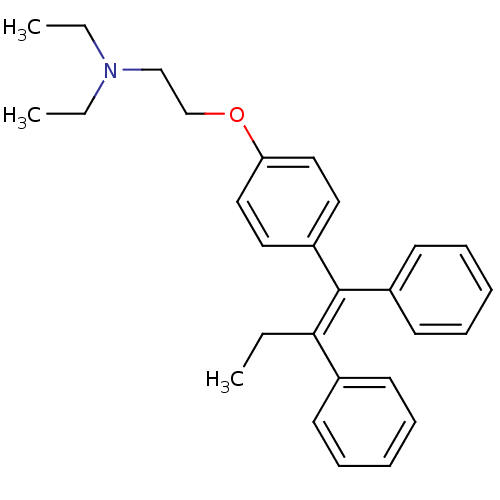
 Tamoxifen, 1 BDBM85467
Tamoxifen, 1 BDBM85467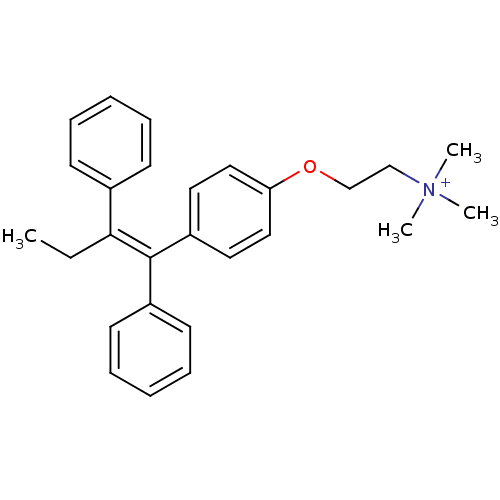 CHEMBL1213783 BDBM50323911 Tamoxifen methyl iodide
CHEMBL1213783 BDBM50323911 Tamoxifen methyl iodide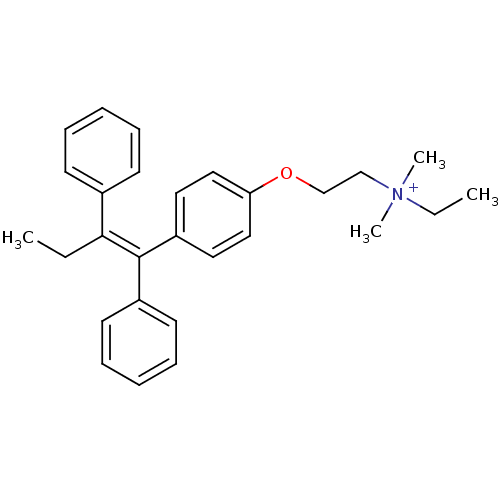 CHEMBL1213784 BDBM50323912 Tamoxifen ethyl bromide
CHEMBL1213784 BDBM50323912 Tamoxifen ethyl bromide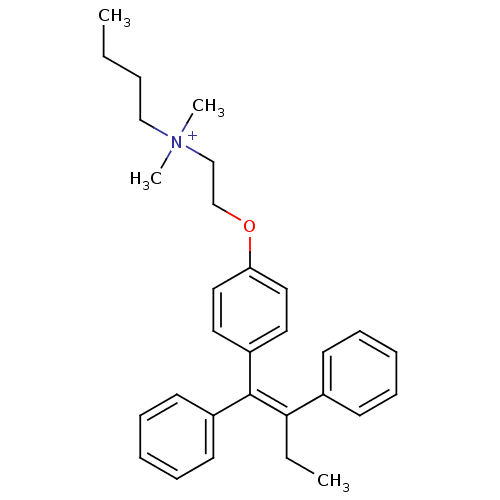 Tamoxifen butyl bromide CHEMBL1213786 BDBM50323914
Tamoxifen butyl bromide CHEMBL1213786 BDBM50323914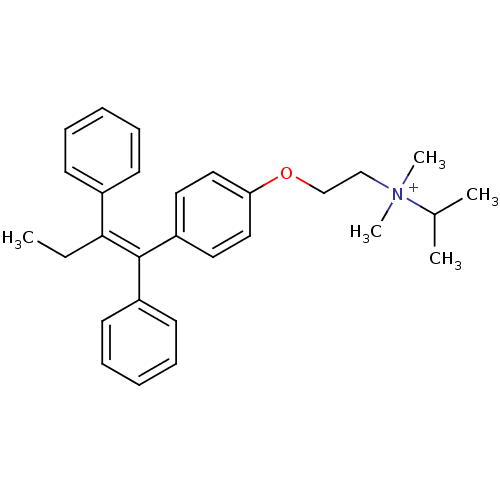 Tamoxifen isopropyl bromide CHEMBL1213785 BDBM50323913
Tamoxifen isopropyl bromide CHEMBL1213785 BDBM50323913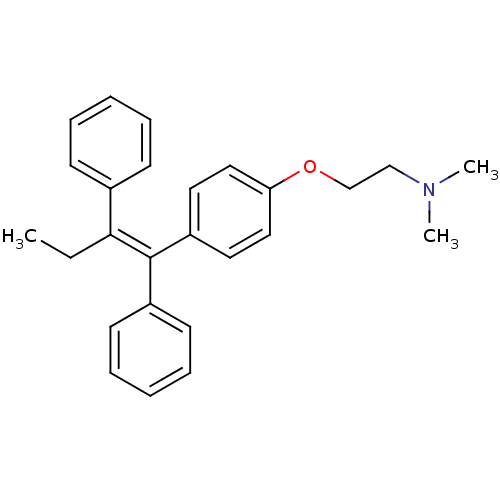 2-{4-[(1Z)-1,2-diphenylbut-1-en-1-yl]phenoxy}-N,N-dimethylethanamine Nolvadex US20240217988, Example Tamoxifen cid_2733526 FOSTRIECIN SODIUM Tamoxifen Tamoxifen (8) CHEMBL83 (2-{4-[(1Z)-1,2-diphenylbut-1-en-1-yl]phenoxy}ethyl)dimethylamine NCGC00024928 Tamoxifen, 7 med.21724, Compound Tamoxifen BDBM20607
2-{4-[(1Z)-1,2-diphenylbut-1-en-1-yl]phenoxy}-N,N-dimethylethanamine Nolvadex US20240217988, Example Tamoxifen cid_2733526 FOSTRIECIN SODIUM Tamoxifen Tamoxifen (8) CHEMBL83 (2-{4-[(1Z)-1,2-diphenylbut-1-en-1-yl]phenoxy}ethyl)dimethylamine NCGC00024928 Tamoxifen, 7 med.21724, Compound Tamoxifen BDBM20607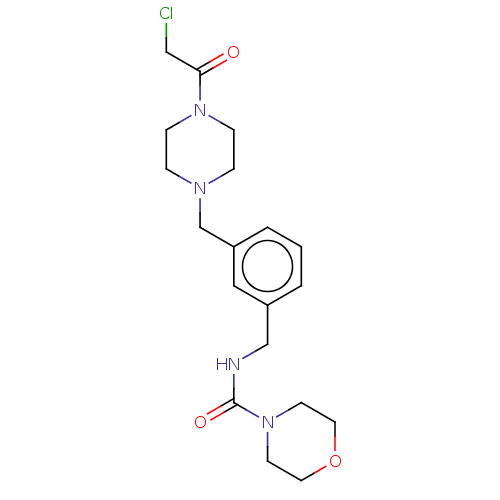 TAM-UNI-d1c3dd9f-12 BDBM496740
TAM-UNI-d1c3dd9f-12 BDBM496740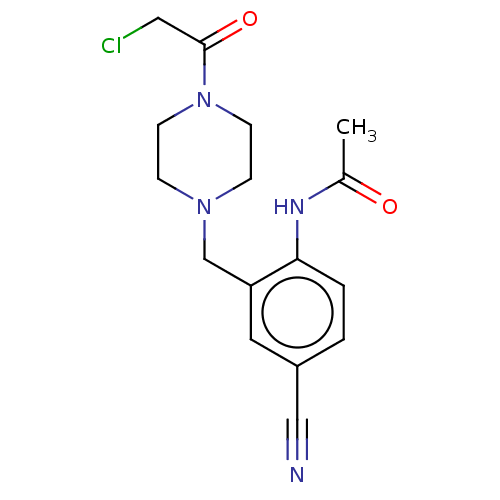 TAM-UNI-d1c3dd9f-18 BDBM496741
TAM-UNI-d1c3dd9f-18 BDBM496741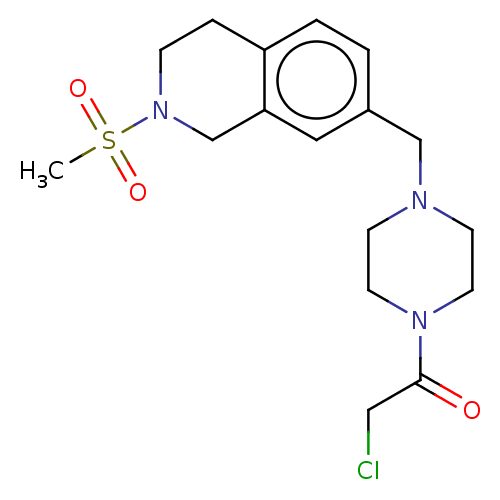 TAM-UNI-d1c3dd9f-5 BDBM496739
TAM-UNI-d1c3dd9f-5 BDBM496739
- Shagufta, na; Ahmad, I Tamoxifen a pioneering drug: An update on the therapeutic potential of tamoxifen derivatives. Eur J Med Chem 143: 515-531 (2018)
- Jia, Z; Wu, Y; Pan, Y; Zhou, J; Li, Q Salts of TAM inhibitors US Patent US11104682 (2021)
- Li, Y; Burns, DM; Feng, H; Glenn, J; He, C; Huang, T; Mei, S; Pan, J; Wang, X; Ye, Y Pyrrolopyrimidine derivatives as TAM inhibitors US Patent US10053465 (2018)
- Li, Y; Burns, DM; Feng, H; Glenn, J; He, C; Mei, S; Pan, J; Wang, X; Ye, Y Pyrrolopyrimidine derivatives as TAM inhibitors US Patent US11136326 (2021)
- Li, Y; Wang, X; He, C Pyrrolotriazine compounds as TAM inhibitors US Patent US10442810 (2019)
- Li, Y; Wang, X; He, C Pyrrolotriazine compounds as tam inhibitors US Patent US9981975 (2018)
- Price, S; Bender, SG; Yahn, R; Till, NA; Varady, S; LaLonde, RL Searching for an ideal SERM: Mining tamoxifen structure-activity relationships. Bioorg Med Chem Lett 52: (2021)
- Li, Y; Burns, DM Bicyclic fused pyrimidine compounds as TAM inhibitors US Patent US10005788 (2018)
- Ahmed, NS; Elghazawy, NH; ElHady, AK; Engel, M; Hartmann, RW; Abadi, AH Design and synthesis of novel tamoxifen analogues that avoid CYP2D6 metabolism. Eur J Med Chem 112: 171-9 (2016)
- Rivera-Guevara, C; Pérez-Alvarez, V; García-Becerra, R; Ordaz-Rosado, D; Morales-Ríos, MS; Hernández-Gallegos, E; Cooney, AJ; Bravo-Gómez, ME; Larrea, F; Camacho, J Genomic action of permanently charged tamoxifen derivatives via estrogen receptor-alpha. Bioorg Med Chem 18: 5593-601 (2010)
- Shagufta, na; Ahmad, I; Nelson, DJ; Hussain, MI; Nasar, NA Potential of covalently linked tamoxifen hybrids for cancer treatment: recent update. RSC Med Chem 15: 1877-1898
- Li, Y; Burns, DM Fused bicyclic 1,2,4-triazine compounds as TAM inhibitors US Patent US9708333 (2017)
- Xiong, R; Patel, HK; Gutgesell, LM; Zhao, J; Delgado-Rivera, L; Pham, TN; Zhao, H; Carlson, K; Martin, T; Katzenellenbogen, JA; Moore, TW; Tonetti, DA; Thatcher, GR Selective Human Estrogen Receptor Partial Agonists (ShERPAs) for Tamoxifen-Resistant Breast Cancer. J Med Chem 59: 219-37 (2016)
- Allen, S; Boys, ML; Cook, A; Gaudino, J; Hinklin, RJ; Laird, E; McNulty, OT; Metcalf, AT; Newhouse, B; Robinson, JE Bicyclic fused pyridine compounds as inhibitors of TAM kinases US Patent US11247990 (2022)
- Hasegawa, M; Yasuda, Y; Tanaka, M; Nakata, K; Umeda, E; Wang, Y; Watanabe, C; Uetake, S; Kunoh, T; Shionyu, M; Sasaki, R; Shiina, I; Mizukami, T A novel tamoxifen derivative, ridaifen-F, is a nonpeptidic small-molecule proteasome inhibitor. Eur J Med Chem 71: 290-305 (2014)
- Bekaii-Saab, TS; Perloff, MD; Weemhoff, JL; Greenblatt, DJ; von Moltke, LL Interactions of tamoxifen, N-desmethyltamoxifen and 4-hydroxytamoxifen with P-glycoprotein and CYP3A. Biopharm Drug Dispos 25: 283-9 (2004)
- Tsukuda, S; Kusayanagi, T; Umeda, E; Watanabe, C; Tosaki, YT; Kamisuki, S; Takeuchi, T; Takakusagi, Y; Shiina, I; Sugawara, F Ridaifen B, a tamoxifen derivative, directly binds to Grb10 interacting GYF protein 2. Bioorg Med Chem 21: 311-20 (2012)
- Chao, EY; Collins, JL; Gaillard, S; Miller, AB; Wang, L; Orband-Miller, LA; Nolte, RT; McDonnell, DP; Willson, TM; Zuercher, WJ Structure-guided synthesis of tamoxifen analogs with improved selectivity for the orphan ERRgamma. Bioorg Med Chem Lett 16: 821-4 (2006)
- Carpenter, C; Sorenson, RJ; Jin, Y; Klossowski, S; Cierpicki, T; Gnegy, M; Showalter, HD Design and synthesis of triarylacrylonitrile analogues of tamoxifen with improved binding selectivity to protein kinase C. Bioorg Med Chem 24: 5495-5504 (2016)
- Palermo, AF; Diennet, M; El Ezzy, M; Williams, BM; Cotnoir-White, D; Mader, S; Gleason, JL Incorporation of histone deacetylase inhibitory activity into the core of tamoxifen - A new hybrid design paradigm. Bioorg Med Chem 26: 4428-4440 (2018)
- Suárez, RM; Chevot, F; Cavagnino, A; Saettel, N; Radvanyi, F; Piguel, S; Bernard-Pierrot, I; Stoven, V; Legraverend, M Inhibitors of the TAM subfamily of tyrosine kinases: synthesis and biological evaluation. Eur J Med Chem 61: 2-25 (2013)
- Hinklin, RJ; Allen, S; Barbour, P; Cook, A; Dahlke, J; Gaudino, J; Laird, E; McNulty, OT; Zhao, Q Pyrazolo[3,4-b]pyridine compounds as inhibitors of TAM and MET kinases US Patent US11104676 (2021)
- Kahraman, M; Govek, SP; Nagasawa, JY; Lai, A; Bonnefous, C; Douglas, K; Sensintaffar, J; Liu, N; Lee, K; Aparicio, A; Kaufman, J; Qian, J; Shao, G; Prudente, R; Joseph, JD; Darimont, B; Brigham, D; Heyman, R; Rix, PJ; Hager, JH; Smith, ND Maximizing ER-α Degradation Maximizes Activity in a Tamoxifen-Resistant Breast Cancer Model: Identification of GDC-0927. ACS Med Chem Lett 10: 50-55 (2019)
- Govek, SP; Bonnefous, C; Julien, JD; Nagasawa, JY; Kahraman, M; Lai, AG; Douglas, KL; Aparicio, AM; Darimont, BD; Grillot, KL; Joseph, JD; Kaufman, JA; Lee, KJ; Lu, N; Moon, MJ; Prudente, RY; Sensintaffar, J; Rix, PJ; Hager, JH; Smith, ND Selective estrogen receptor degraders with novel structural motifs induce regression in a tamoxifen-resistant breast cancer xenograft. Bioorg Med Chem Lett 29: 367-372 (2019)
- Shoda, T; Kato, M; Harada, R; Fujisato, T; Okuhira, K; Demizu, Y; Inoue, H; Naito, M; Kurihara, M Synthesis and evaluation of tamoxifen derivatives with a long alkyl side chain as selective estrogen receptor down-regulators. Bioorg Med Chem 23: 3091-6 (2015)
- Yen, S; Liao, C; Wang, H; Chen, P; Pan, Y; Li, T; Chen, B; Chiou, S Heterocycle compounds as Tyro3, Axl and MerTK (TAM) family of receptor tyrosine kinase inhibitors US Patent US11407757 (2022)
- Paolino, M; Choidas, A; Wallner, S; Pranjic, B; Uribesalgo, I; Loeser, S; Jamieson, AM; Langdon, WY; Ikeda, F; Fededa, JP; Cronin, SJ; Nitsch, R; Schultz-Fademrecht, C; Eickhoff, J; Menninger, S; Unger, A; Torka, R; Gruber, T; Hinterleitner, R; Baier, G; Wolf, D; Ullrich, A; Klebl, BM; Penninger, JM The E3 ligase Cbl-b and TAM receptors regulate cancer metastasis via natural killer cells. Nature 507: 508-12
- Lai, A; Kahraman, M; Govek, S; Nagasawa, J; Bonnefous, C; Julien, J; Douglas, K; Sensintaffar, J; Lu, N; Lee, KJ; Aparicio, A; Kaufman, J; Qian, J; Shao, G; Prudente, R; Moon, MJ; Joseph, JD; Darimont, B; Brigham, D; Grillot, K; Heyman, R; Rix, PJ; Hager, JH; Smith, ND Identification of GDC-0810 (ARN-810), an Orally Bioavailable Selective Estrogen Receptor Degrader (SERD) that Demonstrates Robust Activity in Tamoxifen-Resistant Breast Cancer Xenografts. J Med Chem 58: 4888-904 (2015)
- Nagasawa, J; Govek, S; Kahraman, M; Lai, A; Bonnefous, C; Douglas, K; Sensintaffar, J; Lu, N; Lee, K; Aparicio, A; Kaufman, J; Qian, J; Shao, G; Prudente, R; Joseph, JD; Darimont, B; Brigham, D; Maheu, K; Heyman, R; Rix, PJ; Hager, JH; Smith, ND Identification of an Orally Bioavailable Chromene-Based Selective Estrogen Receptor Degrader (SERD) That Demonstrates Robust Activity in a Model of Tamoxifen-Resistant Breast Cancer. J Med Chem 61: 7917-7928 (2018)
- Baladi, T; Aziz, J; Dufour, F; Abet, V; Stoven, V; Radvanyi, F; Poyer, F; Wu, TD; Guerquin-Kern, JL; Bernard-Pierrot, I; Garrido, SM; Piguel, S Design, synthesis, biological evaluation and cellular imaging of imidazo[4,5-b]pyridine derivatives as potent and selective TAM inhibitors. Bioorg Med Chem 26: 5510-5530 (2018)
- Kraft, KS; Ruenitz, PC; Bartlett, MG Carboxylic acid analogues of tamoxifen: (Z)-2-[p-(1, 2-diphenyl-1-butenyl)phenoxy]-N,N-dimethylethylamine. Estrogen receptor affinity and estrogen antagonist effects in MCF-7 cells. J Med Chem 42: 3126-33 (1999)
- Sahin, C; Magomedova, L; Ferreira, TAM; Liu, J; Tiefenbach, J; Alves, PS; Queiroz, FJG; Oliveira, AS; Bhattacharyya, M; Grouleff, J; Nogueira, PCN; Silveira, ER; Moreira, DC; Leite, JRSA; Brand, GD; Uehling, D; Poda, G; Krause, H; Cummins, CL; Romeiro, LAS J Med Chem 65: 1961-1978 (2022)
- Wágner, G; Mocking, TAM; Arimont, M; Provensi, G; Rani, B; Silva-Marques, B; Latacz, G; Da Costa Pereira, D; Karatzidou, C; Vischer, HF; Wijtmans, M; Kieć-Kononowicz, K; de Esch, IJP; Leurs, R J Med Chem 62: 10848-10866 (2019)
- BENITA, S; TAM, J; NASSAR, T; FREEMAN, N US Patent US20240189287 (2024)
- McDermott, U; Sharma, SV; Dowell, L; Greninger, P; Montagut, C; Lamb, J; Archibald, H; Raudales, R; Tam, A; Lee, D; Rothenberg, SM; Supko, JG; Sordella, R; Ulkus, LE; Iafrate, AJ; Maheswaran, S; Njauw, CN; Tsao, H; Drew, L; Hanke, JH; Ma, XJ; Erlander, MG; Gray, NS; Haber, DA; Settleman, J Proc Natl Acad Sci U S A 104: 19936-41 (2007)
- Le Brazidec, JY; Pasis, A; Tam, B; Boykin, C; Wang, D; Marcotte, DJ; Claassen, G; Chong, JH; Chao, J; Fan, J; Nguyen, K; Silvian, L; Ling, L; Zhang, L; Choi, M; Teng, M; Pathan, N; Zhao, S; Li, T; Taveras, A Bioorg Med Chem Lett 22: 4033-7 (2012)
- Zhang, L; Fan, J; Chong, JH; Cesena, A; Tam, BY; Gilson, C; Boykin, C; Wang, D; Aivazian, D; Marcotte, D; Xiao, G; Le Brazidec, JY; Piao, J; Lundgren, K; Hong, K; Vu, K; Nguyen, K; Gan, LS; Silvian, L; Ling, L; Teng, M; Reff, M; Takeda, N; Timple, N; Wang, Q; Morena, R; Khan, S; Zhao, S; Li, T; Lee, WC; Taveras, AG; Chao, J Bioorg Med Chem Lett 21: 5633-7 (2011)
- Staben, ST; Feng, JA; Lyle, K; Belvin, M; Boggs, J; Burch, JD; Chua, CC; Cui, H; DiPasquale, AG; Friedman, LS; Heise, C; Koeppen, H; Kotey, A; Mintzer, R; Oh, A; Roberts, DA; Rouge, L; Rudolph, J; Tam, C; Wang, W; Xiao, Y; Young, A; Zhang, Y; Hoeflich, KP J Med Chem 57: 1033-45 (2014)
- Tan, L; Gurbani, D; Weisberg, EL; Hunter, JC; Li, L; Jones, DS; Ficarro, SB; Mowafy, S; Tam, CP; Rao, S; Du, G; Griffin, JD; Sorger, PK; Marto, JA; Westover, KD; Gray, NS Bioorg Med Chem 25: 838-846 (2017)
- Franzini, M; Ye, XM; Adler, M; Aubele, DL; Garofalo, AW; Gauby, S; Goldbach, E; Probst, GD; Quinn, KP; Santiago, P; Sham, HL; Tam, D; Truong, A; Ren, Z Bioorg Med Chem Lett 23: 1967-73 (2013)
- Bienaymé, H; Chêne, L; Grisoni, S; Grondin, A; Kaloun, el-B; Poigny, S; Rahali, H; Tam, E Bioorg Med Chem Lett 16: 4830-3 (2006)
- Isabel, E; Black, WC; Bayly, CI; Grimm, EL; Janes, MK; McKay, DJ; Nicholson, DW; Rasper, DM; Renaud, J; Roy, S; Tam, J; Thornberry, NA; Vaillancourt, JP; Xanthoudakis, S; Zamboni, R Bioorg Med Chem Lett 13: 2137-40 (2003)
- Nguyen, PQ; Luu, TT; Bai, Y; Nguyen, GK; Pervushin, K; Tam, JP J Nat Prod 78: 695-704 (2015)
- Foote, KM; Blades, K; Cronin, A; Fillery, S; Guichard, SS; Hassall, L; Hickson, I; Jacq, X; Jewsbury, PJ; McGuire, TM; Nissink, JW; Odedra, R; Page, K; Perkins, P; Suleman, A; Tam, K; Thommes, P; Broadhurst, R; Wood, C J Med Chem 56: 2125-38 (2013)
- Dalal, K; Morin, H; Ban, F; Shepherd, A; Fernandez, M; Tam, KJ; Li, H; LeBlanc, E; Lack, N; Prinz, H; Rennie, PS; Cherkasov, A Eur J Med Chem 157: 1164-1173 (2018)
- Xu, B; Wang, ZP; Liu, Q; Yang, X; Li, X; Huang, D; Qiu, Y; Tam, KY; Zhang, SL; He, Y Eur J Med Chem 214: (2021)
- Xiang, J; Wan, ZK; Li, HQ; Ipek, M; Binnun, E; Nunez, J; Chen, L; McKew, JC; Mansour, TS; Xu, X; Suri, V; Tam, M; Xing, Y; Li, X; Hahm, S; Tobin, J; Saiah, E J Med Chem 51: 4068-71 (2008)
- Gopalsamy, A; Shi, M; Golas, J; Vogan, E; Jacob, J; Johnson, M; Lee, F; Nilakantan, R; Petersen, R; Svenson, K; Chopra, R; Tam, MS; Wen, Y; Ellingboe, J; Arndt, K; Boschelli, F J Med Chem 51: 373-5 (2008)
- Shishodia, S; Demetriades, M; Zhang, D; Tam, NY; Maheswaran, P; Clunie-O'Connor, C; Tumber, A; Leung, IKH; Ng, YM; Leissing, TM; El-Sagheer, AH; Salah, E; Brown, T; Aik, WS; McDonough, MA; Schofield, CJ J Med Chem 64: 16609-16625 (2021)
- El Abdellaoui, H; Varaprasad, CV; Barawkar, D; Chakravarty, S; Maderna, A; Tam, R; Chen, H; Allan, M; Wu, JZ; Appleby, T; Yan, S; Zhang, W; Lang, S; Yao, N; Hamatake, R; Hong, Z Bioorg Med Chem Lett 16: 5561-6 (2006)
- Li, W; Li, J; Wu, Y; Wu, J; Hotchandani, R; Cunningham, K; McFadyen, I; Bard, J; Morgan, P; Schlerman, F; Xu, X; Tam, S; Goldman, SJ; Williams, C; Sypek, J; Mansour, TS J Med Chem 52: 1799-802 (2009)
- Moretto, AF; Kirincich, SJ; Xu, WX; Smith, MJ; Wan, ZK; Wilson, DP; Follows, BC; Binnun, E; Joseph-McCarthy, D; Foreman, K; Erbe, DV; Zhang, YL; Tam, SK; Tam, SY; Lee, J Bioorg Med Chem 14: 2162-77 (2006)
- Cheng, CY; Wu, SC; Hsin, LW; Tam, SW J Med Chem 35: 2243-7 (1992)
- Green, N; Hu, Y; Janz, K; Li, HQ; Kaila, N; Guler, S; Thomason, J; Joseph-McCarthy, D; Tam, SY; Hotchandani, R; Wu, J; Huang, A; Wang, Q; Leung, L; Pelker, J; Marusic, S; Hsu, S; Telliez, JB; Hall, JP; Cuozzo, JW; Lin, LL J Med Chem 50: 4728-45 (2007)
- Krantz, A; Spencer, RW; Tam, TF; Liak, TJ; Copp, LJ; Thomas, EM; Rafferty, SP J Med Chem 33: 464-79 (1990)
- Wang, G; Dyatkina, N; Prhavc, M; Williams, C; Serebryany, V; Hu, Y; Huang, Y; Wan, J; Wu, X; Deval, J; Fung, A; Jin, Z; Tan, H; Shaw, K; Kang, H; Zhang, Q; Tam, Y; Stoycheva, A; Jekle, A; Smith, DB; Beigelman, L J Med Chem 62: 4555-4570 (2019)
- Cain, GA; Christos, TE; Johnson, AL; Pottorf, RS; Confalone, PN; William Tam, S; Schmidt, WK Bioorg Med Chem Lett 3: 2055-2060 (1993)
- Ferdinandy, P; Csont, TB; Csonka, C; Kedvesné Kupai, K; Kovács, L; Kis-Tamás, A; Takács, FT; Kónya, D; Medgyes, G; Cseh, S; Hajdú, I; Lörincz, Z; Dormán, G; Görbe, A US Patent US9487462 (2016)
- ChEBML_35744 Displacement of [3H]tamoxifen from antiestrogen binding site (AEBS)
- ChEMBL_1519534 (CHEMBL3625856) Antagonist activity at ERalpha receptor in human MCF7 cells in presence of 0.25 uM tamoxifen
- ChEMBL_1519535 (CHEMBL3625857) Agonist activity at progesterone receptor in human MCF7 cells in presence of 0.25 uM tamoxifen
- ChEMBL_550766 (CHEMBL1008660) Displacement of [3H]tamoxifen from antiestrogen binding site in Sprague-Dawley rat liver by liquid scintillation counting
- ChEMBL_2022297 (CHEMBL4676110) Induction of ERalpha degradation in human MCF7 cells assessed as decrease in ERalpha expression in presence of tamoxifen by Western blot analysis
- ChEMBL_1649839 (CHEMBL3998973) Induction of ERalpha degradation in tamoxifen-sensitive human MCF7:WS8 cells after 24 hrs by CellTag 700 staining based In-cell western assay
- ChEMBL_1926284 (CHEMBL4429356) Downregulation of ER-alpha expression in human tamoxifen-resistant T47D cells over-expressing PKC-alpha measured after 5 days by Western blot analysis
- ChEMBL_1649840 (CHEMBL3998974) Antagonist activity at ERalpha in tamoxifen-sensitive human MCF7:WS8 cells assessed as inhibition of estradiol-induced response after 18 hrs by luciferase reporter gene assay
- ChEMBL_1869278 (CHEMBL4370344) Induction of ERalpha degradation in human MCF7 cells assessed as reduction in ERalpha protein expression after 5 hrs in absence of 0.25 uM tamoxifen by formaldehyde-staining based assay
- ChEMBL_1869280 (CHEMBL4370346) Induction of ERalpha degradation in human MCF7 cells assessed as reduction in ERalpha protein expression after 5 hrs in presence of 0.25 uM tamoxifen by formaldehyde-staining based assay
- ChEMBL_1837761 (CHEMBL4337894) Inhibition of Influenza A virus RNA-dependent RNA polymerase PA N-terminal endonuclease using [6-FAM]AATCGCAGGCAGCACTC[TAM] substrate and measured over 45 mins by fluorescence based assay
- ChEMBL_2156795 (CHEMBL5041455) Inhibition of TAM-3Y-F2Pmp peptide binding to N-terminal His6-fused STEP32 (282 to 563 residues) (unknown origin) expressed in Escherichia coli BL21(DE3) by SPR analysis
- ChEMBL_2156794 (CHEMBL5041454) Inhibition of TAM-3Y-F2Pmp peptide binding to N-terminal His6-fused STEP32 (282 to 563 residues) (unknown origin) expressed in Escherichia coli BL21(DE3) measured for 30 mins by fluorescence polarization method
- Verification on Inhibitory Activities of TAM Receptor Inhibiting Compounds on Tyro 3, Axl, and Mer For evaluation of inhibitory activities of the compounds according to the present invention on Tyro 3, Axl, and Mer, the following test was carried out.The specific test method followed the method provided by Cisbio. The compounds of the examples were prepared at various concentrations, followed by addition of Tyro 3, Axl, or Mer and substrate peptides, and then ATP was added to initiate a reaction. After 1 hour, the reaction was stopped by addition of a solution containing EDTA. Thereafter, the amount of phosphorylated peptides was measured. Here, the phosphorylated peptides were treated with an europium (Eu)-labeled antibody recognizing phosphorylated peptides, and then after 1 hour, excited by irradiation of the light of a wavelength of 320 or 340 nm using an Envision Reader. The amount of light emitted at 665 nm was measured to investigate the degrees of inhibition of TAM receptors. The TAM receptor inhibitory activities (IC50 values) of the respective compounds were analyzed using the GraphPad Prism program, and tabulated in Tables 1 to 3 below. LDC1267 (CAS no. 1361030-48-9, Calbiochem), which is a TAM receptor inhibitor compound, was used as a positive control.
- Competition Displacement Assay Compound binding to CB1R was assessed in competition displacement assays using [3H]CP-55,940 as the radioligand and crude membranes from mouse brain. See Tam, J., Vemuri, V. K., Liu, J., Batkai, S., Mukhopadhyay, B., Godlewski, G., Osei-Hyiaman, D., Ohnuma, S., Ambudkar, S. V., Pickel, J., et al., J. Clin. Invest. 2010, 120, 2953-2966.
- competition displacement assays Compound binding to CB1R was assessed in competition displacement assays using [3H]CP-55,940 as the radioligand and crude membranes from mouse brain. See Tam, J., Vemuri, V. K., Liu, J., Batkai, S., Mukhopadhyay, B., Godlewski, G., Osei-Hyiaman, D., Ohnuma, S., Ambudkar, S. V., Pickel, J., et al., J. Clin. Invest. 2010, 120, 2953-2966. All data were in triplicates with Ki values determined from three independent experiments.
- Endonuclease Activity Assay Endonuclease activity assays were carried out in Black Costar 96-well plates. Each well contained a total volume of 100 μL comprised of: buffer (20 mM Tris, 150 mM NaCl, 2 mM MnCl2, 10 mM β-mercaptoethanol, 0.2% Triton-X100, pH=8.0), influenza PA endonuclease (25 nM) prepared from Example 1, inhibitor (various concentrations) in buffer, and fluorescent ssDNA-oligo substrate (200 nM). A single-stranded, 17-mer DNA substrate labeled with a 5′-FAM fluorophore and a 3′-TAMRA quencher ([6-FAM]AATCGCAGGCAGCACTC[TAM]) (SEQ ID NO:2) synthesized by Sigma-Aldrich was employed to measure endonucleic cleavage. Upon addition of the substrate, the change in fluorescence was measured over 45 minutes at 37° C. (excitation: 485 nm; emission 528 nm). The positive control wells contained no inhibitor for preliminary screens, and were set as an arbitrary 100% activity. Compounds that exhibited >80% inhibition at a concentration of 200 μM were re-evaluated at a concentration of 50 μM. Confirmation screens employed EGCG (Epigallocatechin 3-gallate), a previously validated inhibitor, as a positive control. The gain was set to 100 and the first 10 data points (the first 10 minutes) were excluded from the activity calculations. Dose-response curves were generated, fitted, and analyzed using Origin8 graphing software. Dose-response curves were compiled and IC50 values determined for compounds exhibiting >50% inhibition at 50 μM.
- TAM Enzymatic Assay The assay buffer contained 50 mM HEPES, pH7.5, 10 mM MgCl2, 1 mM EGTA, 0.01% NP-40 and 2 mM DTT. 0.5 ul test compounds dissolved in DMSO were transferred from compound plates to white 384-well assay plates (Greiner LUMITRAC plates). The final concentration of DMSO was 2.5%. Enzyme solutions of 13.8 nM AXL (Life Technologies, PV4275), or 4.1 nM c-MER (Life Technologies, PV4112), or 0.366 nM TYRO3 (Life Technologies, PR7480A) were prepared in assay buffer. A 1 mM stock solution of peptide substrate Biotin-EQEDEPEGDYFEWLE-amide (Quality Controlled Biochemicals, MA) dissolved in DMSO was diluted to 1 uM in assay buffer containing 100 uM ATP (for AXL and c-MER assays) or 20 uM ATP (for TYRO3 assay). 10 ul enzyme solution (or assay buffer for the enzyme blank) was added to the appropriate wells in each plate, and then 10 ul/well substrate solution was added to initiate the reaction. The plate was protected from light and incubated at room temperature for 60 min. The reaction was stopped by adding 10 ul detection solution containing 50 mM Tris-HCl, pH7.8, 150 mM NaCl, 0.05% BSA, 45 mM EDTA, 180 nM SA-APC (Perkin Elmer, CR130-100) and 3 nM Eu-W1024 anti-phosphotyrosine PY20 (Perkin Elmer, AD0067). The plate was incubated for 1 h at room temperature, and HTRF (homogenous time resolved fluorescence) signal was measured on a PHERAstar FS plate reader (BMG labtech).
- TAM Enzymatic Assay The kinase assay buffer contained 50 mM HEPES, pH7.5, 10 mM MgCl2, 1 mM EGTA, 0.01% NP-40 and 2 mM DTT. 0.1 ul test compounds dissolved in DMSO were transferred from compound plates to white 384-well assay plates (Greiner LUMITRAC plates). The final concentration of DMSO was 1.25%. Enzyme solutions of 5.1 nM phosphor-Axl, or 0.0625 nM c-Mer (Carna Biosciences, 08-108), or 0.366 nM Tyro3 (Life Technologies, PR7480A) were prepared in assay buffer. A 1 mM stock solution of peptide substrate Biotin-EQEDEPEGDYFEWLE-amide SEQ ID NO: 1 (Quality Controlled Biochemicals, MA) dissolved in DMSO was diluted to 1 uM in assay buffer containing 2000 uM ATP. 4 ul enzyme solution (or assay buffer for the enzyme blank) was added to the appropriate wells in each plate, and then 4 ul/well substrate solution was added to initiate the reaction. The plate was protected from light and incubated at room temperature for 60 min. The reaction was stopped by adding 4 ul detection solution containing 50 mM Tris-HCl, pH7.8, 150 mM NaCl, 0.05% BSA, 45 mM EDTA, 180 nM SA-APC (Perkin Elmer, CR130-100) and 3 nM Eu-W1024 anti-phosphotyrosine PY20 (Perkin Elmer, AD0067). The plate was incubated for 1 h at room temperature, and HTRF (homogenous time resolved fluorescence) signal was measured on a PHERAstar FS plate reader (BMG labtech).
- TAM Enzymatic Assay The kinase assay buffer contained 50 mM HEPES, pH7.5, 10 mM MgCl2, 1 mM EGTA, 0.01% NP-40 and 2 mM DTT. 0.1 ul test compounds dissolved in DMSO were transferred from compound plates to white 384-well assay plates (Greiner LLIMITRAC plates). The final concentration of DMSO was 1.25%. Enzyme solutions of 5.1 nM phosphor-Axl, or 0.0625 nM c-Mer (Carna Biosciences, 08-108), or 0.366 nM Tyro3 (Life Technologies, PR7480A) were prepared in assay buffer. A 1 mM stock solution of peptide substrate Biotin-EQEDEPEGDYFEWLE-amide SEQ ID NO: 1 (Quality Controlled Biochemicals, MA) dissolved in DMSO was diluted to 1 uM in assay buffer containing 2000 uM ATP. 4 ul enzyme solution (or assay buffer for the enzyme blank) was added to the appropriate wells in each plate, and then 4 ul/well substrate solution was added to initiate the reaction. The plate was protected from light and incubated at room temperature for 60 min. The reaction was stopped by adding 4 ul detection solution containing 50 mM Tris-HCl, pH7.8, 150 mM NaCl, 0.05% BSA, 45 mM EDTA, 180 nM SA-APC (Perkin Elmer, CR130-100) and 3 nM Eu-W1024 anti-phosphotyrosine PY20 (Perkin Elmer, AD0067). The plate was incubated for 1 h at room temperature, and HTRF (homogenous time resolved fluorescence) signal was measured on a PHERAstar FS plate reader (BMG labtech). Percentage of inhibition was calculated for each concentration and IC50 value was generated from curve fitting with GraphPad Prism software.
- TAM Enzymatic Assay The kinase assay buffer contained 50 mM HEPES, pH7.5, 10 mM MgCl2, 1 mM EGTA, 0.01% NP-40 and 2 mM DTT. 0.1 ul test compounds dissolved in DMSO were transferred from compound plates to white 384-well assay plates (Greiner LUMITRAC plates). The final concentration of DMSO was 1.25%. Enzyme solutions of 5.1 nM phosphor-Axl, or 0.0625 nM c-Mer (Carna Biosciences, 08-108), or 0.366 nM Tyro3 (Life Technologies, PR7480A) were prepared in assay buffer. A 1 mM stock solution of peptide substrate Biotin-EQEDEPEGDYFEWLE-amide SEQ ID NO: 1 (Quality Controlled Biochemicals, MA) dissolved in DMSO was diluted to 1 uM in assay buffer containing 2000 uM ATP. 4 ul enzyme solution (or assay buffer for the enzyme blank) was added to the appropriate wells in each plate, and then 4 ul/well substrate solution was added to initiate the reaction. The plate was protected from light and incubated at room temperature for 60 min. The reaction was stopped by adding 4 ul detection solution containing 50 mM Tris-HCl, pH7.8, 150 mM NaCl, 0.05% BSA, 45 mM EDTA, 180 nM SA-APC (Perkin Elmer, CR130-100) and 3 nM Eu-W1024 anti-phosphotyrosine PY20 (Perkin Elmer, AD0067). The plate was incubated for 1 h at room temperature, and HTRF (homogenous time resolved fluorescence) signal was measured on a PHERAstar FS plate reader (BMG labtech). Percentage of inhibition was calculated for each concentration and IC50 value was generated from curve fitting with GraphPad Prism software.
- TAM Enzymatic Assay The kinase assay buffer contained 50 mM HEPES, pH7.5, 10 mM MgCl2, 1 mM EGTA, 0.01% NP-40 and 2 mM DTT. 0.1 ul test compounds dissolved in DMSO were transferred from compound plates to white 384-well assay plates (Greiner LUMITRAC plates). The final concentration of DMSO was 1.25%. Enzyme solutions of 5.1 nM phosphor-Axl, or 0.0625 nM c-Mer (Carna Biosciences, 08-108), or 0.366 nM Tyro3 (Life Technologies, PR7480A) were prepared in assay buffer. A 1 mM stock solution of peptide substrate Biotin-EQEDEPEGDYFEWLE-amide SEQ ID NO:1 (Quality Controlled Biochemicals, MA) dissolved in DMSO was diluted to 1 uM in assay buffer containing 2000 uM ATP. 4 ul enzyme solution (or assay buffer for the enzyme blank) was added to the appropriate wells in each plate, and then 4 ul/well substrate solution was added to initiate the reaction. The plate was protected from light and incubated at room temperature for 60 min. The reaction was stopped by adding 4 ul detection solution containing 50 mM Tris-HCl, pH7.8, 150 mM NaCl, 0.05% BSA, 45 mM EDTA, 180 nM SA-APC (Perkin Elmer, CR130-100) and 3 nM Eu-W1024 anti-phosphotyrosine PY20 (Perkin Elmer, AD0067). The plate was incubated for 1 h at room temperature, and HTRF (homogenous time resolved fluorescence) signal was measured on a PHERAstar FS plate reader (BMG labtech). Percentage of inhibition was calculated for each concentration and IC50 value was generated from curve fitting with GraphPad Prism software.
- TAM Kinase Assay The kinase assay buffer contained 50 mM HEPES, pH7.5, 10 mM MgCl2, 1 mM EGTA, 0.01% NP-40 and 2 mM DTT. 0.1 ul test compounds dissolved in DMSO were transferred from compound plates to white 384-well assay plates (Greiner LUMITRAC plates). The final concentration of DMSO was 1.25%. Enzyme solutions of 5.1 nM phosphor-Axl (see Axl autophosphorylation assay above), or 0.0625 nM c-Mer (Carna Biosciences, 08-108), or 0.366 nM Tyro3 (Life Technologies, PR7480A) were prepared in assay buffer. A 1 mM stock solution of peptide substrate Biotin-EQEDEPEGDYFEWLE-amide SEQ ID NO: 1 (Quality Controlled Biochemicals, MA) dissolved in DMSO was diluted to 1 uM in assay buffer containing 2000 uM ATP. 4 ul enzyme solution (or assay buffer for the enzyme blank) was added to the appropriate wells in each plate, and then 4 ul/well substrate solution was added to initiate the reaction. The plate was protected from light and incubated at room temperature for 60 min. The reaction was stopped by adding 4 ul detection solution containing 50 mM Tris-HCl, pH7.8, 150 mM NaCl, 0.05% BSA, 45 mM EDTA, 180 nM SA-APC (Perkin Elmer, CR130-100) and 3 nM Eu W1024 anti-phosphotyrosine PY20 (Perkin Elmer, AD0067). The plate was incubated for 1 h at room temperature, and HTRF (homogenous time resolved fluorescence) signal was measured on a PHERAstar FS plate reader (BMG labtech). Percentage of inhibition was calculated for each concentration and IC50 value was generated from curve fitting with GraphPad Prism software.
- TAM Kinase Assay The kinase assay buffer contained 50 mM HEPES, pH7.5, 10 mM MgCl2, 1 mM EGTA, 0.01% NP-40 and 2 mM DTT. 0.1 ul test compounds dissolved in DMSO were transferred from compound plates to white 384-well assay plates (Greiner LUMITRAC plates). The final concentration of DMSO was 1.25%. Enzyme solutions of 5.1 nM phosphor-Axl (see Axl autophosphorylation assay above), or 0.0625 nM c-Mer (Carna Biosciences, 08-108), or 0.366 nM Tyro3 (Life Technologies, PR7480A) were prepared in assay buffer. A 1 mM stock solution of peptide substrate Biotin-EQEDEPEGDYFEWLE-amide SEQ ID NO: 1 (Quality Controlled Biochemicals, MA) dissolved in DMSO was diluted to 1 uM in assay buffer containing 2000 uM ATP. 4 ul enzyme solution (or assay buffer for the enzyme blank) was added to the appropriate wells in each plate, and then 4 ul/well substrate solution was added to initiate the reaction. The plate was protected from light and incubated at room temperature for 60 min. The reaction was stopped by adding 4 ul detection solution containing 50 mM Tris-HCl, pH7.8, 150 mM NaCl, 0.05% BSA, 45 mM EDTA, 180 nM SA-APC (Perkin Elmer, CR130-100) and 3 nM Eu-W1024 anti-phosphotyrosine PY20 (Perkin Elmer, AD0067). The plate was incubated for 1 h at room temperature, and HTRF (homogenous time resolved fluorescence) signal was measured on a PHERAstar FS plate reader (BMG labtech). Percentage of inhibition was calculated for each concentration and IC50 value was generated from curve fitting with GraphPad Prism software.
 Tamoxifen, 1 BDBM85467
Tamoxifen, 1 BDBM85467 CHEMBL1213783 BDBM50323911 Tamoxifen methyl iodide
CHEMBL1213783 BDBM50323911 Tamoxifen methyl iodide CHEMBL1213784 BDBM50323912 Tamoxifen ethyl bromide
CHEMBL1213784 BDBM50323912 Tamoxifen ethyl bromide Tamoxifen butyl bromide CHEMBL1213786 BDBM50323914
Tamoxifen butyl bromide CHEMBL1213786 BDBM50323914 Tamoxifen isopropyl bromide CHEMBL1213785 BDBM50323913
Tamoxifen isopropyl bromide CHEMBL1213785 BDBM50323913 2-{4-[(1Z)-1,2-diphenylbut-1-en-1-yl]phenoxy}-N,N-dimethylethanamine Nolvadex US20240217988, Example Tamoxifen cid_2733526 FOSTRIECIN SODIUM Tamoxifen Tamoxifen (8) CHEMBL83 (2-{4-[(1Z)-1,2-diphenylbut-1-en-1-yl]phenoxy}ethyl)dimethylamine NCGC00024928 Tamoxifen, 7 med.21724, Compound Tamoxifen BDBM20607
2-{4-[(1Z)-1,2-diphenylbut-1-en-1-yl]phenoxy}-N,N-dimethylethanamine Nolvadex US20240217988, Example Tamoxifen cid_2733526 FOSTRIECIN SODIUM Tamoxifen Tamoxifen (8) CHEMBL83 (2-{4-[(1Z)-1,2-diphenylbut-1-en-1-yl]phenoxy}ethyl)dimethylamine NCGC00024928 Tamoxifen, 7 med.21724, Compound Tamoxifen BDBM20607 TAM-UNI-d1c3dd9f-12 BDBM496740
TAM-UNI-d1c3dd9f-12 BDBM496740 TAM-UNI-d1c3dd9f-18 BDBM496741
TAM-UNI-d1c3dd9f-18 BDBM496741 TAM-UNI-d1c3dd9f-5 BDBM496739
TAM-UNI-d1c3dd9f-5 BDBM496739