Reaction Details Report a problem with these data
Report a problem with these data
 Report a problem with these data
Report a problem with these dataTarget
Mitogen-activated protein kinase 14
Ligand
BDBM14836
Substrate
BDBM14832
Meas. Tech.
Fluorescence Exchange Curve Assay
pH
7±n/a
Temperature
296.15±n/a K
Kd
21±n/a nM
Citation
 Kroe, RR; Regan, J; Proto, A; Peet, GW; Roy, T; Landro, LD; Fuschetto, NG; Pargellis, CA; Ingraham, RH Thermal denaturation: a method to rank slow binding, high-affinity P38alpha MAP kinase inhibitors. J Med Chem 46:4669-75 (2003) [PubMed] Article
Kroe, RR; Regan, J; Proto, A; Peet, GW; Roy, T; Landro, LD; Fuschetto, NG; Pargellis, CA; Ingraham, RH Thermal denaturation: a method to rank slow binding, high-affinity P38alpha MAP kinase inhibitors. J Med Chem 46:4669-75 (2003) [PubMed] Article More Info.:
Target
Name:
Mitogen-activated protein kinase 14
Synonyms:
CSAID-binding protein | CSBP | CSBP1 | CSBP2 | CSPB1 | Cytokine suppressive anti-inflammatory drug-binding protein | MAP kinase 14 | MAP kinase MXI2 | MAP kinase p38 alpha | MAPK 14 | MAPK14 | MAX-interacting protein 2 | MK14_HUMAN | MXI2 | Mitogen-activated protein kinase p38 alpha | SAPK2A | Stress-activated protein kinase 2a | p38 MAP kinase alpha/beta
Type:
Serine/threonine-protein kinase
Mol. Mass.:
41286.76
Organism:
Homo sapiens (Human)
Description:
Q16539
Residue:
360
Sequence:
MSQERPTFYRQELNKTIWEVPERYQNLSPVGSGAYGSVCAAFDTKTGLRVAVKKLSRPFQSIIHAKRTYRELRLLKHMKHENVIGLLDVFTPARSLEEFNDVYLVTHLMGADLNNIVKCQKLTDDHVQFLIYQILRGLKYIHSADIIHRDLKPSNLAVNEDCELKILDFGLARHTDDEMTGYVATRWYRAPEIMLNWMHYNQTVDIWSVGCIMAELLTGRTLFPGTDHIDQLKLILRLVGTPGAELLKKISSESARNYIQSLTQMPKMNFANVFIGANPLAVDLLEKMLVLDSDKRITAAQALAHAYFAQYHDPDDEPVADPYDQSFESRDLLIDEWKSLTYDEVISFVPPPLDQEEMES
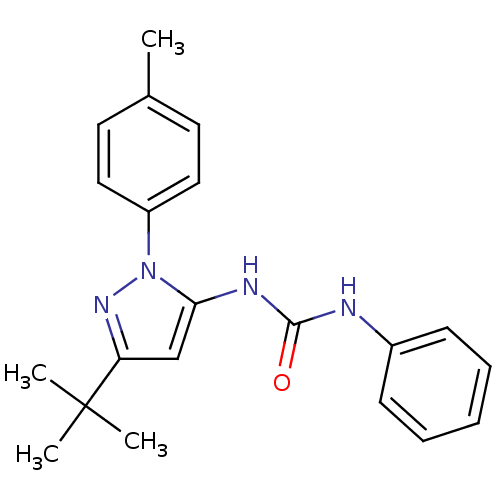
Inhibitor
Name:
BDBM14836
Synonyms:
3-[3-tert-butyl-1-(4-methylphenyl)-1H-pyrazol-5-yl]-1-phenylurea | CHEMBL315100 | Dopamine D2 receptor and serotonin 2a receptor | diaryl urea compound 5
Type:
Small organic molecule
Emp. Form.:
C21H24N4O
Mol. Mass.:
348.4415
SMILES:
Cc1ccc(cc1)-n1nc(cc1NC(=O)Nc1ccccc1)C(C)(C)C
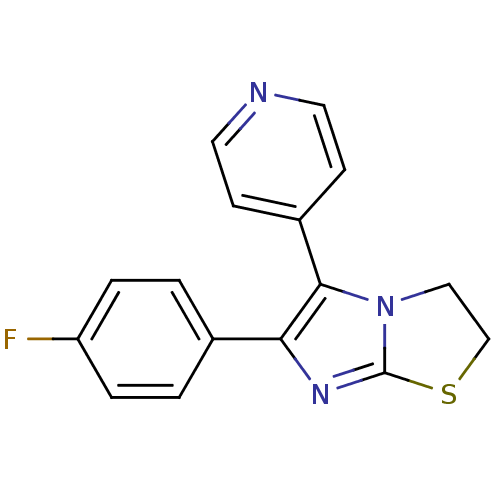
Substrate
Name:
BDBM14832
Synonyms:
3-(4-fluorophenyl)-2-pyridin-4-yl-6-thia-1,4-diazabicyclo[3.3.0]octa-2,4-diene | 4-[6-(4-fluorophenyl)-2H,3H-imidazo[2,1-b][1,3]thiazol-5-yl]pyridine | CHEMBL313417 | D3RKN_48 | SKF 86002
Type:
Small organic molecule
Emp. Form.:
C16H12FN3S
Mol. Mass.:
297.35
SMILES:
Fc1ccc(cc1)-c1nc2SCCn2c1-c1ccncc1