Target (1)
Compound (33)
Article Author (129)
Assay (226)
Sahin, C; Magomedova, L; Ferreira, TAM; Liu, J; Tiefenbach, J; Alves, PS; Queiroz, FJG; Oliveira, AS; Bhattacharyya, M; Grouleff, J; Nogueira, PCN; Silveira, ER; Moreira, DC; Leite, JRSA; Brand, GD; Uehling, D; Poda, G; Krause, H; Cummins, CL; Romeiro, LAS J Med Chem 65: 1961 -1978 (2022) Plantan, I; Selic, L; Mesar, T; Anderluh, PS; Oblak, M; Prezelj, A; Hesse, L; Andrejasic, M; Vilar, M; Turk, D; Kocijan, A; Prevec, T; Vilfan, G; Kocjan, D; Copar, A; Urleb, U; Solmajer, T J Med Chem 50: 4113 -21 (2007) Evans, BE; Rittle, KE; Bock, MG; DiPardo, RM; Freidinger, RM; Whitter, WL; Lundell, GF; Veber, DF; Anderson, PS; Chang, RS J Med Chem 31: 2235 -46 (1989) Peterson, EA; Andrews, PS; Be, X; Boezio, AA; Bush, TL; Cheng, AC; Coats, JR; Colletti, AE; Copeland, KW; DuPont, M; Graceffa, R; Grubinska, B; Harmange, JC; Kim, JL; Mullady, EL; Olivieri, P; Schenkel, LB; Stanton, MK; Teffera, Y; Whittington, DA; Cai, T; La, DS Bioorg Med Chem Lett 21: 2064 -70 (2011) Jochim, AL; Miller, SE; Angelo, NG; Arora, PS Bioorg Med Chem Lett 19: 6023 -6 (2009) Geldenhuys, WJ; Funk, MO; Awale, PS; Lin, L; Carroll, RT Bioorg Med Chem Lett 21: 5498 -501 (2011) Baran, PS; Felding, J; Jin, Y; Yeh, C US Patent US9409931 (2016) Bubenik, M; Mader, P; Mochirian, P; Vallée, F; Clark, J; Truchon, JF; Perryman, AL; Pau, V; Kurinov, I; Zahn, KE; Leclaire, ME; Papp, R; Mathieu, MC; Hamel, M; Duffy, NM; Godbout, C; Casas-Selves, M; Falgueyret, JP; Baruah, PS; Nicolas, O; Stocco, R; Poirier, H; Martino, G; Fortin, AB; Roulston, A; Chefson, A; Dorich, S; St-Onge, M; Patel, P; Pellerin, C; Ciblat, S; Pinter, T; Barabé, F; El Bakkouri, M; Parikh, P; Gervais, C; Sfeir, A; Mamane, Y; Morris, SJ; Black, WC; Sicheri, F; Gallant, M J Med Chem 65: 13198 -13215 (2022) Liu, XH; Zhu, J; Zhou, AN; Song, BA; Zhu, HL; Bai, LS; Bhadury, PS; Pan, CX Bioorg Med Chem 17: 1207 -13 (2009) Reitz, AB; Bennett, DJ; Blum, PS; Codd, EE; Maryanoff, CA; Ortegon, ME; Renzi, MJ; Scott, MK; Shank, RP; Vaught, JL J Med Chem 37: 1060 -2 (1994) Bell, MG; Gernert, DL; Grese, TA; Belvo, MD; Borromeo, PS; Kelley, SA; Kennedy, JH; Kolis, SP; Lander, PA; Richey, R; Sharp, VS; Stephenson, GA; Williams, JD; Yu, H; Zimmerman, KM; Steinberg, MI; Jadhav, PK J Med Chem 50: 6443 -5 (2007) Hylsová, M; Carbain, B; Fanfrlík, J; Musilová, L; Haldar, S; Köprülüoglu, C; Ajani, H; Brahmkshatriya, PS; Jorda, R; Kryštof, V; Hobza, P; Echalier, A; Paruch, K; Lepšík, M Eur J Med Chem 126: 1118 -1128 (2017) Jørgensen, AS; Jacobsen, P; Christiansen, LB; Bury, PS; Kanstrup, A; Thorpe, SM; Naerum, L; Wassermann, K Bioorg Med Chem Lett 10: 2383 -6 (2000) Wichitnithad, W; O'Callaghan, JP; Miller, DB; Train, BC; Callery, PS Bioorg Med Chem 19: 7482 -92 (2011) Borthwick, JA; Ancellin, N; Bertrand, SM; Bingham, RP; Carter, PS; Chung, CW; Churcher, I; Dodic, N; Fournier, C; Francis, PL; Hobbs, A; Jamieson, C; Pickett, SD; Smith, SE; Somers, DO; Spitzfaden, C; Suckling, CJ; Young, RJ J Med Chem 59: 2452 -67 (2016) Levin, JI; Chan, PS; Coupet, J; Thibault, L; Venkatesan, AM; Bailey, TK; Vice, G; Cobuzzi, A; Lai, F; Mellish, N Bioorg Med Chem Lett 4: 1709 -1714 (1994) Flanagan, ME; Blumenkopf, TA; Brissette, WH; Brown, MF; Casavant, JM; Shang-Poa, C; Doty, JL; Elliott, EA; Fisher, MB; Hines, M; Kent, C; Kudlacz, EM; Lillie, BM; Magnuson, KS; McCurdy, SP; Munchhof, MJ; Perry, BD; Sawyer, PS; Strelevitz, TJ; Subramanyam, C; Sun, J; Whipple, DA; Changelian, PS J Med Chem 53: 8468 -84 (2010) Boyd, MJ; Collier, PN; Clark, MP; Deng, H; Kesavan, S; Ronkin, SM; Waal, N; Wang, J; Cao, J; Li, P; Come, J; Davies, I; Duffy, JP; Cochran, JE; Court, JJ; Chandupatla, K; Jackson, KL; Maltais, F; O'Dowd, H; Boucher, C; Considine, T; Taylor, WP; Gao, H; Chakilam, A; Engtrakul, J; Crawford, D; Doyle, E; Phillips, J; Kemper, R; Swett, R; Empfield, J; Bunnage, ME; Charifson, PS; Magavi, SS J Med Chem 64: 17777 -17794 (2021) Guerrero, M; Poddutoori, R; Urbano, M; Peng, X; Spicer, TP; Chase, PS; Hodder, PS; Schaeffer, MT; Brown, S; Rosen, H; Roberts, E Bioorg Med Chem Lett 23: 6346 -9 (2013) Li, J; Rush, TS; Li, W; DeVincentis, D; Du, X; Hu, Y; Thomason, JR; Xiang, JS; Skotnicki, JS; Tam, S; Cunningham, KM; Chockalingam, PS; Morris, EA; Levin, JI Bioorg Med Chem Lett 15: 4961 -6 (2005) Getahun, Z; Jurd, L; Chu, PS; Lin, CM; Hamel, E J Med Chem 35: 1058 -67 (1992) Cogan, PS; Koch, TH J Med Chem 46: 5258 -70 (2003) Selvaraj, R; Liu, S; Hassink, M; Huang, CW; Yap, LP; Park, R; Fox, JM; Li, Z; Conti, PS Bioorg Med Chem Lett 21: 5011 -4 (2011) Salvadori, S; Balboni, G; Guerrini, R; Tomatis, R; Bianchi, C; Bryant, SD; Cooper, PS; Lazarus, LH J Med Chem 40: 3100 -8 (1997) Finke, PE; Shah, SK; Fletcher, DS; Ashe, BM; Brause, KA; Chandler, GO; Dellea, PS; Hand, KM; Maycock, AL; Osinga, DG J Med Chem 38: 2449 -62 (1995) Das, PK; Matada, GSP; Pal, R; Maji, L; Dhiwar, PS; Manjushree, BV; Viji, MP Eur J Med Chem 274: Napper, AD; Hixon, J; McDonagh, T; Keavey, K; Pons, JF; Barker, J; Yau, WT; Amouzegh, P; Flegg, A; Hamelin, E; Thomas, RJ; Kates, M; Jones, S; Navia, MA; Saunders, JO; DiStefano, PS; Curtis, R J Med Chem 48: 8045 -54 (2005) Guzii, AG; Makarieva, TN; Denisenko, VA; Dmitrenok, PS; Popov, RS; Kuzmich, AS; Fedorov, SN; Krasokhin, VB; Kim, NY; Stonik, VA J Nat Prod 81: 2763 -2767 (2018) Jaishankar, P; Hansell, E; Zhao, DM; Doyle, PS; McKerrow, JH; Renslo, AR Bioorg Med Chem Lett 18: 624 -8 (2008) Liang, J; Labadie, S; Zhang, B; Ortwine, DF; Patel, S; Vinogradova, M; Kiefer, JR; Mauer, T; Gehling, VS; Harmange, JC; Cummings, R; Lai, T; Liao, J; Zheng, X; Liu, Y; Gustafson, A; Van der Porten, E; Mao, W; Liederer, BM; Deshmukh, G; An, L; Ran, Y; Classon, M; Trojer, P; Dragovich, PS; Murray, L Bioorg Med Chem Lett 27: 2974 -2981 (2017) Liu, J; Taylor, SF; Dupart, PS; Arnold, CL; Sridhar, J; Jiang, Q; Wang, Y; Skripnikova, EV; Zhao, M; Foroozesh, M J Med Chem 56: 4082 -92 (2013) Quiroz, RV; Reutershan, MH; Schneider, SE; Sloman, D; Lacey, BM; Swalm, BM; Yeung, CS; Gibeau, C; Spellman, DS; Rankic, DA; Chen, D; Witter, D; Linn, D; Munsell, E; Feng, G; Xu, H; Hughes, JME; Lim, J; Saurí, J; Geddes, K; Wan, M; Mansueto, MS; Follmer, NE; Fier, PS; Siliphaivanh, P; Daublain, P; Palte, RL; Hayes, RP; Lee, S; Kawamura, S; Silverman, S; Sanyal, S; Henderson, TJ; Ye, Y; Gao, Y; Nicholson, B; Machacek, MR J Med Chem 64: 3911 -3939 (2021) Arias, DG; Herrera, FE; Garay, AS; Rodrigues, D; Forastieri, PS; Luna, LE; Bürgi, MD; Prieto, C; Iglesias, AA; Cravero, RM; Guerrero, SA Eur J Med Chem 125: 1088 -1097 (2017) Heathcote, DA; Patel, H; Kroll, SH; Hazel, P; Periyasamy, M; Alikian, M; Kanneganti, SK; Jogalekar, AS; Scheiper, B; Barbazanges, M; Blum, A; Brackow, J; Siwicka, A; Pace, RD; Fuchter, MJ; Snyder, JP; Liotta, DC; Freemont, PS; Aboagye, EO; Coombes, RC; Barrett, AG; Ali, S J Med Chem 53: 8508 -22 (2010) Galatin, PS; Abraham, DJ J Med Chem 47: 4163 -5 (2004) Friedman, SH; Ganapathi, PS; Rubin, Y; Kenyon, GL J Med Chem 41: 2424 -9 (1998) Tora, G; Jiang, J; Bostwick, JS; Gargalovic, PS; Onorato, JM; Luk, CE; Generaux, C; Xu, C; Galella, MA; Wang, T; He, Y; Wexler, RR; Finlay, HJ Bioorg Med Chem Lett 50: (2021) De Savi, C; Cox, RJ; Warner, DJ; Cook, AR; Dickinson, MR; McDonough, A; Morrill, LC; Parker, B; Andrews, G; Young, SS; Gilmour, PS; Riley, R; Dearman, MS J Med Chem 57: 4661 -76 (2014) Lipchock, JM; Hendrickson, HP; Douglas, BB; Bird, KE; Ginther, PS; Rivalta, I; Ten, NS; Batista, VS; Loria, JP Biochemistry 56: 96 -106 (2017) Girase, PS; Dhawan, S; Kumar, V; Shinde, SR; Palkar, MB; Karpoormath, R Eur J Med Chem 210: (2021) Grant, PS; Kahlcke, N; Govindpani, K; Hunter, M; MacDonald, C; Brimble, MA; Glass, M; Furkert, DP Bioorg Med Chem Lett 29: (2019) Guerreiro, PS; Estácio, SG; Antunes, F; Fernandes, AS; Pinheiro, PF; Costa, JG; Castro, M; Miranda, JP; Guedes, RC; Oliveira, NG Chem Biol Drug Des 88: 915 -925 (2016) Kasangana, PB; Haddad, PS; Eid, HM; Nachar, A; Stevanovic, T J Nat Prod 81: 2169 -2176 (2018) Faull, AW; Gaskin, H; Hadfield, PS; Jessup, R; Russell, K; Watkins, WJ; Wayne, M Bioorg Med Chem Lett 2: 1181 -1186 (1992) Terai, H; Tan, L; Beauchamp, EM; Hatcher, JM; Liu, Q; Meyerson, M; Gray, NS; Hammerman, PS ACS Chem Biol 10: 2687 -96 (2015) Huang, Y; Hammond, PS; Whirrett, BR; Kuhner, RJ; Wu, L; Childers, SR; Mach, RH J Med Chem 41: 2361 -70 (1998) Page, MI; Hinchliffe, PS; Wood, JM; Harding, LP; Laws, AP Bioorg Med Chem Lett 13: 4489 -92 (2003) Hyohdoh, I; Furuichi, N; Aoki, T; Itezono, Y; Shirai, H; Ozawa, S; Watanabe, F; Matsushita, M; Sakaitani, M; Ho, PS; Takanashi, K; Harada, N; Tomii, Y; Yoshinari, K; Ori, K; Tabo, M; Aoki, Y; Shimma, N; Iikura, H ACS Med Chem Lett 4: 1059 -63 (2013) Guerrero, M; Poddutoori, R; Urbano, M; Peng, X; Spicer, TP; Chase, PS; Hodder, PS; Schaeffer, MT; Brown, S; Rosen, H; Roberts, E Bioorg Med Chem Lett 23: 6346 -9 (2013) Hoffman, PS; Sisson, G; Croxen, MA; Welch, K; Harman, WD; Cremades, N; Morash, MG Antimicrob Agents Chemother 51: 868 -76 (2007) Johansson, L; Sohn, D; Thorberg, SO; Jackson, DM; Kelder, D; Larsson, LG; Rényi, L; Ross, SB; Wallsten, C; Eriksson, H; Hu, PS; Jerning, E; Mohell, N; Westlind-Danielsson, A J Pharmacol Exp Ther 283: 216 -25 (1997) Wood, KW; Lad, L; Luo, L; Qian, X; Knight, SD; Nevins, N; Brejc, K; Sutton, D; Gilmartin, AG; Chua, PR; Desai, R; Schauer, SP; McNulty, DE; Annan, RS; Belmont, LD; Garcia, C; Lee, Y; Diamond, MA; Faucette, LF; Giardiniere, M; Zhang, S; Sun, CM; Vidal, JD; Lichtsteiner, S; Cornwell, WD; Greshock, JD; Wooster, RF; Finer, JT; Copeland, RA; Huang, PS; Morgans, DJ; Dhanak, D; Bergnes, G; Sakowicz, R; Jackson, JR Proc Natl Acad Sci U S A 107: 5839 -44 Humphries, PS; Lafontaine, JA; Agree, CS; Alexander, D; Chen, P; Do, QQ; Li, LY; Lunney, EA; Rajapakse, RJ; Siegel, K; Timofeevski, SL; Wang, T; Wilhite, DM Bioorg Med Chem Lett 19: 2099 -102 (2009) Safina, BS; Baker, S; Baumgardner, M; Blaney, PM; Chan, BK; Chen, YH; Cartwright, MW; Castanedo, G; Chabot, C; Cheguillaume, AJ; Goldsmith, P; Goldstein, DM; Goyal, B; Hancox, T; Handa, RK; Iyer, PS; Kaur, J; Kondru, R; Kenny, JR; Krintel, SL; Li, J; Lesnick, J; Lucas, MC; Lewis, C; Mukadam, S; Murray, J; Nadin, AJ; Nonomiya, J; Padilla, F; Palmer, WS; Pang, J; Pegg, N; Price, S; Reif, K; Salphati, L; Savy, PA; Seward, EM; Shuttleworth, S; Sohal, S; Sweeney, ZK; Tay, S; Tivitmahaisoon, P; Waszkowycz, B; Wei, B; Yue, Q; Zhang, C; Sutherlin, DP J Med Chem 55: 5887 -900 (2012) Hanan, EJ; Eigenbrot, C; Bryan, MC; Burdick, DJ; Chan, BK; Chen, Y; Dotson, J; Heald, RA; Jackson, PS; La, H; Lainchbury, MD; Malek, S; Purkey, HE; Schaefer, G; Schmidt, S; Seward, EM; Sideris, S; Tam, C; Wang, S; Yeap, SK; Yen, I; Yin, J; Yu, C; Zilberleyb, I; Heffron, TP J Med Chem 57: 10176 -91 (2014) Pujala, B; Ansari, A; Sapra, S; Jadhavar, PS; Pendharkar, D; Ramachandran, SA; Saeed, U; Danodia, A; Khan, F; Patni, S; Soni, S; Gupta, A; Chakravarty, S; Sathe, BD US Patent US20240352046 (2024) Jayasekara, PS; Barrett, MO; Ball, CB; Brown, KA; Hammes, E; Balasubramanian, R; Harden, TK; Jacobson, KA J Med Chem 57: 3874 -83 (2014) Lee, YS; Chuang, SH; Huang, LY; Lai, CL; Lin, YH; Yang, JY; Liu, CW; Yang, SC; Lin, HS; Chang, CC; Lai, JY; Jian, PS; Lam, K; Chang, JM; Lau, JY; Huang, JJ J Med Chem 57: 4098 -110 (2014) Mittal, S; Malde, A; Selvam, C; Arun, KH; Johar, PS; Jachak, SM; Ramarao, P; Bharatam, PV; Chawla, HP Bioorg Med Chem Lett 14: 979 -82 (2004) Johnson, PS; Ryckmans, T; Bryans, J; Beal, DM; Dack, KN; Feeder, N; Harrison, A; Lewis, M; Mason, HJ; Mills, J; Newman, J; Pasquinet, C; Rawson, DJ; Roberts, LR; Russell, R; Spark, D; Stobie, A; Underwood, TJ; Ward, R; Wheeler, S Bioorg Med Chem Lett 21: 5684 -7 (2011) Palin, R; Clark, JK; Evans, L; Houghton, AK; Jones, PS; Prosser, A; Wishart, G; Yoshiizumi, K Bioorg Med Chem 16: 2829 -51 (2008) Bissantz, C; Dehmlow, H; Erickson, SD; Karnachi, PS; Kim, K; Martin, RE; Mattei, P; Sander, UO; Pietranico-Cole, SL; Richter, H; Ullmer, C US Patent US10385022 (2019) Jacobsen, EJ; Mitchell, MA; Hendges, SK; Belonga, KL; Skaletzky, LL; Stelzer, LS; Lindberg, TJ; Fritzen, EL; Schostarez, HJ; O'Sullivan, TJ; Maggiora, LL; Stuchly, CW; Laborde, AL; Kubicek, MF; Poorman, RA; Beck, JM; Miller, HR; Petzold, GL; Scott, PS; Truesdell, SE; Wallace, TL; Wilks, JW; Fisher, C; Goodman, LV; Kaytes, PS J Med Chem 42: 1525 -36 (1999) Truong, AP; Aubele, DL; Probst, GD; Neitzel, ML; Semko, CM; Bowers, S; Dressen, D; Hom, RK; Konradi, AW; Sham, HL; Garofalo, AW; Keim, PS; Wu, J; Dappen, MS; Wong, K; Goldbach, E; Quinn, KP; Sauer, JM; Brigham, EF; Wallace, W; Nguyen, L; Hemphill, SS; Bova, MP; Bard, F; Yednock, TA; Basi, G Bioorg Med Chem Lett 19: 4920 -3 (2009) Beaumont, EJ; Devraj, R; Kerry, PS; Kumaravel, G; Loke, PL; Meniconi, M; Palfrey, JJ; North, C; Lecci, C; Tye, H US Patent US10703741 (2020) Silbermann, K; Shah, CP; Sahu, NU; Juvale, K; Stefan, SM; Kharkar, PS; Wiese, M Eur J Med Chem 164: 193 -213 (2019) Fass, DM; Shah, R; Ghosh, B; Hennig, K; Norton, S; Zhao, WN; Reis, SA; Klein, PS; Mazitschek, R; Maglathlin, RL; Lewis, TA; Haggarty, SJ ACS Med Chem Lett 2: 39 -42 (2011) Suresh, L; Onkara, P; Kumar, PS; Pydisetty, Y; Chandramouli, GV Bioorg Med Chem Lett 26: 4007 -14 (2016) Bukhtiyarova, M; Cook, EM; Hancock, PJ; Hruza, AW; Shaw, AW; Adam, GC; Barnard, RJO; McKenna, PM; Holloway, MK; Bell, IM; Carroll, S; Cornella-Taracido, I; Cox, CD; Kutchukian, PS; Powell, DA; Strickland, C; Trotter, BW; Tudor, M; Wolkenberg, S; Li, J; Tellers, DM ACS Med Chem Lett 12: 99 -106 (2021) Braga, SF; Alves, ÉV; Ferreira, RS; Fradico, JR; Lage, PS; Duarte, MC; Ribeiro, TG; Júnior, PA; Romanha, AJ; Tonini, ML; Steindel, M; Coelho, EF; de Oliveira, RB Eur J Med Chem 71: 282 -9 (2014) Lainé, DI; Yan, H; Xie, H; Davis, RS; Dufour, J; Widdowson, KL; Palovich, MR; Wan, Z; Foley, JJ; Schmidt, DB; Hunsberger, GE; Burman, M; Bacon, AM; Webb, EF; Luttmann, MA; Salmon, M; Sarau, HM; Umbrecht, ST; Landis, PS; Peck, BJ; Busch-Petersen, J Bioorg Med Chem Lett 22: 3366 -9 (2012) Qian, X; Liang, GB; Feng, D; Fisher, M; Crumley, T; Rattray, S; Dulski, PM; Gurnett, A; Leavitt, PS; Liberator, PA; Misura, AS; Samaras, S; Tamas, T; Schmatz, DM; Wyvratt, M; Biftu, T Bioorg Med Chem Lett 16: 2817 -21 (2006) Hsu, PH; Chiu, DC; Wu, KL; Lee, PS; Jan, JT; Cheng, YE; Tsai, KC; Cheng, TJ; Fang, JM Eur J Med Chem 154: 314 -323 (2018) Marquez, VE; Liu, PS; Kelley, JA; Driscoll, JS; McCormack, JJ J Med Chem 23: 713 -5 (1980) Rong, SB; Enyedy, IJ; Qiao, L; Zhao, L; Ma, D; Pearce, LL; Lorenzo, PS; Stone, JC; Blumberg, PM; Wang, S; Kozikowski, AP J Med Chem 45: 853 -60 (2002) Venkatesh, C; Doorneweerd, DD; Xia, W; Putt, KS; Low, PS Bioorg Med Chem Lett 42: (2021) da Costa, JS; Lopes, JP; Russowsky, D; Petzhold, CL; Borges, AC; Ceschi, MA; Konrath, E; Batassini, C; Lunardi, PS; Gonçalves, CA Eur J Med Chem 62: 556 -63 (2013) Lyon, RA; Titeler, M; McKenney, JD; Magee, PS; Glennon, RA J Med Chem 29: 630 -4 (1986) Phull, MS; Jadav, SS; Gundla, R; Mainkar, PS Eur J Med Chem 212: (2021) Radwan, AA; Al-Mohanna, F; Alanazi, FK; Manogaran, PS; Al-Dhfyan, A Bioorg Med Chem Lett 26: 1664 -70 (2016) Liu, C; Dunaway-Mariano, D; Mariano, PS Eur J Med Chem 128: 274 -286 (2017) McCarthy, C; Moulton, B; Walker, ER; McMahon, PS US Patent US20230293517 (2023) Drewry, DH; Potjewyd, FM; Bayati, A; Smith, JL; Dickmander, RJ; Howell, S; Taft-Benz, S; Min, SM; Hossain, MA; Heise, M; McPherson, PS; Moorman, NJ; Axtman, AD J Med Chem 65: 12860 -12882 (2022) Lassaux, P; Hamel, M; Gulea, M; Delbrück, H; Mercuri, PS; Horsfall, L; Dehareng, D; Kupper, M; Frère, JM; Hoffmann, K; Galleni, M; Bebrone, C J Med Chem 53: 4862 -76 (2010) Marcin, LR; Warrier, J; Thangathirupathy, S; Shi, J; Karageorge, GN; Pearce, BC; Ng, A; Park, H; Kempson, J; Li, J; Zhang, H; Mathur, A; Reddy, AB; Nagaraju, G; Tonukunuru, G; Gupta, GVRKM; Kamble, M; Mannoori, R; Cheruku, S; Jogi, S; Gulia, J; Bastia, T; Sanmathi, C; Aher, J; Kallem, R; Srikumar, BN; Vijaya, KK; Naidu, PS; Paschapur, M; Kalidindi, N; Vikramadithyan, R; Ramarao, M; Denton, R; Molski, T; Shields, E; Subramanian, M; Zhuo, X; Nophsker, M; Simmermacher, J; Sinz, M; Albright, C; Bristow, LJ; Islam, I; Bronson, JJ; Olson, RE; King, D; Thompson, LA; Macor, JE ACS Med Chem Lett 9: 472 -477 (2018) Lucas, JM; Heinlein, C; Kim, T; Hernandez, SA; Malik, MS; True, LD; Morrissey, C; Corey, E; Montgomery, B; Mostaghel, E; Clegg, N; Coleman, I; Brown, CM; Schneider, EL; Craik, C; Simon, JA; Bedalov, A; Nelson, PS Cancer Discov 4: 1310 -25 (2014) Ng, PS; Manjunatha, UH; Rao, SP; Camacho, LR; Ma, NL; Herve, M; Noble, CG; Goh, A; Peukert, S; Diagana, TT; Smith, PW; Kondreddi, RR Eur J Med Chem 106: 144 -56 (2015) Adams, GL; Pall, PS; Grauer, SM; Zhou, X; Ballard, JE; Vavrek, M; Kraus, RL; Morissette, P; Li, N; Colarusso, S; Bianchi, E; Palani, A; Klein, R; John, CT; Wang, D; Tudor, M; Nolting, AF; Biba, M; Nowak, T; Makarov, AA; Reibarkh, M; Buevich, AV; Zhong, W; Regalado, EL; Wang, X; Gao, Q; Shahripour, A; Zhu, Y; de Simone, D; Frattarelli, T; Pasquini, NM; Magotti, P; Iaccarino, R; Li, Y; Solly, K; Lee, KJ; Wang, W; Chen, F; Zeng, H; Wang, J; Regan, H; Amin, RP; Regan, CP; Burgey, CS; Henze, DA; Sun, C; Tellers, DM J Med Chem 65: 485 -496 (2022) Singh, EK; Ravula, S; Pan, CM; Pan, PS; Vasko, RC; Lapera, SA; Weerasinghe, SV; Pflum, MK; McAlpine, SR Bioorg Med Chem Lett 18: 2549 -54 (2008) Hegde, VR; Chan, TM; Pu, H; Gullo, VP; Patel, MG; Das, P; Wagner, N; Parameswaran, PS; Naik, CG Bioorg Med Chem Lett 12: 3203 -5 (2002) Els, S; Schild, E; Petersen, PS; Kilian, TM; Mokrosinski, J; Frimurer, TM; Chollet, C; Schwartz, TW; Holst, B; Beck-Sickinger, AG J Med Chem 55: 7437 -49 (2012) Portoghese, PS; Sultana, M; Moe, ST; Takemori, AE J Med Chem 37: 579 -85 (1994) Bianchi, BR; El Kouhen, R; Neelands, TR; Lee, CH; Gomtsyan, A; Raja, SN; Vaidyanathan, SN; Surber, B; McDonald, HA; Surowy, CS; Faltynek, CR; Moreland, RB; Jarvis, MF; Puttfarcken, PS J Pharmacol Exp Ther 323: 285 -93 (2007) Leblanc, E; Ban, F; Cavga, AD; Lawn, S; Huang, CF; Mohan, S; Chang, MEK; Flory, MR; Ghaidi, F; Lingadahalli, S; Chen, G; Yu, IPL; Morin, H; Lallous, N; Gleave, ME; Mohammed, H; Young, RN; Rennie, PS; Lack, NA; Cherkasov, A J Med Chem 64: 14968 -14982 (2021) Bellera, CL; Balcazar, DE; Vanrell, MC; Casassa, AF; Palestro, PH; Gavernet, L; Labriola, CA; Gálvez, J; Bruno-Blanch, LE; Romano, PS; Carrillo, C; Talevi, A Eur J Med Chem 93: 338 -48 (2015) FINK, BE; CHERNEY, RJ; NGU, K; VELAPARTHI, U; VACCARO, WD; RUAN, Z; QIN, L; SHIRUDE, PS; RAHAMAN, H US Patent US20240217982 (2024) Santhoshi, A; Mahendar, B; Mattapally, S; Sadhu, PS; Banerjee, SK; Jayathirtha Rao, V Bioorg Med Chem Lett 24: 1952 -7 (2014) Salva, PS; Hite, GJ; Heyman, RA; Gianutsos, G J Med Chem 29: 2111 -3 (1986) Shah, RR; De Vita, E; Sathyamurthi, PS; Conole, D; Zhang, X; Fellows, E; Dickinson, ER; Fleites, CM; Queisser, MA; Harling, JD; Tate, EW J Med Chem 67: 4641 -4654 Changelian, PS; Flanagan, ME; Ball, DJ; Kent, CR; Magnuson, KS; Martin, WH; Rizzuti, BJ; Sawyer, PS; Perry, BD; Brissette, WH; McCurdy, SP; Kudlacz, EM; Conklyn, MJ; Elliott, EA; Koslov, ER; Fisher, MB; Strelevitz, TJ; Yoon, K; Whipple, DA; Sun, J; Munchhof, MJ; Doty, JL; Casavant, JM; Blumenkopf, TA; Hines, M; Brown, MF; Lillie, BM; Subramanyam, C; Shang-Poa, C; Milici, AJ; Beckius, GE; Moyer, JD; Su, C; Woodworth, TG; Gaweco, AS; Beals, CR; Littman, BH; Fisher, DA; Smith, JF; Zagouras, P; Magna, HA; Saltarelli, MJ; Johnson, KS; Nelms, LF; Des Etages, SG; Hayes, LS; Kawabata, TT; Finco-Kent, D; Baker, DL; Larson, M; Si, MS; Paniagua, R; Higgins, J; Holm, B; Reitz, B; Zhou, YJ; Morris, RE; O'Shea, JJ; Borie, DC Science 302: 875 -878 (2003) Jacobsen, EJ; Mitchell, MA; Hendges, SK; Belonga, KL; Skaletzky, LL; Stelzer, LS; Lindberg, TJ; Fritzen, EL; Schostarez, HJ; O'Sullivan, TJ; Maggiora, LL; Stuchly, CW; Laborde, AL; Kubicek, MF; Poorman, RA; Beck, JM; Miller, HR; Petzold, GL; Scott, PS; Truesdell, SE; Wallace, TL; Wilks, JW; Fisher, C; Goodman, LV; Kaytes, PS J Med Chem 42: 1525 -36 (1999) Beseda, I; Czollner, L; Shah, PS; Khunt, R; Gaware, R; Kosma, P; Stanetty, C; Del Ruiz-Ruiz, MC; Amer, H; Mereiter, K; Da Cunha, T; Odermatt, A; Classen-Houben, D; Jordis, U Bioorg Med Chem 18: 433 -54 (2010) Shapiro, PS; MacKerell, Jr., AD; Fletcher, S US Patent US9115122 (2015) Podvinec, M; Lim, SP; Schmidt, T; Scarsi, M; Wen, D; Sonntag, LS; Sanschagrin, P; Shenkin, PS; Schwede, T J Med Chem 53: 1483 -95 (2010) Van Dort, ME; Ciliax, BJ; Gildersleeve, DL; Sherman, PS; Rosenspire, KC; Young, AB; Junck, L; Wieland, DM J Med Chem 31: 2081 -6 (1988) Shirude, PS; Madhavapeddi, P; Naik, M; Murugan, K; Shinde, V; Nandishaiah, R; Bhat, J; Kumar, A; Hameed, S; Holdgate, G; Davies, G; McMiken, H; Hegde, N; Ambady, A; Venkatraman, J; Panda, M; Bandodkar, B; Sambandamurthy, VK; Read, JA J Med Chem 56: 8533 -42 (2013) Sidhu, PS; Zhou, Q; Desai, UR Bioorg Med Chem Lett 24: 5716 -20 (2014) Whittamore, PR; Addie, MS; Bennett, SN; Birch, AM; Butters, M; Godfrey, L; Kenny, PW; Morley, AD; Murray, PM; Oikonomakos, NG; Otterbein, LR; Pannifer, AD; Parker, JS; Readman, K; Siedlecki, PS; Schofield, P; Stocker, A; Taylor, MJ; Townsend, LA; Whalley, DP; Whitehouse, J Bioorg Med Chem Lett 16: 5567 -71 (2006) Rana, D; Kalamuddin, M; Sundriyal, S; Jaiswal, V; Sharma, G; Das Sarma, K; Sijwali, PS; Mohmmed, A; Malhotra, P; Mahindroo, N Bioorg Med Chem 28: (2020) Blaszczyk, R; Brzezinska, J; Dymek, B; Stanczak, PS; Mazurkiewicz, M; Olczak, J; Nowicka, J; Dzwonek, K; Zagozdzon, A; Golab, J; Golebiowski, A ACS Med Chem Lett 11: 433 -438 (2020) Klumb, LA; Chu, V; Stayton, PS Biochemistry 37: 7657 -63 (1998) Yamani, A; Zdżalik-Bielecka, D; Lipner, J; Stańczak, A; Piórkowska, N; Stańczak, PS; Olejkowska, P; Hucz-Kalitowska, J; Magdycz, M; Dzwonek, K; Dubiel, K; Lamparska-Przybysz, M; Popiel, D; Pieczykolan, J; Wieczorek, M Eur J Med Chem 210: (2021) Jarvi, ET; McCarthy, JR; Mehdi, S; Matthews, DP; Edwards, ML; Prakash, NJ; Bowlin, TL; Sunkara, PS; Bey, P J Med Chem 34: 647 -56 (1991) Kosugi, T; Mitchell, DR; Fujino, A; Imai, M; Kambe, M; Kobayashi, S; Makino, H; Matsueda, Y; Oue, Y; Komatsu, K; Imaizumi, K; Sakai, Y; Sugiura, S; Takenouchi, O; Unoki, G; Yamakoshi, Y; Cunliffe, V; Frearson, J; Gordon, R; Harris, CJ; Kalloo-Hosein, H; Le, J; Patel, G; Simpson, DJ; Sherborne, B; Thomas, PS; Suzuki, N; Takimoto-Kamimura, M; Kataoka, K J Med Chem 55: 6700 -15 (2012) Li, YH; Tseng, PS; Evans, KA; Jaworski, JP; Morrow, DM; Fries, HE; Wu, CW; Edwards, RM; Jin, J Bioorg Med Chem Lett 20: 6744 -7 (2010) Zang, J; Peters, F; Cambet, Y; Cifuentes-Pagano, E; Hissabu, MMS; Dustin, CM; Svensson, LH; Olesen, MM; Poulsen, MFL; Jacobsen, S; Tuelung, PS; Narayanan, D; Langkilde, AE; Gajhede, M; Pagano, PJ; Jaquet, V; Vilhardt, F; Bach, A J Med Chem 66: 14963 -15005 (2023) Henke, BR; Consler, TG; Go, N; Hale, RL; Hohman, DR; Jones, SA; Lu, AT; Moore, LB; Moore, JT; Orband-Miller, LA; Robinett, RG; Shearin, J; Spearing, PK; Stewart, EL; Turnbull, PS; Weaver, SL; Williams, SP; Wisely, GB; Lambert, MH J Med Chem 45: 5492 -505 (2002) Hatcher, JM; Vatsan, PS; Wang, E; Jiang, J; Gray, NS ACS Med Chem Lett 12: 1689 -1693 (2021) Charrier, JD; Durrant, SJ; Golec, JM; Kay, DP; Knegtel, RM; MacCormick, S; Mortimore, M; O'Donnell, ME; Pinder, JL; Reaper, PM; Rutherford, AP; Wang, PS; Young, SC; Pollard, JR J Med Chem 54: 2320 -30 (2011) Lampe, JW; Watson, PS; Slade, DJ US Patent US8476295 (2013) Dreyer, MK; Borcherding, DR; Dumont, JA; Peet, NP; Tsay, JT; Wright, PS; Bitonti, AJ; Shen, J; Kim, SH J Med Chem 44: 524 -30 (2001) Moore, WM; Webber, RK; Fok, KF; Jerome, GM; Connor, JR; Manning, PT; Wyatt, PS; Misko, TP; Tjoeng, FS; Currie, MG J Med Chem 39: 669 -72 (1996) Banerjee, A; Yadav, PS; Bajpai, M; Sangana, RR; Gullapalli, S; Gudi, GS; Gharat, LA Bioorg Med Chem Lett 22: 3223 -8 (2012) Bockus, AT; Leung, SF; Earp, DJ; Garcia Dominguez, PS; Spellmeyer, DC; Hernandez, L; Baldomero, MP; Gleason, CE; Walton, BF; Singh, R; Aggen, JB; Dupper, NJ; Shapiro, JA; Kreatsoulas, C; Bambal, RB; Fung, CC; Ramaseshan, M US Patent US20240218021 (2024) de Andrade, P; Mantoani, SP; Gonçalves Nunes, PS; Magadán, CR; Pérez, C; Xavier, DJ; Hojo, ETS; Campillo, NE; Martínez, A; Carvalho, I Bioorg Med Chem 27: 931 -943 (2019) Glunz, PW; Sitkoff, DF; Bodas, MS; Yadav, ND; Patil, S; Maddu Rao, PS US Patent US10112929 (2018) Learmonth, DA; Vieira-Coelho, MA; Benes, J; Alves, PC; Borges, N; Freitas, AP; da-Silva, PS J Med Chem 45: 685 -95 (2002) Zhou, Y; Wei, L; Brady, TP; Murali Mohan Redddy, PS; Nguyen, T; Chen, J; Au, Q; Yoon, IS; Yip, G; Zhang, B; Barber, JR; Ng, SC Bioorg Med Chem Lett 19: 4303 -7 (2009) Zhou, Y; Vu, K; Chen, Y; Pham, J; Brady, T; Liu, G; Chen, J; Nam, J; Murali Mohan Reddy, PS; Au, Q; Yoon, IS; Tremblay, MH; Yip, G; Cher, C; Zhang, B; Barber, JR; Ng, SC Bioorg Med Chem Lett 19: 3128 -35 (2009)
ChEMBL_962568 (CHEMBL2390763) Inhibition of human GSK3beta-mediated YRRAAVPPSPSLSRHSSPHQ(pS)EDEEE phosphorylation ChEMBL_1543460 (CHEMBL3750179) Inhibition of human TBK1 using [KRRRAL[pS]VASLPGL] as substrate ChEMBL_1546481 (CHEMBL3748672) Inhibition of human CK1alpha1 using [KRRRAL[pS]VASLPGL] as substrate ChEMBL_1546482 (CHEMBL3748673) Inhibition of human CK1delta using [KRRRAL[pS]VASLPGL] as substrate ChEMBL_1546483 (CHEMBL3748674) Inhibition of human CK1epsilon using [KRRRAL[pS]VASLPGL] as substrate ChEMBL_1546484 (CHEMBL3748987) Inhibition of human CK1gamma1 using [KRRRAL[pS]VASLPGL] as substrate ChEMBL_1546485 (CHEMBL3748988) Inhibition of human CK1gamma2 using [KRRRAL[pS]VASLPGL] as substrate ChEMBL_1546486 (CHEMBL3748989) Inhibition of human CK1gamma3 using [KRRRAL[pS]VASLPGL] as substrate ChEMBL_1546681 (CHEMBL3748206) Inhibition of human GSK3alpha using [YRRAAVPPSPSLSRHSSPHQ(pS)EDEEE] as substrate ChEMBL_2572610 Inhibition of human PIK3CG using PIP2/PS as substrate by ADP-Glo assay ChEMBL_2572611 Inhibition of human PIK3C2A using PIP2/PS as substrate by ADP-Glo assay ChEMBL_2572612 Inhibition of human PIK3C2B using PIP2/PS as substrate by ADP-Glo assay ChEMBL_2572613 Inhibition of human PIK3C2G using PIP2/PS as substrate by ADP-Glo assay ChEMBL_2572615 Inhibition of human PI4K2B using PI/PS as substrate by ADP-Glo assay ChEMBL_2572616 Inhibition of human PI4K2A using PI/PS as substrate by ADP-Glo assay ChEMBL_2572617 Inhibition of human PIP5K1A using PI/PS as substrate by ADP-Glo assay ChEMBL_2572618 Inhibition of human PIP5K1C using PI/PS as substrate by ADP-Glo assay ChEMBL_2572607 Inhibition of human PIK3CA/PIK3R1 using PIP2/PS as substrate by ADP-Glo assay ChEMBL_2572608 Inhibition of human PIK3CB/PIK3R1 using PIP2/PS as substrate by ADP-Glo assay ChEMBL_2572609 Inhibition of human PIK3CD/PIK3R1 using PIP2/PS as substrate by ADP-Glo assay ChEMBL_967913 (CHEMBL2400729) Inhibition of P110gamma (unknown origin) using PIP2:PS as substrate by TR-FRET assay ChEMBL_967915 (CHEMBL2400731) Inhibition of P110delta (unknown origin) using PIP2:PS as substrate by TR-FRET assay ChEMBL_2328221 Inhibition of N-terminal GST-tagged recombinant human CK1 delta expressed in Escherichia coli BL21 cells using PLSRTL-pS-VA-pS-LPGL as substrate measured after 60 mins in presence of ATP by ADP-Glo assay ChEMBL_1828490 (CHEMBL4328364) Inhibition of human CK1a1 using KRRRAL[pS]VASLPGL as substrate by [gamma-33P]-ATP assay ChEMBL_1828491 (CHEMBL4328365) Inhibition of human CK1a1L using KRRRAL[pS]VASLPGL as substrate by [gamma-33P]-ATP assay ChEMBL_1828492 (CHEMBL4328366) Inhibition of human CK1D using KRRRAL[pS]VASLPGL as substrate by [gamma-33P]-ATP assay ChEMBL_1828493 (CHEMBL4328367) Inhibition of human CK1E using KRRRAL[pS]VASLPGL as substrate by [gamma-33P]-ATP assay ChEMBL_1828494 (CHEMBL4328368) Inhibition of human CK1gamma1 using KRRRAL[pS]VASLPGL as substrate by [gamma-33P]-ATP assay ChEMBL_1828495 (CHEMBL4328369) Inhibition of human CK1gamma2 using KRRRAL[pS]VASLPGL as substrate by [gamma-33P]-ATP assay ChEMBL_1828496 (CHEMBL4328370) Inhibition of human CK1gamma3 using KRRRAL[pS]VASLPGL as substrate by [gamma-33P]-ATP assay ChEMBL_1828565 (CHEMBL4328439) Inhibition of human GSK3A using YRRAAVPPSPSLSRHSSPHQ(pS)EDEEE as substrate by [gamma-33P]-ATP assay ChEMBL_1828566 (CHEMBL4328440) Inhibition of human GSK3B using YRRAAVPPSPSLSRHSSPHQ(pS)EDEEE as substrate by [gamma-33P]-ATP assay ChEMBL_1828754 (CHEMBL4328628) Inhibition of human TBK1 using KRRRAL[pS]VASLPGL as substrate by [gamma-33P]-ATP assay ChEMBL_2065278 (CHEMBL4720531) Inhibition of human TBK1 using KRRRAL[pS]VASLPGL as substrate by [gamma-33P]-ATP assay ChEMBL_2065457 (CHEMBL4720710) Inhibition of human GSK3B using YRRAAVPPSPSLSRHSSPHQ(pS)EDEEE as substrate by [gamma-33P]-ATP assay ChEMBL_2065458 (CHEMBL4720711) Inhibition of human GSK3A using YRRAAVPPSPSLSRHSSPHQ(pS)EDEEE as substrate by [gamma-33P]-ATP assay ChEMBL_2065524 (CHEMBL4720777) Inhibition of human CK1gamma3 using KRRRAL[pS]VASLPGL as substrate by [gamma-33P]-ATP assay ChEMBL_2065525 (CHEMBL4720778) Inhibition of human CK1gamma2 using KRRRAL[pS]VASLPGL as substrate by [gamma-33P]-ATP assay ChEMBL_2065526 (CHEMBL4720779) Inhibition of human CK1gamma1 using KRRRAL[pS]VASLPGL as substrate by [gamma-33P]-ATP assay ChEMBL_2065527 (CHEMBL4720780) Inhibition of human CK1a1L using KRRRAL[pS]VASLPGL as substrate by [gamma-33P]-ATP assay ChEMBL_2065528 (CHEMBL4720781) Inhibition of human CK1a1 using KRRRAL[pS]VASLPGL as substrate by [gamma-33P]-ATP assay ChEMBL_2130712 (CHEMBL4840141) Inhibition of human CK1E using KRRRAL[pS]VASLPGL as substrate by [gamma-33P]-ATP assay ChEMBL_1511998 (CHEMBL3610611) Inhibition of PI3Kgamma (unknown origin) using PI or PIP2:PS as substrate by TR-FRET assay ChEMBL_1511999 (CHEMBL3610612) Inhibition of PI3Kdelta (unknown origin) using PI or PIP2:PS as substrate by TR-FRET assay ChEMBL_1625785 (CHEMBL3868254) Inhibition of human GSK3beta using YRRAAVPPSPSLSRHSSPHQ(pS)EDEEE as substrate in presence of gamma-33P-ATP ChEMBL_1720738 (CHEMBL4135738) Inhibition of human GSK3beta using YRRAAVPPSPSLSRHSSPHQ(pS)EDEEE as substrate in presence of [gamma-33P]-ATP ChEMBL_811985 (CHEMBL2013355) Inhibition of recombinant TBK1 using 5FAM-AhxKRRAL(ps)VASLPGL as substrate by microfluidic mobility shift assay ChEMBL_1666744 (CHEMBL4016540) Inhibition of PI3Kdelta (unknown origin) using PIP2:PS as substrate after 3 hrs by ADP-Glo assay ChEMBL_1666774 (CHEMBL4016570) Inhibition of PI3Kgamma (unknown origin) using PIP2:PS as substrate after 30 mins by ADP-Glo assay ChEMBL_1666775 (CHEMBL4016571) Inhibition of PI3Kalpha (unknown origin) using PIP2:PS as substrate after 30 mins by ADP-Glo assay ChEMBL_1666776 (CHEMBL4016572) Inhibition of PI3Kbeta (unknown origin) using PIP2:PS as substrate after 30 mins by ADP-Glo assay ChEMBL_942558 (CHEMBL2338935) Inhibition of human recombinant GSK3beta using 650HSSPHQ(pS)EDEEE as substrate after 30 mins by luminescence assay ChEMBL_974625 (CHEMBL2410551) Inhibition of full length Pin1 (unknown origin) using WFY(pS)PR-pNA as substrate by spectrophotometric analysis ChEMBL_1496312 (CHEMBL3579761) Inhibition of PI3Kalpha (unknown origin) using PIP2/PS as substrate after 1 hr by luciferase-based luminescence assay ChEMBL_1496315 (CHEMBL3579764) Inhibition of PI3Kbeta (unknown origin) using PIP2/PS as substrate after 1 hr by luciferase-based luminescence assay ChEMBL_1496316 (CHEMBL3579765) Inhibition of PI3Kdelta (unknown origin) using PIP2/PS as substrate after 1 hr by luciferase-based luminescence assay ChEMBL_1496317 (CHEMBL3579766) Inhibition of PI3Kgamma (unknown origin) using PIP2/PS as substrate after 1 hr by luciferase-based luminescence assay ChEMBL_1647788 (CHEMBL3996844) Inhibition of PI3Kdelta (unknown origin) using PIP2:PS as substrate incubated for 3 hrs by ADP-Glo assay ChEMBL_1647796 (CHEMBL3996852) Inhibition of PI3Kalpha (unknown origin) using PIP2:PS as substrate incubated for 30 mins by ADP-Glo assay ChEMBL_1647797 (CHEMBL3996853) Inhibition of PI3Kbeta (unknown origin) using PIP2:PS as substrate incubated for 30 mins by ADP-Glo assay ChEMBL_1647798 (CHEMBL3996854) Inhibition of PI3Kgamma (unknown origin) using PIP2:PS as substrate incubated for 30 mins by ADP-Glo assay ChEMBL_1924910 (CHEMBL4427866) Inhibition of PI3Kdelta (unknown origin) using PIP2:PS as substrate incubated for 3 hrs by ADP-Glo assay ChEMBL_2050709 (CHEMBL4705408) Inhibition of human PI3Kdelta using substrate PIP2:PS and ATP incubated for 1 hr by ADP-Glo assay ChEMBL_2427090 Inhibition of human PI3Kalpha using PI(4,5)P2:PS as substrate in presence of ATP by ADP-Glo assay ChEMBL_2427119 Inhibition of human PI3Kbeta using PI(4,5)P2:PS as substrate in presence of ATP by kinase hotspot assay ChEMBL_2427120 Inhibition of human PI3Kgamma using PI(4,5)P2:PS as substrate in presence of ATP by kinase hotspot assay ChEMBL_2427121 Inhibition of human PI3Kdelta using PI(4,5)P2:PS as substrate in presence of ATP by kinase hotspot assay ChEMBL_880534 (CHEMBL2211469) Inhibition of CK1-alpha using biotin[long chain]-KKRRRAL{pS}VATLPGL substrate in presence of 8 uM ATP ChEMBL_880535 (CHEMBL2211470) Inhibition of CK1-delta using biotin[long chain]-KKRRRAL{pS}VATLPGL substrate in presence of 11 uM ATP ChEMBL_1758865 (CHEMBL4193873) Inhibition of recombinant human PI3K p110beta using PIP2/PS as substrate after 1 hr by ADP-Glo luminescence assay ChEMBL_2121262 (CHEMBL4830409) Inhibition of human GSK3A using YRRAAVPPSPSLSRHSSPHQ(pS)EDEEE as substrate incubated for 2 hrs by [gamma-33P]-ATP assay ChEMBL_2121263 (CHEMBL4830410) Inhibition of human GSK3B using YRRAAVPPSPSLSRHSSPHQ(pS)EDEEE as substrate incubated for 2 hrs by [gamma-33P]-ATP assay ChEMBL_880536 (CHEMBL2211471) Inhibition of CK1-gamma 2 using biotin[long chain]-KKRRRAL{pS}VATLPGL substrate in presence of 32 uM ATP ChEMBL_1289436 (CHEMBL3116932) Inhibition of PI3Kalpha (unknown origin) using PIP2/PS as substrate compound preincubated for 15 mins by luciferase-based luminescence assay ChEMBL_1618552 (CHEMBL3860721) Inhibition of PI3Kdelta (unknown origin) using phosphatidylinositol/PIP2:PS as substrate after 15 to 60 mins by TR-FRET assay ChEMBL_1891209 (CHEMBL4393036) Inhibition of recombinant human GSK3beta using YRRAAVPPSPSLSRHSSPHQ(pS)EDEEE as substrate measured after 60 mins by ADP-Glo luminescence assay ChEMBL_1834774 (CHEMBL4334907) Inhibition of human GSK-3alpha using YRRAAVPPSPSLSRHSSPHQ(pS)EDEEE as substrate measured in presence of [gamma-33P]ATP by radiometric assay ChEMBL_1834775 (CHEMBL4334908) Inhibition of human GSK-3beta using YRRAAVPPSPSLSRHSSPHQ(pS)EDEEE as substrate measured in presence of [gamma-33P]ATP by radiometric assay ChEMBL_2291541 Inhibition of full length GST-tagged human recombinant GSK3-alpha using FL-KRREILSRRP[ps]ERYR-NH2 as substrate incubated for 20 hrs ChEMBL_2355228 Inhibition of PI3Kdelta (unknown origin) using PIP2:PS as substrate in presence of ATP measured after 1 hr by ADP-Glo assay ChEMBL_2464388 Inhibition of PI3Kdelta (unknown origin) using PIP2:PS as substrate measured after 1 hrs in presence of by ADP-Glo Kinase assay ChEMBL_2464391 Inhibition of PI3Kalpha (unknown origin) using PIP2:PS as substrate measured after 1 hrs in presence of by ADP-Glo Kinase assay ChEMBL_2464392 Inhibition of PI3Kbeta (unknown origin) using PIP2:PS as substrate measured after 1 hrs in presence of by ADP-Glo Kinase assay ChEMBL_2464393 Inhibition of PI3Kgamma (unknown origin) using PIP2:PS as substrate measured after 1 hrs in presence of by ADP-Glo Kinase assay ChEMBL_1449888 (CHEMBL3380577) Inhibition of PI3Kalpha (unknown origin) using PIP2/PS as substrate preincubated for 15 mins before substrate addition by luciferase-based luminescence assay ChEMBL_1511295 (CHEMBL3606200) Inhibition of P110gamma (unknown origin) using PI or PIP2:PS as substrate measured for 15 to 60 mins by TR-FRET analysis ChEMBL_1511296 (CHEMBL3606201) Inhibition of P110delta (unknown origin) using PI or PIP2:PS as substrate measured for 15 to 60 mins by TR-FRET analysis ChEMBL_2377779 Inhibition of PI3Kalpha H1047R mutant (unknown origin) using PI(4,5)P2:PS as substrate incubated in presence of ATP by ADP-Glo assay ChEMBL_2482662 Inhibition of PI3Kgamma (unknown origin) using PI3:PS as substrate incubated for 1 hr in presence of ATP by ADP-glo kinase assay ChEMBL_2482663 Inhibition of PI3Kdelta (unknown origin) using PI3:PS as substrate incubated for 1 hr in presence of ATP by ADP-glo kinase assay Inhibition Assay Inhibition of factor VIII ELISA. The interaction between factor VIII and PS was measured by using an enzyme-linked immunoadsorbent assay (ELISA). ChEMBL_1560862 (CHEMBL3778934) Displacement of [125ITyr0-Glu1,Nle17]-PS-Svg in human CRF-R1 expressed in COS-M6 cells after 90 mins by gamma counting assay ChEMBL_2482661 Inhibition of PI3K alpha (unknown origin) using PI3:PS as substrate incubated for 1 hr in presence of ATP by ADP-glo kinase assay ChEMBL_1560863 (CHEMBL3778935) Displacement of [125ITyr0-Glu1,Nle17]-PS-Svg in mouse CRF-R2 beta expressed in COS-M6 cells after 90 mins by gamma counting assay ChEMBL_2115437 (CHEMBL4824378) Inhibition of human full length GSK3beta expressed in baculovirus in Sf9 insect cells using YRRAAVPPSPSLSRHSSPHQ(pS)EDEEE as substrate by ADP-Glo kinase assay ChEMBL_2215274 (CHEMBL5128406) Inhibition of GSK-3beta (unknown origin) using peptide YRRAAVPPSPSLSRHSSPHQ(pS)EDEEE as substrate incubated for 5 to 60 mins by ADP-Glo kinase assay ChEMBL_2072599 (CHEMBL4728133) Inhibition of recombinant human His-tagged PI3Kdelta expressed in baculovirus expression system using PIP2:PS as substrate incubated for 1 hr by invitrogen adapta assay ChEMBL_2072602 (CHEMBL4728136) Inhibition of recombinant human His-tagged PI3Kgamma expressed in baculovirus expression system using PIP2:PS as substrate incubated for 1 hr by invitrogen adapta assay ChEMBL_2230849 (CHEMBL5144621) Inhibition of human recombinant GSK-3 beta using YRRAAVPPSPSLSRHSSPHQ(pS)EDEEE as substrate incubated for 60 mins in presence of ATP by luminescence based analysis ChEMBL_2360058 Competitive inhibition of P13Kdelta (unknown origin) using PIP2:PS as substrate preincubated for 1 hr followed by substrate addition for 1 hr by ADP-Glo assay Inhibition Assay The IC50 value of the inhibition of the cocrystal of I-D1-6 and L-proline prepared in Example 138 on SGLT2 and SGLT1 is measured according to the method recorded in the literature (Meng, W. et al, J. Med. Chem., 2008, 51, 1145-1149). SGLT2 and SGLT1 Inhibition IC50 values of inhibition of some compounds described in the present invention and related compounds on SGLT2 and SGLT1 are determined according to the method similar to that recorded in the document (Meng, W. et al, J. Med. Chem., 2008, 51, 1145-1149). ChEMBL_2072600 (CHEMBL4728134) Inhibition of recombinant human His-tagged PI3Kalpha expressed in insect cell expression system using PIP2:PS as substrate incubated for 1 hr by invitrogen adapta assay ChEMBL_2072601 (CHEMBL4728135) Inhibition of recombinant human His-tagged PI3Kbeta expressed in insect cell expression system using PIP2:PS as substrate incubated for 1 hr by invitrogen adapta assay ChEMBL_2329181 Inhibition of recombinant human Wip1 (1 to 420 residues) using human ATM phosphopeptide (AFEEG-pS-QSTTIGY) as substrate incubated for 7 mins by Bio-mol green assay ChEMBL_1758863 (CHEMBL4193871) Inhibition of recombinant full length His-tagged human PI3K p110gamma expressed in baculovirus expression system using PIP2/PS as substrate after 1 hr by ADP-Glo luminescence assay ChEMBL_2332932 Displacement of FL-KRREILSRRP[ps]ERYR-NH2 from human recombinant full length GST-tagged GSK-3-alpha expressed in insect cells incubated for 20 hrs by fluorescence based analysis ChEBML_1970502 Inhibition of His-tagged recombinant human PI3K p110delta/p85alpha expressed in baculovirus expression system using PIP2:PS lipid as substrate in presence of ATP after 1 hr by Adapta assay ChEMBL_1676639 (CHEMBL4026782) Inhibition of human wild type ALK using YRRAAVPPSPSLSRHSSPHQ(pS)EDEEE as substrate preincubated for 20 mins followed by [gamma-33P]-ATP addition after 2 hrs by filter binding assay ChEMBL_1758866 (CHEMBL4193874) Inhibition of recombinant human full length His-tagged PI3K p110delta/p85alpha expressed in baculovirus expression system using PIP2/PS as substrate after 1 hr by ADP-Glo luminescence assay ChEMBL_2299088 Inhibition of PI3Kalpha (unknown origin) using PIP2:PS as substrate preincubated for 1 hr followed by ATP and substate addition and measured after 1 hr by ADP-glo reagent based assay ChEMBL_2299089 Inhibition of PI3Kdelta (unknown origin) using PIP2:PS as substrate preincubated for 1 hr followed by ATP and substate addition and measured after 1 hr by ADP-glo reagent based assay ChEMBL_2299090 Inhibition of PI3Kbeta (unknown origin) using PIP2:PS as substrate preincubated for 1 hr followed by ATP and substate addition and measured after 1 hr by ADP-glo reagent based assay ChEMBL_2299091 Inhibition of PI3Kgamma (unknown origin) using PIP2:PS as substrate preincubated for 1 hr followed by ATP and substate addition and measured after 1 hr by ADP-glo reagent based assay ChEMBL_2359932 Inhibition of recombinant human GSK-3beta using YRRAAVPPSPSLSRHSSPHQ(pS)EDEE as substrate preincubated for 15 mins followed by substrate addition and measured after 60 mins by Kinase-Glo luminescent kinase assay ChEMBL_2583162 Inhibition of human CK1a1 using KRRRAL[pS]VASLPGL as substrate preincubated for 20 mins followed by [gamma-33P]-ATP addition and measured after 120 mins by radiometric Hot-SpotSM Kinase assay ChEMBL_2583163 Inhibition of human CK1a1L using KRRRAL[pS]VASLPGL as substrate preincubated for 20 mins followed by [gamma-33P]-ATP addition and measured after 120 mins by radiometric Hot-SpotSM Kinase assay ChEMBL_2583164 Inhibition of human CK1g1 using KRRRAL[pS]VASLPGL as substrate preincubated for 20 mins followed by [gamma-33P]-ATP addition and measured after 120 mins by radiometric Hot-SpotSM Kinase assay ChEMBL_2583165 Inhibition of human CK1g2 using KRRRAL[pS]VASLPGL as substrate preincubated for 20 mins followed by [gamma-33P]-ATP addition and measured after 120 mins by radiometric Hot-SpotSM Kinase assay ChEMBL_2583166 Inhibition of human CK1g3 using KRRRAL[pS]VASLPGL as substrate preincubated for 20 mins followed by [gamma-33P]-ATP addition and measured after 120 mins by radiometric Hot-SpotSM Kinase assay ChEMBL_2583232 Inhibition of human GSK3a using YRRAAVPPSPSLSRHSSPHQ(pS)EDEEE as substrate preincubated for 20 mins followed by [gamma-33P]-ATP addition and measured after 120 mins by radiometric Hot-SpotSM Kinase assay ChEMBL_2583233 Inhibition of human GSK3b using YRRAAVPPSPSLSRHSSPHQ(pS)EDEEE as substrate preincubated for 20 mins followed by [gamma-33P]-ATP addition and measured after 120 mins by radiometric Hot-SpotSM Kinase assay ChEMBL_968487 (CHEMBL2400933) Inhibition of recombinant human GSK3 using YRRAAVPPSPSLSRHSSPHQ(pS)EDEEE as substrate preincubated for 10 mins followed by [33P-gamma]-ATP addition measured after 30 mins by liquid scintillation counting analysis ChEBML_1970501 Inhibition of His-tagged recombinant human full length PI3K p110gamma expressed in baculovirus expression system using PIP2:PS lipid as substrate incubated for 1 hr in presence of ATP by Adapta assay ChEMBL_1758864 (CHEMBL4193872) Inhibition of recombinant full length N-terminal His-tagged human PI3K p110alpha/p85alpha expressed in baculovirus expression system using PIP2/PS as substrate after 1 hr by ADP-Glo luminescence assay ChEMBL_2052179 (CHEMBL4707180) Inhibition of human full-length recombinant p110alpha/p85alpha co-expressed in baculovirus expression system using PIP2:PS lipid as substrate incubated for 1 hr in presence of ATP by ADP-Glo assay ChEMBL_2114760 (CHEMBL4823701) Inhibition of full length recombinant GST-tagged human VPS34 expressed in baculovirus expression system using PI/PS as substrate incubated for 60 mins in presence of ATP at Km concentration by luminescence assay ChEMBL_2504753 Inhibition of N-terminal 6His-tagged full-length recombinant human PIP5K1C expressed in baculovirus infected Sf21 cells using PI4P/PS as substrate incubated for 60 mins in presence of ATP by ADP-Glo assay ChEMBL_1741074 (CHEMBL4156824) Inhibition of PI3Kalpha (unknown origin) using PIP2:PS as substrate preincubated for 1 hr followed by substrate addition and measured after 1 hr in presence of 10 uM ATP by ADP-Glo luminescence assay ChEMBL_1741075 (CHEMBL4156825) Inhibition of PI3Kbeta (unknown origin) using PIP2:PS as substrate preincubated for 1 hr followed by substrate addition and measured after 1 hr in presence of 50 uM ATP by ADP-Glo luminescence assay ChEMBL_1741076 (CHEMBL4156826) Inhibition of PI3Kgamma (unknown origin) using PIP2:PS as substrate preincubated for 1 hr followed by substrate addition and measured after 1 hr in presence of 50 uM ATP by ADP-Glo luminescence assay ChEMBL_1741077 (CHEMBL4156827) Inhibition of PI3Kdelta (unknown origin) using PIP2:PS as substrate preincubated for 1 hr followed by substrate addition and measured after 1 hr in presence of 50 uM ATP by ADP-Glo luminescence assay ChEMBL_1928060 (CHEMBL4431132) Inhibition of His-tagged full length human recombinant GSK3beta using [YRRAAVPPSPSLSRHSSPHQ-(pS)EDEEE] substrate and pre-incubarted for 20 mins before [gamma-33P]ATP addition and measured after 2 hrs by radiometric kinase assay ChEMBL_2377743 Inhibition of recombinant N-terminal 6-His tagged full length human PI3Kalpha H1047R mutant using PIP2diC8 and PIP2:PS as substrate incubated for 1 hrs in presence of ATP by fluorescence based plate reader analysis ChEMBL_2122763 (CHEMBL4831996) Inhibition of human recombinant N-terminal His-tagged GSK3beta expressed in Escherichia coli using RRRPASVPPSPSLS RHS(pS)HQRR as substrate incubated for 30 mins in presence of ATP by Kinase-Glo reagent based luminescence assay ChEMBL_2161703 (CHEMBL5046564) Inhibition of recombinant human GSK-3-beta using YRRAAVPPSPSLSRHSSPHQ(pS)EDEEE as substrate preincubated for 10 mins followed by substrate addition and measured after 1 hr in presence of ATP by ADP Glo kinase assay ChEMBL_2291219 Inhibition of human C-terminal his tagged GSK3beta (26 to 383 residues) expressed in Escherichia coli using GYSI(YRRAAVPPSPSLSRHSSPHQ(pS)EDEEE) as peptide substrate incubated for 1 hr in presence of ATP by cook activity assay ChEMBL_2452127 Inhibition of full length GST-tagged human recombinant GSK3-alpha using FL-KRREILSRRP[ps]ERYR-NH2 as substrate preincubated for 30 mins followed by substrate addition and measured after 60 mins by ADP-Glo kinase assay ChEMBL_1893921 (CHEMBL4395842) Inhibition of human recombinant full length GSK3alpha using YRRAAVPPSPSLSRHSSPHQ(pS)EDEEE as substrate preincubated with enzyme for 10 mins followed by substrate and 25 uM ATP addition measured after 60 mins by ADP-Glo luminescence assay ChEMBL_2332931 Displacement of FL-KRREILSRRP[ps]ERYR-NH2 from human recombinant N-terminal His6-tagged GSK-3-beta (14 to 306 residues) H355L mutant expressed in baculovirus-infected Sf21 cells incubated for 20 hrs by fluorescence based analysis ChEMBL_1893920 (CHEMBL4395841) Competitive inhibition of human recombinant full length GSK3beta using YRRAAVPPSPSLSRHSSPHQ(pS)EDEEE as substrate preincubated with enzyme for 10 mins followed by substrate and 25 uM ATP addition measured after 60 mins by ADP-Glo luminescence assay ChEMBL_1893923 (CHEMBL4395844) Competitive inhibition of human recombinant full length GSK3beta using YRRAAVPPSPSLSRHSSPHQ(pS)EDEEE as substrate preincubated with enzyme for 10 mins followed by substrate and 100 uM ATP addition measured after 60 mins by ADP-Glo luminescence assay ChEMBL_1893924 (CHEMBL4395845) Competitive inhibition of human recombinant full length GSK3beta using YRRAAVPPSPSLSRHSSPHQ(pS)EDEEE as substrate preincubated with enzyme for 10 mins followed by substrate and 500 uM ATP addition measured after 60 mins by ADP-Glo luminescence assay ChEMBL_2298676 Inhibition of recombinant human GSK3beta (636 to 661 residues) assessed as reduction in human muscle glycogen synthase type 1 fragment phosphorylation using YRRAAVPPSPSLSRHSSPHQ(pS)EDE as substrate in presence of ATP incubated for 60 mins by ADP-Glo kinase assay ChEMBL_2452126 Inhibition of His-tagged human recombinant GSK3beta expressed in Sf9 cells using biotinylated peptide bio-LC-S-R-H-S-S-P-H-Q-pS-E-D-E-E-E-OH as substrate preincubated for 30 mins followed by substrate addition and measured after 60 mins by ADP-Glo kinase assay Protein Kinase C beta 2 Assay A typical assay was carried out on a 96-well, clear microtiter plate in a Molecular Devices spectrophotometer for 20 minutes at 30° C. in 0.1 mL of assay buffer containing 50 mM HEPES, pH 7.4, 5 nM PKC, 23 units of pyruvate kinase, 33 units of lactate dehydrogenase, 0.15 mM peptide, 0.1 mM ATP, 1 mM DTT, 4 mM PEP, 8 mM MgCl2, 0.3 mM NADH, 60 mM CaCl2, 10 mg/mL PS, 50 ng/mL PMA, 7.5% DMSO and from about 10,000 nM to 0.169 nM compound inhibitor. Stock solutions of 3-sn-phosphatidyl-L-serine (PS) and phorbol-12-myristate-13-acetate (PMA) were sonicated for 30 seconds just prior to addition to assay buffer and assays were initiated by the addition of 100 μM ATP. Mycobacterium tuberculosis Pantothenate Synthetase Assay Southern Research Molecular Libraries Screening Center (SRMLSC) Southern Research Institute (Birmingham, Alabama) NIH Molecular Libraries Screening Centers Network (MLSCN) Award: 1R03MH076412-01 Multi-drug resistant Mycobacterium tuberculosis is becoming an increased health problem, especially in immunocompromised individuals with HIV. This form of TB is more difficult to treat and as a result has a higher mortality rate. Because of this, the discovery of drugs targeting novel pathways such as the synthesis of pantothenate has become increasingly important. Pantothenate synthetase (PS; EC 6.3.2.1), encoded by the panC gene, catalyzes the essential ATP-dependent condensation of D-pantoate and alpha-alanine to form pantothenate in bacteria, yeast and plants; pantothenate is a key precursor for the biosynthesis of coenzyme A (CoA) and acyl carrier protein (ACP). The activity of PS was measured spectrophotometrically by coupling the formation of AMP to the reactions of myokinase, pyr Mycobacterium tuberculosis Pantothenate Synthetase Secondary Assay Southern Research Molecular Libraries Screening Center (SRMLSC) Southern Research Institute (Birmingham, Alabama) NIH Molecular Libraries Screening Centers Network (MLSCN) Award: 1R03MH076412-01 Multi-drug resistant Mycobacterium tuberculosis is becoming an increased health problem, especially in immunocompromised individuals with HIV. This form of TB is more difficult to treat and as a result has a higher mortality rate. Because of this, the discovery of drugs targeting novel pathways such as the synthesis of pantothenate has become increasingly important. Pantothenate synthetase (PS; EC 6.3.2.1), encoded by the panC gene, catalyzes the essential ATP-dependent condensation of D-pantoate and alpha-alanine to form pantothenate in bacteria, yeast and plants; pantothenate is a key precursor for the biosynthesis of coenzyme A (CoA) and acyl carrier protein (ACP). The activity of PS was measured spectrophotometrically by coupling the formation of AMP to the reactions of myokinase, pyruva TR-FRET Adapta Assay PI 3-Kinase Gamma (E), PI 3-Kinase Delta (F):The TR-FRET Adapta Universal Kinase Assay Kit was purchased from Invitrogen Corporation (Carlsbad/CA, USA) (Cat. No. PV5099). The kit contains the following reagents: Adapta Eu-anti-ADP Antibody (Europium labeled anti-ADP antibody in HEPES buffered saline, Cat. No. PV5097), Alexa Fluor 647-labeled ADP tracer (Alexa Fluor 647-labeled ADP tracer in HEPES buffered saline, Cat. No. PV5098), TR-FRET dilution buffer pH 7.5 (Cat. No. PV3574). PIK3CD substrate phosphatidylinositol (PI) was obtained from Invitrogen (vesicules consisting of 2 mM phosphatidylinositol (PI) in 50 mM HEPES pH7.5; Cat. No. PV5371). PIK3CG substrate phosphatidylinositol-4,5-bisphosphate (PIP(4,5)2 was obtained from Invitrogen (PIP2:PS large unilamellar vesicules consisting of 1 mM PIP2: 19 mM PS in 50 mM HEPES pH7.5, 3 mM MgCl2, 1 mM EGTA; Cat. No. PV5100). Biochemical hGSK-3Beta Assay Compounds were tested for their ability to inhibit human Glycogen Synthase Kinase-3 beta (hGSK-3β) to phosphorylate biotin-YRRAAVPPSPSLSRHSSPHQ(pS)EDEEE. Compounds were incubated with 0.5 μCi 33P-ATP, 10 μM ATP, 0.0125 U hGSK-3β (Upstate cell signaling solutions) and 1 μM substrate (biotin-YRRAAVPPSPSLSRHSSPHQ(pS)EDEEE) in 50 mM HEPES, 10 mM MgCl2, 100 mM Na3VO4, 1 mM DTT, 0.0075% Triton, 2% DMSO (total volume 50 μL) for 30 minutes at room temperature. The incubation was stopped by addition of an equal volume of 100 mM EDTA, 4M NaCl. 80 μL of this mixture was added to streptavidin-coated Flash-plates (PerkinElmer). Following a wash step, 33P incorporation was quantified on a MicroBeta microplate liquid scintillation counter (Perkin Elmer). IC50s were determined by fitting a sigmoidal dose-response curve to the counts obtained at the different concentrations in GraphPad Prism. Kinase assay The kinase assay was performed in V-bottom 384-well plates. The final assay volume was 30 μl prepared from 15 μl additions of enzyme, substrates (fluoresceinated peptide FL-KRREILSRRP[ps]ERYR-NH2 and ATP) and test compounds in assay buffer (100 mM HEPES pH 7.4, 10 mM MgCl2, 25 mM Beta-Glycerolphosphate, 0.015% Brij35 and 0.25 mM DTT). The reaction was incubated at room temperature for 20 hours and terminated by adding 45 μl of 35 mM EDTA to each sample. The reaction mixture was analyzed on the Caliper LabChip3000 (Caliper, Hopkinton, Mass.) by electrophoretic separation of the unphosphorylated substrate and phosphorylated product. Inhibition data were calculated by comparison of the no enzyme control reactions for 100% inhibition and vehicle-only reactions for 0% inhibition. The final concentration of reagents in the assay were 250 pM GSK3u or GSK30, 20 uM ATP, 1.5 uM FL-KRREILSRRP[ps]ERYR-NH2, and 1.6% DMSO. Dose response curves were generated to determine the concentration required to inhibit 50% of the kinase activity (IC50). Compounds were dissolved at 10 mM in dimethylsulfoxide (DMSO) and evaluated at eleven concentrations. IC50 values were derived by non-linear regression analysis. Patch clamp assay HEK293 cells were cultured in Dulbecco's Modified Eagle Medium/Nutrient Mixture F-12 (D-MEM/F-12) supplemented with 10% fetal bovine serum, 100 U/mL penicillin G sodium, 100 ug/mL streptomycin sulfate and 500 ug/mL G418. Before testing, cells in culture dishes were washed twice with Hank's Balanced Salt Solution, treated with Accutase and re-suspended in the culture media ( 20 106 cells in 20 mL). Cells in suspension were allowed to recover for 10 minutes in tissue culture incubator set at 37 C. in a humidified 95% air: 5% CO2 atmosphere. Immediately before use in the SyncroPatch 384PE system (SP384PE), the cells were washed twice in extracellular buffer (HB-PS) to remove the culture medium and re-suspended in 20 mL of HB-PS. Extracellular buffer was loaded into the wells of Nanion 384-well Patch Clamp (NPC-384, 4 M) chip (60 uL per well). Then, cell suspension was pipetted into the wells (20 uL per well) of the chip. After establishment of the whole-cell configuration, membrane currents were recorded using patch clamp amplifier in the SP384PE system. Protein Kinase C beta 2 (PKCpII) Assay Protein Kinase C beta 2 (PKCpII) catalyzes the production of ADP from ATP that accompanies the phosphoryl transfer to the PKC Pseudosubstrate peptide (A→S, RFARKGSLRQKNV). This transfer is coupled to the oxidation of β-NADH through the activities of Pyruvate Kinase (PK) and Lactate Dehydrogenase (LDH). β-NADH conversion to NAD+ is monitored by the decrease in absorbance at 340 nm (e=6.22 cm−1 mM−1) using a Molecular Devices SPECTRA max PLUS spectrophotometer.A typical assay was carried out on a 96-well, clear microtiter plate in a Molecular Devices spectrophotometer for 20 minutes at 30° C. in 0.1 mL of assay buffer containing 50 mM HEPES, pH 7.4, 5 nM PKC, 23 units of pyruvate kinase, 33 units of lactate dehydrogenase, 0.15 mM peptide, 0.1 mM ATP, 1 mM DTT, 4 mM PEP, 8 mM MgCl2, 0.3 mM NADH, 60 mM CaCl2), 10 mg/mL PS, 50 ng/mL PMA, 7.5% DMSO and from about 10,000 nM to 0.169 nM compound inhibitor. Stock solutions of 3-sn-phosphatidyl-L-serine (PS) and phorbol-12-myristate-13-acetate (PMA) were sonicated for 30 seconds just prior to addition to assay buffer and assays were initiated by the addition of 100 μM ATP. DGKα Biochemical Activity Assay Table 6: Alternatively, the enzymatic activity of human DGKα was monitored in a biochemical assay in the presence or absence of compounds and using micelles containing 18:1 Diacylglycerol (DAG), 16:0-18:1 PS (POPS) and Octylglucoside as substrate. DGKα activity led to conversion of DAG and ATP to Phosphatidic Acid (PA) and ADP. Levels of ADP were monitored by bioluminescence using the ADP-Glo Kinase Assay (Promega) and were indicative of DGKα activity.Ten nanoliters of test compound dissolved in DMSO at various concentrations were dispensed into a 384-well low volume nonbinding service white plates (Corning #3824) using a Labcyte Echo instrument. Recombinant DGKα (Carna Biosciences) in assay buffer (5 μL in 50 mM MOPS [3-(N-morpholino) propanesulfonic acid], pH 7.2; 0.0025% Triton X-100; 1 mM dithiothreitol; 5 mM MgCl2, 200 μM ATP) was added to the compound-containing plate and was incubated for 15 minutes at 25° C. A substrate solution (5 μL in 1.7 mM 1,2-dioleoyl-sn-glycerol [18:1 DAG], 13.5 mM 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-L-serine [16:0-18:1 PS](POPS), 2 μM CaCl2, 100 mM Octylglucoside (OG), 1 mM DTT) (obtained from Carna Biosciences) diluted in DGK ALPHA assay buffer was then added to start the reaction. Final concentrations were 1 nM DGKα, 100 μM ATP, 1 μM calcium chloride, 0.85 mM 1,2-dioleoyl-sn-glycerol (18:1 DAG), 6.75 mM 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-L-serine (16:0-18:1 PS) (POPS), 1 μM CaCl2, 50 mM Octylglucoside (OG) and 5 mM MgCl2. The reaction mixture was incubated at 25° C. for 1 hour. ADP-Glo Reagent (10 μL, 10 mM Mg added) provided by the kit was added to each well and incubated at 25° C. for 40 minutes. Then, 20 μL of Kinase Detection Reagent was added and incubated at 25° C. for 40 minutes. Luciferase activity of each well was measured via luminescence on an Envision plate reader (PerkinElmer). PREP Enzymatic Activity Assay PREP enzymatic activity assay. To assay baseline prolyl endopeptidase (PREP) activity, 20 ng of recombinant human PREP (rhPREP) (R&S system, #4308-SE) or 20 ng of recombinant mouse PREP (rmPREP) (R&S system, #6339-SE) was incubated with 100 μM of Z-Gly-Pro-AMC peptide (BACHEM, #L-1145) in a PREP assay buffer (25 mM Tris, 250 mM NaCl, 10 mM DTT, pH 7.5) for 30 min at 37° C. protected from light in 96-well black plates (Nunc, #237108). To assay PREP activity inhibition by test compounds, test compounds were pre-incubated with the enzyme for 15 min at 37° C. before starting the reaction by substrate addition in 96-well black plates (Nunc, #237108). 7-Amino-4-Methylcoumarin (AMC) release was detected by measuring fluorescence at Ex/Em 380/460 nm using a Multifunction Microplate Reader (Synergy 4, Biotek). All measurements were carried out in triplicate. Val-boroPro, a non-specific prolyl peptidase inhibitor, was used as a positive control. hPrep Inhibition Assay Table 3: Compounds were tested in a biochemical inhibition assay using Prolyl endopeptidase, Prolyl Oligopeptidase (hPREP) enzyme at 0.6 nM FAC (R&D Systems, 4308-SE) and the substrate Z-Gly-Pro-amino-methylcoumarin (Bachem, I-1145) at 50 μM FAC. 384 Low volume black plates (Greiner #784076) were used. 4 μL, 1.2 nM enzyme solution (25 mM Tris HCl, 250 mM NaCl, 0.01% Triton X-100, 5 mM Glutathione, pH 7.5) was added to 40 nL compounds (in DMSO) at 10 CR, 3-fold dilution series from 50 μM FAC. Plates were incubated for 15 min at rt in dark. 4 μL, 100 μM substrate solution (25 mM Tris HCl, 250 mM NaCl, 0.01% Triton X-100, 5 mM Glutathione, pH 7.5) was added to each well. Plates were centrifuged at 1000 rpm and incubated for 20 min at rt in dark. The plates were read on a PHERAstar reader with excitation 340 nm and emission 460 nm. D Biochemical Assay The TR-FRET Adapta Universal Kinase Assay Kit was purchased from Invitrogen Corporation (Carlsbad/CA, USA) (Cat. No. PV5099). The kit contains the following reagents: Adapta Eu-anti-ADP Antibody (Europium labeled anti-ADP antibody in HEPES buffered saline, Cat. No. PV5097), Alexa Fluor 647-labeled ADP tracer (Alexa Fluor 647-labeled ADP tracer in HEPES buffered saline, Cat. No. PV5098), proprietary TR-FRET dilution buffer pH 7.5 (Cat. No. PV3574).PIK3CD substrate Phosphatidylinositol was obtained from Invitrogen (vesicules consisting of 2 mM PI in 50 mM HEPES pH7.5; Cat. No. PV5371). PIK3CG substrate Phosphatidylinositol-4,5-bisphosphate (PIP(4,5)2 was obtained from Invitrogen (PIP2:PS large unilamellar vesicules consisting of 1 mM PI P2: 19 mM PS in 50 mM HEPES pH7.5, 3 mM MgCl2, 1 mM EGTA; Cat. No. PV5100).Time-Resolved Fluorescence Resonance Energy Transfer (TR-FRET) is a technology based on energy transfer between two adjacent dyes, from an excited electron in one dye (the donor) to an electron of an adjacent dye (the acceptor) through resonance, then released as a photon. This energy transfer is detected by an increase in the fluorescence emission of the acceptor, and a decrease in the fluorescence emission of the donor. TR-FRET assays for protein kinases use a long-lifetime lanthanide Terbium or Europium chelates as the donor species which overcome interference from compound autofluorescence or light scatter from precipitated compounds, by introducing a delay after excitation by a flashlamp excitation source. Inhibition of FAPα In Vitro Enzymatic Activity Assay FAPα enzymatic exopeptidase (dipeptidase) activity assay. To assay baseline FAPa enzymatic exopeptidase activity, 40 ng of recombinant human FAPα (rhFAPα, R&S system, #3715-SE) or 40 ng of recombinant mouse FAPα (rmFAPα, R&S system, #8647-SE) was incubated with 100 μM of Z-Gly-Pro-AMC peptide (BACHEM, #L-1145) in a FAPα assay buffer (50 mM Tris pH 7.4, 100 mM NaCl, 0.1 mg/ml bovine serum albumin) for 1 h at 37° C. protected from light in 96-well black plates (Nunc, #237108). To assay FAPα enzymatic exopeptidase activity inhibition by test compounds, all test compounds were pre-incubated with the enzyme for 15 min at 37° C. before starting the reaction by substrate addition in 96-well black plates (Nunc, #237108). 7-Amino-4-Methylcoumarin (AMC) release was detected by measuring fluorescence at Ex/Em 380/460 nm using a Multifunction Microplate Reader (Synergy 4, Biotek). All measurements were carried out in duplicate. Val-boroPro, a non-specific prolyl peptidase inhibitor, was used as a positive control. hPrep Inhibition Assay Table 3: Compounds were tested in a biochemical inhibition assay using Prolyl endopeptidase, Prolyl Oligopeptidase (hPREP) enzyme at 0.6 nM FAC (R&D Systems, 4308-SE) and the substrate Z-Gly-Pro-amino-methylcoumarin (Bachem, I-1145) at 50 μM FAC. 384 Low volume black plates (Greiner #784076) were used. 4 μL, 1.2 nM enzyme solution (25 mM Tris HCl, 250 mM NaCl, 0.01% Triton X-100, 5 mM Glutathione, pH 7.5) was added to 40 nL compounds (in DMSO) at 10 CR, 3 fold dilution series from 50 μM FAC. Plates were incubated for 15 min at rt in dark. 4 μL, 100 μM substrate solution (25 mM Tris HCl, 250 mM NaCl, 0.01% Triton X-100, 5 mM Glutathione, pH 7.5) was added to each well. Plates were centrifuged at 1000 rpm and incubated for 20 min at rt in dark. The plates were read on a PHERAstar reader with excitation 340 nm and emission 460 nm. Data were analyzed in Genedata Screener . IC50 values were determined by plotting % inhibition versus log compound concentration and using a one site dose response model. DGKα Biochemical Activity Assay Ten nanoliters of test compound dissolved in DMSO at various concentrations were dispensed into a 384-well low volume nonbinding service white plates (Corning #3824) using a Labcyte Echo instrument. Recombinant DGKα (Carna Biosciences) in assay buffer (5 μL in 50 mM MOPS [3-(N-morpholino) propanesulfonic acid], pH 7.2; 0.0025% Triton X-100; 1 mM dithiothreitol; 5 mM MgCl2, 200 μM ATP) was added to the compound-containing plate and was incubated for 15 minutes at 25° C. A substrate solution (5 μL in 1.7 mM 1,2-dioleoyl-sn-glycerol [18:1 DAG], 13.5 mM 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-L-serine [16:0-18:1 PS](POPS), 2 μM CaCl2, 100 mM Octylglucoside (OG), 1 mM DTT) (obtained from Carna Biosciences) diluted in DGK ALPHA assay buffer was then added to start the reaction. Final concentrations were 1 nM DGKα, 100 μM ATP, 1 μM calcium chloride, 0.85 mM 1,2-dioleoyl-sn-glycerol (18:1 DAG), 6.75 mM 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-L-serine (16:0-18:1 PS) (POPS), 1 μM CaCl2), 50 mM Octylglucoside (OG) and 5 mM MgCl2. The reaction mixture was incubated at 25° C. for 1 hour. ADP-Glo Reagent (10 μL, 10 mM Mg added) provided by the kit was added to each well and incubated at 25° C. for 40 minutes. Then, 20 μL of Kinase Detection Reagent was added and incubated at 25° C. for 40 minutes. Luciferase activity of each well was measured via luminescence on an Envision plate reader (PerkinElmer). Inhibition of PI3K alpha Quantification of ATP to ADP conversion as a measure of PI3Kα activity. Active PI3Kα (Life Technologies), in the presence or absence of PI3Kα inhibitor, was reacted with PIP2:PS (Life Technologies), a substrate specifically optimized for use with Class I PI3 kinases, and ultrapure ATP (Promega). The conversion of ATP to ADP by PI3Kα was measured as luminescence signal via Promega ADP-Glo kinase activity assay. Assay was validated using published PI3Kα inhibitors LY294002, PI-103, BYL719, GDC0198 and also DMSO vehicle control.Compounds were prepared at 100× final concentration as a 12-point, 1:3 serial-dilution in DMSO series, with DMSO control as 12th point. Compound was then diluted in (25 mM HEPES pH 7.5, 1 mM EGTA, 0.3% CHAPS) prior to addition to PI3Kα. Active PI3Kα diluted to 0.24 ng/μL (1.1 nM) in (50 mM HEPES pH 7.5, 6 mM MgCl2, 1 mM EGTA, 200 mM NaCl, 0.03% CHAPS, 8 mM DTT) was incubated with compound for 0 hr and 3 hr prior to the start of the reaction. 25 μM PIP2:PS and 60 μM ATP were diluted from stock in (25 mM HEPES pH 7.5, 1 mM EGTA, 0.3% CHAPS) and added to initiate the PI3Kα reaction. Reaction time was 30 minutes. ATP to ADP conversion was measured in Luminescence Counts on DTX880 Plate Reader (Beckman Coulter). Compound IC50s were reported using GraphPad Prism software. Analytical method was non-linear regression, 4-parameter curve fit with bottom fit to validated PI3Kα inhibitor reference controls and no top fit (floating top). PI3KC3 (hVPS34) Kinase Assay The substrate (Phosphatidylinositol (PI): Phosphatidylserine (PS)) was added to freshly prepared reaction buffer (40 mM Tris-HCl (pH7.5), 3 μM Orthovanadate, 20 mM MgCl2, 2 mM DTT, 0.05% CHAPS, 1% DMSO). The PI3KC3 (hVPS34) kinase was delivered into the substrate solution with gentle mixing. Compounds were delivered in 100% DMSO into the kinase reaction mixture by Acoustic technology (Echo550; nanoliter range). The reaction was incubated for 20 min at room temperature. ATP was delivered into the reaction mixture to initiate the reaction. After incubating the mixture for 60 min at 30° C., the reaction was quenched with ADP-Glo reagent (Promega ADP Glo Kinase Assay kit #V9102) and incubated for 40 min at room temperature. Detection mixture (Promega ADP Glo Kinase Assay kit #V9102) was added and the reaction was incubated for 30 minutes. Luminescence was measured. Beta-AR binding assay β1-AR binding was done on rat cortical membrane following a previously described procedure (Beer et al., Biochem. Pharmacol. 37: 1145-1151, 1988). In brief, male Sprague-Dawley rats weighing 250-350 g were decapitated and their brains quickly removed. The cerebral cortices were dissected on ice, weighed and promptly transferred to a 50 ml test tube containing approximately 30 ml of 50 mM Tris-HCl, pH 7.8 (at room temperature). The tissues were homogenized with a polytron and centrifuged at 20,000×g for 12 min at 4° C. The pellet was washed again in the same manner and resuspended at a concentration of 20 mg (original wet wt) per 1 ml in the assay buffer (20 mM Tris-HCl, 10 mM MgCl2, 1 mM EDTA, 0.1 mM ascorbic acid at pH 7.8). To block the β2 sites present in the cortical membrane preparation, 30 nM ICI 118-551 was also added to the assay buffer. To wells containing 100 μl of the test drug and 100 μl of [3H]CGP-12177 (1.4 nM final concentration), 0.8 ml of tissue homogenate was added. After 2 hours at 25° C., the incubation was terminated by rapid filtration. Nonspecific binding was determined by 10 μM propranolol. Beta1-AR Binding Assay Beta1-AR binding was done on rat cortical membrane following a previously described procedure (Beer et al., Biochem. Pharmacol. 37: 1145-1151, 1988). In brief, male Sprague-Dawley rats weighing 250-350 g were decapitated and their brains quickly removed. The cerebral cortices were dissected on ice, weighed and promptly transferred to a 50 ml test tube containing approximately 30 ml of 50 mM Tris-HCl, pH 7.8 (at room temperature). The tissues were homogenized with a polytron and centrifuged at 20,000xg for 12 min at 4° C. The pellet was washed again in the same manner and resuspended at a concentration of 20 mg (original wet wt) per 1 ml in the assay buffer (20 mM Tris-HCl, 10 mM MgCl2, 1 mM EDTA, 0.1 mM ascorbic acid at pH 7.8). To block the beta2 sites present in the cortical membrane preparation, 30 nM ICI 118-551 was also added to the assay buffer. To wells containing 100 ul of the test drug and 100 ul of [3H]CGP-12177 (1.4 nM final concentration), 0.8 ml of tissue homogenate was added. After 2 hours at 25° C., the incubation was terminated by rapid filtration. Nonspecific binding was determined by 10 uM propranolol. Binding Assay For radioligand competition studies, a final concentration of 1x105 THP-1 monocytic leukemia cells are combined with 100 μg of LS WGA PS beads (Amersham, Cat.#: RPNQ 0260) in 40 μL of assay buffer (RPMI 1640 without phenol red, 50 mM HEPES, 5 mM MgCl2, 1 mM CaCl2, 0.1% BSA). The THP-1 cell/bead mixture was added to each well of a 384-well assay plate (PerkinElmer, Cat. #:6007899) containing test compound in 3-fold serial dilution, with final concentrations ranging from 8 μM to 0.14 nM. A final concentration of 0.1 nM [125I]-MIP-1α (PerkinElmer, Cat. #NEX298) in 20 μL assay buffer was added to the reaction. Unlabeled MIP-1α was added in excess to some wells to determine non-specific binding. Sealed assay plates were incubated at room temperature for 12 h then analyzed by LEADseeker. 14-3-3 protein interaction modulators Dose Response Confirmation NIH Molecular Libraries Screening Centers Network [MLSCN] Emory Chemical Biology Discovery Center in MLSCN MLSCN Grant: 1 X01MH78953-01 The 14-3-3 proteins are the prototype for a novel class of protein modules that can recognize phosphoserine/threonine (pS/T)-containing motifs in a variety of signaling proteins. To date, 14-3-3 proteins have been reported to bind more than 200 client proteins. Through these interactions, 14-3-3 proteins play important roles in a wide range of vital regulatory processes, such as Bad-induced apoptosis, Raf-1-mediated cell proliferation, and Cdc25-regulated cell cycle progression. In addition to their participation in diverse physiological processes, 14-3-3 proteins have been implicated in a number of clinically important pathological conditions, such as neurodegenerative disorders and cancers. Thus, such studies on the 14-3-3/client-protein interactions may provide tremendous opportunities for therapeutic interventions. Therefore, chemical tools woul Enzyme Inhibitory Assay The activity of the compounds according to the present invention to inhibit the PARP-1 enzyme was examined using a PARP Assay kit (4671-096-K) purchased from Trevigen. The assay was performed following a modified previously reported method by Lee et al [Methods Find, Exp. Clin. Pharmacol., 27, 617-622, 2005].Histone was coated on 384-well plate, which is a small volume PS plate (784101) of Greiner Bio-One, and left at 25 C. for 2 hours. After that, the plate was rinsed four times with PBS (7.5 mM Na2HPO4, 2.5 mM NaH2PO4, 145 mM NaCl, pH 7.4), and in order to prevent non-specific reaction, the Strep-diluent (provided from kit of Trevigen) was added and left at 25 C. for one hour. After one hour, the plate was again rinsed with PBS four times, and the compounds of the Examples in various concentrations were put into reactant containing PARP-1 enzyme (0.12 unit/well), 2x PARP cocktail. GTPγS Binding The human APJ receptor was cloned by polymerase chain reaction and the gene encoding the receptor was subcloned in pFLAG-CMV-3 expression vector (Sigma, Saint Louis, Mo. USA) in-house at Amgen. A GTPγS binding assay was performed on membranes prepared from CHO cells stably expressing human APJ receptor. The optimum experimental conditions for the concentrations of GDP, MgCl2, and NaCl in the assay buffer were initially determined. The assay was performed in assay buffer [20 mM HEPES, pH 7.5, 5 mM MgCl2, and 0.1% (w/v) BSA with 200 mM NaCl, 3 μM GDP] and membranes expressing human APJ receptor/well along with WGA PS beads. The reaction was initiated by addition of 0.2 nM [35S]GTPγS in the absence or presence of various ligands and incubated at RT for 90 min. Nonspecific binding was determined in the presence of 100 μM GTPγS and was always less than 0.2% of total binding. In Vitro DGK Inhibition Assay First Table: A concentrated liposome solution was prepared in assay buffer without DTT and BSA: 2 mM of DLG in 21 mM of total liposome (2 mM DLG/8 mM PS/11 mM PC). The reaction mixtures contain the assay buffer with a final DLG concentration of 125 uM ATP concentrations of 25 μM (for DGKA assay) or 50 μM (for DGKZ assay). The reactions were started by addition of DGK α and ζ kinases at 4 nM and 2 nM final concentrations, respectively. After 1 hour reaction, the amount of ADP formed was detected with the ADP-Glo kinase assay (Promega) according to the manufacturer instructions. Compounds were added in 11-points dose response, starting at 10 mM, 1:3 dilutions, with a final DMSO concentration of 2%. The multidrop combi was used as a liquid handler and luminescence was read with 0.5 s by the envision reader (PE). Kinase Glo Luminescent Kinase Assay PI 3-Kinase Alpha (A), Pb 3-Kinase Beta (B), Vps34 (C), PI 4-Kinase Beta (D):The luminescence-based ATP detection reagent KinaseGlo was obtained from Promega, (Cat. No. V6714, Lot No. 236161) through Catalys, Wallisellen, Switzerland. L-alpha-phosphatidylinositol (PI, liver, bovine) was obtained from Avanti Polar Lipid (Cat. No. 840042C, Lot#LPI-274), Phosphatidylinositol-4,5-bisphosphate (PIP(4,5)2) was also obtained from Avanti Polar Lipid (Cat. No. 840046X). L-α-Phosphatidylserine (PS) was obtained from Avanti Polar Lipid (Cat. No. 840032C), n-Octylglucoside from Avanti Polar Lipid (Cat. No. 10634425001). Luminescence is a well established readout to determine ATP concentrations and can thus be used to follow the activity of many kinases regardless of their substrate. The Kinase Glo Luminescent Kinase Assay (Promega, Madison/WI, USA) is a homogeneous HTS method of measuring kinase activity by quantifying the amount of ATP remaining in solution following a kinase reaction. VPS34 Enzyme Assays 100 nL compounds in DMSO are added to wells of a 384 well microtitre plate (Greiner 780076). At room temperature: 5 ul VPS34 reaction buffer (Invitrogen Assay Buffer Q (diluted 1 in 5 with nanopure water) plus 2 mM DTT and 2 mM MnCl2) containing ATP (20 uM, Promega) and 200 uM PI-PS substrate (Invitrogen PV5122) is added followed immediately by 5 ul VPS34 reaction buffer (as above) containing VPS34 (5 nM, Millennium Protein Sciences Group) and the mixture is incubated with shaking at room temperature for 1 hour. Then 5 ul VPS34 stop-detect mix (as per Invitrogen Adapta Assay kit (PV5009) instructions (contains kinase quench buffer, TR-FRET buffer, Adapta Eu anti-ADP antibody and Alexa Fluor 647 ADP tracer)) is added to quench the reaction. The plates are then incubated for 30 minutes at room temperature with shaking and then read on a BMG PheraStar Plus reader. In Vitro DGK Inhibition Assay The DGKα and DGKζ reactions were performed using either extruded liposome (DGKα and DGKζ LIPGLO assays) or detergent/lipid micelle substrate (DGKα and DGKζ assays). The reactions were carried out in 50 mM MOPS pH 7.5, 100 mM NaCl, 10 mM MgCl2, 1 μM CaCl2), and 1 mM DTT (assay buffer). The reactions using a detergent/lipid micelle substrate also contained 50 mM octyl B-D-glucopyranoside. The lipid substrate concentrations were 11 mM PS and 1 mM DAG for the detergent/lipid micelle reactions. The lipid substrate concentrations were 2 mM PS, 0.25 mM DAG, and 2.75 mM PC for the extruded liposome reactions. The reactions were carried out in 150 μM ATP. The enzyme concentrations for the DGKα and DGKζ were 5 nM. The compound inhibition studies were carried out as follows: 50 nL droplets of each test compound (top concentration 10 mM with 11 point, 3-fold dilution series for each compound) solubilized in DMSO were transferred to wells of a white 1536 well plate (Corning 3725). A 5 mL enzyme/substrate solution at 2× final reaction concentration was prepared by combining 2.5 mL 4× enzyme solution (20 nM DGKα or DGKζ (prepared as described below) in assay buffer) and 2.5 mL of either 4× liposome or 4× detergent/lipid micelle solution (compositions described below) and incubated at room temperature for 10 minutes. Next, 1 μL 2× enzyme/substrate solution was added to wells containing the test compound and reactions were initiated with the addition of 1 μL 300 uM ATP. The reactions were allowed to proceed for 1 hr, after which 2 μL Glo Reagent (Promega V9101) was added and incubated for 40 minutes. Next, 4 μL Kinase Detection Reagent was added and incubated for 30 minutes. Luminescence was recorded using an EnVision microplate reader. In Vitro DGK Inhibition Assay The reactions were carried out in 50 mM MOPS pH 7.5, 100 mM NaCl, 10 mM MgCl2, 1 μM CaCl2, and 1 mM DTT (assay buffer). The reactions using a detergent/lipid micelle substrate also contained 50 mM octyl B-D-glucopyranoside. The lipid substrate concentrations were 11 mM PS and 1 mM DAG for the detergent/lipid micelle reactions. The lipid substrate concentrations were 2 mM PS, 0.25 mM DAG, and 2.75 mM PC for the extruded liposome reactions (5 mM total lipid). The reactions were carried out in 150 μM ATP. The enzyme concentrations for the DGKα and DGKζ were 5 nM. The compound inhibition studies were carried out as follows: 25 nL (ADPGLO assay) or 50 nL (LIPGLO assay) droplets of each test compound (top concentration 10 mM with 11 point, 3-fold dilution series for each compound) solubilized in DMSO were transferred to wells of a white 1536 well plate (Corning 3725). A 5 mL enzyme/lipid substrate solution at 2× final reaction concentration was prepared by combining 2.5 mL 4× enzyme solution (20 nM DGKα or DGKζ (prepared as described below) in assay buffer) and 2.5 mL of either 4× liposome or 4× detergent/lipid micelle solution (compositions described below) and incubated at room temperature for 10 minutes. Next, 1 μL 2× enzyme/lipid substrate solution was added to wells containing the test compound and reactions were initiated with the addition of 1 μL 300 uM ATP. The reactions were allowed to proceed for 2 hr (ADPGLO assay) or 1 hr (LIPGLO assay), after which 2 μL Glo Reagent (Promega V9101) was added and incubated for 40 minutes. Next, 4 μL Kinase Detection Reagent was added and incubated for 30 minutes. Luminescence was recorded using an EnVision microplate reader. The percent inhibition was calculated from the ATP conversion generated by no enzyme control reactions for 100% inhibition and vehicle-only reactions for 0% inhibition. Inhibition Assay Protein Kinase C beta 2 (PKCβII) catalyzes the production of ADP from ATP that accompanies the phosphoryl transfer to the PKC Pseudosubstrate peptide (A→S, RFARKGSLRQKNV). This transfer is coupled to the oxidation of β-NADH through the activities of Pyruvate Kinase (PK) and Lactate Dehydrogenase (LDH). β-NADH conversion to NAD+ is monitored by the decrease in absorbance at 340 nm (e=6.22 cm−1 mM−1) using a Molecular Devices SPECTRA max PLUS spectrophotometer.A typical assay was carried out on a 96-well, clear microtiter plate in a Molecular Devices spectrophotometer for 20 minutes at 30° C. in 0.1 mL of assay buffer containing 50 mM HEPES, pH 7.4, 5 nM PKC, 23 units of pyruvate kinase, 33 units of lactate dehydrogenase, 0.15 mM peptide, 0.1 mM ATP, 1 mM DTT, 4 mM PEP, 8 mM MgCl2, 0.3 mM NADH, 60 mM CaCl2, 10 mg/mL PS, 50 ng/mL PMA, 7.5% DMSO and from about 10,000 nM to 0.169 nM compound inhibitor. Stock solutions of 3-sn-phosphatidyl-L-serine (PS) and phorbol-12-myristate-13-acetate (PMA) were sonicated for 30 seconds just prior to addition to assay buffer and assays were initiated by the addition of 100 μM ATP.Steady-state kinetic parameters for the bi-bi kinase reaction were determined at saturating phospho-acceptor peptide substrate concentration (0.15 mM) by fitting initial velocity data to the Michaelis-Menten equation, v=V max [S]/(K M +[S]) where v is the measured initial velocity, Vmax is the maximal enzyme velocity, [S] is the ATP substrate concentration, and KM is the Michealis constant for ATP. Enzyme turnover values (kcat) were calculated according to kcat=Vmax[E], where [E] is the total enzyme concentration. Enzyme inhibition constants (apparent Ki values) were determined by fitting initial velocities at variable inhibitor concentrations to a model for ATP competitive inhibition based on the Morrison equation). Morrison, J. F., Biochim. Biophys Acta 185: 269-286 (1969). PI3K-Alpha Kinase (PIK3CA) Activity, Wild-Type and H1047R Mutant and Determining IC50 Values for Inhibitors Recombinant, catalytically active human full length PIK3KA Wild-type and H1047R mutant were purchased as 1:1 complex of N-terminal 6× his tagged p110Figure US20230286960A1-20230914-P00002(catalytic) and untagged p85Figure US20230286960A1-20230914-P00002(regulatory subunit) from EMD Millipore Sigma (cat. no. 14-602M and 14-792M, respectively). The enzyme stocks were diluted to 5× stocks in buffer (20 mM HEPES pH 7.4, 100 mM NaCl, 0.5 mM EGTA, 0.01% triton-x-100) just before use. PIP2diC8 (Avanti Polar Lipids Inc., cat. no. 850185) or phosphoinositol-4,5-bisphosphate with phosphoserine (PIP2:PS) membrane (Thermo Fisher Scientific, cat. no. PV5100) was used as lipid substrates. PIP2diC8 lyophilized powder and PIP2:PS (1:19) membrane stock (1 mM in PIP2) were separately dissolved in milliQ water to a concentration of 250 uM and stored in −20° C. 10 mM stocks of compounds were serially diluted (3×) in neat DMSO and stored in a dessicator at room temperature. 5× compound stocks in 25% DMSO were prepared fresh from neat DMSO stocks. Wild-type (WT) and H1047R mutant protein, along with buffer components (except ATP), were incubated with or without compound at 27° C. for 1 h. After incubation, the reaction was initiated by the addition of 5 uL of 125 uM ATP. A typical assay mixture (25 uL) comprised 40 mM HEPES buffer, pH 7.4, 25 mM MgCl2, 0.01% v/v triton-X-100, 5% v/v DMSO, 20 mM NaCl, 1-5 nM wt or H1047R, 25 uM ATP, and 50 uM PIP2diC8 or PIP2 in membrane. The reaction was allowed to proceed until about 1000 conversion (2.5 uM ADP) after which time, 10 uL of reaction mixture was quenched with 25 uL of transcreener reagent (Transcreener ADP2 FI assay kit, BellBrook labs, Cat. No. 3013). The contents were incubated at rt for 1 h and fluorescence was measured using a plate reader (Paradigm, Molecular Devices). FlashPlate™ Assay DGAT activity in membrane preparations was assayed in 50 mM Tris-HCl (pH 7.4), 150 mM MgCl2, 1 mM EDTA and 0.2% BSA, containing 50 μM DAG, 32 μg/ml PC/PS and 8.4 μM [3H]-oleoylCoA (at a specific activity of 30 nCi/well) in a final volume of 50 μl in 384-well format using the red shifted Basic Image FlashPlate™ (Perkin Elmer Cat.No. SMP400). In detail, 10 μl enzyme mix and 10 μl substrate mix were added to 30 μl of assay buffer, optionally in the presence of 1 μl DMSO (blank and controls) or 1 μl of the compound to be tested. This reaction mixture was incubated for 120 minutes at 37° C. and the enzymatic reaction stopped by adding 20 μl of the stop mix. The plates were sealed and the vesicles allowed to settle overnight at room temperature. Plates were centrifuged for 5 minutes at 1500 rpm and measured in Leadseeker. Inhibition Assay Protein Kinase C beta 2 (PKC beta II) catalyzes the production of ADP from ATP that accompanies the phosphoryl transfer to the PKC Pseudosubstrate peptide (A->S, RFARKGSLRQKNV). This transfer is coupled to the oxidation of p-NADH through the activities of Pyruvate Kinase (PK) and Lactate Dehydrogenase (LDH). (3-NADH conversion to NAD+ is monitored by the decrease in absorbance at 340 nm (e=6.22 cm-1 mM-1) using a Molecular Devices SPECTRA max PLUS spectrophotometer.A typical assay was carried out on a 96-well, clear microtiter plate in a Molecular Devices spectrophotometer for 20 minutes at 30 C. in 0.1 mL of assay buffer containing 50 mM HEPES, pH 7.4, 5 nM PKC, 23 units of pyruvate kinase, 33 units of lactate dehydrogenase, 0.15 mM peptide, 0.1 mM ATP, 1 mM DTT, 4 mM PEP, 8 mM MgCl2, 0.3 mM NADH, 60 mM CaCl2, 10 mg/mL PS, 50 ng/mL PMA, 7.5% DMSO and from about 10,000 nM to 0.169 nM compound inhibitor. TEAD4 FRET Assay Compound IC50 values were assessed following a 10-point, half-log10 dilution schema starting at 100 μM compound concentration. Specifically, human TEAD protein from TEAD4(217-434) was cloned into an overexpression vector, expressed as an N-terminal HIS-TEV-Avi-tagged fusion protein in E coli, and subsequently purified, then protein was chemically depalmitoylated & biotinylated. The assay was performed in 384-well LV plates (384-well black, medium binding, PS, HIBASE, GREINER #784076) and run in the presence and absence of the compound of interest. Each well of 5 μL assay mixture contained 10 mM Tris-HCl (pH 7.5), 100 mM NaCl, 0.05 mM EDTA, 1 mM TECP, 1% DMSO, 0.03% Pluronic acid F127, 20 nM dePal Avi TEAD4 (217-434)-depalmitoylated & biotinylated protein, 0.8 nM Streptavidin Terbium cryptate (CisBio #610SATLB), 625 nM FAM labelled Probe A. Reactions were incubated at 25° C. for 120 min before reading on a PHERASTAR FSX Plate Reader (337 520 490 HTRF module required) (Supplier BMP). PI3K-Alpha kinase (PIK3CA) activity, wild-type and H1047R mutant and determining IC50 values for inhibitors Recombinant, catalytically active human full length PIK3KA Wild-type and H1047R mutant were purchased as 1:1 complex of N-terminal 6×his tagged p110<(catalytic) and untagged p85<(regulatory subunit) from EMD Millipore Sigma (cat. no. 14-602M and 14-792M, respectively). The enzyme stocks were diluted to 5× stocks in buffer (20 mM HEPES pH 7.4, 100 mM NaCl, 0.5 mM EGTA, 0.01% triton-x-100) just before use. PIP2diC8 (Avanti Polar Lipids Inc., cat. no. 850185) or phosphoinositol-4,5-bisphosphate with phosphoserine (PIP2:PS) membrane (Thermo Fisher Scientific, cat. no. PV5100) was used as lipid substrates. PIP2diC8 lyophilized powder and PIP2:PS (1:19) membrane stock (1 mM in PIP2) were separately dissolved in milliQ water to a concentration of 250 uM and stored in −20° C. 10 mM stocks of compounds were serially diluted (3×) in neat DMSO and stored in a dessicator at room temperature. 5× compound stocks in 25% DMSO were prepared fresh from neat DMSO stocks. Wild-type (WT) and H1047R mutant protein, along with buffer components (except ATP), were incubated with or without compound at 27° C. for 1h. After incubation, the reaction was initiated by the addition of 5 uL of 125 uM ATP. A typical assay mixture (25 uL) comprised 40 mM HEPES buffer, pH 7.4, 25 mM MgCl2, 0.01% v/v triton-X-100, 5% v/v DMSO, 20 mM NaCl, 1-5 nM wt or H1047R, 25 uM ATP, and 50 uM PTP2diC8 or PTP2 in membrane. The reaction was allowed to proceed until about 1000 conversion (2.5 uM ADP) after which time, 10 uL of reaction mixture was quenched with 25 uL of transcreener reagent (Transcreener ADP2 FI assay kit, BellBrook labs, Cat. No. 3013). The contents were incubated at rt for 1h and fluorescence was measured using a plate reader (Paradigm, Molecular Devices). The same assay was also run at pH 6.0 or 6.4 using MOPS buffer (Fisher BioReagents, CAS 1132-61-2). A calibration curve was generated under identical buffer conditions with varying ADP amounts. Using that, the observed fluorescence was converted to uM ADP. Biochemical Assay For the biochemical assay panel, 50 nl of the test compounds, reference compounds and buffer/DMSO control are transferred to the respective wells of a 384-well white GREINER "SMALL VOLUME" PS plate. The assay panel is run at room temperature on a Biomek FX liquid handling workstation. To the assay plates containing 50 nl compound or control solutions in 90% DMSO, 4.5 ul of E3 ligase solution were added per well, followed by 4.5 ul of the pre-incubated E1/E2/Ub mix or the pre-diluted ubiquitin (LOW control). Plates are shaken vigorously after each addition. In this assay the compound concentrations range from 3 nM to 10 uM in an 8-point dose-response curve. After 45 min of incubation the ubiquitinylation reactions were stopped by adding 4.5 ul 2 mM NEM, immediately followed by 4.5 ul of a detection solution including the XL665-labeled antibody and the streptavidin-coupled europium to give a total volume of 18 ul. After an incubation time of 45 min in the dark, the plates are transferred into the Pherastar fluorescence reader to measure the TR-FRET signal. Inhibition of PI3Kα Compounds were prepared at 100× final concentration using a 12-point, 1:3 serial-dilution in DMSO, with DMSO control as the 12th point. Compound was then diluted in HEPES buffer (25 mM HEPES pH 7.5, 1 mM EGTA, 0.3% CHAPS) prior to addition to PI3Kα. Active PI3Kα diluted to 0.24 ng/μL (1.1 nM) in (50 mM HEPES pH 7.5, 6 mM MgCl2, 1 mM EGTA, 200 mM NaCl, 0.03% CHAPS, 8 mM DTT) was incubated with compound for 0 hr and 3 hr prior to the start of the reaction. 25 μM PIP2:PS and 60 μM ATP were diluted from stock solution (25 mM HEPES pH 7.5, 1 mM EGTA, 0.3% CHAPS) and added to initiate the PI3Kα reaction. Reaction time was 30 minutes. ATP to ADP conversion was measured in Luminescence Counts on DTX880 Plate Reader (Beckman Coulter). The IC50 of the compounds were reported using the GraphPad Prism software. Analytical method was non-linear regression, 4-parameter curve fit with bottom fit to validated PI3Kα inhibitor reference controls and no top fit (floating top). MT-4 HIV Wild Type Virus Infection Assay (IIIB Virus) Test compounds and controls were serially diluted and spotted in replicate into 384 well black assay plates via acoustic transfer (Echo). MT-4 cells were grown in batch, centrifuged and resuspended into fresh CCM media (RPMI w/10% FBS, 1% PS) at 2×106 cells/ml. MT-4 cells were acutely infected with HIV-1 IIIB strain. The size of each infection mix was scaled by the number of sample plates to be tested. Each infection mix was transferred into 5 mL closed tubes and nutated rapidly on a shaker at 37° C. incubator for 1 hour. The infection mixes were then diluted 25× in fresh cell culture media and then added to assay plates at 40 μL per well using a ViaFlo 384 pipettor. After 5 day incubation at 37° C. in a CO2 incubator, assay plates were processed with Cell-titer glo reagent using a ViaFlo 384 with an addition/mixing program. Plates were read immediately on Envision reader. Assay signals were plotted and dose response curves generated to determine individual compound EC50s. [35S]GTPgammaS Binding The human APJ receptor was cloned by polymerase chain reaction and the gene encoding the receptor was subcloned in pFLAG-CMV-3 expression vector (Sigma, Saint Louis, Mo. USA) in-house at Amgen. A GTPγS binding assay was performed on membranes prepared from CHO cells stably expressing human APJ receptor. The optimum experimental conditions for the concentrations of GDP, MgCl2, and NaCl in the assay buffer were initially determined. The assay was performed in assay buffer [20 mM HEPES, pH 7.5, 5 mM MgCl2, and 0.1% (w/v) BSA with 200 mM NaCl, 3 μM GDP] and membranes expressing human APJ receptor/well along with WGA PS beads. The reaction was initiated by addition of 0.2 nM [35S]GTPγS (Perkin Elmer Life and Analytical Sciences, Waltham USA) in the absence or presence of various ligands and incubated at RT for 90 min. Nonspecific binding was determined in the presence of 100 μM GTPγS and was always less than 0.2% of total binding. All the results presented are means of several independent experiments and analyzed by non-linear regression methods using commercially available program Prism (GraphPad, San Diego, Calif.). Enzyme Assay 100 nL compounds in DMSO are added to wells of a 384 well microtitre plate (Greiner 780076). At room temperature: 5 ul VPS34 reaction buffer (Invitrogen Assay Buffer Q (diluted 1 in 5 with nanopure water) plus 2 mM DTT and 2 mM MnCl2) containing ATP (20 uM, Promega) and 200 uM PI-PS substrate (Invitrogen PV5122) is added followed immediately by 5 ul VPS34 reaction buffer (as above) containing VPS34 (5 nM, Millennium Protein Sciences Group) and the mixture is incubated with shaking at room temperature for 1 hour. Then 5 ul VPS34 stop-detect mix (as per Invitrogen Adapta Assay kit (PV5009) instructions (contains kinase quench buffer, TR-FRET buffer, Adapta Eu anti-ADP antibody and Alexa Fluor 647 ADP tracer)) is added to quench the reaction. The plates are then incubated for 30 minutes at room temperature with shaking and then read on a BMG PheraStar Plus reader.For the assay methods described above, test compound percent inhibition, at various concentrations, is calculated relative to control (DMSO and EDTA) treated samples. Compound concentration versus percent inhibition curves are fitted to generate IC50 values. One skilled in the art will appreciate that the values generated either as percentage inhibition at a single concentration or IC50 values are subject to experimental variation. [35S]GTPγS Binding The human APJ receptor was cloned by polymerase chain reaction and the gene encoding the receptor was subcloned in pFLAG-CMV-3 expression vector (Sigma, Saint Louis, Mo. USA) in-house at Amgen. A GTPγS binding assay was performed on membranes prepared from CHO cells stably expressing human APJ receptor. The optimum experimental conditions for the concentrations of GDP, MgCl2, and NaCl in the assay buffer were initially determined. The assay was performed in assay buffer [20 mM HEPES, pH 7.5, 5 mM MgCl2, and 0.1% (w/v) BSA with 200 mM NaCl, 3 M GDP] and membranes expressing human APJ receptor/well along with WGA PS beads. The reaction was initiated by addition of 0.2 nM [35S]GTPγS (Perkin Elmer Life and Analytical Sciences, Waltham USA) in the absence or presence of various ligands and incubated at RT for 90 min. Nonspecific binding was determined in the presence of 100 M GTPγS and was always less than 0.2% of total binding. All the results presented are means of several independent experiments and analyzed by non-linear regression methods using commercially available program Prism (GraphPad, San Diego, Calif.) to obtain EC50. [35S]GTPgammaS Binding Assay The human APJ receptor was cloned by polymerase chain reaction and the gene encoding the receptor was subcloned in pFLAG-CMV -3 expression vector (Sigma, Saint Louis, Mo. USA) in-house at Amgen. A GTPγS binding assay was performed on membranes prepared from CHO cells stably expressing human APJ receptor. The optimum experimental conditions for the concentrations of GDP, MgCl2, and NaCl in the assay buffer were initially determined. The assay was performed in assay buffer [20 mM HEPES, pH 7.5, 5 mM MgCl2, and 0.1% (w/v) BSA with 200 mM NaCl, 3 μM GDP] and membranes expressing human APJ receptor/well along with WGA PS beads. The reaction was initiated by addition of 0.2 nM [35S]GTPγS (Perkin Elmer Life and Analytical Sciences, Waltham USA) in the absence or presence of various ligands and incubated at RT for 90 min. Nonspecific binding was determined in the presence of 100 μM GTPγS and was always less than 0.2% of total binding. All the results presented are means of several independent experiments and analyzed by non-linear regression methods using commercially available program Prism (GraphPad, San Diego, Calif.) [35S]GTPgammaS Binding Assay The human APJ receptor was cloned by polymerase chain reaction and the gene encoding the receptor was subcloned in pFLAG-CMV -3 expression vector (Sigma, Saint Louis, Mo. USA) in-house at Amgen. A GTPγS binding assay was performed on membranes prepared from CHO cells stably expressing human APJ receptor. The optimum experimental conditions for the concentrations of GDP, MgCl2, and NaCl in the assay buffer were initially determined. The assay was performed in assay buffer [20 mM HEPES, pH 7.5, 5 mM MgCl2, and 0.1% (w/v) BSA with 200 mM NaCl, 3 μM GDP] and membranes expressing human APJ receptor/well along with WGA PS beads. The reaction was initiated by addition of 0.2 nM [35S]GTPγS (Perkin Elmer Life and Analytical Sciences, Waltham USA) in the absence or presence of various ligands and incubated at RT for 90 min. Nonspecific binding was determined in the presence of 100 μM GTPγS and was always less than 0.2% of total binding. All the results presented are means of several independent experiments and analyzed by non-linear regression methods using commercially available program Prism (GraphPad, San Diego, Calif.) to obtain EC50. AOC1 Biochemical Assay For the determination of AOC1 activity or compound AOC1 inhibition potency, the compound inhibitors are dissolved in DMSO and adjusted to the respective assay concentration with reaction buffer (100 mM sodiumphosphate, 0.05% Pluronic F-127 (#P3000MP Sigma-Aldrich), pH 7.4). An aliquot of 3 μL of the compound dilution is added to a 384 well plate (Optiplate, PS, flat bottom F, black, PERKIN ELMER, #6007270) in a DMSO concentration of 6.6%.An AOC1 enzyme aliquot (#8297-AO-010, R&D Systems) is thawed on ice, diluted in reaction buffer and added in a volume of 7 μL to the wells to give a final assay concentration of 1 ng/well. After incubation of inhibitor and enzyme for 30 minutes at 37° C., the enzymatic reaction is started with the addition of 10 μL of Amplex® Red reaction mix (final assay concentration: 100 mM sodiumphosphate, 120 μM Amplex® Red reagent (#A22177 Molecular Probes), 1.5 U/mL Horseradish Peroxidase (#P8375 Sigma-Aldrich), 200 μM putrescine (#P7505 Sigma-Alrdich), 0.05% Pluronic F-127 (#P3000MP Sigma-Aldrich), pH 7.4, 37° C.).After an incubation for 30 minutes at 37° C. the turnover of the substrate is determined directly (or after the addition of an excess of an amine-oxidase inhibitor) with a fluorescence reader (Ex 540 nm/Em 590 nm) like Envision 2104 Multilabel Reader (PERKIN ELMER). AOC3 Biochemical Assay For the determination of AOC3 activity or compound inhibition potency, the compound inhibitors are dissolved in DMSO and adjusted to the respective assay concentration with reaction buffer (50 mM HEPES, 5 mM KCl, 2 mM CaCl2, 1.4 mM MgCl2, 120 mM NaCl, 0.001% (v/v) Tween 20, 100 μM TCEP, pH 7.4). An aliquot of 3 μL of the compound dilution is added to a 384 well plate (Optiplate, PS, flat bottom, white, PERKIN ELMER, #6007290) with a final DMSO concentration of 6.6%. Recombinant CHO cells, overexpressing the human (1500 cells/well), mouse (1000 cells/well) or rat (500 cells/well) AOC3 enzyme are diluted in reaction buffer and added in a volume of 15 μL to the wells. After incubation for 20 minutes at 37° C., 2 μL of MAO substrate (dissolved in DMSO at 16 mM, adjusted to assay concentration in reaction buffer to a final assay concentration of 20 μM) is added and further incubated for 60 minutes at 37° C. The turnover of the substrate is determined by the addition of 20 μL of the detection-mix which was generated by the addition of reconstitution buffer with esterase (PROMEGA, #V1402) to the luciferine detection reagent (PROMEGA, #V1402). After an incubation period of 20 minutes, the luminescent signal is measured with Envision 2104 Multilabel Reader (PERKIN ELMER). Competitive Delta Receptor Binding Assay Step 1: 50 μL of a vehicle (1% DMSO) was added into a total binding tube (TB), 50 μL of DADLE (with a final concentration of 1.0×10−5 M) was added into a non-specific binding tube (NB), and 50 μL of a test compound was added into each compound binding tube (CB).Step 2: 100 μL of a buffer (homogenate A) was added into each reaction tube.Step 3: the prepared membrane was suspended in the homogenate A to obtain a 10 mg/mL membrane suspension for later use.Step 4: 50 μL of radioligand 3H-DADLE was added into each reaction tube to reach a final concentration of 4 nM.Step 5: 50 μL of the membrane solution was added into each reaction tube.Step 6: each reaction tube was incubated at 25° C. for 90 mp. After the reaction was completed, the binding ligands were rapidly filtered under reduced pressure. The UniFilter GF/C plate was saturated with 0.5% PEI solution 1 h in advance, fully washed with cold Tris buffer, filtered under vacuum, then placed into a thermostatic dryer, and dried for 30 min. The filter plate was taken out, and MICROSCINT PS scintillation solution was added at 40 μL/well.Step 7: the filter plate was put into a liquid scintillation counter for counting. Competitive Kappa Receptor Binding Assay Step 1: 50 μL of a vehicle (1% DMSO) was added into a total binding tube (TB), 50 μL of U69593 (with a final concentration of 1.0×10−5 M) was added into a non-specific binding tube (NB), and 50 μL of a test compound was added into each compound binding tube (CB).Step 2: 100 μL of a buffer (homogenate A) was added into each reaction tube.Step 3: the prepared membrane was suspended in the homogenate A to obtain a 15 mg/mL membrane suspension for later use.Step 4: 50 μL of radioligand 3H-U69593 was added into each reaction tube to reach a final concentration of 2 nM.Step 5: 50 μL of the membrane solution was added into each reaction tube.Step 6: each reaction tube was incubated at 25° C. for 90 min. After the reaction was completed, the binding ligands were rapidly filtered under reduced pressure. The UniFilter GF/C plate was saturated with 0.5% PEI solution 1 h in advance, fully washed with cold Tris buffer, filtered under vacuum, then placed into a thermostatic dryer, and dried for 30 min. The filter plate was taken out, and MICROSCINT PS scintillation solution was added at 40 μL/well.Step 7: the filter plate was put into a liquid scintillation counter for counting. Enzyme Assay In this assay, CSNK1D phosphorylates a substrate peptide PLSRTL-pS-VASLPGL in the presence of ATP. This substrate peptide has been modeled after the sequences surrounding three main cyclic AMP-dependent protein kinase sites of glycogen synthase. This assay monitors CSNK1D kinase activity by measuring the amount of ADP produced in the assay. A substrate mix is prepared by diluting peptide substrate (final concentration 150 μM) with ATP (final concentration 20 μM) in assay buffer (50 mM Tris/HCl pH 7.4+10 mM MgCl2+1 mM DTT+0.1% BSA). The substrate mix is added to each well of a low volume, 384-well, white opaque plate. Test compounds were diluted in HBSS and added in a dose-response to the plate. To start the reaction, 2 nM of constitutively active human recombinant GST cleaved CSNK1D (University of Dundee, clone DU 19064, stored at 0.28 mg/mL in 50 mM Tris/HCl pH 7.5, 150 mM NaCl, 270 mM Sucrose, 0.1 mM EGTA, 0.1% 2-mercaptoethanol, 0.02% Brij-35.1 mM benzamidine, 0.2 mM PMSF) was added to each well and the plate centrifuged for 5 minutes at 1500 rpm. The total volume of each reaction is 5 ul (2 μL of substrate mix, 1 μL of diluted compounds, and 2 μL of human recombinant CSNK1D). The plates are incubated for 45 minutes at room temperature. Human VPS34 Enzyme Assay 100 nL compounds in DMSO are added to wells of a 384 well microtitre plate (Greiner 780076). At room temperature: 5 ul VPS34 reaction buffer (Invitrogen Assay Buffer Q (diluted 1 in 5 with nanopure water) plus 2 mM DTT and 2 mM MnCl2) containing ATP (20 uM, Promega) and 200 uM PI-PS substrate (Invitrogen PV5122) is added followed immediately by 5 ul VPS34 reaction buffer (as above) containing VPS34 (5 nM, Millennium Protein Sciences Group) and the mixture is incubated with shaking at room temperature for 1 hour. Then 5 ul VPS34 stop-detect mix (as per Invitrogen Adapta Assay kit (PV5009) instructions (contains kinase quench buffer, TR-FRET buffer, Adapta Eu anti-ADP antibody and Alexa Fluor 647 ADP tracer)) is added to quench the reaction. The plates are then incubated for 30 minutes at room temperature with shaking and then read on a BMG PheraStar Plus reader.For the assay methods described above, test compound percent inhibition, at various concentrations, is calculated relative to control (DMSO and EDTA) treated samples. Compound concentration versus percent inhibition curves are fitted to generate IC50 values. One skilled in the art will appreciate that the values generated either as percentage inhibition at a single concentration or IC50 values are subject to experimental variation. Mu Receptor Affinity Assay Step 1: 50 μL of a vehicle (1% DMSO) was added into a total binding tube (TB), 50 μL of DAMGO (with a final concentration of 1.0×10−5 M) was added into a non-specific binding tube (NB), and 50 μL of a test compound was added into each compound binding tube (CB).Step 2: 100 μL of a buffer (homogenate A) was added into each reaction tube.Step 3: the prepared membrane was suspended in the homogenate A to obtain a 10 mg/mL membrane suspension for later use.Step 4: 50 μL of radioligand [3H] DAMGO was added into each reaction tube to reach a final concentration of 2 nM.Step 5: 50 μL of the membrane solution was added into each reaction tube.Step 6: each reaction tube was incubated at 25° C. for 90 min. After the reaction was completed, the binding ligands were rapidly filtered under reduced pressure. The UniFilter GF/C plate was saturated with 0.5% PEI solution 1 h in advance, fully washed with cold Tris buffer, filtered under vacuum, then placed into a thermostatic dryer, and dried for 30 min. The filter plate was taken out, and MICROSCINT PS scintillation solution was added at 40 μL/well.Step 7: the scintillation vial was put into a liquid scintillation counter for counting. In Vitro DGK Inhibition Assays DGK α and ζ kinase use ATP to phosphorylate the substrate 1,2-dilauroyl-sn-glycerol (DLG, incorporated in the liposomes). ATP is converted to ADP as a result of this enzymatic reaction. After the kinase reaction, an ATP-depletion reagent is added to terminate the kinase reaction and deplete any remaining ATP, leaving only ADP. Second, a detection reagent is added to simultaneously convert ADP to ATP and allow the newly synthesized ATP to be converted to light using a coupled luciferase/luciferin reaction. A concentrated liposome solution was prepared in assay buffer without DTT and BSA: 2 mM of DLG in 21 mM of total liposome (2 mM DLG/8 mM PS/11 mM PC). The reaction mixtures contain the assay buffer with a final DLG concentration of 125 uM ATP concentrations of 25 μM (for DGKA assay) or 50 μM (for DGKZ assay). The reactions were started by addition of DGK α and ζ kinases at 4 nM and 2 nM final concentrations, respectively. After 1 hour reaction, the amount of ADP formed was detected with the ADP-Glo kinase assay (Promega) according to the manufacturer instructions. Compounds were added in 11-points dose response, starting at 10 mM, 1:3 dilutions, with a final DMSO concentration of 2%. The multidrop combi was used as a liquid handler and luminescence was read with 0.5 s by the envision reader (PE). TR-FRET Adapta Assay for PI 3-Kinase Gamma (E), PI 3-Kinase Delta (F) The TR-FRET Adapta Universal Kinase Assay Kit was purchased from Invitrogen Corporation (Carlsbad, Calif., USA) (Cat. No. PV5099). The kit contains the following reagents: Adapta Eu-anti-ADP Antibody (Europium labeled anti-ADP antibody in HEPES buffered saline, Cat. No. PV5097), Alexa Fluor 647-labeled ADP tracer (Alexa Fluor 647-labeled ADP tracer in HEPES buffered saline, Cat. No. PV5098), TR-FRET dilution buffer pH 7.5 (Cat. No. PV3574). PIK3CD substrate phosphatidylinositol (PI) was obtained from Invitrogen (vesicules consisting of 2 mM phosphatidylinositol (PI) in 50 mM HEPES pH7.5; Cat. No. PV5371). PIK3CG substrate phosphatidylinositol-4,5-bisphosphate (PIP(4,5)2 was obtained from Invitrogen (PIP2:PS large unilamellar vesicules consisting of 1 mM PIP2:19 mM PS in 50 mM HEPES pH7.5, 3 mM MgCl2, 1 mM EGTA; Cat. No. PV5100). Time-Resolved Fluorescence Resonance Energy Transfer (TR-FRET) is a technology based on energy transfer between two adjacent dyes, from an excited electron in one dye (the donor) to an electron of an adjacent dye (the acceptor) through resonance, then released as a photon. This energy transfer is detected by an increase in the fluorescence emission of the acceptor, and a decrease in the fluorescence emission of the donor. TR-FRET assays for protein kinases use a long-lifetime lanthanide Terbium or Europium chelates as the donor species which overcome interference from compound autofluorescence or light scatter from precipitated compounds, by introducing a delay after excitation by a flashlamp excitation source. Results are often expressed as a ratio of the intensities of the acceptor and donor fluorophores. The ratiometric nature of such a value corrects for differences in assay volumes between wells, as well as corrects for quenching effects due to colored compounds. The Adapta assay can be divided into two phases: a kinase reaction phase and an ADP detection phase. In the kinase reaction phase, all kinase reaction components are added to the well and the reaction is allowed to incubate for a set period of time specific for each kinase. After the reaction, a detection solution of Eu-labeled anti-ADP antibody, Alexa Fluor 647-labeled ADP tracer, and EDTA (to stop the kinase reaction) are added to the assay well. ADP formed by the kinase reaction will displace the Alexa Fluor 647-labeled ADP tracer from the antibody, resulting in a decrease in TR-FRET signal. In the presence of an inhibitor, the amount of ADP formed by the kinase reaction is reduced, and the resulting intact antibody-tracer interaction maintains a high TR-FRET signal. In the Adapta assay, the donor (Europium-anti-ADP antibody) is excited at 340 nm and will transfer its energy to the acceptor (Alexa Fluor 647-labeled ADP tracer). The emission from the Alexa Fluor 647 can be monitored with a filter centered at 665 nm because it is located between the emission peaks of the donor, which is measured at 615/620 nm. GTPγS Binding The human APJ receptor was cloned by polymerase chain reaction and the gene encoding the receptor was subcloned in pFLAG-CMV -3 expression vector (Sigma, Saint Louis, Mo. USA) in-house at Amgen. A GTPγS binding assay was performed on membranes prepared from CHO cells stably expressing human APJ receptor. The optimum experimental conditions for the concentrations of GDP, MgCl2, and NaCl in the assay buffer were initially determined. The assay was performed in 9 μL assay buffer [20 mM HEPES, pH 7.5, 5 mM MgCl2, 100 mM NaCl and 0.1% (w/v) BSA], 1 μL of diluted test compound (starting with 0.75 mM, 2-fold serial dilution with DMSO, total 22 points), 10 μL of 18 μM GDP (final concentration of 3 μM GDP), 20 μL of 0.25 μg/mL membrane protein expressing human APJ receptor captured with WGA PS beads (final concentration of 5 μg per well), and 20 μL of 0.3 nM [35S]GTPγS (final concentration is 0.1 nM [35S]GTPγS)(Perkin Elmer Life and Analytical Sciences, Waltham USA). One column of the plate was 1 μL of DMSO as background and another column of the plate was 1 μL of 180 μM Pyr-Apelin-13 which was used as control at a final concentration of 3 μM. Incubation was at RT for 90 min and the microplate was read using a ViewLux ultra HTS Microplate Imager. [35S]GTPγS Binding Assay The human APJ receptor was cloned by polymerase chain reaction and the gene encoding the receptor was subcloned in pFLAG-CMV -3 expression vector (Sigma, Saint Louis, Mo. USA) in-house at Amgen. A GTPγS binding assay was performed on membranes prepared from CHO cells stably expressing human APJ receptor. The optimum experimental conditions for the concentrations of GDP, MgCl2, and NaCl in the assay buffer were initially determined. The assay was performed in 9 μL assay buffer [20 mM HEPES, pH 7.5, 5 mM MgCl2, 100 mM NaCl and 0.1% (w/v) BSA], 1 μL of diluted test compound (starting with 0.75 mM, 2-fold serial dilution with DMSO, total 22 points), 10 μL of 18 μM GDP (final concentration of 3 μM GDP), 20 μL of 0.25 μg/mL membrane protein expressing human APJ receptor captured with WGA PS beads (final concentration of 5 μg per well), and 20 μL of 0.3 nM [35S]GTPγS (final concentration is 0.1 nM [35S]GTPγS)(Perkin Elmer Life and Analytical Sciences, Waltham USA). One column of the plate was 1 μL of DMSO as background and another column of the plate was 1 μL of 180 μM Pyr-Apelin-13 which was used as control at a final concentration of 3 μM. Incubation was at RT for 90 min and the microplate was read using a ViewLux ultra HTS Microplate Imager (PerkinElmer, Inc.). [35S]GTPγS Binding The human APJ receptor was cloned by polymerase chain reaction and the gene encoding the receptor was subcloned in pFLAG-CMV -3 expression vector (Sigma, Saint Louis, Mo. USA) in-house at Amgen. A GTPγS binding assay was performed on membranes prepared from CHO cells stably expressing human APJ receptor. The optimum experimental conditions for the concentrations of GDP, MgCl2, and NaCl in the assay buffer were initially determined. The assay was performed in 9 μL assay buffer [20 mM HEPES, pH 7.5, 5 mM MgCl2, 100 mM NaCl and 0.1% (w/v) BSA], 1 μL of diluted test compound (starting with 0.75 mM, 2-fold serial dilution with DMSO, total 22 points), 10 μL of 18 M GDP (final concentration of 3 μM GDP), 20 μL of 0.25 μg/mL membrane protein expressing human APJ receptor captured with WGA PS beads (final concentration of 5 μg per well), and 20 μL of 0.3 nM [35S]GTPγS (final concentration is 0.1 nM [35S]GTPγS)(Perkin Elmer Life and Analytical Sciences, Waltham USA). One column of the plate was 1 μL of DMSO as background and another column of the plate was 1 μL of 180 M Pyr-Apelin-13 which was used as control at a final concentration of 3 M. Incubation was at RT for 90 min and the microplate was read using a ViewLux ultra HTS Microplate Imager (PerkinElmer, Inc.). ADP-Glo Format PI3K Assay The ADP-Glo format PI3K assays were performed in Proxiplate 384-well plates (Perkin Elmer #6008280). The final assay volume was 2 μl prepared from 1 μl additions of enzyme/PIP2:PS lipid (Invitrogen #PV5100) mixture and 1 μl ATP (provided in kit, Promega #V9101) and test compounds in assay buffer (50 mM HEPES pH 7.5, 3 mM MgCl2, 100 mM NaCl, 0.5 mM EGTA, 2 mM DTT, 0.03% CHAPS). The reaction was initiated by the combination of enzyme/lipid, ATP, and test compounds. The reaction mixture was incubated at room temperature for 30 minutes (PI3K Alpha, Beta, Gamma) or 3 hours for PI3K Delta. ADP-Glo (2 μl), followed by Kinase Detection reagent (4 μl), were added to reactions following the initial incubation and allowed to incubate 40 minutes at room temperature. The reaction mixture was analyzed on the TOPCOUNT (Perkin Elmer). Inhibition data were calculated by comparison to no enzyme control reactions for 100% inhibition and vehicle-only reactions for 0% inhibition. The final concentration of enzyme in the assays are PI3K Alpha [0.5 nM], PI3K Beta [2 nM], PI3K Gamma [20 nM], PI3K Delta [0.5 nM]. ATP final concentrations are as follows: for Alpha [10 μM], for Beta [12.5 μM], for Gamma [6.5 μM], for Delta [100 μM]. Lipid final concentration was the same for all enzymes, [25 μM]. Dose response curves were generated to determine the concentration required to inhibit 50% of activity. Compounds were dissolved at 0.12 mM in dimethylsulfoxide (DMSO) and evaluated at eleven concentrations. The IC50 values were derived by non-linear regression analysis. ADP-Glo Format PI3K Assays The ADP-Glo format PI3K assays were performed in Proxiplate 384-well plates (Perkin Elmer #6008280). The final assay volume was 2 μl prepared from 1 μl additions of enzyme/PIP2:PS lipid (Invitrogen #PV5100) mixture and 1 μl ATP (provided in kit, Promega #V9101) and test compounds in assay buffer (50 mM HEPES pH 7.5, 3 mM MgCl2, 100 mM NaCl, 0.5 mM EGTA, 2 mM DTT, 0.03% CHAPS). The reaction was initiated by the combination of enzyme/lipid, ATP, and test compounds. The reaction mixture was incubated at room temperature for 30 minutes (PI3K Alpha, Beta, Gamma) or 3 hours for PI3K Delta. ADP-Glo (2 μl), followed by Kinase Detection reagent (4 μl), were added to reactions following the initial incubation and allowed to incubate 40 minutes at room temperature. The reaction mixture was analyzed on the TOPCOUNT (Perkin Elmer). Inhibition data were calculated by comparison to no enzyme control reactions for 100% inhibition and vehicle-only reactions for 0% inhibition. The final concentration of enzyme in the assays are PI3K Alpha [0.5 nM], PI3K Beta [2 nM], PI3K Gamma [20 nM], PI3K Delta [0.5 nM]. ATP final concentrations are as follows: for Alpha [10 μM], for Beta [12.5 μM], for Gamma [6.5 μM], for Delta [100 μM]. Lipid final concentration was the same for all enzymes, [25 μM]. Dose response curves were generated to determine the concentration required to inhibit 50% of activity. Compounds were dissolved at 0.12 mM in dimethylsulfoxide (DMSO) and evaluated at eleven concentrations. In Vitro DGK Inhibition Assay The DGKα and DGKζ reactions were performed using extruded liposomes (DGKα and DGKζ LIPGLO assays). The reactions were carried out in 50 mM MOPS pH 7.5, 100 mM NaCl, 10 mM MgCl2, 1 μM CaCl2), and 1 mM DTT (assay buffer). The lipid substrate concentrations were 2 mM PS, 0.25 mM DAG, and 2.75 mM PC for the extruded liposome reactions. The reactions were carried out in 150 μM ATP. The enzyme concentrations for the DGKα and DGKζ were 5 nM.The compound inhibition studies were carried out as follows: 50 nL droplets of each test compound (top concentration 10 mM with 11 point, 3-fold dilution series for each compound) solubilized in DMSO were transferred to wells of a white 1536 well plate (Corning 3725). A 5 mL enzyme/substrate solution at 2× final reaction concentration was prepared by combining 2.5 mL 4× enzyme solution (20 nM DGKα or DGKζ (prepared as described below) in assay buffer) and 2.5 mL of 4× liposome solution (compositions described below) and incubated at room temperature for 10 minutes. Next, 1 μL 2× enzyme/substrate solution was added to wells containing the test compound and reactions were initiated with the addition of 1 μL 300 uM ATP. The reactions were allowed to proceed for 1 hr, after which 2 μL Glo Reagent (Promega V9101) was added and incubated for 40 minutes. Next, 4 μL Kinase Detection Reagent was added and incubated for 30 minutes. Luminescence was recorded using an EnVision microplate reader. The compounds were evaluated at 11 concentrations to determine IC50. Inhibition Assay Inhibition assay of FAP. The compounds were precisely weighed (range between 20-1000 pg) on a micro-scale (HuberLab, Sartorius QUINTIX35-1S) and dissolved in DMSO to prepare stock solutions at 1 mM. Stock solutions were further diluted with FAP activation buffer (50 mM Tris, 100 mM NaCl, 1 mM EDTA, pH=7.4) to a final DMSO concentration of 10% v/v. 10 μL of each inhibitor were serially diluted in a 384 well plate (grenier-bio one, PS, F-bottom, Black, non-binding) with 10 μL of 10% DMSO FAP buffer (1:1 dilution). FAP (human and murine) was diluted to the desired concentration with FAP activation buffer and added to the serial dilution of inhibitor (10 μL for each well). Each protein-inhibitor solution was incubated for 30 minutes at room temperature. Each measurement was performed with an enzyme concentration (FAP) lower than the expected IC50. A 60 μM solution of substrate (Z-Gly-Pro-AMC) in FAP activation buffer (<1% DMSO) was added to the plate (10 μL for each well). The reaction was incubated from 1 minute to 48 hours at room temperature (22° C.-25° C.). The emission was read at 465 nm (excitation wavelength 360 nm) for 40 μs by Tecan Spark (s.n. 1808004082). The following controls were added to each row: positive control (no inhibitor control, 10 μL of 10% DMSO FAP buffer+10 μL of FAP solution+10 μL of substrate solution), negative control (no protein control, 10 μL of 10% DMSO FAP buffer+10 μL of FAP activation buffer+10 μL of substrate solution). Scintillation Proximity Assay (SPA) for Detection of SMYD2 Enzymatic Inhibition SMYD2 inhibitory activities of the compounds described in the present invention were quantified using a scintillation proximity assay (SPA) which measures methylation by the enzyme of the synthetic, biotinylated peptide Btn-Ahx GSRAHSSHLKSKKGQSTSRH (SEQ ID NO: 3) Amid x TFA (Biosyntan) derived from p53 and referred to from here on as p53 Peptide. The SMYD2 full length enzyme was produced in-house by expression (with an N-terminal 6×His tag) in E. coli and purification by affinity chromatography on a Ni-NTA Sepharose column followed by a size exclusion chromatography step on a Superdex 200 16/60 column (GE Healthcare).In a typical assay 11 different concentrations of each compound (0.1 nM, 0.33 nM, 1.1 nM, 3.8 nM, 13 nM, 44 nM, 0.15 μM, 0.51 μM, 1.7 μM, 5.9 μM and 20 μM) were tested in duplicate within the same microtiter plate. To this end, 100-fold concentrated compound solutions (in DMSO) were previously prepared by serial dilution (1:3.4) of 2 mM stocks in a clear low volume 384-well source microtiter plate (Greiner Bio-One), from which 50 nl of compound solutions were transferred into a white low volume test microtiter plate from the same supplier. Subsequently, 2.5 μl SMYD2 in aqueous assay buffer [50 mM Tris/HCl pH 9.0 (AppliChem), 1 mM dithiothreitol (DTT, Sigma), 0.01% (w/v) bovine serum albumine (BSA, Sigma), 0.0022% (v/v) Pluronic (Sigma)] were added to the compounds in the test plate to a final enzyme concentration of typically 3 nM (this parameter was adjusted depending on the activity of the enzyme lot in order to be within the linear dynamic range of the assay). The samples were then incubated for 15 min at 22° C. to allow pre-equilibration of the putative enzyme-inhibitor complexes before the start of the methylation reaction, which was initiated by the addition of 2.5 μl 2-fold concentrated solution (in assay buffer) of titrated S-Adenosyl-L-Methionine (3H-SAM, Perkin Elmer, final concentration: 60 nM) and p53 Peptide substrate (final concentration: 1.0 μM). The resulting mixture (5 μl final volume) was shortly centrifuged (2 min., 1500 rpm) and incubated at 22° C. during 30 min. Thereupon the reaction was stopped by adding 3 μl of Streptavidin PS SPA imaging beads (Perkin Elmer, final concentration of 3.12 μg/μl) and cold SAM (AK Scientific, 25 μM final concentration) for non-specific binding reduction. Plates containing the stopped reaction were sealed with transparent adhesive foil (Perkin Elmer), centrifuged (2 min., 1500 rpm), and further incubated for at least 1 h at RT (or overnight at 4° C.) in order to allow the SPA signals to develop. Subsequently, the amount of product was evaluated by measuring the energy transfer from the β-particles emitted by the 3H-labeled substrate to the Europium scintillator co-polymerized in the polystyrene matrix of the PS imaging beads, using the standard settings for this purpose of a Viewlux (Perkin-Elmer) CCD plate imaging device (emission filter 613/55 (IFP). The resulting scintillation counts were taken as indicator for the amount of methylated peptide per well. The data were normalised using two sets of control wells (typically 16 each) for high- (=enzyme reaction with DMSO instead of test compound=0%=Minimum inhibition) and low- (=all assay components without enzyme=100%=Maximum inhibition) SMYD2 activity. IC50 values were calculated by fitting the normalized inhibition data to a 4-parameter logistic equation using the Screener analysis software from Genedata. DGKalpha and DGKzeta Biochemical Assays Compounds of the present invention were prepared into 10 mM DMSO solution and 10 nL of stock was transferred into 384 plates (Optiplate 384 plate) using Echo550. DMSO was used as high control, and ATP substrate buffer was used as a low control. A 1× enzyme assay buffer was prepared (Hepes, pH 7.0 25 mM, BSA 0.05%, Triton-X100 0.002%, CaCl2) 1 μM, MgCl2 10 mM, DTT 2 mM). The enzyme assay was performed by diluting enzyme DGKα (1 μg/μL DGKα, Carna12-101, SEQ ID NO: 3) or DGKζ (1μ/μL DGKζ, Carna 12-110, SEQ ID NO: 4) using 1× assay buffer. OAG (1-oleoyl-2-acetyl-sn-glycerol, 25 mg/ml, Avanti 8001000) and PS (10 mg/ml, Avanti 840032P) were mixed at the ratio of 1:2. A 1× substrate solution was prepared with 1× assay buffer by 100-fold dilution. The substrate solution was sonicated on ice for 1 min. The pure ATP was added to the substrate solution (DGKα:400 μM). 5 μL of the enzyme solution were added to the 384 well plate, and the plate was spun for 1 min at 1000 rpm and incubated for 30 mins at RT. 5 μL of 1× substrate solution were added to the 384 well plate, the plate was spun and then incubated for 45 mins at RT. 10 μL ADP-Glo detergent was added to stop the assay. After 60 mins at RT, 20 μL ADP-Glo Detection buffer was added as the final step. Plate was read after 45 min incubation at RT. IL-1beta Release Assay The activation of P2X7 by ATP leads to a fast transient activation of cells resulting in influx of Ca2+ followed by conversion of pro-IL-1β to active IL-1β. The functional activity of P2X7 compounds was measured by the release of mature IL-1β in the culture medium of THP-1 cells, detected by sandwich ELISA. Cells were maintained in complete growth medium (RPMI 1640+10% HI-FCS+2 mM L-glutamine+1×PS). Every 3 days, the medium was renewed by diluting the cells 1/3 to 1/4 as cell density did not exceed 0.5 million cells per ml (seeding cell density @ 1×105/ml). THP-1 cells were harvested from the flask in 50 ml by centrifugation for 3 min at 100 g. The cells were resuspended to 2×105 cells/ml in medium supplemented with 0.5 μM PMA and incubated. The cells were washed and resuspended to 1.5×105 cells/ml in medium complemented with 10 ng/ml LPS, and the cells were primed for 4 h at 37° C., 5% CO2. After addition of 20 μL of prediluted test compounds, blank, standard and control reagents, cells were incubated for a further 20 min at 37° C. and stimulated with 0.8 mM BzATP for 30 minutes. The cells were centrifuged, supernatant was collected and the presence of mature IL-1β was detected using Dual human IL-1b kit following manufacturer's instruction. The tetrahydrobenzodiazepine analogs effectively modulated the activity of P2X7 in the cells as measured by the levels of pro-inflammatory cytokine IL-1β, which is released by the activation of P2X7 receptor. IL-1beta release assay The activation of P2X7 by ATP leads to a fast transient activation of cells resulting in influx of Ca2+ followed by conversion of pro-IL-1β to active IL-1β. The functional activity of P2X7 compounds was measured by the release of mature IL-13 in the culture medium of THP-1 cells, detected by sandwich ELISA. Cells were maintained in complete growth medium (RPMI 1640+10% HI-FCS+2 mM L-glutamine+1×PS). Every 3 days, the medium was renewed by diluting the cells 1/3 to 1/4 as cell density did not exceed 0.5 million cells per ml (seeding cell density @1×105/ml). THP-1 cells were harvested from the flask in 50 ml by centrifugation for 3 min at 100 g. The cells were resuspended to 2×105 cells/ml in medium supplemented with 0.5 μM PMA and incubated. The cells were washed and resuspended to 1.5×105 cells/ml in medium complemented with 10 ng/ml LPS, and the cells were primed for 4 h at 37° C., 5% CO2. After addition of 20 μL of prediluted test compounds, blank, standard and control reagents, cells were incubated for a further 20 min at 37° C. and stimulated with 0.8 mM BzATP for 30 minutes. The cells were centrifuged, supernatant was collected and the presence of mature IL-1β was detected using Dual human IL-1b kit following manufacturer's instruction. The tetrahydrobenzodiazepine analogs effectively modulated the activity of P2X7 in the cells as measured by the levels of pro-inflammatory cytokine IL-1β, which is released by the activation of P2X7 receptor. In Vitro DGK Inhibition Assay The DGKα and DGKζ reactions were performed using either extruded liposome (DGKα and DGKζ LIPGLO assays). The reactions were carried out in 50 mM MOPS pH 7.5, 100 mM NaCl, 10 mM MgCl2, 1 μM CaCl2), and 1 mM DTT (assay buffer). The lipid substrate concentrations were 2 mM PS, 0.25 mM DAG, and 2.75 mM PC for the extruded liposome reactions (5 mM total lipid). The reactions were carried out in 150 μM ATP. The enzyme concentrations for the DGKα and DGKζ were 5 nM.The compound inhibition studies were carried out as follows: 25 nL droplets of each test compound (top concentration 10 mM with 11 point, 3-fold dilution series for each compound) solubilized in DMSO were transferred to wells of a white 1536 well plate (Corning 3725). A 5 mL enzyme/lipid substrate solution at 2× final reaction concentration was prepared by combining 2.5 mL 4× enzyme solution (20 nM DGKα or DGKζ (prepared as described below) in assay buffer) and 2.5 mL of 4× detergent/lipid micelle solution (compositions described below) and incubated at room temperature for 10 minutes. Next, 1 μL 2× enzyme/lipid substrate solution was added to wells containing the test compound and reactions were initiated with the addition of 1 μL 300 uM ATP. The reactions were allowed to proceed for 2 hr, after which 2 μL Glo Reagent (Promega V9101) was added and incubated for 40 minutes. Next, 4 μL Kinase Detection Reagent was added and incubated for 30 minutes. Luminescence was recorded using an EnVision microplate reader. The percent inhibition was calculated from the ATP conversion generated by no enzyme control reactions for 100% inhibition and vehicle-only reactions for 0% inhibition. The compounds were evaluated at 11 concentrations to determine IC50. [35S]GTPγS Binding The human APJ receptor was cloned by polymerase chain reaction and the gene encoding the receptor was subcloned in pFLAG-CMV -3 expression vector (Sigma, Saint Louis, Mo. USA) in-house at Amgen. A GTPγS binding assay was performed on membranes prepared from CHO cells stably expressing human APJ receptor. The optimum experimental conditions for the concentrations of GDP, MgCl2, and NaCl in the assay buffer were initially determined. The assay was performed in 9 μL assay buffer [20 mM HEPES, pH 7.5, 5 mM MgCl2, 100 mM NaCl and 0.1% (w/v) BSA], 1 μL of diluted test compound (starting with 0.75 mM, 2-fold serial dilution with DMSO, total 22 points), 10 μL of 18 μM GDP (final concentration of 3 μM GDP), 20 μL of 0.25 μg/m L membrane protein expressing human APJ receptor captured with WGA PS beads (final concentration of 5 μg per well), and 20 μL of 0.3 nM [35S]GTPγS (final concentration is 0.1 nM [35S]GTPγS)(Perkin Elmer Life and Analytical Sciences, Waltham USA). One column of the plate was 1 μL of DMSO as background and another column of the plate was 1 μL of 180 μM Pyr-Apelin-13 which was used as control at a final concentration of 3 μM. Incubation was at RT for 90 min and the microplate was read using a ViewLux ultra HTS Microplate Imager (PerkinElmer, Inc.). All the results presented are means of several independent experiments and analyzed by non-linear regression methods using the commercially available program Prism (GraphPad, San Diego, Calif.) providing the EC50 values detailed in Table 3. [35S]GTPgammaS Binding The human APJ receptor was cloned by polymerase chain reaction and the gene encoding the receptor was subcloned in pFLAG-CMV -3 expression vector (Sigma, Saint Louis, Mo. USA) in-house at Amgen. A GTPγS binding assay was performed on membranes prepared from CHO cells stably expressing human APJ receptor. The optimum experimental conditions for the concentrations of GDP, MgCl2, and NaCl in the assay buffer were initially determined. The assay was performed in 9 μL assay buffer [20 mM HEPES, pH 7.5, 5 mM MgCl2, 100 mM NaCl and 0.1% (w/v) BSA], 1 μL of diluted test compound (starting with 0.75 mM, 2-fold serial dilution with DMSO, total 22 points), 10 μL of 18 μM GDP (final concentration of 3 μM GDP), 20 μL of 0.25 μg/mL membrane protein expressing human APJ receptor captured with WGA PS beads (final concentration of 5 μg per well), and 20 μL of 0.3 nM [35S]GTPγS (final concentration is 0.1 nM [35S]GTPγS)(Perkin Elmer Life and Analytical Sciences, Waltham USA). One column of the plate was 1 μL of DMSO as background and another column of the plate was 1 μL of 180 μM Pyr-Apelin-13 which was used as control at a final concentration of 3 μM. Incubation was at RT for 90 min and the microplate was read using a ViewLux ultra HTS Microplate Imager (PerkinElmer, Inc.). All the results presented are means of several independent experiments and analyzed by non-linear regression methods using the commercially available program Prism (GraphPad, San Diego, Calif.) providing the EC50 values detailed in Table 13. [35S]GTPgammaS Binding The human APJ receptor was cloned by polymerase chain reaction and the gene encoding the receptor was subcloned in pFLAG-CMV -3 expression vector (Sigma, Saint Louis, Mo. USA) in-house at Amgen. A GTPγS binding assay was performed on membranes prepared from CHO cells stably expressing human APJ receptor. The optimum experimental conditions for the concentrations of GDP, MgCl2, and NaCl in the assay buffer were initially determined. The assay was performed in 9 μL assay buffer [20 mM HEPES, pH 7.5, 5 mM MgCl2, 100 mM NaCl and 0.1% (w/v) BSA], 1 μL of diluted test compound (starting with 0.75 mM, 2-fold serial dilution with DMSO, total 22 points), 10 μL of 18 μM GDP (final concentration of 3 μM GDP), 20 μL of 0.25 μg/mL membrane protein expressing human APJ receptor captured with WGA PS beads (final concentration of 5 μg per well), and 20 μL of 0.3 nM [35S]GTPγS (final concentration is 0.1 nM [35S]GTPγS)(Perkin Elmer Life and Analytical Sciences, Waltham USA). One column of the plate was 1 μL of DMSO as background and another column of the plate was 1 μL of 180 μM Pyr-Apelin-13 which was used as control at a final concentration of 3 μM. Incubation was at RT for 90 min and the microplate was read using a ViewLux ultra HTS Microplate Imager (PerkinElmer, Inc.). All the results presented are means of several independent experiments and analyzed by non-linear regression methods using the commercially available program Prism (GraphPad, San Diego, Calif.) providing the EC50 values detailed in Table 40. [35S]GTPgammaS Binding The human APJ receptor was cloned by polymerase chain reaction and the gene encoding the receptor was subcloned in pFLAG-CMV -3 expression vector (Sigma, Saint Louis, Mo. USA) in-house at Amgen. A GTPγS binding assay was performed on membranes prepared from CHO cells stably expressing human APJ receptor. The optimum experimental conditions for the concentrations of GDP, MgCl2, and NaCl in the assay buffer were initially determined. The assay was performed in 9 μL assay buffer [20 mM HEPES, pH 7.5, 5 mM MgCl2, 100 mM NaCl and 0.1% (w/v) BSA], 1 μL of diluted test compound (starting with 0.75 mM, 2-fold serial dilution with DMSO, total 22 points), 10 μL of 18 μM GDP (final concentration of 3 μM GDP), 20 μL of 0.25 μg/mL membrane protein expressing human APJ receptor captured with WGA PS beads (final concentration of 5 μg per well), and 20 μL of 0.3 nM [35S]GTPγS (final concentration is 0.1 nM [35S]GTPγS)(Perkin Elmer Life and Analytical Sciences, Waltham USA). One column of the plate was 1 μL of DMSO as background and another column of the plate was 1 μL of 180 μM Pyr-Apelin-13 which was used as control at a final concentration of 3 μM. Incubation was at RT for 90 min and the microplate was read using a ViewLux ultra HTS Microplate Imager (PerkinElmer, Inc.). All the results presented are means of several independent experiments and analyzed by non-linear regression methods using the commercially available program Prism (GraphPad, San Diego, Calif.) providing the EC50 values. Biochemical Assay The ingredients required for this purpose are mixed in a black 384-well microtiter plate with transparent base (from Greiner, catalog number 781092). The requirements in this connection for each well of the 384-well microtiter plate are 5 nM GSK3β (from Upstate, catalog number xy), 40 μM GSK3β substrate GSM (sequence H-RRRPASVPPSPSLSRHS-(pS)-HQRR, from Upstate, catalog number 2-533), 30 μM nicotinamide adenine dinucleotide NADH (Roche Diagnostics, catalog number 10107735), 50 μM adenosine triphosphate ATP (from Sigma, catalog number A7966) and 2 mM phosphoenolpyruvate (from Roche, catalog number 128112). The required reaction buffer in which the biochemical reaction takes place consists of 50 mM Trizma hydrochloride Tris-HCl pH: 7.5 (from Sigma, catalog number T3253), 5 mM magnesium chloride MgCl2 (from Sigma, catalog number M8266), 0.2 mM DL-dithiothreitol DTT (from Sigma, catalog number D9779), 2 mM ethylenediaminetetraacetic acid EDTA (from Sigma, catalog number E6758), 0.01% Triton X-100 (from Sigma, catalog number T8787) and 0.05% bovine serum albumin BSA (from Sigma, catalog number B4287). Active substances are dissolved in dimethyl sulfoxide DMSO (from Sigma, catalog number D8418) in a concentration of 10 mM. Active substances are added in serial concentrations of 10 μM, 1 μM, 0.1 μM, 0.01 μM, 0.001 μM, 0.0001 μM, 0.00001 μM, 0.000001 μM to the mixtures of the biochemical reaction. As control, dimethyl sulfoxide is added instead of substance in a final concentration of 0.1%. The reaction is incubated at 30° C. for 2 hours and then the resulting fluorescence is measured in a Tecan Safire-XFLUOR4 instrument, version V4.50 (serial number 12901300283) with the specifications: measurement mode−fluorescence measured from below, extinction wavelength 340 nm, emission wavelength 465 nm, slit width extinction 5 nm, slit width emission 5 nm, gain mode 120, delay 0 μs, number of light flashes per measurement 3, and an integration time of 40 μs. Biochemical Assay The MAO-Glo Assay (commercial available from PROMEGA, #V1402) provides a sensitive method for the measurement of monoamine oxidase (MAO) activity (Valley, M. P. et al., 2006, Anal. Biochem. 359: 238-246) from a variety of tissues, biofluids or recombinant expressed or purified enzymes. As substrate a derivate of the beetle luciferin ((4S)-4,5-dihydro-2-(6-hydroxybenzothiazolyl)-4-thiazole-carboxylic acid) is used, which is oxidized at a primary amine moiety. After a spontaneous elimination and a catalyzed esterase reaction, the turnover of the luciferine by the luciferase is recorded as a signal of AOC3 activity.For the determination of AOC3 activity or compound inhibition potency, the compound inhibitors are dissolved in DMSO and adjusted to the respective assay concentration with reaction buffer (50 mM HEPES, 5 mM KCl, 2 mM CaCl2), 1.4 mM MgCl2, 120 mM NaCl, 0.001% (v/v) Tween 20, 100 μM TCEP, pH 7.4). An aliquot of 3 μL of the compound dilution is added to a 384 well plate (Optiplate, PS, flat bottom, white, PERKIN ELMER, #6007290) with a final DMSO concentration of 6.6%. Recombinant CHO cells, overexpressing the human (1500 cells/well), mouse (1000 cells/well) or rat (500 cells/well) AOC3 enzyme are diluted in reaction buffer and added in a volume of 15 μL to the wells. After incubation for 20 minutes at 37° C., 2 μL of MAO substrate (dissolved in DMSO at 16 mM, adjusted to assay concentration in reaction buffer to a final assay concentration of 20 μM) is added and further incubated for 60 minutes at 37° C. The turnover of the substrate is determined by the addition of 20 μL of the detection-mix which was generated by the addition of reconstitution buffer with esterase (PROMEGA, #V1402) to the luciferine detection reagent (PROMEGA, #V1402). After an incubation period of 20 minutes, the luminescent signal is measured with Envision 2104 Multilabel Reader (PERKIN ELMER).Alternative assays for the determination of the AOC3 enzymatic activity could be the extraction of 14C-labelled benzylamine reaction product or the Amplex Red Monoamine Oxidase reaction (Molecular Probes, Netherlands) as described in Gella et al. (Gella, A. et al., 2013, J. Neural Transm. 120: 1015-1018). CSNK1A1 1uM ATP Assay Recombinant fusion protein of N-terminal Glutathion-S-Transferase (GST) and full-length human CSNK1A1, expressed by baculovirus infected insect cells and purified via Glutathion affinity chromatography, was purchased from Life Technologies (product no. PV4174) and used as enzyme. As substrate for the kinase reaction the biotinylated peptide biotin-Ahx-KRRRAL-pS-VASLPGL (C-terminus in amide form) was used which can be purchased e.g. from the company Biosyntan (Berlin-Buch, Germany).[2164]For the assay 50 nl of a 100 fold concentrated solution of the test compound in DMSO was pipetted into a white low volume 384-well microtiter plate (Greiner Bio-One, Frickenhausen, Germany), 2 μL of a solution of CSNK1A1 in aqueous assay buffer [50 mM HEPES pH 7.5, 10% (v/v) glycerol, 10 mM MgCl2, 50 mM NaCl, 1 mM dithiothreitol, 0.01% (w/v) bovine serum albumin, 0.01% (v/v) Triton X-100] were added and the mixture was incubated for 15 min at 22° C. to allow pre-binding of the test compounds to the enzyme before the start of the kinase reaction. Then the kinase reaction was started by the addition of 3 μL of a solution of ATP (1.67 uM=>final conc. in the 5 μL assay volume is 1 mM) and peptide substrate (167 μM=>final conc. in the 5 μL assay volume is 100 μM) in assay buffer and the resulting mixture was incubated for a reaction time of 30 min at 22° C. The concentration of CSNK1A1 was adjusted depending of the activity of the enzyme lot and was chosen appropriate to have the assay in the linear range, a typical concentration is about 0.4 ng/μL. The reaction was stopped by the addition of 2.5 μL of ADP-Glo-reagent (1:1.5 fold diluted with water) and the resulting mixture was incubated at 22° C. for 1 h to convert the ATP not consumed in the kinase reaction completely to cAMP. Subsequently 2.5 μL of the kinase detection reagent (1.2 fold more concentrated than recommended by the producer) were added, the resulting mixture was incubated at 22° C. for 1 h and then the luminescence measured with a suitable measurement instrument (e.g. Viewlux™ from Perkin-Elmer). The amount of emitted light was taken as a measure for the amount of ADP generated and thereby for the activity of the CSNK1A1. CSNK1A1 High ATP Assay Recombinant fusion protein of N-terminal Glutathion-S-Transferase (GST) and full-length human CSNK1A1, expressed by baculovirus infected insect cells and purified via Glutathion affinity chromatography, was purchased from Life Technologies (product no. PV4174) and used as enzyme. As substrate for the kinase reaction the biotinylated peptide biotin-Ahx-KRRRAL-pS-VASLPGL (C-terminus in amide form) was used which can be purchased e.g. from the company Biosyntan (Berlin-Buch, Germany). For the assay 50 nl of a 100 fold concentrated solution of the test compound in DMSO was pipetted into a white low volume 384-well microtiter plate (Greiner Bio-One, Frickenhausen, Germany), 2 μL of a solution of CSNK1A1 in aqueous assay buffer [50 mM HEPES pH 7.5, 10% (v/v) glycerol, 10 mM MgCl2, 50 mM NaCl, 1 mM dithiothreitol, 0.01% (w/v) bovine serum albumin, 0.01% (v/v) Triton X-100] were added and the mixture was incubated for 15 min at 22 C. to allow pre-binding of the test compounds to the enzyme before the start of the kinase reaction. Then the kinase reaction was started by the addition of 3 μL of a solution of ATP (1.67 mM=>final conc. in the 5 μL assay volume is 1 mM) and peptide substrate (167 μM=>final conc. in the 5 μL assay volume is 100 μM) in assay buffer and the resulting mixture was incubated for a reaction time of 30 min at 22 C. The concentration of CSNK1A1 was adjusted depending of the activity of the enzyme lot and was chosen appropriate to have the assay in the linear range, a typical concentration is about 0.4 ng/μL. The reaction was stopped by the addition of 2.5 μL of ADP-Glo-reagent (1:1.5 fold diluted with water) and the resulting mixture was incubated at 22 C. for 1 h to convert the ATP not consumed in the kinase reaction completely to cAMP. Subsequently 2.5 μL of the kinase detection reagent (1.2 fold more concentrated than recommended by the producer) were added, the resulting mixture was incubated at 22 C. for 1 h and then the luminescence measured with a suitable measurement instrument (e.g. Viewlux from Perkin-Elmer). CSNK1A1 High ATP Assay Recombinant fusion protein of N-terminal Glutathion-S-Transferase (GST) and full-length human CSNK1A1, expressed by baculovirus infected insect cells and purified via Glutathion affinity chromatography, was purchased from Life Technologies (product no. PV4174) and used as enzyme. As substrate for the kinase reaction the biotinylated peptide biotin-Ahx-KRRRAL-pS-VASLPGL (C-terminus in amide form) was used which can be purchased e.g. from the company Biosyntan (Berlin-Buch, Germany).[2164]For the assay 50 nl of a 100 fold concentrated solution of the test compound in DMSO was pipetted into a white low volume 384-well microtiter plate (Greiner Bio-One, Frickenhausen, Germany), 2 μL of a solution of CSNK1A1 in aqueous assay buffer [50 mM HEPES pH 7.5, 10% (v/v) glycerol, 10 mM MgCl2, 50 mM NaCl, 1 mM dithiothreitol, 0.01% (w/v) bovine serum albumin, 0.01% (v/v) Triton X-100] were added and the mixture was incubated for 15 min at 22° C. to allow pre-binding of the test compounds to the enzyme before the start of the kinase reaction. Then the kinase reaction was started by the addition of 3 μL of a solution of ATP (1.67 mM=>final conc. in the 5 μL assay volume is 1 mM) and peptide substrate (167 μM=>final conc. in the 5 μL assay volume is 100 μM) in assay buffer and the resulting mixture was incubated for a reaction time of 30 min at 22° C. The concentration of CSNK1A1 was adjusted depending of the activity of the enzyme lot and was chosen appropriate to have the assay in the linear range, a typical concentration is about 0.4 ng/μL. The reaction was stopped by the addition of 2.5 μL of ADP-Glo-reagent (1:1.5 fold diluted with water) and the resulting mixture was incubated at 22° C. for 1 h to convert the ATP not consumed in the kinase reaction completely to cAMP. Subsequently 2.5 μL of the kinase detection reagent (1.2 fold more concentrated than recommended by the producer) were added, the resulting mixture was incubated at 22° C. for 1 h and then the luminescence measured with a suitable measurement instrument (e.g. Viewlux™ from Perkin-Elmer). The amount of emitted light was taken as a measure for the amount of ADP generated and thereby for the activity of the CSNK1A1. CSNK1A1 Inhibitory Activity Asaay 1 Recombinant fusion protein of N-terminal Glutathion-S-Transferase (GST) and full-length human CSNK1A1, expressed by baculovirus infected insect cells and purified via Glutathion affinity chromatography, was purchased from Life Technologies (product no. PV4174) and used as enzyme. As substrate for the kinase reaction the biotinylated peptide biotin-Ahx-KRRRAL-pS-VASLPGL (C-terminus in amide form) was used which can be purchased e.g. from the company Biosyntan (Berlin-Buch, Germany). For the assay 50 nl of a 100 fold concentrated solution of the test compound in DMSO was pipetted into a white low volume 384-well microtiter plate (Greiner Bio-One, Frickenhausen, Germany), 2 μL of a solution of CSNK1A1 in aqueous assay buffer [50 mM HEPES pH 7.5, 10% (v/v) glycerol, 10 mM MgCl2, 50 mM NaCl, 1 mM dithiothreitol, 0.01% (w/v) bovine serum albumin, 0.01% (v/v) Triton X-100] were added and the mixture was incubated for 15 min at 22 C. to allow pre-binding of the test compounds to the enzyme before the start of the kinase reaction. Then the kinase reaction was started by the addition of 3 μL of a solution of ATP (1.67 mM=>final conc. in the 5 μL assay volume is 1 mM) and peptide substrate (167 μM=>final conc. in the 5 μL assay volume is 100 μM) in assay buffer and the resulting mixture was incubated for a reaction time of 30 min at 22 C. The concentration of CSNK1A1 was adjusted depending of the activity of the enzyme lot and was chosen appropriate to have the assay in the linear range, a typical concentration is about 0.4 ng/μL. The reaction was stopped by the addition of 2.5 μL of ADP-Glo-reagent (1:1.5 fold diluted with water) and the resulting mixture was incubated at 22 C. for 1 h to convert the ATP not consumed in the kinase reaction completely to cAMP. Subsequently 2.5 μL of the kinase detection reagent (1.2 fold more concentrated than recommended by the producer) were added, the resulting mixture was incubated at 22 C. for 1 h and then the luminescence measured with a suitable measurement instrument (e.g. Viewlux from Perkin-Elmer). CSNK1A1 Inhibitory activity Assay 2 Recombinant fusion protein of N-terminal Glutathion-S-Transferase (GST) and full-length human CSNK1A1, expressed by baculovirus infected insect cells and purified via Glutathion affinity chromatography, was purchased from Life Technologies (product no. PV4174) and used as enzyme. As substrate for the kinase reaction the biotinylated peptide biotin-Ahx-KRRRAL-pS-VASLPGL (C-terminus in amide form) was used which can be purchased e.g. from the company Biosyntan (Berlin-Buch, Germany). For the assay 50 nl of a 100 fold concentrated solution of the test compound in DMSO was pipetted into a white low volume 384-well microtiter plate (Greiner Bio-One, Frickenhausen, Germany), 2 μL of a solution of CSNK1A1 in aqueous assay buffer [50 mM HEPES pH 7.5, 10% (v/v) glycerol, 10 mM MgCl2, 50 mM NaCl, 1 mM dithiothreitol, 0.01% (w/v) bovine serum albumin, 0.01% (v/v) Triton X-100] were added and the mixture was incubated for 15 min at 22 C. to allow pre-binding of the test compounds to the enzyme before the start of the kinase reaction. Then the kinase reaction was started by the addition of 3 μL of a solution of ATP (1.67 mM=>final conc. in the 5 μL assay volume is 1 mM) and peptide substrate (167 μM=>final conc. in the 5 μL assay volume is 100 μM) in assay buffer and the resulting mixture was incubated for a reaction time of 30 min at 22 C. The concentration of CSNK1A1 was adjusted depending of the activity of the enzyme lot and was chosen appropriate to have the assay in the linear range, a typical concentration is about 0.4 ng/μL. The reaction was stopped by the addition of 2.5 μL of ADP-Glo-reagent (1:1.5 fold diluted with water) and the resulting mixture was incubated at 22 C. for 1 h to convert the ATP not consumed in the kinase reaction completely to cAMP. Subsequently 2.5 μL of the kinase detection reagent (1.2 fold more concentrated than recommended by the producer) were added, the resulting mixture was incubated at 22 C. for 1 h and then the luminescence measured with a suitable measurement instrument (e.g. Viewlux from Perkin-Elmer). Biochemical Assays for PI3Kalpha, PI3Kbeta The luminescence-based ATP detection reagent KinaseGlo was obtained from Promega, (Cat. No. V6714, Lot No. 236161) through Catalys, Wallisellen, Switzerland. (L-alpha-phosphatidylinositol (PI), Liver, Bovine) were obtained from Avanti Polar Lipid (Cat. No. 840042C, Lot#LPI-274), Phosphatidylinositol-4,5-bisphosphate (PIP(4,5)2(Avanti, Cat. No. 840046X) or L-α-phosphatidylinositol (PI) was obtained from Avanti Polar Lipid (Cat. No. 840042C, Lot#LPI-274). L-α-Phosphatidylserine (PS) was from Avanti Polar Lipid (Cat. No. 840032C), n-Octylglucoside Avanti Polar Lipid (Cat. No. 10634425001). Luminescence is a well established readout to determine ATP concentrations and can thus be used to follow the activity of many kinases regardless of their substrate. The Kinase Glo Luminescent Kinase Assay (Promega, Madison/WI, USA) is a homogeneous HTS method of measuring kinase activity by quantifying the amount of ATP remaining in solution following a kinase reaction.50 nL of compound dilutions were dispensed onto black 384-well low volume Non Binding Styrene (NBS) plates (Costar Cat. No. NBS#3676) as described in section 8.2. L-α-phosphatidylinositol (PI), provided as 10 mg/ml solution in methanol, was transferred into a glass tube and dried under nitrogen beam. It was then resuspended in 3% OctylGlucoside by vortexing and stored at 4° C. 5 μL of a mix of PI/OG with the PI3Ka and Pi3Kb subtypes were added. Kinase reactions were started by addition of 5 μl of ATP-mix containing in a final volume 10 μL 10 mM TRIS-HCl pH 7.5, 3 mM MgCl2, 50 mM NaCl, 0.05% CHAPS, 1 mM DTT and 1 μM ATP, and occurred at room temperature. Reactions were stopped with 10 μl of KinaseGlo and plates were read 10 mins later in a Synergy2 reader using an integration time of 0.1 seconds per well. 2.5 μM of NVP-BGT226 (standard) was added to the assay plates to generate the 100% inhibition of the kinase reaction, and the 0% inhibition was given by the solvent vehicle (90% DMSO in water). NVP-BGT226 was used as a reference compound and included in all assay plates in the form of 16 dilution points in duplicate.IC50 values of the percentage inhibition of each compound at 8 concentrations (usually 10, 3.0, 1.0, 0.3, 0.1, 0.030, 0.010 and 0.003 μM) n=2 were derived by fitting a sigmoidal dose-response curve to a plot of assay readout over inhibitor concentration as described. All fits were performed with the program XLfit4 (ID Business Solutions, Guildford, UK). CSNK1A1 Assay 2 CSNK1A1-inhibitory activity of compounds of the present invention in presence of 1 μM adenosine-tri-phosphate (ATP) was quantified employing the CSNK1A1 assay as described in the following paragraphs. In essence, the enzyme activity is measured by quantification of the adenosine-di-phosphate (ADP), which is generated as a co-product of the enzyme reaction, via the ADP-Glo Kinase Assay kit from the company Promega. This detection system works as follows: In a first step the ATP not consumed in the kinase reaction is quantitatively converted to cAMP employing an adenylate cyclase ( ADP-Glo-reagent ), then the adenylate cyclase is stopped and the ADP generated in the kinase reaction converted to ATP which generates in a luciferase-based reaction a glow-luminescence signal ( Kinase Detection Reagent ). Recombinant fusion protein of N-terminal Glutathion-S-Transferase (GST) and full-length human CSNK1A1, expressed by baculovirus infected insect cells and purified via Glutathion affinity chromatography, was purchased from Life Technologies (product no. PV4174) and used as enzyme. As substrate for the kinase reaction the biotinylated peptide biotin-Ahx-KRRRAL-pS-VASLPGL (C-terminus in amide form) was used which can be purchased e.g. from the company Biosyntan (Berlin-Buch, Germany).For the assay 50 nl of a 100 fold concentrated solution of the test compound in DMSO was pipetted into a white 1536-well microtiter plate (Greiner Bio-One, Frickenhausen, Germany), 2 μL of a solution of CSNK1A1 in aqueous assay buffer [50 mM HEPES pH 7.5, 10% (v/v) glycerol, 10 mM MgCl2, 50 mM NaCl, 1 mM dithiothreitol, 0.01% (w/v) bovine serum albumin, 0.01% (v/v) Triton X-100] were added and the mixture was incubated for 15 min at 22 C. to allow pre-binding of the test compounds to the enzyme before the start of the kinase reaction. Then the kinase reaction was started by the addition of 3 μL of a solution of ATP (1.67 μM=>final conc. in the 5 μL assay volume is 1 μM) and peptide substrate (50 μM=>final conc. in the 5 μL assay volume is 30 μM) in assay buffer and the resulting mixture was incubated for a reaction time of 30 min at 22 C. The concentration of CSNK1A1 was adjusted depending of the activity of the enzyme lot and was chosen appropriate to have the assay in the linear range, a typical concentrations are about 0.0375 ng/μL. Scintillation Proximity Assay (SPA) Assay B (Human Truncated DNMT1(601-1600)) This assay used Scintillation Proximity technology in a signal increase format to evaluate the potency of compounds. Human truncated DNMT1(601-1600), single hemi-methylated CpG site oligonucleotide, and Tritiated SAM were utilized to monitor activity. Assay plate creation consisted of the following parameters: 10 mM Compounds (11-point, 3-fold serial dilution) were stamped at 100 nL per well (100× in 100% DMSO) into a Griener white LV 384 well plate (#784075). Assay buffer mix was made on the day of assay consisting of: base buffer: (500 mM Hepes, ph 8, 1M MgCl2 made in advance, stored at room temp as a stock), 10% NP40-Surfact AMPS, 10% Ultrapure BSA 50 mg/ml, and 2M DTT (DL-Dithiolthreitol). The 2× enzyme mix was then prepared consisting of: DNMT1 protein (truncate human DNMT1—601-1600, made in house at 16.876 uM stock concentration) added to the assay buffer mix. The 2× substrate mix was made last, and consists of: 1 mM 40-mer hemi-methylated DNA Oligonucleotide, 12.5 uM 3H-SAM (Adenosyl-L-Methionine-S-[methyl-3H] Specific Activity 55-85 Ci/mmol) and 32 mM solution of S-Adenosyl-L-Methionine (this was diluted to 1 mM in Nuclease Free-H2O before adding to substrate mix) added into the assay buffer mix (3H-SAM is added last). Five uL of the assay buffer mix was dispensed into column 18 ONLY using a Thermo Multidrop combi. Next, 5 uL of the 2× Enzyme mix was dispensed to columns 1-17, 19-24 using a Thermo Multidrop combi. Then 5 uL of the 2× Substrate mix was dispensed to the full plate using a Thermo Multidrop combi. Plates were stacked and incubated for 40 minutes with a cover plate over the top plate. The quench mix was made around the 25 minute mark of the incubation step, which consisted of: 32 mM solution of S-Adenosyl-L-Methionine & PerkinElmer PEI PS Imaging Beads (Cat. #RPNQ0098)(10 mg/ml) into Nuclease Free-H2O. The quench mix was vortexed prior to use to get the beads in solution. After the 40 minute incubation, 10 uL of the quench mix was dispensed to the full plate using a Thermo Multidrop combi. Plates were sealed with a clear seal and centrifuged at 1000 rpm/1 min and dark adapted for 30 minutes. Plates were read on a Viewlux (PerkinElmer, 613 nm emission filter, 300 sec dual-exposure, (10 min. total read time)). Agonistic Activity of Polypeptide Compounds for GHSR-1a The activity of GHSR-1a can be measured using different techniques, for example, by detecting the change in the intracellular conformation of GHSR, the change in G-protein coupling activity, and/or the change in intracellular messengers. Techniques such as measuring intracellular Ca2+ are preferably used to measure the activity of GHSR-1a. Examples of techniques known in the art that can be used to measure Ca2+ include the use of FLIPR calcium ion assay kits, among others. The FLIPR calcium ion assay kits use a calcium ion sensitive indicator and a masking dye to ensure that a researcher carries out high-sensitivity fluorescent screening for G protein-coupled receptors, ion channels and other calcium ion sensitive targets. This experiment used FLIPR calcium 6 assay kits and FLIPR calcium 6-QF assay kits.1. Process1.1. Cell Culture and Reagent Preparationa) Cell line: Flp In-CHO-GHSR Stable Pool;b) Complete medium: F12K+10% fetal bovine serum+1×penicillin-streptomycin (PS)+600 μg/mL hygromycin B;c) Cell seeding medium: F12K+10% fetal bovine serum.d) Assay buffer: 1× HBSS+20 mM HEPES.e) 10× component A: Assay buffer and component A were left at room temperature (RT), 10 mL of buffer was added to component A, and the mixture was vortexed for 1-2 min and stored at −20° C.1.2. Compound Managementa) Compound stock solutions: the powders from in-house synthesis were made into 10 mM stock solutions in DMSO according to the standard protocol.b) Compound storage: all compounds in DMSO were stored in room temperature desiccators for short-term storage (at most 4 months). The remaining compounds were left at −20 ° C. for long-term storage.1.3. Agonist Activity Assaya) Flp In-CHO-GHSR Stable Pool cells were cultured in complete medium.b) The cells were placed in 25 lbs/inch cell seeding medium in a 384-well cell culture plate (Corning, 3764) at 7k cells/well and cultured overnight at 37° C. with 5% CO2.c) 20× component A was thawed at room temperature, diluted with assay buffer to 2×, and left at RT.d) The Petri dish was taken out of the incubator and equilibrated at room temperature for 10 min. The medium was changed to apricot buffer. After the final wash, 20 μL of buffer was kept in each well, 20 μL of 2× component A was then added to each well, and the plate was incubated at 37° C. for 3-5 s.e) 10 μL of 5× compound was added to the 384-well cell culture plate, and data collection was immediately performed using FLIPR Tetra. Kinase Glo Luminescent Kinase Assay (Kglo) for PI 3-Kinase Alpha (A), PI 3-Kinase Beta (B), Vps34 (C), PI 4-Kinase Beta (D) The luminescence-based ATP detection reagent KinaseGlo was obtained from Promega, (Cat. No. V6714, Lot No. 236161) through Catalys, Wallisellen, Switzerland. L-alpha-phosphatidylinositol (PI, liver, bovine) was obtained from Avanti Polar Lipid (Cat. No. 840042C, Lot#LPI-274), phosphatidylinositol-4,5-bisphosphate (PIP(4,5)2) was obtained from Avanti Polar Lipid (Cat. No. 840046X). L-α-phosphatidylserine (PS) was obtained from Avanti Polar Lipid (Cat. No. 840032C), n-octylglucoside from Avanti Polar Lipid (Cat. No. 10634425001). Luminescence is a well established readout to determine ATP concentrations and can thus be used to follow the activity of many kinases regardless of their substrate. The Kinase Glo Luminescent Kinase Assay (Promega, Madison, Wis., USA) is a homogeneous HTS method of measuring kinase activity by quantifying the amount of ATP remaining in solution following a kinase reaction.50 nL of compound dilutions were dispensed onto black 384-well low volume Non Binding Styrene (NBS) plates (Costar Cat. No. NBS#3676). L-α-phosphatidylinositol (PI), provided as 10 mg/ml solution in methanol, was transferred into a glass tube and dried under a nitrogen beam. It was then resuspended in 3% OctylGlucoside (1-O-n-octyl-beta-D-glucopyranoside) by vortexing and stored at 4° C. 5 μl of a mix of PI/OctylGlucoside with the PI 3-kinase alpha and PI 3-kinase beta subtypes, or Vps34 or PI 4-kinase beta were added. Kinase reactions were started by the addition of 5 μl of an ATP-mix containing in a final volume 10 μl 10 mM TRIS-HCl pH 7.5, 3 mM MgCl2, 50 mM NaCl, 0.05% CHAPS, 1 mM DTT and 1 μM ATP at room temperature. Reactions were stopped with 10 μl of KinaseGlo and plates were read 10 mins later in a Synergy2 reader using an integration time of 0.1 seconds per well. 2.5 μM of NVP-BGT226 (1-(3-(trifluoromethyl)-4-(piperazin-1-yl)phenyl)-8-(6-methoxypyridin-3-yl)-3-methyl-1H-imidazo[4,5-c]quinolin-2(3H)-one) was added to the assay plates to generate the 100% inhibition of the kinase reaction, and the 0% inhibition was given by the solvent vehicle (90% DMSO in water). (1-(3-(trifluoromethyl)-4-(piperazin-1-yl)phenyl)-8-(6-methoxypyridin-3-yl)-3-methyl-1H-imidazo[4,5-c]quinolin-2(3H)-one) was used as a reference compound and included in all assay plates in the form of 16 dilution points in duplicate.IC50 values of the percentage inhibition of each compound at 8 concentrations (10, 3.0, 1.0, 0.3, 0.1, 0.030, 0.010 and 0.003 μM) n=2 were derived by fitting a sigmoidal dose-response curve to a plot of assay readout over inhibitor concentration as described. All fits were performed with the program XLfit4 (ID Business Solutions, Guildford, UK). Scintillation Proximity Assay (SPA) Compounds contained herein were evaluated for their ability to inhibit the methyltransferase activity of EZH2 within the PRC2 complex. Human PRC2 complex was prepared by co-expressing each of the 5 member proteins (FLAG-EZH2, EED, SUZ12, RbAp48, AEBP2) in Sf9 cells followed by co-purification. Enzyme activity was measured in a scintillation proximity assay (SPA) where a tritiated methyl group is transferred from 3H-SAM to a lysine residue on Histone H3 of a mononucleosome, purified from HeLa cells. Mononucleosomes were captured on SPA beads and the resulting signal is read on a ViewLux plate reader. Compound Preparation 1. Prepare 10 mM stock of compounds from solid in 100% DMSO. 2. Set up an 11-point serial dilution (1:3 dilution, top concentration 10 mM) in 100% DMSO for each test compound in a 384 well plate leaving columns 6 and 18 for DMSO controls. 3. Dispense 100 nL of compound from the dilution plate into reaction plates (Grenier Bio-One, 384-well, Cat#784075). Part B. Reagent Preparation Prepare the following solutions: 1. 50 mM Tris-HCl, pH 8: Per 1 L of base buffer, combine 1 M Tris-HCl, pH 8 (50 mL) and distilled water (950 mL). 2. 1× Assay Buffer: Per 10 mL of 1× Assay Buffer, combine 50 mM Tris-HCl, pH 8 (9958 uL), 1 M MgCl2 (20 uL), 2 M DTT (20 uL), and 10% Tween-20 (2 uL) to provide a final concentration of 50 mM Tris-HCl, pH 8, 2 mM MgCl2, 4 mM DTT, 0.002% Tween-20. 3. 2× Enzyme Solution: Per 10 mL of 2× Enzyme Solution, combine 1× Assay Buffer and PRC2 complex to provide a final enzyme concentration of 10 nM. 4. SPA Bead Suspension: Per 1 mL of SPA Bead Suspension, combine PS-PEI coated LEADSeeker beads (40 mg) and H2O (1 mL) to provide a final concentration of 40 mg/mL. 5. 2× Substrate Solution: Per 10 mL of 2× Substrate Solution, combine 1× Assay Buffer (9728.55 uL), 800 ug/mL mononucleosomes (125 uL), 1 mM cold SAM (4 uL), and 7.02 uM 3H-SAM (142.45 uL; 0.55 mCi/mL) to provide a final concentration of 5 ug/mL nucleosomes, 0.2 uM cold SAM, and 0.05 uM 3H-SAM. 6. 2.67× Quench/Bead Mixture: Per 10 mL of 2.67× Quench/Bead Mixture, combine ddH2O (9358 uL), 10 mM cold SAM (267 uL), 40 mg/mL Bead Suspension (375 uL) to provide a final concentration of 100 uM cold SAM and 0.5 mg/mL SPA beads. Part C. Assay Reaction in 384-well Grenier Bio-One Plates Compound Addition 1. Dispense 100 nL/well of 100× Compound to test wells (as noted above). 2. Dispense 100 nL/well of 100% DMSO to columns 6 & 18 for high and low controls, respectively. Assay 1. Dispense 5 uL/well of 1× Assay Buffer to column 18 (low control reactions). 2. Dispense 5 uL/well of 2× Enzyme Solution to columns 1-17, 19-24. 3. Spin assay plates for 1 min at 500 rpm. 4. Stack the assay plates, covering the top plate. 5. Incubate the compound/DMSO with the enzyme for 30 min at room temperature. 6. Dispense 5 uL/well of 2× Substrate Solution to columns 1-24. 7. Spin assay plates for 1 min at 500 rpm. 8. Stack the assay plates, covering the top plate. 9. Incubate the assay plates at room temperature for 1 hour. Quench/Bead Addition 1. Dispense 5 uL/well of the 3× Quench/Bead Mixture to columns 1-24. 2. Seal the top of each assay plate with adhesive TopSeal. 3. Spin assay plates for 1 min at 500 rpm. 4. Equilibrate the plates for >20 min. Read plates 1. Read the assay plates on the Viewlux Plate Reader utilizing the 613 nm emission filter with a 300 s read time. Reagent addition can be done manually or with automated liquid handler. Scintillation Proximity Assay (SPA), Protocol 1 Compounds contained herein were evaluated for their ability to inhibit the methyltransferase activity of EZH2 within the PRC2 complex. Human PRC2 complex was prepared by co-expressing each of the 5 member proteins (FLAG-EZH2, EED, SUZ12, RbAp48, AEBP2) in Sf9 cells followed by co-purification. Enzyme activity was measured in a scintillation proximity assay (SPA) where a tritiated methyl group is transferred from 3H-SAM to a lysine residue on Histone H3 of a mononucleosome, purified from HeLa cells. Mononucleosomes were captured on SPA beads and the resulting signal is read on a ViewLux plate reader.Part A. Compound Preparation 1. Prepare 10 mM stock of compounds from solid in 100% DMSO. 2. Set up an 11-point serial dilution (1:3 dilution, top concentration 10 mM) in 100% DMSO for each test compound in a 384 well plate leaving columns 6 and 18 for DMSO controls. 3. Dispense 100 nL of compound from the dilution plate into reaction plates (Grenier Bio-One, 384-well, Cat#784075). Part B. Reagent Preparation Prepare the following solutions: 1. 50 mM Tris-HCl, pH 8: Per 1 L of base buffer, combine 1 M Tris-HCl, pH 8 (50 mL) and distilled water (950 mL). 2. 1× Assay Buffer: Per 10 mL of 1× Assay Buffer, combine 50 mM Tris-HCl, pH 8 (9958 uL), 1 M MgCl2 (20 uL), 2 M DTT (20 uL), and 10% Tween-20 (2 uL) to provide a final concentration of 50 mM Tris-HCl, pH 8, 2 mM MgCl2, 4 mM DTT, 0.002% Tween-20. 3. 2× Enzyme Solution: Per 10 mL of 2× Enzyme Solution, combine 1× Assay Buffer and PRC2 complex to provide a final enzyme concentration of 10 nM. 4. SPA Bead Suspension: Per 1 mL of SPA Bead Suspension, combine PS-PEI coated LEADSeeker beads (40 mg) and H2O (1 mL) to provide a final concentration of 40 mg/mL. 5. 2× Substrate Solution: Per 10 mL of 2× Substrate Solution, combine 1× Assay Buffer (9728.55 uL), 800 ug/mL mononucleosomes (125 uL), 1 mM cold SAM (4 uL), and 7.02 uM 3H-SAM (142.45 uL; 0.55 mCi/mL) to provide a final concentration of 5 ug/mL nucleosomes, 0.2 uM cold SAM, and 0.05 uM 3H-SAM. 6. 2.67× Quench/Bead Mixture: Per 10 mL of 2.67× Quench/Bead Mixture, combine ddH2O (9358 uL), 10 mM cold SAM (267 uL), 40 mg/mL Bead Suspension (375 uL) to provide a final concentration of 100 uM cold SAM and 0.5 mg/mL SPA beads. Part C. Assay Reaction in 384-well Grenier Bio-One Plates Compound Addition 1. Dispense 100 nL/well of 100× Compound to test wells (as noted above). 2. Dispense 100 nL/well of 100% DMSO to columns 6 & 18 for high and low controls, respectively. Assay 1. Dispense 5 uL/well of 1× Assay Buffer to column 18 (low control reactions). 2. Dispense 5 uL/well of 2× Enzyme Solution to columns 1-17, 19-24. 3. Spin assay plates for 1 min at 500 rpm. 4. Stack the assay plates, covering the top plate. 5. Incubate the compound/DMSO with the enzyme for 30 min at room temperature. 6. Dispense 5 uL/well of 2× Substrate Solution to columns 1-24. 7. Spin assay plates for 1 min at 500 rpm. 8. Stack the assay plates, covering the top plate. 9. Incubate the assay plates at room temperature for 1 hour. Quench/Bead Addition 1. Dispense 5 uL/well of the 3× Quench/Bead Mixture to columns 1-24. 2. Seal the top of each assay plate with adhesive TopSeal. 3. Spin assay plates for 1 min at 500 rpm. 4. Equilibrate the plates for >20 min. Read plates 1. Read the assay plates on the Viewlux Plate Reader utilizing the 613 nm emission filter with a 300 s read time. Reagent addition can be done manually or with automated liquid handler. *The final DMSO concentration in this assay is 1%. *The positive control is in column 6; negative control is in column 18. *Final starting concentration of compounds is 100 μM.
 BDBM88108 US9694016, 1145 US10709712, Example 1145 US10245267, Example 1145
BDBM88108 US9694016, 1145 US10709712, Example 1145 US10245267, Example 1145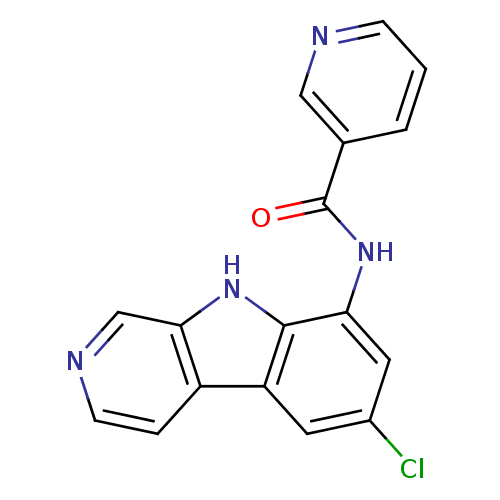 CHEMBL79004 BDBM50130248 PS-1145 N-(6-Chloro-9H-beta-carbolin-8-yl)-nicotinamide
CHEMBL79004 BDBM50130248 PS-1145 N-(6-Chloro-9H-beta-carbolin-8-yl)-nicotinamide US12195427, Compound 1145 US11584714, Compound 1145 BDBM595117 US12351648, Compound 1145
US12195427, Compound 1145 US11584714, Compound 1145 BDBM595117 US12351648, Compound 1145 US10774051, Compound I-1145 US11124486, Compound I-1145 BDBM460747
US10774051, Compound I-1145 US11124486, Compound I-1145 BDBM460747 BDBM149111 US8962648, 1145
BDBM149111 US8962648, 1145 US9302989, 1145 BDBM216240
US9302989, 1145 BDBM216240 BDBM624125 US11780845, Example 1145
BDBM624125 US11780845, Example 1145 BDBM643707 US20240002391, Compound 1145
BDBM643707 US20240002391, Compound 1145 US11286268, Compound 1145 BDBM545654
US11286268, Compound 1145 BDBM545654 BDBM218114 US12240839, Compound X US9212182, 1145
BDBM218114 US12240839, Compound X US9212182, 1145 Roche-Dataset for PDE10A, Compound 1145 BDBM564831
Roche-Dataset for PDE10A, Compound 1145 BDBM564831 N-(6-Chloro-9H-beta-carbolin-8-yl)-benzamide N-(6-chloro-9H-pyrido[3,4-b]indol-8-yl)benzamide BDBM50130230 CHEMBL78862 PS-1145
N-(6-Chloro-9H-beta-carbolin-8-yl)-benzamide N-(6-chloro-9H-pyrido[3,4-b]indol-8-yl)benzamide BDBM50130230 CHEMBL78862 PS-1145 PS-1 BDBM32576
PS-1 BDBM32576 BDBM32577 ent-PS-1
BDBM32577 ent-PS-1 BDBM50495956 CHEMBL223482 PS-6130
BDBM50495956 CHEMBL223482 PS-6130 Thymeleatoxin (low PS) BDBM86439
Thymeleatoxin (low PS) BDBM86439 (-)-Indolactam V (low PS) BDBM86437
(-)-Indolactam V (low PS) BDBM86437 B8-DL-B8 (low PS) BDBM86438
B8-DL-B8 (low PS) BDBM86438 PS-2383 ABBOTT-22370 BDBM50240100 Trimetozine Trimolide
PS-2383 ABBOTT-22370 BDBM50240100 Trimetozine Trimolide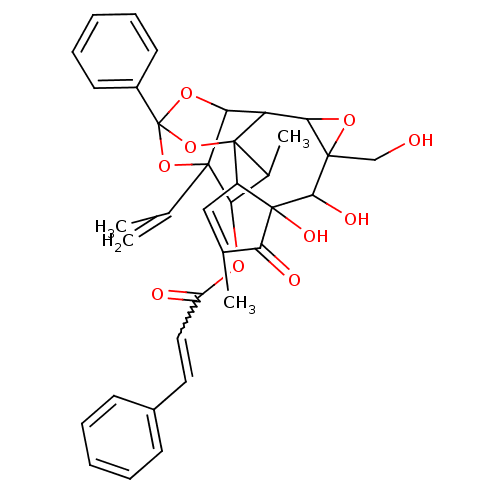 Thymeleatoxin (low PS) BDBM86429 NSC_6437389 CAS_6437389 Thymeleatoxin
Thymeleatoxin (low PS) BDBM86429 NSC_6437389 CAS_6437389 Thymeleatoxin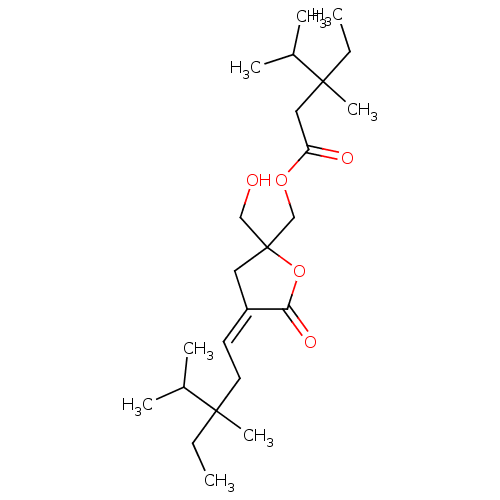 B8-DL-B8 B8-DL-B8 (low PS) BDBM86435
B8-DL-B8 B8-DL-B8 (low PS) BDBM86435 CAS_105000 NSC_105000 (-)-Indolactam V (low PS) BDBM86430 (-)-Indolactam V
CAS_105000 NSC_105000 (-)-Indolactam V (low PS) BDBM86430 (-)-Indolactam V CHEMBL309056 PS-154636-2 BDBM50071549 PS-154636-1 N-[(S)-(1-Benzyl-3-butylcarbamoyl-2-hydroxy-propylcarbamoyl)-phenyl-methyl]-2,4-dimethoxy-benzamide
CHEMBL309056 PS-154636-2 BDBM50071549 PS-154636-1 N-[(S)-(1-Benzyl-3-butylcarbamoyl-2-hydroxy-propylcarbamoyl)-phenyl-methyl]-2,4-dimethoxy-benzamide (S)-1-(4-(2-chloro-3-(2,3-dihydrobenzo[b][1,4]dioxin-6-yl)benzyloxy)-2-(3-cyanobenzyloxyl)benzyl)piperidine-2-carboxylic acid BDBM363222 US9850225, Example 1145
(S)-1-(4-(2-chloro-3-(2,3-dihydrobenzo[b][1,4]dioxin-6-yl)benzyloxy)-2-(3-cyanobenzyloxyl)benzyl)piperidine-2-carboxylic acid BDBM363222 US9850225, Example 1145 2-{4-[4-(5-fluoro-2-methoxyphenyl)-1H-pyrrolo[2,3-b]pyridin-2-yl]-6,6-dimethyl-3,6-dihydropyridin-1(2H)-yl}-N,N-dimethylacetamide BDBM353913 US9796708, Example 1145
2-{4-[4-(5-fluoro-2-methoxyphenyl)-1H-pyrrolo[2,3-b]pyridin-2-yl]-6,6-dimethyl-3,6-dihydropyridin-1(2H)-yl}-N,N-dimethylacetamide BDBM353913 US9796708, Example 1145 5-{4-amino-5-[(4,4- difluoropiperidin-1- yl)methyl]pyrrolo[2,1- f][1,2,4]triazin-7-yl}-2- chloro-N-[(3R,4S)-4- fluoro-1-(3,3,3- trifluoropropanoyl) pyrrolidin-3-yl]benzamide BDBM599909 US11618753, Example 1145
5-{4-amino-5-[(4,4- difluoropiperidin-1- yl)methyl]pyrrolo[2,1- f][1,2,4]triazin-7-yl}-2- chloro-N-[(3R,4S)-4- fluoro-1-(3,3,3- trifluoropropanoyl) pyrrolidin-3-yl]benzamide BDBM599909 US11618753, Example 1145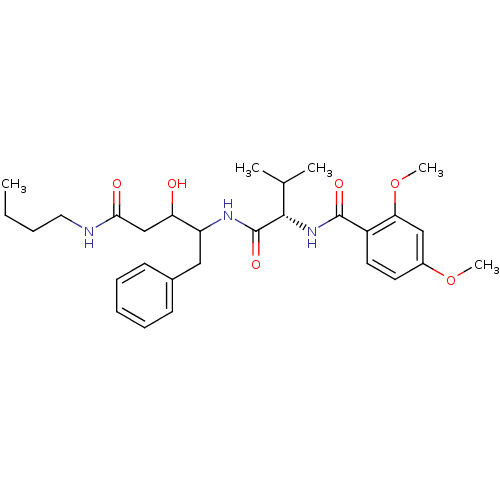 CHEMBL427946 N-[(S)-1-(1-Benzyl-3-butylcarbamoyl-2-hydroxy-propylcarbamoyl)-2-methyl-propyl]-2,4-dimethoxy-benzamide PS-725074 BDBM50071551
CHEMBL427946 N-[(S)-1-(1-Benzyl-3-butylcarbamoyl-2-hydroxy-propylcarbamoyl)-2-methyl-propyl]-2,4-dimethoxy-benzamide PS-725074 BDBM50071551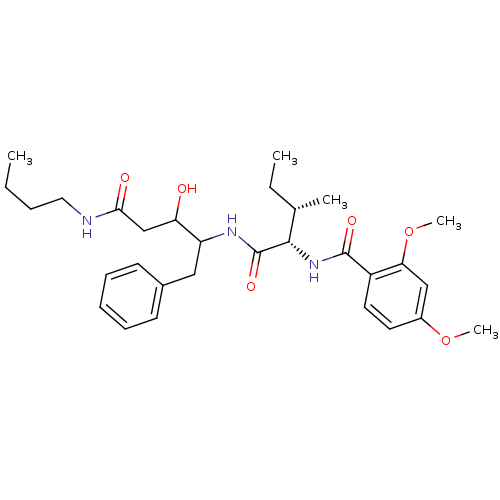 CHEMBL74562 N-[(1S,2S)-1-(1-Benzyl-3-butylcarbamoyl-2-hydroxy-propylcarbamoyl)-2-methyl-butyl]-2,4-dimethoxy-benzamide BDBM50071550 PS-662477
CHEMBL74562 N-[(1S,2S)-1-(1-Benzyl-3-butylcarbamoyl-2-hydroxy-propylcarbamoyl)-2-methyl-butyl]-2,4-dimethoxy-benzamide BDBM50071550 PS-662477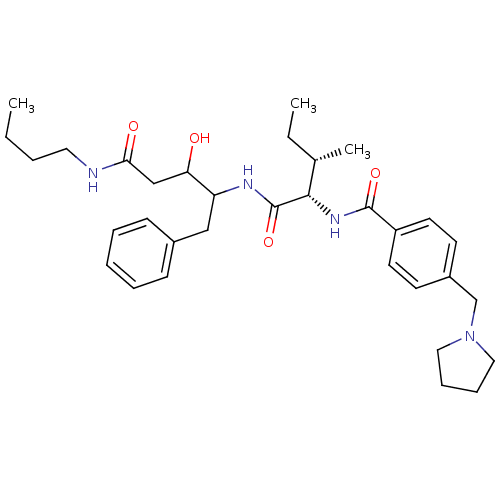 BDBM50071556 PS-444035 N-[(1S,2S)-1-(1-Benzyl-3-butylcarbamoyl-2-hydroxy-propylcarbamoyl)-2-methyl-butyl]-4-pyrrolidin-1-ylmethyl-benzamide CHEMBL74943
BDBM50071556 PS-444035 N-[(1S,2S)-1-(1-Benzyl-3-butylcarbamoyl-2-hydroxy-propylcarbamoyl)-2-methyl-butyl]-4-pyrrolidin-1-ylmethyl-benzamide CHEMBL74943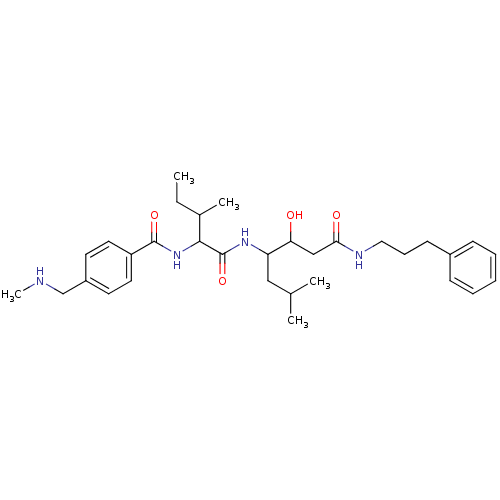 CHEMBL94646 N-(1-{1-[1-Hydroxy-2-(3-phenyl-propylcarbamoyl)-ethyl]-3-methyl-butylcarbamoyl}-2-methyl-butyl)-4-methylaminomethyl-benzamide BDBM50146770 PS-222036
CHEMBL94646 N-(1-{1-[1-Hydroxy-2-(3-phenyl-propylcarbamoyl)-ethyl]-3-methyl-butylcarbamoyl}-2-methyl-butyl)-4-methylaminomethyl-benzamide BDBM50146770 PS-222036 BDBM671979 US20240148821, Compound 1145 (5S,8S,11S,15S,18S,23aS,29S,35S,37aS)-8-((S)-sec-butyl)-29-(3-chloro-4-(trifluoromethyl)phenethyl)-18,35-dicyclopentyl-11-(cyclopentylmethyl)-5,12,16,19,33,36-hexamethyl-15-(morpholine-4-carbonyl)docosahydro-2H,4H-spiro[azeto[2,1-u]pyrrolo[2,1-i][1,4,7,10,13,16,19,22,25,28,31]undecaazacyclotetratriacontine-21,1'-cyclopentan]-4,7,10,13,17,20,23,28,31,34,37(14H,22H)-undecaone
BDBM671979 US20240148821, Compound 1145 (5S,8S,11S,15S,18S,23aS,29S,35S,37aS)-8-((S)-sec-butyl)-29-(3-chloro-4-(trifluoromethyl)phenethyl)-18,35-dicyclopentyl-11-(cyclopentylmethyl)-5,12,16,19,33,36-hexamethyl-15-(morpholine-4-carbonyl)docosahydro-2H,4H-spiro[azeto[2,1-u]pyrrolo[2,1-i][1,4,7,10,13,16,19,22,25,28,31]undecaazacyclotetratriacontine-21,1'-cyclopentan]-4,7,10,13,17,20,23,28,31,34,37(14H,22H)-undecaone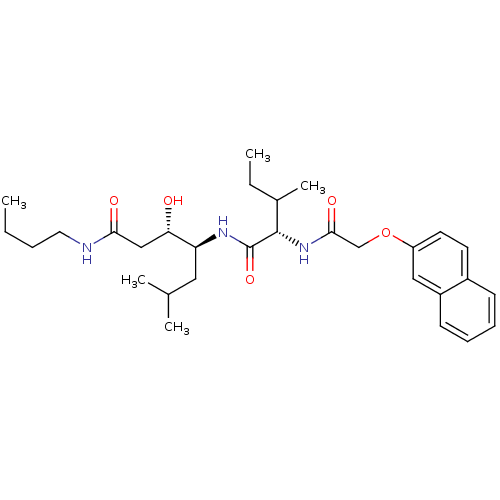 PS-777621 CHEMBL318055 BDBM50146527 (S)-3-(S)-Hydroxy-6-methyl-4-{(S)-3-methyl-2-[2-(naphthalen-2-yloxy)-acetylamino]-pentanoylamino}-heptanoic acid butylamide (3S,4S)-N-butyl-3-hydroxy-6-methyl-4-((2S,3S)-3-methyl-2-(2-(naphthalen-2-yloxy)acetamido)pentanamido)heptanamide
PS-777621 CHEMBL318055 BDBM50146527 (S)-3-(S)-Hydroxy-6-methyl-4-{(S)-3-methyl-2-[2-(naphthalen-2-yloxy)-acetylamino]-pentanoylamino}-heptanoic acid butylamide (3S,4S)-N-butyl-3-hydroxy-6-methyl-4-((2S,3S)-3-methyl-2-(2-(naphthalen-2-yloxy)acetamido)pentanamido)heptanamide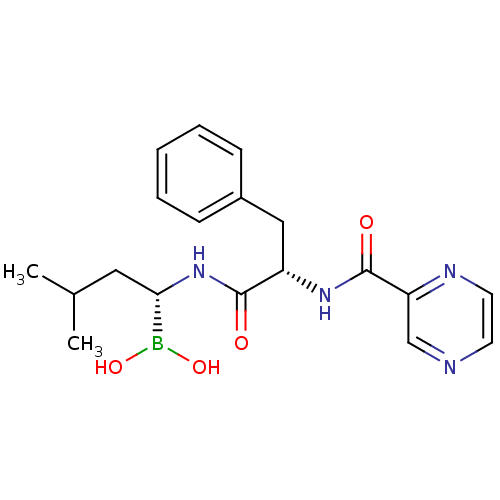 US11542283, Compound Velcade Dipeptidyl boronic acid derivative LDP-341 BORTEZOMIB Peptidyl boronic acid derivative PS-341 Velcade cid_387447 N-[(1R)-1-(DIHYDROXYBORYL)-3-METHYLBUTYL]-N-(PYRAZIN-2-YLCARBONYL)-L-PHENYLALANINAMIDE CHEMBL325041 (R)-3-methyl-1-((S)-3-phenyl-2-(pyrazine-6-carboxamido)propanamido)butylboronic acid BDBM50069989 (R)-3-methyl-1-((S)-3-phenyl-2-(pyrazine-2-carboxamido)propanamido)butylboronic acid
US11542283, Compound Velcade Dipeptidyl boronic acid derivative LDP-341 BORTEZOMIB Peptidyl boronic acid derivative PS-341 Velcade cid_387447 N-[(1R)-1-(DIHYDROXYBORYL)-3-METHYLBUTYL]-N-(PYRAZIN-2-YLCARBONYL)-L-PHENYLALANINAMIDE CHEMBL325041 (R)-3-methyl-1-((S)-3-phenyl-2-(pyrazine-6-carboxamido)propanamido)butylboronic acid BDBM50069989 (R)-3-methyl-1-((S)-3-phenyl-2-(pyrazine-2-carboxamido)propanamido)butylboronic acid