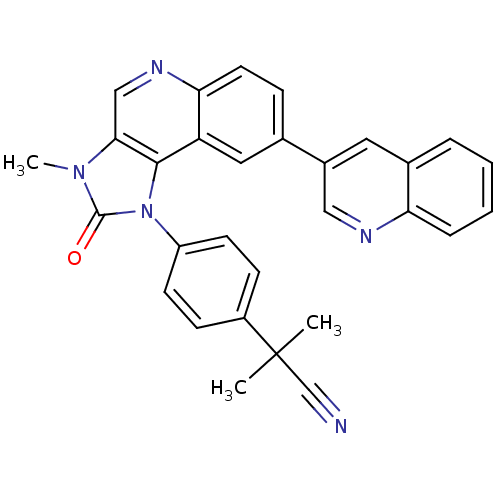
 BDBM92862 US9284315, BEZ-235 mTOR Inhibitor, BEZ235
BDBM92862 US9284315, BEZ-235 mTOR Inhibitor, BEZ235 NVP-DPP728 BDBM11594
NVP-DPP728 BDBM11594 Leq506 LEQ-506 BDBM50587737 Nvp-leq-506 Leq 506 NVP-LEQ506
Leq506 LEQ-506 BDBM50587737 Nvp-leq-506 Leq 506 NVP-LEQ506 BDBM142096 US8933056, NVP-LAF237 analogue
BDBM142096 US8933056, NVP-LAF237 analogue Fevipiprant BDBM50233520 QAW039 NVP-QAW039
Fevipiprant BDBM50233520 QAW039 NVP-QAW039 US8933056, NVP-LAF237 analogue Thioxamide BDBM142097
US8933056, NVP-LAF237 analogue Thioxamide BDBM142097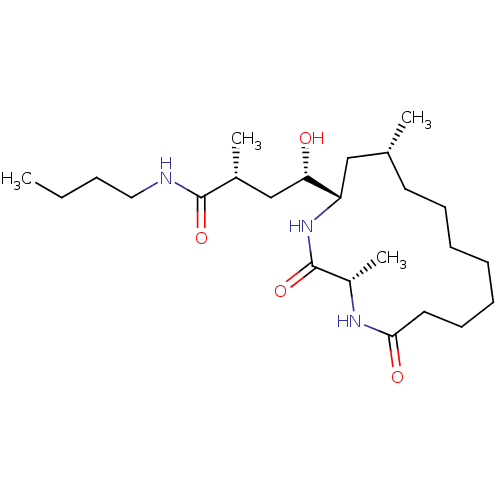 macrocyclic peptidomimetic, 3b NVP-ARV99 BDBM29748
macrocyclic peptidomimetic, 3b NVP-ARV99 BDBM29748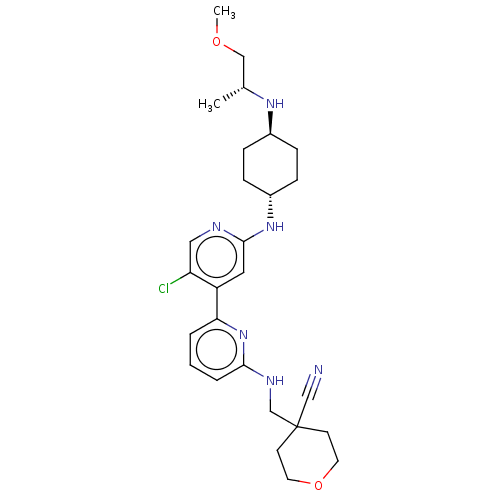 US8778951, 310 US11591322, Compound NVP-2 BDBM126500
US8778951, 310 US11591322, Compound NVP-2 BDBM126500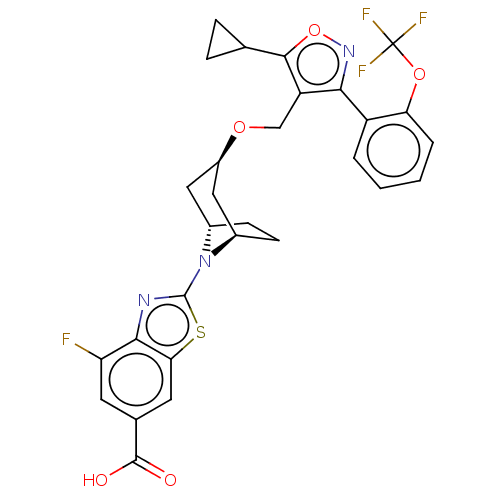 LJN-452 LJN452 Tropifexor NVP-LJN452-NXA BDBM50527040
LJN-452 LJN452 Tropifexor NVP-LJN452-NXA BDBM50527040 CGM 097 Cgm-097 Cgm097 NVP-CGM-097 BDBM50637898
CGM 097 Cgm-097 Cgm097 NVP-CGM-097 BDBM50637898 Jdq-443 Jdq 443 US11702409, Example 1b JDQ443 BDBM50579985 Nvp-jdq-443
Jdq-443 Jdq 443 US11702409, Example 1b JDQ443 BDBM50579985 Nvp-jdq-443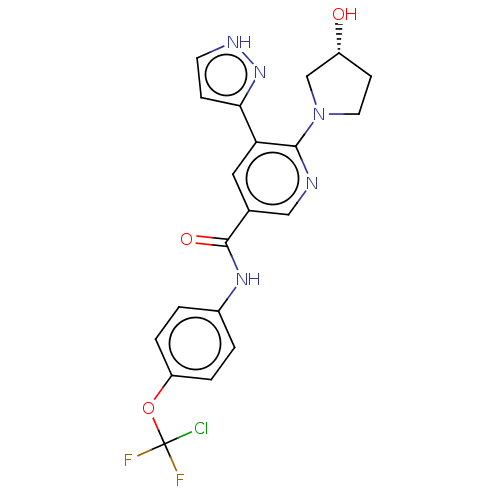 BDBM50459091 ABL-001 ABL001 ABL001-NX US11649218, Example ABL-001 Asciminib NVP-ABL001
BDBM50459091 ABL-001 ABL001 ABL001-NX US11649218, Example ABL-001 Asciminib NVP-ABL001 NVP-LAF237 CHEMBL142703 (2S)-1-{2-[(3-hydroxyadamantan-1-yl)amino]acetyl}pyrrolidine-2-carbonitrile BDBM11695
NVP-LAF237 CHEMBL142703 (2S)-1-{2-[(3-hydroxyadamantan-1-yl)amino]acetyl}pyrrolidine-2-carbonitrile BDBM11695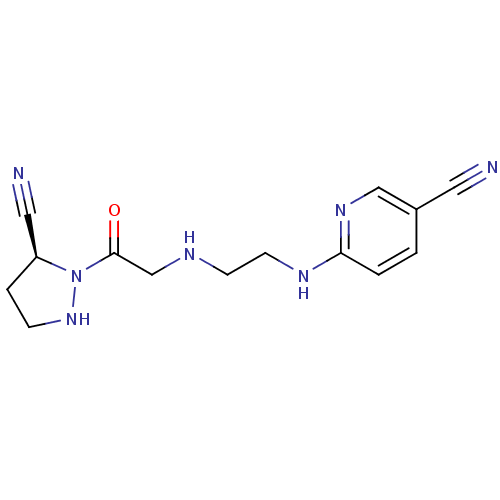 6-{2-[2-((S)-5-Cyano-pyrazolidin-1-yl)-2-oxo-ethylamino]-ethylamino}-nicotinonitrile BDBM50150870 NVP-728 CHEMBL184335
6-{2-[2-((S)-5-Cyano-pyrazolidin-1-yl)-2-oxo-ethylamino]-ethylamino}-nicotinonitrile BDBM50150870 NVP-728 CHEMBL184335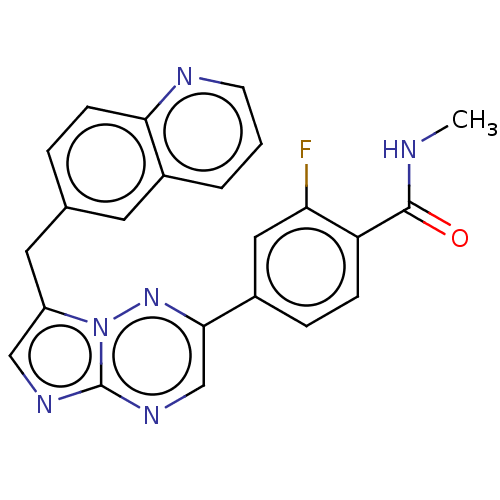 NVP-INC280 US11261191, Example 7 INCB-28060 US20240294535, Compound Capmatinib Capmatinib BDBM50146167 US10738052, Example 7 INC-280 US9944645, 7
NVP-INC280 US11261191, Example 7 INCB-28060 US20240294535, Compound Capmatinib Capmatinib BDBM50146167 US10738052, Example 7 INC-280 US9944645, 7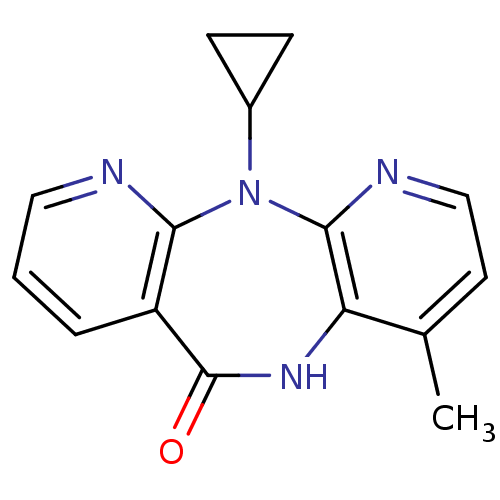 CHEMBL57 Nevirapine Viramune BDBM1434 2-cyclopropyl-7-methyl-2,4,9,15-tetraazatricyclo[9.4.0.0^{3,8}]pentadeca-1(11),3,5,7,12,14-hexaen-10-one Nevirapine (NVP) US11420959, Example NVP 11-cyclopropyl-5,11-dihydro-4-methyl-6H-dipyrido[3,2-b:2 ,3 -e][1,4]diazepin-6-one BI-RG-587
CHEMBL57 Nevirapine Viramune BDBM1434 2-cyclopropyl-7-methyl-2,4,9,15-tetraazatricyclo[9.4.0.0^{3,8}]pentadeca-1(11),3,5,7,12,14-hexaen-10-one Nevirapine (NVP) US11420959, Example NVP 11-cyclopropyl-5,11-dihydro-4-methyl-6H-dipyrido[3,2-b:2 ,3 -e][1,4]diazepin-6-one BI-RG-587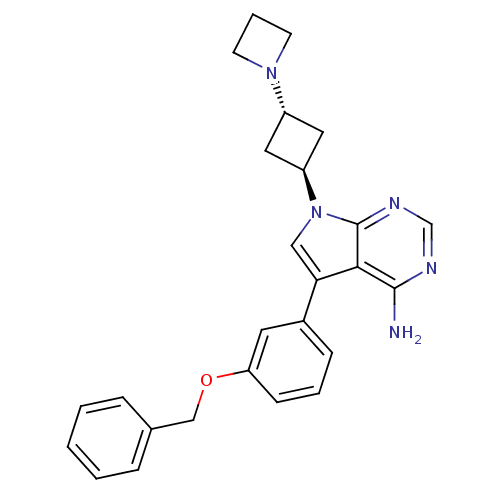 7-(3-(azetidin-1-yl)cyclobutyl)-5-(3-(benzyloxy)phenyl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine NVP-AEW541 BDBM50296348 CHEMBL551064
7-(3-(azetidin-1-yl)cyclobutyl)-5-(3-(benzyloxy)phenyl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine NVP-AEW541 BDBM50296348 CHEMBL551064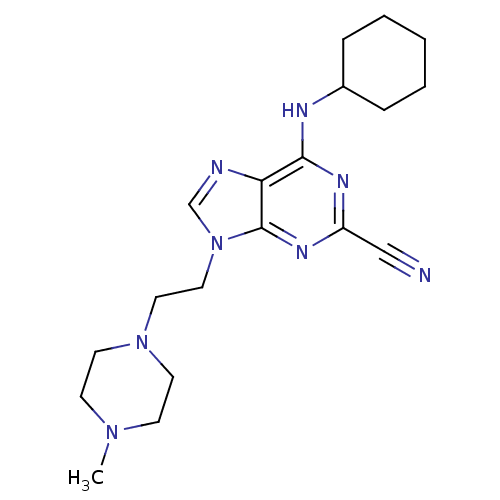 CHEMBL363847 BDBM19746 NVP-ABE854 Purine lead structure, 1 6-(cyclohexylamino)-9-[2-(4-methylpiperazin-1-yl)ethyl]-9H-purine-2-carbonitrile
CHEMBL363847 BDBM19746 NVP-ABE854 Purine lead structure, 1 6-(cyclohexylamino)-9-[2-(4-methylpiperazin-1-yl)ethyl]-9H-purine-2-carbonitrile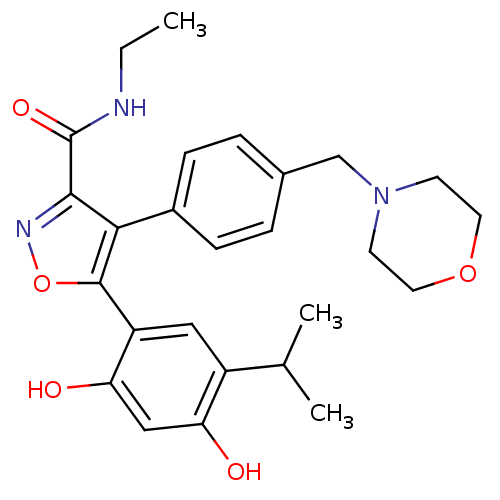 5-[2,4-dihydroxy-5-(propan-2-yl)phenyl]-N-ethyl-4-[4-(morpholin-4-ylmethyl)phenyl]-1,2-oxazole-3-carboxamide Isoxazole, 40f VER-52296/NVP-AUY922 BDBM20926
5-[2,4-dihydroxy-5-(propan-2-yl)phenyl]-N-ethyl-4-[4-(morpholin-4-ylmethyl)phenyl]-1,2-oxazole-3-carboxamide Isoxazole, 40f VER-52296/NVP-AUY922 BDBM20926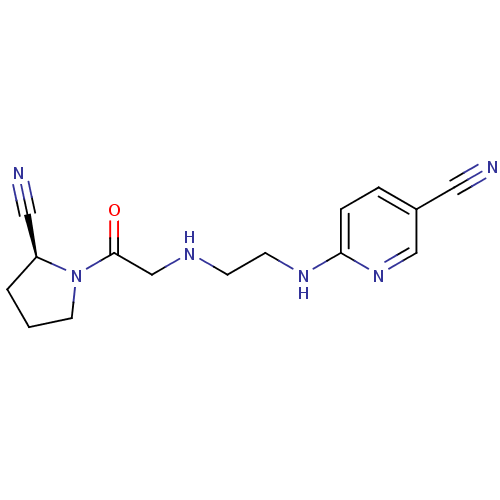 6-{[2-({2-[(2S)-2-cyanopyrrolidin-1-yl]-2-oxoethyl}amino)ethyl]amino}pyridine-3-carbonitrile NVP-DPP728 BDBM11113 CHEMBL77538 cyanopyrrolidine derivative 2 BMCL15687 Compound 2 cyano-(S)-pyrrolidine deriv. 1
6-{[2-({2-[(2S)-2-cyanopyrrolidin-1-yl]-2-oxoethyl}amino)ethyl]amino}pyridine-3-carbonitrile NVP-DPP728 BDBM11113 CHEMBL77538 cyanopyrrolidine derivative 2 BMCL15687 Compound 2 cyano-(S)-pyrrolidine deriv. 1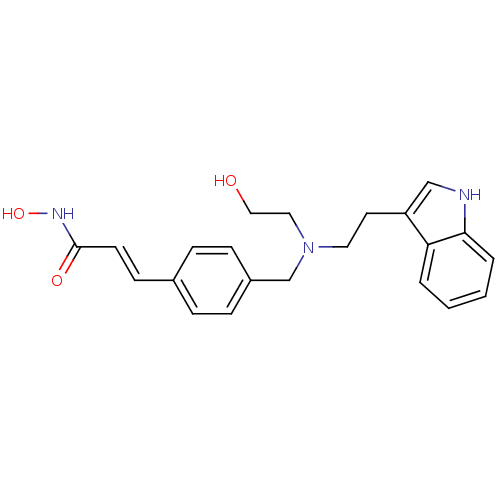 CHEMBL356066 Dacinostat (2E)-N-hydroxy-3-(4-{[(2-hydroxyethyl)[2-(1H-indol-3-yl)ethyl]amino]methyl}phenyl)prop-2-enamide LAQ-824 BDBM19428 (E)-N-hydroxy-3-[4-[[2-hydroxyethyl-[2-(1H-indol-3-yl)ethyl]amino]methyl]phenyl]prop-2-enamide NVP-LAQ824
CHEMBL356066 Dacinostat (2E)-N-hydroxy-3-(4-{[(2-hydroxyethyl)[2-(1H-indol-3-yl)ethyl]amino]methyl}phenyl)prop-2-enamide LAQ-824 BDBM19428 (E)-N-hydroxy-3-[4-[[2-hydroxyethyl-[2-(1H-indol-3-yl)ethyl]amino]methyl]phenyl]prop-2-enamide NVP-LAQ824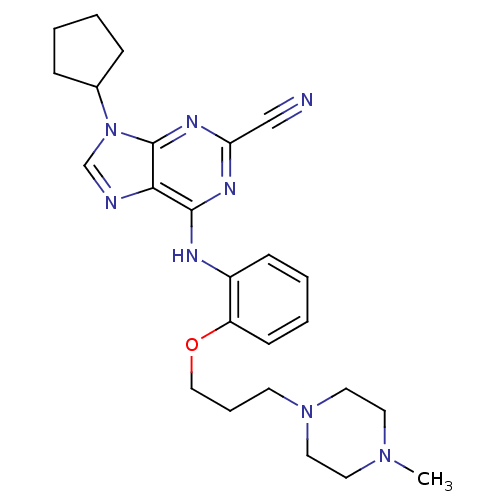 NVP-ABJ688 CHEMBL188139 9-CYCLOPENTYL-6-{2-[3-(4-METHYL-PIPERAZIN-1-YL)-PROPOXY]-PHENYLAMINO}-9H-PURINE-2-CARBONITRILE 9-cyclopentyl-6-{2-[3-(4-methylpiperazin-1-yl)propoxy]phenylamino}-9H-purine-2-carbonitrile 9-cyclopentyl-6-({2-[3-(4-methylpiperazin-1-yl)propoxy]phenyl}amino)-9H-purine-2-carbonitrile BDBM50156087
NVP-ABJ688 CHEMBL188139 9-CYCLOPENTYL-6-{2-[3-(4-METHYL-PIPERAZIN-1-YL)-PROPOXY]-PHENYLAMINO}-9H-PURINE-2-CARBONITRILE 9-cyclopentyl-6-{2-[3-(4-methylpiperazin-1-yl)propoxy]phenylamino}-9H-purine-2-carbonitrile 9-cyclopentyl-6-({2-[3-(4-methylpiperazin-1-yl)propoxy]phenyl}amino)-9H-purine-2-carbonitrile BDBM50156087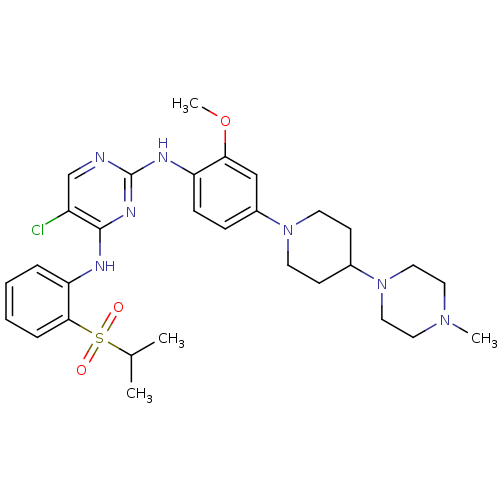 CHEMBL509032 NVP-TAE684 TAE684 US10647694, Example 36 TAE-684 5-chloro-N4-(2-(isopropylsulfonyl)phenyl)-N2-(2-methoxy-4-(4-(4-methylpiperazin-1-yl)piperidin-1-yl)phenyl)pyrimidine-2,4-diamine 5-CHLORO-N-[2-METHOXY-4-[4-(4-METHYLPIPERAZIN-1-YL)PIPERIDIN-1-YL]PHENYL]-N'-(2-PROPAN-2-YLSULFONYLPHENYL)PYRIMIDINE-2,4-DIAMINE BDBM50242742
CHEMBL509032 NVP-TAE684 TAE684 US10647694, Example 36 TAE-684 5-chloro-N4-(2-(isopropylsulfonyl)phenyl)-N2-(2-methoxy-4-(4-(4-methylpiperazin-1-yl)piperidin-1-yl)phenyl)pyrimidine-2,4-diamine 5-CHLORO-N-[2-METHOXY-4-[4-(4-METHYLPIPERAZIN-1-YL)PIPERIDIN-1-YL]PHENYL]-N'-(2-PROPAN-2-YLSULFONYLPHENYL)PYRIMIDINE-2,4-DIAMINE BDBM50242742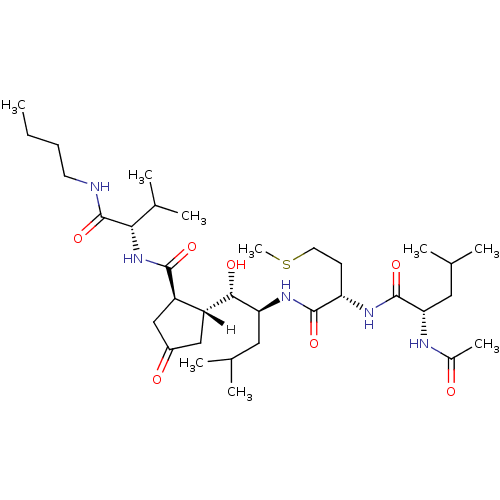 BDBM16057 N-acetyl-L-leucyl-N-{(1S)-1-[(S)-[(1R,2R)-2-{[(1S)-1-(butylcarbamoyl)-2-methylpropyl]carbamoyl}-4-oxocyclopentyl](hydroxy)methyl]-3-methylbutyl}-L-methioninamide (2S)-N-[(1S)-1-{[(1S,2S)-1-[(1R,2R)-2-{[(1S)-1-(butylcarbamoyl)-2-methylpropyl]carbamoyl}-4-oxocyclopentyl]-1-hydroxy-4-methylpentan-2-yl]carbamoyl}-3-(methylsulfanyl)propyl]-2-acetamido-4-methylpentanamide NVP-AUR200 Val-NHBu deriv. 45
BDBM16057 N-acetyl-L-leucyl-N-{(1S)-1-[(S)-[(1R,2R)-2-{[(1S)-1-(butylcarbamoyl)-2-methylpropyl]carbamoyl}-4-oxocyclopentyl](hydroxy)methyl]-3-methylbutyl}-L-methioninamide (2S)-N-[(1S)-1-{[(1S,2S)-1-[(1R,2R)-2-{[(1S)-1-(butylcarbamoyl)-2-methylpropyl]carbamoyl}-4-oxocyclopentyl]-1-hydroxy-4-methylpentan-2-yl]carbamoyl}-3-(methylsulfanyl)propyl]-2-acetamido-4-methylpentanamide NVP-AUR200 Val-NHBu deriv. 45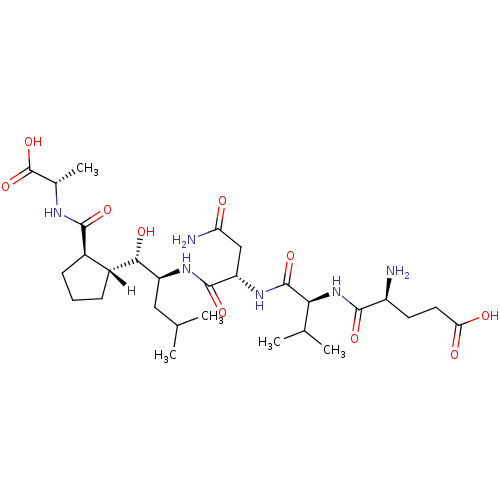 truncated OM99-2 cyclopentano analog 5 NVP-AMK640 (4S)-4-amino-4-{[(1S)-1-{[(1S)-2-carbamoyl-1-{[(1S,2S)-1-[(1R,2R)-2-{[(1S)-1-carboxyethyl]carbamoyl}cyclopentyl]-1-hydroxy-4-methylpentan-2-yl]carbamoyl}ethyl]carbamoyl}-2-methylpropyl]carbamoyl}butanoic acid L-alpha-glutamyl-L-valyl-N1-{(1S)-1-[(S)-[(1R,2R)-2-{[(1S)-1-carboxyethyl]carbamoyl}cyclopentyl](hydroxy)methyl]-3-methylbutyl}-L-aspartamide BDBM16051
truncated OM99-2 cyclopentano analog 5 NVP-AMK640 (4S)-4-amino-4-{[(1S)-1-{[(1S)-2-carbamoyl-1-{[(1S,2S)-1-[(1R,2R)-2-{[(1S)-1-carboxyethyl]carbamoyl}cyclopentyl]-1-hydroxy-4-methylpentan-2-yl]carbamoyl}ethyl]carbamoyl}-2-methylpropyl]carbamoyl}butanoic acid L-alpha-glutamyl-L-valyl-N1-{(1S)-1-[(S)-[(1R,2R)-2-{[(1S)-1-carboxyethyl]carbamoyl}cyclopentyl](hydroxy)methyl]-3-methylbutyl}-L-aspartamide BDBM16051
- Seixas, JD; Luengo-Arratta, SA; Diaz, R; Saldivia, M; Rojas-Barros, DI; Manzano, P; Gonzalez, S; Berlanga, M; Smith, TK; Navarro, M; Pollastri, MP Establishment of a structure-activity relationship of 1H-imidazo[4,5-c]quinoline-based kinase inhibitor NVP-BEZ235 as a lead for African sleeping sickness. J Med Chem 57: 4834-48 (2014)
- Sandham, DA; Barker, L; Brown, L; Brown, Z; Budd, D; Charlton, SJ; Chatterjee, D; Cox, B; Dubois, G; Duggan, N; Hall, E; Hatto, J; Maas, J; Manini, J; Profit, R; Riddy, D; Ritchie, C; Sohal, B; Shaw, D; Stringer, R; Sykes, DA; Thomas, M; Turner, KL; Watson, SJ; West, R; Willard, E; Williams, G; Willis, J Discovery of Fevipiprant (NVP-QAW039), a Potent and Selective DP ACS Med Chem Lett 8: 582-586 (2017)
- Galkin, AV; Melnick, JS; Kim, S; Hood, TL; Li, N; Li, L; Xia, G; Steensma, R; Chopiuk, G; Jiang, J; Wan, Y; Ding, P; Liu, Y; Sun, F; Schultz, PG; Gray, NS; Warmuth, M Identification of NVP-TAE684, a potent, selective, and efficacious inhibitor of NPM-ALK. Proc Natl Acad Sci U S A 104: 270-5 (2007)
- McBride, CM; Levine, B; Xia, Y; Bellamacina, C; Machajewski, T; Gao, Z; Renhowe, P; Antonios-McCrea, W; Barsanti, P; Brinner, K; Costales, A; Doughan, B; Lin, X; Louie, A; McKenna, M; Mendenhall, K; Poon, D; Rico, A; Wang, M; Williams, TE; Abrams, T; Fong, S; Hendrickson, T; Lei, D; Lin, J; Menezes, D; Pryer, N; Taverna, P; Xu, Y; Zhou, Y; Shafer, CM Design, structure-activity relationship, and in vivo characterization of the development candidate NVP-HSP990. J Med Chem 57: 9124-9 (2014)
- NVP-BHG712: Effects of Regioisomers on the Affinity and Selectivity toward the EPHrin Family.
- Discovery of Darovasertib (NVP-LXS196), a Pan-PKC Inhibitor for the Treatment of Metastatic Uveal Melanoma.
- Furet, P; Guagnano, V; Fairhurst, RA; Imbach-Weese, P; Bruce, I; Knapp, M; Fritsch, C; Blasco, F; Blanz, J; Aichholz, R; Hamon, J; Fabbro, D; Caravatti, G Discovery of NVP-BYL719 a potent and selective phosphatidylinositol-3 kinase alpha inhibitor selected for clinical evaluation. Bioorg Med Chem Lett 23: 3741-8 (2013)
- Burger, MT; Pecchi, S; Wagman, A; Ni, ZJ; Knapp, M; Hendrickson, T; Atallah, G; Pfister, K; Zhang, Y; Bartulis, S; Frazier, K; Ng, S; Smith, A; Verhagen, J; Haznedar, J; Huh, K; Iwanowicz, E; Xin, X; Menezes, D; Merritt, H; Lee, I; Wiesmann, M; Kaufman, S; Crawford, K; Chin, M; Bussiere, D; Shoemaker, K; Zaror, I; Maira, SM; Voliva, CF Identification of NVP-BKM120 as a Potent, Selective, Orally Bioavailable Class I PI3 Kinase Inhibitor for Treating Cancer. ACS Med Chem Lett 2: 774-779 (2011)
- Fairhurst, RA; Furet, P; Imbach-Weese, P; Stauffer, F; Rueeger, H; McCarthy, C; Ripoche, S; Oswald, S; Arnaud, B; Jary, A; Maira, M; Schnell, C; Guthy, DA; Wartmann, M; Kiffe, M; Desrayaud, S; Blasco, F; Widmer, T; Seiler, F; Gutmann, S; Knapp, M; Caravatti, G Identification of NVP-CLR457 as an Orally Bioavailable Non-CNS-Penetrant pan-Class IA Phosphoinositol-3-Kinase Inhibitor. J Med Chem 65: 8345-8379 (2022)
- In vivo antitumor activity of NVP-AEW541-A novel, potent, and selective inhibitor of the IGF-IR kinase.
- Sandham, DA; Arnold, N; Aschauer, H; Bala, K; Barker, L; Brown, L; Brown, Z; Budd, D; Cox, B; Docx, C; Dubois, G; Duggan, N; England, K; Everatt, B; Furegati, M; Hall, E; Kalthoff, F; King, A; Leblanc, CJ; Manini, J; Meingassner, J; Profit, R; Schmidt, A; Simmons, J; Sohal, B; Stringer, R; Thomas, M; Turner, KL; Walker, C; Watson, SJ; Westwick, J; Willis, J; Williams, G; Wilson, C Discovery and characterization of NVP-QAV680, a potent and selective CRTh2 receptor antagonist suitable for clinical testing in allergic diseases. Bioorg Med Chem 21: 6582-91 (2013)
- Shultz, MD; Cheung, AK; Kirby, CA; Firestone, B; Fan, J; Chen, CH; Chen, Z; Chin, DN; Dipietro, L; Fazal, A; Feng, Y; Fortin, PD; Gould, T; Lagu, B; Lei, H; Lenoir, F; Majumdar, D; Ochala, E; Palermo, MG; Pham, L; Pu, M; Smith, T; Stams, T; Tomlinson, RC; Touré, BB; Visser, M; Wang, RM; Waters, NJ; Shao, W Identification of NVP-TNKS656: the use of structure-efficiency relationships to generate a highly potent, selective, and orally active tankyrase inhibitor. J Med Chem 56: 6495-511 (2013)
- Holzer, P; Masuya, K; Furet, P; Kallen, J; Valat-Stachyra, T; Ferretti, S; Berghausen, J; Bouisset-Leonard, M; Buschmann, N; Pissot-Soldermann, C; Rynn, C; Ruetz, S; Stutz, S; Chène, P; Jeay, S; Gessier, F Discovery of a Dihydroisoquinolinone Derivative (NVP-CGM097): A Highly Potent and Selective MDM2 Inhibitor Undergoing Phase 1 Clinical Trials in p53wt Tumors. J Med Chem 58: 6348-58 (2015)
- Guagnano, V; Furet, P; Spanka, C; Bordas, V; Le Douget, M; Stamm, C; Brueggen, J; Jensen, MR; Schnell, C; Schmid, H; Wartmann, M; Berghausen, J; Drueckes, P; Zimmerlin, A; Bussiere, D; Murray, J; Graus Porta, D Discovery of 3-(2,6-dichloro-3,5-dimethoxy-phenyl)-1-{6-[4-(4-ethyl-piperazin-1-yl)-phenylamino]-pyrimidin-4-yl}-1-methyl-urea (NVP-BGJ398), a potent and selective inhibitor of the fibroblast growth factor receptor family of receptor tyrosine kinase. J Med Chem 54: 7066-83 (2011)
- Remiszewski, SW; Sambucetti, LC; Bair, KW; Bontempo, J; Cesarz, D; Chandramouli, N; Chen, R; Cheung, M; Cornell-Kennon, S; Dean, K; Diamantidis, G; France, D; Green, MA; Howell, KL; Kashi, R; Kwon, P; Lassota, P; Martin, MS; Mou, Y; Perez, LB; Sharma, S; Smith, T; Sorensen, E; Taplin, F; Trogani, N; Versace, R; Walker, H; Weltchek-Engler, S; Wood, A; Wu, A; Atadja, P N-hydroxy-3-phenyl-2-propenamides as novel inhibitors of human histone deacetylase with in vivo antitumor activity: discovery of (2E)-N-hydroxy-3-[4-[[(2-hydroxyethyl)[2-(1H-indol-3-yl)ethyl]amino]methyl]phenyl]-2-propenamide (NVP-LAQ824). J Med Chem 46: 4609-24 (2003)
- ChEMBL_1650982 (CHEMBL4000116) Displacement of [3H]NVP-QAW039 from human DP2 receptor expressed in CHO cell membranes by TopCount scintillation assay
- HIV-1 RT Inhibitory Assay The NNRTI compound to be evaluated (NVP) was serially diluted in 50% DMSO. The reaction mixtures containing 150 nM labeled primer−template hybrid, 100 μM dATP, 0.25 or 6 mM free Mg2+ (adjusted according to the concentration of dATP), and 5% DMSO were preincubated at 37 °C for 3 min in a total volume of 8.5 μL of reaction buffer (see above). Twenty-five nM HIV-1 or K103N RT (at similar activities of primer extension) was preincubated with different dilutions of NVP (as noted in the figure legends) for 10 min at room temperature. Reaction was then initiated by adding the NVP/ RT mix to the primer−template hybrid. The final pH of the reactions was 7.7. After extension for 3 min, the reactions were stopped by adding 12.5 μL of 2× gel loading buffer and the samples were resolved in a 16% denaturing polyacrylamide−7 M urea gel.
- Fluorogenic Assay DPP-IV inhibitors were measured for their ability to inhibit DPP-IV mediated cleavage of Ala-Pro-7-amido-4-trifluoromethylcoumarin in a fluorogenic assay. The compounds were measured in triplicate at 5 to 7 concentrations in the range of 100 uM to 100 pM. IC50 values were calculated with a nonlinear best-fit regression model. All assays were calibrated with NVP-DPP728 as internal standard inhibitor.
- Anti-HIV-1 Activity Test C8166 cells infected with HIV-1 were used for determining the anti-HIV biological activity at the cellular level. The specific method was described below.Cytotoxicity experiment: The toxicity of the compounds on C8166 cells was determined by MTT method. In a 96-well cell culture plate, the compounds were subjected to 5-fold serial dilution and 100 μL of C8166 cell suspension (4×105/mL) was added into each well. Three replicate wells were set for each concentration. At the same time, a cell control group without drugs and drug control groups with Zidovudine (AZT) or Nevirapine (NVP) were set. The cells were incubated at 37° C. in a 5% CO2 incubator for three days, followed by the addition of MTT solution into each well, and then the cells were incubated at 37° C. for 4 hours. 15% SDS-50% DMF was added to each well and the cells were incubated at 37° C. in a 5% CO2 incubator overnight. After mixing evenly, the OD values were measured by BIO-TEK ELx800 ELISA instrument (determination wavelength: 570 nm; reference wavelength: 630 nm). The dose-response curve was graphed according to the experimental results, and the CC50 was calculated (the concentrations of the compounds required to produce toxicity on 50% cells).Syncytium inhibition experiment: 100 μL of C8166 cell suspension (4×105/mL) was inoculated into each well of a 96-well cell culture plate containing 5-fold serial dilutions of the compounds, followed by addition of HIV-1IIIB diluted supernatant (MOI=0.04). Three replicate wells were set for each serial concentration. At the same time, negative control wells of HIV-1IIIB infection without compounds and positive control wells with Zidovudine (AZT) or Nevirapine (NVP) were set. The cells were incubated at 37° C. in a 5% CO2 incubator for three days. The number of the syncytia was counted in five non-overlapping fields of view by using an inverted microscope (100×). The dose-response curves were graphed according to the experimental results, and the 50% effective concentrations of the compounds for inhibiting the virus (EC50, 50% effective concentration) were calculated according to Reed & Muench method. Calculation formula: cytopathic inhibition rate (%)=(1−number of syncytia in experimental wells/number of syncytia in control well)×100%.
- Biochemical Assays for PI3Kalpha, PI3Kbeta The luminescence-based ATP detection reagent KinaseGlo was obtained from Promega, (Cat. No. V6714, Lot No. 236161) through Catalys, Wallisellen, Switzerland. (L-alpha-phosphatidylinositol (PI), Liver, Bovine) were obtained from Avanti Polar Lipid (Cat. No. 840042C, Lot#LPI-274), Phosphatidylinositol-4,5-bisphosphate (PIP(4,5)2(Avanti, Cat. No. 840046X) or L-α-phosphatidylinositol (PI) was obtained from Avanti Polar Lipid (Cat. No. 840042C, Lot#LPI-274). L-α-Phosphatidylserine (PS) was from Avanti Polar Lipid (Cat. No. 840032C), n-Octylglucoside Avanti Polar Lipid (Cat. No. 10634425001). Luminescence is a well established readout to determine ATP concentrations and can thus be used to follow the activity of many kinases regardless of their substrate. The Kinase Glo Luminescent Kinase Assay (Promega, Madison/WI, USA) is a homogeneous HTS method of measuring kinase activity by quantifying the amount of ATP remaining in solution following a kinase reaction.50 nL of compound dilutions were dispensed onto black 384-well low volume Non Binding Styrene (NBS) plates (Costar Cat. No. NBS#3676) as described in section 8.2. L-α-phosphatidylinositol (PI), provided as 10 mg/ml solution in methanol, was transferred into a glass tube and dried under nitrogen beam. It was then resuspended in 3% OctylGlucoside by vortexing and stored at 4° C. 5 μL of a mix of PI/OG with the PI3Ka and Pi3Kb subtypes were added. Kinase reactions were started by addition of 5 μl of ATP-mix containing in a final volume 10 μL 10 mM TRIS-HCl pH 7.5, 3 mM MgCl2, 50 mM NaCl, 0.05% CHAPS, 1 mM DTT and 1 μM ATP, and occurred at room temperature. Reactions were stopped with 10 μl of KinaseGlo and plates were read 10 mins later in a Synergy2 reader using an integration time of 0.1 seconds per well. 2.5 μM of NVP-BGT226 (standard) was added to the assay plates to generate the 100% inhibition of the kinase reaction, and the 0% inhibition was given by the solvent vehicle (90% DMSO in water). NVP-BGT226 was used as a reference compound and included in all assay plates in the form of 16 dilution points in duplicate.IC50 values of the percentage inhibition of each compound at 8 concentrations (usually 10, 3.0, 1.0, 0.3, 0.1, 0.030, 0.010 and 0.003 μM) n=2 were derived by fitting a sigmoidal dose-response curve to a plot of assay readout over inhibitor concentration as described. All fits were performed with the program XLfit4 (ID Business Solutions, Guildford, UK).
- Kinase Glo Luminescent Kinase Assay (Kglo) for PI 3-Kinase Alpha (A), PI 3-Kinase Beta (B), Vps34 (C), PI 4-Kinase Beta (D) The luminescence-based ATP detection reagent KinaseGlo was obtained from Promega, (Cat. No. V6714, Lot No. 236161) through Catalys, Wallisellen, Switzerland. L-alpha-phosphatidylinositol (PI, liver, bovine) was obtained from Avanti Polar Lipid (Cat. No. 840042C, Lot#LPI-274), phosphatidylinositol-4,5-bisphosphate (PIP(4,5)2) was obtained from Avanti Polar Lipid (Cat. No. 840046X). L-α-phosphatidylserine (PS) was obtained from Avanti Polar Lipid (Cat. No. 840032C), n-octylglucoside from Avanti Polar Lipid (Cat. No. 10634425001). Luminescence is a well established readout to determine ATP concentrations and can thus be used to follow the activity of many kinases regardless of their substrate. The Kinase Glo Luminescent Kinase Assay (Promega, Madison, Wis., USA) is a homogeneous HTS method of measuring kinase activity by quantifying the amount of ATP remaining in solution following a kinase reaction.50 nL of compound dilutions were dispensed onto black 384-well low volume Non Binding Styrene (NBS) plates (Costar Cat. No. NBS#3676). L-α-phosphatidylinositol (PI), provided as 10 mg/ml solution in methanol, was transferred into a glass tube and dried under a nitrogen beam. It was then resuspended in 3% OctylGlucoside (1-O-n-octyl-beta-D-glucopyranoside) by vortexing and stored at 4° C. 5 μl of a mix of PI/OctylGlucoside with the PI 3-kinase alpha and PI 3-kinase beta subtypes, or Vps34 or PI 4-kinase beta were added. Kinase reactions were started by the addition of 5 μl of an ATP-mix containing in a final volume 10 μl 10 mM TRIS-HCl pH 7.5, 3 mM MgCl2, 50 mM NaCl, 0.05% CHAPS, 1 mM DTT and 1 μM ATP at room temperature. Reactions were stopped with 10 μl of KinaseGlo and plates were read 10 mins later in a Synergy2 reader using an integration time of 0.1 seconds per well. 2.5 μM of NVP-BGT226 (1-(3-(trifluoromethyl)-4-(piperazin-1-yl)phenyl)-8-(6-methoxypyridin-3-yl)-3-methyl-1H-imidazo[4,5-c]quinolin-2(3H)-one) was added to the assay plates to generate the 100% inhibition of the kinase reaction, and the 0% inhibition was given by the solvent vehicle (90% DMSO in water). (1-(3-(trifluoromethyl)-4-(piperazin-1-yl)phenyl)-8-(6-methoxypyridin-3-yl)-3-methyl-1H-imidazo[4,5-c]quinolin-2(3H)-one) was used as a reference compound and included in all assay plates in the form of 16 dilution points in duplicate.IC50 values of the percentage inhibition of each compound at 8 concentrations (10, 3.0, 1.0, 0.3, 0.1, 0.030, 0.010 and 0.003 μM) n=2 were derived by fitting a sigmoidal dose-response curve to a plot of assay readout over inhibitor concentration as described. All fits were performed with the program XLfit4 (ID Business Solutions, Guildford, UK).
 BDBM92862 US9284315, BEZ-235 mTOR Inhibitor, BEZ235
BDBM92862 US9284315, BEZ-235 mTOR Inhibitor, BEZ235 NVP-DPP728 BDBM11594
NVP-DPP728 BDBM11594 Leq506 LEQ-506 BDBM50587737 Nvp-leq-506 Leq 506 NVP-LEQ506
Leq506 LEQ-506 BDBM50587737 Nvp-leq-506 Leq 506 NVP-LEQ506 BDBM142096 US8933056, NVP-LAF237 analogue
BDBM142096 US8933056, NVP-LAF237 analogue Fevipiprant BDBM50233520 QAW039 NVP-QAW039
Fevipiprant BDBM50233520 QAW039 NVP-QAW039 US8933056, NVP-LAF237 analogue Thioxamide BDBM142097
US8933056, NVP-LAF237 analogue Thioxamide BDBM142097 macrocyclic peptidomimetic, 3b NVP-ARV99 BDBM29748
macrocyclic peptidomimetic, 3b NVP-ARV99 BDBM29748 US8778951, 310 US11591322, Compound NVP-2 BDBM126500
US8778951, 310 US11591322, Compound NVP-2 BDBM126500 LJN-452 LJN452 Tropifexor NVP-LJN452-NXA BDBM50527040
LJN-452 LJN452 Tropifexor NVP-LJN452-NXA BDBM50527040 CGM 097 Cgm-097 Cgm097 NVP-CGM-097 BDBM50637898
CGM 097 Cgm-097 Cgm097 NVP-CGM-097 BDBM50637898 Jdq-443 Jdq 443 US11702409, Example 1b JDQ443 BDBM50579985 Nvp-jdq-443
Jdq-443 Jdq 443 US11702409, Example 1b JDQ443 BDBM50579985 Nvp-jdq-443 BDBM50459091 ABL-001 ABL001 ABL001-NX US11649218, Example ABL-001 Asciminib NVP-ABL001
BDBM50459091 ABL-001 ABL001 ABL001-NX US11649218, Example ABL-001 Asciminib NVP-ABL001 NVP-LAF237 CHEMBL142703 (2S)-1-{2-[(3-hydroxyadamantan-1-yl)amino]acetyl}pyrrolidine-2-carbonitrile BDBM11695
NVP-LAF237 CHEMBL142703 (2S)-1-{2-[(3-hydroxyadamantan-1-yl)amino]acetyl}pyrrolidine-2-carbonitrile BDBM11695 6-{2-[2-((S)-5-Cyano-pyrazolidin-1-yl)-2-oxo-ethylamino]-ethylamino}-nicotinonitrile BDBM50150870 NVP-728 CHEMBL184335
6-{2-[2-((S)-5-Cyano-pyrazolidin-1-yl)-2-oxo-ethylamino]-ethylamino}-nicotinonitrile BDBM50150870 NVP-728 CHEMBL184335 NVP-INC280 US11261191, Example 7 INCB-28060 US20240294535, Compound Capmatinib Capmatinib BDBM50146167 US10738052, Example 7 INC-280 US9944645, 7
NVP-INC280 US11261191, Example 7 INCB-28060 US20240294535, Compound Capmatinib Capmatinib BDBM50146167 US10738052, Example 7 INC-280 US9944645, 7 CHEMBL57 Nevirapine Viramune BDBM1434 2-cyclopropyl-7-methyl-2,4,9,15-tetraazatricyclo[9.4.0.0^{3,8}]pentadeca-1(11),3,5,7,12,14-hexaen-10-one Nevirapine (NVP) US11420959, Example NVP 11-cyclopropyl-5,11-dihydro-4-methyl-6H-dipyrido[3,2-b:2 ,3 -e][1,4]diazepin-6-one BI-RG-587
CHEMBL57 Nevirapine Viramune BDBM1434 2-cyclopropyl-7-methyl-2,4,9,15-tetraazatricyclo[9.4.0.0^{3,8}]pentadeca-1(11),3,5,7,12,14-hexaen-10-one Nevirapine (NVP) US11420959, Example NVP 11-cyclopropyl-5,11-dihydro-4-methyl-6H-dipyrido[3,2-b:2 ,3 -e][1,4]diazepin-6-one BI-RG-587 7-(3-(azetidin-1-yl)cyclobutyl)-5-(3-(benzyloxy)phenyl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine NVP-AEW541 BDBM50296348 CHEMBL551064
7-(3-(azetidin-1-yl)cyclobutyl)-5-(3-(benzyloxy)phenyl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine NVP-AEW541 BDBM50296348 CHEMBL551064 CHEMBL363847 BDBM19746 NVP-ABE854 Purine lead structure, 1 6-(cyclohexylamino)-9-[2-(4-methylpiperazin-1-yl)ethyl]-9H-purine-2-carbonitrile
CHEMBL363847 BDBM19746 NVP-ABE854 Purine lead structure, 1 6-(cyclohexylamino)-9-[2-(4-methylpiperazin-1-yl)ethyl]-9H-purine-2-carbonitrile 5-[2,4-dihydroxy-5-(propan-2-yl)phenyl]-N-ethyl-4-[4-(morpholin-4-ylmethyl)phenyl]-1,2-oxazole-3-carboxamide Isoxazole, 40f VER-52296/NVP-AUY922 BDBM20926
5-[2,4-dihydroxy-5-(propan-2-yl)phenyl]-N-ethyl-4-[4-(morpholin-4-ylmethyl)phenyl]-1,2-oxazole-3-carboxamide Isoxazole, 40f VER-52296/NVP-AUY922 BDBM20926 6-{[2-({2-[(2S)-2-cyanopyrrolidin-1-yl]-2-oxoethyl}amino)ethyl]amino}pyridine-3-carbonitrile NVP-DPP728 BDBM11113 CHEMBL77538 cyanopyrrolidine derivative 2 BMCL15687 Compound 2 cyano-(S)-pyrrolidine deriv. 1
6-{[2-({2-[(2S)-2-cyanopyrrolidin-1-yl]-2-oxoethyl}amino)ethyl]amino}pyridine-3-carbonitrile NVP-DPP728 BDBM11113 CHEMBL77538 cyanopyrrolidine derivative 2 BMCL15687 Compound 2 cyano-(S)-pyrrolidine deriv. 1 CHEMBL356066 Dacinostat (2E)-N-hydroxy-3-(4-{[(2-hydroxyethyl)[2-(1H-indol-3-yl)ethyl]amino]methyl}phenyl)prop-2-enamide LAQ-824 BDBM19428 (E)-N-hydroxy-3-[4-[[2-hydroxyethyl-[2-(1H-indol-3-yl)ethyl]amino]methyl]phenyl]prop-2-enamide NVP-LAQ824
CHEMBL356066 Dacinostat (2E)-N-hydroxy-3-(4-{[(2-hydroxyethyl)[2-(1H-indol-3-yl)ethyl]amino]methyl}phenyl)prop-2-enamide LAQ-824 BDBM19428 (E)-N-hydroxy-3-[4-[[2-hydroxyethyl-[2-(1H-indol-3-yl)ethyl]amino]methyl]phenyl]prop-2-enamide NVP-LAQ824 NVP-ABJ688 CHEMBL188139 9-CYCLOPENTYL-6-{2-[3-(4-METHYL-PIPERAZIN-1-YL)-PROPOXY]-PHENYLAMINO}-9H-PURINE-2-CARBONITRILE 9-cyclopentyl-6-{2-[3-(4-methylpiperazin-1-yl)propoxy]phenylamino}-9H-purine-2-carbonitrile 9-cyclopentyl-6-({2-[3-(4-methylpiperazin-1-yl)propoxy]phenyl}amino)-9H-purine-2-carbonitrile BDBM50156087
NVP-ABJ688 CHEMBL188139 9-CYCLOPENTYL-6-{2-[3-(4-METHYL-PIPERAZIN-1-YL)-PROPOXY]-PHENYLAMINO}-9H-PURINE-2-CARBONITRILE 9-cyclopentyl-6-{2-[3-(4-methylpiperazin-1-yl)propoxy]phenylamino}-9H-purine-2-carbonitrile 9-cyclopentyl-6-({2-[3-(4-methylpiperazin-1-yl)propoxy]phenyl}amino)-9H-purine-2-carbonitrile BDBM50156087 CHEMBL509032 NVP-TAE684 TAE684 US10647694, Example 36 TAE-684 5-chloro-N4-(2-(isopropylsulfonyl)phenyl)-N2-(2-methoxy-4-(4-(4-methylpiperazin-1-yl)piperidin-1-yl)phenyl)pyrimidine-2,4-diamine 5-CHLORO-N-[2-METHOXY-4-[4-(4-METHYLPIPERAZIN-1-YL)PIPERIDIN-1-YL]PHENYL]-N'-(2-PROPAN-2-YLSULFONYLPHENYL)PYRIMIDINE-2,4-DIAMINE BDBM50242742
CHEMBL509032 NVP-TAE684 TAE684 US10647694, Example 36 TAE-684 5-chloro-N4-(2-(isopropylsulfonyl)phenyl)-N2-(2-methoxy-4-(4-(4-methylpiperazin-1-yl)piperidin-1-yl)phenyl)pyrimidine-2,4-diamine 5-CHLORO-N-[2-METHOXY-4-[4-(4-METHYLPIPERAZIN-1-YL)PIPERIDIN-1-YL]PHENYL]-N'-(2-PROPAN-2-YLSULFONYLPHENYL)PYRIMIDINE-2,4-DIAMINE BDBM50242742 BDBM16057 N-acetyl-L-leucyl-N-{(1S)-1-[(S)-[(1R,2R)-2-{[(1S)-1-(butylcarbamoyl)-2-methylpropyl]carbamoyl}-4-oxocyclopentyl](hydroxy)methyl]-3-methylbutyl}-L-methioninamide (2S)-N-[(1S)-1-{[(1S,2S)-1-[(1R,2R)-2-{[(1S)-1-(butylcarbamoyl)-2-methylpropyl]carbamoyl}-4-oxocyclopentyl]-1-hydroxy-4-methylpentan-2-yl]carbamoyl}-3-(methylsulfanyl)propyl]-2-acetamido-4-methylpentanamide NVP-AUR200 Val-NHBu deriv. 45
BDBM16057 N-acetyl-L-leucyl-N-{(1S)-1-[(S)-[(1R,2R)-2-{[(1S)-1-(butylcarbamoyl)-2-methylpropyl]carbamoyl}-4-oxocyclopentyl](hydroxy)methyl]-3-methylbutyl}-L-methioninamide (2S)-N-[(1S)-1-{[(1S,2S)-1-[(1R,2R)-2-{[(1S)-1-(butylcarbamoyl)-2-methylpropyl]carbamoyl}-4-oxocyclopentyl]-1-hydroxy-4-methylpentan-2-yl]carbamoyl}-3-(methylsulfanyl)propyl]-2-acetamido-4-methylpentanamide NVP-AUR200 Val-NHBu deriv. 45 truncated OM99-2 cyclopentano analog 5 NVP-AMK640 (4S)-4-amino-4-{[(1S)-1-{[(1S)-2-carbamoyl-1-{[(1S,2S)-1-[(1R,2R)-2-{[(1S)-1-carboxyethyl]carbamoyl}cyclopentyl]-1-hydroxy-4-methylpentan-2-yl]carbamoyl}ethyl]carbamoyl}-2-methylpropyl]carbamoyl}butanoic acid L-alpha-glutamyl-L-valyl-N1-{(1S)-1-[(S)-[(1R,2R)-2-{[(1S)-1-carboxyethyl]carbamoyl}cyclopentyl](hydroxy)methyl]-3-methylbutyl}-L-aspartamide BDBM16051
truncated OM99-2 cyclopentano analog 5 NVP-AMK640 (4S)-4-amino-4-{[(1S)-1-{[(1S)-2-carbamoyl-1-{[(1S,2S)-1-[(1R,2R)-2-{[(1S)-1-carboxyethyl]carbamoyl}cyclopentyl]-1-hydroxy-4-methylpentan-2-yl]carbamoyl}ethyl]carbamoyl}-2-methylpropyl]carbamoyl}butanoic acid L-alpha-glutamyl-L-valyl-N1-{(1S)-1-[(S)-[(1R,2R)-2-{[(1S)-1-carboxyethyl]carbamoyl}cyclopentyl](hydroxy)methyl]-3-methylbutyl}-L-aspartamide BDBM16051