Target (9)
Compound (27)
Article Title (89)
Assay (199)
Hossain, MA; Bathgate, RAD Challenges in the design of insulin and relaxin/insulin-like peptide mimetics. Bioorg Med Chem 26: 2827 -2841 (2018) Collier, GR; Walder, KR; Campbell, JA; Molero-Navajas, J; Konstantopoulos, N; Krippner, GY Insulin sensitisers and methods of treatment US Patent US10172837 (2019) Cantin, LD; Magnuson, S; Gunn, D; Barucci, N; Breuhaus, M; Bullock, WH; Burke, J; Claus, TH; Daly, M; Decarr, L; Gore-Willse, A; Hoover-Litty, H; Kumarasinghe, ES; Li, Y; Liang, SX; Livingston, JN; Lowinger, T; Macdougall, M; Ogutu, HO; Olague, A; Ott-Morgan, R; Schoenleber, RW; Tersteegen, A; Wickens, P; Zhang, Z; Zhu, J; Zhu, L; Sweet, LJ PDE-10A inhibitors as insulin secretagogues. Bioorg Med Chem Lett 17: 2869 -73 (2007) Wickens, P; Zhang, C; Ma, X; Zhao, Q; Amatruda, J; Bullock, W; Burns, M; Cantin, LD; Chuang, CY; Claus, T; Dai, M; Dela Cruz, F; Dickson, D; Ehrgott, FJ; Fan, D; Heald, S; Hentemann, M; Iwuagwu, CI; Johnson, JS; Kumarasinghe, E; Ladner, D; Lavoie, R; Liang, S; Livingston, JN; Lowe, D; Magnuson, S; Mannelly, G; Mugge, I; Ogutu, H; Pleasic-Williams, S; Schoenleber, RW; Shapiro, J; Shelekhin, T; Sweet, L; Town, C; Tsutsumi, M Indanylacetic acids as PPAR-delta activator insulin sensitizers. Bioorg Med Chem Lett 17: 4369 -73 (2007) Yang, D; Qin, W; Shi, X; Zhu, B; Xie, M; Zhao, H; Teng, B; Wu, Y; Zhao, R; Yin, F; Ren, P; Liu, L; Li, Z Stabilized β-Hairpin Peptide Inhibits Insulin Degrading Enzyme. J Med Chem 61: 8174 -8185 (2018) Kraupner, N; Dinh, CP; Wen, X; Landry, V; Herledan, A; Leroux, F; Bosc, D; Charton, J; Maillard, C; Warenghem, S; Duplan, I; Piveteau, C; Hennuyer, N; Staels, B; Deprez, B; Deprez-Poulain, R Identification of indole-based activators of insulin degrading enzyme. Eur J Med Chem 228: (2022) Liu, DR; Maianti, JP; Saghatelian, A; Kleiner, RE Macrocyclic insulin-degrading enzyme (IDE) inhibitors and uses thereof US Patent US9243038 (2016) Barlow, N; Vanga, SR; Sävmarker, J; Sandström, A; Burns, P; Hallberg, A; Åqvist, J; Gutiérrez-de-Terán, H; Hallberg, M; Larhed, M; Chai, SY; Thompson, PE Macrocyclic peptidomimetics as inhibitors of insulin-regulated aminopeptidase (IRAP). RSC Med Chem 11: 234 -244 (2020) Bell, IM; Stirdivant, SM; Ahern, J; Culberson, JC; Darke, PL; Dinsmore, CJ; Drakas, RA; Gallicchio, SN; Graham, SL; Heimbrook, DC; Hall, DL; Hua, J; Kett, NR; Kim, AS; Kornienko, M; Kuo, LC; Munshi, SK; Quigley, AG; Reid, JC; Trotter, BW; Waxman, LH; Williams, TM; Zartman, CB Biochemical and structural characterization of a novel class of inhibitors of the type 1 insulin-like growth factor and insulin receptor kinases. Biochemistry 44: 9430 -40 (2005) Eliahu, S; Barr, HM; Camden, J; Weisman, GA; Fischer, B A novel insulin secretagogue based on a dinucleoside polyphosphate scaffold. J Med Chem 53: 2472 -81 (2010) Tang, YB; Lu, D; Chen, Z; Hu, C; Yang, Y; Tian, JY; Ye, F; Wu, L; Zhang, ZY; Xiao, Z Design, synthesis and insulin-sensitising effects of novel PTP1B inhibitors. Bioorg Med Chem Lett 23: 2313 -8 (2013) A highly selective dual insulin receptor (IR)/insulin-like growth factor 1 receptor (IGF-1R) inhibitor derived from an extracellular signal-regulated kinase (ERK) inhibitor. Zhang, J; Wang, J; Wu, H; He, Y; Zhu, G; Cui, X; Tang, L Design, synthesis and insulin-sensitizing activity of indomethacin and diclofenac derivatives. Bioorg Med Chem Lett 19: 3324 -7 (2009) Patil, NA; Hughes, RA; Rosengren, KJ; Kocan, M; Ang, SY; Tailhades, J; Separovic, F; Summers, RJ; Grosse, J; Wade, JD; Bathgate, RA; Hossain, MA Engineering of a Novel Simplified Human Insulin-Like Peptide 5 Agonist. J Med Chem 59: 2118 -25 (2016) Taygerly, JP; McGee, LR; Rubenstein, SM; Houze, JB; Cushing, TD; Li, Y; Motani, A; Chen, JL; Frankmoelle, W; Ye, G; Learned, MR; Jaen, J; Miao, S; Timmermans, PB; Thoolen, M; Kearney, P; Flygare, J; Beckmann, H; Weiszmann, J; Lindstrom, M; Walker, N; Liu, J; Biermann, D; Wang, Z; Hagiwara, A; Iida, T; Aramaki, H; Kitao, Y; Shinkai, H; Furukawa, N; Nishiu, J; Nakamura, M Discovery of INT131: a selective PPAR¿ modulator that enhances insulin sensitivity. Bioorg Med Chem 21: 979 -92 (2013) Min, J; Kyung Kim, Y; Cipriani, PG; Kang, M; Khersonsky, SM; Walsh, DP; Lee, JY; Niessen, S; Yates, JR; Gunsalus, K; Piano, F; Chang, YT Forward chemical genetic approach identifies new role for GAPDH in insulin signaling. Nat Chem Biol 3: 55 -9 (2007) Pasternak, A; Feng, Z; de Jesus, R; Ye, Z; He, S; Dobbelaar, P; Bradley, SA; Chicchi, GG; Tsao, KL; Trusca, D; Eiermann, GJ; Li, C; Feng, Y; Wu, M; Shao, Q; Zhang, BB; Nargund, R; Mills, SG; Howard, AD; Yang, L; Zhou, YP Stimulation of Glucose-Dependent Insulin Secretion by a Potent, Selective sst3 Antagonist. ACS Med Chem Lett 3: 289 -293 (2012) BMS-754807, a small molecule inhibitor of insulin-like growth factor-1R/IR. Lum, RT; Cheng, M; Cristobal, CP; Goldfine, ID; Evans, JL; Keck, JG; Macsata, RW; Manchem, VP; Matsumoto, Y; Park, SJ; Rao, SS; Robinson, L; Shi, S; Spevak, WR; Schow, SR Design, synthesis, and structure-activity relationships of novel insulin receptor tyrosine kinase activators. J Med Chem 51: 6173 -87 (2008) Abdul-Hay, SO; Lane, AL; Caulfield, TR; Claussin, C; Bertrand, J; Masson, A; Choudhry, S; Fauq, AH; Maharvi, GM; Leissring, MA Optimization of peptide hydroxamate inhibitors of insulin-degrading enzyme reveals marked substrate-selectivity. J Med Chem 56: 2246 -55 (2013) Andersson, H; Demaegdt, H; Johnsson, A; Vauquelin, G; Lindeberg, G; Hallberg, M; Erdelyi, M; Karlen, A; Hallberg, A Potent macrocyclic inhibitors of insulin-regulated aminopeptidase (IRAP) by olefin ring-closing metathesis. J Med Chem 54: 3779 -92 (2011) Mpakali, A; Saridakis, E; Giastas, P; Maben, Z; Stern, LJ; Larhed, M; Hallberg, M; Stratikos, E Structural Basis of Inhibition of Insulin-Regulated Aminopeptidase by a Macrocyclic Peptidic Inhibitor. ACS Med Chem Lett 11: 1429 -1434 (2020) Huang, C; Palani, A; Yang, Z; Deng, Q; Reddy, V; Nargund, RP; Lin, S; Altezza, S; Bianchi, E; Orvieto, F; Carrington, P Discovery of Insulin/GLP-1/Glucagon Triagonists for the Treatment of Diabetes and Obesity. ACS Med Chem Lett 13: 1255 -1261 (2022) Kim, H; Deng, L; Xiong, X; Hunter, WD; Long, MC; Pirrung, MC Glyceraldehyde 3-phosphate dehydrogenase is a cellular target of the insulin mimic demethylasterriquinone B1. J Med Chem 50: 3423 -6 (2007) Nguyen, PH; Zhao, BT; Ali, MY; Choi, JS; Rhyu, DY; Min, BS; Woo, MH Insulin-mimetic selaginellins from Selaginella tamariscina with protein tyrosine phosphatase 1B (PTP1B) inhibitory activity. J Nat Prod 78: 34 -42 (2015) Kennedy, BJ; Lato, AM; Fisch, AR; Burke, SJ; Kirkland, JK; Prevatte, CW; Dunlap, LE; Smith, RT; Vogiatzis, KD; Collier, JJ; Campagna, SR Potent Anti-Inflammatory, Arylpyrazole-Based Glucocorticoid Receptor Agonists That Do Not Impair Insulin Secretion. ACS Med Chem Lett 12: 1568 -1577 (2021) Giordanetto, F; Revell, JD; Knerr, L; Hostettler, M; Paunovic, A; Priest, C; Janefeldt, A; Gill, A Stapled Vasoactive Intestinal Peptide (VIP) Derivatives Improve VPAC2 Agonism and Glucose-Dependent Insulin Secretion. ACS Med Chem Lett 4: 1163 -8 (2013) Momose, Y; Maekawa, T; Yamano, T; Kawada, M; Odaka, H; Ikeda, H; Sohda, T Novel 5-substituted 2,4-thiazolidinedione and 2,4-oxazolidinedione derivatives as insulin sensitizers with antidiabetic activities. J Med Chem 45: 1518 -34 (2002) Mascarello, A; Frederico, MJ; Castro, AJ; Mendes, CP; Dutra, MF; Woehl, VM; Yunes, RA; Mena Barreto Silva, FR; Nunes, RJ Novel sulfonyl(thio)urea derivatives act efficiently both as insulin secretagogues and as insulinomimetic compounds. Eur J Med Chem 86: 491 -501 (2014) Tanis, SP; Colca, JR; Parker, TT; Artman, GD; Larsen, SD; McDonald, WG; Gadwood, RC; Kletzien, RF; Zeller, JB; Lee, PH; Adams, WJ PPARγ-sparing thiazolidinediones as insulin sensitizers. Design, synthesis and selection of compounds for clinical development. Bioorg Med Chem 26: 5870 -5884 (2018) Nguyen, PH; Yang, JL; Uddin, MN; Park, SL; Lim, SI; Jung, DW; Williams, DR; Oh, WK Protein tyrosine phosphatase 1B (PTP1B) inhibitors from Morinda citrifolia (Noni) and their insulin mimetic activity. J Nat Prod 76: 2080 -7 (2013) Zheng, N; Karra, P; VandenBerg, MA; Kim, JH; Webber, MJ; Holland, WL; Chou, DH Synthesis and Characterization of an A6-A11 Methylene Thioacetal Human Insulin Analogue with Enhanced Stability. J Med Chem 62: 11437 -11443 (2019) Targeting insulin-like growth factor 2 mRNA-binding proteins (IGF2BPs) for the treatment of cancer. Mulvihill, MJ; Ji, QS; Werner, D; Beck, P; Cesario, C; Cooke, A; Cox, M; Crew, A; Dong, H; Feng, L; Foreman, KW; Mak, G; Nigro, A; O'Connor, M; Saroglou, L; Stolz, KM; Sujka, I; Volk, B; Weng, Q; Wilkes, R 1,3-Disubstituted-imidazo[1,5-a]pyrazines as insulin-like growth-factor-I receptor (IGF-IR) inhibitors. Bioorg Med Chem Lett 17: 1091 -7 (2007) Fischer, B; Chulkin, A; Boyer, JL; Harden, KT; Gendron, FP; Beaudoin, AR; Chapal, J; Hillaire-Buys, D; Petit, P 2-thioether 5'-O-(1-thiotriphosphate)adenosine derivatives as new insulin secretagogues acting through P2Y-Receptors. J Med Chem 42: 3636 -46 (1999) Liu, K; Xu, L; Szalkowski, D; Li, Z; Ding, V; Kwei, G; Huskey, S; Moller, DE; Heck, JV; Zhang, BB; Jones, AB Discovery of a potent, highly selective, and orally efficacious small-molecule activator of the insulin receptor. J Med Chem 43: 3487 -94 (2000) Jin, M; Petronella, BA; Cooke, A; Kadalbajoo, M; Siu, KW; Kleinberg, A; May, EW; Gokhale, PC; Schulz, R; Kahler, J; Bittner, MA; Foreman, K; Pachter, JA; Wild, R; Epstein, D; Mulvihill, MJ Discovery of novel insulin-like growth factor-1 receptor inhibitors with unique time-dependent binding kinetics. ACS Med Chem Lett 4: 627 -31 (2013) Zhang, X; Bathgate, RAD; Hossain, MA Human Insulin-like Peptide 5 (INSL5). Identification of a Simplified Version of Two-Chain Analog A13. ACS Med Chem Lett 11: 2455 -2460 (2020) Leroux, F; Bosc, D; Beghyn, T; Hermant, P; Warenghem, S; Landry, V; Pottiez, V; Guillaume, V; Charton, J; Herledan, A; Urata, S; Liang, W; Sheng, L; Tang, WJ; Deprez, B; Deprez-Poulain, R Identification of ebselen as a potent inhibitor of insulin degrading enzyme by a drug repurposing screening. Eur J Med Chem 179: 557 -566 (2019) Mpakali, A; Saridakis, E; Harlos, K; Zhao, Y; Kokkala, P; Georgiadis, D; Giastas, P; Papakyriakou, A; Stratikos, E Ligand-Induced Conformational Change of Insulin-Regulated Aminopeptidase: Insights on Catalytic Mechanism and Active Site Plasticity. J Med Chem 60: 2963 -2972 (2017) Hubbard, RD; Bamaung, NY; Palazzo, F; Zhang, Q; Kovar, P; Osterling, DJ; Hu, X; Wilsbacher, JL; Johnson, EF; Bouska, J; Wang, J; Bell, RL; Davidsen, SK; Sheppard, GS Pyrazolo[3,4-d]pyrimidines as potent inhibitors of the insulin-like growth factor receptor (IGF-IR). Bioorg Med Chem Lett 17: 5406 -9 (2007) Feng, B; Zhang, J; Liu, Z; Xu, Y; Hu, H Discovery and biological evaluation of novel dual PTP1B and ACP1 inhibitors for the treatment of insulin resistance. Bioorg Med Chem 97: (2024) Buchanan, JL; Newcomb, JR; Carney, DP; Chaffee, SC; Chai, L; Cupples, R; Epstein, LF; Gallant, P; Gu, Y; Harmange, JC; Hodge, K; Houk, BE; Huang, X; Jona, J; Joseph, S; Jun, HT; Kumar, R; Li, C; Lu, J; Menges, T; Morrison, MJ; Novak, PM; van der Plas, S; Radinsky, R; Rose, PE; Sawant, S; Sun, JR; Surapaneni, S; Turci, SM; Xu, K; Yanez, E; Zhao, H; Zhu, X Discovery of 2,4-bis-arylamino-1,3-pyrimidines as insulin-like growth factor-1 receptor (IGF-1R) inhibitors. Bioorg Med Chem Lett 21: 2394 -9 (2011) Discovery of aurones bearing two amine functionalities as SHIP2 inhibitors with insulin-sensitizing effect in rat myotubes. Andersson, H; Demaegdt, H; Vauquelin, G; Lindeberg, G; Karlén, A; Hallberg, M; Erdélyi, M; Hallberg, A Disulfide cyclized tripeptide analogues of angiotensin IV as potent and selective inhibitors of insulin-regulated aminopeptidase (IRAP). J Med Chem 53: 8059 -71 (2010) Toda, N; Hao, X; Ogawa, Y; Oda, K; Yu, M; Fu, Z; Chen, Y; Kim, Y; Lizarzaburu, M; Lively, S; Lawlis, S; Murakoshi, M; Nara, F; Watanabe, N; Reagan, JD; Tian, H; Fu, A; Motani, A; Liu, Q; Lin, YJ; Zhuang, R; Xiong, Y; Fan, P; Medina, J; Li, L; Izumi, M; Okuyama, R; Shibuya, S Potent and Orally Bioavailable GPR142 Agonists as Novel Insulin Secretagogues for the Treatment of Type 2 Diabetes. ACS Med Chem Lett 4: 790 -4 (2013) Jin, M; Kleinberg, A; Cooke, A; Gokhale, PC; Foreman, K; Dong, H; Siu, KW; Bittner, MA; Mulvihill, KM; Yao, Y; Landfair, D; O'Connor, M; Mak, G; Pachter, JA; Wild, R; Rosenfeld-Franklin, M; Ji, Q; Mulvihill, MJ Potent and selective cyclohexyl-derived imidazopyrazine insulin-like growth factor 1 receptor inhibitors with in vivo efficacy. Bioorg Med Chem Lett 21: 1176 -80 (2011) Bleasdale, JE; Ogg, D; Palazuk, BJ; Jacob, CS; Swanson, ML; Wang, XY; Thompson, DP; Conradi, RA; Mathews, WR; Laborde, AL; Stuchly, CW; Heijbel, A; Bergdahl, K; Bannow, CA; Smith, CW; Svensson, C; Liljebris, C; Schostarez, HJ; May, PD; Stevens, FC; Larsen, SD Small molecule peptidomimetics containing a novel phosphotyrosine bioisostere inhibit protein tyrosine phosphatase 1B and augment insulin action. Biochemistry 40: 5642 -54 (2001) Antitumor activity of GSK1904529A, a small-molecule inhibitor of the insulin-like growth factor-I receptor tyrosine kinase. Emmitte, KA; Wilson, BJ; Baum, EW; Emerson, HK; Kuntz, KW; Nailor, KE; Salovich, JM; Smith, SC; Cheung, M; Gerding, RM; Stevens, KL; Uehling, DE; Mook, RA; Moorthy, GS; Dickerson, SH; Hassell, AM; Leesnitzer, MA; Shewchuk, LM; Groy, A; Rowand, JL; Anderson, K; Atkins, CL; Yang, J; Sabbatini, P; Kumar, R Discovery and optimization of imidazo[1,2-a]pyridine inhibitors of insulin-like growth factor-1 receptor (IGF-1R). Bioorg Med Chem Lett 19: 1004 -8 (2009) Fromont, C; Atzori, A; Kaur, D; Hashmi, L; Greco, G; Cabanillas, A; Nguyen, HV; Jones, DH; Garzón, M; Varela, A; Stevenson, B; Iacobini, GP; Lenoir, M; Rajesh, S; Box, C; Kumar, J; Grant, P; Novitskaya, V; Morgan, J; Sorrell, FJ; Redondo, C; Kramer, A; Harris, CJ; Leighton, B; Vickers, SP; Cheetham, SC; Kenyon, C; Grabowska, AM; Overduin, M; Berditchevski, F; Weston, CJ; Knapp, S; Fischer, PM; Butterworth, S Discovery of Highly Selective Inhibitors of Calmodulin-Dependent Kinases That Restore Insulin Sensitivity in the Diet-Induced Obesity J Med Chem 63: 6784 -6801 (2020) Admyre, T; Amrot-Fors, L; Andersson, M; Bauer, M; Bjursell, M; Drmota, T; Hallen, S; Hartleib-Geschwindner, J; Lindmark, B; Liu, J; Löfgren, L; Rohman, M; Selmi, N; Wallenius, K Inhibition of AMP deaminase activity does not improve glucose control in rodent models of insulin resistance or diabetes. Chem Biol 21: 1486 -96 (2014) Li, R; Pourpak, A; Morris, SW Inhibition of the insulin-like growth factor-1 receptor (IGF1R) tyrosine kinase as a novel cancer therapy approach. J Med Chem 52: 4981 -5004 (2010) Yang, JL; Ha, TKQ; Lee, BW; Kim, J; Oh, WK PTP1B inhibitors from the seeds of Iris sanguinea and their insulin mimetic activities via AMPK and ACC phosphorylation. Bioorg Med Chem Lett 27: 5076 -5081 (2017) Mountford, SJ; Albiston, AL; Charman, WN; Ng, L; Holien, JK; Parker, MW; Nicolazzo, JA; Thompson, PE; Chai, SY Synthesis, structure-activity relationships and brain uptake of a novel series of benzopyran inhibitors of insulin-regulated aminopeptidase. J Med Chem 57: 1368 -77 (2014) Discovery of OSI-906: a selective and orally efficacious dual inhibitor of the IGF-1 receptor and insulin receptor. Vourloumis, D; Mavridis, I; Athanasoulis, A; Temponeras, I; Koumantou, D; Giastas, P; Mpakali, A; Magrioti, V; Leib, J; van Endert, P; Stratikos, E; Papakyriakou, A Discovery of Selective Nanomolar Inhibitors for Insulin-Regulated Aminopeptidase Based on α-Hydroxy-β-amino Acid Derivatives of Bestatin. J Med Chem 65: 10098 -10117 (2022) Ducray, R; Simpson, I; Jung, FH; Nissink, JW; Kenny, PW; Fitzek, M; Walker, GE; Ward, LT; Hudson, K Discovery of novel imidazo[1,2-a]pyridines as inhibitors of the insulin-like growth factor-1 receptor tyrosine kinase. Bioorg Med Chem Lett 21: 4698 -701 (2011) PubChem, PC Dose Response: Fluorescence polarization-based cell-based high throughput dose response assay for inhibitors of insulin-degrading enzyme (IDE) PubChem Bioassay (2010) Plisson, F; Hill, TA; Mitchell, JM; Hoang, HN; de Araujo, AD; Xu, W; Cotterell, A; Edmonds, DJ; Stanton, RV; Derksen, DR; Loria, PM; Griffith, DA; Price, DA; Liras, S; Fairlie, DP Helixconstraints and amino acid substitution in GLP-1 increase cAMP and insulin secretion but not beta-arrestin 2 signaling. Eur J Med Chem 127: 703 -714 (2017) Charton, J; Gauriot, M; Guo, Q; Hennuyer, N; Marechal, X; Dumont, J; Hamdane, M; Pottiez, V; Landry, V; Sperandio, O; Flipo, M; Buee, L; Staels, B; Leroux, F; Tang, WJ; Deprez, B; Deprez-Poulain, R Imidazole-derived 2-[N-carbamoylmethyl-alkylamino]acetic acids, substrate-dependent modulators of insulin-degrading enzyme in amyloid-ß hydrolysis. Eur J Med Chem 79: 184 -93 (2014) Wittman, MD; Balasubramanian, B; Stoffan, K; Velaparthi, U; Liu, P; Krishnanathan, S; Carboni, J; Li, A; Greer, A; Attar, R; Gottardis, M; Chang, C; Jacobson, B; Sun, Y; Hansel, S; Zoeckler, M; Vyas, DM Novel 1H-(benzimidazol-2-yl)-1H-pyridin-2-one inhibitors of insulin-like growth factor I (IGF-1R) kinase. Bioorg Med Chem Lett 17: 974 -7 (2007) Mayer, SC; Banker, AL; Boschelli, F; Di, L; Johnson, M; Kenny, CH; Krishnamurthy, G; Kutterer, K; Moy, F; Petusky, S; Ravi, M; Tkach, D; Tsou, HR; Xu, W Lead identification to generate isoquinolinedione inhibitors of insulin-like growth factor receptor (IGF-1R) for potential use in cancer treatment. Bioorg Med Chem Lett 18: 3641 -5 (2008) Zhu, YF; Wang, XC; Connors, P; Wilcoxen, K; Gao, Y; Gross, R; Strack, N; Gross, T; McCarthy, JR; Xie, Q; Ling, N; Chen, C Quinoline-carboxylic acids are potent inhibitors that inhibit the binding of insulin-like growth factor (IGF) to IGF-binding proteins. Bioorg Med Chem Lett 13: 1931 -4 (2003) Human target validation of phosphoinositide 3-kinase (PI3K)β: effects on platelets and insulin sensitivity, using AZD6482 a novel PI3Kβ inhibitor. Miller, LM; Mayer, SC; Berger, DM; Boschelli, DH; Boschelli, F; Di, L; Du, X; Dutia, M; Floyd, MB; Johnson, M; Kenny, CH; Krishnamurthy, G; Moy, F; Petusky, S; Tkach, D; Torres, N; Wu, B; Xu, W Lead identification to generate 3-cyanoquinoline inhibitors of insulin-like growth factor receptor (IGF-1R) for potential use in cancer treatment. Bioorg Med Chem Lett 19: 62 -6 (2009) Mulvihill, MJ; Ji, QS; Coate, HR; Cooke, A; Dong, H; Feng, L; Foreman, K; Rosenfeld-Franklin, M; Honda, A; Mak, G; Mulvihill, KM; Nigro, AI; O'Connor, M; Pirrit, C; Steinig, AG; Siu, K; Stolz, KM; Sun, Y; Tavares, PA; Yao, Y; Gibson, NW Novel 2-phenylquinolin-7-yl-derived imidazo[1,5-a]pyrazines as potent insulin-like growth factor-I receptor (IGF-IR) inhibitors. Bioorg Med Chem 16: 1359 -75 (2008) Liu, X; Xie, H; Luo, C; Tong, L; Wang, Y; Peng, T; Ding, J; Jiang, H; Li, H Discovery and SAR of thiazolidine-2,4-dione analogues as insulin-like growth factor-1 receptor (IGF-1R) inhibitors via hierarchical virtual screening. J Med Chem 53: 2661 -5 (2010) Chen, C; Zhu, YF; Liu, XJ; Lu, ZX; Xie, Q; Ling, N Discovery of a series of nonpeptide small molecules that inhibit the binding of insulin-like growth factor (IGF) to IGF-binding proteins. J Med Chem 44: 4001 -10 (2001) Wilson, KJ; Xiao, J; Chen, CZ; Huang, Z; Agoulnik, IU; Ferrer, M; Southall, N; Hu, X; Zheng, W; Xu, X; Wang, A; Myhr, C; Barnaeva, E; George, ER; Agoulnik, AI; Marugan, JJ Optimization of the first small-molecule relaxin/insulin-like family peptide receptor (RXFP1) agonists: Activation results in an antifibrotic gene expression profile. Eur J Med Chem 156: 79 -92 (2018) Engen, W; O'Brien, TE; Kelly, B; Do, J; Rillera, L; Stapleton, LK; Youngren, JF; Anderson, MO Synthesis of aryl-heteroaryl ureas (AHUs) based on 4-aminoquinoline and their evaluation against the insulin-like growth factor receptor (IGF-1R). Bioorg Med Chem 18: 5995 -6005 (2010) Sblano, S; Cerchia, C; Laghezza, A; Piemontese, L; Brunetti, L; Leuci, R; Gilardi, F; Thomas, A; Genovese, M; Santi, A; Tortorella, P; Paoli, P; Lavecchia, A; Loiodice, F A chemoinformatics search for peroxisome proliferator-activated receptors ligands revealed a new pan-agonist able to reduce lipid accumulation and improve insulin sensitivity. Eur J Med Chem 235: (2022) Patnaik, S; Stevens, KL; Gerding, R; Deanda, F; Shotwell, JB; Tang, J; Hamajima, T; Nakamura, H; Leesnitzer, MA; Hassell, AM; Shewchuck, LM; Kumar, R; Lei, H; Chamberlain, SD Discovery of 3,5-disubstituted-1H-pyrrolo[2,3-b]pyridines as potent inhibitors of the insulin-like growth factor-1 receptor (IGF-1R) tyrosine kinase. Bioorg Med Chem Lett 19: 3136 -40 (2009) Wittman, MD; Carboni, JM; Yang, Z; Lee, FY; Antman, M; Attar, R; Balimane, P; Chang, C; Chen, C; Discenza, L; Frennesson, D; Gottardis, MM; Greer, A; Hurlburt, W; Johnson, W; Langley, DR; Li, A; Li, J; Liu, P; Mastalerz, H; Mathur, A; Menard, K; Patel, K; Sack, J; Sang, X; Saulnier, M; Smith, D; Stefanski, K; Trainor, G; Velaparthi, U; Zhang, G; Zimmermann, K; Vyas, DM Discovery of a 2,4-disubstituted pyrrolo[1,2-f][1,2,4]triazine inhibitor (BMS-754807) of insulin-like growth factor receptor (IGF-1R) kinase in clinical development. J Med Chem 52: 7360 -3 (2009) Cheng, X; Merz, KH; Vatter, S; Zeller, J; Muehlbeyer, S; Thommet, A; Christ, J; Wölfl, S; Eisenbrand, G Identification of a Water-Soluble Indirubin Derivative as Potent Inhibitor of Insulin-like Growth Factor 1 Receptor through Structural Modification of the Parent Natural Molecule. J Med Chem 60: 4949 -4962 (2017) Hubbard, RD; Bamaung, NY; Fidanze, SD; Erickson, SA; Palazzo, F; Wilsbacher, JL; Zhang, Q; Tucker, LA; Hu, X; Kovar, P; Osterling, DJ; Johnson, EF; Bouska, J; Wang, J; Davidsen, SK; Bell, RL; Sheppard, GS Development of multitargeted inhibitors of both the insulin-like growth factor receptor (IGF-IR) and members of the epidermal growth factor family of receptor tyrosine kinases. Bioorg Med Chem Lett 19: 1718 -21 (2009) Degorce, SL; Boyd, S; Curwen, JO; Ducray, R; Halsall, CT; Jones, CD; Lach, F; Lenz, EM; Pass, M; Pass, S; Trigwell, C Discovery of a Potent, Selective, Orally Bioavailable, and Efficacious Novel 2-(Pyrazol-4-ylamino)-pyrimidine Inhibitor of the Insulin-like Growth Factor-1 Receptor (IGF-1R). J Med Chem 59: 4859 -66 (2016) Fidanze, SD; Erickson, SA; Wang, GT; Mantei, R; Clark, RF; Sorensen, BK; Bamaung, NY; Kovar, P; Johnson, EF; Swinger, KK; Stewart, KD; Zhang, Q; Tucker, LA; Pappano, WN; Wilsbacher, JL; Wang, J; Sheppard, GS; Bell, RL; Davidsen, SK; Hubbard, RD Imidazo[2,1-b]thiazoles: multitargeted inhibitors of both the insulin-like growth factor receptor and members of the epidermal growth factor family of receptor tyrosine kinases. Bioorg Med Chem Lett 20: 2452 -5 (2010) Velaparthi, U; Wittman, M; Liu, P; Stoffan, K; Zimmermann, K; Sang, X; Carboni, J; Li, A; Attar, R; Gottardis, M; Greer, A; Chang, CY; Jacobsen, BL; Sack, JS; Sun, Y; Langley, DR; Balasubramanian, B; Vyas, D Discovery and initial SAR of 3-(1H-benzo[d]imidazol-2-yl)pyridin-2(1H)-ones as inhibitors of insulin-like growth factor 1-receptor (IGF-1R). Bioorg Med Chem Lett 17: 2317 -21 (2007) Wittman, M; Carboni, J; Attar, R; Balasubramanian, B; Balimane, P; Brassil, P; Beaulieu, F; Chang, C; Clarke, W; Dell, J; Eummer, J; Frennesson, D; Gottardis, M; Greer, A; Hansel, S; Hurlburt, W; Jacobson, B; Krishnananthan, S; Lee, FY; Li, A; Lin, TA; Liu, P; Ouellet, C; Sang, X; Saulnier, MG; Stoffan, K; Sun, Y; Velaparthi, U; Wong, H; Yang, Z; Zimmermann, K; Zoeckler, M; Vyas, D Discovery of a (1H-benzoimidazol-2-yl)-1H-pyridin-2-one (BMS-536924) inhibitor of insulin-like growth factor I receptor kinase with in vivo antitumor activity. J Med Chem 48: 5639 -43 (2005) Lipinski, CA; Reaume, AG High throughput in vivo phenotypic screening for drug repurposing: Discovery of MLR-1023 a novel insulin sensitizer and novel Lyn kinase activator with clinical proof of concept. Bioorg Med Chem 28: (2020) Wang, GT; Mantei, RA; Hubbard, RD; Wilsbacher, JL; Zhang, Q; Tucker, L; Hu, X; Kovar, P; Johnson, EF; Osterling, DJ; Bouska, J; Wang, J; Davidsen, SK; Bell, RL; Sheppard, GS Substituted 4-amino-1H-pyrazolo[3,4-d]pyrimidines as multi-targeted inhibitors of insulin-like growth factor-1 receptor (IGF1R) and members of ErbB-family receptor kinases. Bioorg Med Chem Lett 20: 6067 -71 (2010) Yamazaki, H; Kanno, SI; Abdjul, DB; Namikoshi, M A bromopyrrole-containing diterpene alkaloid from the Okinawan marine sponge Agelas nakamurai activates the insulin pathway in Huh-7 human hepatoma cells by inhibiting protein tyrosine phosphatase 1B. Bioorg Med Chem Lett 27: 2207 -2209 (2017) Zhu, YF; Wilcoxen, K; Gross, T; Connors, P; Strack, N; Gross, R; Huang, CQ; McCarthy, JR; Xie, Q; Ling, N; Chen, C 6,7-dihydroxyisoquinoline-3-carboxylic acids are potent inhibitors on the binding of insulin-like growth factor (IGF) to IGF-binding proteins: optimization of the 1-position benzoyl side chain. Bioorg Med Chem Lett 13: 1927 -30 (2003) Velaparthi, U; Liu, P; Balasubramanian, B; Carboni, J; Attar, R; Gottardis, M; Li, A; Greer, A; Zoeckler, M; Wittman, MD; Vyas, D Imidazole moiety replacements in the 3-(1H-benzo[d]imidazol-2-yl)pyridin-2(1H)-one inhibitors of insulin-like growth factor receptor-1 (IGF-1R) to improve cytochrome P450 profile. Bioorg Med Chem Lett 17: 3072 -6 (2007) Velaparthi, U; Saulnier, MG; Wittman, MD; Liu, P; Frennesson, DB; Zimmermann, K; Carboni, JM; Gottardis, M; Li, A; Greer, A; Clarke, W; Yang, Z; Menard, K; Lee, FY; Trainor, G; Vyas, D Insulin-like growth factor-1 receptor (IGF-1R) kinase inhibitors: SAR of a series of 3-[6-(4-substituted-piperazin-1-yl)-4-methyl-1H-benzimidazol-2-yl]-1H-pyridine-2-one. Bioorg Med Chem Lett 20: 3182 -5 (2010) Iqbal, Z; Morahan, G; Arooj, M; Sobolev, AN; Hameed, S Synthesis of new arylsulfonylspiroimidazolidine-2',4'-diones and study of their effect on stimulation of insulin release from MIN6 cell line, inhibition of human aldose reductase, sorbitol accumulations in various tissues and oxidative stress. Eur J Med Chem 168: 154 -175 (2019) Giordanetto, F; Wållberg, A; Ghosal, S; Iliefski, T; Cassel, J; Yuan, ZQ; von Wachenfeldt, H; Andersen, SM; Inghardt, T; Tunek, A; Nylander, S Discovery of phosphoinositide 3-kinases (PI3K) p110ß isoform inhibitor 4-[2-hydroxyethyl(1-naphthylmethyl)amino]-6-[(2S)-2-methylmorpholin-4-yl]-1H-pyrimidin-2-one, an effective antithrombotic agent without associated bleeding and insulin resistance. Bioorg Med Chem Lett 22: 6671 -6 (2012) Velaparthi, U; Wittman, M; Liu, P; Carboni, JM; Lee, FY; Attar, R; Balimane, P; Clarke, W; Sinz, MW; Hurlburt, W; Patel, K; Discenza, L; Kim, S; Gottardis, M; Greer, A; Li, A; Saulnier, M; Yang, Z; Zimmermann, K; Trainor, G; Vyas, D Discovery and evaluation of 4-(2-(4-chloro-1H-pyrazol-1-yl)ethylamino)-3-(6-(1-(3-fluoropropyl)piperidin-4-yl)-4-methyl-1H-benzo[d]imidazol-2-yl)pyridin-2(1H)-one (BMS-695735), an orally efficacious inhibitor of insulin-like growth factor-1 receptor kinase with broad spectrum in vivo antitumor acti J Med Chem 51: 5897 -900 (2008)
Insulin reductase assay Insulin reductase assay. ChEBML_90257 Inhibition of insulin receptor ChEBML_90271 Inhibition of Insulin receptor ChEMBL_759533 (CHEMBL1811059) Inhibition of insulin-induced autophosphorylation of human insulin receptor expressed in CHO cells ChEBML_90277 Inhibition of insulin receptor autophosphorylation ChEMBL_334490 (CHEMBL862525) Inhibition of insulin receptor ChEMBL_377345 (CHEMBL865408) Inhibition of Insulin receptor ChEMBL_506205 (CHEMBL941409) Inhibition of insulin receptor ChEMBL_581990 (CHEMBL1058970) Inhibition of insulin receptor ChEMBL_635332 (CHEMBL1119645) Inhibition of insulin receptor ChEMBL_698757 (CHEMBL1648482) Inhibition of insulin receptor ChEMBL_759532 (CHEMBL1811058) Inhibition of insulin receptor ChEMBL_794804 (CHEMBL1935987) Inhibition of insulin receptor ChEMBL_825876 (CHEMBL2044753) Inhibition of insulin receptor ChEMBL_90267 (CHEMBL699132) Inhibition of Insulin receptor ChEMBL_90276 (CHEMBL699141) Inhibition of Insulin receptor ChEBML_90279 Inhibition of Insulin receptor kinase-beta ChEMBL_2544064 Inhibition of insulin receptor (unknown origin) ChEMBL_563807 (CHEMBL994319) Inhibition of human insulin receptor ChEMBL_655044 (CHEMBL1244088) Inhibition of human insulin receptor ChEMBL_741336 (CHEMBL1764805) Inhibition of insulin receptor kinase ChEMBL_1363800 (CHEMBL3294217) Inhibition of insulin receptor (unknown origin) ChEMBL_215892 (CHEMBL820824) Inhibition of beta-insulin receptor kinase ChEMBL_2251074 (CHEMBL5165284) Inhibition of Insulin receptor (unknown origin) ChEBML_90259 Inhibition of insulin beta-R in HepG2 cells ChEBML_90397 Inhibition of Insulin-like growth factor I receptor ChEMBL_1460545 (CHEMBL3395577) Inhibition of recombinant insulin receptor (unknown origin) ChEMBL_2267608 Inhibition of Insulin receptor tyrosine kinase (unknown origin) ChEMBL_2297251 Binding affinity to insulin regulated aminopeptidase (unknown origin) ChEMBL_614233 (CHEMBL1106061) Inhibition of insulin receptor by HTRF assay ChEMBL_699678 (CHEMBL1648321) Inhibition of insulin receptor after 1 hr ChEMBL_885942 (CHEMBL2215704) Inhibition of insulin receptor by AlphaScreen analysis ChEMBL_90266 (CHEMBL699131) Inhibition of insulin receptor (InsR) tyrosine kinase ChEBML_90282 Inhibition of human insulin-like growth factor I receptor ChEMBL_305383 (CHEMBL832897) Inhibition of Insulin receptor kinase in P19 cells ChEMBL_566615 (CHEMBL960141) Inhibition of insulin receptor by virtual HTS assay ChEMBL_581979 (CHEMBL1058959) Inhibition of insulin receptor by cell based assay ChEMBL_88875 (CHEMBL698212) Activation of human insulin receptor tyrosine kinase (IRTK) ChEMBL_90285 (CHEMBL697504) Inhibition of Insulin-like growth factor I receptor ChEMBL_90396 (CHEMBL698351) Inhibition of Insulin-like growth factor I receptor ChEMBL_941897 (CHEMBL2329963) Inhibition of human recombinant IDE-mediated insulin degradation ChEBML_90404 Ability to displace Insulin-Like Growth Factor (IGF-I) from its binding to human insulin-like growth factor binding protein 3 (hIGFBP-3) ChEMBL_2527721 Inhibition of Insulin receptor expressed in HEK293 cells assessed as reduction insulin-stimulated INSR phosphorylation incubated for 1 hr by sandwich ELISA analysis ChEBML_90403 Ability of compound to displace Insulin-Like Growth Factor (IGF-I) from its binding to Insulin-like growth factor binding protein 3 (IGFBP-3) ChEBML_90543 Ability of compound to displace Insulin-Like Growth Factor (IGF-I) from its binding to Insulin-like growth factor binding protein 5 (IGFBP-5) ChEBML_88868 Inhibition of insulin receptor kinase (Inactive at 1 mM ATP) ChEMBL_1500622 (CHEMBL3587752) Inhibition of HSP27 (unknown origin) by insulin aggregation assay ChEMBL_2377430 Covalent inhibition of PDI (unknown origin) by insulin aggregation assay ChEMBL_2525422 Inhibition of insulin-R (unknown origin) in presence of ATP ChEMBL_305115 (CHEMBL831583) Inhibition of human insulin-like growth factor I receptor ChEMBL_306046 (CHEMBL874552) Inhibition of human Insulin receptor expressed in CHO cells ChEMBL_519660 (CHEMBL947811) Binding affinity to insulin receptor by liquid scintillation counting ChEMBL_90265 (CHEMBL699130) Inhibition of insulin receptor mediated mitogenesis of NIH3T3 cells ChEMBL_971459 (CHEMBL2406337) Inhibition of insulin receptor (unknown origin) after 60 mins ChEMBL_1869997 (CHEMBL4371164) Inhibition of human placental insulin receptor expressed in CHO cells assessed as decrease in insulin-stimulated [3H]-2-deoxyglucose uptake preincubated for 1 hr ChEMBL_950881 (CHEMBL2349705) Inhibition of Akt phosphorylation in human insulin-stimulated A549 cells incubated for 2 hrs prior to insulin-induction measured after 30 mins by ELISA ChEMBL_1767895 (CHEMBL4220007) Displacement of [TyrA14-125I]-human insulin from human insulin receptor isoform A expressed in baby hamster kidney cells after 2 days by gamma counting method ChEMBL_538724 (CHEMBL1035035) Activation of insulin receptor tyrosine kinase in mouse 3T3-L1 cells assessed as increase in 2-deoxy-D-[14C]glucose transport in presence of insulin ChEMBL_210560 (CHEMBL816512) Inhibition of thioredoxin reductase in the presence of thioredoxinand insulin ChEMBL_2377431 Non-covalent inhibition of PDI (unknown origin) by insulin aggregation assay ChEMBL_471122 (CHEMBL921275) Inhibition of human purified insulin receptor expressed in HepG2 cells ChEMBL_807032 (CHEMBL1959424) Inhibition of insulin receptor using fluorescent substrate by IMAP assay ChEMBL_90405 (CHEMBL698360) Inhibitory activity against Insulin-like growth factor binding protein 3 ChEMBL_1706543 (CHEMBL4057776) Inhibition of insulin stimulated INSR phosphorylation in human HeLa cells preincubated for 1 hr followed by insulin addition measured after 5 mins by Western blot analysis ChEMBL_1570092 (CHEMBL3789694) Inhibition of insulin receptor (unknown origin) in presence of [gamma33P]ATP ChEMBL_519651 (CHEMBL946778) Inhibition of human insulin receptor autophosphorylation in transfected mouse NIH3T3 cells ChEMBL_591804 (CHEMBL1041544) Inhibition of insulin receptor by [gamma-33-P]ATP based assay ChEMBL_654586 (CHEMBL1243630) Inhibition of human insulin receptor-mediated Poly Glu4Tyr phosphorylation by ELISA ChEMBL_873671 (CHEMBL2188787) Agonist activity at rat P2Y1R assessed as glucose-dependent insulin secretion Reductase Activity Assay Reductase activity was assayed by measuring the PDI-catalyzed reduction of insulin in the presence of DTT, thus measuring the aggregation of reduced insulin chains at 650nm. ChEMBL_2567995 Inhibition of INSR phosphorylation in insulin-stimulated human H4-II-E cells preincubated with compound for 30 mins followed by insulin stimulation and measured after 10 mins by immunoblot analysis ChEMBL_1513260 (CHEMBL3610968) Inhibition of insulin receptor (unknown origin) by homogeneous time resolved fluorescence assay ChEMBL_745322 (CHEMBL1775397) Activation of FFA1 in mouse islets assessed as glucose-dependent insulin secretion ChEMBL_796984 (CHEMBL1944059) Inhibition of insulin receptor after 20 mins by time-resolved fluorescence assay ChEMBL_162259 (CHEMBL772438) Inhibitory activity against Protein-tyrosine phosphatase 1B-mediated dephosphorylation of phosphorylated insulin receptor using (CHO) cell line transfected with an expression plasmid encoding the normal human insulin receptor [CHO/HIRc] ChEMBL_2542048 Inhibition of human IR autophosphorylation in insulin- stimulated mouse NIH-3T3-hIR cells pre incubated for 6 hr followed by insulin stimulation and measured after 15 mins by Western blot analysis ChEMBL_162252 (CHEMBL770119) Inhibition of human Protein-tyrosine phosphatase 1B (PTP1B) dephosphorylation of insulin receptor peptide ChEMBL_521715 (CHEMBL1001626) Agonist activity at rat pancreas P2Y1 receptor assessed as enhancement of insulin secretion ChEMBL_801839 (CHEMBL1947451) Displacement of thioflavin T from insulin receptor by thioflavin-T fluorescent dye assay ChEMBL_90284 (CHEMBL697349) In vitro inhibition of Insulin-like growth factor I receptor expressed in baculovirus ChEMBL_1279808 (CHEMBL3095509) Activation of glucokinase in rat INS-1 cells assessed as glucose-stimulated insulin secretion ChEMBL_1672669 (CHEMBL4022698) Inhibition of human insulin receptor tyrosine kinase after 1 hr by scintillation counting method ChEMBL_2266110 Inhibition of Escherichia coil TrxR incubated for 2 hrs in presence of DTT and insulin ChEMBL_519642 (CHEMBL946769) Inhibition of GST-tagged insulin receptor expressed in baculovirus by time-resolved fluorescence assay ChEMBL_884453 (CHEMBL2212215) Inhibition of human recombinant TRX-2 up to 60 mins by insulin reduction assay ChEMBL_884454 (CHEMBL2212216) Inhibition of human recombinant TRX-1 up to 60 mins by insulin reduction assay ChEMBL_1909270 (CHEMBL4411716) Activation of insulin-stimulated glycogen synthase in human HepG2 cells using [U-14C]UDP glucose as substrate preincubated for 18 hrs followed by insulin stimulation and measured after 15 mins by beta-counting method ChEMBL_2545794 Inhibition of PI3Kbeta in human adipocytes assessed as reduction in insulin-induced 2-deoxy-[U-14C]-glucose uptake preincubated for 3 hrs followed by insulin addition and measured after 30 mins by microbeta scintillation counting analysis ChEMBL_800636 (CHEMBL1947668) Activation of SUR1 in rat pancreatic islets assessed as inhibition of glucose-induced insulin secretion ChEMBL_90401 (CHEMBL698356) The compound was tested for binding affinity against insulin-like growth factor binding protein 1 ChEMBL_90402 (CHEMBL698357) The compound was tested for binding affinity against insulin-like growth factor binding protein 2 ChEMBL_90406 (CHEMBL698361) The compound was tested for binding affinity against Insulin-like growth factor binding protein 4 ChEMBL_90407 (CHEMBL698362) The compound was tested for binding affinity against Insulin-like growth factor binding protein 4 ChEMBL_90544 (CHEMBL700614) The compound was tested for binding affinity against Insulin-like growth factor binding protein 5 ChEMBL_90545 (CHEMBL700615) The compound was tested for binding affinity against Insulin-like growth factor binding protein 6 ChEMBL_959585 (CHEMBL2382530) Agonist activity at GPR40 in rat INS-1 cells assessed as glucose-stimulated insulin secretion ChEMBL_776271 (CHEMBL1914068) Inhibition of PIK3CA-mediated cell signaling in PTEN-deficient human U87MG cells assessed as inhibition of insulin-induced pAkt/PKB phosphorylation at Thr308 treated for 15 mins before insulin challenge measured after 5 mins by immunoblotting ChEMBL_776272 (CHEMBL1914069) Inhibition of PIK3CA-mediated cell signaling in PTEN-deficient human U87MG cells assessed as inhibition of insulin-induced pAkt/PKB phosphorylation at Ser473 treated for 15 mins before insulin challenge measured after 5 mins by immunoblotting ChEMBL_1719739 (CHEMBL4134739) Inhibition of TrxR1 in human HL60 cells incubated for 24 hrs by endpoint insulin reduction assay ChEMBL_858381 (CHEMBL2168596) Activation of glucokinase in rat INS-1 cells assessed as stimulation of glucose-induced insulin secretion ChEMBL_90408 (CHEMBL698363) Dissociation constant binding to Insulin-like growth factor binding protein 5 at the residue L-81 ChEMBL_90424 (CHEMBL698181) Dissociation constant in binding to Insulin-like growth factor binding protein 5 at the residue L73 ChEMBL_90425 (CHEMBL698182) Dissociation constant in binding to Insulin-like growth factor binding protein 5 at the residue L81 ChEMBL_90426 (CHEMBL697415) Dissociation constant in binding to Insulin-like growth factor binding protein 5 at the residue R87 ChEMBL_90427 (CHEMBL697416) Dissociation constant in binding to Insulin-like growth factor binding protein 5 at the residue S85 ChEMBL_90428 (CHEMBL697417) Dissociation constant in binding to Insulin-like growth factor binding protein 5 at the residue Y50 ChEMBL_90429 (CHEMBL697418) Dissociation constant in binding to Insulin-like growth factor binding protein 5 at the residue Y86 ChEMBL_1570094 (CHEMBL3789696) Inhibition of full length insulin receptor (unknown origin) transfected in Ba/F3 cells assessed as cell proliferation ChEMBL_1667905 (CHEMBL4017793) Inhibition of insulin receptor kinase (unknown origin) using TK as substrate after 30 mins by HTRF assay ChEMBL_1897038 (CHEMBL4399073) Inhibition of human recombinant IDE expressed in CHO cells in presence of [125I]-insulin by HTRF assay ChEMBL_581975 (CHEMBL1058955) Inhibition of insulin receptor-mediated proliferation of mouse NIH/3T3 cells after 48 hrs by MTT assay ChEMBL_90281 (CHEMBL697346) Concentration required to achieve 50% inhibition of tyrosine phosphorylation on human Insulin-like growth factor I receptor ChEMBL_90409 (CHEMBL698364) Dissociation constant in BS binding to Insulin-like growth factor binding protein 5 at the residue K91 ChEMBL_90410 (CHEMBL698365) Dissociation constant in DMSO binding to Insulin-like growth factor binding protein 5 at the residue K91 ChEMBL_90411 (CHEMBL698366) Dissociation constant in DMSO binding to Insulin-like growth factor binding protein 5 at the residue L73 ChEMBL_90412 (CHEMBL698367) Dissociation constant in DMSO binding to Insulin-like growth factor binding protein 5 at the residue L81 ChEMBL_90413 (CHEMBL698368) Dissociation constant in DMSO binding to Insulin-like growth factor binding protein 5 at the residue R87 ChEMBL_90414 (CHEMBL698369) Dissociation constant in DMSO binding to Insulin-like growth factor binding protein 5 at the residue S85 ChEMBL_90415 (CHEMBL698218) Dissociation constant in DMSO binding to Insulin-like growth factor binding protein 5 at the residue Y50 ChEMBL_90416 (CHEMBL698219) Dissociation constant in DMSO binding to Insulin-like growth factor binding protein 5 at the residue Y86 ChEMBL_90417 (CHEMBL698220) Dissociation constant in PBS binding to Insulin-like growth factor binding protein 5 at the residue K91 ChEMBL_90418 (CHEMBL698221) Dissociation constant in PBS binding to Insulin-like growth factor binding protein 5 at the residue L73 ChEMBL_90419 (CHEMBL698222) Dissociation constant in PBS binding to Insulin-like growth factor binding protein 5 at the residue L81 ChEMBL_90420 (CHEMBL698223) Dissociation constant in PBS binding to Insulin-like growth factor binding protein 5 at the residue R87 ChEMBL_90421 (CHEMBL698838) Dissociation constant in PBS binding to Insulin-like growth factor binding protein 5 at the residue S85 ChEMBL_90422 (CHEMBL698839) Dissociation constant in PBS binding to Insulin-like growth factor binding protein 5 at the residue Y50 ChEMBL_90423 (CHEMBL698180) Dissociation constant in PBS binding to Insulin-like growth factor binding protein 5 at the residue Y86 ChEMBL_631420 (CHEMBL1111957) Inhibition of insulin receptor assessed as [33P]gamma-ATP incorporation into substrate after 60 mins by gamma counting ChEMBL_1339905 (CHEMBL3243742) Induction of human recombinant IDE-mediated insulin hydrolysis preincubated for 10 mins measured after 30 mins by spectrophotometer analysis ChEMBL_2524990 Inhibition of GST-tagged human Insulin receptor expressed in baculovirus infected Sf9 cells using retinoblastoma protein in presence of ATP ChEMBL_675743 (CHEMBL1272377) Inhibition of Pin1 in serum starved human PC3 cells assessed as reduction in insulin-induced p7)S6 kinase phosphorylation ChEMBL_1869996 (CHEMBL4371163) Non-competitive inhibition of human placental insulin receptor expressed in CHO cell membrane assessed as decrease in insulin-stimulated A2 phosphorylation preincubated for 15 mins followed by substrate addition and measured after 15 mins in presence of [gamma-32P]ATP by Dixon plot analysis ChEMBL_1455264 (CHEMBL3362270) Agonist activity at mouse FFA4 receptor in mouse MIN6 cells assessed as induction of glucose-induced insulin secretion by AlphaLISA ChEMBL_1565351 (CHEMBL3782745) Inhibition of human recombinant PTP-1B using IR5 insulin receptor residues as substrate after 30 mins by malachite green assay ChEMBL_1623252 (CHEMBL3865604) Agonist activity at GPR40 in rat RINm cells assessed as increase glucose-stimulated insulin secretion after 1 hr by ELISA ChEMBL_1850470 (CHEMBL4351094) Inhibition of PI3Kdelta (unknown origin) preincubated for 30 mins followed by insulin stimulation for 5 mins by Western blot analysis ChEMBL_1922001 (CHEMBL4424846) Inhibition of human N-terminal His6-tagged insulin receptor (1005 to 1310 residues) expressed in baculovirus infected Sf21 insect cells ChEMBL_1775688 (CHEMBL4232680) Inhibition of human IDE using insulin as substrate preincubated for 10 mis followed by substrate addition and measured after 30 mins ChEMBL_1883833 (CHEMBL4385332) Agonist activity at GPR40 in human islets assessed as induction of glucose-stimulated insulin secretion after 1 hr by HTRF assay ChEMBL_2028620 (CHEMBL4682778) Inhibition of PDIA3 (unknown origin) expressed in Escherichia coli BL21 (DE3) incubated for 1 hr and measured by insulin reduction assay ChEMBL_2028621 (CHEMBL4682779) Inhibition of PDIA4 (unknown origin) expressed in Escherichia coli BL21 (DE3) incubated for 1 hr and measured by insulin reduction assay ChEMBL_2028623 (CHEMBL4682781) Inhibition of PDIA6 (unknown origin) expressed in Escherichia coli BL21 (DE3) incubated for 1 hr and measured by insulin reduction assay ChEBML_1645791 Agonist activity at GLP1R in rat INS-1 cells assessed as increase in glucose-stimulated insulin secretion after 1 hr by HTRF assay ChEMBL_1897041 (CHEMBL4399076) Inhibition of recombinant IDE exosite (unknown origin) expressed in Escherichia coli using insulin as substrate incubated for 4 hrs by AlphaLisa assay ChEMBL_1296283 (CHEMBL3132611) Inhibition of Insulin receptor (unknown origin) using biotinylated poly-Glu-Tyr (4:1) as substrate preincubated for 5 mins measured after 12 hrs ChEMBL_1645791 (CHEMBL3994847) Agonist activity at GLP1R in rat INS-1 cells assessed as increase in glucose-stimulated insulin secretion after 1 hr by HTRF assay ChEMBL_1861481 (CHEMBL4362337) Inhibition of TrxR1 (unknown origin) using DTNB as substrate preincubated for 30 mins in presence of insulin followed by substrate addition by colorimetry ChEMBL_2069558 (CHEMBL4724811) Agonist activity at GPR119 in glucose-induced golden hamster HIT-T15 cells assessed as increase in insulin secretion after 2 hrs by A1phaLISA ChEMBL_971485 (CHEMBL2406363) Inhibition of TEL-fused insulin receptor (unknown origin) expressed in mouse BAF3 cells after 2 to 3 days by luciferase reporter gene assay ChEMBL_1784836 (CHEMBL4256353) Inhibition of NADPH-reduced recombinant rat TrxR1 expressed in Escherichia coli harboring gor mutant assessed as suppression of insulin reduction using Trx as substrate pretreated for 30 mins followed by Trx addition for 30 mins and subsequent eosin-labelled bovine insulin addition measured every 5 mins for 1 hr by fluorescence assay ChEMBL_2104460 (CHEMBL4812963) Agonist activity at rat GPR119 in golden hamster HIT-T15 cells assessed as induction of glucose-induced insulin secretion after 2 hrs by A1phaLISA ChEMBL_1661563 (CHEMBL4011175) Induction of ERalpha degradation in human MCF7 cells assessed as inhibition of insulin-mediated cell proliferation after 6 days by Hoechst 33258 dye-based assay ChEMBL_1888984 (CHEMBL4390738) Inhibition of recombinant human N-terminal His-tagged PDI expressed in Escherichia coli BL21 (DE3) pLysS assessed as reduction in enzyme-mediated bovine insulin aggregation ChEMBL_863868 (CHEMBL2176258) Inhibition of INSR expressed in CHO cells assessed as inhibition of receptor phosphorylation pre-incubated before insulin stimulation by Meso-Scale Discovery pY-IR assay ChEMBL_218251 (CHEMBL819219) In vitro beta-adrenergic activity against beta-1 adrenergic receptor by the inhibition of insulin stimulated [14C]- glucose incorporation into glycogen in isolated rat soleus muscle ChEMBL_1933011 (CHEMBL4478663) Inhibition of AKT1 phosphorylation at Ser473 in EGF/insulin-stimulated human AN3CA cells incubated for 1 hr followed by stimulation for 15 mins by Western blot analysis ChEMBL_1933012 (CHEMBL4478664) Inhibition of AKT1 phosphorylation at Ser473 in EGF/insulin-stimulated human A2780 cells incubated for 1 hr followed by stimulation for 15 mins by Western blot analysis ChEMBL_1933013 (CHEMBL4478665) Inhibition of AKT1 phosphorylation at Thr308 in EGF/insulin-stimulated human AN3CA cells incubated for 1 hr followed by stimulation for 15 mins by Western blot analysis ChEMBL_1933014 (CHEMBL4478666) Inhibition of AKT1 phosphorylation at Thr308 in EGF/insulin-stimulated human A2780 cells incubated for 1 hr followed by stimulation for 15 mins by Western blot analysis ChEMBL_1897052 (CHEMBL4399087) Inhibition of wild type human IDE catalytic site using insulin as substrate preincubated for 10 mins followed by substrate addition and measured after 10 mins by Luminex kit method ChEMBL_1988385 (CHEMBL4621932) Inhibition of PI3Kalpha in human BT474 cells expressing P3KCA mutant assessed as reduction in AKT phosphorylation at residue S473 incubated for 2 hrs followed by insulin addition by ELISA Metabolic Measurements Glucose in tail blood was measured using a glucometer (One-Touch Basic; Lifescan, CA). For glucose tolerance tests (GTTs), mice were fasted for 10 hours and then injected with 20% D-glucose (2 mg/g body weight) and the blood glucose was monitored immediately before and at 15, 30, 60 and 120 mins following the injection. For insulin tolerance tests (ITTs), 4-h fasted animals were given insulin (0.75 mU/g) and blood glucose was measured immediately before and at 30, 60 and 120 minutes postinjection. Serum insulin, cholesterol, triglycerides (Stanbio Labs, TX), BDNF (Abnova), IGF1 and IGFBPs (R&D Systems) were determined by enzyme-linked immunosorbent assay. ChEMBL_1570093 (CHEMBL3789695) Inhibition of full length insulin receptor (unknown origin) autophosphorylation transfected in HEK293 cells pretreated for 60 mins followed by IGF-1 stimulation measured after 10 mins by quantitative Western blot analysis ChEMBL_1933015 (CHEMBL4478667) Inhibition of AKT1 in EGF/insulin-stimulated human AN3CA cells assessed as reduction in phosphorylation at Thr246 incubated for 1 hr followed by stimulation for 15 mins by Western blot analysis ChEMBL_1933016 (CHEMBL4478668) Inhibition of AKT1 in EGF/insulin-stimulated human A2780 cells assessed as reduction in phosphorylation at Thr246 incubated for 1 hr followed by stimulation for 15 mins by Western blot analysis ChEMBL_2074276 (CHEMBL4729810) Inhibition of recombinant human wild-type N-terminal His-tagged PDIA1 expressed in Escherichia coli strain BL21(DE3) using bovine insulin as substrate preincubated for 1 hr followed by substrate addition ChEMBL_2516987 Inhibition of Ins-R in A14 [Human melanoma] cells assessed as inhibition of autophosphorylation of Ins-R incubated for 90 mins followed by insulin stimulation measured after 10 mins by ELISA analysis ChEMBL_901085 (CHEMBL3062690) Agonist activity at Homo sapiens (human) PPARgamma expressed in mouse 3T3-L1 cells incubated for 2 days followed by compound wash out measured after 4 days by insulin receptor binding assay ChEMBL_1767894 (CHEMBL4220006) Agonist activity at B6D2F1 mouse insulin receptor assessed as increase in glucose incorporation into lipid phase after 2 hrs in presence of D-[3-3H]glucose by TopCount microplate scintillation counting method ChEMBL_1897053 (CHEMBL4399088) Inhibition of human recombinant IDE expressed in Escherichia coli BL21 (DE3) cells using insulin as substrate preincubated for 10 mins followed by substrate addition and measured after 15 mins by fluorescence based assay ChEMBL_2264503 Inhibition of GST-tagged recombinant human IR ( 919 to 1343 residues) autophosphorylation expressed in mouse NIH-3T3-A14 cells preincubated for 90 mins followed by insulin stimulation and measured after 10 mins by ELISA ChEMBL_2248960 (CHEMBL5163170) Positive allosteric modulator activity at GLP-1R in rat INS1 beta-cells assessed potentiation of GLP-1(7-36)NH2 induced insulin secretion incubated for 30 mins in presence of high glucose condition by ELISA ChEMBL_1717891 (CHEMBL4132891) Antagonist activity at GLUT4 in rat adipocytes assessed as reduction in insulin-stimulated [3H]2-deoxyglucose uptake preincubated for 1 min followed by [3H]2-deoxyglucose addition measured after 1 min by liquid scintillation counting analysis ChEMBL_2248961 (CHEMBL5163171) Positive allosteric modulator activity at GLP-1R in rat INS1 beta-cells assessed potentiation of GLP-1(7-36)NH2 induced insulin secretion incubated for 30 mins in presence of 0.2 nM of GLP-1 by ELISA ChEMBL_2248962 (CHEMBL5163172) Positive allosteric modulator activity at GLP-1R in rat INS1 beta-cells assessed potentiation of GLP-1(7-36)NH2 induced insulin secretion at 0.1 nM incubated for 30 mins in presence of exendin(9-39)NH2 by ELISA ChEMBL_1661323 (CHEMBL4010935) Agonist activity at TGR5 in STC1 cells derived from double transgenic mouse expressing rat insulin promoters linked to SV40 large T antigen and to polyomavirus small T antigen assessed as increase in cAMP level after 40 mins by mass spectrometric method ChEMBL_662256 (CHEMBL1252348) Inhibition of PKB/Akt activation in human overnight starved MCF7 cells assessed as phosphorylated Akt at Ser473 level at 50 ug/ml after 1 hr followed by treated with 1 ug/mL of insulin for 15 mins by Western blotting cAMP Assays Activation of GLP-1 receptor is known to stimulate cyclic AMP (cAMP) production in cells which indicates primary coupling to the G as subunit of the G protein heterotrimeric complex. Evidence suggests signaling through G as induced cAMP stimulation elicits the desired pharmacological response regarding insulin release from pancreatic β-cells. Inhibition of ALK IR Anaplastic Lymphoma Kinase Activity To measure the activity of the N2-(2-methoxyphenyl)pyrimidine derivative represented by formula 1 of the present invention to inhibit anaplastic lymphoma kinase (ALK) activity at enzyme level, the following experiment was performed by the same manner as described in experimental example 1 except that IR (Insulin Receptor) protein was used instead of ALK WT protein. In vitro Assay The method of determining thioredoxin activity is based on the reduction of insulin by thioredoxin. Thioredoxin is reconstituted is by thioredoxin reductase, with NADPH. The resulting free thiol -SH groups react with 5,5′-dithiobis-2-nitrobenzoic acid (DTNB). The termination of the reaction results in the formation of a red colour. The intensity of colouration is determined spectrophotometrically at 412 nm and this corresponds to the number of reduced sulfahydryl groups. The reaction was performed at 37° C. over 30 min. in a buffer containing 50 mM Tris-HCl and 20 mM EDTA at a pH of 7.6. The substrate concentrations used were: 0.25 μM human recombinant thioredoxin and 0.325 μM rat recombinant thioredoxin reductase (IMCO Corporation Ltd AB, Sweden) and 316 μM of insulin, 0.8 μM NADPH, 8 mM DTNB (Sigma Aldrich). Dose Response: Fluorescence polarization-based cell-based high throughput dose response assay for inhibitors of insulin-degrading enzyme (IDE) Source (MLPCN Center Name): The Scripps Research Institute Molecular Screening Center (SRIMSC) Center Affiliation: The Scripps Research Institute (TSRI) Assay Provider: Malcolm Leissring, Mayo Clinic College of Medicine Network: Molecular Libraries Probe Production Centers Network (MLPCN) Grant Proposal Number: 1 R03 DA024888-01 Grant Proposal PI: Malcolm Leissring, Mayo Clinic College of Medicine External Assay ID: IDE_INH_FP_1536_3XIC50 DRUN Name: Dose Response: Fluorescence polarization-based cell-based high throughput dose response assay for inhibitors of insulin-degrading enzyme (IDE). Description: Alzheimer's disease (AD) is characterized by accumulation of amyloid beta-protein (A-beta; Abeta) in brain regions involved in memory and cognition (1). The steady-state levels of AB reflect a balance between its production via beta- and gamma-secretases and its catabolism by proteolytic degradation (2-4). Because Abeta cleavage products are less neurotoxic than intact Abeta, e PDI Inhibition Assay The assay was carried out in 384-well plates according to literature procedures [46]. Each well contained 100mM sodium phosphate and 0.2mM EDTA pH 7.0. 10ng PDI was pre-incubated with various concentration of probes (4% DMSO) in 20uL buffer for 30 min at 37 degrees Celsius, followed by the addition of insulin (0.16mM final concentration) and DDT (1mM final concentration). The enzyme reaction was monitored at 650nm on a Bioteck microplate reader. DNA Methylation Assay Assays (0.1 ml) were conducted in 96-well black half-area plates in either a Wallac VICTOR2 or Biotek Synergy Neo plate reader at 37 °C in 10 mM Tris, pH 7.5, 100 mM potassium glutamate, 1 mM MgCl2, 1 mM DTT, 0.1 mg/ml BSA, and 5% glycerol. Assays contained varying amounts of oligonucleotide 8006 (5'-FAM-CCTATGCGmCATCAGTTTTCTGATGmCGmCATAGG-3'-Iowa Black, in which mC denotes 5-methyldeoxycytidylate residues (Integrated DNA Technologies, Coralville, IA), AdoMet (HPLC-purified, Sigma), and small molecule inhibitors (5-azaC, Sigma; SGI-1027, generous gift of Dr. Jian Jin, University of North Carolina; LCA, TCI America). Other anthraquinone compounds examined were purchased from ChemBridge Corp, San Diego, CA (UI1055 is N,N-diethyl-1-nitro-9,10-dioxo-9,10-dihydroanthracene-2-carboxamide; UI1060 is ethyl N-{[1-(butylthio)-9,10-dioxo-9,10-dihydroanthra-cen-2-yl] carbonyl}glycinate; UI1061 is 1-(butylsulfonyl)-9,10-dioxo-9,10-dihydroanthracene-2-carboxylic acid) and Sigma (anthraquinon and anthraquinone 2-carboxylic acid). All assays were conducted in triplicate and contained either human Dnmt1 (621-1600), human Dnmt1 (351-1600), human Dnmt3a (590-912), or M.SssI methyltransferase (New England Biolabs) and 0.8 units of Gla I (Sibenzyme, West RoxRoxbury, MA), except for the Gla I control assay, which did not contain a methyltransferase. Data were fitted using Prism (GraphPad Software, Inc). In Vitro Kinase Assay Inhibition of kinase activity against a variety of recombinant kinases [FLT3, FLT3 D835Y, anaplastic lymphoma kinase (ALK), insulin receptor, and epidermal growthfactor receptor (EGFR)] was measured using homogeneous time-resolved fluorescence (HTRF) assays [Choi et al., Bioorg. Med. Chem. Lett., 20:2033-2037]. Assays consist of enzymes mixed with serially diluted compounds and peptide substrates in a kinase reaction buffer (250 mM HEPES (pH 7.0), 0.5 mM orthovanadate, 0.05% bovine serum albumin, 0.1% NaN3). Following the addition of reagents for detection, the TR-FRET signal was measured using an EnVision multi-label reader (Perkin Elmer, Waltham, MA). In vitro degradation assay In vitro degradation assay protocol for compounds 3-33: Panc02.13 cells were purchased from ATCC and cultured in RPMI-1640 (Gibco), supplemented with 15% FBS (ATCC) and 10 Units/mL human recombinant insulin (Gibco). PROTAC treatments were carried out in 12-well plates for 16 h. TLR3 agonist Poly I:C (Invivogen; tlr1-pic) was added for the final 3 h. Cells were harvested, and lysed in RIPA buffer (50 mM Tris pH8, 150 mM NaCl, 1% Tx-100, 0.1% SDS, 0.5% Sodium Deoxycholate) supplemented with protease and phosphatase inhibitors. Lysates were clarified at 16,000 g for 10 minutes, and supernatants were separated by SDS-PAGE. Immunoblotting was performed using standard protocols. The antibodies used were TBK1 (Cell Signaling#3504), pIRF3 (abcam#ab76493), and GAPDH (Cell Signaling#5174). SAR analysis for the identification of translation initiation inhibitors (PABP) Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute(SBMRI, San Diego, CA) Network: NIH Molecular Libraries Probe Production Centers Network (MLPCN) Grant Number: 1R03MH084835-01 Assay Provider: Jerry Pelletier, Ph.D, McGill University, Montreal, Canada Translation is an essential cellular process whose deregulation is associated with alterations in cell growth, cell cycle progression, and cell death responses. The initiation phase of translation is a key target for regulation when cells are exposed to various environmental cues (e.g. insulin, amino acid starvation, mitogenic stimulation, hypoxia, etc). As well, translation initiation control is usurped upon viral infection and is deregulated in many human cancers. Over-expression of certain translation factors can lead to malignant transformation and many of the components of the translational apparatus are over-expressed in human cancers. Several tumor sup SAR analysis for the identification of translation initiation inhibitors (eIF4H) Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBIMR, San Diego, CA) Network: NIH Molecular Libraries Probe Production Centers Network (MLPCN) Grant Number: 1R03MH084835-01 Assay Provider: Jerry Pelletier, Ph.D, McGill University, Montreal, Canada Translation is an essential cellular process whose deregulation is associated with alterations in cell growth, cell cycle progression, and cell death responses. The initiation phase of translation is a key target for regulation when cells are exposed to various environmental cues (e.g. insulin, amino acid starvation, mitogenic stimulation, hypoxia, etc). As well, translation initiation control is usurped upon viral infection and is deregulated in many human cancers. Over-expression of certain translation factors can lead to malignant transformation and many of the components of the translational apparatus are over-expressed in human cancers. Several tumor su uHTS fluorescence polarization assay for the identification of translation initiation inhibitors (PABP) Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Production Centers Network (MLPCN) Grant Number: 1R03MH084835-01 Assay Provider: Jerry Pelletier, Ph.D, McGill University, Montreal, Canada Translation is an essential cellular process whose deregulation is associated with alterations in cell growth, cell cycle progression, and cell death responses. The initiation phase of translation is a key target for regulation when cells are exposed to various environmental cues (e.g. insulin, amino acid starvation, mitogenic stimulation, hypoxia, etc). As well, translation initiation control is usurped upon viral infection and is deregulated in many human cancers. Over-expression of certain translation factors can lead to malignant transformation and many of the components of the translational apparatus are over-expressed in human cancers. Several tumor suppresso uHTS fluorescence polarization assay for the identification of translation initiation inhibitors (eIF4H) Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Production Centers Network (MLPCN) Grant Number: 1R03MH084835-01 Assay Provider: Jerry Pelletier, Ph.D, McGill University, Montreal, Canada Translation is an essential cellular process whose deregulation is associated with alterations in cell growth, cell cycle progression, and cell death responses. The initiation phase of translation is a key target for regulation when cells are exposed to various environmental cues (e.g. insulin, amino acid starvation, mitogenic stimulation, hypoxia, etc). As well, translation initiation control is usurped upon viral infection and is deregulated in many human cancers. Over-expression of certain translation factors can lead to malignant transformation and many of the components of the translational apparatus are over-expressed in human cancers. Several tumor suppresso CAMP Assays Activation of GLP-1 receptor is known to stimulate cyclic AMP (cAMP) production in cells which indicates primary coupling to the Gαs subunit of the G protein heterotrimeric complex. Evidence suggests signaling through Gαs induced CAMP stimulation elicits the desired pharmacological response regarding insulin release from pancreatic β-cells. To optimize functional activity directed toward Gαs coupling, a HEK293/CRELuc cell line developed by HDB stably expressing the GLP-1 Receptor was used. 200× concentration of compound working solutions were prepared (Agilent Technologies Bravo) with 1/2 log serial dilution in 384-well Echo LDV plate (Labcyte, Cat #LP-0200). 50 nL/well 200× concentration of compound working solutions were moved to 384-well white low volume plate (Greiner, Cat #784075) using Labcyte ECHO550. 1×105 cells/mL HEK293/GLP1R/CRE-LUC (HD Biosciences) cell suspensions prepared with assay buffer [DPBS containing 0.5 mM IBMX (Sigma,Cat #I5879) and 0.1% BSA (GENVIEW, Cat #FA016-100 g)], 10 uL cell suspensions were added to each well of previous generated assay plate which already contains 50 nl compound at 200× concentration using ThermoFisher Multidrop Combi (1000cells/well). Seal the plate and incubate at 37° C. with 5% CO2 for 30 min. Fluorescence Intensity-Based Assay To test the synthesized compounds for their potential to inhibit SHP2 activity, a fluorescence intensity-based assay using 6,8-difluoro-4-methylumbelliferyl phosphate (DiFMUP) as the substrate was adapted for three recombinant SHP2 constructs: 1) the SHP2 catalytic domain (SHP2cat; residues 248-527), 2) the full-length SHP2-E76K oncogenic mutant, and 3) the full-length SHP2 wild-type (SHP2-WT). The recombinant SHP2 proteins were expressed and purified. A dually phosphorylated peptide derived from the insulin receptor substrate 1 (IRS-1) served as a surrogate binding protein and was used to activate SHP2-WT. The constitutively active E76K mutant did not require activation. Similarly, the SHP2cat construct, which lacks the SH2 domains, did not need to be activated. Michaelis-Menten experiments to determine the DiFMUP Michaelis-Menten constant (Kmn) for each SHP2 construct were performed and yielded the following values: SHP2-WT, Km=60 μM; SHP2-E76K, Km=20 μM; SHP2cat, Km=20 μM. Relative maximum rates (Vmax) of the DiFMUP reactions were as follows: SHP2-WT, Vmax=871 AFU/min; SHP2-E76K, Vmax, =2730 AFU/min; SHP2cat, Vmax=2912 AFU/min. IC50 values for each compound were determined from initial rates in 10-point dose-response assays using DiFMUP at a concentration corresponding to its Km value for the respective SHP2 construct. HepaRG-CAR Cell-Based Assay for Quantitation of Glycolate Oxidase Inhibition A HepaRG human hepatic cell line was transfected for stable overexpression of the constitutive androstane receptor (i.e., HepaRG-CAR cells), as reported by van der Mark et al. (Drug Metab. Dispos., 2017, 45:56-67. Overexpression of CAR in these cells resulted in higher levels of glycolate oxidase (GOX) expression compared to the parental HepaRG cells. HepaRG-CAR cells were plated in a 12-wells plate and incubated for 4 weeks until fully differentiated.To measure cellular glycolate flux, the HepaRG-CAR cells were incubated in Williams medium supplemented with 10% fetal bovine serum (FBS), 5 μg/mL insulin, 50 μM hydrocortisone hemisuccinate, 2 mM glutamine, 5000 U/mL penicillin and 5 mg/mL streptomycin. Test compounds were added to the medium at 0, 0.3, 1, 3 or 10 μM and incubated for 30 minutes, after which 500 μM glycolate was also added. After incubation for 48 hours, 400 μL medium was taken from the culture plate and added to 60 μL 37% HCl.Internal standards (2,2-d2 glycolate, 1,2-13C2 oxalate and 13C2-glyoxylate) and hydroxylamine were added followed by another 30 minute incubation at 80° C. The acids were extracted using ethyl acetate with NaCl. The organic phase was dried under nitrogen and derivatized with N-tert-butyldimethylsilyl-N-methyl trifluoroacetamide (MTBSTFA) for 30 minutes at 80° C. The amounts of glycolate, glyoxylate and oxalate were determined by gas chromatography-mass spectrometry (GC-MS) analysis, using a 25 meter CP-Sil 5 CB low bleed column. A standard curve was used to calculate the concentrations of each acid in the culture medium. Mobility Shift Assay Compound activity was determined using in house His tagged full-length PTPN1 protein (SEQ ID NO: 3) in an in vitro enzymatic reaction. The enzymatic assay used to determine activity is a mobility shift assay using a LabChip EZ Reader by Caliper Life Sciences. The enzymatic reaction was carried out in assay buffer (50 mM HEPES pH 7.5, 1 mM EGTA, 10 mM EDTA, 0.01% Tween 20, and 2 mM DTT). The compounds were dispensed on a white 384 well ProxiPlate™ (PerkinElmer Cat #6008289) plate using a Labcyte Echo liquid handler at varying concentrations (12 point, 1:3 dilution). The enzyme (at 0.5 nM) was incubated with compound for 10 minutes at room temperature. Thereafter, the substrate (phosphorylated insulin receptor probe sequence: ((OG488)-(NH-CH2-CH2-O-CH2-CH2-O-CH2-CO)-T-R-D-I-(PY)-E-T-D-Y-Y-R-K-K-NH2) (SEQ ID NO: 2) was added at 2 μM to the plates and incubated for another 10 minutes at room temperature. Finally, a quench solution (water and 4-bromo-3-(2-oxo-2-propoxyethoxy)-5-(3-{[1-(phenylmethanesulfonyl)piperidin-4-yl]amino}phenyl)thiophene-2-carboxylic acid) was added to the plates, which were then run on the EZ Reader (excitation 488 nm, emission 530 nm) to measure % conversion (the amount of phosphorylated substrate which was de-phosphorylated by PTPN1). Each plate had a 100% control (inhibitor: 4-bromo-3-(2-oxo-2-propoxyethoxy)-5-(3-{[1-(phenylmethanesulfonyl)piperidin-4-yl]amino}phenyl)thiophene-2-carboxylic acid) and 0% control (DMSO), which were used to calculate % inhibition. The % inhibition was then used to calculate the IC50 values. pAKT Protocol Inhibition of the PI3K-AKT-mTOR pathway was measured by quantifying the loss of (Ser-473) pAKT using AlphaScreen (Perkin Elmer). B103 (Rat Neuroblastoma) cells were seeded in serum containing medium (High Glucose DMEM (-Phenol Red)+10% FBS+2× Glutamax+1 mM Sodium Pyruvate+10 mM HEPES+1× Non-Essential Amino Acids+1× Pen/Strep) on a 96-well tissue culture treated plate and grown for 20 hours. Cells were then serum starved in serum free medium (High Glucose DMEM (-Phenol Red)+1× Glutamax+1 mM Sodium Pyruvate+1× Pen/Strep) for 6 hours prior to a 2-hour pretreatment with inhibitors of the pathway, including reference inhibitor LY294002. These inhibitors were prepared at a 200× final concentration as a 6-point, 1:3 serial dilution in DMSO series, with DMSO as the 7th point. The inhibitors were then diluted in experimental medium (High Glucose DMEM (-Phenol Red)+1× Glutamax+1 mM Sodium Pyruvate+1× Pen/Strep+25 mM HEPES+0.1% BSA) and combined with the cells at 1× final concentration in 0.5% DMSO. The cells were then stimulated for 20 minutes with (2.5 μg/mL) insulin, an activator of the PI3K-AKT-mTOR pathway and a demonstrated (Ser-473) pAKT agonist. Cells were promptly lysed using Perkin Elmer proprietary lysis buffer and the (Ser-473) pAKT and total AKT contained in the lysate was measured by AlphaScreen. In AlphaScreen, donor beads were coated with streptavidin to capture one of the antibodies, which is biotinylated. Acceptor beads were coated with Protein A to immobilize the other antibody. In the presence of target protein, the two antibodies bring the donor and acceptor beads close together, generating signal. The amount of light emission is directly proportional to the amount of target protein present in the sample. For each inhibitor tested: the ratio of measured (Ser-473) pAKT/totalAKT was plotted in GraphPad Prism as a 7-point, non-linear regression, 4-parameter curve with bottom constrained to reference control bottom and unconstrained top anchored to DMSO. PHH Natural Infection Assay Detailed procedures regarding primary human hepatocyte (PHH) HBV natural infection assay are described as below. One tube of frozen PHH (10 million cells) is thawed in 37 oC water bath and then transferred to 20 mL of PHH thawing medium (Sigma, InVitroGRO HT Medium, Cat. S03319) with gently mixing. The cells were then centrifuged at 80 g/min for 5 min, the supernatant was discarded and the tube was refilled with 25 mL of PHH plating medium (Sigma , InVitroGRO CP Medium, Cat. S03317). The tube was shaken very gently to re-suspend all cells, and then 50 µl of cells were transferred to each well 384-well collagen I coated plate withappropriate liquid handling equipment, e.g. Integra VI AFL0384 or Agilent Bravo. The cells were then cultured for 24 hours in a cell incubator. For HBV infection, after PHH attachment on the culture plate, the plating medium was removed and replenished with PHH culture medium containing HBV virus. The PHH culture medium was prepared with Dulbecco's Modified Eagle Medium (DMEM)/F12 (1: 1 in volume ratio) containing 10% fetal bovine serum (Gibco, Cat.10099141), 5 ng/mL human epidermal growth factor (Gibco, Cat.PHG0311L), 20 ng/mL dexamethasone (Sigma, Cat.D4902-100mg), 250 ng/mL human recombinant insulin (Gibco, Cat.41400045) and 100 U/mL penicillin. HBV virus at 200 genome equivalent (GE) per cell with 4% PEG8000 (Sigma, Cat.P1458) containing culture medium were added to the PHH culture medium for infection. The cells were then cultured for 24 hours in cell incubator. Then the cell culture supernatant was removed. The HBV-infected PHH were cultured with sandwich culture method with PHH culture medium containing 1% DMSO and 0.25 mg/mL matrix gel for 72 hours. The supernatant was then refreshed with PHH culture medium containing different concentrations of testing compounds for two times with 72-hour interval. At the end of treatment, the supernatant was collected for viral markers measurements, including HBsAg, HBeAg, HBV DNA and cytotoxicity. HBsAg and HBeAg were detected using alphalisa method using their specific antibodies. For HBV DNA detection, HBV DNA Quantitative Fluorescence Diagnostic Kit (Sansure Biotech Inc.) was used following the manufacture s protocol. PHH Natural Infection Assay Detailed procedures regarding primary human hepatocyte (PHH) HBV natural infection assay are described as below. One tube of frozen PHH (10 million cells) is thawed in 37 oC water bath and then transferred to 20 mL of PHH thawing medium (Sigma , InVitroGRO HT Medium, Cat. S03319) with gently mixing. The cells were then centrifuged at 80 g/min for 5 min, the supernatant was discarded and the tube was refilled with 25 mL of PHH plating medium (Sigma , InVitroGRO CP Medium, Cat. S03317). The tube was shaken very gently to re-suspend all cells.50 μl of cells were transferred to each well 384-well collagen I coated plate with appropriate liquid handling equipment, e.g. Integra VIAFLO384 or Agilent Bravo. The cells were then cultured for 24 hours in a cell incubator. For HBV infection, after PHH attachment on the culture plate, the plating medium was removed and replenished with PHH culture medium containing HBV virus. The PHH culture medium was prepared with Dulbecco's Modified Eagle Medium (DMEM)/F12 (1: 1 in volume ratio) containing 10% fetal bovine serum (Gibco, Cat.10099141), 5 ng/mL human epidermal growth factor (Gibco, Cat.PHG0311L), 20 ng/mL dexamethasone (Sigma, Cat.D4902-100mg), 250 ng/mL human recombinant insulin (Gibco, Cat.41400045) and 100 U/mL penicillin. HBV virus at 200 genome equivalent (GE) per cell with 4% PEG8000 (Sigma, Cat.P1458) containing culture medium were added to the PHH culture medium for infection. The cells were then cultured for 24 hours in cell incubator. Then the cell culture supernatant was removed. The HBV-infected PHH were cultured with sandwich culture method with PHH culture medium containing 1% DMSO and 0.25 mg/mL matrix gel for 72 hours. The supernatant was then refreshed with PHH culture medium containing different concentrations of testing compounds for two times with 72-hour interval. At the end of treatment, the supernatant was collected for viral markers measurements, including HBsAg, HBeAg, HBV DNA and cytotoxicity. HBsAg and HBeAg were detected using alphalisa method using their specific antibodies. For HBV DNA detection, HBV DNA Quantitative Fluorescence Diagnostic Kit (Sansure Biotech Inc.) was used following the manufactures protocol. Cytotoxicity was determined using Cell Counting Kit-8 (CCK8, Dojindo Molecular Technologies, Inc.). PHH Natural Infection Assay Detailed procedures regarding primary human hepatocyte (PHH) HBV natural infection assay are described as below. One tube of frozen PHH (10 million cells) is thawed in 37 oC water bath and then transferred to 20 mL of PHH thawing medium (Sigma , InVitroGRO HT Medium, Cat. S03319) with gently mixing. The cells were then centrifuged at 80 g/min for 5 min, the supernatant was discarded and the tube was refilled with 25 mL of PHH plating medium (Sigma , InVitroGRO CP Medium, Cat. S03317). The tube was shaken very gently to re-suspend all cells.50 μl of cells were transferred to each well 384-well collagen I coated plate with appropriate liquid handling equipment, e.g. Integra VIAFLO384 or Agilent Bravo. The cells were then cultured for 24 hours in a cell incubator. For HBV infection, after PHH attachment on the culture plate, the plating medium was removed and replenished with PHH culture medium containing HBV virus. The PHH culture medium was prepared with Dulbecco's Modified Eagle Medium (DMEM)/F12 (1: 1 in volume ratio) containing 10% fetal bovine serum (Gibco, Cat.10099141), 5 ng/mL human epidermal growth factor (Gibco, Cat.PHG0311L), 20 ng/mL dexamethasone (Sigma, Cat.D4902-100mg), 250 ng/mL human recombinant insulin (Gibco, Cat.41400045) and 100 U/mL penicillin. HBV virus at 200 genome equivalent (GE) per cell with 4% PEG8000 (Sigma, Cat.P1458) containing culture medium were added to the PHH culture medium for infection. The cells were then cultured for 24 hours in cell incubator. Then the cell culture supernatant was removed. The HBV-infected PHH were cultured with sandwich culture method with PHH culture medium containing 1% DMSO and 0.25 mg/mL matrix gel for 72 hours. The supernatant was then refreshed with PHH culture medium containing different concentrations of testing compounds for two times with 72-hour interval. At the end of treatment, the supernatant was collected for viral markers measurements, including HBsAg, HBeAg, HBV DNA and cytotoxicity. HBsAg and HBeAg were detected using alphalisa method using their specific antibodies. For HBV DNA detection, HBV DNA Quantitative Fluorescence Diagnostic Kit (Sansure Biotech Inc.) was used following the manufacture s protocol. Cytotoxicity was determined using Cell Counting Kit-8 (CCK8, Dojindo Molecular Technologies, Inc.). PHH Natural Infection Assay Detailed procedures regarding primary human hepatocyte (PHH) HBV natural infection assay are described as below. One tube of frozen PHH (10 million cells) is thawed in 37 °C water bath and then transferred to 20 mL of PHH thawing medium (Sigma, InVitroGRO HT Medium, Cat. S03319) with gently mixing. The cells were then centrifuged at 80 g/min for 5 min, the supernatant was discarded and the tube was refilled with 25 mL of PHH plating medium (Sigma , InVitroGRO CP Medium, Cat. S03317). The tube was shaken very gently to re-suspend all cells, and then 50 mΐ of cells were transferred to each well 384-well collagen I coated plate with appropriate liquid handling equipment, e.g. Integra VIAFL0384 or Agilent Bravo. The cells were then cultured for 24 hours in a cell incubator. For HBV infection, after PHH attachment on the culture plate, the plating medium was removed and replenished with PHH culture medium containing HBV virus. The PHH culture medium was prepared with Dulbecco's Modified Eagle Medium (DMEM)/F12 (1: 1 in volume ratio) containing 10% fetal bovine serum (Gibco, Cat.10099141), 5 ng/mL human epidermal growth factor (Gibco, Cat.PHG0311L), 20 ng/mL dexamethasone (Sigma, Cat.D4902- 1 OOmg), 250 ng/mL human recombinant insulin (Gibco, Cat.41400045) and 100 U/mL penicillin. HBV virus at 200 genome equivalent (GE) per cell with 4% PEG8000 (Sigma, Cat.P1458) containing culture medium were added to the PHH culture medium for infection. The cells were then cultured for 24 hours in cell incubator. Then the cell culture supernatant was removed. The HBV-infected PHH were cultured with sandwich culture method with PHH culture medium containing 1% DMSO and 0.25 mg/mL matrix gel for 72 hours. The supernatant was then refreshed with PHH culture medium containing different concentrations of testing compounds for two times with 72 -hour interval. At the end of treatment, the supernatant was collected for viral markers measurements, including HBsAg, HBeAg, HBV DNA and cytotoxicity. HBsAg and HBeAg were detected using alphalisa method using their specific antibodies. For HBV DNA detection, HBV DNA Quantitative Fluorescence Diagnostic Kit (Sansure Biotech Inc.) was used following the manufacture s protocol. Cytotoxicity was determined using Cell Counting Kit-8 (CCK8, Dojindo Molecular Technologies, Inc.). PHH Natural Infection Assay Detailed procedures regarding primary human hepatocyte (PHH) HBV natural infection assay are described as below. One tube of frozen PHH (10 million cells) is thawed in 37 °C water bath and then transferred to 20 mL of PHH thawing medium (Sigma, InVitroGRO HT Medium, Cat. S03319) with gently mixing. The cells were then centrifuged at 80 g/min for 5 min, the supernatant was discarded and the tube was refilled with 25 mL of PHH plating medium (Sigma, InVitroGRO CP Medium, Cat. S03317). The tube was shaken very gently to re-suspend all cells, and then 50 μL of cells were transferred to each well 384-well collagen I coated plate with appropriate liquid handling equipment, e.g. Integra VIAFL0384 or Agilent Bravo. The cells were then cultured for 24 hours in a cell incubator. For HBV infection, after PHH attachment on the culture plate, the plating medium was removed and replenished with PHH culture medium containing HBV virus. The PHH culture medium was prepared with Dulbecco's Modified Eagle Medium (DMEM)/F12 (1: 1 in volume ratio) containing 10% fetal bovine serum (Gibco,Cat.10099141), 5 ng/mL human epidermal growth factor (Gibco, Cat.PHG0311L), 20 ng/mL dexamethasone (Sigma, Cat.D4902-100mg), 250 ng/mL human recombinant insulin (Gibco,Cat.41400045) and 100 U/mL penicillin. HBV virus at 200 genome equivalent (GE) per cell with 4% PEG8000 (Sigma, Cat.P1458) containing culture medium were added to the PHH culture medium for infection. The cells were then cultured for 24 hours in cell incubator. Then the cell culture supernatant was removed. The HBV-infected PHH were cultured with sandwichculture method with PHH culture medium containing 1% DMSO and 0.25 mg/mL matrix gel for 72 hours. The supernatant was then refreshed with PHH culture medium containing different concentrations of testing compounds for two times with 72-hour interval. At the end of treatment, the supernatant was collected for viral markers measurements, including HBsAg, HBeAg, HBV DNA and cytotoxicity. HBsAg and HBeAg were detected using alphalisa method using their specific antibodies. For HBV DNA detection, HBV DNA Quantitative Fluorescence Diagnostic Kit (Sansure Biotech Inc.) was used following the manufacture s protocol. Cytotoxicity was determined using Cell Counting Kit-8 (CCK8, Dojindo Molecular Technologies, Inc.).
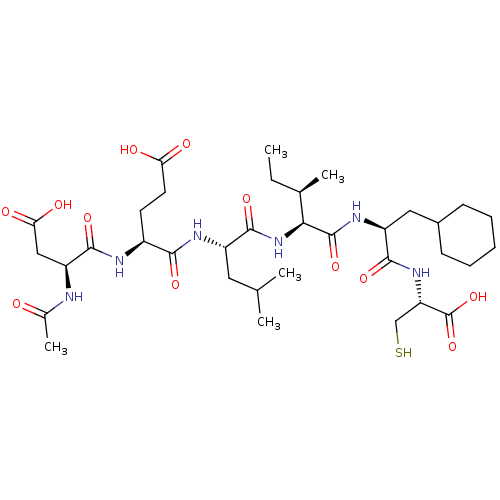 CHEMBL179084 BDBM50158804 AcAsp-D-Gla-Leu-Ile-Cha-Cys
CHEMBL179084 BDBM50158804 AcAsp-D-Gla-Leu-Ile-Cha-Cys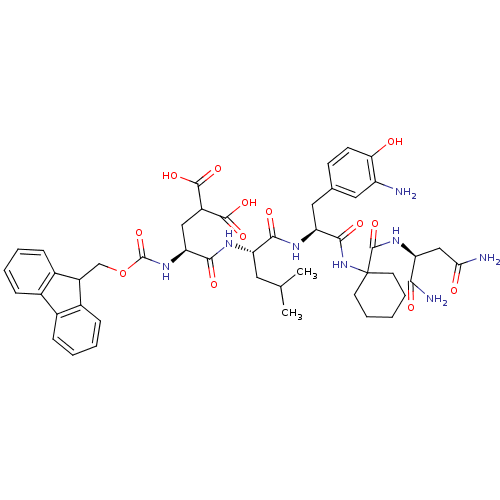 BDBM50264466 CHEMBL444948 N-Fmoc-Gla-Leu-(3'-NH2-Tyr)-Ac6c-Glu-Asn-amide
BDBM50264466 CHEMBL444948 N-Fmoc-Gla-Leu-(3'-NH2-Tyr)-Ac6c-Glu-Asn-amide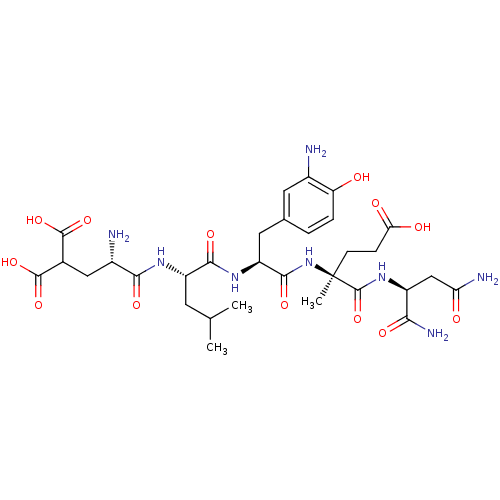 H-Gla-Leu-(3'-NH2-Tyr)-(alpha-Me)Glu-Asn-amide BDBM50264460 CHEMBL507364
H-Gla-Leu-(3'-NH2-Tyr)-(alpha-Me)Glu-Asn-amide BDBM50264460 CHEMBL507364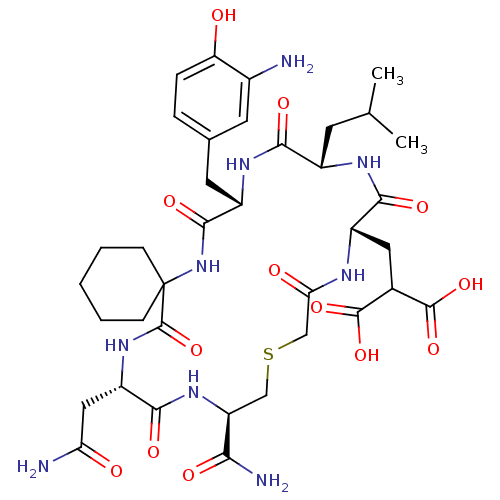 BDBM50183416 cyclo-(CH2-CO-Gla-Leu-(3'-NH2-Tyr)-Ac6c-Asn-Cys)-amide CHEMBL202731
BDBM50183416 cyclo-(CH2-CO-Gla-Leu-(3'-NH2-Tyr)-Ac6c-Asn-Cys)-amide CHEMBL202731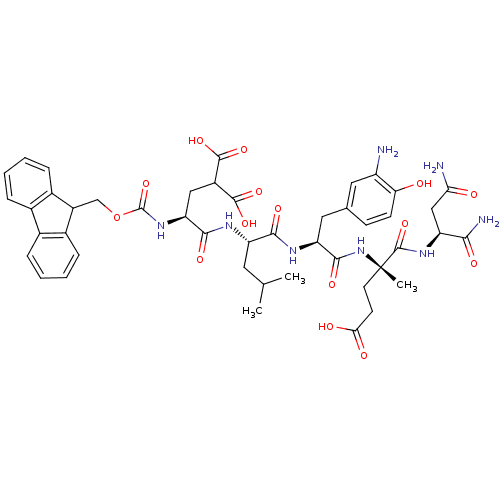 N-Fmoc-Gla-Leu-(3'-NH2-Tyr)-(alpha-Me)Glu-Asn-amide BDBM50264461 CHEMBL508925
N-Fmoc-Gla-Leu-(3'-NH2-Tyr)-(alpha-Me)Glu-Asn-amide BDBM50264461 CHEMBL508925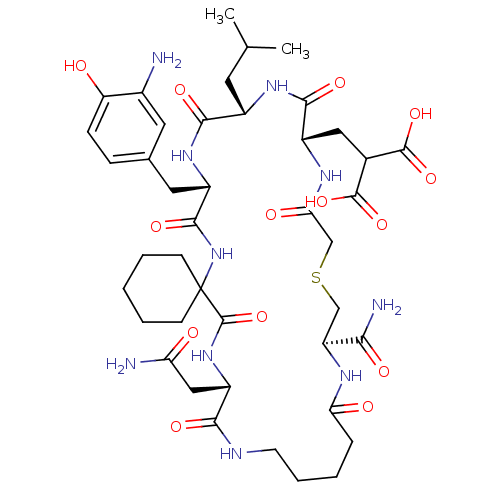 BDBM50183426 cyclo-(CH2-CO-Gla-Leu-(3'-NH2-Tyr)-Ac6c-Asn-Ava-Cys)-amide CHEMBL262988
BDBM50183426 cyclo-(CH2-CO-Gla-Leu-(3'-NH2-Tyr)-Ac6c-Asn-Ava-Cys)-amide CHEMBL262988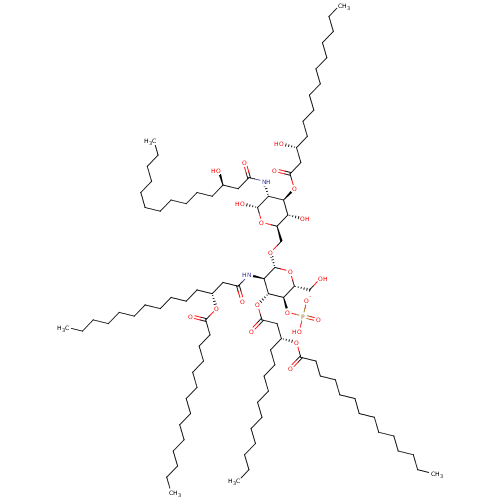 US11458151, Compound MPL Monophosphoryl Lipid A Monophosphoryl hexaacyl disaccharide MPLA GLA Glycopyranoside Lipid A BDBM633255
US11458151, Compound MPL Monophosphoryl Lipid A Monophosphoryl hexaacyl disaccharide MPLA GLA Glycopyranoside Lipid A BDBM633255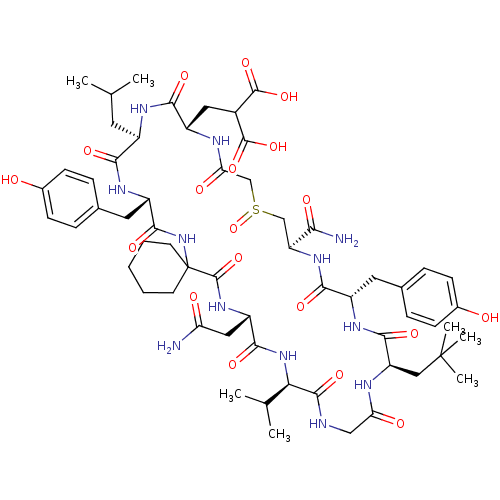 (R)-cyclic ((N-AC)Gla Leu Tyr Ach Asn Val Gly NPG Tyr Cys(O) NH2) (S)-cyclic ((N-AC)Gla Leu Tyr Ach Asn Val Gly NPG Tyr Cys(O) NH2) BDBM50129417 CHEMBL437456
(R)-cyclic ((N-AC)Gla Leu Tyr Ach Asn Val Gly NPG Tyr Cys(O) NH2) (S)-cyclic ((N-AC)Gla Leu Tyr Ach Asn Val Gly NPG Tyr Cys(O) NH2) BDBM50129417 CHEMBL437456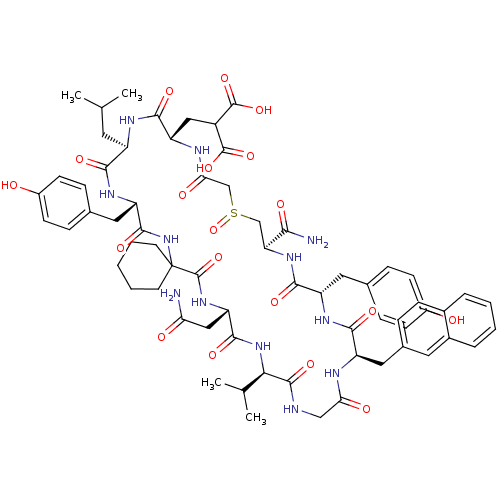 (S)-cyclic ((N-AC)Gla Leu Tyr Ach Asn Val Gly Nal Tyr Cys(O) NH2) BDBM50129418 (R)-cyclic ((N-AC)Gla Leu Tyr Ach Asn Val Gly Nal Tyr Cys(O) NH2) CHEMBL411790
(S)-cyclic ((N-AC)Gla Leu Tyr Ach Asn Val Gly Nal Tyr Cys(O) NH2) BDBM50129418 (R)-cyclic ((N-AC)Gla Leu Tyr Ach Asn Val Gly Nal Tyr Cys(O) NH2) CHEMBL411790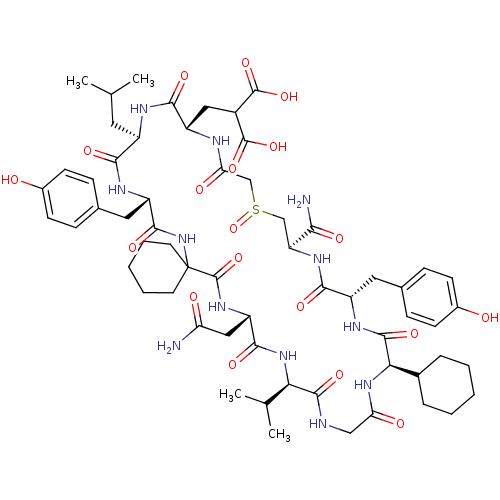 BDBM50129420 (R)-cyclic ((N-AC)Gla Leu Tyr Ach Asn Val Gly Cha Tyr Cys(O) NH2) CHEMBL406289 (S)-cyclic ((N-AC)Gla Leu Tyr Ach Asn Val Gly Cha Tyr Cys(O) NH2)
BDBM50129420 (R)-cyclic ((N-AC)Gla Leu Tyr Ach Asn Val Gly Cha Tyr Cys(O) NH2) CHEMBL406289 (S)-cyclic ((N-AC)Gla Leu Tyr Ach Asn Val Gly Cha Tyr Cys(O) NH2)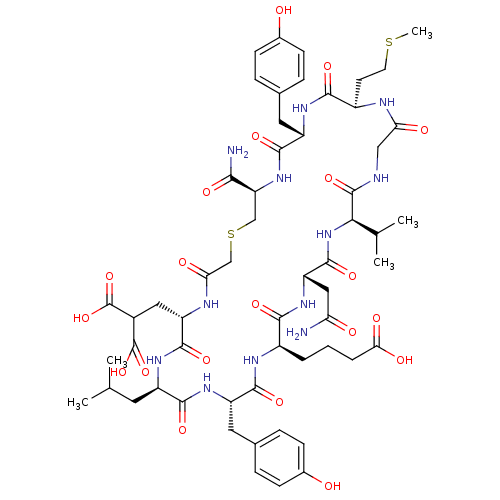 BDBM50129413 CHEMBL405191 cyclic ((N-AC)Gla Leu Tyr Adi Asn Val Gly Met Tyr Cys NH2)
BDBM50129413 CHEMBL405191 cyclic ((N-AC)Gla Leu Tyr Adi Asn Val Gly Met Tyr Cys NH2)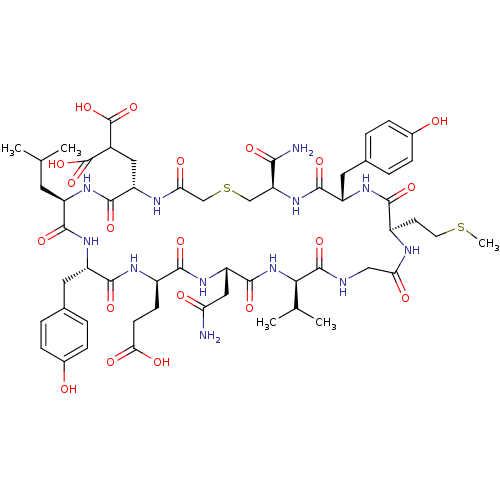 BDBM50129421 CHEMBL217433 cyclic ((N-AC)Gla Leu Tyr Pro Asn Val Gly Met Tyr Cys NH2)
BDBM50129421 CHEMBL217433 cyclic ((N-AC)Gla Leu Tyr Pro Asn Val Gly Met Tyr Cys NH2)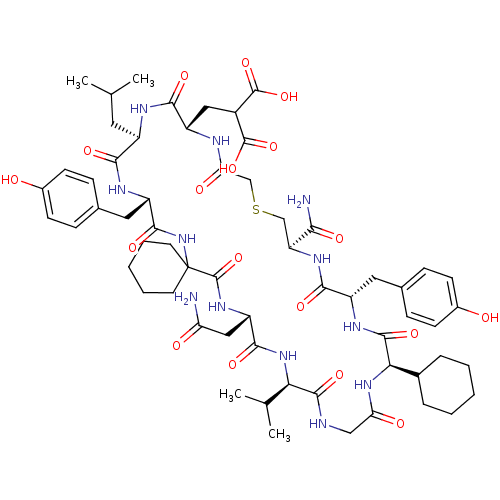 BDBM50129424 CHEMBL410772 cyclic ((N-AC)Gla Leu Tyr Ach Asn Val Gly Cha Tyr Cys NH2)
BDBM50129424 CHEMBL410772 cyclic ((N-AC)Gla Leu Tyr Ach Asn Val Gly Cha Tyr Cys NH2)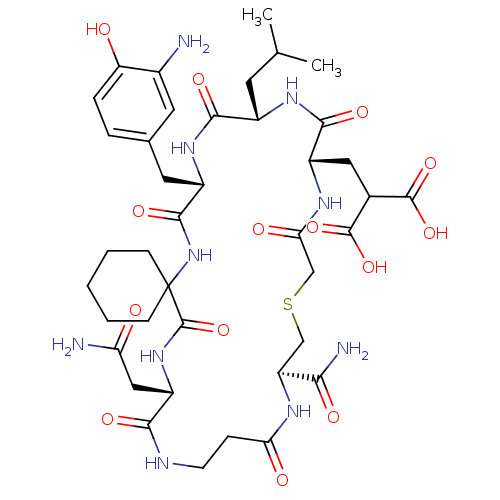 CHEMBL204237 BDBM50183423 cyclo-(CH2-CO-Gla-Leu-(3'-NH2-Tyr)-Ac6c-Asn-beta-Ala-Cys)-amide
CHEMBL204237 BDBM50183423 cyclo-(CH2-CO-Gla-Leu-(3'-NH2-Tyr)-Ac6c-Asn-beta-Ala-Cys)-amide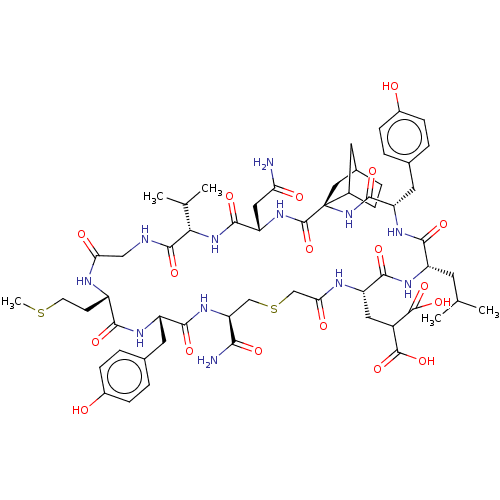 CHEMBL2369467 cyclic ((N-AC)Gla Leu Tyr BCH Asn Val Gly Met Tyr Cys NH2) BDBM50129419
CHEMBL2369467 cyclic ((N-AC)Gla Leu Tyr BCH Asn Val Gly Met Tyr Cys NH2) BDBM50129419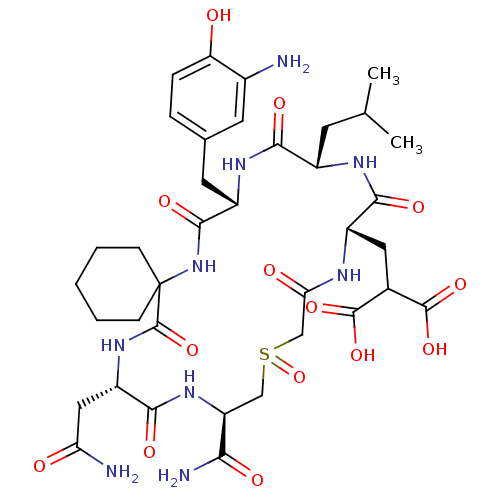 CHEMBL381980 cyclo-(CH2-CO-Gla-Leu-(3'-NH2-Tyr)-Ac6c-Asn-Cys (O))-amide (R) BDBM50183431
CHEMBL381980 cyclo-(CH2-CO-Gla-Leu-(3'-NH2-Tyr)-Ac6c-Asn-Cys (O))-amide (R) BDBM50183431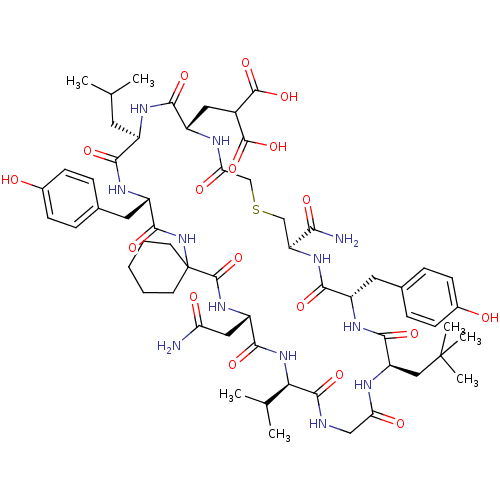 CHEMBL437329 BDBM50129415 cyclic ((N-AC)Gla Leu Tyr Ach Asn Val Gly NPG Tyr Cys NH2)
CHEMBL437329 BDBM50129415 cyclic ((N-AC)Gla Leu Tyr Ach Asn Val Gly NPG Tyr Cys NH2) cyclic ((N-AC)Gla Leu Tyr Ach Asn Val Gly Bip Tyr Cys NH2) BDBM50129411 CHEMBL266273
cyclic ((N-AC)Gla Leu Tyr Ach Asn Val Gly Bip Tyr Cys NH2) BDBM50129411 CHEMBL266273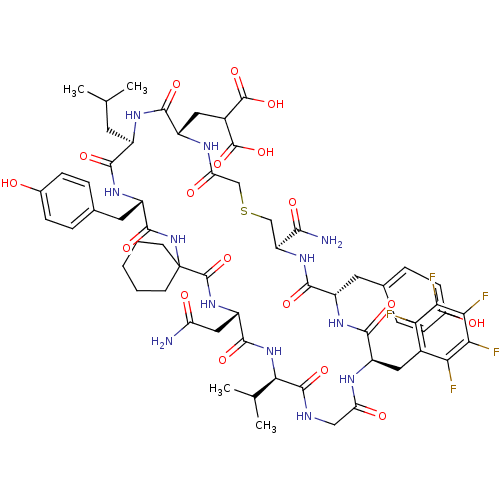 cyclic ((N-AC)Gla Leu Tyr Ach Asn Val Gly F5Phe Tyr Cys NH2) BDBM50129412 CHEMBL413683
cyclic ((N-AC)Gla Leu Tyr Ach Asn Val Gly F5Phe Tyr Cys NH2) BDBM50129412 CHEMBL413683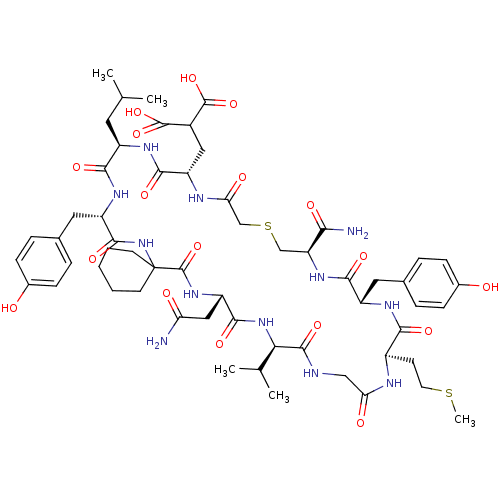 cyclic ((N-AC)Gla Leu Tyr Ach Asn Val Gly Met Tyr Cys NH2) BDBM50129423 CHEMBL407241
cyclic ((N-AC)Gla Leu Tyr Ach Asn Val Gly Met Tyr Cys NH2) BDBM50129423 CHEMBL407241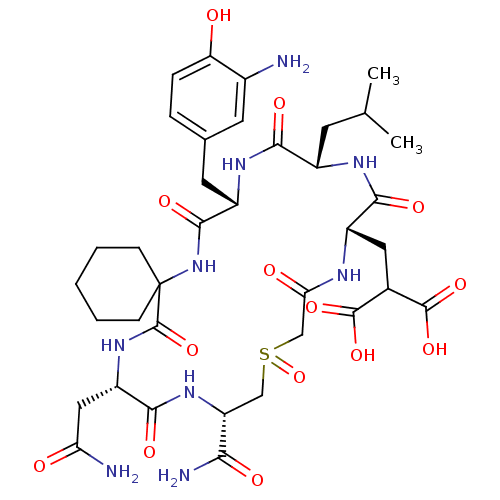 cyclo-(CH2-CO-Gla-Leu-(3'-NH2-Tyr)-Ac6c-Asn-Cys (O))-amide (S) BDBM50183421 CHEMBL204668
cyclo-(CH2-CO-Gla-Leu-(3'-NH2-Tyr)-Ac6c-Asn-Cys (O))-amide (S) BDBM50183421 CHEMBL204668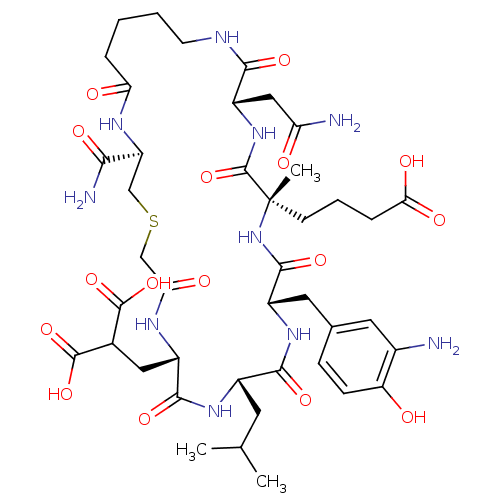 BDBM50264458 CHEMBL452659 Cyclo-[CH2-CO-Gla-Leu-(3'-NH2-Tyr)-(alpha-Me)Adi-Asn-Ava-Cys]-amide
BDBM50264458 CHEMBL452659 Cyclo-[CH2-CO-Gla-Leu-(3'-NH2-Tyr)-(alpha-Me)Adi-Asn-Ava-Cys]-amide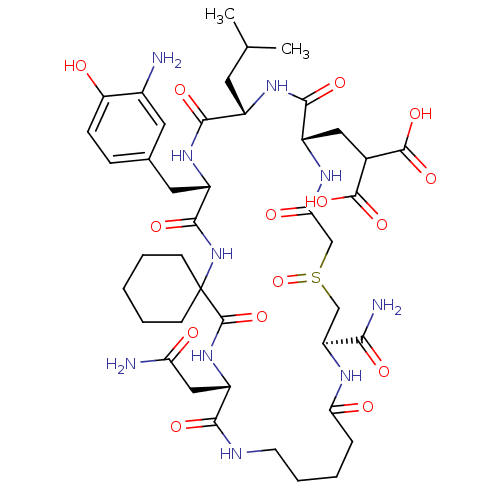 CHEMBL203761 BDBM50183425 cyclo-(CH2-CO-Gla-Leu-(3'-NH2-Tyr)-Ac6c-Asn-Ava-Cys (O))-amide (R)
CHEMBL203761 BDBM50183425 cyclo-(CH2-CO-Gla-Leu-(3'-NH2-Tyr)-Ac6c-Asn-Ava-Cys (O))-amide (R)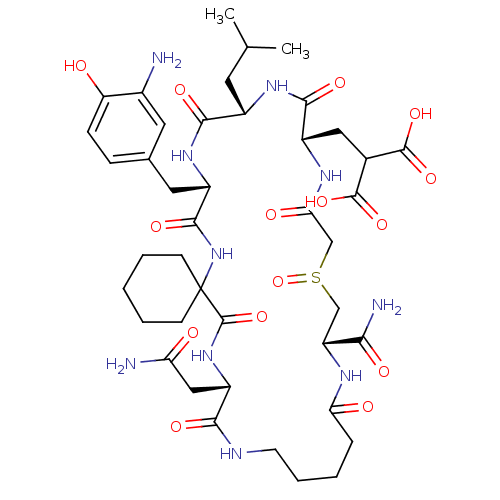 CHEMBL205022 cyclo-(CH2-CO-Gla-Leu-(3'-NH2-Tyr)-Ac6c-Asn-Ava-Cys (O))-amide (S) BDBM50183430
CHEMBL205022 cyclo-(CH2-CO-Gla-Leu-(3'-NH2-Tyr)-Ac6c-Asn-Ava-Cys (O))-amide (S) BDBM50183430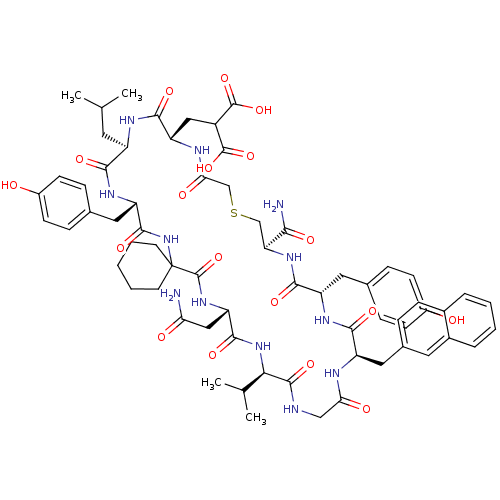 CHEMBL411181 BDBM50129416 cyclic ((N-AC)Gla Leu Tyr Ach Asn Val Gly 2-Nal Tyr Cys NH2)
CHEMBL411181 BDBM50129416 cyclic ((N-AC)Gla Leu Tyr Ach Asn Val Gly 2-Nal Tyr Cys NH2)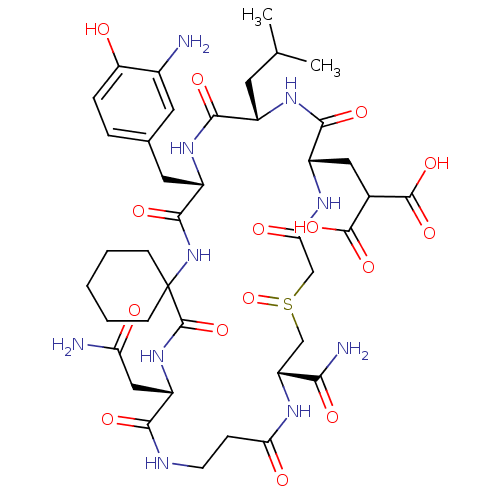 BDBM50183429 cyclo-(CH2-CO-Gla-Leu-(3'-NH2-Tyr)-Ac6c-Asn-beta-Ala-Cys (O))-amide (S) CHEMBL203512
BDBM50183429 cyclo-(CH2-CO-Gla-Leu-(3'-NH2-Tyr)-Ac6c-Asn-beta-Ala-Cys (O))-amide (S) CHEMBL203512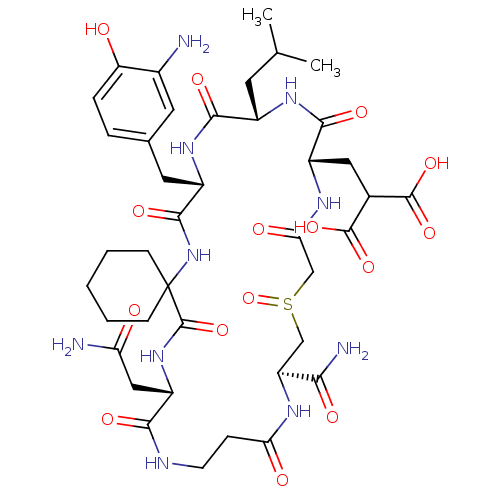 cyclo-(CH2-CO-Gla-Leu-(3'-NH2-Tyr)-Ac6c-Asn-beta-Ala-Cys-(O))-amide (R) BDBM50183417 CHEMBL383709
cyclo-(CH2-CO-Gla-Leu-(3'-NH2-Tyr)-Ac6c-Asn-beta-Ala-Cys-(O))-amide (R) BDBM50183417 CHEMBL383709