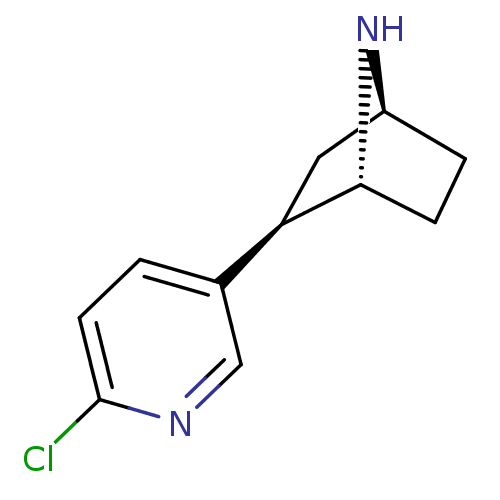
BDBM50143314 (+)-Epibatidine::(-)-epibatidine::(1R,2R,4S)-2-(6-Chloro-pyridin-3-yl)-7-aza-bicyclo[2.2.1]heptane::2-(6-Chloro-pyridin-3-yl)-7-aza-bicyclo[2.2.1]heptane (epibatidine)::CHEMBL298826::EPIBATIDINE
SMILES Clc1ccc(cn1)[C@H]1C[C@@H]2CC[C@H]1N2
InChI Key InChIKey=NLPRAJRHRHZCQQ-IVZWLZJFSA-N
Activity Spreadsheet -- Enzyme Inhibition Constant Data from BindingDB
 Found 2 hits for monomerid = 50143314
Found 2 hits for monomerid = 50143314
Affinity DataKi: 7.40nM ΔG°: -10.9kcal/molepH: 7.4 T: 2°CAssay Description:Assay Description 2 Assays used to generate Ki or EC50 values. 1) SPA Assay - Quick screen binding assays were performed using 100 ul of 0.2 mg/ml an...More data for this Ligand-Target Pair
Affinity DataEC50: 28nMpH: 7.4 T: 2°CAssay Description:Assay Description 2 Assays used to generate Ki or EC50 values. 1) SPA Assay - Quick screen binding assays were performed using 100 ul of 0.2 mg/ml an...More data for this Ligand-Target Pair
 3D Structure (crystal)
3D Structure (crystal)