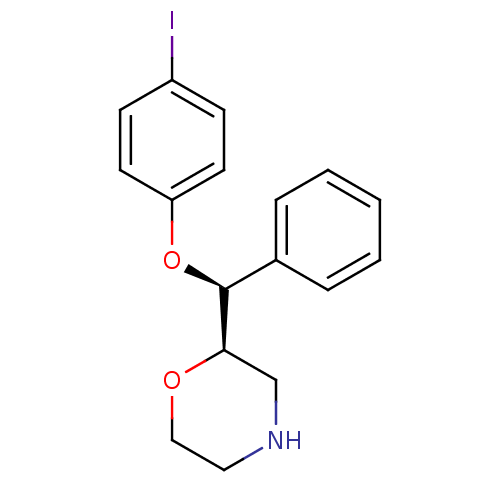
BDBM50301213 (2R,3S)-2-[(4-Iodophenoxy)phenylmethyl]morpholine::CHEMBL579221
SMILES Ic1ccc(O[C@H]([C@H]2CNCCO2)c2ccccc2)cc1
InChI Key InChIKey=SFIZDMBLVRMOCD-SJORKVTESA-N
Data 3 KI
Activity Spreadsheet -- Enzyme Inhibition Constant Data from BindingDB
 Found 3 hits for monomerid = 50301213
Found 3 hits for monomerid = 50301213
TargetSodium-dependent serotonin transporter(Rattus norvegicus (rat))
University Of Glasgow
Curated by ChEMBL
University Of Glasgow
Curated by ChEMBL
Affinity DataKi: 34.5nMAssay Description:Displacement of [3H]citalopram from SERT in Sprague-Dawley rat brain after 2 hrs by liquid scintillation analysisMore data for this Ligand-Target Pair
Affinity DataKi: 1.10E+3nMAssay Description:Displacement of [3H]nisoxetine from NAT in Sprague-Dawley rat brain after 4 hrs by liquid scintillation analysisMore data for this Ligand-Target Pair
TargetSodium-dependent dopamine transporter(Rattus norvegicus (rat))
University Of Glasgow
Curated by ChEMBL
University Of Glasgow
Curated by ChEMBL
Affinity DataKi: 2.10E+3nMAssay Description:Displacement of [3H]WIN-35,428 from DAT in Sprague-Dawley rat brain after 2 hrs by liquid scintillation analysisMore data for this Ligand-Target Pair