Reaction Details Report a problem with these data
Report a problem with these data
 Report a problem with these data
Report a problem with these dataTarget
MAP kinase-activated protein kinase 2
Ligand
BDBM50395280
Substrate
n/a
Meas. Tech.
ChEMBL_860552 (CHEMBL2167126)
IC50
600±n/a nM
Citation
 Kosugi, T; Mitchell, DR; Fujino, A; Imai, M; Kambe, M; Kobayashi, S; Makino, H; Matsueda, Y; Oue, Y; Komatsu, K; Imaizumi, K; Sakai, Y; Sugiura, S; Takenouchi, O; Unoki, G; Yamakoshi, Y; Cunliffe, V; Frearson, J; Gordon, R; Harris, CJ; Kalloo-Hosein, H; Le, J; Patel, G; Simpson, DJ; Sherborne, B; Thomas, PS; Suzuki, N; Takimoto-Kamimura, M; Kataoka, K Mitogen-activated protein kinase-activated protein kinase 2 (MAPKAP-K2) as an antiinflammatory target: discovery and in vivo activity of selective pyrazolo[1,5-a]pyrimidine inhibitors using a focused library and structure-based optimization approach. J Med Chem 55:6700-15 (2012) [PubMed] Article
Kosugi, T; Mitchell, DR; Fujino, A; Imai, M; Kambe, M; Kobayashi, S; Makino, H; Matsueda, Y; Oue, Y; Komatsu, K; Imaizumi, K; Sakai, Y; Sugiura, S; Takenouchi, O; Unoki, G; Yamakoshi, Y; Cunliffe, V; Frearson, J; Gordon, R; Harris, CJ; Kalloo-Hosein, H; Le, J; Patel, G; Simpson, DJ; Sherborne, B; Thomas, PS; Suzuki, N; Takimoto-Kamimura, M; Kataoka, K Mitogen-activated protein kinase-activated protein kinase 2 (MAPKAP-K2) as an antiinflammatory target: discovery and in vivo activity of selective pyrazolo[1,5-a]pyrimidine inhibitors using a focused library and structure-based optimization approach. J Med Chem 55:6700-15 (2012) [PubMed] Article More Info.:
Target
Name:
MAP kinase-activated protein kinase 2
Synonyms:
MAP kinase-activated protein kinase 2 (MAPKAPK2) | MAP kinase-activated protein kinase 2 (MK2) | MAP kinase-activated protein kinase 2 (p38/MK2) | MAPK-Activated Protein Kinase 2 (MK2) | MAPK-activated protein kinase 2 | MAPK2_HUMAN | MAPKAP kinase 2 | MAPKAPK-2 | MAPKAPK2 | MK2
Type:
Serine/threonine-protein kinase
Mol. Mass.:
45579.87
Organism:
Homo sapiens (Human)
Description:
P49137
Residue:
400
Sequence:
MLSNSQGQSPPVPFPAPAPPPQPPTPALPHPPAQPPPPPPQQFPQFHVKSGLQIKKNAIIDDYKVTSQVLGLGINGKVLQIFNKRTQEKFALKMLQDCPKARREVELHWRASQCPHIVRIVDVYENLYAGRKCLLIVMECLDGGELFSRIQDRGDQAFTEREASEIMKSIGEAIQYLHSINIAHRDVKPENLLYTSKRPNAILKLTDFGFAKETTSHNSLTTPCYTPYYVAPEVLGPEKYDKSCDMWSLGVIMYILLCGYPPFYSNHGLAISPGMKTRIRMGQYEFPNPEWSEVSEEVKMLIRNLLKTEPTQRMTITEFMNHPWIMQSTKVPQTPLHTSRVLKEDKERWEDVKEEMTSALATMRVDYEQIKIKKIEDASNPLLLKRRKKARALEAAALAH
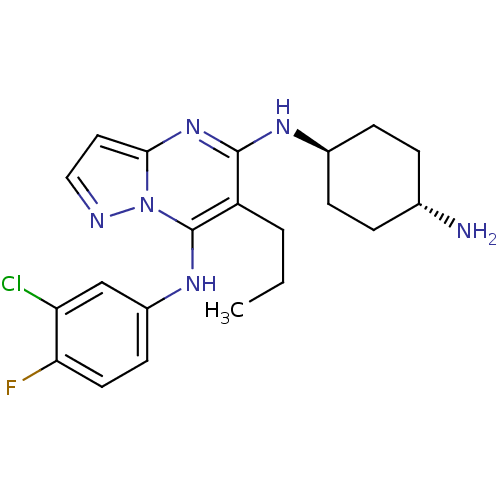
Inhibitor
Name:
BDBM50395280
Synonyms:
CHEMBL2163992
Type:
Small organic molecule
Emp. Form.:
C21H26ClFN6
Mol. Mass.:
416.923
SMILES:
CCCc1c(N[C@H]2CC[C@H](N)CC2)nc2ccnn2c1Nc1ccc(F)c(Cl)c1 |r,wU:6.5,wD:9.9,(41.91,-26.84,;40.57,-27.61,;40.57,-29.15,;39.23,-29.91,;39.23,-31.45,;40.57,-32.23,;41.9,-31.46,;41.9,-29.93,;43.24,-29.16,;44.58,-29.94,;45.91,-29.18,;44.57,-31.48,;43.23,-32.24,;37.91,-32.22,;36.58,-31.45,;35.1,-31.93,;34.19,-30.68,;35.1,-29.43,;36.58,-29.91,;37.91,-29.14,;37.91,-27.6,;36.58,-26.82,;36.59,-25.29,;35.26,-24.51,;33.92,-25.28,;32.59,-24.51,;33.92,-26.83,;32.59,-27.6,;35.25,-27.59,)|