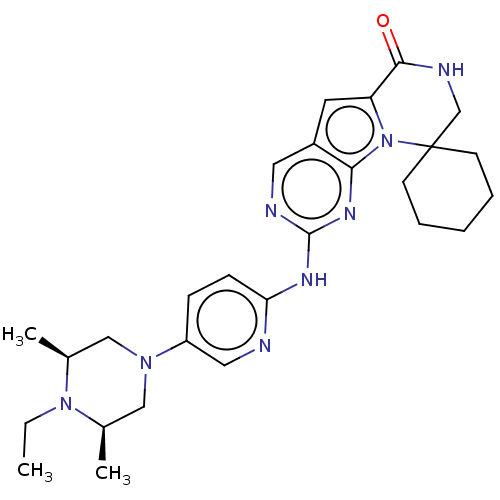
 US9464092, FF BDBM253940 US9527857, FF
US9464092, FF BDBM253940 US9527857, FF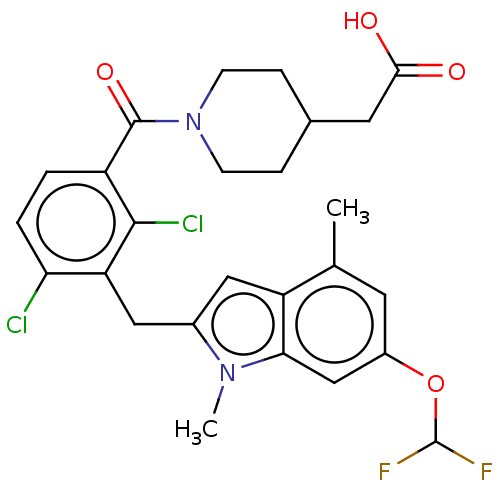 BDBM292830 US10106501, Example FF
BDBM292830 US10106501, Example FF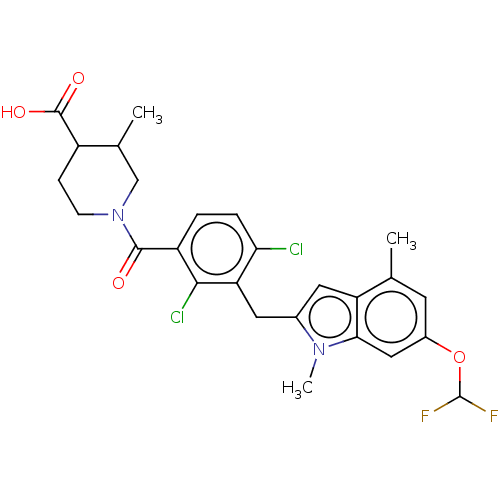 US10106501, Example FF-1 BDBM292831
US10106501, Example FF-1 BDBM292831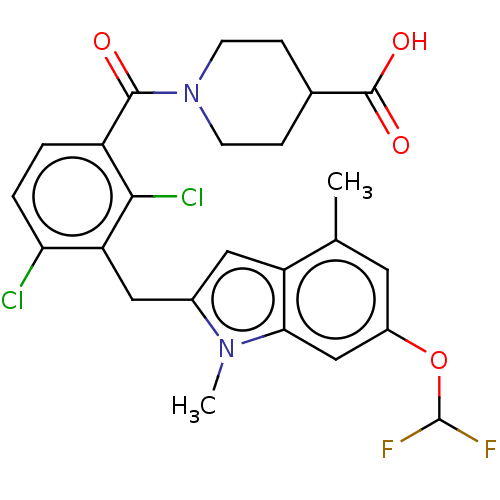 US10106501, Example FF-2 BDBM292832
US10106501, Example FF-2 BDBM292832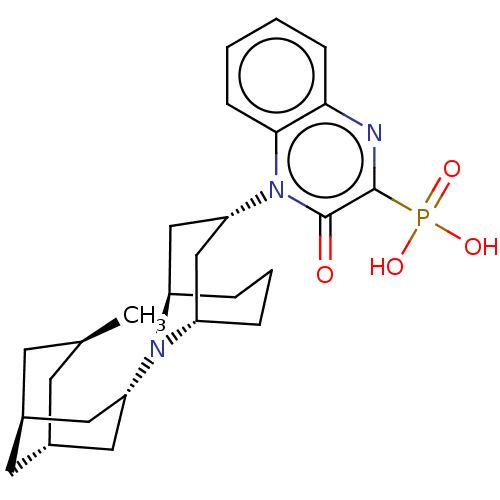 US9598447, Ref No. FF BDBM303064
US9598447, Ref No. FF BDBM303064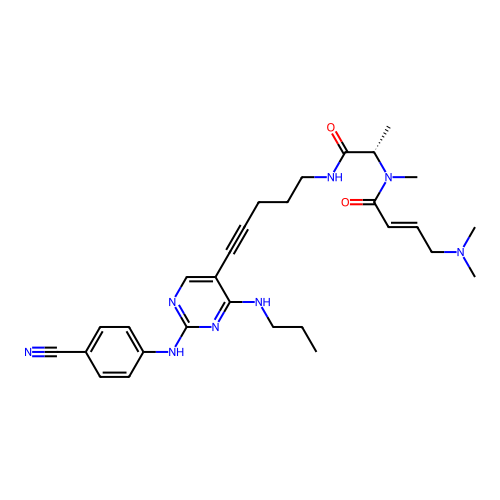 Ff-10101 BDBM50647812 FLT3-IN-1
Ff-10101 BDBM50647812 FLT3-IN-1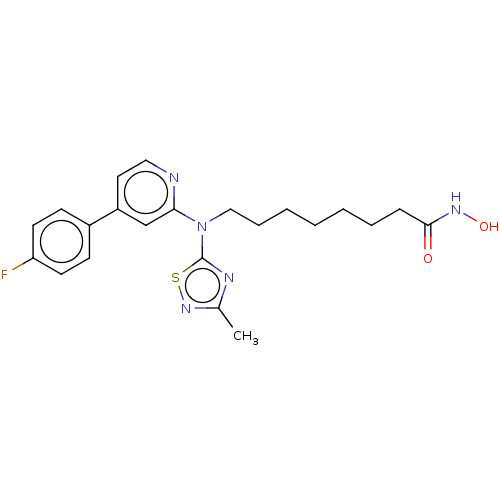 US10150763, Example FF BDBM310732 8-((4-(4-Fluorophenyl)-pyridin-2-yl)(3-methyl-1,2,4-thiadiazol-5-yl)amino)-N-hydroxyoctanamide
US10150763, Example FF BDBM310732 8-((4-(4-Fluorophenyl)-pyridin-2-yl)(3-methyl-1,2,4-thiadiazol-5-yl)amino)-N-hydroxyoctanamide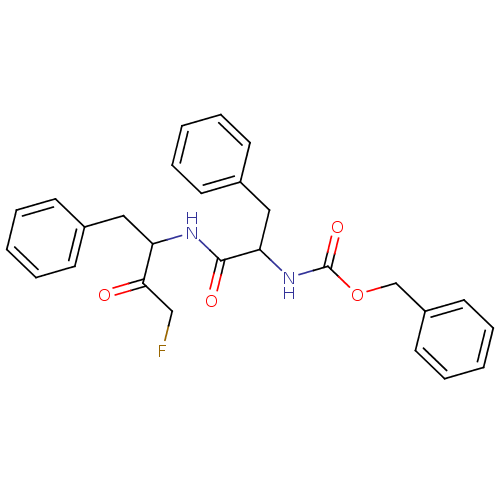 BDBM110185 benzyl N-[1-[(4-fluoro-3-oxo-1-phenylbutan-2-yl)amino]-1-oxo-3-phenylpropan-2-yl]carbamate Z-FF-FMK
BDBM110185 benzyl N-[1-[(4-fluoro-3-oxo-1-phenylbutan-2-yl)amino]-1-oxo-3-phenylpropan-2-yl]carbamate Z-FF-FMK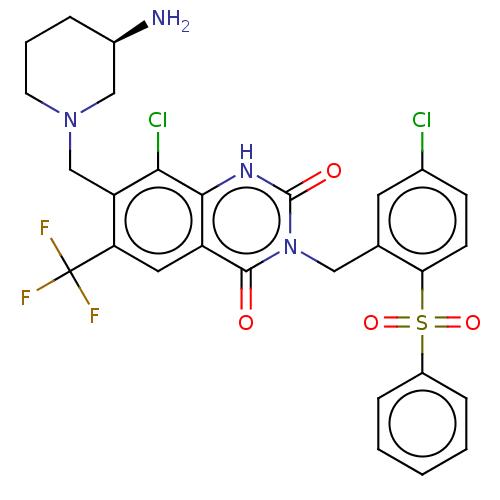 7-[[(3R)-3-Aminopiperidin-1-yl]methyl]-3-[[2-(benzenesulfonyl)-5-chlorophenyl]methyl]-8-chloro-6-(trifluoromethyl)-1H-quinazoline-2,4-dione BDBM286935 US9567304, Compound FF-2
7-[[(3R)-3-Aminopiperidin-1-yl]methyl]-3-[[2-(benzenesulfonyl)-5-chlorophenyl]methyl]-8-chloro-6-(trifluoromethyl)-1H-quinazoline-2,4-dione BDBM286935 US9567304, Compound FF-2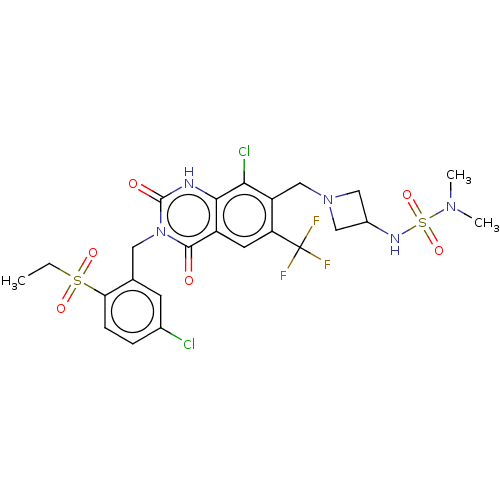 8-Chloro-3-[(5-chloro-2-ethylsulfonylphenyl)methyl]-7-[[3-(dimethylsulfamoylamino)azetidin-1-yl]methyl]-2,4-dioxo-6-(trifluoromethyl)-1H-quinazoline BDBM286940 US9567304, Compound FF-7
8-Chloro-3-[(5-chloro-2-ethylsulfonylphenyl)methyl]-7-[[3-(dimethylsulfamoylamino)azetidin-1-yl]methyl]-2,4-dioxo-6-(trifluoromethyl)-1H-quinazoline BDBM286940 US9567304, Compound FF-7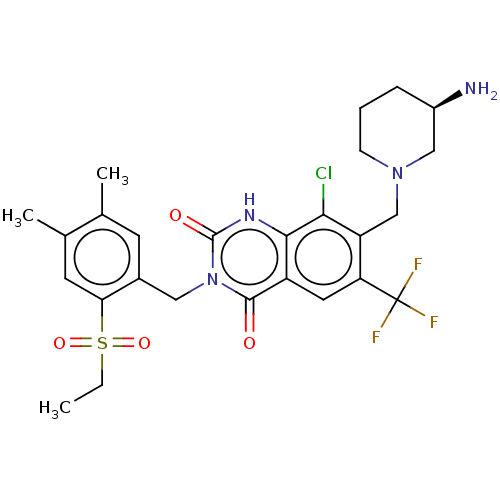 BDBM286938 7-[[(3R)-3-Aminopiperidin-1-yl]methyl]-8-chloro-3-[(2-ethylsulfonyl-4,5-dimethylphenyl)methyl]-6-(trifluoromethyl)-1H-quinazoline-2,4-dione US9567304, Compound FF-5
BDBM286938 7-[[(3R)-3-Aminopiperidin-1-yl]methyl]-8-chloro-3-[(2-ethylsulfonyl-4,5-dimethylphenyl)methyl]-6-(trifluoromethyl)-1H-quinazoline-2,4-dione US9567304, Compound FF-5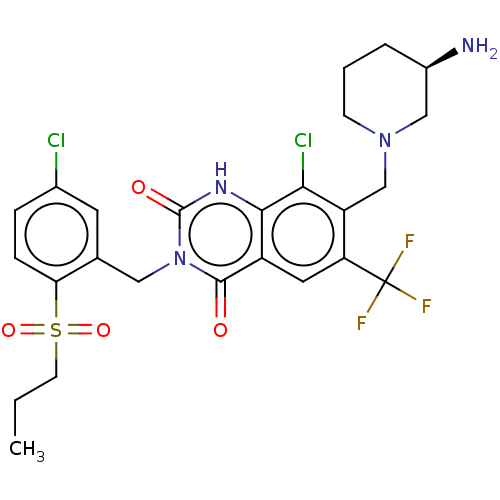 US9567304, Compound FF-1 7-[[(3R)-3-Aminopiperidin-1-yl]methyl]-8-chloro-3-[(5-chloro-2-propylsulfonylphenyl)methyl]-6-(trifluoromethyl)-1H-quinazoline-2,4-dione BDBM286934
US9567304, Compound FF-1 7-[[(3R)-3-Aminopiperidin-1-yl]methyl]-8-chloro-3-[(5-chloro-2-propylsulfonylphenyl)methyl]-6-(trifluoromethyl)-1H-quinazoline-2,4-dione BDBM286934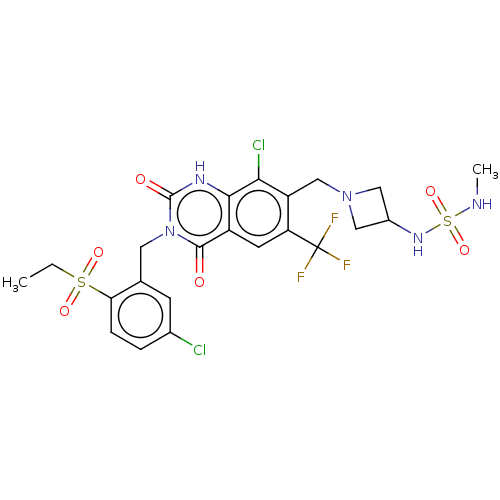 US9567304, Compound FF-8 8-Chloro-3-[(5-chloro-2-ethylsulfonylphenyl)methyl]-7-[[3-(methylsulfamoylamino)azetidin-1-yl]methyl]-6-(trifluoromethyl)-1H-quinazoline-2,4-dione BDBM286941
US9567304, Compound FF-8 8-Chloro-3-[(5-chloro-2-ethylsulfonylphenyl)methyl]-7-[[3-(methylsulfamoylamino)azetidin-1-yl]methyl]-6-(trifluoromethyl)-1H-quinazoline-2,4-dione BDBM286941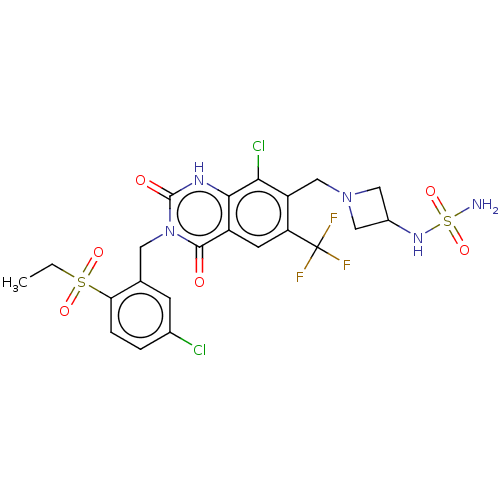 US9567304, Compound FF-9 8-Chloro-3-[(5-chloro-2-ethylsulfonylphenyl)methyl]-2,4-dioxo-7-[[3-(sulfamoylamino)azetidin-1-yl]methyl]-6-(trifluoromethyl)-1H-quinazoline BDBM286942
US9567304, Compound FF-9 8-Chloro-3-[(5-chloro-2-ethylsulfonylphenyl)methyl]-2,4-dioxo-7-[[3-(sulfamoylamino)azetidin-1-yl]methyl]-6-(trifluoromethyl)-1H-quinazoline BDBM286942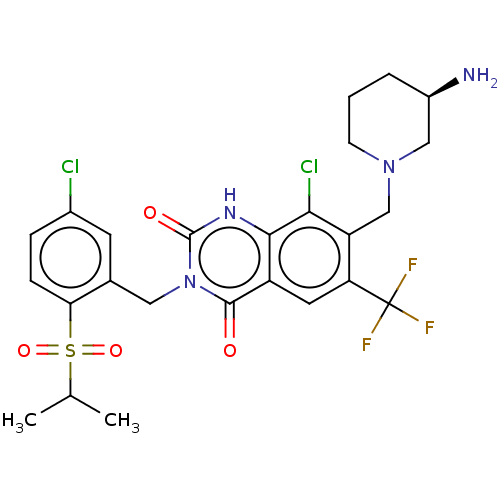 BDBM286936 US9567304, Compound FF-3 7-[[(3R)-3-Aminopiperidin-1-yl]methyl]-8-chloro-3-[(5-chloro-2-propan-2-ylsulfonylphenyl)methyl]-6-(trifluoromethyl)-1H-quinazoline-2,4-dione
BDBM286936 US9567304, Compound FF-3 7-[[(3R)-3-Aminopiperidin-1-yl]methyl]-8-chloro-3-[(5-chloro-2-propan-2-ylsulfonylphenyl)methyl]-6-(trifluoromethyl)-1H-quinazoline-2,4-dione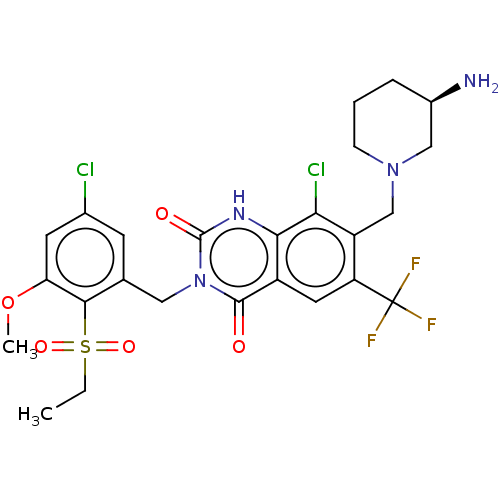 BDBM286937 7-[[(3R)-3-Aminopiperidin-1-yl]methyl]-8-chloro-3-[(5-chloro-2-ethylsulfonyl-3-methoxyphenyl)methyl]-6-(trifluoromethyl)-1H-quinazoline-2,4-dione US9567304, Compound FF-4
BDBM286937 7-[[(3R)-3-Aminopiperidin-1-yl]methyl]-8-chloro-3-[(5-chloro-2-ethylsulfonyl-3-methoxyphenyl)methyl]-6-(trifluoromethyl)-1H-quinazoline-2,4-dione US9567304, Compound FF-4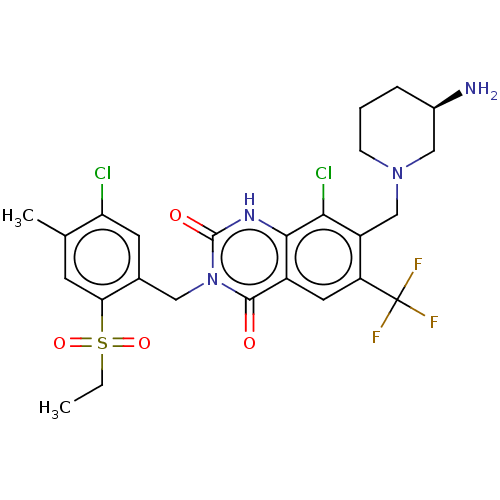 BDBM286939 7-[[(3R)-3-Aminopiperidin-1-yl]methyl]-8-chloro-3-[(5-chloro-2-ethylsulfonyl-4-methylphenyl)methyl]-6-(trifluoromethyl)-1H-quinazoline-2,4-dione US9567304, Compound FF-6
BDBM286939 7-[[(3R)-3-Aminopiperidin-1-yl]methyl]-8-chloro-3-[(5-chloro-2-ethylsulfonyl-4-methylphenyl)methyl]-6-(trifluoromethyl)-1H-quinazoline-2,4-dione US9567304, Compound FF-6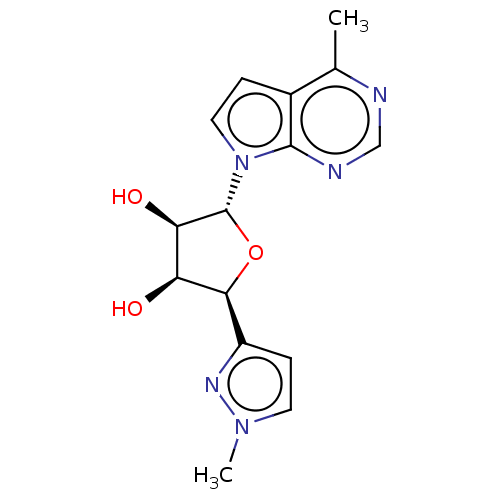 (Scheme FF) (2S,3S,4R,5R)-2-(1-methyl-1H-pyrazol-3-yl)-5-(4-methyl-7H-pyrrolo[2,3-d]pyrimidin-7-yl)tetrahydrofuran-3,4-diol (EE-2) US10428104, Example 77 BDBM415546
(Scheme FF) (2S,3S,4R,5R)-2-(1-methyl-1H-pyrazol-3-yl)-5-(4-methyl-7H-pyrrolo[2,3-d]pyrimidin-7-yl)tetrahydrofuran-3,4-diol (EE-2) US10428104, Example 77 BDBM415546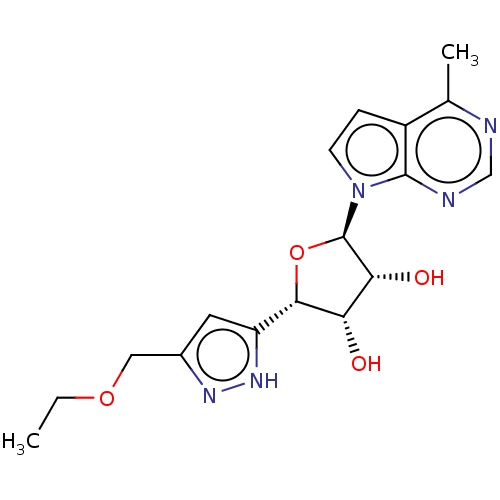 US10428104, Example 78 (Scheme GG) (2S,3S,4R,5R)-2-(3-(ethoxymethyl)-1H-pyrazol-5-yl)-5-(4-methyl-7H-pyrrolo[2,3-d]pyrimidin-7-yl)tetrahydrofuran-3,4-diol (FF-6) BDBM415547
US10428104, Example 78 (Scheme GG) (2S,3S,4R,5R)-2-(3-(ethoxymethyl)-1H-pyrazol-5-yl)-5-(4-methyl-7H-pyrrolo[2,3-d]pyrimidin-7-yl)tetrahydrofuran-3,4-diol (FF-6) BDBM415547
- Prokai, L; Prokai-Tatrai, K; Zharikova, A; Li, X; Rocca, JR Combinatorial lead optimization of a neuropeptide FF antagonist. J Med Chem 44: 1623-6 (2001)
- Oishi, S; Misu, R; Tomita, K; Setsuda, S; Masuda, R; Ohno, H; Naniwa, Y; Ieda, N; Inoue, N; Ohkura, S; Uenoyama, Y; Tsukamura, H; Maeda, K; Hirasawa, A; Tsujimoto, G; Fujii, N Activation of Neuropeptide FF Receptors by Kisspeptin Receptor Ligands. ACS Med Chem Lett 2: 53-57 (2011)
- Gealageas, R; Schneider, S; Humbert, JP; Bertin, I; Schmitt, M; Laboureyras, E; Dugave, C; Mollereau, C; Simonnet, G; Bourguignon, JJ; Simonin, F; Bihel, F Development of sub-nanomolar dipeptidic ligands of neuropeptide FF receptors. Bioorg Med Chem Lett 22: 7471-4 (2012)
- Nguyen, T; Marusich, J; Li, JX; Zhang, Y Neuropeptide FF and Its Receptors: Therapeutic Applications and Ligand Development. J Med Chem 63: 12387-12402 (2020)
- Naef, R; Tenor, H 2-phenyl-3,4-dihydropyrrolo[2,1-Ff] [1,2,4]triazinone derivatives as phosphodiesterase inhibitors and uses thereof US Patent US11242347 (2022)
- Gicquel, S; Mazarguil, H; Desprat, C; Allard, M; Devillers, JP; Simonnet, G; Zajac, JM Structure-activity study of neuropeptide FF: contribution of N-terminal regions to affinity and activity. J Med Chem 37: 3477-81 (1994)
- De Neve, J; Elhabazi, K; Gonzalez, S; Herby, C; Schneider, S; Utard, V; Fellmann-Clauss, R; Petit-Demouliere, N; Lecat, S; Kremer, M; Ces, A; Daubeuf, F; Martin, C; Ballet, S; Bihel, F; Simonin, F Multitarget μ-Opioid Receptor Agonists─Neuropeptide FF Receptor Antagonists Induce Potent Antinociception with Reduced Adverse Side Effects. J Med Chem 67: 7603-7619
- Wang, ZL; Pan, JX; Song, JJ; Tang, HH; Yu, HP; Li, XH; Li, N; Zhang, T; Zhang, R; Zhang, MN; Xu, B; Fang, Q; Wang, R Structure-Based Optimization of Multifunctional Agonists for Opioid and Neuropeptide FF Receptors with Potent Nontolerance Forming Analgesic Activities. J Med Chem 59: 10198-10208 (2016)
- Zhang, M; Xu, B; Li, N; Liu, H; Shi, X; Zhang, Q; Shi, Y; Xu, K; Xiao, J; Chen, D; Zhu, H; Sun, Y; Zhang, T; Zhang, R; Fang, Q Synthesis and Biological Characterization of Cyclic Disulfide-Containing Peptide Analogs of the Multifunctional Opioid/Neuropeptide FF Receptor Agonists That Produce Long-Lasting and Nontolerant Antinociception. J Med Chem 63: 15709-15725 (2020)
- Zhang, M; Xu, B; Li, N; Zhang, R; Zhang, Q; Shi, X; Xu, K; Xiao, J; Chen, D; Niu, J; Shi, Y; Fang, Q Development of Multifunctional and Orally Active Cyclic Peptide Agonists of Opioid/Neuropeptide FF Receptors that Produce Potent, Long-Lasting, and Peripherally Restricted Antinociception with Diminished Side Effects. J Med Chem 64: 13394-13409 (2021)
- Keller, M; Kuhn, KK; Einsiedel, J; Hübner, H; Biselli, S; Mollereau, C; Wifling, D; Svobodová, J; Bernhardt, G; Cabrele, C; Vanderheyden, PM; Gmeiner, P; Buschauer, A Mimicking of Arginine by Functionalized N(¿)-Carbamoylated Arginine As a New Broadly Applicable Approach to Labeled Bioactive Peptides: High Affinity Angiotensin, Neuropeptide Y, Neuropeptide FF, and Neurotensin Receptor Ligands As Examples. J Med Chem 59: 1925-45 (2016)
- Albelwi, FF; Nafie, MS; Albujuq, NR; Hourani, W; Aljuhani, A; Darwish, KM; Tawfik, MM; Rezki, N; Aouad, MR RSC Med Chem 15: 2440-2461
- Amin, KM; Barsoum, FF; Awadallah, FM; Mohamed, NE Eur J Med Chem 123: 191-201 (2016)
- Byk, G; Duchesne, M; Parker, F; Lelievre, Y; Guitton, JD; Clerc, FF; Becquart, J; Tocque, B; Scherman, D Bioorg Med Chem Lett 5: 2677-2682 (1995)
- Verschueren, WG; Dierynck, I; Amssoms, KI; Hu, L; Boonants, PM; Pille, GM; Daeyaert, FF; Hertogs, K; Surleraux, DL; Wigerinck, PB J Med Chem 48: 1930-40 (2005)
- Smith, AL; DeMorin, FF; Paras, NA; Huang, Q; Petkus, JK; Doherty, EM; Nixey, T; Kim, JL; Whittington, DA; Epstein, LF; Lee, MR; Rose, MJ; Babij, C; Fernando, M; Hess, K; Le, Q; Beltran, P; Carnahan, J J Med Chem 52: 6189-92 (2009)
- Messore, A; Madia, VN; Pescatori, L; Saccoliti, F; Tudino, V; De Leo, A; Bortolami, M; De Vita, D; Scipione, L; Pepi, F; Costi, R; Rivara, S; Scalvini, L; Mor, M; Ferrara, FF; Pavoni, E; Roscilli, G; Cassinelli, G; Milazzo, FM; Battistuzzi, G; Di Santo, R; Giannini, G J Med Chem 61: 10834-10859 (2018)
- Dias Viegas, FP; de Freitas Silva, M; Divino da Rocha, M; Castelli, MR; Riquiel, MM; Machado, RP; Vaz, SM; Simões de Lima, LM; Mancini, KC; Marques de Oliveira, PC; Morais, ÉP; Gontijo, VS; da Silva, FMR; D'Alincourt da Fonseca Peçanha, D; Castro, NG; Neves, GA; Giusti-Paiva, A; Vilela, FC; Orlandi, L; Camps, I; Veloso, MP; Leomil Coelho, LF; Ionta, M; Ferreira-Silva, GÁ; Pereira, RM; Dardenne, LE; Guedes, IA; de Oliveira Carneiro Junior, W; Quaglio Bellozi, PM; Pinheiro de Oliveira, AC; Ferreira, FF; Pruccoli, L; Tarozzi, A; Viegas, C Eur J Med Chem 147: 48-65 (2018)
- Fleming, FF; Yao, L; Ravikumar, PC; Funk, L; Shook, BC J Med Chem 53: 7902-17 (2010)
- Li, JH; Han, SJ; Hamdan, FF; Kim, SK; Jacobson, KA; Bloodworth, LM; Zhang, X; Wess, J J Biol Chem 282: 26284-93 (2007)
- Banzato, TP; Gubiani, JR; Bernardi, DI; Nogueira, CR; Monteiro, AF; Juliano, FF; de Alencar, SM; Pilli, RA; Lima, CA; Longato, GB; Ferreira, AG; Foglio, MA; Carvalho, JE; Vendramini-Costa, DB; Berlinck, RGS J Nat Prod 83: 1784-1793 (2020)
- McPherson, DW; Lambert, CR; Jahn, K; Sood, V; McRee, RC; Zeeberg, B; Reba, RC; Knapp, FF J Med Chem 38: 3908-17 (1995)
- Daniels, BJ; Li, FF; Furkert, DP; Brimble, MA J Nat Prod 82: 2054-2065 (2019)
- Doppalapudi, VR; Tryder, N; Li, L; Aja, T; Griffith, D; Liao, FF; Roxas, G; Ramprasad, MP; Bradshaw, C; Barbas, CF Bioorg Med Chem Lett 17: 501-6 (2007)
- Schierle, S; Chaikuad, A; Lillich, FF; Ni, X; Woltersdorf, S; Schallmayer, E; Renelt, B; Ronchetti, R; Knapp, S; Proschak, E; Merk, D J Med Chem 64: 5123-5136 (2021)
- Cheng, Y; Albrecht, BK; Brown, J; Buchanan, JL; Buckner, WH; DiMauro, EF; Emkey, R; Fremeau, RT; Harmange, JC; Hoffman, BJ; Huang, L; Huang, M; Lee, JH; Lin, FF; Martin, MW; Nguyen, HQ; Patel, VF; Tomlinson, SA; White, RD; Xia, X; Hitchcock, SA J Med Chem 51: 5019-34 (2008)
- Knubel, CP; Insfran, C; Martinez, FF; Diaz Lujan, C; Fretes, RE; Theumer, MG; Cervi, L; Motran, CC ACS Med Chem Lett 8: 757-761 (2017)
- Mattson, RJ; Catt, JD; Sloan, CP; Gao, Q; Carter, RB; Gentile, A; Mahle, CD; Matos, FF; McGovern, R; VanderMaelen, CP; Yocca, FD Bioorg Med Chem Lett 13: 285-8 (2002)
- Laggner, C; Schieferer, C; Fiechtner, B; Poles, G; Hoffmann, RD; Glossmann, H; Langer, T; Moebius, FF J Med Chem 48: 4754-64 (2005)
- Abd El-Karim, SS; Anwar, MM; Ahmed, NS; Syam, YM; Elseginy, SA; Aly, HF; Younis, EA; Khalil, WKB; Ahmed, KA; Mohammed, FF; Rizk, M Eur J Med Chem 260:
- Lazareno, S; Buckley, NJ; Roberts, FF Mol Pharmacol 38: 805-15 (1990)
- Shang, FF; Lu, Q; Lin, T; Pu, M; Xiao, R; Liu, W; Deng, H; Guo, H; Quan, ZS; Ding, C; Shen, QK J Med Chem 66: 12931-12949 (2023)
- Casimiro-Garcia, A; Allais, C; Brennan, A; Choi, C; Dower, G; Farley, KA; Fleming, M; Flick, A; Frisbie, RK; Hall, J; Hepworth, D; Jones, H; Knafels, JD; Kortum, S; Lovering, FE; Mathias, JP; Mohan, S; Morgan, PM; Parng, C; Parris, K; Pullen, N; Schlerman, F; Stansfield, J; Strohbach, JW; Vajdos, FF; Vincent, F; Wang, H; Wang, X; Webster, R; Wright, SW J Med Chem 65: 757-784 (2022)
- Wagner, FF; Lundh, M; Kaya, T; McCarren, P; Zhang, YL; Chattopadhyay, S; Gale, JP; Galbo, T; Fisher, SL; Meier, BC; Vetere, A; Richardson, S; Morgan, NG; Christensen, DP; Gilbert, TJ; Hooker, JM; Leroy, M; Walpita, D; Mandrup-Poulsen, T; Wagner, BK; Holson, EB ACS Chem Biol 11: 363-74 (2016)
- Li, SM; Chou, JY; Tsai, SE; Tseng, CC; Chung, CY; Zeng, WZ; Hu, YP; Uramaru, N; Huang, GJ; Wong, FF Eur J Med Chem 257:
- Shen, XB; Chen, X; Zhang, ZY; Wu, FF; Liu, XH Eur J Med Chem 225: (2021)
- Yang, FF; Xu, XL; Hu, T; Liu, JQ; Zhou, JZ; Ma, LY; Liu, HM J Med Chem 66: 4275-4293 (2023)
- Sayago, C; Gonzalez Valcarcel, IC; Qian, Y; Lee, J; Alsina-Fernandez, J; Fite, NC; Carrillo, JJ; Zhang, FF; Chalmers, MJ; Dodge, JA; Broughton, H; Espada, A ACS Med Chem Lett 9: 912-916 (2018)
- Kuduk, SD; Zheng, FF; Sepp-Lorenzino, L; Rosen, N; Danishefsky, SJ Bioorg Med Chem Lett 9: 1233-8 (1999)
- Marinozzi, M; Castro Navas, FF; Maggioni, D; Carosati, E; Bocci, G; Carloncelli, M; Giorgi, G; Cruciani, G; Fontana, R; Russo, V J Med Chem 60: 6548-6562 (2017)
- Allen, JR; Biswas, K; Chavez, Jr., F; Chen, N; De Morin, FF; Falsey, JR; Frohn, MJ; Harrington, PE; Home, DB; Harrington, EH; Kaller, MR; Kunz, RK; Monenschein, H; Nguyen, TT; Pickrell, AJ; Reichelt, A; Rumfelt, S; Rzasa, RM; Sham, K; Yao, G US Patent US8759532 (2014)
- Hadida Ruah, SS; Grootenhuis, PD; Van Goor, FF; Zhou, J; Bear, BR; Miller, MT; McCartney, J; Numa, MM US Patent US10022352 (2018)
- RORgamma Gal4 Reporter Gene Assay (FF) Cells were incubated for additional 16 h before firefly (FF) luciferase activities were measured sequentially in the same cell extract using a Dual-Light-Luciferase-Assay system (Dyer et al., Anal. Biochem. 2000, 282:158). All experiments were done at least in triplicates.
- ChEMBL_616836 (CHEMBL1100220) Inhibition of human mPGES1 in IL-1beta treated human FF cells assessed as blockade of PGH2 to PGE2 conversion after 50 mins by ELISA
- ChEMBL_653215 (CHEMBL1226418) Inhibition of eIF4A-mediated cap-dependent protein synthesis in FF-HCV-Ren mRNA transfected Swiss mouse Krebs2 cell extract by [35S]methionine metabolic labeling study
- ChEMBL_616837 (CHEMBL1100221) Inhibition of human COX2 in IL-1beta treated human FF cells assessed as blockade of SnCl2-mediated PGH2 to PGF2alpha conversion after 50 mins by ELISA
- ChEMBL_2093731 (CHEMBL4774994) Inverse agonist activity at human LXRalpha transfected in human HEK293T cells co-transfected with pSG5 and pGL3/(DR-4)-c-fos-FF-luc/pCMV/Renilla-luc assessed as receptor transactivation incubated for 20 hrs by dual luciferase reporter gene assay
- ChEMBL_2093733 (CHEMBL4774996) Inverse agonist activity at human LXRbeta transfected in human HEK293T cells co-transfected with pSG5 and pGL3/(DR-4)-c-fos-FF-luc/pCMV/Renilla-luc assessed as receptor transactivation incubated for 20 hrs by dual luciferase reporter gene assay
- Cathepsin L Inhibition Assay Inhibition of cathepsin L was assayed in reaction buffer (0.1M NaOAc-HCl, 1 mM EDTA, 0.1% β-mercaptoethanol, pH 5.5) containing 20 µM substrate and 4 nM cathepsin L. Z-FR-AMC was used as a substrate for cathepsin B. Cathepsin L was activated by incubation in assay buffer at 37 °C for 30 min prior to initiation of the reaction by addition of the substrate and test compound. The reaction mixture was then incubated at room temperature for 30 min with shaking. Fluorescence intensities were measured at 360 nm excitation and 450 nm emission wavelengths. Z-FF-FMK was used as a positive control for inhibition of cathepsin L.
- Dose-response biochemical assay of inhibitors of Rho kinase 2 (Rock2) Source (MLSCN Center Name): The Scripps Research Institute Molecular Screening Center Center Affiliation: The Scripps Research Institute, TSRI Assay Provider: Scripps Florida Network: Molecular Library Screening Center Network (MLSCN) Proposal Number: None External Assay ID: Rock2_INH_ LUMI_1536_ IC50 Name: Dose-response biochemical assay of inhibitors of Rho kinase 2 (Rock2) Description: Rho-Kinase is a serine/threonine kinase involved in the regulation of smooth muscle contraction and cytoskeletal reorganization of nonmuscle cells (1). Its inhibition is known to promote the smooth muscle relaxation. Thus, small-molecule inhibitors of Rho-Kinase may be effective probes for treatment of cerebral vasospasm (2) and potentially effective for treatment of angina (3), hypertension (4), arteriosclerosis (5), and erectile dysfunction (6). References: [1] Trauger JW, Lin FF, Turner MS, Stephens J, LoGrasso PV. Kinetic mechanism for human Rho-Kinase II (ROCK-II).Biochemistry. 2002 J
- Inhibition Assay Preparation of rhACC1. Two liters of SF9 cells, infected with recombinant baculovirus containing full length human ACC1 cDNA, were suspended in ice-cold lysis buffer (25 mM Tris, pH 7.5; 150 mM NaCl; 10% glycerol; 5 mM imidazole (EMD Bioscience; Gibbstown, N.J.); 2 mM TCEP (BioVectra; Charlottetown, Canada); Benzonase nuclease (10000 U/100 g cell paste; Novagen; Madison, Wis.); EDTA-free protease inhibitor cocktail (1 tab/50 mL; Roche Diagnostics; Mannheim, Germany). Cells were lysed by 3 cycles of freeze-thaw and centrifuged at 40,000xg for 40 minutes (4 C.). Supernatant was directly loaded onto a HisTrap FF crude column (GE Healthcare; Piscataway, N.J.) and eluted with an imidazole gradient up to 0.5 M over 20 column volumes (CV). ACC1-containing fractions were pooled and diluted 1:5 with 25 mM Tris, pH 7.5, 2 mM TCEP, 10% glycerol and direct loaded onto a CaptoQ (GE Healthcare) column and eluted with an NaCl gradient up to 1 M over 20 CV's.
- RORgamma Gal4 Reporter Gene Assay (FRET) Determination of a ligand mediated Gal4 promoter driven transactivation to quantify ligand binding to RORγ was performed as follows: DNA encoding three different RORγ protein fragments was cloned into vector pCMV-BD (Stratagene). Expression was under control of a CMV promoter and as fusion to the DNA-binding domain of the yeast protein GAL4. The amino acid boundaries of the three proteins and the respective database entries are listed in Table 2. Other vectors used were pFR-Luc (Stratagene) as regulated reporter plasmid. pFR-Luc contains a synthetic promoter with five tandem repeats of the yeast GAL4 binding sites that control expression of the Photinus pyralis (American firefly) luciferase gene. In order to improve experimental accuracy the plasmid pRL-CMV was cotransfected. pRL-CMV contains the constitutive CMV promoter, controlling the expression of the Renilla reniformis luciferase. All Gal4 reporter gene assays were done in 293T cells (DSMZ (German Collection of Microorganisms and Cell Cultures), Braunschweig, Germany, ACC635) grown in Minimum Essential Medium (MEM) with Phenol Red. The medium is supplemented with 10% fetal bovine serum, 0.1 mM nonessential amino acids, 1 mM sodium pyruvate, 1% Glutamax and 100 units Penicilin/Streptavidin per mL at 37° C. in 5% CO2.For the assay, 5×105 cells were plated per well in 96 well plates in 100 μL per well, incubated over night at 37° C. in 5% CO2. The following day, medium was discarded and the cells were transiently transfected using 20 μL per well of a OptiMEM PEI-based transfection-reagent (Sigma-Aldrich, 408727) including the three plasmids described above. About 4 h after addition of the transfection solution, fresh Minimal Essential Medium (MEM, same composition as used for plating cells, but without serum) was added. Then compound stocks, prediluted in MEM (same composition as used for plating cells) were added (final vehicle concentration not exceeding 0.1%).Cells were incubated for additional 16 h before firefly (FF) and renilla (REN) luciferase activities were measured sequentially in the same cell extract using a Dual-Light-Luciferase-Assay system (Dyer et al., Anal. Biochem. 2000, 282:158). All experiments were done at least in triplicates.Applying the Gal4 reporter gene assay as described above, the Examples of the present invention usually have an inhibition activity (IC50 FF resp. IC50 RENnorm) ranging from below 10 nM to about 20 μM, and typically, from about 10 nM to about 1 μM. The RORγ modulating compounds of the invention desirably have an inhibition in the Gal4 reporter gene assay ranging from below 10 nM to about 1 μM. Table 3 list the pIC50-value of typical examples of compounds of the invention that have an RORγ activity in the Gal4 reporter gene assay for firefly (FF) and renilla normalised (RENnorm) luciferase measurements (nt=not tested). It is understood that the data illustrated below may have reasonable variation depending on the specific conditions and procedures used by the person conducting the test. The efficacy was determined in comparison to the RORγt inhibitor T0901317 (equals 100%) and the pIC50-value is underlined, when the efficacy of the compound is below 50% of the reference.
- Binding Assay Human eIF4E (aa 28-217) with a C-terminal His tag (HH-eIF4E) was expressed in E. coli in inclusion bodies. The protein was solubilized with 8 M urea and purified under denaturing conditions using nickel-charged HisTrap HP columns (GE Healthcare). The protein was refolded by diluting to approximately 0.25 mg/mL with 6 M urea, 20 mM Hepes pH 7.0, 500 mM NaCl, 1 mM DTT, 1 mM EDTA, and 0.5 M arginine HCl, and then dialyzing overnight into the same buffer without the urea. The protein was further dialyzed into 20 mM Hepes pH 6.5, 50 mM NaCl, 1 mM EDTA, and 1 mM DTT, filtered, and then concentrated using Hitrap SP sepharose FF columns (GE Healthcare). The protein was dialyzed into 20 mM Hepes pH 7.0, 500 mM NaCl, 5 mM DTT, and 10% glycerol and stored at -80 °C until use. Test compounds (1.6 mM stock in DMSO) were diluted 3 fold in series in DMSO. Compound solutions were diluted 4 fold in Assay Buffer (50 mM sodium phosphate, pH 6.5, 50 mM KC1, 1 mM DTT and 0.5 mg/mL gammaglobulin).
- Inhibition of the Activity of ACC1 The ACC inhibitory activity of the compounds of the present invention was demonstrated by methods based on standard procedures. The direct inhibition of ACC1 for the compounds of the present invention was determined using preparations of recombinant human ACC1 (rhACC1) (SEQ ID NO. 1). Preparation of rhACC1: Two liters of SF9 cells, infected with recombinant baculovirus containing full length human ACC1 cDNA, were suspended in ice-cold lysis buffer (25 mM Tris, pH 7.5; 150 mM NaCl; 10% glycerol; 5 mM imidazole (EMD Bioscience; Gibbstown, NJ); 2 mM TCEP (BioVectra; Charlottetown, Canada); Benzonase nuclease (10000 U/100 g cell paste; Novagen; Madison, WI); EDTA-free protease inhibitor cocktail (1 tab/50 ml; Roche Diagnostics; Mannheim, Germany). Cells were lysed by 3 cycles of freeze-thaw and centrifuged at 40,000×g for 40 minutes (4° C.). Supernatant was directly loaded onto a HisTrap FF crude column (GE Healthcare; Piscataway, NJ) and eluted with an imidazole gradient up to 0.5 M over 20 column volumes (CV). ACC1-containing fractions were pooled and diluted 1:5 with 25 mM Tris, pH 7.5, 2 mM TCEP, 10% glycerol and direct loaded onto a CaptoQ (GE Healthcare) column and eluted with an NaCl gradient up to 1 M over 20 CV's. Phosphate groups were removed from purified ACC1 by incubation with lambda phosphatase (100 U/10 μM target protein; New England Biolabs; Beverly, MA) for 14 hours at 4° C.; okadaic acid was added (1 μM final concentration; Roche Diagnostics) to inhibit the phosphatase. Purified ACC1 was exchanged into 25 mM Tris, pH 7.5, 2 mM TCEP, 10% glycerol, 0.5 M NaCl by 6 hour dialysis at 4° C. Aliquots were prepared and frozen at −80° C.
- Inhibition of Protease Activity Assay Full-length cDNA of human MALT1 gene (GenBank accession No: AB026118.1) amplified by PCR was inserted in flame to a SalI site located downstream of GST gene in a pGEX6P3 vector (GE Healthcare Japan Corp.) to prepare a vector (hereinafter, referred to as a pGEX6P3-MALT1 vector). Subsequently, E. coli for protein expression (BL21-RIL-codon plus-DE3, Agilent Technologies, Inc.) was transformed with the pGEX6P3-MALT1 vector and then analyzed by ampicillin resistance screening and colony PCR to obtain an E. coli strain expressing recombinant GST fusion MALT1. Protein expression was induced with isopropyl-(3-thiogalactopyranoside. After the expression induction, E. coli precipitates were recovered by centrifugation from the E. coli culture solution, and the E. coli precipitates were homogenized and then centrifuged to obtain a supernatant. The supernatant was purified using GSTrap FF column (GE Healthcare Japan Corp.) to obtain recombinant GST fusion MALT1.B) Evaluation of Inhibition of Protease Activity of MALT1:To 89 μL of an enzyme solution (4.8 g/mL GST fusion MALT1, 50 mmol/L MES, 150 mmol/L NaCl, 10% sucrose, 0.1% CHAPS, 10 mmol/L dithiothreitol, and 1 mol/L tri-ammonium citrate) per specimen, 1 μL of a test compound (DMSO-diluted solution) of each concentration was added to prepare a mixed solution. The mixed solution was incubated at room temperature for 30 minutes, followed by the measurement of the fluorescence value of the mixed solution (fluorescence value of the first measurement) (Ex: 380 nm, Em: 460 nm; Envision (Perkin Elmer Inc.)). Next, 10 μL of 200 μmol/L substrate (Ac-LRSR-AMC, SM Biochemicals LLC) was added (final concentration: 20 μmol/L) to the mixed solution, and the mixture was reacted by incubation at 30° C. for 80 minutes, followed by the measurement of the fluorescence value of the reaction solution (fluorescence value of the second measurement) (Ex: 380 nm, Em: 460 nm; Envision (Perkin Elmer Inc.)).
- In Vitro Assays for IDH1m (R132H or R132C) Inhibitors In the primary reaction, the reduction of α-KG acid to 2-HG is accompanied by a concomitant oxidation of NADPH to NADP. The amount of NADPH remaining at the end of the reaction time is measured in a secondary diaphorase/resazurin reaction in which the NADPH is consumed in a 1:1 molar ratio with the conversion of resazurin to the highly fluorescent resorufin. Uninhibited reactions exhibit a low fluorescence at the end of the assay, while reactions in which the consumption of NADPH by R132H IDH1 has been inhibited by a small molecule show a high fluorescence.The primary reaction is performed in a volume of 50 μL 1× Buffer (150 mM NaCl, 20 mM Tris 7.5, 10 mM MgCl2, 0.05% (w/v) bovine serum albumin), contained 0.25 ug/mL (2.7 nM) IDH1 wt/IDH1 R132H heterodimer, 0.3 mM alpha-ketoglutarate, 4 μM NADPH, and either 300 μM NADP (saturated) or 30 μM NADP (without saturation), and 1 uL of 50× compound in DMSO. The mixture of compound, enzyme, and cofactor is pre-incubated at room temperature for 1 hr prior to the addition of alpha-ketoglutarate. To perform the secondary reaction, 10 uL of 1× buffer containing 36 μg/ml diaphorase and 30 mM resazurin is added to the primary reaction and incubated for a further 5 minutes at 25° C. Florescence is read on a Spectramax platereader at Ex 544 Em 590. Compounds or compound dilutions are prepared in 100% DMSO concentration and diluted 1:50 into the final reaction. IDH1 wt/IDH1 R132C is assayed under similar conditions except that 1X Buffer is 50 mM K2HPO4, pH 6.5; 10 mM MgCl2; 10% glycerol; 0.03% (w/v) bovine serum albumin and final concentrations are 0.4 ug/mL (4.3 nM) IDH1 wt/IDH1 R132C heterodimer, 0.02 mM alpha-ketoglutarate, 4 uM NADPH, and either 300 μM NADP (saturated) or 30 μM NADP (without saturation). IC50s are determined.IDH1 or IDH2 wildtype (wt) and mutant heterodimers are expressed and purified by methods known in the art. For example, IDH1 wt/R132m heterodimer is expressed and purified as follows. Co-expression of IDH1 wt-his and IDH1R132C-flag is carried out in sf9 insect cells. Cells (25 g) are resuspended in 250 ml of 50 mM Tris, 500 mM NaCl, pH7.4, at 4° C. with stirring. Cells are disrupted with 4 passes through an M-Y110 Micro fluidizer (Microfluidics) set to 500 psi, and then centrifuged at 22,000 ref for 20 min at 4° C. The supernatant is harvested and loaded at 15 cm/h on a Histrap FF 5*1 ml column (GE) which is equilibrated with 50 mM Tris, 500 mM NaCl, pH7.4. Host cell contaminants are removed by washing the column with equilibration buffer followed by equilibration buffer containing 20 mM imidazole and 60 mM imidazole to baseline. IDH1 wt-his homodimer and IDH1 wt-his/IDH1R132C-flag are eluted by equilibration buffer containing 250 mM imidazole. Fractions eluted by 250 mM imidazole are pooled together and loaded at 15 cm/h onto a column pre-packed with 10 ml ANTI-FLAG M2 Affinity Gel (Sigma), the column is equilibrated with 50 mM Tris, 500 mM NaCl, pH7.4. After washing with equilibration buffer, IDH1 wt-his/IDH1R132C-flag heterodimer is eluted by equilibration buffer containing flag peptide (0.2 mg/ml). Aliquots of IDH1 wt-his/IDH1R132C-flag are flash frozen in liquid N2 and stored at −80° C. Same conditions are used for the purification of IDH1 wt-his/IDH1R132H-flag.
- In Vitro Assays for IDH1m (R132H or R132C) Inhibitors In the primary reaction, the reduction of α-KG acid to 2-HG is accompanied by a concomitant oxidation of NADPH to NADP. The amount of NADPH remaining at the end of the reaction time is measured in a secondary diaphorase/resazurin reaction in which the NADPH is consumed in a 1:1 molar ratio with the conversion of resazurin to the highly fluorescent resorufin. Uninhibited reactions exhibit a low fluorescence at the end of the assay, while reactions in which the consumption of NADPH by R132H IDH1 has been inhibited by a small molecule show a high fluorescence.The primary reaction is performed in a volume of 50 μL 1× Buffer (150 mM NaCl, 20 mM Tris 7.5, 10 mM MgCl2, 0.05% (w/v) bovine serum albumin), contained 0.25 ug/mL (2.7 nM) IDH1 wt/IDH1 R132H heterodimer, 0.3 mM alpha-ketoglutarate, 4 μM NADPH, and either 300 μM NADP (saturated) or 30 μM NADP (without saturation), and 1 uL of 50× compound in DMSO. The mixture of compound, enzyme, and cofactor is pre-incubated at room temperature for 1 hr prior to the addition of alpha-ketoglutarate. To perform the secondary reaction, 10 uL of 1× buffer containing 36 μg/ml diaphorase and 30 mM resazurin is added to the primary reaction and incubated for a further 5 minutes at 25° C. Florescence is read on a Spectramax platereader at Ex 544 Em 590. Compounds or compound dilutions are prepared in 100% DMSO concentration and diluted 1:50 into the final reaction. IDH1 wt/IDH1 R132C is assayed under similar conditions except that 1× Buffer is 50 mM K2HP04, pH 6.5; 10 mM MgCl2; 10% glycerol; 0.03% (w/v) bovine serum albumin and final concentrations are 0.4 ug/mL (4.3 nM) IDH1 wt/IDH1 R132C heterodimer, 0.02 mM alpha-ketoglutarate, 4 uM NADPH, and either 300 μM NADP (saturated) or 30 μM NADP (without saturation). IC50s are determined.IDH1 or IDH2 wildtype (wt) and mutant heterodimers are expressed and purified by methods known in the art. For example, IDH1wt/R132m heterodimer is expressed and purified as follows. Co-expression of IDH1wt-his and IDH1R132C-flag is carried out in sf9 insect cells. Cells (25 g) are resuspended in 250 ml of 50 mM Tirs, 500 mM NaCl, pH7.4, at 4° C. with stirring. Cells are disrupted with 4 passes through an M-Y110 Micro fluidizer (Microfluidics) set to 500 psi, and then centrifuged at 22,000 rcf for 20 min at 4° C. The supernatant is harvested and loaded at 15 cm/h on a Histrap FF 5*1 ml column (GE) which is equilibrated with 50 mM Tirs, 500 mM NaCl, pH7.4. Host cell contaminants are removed by washing the column with equilibration buffer followed by equilibration buffer containing 20 mM imidazole and 60 mM imidazole to baseline. IDH1wt-his homodimer and IDH1wt-his/IDH1R132C-flag are eluted by equilibration buffer containing 250 mM imidazole. Fractions eluted by 250 mM imidazole are pooled together and loaded at 15 cm/h onto a column pre-packed with 10 ml ANTI-FLAG M2 Affinity Gel (Sigma), the column is equilibrated with 50 mM Tris, 500 mM NaCl, pH7.4. After washing with equilibration buffer, IDH1wt-his/IDH1R132C-flag heterodimer is eluted by equilibration buffer containing flag peptide (0.2 mg/ml). Aliquots of IDH1wt-his/IDH1R132C-flag are flash frozen in liquid N2 and stored at −80° C. Same conditions are used for the purification of IDH1wt-his/IDH1R132H-flag.
- In Vitro Assays for IDH1m (R132H or R132C) Inhibitors In the primary reaction, the reduction of α-KG acid to 2-HG is accompanied by a concomitant oxidation of NADPH to NADP. The amount of NADPH remaining at the end of the reaction time is measured in a secondary diaphorase/resazurin reaction in which the NADPH is consumed in a 1:1 molar ratio with the conversion of resazurin to the highly fluorescent resorufin. Uninhibited reactions exhibit a low fluorescence at the end of the assay, while reactions in which the consumption of NADPH by R132H IDH1 has been inhibited by a small molecule show a high fluorescence.The primary reaction is performed in a volume of 50 μL 1× Buffer (150 mM NaCl, 20 mM Tris 7.5, 10 mM MgCl2, 0.05% (w/v) bovine serum albumin), contained 0.25 ug/mL (2.7 nM) IDH1 wt/IDH1 R132H heterodimer, 0.3 mM alpha-ketoglutarate, 4 μM NADPH, and either 300 μM NADP (saturated) or 30 μM NADP (without saturation), and 1 uL of 50× compound in DMSO. The mixture of compound, enzyme, and cofactor is pre-incubated at room temperature for 1 hr prior to the addition of alpha-ketoglutarate. To perform the secondary reaction, 10 uL of 1× buffer containing 36 μg/ml diaphorase and 30 mM resazurin is added to the primary reaction and incubated for a further 5 minutes at 25° C. Florescence is read on a Spectramax platereader at Ex 544 Em 590. Compounds or compound dilutions are prepared in 100% DMSO concentration and diluted 1:50 into the final reaction. IDH1 wt/IDH1 R132C is assayed under similar conditions except that 1× Buffer is 50 mM K2HPO4, pH 6.5; 10 mM MgCl2; 10% glycerol; 0.03% (w/v) bovine serum albumin and final concentrations are 0.4 ug/mL (4.3 nM) IDH1 wt/IDH1 R132C heterodimer, 0.02 mM alpha-ketoglutarate, 4 uM NADPH, and either 300 μM NADP (saturated) or 30 μM NADP (without saturation). IC50s are determined.IDH1 or IDH2 wildtype (wt) and mutant heterodimers are expressed and purified by methods known in the art. For example, IDH1wt/R132m heterodimer is expressed and purified as follows. Co-expression of IDH1wt-his and IDH1R132C-flag is carried out in sf9 insect cells. Cells (25 g) are resuspended in 250 ml of 50 mM Tris, 500 mM NaCl, pH7.4, at 4° C. with stirring. Cells are disrupted with 4 passes through an M-Y110 Micro fluidizer (Microfluidics) set to 500 psi, and then centrifuged at 22,000 rcf for 20 min at 4° C. The supernatant is harvested and loaded at 15 cm/h on a Histrap FF 5*1 ml column (GE) which is equilibrated with 50 mM Tris, 500 mM NaCl, pH7.4. Host cell contaminants are removed by washing the column with equilibration buffer followed by equilibration buffer containing 20 mM imidazole and 60 mM imidazole to baseline. IDH1wt-his homodimer and IDH1wt-his/IDH1R132C-flag are eluted by equilibration buffer containing 250 mM imidazole. Fractions eluted by 250 mM imidazole are pooled together and loaded at 15 cm/h onto a column pre-packed with 10 ml ANTI-FLAG M2 Affinity Gel (Sigma), the column is equilibrated with 50 mM Tris, 500 mM NaCl, pH7.4. After washing with equilibration buffer, IDH1wt-his/IDH1R132C-flag heterodimer is eluted by equilibration buffer containing flag peptide (0.2 mg/ml). Aliquots of IDH1wt-his/IDH1R132C-flag are flash frozen in liquid N2 and stored at −80° C. Same conditions are used for the purification of IDH1wt-his/IDH1R132H-flag.
- kinetic binding analysis Dual histidine and Avi tagged human EIF4E (His6-3C-avi-eIF4E) was expressed in 8 L of TB Media. Induction by 0.4 mM IPTG occurred at 2.0 OD600, and cells were harvested at 15 OD600. 225 gram pellet was diluted in buffer (50 mM Tris, 500 mM NaCl, 2 mM MgCl2, 1 mM TCEP, pH 7.5 containing 10% glycerol, protease inhibitors, and DNase) to volume of 600 mL and passed once over the microfluidizer. Sample was run on 5 mL HisTrap HP IMAC at 4.0 mL/min IMAC with a 25 mM to 500 mM imidazole gradient for one column volume. GST-PreScission Protease (2 mg, made in-house) was added to sample and allowed to react overnight at 4° C. The cleaved pool was passed over 0.5 mL GST and 0.5 IMAC resin in a gravity column. Sample volume was increased to 800 mL using 50 mM Tris pH 7.5, 1 mM TCEP and passed over a 5 mL hiTrap SP FF at 4.0 mL/min with a 0 to 1 M NaCl gradient over 20 column volumes. Sample was then injected onto a 124 mL S75 Gel Filtration Column at 20 mg/mL. Final Avi-eIF4E (26931 Da) was diluted to 1 mg/mL in 1× Bicine buffer to a volume of 2.5 mg and mixed with ATP/Biotin Mix (10 mM ATP, 10 mM Mg(OAc)2, 50 μM d-biotin final). Biotin Ligase (25 μg BirA produced in house) was added to reaction. Reactions were performed with mixing (500 rpm) on Eppendorf ThermoMixer R at 30° C. for 60 minutes and checked for completeness using LC-MS. To the sample, 100 μl of immobilized glutathione (1:1 with buffer) was add and mixed for 15 min at 4° C. to bind C3 and Bir3 and removed by centrifugation. The sample was buffer exchanged using two consecutive PD-10 columns equilibrated with 20 mM HEPES, 100 mM KCl, 1 mM DTT, pH 7.5.Due to low eIF4E stability, the streptavidin coated chip was prepared and run at 10° C. on the Biacore T200. The eIF4E (0.04 mg/mL, 150 μl) was bound to the sample channel of Series S Sensor Chip SA (GE Life Sciences, BR-1005-31) to surface density of 5000 to 7000 RU (Response Units). Buffer flowed over the chip at 30 μL/min, using 1×PBS, 50 mM NaCl, 0.1% Glycerol, 0.1% CHAPS, and 1% DMSO. Samples of various cap analogs were diluted to various concentrations in a range of 100 μM to less than 1 nM. Samples were injected into the Biacore chip with a two minute association time and a five minute dissociation time. Several buffer injections were done for each sample for blank subtraction.Analysis was done for all sets using Biacore T200 evaluation software. Binding analysis for all compounds is reported as response units at 1 micromolar compound where a higher value for RU is interpreted as greater ligand binding to the surface immobilized eIF4E protein. A subset of compounds was further characterized to determine dissociation constants using kinetic binding. Steady State Affinity fits were done with default settings (4 seconds before injection stop with 5 second window). Kinetic fits were normally done with 1:1 binding model, with constant RI=0 and all other variables set to fit globally.
 US9464092, FF BDBM253940 US9527857, FF
US9464092, FF BDBM253940 US9527857, FF BDBM292830 US10106501, Example FF
BDBM292830 US10106501, Example FF US10106501, Example FF-1 BDBM292831
US10106501, Example FF-1 BDBM292831 US10106501, Example FF-2 BDBM292832
US10106501, Example FF-2 BDBM292832 US9598447, Ref No. FF BDBM303064
US9598447, Ref No. FF BDBM303064 Ff-10101 BDBM50647812 FLT3-IN-1
Ff-10101 BDBM50647812 FLT3-IN-1 US10150763, Example FF BDBM310732 8-((4-(4-Fluorophenyl)-pyridin-2-yl)(3-methyl-1,2,4-thiadiazol-5-yl)amino)-N-hydroxyoctanamide
US10150763, Example FF BDBM310732 8-((4-(4-Fluorophenyl)-pyridin-2-yl)(3-methyl-1,2,4-thiadiazol-5-yl)amino)-N-hydroxyoctanamide BDBM110185 benzyl N-[1-[(4-fluoro-3-oxo-1-phenylbutan-2-yl)amino]-1-oxo-3-phenylpropan-2-yl]carbamate Z-FF-FMK
BDBM110185 benzyl N-[1-[(4-fluoro-3-oxo-1-phenylbutan-2-yl)amino]-1-oxo-3-phenylpropan-2-yl]carbamate Z-FF-FMK 7-[[(3R)-3-Aminopiperidin-1-yl]methyl]-3-[[2-(benzenesulfonyl)-5-chlorophenyl]methyl]-8-chloro-6-(trifluoromethyl)-1H-quinazoline-2,4-dione BDBM286935 US9567304, Compound FF-2
7-[[(3R)-3-Aminopiperidin-1-yl]methyl]-3-[[2-(benzenesulfonyl)-5-chlorophenyl]methyl]-8-chloro-6-(trifluoromethyl)-1H-quinazoline-2,4-dione BDBM286935 US9567304, Compound FF-2 8-Chloro-3-[(5-chloro-2-ethylsulfonylphenyl)methyl]-7-[[3-(dimethylsulfamoylamino)azetidin-1-yl]methyl]-2,4-dioxo-6-(trifluoromethyl)-1H-quinazoline BDBM286940 US9567304, Compound FF-7
8-Chloro-3-[(5-chloro-2-ethylsulfonylphenyl)methyl]-7-[[3-(dimethylsulfamoylamino)azetidin-1-yl]methyl]-2,4-dioxo-6-(trifluoromethyl)-1H-quinazoline BDBM286940 US9567304, Compound FF-7 BDBM286938 7-[[(3R)-3-Aminopiperidin-1-yl]methyl]-8-chloro-3-[(2-ethylsulfonyl-4,5-dimethylphenyl)methyl]-6-(trifluoromethyl)-1H-quinazoline-2,4-dione US9567304, Compound FF-5
BDBM286938 7-[[(3R)-3-Aminopiperidin-1-yl]methyl]-8-chloro-3-[(2-ethylsulfonyl-4,5-dimethylphenyl)methyl]-6-(trifluoromethyl)-1H-quinazoline-2,4-dione US9567304, Compound FF-5 US9567304, Compound FF-1 7-[[(3R)-3-Aminopiperidin-1-yl]methyl]-8-chloro-3-[(5-chloro-2-propylsulfonylphenyl)methyl]-6-(trifluoromethyl)-1H-quinazoline-2,4-dione BDBM286934
US9567304, Compound FF-1 7-[[(3R)-3-Aminopiperidin-1-yl]methyl]-8-chloro-3-[(5-chloro-2-propylsulfonylphenyl)methyl]-6-(trifluoromethyl)-1H-quinazoline-2,4-dione BDBM286934 US9567304, Compound FF-8 8-Chloro-3-[(5-chloro-2-ethylsulfonylphenyl)methyl]-7-[[3-(methylsulfamoylamino)azetidin-1-yl]methyl]-6-(trifluoromethyl)-1H-quinazoline-2,4-dione BDBM286941
US9567304, Compound FF-8 8-Chloro-3-[(5-chloro-2-ethylsulfonylphenyl)methyl]-7-[[3-(methylsulfamoylamino)azetidin-1-yl]methyl]-6-(trifluoromethyl)-1H-quinazoline-2,4-dione BDBM286941 US9567304, Compound FF-9 8-Chloro-3-[(5-chloro-2-ethylsulfonylphenyl)methyl]-2,4-dioxo-7-[[3-(sulfamoylamino)azetidin-1-yl]methyl]-6-(trifluoromethyl)-1H-quinazoline BDBM286942
US9567304, Compound FF-9 8-Chloro-3-[(5-chloro-2-ethylsulfonylphenyl)methyl]-2,4-dioxo-7-[[3-(sulfamoylamino)azetidin-1-yl]methyl]-6-(trifluoromethyl)-1H-quinazoline BDBM286942 BDBM286936 US9567304, Compound FF-3 7-[[(3R)-3-Aminopiperidin-1-yl]methyl]-8-chloro-3-[(5-chloro-2-propan-2-ylsulfonylphenyl)methyl]-6-(trifluoromethyl)-1H-quinazoline-2,4-dione
BDBM286936 US9567304, Compound FF-3 7-[[(3R)-3-Aminopiperidin-1-yl]methyl]-8-chloro-3-[(5-chloro-2-propan-2-ylsulfonylphenyl)methyl]-6-(trifluoromethyl)-1H-quinazoline-2,4-dione BDBM286937 7-[[(3R)-3-Aminopiperidin-1-yl]methyl]-8-chloro-3-[(5-chloro-2-ethylsulfonyl-3-methoxyphenyl)methyl]-6-(trifluoromethyl)-1H-quinazoline-2,4-dione US9567304, Compound FF-4
BDBM286937 7-[[(3R)-3-Aminopiperidin-1-yl]methyl]-8-chloro-3-[(5-chloro-2-ethylsulfonyl-3-methoxyphenyl)methyl]-6-(trifluoromethyl)-1H-quinazoline-2,4-dione US9567304, Compound FF-4 BDBM286939 7-[[(3R)-3-Aminopiperidin-1-yl]methyl]-8-chloro-3-[(5-chloro-2-ethylsulfonyl-4-methylphenyl)methyl]-6-(trifluoromethyl)-1H-quinazoline-2,4-dione US9567304, Compound FF-6
BDBM286939 7-[[(3R)-3-Aminopiperidin-1-yl]methyl]-8-chloro-3-[(5-chloro-2-ethylsulfonyl-4-methylphenyl)methyl]-6-(trifluoromethyl)-1H-quinazoline-2,4-dione US9567304, Compound FF-6 (Scheme FF) (2S,3S,4R,5R)-2-(1-methyl-1H-pyrazol-3-yl)-5-(4-methyl-7H-pyrrolo[2,3-d]pyrimidin-7-yl)tetrahydrofuran-3,4-diol (EE-2) US10428104, Example 77 BDBM415546
(Scheme FF) (2S,3S,4R,5R)-2-(1-methyl-1H-pyrazol-3-yl)-5-(4-methyl-7H-pyrrolo[2,3-d]pyrimidin-7-yl)tetrahydrofuran-3,4-diol (EE-2) US10428104, Example 77 BDBM415546 US10428104, Example 78 (Scheme GG) (2S,3S,4R,5R)-2-(3-(ethoxymethyl)-1H-pyrazol-5-yl)-5-(4-methyl-7H-pyrrolo[2,3-d]pyrimidin-7-yl)tetrahydrofuran-3,4-diol (FF-6) BDBM415547
US10428104, Example 78 (Scheme GG) (2S,3S,4R,5R)-2-(3-(ethoxymethyl)-1H-pyrazol-5-yl)-5-(4-methyl-7H-pyrrolo[2,3-d]pyrimidin-7-yl)tetrahydrofuran-3,4-diol (FF-6) BDBM415547