 US8889724, EC BDBM139535
US8889724, EC BDBM139535 BDBM292781 US10106501, Example EC
BDBM292781 US10106501, Example EC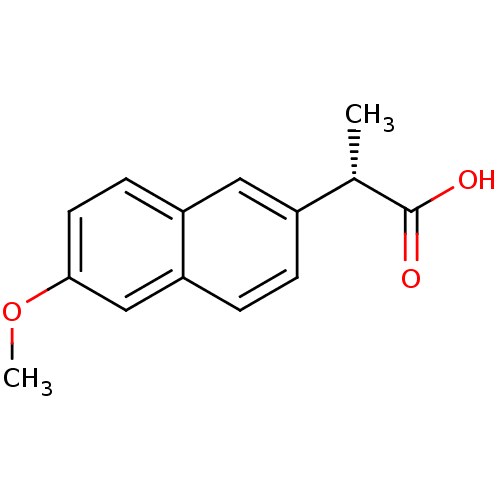 2-(6-Methoxy-naphthalen-2-yl)-propionic acid (S)-2-(6-Methoxy-naphthalen-2-yl)-propionic acid US11459295, Compound S-Naproxen (1) Equiproxen Naprelan (S)-2-(6-methoxynaphthalen-2-yl)propanoic acid Naproxen2-(6-Methoxy-naphthalen-2-yl)-propionic acid Anaprox 2-(6-Methoxy-naphthalen-2-yl)-propionic acid(naproxen) (2S)-2-(6-methoxynaphthalen-2-yl)propanoic acid naproxene Aleve CHEMBL154 2-(6-methoxy-2-naphthyl)propanoic acid Ec-naprosyn S-NAPROXEN Naproxen 2-(6-methoxynaphthalen-2-yl)propanoic acid BDBM50339185 RS-3540 (S)-naproxen Naprosyn
2-(6-Methoxy-naphthalen-2-yl)-propionic acid (S)-2-(6-Methoxy-naphthalen-2-yl)-propionic acid US11459295, Compound S-Naproxen (1) Equiproxen Naprelan (S)-2-(6-methoxynaphthalen-2-yl)propanoic acid Naproxen2-(6-Methoxy-naphthalen-2-yl)-propionic acid Anaprox 2-(6-Methoxy-naphthalen-2-yl)-propionic acid(naproxen) (2S)-2-(6-methoxynaphthalen-2-yl)propanoic acid naproxene Aleve CHEMBL154 2-(6-methoxy-2-naphthyl)propanoic acid Ec-naprosyn S-NAPROXEN Naproxen 2-(6-methoxynaphthalen-2-yl)propanoic acid BDBM50339185 RS-3540 (S)-naproxen Naprosyn
- Chen, H; Roques, BP; Fournié-Zaluski, MC Design of the first highly potent and selective aminopeptidase N (EC 3.4.11.2) inhibitor. Bioorg Med Chem Lett 9: 1511-6 (1999)
- Gomez-Monterrey, I; Beaumont, A; Nemecek, P; Roques, BP; Fournie-Zaluski, MC New thiol inhibitors of neutral endopeptidase EC 3.4.24.11: synthesis and enzyme active-site recognition. J Med Chem 37: 1865-73 (1994)
- Fink, CA; Qiao, Y; Berry, CJ; Sakane, Y; Ghai, RD; Trapani, AJ New alpha-thiol dipeptide dual inhibitors of angiotensin-I converting enzyme and neutral endopeptidase EC 3.4.24.11. J Med Chem 38: 5023-30 (1996)
- De Lombaert, S; Erion, MD; Tan, J; Blanchard, L; el-Chehabi, L; Ghai, RD; Sakane, Y; Berry, C; Trapani, AJ N-Phosphonomethyl dipeptides and their phosphonate prodrugs, a new generation of neutral endopeptidase (NEP, EC 3.4.24.11) inhibitors. J Med Chem 37: 498-511 (1994)
- Danvy, D; Monteil, T; Plaquevent, JC; Duhamel, L; Duhamel, P; Gros, C; Noël, N; Schwartz, JC; Lecomte, JM Studies on the structural feature of S'1 subsite of neprilysin (EC.3.4.24.11): Stereochemical requirement for the enzyme-inhibitor docking process Bioorg Med Chem Lett 6: 2437-2440 (1996)
- David, C; Bischoff, L; Meudal, H; Mothé, A; De Mota, N; DaNascimento, S; Llorens-Cortes, C; Fournié-Zaluski, MC; Roques, BP Investigation of subsite preferences in aminopeptidase A (EC 3.4.11.7) led to the design of the first highly potent and selective inhibitors of this enzyme. J Med Chem 42: 5197-211 (2000)
- Dominguez, C; Duffy, DE; Han, Q; Alexander, RS; Galemmo, RA; Park, JM; Wong, PC; Amparo, EC; Knabb, RM; Luettgen, J; Wexler, RR Bioorg Med Chem Lett 9: 925-30 (1999)
- Adams, J; Anderson, EC; Blackham, EE; Chiu, YW; Clarke, T; Eccles, N; Gill, LA; Haye, JJ; Haywood, HT; Hoenig, CR; Kausas, M; Le, J; Russell, HL; Smedley, C; Tipping, WJ; Tongue, T; Wood, CC; Yeung, J; Rowedder, JE; Fray, MJ; McInally, T; Macdonald, SJ ACS Med Chem Lett 5: 1207-12 (2014)
- Decicco, CP; Seng, JL; Kennedy, KE; Covington, MB; Welch, PK; Arner, EC; Magolda, RL; Nelson, DJ Bioorg Med Chem Lett 7: 2331-2336 (1997)
- Birkinshaw, TN; Teague, SJ; Beech, C; Bonnert, RV; Hill, S; Patel, A; Reakes, S; Sanganee, H; Dougall, IG; Phillips, TT; Salter, S; Schmidt, J; Arrowsmith, EC; Carrillo, JJ; Bell, FM; Paine, SW; Weaver, R Bioorg Med Chem Lett 16: 4287-90 (2006)
- Kamble, SH; Berthold, EC; King, TI; Raju Kanumuri, SR; Popa, R; Herting, JR; León, F; Sharma, A; McMahon, LR; Avery, BA; McCurdy, CR J Nat Prod 84: 1104-1112 (2021)
- Speake, JD; Navas, F; Bishop, MJ; Garrison, DT; Bigham, EC; Hodson, SJ; Saussy, DL; Liacos, JA; Irving, PE; Sherman, BW Bioorg Med Chem Lett 13: 1183-6 (2003)
- Rana, S; Blowers, EC; Natarajan, A J Med Chem 58: 2-29 (2015)
- Alexander, RP; Warrellow, GJ; Eaton, MA; Boyd, EC; Head, JC; Porter, JR; Brown, JA; Reuberson, JT; Hutchinson, B; Turner, P; Boyce, B; Barnes, D; Mason, B; Cannell, A; Taylor, RJ; Zomaya, A; Millican, A; Leonard, J; Morphy, R; Wales, M; Perry, M; Allen, RA; Gozzard, N; Hughes, B; Higgs, G Bioorg Med Chem Lett 12: 1451-6 (2002)
- Argiriadi, MA; Breinlinger, EC; Chien, EY; Cowart, MD; Frank, KE; Friedman, MM; Hardee, DJ; Herold, JM; Liu, H; Qiu, W; Scanio, MJ; Schrimpf, MR; Vargo, TR; Van Epps, SA; Webster, MP; Little, AJ; Katcher, MH US Patent US20190276450 (2019)
- Tadesse, S; Caldon, EC; Tilley, W; Wang, S J Med Chem 62: 4233-4251 (2019)
- Granchi, C; Roy, S; Giacomelli, C; Macchia, M; Tuccinardi, T; Martinelli, A; Lanza, M; Betti, L; Giannaccini, G; Lucacchini, A; Funel, N; León, LG; Giovannetti, E; Peters, GJ; Palchaudhuri, R; Calvaresi, EC; Hergenrother, PJ; Minutolo, F J Med Chem 54: 1599-612 (2011)
- Chan, EC; New, LS; Chua, TB; Yap, CW; Ho, HK; Nelson, SD Drug Metab Dispos 40: 1414-22 (2012)
- Chen, EC; Khuri, N; Liang, X; Stecula, A; Chien, HC; Yee, SW; Huang, Y; Sali, A; Giacomini, KM J Med Chem 60: 2685-2696 (2017)
- Cherney, EC; Zhang, L; Nara, S; Zhu, X; Gullo-Brown, J; Maley, D; Lin, TA; Hunt, JT; Huang, C; Yang, Z; Darienzo, C; Discenza, L; Ranasinghe, A; Grubb, M; Ziemba, T; Traeger, SC; Li, X; Johnston, K; Kopcho, L; Fereshteh, M; Foster, K; Stefanski, K; Fargnoli, J; Swanson, J; Brown, J; Delpy, D; Seitz, SP; Borzilleri, R; Vite, G; Balog, A ACS Med Chem Lett 12: 288-294 (2021)
- Harris, NV; Smith, C; Ashton, MJ; Bridge, AW; Bush, RC; Coffee, EC; Dron, DI; Harper, MF; Lythgoe, DJ; Newton, CG J Med Chem 35: 4384-92 (1992)
- Edmonds, DJ; Kung, DW; Kalgutkar, AS; Filipski, KJ; Ebner, DC; Cabral, S; Smith, AC; Aspnes, GE; Bhattacharya, SK; Borzilleri, KA; Brown, JA; Calabrese, MF; Caspers, NL; Cokorinos, EC; Conn, EL; Dowling, MS; Eng, H; Feng, B; Fernando, DP; Genung, NE; Herr, M; Kurumbail, RG; Lavergne, SY; Lee, EC; Li, Q; Mathialagan, S; Miller, RA; Panteleev, J; Polivkova, J; Rajamohan, F; Reyes, AR; Salatto, CT; Shavnya, A; Thuma, BA; Tu, M; Ward, J; Withka, JM; Xiao, J; Cameron, KO J Med Chem 61: 2372-2383 (2018)
- Samal, RP; Khedkar, VM; Pissurlenkar, RR; Bwalya, AG; Tasdemir, D; Joshi, RA; Rajamohanan, PR; Puranik, VG; Coutinho, EC Chem Biol Drug Des 81: 715-29 (2013)
- Colletti, SL; Frie, JL; Dixon, EC; Singh, SB; Choi, BK; Scapin, G; Fitzgerald, CE; Kumar, S; Nichols, EA; O'Keefe, SJ; O'Neill, EA; Porter, G; Samuel, K; Schmatz, DM; Schwartz, CD; Shoop, WL; Thompson, CM; Thompson, JE; Wang, R; Woods, A; Zaller, DM; Doherty, JB J Med Chem 46: 349-52 (2003)
- Giannetti, AM; Wong, H; Dijkgraaf, GJ; Dueber, EC; Ortwine, DF; Bravo, BJ; Gould, SE; Plise, EG; Lum, BL; Malhi, V; Graham, RA J Med Chem 54: 2592-601 (2011)
- Ordonez-Rubiano, SC; Maschinot, CA; Wang, S; Sood, S; Baracaldo-Lancheros, LF; Strohmier, BP; McQuade, AJ; Smith, BC; Dykhuizen, EC J Med Chem 66: 11250-11270 (2023)
- Lenger, J; Schröder, M; Ennemann, EC; Müller, B; Wong, CH; Noll, T; Dierks, T; Hanson, SR; Sewald, N Bioorg Med Chem 20: 622-7 (2012)
- Erigür, EC; Altuğ, C; Angeli, A; Supuran, CT Bioorg Med Chem Lett 59: (2022)
- Severance, ZC; Nuñez, JI; Le-McClain, AT; Malinky, CA; Bensen, RC; Fogle, RS; Manginelli, GW; Sakers, SH; Falcon, EC; Bui, RH; Snead, KJ; Bourne, CR; Burgett, AWG J Med Chem 66: 3866-3875 (2023)
- Traynelis, SF; Mullasseril, P; Garnier, EC; Liotta, DC; Zimmerman, S US Patent US9737522 (2017)
- Kreutter, KD; Lu, T; Lee, L; Giardino, EC; Patel, S; Huang, H; Xu, G; Fitzgerald, M; Haertlein, BJ; Mohan, V; Crysler, C; Eisennagel, S; Dasgupta, M; McMillan, M; Spurlino, JC; Huebert, ND; Maryanoff, BE; Tomczuk, BE; Damiano, BP; Player, MR Bioorg Med Chem Lett 18: 2865-70 (2008)
- Hachey, AC; Fenton, AD; Heidary, DK; Glazer, EC J Med Chem 66: 398-412 (2023)
- Cui, H; Divakaran, A; Hoell, ZJ; Ellingson, MO; Scholtz, CR; Zahid, H; Johnson, JA; Griffith, EC; Gee, CT; Lee, AL; Khanal, S; Shi, K; Aihara, H; Shah, VH; Lee, RE; Harki, DA; Pomerantz, WCK J Med Chem 65: 2342-2360 (2022)
- Bellenie, BR; Bloomfield, GC; Bruce, I; Culshaw, AJ; Hall, EC; Hollingworth, GJ; Neef, J; Spendiff, M; Watson, SJ US Patent US10112926 (2018)
- Hank EC
- Tan, Q; Blizzard, TA; Morgan, JD; Birzin, ET; Chan, W; Yang, YT; Pai, LY; Hayes, EC; DaSilva, CA; Warrier, S; Yudkovitz, J; Wilkinson, HA; Sharma, N; Fitzgerald, PM; Li, S; Colwell, L; Fisher, JE; Adamski, S; Reszka, AA; Kimmel, D; DiNinno, F; Rohrer, SP; Freedman, LP; Schaeffer, JM; Hammond, ML Bioorg Med Chem Lett 15: 1675-81 (2005)
- Bhat, R; Adam, AT; Lee, JJ; Gasiewicz, TA; Henry, EC; Rotella, DP Bioorg Med Chem Lett 24: 2263-6 (2014)
- Hill, MD; Blanco, MJ; Salituro, FG; Bai, Z; Beckley, JT; Ackley, MA; Dai, J; Doherty, JJ; Harrison, BL; Hoffmann, EC; Kazdoba, TM; Lanzetta, D; Lewis, M; Quirk, MC; Robichaud, AJ J Med Chem 65: 9063-9075 (2022)
- Lee, SL; Hsu, EC; Chou, CC; Chuang, HC; Bai, LY; Kulp, SK; Chen, CS J Med Chem 54: 6364-74 (2011)
- Hulme, EC; Birdsall, NJ; Buckley, NJ Annu Rev Pharmacol Toxicol 30: 633-73 (1990)
- Teichert, RW; Jimenez, EC; Twede, V; Watkins, M; Hollmann, M; Bulaj, G; Olivera, BM J Biol Chem 282: 36905-13 (2007)
- Varney, MA; Rao, SP; Jachec, C; Deal, C; Hess, SD; Daggett, LP; Lin, F; Johnson, EC; Veliçelebi, G J Pharmacol Exp Ther 285: 358-70 (1998)
- Furber, M; Alcaraz, L; Bent, JE; Beyerbach, A; Bowers, K; Braddock, M; Caffrey, MV; Cladingboel, D; Collington, J; Donald, DK; Fagura, M; Ince, F; Kinchin, EC; Laurent, C; Lawson, M; Luker, TJ; Mortimore, MM; Pimm, AD; Riley, RJ; Roberts, N; Robertson, M; Theaker, J; Thorne, PV; Weaver, R; Webborn, P; Willis, P J Med Chem 50: 5882-5 (2007)
- Beukers, MW; Wanner, MJ; Von Frijtag Drabbe Künzel, JK; Klaasse, EC; IJzerman, AP; Koomen, GJ J Med Chem 46: 1492-503 (2003)
- Titus, RD; Kornfeld, EC; Jones, ND; Clemens, JA; Smalstig, EB; Fuller, RW; Hahn, RA; Hynes, MD; Mason, NR; Wong, DT; Foreman, MM J Med Chem 26: 1112-6 (1983)
- Sundaramoorthi, R; Siedem, C; Vu, CB; Dalgarno, DC; Laird, EC; Botfield, MC; Combs, AB; Adams, SE; Yuan, RW; Weigele, M; Narula, SS Bioorg Med Chem Lett 11: 1665-9 (2001)
- Lawson, EC; Hoekstra, WJ; Addo, MF; Andrade-Gordon, P; Damiano, BP; Kauffman, JA; Mitchell, JA; Maryanoff, BE Bioorg Med Chem Lett 11: 2619-22 (2001)
- Filipski, KJ; Bian, J; Ebner, DC; Lee, EC; Li, JC; Sammons, MF; Wright, SW; Stevens, BD; Didiuk, MT; Tu, M; Perreault, C; Brown, J; Atkinson, K; Tan, B; Salatto, CT; Litchfield, J; Pfefferkorn, JA; Guzman-Perez, A Bioorg Med Chem Lett 22: 415-20 (2011)
- Emert-Sedlak, L; Kodama, T; Lerner, EC; Dai, W; Foster, C; Day, BW; Lazo, JS; Smithgall, TE ACS Chem Biol 4: 939-47 (2009)
- Shreder, KR; Lin, EC; Wu, J; Cajica, J; Amantea, CM; Hu, Y; Okerberg, E; Brown, HE; Pham, LM; Chung, de M; Fraser, AS; McGee, E; Rosenblum, JS; Kozarich, JW Bioorg Med Chem Lett 22: 5748-51 (2012)
- Murugesan, N; Gu, Z; Spergel, S; Young, M; Chen, P; Mathur, A; Leith, L; Hermsmeier, M; Liu, EC; Zhang, R; Bird, E; Waldron, T; Marino, A; Koplowitz, B; Humphreys, WG; Chong, S; Morrison, RA; Webb, ML; Moreland, S; Trippodo, N; Barrish, JC J Med Chem 46: 125-37 (2002)
- Bock, MG; DiPardo, RM; Mellin, EC; Newton, RC; Veber, DF; Freedman, SB; Smith, AJ; Patel, S; Kemp, JA; Marshall, GR J Med Chem 37: 722-4 (1994)
- Peet, NP; Lentz, NL; Meng, EC; Dudley, MW; Ogden, AM; Demeter, DA; Weintraub, HJ; Bey, P J Med Chem 33: 3127-30 (1991)
- Davis, GC; Kong, Y; Paige, M; Li, Z; Merrick, EC; Hansen, T; Suy, S; Wang, K; Dakshanamurthy, S; Cordova, A; McManus, OB; Williams, BS; Chruszcz, M; Minor, W; Patel, MK; Brown, ML Bioorg Med Chem 20: 2180-8 (2012)
- Candito, DA; Simov, V; Gulati, A; Kattar, S; Chau, RW; Lapointe, BT; Methot, JL; DeMong, DE; Graham, TH; Kurukulasuriya, R; Keylor, MH; Tong, L; Morriello, GJ; Acton, JJ; Pio, B; Liu, W; Scott, JD; Ardolino, MJ; Martinot, TA; Maddess, ML; Yan, X; Gunaydin, H; Palte, RL; McMinn, SE; Nogle, L; Yu, H; Minnihan, EC; Lesburg, CA; Liu, P; Su, J; Hegde, LG; Moy, LY; Woodhouse, JD; Faltus, R; Xiong, T; Ciaccio, P; Piesvaux, JA; Otte, KM; Kennedy, ME; Bennett, DJ; DiMauro, EF; Fell, MJ; Neelamkavil, S; Wood, HB; Fuller, PH; Ellis, JM J Med Chem 65: 16801-16817 (2022)
- Naumann, EC; Göring, S; Ogorek, I; Weggen, S; Schmidt, B Bioorg Med Chem Lett 23: 3852-6 (2013)
- Pan, W; Miao, HQ; Xu, YJ; Navarro, EC; Tonra, JR; Corcoran, E; Lahiji, A; Kussie, P; Kiselyov, AS; Wong, WC; Liu, H Bioorg Med Chem Lett 16: 409-12 (2005)
- Dhar, A; Liu, S; Klucik, J; Berlin, KD; Madler, MM; Lu, S; Ivey, RT; Zacheis, D; Brown, CW; Nelson, EC; Birckbichler, PJ; Benbrook, DM J Med Chem 42: 3602-14 (1999)
- Reid, M; Carlyle, I; Caulfield, WL; Clarkson, TR; Cusick, F; Epemolu, O; Gilfillan, R; Goodwin, R; Jaap, D; O'Donnell, EC; Presland, J; Rankovic, Z; Spinks, D; Spinks, G; Thomson, AM; Thomson, F; Strain, J; Wishart, G Bioorg Med Chem Lett 20: 3713-6 (2010)
- Thakur, A; Tawa, GJ; Henderson, MJ; Danchik, C; Liu, S; Shah, P; Wang, AQ; Dunn, G; Kabir, M; Padilha, EC; Xu, X; Simeonov, A; Kharbanda, S; Stone, R; Grewal, G J Med Chem 63: 4256-4292 (2020)
- Song, D; Lee, JY; Park, EC; Choi, NE; Nam, HY; Seo, J; Lee, J Bioorg Med Chem Lett 87: (2023)
- Ji, C; Sharma, I; Pratihar, D; Hudson, LL; Maura, D; Guney, T; Rahme, LG; Pesci, EC; Coleman, JP; Tan, DS ACS Chem Biol 11: 3061-3067 (2016)
- Zhang, Q; Major, MB; Takanashi, S; Camp, ND; Nishiya, N; Peters, EC; Ginsberg, MH; Jian, X; Randazzo, PA; Schultz, PG; Moon, RT; Ding, S Proc Natl Acad Sci U S A 104: 7444-8 (2007)
- Grasberger, BL; Lu, T; Schubert, C; Parks, DJ; Carver, TE; Koblish, HK; Cummings, MD; LaFrance, LV; Milkiewicz, KL; Calvo, RR; Maguire, D; Lattanze, J; Franks, CF; Zhao, S; Ramachandren, K; Bylebyl, GR; Zhang, M; Manthey, CL; Petrella, EC; Pantoliano, MW; Deckman, IC; Spurlino, JC; Maroney, AC; Tomczuk, BE; Molloy, CJ; Bone, RF J Med Chem 48: 909-12 (2005)
- DeRatt, LG; Pietsch, EC; Cisar, JS; Jacoby, E; Kazmi, F; Matico, R; Shaffer, P; Tanner, A; Wang, W; Attar, R; Edwards, JP; Kuduk, SD ACS Med Chem Lett 15: 381-387 (2024)
- Das, BC; Madhukumar, AV; Anguiano, J; Kim, S; Sinz, M; Zvyaga, TA; Power, EC; Ganellin, CR; Mani, S Bioorg Med Chem Lett 18: 3974-7 (2008)
- Kalisiak, J; Ralph, EC; Cashman, JR J Med Chem 55: 465-74 (2012)
- Yamazaki, DAS; Rozada, AMF; Baréa, P; Reis, EC; Basso, EA; Sarragiotto, MH; Seixas, FAV; Gauze, GF Bioorg Med Chem 32: (2021)
- Wissner, A; Overbeek, E; Reich, MF; Floyd, MB; Johnson, BD; Mamuya, N; Rosfjord, EC; Discafani, C; Davis, R; Shi, X; Rabindran, SK; Gruber, BC; Ye, F; Hallett, WA; Nilakantan, R; Shen, R; Wang, YF; Greenberger, LM; Tsou, HR J Med Chem 46: 49-63 (2002)
- Scott EC
- Zhang, T; Chen, Y; Guo, L; Hruza, A; Jian, T; Li, B; Meng, D; Parker, DL; Sherer, EC; Wood, HB; Sakurada, I US Patent US10189819 (2019)
- Hazai, E; Joshi, P; Skoviak, EC; Suryanarayanan, A; Schulte, MK; Bikadi, Z Bioorg Med Chem 17: 5796-805 (2009)
- Smith, EC; McQuaid, LA; Calligaro, DO; O'Malley, PJ Bioorg Med Chem Lett 3: 81-84 (1993)
- Wipf, P; Hopkins, TD; Jung, JK; Rodriguez, S; Birmingham, A; Southwick, EC; Lazo, JS; Powis, G Bioorg Med Chem Lett 11: 2637-41 (2001)
- Akkari, R; Alvey, LJ; Bock, XM; Brown, BS; Claes, PI; Cowart, MD; Conrath, KE; Cyr, D; De Lemos, E; De Wilde, GJ; Desroy, N; Duthion, B; Gfesser, GA; Gosmini, RL; Housseman, CG; Jansen, KK; Ji, J; Kym, PR; Lefrancois, J; Mammoliti, O; Menet, CJ; Merayo, NM; Newsome, GJ; Palisse, AM; Patel, SV; Pizzonero, MR; Shrestha, A; Swift, EC; Tse, C; Van der Plas, SE; Wang, X US Patent US10130622 (2018)
- Yao, L; Mustafa, N; Tan, EC; Poulsen, A; Singh, P; Duong-Thi, MD; Lee, JXT; Ramanujulu, PM; Chng, WJ; Yen, JJY; Ohlson, S; Dymock, BW J Med Chem 60: 8336-8357 (2017)
- Taylor, EC; Dowling, JE Bioorg Med Chem Lett 7: 453-456 (1997)
- Nguyen, T; Decker, AM; Snyder, RW; Tonetti, EC; Gamage, TF; Zhang, Y Eur J Med Chem 231: (2022)
- Karatas, H; Townsend, EC; Bernard, D; Dou, Y; Wang, S J Med Chem 53: 5179-85 (2010)
- Truong, EC; Phuan, PW; Reggi, AL; Ferrera, L; Galietta, LJV; Levy, SE; Moises, AC; Cil, O; Diez-Cecilia, E; Lee, S; Verkman, AS; Anderson, MO J Med Chem 60: 4626-4635 (2017)
- Debenham, JS; Graham, TH; Verras, A; Zhang, Y; Clements, MJ; Kuethe, JT; Madsen-Duggan, C; Liu, W; Bhatt, UR; Chen, D; Chen, Q; Garcia-Calvo, M; Geissler, WM; He, H; Li, X; Lisnock, J; Shen, Z; Tong, X; Tung, EC; Wiltsie, J; Xu, S; Hale, JJ; Pinto, S; Shen, DM Bioorg Med Chem Lett 23: 6228-33 (2013)
- Wilkening, RR; Ratcliffe, RW; Tynebor, EC; Wildonger, KJ; Fried, AK; Hammond, ML; Mosley, RT; Fitzgerald, PM; Sharma, N; McKeever, BM; Nilsson, S; Carlquist, M; Thorsell, A; Locco, L; Katz, R; Frisch, K; Birzin, ET; Wilkinson, HA; Mitra, S; Cai, S; Hayes, EC; Schaeffer, JM; Rohrer, SP Bioorg Med Chem Lett 16: 3489-94 (2006)
- Parr, BT; Pastor, R; Sellers, BD; Pei, Z; Jaipuri, FA; Castanedo, GM; Gazzard, L; Kumar, S; Li, X; Liu, W; Mendonca, R; Pavana, RK; Potturi, H; Shao, C; Velvadapu, V; Waldo, JP; Wu, G; Yuen, PW; Zhang, Z; Zhang, Y; Harris, SF; Oh, AJ; DiPasquale, A; Dement, K; La, H; Goon, L; Gustafson, A; VanderPorten, EC; Mautino, MR; Liu, Y ACS Med Chem Lett 11: 541-549 (2020)
- Ma, Y; Yang, KS; Geng, ZZ; Alugubelli, YR; Shaabani, N; Vatansever, EC; Ma, XR; Cho, CC; Khatua, K; Xiao, J; Blankenship, LR; Yu, G; Sankaran, B; Li, P; Allen, R; Ji, H; Xu, S; Liu, WR Eur J Med Chem 240: (2022)
- Jungheim, LN; Cohen, JD; Johnson, RB; Villarreal, EC; Wakulchik, M; Loncharich, RJ; Wang, QM Bioorg Med Chem Lett 7: 1589-1594 (1997)
- Spiliotopoulos, D; Zhu, J; Wamhoff, EC; Deerain, N; Marchand, JR; Aretz, J; Rademacher, C; Caflisch, A Bioorg Med Chem Lett 27: 2472-2478 (2017)
- Bar-Tana, J; Ben-Shoshan, S; Blum, J; Migron, Y; Hertz, R; Pill, J; Rose-Khan, G; Witte, EC J Med Chem 32: 2072-84 (1989)
- Inglis, SR; Zervosen, A; Woon, EC; Gerards, T; Teller, N; Fischer, DS; Luxen, A; Schofield, CJ J Med Chem 52: 6097-106 (2009)
- Liu, Y; Yang, EC; Yang, YW; Zhang, HY; Fan, Z; Ding, F; Cao, R J Org Chem 69: 173-80 (2004)
- Cheltsov, AV; Aoyagi, M; Aleshin, A; Yu, EC; Gilliland, T; Zhai, D; Bobkov, AA; Reed, JC; Liddington, RC; Abagyan, R J Med Chem 53: 3899-906 (2010)
- Dorn, CP; Zimmerman, M; Yang, SS; Yurewicz, EC; Ashe, BM; Frankshun, R; Jones, H J Med Chem 20: 1464-8 (1977)
- Zimmerman, SS; Khatri, A; Garnier-Amblard, EC; Mullasseril, P; Kurtkaya, NL; Gyoneva, S; Hansen, KB; Traynelis, SF; Liotta, DC J Med Chem 57: 2334-56 (2014)
- CECERE, G; GOBBI, L; HERNANDEZ, M; KNOFLACH, F; KOBLET, A; O''CONNOR, EC; OLIVARES MORALES, AM; REUTLINGER, M; RUNTZ-SCHMITT, V; WAMSTEEKER CUSULIN, JI; ZORN, N US Patent US20240002393 (2024)
- ChEMBL_54739 (CHEMBL667190) Inhibitory activity against Escherichia coli (ec) Dihydrofolate reductase
- Enzyme Inhibition Assay Enzyme inhibition assay using neutrol endopeptiase EC 3.4.24.11 (NEP).
- ChEMBL_54740 (CHEMBL667191) Inhibitory activity against recombinant ec dihydrofolate reductase(in 50 uM dihydrofolic acid)
- LPO/TPO Selectivity Assay The halogenation activities of LPO (EC 1.11.1.7) and TPO (EC 1.11.1.8) were determined using a modified method of that described previously [Ferrari et al., J. Inorg. Biochem., 68:61-69].
- ChEBML_144313 The compound was tested for its inhibitory activity against neutral endopeptidase (EC.3.4.24.111) in rat kidney
- ChEMBL_47172 (CHEMBL883496) In vitro concentration that causes 50% inhibition of carbonic anhydrase carbonate hydro-lyase (EC.4.2.1.1) isolated from bovine erythrocyte
- ChEMBL_87236 (CHEMBL696456) Compound was tested against histamine N-methyl-transferase in rat brain [EC 2.1.1.8](relative weak affinity on rat enzyme)
- ChEMBL_1546959 (CHEMBL3748062) Inhibition of 6His-tagged human recombinant IDO1 expressed in Escherichia coli EC 539 using L-tryptophan as substrate after 1 hr by plate reader analysis
- ChEMBL_2235845 (CHEMBL5149617) Agonist activity at human CB2 receptor expressed in CHO-K1 cells assessed as inhibition of froskolin-stimulated cAMP accumulation incubated for 90 mins in presence of EC-21a by HitHunter chemiluminescence based assay
- Inhibition of A-beta Fibril Formation EC/IC50 was the effective concentration of agonist inhibiting the formation of A-beta fibrils to 50% of the control value. Three independent measurements were made for all cases.
- ChEMBL_2235846 (CHEMBL5149618) Agonist activity at human CB2 receptor expressed in CHO-K1 cells assessed as inhibition of froskolin-stimulated cAMP accumulation at 50 nM incubated for 90 mins in presence of EC-21a by HitHunter chemiluminescence based assay relative to CP55,940
- In Vitro MKP-3 Dose Response Assay for SAR Study Data Source: Burnham Center for Chemical Genomics (BCCG) Source Affiliation: Burnham Institute for Medical Research (BIMR, La Jolla, CA) Network: NIH Molecular Libraries Screening Centers Network (MLSCN) Grant Number: XO1 MH076390-01 Assay Provider: Dr. John Lazo, University of Pittsburg This MKP-3 dose response assay is developed and performed for the purpose of SAR study on analogs of hits originally identified in the MKP-3 in vitro HTS assay (AID 425) MKP-3 (mitogen-activated protein kinase phosphatase-3; EC 3.1.3.48, EC 3.1.3.16), a dual specificity phosphatase negatively regulates ERK1/2 by catalyzing the removal of a phosphoryl group from Thr(P) and Tyr(P) in the activation loop consensus motif -pTXpY. MKP-3 screening was performed using a biochemical assay developed at the laboratory of Prof. John Lazo (University of Pittsburg). The assay was optimized and run at the Burnham Center for Chemical Genomics (BCCG) as part of the Molecular Library Screening Center Network (MLSCN
- MKP-3 in vitro HTS assay Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Screening Centers Network (MLSCN) Grant Number: XO1 MH076390-01 Assay Provider: Dr. John Lazo, University of Pittsburg MKP-3 (mitogen-activated protein kinase phosphatase-3; EC 3.1.3.48, EC 3.1.3.16), a dual specificity phosphatase negatively regulates ERK1/2 by catalyzing the removal of a phosphoryl group from Thr(P) and Tyr(P) in the activation loop consensus motif -pTXpY. MKP-3 screening was performed using a biochemical assay developed at the laboratory of Prof. John Lazo (University of Pittsburg). The assay was optimized and run at the Sanford-Burnham Center for Chemical Genomics (SBCCG) as part of the Molecular Library Screening Center Network (MLSCN). Enzyme activity and its inhibition by screened compounds was measured in an end-point assay based on hydrolysis of 3-O-methylfluorescein phosphate
- MKP-3 in vitro secondary assay for identification of irreversible and slow-binding inhibitors Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute(SBMRI, San Diego, CA) Network: NIH Molecular Libraries Screening Centers Network (MLSCN) Grant Number: XO1 MH076390-01 Assay Provider: Dr. John Lazo, University of Pittsburg This assay is designed as a counter-screen for the MKP-3 in vitro HTS assay (AID 425) aimed at identification of compounds with time-dependent behavior. MKP-3 (mitogen-activated protein kinase phosphatase-3; EC 3.1.3.48, EC 3.1.3.16), a dual specificity phosphatase negatively regulates ERK1/2 by catalyzing the removal of a phosphoryl group from Thr(P) and Tyr(P) in the activation loop consensus motif -pTXpY. MKP-3 has a labile cysteine in its active site that is crucial for the catalysis. As a result compounds capable of cysteine oxidation or modification are likely to appear as screening hits. The current biochemical assay is designed as a rapid diagnostic tool for identification of
- Competition Binding Assay and Inhibition of Ca2+ Uptake Assay Binding assay mixtures were set up and contained [3H] RTX, various concentrations of competing ligands, and rVR1-transfected CHO cells. Nonspecific binding was determined in the presence of 100 nM nonradioactive RTX. After the binding reaction, membrane-bound RTX was separated from the free by centrifugation, and the radioactivity was determined by scintillation counting. Equilibrium binding parameters (Ki, Bmax, and cooperativity) were determined by fitting the Hill equation to the measured values with the aid of the program Origin 6.0. EC/IC50 is the functional potencies of TRPV1 antagonists against 50 nM capsaicin at the CHO/rVR1 cells, as determined in a Ca2+ uptake assay.
- Competition Binding Assay and Inhibition of Ca2+ Uptake Assay Binding assay mixtures were set up and contained [3H] RTX, various concentrations of competing ligands, and rVR1-transfected CHO cells. Nonspecific binding was determined in the presence of 100 nM nonradioactive RTX. After the binding reaction, membrane-bound RTX was separated from the free by centrifugation, and the radioactivity was determined by scintillation counting. Equilibrium binding parameters (Ki, Bmax, and cooperativity) were determined by fitting the Hill equation to the measured values with the aid of the program Origin 6.0. EC/IC50 is the functional potencies of TRPV1 antagonists against 50 nM capsaicin at the CHO/rVR1 cells, as determined in a Ca2+ uptake assay.
- Radioligand Binding Assay and Agonist Ca2+ Influx Functional Assay The hTRPV1-expressing CHO cell membranes were incubated with [3H]A-778317 and test compounds to establish equilibrium. After incubation was terminated, bound tritium radioactivity (dpm) was counted in a scintillation counter. The Ki values were determined by the application of the Cheng-Prusoff equation: Ki = (IC50 x Kd)/(Kd+[L]) where [L] is the concentration of [3H]A-778317 (2 nM ) and Kd is the dissociation constant of the [3H]A-778317 determined in the saturation analysis (Kd=6.2 nM for A-778317). EC/IC50 is the functional potencies of TRPV1 agonist at the hTRPV1 receptor, as determined in a Ca2+ influx assay.
- Radioligand Binding Assay and Antagonist Ca2+ Influx Functional Assay The hTRPV1-expressing CHO cell membranes were incubated with [3H]A-778317 and test compounds to establish equilibrium. After incubation was terminated, bound tritium radioactivity (dpm) was counted in a scintillation counter. The Ki values were determined by the application of the Cheng-Prusoff equation: Ki = (IC50 x Kd)/(Kd+[L]) where [L] is the concentration of [3H]A-778317 (2 nM ) and Kd is the dissociation constant of the [3H]A-778317 determined in the saturation analysis (Kd=6.2 nM for A-778317). EC/IC50 is the functional potencies of TRPV1 antagonists against 50 nM CAP at the hTRPV1 receptor, as determined in a Ca2+ influx assay.
- BCA Protein Assay Escherichia coli BL21*DE3 pET30a-Ec yeaWX #1 (Ec YeaWX) strain was generated as described below. The contiguous Escherichia coli coding sequence yeaW (equivalent to uniprot ID P0ABR7.1 (YeaW) (SEQ ID NO: 2)) and yeaX (equivalent to uniprot ID P76254.1 (YeaX) (SEQ ID NO: 3)) were PCR amplified from Escherichia coli strain K-12 substr. BW25113 genomic DNA. PCR primers (YeaW_Nde I_fwd2-SEQ ID NO: 4; YeaX_rev2-SEQ ID NO: 5) were designed to create a 5′ NdeI restriction site including the ATG start codon of yeaW and create a PstI restriction site just 3′ of the yeaX TAG stop codon.The bacteria were grown aerobically in 50 mL LB broth (Difco #244620; 10 g/L Tryptone, 5 g/L yeast extract, 10 g/L NaCl, 50 μg/mL kanamycin), in a 500 mL Erlenmeyer flask. The cultures were inoculated from glycerol stock of BL21*DE3 pET30a-Ec yeaWX #1 strain. Strains were cultured all day at 37° C. with 250 rpm shaking. Two 300 mL Minimal M9 Medium (6 g/L Na2HPO4, 3 g/L KH2PO4, 0.5 g/L NaCl, 1 g/L NH4Cl, 0.1 mM CaCl2, 1 mM MgSO4, 0.2% Dextrose, 1 mg/L Thiamine, 50 μg/mL kanamycin), in 1 L Erlenmeyer flasks, were inoculated with 5 mL of the LB broth day culture and cultured overnight at 37° C. with 250 rpm shaking. The overnight cultures were used to inoculate twelve 1 L cultures of Minimal M9 media in 2.8 L fluted Erlenmeyer flasks to an OD 600 nm of 0.05 (typically approximately 28 mLs), which were grown at 37° C. with 250 rpm shaking until an OD600 of approximately 0.4 was reached. Expression of YeaWX was induced with 1 mM IPTG and the induced cultures were further grown overnight at 37° C. with 250 rpm shaking. The biomass was pelleted by centrifugation at 6000×g for 12 minutes at 4° C. The cell pellet was suspended in 240 mL of ice-cold 1× Phosphate Buffered Saline (Ca2+ and Mg2+ free). Ninety micrograms of Lysozyme (Sigma #L6876 Lot #SLBG8654V; Sigma-Aldrich Corp., St. Louis, Mo.) was added and incubated with 320 rpm shaking for 30 minutes at 4° C. Lysis was achieved via French press with a 4° C. prechilled 1″ diameter chamber at 1000 psi (high ratio; internal PSI equivalent 16000). The lysate was centrifuged at 6,000×g for 12 minutes at 4° C. to pellet extra debris. Glycerol was added to the centrifuged lysate supernatant at a final concentration of 15% A protein concentration of the centrifuged lysate supernatant was determined by a BCA Protein Assay Kit (Pierce #23225), typically in the 2.5 to 4.5 mg/ml range. The centrifuged Ec YeaWX lysate supernatant was aliquoted into 20 mL volumes and stored frozen at −80° C.Ec YeaWX lysate was diluted to 2.0 mg/mL protein with 1× Dulbecco's phosphate buffered saline (DPBS) plus 15% glycerol. Nicotinamide adenine dinucleotide phosphate (NADPH) was added to 250 μM. One hundred and fifty microliters of Ec YeaWX lysate was dispensed into a deep-well plate (polypropylene, 2 mL volume, Corning Axygen catalogue #P-DW-20-C). Candidate IC50 compounds from TABLE 1 and vehicle control (respective vehicle control of DMSO or water), or control compounds (IC50 control, 8-Quinolinol hemisulfate salt (Sigma Catalog #55100)) were added at a 1:100 dilution (e.g., 1.5 μL per well). The plates were agitated on a plate shaker for 1 minute. d9-carnitine chloride (1.5 μL of 5 mM) was added to all wells to reach a final d9-carnitine chloride concentration of 50 μM.
- Lipoxygenase Inhibition Assay A mixture of test compound (10 μL; 1 mM) in MeOH, type-1B lipoxygenase (EC 1.13.11.12; from soyabean) (20 μL; 70 units) in 0.1 M aqueous phosphate buffer (pH 8.0) in a total volume of 160 μL was incubated for 10 min at 25°C. Then the reaction was initiated by the addition of a solution of linoleic acid as substrate (10 μL; 20 μM), resulting in the formation of (9Z,11E,13S)-13-hydroperoxyoctadeca-9,11-dienoate. The change in UV absorbance at 234 nm was followed over a period of 6 min. All reactions were performed in triplicate, and analyzed with a 96-well plate reader (SpectraMax Plus 384; Molecular Devices).
- In Vitro α-Glucosidase Inhibitory Assay The α-glucosidase inhibition assay had been carried out using baker's yeast α-glucosidase (EC 3.2.1.20) and p-nitrophenyl α-d-glucopyranoside. The test compounds were dissolved in DMSO to prepare the required distributing concentration. α-Glucosidase inhibitory activity was assayed by using 0.1 M phosphate buffer (pH 6.8) at 37 °C. The enzyme (0.1 U/mL) in phosphate buffer saline was incubated with various concentrations of test compounds at 37 °C for 15 min. Then 1.25 mM p-nitrophenyl α-d-glucopyranoside was added to the mixture as a substrate. After further incubation at 37 °C for 30 min. The absorbance was measured spectrophotometrically at 405 nm. The sample solution was replaced by DMSO as a control. Acarbose was used as a positive control. All experiments were carried out in triplicates.
- Inhibitory Effect of Compounds on Cat K Cathepsin K (CTSK, EC 3.4.22.38) is a lysosomal cysteine protease, participates in osteoclastic bone remodeling and resorption, and can further degrade collagen, gelatin and elastin. The cathepsin K inhibitor screening kit from Biovision uses the ability of the active cathepsin K to cleave a synthetic AFC-based peptide substrate to release AFC and the AFC can be easily quantified by using a fluorometer or fluorescent microplate reader. In the presence of a specific cathepsin K specific inhibitor, the cleavage of the substrate is reduced/eliminated, resulting in a reduction or complete loss of AFC fluorescence. The simple and high-throughput adaptive assay kit can be used to screen/study/characterize a potential inhibitor of the cathepsin K.Figure US20250145576A1-20250508-C00016[0223]20 μL of a buffer, a Cat K inhibitor and test compounds (1 μM and 10 μM) were added to 96-well plates to serve as an EC well, an IC well and an S well, respectively; 50 μL of a Cat K enzyme solution was added into all the wells respectively and the mixture was incubated for 10-15 min at room temperature to establish an enzyme inhibitor complex; 30 μL of a Cat K substrate solution was added into all the wells and the mixture was incubated for 30-60 min at room temperature; and the final volume of the reaction was 100 μL. At 30-60 min, two time points T1 and T2 were selected to detect fluorescence absorption values (Ex/Em=400/505 nm), the fluorescence absorption values were recorded as RFU1 and RFU2, and the inhibition rates (%) of the test compounds on the Cat K were calculated.Slope=(RFU2-RFU1)/(T2-T1)Inhibitionrate(%)=(ECslope-Sslope)/ECslope×100
- Patch Clamp Assay The patch-clamp measurement was carried out at room temperature in whole-cell configuration on human embryonic kidney cells (HEK293) which have been transfected in a stable manner with the hERG gene.The whole-cell configurations were carried out using an automated patch clamp device (Patchliner, Nanion Technologies, Munich). This is a glass chip-based system with which automated whole-cell measurements on up to 8 cells simultaneously are possible. The glass chip has a hole of defined size to which the cell is transferred into the Gigaseal by application of a reduced pressure and brought into the whole-cell configuration. Buffer, cell suspension and test substances were added to microchannels of the chip using a Teflon-coated pipette. The cells were clamped to a holding potential of -80 mV. For measurement of substance-promoted inhibition of the Kv11.1 channel, the following voltage protocol was applied at 10-second intervals: 51 ms/-80 mV, 500 ms/+40 mV, 500 ms/-40 mV, 200 ms/-80 mV. The leakage current is subtracted by means of the P4 method. The cells were resuspended in extracellular buffer (EC) and applied to the chip. After the cell had been collected, the seal was improved by addition of a seal enhancer buffer. As soon as the whole-cell configuration had been reached, the seal enhancer buffer was washed out and replaced by extracellular buffer. The measurement started in EC for 1.5 min. DMSO (vehicle control, 0.1% of DMSO) was then applied, and the control current was recorded for 3 min. The test substance was subsequently added twice in the same concentration, and the potassium current was measured for 3.5 min in each case.
- Activity Inhibition Assay Specifically, in order to determine IC50 of the compounds for neuraminidase, 0.01 U/ml of neuraminidase (EC. 3.2.1.8, C. perfringens, SIGMA, N2876) as an enzyme, 4-methylumbelliferyl-a-D-N-acetylneuraminic acid (SIGMA, M8639) mixed in 0.1 mM acetate buffer [50 mM sodium acetate (pH 5.0)] as a substrate, and inhibitors dissolved in methanol in each concentration were prepared. Fluorescence analysis was observed by SpectraMax M3 through excitation at 365 nm and emission at 450 nm (gain 40 nm). The activity analysis was performed on a 96 well plate (SPL Life Sciences, Korea) by providing total 200 μl of a solution comprising an enzyme (10 μl), a substrate (20 μl), an inhibitor (10 μl), and an acetate buffer (160 μl). By plotting initial rates of inhibition reactions occurring at different concentrations of inhibitors, the inhibition capability was evaluated.
- Cathepsin B dose-response confirmation Screening Center: Penn Center for Molecular Discovery Center Affiliation: University of Pennsylvania Network: Molecular Library Screening Center Network (MLSCN) Assay Provider: Scott Diamond, University of Pennsylvania Grant number: MH076406-01 Human liver cathepsin B (EC 3.4.22.1) is a lysosomal cysteine protease. There has been a recent resurgence of interest in cathepsin B due to research showing that proteolysis by this enzyme is required for the entry and replication of the Ebola and SARS viruses in human cells. Thus cathepsin B inhibitors have potential as novel anti-viral agents. Cathepsin B is also implicated in cancer progression. Upregulation and secretion of this enzyme occurs in many types of tumors and correlates positively with their invasive and metastatic capabilities. Cathepsin B facilitates tumor invasion by dissolving extracellular barriers. Inhibitors of cathepsin B thus have been studied as potential anti-cancer agents. A high-throughput screen for cathepsin B
- Cathepsin G dose-response confirmation Screening Center: Penn Center for Molecular Discovery Center Affiliation: University of Pennsylvania Network: Molecular Library Screening Center Network (MLSCN) Assay Provider: Scott Diamond, University of Pennsylvania Grant number: MH076406-01 Cathepsin G (EC 3.4.21.20) is a chymotrypsin-like serine protease that is secreted from neutrophils. Disregulated cathepsin G activity is implicated in the progression of various chronic inflammatory diseases such as asthma and chronic pulmonary obstructive disease. Thus cathepsin G inhibitors represent useful probes to further elucidate the role of this enzyme in inflammation and may provide a starting point for the development of novel therapeutic agents. A high-throughput screen for cathepsin G inhibitors was designed as an end-point assay monitoring the release of the fluorophore aminomethyl coumarin (AMC) upon enzymatic hydrolysis of an AMC-labeled dipeptide. Primary HTS results have been reported previously (AID 581). Compounds identifi
- Cathepsin L dose-response confirmation Screening Center: Penn Center for Molecular Discovery Center Affiliation: University of Pennsylvania Network: Molecular Library Screening Center Network (MLSCN) Assay Provider: Scott Diamond, University of Pennsylvania Grant number: MH076406-01 Human liver cathepsin L (EC 3.4.22.15) is a lysosomal cysteine protease. Recent interest in cathepsin L has been generated by research showing that proteolysis by this enzyme is required for the entry and replication of the SARS and Ebola viruses in human cells. Thus cathepsin L inhibitors have potential as novel anti-viral agents. Cathepsin L inhibitors may also be active against Plasmodium falciparum, the parasite responsible for human malaria. Plasmodium contains cathepsin L-like cysteine proteases known as falcipains that appear to promote virulence of the parasite through haemoglobin digestion and erythrocyte invasion. A high-throughput screen for cathepsin L inhibitors was designed as an end-point assay monitoring the release of the fl
- Cathepsin S dose-response confirmation Screening Center: Penn Center for Molecular Discovery Center Affiliation: University of Pennsylvania Network: Molecular Library Screening Center Network (MLSCN) Assay Provider: Scott Diamond, University of Pennsylvania Grant number: MH076406-01 Human cathepsin S (EC 3.4.22.27) is a lysosomal cysteine protease that is expressed in antigen-presenting cells, especially dendritic cells, B-cells and macrophages. Cathepsin S plays a key role in the processing of antigenic peptides for presentation by MHC Class II molecules on the surface of antigen-presenting cells. Thus inhibitors of cathepsin S may be immunomodulators effective in the treatment of autoimmune diseases. A high-throughput screen for cathepsin S inhibitors was designed as an end-point assay monitoring the release of the fluorophore aminomethyl coumarin (AMC) upon enzymatic hydrolysis of an AMC-labeled dipeptide. Primary HTS results have been reported previously (AID 501). Compounds identified as hits in HTS of mixtures of 1
- GAPDH Dose Response Colorimetric Assay Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Screening Centers Network (MLSCN) Grant Number: None This glyceraldehydes-3-phosphate dehydrogenase (GAPDH; EC 1.2.1.12) dose-response assay is developed and performed at the Sanford-Burnham Center for Chemical Genomics for characterization of the hits of biochemical assays. GAPDH is found in all mammalian tissues, and is considered a "housekeeping enzyme" unaffected by most physiological, hormonal, and metabolic changes. Therefore, compounds that inhibit GAPDH are expected to have pronounced cytotoxic effect and would be unfavorable as chemical probes. In addition, GAPDH has a labile cysteine in its active site that is crucial for the catalysis, resembling several other enzyme groups, such as cysteine-based proteases and phosphatases. As a result, the compounds capable of cysteine oxidation or mod
- High Throughput Screen to Identify Compounds that Inhibit Class II HMG-CoA Reductases - Confirmatory Screen Southern Research Molecular Libraries Screening Center (SRMLSC) Southern Research Institute (Birmingham, Alabama) NIH Molecular Libraries Screening Centers Network (MLSCN) Submitted by Dr. Cynthia Stauffacher (Purdue University) Award: 1-R03-MH082373-01 A number of common human pathogens, including Enterococcus faecalis, Streptococcus pneumoniae, and Staphylococcus aureus, are becoming progressively more resistant to antibiotics and pose a serious public health threat, especially to post-surgical and trauma patients. Therefore, the discovery of drugs targeting novel pathways in these pathogens has become increasingly important. The synthesis of isoprenoids, which in these gram-positive pathogenic bacteria (Hedl, 2002) occurs exclusively via the mevalonate pathway, is essential for their survival. The central mevalonate pathway enzyme, 3-hydroxy-3-methylglutaryl coenzyme A reductase (HMGR; EC 1.1.1.34), reduces HMG-CoA to mevalonate using NADPH (Hedl, 2004). Bacterial HMG-CoA redu
- In Vitro Inhibition Assay The in vitro inhibition of recombinant human neutral endopeptidase (NEP, EC 3.424.11) can be determined as follows:Recombinant human neutral endopeptidase (expressed in insect cells and purified using standard methods, final concentration 7 pM) is pre-incubated with test compounds at various concentrations for 1 hour at room temperature in 10 mM sodium phosphate buffer at pH 7.4, containing 150 mM NaCl and 0.05% (w/v) CHAPS. The enzymatic reaction is started by the addition of a synthetic peptide substrate Cys(PT14)-Arg-Arg-Leu-Trp-OH to a final concentration of 0.7 uM. Substrate hydrolysis leads to an increase fluorescence lifetime (FLT) of PT14 measured by the means of a FLT reader as described by Doering et al. (2009). The effect of the compound on the enzymatic activity was determined after 1 hour (t=60 min) incubation at room temperature.
- Luminescent assay for HTS discovery of chemical activators of placental alkaline phosphatase Sanford-Burnham Center for Chemical Genomics (SBCCG) Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) NIH Molecular Libraries Screening Centers Network (MLSCN) Alkaline phosphatase (EC 3.1.3.1) (APs) catalyze the hydrolysis of phosphomonoesters, releasing phosphate and alcohol. APs are dimeric enzymes found in most organisms. In human, four isozymes of APs have been identified: three isozymes are tissue-specific and the fourth one is tissue-nonspecific. Placental alkaline phosphatase (PLAP) is highly expressed in primate placental tissue. Its biological function is unknown. Identification of PLAP-specific inhibitors should provide necessary tools for characterization of its biological role. PLAP screening was developed and performed at the Sanford-Burnham Center for Chemical Genomics (SBCCG) as part of the Molecular Library Screening Center Network (MLSCN). This assay represents a selectivity screening for tissue nonspecific alkaline phosphatase (TNAP) screened at BCC
- Luminescent assay for HTS discovery of chemical inhibitors of placental alkaline phosphatase Sanford-Burnham Center for Chemical Genomics (SBCCG) Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) NIH Molecular Libraries Screening Centers Network (MLSCN) Alkaline phosphatase (EC 3.1.3.1) (APs) catalyze the hydrolysis of phosphomonoesters, releasing phosphate and alcohol. APs are dimeric enzymes found in most organisms. In human, four isozymes of APs have been identified: three isozymes are tissue-specific and the fourth one is tissue-nonspecific. Placental alkaline phosphatase (PLAP) is highly expressed in primate placental tissue. Its biological function is unknown. Identification of PLAP-specific inhibitors should provide necessary tools for characterization of its biological role. PLAP screening was developed and performed at the Sanford-Burnham Center for Chemical Genomics (SBCCG) as part of the Molecular Library Screening Center Network (MLSCN). This assay represents a selectivity screening for tissue nonspecific alkaline phosphatase (TNAP) screened at B
- Mycobacterium tuberculosis Pantothenate Synthetase Assay Southern Research Molecular Libraries Screening Center (SRMLSC) Southern Research Institute (Birmingham, Alabama) NIH Molecular Libraries Screening Centers Network (MLSCN) Award: 1R03MH076412-01 Multi-drug resistant Mycobacterium tuberculosis is becoming an increased health problem, especially in immunocompromised individuals with HIV. This form of TB is more difficult to treat and as a result has a higher mortality rate. Because of this, the discovery of drugs targeting novel pathways such as the synthesis of pantothenate has become increasingly important. Pantothenate synthetase (PS; EC 6.3.2.1), encoded by the panC gene, catalyzes the essential ATP-dependent condensation of D-pantoate and alpha-alanine to form pantothenate in bacteria, yeast and plants; pantothenate is a key precursor for the biosynthesis of coenzyme A (CoA) and acyl carrier protein (ACP). The activity of PS was measured spectrophotometrically by coupling the formation of AMP to the reactions of myokinase, pyr
- Mycobacterium tuberculosis Pantothenate Synthetase Secondary Assay Southern Research Molecular Libraries Screening Center (SRMLSC) Southern Research Institute (Birmingham, Alabama) NIH Molecular Libraries Screening Centers Network (MLSCN) Award: 1R03MH076412-01 Multi-drug resistant Mycobacterium tuberculosis is becoming an increased health problem, especially in immunocompromised individuals with HIV. This form of TB is more difficult to treat and as a result has a higher mortality rate. Because of this, the discovery of drugs targeting novel pathways such as the synthesis of pantothenate has become increasingly important. Pantothenate synthetase (PS; EC 6.3.2.1), encoded by the panC gene, catalyzes the essential ATP-dependent condensation of D-pantoate and alpha-alanine to form pantothenate in bacteria, yeast and plants; pantothenate is a key precursor for the biosynthesis of coenzyme A (CoA) and acyl carrier protein (ACP). The activity of PS was measured spectrophotometrically by coupling the formation of AMP to the reactions of myokinase, pyruva
- RNA polymerase SAR Screening Center: Penn Center for Molecular Discovery Center Affiliation: University of Pennsylvania Network: Molecular Library Screening Center Network (MLSCN) Assay Provider: Arkady Mustaev, Public Health Research Institute, Newark, NJ Grant number: MH076325-01 DNA-directed RNA polymerase (EC 2.7.7.6) is responsible for bacterial RNA synthesis and as such is essential for bacterial gene expression. Owing to its central role in DNA transcription, the enzyme RNA polymerase is the target of various natural antibiotics. The best known is rifampicin, a potent and broad-spectrum anti-infective agent that is particularly effective against intracellular pathogens, such as Mycobacterium tuberculosis, for which it is one of the most widely used chemotherapeutic agents. However, the emergence of drug-resistant bacteria has become a major public health problem, so the discovery of novel RNA polymerase inhibitors is an important goal. A high-throughput screen was designed to discover novel in
- RNA polymerase dose-response confirmation Screening Center: Penn Center for Molecular Discovery Center Affiliation: University of Pennsylvania Network: Molecular Library Screening Center Network (MLSCN) Assay Provider: Arkady Mustaev, Public Health Research Institute, Newark, NJ Grant number: MH076325-01 DNA-directed RNA polymerase (EC 2.7.7.6) is responsible for bacterial RNA synthesis and as such is essential for bacterial gene expression. Owing to its central role in DNA transcription, the enzyme RNA polymerase is the target of various natural antibiotics. The best known is rifampicin, a potent and broad-spectrum anti-infective agent that is particularly effective against intracellular pathogens, such as Mycobacterium tuberculosis, for which it is one of the most widely used chemotherapeutic agents. However, the emergence of drug-resistant bacteria has become a major public health problem, so the discovery of novel RNA polymerase inhibitors is an important goal. A high-throughput screen was designed to discover novel in
- SAR analysis of an In Vitro TNAP Dose Response Luminescent Assay Sanford-Burnham Center for Chemical Genomics (SBCCG) Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) NIH Molecular Libraries Screening Centers Network (MLSCN) Grant Number: MH077602-01 Assay Provider: Dr. Jose Luis Millan, Sanford-Burnham Medical Research Institute, San Diego, CA This TNAP dose response assay is developed and performed for the purpose of SAR study on analogs of hits originally identified in the TNAP luminescent HTS assay (AID 518) Alkaline phosphatase (EC 3.1.3.1) (APs) catalyze the hydrolysis of phosphomonoesters, releasing phosphate and alcohol. APs are dimeric enzymes found in the most organism. In human, four isozymes of APs have been identified. Three isozymes are tissue-specific and the fourth one is tissue-nonsepecifc, named TNAP. TNAP deficiency is associated with defective bone mineralization in the form of rickets and osteomalacia. Therefore, there are therapeutic potentials of inhibiting TNAP activity
- TNAP luminescent HTS assay Sanford-Burnham Center for Chemical Genomics (SBCCG) Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) NIH Molecular Libraries Screening Centers Network (MLSCN) Grant Number: MH077602-01 Alkaline phosphatase (EC 3.1.3.1) (APs) catalyze the hydrolysis of phosphomonoesters, releasing phosphate and alcohol. APs are dimeric enzymes found in the most organism. In human, four isozymes of APs have been identified. Three isozymes are tissue-specific and the fourth one is tissue-nonsepecifc, named TNAP. TNAP deficiency is associated with defective bone mineralization in the form of rickets and osteomalacia. Therefore, there are therapeutic potentials of inhibiting TNAP activity. The goal of this HTS is to identify novel and specific inhibitors of TNAP. TNAP screening was developed and performed at the Sanford-Burnham Center for Chemical Genomics (SBCCG) as part of the Molecular Library Screening Center Network (MLSCN). XO1 submission, MH077602-01, Pharmacological inhibitors o
- alpha-Glucosidase Inhibition Assay The alpha-glucosidase inhibition assay had been carried out using baker's yeast alpha-glucosidase (EC 3.2.1.20) and p-nitrophenyl-alpha-D-glucopyranoside (46 50). The samples (5 ug/mL) were prepared by dissolving the compounds 1-15 in DMSO. Test samples (10 uL) which had been prepared were reconstituted in 100 uL of phosphate buffer (100 mM) at pH 6.8 in 96-well microplate and incubated with 50 uL of baker's yeast alpha-glucosidase for 5 min before 50 uL of p-nitrophenyl-alpha-D-glucopyranoside (5 mM) was added. After incubating for 5 min, the absorbance was measured at 405 nm using SpectraMax plus384 (Molecular Devices Corporation, Sunnyvale, CA, USA). Blank in which the substrate was changed with 50 uL of buffer was analysed to accurately determine the background absorbance. Control sample was prepared to contain 10 uL DMSO instead of test samples.
- Antiviral Assay To evaluate the anti-HSV activity of 1, 17 or Brequinar by plaque reduction assays (PRA), Vero cells were seeded in 24-well plates at a density of 70×103 cells. After 24 h, cells were treated with different concentrations of 17, 1 or Brequinar 1 h prior to infection, and then infected with HSV-1 or HSV-2 (50 PFU/well). Following virus adsorption (2 h at 37 C), cultures were maintained in medium-containing 0.8% methylcellulose (Sigma) plus compounds. At 48 h post infection (h.p.i.), cells were fixed and stained by using 20% ethanol and 1% crystal violet. Plaques were microscopically counted, and the mean plaque counts for each concentration expressed as a percentage of the mean plaque count for the control virus. The number of plaques was plotted as a function of drug concentration; concentration producing 50% reduction in plaque formation (EC 50) was determined as described by Terlizzi et al. (Antiviral Research 132, 154-164, 2016).
- Cathepsin B Inhibitor Series SAR Study Human liver cathepsin B (EC 3.4.22.1) is a lysosomal cysteine protease. There has been a recent resurgence of interest in cathepsin B due to research showing that proteolysis by this enzyme is required for the entry and replication of the Ebola and SARS viruses in human cells. Thus cathepsin B inhibitors have potential as novel anti-viral agents. Cathepsin B is also implicated in cancer progression. Upregulation and secretion of this enzyme occurs in many types of tumors and correlates positively with their invasive and metastatic capabilities. Cathepsin B facilitates tumor invasion by dissolving extracellular barriers. Inhibitors of cathepsin B thus have been studied as potential anti-cancer agents. Two high-throughput screens against cathepsin B reported previously under AID 453 and AID 488 each independently revealed the same series of aminopyrazole inhibitors. Six compounds active in the original HTS were resynthesized, and 21 new compounds were made to explore SAR. As in the o
- Cathepsin L dose-response testing in the presence of cysteine Screening Center: Penn Center for Molecular Discovery Center Affiliation: University of Pennsylvania Network: Molecular Library Screening Center Network (MLSCN) Assay Provider: Scott Diamond, University of Pennsylvania Grant number: MH076406-01 Human liver cathepsin L (EC 3.4.22.15) is a lysosomal cysteine protease. Recent interest in cathepsin L has been generated by research showing that proteolysis by this enzyme is required for the entry and replication of the SARS and Ebola viruses in human cells. Thus cathepsin L inhibitors have potential as novel anti-viral agents. Cathepsin L inhibitors may also be active against Plasmodium falciparum, the parasite responsible for human malaria. Plasmodium contains cathepsin L-like cysteine proteases known as falcipains that appear to promote virulence of the parasite through haemoglobin digestion and erythrocyte invasion. A high-throughput screen for cathepsin L inhibitors was designed as an end-point assay monitoring the release of the fl
- Cathepsin L probe #2 dose-response testing Screening Center: Penn Center for Molecular Discovery Center Affiliation: University of Pennsylvania Network: Molecular Library Screening Center Network (MLSCN) Assay Provider: Scott Diamond, University of Pennsylvania Grant number: MH076406-01 Human liver cathepsin L (EC 3.4.22.15) is a lysosomal cysteine protease. Recent interest in cathepsin L has been generated by research showing that proteolysis by this enzyme is required for the entry and replication of the SARS and Ebola viruses in human cells. Thus cathepsin L inhibitors have potential as novel anti-viral agents. Cathepsin L inhibitors may also be active against Plasmodium falciparum, the parasite responsible for human malaria. Plasmodium contains cathepsin L-like cysteine proteases known as falcipains that appear to promote virulence of the parasite through haemoglobin digestion and erythrocyte invasion. A high-throughput screen for cathepsin L inhibitors was designed as an end-point assay monitoring the release of the fl
- Complement factor C1s IC50 from mixture screen Molecular Library Screening Center Network (MLSCN) Penn Center for Molecular Discovery (PCMD) Assay Provider: Dr. Scott L. Diamond, University of Pennsylvania MLSCN Grant: X01-MH076406-01 Complement factor C1s (EC 3.4.21.42) is a trypsin-like serine protease that is activated in one of the first steps in the classical complement cascade. Despite the essential role for the complement cascade in immune defense, unregulated activation leading to acute inflammation and tissue damage has been implicated in many disease states. Under normal conditions the activity of C1s is modulated by its endogenous inhibitor, C1 esterase inhibitor. Pathological conditions lead to excessive activation of C1s; thus a small molecule inhibitor would be useful in the treatment of ischemia-reperfusion injury and other complement-mediated diseases. Assay The high-throughput screen for complement factor C1s inhibitors has been reported earlier (AID 538). The assay consisted of an end-point assay monitoring th
- Dose Response concentration confirmation of uHTS hits from a small molecule activators of human intestinal alkaline phosphatase via a luminescent assay - Set 2 Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Production Centers Network (MLPCN) Grant Proposal Number: X01-MH077602-01 Assay Provider Dr. Jose Luis Millan, Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Alkaline phosphatase (EC 3.1.3.1) (APs) catalyze the hydrolysis of phosphomonoesters, releasing inorganic phosphate and alcohol. APs are dimeric enzymes found in most organisms. In human, four isozymes of APs have been identified. One isozyme is tissue-nonspecific (designated TNAP) and three other isozymes are tissue-specific and named according to the tissue of their predominant expression: intestinal (IAP), placental (PLAP) and germ cell (GCAP) alkaline phosphatases. IAP expression is largely restricted to the gut, especially to the epithelial cells (enterocytes) of the small intestinal mucosa. The exact biological function of
- Dose Response concentration confirmation of uHTS hits from a small molecule activators of human intestinal alkaline phosphatase via a luminescent assay Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Production Centers Network (MLPCN) Grant Proposal Number: X01-MH077602-01 Assay Provider Dr. Jose Luis Millan, Sanford-Burnham Medical Research Institute(SBMRI, San Diego, CA) Alkaline phosphatase (EC 3.1.3.1) (APs) catalyze the hydrolysis of phosphomonoesters, releasing inorganic phosphate and alcohol. APs are dimeric enzymes found in most organisms. In human, four isozymes of APs have been identified. One isozyme is tissue-nonspecific (designated TNAP) and three other isozymes are tissue-specific and named according to the tissue of their predominant expression: intestinal (IAP), placental (PLAP) and germ cell (GCAP) alkaline phosphatases. IAP expression is largely restricted to the gut, especially to the epithelial cells (enterocytes) of the small intestinal mucosa. The exact biological function of
- Dose Response concentration confirmation of uHTS hits from a small molecule activators of mouse intestinal alkaline phosphatase via a luminescent assay Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Production Centers Network (MLPCN) Grant Proposal Number: X01-MH077602-01 Alkaline phosphatase (EC 3.1.3.1) (APs) catalyze the hydrolysis of phosphomonoesters, releasing inorganic phosphate and alcohol. APs are dimeric enzymes found in most organisms. In human, four isozymes of APs have been identified. One isozyme is tissue-nonspecific (designated TNAP) and three other isozymes are tissue-specific and named according to the tissue of their predominant expression: intestinal (IAP), placental (PLAP) and germ cell (GCAP) alkaline phosphatases. IAP expression is largely restricted to the gut, especially to the epithelial cells (enterocytes) of the small intestinal mucosa. The exact biological function of IAP is unknown. The goal of this HTS is to confirm hits in "uHTS Luminescent assay for identificatio
- Dose Response confirmation of uHTS activators of Human Intestinal Alkaline Phosphatase using Mouse Intestinal Alkaline Phosphatase Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Production Centers Network (MLPCN) Grant Proposal Number: X01-MH077602-01 Alkaline phosphatase (EC 3.1.3.1) (APs) catalyze the hydrolysis of phosphomonoesters, releasing inorganic phosphate and alcohol. APs are dimeric enzymes found in most organisms. In human, four isozymes of APs have been identified. One isozyme is tissue-nonspecific (designated TNAP) and three other isozymes are tissue-specific and named according to the tissue of their predominant expression: intestinal (IAP), placental (PLAP) and germ cell (GCAP) alkaline phosphatases. IAP expression is largely restricted to the gut, especially to the epithelial cells (enterocytes) of the small intestinal mucosa. The exact biological function of IAP is unknown. The goal of this MLPCN probe project is to identify novel and specific activators of
- Dose Response confirmation of uHTS activators of Human Intestinal Alkaline Phosphatase using Placental Alkaline Phosphatase Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Production Centers Network (MLPCN) Grant Proposal Number: X01-MH077602-01 Assay Provider Dr. Jose Luis Millan, Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Alkaline phosphatase (EC 3.1.3.1) (APs) catalyze the hydrolysis of phosphomonoesters, releasing inorganic phosphate and alcohol. APs are dimeric enzymes found in most organisms. In human, four isozymes of APs have been identified. One isozyme is tissue-nonspecific (designated TNAP) and three other isozymes are tissue-specific and named according to the tissue of their predominant expression: intestinal (IAP), placental (PLAP) and germ cell (GCAP) alkaline phosphatases. IAP expression is largely restricted to the gut, especially to the epithelial cells (enterocytes) of the small intestinal mucosa. The exact biological function of
- Dose Response confirmation of uHTS activators of Human Intestinal Alkaline Phosphatase using Tissue Nonspecific Alkaline Phosphatase. Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Production Centers Network (MLPCN) Grant Proposal Number: X01-MH077602-01 Assay Provider Dr. Jose Luis Millan, Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Alkaline phosphatase (EC 3.1.3.1) (APs) catalyze the hydrolysis of phosphomonoesters, releasing inorganic phosphate and alcohol. APs are dimeric enzymes found in most organisms. In human, four isozymes of APs have been identified. One isozyme is tissue-nonspecific (designated TNAP) and three other isozymes are tissue-specific and named according to the tissue of their predominant expression: intestinal (IAP), placental (PLAP) and germ cell (GCAP) alkaline phosphatases. IAP expression is largely restricted to the gut, especially to the epithelial cells (enterocytes) of the small intestinal mucosa. The exact biological function of
- Dose Response confirmation of uHTS activators of Mouse Intestinal Alkaline Phosphatase using Human Intestinal Alkaline Phosphatase Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Production Centers Network (MLPCN) Grant Proposal Number: X01-MH077602-01 Assay Provider Dr. Jose Luis Millan, Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Alkaline phosphatase (EC 3.1.3.1) (APs) catalyze the hydrolysis of phosphomonoesters, releasing inorganic phosphate and alcohol. APs are dimeric enzymes found in most organisms. In human, four isozymes of APs have been identified. One isozyme is tissue-nonspecific (designated TNAP) and three other isozymes are tissue-specific and named according to the tissue of their predominant expression: intestinal (IAP), placental (PLAP) and germ cell (GCAP) alkaline phosphatases. IAP expression is largely restricted to the gut, especially to the epithelial cells (enterocytes) of the small intestinal mucosa. The exact biological function of
- Dose Response confirmation of uHTS activators of Mouse Intestinal Alkaline Phosphatase using Placental Alkaline Phosphatase Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Production Centers Network (MLPCN) Grant Proposal Number: X01-MH077602-01 Alkaline phosphatase (EC 3.1.3.1) (APs) catalyze the hydrolysis of phosphomonoesters, releasing inorganic phosphate and alcohol. APs are dimeric enzymes found in most organisms. In human, four isozymes of APs have been identified. One isozyme is tissue-nonspecific (designated TNAP) and three other isozymes are tissue-specific and named according to the tissue of their predominant expression: intestinal (IAP), placental (PLAP) and germ cell (GCAP) alkaline phosphatases. IAP expression is largely restricted to the gut, especially to the epithelial cells (enterocytes) of the small intestinal mucosa. The exact biological function of IAP is unknown. The goal of this HTS is to confirm hits in "uHTS Luminescent assay for identificatio
- Dose Response confirmation of uHTS activators of Mouse Intestinal Alkaline Phosphatase using Tissue Nonspecific Alkaline Phosphatase. Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Production Centers Network (MLPCN) Grant Proposal Number: X01-MH077602-01 Alkaline phosphatase (EC 3.1.3.1) (APs) catalyze the hydrolysis of phosphomonoesters, releasing inorganic phosphate and alcohol. APs are dimeric enzymes found in most organisms. In human, four isozymes of APs have been identified. One isozyme is tissue-nonspecific (designated TNAP) and three other isozymes are tissue-specific and named according to the tissue of their predominant expression: intestinal (IAP), placental (PLAP) and germ cell (GCAP) alkaline phosphatases. IAP expression is largely restricted to the gut, especially to the epithelial cells (enterocytes) of the small intestinal mucosa. The exact biological function of IAP is unknown. The goal of this HTS is to confirm hits in "uHTS Luminescent assay for identificatio
- Dose Response confirmation of uHTS hits from a small molecule inhibitors of human intestinal alkaline phosphatase via a luminescent assay - Set 2 Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute(SBMRI, San Diego, CA) Network: NIH Molecular Libraries Probe Production Centers Network (MLPCN) Grant Proposal Number: X01-MH077602-01 Assay Provider Dr. Jose Luis Millan, Sanford-Burnham Medical Research Institute(SBMRI, San Diego, CA) Alkaline phosphatase (EC 3.1.3.1) (APs) catalyze the hydrolysis of phosphomonoesters, releasing inorganic phosphate and alcohol. APs are dimeric enzymes found in most organisms. In human, four isozymes of APs have been identified. One isozyme is tissue-nonspecific (designated TNAP) and three other isozymes are tissue-specific and named according to the tissue of their predominant expression: intestinal (IAP), placental (PLAP) and germ cell (GCAP) alkaline phosphatases. IAP expression is largely restricted to the gut, especially to the epithelial cells (enterocytes) of the small intestinal mucosa. The exact biological functio
- Dose Response confirmation of uHTS hits from a small molecule inhibitors of human intestinal alkaline phosphatase via a luminescent assay Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Production Centers Network (MLPCN) Grant Proposal Number: X01-MH077602-01 Assay Provider Dr. Jose Luis Millan, Sanford-Burnham Medical Research Institute(SBMRI, San Diego, CA) Alkaline phosphatase (EC 3.1.3.1) (APs) catalyze the hydrolysis of phosphomonoesters, releasing inorganic phosphate and alcohol. APs are dimeric enzymes found in most organisms. In human, four isozymes of APs have been identified. One isozyme is tissue-nonspecific (designated TNAP) and three other isozymes are tissue-specific and named according to the tissue of their predominant expression: intestinal (IAP), placental (PLAP) and germ cell (GCAP) alkaline phosphatases. IAP expression is largely restricted to the gut, especially to the epithelial cells (enterocytes) of the small intestinal mucosa. The exact biological function of
- Dose Response confirmation of uHTS hits from a small molecule inhibitors of mouse intestinal alkaline phosphatase via a luminescent assay Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Production Centers Network (MLPCN) Grant Proposal Number: X01-MH077602-01 Assay Provider Dr. Jose Luis Millan, Sanford-Burnham Medical Research Institute(SBMRI, San Diego, CA) Alkaline phosphatase (EC 3.1.3.1) (APs) catalyze the hydrolysis of phosphomonoesters, releasing inorganic phosphate and alcohol. APs are dimeric enzymes found in most organisms. In human, four isozymes of APs have been identified. One isozyme is tissue-nonspecific (designated TNAP) and three other isozymes are tissue-specific and named according to the tissue of their predominant expression: intestinal (IAP), placental (PLAP) and germ cell (GCAP) alkaline phosphatases. IAP expression is largely restricted to the gut, especially to the epithelial cells (enterocytes) of the small intestinal mucosa. The exact biological function of
- Dose Response confirmation of uHTS inhibitors of Human Intestinal Alkaline Phosphatase using Mouse Intestinal Alkaline Phosphatase Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Production Centers Network (MLPCN) Grant Proposal Number: X01-MH077602-01 Assay Provider Dr. Jose Luis Millan, Sanford-Burnham Medical Research Institute(SBMRI, San Diego, CA) Alkaline phosphatase (EC 3.1.3.1) (APs) catalyze the hydrolysis of phosphomonoesters, releasing inorganic phosphate and alcohol. APs are dimeric enzymes found in most organisms. In human, four isozymes of APs have been identified. One isozyme is tissue-nonspecific (designated TNAP) and three other isozymes are tissue-specific and named according to the tissue of their predominant expression: intestinal (IAP), placental (PLAP) and germ cell (GCAP) alkaline phosphatases. IAP expression is largely restricted to the gut, especially to the epithelial cells (enterocytes) of the small intestinal mucosa. The exact biological function of
- Dose Response confirmation of uHTS inhibitors of Human Intestinal Alkaline Phosphatase using Placental Alkaline Phosphatase Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute(SBMRI, San Diego, CA) Network: NIH Molecular Libraries Probe Production Centers Network (MLPCN) Grant Proposal Number: X01-MH077602-01 Assay Provider Dr. Jose Luis Millan, Sanford-Burnham Medical Research Institute, San Diego, CA) Alkaline phosphatase (EC 3.1.3.1) (APs) catalyze the hydrolysis of phosphomonoesters, releasing inorganic phosphate and alcohol. APs are dimeric enzymes found in most organisms. In human, four isozymes of APs have been identified. One isozyme is tissue-nonspecific (designated TNAP) and three other isozymes are tissue-specific and named according to the tissue of their predominant expression: intestinal (IAP), placental (PLAP) and germ cell (GCAP) alkaline phosphatases. IAP expression is largely restricted to the gut, especially to the epithelial cells (enterocytes) of the small intestinal mucosa. The exact biological function of I
- Dose Response confirmation of uHTS inhibitors of Human Intestinal Alkaline Phosphatase using Tissue Nonspecific Alkaline Phosphatase. Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute(SBMRI, San Diego, CA) Network: NIH Molecular Libraries Probe Production Centers Network (MLPCN) Grant Proposal Number: X01-MH077602-01 Assay Provider Dr. Jose Luis Millan, Sanford-Burnham Medical Research Institute(SBMRI, San Diego, CA) Alkaline phosphatase (EC 3.1.3.1) (APs) catalyze the hydrolysis of phosphomonoesters, releasing inorganic phosphate and alcohol. APs are dimeric enzymes found in most organisms. In human, four isozymes of APs have been identified. One isozyme is tissue-nonspecific (designated TNAP) and three other isozymes are tissue-specific and named according to the tissue of their predominant expression: intestinal (IAP), placental (PLAP) and germ cell (GCAP) alkaline phosphatases. IAP expression is largely restricted to the gut, especially to the epithelial cells (enterocytes) of the small intestinal mucosa. The exact biological functio
- Dose Response confirmation of uHTS inhibitors of Mouse Intestinal Alkaline Phosphatase using Human Intestinal Alkaline Phosphatase Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute(SBMRI, San Diego, CA) Network: NIH Molecular Libraries Probe Production Centers Network (MLPCN) Grant Proposal Number: X01-MH077602-01 Assay Provider Dr. Jose Luis Millan, Sanford-Burnham Medical Research Institute(SBMRI, San Diego, CA) Alkaline phosphatase (EC 3.1.3.1) (APs) catalyze the hydrolysis of phosphomonoesters, releasing inorganic phosphate and alcohol. APs are dimeric enzymes found in most organisms. In human, four isozymes of APs have been identified. One isozyme is tissue-nonspecific (designated TNAP) and three other isozymes are tissue-specific and named according to the tissue of their predominant expression: intestinal (IAP), placental (PLAP) and germ cell (GCAP) alkaline phosphatases. IAP expression is largely restricted to the gut, especially to the epithelial cells (enterocytes) of the small intestinal mucosa. The exact biological functio
- Dose Response confirmation of uHTS inhibitors of Mouse Intestinal Alkaline Phosphatase using Placental Alkaline Phosphatase Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Production Centers Network (MLPCN) Grant Proposal Number: X01-MH077602-01 Assay Provider Dr. Jose Luis Millan, Sanford-Burnham Medical Research Institute(SBMRI, San Diego, CA) Alkaline phosphatase (EC 3.1.3.1) (APs) catalyze the hydrolysis of phosphomonoesters, releasing inorganic phosphate and alcohol. APs are dimeric enzymes found in most organisms. In human, four isozymes of APs have been identified. One isozyme is tissue-nonspecific (designated TNAP) and three other isozymes are tissue-specific and named according to the tissue of their predominant expression: intestinal (IAP), placental (PLAP) and germ cell (GCAP) alkaline phosphatases. IAP expression is largely restricted to the gut, especially to the epithelial cells (enterocytes) of the small intestinal mucosa. The exact biological function of
- Dose Response confirmation of uHTS inhibitors of Mouse Intestinal Alkaline Phosphatase using Tissue Nonspecific Alkaline Phosphatase. Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Production Centers Network (MLPCN) Grant Proposal Number: X01-MH077602-01 Assay Provider Dr. Jose Luis Millan, Sanford-Burnham Medical Research Institute(SBMRI, San Diego, CA) Alkaline phosphatase (EC 3.1.3.1) (APs) catalyze the hydrolysis of phosphomonoesters, releasing inorganic phosphate and alcohol. APs are dimeric enzymes found in most organisms. In human, four isozymes of APs have been identified. One isozyme is tissue-nonspecific (designated TNAP) and three other isozymes are tissue-specific and named according to the tissue of their predominant expression: intestinal (IAP), placental (PLAP) and germ cell (GCAP) alkaline phosphatases. IAP expression is largely restricted to the gut, especially to the epithelial cells (enterocytes) of the small intestinal mucosa. The exact biological function of
- Luminescent assay for HTS discovery of chemical inhibitors of placental alkaline phosphatase confirmation Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Screening Centers Network (MLSCN) Grant Proposal Number: MH077602-01 Assay Provider: Dr. Jose Luis Millan, Sanford-Burnham Medical Research Institute, San Diego, CA. This PLAP dose response assay is developed and performed to confirm hits originally identified in the PLAP Luminescent HTS assay (AID 690) and to study the structure-activity relationship on analogs of the confirmed hits. Compounds are either acquired from commercial sources or synthesized internally. Alkaline phosphatase (EC 3.1.3.1) (APs) catalyze the hydrolysis of phosphomonoesters, releasing phosphate and alcohol. APs are dimeric enzymes found in most organisms. In human, four isozymes of APs have been identified: three isozymes are tissue-specific and the fourth one is tissue-nonspecific. Placental alkaline phosphatase (PLAP) is high
- Luminescent assay for identification of inhibitors of human intestinal alkaline phosphatase Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Screening Centers Network (MLSCN) Grant Proposal Number: XO1 MH077602-01 Alkaline phosphatase (EC 3.1.3.1) (APs) catalyze the hydrolysis of phosphomonoesters, releasing inorganic phosphate and alcohol. APs are dimeric enzymes found in most organisms. In human, four isozymes of APs have been identified. One isozyme is tissue-nonspecific (designated TNAP) and three other isozymes are tissue-specific and named according to the tissue of their predominant expression: intestinal (IAP), placental (PLAP) and germ cell (GCAP) alkaline phosphatases. IAP expression is largely restricted to the gut, especially to the epithelial cells (enterocytes) of the small intestinal mucosa. IAP is inhibited by a number of inhibitors (1). They include L-phenylalanine, (2, 3), L-tryptophan (4), L-leucine and phenylalanine-g
- SAR assay for compounds activating TNAP in the absence of phosphate acceptor performed in a luminescent assay Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Screening Centers Network (MLSCN) Grant Proposal Number: 1R03 MH082385-01 This TNAP dose response assay is developed and performed to confirm hits originally identified in the TNAP luminescent HTS assay (AID 1001) and to study the structure-activity relationship on analogs of the confirmed hits. Compounds are either acquired from commercial sources or synthesized internally. Alkaline phosphatases (EC 3.1.3.1) (APs) catalyze the hydrolysis of phosphomonoesters, releasing phosphate and alcohol. APs are dimeric enzymes found in the most organisms. In human, four isozymes of APs have been identified. Three isozymes are tissue-specific and the fourth one is tissue-non specific, named TNAP. TNAP deficiency is associated with defective bone mineralization in the form of rickets and osteomalacia. Therefore, t
- SAR assay for compounds activating TNAP in the presence of 100 mM DEA performed in a luminescence assay Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Screening Centers Network (MLSCN) Grant Proposal Number: 1R03 MH082385-01 This TNAP dose response assay is developed and performed to confirm hits originally identified in the TNAP luminescent HTS assay (AID 1001) and to study the structure-activity relationship on analogs of the confirmed hits. Compounds are either acquired from commercial sources or synthesized internally. Alkaline phosphatases (EC 3.1.3.1) (APs) catalyze the hydrolysis of phosphomonoesters, releasing phosphate and alcohol. APs are dimeric enzymes found in the most organisms. In human, four isozymes of APs have been identified. Three isozymes are tissue-specific and the fourth one is tissue-non specific, named TNAP. TNAP deficiency is associated with defective bone mineralization in the form of rickets and osteomalacia. Therefore, t
- SAR assay for compounds inhibiting TNAP in the absence of phosphate acceptor performed in a luminescent assay Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Screening Centers Network (MLSCN) Grant Number: MH077602-01 Assay Provider: Dr. Jose Luis Millan, Sanford-Burnham Medical Research Institute, San Diego, CA This TNAP dose response assay is developed and performed for the purpose of SAR study on analogs of hits originally identified in the TNAP luminescent HTS assay (AID 518) Alkaline phosphatase (EC 3.1.3.1) (APs) catalyze the hydrolysis of phosphomonoesters, releasing phosphate and alcohol. APs are dimeric enzymes found in the most organism. In human, four isozymes of APs have been identified. Three isozymes are tissue-specific and the fourth one is tissue-nonsepecifc, named TNAP. TNAP overexpression is associated with excessive calcification observed in different tissues. Therefore, there are therapeutic potentials of in
- uHTS identification of compounds activating TNAP in the absence of phosphate acceptor performed in luminescent assay Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Screening Centers Network (MLSCN) Grant Proposal Number: 1R03 MH082385-01 Alkaline phosphatases (EC 3.1.3.1) (APs) catalyze the hydrolysis of phosphomonoesters, releasing phosphate and alcohol. APs are dimeric enzymes found in the most organisms. In human, four isozymes of APs have been identified. Three isozymes are tissue-specific and the fourth one is tissue-non specific, named TNAP. TNAP deficiency is associated with defective bone mineralization in the form of rickets and osteomalacia. Therefore, there is therapeutic potential of modulating TNAP activity. The goal of this HTS is to identify novel and specific activators of TNAP. The only known to date class of alkaline phosphatases activators are amino-containing alcohols, such as diethanolamine (DEA), that act as phosphoacceptor substrate and exh
- Lipo-oxygenase Inhibitory Assay Solution A was 50 mm DMAB in an 100 mm phosphate buffer (pH 7.0). Solution B was a mixture of 10 mm MBTH (3 mL) and hemoglobin (5 mg/mL, 3 mL) in 50 mm phosphatebuffer at pH 5.0 (25 mL). A linoleic acid solution was prepared by mixing 5 mg of linoleic acid with 50 mg Tween-20 in 3 mL water and then diluted with KOH 100 mmto a final volume of 5 mL. In the standard assay, the sample in ethanol (25 uL), SLO (EC 1,13,11,12; 4000 units/mL; 25 uL) and phosphate buffer, pH 7.0 (50 mm; 900 uL), were mixed in a test-tube and preincubation was carried out for 5 min at room temperature. A control test was done with the same volume of ethanol. Afterthe preincubation, linoleic acid solution (50 uL) was added to start the peroxidation reaction, and, 7 min later, solution A (270 uL) and then solution B (130 uL) were added to start the color formation. Further, 5 min later, 200 uL of a 2% SDS solution was added to terminate the reaction.
- Fluorescence Lifetime-Based Assay The activity of a compound according to the present invention can be assessed by the following in vitro & in vivo methods and/or by the following in vitro & in vivo methods well-described in the art. See A fluorescence lifetime-based assay for protease inhibitor profiling on human kallikrein 7 Doering K, Meder G, Hinnenberger M, Woelcke J, Mayr L M, Hassiepen U Biomol Screen. 2009 January; 14(1):1-9.In particular, the in vitro inhibition of recombinant human neutral endopeptidase (NEP, EC 3.4.24.11) can be determined as follows:Recombinant human neutral endopeptidase (expressed in insect cells and purified using standard methods, final concentration 7 pM) is pre-incubated with test compounds at various concentrations for 1 hour at room temperature in 10 mM sodium phosphate buffer at pH 7.4, containing 150 mM NaCl and 0.05% (w/v) CHAPS. The enzymatic reaction is started by the addition of a synthetic peptide substrate Cys(PT14)-Arg-Arg-Leu-Trp-OH to a final concentration of 0.7 μM. Substrate hydrolysis leads to an increase fluorescence lifetime (FLT) of PT14 measured by the means of a FLT reader as described by Doering et al. (2009). The effect of the compound on the enzymatic activity was determined after 1 hour (t=60 min) incubation at room temperature.
- Kv11.1 (hERG) ION CHANNEL ACTIVITY (Patch Clamp Assay) Method for the detection and characterisation of test substances which interfere with the Kv11.1 (hERG) channel: Kv11.1 (hERG, human ether a-go-go related gene) is a potassium channel which plays a central role for repolarisation of the cells in the ventricular cardiomyocytes.The patch-clamp measurement was carried out at room temperature in whole-cell configuration on human embryonic kidney cells (HEK293) which have been transfected in a stable manner with the hERG gene.The whole-cell configurations were carried out using an automated patch clamp device (Patchliner, Nanion Technologies, Munich). This is a glass chip-based system with which automated whole-cell measurements on up to 8 cells simultaneously are possible. The glass chip has a hole of defined size to which the cell is transferred into the Gigaseal by application of a reduced pressure and brought into the whole-cell configuration. Buffer, cell suspension and test substances were added to microchannels of the chip using a Teflon-coated pipette. The cells were clamped to a holding potential of −80 mV. For measurement of substancepromoted inhibition of the Kv11.1 channel, the following voltage protocol was applied at 10-second intervals: 51 ms/−80 mV, 500 ms/+40 mV, 500 ms/−40 mV, 200 ms/−80 mV. The leakage current is subtracted by means of the P4 method. The cells were resuspended in extracellular buffer (EC) and applied to the chip. After the cell had been collected, the seal was improved by addition of a seal enhancer buffer. As soon as the whole-cell configuration had been reached, the seal enhancer buffer was washed out and replaced by extracellular buffer. The measurement started in EC for 1.5 min. DMSO (vehicle control, 0.1% of DMSO) was then applied, and the control current was recorded for 3 min. The test substance was subsequently added twice in the same concentration, and the potassium current was measured for 3.5 min in each case.If the measurement result of a test substance at an initial concentration of 10 μM was smaller than (−)50% effect (threshold value) (for example (−)60% effect), the test substance was, in order to determine a dose/action relationship, added cumulatively in increasing concentration, where each concentration was measured for 5 min.The reference substance used was the Kv11.1 (hERG) ion channel blocker quinidine. The effects of test substances and quinidine were standardised to the associated vehicle control. The effect on the Kv11.1 (hERG) channel activity was assessed from the potassium current at −40 mV. For the calculation, the current was evaluated for the respective final trace. A test-substance-induced inhibition of the Kv11.1(hERG) channel was standardised to the vehicle control (0.1% of DMSO).During the measurement, an aliquot of the test substance was taken for concentration determination. The sample was measured immediately by HPLC, and the final concentration was determined from a calibration curve.
- n Vitro Testing of DPP IV Inhibitory Activity itagliptin phosphate monohydrate and the different tested compounds were dissolved in dimethyl sulphoxide (DMSO) and diluted with tris buffer (pH 8.0, 50 mM) to achieve the required concentrations. DPP-IV inhibition assay was conducted using a kit purchased from Biovision (Milpitas, Calif., USA). Briefly, DPP-IV enzyme was diluted with tris buffer and pipetted 50 μl into glass mini-cells. Subsequently the 25 μl of tris buffer, sitagliptin phosphate monohydrate or the tested compounds was added and incubated at 37° C. for 20 min protected from light. Finally, a volume of 25 μl of DPP-IV substrate was added into each tube. The fluorescence generated from hydrolysis of the substrate was read at Ex/Em=360/460 nm using Modulus single tube multimode reader (Promega, USA). Baseline fluorescence was recorded before incubation (R1) and 20 min after incubation (R2). Percentage inhibition was calculated according to the following equation:% inhibition=(ΔRFU test/ΔRFU E C)*100where, ΔRFU=R2−R1 and EC is the enzyme control (enzyme solution without inhibitor) which is considered 100% activity.The standard DPP-IV inhibitor drug sitagliptin was employed as positive control. Screening was done of all the synthesized compounds at 100 nM then a dose-response study was conducted of the compounds that showed a promising inhibitory effect on DPP-IV activity (>80% inhibition which was comparable to that of sitalgliptin). Sigmoidal dose-response curves for DPP-IV % inhibition versus log concentrations were plotted using Graphpad prism software, version 5.00 (GraphPad Software, Inc. La Jolla, Calif., USA).
- OPR Mu Antagonist Assay The purpose of this assay is to confirm the potency and selectivity of compounds synthesized to be OPRK1 Antagonists. This assay monitors the OPRMu1 activation, in membrane recruitment of β-arrestin. The assay monitors GPCR-β-arrestin proximity using low affinity fragment complementation of beta-galactosidase (beta-gal). It employs U20S cells which express OPRMu1 fused to the complementary beta-gal fragment (enzyme acceptor). As designed, compounds that act as antagonists will prevent receptor activation resulting in reduce well luminescence. Compounds were tested in triplicate using a 10-point, 1:3 dilution series starting at a nominal concentration of 10 micromolar.The Discover X OPRMu1-U20S cell line was routinely cultured in T175 flasks at 37° C., 5% CO2 and 95% relative humidity (RH). The growth media consisted of DMEM/F12 1:1 Media supplemented with 10% v/v heat inactivated fetal bovine serum, 25 mM HEPES, Non-essential amino acids, 1 mM Sodium Pyruvate, 1× antibiotic mix (penicillin streptomycin) plus 500 ug/mL Geneticin and 300 ug/mL Hygromycin (selection antibiotics).On Day 1 of the assay, 5000 cells in 20 ul of assay buffer (Discover X's Cell Plating Reagent 5) were seeded into each well of a 384 Corning 3570 standard white plate, and incubated 16-24 hours at 37° C., 5% CO2 and 95% (RH). On Day 2, 100 nl of test compound in DMSO was added to the appropriate wells, 100 nl of DMSO added to control wells and plates were incubated for 30 min at 37° C., 5% CO2 and 95% (RH). Next 100 nl of DAMGO OPRMu1 or DMSO in assay media (EC80 Challenge consists of 100 nl of 50 uM DAMGO, final assay concentration=250 nM, 100% Response wells receive 100 nl 200 uM DAMGO). After incubation for 3 hours at 37° C., 5% CO2 and 95% (RH), 10 ul of Path Hunter Detection Mix is added to each well, plate placed on a plate rotator/mixer for 10 minutes and then incubated at room temperature, in the dark for 1 hour. Well luminescence was measured on Perkin Elmer's Envision.The Percent Inhibition was calculated from the median ratio as follows:% Inhibition = 100 - ( 100 ( Compound Well - 0 % Response Well EC 80 Control Well - 0 % Response Well ) )where: COMPOUND WELL is defined as the well containing test compound; EC80 CONTROL WELL is defined as wells containing DAMGO challenge (250 nM final)=0% inhibition; and 0% RESPONSE WELL is defined as wells containing DMSO=100% inhibition. IC50 is defined as the concentration of compound required to achieve 50% inhibition.
 US8889724, EC BDBM139535
US8889724, EC BDBM139535 BDBM292781 US10106501, Example EC
BDBM292781 US10106501, Example EC 2-(6-Methoxy-naphthalen-2-yl)-propionic acid (S)-2-(6-Methoxy-naphthalen-2-yl)-propionic acid US11459295, Compound S-Naproxen (1) Equiproxen Naprelan (S)-2-(6-methoxynaphthalen-2-yl)propanoic acid Naproxen2-(6-Methoxy-naphthalen-2-yl)-propionic acid Anaprox 2-(6-Methoxy-naphthalen-2-yl)-propionic acid(naproxen) (2S)-2-(6-methoxynaphthalen-2-yl)propanoic acid naproxene Aleve CHEMBL154 2-(6-methoxy-2-naphthyl)propanoic acid Ec-naprosyn S-NAPROXEN Naproxen 2-(6-methoxynaphthalen-2-yl)propanoic acid BDBM50339185 RS-3540 (S)-naproxen Naprosyn
2-(6-Methoxy-naphthalen-2-yl)-propionic acid (S)-2-(6-Methoxy-naphthalen-2-yl)-propionic acid US11459295, Compound S-Naproxen (1) Equiproxen Naprelan (S)-2-(6-methoxynaphthalen-2-yl)propanoic acid Naproxen2-(6-Methoxy-naphthalen-2-yl)-propionic acid Anaprox 2-(6-Methoxy-naphthalen-2-yl)-propionic acid(naproxen) (2S)-2-(6-methoxynaphthalen-2-yl)propanoic acid naproxene Aleve CHEMBL154 2-(6-methoxy-2-naphthyl)propanoic acid Ec-naprosyn S-NAPROXEN Naproxen 2-(6-methoxynaphthalen-2-yl)propanoic acid BDBM50339185 RS-3540 (S)-naproxen Naprosyn