 Lactate Lactic Acid BDBM23233 2-hydroxypropanoic acid
Lactate Lactic Acid BDBM23233 2-hydroxypropanoic acid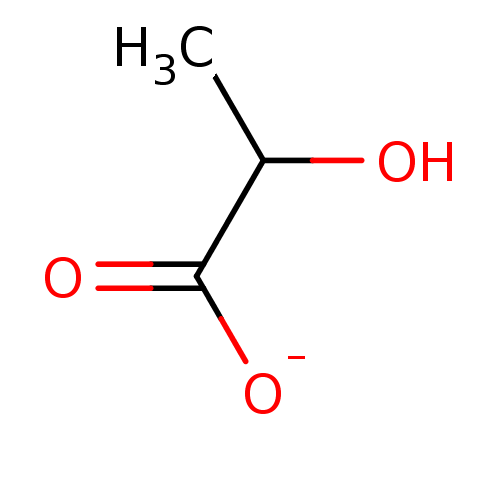 SODIUM LACTATE CHEMBL1357 Sodium; 2-hydroxy-propionate BDBM50159794
SODIUM LACTATE CHEMBL1357 Sodium; 2-hydroxy-propionate BDBM50159794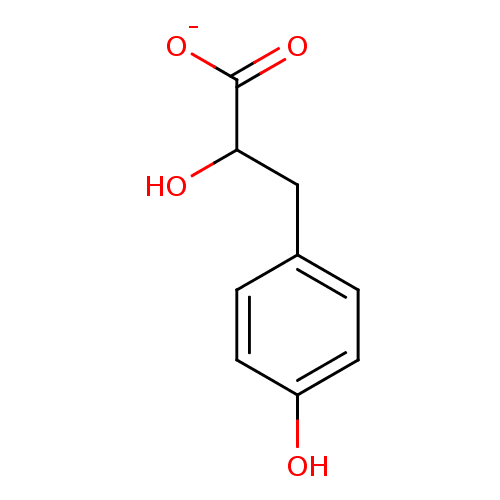 3-(4-Hydroxyphenyl)lactate 4-Hydroxyphenyllactate BDBM50269986 p-Hydroxyphenyllactate
3-(4-Hydroxyphenyl)lactate 4-Hydroxyphenyllactate BDBM50269986 p-Hydroxyphenyllactate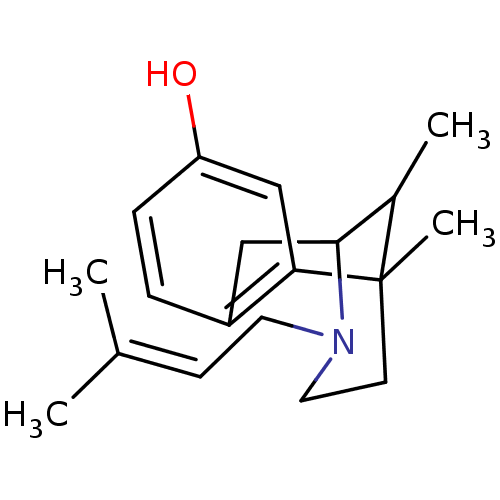 6,11-Dimethyl-3-(3-methyl-but-2-enyl)-1,2,3,4,5,6-hexahydro-2,6-methano-benzo[d]azocin-8-ol; lactate PENTAZOCINE (+) (Pentazocine) 6,11-Dimethyl-3-(3-methyl-but-2-enyl)-1,2 ,3,4,5,6-hexahydro-2,6-methano-benzo[d]azocin-8-ol PENTAZOCINE (-) 2-[3-cyclopropylmethyl-11-hydroxy-15-methoxy-(15S)-13-oxa-3-azahexacyclo[13.2.2.12,8.01,6.06,14.07,12]icosa-7,9,11-trien-16-yl]-3,3-dimethyl-2-butanol(Pentazocine) CHEMBL100116 6,11-Dimethyl-3-(3-methyl-but-2-enyl)-1,2,3,4,5,6-hexahydro-2,6-methano-benzo[d]azocin-8-ol (Pentazocine) 6,11-Dimethyl-3-(3-methyl-but-2-enyl)-1,2,3,4,5,6-hexahydro-2,6-methano-benzo[d]azocin-8-ol (pentazocine)2-{4-[(2,4-Diamino-pteridin-6-ylmethyl)-methyl-amino]-benzoylamino}-pentanedioic acid BDBM50032403 PENTAZOCINE 6,11-Dimethyl-3-(3-methyl-but-2-enyl)-1,2,3,4,5,6-hexahydro-2,6-methano-benzo[d]azocin-8-ol (pentazocine) 1,13-dimethyl-10-(3-methyl-2-butenyl)-(1R,9R,13R)-10-azatricyclo[7.3.1.02,7]trideca-2(7),3,5-trien-4-ol(Pentazocine)
6,11-Dimethyl-3-(3-methyl-but-2-enyl)-1,2,3,4,5,6-hexahydro-2,6-methano-benzo[d]azocin-8-ol; lactate PENTAZOCINE (+) (Pentazocine) 6,11-Dimethyl-3-(3-methyl-but-2-enyl)-1,2 ,3,4,5,6-hexahydro-2,6-methano-benzo[d]azocin-8-ol PENTAZOCINE (-) 2-[3-cyclopropylmethyl-11-hydroxy-15-methoxy-(15S)-13-oxa-3-azahexacyclo[13.2.2.12,8.01,6.06,14.07,12]icosa-7,9,11-trien-16-yl]-3,3-dimethyl-2-butanol(Pentazocine) CHEMBL100116 6,11-Dimethyl-3-(3-methyl-but-2-enyl)-1,2,3,4,5,6-hexahydro-2,6-methano-benzo[d]azocin-8-ol (Pentazocine) 6,11-Dimethyl-3-(3-methyl-but-2-enyl)-1,2,3,4,5,6-hexahydro-2,6-methano-benzo[d]azocin-8-ol (pentazocine)2-{4-[(2,4-Diamino-pteridin-6-ylmethyl)-methyl-amino]-benzoylamino}-pentanedioic acid BDBM50032403 PENTAZOCINE 6,11-Dimethyl-3-(3-methyl-but-2-enyl)-1,2,3,4,5,6-hexahydro-2,6-methano-benzo[d]azocin-8-ol (pentazocine) 1,13-dimethyl-10-(3-methyl-2-butenyl)-(1R,9R,13R)-10-azatricyclo[7.3.1.02,7]trideca-2(7),3,5-trien-4-ol(Pentazocine)
- Deck, LM; Royer, RE; Chamblee, BB; Hernandez, VM; Malone, RR; Torres, JE; Hunsaker, LA; Piper, RC; Makler, MT; Vander Jagt, DL Selective inhibitors of human lactate dehydrogenases and lactate dehydrogenase from the malarial parasite Plasmodium falciparum. J Med Chem 41: 3879-87 (1998)
- Thabault, L; Brisson, L; Brustenga, C; Martinez Gache, SA; Prévost, JRC; Kozlova, A; Spillier, Q; Liberelle, M; Benyahia, Z; Messens, J; Copetti, T; Sonveaux, P; Frédérick, R Interrogating the Lactate Dehydrogenase Tetramerization Site Using (Stapled) Peptides. J Med Chem 63: 4628-4643 (2020)
- Ding, J; Gumpena, R; Boily, MO; Caron, A; Chong, O; Cox, JH; Dumais, V; Gaudreault, S; Graff, AH; King, A; Knight, J; Oballa, R; Surendradoss, J; Tang, T; Wu, J; Lowther, WT; Powell, DA Dual Glycolate Oxidase/Lactate Dehydrogenase A Inhibitors for Primary Hyperoxaluria. ACS Med Chem Lett 12: 1116-1123 (2021)
- Christov, PP; Kim, K; Jana, S; Romaine, IM; Rai, G; Mott, BT; Allweil, AA; Lamers, A; Brimacombe, KR; Urban, DJ; Lee, TD; Hu, X; Lukacs, CM; Davies, DR; Jadhav, A; Hall, MD; Green, N; Moore, WJ; Stott, GM; Flint, AJ; Maloney, DJ; Sulikowski, GA; Waterson, AG Optimization of ether and aniline based inhibitors of lactate dehydrogenase. Bioorg Med Chem Lett 41: (2021)
- Choi, SR; Pradhan, A; Hammond, NL; Chittiboyina, AG; Tekwani, BL; Avery, MA Design, synthesis, and biological evaluation of Plasmodium falciparum lactate dehydrogenase inhibitors. J Med Chem 50: 3841-50 (2007)
- Kohlmann, A; Zech, SG; Li, F; Zhou, T; Squillace, RM; Commodore, L; Greenfield, MT; Lu, X; Miller, DP; Huang, WS; Qi, J; Thomas, RM; Wang, Y; Zhang, S; Dodd, R; Liu, S; Xu, R; Xu, Y; Miret, JJ; Rivera, V; Clackson, T; Shakespeare, WC; Zhu, X; Dalgarno, DC Fragment growing and linking lead to novel nanomolar lactate dehydrogenase inhibitors. J Med Chem 56: 1023-40 (2013)
- Fauber, BP; Dragovich, PS; Chen, J; Corson, LB; Ding, CZ; Eigenbrot, C; Labadie, S; Malek, S; Peterson, D; Purkey, HE; Robarge, K; Sideris, S; Ultsch, M; Wei, B; Yen, I; Yue, Q; Zhou, A Identification of 3,6-disubstituted dihydropyrones as inhibitors of human lactate dehydrogenase. Bioorg Med Chem Lett 24: 5683-7 (2014)
- Rupiani, S; Buonfiglio, R; Manerba, M; Di Ianni, L; Vettraino, M; Giacomini, E; Masetti, M; Falchi, F; Di Stefano, G; Roberti, M; Recanatini, M Identification of N-acylhydrazone derivatives as novel lactate dehydrogenase A inhibitors. Eur J Med Chem 101: 63-70 (2015)
- Maloney, DJ; Waterson, AG; Bantukallu, GR; Brimacombe, KR; Christov, P; Dang, CV; Darley-Usmar, V; Hu, X; Jadhav, A; Jana, S; Kim, K; Kouznetsova, JL; Moore, WJ; Mott, BT; Neckers, LM; Simeonov, A; Sulikowski, GA; Urban, DJ; Yang, SM Small molecule inhibitors of lactate dehydrogenase and methods of use thereof US Patent US10961200 (2021)
- Rai, G; Urban, DJ; Mott, BT; Hu, X; Yang, SM; Benavides, GA; Johnson, MS; Squadrito, GL; Brimacombe, KR; Lee, TD; Cheff, DM; Zhu, H; Henderson, MJ; Pohida, K; Sulikowski, GA; Dranow, DM; Kabir, M; Shah, P; Padilha, E; Tao, D; Fang, Y; Christov, PP; Kim, K; Jana, S; Muttil, P; Anderson, T; Kunda, NK; Hathaway, HJ; Kusewitt, DF; Oshima, N; Cherukuri, M; Davies, DR; Norenberg, JP; Sklar, LA; Moore, WJ; Dang, CV; Stott, GM; Neckers, L; Flint, AJ; Darley-Usmar, VM; Simeonov, A; Waterson, AG; Jadhav, A; Hall, MD; Maloney, DJ Pyrazole-Based Lactate Dehydrogenase Inhibitors with Optimized Cell Activity and Pharmacokinetic Properties. J Med Chem 63: 10984-11011 (2020)
- Hall, M; Urban, DJ; Knight, J; Holmes, R; Wood, KD; Waterson, A; Darley-Usmar, VM; Neckers, LM Treating primary or idiopathic hyperoxaluria with small molecule inhibitors of lactate dehydrogenase US Patent US11752138 (2023)
- Purkey, HE; Robarge, K; Chen, J; Chen, Z; Corson, LB; Ding, CZ; DiPasquale, AG; Dragovich, PS; Eigenbrot, C; Evangelista, M; Fauber, BP; Gao, Z; Ge, H; Hitz, A; Ho, Q; Labadie, SS; Lai, KW; Liu, W; Liu, Y; Li, C; Ma, S; Malek, S; O'Brien, T; Pang, J; Peterson, D; Salphati, L; Sideris, S; Ultsch, M; Wei, B; Yen, I; Yue, Q; Zhang, H; Zhou, A Cell Active Hydroxylactam Inhibitors of Human Lactate Dehydrogenase with Oral Bioavailability in Mice. ACS Med Chem Lett 7: 896-901 (2016)
- Choi, SR; Beeler, AB; Pradhan, A; Watkins, EB; Rimoldi, JM; Tekwani, B; Avery, MA Generation of oxamic acid libraries: antimalarials and inhibitors of Plasmodium falciparum lactate dehydrogenase. J Comb Chem 9: 292-300 (2007)
- Fauber, BP; Dragovich, PS; Chen, J; Corson, LB; Ding, CZ; Eigenbrot, C; Giannetti, AM; Hunsaker, T; Labadie, S; Liu, Y; Liu, Y; Malek, S; Peterson, D; Pitts, K; Sideris, S; Ultsch, M; VanderPorten, E; Wang, J; Wei, B; Yen, I; Yue, Q Identification of 2-amino-5-aryl-pyrazines as inhibitors of human lactate dehydrogenase. Bioorg Med Chem Lett 23: 5533-9 (2013)
- Dvorak, CA; Liu, C; Shelton, J; Kuei, C; Sutton, SW; Lovenberg, TW; Carruthers, NI Identification of Hydroxybenzoic Acids as Selective Lactate Receptor (GPR81) Agonists with Antilipolytic Effects. ACS Med Chem Lett 3: 637-639 (2012)
- Wei, B; Robarge, K; Labadie, SS; Chen, J; Corson, LB; DiPasquale, A; Dragovich, PS; Eigenbrot, C; Evangelista, M; Fauber, BP; Hitz, A; Hong, R; Lai, KW; Liu, W; Ma, S; Malek, S; O'Brien, T; Pang, J; Peterson, D; Salphati, L; Sampath, D; Sideris, S; Ultsch, M; Xu, Z; Yen, I; Yu, D; Yue, Q; Zhou, A; Purkey, HE Structure-based optimization of hydroxylactam as potent, cell-active inhibitors of lactate dehydrogenase. Bioorg Med Chem Lett 59: (2022)
- Ward, RA; Brassington, C; Breeze, AL; Caputo, A; Critchlow, S; Davies, G; Goodwin, L; Hassall, G; Greenwood, R; Holdgate, GA; Mrosek, M; Norman, RA; Pearson, S; Tart, J; Tucker, JA; Vogtherr, M; Whittaker, D; Wingfield, J; Winter, J; Hudson, K Design and synthesis of novel lactate dehydrogenase a inhibitors by fragment-based lead generation. J Med Chem 55: 3285-306 (2012)
- Rai, G; Brimacombe, KR; Mott, BT; Urban, DJ; Hu, X; Yang, SM; Lee, TD; Cheff, DM; Kouznetsova, J; Benavides, GA; Pohida, K; Kuenstner, EJ; Luci, DK; Lukacs, CM; Davies, DR; Dranow, DM; Zhu, H; Sulikowski, G; Moore, WJ; Stott, GM; Flint, AJ; Hall, MD; Darley-Usmar, VM; Neckers, LM; Dang, CV; Waterson, AG; Simeonov, A; Jadhav, A; Maloney, DJ Discovery and Optimization of Potent, Cell-Active Pyrazole-Based Inhibitors of Lactate Dehydrogenase (LDH). J Med Chem 60: 9184-9204 (2017)
- Chan, BS; Endo, S; Kanai, N; Schuster, VL Identification of lactate as a driving force for prostanoid transport by prostaglandin transporter PGT. Am J Physiol Renal Physiol 282: 1097-102 (2002)
- De Leo, M; Peruzzi, L; Granchi, C; Tuccinardi, T; Minutolo, F; De Tommasi, N; Braca, A Constituents of Polygala flavescens ssp. flavescens and Their Activity as Inhibitors of Human Lactate Dehydrogenase. J Nat Prod 80: 2077-2087 (2017)
- Zhou, Y; Tao, P; Wang, M; Xu, P; Lu, W; Lei, P; You, Q Development of novel human lactate dehydrogenase A inhibitors: High-throughput screening, synthesis, and biological evaluations. Eur J Med Chem 177: 105-115 (2019)
- Di Magno, L; Coluccia, A; Bufano, M; Ripa, S; La Regina, G; Nalli, M; Di Pastena, F; Canettieri, G; Silvestri, R; Frati, L Discovery of novel human lactate dehydrogenase inhibitors: Structure-based virtual screening studies and biological assessment. Eur J Med Chem 240: (2022)
- Dragovich, PS; Fauber, BP; Corson, LB; Ding, CZ; Eigenbrot, C; Ge, H; Giannetti, AM; Hunsaker, T; Labadie, S; Liu, Y; Malek, S; Pan, B; Peterson, D; Pitts, K; Purkey, HE; Sideris, S; Ultsch, M; VanderPorten, E; Wei, B; Xu, Q; Yen, I; Yue, Q; Zhang, H; Zhang, X Identification of substituted 2-thio-6-oxo-1,6-dihydropyrimidines as inhibitors of human lactate dehydrogenase. Bioorg Med Chem Lett 23: 3186-94 (2013)
- Sharma, H; Mondal, S; Urquiza, U; Esparza, C; Bartlett, S; Santa-Pinter, L; Hill, H; White, M; Sharma, P; Luckett-Chastain, L; Cooper, A; Rasel, M; Gao, P; Battaile, KP; Shukla, SK; Lovell, S; Ihnat, MA Synthesis and biological characterization of an orally bioavailable lactate dehydrogenase-A inhibitor against pancreatic cancer. Eur J Med Chem 275:
- Maloney, DJ; Waterson, AG; Bantukallu, GR; Brimacombe, KR; Christov, P; Dang, CV; Darley-Usmar, VM; Hall, M; Hu, X; Jadhav, A; Jana, S; Kim, K; Moore, WJ; Mott, BT; Neckers, LM; Simeonov, A; Sulikowski, GA; Urban, DJ; Yang, SM 1 H-pyrazol-1-yl-thiazoles as inhibitors of lactate dehydrogenase and methods of use thereof US Patent US10954228 (2021)
- Cao, W; Fang, L; Teng, S; Chen, H; Wang, Z Computer-aided discovery and biological characterization of human lactate dehydrogenase 5 inhibitors with anti-osteosarcoma activity. Bioorg Med Chem Lett 28: 2229-2233 (2018)
- Sun, N; Kabir, M; Lee, Y; Xie, L; Hu, X; Velez, J; Chen, X; Kaniskan, HÜ; Jin, J Discovery of the First Lactate Dehydrogenase Proteolysis Targeting Chimera Degrader for the Treatment of Pancreatic Cancer. J Med Chem 66: 596-610 (2023)
- Wang, F; Zhao, Q; Liu, J; Wang, Z; Kong, D Identification of human lactate dehydrogenase A inhibitors with anti-osteosarcoma activity through cell-based phenotypic screening. Bioorg Med Chem Lett 30: (2020)
- Dragovich, PS; Fauber, BP; Boggs, J; Chen, J; Corson, LB; Ding, CZ; Eigenbrot, C; Ge, H; Giannetti, AM; Hunsaker, T; Labadie, S; Li, C; Liu, Y; Liu, Y; Ma, S; Malek, S; Peterson, D; Pitts, KE; Purkey, HE; Robarge, K; Salphati, L; Sideris, S; Ultsch, M; VanderPorten, E; Wang, J; Wei, B; Xu, Q; Yen, I; Yue, Q; Zhang, H; Zhang, X; Zhou, A Identification of substituted 3-hydroxy-2-mercaptocyclohex-2-enones as potent inhibitors of human lactate dehydrogenase. Bioorg Med Chem Lett 24: 3764-71 (2014)
- Rani, R; Kumar, V Recent Update on Human Lactate Dehydrogenase Enzyme 5 (hLDH5) Inhibitors: A Promising Approach for Cancer Chemotherapy. J Med Chem 59: 487-96 (2016)
- Cameron, A; Read, J; Tranter, R; Winter, VJ; Sessions, RB; Brady, RL; Vivas, L; Easton, A; Kendrick, H; Croft, SL; Barros, D; Lavandera, JL; Martin, JJ; Risco, F; García-Ochoa, S; Gamo, FJ; Sanz, L; Leon, L; Ruiz, JR; Gabarró, R; Mallo, A; Gómez de las Heras, F Identification and activity of a series of azole-based compounds with lactate dehydrogenase-directed anti-malarial activity. J Biol Chem 279: 31429-39 (2004)
- Draoui, N; Schicke, O; Fernandes, A; Drozak, X; Nahra, F; Dumont, A; Douxfils, J; Hermans, E; Dogné, JM; Corbau, R; Marchand, A; Chaltin, P; Sonveaux, P; Feron, O; Riant, O Synthesis and pharmacological evaluation of carboxycoumarins as a new antitumor treatment targeting lactate transport in cancer cells. Bioorg Med Chem 21: 7107-17 (2013)
- Chen, CY; Feng, Y; Chen, JY; Deng, H Identification of a potent inhibitor targeting human lactate dehydrogenase A and its metabolic modulation for cancer cell line. Bioorg Med Chem Lett 26: 72-5 (2016)
- Moya-Garzon, MD; Rodriguez-Rodriguez, B; Martin-Higueras, C; Franco-Montalban, F; Fernandes, MX; Gomez-Vidal, JA; Pey, AL; Salido, E; Diaz-Gavilan, M New salicylic acid derivatives, double inhibitors of glycolate oxidase and lactate dehydrogenase, as effective agents decreasing oxalate production. Eur J Med Chem 237: (2022)
- Labadie, S; Dragovich, PS; Chen, J; Fauber, BP; Boggs, J; Corson, LB; Ding, CZ; Eigenbrot, C; Ge, H; Ho, Q; Lai, KW; Ma, S; Malek, S; Peterson, D; Purkey, HE; Robarge, K; Salphati, L; Sideris, S; Ultsch, M; VanderPorten, E; Wei, B; Xu, Q; Yen, I; Yue, Q; Zhang, H; Zhang, X; Zhou, A Optimization of 5-(2,6-dichlorophenyl)-3-hydroxy-2-mercaptocyclohex-2-enones as potent inhibitors of human lactate dehydrogenase. Bioorg Med Chem Lett 25: 75-82 (2015)
- Granchi, C; Roy, S; Giacomelli, C; Macchia, M; Tuccinardi, T; Martinelli, A; Lanza, M; Betti, L; Giannaccini, G; Lucacchini, A; Funel, N; León, LG; Giovannetti, E; Peters, GJ; Palchaudhuri, R; Calvaresi, EC; Hergenrother, PJ; Minutolo, F Discovery of N-hydroxyindole-based inhibitors of human lactate dehydrogenase isoform A (LDH-A) as starvation agents against cancer cells. J Med Chem 54: 1599-612 (2011)
- Nair, RN; Mishra, JK; Li, F; Tortosa, M; Yang, C; Doherty, JR; Cameron, M; Cleveland, JL; Roush, WR; Bannister, TD Exploiting the co-reliance of tumours upon transport of amino acids and lactate: Gln and Tyr conjugates of MCT1 inhibitors. Medchemcomm 7: 900-905 (2016)
- Xiang, S; Huang, D; He, Q; Li, J; Tam, KY; Zhang, SL; He, Y Development of dual inhibitors targeting pyruvate dehydrogenase kinases and human lactate dehydrogenase A: High-throughput virtual screening, synthesis and biological validation. Eur J Med Chem 203: (2020)
- He, S; Wang, Q Discovery of human lactate dehydrogenase 5 inhibitors (hLDH5) with anti-lung cancer activity through an in silico method and biological validation. Bioorg Med Chem Lett 29: 2459-2463 (2019)
- Sova, M; Cadez, G; Turk, S; Majce, V; Polanc, S; Batson, S; Lloyd, AJ; Roper, DI; Fishwick, CW; Gobec, S Design and synthesis of new hydroxyethylamines as inhibitors of D-alanyl-D-lactate ligase (VanA) and D-alanyl-D-alanine ligase (DdlB). Bioorg Med Chem Lett 19: 1376-9 (2009)
- Bai, Y; He, X; Bai, Y; Sun, Y; Zhao, Z; Chen, X; Li, B; Xie, J; Li, Y; Jia, P; Meng, X; Zhao, Y; Ding, Y; Xiao, C; Wang, S; Yu, J; Liao, S; Zhang, Y; Zhu, Z; Zhang, Q; Zhao, Y; Qin, F; Zhang, Y; Wei, X; Zeng, M; Liang, J; Cuan, Y; Shan, G; Fan, TP; Wu, B; Zheng, X Polygala tenuifolia-Acori tatarinowii herbal pair as an inspiration for substituted cinnamic α-asaronol esters: Design, synthesis, anticonvulsant activity, and inhibition of lactate dehydrogenase study. Eur J Med Chem 183: (2019)
- Cui, W; Lv, W; Qu, Y; Ma, R; Wang, YW; Xu, YJ; Wu, D; Chen, X Discovery of 2-((3-cyanopyridin-2-yl)thio)acetamides as human lactate dehydrogenase A inhibitors to reduce the growth of MG-63 osteosarcoma cells: Virtual screening and biological validation. Bioorg Med Chem Lett 26: 3984-7 (2016)
- ChEMBL_835931 (CHEMBL2077069) TP_TRANSPORTER: inhibition of L-Lactate uptake (L-Lactate:30mM) in Xenopus laevis oocytes
- ChEMBL_989588 (CHEMBL2444903) Inhibition of MCT4-mediated [14C]-lactate uptake in human SiHa cells in lactate-containing medium assessed as remaining lactate concentration in culture medium after 12 mins by liquid scintillation counting
- ChEMBL_989589 (CHEMBL2444904) Inhibition of MCT4-mediated [14C]-lactate uptake in human SiHa cells in lactate-containing medium assessed as remaining lactate concentration in culture medium after 24 hrs by liquid scintillation counting
- ChEMBL_1498543 (CHEMBL3584796) Inhibition of MCT1-mediated lactate transport in rat RBE4 cells incubated for 15 mins by [14C]-lactate uptake assay
- ChEMBL_1513323 (CHEMBL3611227) Inhibition of lactate dehydrogenase-A in human Raji cells assessed as reduction of intracellular lactate level after 3 hrs
- Enzyme Assay Enzyme assay using lactate dehydrogenase A (LDHA).
- ChEMBL_96432 (CHEMBL706379) inhibitory activity against Human Lactate Dehydrogenase (LDH-H)
- ChEMBL_96433 (CHEMBL706380) inhibitory activity against Human Lactate Dehydrogenase (LDH-M)
- ChEMBL_96560 (CHEMBL709293) Apparent inhibition constant against mammalian lactate dehydrogenase (LDH)
- ChEBML_96562 In vitro inhibitory activity against pig heart lactate dehydrogenase (LDH)
- ChEMBL_838262 (CHEMBL2076210) TP_TRANSPORTER: inhibition of lactate uptake in Xenopus laevis oocytes
- ChEMBL_838272 (CHEMBL2076220) TP_TRANSPORTER: inhibition of lactate uptake in Xenopus laevis oocytes
- ChEMBL_2017128 (CHEMBL4670706) Inhibition of LDHA in human A673 cells assessed as decrease in lactate production incubated for 2 hrs by lactate oxidase-coupled fluorescence based assay
- ChEMBL_2434372 Inhibition of MCT1 in human SiHa cells by lactate reporter assay
- ChEMBL_2434387 Inhibition of MCT1 in human HAP1 cells by lactate transport assay
- ChEMBL_2434388 Inhibition of MCT4 in human HAP1 cells by lactate transport assay
- ChEMBL_2017130 (CHEMBL4670708) Inhibition of LDHA in human MIA PaCa-2 cells assessed as decrease in lactate production incubated for 2 hrs by lactate oxidase-coupled fluorescence based assay
- ChEMBL_160093 (CHEMBL767695) In vivo inhibion of pyruvate dehydrogenase kinase, increased oxidation of lactate
- ChEMBL_2434376 Inhibition of MCT4 in human EVSA-T cells by lactate transport assay
- ChEMBL_2434384 Inhibition of MCT1 in human Hs-578T cells by lactate transport assay
- ChEMBL_2113514 (CHEMBL4822364) Activation of PKM2 (unknown origin) by pyruvate kinase-lactate dehydrogenase coupled assay
- ChEMBL_2434371 Inhibition of MCT4 in human MDA-MB-231 cells by lactate transport assay
- ChEMBL_566219 (CHEMBL963300) Inhibition of Enterococcus faecium VanA by pyruvate kinase/lactate dehydrogenase coupled assay
- ChEMBL_818701 (CHEMBL2033190) Inhibition of Eg5 ATPase activity by pyruvate kinase/lactate dehydrogenase-linked assay
- ChEBML_1448600 Inhibition of methicillin-resistant Staphylococcus aureus pyruvate kinase by coupled lactate dehydrogenase continuous assay
- ChEMBL_1679402 (CHEMBL4029679) Inhibition of recombinant PIM1 (unknown origin) by pyruvate kinase/lactate dehydrogenase-coupled assay
- ChEMBL_1928158 (CHEMBL4431230) Inhibition of LDHA in human MiPaca2 cells assessed as inhibition of lactate production
- ChEMBL_483709 (CHEMBL954608) Inhibition of HIV1 protease-mediated proteolysis of rabbit muscle lactate dehydrogenase by spectrophotometry
- ChEMBL_818702 (CHEMBL2033191) Inhibition of human Kif5A ATPase activity by pyruvate kinase/lactate dehydrogenase-linked assay
- ChEMBL_818703 (CHEMBL2033192) Inhibition of human Kif5C ATPase activity by pyruvate kinase/lactate dehydrogenase-linked assay
- ChEMBL_1461346 (CHEMBL3396196) Inhibition of Mycobacterium tuberculosis thymidylate kinase activity by pyruvate kinase-lactate dehydrogenase coupled assay
- ChEMBL_824371 (CHEMBL2044661) Inhibition of basal ATPase activity of Eg5 by coupled pyruvate kinase/lactate dehydrogenase assay
- ChEMBL_1784112 (CHEMBL4255629) Inhibition of LDH (unknown origin) assessed as reduction in lactate production by cell-based assay
- ChEMBL_65923 (CHEMBL678318) Inhibitory concentration towards rat mitochondrial F1F0 ATP hydrolase using a pyruvate kinase / lactate dehydrogenase system
- Enzyme Assay The enzyme was assayed at pH 7.0 using the pyruvate kinase/lactate dehydroenase coupling system.
- ChEMBL_1475335 (CHEMBL3425232) Inhibition of PFKFB3 in human A549 cells assessed as decrease in lactate secretion after 4 hrs
- ChEMBL_1500621 (CHEMBL3587751) Binding affinity to rabbit hind leg muscle SERCA1a by pyruvate kinase and lactate dehydrogenase coupled assay
- ChEMBL_1618019 (CHEMBL3860188) Inhibition of basal ATPase activity of human CENP-E by pyruvate kinase/lactate dehydrogenase-linked assay
- ChEMBL_817613 (CHEMBL2027261) Inhibition of p38beta using KRELVEPLTPSGEAPNQALLR as substrate for 10 mins by lactate dehydrogenase-coupled spectrophotometric assay
- ChEMBL_1499594 (CHEMBL3584540) Inhibition of PAK1 (unknown origin) using Syntide2 peptide substrate by pyruvate kinase and lactate dehydrogenase coupled assay
- ChEMBL_1513380 (CHEMBL3611420) Inhibition of PAK1 (unknown origin) using Syntide2 peptide as substrate by pyruvate kinase/lactate dehydrogenase coupled assay
- ChEMBL_1513384 (CHEMBL3611424) Inhibition of PAK5 (unknown origin) using peptide 7 as substrate by pyruvate kinase/lactate dehydrogenase coupled assay
- ChEMBL_1513385 (CHEMBL3611425) Inhibition of PAK6 (unknown origin) using peptide 7 as substrate by pyruvate kinase/lactate dehydrogenase coupled assay
- ChEMBL_1618032 (CHEMBL3860201) Inhibition of microtubule-stimulated ATPase activity of human CENP-E by pyruvate kinase/lactate dehydrogenase-linked assay
- ChEMBL_1721609 (CHEMBL4136609) Inhibition of recombinant SRC (unknown origin) using polyE4Y as substrate by pyruvate kinase-lactate dehydrogenase coupled assay
- ChEMBL_817607 (CHEMBL2027255) Inhibition of p38alpha kinase using KRELVEPLTPSGEAPNQALLR as substrate for 20 mins by lactate dehydrogenase-coupled spectrophotometric assay
- ChEMBL_1513325 (CHEMBL3611229) Inhibition of human liver purified lactate dehydrogenase-A using pyruvate as substrate assessed as disappearance of NADH fluorescence
- ChEMBL_1513326 (CHEMBL3611230) Inhibition of human liver purified lactate dehydrogenase-A using NADH as substrate assessed as disappearance of NADH fluorescence
- ChEMBL_1777996 (CHEMBL4234988) Inhibition of MT-stimulated EG5 ATPase activity (unknown origin) by pyruvate kinase/lactate dehydrogenase enzyme coupled photometric assay
- ChEMBL_1721607 (CHEMBL4136607) Inhibition of recombinant GSK3beta (unknown origin) using HSSPHQ(Sp)EDEEE as substrate by pyruvate kinase-lactate dehydrogenase coupled assay
- ChEMBL_1721608 (CHEMBL4136608) Inhibition of recombinant CDK2/cyclin A (unknown origin) using MAHHRSPRKRAKKK as substrate by pyruvate kinase-lactate dehydrogenase coupled assay
- ChEMBL_2270628 Inhibition of PFKFB3 in human HCT-116 cells assessed as inhibition of glucose induced lactate production by absorbance based analysis
- ChEMBL_2306764 Inhibition of MCT1 in human K562 cells assessed as inhibition of lactate efflux incubated for 1 hr by FLIPR method
- ChEMBL_616299 (CHEMBL1102102) Inhibition of GSK3-beta assessed as NADH level after 10 mins by pyruvate kinase/lactate dehydrogenase coupled spectrophotometric assay
- ChEMBL_617338 (CHEMBL1101849) Inhibition of ATPase activity of human recombinant EG5 assessed as ATP hydrolysis by pyruvate kinase-lactate dehydrogenase coupled assay
- ChEMBL_877484 (CHEMBL2183638) Inhibition of recombinant Plasmodium falciparum TMPK using TMP as substrate by pyruvate kinase and lactate dehydrogenase enzyme coupled assay
- Assay 4: MCT1-Mediated Lactate Transport in BT20 Breast Cancer Cells MCT1 activity may be measured using BT-20 breast cancer cells that express high native levels of MCT1, but do not express MCT4 and are known to those with skill in the art. Preparation of BCECF loaded cells are as described for Assay 1. Lactate transport assay is as described for Assay 1, except 10 mM L-lactate (rather than 50 mM) is added.
- MCT1-Mediated Lactate Transport in BT20 Breast Cancer Cells (Assay 4) MCT1 activity may be measured using BT-20 breast cancer cells that express high native levels of MCT1, but do not express MCT4 and are known to those with skill in the art. Preparation of BCECF loaded cells are as described for Assay 1. Lactate transport assay is as described for Assay 1, except 10 mM L-lactate (rather than 50 mM) is added.
- MCT1-Mediated Lactate Transport in BT20 Breast Cancer Cells MCT1 activity may be measured using BT-20 breast cancer cells that express high native levels of MCT1, but do not express MCT4 and are known to those with skill in the art. Preparation of BCECF loaded cells are as described for Assay 1. Lactate transport assay is as described for Assay 1, except 10 mM L-lactate (rather than 50 mM) is added.
- ChEMBL_2274354 Inhibition of MCT1 (unknown origin) assessed as reduction in [14C]lactate uptake incubated for 20 mins by liquid scintillation counting analysis
- ChEMBL_2274355 Inhibition of MCT4 (unknown origin) assessed as reduction in [14C]lactate uptake incubated for 20 mins by liquid scintillation counting analysis
- ChEMBL_2306763 Inhibition of MCT4 in human NCI-H358 cells assessed as inhibition of lactate efflux incubated for 1 hr by FLIPR method
- Lactate Dehydrogenase Inhibition Assay An LDH enzymatic assay developed for high throughput format was used. The dehydrogenase reaction was run in the lactate to pyruvate direction and coupled with the ability of diaphorase to reduce p-iodonitrotetrazolium violet using the NADH generated in the conversion of lactate to pyruvate. The progression of the coupling reaction was monitored as the increase of absorbance at 492 nm. Positive hits were subjected to additional analysis to determine IC50 values.
- ChEMBL_1849700 (CHEMBL4350241) Inhibition of recombinant human LDHA expressed in Escherichia coli using lactate as substrate incubated for 30 mins followed by substrate addition
- ChEMBL_1933556 (CHEMBL4479208) Inhibition of human LDHA assessed as conversion of pyruvate to lactate measuring disappearance of NADH after 10 mins by fluorescence assay
- ChEMBL_2109294 (CHEMBL4817969) Inhibition of LDHA in human A673 cells assessed as reduction in lactate production incubated for 2 hrs by fluorescence based assay
- ChEMBL_2274356 Inhibition of MCT1 in rat Brain-like endothelial cells incubated for 15 mins in presence of [14C]lactate by BCA protein assay
- ChEMBL_2274367 Binding affinity to human MCT4 expressed in Xenopus laevis oocytes assessed as inhibition constant in presence of [14C]lactate by fluorescence assay
- ChEMBL_1513322 (CHEMBL3611226) Inhibition of human liver purified lactate dehydrogenase-A using pyruvate as substrate assessed as NADH oxidation for 3 mins by fluorimetric method
- ChEMBL_1581840 (CHEMBL3816459) Inhibition of MCT1 in rat brain endothelial 4 cells assessed as L-[14C]-Lactate Uptake after 15 mins by scintillation spectrometric analysis
- ChEMBL_2109291 (CHEMBL4817966) Inhibition of LDHA in human MIA PaCa2 cells assessed as reduction in lactate production incubated for 2 hrs by fluorescence based assay
- ChEMBL_1499769 (CHEMBL3585218) Inhibition of human leukocytic ROCK1 expressed in insect cells using KKRNRTLSV as substrate after 10 mins by pyruvate kinase/lactate dehydrogenase coupled assay
- ChEMBL_1653989 (CHEMBL4003355) Inhibition of human DYRK1A preincubated for 10 mins followed by YRASPSRPESPRPPA-amide substrate addition by pyruvate kinase and lactate dehydrogenase coupled enzyme assay
- ChEMBL_2261029 (CHEMBL5216040) Inhibition of MCT1 in rat RBE4 cells assessed as reduction in [14C]lactate uptake measured after 15 mins by liquid scintillation counting analysis
- ChEMBL_2306754 Inhibition of MCT4 in human SK-BR-3 cells assessed as inhibition of lactate efflux incubated for 4 hrs by automated microplate reader analysis
- ChEMBL_1451367 (CHEMBL3363651) Inhibition of MCT1 (unknown origin) overexpressed in human MCF7 cells assessed as inhibition of transporter-mediated [14C]lactate transport after 10 mins by scintillation counting
- ChEMBL_2132824 (CHEMBL4842434) Non-competitive inhibition of human LDHA assessed as reduction in lactate production using pyruvate as substrate in presence of NADH by Lineweaver-Burk plot analysis
- ChEMBL_1499595 (CHEMBL3584541) Inhibition of N-terminal His6-tagged human recombinant PAK4 (300 to 591 amino acids) using peptide-7 substrate by pyruvate kinase and lactate dehydrogenase coupled assay
- ChEMBL_1513383 (CHEMBL3611423) Inhibition of N-terminal His6-tagged recombinant human PAK4 kinase domain (300 to 591) using peptide 7 as substrate by pyruvate kinase/lactate dehydrogenase coupled assay
- ChEMBL_1548834 (CHEMBL3755390) Inhibition of N-terminal MKLP2 (56 to 505 residues) ATPase basal activity isolated from human hepatocellular carcinoma cells by pyruvate kinase/lactate dehydrogenase enzyme linked assay
- ChEMBL_1682047 (CHEMBL4032324) Inhibition of LDH in human MIAPaCa2 cells assessed as reduction in lactate production preincubated for 2 hrs measured after 30 mins by high throughput fluorescence assay
- ChEMBL_1682057 (CHEMBL4032334) Inhibition of LDH in human A673 cells assessed as reduction in lactate production preincubated for 2 hrs measured after 30 mins by high throughput fluorescence assay
- ChEMBL_1776287 (CHEMBL4233279) Inhibition of PKAalpha (1 to 351 residues) (unknown origin) using Leu-Arg-Arg-Ala-Ser-Leu-Gly as substrate by pyruvate kinase-lactate dehydrogenase coupled assay
- MCT4-Mediated Lactate Transport in NCI-H358 Lung Adenocarcinoma Cell NCI-H358 lung adenocarcinoma cells may be used to measure MCT4 activity in cells with high native levels of MCT4 and low levels of MCT1 and are known to those with skill in the art. Preparation of BCECF-loaded cells and lactate transport activity may be determined as described for Assay 1.
- ChEMBL_1499749 (CHEMBL3585198) Inhibition of PKAalpha (1 to 351 amino acids) (unknown origin) using Leu-Arg-Arg-Ala-Ser-Leu-Gly as substrate by pyruvate kinase/lactate dehydrogenase coupled assay
- ChEMBL_1495598 (CHEMBL3578336) Inhibition of wild type BRAF (unknown origin) assessed as ADP formation measured for 5 hrs by pyruvate kinase/lactate dehydrogenase coupled assay in presence of ATP, MEK1, NADH
- ChEMBL_1548836 (CHEMBL3755392) Inhibition of microtubule-stimulated N-terminal MKLP-2 (56 to 505 residues) ATPase activity isolated from human hepatocellular carcinoma cells by pyruvate kinase/lactate dehydrogenase enzyme linked assay
- ChEMBL_1776286 (CHEMBL4233278) Inhibition of ROCK1 (6 to 553 residues) (unknown origin) using Lys-Lys-Arg-Asn-Arg-Thr-Leu-Ser-Val as substrate by pyruvate kinase-lactate dehydrogenase coupled assay
- ChEMBL_2118570 (CHEMBL4827636) Inhibition of LDHA in wild type CD1 mouse primary hepatocyte assessed as inhibition of lactate production using pyruvate as substrate measured after 30 mins by LC-MS analysis
- Assay 2: MCT4-Mediated Lactate Transport in NCI-H358 Lung Adenocarcinoma Cells NCI-H358 lung adenocarcinoma cells may be used to measure MCT4 activity in cells with high native levels of MCT4 and low levels of MCT1 and are known to those with skill in the art. Preparation of BCECF-loaded cells and lactate transport activity may be determined as described for Assay 1.
- Assay 3: MCT4-Mediated Lactate Transport in MDA-MB-231 Breast Cancer Cells MDA-MB-231 breast cancer cells may be used to measure MCT4 activity in cells with high native levels of MCT4 and low levels of MCT1 and are known to those with skill in the art. Preparation of BCECF-loaded cells and lactate transport activity may be determined as described for Assay 1.
- MCT4-Mediated Lactate Transport in MDA-MB-231 Breast Cancer Cells (Assay 3) MDA-MB-231 breast cancer cells may be used to measure MCT4 activity in cells with high native levels of MCT4 and low levels of MCT1 and are known to those with skill in the art. Preparation of BCECF-loaded cells and lactate transport activity may be determined as described for Assay 1.
- MCT4-Mediated Lactate Transport in NCI-H358 Lung Adenocarcinoma Cells (Assay 2) NCI-H358 lung adenocarcinoma cells may be used to measure MCT4 activity in cells with high native levels of MCT4 and low levels of MCT1 and are known to those with skill in the art. Preparation of BCECF-loaded cells and lactate transport activity may be determined as described for Assay 1.
- ChEMBL_1495597 (CHEMBL3578335) Inhibition of recombinant BRAF V600E mutant (unknown origin) assessed as ADP formation measured for 5 hrs by pyruvate kinase/lactate dehydrogenase coupled assay in presence of ATP, MEK1, NADH
- ChEMBL_1495599 (CHEMBL3578337) Inhibition of wild type CRAF (unknown origin) assessed as ADP formation measured for 5 hrs by pyruvate kinase/lactate dehydrogenase coupled assay in presence of ATP, MEK1, PEP, NADH
- ChEMBL_1931161 (CHEMBL4434412) Antagonist activity at integrin alpha4beta7 expressed in human RPMI8866 cells assessed as reduction in cell adhesion to rat MadCAM incubated for 40 to 60 mins by lactate dehydrogenase assay
- ChEMBL_2060300 (CHEMBL4715301) Inhibition of c-Met (unknown origin) using poly Glu-Tyr as substrate preincubated for 10 mins followed by ATP addition by phosphoenolpyruvate/pyruvate kinase/lactate dehydrogenase coupled radiometric assay
- ChEBML_1654001 Inhibition of human CLK3 (275 to 632 residues) expressed in Escherichia coli BL21(DE3) preincubated for 10 mins followed by substrate addition by pyruvate kinase and lactate dehydrogenase coupled enzyme assay
- ChEMBL_1586646 (CHEMBL3822136) Inhibition of Helicobacter pylori shikimate kinase using shikimic acid substrate by measuring oxidation of NADH to NAD by pyruvate kinase and lactate dehydrogenase coupled enzyme assay based Dixon plot analysis
- ChEMBL_1931162 (CHEMBL4434413) Antagonist activity at integrin alpha4beta1 expressed in human Jurkat cells assessed as reduction in cell adhesion to human VCAM-1 incubated for 40 to 60 mins by lactate dehydrogenase assay
- Biological Activity Assay 4 MCT1 activity may be measured using BT-20 breast cancer cells that express high native levels of MCT1, but do not express MCT4 and are known to those with skill in the art. Preparation of BCECF loaded cells are as described for Assay 1. Lactate transport assay is as described for Assay 1, except 10 mM L-lactate (rather than 50 mM) is added.
- ChEBML_1654000 Inhibition of human CLK1 (148 to 484 residues) expressed in Escherichia coli BL21(DE3) preincubated for 10 mins followed by AFRREWSPGKEAKK substrate addition by pyruvate kinase and lactate dehydrogenase coupled enzyme assay
- ChEMBL_1654001 (CHEMBL4003367) Inhibition of human CLK3 (275 to 632 residues) expressed in Escherichia coli BL21(DE3) preincubated for 10 mins followed by substrate addition by pyruvate kinase and lactate dehydrogenase coupled enzyme assay
- ChEMBL_1801095 (CHEMBL4273387) Inhibition of basal ATPase activity of N-terminal His6-tagged human Eg5 motor domain (1 to 368 residues) expressed in Escherichia coli BL21 (DE3) by pyruvate kinase-lactate dehydrogenase coupled assay
- ChEMBL_977859 (CHEMBL2421461) Inhibition of microtubule-stimulated ATPase activity of N-terminal His-6-tagged human wild type Eg5 (1 to 368) expressed in Escherichia coli BL21 by pyruvate kinase/lactate dehydrogenase-linked assay
- ChEMBL_1801093 (CHEMBL4273385) Inhibition of N-terminal His6-tagged microtubule-stimulated ATPase activity of human Eg5 motor domain (1 to 368 residues) expressed in Escherichia coli BL21 (DE3) by pyruvate kinase-lactate dehydrogenase coupled assay
- ChEMBL_1892980 (CHEMBL4394901) Inhibition of recombinant human adenosine kinase expressed in Escherichia coli BL21 (DE3) using adenosine as substrate in presence of ATP and PEP by pyruvate kinase-lactate dehydrogenase-coupled UV-vis spectrophotometric assay
- ChEMBL_940910 (CHEMBL2330556) Inhibition of human N-terminal His6-tagged Eg5 (1 to 368 amino acid residues) motor domain basal ATPase activity expressed in Escherichia coli BL21 (DE3) by pyruvate kinase/lactate dehydrogenase-coupled assay
- ChEMBL_1617999 (CHEMBL3860168) Inhibition of basal ATPase activity of N-terminal His6-tagged/SUMO-fused human MPP1 motor domain (57 to 491 residues) expressed in Escherichia coli BL21 CodonPlus by pyruvate kinase/lactate dehydrogenase-linked assay
- ChEMBL_1618001 (CHEMBL3860170) Inhibition of basal ATPase activity of N-terminal His6-tagged/SUMO-fused human MPP1 motor domain (2 to 477 residues) expressed in Escherichia coli BL21 CodonPlus by pyruvate kinase/lactate dehydrogenase-linked assay
- ChEMBL_1733255 (CHEMBL4148791) Inhibition of human mitotic kinesin Eg5 motor domain (1 to 368 residues) assessed as reduction in steady-state basal ATPase activity using ATP as substrate by pyruvate kinase/lactate dehydrogenase enzyme coupled assay
- ChEMBL_1886104 (CHEMBL4387686) Non-competitive inhibition of C-terminally His6-tagged human UCK2 expressed in Escherichia coli BL21(DE3) cells using phosphoenolpyruvate, NADH, uridine level by spectrophotometry based pyruvate kinase and lactate dehydrogenase coupled enzyme assay
- ChEMBL_772123 (CHEMBL1838042) Inhibition of His6-tagged Yersinia pestis KIM6 CDP-ME kinase assessed as ADP production after 30 mins by Kinase Glo luminescence-based assay or standard pyruvate kinase/lactate dehydrogenase-coupled absorbance-based assay
- ChEMBL_938549 (CHEMBL2327388) Inhibition of MT-stimulated human N-terminal His6-tagged Eg5 (1 to 368 amino acid residues) motor domain ATPase activity expressed in Escherichia coli BL21 (DE3) by pyruvate kinase/lactate dehydrogenase-coupled assay
- ChEMBL_1618000 (CHEMBL3860169) Inhibition of microtubule-stimulated ATPase activity of N-terminal His6-tagged/SUMO-fused human MPP1 motor domain (57 to 491 residues) expressed in Escherichia coli BL21 CodonPlus by pyruvate kinase/lactate dehydrogenase-linked assay
- ChEMBL_1618002 (CHEMBL3860171) Inhibition of microtubule-stimulated ATPase activity of N-terminal His6-tagged/SUMO-fused human MPP1 motor domain (2 to 477 residues) expressed in Escherichia coli BL21 CodonPlus by pyruvate kinase/lactate dehydrogenase-linked assay
- Enzyme Inhibition Assay A coupled spectrophotometric assay was used in which ADP generated by ERK2 was converted to ATP by pyruvate kinase with the production of pyruvate from phosphoenol pyruvate. Lactate dehydrogenase reduces pyruvate to lactate with the oxidation of NADH. NADH depletion was monitored at 340 nm using a microplate reader. The decrease of absorbance was monitored as a function of time and the resulting data was fitted to a competitive inhibition kinetic model to determine the Ki.
- ChEMBL_1667053 (CHEMBL4016849) Inhibition of human C-terminus CLK1 (148 to 484 residues) expressed in Escherichia coli BL21(DE3) using AFRREWSPGKEAKK as substrate preincubated for 10 mins followed by ATP addition by pyruvate kinase-lactate dehydrogenase coupled assay
- ChEMBL_1886100 (CHEMBL4387682) Non-competitive inhibition of C-terminally His6-tagged human UCK2 expressed in Escherichia coli BL21(DE3) cells using phosphoenolpyruvate, NADH and varying uridine level by spectrophotometry based pyruvate kinase and lactate dehydrogenase coupled enzyme assay
- ChEMBL_816345 (CHEMBL2024743) Inhibition of N-terminal hexa-histidine tagged human cloned Eg5 (1 to 368 amino acids) expressed in Escherichia coli BL21 (DE3) assessed as reduction in basal ATPase activity by pyruvate kinase/lactate dehydrogenase-linked assay
- ChEMBL_1886101 (CHEMBL4387683) Non-competitive inhibition of C-terminally His6-tagged human UCK2 expressed in Escherichia coli BL21(DE3) cells using uridine, phosphoenolpyruvate, NADH and varying ATP level by spectrophotometry based pyruvate kinase and lactate dehydrogenase coupled enzyme assay
- ChEMBL_2282176 Inhibition of human MCT4 in human SNU-398 cells assessed as inhibition of lactate efflux preincubated for 30 mins followed by D(+)glucose and measured after 4 hrs by dialysis based UHPLC-ESI-Q-Orbitrap-MS analysis
- ChEMBL_2282177 Inhibition of human MCT4 in human NCI-H358 cells assessed as inhibition of lactate efflux preincubated for 30 mins followed by D(+)glucose and measured after 4 hrs by dialysis based UHPLC-ESI-Q-Orbitrap-MS analysis
- ChEMBL_2282178 Inhibition of human MCT4 in human NCI-H441 cells assessed as inhibition of lactate efflux preincubated for 30 mins followed by D(+)glucose and measured after 4 hrs by dialysis based UHPLC-ESI-Q-Orbitrap-MS analysis
- ChEMBL_2282192 Inhibition of human MCT4 in human RT-4 cells assessed as inhibition of lactate efflux preincubated for 30 mins followed by D(+)glucose and measured after 4 hrs by dialysis based UHPLC-ESI-Q-Orbitrap-MS analysis
- ChEMBL_816346 (CHEMBL2024744) Inhibition of N-terminal hexa-histidine tagged human cloned Eg5 (1 to 368 amino acids) expressed in Escherichia coli BL21 (DE3) assessed as reduction of MT-stimulated ATPase activity by pyruvate kinase/lactate dehydrogenase-linked assay
- ChEMBL_1499748 (CHEMBL3585197) Inhibition of ROCK1 (6 to 553 amino acids) (unknown origin) using Lys-Lys-Arg-Asn-Arg-Thr-Leu-Ser-Val as substrate preincubated for 15 mins followed by ATP addition by pyruvate kinase/lactate dehydrogenase coupled assay
- ChEMBL_2282159 Inhibition of human MCT4 in human MDA-MB-231 cells assessed as inhibition of lactate efflux preincubated for 30 mins followed by D(+)glucose and measured after 4 hrs by dialysis based UHPLC-ESI-Q-Orbitrap-MS analysis
- ChEMBL_2282175 Inhibition of human MCT4 in human SNU-398 cells assessed as inhibition of radioactive lactate efflux preincubated for 30 mins followed by D(+)glucose and measured after 4 hrs by dialysis based UHPLC-ESI-Q-Orbitrap-MS analysis
- ChEMBL_2282191 Inhibition of human MCT4 in human MIA PaCa-2 cells assessed as inhibition of lactate efflux preincubated for 30 mins followed by D(+)glucose and measured after 4 hrs by dialysis based UHPLC-ESI-Q-Orbitrap-MS analysis
- ChEMBL_1669644 (CHEMBL4019532) Inhibition of recombinant human N-terminal His-tagged KHK expressed in Escherichia coli BL21(DE3) using fructose as substrate incubated for 30 mins followed by ATP addition measured for 30 mins by pyruvate kinase-lactate dehydrogenase coupled enzyme assay
- ChEMBL_1669645 (CHEMBL4019533) Inhibition of recombinant rat N-terminal His-tagged KHK expressed in Escherichia coli BL21(DE3) using fructose as substrate incubated for 30 mins followed by ATP addition measured for 30 mins by pyruvate kinase-lactate dehydrogenase coupled enzyme assay
- ChEMBL_2282174 Inhibition of human MCT4 in human MDA-MB-231 cells assessed as inhibition of radioactive lactate efflux preincubated for 30 mins followed by D(+)glucose and measured after 4 hrs by dialysis based UHPLC-ESI-Q-Orbitrap-MS analysis
- ChEMBL_1669642 (CHEMBL4019530) Inhibition of recombinant human N-terminal His-tagged KHK-C expressed in Escherichia coli BL21(DE3) using fructose as substrate incubated for 30 mins followed by ATP addition measured for 30 mins by pyruvate kinase-lactate dehydrogenase coupled enzyme assay
- ChEMBL_1669670 (CHEMBL4019558) Inhibition of recombinant human N-terminal His-tagged KHK-A expressed in Escherichia coli BL21(DE3) using fructose as substrate incubated for 30 mins followed by ATP addition measured for 30 mins by pyruvate kinase-lactate dehydrogenase coupled enzyme assay
- ChEMBL_2022241 (CHEMBL4676054) Inhibition of 10 nM recombinant human N-terminal His-tagged KHKC expressed in Escherichia coli BL21 (DE3) using fructose as substrate preincubated for 30 mins followed by ATP addition and measured for 30 mins by pyruvate kinase-lactate dehydrogenase coupled assay
- ChEMBL_2022242 (CHEMBL4676055) Inhibition of 1 nM recombinant human N-terminal His-tagged KHKC expressed in Escherichia coli BL21 (DE3) using fructose as substrate preincubated for 30 mins followed by ATP addition and measured for 3 hrs by pyruvate kinase-lactate dehydrogenase coupled assay
- Lactate Transport in MCT4-Expressing MDA-MB-453 Breast Cancer Cells MCT4 may be stably expressed in MDA-MB-453 breast cancer cells that do not express native MCT1 or MCT4. MCT4 activity may be assessed by monitoring the intracellular pH change that accompanies lactate/proton symport, using the pH-sensitive fluorescent dye 2',7'-bis-(carboxyethyl)-5(6)-carboxyfluorescein (BCECF), in a manner similar to that previously reported for MCT1 and MCT4. The following is an exemplary procedure for assaying MCT4 activity of the compounds of Formula (I).
- ChEMBL_1892973 (CHEMBL4394894) Inhibition of wild-type N-terminal TEV cleavage site-fused/His-tagged Mycobacterium tuberculosis H37Rv adenosine kinase expressed in Escherichia coli BL21 (DE3) using adenosine as substrate in presence of ATP and PEP by pyruvate kinase-lactate dehydrogenase coupled UV-vis spectrophotometric method
- ChEMBL_1895210 (CHEMBL4397245) Inhibition of recombinant human His6-tagged PDE4D2 expressed in baculovirus infected Sf9 insect cells using cAMP as substrate preincubated for 5 to 10 mins followed by substrate addition and measured for 10 mins by yeast myokinase/pyruvate kinase/lactate dehydrogenase-coupled fluorescence assay
- ChEMBL_2022251 (CHEMBL4676064) Inhibition of recombinant human N-terminal His-tagged KHKC expressed in Escherichia coli BL21 (DE3) using 8 mM fructose as substrate preincubated for 30 mins followed by 2 mM ATP addition and measured for 30 min by pyruvate kinase-lactate dehydrogenase coupled assay
- ChEMBL_2022252 (CHEMBL4676065) Inhibition of recombinant human N-terminal His-tagged KHKA expressed in Escherichia coli BL21 (DE3) using 8 mM fructose as substrate preincubated for 30 mins followed by 2 mM ATP addition and measured for 30 min by pyruvate kinase-lactate dehydrogenase coupled assay
- ChEMBL_2022253 (CHEMBL4676066) Inhibition of recombinant rat N-terminal His-tagged KHK expressed in Escherichia coli BL21 (DE3) using 8 mM fructose as substrate preincubated for 30 mins followed by 2 mM ATP addition and measured for 30 min by pyruvate kinase-lactate dehydrogenase coupled assay
- Enzyme Inhibition Assay The phosphorylation of L-Hse was monitored by coupling the formation of ADP with pyruvate kinase and lactate dehydrogenase (PK/LDH). The resulting oxidation of NADH was monitored at 340 nm by using a SpectraMax plate reader in a 96-well format.
- Enzyme Inhibition Assay The phosphorylation of L-Hse was monitored by coupling the formation of ADP with pyruvate kinase and lactate dehydrogenase (PK/LDH). The resulting oxidation of NADH was monitored at 340 nm by using a SpectraMax plate reader in a 96-well format.
- ChEMBL_1895162 (CHEMBL4397197) Inhibition of recombinant human His6-tagged PDE4B1 UCR1 S133D mutant expressed in baculovirus infected Sf9 insect cells using cAMP as substrate preincubated for 5 to 10 mins followed by substrate addition and measured for 10 mins by yeast myokinase/pyruvate kinase/lactate dehydrogenase-coupled fluorescence assay
- Inhibition Assay Primary screening was performed using PKM2 in the presence of FBP. Compounds were chosen as potential "hits" if the compound demonstrated inhibition of PKM2 activity greater than 50%. Activity was measured by monitoring the concentration of NADH. Pyruvate, produced by the enzymatic activity of pyruvate kinase, is converted into lactate by lactate dehydrogenase, which requires the consumption of NADH (NADH→NAD+). Thus, the activity of PKM2 was indirectly measured by monitoring the consumption of NADH through fluorescence assays. Additionally, the activity of the PKM2 enzyme was directly monitored by measuring the production of ATP, as ATP is produced when phosphoenolpyruvate is converted to pyruvate by PKM2.
- MCT4-Mediated Lactate Transport in MDA-MB-231 Breast Cancer Cells MDA-MB-231 breast cancer cells may be used to measure MCT4 activity in cells with high native levels of MCT4 and low levels of MCT1 and are known to those with skill in the art. MCT4 activity may be assessed by monitoring the intracellular pH change that accompanies lactate/proton symport, using the pH-sensitive fluorescent dye 2′,7′-bis-(carboxyethyl)-5(6)-carboxyfluorescein (BCECF), in a manner similar to that previously reported for MCT1 and MCT4. The following is an exemplary procedure for assaying MCT4 activity of the compounds of Formula (I).
- Lactate Transport in MCT4-Expressing MDA-MB-453 Breast Cancer Cells MCT4 may be stably expressed in MDA-MB-453 breast cancer cells that do not express native MCT1 or MCT4. MCT4 activity may be assessed by monitoring the intracellular pH change that accompanies lactate/proton symport, using the pH-sensitive fluorescent dye 2′,7′-bis-(carboxyethyl)-5(6)-carboxyfluorescein (BCECF), in a manner similar to that previously reported for MCT1 and MCT4. The following is an exemplary procedure for assaying MCT4 activity of the compounds of Formula (I). 2.5 μL BCECF-loaded cells, along with either 10 μL DMSO or 100×compound in DMSO, are added to 937.5 μL of Tyrode's Solution in a quartz 1.0 mL cuvette (PerkinElmer, B0631116). Fluorescence measurements are performed on a PerkinElmer LS55 fluorescence spectrometer with dual excitation wavelengths achieved using a filter wheel (FL Winlab program: Fast Filter; Excitation 490/440; Emission 535). After establishing baseline BCECF fluorescence (around 10-20 s), 50 μL of 1 M sodium L-lactate (Sigma-Aldrich) is added to the cuvette (final concentration: 50 mM) and rapidly mixed. The time-dependent decrease in BCECF fluorescence (490/440 ratio) may be fit to an exponential decay curve (Prism GraphPad) to determine the rate of lactate transport.
- Enzyme Inhibition Assay The assay phosphotransferase activity was followed spectrophotometrically b reduction of NADP in the presence of an excess of glucose-6-phosphate-dehydrogenase (method A). ATPase activity was measured by spectrophotometric measurement of the rate of oxidation of NADH in the presence of phospho-enol-pyruvate, pyruvated-kinase and lactate-dehydrogenase (method B).
- Kinase Inhibition Assay Phosphorylation reactions were monitored using a coupled-enzyme assay in which ADP production was coupled to NADH oxidation by pyruvate kinase and lactate dehydrogenase. The assay was carried out in a buffer containing phosphoenolpyruvate, NADH, pyruvate kinase, lactate dehydrogenase, and PIM kinase. The reaction was monitored at 340 nm at 25 deg C on a Spectramax spectrophotometer (Molecular Devices) and started by addition ATP after a 10-min preincubation at 25 deg C. A recognition peptide of the PIM1 substrate p21 (RKRRQTSMTD) was used. DMSO-dissolved inhibitors were added at the preincubation period resulting in a 2% final DMSO. Kinetic analysis was done by nonlinear regression fitting using the program KaleidaGraph (Synergy Software).
- Biological Activity Assay 2 NCI-H358 lung adenocarcinoma cells may be used to measure MCT4 activity in cells with high native levels of MCT4 and low levels of MCT1 and are known to those with skill in the art. Preparation of BCECF-loaded cells and lactate transport activity may be determined as described for Assay 1.
- Intracellular ZAP-70 Kinase Inhibition Assay A coupled spectrophotometric assay was used wherein ADP generated by ZAP-70 kinase was converted to ATP by pyruvate kinase (PK), with concomitant production of pyruvate from phosphoenolpyruvate (PEP). LDH reduces pyruvate to lactate by oxidizing NADH. NADH depletion was monitored at 340 nm using a microplate reader (Spectra Max 250, Molecular Device).
- ACCase Enzymatic Assay In order to effectively screen out non-specific modulators of pyruvate kinase and lactate dehydrogenase (the coupled portion of the reaction), a PK/LDH inhibition test was developed. The complete 200 μl reaction mixture contained 52.5 mM HEPES (pH8), 2.625 mM MgCl2, 0.525 mM DTT, 11 mM NaHCO3, 1% DMSO with or without inhibitor, 1× pyruvate kinase/lactate dehydrogenase (PK/LDH), 0.3 mM NADH, and 0.5 mM PEP. The reactions were incubated at 30° C. for 10 minutes and then initiated by the addition of 66 μM ADP. The initiated reactions were read immediately via plate reader at OD340 and kinetic readings were acquired every 20 s for 15 minutes while remaining at 30° C.
- Lactate Transport in MCT4-Expressing MDA-MB-453 Breast Cancer Cells (Assay 1) MCT4 may be stably expressed in MDA-MB-453 breast cancer cells that do not express native MCT1 or MCT4. MCT4 activity may be assessed by monitoring the intracellular pH change that accompanies lactate/proton symport, using the pH-sensitive fluorescent dye 2′,7′-bis-(carboxyethyl)-5(6)-carboxyfluorescein (BCECF), in a manner similar to that previously reported for MCT1 and MCT4. The following is an exemplary procedure for assaying MCT4 activity of the compounds of Formula (I).Preparing BCECF-Loaded Cells:Cells (˜7×106) are trypsinized (0.05% Trypsin-EDTA), pelleted (300 g, 5 min), and resuspended in 1 mL Tyrode's Solution, pH 7.4 (119 mM NaCl, 5 mM KCl, 25 mM HEPES, pH 7.4, 2 mM CaCl2), 2 mM MgCl2, 6 g/L glucose). 10 μL of a 30 mM DMSO stock of BCECF-AM ester (Life Technologies) is added and the cells are incubated at 37° C. for 5 min. The cells are pelleted (300 g, 5 min), washed once with 1 mL Tyrode's Solution, pH 7.4, re-pelleted (300 g, 5 min), and resuspended in 1 mL Tyrode's Solution, pH 7.4.Lactate Transport Assay:2.5 μL BCECF-loaded cells, along with either 10 μL DMSO or 100×compound in DMSO, are added to 937.5 μL of Tyrode's Solution in a quartz 1.0 mL cuvette (PerkinElmer, B0631116). Fluorescence measurements are performed on a PerkinElmer LS55 fluorescence spectrometer with dual excitation wavelengths achieved using a filter wheel (FL Winlab program: Fast Filter; Excitation 490/440; Emission 535). After establishing baseline BCECF fluorescence (around 10-20 s), 50 μL of 1 M sodium L-lactate (Sigma-Aldrich) is added to the cuvette (final concentration: 50 mM) and rapidly mixed. The time-dependent decrease in BCECF fluorescence (490/440 ratio) may be fit to an exponential decay curve (Prism GraphPad) to determine the rate of lactate transport.
- Coupled Diaphorase Assay The inhibitory properties of the compounds were investigated using a coupled enzyme assay that links the lactate dehydrogenase (LDH) reaction to the production of fluorescent resorutin by diaphorase.Human lactate dehydrogenases (LDH) catalyze the reversible interconversion between pyruvate and lactate. LDH is capable of catalyzing both the forward (pyruvate to lactate) and the reverse (lactate to pyruvate) reaction, using either NADH or NAD+ as a cofactor. The reaction proceeds in either direction dependent on various factors, such as substrate availability, the presence of necessary cofactors, temperature and pH. Different isoforms (LDH A, B, and C) of the enzyme favor different reaction directions LDHA prefers the conversion from pyruvate to lactate, whereas LDHB preferentially oxidizes lactate to pyruvate.The coupled assay relies on the oxidation of NAD+ to NADH throughout the conversion of lactate to pyruvate by LDH (isoforms A, B and C). The produced NADH serves as cofactor in the diaphorase reaction, which reduces non-fluorescent resazurin to fluorescent resorufin. Therefore, the assay indirectly monitors the rate of pyruvate production. Although the consumption of NADH can be directly monitored due to the intrinsic fluorescence of the molecule (excitation: 340 nm, emission: 460 nm) there are problems linked to the direct readout method. It has been shown that many compounds in chemical libraries interfere with the assay due to fluorescent properties similar to NADH. Shifting the assay to longer wavelengths by coupling the LDH reaction to the conversion of resazurin to fluorescent resorufin by diaphorase reduces this compound interference. The assay direction was thus chosen to provide a robust and reliable assay.Applying the LDHA reaction in the preferred direction for the conversion of pyruvate to lactate under oxidation of NADH to NAD+ would necessitate running the LDHA reaction to about 80% completion and adding the diaphorase assay reagents afterwards in order to avoid enzyme competition for NADH. As a result, such a method would be expected to be more prone to errors, since too high conversion rates will lead to extenuation of the IC50 values obtained (Davis et al., ASSAY and Drug Dev. Tech. 14 (3): 175-179, 2016). When not running the assay in the preferred direction for LDHA, more conservative IC50 values would be expected to be obtained compared to earlier published results for other LDHA inhibitor compounds. Therefore, actual IC50 values could thus be expected to be lower.For the determination of IC50 values a coupled diaphorase assay was adopted from Bembenek et al. (A Fluorescence-Based Coupling Reaction for Monitoring the Activity of Recombinant Human NAD Synthetase. ASSAY and Drug Development Technologies, 2005. 3(5): 533-541). Compounds were tested in duplicates using 2-fold, 3-fold or 4-fold serial dilutions including 11 individual concentrations, starting from 5000 μM to 30 μM. A no-substrate control representing 100% inhibition or oxamate-inhibition controls (28.7 mM final oxamate concentration in assay) and a control containing the complete substrate solution as well as DMSO representing the fully uninhibited reaction were added. Oxamate is a well characterized inhibitor of LDH that inhibits LDH enzyme activity in the mM range in vitro with high specificity (Papacostantinou el al., J. Biol. Chem. 236: 278-284, 1961). The controls allowed for the calculation of the percentage inhibition for each data point. The assay buffer consisted of 50 mM HEPES pH 7.4, 5 mM MgCl2 and 0.05% pluronic acid F-127. Enzyme solution leading to final concentrations of 4-7 nM LDHA or 6 nM LDHB, as well as 0.2 U/ml diaphorase in the reaction well was dispensed into 384-well plates (Greiner bio-one) using a CyBi -SELMA robotic pipettor. Compound dilutions and the enzyme were incubated for at least 20 min at room temperature. Thereafter, the substrate solution was added.
- Lactate Dehydrogenase Inhibition Assay LDH was assayed spectrophotometrically for reduction of pyruvate using NADH by recording the changed in absorbance at 340 nm. The reaction was initiated by the addition of NADH, and the decrease in absorbance at 340 nm was monitored for 5 min. The compounds showing inhibition of more than 50% at 50 ug/mL concentration were retested and their IC50 values were determined.
- Lactate Excretion Assays Parasite cultures were seeded in 96-well plates (1% hematocrit, 1-2% starting parasitemia, 100 μL total volume) with varying concentrations of the test compounds in a final concentration of 0.1% DMSO. Plates were cultured for 48 hours before harvesting the culture medium for LC-MS analysis. At harvest, thin smears were taken and examined microscopically to observe any alterations in parasite morphology.
- Ki Determination A coupled spectrophotometric assay was used in which ADP generated by kinase was converted to ATP with the concomitant production of pyruvate from PEP. LDH reduces pyruvate to lactate with the oxidation of NADH. NADH depletion was monitored at 340 nm using a microplate reader. The IC50 was evaluated from the data as a function of inhibitor concentration. The Ki value was calculated according to the Cheng-Prusoff approximation.
- ATPase Assay The ATP hydrolysis activity of S. aureus DNA gyrase is measured by coupling the production of ADP through pyruvate kinase/lactate dehydrogenase to the oxidation of NADH. This method has been described previously (Tamura and Gellert, 1990, J. Biol. Chem., 265, 21342).ATPase assays are carried out at 30° C. in buffered solutions containing 100 mM TRIS pH 7.6, 1.5 mM MgCl2, 150 mM KCl. The coupling system contains final concentrations of 2.5 mM phosphoenol pyruvate, 200 μM nicotinamide adenine dinucleotide (NADH), 1 mM DTT, 30 ug/ml pyruvate kinase, and 10 ug/ml lactate dehydrogenase. The enzyme (90 nM final concentration) and a DMSO solution (3% final concentration) of a compound is added. The reaction mixture is allowed to incubate for 10 minutes at 30° C. The reaction is initiated by the addition of ATP to a final concentration of 0.9 mM, and the rate of NADH disappearance is monitored at 340 nanometers over the course of 10 minutes.
- Enzymatic Assay Activity of unphosphorylated c-FMS kinase (uFMS, Seq. ID no. 1) was determined by following the production of ADP from the FMS kinase reaction with ATP and poly E4Y as substrates through coupling with the pyruvate kinase/lactate dehydrogenase system (e.g., Schindler et al. Science (2000) 289: 1938-1942). In this assay, the oxidation of NADH (thus the decrease at A340 nm) was continuously monitored spectrophometrically. The reaction mixture (100 .mu.L) contained FMS (purchased from Millipore) (10 nM), polyE4Y (1 mg/mL), MgCl.sub.2 (10 mM), pyruvate kinase (4 units), lactate dehydrogenase (0.7 units), phosphoenol pyruvate (1 mM), NADH (0.28 mM) and ATP (500 .mu.M) in 90 mM Tris buffer containing 0.2% octyl-glucoside and 1% DMSO, pH 7.5. The inhibition reaction was started by mixing serial diluted test compound with the above reaction mixture. The absorption at 340 nm was monitored continuously for 4 hours at 30.degree. C. on Synergy 2 plate reader.
- Spectrophotometric 384 Well Assay Malonyl CoA formation by acetyl CoA carboxylases is stoichometrically linked to the consumption of ATP. ACC2 activity is measured in a NADH-linked kinetic method measuring ADP generated during the ACC reaction using a coupled lactate dehydrogenase/pyruvate kinase reaction. Assay reactions are then carried out in 384-well plates, with hACC2 in an appropriate dilution and at final assay concentrations (f.c.) of 100 mM Tris (pH 7.5), 10 mM trisodium citrate, 25 mM KHCO3, 10 mM MgCl2, 0.5 mg/ml BSA, 3.75 mM reduced L-glutathione, 15 U/ml lactate dehydrogenase, 0.5 mM phosphoenolpyruvate, 15 U/ml pyruvate kinase, compounds at different concentrations at final DMSO concentrations of 1%.The enzymatic reaction is then started by addition of a mixture of NADH, acetylCoenzyme A (both 2000 f.c.) and ATP (500 uM f.c.). The decrease of the optical density (slope S) is then determined at 25° C. at a wavelength of 340 nm over 15 minutes in a spectrophotometric reader.
- Spectrophotometric 384 Well Assay Malonyl CoA formation by acetyl CoA carboxylases is stoichometrically linked to the consumption of ATP. ACC2 activity is measured in a NADH-linked kinetic method measuring ADP generated during the ACC reaction using a coupled lactate dehydrogenase/pyruvate kinase reaction.For biological testing, a human ACC2 construct which lacks the 128 amino acids at the N-terminus for increased solubility (nt 385-6966 in Genbank entry AJ575592) is cloned. The protein is then expressed in insect cells using a baculoviral expression system. Protein purification is performed by anion exchange.All compounds are dissolved in dimethyl sulfoxide (DMSO) to a concentration of 10 mM.Assay reactions are then carried out in 384-well plates, with hACC2 in an appropriate dilution and at final assay concentrations (f.c.) of 100 mM Tris (pH 7.5), 10 mM trisodium citrate, 25 mM KHCO3, 10 mM MgCl2, 0.5 mg/mL BSA, 3.75 mM reduced L-glutathione, 15 U/mL lactate dehydrogenase, 0.5 mM phosphoenolpyruvate.
- Spectrophotometric 384 Well Assay Malonyl CoA formation by acetyl CoA carboxylases is stoichometrically linked to the consumption of ATP. ACC2 activity is measured in a NADH-linked kinetic method measuring ADP generated during the ACC reaction using a coupled lactate dehydrogenase/pyruvate kinase reaction.For biological testing, a human ACC2 construct which lacks the 128 amino acids at the N-terminus for increased solubility (nt 385-6966 in Genbank entry AJ575592) is cloned. The protein is then expressed in insect cells using a baculoviral expression system. Protein purification is performed by anion exchange.All compounds are dissolved in dimethyl sulfoxide (DMSO) to a concentration of 10 mM.Assay reactions are then carried out in 384-well plates, with hACC2 in an appropriate dilution and at final assay concentrations (f.c.) of 100 mM Tris (pH 7.5), 10 mM trisodium citrate, 25 mM KHCO3, 10 mM MgCl2, 0.5 mg/ml BSA, 3.75 mM reduced L-glutathione, 15 U/ml lactate dehydrogenase, 0.5 mM phosphoenolpyruvate.
- Inhibition of Eg5 ATPase Activity The microtubule-activated ATPase rates were measured using the pyruvate kinase/lactate dehydrogenase-linked assay. To optimize the signal for basal Eg5 activity at low protein concentration, measurements were performed in the presence of 300 mM NaCl. Eg5 at seven different concentrations was incubated at room temperature for 25 min with test compounds from 0 to 5 uM. After the reaction, the resulting decrease in absorbance at 340 nm was measured using the 96-well Sunrise photometer.
- Inhibition Assay Protein Kinase C beta 2 (PKC beta II) catalyzes the production of ADP from ATP that accompanies the phosphoryl transfer to the PKC Pseudosubstrate peptide (A->S, RFARKGSLRQKNV). This transfer is coupled to the oxidation of p-NADH through the activities of Pyruvate Kinase (PK) and Lactate Dehydrogenase (LDH). (3-NADH conversion to NAD+ is monitored by the decrease in absorbance at 340 nm (e=6.22 cm-1 mM-1) using a Molecular Devices SPECTRA max PLUS spectrophotometer.A typical assay was carried out on a 96-well, clear microtiter plate in a Molecular Devices spectrophotometer for 20 minutes at 30 C. in 0.1 mL of assay buffer containing 50 mM HEPES, pH 7.4, 5 nM PKC, 23 units of pyruvate kinase, 33 units of lactate dehydrogenase, 0.15 mM peptide, 0.1 mM ATP, 1 mM DTT, 4 mM PEP, 8 mM MgCl2, 0.3 mM NADH, 60 mM CaCl2, 10 mg/mL PS, 50 ng/mL PMA, 7.5% DMSO and from about 10,000 nM to 0.169 nM compound inhibitor.
- Inhibition Assay Specific high-throughput screening assays were developed for KHK-C and KHK-A using recombinant proteins. Purified human recombinant KHK-C and KHK-A were produced using the Bio-Rad Profinity eXact Fusion-Tag System and Profinia protein purification instrument. The assays consist of a 3-step, coupled-enzyme process involving fructokinase (KHK), pyruvate kinase (PK) and lactate dehydrogenase (LDH). 1,2 The disappearance of NADH is measured kinetically by A340 at 37 ° C. The enzymatic assay was carried out in a total reaction volume of 200 ul containing 50 mM PIPES, 6 mM MgCl2, 100 mM KCl, 100 uM-5 mM ATP, 2 mM phosphoenolpyruvate, 0.3 mM NADH, 15 U of pyruvate kinase, 15 U of lactate dehydrogenase, and 75-1000 ng KHKC. 1 mM fructose was added to the reactions, except for the no fructose controls which utilized water. The high-throughput assay was used to identify inhibitors that have an IC50 value <5 M for KHK-C.5 The '559 pub also sets forth a KHK assay for testing inhibition activity of potential KHK inhibitors.
- Biological Activity Assay 3 MDA-MB-231 breast cancer cells may be used to measure MCT4 activity in cells with high native levels of MCT4 and low levels of MCT1 and are known to those with skill in the art. MCT4 activity may be assessed by monitoring the intracellular pH change that accompanies lactate/proton symport, using the pH-sensitive fluorescent dye 2′,7′-bis-(carboxyethyl)-5(6)-carboxyfluorescein (BCECF), in a manner similar to that previously reported for MCT1 and MCT4.
- Spectrophotometric 384 Well Assay Malonyl CoA formation by acetyl CoA carboxylases is stoichometrically linked to the consumption of ATP. ACC2 activity is measured in a NADH-linked kinetic method measuring ADP generated during the ACC reaction using a coupled lactate dehydrogenase/pyruvate kinase reaction. For biological testing, a human ACC2 construct which lacks the 128 amino acids at the N-terminus for increased solubility (nt 385-6966 in Genbank entry AJ575592) is cloned. The protein is then expressed in insect cells using a baculoviral expression system. Protein purification is performed by anion exchange.
- Biochemical Assay for PERK Activity of PERK kinase was determined spectroscopically using a coupled pyruvate kinase/lactate dehydrogenase assay that continuously monitors the ATP hydrolysis-dependent oxidation of NADH (e.g., Schindler et al. Science (2000) 289: 1938-1942). Assays were conducted in 384-well plates (100 μL final volume) using 10 nM PERK (from Beryllium), 0.25 mg/mL Myelin Basic Protein substrate, 1.5 units pyruvate kinase, 2.1 units lactate dehydrogenase, 1 mM phosphoenol pyruvate, 0.28 mM NADH and 1 mM ATP in assay buffer (100 mM Tris, pH 7.5, 15 mM MgCl2, 0.5 mM DTT, 0.004% (w/v) BSA, and 0.004% Triton X-100). Inhibition of PERK was measured by adding serial diluted test compound (final assay concentration of 1% DMSO). A decrease in absorption at 340 nm was monitored continuously for 6 hours at 30° C. on a multi-mode microplate reader (BioTek). The reaction rate was calculated using the 2-3 h time frame. The reaction rate at each concentration of compound was converted to percent inhibition using controls (i.e. reaction with no test compound and reaction with a known inhibitor) and IC50 values were calculated using software routines in Prism (GraphPad software).
- Biochemical Assay for PERK Activity of PERK kinase was determined spectroscopically using a coupled pyruvate kinase/lactate dehydrogenase assay that continuously monitors the ATP hydrolysis-dependent oxidation of NADH (e.g., Schindler et al. Science (2000) 289: 1938-1942). Assays were conducted in 384-well plates (100 μt final volume) using 10 nM PERK (from Beryllium), 0.25 mg/mL Myelin Basic Protein substrate, 1.5 units pyruvate kinase, 2.1 units lactate dehydrogenase, 1 mM phosphoenol pyruvate, 0.28 mM NADH and 1 mM ATP in assay buffer (100 mM Tris, pH 7.5, 15 mM MgCl2, 0.5 mM DTT, 0.004% (w/v) BSA, and 0.004% Triton X-100). Inhibition of PERK was measured by adding serial diluted test compound (final assay concentration of 1% DMSO). A decrease in absorption at 340 nm was monitored continuously for 6 hours at 30° C. on a multi-mode microplate reader (BioTek). The reaction rate was calculated using the 2-3 h time frame. The reaction rate at each concentration of compound was converted to percent inhibition using controls (i.e., reaction with no test compound and reaction with a known inhibitor) and IC50 values were calculated using software routines in Prism (GraphPad software).
- Inhibition Assay The entire lactate uptake studies for the inhibition of MCT1 were carried out on RBE4 (Rat Brain Endothelial 4) cells. The expression of MCT1 on these cells was confirmed by Western Blotting. The cells were plated approximately 20-24 hours before the experiment, the number of cells being approximately 105 cells per well. The test compounds were dissolved in DMSO and diluted 1000 times using a solution of HEPES buffer (pH 7.43) which consists of 3 uM 14C-Lactate and 2 uM L-Lactate. The cells were washed twice with 500 uL of HEPES buffer and the cells were allowed to equilibrate for 15 minutes at 37° C. The HEPES buffer was removed and 250 uL of the test sample was added in triplicates. This was repeated for all the compounds, including the controls (CHC and DMSO). After 15 minutes, the compounds were removed from the well and 500 uL of ice-cold stop buffer (0.1 mM CHC solution in HEPES buffer) was added. The plate was kept on ice. Now, the HEPES buffer in one triplicate was removed and DMSO solution was added and immediately removed and ice-cold stop buffer was added. This was considered as "Zero". One triplicate was left blank, which was used for protein assay after lysing the cells. The cells were washed twice with ice-cold HEPES buffer and then 250 uL of 0.1M NaOH in 5% Triton-X solution was added and the plate is kept on a shaker for 40 minutes to lyse the cells. 150 uL of the lysed cells was added into 4 mL of the scintillation fluid in a scintillation vial and scintillation count was obtained in disintegrations per minute (dpm). The percent inhibition values were calculated by taking DMSO as minimum. Concentration study (usually 9-12 dilutions) was done to determine the IC50 of each compound.
- Biochemical Assay for A-Raf (1) Activity of A-Raf kinase (SEQ. ID No: 1) was determined spectroscopically using a coupled pyruvate kinase/lactate dehydrogenase assay that continuously monitors the ATP hydrolysis-dependent oxidation of NADH (e.g., Schindler et al., Science, 2000, 289, 1938-1942). Assays were conducted in 384-well plates (100 μL final volume) using 5.55 nM A-Raf (Sigma), 1.5 units pyruvate kinase, 2.1 units lactate dehydrogenase, 1 mM phosphoenol pyruvate, 0.28 mM NADH, 30.1 nM MEK (SignalChem), and 1 mM ATP in assay buffer (100 mM Tris, pH 7.5, 15 mM MgCl2, 0.5 mM DTT, 0.1% octyl-glucoside, 0.002% (w/v) BSA, and 0.002% Triton X-100). Inhibition of A-Raf was measured by adding serial diluted test compound (final assay concentration of 1% DMSO). A decrease in absorption at 340 nm was monitored continuously for 6 h at 30° C. on a multi-mode microplate reader (BioTek). The reaction rate was calculated using the 4 to 5 h time frame. The reaction rate at each concentration of compound was converted to percent inhibition using controls (i.e., reaction with no test compound and reaction with a known inhibitor) and IC50 values were calculated by fitting a four-parameter sigmoidal curve to the data using Prism (GraphPad software).
- Biochemical Assay for A-Raf (2) Activity of A-Raf kinase (SEQ. ID No: 1) was determined spectroscopically using a coupled pyruvate kinase/lactate dehydrogenase assay that continuously monitors the ATP hydrolysis-dependent oxidation of NADH (e.g., Schindler et al., Science, 2000, 289, 1938-1942). Assays were conducted in 384-well plates (25 μL final volume) using 20 nM A-Raf (Eurofins), 1.5 units pyruvate kinase, 2.1 units lactate dehydrogenase, 1 mM phosphoenol pyruvate, 0.7 mM NADH, 100 nM MEK (SignalChem), and 1 mM ATP in assay buffer (100 mM Tris, pH 7.5, 15 mM MgCl2, 0.5 mM DTT, 0.1% octyl-glucoside, 0.002% (w/v) BSA, and 0.002% Triton X-100). Inhibition of A-Raf was measured by adding serial diluted test compound (final assay concentration of 1% DMSO). A decrease in absorption at 340 nm was monitored hourly for 4 h at 30° C. on a multi-mode microplate reader (BioTek). The reaction rate was calculated using the 3 to 4 h time frame. The reaction rate at each concentration of compound was converted to percent inhibition using controls (i.e., reaction with no test compound and reaction with a known inhibitor) and IC50 values were calculated by fitting a four-parameter sigmoidal curve to the data using Prism (GraphPad software).
- Biochemical Assay for B-Raf (1) Activity of B-Raf kinase (SEQ. ID NO: 2) was determined spectroscopically using a coupled pyruvate kinase/lactate dehydrogenase assay that continuously monitors the ATP hydrolysis-dependent oxidation of NADH (e.g., Schindler et al., Science, 2000, 289, 1938-1942). Assays were conducted in 384-well plates (100 μL final volume) using 0.13 nM B-Raf (Sigma), 1.5 units pyruvate kinase, 2.1 units lactate dehydrogenase, 1 mM phosphoenol pyruvate, 0.28 mM NADH, 30.1 nM MEK (SignalChem), and 1 mM ATP in assay buffer (100 mM Tris, Ph 7.5, 15 mM MgCl2, 0.5 mM DTT, 0.1% octyl-glucoside, 0.002% (w/v) BSA, and 0.002% Triton X-100). Inhibition of B-Raf was measured by adding serial diluted test compound (final assay concentration of 1% DMSO). A decrease in absorption at 340 nm was monitored continuously for 6 h at 30° C. on a multi-mode microplate reader (BioTek). The reaction rate was calculated using the 4 to 5 h time frame. The reaction rate at each concentration of compound was converted to percent inhibition using controls (i.e., reaction with no test compound and reaction with a known inhibitor) and IC50 values were calculated by fitting a four-parameter sigmoidal curve to the data using Prism (GraphPad software).
- Biochemical Assay for B-Raf (2) Activity of B-Raf kinase (SEQ. ID NO: 2) was determined spectroscopically using a coupled pyruvate kinase/lactate dehydrogenase assay that continuously monitors the ATP hydrolysis-dependent oxidation of NADH (e.g., Schindler et al., Science, 2000, 289, 1938-1942). Assays were conducted in 384-well plates (25 μL final volume) using 2 nM B-Raf (Sigma), 1.5 units pyruvate kinase, 2.1 units lactate dehydrogenase, 1 mM phosphoenol pyruvate, 0.7 mM NADH, 50 nM MEK (SignalChem), and 1 mM ATP in assay buffer (100 mM Tris, pH 7.5, 15 mM MgCl2, 0.5 mM DTT, 0.1% octyl-glucoside, 0.002% (w/v) BSA, and 0.002% Triton X-100). Inhibition of B-Raf was measured by adding serial diluted test compound (final assay concentration of 1% DMSO). A decrease in absorption at 340 nm was monitored hourly for 4 h at 30° C. on a multi-mode microplate reader (BioTek). The reaction rate was calculated using the 2 to 3 h time frame. The reaction rate at each concentration of compound was converted to percent inhibition using controls (i.e., reaction with no test compound and reaction with a known inhibitor) and IC50 values were calculated by fitting a four-parameter sigmoidal curve to the data using Prism (GraphPad software).
- Biochemical Assay for B-Raf (V600E) (1) Activity of B-Raf (V600E) (SEQ. ID NO: 4) kinase was determined spectroscopically using a coupled pyruvate kinase/lactate dehydrogenase assay that continuously monitors the ATP hydrolysis-dependent oxidation of NADH (e.g., Schindler et al., Science, 2000, 289, 1938-1942). Assays were conducted in 384-well plates (100 μL final volume) using 0.03 nM B-Raf (SignalChem), 1.5 units pyruvate kinase, 2.1 units lactate dehydrogenase, 1 mM phosphoenol pyruvate, 0.28 mM NADH, 30.1 nM MEK (SignalChem), and 1 mM ATP in assay buffer (100 mM Tris, Ph 7.5, 15 mM MgCl2, 0.5 mM DTT, 0.1% octyl-glucoside, 0.002% (w/v) BSA, and 0.002% Triton X-100). Inhibition of B-Raf (V600E) was measured by adding serial diluted test compound (final assay concentration of 1% DMSO). A decrease in absorption at 340 nm was monitored continuously for 6 h at 30° C. on a multi-mode microplate reader (BioTek). The reaction rate was calculated using the 3 to 4 h time frame. The reaction rate at each concentration of compound was converted to percent inhibition using controls (i.e., reaction with no test compound and reaction with a known inhibitor) and IC50 values were calculated by fitting a four-parameter sigmoidal curve to the data using Prism (GraphPad software).
- Biochemical Assay for B-Raf (V600E) (2) Activity of B-Raf (V600E) (SEQ. ID NO: 4) kinase was determined spectroscopically using a coupled pyruvate kinase/lactate dehydrogenase assay that continuously monitors the ATP hydrolysis-dependent oxidation of NADH (e.g., Schindler et al., Science, 2000, 289, 1938-1942). Assays were conducted in 384-well plates (25 μL final volume) using 0.5 nM B-Raf (deCode), 1.5 units pyruvate kinase, 2.1 units lactate dehydrogenase, 1 mM phosphoenol pyruvate, 0.7 mM NADH, 100 nM MEK (SignalChem), and 1 mM ATP in assay buffer (100 mM Tris, pH 7.5, 15 mM MgCl2, 0.5 mM DTT, 0.1% octyl-glucoside, 0.002% (w/v) BSA, and 0.002% Triton X-100). Inhibition of B-Raf (V600E) was measured by adding serial diluted test compound (final assay concentration of 1% DMSO). A decrease in absorption at 340 nm was monitored hourly for 4 h at 30° C. on a multi-mode microplate reader (BioTek). The reaction rate was calculated using the 3 to 4 h time frame. The reaction rate at each concentration of compound was converted to percent inhibition using controls (i.e., reaction with no test compound and reaction with a known inhibitor) and IC50 values were calculated by fitting a four-parameter sigmoidal curve to the data using Prism (GraphPad software).
- Biochemical Assay for C-Raf (1) Activity of C-Raf kinase (SEQ. ID NO: 3) was determined spectroscopically using a coupled pyruvate kinase/lactate dehydrogenase assay that continuously monitors the ATP hydrolysis-dependent oxidation of NADH (e.g., Schindler et al., Science, 2000, 289, 1938-1942). Assays were conducted in 384-well plates (100 μL final volume) using 0.43 nM C-Raf (Sigma), 1.5 units pyruvate kinase, 2.1 units lactate dehydrogenase, 1 mM phosphoenol pyruvate, 0.28 mM NADH, 30.1 nM MEK (SignalChem), and 1 mM ATP in assay buffer (100 mM Tris, Ph 7.5, 15 mM MgCl2, 0.5 mM DTT, 0.1% octyl-glucoside, 0.002% (w/v) BSA, and 0.002% Triton X-100). Inhibition of C-Raf was measured by adding serial diluted test compound (final assay concentration of 1% DMSO). A decrease in absorption at 340 nm was monitored continuously for 6 h at 30° C. on a multi-mode microplate reader (BioTek). The reaction rate was calculated using the 4 to 5 h time frame. The reaction rate at each concentration of compound was converted to percent inhibition using controls (i.e., reaction with no test compound and reaction with a known inhibitor) and IC50 values were calculated by fitting a four-parameter sigmoidal curve to the data using Prism (GraphPad software).
- Biochemical Assay for C-Raf (2) Activity of C-Raf kinase (SEQ. ID NO: 3) was determined spectroscopically using a coupled pyruvate kinase/lactate dehydrogenase assay that continuously monitors the ATP hydrolysis-dependent oxidation of NADH (e.g., Schindler et al., Science, 2000, 289, 1938-1942). Assays were conducted in 384-well plates (25 μL final volume) using 3.84 nM C-Raf (Eurofins), 1.5 units pyruvate kinase, 2.1 units lactate dehydrogenase, 1 mM phosphoenol pyruvate, 0.7 mM NADH, 50 nM MEK (SignalChem), and 1 mM ATP in assay buffer (100 mM Tris, pH 7.5, 15 mM MgCl2, 0.5 mM DTT, 0.1% octyl-glucoside, 0.002% (w/v) BSA, and 0.002% Triton X-100). Inhibition of C-Raf was measured by adding serial diluted test compound (final assay concentration of 1% DMSO). A decrease in absorption at 340 nm was monitored hourly for 4 h at 30° C. on a multi-mode microplate reader (BioTek). The reaction rate was calculated using the 2 to 3 h time frame. The reaction rate at each concentration of compound was converted to percent inhibition using controls (i.e., reaction with no test compound and reaction with a known inhibitor) and IC50 values were calculated by fitting a four-parameter sigmoidal curve to the data using Prism (GraphPad software).
- CSF1R Kinase Assay The activity of CSF1R kinase (CSF1R, SEQ ID NO: 1) was determined spectroscopically using a coupled pyruvate kinase/lactate dehydrogenase assay that continuously monitors the ATP hydrolysis-dependent oxidation of NADH (e.g., Schindler et al. Science (2000) 289: 1938-1942 Assays were conducted in 384-well plates (100 μL final volume) using 10 nM CSF1R (Eurofins), 1.5 units pyruvate kinase, 2.1 units lactate dehydrogenase, 1 mM phosphoenol pyruvate, 0.28 mM NADH, 0.7 mg/mL PolyEY and 1 mM ATP in assay buffer (100 mM Tris, pH 7.5, 15 mM MgCl2, 0.5 mM DTT, 0.1% octyl-glucoside, 0.002% (w/v) BSA, and 0.002% Triton X-100). Inhibition of CSF1R was measured by adding serial diluted test compound (final assay concentration of 1% DMSO). A decrease in absorption at 340 nm was monitored continuously for 4 hours at 30° C. on a multi-mode microplate reader (BioTek). The reaction rate was calculated using the 2-3 h time frame. The reaction rate at each concentration of compound was converted to percent inhibition using controls (i.e. reaction with no test compound and reaction with a known inhibitor) and IC50values were calculated by fitting a four-parameter sigmoidal curve to the data using Prism (GraphPad software).
- FLT3 Kinase Assay The activity of FLT3 kinase was determined spectroscopically using a coupled pyruvate kinase/lactate dehydrogenase assay that continuously monitors the ATP hydrolysis-dependent oxidation of NADH (e.g., Schindler et al. Science (2000) 289: 1938-1942). Assays were conducted in 384-well plates (100 μL final volume) using 1.6 nM FLT3 (Invitrogen), 1.5 units pyruvate kinase, 2.1 units lactate dehydrogenase, 1 mM phosphoenol pyruvate, 0.28 mM NADH, 0.7 mg/mL PolyEY and 1 mM ATP in assay buffer (100 mM Tris, pH 7.5, 15 mM MgCl2, 0.5 mM DTT, 0.1% octyl-glucoside, 0.002% (w/v) BSA, and 0.002% Triton X-100). Inhibition of FLT3 was measured by adding serial diluted test compound (final assay concentration of 1% DMSO). A decrease in absorption at 340 nm was monitored continuously for 6 hours at 30° C. on a multi-mode microplate reader (BioTek). The reaction rate was calculated using the 3-4 h time frame. The reaction rate at each concentration of compound was converted to percent inhibition using controls (i.e. reaction with no test compound and reaction with a known inhibitor) and IC50 values were calculated by fitting a four-parameter sigmoidal curve to the data using Prism (GraphPad software).
- PDGFRα Kinase Assay The activity of unphosphorylated PDGFRα kinase was determined spectroscopically using a coupled pyruvate kinase/lactate dehydrogenase assay that continuously monitors the ATP hydrolysis-dependent oxidation of NADH (e.g., Schindler et al. Science (2000) 289: 1938-1942). Assays were conducted in 384-well plates (100 μL final volume) using 11.7 nM PDGFRα (DeCode Biostructures), 1.5 units pyruvate kinase, 2.1 units lactate dehydrogenase, 1 mM phosphoenol pyruvate, 0.28 mM NADH, 0.7 mg/mL PolyEY and 1 mM ATP in assay buffer (100 mM Tris, pH 7.5, 15 mM MgCl2, 0.5 mM DTT, 0.1% octyl-glucoside, 0.002% (w/v) BSA, and 0.002% Triton X-100). Inhibition of PDGFRα was measured by adding serial diluted test compound (final assay concentration of 1% DMSO). A decrease in absorption at 340 nm was monitored continuously for 6 hours at 30° C. on a multi-mode microplate reader (BioTek). The reaction rate was calculated using the 2-3 h time frame. The reaction rate at each concentration of compound was converted to percent inhibition using controls (i.e. reaction with no test compound and reaction with a known inhibitor) and IC50values were calculated by fitting a four-parameter sigmoidal curve to the data using Prism (GraphPad software).
- Protein Kinase C beta 2 (PKCpII) Assay Protein Kinase C beta 2 (PKCpII) catalyzes the production of ADP from ATP that accompanies the phosphoryl transfer to the PKC Pseudosubstrate peptide (A→S, RFARKGSLRQKNV). This transfer is coupled to the oxidation of β-NADH through the activities of Pyruvate Kinase (PK) and Lactate Dehydrogenase (LDH). β-NADH conversion to NAD+ is monitored by the decrease in absorbance at 340 nm (e=6.22 cm−1 mM−1) using a Molecular Devices SPECTRA max PLUS spectrophotometer.A typical assay was carried out on a 96-well, clear microtiter plate in a Molecular Devices spectrophotometer for 20 minutes at 30° C. in 0.1 mL of assay buffer containing 50 mM HEPES, pH 7.4, 5 nM PKC, 23 units of pyruvate kinase, 33 units of lactate dehydrogenase, 0.15 mM peptide, 0.1 mM ATP, 1 mM DTT, 4 mM PEP, 8 mM MgCl2, 0.3 mM NADH, 60 mM CaCl2), 10 mg/mL PS, 50 ng/mL PMA, 7.5% DMSO and from about 10,000 nM to 0.169 nM compound inhibitor. Stock solutions of 3-sn-phosphatidyl-L-serine (PS) and phorbol-12-myristate-13-acetate (PMA) were sonicated for 30 seconds just prior to addition to assay buffer and assays were initiated by the addition of 100 μM ATP.
- The Alizarin Assay (ARS) (Lactate) The alizarin-red binding assay is a colorimetric assay used to determine the inhibition affinity of boronate compounds to glucose. The assay is based on a colour shift of alizarin-red upon binding to boronate, which shift can be followed by change in absorbance in the 330-340 nm region. For determination of the inhibitory constant (Ki) between the boronate and the carbohydrate, 400 μM of boronic acids is dissolved in a 20 mM phosphate buffer pH 7.4 under gentle stirring. Upon complete dissolution of the compound, 200 μM of Alizarin red (ARS) is added to the solution. The ARS-boronate solution is then aliquoted into a 96 multiwell plate (black, flat and clear bottom) 1:1 with appropriate carbohydrate. In particular, L-lactate solutions are prepared in a 20 mM phosphate buffer pH 7.4 at these concentrations respectively: 1000, 500, 250, 100, 50, 25, 10, 5, 2.5, 1, 0.25, 0.1 mM and 2500, 1000, 500, 100, 50, 10, 5, 1, 0.5, 0.1, 0.05, 0.01 mM. The plate with ARS-boronate mixed with carbohydrate is incubated 20 minutes at room temperature. After 5 minutes of centrifugation at 4000 rpm the plate is placed in a multiwell spectrometer (Spectra Max, Molecular Devices) for absorption detection.
- Biochemical Assay for uKIT Activity of c-KIT kinase (SEQ. ID NO. 1 or SEQ. ID NO. 2) was determined spectroscopically using a coupled pyruvate kinase/lactate dehydrogenase assay that continuously monitors the ATP hydrolysis-dependent oxidation of NADH (e.g., Schindler et al. Science (2000) 289: 1938-1942). Assays were conducted in 384-well plates (100 μL final volume) using 16 nM (Decode, SEQ ID No. 1) or 4.36 nM (Signal Chem, SEQ. ID No. 2), 0.7 mg/mL PE4Y substrate, 1.5 units pyruvate kinase, 2.1 units lactate dehydrogenase, 1 mM phosphoenol pyruvate, 0.28 mM NADH and 1 mM ATP in assay buffer (100 mM Tris, pH 7.5, 15 mM MgCl2, 0.5 mM DTT, 0.1% octyl-glucoside, 0.002% (w/v) BSA, and 0.002% Triton X-100). Inhibition of KIT was measured by adding serial diluted test compound (final assay concentration of 1% DMSO). A decrease in absorption at 340 nm was monitored continuously for 6 h at 30° C. on a multi-mode microplate reader (BioTek). The reaction rate was calculated using the 2-3 h time frame. The reaction rate at each concentration of compound was converted to percent inhibition using controls (i.e., reaction with no test compound and reaction with a known inhibitor) and IC50 values were calculated using software routines in Prism (GraphPad software).
- c-Kit Kinase Assay The activity of unphosphorylated c-KIT kinase (c-KIT, SEQ ID NO: 2) was determined spectroscopically using a coupled pyruvate kinase/lactate dehydrogenase assay that continuously monitors the ATP hydrolysis-dependent oxidation of NADH (e.g., Schindler et al. Science (2000) 289: 1938-1942). Assays were conducted in 384-well plates (100 μL final volume) using 16 nM c-KIT (DeCode Biostructures), 1.5 units pyruvate kinase, 2.1 units lactate dehydrogenase, 1 mM phosphoenol pyruvate, 0.28 mM NADH, 0.7 mg/mL PolyEY and 1 mM ATP in assay buffer (100 mM Tris, pH 7.5, 15 mM MgCl2, 0.5 mM DTT, 0.1% octyl-glucoside, 0.002% (w/v) BSA, and 0.002% Triton X-100). Inhibition of c-KIT was measured by adding serial diluted test compound (final assay concentration of 1% DMSO). A decrease in absorption at 340 nm was monitored continuously for 6 hours at 30° C. on a multi-mode microplate reader (BioTek). The reaction rate was calculated using the 2-3 h time frame. The reaction rate at each concentration of compound was converted to percent inhibition using controls (i.e. reaction with no test compound and reaction with a known inhibitor) and IC50 values were calculated by fitting a four-parameter sigmoidal curve to the data using Prism (GraphPad software). as implemented in the GraphPad Prism software package.
- ATPase Assay The conversion of ATP to ADP by S. aureus TopoIV enzyme is coupled to the conversion of NADH to NAD+, and the progress of the reaction is measured by the change in absorbance at 340 nm. TopoIV (64 nM) is incubated with the selected compound (3% DMSO final) in buffer for 10 minutes at 30 ° C. The buffer consists of 100 mM Tris 7.5, 1.5 mM MgCl2, 200 mM K.Glutamate, 2.5 mM phosphoenol pyruvate, 0.2 mM NADH, 1 mM DTT, 5 g/mL linearized DNA, 50 g/mL BSA, 30 g/mL pyruvate kinase, and 10 g/mL lactate dehyrodgenase (LDH). The reaction is initiated with ATP, and rates are monitored continuously for 20 minutes at 30 ° C. on a Molecular Devices SpectraMAX plate reader.
- LDH Activity Assay Lactate dehydrogenase activity was determined by recording the absorbance change at 340 nm produced by the oxidation of NADH. Assays were performed at 37°C. The reagent mixture contained 0.115 mM NADH, 50 mM sodium phosphate buffer, pH 7.4, sodium pyruvate (0.31 mM for LDH-C4 and LDH-B4 and 1.25 mM for LDH-A4) and the enzyme preparation, diluted with phosphate buffer, pH 7.4, to provide a ΔE340 of 0.06-0.07 per minute when the activity was assayed in a 1-cm light path. For determination of dissociation constants, the isozymes, the inhibitors (oxamate or oxamate derivatives) and the coenzyme were incubated with the buffer used in the assay for 10 min at 37°C before adding the substrate.
- Spectrophotometric 384 Well Assay Assay reactions are then carried out in 384-well plates, with hACC2 in an appropriate dilution and at final assay concentrations (f.c.) of 100 mM Tris (pH 7.5), 10 mM trisodium citrate, 25 mM KHCO3, 10 mM MgCl2, 0.5 mg/ml BSA, 3.75 mM reduced L-glutathione, 15 U/ml lactate dehydrogenase, 0.5 mM phosphoenolpyruvate, 15 U/ml pyruvate kinase, compounds at different concentrations at final DMSO concentrations of 1%.The enzymatic reaction is then started by addition of a mixture of NADH, acetyl Coenzyme A (both 200 μM f.c.) and ATP (500 uM f.c.). The decrease of the optical density (slope S) is then determined at 25° C. at a wavelength of 340 nm over 15 minutes in a spectrophotometric reader.
- Inhibition Assay Compounds of the present invention were screened for their ability to inhibit GSK-3β (AA 1-420) activity using a standard coupled enzyme system (Fox et al., Protein Sci. 1998, 7, 2249). Reactions were carried out in a solution containing 100 mM HEPES (pH 7.5), 10 mM MgCl2, 25 mM NaCl, 300 μM NADH, 1 mM DTT and 1.5% DMSO. Final substrate concentrations in the assay were 20 μM ATP (Sigma Chemicals, St Louis, Mo.) and 300 μM peptide (American Peptide, Sunnyvale, Calif.). Reactions were carried out at 30 C. and 20 nM GSK-3β. Final concentrations of the components of the coupled enzyme system were 2.5 mM phosphoenolpyruvate, 300 μM NADH, 30 μg/ml pyruvate kinase and 10 μg/ml lactate dehydrogenase.
- Protein Kinase C beta 2 Assay A typical assay was carried out on a 96-well, clear microtiter plate in a Molecular Devices spectrophotometer for 20 minutes at 30° C. in 0.1 mL of assay buffer containing 50 mM HEPES, pH 7.4, 5 nM PKC, 23 units of pyruvate kinase, 33 units of lactate dehydrogenase, 0.15 mM peptide, 0.1 mM ATP, 1 mM DTT, 4 mM PEP, 8 mM MgCl2, 0.3 mM NADH, 60 mM CaCl2, 10 mg/mL PS, 50 ng/mL PMA, 7.5% DMSO and from about 10,000 nM to 0.169 nM compound inhibitor. Stock solutions of 3-sn-phosphatidyl-L-serine (PS) and phorbol-12-myristate-13-acetate (PMA) were sonicated for 30 seconds just prior to addition to assay buffer and assays were initiated by the addition of 100 μM ATP.
- Myofibril Assays To evaluate the effect of compounds on the ATPase activity of full-length cardiac myosin in the context of the native sarcomere, skinned myofibril assays were performed. Bovine cardiac myofibrils were obtained by homogenizing bovine cardiac left ventricular tissue in the presence of a detergent such as triton X-100. Such treatment removes membranes and a majority of the soluble cytoplasmic proteins but leaves intact the cardiac sarcomeric acto-myosin apparatus. Myofibril preparations retain the ability to hydrolyze ATP in an Ca2+ regulated manner. ATPase activities of such myofibril preparations in the presence and absence of compounds were assayed at Ca2+ concentrations activating to a defined fraction of the maximal rate (i.e., 25%, 75%). Small molecule agents were assessed for their ability to inhibit the steady-state ATPase activity of bovine cardiac myofibrils using pyruvate kinase and lactate dehydrogenase (PK/LDH)-coupled enzyme system. This assay regenerates myosin-produced ADP into ATP by oxidizing NADH, producing an absorbance change at 340 nm. Prior to testing small molecule agents, the bovine cardiac myofibrils were assessed for their calcium responsiveness and the calcium concentration that achieves either a 50% (pCa50) or 75% (pCa75) activation of the myofibril system was chosen as the final condition for assessing the inhibitory activity of the small molecule agents. All enzymatic activity was measured in a buffered solution containing 12 mM PIPES (piperazine-N,N′-bis(2-ethanesulfonic acid), 2 mM magnesium chloride at pH 6.8 (PM 12 buffer). Final assay conditions were 1 mg/mL of bovine cardiac myofibrils, 4 U/mL pyruvate kinase, 6 U/mL lactate dehydrogenase, 50 μM ATP, 0.1 mg/mL BSA (bovine serum albumin), 10 ppm antifoam, 1 mM DTT, 0.5 mM NADH, 1.5 mM PEP, 0.6 mM EGTA, and an amount of CaCl2) sufficient to achieve either 50% or 75% activation of the myofibril ATPase activity.
- n Vitro CDK7 (5 nM) Assay Inhibition potencies of compounds were studied using an absorbance kinetic assay as described below. Absorbance kinetic assay (5 nM CDK7/Cyclin H/MAT-1 complex}CDK7 complex catalyzes the ATP-dependent phosphorylation of a peptide substrate CDK7/9-tide that is derived from RNA Pol II to produce phosphorylated peptide and ADP. The kinase reaction product ADP was converted to lactate and NAD+ in the presence of phosphoenol pyruvate (PEP), NADH and coupling enzymes lactate dehydrogenase (LDH) and pyruvate kinase (PK). CDK7 complex catalytic activity was measured by following the absorbance intensity continuously at 340 nm that corresponds with the depletion of NADH.Compound potencies were measured by a 12-point dose response manner under the assay conditions of 300 μM CDK7/9 tide (KMpeptide =140.5 ± 18.5 μM), 500μM ATP (KMATP = 27.8 ± 4.1 μM), 500 μM PEP, 100 μM NADH, 0.6-lunit PK/0.9-1.4 unit and 20 nM CDK7/Cyclin H/MAT-1 complex in a buffer containing 20 mM Tris, pH 7.4, 10 mM MgCF and 0.004% Triton X-100. Absorbance at 340 nm was followed kinetically at an interval of 2 minutes for 8 hours.The assay was carried out with 100 μl reaction volume per well in a 384-well plate that was pre-spotted nanoliter volume of compounds by LabCyte Echo 555. Compound dilution plates were made by 2-fold (could vary upon necessity) dilution in DMSO for 11 concentrations plus a DMSO control of uninhibited reaction. 2x substrate and coupling reagent mixture was added to the assay plate followed by an addition of an equal volume of 40 nM CDK7/Cyclin H/MAT-1 complex. After mixing, assay plates were spun at 2000 rpm for 3 minutes and then transferred to the plate reader for data collection.
- Determination of Inhibitory Potencies by Coupled ATPase Activity Assay Inhibitory potencies of compounds were determined in a coupled ATPase activity assay using SERCA microsomes at 14 different inhibitor concentrations. In a plastic cuvette, the enzyme and the inhibitor were incubated 37 deg C in the assay buffer containing the enzyme pyruvate kinase and lactate dehydrogenase. After checking for potential background rates of NADH oxidation, the reaction was started by adding ATP. Rates of the rate-limiting NADH oxidation were determined spectroscopically for 5 min with a spectrophotometer operating at a wavelength of 340 nm. ATP hydrolysis rates as a function of inhibitor concentration were fit to a three-parameter logistic equation (amplitude, offset, and IC50). The IC50 was the concentration of inhibitor that caused inhibition of half of the ATPase activity observed in the absence of inhibitor. Reported IC50 values were the average of at least three independent trials.
- Biological Assay Assay A, a 384-well format on a Corning 3653 assay plate is used, and monitored by UV-vis spectroscopy in continuous mode at rt. Compounds were prepared in DMSO as 4 mM stocks, diluted using an 11-point half-log scheme on a Biomek FX (Beckman Coulter), and incubated at rt for 30 minutes with the reaction mixture containing 50 mM HEPES, pH 7.4, 140 mM KCl, 3.5 mM MgCl2, 0.8 mM fructose, 2 mM TCEP, 0.8 mM PEP, 0.7 mM NADH, 0.01% Triton X-100, 30 U/mL pyruvate kinase-lactate dehydrogenase, and 10 nM purified KHK-C. The compound concentration in each well ranged from 1 nM to 100 μM. The reaction was initiated with the addition of 0.2 mM ATP. The absorbance was measured for 30 minutes on a SpectraMax reader (Molecular Devices) after ATP was added. The concentrations provided are based on the final mixture volume of 40 μL (referred to as the final concentration).
- Biological Assay Assay A, a 384-well format on a Corning 3653 assay plate is used, and monitored by UV-vis spectroscopy in continuous mode at rt. Compounds were prepared in DMSO as 4 mM stocks, diluted using an 11-point half-log scheme on a Biomek FX (Beckman Coulter), and incubated at rt for 30 minutes with the reaction mixture containing 50 mM HEPES, pH 7.4, 140 mM KCl, 3.5 mM MgCl2, 0.8 mM fructose, 2 mM TCEP, 0.8 mM PEP, 0.7 mM NADH, 0.01% Triton X-100, 30 U/mL pyruvate kinase-lactate dehydrogenase, and 10 nM purified KHK-C. The compound concentration in each well ranged from 1 nM to 100 uM. The reaction was initiated with the addition of 0.2 mM ATP. The absorbance was measured for 30 minutes on a SpectraMax reader (Molecular Devices) after ATP was added. The concentrations provided are based on the final mixture volume of 40 uL (referred to as the final concentration).
- KHK Inhibition Assay Determination of Human and Rat recombinant KHK-A and KHK-C isozyme IC50 ValuesAn assay reagent cocktail was prepared by combining NADH, water, TEA, KCl, MgCl2, PEP, ATP, DTT, coupling enzymes (pyruvate kinase and lactate dehydrogenase, LDH) to final concentrations as shown in Table B.TABLE BReagent Final ConcentrationNADH 300 μMWater —TEA 33 mMKCl 100 mMMgCl2 6 mMPEP 1.33 mMATP 0.1 mMDTT 12 mMPyruvate kinase 1.0 U/mLLDH 1.0 U/mL[0367]To this was added the relevant KHK isozyme to a final concentration of 6 nM. Aliquots of each inhibitor compound were diluted via 5-fold serial dilutions to produce final concentrations ranging from 1000 nM to 0.064 nM. The inhibitor aliquots were added to the assay reagent cocktail containing KHK with fructose (at a concentration of 2 mM) in a 96-well plate. The absorbance at 340 nm was measured via spectrophotometry and inhibition was analyzed using non-linear regression.
- Receptor Tyrosine Kinase Activity Assay Recombinant human c-Met protein (Invitrogen, Carlsbad, Calif., USA) is used. As substrate for the kinase reaction the peptide KKKSPGEYVNIEFG (JPT, Germany) is used. For the assay, 1 uL of a 51-fold concentrated solution of the test compound in DMSO is pipetted into a white 384-well microtiter plate (Greiner Bio-One, Frickenhausen, Germany). 25 uL of a solution of c-Met (final concentration 30 nM) and pyruvate kinase/lactate dehydrogenase (Roche Diagnostics, Mannheim, Germany; final concentration 8 mg/L) in assay buffer [3-(N-morpholino)propanesulfonic acid (MOPS), 50 mM, pH 7; MgCl2, 10 mM; bovine serum albumin (BSA), 0.01%; Triton X 100, 0.01%; DTT, 2 mM] are added, and the mixture is incubated for 5 min at room temperature. Then, the kinase reaction is started by the addition of 25 uL of a solution of adenosine triphosphate (ATP, final concentration 30 uM), substrate (final concentration 100 uM), nicotinamide adenine dinucleotide.
- Spectrophotometric 384 Well Assay Assay reactions are then carried out in 384-well plates, with hACC2 in an appropriate dilution and at final assay concentrations (f.c.) of 100 mM Tris (pH 7.5), 10 mM trisodium citrate, 25 mM KHCO3, 10 mM MgCl2, 0.5 mg/ml BSA, 3.75 mM reduced L-glutathione, 15 U/ml lactate dehydrogenase, 0.5 mM phosphoenolpyruvate, 15 U/ml pyruvate kinase, compounds at different concentrations at final DMSO concentrations of 1%.The enzymatic reaction is then started by addition of a mixture of NADH, acetyl Coenzyme A (both 2000 f.c.) and ATP (500 uM f.c.). The decrease of the optical density (slope S) is then determined at 25° C. at a wavelength of 340 nm over 15 minutes in a spectrophotometric reader.Each assay microtiter plate contains wells with vehicle instead of compound as controls for the non-inhibited enzyme (100% CTL; HIGH) and wells without acetyl-CoA as controls for non-specific NADH degradation (0% CTL; LOW).
- Tyrosine Kinase Activity Assay Recombinant human c-Met protein (Invitrogen, Carlsbad, Calif., USA) is used. As substrate for the kinase reaction the peptide KKKSPGEYVNIEFG (JPT, Germany) is used. For the assay, 1 uL of a 51-fold concentrated solution of the test compound in DMSO is pipetted into a white 384-well microtiter plate (Greiner Bio-One, Frickenhausen, Germany). 25 uL of a solution of c-Met (final concentration 30 nM) and pyruvate kinase/lactate dehydrogenase (Roche Diagnostics, Mannheim, Germany; final concentration 8 mg/L) in assay buffer [3-(N-morpholino)propane-sulfonic acid (MOPS), 50 mM, pH 7; MgCl2, 10 mM; bovine serum albumin (BSA), 0.01%; Triton X 100, 0.01%; DTT, 2 mM] are added, and the mixture is incubated for 5 min at room temperature. Then, the kinase reaction is started by the addition of 25 uL of a solution of adenosine triphosphate (ATP, final concentration 30 uM), substrate (final concentration 100 uM), nicotinamide adenine dinucleotide.
- Tyrosine Kinase Activity Assay Recombinant human c-Met protein (Invitrogen, Carlsbad, Calif., USA) is used. As substrate for the kinase reaction the peptide KKKSPGEYVNIEFG (JPT, Germany) is used. For the assay, 1 uL of a 51-fold concentrated solution of the test compound in DMSO is pipetted into a white 384-well microtiter plate (Greiner Bio-One, Frickenhausen, Germany). 25 uL of a solution of c-Met (final concentration 30 nM) and pyruvate kinase/lactate dehydrogenase (Roche Diagnostics, Mannheim, Germany; final concentration 8 mg/L) in assay buffer [3-(N-morpholino)propanesulfonic acid (MOPS), 50 mM, pH 7; MgCl2, 10 mM; bovine serum albumin (BSA), 0.01%; Triton X 100, 0.01%; DTT, 2 mM] are added, and the mixture is incubated for 5 min at room temperature. Then, the kinase reaction is started by the addition of 25 uL of a solution of adenosine triphosphate (ATP, final concentration 30 uM), substrate (final concentration 100 uM), nicotinamide adenine dinucleotide.
- Inhibition Assay Protein Kinase C beta 2 (PKCβII) catalyzes the production of ADP from ATP that accompanies the phosphoryl transfer to the PKC Pseudosubstrate peptide (A→S, RFARKGSLRQKNV). This transfer is coupled to the oxidation of β-NADH through the activities of Pyruvate Kinase (PK) and Lactate Dehydrogenase (LDH). β-NADH conversion to NAD+ is monitored by the decrease in absorbance at 340 nm (e=6.22 cm−1 mM−1) using a Molecular Devices SPECTRA max PLUS spectrophotometer.A typical assay was carried out on a 96-well, clear microtiter plate in a Molecular Devices spectrophotometer for 20 minutes at 30° C. in 0.1 mL of assay buffer containing 50 mM HEPES, pH 7.4, 5 nM PKC, 23 units of pyruvate kinase, 33 units of lactate dehydrogenase, 0.15 mM peptide, 0.1 mM ATP, 1 mM DTT, 4 mM PEP, 8 mM MgCl2, 0.3 mM NADH, 60 mM CaCl2, 10 mg/mL PS, 50 ng/mL PMA, 7.5% DMSO and from about 10,000 nM to 0.169 nM compound inhibitor. Stock solutions of 3-sn-phosphatidyl-L-serine (PS) and phorbol-12-myristate-13-acetate (PMA) were sonicated for 30 seconds just prior to addition to assay buffer and assays were initiated by the addition of 100 μM ATP.Steady-state kinetic parameters for the bi-bi kinase reaction were determined at saturating phospho-acceptor peptide substrate concentration (0.15 mM) by fitting initial velocity data to the Michaelis-Menten equation, v=V max [S]/(K M +[S]) where v is the measured initial velocity, Vmax is the maximal enzyme velocity, [S] is the ATP substrate concentration, and KM is the Michealis constant for ATP. Enzyme turnover values (kcat) were calculated according to kcat=Vmax[E], where [E] is the total enzyme concentration. Enzyme inhibition constants (apparent Ki values) were determined by fitting initial velocities at variable inhibitor concentrations to a model for ATP competitive inhibition based on the Morrison equation). Morrison, J. F., Biochim. Biophys Acta 185: 269-286 (1969).
- n Vitro CDK7 (20 nM) Assay Inhibition potencies of compounds were studied using an absorbance kinetic assay as described below. Compounds with potencies approaching the limit of detection of the assay (IC50 < 10 nM) were further assessed in a more sensitive fluorescence end-point assay. Absorbance kinetic assay (20 nM CDK7/Cyclin H/MAT-1 complex}CDK7 complex catalyzes the ATP-dependent phosphorylation of a peptide substrate CDK7/9-tide that is derived from RNA Pol II to produce phosphorylated peptide and ADP. The kinase reaction product ADP was converted to lactate and NAD+ in the presence of phosphoenol pyruvate (PEP), NADH and coupling enzymes lactate dehydrogenase (LDH) and pyruvate kinase (PK). CDK7 complex catalytic activity was measured by following the absorbance intensity continuously at 340 nm that corresponds with the depletion of NADH.Compound potencies were measured by a 12-point dose response manner under the assay conditions of 300 μM CDK7/9 tide (KMpeptide =140.5 ± 18.5 μM), 500μM ATP (KMATP = 27.8 ± 4.1 μM), 500 μM PEP, 100 μM NADH, 0.6-lunit PK/0.9-1.4 unit and 20 nM CDK7/Cyclin H/MAT-1 complex in a buffer containing 20 mM Tris, pH 7.4, 10 mM MgCF and 0.004% Triton X-100. Absorbance at 340 nm was followed kinetically at an interval of 2 minutes for 8 hours.The assay was carried out with 100 μl reaction volume per well in a 384-well plate that was pre-spotted nanoliter volume of compounds by LabCyte Echo 555. Compound dilution plates were made by 2-fold (could vary upon necessity) dilution in DMSO for 11 concentrations plus a DMSO control of uninhibited reaction. 2x substrate and coupling reagent mixture was added to the assay plate followed by an addition of an equal volume of 40 nM CDK7/Cyclin H/MAT-1 complex. After mixing, assay plates were spun at 2000 rpm for 3 minutes and then transferred to the plate reader for data collection.
- ACCase Enzymatic Assay Ustilago maydis acetyl CoA carboxylase (ACCase) was cloned, expressed, and purified as described (Weatherly et al, Biochem. J., 2004) and the test compounds were tested in a 96-well plate format. Primary in vitro screening consisted of obtaining dose response data at 100, 33, 10, and 1 μM inhibitor. Actives in the primary screen were re-tested to establish IC50 values.Direct detection of the conversion of acetyl CoA to malonyl CoA by ACCase was not feasible, but during this process ATP is converted to ADP which allowed for detection through a standard reaction coupling with ADP recycling to the oxidation of NADH. Thus, ACCase activity was measured via kinetic OD340 measurements of the conversion of NADH to NAD in a coupled reaction involving the conversion of phosphoenolpyruvate (PEP) to lactate.The complete 200 ul reaction mixture contained 52.5 mM HEPES (pH8), 2.625 mM MgCl2, 1 mM ATP, 0.525 mM DTT, 11 mM NaHCO3, 1% DMSO with or without inhibitor, 1× pyruvate kinase/lactate dehydrogenase (PK/LDH), 0.3 mM NADH, 0.5 mM PEP, and 5 μg ACCase. The reactions were incubated at 30° C. for 10 minutes and then initiated by the addition of 0.33 mM acetyl CoA. The initiated reactions were read immediately via plate reader at OD340 and kinetic readings were acquired every 20 s for 15 minutes while keeping the temperature at 30° C.A slope of the kinetic curve was determined by using the 2 to 7 minute data which was then calculated as percent inhibition relative to the no inhibitor control.The primary screens were conducted in duplicate and the IC50's conducted in triplicate. Averages were reported along with standard deviation calculation to generate error bars.Each plate contained its own controls and consisted of a DMSO only control, 5-fold titration series of soraphen from 2 μM to 3.2 nM, and an ADP coupled reaction control.In order to effectively screen out non-specific modulators of pyruvate kinase and lactate dehydrogenase (the coupled portion of the reaction), a PK/LDH inhibition test was developed. The complete 200 μl reaction mixture contained 52.5 mM HEPES (pH8), 2.625 mM MgCl2, 0.525 mM DTT, 11 mM NaHCO3, 1% DMSO with or without inhibitor, 1× pyruvate kinase/lactate dehydrogenase (PK/LDH), 0.3 mM NADH, and 0.5 mM PEP. The reactions were incubated at 30° C. for 10 minutes and then initiated by the addition of 66 μM ADP. The initiated reactions were read immediately via plate reader at OD340 and kinetic readings were acquired every 20 s for 15 minutes while remaining at 30° C.A slope of the kinetic curve was determined by using the 2 to 7 minute data which was then calculated as percent inhibition relative to the no inhibitor control. Those compounds which had no significant PK/LDH inhibition at or above the IC50 in the ACCase assay, were considered to be valid modulators of only ACCase.
- ACCase Enzymatic Assay Ustilago maydis acetyl CoA carboxylase (ACCase) was cloned, expressed, and purified as described (Weatherly et al, Biochem. J., 2004) and the test compounds were tested in a 96-well plate format. Primary in vitro screening consisted of obtaining dose response data at 100, 33, 10, and 1 μM inhibitor. Actives in the primary screen were re-tested to establish IC50 values.Direct detection of the conversion of acetyl CoA to malonyl CoA by ACCase was not feasible, but during this process ATP is converted to ADP which allowed for detection through a standard reaction coupling with ADP recycling to the oxidation of NADH. Thus, ACCase activity was measured via kinetic OD340 measurements of the conversion of NADH to NAD in a coupled reaction involving the conversion of phosphoenolpyruvate (PEP) to lactate.The complete 200 ul reaction mixture contained 52.5 mM HEPES (pH8), 2.625 mM MgCl2, 1 mM ATP, 0.525 mM DTT, 11 mM NaHCO3, 1% DMSO with or without inhibitor, 1× pyruvate kinase/lactate dehydrogenase (PK/LDH), 0.3 mM NADH, 0.5 mM PEP, and 5 μg ACCase. The reactions were incubated at 30° C. for 10 minutes and then initiated by the addition of 0.33 mM acetyl CoA. The initiated reactions were read immediately via plate reader at OD340 and kinetic readings were acquired every 20 s for 15 minutes while keeping the temperature at 30° C.A slope of the kinetic curve was determined by using the 2 to 7 minute data which was then calculated as percent inhibition relative to the no inhibitor control.The primary screens were conducted in duplicate and the IC50's conducted in triplicate. Averages were reported along with standard deviation calculation to generate error bars.Each plate contained its own controls and consisted of a DMSO only control, 5-fold titration series of soraphen from 2 μM to 3.2 nM, and an ADP coupled reaction control.In order to effectively screen out non-specific modulators of pyruvate kinase and lactate dehydrogenase (the coupled portion of the reaction), a PK/LDH inhibition test was developed. The complete 200 μl reaction mixture contained 52.5 mM HEPES (pH8), 2.625 mM MgCl2, 0.525 mM DTT, 11 mM NaHCO3, 1% DMSO with or without inhibitor, 1× pyruvate kinase/lactate dehydrogenase (PK/LDH), 0.3 mM NADH, and 0.5 mM PEP. The reactions were incubated at 30° C. for 10 minutes and then initiated by the addition of 66 μM ADP. The initiated reactions were read immediately via plate reader at OD340 and kinetic readings were acquired every 20 s for 15 minutes while remaining at 30° C.A slope of the kinetic curve was determined by using the 2 to 7 minute data which was then calculated as percent inhibition relative to the no inhibitor control. Those compounds which had no significant PK/LDH inhibition at or above the IC50 in the ACCase assay, were considered to be valid modulators of only ACCase.
- Enolase Enzymatic Assays Enolase activity was measured by two different methods: a fluorometric NADH-linked assay or adirect spectrophotometric assay via formation of PEP. In the fluorescent assay,enolase activity was measured via NADH oxidation in a pyruvate kinase-dehydrogenase coupled assay. The assay is conducted in 10 mM KCl, 5 mM MgSO4, and 100 mM triethanolamine at pH 7.4, with 400 uM NADH and 2 mM ADP. 2-PGA, pyruvate kinase (PK) and lactate dehydrogenase(LDH) are provided in excess, with conversion of 2-PGA to PEP by enolase being rate limiting. PEP (with ADP) is substrate of PK; pyruvate formed by thisreaction is linked to NADH oxidation by LDH. Enolase activity is determinedby fluorescence measurement of oxidation of NADH by excitation at 340 nmand emission at 460 nm. The substrate concentration was 5 mM 2-PGA unlessotherwise indicated. Fluorescence was measured using an Omega FluorescencePlate Reader (BMG Labtech). Alternatively, enolase activity was measured directly by the appearance of PEP from 2-PGA via absorption at 240 nm. The assay medium was the same, except that all the auxiliary reagents (PK or LDH, NADH and ADP) are omitted. Both assays were conducted in a 96-well plate format, with the direct assay performed in UV-transmissible plates.
- Glycogen Synthase Activity Assay To a polystyrene 96-well plate, a solution containing 30 mM glycylglycine (pH 7.3), 40 mM KCl, 20 mM MgCl2, 9.2% DMSO containing one of the test compounds at various concentrations, and 10 mM glucose-6-phosphate (Sigma-Aldrich Corporation, G7879) was added by 12 uL/well. Next, a substrate solution containing 30 mM glycylglycine (pH 7.3), 4.3 mg/mL of glycogen (Sigma-Aldrich Corporation, G8876), 21.6 mM UDP-glucose (Sigma-Aldrich Corporation, U4625), 21.6 mM phosphoenolpyruvic acid (Sigma-Aldrich Corporation, P0564), and 4.05 mM NADH (Sigma-Aldrich Corporation, N8129) was added by 18 uL/well. Further, an enzyme solution containing 50 mM Tris-HCl (pH 8.0), 27 mM DTT (Nacalai Tesque, Inc., 14128-04), 0.2 mg/mL of bovine serum albumin, 0.17 mg/mL of the glycogen synthase, 1.5 uL of a pyruvate kinase/lactate dehydrogenase solution (Sigma-Aldrich Corporation, P0294) was added by 18 uL/well to prepare a reaction solution. After the reaction solution was incubated (at 30° C. for 25 minutes for Examples 1 to 10, at 37° C. for 20 minutes for Examples 11 to 75), the absorbance at 340 nm was measured using Benchmark Plus (Bio-Rad Laboratories, Inc.).
- Assaying Cardiac Muscle Myosin II The cardiac muscle myosin II actin-activated ATPase assay is a biochemical assay. Specifically, it is an NADH (nicotinamide adenine dinucleotide)-coupled ATPase assay that relies on NADH fluorescence as a readout. Cardiac myosin is a mechanochemical energy transducer that hydrolyzes ATP to generate force in the presence of its activator, F-actin. The resulting ADP is regenerated to ATP by pyruvate kinase (PK) that transforms one molecule of phosphoenolpyruvate (PEP) to pyruvate in parallel. Subsequently, pyruvate is reduced to lactate by lactate dehydrogenase (LDH) that, in turn, oxidizes one molecule of NADH to NAD. Therefore, the decrease in NADH concentration as a function of time equals the ATP hydrolysis rate. Bovine cardiac myosin is obtained from a commercial source, Cytoskeleton. PK, LDH, ATP, PEP, and NADH are obtained from Sigma. F-actin is prepared in house from Rabbit Muscle Acetone Powder. The assay is run at 25° C. in 384 well black-wall polystyrene microplates with a total volume of 20 μl per well. NADH fluorescence is monitored for 30 minutes with an EnVision Multimode Plate Reader. The slope of the fluorescence response, which is proportional to the reaction rate, is determined by simple linear regression. Final assay conditions are 300 nM cardiac myosin, 10 μM actin, 40 U/ml LDH, 200 U/ml PK, 220 μM NADH, 1 mM PEP, 1 mM ATP in a buffer containing 10 mM 3-(N-morpholino)propanesulfonic acid (pH=7.0), 2 mM MgCl2, 0.15 mM ethylene glycol-bis(p-aminoethyl ether)-N,N,N′,N′-tetraacetic acid, 0.1 mg/mL bovine serum albumin, 0.5% (V/V) dimethyl sulfoxide (DMSO) and 1 mM dithiothreitol. Prior to testing the inhibitory activity of the compounds, a two-fold dilution series starting at 10 mM compound concentration is prepared in DMSO. Subsequently, 100 nl is transferred to each well of the measuring plate containing a mixture of myosin, LDH and PK. The enzymatic reaction is started with the addition of a mixture containing ATP, PEP, NADH and actin. The highest final compound concentration is 50 uM. 20 μM para-aminoblebbistatin in 0.5% DMSO serves as the positive control and 0.5% DMSO alone is the negative control. Reaction rates are determined by using the fluorescence responses of a dilution series of NADH included in all plates and plotted as a function of inhibitor concentration. All measurements are carried out in triplicate. Inhibitory constants are determined by fitting the 16 point dose-response data to a quadratic equation corresponding to a simple one-to-one binding model. Small molecules showing no observable inhibition at or below their solubility are reported as inactive.
- Kinase Activity Assay (NADH Read-Out) Recombinant human c-Met protein (Invitrogen, Carlsbad, Calif., USA) is used. As substrate for the kinase reaction the peptide KKKSPGEYVNIEFG (JPT, Germany) is used. For the assay, 1 μL of a 51-fold concentrated solution of the test compound in DMSO is pipetted into a white 384-well microtiter plate (Greiner Bio-One, Frickenhausen, Germany). 25 μL of a solution of c-Met (final concentration 30 nM) and pyruvate kinase/lactate dehydrogenase (Roche Diagnostics, Mannheim, Germany; final concentration 8 mg/L) in assay buffer [3-(N-morpholino)propanesulfonic acid (MOPS), 50 mM, pH 7; MgCl2, 10 mM; bovine serum albumin (BSA), 0.01%; Triton X 100, 0.01%; DTT, 2 mM] are added, and the mixture is incubated for 5 min at room temperature. Then, the kinase reaction is started by the addition of 25 μL of a solution of adenosine triphosphate (ATP, final concentration 30 μM), substrate (final concentration 100 μM), nicotinamide adenine dinucleo-tide (NADH, final concentration 50 μM) and dithiothreitol (DTT, final concentration 2 mM) in assay buffer, and the resulting mixture is incubated for a reaction time of 100 min at 32° C. Subsequently, the amount of phosphorylated substrate is evaluated by measurement of the decrease of NADH fluorescence. Therefore, the fluorescence emissions at 465 nm after excitation at 340 nm is measured in a fluorescence reader, e.g. Tecan Ultra (Tecan, Männedorf, Switzerland).
- KHK-C Assay A A 384-well format on a Corning 3653 assay plate is used, and monitored by UV-vis spectroscopy in continuous mode at rt. Compounds were prepared in DMSO as 4 mM stocks, diluted using an 11-point half-log scheme on a Biomek FX (Beckman Coulter), and incubated at rt for 30 minutes with the reaction mixture containing 50 mM HEPES, pH 7.4, 140 mM KCl, 3.5 mM MgCl2, 0.8 mM fructose, 2 mM TCEP, 0.8 mM PEP, 0.7 mM NADH, 0.01% Triton X-100, 30 U/mL pyruvate kinase-lactate dehydrogenase, and 10 nM purified KHK-C. The compound concentration in each well ranged from 1 nM to 100 μM. The reaction was initiated with the addition of 0.2 mM ATP. The absorbance was measured for 30 minutes on a SpectraMax reader (Molecular Devices) after ATP was added. The concentrations provided are based on the final mixture volume of 40 μL (referred to as the final concentration). Controls: N8-(cyclopropylmethyl)-N4-(2-(methylthio)phenyl)-2-(piperazin-1-yl)pyrimido[5,4-d]pyrimidine-4,8-diamine at 2 μM final concentration was used as high percent effect (HPE) control, and 2.5% DMSO which was present in all reaction wells was used as zero percent effect (ZPE) control. Reaction rates were obtained for 300-1800 seconds time window in units of 1000*AU/min (absorbance unit per minute), and average values for ZPE and HPE controls from 16 wells each were calculated, AveZPE and AveHPE, respectively.
- Kinetic Assay Assays are performed in 96-well plates in a total volume of 200 μl/well. Compounds are dissolved in dimethylsulfoxide (DMSO) and added to plates in a volume of 10 μl followed by addition of 165 μl of assay mix. Plates are pre-incubated at 25° C. for 15 min and the reactions are initiated by the addition of 25 μl of cAMP followed by thorough mixing. Reaction rates are measured by monitoring the decrease in fluorescence using excitation at 355 nm and emission at 460 nm for a period of 10 min in a fluorescence plate reader. Initial rates (slopes) are determined from linear portions of the progress curves. Final concentrations of assay components are as follows: 50 mM Tris, pH 8, 10 mM MgCl2, 50 mM KCl, 2% DMSO, 5 mM tris(2-carboxyethyl)phosphine (TCEP), 0.4 mM phosphenolpyruvate (PEP), 0.01 mM NADH, 0.04 mM adenosine triphosphate (ATP), 0.004 mM cAMP, 7.5 units myokinase from yeast, 1.6 units pyruvate kinase, 2 units lactate dehydrogenase, and either 0.5 nM human PDE4D7 or 10 nM human PDE4B1. All data are percent normalized relative to controls and are presented as percent inhibition. An inhibitory concentration 50% (IC50) value is calculated by fitting of a sigmoidal dose response curve. Human PDE4D7 contained a mutation of serine 54 to aspartic acid to mimic activation by cAMP-dependent protein kinase A (PKA). Human PDE4B1 contained a corresponding mutation of serine 133 to aspartic acid to also mimic PKA activation.
- In Vitro CDK7 Assay CDK7 activity is measured by following the production of ADP generated from ATP-dependent phosphorylation of the peptide substrate derived from RNA Pol II (CDK7/9 tide) by CDK7. Pyruvate kinase converts ADP and phosphoenolpyruvate (PEP) to ATP and pyruvate. Lactase dehydrogenase catalyzes pyruvate to lactate with a concomitant conversion of NADH to oxidized form NAD+, which is spectrophotometrically measured at 340 nm. The CDK7 assay was performed in 384-well microplates with a final volume of 100 μL. Inhibitor serial dilutions and liquid handing for the assay were performed by using Janus from PerkinElmer (Downers Grove, IL) and Tempest from Formulatrix (Bedford, MA), respectively. To determine inhibitor potency of irreversible covalent inhibitors (kinact/K1 ratios), 500 nL of inhibitor in DMSO (or DMSO for controls) was added to the assay plate using Echo 555 from Labcyte (San Jose, CA) followed by 50 μL of assay mixture consisting of 600 μM peptide substrate (CDK7/9 tide, YSPTSPSYSPTSPSYSPTSPSKKKK), 1 mM ATP, 1 mM PEP, 200 μM NADH, 1.2-2 units of PK, 1.8-2.8 units of LDH, 20 mM Tris-HCl (pH 7.4), 10 mM MgCl2, and 0.004% Triton X-100. Reactions were initiated by the addition of 50 μL of 40 nM CDK7/cyclinH/MAT1 trimeric complex in 20 mM Tri-HCl (pH 7.4), 10 mM MgCl2, and 0.004% Triton X-100. The assay plates were centrifuged at 3220 g for 5 min using Centrifuge 5810 from Eppendorf (Hauppauge, NY) and then the absorbance changes were read at 340 nm at room temperature using Infinite M1000 from Tecan (Mannedorf, Switzerland) every 2 min for 8 hours.
- Kinetic Biochemical Assay Compounds that inhibit the hGYS1 enzyme and, subsequently, the downstream conversion of NADH to NAD+, were tested using assay ready plates (black, clear bottom 384 well plates) in a final DMSO reaction volume of 2.5% DMSO. The Assay Buffer contained 50 mM Tris pH 7.5, 2 mM MgCl2, and 100 mM KCl. Fresh stocks of BSA at a final concentration of 0.02% and TCEP at 1 mM were added before splitting buffer into hGYS1 buffer and substrate buffer. To the hGYS1 buffer, rabbit liver glycogen was added at a final concentration of 0.2% glycogen. Glucose-6-Phosphate was added at 1 mM, recombinant hGYS1/GN1 protein was added at 50 nM to the substrate buffer, phosphoenolpyruvate (PEP) was added at 2 mM, UDP-Glucose was added at 0.8 mM, NADH) was added at 0.6 mM, and Pyruvate Kinase/Lactate Dehydrogenase was added at 20 units/mL. The reaction was initiated by mixing hGYS1 buffer and substrate buffer at a 1:1 ratio. Both buffers were plated using a liquid dispensing device with hGYS1 buffer plated first followed by the substrate buffer. Plates were spun briefly to eliminate air bubbles and are immediately read in continuous mode at an absorbance of 340 nm, for 10 time points in one-minute increments, for a total of 10 minutes. The slope from these 10 time points was normalized to the positive and negative control wells. The duplicate % inhibition values are then averaged and fit to a Hill equation for dose response according to the Levenberg-Marquardt algorithm with the Hill equation maximum set to 100 and the minimum set to 0.
- DYRK1A activity assay SH-SY5Y cells are cultured in DMEM/F-12 medium supplemented with 15% FBS, Non-essential Amino Acid and Penicillin/Streptamycin. Two days before treatment, cells are seeded onto 96 well plates at 20e5 cells/well.DMSO-resuspended compounds are dispensed to 8 wells as a serial titration from 10 μM to 4.6 nM final and cells are exposed overnight (16-18 h) before harvest. Wells are visualized checked for cell death or change in morphology and supernatants are tested for cytotoxicity by measurement of lactate dehydrogenase release (LDH, CytoToxOne kit, Progema) if necessary. As controls, commercially available DYRK1A inhibitors, Harmine and Indy were shown to have good DYRK1A inhibition in the kinase assay with no CDK1 activity (EC50 18 and 53n M respectively, 6 μM for CDK1) but weak EC50 in the Tau assay: about 10 μM for Harmine and 30 μM for Indy.Cells are lyzed with RIPA buffer complemented with phosphatase and protease inhibitors (Thermo Scientific) then lysates are sonicated and spun down at 12,000 g for 10 min to remove any cellular debris. Lysates are then either directly tested for pSer396 by ELISA (Life Technology, Kit KHB7031) or loaded on NuPage Bis-Tris gels for western blot analysis. Colorimetric detection of ELISA signal is performed by Cytation3 plate reader (Biotek) and the chemoluminecence signal for HRP-linked antibodies used in western blotting is detected using a Carestream Image Station. The same pSer396 antibody is used for detection of pTau in both assays.Blot densitometry for pSer396 and beta-actin are analyzed using ImageJ (NIH) and pSer396/Total Tau ELISA signal was used to plot, draw the curve fitting, and determine each compounds EC50 in Prism (GraphPad).
- The GYS2 coupled enzyme assay Compounds that inhibit the hGYS2 enzyme and, subsequently, the downstream conversion of NADH to NAD+, were tested using assay ready plates (black, clear bottom 384 well plates) in a final DMSO reaction volume of 2.5% DMSO. The Assay Buffer contained 50 mM Tris pH 7.5, 2 mM MgCl2, and 100 mM KCl. Fresh stocks of BSA at a final concentration of 0.02% and TCEP 1 mM were added before splitting buffer into hGYS2 buffer and substrate buffer. To the hGYS2 buffer, rabbit liver glycogen was added at a final concentration of 0.2% glycogen. Glucose-6-Phosphate was added at 2 mM, recombinant hGYS2/GN1 protein was added at 200 nM to the substrate buffer, phosphoenolpyruvate (PEP) was added at 2 mM, UDP-Glucose was added at 2 mM, NADH was added at 0.6 mM, and Pyruvate Kinase/Lactate Dehydrogenase was added at 20 units/mL. The reaction was initiated by mixing hGYS2 buffer and substrate buffer at a 1:1 ratio. Both buffers were plated using a liquid dispensing device with hGYS2 buffer plated first followed by the substrate buffer. Plates were spun briefly to eliminate air bubbles and are immediately read in continuous mode at an absorbance of 340 nm, for 10 time points in one-minute increments, for a total of 10 minutes. The slope from these 10 time points was normalized to the positive and negative control wells. The duplicate % inhibition values are then averaged and fit to a Hill equation for dose response according to the Levenberg-Marquardt algorithm with the Hill equation maximum set to 100 and the minimum set to 0.The results are shown in Table 4 below
- KHK Assay A A 384-well format on a Corning 3653 assay plate is used, and monitored by UV-vis spectroscopy in continuous mode at rt. Compounds were prepared in DMSO as 4 mM stocks, diluted using an 11-point half-log scheme on a Biomek FX (Beckman Coulter), and incubated at rt for 30 minutes with the reaction mixture containing 50 mM HEPES, pH 7.4, 140 mM KCl, 3.5 mM MgCl2, 0.8 mM fructose, 2 mM TCEP, 0.8 mM PEP, 0.7 mM NADH, 0.01% Triton X-100, 30 U/mL pyruvate kinase-lactate dehydrogenase, and 10 nM purified KHK-C. The compound concentration in each well ranged from 1 nM to 100 μM. The reaction was initiated with the addition of 0.2 mM ATP. The absorbance was measured for 30 minutes on a SpectraMax reader (Molecular Devices) after ATP was added. The concentrations provided are based on the final mixture volume of 40 μL (referred to as the final concentration).Controls: N8-(cyclopropylmethyl)-N4-(2-(methylthio)phenyl)-2-(piperazin-1-yl)pyrimido[5,4-d]pyrimidine-4,8-diamine at 2 μM final concentration was used as high percent effect (HPE) control, and 2.5% DMSO which was present in all reaction wells was used as zero percent effect (ZPE) control. Reaction rates were obtained for 300-1800 seconds time window in units of 1000*AU/min (absorbance unit per minute), and average values for ZPE and HPE controls from 16 wells each were calculated, AveZPE and AveHPE, respectively.Percent inhibition (% inhibition) was calculated for each well using this equation:100 - 100 × ( Compound absorbance rate value - Ave HPE ) ( Ave ZPE - Ave HPE )The % inhibition was then plotted against the log of compound concentration using GraphPad Prism, and the data was fit to the equation log [compound] vs. response variable slope using nonlinear regression analysis to give IC50 values. For each compound tested, the IC50 provided is the average based on at least two separate assays conducted on separate days.
- Binding Inhibition Assay The ability of test substances to inhibit the binding of a human B cell line, RPMI-8866, which is known to express α4β7 integrin, to MAdCAM-1 was measured. To 96-well microtiter plates, recombinant mouse MAdCAM-1/Fc (R&D systems) solution (1 ug/mL) diluted with buffer A (carbonate buffer, pH 9.6) was added at 50 uL/well, followed by incubation at 4° C. overnight. After washing once with PBS, Block Ace (Snow Brand Milk Products Company, Limited) was added at 150 uL/well, followed by incubation at room temperature for 2 hours. After removal, washing was conducted once with PBS.100 uL of each test substance diluted with a binding buffer (DMEM containing 40 mM HEPES, 0.2% BSA, and 4 mM MnCl2) to various concentrations and 100 uL of RPMI-8866 cells (2x106 cell/mL) were added to plates coated with MAdCAM-1/Fc (5x105 cells/well) with human serum being contained at a final concentration of 50%, followed by incubation at 30° C. for 15 minutes to 60 minutes. After the cells were bound to each well, unbound cells were removed by washing with PBS. To the plates, buffer C (PBS containing 1.5% Triton X-100) was added at 50 uL/well to lyse the bound RPMI-8866 cells. To 30 uL of the cell lysate, 30 uL of Substrate Buffer (Promega, CytoTox 96 Non-Radioactive Cytotoxicity Assay) was added, and the reaction was allowed to proceed at room temperature in a dark place for 30 minutes. To each well, 30 uL of Stop Solution (Promega, CytoTox 96 Non-Radioactive Cytotoxicity Assay) was added, and the absorbance at 490 nm was measured by using a plate reader. Here, by the obtained absorbance, the activity of the lactate dehydrogenase (LDH) dissolved into the supernatant of each well was detected. In other words, the absorbance was proportional to the number of the RPMI-8866 cells bound to MAdCAM-1 and remaining on the plate. The test was duplicated. The binding ratios of the cells at various concentrations were determined, with the absorbance of a well containing no test substance taken as 100%. Then, the concentration IC50, at which 50% binding inhibition was achieved, was calculated.
- Binding Inhibition Assay The ability of test substances to inhibit the binding of a human B cell line, RPMI-8866, which is known to express α4β7 integrin, to MAdCAM-1 was measured.To 96-well microtiter plates, recombinant mouse MAdCAM-1/Fc (R&D systems) solution (0.75 ug/mL) diluted with buffer A (carbonate buffer solution, pH 9.6) was added at 50 uL/well, followed by incubation at 4° C. overnight. After washing once with PBS, Block Ace (Snow Brand Milk Products Company, Limited) was added at 150 uL/well, followed by incubation at room temperature for 2 hours. After removal, washing was conducted once with PBS.100 uL of each test substance diluted with binding buffer (DMEM containing 40 mM HEPES, 0.2% BSA, and 4 mM MnCl2) to various concentrations and 100 uL of RPMI-8866 cells (2x106 cell/mL) were added to the plates coated with MAdCAM-1/Fc (5x105 cells/well), followed by incubation at 30° C. for 15 minutes to 60 minutes. After the cells were bound to each well, unbound cells were removed by washing with PBS. To the plates, buffer C (PBS containing 1.5% Triton X-100) was added at 50 uL/well to lyse the bound RPMI-8866 cells. To 30 uL of the cell lysate, 30 uL of Substrate Buffer (Promega, CytoTox 96 Non-Radioactive Cytotoxicity Assay) was added, and the reaction was allowed to proceed at room temperature in a dark place for 30 minutes. To each well, 30 uL of Stop Solution (Promega, CytoTox 96 Non-Radioactive Cytotoxicity Assay) was added, and the absorbance at 490 nm was measured by using a plate reader. Here, by the obtained absorbance, the activity of the lactate dehydrogenase (LDH) dissolved into the supernatant of each well was detected. In other words, the absorbance was proportional to the number of the RPMI-8866 cells bound to MAdCAM-1 and remaining on the plate. The test was duplicated. The binding ratios of the cells at various concentrations were determined, with the absorbance of a well containing no test substance taken as 100%. Then, the concentration IC50, at which 50% binding inhibition was achieved, was calculated.
- POLQ Enzyme Activity Inhibition Assay Brief introduction to the experimental principle: The N-terminal active peptide segment (MI-N899) of POLQ with ATPase activity was co-incubated with compounds, and then reacted with substrate dT50 under the action of ATP to generate ADP, which participated in the subsequent NADH oxidative coupling enzymatic reaction and catalysed the NADH reaction to generate NAD+. The Envision microplate reader from Perkin Elmer Inc was used to measure the reduction of OD value of NADH at 340 nm, thus reflecting the enzyme activity.Experimental instruments: Echo 650 pipetting system from Labcyte Inc, Envision microplate reader from Perkin Elmer Inc; 5810R centrifuge from Eppendorf Inc, and BSD-YX3400 thermostatic shaker from Boxun Inc.Experimental MaterialsReagent BrandPOLQ (N) Synthesized by Pharmaron IncATP SigmadT50 Synthesized by Genscript IncNADH RochePEP SigmaLactate dehydrogenase SigmaPyruvate kinase Sigma384-well plate Greiner Bio-OneExperimental method: The POLQ enzyme was diluted to 100 nM with a reaction buffer (20 mM Tris HCl (pH 7.80), 80 mM KCl, 10 mM MgCl2, 1 mM DTT, 0.01% w/v bovine serum albumin, 0.01% v/v Tween-20, and 5% v/v glycerol). The compounds to be tested were diluted to different concentrations in dimethyl sulfoxide (DMSO) using the Echo 650 pipetting system, and transferred to a 384-well plate, 20 μL/well of 100 nM POLQ was added, and the mixture was incubated at room temperature for 15 minutes. A reaction mixture solution was formulated, and the concentration of each component in the reaction mixture solution was: 100 μM ATP, 300 nM dT50, 300 μM NADH, 6 mM PEP, 10 U/mL lactate dehydrogenase and 20 U/mL pyruvate kinase. 20 μL/well of reaction mixture solution was added to start the enzyme reaction. The final concentration of the compounds in the reaction system started from 10 μM, and was subjected to 3-fold gradient dilution. The concentration range was from 10 μM to 0.0005 μM, and the final concentration of DMSO in the system was 0.2% v/v. The 384-well plate was reacted at room temperature for 20 minutes, and then the OD value at 340 nm was read with an Envision microplate reader.
- Binding Inhibition Assay The ability of test substances to inhibit the binding of a human T-cell line, Jurkat, which is known to express α4β1 integrin, to VCAM-1 was measured.To 96-well microtiter plates, recombinant human VCAM-1/Fc (R&D systems) solution (1 ug/mL) diluted with buffer A (carbonate buffer, pH 9.6) was added at 50 uL/well, followed by incubation at 4° C. overnight. After washing once with PBS, Block Ace (Snow Brand Milk Products Company, Limited) was added at 150 uL/well, followed by incubation at room temperature for 2 hours. After removal, washing was conducted once with PBS. 100 uL of each test substance diluted with a binding buffer (DMEM containing 40 mM HEPES, 0.2% BSA, and 4 mM MnCl2) to various concentrations and 100 uL of the Jurkat cells (2x106 cell/mL) were added to the plates coated with VCAM-1/Fc (5x105 cells/well), followed by incubation at 30° C. for 15 minutes to 60 minutes. After the cells were bound to each well, unbound cells were removed by washing with PBS. To the plates, buffer C (PBS containing 1.5% Triton X-100) was added at 50 uL/well to lyse the bound Jurkat cells. To 30 uL of the cell lysate, 30 uL of Substrate Buffer (Promega, CytoTox 96 Non-Radioactive Cytotoxicity Assay) was added, and the reaction was allowed to proceed at room temperature in a dark place for 30 minutes. To each well, 30 uL of Stop Solution (Promega, CytoTox 96 Non-Radioactive Cytotoxicity Assay) was added, and the absorbance at 490 nm was measured by using a plate reader. Here, by the obtained absorbance, the activity of the lactate dehydrogenase (LDH) dissolved into the supernatant of each well was detected. In other words, the absorbance was proportional to the number of the Jurkat cells bound to VCAM-1 and remaining on the plate. The test was duplicated. The binding ratios of the cells at various concentrations were determined, with the absorbance of a well containing no test substances taken as 100%. Then, the concentration IC50, at which 50% binding inhibition was achieved, was calculated. Table 1 collectively shows the obtained results. Note that, among the compounds synthesized in the corresponding Examples, the compounds in the free form (Compounds A-1 to A-39) were used as the test compounds. Hereinafter, the same shall apply.
 Lactate Lactic Acid BDBM23233 2-hydroxypropanoic acid
Lactate Lactic Acid BDBM23233 2-hydroxypropanoic acid SODIUM LACTATE CHEMBL1357 Sodium; 2-hydroxy-propionate BDBM50159794
SODIUM LACTATE CHEMBL1357 Sodium; 2-hydroxy-propionate BDBM50159794 3-(4-Hydroxyphenyl)lactate 4-Hydroxyphenyllactate BDBM50269986 p-Hydroxyphenyllactate
3-(4-Hydroxyphenyl)lactate 4-Hydroxyphenyllactate BDBM50269986 p-Hydroxyphenyllactate 6,11-Dimethyl-3-(3-methyl-but-2-enyl)-1,2,3,4,5,6-hexahydro-2,6-methano-benzo[d]azocin-8-ol; lactate PENTAZOCINE (+) (Pentazocine) 6,11-Dimethyl-3-(3-methyl-but-2-enyl)-1,2 ,3,4,5,6-hexahydro-2,6-methano-benzo[d]azocin-8-ol PENTAZOCINE (-) 2-[3-cyclopropylmethyl-11-hydroxy-15-methoxy-(15S)-13-oxa-3-azahexacyclo[13.2.2.12,8.01,6.06,14.07,12]icosa-7,9,11-trien-16-yl]-3,3-dimethyl-2-butanol(Pentazocine) CHEMBL100116 6,11-Dimethyl-3-(3-methyl-but-2-enyl)-1,2,3,4,5,6-hexahydro-2,6-methano-benzo[d]azocin-8-ol (Pentazocine) 6,11-Dimethyl-3-(3-methyl-but-2-enyl)-1,2,3,4,5,6-hexahydro-2,6-methano-benzo[d]azocin-8-ol (pentazocine)2-{4-[(2,4-Diamino-pteridin-6-ylmethyl)-methyl-amino]-benzoylamino}-pentanedioic acid BDBM50032403 PENTAZOCINE 6,11-Dimethyl-3-(3-methyl-but-2-enyl)-1,2,3,4,5,6-hexahydro-2,6-methano-benzo[d]azocin-8-ol (pentazocine) 1,13-dimethyl-10-(3-methyl-2-butenyl)-(1R,9R,13R)-10-azatricyclo[7.3.1.02,7]trideca-2(7),3,5-trien-4-ol(Pentazocine)
6,11-Dimethyl-3-(3-methyl-but-2-enyl)-1,2,3,4,5,6-hexahydro-2,6-methano-benzo[d]azocin-8-ol; lactate PENTAZOCINE (+) (Pentazocine) 6,11-Dimethyl-3-(3-methyl-but-2-enyl)-1,2 ,3,4,5,6-hexahydro-2,6-methano-benzo[d]azocin-8-ol PENTAZOCINE (-) 2-[3-cyclopropylmethyl-11-hydroxy-15-methoxy-(15S)-13-oxa-3-azahexacyclo[13.2.2.12,8.01,6.06,14.07,12]icosa-7,9,11-trien-16-yl]-3,3-dimethyl-2-butanol(Pentazocine) CHEMBL100116 6,11-Dimethyl-3-(3-methyl-but-2-enyl)-1,2,3,4,5,6-hexahydro-2,6-methano-benzo[d]azocin-8-ol (Pentazocine) 6,11-Dimethyl-3-(3-methyl-but-2-enyl)-1,2,3,4,5,6-hexahydro-2,6-methano-benzo[d]azocin-8-ol (pentazocine)2-{4-[(2,4-Diamino-pteridin-6-ylmethyl)-methyl-amino]-benzoylamino}-pentanedioic acid BDBM50032403 PENTAZOCINE 6,11-Dimethyl-3-(3-methyl-but-2-enyl)-1,2,3,4,5,6-hexahydro-2,6-methano-benzo[d]azocin-8-ol (pentazocine) 1,13-dimethyl-10-(3-methyl-2-butenyl)-(1R,9R,13R)-10-azatricyclo[7.3.1.02,7]trideca-2(7),3,5-trien-4-ol(Pentazocine)