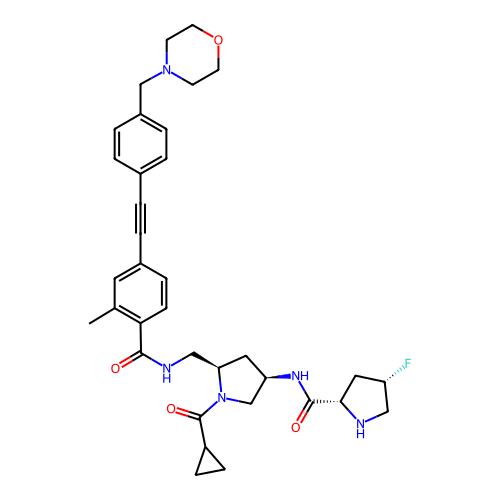
BDBM744659 (2S,4S)—N-((3R,5R)-1-(Cyclopropanecarbonyl)-5-((2-methyl-4-((4-(morpholinomethyl)phenyl)ethynyl)benzamido)methyl)pyrrolidin-3-yl)-4-fluoropyrrolidine-2-carboxamide::US20250171400, Example 7
SMILES Cc1cc(C#Cc2ccc(CN3CCOCC3)cc2)ccc1C(=O)NC[C@H]1C[C@@H](NC(=O)[C@@H]2C[C@H](F)CN2)CN1C(=O)C1CC1
InChI Key
PDB links: 1 PDB ID matches this monomer.
Activity Spreadsheet -- Enzyme Inhibition Constant Data from BindingDB
 Found 2 hits for monomerid = 744659
Found 2 hits for monomerid = 744659
TargetUDP-3-O-acyl-N-acetylglucosamine deacetylase(Pseudomonas aeruginosa (strain ATCC 15692 / PAO1 /...)TBA
Affinity DataIC50: 75nMAssay Description:Dilutions of test compound were pre-incubated with 5 nM P. aeruginosa or E. coli LpxC for 10 minutes at room temperature in 50 mM NaH2PO4, 500 mM suc...More data for this Ligand-Target Pair
Affinity DataIC50: 500nMAssay Description:Dilutions of test compound were pre-incubated with 5 nM P. aeruginosa or E. coli LpxC for 10 minutes at room temperature in 50 mM NaH2PO4, 500 mM suc...More data for this Ligand-Target Pair
 3D Structure (crystal)
3D Structure (crystal)