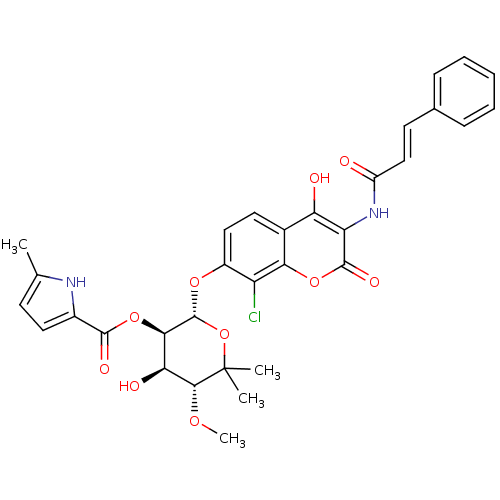
BDBM50330297 (2S,3R,4R,5R)-2-(8-chloro-3-cinnamamido-4-hydroxy-2-oxo-2H-chromen-7-yloxy)-4-hydroxy-5-methoxy-6,6-dimethyltetrahydro-2H-pyran-3-yl 5-methyl-1H-pyrrole-2-carboxylate::CHEMBL1275896
SMILES CO[C@@H]1[C@@H](O)[C@@H](OC(=O)c2ccc(C)[nH]2)[C@H](Oc2ccc3c(O)c(NC(=O)\C=C\c4ccccc4)c(=O)oc3c2Cl)OC1(C)C
InChI Key InChIKey=OHUMOGWHGJTPKY-ULGXMYAKSA-N
Data 2 IC50
Activity Spreadsheet -- Enzyme Inhibition Constant Data from BindingDB
 Found 2 hits for monomerid = 50330297
Found 2 hits for monomerid = 50330297
Affinity DataIC50: 1.20E+3nMAssay Description:Inhibition of Escherichia coli JM109 DNA gyrase subunit B assessed as DNA triple helix formation by supercoiling assayMore data for this Ligand-Target Pair
Affinity DataIC50: 598nMAssay Description:Inhibition of Escherichia coli JM109 DNA gyrase subunit B assessed as ATP hydrolysis by ATPase assayMore data for this Ligand-Target Pair