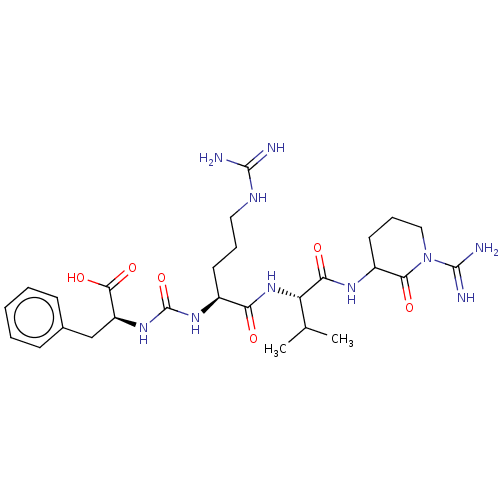
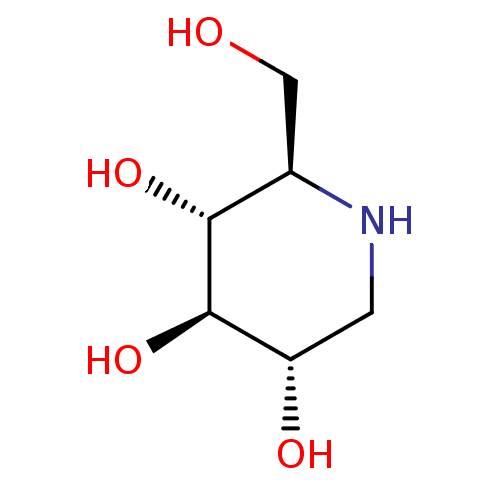
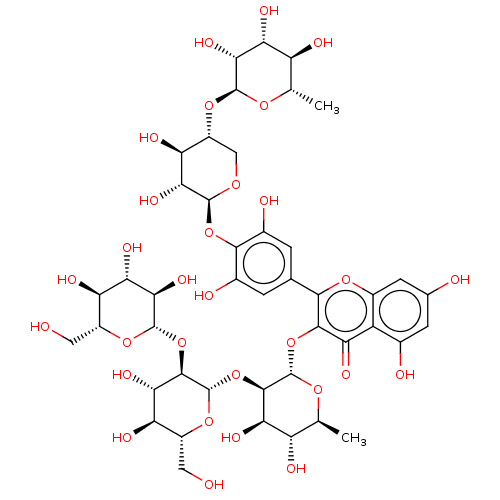
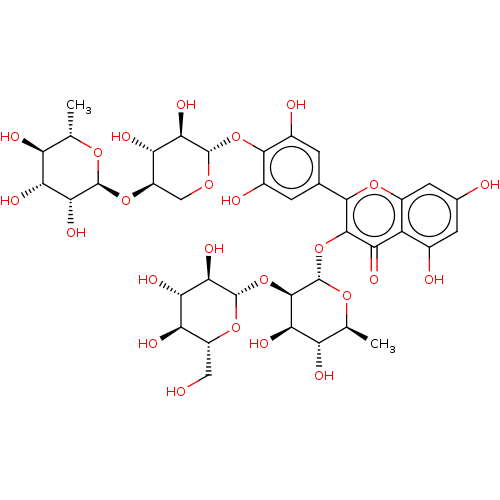
Affinity DataKi: 1nMpH: 7.0Assay Description:The release of 2-chloro-4-nitrophenol resulting from the HPA-catalyzed hydrolysis of 2-chloro-4-nitrophenyl a-maltotrioside (CNPG3) was monitored at ...More data for this Ligand-Target Pair
Affinity DataKi: 2.70nMpH: 7.0Assay Description:The release of 2-chloro-4-nitrophenol resulting from the HPA-catalyzed hydrolysis of 2-chloro-4-nitrophenyl a-maltotrioside (CNPG3) was monitored at ...More data for this Ligand-Target Pair
Affinity DataKi: 7nMpH: 7.0Assay Description:The release of 2-chloro-4-nitrophenol resulting from the HPA-catalyzed hydrolysis of 2-chloro-4-nitrophenyl a-maltotrioside (CNPG3) was monitored at ...More data for this Ligand-Target Pair
Affinity DataKi: 8.10nM ΔG°: -47.0kJ/moleT: 2°CAssay Description:Inhibitor concentrations varied from 0.25 to 5 times the Ki value in 50 mM sodium phosphate, 100 mM sodium chloride buffer, pH 7.0, at 30 °C. 2-Chlor...More data for this Ligand-Target Pair
Affinity DataKi: 9.10nM ΔG°: -46.7kJ/moleT: 2°CAssay Description:Inhibitor concentrations varied from 0.25 to 5 times the Ki value in 50 mM sodium phosphate, 100 mM sodium chloride buffer, pH 7.0, at 30 °C. 2-Chlor...More data for this Ligand-Target Pair
Affinity DataKi: 11nMAssay Description:Competitive inhibition of human recombinant cathepsin K using Z-FR-MCA fluorogenic substrate by Handerson plot analysisMore data for this Ligand-Target Pair
Affinity DataKi: 21.3nM ΔG°: -44.5kJ/moleT: 2°CAssay Description:Inhibitor concentrations varied from 0.25 to 5 times the Ki value in 50 mM sodium phosphate, 100 mM sodium chloride buffer, pH 7.0, at 30 °C. 2-Chlor...More data for this Ligand-Target Pair
Affinity DataKi: 41nMAssay Description:Competitive inhibition of human recombinant cathepsin K using Z-FR-MCA fluorogenic substrate by Handerson plot analysisMore data for this Ligand-Target Pair
Affinity DataKi: 42.4nM ΔG°: -42.8kJ/moleT: 2°CAssay Description:Inhibitor concentrations varied from 0.25 to 5 times the Ki value in 50 mM sodium phosphate, 100 mM sodium chloride buffer, pH 7.0, at 30 °C. 2-Chlor...More data for this Ligand-Target Pair
Affinity DataKi: 79.3nM ΔG°: -41.2kJ/moleT: 2°CAssay Description:Inhibitor concentrations varied from 0.25 to 5 times the Ki value in 50 mM sodium phosphate, 100 mM sodium chloride buffer, pH 7.0, at 30 °C. 2-Chlor...More data for this Ligand-Target Pair
Affinity DataKi: 93.3nM ΔG°: -40.8kJ/moleT: 2°CAssay Description:Inhibitor concentrations varied from 0.25 to 5 times the Ki value in 50 mM sodium phosphate, 100 mM sodium chloride buffer, pH 7.0, at 30 °C. 2-Chlor...More data for this Ligand-Target Pair
Affinity DataKi: 105nMAssay Description:Competitive inhibition of human recombinant cathepsin K using Z-FR-MCA fluorogenic substrate by Dixon plot analysisMore data for this Ligand-Target Pair
Affinity DataKi: 105nMAssay Description:Competitive inhibition of human recombinant cathepsin K using Z-FR-MCA fluorogenic substrate by Dixon plot analysisMore data for this Ligand-Target Pair
Affinity DataKi: 163nMAssay Description:Competitive inhibition of human recombinant cathepsin K using Z-FR-MCA fluorogenic substrate by Dixon plot analysisMore data for this Ligand-Target Pair
Affinity DataKi: 250nMAssay Description:The inhibition of TreS by a range of known alpha-glucosidase inhibitor was assayed.More data for this Ligand-Target Pair
Affinity DataKi: 393nMAssay Description:Competitive inhibition of human recombinant cathepsin K using Z-FR-MCA fluorogenic substrate by Dixon plot analysisMore data for this Ligand-Target Pair
Affinity DataKi: 730nM ΔG°: -35.6kJ/moleT: 2°CAssay Description:Inhibitor concentrations varied from 0.25 to 5 times the Ki value in 50 mM sodium phosphate, 100 mM sodium chloride buffer, pH 7.0, at 30 °C. 2-Chlor...More data for this Ligand-Target Pair
Affinity DataKi: 2.24E+3nM ΔG°: -32.8kJ/moleT: 2°CAssay Description:Inhibitor concentrations varied from 0.25 to 5 times the Ki value in 50 mM sodium phosphate, 100 mM sodium chloride buffer, pH 7.0, at 30 °C. 2-Chlor...More data for this Ligand-Target Pair
Affinity DataKi: 2.50E+3nMAssay Description:The inhibition of TreS by a range of known alpha-glucosidase inhibitor was assayed.More data for this Ligand-Target Pair
Affinity DataKi: 1.39E+4nM ΔG°: -28.2kJ/moleT: 2°CAssay Description:Inhibitor concentrations varied from 0.25 to 5 times the Ki value in 50 mM sodium phosphate, 100 mM sodium chloride buffer, pH 7.0, at 30 °C. 2-Chlor...More data for this Ligand-Target Pair
Affinity DataKi: 4.68E+4nM ΔG°: -25.1kJ/moleT: 2°CAssay Description:Inhibitor concentrations varied from 0.25 to 5 times the Ki value in 50 mM sodium phosphate, 100 mM sodium chloride buffer, pH 7.0, at 30 °C. 2-Chlor...More data for this Ligand-Target Pair
Affinity DataKi: 1.28E+5nM ΔG°: -22.6kJ/moleT: 2°CAssay Description:Inhibitor concentrations varied from 0.25 to 5 times the Ki value in 50 mM sodium phosphate, 100 mM sodium chloride buffer, pH 7.0, at 30 °C. 2-Chlor...More data for this Ligand-Target Pair
Affinity DataKi: 1.40E+5nMAssay Description:The inhibition of TreS by a range of known alpha-glucosidase inhibitor was assayed.More data for this Ligand-Target Pair
Affinity DataIC50: 8.5nMpH: 7.0Assay Description:After kinetic analysis with HPA, piHA-Dm was tested as an inhibitor of ten additional enzymes. The conditions are listed as follows: Agrobacterium fa...More data for this Ligand-Target Pair
Affinity DataIC50: 34nMpH: 7.0Assay Description:After kinetic analysis with HPA, piHA-Dm was tested as an inhibitor of ten additional enzymes. The conditions are listed as follows: Agrobacterium fa...More data for this Ligand-Target Pair
Affinity DataIC50: 520nMpH: 7.0Assay Description:After kinetic analysis with HPA, piHA-Dm was tested as an inhibitor of ten additional enzymes. The conditions are listed as follows: Agrobacterium fa...More data for this Ligand-Target Pair
Affinity DataIC50: 990nMpH: 7.0Assay Description:After kinetic analysis with HPA, piHA-Dm was tested as an inhibitor of ten additional enzymes. The conditions are listed as follows: Agrobacterium fa...More data for this Ligand-Target Pair

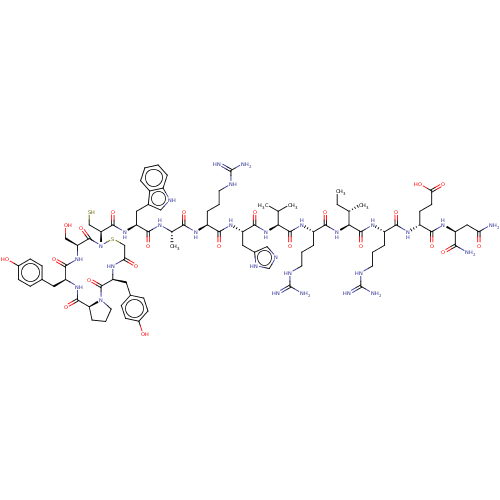
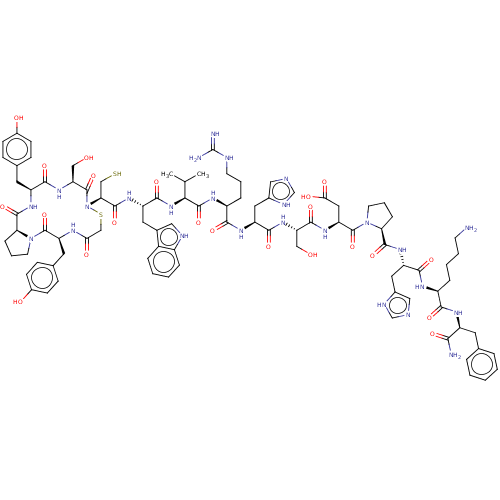


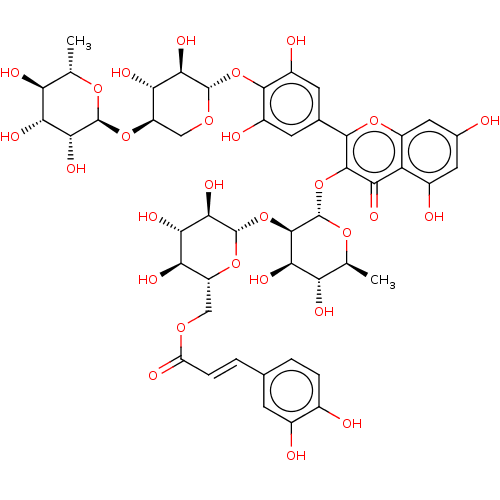
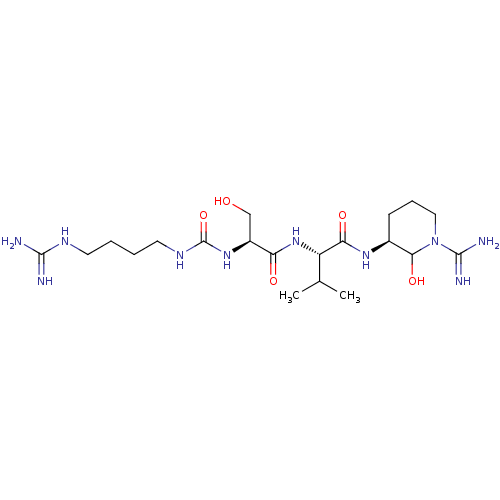

 3D Structure (crystal)
3D Structure (crystal)