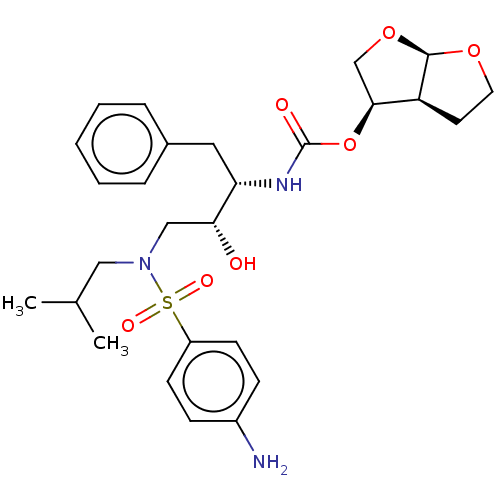
BDBM8125 (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl N-[(2S,3R)-4-[(4-aminobenzene)(2-methylpropyl)sulfonamido]-3-hydroxy-1-phenylbutan-2-yl]carbamate::CHEMBL1323::Darunavir::Darunavir (DRV)::TMC-114::UIC-94017::US10806794, Compound Darunavir
SMILES [H][C@@]1(CO[C@@]2([H])OCC[C@@]12[H])OC(=O)N[C@@H](Cc1ccccc1)[C@H](O)CN(CC(C)C)S(=O)(=O)c1ccc(N)cc1
InChI Key InChIKey=CJBJHOAVZSMMDJ-HEXNFIEUSA-N
Activity Spreadsheet -- Enzyme Inhibition Constant Data from BindingDB
 Found 14 hits for monomerid = 8125
Found 14 hits for monomerid = 8125
TargetDimer of Gag-Pol polyprotein [501-599,Q508K,L534I,L564I,C568A,C596A](Human immunodeficiency virus type 1)
Purdue University
Purdue University
Affinity DataKi: 0.0140nM ΔG°: -14.8kcal/molepH: 6.4 T: 2°CAssay Description:The Ki values were determined by substrate cleavage assay using fluorogenic substrate, 2-(aminobenzoyl)-Thr-Ile-Nle-Phe(p-NO2)-Gln-Arg-NH2. A standar...More data for this Ligand-Target Pair
Affinity DataKi: 0.0160nMAssay Description:Inhibition of HIV1 proteaseMore data for this Ligand-Target Pair
Affinity DataKi: 0.0160nMAssay Description:Inhibition of HIV1 proteaseMore data for this Ligand-Target Pair
Affinity DataKi: 0.0160nMAssay Description:Inhibition of HIV1 wild type NL4-3 protease expressed in Escherichia coli Rosetta (DE3) pLysS using Abz-Thr-Ile-Nle-Phe-(pNO2)-Gln-Arg-NH2 as substra...More data for this Ligand-Target Pair
Affinity DataKi: 0.0160nMAssay Description:Inhibition of HIV-1 protease by fluorometric assayMore data for this Ligand-Target Pair
TargetDimer of Gag-Pol polyprotein [501-599,Q508K,L534I,L564I,C568A,C596A](Human immunodeficiency virus type 1)
Purdue University
Purdue University
Affinity DataKi: 0.0160nM ΔG°: -14.7kcal/mole EC50: 1.60nMpH: 6.4 T: 2°CAssay Description:The Ki values were determined by substrate cleavage assay using fluorogenic substrate, 2-(aminobenzoyl)-Thr-Ile-Nle-Phe(p-NO2)-Gln-Arg-NH2. A standar...More data for this Ligand-Target Pair
Affinity DataKi: 0.0160nMAssay Description:Inhibition of wild type HIV-1 NL4-3 protease expressed in Escherichia coli Rosetta (DE3)pLysS using Ac-Thr-Ile-Nle-Nle-Gln-Arg-NH2 as substrate by fl...More data for this Ligand-Target Pair
Affinity DataKi: 0.0160nMAssay Description:Inhibition of HIV1 proteaseMore data for this Ligand-Target Pair
Affinity DataKi: 0.0160nMAssay Description:Inhibition of HIV1 protease using Ac-Thr-Ile-Nle-Nle-Gln-Arg-NH2 substrate by continuous fluorometric assayMore data for this Ligand-Target Pair
Affinity DataKi: 0.0160nMAssay Description:Inhibition of HIV-1 proteaseMore data for this Ligand-Target Pair
Affinity DataIC50: 1.54E+4nMAssay Description:Inhibition of P-gp in human CMEC/D3 cells using NBD-Aba as substrate after 30 mins by flow cytometric analysisMore data for this Ligand-Target Pair
Affinity DataIC50: 6.60E+3nMAssay Description:Inhibition of P-gp in human CMEC/D3 cells using calcein-AM as substrate after 30 mins by flow cytometric analysisMore data for this Ligand-Target Pair
Affinity DataIC50: 6.00E+4nMAssay Description:Inhibition of P-gp in human 12D7-MDR cells using NBD-Aba as substrate after 30 mins by flow cytometric analysisMore data for this Ligand-Target Pair
Affinity DataIC50: 3.32E+4nMAssay Description:Inhibition of P-gp in human 12D7-MDR cells using calcein-AM as substrate after 30 mins by flow cytometric analysisMore data for this Ligand-Target Pair
 3D Structure (crystal)
3D Structure (crystal)