Reaction Details Report a problem with these data
Report a problem with these data
 Report a problem with these data
Report a problem with these dataTarget
cGMP-dependent protein kinase
Ligand
BDBM50182355
Substrate
n/a
Meas. Tech.
ChEMBL_349021 (CHEMBL865064)
IC50
0.13±n/a nM
Citation
 Biftu, T; Feng, D; Fisher, M; Liang, GB; Qian, X; Scribner, A; Dennis, R; Lee, S; Liberator, PA; Brown, C; Gurnett, A; Leavitt, PS; Thompson, D; Mathew, J; Misura, A; Samaras, S; Tamas, T; Sina, JF; McNulty, KA; McKnight, CG; Schmatz, DM; Wyvratt, M Synthesis and SAR studies of very potent imidazopyridine antiprotozoal agents. Bioorg Med Chem Lett 16:2479-83 (2006) [PubMed] Article
Biftu, T; Feng, D; Fisher, M; Liang, GB; Qian, X; Scribner, A; Dennis, R; Lee, S; Liberator, PA; Brown, C; Gurnett, A; Leavitt, PS; Thompson, D; Mathew, J; Misura, A; Samaras, S; Tamas, T; Sina, JF; McNulty, KA; McKnight, CG; Schmatz, DM; Wyvratt, M Synthesis and SAR studies of very potent imidazopyridine antiprotozoal agents. Bioorg Med Chem Lett 16:2479-83 (2006) [PubMed] Article More Info.:
Target
Name:
cGMP-dependent protein kinase
Synonyms:
n/a
Type:
PROTEIN
Mol. Mass.:
112911.27
Organism:
Eimeria tenella
Description:
ChEMBL_469577
Residue:
1003
Sequence:
MGACSSKAQHQTRDPEPREQQAAQEQKSTGPSGAPNDAPAPAEAERKMSGSSATAPKGEMPTASTGTPEQQQQQQQQQQQQQEQQQHPEHQQSEKQQQHGEEQQQERKPSQQQQNEEAAAPHKHGGERKVQKAIKQQEDTQAEDARLLGHLEKREKTPSDLSLIRDSLSTNLVCSSLNDAEVEALANAVEFFTFKKGDVVTKQGESGSYFFIVHSGEFEVIVNDKVVNKILTGQAFGEISLIHNSARTATIKTLSEDAALWGVQRQVFRETLKQLSSRNFAENRQFLASVKFFEMLTEAQKNVITNALVVQSFQPGQAIVKEGEKGDVLYILKSGKALVSIKNKEVRVLQRGEYFGERALLYDEPRSATITAEEPTVCVSIGRDLLDRVLGNLQHVLFRNIMLEALQQSKVFASFPTEQLSRLIGSVVVKDYPENYIILDRENRTRASASALFSAQGVRFFFVLEGEVSVFAYKDKSSSSSSSSSSSSSSSSAEGEMELHLIDTLKRGQAFGDEYVLSPNKPFAHCVKSNGPTKLALLTASALTATLGGQDIDETLDYNNKLAITKKMYIFRYLSEQQTQTLIKAFKTVRYTQGESIIREGEIGSRFFIIKLGEVVILKGEKRVRTLGRHDYFGERALLHDERRSATVAANSPEVDLWVVDKDVFLQIVKGPMLTHLEERIRMQDTKVEFKDLNVVRVVGRGTFGTVKLVQHIPTQMRYALKCVSRKSVVALNQQDHIRLEREIMAENDHPFIIRLVRTFRDKEFLYFLTELVTGGELYDAIRKLGLLGRYQAQFYLASIVLAIEYLHERNIAYRDLKPENILLDSQGYVKLIDFGCAKKMQGRAYTLVGTPHYMAPEVILGKGYTLTADTWAFGVCLYEFMCGPLPFGNDAEDQLEIFRDILAGKLIFPHYVTDQDAINLMKRLLCRLPEVRIGCSINGYKDIKEHAFFSDFDWDRLAGRDLSPPLLPKGEIYAEDAEEGGLDIEEDEGIELEDEYEWDKDF
Inhibitor
Name:
BDBM50182355
Synonyms:
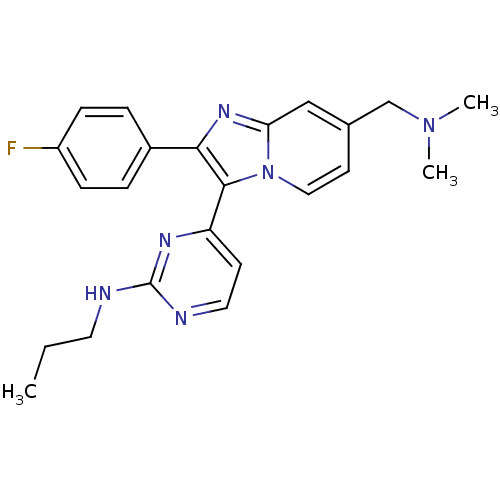
4-(7-((dimethylamino)methyl)-2-(4-fluorophenyl)H-imidazo[1,2-a]pyridin-3-yl)-N-propylpyrimidin-2-amine | CHEMBL205377 | {4-[7-dimethylaminomethyl-2-(4-fluorophenyl)imidazo[1,2-a]pyridin-3-yl]pyrimidin-2-yl}propylamine
Type:
Small organic molecule
Emp. Form.:
C23H25FN6
Mol. Mass.:
404.4832
SMILES:
CCCNc1nccc(n1)-c1c(nc2cc(CN(C)C)ccn12)-c1ccc(F)cc1