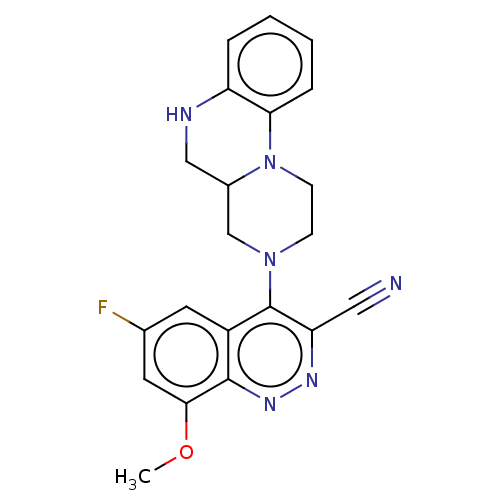
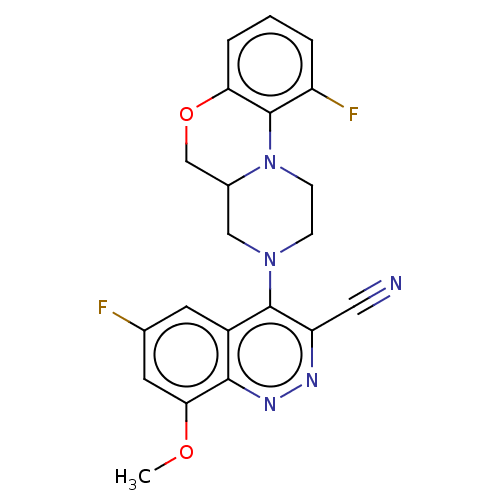
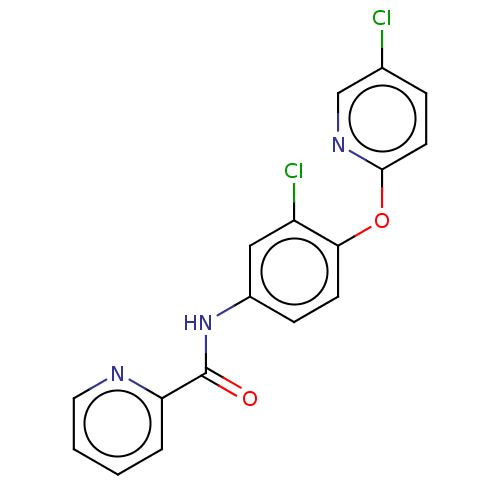
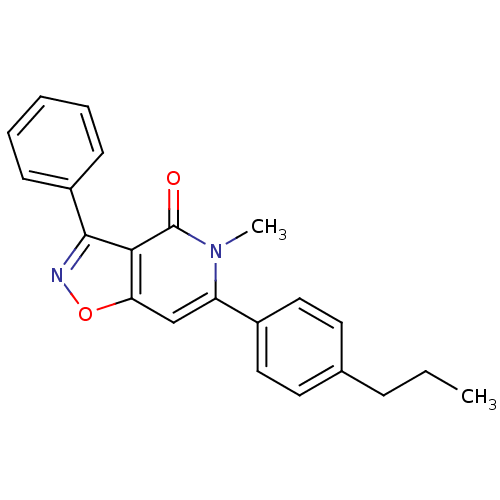
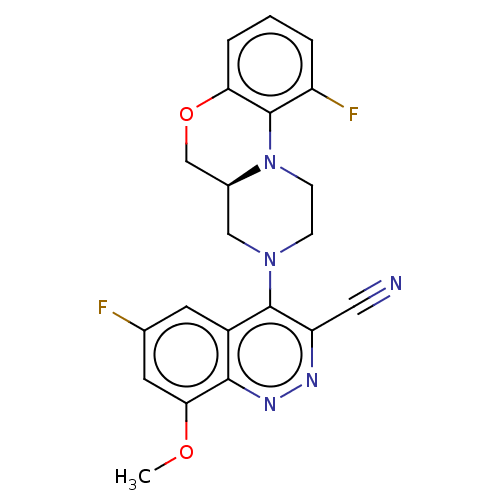
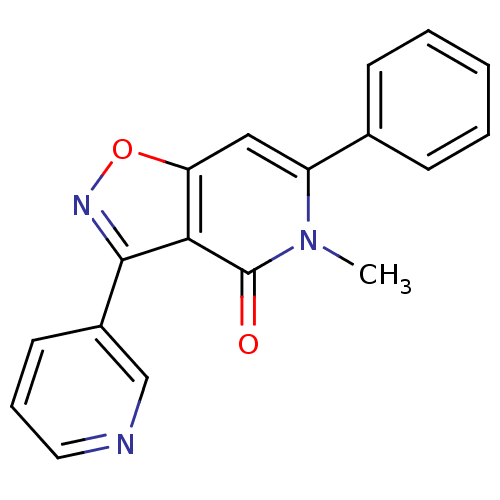
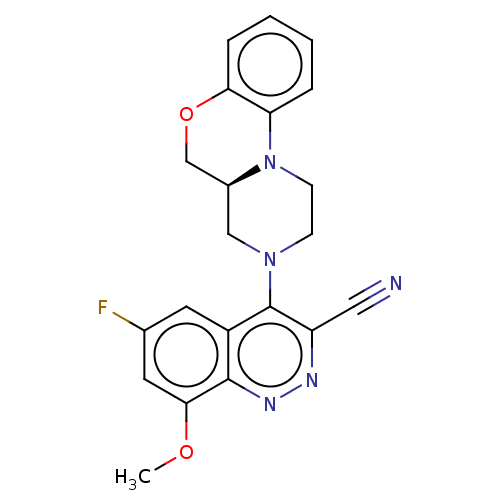
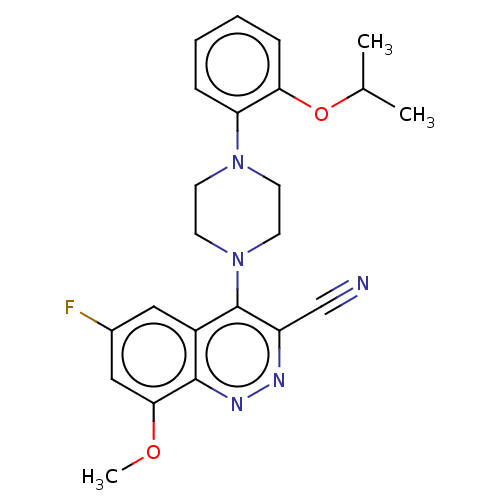
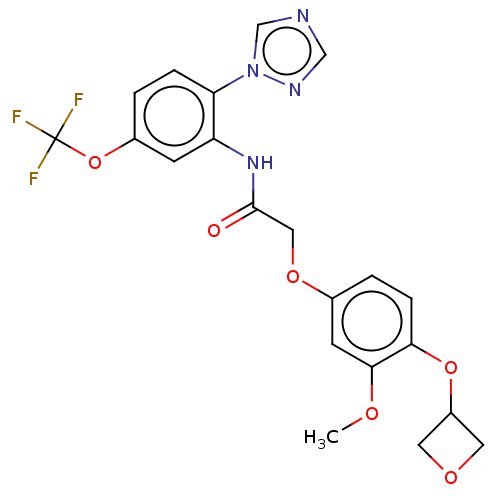
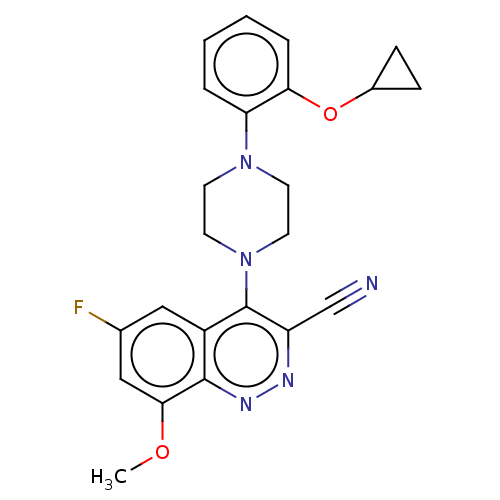
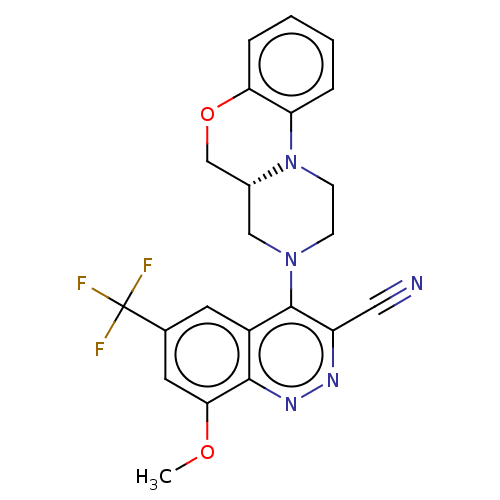
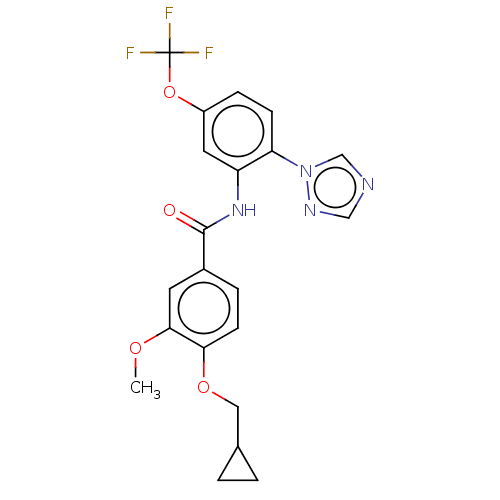
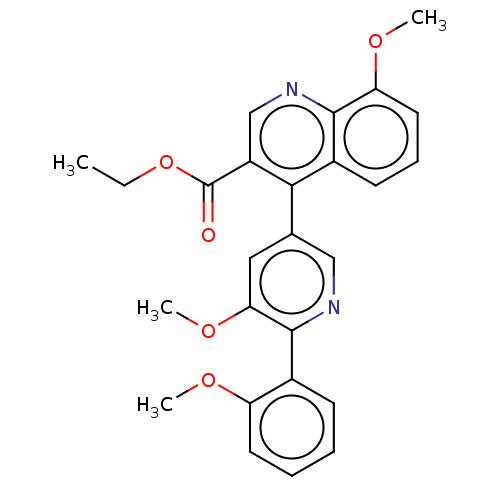
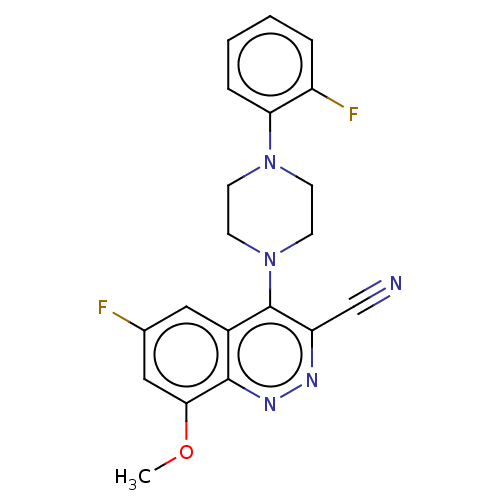
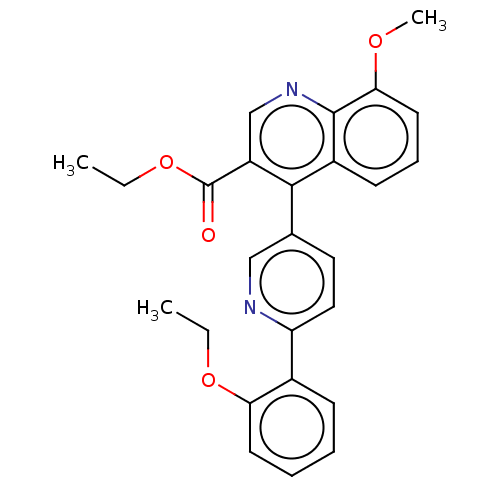
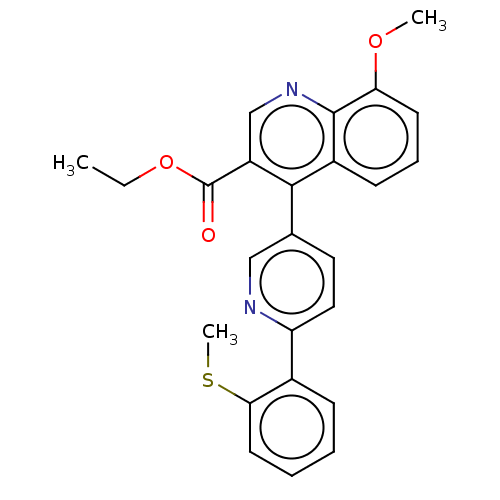
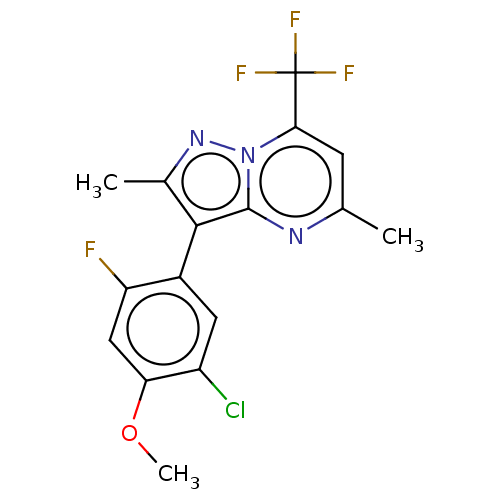
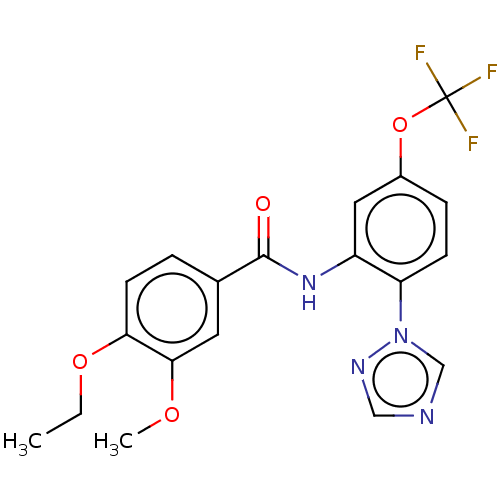
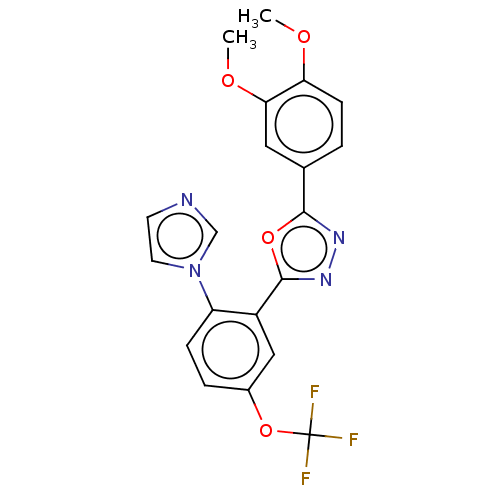
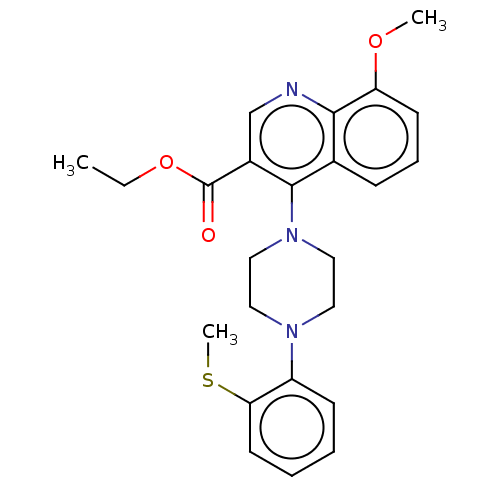
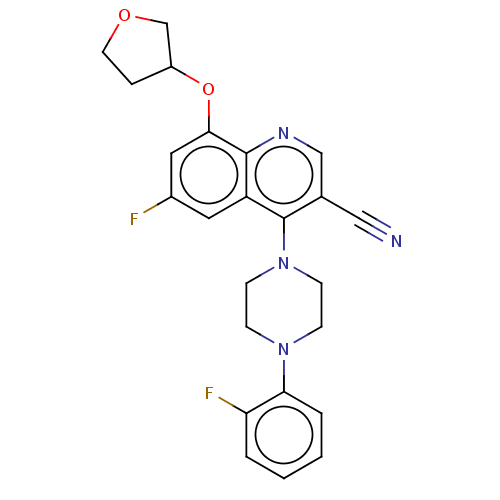
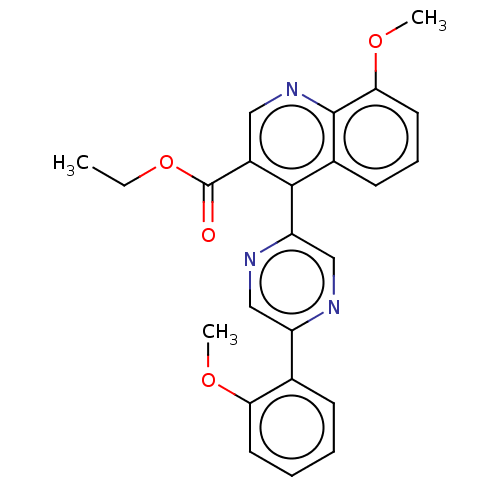
Affinity DataIC50: 12nMAssay Description:Antagonist activity at rat mGluR7 expressed in CHO cells assessed as inhibition of LAP-induced intracellular calcium mobilization by FLIPRMore data for this Ligand-Target Pair
Affinity DataIC50: 20nMAssay Description:Antagonist activity at rat mGluR7 expressed in CHO cells assessed as inhibition of LAP-induced intracellular calcium mobilization by FLIPRMore data for this Ligand-Target Pair
Affinity DataIC50: 21nMAssay Description:Antagonist activity at rat mGluR7 expressed in CHO cells assessed as inhibition of LAP-induced intracellular calcium mobilization by FLIPRMore data for this Ligand-Target Pair
Affinity DataIC50: 22nMAssay Description:Antagonist activity at rat mGluR7 expressed in CHO cells assessed as inhibition of LAP-induced intracellular calcium mobilization by FLIPRMore data for this Ligand-Target Pair
Affinity DataIC50: 26nMAssay Description:Antagonist activity at rat mGluR7 expressed in CHO cells assessed as inhibition of LAP-induced intracellular calcium mobilization by FLIPRMore data for this Ligand-Target Pair
Affinity DataIC50: 26nMAssay Description:Antagonist activity at rat mGluR7 expressed in CHO cells assessed as inhibition of LAP-induced intracellular calcium mobilization by FLIPRMore data for this Ligand-Target Pair
Affinity DataIC50: 29nMAssay Description:Antagonist activity at rat mGluR7 expressed in CHO cells assessed as inhibition of LAP-induced intracellular calcium mobilization by FLIPRMore data for this Ligand-Target Pair
Affinity DataIC50: 31nMAssay Description:Antagonist activity at rat mGluR7 expressed in CHO cells assessed as inhibition of LAP-induced intracellular calcium mobilization by FLIPRMore data for this Ligand-Target Pair
Affinity DataEC50: 43nMAssay Description:Positive allosteric modulation of rat mGluR7More data for this Ligand-Target Pair
Affinity DataIC50: 51nMAssay Description:Antagonist activity at rat mGluR7 expressed in CHO cells assessed as inhibition of LAP-induced intracellular calcium mobilization by FLIPRMore data for this Ligand-Target Pair
Affinity DataEC50: 56nMAssay Description:Positive allosteric modulation of rat mGluR7More data for this Ligand-Target Pair
Affinity DataIC50: 72nMAssay Description:Antagonist activity at rat mGluR7 expressed in CHO cells assessed as inhibition of LAP-induced intracellular calcium mobilization by FLIPRMore data for this Ligand-Target Pair
Affinity DataIC50: 80nMAssay Description:Inhibition of [3H] -LY341495 binding to rat mGlu7 expressed in CHO cell membrane incubated for 30 mins in presence of LY341495 by liquid scintillatio...More data for this Ligand-Target Pair
Affinity DataIC50: 96nMAssay Description:Antagonist activity at rat mGluR7 expressed in CHO cells assessed as inhibition of LAP-induced intracellular calcium mobilization by FLIPRMore data for this Ligand-Target Pair
Affinity DataIC50: 101nMAssay Description:Antagonist activity at rat mGluR7 expressed in CHO cells assessed as inhibition of LAP-induced intracellular calcium mobilization by FLIPRMore data for this Ligand-Target Pair
Affinity DataKi: 110nMAssay Description:Binding affinity towards metabotropic glutamate receptor 7 of rat expressed in CHO cells was determined by using [3H]MGS-0008More data for this Ligand-Target Pair
Affinity DataEC50: 120nMAssay Description:Positive allosteric modulation of rat mGluR7More data for this Ligand-Target Pair
Affinity DataEC50: 130nMAssay Description:Positive allosteric modulation of rat mGluR7More data for this Ligand-Target Pair
Affinity DataIC50: 140nMAssay Description:Antagonist activity at rat mGluR7 expressed in CHO cells assessed as inhibition of LAP-induced intracellular calcium mobilization by FLIPRMore data for this Ligand-Target Pair
Affinity DataEC50: 146nMAssay Description:Positive allosteric modulation of rat mGlu7 receptor expressed in HEK cells co-expressing Galphai5 assessed as increase in glutamate-induced calcium ...More data for this Ligand-Target Pair
Affinity DataIC50: 150nMAssay Description:Antagonist activity at rat mGluR7 expressed in CHO cells assessed as inhibition of LAP-induced intracellular calcium mobilization by FLIPRMore data for this Ligand-Target Pair
Affinity DataEC50: 150nMAssay Description:Positive allosteric modulation of rat mGluR7More data for this Ligand-Target Pair
Affinity DataIC50: 190nMAssay Description:Inhibition of [3H] -LY341495 binding to rat mGlu7 expressed in CHO cell membrane incubated for 30 mins by liquid scintillation analysisMore data for this Ligand-Target Pair
Affinity DataIC50: 210nMAssay Description:Antagonist activity at rat mGluR7 expressed in CHO cells assessed as inhibition of LAP-induced intracellular calcium mobilization by FLIPRMore data for this Ligand-Target Pair
Affinity DataIC50: 240nMAssay Description:Antagonist activity at rat mGluR7 expressed in CHO cells assessed as inhibition of LAP-induced intracellular calcium mobilization by FLIPRMore data for this Ligand-Target Pair
Affinity DataEC50: 260nMAssay Description:Positive allosteric modulation of rat mGluR7More data for this Ligand-Target Pair
Affinity DataEC50: 260nMAssay Description:Positive allosteric modulation of rat mGluR7More data for this Ligand-Target Pair
Affinity DataIC50: 263nMAssay Description:Negative allosteric modulation of rat mGlu7 in HEK cells expressing Galpha15 assessed as inhibition of L-AP4-induced response measured by calcium mob...More data for this Ligand-Target Pair
Affinity DataIC50: 263nMAssay Description:Negative allosteric modulation of rat mGlu7 in HEK cells expressing Galpha15 assessed as inhibition of L-AP4-induced response measured by calcium mob...More data for this Ligand-Target Pair
Affinity DataEC50: 310nMAssay Description:Positive allosteric modulation of rat mGluR7More data for this Ligand-Target Pair
Affinity DataEC50: 340nMAssay Description:Positive allosteric modulation of rat mGluR7More data for this Ligand-Target Pair
Affinity DataIC50: 347nMAssay Description:Negative allosteric modulation of rat mGlu7 receptor expressed in HEK cells harboring Ga15 assessed as reduction in L-AP4-induced calcium flux preinc...More data for this Ligand-Target Pair
Affinity DataIC50: 347nMAssay Description:Negative allosteric modulation of rat mGlu7 in HEK cells expressing Galpha15 assessed as inhibition of L-AP4-induced response measured by calcium mob...More data for this Ligand-Target Pair
Affinity DataIC50: 350nMAssay Description:Negative allosteric modulation of rat mGlu7 receptor expressed in HEK cells co-expressing Galpha15 assessed as reduction in calcium mobilization incu...More data for this Ligand-Target Pair
Affinity DataIC50: 350nMAssay Description:Negative allosteric modulation of rat mGlu7 receptor expressed in HEK cells harboring Ga15 assessed as reduction in L-AP4-induced calcium flux preinc...More data for this Ligand-Target Pair
Affinity DataEC50: 360nMAssay Description:Positive allosteric modulation of rat mGluR7More data for this Ligand-Target Pair
Affinity DataIC50: 370nMAssay Description:Negative allosteric modulation of rat mGlu7 receptor expressed in HEK cells co-expressing Galpha15 assessed as reduction in calcium mobilization incu...More data for this Ligand-Target Pair
Affinity DataIC50: 410nMAssay Description:Negative allosteric modulation of rat mGlu7 receptor expressed in HEK cells co-expressing Galpha15 assessed as reduction in calcium mobilization incu...More data for this Ligand-Target Pair
Affinity DataEC50: 479nMAssay Description:Positive allosteric modulation of rat mGlu7 receptor expressed in HEK cells co-expressing Galphai5 assessed as increase in glutamate-induced calcium ...More data for this Ligand-Target Pair
Affinity DataEC50: 480nMAssay Description:Positive allosteric modulation of rat mGlu7 receptor expressed in HEK cells co-expressing Galphai5 assessed as increase in glutamate-induced calcium ...More data for this Ligand-Target Pair
Affinity DataIC50: 501nMAssay Description:Negative allosteric modulation of rat mGlu7 in HEK cells expressing Galpha15 assessed as inhibition of L-AP4-induced response measured by calcium mob...More data for this Ligand-Target Pair
Affinity DataIC50: 501nMAssay Description:Negative allosteric modulation of rat mGlu7 in HEK cells expressing Galpha15 assessed as inhibition of L-AP4-induced response measured by calcium mob...More data for this Ligand-Target Pair
Affinity DataIC50: 501nMAssay Description:Negative allosteric modulation of rat mGlu7 in HEK cells expressing Galpha15 assessed as inhibition of L-AP4-induced response measured by calcium mob...More data for this Ligand-Target Pair
Affinity DataIC50: 510nMAssay Description:Negative allosteric modulation of rat mGlu7 receptor expressed in HEK cells co-expressing Galpha15 assessed as reduction in calcium mobilization incu...More data for this Ligand-Target Pair
Affinity DataEC50: 510nMAssay Description:Positive allosteric modulation of rat mGlu7 receptor expressed in HEK293 cells co-expressing Gai5 assessed as potentiation of L-AP4-induced calcium m...More data for this Ligand-Target Pair
Affinity DataIC50: 510nMAssay Description:Negative allosteric modulation of rat mGlu7 expressed in HEK cells co-expressing Galpha15 assessed as reduction in L-AP4-induced intracellular calciu...More data for this Ligand-Target Pair
Affinity DataIC50: 513nMAssay Description:Negative allosteric modulation of rat mGlu7 expressed in HEK cells co-expressing Galpha15 assessed as reduction in L-AP4-induced intracellular calciu...More data for this Ligand-Target Pair
Affinity DataIC50: 540nMAssay Description:Negative allosteric modulation of rat mGlu7 receptor expressed in HEK cells co-expressing Galpha15 assessed as reduction in calcium mobilization incu...More data for this Ligand-Target Pair
Affinity DataIC50: 550nMAssay Description:Negative allosteric modulation of rat mGlu7 in HEK cells expressing Galpha15 assessed as inhibition of L-AP4-induced response measured by calcium mob...More data for this Ligand-Target Pair
Affinity DataIC50: 570nMAssay Description:Negative allosteric modulation of rat mGlu7 receptor expressed in HEK cells co-expressing Galpha15 assessed as reduction in calcium mobilization incu...More data for this Ligand-Target Pair
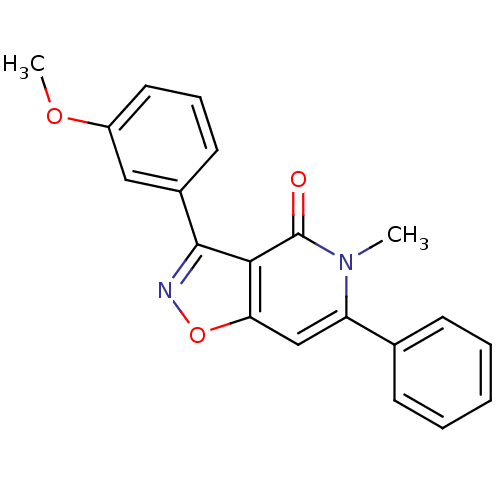

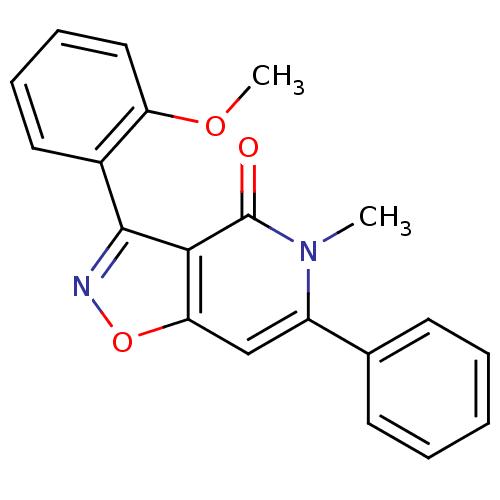
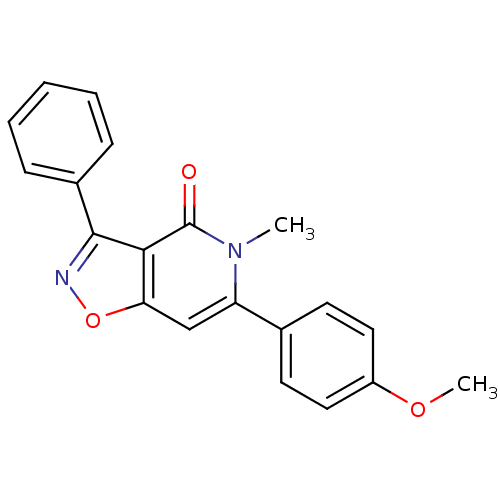

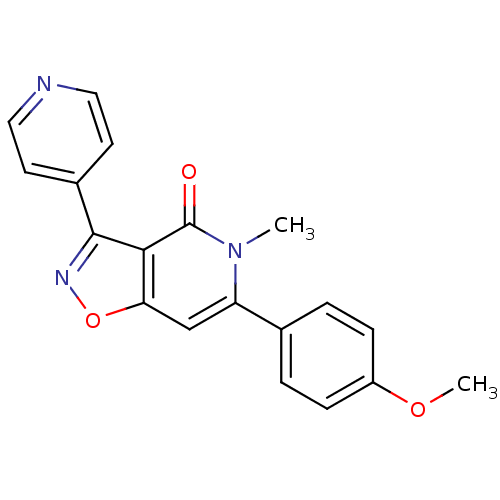
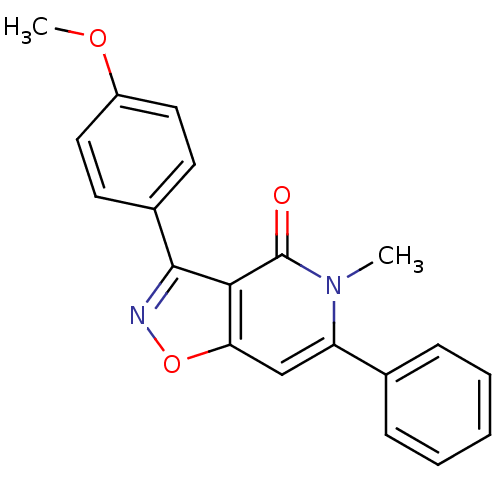
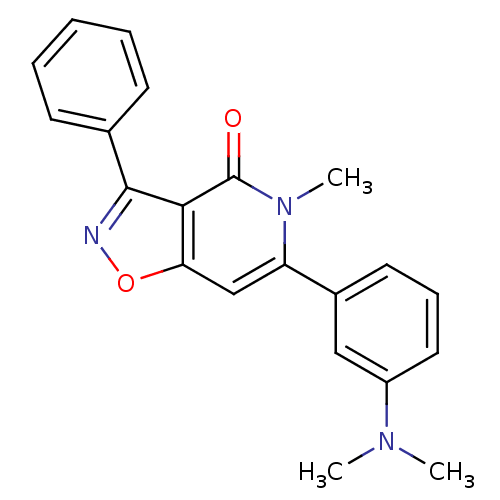
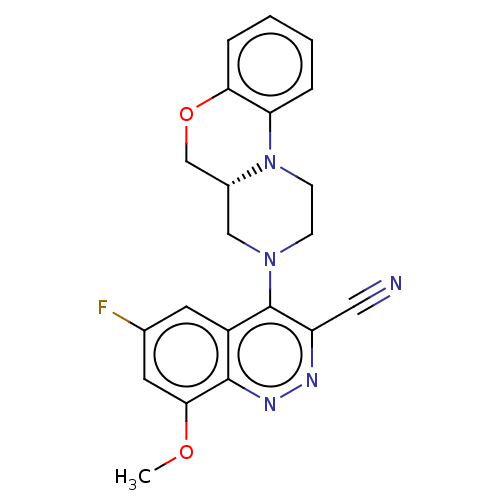
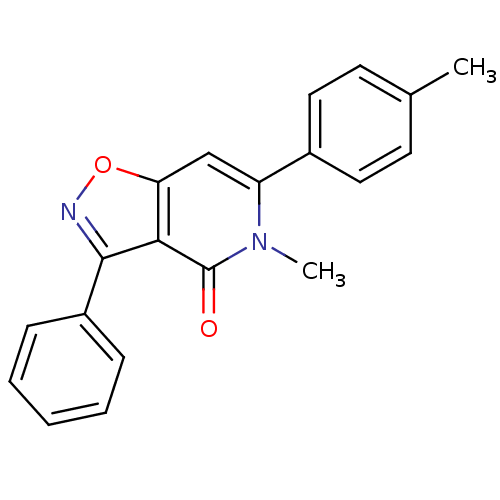
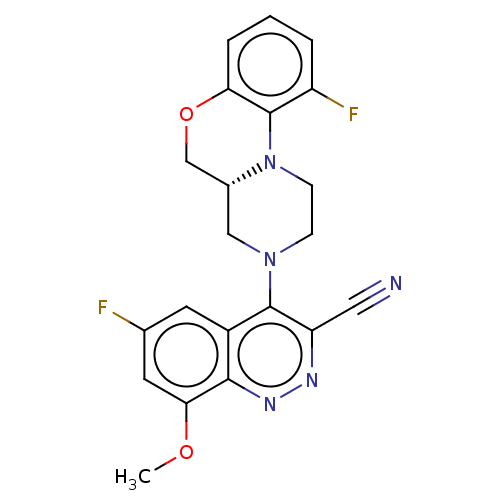
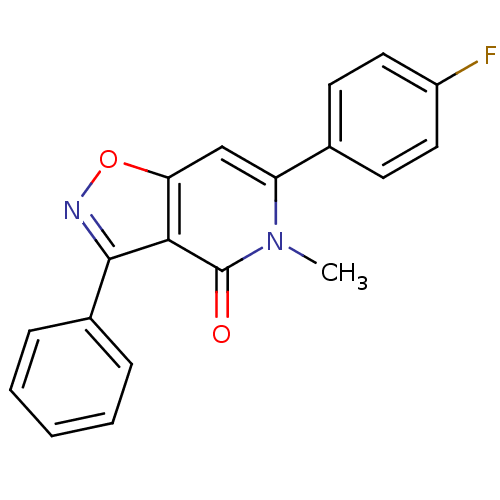
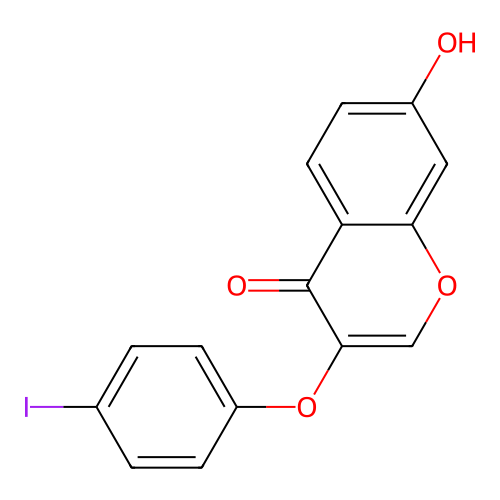
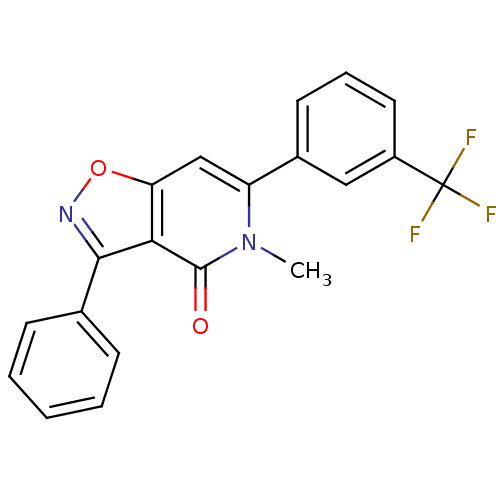
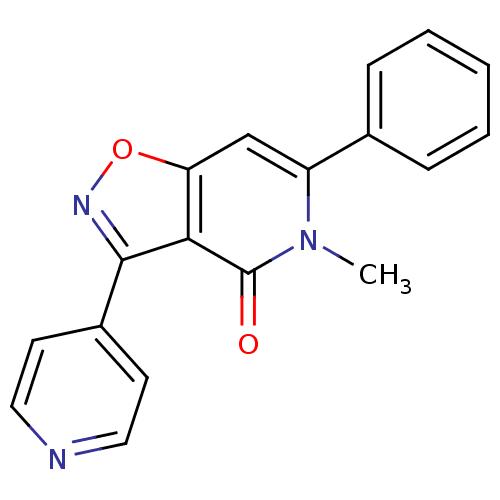
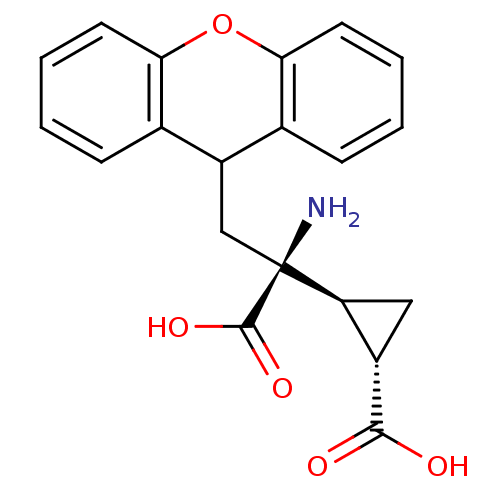
 3D Structure (crystal)
3D Structure (crystal)