Reaction Details Report a problem with these data
Report a problem with these data
 Report a problem with these data
Report a problem with these dataTarget
Sodium- and chloride-dependent glycine transporter 1
Ligand
BDBM50460351
Substrate
n/a
Meas. Tech.
ChEMBL_1769377 (CHEMBL4221489)
IC50
7.0±n/a nM
Citation
 Varnes, JG; Xiong, H; Forst, JM; Holmquist, CR; Ernst, GE; Frietze, W; Dembofsky, B; Andisik, DW; Palmer, WE; Hinkley, L; Steelman, GB; Wilkins, DE; Tian, G; Jonak, G; Potts, WM; Wang, X; Brugel, TA; Alhambra, C; Wood, MW; Veale, CA; Albert, JS Bicyclo((aryl)methyl)benzamides as inhibitors of GlyT1. Bioorg Med Chem Lett 28:1043-1049 (2018) [PubMed] Article
Varnes, JG; Xiong, H; Forst, JM; Holmquist, CR; Ernst, GE; Frietze, W; Dembofsky, B; Andisik, DW; Palmer, WE; Hinkley, L; Steelman, GB; Wilkins, DE; Tian, G; Jonak, G; Potts, WM; Wang, X; Brugel, TA; Alhambra, C; Wood, MW; Veale, CA; Albert, JS Bicyclo((aryl)methyl)benzamides as inhibitors of GlyT1. Bioorg Med Chem Lett 28:1043-1049 (2018) [PubMed] Article More Info.:
Target
Name:
Sodium- and chloride-dependent glycine transporter 1
Synonyms:
GlyT-1 | GlyT1 | Glycine Transporters (GlyT1c) | Glycine transporter 1 | SC6A9_HUMAN | SLC6A9 | Sodium- and chloride-dependent glycine transporter 1 | Sodium- and chloride-dependent glycine transporter 1 (GlyT1) | Sodium- and chloride-dependent glycine transporter 1 (GlyT1c) | Sodium-and chloride-dependent glycine transporter 1 (GlyT-1c) | Solute carrier family 6 member 9
Type:
Enzyme
Mol. Mass.:
78270.54
Organism:
Homo sapiens (Human)
Description:
P48067
Residue:
706
Sequence:
MSGGDTRAAIARPRMAAAHGPVAPSSPEQVTLLPVQRSFFLPPFSGATPSTSLAESVLKVWHGAYNSGLLPQLMAQHSLAMAQNGAVPSEATKRDQNLKRGNWGNQIEFVLTSVGYAVGLGNVWRFPYLCYRNGGGAFMFPYFIMLIFCGIPLFFMELSFGQFASQGCLGVWRISPMFKGVGYGMMVVSTYIGIYYNVVICIAFYYFFSSMTHVLPWAYCNNPWNTHDCAGVLDASNLTNGSRPAALPSNLSHLLNHSLQRTSPSEEYWRLYVLKLSDDIGNFGEVRLPLLGCLGVSWLVVFLCLIRGVKSSGKVVYFTATFPYVVLTILFVRGVTLEGAFDGIMYYLTPQWDKILEAKVWGDAASQIFYSLGCAWGGLITMASYNKFHNNCYRDSVIISITNCATSVYAGFVIFSILGFMANHLGVDVSRVADHGPGLAFVAYPEALTLLPISPLWSLLFFFMLILLGLGTQFCLLETLVTAIVDEVGNEWILQKKTYVTLGVAVAGFLLGIPLTSQAGIYWLLLMDNYAASFSLVVISCIMCVAIMYIYGHRNYFQDIQMMLGFPPPLFFQICWRFVSPAIIFFILVFTVIQYQPITYNHYQYPGWAVAIGFLMALSSVLCIPLYAMFRLCRTDGDTLLQRLKNATKPSRDWGPALLEHRTGRYAPTIAPSPEDGFEVQPLHPDKAQIPIVGSNGSSRLQDSRI
Inhibitor
Name:
BDBM50460351
Synonyms:
CHEMBL4225053
Type:
Small organic molecule
Emp. Form.:
C22H26N2O
Mol. Mass.:
334.4546
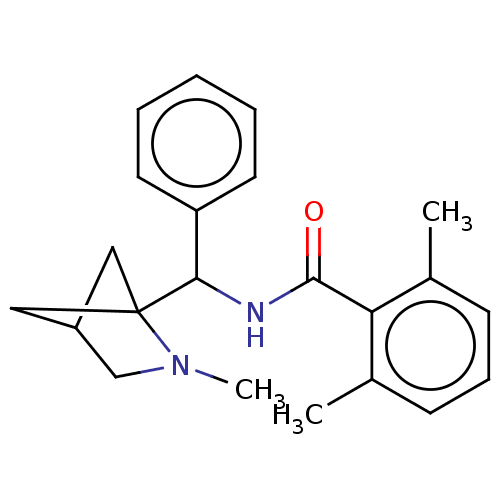
SMILES:
CN1CC2CC1(C2)C(NC(=O)c1c(C)cccc1C)c1ccccc1 |(27.16,-27.5,;27.49,-25.97,;26.44,-24.81,;27.22,-23.46,;28.75,-23.78,;28.91,-25.33,;27.62,-24.96,;30.27,-26.11,;30.27,-27.64,;31.6,-28.42,;32.92,-27.65,;31.6,-29.96,;30.26,-30.71,;28.91,-29.93,;30.26,-32.25,;31.59,-33.03,;32.92,-32.25,;32.92,-30.71,;34.27,-29.93,;31.59,-25.34,;31.59,-23.8,;32.92,-23.04,;34.26,-23.81,;34.26,-25.34,;32.92,-26.11,)|