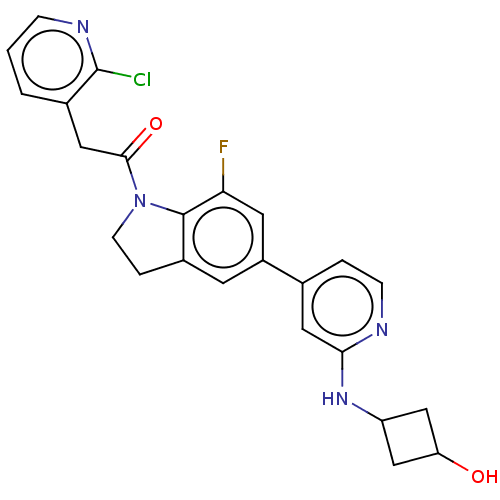
 US11465984, Example P53 BDBM575772 2-(2- chloropyridin-3- yl)-1-(7-fluoro- 5-(2-((3- hydroxylcyclobutyl) amino)pyridin-4- yl)indolin-1-yl) ethan-1-one
US11465984, Example P53 BDBM575772 2-(2- chloropyridin-3- yl)-1-(7-fluoro- 5-(2-((3- hydroxylcyclobutyl) amino)pyridin-4- yl)indolin-1-yl) ethan-1-one
- Lain, S; Lane, DP; Stark, MJ; McCarthy, AR; Hollick, JJ; Westwood, NJ P53 activating compounds US Patent US8501991 (2013)
- Doemling, A p53-Mdm2 antagonists US Patent US9187441 (2015)
- Espadinha, M; Lopes, EA; Marques, V; Amaral, JD; Dos Santos, DJVA; Mori, M; Daniele, S; Piccarducci, R; Zappelli, E; Martini, C; Rodrigues, CMP; Santos, MMM Discovery of MDM2-p53 and MDM4-p53 protein-protein interactions small molecule dual inhibitors. Eur J Med Chem 241: (2022)
- An overview of the functions of p53 and drugs acting either on wild- or mutant-type p53.
- Gomha, SM; Eldebss, TM; Abdulla, MM; Mayhoub, AS Diphenylpyrroles: Novel p53 activators. Eur J Med Chem 82: 472-9 (2014)
- Niazi, S; Purohit, M; Niazi, JH Role of p53 circuitry in tumorigenesis: A brief review. Eur J Med Chem 158: 7-24 (2018)
- Ishiba, H; Noguchi, T; Shu, K; Ohno, H; Honda, K; Kondoh, Y; Osada, H; Fujii, N; Oishi, S Investigation of the inhibitory mechanism of apomorphine against MDM2-p53 interaction. Bioorg Med Chem Lett 27: 2571-2574 (2017)
- Rew, Y; Sun, D; Gonzalez-Lopez De Turiso, F; Bartberger, MD; Beck, HP; Canon, J; Chen, A; Chow, D; Deignan, J; Fox, BM; Gustin, D; Huang, X; Jiang, M; Jiao, X; Jin, L; Kayser, F; Kopecky, DJ; Li, Y; Lo, MC; Long, AM; Michelsen, K; Oliner, JD; Osgood, T; Ragains, M; Saiki, AY; Schneider, S; Toteva, M; Yakowec, P; Yan, X; Ye, Q; Yu, D; Zhao, X; Zhou, J; Medina, JC; Olson, SH Structure-based design of novel inhibitors of the MDM2-p53 interaction. J Med Chem 55: 4936-54 (2012)
- Galatin, PS; Abraham, DJ A nonpeptidic sulfonamide inhibits the p53-mdm2 interaction and activates p53-dependent transcription in mdm2-overexpressing cells. J Med Chem 47: 4163-5 (2004)
- Dickens, MP; Roxburgh, P; Hock, A; Mezna, M; Kellam, B; Vousden, KH; Fischer, PM 5-Deazaflavin derivatives as inhibitors of p53 ubiquitination by HDM2. Bioorg Med Chem 21: 6868-77 (2013)
- Yagi, H; Chuman, Y; Kozakai, Y; Imagawa, T; Takahashi, Y; Yoshimura, F; Tanino, K; Sakaguchi, K A small molecule inhibitor of p53-inducible protein phosphatase PPM1D. Bioorg Med Chem Lett 22: 729-32 (2011)
- Chessari, G; Howard, S; Buck, IM; Cons, BD; Johnson, CN; Holvey, RS; Rees, DC; St. Denis, JD; Tamanini, E; Golding, BT; Hardcastle, IR; Cano, CF; Miller, DC; Noble, ME; Griffin, RJ; Osborne, JD; Peach, J; Lewis, A; Hirst, KL; Whittaker, BP; Watson, DW; Mitchell, DR Isoindolinone inhibitors of the MDM2-p53 interaction having anticancer activity US Patent US10544132 (2020)
- Hayashi, R; Wang, D; Hara, T; Iera, JA; Durell, SR; Appella, DH N-acylpolyamine inhibitors of HDM2 and HDMX binding to p53. Bioorg Med Chem 17: 7884-93 (2009)
- Gonzalez-Lopez de Turiso, F; Sun, D; Rew, Y; Bartberger, MD; Beck, HP; Canon, J; Chen, A; Chow, D; Correll, TL; Huang, X; Julian, LD; Kayser, F; Lo, MC; Long, AM; McMinn, D; Oliner, JD; Osgood, T; Powers, JP; Saiki, AY; Schneider, S; Shaffer, P; Xiao, SH; Yakowec, P; Yan, X; Ye, Q; Yu, D; Zhao, X; Zhou, J; Medina, JC; Olson, SH Rational design and binding mode duality of MDM2-p53 inhibitors. J Med Chem 56: 4053-70 (2013)
- Estrada-Ortiz, N; Neochoritis, CG; Twarda-Clapa, A; Musielak, B; Holak, TA; Dömling, A Artificial Macrocycles as Potent p53-MDM2 Inhibitors. ACS Med Chem Lett 8: 1025-1030 (2017)
- Baud, MGJ; Bauer, MR; Verduci, L; Dingler, FA; Patel, KJ; Horil Roy, D; Joerger, AC; Fersht, AR Aminobenzothiazole derivatives stabilize the thermolabile p53 cancer mutant Y220C and show anticancer activity in p53-Y220C cell lines. Eur J Med Chem 152: 101-114 (2018)
- Miyazaki, M; Naito, H; Sugimoto, Y; Kawato, H; Okayama, T; Shimizu, H; Miyazaki, M; Kitagawa, M; Seki, T; Fukutake, S; Aonuma, M; Soga, T Lead optimization of novel p53-MDM2 interaction inhibitors possessing dihydroimidazothiazole scaffold. Bioorg Med Chem Lett 23: 728-32 (2013)
- Cominetti, MM; Goffin, SA; Raffel, E; Turner, KD; Ramoutar, JC; O'Connell, MA; Howell, LA; Searcey, M Identification of a new p53/MDM2 inhibitor motif inspired by studies of chlorofusin. Bioorg Med Chem Lett 25: 4878-80 (2015)
- Carry, JC; Garcia-Echeverria, C Inhibitors of the p53/hdm2 protein-protein interaction-path to the clinic. Bioorg Med Chem Lett 23: 2480-5 (2013)
- Yu, S; Qin, D; Shangary, S; Chen, J; Wang, G; Ding, K; McEachern, D; Qiu, S; Nikolovska-Coleska, Z; Miller, R; Kang, S; Yang, D; Wang, S Potent and orally active small-molecule inhibitors of the MDM2-p53 interaction. J Med Chem 52: 7970-3 (2009)
- Gonzalez, AZ; Eksterowicz, J; Bartberger, MD; Beck, HP; Canon, J; Chen, A; Chow, D; Duquette, J; Fox, BM; Fu, J; Huang, X; Houze, JB; Jin, L; Li, Y; Li, Z; Ling, Y; Lo, MC; Long, AM; McGee, LR; McIntosh, J; McMinn, DL; Oliner, JD; Osgood, T; Rew, Y; Saiki, AY; Shaffer, P; Wortman, S; Yakowec, P; Yan, X; Ye, Q; Yu, D; Zhao, X; Zhou, J; Olson, SH; Medina, JC; Sun, D Selective and potent morpholinone inhibitors of the MDM2-p53 protein-protein interaction. J Med Chem 57: 2472-88 (2014)
- Pan, W; Lahue, BR; Ma, Y; Nair, LG; Shipps, GW; Wang, Y; Doll, R; Bogen, SL Core modification of substituted piperidines as novel inhibitors of HDM2-p53 protein-protein interaction. Bioorg Med Chem Lett 24: 1983-6 (2014)
- Vaupel, A; Bold, G; De Pover, A; Stachyra-Valat, T; Hergovich-Lisztwan, J; Kallen, J; Masuya, K; Furet, P Tetra-substituted imidazoles as a new class of inhibitors of the p53-MDM2 interaction. Bioorg Med Chem Lett 24: 2110-4 (2014)
- Chen, KX; Yan, X; Huang, J; Nie, Y; Hu, G; Li, J; Chen, S; Dong, J; Wang, T Substituted pyrrolo[3,4-d]imidazoles as MDM2-p53 inhibitors US Patent US11639355 (2023)
- Grasberger, BL; Lu, T; Schubert, C; Parks, DJ; Carver, TE; Koblish, HK; Cummings, MD; LaFrance, LV; Milkiewicz, KL; Calvo, RR; Maguire, D; Lattanze, J; Franks, CF; Zhao, S; Ramachandren, K; Bylebyl, GR; Zhang, M; Manthey, CL; Petrella, EC; Pantoliano, MW; Deckman, IC; Spurlino, JC; Maroney, AC; Tomczuk, BE; Molloy, CJ; Bone, RF Discovery and cocrystal structure of benzodiazepinedione HDM2 antagonists that activate p53 in cells. J Med Chem 48: 909-12 (2005)
- Ding, Q; Zhang, Z; Liu, JJ; Jiang, N; Zhang, J; Ross, TM; Chu, XJ; Bartkovitz, D; Podlaski, F; Janson, C; Tovar, C; Filipovic, ZM; Higgins, B; Glenn, K; Packman, K; Vassilev, LT; Graves, B Discovery of RG7388, a potent and selective p53-MDM2 inhibitor in clinical development. J Med Chem 56: 5979-83 (2014)
- Mizuno-Kaneko, M; Hashimoto, I; Miyahara, K; Kochi, M; Ohashi, N; Tsumura, K; Suzuki, K; Tamura, T Molecular Design of Cyclic Peptides with Cell Membrane Permeability and Development of MDMX-p53 Inhibitor. ACS Med Chem Lett 14: 1174-1178 (2023)
- Li, C; Zhan, C; Zhao, L; Chen, X; Lu, WY; Lu, W Functional consequences of retro-inverso isomerization of a miniature protein inhibitor of the p53-MDM2 interaction. Bioorg Med Chem 21: 4045-50 (2013)
- Neochoritis, CG; Wang, K; Estrada-Ortiz, N; Herdtweck, E; Kubica, K; Twarda, A; Zak, KM; Holak, TA; Dömling, A 2,30-Bis(10H-indole) heterocycles: New p53/MDM2/MDMX antagonists. Bioorg Med Chem Lett 25: 5661-6 (2015)
- Zhang, R; Mayhood, T; Lipari, P; Wang, Y; Durkin, J; Syto, R; Gesell, J; McNemar, C; Windsor, W Fluorescence polarization assay and inhibitor design for MDM2/p53 interaction. Anal Biochem 331: 138-46 (2004)
- Silvestri, AP; Zhang, Q; Ping, Y; Muir, EW; Zhao, J; Chakka, SK; Wang, G; Bray, WM; Chen, W; Fribourgh, JL; Tripathi, S; He, Y; Rubin, SM; Satz, AL; Pye, CR; Kuai, L; Su, W; Schwochert, JA DNA-Encoded Macrocyclic Peptide Libraries Enable the Discovery of a Neutral MDM2-p53 Inhibitor. ACS Med Chem Lett 14: 820-826 (2023)
- Neochoritis, CG; Atmaj, J; Twarda-Clapa, A; Surmiak, E; Skalniak, L; Köhler, LM; Muszak, D; Kurpiewska, K; Kalinowska-Tłuścik, J; Beck, B; Holak, TA; Dömling, A Hitting on the move: Targeting intrinsically disordered protein states of the MDM2-p53 interaction. Eur J Med Chem 182: (2019)
- Vaupel, A; Holzer, P; Ferretti, S; Guagnano, V; Kallen, J; Mah, R; Masuya, K; Ruetz, S; Rynn, C; Schlapbach, A; Stachyra, T; Stutz, S; Todorov, M; Jeay, S; Furet, P In vitro and in vivo characterization of a novel, highly potent p53-MDM2 inhibitor. Bioorg Med Chem Lett 28: 3404-3408 (2018)
- Zhang, S; Lou, J; Li, Y; Zhou, F; Yan, Z; Lyu, X; Zhao, Y Recent Progress and Clinical Development of Inhibitors that Block MDM4/p53 Protein-Protein Interactions. J Med Chem 64: 10621-10640 (2021)
- Markowitz, J; Chen, I; Gitti, R; Baldisseri, DM; Pan, Y; Udan, R; Carrier, F; MacKerell, AD; Weber, DJ Identification and characterization of small molecule inhibitors of the calcium-dependent S100B-p53 tumor suppressor interaction. J Med Chem 47: 5085-93 (2004)
- Surmiak, E; Neochoritis, CG; Musielak, B; Twarda-Clapa, A; Kurpiewska, K; Dubin, G; Camacho, C; Holak, TA; Dömling, A Rational design and synthesis of 1,5-disubstituted tetrazoles as potent inhibitors of the MDM2-p53 interaction. Eur J Med Chem 126: 384-407 (2017)
- Tian, Y; Ma, Y; Gibeau, CR; Lahue, BR; Shipps, GW; Strickland, C; Bogen, SL Structure-activity relationship study of 4-substituted piperidines at Leu26 moiety of novel p53-hDM2 inhibitors. Bioorg Med Chem Lett 26: 2735-8 (2016)
- Li, X; Liu, C; Chen, S; Hu, H; Su, J; Zou, Y d-Amino acid mutation of PMI as potent dual peptide inhibitors of p53-MDM2/MDMX interactions. Bioorg Med Chem Lett 27: 4678-4681 (2017)
- Wang, W; Cao, H; Wolf, S; Camacho-Horvitz, MS; Holak, TA; Dömling, A Benzimidazole-2-one: a novel anchoring principle for antagonizing p53-Mdm2. Bioorg Med Chem 21: 3982-95 (2013)
- Stoll, R; Renner, C; Hansen, S; Palme, S; Klein, C; Belling, A; Zeslawski, W; Kamionka, M; Rehm, T; Mühlhahn, P; Schumacher, R; Hesse, F; Kaluza, B; Voelter, W; Engh, RA; Holak, TA Chalcone derivatives antagonize interactions between the human oncoprotein MDM2 and p53. Biochemistry 40: 336-44 (2001)
- Li, J; Wu, Y; Guo, Z; Zhuang, C; Yao, J; Dong, G; Yu, Z; Min, X; Wang, S; Liu, Y; Wu, S; Zhu, S; Sheng, C; Miao, Z; Zhang, W Discovery of 1-arylpyrrolidone derivatives as potent p53-MDM2 inhibitors based on molecule fusing strategy. Bioorg Med Chem Lett 24: 2648-50 (2014)
- Bogen, SL; Pan, W; Gibeau, CR; Lahue, BR; Ma, Y; Nair, LG; Seigel, E; Shipps, GW; Tian, Y; Wang, Y; Lin, Y; Liu, M; Liu, S; Mirza, A; Wang, X; Lipari, P; Seidel-Dugan, C; Hicklin, DJ; Bishop, WR; Rindgen, D; Nomeir, A; Prosise, W; Reichert, P; Scapin, G; Strickland, C; Doll, RJ Discovery of Novel 3,3-Disubstituted Piperidines as Orally Bioavailable, Potent, and Efficacious HDM2-p53 Inhibitors. ACS Med Chem Lett 7: 324-9 (2016)
- Datta, S; Bucks, ME; Koley, D; Lim, PX; Savinov, SN Functional profiling of p53-binding sites in Hdm2 and Hdmx using a genetic selection system. Bioorg Med Chem 18: 6099-108 (2010)
- Gonzalez, AZ; Li, Z; Beck, HP; Canon, J; Chen, A; Chow, D; Duquette, J; Eksterowicz, J; Fox, BM; Fu, J; Huang, X; Houze, J; Jin, L; Li, Y; Ling, Y; Lo, MC; Long, AM; McGee, LR; McIntosh, J; Oliner, JD; Osgood, T; Rew, Y; Saiki, AY; Shaffer, P; Wortman, S; Yakowec, P; Yan, X; Ye, Q; Yu, D; Zhao, X; Zhou, J; Olson, SH; Sun, D; Medina, JC Novel inhibitors of the MDM2-p53 interaction featuring hydrogen bond acceptors as carboxylic acid isosteres. J Med Chem 57: 2963-88 (2014)
- Tian, Y; Lahue, BR; Ma, Y; Nair, LG; Pan, W; Doll, RJ; Guzi, T; Ma, Y; Wang, Y; Bogen, SL Phe19 modification of HDM2-p53 PPI inhibitors to alleviate CYP3A4 metabolism/mechanism-based inhibition liability. Bioorg Med Chem Lett 61: (2022)
- Hardcastle, IR; Ahmed, SU; Atkins, H; Farnie, G; Golding, BT; Griffin, RJ; Guyenne, S; Hutton, C; Källblad, P; Kemp, SJ; Kitching, MS; Newell, DR; Norbedo, S; Northen, JS; Reid, RJ; Saravanan, K; Willems, HM; Lunec, J Small-molecule inhibitors of the MDM2-p53 protein-protein interaction based on an isoindolinone scaffold. J Med Chem 49: 6209-21 (2006)
- Liao, G; Yang, D; Ma, L; Li, W; Hu, L; Zeng, L; Wu, P; Duan, L; Liu, Z The development of piperidinones as potent MDM2-P53 protein-protein interaction inhibitors for cancer therapy. Eur J Med Chem 159: 1-9 (2018)
- Noguchi, T; Oishi, S; Honda, K; Kondoh, Y; Saito, T; Kubo, T; Kaneda, M; Ohno, H; Osada, H; Fujii, N Affinity-based screening of MDM2/MDMX-p53 interaction inhibitors by chemical array: identification of novel peptidic inhibitors. Bioorg Med Chem Lett 23: 3802-5 (2013)
- Rew, Y; Sun, D; Yan, X; Beck, HP; Canon, J; Chen, A; Duquette, J; Eksterowicz, J; Fox, BM; Fu, J; Gonzalez, AZ; Houze, J; Huang, X; Jiang, M; Jin, L; Li, Y; Li, Z; Ling, Y; Lo, MC; Long, AM; McGee, LR; McIntosh, J; Oliner, JD; Osgood, T; Saiki, AY; Shaffer, P; Wang, YC; Wortman, S; Yakowec, P; Ye, Q; Yu, D; Zhao, X; Zhou, J; Medina, JC; Olson, SH Discovery of AM-7209, a potent and selective 4-amidobenzoic acid inhibitor of the MDM2-p53 interaction. J Med Chem 57: 10499-511 (2014)
- Ding, K; Lu, Y; Nikolovska-Coleska, Z; Wang, G; Qiu, S; Shangary, S; Gao, W; Qin, D; Stuckey, J; Krajewski, K; Roller, PP; Wang, S Structure-based design of spiro-oxindoles as potent, specific small-molecule inhibitors of the MDM2-p53 interaction. J Med Chem 49: 3432-5 (2006)
- Synthesis and biological evaluation of 4-imidazolidinone-containing compounds as potent inhibitors of the MDM2/p53 interaction.
- Madden, MM; Muppidi, A; Li, Z; Li, X; Chen, J; Lin, Q Synthesis of cell-permeable stapled peptide dual inhibitors of the p53-Mdm2/Mdmx interactions via photoinduced cycloaddition. Bioorg Med Chem Lett 21: 1472-5 (2011)
- Ji, C; Wang, S; Chen, S; He, S; Jiang, Y; Miao, Z; Li, J; Sheng, C Design, synthesis and biological evaluation of novel antitumor spirotetrahydrothiopyran-oxindole derivatives as potent p53-MDM2 inhibitors. Bioorg Med Chem 25: 5268-5277 (2017)
- Sun, D; Li, Z; Rew, Y; Gribble, M; Bartberger, MD; Beck, HP; Canon, J; Chen, A; Chen, X; Chow, D; Deignan, J; Duquette, J; Eksterowicz, J; Fisher, B; Fox, BM; Fu, J; Gonzalez, AZ; Gonzalez-Lopez De Turiso, F; Houze, JB; Huang, X; Jiang, M; Jin, L; Kayser, F; Liu, JJ; Lo, MC; Long, AM; Lucas, B; McGee, LR; McIntosh, J; Mihalic, J; Oliner, JD; Osgood, T; Peterson, ML; Roveto, P; Saiki, AY; Shaffer, P; Toteva, M; Wang, Y; Wang, YC; Wortman, S; Yakowec, P; Yan, X; Ye, Q; Yu, D; Yu, M; Zhao, X; Zhou, J; Zhu, J; Olson, SH; Medina, JC Discovery of AMG 232, a potent, selective, and orally bioavailable MDM2-p53 inhibitor in clinical development. J Med Chem 57: 1454-72 (2014)
- Zhang, Z; Chu, XJ; Liu, JJ; Ding, Q; Zhang, J; Bartkovitz, D; Jiang, N; Karnachi, P; So, SS; Tovar, C; Filipovic, ZM; Higgins, B; Glenn, K; Packman, K; Vassilev, L; Graves, B Discovery of Potent and Orally Active p53-MDM2 Inhibitors RO5353 and RO2468 for Potential Clinical Development. ACS Med Chem Lett 5: 124-7 (2014)
- Guo, Z; Zhuang, C; Zhu, L; Zhang, Y; Yao, J; Dong, G; Wang, S; Liu, Y; Chen, H; Sheng, C; Miao, Z; Zhang, W Structure-activity relationship and antitumor activity of thio-benzodiazepines as p53-MDM2 protein-protein interaction inhibitors. Eur J Med Chem 56: 10-16 (2012)
- Lawrence, HR; Li, Z; Yip, ML; Sung, SS; Lawrence, NJ; McLaughlin, ML; McManus, GJ; Zaworotko, MJ; Sebti, SM; Chen, J; Guida, WC Identification of a disruptor of the MDM2-p53 protein-protein interaction facilitated by high-throughput in silico docking. Bioorg Med Chem Lett 19: 3756-9 (2009)
- Yoshimura, C; Miyafusa, T; Tsumoto, K Identification of small-molecule inhibitors of the human S100B-p53 interaction and evaluation of their activity in human melanoma cells. Bioorg Med Chem 21: 1109-15 (2013)
- Miyazaki, M; Uoto, K; Sugimoto, Y; Naito, H; Yoshida, K; Okayama, T; Kawato, H; Miyazaki, M; Kitagawa, M; Seki, T; Fukutake, S; Aonuma, M; Soga, T Discovery of DS-5272 as a promising candidate: A potent and orally active p53-MDM2 interaction inhibitor. Bioorg Med Chem 23: 2360-7 (2015)
- Tsukamoto, S; Yoshida, T; Hosono, H; Ohta, T; Yokosawa, H Hexylitaconic acid: a new inhibitor of p53-HDM2 interaction isolated from a marine-derived fungus, Arthrinium sp. Bioorg Med Chem Lett 16: 69-71 (2006)
- Cummings, MD; Schubert, C; Parks, DJ; Calvo, RR; LaFrance, LV; Lattanze, J; Milkiewicz, KL; Lu, T Substituted 1,4-benzodiazepine-2,5-diones as alpha-helix mimetic antagonists of the HDM2-p53 protein-protein interaction. Chem Biol Drug Des 67: 201-5 (2006)
- Mularski, J; Malarz, K; Pacholczyk, M; Musiol, R The p53 stabilizing agent CP-31398 and multi-kinase inhibitors. Designing, synthesizing and screening of styrylquinazoline series. Eur J Med Chem 163: 610-625 (2019)
- Liu, Y; Wang, X; Wang, G; Yang, Y; Yuan, Y; Ouyang, L The past, present and future of potential small-molecule drugs targeting p53-MDM2/MDMX for cancer therapy. Eur J Med Chem 176: 92-104 (2019)
- Luo, HJ; Si, DJ; Sun, XJ; Wang, MY; Yang, YB; Wang, B; Wen, HM; Li, W; Liu, J Structure-based discovery of novel α-aminoketone derivatives as dual p53-MDM2/MDMX inhibitors for the treatment of cancer. Eur J Med Chem 252: (2023)
- HDAC/NAMPT dual inhibitors overcome initial drug-resistance in p53-null leukemia cells.
- Clement, JA; Kitagaki, J; Yang, Y; Saucedo, CJ; O'Keefe, BR; Weissman, AM; McKee, TC; McMahon, JB Discovery of new pyridoacridine alkaloids from Lissoclinum cf. badium that inhibit the ubiquitin ligase activity of Hdm2 and stabilize p53. Bioorg Med Chem 16: 10022-8 (2008)
- Furet, P; Chène, P; De Pover, A; Valat, TS; Lisztwan, JH; Kallen, J; Masuya, K The central valine concept provides an entry in a new class of non peptide inhibitors of the p53-MDM2 interaction. Bioorg Med Chem Lett 22: 3498-502 (2012)
- Li, X; Jiang, Y; Peterson, YK; Xu, T; Himes, RA; Luo, X; Yin, G; Inks, ES; Dolloff, N; Halene, S; Chan, SSL; Chou, CJ Design of Hydrazide-Bearing HDACIs Based on Panobinostat and Their p53 and FLT3-ITD Dependency in Antileukemia Activity. J Med Chem 63: 5501-5525 (2020)
- Zhao, Y; Aguilar, A; Bernard, D; Wang, S Small-molecule inhibitors of the MDM2-p53 protein-protein interaction (MDM2 Inhibitors) in clinical trials for cancer treatment. J Med Chem 58: 1038-52 (2015)
- Sang, P; Shi, Y; Lu, J; Chen, L; Yang, L; Borcherds, W; Abdulkadir, S; Li, Q; Daughdrill, G; Chen, J; Cai, J α-Helix-Mimicking Sulfono-γ-AApeptide Inhibitors for p53-MDM2/MDMX Protein-Protein Interactions. J Med Chem 63: 975-986 (2020)
- Surmiak, E; Twarda-Clapa, A; Zak, KM; Musielak, B; Tomala, MD; Kubica, K; Grudnik, P; Madej, M; Jablonski, M; Potempa, J; Kalinowska-Tluscik, J; Dömling, A; Dubin, G; Holak, TA A Unique Mdm2-Binding Mode of the 3-Pyrrolin-2-one- and 2-Furanone-Based Antagonists of the p53-Mdm2 Interaction. ACS Chem Biol 11: 3310-3318 (2016)
- Parks, DJ; LaFrance, LV; Calvo, RR; Milkiewicz, KL; Marugán, JJ; Raboisson, P; Schubert, C; Koblish, HK; Zhao, S; Franks, CF; Lattanze, J; Carver, TE; Cummings, MD; Maguire, D; Grasberger, BL; Maroney, AC; Lu, T Enhanced pharmacokinetic properties of 1,4-benzodiazepine-2,5-dione antagonists of the HDM2-p53 protein-protein interaction through structure-based drug design. Bioorg Med Chem Lett 16: 3310-4 (2006)
- Huart, AS; Saxty, B; Merritt, A; Nekulova, M; Lewis, S; Huang, Y; Vojtesek, B; Kettleborough, C; Hupp, TR A Casein kinase 1/Checkpoint kinase 1 pyrazolo-pyridine protein kinase inhibitor as novel activator of the p53 pathway. Bioorg Med Chem Lett 23: 5578-85 (2013)
- Miyazaki, M; Kawato, H; Naito, H; Ikeda, M; Miyazaki, M; Kitagawa, M; Seki, T; Fukutake, S; Aonuma, M; Soga, T Discovery of novel dihydroimidazothiazole derivatives as p53-MDM2 protein-protein interaction inhibitors: synthesis, biological evaluation and structure-activity relationships. Bioorg Med Chem Lett 22: 6338-42 (2012)
- Teng, J; Chen, Y; Xiao, S; Li, T; Su, G; Wang, G; Zhao, Y Novel ginsenoside derivatives induce apoptosis in HepG-2 cells via the MDM2-p53 signaling pathway. Bioorg Med Chem Lett 78: (2022)
- Tatsuda, D; Momose, I; Someno, T; Sawa, R; Kubota, Y; Iijima, M; Kunisada, T; Watanabe, T; Shibasaki, M; Nomoto, A Quinofuracins A-E, produced by the fungus Staphylotrichum boninense PF1444, show p53-dependent growth suppression. J Nat Prod 78: 188-95 (2015)
- Lu, Y; Nikolovska-Coleska, Z; Fang, X; Gao, W; Shangary, S; Qiu, S; Qin, D; Wang, S Discovery of a nanomolar inhibitor of the human murine double minute 2 (MDM2)-p53 interaction through an integrated, virtual database screening strategy. J Med Chem 49: 3759-62 (2006)
- Zhao, Y; Yu, S; Sun, W; Liu, L; Lu, J; McEachern, D; Shargary, S; Bernard, D; Li, X; Zhao, T; Zou, P; Sun, D; Wang, S A potent small-molecule inhibitor of the MDM2-p53 interaction (MI-888) achieved complete and durable tumor regression in mice. J Med Chem 56: 5553-61 (2014)
- Hardcastle, IR; Liu, J; Valeur, E; Watson, A; Ahmed, SU; Blackburn, TJ; Bennaceur, K; Clegg, W; Drummond, C; Endicott, JA; Golding, BT; Griffin, RJ; Gruber, J; Haggerty, K; Harrington, RW; Hutton, C; Kemp, S; Lu, X; McDonnell, JM; Newell, DR; Noble, ME; Payne, SL; Revill, CH; Riedinger, C; Xu, Q; Lunec, J Isoindolinone inhibitors of the murine double minute 2 (MDM2)-p53 protein-protein interaction: structure-activity studies leading to improved potency. J Med Chem 54: 1233-43 (2011)
- Day, CL; Smits, C; Fan, FC; Lee, EF; Fairlie, WD; Hinds, MG Structure of the BH3 domains from the p53-inducible BH3-only proteins Noxa and Puma in complex with Mcl-1. J Mol Biol 380: 958-71 (2008)
- Wang, Y; Zhu, J; Liu, JJ; Chen, X; Mihalic, J; Deignan, J; Yu, M; Sun, D; Kayser, F; McGee, LR; Lo, MC; Chen, A; Zhou, J; Ye, Q; Huang, X; Long, AM; Yakowec, P; Oliner, JD; Olson, SH; Medina, JC Optimization beyond AMG 232: discovery and SAR of sulfonamides on a piperidinone scaffold as potent inhibitors of the MDM2-p53 protein-protein interaction. Bioorg Med Chem Lett 24: 3782-5 (2014)
- Twarda-Clapa, A; Krzanik, S; Kubica, K; Guzik, K; Labuzek, B; Neochoritis, CG; Khoury, K; Kowalska, K; Czub, M; Dubin, G; Dömling, A; Skalniak, L; Holak, TA 1,4,5-Trisubstituted Imidazole-Based p53-MDM2/MDMX Antagonists with Aliphatic Linkers for Conjugation with Biological Carriers. J Med Chem 60: 4234-4244 (2017)
- Weinstabl, H; Gollner, A; Ramharter, J; Wunberg, T Spiro[3H-indole-3,2′-pyrrolidin]-2(1H)-one compounds and derivatives as MDM2-P53 inhibitors US Patent US10576064 (2020)
- Ramharter, J; Broeker, J; Gille, A; Gollner, A; Henry, M; Kerres, N; Weinstabl, H Spiro[3H-indole-3,2′-pyrrolidin]-2(1H)-one compounds and derivatives as MDM2-p53 inhibitors US Patent US10919913 (2021)
- Cheng, J; Yan, Z; Jiang, K; Liu, C; Xu, D; Lyu, X; Hu, X; Zhang, S; Zhou, Y; Li, J; Zhao, Y Discovery of JN122, a Spiroindoline-Containing Molecule that Inhibits MDM2/p53 Protein-Protein Interaction and Exerts Robust In Vivo Antitumor Efficacy. J Med Chem 66: 16991-17025 (2023)
- He, S; Dong, G; Wu, S; Fang, K; Miao, Z; Wang, W; Sheng, C Small Molecules Simultaneously Inhibiting p53-Murine Double Minute 2 (MDM2) Interaction and Histone Deacetylases (HDACs): Discovery of Novel Multitargeting Antitumor Agents. J Med Chem 61: 7245-7260 (2018)
- Liu, Q; Zhang, B; Wang, Y; Wang, X; Gou, S Discovery of phthalazino[1,2-b]-quinazolinone derivatives as multi-target HDAC inhibitors for the treatment of hepatocellular carcinoma via activating the p53 signal pathway. Eur J Med Chem 229: (2022)
- RAMHARTER, J; BROEKER, J; GILLE, A; GOLLNER, A; HENRY, M; KERRES, N; WEINSTABL, H NEW SPIRO[3H-INDOLE-3,2'-PYRROLIDIN]-2(1H)-ONE COMPOUNDS AND DERIVATIVES AS MDM2-P53 INHIBITORS US Patent US20240109913 (2024)
- Activation of the p53-MDM4 regulatory axis defines the anti-tumour response to PRMT5 inhibition through its role in regulating cellular splicing.
- Design, synthesis, biological evaluation and in silico studies of novel quinoline derivatives as potential radioprotective molecules targeting the TLR2 and p53 pathways.
- Hoffmann, G; Breitenbücher, F; Schuler, M; Ehrenhofer-Murray, AE A novel sirtuin 2 (SIRT2) inhibitor with p53-dependent pro-apoptotic activity in non-small cell lung cancer. J Biol Chem 289: 5208-16 (2014)
- Stahlecker, J; Klett, T; Schwer, M; Jaag, S; Dammann, M; Ernst, LN; Braun, MB; Zimmermann, MO; Kramer, M; Lämmerhofer, M; Stehle, T; Coles, M; Boeckler, FM Revisiting a challenging p53 binding site: a diversity-optimized HEFLib reveals diverse binding modes in T-p53C-Y220C. RSC Med Chem 13: 1575-1586 (2022)
- Kargbo, RB Modulators of the Cystic Fibrosis Transmembrane Conductance Regulator Protein. ACS Med Chem Lett 9: 592-593 (2018)
- Ling, Y; Xu, C; Luo, L; Cao, J; Feng, J; Xue, Y; Zhu, Q; Ju, C; Li, F; Zhang, Y; Zhang, Y; Ling, X Novel β-Carboline/Hydroxamic Acid Hybrids Targeting Both Histone Deacetylase and DNA Display High Anticancer Activity via Regulation of the p53 Signaling Pathway. J Med Chem 58: 9214-27 (2015)
- Turner, EM; Blazer, LL; Neubig, RR; Husbands, SM Small Molecule Inhibitors of Regulator of G Protein Signalling (RGS) Proteins. ACS Med Chem Lett 3: 146-150 (2012)
- Zheng, GH; Shen, JJ; Zhan, YC; Yi, H; Xue, ST; Wang, Z; Ji, XY; Li, ZR Design, synthesis and in vitro and in vivo antitumour activity of 3-benzylideneindolin-2-one derivatives, a novel class of small-molecule inhibitors of the MDM2-p53 interaction. Eur J Med Chem 81: 277-88 (2014)
- Li, X; Peterson, YK; Inks, ES; Himes, RA; Li, J; Zhang, Y; Kong, X; Chou, CJ Class I HDAC Inhibitors Display Different Antitumor Mechanism in Leukemia and Prostatic Cancer Cells Depending on Their p53 Status. J Med Chem 61: 2589-2603 (2018)
- Horswill, AR; Gordon, CP Structure-Activity Relationship Studies of Small Molecule Modulators of the Staphylococcal Accessory Gene Regulator. J Med Chem 63: 2705-2730 (2020)
- Kulén, M; Lindgren, M; Hansen, S; Cairns, AG; Grundström, C; Begum, A; van der Lingen, I; Brännström, K; Hall, M; Sauer, UH; Johansson, J; Sauer-Eriksson, AE; Almqvist, F Structure-Based Design of Inhibitors Targeting PrfA, the Master Virulence Regulator of Listeria monocytogenes. J Med Chem 61: 4165-4175 (2018)
- Lorca, GL; Ezersky, A; Lunin, VV; Walker, JR; Altamentova, S; Evdokimova, E; Vedadi, M; Bochkarev, A; Savchenko, A Glyoxylate and pyruvate are antagonistic effectors of the Escherichia coli IclR transcriptional regulator. J Biol Chem 282: 16476-91 (2007)
- Shoda, T; Kato, M; Fujisato, T; Misawa, T; Demizu, Y; Inoue, H; Naito, M; Kurihara, M Synthesis and evaluation of raloxifene derivatives as a selective estrogen receptor down-regulator. Bioorg Med Chem 24: 2914-2919 (2016)
- Frezza, M; Soulère, L; Reverchon, S; Guiliani, N; Jerez, C; Queneau, Y; Doutheau, A Synthetic homoserine lactone-derived sulfonylureas as inhibitors of Vibrio fischeri quorum sensing regulator. Bioorg Med Chem 16: 3550-6 (2008)
- PRMT5: A novel regulator of Hepatitis B virus replication and an arginine methylase of HBV core.
- Laaroussi, H; Ding, Y; Teng, Y; Deschamps, P; Vidal, M; Yu, P; Broussy, S Synthesis of indole inhibitors of silent information regulator 1 (SIRT1), and their evaluation as cytotoxic agents. Eur J Med Chem 202: (2020)
- Tang, YT; Gao, R; Havranek, JJ; Groisman, EA; Stock, AM; Marshall, GR Inhibition of bacterial virulence: drug-like molecules targeting the Salmonella enterica PhoP response regulator. Chem Biol Drug Des 79: 1007-17 (2012)
- Rose, NR; Woon, EC; Tumber, A; Walport, LJ; Chowdhury, R; Li, XS; King, ON; Lejeune, C; Ng, SS; Krojer, T; Chan, MC; Rydzik, AM; Hopkinson, RJ; Che, KH; Daniel, M; Strain-Damerell, C; Gileadi, C; Kochan, G; Leung, IK; Dunford, J; Yeoh, KK; Ratcliffe, PJ; Burgess-Brown, N; von Delft, F; Muller, S; Marsden, B; Brennan, PE; McDonough, MA; Oppermann, U; Klose, RJ; Schofield, CJ; Kawamura, A Plant growth regulator daminozide is a selective inhibitor of human KDM2/7 histone demethylases. J Med Chem 55: 6639-43 (2012)
- Small molecule modulators of cystic fibrosis transmembrane conductance regulator (CFTR): Structure, classification, and mechanisms.
- The "Doorstop Pocket" In Thioredoxin Reductases─An Unexpected Druggable Regulator of the Catalytic Machinery.
- Abdel-Magid, AF Targeting the Cystic Fibrosis Transmembrane Conductance Regulator (CFTR) Protein for the Treatment of Cystic Fibrosis. ACS Med Chem Lett 7: 725-7 (2016)
- Mirguet, O; Gosmini, R; Toum, J; Clément, CA; Barnathan, M; Brusq, JM; Mordaunt, JE; Grimes, RM; Crowe, M; Pineau, O; Ajakane, M; Daugan, A; Jeffrey, P; Cutler, L; Haynes, AC; Smithers, NN; Chung, CW; Bamborough, P; Uings, IJ; Lewis, A; Witherington, J; Parr, N; Prinjha, RK; Nicodème, E Discovery of epigenetic regulator I-BET762: lead optimization to afford a clinical candidate inhibitor of the BET bromodomains. J Med Chem 56: 7501-15 (2013)
- Alcacio, T; Baek, M; Grootenhuis, P; Hadida Ruah, SS; Hughes, RM; Keshavarz-Shokri, A; McAuley-Aoki, R; McCartney, J; Miller, MT; Van Goor, F; Zhang, B; Anderson, C; Cleveland, T; Frieman, BA; Khatuya, H; Joshi, PV; Krenitsky, PJ; Melillo, V; Pierre, FJ; Termin, AP; Uy, J; Zhou, J; Abela, AR; Busch, BB; Paraselli, P Modulator of cystic fibrosis transmembrane conductance regulator, pharmaceutical compositions, methods of treatment, and process for making the modulator US Patent US11186566 (2021)
- Clemens, JJ; Abela, AR; Anderson, CD; Busch, BB; Chen, WG; Cleveland, T; Coon, TR; Frieman, B; Ghirmai, SG; Grootenhuis, P; Gulevich, AV; Hadida Ruah, SS; Hsia, CK; Kang, P; Khatuya, H; McCartney, J; Miller, MT; Paraselli, P; Pierre, F; Swift, SE; Termin, A; Uy, J; Vogel, CV; Zhou, J Modulators of Cystic Fibrosis Transmembrane Conductance regulator, pharmaceutical compositions, methods of treatment, and process for making the modulators US Patent US11866450 (2024)
- Zender, M; Klein, T; Henn, C; Kirsch, B; Maurer, CK; Kail, D; Ritter, C; Dolezal, O; Steinbach, A; Hartmann, RW Discovery and biophysical characterization of 2-amino-oxadiazoles as novel antagonists of PqsR, an important regulator of Pseudomonas aeruginosa virulence. J Med Chem 56: 6761-74 (2013)
- J., CJ; Schulz, BW; C., BB; Thomas, C; Timothy, C; Michel, G; Jan, GP; Sabina, HR; Julie, L; Thomas, MM; Prasuna, P; K., RY; Jagadeeswar, RT; Claudio, S; Lino, V; Jinglan, Z MACROCYCLES CONTAINING A 1,3,4-OXADIAZOLE RING FOR USE AS MODULATORS OF CYSTIC FIBROSIS TRANSMEMBRANE CONDUCTANCE REGULATOR WIPO WO2022109573 (2022)
- Synthesis and Biological Evaluation of Pyrazole-Pyrimidones as a New Class of Correctors of the Cystic Fibrosis Transmembrane Conductance Regulator (CFTR).
- Cottet, K; Xu, B; Coric, P; Bouaziz, S; Michel, S; Vidal, M; Lallemand, MC; Broussy, S Guttiferone A Aggregates Modulate Silent Information Regulator 1 (SIRT1) Activity. J Med Chem 59: 9560-9566 (2016)
- Son, JH; Zhu, JS; Phuan, PW; Cil, O; Teuthorn, AP; Ku, CK; Lee, S; Verkman, AS; Kurth, MJ High-Potency Phenylquinoxalinone Cystic Fibrosis Transmembrane Conductance Regulator (CFTR) Activators. J Med Chem 60: 2401-2410 (2017)
- Knapp, JM; Wood, AB; Phuan, PW; Lodewyk, MW; Tantillo, DJ; Verkman, AS; Kurth, MJ Structure-activity relationships of cyanoquinolines with corrector-potentiator activity in ΔF508 cystic fibrosis transmembrane conductance regulator protein. J Med Chem 55: 1242-51 (2012)
- Gosmini, R; Nguyen, VL; Toum, J; Simon, C; Brusq, JM; Krysa, G; Mirguet, O; Riou-Eymard, AM; Boursier, EV; Trottet, L; Bamborough, P; Clark, H; Chung, CW; Cutler, L; Demont, EH; Kaur, R; Lewis, AJ; Schilling, MB; Soden, PE; Taylor, S; Walker, AL; Walker, MD; Prinjha, RK; Nicodème, E The discovery of I-BET726 (GSK1324726A), a potent tetrahydroquinoline ApoA1 up-regulator and selective BET bromodomain inhibitor. J Med Chem 57: 8111-31 (2014)
- Greco, MN; Connelly, MA; Leo, GC; Olson, MW; Powell, E; Huang, Z; Hawkins, M; Smith, C; Schalk-Hihi, C; Darrow, AL; Xin, H; Lang, W; Damiano, BP; Hlasta, DJ A thiocarbamate inhibitor of endothelial lipase raises HDL cholesterol levels in mice. Bioorg Med Chem Lett 23: 2595-7 (2013)
- Koch, S; Hemrick-Luecke, SK; Thompson, LK; Evans, DC; Threlkeld, PG; Nelson, DL; Perry, KW; Bymaster, FP Comparison of effects of dual transporter inhibitors on monoamine transporters and extracellular levels in rats. Neuropharmacology 45: 935-44 (2003)
- Marivingt-Mounir, C; Norez, C; Dérand, R; Bulteau-Pignoux, L; Nguyen-Huy, D; Viossat, B; Morgant, G; Becq, F; Vierfond, JM; Mettey, Y Synthesis, SAR, crystal structure, and biological evaluation of benzoquinoliziniums as activators of wild-type and mutant cystic fibrosis transmembrane conductance regulator channels. J Med Chem 47: 962-72 (2004)
- Chi, YH; Yeh, TK; Ke, YY; Lin, WH; Tsai, CH; Wang, WP; Chen, YT; Su, YC; Wang, PC; Chen, YF; Wu, ZW; Yeh, JY; Hung, MC; Wu, MH; Wang, JY; Chen, CP; Song, JS; Shih, C; Chen, CT; Chang, CP Discovery and Synthesis of a Pyrimidine-Based Aurora Kinase Inhibitor to Reduce Levels of MYC Oncoproteins. J Med Chem 64: 7312-7330 (2021)
- Horie, K; Tang, F; Borchardt, RT Isolation and characterization of Caco-2 subclones expressing high levels of multidrug resistance protein efflux transporter. Pharm Res 20: 161-8 (2003)
- Choi, D; Piao, YL; Wu, Y; Cho, H Control of the intracellular levels of prostaglandin E2 through inhibition of the 15-hydroxyprostaglandin dehydrogenase for wound healing. Bioorg Med Chem 21: 4477-84 (2013)
- Lee, S; Phuan, PW; Felix, CM; Tan, JA; Levin, MH; Verkman, AS Nanomolar-Potency Aminophenyl-1,3,5-triazine Activators of the Cystic Fibrosis Transmembrane Conductance Regulator (CFTR) Chloride Channel for Prosecretory Therapy of Dry Eye Diseases. J Med Chem 60: 1210-1218 (2017)
- McConnell, JR; Buckton, LK; McAlpine, SR Regulating the master regulator: Controlling heat shock factor 1 as a chemotherapy approach. Bioorg Med Chem Lett 25: 3409-14 (2015)
- Mattmann, ME; Geske, GD; Worzalla, GA; Chandler, JR; Sappington, KJ; Greenberg, EP; Blackwell, HE Synthetic ligands that activate and inhibit a quorum-sensing regulator in Pseudomonas aeruginosa. Bioorg Med Chem Lett 18: 3072-5 (2008)
- Holmes, IP; Blunt, RJ; Lorthioir, OE; Blowers, SM; Gribble, A; Payne, AH; Stansfield, IG; Wood, M; Woollard, PM; Reavill, C; Howes, CM; Micheli, F; Di Fabio, R; Donati, D; Terreni, S; Hamprecht, D; Arista, L; Worby, A; Watson, SP The identification of a selective dopamine D2 partial agonist, D3 antagonist displaying high levels of brain exposure. Bioorg Med Chem Lett 20: 2013-6 (2010)
- Shaw, SJ; Goff, DA; Lin, N; Singh, R; Li, W; McLaughlin, J; Baltgalvis, KA; Payan, DG; Kinsella, TM Developing DYRK inhibitors derived from the meridianins as a means of increasing levels of NFAT in the nucleus. Bioorg Med Chem Lett 27: 2617-2621 (2017)
- Shi, ZC; Devasagayaraj, A; Gu, K; Jin, H; Marinelli, B; Samala, L; Scott, S; Stouch, T; Tunoori, A; Wang, Y; Zang, Y; Zhang, C; Kimball, SD; Main, AJ; Sun, W; Yang, Q; Nouraldeen, A; Yu, XQ; Buxton, E; Patel, S; Nguyen, N; Swaffield, J; Powell, DR; Wilson, A; Liu, Q Modulation of peripheral serotonin levels by novel tryptophan hydroxylase inhibitors for the potential treatment of functional gastrointestinal disorders. J Med Chem 51: 3684-7 (2008)
- Chen, H; Liang, B P-phenylenediamine derivative as potassium channel regulator and preparation method and medical application thereof US Patent US11365181 (2022)
- Kuttruff, CA; Ferrara, M; Bretschneider, T; Hoerer, S; Handschuh, S; Nosse, B; Romig, H; Nicklin, P; Roth, GJ Discovery of BI-2545: A Novel Autotaxin Inhibitor That Significantly Reduces LPA Levels in Vivo. ACS Med Chem Lett 8: 1252-1257 (2017)
- Bellotti, M; Salis, A; Grozio, A; Damonte, G; Vigliarolo, T; Galatini, A; Zocchi, E; Benatti, U; Millo, E Synthesis, structural characterization and effect on human granulocyte intracellular cAMP levels of abscisic acid analogs. Bioorg Med Chem 23: 22-32 (2014)
- Grand, DL; Gosling, M; Baettig, U; Bahra, P; Bala, K; Brocklehurst, C; Budd, E; Butler, R; Cheung, AK; Choudhury, H; Collingwood, SP; Cox, B; Danahay, H; Edwards, L; Everatt, B; Glaenzel, U; Glotin, AL; Groot-Kormelink, P; Hall, E; Hatto, J; Howsham, C; Hughes, G; King, A; Koehler, J; Kulkarni, S; Lightfoot, M; Nicholls, I; Page, C; Pergl-Wilson, G; Popa, MO; Robinson, R; Rowlands, D; Sharp, T; Spendiff, M; Stanley, E; Steward, O; Taylor, RJ; Tranter, P; Wagner, T; Watson, H; Williams, G; Wright, P; Young, A; Sandham, DA Discovery of Icenticaftor (QBW251), a Cystic Fibrosis Transmembrane Conductance Regulator Potentiator with Clinical Efficacy in Cystic Fibrosis and Chronic Obstructive Pulmonary Disease. J Med Chem 64: 7241-7260 (2021)
- Ye, L; Knapp, JM; Sangwung, P; Fettinger, JC; Verkman, AS; Kurth, MJ Pyrazolylthiazole as DeltaF508-cystic fibrosis transmembrane conductance regulator correctors with improved hydrophilicity compared to bithiazoles. J Med Chem 53: 3772-81 (2010)
- Pesci, E; Bettinetti, L; Fanti, P; Galietta, LJ; La Rosa, S; Magnoni, L; Pedemonte, N; Sardone, GL; Maccari, L Novel Hits in the Correction of ΔF508-Cystic Fibrosis Transmembrane Conductance Regulator (CFTR) Protein: Synthesis, Pharmacological, and ADME Evaluation of Tetrahydropyrido[4,3-d]pyrimidines for the Potential Treatment of Cystic Fibrosis. J Med Chem 58: 9697-711 (2015)
- Warrell, Jr., RP; Piwinski, JJ Bifunctional compounds and use for reducing uric acid levels US Patent US10093658 (2018)
- Piwinski, JJ; Buckle, RN; Larivée, A; Siddiqui, A; Warrell, Jr., RP Compounds and their use for reducing uric acid levels US Patent US11168075 (2021)
- Warrell, Jr., RP Methods for reducing uric acid levels using barbiturate derivatives US Patent US9428466 (2016)
- Green, J; Tinson, RAJ; Betts, JHJ; Piras, M; Pelut, A; Steverding, D; Wren, SP; Searcey, M; Troeberg, L Suramin analogues protect cartilage against osteoarthritic breakdown by increasing levels of tissue inhibitor of metalloproteinases 3 (TIMP-3) in the tissue. Bioorg Med Chem 92: (2023)
- PubChem, PC Luminescence-based cell-based high throughput dose response assay to identify activators of the function of SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 (SMARCA2, BRM) PubChem Bioassay (2013)
- Cussac, D; Newman-Tancredi, A; Quentric, Y; Carpentier, N; Poissonnet, G; Parmentier, JG; Goldstein, S; Millan, MJ Characterization of phospholipase C activity at h5-HT2C compared with h5-HT2B receptors: influence of novel ligands upon membrane-bound levels of [3H]phosphatidylinositols. Naunyn Schmiedebergs Arch Pharmacol 365: 242-52 (2002)
- Bengtsson, C; Blaho, S; Saitton, DB; Brickmann, K; Broddefalk, J; Davidsson, O; Drmota, T; Folmer, R; Hallberg, K; Hallén, S; Hovland, R; Isin, E; Johannesson, P; Kull, B; Larsson, LO; Löfgren, L; Nilsson, KE; Noeske, T; Oakes, N; Plowright, AT; Schnecke, V; Ståhlberg, P; Sörme, P; Wan, H; Wellner, E; Oster, L Design of small molecule inhibitors of acetyl-CoA carboxylase 1 and 2 showing reduction of hepatic malonyl-CoA levels in vivo in obese Zucker rats. Bioorg Med Chem 19: 3039-53 (2011)
- Nomura, DK; Hudak, CS; Ward, AM; Burston, JJ; Issa, RS; Fisher, KJ; Abood, ME; Wiley, JL; Lichtman, AH; Casida, JE Monoacylglycerol lipase regulates 2-arachidonoylglycerol action and arachidonic acid levels. Bioorg Med Chem Lett 18: 5875-8 (2008)
- Bymaster, FP; Katner, JS; Nelson, DL; Hemrick-Luecke, SK; Threlkeld, PG; Heiligenstein, JH; Morin, SM; Gehlert, DR; Perry, KW Atomoxetine increases extracellular levels of norepinephrine and dopamine in prefrontal cortex of rat: a potential mechanism for efficacy in attention deficit/hyperactivity disorder. Neuropsychopharmacology 27: 699-711 (2002)
- Wang, C; Zhang, YF; Guo, S; Zhao, Q; Zeng, Y; Xie, Z; Xie, X; Lu, B; Hu, Y GPR52 Antagonist Reduces Huntingtin Levels and Ameliorates Huntington's Disease-Related Phenotypes. J Med Chem 64: 941-957 (2021)
- Wortmann, L; Lindenthal, B; Muhn, P; Walter, A; Nubbemeyer, R; Heldmann, D; Sobek, L; Morandi, F; Schrey, AK; Moosmayer, D; Günther, J; Kuhnke, J; Koppitz, M; Lücking, U; Röhn, U; Schäfer, M; Nowak-Reppel, K; Kühne, R; Weinmann, H; Langer, G Discovery of BAY-298 and BAY-899: Tetrahydro-1,6-naphthyridine-Based, Potent, and Selective Antagonists of the Luteinizing Hormone Receptor Which Reduce Sex Hormone Levels in Vivo. J Med Chem 62: 10321-10341 (2019)
- Boucherle, B; Bertrand, J; Maurin, B; Renard, BL; Fortuné, A; Tremblier, B; Becq, F; Norez, C; Décout, JL A new 9-alkyladenine-cyclic methylglyoxal diadduct activates wt- and F508del-cystic fibrosis transmembrane conductance regulator (CFTR) in vitro and in vivo. Eur J Med Chem 83: 455-65 (2014)
- Seeman, P; Tallerico, T Antipsychotic drugs which elicit little or no parkinsonism bind more loosely than dopamine to brain D2 receptors, yet occupy high levels of these receptors. Mol Psychiatry 3: 123-34 (1998)
- PubChem, PC Late stage assay provider counterscreen results from the probe development effort to identify inhibitors of kruppel-like factor 5 (KLF5): chemiluminescence-based western blot assay for inhibitors of KLF5 protein levels PubChem Bioassay (2010)
- ChEMBL_328243 (CHEMBL869434) Inhibition of p53-HDM2 interaction
- ChEMBL_852441 (CHEMBL2156544) Inhibition of MDM2 binding to p53
- ChEMBL_861666 (CHEMBL2173003) Inhibition of HDM2 binding to p53
- ChEMBL_969084 (CHEMBL2400965) Antagonist activity at MDM2 (unknown origin)-p53 complex assessed as release of p53 by NMR spectroscopic analysis
- ChEMBL_1980318 (CHEMBL4613580) Activation of p53 in human SMMC7721 cells
- ChEMBL_1719235 (CHEMBL4134235) Inhibition of p53 in human HCT116 cells assessed as reduction in nutlin-induced p53 transcriptional response by FRET assay
- ChEMBL_2264535 Displacement of p53 from human MDM2 by HTRF assay
- ChEMBL_736488 (CHEMBL1694433) Inhibition of Mdm2 -p53 protein interaction by ELISA
- ChEMBL_1778139 (CHEMBL4235131) Activation of p53 in human MCF7 cell lysates assessed as increase in total p53 protein level by Western blot analysis
- ChEMBL_951257 (CHEMBL2353483) Inhibition of MDM2 (unknown origin) assessed as activation of p53
- ChEMBL_1852372 (CHEMBL4352996) Inhibition of p53 protein binding to MDM2 (unknown origin)
- ChEMBL_1852380 (CHEMBL4353004) Inhibition of p53 protein binding to MDMX (unknown origin)
- ChEMBL_653502 (CHEMBL1226705) Inhibition of Rad3-related (ATR) protein-mediated p53 phosphorylation
- ChEMBL_861560 (CHEMBL2174501) Inhibition of SIRT1 using p53-based 18mer as substrate
- Dose Response confirmation of Inhibitors of Mdm2/MdmX interaction in luminescent format Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego CA) Network: NIH Molecular Libraries Probe Production Centers Network (MLPCN) Grant Number: R03 MH089489-01 Assay Provider: Dr. Geoffrey M. Wahl, Salk Institute for Biological Studies, San Diego, CA A wild type but attenuated p53 is retained in approximately 50% of human tumors, and reactivation of p53 in such tumors is an attractive chemotherapeutic strategy. p53 activity is restricted in vivo by mdm2 and mdmx, and knockout of either of these proteins is embryonic lethal in a p53-dependent manner (1, 2). Both proteins bind to p53 via a hydrophobic N-terminal pocket and block p53-dependent transcription of genes required for tumor suppression. Efforts to reactivate p53 with small molecules have focused on inhibition of the mdm2/p53 interaction, which leads to increased p53 levels and activity. However, recent reports indicate that targetin
- ChEMBL_735889 (CHEMBL1694393) Inhibition of human GST-tagged Mdm2 expressed in Escherichia coli harboring integrated p53-Mmd2 protein assessed as blockade of enzyme-p53 interaction by ELISA assay
- ChEMBL_735890 (CHEMBL1694394) Inhibition of human GST-tagged MDMX expressed in Escherichia coli harboring integrated p53-Hmd2 protein assessed as blockade of enzyme-p53 interaction by ELISA assay
- ChEMBL_873189 (CHEMBL2183420) Inhibition of DNA-PK assessed as inhibition of p53 peptide phosphorylation
- ChEMBL_1796587 (CHEMBL4268704) Inhibition of p53-MDM2 interaction (unknown origin) by HTRF assay
- ChEMBL_664220 (CHEMBL1261727) Inhibition of GST-tagged p53 binding to Hdm2 by ELISA
- ChEMBL_664222 (CHEMBL1261729) Inhibition of GST-tagged p53 binding to MDMX by ELISA
- ChEMBL_852440 (CHEMBL2156543) Inhibition of GST-tagged MDM2 binding to p53 by ELISA
- ChEMBL_2537789 Inhibition of FLAG-tagged SMYD2 methylation co-transfected in human HEK293 cells assessed as decrease in levels of p53 Lys370 me1 by Western blot analysis
- ChEMBL_874350 (CHEMBL2182648) Activity at p53 in human HCT116 cells expressing wild type p53 assessed as induction of p21 mRNA expression after 7 hrs by RT-PCR analysis
- ChEMBL_1852382 (CHEMBL4353006) Inhibition of p53 protein binding to MDM2 in human SJSA1 cells
- ChEMBL_1904689 (CHEMBL4407047) Inhibition of P300 (unknown origin) assessed as reduction in p53 acetylation
- ChEMBL_2332806 Inhibition of human ATM kinase using p53 as substrate by FRET assay
- ChEMBL_72328 (CHEMBL685228) Inhibition of p53 binding to Glutathione S-transferase 2 (hdm2-GST)
- Dose Response confirmation of Inhibitors of Mdm2/MdmX interaction using a Brca1/Bard1 BiLC Counterscreen assay Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego CA) Network: NIH Molecular Libraries Probe Production Centers Network (MLPCN) Grant Number: R03 MH089489-01 Assay Provider: Dr. Geoffrey M. Wahl, Salk Institute for Biological Studies, San Diego, CA A wild type but attenuated p53 is retained in approximately 50% of human tumors, and reactivation of p53 in such tumors is an attractive chemotherapeutic strategy. p53 activity is restricted in vivo by mdm2 and mdmx, and knockout of either of these proteins is embryonic lethal in a p53-dependent manner (1, 2). Both proteins bind to p53 via a hydrophobic N-terminal pocket and block p53-dependent transcription of genes required for tumor suppression. Efforts to reactivate p53 with small molecules have focused on inhibition of the mdm2/p53 interaction, which leads to increased p53 levels and activity. However, recent reports indicate that targetin
- ChEMBL_1852381 (CHEMBL4353005) Inhibition of p53 derived Cy5-p53 peptide (18 to 26 residues) binding to human MDM2 (2 to 188 residues) after 30 mins by TR-FRET assay
- ChEMBL_2205941 (CHEMBL5118649) Inhibition of FLAG-tagged SMYD2 (unknown origin) expressed in human U2OS cells mediated intracellular P53 methylation incubated for 15 hrs using p53 as substrate by ELISA
- ChEMBL_1738283 (CHEMBL4154033) Binding affinity to p53 Y220C mutant (unknown origin)
- ChEMBL_2205938 (CHEMBL5118646) Inhibition of FLAG-tagged SMYD2 (unknown origin) expressed in human U2OS cells mediated intracellular p53 methylation incubated for 24 hrs using p53 as substrate by immunofluorescence analysis
- ChEMBL_1576259 (CHEMBL3801205) Inhibition of MDM2 in human HCT116 p53+/+ cells assessed as growth inhibition
- ChEMBL_2559191 Inhibition of human SMG1 using glutathione S-transferase-p53 as substrate by DELFIA
- ChEMBL_466592 (CHEMBL930243) Binding affinity to human HDM2 assessed as inhibition of HDM2-p53 interaction
- ChEMBL_589613 (CHEMBL1052166) Induction of p53-Ser15 phosphorylation in human HCT116 cells after 24 hrs
- ChEMBL_811269 (CHEMBL2015221) Displacement of fluorescein-labeled p53 peptide from HDM2 by fluorescence polarization assay
- ChEMBL_967889 (CHEMBL2400705) Inhibition of p53 (unknown origin) phosphorylation at Ser15 by cell-based assay
- ChEMBL_977770 (CHEMBL2424317) Inhibition of HDAC3/NcoR2 (unknown origin) using RHKKAc from p53 as substrate
- ChEMBL_1363438 (CHEMBL3295827) Inhibition of GST-tagged HDM2 (unknown origin) expressed in Escherichia coli BL21 cells assessed as p53 activation by measuring inhibition of His-p53 ubiquitination after 1 hr by fluorescence spectroscopy
- ChEMBL_2027563 (CHEMBL4681721) Inhibition of SMYD2 (unknown origin) expressed in in human U2OS cells assessed as reduction in monomethylated p53 levels incubated for 24 hrs by immunofluorescence assay
- ChEMBL_2537790 Inhibition of FLAG-tagged SMYD2 methylation co-transfected in human U2OS cells assessed as decrease in levels of p53 by measured after 15 hrs by cell-based ELISA analysis
- ChEMBL_1973136 (CHEMBL4605954) Inhibition of USP7 in human RKO cells transfected with p53 luciferase reporter vector assessed as increase in p53-dependent luciferase activity measured after 18 hrs by BrightGlo luciferase assay
- ChEMBL_876865 (CHEMBL2184459) Binding affinity to p53 Y220C mutant by NMR spectroscopy
- ChEBML_1696268 Inhibition of p53 binding to MDM2 (unknown origin) after 30 mins by SPR analysis
- ChEBML_1696269 Inhibition of p53 binding to MDMX (unknown origin) after 30 mins by SPR analysis
- ChEMBL_1350282 (CHEMBL3267620) Binding affinity to MDM2 (unknown origin) assessed as inhibition of interaction with p53
- ChEMBL_1556177 (CHEMBL3768631) Inhibition of flag-tagged ATM (unknown origin) using p53 as substrate by ELISA
- ChEMBL_1852374 (CHEMBL4352998) Inhibition of p53 protein binding to MDM2 (unknown origin) by western blot analysis
- ChEMBL_2488072 Inhibition of HDAC1 (unknown origin) using biotinylated P53 peptide as substrate by HTRF assay
- ChEMBL_2559192 Inhibition of ATR (unknown origin) using glutathione S-transferase-p53 as substrate by DELFIA
- ChEMBL_874011 (CHEMBL2186136) Inhibition of HDAC1 using p53 (379 to 382 residues) based fluorogenic peptide substrate
- ChEMBL_874013 (CHEMBL2186138) Inhibition of HDAC3 using p53 (379 to 382 residues) based fluorogenic peptide substrate
- ChEMBL_874014 (CHEMBL2186139) Inhibition of HDAC2 using p53 (379 to 382 residues) based fluorogenic peptide substrate
- ChEMBL_876028 (CHEMBL2182319) Inhibition of HDAC11 using p53 (379 to 382 residues) based fluorogenic peptide substrate
- ChEMBL_876029 (CHEMBL2182320) Inhibition of HDAC10 using p53 (379 to 382 residues) based fluorogenic peptide substrate
- ChEMBL_876030 (CHEMBL2182321) Inhibition of HDAC9 using p53 (379 to 382 residues) based fluorogenic peptide substrate
- ChEMBL_876031 (CHEMBL2182322) Inhibition of HDAC8 using p53 (379 to 382 residues) based fluorogenic peptide substrate
- ChEMBL_876032 (CHEMBL2182323) Inhibition of HDAC7 using p53 (379 to 382 residues) based fluorogenic peptide substrate
- ChEMBL_876033 (CHEMBL2182324) Inhibition of HDAC6 using p53 (379 to 382 residues) based fluorogenic peptide substrate
- ChEMBL_876034 (CHEMBL2182325) Inhibition of HDAC5 using p53 (379 to 382 residues) based fluorogenic peptide substrate
- ChEMBL_876035 (CHEMBL2182326) Inhibition of HDAC4 using p53 (379 to 382 residues) based fluorogenic peptide substrate
- ChEMBL_977174 (CHEMBL2416767) Inhibition of human recombinant HDAC6 using p53 residues 379-382 (RHKKAc) as substrate
- ChEMBL_977175 (CHEMBL2416768) Inhibition of human recombinant HDAC4 using p53 residues 379-382 (RHKKAc) as substrate
- ChEMBL_977176 (CHEMBL2416769) Inhibition of human recombinant HDAC1 using p53 residues 379-382 (RHKKAc) as substrate
- ChEMBL_2205942 (CHEMBL5118650) Inhibition of FLAG-tagged SMYD2 (unknown origin) expressed in human KYSE-150 cells mediated intracellular P53 methylation at lysine 370 incubated for 15 hrs using p53 as substrate by ELISA
- ChEMBL_1444859 (CHEMBL3379629) Inhibition of ATM (unknown origin) using p53-Q10-K17 peptide substrate by alphascreen assay
- ChEMBL_1556174 (CHEMBL3768628) Inhibition of human ATM using p53 as substrate preincubated for 10 mins by ELISA
- ChEMBL_1718864 (CHEMBL4133864) Inhibition of human HDAC1 using p53 (379 to 382 residues) derived fluorogenic peptide RHKKAc
- ChEMBL_1718865 (CHEMBL4133865) Inhibition of human HDAC2 using p53 (379 to 382 residues) derived fluorogenic peptide RHKKAc
- ChEMBL_947124 (CHEMBL2341094) Inhibition of human S100B/human TAMRA-p53(367-388) interaction by fluorescence polarization assay
- ChEMBL_1504432 (CHEMBL3592997) Inhibition of SMYD2 (unknown origin) using p53 (361 to 380) as substrate assessed as incorporation of tritium labeled methyl group from [3H]-SAM to p53 peptide substrate after 1 hr by scintillation proximity assay
- ChEMBL_2114758 (CHEMBL4823699) Inhibition of full length recombinant FLAG-tagged human ATM assessed as decrease in p53 S15 phosphorylation using full length myc-tagged p53 as substrate incubated for 30 mins in presence of ATP by ELISA
- ChEMBL_1474783 (CHEMBL3424240) Induction of p53-dependent growth suppression of human LNZTA3 cells after 72 hrs by MTT assay
- ChEMBL_1556175 (CHEMBL3768629) Inhibition of ATM in human U2OS cells assessed as inhibition of p53 phosphorylation at Ser15 residue
- ChEMBL_665716 (CHEMBL1261685) Inhibition of mTOR in p53-deficient MEF assessed as phosphorylation of S6K1 at Thr389 by immunoblotting
- ChEMBL_741994 (CHEMBL1768754) Inhibition of mTOR in p53-deficient MEF assessed as phosphorylation of S6K1 at Thr389 by immunoblotting
- ChEMBL_1336809 (CHEMBL3241500) Binding affinity to MDMX (unknown origin) assessed as inhibition of interaction with p53 in serum free buffer incubated for 20 mins prior to p53 addition measured after 60 mins by HTRF assay
- ChEMBL_1339943 (CHEMBL3243813) Binding affinity to MDMX (unknown origin) assessed as inhibition of interaction with p53 in serum free buffer incubated for 20 mins prior to p53 addition measured after 60 mins by HTRF assay
- ChEMBL_1347756 (CHEMBL3271857) Binding affinity to MDM2 in human MCF7 cells assessed as inhibition of MDM2-p53 interaction
- ChEMBL_1718866 (CHEMBL4133866) Inhibition of human HDAC3/NCOR2 using p53 (379 to 382 residues) derived fluorogenic peptide RHKKAc
- ChEMBL_1795357 (CHEMBL4267474) Inhibition of recombinant p53 protein binding to recombinant human MDM2 by surface plasmon resonance method
- ChEMBL_2345087 Inhibition of human recombinant HDAC1 using fluorogenic p53 (379-382 residues) (RHKK(Ac)AMC) as substrate
- ChEMBL_2345088 Inhibition of human recombinant HDAC3 using fluorogenic p53 (379-382 residues) (RHKK(Ac)AMC) as substrate
- ChEMBL_2345090 Inhibition of human recombinant HDAC6 using fluorogenic p53 (379-382 residues) (RHKK(Ac)AMC) as substrate
- ChEMBL_2345092 Inhibition of human recombinant HDAC2 using fluorogenic p53 (379-382 residues) (RHKK(Ac)AMC) as substrate
- ChEMBL_2345096 Inhibition of human recombinant HDAC10 using fluorogenic p53 (379-382 residues) (RHKK(Ac)AMC) as substrate
- ChEMBL_2427094 Inhibition of human full length recombinant ATM using GST-cMyc-p53 as substrate by ELISA method
- ChEMBL_2469101 Inhibition of HDAC1 (unknown origin) using p53 residues 379 to 382 [RHKK(Ac)] as fluorogenic substrate
- ChEMBL_2469103 Inhibition of HDAC3 (unknown origin) using p53 residues 379 to 382 [RHKK(Ac)] as fluorogenic substrate
- ChEMBL_989970 (CHEMBL2447327) Inhibition of GST-tagged HDM2 (unknown origin) assessed as p53 ubiquitination by Western blot analysis
- ChEMBL_951641 (CHEMBL2351555) Inhibition of human recombinant GST-thrombin-MDM2 (1 to 188) expressed in Escherichia coli assessed as reduction of MDM2-p53 interaction incubated for 20 mins followed by p53 addition measured after 60 mins by HTRF assay
- ChEMBL_1874860 (CHEMBL4376149) Inhibition of V5-tagged SMYD2 (unknown origin) transfected in HEK293 cells co-transfected with FLAG-tagged p53 assessed as reduction in p53 methylation at K370 residue measured after 20 hrs by immunoblot analysis
- ChEMBL_306060 (CHEMBL874553) Binding affinity between MDM2 and p53 protein in fluorescence peptide assay
- ChEMBL_1874857 (CHEMBL4376146) Inhibition of V5-tagged SMYD2 (unknown origin) transfected in human MCF7 cells co-transfected with FLAG-tagged p53 assessed as reduction in p53 methylation at K370 residue measured after 20 hrs by immunoblot analysis
- ChEMBL_1874859 (CHEMBL4376148) Inhibition of V5-tagged SMYD2 (unknown origin) transfected in human HeLa cells co-transfected with FLAG-tagged p53 assessed as reduction in p53 methylation at K370 residue measured after 20 hrs by immunoblot analysis
- ChEMBL_1874861 (CHEMBL4376150) Inhibition of V5-tagged SMYD2 (unknown origin) transfected in human CAL120 cells co-transfected with FLAG-tagged p53 assessed as reduction in p53 methylation at K370 residue measured after 20 hrs by immunoblot analysis
- ChEMBL_1874862 (CHEMBL4376151) Inhibition of V5-tagged SMYD2 (unknown origin) transfected in human U2OS cells co-transfected with FLAG-tagged p53 assessed as reduction in p53 methylation at K370 residue measured after 20 hrs by immunoblot analysis
- ChEMBL_951640 (CHEMBL2351554) Inhibition of human recombinant GST-thrombin-MDM2 (1 to 188) expressed in Escherichia coli assessed as reduction of MDM2-p53 interaction incubated for 20 mins followed by p53 addition measured after 60 mins by HTRF assay in presence of 15% serum
- ChEBML_1681049 Inhibition of human HDAC10 using fluorogenic peptide from p53 residues 379 to 382 as substrate in presence of ATP
- ChEMBL_961694 (CHEMBL2389364) Inhibition of N-terminal human recombinant MDM2 assessed as inhibition of protein interaction with p53 by HTRF assay
- ChEMBL_1574245 (CHEMBL3802428) Binding affinity to human MDM2 assessed as inhibition of interaction with p53 by fluorescence polarization assay
- ChEMBL_1776431 (CHEMBL4233423) Inhibition of human HDAC1 using p53 (379 to 382 residues) derived fluorogenic peptide RHKKAc as substrate
- ChEMBL_1776433 (CHEMBL4233425) Inhibition of human HDAC6 using p53 (379 to 382 residues) derived fluorogenic peptide RHKKAc as substrate
- ChEMBL_1852373 (CHEMBL4352997) Inhibition of wild type p53 protein binding to MDM2 in human HCT116 cells by immunoblot analysis
- ChEMBL_2093853 (CHEMBL4775116) Inhibition of ATM (unknown origin) assessed as ATM-dependent phosphorylation using GST-p53 ser15 as substrate
- ChEMBL_2525403 Inhibition of human HeLa cell nuclear extract purified DNA-PK using p53 peptide as substrate by ELISA
- ChEMBL_815413 (CHEMBL2027065) Inhibition of p53 derived peptide binding to MDM4 using fluorescent dye Cy5 by TR-FRET assay
- ChEMBL_959852 (CHEMBL2383516) Antagonist activity at human GST-tagged MDM2 assessed as inhibition of binding to full length p53
- ChEMBL_1473619 (CHEMBL3420958) Inhibition of SMYD2 in human KYSE-150 cells assessed as reduction of monomethylation of p53 K370 by sandwich ELISA method
- ChEMBL_1874858 (CHEMBL4376147) Inhibition of V5-tagged SMYD2 (unknown origin) transfected in human MDA-MB-231 cells co-transfected with FLAG-tagged p53 assessed as reduction in p53 methylation at K370 residue measured after 20 hrs by immunoblot analysis
- Time Resolved Fluorescence Energy Transfer (TR-FRET) Assay The inhibition of p53-Hdm2 and p53-Hdm4 interactions is measured by time resolved fluorescence energy transfer (TR-FRET). Fluorescence energy transfer (or Foerster resonance energy transfer) describes an energy transfer between donor and acceptor fluorescent molecules. For this assay, MDM2 protein (amino acids 2-188) and MDM4 protein (amino acids 2-185), tagged with a C-terminal Biotin moiety, are used in combination with a Europium labeled streptavidin (Perkin Elmer, Inc., Waltham, Mass., USA) serving as the donor fluorophore. The p53 derived, Cy5 labeled peptide Cy5-TFSDLWKLL (p53 aa 18-26) is the energy acceptor. Upon excitation of the donor molecule at 340 nm, binding interaction between MDM2 or MDM4 and the p53 peptide induces energy transfer and enhanced response at the acceptor emission wavelength at 665 nm. Disruption of the formation of the p53-MDM2 or p53-MDM4 complex due to an inhibitor molecule binding to the p53 binding site of MDM2 or MDM4 results in increased donor emission.
- ChEMBL_1351457 (CHEMBL3267322) Inhibition of MDM2 (unknown origin) (2 to 188) assessed as inhibition of p53-MDM2 interaction by TR-FRET assay
- ChEMBL_2214602 (CHEMBL5127734) Inhibition of human DNA-PK using p53 as substrate incubated for 30 mins in presence of by HTRF assay
- ChEMBL_2427955 Inhibition of human ATR/ATRIP using GST-tagged p53 as substrate in presence of 3 uM ATP by AlphaScreen assay
- ChEBML_1649741 Inhibition of human KDAC3 using FITC-labeled p53 acetylated peptide as substrate after 60 mins by fluorescence assay
- ChEMBL_1556907 (CHEMBL3771932) Inhibition of recombinant human KDAC1 using acetylated p53 (379 to 382 residues) as substrate by fluorescence assay
- ChEMBL_1556992 (CHEMBL3772323) Inhibition of recombinant human KDAC6 using acetylated p53 (379 to 382 residues) as substrate by fluorescence assay
- ChEMBL_1556993 (CHEMBL3772324) Inhibition of recombinant human KDAC8 using diacetylated p53 (379 to 382 residues) as substrate by fluorescence assay
- ChEMBL_1729768 (CHEMBL4145046) Displacement of p53 from MDM2 in human U87MG cells incubated for 10 mins by sandwich ELISA method
- ChEMBL_1847051 (CHEMBL4347592) Inhibition of recombinant human HDAC6 using p53 (379 to 382 residues) derived fluorogenic peptide RHKKAc as substrate
- ChEMBL_2020103 (CHEMBL4673916) Inhibition of HDAC6 (unknown origin) using (AMC)-labelled p53 peptide( 379 to 382 residues) RHKKAc as substrate
- ChEMBL_2020104 (CHEMBL4673917) Inhibition of HDAC1 (unknown origin) using (AMC)-labelled p53 peptide( 379 to 382 residues) RHKKAc as substrate
- ChEMBL_841972 (CHEMBL2091389) Inhibition of MDM2 binding domain assessed as inhibition of p53 binding to MDM2 after 1 hr by fluorescence polarization assay
- ChEBML_1553163 Inhibition of recombinant human CK1epsilon using GST-p53 (1 to 64 residues) as substrate by SDS-PAGE based autoradiography
- ChEMBL_1349594 (CHEMBL3265663) Inhibition of HDAC1 (unknown origin) using RHKK(Ac) (379 to 382) p53 peptide as substrate by fluorescence assay
- ChEMBL_1349595 (CHEMBL3265664) Inhibition of HDAC2 (unknown origin) using RHKK(Ac) (379 to 382) p53 peptide as substrate by fluorescence assay
- ChEMBL_1349596 (CHEMBL3265665) Inhibition of HDAC6 (unknown origin) using RHKK(Ac) (379 to 382) p53 peptide as substrate by fluorescence assay
- ChEMBL_1556991 (CHEMBL3772322) Inhibition of recombinant human KDAC3/NcoR2 using acetylated p53 (379 to 382 residues) as substrate by fluorescence assay
- ChEMBL_1649741 (CHEMBL3998875) Inhibition of human KDAC3 using FITC-labeled p53 acetylated peptide as substrate after 60 mins by fluorescence assay
- ChEMBL_1729769 (CHEMBL4145047) Displacement of p53 from MDM4 in human SHSY-5Y cells incubated for 10 mins by sandwich ELISA method
- ChEMBL_1932538 (CHEMBL4478190) Inhibition of DNAPK (unknown origin) using p53-based peptide substrate preincubated for 5 min prior to ATP addition
- ChEMBL_2345091 Inhibition of human recombinant HDAC8 using fluorogenic diacetylated p53 (379-382 residues) (RHKK(Ac)K(Ac)AMC) as substrate
- ChEMBL_593610 (CHEMBL1039715) Inhibition of his-tagged human recombinant MDM2 binding to p53-based peptide PMDM6-F by fluorescence polarization assay
- ChEMBL_786126 (CHEMBL1919869) Inhibition of DNA-PK using fluorescein-p53 peptide as substrate after 1 hr by TR-FRET enzyme assay
- ChEMBL_873187 (CHEMBL2183418) Inhibition of DNA-PK isolated from human HeLa cell extract assessed as inhibition of p53 peptide fragment phosphorylation after 10 mins
- Method of Inhibiting HDAC8 Further provided herein are methods of inhibiting HDAC8 mediated deacetylation of p53. In one aspect, the method includes contacting HDAC8 with a HDAC8 inhibitor as described herein, thereby inhibiting HDAC8 mediated deacetylation of p53. The inhibition of HDAC8 may allow for acetylation and activation of p53, thereby mediating cell apoptosis. The contacting may be performed in vitro or in vivo. The contacting may be performed in vitro. The contacting may be performed in vivo. The contacting may be performed in an organism. The inhibition of HDAC8 mediated deacetylation of p53 may be monitored by techniques known in the art, including for example, fluorescent and colorimetric assays.
- HDM2 Binding Assay Binding assay using 1,4-benzodiazepine-2,5-dione (BZD) scaffold and HDM2-p53 protein.
- ChEMBL_1336775 (CHEMBL3241171) Binding affinity to GST-thrombin-tagged human MDM2 (1 to 188) expressed in Escherichia coli assessed as inhibition of interaction with p53 in serum free buffer incubated for 20 mins prior to p53 addition measured after 60 mins by HTRF assay
- ChEMBL_1339578 (CHEMBL3243058) Binding affinity to GST-thrombin-tagged human MDM2 (1 to 188) expressed in Escherichia coli assessed as inhibition of interaction with p53 in serum free buffer incubated for 20 mins prior to p53 addition measured after 60 mins by HTRF assay
- ChEMBL_1668048 (CHEMBL4017936) Inhibition of biotin-labelled p53 binding to MDM2 (25 to 109 residues) (unknown origin) by surface plasmon resonance method
- ChEMBL_1852378 (CHEMBL4353002) Inhibition of p53 derived Cy5-TFSDLWKLL peptide binding to human MDM2 (2 to 188 residues) by TR-FRET assay
- ChEMBL_1852379 (CHEMBL4353003) Inhibition of p53 derived Cy5-TFSDLWKLL peptide binding to human MDMX (2 to 185 residues) by TR-FRET assay
- ChEMBL_1875130 (CHEMBL4376419) Inhibition of human HDAC10 using fluorescent-labelled p53 residues 379 to 382 (RHKKAc) peptide as substrate by fluorescence assay
- ChEMBL_1878101 (CHEMBL4379495) Inhibition of recombinant full-length human ATM using GST-cMyc-p53 as substrate measured after 40 mins by ELISA
- ChEMBL_2020095 (CHEMBL4673908) Inhibition of human HDAC2 using p53 fluorogenic peptide (379 to 382 residues) RHKKAc as substrate by fluorescence-based assay
- ChEMBL_2020096 (CHEMBL4673909) Inhibition of human HDAC6 using p53 fluorogenic peptide (379 to 382 residues) RHKKAc as substrate by fluorescence-based assay
- ChEMBL_2020097 (CHEMBL4673910) Inhibition of human HDAC1 using p53 fluorogenic peptide (379 to 382 residues) RHKKAc as substrate by fluorescence-based assay
- ChEMBL_2028076 (CHEMBL4682234) Inhibition of DNA-PK isolated from human HeLa cells assessed as reduction in p53 Ser15 phosphorylation by HTRF assay
- ChEMBL_2035574 (CHEMBL4689732) Inhibition of His-tagged SIRT1 (unknown origin) expressed in Escherichia coli codon plus (DE3) using acetylated p53 as substrate
- ChEMBL_2090166 (CHEMBL4771429) Inhibition of HDAC1 (unknown origin) using p53 residues (379 to 382)(RHKK(Ac)-AMC) as substrate by fluorescent method
- ChEMBL_2090167 (CHEMBL4771430) Inhibition of HDAC2 (unknown origin) using p53 residues (379 to 382)(RHKK(Ac)-AMC) as substrate by fluorescent method
- ChEMBL_2090168 (CHEMBL4771431) Inhibition of HDAC3 (unknown origin) using p53 residues (379 to 382)(RHKK(Ac)-AMC) as substrate by fluorescent method
- ChEMBL_2090174 (CHEMBL4771437) Inhibition of HDAC6 (unknown origin) using p53 residues (379 to 382)(RHKK(Ac)-AMC) as substrate by fluorescent method
- ChEMBL_2090175 (CHEMBL4771438) Inhibition of HDAC10 (unknown origin) using p53 residues (379 to 382)(RHKK(Ac)-AMC) as substrate by fluorescent method
- ChEMBL_2090176 (CHEMBL4771439) Inhibition of HDAC11 (unknown origin) using p53 (379 to 382 residues)(RHKK(Ac)-AMC) as substrate by fluorescent method
- ChEMBL_768516 (CHEMBL1832438) Inhibition of G9a in p53-deficient human HCT116 cells assessed as reduction of H3K9me2 after 48 hrs by In-Cell Western assay
- ChEMBL_1336776 (CHEMBL3241172) Binding affinity to GST-thrombin-tagged human MDM2 (1 to 188) expressed in Escherichia coli assessed as inhibition of interaction with p53 in buffer containing 15% human serum incubated for 20 mins prior to p53 addition measured after 60 mins by HTRF assay
- ChEMBL_1339579 (CHEMBL3243059) Binding affinity to GST-thrombin-tagged human MDM2 (1 to 188) expressed in Escherichia coli assessed as inhibition of interaction with p53 in buffer containing 15% human serum incubated for 20 mins prior to p53 addition measured after 60 mins by HTRF assay
- ChEMBL_1556910 (CHEMBL3771935) Inhibition of recombinant human KDAC3 using FITC-p53 acetylated peptide substrate incubated for 60 mins by microfluidic chip-based assay
- ChEMBL_1585282 (CHEMBL3821545) Inhibition of SMYD2 (unknown origin) expressed in human U2OS cells assessed as reduction in p53 methylation by Western blot analysis
- ChEMBL_1983107 (CHEMBL4616369) Binding affinity to recombinant human MDM2 expressed in Escherichia coli DH5alpha assessed as inhibition of MDM2/p53 protein-protein interaction
- ChEMBL_2048962 (CHEMBL4703661) Binding affinity to human MDM2 assessed as dissociation constant in presence of 50 uM p53 peptide by fluorescence polarization assay
- ChEMBL_2090260 (CHEMBL4771523) Inhibition of human HDAC6 using p53 residues 379-382 (RHKKAc) as substrate measured after 2 hrs by Cheng-Prusoff analysis
- ChEMBL_2311255 Displacement of FAM labeled p53 fragment from human MDM2 (1 to 116 residues) incubated for 1 hrs by fluorescence polarization assay
- ChEMBL_2443957 Inhibition of human recombinant HDAC1 using fluorescent peptide p53 (379 to 382 residues) RHKK(Ac) as substrate by fluorescence based analysis
- ChEMBL_2443958 Inhibition of human recombinant HDAC2 using fluorescent peptide p53 (379 to 382 residues) RHKK(Ac) as substrate by fluorescence based analysis
- ChEMBL_2443959 Inhibition of human recombinant HDAC3 using fluorescent peptide p53 (379 to 382 residues) RHKK(Ac) as substrate by fluorescence based analysis
- ChEMBL_2443962 Inhibition of human recombinant HDAC6 using fluorescent peptide p53 (379 to 382 residues) RHKK(Ac) as substrate by fluorescence based analysis
- ChEMBL_2443966 Inhibition of human recombinant HDAC10 using fluorescent peptide p53 (379 to 382 residues) RHKK(Ac) as substrate by fluorescence based analysis
- ChEMBL_2443967 Inhibition of human recombinant HDAC11 using fluorescent peptide p53 (379 to 382 residues) RHKK(Ac) as substrate by fluorescence based analysis
- ChEMBL_2537787 Inhibition of SMYD2 (unknown origin) using p53(361-380) peptide as a substrate incubated for 1 hr by scintillation proximity assay
- ChEMBL_2214589 (CHEMBL5127721) Inhibition of full-length recombinant human ATR/ATRIP using p53 as substrate incubated for 30 mins in presence of ATP by HTRF assay
- ChEMBL_2355383 Inhibition of HDAC in human HeLa cell nuclear extract using Kac fluorogenic peptide as substrate containing residues 379-382 of p53 by fluorescence assay
- ChEMBL_1362099 (CHEMBL3294652) Binding affinity to MDM2 (unknown origin) assessed as inhibition of interaction with p53 after 1 hr by fluorescence polarization binding assay
- ChEMBL_1440401 (CHEMBL3386214) Inhibition of human recombinant CK1epsilon expressed in Escherichia coli using GST-p53 (1 to 64 residues) by SDS-PAGE based autoradiography
- ChEMBL_1878103 (CHEMBL4379497) Inhibition of recombinant full length human DNA-PK using GST-cMyc-p53 as substrate incubated for 40 mins by fluorescence assay
- ChEMBL_2090129 (CHEMBL4771392) Inhibition of human HDAC3/NcoR2 expressed in sf9 cells using p53 residues 379-382 (RHKKAc) as substrate by fluorescence-based assay
- ChEMBL_2090169 (CHEMBL4771432) Inhibition of HDAC8 (unknown origin) using p53 (379 to 382 residues)(RHK(Ac)K(ac)AMC) as substrate by fluorescent method
- ChEMBL_2090256 (CHEMBL4771519) Inhibition of human HDAC1 using p53 (379 to 382 residues) (RHKKAc) as substrate measured after 2 hrs by cheng-Prusoff analysis
- ChEMBL_2090257 (CHEMBL4771520) Inhibition of human HDAC2 using p53 (379 to 382 residues) (RHKKAc) as substrate measured after 2 hrs by Cheng-Prusoff analysis
- ChEMBL_2090258 (CHEMBL4771521) Inhibition of human HDAC3/NcoR2 using p53 residues 379-382 (RHKKAc) as substrate measured after 2 hrs by Cheng-Prusoff analysis
- ChEMBL_2488435 Inhibition of HDAC7 (unknown origin) using fluorescent peptide p53 (379 to 382 residues) RHKK(Ac)AMC as substrate by fluorescence based analysis
- ChEMBL_2488436 Inhibition of HDAC6 (unknown origin) using fluorescent peptide p53 (379 to 382 residues) RHKK(Ac)AMC as substrate by fluorescence based analysis
- ChEMBL_2488437 Inhibition of HDAC3 (unknown origin) using fluorescent peptide p53 (379 to 382 residues) RHKK(Ac)AMC as substrate by fluorescence based analysis
- ChEMBL_2488444 Inhibition of HDAC1 (unknown origin) using fluorescent peptide p53 (379 to 382 residues) RHKK(Ac)AMC as substrate by fluorescence based analysis
- ChEMBL_856497 (CHEMBL2161543) Inhibition of human recombinant SMG1 expressed in HEK293 cells using GST-tagged p53 as substrate after 1 hr by DELFIA assay
- ChEMBL_968096 (CHEMBL2398954) Inhibition of TMRA-p53 binding to MDM2 (25 to 109 amino acids) (unknown origin) after 30 mins by fluorescence polarization assay
- ChEMBL_969491 (CHEMBL2406002) Binding affinity to GST-tagged MDM2 (unknown origin) assessed as inhibition of interaction with p53 after 1 hr by HTRF assay
- ChEMBL_972652 (CHEMBL2410632) Inhibition of HDAC11 (unknown origin) using fluorogenic peptide from p53 residues (379 to 382) (RHKK(Ac)) as substrate by fluorescence assay
- ChEMBL_972653 (CHEMBL2410633) Inhibition of HDAC10 (unknown origin) using fluorogenic peptide from p53 residues (379 to 382) (RHKK(Ac)) as substrate by fluorescence assay
- ChEMBL_972657 (CHEMBL2410760) Inhibition of HDAC6 (unknown origin) using fluorogenic peptide from p53 residues (379 to 382) (RHKK(Ac)) as substrate by fluorescence assay
- ChEMBL_972660 (CHEMBL2410763) Inhibition of HDAC3 (unknown origin) using fluorogenic peptide from p53 residues (379 to 382) (RHKK(Ac)) as substrate by fluorescence assay
- ChEMBL_972661 (CHEMBL2410764) Inhibition of HDAC2 (unknown origin) using fluorogenic peptide from p53 residues (379 to 382) (RHKK(Ac)) as substrate by fluorescence assay
- ChEMBL_972662 (CHEMBL2410765) Inhibition of HDAC1 (unknown origin) using fluorogenic peptide from p53 residues (379 to 382) (RHKK(Ac)) as substrate by fluorescence assay
- ChEMBL_2245201 (CHEMBL5159411) Inhibition of human recombinant full length ATR using GST-cMyc-p53 as substrate incubated for 40 mins in presence of ATP by HTRF assay
- ChEMBL_2311265 Inhibition of ATM (unknown origin) using GST-cMyc-p53 as substrate incubated for 30 mins in presence of Mg/ATP mix by HTRF-based analysis
- ChEMBL_2048576 (CHEMBL4703275) Inhibition of SMYD2 (unknown origin) transfected in human U2OS cells assessed as inhibition of methylation of monomethyl p53 peptide incubated for 24 hrs by immunofluorescence assay
- ChEMBL_1288104 (CHEMBL3110643) Binding affinity to GST-tagged MDM2 (unknown origin) assessed as inhibition of interaction with biotinylated p53 after 1 hr by HTRF assay
- ChEMBL_1296787 (CHEMBL3132470) Binding affinity to human GST-thrombin-tagged MDM2 assessed as inhibition of interaction with human p53 after 1 hr by HTRF assay
- ChEMBL_1440399 (CHEMBL3386212) Inhibition of rat CK1delta kinetic domain expressed in Escherichia coli using GST-p53 (1 to 64 residues) by SDS-PAGE based autoradiography
- ChEMBL_1440400 (CHEMBL3386213) Inhibition of GST-tagged rat CK1delta expressed in Escherichia coli using GST-p53 (1 to 64 residues) by SDS-PAGE based autoradiography
- ChEMBL_2020108 (CHEMBL4673921) Inhibition of human full-length recombinant HDAC1 using p53 fluorogenic peptide (79 to 382 residues) (RHKK(Ac)) as substrate by fluorescence method
- ChEMBL_2028066 (CHEMBL4682224) Inhibition of recombinant full-length human FLAG-tagged ATM assessed as reduction in p53 S15 phosphorylation incubated for 30 mins by ELISA
- ChEMBL_2090127 (CHEMBL4771390) Inhibition of human HDAC1 expressed in baculovirus infected sf9 cells using p53 residues 379-382 (RHKKAc) as substrate by fluorescence-based assay
- ChEMBL_2090128 (CHEMBL4771391) Inhibition of human HDAC2 expressed in baculovirus infected sf9 cells using p53 residues 379-382 (RHKKAc) as substrate by fluorescence-based assay
- ChEMBL_2090130 (CHEMBL4771393) Inhibition of human HDAC8 expressed in baculovirus infected sf9 cells using p53 residues 379-382 (RHKAcKAc) as substrate by fluorescence-based assay
- ChEMBL_2090135 (CHEMBL4771398) Inhibition of human HDAC6 expressed in baculovirus infected sf9 cells using p53 residues 379-382 (RHKKAc) as substrate by fluorescence-based assay
- ChEMBL_2090183 (CHEMBL4771446) Inhibition of HDAC1 (unknown origin) using p53 (379 to 382 residues) (Arg-His-Lys(Ac)-Lys(Ac)) as substrate by fluorimetric assay
- ChEMBL_2090191 (CHEMBL4771454) Inhibition of HDAC6 (unknown origin) using p53 (379 to 382 residues) (Arg-His-Lys(Ac)-Lys(Ac)) as substrate by fluorimetric assay
- ChEMBL_2090259 (CHEMBL4771522) Inhibition of human HDAC8 using p53 residues 379-382 (RHKAcKAc) as substrate as substrate measured after 2 hrs by Cheng-Prusoff analysis
- ChEMBL_2093851 (CHEMBL4775114) Inhibition of full length Flag-tagged ATM (unknown origin) using GST-p53(1 to 101 residues) as substrates incubated for 90 mins
- ChEMBL_2443964 Inhibition of human recombinant HDAC8 using fluorescent peptide p53 (379 to 382 residues) (RHK(Ac)K(Ac)) as substrate by fluorescence based analysis
- ChEMBL_2461916 Inhibition of human ATR/ATRIP complex using GST-cMyc-p53 substrate measured after 30 mins by HTRF based non-radioactive in vitro assay
- ChEMBL_1825984 (CHEMBL4325748) Binding affinity to MDMX (unknown origin) by using (FITC)-labeled p53 peptide based fluorescence polarization competitive assay
- ChEMBL_2311269 Inhibition of ATR (unknown origin) using GST-cMyc-p53 as substrate incubated for 30 mins in the presence of Mg/ATP mix by HTRF-based analysis
- ChEMBL_2427951 Inhibition of human full length recombinant ATM using GST-c-Myc-p53 as substrate measured after 30 mins in presence of Mg/ATP mix by ELISA
- ChEMBL_589609 (CHEMBL1052162) Induction of p53 in human HCT116 cells coexpressing pp53TA-luc assessed as inhibition of cell proliferation after 8 hrs by firefly/renilla luciferase reporter assay
- ChEMBL_1466806 (CHEMBL3405562) Binding affinity to GST-tagged MDM2 (unknown origin) assessed as inhibition of interaction with His-tagged p53 after 1 hr by HTRF assay
- ChEMBL_1991282 (CHEMBL4625017) Inhibition of His-tagged human SIRT6 expressed in Escherichia coli BL21(DE3) using Ac-p53 as substrate by fluorescence-based coupled-enzyme assay
- ChEMBL_2087781 (CHEMBL4769044) Inhibition of human recombinant HDAC1 expressed in baculovirus using fluorogenic peptide p53 residues 379-382 (RHKK(Ac)AMC) as substrate by Fluorescence analysis
- ChEMBL_2087782 (CHEMBL4769045) Inhibition of human recombinant HDAC2 expressed in baculovirus using fluorogenic peptide p53 residues 379-382 (RHKK(Ac)AMC) as substrate by Fluorescence analysis
- ChEMBL_2087783 (CHEMBL4769046) Inhibition of human recombinant HDAC6 expressed in baculovirus using fluorogenic peptide p53 residues 379-382 (RHKK(Ac)AMC) as substrate by Fluorescence analysis
- ChEMBL_2087784 (CHEMBL4769047) Inhibition of human recombinant HDAC8 expressed in baculovirus using fluorogenic peptide p53 residues 379-382 (RHKK(Ac)AMC) as substrate by Fluorescence analysis
- ChEMBL_2087785 (CHEMBL4769048) Inhibition of human recombinant HDAC3 expressed in baculovirus using fluorogenic peptide p53 residues 379-382 (RHKK(Ac)AMC) as substrate by Fluorescence analysis
- ChEMBL_2087786 (CHEMBL4769049) Inhibition of human recombinant HDAC4 expressed in baculovirus using fluorogenic peptide p53 residues 379-382 (RHKK(Ac)AMC) as substrate by Fluorescence analysis
- ChEMBL_2087787 (CHEMBL4769050) Inhibition of human recombinant HDAC5 expressed in baculovirus using fluorogenic peptide p53 residues 379-382 (RHKK(Ac)AMC) as substrate by Fluorescence analysis
- ChEMBL_2087788 (CHEMBL4769051) Inhibition of human recombinant HDAC7 expressed in baculovirus using fluorogenic peptide p53 residues 379-382 (RHKK(Ac)AMC) as substrate by Fluorescence analysis
- ChEMBL_2087789 (CHEMBL4769052) Inhibition of human recombinant HDAC9 expressed in baculovirus using fluorogenic peptide p53 residues 379-382 (RHKK(Ac)AMC) as substrate by Fluorescence analysis
- ChEMBL_2087790 (CHEMBL4769053) Inhibition of human recombinant HDAC10 expressed in baculovirus using fluorogenic peptide p53 residues 379-382 (RHKK(Ac)AMC) as substrate by Fluorescence analysis
- ChEMBL_2205937 (CHEMBL5118645) Inhibition of SMYD3 (unknown origin) using p53 (361 to 380 residues) and [3H]-SAM as substrate incubated for 90 mins by SPA assay
- ChEMBL_2358003 Inhibition of N-terminal biotin-his-tagged MDMX (24 to 108 residues)(unknown origin) binding to p53 incubated for 1 hr by Alphascreen assay
- ChEMBL_2538642 Inhibition of BRD4 in mouse primary skin-derived precursors (Nf1-/-, P53-/-) assessed as reduction in cell viability incubated for 72 hrs by SRB assay
- ChEMBL_972655 (CHEMBL2410635) Inhibition of HDAC8 (unknown origin) using fluorogenic peptide from p53 residues (379 to 382) (RHK(Ac)K(Ac)) as substrate by fluorescence assay
- ChEMBL_1467914 (CHEMBL3413118) Inhibition of human recombinant SIRT1 assessed as deacetylation of N-acetyl lysine residue in p53 (317 to 320) after 30 mins by luciferase-mediated luminescence assay
- ChEMBL_1467916 (CHEMBL3413120) Inhibition of human recombinant SIRT2 assessed as deacetylation of N-acetyl lysine residue in p53 (317 to 320) after 30 mins by luciferase-mediated luminescence assay
- ChEMBL_1806332 (CHEMBL4305691) Inhibition of recombinant human SIRT2 demyristoylase activity using p53(Myr)-AMC as substrate measured after 3 hrs in presence of NAD+ and trypsin by fluorescence assay
- ChEMBL_1899166 (CHEMBL4401281) Inhibition of SMYD2 (unknown origin) using [3H]-p53 (361 to 380 residues) as substrate after 1 hr in presence of [3H]-SAM by scintillation proximity assay
- ChEMBL_2214615 (CHEMBL5127747) Inhibition of full-length human DNA-PK expressed in baculovirus expression system using p53 as substrate incubated for 1 hr in presence of ATP by ELISA
- ChEMBL_2311268 Inhibition of DNA-PK (unknown origin) using GST-cMyc-p53 as substrate incubated for 30 mins in the presence of Mg/ATP mix by HTRF-based analysis
- ChEMBL_2427952 Inhibition of human full length recombinant DNA-PK using GST-c-Myc-p53 as substrate measured after 30 mins in presence of Mg/ATP mix by ELISA
- ChEBML_1553162 Inhibition of recombinant rat GST-tagged CK1delta expressed in Escherichia coli using GST-p53 (1 to 64 residues) as substrate by SDS-PAGE based autoradiography
- ChEMBL_1585034 (CHEMBL3820275) Binding affinity to MDM2 in human U87MG cells assessed as inhibition of MDM2/p53 protein interaction after 10 mins by quantitative sandwich immuno assay
- ChEMBL_1614263 (CHEMBL3856332) Inhibition of recombinant SIRT1 (unknown origin) assessed as deacetylation activity using acetylated p53 as substrate measured after 2 hrs by Fluor de Lys assay
- ChEMBL_1670058 (CHEMBL4019946) Inhibition of MDM2 in human U87MG cells assessed as reduction in MDM2 interaction with p53 after 10 mins by quantitative sandwich immune-enzymatic assay
- ChEMBL_1795643 (CHEMBL4267760) Inhibition of Cy5-labeled p53 derived TFSDLWKLL peptide binding to C-terminal biotin-labelled human MDM2 (2 to 188 residues) by TR-FRET assay
- ChEMBL_1754484 (CHEMBL4189244) Inhibition of rat recombinant GST-tagged CK1delta using GST-tagged mouse p53 (1 to 64 residues) as substrate in presence of radiolabelled-ATP by Cherenkov counting method
- ChEMBL_1754485 (CHEMBL4189245) Inhibition of human CK1delta transcription variant 1 using GST-tagged mouse p53 (1 to 64 residues) as substrate in presence of radiolabelled-ATP by Cherenkov counting method
- ChEMBL_1754486 (CHEMBL4189246) Inhibition of human CK1delta transcription variant 2 using GST-tagged mouse p53 (1 to 64 residues) as substrate in presence of radiolabelled-ATP by Cherenkov counting method
- ChEMBL_874346 (CHEMBL2182644) Activity at p53 in human SJSA1 cells assessed as induction of p21 mRNA expression after 7 hrs by qRT-PCR analysis in presence of 10% human serum
- ChEMBL_1539473 (CHEMBL3737093) Competition binding affinity to MDM2 (unknown origin) using p53 mimicking peptide TSFAEYWNLLSP after 30 mins by fluorescence polarization assay
- ChEMBL_1539474 (CHEMBL3737264) Competition binding affinity to MDMX (unknown origin) using p53 mimicking peptide TSFAEYWNLLSP after 30 mins by fluorescence polarization assay
- Time Resolved Fluorescence Energy Transfer (TR-FRET) Assay The inhibition of p53-MDM2 and p53-MDM4 interactions is measured by time resolved fluorescence energy transfer (TR-FRET). Fluorescence energy transfer (or Foerster resonance energy transfer) describes an energy transfer between donor and acceptor fluorescent molecules. For this assay, human MDM2 protein (amino acids 2-188) and human MDM4 protein (amino acids 2-185), tagged with a C-terminal biotin moiety, are used in combination with a Europium labeled streptavidin (Perkin Elmer, Inc., Waltham, MA, USA) serving as the donor fluorophore. The p53 derived, Cy5 labeled peptide Cy5-TFSDLWKLL (p53 aa18-26) is the energy acceptor. Upon excitation of the donor molecule at 340nm, binding interaction between MDM2 or MDM4 and the p53 peptide induces energy transfer and enhanced response at the acceptor emission wavelength at 665 nm.
- Time Resolved Fluorescence Energy Transfer (TR-FRET) Assay The inhibition of p53-MDM2 and p53-MDM4 interactions is measured by time resolved fluorescence energy transfer (TR-FRET). Fluorescence energy transfer (or Foerster resonance energy transfer) describes an energy transfer between donor and acceptor fluorescent molecules. For this assay, human MDM2 protein (amino acids 2-188) and human MDM4 protein (amino acids 2-185), tagged with a C-terminal biotin moiety, are used in combination with a Europium labeled streptavidin (Perkin Elmer, Inc., Waltham, Mass., USA) serving as the donor fluorophore. The p53 derived, Cy5 labeled peptide Cy5-TFSDLWKLL (p53 aa18-26) is the energy acceptor. Upon excitation of the donor molecule at 340 nm, binding interaction between MDM2 or MDM4 and the p53 peptide induces energy transfer and enhanced response at the acceptor emission wavelength at 665 nm.
- ChEMBL_1296786 (CHEMBL3132469) Binding affinity to human GST-thrombin-tagged MDM2 assessed as inhibition of interaction with human p53 after 1 hr by HTRF assay in presence of 15% human serum
- ChEMBL_1458455 (CHEMBL3370341) Binding affinity to human GST-thrombin-tagged MDM2 assessed as inhibition of interaction with human p53 after 1 hr by HTRF assay in presence of 15% human serum
- ChEMBL_1569637 (CHEMBL3790863) Inhibition of doxorubicin-stimulated p53 (unknown origin) expressed in human RKO cells preincubated for 24 hrs followed by addition of doxorubicin for 16 hrs by luciferase reporter assay
- ChEMBL_1932514 (CHEMBL4478166) Inhibition of ATM (unknown origin) using p53 as substrate incubated for 30 mins followed by substrate addition measured after 2 hrs in presence of ATP by HTRF method
- ChEMBL_2214588 (CHEMBL5127720) Inhibition of full-length human recombinant FLAG-tagged ATR/c-Myc-tagged ATRIP using p53 as substrate incubated for 30 mins in presence of ATP by HTRF assay
- ChEMBL_1336790 (CHEMBL3241329) Binding affinity to MDM2 in human HCT116 cells expressing p53 assessed as upregulation of p21 mRNA expression after 7 hrs by quantitative RT-PCR analysis
- ChEMBL_1668047 (CHEMBL4017935) Inhibition of biotin-labelled L-p53 binding to L-MDM2 (25 to 109 residues) (unknown origin) by FAM labeled P4 peptide based fluorescence polarization assay
- ChEMBL_1668053 (CHEMBL4017941) Inhibition of biotin-labelled D-p53 binding to D-MDM2 (25 to 109 residues) (unknown origin) by FAM labeled P4 peptide based fluorescence polarization assay
- ChEMBL_1670059 (CHEMBL4019947) Inhibition of MDM4 in human SH-SY5Y cells assessed as reduction in MDM4 interaction with p53 after 10 mins by quantitative sandwich immune-enzymatic assay
- ChEMBL_1738061 (CHEMBL4153811) Inhibition of ATM (unknown origin) using p53 as substrate preincubated for 30 mins followed by substrate addition and measured after 2 hrs by HTRF assay
- ChEMBL_1765222 (CHEMBL4200469) Inhibition of ATM (unknown origin) using p53 as substrate pretreated for 30 mins followed by substrate addition and measured after 2 hrs by HTRF assay
- ChEMBL_1778144 (CHEMBL4235136) Induction of p53 transcriptional activity (unknown origin) expressed in HEK293 cells coexpressing TK-driven Rluc after 20 to 22 hrs by luciferase reporter gene assay
- ChEMBL_1795364 (CHEMBL4267481) Inhibition of FAM-tagged p53-based fluorescent probe binding to human His-tagged MDM2 (1 to 118 residues) after 15 mins by fluorescence polarization assay
- ChEMBL_2090184 (CHEMBL4771447) Inhibition of human HDAC2 expressed in Sf9 cells using p53 (379 to 382 residues) (Arg-His-Lys(Ac)-Lys(Ac)) as substrate by fluorimetric assay
- ChEMBL_2090240 (CHEMBL4771503) Inhibition of HDAC1 (unknown origin) using p53 (379 to 382 residues) (Arg-His-Lys-Lys(Ac)) as substrate incubated for 30 mins by fluorescence method
- ChEMBL_2090241 (CHEMBL4771504) Inhibition of HDAC2 (unknown origin) using p53 (379 to 382 residues) (Arg-His-Lys-Lys(Ac)) as substrate incubated for 30 mins by fluorescence method
- ChEMBL_2090245 (CHEMBL4771508) Inhibition of HDAC10 (unknown origin) using p53 (379 to 382 residues) (Arg-His-Lys-Lys(Ac)) as substrate incubated for 30 mins by fluorescence method
- ChEMBL_2090246 (CHEMBL4771509) Inhibition of HDAC11 (unknown origin) using p53 (379 to 382 residues) (Arg-His-Lys-Lys(Ac)) as substrate incubated for 30 mins by fluorescence method
- ChEMBL_2163026 (CHEMBL5047887) Inhibition of HDAC1 (unknown origin) using p53 (379 to 382 residues) (RHKK(Ac-AMC) as fluorogenic substrate incubated for 1 hrs by fluorescence based assay
- ChEMBL_2163090 (CHEMBL5047951) Inhibition of HDAC8 (unknown origin) using p53 (379 to 382 residues) (RHKK(Ac-AMC) as fluorogenic substrate incubated for 2 hrs by fluorescence based assay
- ChEMBL_2205939 (CHEMBL5118647) Inhibition of SMYD2 (unknown origin) using [H3]-SAM and p53 (361 to 380 residues) as peptide substrate incubated for 1 hr by scintillation proximity assay
- ChEMBL_305973 (CHEMBL832959) Antagonistic concentration for accessory gene regulator C1 of Staphylococcus aureus
- ChEMBL_2114766 (CHEMBL4823707) Inhibition of DNA-PK isolated from human HeLa nuclear extract using full length His-tagged p53 as substrate measured after 75 mins in presence of ATP by HTRF assay
- ChEMBL_2214583 (CHEMBL5127715) Inhibition of full-length human recombinant FLAG-tagged ATR/c-Myc-tagged ATRIP using p53 as substrate incubated for 30 mins in presence of ATP by fluorescence based assay
- ChEBML_1661768 Inhibition of FAM-tagged p53-based PMDM6-F peptide binding to human recombinant His-tagged MDM2 (1 to 118 residues) after 30 mins by fluorescence polarization assay
- ChEMBL_1795366 (CHEMBL4267483) Inhibition of p53 protein binding to recombinant human GST-tagged MDM2 (1 to 118 residues) expressed in Escherichia coli BL21 (DE3) after 30 mins by ELISA
- ChEMBL_1798009 (CHEMBL4270126) Inhibition of human recombinant His-tagged full-length Sirt1 expressed in Escherichia coli BL21 DE3 Rosetta2 using acetyl-p53-peptide as substrate by coupled deacetylation assay
- ChEMBL_1902914 (CHEMBL4405136) Inhibition of HDAC10 (unknown origin) using fluorogenic peptide p53 residues 379-382 (RHKK(Ac)AMC) as substrate measured after 1 to 2 hrs by fluorescence assay
- ChEMBL_2090242 (CHEMBL4771505) Inhibition of HDAC3/NCoR2 (unknown origin) using p53 (379 to 382 residues) (Arg-His-Lys-Lys(Ac)) as substrate incubated for 30 mins by fluorescence method
- ChEMBL_2021475 (CHEMBL4675288) Competitive inhibition of human CSE using varying levels of L-Cys as substrate and fixed PLP levels
- ChEMBL_1339586 (CHEMBL3243066) Binding affinity to MDM2 in human HCT116 cells expressing p53 assessed as upregulation of p21 mRNA expression after 7 hrs by qRT-PCR analysis in presence of 10% human serum
- ChEMBL_1661768 (CHEMBL4011380) Inhibition of FAM-tagged p53-based PMDM6-F peptide binding to human recombinant His-tagged MDM2 (1 to 118 residues) after 30 mins by fluorescence polarization assay
- ChEMBL_1983108 (CHEMBL4616370) Binding affinity to N-terminal 6xHis-tagged MDM2 (unknown origin) (1 to 126 residues) assessed as inhibition of MDM2/p53 protein-protein interaction by fluorescence anisotrophy assay
- ChEMBL_2214586 (CHEMBL5127718) Inhibition of human FLAG-tagged ATR/c-Myc-tagged ATRIP expressed in mammalian expression system using p53 as substrate incubated for 2 hrs by fluorescence based assay
- ChEMBL_933815 (CHEMBL2321369) Inhibition of human His-tagged p53 (1 to 132 amino acid residues)/GST-tagged MDM2 L33E mutant (25 to 108 amino acid residues) interaction by HTRF assay
- ChEMBL_304175 (CHEMBL829171) Effective concentration to antagonise accessory gene regulator C2 of Staphylococcus aureus
- ChEMBL_304176 (CHEMBL829172) Effective concentration to antagonise accessory gene regulator C3 of Staphylococcus aureus
- ChEMBL_304177 (CHEMBL829173) Effective concentration to antagonise accessory gene regulator C4 of Staphylococcus aureus
- ChEMBL_305974 (CHEMBL832960) Concentration required to antagonise accessory gene regulator C2 of Staphylococcus aureus
- ChEMBL_305975 (CHEMBL832961) Concentration required to antagonise accessory gene regulator C3 of Staphylococcus aureus
- ChEMBL_305976 (CHEMBL832141) Concentration required to antagonise accessory gene regulator C4 of Staphylococcus aureus
- ChEMBL_99545 (CHEMBL707778) Concentration at which one-half of the maximum Low density lipoprotein receptor is upregulated in HepG2 cells
- ChEMBL_1561479 (CHEMBL3779616) Inhibition of p53-CBP bromodomain interaction in human U2OS cells assessed as inhibition of p21 activation incubated overnight followed by doxorubicin addition measured after 24 hrs by luciferase reporter gene assay
- ChEMBL_1614254 (CHEMBL3856323) Inhibition of recombinant human SIRT1 (193 to 747 residues) expressed in Escherichia coli BL21 (DE3) using AMC-labeled p53-derived acetylated lysine as substrate in presence of NAD+ by HTS assay
- ChEMBL_1823456 (CHEMBL4323220) Binding affinity to cereblon in human RV4:11 cells expressing wild type p53 assessed as cell growth inhibition measured after 4 days
- ChEMBL_1825972 (CHEMBL4325736) Binding affinity to human MDM2 expressed in Escherichia coli expression system by using (FITC)-labeled p53 peptide based fluorescence polarization competitive assay
- ChEMBL_87376 (CHEMBL694305) Inhibition of histidine protein kinase (KinA) phosphorylation in the presence of response regulator (Spo0F)
- ChEMBL_1695931 (CHEMBL4046821) Inhibition of DNA-PK in human HeLa nuclear extracts incubated for 40 mins using calf thymus DNA and fluoroscein-p53[Ser15]-peptide by Lanthascreen time resolved fluorescence assay
- ChEMBL_1806362 (CHEMBL4305721) Inhibition of recombinant human SIRT2 deacylation activity using fluorogenic p53 (Arg-His-Lys-Lys(Ac)) (379 to 382 residues) as substrate measured after 1 hr by fluorescence assay
- ChEMBL_2090193 (CHEMBL4771456) Inhibition of recombinant human HDAC11 expressed in baculovirus infected sf9 cells using p53 (379 to 382 residues) (Arg-His-Lys(Ac)-Lys(Ac)) as substrate by fluorimetric assay
- ChEMBL_2488455 Inhibition of HDAC6 (unknown origin) extracted from baculovirus infected Sf9 insect cells using fluorescent peptide p53 (379 to 382 residues) RHKK(Ac)AMC as substrate by fluorescence based analysis
- ChEMBL_1469499 (CHEMBL3412296) Binding affinity to human GST-thrombin-tagged MDM2 ( 1 to 188 aa) assessed as inhibition of interaction with human p53 preincubated with compound for 20 mins by HTRF assay
- ChEMBL_850546 (CHEMBL2157768) Antagonist activity at MDM2 (residues 25 to 109) assessed as inhibition of binding to immobilized p53 (residues 15 to 29) preincubated for 30 mins by surface plasmon resonance method
- ChEMBL_2355399 Inhibition of HDAC1 (unknown origin) using fluorogenic peptide as the substrate containing residues 379-382 of p53 pre-incubated for 15 mins followed by substrate addition incubated for 30 mins by multimode plate reader
- ChEMBL_1823460 (CHEMBL4323224) Binding affinity to cereblon in human RV4:11 cells expressing wild type p53 assessed as reduction in cell growth inhibition measured after 4 days in presence of 10 uM lenalidomide
- ChEMBL_1823461 (CHEMBL4323225) Binding affinity to cereblon in human RV4:11 cells expressing wild type p53 assessed as reduction in cell growth inhibition measured after 4 days in presence of 30 uM lenalidomide
- ChEMBL_1823462 (CHEMBL4323226) Binding affinity to cereblon in human RV4:11 cells expressing wild type p53 assessed as reduction in cell growth inhibition measured after 4 days in presence of 100 uM lenalidomide
- ChEMBL_2033595 (CHEMBL4687753) Inhibition of FAM tagged p53-based peptide binding to recombinant human His-tagged MDM2 protein (residues 1 to 118 residues) incubated for 15 mins by fluorescence-polarization-based binding assay
- ChEMBL_2090190 (CHEMBL4771453) Inhibition of recombinant human C-terminal His-tagged HDAC8 expressed in baculovirus infected sf9 cells using p53 (379 to 382 residues) (Arg-His-Lys(Ac)-Lys(Ac)) by fluorimetric assay
- ChEMBL_2239666 (CHEMBL5153562) Inhibition of recombinant human HDAC1 using p53 (Ac-RHKK(Acetyl)-AMC) as substrate preincubated for 5 mins followed by substrate addition and measured after 90 mins by fluorescence based analysis
- ChEMBL_2239667 (CHEMBL5153563) Inhibition of recombinant human HDAC2 using p53 (Ac-RHKK(Acetyl)-AMC) as substrate preincubated for 5 mins followed by substrate addition and measured after 30 mins by fluorescence based analysis
- ChEMBL_2239668 (CHEMBL5153564) Inhibition of recombinant human HDAC3 using p53 (Ac-RHKK(Acetyl)-AMC) as substrate preincubated for 5 mins followed by substrate addition and measured after 30 mins by fluorescence based analysis
- ChEMBL_2367190 Inhibition of MDM2 (1 to 118 residues)/p53 (unknown origin) interaction expressed in Escherichia coli BL21(DE3) cells assessed as inhibition constant incubated for 1.5 hrs by competitive fluorescence polarization assay
- ChEMBL_2367191 Inhibition of MDM4 (14 to 111 residues)/p53 (unknown origin) interaction expressed in Escherichia coli BL21(DE3) cells assessed as inhibition constant incubated for 1.5 hrs by competitive fluorescence polarization assay
- ChEMBL_2486403 Inhibition of purified recombinant human HDAC2 assessed as inhibition constant using residues 379-382 (RHKK(Ac)AMC) fluorogenic substrate p53 fluorogenic peptide as substrate measured after 90 mins by fluorescence assay
- ChEMBL_2486405 Inhibition of purified recombinant human HDAC6 assessed as inhibition constant using residues 379-382 (RHKK(Ac)AMC) fluorogenic substrate p53 fluorogenic peptide as substrate measured after 90 mins by fluorescence assay
- ChEMBL_2486406 Inhibition of purified recombinant human HDAC8 assessed as inhibition constant using residues 379-382 (RHKK(Ac)AMC) fluorogenic substrate p53 fluorogenic peptide as substrate measured after 90 mins by fluorescence assay
- ChEMBL_2486407 Inhibition of purified recombinant human HDAC10 assessed as inhibition constant using residues 379-382 (RHKK(Ac)AMC) fluorogenic substrate p53 fluorogenic peptide as substrate measured after 90 mins by fluorescence assay
- ChEMBL_1445462 (CHEMBL3380268) Binding affinity to human His-tagged MDM2 (1 to 118 residues) using FAM tagged p53-based peptide by fluorescence prolarization based protein binding assay
- ChEMBL_1504425 (CHEMBL3592990) Inhibition of recombinant full-length human SMYD2 (1 to 433) expressed in Escherichia coli BL21Star(DE3) cells using p53 peptide as substrate after 90 mins by scintillation proximity assay in presence of [3H]-SAM
- ChEMBL_1806350 (CHEMBL4305709) Inhibition of recombinant human SIRT3 (118 to 399 residues) demyristoylase activity expressed in Escherichia coli using p53(Myr)-AMC as substrate measured after 3 hrs in presence of NAD+ and trypsin by fluorescence assay
- ChEMBL_2059819 (CHEMBL4714820) Inhibition of p53(Myr)-AMC binding to SIRT2 (unknown origin) incubated for 3 hrs in presence of NAD+ followed by trypsin addition and measured at 5 mins interval for 20 mins by fluorometric method
- ChEMBL_1294426 (CHEMBL3129252) Inhibition of NTRK3 (unknown origin) using Km levels of ATP
- ChEMBL_1294427 (CHEMBL3129253) Inhibition of NTRK1 (unknown origin) using Km levels of ATP
- ChEMBL_1294428 (CHEMBL3129254) Inhibition of PTK2 (unknown origin) using Km levels of ATP
- ChEMBL_1294429 (CHEMBL3129255) Inhibition of TNK2 (unknown origin) using Km levels of ATP
- ChEMBL_1294430 (CHEMBL3129256) Inhibition of PTK2B (unknown origin) using Km levels of ATP
- ChEMBL_1294431 (CHEMBL3129257) Inhibition of JAK2 (unknown origin) using Km levels of ATP
- ChEMBL_1294432 (CHEMBL3129258) Inhibition of FES (unknown origin) using Km levels of ATP
- ChEMBL_1294433 (CHEMBL3129259) Inhibition of NTRK2 (unknown origin) using Km levels of ATP
- ChEMBL_1294434 (CHEMBL3129260) Inhibition of LTK (unknown origin) using Km levels of ATP
- ChEMBL_1294435 (CHEMBL3129261) Inhibition of FER (unknown origin) using Km levels of ATP
- ChEMBL_812462 (CHEMBL2014248) Competitive inhibition of Chk1 in presence of higher ATP levels
- Time Resolved Fluorescence Energy Transfer (TR-FRET) Assay Assay 1: This assay was used to evaluate compounds displaying inhibition of p53-MDM2 interaction and p53-MDM4 interaction at IC50s of 0.005 to 50 μM (p53-MDM2 Assay 1 and p53-MDM4 Assay 1, respectively). The test is performed in white 384-well plates (Greiner Bio-One, reference 781207) in a total volume of 60 μL by adding 1 μL of compounds tested at different concentrations diluted in 100% DMSO (1.7% final DMSO concentration) in reaction buffer (PBS, 125 mM NaCl, 0.001% Novexin (consists of carbohydrate polymers), designed to increase the solubility and stability of proteins; Expedeon Ltd., Cambridgeshire, United Kingdom), 0.01% Gelatin, 0.01% 0.2%, Pluronic F-127 (block copolymer from ethylenoxide and propyleneoxide), 1 mM DTT). After addition of 1.25 nM MDM2-biotinylated or 2.5 nM MDM4-biotinylated (internal preparations, both MDM2 and MDM4 are biotinylated at the C-terminal of the peptide construct), and 0.625 nM Europium labeled streptavidin (Perkin Elmer), the solution is pre-incubated for 15 minutes at room temperature, then 10 nM Cy5-p53 peptide (internal preparation, the Cy5 dye is directly bound to the N-terminal part of p53 peptide construct) is added before an incubation at room temperature for 15 minutes prior to reading the plate. For measurement of samples, a Victor II microplate reader (Perkin Elmer) is used with the following settings: Excitation 340 nm, Emission Donor 620 nm and Emission Acceptor 665 nm.
- ChEMBL_2019740 (CHEMBL4673553) Inhibition of C-terminal His/FLAG-tagged human HDAC1 expressed in Sf9 insect cells using acetylated P53(379-382 residues) (RHKK(Ac)) as substrate incubated for 30 mins by fluorescence method
- ChEMBL_1469730 (CHEMBL3413418) Binding affinity to human GST-thrombin-tagged MDM2 ( 1 to 188 aa) assessed as inhibition of interaction with human p53 preincubated with compound for 20 mins by HTRF assay in presence of 15% human serum
- ChEMBL_1750376 (CHEMBL4185136) Binding affinity to MDM2 (unknown origin) in assessed as reduction in MDM2 binding to Fl-p53-TAD after 30 mins by fluorescence polarization competition assay
- ChEMBL_1440407 (CHEMBL3386220) Inhibition of GST-tagged mouse CK1delta transcription variant 1 mutant expressed in Escherichia coli at 15 degC for 14 hrs using GST-p53 (1 to 64 residues) by SDS-PAGE based autoradiography
- ChEMBL_1440408 (CHEMBL3386221) Inhibition of GST-tagged mouse CK1delta transcription variant 1 mutant expressed in Escherichia coli at 37 degC for 2 hrs using GST-p53 (1 to 64 residues) by SDS-PAGE based autoradiography
- ChEMBL_1440409 (CHEMBL3386222) Inhibition of GST-tagged mouse CK1delta transcription variant 2 mutant expressed in Escherichia coli at 15 degC for 14 hrs using GST-p53 (1 to 64 residues) by SDS-PAGE based autoradiography
- ChEMBL_1440410 (CHEMBL3386223) Inhibition of GST-tagged mouse CK1delta transcription variant 2 mutant expressed in Escherichia coli at 37 degC for 2 hrs using GST-p53 (1 to 64 residues) by SDS-PAGE based autoradiography
- ChEMBL_861196 (CHEMBL2173264) Competitive binding affinity to full length human SMYD2 amino acid 1 to 433 expressed in Escherichia coli BL21 (DE3) after 90 mins by radioactive filter-binding assay in presence of P53 peptide
- Binding Assay The binding affinity of the MDM2 inhibitors disclosed herein was determined using a fluorescence polarization-based (FP-based) binding assay using a recombinant human His-tagged MDM2 protein (residues 1-118) and a fluorescently tagged p53-based peptide. The design of the fluorescence probe was based upon a previously reported high-affinity p53-based peptidomimetic compound called PMDM6-F (García-Echeverria et al., J. Med. Chem. 43: 3205-3208 (2000)).
- ChEMBL_457331 (CHEMBL941864) Inhibition of Plasmodium falciparum FabI in presence of variable NADH levels
- ChEMBL_842366 (CHEMBL2091983) Activation of human TRPA1 assessed as stimulation of intracellular Ca2+ levels
- ChEMBL_1738289 (CHEMBL4154039) Binding affinity to N-terminal His6-tagged p53 DNA binding domain Y220C mutant (94 to 312 residues) (unknown origin) expressed in Escherichia coli C41 by ITC
- ChEMBL_1823457 (CHEMBL4323221) Binding affinity to cereblon in human RV4:11/IRMI-2 cells harboring p53 hotspot mutation Y236H/R249G assessed as cell growth inhibition measured after 4 days
- ChEMBL_967970 (CHEMBL2401074) Binding affinity to recombinant human His-tagged MDM2 (1 to 118 amino acids) using p53-based PMDM6-F as probe after 15 mins by competition assay
- ChEMBL_863264 (CHEMBL2176207) Inhibition of human recombinant SIRT2 expressed in Escherichia coli cells using acetylated Lys side chain amino acids 379-382 (Arg-His-Lys-Lys(Ac)) p53 conjugated with aminomethylcoumarin as substrate by fluorescence assay
- ChEMBL_863265 (CHEMBL2176208) Inhibition of human recombinant SIRT1 expressed in Escherichia coli cells using acetylated Lys side chain amino acids 379-382 (Arg-His-Lys-Lys(Ac)) p53 conjugated with aminomethylcoumarin as substrate by fluorescence assay
- FRET Assay Methods: An HDM2 FRET assay was developed to assess the compounds' inhibitory activity towards binding of p53 protein. A truncated version of HDM2 with residues 17 to 125 (containing p53 binding surface, Science (1994) 265, 346-355), with N-terminal His and Thioredoxin tag was generated in pET32a expression vector and expressed in E. coli strain BL21(DE3)Rosetta. Protein was purified using Ni-affinity chromatography, followed by size exclusion chromatography using Superdex 75 26/60 column. To assess inhibition of p53 binding to HDM2, a FITC labeled 8-mer peptide (SEQ ID NO:1: Ac-Phe-Arg-Dpr-Ac6c-(6-Br)Trp-Glu-Glu-Leu-NH2; Anal Biochem. 2004 Aug. 1; 331(1):138-46) with strong affinity towards the p53 binding pocket of HDM2 was used. The HDM2 assay buffer contained 1x Phosphate Buffered Saline (Invitrogen, Cat#14190), 0.01% BSA (Jackson ImmunoResearch, Cat#001-000-162), 0.01% Tween-20.
- ChEMBL_304243 (CHEMBL841798) Effective concentration to antagonise accessory gene regulator C1 of Staphylococcus aureus; range = 9-11 uM
- ChEMBL_1738290 (CHEMBL4154040) Binding affinity to 15N-labeled T-p53 DNA binding domain Y220C mutant (94 to 312 residues) (unknown origin) expressed in Escherichia coli C41 by HSQC-NMR analysis
- ChEMBL_1803480 (CHEMBL4275772) Non-competitive inhibition of rat liver ACLY using varying levels of citrate
- ChEMBL_457861 (CHEMBL925192) Inhibition of gamma-secretase assessed as reduction of amyloid-beta 42 levels
- ChEMBL_1806346 (CHEMBL4305705) Inhibition of recombinant human C-terminal His6-tagged SIRT1 (Met1 to Ser747 residues) demyristoylase activity expressed in Escherichia coli using p53(Myr)-AMC as substrate measured after 3 hrs in presence of NAD+ and trypsin by fluorescence assay
- ChEMBL_1823463 (CHEMBL4323227) Binding affinity to cereblon in human RV4:11/IRMI-2 cells harboring p53 hotspot mutation Y236H/R249G assessed as reduction in cell growth inhibition measured after 4 days in presence of 10 uM lenalidomide
- ChEMBL_1823464 (CHEMBL4323228) Binding affinity to cereblon in human RV4:11/IRMI-2 cells harboring p53 hotspot mutation Y236H/R249G assessed as reduction in cell growth inhibition measured after 4 days in presence of 30 uM lenalidomide
- ChEMBL_1823465 (CHEMBL4323229) Binding affinity to cereblon in human RV4:11/IRMI-2 cells harboring p53 hotspot mutation Y236H/R249G assessed as reduction in cell growth inhibition measured after 4 days in presence of 100 uM lenalidomide
- Mdm2-p53 Inhibition AlphaScreen Assay This assay is used to determine whether the compounds inhibit the p53-MDM2 interaction and thus restore p53 function.15 μL of compound in 20% DMSO (serial pre-dilutions of compound are done in 100% DMSO) is pipetted to the wells of a white OptiPlate-96 (PerkinElmer). A mix consisting of 20 nM GST-MDM2 protein (aa 23-117) and 20 nM biotinylated p53 wt peptide (encompassing aa 16-27 of wt human p53, amino acid sequence QETFSDLWKLLP-Ttds-Lys-Biotin, molecular weight 2132.56 g/mol) is prepared in assay buffer (50 mM Tris/HCl pH 7.2; 120 mM NaCl; 0.1% bovine serum albumin (BSA); 5 mM dithiothreitol (DTT); 1 mM ethylenediaminetetraacetic acid (EDTA); 0.01% Tween 20). 30 μL of the mix is added to the compound dilutions and incubated for 15 min at rt while gently shaking the plate at 300 rounds per minute (rpm). Subsequently, 15 μL of premixed AlphaLISA Glutathione Acceptor Beads and AlphaScreen Streptavidin Donor Beads from PerkinElmer (in assay buffer at a concentration of 10 μg/mL each) are added and the samples are incubated for 30 min at rt in the dark (shaking 300 rpm). Afterwards, the signal is measured in a PerkinElmer Envision HTS Multilabel Reader using the AlphaScreen protocol from PerkinElmer.Each plate contains negative controls where biotinylated p53-peptide and GST-MDM2 are left out and replaced by assay buffer. Negative control values are entered as low basis value when using the software GraphPad Prism for calculations.
- Homogenous Time-Resolved Fluorescence Assay (HTRF) The standard assay conditions for the in vitro HTRF assay consisted of a 50 ul total reaction volume in black 384-well Costar polypropylene plates in 1PBS buffer pH 7.4, 1 mM DTT, 0.1% BSA, 2.5 nM GST-hMDM2 (aa 1-188), 5 nM biotinylated-p53 (aa 1-83), 1.8 nM SA-XLent (Cisbio; Bedford, Mass.), 0.6 nM anti-GST cryptate monoclonal antibody (Cisbio; Bedford, Mass.) and 200 mM KF. Amino acid residues 1-188 of human MDM2 were expressed as an amino-terminal glutathione S-transferase (GST) fusion protein (GST-hMDM2) in Escherichia coli. Residues 1-83 of human p53 were expressed as an amino-terminal AviTag-TrxA-6His fusion protein (biotinylated p53) in E. coli. Each protein was purified from cell paste by affinity chromatography.Specifically, 10 uL of GST-hMDM2 was incubated with 10 ul of diluted compound (various concentrations, serially diluted) in 10% DMSO for 20 minutes at room temperature. 20 uL of biotinylated-p53 was added to the GST-hMDM2+compound mixture.
- ChEBML_44471 Kd (half effective concentration) of compound against CFTR (cystic fibrosis transmembrane conductance regulator) in rat was determined
- ChEMBL_2205936 (CHEMBL5118644) Inhibition of full length human SMYD2 (1 to 433 residues) expressed in Escherichia coli strain BL21 (DE3) using p53 (361 to 380 residues) and [3H]-SAM as substrate incubated for 90 mins by SPA assay
- ChEMBL_1801960 (CHEMBL4274252) Competitive inhibition of recombinant human AChE using varying levels of acetylthiocholine as substrate
- ChEMBL_1803477 (CHEMBL4275769) Competitive inhibition of human liver ACLY using varying levels of citrate as substrate
- ChEMBL_1874852 (CHEMBL4376141) Inhibition of recombinant human SMYD2 (1 to 433 residues) expressed in baculovirus infected Sf9 insect cells using biotinylated-p53 peptide (361 to 380 residues) as substrate measured after 50 mins in presence of 70 nM [3H]-SAM by scintillation proximity assay
- ChEMBL_1874854 (CHEMBL4376143) Inhibition of recombinant human SMYD2 (1 to 433 residues) expressed in baculovirus infected Sf9 insect cells using biotinylated-p53 peptide (361 to 380 residues) as substrate measured after 120 mins in presence of 20 uM [3H]-SAM by scintillation proximity assay
- ChEMBL_967971 (CHEMBL2401075) Binding affinity to recombinant human His-tagged MDM2 (1 to 118 amino acids) using p53-based PMDM6-F as probe after 15 to 30 mins by fluorescence polarization assay
- ChEMBL_2090192 (CHEMBL4771455) Inhibition of recombinant human N-terminal FLAG-tagged HDAC10 (2 to 631 residues) expressed in baculovirus infected sf9 cells using p53 (379 to 382 residues) (Arg-His-Lys(Ac)-Lys(Ac)) as substrate by fluorimetric assay
- ChEMBL_2090223 (CHEMBL4771486) Inhibition of human full length recombinant HDAC1 using p53 (379 to 382 residues) (RHKK(Ac)AMC) as substrate preincubated for 5 to 10 mins followed by substrate addition and measured after 2 hrs by fluorescence method
- ChEMBL_2090224 (CHEMBL4771487) Inhibition of human full length recombinant HDAC2 using p53 (379 to 382 residues) (RHKK(Ac)AMC) as substrate preincubated for 5 to 10 mins followed by substrate addition and measured after 2 hrs by fluorescence method
- ChEMBL_2090231 (CHEMBL4771494) Inhibition of human full length recombinant HDAC10 using p53 (379 to 382 residues) (RHKK(Ac)AMC) as substrate preincubated for 5 to 10 mins followed by substrate addition and measured after 2 hrs by fluorescence method
- ChEMBL_2090232 (CHEMBL4771495) Inhibition of human full length recombinant HDAC11 using p53 (379 to 382 residues) (RHKK(Ac)AMC) as substrate preincubated for 5 to 10 mins followed by substrate addition and measured after 2 hrs by fluorescence method
- ChEMBL_2090233 (CHEMBL4771496) Inhibition of human full length recombinant HDAC6 using p53 (379 to 382 residues) (RHKK(Ac)AMC) as substrate preincubated for 5 to 10 mins followed by substrate addition and measured after 2 hrs by fluorescence method
- ChEMBL_1866658 (CHEMBL4367633) Selective estrogen receptor down regulator activity at ERalpha (unknown origin)
- ChEMBL_1801961 (CHEMBL4274253) Non-competitive inhibition of recombinant human AChE using varying levels of acetylthiocholine as substrate
- ChEMBL_547031 (CHEMBL1036029) Activity of rat recombinant DNA polymerase beta using variable levels of DNA template-primer
- ChEMBL_455074 (CHEMBL887103) Inhibition of p38-alpha in THP1 cells assessed as TNFalpha levels
- Homogenous Time-Resolved Fluorescence Assay The standard assay conditions for the in vitro HTRF assay consisted of a 50 ul total reaction volume in black 384-well Costar polypropylene plates in 1×PBS buffer pH 7.4, 1 mM DTT, 0.1% BSA, 2.5 nM GST-hMDM2 (aa 1-188), 5 nM biotinylated-p53 (aa 1-83), 1.8 nM SA-XLent (Cisbio; Bedford, Mass.), 0.6 nM anti-GST cryptate monoclonal antibody (Cisbio; Bedford, Mass.) and 200 mM KF. Amino acid residues 1-188 of human MDM2 were expressed as an amino-terminal glutathione-S-transferase (GST) fusion protein (GST-hMDM2) in Escherichia coli. Residues 1-83 of human p53 were expressed as an amino-terminal AviTag -TrxA-6×His fusion protein (biotinylated p53) in E. coli. Each protein was purified from cell paste by affinity chromatography.Specifically, 10 uL of GST-hMDM2 was incubated with 10 ul of diluted compound (various concentrations, serially diluted) in 10% DMSO for 20 minutes at room temperature. 20 uL of biotinylated-p53 was added to the GST-hMDM2+compound mixture, and then incubated at room temperature for 60 min. 10 uL of detection buffer consisting of SA-XLent, anti-GST cryptate antibody and KF was added to GST-hMDM2, biotinylated-p53 and compound reaction and left at room temperature to reach equilibrium for >4 hrs. The final concentration of DMSO in the reaction was 2%. Time-resolved fluorescence readings were measured on a microplate multilabel reader. Percentage of inhibition was calculated relative to nutlin-3.As the potencies of the HDM2 inhibitors increased, an improved HTRF assay (HTRF2 assay) was developed. All assay conditions remained the same as described above, with the exception of the following changes in reagent concentrations: 0.2 nM GST-hMDM2 (1-188), 0.5 nM biotinylated-p53 (1-83), 0.18 nM SA-XLent, and 100 mM KF.
- ChEMBL_1852978 (CHEMBL4353602) Inhibition of human N-terminal His-tagged SIRT1 expressed in Escherichia coli using p53 (Arg-His-Lys-Lys(Ac)) (379 to 382 residues) as substrate incubated for 3 hrs measured up to 30 mins by fluorescence assay
- ChEMBL_1852980 (CHEMBL4353604) Inhibition of human N-terminal His-tagged SIRT2 expressed in Escherichia coli using p53 (Gln-Pro-Lys-Lys(Ac)) (317 to 320 residues) as substrate incubated for 3 hrs measured up to 30 mins by fluorescence assay
- ChEMBL_2090225 (CHEMBL4771488) Inhibition of human full length recombinant HDAC3/ NcoR2 using p53 (379 to 382 residues) (RHKK(Ac)AMC) as substrate preincubated for 5 to 10 mins followed by substrate addition and measured after 2 hrs by fluorescence method
- ChEMBL_2090227 (CHEMBL4771490) Inhibition of human full length recombinant HDAC4 using p53 (379 to 382 residues) Ac-LGK(TFA)-AMC as substrate preincubated for 5 to 10 mins followed by substrate addition and measured after 2 hrs by fluorescence method
- ChEMBL_2090228 (CHEMBL4771491) Inhibition of human full length recombinant HDAC5 using p53 (379 to 382 residues) Ac-LGK(TFA)-AMC as substrate preincubated for 5 to 10 mins followed by substrate addition and measured after 2 hrs by fluorescence method
- ChEMBL_2090229 (CHEMBL4771492) Inhibition of human full length recombinant HDAC7 using p53 (379 to 382 residues) Ac-LGK(TFA)-AMC as substrate preincubated for 5 to 10 mins followed by substrate addition and measured after 2 hrs by fluorescence method
- ChEMBL_2090230 (CHEMBL4771493) Inhibition of human full length recombinant HDAC9 using p53 (379 to 382 residues) Ac-LGK(TFA)-AMC as substrate preincubated for 5 to 10 mins followed by substrate addition and measured after 2 hrs by fluorescence method
- FRET Assay Methods: An HDM2 FRET assay was developed to assess the compounds' inhibitory activity towards binding of p53 protein. A truncated version of HDM2 with residues 17 to 125 (containing p53 binding surface, Science (1994) 265, 346-355), with N-terminal His and Thioredoxin tag was generated in pET32a expression vector and expressed in E. coli strain BL21(DE3)Rosetta. Protein was purified using Ni-affinity chromatography, followed by size exclusion chromatography using Superdex 75 26/60 column. To assess inhibition of p53 binding to HDM2, a FITC labeled 8-mer peptide (SEQ ID NO:1: Ac-Phe-Arg-Dpr-Ac6c-(6-Br)Trp-Glu-Glu-Leu-NH2; Anal Biochem. 2004 Aug. 1; 331(1):138-46) with strong affinity towards the p53 binding pocket of HDM2 was used. The HDM2 assay buffer contained 1x Phosphate Buffered Saline (Invitrogen, Cat#14190), 0.01% BSA (Jackson ImmunoResearch, Cat#001-000-162), 0.01% Tween-20. In the 1x assay buffer recombinant HDM2 protein, peptide and Lumi-4-Tb Cryptate-conjugate mouse.
- Homogenous Time-Resolved Fluorescence Assay (HTRF2 Assay) The standard assay conditions for the in vitro HTRF assay consisted of a 50 ul total reaction volume in black 384-well Costar polypropylene plates in 1×PBS buffer pH 7.4, 1 mM DTT, 0.1% BSA, 2.5 nM GST-hMDM2 (aa 1-188), 5 nM biotinylated-p53 (aa 1-83), 1.8 nM SA-XLent (Cisbio; Bedford, Mass.), 0.6 nM anti-GST cryptate monoclonal antibody (Cisbio; Bedford, Mass.) and 200 mM KF. Amino acid residues 1-188 of human MDM2 were expressed as an amino-terminal glutathione-S-transferase (GST) fusion protein (GST-hMDM2) in Escherichia coli. Residues 1-83 of human p53 were expressed as an amino-terminal AviTag™-TrxA-6×His fusion protein (biotinylated p53) in E. coli. Each protein was purified from cell paste by affinity chromatography.Specifically, 10 uL of GST-hMDM2 was incubated with 10 ul of diluted compound (various concentrations, serially diluted) in 10% DMSO for 20 minutes at room temperature. 20 uL of biotinylated-p53 was added to the GST-hMDM2+compound mixture, and then incubated at room temperature for 60 min. 10 uL of detection buffer consisting of SA-XLent, anti-GST cryptate antibody and KF was added to GST-hMDM2, biotinylated-p53 and compound reaction and left at room temperature to reach equilibrium for >4 hrs. The final concentration of DMSO in the reaction was 2%. Time-resolved fluorescence readings were measured on a microplate multilabel reader. Percentage of inhibition was calculated relative to nutlin-3. All assay conditions remained the same as described above, with the exception of the following changes in reagent concentrations: 0.2 nM GST-hMDM2 (1-188), 0.5 nM biotinylated-p53 (1-83), 0.18 nM SA-XLent, and 100 mM KF.
- ChEMBL_1664928 (CHEMBL4014724) Inhibition of full length human C-terminal His-tagged KDAC3/N-terminal GST-tagged human NCOR2 (395 to 489 residues) expressed in baculovirus expression system using FITC-p53 acetylated peptide as substrate after 60 mins by microfluidic assay
- ChEMBL_1784704 (CHEMBL4256221) Inhibition of recombinant human full length ATR/ATRIP expressed in mammalian expression system using c-Myc-tagged p53 as substrate incubated for 15 mins followed by substrate addition measured after 25 to 35 mins by TR-FRET assay
- ChEMBL_1902906 (CHEMBL4405128) Inhibition of recombinant human GST-tagged HDAC2 (1 to 488 residues) expressed in baculovirus infected sf21 cells using fluorogenic peptide p53 residues 379-382 (RHKK(Ac)AMC) as substrate measured after 1 to 2 hrs by fluorescence assay
- ChEMBL_2090226 (CHEMBL4771489) Inhibition of human full length recombinant HDAC8 using p53 (379 to 382 residues) (RHK(Ac)K(Ac)AMC as substrate preincubated for 5 to 10 mins followed by substrate addition and measured after 2 hrs by fluorescence method
- ChEMBL_1431546 (CHEMBL3390467) Competitive inhibition of human recombinant ribonucleotide reductase in presence of varying levels of ATP by Dixon plot
- MDM2-p53 Inhibition AlphaScreen This assay is used to determine whether the compounds inhibit the p53-MDM2 interaction and thus restore p53 function.15 μL of compound in 20% DMSO (serial pre-dilutions of compound are done in 100% DMSO) is pipetted to the wells of a white OptiPlate-96 (PerkinElmer). A mix consisting of 20 nM GST-MDM2 protein (aa 23-117) and 20 nM biotinylated p53 wt peptide (encompassing aa 16-27 of wt human p53, amino acid sequence QETFSDLWKLLP-Ttds-Lys-Biotin, molecular weight 2132.56 g/mol) is prepared in assay buffer (50 mM Tris/HCl pH 7.2; 120 mM NaCl; 0.1% bovine serum albumin (BSA); 5 mM dithiothreitol (DTT); 1 mM ethylenediaminetetraacetic acid (EDTA); 0.01% Tween 20). 30 μL of the mix is added to the compound dilutions and incubated for 15 min at rt while gently shaking the plate at 300 rounds per minute (rpm). Subsequently, 15 μL of premixed AlphaLISA Glutathione Acceptor Beads and AlphaScreen Streptavidin Donor Beads from PerkinElmer (in assay buffer at a concentration of 10 μg/mL each) are added and the samples are incubated for 30 min at rt in the dark (shaking 300 rpm). Afterwards, the signal is measured in a PerkinElmer Envision HTS Multilabel Reader using the AlphaScreen protocol from PerkinElmer.Each plate contains negative controls where biotinylated p53-peptide and GST-MDM2 are left out and replaced by assay buffer. Negative control values are entered as low basis value when using the software GraphPad Prism for calculations. Furthermore, a positive control (5% DMSO instead of test compound; with protein/peptide mix) is pipetted. Determination of IC50 values are carried out using GraphPad Prism 3.03 software (or updates thereof).
- Mdm2-p53 Inhibition AlphaScreen This assay is used to determine whether the compounds inhibit the p53-MDM2 interaction and thus restore p53 function.15 μL of compound in 20% DMSO (serial pre-dilutions of compound are done in 100% DMSO) is pipetted to the wells of a white OptiPlate-96 (PerkinElmer). A mix consisting of 20 nM GST-MDM2 protein (aa 23-117) and 20 nM biotinylated p53 wt peptide (encompassing aa 16-27 of wt human p53, amino acid sequence QETFSDLWKLLP-Ttds-Lys-Biotin, molecular weight 2132.56 g/mol) is prepared in assay buffer (50 mM Tris/HCl pH 7.2; 120 mM NaCl; 0.1% bovine serum albumin (BSA); 5 mM dithiothreitol (DTT); 1 mM ethylenediaminetetraacetic acid (EDTA); 0.01% Tween 20). 30 μL of the mix is added to the compound dilutions and incubated for 15 min at rt while gently shaking the plate at 300 rounds per minute (rpm). Subsequently, 15 μL of premixed AlphaLISA Glutathione Acceptor Beads and AlphaScreen Streptavidin Donor Beads from PerkinElmer (in assay buffer at a concentration of 10 μg/mL each) are added and the samples are incubated for 30 min at rt in the dark (shaking 300 rpm). Afterwards, the signal is measured in a PerkinElmer Envision HTS Multilabel Reader using the AlphaScreen protocol from PerkinElmer.Each plate contains negative controls where biotinylated p53-peptide and GST-MDM2 are left out and replaced by assay buffer. Negative control values are entered as low basis value when using the software GraphPad Prism for calculations. Furthermore, a positive control (5% DMSO instead of test compound; with protein/peptide mix) is pipetted. Determination of IC50 values are carried out using GraphPad Prism 3.03 software (or updates thereof).
- Mdm2-p53 Inhibition AlphaScreen This assay is used to determine whether the compounds inhibit the p53-MDM2 interaction and thus restore p53 function.15 μL of compound in 20% DMSO (serial pre-dilutions of compound are done in 100% DMSO) is pipetted to the wells of a white OptiPlate-96 (PerkinElmer). A mix consisting of nM GST-MDM2 protein (aa 23-117) and 20 nM biotinylated p53 wt peptide (encompassing aa 16-27 of wt human p53, amino acid sequence QETFSDLWKLLP-Ttds-Lys-Biotin, molecular weight 2132.56 g/mol) is prepared in assay buffer (50 mM Tris/HCl pH 7.2; 120 mM NaCl; 0.1% bovine serum albumin (BSA); 5 mM dithiothreitol (DTT); 1 mM ethylenediaminetetraacetic acid (EDTA); 0.01% Tween 20). 30 μL of the mix is added to the compound dilutions and incubated for 15 min at rt while gently shaking the plate at 300 rounds per minute (rpm). Subsequently, 15 μL of premixed AlphaLISA Glutathione Acceptor Beads and AlphaScreen Streptavidin Donor Beads from PerkinElmer (in assay buffer at a concentration of 10 μg/mL each) are added and the samples are incubated for 30 min at rt in the dark (shaking 300 rpm). Afterwards, the signal is measured in a PerkinElmer Envision HTS Multilabel Reader using the AlphaScreen protocol from PerkinElmer.Each plate contains negative controls where biotinylated p53-peptide and GST-MDM2 are left out and replaced by assay buffer. Negative control values are entered as low basis value when using the software GraphPad Prism for calculations. Furthermore, a positive control (5% DMSO instead of test compound; with protein/peptide mix) is pipetted. Determination of IC50 values are carried out using GraphPad Prism 3.03 software (or updates thereof).
- ChEMBL_1672388 (CHEMBL4022417) Inhibition of LSD1 in human THP1 cells assessed as upregulation of CD86 levels by ELISA
- ChEMBL_1803481 (CHEMBL4275773) Non-competitive inhibition of rat liver ACLY using citrate substrate and varying levels of ATP
- ChEMBL_1890270 (CHEMBL4392024) Competitive inhibition of human placental microsome aromatase using varying levels of [1beta2beta3H]-androstenedione as substrate
- ChEMBL_2443720 Inhibition of mushroom tyrosinase using varying levels of L-DOPA as substrate by Dixon plot analysis
- ChEMBL_457860 (CHEMBL925191) Inhibition of gamma-secretase assessed as reduction of amyloid-beta 40 levels in H4 cells
- ChEMBL_842220 (CHEMBL2091725) Activation of rat TRPA1 expressed in HEK293 cells assessed as stimulation of intracellular Ca2+ levels
- ChEMBL_842223 (CHEMBL2091728) Activation of rat TRPM8 expressed in HEK293 cells assessed as stimulation of intracellular Ca2+ levels
- ChEMBL_855972 (CHEMBL2161973) Inhibition of LYP expressed in Escherichia coli BL21 using increasing levels of ARLIED-NEpYTAREG substrate
- Biochemical Kinase Assay The IC50 value was determined with the aid of a biochemical ATM kinase assay. The assay consists of two steps: the enzymatic reaction and the detection step. Firstly, ATM (ataxia telangiectasia mutated) protein and the test substance are incubated at different concentrations with addition of substrate protein p53 and ATP. ATM mediates the phosphorylation of p53 at several positions, including at amino acid S15. The amount of phosphorylated p53 is determined with the aid of specific antibodies and the TR-FRET technique. The enzymatic ATM assay is carried out as TR-FRET (HTRF , Cisbio Bioassays) based 384-well assay. In the first step, purified human recombinant ATM (human ATM, full length, GenBank ID NM_000051, expressed in a mammal cell line) is incubated in assay buffer for 15 minutes with the ATM inhibitor in various concentrations and without test substance as negative or neutral control. The assay buffer comprises 25 mM HEPES pH 8.0, 10 mM Mg(CH3COO)2, 1 mM MnCl2, 0.1% BSA and 0.01% Brij 35, 5 mM dithiothreitol (DTT). The test-substance solutions were dispensed into the microtitre plates using an ECHO 555 (Labcyte). In the second step, purified human recombinant cmyc-labelled p53 (human p53, full length, GenBank ID BC003596, expressed in Sf21 insect cells) and ATP are added, and the reaction mixture is incubated at 22° C. for 30-35 minutes. The pharmacologically relevant assay volume is 5 μl. The final concentrations in the assay during incubation of the reaction mixture are 0.3-0.4 nM ATM, 50-75 nM p53 and 10 μM ATP. The enzymatic reaction is stopped by addition of EDTA. The formation of phosphorylated p53 as the result of the ATM-mediated reaction in the presence of ATP is detected via specific antibodies[labelled with the fluorophorene europium (Eu) as donor and d2 as acceptor (Cisbio Bioassays)] which enable FRET. 2 μl of antibody-containing stop solution (12.5 mM HEPES pH 8.0, 125 mM EDTA, 30 mM sodium chloride, 300 mM potassium fluoride, 0.1006% Tween-20, 0.005% Brij 35, 0.21 nM anti-phospho-p53(ser15)-Eu antibody and 15 nM anti-cmyc-d2 antibody) are added to the reaction mixture. After incubation, usually for 2 hours (between 1.5 and 15 h), for signal development, the plates are analysed in a plate reader (EnVision, PerkinElmer) using TRF mode (and with laser excitation). After excitation of the donor europium at a wavelength of 340 nm, the emitted fluorescence light both of the acceptor d2 at 665 nm and also of the donor Eu at 615 nm is measured. The amount of phosphorylated p53 is directly proportional to the quotient of the amounts of light emitted, i.e. the relative fluorescence units (RFU) at 665 nm and 615 nm.
- ChEMBL_2208428 (CHEMBL5121377) Inhibition of gamma-secretase (unknown origin) assessed as decrease in Abeta42 levels
- ChEMBL_90338 (CHEMBL696391) Inhibition of 1 uM NECA-stimulated cyclic AMP levels in human platelets
- ChEMBL_1656121 (CHEMBL4005591) Inhibition of recombinant human N-terminal GST-tagged SIRT3 (101 to 399 residues) expressed in Escherichia coli using p53 derived (379 to 382 residues) fluorogenic peptide substrate RHKK(Ac)-AMC) preincubated for 10 mins followed by substrate addition in presence of NAD+ by fluorescence assay
- ChEMBL_1852989 (CHEMBL4353613) Inhibition of human N-terminal His-tagged SIRT2 expressed in Escherichia coli using p53 (Gln-Pro-Lys-Lys(Ac)) (317 to 320 residues) as substrate incubated for 3 hrs measured up to 30 mins in presence of NAD+ by fluorescence-based Michaelis-Menten plot analysis
- Homogenous Time-Resolved Fluorescence Assay (HTRF1 Assay) The standard assay conditions for the in vitro HTRF assay consisted of a 50 ul total reaction volume in black 384-well Costar polypropylene plates in 1xPBS buffer pH 7.4, 1 mM DTT, 0.1% BSA, 2.5 nM GST-hMDM2 (aa 1-188), 5 nM biotinylated-p53 (aa 1-83), 1.8 nM SA-XLent (Cisbio; Bedford, Mass.), 0.6 nM anti-GST cryptate monoclonal antibody (Cisbio; Bedford, Mass.) and 200 mM KF. Amino acid residues 1-188 of human MDM2 were expressed as an amino-terminal glutathione-5-transferase (GST) fusion protein (GST-hMDM2) in Escherichia coli. Residues 1-83 of human p53 were expressed as an amino-terminal AviTag-TrxA-6×His fusion protein (biotinylated p53) in E. coli. Each protein was purified from cell paste by affinity chromatography. Specifically, 10 uL of GST-hMDM2 was incubated with 10 ul of diluted compound (various concentrations, serially diluted) in 10% DMSO for 20 minutes at room temperature. 20 uL of biotinylated-p53 was added to the GST-hMDM2+compound mixture, and then incubated at room temperature for 60 min. 10 uL of detection buffer consisting of SA-XLent, anti-GST cryptate antibody and KF was added to GST-hMDM2, biotinylated-p53 and compound reaction and left at room temperature to reach equilibrium for >4 hrs. The final concentration of DMSO in the reaction was 2%. Time-resolved fluorescence readings were measured on a microplate multilabel reader. Percentage of inhibition was calculated relative to nutlin-3.
- Homogenous Time-Resolved Fluorescence Assay (HTRF1 Assay) The standard assay conditions for the in vitro HTRF assay consisted of a 50 ul total reaction volume in black 384-well Costar polypropylene plates in 1xPBS buffer pH 7.4, 1 mM DTT, 0.1% BSA, 2.5 nM GST-hMDM2 (aa 1-188), 5 nM biotinylated-p53 (aa 1-83), 1.8 nM SA-XLent (Cisbio; Bedford, Mass.), 0.6 nM anti-GST cryptate monoclonal antibody (Cisbio; Bedford, Mass.) and 200 mM KF. Amino acid residues 1-188 of human MDM2 were expressed as an amino-terminal glutathione-S-transferase (GST) fusion protein (GST-hMDM2) in Escherichia coli. Residues 1-83 of human p53 were expressed as an amino-terminal AviTag-TrxA-6-His fusion protein (biotinylated p53) in E. coli. Each protein was purified from cell paste by affinity chromatography.Specifically, 10 uL of GST-hMDM2 was incubated with 10 ul of diluted compound (various concentrations, serially diluted) in 10% DMSO for 20 minutes at room temperature. 20 uL of biotinylated-p53 was added to the GST-hMDM2+compound mixture, and then incubated at room temperature for 60 min. 10 uL of detection buffer consisting of SA-XLent, anti-GST cryptate antibody and KF was added to GST-hMDM2, biotinylated-p53 and compound reaction and left at room temperature to reach equilibrium for >4 hrs. The final concentration of DMSO in the reaction was 2%. Time-resolved fluorescence readings were measured on a microplate multilabel reader. Percentage of inhibition was calculated relative to nutlin-3.
- Homogenous Time-Resolved Fluorescence Assay (HTRF1 Assay) The standard assay conditions for the in vitro HTRF assay consisted of a 50 ul total reaction volume in black 384-well Costar polypropylene plates in 1×PBS buffer pH 7.4, 1 mM DTT, 0.1% BSA, 2.5 nM GST-hMDM2 (aa 1-188), 5 nM biotinylated-p53 (aa 1-83), 1.8 nM SA-XLent (Cisbio; Bedford, Mass.), 0.6 nM anti-GST cryptate monoclonal antibody (Cisbio; Bedford, Mass.) and 200 mM KF. Amino acid residues 1-188 of human MDM2 were expressed as an amino-terminal glutathione-S-transferase (GST) fusion protein (GST-hMDM2) in Escherichia coli. Residues 1-83 of human p53 were expressed as an amino-terminal AviTag-TrxA-6×His fusion protein (biotinylated p53) in E. coli. Each protein was purified from cell paste by affinity chromatography.Specifically, 10 uL of GST-hMDM2 was incubated with 10 ul of diluted compound (various concentrations, serially diluted) in 10% DMSO for 20 minutes at room temperature. 20 uL of biotinylated-p53 was added to the GST-hMDM2+ compound mixture, and then incubated at room temperature for 60 min. 10 uL of detection buffer consisting of SA-XLent, anti-GST cryptate antibody and KF was added to GST-hMDM2, biotinylated-p53 and compound reaction and left at room temperature to reach equilibrium for >4 hrs. The final concentration of DMSO in the reaction was 2%. Time-resolved fluorescence readings were measured on a microplate multilabel reader. Percentage of inhibition was calculated relative to nutlin-3.
- Homogenous Time-Resolved Fluorescence Assay (HTRF2 Assay) The standard assay conditions for the in vitro HTRF assay consisted of a 50 ul total reaction volume in black 384-well Costar polypropylene plates in 1xPBS buffer pH 7.4, 1 mM DTT, 0.1% BSA, 0.2 nM GST-hMDM2 (aa 1-188), 0.5 nM biotinylated-p53 (aa 1-83), 0.18 nM SA-XLent (Cisbio; Bedford, Mass.), 0.6 nM anti-GST cryptate monoclonal antibody (Cisbio; Bedford, Mass.) and 100 mM KF. Amino acid residues 1-188 of human MDM2 were expressed as an amino-terminal glutathione-5-transferase (GST) fusion protein (GST-hMDM2) in Escherichia coli. Residues 1-83 of human p53 were expressed as an amino-terminal AviTag-TrxA-6×His fusion protein (biotinylated p53) in E. coli. Each protein was purified from cell paste by affinity chromatography. Specifically, 10 uL of GST-hMDM2 was incubated with 10 ul of diluted compound (various concentrations, serially diluted) in 10% DMSO for 20 minutes at room temperature. 20 uL of biotinylated-p53 was added to the GST-hMDM2+compound mixture, and then incubated at room temperature for 60 min. 10 uL of detection buffer consisting of SA-XLent, anti-GST cryptate antibody and KF was added to GST-hMDM2, biotinylated-p53 and compound reaction and left at room temperature to reach equilibrium for >4 hrs. The final concentration of DMSO in the reaction was 2%. Time-resolved fluorescence readings were measured on a microplate multilabel reader. Percentage of inhibition was calculated relative to nutlin-3.
- ChEBML_210283 In vitro inhibition of thromboxane synthase (TSI) activity was determined by using human serum levels of TXB2
- ChEMBL_450439 (CHEMBL900723) Inhibition of COX2 in LPS-stimulated J774 cells assessed as inhibition of PGE2 levels by radioimmunoassay
- ChEMBL_461557 (CHEMBL928698) Inhibition of human gamma secretase in H4 cells assessed as reduction of amyloid beta-40 levels
- ChEMBL_855966 (CHEMBL2161840) Noncompetitive inhibition of LYP expressed in Escherichia coli BL21 using increasing levels of ARLIED-NEpYTAREG substrate
- ChEMBL_878322 (CHEMBL2185950) Noncompetitive inhibition of Trypanosoma brucei TR using varying levels of trypanothione disulfide by Lineweaver-Burk plot
- ChEMBL_1720538 (CHEMBL4135538) Inhibition of recombinant human full length HDAC1 expressed in baculovirus infected Sf9 insect cells using p53 (379 to 382 residues) derived RHKKAc as substrate preincubated for 5 to 10 mins followed by substrate addition measured after 2 hrs by fluorescence assay
- ChEMBL_1720539 (CHEMBL4135539) Inhibition of recombinant human full length HDAC6 expressed in baculovirus infected Sf9 insect cells using p53 (379 to 382 residues) derived RHKKAc as substrate preincubated for 5 to 10 mins followed by substrate addition measured after 2 hrs by fluorescence assay
- ChEMBL_1852982 (CHEMBL4353606) Inhibition of human N-terminal His-tagged SIRT3 (102 to 399 residues) expressed in Escherichia coli using p53 (Gln-Pro-Lys-Lys(Ac)) (317 to 320 residues) as substrate incubated for 3 hrs measured up to 30 mins by fluorescence assay
- ChEMBL_1902910 (CHEMBL4405132) Inhibition of recombinant human N-terminal GST-tagged HDAC6 (1 to 1125 residues) expressed in baculovirus infected insect cells using fluorogenic peptide p53 residues 379-382 (RHKK(Ac)AMC) as substrate measured after 1 to 2 hrs by fluorescence assay
- ATM Kinase Assay Determination of ATM Inhibition (IC50 ATM) The IC50 value was determined with the aid of a biochemical ATM kinase assay. The assay consists of two steps: the enzymatic reaction and the detection step. Firstly, ATM (ataxia telangiectasia mutated) protein and the test substance are incubated at different concentrations with addition of substrate protein p53 and ATP. ATM mediates the phosphorylation of p53 at several positions, including at amino acid S15. The amount of phosphorylated p53 is determined with the aid of specific antibodies and the TR-FRET technique. The enzymatic ATM assay is carried out as TR-FRET (HTRF , Cisbio Bioassays) based 384-well assay. In the first step, purified human recombinant ATM (human ATM, full length, GenBank ID NM_000051, expressed in a mammal cell line) is incubated in assay buffer for 15 minutes with the ATM inhibitor in various concentrations and without test substance as negative or neutral control. The assay buffer comprises 25 mM HEPES pH 8.0, 10 mM Mg(CH3COO)2, 1 mM MnCl2, 0.1% BSA and 0.01% Brij 35, 5 mM dithiothreitol (DTT). The test-substance solutions were dispensed into the microtitre plates using an ECHO 555 (Labcyte). In the second step, purified human recombinant cmyc-labelled p53 (human p53, full length, GenBank ID BC003596, expressed in Sf21 insect cells) and ATP are added, and the reaction mixture is incubated at 22° C. for 30-35 minutes. The pharmacologically relevant assay volume is 5 μl. The final concentrations in the assay during incubation of the reaction mixture are 0.3-0.4 nM ATM, 50-75 nM p53 and 10 μM ATP. The enzymatic reaction is stopped by addition of EDTA. The formation of phosphorylated p53 as the result of the ATM-mediated reaction in the presence of ATP is detected via specific antibodies [labelled with the fluorophorene europium (Eu) as donor and d2 as acceptor (Cisbio Bioassays)] which enable FRET. 2 μl of antibody-containing stop solution (12.5 mM HEPES pH 8.0, 125 mM EDTA, 30 mM sodium chloride, 300 mM potassium fluoride, 0.1006% Tween-20, 0.005% Brij 35, 0.21 nM anti-phospho-p53(ser15)-Eu antibody and 15 nM anti-cmyc-d2 antibody) are added to the reaction mixture. After incubation, usually for 2 hours (between 1.5 and 15 h), for signal development, the plates are analysed in a plate reader (EnVision, PerkinElmer) using TRF mode (and with laser excitation). After excitation of the donor europium at a wavelength of 340 nm, the emitted fluorescence light both of the acceptor d2 at 665 nm and also of the donor Eu at 615 nm is measured. The amount of phosphorylated p53 is directly proportional to the quotient of the amounts of light emitted, i.e. the relative fluorescence units (RFU) at 665 nm and 615 nm. The measurement data were processed by means of Genedata Screener software. IC50 determinations are carried out, in particular, by fitting a dose/action curve to the data points by means of nonlinear regression analysis.
- ATM kinase assay—determination of ATM inhibition (IC50 ATM) The IC50 value was determined with the aid of a biochemical ATM kinase assay. The assay consists of two steps: the enzymatic reaction and the detection step. Firstly, ATM (ataxia telangiectasia mutated) protein and the test substance are incubated at different concentrations with addition of substrate protein p53 and ATP. ATM mediates the phosphorylation of p53 at several positions, including at amino acid S15. The amount of phosphorylated p53 is determined with the aid of specific antibodies and the TR-FRET technique. The enzymatic ATM assay is carried out as TR-FRET (HTRF™, Cisbio Bioassays) based 384-well assay. In the first step, purified human recombinant ATM (human ATM, full length, GenBank ID NM_000051, expressed in a mammal cell line) is incubated in assay buffer for 15 minutes with the ATM inhibitor in various concentrations and without test substance as negative or neutral control. The assay buffer comprises 25 mM HEPES pH 8.0, 10 mM Mg(CH3COO)2, 1 mM MnCl2, 0,1% BSA and 0,01% Brij 35, 5 mM dithiothreitol (DTT). The test-substance solutions were dispensed into the microtitre plates using an ECHO 555 (Labcyte). In the second step, purified human recombinant cmyc-labelled p53 (human p53, full length, GenBank ID BC003596, expressed in Sf21 insect cells) and ATP are added, and the reaction mixture is incubated at 22° C. for 30-35 minutes. The pharmacologically relevant assay volume is 5 μl. The final concentrations in the assay during incubation of the reaction mixture are 0.3-0.4 nM ATM, 50-75 nM p53 and 10 μM ATP. The enzymatic reaction is stopped by addition of EDTA. The formation of phosphorylated p53 as the result of the ATM-mediated reaction in the presence of ATP is detected via specific antibodies [labelled with the fluorophorene europium (Eu) as donor and d2 as acceptor (Cisbio Bio-assays)] which enable FRET. 2 μl of antibody-containing stop solution (12.5 mM HEPES pH 8.0, 125 mM EDTA, 30 mM sodium chloride, 300mM potassium fluoride, 0.1006% Tween-20, 0.005% Brij 35, 0.21 nM anti-phospho-p53(ser15)-Eu antibody and 15 nM anti-cmyc-d2 antibody) are added to the reaction mixture. After incubation, usually for 2 hours (between 1.5 and 15h), for signal development, the plates are analysed in a plate reader (EnVision, PerkinElmer) using TRF mode (and with laser excitation). After excitation of the donor europium at a wavelength of 340 nm, the emitted fluorescence light both of the acceptor d2 at 665 nm and also of the donor Eu at 615 nm is measured. The amount of phosphorylated p53 is directly proportional to the quotient of the amounts of light emitted, i.e. the relative fluorescence units (RFU) at 665 nm and 615 nm. The measurement data were processed by means of Genedata Screener software. IC50 determinations are carried out, in particular, by fitting a dose/action curve to the data points by means of nonlinear regression analysis.
- ChEMBL_1794156 (CHEMBL4266273) Competitive inhibition of Aurora A kinase in human cells using varying ATP levels
- ChEMBL_1882462 (CHEMBL4383961) Inhibition of PCSK9 in human HepG2 cells assessed as increase in LDLR levels
- ChEMBL_1925284 (CHEMBL4428356) Modulation of gamma secretase (unknown origin) assessed as reduction in amyloid beta levels
- ChEMBL_2208420 (CHEMBL5121369) Modulation of gamma-secretase (unknown origin) assessed as decrease in total Abeta42 levels
- ChEMBL_2208421 (CHEMBL5121370) Modulation of gamma-secretase (unknown origin) assessed as decrease in free Abeta42 levels
- ATM Kinase Assay The IC50 value was determined with the aid of a biochemical ATM kinase assay. The assay consists of two steps: the enzymatic reaction and the detection step. Firstly, ATM (ataxia telangiectasia mutated) protein and the test substance are incubated at different concentrations with addition of substrate protein p53 and ATP. ATM mediates the phosphorylation of p53 at several positions, including at amino acid S15. The amount of phosphorylated p53 is determined with the aid of specific antibodies and the TR-FRET technique. The enzymatic ATM assay is carried out as TR-FRET (HTRF , Cisbio Bioassays) based 384-well assay. In the first step, purified human recombinant ATM (human ATM, full length, GenBank ID NM_000051, expressed in a mammal cell line) is incubated in assay buffer for 15 minutes with the ATM inhibitor in various concentrations and without test substance as negative or neutral control. The assay buffer comprises 25 mM HEPES pH 8.0, 10 mM Mg(CH3COO)2, 1 mM MnCl2, 0.1% BSA and 0.01% Brij 35, 5 mM dithiothreitol (DTT). The test-substance solutions were dispensed into the microtitre plates using an ECHO 555 (Labcyte). In the second step, purified human recombinant cmyc-labelled p53 (human p53, full length, GenBank ID BC003596, expressed in Sf21 insect cells) and ATP are added, and the reaction mixture is incubated at 22° C. for 30-35 minutes. The pharmacologically relevant assay volume is 5 μl. The final concentrations in the assay during incubation of the reaction mixture are 0.3-0.4 nM ATM, 50-75 nM p53 and 10 μM ATP. The enzymatic reaction is stopped by addition of EDTA. The formation of phosphorylated p53 as the result of the ATM-mediated reaction in the presence of ATP is detected via specific antibodies [labelled with the fluorophorene europium (Eu) as donor and d2 as acceptor (Cisbio Bioassays)] which enable FRET. 2 μl of antibody-containing stop solution (12.5 mM HEPES pH 8.0, 125 mM EDTA, 30 mM sodium chloride, 300 mM potassium fluoride, 0.1006% Tween-20, 0.005% Brij 35, 0.21 nM anti-phospho-p53(ser15)-Eu antibody and 15 nM anti-cmyc-d2 antibody) are added to the reaction mixture. After incubation, usually for 2 hours (between 1.5 and 15 h), for signal development, the plates are analysed in a plate reader (EnVision, PerkinElmer) using TRF mode (and with laser excitation). After excitation of the donor europium at a wavelength of 340 nm, the emitted fluorescence light both of the acceptor d2 at 665 nm and also of the donor Eu at 615 nm is measured. The amount of phosphorylated p53 is directly proportional to the quotient of the amounts of light emitted, i.e. the relative fluorescence units (RFU) at 665 nm and 615 nm. The measurement data were processed by means of Genedata Screener software. IC50 determinations are carried out, in particular, by fitting a dose/action curve to the data points by means of nonlinear regression analysis.
- ChEMBL_1656115 (CHEMBL4005585) Inhibition of recombinant human C-terminal His-tagged SIRT2 (50 to 389 end residues) expressed in Escherichia coli using p53 derived (379 to 382 residues) fluorogenic peptide substrate RHKK(Ac)-AMC preincubated for 10 mins followed by substrate addition in presence of NAD+ by fluorescence assay
- ChEMBL_1656119 (CHEMBL4005589) Inhibition of recombinant human N-terminal His-tagged SIRT1 (1 to 747 end residues) expressed in Escherichia coli using p53 derived (379 to 382 residues) fluorogenic peptide substrate RHKK(Ac)-AMC preincubated for 10 mins followed by substrate addition in presence of NAD+ by fluorescence assay
- Inhibition AlphaScreen Assay This assay is used to determine whether the compounds inhibit the p53-MDM2 interaction and thus restore p53 function.15 μL of compound in 20% DMSO (serial pre-dilutions of compound are done in 100% DMSO) is pipetted to the wells of a white OptiPlate-96 (PerkinElmer). A mix consisting of 20 nM GST-MDM2 protein (aa 23-117) and 20 nM biotinylated p53 wt peptide (encompassing aa 16-27 of wt human p53, amino acid sequence QETFSDLWKLLP-Ttds-Lys-Biotin, molecular weight 2132.56 g/mol) is prepared in assay buffer (50 mM Tris/HCl pH 7.2; 120 mM NaCl; 0.1% bovine serum albumin (BSA); 5 mM dithiothreitol (DTT); 1 mM ethylenediaminetetraacetic acid (EDTA); 0.01% Tween 20). 30 μL of the mix is added to the compound dilutions and incubated for 15 min at rt while gently shaking the plate at 300 rounds per minute (rpm). Subsequently, 15 μL of premixed AlphaLISA Glutathione Acceptor Beads and AlphaScreen Streptavidin Donor Beads from PerkinElmer (in assay buffer at a concentration of 10 μg/mL each) are added and the samples are incubated for 30 min at rt in the dark (shaking 300 rpm). Afterwards, the signal is measured in a PerkinElmer Envision HTS Multilabel Reader using the AlphaScreen protocol from PerkinElmer.Each plate contains negative controls where biotinylated p53-peptide and GST-MDM2 are left out and replaced by assay buffer. Negative control values are entered as low basis value when using the software GraphPad Prism for calculations.
- Enzymatic Assay The IC50 values for aforementioned compounds against HDACs were determined. HDAC 1 to 11 can be assayed by using acetylated AMC-labeled peptide substrate. The Substrate 1, a fluorogenic peptide from p53 residues 379-382 (RHKKAc) is used for all MAC 1 to 11 but HDAC8, which has a substrate II (RHKAcKAc), a fluorogenic diacyl peptide based on residues 179-382 of p53. Compounds were tested in 10-dose 1050 mode in duplicate with 3-fold serial dilution starting at 10 uM.
- ChEMBL_2062311 (CHEMBL4717564) Competitive inhibition of sEH (unknown origin) using varying levels of PHOME as substrate by Dixon plot analysis
- ChEMBL_2063497 (CHEMBL4718750) Competitive inhibition of human S1PL using varying levels of RBM13 as substrate by Lineweaver-Burk plot analysis
- ChEMBL_210283 (CHEMBL808529) In vitro inhibition of thromboxane synthase (TSI) activity was determined by using human serum levels of TXB2
- ChEMBL_210291 (CHEMBL808537) In vitro inhibition of thromboxane synthase (TSI) activity was determined by using human serum levels of TXB2
- ChEMBL_306686 (CHEMBL830020) Inhibition of 100 nM-NECA stimulation of cAMP levels in human adenosine A2b receptor expressing CHO cells
- AlphaScreen Assay The following example describes an assay that measured the ability of compounds to inhibit the binding of p53 to MDM2 using the AlphaScreen assay technology (PerkinElmer). The following protocol is an adaptation of the method described by H. R. Lawrence et al. (Bioorg. Med. Chem. Lett. 19 (2009) 3756-3759). Recombinant, truncated, human, N-terminal GST-MDM2 (aa 1-150) was obtained from GeneScript. Wild-type, full length human N-terminal 6-his p53 was purchased from SignalChem.Resulting in a final reaction volume of 24 μl PBS, 0.1% Tween-20, and 10% glycerol, 30 ng of MDM2 was added, followed by the addition of 10 of compound diluted in 100% DMSO that provided a final DMSO concentration of 4%. 30 ng of p53 was then added, mixed, and incubated at room temperature for 1 hour. Glutathione donor beads and Nickel acceptor beads (0.5 μg each; PerkinElmer) were added under subdued lighting conditions to a final reaction volume of 30 μl/well in a 96 well, volume Proxima plate.
- ChEMBL_1902907 (CHEMBL4405129) Inhibition of recombinant full length human C-terminal His-tagged HDAC3 (1 to 428 residues) expressed in baculovirus infected insect cells using fluorogenic peptide p53 residues 379-382 (RHKK(Ac)AMC) as substrate measured after 1 to 2 hrs by fluorescence assay
- ChEMBL_2020056 (CHEMBL4673869) Inhibition of human recombinant full length HDAC6 expressed in baculovirus infected sf9 insect cells using p53 fluorogenic peptide (379 to 382 residues) (RHKKAc) as substrate preincubated for 5 to 10 mins followed by substrate addition and measured after 2 hrs by fluorescence method
- ChEMBL_2020057 (CHEMBL4673870) Inhibition of human recombinant full length HDAC2 expressed in baculovirus infected sf9 insect cells using p53 fluorogenic peptide (379 to 382 residues) (RHKKAc) as substrate preincubated for 5 to 10 mins followed by substrate addition and measured after 2 hrs by fluorescence method
- ChEMBL_2020058 (CHEMBL4673871) Inhibition of human recombinant full length HDAC1 expressed in baculovirus infected sf9 insect cells using p53 fluorogenic peptide (379 to 382 residues) (RHKKAc) as substrate preincubated for 5 to 10 mins followed by substrate addition and measured after 2 hrs by fluorescence method
- ChEMBL_2090185 (CHEMBL4771448) Inhibition of human C-terminal His-tagged HDAC3 (1 to 428 residues)/human N-terminal GST-tagged NcoR2 (395 to 489 residues) expressed in sf9 cells using p53 (379 to 382 residues) (Arg-His-Lys(Ac)-Lys(Ac)) as substrate by fluorimetric assay
- ChEMBL_1634999 (CHEMBL3877897) Competitive inhibition of horse serum BChE in presence of varying levels of butyrylthiocholine iodide substrate by Lineweaver-burk plot method
- ChEMBL_1766754 (CHEMBL4202001) Non-competitive inhibition of human recombinant ALDH1A1 assessed as reduction of NAD(P)H formation using varying levels of NAD+
- ChEBML_38924 Stimulation of cAMP levels in CHO cells expressing the recombinant human beta-3 adrenergic receptor
- ChEMBL_106195 (CHEMBL714613) Intracellular levels of cAMP in HEK 293 cells expressing human melanocortin 4 receptor (hMC4R)
- ChEMBL_1560971 (CHEMBL3779604) Antagonist activity at human somatostatin receptor type 3 assessed as inhibition of cAMP levels
- ChEMBL_1658851 (CHEMBL4008463) Inhibition of NAMPT in human Jurkat T cells assessed as reduction in NAD levels
- ChEMBL_2266257 Inhibition of GCS in human iPSC derived neurons assessed as reduction in brain GlcCer levels
- ChEMBL_2538822 Inhibition of SMYD2 in human HEK293T cells assessed as BTF3me1 levels by Western blot analysis
- Homogeneous Time-Resolved Fluorescence (HTRF) Assay The ability of the compounds to inhibit the interaction between p53 and MDM2 proteins was measured by an HTRF (homogeneous time-resolved fluorescence) assay in which recombinant GST-tagged MDM2 binds to a peptide that resembles the MDM2-interacting region of p53 (Lane et al.). Binding of GST-MDM2 protein and p53-peptide (biotinylated on its N-terminal end) is registered by the FRET (fluorescence resonance energy transfer) between Europium (Eu)-labeled anti-GST antibody and streptavidin-conjugated Allophycocyanin (APC).Test is performed in black flat-bottom 384-well plates (Costar) in a total volume of 40 uL containing 90 nM biotinylate peptide, 160 ng/ml GST-MDM2, 20 nM streptavidin-APC (PerkinElmerWallac), 2 nM Eu-labeled anti-GST-antibody (PerkinElmerWallac), 0.02% bovine serum albumin (BSA), 1 mM dithiothreitol (DTT) and 20 mM Tris-borate saline (TBS) buffer as follows: Add 10 uL of GST-MDM2 (640 ng/ml working solution) in reaction buffer to each well.
- ChEMBL_1630545 (CHEMBL3873251) Competitive inhibition of rabbit muscle glycogen phosphorylase-b in presence of varying glucose-1-phosphate levels and NADP
- ChEMBL_2014475 (CHEMBL4668053) Mixed type inhibition of human erythrocytes AChE using varying levels of acetylthiocholine iodide substrate by Dixon plot analysis
- ChEMBL_2062309 (CHEMBL4717562) Mixed type inhibition of sEH (unknown origin) using varying levels of PHOME as substrate by Dixon plot analysis
- ChEMBL_2062310 (CHEMBL4717563) Non competitive inhibition of sEH (unknown origin) using varying levels of PHOME as substrate by Dixon plot analysis
- ELISA Assay Streptavidin-coated 96-well plates are used to immobilise a biotin-tagged IP3 p53-derived peptide (MPRFMDYWEGLN). This is a peptide analogue derived from the p53 binding site for MDM2 (QETFSDLWKLLP). IP3 has a higher affinity for MDM2 than the native peptide and has been used elsewhere to identify antagonists of the binding between MDM2 and p53 (Stoll et al 2001). Aliquots of MDM2 generated by in vitro translation are pre-incubated for 20 minutes at room temperature (i.e. 20-25C.) with test compounds and controls, before transfer into the IP3-coated 96-well plates. Following a further incubation period of 90 minutes at 4C., the plates are washed to remove unbound MDM2 and the residual bound MDM2 is detected using a primary monoclonal antibody (MDM2 Ab-1, clone IF2, Oncogene Research Products) and HRP-conjugated secondary antibody (Goat anti-mouse, Dako PO447).
- ChEMBL_1902905 (CHEMBL4405127) Inhibition of full length recombinant human C-terminal FLAG/His-tagged HDAC1 (1 to 482 residues) expressed in baculovirus infected Sf21 insect cells using fluorogenic peptide p53 residues 379-382 (RHKK(Ac)AMC) as substrate measured after 1 to 2 hrs by fluorescence assay
- ChEMBL_2047186 (CHEMBL4701885) Inhibition of EGFR in human KYSE-520 cells assessed as reduction in p-ERK levels
- ChEMBL_2047188 (CHEMBL4701887) Inhibition of RAF in human A-375 cells assessed as reduction in p-ERK levels
- ChEMBL_2047199 (CHEMBL4701898) Inhibition of MEK in human KYSE-520 cells assessed as reduction in p-ERK levels
- ChEMBL_218113 (CHEMBL822475) Increased cAMP levels of Chinese hamster ovary (CHO) cells expressing human beta 1 Adrenergic receptor
- ChEMBL_218117 (CHEMBL821784) Increased cAMP levels of Chinese hamster ovary (CHO) cells expressing human beta 2 adrenergic receptors
- ChEMBL_3905 (CHEMBL619910) Inhibition of 5-lipoxygenase by measuring 5-HETE levels in RBL-1 cell-free supernatant
- ChEMBL_548295 (CHEMBL1028365) Inhibition of rat fetal brain CDK5 assessed as phosphorylated histone H1 levels by immuno-precipitation
- ChEMBL_633015 (CHEMBL1118240) Activation of TRPA1 channel expressed in CHO cells assessed as increase in intracellular calcium levels
- ChEMBL_970232 (CHEMBL2406499) Inhibition of cMET in human MKN45 cells assessed as phosphorylated MET levels after 4 hrs
- ChEMBL_1850314 (CHEMBL4350855) Uncompetitive inhibition of electric eel BChE using varying levels of butyrylcholine chloride as substrate by Lineweaver-Burk plot analysis
- ChEMBL_1850315 (CHEMBL4350856) Mixed inhibition of equine serum BChE using varying levels of butyrylcholine chloride as substrate by Lineweaver-Burk plot analysis
- ATR/ATRIP Enzymatic Assay Detection of ATR kinase activity utilized the AlphaScreen system to measure the phosphorylation of the substrate protein p53. Recombinant purified ATR/ATRIP (Eurofins cat #14-953) at a final concentration of 0.63 nM in assay buffer (50 mM Hepes pH 7.4, 0.1 mM vanadate, 0.5 mM DTT, 0.1 mM EGTA, 5 mM MnCl2, 0.01% Brij-30, 1% glycerol, 0.05% BSA) was mixed with compound serially diluted in 10% DMSO. The final DMSO concentration was 1.25%. A pre-mix of GST-tagged p53 (full length, Enzo Life Sciences cat # BML-FW9370) and adenosine 5′-triphosphate, ATP (Sigma-Aldrich cat #10519979001, Roche Diagnostic) in assay buffer was added to the enzyme:compound mix for a final concentration of 25 nM GST-p53 and 3 μM ATP. The reaction was allowed to proceed at room temperature for 1 hour then stopped by the addition of a pre-mix of phospho-p53 (Ser 15) antibody (New England Biolabs cat #9284S) at 1:3000 final dilution, 14.3 pg/mL glutathione donor beads (PerkinElmer Life Sciences cat #6765301) and 14.3 pg/mL protein A acceptor beads (PerkinElmer Life Sciences cat #670137) final bead concentration in buffer (60 mM EDTA in 50 mM Tris, pH 7.4 and 0.1% BSA). Plates were incubated at room temperature in the dark for 4 hours and read on a BMG Polarstar using AlphaScreen dedicated filters.
- Fluorescence Anisotropy Competition Binding Assay Inhibitory activities of NAPAs were measured by fluorescence anisotropy competition experiments. Protein was incubated with fluorescein-labeled human p53 peptide in the presence of test compound. Experiments were performed with a 70-fold molar excess of protein over labeled p53-peptide to ensure that the peptide is completely bound by the protein. Under these conditions, there is no difference in the minimum fluorescence polarization values between direct and competition assays. Fluorescence anisotropy was measured at 25 deg C on a Beacon 2000 (PanVera, Madison, WI). Data points were measured in triplicate. IC50 values were calculated by nonlinear regression curve fitting.
- ChEMBL_1705921 (CHEMBL4057154) Competitive inhibition of Trypanosoma cruzi trypanothione reductase using varying levels of trypanothione disulfide as substrate by Lineweaver--burk plot analysis
- ChEMBL_1739706 (CHEMBL4155456) Mixed-type inhibition of equine serum BChE using varying levels of butyrylthiocholine iodide as substrate by Lineweaver-burk plot analysis
- ChEMBL_1906045 (CHEMBL4408403) Inhibition of equine serum BChE using varying levels of butyrylthiocholine iodide as substrate by Line weaver Burk reciprocal plot analysis
- ChEMBL_2087001 (CHEMBL4768264) Non competitive inhibition of human erythrocyte AChE using varying levels of acetylthiocholine iodide as substrate by Lineweaver-Burk plot analysis
- ChEMBL_2087002 (CHEMBL4768265) Non competitive inhibition of human plasma BuChE using varying levels of butyrylthiocholine iodide as substrate by Lineweaver-Burk plot analysis
- ChEMBL_2087003 (CHEMBL4768266) Mixed type inhibition of human erythrocyte AChE using varying levels of acetylthiocholine iodide as substrate by Lineweaver-Burk plot analysis
- ChEMBL_970209 (CHEMBL2406476) Inhibition of cMET in human MKN45 cells assessed as phosphorylated MET levels after 4 hrs in presence of mouse plasma
- ChEMBL_1902912 (CHEMBL4405134) Inhibition of recombinant C-terminal His-fusion tagged and N-terminal Streptavidin2-tagged human HDAC8 (1 to 377 residues) expressed in insect cells using fluorogenic peptide p53 residues 379-382 (RHK(Ac)K(Ac)AMC) as substrate measured after 1 to 2 hrs by fluorescence assay
- ChEMBL_1809878 (CHEMBL4309338) Inhibition of IDH1 R132H mutant in human U87MG cells assessed as decrease in 2-HG levels
- ChEMBL_1925277 (CHEMBL4428349) Inhibition of gamma secretase (unknown origin) assessed as reduction in amyloid beta levels by HTRF assay
- ChEMBL_1925281 (CHEMBL4428353) Inhibition of gamma secretase in HEK293 cells expressing APP assessed as reduction in amyloid beta levels
- ChEMBL_1925289 (CHEMBL4428361) Inhibition of gamma secretase (unknown origin) assessed as reduction in amyloid beta levels by sandwich ELISA
- ChEMBL_2081278 (CHEMBL4737069) Inhibition of wild type EZH2 in human KARPAS-422 cells assessed as effect on H3K27Me3 levels
- ChEMBL_313611 (CHEMBL835018) Inhibition of forskolin stimulated cAMP levels in CHO cell membranes expressing the human NOP receptor (CHOhNOP)
- ChEMBL_423827 (CHEMBL911272) Activity at human adenosine A2B receptor expressed in CHO cells assessed as stimulation of cAMP levels
- ChEMBL_1650525 (CHEMBL3999659) Competitive inhibition of recombinant Trypanosoma cruzi cruzain in presence of varying levels of Z-phe-Arg-amidomethylcoumarin substrate by Lineweaver-Burk plot analysis
- ChEMBL_1630547 (CHEMBL3873253) Competitive inhibition of rabbit muscle glycogen phosphorylase-a in presence of varying levels of glucose-1-phosphate and constant concentration of glycogen by Hill plot analysis
- Inhibitory Effect of Compounds Disclosed Herein on ATR Enzyme The following method was used to determine the inhibitory effect of the compounds disclosed herein on ATR enzyme. The experimental method was briefly described as follows:I. Experimental Materials and Instruments1. ATR enzyme (Eurofins Pharma Discovery Services, 14-953-M)2. GST-tag P53 protein (Eurofins Pharma Discovery Services, 14-952-M)3. 384-well plate (Thermo Scientific, 267462)4. U-shaped bottom 96-well plate (Corning, 3795)5. MAb Anti-phospho p53-Eu cryptate (Cisbio, 61P08KAE)6. MAb Anti GST-d2 (Cisbio, 61GSTDLF)7. ATP solution (Promega, V916B)8. EDTA (Thermo Scientific, AM9260G)9. HEPES (Gibco, 15630-080)10. Microplate reader (BMG, PHERAsta)II. Experimental Procedures1 nM ATR enzyme, 50 nM P53 protein, 7.435 μM ATP and small molecule compounds of different concentrations (serially 3-fold diluted from 1 μM to the 11th concentration) were mixed and incubated at room temperature for 2 h. A terminating buffer (12.5 mM HEPES, 250 mM EDTA) was added. The mixture was well mixed before 0.42 ng/well of mAb anti-phospho p53-Eu cryptate and 25 ng/well of mAb anti GST-d2 were added. The mixture was incubated overnight at room temperature, and the fluorescence signals at 620 nm and 665 nm were detected using a PHERAstar system. Data were processed using GraphPad software.
- ChEMBL_1650377 (CHEMBL3999511) Competitive inhibition of recombinant human MAO-A using varying levels of kynuramine substrate after 5 mins by Lineweaver-Burk plot analysis
- ChEMBL_1730448 (CHEMBL4145984) Reversible-competitive inhibition of human MAO-A using varying levels of tyramine as substrate after 30 mins by Lineweaver-Burk plot
- ChEMBL_787208 (CHEMBL1918878) Inhibition of HSP90beta in human SKBR3 cells assessed as down regulation of HER2 expression levels after 24 hrs by FACS analysis
- ChEMBL_1476665 (CHEMBL3428750) Inhibition of LPS-induced COX2 in human whole blood assessed as reduction in PGH2 levels by radioimmunoassay
- ChEMBL_1476666 (CHEMBL3428751) Inhibition of LPS-induced COX1 in human whole blood assessed as reduction in TXB2 levels by radioimmunoassay
- ChEMBL_2047195 (CHEMBL4701894) Inhibition of KRAS G12C mutant in human NCI-H358 cells assessed as reduction in p-ERK levels
- ChEMBL_2165240 (CHEMBL5050101) Inhibition of EZH2 in HEK293T cells assessed as H3K27me3 levels incubated for 48 hrs by flow cytometry
- ChEMBL_2353755 Inhibition of NSD3 degradation in human KMS-11 cells assessed as reduction in H3K36me2 levels by FRET assay
- ChEMBL_450438 (CHEMBL900722) Inhibition of COX1 in mouse J774 cells assessed as arachidonic acid-induced PGE2 levels by radio immunoassay
- ChEMBL_511445 (CHEMBL1003926) Binding affinity to Mycobacterium tuberculosis recombinant MbtA using saturated levels of salicylic acid substrate by isothermal calorimetry
- ChEMBL_863278 (CHEMBL2176221) Inhibition of AGT in human HL60 cells assessed as AGT levels using [3H]AGT after 2 hrs
- HTRF2 Assay As the potencies of the HDM2 inhibitors increased, an improved HTRF assay (HTRF2 assay) was developed. All assay conditions remained the same as described above, with the exception of the following changes in reagent concentrations: 0.2 nM GST-hMDM2 (1-188), 0.5 nM biotinylated-p53 (1-83), 0.18 nM SA-XLent, and 100 mM KF.
- ChEMBL_1669318 (CHEMBL4019206) Competitive inhibition of AEP (unknown origin) using varying levels of Z-AAN-AMC substrate after 10 mins by Lineweaver-Burk plot analysis
- ChEMBL_1800952 (CHEMBL4273244) Inhibition of recombinant human AChE expressed in HEK293 cells using varying levels of acetylthiocholine iodide as substrate by Lineweaver-burk plot analysis
- ChEBML_143203 Agonistic activity was determined against human Neuromedin B receptor transfected in HEK293 cells as accumulation levels of inositol phosphate
- ChEBML_1669766 Inhibition of CrtN in Staphylococcus aureus Newman assessed as reduction in staphyloxanthin levels after 48 hrs by spectrophotometric analysis
- ChEBML_216183 Agonistic activity was assessed by measurement of cAMP accumulation levels in CHO cells expressing human beta-2-adrenergic receptor
- ChEMBL_1615982 (CHEMBL3858051) Inhibition of EZH2 in human KARPAS422 cells assessed as reduction in H3K27Me3 levels after 72 hrs by ELISA
- ChEMBL_1677652 (CHEMBL4027795) Inhibition of CrtM in Staphylococcus aureus assessed as reduction in staphyloxanthin levels after 72 hrs by spectrophotometric method
- ChEMBL_1700376 (CHEMBL4051358) Inhibition of ERK2 in human A375 cells harboring BRAF V600E mutant assessed as decrease in phosphorylated ERK2 levels
- ChEMBL_1700377 (CHEMBL4051359) Inhibition of ERK2 in human A375 cells harboring BRAF V600E mutant assessed as decrease in phosphorylated RSK levels
- ChEMBL_1929600 (CHEMBL4432776) Inhibition of PI3Kdelta in human KARPAS422 cells assessed as reduction in AktS473 phosphorylation levels by Western blot analysis
- ChEMBL_1929603 (CHEMBL4432779) Inhibition of PI3Kdelta in human KARPAS422 cells assessed as reduction in S6 phosphorylation levels by Western blot analysis
- ChEMBL_2016282 (CHEMBL4669860) Agonist activity at human SST2 expressed in CHOK1 cells assessed as inhibition of NKH477-stimulated intracellular cAMP levels
- ChEMBL_2047197 (CHEMBL4701896) Inhibition of KRAS G12C mutant in human MIA PaCa-2 cells assessed as reduction in p-ERK levels
- ChEMBL_2363563 Inhibition of THP1 in human serotonergic BON cells assessed as reduced the intracellular serotonin levels incubated for 72 hrs
- ChEMBL_1854945 (CHEMBL4355674) Inhibition of recombinant N-terminal GST-tagged human HDAC6 (1 to 1215 residues) expressed in baculovirus infected sf9 insect cells using p53 (379 to 382 residues) derived fluorogenic peptide RHKKAc as substrate preincubated for 5 to 10 mins followed by substrate addition and further incubated for 2 hrs by fluorescence assay
- ChEMBL_1801926 (CHEMBL4274218) Competitive inhibition of AChE in human erythrocytes using varying levels of acetylthiocholine as substrate measured for 1 min by Lineweaver-burk plot analysis
- ChEMBL_1862249 (CHEMBL4363105) Competitive inhibition of red kidney bean PAP using varying levels of pNPP as substrate measured at pH 6.2 by UV-vis spectrophotometric analysis
- ChEMBL_1921374 (CHEMBL4424219) Competitive inhibition of recombinant human PTP1B expressed in Escherichia coli using varying levels of p-nitrophenol as substrate by Lineweaver-burk plot analysis
- ChEMBL_2123834 (CHEMBL4833067) Competitive inhibition of Trypanosoma cruzi cruzain assessed as inhibition constant using varying levels of Z-FR-AMC as substrate by Michaelis-Menten analysis
- ChEMBL_2129834 (CHEMBL4839263) Inhibition of human soluble adenylyl cyclase assessed as reduction in cAMP levels in the presence of alpha-32p labeled ATP by biochemical assay
- ChEMBL_2261996 (CHEMBL5217007) Competitive inhibition of human recombinant MAO-A assessed as inhibitory constant using varying levels of kynuramine as substrate by Lineweaver-burk plot analysis
- ChEMBL_2261997 (CHEMBL5217008) Competitive inhibition of human recombinant MAO-B assessed as inhibitory constant using varying levels of kynuramine as substrate by Lineweaver-burk plot analysis
- ChEMBL_2262985 (CHEMBL5217996) Inhibition of CBP/p300 in human COLO 320HSR cells assessed as reduction of C-myc levels measured after 24 hrs by HTRF assay
- ChEMBL_432673 (CHEMBL919244) Inhibition of Helicobacter pylori SS1 recombinant shikimate kinase expressed in BL21 (DE3) cells in presence of varying shikimate levels by double coupled assay
- ChEMBL_432674 (CHEMBL920648) Inhibition of Helicobacter pylori SS1 recombinant shikimate kinase expressed in BL21 (DE3) cells in presence of varying MgATP levels by double coupled assay
- ChEMBL_1630555 (CHEMBL3873261) Competitive inhibition of rabbit muscle glycogen phosphorylase-b in presence of varying levels of glucose-1-phosphate and constant concentration of glycogen and AMP by double reciprocal plot method
- p53-MDM2 Binding ELISA Interference of the p53-MDM2 binding by test compounds was measured in a 96-well polypropylene round-bottom microtiter plate (Costar, Serocluster). Human MDM2 (amino acids 1-118) was preincubated with 10% DMSO or compounds in assay buffer. After 15-min incubation of the sample, a biotinylated p53-derived peptide was added. As a negative control, buffer only was added into separate wells (blanks). After another 30 min, the incubation mixture was added to 96-well plates coated with streptavidin. After 1 h, the wells were extensively washed. Then, the MDM2 specific antibody was added. After 1 h, the wells were thoroughly washed and the secondary antibody (anti-rabbit-IgG-POD) was added. After another hour, the wells were washed, and the peroxidase subtrate ABTS was added. After 30 min, OD 405/490 nm was determined with a Dynatech MR 7000 ELISA reader. Compounds were titrated to determine IC50 values twice (range of compound concentration: 0.5, 1, 5, 10, 25, 50, 125, 250 uM).
- ChEMBL_1892974 (CHEMBL4394895) Inhibition of purified human adenosine kinase using varying levels of [3H]Ado as substrate in presence of adenosine deaminase inhibitor deoxycoformycin by Line-weaver burk plot analysis
- ChEMBL_143203 (CHEMBL752220) Agonistic activity was determined against human Neuromedin B receptor transfected in HEK293 cells as accumulation levels of inositol phosphate
- ChEMBL_1650851 (CHEMBL3999985) Inhibition of IDO1 in IFN-gamma-stimulated human HeLa cells assessed as decrease in kynurenine levels after 48 hrs
- ChEMBL_1669766 (CHEMBL4019654) Inhibition of CrtN in Staphylococcus aureus Newman assessed as reduction in staphyloxanthin levels after 48 hrs by spectrophotometric analysis
- ChEMBL_1669774 (CHEMBL4019662) Inhibition of CrtN in Staphylococcus aureus Mu50 assessed as reduction in staphyloxanthin levels after 48 hrs by spectrophotometric analysis
- ChEMBL_1779341 (CHEMBL4236333) Agonist activity at recombinant rat Cb1 receptor expressed in HEK293 cells assessed as inhibition of forskolin-stimulated cAMP levels
- ChEMBL_1779342 (CHEMBL4236334) Agonist activity at recombinant human Cb2 receptor expressed in HEK293 cells assessed as inhibition of forskolin-stimulated cAMP levels
- ChEMBL_1784807 (CHEMBL4256324) Inhibition of IDO1 in IFN-gamma-stimulated human HeLa cells assessed as decrease in kynurenine levels after 48 hrs
- ChEMBL_1913238 (CHEMBL4415821) Inhibition of IDO1 in IFN-gamma-stimulated human HeLa cells assessed as decrease in kynurenine levels after 48 hrs
- ChEMBL_1924844 (CHEMBL4427800) Inhibition of CENP-E in human HeLa cells assessed as HH3 phosphorylated levels after 24 hrs by FACS analysis
- ChEMBL_1929601 (CHEMBL4432777) Inhibition of PI3Kdelta in human CCRF-SB cells assessed as reduction in AktS473 phosphorylation levels by Western blot analysis
- ChEMBL_1929604 (CHEMBL4432780) Inhibition of PI3Kdelta in human CCRF-SB cells assessed as reduction in S6 phosphorylation levels by Western blot analysis
- ChEMBL_473541 (CHEMBL939195) Antagonist activity at human adenosine A2B receptor expressed in CHO cells assessed as inhibition of NECA-stimulated cAMP levels
- ChEMBL_520157 (CHEMBL947845) Binding affinity to human recombinant BHMT expressed in Escherichia coli using variable levels of betaine substrate by Dixon plot
- ChEMBL_1891476 (CHEMBL4393303) Selective estrogen receptor down-regulator activity at FLAG-tagged ERalpha (unknown origin) expressed in HEK293 cells assessed as induction of ERalpha degradation by luciferase reporter gene assay
- ChEMBL_1557609 (CHEMBL3772526) Inhibition of PAK1 in human EBC1 cells assessed as reduction in levels of MEK phosphorylation at S298 residue after 2 hrs by HTRF assay
- ChEMBL_1664063 (CHEMBL4013744) Downregulation of ERalpha in human MCF-7 cells assessed as reduction of estradiol-induced GREB1 mRNA levels after 24 hrs by RT-PCR method
- ChEMBL_1664064 (CHEMBL4013745) Downregulation of ERalpha in human MCF-7 cells assessed as reduction of estradiol-induced PgR mRNA levels after 24 hrs by RT-PCR method
- ChEMBL_1664065 (CHEMBL4013746) Downregulation of ERalpha in human MCF-7 cells assessed as reduction of estradiol-induced pS2 mRNA levels after 24 hrs by RT-PCR method
- ChEMBL_1704370 (CHEMBL4055603) Mixed-type inhibition of electric eel AChE using varying levels of acetylthiocholine iodide as substrate measured for 2 mins by Lineweaver-Burk plot analysis
- ChEMBL_1801927 (CHEMBL4274219) Non-competitive inhibition of AChE in human erythrocytes using varying levels of acetylthiocholine as substrate measured for 1 min by Lineweaver-burk plot analysis
- ChEMBL_1843984 (CHEMBL4344411) Inhibition of human alpha-thrombin using varying levels of Tos-Gly-Pro-Arg-AMC-TFA as substrate by fluorescence based Michaelis-Menten equation analysis
- ChEMBL_2018481 (CHEMBL4672059) Non-competitive inhibition of recombinant human MIF assessed as inhibition constant using varying levels of p-hydroxyphenylpyruvate as substrate by Lineweaver-Burk plot analysis
- ChEMBL_2072564 (CHEMBL4728098) Mixed type inhibition of electric eel AChE assessed as inhibition constant using varying levels of acetylthiocholine as substrate by reciprocal Lineweaver-Burk plot analysis
- ChEMBL_2196078 (CHEMBL5108594) Non competitive type inhibition of human TS assessed as inhibition constant using varying levels of mTHF as substrate by UV-Vis spectroscopy based analysis
- AlphaScreen Assay When two proteins interact (one of which is conjugated to the donor bead and the other to the acceptor bead) the beads are brought into close proximity. Excitation of a photosensitizer in the donor bead results in the production of singlet oxygen that diffuses to and reacts with a chemiluminescent molecule in the acceptor bead. This results in the activation of fluorophores in the acceptor bead which results emitted light that can be detected. In this assay, GST-MDM2 (1-150) and full-length His6-wt-p53 were expressed in E. coli and affinity purified under non-denaturing conditions. To detect the MDM2-p53 interaction by the AlphaScreen assay (Perkin Elmer), GST-MDM2, His6-p53, and potential inhibitors were mixed in binding buffer, and incubated for 1 h at 24 deg C. Nickel acceptor beads and glutathione donor beads were added. Following 1 h incubation at 24 deg C, the mixture was analyzed in a fluorometer at an excitation wavelength of 680 nm. Nutlin-3a obtained from Cayman Chemical was used as a control. The IC50 values reported are from runs repeated four times.
- ChEMBL_1890267 (CHEMBL4392021) Competitive reversible inhibition of human placental microsome aromatase using varying levels of [1beta3H]-androstenedione as substrate measured after 5 mins in presence of NADPH by Dixon-plot analysis
- ChEMBL_1890268 (CHEMBL4392022) Competitive irreversible inhibition of human placental microsome aromatase using varying levels of [1beta3H]-androstenedione as substrate measured after 5 mins in presence of NADPH by Dixon-plot analysis
- ChEMBL_1630552 (CHEMBL3873258) Inhibition of rabbit muscle glycogen phosphorylase-b after 1 min in presence of varying levels of glucose-1-phosphate and constant concentration of glycogen and AMP by double reciprocal plot method
- ChEMBL_1511674 (CHEMBL3607427) Inhibition of EZH2 in human HeLa cells assessed as reduction in H3K27me3 levels incubated for 72 hrs by ELISA method
- ChEMBL_1929599 (CHEMBL4432775) Inhibition of PI3Kdelta in human SU-DHL-5 cells assessed as reduction in AktS473 phosphorylation levels by Western blot analysis
- ChEMBL_1929602 (CHEMBL4432778) Inhibition of PI3Kdelta in human SU-DHL-5 cells assessed as reduction in S6 phosphorylation levels by Western blot analysis
- ChEMBL_2538527 Inhibition of G9a in human PC-3 cells assessed as decrease in H3k2me2 levels after 72 hrs by Western blot analysis
- ChEMBL_436392 (CHEMBL904700) Activity at human A2B receptor expressed in HEK293 cells assessed as inhibition of NECA-induced intracellular cAMP levels by ELISA
- ChEMBL_436393 (CHEMBL904701) Activity at mouse A2B receptor expressed in CHO cells assessed as inhibition of NECA-induced intracellular cAMP levels by ELISA
- ChEMBL_458595 (CHEMBL942878) Antagonist activity at human vasopressin V2 receptor expressed in HEK293 cells assessed as inhibition of Arg-vasopressin-induced cAMP levels
- ChEMBL_458598 (CHEMBL942881) Antagonist activity at rat vasopressin V2 receptor expressed in HEK293 cells assessed as inhibition of Arg-vasopressin-induced cAMP levels
- ChEMBL_970231 (CHEMBL2406498) Inhibition of short-form RON (unknown origin) expressed in human HeLa cells assessed as phosphorylated RON levels after 4 hrs
- ChEMBL_1654016 (CHEMBL4003382) Mixed-type inhibition of Electrophorus electricus AChE pretreated for 15 mins followed by varying levels of acetylthiocholine iodide substrate addition by Lineweaver-Burk plot analysis
- ChEMBL_1720207 (CHEMBL4135207) Inhibition of Trypanosoma cruzi trypanothione reductase assessed as reduction in NADPH consumption using varying levels of trypanothione disulfide as substrate by Lineweaver-Burk plot analysis
- ChEMBL_1748392 (CHEMBL4182902) Inhibition of Ostrinia furnacalis Hex1 expressed in Pichia pastoris using varying levels of 4-MU-GlcNAc as substrate measured after 30 mins by fluorescence assay
- ChEMBL_1809917 (CHEMBL4309377) Inhibition of IDH1 R132C mutant (unknown origin) assessed as reduction in 2-HG levels after 90 mins in presence of NADPH by resazurin-based assay
- ChEMBL_1809918 (CHEMBL4309378) Inhibition of IDH1 R132G mutant (unknown origin) assessed as reduction in 2-HG levels after 90 mins in presence of NADPH by resazurin-based assay
- ChEMBL_1809919 (CHEMBL4309379) Inhibition of IDH1 R132H mutant (unknown origin) assessed as reduction in 2-HG levels after 90 mins in presence of NADPH by resazurin-based assay
- ChEMBL_1809920 (CHEMBL4309380) Inhibition of IDH1 R132S mutant (unknown origin) assessed as reduction in 2-HG levels after 90 mins in presence of NADPH by resazurin-based assay
- ChEMBL_1809921 (CHEMBL4309381) Inhibition of IDH2 R172K mutant (unknown origin) assessed as reduction in 2-HG levels after 90 mins in presence of NADPH by resazurin-based assay
- ChEMBL_1809922 (CHEMBL4309382) Inhibition of IDH2 R140Q mutant (unknown origin) assessed as reduction in 2-HG levels after 90 mins in presence of NADPH by resazurin-based assay
- ChEMBL_1830127 (CHEMBL4330001) Inhibition of recombinant human OGA expressed in Escherichia coli BL21 (DE3) using varying levels of pNP-beta-GlcNAc as substrate by Lineweaver-burk plot analysis
- ChEMBL_1891487 (CHEMBL4393314) Competitive-inhibition of MERS-CoV papain-like protease using varying levels of Z-Arg-Leu-Arg-Gly-Gly-AMC as substrate by Dixon-plot analysis
- ChEMBL_2114068 (CHEMBL4822918) Non competitive type inhibition of human AChE assessed as inhibition constant using varying levels of acetylthiocholine as substrate by double reciprocal Lineweaver-Burk plot analysis
- ChEMBL_878321 (CHEMBL2185949) Mixed type inhibition of Trypanosoma brucei TR using varying levels of trypanothione disulfide assessed as inhibition constant for enzyme-inhibitor complex by Lineweaver-Burk plot
- ChEMBL_1854944 (CHEMBL4355673) Inhibition of recombinant full length C-terminal His/FLAG tagged human HDAC1 (1 to 482 residues) expressed in baculovirus infected sf9 insect cells using p53 (379 to 382 residues) derived fluorogenic peptide RHKKAc as substrate preincubated for 5 to 10 mins followed by substrate addition and further incubated for 2 hrs by fluorescence assay
- ChEMBL_1774350 (CHEMBL4231342) Mixed-type inhibition of Trypanosoma brucei trypanothione reductase assessed as enzyme-inhibitor complex using varying levels of TS2 as substrate in presence of NADPH by Lineweaver-Burk plot analysis
- ChEMBL_2526304 Competitive inhibition of recombinant human GST-fused JAK1 (852 to 1142 residues) using 5-FAM-KKSRGDYMTMQIG as substrate in presence of varying levels of MgATP by Lineweaver-Burk plot analysis
- ChEMBL_2035579 (CHEMBL4689737) Antagonist activity at recombinant GST-tagged FXR LBD (unknown origin) assessed as inhibition of GW4064-induced Fluorecein-SRC2-2 coactivator peptide recruitment by LanthaScreen TR-FRET co-regulator assay
- ATR/ATRIP Kinase Assay The ATR/ATRIP enzymatic assay is performed as a TR-FRET-(HTRF™, Cisbio Bioassays) based 384-well assay. In a first step, purified human recombinant ATR/ATRIP (human ATR, full length, GenBank ID: NM_001184.3, and human ATRIP, full length, GenBank ID AF451323.1, co-expressed in a mammalian cell line) is incubated in assay buffer for 15 minutes at 22° C. with test compound at different concentrations or without test compound (as a negative control). The assay buffer contains 25 mM HEPES pH 8.0, 10 mM Mg(CH3COO)2, 1 mM MnCl2, 0.1% BSA, 0.01% Brij® 35, and 5 mM dithiothreitol (DTT). An Echo 555 (Labcyte) is used for dispensing of compound solutions. Then, in a second step, purified human recombinant cmyc-tagged p53 (human p53, full length, GenBank ID: BC003596, expressed in Sf21 insect cells) and ATP are added and the reaction mixture is incubated for 25-35 minutes, typically 25 minutes, at 22° C. The pharmacologically relevant assay volume is 5 μl. The final concentrations in the assay during incubation of the reaction mixture are 0.3-0.5 nM, typically 0.3 nM, ATR/ATRIP, 50 nM p53, and 0.5 μM ATP. The enzymatic reaction is stopped by the addition of EDTA. The generation of phosphorylated p53 as a result of the ATR mediated reaction in the presence of ATP is detected by using specific antibodies [labeled with the fluorophores europium (Eu) as donor and d2 as acceptor (Cisbio Bioassays)] enabling FRET. For this purpose, 2 μl of antibody-containing stop solution (12.5 mM HEPES pH 8.0, 125 mM EDTA, 30 mM sodium chloride, 300 mM potassium fluoride, 0.006% Tween-20, 0.005% Brij® 35, 0.21 nM anti-phospho-p53(Ser15)-Eu antibody, 15 nM anti-cmyc-d2 antibody) are added to the reaction mixture. Following signal development for 2 h the plates are analyzed in an EnVision (PerkinElmer) microplate reader using the TRF mode with laser excitation. Upon excitation of the donor europium at 340 nm the emitted fluorescence light of the acceptor d2 at 665 nm as well as from the donor Eu at 615 nm are measured. The amount of phosphorylated p53 is directly proportional to the ratio of the amounts of emitted light i.e. the ratio of the relative fluorescence units (rfu) at 665 nm and 615 nm. Data are processed employing the Genedata Screener software. In particular, IC50 values are determined in the usual manner by fitting a dose-response curve to the data points using nonlinear regression analysis.
- ChEBML_1685824 Mixed-type inhibition of bovine milk xanthine oxidase assessed as enzyme-substrate-inhibitor complex using varying levels of xanthine as substrate by Line-weaver burk plot analysis
- ChEMBL_1636689 (CHEMBL3879587) Uncompetitive inhibition of electric eel AChE using varying levels of acetylthiocholine iodide substrate preincubated for 20 mins followed by substrate addition by Lineweaver-Burk plot analysis
- ChEMBL_1653320 (CHEMBL4002686) Non-competitive inhibition of recombinant human alpha GAL-A using varying levels of 4-methylumbelliferyl alpha-D-galactopyranoside substrate at pH 4.6 Lineweaver-burk plot analysis
- ChEMBL_1653321 (CHEMBL4002687) Non-competitive inhibition of recombinant human alpha GAL-A using varying levels of 4-methylumbelliferyl alpha-D-galactopyranoside substrate at pH 7 Lineweaver-burk plot analysis
- ChEMBL_1685823 (CHEMBL4036302) Mixed-type inhibition of bovine milk xanthine oxidase assessed as enzyme-inhibitor complex using varying levels of xanthine as substrate by Line-weaver burk plot analysis
- ChEMBL_1719626 (CHEMBL4134626) Noncompetitive inhibition of human neutrophil elastase using varying levels of MeOSuc-AAPV-pNA as substrate measured after 30 mins by double-reciprocal Lineweaver-Burk plot analysis
- ChEMBL_1793338 (CHEMBL4265257) Activation of human Y1R expressed in HEK293 cells assessed as inhibition of isoproterenol-induced increase in intracellular cAMP levels by calcium 5 dye-based FLIPR assay
- ChEMBL_1793339 (CHEMBL4265258) Activation of human Y2R expressed in HEK293 cells assessed as inhibition of isoproterenol-induced increase in intracellular cAMP levels by calcium 5 dye-based FLIPR assay
- ChEMBL_1793340 (CHEMBL4265259) Activation of human Y4R expressed in HEK293 cells assessed as inhibition of isoproterenol-induced increase in intracellular cAMP levels by calcium 5 dye-based FLIPR assay
- ChEMBL_1793341 (CHEMBL4265260) Activation of human Y5R expressed in HEK293 cells assessed as inhibition of isoproterenol-induced increase in intracellular cAMP levels by calcium 5 dye-based FLIPR assay
- ChEMBL_1811410 (CHEMBL4310870) Competitive inhibition of human MAOB expressed in insect cell microsomes using varying levels of kynuramine as substrate measured after 20 mins by Lineweaver-burk plot analysis
- ChEMBL_1825766 (CHEMBL4325530) Reversible inhibition of recombinant human MAOA expressed in Pichia pastoris using varying levels of kynuramine as substrate measured after 5 mins by Michaelis-Menten equation analysis
- ChEMBL_1825767 (CHEMBL4325531) Reversible inhibition of recombinant human MAOB expressed in Pichia pastoris using varying levels of kynuramine as substrate measured after 5 mins by Michaelis-Menten equation analysis
- ChEMBL_1825768 (CHEMBL4325532) Irreversible inhibition of recombinant human MAOA expressed in Pichia pastoris using varying levels of kynuramine as substrate measured after 5 mins by Michaelis-Menten equation analysis
- ChEMBL_1825769 (CHEMBL4325533) Irreversible inhibition of recombinant human MAOB expressed in Pichia pastoris using varying levels of kynuramine as substrate measured after 5 mins by Michaelis-Menten equation analysis
- ChEMBL_878320 (CHEMBL2185948) Mixed type inhibition of Trypanosoma brucei TR using varying levels of trypanothione disulfide assessed as inhibition constant for enzyme-substrate-inhibitor complex by Lineweaver-Burk plot
- ATR Enzyme Assay I. Experimental Materials and Instruments1. ATR enzyme (Eurofins Pharma Discovery Services, 14-953M)2. GST-tagged P53 protein (Eurofins Pharma Discovery Services, 14-952M)3. 384-well plate (Geriner bio-one, 784075)4. U-bottom 96-well plate (Geriner bio-one, 651201)5. Anti-phospho-P53 protein antibody labeled with europium cryptate (cisbio, 61P08KAZ)6. Anti-GST antibody linked to d2 (cisbio, 61GSTDLB)7. ATP solution (Sigma, R0441)8. DTT (Sigma, D0632-259)9. HEPES (Sigma, 15630080)10. Microplate reader (Envision 2104 Multilabel Reader)II. Experimental Steps15 nM ATR enzyme, 80 nM P53 protein, 300 nM ATP (the final concentrations were 40 nM and 150 nM, respectively), and small molecule compounds of various concentrations (the final concentrations (nM) of the ten points were 2985.0, 895.5, 298.5, 110.56, 33.17, 11.06, 4.09, 1.23, 0.41 and 0.15, respectively, and the final dimethyl sulfoxide concentration was 0.498%) were mixed and incubated at room temperature for 90 minutes. 10 μL of 2× cocktail buffer was added to the mixture of ATR, compound and substrate in the assay plate (anti-phospho-p53-Eu and anti-GST-d2 were diluted in the assay buffer). The resulting mixture was centrifuged at 1000 rpm for 30 seconds, and incubated overnight at 4° C. in the dark (a total of 20 μl in each well). The FRET signal (endpoint) was measured in the Envision instrument (HTRF 665/612 ratio was calculated at 665 nm emission and 612 nm emission). Data were processed using GraphPad software.
- In Vitro MDM2 Assay The ability of the compounds to inhibit the interaction between p53 and MDM2 proteins was measured by an HTRF (homogeneous time-resolved fluorescence) assay in which recombinant GST-tagged MDM2 binds to a peptide that resembles the MDM2-interacting region of p53 (Lane et al). Binding of GST-MDM2 protein and p53-peptide (biotinyiated on its N-terminal) is registered by the FRET (fluorescence resonance energy transfer) between Europium (Eu)-labeled anti-GST antibody and streptavidin-conjugated Allophycocyanin (APC). Test is performed in black flat-bottom 384-well plates (Costar) in a total volume of 40 uL containing:90 nM biotinylate peptide, 160 ng/ml GST-MDM2, 20 nM streptavidin-APC (PerkinElmer Wallac), 2 nM Eu-labeled anti-GST-antibody (PerkmElmerWallac), 0.02% bovine serum albumin (BSA), 1 mM dithiothreitol (DTT) and 20 mM Tris-borate saline (TBS) buffer as follows: Add 10 ul. of GST-MDM2 (640 ng/ml working solution) in reaction buffer to each well. Add 10 uL diluted compounds (1:5 dilution in reaction buffer) to each well, mix by shaking. Add 20 uL biotinyiated p53 peptide (180 nM working solution) in reaction buffer to each well and mix on shaker. Incubate at 37° C. for 1 h. Add 20 11 L streptavidin-APC and Eii-anti-GST antibody mixture (6 nM Eu-anti-GST and 60 nM streptavidin-APC working solution) in TBS buffer with 0.02% BSA, shake at room temperature for 30 minutes and read using a TRF-capable plate reader at 665 and 615 nm (Victor 5, Perk in ElmcrWallac).
- ChEMBL_1581012 (CHEMBL3813017) Inhibition of Staphylococcus aureus PDF using formyl-methionine-alanine-serine as substrate assessed as increase in NADH levels by microtiter ELISA
- ChEMBL_1621818 (CHEMBL3864101) Noncompetitive inhibition of OGT (unknown origin) using variable UDP-GlcNAc levels and fixed CKII peptide level by UDP-Glo glycosyltransferase assay
- ChEMBL_1621819 (CHEMBL3864102) Noncompetitive inhibition of OGT (unknown origin) using fixed UDP-GlcNAc levels and variable CKII peptide level by UDP-Glo glycosyltransferase assay
- ChEMBL_1652997 (CHEMBL4002252) Inhibition of bovine xanthine oxidase assessed as reduction in uric acid levels using xanthine as substrate after 120 mins by spectrophotometry
- ChEMBL_1698421 (CHEMBL4049311) Inhibition of ATX in human whole blood assessed as decrease in LPA levels after 2 hrs by LC-MS/MS method
- ChEMBL_1729041 (CHEMBL4144319) Inhibition of p300/CBP in human PC3 cells assessed as reduction in acetylated H3K27 levels after 3 hrs by fluorescence assay
- ChEMBL_1735605 (CHEMBL4151141) Inhibition of PRMT5 in human MCF7 cells assessed as reduction in SmBB'-Rme2s levels after 48 hrs by Western blot analysis
- ChEMBL_1780211 (CHEMBL4237203) Activation of SOS1 in human HeLa cells assessed as increase in RAS-GTP levels after 30 mins by pull-down assay
- ChEMBL_1846170 (CHEMBL4346597) Inhibition of Flag-tagged PRMT6 (unknown origin) expressed in HEK293 cells incubated for 20 hrs assessed as reduction in H3R2me2a levels
- ChEMBL_1846173 (CHEMBL4346600) Inhibition of PRMT1 in human MCF7 cells assessed as reduction in H4R3me2a levels incubated for 2 days by fluorescence based assay
- ChEMBL_1925282 (CHEMBL4428354) Modulation of gamma secretase in HEK cells expressing APP Swedish mutant assessed as reduction in amyloid beta levels after 5 hrs
- ChEMBL_215802 (CHEMBL820091) Antagonistic activity towards human vanilloid receptor subtype 1 expressed in HEK293 cell membrane, as inhibition of agonist-induced intracellular [Ca2+] levels.
- ChEMBL_616846 (CHEMBL1100230) Inhibition of human COX2 in IL-1-beta-stimulated human RASF cells assessed as PGF2alpha levels after 50 mins by ELISA
- ChEMBL_686303 (CHEMBL1293059) Antagonist activity at human recombinant TRPV1 assessed as inhibition of capsaicin-induced in intracellular calcium levels by cell-based FLIPR assay
- ChEMBL_842361 (CHEMBL2091978) Inhibition of rat TRPM8 expressed in HEK293 cells assessed as reduction in 0.25 uM icilin-stimulated increase in intracellular Ca2+ levels
- ChEMBL_842362 (CHEMBL2091979) Inhibition of rat TRPM8 expressed in HEK293 cells assessed as reduction in 20 uM menthol-stimulated increase in intracellular Ca2+ levels
- ChEMBL_1774352 (CHEMBL4231344) Mixed-type inhibition of Trypanosoma brucei trypanothione reductase assessed as enzyme-inhibitor-substrate complex using varying levels of TS2 as substrate in presence of NADPH by Lineweaver-Burk plot analysis
- ChEMBL_1892975 (CHEMBL4394896) Inhibition of Mycobacterium tuberculosis H37Ra ATCC 25177 adenosine kinase using varying levels of [3H]Ado as substrate in presence of adenosine deaminase inhibitor deoxycoformycin by Line-weaver burk plot analysis
- Biological Activity Assay 2 NCI-H358 lung adenocarcinoma cells may be used to measure MCT4 activity in cells with high native levels of MCT4 and low levels of MCT1 and are known to those with skill in the art. Preparation of BCECF-loaded cells and lactate transport activity may be determined as described for Assay 1.
- ChEMBL_1685824 (CHEMBL4036303) Mixed-type inhibition of bovine milk xanthine oxidase assessed as enzyme-substrate-inhibitor complex using varying levels of xanthine as substrate by Line-weaver burk plot analysis
- ChEMBL_1715070 (CHEMBL4125119) Mixed-type inhibition of calf intestinal alkaline phosphatase assessed as enzyme-inhibitor dissociation constant using varying levels of p-NPP as substrate by Lineweaver-Burk plot analysis
- ChEMBL_1720205 (CHEMBL4135205) Non-competitive inhibition of Trypanosoma cruzi trypanothione reductase assessed as reduction in NADPH consumption using varying levels of trypanothione disulfide as substrate by Lineweaver-Burk plot analysis
- ChEMBL_1720206 (CHEMBL4135206) Mixed-type inhibition of Trypanosoma cruzi trypanothione reductase assessed as reduction in NADPH consumption using varying levels of trypanothione disulfide as substrate by Lineweaver-Burk plot analysis
- ChEMBL_1748391 (CHEMBL4182901) Inhibition of recombinant human OGA expressed in Escherichia coli BL21(DE3) using varying levels of 4-MU-GlcNAc as substrate measured after 30 mins by fluorescence assay
- ChEMBL_1906171 (CHEMBL4408529) Non-competitive inhibition of electric eel AChE using varying levels of acetylthiocholine iodide as substrate measured at 2 mins interval for 10 mins by Dixon plot analysis
- ChEMBL_1906172 (CHEMBL4408530) Non-competitive inhibition of equine serum BuChE using varying levels of butyrylthiocholine iodide as substrate measured at 2 mins interval for 10 mins by Dixon plot analysis
- ChEMBL_2526306 Inhibition of recombinant human GST-tagged JAK3 (781 to 1124 residues) expressed in insect cells using FITC-KGGEEEEYFE as substrate in presence of varying levels of MgATP by Lineweaver-Burk plot analysis
- ChEMBL_1472824 (CHEMBL3419486) Agonist activity at human adenosine A2A receptor expressed in CHO cells assessed as stimulation of [3H]cAMP levels by scintillation counting method
- ChEMBL_1472825 (CHEMBL3419487) Agonist activity at human adenosine A2B receptor expressed in CHO cells assessed as stimulation of [3H]cAMP levels by scintillation counting method
- ChEMBL_1579150 (CHEMBL3811870) Inhibition of GLS1 in human PC3 cells assessed as cellular glutamate depletion levels after 6 hrs by Glutamate Oxidase/ AmplexRed coupled assay
- ChEMBL_1713890 (CHEMBL4123939) Inhibition of ATX in human serum assessed as reduction in LPA 18:1 levels after 3 hrs by LC-MS/MS analysis
- ChEMBL_1779720 (CHEMBL4236712) Agonist activity at recombinant human TRPV1 expressed in HEK293 cells assessed as induction of Ca2+ levels by Fuo-4-AM based spectrofluorimetry
- ChEMBL_1809898 (CHEMBL4309358) Inhibition of IDH1 in human HT1080 cells assessed as decrease in 2-HG levels after 48 hrs by LC-MS/MS analysis
- ChEMBL_1848554 (CHEMBL4349095) Inhibition of TTK in human CAL51 cells assessed as decrease in phosphorylated TTK T686 levels after 1 hr by Western blot analysis
- ChEMBL_1977334 (CHEMBL4610469) Modulatory activity of gamma secretase in human SKNBE2 cells assessed as decrease in Abeta42 levels incubated for 6 hrs by sandwich ELISA
- ChEMBL_2359400 Inhibition of human EZH2 in human MCF-10A cells assessed as reduction in H3K27me3 levels incubated for 72 hrs by Western blot analysis
- ChEMBL_616845 (CHEMBL1100229) Inhibition of human prostacyclin synthase in IL-1-beta-stimulated human RASF cells assessed as PGF1alpha levels after 50 mins by ELISA
- ChEMBL_842221 (CHEMBL2091726) Inhibition of rat TRPA1 expressed in HEK293 cells assessed as reduction in 100 uM allyl isothiocyanate-stimulated increase in intracellular Ca2+ levels
- Ataxia-Telangiectasia Mutated (ATM) Biochemical Assay Using Reaction Biology's HTRF Kinase Assay Protocol The assay detects kinase activity by antibody based HTRF assay. The GST-tagged p53 protein serves as substrate and Eu-labeled-antibody with specific phosphorylation site binds upon phosphorylation which is monitored by HTRF from the anti-GST-d2.Reagent: Reaction buffer; 50 mM Hepes (pH 7.5), 150 mM NaCl, 10 mM MnCl2, 1% Glycerol, 0.01% Brij35, 1 mM DTT, 1% DMSO.Substrate: Human p53 (aa2-393), RBC produced in E. coli, N-terminal GST-Tag, MW=70.42 kDa.[0429]Detection; MAb Anti-p53-pS15-Eu cryptate, CisBio cat #61P08KAZ; MAb Anti GST-d2Reaction Procedure: 1. Deliver 2× Enzyme in wells of reaction plate except No E control wells. Add buffer instead; 2. Deliver compounds in 100% DMSO into the kinase reaction mixture by Acoustic technology (Echo550; nanoliter range), incubate for 30 min at room temperature; 3. Deliver 2× substrate+ATP mixture into the reaction mixture to initiate the reaction; 4. Incubate for 2 hours at room temperature.
- FRET Assay An HDM2 FRET assay was developed to assess the compounds' inhibitory activity towards binding of p53 protein. A truncated version of HDM2 with residues 17 to 125 (containing p53 binding surface, Science (1994) 265, 346-355), with N-terminal His and Thioredoxin tag was generated in pET32a expression vector and expressed in E. coli strain BL21(DE3)Rosetta. Protein was purified using Ni-affinity chromatography, followed by size exclusion chromatography using Superdex 75 26/60 column. To assess inhibition of p53 binding to HDM2, a FITC labeled 8-mer peptide (sequence: Ac-Phe-Arg-Dpr-Ac6c-(6-Br)Trp-Glu-Glu-Leu-NH2; Anal Biochem. 2004 Aug. 1; 331(1):138-46) with strong affinity towards p53 binding pocket of HDM2 was used. The HDM2 assay buffer contained 1× Phosphate Buffered Saline (Invitrogen, Cat#14190), 0.01% BSA (Jackson ImmunoResearch, Cat#001-000-162), 0.01% Tween-20. In the 1× assay buffer recombinant HDM2 protein, peptide and Lumi4-Tb Cryptate-conjugate mouse anti-6×His antibody (cisbio, Cat# Tb61HISTLB) were added and transferred to ProxiPlate PLUS (PerkinElmer, Cat#6008269), containing compounds so that final DMSO concentration is 0.1%. Final concentrations of reagents in the assay wells are 0.5 nM HDM2, 0.25 nM anti HIS (Tb label) antibody and 3 nM peptide. After two hour incubation at room temperature in a humidified chamber plates were read on EnVision plate reader with the following settings: excitation: UV, 340 nM, two emission filters: 520 nm and 495 nm respectively. Ratio of em520/em495 was used to calculate % inhibition and to obtain IC50 with 4-parameter logistic equation.
- FRET Assay Methods: An HDM2 FRET assay was developed to assess the compounds' inhibitory activity towards binding of p53 protein. A truncated version of HDM2 with residues 17 to 125 (containing p53 binding surface, Science (1994) 265, 346-355), with N-terminal His and Thioredoxin tag was generated in pET32a expression vector and expressed in E. coli strain BL21(DE3)Rosetta. Protein was purified using Ni-affinity chromatography, followed by size exclusion chromatography using Superdex 75 26/60 column. To assess inhibition of p53 binding to HDM2, a FITC labeled 8-mer peptide (SEQ ID NO:1: Ac-Phe-Arg-Dpr-Ac6c-(6-Br)Trp-Glu-Glu-Leu-NH2; Anal Biochem. 2004 Aug. 1; 331(1):138-46) with strong affinity towards the p53 binding pocket of HDM2 was used. The HDM2 assay buffer contained 1× Phosphate Buffered Saline (Invitrogen, Cat#14190), 0.01% BSA (Jackson ImmunoResearch, Cat#001-000-162), 0.01% Tween-20. In the 1× assay buffer recombinant HDM2 protein, peptide and Lumi4-Tb Cryptate-conjugate mouse anti-6×His antibody (cisbio, Cat#Tb61HISTLB) were added and transferred to ProxiPlate PLUS (PerkinElmer, Cat#6008269), containing compounds so that final DMSO concentration is 0.1%. Final concentrations of reagents in the assay wells are 0.5 nM HDM2, 0.25 nM anti HIS (Tb label) antibody and 3 nM peptide. After two hour incubation at room temperature in a humidified chamber plates were read on EnVision plate reader with the following settings: excitation: UV, 340 nM, two emission filters: 520 nm and 495 nm respectively. Ratio of em520/em495 was used to calculate % inhibition and to obtain IC50 with 4-parameter logistic equation.
- In Vitro Evaluation TR/ATRIP(h) was incubated in assay buffer containing 50 nM GST-cMyc-p53 and Mg/ATP (according to concentration required). The reaction was initiated by adding Mg/ATP mixture. After incubating for 30 min at room temperature, a stop solution containing EDTA to was added to terminate the reaction. Finally, detecting buffer containing d2-labeled anti-GST monoclonal antibody and europium-labeled anti-phospho Ser15 antibody against phosphorylated p53 were added. Then the plate was read in time-resolved fluorescence mode and homogeneous time resolution was performed.The fluorescence (HTRF) signal was determined according to the formula HTRF=10000×(Em665 nm/Em620 nm).
- ChEMBL_1623122 (CHEMBL3865474) Competitive inhibition of SETD8 (unknown origin) using biotin-labeled H4 (1 to 24 residues) as substrate after 1 hr in presence of varying levels of [3H]SAM by Lineweaver-Burk plot analysis
- ChEMBL_1715560 (CHEMBL4125609) Competitive inhibition of recombinant Bacillus cereus BC1974 expressed in Escherichia coli using varying levels of GlcNAc5 as substrate measured after 5 mins in presence of fluorescamine by Line-weaver burk plot analysis
- ChEMBL_1715561 (CHEMBL4125610) Competitive inhibition of recombinant Bacillus cereus BC1960 expressed in Escherichia coli using varying levels of GlcNAc5 as substrate measured after 10 mins in presence of fluorescamine by Line-weaver burk plot analysis
- ChEMBL_2526305 Inhibition of recombinant human GST-tagged JAK2 (809 to 1153+9 residues) expressed in insect cells using FITC-KGGEEEEYFE as substrate in presence of varying levels of MgATP by Lineweaver-Burk plot analysis
- ChEMBL_1704274 (CHEMBL4055507) Competitive inhibition of porcine liver carboxylesterase using varying levels of 4-nitrophenol acetate as substrate preincubated for 10 mins followed by substrate addition by Lineweaver-Burk plot analysis
- ChEMBL_1715071 (CHEMBL4125120) Mixed-type inhibition of calf intestinal alkaline phosphatase assessed as enzyme-substrate-inhibitor dissociation constant using varying levels of p-NPP as substrate by Lineweaver-Burk plot analysis
- ChEMBL_1723560 (CHEMBL4138838) Mixed-type inhibition of electric eel AChE using varying levels of acetylthiocholine iodide as substrate preincubated for 15 mins followed by substrate addition by Lineweaver-Burk plot analysis
- ChEMBL_1739235 (CHEMBL4154985) Non-competitive inhibition of electric eel AChE using varying levels of acetylthiocholine iodide as substrate pretreated for 5 mins followed by substrate addition by Lineweaver-Burk plot analysis
- ChEMBL_1740535 (CHEMBL4156285) Non-competitive inhibition of recombinant human AChE expressed in HEK293 cells assessed as enzyme-substrate-inhibitor complex using varying levels of acetylthiocholine as substrate by Dixon plot analysis
- ChEMBL_2063498 (CHEMBL4718751) Competitive inhibition of rat liver microsomal S1PL using varying levels of [3-3H]-D(+) erythro-sphinganine-1-phosphate as substrate incubated for 15 mins by Dixon plot analysis
- ChEMBL_2520733 Competitive inhibition of human neutrophil elastase using varying levels of MeOSuc-AAPV-MCA as substrate measured at 60 mins interval for up to several hrs by Dixon plot analysis
- ChEMBL_2520735 Competitive inhibition of mouse neutrophil elastase using varying levels of MeOSuc-AAPV-MCA as substrate measured at 60 mins interval for up to several hrs by Dixon plot analysis
- ATR Kinase Activity Assay The amount of phosphorylated P53 protein was determined using a time-resolved fluorescence system. Before the starting of the experiment, the following working solutions were prepared as needed: 1× reaction buffer (20 mM HEPES PH8.0, 1% glycerol, 0.01% Brij-35), dilution buffer (20 mM HEPES PH8.0, 1% glycerol, 0.01% Brij-35, 5 mM DTT and 1% BSA), stop buffer (20 mM HEPES PH8.0, 1% glycerol, 0.01% Brij-35, 250 mM EDTA), detection buffer (50 mM HEPES PH7.0, 150 mM NaCl, 267 mM KF, 0.1% sodium cholate, 0.01% Tween-20, 0.0125% sodium azide). The clinically investigational drug AZD6738 (purchased from Selleck) was used as a positive control.The specific steps of the assay are as follows:4× gradient dilution compound solutions were prepared with the 1× reaction buffer to obtain 9 different compound concentrations, and 2.5 μL of the 4× gradient dilution compound solutions are added to a 384-well assay plate (784075, Greiner). 4× p53 substrate working solution (40 nM) was prepared with the 1× reaction buffer and 2.5 μL of the 4× p53 substrate working solution was added to the 384-well assay plate. 4×ATR/ATRIP working solution (12.8 ng/μL) was prepared with the dilution buffer and 2.5 μL of the 4×ATR/ATRIP working solution was added to the 384-well assay plate. 4×ATP working solution (2 mM) was prepared with deionized water, and 2.5 μL of the 4×ATP working solution was added to the 384-well assay plate. The plate was incubated at room temperature for 30 minutes in the dark. 5 μL of the stop solution was added to the 384 assay plate. Finally, 5 μL of the detection mixture (0.09 ng/μL of Anti-phospho-p53(ser15)-K and 6 ng/μL of Anti-GST-d2) to the 384-well assay plate. After incubation at room temperature overnight, the fluorescence signals were detected using an M5e (Molecular Device) instrument (excitation wavelength of 320 nm, emission wavelengths of 620 nm and 665 nm).
- ChEBML_1690006 Inhibition of ATX in rat plasma assessed as reduction in plasma lysophosphatidic acid 18:2 levels after 2 hrs by LC-MS/MS method
- ChEMBL_1677511 (CHEMBL4027654) Inhibition of CrtN in Staphylococcus aureus Newman assessed as reduction in staphyloxanthin levels after 48 hrs by spectrophotometric method-based pigment inhibition assay
- ChEMBL_1731875 (CHEMBL4147411) Inhibition of recombinant human PDE4D expressed in Escherichia coli assessed as increase in cAMP levels after 60 mins by BIOMOLGREEN dye-based assay
- ChEMBL_1784286 (CHEMBL4255803) Inhibition of CrtN in Staphylococcus aureus Newman assessed as reduction in staphyloxanthin levels after 48 hrs by spectrophotometric method-based pigment inhibition assay
- ChEMBL_1789224 (CHEMBL4260958) Agonist activity at human dopamine D1 receptor expressed in HEK293T cells assessed as induction of cAMP levels after 30 mins by HTRF assay
- ChEMBL_1848553 (CHEMBL4349094) Inhibition of TTK in human CAL51 cells assessed as decrease in phosphorylated KNL-1 T943 levels after 1 hr by Western blot analysis
- ChEMBL_2268607 Inhibition of human EZH2 in human MCF-10A cells assessed as reduction in H3K27me3 levels incubated for 72 hrs by In-cell western assay
- ChEMBL_2571750 Inhibition of human EZH2 in human MCF-10A cells assessed as reduction in H3K27me3 levels incubated for 72 hrs by In-cell western assay
- ChEMBL_616847 (CHEMBL1100231) Inhibition of human thromboxane A synthase in IL-1-beta-stimulated human RASF cells assessed as TXB2 levels after 50 mins by ELISA
- ChEMBL_629245 (CHEMBL1121359) Antagonist activity at human TRPV1 expressed in tetracycline-stimulated HEK293 cells assessed as inhibition of capsaicin-induced intracellular calcium levels by fluorimetric assay
- ChEMBL_877077 (CHEMBL2186813) Inhibition of human thymidylate synthase expressed in Escherichia coli incubated for 1 hr using varying dUMP levels by spectrophotometry based Lineweaver-Burk method
- ChEMBL_877078 (CHEMBL2186814) Inhibition of human thymidylate synthase expressed in Escherichia coli incubated for 1 hr using varying mTHF levels by spectrophotometry based Lineweaver-Burk method
- ChEMBL_946397 (CHEMBL2339174) Inhibition of Escherichia coli lipopolysaccharide-induced COX2 activity in mouse J774 cells assessed as decrease in PGE2 levels after 24 hrs by RIA
- ChEMBL_1630550 (CHEMBL3873256) Competitive inhibition of rabbit muscle glycogen phosphorylase-a assessed as release of inorganic phosphate using varying levels of glucose-1-phosphate and constant concentration of AMP and glycogen measured after 1 min by Dixon plot method
- MCT4-Mediated Lactate Transport in NCI-H358 Lung Adenocarcinoma Cell NCI-H358 lung adenocarcinoma cells may be used to measure MCT4 activity in cells with high native levels of MCT4 and low levels of MCT1 and are known to those with skill in the art. Preparation of BCECF-loaded cells and lactate transport activity may be determined as described for Assay 1.
- ChEMBL_1782681 (CHEMBL4254198) ATP-competitive inhibition of human CK2-alpha/beta expressed in Escherichia coli BL21(DE3) using RRRDDDSDDD peptide as substrate after 15 mins in presence of varying levels of ATP by capillary electrophoresis method
- ChEBML_1691469 Mixed-type inhibition of recombinant human MAO-A assessed as reduction in 4-hydroxyquinoline formation using varying levels of kynuramine as substrate after 20 mins by Lineweaver-Burk plot analysis
- ChEMBL_1659738 (CHEMBL4009350) Uncompetitive inhibition of full length recombinant human ALDH1A1 expressed in Escherichia coli BL21(DE3) using propionaldehyde as substrate in presence of varying levels NAD+ by Lineweaver-Burk plot analysis
- ChEMBL_1691468 (CHEMBL4042117) Competitive inhibition of recombinant human MAO-A assessed as reduction in 4-hydroxyquinoline formation using varying levels of kynuramine as substrate after 20 mins by Lineweaver-Burk plot analysis
- ChEMBL_1795339 (CHEMBL4267456) Non-competitive inhibition of recombinant HisA-tagged Trypanosoma cruzi trans-sialidase expressed in Escherichia coli BL21 (DE3) using varying levels of MuNANA as substrate by Lineweaver-burk plot analysis
- ChEMBL_2058591 (CHEMBL4713592) Covalent inhibition of N-terminal GST tagged human ALK cytoplasmic domain (1058 to 1620 residues) expressed in Sf21 cells incubated for 2 hrs in presence of high ATP levels
- ChEMBL_2124395 (CHEMBL4833628) Inhibition of gamma secretase (unknown origin) expressed in HEK293 cells assessed as reduction of A-beta40 levels using C100-FLAG as substrate incubated for 2 hrs by sandwich ELISA
- ATR Enzyme Assay In this experiment, the phosphorylation level of substrate protein P53 (Eurofins, 14-952) was detected by HTRF technology to measure the activity of ATR/ATRIP (Eurofins, 14-953) kinase. Reaction buffer (25 mM HEPES pH 8.0, 0.01% Brij-35, 1% Glycerol, 5 mM DTT, 1 mg/mL BSA), termination buffer (12.5 mM HEPES pH 8.0, 0.005% Brij-35, 0.5% Glycerol, 250 mM EDTA) and assay buffer (50 mM HEPES pH 7.0, 150 mM NaCl, 267 mM KF, 0.1% sodium cholate, 0.01% Tween 20) were prepared in advance. ATR/ATRIP was diluted with reaction buffer to a working solution of 2 ng/μL, and the substrate protein P53 was diluted with reaction buffer to a working solution of 80 nM, and 4 nM of ATP (Sigma, A2383) working solution (containing 40 mM MnCl2) was prepared with reaction buffer. The compound was 3-fold diluted with DMSO, then diluted with reaction buffer into a working solution, then added to a 384-well plate at 2.5 μL/well, and centrifuged at 1500 rpm for 40 s. Then 2.5 μL of ATR/ATRIP working solution, P53 working solution and ATP working solution were added into the 384-well plate, centrifuged at 1500 rpm for 40 s and reacted at room temperature for 30 minutes.
- ChEMBL_2281749 Binding affinity to human Gal-1 assessed as decrease in Gal-1 levels by ELISA
- ChEMBL_1613481 (CHEMBL3855281) Activity at human muscarinic acetylcholine receptor M1 transfected in CHO-K1 cells assessed as intracellular calcium levels in presence of acetylcholine by FLIPR assay
- ChEMBL_1613482 (CHEMBL3855282) Activity at human muscarinic acetylcholine receptor M3 transfected in CHO-K1 cells assessed as intracellular calcium levels in presence of acetylcholine by FLIPR assay
- ChEMBL_1622747 (CHEMBL3865099) Inhibition of ATX in human plasma assessed as decrease in LPA C18:1 levels after 24 hrs by horseradish peroxidase/choline oxidase-coupled assay
- ChEMBL_1657189 (CHEMBL4006659) Mixed-type inhibition of electric eel AChE preincubated for 5 mins followed by varying levels acetylthiocholine iodide substrate addition by Lineweaver-Burk plot analysis
- ChEMBL_1666008 (CHEMBL4015804) Inhibition of PI3Kdelta in human PBMC assessed as reduction in anti-human CD28 antibody stimulated IL-6 secretion levels after 72 hrs by ELISA
- ChEMBL_1809881 (CHEMBL4309341) Inhibition of IDH1 RI32C mutant in human HT1080 cells assessed as reduction in intracellular 2-HG levels after 24 hrs by LC-MS analysis
- ChEMBL_1887245 (CHEMBL4388922) Inhibition of gamma secretase in human SKNBE2 cells expressing human APP695 assessed as increase in Abeta38 levels incubated for 18 hrs by sandwich ELISA
- ChEMBL_1898208 (CHEMBL4400243) Agonist activity at human GLP1 receptor expressed in HEK293 cells assessed as induction of cAMP levels after 20 mins by time-resolved fluorescence analysis
- ChEMBL_1924989 (CHEMBL4427945) Inhibition of KDM5A in human PC9 cells assessed as increase in H3K4me3 levels preincubated for 4 days measured up to 24 hrs by ELISA
- ChEMBL_1925279 (CHEMBL4428351) Inhibition of gamma secretase in HEK293 cells expressing APP 695 assessed as reduction in amyloid beta levels after 5 hrs by Western blot analysis
- ChEMBL_38169 (CHEMBL650240) Beta-2 adrenergic receptor agonist activity was determined by a measurement of increased cAMP levels in CHO cells expressing human Beta-2 adrenergic receptor
- ChEMBL_629502 (CHEMBL1120791) Antagonist activity at rat NR1/NR2A receptor expressed in BHK21 cells assessed as inhibition of (S)-Glu-induced intracellular calcium levels by FLIPR assay
- ChEMBL_1691975 (CHEMBL4042624) Inhibition of quorum sensing regulator protein RhlR in Pseudomonas aeruginosa PAO1 harboring reporter plasmid rhlA-gfp assessed as reduction in rhlA expression measured every 15 mins for 12 hrs by GFP-fluorescence assay
- ChEMBL_1691469 (CHEMBL4042118) Mixed-type inhibition of recombinant human MAO-A assessed as reduction in 4-hydroxyquinoline formation using varying levels of kynuramine as substrate after 20 mins by Lineweaver-Burk plot analysis
- ChEMBL_1691471 (CHEMBL4042120) Mixed-type inhibition of recombinant human MAO-B assessed as reduction in 4-hydroxyquinoline formation using varying levels of kynuramine as substrate after 20 mins by Lineweaver-Burk plot analysis
- ChEMBL_1992584 (CHEMBL4626319) Competitive inhibition of Mycobacterium tuberculosis PTPB expressed in Escherichia coli BL21 (DE3) using varying levels of p-nitrophenyl phosphate as substrate measured after 30 mins by Lineweaver-Burk plot analysis
- ChEMBL_1659745 (CHEMBL4009357) Non-competitive inhibition of full length recombinant human ALDH2 expressed in Escherichia coli assessed as reduction in dehydrogenase activity using varying levels of propionaldehyde as substrate in presence of NAD+ by Lineweaver-Burk plot analysis
- ChEMBL_2124396 (CHEMBL4833629) Noncompetitive inhibition of gamma secretase (unknown origin) expressed in HEK293 cells assessed as reduction of A-beta40 levels using varying level of C100-FLAG as substrate incubated for 2 hrs by double reciprocal plot analysis
- Assay 2: MCT4-Mediated Lactate Transport in NCI-H358 Lung Adenocarcinoma Cells NCI-H358 lung adenocarcinoma cells may be used to measure MCT4 activity in cells with high native levels of MCT4 and low levels of MCT1 and are known to those with skill in the art. Preparation of BCECF-loaded cells and lactate transport activity may be determined as described for Assay 1.
- Assay 3: MCT4-Mediated Lactate Transport in MDA-MB-231 Breast Cancer Cells MDA-MB-231 breast cancer cells may be used to measure MCT4 activity in cells with high native levels of MCT4 and low levels of MCT1 and are known to those with skill in the art. Preparation of BCECF-loaded cells and lactate transport activity may be determined as described for Assay 1.
- MCT4-Mediated Lactate Transport in MDA-MB-231 Breast Cancer Cells (Assay 3) MDA-MB-231 breast cancer cells may be used to measure MCT4 activity in cells with high native levels of MCT4 and low levels of MCT1 and are known to those with skill in the art. Preparation of BCECF-loaded cells and lactate transport activity may be determined as described for Assay 1.
- MCT4-Mediated Lactate Transport in NCI-H358 Lung Adenocarcinoma Cells (Assay 2) NCI-H358 lung adenocarcinoma cells may be used to measure MCT4 activity in cells with high native levels of MCT4 and low levels of MCT1 and are known to those with skill in the art. Preparation of BCECF-loaded cells and lactate transport activity may be determined as described for Assay 1.
- ChEMBL_1672745 (CHEMBL4022774) Inhibition of AICARFT in human NCI-H460 cells assessed as increase in ZMP levels using low folate media after 16 hrs by LC-MS method
- ChEMBL_1672757 (CHEMBL4022786) Inhibition of AICARFT in human NCI-H460 cells assessed as increase in ZMP levels using regular folate media after 16 hrs by LC-MS method
- ChEMBL_1677514 (CHEMBL4027657) Inhibition of CrtN in methicillin-resistant Staphylococcus aureus Mu50 assessed as reduction in staphyloxanthin levels after 48 hrs by spectrophotometric method-based pigment inhibition assay
- ChEMBL_1784288 (CHEMBL4255805) Inhibition of CrtN in vancomycin-intermediate Staphylococcus aureus Mu50 assessed as reduction in staphyloxanthin levels after 48 hrs by spectrophotometric method-based pigment inhibition assay
- ChEMBL_1784289 (CHEMBL4255806) Inhibition of CrtN in linezolid-resistant Staphylococcus aureus NRS271 assessed as reduction in staphyloxanthin levels after 48 hrs by spectrophotometric method-based pigment inhibition assay
- ChEMBL_1809899 (CHEMBL4309359) Inhibition of IDH1 RI32H mutant in human HT1080 cells assessed as reduction in intracellular 2-HG levels after 24 hrs by resazurin fluorescence based assay
- ChEMBL_1809900 (CHEMBL4309360) Inhibition of IDH1 RI32C mutant in human U87 cells assessed as reduction in intracellular 2-HG levels after 24 hrs by resazurin fluorescence based assay
- ChEMBL_1821297 (CHEMBL4320957) Inhibition of 5'-biotin labeled duplex forked-DNA binding to BLM (unknown origin) assessed as increase in unbound DNA levels after 1 hr by ELISA
- ChEMBL_1827362 (CHEMBL4327236) Agonist activity at recombinant human CB2R expressed in CHOK1 cells assessed as inhibition of NKH477-stimulated intracellular cAMP levels after 30 mins by chemiluminescent assay
- ChEMBL_1880797 (CHEMBL4382191) Induction of ERalpha protein degradation in human MCF7 cells assessed as reduction in ERalpha protein levels incubated for 24 hrs by In-cell western assay
- ChEMBL_2057905 (CHEMBL4712906) Inhibition of OGA in human SH-SY5Y cells assessed as increase in O-GIcNAcylated protein levels incubated for 48 hrs by in-cell Western assay
- ChEMBL_2094590 (CHEMBL4775853) Inhibition of A2AR (unknown origin) expressed in CHO cells assessed as reduction in NECA-stimulated cellular cAMP levels incubated for 90 mins by FRET assay
- ChEMBL_2110462 (CHEMBL4819312) Inhibition of G9a in human MDA-MB-231 cells assessed as effect on H3K9Me2 levels incubated for 48 hrs by immunofluorescence in-cell western assay
- ChEMBL_215804 (CHEMBL820093) Functional antagonistic activity against human vanilloid receptor subtype 1 in HEK293 cell membranes was determined as inhibition of agonist-induced increases in intracellular [Ca2+] levels
- ChEMBL_497565 (CHEMBL998622) Antagonist activity at human CXCR3 expressed in HEK293T cells coexpressing Galphaqi5 assessed as inhibition of CXCL10-induced [3H]inositol phosphate levels by liquid scintillation counting
- ChEMBL_641610 (CHEMBL1176312) Antagonist activity at human recombinant TRPV1 receptor expressed in human 1321 cells assessed as inhibition of capsaicin-induced in intracellular calcium levels by FLIPR assay
- ChEBML_1624058 Inhibition of GST-tagged c-Met (unknown origin) assessed as phosphotyrosine levels preincubated foe 15 mins followed by addition of poly(glutamic acid-tyrosine (4:1)) as substrate and ATP by ELISA
- ChEMBL_1714535 (CHEMBL4124584) Mixed-type inhibition of equine serum BChE using varying levels of butyrylthiocholine iodide as substrate pretreated for 10 mins followed by substrate addition measured by Lineweaver-Burk double reciprocal plot analysis
- ChEMBL_1824460 (CHEMBL4324224) Inhibition of NS5A in HCV genotype 2a infected in human Huh7.5/J6/JFH1/EMCVIRES/hRlucNeo cells assessed as inhibition of replicon levels incubated for 72 hrs by luciferase reporter gene assay
- ChEMBL_1830125 (CHEMBL4329999) Inhibition of recombinant human OGA expressed in Escherichia coli BL21 (DE3) using varying levels of 4-Mu-GlcNAc as substrate measured after 30 mins by fluorescence assay by dixon plot analysis
- ChEMBL_424171 (CHEMBL907862) Agonist activity at human beta3 adrenergic receptor expressed in CHO cells assessed as cAMP levels
- ChEMBL_728638 (CHEMBL1685768) Agonist activity at human S1P1 receptor expressed in CHO cells assessed as intracellular calcium levels
- ChEMBL_728639 (CHEMBL1685769) Agonist activity at human S1P2 receptor expressed in CHO cells assessed as intracellular calcium levels
- ChEMBL_728640 (CHEMBL1685770) Agonist activity at human S1P3 receptor expressed in CHO cells assessed as intracellular calcium levels
- ChEMBL_728641 (CHEMBL1685771) Agonist activity at human S1P5 receptor expressed in CHO cells assessed as intracellular calcium levels
- ChEMBL_753427 (CHEMBL1799857) Agonist activity at human S1P1 receptor expressed in CHO cells assessed as intracellular calcium levels
- ChEMBL_753428 (CHEMBL1799858) Agonist activity at human S1P3 receptor expressed in CHO cells assessed as intracellular calcium levels
- ChEMBL_875033 (CHEMBL2184351) Antagonist activity at human 5HT4ER expressed in CHO cells assessed as reduction in cAMP levels
- ChEMBL_1624058 (CHEMBL3866470) Inhibition of GST-tagged c-Met (unknown origin) assessed as phosphotyrosine levels preincubated foe 15 mins followed by addition of poly(glutamic acid-tyrosine (4:1)) as substrate and ATP by ELISA
- ChEMBL_1662598 (CHEMBL4012279) Competitive inhibition of bovine xanthine oxidase using varying levels of xanthine as substrate preincubated for 10 mins followed by substrate addition measured for 15 mins by Lineweaver-Burk plot/Dixon plot analysis
- ChEMBL_1779756 (CHEMBL4236748) Mixed-type inhibition of equine serum BuChE using varying levels of butyrylthiocholine iodide as substrate preincubated for 10 mins followed by substrate addition measured for 5 mins by Lineweaver-Burk plot analysis
- ChEMBL_1824459 (CHEMBL4324223) Inhibition of NS5A in HCV genotype 1b infected in human HuH7.5/Con1/SG-Neo(I)-hRluc2aUb cells assessed as inhibition of replicon levels incubated for 72 hrs by luciferase reporter gene assay
- ChEMBL_1824494 (CHEMBL4324258) Inhibition of NS5A in HCV genotype 3a infected in human HuH7.5/Con1/SG-Neo(I)-hRluc2aUb cells assessed as inhibition of replicon levels incubated for 72 hrs by luciferase reporter gene assay
- ChEMBL_1824509 (CHEMBL4324273) Inhibition of NS5A in HCV genotype 1a infected in human HuH7.5/Con1/SG-Neo(I)-hRluc2aUb cells assessed as inhibition of replicon levels incubated for 72 hrs by luciferase reporter gene assay
- ChEMBL_1824512 (CHEMBL4324276) Inhibition of NS5A in HCV genotype 4a infected in human HuH7.5/Con1/SG-Neo(I)-hRluc2aUb cells assessed as inhibition of replicon levels incubated for 72 hrs by luciferase reporter gene assay
- ChEMBL_1824513 (CHEMBL4324277) Inhibition of NS5A in HCV genotype 6a infected in human HuH7.5/Con1/SG-Neo(I)-hRluc2aUb cells assessed as inhibition of replicon levels incubated for 72 hrs by luciferase reporter gene assay
- ChEMBL_1830126 (CHEMBL4330000) Inhibition of recombinant human His6-tagged HexB expressed in Pichia pastoris using varying levels of 4-Mu-GlcNAc as substrate measured after 30 mins by fluorescence assay by dixon plot analysis a
- ChEMBL_2080389 (CHEMBL4736180) Competitive-inhibition of recombinant human C-terminal His6-tagged Sphk1 expressed in baculovirus infected Sf21 insect cells using varying levels of sphingosine as substrate measured after 30 mins by LC-MS analysis
- ChEMBL_813546 (CHEMBL2019655) Inhibition of ketokinase isoform C in human HepG2 cells assessed as levels of fructose-1-phosphate preincubated for 30 mins followed by substrate addition measured after 3 hrs by LC-MS analysis
- ChEMBL_1672758 (CHEMBL4022787) Inhibition of AICARFT in human MDA-MB-231 cells assessed as increase in ZMP levels using low folate media after 16 hrs by LC-MS method
- ChEMBL_1672759 (CHEMBL4022788) Inhibition of AICARFT in human MDA-MB-231 cells assessed as increase in ZMP levels using regular folate media after 16 hrs by LC-MS method
- ChEMBL_1677512 (CHEMBL4027655) Inhibition of CrtN in methicillin-resistant Staphylococcus aureus USA400 MW2 assessed as reduction in staphyloxanthin levels after 48 hrs by spectrophotometric method-based pigment inhibition assay
- ChEMBL_1677513 (CHEMBL4027656) Inhibition of CrtN in methicillin-resistant Staphylococcus aureus LAC USA300 assessed as reduction in staphyloxanthin levels after 48 hrs by spectrophotometric method-based pigment inhibition assay
- ChEMBL_1752869 (CHEMBL4187629) Competitive inhibition of human recombinant soluble epoxide hydrolase expressed in baculovirus-infected High Five cells S9 fraction using varying CMNPC substrate levels by fluorescence based assay
- ChEMBL_1764672 (CHEMBL4199919) Inhibition of IDH1 R132H mutant (unknown origin) expressed in HCT116 cells assessed as reduction in 2-HG levels after 48 hrs by LC-MS/MS analysis
- ChEMBL_1784290 (CHEMBL4255807) Inhibition of CrtN in methicillin-resistant Staphylococcus aureus USA400 MW2 assessed as reduction in staphyloxanthin levels after 48 hrs by spectrophotometric method-based pigment inhibition assay
- ChEMBL_1784291 (CHEMBL4255808) Inhibition of CrtN in methicillin-resistant Staphylococcus aureus LAC USA300 assessed as reduction in staphyloxanthin levels after 48 hrs by spectrophotometric method-based pigment inhibition assay
- ChEMBL_1825945 (CHEMBL4325709) Inhibition of IDH1 R132H mutant in human HCT116 cells assessed as reduction in 2-HG levels after 24 hrs by RapidFire high-throughput mass spectrometry assay
- ChEMBL_1825946 (CHEMBL4325710) Inhibition of IDH1 R132C mutant in human HCT116 cells assessed as reduction in 2-HG levels after 24 hrs by RapidFire high-throughput mass spectrometry assay
- ChEMBL_1859392 (CHEMBL4360248) Inhibition of IDH1 R132H mutant in human HCT116 cells assessed as reduction in 2-HG levels after 24 hrs by RapidFire high-throughput mass spectrometry assay
- ChEMBL_1859393 (CHEMBL4360249) Inhibition of IDH1 R132C mutant in human HCT116 cells assessed as reduction in 2-HG levels after 24 hrs by RapidFire high-throughput mass spectrometry assay
- ChEMBL_1925278 (CHEMBL4428350) Inhibition of full-length human gamma secretase expressed in SH-SY5Y spbetaA4CTF cells assessed as reduction in amyloid beta levels after 90 mins by HTRF assay
- ChEMBL_2059132 (CHEMBL4714133) Stabilization of human JAK2 KD expressed in Expi293F (HEK293F) cells assessed as increase in JAK2 protein levels incubated for 3 mins by isothermal dose response assay
- ChEMBL_2217502 (CHEMBL5130634) Inhibition of PRMT5 methyltransferase activity in human MTAP -/- Calu-6 cells assessed as reduction in SmB SDMA levels measured after 2 days by HTRF based assay
- ChEMBL_878952 (CHEMBL2185515) Inhibition of BACE1 expressed in CHO 2B7 cells expressing wild type APP695 assessed as reduction in Abeta40/Abeta42 levels incubated for 16 hrs by sandwich ELISA
- ChEMBL_2027414 (CHEMBL4681572) Binding affinity to GST-tagged ERRgamma LBD (unknown origin) using fluorescein PGC1alpha as coactivator incubated for 1 hr by LanthaScreen TR-FRET co-regulator assay
- ChEMBL_2027416 (CHEMBL4681574) Binding affinity to GST-tagged ERRalpha LBD (unknown origin) using fluorescein PGC1alpha as coactivator incubated for 1 hr by LanthaScreen TR-FRET co-regulator assay
- ChEMBL_2027417 (CHEMBL4681575) Binding affinity to GST-tagged ERRbeta LBD (unknown origin) using fluorescein PGC1alpha as coactivator incubated for 1 hr by LanthaScreen TR-FRET co-regulator assay
- ChEMBL_2027418 (CHEMBL4681576) Binding affinity to GST-tagged ERalpha LBD (unknown origin) using fluorescein PGC1alpha as coactivator incubated for 1 hr by LanthaScreen TR-FRET co-regulator assay
- ChEMBL_1585294 (CHEMBL3821557) Competitive inhibition of full length 6xHis-tagged SMYD2 (unknown origin) expressed in Escherichia coli using varying levels of Btn-Ahx-GSRAHSSHLKSKKGQSTSRH-amide substrate after 30 mins in presence of fixed 3H-SAM level by scintillation proximity assay
- DNA-PK Enzyme-Linked Immunosorbent Assay On day one, coat 96-well plate (ThermoFisher. Cat #: 442404) with GST-p53 (1-101) peptide (purified by Pharmaron, BCS department) by diluting 3 μg of GST-p53 each well with 0.1 M Na2CO3/NaHCO3, pH 9.6. Incubate the plate overnight at 4° C. The second day, remove coating buffer, wash 2× with PBST (1×PBS containing 0.1% Tween-20). Then add DNA-PK enzyme solution (Invitrogen, #PR9107A; the final DNA-PK concentration is 0.1 μg/mL), series dilution compounds (the final top concentration is 100 nM, 3 fold series dilution, with total 10 doses) and ATP solution (the final ATP concentration is 20 μM) to the 96-well plate. Incubate the plate at 25° C. for 1 hour. Then wash 3× with PBST (1×PBS containing 0.1% Tween-20). Block the plate with PBST+ 1% BSA at 4° C. overnight. The third day, wash 4× with PBST (1×PBS containing 0.1% Tween-20). Then add Phospho-p53 primary antibody (cell signaling Technology, #9286, Phospho-p53 (Ser15) (16G8) Mouse mAb) (1/1000) to each well. Seal with plate and incubate the plate for 1 h at 37° C. Wash 4× with PBST (1×PBS containing 0.1% Tween-20), add 100 μL of HRP-linked secondary antibody (Cell signaling Technology, #7076, Anti-mouse IgG, HRP-linked Antibody) (1/1000) to each well. Seal with tape and incubate the plate for 30 min at 37° C. Wash 4× with PBST (1×PBS containing 0.1% Tween-20), add 100 μL of TMB (Cell signaling Technology, #7004) substrate to each well. Seal with tape and incubate the plate for 10 min at 37° C. Then add 100 μL of Stop solution (Cell signaling Technology, #7002) to each well. Read the plate at 450 nm to detect absorption.
- ChEBML_158586 Biochemical index for Prostaglandin G/H synthase 1 measured as, thromboxane 2 (TXB2) levels following blood coagulation
- ChEMBL_1580708 (CHEMBL3811154) Agonist activity at human GPR119 receptor assessed as increase in cellular cAMP levels by HTRF assay
- ChEMBL_424173 (CHEMBL906194) Agonist activity at rat beta-3 adrenergic receptor expressed in CHO cells assessed as cAMP levels
- ChEMBL_424175 (CHEMBL907249) Agonist activity at human beta-2 adrenergic receptor expressed in CHO cells assessed as cAMP levels
- ChEMBL_424177 (CHEMBL907251) Agonist activity at human beta-1 adrenergic receptor expressed in CHO cells assessed as cAMP levels
- Time Resolved Fluorescence Energy Transfer (TR-FRET) Assay Assay 2: For selected compounds displaying IC50s between 0.05 and 5 nM on MDM2, a slightly modified assay is used with the following adaptations: 0.1 nM MDM2, 0.1 nM Europium labeled streptavidin and Tecan genios Pro is used as a microplate reader for the fluorescence measurements (p53-MDM2 Assay 2). The test is performed in white 384-well plates (Greiner Bio-One, reference 781207) in a total volume of 60 μL by adding 1 μL of compounds tested at different concentrations diluted in 100% DMSO (1.7% final DMSO concentration) in reaction buffer (PBS, 125 mM NaCl, 0.001% Novexin (consists of carbohydrate polymers), designed to increase the solubility and stability of proteins; Expedeon Ltd., Cambridgeshire, United Kingdom), 0.01% Gelatin, 0.01% 0.2%, Pluronic F-127 (block copolymer from ethylenoxide and propyleneoxide), 1 mM DTT). After addition of 1.25 nM MDM2-biotinylated or 2.5 nM MDM4-biotinylated (internal preparations, both MDM2 and MDM4 are biotinylated at the C-terminal of the peptide construct), and 0.625 nM Europium labeled streptavidin (Perkin Elmer), the solution is pre-incubated for 15 minutes at room temperature, then 10 nM Cy5-p53 peptide (internal preparation, the Cy5 dye is directly bound to the N-terminal part of p53 peptide construct) is added before an incubation at room temperature for 15 minutes prior to reading the plate. For measurement of samples, a Victor II microplate reader (Perkin Elmer) is used with the following settings: Excitation 340 nm, Emission Donor 620 nm and Emission Acceptor 665 nm.
- ChEMBL_1659742 (CHEMBL4009354) Competitive inhibition of full length recombinant human ALDH2 expressed in Escherichia coli assessed as reduction in dehydrogenase activity using propionaldehyde as substrate in presence of varying levels NAD+ by Lineweaver-Burk plot analysis
- ChEMBL_1763451 (CHEMBL4198698) Mixed type inhibition of human erythrocyte AChE assessed as enzyme-inhibitor complex using varying levels of acetylthiocholine iodide as substrate preincubated for 5 mins followed by substrate addition by Lineweaver-Burk plot analysis
- ChEMBL_1871707 (CHEMBL4372874) Competitive inhibition of Mycobacterium tuberculosis PTPB using varying levels of pNPP as substrate preincubated for 15 mins followed by substrate addition and measured after 2 to 6 mins by Lineweaver-burk plot analysis
