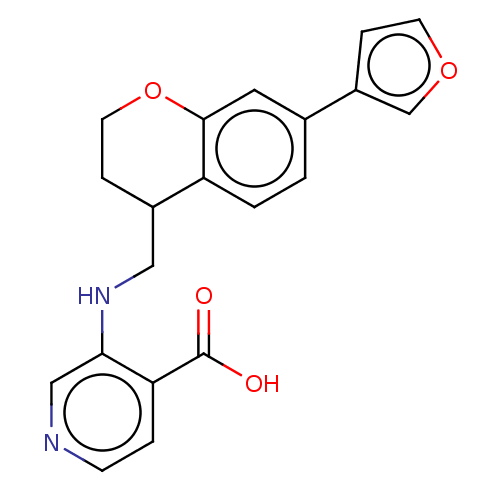
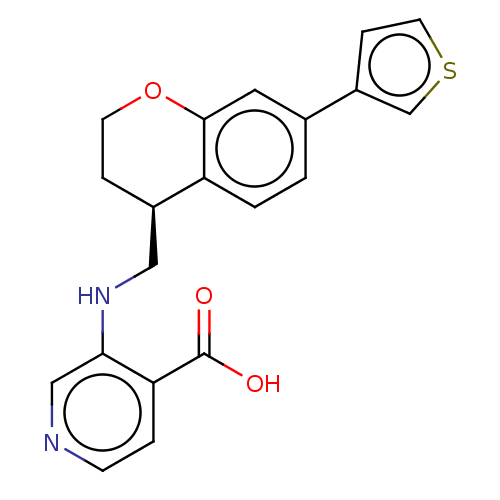
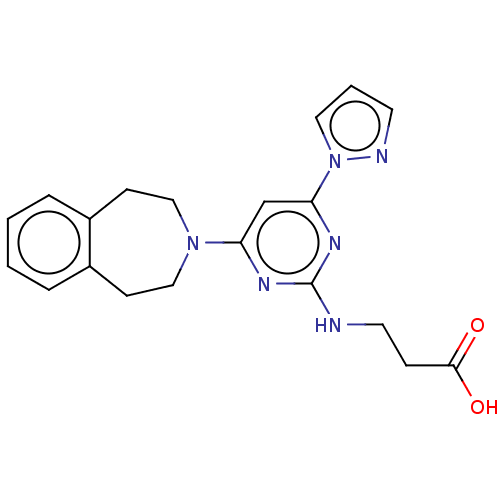
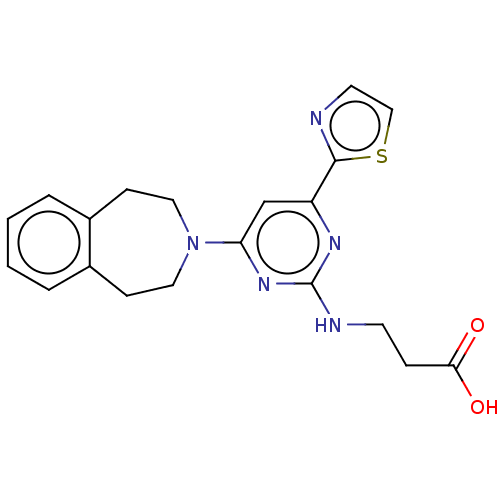
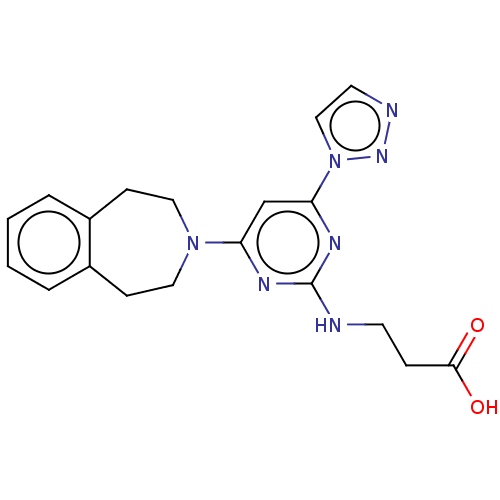
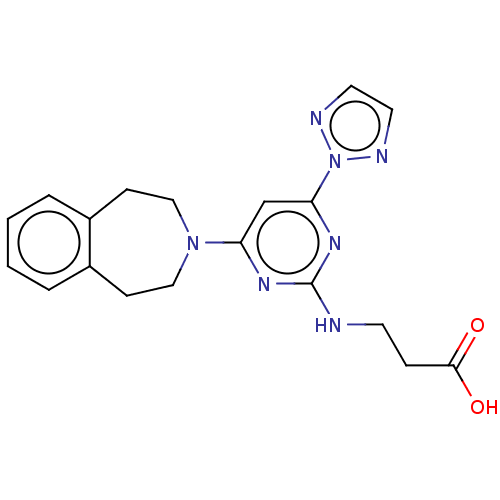
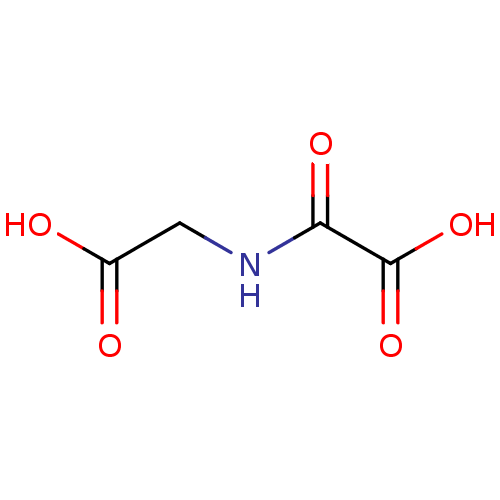
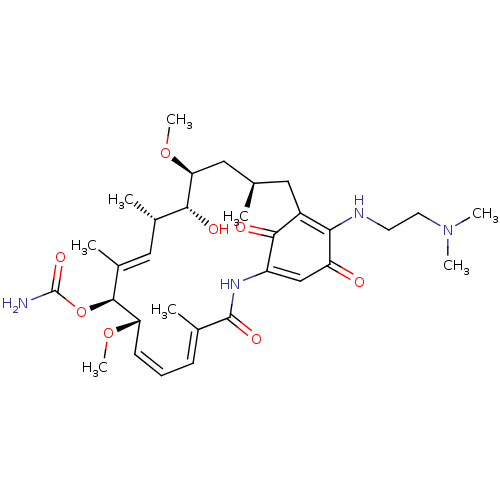
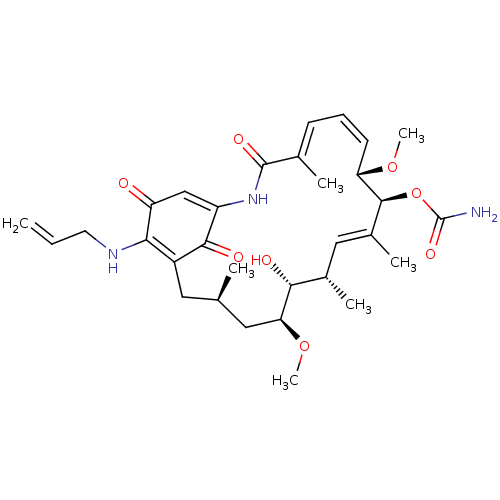
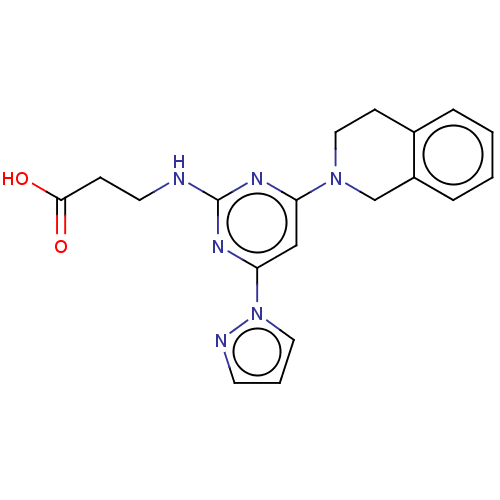
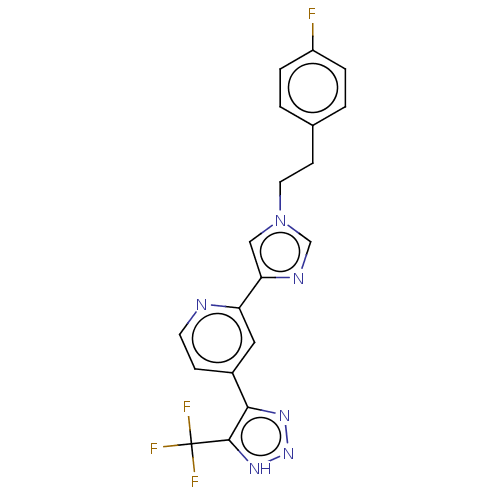
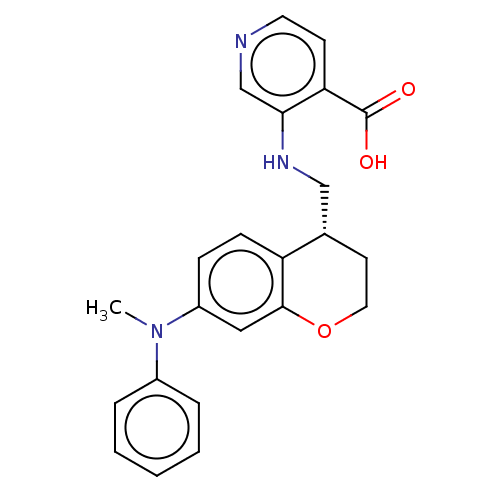
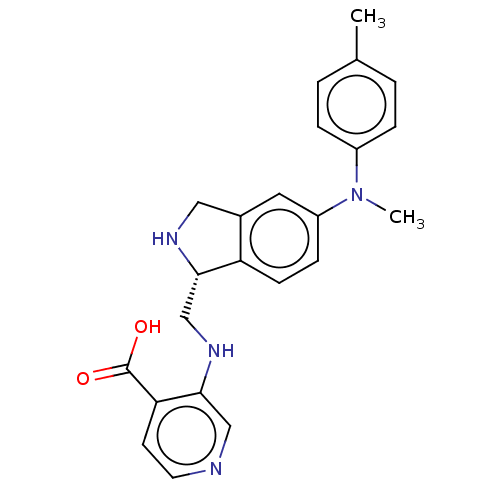
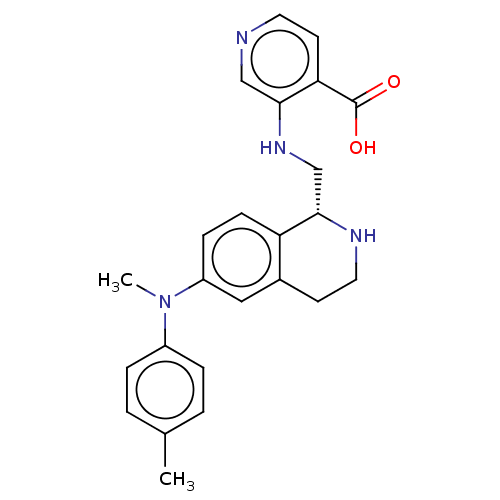
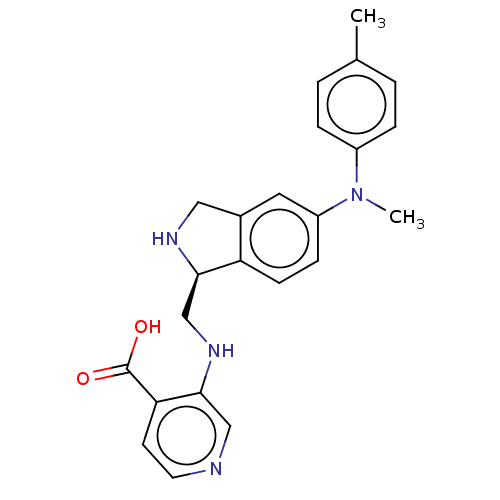
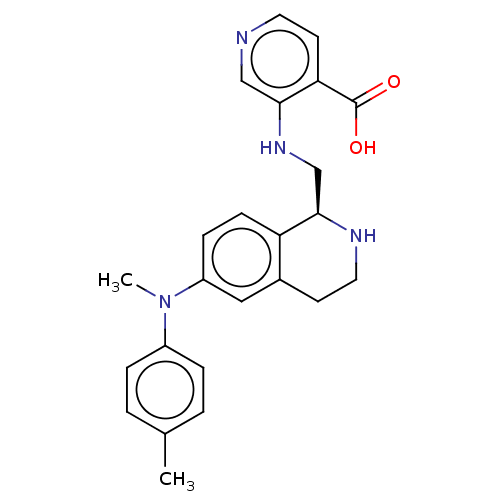
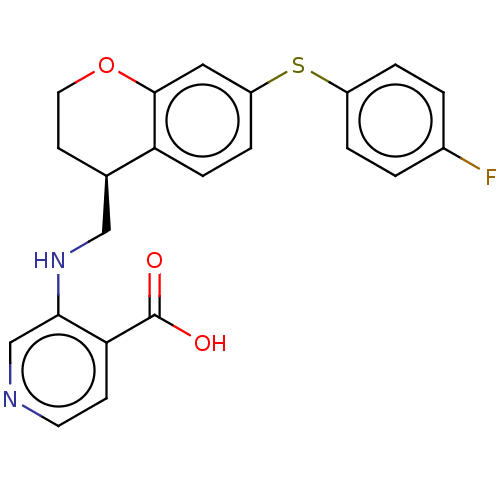
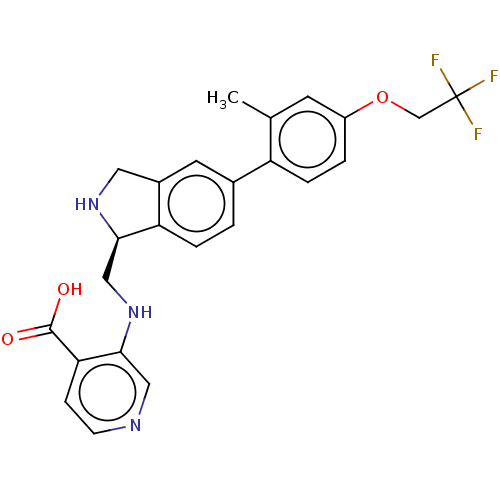
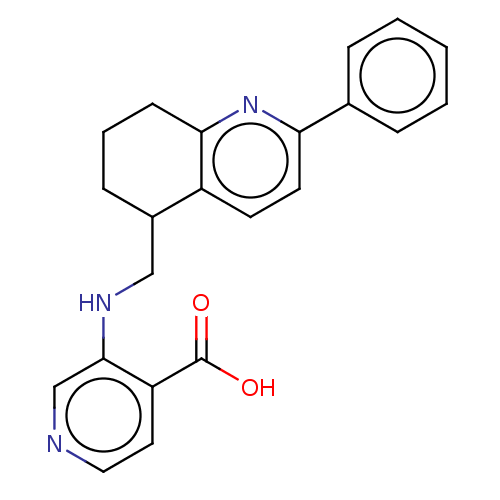
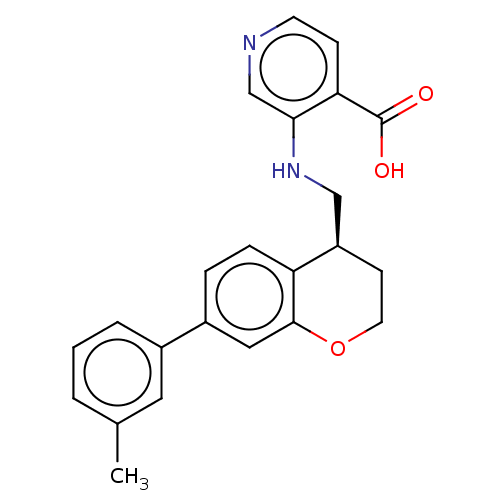
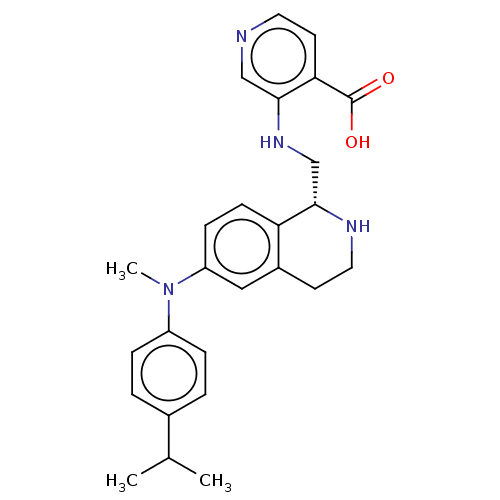
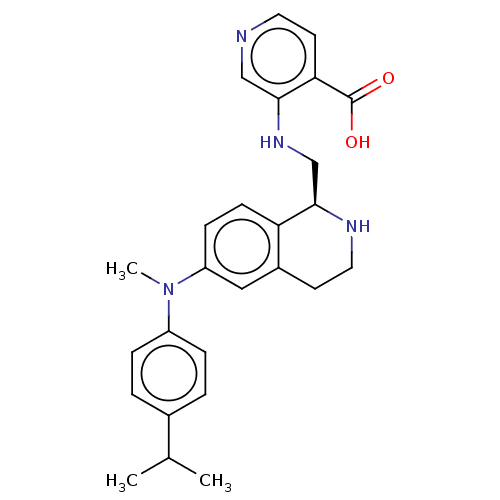
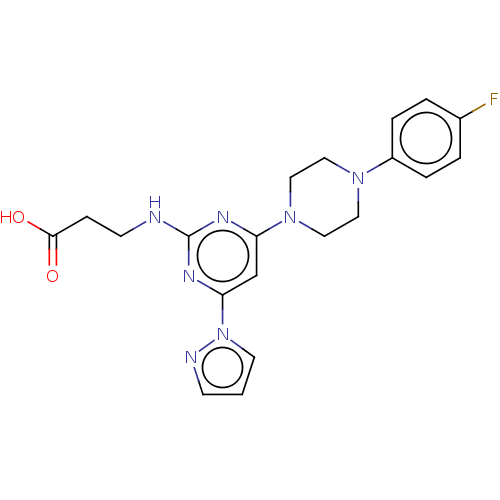
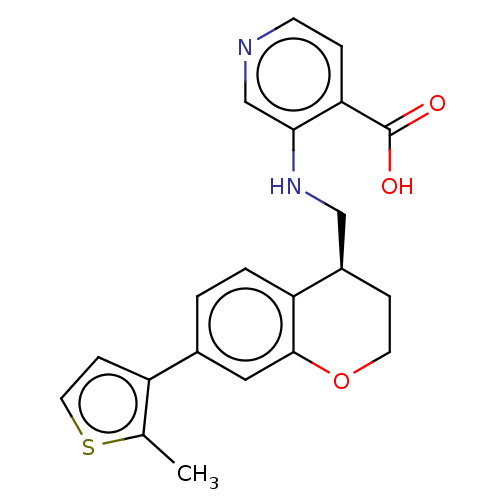
Report error Found 443 Enz. Inhib. hit(s) with Target = 'Lysine-specific demethylase 6B'
Affinity DataIC50: 1nMAssay Description:Inhibition of KDM6B (1043 to 1643 residues)(unknown origin) by Alphalisa assayMore data for this Ligand-Target Pair
Affinity DataIC50: 1nMAssay Description:Inhibition of epitope-tagged KDM6B (1026 to 1682 residues)(unknown origin) transfected in human U2OS cells using H3K27me2 peptide as substrateMore data for this Ligand-Target Pair
Affinity DataIC50: 1.70nMAssay Description:Inhibition of epitope-tagged KDM6B (1026 to 1682 residues)(unknown origin) transfected in human U2OS cells using H3K27me2 peptide as substrateMore data for this Ligand-Target Pair
Ligand InfoSimilars
Affinity DataIC50: 2.5nMAssay Description:Inhibition of epitope-tagged KDM6B (1026 to 1682 residues)(unknown origin) transfected in human U2OS cells using H3K27me2 peptide as substrateMore data for this Ligand-Target Pair
Affinity DataIC50: 2.80nMAssay Description:Inhibition of KDM6B (1043 to 1643 residues)(unknown origin) by Alphalisa assayMore data for this Ligand-Target Pair
Ligand InfoSimilars
Affinity DataIC50: 3.10nMAssay Description:Inhibition of epitope-tagged KDM6B (1026 to 1682 residues)(unknown origin) transfected in human U2OS cells using H3K27me2 peptide as substrateMore data for this Ligand-Target Pair
Affinity DataIC50: 4.90nMAssay Description:Inhibition of KDM6B (1043 to 1643 residues)(unknown origin) by Alphalisa assayMore data for this Ligand-Target Pair
Affinity DataIC50: 8.60nMAssay Description:Inhibition of KDM6B (1043 to 1643 residues)(unknown origin) by Alphalisa assayMore data for this Ligand-Target Pair
Affinity DataIC50: 16nMAssay Description:Inhibition of C-terminal FLAG-tagged human KDM6B expressed in expressed in Sf9 cells pre-incubated for 4 hrs before substrate addition and measured a...More data for this Ligand-Target Pair
Affinity DataIC50: 28nMAssay Description:Inhibition of KDM6B (unknown origin ) by TR-FRET assayMore data for this Ligand-Target Pair
Affinity DataIC50: 60nMAssay Description:Inhibition of JMJD3 (unknown origin) by Alphascreen assayMore data for this Ligand-Target Pair
Affinity DataIC50: 60nMAssay Description:Competitive inhibition of KDM6B (unknown origin) using biotinylated H3K27me3 peptide as substrate preincubated for 15 mins followed by substrate addi...More data for this Ligand-Target Pair
Ligand InfoSimilars
Affinity DataIC50: 60nMAssay Description:Inhibition of KDM6B (unknown origin)More data for this Ligand-Target Pair
Affinity DataIC50: 60nMAssay Description:Inhibition of recombinant human N-terminal His6/Flag-tagged TEV-protease cleavage site fused JMJD3 1637-1675 deletion mutant (1141 to 1682 residues) ...More data for this Ligand-Target Pair
Ligand InfoSimilars
Affinity DataIC50: 63nMAssay Description:Inhibition of KDM6B (unknown origin)More data for this Ligand-Target Pair
Affinity DataIC50: 100nMAssay Description:Inhibition of KDM6B (unknown origin)More data for this Ligand-Target Pair
Affinity DataIC50: 100nMT: 2°CAssay Description:The enzymatic assay of JMJD3 activity is based upon Time Resolved-Fluorescence Resonance Energy Transfer (TR-FRET) detection. The ability of test com...More data for this Ligand-Target Pair
Affinity DataIC50: 100nMT: 2°CAssay Description:The enzymatic assay of JMJD3 activity is based upon Time Resolved-Fluorescence Resonance Energy Transfer (TR-FRET) detection. The ability of test com...More data for this Ligand-Target Pair
Affinity DataIC50: 140nMAssay Description:Inhibition of recombinant JMJD3 (unknown origin) incubated for 15 mins prior to substrate addition by AlphaScreen methodMore data for this Ligand-Target Pair
Affinity DataIC50: 150nMAssay Description:Inhibition of JMJD3 (unknown origin) using biotinylated H3K27me3 peptide as substrate after 1 hr by AlphaLISA assayMore data for this Ligand-Target Pair
Affinity DataIC50: 150nMAssay Description:Inhibition of JMJD3 (unknown origin) using biotinylated H3K27me3 peptide as substrate after 1 hr by AlphaLISA assayMore data for this Ligand-Target Pair
Affinity DataIC50: 160nMpH: 7.5Assay Description:The demethylase AlphaScreen assay was performed in 384-well plate format using white proxiplates (PerkinElmer), and transfer of compound (100 nl) was...More data for this Ligand-Target Pair
Affinity DataIC50: 200nMAssay Description:Inhibition of JMJD3 (unknown origin) using biotinylated H3K27me3 peptide as substrate after 1 hr by AlphaLISA assayMore data for this Ligand-Target Pair
Affinity DataIC50: 200nMAssay Description:Inhibition of KDM6B (unknown origin) expressed in Escherichia coli and measured after 1 hrs by alpha screen assayMore data for this Ligand-Target Pair
Affinity DataIC50: 210nMAssay Description:Inhibition of JMJD3 (unknown origin) using biotinylated H3K27me3 peptide as substrate after 1 hr by AlphaLISA assayMore data for this Ligand-Target Pair
Affinity DataIC50: 270nMAssay Description:Inhibition of JMJD3 (unknown origin) using biotinylated H3K27me3 peptide as substrate after 1 hr by AlphaLISA assayMore data for this Ligand-Target Pair
Affinity DataIC50: 316nMAssay Description:Inhibition of KDM6B (unknown origin)More data for this Ligand-Target Pair
Affinity DataIC50: 400nMAssay Description:Inhibition of KDM6B (unknown origin) measured by AlphaScreen assayMore data for this Ligand-Target Pair
Affinity DataIC50: 400nMAssay Description:Inhibition of KDM6B (unknown origin) measured by AlphaScreen assayMore data for this Ligand-Target Pair
Affinity DataIC50: 400nMAssay Description:Inhibition of KDM6B (unknown origin) measured by AlphaScreen assayMore data for this Ligand-Target Pair
Affinity DataIC50: 490nMAssay Description:Inhibition of JMJD3 (unknown origin) using biotinylated H3K27me3 peptide as substrate after 1 hr by AlphaLISA assayMore data for this Ligand-Target Pair
Affinity DataIC50: 500nMAssay Description:The ability of test compounds to inhibit the activity of JMJD3 was determined in 384-well plate format under the following reaction conditions: 1 nM ...More data for this Ligand-Target Pair
Affinity DataIC50: 500nMAssay Description:The ability of test compounds to inhibit the activity of JMJD3 was determined in 384-well plate format under the following reaction conditions: 1 nM ...More data for this Ligand-Target Pair
Affinity DataIC50: 550nMT: 2°CAssay Description:The enzymatic assay of JMJD3 activity is based upon Time Resolved-Fluorescence Resonance Energy Transfer (TR-FRET) detection. The ability of test com...More data for this Ligand-Target Pair
Affinity DataIC50: 550nMAssay Description:The enzymatic assay of JMJD3 activity is based upon Time Resolved-Fluorescence Resonance Energy Transfer (TR-FRET) detection. The ability of test com...More data for this Ligand-Target Pair
Affinity DataIC50: 550nMAssay Description:JMJD3: The enzymatic assay of JMJD3 activity is based upon Time Resolved-Fluorescence Resonance Energy Transfer (TR-FRET) detection. The ability of t...More data for this Ligand-Target Pair
Affinity DataIC50: 550nMAssay Description:The enzymatic assay of JMJD3 activity is based upon Time Resolved-Fluorescence Resonance Energy Transfer (TR-FRET) detection. The ability of test com...More data for this Ligand-Target Pair
Affinity DataIC50: 550nMAssay Description:JMJD3: The enzymatic assay of JMJD3 activity is based upon Time Resolved-Fluorescence Resonance Energy Transfer (TR-FRET) detection. The ability of t...More data for this Ligand-Target Pair
Affinity DataIC50: 550nMT: 2°CAssay Description:The enzymatic assay of JMJD3 activity is based upon Time Resolved-Fluorescence Resonance Energy Transfer (TR-FRET) detection. The ability of test com...More data for this Ligand-Target Pair
Affinity DataIC50: 550nMAssay Description:JMJD3: The enzymatic assay of JMJD3 activity is based upon Time Resolved-Fluorescence Resonance Energy Transfer (TR-FRET) detection. The ability of t...More data for this Ligand-Target Pair
Affinity DataIC50: 550nMAssay Description:JMJD3: The enzymatic assay of JMJD3 activity is based upon Time Resolved-Fluorescence Resonance Energy Transfer (TR-FRET) detection. The ability of t...More data for this Ligand-Target Pair
Affinity DataIC50: 550nMT: 2°CAssay Description:The enzymatic assay of JMJD3 activity is based upon Time Resolved-Fluorescence Resonance Energy Transfer (TR-FRET) detection. The ability of test com...More data for this Ligand-Target Pair
Affinity DataIC50: 550nMAssay Description:JMJD3: The enzymatic assay of JMJD3 activity is based upon Time Resolved-Fluorescence Resonance Energy Transfer (TR-FRET) detection. The ability of t...More data for this Ligand-Target Pair
Affinity DataIC50: 550nMAssay Description:JMJD3: The enzymatic assay of JMJD3 activity is based upon Time Resolved-Fluorescence Resonance Energy Transfer (TR-FRET) detection. The ability of t...More data for this Ligand-Target Pair
Affinity DataIC50: 550nMT: 2°CAssay Description:The enzymatic assay of JMJD3 activity is based upon Time Resolved-Fluorescence Resonance Energy Transfer (TR-FRET) detection. The ability of test com...More data for this Ligand-Target Pair
Affinity DataIC50: 550nMAssay Description:The enzymatic assay of JMJD3 activity is based upon Time Resolved-Fluorescence Resonance Energy Transfer (TR-FRET) detection. The ability of test com...More data for this Ligand-Target Pair
Affinity DataIC50: 550nMAssay Description:JMJD3: The enzymatic assay of JMJD3 activity is based upon Time Resolved-Fluorescence Resonance Energy Transfer (TR-FRET) detection. The ability of t...More data for this Ligand-Target Pair
Affinity DataIC50: 550nMAssay Description:JMJD3: The enzymatic assay of JMJD3 activity is based upon Time Resolved-Fluorescence Resonance Energy Transfer (TR-FRET) detection. The ability of t...More data for this Ligand-Target Pair
Affinity DataIC50: 550nMAssay Description:Inhibition of JMJD3 (unknown origin) using biotinylated H3K27me3 peptide as substrate after 1 hr by AlphaLISA assayMore data for this Ligand-Target Pair
Affinity DataIC50: 550nMT: 2°CAssay Description:The enzymatic assay of JMJD3 activity is based upon Time Resolved-Fluorescence Resonance Energy Transfer (TR-FRET) detection. The ability of test com...More data for this Ligand-Target Pair

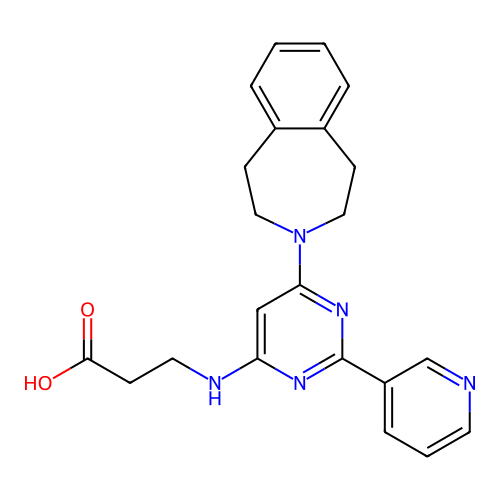


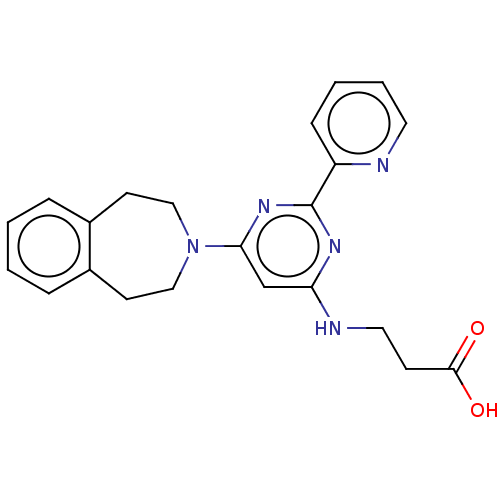
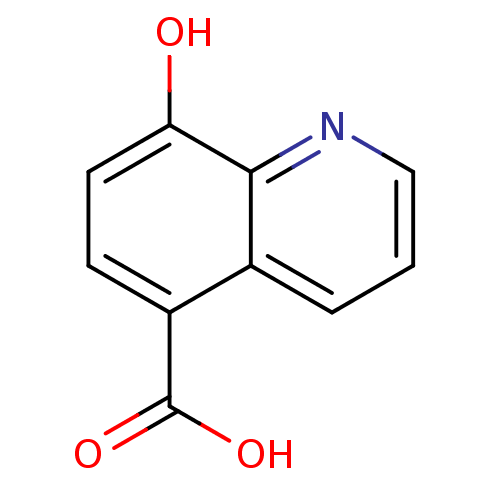
 3D Structure (crystal)
3D Structure (crystal)