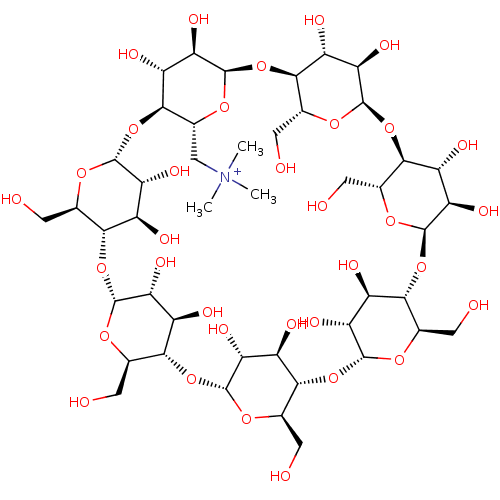
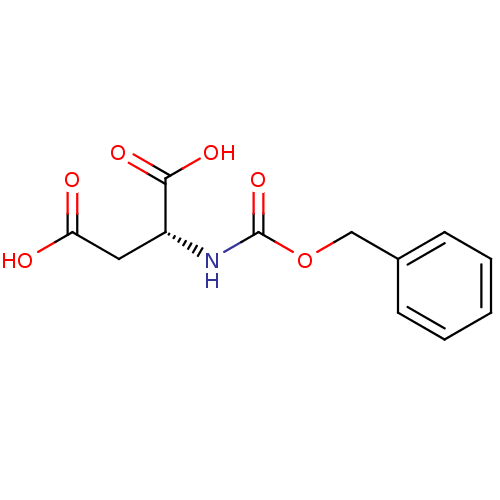
BDBM36136 TMA-beta-cyclodextrin
SMILES C[N+](C)(C)C[C@H]1O[C@@H]2O[C@@H]3[C@@H](CO)O[C@H](O[C@@H]4[C@@H](CO)O[C@H](O[C@@H]5[C@@H](CO)O[C@H](O[C@@H]6[C@@H](CO)O[C@H](O[C@@H]7[C@@H](CO)O[C@H](O[C@@H]8[C@@H](CO)O[C@H](O[C@H]1[C@H](O)[C@H]2O)[C@H](O)[C@H]8O)[C@H](O)[C@H]7O)[C@H](O)[C@H]6O)[C@H](O)[C@H]5O)[C@H](O)[C@H]4O)[C@H](O)[C@H]3O
InChI Key InChIKey=YPNPGLJLLOPIHV-HCHLQLBUSA-N
Data 4 ITC
Activity Spreadsheet -- ITC Data from BindingDB
 Found 4 hits for monomerid = 36136
Found 4 hits for monomerid = 36136
ITC Data−TΔS°: -0.385kcal/mole ΔH°: -2.41kcal/mole logk: 114
pH: 6.9 T: 25.00°C
pH: 6.9 T: 25.00°C
In DepthDetails
ITC Data−TΔS°: -0.242kcal/mole ΔH°: -2.60kcal/mole logk: 119
pH: 6.9 T: 25.00°C
pH: 6.9 T: 25.00°C
In DepthDetails
ITC Data−TΔS°: -0.164kcal/mole ΔH°: -2.63kcal/mole logk: 111
pH: 6.9 T: 25.00°C
pH: 6.9 T: 25.00°C
In DepthDetails
ITC Data−TΔS°: -0.263kcal/mole ΔH°: -2.53kcal/mole logk: 111
pH: 6.9 T: 25.00°C
pH: 6.9 T: 25.00°C
In DepthDetails