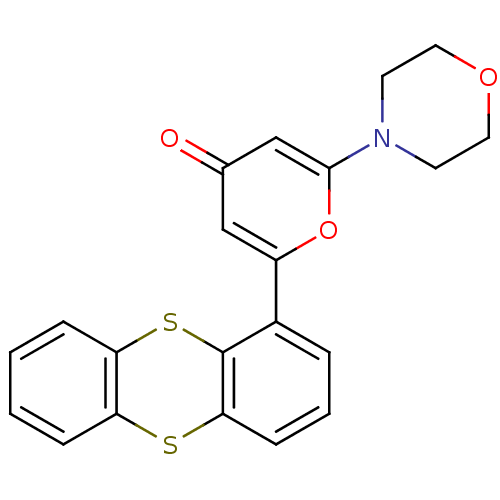
 KU-55933 BDBM50208517 2-morpholin-4-yl-6-thianthren-1-yl-pyran-4-one CHEMBL222102
KU-55933 BDBM50208517 2-morpholin-4-yl-6-thianthren-1-yl-pyran-4-one CHEMBL222102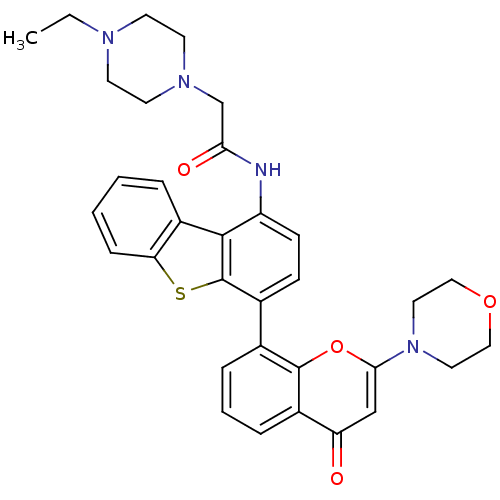 BDBM50319926 KU-0060648 CHEMBL1086377 2-(4-ethylpiperazin-1-yl)-N-(4-(2-morpholino-4-oxo-4H-chromen-8-yl)dibenzo[b,d]thiophen-1-yl)acetamide
BDBM50319926 KU-0060648 CHEMBL1086377 2-(4-ethylpiperazin-1-yl)-N-(4-(2-morpholino-4-oxo-4H-chromen-8-yl)dibenzo[b,d]thiophen-1-yl)acetamide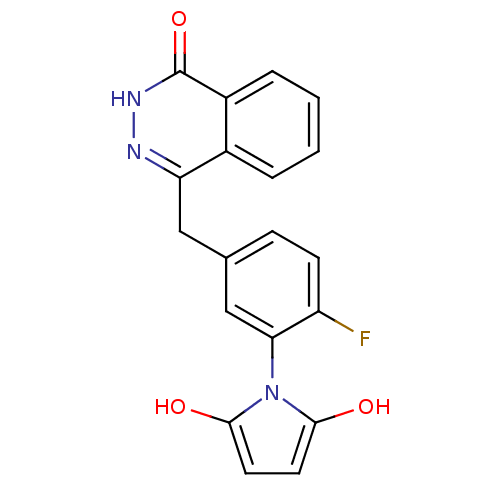 1-[2-Fluoro-5-(4-oxo-3,4-dihydro-phthalazin-1-ylmethyl)-phenyl]-pyrrolidine-2,5-dione 1-(2-fluoro-5-((4-oxo-3,4-dihydrophthalazin-1-yl)methyl)phenyl)pyrrolidine-2,5-dione BDBM50165486 KU-58684 CHEMBL196444
1-[2-Fluoro-5-(4-oxo-3,4-dihydro-phthalazin-1-ylmethyl)-phenyl]-pyrrolidine-2,5-dione 1-(2-fluoro-5-((4-oxo-3,4-dihydrophthalazin-1-yl)methyl)phenyl)pyrrolidine-2,5-dione BDBM50165486 KU-58684 CHEMBL196444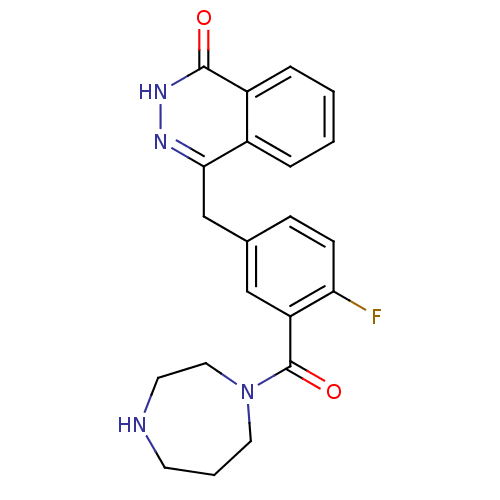 4-{[3-(1,4-diazepan-1-ylcarbonyl)-4-fluorophenyl]methyl}-1,2-dihydrophthalazin-1-one 4-(3-(1,4-diazepane-1-carbonyl)-4-fluorobenzyl)phthalazin-1(2H)-one 4-[[3-(1,4-Diazepane-1-carbonyl)-4-fluorophenyl]methyl]-2Hphthalazin-1-one BDBM27533 CHEMBL380648 KU-0058948 4-[3-(1,4-diazepan-1-ylcarbonyl)-4-fluorobenzyl]phthalazin-1(2H)-one Homopiperazine analogue, 14
4-{[3-(1,4-diazepan-1-ylcarbonyl)-4-fluorophenyl]methyl}-1,2-dihydrophthalazin-1-one 4-(3-(1,4-diazepane-1-carbonyl)-4-fluorobenzyl)phthalazin-1(2H)-one 4-[[3-(1,4-Diazepane-1-carbonyl)-4-fluorophenyl]methyl]-2Hphthalazin-1-one BDBM27533 CHEMBL380648 KU-0058948 4-[3-(1,4-diazepan-1-ylcarbonyl)-4-fluorobenzyl]phthalazin-1(2H)-one Homopiperazine analogue, 14
- Ahn, BZ; Baik, KU; Kweon, GR; Lim, K; Hwang, BD J Med Chem 38: 1044-7 (1995)
- Kling, A; Lange, UE; Mack, H; Bakker, MH; Drescher, KU; Hornberger, W; Hutchins, CW; Möller, A; Müller, R; Schmidt, M; Unger, L; Wicke, K; Schellhaas, K; Steiner, G Bioorg Med Chem Lett 15: 5567-73 (2005)
- Kubas, H; Meyer, U; Hechenberger, M; Klein, KU; Plitt, P; Zemribo, R; Spexgoor, HW; van Assema, SG; Abel, U Bioorg Med Chem Lett 23: 6370-6 (2013)
- Thompson, JC; Dao, WT; Ku, A; Rodriguez-Beltran, SL; Amezcua, M; Palomino, AY; Lien, T; Salzameda, NT Bioorg Med Chem 28: (2020)
- Lee, S; Ku, AF; Vippila, MR; Wang, Y; Zhang, M; Wang, X; Hedstrom, L; Cuny, GD Bioorg Med Chem Lett 30: (2020)
- Naik, R; Ban, HS; Jang, K; Kim, I; Xu, X; Harmalkar, D; Shin, SA; Kim, M; Kim, BK; Park, J; Ku, B; Oh, S; Won, M; Lee, K J Med Chem 60: 8631-8646 (2017)
- Son, JH; Zhu, JS; Phuan, PW; Cil, O; Teuthorn, AP; Ku, CK; Lee, S; Verkman, AS; Kurth, MJ J Med Chem 60: 2401-2410 (2017)
- Vlattas, I; Dellureficio, J; Ku, E; Bohacek, R; Xiaolu, Zhang Bioorg Med Chem Lett 6: 2091-2096 (1996)
- Chun, K; Park, JS; Lee, HC; Kim, YH; Ye, IH; Kim, KJ; Ku, IW; Noh, MY; Cho, GW; Kim, H; Kim, SH; Kim, J Bioorg Med Chem Lett 23: 3983-7 (2013)
- Pativada, T; Kim, MH; Lee, JH; Hong, SS; Choi, CW; Choi, YH; Kim, WJ; Song, DW; Park, SI; Lee, EJ; Seo, BY; Kim, H; Kim, HK; Lee, KH; Ahn, SK; Ku, JM; Park, GH J Med Chem 62: 6063-6082 (2019)
- Lee, KL; Foley, MA; Chen, L; Behnke, ML; Lovering, FE; Kirincich, SJ; Wang, W; Shim, J; Tam, S; Shen, MW; Khor, S; Xu, X; Goodwin, DG; Ramarao, MK; Nickerson-Nutter, C; Donahue, F; Ku, MS; Clark, JD; McKew, JC J Med Chem 50: 1380-400 (2007)
- Drilon, A; Nagasubramanian, R; Blake, JF; Ku, N; Tuch, BB; Ebata, K; Smith, S; Lauriault, V; Kolakowski, GR; Brandhuber, BJ; Larsen, PD; Bouhana, KS; Winski, SL; Hamor, R; Wu, WI; Parker, A; Morales, TH; Sullivan, FX; DeWolf, WE; Wollenberg, LA; Gordon, PR; Douglas-Lindsay, DN; Scaltriti, M; Benayed, R; Raj, S; Hanusch, B; Schram, AM; Jonsson, P; Berger, MF; Hechtman, JF; Taylor, BS; Andrews, S; Rothenberg, SM; Hyman, DM Cancer Discov 7: 963-972
- Kim, M; Kim, S; Ku, S; Park, C; Joe, B; Chun, K; Ye, I; Choi, J; Ryu, D; Park, J; Lee, H; Choi, J; Kim, Y US Patent US8815891 (2014)
- Giancola, JB; Bonifazi, A; Cao, J; Ku, T; Haraczy, AJ; Lam, J; Rais, R; Coggiano, MA; Tanda, G; Newman, AH Eur J Med Chem 208: (2020)
- Camacho-Hernandez, GA; Casiraghi, A; Rudin, D; Luethi, D; Ku, TC; Guthrie, DA; Straniero, V; Valoti, E; Schütz, GJ; Sitte, HH; Newman, AH RSC Med Chem 12: 1174-1186 (2021)
- Gleason, JG; Hall, RF; Perchonock, CD; Erhard, KF; Frazee, JS; Ku, TW; Kondrad, K; McCarthy, ME; Mong, S; Crooke, ST J Med Chem 30: 959-61 (1987)
- Zhou, W; Liu, X; Tu, Z; Zhang, L; Ku, X; Bai, F; Zhao, Z; Xu, Y; Ding, K; Li, H J Med Chem 56: 7821-37 (2013)
- Kusunoki, N; Takara, K; Tanigawara, Y; Yamauchi, A; Ueda, K; Komada, F; Ku, Y; Kuroda, Y; Saitoh, Y; Okumura, K Jpn J Cancer Res 89: 1220-8 (1999)
- Black, LA; Nersesian, DL; Sharma, P; Ku, YY; Bennani, YL; Marsh, KC; Miller, TR; Esbenshade, TA; Hancock, AA; Cowart, M Bioorg Med Chem Lett 17: 1443-6 (2007)
- Aarhus, TI; Bjørnstad, F; Wolowczyk, C; Larsen, KU; Rognstad, L; Leithaug, T; Unger, A; Habenberger, P; Wolf, A; Bjørkøy, G; Pridans, C; Eickhoff, J; Klebl, B; Hoff, BH; Sundby, E J Med Chem 66: 6959-6980 (2023)
- Tewes, B; Frehland, B; Schepmann, D; Schmidtke, KU; Winckler, T; Wünsch, B Bioorg Med Chem 18: 8005-15 (2010)
- Matter, H; Schwab, W; Barbier, D; Billen, G; Haase, B; Neises, B; Schudok, M; Thorwart, W; Schreuder, H; Brachvogel, V; Lönze, P; Weithmann, KU J Med Chem 42: 1908-20 (1999)
- Klabunde, T; Wendt, KU; Kadereit, D; Brachvogel, V; Burger, HJ; Herling, AW; Oikonomakos, NG; Kosmopoulou, MN; Schmoll, D; Sarubbi, E; von Roedern, E; Schönafinger, K; Defossa, E J Med Chem 48: 6178-93 (2005)
- Mederski, WW; Cezanne, B; van Amsterdam, C; Bühring, KU; Dorsch, D; Gleitz, J; März, J; Tsaklakidis, C Bioorg Med Chem Lett 14: 5817-22 (2004)
- Additional SAR compounds tested via Multiplex dose response to identify specific small molecule inhibitors of Ras and Ras-related GTPases specifically Cdc42 wildtype University of New Mexico Assay Overview: Assay Support: NIH I RO3 MH081231-01 HTS to identify specific small molecule inhibitors of Ras and Ras-related GTPases PI: Angela Wandinger-Ness, Ph.D. Co-PI: Larry Sklar, Ph.D. Assay Development: Zurab Surviladze, Ph.D. Assay Implementation: Zurab Surviladze, Danuta Wlodek, Terry Foutz, Mark Carter, Anna Waller Target Team Leader for the Center: Larry Sklar (lsklar@salud.unm.edu) UNM Cheminformatics: Cristian Bologa, Ph.D., Fabiola Miscioscia, Ph.D., Ramona Curpan, Ph.D., Oleg Ursu, Ph.D. Chemistry: University of Kansas Specialized Chemistry Center KU Specialized Chemistry Center PI: Jeff Aube, Ph.D. KU SCC Project Manager: Jennifer E. Golden. Ph.D. KU SCC Chemists on this project: Chad Schroeder, M.S., Denise Simpson, Ph.D., Julica Noeth, B.S. Dose Response Assay Background and Significance: Ras and related small molecular weight GTPases function in the regulation of signaling and cell growth, and collectively serve to control cell proli
- Additional SAR compounds tested via Multiplex dose response to identify specific small molecule inhibitors of Ras and Ras-related GTPases specifically Rab2 wildtype University of New Mexico Assay Overview: Assay Support: NIH I RO3 MH081231-01 HTS to identify specific small molecule inhibitors of Ras and Ras-related GTPases PI: Angela Wandinger-Ness, Ph.D. Co-PI: Larry Sklar, Ph.D. Assay Development: Zurab Surviladze, Ph.D. Assay Implementation: Zurab Surviladze, Danuta Wlodek, Terry Foutz, Mark Carter, Anna Waller Target Team Leader for the Center: Larry Sklar (lsklar@salud.unm.edu) UNM Cheminformatics: Cristian Bologa, Ph.D., Fabiola Miscioscia, Ph.D., Ramona Curpan, Ph.D., Oleg Ursu, Ph.D. Chemistry: University of Kansas Specialized Chemistry Center KU Specialized Chemistry Center PI: Jeff Aube, Ph.D. KU SCC Project Manager: Jennifer E. Golden. Ph.D. KU SCC Chemists on this project: Chad Schroeder, M.S., Denise Simpson, Ph.D., Julica Noeth, B.S. Dose Response Assay Background and Significance: Ras and related small molecular weight GTPases function in the regulation of signaling and cell growth, and collectively serve to control cell proli
- Additional SAR compounds tested via Multiplex dose response to identify specific small molecule inhibitors of Ras and Ras-related GTPases specifically Rab7 wildtype University of New Mexico Assay Overview: Assay Support: NIH I RO3 MH081231-01 HTS to identify specific small molecule inhibitors of Ras and Ras-related GTPases PI: Angela Wandinger-Ness, Ph.D. Co-PI: Larry Sklar, Ph.D. Assay Development: Zurab Surviladze, Ph.D. Assay Implementation: Zurab Surviladze, Danuta Wlodek, Terry Foutz, Mark Carter, Anna Waller Target Team Leader for the Center: Larry Sklar (lsklar@salud.unm.edu) UNM Cheminformatics: Cristian Bologa, Ph.D., Fabiola Miscioscia, Ph.D., Ramona Curpan, Ph.D., Oleg Ursu, Ph.D. Chemistry: University of Kansas Specialized Chemistry Center KU Specialized Chemistry Center PI: Jeff Aube, Ph.D. KU SCC Project Manager: Jennifer E. Golden. Ph.D. KU SCC Chemists on this project: Chad Schroeder, M.S., Denise Simpson, Ph.D., Julica Noeth, B.S. Dose Response Assay Background and Significance: Ras and related small molecular weight GTPases function in the regulation of signaling and cell growth, and collectively serve to control cell proli
- Additional SAR compounds tested via Multiplex dose response to identify specific small molecule inhibitors of Ras and Ras-related GTPases specifically Rac1 wildtype University of New Mexico Assay Overview: Assay Support: NIH I RO3 MH081231-01 HTS to identify specific small molecule inhibitors of Ras and Ras-related GTPases PI: Angela Wandinger-Ness, Ph.D. Co-PI: Larry Sklar, Ph.D. Assay Development: Zurab Surviladze, Ph.D. Assay Implementation: Zurab Surviladze, Danuta Wlodek, Terry Foutz, Mark Carter, Anna Waller Target Team Leader for the Center: Larry Sklar (lsklar@salud.unm.edu) UNM Cheminformatics: Cristian Bologa, Ph.D., Fabiola Miscioscia, Ph.D., Ramona Curpan, Ph.D., Oleg Ursu, Ph.D. Chemistry: University of Kansas Specialized Chemistry Center KU Specialized Chemistry Center PI: Jeff Aube, Ph.D. KU SCC Project Manager: Jennifer E. Golden. Ph.D. KU SCC Chemists on this project: Chad Schroeder, M.S., Denise Simpson, Ph.D., Julica Noeth, B.S. Dose Response Assay Background and Significance: Ras and related small molecular weight GTPases function in the regulation of signaling and cell growth, and collectively serve to control cell proli
- Additional SAR compounds tested via Multiplex dose response to identify specific small molecule inhibitors of Ras and Ras-related GTPases specifically Ras wildtype University of New Mexico Assay Overview: Assay Support: NIH I RO3 MH081231-01 HTS to identify specific small molecule inhibitors of Ras and Ras-related GTPases PI: Angela Wandinger-Ness, Ph.D. Co-PI: Larry Sklar, Ph.D. Assay Development: Zurab Surviladze, Ph.D. Assay Implementation: Zurab Surviladze, Danuta Wlodek, Terry Foutz, Mark Carter, Anna Waller Target Team Leader for the Center: Larry Sklar (lsklar@salud.unm.edu) UNM Cheminformatics: Cristian Bologa, Ph.D., Fabiola Miscioscia, Ph.D., Ramona Curpan, Ph.D., Oleg Ursu, Ph.D. Chemistry: University of Kansas Specialized Chemistry Center KU Specialized Chemistry Center PI: Jeff Aube, Ph.D. KU SCC Project Manager: Jennifer E. Golden. Ph.D. KU SCC Chemists on this project: Chad Schroeder, M.S., Denise Simpson, Ph.D., Julica Noeth, B.S. Dose Response Assay Background and Significance: Ras and related small molecular weight GTPases function in the regulation of signaling and cell growth, and collectively serve to control cell proli
- Oxadiazole SAR compounds tested via Multiplex dose response to identify specific small molecule inhibitors of Ras and Ras-related GTPases specifically Cdc42 wildtype University of New Mexico Assay Overview: Assay Support: NIH I RO3 MH081231-01 HTS to identify specific small molecule inhibitors of Ras and Ras-related GTPases PI: Angela Wandinger-Ness, Ph.D. Co-PI: Larry Sklar, Ph.D. Assay Development: Zurab Surviladze, Ph.D. Assay Implementation: Zurab Surviladze, Danuta Wlodek, Terry Foutz, Mark Carter, Anna Waller Target Team Leader for the Center: Larry Sklar (lsklar@salud.unm.edu) UNM Cheminformatics: Cristian Bologa, Ph.D., Fabiola Miscioscia, Ph.D., Ramona Curpan, Ph.D., Oleg Ursu, Ph.D. Chemistry: University of Kansas Specialized Chemistry Center KU Specialized Chemistry Center PI: Jeff Aube, Ph.D. KU SCC Project Manager: Jennifer E. Golden. Ph.D. KU SCC Chemists on this project: Chad Schroeder, M.S., Denise Simpson, Ph.D., Julica Noeth, B.S. Dose Response Assay Background and Significance: Ras and related small molecular weight GTPases function in the regulation of signaling and cell growth, and collectively serve to control cell proli
- Oxadiazole SAR compounds tested via Multiplex dose response to identify specific small molecule inhibitors of Ras and Ras-related GTPases specifically Rab2 wildtype University of New Mexico Assay Overview: Assay Support: NIH I RO3 MH081231-01 HTS to identify specific small molecule inhibitors of Ras and Ras-related GTPases PI: Angela Wandinger-Ness, Ph.D. Co-PI: Larry Sklar, Ph.D. Assay Development: Zurab Surviladze, Ph.D. Assay Implementation: Zurab Surviladze, Danuta Wlodek, Terry Foutz, Mark Carter, Anna Waller Target Team Leader for the Center: Larry Sklar (lsklar@salud.unm.edu) UNM Cheminformatics: Cristian Bologa, Ph.D., Fabiola Miscioscia, Ph.D., Ramona Curpan, Ph.D., Oleg Ursu, Ph.D. Chemistry: University of Kansas Specialized Chemistry Center KU Specialized Chemistry Center PI: Jeff Aube, Ph.D. KU SCC Project Manager: Jennifer E. Golden. Ph.D. KU SCC Chemists on this project: Chad Schroeder, M.S., Denise Simpson, Ph.D., Julica Noeth, B.S. Dose Response Assay Background and Significance: Ras and related small molecular weight GTPases function in the regulation of signaling and cell growth, and collectively serve to control cell proli
- Oxadiazole SAR compounds tested via Multiplex dose response to identify specific small molecule inhibitors of Ras and Ras-related GTPases specifically Rab7 wildtype University of New Mexico Assay Overview: Assay Support: NIH I RO3 MH081231-01 HTS to identify specific small molecule inhibitors of Ras and Ras-related GTPases PI: Angela Wandinger-Ness, Ph.D. Co-PI: Larry Sklar, Ph.D. Assay Development: Zurab Surviladze, Ph.D. Assay Implementation: Zurab Surviladze, Danuta Wlodek, Terry Foutz, Mark Carter, Anna Waller Target Team Leader for the Center: Larry Sklar (lsklar@salud.unm.edu) UNM Cheminformatics: Cristian Bologa, Ph.D., Fabiola Miscioscia, Ph.D., Ramona Curpan, Ph.D., Oleg Ursu, Ph.D. Chemistry: University of Kansas Specialized Chemistry Center KU Specialized Chemistry Center PI: Jeff Aube, Ph.D. KU SCC Project Manager: Jennifer E. Golden. Ph.D. KU SCC Chemists on this project: Chad Schroeder, M.S., Denise Simpson, Ph.D., Julica Noeth, B.S. Dose Response Assay Background and Significance: Ras and related small molecular weight GTPases function in the regulation of signaling and cell growth, and collectively serve to control cell proli
- Oxadiazole SAR compounds tested via Multiplex dose response to identify specific small molecule inhibitors of Ras and Ras-related GTPases specifically Rac1 wildtype University of New Mexico Assay Overview: Assay Support: NIH I RO3 MH081231-01 HTS to identify specific small molecule inhibitors of Ras and Ras-related GTPases PI: Angela Wandinger-Ness, Ph.D. Co-PI: Larry Sklar, Ph.D. Assay Development: Zurab Surviladze, Ph.D. Assay Implementation: Zurab Surviladze, Danuta Wlodek, Terry Foutz, Mark Carter, Anna Waller Target Team Leader for the Center: Larry Sklar (lsklar@salud.unm.edu) UNM Cheminformatics: Cristian Bologa, Ph.D., Fabiola Miscioscia, Ph.D., Ramona Curpan, Ph.D., Oleg Ursu, Ph.D. Chemistry: University of Kansas Specialized Chemistry Center KU Specialized Chemistry Center PI: Jeff Aube, Ph.D. KU SCC Project Manager: Jennifer E. Golden. Ph.D. KU SCC Chemists on this project: Chad Schroeder, M.S., Denise Simpson, Ph.D., Julica Noeth, B.S. Dose Response Assay Background and Significance: Ras and related small molecular weight GTPases function in the regulation of signaling and cell growth, and collectively serve to control cell proli
- Oxadiazole SAR compounds tested via Multiplex dose response to identify specific small molecule inhibitors of Ras and Ras-related GTPases specifically Ras wildtype University of New Mexico Assay Overview: Assay Support: NIH I RO3 MH081231-01 HTS to identify specific small molecule inhibitors of Ras and Ras-related GTPases PI: Angela Wandinger-Ness, Ph.D. Co-PI: Larry Sklar, Ph.D. Assay Development: Zurab Surviladze, Ph.D. Assay Implementation: Zurab Surviladze, Danuta Wlodek, Terry Foutz, Mark Carter, Anna Waller Target Team Leader for the Center: Larry Sklar (lsklar@salud.unm.edu) UNM Cheminformatics: Cristian Bologa, Ph.D., Fabiola Miscioscia, Ph.D., Ramona Curpan, Ph.D., Oleg Ursu, Ph.D. Chemistry: University of Kansas Specialized Chemistry Center KU Specialized Chemistry Center PI: Jeff Aube, Ph.D. KU SCC Project Manager: Jennifer E. Golden. Ph.D. KU SCC Chemists on this project: Chad Schroeder, M.S., Denise Simpson, Ph.D., Julica Noeth, B.S. Dose Response Assay Background and Significance: Ras and related small molecular weight GTPases function in the regulation of signaling and cell growth, and collectively serve to control cell proli
- Pyrazoline SAR compounds tested via Multiplex dose response to identify specific small molecule inhibitors of Ras and Ras-related GTPases specifically Rab2 wildtype University of New Mexico Assay Overview: Assay Support: NIH I RO3 MH081231-01 HTS to identify specific small molecule inhibitors of Ras and Ras-related GTPases PI: Angela Wandinger-Ness, Ph.D. Co-PI: Larry Sklar, Ph.D. Assay Development: Zurab Surviladze, Ph.D. Assay Implementation: Zurab Surviladze, Danuta Wlodek, Terry Foutz, Mark Carter, Anna Waller Target Team Leader for the Center: Larry Sklar (lsklar@salud.unm.edu) UNM Cheminformatics: Cristian Bologa, Ph.D., Fabiola Miscioscia, Ph.D., Ramona Curpan, Ph.D., Oleg Ursu, Ph.D. Chemistry: University of Kansas Specialized Chemistry Center KU Specialized Chemistry Center PI: Jeff Aube, Ph.D. KU SCC Project Manager: Jennifer E. Golden. Ph.D. KU SCC Chemists on this project: Chad Schroeder, M.S., Denise Simpson, Ph.D., Julica Noeth, B.S. Dose Response Assay Background and Significance: Ras and related small molecular weight GTPases function in the regulation of signaling and cell growth, and collectively serve to control cell proli
- Pyrazoline SAR compounds tested via Multiplex dose response to identify specific small molecule inhibitors of Ras and Ras-related GTPases specifically Rab7 wildtype University of New Mexico Assay Overview: Assay Support: NIH I RO3 MH081231-01 HTS to identify specific small molecule inhibitors of Ras and Ras-related GTPases PI: Angela Wandinger-Ness, Ph.D. Co-PI: Larry Sklar, Ph.D. Assay Development: Zurab Surviladze, Ph.D. Assay Implementation: Zurab Surviladze, Danuta Wlodek, Terry Foutz, Mark Carter, Anna Waller Target Team Leader for the Center: Larry Sklar (lsklar@salud.unm.edu) UNM Cheminformatics: Cristian Bologa, Ph.D., Fabiola Miscioscia, Ph.D., Ramona Curpan, Ph.D., Oleg Ursu, Ph.D. Chemistry: University of Kansas Specialized Chemistry Center KU Specialized Chemistry Center PI: Jeff Aube, Ph.D. KU SCC Project Manager: Jennifer E. Golden. Ph.D. KU SCC Chemists on this project: Chad Schroeder, M.S., Denise Simpson, Ph.D., Julica Noeth, B.S. Dose Response Assay Background and Significance: Ras and related small molecular weight GTPases function in the regulation of signaling and cell growth, and collectively serve to control cell proli
- Pyrazoline SAR compounds tested via Multiplex dose response to identify specific small molecule inhibitors of Ras and Ras-related GTPases specifically Rac1 wildtype University of New Mexico Assay Overview: Assay Support: NIH I RO3 MH081231-01 HTS to identify specific small molecule inhibitors of Ras and Ras-related GTPases PI: Angela Wandinger-Ness, Ph.D. Co-PI: Larry Sklar, Ph.D. Assay Development: Zurab Surviladze, Ph.D. Assay Implementation: Zurab Surviladze, Danuta Wlodek, Terry Foutz, Mark Carter, Anna Waller Target Team Leader for the Center: Larry Sklar (lsklar@salud.unm.edu) UNM Cheminformatics: Cristian Bologa, Ph.D., Fabiola Miscioscia, Ph.D., Ramona Curpan, Ph.D., Oleg Ursu, Ph.D. Chemistry: University of Kansas Specialized Chemistry Center KU Specialized Chemistry Center PI: Jeff Aube, Ph.D. KU SCC Project Manager: Jennifer E. Golden. Ph.D. KU SCC Chemists on this project: Chad Schroeder, M.S., Denise Simpson, Ph.D., Julica Noeth, B.S. Dose Response Assay Background and Significance: Ras and related small molecular weight GTPases function in the regulation of signaling and cell growth, and collectively serve to control cell proli
- Pyrazoline SAR compounds tested via Multiplex dose response to identify specific small molecule inhibitors of Ras and Ras-related GTPases specifically Ras wildtype University of New Mexico Assay Overview: Assay Support: NIH I RO3 MH081231-01 HTS to identify specific small molecule inhibitors of Ras and Ras-related GTPases PI: Angela Wandinger-Ness, Ph.D. Co-PI: Larry Sklar, Ph.D. Assay Development: Zurab Surviladze, Ph.D. Assay Implementation: Zurab Surviladze, Danuta Wlodek, Terry Foutz, Mark Carter, Anna Waller Target Team Leader for the Center: Larry Sklar (lsklar@salud.unm.edu) UNM Cheminformatics: Cristian Bologa, Ph.D., Fabiola Miscioscia, Ph.D., Ramona Curpan, Ph.D., Oleg Ursu, Ph.D. Chemistry: University of Kansas Specialized Chemistry Center KU Specialized Chemistry Center PI: Jeff Aube, Ph.D. KU SCC Project Manager: Jennifer E. Golden. Ph.D. KU SCC Chemists on this project: Chad Schroeder, M.S., Denise Simpson, Ph.D., Julica Noeth, B.S. Dose Response Assay Background and Significance: Ras and related small molecular weight GTPases function in the regulation of signaling and cell growth, and collectively serve to control cell prol
- Additional SAR compounds tested by Multiplex dose response to identify specific small molecule inhibitors of Ras and Ras-related GTPases specifically Cdc42 activated mutant University of New Mexico Assay Overview: Assay Support: NIH I RO3 MH081231-01 HTS to identify specific small molecule inhibitors of Ras and Ras-related GTPases PI: Angela Wandinger-Ness, Ph.D. Co-PI: Larry Sklar, Ph.D. Assay Development: Zurab Surviladze, Ph.D. Assay Implementation: Zurab Surviladze, Danuta Wlodek, Terry Foutz, Mark Carter, Anna Waller Target Team Leader for the Center: Larry Sklar (lsklar@salud.unm.edu) UNM Cheminformatics: Cristian Bologa, Ph.D., Fabiola Miscioscia, Ph.D., Ramona Curpan, Ph.D., Oleg Ursu, Ph.D. Chemistry: University of Kansas Specialized Chemistry Center KU Specialized Chemistry Center PI: Jeff Aube, Ph.D. KU SCC Project Manager: Jennifer E. Golden. Ph.D. KU SCC Chemists on this project: Chad Schroeder, M.S., Denise Simpson, Ph.D., Julica Noeth, B.S. Dose Response Assay Background and Significance: Ras and related small molecular weight GTPases function in the regulation of signaling and cell growth, and collectively serve to control cell proli
- Additional SAR compounds tested via Multiplex dose response to identify specific small molecule inhibitors of Ras and Ras-related GTPases specifically Rac1 activated mutant University of New Mexico Assay Overview: Assay Support: NIH I RO3 MH081231-01 HTS to identify specific small molecule inhibitors of Ras and Ras-related GTPases PI: Angela Wandinger-Ness, Ph.D. Co-PI: Larry Sklar, Ph.D. Assay Development: Zurab Surviladze, Ph.D. Assay Implementation: Zurab Surviladze, Danuta Wlodek, Terry Foutz, Mark Carter, Anna Waller Target Team Leader for the Center: Larry Sklar (lsklar@salud.unm.edu) UNM Cheminformatics: Cristian Bologa, Ph.D., Fabiola Miscioscia, Ph.D., Ramona Curpan, Ph.D., Oleg Ursu, Ph.D. Chemistry: University of Kansas Specialized Chemistry Center KU Specialized Chemistry Center PI: Jeff Aube, Ph.D. KU SCC Project Manager: Jennifer E. Golden. Ph.D. KU SCC Chemists on this project: Chad Schroeder, M.S., Denise Simpson, Ph.D., Julica Noeth, B.S. Dose Response Assay Background and Significance: Ras and related small molecular weight GTPases function in the regulation of signaling and cell growth, and collectively serve to control cell proli
- Additional SAR compounds tested via Multiplex dose response to identify specific small molecule inhibitors of Ras and Ras-related GTPases specifically Ras activated mutant University of New Mexico Assay Overview: Assay Support: NIH I RO3 MH081231-01 HTS to identify specific small molecule inhibitors of Ras and Ras-related GTPases PI: Angela Wandinger-Ness, Ph.D. Co-PI: Larry Sklar, Ph.D. Assay Development: Zurab Surviladze, Ph.D. Assay Implementation: Zurab Surviladze, Danuta Wlodek, Terry Foutz, Mark Carter, Anna Waller Target Team Leader for the Center: Larry Sklar (lsklar@salud.unm.edu) UNM Cheminformatics: Cristian Bologa, Ph.D., Fabiola Miscioscia, Ph.D., Ramona Curpan, Ph.D., Oleg Ursu, Ph.D. Chemistry: University of Kansas Specialized Chemistry Center KU Specialized Chemistry Center PI: Jeff Aube, Ph.D. KU SCC Project Manager: Jennifer E. Golden. Ph.D. KU SCC Chemists on this project: Chad Schroeder, M.S., Denise Simpson, Ph.D., Julica Noeth, B.S. Dose Response Assay Background and Significance: Ras and related small molecular weight GTPases function in the regulation of signaling and cell growth, and collectively serve to control cell proli
- Oxadiazole SAR compounds tested via Multiplex dose response to identify specific small molecule inhibitors of Ras and Ras-related GTPases specifically Rac1 activated mutant University of New Mexico Assay Overview: Assay Support: NIH I RO3 MH081231-01 HTS to identify specific small molecule inhibitors of Ras and Ras-related GTPases PI: Angela Wandinger-Ness, Ph.D. Co-PI: Larry Sklar, Ph.D. Assay Development: Zurab Surviladze, Ph.D. Assay Implementation: Zurab Surviladze, Danuta Wlodek, Terry Foutz, Mark Carter, Anna Waller Target Team Leader for the Center: Larry Sklar (lsklar@salud.unm.edu) UNM Cheminformatics: Cristian Bologa, Ph.D., Fabiola Miscioscia, Ph.D., Ramona Curpan, Ph.D., Oleg Ursu, Ph.D. Chemistry: University of Kansas Specialized Chemistry Center KU Specialized Chemistry Center PI: Jeff Aube, Ph.D. KU SCC Project Manager: Jennifer E. Golden. Ph.D. KU SCC Chemists on this project: Chad Schroeder, M.S., Denise Simpson, Ph.D., Julica Noeth, B.S. Dose Response Assay Background and Significance: Ras and related small molecular weight GTPases function in the regulation of signaling and cell growth, and collectively serve to control cell proli
- Oxadiazole SAR compounds tested via Multiplex dose response to identify specific small molecule inhibitors of Ras and Ras-related GTPases specifically Ras activated mutant University of New Mexico Assay Overview: Assay Support: NIH I RO3 MH081231-01 HTS to identify specific small molecule inhibitors of Ras and Ras-related GTPases PI: Angela Wandinger-Ness, Ph.D. Co-PI: Larry Sklar, Ph.D. Assay Development: Zurab Surviladze, Ph.D. Assay Implementation: Zurab Surviladze, Danuta Wlodek, Terry Foutz, Mark Carter, Anna Waller Target Team Leader for the Center: Larry Sklar (lsklar@salud.unm.edu) UNM Cheminformatics: Cristian Bologa, Ph.D., Fabiola Miscioscia, Ph.D., Ramona Curpan, Ph.D., Oleg Ursu, Ph.D. Chemistry: University of Kansas Specialized Chemistry Center KU Specialized Chemistry Center PI: Jeff Aube, Ph.D. KU SCC Project Manager: Jennifer E. Golden. Ph.D. KU SCC Chemists on this project: Chad Schroeder, M.S., Denise Simpson, Ph.D., Julica Noeth, B.S. Dose Response Assay Background and Significance: Ras and related small molecular weight GTPases function in the regulation of signaling and cell growth, and collectively serve to control cell proli
- Pyrazoline SAR compounds tested via Multiplex dose response to identify specific small molecule inhibitors of Ras and Ras-related GTPases specifically Cdc42 activated mutant University of New Mexico Assay Overview: Assay Support: NIH I RO3 MH081231-01 HTS to identify specific small molecule inhibitors of Ras and Ras-related GTPases PI: Angela Wandinger-Ness, Ph.D. Co-PI: Larry Sklar, Ph.D. Assay Development: Zurab Surviladze, Ph.D. Assay Implementation: Zurab Surviladze, Danuta Wlodek, Terry Foutz, Mark Carter, Anna Waller Target Team Leader for the Center: Larry Sklar (lsklar@salud.unm.edu) UNM Cheminformatics: Cristian Bologa, Ph.D., Fabiola Miscioscia, Ph.D., Ramona Curpan, Ph.D., Oleg Ursu, Ph.D. Chemistry: University of Kansas Specialized Chemistry Center KU Specialized Chemistry Center PI: Jeff Aube, Ph.D. KU SCC Project Manager: Jennifer E. Golden. Ph.D. KU SCC Chemists on this project: Chad Schroeder, M.S., Denise Simpson, Ph.D., Julica Noeth, B.S. Dose Response Assay Background and Significance: Ras and related small molecular weight GTPases function in the regulation of signaling and cell growth, and collectively serve to control cell proli
- Pyrazoline SAR compounds tested via Multiplex dose response to identify specific small molecule inhibitors of Ras and Ras-related GTPases specifically Cdc42 wildtype protein University of New Mexico Assay Overview: Assay Support: NIH I RO3 MH081231-01 HTS to identify specific small molecule inhibitors of Ras and Ras-related GTPases PI: Angela Wandinger-Ness, Ph.D. Co-PI: Larry Sklar, Ph.D. Assay Development: Zurab Surviladze, Ph.D. Assay Implementation: Zurab Surviladze, Danuta Wlodek, Terry Foutz, Mark Carter, Anna Waller Target Team Leader for the Center: Larry Sklar (lsklar@salud.unm.edu) UNM Cheminformatics: Cristian Bologa, Ph.D., Fabiola Miscioscia, Ph.D., Ramona Curpan, Ph.D., Oleg Ursu, Ph.D. Chemistry: University of Kansas Specialized Chemistry Center KU Specialized Chemistry Center PI: Jeff Aube, Ph.D. KU SCC Project Manager: Jennifer E. Golden. Ph.D. KU SCC Chemists on this project: Chad Schroeder, M.S., Denise Simpson, Ph.D., Julica Noeth, B.S. Dose Response Assay Background and Significance: Ras and related small molecular weight GTPases function in the regulation of signaling and cell growth, and collectively serve to control cell prol
- Pyrazoline SAR compounds tested via Multiplex dose response to identify specific small molecule inhibitors of Ras and Ras-related GTPases specifically Rac1 activated mutant University of New Mexico Assay Overview: Assay Support: NIH I RO3 MH081231-01 HTS to identify specific small molecule inhibitors of Ras and Ras-related GTPases PI: Angela Wandinger-Ness, Ph.D. Co-PI: Larry Sklar, Ph.D. Assay Development: Zurab Surviladze, Ph.D. Assay Implementation: Zurab Surviladze, Danuta Wlodek, Terry Foutz, Mark Carter, Anna Waller Target Team Leader for the Center: Larry Sklar (lsklar@salud.unm.edu) UNM Cheminformatics: Cristian Bologa, Ph.D., Fabiola Miscioscia, Ph.D., Ramona Curpan, Ph.D., Oleg Ursu, Ph.D. Chemistry: University of Kansas Specialized Chemistry Center KU Specialized Chemistry Center PI: Jeff Aube, Ph.D. KU SCC Project Manager: Jennifer E. Golden. Ph.D. KU SCC Chemists on this project: Chad Schroeder, M.S., Denise Simpson, Ph.D., Julica Noeth, B.S. Dose Response Assay Background and Significance: Ras and related small molecular weight GTPases function in the regulation of signaling and cell growth, and collectively serve to control cell proli
- Pyrazoline SAR compounds tested via Multiplex dose response to identify specific small molecule inhibitors of Ras and Ras-related GTPases specifically Ras activated mutant University of New Mexico Assay Overview: Assay Support: NIH I RO3 MH081231-01 HTS to identify specific small molecule inhibitors of Ras and Ras-related GTPases PI: Angela Wandinger-Ness, Ph.D. Co-PI: Larry Sklar, Ph.D. Assay Development: Zurab Surviladze, Ph.D. Assay Implementation: Zurab Surviladze, Danuta Wlodek, Terry Foutz, Mark Carter, Anna Waller Target Team Leader for the Center: Larry Sklar (lsklar@salud.unm.edu) UNM Cheminformatics: Cristian Bologa, Ph.D., Fabiola Miscioscia, Ph.D., Ramona Curpan, Ph.D., Oleg Ursu, Ph.D. Chemistry: University of Kansas Specialized Chemistry Center KU Specialized Chemistry Center PI: Jeff Aube, Ph.D. KU SCC Project Manager: Jennifer E. Golden. Ph.D. KU SCC Chemists on this project: Chad Schroeder, M.S., Denise Simpson, Ph.D., Julica Noeth, B.S. Dose Response Assay Background and Significance: Ras and related small molecular weight GTPases function in the regulation of signaling and cell growth, and collectively serve to control cell proli
- Dose Response of Flow Cytometric HTS Screen for inhibitors of the ABC transporter ABCB6 for Cherry Pick01 University of New Mexico Assay Overview: Assay Support: NIH 1 R03 MH093193-01A1 High Throughput Screening for inhibitors of the ABC transporter ABCB6 PI: Partha Krishnamurthy, Ph.D. Screening Center PI: Larry Sklar, Ph.D. / UNMCMD Screening Lead: Mohiuddin Khan, Ph.D. Assay Implementation: Stephanie Chavez, Dominique Perez, Matthew Garcia, J. Jacob Strouse, Ph.D., Mark Carter, M.S., Anna Waller, Ph.D. UNM Cheminformatics: Cristian Bologa, Ph.D., Oleg Ursu, Ph.D. Chemistry: University of Kansas Specialized Chemistry Center KU Specialized Chemistry Center PI: Jeff Aube, Ph.D. KU SCC Project Manager: Jennifer E. Golden. Ph.D. Assay Background and Significance: The goal of this project is to implement a high throughput screening (HTS) assay to identify and develop selective chemical probes that regulate ABCB6 expression or function. The long-term objective is to use these novel pharmacological tools to understand not only the significance of ABCB6 in tumor growth and proliferation a
- Diaxonhit Phosphodiesterase Assay The phosphodiesterase assay was developed using the LANCE cAMP kit (PerkinElmer). The assay buffer contained HBSS with 5 mM HEPES, 0.1% BSA, and 1.5 mM MgCl2, pH 7.4. PDE10A (BPS Bioscience) was used at 200 pg/well (with a specific activity of 3200 pmole/min/ug with assay conditions: 10 mM Tris-HCl, pH 7.4, 10 mM MgCl2, 1 mM MnCl2, 200 uM cAMP, 2.5 kU 5' nucleotidase, 37° C., 20 min) and PDE4D3 (BPS Bioscience) was used at 100 pg/well (with a specific activity of 32713 pmole/min/ug with assay conditions: 10 mM Tris-HCl, pH 7.4, 10 mM MgCl2, 1 mM MnCl2, 200 uM cAMP, 2.5 kU 5' nucleotidase, 37° C., 20 min). The Biotin-cAMP tracer, supplied in 10 mmol/L Tris-HCl buffered (pH 8.0) salt solution with 1 mmol/L ethylenediaminetetraacetic acid (EDTA), 0.1% bovine serum albumin (BSA), and 0.05% sodium azide, is used at a dilution of 1/375. The assay detection mixture contained the LANCE Eu-W8044 labeled streptavidin 1/2250 (supplied in 50 mmol/L Tris-HCl buffer).
- Homogenous Time-Resolved Fluorescence Resonance Energy Transfer (TR-FRET) Technology The phosphodiesterase assay was developed using the LANCE cAMP kit (PerkinElmer). The assay buffer contained HBSS with 5 mM HEPES, 0.1% BSA, and 1.5 mM MgCl2, pH 7.4. PDE10A (BPS Bioscience) was used at 200 pg/well (with a specific activity of 3200 pmole/min/μg with assay conditions: 10 mM Tris-HCl, pH 7.4, 10 mM MgCl2, 1 mM MnCl2, 200 μM cAMP, 2.5 kU 5′ nucleotidase, 37° C., 20 min) and PDE4D3 (BPS Bioscience) was used at 100 pg/well (with a specific activity of 32713 pmole/min/μg with assay conditions: 10 mM Tris-HCl, pH 7.4, 10 mM MgCl2, 1 mM MnCl2, 200 μM cAMP, 2.5 kU 5′ nucleotidase, 37° C., 20 min). The Biotin-cAMP tracer, supplied in 10 mmol/L Tris-HCl buffered (pH 8.0) salt solution with 1 mmol/L ethylenediaminetetraacetic acid (EDTA), 0.1% bovine serum albumin (BSA), and 0.05% sodium azide, is used at a dilution of 1/375. The assay detection mixture contained the LANCE Eu-W8044 labeled streptavidin 1/2250 (supplied in 50 mmol/L Tris-HCl buffered (pH 7.8) salt solution with 0.9% sodium chloride (NaCl), 0.1% BSA, and 0.05% sodium azide) and the Alexa Fluor 647-anti cAMP antibody 1/200 (supplied in 50 mmol/L Tris-HCl buffered (pH 7.8) salt solution with 0.9% NaCl, 0.1% BSA, and 0.05% sodium azide). Chemical compounds were dissolved in DMSO (final concentration 2% (v/v)).
- Activity Assay Inhibitory activity-1 assay: To a solution containing 20 mM Tris-HCl (pH 7.5), 1 mM MgCl2, 100 μM EDTA, 330 μg/ml bovine serum albumin, 50 kU/ml 5′-nucleotidase, 0.1 μCi 3H-cAMP (64 nM cAMP), and PDE10A (H-PDE10A2, Human Phosphodiesterase 10A2, Scottish Biomedical), a test compound was added, and the mixture was reacted at 25° C. for 2 hours. To the reaction solution, QAE-Sephadex (17-0190-01, GE Healthcare Japan Corp.) suspended in 10 mM HEPES-Na (pH 7.0) (hereinafter, also referred to as a “QAE-Sephadex suspension”) was added, and the mixture was shaken for 1 minute and left standing for 5 minutes to obtain a supernatant. To the supernatant, a QAE-Sephadex suspension was further added, and the mixture was shaken for 1 minute and left standing for 5 minutes. Then, the obtained supernatant was transferred to LumaPlate (PerkinElmer, Inc.) and assayed using a radiation counter (TopCount NXT, PerkinElmer, Inc.).
- Homogenous Time-Resolved Fluorescence Resonance Energy Transfer Assay The PDE assay is based on the homogenous time-resolved fluorescence resonance energy transfer (TR-FRET) technology (LANCE from Perkin Elmer). This competition based assay is formatted using a cAMP specific antibody labeled with the dye, Alexa Fluor 647, biotin-cAMP and streptavidin labeled with Europium (Eu-SA). As the complex of Eu-SA/biotin-cAMP/Alexa Fluor 647 labeled antibody is formed, an increase in signal is generated. When there is PDE activity, resulting in the degradation of the cyclic nucleotide, the complex is not formed and a decrease in signal is observed. The phosphodiesterase assay was developed using the LANCE cAMP kit (PerkinElmer). The assay buffer contained HBSS with 5 mM HEPES, 0.1% BSA, and 1.5 mM MgCl2, pH 7.4. PDE10A (BPS Bioscience) was used at 200 pg/well (with a specific activity of 3200 pmole/min/ug with assay conditions: 10 mM Tris-HCl, pH7.4, 10 mM MgCl2, 1 mM MnCl2, 200 uM cAMP, 2.5 kU 5'nucleotidase, 37 C, 20 min).
- ENPP1 Enzyme Activity Assay 3 nM mouse ENPP1 was incubated with 5 uM cGAMP and 5-fold serial dilutions of compounds in buffer containing 50 mM Tris pH 7.6, 250 nM NaCl, 500 uM CaCl2, and 1 uM ZnCl2 (total reaction volume=10 μL) at room temperature for 3 hours, after which the reactions were heat inactivated at 95° C. for 10 minutes. The AMP degradation product was converted to ATP, which was detected using luciferase. To achieve this, an enzyme mixture of polyphosphate:AMP phosphotransferase (PAP) and myokinase was prepared according to Goueli et al. in EP2771480. Briefly, PAP was diluted to 2 mg/mL in buffer containing 50 mM Tris pH 7.5, 0.1% NP-40. Myokinase was diluted to 2 KU/mL in buffer containing 3.2 mM ammonium sulfate pH 6.0, 1 mM EDTA, and 4 mM polyphosphate. The heat-inactivated ENPP1 reaction was incubated with PAP (0.01 μg/μL) and myokinase (0.0075 U/μL) in buffer containing 40 mM Tris pH 7.5, 0.05 mg/mL Prionex, 5 mM MgCl2, 20 μM polyphosphate, and 0.15 g/L phenol red (for ease of pipetting) for 3 hours (total reaction volume=20 μL). CellTiterGlo (20 uL) was added to the reaction according to manufacturer's protocol and luminescence was measured. Data were normalized to 100% enzyme activity (no compound) and 0% enzyme activity (no enzyme) before being fit to the function 100/(1+([compound]/IC50)).
- Biological Assay IRAK4 enzyme (Carna Biosciences, Chuo-ku, Kobe, Japan) activity was measured by detecting phosphorylated peptide substrate formation using an antibody against the phosphorylated peptide substrate. This is a time-resolved fluorescence resonance energy transfer (TR-FRET) immunoassay, based on the STK1 KinEASE Assay (Cisbio, Bedford, Mass.). The assay was designed as a simple two-step, endpoint assay (a 5 μl enzyme reaction followed by 5 μl stop and detect Solution) performed in ProxiPlate-384 Plus plates (Perkin Elmer, Waltham, Mass.). Staurosporine, a non-selective kinase inhibitor was used as a positive control. Compounds diluted in DMSO were spotted into 384 well plates using a Labcyte Echo 550 Liquid Handling System prior to addition of IRAK4 enzyme and peptide substrate. Reaction solutions were delivered using a Multi-Flo (Bio-Tek Instruments). The enzyme and peptide solution was incubated with compound for 15 minutes at room temp before the reaction was initiated by the addition of ATP. The standard 5 μl reaction mixture contained 500 μM ATP, 2 μM peptide (STK1 Peptide), 0.75 nM of IRAK4 in reaction buffer (50 mM HEPES, pH 7.0, 0.02% NaN3, 0.01% BSA, 0.1 mM Orthovanadate, 5 mM MgCl2, 0.025% NP-40, 1 mM DTT). After 120 min of incubation at room temperature, 5 μl of Stop and Detect Solution (1:100 Cryptate labeled anti-phosphorylated peptide antibody solution and 125 nM Tracer in a 50 mM HEPES pH 7.0 detection buffer containing sufficient EDTA) was added. The plate was then further incubated for 60 minutes at room temperature and read on Envision 2103 Multilabeled reader (PerkinElmer) with excitation/emission/FRET emission at 340 nm/615 nm/665 nm, respectively. Fluorescence intensities at 615 nm and 665 nm emission wavelengths were expressed as a ratio (665 nm/615 nm).
- IRAK4 Biochemical Assay IRAK4 enzyme (Carna Biosciences, Chuo-ku, Kobe, Japan) activity was measured by detecting phosphorylated peptide substrate formation using an antibody against the phosphorylated peptide substrate. This is a time-resolved fluorescence resonance energy transfer (TR-FRET) immunoassay, based on the STK1 KinEASE Assay (Cisbio, Bedford, Mass.). The assay was designed as a simple two-step, endpoint assay (a 5 μl enzyme reaction followed by 5 μl stop and detect Solution) performed in ProxiPlate-384 Plus plates (Perkin Elmer, Waltham, Mass.). Staurosporine, a non-selective kinase inhibitor was used as a positive control. Compounds diluted in DMSO were spotted into 384 well plates using a Labcyte Echo 550 Liquid Handling System prior to addition of IRAK4 enzyme and peptide substrate. Reaction solutions were delivered using a Multi-Flo (Bio-Tek Instruments). The enzyme and peptide solution was incubated with compound for 15 minutes at room temp before the reaction was initiated by the addition of ATP. The standard 5 μl reaction mixture contained 500 μM ATP, 2 M peptide (STK1 Peptide), 0.75 nM of IRAK4 in reaction buffer (50 mM HEPES, pH 7.0, 0.02% NaN3, 0.01% BSA, 0.1 mM Orthovanadate, 5 mM MgC2, 0.025% NP-40, 1 mM DTT). After 120 min of incubation at room temperature, 5 μl of Stop and Detect Solution (1:100 Cryptate labeled anti-phosphorylated peptide antibody solution and 125 nM Tracer in a 50 mM HEPES pH 7.0 detection buffer containing sufficient EDTA) was added. The plate was then further incubated for 60 minutes at room temperature and read on Envision 2103 Multilabeled reader (PerkinElmer) with excitation/emission/FRET emission at 340 nm/615 nm665 nm, respectively. Fluorescence intensities at 615 nm and 665 nm emission wavelengths were expressed as aratio (665 nm/615 nm). Percentage of inhibition was calculated as below:% Inhibition=100×(Ratiosample−Ratio0% Inhibition)/(Ratio100% Inhibition−Ratio0% Inhibition)The 0% inhibition value comes from control wells lacking inhibitor. The 100% inhibition value comes from control wells containing a saturating amount of known inhibitor staurosporine.
- Method of Biotinylated Wild-Type STING Protein Into pRSF1b (Novagen) having altered multiple cloning site was inserted Escherichia coli BirA, and transfected to ECOS JM109, whereby pRH8/FLAG-BirA was constructed. pET21HH/His-Avi-SUMO-FLAG-hTMEM173(139-379XH232R) (which was constructed by the method mentioned in the Example 36) and pRH8/FLAG-BirA for Avi tag biotinylation were simultaneously transformed to ECO (trade name) Competent E. coli BL21(DE3) to prepare His-Avi-SUMO-FLAG-hSTING (139-379, H232R)-expressing cell line. The expressing cell line was added to LB medium (10 g/L Tryptone, 5 g/L Yeast Extract, 5 g/L NaCl) containing ampicillin (100 μg/L) and kanamycin (50 μg/L), and the mixture was pre-cultured at 30° C., and expanded to TB medium (12 g/L Tryptone, 24 g/L Yeast Extract, 4 mL/L Glycerol, 2.3 g/L KH2PO4, 12.5 g/L K2HPO4) containing the same antibiotics, and the mixture was cultured at 37° C. When the turbidity of the culture solution reached 500 KU, the culture temperature was reduced to 16° C., 0.1 mM isopropylthiogalactoside and 50 μM (+)-biotin were added thereto, and the mixture was cultured for additional 16 hr.The culture solution was centrifuged, the obtained fungus bodies were suspended in Lysis Buffer (50 mM TrisHCl, 150 mM NaCl, 20 mM Imidazole, 1 mg/mL Lysozyme, 5 U/mL SEM Nuclease, recombinant, Complete EDTA-free, pH7.6), and the protein was extracted by ultrasonic fragmentation. The reagent was added thereto so that the salt concentration of the extract was adjusted to 300 mM NaCl, and the supernatant was collected by centrifugation. The obtained supernatant was passed through NiNTA superflow Cartridge equilibrated with Wash Buffer (50 mM TrisHCl, 300 mM NaCl, 20 mM Imidazole, pH7.6), and the Cartridge was washed with Wash Buffer, and eluted with Elution Buffer (50 mM TrisHCl, 300 mM NaCl, 250 mM Imidazole, pH7.6).
- IRAK4 Biochemical Assay IRAK4 enzyme (Carna Biosciences, Chuo-ku, Kobe, Japan) activity was measured by detecting phosphorylated peptide substrate formation using an antibody against the phosphorylated peptide substrate. This is a time-resolved fluorescence resonance energy transfer (TR-FRET) immunoassay, based on the STK1 KinEASE Assay (Cisbio, Bedford, Mass.). The assay was designed as a simple two-step, endpoint assay (a 5 μl enzyme reaction followed by 5 μl stop and detect Solution) performed in ProxiPlate-384 Plus plates (Perkin Elmer, Waltham, Mass.). Staurosporine, a non-selective kinase inhibitor was used as a positive control. Compounds diluted in DMSO were spotted into 384 well plates using a Labcyte Echo 550 Liquid Handling System prior to addition of IRAK4 enzyme and peptide substrate. Reaction solutions were delivered using a Multi-Flo (Bio-Tek Instruments). The enzyme and peptide solution was incubated with compound for 15 minutes at room temp before the reaction was initiated by the addition of ATP. The standard 5 μl reaction mixture contained 500 μM ATP, 2 μM peptide (STK1 Peptide), 0.75 nM of IRAK4 in reaction buffer (50 mM HEPES, pH 7.0, 0.02% NaN3, 0.01% BSA, 0.1 mM Orthovanadate, 5 mM MgCl2, 0.025% NP-40, 1 mM DTT). After 120 min of incubation at room temperature, 5 μl of Stop and Detect Solution (1:100 Cryptate labeled anti-phosphorylated peptide antibody solution and 125 nM Tracer in a 50 mM HEPES pH 7.0 detection buffer containing sufficient EDTA) was added. The plate was then further incubated for 60 minutes at room temperature and read on Envision 2103 Multilabeled reader (PerkinElmer) with excitation/emission/FRET emission at 340 nm/615 nm/665 nm, respectively. Fluorescence intensities at 615 nm and 665 nm emission wavelengths were expressed as a ratio (665 nm/615 nm). Percentage of inhibition was calculated as below:% Inhibition=100×(RatioSample−Ratio0% Inhibition)/(Ratio100% Inhibition−Ratio0% Inhibition)The 0% inhibition value comes from control wells lacking inhibitor. The 100% inhibition value comes from control wells containing a saturating amount of known inhibitor staurosporine.
- Phosphodiesterase Assay The PDE assay is based on the homogenous time-resolved fluorescence resonance energy transfer (TR-FRET) technology (LANCE from Perkin Elmer). This competition based assay is formatted using a cAMP specific antibody labeled with the dye, Alexa Fluor 647, biotin-cAMP and streptavidin labeled with Europium (Eu-SA). As the complex of Eu-SA/biotin-cAMP/Alexa Fluor 647 labeled antibody is formed, an increase in signal is generated. When there is PDE activity, resulting in the degradation of the cyclic nucleotide, the complex is not formed and a decrease in signal is observed. The phosphodiesterase assay was developed using the LANCE cAMP kit (PerkinElmer). The assay buffer contained HBSS with 5 mM HEPES, 0.1% BSA, and 1.5 mM MgCl2, pH 7.4. PDE10A (BPS Bioscience) was used at 200 pg/well (with a specific activity of 3200 μmole/min/μg with assay conditions: 10 mM Tris-HCl, pH7.4, 10 mM MgCl2, 1 mM MnCl2, 200 μM cAMP, 2.5 kU 5′ nucleotidase, 37° C., 20 min). The Biotin-cAMP tracer, supplied in 10 mmol/L Tris-HCl buffered (pH 8.0) salt solution with 1 mmol/L ethylenediaminetetraacetic acid (EDTA), 0.1% bovine serum albumin (BSA), and 0.05% sodium azide, is used at a dilution of 1/375. The assay detection mixture contained the LANCE Eu-W8044 labeled streptavidin 1/2250 (supplied in 50 mmol/L Tris-HCl buffered (pH 7.8) salt solution with 0.9% sodium chloride (NaCl), 0.1% BSA, and 0.05% sodium azide) and the Alexa Fluor 647-anti cAMP antibody 1/200 (supplied in 50 mmol/L Tris-HCl buffered (pH 7.8) salt solution with 0.9% NaCl, 0.1% BSA, and 0.05% sodium azide). Chemical compounds were dissolved in DMSO (final concentration 2% (v/v)). In a 384-well plate, 2 μL inhibitor and 3 μL PDE were added to the well, followed by the addition of 5 μl substrate biotinylated cAMP (1:5). After 60 min incubation at room temperature, 10 μL of assay detection mixture was added to the assay plate. After 1 h at room temperature, the signal was measured on EnVision (Perkin Elmer).
- IRAK4 Monocyte TNFalpha Cell Based Assay Cryopreserved human monocytes (Stem Cell Technologies) were thawed, diluted in RPMI with GlutaMAX (Gibco 200 mM L-alanyl-L-glutamine) (10 mM HEPES, 1× Pen-Strep, 55 μM -mercaptoethanol, 1 mM Sodium pyruvate) media containing 10% FBS to 0.125×106 cells/ml and recovered at 37° C. for 2 hours. The cell suspension was then plated at a density of 5,000 cells/well onto black 384 well Greiner clear bottom plates. Plates were pre-spotted with test compounds and serially diluted in DMSO where 200 nL/well were delivered using the Echo 550 acoustic liquid dispenser (Labcyte ) for a final DMSO concentration of 0.5%. Plated cells were treated with compound for 1 hour at 37° C. Cells were then stimulated with 50 pg/ml of LPS (Sigma) excluding outside columns of plate used for unstimulated cell control wells. Cells were incubated for an additional 4 hours at 37° C. Cells were then spun out of the media and 5 μl of sample were taken and analyzed for total TNFα content using the TR-FRET Human TNFα detection system (CisBio). This system utilizes two labeled antibodies (cryptate and XL665) that bind to two different epitopes of the TNFα molecule and produce FRET signal proportional to the concentration of TNFα in the sample. Detection antibodies are mixed 50:50 and 5 μL were dispensed into each well. Plates were covered with clear seals and incubated at room temp overnight. The following morning plates were read using an Envision 2103 Multilabeled reader (PerkinElmer) with excitation/emission/FRET emission at 340 nm/615 nm/665 nm, respectively. Fluorescence intensities at 615 nm and 665 nm emission wavelengths were expressed as a ratio (665 nm/615 nm). Percent of control was calculated as follows:% Control=100×(RatioSample−Ratio0% stimulation)/(Ratio100% Stimulation−Ratio0% Stimulation)where unstimulated cells (0% stimulation) were the negative control and stimulated cells (100% stimulation) were used as the positive control.IRAK4 Biochemical Assay Procedure:IRAK4 enzyme (Carna Biosciences, Chuo-ku, Kobe, Japan) activity was measured by detecting phosphorylated peptide substrate formation using an antibody against the phosphorylated peptide substrate. This is a time-resolved fluorescence resonance energy transfer (TR-FRET) immunoassay, based on the STK1 KinEASE Assay (Cisbio, Bedford, Mass.). The assay was designed as a simple two-step, endpoint assay (a 5 μl enzyme reaction followed by 5 μl stop and detect Solution) performed in ProxiPlate-384 Plus plates (Perkin Elmer, Waltham, Mass.). Staurosporine, a non-selective kinase inhibitor was used as a positive control. Compounds diluted in DMSO were spotted into 384 well plates using a Labcyte Echo 550 Liquid Handling System prior to addition of IRAK4 enzyme and peptide substrate. Reaction solutions were delivered using a Multi-Flo (Bio-Tek Instruments). The enzyme and peptide solution was incubated with compound for 15 minutes at room temp before the reaction was initiated by the addition of ATP. The standard 5 μl reaction mixture contained 500 □M ATP, 2 □M peptide (STK1 Peptide), 0.75 nM of IRAK4 in reaction buffer (50 mM HEPES, pH 7.0, 0.02% NaN3, 0.01% BSA, 0.1 mM Orthovanadate, 5 mM MgCl2, 0.025% NP-40, 1 mM DTT). After 120 min of incubation at room temperature, 5 □l of Stop and Detect Solution (1:100 Cryptate labeled anti-phosphorylated peptide antibody solution and 125 nM Tracer in a 50 mM HEPES pH 7.0 detection buffer containing sufficient EDTA) was added. The plate was then further incubated for 60 minutes at room temperature and read on Envision 2103 Multilabeled reader (PerkinElmer) with excitation/emission/FRET emission at 340 nm/615 nm/665 nm, respectively. Fluorescence intensities at 615 nm and 665 nm emission wavelengths were expressed as a ratio (665 nm/615 nm).
 KU-55933 BDBM50208517 2-morpholin-4-yl-6-thianthren-1-yl-pyran-4-one CHEMBL222102
KU-55933 BDBM50208517 2-morpholin-4-yl-6-thianthren-1-yl-pyran-4-one CHEMBL222102 BDBM50319926 KU-0060648 CHEMBL1086377 2-(4-ethylpiperazin-1-yl)-N-(4-(2-morpholino-4-oxo-4H-chromen-8-yl)dibenzo[b,d]thiophen-1-yl)acetamide
BDBM50319926 KU-0060648 CHEMBL1086377 2-(4-ethylpiperazin-1-yl)-N-(4-(2-morpholino-4-oxo-4H-chromen-8-yl)dibenzo[b,d]thiophen-1-yl)acetamide 1-[2-Fluoro-5-(4-oxo-3,4-dihydro-phthalazin-1-ylmethyl)-phenyl]-pyrrolidine-2,5-dione 1-(2-fluoro-5-((4-oxo-3,4-dihydrophthalazin-1-yl)methyl)phenyl)pyrrolidine-2,5-dione BDBM50165486 KU-58684 CHEMBL196444
1-[2-Fluoro-5-(4-oxo-3,4-dihydro-phthalazin-1-ylmethyl)-phenyl]-pyrrolidine-2,5-dione 1-(2-fluoro-5-((4-oxo-3,4-dihydrophthalazin-1-yl)methyl)phenyl)pyrrolidine-2,5-dione BDBM50165486 KU-58684 CHEMBL196444 4-{[3-(1,4-diazepan-1-ylcarbonyl)-4-fluorophenyl]methyl}-1,2-dihydrophthalazin-1-one 4-(3-(1,4-diazepane-1-carbonyl)-4-fluorobenzyl)phthalazin-1(2H)-one 4-[[3-(1,4-Diazepane-1-carbonyl)-4-fluorophenyl]methyl]-2Hphthalazin-1-one BDBM27533 CHEMBL380648 KU-0058948 4-[3-(1,4-diazepan-1-ylcarbonyl)-4-fluorobenzyl]phthalazin-1(2H)-one Homopiperazine analogue, 14
4-{[3-(1,4-diazepan-1-ylcarbonyl)-4-fluorophenyl]methyl}-1,2-dihydrophthalazin-1-one 4-(3-(1,4-diazepane-1-carbonyl)-4-fluorobenzyl)phthalazin-1(2H)-one 4-[[3-(1,4-Diazepane-1-carbonyl)-4-fluorophenyl]methyl]-2Hphthalazin-1-one BDBM27533 CHEMBL380648 KU-0058948 4-[3-(1,4-diazepan-1-ylcarbonyl)-4-fluorobenzyl]phthalazin-1(2H)-one Homopiperazine analogue, 14