Reaction Details Report a problem with these data
Report a problem with these data
 Report a problem with these data
Report a problem with these dataTarget
Oxysterols receptor LXR-beta
Ligand
BDBM382353
Substrate
n/a
Meas. Tech.
ChEMBL_1837940 (CHEMBL4338073)
IC50
>10000±n/a nM
Citation
 Marcoux, D; Duan, JJ; Shi, Q; Cherney, RJ; Srivastava, AS; Cornelius, L; Batt, DG; Liu, Q; Beaudoin-Bertrand, M; Weigelt, CA; Khandelwal, P; Vishwakrishnan, S; Selvakumar, K; Karmakar, A; Gupta, AK; Basha, M; Ramlingam, S; Manjunath, N; Vanteru, S; Karmakar, S; Maddala, N; Vetrichelvan, M; Gupta, A; Rampulla, RA; Mathur, A; Yip, S; Li, P; Wu, DR; Khan, J; Ruzanov, M; Sack, JS; Wang, J; Yarde, M; Cvijic, ME; Li, S; Shuster, DJ; Borowski, V; Xie, JH; McIntyre, KW; Obermeier, MT; Fura, A; Stefanski, K; Cornelius, G; Hynes, J; Tino, JA; Macor, JE; Salter-Cid, L; Denton, R; Zhao, Q; Carter, PH; Dhar, TGM Rationally Designed, Conformationally Constrained Inverse Agonists of ROR?t-Identification of a Potent, Selective Series with Biologic-Like in Vivo Efficacy. J Med Chem 62:9931-9946 (2019) [PubMed] Article
Marcoux, D; Duan, JJ; Shi, Q; Cherney, RJ; Srivastava, AS; Cornelius, L; Batt, DG; Liu, Q; Beaudoin-Bertrand, M; Weigelt, CA; Khandelwal, P; Vishwakrishnan, S; Selvakumar, K; Karmakar, A; Gupta, AK; Basha, M; Ramlingam, S; Manjunath, N; Vanteru, S; Karmakar, S; Maddala, N; Vetrichelvan, M; Gupta, A; Rampulla, RA; Mathur, A; Yip, S; Li, P; Wu, DR; Khan, J; Ruzanov, M; Sack, JS; Wang, J; Yarde, M; Cvijic, ME; Li, S; Shuster, DJ; Borowski, V; Xie, JH; McIntyre, KW; Obermeier, MT; Fura, A; Stefanski, K; Cornelius, G; Hynes, J; Tino, JA; Macor, JE; Salter-Cid, L; Denton, R; Zhao, Q; Carter, PH; Dhar, TGM Rationally Designed, Conformationally Constrained Inverse Agonists of ROR?t-Identification of a Potent, Selective Series with Biologic-Like in Vivo Efficacy. J Med Chem 62:9931-9946 (2019) [PubMed] Article More Info.:
Target
Name:
Oxysterols receptor LXR-beta
Synonyms:
LXRB | Liver X receptor beta (NR1H2) | Liver X, LXR beta | NER | NR1H2 | NR1H2_HUMAN | Nuclear receptor NER | UNR | Ubiquitously-expressed nuclear receptor
Type:
Enzyme Catalytic Domain
Mol. Mass.:
50978.79
Organism:
Homo sapiens (Human)
Description:
P55055
Residue:
460
Sequence:
MSSPTTSSLDTPLPGNGPPQPGAPSSSPTVKEEGPEPWPGGPDPDVPGTDEASSACSTDWVIPDPEEEPERKRKKGPAPKMLGHELCRVCGDKASGFHYNVLSCEGCKGFFRRSVVRGGARRYACRGGGTCQMDAFMRRKCQQCRLRKCKEAGMREQCVLSEEQIRKKKIRKQQQESQSQSQSPVGPQGSSSSASGPGASPGGSEAGSQGSGEGEGVQLTAAQELMIQQLVAAQLQCNKRSFSDQPKVTPWPLGADPQSRDARQQRFAHFTELAIISVQEIVDFAKQVPGFLQLGREDQIALLKASTIEIMLLETARRYNHETECITFLKDFTYSKDDFHRAGLQVEFINPIFEFSRAMRRLGLDDAEYALLIAINIFSADRPNVQEPGRVEALQQPYVEALLSYTRIKRPQDQLRFPRMLMKLVSLRTLSSVHSEQVFALRLQDKKLPPLLSEIWDVHE
Inhibitor
Name:
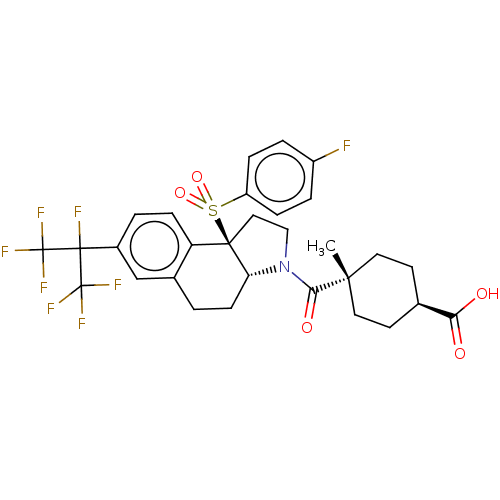
BDBM382353
Synonyms:
US10273259, Example 66 | US10711020, Example 66
Type:
Small organic molecule
Emp. Form.:
C30H29F8NO5S
Mol. Mass.:
667.607
SMILES:
C[C@@]1(CC[C@@H](CC1)C(O)=O)C(=O)N1CC[C@@]2([C@H]1CCc1cc(ccc21)C(F)(C(F)(F)F)C(F)(F)F)S(=O)(=O)c1ccc(F)cc1 |r,wU:16.17,4.7,1.0,wD:1.10,15.38,(-5.67,-3.47,;-5.41,-1.95,;-4.64,-.61,;-5.42,.72,;-6.96,.71,;-7.72,-.62,;-6.95,-1.95,;-7.73,2.04,;-9.27,2.04,;-6.97,3.38,;-3.96,-2.47,;-3.69,-3.98,;-2.78,-1.47,;-2.9,.06,;-1.47,.64,;-.48,-.53,;-1.29,-1.84,;-.56,-3.2,;.98,-3.25,;1.79,-1.94,;3.33,-1.99,;4.14,-.68,;3.41,.68,;1.87,.73,;1.06,-.58,;5.68,-.73,;5.73,.81,;7.22,-.77,;8.76,-.82,;7.17,-2.31,;7.27,.77,;5.63,-2.26,;4.28,-2.99,;6.94,-3.08,;5.58,-3.8,;.19,.85,;1.6,1.48,;.51,2.36,;-.67,2.13,;-2.21,2.01,;-3.07,3.29,;-2.4,4.67,;-3.27,5.95,;-.87,4.79,;-0,3.51,)|