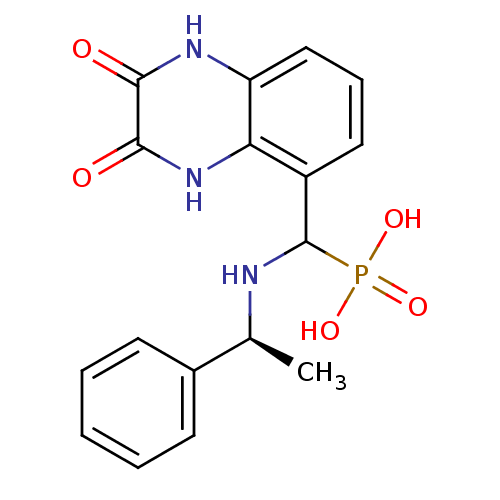
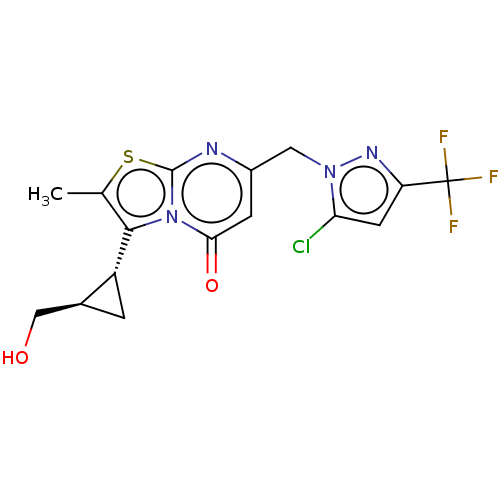
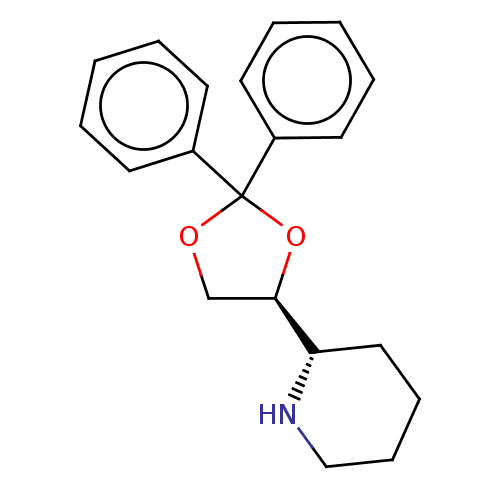
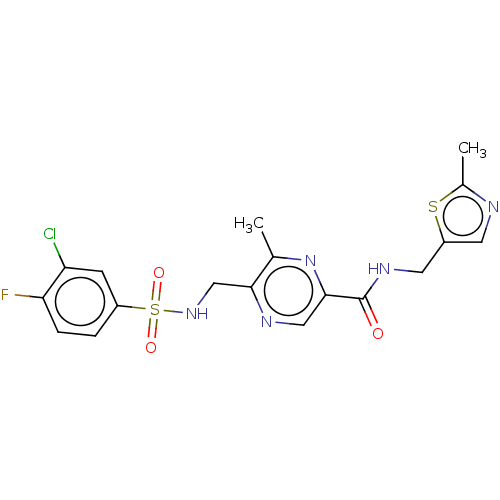
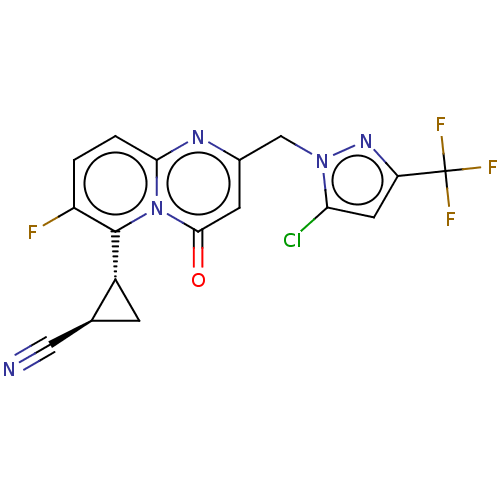
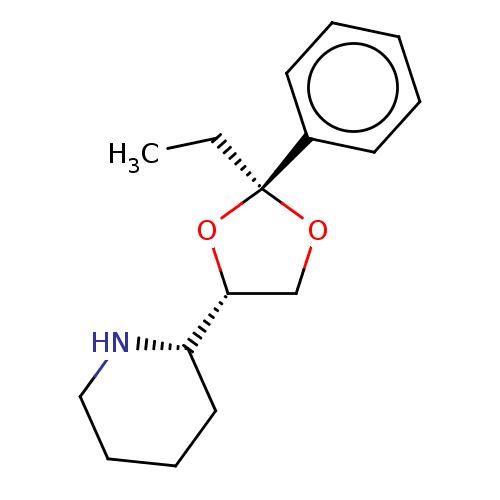
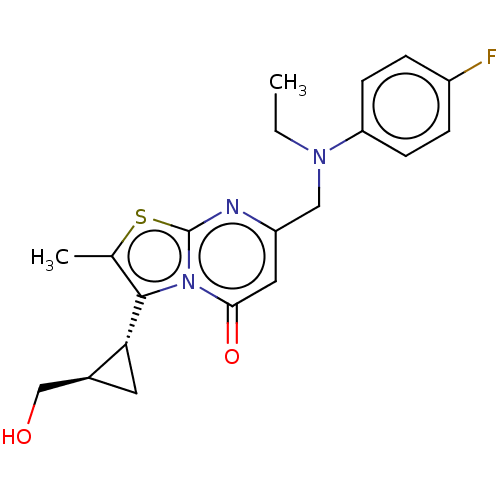
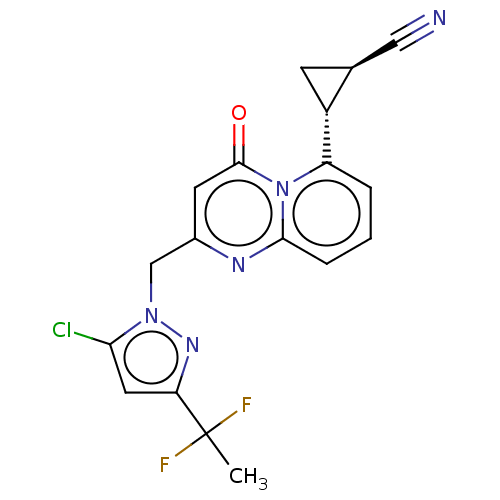
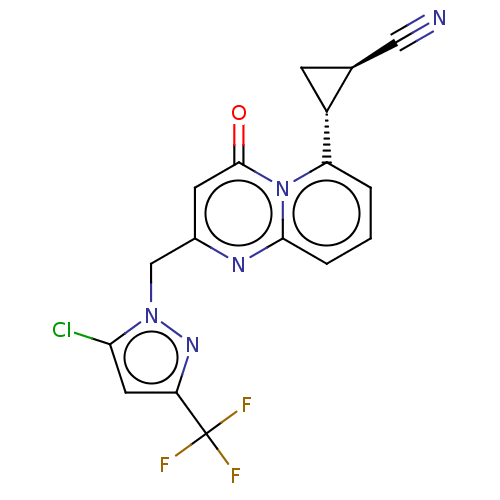
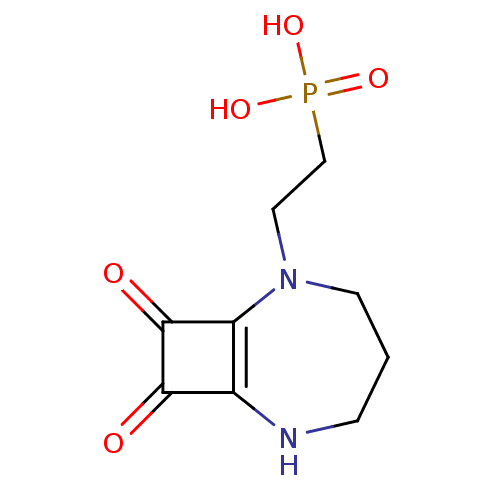
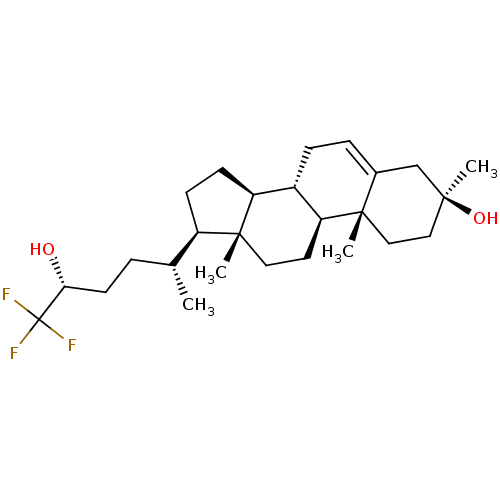
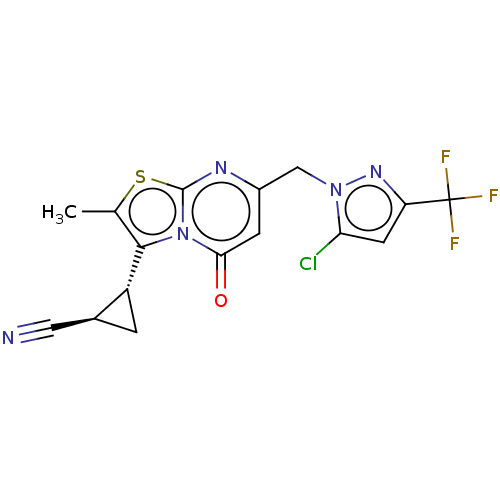
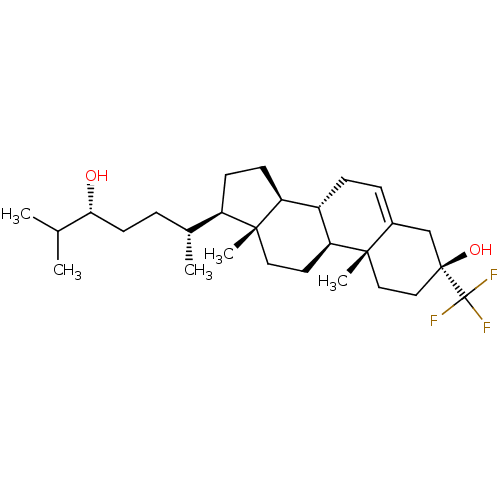
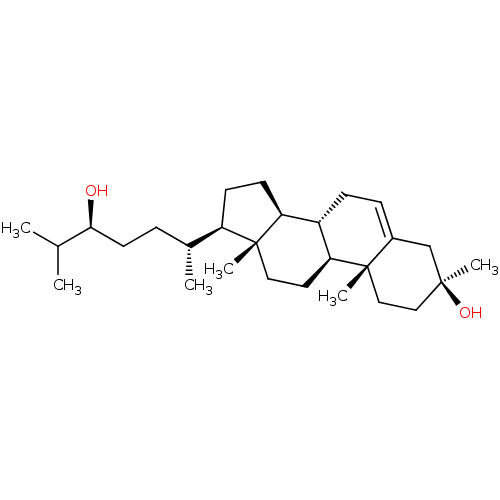
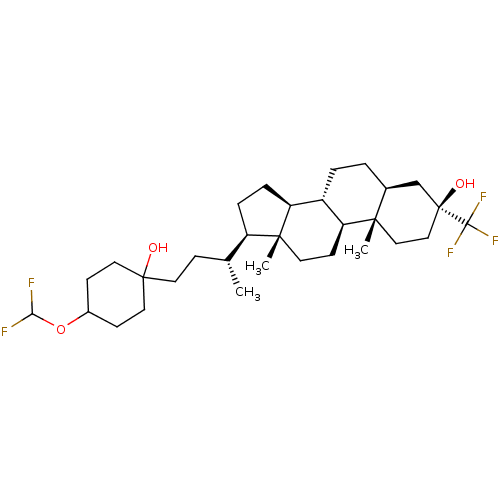
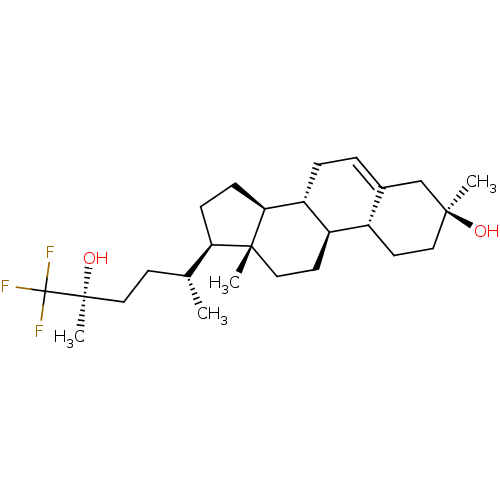
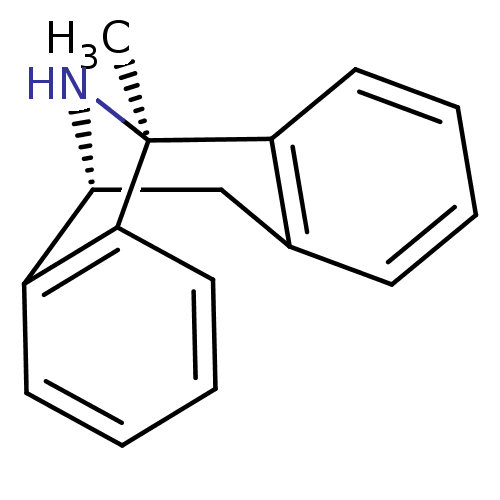Affinity DataIC50: 0.200nMAssay Description:Inhibitory activity against Xenopus laevis oocyte expressing 1A/2A heteromeric human NMDA (hNMDA) receptorMore data for this Ligand-Target Pair
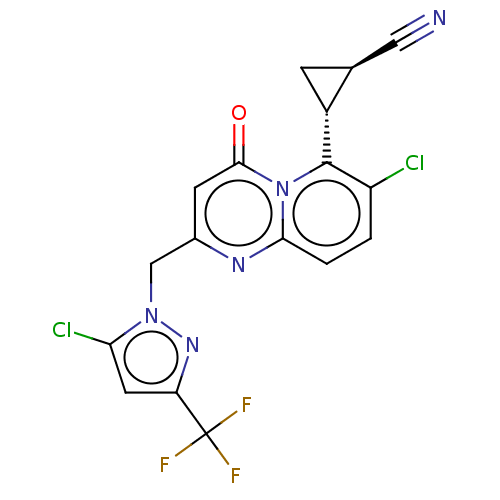
Affinity DataIC50: 0.950nMAssay Description:Antagonist activity at recombinant GluN1/GluN2A receptor (unknown origin) expressed in Xenopus laevis oocytes assessed as inhibition of glycine/gluta...More data for this Ligand-Target Pair
Affinity DataKi: 0.960nMAssay Description:Displacement of [3H]DCKA from NMDA receptor (unknown origin)More data for this Ligand-Target Pair
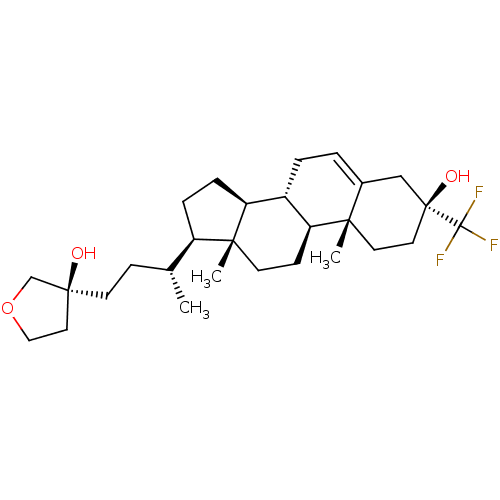
Affinity DataIC50: 1.10nMAssay Description:Inhibitory activity against Xenopus laevis oocyte expressing 1A/2A heteromeric human NMDA (hNMDA) receptorMore data for this Ligand-Target Pair
Affinity DataKi: 1.40nMAssay Description:Displacement of [3H]DCKA from NMDA receptor (unknown origin)More data for this Ligand-Target Pair
Affinity DataKi: 2.30nMAssay Description:Displacement of [3H]DCKA from NMDA receptor (unknown origin)More data for this Ligand-Target Pair
Affinity DataIC50: 2.60nMAssay Description:Displacement of [3H]L-689560 from NMDA receptor (unknown origin)More data for this Ligand-Target Pair
Affinity DataKi: 2.60nMAssay Description:Displacement of [3H]DCKA from NMDA receptor (unknown origin)More data for this Ligand-Target Pair
Affinity DataKi: 3nMAssay Description:Displacement of [3H]DCKA from NMDA receptor (unknown origin)More data for this Ligand-Target Pair
Affinity DataKi: 3.10nMAssay Description:Displacement of [3H]DCKA from NMDA receptor (unknown origin)More data for this Ligand-Target Pair
Affinity DataKi: 3.30nMAssay Description:Displacement of [3H]DCKA from NMDA receptor (unknown origin)More data for this Ligand-Target Pair
Affinity DataIC50: 4.20nMAssay Description:Inhibitory activity against Xenopus laevis oocyte expressing 1A/2A heteromeric human NMDA (hNMDA) receptorMore data for this Ligand-Target Pair
Affinity DataIC50: 4.20nMAssay Description:Antagonist activity at recombinant GluN1/GluN2A receptor (unknown origin) expressed in Xenopus laevis oocytes assessed as inhibition of glycine/gluta...More data for this Ligand-Target Pair
Affinity DataKi: 4.40nMAssay Description:Displacement of [3H]DCKA from NMDA receptor (unknown origin)More data for this Ligand-Target Pair
Affinity DataIC50: 4.40nMAssay Description:Displacement of [3H]L-689560 from NMDA receptor (unknown origin)More data for this Ligand-Target Pair
Affinity DataKi: 4.5nMAssay Description:Antagonist activity at NMDA receptor (unknown origin) assessed as inhibition constantMore data for this Ligand-Target Pair
Affinity DataKi: 5.10nMAssay Description:Displacement of [3H]DCKA from NMDA receptor (unknown origin)More data for this Ligand-Target Pair
Affinity DataKi: 6.60nMAssay Description:Displacement of [3H]DCKA from NMDA receptor (unknown origin)More data for this Ligand-Target Pair
Affinity DataKi: 9.60nMAssay Description:Displacement of [3H]DCKA from NMDA receptor (unknown origin)More data for this Ligand-Target Pair
Affinity DataKi: 9.90nMAssay Description:Displacement of [3H]DCKA from NMDA receptor (unknown origin)More data for this Ligand-Target Pair
Affinity DataIC50: 10nMAssay Description:Displacement of [3H]MDL from NMDA receptor (unknown origin)More data for this Ligand-Target Pair
Affinity DataIC50: 10nMAssay Description:Displacement of [3H]MDL from NMDA receptor (unknown origin)More data for this Ligand-Target Pair
Affinity DataIC50: 10nMAssay Description:Displacement of [3H]MDL from NMDA receptor (unknown origin)More data for this Ligand-Target Pair
Affinity DataEC50: 11nMAssay Description:Positive allosteric modulator activity at GluN2A receptor (unknown origin) by FLIPR assayMore data for this Ligand-Target Pair
Affinity DataIC50: 11nMAssay Description:Antagonist activity at recombinant GluN1/GluN2A receptor (unknown origin) expressed in Xenopus laevis oocytes assessed as inhibition of glycine/gluta...More data for this Ligand-Target Pair
Affinity DataEC50: 13nMAssay Description:Cells are transferred as suspension in serum-free medium to the QPatch HTX system and kept in the cell storage tank/stirrer during experiments. All s...More data for this Ligand-Target Pair
Affinity DataIC50: 16.2nMAssay Description:Cell Culture and plating: HEK293 cells expressing NR1/NR2A (Chantest, Cleveland, Ohio) were grown to 70-80% confluency as an adherent monolayer in st...More data for this Ligand-Target Pair
Affinity DataIC50: 18nMAssay Description:Inhibitory activity against Xenopus laevis oocyte expressing 1A/2A heteromeric human NMDA (hNMDA) receptorMore data for this Ligand-Target Pair
Affinity DataEC50: 18nMAssay Description:Positive allosteric modulation of GluN1/GluN2A NMDAR (unknown origin) expressed in Dox-inducible cells measured every 5 mins by BD calcium indicator ...More data for this Ligand-Target Pair
Affinity DataKi: 19nMAssay Description:Binding affinity to NMDA receptor (unknown origin)More data for this Ligand-Target Pair
Affinity DataIC50: 19.6nMAssay Description:Cell Culture and plating: HEK293 cells expressing NR1/NR2A (Chantest, Cleveland, Ohio) were grown to 70-80% confluency as an adherent monolayer in st...More data for this Ligand-Target Pair
Affinity DataEC50: 21nMAssay Description:Positive allosteric modulation of GluN1/GluN2A receptor (unknown origin) expressed in CHO cells assessed as increase in glutamate-induced calcium flu...More data for this Ligand-Target Pair
Affinity DataEC50: 21nMAssay Description:Positive allosteric modulation of GluN1/GluN2A NMDAR (unknown origin) expressed in Dox-inducible cells measured every 5 mins by BD calcium indicator ...More data for this Ligand-Target Pair
Affinity DataKi: 22nMAssay Description:Binding affinity to NMDA receptor (unknown origin)More data for this Ligand-Target Pair
Affinity DataEC50: 24nMAssay Description:Positive allosteric modulation of GluN1/GluN2A NMDAR (unknown origin) expressed in Dox-inducible cells measured every 5 mins by BD calcium indicator ...More data for this Ligand-Target Pair
Affinity DataEC50: 24nMAssay Description:Positive allosteric modulation of GluN1/GluN2A receptor (unknown origin) expressed in CHO cells assessed as increase in glutamate-induced calcium flu...More data for this Ligand-Target Pair
Affinity DataIC50: 29nMAssay Description:Antagonist activity at NR2A transfected in oocytesMore data for this Ligand-Target Pair
Affinity DataIC50: 30nMAssay Description:Binding affinity to NMDA receptor (unknown origin)More data for this Ligand-Target Pair
Affinity DataEC50: 30nMAssay Description:Positive allosteric modulation of recombinant human GluN1/GluN2A receptor stably expressed in HEK293 cells assessed as increase in glycine/L-glutamat...More data for this Ligand-Target Pair
Affinity DataEC50: 30nMAssay Description:Positive allosteric modulation of GluN1/GluN2A NMDAR (unknown origin) expressed in Dox-inducible cells measured every 5 mins by BD calcium indicator ...More data for this Ligand-Target Pair
Affinity DataEC50: 30nMAssay Description:Positive allosteric modulation of recombinant human GluN1/GluN2A receptor stably expressed in HEK293 cells assessed as increase in glycine/L-glutamat...More data for this Ligand-Target Pair
Affinity DataEC50: 30nMAssay Description:Positive allosteric modulation of GluN1/GluN2A receptor (unknown origin) expressed in CHO cells assessed as increase in glutamate-induced calcium flu...More data for this Ligand-Target Pair
Affinity DataEC50: 31.6nMAssay Description:The whole-cell patch-clamp technique was used to investigate the effects of positive allosteric modulating activity of test compounds on GlunN1/GluN2...More data for this Ligand-Target Pair
Affinity DataEC50: 33nMAssay Description:Positive allosteric modulation of recombinant human GluN1/GluN2A receptor stably expressed in HEK293 cells assessed as increase in glycine/L-glutamat...More data for this Ligand-Target Pair
Affinity DataEC50: 33.6nMAssay Description:The whole-cell patch-clamp technique was used to investigate the effects of positive allosteric modulating activity of test compounds on GlunN1/GluN2...More data for this Ligand-Target Pair
Affinity DataEC50: 34.5nMAssay Description:The whole-cell patch-clamp technique was used to investigate the effects of positive allosteric modulating activity of test compounds on GlunN1/GluN2...More data for this Ligand-Target Pair
Affinity DataEC50: 35.1nMAssay Description:The whole-cell patch-clamp technique was used to investigate the effects of positive allosteric modulating activity of test compounds on GlunN1/GluN2...More data for this Ligand-Target Pair
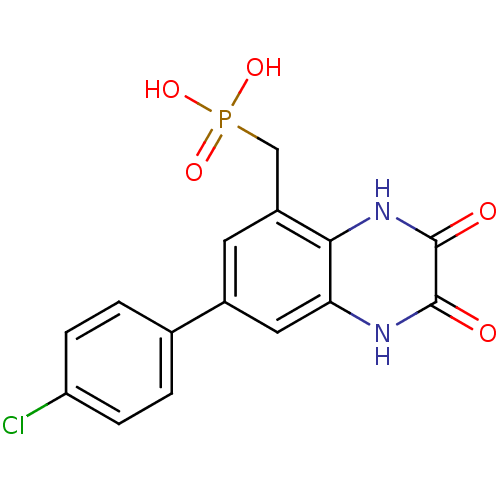

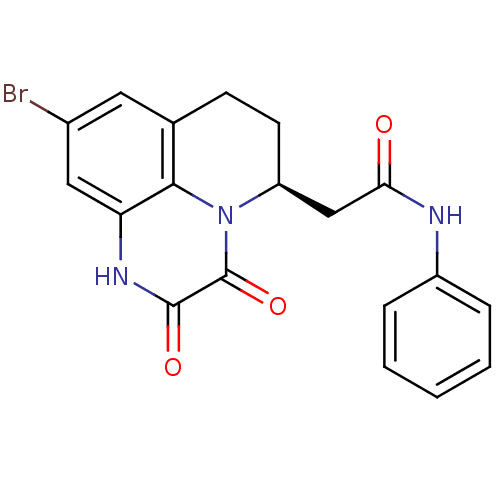
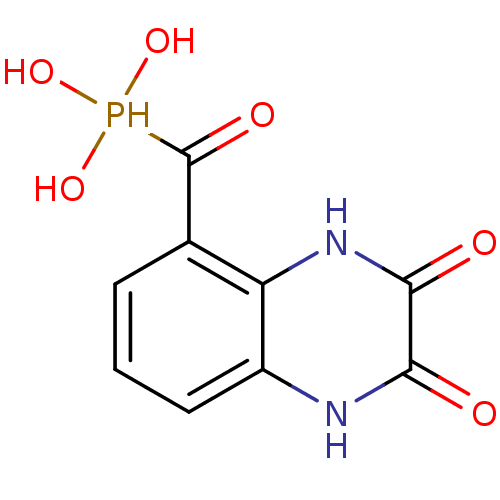
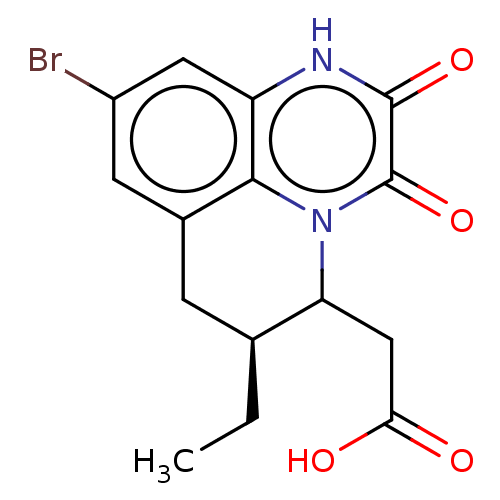

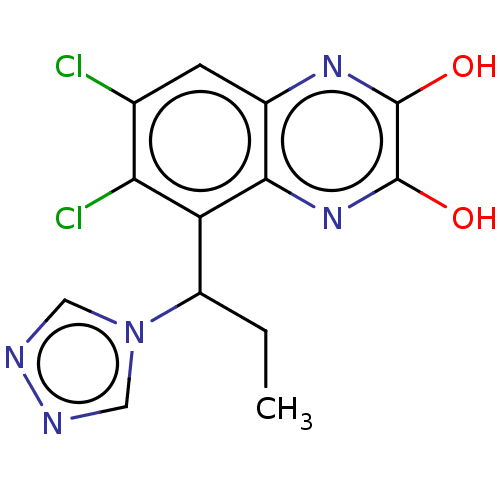

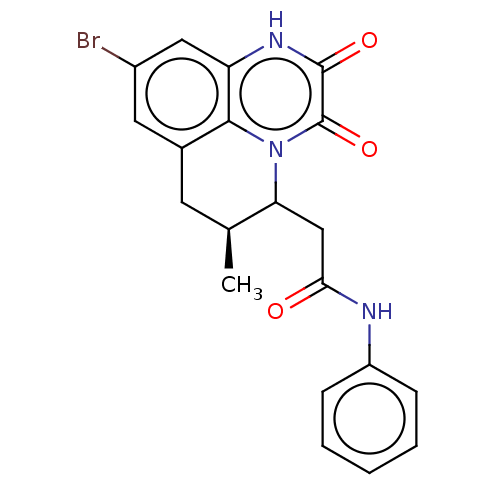
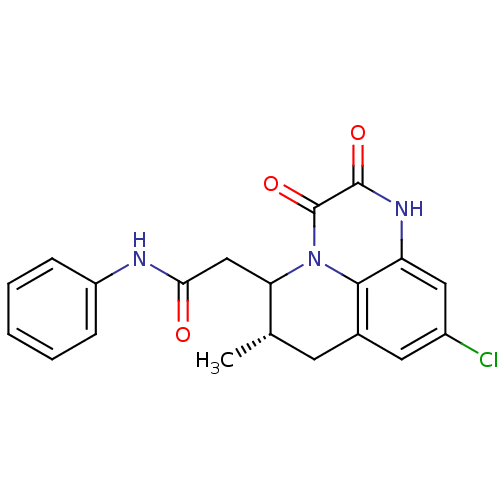
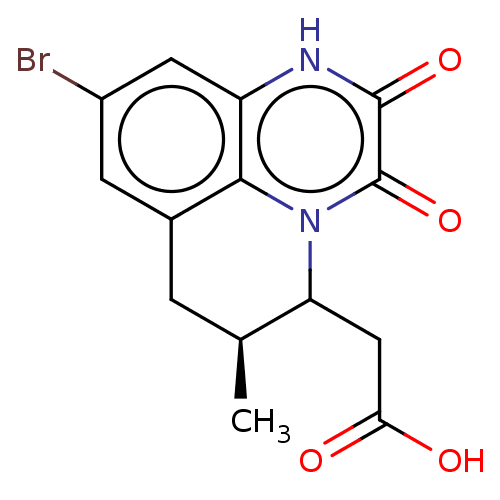
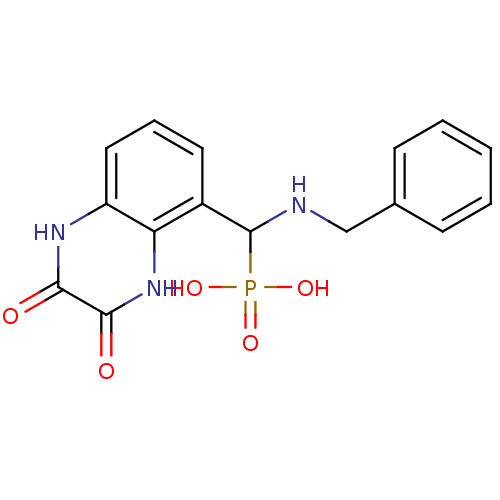

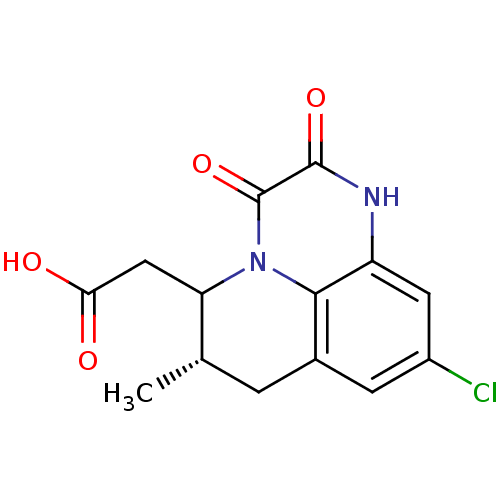
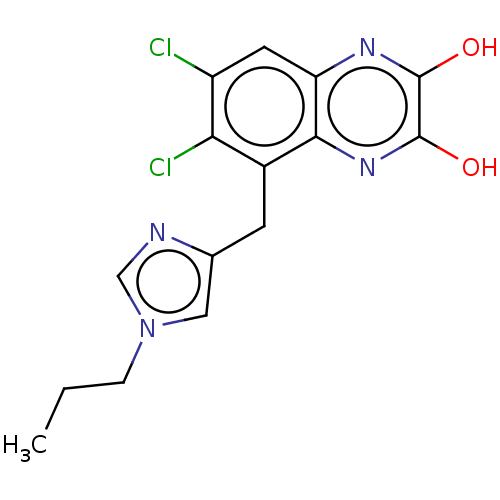
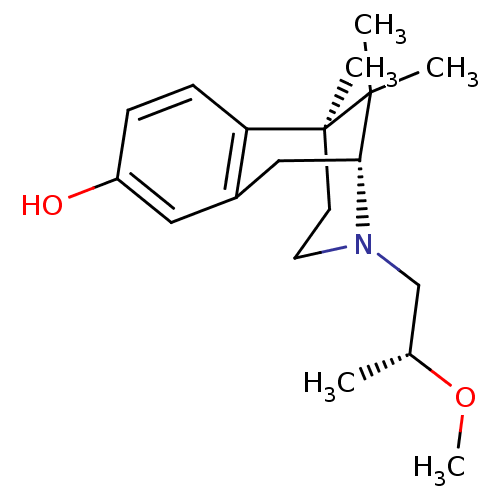
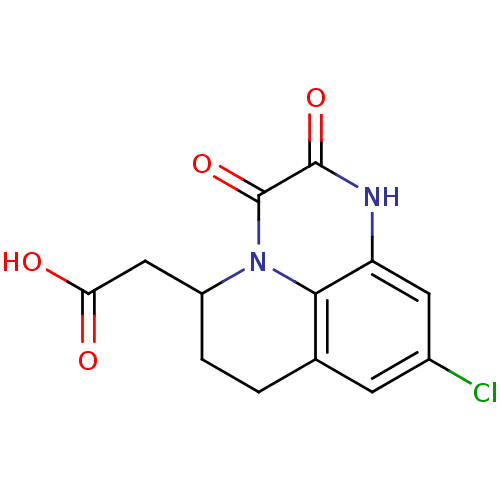
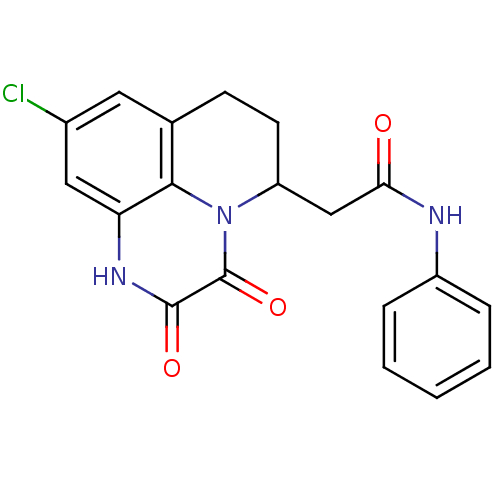
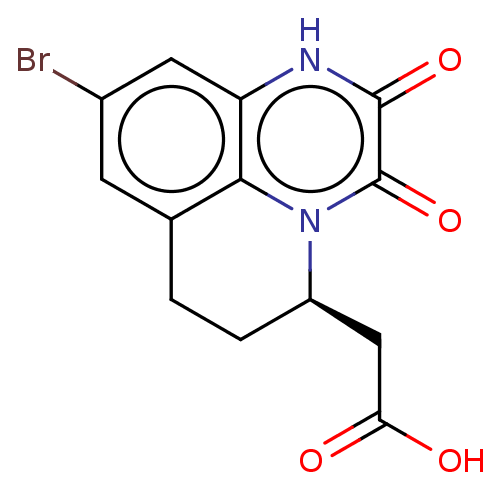
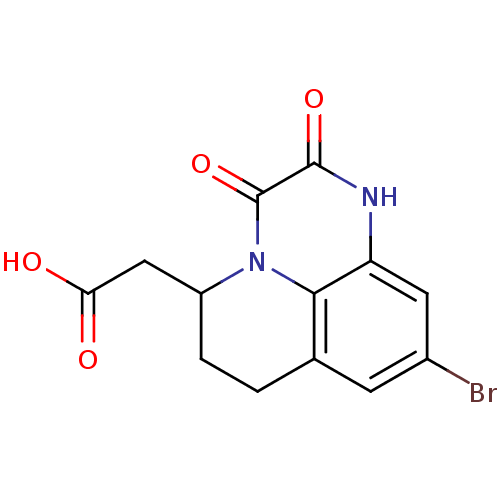
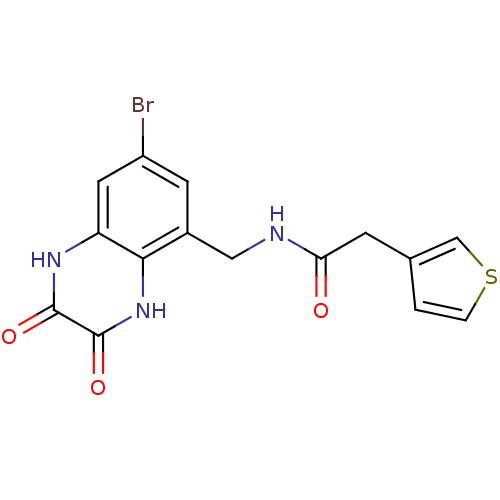
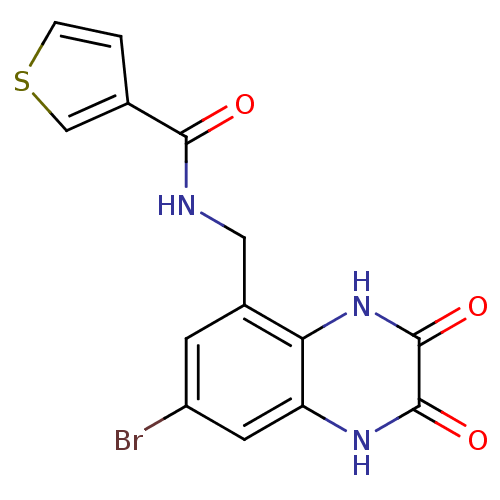
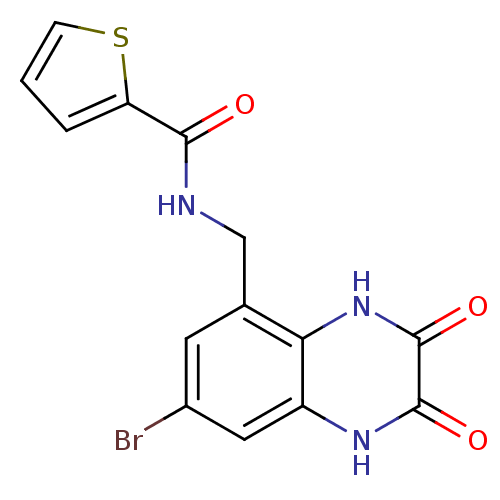
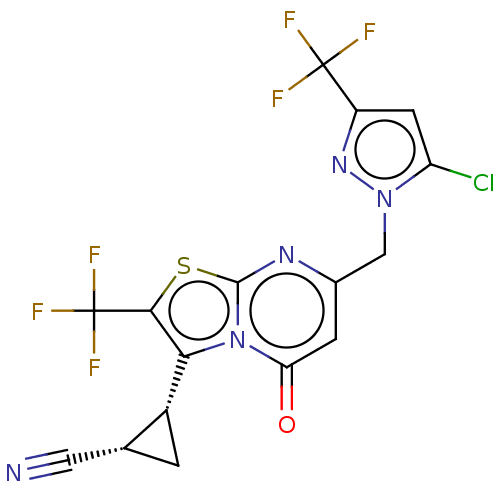

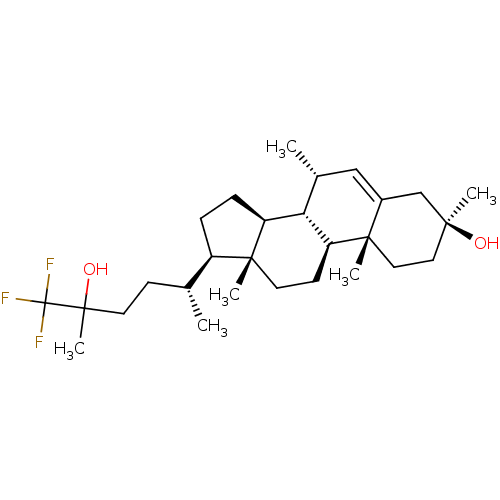
 3D Structure (crystal)
3D Structure (crystal)