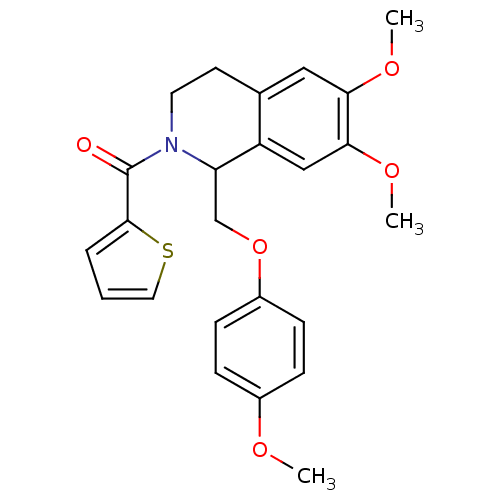
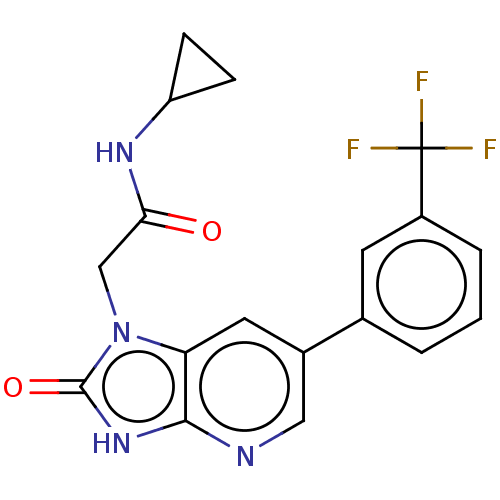
Affinity DataKi: 63nMAssay Description:To determine the affinity of the compounds of the present invention a SPA is used. The assay is run in a 384-plate format (OptiPlate-384) where each ...More data for this Ligand-Target Pair
Affinity DataKi: 96nMAssay Description:To determine the affinity of the compounds of the present invention a SPA is used. The assay is run in a 384-plate format (OptiPlate-384) where each ...More data for this Ligand-Target Pair
Affinity DataKi: 140nMAssay Description:To determine the affinity of the compounds of the present invention a SPA is used. The assay is run in a 384-plate format (OptiPlate-384) where each ...More data for this Ligand-Target Pair
Affinity DataEC50: 150nMAssay Description:Positive allosteric modulation of recombinant human GluN1/GluN2C receptor stably expressed in HEK293 cells assessed as increase in glycine/L-glutamat...More data for this Ligand-Target Pair
Affinity DataKi: 170nMAssay Description:To determine the affinity of the compounds of the present invention a SPA is used. The assay is run in a 384-plate format (OptiPlate-384) where each ...More data for this Ligand-Target Pair
Affinity DataKi: 180nMAssay Description:To determine the affinity of the compounds of the present invention a SPA is used. The assay is run in a 384-plate format (OptiPlate-384) where each ...More data for this Ligand-Target Pair
Affinity DataKi: 220nMAssay Description:To determine the affinity of the compounds of the present invention a SPA is used. The assay is run in a 384-plate format (OptiPlate-384) where each ...More data for this Ligand-Target Pair
Affinity DataKi: 220nMAssay Description:To determine the affinity of the compounds of the present invention a SPA is used. The assay is run in a 384-plate format (OptiPlate-384) where each ...More data for this Ligand-Target Pair
Affinity DataKi: 260nMAssay Description:To determine the affinity of the compounds of the present invention a SPA is used. The assay is run in a 384-plate format (OptiPlate-384) where each ...More data for this Ligand-Target Pair
Affinity DataKi: 270nMAssay Description:To determine the affinity of the compounds of the present invention a SPA is used. The assay is run in a 384-plate format (OptiPlate-384) where each ...More data for this Ligand-Target Pair
Affinity DataKi: 360nMAssay Description:To determine the affinity of the compounds of the present invention a SPA is used. The assay is run in a 384-plate format (OptiPlate-384) where each ...More data for this Ligand-Target Pair
Affinity DataIC50: 400nMAssay Description:Inhibition of recombinant GluN1/GluN2C receptor (unknown origin) expressed in HEK293 cells assessed as inhibition of glutamate-evoked current by whol...More data for this Ligand-Target Pair
Affinity DataKi: 490nMAssay Description:To determine the affinity of the compounds of the present invention a SPA is used. The assay is run in a 384-plate format (OptiPlate-384) where each ...More data for this Ligand-Target Pair
Affinity DataKi: 690nMAssay Description:To determine the affinity of the compounds of the present invention a SPA is used. The assay is run in a 384-plate format (OptiPlate-384) where each ...More data for this Ligand-Target Pair
Affinity DataIC50: 800nMAssay Description:The assay involves using a cell line that expresses the NR1 subunit together with either NR2C or NR2D. These cell lines can be prepared by transfecti...More data for this Ligand-Target Pair
Affinity DataKi: 860nMAssay Description:To determine the affinity of the compounds of the present invention a SPA is used. The assay is run in a 384-plate format (OptiPlate-384) where each ...More data for this Ligand-Target Pair
Affinity DataKi: 870nMAssay Description:To determine the affinity of the compounds of the present invention a SPA is used. The assay is run in a 384-plate format (OptiPlate-384) where each ...More data for this Ligand-Target Pair
Affinity DataIC50: 1.00E+3nMAssay Description:The assay involves using a cell line that expresses the NR1 subunit together with either NR2C or NR2D. These cell lines can be prepared by transfecti...More data for this Ligand-Target Pair
Affinity DataIC50: 1.00E+3nMAssay Description:The assay involves using a cell line that expresses the NR1 subunit together with either NR2C or NR2D. These cell lines can be prepared by transfecti...More data for this Ligand-Target Pair
Affinity DataIC50: 1.20E+3nMAssay Description:The assay involves using a cell line that expresses the NR1 subunit together with either NR2C or NR2D. These cell lines can be prepared by transfecti...More data for this Ligand-Target Pair
Affinity DataEC50: 1.90E+3nMAssay Description:Positive allosteric modulator activity at GluN2C NMDA receptor (unknown origin) expressed in CHO cells co-expressing GluN1a in the presence of L-glut...More data for this Ligand-Target Pair
Affinity DataIC50: 2.00E+3nMAssay Description:The assay involves using a cell line that expresses the NR1 subunit together with either NR2C or NR2D. These cell lines can be prepared by transfecti...More data for this Ligand-Target Pair
Affinity DataIC50: 2.00E+3nMAssay Description:Negative allosteric modulation of GluN2C receptor (unknown origin) expressed in xenopus laevis oocytes assessed as reduction in glycine-induced chann...More data for this Ligand-Target Pair
Affinity DataIC50: 2.00E+3nMAssay Description:The assay involves using a cell line that expresses the NR1 subunit together with either NR2C or NR2D. These cell lines can be prepared by transfecti...More data for this Ligand-Target Pair
Affinity DataIC50: 2.40E+3nMAssay Description:The assay involves using a cell line that expresses the NR1 subunit together with either NR2C or NR2D. These cell lines can be prepared by transfecti...More data for this Ligand-Target Pair
Affinity DataIC50: 2.60E+3nMAssay Description:Negative allosteric modulation of GluN2C receptor (unknown origin) expressed in xenopus laevis oocytes assessed as reduction in glycine-induced chann...More data for this Ligand-Target Pair
Affinity DataEC50: 2.70E+3nMAssay Description:Positive allosteric modulation of GluN2C receptor (unknown origin) expressed in xenopus laevis oocytes assessed as increase in glycine-induced channe...More data for this Ligand-Target Pair
Affinity DataIC50: 3.00E+3nMAssay Description:The assay involves using a cell line that expresses the NR1 subunit together with either NR2C or NR2D. These cell lines can be prepared by transfecti...More data for this Ligand-Target Pair
Affinity DataIC50: 3.00E+3nMAssay Description:The assay involves using a cell line that expresses the NR1 subunit together with either NR2C or NR2D. These cell lines can be prepared by transfecti...More data for this Ligand-Target Pair
Affinity DataIC50: 3.00E+3nMAssay Description:The assay involves using a cell line that expresses the NR1 subunit together with either NR2C or NR2D. These cell lines can be prepared by transfecti...More data for this Ligand-Target Pair
Affinity DataKi: 3.90E+3nMAssay Description:To determine the affinity of the compounds of the present invention a SPA is used. The assay is run in a 384-plate format (OptiPlate-384) where each ...More data for this Ligand-Target Pair
Affinity DataIC50: 4.00E+3nMAssay Description:The assay involves using a cell line that expresses the NR1 subunit together with either NR2C or NR2D. These cell lines can be prepared by transfecti...More data for this Ligand-Target Pair
Affinity DataEC50: 4.70E+3nMAssay Description:Positive allosteric modulation of GluN1/GluN2C receptor (unknown origin) assessed as increase in glutamate-induced calcium flux measured at time inte...More data for this Ligand-Target Pair
Affinity DataIC50: 5.00E+3nMAssay Description:The assay involves using a cell line that expresses the NR1 subunit together with either NR2C or NR2D. These cell lines can be prepared by transfecti...More data for this Ligand-Target Pair
Affinity DataIC50: 5.40E+3nMAssay Description:Negative allosteric modulation of GluN2C receptor (unknown origin) expressed in xenopus laevis oocytes assessed as reduction in glycine-induced chann...More data for this Ligand-Target Pair
Affinity DataIC50: 6.00E+3nMAssay Description:The assay involves using a cell line that expresses the NR1 subunit together with either NR2C or NR2D. These cell lines can be prepared by transfecti...More data for this Ligand-Target Pair
Affinity DataIC50: 6.00E+3nMAssay Description:The assay involves using a cell line that expresses the NR1 subunit together with either NR2C or NR2D. These cell lines can be prepared by transfecti...More data for this Ligand-Target Pair
Affinity DataEC50: 6.60E+3nMAssay Description:Positive allosteric modulation of GluN2C receptor (unknown origin) expressed in xenopus laevis oocytes assessed as increase in glycine-induced channe...More data for this Ligand-Target Pair
Affinity DataIC50: 7.00E+3nMAssay Description:Negative allosteric modulation of GluN2C receptor (unknown origin) expressed in xenopus laevis oocytes assessed as reduction in glycine-induced chann...More data for this Ligand-Target Pair
Affinity DataIC50: 7.00E+3nMAssay Description:The assay involves using a cell line that expresses the NR1 subunit together with either NR2C or NR2D. These cell lines can be prepared by transfecti...More data for this Ligand-Target Pair
Affinity DataIC50: 7.00E+3nMAssay Description:The assay involves using a cell line that expresses the NR1 subunit together with either NR2C or NR2D. These cell lines can be prepared by transfecti...More data for this Ligand-Target Pair
Affinity DataIC50: 7.10E+3nMAssay Description:Negative allosteric modulation of GluN2C receptor (unknown origin) expressed in xenopus laevis oocytes assessed as reduction in glycine-induced chann...More data for this Ligand-Target Pair
Affinity DataKi: 7.40E+3nMAssay Description:To determine the affinity of the compounds of the present invention a SPA is used. The assay is run in a 384-plate format (OptiPlate-384) where each ...More data for this Ligand-Target Pair
Affinity DataEC50: 7.40E+3nMAssay Description:Positive allosteric modulation of GluN1/GluN2C NMDAR (unknown origin) expressed in Dox-inducible cells by BD calcium indicator dye based-fluorescence...More data for this Ligand-Target Pair
Affinity DataIC50: 9.00E+3nMAssay Description:The assay involves using a cell line that expresses the NR1 subunit together with either NR2C or NR2D. These cell lines can be prepared by transfecti...More data for this Ligand-Target Pair
Affinity DataIC50: 1.00E+4nMAssay Description:Inhibition of human GluN2C receptor by FLIPR assayMore data for this Ligand-Target Pair
Affinity DataIC50: 1.00E+4nMAssay Description:Inhibition of human GluN2C receptor by FLIPR assayMore data for this Ligand-Target Pair
Affinity DataIC50: 1.00E+4nMAssay Description:The assay involves using a cell line that expresses the NR1 subunit together with either NR2C or NR2D. These cell lines can be prepared by transfecti...More data for this Ligand-Target Pair
Affinity DataIC50: 1.00E+4nMAssay Description:Negative allosteric modulation of recombinant human GluN1a/GluN2C expressed in CHO-T-REx cells assessed as inhibition of glutamate/glycine-induced re...More data for this Ligand-Target Pair
Affinity DataIC50: 1.00E+4nMAssay Description:Inhibition of human GluN2C receptor by FLIPR assayMore data for this Ligand-Target Pair

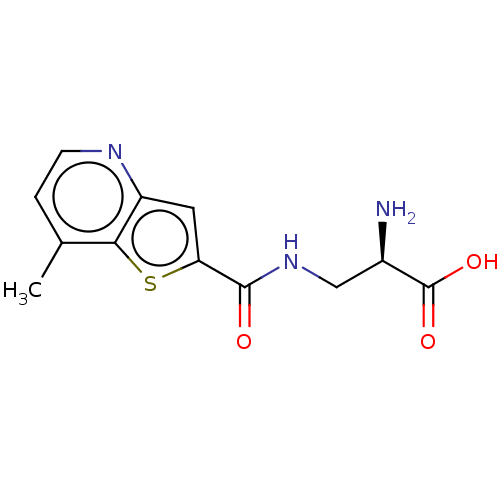

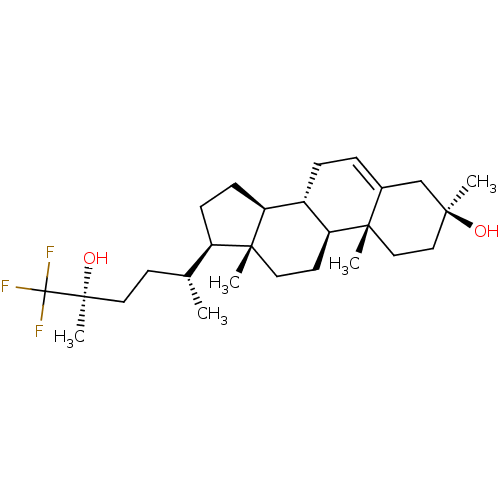
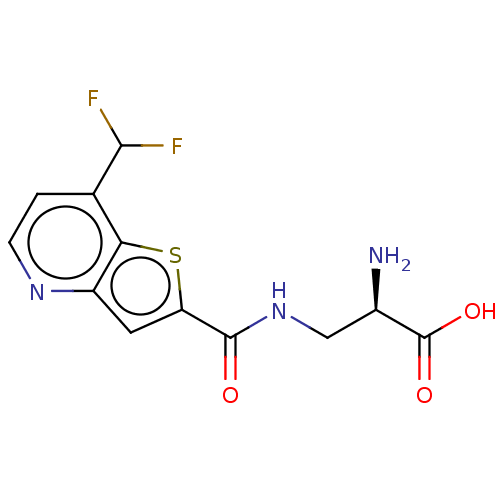
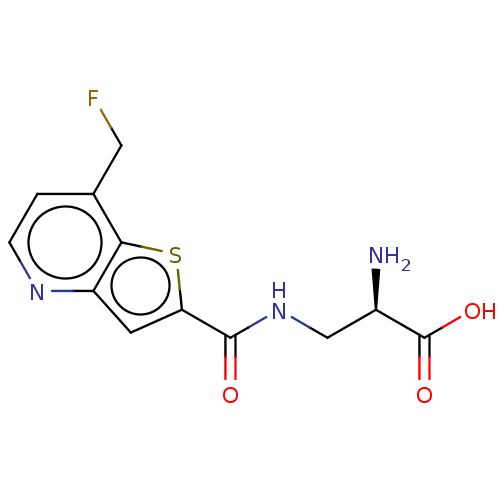
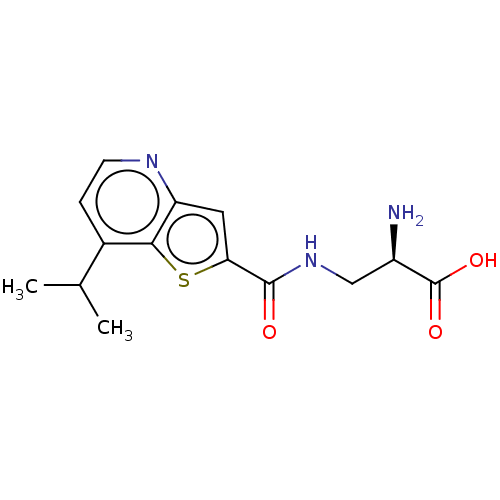
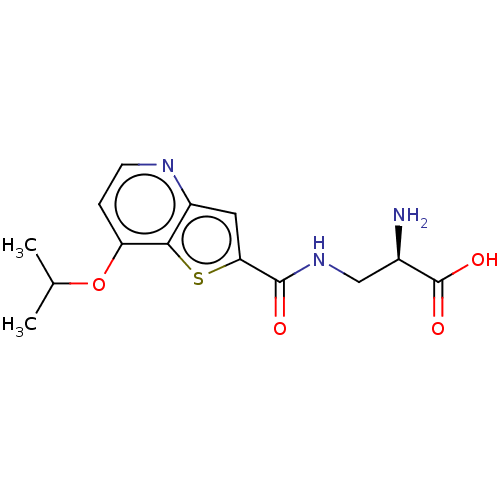
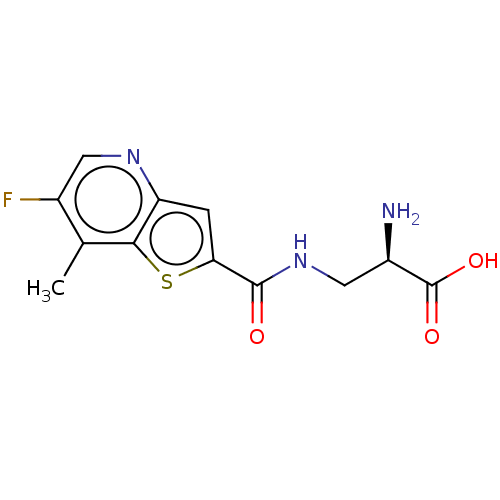
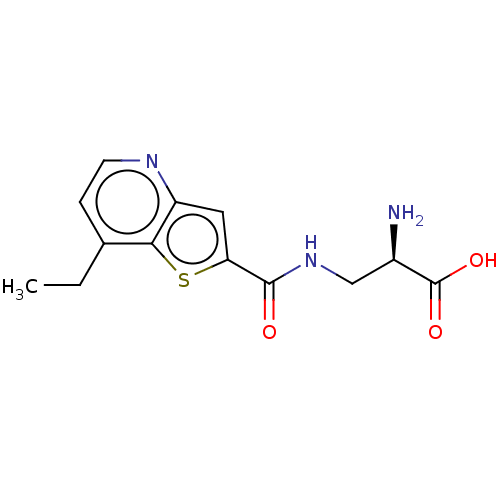

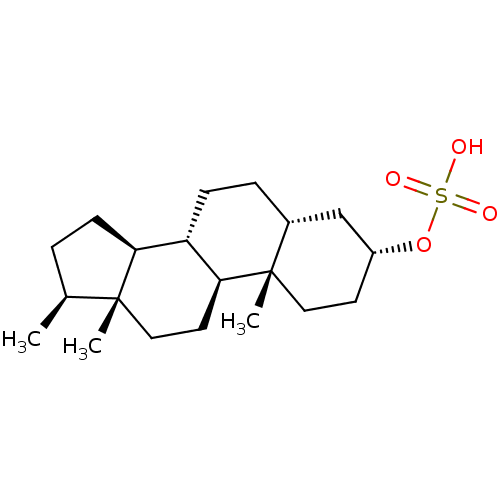
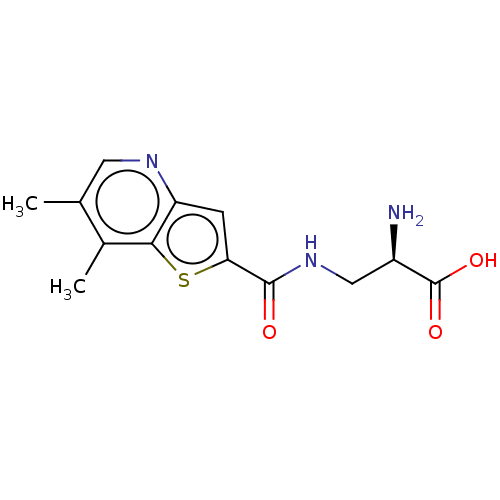
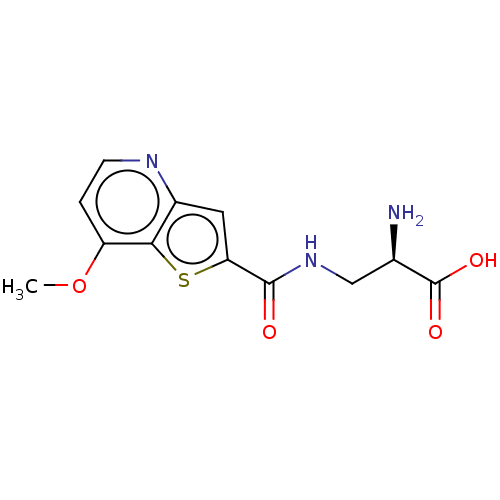
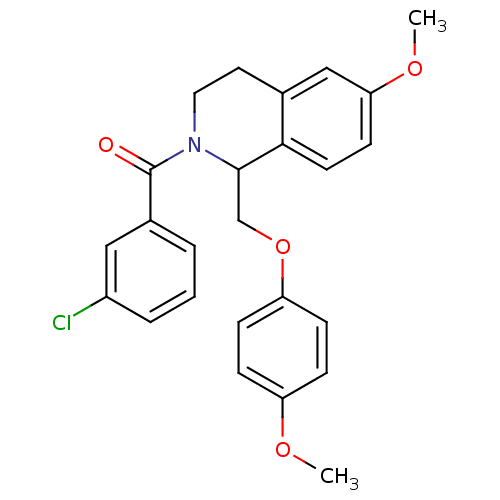
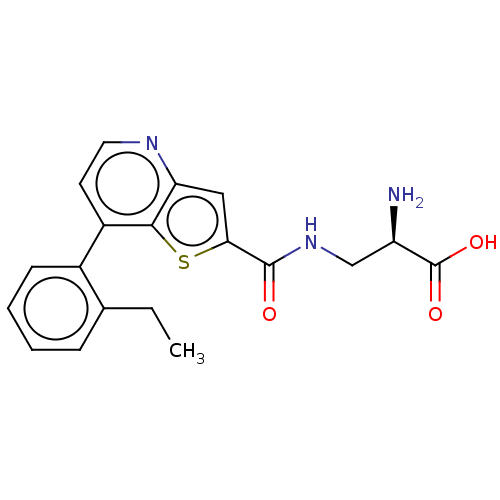
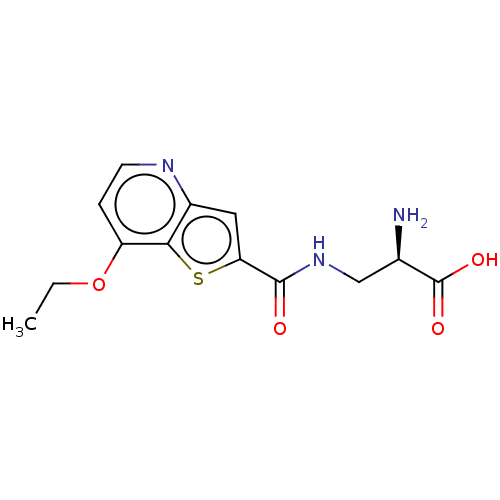
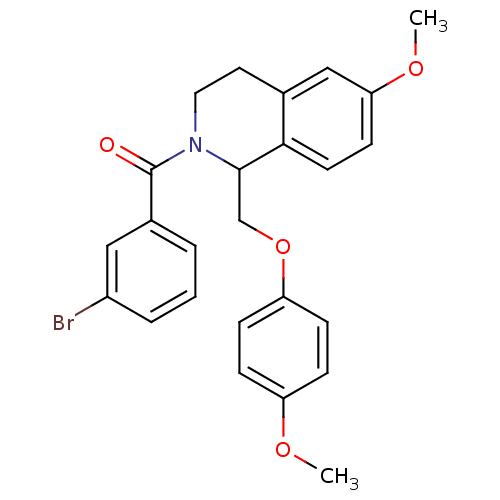
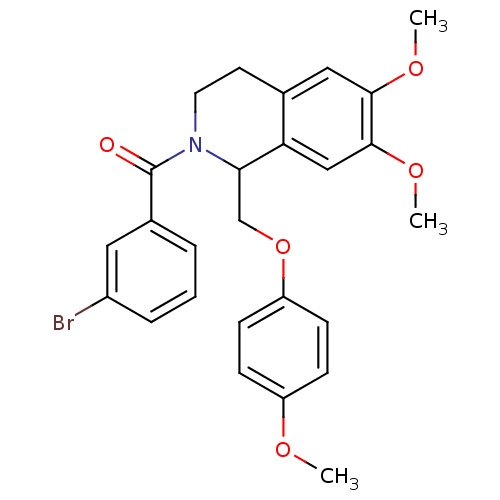
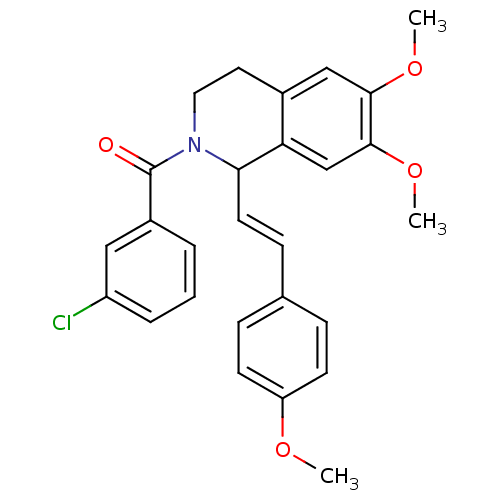
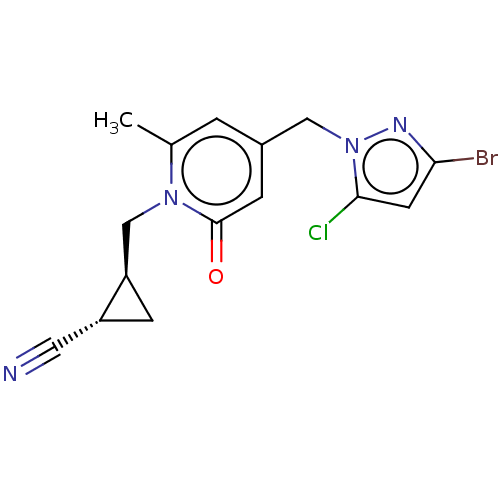


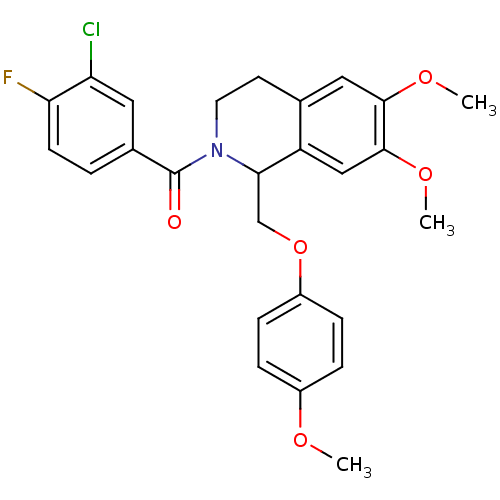
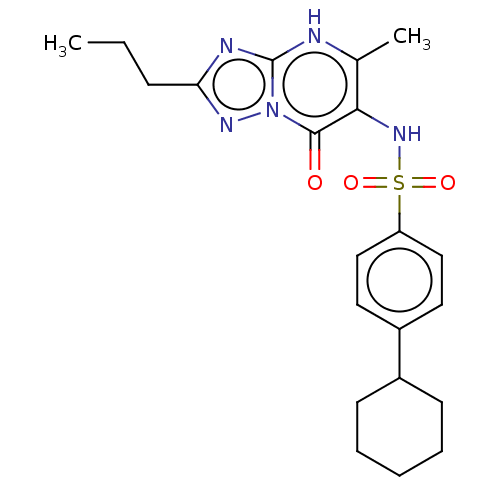
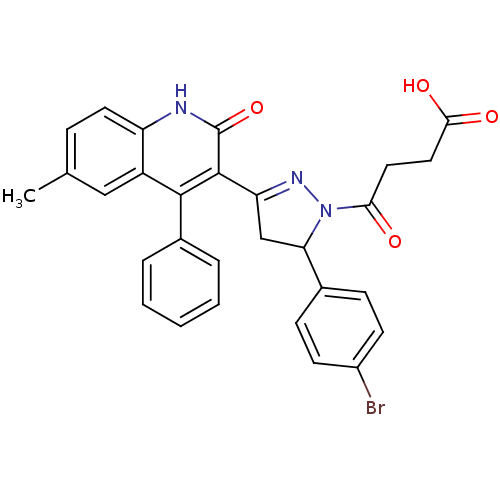
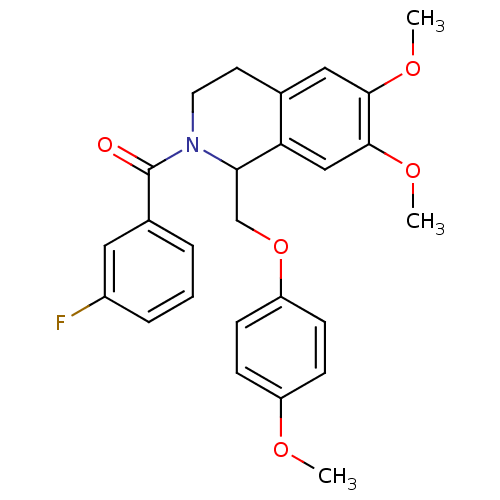
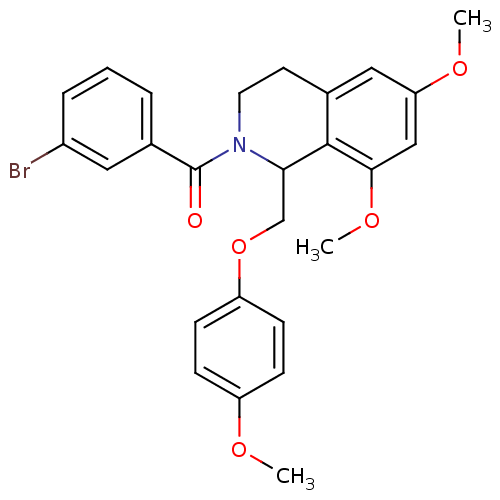
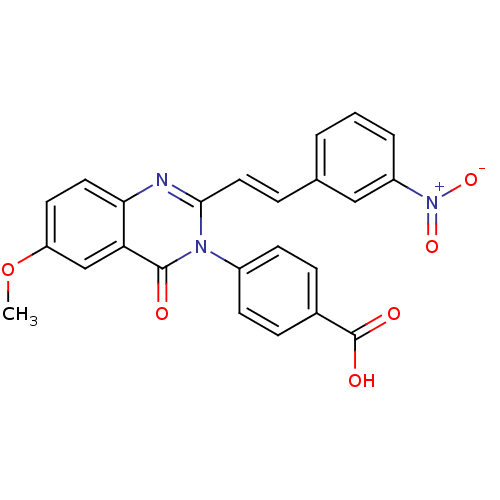
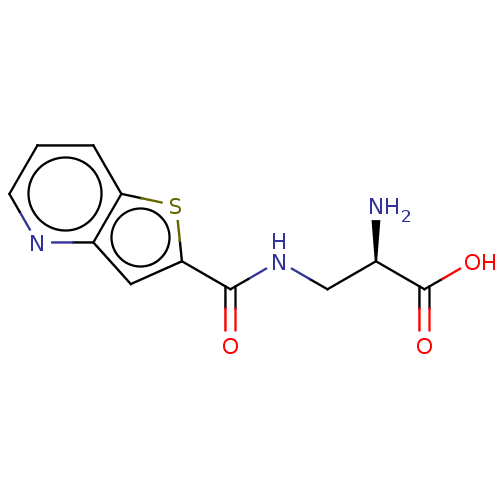

 3D Structure (crystal)
3D Structure (crystal)