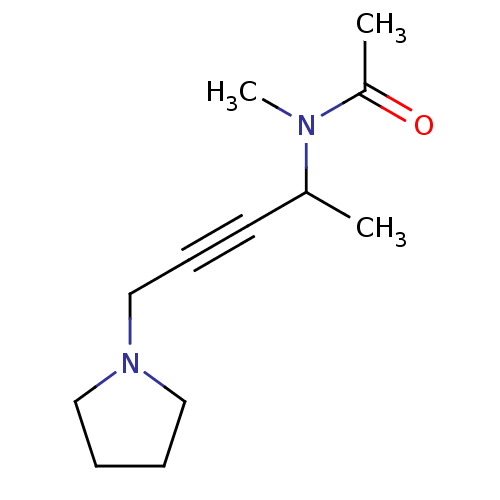
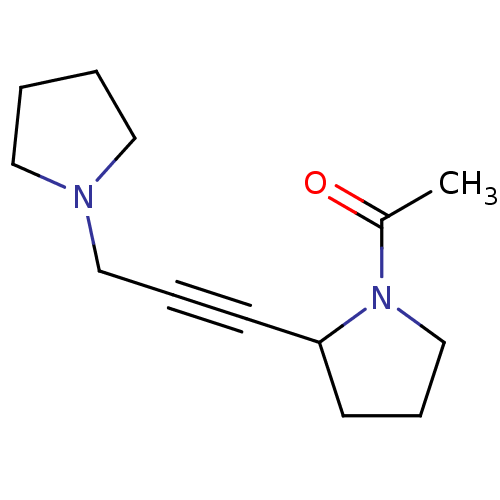
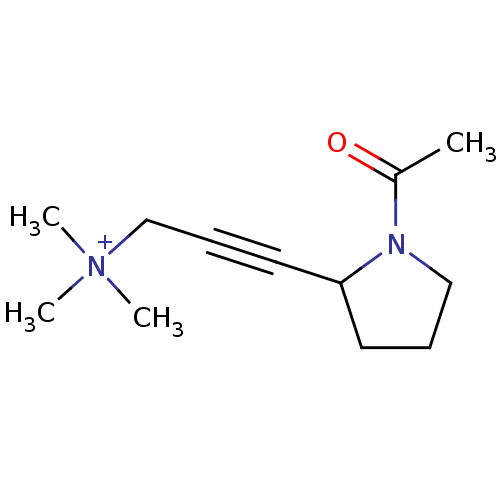
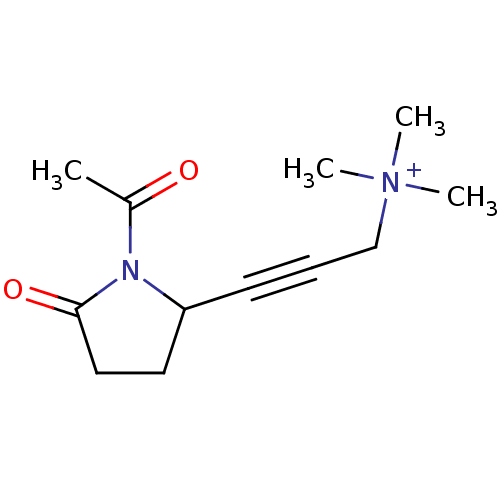
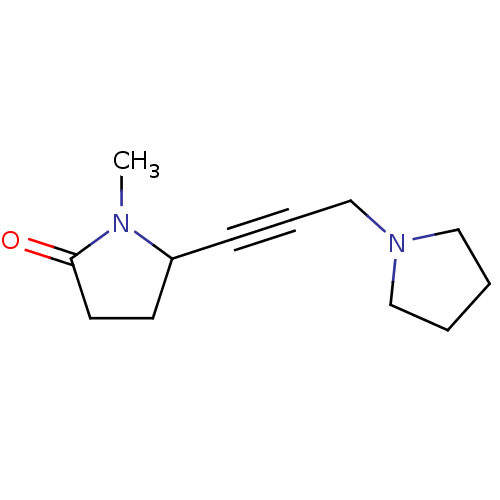
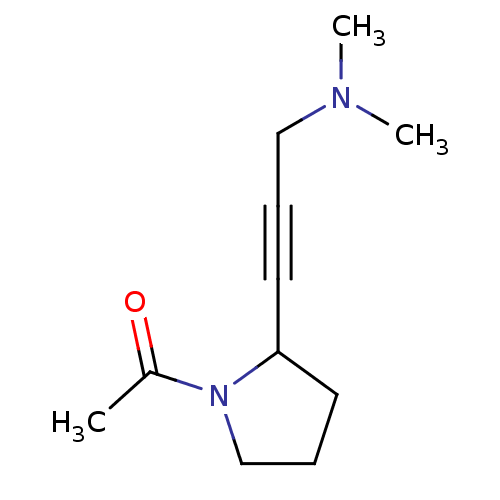
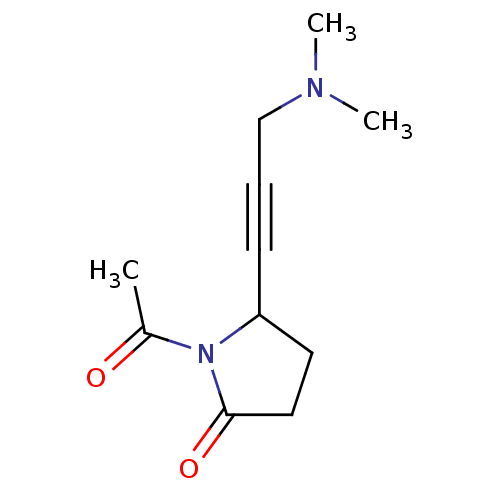
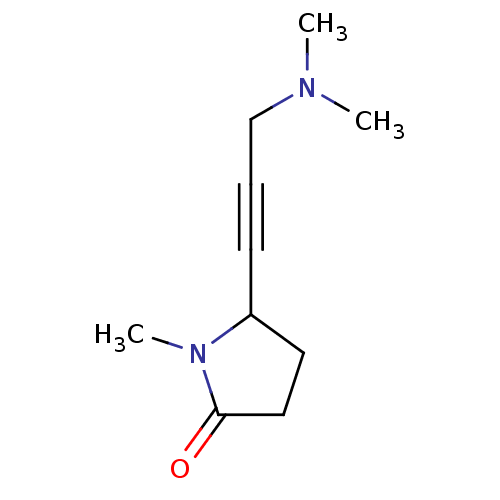
Affinity DataKi: 64nMAssay Description:Compound was tested for the concentration that inhibited the specific binding of (-)-[3H]-NMS to muscarinic receptor in the rat cerebral cortex.More data for this Ligand-Target Pair
Affinity DataKi: 670nMAssay Description:Compound was tested for the concentration that inhibited the specific binding of (-)-[3H]-NMS to muscarinic receptor in the rat cerebral cortex.More data for this Ligand-Target Pair
Affinity DataKi: 780nMAssay Description:Compound was tested for the concentration that inhibited the specific binding of (-)-[3H]-NMS to muscarinic acetylcholine receptor in the rat cerebra...More data for this Ligand-Target Pair
Affinity DataKi: 6.10E+3nMAssay Description:Compound was tested for the concentration that inhibited the specific binding of (-)-[3H]-NMS to muscarinic acetylcholine receptor in the rat cerebra...More data for this Ligand-Target Pair
Affinity DataKi: 6.40E+3nMAssay Description:Compound was tested for the concentration that inhibited the specific binding of (-)-[3H]-NMS to muscarinic acetylcholine receptor in the rat cerebra...More data for this Ligand-Target Pair
Affinity DataKi: 1.83E+4nMAssay Description:Compound was tested for the concentration that inhibited the specific binding of (-)-[3H]-NMS to muscarinic receptor in the rat cerebral cortex.More data for this Ligand-Target Pair
Affinity DataKi: 2.54E+4nMAssay Description:Compound was tested for the concentration that inhibited the specific binding of (-)-[3H]-NMS to muscarinic receptor in the rat cerebral cortexMore data for this Ligand-Target Pair
Affinity DataKi: 5.32E+4nMAssay Description:Compound was tested for the concentration that inhibited the specific binding of (-)-[3H]-NMS to muscarinic acetylcholine receptor in the rat cerebra...More data for this Ligand-Target Pair
Affinity DataKi: 1.45E+5nMAssay Description:Compound was tested for the concentration that inhibited the specific binding of (-)-[3H]-NMS to muscarinic acetylcholine receptor in the rat cerebra...More data for this Ligand-Target Pair
Affinity DataKi: 2.04E+5nMAssay Description:Compound was tested for the concentration that inhibited the specific binding of (-)-[3H]-NMS to muscarinic acetylcholine receptor in the rat cerebra...More data for this Ligand-Target Pair