 BDBM86673 CAS_510732-84-0 ATC 0065 ATC0065
BDBM86673 CAS_510732-84-0 ATC 0065 ATC0065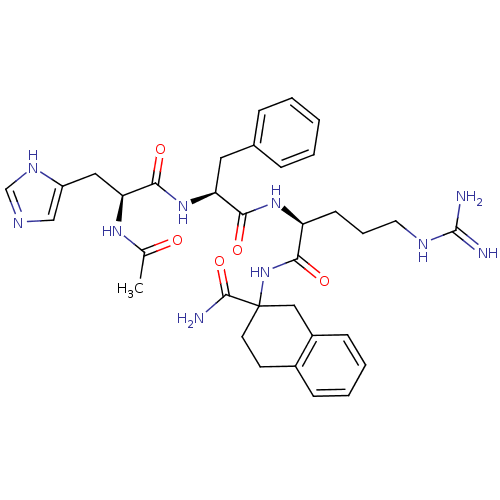 CHEMBL153420 Ac-His-D-Phe-Arg-Atc-NH2 BDBM50122062
CHEMBL153420 Ac-His-D-Phe-Arg-Atc-NH2 BDBM50122062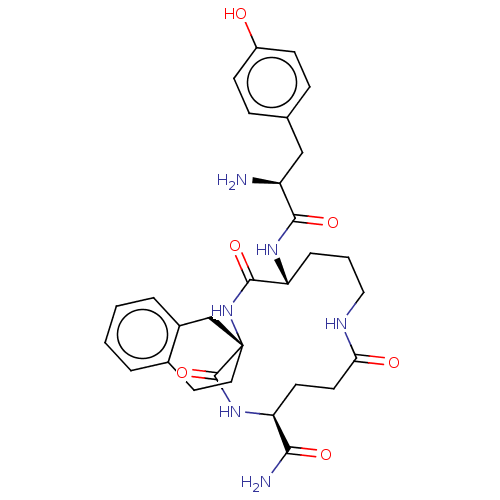 CHEMBL3037875 BDBM50007326 H-Tyr-c[D-Orn-(D or L)Atc-Glu]-NH2
CHEMBL3037875 BDBM50007326 H-Tyr-c[D-Orn-(D or L)Atc-Glu]-NH2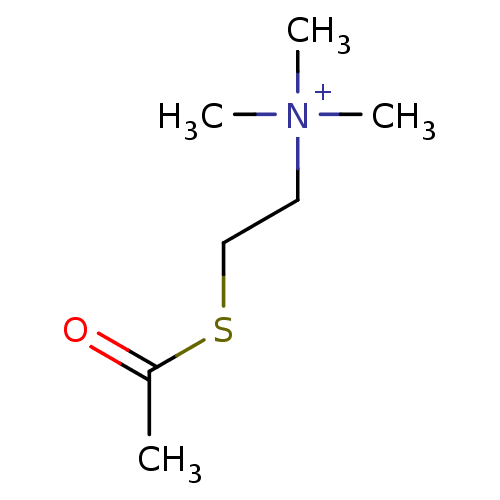 [2-(acetylsulfanyl)ethyl]trimethylazanium iodide ATC acetylthiocholine chloride Acetylthiocholine acetylthiocholine iodide (2-Mercaptoethyl)trimethylammonium iodide acetate BDBM8959
[2-(acetylsulfanyl)ethyl]trimethylazanium iodide ATC acetylthiocholine chloride Acetylthiocholine acetylthiocholine iodide (2-Mercaptoethyl)trimethylammonium iodide acetate BDBM8959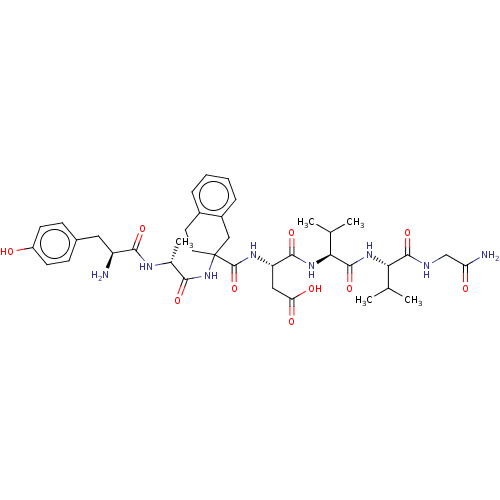 BDBM50001459 H-Tyr-D-Ala-(R or S)Atc-Asp-Val-Val-Gly-NH2 3-[(2-{2-[2-Amino-3-(4-hydroxy-phenyl)-propionylamino]-propionylamino}-1,2,3,4-tetrahydro-naphthalene-2-carbonyl)-amino]-N-{1-[1-(carbamoylmethyl-carbamoyl)-2-methyl-propylcarbamoyl]-2-methyl-propyl}-succinamic acid CHEMBL124941
BDBM50001459 H-Tyr-D-Ala-(R or S)Atc-Asp-Val-Val-Gly-NH2 3-[(2-{2-[2-Amino-3-(4-hydroxy-phenyl)-propionylamino]-propionylamino}-1,2,3,4-tetrahydro-naphthalene-2-carbonyl)-amino]-N-{1-[1-(carbamoylmethyl-carbamoyl)-2-methyl-propylcarbamoyl]-2-methyl-propyl}-succinamic acid CHEMBL124941
- ChEMBL_1511325 (CHEMBL3606230) Inhibition of electric eel AChE using ATC iodide substrate by Ellman's assay
- ChEMBL_1511326 (CHEMBL3606231) Inhibition of equine serum BChE using ATC iodide substrate by Ellman's assay
- ChEMBL_1547639 (CHEMBL3755509) Mixed-type inhibition of electric eel AChE using ATC as substrate assessed as hydrolysis of ATC preincubated with protein for 15 mins followed by substrate addition by Lineweaver-Burk plot analysis
- ChEMBL_1458502 (CHEMBL3370619) Inhibition of electric eel AChE using ATC iodide substrate by DTNB based assay
- ChEMBL_1458503 (CHEMBL3370620) Inhibition of equine serum BChE using ATC iodide substrate by DTNB based assay
- ChEMBL_1854516 (CHEMBL4355245) Inhibition of electric eel AChE using ATC as substrate after 15 mins by Ellman's method
- ChEMBL_2073679 (CHEMBL4729213) Inhibition of recombinant human AChE using ATC as substrate by DTNB-reagent based Ellman's method
- ChEMBL_2424348 Inhibition of electric eel AChE using ATC as substrate incubated for 2 mins by Ellman's method
- ChEMBL_2329087 Inhibition of human erythrocyte AChE using ATC iodide as substrate preincubated for 5 mins followed by substrate addition measured Ellmans method
- ChEMBL_1621946 (CHEMBL3864298) Inhibition of electric eel AChE using ATC as substrate preincubated for 15 mins followed by substrate addition by UV-spectrophotometric analysis
- ChEMBL_1549348 (CHEMBL3755054) Inhibition of human AChE using ATC as substrate preincubated for 5 mins followed by substrate addition measured after 2 mins by Ellman's assay
- ChEMBL_1549346 (CHEMBL3755052) Inhibition of electric eel AChE using ATC as substrate preincubated for 5 mins followed by substrate addition measured after 2 mins by Ellman's assay
- ChEMBL_1547640 (CHEMBL3755510) Inhibition of electric eel AChE using ATC as substrate preincubated for 15 mins followed by substrate addition measured at 1 min intervals by Ellman method
- ChEMBL_1673532 (CHEMBL4023561) Inhibition of human erythrocyte AChE using ATC iodide as substrate preincubated for 4.5 mins followed by substrate addition measured after 2.5 mins by Ellman's method
- ChEMBL_1673537 (CHEMBL4023566) Inhibition of electric eel AChE using ATC iodide as substrate preincubated for 4.5 mins followed by substrate addition measured after 2.5 mins by Ellman's method
- ChEMBL_2121991 (CHEMBL4831138) Inhibition of electric eel AchE using ATC as substrate preincubated with enzyme for 5 mins followed by substrate addition for 5 mins by Ellman's method
- ChEMBL_2054929 (CHEMBL4709930) Inhibition of human erythrocyte AChE using ATC iodide as substrate preincubated with enzyme for 4.5 mins followed by substrate addition and measured after 2.5 mins by Ellman's method
- ChEMBL_2054930 (CHEMBL4709931) Inhibition of Electric eel AChE using ATC iodide as substrate preincubated with enzyme for 4.5 mins followed by substrate addition and measured after 2.5 mins by Ellman's method
- ChEMBL_2288317 Inhibition of human AChE using ATC and BTC iodide as substrate preincubated for 5 mins followed by substrate addition and measured after 2 mins by DTNB reagent based assay
- ChEMBL_1816033 (CHEMBL4315607) Reactivation of POX-inhibited human recombinant AChE assessed as dissociation rate constant using ATC as substrate incubated for 60 mins followed by substrate addition by Ellman's method based spectrophotometry
- ChEMBL_2288315 Inhibition of electric eel AChE using ATC and BTC iodide as substrate preincubated for 5 mins followed by substrate addition and measured after 2 mins by DTNB reagent based assay
- ChEMBL_2288316 Inhibition of equine serum BuChE using ATC and BTC iodide as substrate preincubated for 5 mins followed by substrate addition and measured after 2 mins by DTNB reagent based assay
- Enzyme Inhibition Assay All the assays were under 0.1 M KH2PO4/K2HPO4 buffer, pH 8.0, using a Shimadzu 2450 Spectrophotometer. Enzyme solutions were prepared to give 2.0 units/mL in 2 mL aliquots. The assay medium contained phosphate buffer, pH 8.0 (1 mL), 50 μL of 0.01 M DTNB, 10 μL of enzyme, and 50 μL of 0.01 M substrate (ATC). The substrate was added to the assay medium containing enzyme, buffer, and DTNB with inhibitor after 15 min of incubation time. The activity was determined by measuring the increase in absorbance at 412 nm at 1 min intervals at 37°C. In vitro BuChE assay use the similar method described above.
- Enzymatic Assay Enzyme solutions were prepared in gelatine solution (1%), at a concentration of 2.5 units/mL. AChE or BChE solution (50 μL) and compound solution (50 μL), which is prepared in 2% DMSO at a concentration range of 10^−1-10^−6 mM, were added to 3.0 mL phosphate buffer (pH8 ± 0.1) and incubated at 25°C for 5 min. The reaction was started by adding (DTNB) (50 μL) and ATC (10 μL) to the enzyme-inhibitor mixture. The production of the yellow anion was recorded for 10 min at 412 nm. As a control, an identical solution of the enzyme without the inhibitor was processed following the same protocol. The blank reading contained 3.0 mL buffer, 50 μL 2% DMSO, 50 μL DTNB and 10 μL substrate. All processes were assayed in triplicate.
- RNase H (RH) Assay HIV RT-associated RNase H activity was measured as previously described.[J. Med. Chem. 2014, 57:3223-3234] Briefly, the reaction mixture comprised Tris HCl (100 mL, 50 mm, pH 7.8) containingMgCl2 (6 mm), dithiothreitol (1 mm), KCl (80 mm), hybrid RNA/DNA 5'-GAUCU GAGCC UGGGA GCU-Fluorescin-3' (0.25 mm; HPLC, dry, QC: Mass Check; Metabion, Steinkirchen, Germany), 5'-Dabcyl-AGCTC CCAGG CTCAG ATC-3' (HPLC, dry, QC: Mass Check), and HIV-1 RT (20 ng, according to a linear range of dose-activity curve). The reaction mixture was incubated for 1 h at 37 °C, and the reaction was stopped by addition of EDTA; products were measured with a Victor 3 multilabel counter plate reader (model 1420-051; PerkinElmer) equipped with filters (lex=490 nm, lem=528 nm).
- AChE and BuChE Inhibition Assay This spectrophotometric assay is based on the reaction of 5,5-dithio-bis(2-nitrobenzoic)acid (DTNB) with thiocholine to yield a colored product. Shimadzu (Kyoto, Japan) UV-1700 UV-vis spectrophotometer was used to record the measurements. The enzyme solutions were prepared at 2.5 units/mL concentrationin 1% gelatin solution. BChE/AChE and compound solutions (50 μL each) in 2% DMSO at 10^-1-10^-6 mM concentration were added to 3 mL phosphate buffer (pH 8 &plusnm; 0.1)followed by incubation for 5 min at 25 °C. The reaction was commenced by the addition of acetylthiocholine iodide (ATC) (10 μL) and DTNB (50 μL) to the enzyme-inhibitor mixture. The yellow anion production was measured at 412 nm for 10 min. Using similar method, same enzyme solution without the inhibitor was processed which served as a control. The blank reading contained 10 μL substrate, 50 μL DTNB, 50 μL (2%) DMSO, and 3 mL buffer. The reaction was performed in triplicate.
- Enzymatic Assay Fluorogenic biochemical assays used recombinant full-length FEN-1 or catalytic domains of Exo1, XPG or GEN1 and 200 nM of a DNA substrate (formed by annealing three oligonucleotides quencher (5′-CAC GTT GAC TAC CGC TCA ATC CTG ACG AAC ACA TC-BHQ2), flap (5′-TAMRA-GA TGT CAA GCA GTC CTA ACT TTG AGG CAG AGT CCG C) and template (5′-GC GGA CTC TGC CTC AAG ACG GTA GTC AAC GTG-3′) (PMID: 21062821)) in 50 mM Tris pH 8.0, 10 mM MgCl2, 1 mM DTT, and 0.01% Tween-20. Inhibitors arrayed in dose response were added from DMSO stocks (using a V&P 384-pintool head) to the enzyme solution in low volume, black polystyrene 384-well plates (Corning #3677) 15 minutes prior to the addition of the DNA substrate. Reactions were allowed to proceed for 2-4 hours at 25° C. or 37° C. Fluorescence data (Excitation: 530 nm/10 nm bandwidth; Emission: 590 nm/20 nm bandwith) were measured with an Infinite M1000 plate reader (Tecan).
- HIV RT Filter Binding IC50 Assay The RT IC50 filter binding assay utilized a synthetic D19/D78mer annealed primer template complex prepared by incubating 50 μM of 19-mer [5′-G TCC CTG TTC GGG CGC CAC-3′] with 50 μM of 78-mer [3-CGA CCG TCC AGG GAC AAG CCC GCG GTG GCG ATC TCT TGA CAT TCC GTA ACC TTC GTA TTT TAA GCA TCA TAG TAC ACA-5] in 50 mM Tris pH 7.8 at 95° C. for 5 minutes, 55° C. for 15 minutes, and 37° C. for 10 minutes. 22.5 nM HIV RT was added to reaction mix containing 150 mM Tris-Cl pH 7.8, 180 mM KCl, 0.33 mM DTT, 0.9 mg/mL bSA, 12.6% glycerol, 3 uM D19/D78 mer. Serial dilutions of compounds were added at equal volumes and reactions were heated to 37° C. The reactions were initiated with equal volumes of dNTP mix containing 150 μM each of noncompeting dNTP and 1.2 μM of competing dNTP, and 0.1 μCi/μL competing dNTP. After 5 minutes at 37° C., 5 μL reactions were spotted on DE81 paper, washed three times in 0.125M Na2HPO4, dried, and quantified by autoradiography. IC50 values were calculated in Prism by non-linear regression analysis using the dose-response (variable slope) equation (four-parameter logistic equation): Y=Bottom+(Top-Bottom)/(1+10{circumflex over ( )}((Log IC50−X)*HillSlope)).
- Antiviral assay VeroE6 cells and TMPRSS2-overexpressing VeroE6 (VeroE6TMPRSS2) cells were obtained from the Japanese Collection of Research Bioresources (JCRB) Cell Bank (Osaka, Japan). VeroE6 cells were maintained in Dulbecco s modified Eagle s medium (d-MEM) supplemented with 10% fetal bovine serum (FCS), 100 μg/ml of penicillin, and 100 μg/ml of streptomycin. VeroE6TMPRSS2 cells were maintained in d-MEM as reported (ref.1) in the presence of 1 mg/ml of G418. SARS-CoV-2 strain JPN/TY/WK-521 (SARS-CoV-2WK-521) was obtained from the National Institute of Infectious Diseases (Tokyo, Japan).Antiviral assay was carried out as described recently (ref 1): Cells were seeded in a 96-well plate (2x104 cells/well) and incubated. After 24 h, virus was inoculated into cells at multiplicity of infection (MOI) of 0.05. After an additional 72 h, cell culture supernatants were harvested and viral RNA was extracted using a QIAamp viral RNA minikit (Qiagen, Hilden, Germany), and quantitative RT-PCR (RT-qPCR) was then performed using One Step PrimeScript III RT-qPCR mix (TaKaRa Bio, Shiga, Japan) following the instructions of the manufacturers. The primers and probe used for detecting SARS-CoV-2 envelope (6) were 5=-ACT TCT TTT TCT TGC TTT CGT GGT-3= (forward), 5=-GCA GCA GTA CGC ACA CAA TC-3= (reverse), and 5=-FAM-CTA GTT ACA CTA GCC ATC CTT ACT GC-black hole quencher 1 (BHQ1)-3= (probe). To determine the cytotoxicity of each compound, cells were seeded in a 96-well plate (2_104 cells/well). One day later, various concentrations of each compound were added, and cells were incubated for additional 3 days. The 50% cytotoxic concentrations (CC50) values were determined using the WST-8 assay and Cell Counting Kit-8 (Dojindo, Kumamoto, Japan).
 BDBM86673 CAS_510732-84-0 ATC 0065 ATC0065
BDBM86673 CAS_510732-84-0 ATC 0065 ATC0065 CHEMBL153420 Ac-His-D-Phe-Arg-Atc-NH2 BDBM50122062
CHEMBL153420 Ac-His-D-Phe-Arg-Atc-NH2 BDBM50122062 CHEMBL3037875 BDBM50007326 H-Tyr-c[D-Orn-(D or L)Atc-Glu]-NH2
CHEMBL3037875 BDBM50007326 H-Tyr-c[D-Orn-(D or L)Atc-Glu]-NH2 [2-(acetylsulfanyl)ethyl]trimethylazanium iodide ATC acetylthiocholine chloride Acetylthiocholine acetylthiocholine iodide (2-Mercaptoethyl)trimethylammonium iodide acetate BDBM8959
[2-(acetylsulfanyl)ethyl]trimethylazanium iodide ATC acetylthiocholine chloride Acetylthiocholine acetylthiocholine iodide (2-Mercaptoethyl)trimethylammonium iodide acetate BDBM8959 BDBM50001459 H-Tyr-D-Ala-(R or S)Atc-Asp-Val-Val-Gly-NH2 3-[(2-{2-[2-Amino-3-(4-hydroxy-phenyl)-propionylamino]-propionylamino}-1,2,3,4-tetrahydro-naphthalene-2-carbonyl)-amino]-N-{1-[1-(carbamoylmethyl-carbamoyl)-2-methyl-propylcarbamoyl]-2-methyl-propyl}-succinamic acid CHEMBL124941
BDBM50001459 H-Tyr-D-Ala-(R or S)Atc-Asp-Val-Val-Gly-NH2 3-[(2-{2-[2-Amino-3-(4-hydroxy-phenyl)-propionylamino]-propionylamino}-1,2,3,4-tetrahydro-naphthalene-2-carbonyl)-amino]-N-{1-[1-(carbamoylmethyl-carbamoyl)-2-methyl-propylcarbamoyl]-2-methyl-propyl}-succinamic acid CHEMBL124941