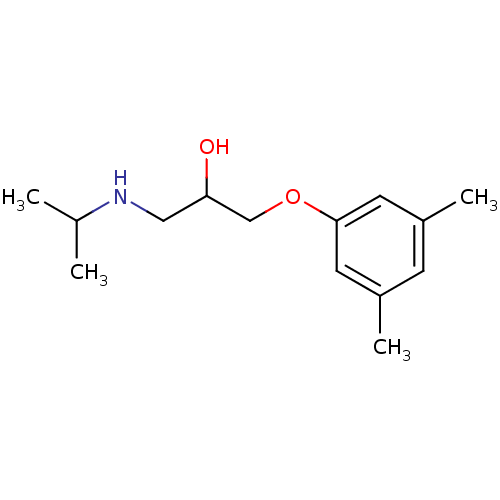
 CHEMBL1159714 Ko 707 BDBM50421719
CHEMBL1159714 Ko 707 BDBM50421719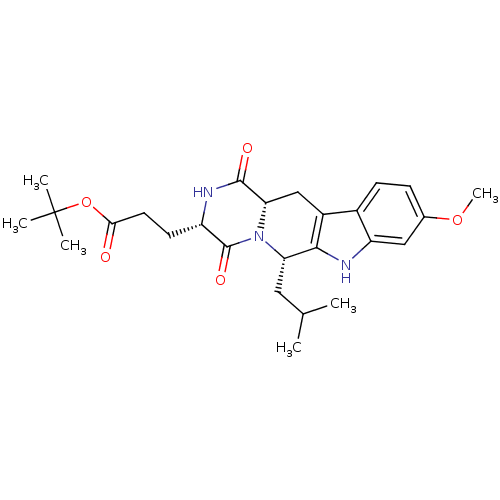 3-((3S,6S)-6-Isobutyl-9-methoxy-1,4-dioxo-1,2,3,4,6,7,12,12a-octahydro-pyrazino[1',2':1,6]pyrido[3,4-b]indol-3-yl)-propionic acid tert-butyl ester BDBM50305083 3-((3S,6S,12aS)-6-Isobutyl-9-methoxy-1,4-dioxo-1,2,3,4,6,7,12,12a-octahydro-pyrazino[1',2':1,6]pyrido[3,4-b]indol-3-yl)-propionic acid tert-butyl ester Ko143 Ko-143 US9695174, Ko143 CHEMBL488910
3-((3S,6S)-6-Isobutyl-9-methoxy-1,4-dioxo-1,2,3,4,6,7,12,12a-octahydro-pyrazino[1',2':1,6]pyrido[3,4-b]indol-3-yl)-propionic acid tert-butyl ester BDBM50305083 3-((3S,6S,12aS)-6-Isobutyl-9-methoxy-1,4-dioxo-1,2,3,4,6,7,12,12a-octahydro-pyrazino[1',2':1,6]pyrido[3,4-b]indol-3-yl)-propionic acid tert-butyl ester Ko143 Ko-143 US9695174, Ko143 CHEMBL488910
- Fuchss, T; Schiemann, K Imidazolonylquinolines and the use thereof as ATM kinase inhibitors US Patent US10457677 (2019)
- Toledo-Sherman, LM; Dominguez, C; Breccia, P; Van De Poël, AJ; Wishart, G; Vater, HD; Esmieu, WR; Clissold, C; Blackaby, WP ATM kinase inhibitors and compositions and methods of use thereof US Patent US11685734 (2023)
- ZHOU, D; CHENG, Z; WANG, Z SELECTIVE MODULATORS OF ATAXIA TELANGIECTASIA MUTATED (ATM) KINASE AND USES THEREOF US Patent US20240207263 (2024)
- Van de Poël, A; Toledo-Sherman, L; Breccia, P; Cachope, R; Bate, JR; Angulo-Herrera, I; Wishart, G; Matthews, KL; Martin, SL; Peacock, M; Barnard, A; Cox, HC; Jones, G; McAllister, G; Vater, H; Esmieu, W; Clissold, C; Lamers, M; Leonard, P; Jarvis, RE; Blackaby, W; Eznarriaga, M; Lazari, O; Yates, D; Rose, M; Jang, SW; Muñoz-Sanjuan, I; Dominguez, C Structure-Based Exploration of Selectivity for ATM Inhibitors in Huntington's Disease. J Med Chem 64: 5018-5036 (2021)
- Fu, J; Wang, Y; Sun, Y; Wu, G; Lu, A; Zhang, S; Goodnow, R; Gilmer, T; Kastan, M; Kirsch, D Dual ATM and DNA-PK inhibitors for use in anti-tumor therapy US Patent US12187742 (2025)
- Laufer, S; Forster, M; Dimitrov, T; Zender, L; Moschopoulou, A IMIDAZO[4,5-C]QUINOLINE COMPOUNDS AND THEIR USE AS ATM KINASE INHIBITORS US Patent US20230348462 (2023)
- Small Molecular Inhibitors That Target ATM for Drug Discovery: Current Research and Potential Prospective.
- Dimitrov, T; Anli, C; Moschopoulou, AA; Kronenberger, T; Kudolo, M; Geibel, C; Schwalm, MP; Knapp, S; Zender, L; Forster, M; Laufer, S Development of novel urea-based ATM kinase inhibitors with subnanomolar cellular potency and high kinome selectivity. Eur J Med Chem 235: (2022)
- Barlaam, B; Cadogan, E; Campbell, A; Colclough, N; Dishington, A; Durant, S; Goldberg, K; Hassall, LA; Hughes, GD; MacFaul, PA; McGuire, TM; Pass, M; Patel, A; Pearson, S; Petersen, J; Pike, KG; Robb, G; Stratton, N; Xin, G; Zhai, B Discovery of a Series of 3-Cinnoline Carboxamides as Orally Bioavailable, Highly Potent, and Selective ATM Inhibitors. ACS Med Chem Lett 9: 809-814 (2018)
- Dou, X; Sun, X; Huang, H; Jiang, L; Jin, Z; Liu, Y; Zou, Y; Li, Z; Zhu, G; Jin, H; Jiao, N; Zhang, L; Liu, Z; Zhang, L Discovery of novel ataxia telangiectasia mutated (ATM) kinase modulators: Computational simulation, biological evaluation and cancer combinational chemotherapy study. Eur J Med Chem 233: (2022)
- Degorce, SL; Barlaam, B; Cadogan, E; Dishington, A; Ducray, R; Glossop, SC; Hassall, LA; Lach, F; Lau, A; McGuire, TM; Nowak, T; Ouvry, G; Pike, KG; Thomason, AG Discovery of Novel 3-Quinoline Carboxamides as Potent, Selective, and Orally Bioavailable Inhibitors of Ataxia Telangiectasia Mutated (ATM) Kinase. J Med Chem 59: 6281-92 (2016)
- Rescue of fragile X syndrome phenotypes in Fmr1 KO mice by the small-molecule PAK inhibitor FRAX486.
- Discovery of a Meisoindigo-Derived PROTAC as the ATM Degrader: Revolutionizing Colorectal Cancer Therapy via Synthetic Lethality with ATR Inhibitors.
- Zhou, D; Cheng, Z 1-isopropyl-3-methyl-8-(pyridin-3-yl)-1,3-dihydro-2H-imidazo[4,5-c] cinnolin-2-one as selective modulators of ataxia telangiectasia mutated (ATM) kinase and uses thereof US Patent US12226414 (2025)
- Deng, D; Yang, Y; Zou, Y; Liu, K; Zhang, C; Tang, M; Yang, T; Chen, Y; Yuan, X; Guo, Y; Zhang, S; Si, W; Peng, B; Xu, Q; He, W; Xu, D; Xiang, M; Chen, L Discovery and Evaluation of 3-Quinoxalin Urea Derivatives as Potent, Selective, and Orally Available ATM Inhibitors Combined with Chemotherapy for the Treatment of Cancer via Goal-Oriented Molecule Generation and Virtual Screening. J Med Chem 66: 9495-9518 (2023)
- Pike, KG; Barlaam, B; Cadogan, E; Campbell, A; Chen, Y; Colclough, N; Davies, NL; de-Almeida, C; Degorce, SL; Didelot, M; Dishington, A; Ducray, R; Durant, ST; Hassall, LA; Holmes, J; Hughes, GD; MacFaul, PA; Mulholland, KR; McGuire, TM; Ouvry, G; Pass, M; Robb, G; Stratton, N; Wang, Z; Wilson, J; Zhai, B; Zhao, K; Al-Huniti, N The Identification of Potent, Selective, and Orally Available Inhibitors of Ataxia Telangiectasia Mutated (ATM) Kinase: The Discovery of AZD0156 (8-{6-[3-(Dimethylamino)propoxy]pyridin-3-yl}-3-methyl-1-(tetrahydro-2 H-pyran-4-yl)-1,3-dihydro-2 H-imidazo[4,5- c]quinolin-2-one). J Med Chem 61: 3823-3841 (2018)
- Edmonds, DJ; Kung, DW; Kalgutkar, AS; Filipski, KJ; Ebner, DC; Cabral, S; Smith, AC; Aspnes, GE; Bhattacharya, SK; Borzilleri, KA; Brown, JA; Calabrese, MF; Caspers, NL; Cokorinos, EC; Conn, EL; Dowling, MS; Eng, H; Feng, B; Fernando, DP; Genung, NE; Herr, M; Kurumbail, RG; Lavergne, SY; Lee, EC; Li, Q; Mathialagan, S; Miller, RA; Panteleev, J; Polivkova, J; Rajamohan, F; Reyes, AR; Salatto, CT; Shavnya, A; Thuma, BA; Tu, M; Ward, J; Withka, JM; Xiao, J; Cameron, KO J Med Chem 61: 2372-2383 (2018)
- Mandadapu, SR; Weerawarna, PM; Prior, AM; Uy, RA; Aravapalli, S; Alliston, KR; Lushington, GH; Kim, Y; Hua, DH; Chang, KO; Groutas, WC Bioorg Med Chem Lett 23: 3709-12 (2013)
- Clausen, JD; Kjellerup, L; Cohrt, KO; Hansen, JB; Dalby-Brown, W; Winther, AL Bioorg Med Chem Lett 27: 4564-4570 (2017)
- Wang, F; Jeon, KO; Salovich, JM; Macdonald, JD; Alvarado, J; Gogliotti, RD; Phan, J; Olejniczak, ET; Sun, Q; Wang, S; Camper, D; Yuh, JP; Shaw, JG; Sai, J; Rossanese, OW; Tansey, WP; Stauffer, SR; Fesik, SW J Med Chem 61: 5623-5642 (2018)
- Zhao, B; Bower, MJ; McDevitt, PJ; Zhao, H; Davis, ST; Johanson, KO; Green, SM; Concha, NO; Zhou, BB J Biol Chem 277: 46609-15 (2002)
- Zankel, TC; Isbell, SL; Ko, AA US Patent US10308607 (2019)
- Ko, B; Jang, Y; Kwak, SH; You, H; Kim, JH; Lee, JE; Park, HD; Kim, SK; Goddard, WA; Han, JH; Kim, YC J Med Chem 66: 14564-14582 (2023)
- Li, Z; Liao, C; Ko, BC; Shan, S; Tong, EH; Yin, Z; Pan, D; Wong, VK; Shi, L; Ning, ZQ; Hu, W; Zhou, J; Chung, SS; Lu, XP Bioorg Med Chem Lett 14: 3507-11 (2004)
- Ko, CC; Chen, YJ; Chen, CT; Liu, YC; Cheng, FC; Hsu, KC; Chow, LP J Biol Chem 289: 22078-89 (2014)
- Chen, CH; Lee, O; Yao, CN; Chuang, MY; Chang, YL; Chang, MH; Wen, YF; Yang, WH; Ko, CH; Chou, NT; Lin, MW; Lai, CP; Sun, CY; Wang, LM; Chen, YC; Hseu, TH; Chang, CN; Hsu, HC; Lin, HC; Chang, YL; Shih, YC; Chou, SH; Hsu, YL; Tseng, HW; Liu, CP; Tu, CM; Hu, TL; Tsai, YJ; Chen, TS; Lin, CL; Chiou, SJ; Liu, CC; Hwang, CS Bioorg Med Chem Lett 20: 6129-32 (2010)
- Ratni, H; Ebeling, M; Baird, J; Bendels, S; Bylund, J; Chen, KS; Denk, N; Feng, Z; Green, L; Guerard, M; Jablonski, P; Jacobsen, B; Khwaja, O; Kletzl, H; Ko, CP; Kustermann, S; Marquet, A; Metzger, F; Mueller, B; Naryshkin, NA; Paushkin, SV; Pinard, E; Poirier, A; Reutlinger, M; Weetall, M; Zeller, A; Zhao, X; Mueller, L J Med Chem 61: 6501-6517 (2018)
- Nam, Y; Ryu, KD; Jang, C; Moon, YH; Kim, M; Ko, D; Chung, KS; Gandini, MA; Lee, KT; Zamponi, GW; Lee, JY Bioorg Med Chem 28: (2020)
- Kim, YK; Kwon, O; Park, H; Park, J; Choi, HG; Son, JB; Ko, E; Kim, SY; Lee, S; Kang, SY; Ko, YK; Park, J US Patent US11447480 (2022)
- Brand, S; Ko, EJ; Viayna, E; Thompson, S; Spinks, D; Thomas, M; Sandberg, L; Francisco, AF; Jayawardhana, S; Smith, VC; Jansen, C; De Rycker, M; Thomas, J; MacLean, L; Osuna-Cabello, M; Riley, J; Scullion, P; Stojanovski, L; Simeons, FRC; Epemolu, O; Shishikura, Y; Crouch, SD; Bakshi, TS; Nixon, CJ; Reid, IH; Hill, AP; Underwood, TZ; Hindley, SJ; Robinson, SA; Kelly, JM; Fiandor, JM; Wyatt, PG; Marco, M; Miles, TJ; Read, KD; Gilbert, IH J Med Chem 60: 7284-7299 (2017)
- Cheng, MC; Li, CY; Ko, HC; Ko, FN; Lin, YL; Wu, TS J Nat Prod 69: 1305-9 (2006)
- Hillmann, P; Ko, GY; Spinrath, A; Raulf, A; von Kügelgen, I; Wolff, SC; Nicholas, RA; Kostenis, E; Höltje, HD; Müller, CE J Med Chem 52: 2762-75 (2009)
- Lee, WG; Lee, SD; Cho, JH; Jung, Y; Kim, JH; Hien, TT; Kang, KW; Ko, H; Kim, YC J Med Chem 55: 3687-98 (2012)
- Cheng, MC; Li, CY; Ko, HC; Ko, FN; Lin, YL; Wu, TS J Nat Prod 69: 1305-9 (2006)
- Ko, HH; Weng, JR; Tsao, LT; Yen, MH; Wang, JP; Lin, CN Bioorg Med Chem Lett 14: 1011-4 (2004)
- Kwon, SH; Kim, S; Park, AY; Lee, S; Gadhe, CG; Seo, BA; Park, JS; Jo, S; Oh, Y; Kweon, SH; Ma, SX; Kim, WR; Kim, M; Kim, H; Kim, JE; Lee, S; Lee, J; Ko, HS J Med Chem 64: 15091-15110 (2021)
- Ko HY
- Ko, J; Hwang, H; Chin, J; Hahn, D; Lee, J; Yang, I; Shin, K; Ham, J; Kang, H Bioorg Med Chem Lett 20: 6017-9 (2010)
- Bhattarai, BR; Ko, JH; Shrestha, S; Kafle, B; Cho, H; Kang, JH; Cho, H Bioorg Med Chem Lett 20: 1075-7 (2010)
- Ko, K; Kim, HJ; Ho, PS; Lee, SO; Lee, JE; Min, CR; Kim, YC; Yoon, JH; Park, EJ; Kwon, YJ; Yun, JH; Yoon, DO; Kim, JS; Park, WS; Oh, SS; Song, YM; Cho, WK; Morikawa, K; Lee, KJ; Park, CH J Med Chem 61: 2949-2961 (2018)
- Lee, W; Ko, KR; Kim, HK; Lee, DS; Nam, IJ; Lim, S; Kim, S J Nat Prod 81: 1343-1356 (2018)
- Ko, KS; Steffey, ME; Brandvold, KR; Soellner, MB ACS Med Chem Lett 4: 779-783 (2013)
- Yang, HY; Tae, J; Seo, YW; Kim, YJ; Im, HY; Choi, GD; Cho, H; Park, WK; Kwon, OS; Cho, YS; Ko, M; Jang, H; Lee, J; Choi, K; Kim, CH; Lee, J; Pae, AN Eur J Med Chem 63: 558-69 (2013)
- Nam, M; Kim, T; Kwak, J; Seo, SH; Ko, MK; Lim, EJ; Min, SJ; Cho, YS; Keum, G; Baek, DJ; Lee, J; Pae, AN Eur J Med Chem 97: 245-58 (2015)
- Jackson, JJ; Shibuya, GM; Ravishankar, B; Adusumilli, L; Bradford, D; Brockstedt, DG; Bucher, C; Bui, M; Cho, C; Colas, C; Cutler, G; Dukes, A; Han, X; Hu, DX; Jacobson, S; Kassner, PD; Katibah, GE; Ko, MYM; Kolhatkar, U; Leger, PR; Ma, A; Marshall, L; Maung, J; Ng, AA; Okano, A; Pookot, D; Poon, D; Ramana, C; Reilly, MK; Robles, O; Schwarz, JB; Shakhmin, AA; Shunatona, HP; Sreenivasan, R; Tivitmahaisoon, P; Xu, M; Zaw, T; Wustrow, DJ; Zibinsky, M J Med Chem 65: 12895-12924 (2022)
- Srivastava, AS; Ko, S; Watterson, SH; Pattoli, MA; Skala, S; Cheng, L; Obermeier, MT; Vickery, R; Discenza, LN; D'Arienzo, CJ; Gillooly, KM; Taylor, TL; Pulicicchio, C; McIntyre, KW; Yip, S; Li, P; Sun, D; Wu, DR; Dai, J; Wang, C; Zhang, Y; Wang, B; Pawluczyk, J; Kempson, J; Zhao, R; Hou, X; Rampulla, R; Mathur, A; Galella, MA; Salter-Cid, L; Barrish, JC; Carter, PH; Fura, A; Burke, JR; Tino, JA ACS Med Chem Lett 11: 2195-2203 (2020)
- Lim, CJ; Woo, SE; Ko, SI; Lee, BH; Oh, KS; Yi, KY Bioorg Med Chem Lett 26: 4684-4686 (2016)
- Hwang, GJ; Jang, M; Son, S; Lee, B; Jang, JP; Lee, JS; Ko, SK; Hong, YS; Ahn, JS; Jang, JH J Nat Prod 84: 2420-2426 (2021)
- Batt, DG; Bertrand, MB; Delucca, G; Galella, MA; Ko, SS; Langevine, CM; Liu, Q; Shi, Q; Srivastava, AS; Tino, JA; Watterson, SH US Patent US9334290 (2016)
- Walpole, C; Ko, SY; Brown, M; Beattie, D; Campbell, E; Dickenson, F; Ewan, S; Hughes, GA; Lemaire, M; Lerpiniere, J; Patel, S; Urban, L J Med Chem 41: 3159-73 (1998)
- K-M Chen, C; Hudock, MP; Zhang, Y; Guo, RT; Cao, R; No, JH; Liang, PH; Ko, TP; Chang, TH; Chang, SC; Song, Y; Axelson, J; Kumar, A; Wang, AH; Oldfield, E J Med Chem 51: 5594-607 (2008)
- Quang, TH; Ngan, NT; Ko, W; Kim, DC; Yoon, CS; Sohn, JH; Yim, JH; Kim, YC; Oh, H Bioorg Med Chem Lett 24: 5787-91 (2014)
- Ryu, CK; Kang, HY; Lee, SK; Nam, KA; Hong, CY; Ko, WG; Lee, BH Bioorg Med Chem Lett 10: 461-4 (2000)
- Ko Y
- Jeon, WS; Moon, K; Park, SH; Chun, H; Ko, YH; Lee, JY; Lee, ES; Samal, S; Selvapalam, N; Rekharsky, MV; Sindelar, V; Sobransingh, D; Inoue, Y; Kaifer, AE; Kim, K J Am Chem Soc 127: 12984-9 (2005)
- Kim, YK; Kwon, O; Park, H; Park, J; Choi, HG; Son, JB; Ko, E; Kim, SY; Lee, S; Kang, SY; Ko, YK; Park, J US Patent US11447480 (2022)
- Jackson, PF; Tays, KL; Maclin, KM; Ko, YS; Li, W; Vitharana, D; Tsukamoto, T; Stoermer, D; Lu, XC; Wozniak, K; Slusher, BS J Med Chem 44: 4170-5 (2001)
- Kim, NJ; Lee, KO; Koo, BW; Li, F; Yoo, JK; Park, HJ; Min, KH; Lim, JI; Kim, MK; Kim, JK; Suh, YG Bioorg Med Chem Lett 17: 3595-8 (2007)
- Mederski, WW; Dorsch, D; Osswald, M; Beier, N; Lues, I; Minck, KO; Schelling, P; Ladstetter, BJ Bioorg Med Chem Lett 5: 2665-2670 (1995)
- Shaaban, MA; Elshaier, YAMM; Hammad, AH; Farag, NA; Hassan Haredy, H; AbdEl-Ghany, AA; Mohamed, KO Bioorg Med Chem Lett 30: (2020)
- Mohammed, KO; Nissan, YM Chem Biol Drug Des 84: 473-88 (2014)
- Bugge, S; Moen, IU; Sylte, KO; Sundby, E; Hoff, BH Eur J Med Chem 94: 175-94 (2015)
- Narayanan, D; Tran, KT; Pallesen, JS; Solbak, SMØ; Qin, Y; Mukminova, E; Luchini, M; Vasilyeva, KO; González Chichón, D; Goutsiou, G; Poulsen, C; Haapanen, N; Popowicz, GM; Sattler, M; Olagnier, D; Gajhede, M; Bach, A J Med Chem 65: 14481-14526 (2022)
- Yerdelen, KO; Koca, M; Anil, B; Sevindik, H; Kasap, Z; Halici, Z; Turkaydin, K; Gunesacar, G Bioorg Med Chem Lett 25: 5576-82 (2015)
- Pontius, A; Krick, A; Mesry, R; Kehraus, S; Foegen, SE; Mu¨ller, M; Klimo, K; Gerha¨user, C; Ko¨nig, GM J Nat Prod 71: 1793-1799 (2008)
- Mattei, P; Boehringer, M; Di Giorgio, P; Fischer, H; Hennig, M; Huwyler, J; Koçer, B; Kuhn, B; Loeffler, BM; Macdonald, A; Narquizian, R; Rauber, E; Sebokova, E; Sprecher, U Bioorg Med Chem Lett 20: 1109-13 (2010)
- Bosnar, M; Kragol, G; Koštrun, S; Vujasinovic, I; Bošnjak, B; Bencetic Mihaljevic, V; Marušic Ištuk, Z; Kapic, S; Hrvacic, B; Brajša, K; Tavcar, B; Jelic, D; Glojnaric, I; Verbanac, D; Culic, O; Padovan, J; Alihodžic, S; Erakovic Haber, V; Spaventi, R J Med Chem 55: 6111-23 (2012)
- Schwardt, O; Rabbani, S; Hartmann, M; Abgottspon, D; Wittwer, M; Kleeb, S; Zalewski, A; Smieško, M; Cutting, B; Ernst, B Bioorg Med Chem 19: 6454-73 (2011)
- ZakoSek, M; Mihevc, SP; Majdic, G; Ko{hacek over (s)}ak, U; Gobec, S US Patent US20230331674 (2023)
- ChEMBL_2447239 Inhibition of human ATM
- ChEMBL_473775 (CHEMBL937043) Inhibition of ATM
- ChEMBL_596682 (CHEMBL1049652) Inhibition of ATM
- ChEMBL_660433 (CHEMBL1250023) Inhibition of ATM
- ChEMBL_880958 (CHEMBL2216370) Inhibition of ATM
- ChEMBL_2265605 Inhibition of ATM (unknown origin)
- ChEMBL_2340346 Inhibition of ATM (unknown origin)
- ChEMBL_2426873 Inhibition of ATM (unknown origin)
- ChEMBL_2482440 Inhibition of ATM (unknown origin)
- ChEMBL_2545615 Inhibition of ATM (unknown origin)
- ChEMBL_429365 (CHEMBL916757) Inhibition of ATM kinase
- ChEMBL_1445018 (CHEMBL3372426) Inhibition of ATM (unknown origin)
- ChEMBL_1450109 (CHEMBL3374531) Inhibition of ATM (unknown origin)
- ChEMBL_1501974 (CHEMBL3588381) Inhibition of ATM (unknown origin)
- ChEMBL_1556176 (CHEMBL3768630) Inhibition of ATM (unknown origin)
- ChEMBL_2093846 (CHEMBL4775109) Inhibition of ATM (unknown origin)
- ChEMBL_2214591 (CHEMBL5127723) Inhibition of ATM (unknown origin)
- ChEMBL_2265604 Binding affinity to ATM (unknown origin)
- ChEMBL_978828 (CHEMBL2423430) Inhibition of ATM (unknown origin)
- ChEMBL_2093853 (CHEMBL4775116) Inhibition of ATM (unknown origin) assessed as ATM-dependent phosphorylation using GST-p53 ser15 as substrate
- ChEMBL_2229376 (CHEMBL5142889) Inhibition of ATM in human HT-29 cells assessed as reduction in ATM autophosphorylation at S1981 residue
- ChEMBL_2430628 Inhibition of ATM (unknown origin) HTRF assay
- ChEMBL_1672997 (CHEMBL4023026) Inhibition of full length recombinant human ATM
- ChEMBL_2340345 Inhibition of ATM (unknown origin) by ELISA method
- ChEMBL_1460642 (CHEMBL3395883) Inhibition of ATM kinase in human glioma cells
- ChEMBL_2028061 (CHEMBL4682219) Inhibition of ATM (unknown origin) by biochemical assay
- ChEMBL_2248170 (CHEMBL5162380) Inhibition of ATM (unknown origin) by FRET assay
- ChEMBL_305596 (CHEMBL828043) Inhibition of ATM kinase using rabbit polyclonal antisera
- ChEMBL_2482434 Binding affinity to ATM (unknown origin) assessed as inhibition constant
- ChEMBL_305814 (CHEMBL829467) Inhibition of Mutated in ataxia telangiectasia protein ATM kinase
- ChEMBL_2332806 Inhibition of human ATM kinase using p53 as substrate by FRET assay
- ChEMBL_2246001 (CHEMBL5160211) Competitive inhibition of ATM (unknown origin) measured by ATP-competitive binding assay
- ChEMBL_940917 (CHEMBL2330563) Inhibition of ATM-mediated autophosphorylation at serine 1981 in human HT29 cells
- ChEMBL_967888 (CHEMBL2400704) Inhibition of ATM (unknown origin) phosphorylation at Ser1981 by cell-based assay
- ChEMBL_1556177 (CHEMBL3768631) Inhibition of flag-tagged ATM (unknown origin) using p53 as substrate by ELISA
- ChEMBL_2028062 (CHEMBL4682220) Inhibition of ATM in human MCF7 cells assessed as reduction in KAP1 phosphorylation
- ChEMBL_1444859 (CHEMBL3379629) Inhibition of ATM (unknown origin) using p53-Q10-K17 peptide substrate by alphascreen assay
- ChEMBL_1556115 (CHEMBL3768359) Inhibition of ATM kinase in human MCF7 cells after 1 hr by immunofluorescence assay
- ChEMBL_1556174 (CHEMBL3768628) Inhibition of human ATM using p53 as substrate preincubated for 10 mins by ELISA
- ChEMBL_2332816 Inhibition of ATM kinase in human A549 cells in presence of etoposide by ICW assay
- ChEMBL_2514127 Binding affinity to human ATM incubated for 45 mins by Kinobead based pull down assay
- ChEMBL_743383 (CHEMBL1767682) Inhibition of full length recombinant ATM after 24 hrs by radiometric phosphate incorporation assay
- ChEMBL_1797586 (CHEMBL4269703) Inhibition of ATM phosphorylation at Ser1981 residue in human HT-29 cells using ionizing radiation
- ChEMBL_1880181 (CHEMBL4381575) Inhibition of ATM (unknown origin) assessed as reduction in CHK2 phosphorylation by cell based assay
- ChEMBL_2427094 Inhibition of human full length recombinant ATM using GST-cMyc-p53 as substrate by ELISA method
- ChEMBL_2430627 Binding affinity to recombinant ATM (unknown origin) assessed as dissociation constant by surface plasmon resonance assay
- ChEMBL_1556175 (CHEMBL3768629) Inhibition of ATM in human U2OS cells assessed as inhibition of p53 phosphorylation at Ser15 residue
- ChEMBL_1809509 (CHEMBL4308969) Inhibition of ATM in human HCT116 cells assessed as after 1 hr by Western blot analysis
- ChEMBL_2476844 Inhibition of tetracycline-inducible FLAG-tagged human PARL stably transfected in HEK293T harboring FITR/PARL KO
- ChEMBL_873195 (CHEMBL2183783) Inhibition of ATM isolated from human HeLa cell extract using glutathione S-transferase-p53N66 as substrate by ELISA
- ChEMBL_480051 (CHEMBL927990) Inhibition of mouse PKCtheta in KO cells assessed as blockade of anti CD28-stimulated IL2 production
- ChEMBL_1878101 (CHEMBL4379495) Inhibition of recombinant full-length human ATM using GST-cMyc-p53 as substrate measured after 40 mins by ELISA
- ChEMBL_2214616 (CHEMBL5127748) Inhibition of ATM in human MCF7 cells using etoposide as substrate incubated for 1 hr by Cell Western assay
- ChEMBL_2340352 Inhibition of flag tagged- ATM (unknown origin) expressed in Escherichia coli BL21 incubated for 20 mins in presence of ATP
- ChEMBL_2246002 (CHEMBL5160212) Inhibition of ATM in human MCF7 cells assessed as decrease in pKAP-1 level measured by In-cell western assay
- ChEMBL_2248172 (CHEMBL5162382) Inhibition of ATM in human A549 cells assessed as reduction in etoposide-stimulated KAP1 phosphorylation by In-Cell-Western assay
- ChEMBL_2322510 Inhibition of ATM (unknown origin) using peptide as substrate preincubated for 10 mins followed by substrate addition by microplate reader analysis
- ChEMBL_2449217 Inhibition of ATM in human HT-29 cells incubated for 2 hrs by Alexa Fluor 488/Hoeschst staining based plate reader assay
- ChEMBL_2154863 (CHEMBL5039523) Agonist activity at STING KO human THP-1 dual cells incubated for 20 hrs by luciferase reporter gene assay
- ChEMBL_1765209 (CHEMBL4200456) Inhibition of ATM autophosphorylation at Ser1981 residue in human HT-29 cells measured after 1 hr by Hoechst staining based fluorescence assay
- ChEMBL_2028066 (CHEMBL4682224) Inhibition of recombinant full-length human FLAG-tagged ATM assessed as reduction in p53 S15 phosphorylation incubated for 30 mins by ELISA
- ChEMBL_2028067 (CHEMBL4682225) Inhibition of ATM in human MCF7 cells assessed as reduction in etoposide-stimulated KAP1 phosphorylation incubated for 1 hr by ICW assay
- ChEMBL_2093851 (CHEMBL4775114) Inhibition of full length Flag-tagged ATM (unknown origin) using GST-p53(1 to 101 residues) as substrates incubated for 90 mins
- ChEMBL_2114759 (CHEMBL4823700) Inhibition of ATM in human U2OS cells assessed as reduction in etoposide-stimulated KAP1 phosphorylation incubated for 60 mins by immunoreactivity assay
- ChEMBL_2340254 Inhibition of ATM (unknown origin) preincubated with compound for 30 mins followed by substrate addition and measured after 2 hrs by HTRF method
- ChEMBL_2322358 Agonist activity at STING in human STHP1-Dual KO-STING cells incubated for 20 hrs by Quanti-luc reagent based assay
- ChEMBL_2354137 Agonist activity at STING in PMA-differentiated human THP1-Dual KO-STING cells incubated for 24 hrs by QUANTI-Blue assay
- ChEMBL_2311265 Inhibition of ATM (unknown origin) using GST-cMyc-p53 as substrate incubated for 30 mins in presence of Mg/ATP mix by HTRF-based analysis
- ChEMBL_1738061 (CHEMBL4153811) Inhibition of ATM (unknown origin) using p53 as substrate preincubated for 30 mins followed by substrate addition and measured after 2 hrs by HTRF assay
- ChEMBL_1765222 (CHEMBL4200469) Inhibition of ATM (unknown origin) using p53 as substrate pretreated for 30 mins followed by substrate addition and measured after 2 hrs by HTRF assay
- ChEMBL_2427951 Inhibition of human full length recombinant ATM using GST-c-Myc-p53 as substrate measured after 30 mins in presence of Mg/ATP mix by ELISA
- ChEMBL_2473762 Inhibition of PRMT5 in human HCT-116 cells with MTAP KO assessed as decrease in SMDA level incubated for 48 hrs by immunofluorescence analysis
- ChEMBL_2271781 Inhibition of ATM in human HCT-116 cells preincubated for 1 hr followed by UV radiation and measured after 1 hr by Bradford assay based immunoblotting analysis
- ChEMBL_2329181 Inhibition of recombinant human Wip1 (1 to 420 residues) using human ATM phosphopeptide (AFEEG-pS-QSTTIGY) as substrate incubated for 7 mins by Bio-mol green assay
- ChEMBL_1852127 (CHEMBL4352751) Inhibition of full-length ATM (unknown origin) using DPSVEPPLSQETFSDKKK peptide as substrate measured after 24 hrs in presence of [gamma-33P] ATP by microbeta liquid scintillation counting analysis
- ChEMBL_1932514 (CHEMBL4478166) Inhibition of ATM (unknown origin) using p53 as substrate incubated for 30 mins followed by substrate addition measured after 2 hrs in presence of ATP by HTRF method
- ChEMBL_2093845 (CHEMBL4775108) Inhibition of human HeLa nuclear extract derived ATM using glutathioneS-transferase-p53N66 as substrate preincubated for 10 mins followed by ATP addition and measured after 1 hr by ELISA
- ChEMBL_2526498 Inhibition of ATM in human HeLa cells lysate pre incubated for 15 mins followed by ADP acyl phosphate probe addition and measured after 10 mins by LC-MS/MS analysis
- ChEMBL_2160781 (CHEMBL5045531) Agonist activity at STING in human THP1 Dual KO-STING cells assessed as IRF reporter activation incubated for 20 hrs by quanti-blue SEAP reporter gene assay
- ChEMBL_1932515 (CHEMBL4478167) Inhibition of ATM phosphorylation at Ser-1981 residue in human HT-29 cells incubated for 1 hr followed by X-ray irradiation by Hoechst staining based fluorescence plate reader analysis
- ChEMBL_2160782 (CHEMBL5045532) Agonist activity at STING in mouse RAW-Lucia ISG-KO-STING cells assessed as IRF reporter activation incubated for 20 hrs by quanti-blue SEAP reporter gene assay
- ChEMBL_2160784 (CHEMBL5045534) Agonist activity at STING in human THP1-Dual KO-STING cells assessed as NF-kappaB reporter activation incubated for 20 hrs by quanti-blue SEAP reporter gene assay
- ChEMBL_1738020 (CHEMBL4153770) Inhibition of ATM autophosphorylation at Ser1981 in human HT29 cells preincubated for 1 hr followed by X ray irradiation and measured after 1 hr by Hoechst 33342 dye-based immunofluorescence assay
- Enzymatic ATM Assay The inhibitory activity on the isolated ATM kinase was determined by a commercial supplier (Reaction Biology Corp., PA, US) using an activity based FRET assay. Compounds were therefore first pre-incubated with human ATM before substrate (p53) and ATP were added to initiate the phosphorylation reaction. After a given time, the kinase reaction was stopped and the amount of phosphorylated substrate was quantified using standard FRET detection methods. Based on a 5-point dilution row starting at 10 μM with a 10-fold dilution factor, the half-maximal inhibitory concentration (IC50 value) was calculated for each final compound. Detailed assay conditions and parameters can be requested from the commercial supplier.
- ATM Kinase Assay Determination of ATM Inhibition (IC50 ATM) The IC50 value was determined with the aid of a biochemical ATM kinase assay. The assay consists of two steps: the enzymatic reaction and the detection step. Firstly, ATM (ataxia telangiectasia mutated) protein and the test substance are incubated at different concentrations with addition of substrate protein p53 and ATP. ATM mediates the phosphorylation of p53 at several positions, including at amino acid S15. The amount of phosphorylated p53 is determined with the aid of specific antibodies and the TR-FRET technique. The enzymatic ATM assay is carried out as TR-FRET (HTRF , Cisbio Bioassays) based 384-well assay. In the first step, purified human recombinant ATM (human ATM, full length, GenBank ID NM_000051, expressed in a mammal cell line) is incubated in assay buffer for 15 minutes with the ATM inhibitor in various concentrations and without test substance as negative or neutral control. The assay buffer comprises 25 mM HEPES pH 8.0, 10 mM Mg(CH3COO)2, 1 mM MnCl2, 0.1% BSA and 0.01% Brij 35, 5 mM dithiothreitol (DTT). The test-substance solutions were dispensed into the microtitre plates using an ECHO 555 (Labcyte). In the second step, purified human recombinant cmyc-labelled p53 (human p53, full length, GenBank ID BC003596, expressed in Sf21 insect cells) and ATP are added, and the reaction mixture is incubated at 22° C. for 30-35 minutes. The pharmacologically relevant assay volume is 5 μl. The final concentrations in the assay during incubation of the reaction mixture are 0.3-0.4 nM ATM, 50-75 nM p53 and 10 μM ATP. The enzymatic reaction is stopped by addition of EDTA. The formation of phosphorylated p53 as the result of the ATM-mediated reaction in the presence of ATP is detected via specific antibodies [labelled with the fluorophorene europium (Eu) as donor and d2 as acceptor (Cisbio Bioassays)] which enable FRET. 2 μl of antibody-containing stop solution (12.5 mM HEPES pH 8.0, 125 mM EDTA, 30 mM sodium chloride, 300 mM potassium fluoride, 0.1006% Tween-20, 0.005% Brij 35, 0.21 nM anti-phospho-p53(ser15)-Eu antibody and 15 nM anti-cmyc-d2 antibody) are added to the reaction mixture. After incubation, usually for 2 hours (between 1.5 and 15 h), for signal development, the plates are analysed in a plate reader (EnVision, PerkinElmer) using TRF mode (and with laser excitation). After excitation of the donor europium at a wavelength of 340 nm, the emitted fluorescence light both of the acceptor d2 at 665 nm and also of the donor Eu at 615 nm is measured. The amount of phosphorylated p53 is directly proportional to the quotient of the amounts of light emitted, i.e. the relative fluorescence units (RFU) at 665 nm and 615 nm. The measurement data were processed by means of Genedata Screener software. IC50 determinations are carried out, in particular, by fitting a dose/action curve to the data points by means of nonlinear regression analysis.
- ATM kinase assay—determination of ATM inhibition (IC50 ATM) The IC50 value was determined with the aid of a biochemical ATM kinase assay. The assay consists of two steps: the enzymatic reaction and the detection step. Firstly, ATM (ataxia telangiectasia mutated) protein and the test substance are incubated at different concentrations with addition of substrate protein p53 and ATP. ATM mediates the phosphorylation of p53 at several positions, including at amino acid S15. The amount of phosphorylated p53 is determined with the aid of specific antibodies and the TR-FRET technique. The enzymatic ATM assay is carried out as TR-FRET (HTRF™, Cisbio Bioassays) based 384-well assay. In the first step, purified human recombinant ATM (human ATM, full length, GenBank ID NM_000051, expressed in a mammal cell line) is incubated in assay buffer for 15 minutes with the ATM inhibitor in various concentrations and without test substance as negative or neutral control. The assay buffer comprises 25 mM HEPES pH 8.0, 10 mM Mg(CH3COO)2, 1 mM MnCl2, 0,1% BSA and 0,01% Brij 35, 5 mM dithiothreitol (DTT). The test-substance solutions were dispensed into the microtitre plates using an ECHO 555 (Labcyte). In the second step, purified human recombinant cmyc-labelled p53 (human p53, full length, GenBank ID BC003596, expressed in Sf21 insect cells) and ATP are added, and the reaction mixture is incubated at 22° C. for 30-35 minutes. The pharmacologically relevant assay volume is 5 μl. The final concentrations in the assay during incubation of the reaction mixture are 0.3-0.4 nM ATM, 50-75 nM p53 and 10 μM ATP. The enzymatic reaction is stopped by addition of EDTA. The formation of phosphorylated p53 as the result of the ATM-mediated reaction in the presence of ATP is detected via specific antibodies [labelled with the fluorophorene europium (Eu) as donor and d2 as acceptor (Cisbio Bio-assays)] which enable FRET. 2 μl of antibody-containing stop solution (12.5 mM HEPES pH 8.0, 125 mM EDTA, 30 mM sodium chloride, 300mM potassium fluoride, 0.1006% Tween-20, 0.005% Brij 35, 0.21 nM anti-phospho-p53(ser15)-Eu antibody and 15 nM anti-cmyc-d2 antibody) are added to the reaction mixture. After incubation, usually for 2 hours (between 1.5 and 15h), for signal development, the plates are analysed in a plate reader (EnVision, PerkinElmer) using TRF mode (and with laser excitation). After excitation of the donor europium at a wavelength of 340 nm, the emitted fluorescence light both of the acceptor d2 at 665 nm and also of the donor Eu at 615 nm is measured. The amount of phosphorylated p53 is directly proportional to the quotient of the amounts of light emitted, i.e. the relative fluorescence units (RFU) at 665 nm and 615 nm. The measurement data were processed by means of Genedata Screener software. IC50 determinations are carried out, in particular, by fitting a dose/action curve to the data points by means of nonlinear regression analysis.
- ChEMBL_2114758 (CHEMBL4823699) Inhibition of full length recombinant FLAG-tagged human ATM assessed as decrease in p53 S15 phosphorylation using full length myc-tagged p53 as substrate incubated for 30 mins in presence of ATP by ELISA
- ChEMBL_1738015 (CHEMBL4153765) Inhibition of ATM derived from human HeLa cell nuclear extract using glutathione S-transferase p53N66 as substrate preincubated for 10 mins followed by ATP addition and subsequent incubation for 1 hr measured after 1.5 hrs by ELISA
- ChEMBL_1901760 (CHEMBL4403982) Inhibition of ATM in human HT-29 cells assessed as reduction in irradiation-induced autophosphorylation at Ser1981 residue preincubated for 1 hr followed by 6 Gy irradiation and measured after 1 hr by Hoechst staining-based imaging analysis
- ChEMBL_1979591 (CHEMBL4612726) Inhibition of N-terminally FLAG-tagged recombinant full-length ATM (unknown origin) expressed in HEK239-6E cells using biotin-PEG2-SVEPPLSQETFSD as substrate preincubated for 15 mins followed by substrate addition and measured after 90 mins by TR-FRET assay
- ATM Kinase Assay The IC50 value was determined with the aid of a biochemical ATM kinase assay. The assay consists of two steps: the enzymatic reaction and the detection step. Firstly, ATM (ataxia telangiectasia mutated) protein and the test substance are incubated at different concentrations with addition of substrate protein p53 and ATP. ATM mediates the phosphorylation of p53 at several positions, including at amino acid S15. The amount of phosphorylated p53 is determined with the aid of specific antibodies and the TR-FRET technique. The enzymatic ATM assay is carried out as TR-FRET (HTRF , Cisbio Bioassays) based 384-well assay. In the first step, purified human recombinant ATM (human ATM, full length, GenBank ID NM_000051, expressed in a mammal cell line) is incubated in assay buffer for 15 minutes with the ATM inhibitor in various concentrations and without test substance as negative or neutral control. The assay buffer comprises 25 mM HEPES pH 8.0, 10 mM Mg(CH3COO)2, 1 mM MnCl2, 0.1% BSA and 0.01% Brij 35, 5 mM dithiothreitol (DTT). The test-substance solutions were dispensed into the microtitre plates using an ECHO 555 (Labcyte). In the second step, purified human recombinant cmyc-labelled p53 (human p53, full length, GenBank ID BC003596, expressed in Sf21 insect cells) and ATP are added, and the reaction mixture is incubated at 22° C. for 30-35 minutes. The pharmacologically relevant assay volume is 5 μl. The final concentrations in the assay during incubation of the reaction mixture are 0.3-0.4 nM ATM, 50-75 nM p53 and 10 μM ATP. The enzymatic reaction is stopped by addition of EDTA. The formation of phosphorylated p53 as the result of the ATM-mediated reaction in the presence of ATP is detected via specific antibodies [labelled with the fluorophorene europium (Eu) as donor and d2 as acceptor (Cisbio Bioassays)] which enable FRET. 2 μl of antibody-containing stop solution (12.5 mM HEPES pH 8.0, 125 mM EDTA, 30 mM sodium chloride, 300 mM potassium fluoride, 0.1006% Tween-20, 0.005% Brij 35, 0.21 nM anti-phospho-p53(ser15)-Eu antibody and 15 nM anti-cmyc-d2 antibody) are added to the reaction mixture. After incubation, usually for 2 hours (between 1.5 and 15 h), for signal development, the plates are analysed in a plate reader (EnVision, PerkinElmer) using TRF mode (and with laser excitation). After excitation of the donor europium at a wavelength of 340 nm, the emitted fluorescence light both of the acceptor d2 at 665 nm and also of the donor Eu at 615 nm is measured. The amount of phosphorylated p53 is directly proportional to the quotient of the amounts of light emitted, i.e. the relative fluorescence units (RFU) at 665 nm and 615 nm. The measurement data were processed by means of Genedata Screener software. IC50 determinations are carried out, in particular, by fitting a dose/action curve to the data points by means of nonlinear regression analysis.
- ChEMBL_2129936 (CHEMBL4839365) Inhibition of full-length N-terminal FLAG-tagged human ATM expressed in HEK293-6E cells using biotin-PEG2-SVEPPLSQETFSD as substrate preincubated for 15 mins followed by substrate addition and measured after 90 mins in presence of ATP by TR-FRET assay
- Biochemical Kinase Assay The IC50 value was determined with the aid of a biochemical ATM kinase assay. The assay consists of two steps: the enzymatic reaction and the detection step. Firstly, ATM (ataxia telangiectasia mutated) protein and the test substance are incubated at different concentrations with addition of substrate protein p53 and ATP. ATM mediates the phosphorylation of p53 at several positions, including at amino acid S15. The amount of phosphorylated p53 is determined with the aid of specific antibodies and the TR-FRET technique. The enzymatic ATM assay is carried out as TR-FRET (HTRF , Cisbio Bioassays) based 384-well assay. In the first step, purified human recombinant ATM (human ATM, full length, GenBank ID NM_000051, expressed in a mammal cell line) is incubated in assay buffer for 15 minutes with the ATM inhibitor in various concentrations and without test substance as negative or neutral control. The assay buffer comprises 25 mM HEPES pH 8.0, 10 mM Mg(CH3COO)2, 1 mM MnCl2, 0.1% BSA and 0.01% Brij 35, 5 mM dithiothreitol (DTT). The test-substance solutions were dispensed into the microtitre plates using an ECHO 555 (Labcyte). In the second step, purified human recombinant cmyc-labelled p53 (human p53, full length, GenBank ID BC003596, expressed in Sf21 insect cells) and ATP are added, and the reaction mixture is incubated at 22° C. for 30-35 minutes. The pharmacologically relevant assay volume is 5 μl. The final concentrations in the assay during incubation of the reaction mixture are 0.3-0.4 nM ATM, 50-75 nM p53 and 10 μM ATP. The enzymatic reaction is stopped by addition of EDTA. The formation of phosphorylated p53 as the result of the ATM-mediated reaction in the presence of ATP is detected via specific antibodies[labelled with the fluorophorene europium (Eu) as donor and d2 as acceptor (Cisbio Bioassays)] which enable FRET. 2 μl of antibody-containing stop solution (12.5 mM HEPES pH 8.0, 125 mM EDTA, 30 mM sodium chloride, 300 mM potassium fluoride, 0.1006% Tween-20, 0.005% Brij 35, 0.21 nM anti-phospho-p53(ser15)-Eu antibody and 15 nM anti-cmyc-d2 antibody) are added to the reaction mixture. After incubation, usually for 2 hours (between 1.5 and 15 h), for signal development, the plates are analysed in a plate reader (EnVision, PerkinElmer) using TRF mode (and with laser excitation). After excitation of the donor europium at a wavelength of 340 nm, the emitted fluorescence light both of the acceptor d2 at 665 nm and also of the donor Eu at 615 nm is measured. The amount of phosphorylated p53 is directly proportional to the quotient of the amounts of light emitted, i.e. the relative fluorescence units (RFU) at 665 nm and 615 nm.
- Inhibition on the Kinases (ATM, DNA-PK) Following the experimental methods reported in the reference documents, the test compounds were diluted in a 3-fold series to 0.51 nM (a total of 10 concentrations) starting from 10 μM, and their inhibitory activity against the kinases ATM1 and DNA-PK2 were measured respectively.
- ATM Biochemical Assay ATM (Millipore, Cat. No. 14-933) enzyme solution was prepared in 1x kinase base buffer. 10 ul of 2x enzyme solution was transferred to each well of the 384-well assay plate containing 100 nl compounds added by Echo. The plate was incubated at room temperature for 10 minutes. 2x peptide solution was prepared with FAM-labeled peptide and ATP in the 1x kinase base buffer (final concentration: 1.5 nM). 10 ul of 2x peptide solution was added to each well of the 384-well assay plate which was incubated at 37 C. for 210 min before 40 ul stop buffer was added to stop reaction. Data was collected by Caliper.
- ATM Biochemical Potency Assay ATM (Millipore, Cat. No. 14-933) enzyme solution was prepared in 1× kinase base buffer. 10 μl of 2× enzyme solution was transferred to each well of the 384-well assay plate containing 100 nl compounds added by Echo. The plate was incubated at room temperature for 10 minutes. 2× peptide solution was prepared with FAM-labeled peptide and ATP in the 1× kinase base buffer (final concentration: 1.5 nM). 10 μl of 2× peptide solution was added to each well of the 384-well assay plate which was incubated at 37° C. for 210 min before 40 μl stop buffer was added to stop reaction. Data was collected by Caliper.
- Cellular Assay ATM and ATR have distinct and overlapping responses to DNA damage. They must participate together and responses must be co-ordinated. Both pathways may be activated by ionising radiation, however only ATR is activated by UV. Since UV treatment is not practical for use in a high throught-put cell assay, the UV mimetic 4NQ0 (Sigma) was chosen to activate the ATR DNA damage response pathway.
- Biological Assay ATM: Recombinant, full length human FLAG-tagged ATM activity (Eurofins 14-933) was measured in an ELISA for p53 S15 phosphorylation, in a 384 well V-bottom polypropylene plate (Greiner Bio One 781280). Reactions contained 0.75 nM ATM, 25 nM full length myc tagged p53 (Eurofins 23-034), 10 μM UltraPure ATP (Promega V915B) and serial dilutions of inhibitors (0.1% DMSO final) in 25 mM HEPES, pH 7.5. 5 mM MgCl2, 2.5 mM MnCl2, 0.006% Brij-35, 0.5% glycerol, 0.1 mg/ml BSA, 0.5 mM DTT and were allowed to proceed for 30 min at 20° C. 70 mM EDTA final terminated the kinase reactions. An aliquot of the terminated reaction (25 μl) was transferred to a 384-well, high binding microplate (Greiner Bio One 781061), precoated with anti-myc capture antibody overnight (Millipore 05-724) diluted in 1:1000 in PBS, then blocked with Odyssey blocking buffer (LI-COR Biosciences 92740000) for 60 min at 20° C. The terminated reaction was allowed to capture for 2 h at 20° C. Phosphorylation was measured using an antibody specific to p53 S15 phosphorylation (1:5000 Abcam ab38497), anti-rabbit HRP (1:1000 Cell Signalling Technology, 7074) and TMB (ab171522). Antibodies were diluted in Odyssey blocking buffer and incubated for 60 min at 20° C. Three washes with PBS containing 0.05% (v/v) Tween 20 (PBS-T) were performed between each incubation. The TMB reaction was stopped with an equal volume of 0.2 M sulfuric acid and absorbance read at 450 nm on the Perkin Elmer Envision within 30 min of terminating the reaction. The percentage of inhibition was calculated for each concentration of compound using 0.1% DMSO control wells as 0% inhibition and 1 μM KU60019 as 100% inhibition.
- ATM Kinase Assay PI3K (p120γ) (human) was incubated in an assay buffer comprising 10 μM phosphatidylinositol 4,5-bisphosphate and MgATP (concentration as necessary). The reaction was initiated by addition of MgATP solution. After an incubation time of 30 min at room temperature, the reaction was stopped by addition of a solution consisting of EDTA and biotinylated phosphatidylinositol 3,4,5-trisphosphate. Finally, the detection buffer, consisting of a europium-labelled anti-GST monoclonal antibody, a GST-labelled GRP1 PH domain and streptavidin allophycocyanin, was added. The plate was read out via homogeneous time-resolved fluorescence (HTRF) and the corresponding signals were evaluated via the formula HTRF=10000×(Em665 nm/Em620 nm).
- ATM Kinase Assay PI3K p110α/p85α (human) was incubated in an assay buffer comprising 10 μM phosphatidylinositol 4,5-bisphosphate and MgATP (concentration as necessary). The reaction was initiated by addition of MgATP solution. After incubation at room temperature for 40 minutes, the reaction was stopped by addition of a solution consisting of EDTA and biotinylated phosphatidylinositol 3,4,5-trisphosphate. Finally, the detection buffer, consisting of a europium-labelled anti-GST monoclonal antibody, a GST-labelled GRP1 PH domain and streptavidin allophycocyanin, was added. The plate was read out via homogeneous time-resolved fluorescence (HTRF) and the corresponding signals were evaluated via the formula HTRF=10000×(Em665 nm/Em620 nm).
- ATM Kinase Assay mTOR (human) was incubated with 50 mM HEPES pH 7.5, 1 mM EGTA, 0.01% Tween 20, 2 mg/ml of the substrate, 3 mM MnCl2 and [γ-33P-ATP] (specific activity approximately 500 cpm/pmol, concentration as necessary). The reaction was initiated by addition of MgATP solution. After incubation at room temperature for 40 minutes, the reaction was stopped by addition of 3% phosphoric acid. 10 μl of the reaction solution were then transferred dropwise onto a P30 filtermat and washed three times for 5 min in 75 mM phosphoric acid and once in methanol, dried and evaluated by means of liquid scintillation counting.
- ATM Kinase Assay PI3K p110β/p85α (human) was incubated in an assay buffer comprising 10 μM phosphatidylinositol 4,5-bisphosphate and MgATP (concentration as necessary). The reaction was initiated by addition of MgATP solution. After an incubation time of 30 min at room temperature, the reaction was stopped by addition of a solution consisting of EDTA and biotinylated phosphatidylinositol 3,4,5-trisphosphate. Finally, the detection buffer, consisting of a europium-labelled anti-GST monoclonal antibody, a GST-labelled GRP1 PH domain and streptavidin allophycocyanin, was added. The plate was read out via homogeneous time-resolved fluorescence (HTRF) and the corresponding signals were evaluated via the formula HTRF=10000×(Em665 nm/Em620 nm).
- ATM Kinase Assay PI3K p110δ/p85α (human) was incubated in an assay buffer comprising 10 μM phosphatidylinositol 4,5-bisphosphate and MgATP (concentration as necessary). The reaction was initiated by addition of MgATP solution. After an incubation time of 30 min at room temperature, the reaction was stopped by addition of a solution consisting of EDTA and biotinylated phosphatidylinositol 3,4,5-trisphosphate. Finally, the detection buffer, consisting of a europium-labelled anti-GST monoclonal antibody, a GST-labelled GRP1 PH domain and streptavidin allophycocyanin, was added. The plate was read out via homogeneous time-resolved fluorescence (HTRF) and the corresponding signals were evaluated via the formula HTRF=10000×(Em665 nm/Em620 nm).
- Determination of the value of binding affinity constant (KO between the compound and BRD4 BD2 protein The purity of BRD4 BD2 protein used in the experiment was greater than 95%, and the protein concentration was 46.33 uM. The 96-well plate was purchased from Corning (black, #3694). The multifunctional microplate reader was a product of TECAN, model: SPARK 10M. Buffer: 100 mM potassium phosphate (pH 6.5), 2% ethylene glycol (Sigma) and 0.01% Trition X-100 (Sigma). The experimental water was Millipore-Q pure water.The Ki value of the compound and BRD4 BD2 protein was measured according to the FP test procedures for detecting the Ki value of the compound and BRD4 BD1 protein except that the BRD4 BD1 protein was replaced with the BRD4 BD2 protein.
- Determination of the value of binding affinity constant (KO between the compound and BRD4 BD1 protein The purity of BRD4 BD1 protein used in the experiment was greater than 95%, and the protein concentration was 43.4 uM. The 96-well plate was purchased from Corning (black, #3694). The multifunctional microplate reader was a product of TECAN, model: SPARK 10M. Buffer: 100 mM potassium phosphate (pH 6.5), 2% ethylene glycol (Sigma) and 0.01% Trition X-100 (Sigma). The experimental water was Millipore-Q pure water.The specific experimental steps were as follows.First, the compound to be tested was dissolved in ethylene glycol to prepare into a 10 mM standard stock solution. Subsequently, the standard stock solution of the compound to be tested was diluted into a working sample solution with the buffer in an EP tube and ready for use. The concentration of the prepared working sample solution was 5 times of the highest sample concentration required on the test plate (5×test compound solution).40 λL of a 5× test compound solution of a sample A was added to wells B1-B3 of a 96-well plate, and 40 μL of a 5× test compound solution of a sample B was added to wells B7-B9 of the 96-well plate, respectively. 20 uL of the buffer was added to the remaining wells, except for wells B1-B3 and B7-B9. Then, 20 uL of a solution was taken from wells B1-B3 to C1-C3, and this 2-fold dilution was repeated from C1-C3 until H4-H6; in the same way, 20 uL of a solution was taken from B7-B9 to C7-C9, this 2-fold dilution was repeated from C7-C9 until H10-H12. Finally, 80 uL of a mixed solution containing 2.5 nM Tracer and 37.5 nM BRD4 BD1 protein was added to each well.
- Cellular Assay Assay a) HT29 cells (ECACC #85061109) were seeded into 384 well assay plates (Costar #3712) at a density of 3500 cells/well in 40 μl EMEM medium containing 1% L glutamine and 10% FBS and allowed to adhere overnight. The following morning compounds of Formula (I) in 100% DMSO were added to assay plates by acoustic dispensing. After 1 h incubation at 37° C. and 5% CO2 plates (up to 6 at a time) were irradiated using the X-RAD 320 instrument (PXi) with equivalent to 600 cGy. Plates were returned to the incubator for a further 1 h. Then cells were fixed by adding 20 μl of 3.7% formaldehyde in PBS solution and incubating for 20 minutes at r.t. before being washed with 50 μl/well PBS, using a Biotek EL405 plate washer. Then 20 μ 0.1% Triton X100 in PBS was added and incubated for 20 minutes at r.t., to permeabalise cells. Then the plates were washed once with 50 μl/well PBS, using a Biotek EL405 plate washer. Phospho-ATM Ser1981 antibody (Millipore #MAB3806) was diluted 10000 fold in PBS containing 0.05% polysorbate/Tween and 3% BSA and 20 μl was added to each well and incubated over night at r.t. The next morning plates were washed three times with 50 μl/well PBS, using a Biotek EL405 plate washer, and then 20 μl of secondary Ab solution, containing 500 fold diluted Alexa Fluor® 488 Goat anti-rabbit IgG (Life Technologies, A11001) and 0.002 mg/ml Hoeschst dye (Life technologies #H-3570), in PBS containing 0.05% polysorbate/Tween and 3% BSA, was added. After 1 h incubation at r.t., the plates were washed three times with 50 μl/well PBS, using a Biotek EL405 plate washer, and plates were sealed and kept in PBS at 4° C. until read. Plates were read using an ArrayScan VTI instrument, using an XF53 filter with 10× objective. A two laser set up was used to analyse nuclear staining with Hoeschst (405 nm) and secondary antibody staining of pSer1981 (488 nm).
- Inhibitory Activities of Compounds on ATM (1) Preparation of 1× kinase basal buffer and reaction termination solution1) 1× kinase basal buffer50 mM HEPES, pH 7.50.0015% Brij-35 (polyoxyethylene lauryl ether)100 mM Na3VO45 M NaCl1 M MgCl21 M MnCl22) Reaction termination solution100 mM HEPES, pH 7.50.015% Brij-350.2% Coating Reagent #350 mM EDTA(2) Preparation of test compound1) Dissolution and dilution of compound: a compound was dissolved into DMSO to yield a 10 mM or 5 mM stock solution. To a 96-well plate, 98 μL of DMSO and 2 μL of the 10 mM stock solution were added and mixed evenly so that the concentration of the solution was 200 μM. To another 96-well plate, 45 μL of DMSO and 5 μL of the 200 μM solution were added to yield a 20 μM working solution.2) The working solution of the compound was sequentially diluted in the 96-well plate by the way of taking 10 μL of a solution at a higher concentration to 30 μL of DMSO to yield a mixed solution at a lower concentration and transferring the mixed solution to the next well, and so on, in order to set up 10 concentration gradients.3) 100 μL of DMSO was added to the blank well and served as a blank control without compound or enzyme.4) Preparation of intermediate sample plate: 40 μL of each of the solutions with gradient concentrations prepared in the 96-well plate was taken and transferred to a new 384-well plate as an intermediate sample plate.(3) Preparation of test plate100 nL of the compound solution was taken from each well of the intermediate sample plate to a 384-well plate as a test plate.(4) Kinase reaction1) The kinase was dissolved in the 1× kinase basal buffer to yield a 2× enzyme solution.2) 10 μL of the 2× enzyme solution was taken to the 384-well test plate.3) The 384-well test plate was incubated at room temperature for 10 min.4) FAM-labeled polypeptide substrate and ATP were dissolved in the 1× kinase basal buffer to yield a 2× substrate peptide solution.5) 10 μL of the 2× substrate peptide solution was taken to each well of the 384-well test plate, respectively.6) Progress and termination of enzymatic reaction: the test plate, to which the enzyme solution and the substrate peptide solution were added, was incubated at 37° C. for a while, and then 35 μL of the reaction termination solution was added to terminate the reaction.(5) Reading of the reaction wells(6) Calculation of inhibitory rates by means of curve fitting to the read values
 CHEMBL1159714 Ko 707 BDBM50421719
CHEMBL1159714 Ko 707 BDBM50421719 3-((3S,6S)-6-Isobutyl-9-methoxy-1,4-dioxo-1,2,3,4,6,7,12,12a-octahydro-pyrazino[1',2':1,6]pyrido[3,4-b]indol-3-yl)-propionic acid tert-butyl ester BDBM50305083 3-((3S,6S,12aS)-6-Isobutyl-9-methoxy-1,4-dioxo-1,2,3,4,6,7,12,12a-octahydro-pyrazino[1',2':1,6]pyrido[3,4-b]indol-3-yl)-propionic acid tert-butyl ester Ko143 Ko-143 US9695174, Ko143 CHEMBL488910
3-((3S,6S)-6-Isobutyl-9-methoxy-1,4-dioxo-1,2,3,4,6,7,12,12a-octahydro-pyrazino[1',2':1,6]pyrido[3,4-b]indol-3-yl)-propionic acid tert-butyl ester BDBM50305083 3-((3S,6S,12aS)-6-Isobutyl-9-methoxy-1,4-dioxo-1,2,3,4,6,7,12,12a-octahydro-pyrazino[1',2':1,6]pyrido[3,4-b]indol-3-yl)-propionic acid tert-butyl ester Ko143 Ko-143 US9695174, Ko143 CHEMBL488910