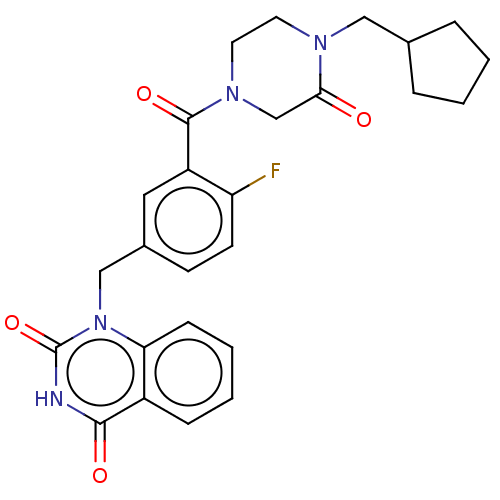
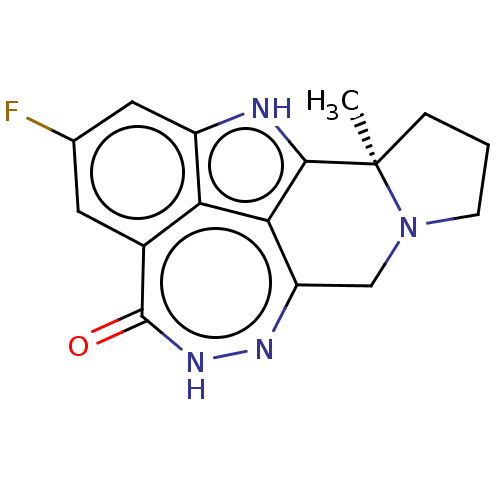
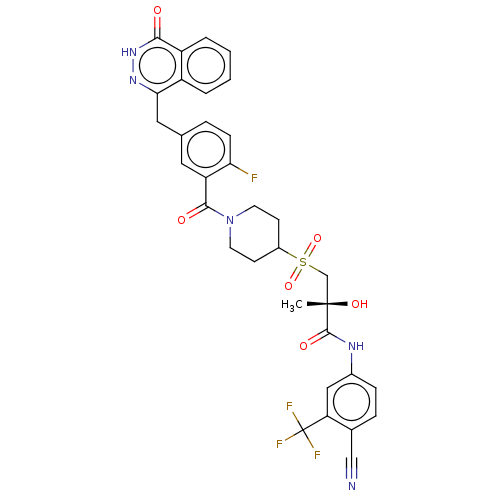
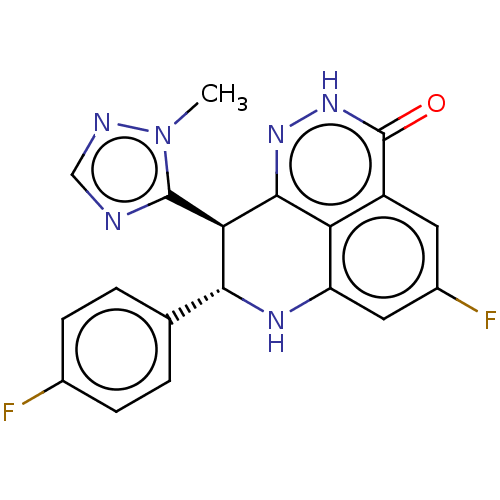
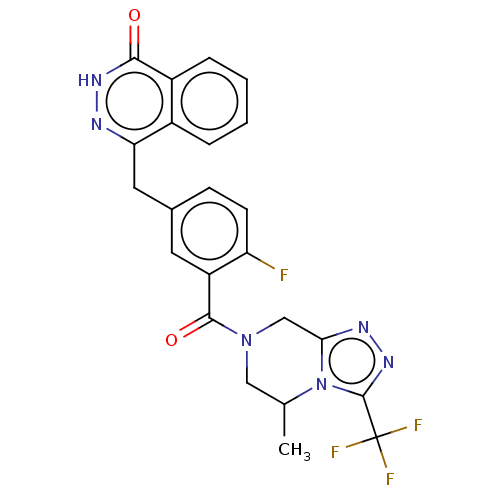
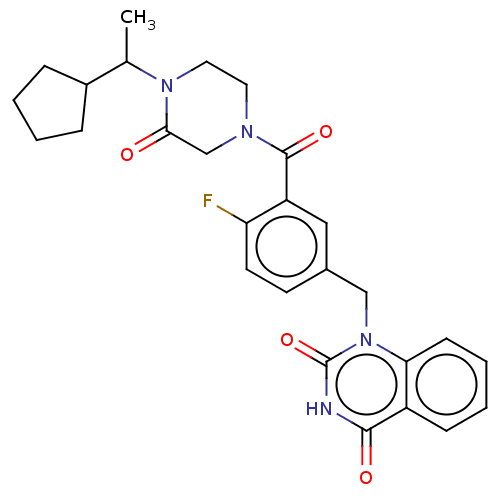
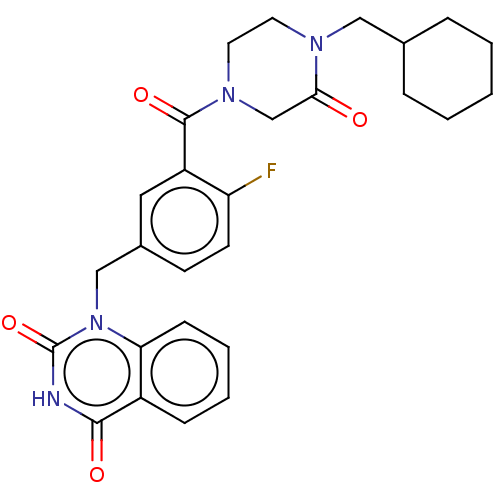
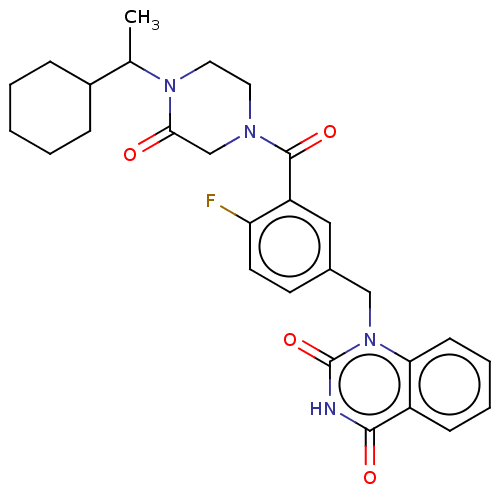
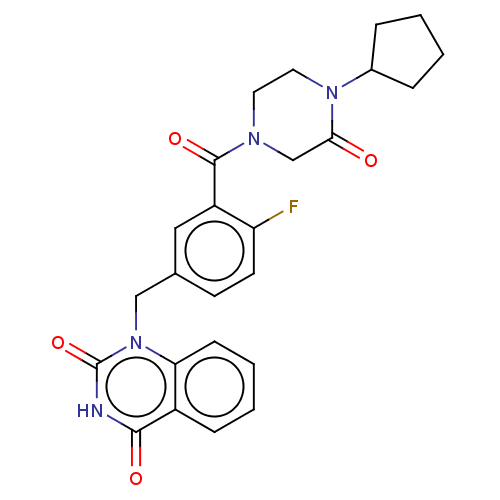
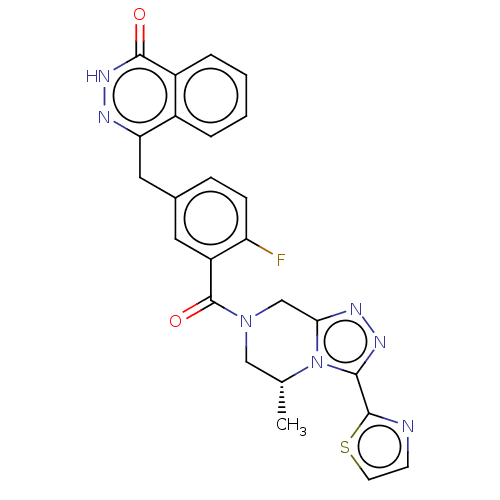
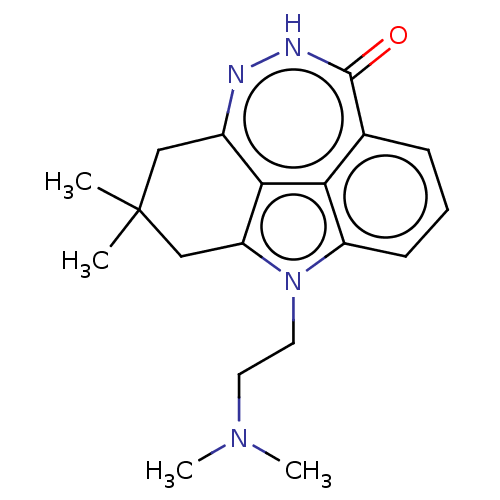
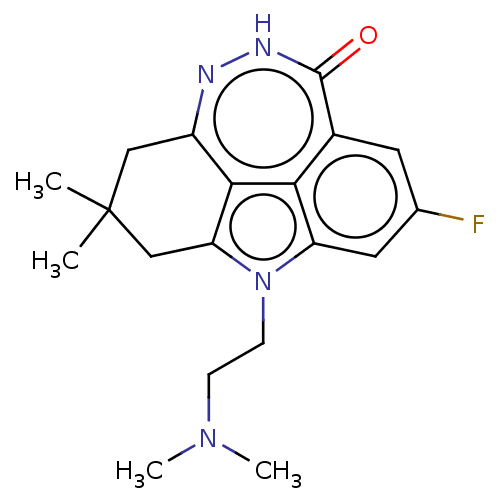
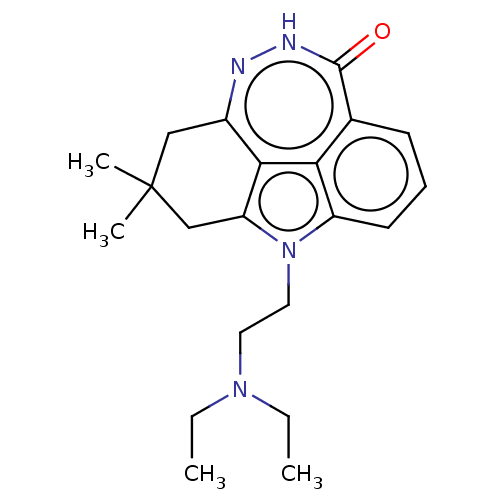
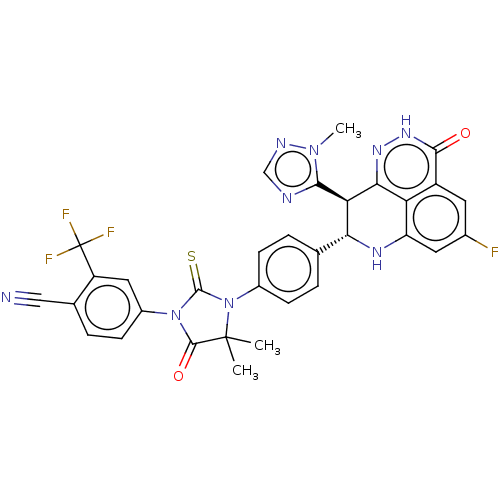
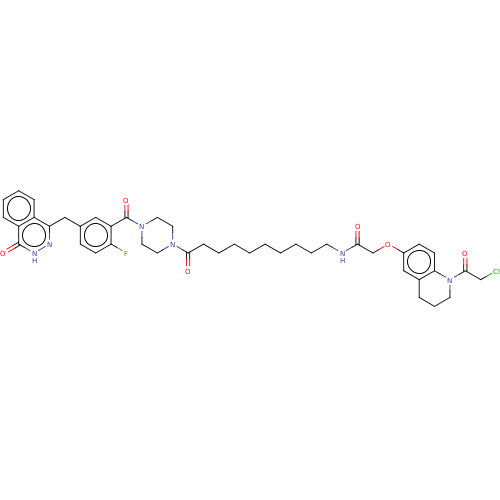
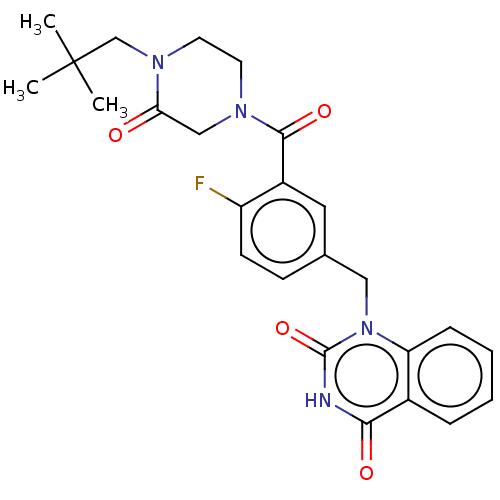
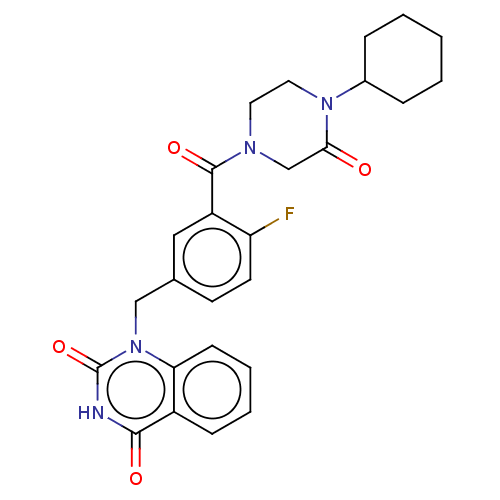
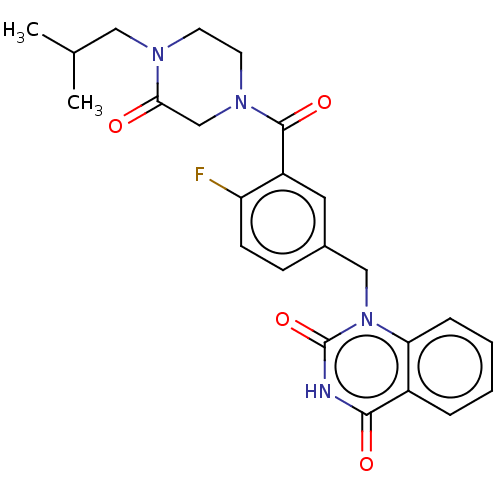
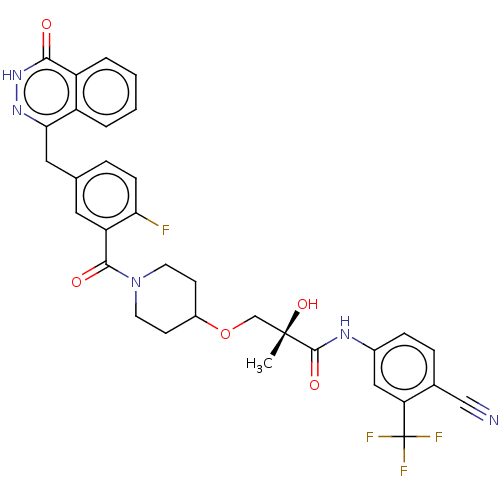
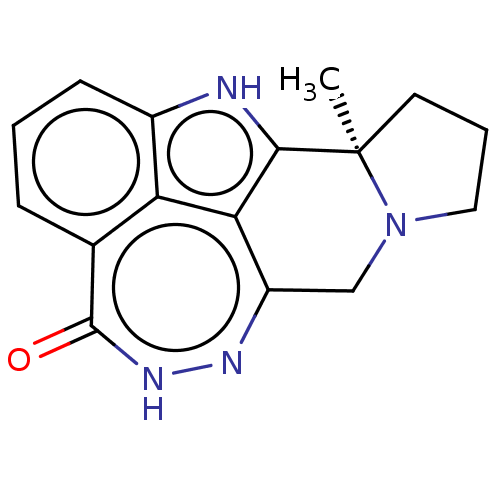
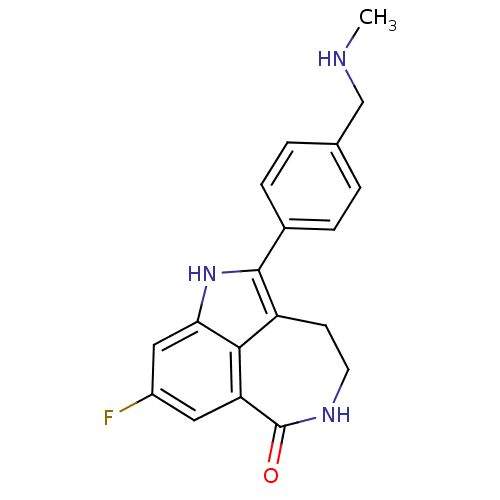
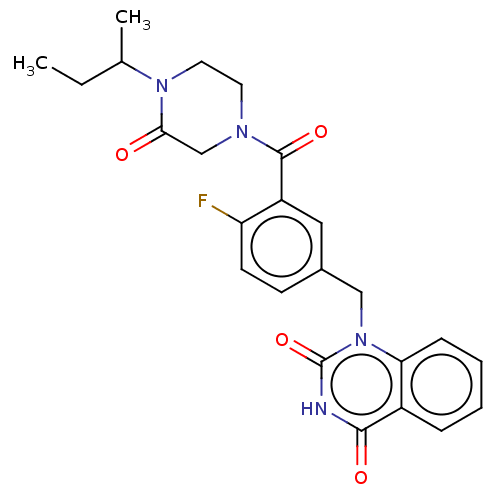
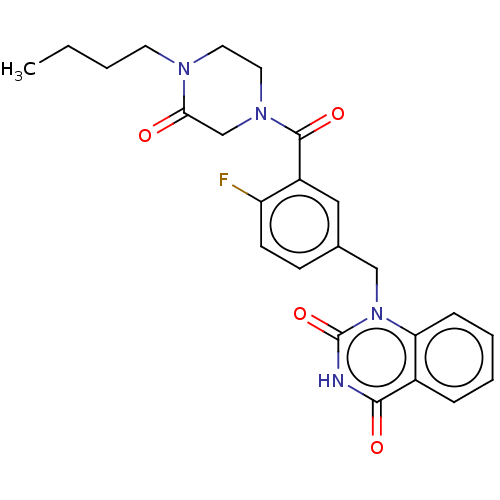
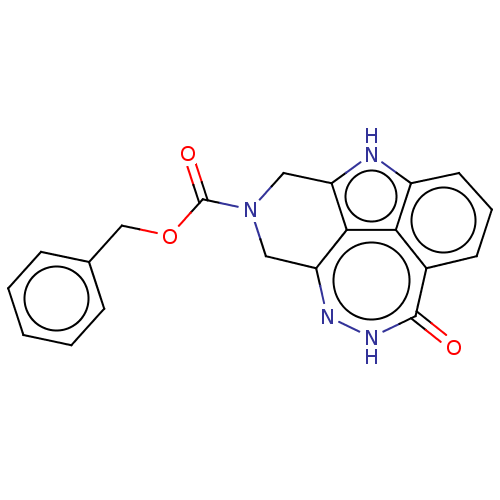
Affinity DataIC50: 0.00100nMAssay Description:Inhibition of PARP2 (unknown origin) using histone as substrate by ELISAMore data for this Ligand-Target Pair
Affinity DataIC50: 0.00100nMAssay Description:Inhibition of PARP2 (unknown origin)More data for this Ligand-Target Pair
Affinity DataIC50: 0.0500nMAssay Description:Inhibition of PARP2 (unknown origin)More data for this Ligand-Target Pair
Affinity DataIC50: 0.100nMAssay Description:Inhibition of PARP-2 (unknown origin)More data for this Ligand-Target Pair
Affinity DataIC50: 0.100nMAssay Description:Inhibition of human recombinant PARP2 expressed in Escherichia coli BL21(DE3) incubated for 1 hr by ELISA assayMore data for this Ligand-Target Pair
Affinity DataIC50: 0.110nMAssay Description:Inhibition of PARP-2 (unknown origin)More data for this Ligand-Target Pair
Affinity DataIC50: 0.110nMAssay Description:Inhibition of PARP2 (unknown origin) using histone/NAD+ as substrates by NAD/NADH-Glo assayMore data for this Ligand-Target Pair
Affinity DataIC50: 0.130nMAssay Description:Inhibition of N-terminal GST-tagged human PARP2 (2 to 583 residues) expressed in baculovirus-infected Sf9 cells using histone as substrate incubated ...More data for this Ligand-Target Pair
Affinity DataIC50: 0.200nMAssay Description:i. PARP1 assay: Human recombinant PARP1 at a final concentration of 2.5 nM was combined with histone H4 (20 μM) and test articles at various con...More data for this Ligand-Target Pair
Affinity DataIC50: 0.200nMAssay Description:PARP-2 and PARP-3 enzymatic assays were conducted using commercial PARP-2/PARP-3 Chemiluminescent Assay Kit (BPS Biosciences) and the protocols with ...More data for this Ligand-Target Pair
Affinity DataIC50: 0.200nMAssay Description:Inhibition of PARP-2 (unknown origin) pre-incubated for 30 mins before addition of activated DNA and NAD by chemiluminescent assayMore data for this Ligand-Target Pair
Affinity DataIC50: 0.200nMAssay Description:Inhibition of PARP2 (unknown origin) using NAD as substrate incubated for 1 minMore data for this Ligand-Target Pair
Affinity DataIC50: 0.220nMAssay Description:In order to test the selectivity of substituents on piperazinotriazole ring within the PARP family, the selectivity of compound S3 and positive compo...More data for this Ligand-Target Pair
Affinity DataIC50: 0.220nMAssay Description:Inhibition of recombinant human PARP2 using histone as substrate after 1 hr in presence of NAD+ by ELISAMore data for this Ligand-Target Pair
Affinity DataIC50: 0.220nMAssay Description:Inhibition of human recombinant PARP2 expressed in Escherichia coli BL21(DE3) incubated for 1 hr by ELISA assayMore data for this Ligand-Target Pair
Affinity DataIC50: 0.240nMAssay Description:Inhibition of human recombinant PARP2 expressed in Escherichia coli BL21(DE3) incubated for 1 hr by ELISA assayMore data for this Ligand-Target Pair
Affinity DataIC50: 0.260nMAssay Description:Inhibition of human recombinant PARP2 expressed in Escherichia coli BL21(DE3) incubated for 1 hr by ELISA assayMore data for this Ligand-Target Pair
Affinity DataKd: 0.270nMAssay Description:Binding affinity to recombinant human PARP-2 by SPR analysisMore data for this Ligand-Target Pair
Affinity DataKd: 0.280nMAssay Description:Binding affinity to recombinant human HisGST-tagged PARP-2 catalytic domain by surface plasmon resonance analysisMore data for this Ligand-Target Pair
Affinity DataIC50: 0.290nMAssay Description:Inhibition of human recombinant PARP2 expressed in Escherichia coli BL21(DE3) incubated for 1 hr by ELISA assayMore data for this Ligand-Target Pair
Affinity DataIC50: 0.300nMAssay Description:Inhibition of PARP2 (unknown origin) by ELISA assayMore data for this Ligand-Target Pair
Affinity DataIC50: 0.300nMAssay Description:PARP-2 and PARP-3 enzymatic assays were conducted using commercial PARP-2/PARP-3 Chemiluminescent Assay Kit (BPS Biosciences) and the protocols with ...More data for this Ligand-Target Pair
Affinity DataIC50: 0.300nMAssay Description:Inhibition of PARP-2 (unknown origin)More data for this Ligand-Target Pair
Affinity DataIC50: 0.300nMAssay Description:Inhibition of PARP-2 (unknown origin) pre-incubated for 30 mins before addition of activated DNA and NAD by chemiluminescent assayMore data for this Ligand-Target Pair
Affinity DataIC50: 0.300nMAssay Description:Inhibition of PARP-2 (unknown origin) pre-incubated for 30 mins before addition of activated DNA and NAD by chemiluminescent assayMore data for this Ligand-Target Pair
Affinity DataIC50: 0.300nMAssay Description:PARP-2 and PARP-3 enzymatic assays were conducted using commercial PARP-2/PARP-3 Chemiluminescent Assay Kit (BPS Biosciences) and the protocols with ...More data for this Ligand-Target Pair
Affinity DataIC50: 0.300nMAssay Description:Inhibition of PARP-2 (unknown origin) pre-incubated for 30 mins before addition of activated DNA and NAD by chemiluminescent assayMore data for this Ligand-Target Pair
Affinity DataIC50: 0.300nMAssay Description:Inhibition of PARP2 (unknown origin)More data for this Ligand-Target Pair
Affinity DataIC50: 0.300nMAssay Description:Inhibition of PARP2 (unknown origin)More data for this Ligand-Target Pair
Affinity DataIC50: 0.300nMAssay Description:Inhibition of PARP2 (unknown origin)More data for this Ligand-Target Pair
Affinity DataIC50: 0.300nMAssay Description:i. PARP1 assay: Human recombinant PARP1 at a final concentration of 2.5 nM was combined with histone H4 (20 μM) and test articles at various con...More data for this Ligand-Target Pair
Affinity DataIC50: 0.300nMAssay Description:PARP-2 and PARP-3 enzymatic assays were conducted using commercial PARP-2/PARP-3 Chemiluminescent Assay Kit (BPS Biosciences) and the protocols with ...More data for this Ligand-Target Pair
Affinity DataIC50: 0.320nMAssay Description:Inhibition of PARP2 (unknown origin)More data for this Ligand-Target Pair
Affinity DataIC50: 0.330nMAssay Description:Inhibition of human recombinant PARP2 expressed in Escherichia coli BL21(DE3) incubated for 1 hr by ELISA assayMore data for this Ligand-Target Pair
Affinity DataIC50: 0.330nMAssay Description:Inhibition of human recombinant PARP2 expressed in Escherichia coli BL21(DE3) incubated for 1 hr by ELISA assayMore data for this Ligand-Target Pair
Affinity DataIC50: 0.380nMAssay Description:Inhibition of human recombinant PARP2 expressed in Escherichia coli BL21(DE3) incubated for 1 hr by ELISA assayMore data for this Ligand-Target Pair
Affinity DataIC50: 0.400nMAssay Description:i. PARP1 assay: Human recombinant PARP1 at a final concentration of 2.5 nM was combined with histone H4 (20 μM) and test articles at various con...More data for this Ligand-Target Pair
Affinity DataIC50: 0.400nMAssay Description:PARP-2 and PARP-3 enzymatic assays were conducted using commercial PARP-2/PARP-3 Chemiluminescent Assay Kit (BPS Biosciences) and the protocols with ...More data for this Ligand-Target Pair
Affinity DataIC50: 0.400nMAssay Description:Inhibition of PARP2 (unknown origin)More data for this Ligand-Target Pair
Affinity DataIC50: 0.400nMAssay Description:Inhibition of PARP-2 (unknown origin) pre-incubated for 30 mins before addition of activated DNA and NAD by chemiluminescent assayMore data for this Ligand-Target Pair
Affinity DataIC50: 0.440nMAssay Description:Inhibition of human recombinant PARP2 expressed in Escherichia coli BL21(DE3) incubated for 1 hr by ELISA assayMore data for this Ligand-Target Pair
Affinity DataIC50: 0.450nMAssay Description:In order to test the selectivity of substituents on piperazinotriazole ring within the PARP family, the selectivity of compound S3 and positive compo...More data for this Ligand-Target Pair
Affinity DataIC50: 0.490nMAssay Description:Inhibition of human recombinant PARP2 expressed in Escherichia coli BL21(DE3) incubated for 1 hr by ELISA assayMore data for this Ligand-Target Pair
Affinity DataIC50: 0.5nMAssay Description:Inhibition of PARP2 (unknown origin)More data for this Ligand-Target Pair
Affinity DataIC50: 0.5nMAssay Description:Inhibition of PARP-2 (unknown origin)More data for this Ligand-Target Pair
Affinity DataIC50: 0.5nMAssay Description:Inhibition of human N-terminal GST-tagged PARP2 (2 to 583 residues) expressed in baculovirus infected Sf9 insect cells using histone as substrate mea...More data for this Ligand-Target Pair
Affinity DataIC50: 0.5nMAssay Description:Inhibition of human N-terminal GST-tagged PARP2 (2 to 583 residues) expressed in baculovirus infected Sf9 insect cells using histone as substrate mea...More data for this Ligand-Target Pair
Affinity DataIC50: 0.5nMAssay Description:PARP2 and PARP3 enzymatic assays were conducted using commercial PARP-2/PARP-3 Chemiluminescent Assay Kit (BPS Biosciences) and the protocols with th...More data for this Ligand-Target Pair
Affinity DataIC50: 0.5nMAssay Description:Inhibition of PARP2 (unknown origin)More data for this Ligand-Target Pair
Affinity DataIC50: 0.5nMAssay Description:PARP-2 and PARP-3 enzymatic assays were conducted using commercial PARP-2/PARP-3 Chemiluminescent Assay Kit (BPS Biosciences) and the protocols with ...More data for this Ligand-Target Pair


 3D Structure (crystal)
3D Structure (crystal)