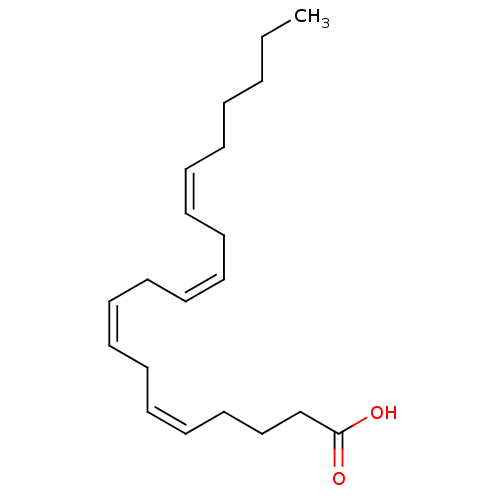
BDBM22319 (5Z,8Z,11Z,14Z)-icosa-5,8,11,14-tetraenoic acid::Arachidonic acid::Arachidonic acid (AA)::CHEMBL15594::[1-14C]Arachidonic acid
SMILES CCCCC\C=C/C\C=C/C\C=C/C\C=C/CCCC(=O)O
InChI Key InChIKey=YZXBAPSDXZZRGB-UHFFFAOYSA-N
Activity Spreadsheet -- Enzyme Inhibition Constant Data from BindingDB
 Found 35 hits for monomerid = 22319
Found 35 hits for monomerid = 22319
Affinity DataEC50: 7.80nMAssay Description:Activation of recombinant human PP5 (16 to 499 residues) Escherichia coli BL21 (DE3) using pNPP incubated for 60 minsMore data for this Ligand-Target Pair
Affinity DataKd: 42.1nMAssay Description:The method entails two steps [Lin et al., Biochemistry, 38:185-190]. In the first step, the Kd for association of FABP5 with the fluorescent lipid pr...More data for this Ligand-Target Pair
Affinity DataKd: 180nMAssay Description:Binding affinity against Adipocyte lipid binding proteinMore data for this Ligand-Target Pair
Affinity DataKi: 330nMAssay Description:Ki values for sodium fluorescein (10 uM) uptake in OATP1B1-transfected CHO cellsMore data for this Ligand-Target Pair
Affinity DataIC50: 603nMAssay Description:pIC50 values for sodium fluorescein (10 uM) uptake in OATP1B1-transfected CHO cellsMore data for this Ligand-Target Pair
Affinity DataKi: 820nMAssay Description:Ki values for sodium fluorescein (10 uM) uptake in OATP1B3-transfected CHO cellsMore data for this Ligand-Target Pair
Affinity DataIC50: 1.07E+3nMAssay Description:pIC50 values for sodium fluorescein (10 uM) uptake in OATP1B3-transfected CHO cellsMore data for this Ligand-Target Pair
Affinity DataIC50: 1.20E+3nMAssay Description:Inhibition of PPAR-alpha (unknown origin) by SPA assayMore data for this Ligand-Target Pair
Affinity DataIC50: 1.20E+3nMAssay Description:Displacement of [3H]GW2331 from Homo sapiens (human) PPARalpha receptor by scintillation proximity assayMore data for this Ligand-Target Pair
Affinity DataIC50: 1.60E+3nMAssay Description:Displacement of [3H]BRL49653 from Homo sapiens (human) PPARgamma receptor by scintillation proximity assayMore data for this Ligand-Target Pair
Affinity DataKi: 2.59E+3nM ΔG°: -7.74kcal/molepH: 8.2 T: 2°CAssay Description:In brief, both wild-type and mutant hFABP5 were expressed and purified to homogeneity as described above and dialyzed in PBS (pH 8.2). Binding affini...More data for this Ligand-Target Pair
Affinity DataIC50: 2.75E+3nMAssay Description:Inhibition of FABP4 (unknown origin) incubated for 10 mins by fluorescence based analysisMore data for this Ligand-Target Pair
Affinity DataIC50: 3.10E+3nMAssay Description:Displacement of [3H]GW2433 from Homo sapiens (human) PPARdelta receptor by scintillation proximity assayMore data for this Ligand-Target Pair
Target60 kDa heat shock protein, mitochondrial(Human)
Indiana University School of Medicine
Curated by ChEMBL
Indiana University School of Medicine
Curated by ChEMBL
Affinity DataIC50: 4.50E+3nMAssay Description:Inhibition of human N-terminal octa-His-tagged HSP60 expressed in Escherichia coli Rosetta(DE3) pLysS/human HSP10 expressed in Escherichia coli Roset...More data for this Ligand-Target Pair
Affinity DataKi: 5.01E+3nM ΔG°: -7.35kcal/molepH: 8.2 T: 2°CAssay Description:In brief, both wild-type and mutant hFABP5 were expressed and purified to homogeneity as described above and dialyzed in PBS (pH 8.2). Binding affini...More data for this Ligand-Target Pair
Affinity DataIC50: 5.13E+3nMAssay Description:Inhibition of FABP4 (unknown origin)More data for this Ligand-Target Pair
TargetVoltage-dependent L-type calcium channel subunit alpha-1C(Rat)
Jagiellonian University
Curated by ChEMBL
Jagiellonian University
Curated by ChEMBL
Affinity DataIC50: 5.80E+3nMAssay Description:Inhibition of L-type calcium channel measured using whole-cell patch clamp in rat ventricular myocytesMore data for this Ligand-Target Pair
Affinity DataIC50: 7.30E+3nMAssay Description:Inhibition of Escherichia coli GroEL expressed in Escherichia coliDH5alpha/Escherichia coli GroES expressed in Escherichia coli BL21 (DE3) assessed a...More data for this Ligand-Target Pair
Affinity DataKd: 8.07E+3nMAssay Description:Inhibitory activity of the synthesized compounds on COX-1 and COX-2 enzymes were evaluated at 40 μm using the COX Inhibitor Screening Assay Kit ...More data for this Ligand-Target Pair
TargetVoltage-dependent L-type calcium channel subunit alpha-1C(Rat)
Jagiellonian University
Curated by ChEMBL
Jagiellonian University
Curated by ChEMBL
Affinity DataIC50: 8.50E+3nMAssay Description:Inhibition of L-type calcium channel measured using whole-cell patch clamp in rat ventricular myocytesMore data for this Ligand-Target Pair
Affinity DataKi: 9.37E+3nM ΔG°: -6.97kcal/molepH: 8.2 T: 2°CAssay Description:In brief, both wild-type and mutant hFABP5 were expressed and purified to homogeneity as described above and dialyzed in PBS (pH 8.2). Binding affini...More data for this Ligand-Target Pair
TargetTransient receptor potential cation channel subfamily V member 2(Rat)
National Research Council (Cnr)
Curated by ChEMBL
National Research Council (Cnr)
Curated by ChEMBL
Affinity DataIC50: 1.00E+4nMAssay Description:Antagonist activity at recombinant rat TRPV2 expressed in HEK293 cells assessed as inhibition of LPC-induced Ca2+ levels preincubated for 5 mins foll...More data for this Ligand-Target Pair
TargetTransient receptor potential cation channel subfamily V member 2(Rat)
National Research Council (Cnr)
Curated by ChEMBL
National Research Council (Cnr)
Curated by ChEMBL
Affinity DataIC50: 1.00E+4nMAssay Description:Antagonist activity at recombinant rat TRPV2 expressed in HEK293 cells assessed as inhibition of CBD-induced Ca2+ levels preincubated for 5 mins foll...More data for this Ligand-Target Pair
Affinity DataIC50: 1.80E+4nMAssay Description:Inhibition of Escherichia coli GroEL expressed in Escherichia coli DH5alpha/Escherichia coli GroES expressed in Escherichia coli BL21 (DE3) assessed ...More data for this Ligand-Target Pair
Affinity DataIC50: 2.82E+4nMAssay Description:Inhibition of aromatase in human placental microsomes by radiometric methodMore data for this Ligand-Target Pair
TargetNuclear receptor subfamily 4 group A member 2(Human)
Goethe University Frankfurt
Curated by ChEMBL
Goethe University Frankfurt
Curated by ChEMBL
Affinity DataKd: 5.00E+4nMAssay Description:Binding affinity towards Nurr1 ligand binding domain (unknown origin) measured by tryptophan fluorescence spectroscopyMore data for this Ligand-Target Pair
Affinity DataEC50: 7.76E+4nMAssay Description:Activation of PPP5C (unknown origin) TPR domainMore data for this Ligand-Target Pair
Affinity DataIC50: 1.00E+5nMAssay Description:Inhibition of native rhodanese (unknown origin) assessed as reduction in rhodanese enzyme activity after 45 mins by Fe(SCN)3 dye based spectrometric ...More data for this Ligand-Target Pair
Affinity DataIC50: 1.00E+5nMAssay Description:Concentration required to inhibit human Cytosolic phospholipase A2More data for this Ligand-Target Pair
TargetFatty-acid amide hydrolase 1(Human)
Johnson & Johnson Pharmaceutical Research and Development
Curated by ChEMBL
Johnson & Johnson Pharmaceutical Research and Development
Curated by ChEMBL
Affinity DataIC50: 1.00E+5nMAssay Description:Inhibition of FAAHMore data for this Ligand-Target Pair
Affinity DataIC50: 1.05E+5nMAssay Description:Inhibition of amidolytic activity of human tissue factor/human factor 7aMore data for this Ligand-Target Pair
Affinity DataIC50: 2.00E+5nMAssay Description:Inhibition of human CYP51 expressed in Topp 3 cells by lanosterol demethylase assayMore data for this Ligand-Target Pair
Affinity DataIC50: 2.00E+5nMAssay Description:Inhibition of pig pancreatic trypsin after 15 minsMore data for this Ligand-Target Pair
Affinity DataIC50: 2.50E+5nMAssay Description:Inhibition of ATPase activity of Escherichia coli GroEL expressed in Escherichia coliDH5alpha incubated for 60 mins using ATP by spectrometric analys...More data for this Ligand-Target Pair
Activity Spreadsheet -- ITC Data from BindingDB
 Found 4 hits for monomerid = 22319
Found 4 hits for monomerid = 22319
CellMannoprotein ligand-binding domain 2 (Mp1p-LBD2)(Talaromyces marneffei)
The University of Hong Kong
The University of Hong Kong
ITC DataΔG°: -11.2kcal/mole −TΔS°: 3.33kcal/mole ΔH°: -14.2kcal/mole logk: 7.69E+7
pH: 8.0 T: 37.00°C
pH: 8.0 T: 37.00°C
CellMannoprotein ligand-binding domain 2 (Mp1p-LBD2)(Talaromyces marneffei)
The University of Hong Kong
The University of Hong Kong
ITC DataΔG°: -12.3kcal/mole −TΔS°: 2.30kcal/mole ΔH°: -11.0kcal/mole logk: 4.35E+8
pH: 8.0 T: 37.00°C
pH: 8.0 T: 37.00°C
CellMannoprotein ligand-binding domain 2 (Mp1p-LBD2 I332A)(Talaromyces marneffei)
The University of Hong Kong
The University of Hong Kong
ITC DataΔG°: -8.18kcal/mole −TΔS°: 6.44kcal/mole ΔH°: -14.5kcal/mole logk: 5.88E+5
pH: 8.0 T: 37.00°C
pH: 8.0 T: 37.00°C
CellMannoprotein ligand-binding domain 2 (Mp1p-LBD2 V309D)(Talaromyces marneffei)
The University of Hong Kong
The University of Hong Kong
ITC DataΔG°: -7.69kcal/mole −TΔS°: 1.93kcal/mole ΔH°: -9.31kcal/mole logk: 2.63E+5
pH: 8.0 T: 37.00°C
pH: 8.0 T: 37.00°C
 3D Structure (crystal)
3D Structure (crystal)