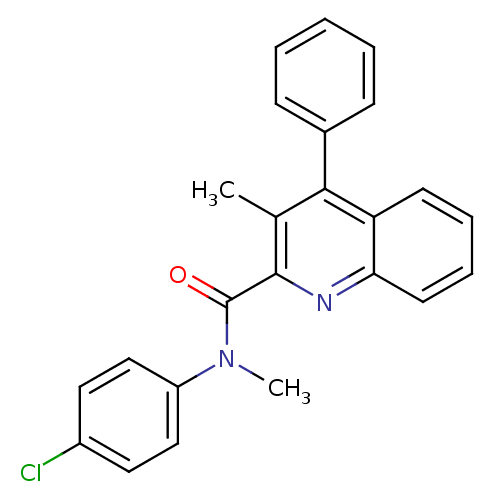
BDBM50059738 3-Methyl-4-phenyl-quinoline-2-carboxylic acid (4-chloro-phenyl)-methyl-amide::CHEMBL275410
SMILES CN(C(=O)c1nc2ccccc2c(c1C)-c1ccccc1)c1ccc(Cl)cc1
InChI Key InChIKey=VITHAIPHWHWXDX-UHFFFAOYSA-N
Data 4 IC50
Activity Spreadsheet -- Enzyme Inhibition Constant Data from BindingDB
 Found 4 hits for monomerid = 50059738
Found 4 hits for monomerid = 50059738
Affinity DataIC50: 6.5nMAssay Description:In vitro binding affinity (pIC50) was tested against peripheral benzodiazepine receptor (PBR) in rat.More data for this Ligand-Target Pair
Affinity DataIC50: 6.40nMAssay Description:Compound was tested for inhibitory activity against Dihydrofolate Reductase(DHFR) of rat liverMore data for this Ligand-Target Pair
Affinity DataIC50: 6.40nMAssay Description:Test in vitro, for potential ability to displace [3H]1 from PBR in rat brain cortexMore data for this Ligand-Target Pair
Affinity DataIC50: 6.40nMAssay Description:In vitro displacement of [3H]PK11195 from the peripheral benzodiazepine receptor (PBR) in rat cortexMore data for this Ligand-Target Pair