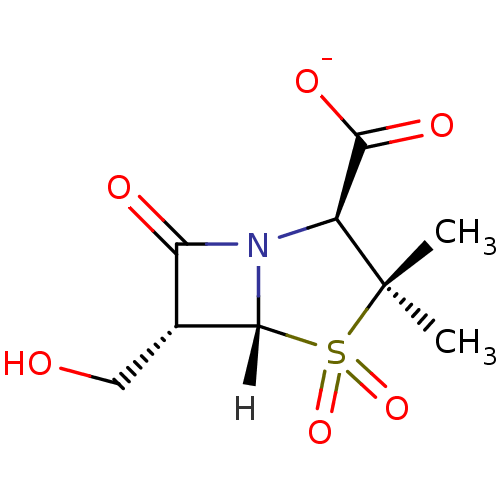
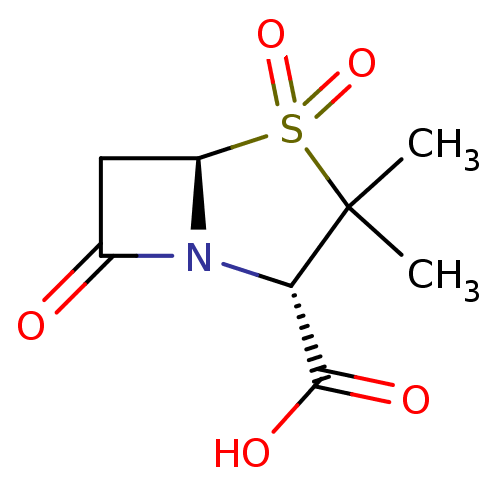
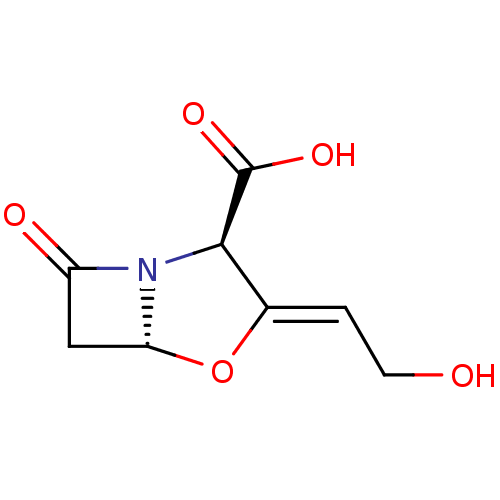
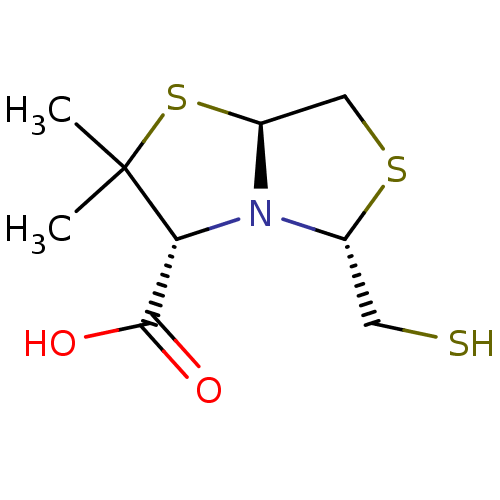
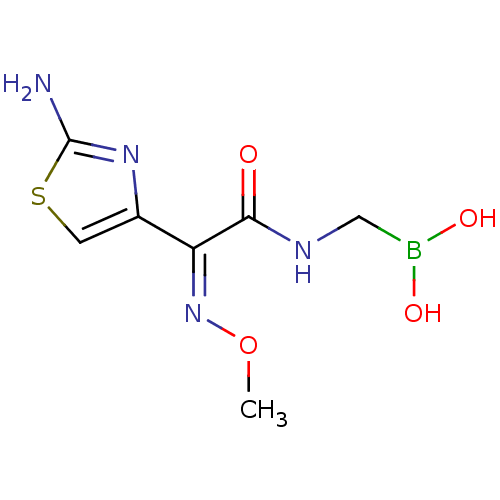
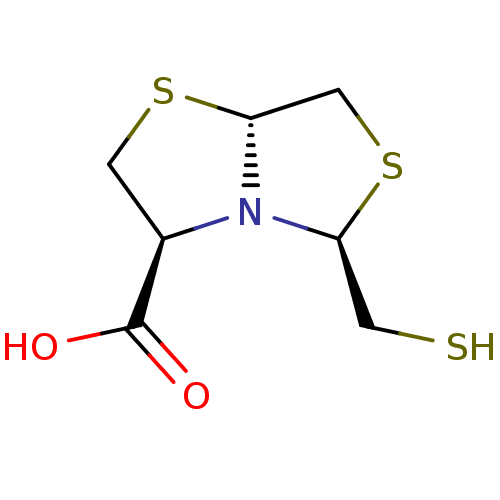
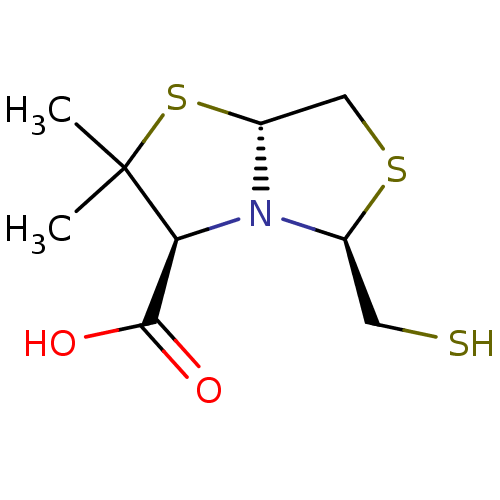
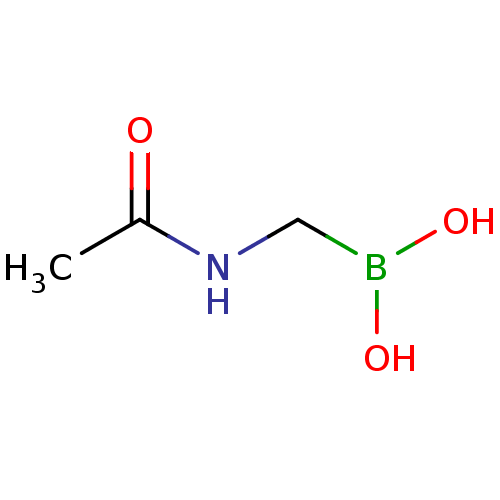
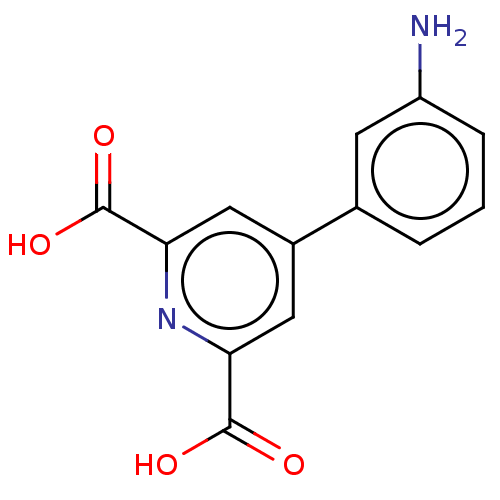
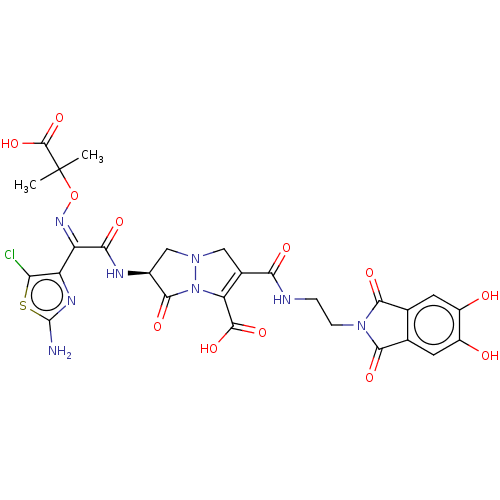
Affinity DataKi: 30nM IC50: 150nMpH: 7.2Assay Description:Steady-state kinetics were performed on an Agilent 8453 diode array spectrophotometer (Palo Alto, CA) in 50 mM sodium phosphate buffer (pH 7.2) suppl...More data for this Ligand-Target Pair
Affinity DataKi: 50nM IC50: 30nMpH: 7.2Assay Description:Steady-state kinetics were performed on an Agilent 8453 diode array spectrophotometer (Palo Alto, CA) in 50 mM sodium phosphate buffer (pH 7.2) suppl...More data for this Ligand-Target Pair
Affinity DataKi: 150nM IC50: 160nMpH: 7.2Assay Description:Steady-state kinetics were performed on an Agilent 8453 diode array spectrophotometer (Palo Alto, CA) in 50 mM sodium phosphate buffer (pH 7.2) suppl...More data for this Ligand-Target Pair
Affinity DataKi: 220nM IC50: 30nMpH: 7.4Assay Description:Steady state kinetics were performed on an Agilent 8453 diode array spectrophotometer in 10 mM phosphate-buffered saline (pH 7.4). IC50, defined as t...More data for this Ligand-Target Pair
Affinity DataKi: 380nM IC50: 60nMpH: 7.2Assay Description:Steady-state kinetics were performed on an Agilent 8453 diode array spectrophotometer (Palo Alto, CA) in 50 mM sodium phosphate buffer (pH 7.2) suppl...More data for this Ligand-Target Pair
Affinity DataKi: 560nM IC50: 320nMpH: 7.4Assay Description:Steady state kinetics were performed on an Agilent 8453 diode array spectrophotometer in 10 mM phosphate-buffered saline (pH 7.4). IC50, defined as t...More data for this Ligand-Target Pair
Affinity DataKi: 1.50E+3nM IC50: 3.40E+3nMpH: 7.4Assay Description:Steady state kinetics were performed on an Agilent 8453 diode array spectrophotometer in 10 mM phosphate-buffered saline (pH 7.4). IC50, defined as t...More data for this Ligand-Target Pair
Affinity DataKi: 2.00E+3nM IC50: 100nMpH: 7.4Assay Description:Steady state kinetics were performed on an Agilent 8453 diode array spectrophotometer in 10 mM phosphate-buffered saline (pH 7.4). IC50, defined as t...More data for this Ligand-Target Pair
Affinity DataKi: 2.20E+3nMAssay Description:Binding affinity at Beta-lactamase SHV1 expressed in Escherichia coli DH10B cellsMore data for this Ligand-Target Pair
TargetMetallo-beta-lactamase VIM-2(Pseudomonas aeruginosa (g-Proteobacteria))
Louis Stokes Cleveland Veterans Affairs Medical Center
Louis Stokes Cleveland Veterans Affairs Medical Center
Affinity DataKi: 3.70E+3nMAssay Description:The Ki for each inhibitor was determined by direct competition assays under steady-state conditions. The initial velocity was measured in the presenc...More data for this Ligand-Target Pair
TargetMetallo-beta-lactamase VIM-2(Pseudomonas aeruginosa (g-Proteobacteria))
Louis Stokes Cleveland Veterans Affairs Medical Center
Louis Stokes Cleveland Veterans Affairs Medical Center
Affinity DataKi: 3.80E+3nMAssay Description:The Ki for each inhibitor was determined by direct competition assays under steady-state conditions. The initial velocity was measured in the presenc...More data for this Ligand-Target Pair
Affinity DataKi: 4.50E+3nMAssay Description:Binding affinity at Beta-lactamase SHV5 expressed in Escherichia coli DH10B cellsMore data for this Ligand-Target Pair
TargetVIM-24(Klebsiella pneumoniae (Enterobacteria))
Louis Stokes Cleveland Veterans Affairs Medical Center
Louis Stokes Cleveland Veterans Affairs Medical Center
Affinity DataKi: 4.90E+3nMAssay Description:The Ki for each inhibitor was determined by direct competition assays under steady-state conditions. The initial velocity was measured in the presenc...More data for this Ligand-Target Pair
Affinity DataKi: 5.00E+3nMAssay Description:Binding affinity at Beta-lactamase SHV5 expressed in Escherichia coli DH10B cellsMore data for this Ligand-Target Pair
TargetMetallo-beta-lactamase VIM-2(Pseudomonas aeruginosa (g-Proteobacteria))
Louis Stokes Cleveland Veterans Affairs Medical Center
Louis Stokes Cleveland Veterans Affairs Medical Center
Affinity DataKi: 5.40E+3nMAssay Description:The Ki for each inhibitor was determined by direct competition assays under steady-state conditions. The initial velocity was measured in the presenc...More data for this Ligand-Target Pair
TargetVIM-24(Klebsiella pneumoniae (Enterobacteria))
Louis Stokes Cleveland Veterans Affairs Medical Center
Louis Stokes Cleveland Veterans Affairs Medical Center
Affinity DataKi: 6.00E+3nMAssay Description:The Ki for each inhibitor was determined by direct competition assays under steady-state conditions. The initial velocity was measured in the presenc...More data for this Ligand-Target Pair
Affinity DataKi: 8.90E+3nMAssay Description:Binding affinity at Beta-lactamase SHV1 expressed in Escherichia coli DH10B cellsMore data for this Ligand-Target Pair
TargetVIM-24(Klebsiella pneumoniae (Enterobacteria))
Louis Stokes Cleveland Veterans Affairs Medical Center
Louis Stokes Cleveland Veterans Affairs Medical Center
Affinity DataKi: 1.10E+4nMAssay Description:The Ki for each inhibitor was determined by direct competition assays under steady-state conditions. The initial velocity was measured in the presenc...More data for this Ligand-Target Pair
TargetVIM-24(Klebsiella pneumoniae (Enterobacteria))
Louis Stokes Cleveland Veterans Affairs Medical Center
Louis Stokes Cleveland Veterans Affairs Medical Center
Affinity DataKi: 1.20E+4nMAssay Description:The Ki for each inhibitor was determined by direct competition assays under steady-state conditions. The initial velocity was measured in the presenc...More data for this Ligand-Target Pair
TargetMetallo-beta-lactamase VIM-2(Pseudomonas aeruginosa (g-Proteobacteria))
Louis Stokes Cleveland Veterans Affairs Medical Center
Louis Stokes Cleveland Veterans Affairs Medical Center
Affinity DataKi: 1.40E+4nMAssay Description:The Ki for each inhibitor was determined by direct competition assays under steady-state conditions. The initial velocity was measured in the presenc...More data for this Ligand-Target Pair
Affinity DataKi: 3.00E+5nMAssay Description:Binding affinity at Beta-lactamase SHV1 expressed in Escherichia coli DH10B cellsMore data for this Ligand-Target Pair
Affinity DataKi: 3.60E+5nMAssay Description:Binding affinity at Beta-lactamase SHV5 expressed in Escherichia coli DH10B cellsMore data for this Ligand-Target Pair
TargetMetallo-beta-lactamase VIM-2(Pseudomonas aeruginosa (g-Proteobacteria))
Louis Stokes Cleveland Veterans Affairs Medical Center
Louis Stokes Cleveland Veterans Affairs Medical Center
Affinity DataIC50: 210nMAssay Description:Binding affinity at 5-hydroxytryptamine 1A receptor by the displacement of [3H]8-OH-DPAT in rat brain.More data for this Ligand-Target Pair
TargetMetallo-beta-lactamase type 2(Serratia marcescens)
University of California San Diego
Curated by ChEMBL
University of California San Diego
Curated by ChEMBL
Affinity DataIC50: 240nMAssay Description:Inhibition of Serratia marcescens IMP1 expressed in Escherichia coli BL21(DE3) using chromacef as substrate preincubated for 10 mins followed by subs...More data for this Ligand-Target Pair
TargetMetallo-beta-lactamase VIM-2(Pseudomonas aeruginosa (g-Proteobacteria))
Louis Stokes Cleveland Veterans Affairs Medical Center
Louis Stokes Cleveland Veterans Affairs Medical Center
Affinity DataIC50: 1.66E+3nMAssay Description:Inhibition of Pseudomonas aeruginosa VIM2 expressed in Escherichia coli BL21(DE3) using chromacef as substrate preincubated for 10 mins followed by s...More data for this Ligand-Target Pair
TargetPeptidoglycan D,D-transpeptidase FtsI(Pseudomonas aeruginosa)
Louis Stokes Cleveland Department Of Veterans Affairs Medical Center
Curated by ChEMBL
Louis Stokes Cleveland Department Of Veterans Affairs Medical Center
Curated by ChEMBL
Affinity DataIC50: 2.50E+3nMAssay Description:Inhibition of Pseudomonas aeruginosa PBP3 assessed as reduction in fluorescence intensity using Bocillin-protein as fluorescent substrate preincubate...More data for this Ligand-Target Pair
TargetPeptidoglycan D,D-transpeptidase FtsI(Pseudomonas aeruginosa)
Louis Stokes Cleveland Department Of Veterans Affairs Medical Center
Curated by ChEMBL
Louis Stokes Cleveland Department Of Veterans Affairs Medical Center
Curated by ChEMBL
Affinity DataIC50: 2.50E+3nMAssay Description:Inhibition of Pseudomonas aeruginosa PBP3 assessed as reduction in fluorescence intensity of bocillin labeled protein pre-incubated for 20 mins befor...More data for this Ligand-Target Pair
TargetMetallo-beta-lactamase type 2(Serratia marcescens)
University of California San Diego
Curated by ChEMBL
University of California San Diego
Curated by ChEMBL
Affinity DataIC50: 3.03E+3nMAssay Description:Binding affinity at 5-hydroxytryptamine 1A receptor by the displacement of [3H]8-OH-DPAT in rat brain.More data for this Ligand-Target Pair
TargetPeptidoglycan D,D-transpeptidase FtsI(Pseudomonas aeruginosa)
Louis Stokes Cleveland Department Of Veterans Affairs Medical Center
Curated by ChEMBL
Louis Stokes Cleveland Department Of Veterans Affairs Medical Center
Curated by ChEMBL
Affinity DataIC50: 5.00E+3nMAssay Description:Inhibition of Pseudomonas aeruginosa PBP3 assessed as reduction in fluorescence intensity using Bocillin-protein as fluorescent substrate preincubate...More data for this Ligand-Target Pair


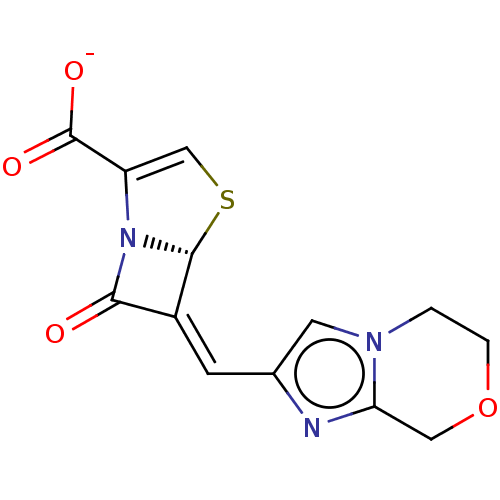
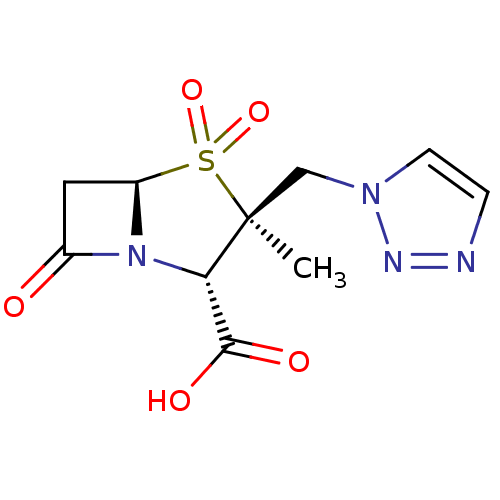
 3D Structure (crystal)
3D Structure (crystal)