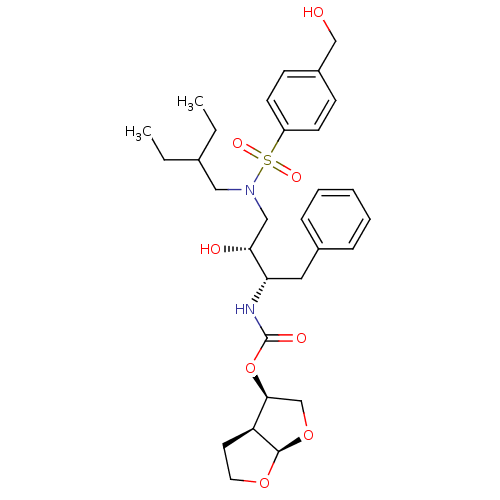
BDBM112661 (3R,3aS,6aR)-Hexahydrofuro[2,3-b]furan-3-yl ((2S,3R)-4-(N-(2-ethylbutyl)-4-(hydroxymethyl)phenylsulfonamido)-3-hydroxy-1-phenylbutan-2-yl)carbamate (11c)
SMILES CCC(CC)CN(C[C@@H](O)[C@H](Cc1ccccc1)NC(=O)O[C@H]1CO[C@H]2OCC[C@@H]12)S(=O)(=O)c1ccc(CO)cc1
InChI Key InChIKey=KOYHHSRUNXTXFB-WNJKUOTESA-N
Activity Spreadsheet -- Enzyme Inhibition Constant Data from BindingDB
 Found 6 hits for monomerid = 112661
Found 6 hits for monomerid = 112661
TargetGag-Pol polyprotein [484-582,Q491K,I494L,I497V,V499I,E519D,R541K,D544E,Q553K,L573M](Human immunodeficiency virus)
University Of Massachusetts
University Of Massachusetts
Affinity DataKi: 0.000500nM ΔG°: -16.8kcal/molepH: 4.7 T: 2°CAssay Description:The reaction mixture contained 2 µL of protease and 2 µL of inhibitor (or DMSO as a control) and was incubated for 20-30 min at room temper...More data for this Ligand-Target Pair
TargetGag-Pol polyprotein [484-582,Q491K,I497V,V499I,E519D,G532V,I538V,R541K,D544E,L547P,Q553K,V566A,L573M](Human immunodeficiency virus)
University Of Massachusetts
University Of Massachusetts
Affinity DataKi: 0.00500nM ΔG°: -15.4kcal/molepH: 4.7 T: 2°CAssay Description:The reaction mixture contained 2 µL of protease and 2 µL of inhibitor (or DMSO as a control) and was incubated for 20-30 min at room temper...More data for this Ligand-Target Pair
TargetProtease(Human immunodeficiency virus 1 (HIV-1))
University Of Massachusetts Medical School
Curated by ChEMBL
University Of Massachusetts Medical School
Curated by ChEMBL
Affinity DataKi: 0.00500nMAssay Description:Inhibition of HIV1 NL4-3 protease I84V mutant expressed in Escherichia coli TAP-106 cells using EDANS/DABCYL-labelled 10-amino acid containing natura...More data for this Ligand-Target Pair
TargetGag-Pol polyprotein [484-582,Q491K,I494L,I497V,V499I,E519D,I534V,R541K,D544E,Q553K,A555V,L573M](Human immunodeficiency virus)
University Of Massachusetts
University Of Massachusetts
Affinity DataKi: 0.0260nM ΔG°: -14.4kcal/molepH: 4.7 T: 2°CAssay Description:The reaction mixture contained 2 µL of protease and 2 µL of inhibitor (or DMSO as a control) and was incubated for 20-30 min at room temper...More data for this Ligand-Target Pair
TargetProtease(Human immunodeficiency virus 1 (HIV-1))
University Of Massachusetts Medical School
Curated by ChEMBL
University Of Massachusetts Medical School
Curated by ChEMBL
Affinity DataKi: 0.0550nMAssay Description:Inhibition of HIV1 NL4-3 protease I50V/A71V mutant expressed in Escherichia coli TAP-106 cells using EDANS/DABCYL-labelled 10-amino acid containing n...More data for this Ligand-Target Pair
TargetGag-Pol polyprotein [484-582,Q491K,I497V,V499I,E519D,R541K,D544E,L547P,Q553K,A555V,G557S,I568V,L573M,L574M](Human immunodeficiency virus)
University Of Massachusetts
University Of Massachusetts
Affinity DataKi: 0.0700nM ΔG°: -13.8kcal/molepH: 4.7 T: 2°CAssay Description:The reaction mixture contained 2 µL of protease and 2 µL of inhibitor (or DMSO as a control) and was incubated for 20-30 min at room temper...More data for this Ligand-Target Pair
 3D Structure (crystal)
3D Structure (crystal)