 BDBM356619 US10214478, Compound NM-001
BDBM356619 US10214478, Compound NM-001 BDBM356624 US10214478, Compound NM-008
BDBM356624 US10214478, Compound NM-008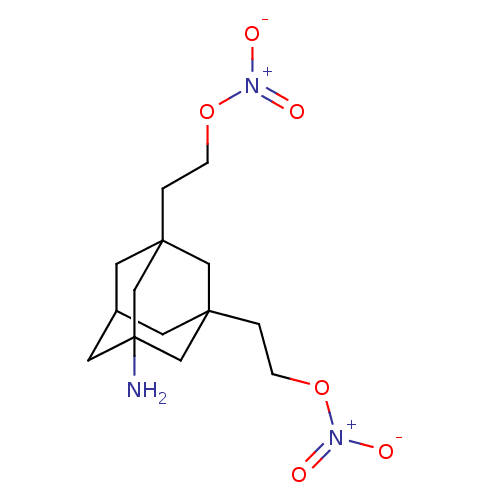 BDBM356628 US10214478, Compound NM-009
BDBM356628 US10214478, Compound NM-009 BDBM356629 US10214478, Compound NM-011
BDBM356629 US10214478, Compound NM-011 BDBM356630 US10214478, Compound NM-012
BDBM356630 US10214478, Compound NM-012 US10214478, Compound NM-002 BDBM356620
US10214478, Compound NM-002 BDBM356620 US10214478, Compound NM-003 BDBM356621
US10214478, Compound NM-003 BDBM356621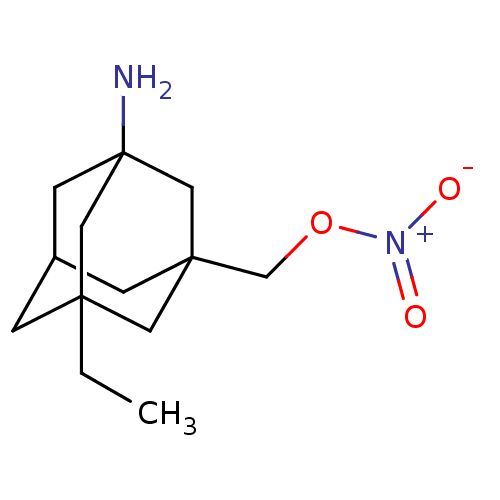 US10214478, Compound NM-004 BDBM356622
US10214478, Compound NM-004 BDBM356622 US10214478, Compound NM-005 BDBM356623
US10214478, Compound NM-005 BDBM356623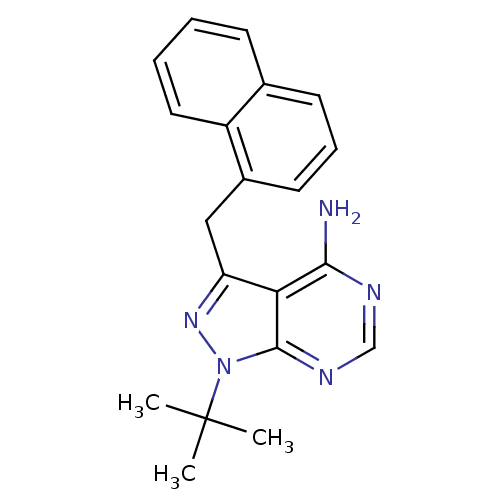 US10544104, Compound 5d CHEMBL573578 US9765037, Compound 5d BDBM50298225 US11247972, Compound 5d CHEMBL2069955 NM-PP1
US10544104, Compound 5d CHEMBL573578 US9765037, Compound 5d BDBM50298225 US11247972, Compound 5d CHEMBL2069955 NM-PP1
- Ménard, D; Niculescu-Duvaz, I; Dijkstra, HP; Niculescu-Duvaz, D; Suijkerbuijk, BM; Zambon, A; Nourry, A; Roman, E; Davies, L; Manne, HA; Friedlos, F; Kirk, R; Whittaker, S; Gill, A; Taylor, RD; Marais, R; Springer, CJ Novel potent BRAF inhibitors: toward 1 nM compounds through optimization of the central phenyl ring. J Med Chem 52: 3881-91 (2009)
- Peterson, K; Kumar, R; Stenström, O; Verma, P; Verma, PR; Håkansson, M; Kahl-Knutsson, B; Zetterberg, F; Leffler, H; Akke, M; Logan, DT; Nilsson, UJ Systematic Tuning of Fluoro-galectin-3 Interactions Provides Thiodigalactoside Derivatives with Single-Digit nM Affinity and High Selectivity. J Med Chem 61: 1164-1175 (2018)
- Ahmed, RF; Mahmoud, WR; Abdelgawad, NM; Fouad, MA; Said, MF Eur J Med Chem 261:
- Durrant, SJ; Ahmad, NM; Beck, EM; Carvalho Meireles, LM; Chudyk, EI; Etxebarria Jardi, G; Galan, B; Hadida Ruah, SS; Hurley, DJ; Knegtel, RM; Neubert, TD; Pinder, JL; Pontillo, J; Pullin, R; Schmidt, Y; Shaw, DM; Skerratt, S; Stamos, D; Thomson, SA; Virani, AN; Wray, C US Patent US11919887 (2024)
- Manley, PW; Tuffin, DP; Allanson, NM; Buckle, PE; Lad, N; Lai, SM; Lunt, DO; Porter, RA; Wade, PJ J Med Chem 30: 1812-8 (1987)
- Afifi, AH; Kagiyama, I; El-Desoky, AH; Kato, H; Mangindaan, REP; de Voogd, NJ; Ammar, NM; Hifnawy, MS; Tsukamoto, S J Nat Prod 80: 2045-2050 (2017)
- Paudel, P; Pandey, P; Paris, JJ; Ashpole, NM; Mahdi, F; Tian, JM; Lee, J; Wang, M; Xu, M; Chittiboyina, AG; Khan, IA; Ross, SA; Li, XC J Nat Prod 86: 1786-1792 (2023)
- Aston, NM; Bamborough, P; Buckton, JB; Edwards, CD; Holmes, DS; Jones, KL; Patel, VK; Smee, PA; Somers, DO; Vitulli, G; Walker, AL J Med Chem 52: 6257-69 (2009)
- Barkey, NM; Tafreshi, NK; Josan, JS; De Silva, CR; Sill, KN; Hruby, VJ; Gillies, RJ; Morse, DL; Vagner, J J Med Chem 54: 8078-84 (2011)
- Manning, DD; Cioffi, CL; Usyatinsky, A; Fitzpatrick, K; Masih, L; Guo, C; Zhang, Z; Choo, SH; Sikkander, MI; Ryan, KN; Naginskaya, J; Hassler, C; Dobritsa, S; Wierschke, JD; Earley, WG; Butler, AS; Brady, CA; Barnes, NM; Cohen, ML; Guzzo, PR Bioorg Med Chem Lett 21: 58-61 (2010)
- Deekonda, S; Cole, J; Sunna, S; Rankin, D; Largent-Milnes, TM; Davis, P; BassiriRad, NM; Lai, J; Vanderah, TW; Porecca, F; Hruby, VJ Bioorg Med Chem Lett 26: 222-7 (2016)
- Deekonda, S; Wugalter, L; Rankin, D; Largent-Milnes, TM; Davis, P; Wang, Y; Bassirirad, NM; Lai, J; Kulkarni, V; Vanderah, TW; Porreca, F; Hruby, VJ Bioorg Med Chem Lett 25: 4683-8 (2015)
- Van Calenbergh, S; von Frijtag Drabbe Künzel, JK; Blaton, NM; Peeters, OM; Rozenski, J; Van Aerschot, A; De Bruyn, A; De Keukeleire, D; IJzerman, AP; Herdewijn, P J Med Chem 40: 3765-72 (1997)
- Hulten, J; Bonham, NM; Nillroth, U; Hansson, T; Zuccarello, G; Bouzide, A; Aqvist, J; Classon, B; Danielson, UH; Karlen, A; Kvarnstrom, I; Samuelsson, B; Hallberg, A J Med Chem 40: 885-97 (1997)
- Lee, SY; Namasivayam, V; Boshta, NM; Perotti, A; Mirza, S; Bua, S; Supuran, CT; Müller, CE RSC Med Chem 12: 1187-1206 (2021)
- Scheuplein, NJ; Bzdyl, NM; Kibble, EA; Lohr, T; Holzgrabe, U; Sarkar-Tyson, M J Med Chem 63: 13355-13388 (2020)
- Sanabria-Ríos, DJ; Rivera-Torres, Y; Rosario, J; Gutierrez, R; Torres-García, Y; Montano, N; Ortíz-Soto, G; Ríos-Olivares, E; Rodríguez, JW; Carballeira, NM Bioorg Med Chem Lett 25: 5067-71 (2015)
- Furuya, T; Shapiro, AB; Comita-Prevoir, J; Kuenstner, EJ; Zhang, J; Ribe, SD; Chen, A; Hines, D; Moussa, SH; Carter, NM; Sylvester, MA; Romero, JAC; Vega, CV; Sacco, MD; Chen, Y; O'Donnell, JP; Durand-Reville, TF; Miller, AA; Tommasi, RA Bioorg Med Chem 28: (2020)
- Dias, RFC; Ribeiro, BMRM; Cassani, NM; Farago, DN; Antoniucci, GA; de Oliveira Rocha, RE; de Oliveira Souza, F; Pilau, EJ; Jardim, ACG; Ferreira, RS; de Oliveira Rezende Júnior, C Bioorg Med Chem 95: (2023)
- Gupta, S; Rodrigues, LM; Esteves, AP; Oliveira-Campos, AM; Nascimento, MS; Nazareth, N; Cidade, H; Neves, MP; Fernandes, E; Pinto, M; Cerqueira, NM; Brás, N Eur J Med Chem 43: 771-80 (2008)
- Giblin, GM; O'Shaughnessy, CT; Naylor, A; Mitchell, WL; Eatherton, AJ; Slingsby, BP; Rawlings, DA; Goldsmith, P; Brown, AJ; Haslam, CP; Clayton, NM; Wilson, AW; Chessell, IP; Wittington, AR; Green, R J Med Chem 50: 2597-600 (2007)
- Cuong, NM; Khanh, PN; Huyen, PT; Duc, HV; Huong, TT; Ha, VT; Durante, M; Sgaragli, G; Fusi, F J Nat Prod 77: 1586-93 (2014)
- Miyamoto, DK; Curnutt, NM; Park, SM; Stavropoulos, A; Kharas, MG; Woo, CM J Med Chem 66: 16953-16979 (2023)
- Balachandran, S; Rodge, A; Gadekar, PK; Yadav, VN; Kamath, D; Chetrapal-Kunwar, A; Bhatt, P; Srinivasan, S; Sharma, S; Vishwakarma, RA; Dagia, NM Bioorg Med Chem Lett 19: 4773-6 (2009)
- Báez-Santos, YM; Barraza, SJ; Wilson, MW; Agius, MP; Mielech, AM; Davis, NM; Baker, SC; Larsen, SD; Mesecar, AD J Med Chem 57: 2393-412 (2014)
- Dolbois, A; Batiste, L; Wiedmer, L; Dong, J; Brütsch, M; Huang, D; Deerain, NM; Spiliotopoulos, D; Cheng-Sánchez, I; Laul, E; Nevado, C; Śledź, P; Caflisch, A ACS Med Chem Lett 11: 1573-1580 (2020)
- Bubenik, M; Mader, P; Mochirian, P; Vallée, F; Clark, J; Truchon, JF; Perryman, AL; Pau, V; Kurinov, I; Zahn, KE; Leclaire, ME; Papp, R; Mathieu, MC; Hamel, M; Duffy, NM; Godbout, C; Casas-Selves, M; Falgueyret, JP; Baruah, PS; Nicolas, O; Stocco, R; Poirier, H; Martino, G; Fortin, AB; Roulston, A; Chefson, A; Dorich, S; St-Onge, M; Patel, P; Pellerin, C; Ciblat, S; Pinter, T; Barabé, F; El Bakkouri, M; Parikh, P; Gervais, C; Sfeir, A; Mamane, Y; Morris, SJ; Black, WC; Sicheri, F; Gallant, M J Med Chem 65: 13198-13215 (2022)
- Ford, DJ; Duggan, NM; Fry, SE; Ripoll-Rozada, J; Agten, SM; Liu, W; Corcilius, L; Hackeng, TM; van Oerle, R; Spronk, HMH; Ashhurst, AS; Mini Sasi, V; Kaczmarski, JA; Jackson, CJ; Pereira, PJB; Passioura, T; Suga, H; Payne, RJ J Med Chem 64: 7853-7876 (2021)
- Wang, P; Wadsworth, PA; Dvorak, NM; Singh, AK; Chen, H; Liu, Z; Zhou, R; Holthauzen, LMF; Zhou, J; Laezza, F J Med Chem 63: 11522-11547 (2020)
- Sun, H; Monenschein, H; Schiffer, HH; Reichard, HA; Kikuchi, S; Hopkins, M; Macklin, TK; Hitchcock, S; Adams, M; Green, J; Brown, J; Murphy, ST; Kaushal, N; Collia, DR; Moore, S; Ray, WJ; English, NM; Carlton, MBL; Brice, NL J Med Chem 64: 9875-9890 (2021)
- Berhe, S; Slupe, A; Luster, C; Charlier, HA; Warner, DL; Zalkow, LH; Burgess, EM; Enwerem, NM; Bakare, O Bioorg Med Chem 18: 134-41 (2010)
- Schepetkin, IA; Khlebnikov, AI; Potapov, AS; Kovrizhina, AR; Matveevskaya, VV; Belyanin, ML; Atochin, DN; Zanoza, SO; Gaidarzhy, NM; Lyakhov, SA; Kirpotina, LN; Quinn, MT Eur J Med Chem 161: 179-191 (2019)
- Ganta, NM; Gedda, G; Rathnakar, B; Satyanarayana, M; Yamajala, B; Ahsan, MJ; Jadav, SS; Balaraju, T Eur J Med Chem 164: 576-601 (2019)
- Nieto, CT; Gonzalez-Nunez, V; Rodríguez, RE; Diez, D; Garrido, NM Eur J Med Chem 101: 150-62 (2015)
- Mohamed, HA; Girgis, NM; Wilcken, R; Bauer, MR; Tinsley, HN; Gary, BD; Piazza, GA; Boeckler, FM; Abadi, AH J Med Chem 54: 495-509 (2011)
- Goudgaon, NM; Schinazi, RF J Med Chem 34: 3305-9 (1991)
- de Costa, BR; Rice, KC; Bowen, WD; Thurkauf, A; Rothman, RB; Band, L; Jacobson, AE; Radesca, L; Contreras, PC; Gray, NM J Med Chem 33: 3100-10 (1990)
- Grob, NM; Häussinger, D; Deupi, X; Schibli, R; Behe, M; Mindt, TL J Med Chem 63: 4484-4495 (2020)
- Bohmann, JA; Gutierrez, NM; Mcdonough, JA; Campbell, RF; Joyce, MG; Panchal, R; Sankhala, R; Duplantier, A US Patent US11963959 (2024)
- McGonagle, AE; Jordan, A; Waszkowycz, B; Hutton, C; Waddell, I; Hitchin, JR; Smith, KM; Hamilton, NM US Patent US10239843 (2019)
- Lentini, NA; Schroeder, CM; Harmon, NM; Huang, X; Schladetsch, MA; Foust, BJ; Poe, MM; Hsiao, CC; Wiemer, AJ; Wiemer, DF ACS Med Chem Lett 12: 136-142 (2021)
- Schneider, NO; Gilreath, K; Henriksen, NM; Donaldson, WA; Chaudhury, S; St Maurice, M ACS Med Chem Lett 15: 1088-1093
- Heron, NM; Anderson, M; Blowers, DP; Breed, J; Eden, JM; Green, S; Hill, GB; Johnson, T; Jung, FH; McMiken, HH; Pannifer, AD; Pauptit, RA; Pink, J; Roberts, NJ Bioorg Med Chem Lett 16: 1320-23 (2006)
- Yi Mok, N; Chadwick, J; Kellett, KA; Hooper, NM; Johnson, AP; Fishwick, CW Bioorg Med Chem Lett 19: 6770-4 (2009)
- Howarth, NM; Purohit, A; Reed, MJ; Potter, BV J Med Chem 37: 219-21 (1994)
- Zorbaz, T; Malinak, D; Hofmanova, T; Maraković, N; Žunec, S; Hrvat, NM; Andrys, R; Psotka, M; Zandona, A; Svobodova, J; Prchal, L; Fingler, S; Katalinić, M; Kovarik, Z; Musilek, K Eur J Med Chem 238: (2022)
- Ibrahim, NM; Fahim, SH; Hassan, M; Farag, AE; Georgey, HH Eur J Med Chem 228: (2022)
- Meier, JC; Tallant, C; Fedorov, O; Witwicka, H; Hwang, SY; van Stiphout, RG; Lambert, JP; Rogers, C; Yapp, C; Gerstenberger, BS; Fedele, V; Savitsky, P; Heidenreich, D; Daniels, DL; Owen, DR; Fish, PV; Igoe, NM; Bayle, ED; Haendler, B; Oppermann, UCT; Buffa, F; Brennan, PE; Müller, S; Gingras, AC; Odgren, PR; Birnbaum, MJ; Knapp, S ACS Chem Biol 12: 2619-2630 (2017)
- Islam, NM; Kato, T; Nishino, N; Kim, HJ; Ito, A; Yoshida, M Bioorg Med Chem Lett 20: 997-9 (2010)
- Richardson, NL; O'Malley, LJ; Weissberger, D; Tumber, A; Schofield, CJ; Griffith, R; Jones, NM; Hunter, L Bioorg Med Chem 38: (2021)
- Yang, Q; Lachapelle, EA; Kablaoui, NM; Webb, D; Popiolek, M; Grimwood, S; Kozak, R; O'Connor, RE; Lazzaro, JT; Butler, CR; Zhang, L ACS Med Chem Lett 10: 941-948 (2019)
- Kelly, NM; Wellejus, A; Elbrønd-Bek, H; Weidner, MS; Jørgensen, SH Bioorg Med Chem 21: 3334-47 (2013)
- Kennedy, NM; Schmid, CL; Ross, NC; Lovell, KM; Yue, Z; Chen, YT; Cameron, MD; Bohn, LM; Bannister, TD J Med Chem 61: 8895-8907 (2018)
- Shore, ER; Awais, M; Kershaw, NM; Gibson, RR; Pandalaneni, S; Latawiec, D; Wen, L; Javed, MA; Criddle, DN; Berry, N; O'Neill, PM; Lian, LY; Sutton, R J Med Chem 59: 2596-611 (2016)
- Ngoc, TM; Khoi, NM; Ha, do T; Nhiem, NX; Tai, BH; Don, DV; Luong, HV; Son, DC; Bae, K Bioorg Med Chem Lett 22: 4625-8 (2012)
- Surleraux, DL; de Kock, HA; Verschueren, WG; Pille, GM; Maes, LJ; Peeters, A; Vendeville, S; De Meyer, S; Azijn, H; Pauwels, R; de Bethune, MP; King, NM; Prabu-Jeyabalan, M; Schiffer, CA; Wigerinck, PB J Med Chem 48: 1965-73 (2005)
- Jayatilake, GS; Jayasuriya, H; Lee, ES; Koonchanok, NM; Geahlen, RL; Ashendel, CL; McLaughlin, JL; Chang, CJ J Nat Prod 56: 1805-10 (1993)
- Fougiaxis, V; He, B; Khan, T; Vatinel, R; Koutroumpa, NM; Afantitis, A; Lesire, L; Sierocki, P; Deprez, B; Deprez-Poulain, R J Med Chem 67: 11597-11621
- Lawande, PP; Sontakke, VA; Kumbhar, NM; Bhagwat, TR; Ghosh, S; Shinde, VS Bioorg Med Chem Lett 27: 5291-5295 (2017)
- Hennessy, EJ; Adam, A; Aquila, BM; Castriotta, LM; Cook, D; Hattersley, M; Hird, AW; Huntington, C; Kamhi, VM; Laing, NM; Li, D; MacIntyre, T; Omer, CA; Oza, V; Patterson, T; Repik, G; Rooney, MT; Saeh, JC; Sha, L; Vasbinder, MM; Wang, H; Whitston, D J Med Chem 56: 9897-919 (2013)
- Wang, L; Wang, GT; Wang, X; Tong, Y; Sullivan, G; Park, D; Leonard, NM; Li, Q; Cohen, J; Gu, WZ; Zhang, H; Bauch, JL; Jakob, CG; Hutchins, CW; Stoll, VS; Marsh, K; Rosenberg, SH; Sham, HL; Lin, NH J Med Chem 47: 612-26 (2004)
- Faber, EB; Wang, N; John, K; Sun, L; Wong, HL; Burban, D; Francis, R; Tian, D; Hong, KH; Yang, A; Wang, L; Elsaid, M; Khalid, H; Levinson, NM; Schönbrunn, E; Hawkinson, JE; Georg, GI J Med Chem 66: 1928-1940 (2023)
- Shiozaki, M; Maeda, K; Miura, T; Kotoku, M; Yamasaki, T; Matsuda, I; Aoki, K; Yasue, K; Imai, H; Ubukata, M; Suma, A; Yokota, M; Hotta, T; Tanaka, M; Hase, Y; Haas, J; Fryer, AM; Laird, ER; Littmann, NM; Andrews, SW; Josey, JA; Mimura, T; Shinozaki, Y; Yoshiuchi, H; Inaba, T J Med Chem 54: 2839-63 (2011)
- Abdel-Hamid, MK; Abdel-Hafez, AA; El-Koussi, NA; Mahfouz, NM; Innocenti, A; Supuran, CT Bioorg Med Chem 15: 6975-84 (2007)
- Loh, VM; Cockcroft, XL; Dillon, KJ; Dixon, L; Drzewiecki, J; Eversley, PJ; Gomez, S; Hoare, J; Kerrigan, F; Matthews, IT; Menear, KA; Martin, NM; Newton, RF; Paul, J; Smith, GC; Vile, J; Whittle, AJ Bioorg Med Chem Lett 15: 2235-8 (2005)
- Boskovic, ZV; Kemp, MM; Freedy, AM; Viswanathan, VS; Pop, MS; Fuller, JH; Martinez, NM; Figueroa Lazú, SO; Hong, JA; Lewis, TA; Calarese, D; Love, JD; Vetere, A; Almo, SC; Schreiber, SL; Koehler, AN ACS Chem Biol 11: 1844-51 (2016)
- Lavecchia, A; Di Giovanni, C; Pesapane, A; Montuori, N; Ragno, P; Martucci, NM; Masullo, M; De Vendittis, E; Novellino, E J Med Chem 55: 4142-58 (2012)
- Mascarenhas, NM; Ghoshal, N Eur J Med Chem 43: 2807-18 (2008)
- Akkari, R; Alvey, LJ; Bock, XM; Brown, BS; Claes, PI; Cowart, MD; Conrath, KE; Cyr, D; De Lemos, E; De Wilde, GJ; Desroy, N; Duthion, B; Gfesser, GA; Gosmini, RL; Housseman, CG; Jansen, KK; Ji, J; Kym, PR; Lefrancois, J; Mammoliti, O; Menet, CJ; Merayo, NM; Newsome, GJ; Palisse, AM; Patel, SV; Pizzonero, MR; Shrestha, A; Swift, EC; Tse, C; Van der Plas, SE; Wang, X US Patent US10130622 (2018)
- Schüß, C; Vu, O; Schubert, M; Du, Y; Mishra, NM; Tough, IR; Stichel, J; Weaver, CD; Emmitte, KA; Cox, HM; Meiler, J; Beck-Sickinger, AG J Med Chem 64: 2801-2814 (2021)
- El Sayed, MT; El-Sharief, MAMS; Zarie, ES; Morsy, NM; Elsheakh, AR; Voronkov, A; Berishvili, V; Hassan, GS Bioorg Med Chem Lett 28: 952-957 (2018)
- Allerton, CM; Barber, CG; Beaumont, KC; Brown, DG; Cole, SM; Ellis, D; Lane, CA; Maw, GN; Mount, NM; Rawson, DJ; Robinson, CM; Street, SD; Summerhill, NW J Med Chem 49: 3581-94 (2006)
- Teng, YH; Berger, WT; Nesbitt, NM; Kumar, K; Balius, TE; Rizzo, RC; Tonge, PJ; Ojima, I; Swaminathan, S Bioorg Med Chem 23: 5489-95 (2015)
- O'Boyle, NM; Barrett, I; Greene, LM; Carr, M; Fayne, D; Twamley, B; Knox, AJS; Keely, NO; Zisterer, DM; Meegan, MJ J Med Chem 61: 514-534 (2018)
- Adams, GL; Pall, PS; Grauer, SM; Zhou, X; Ballard, JE; Vavrek, M; Kraus, RL; Morissette, P; Li, N; Colarusso, S; Bianchi, E; Palani, A; Klein, R; John, CT; Wang, D; Tudor, M; Nolting, AF; Biba, M; Nowak, T; Makarov, AA; Reibarkh, M; Buevich, AV; Zhong, W; Regalado, EL; Wang, X; Gao, Q; Shahripour, A; Zhu, Y; de Simone, D; Frattarelli, T; Pasquini, NM; Magotti, P; Iaccarino, R; Li, Y; Solly, K; Lee, KJ; Wang, W; Chen, F; Zeng, H; Wang, J; Regan, H; Amin, RP; Regan, CP; Burgey, CS; Henze, DA; Sun, C; Tellers, DM J Med Chem 65: 485-496 (2022)
- Patel, NM; Bennett, F; Girijavallabhan, VM; Dasmahapatra, B; Butkiewicz, N; Hart, A Bioorg Med Chem Lett 8: 931-4 (1999)
- Paul, NM; Taylor, M; Kumar, R; Deschamps, JR; Luedtke, RR; Newman, AH J Med Chem 51: 6095-109 (2008)
- DiRaimondo, TR; Plugis, NM; Jin, X; Khosla, C J Med Chem 56: 1301-10 (2013)
- Costa, TEMM; Raghavendra, NM; Penido, C Eur J Med Chem 189: (2020)
- Heck, MC; Wagner, CE; Shahani, PH; MacNeill, M; Grozic, A; Darwaiz, T; Shimabuku, M; Deans, DG; Robinson, NM; Salama, SH; Ziller, JW; Ma, N; van der Vaart, A; Marshall, PA; Jurutka, PW J Med Chem 59: 8924-8940 (2016)
- Harrison, T; Owens, AP; Williams, BJ; Swain, CJ; Williams, A; Carlson, EJ; Rycroft, W; Tattersall, FD; Cascieri, MA; Chicchi, GG; Sadowski, S; Rupniak, NM; Hargreaves, RJ J Med Chem 44: 4296-9 (2001)
- Elshemy, HAH; Abdelall, EKA; Azouz, AA; Moawad, A; Ali, WAM; Safwat, NM Eur J Med Chem 127: 10-21 (2017)
- Letribot, B; Akué-Gédu, R; Santio, NM; El-Ghozzi, M; Avignant, D; Cisnetti, F; Koskinen, PJ; Gautier, A; Anizon, F; Moreau, P Eur J Med Chem 50: 304-10 (2012)
- Cai, L; Liow, JS; Zoghbi, SS; Cuevas, J; Baetas, C; Hong, J; Shetty, HU; Seneca, NM; Brown, AK; Gladding, R; Temme, SS; Herman, MM; Innis, RB; Pike, VW J Med Chem 51: 148-58 (2008)
- Beesu, M; Caruso, G; Salyer, AC; Shukla, NM; Khetani, KK; Smith, LJ; Fox, LM; Tanji, H; Ohto, U; Shimizu, T; David, SA J Med Chem 59: 3311-30 (2016)
- da Silva AJM, na; Melo, PA; Silva, NM; Brito, FV; Buarque, CD; de Souza, DV; Rodrigues, VP; Poças, ES; Noël, F; Albuquerque, EX; Costa, PR Bioorg Med Chem Lett 11: 283-6 (2001)
- Burks, HE; Dechantsreiter, MA; He, G; Nunez, J; Peukert, S; Springer, C; Sun, Y; Thomsen, NM; Tria, GS; Yu, B US Patent US10058534 (2018)
- Tormählen, NM; Martorelli, M; Kuhn, A; Maier, F; Guezguez, J; Burnet, M; Albrecht, W; Laufer, SA; Koch, P J Med Chem 65: 1225-1242 (2022)
- Abramovitz, M; Adam, M; Boie, Y; Carrière, M; Denis, D; Godbout, C; Lamontagne, S; Rochette, C; Sawyer, N; Tremblay, NM; Belley, M; Gallant, M; Dufresne, C; Gareau, Y; Ruel, R; Juteau, H; Labelle, M; Ouimet, N; Metters, KM Biochim Biophys Acta 1483: 285-93 (2000)
- Thanh, ND; Lan, PH; Hai, DS; Anh, HH; Giang, NTK; Van, HTK; Toan, VN; Tri, NM; Toan, DN RSC Med Chem 14: 1114-1130 (2023)
- Caillé, F; Morley, TJ; Tavares, AA; Papin, C; Twardy, NM; Alagille, D; Lee, HS; Baldwin, RM; Seibyl, JP; Barret, O; Tamagnan, GD Bioorg Med Chem Lett 23: 6243-7 (2013)
- Park, H; Urs, AN; Zimmerman, J; Liu, C; Wang, Q; Urs, NM ACS Med Chem Lett 11: 385-392 (2020)
- Cortez, A; Li, Y; Miller, AT; Zhang, X; Yue, K; Maginnis, J; Hampton, J; Hall, de S; Shapiro, M; Nayak, B; D'Oro, U; Li, C; Skibinski, D; Mbow, ML; Singh, M; O'Hagan, DT; Cooke, MP; Valiante, NM; Wu, TY J Med Chem 59: 5868-78 (2016)
- Burns, MR; Jenkins, SA; Vermeulen, NM; Balakrishna, R; Nguyen, TB; Kimbrell, MR; David, SA Bioorg Med Chem Lett 16: 6209-12 (2006)
- Camps, P; El Achab, R; Görbig, DM; Morral, J; Muñoz-Torrero, D; Badia, A; Eladi Baños, J; Vivas, NM; Barril, X; Orozco, M; Luque, FJ J Med Chem 42: 3227-42 (1999)
- Walter, NM; Wentsch, HK; Bührmann, M; Bauer, SM; Döring, E; Mayer-Wrangowski, S; Sievers-Engler, A; Willemsen-Seegers, N; Zaman, G; Buijsman, R; Lämmerhofer, M; Rauh, D; Laufer, SA J Med Chem 60: 8027-8054 (2017)
- Youngman, MA; McNally, JJ; Lovenberg, TW; Reitz, AB; Willard, NM; Nepomuceno, DH; Wilson, SJ; Crooke, JJ; Rosenthal, D; Vaidya, AH; Dax, SL J Med Chem 43: 346-50 (2000)
- Zhou, B; Shetye, G; Yu, Y; Santarsiero, BD; Klein, LL; Abad-Zapatero, C; Wolf, NM; Cheng, J; Jin, Y; Lee, H; Suh, JW; Lee, H; Bisson, J; McAlpine, JB; Chen, SN; Cho, SH; Franzblau, SG; Pauli, GF J Nat Prod 83: 657-667 (2020)
- Moreau, F; Atamanyuk, D; Blaukopf, M; Barath, M; Herczeg, M; Xavier, NM; Monbrun, J; Airiau, E; Henryon, V; Leroy, F; Floquet, S; Bonnard, D; Szabla, R; Brown, C; Junop, MS; Kosma, P; Gerusz, V J Med Chem 67: 6610-6623
- Malamas, M; Makriyannis, A; Subramanian, KV; Whitten, KM; Zvonok, NM; West, JM; Mccormack, M; Pavlopoulos, S US Patent US9963444 (2018)
- Mohamed, AR; Mostafa, A; El Hassab, MA; Hedeab, GM; Mahmoud, SH; George, RF; Georgey, HH; Abdel Gawad, NM; El-Ashrey, MK RSC Med Chem 14: 899-920 (2023)
- Furqan, M; Fayyaz, A; Firdous, F; Raza, H; Bilal, A; Saleem, RSZ; Shahzad-Ul-Hussan, S; Wang, D; Youssef, FS; Al Musayeib, NM; Ashour, ML; Hussain, H; Faisal, A J Nat Prod 85: 1503-1513 (2022)
- Ibrahim, MK; Taghour, MS; Metwaly, AM; Belal, A; Mehany, ABM; Elhendawy, MA; Radwan, MM; Yassin, AM; El-Deeb, NM; Hafez, EE; ElSohly, MA; Eissa, IH Eur J Med Chem 155: 117-134 (2018)
- Assis, DM; Gontijo, VS; de Oliveira Pereira, I; Santos, JA; Camps, I; Nagem, TJ; Ellena, J; Izidoro, MA; dos Santos Tersariol, IL; de Barros, NM; Doriguetto, AC; dos Santos, MH; Juliano, MA J Enzyme Inhib Med Chem 28: 661-70 (2013)
- van Loevezijn, A; Venhorst, J; Iwema Bakker, WI; Lange, JH; de Looff, W; Henzen, R; de Vries, J; van de Woestijne, RP; den Hartog, AP; Verhoog, S; van der Neut, MA; de Bruin, NM; Kruse, CG Bioorg Med Chem Lett 26: 1605-11 (2016)
- Lange, JH; Coolen, HK; van Stuivenberg, HH; Dijksman, JA; Herremans, AH; Ronken, E; Keizer, HG; Tipker, K; McCreary, AC; Veerman, W; Wals, HC; Stork, B; Verveer, PC; den Hartog, AP; de Jong, NM; Adolfs, TJ; Hoogendoorn, J; Kruse, CG J Med Chem 47: 627-43 (2004)
- Muñoz-Torrero López-Ibarra, D; Inestrosa Cantín, NM; Viayna Gaza, E; Sola Lao, I; Vázquez Cruz, S US Patent US9238626 (2016)
- NAV1.7 INWARD CURRENT BLOCK (nM) (Automated Patchclamp) NAV1.7 INWARD CURRENT BLOCK (nM) (Automated Patchclamp).
- NAV1.7 SLOW INACTIVATION BLOCK (nM) (Automated Patchclamp) NAV1.7 SLOW INACTIVATION BLOCK (nM) (Automated Patchclamp).
- NAV1.7 SLOW INACTIVATION BLOCK (nM) (Manual Patchclamp) NAV1.7 SLOW INACTIVATION BLOCK (nM) (Manual Patchclamp).
- ChEBML_145204 Displacement of 0.5 nM [3H]bremazocine from guinea pig brain membrane opioid receptor kappa with 100 nM DAGO and 100 nM DPDPE
- ChEBML_62108 Displacement of [3H]spiperone [0.5 nM (Kd=0.1 nM)] from recombinant human dopamine receptor D3 expressed in CHO cells at 0.5 nM concentration
- ChEMBL_62108 (CHEMBL674986) Displacement of [3H]spiperone [0.5 nM (Kd=0.1 nM)] from recombinant human dopamine receptor D3 expressed in CHO cells at 0.5 nM concentration
- ChEMBL_141127 (CHEMBL746618) Inhibitory activity against Candida albicans (Nmt) assessed as inhibitory concentration (nM); 3.7-9.4 nM
- NAV1.7 INWARD CURRENT BLOCK (nM) (0.25HZ Automated Patchclamp) NAV1.7 INWARD CURRENT BLOCK (nM) (0.25HZ Automated Patchclamp).
- NAV1.7 INWARD CURRENT BLOCK (nM) (7HZ Manual Patchclamp) NAV1.7 INWARD CURRENT BLOCK (nM) (7HZ Manual Patchclamp).
- ChEBML_61801 Ability to displace [3H]spiperone [0.5 nM (Kd=0.2-0.45 nM)] from human cloned dopamine receptor D2S expressed in CHO cells at 0.5 nM concentration
- ChEMBL_60054 (CHEMBL872881) Displacement of [3H]-spiperone [0.5 nM (Kd=0.2-0.45 nM)] from human recombinant dopamine receptor D2S expressed in CHO cells at 0.5 nM concentration
- ChEMBL_61801 (CHEMBL670554) Ability to displace [3H]spiperone [0.5 nM (Kd=0.2-0.45 nM)] from human cloned dopamine receptor D2S expressed in CHO cells at 0.5 nM concentration
- ChEMBL_146672 (CHEMBL753164) In vivo binding affinity against kappa opioid receptor was measured by using labeled ligand [3H]ethylketocyclazocine (1 nM) with 500 nM DADLE and 20 nM sufentanil
- ChEBML_60196 Displacement of [3H]SCH-23390 [0.3 nM (Kd=0.35 nM)] from dopamine receptor D1 in bovine striatal membranes
- ChEMBL_60196 (CHEMBL674339) Displacement of [3H]SCH-23390 [0.3 nM (Kd=0.35 nM)] from dopamine receptor D1 in bovine striatal membranes
- ChEMBL_60053 (CHEMBL671368) Displacement of [3H]spiperone [0.5 nM (Kd=0.1 nM)] from human recombinant dopamine receptor D2L expressed in CHO cells
- Anti-HIV Cytoprotection Assay The in vitro antiviral activity profile for CMX157 was evaluated for cell-type effects and HIV strain effects. It is active against all major subtypes of HIV-1 in PBMCs with EC50 values ranging between 0.20 and 7.18 nanomolar (nM). In a PHENOSENSE assay, EC50s for CMX157 ranged from 0.66 nM for 74V/184V to 57 nM for 62V/69SVG/75I/215I; corresponding EC50s for tenofovir were 227 nM and 16,959 nM respectively (see FIG. 3). CMX157 IC50s for 41L/210W/215Y averaged 6.3 nM without 184V and 2.2 nM with 184V (2,240 and 770 nM for tenofovir respectively).
- ChEMBL_473654 (CHEMBL937672) Inhibition of human EGFR at 10000 nM
- ChEBML_201714 Binding affinity against sigma receptor in bovine cerebellum using 2.0 nM [3H]haloperidol in the presence of 25 nM unlabeled spiperone
- ChEBML_63102 Ability to displace [3H]spiperone [0.5 nM (Kd=0.1-0.45 nM)] from human recombinant dopamine receptor D4 expressed in CHO cells
- In vitro Assay (1 nM) Each compound for inhibiting enzymatic activity of recombinant 15-PGDH (1 nM) in an in vitro assay.
- In vitro Assay (5 nM) Each compound for inhibiting enzymatic activity of recombinant 15-PGDH (5 nM) in an in vitro assay.
- HDAC Enzyme Activity Inhibition (In Vitro) An HDAC enzyme inhibitory capacity of test material was measured by using HDAC1 Fluorimetric Drug Discovery Assay Kit (Enzolifesciences: BML-AK511) and HDAC6 human recombinant (Calbiochem: 382180). For a HDAC1 assay, samples were treated at a concentration of 100 nM, 1000 nM and 10000 nM. For an HDAC6 assay, samples were treated at a concentration of 0.1 nM, 1 nM, 10 nM, 100 nM and 1000 nM. After the sample treatment, a reaction was continued at 37° C. for 60 minutes, treated with a developer, and subjected to reaction at 37° C. for 30 minutes, after which fluorescence intensity (Ex 390 nm, Em 460 nm) was measured by using FlexStation3 (Molecular device). For final result values, each IC51 value was calculated with GraphPad Prism 4.0 program.
- In Vitro Assay All of the compounds of Examples 1 to 16 and Tables 1 to 19 were tested in one or more of the assays described above. In the following tables, for the JAK1, JAK 2, JAK3, and TYK2 enzyme assays, A represents a pKi value≥10 (Ki≤0.1 nM), B represents a pKi value between 9 and 10 (Ki between 1 nM and 0.1 nM), C represents a pKi value between 9 and 9.5 (Ki between 1 nM and 0.32 nM), and D represents a pKi value between 8.5 and 9 (Ki between 32 nM and 1 nM). For the BEAS-2B cell potency assay, A represents a pIC50 value≥8 (IC50≤10 nM) and B represents a pIC50 value between 7.4 and 8 (IC50 between 40 nM and 10 nM).
- VEGFA ELISA Assay 786-O cells in the logarithmic growth phase were inoculated into a 96-well plate at a cell concentration of 65,000 cells per ml of culture liquid, 180 μL per well. The compounds were diluted to the corresponding concentrations, and 20 μL of compound solutions of various concentrations were added to the corresponding cell wells, so that the final concentrations of the compounds were 1.5 nM, 4.6 nM, 13.7 nM, 41.2 nM, 123.5 nM, 370.4 nM, 1111.1 nM, 3333.3 nM, and 10000 nM, respectively. After culturing for 24 h, the cell culture supernatant was collected and the VEGFA concentration was determined using an ELISA kit (purchased from Abeam). Finally, the reaction was terminated and the absorbance value of each well was measured at a wavelength of 450 nm using an ELISA reader.
- ChEMBL_201714 (CHEMBL803751) Binding affinity against sigma receptor in bovine cerebellum using 2.0 nM [3H]haloperidol in the presence of 25 nM unlabeled spiperone
- ChEMBL_226542 (CHEMBL846482) Binding affinity against sigma receptor in bovine cerebellum using 2.0 nM [3H]haloperidol in the presence of 25 nM unlabeled spiperone
- ChEMBL_63102 (CHEMBL674495) Ability to displace [3H]spiperone [0.5 nM (Kd=0.1-0.45 nM)] from human recombinant dopamine receptor D4 expressed in CHO cells
- ChEBML_142953 Inhibition of 0.5 nM [3H]nisoxetine binding toNorepinephrine transporter
- ChEBML_48367 Inhibitory activity against recombinant human cathepsin L (1.2 nM)
- ChEMBL_467649 (CHEMBL936672) Inhibition of 100 nM Mycobacterium tuberculosis H37RV InhA
- ChEMBL_467650 (CHEMBL936673) Inhibition of 1 nM Mycobacterium tuberculosis H37RV InhA
- ChEMBL_70144 (CHEMBL685967) Inhibition of bovine brain Farnesyltransferase at 1 nM
- ChEMBL_147277 (CHEMBL755199) In vivo binding affinity against delta Opioid receptor was measured by using labeled ligand [3H]DADLE (1 nM) with 4 nM sufentanil
- Fluorescence Polarization Assay Polarization for the fluorescein-UDP conjugated probe in the presence of inhibitors was measured on a Tecan M1000 microplate reader, exciting with 470 nm light and monitoring emission at 525 nm. Each condition was tested in triplicate in the presence of 220 nM K. pneumoniae UGM and 23.3 nM of the probe.
- ChEBML_162430 Inhibition of Protein-tyrosine phosphatase 1B in 300 nM DTT
- ChEBML_201999 Inhibition of 0.2 nM [3H]paroxetine binding to Serotonin transporter
- ChEBML_62004 Displacement of 0.5 nM [3H]WIN-35248 from Dopamine transporter
- ChEMBL_142953 (CHEMBL750805) Inhibition of 0.5 nM [3H]nisoxetine binding toNorepinephrine transporter
- ChEMBL_161572 (CHEMBL769089) Inhibition of Trypanosoma brucei protein farnesyltransferase at 50 nM
- ChEMBL_29409 (CHEMBL643382) IC50 against acetylcholinesterase; value ranges from 1-4900 nM.
- ChEMBL_29410 (CHEMBL643383) IC50 against acetylcholinesterase; value ranges from 1.3-380 nM.
- ChEMBL_311781 (CHEMBL826403) Inhibitory concentration required against Escherichia coli MetAP1 (150 nM)
- ChEMBL_311942 (CHEMBL833409) Inhibitory concentration required against Saccharomyces cerevisiae MetAP1 (330 nM)
- ChEMBL_311969 (CHEMBL834386) Inhibitory concentration required against Saccharomyces cerevisiae MetAP1 (330 nM)
- ChEMBL_313633 (CHEMBL835777) pIC50 for 1 nM estradiol-induced Ishikawa cell proliferation
- ChEMBL_323053 (CHEMBL857324) Agonist activity against human orexin 1 receptor; EC50; nM
- ChEMBL_323054 (CHEMBL857325) Agonist activity against human orexin 2 receptor; EC50; nM
- ChEMBL_48367 (CHEMBL661683) Inhibitory activity against recombinant human cathepsin L (1.2 nM)
- ChEBML_146568 In vito concentration required to displace [3H]cyclofoxy (Kd = 0.8 nM and concentration is 1.3 nM) from mu and kappa2 receptor in rat brain membranes.
- ChEMBL_145925 (CHEMBL857682) In vito concentration required to displace 9 (Kd = 1.6 nM and concentration is 1.8 nM) from opioid receptor kappa 1 in guinea brain membranes.
- ChEMBL_146422 (CHEMBL757174) In vito concentration required to displace [3H]DAGO (Kd = 0.7 nM and concentration is 1.7 nM) from opioid receptor mu in rat brain membranes.
- ChEMBL_146547 (CHEMBL754966) In vito concentration required to displace [3H]DAGO (Kd = 0.7 nM and concentration is 1.7 nM) from opioid receptor mu in rat brain membranes.
- ChEMBL_146667 (CHEMBL755870) In vito concentration required to displace 9 (Kd = 1.6 nM and concentration is 1.8 nM) from opioid receptor kappa 1 in guinea brain membranes.
- ChEMBL_147215 (CHEMBL873067) In vito concentration required to displace 9 (Kd = 1.6 nM and concentration is 1.8 nM) from opioid receptor kappa 1 in guinea brain membranes.
- ChEMBL_147216 (CHEMBL755421) In vito concentration required to displace 9 (Kd = 1.6 nM and concentration is 1.8 nM) from opioid receptor kappa 2 in guinea brain membranes.
- ChEMBL_151865 (CHEMBL759579) Binding affinity against Protease-activated receptor (PAR-1) using [3H]-s-(p-F-Phe)-homoarginine-L-homoarginine-KY-NH2, 10 nM (Kd= 15 nM)
- ChEMBL_151866 (CHEMBL759580) Binding affinity against Protease-activated receptor (PAR-1) using [3H]-s-(p-F-Phe)-homoarginine-L-homoarginine-KY-NH2, 10 nM (Kd= 15 nM)
- ChEMBL_42209 (CHEMBL658310) Agonist effect on 45 [Ca2+] influx in vanilloid receptor expressing CHO cells relative to maximal capsaicin (300 nM) response, weak effect at 30 nM
- In Vitro Enzyme Assay In vitro enzyme assays were conducted via the Ba(OH)2 precipitation method of Wang et al. (32) using recombinant human PDE11A4, PDE10A1, PDE5A1, PDE6C, PDE8A (BPS Bioscience Inc.), PDE7A (BIOMOL International), and PDE4A10 enzymes (gift from HengmingKe), in the presence of 100 nM cGMP, 30 nM cAMP, 500 nM cGMP, 1.7 μM cGMP, 10 nM cAMP, 15 nM cAMP, and 625 nM cAMP, respectively. Inhibitor concentrations that reduce enzyme activity by 50% (IC50) are presented. The values are means of at least three independent experiments.
- ChEBML_154360 Binding affinity was determined by displacement of 20 nM [3H]- thiazolidinedione from 4 nM biotinylated human Peroxisome proliferator activated receptor gamma (PPAR gamma) ligand binding domain
- ChEMBL_145995 (CHEMBL750589) In vito concentration required to displace [3H]BRM (Kd = 1.0 nM and concentration is 1.8 nM) from opioid receptor kappa 2 in guinea brain membranes.
- ChEMBL_146096 (CHEMBL753324) In vito concentration required to displace [3H]BRM (Kd = 1.0 nM and concentration is 1.8 nM) from opioid receptor kappa 2 in guinea brain membranes.
- ChEMBL_146098 (CHEMBL753326) In vito concentration required to displace [3H]-BRM (Kd = 1.0 nM and concentration is 1.8 nM) from opioid receptor kappa 2 in guinea brain membranes.
- ChEMBL_146567 (CHEMBL755056) In vito concentration required to displace [3H]cyclofoxy (Kd = 0.8 nM and concentration is 1.3 nM) from mu and kappa2 receptor in rat brain membranes.
- ChEMBL_146568 (CHEMBL755057) In vito concentration required to displace [3H]cyclofoxy (Kd = 0.8 nM and concentration is 1.3 nM) from mu and kappa2 receptor in rat brain membranes.
- ChEMBL_146584 (CHEMBL754818) In vito concentration required to displace [3H]DADLE (Kd = 1.6 nM and concentration is 1.9 nM) from high affinity delta-site in rat brain membranes.
- ChEMBL_146585 (CHEMBL754819) In vito concentration required to displace [3H]-DADLE (Kd = 12.2 nM and concentration is 2.1 nM) from low affinity delta-site in rat brain membranes.
- ChEMBL_146593 (CHEMBL752697) In vito concentration required to displace [3H]DADLE (Kd = 1.6 nM and concentration is 1.9 nM) from high affinity delta-site in rat brain membranes.
- ChEMBL_146594 (CHEMBL752698) In vito concentration required to displace [3H]DADLE (Kd = 12.2 nM and concentration is 2.1 nM) from low affinity delta-site in rat brain membranes.
- DDR1 and DDR2 binding assays DDR1 and DDR2 binding assays were performed using Life Technologies LanthaScreen Europium Kinase Binding assay. The compounds were incubated with 5 nM DDR1 (Carna Biosciences) or 5 nM DDR2 (Life Technologies) for 1 hour at room temperature in white 384-well OptiPlate (PerkinElmer), containing 20 nM or 10 nM Kinase Tracer 178 respectively and 2 nM Europium labelled anti-GST antibody (Life Technologies) in assay buffer (50 mM HEPES pH 7.5, 10 mM MgCl2, 1 mM EGTA and 0.01% BRIJ35). The ratio of fluorescence emission 665 nm/615 nm after excitation at 340 nm was obtained using the Tecan Spark 20M plate reader. IC50 values were determined in GraphPad Prism 8.0 software, using 4 parameter model: log(inhibitor) vs. response.
- TR-FRET cereblon binding assay The 6XHis-tagged full length human CRBN bound to full length human DDB1 used in the assay was purified as described elsewhere with the exception that the thrombin cleavage/ortho nickle step was removed.8b In the assay, 60 nM 6Xhis-tagged CRBN-DDB1 was combined with 30 nM cy5-conjugated cereblon modulator (compound 7) and 3 nM LanthaScreen Eu-anti-His Tag antibody (ThermoFisher catalogue no. PV5596) in 20 mM HEPES pH 7, 150 mM NaCl, 0.005% Tween-20 assay buffer. FRET was observed by exciting at 340 nm and monitoring emission at 615 nm (non-FRET emission) and 665 nm (FRET emission), and FRET efficiency was determined by the ratio of FRET to non-FRET emission (665 nm/615 nm).
- ChEBML_306356 Irreversible inhibition of fatty acid amide hydrolase; range=. 5-6 nM
- ChEBML_54965 Inhibition of rat liver dihydrofolate reductase assayed spectrophotometrically at 340 nM
- ChEMBL_142961 (CHEMBL750813) Inhibitory activity against Norepinephrine transporter using 0.5 nM [3H]-radioligand
- ChEMBL_142963 (CHEMBL750815) Inhibitory concentration binding to Norepinephrine transporter using 0.2 nM paroxetine
- ChEMBL_143122 (CHEMBL748004) Inhibition of 0.5 nM [3H]nisoxetine binding to Norepinephrine transporter
- ChEMBL_144837 (CHEMBL750463) Inhibition of [3H]nisoxetine (0.5 nM) binding to Noradrenaline transporter
- ChEMBL_144982 (CHEMBL754324) Inhibitory concentration binding to Norepinephrine transporter using 0.2 nM paroxetine
- ChEMBL_162430 (CHEMBL771833) Inhibition of Protein-tyrosine phosphatase 1B in 300 nM DTT
- ChEMBL_201650 (CHEMBL806551) Inhibition of 0.2 nM [3H]paroxetine binding to Serotonin transporter
- ChEMBL_201665 (CHEMBL803039) Inhibitory concentration binding to Serotonin transporter using 0.5 nM Nisoxetine
- ChEMBL_201999 (CHEMBL809444) Inhibition of 0.2 nM [3H]paroxetine binding to Serotonin transporter
- ChEMBL_2232253 (CHEMBL5146025) Inhibition of JNK3 (unknown origin) irradiated with 400 nm light
- ChEMBL_305286 (CHEMBL832835) Inhibition of human Phosphodiesterase 5; IC50 range 0.03-0.3 nM
- ChEMBL_311943 (CHEMBL833410) Maximal response (3000 nM) at human cannabinoid receptor 1 (hCB1)
- ChEMBL_37258 (CHEMBL650173) Percent adenylyl cyclase activation at 10000 nM of compound concentration
- ChEMBL_38297 (CHEMBL647101) Percent adenylyl cyclase activation at 10000 nM of compound concentration
- ChEMBL_54963 (CHEMBL666694) Inhibition of rat liver DHFR assayed spectrophotometrically at 340 nM
- ChEMBL_61833 (CHEMBL673203) Displacement of [3H]WIN-35428(0.5 nM) from Dopamine transporter
- ChEMBL_62004 (CHEMBL670187) Displacement of 0.5 nM [3H]WIN-35248 from Dopamine transporter
- ChEMBL_69685 (CHEMBL682021) Inhibitory activity against factor Xa, activity expressed as Ki nM
- ChEMBL_71395 (CHEMBL681747) Displacement of 10 nM [3H]dexamethasone from human Glucocorticoid receptor
- ChEMBL_71396 (CHEMBL681748) Displacement of 10 nM [3H]dexamethasone from human Glucocorticoid receptor
- ChEMBL_1486535 (CHEMBL3532204) Binding affinity to CYP2A6 (unknown origin) assessed as type 2 interaction as increase in absorbance 431 to 432 nm and decrease in 406 to 412 nm
- ChEMBL_1486536 (CHEMBL3532205) Binding affinity to CYP2A6 (unknown origin) assessed as type 1 interaction as increase in absorbance 379 to 387 nm and decrease in 414 to 420 nm
- ChEMBL_1488921 (CHEMBL3535230) Binding affinity to CYP2A13 (unknown origin) assessed as type 2 interaction as increase in absorbance 431 to 432 nm and decrease in 406 to 412 nm
- ChEMBL_1488922 (CHEMBL3535231) Binding affinity to CYP2A13 (unknown origin) assessed as type 1 interaction as increase in absorbance 379 to 387 nm and decrease in 414 to 420 nm
- ChEMBL_151864 (CHEMBL759578) Binding Affinity of ligand against Protease-activated Receptor (PAR-1) using [3H]-s-(p-F-Phe)-homoarginine-L-homoarginine-KY-NH2, 10 nM (Kd= 15 nM)
- ChEMBL_1541737 (CHEMBL3745245) Antagonist activity against human D2 receptor expressed in CHO cells assessed as dopamine EC50 at 50 nM by [35S]GTPgammaS binding assay (Rvb = 225 +/- 45 nM)
- ChEMBL_1541738 (CHEMBL3745246) Antagonist activity against human D2 receptor expressed in CHO cells assessed as dopamine EC50 at 300 nM by [35S]GTPgammaS binding assay (Rvb = 225 +/- 45 nM)
- ChEMBL_1541740 (CHEMBL3745248) Antagonist activity against human D2 receptor expressed in CHO cells assessed as dopamine EC50 at 200 nM by [35S]GTPgammaS binding assay (Rvb = 225 +/- 45 nM)
- ChEMBL_154360 (CHEMBL756622) Binding affinity was determined by displacement of 20 nM [3H]- thiazolidinedione from 4 nM biotinylated human Peroxisome proliferator activated receptor gamma (PPAR gamma) ligand binding domain
- In Vitro BRCC36 Activity Assay BRCC36 activity was measured using 1 nM purified BRISC complex and 80 nM di-ubiquitin substrate DiUbK63TAMRA (UF-310, Boston Biochem). TAMRA fluorescence intensity was monitored using a PHERAstar plate reader with excitation at 540 nm and emission at 590 nm. Reactions were performed in black low-volume 384-well plates and analyzed as described above.
- AMPD Enzymatic Activity Assay AMPD1 was added to buffer A (50 mM HEPES, 150 mM KCl, 5 mM MgCl2, and 0.5 mMglutathione [pH 7.4]) to a concentration of 10 nM. A substrate mix consistingof 2mMNADPH, 6mMa-ketoglutarate, 8mMAMP, and 15 U GDH was prepared in buffer A. Substrate mix (4 ml) was added to the plate, followed by centrifugation and incubation for 1 hr. Detection was by absorbance at 340 nm following the conversion of NADPH to NADP+. Recombinant AMPD enzyme activity assays were performed in a 384-well format using the same conditions as described above, except the assay volume was 80 ml and the AMP concentration for the AMPD2 assay was 8 mM. Different AMPD enzyme concentrations were used to have linear reactions: hAMPD1 (4 nM), hAMPD2 (38 nM), hAMPD3 (1 nM), rAMPD1 (116 nM), rAMPD2 (53 nM), rAMPD3 (31 nM), and mAMPD1 (816 nM).
- ChEMBL_1982181 (CHEMBL4615443) Competitive inhibition of human recombinant MAGL at 31.25 nM to 125 nM pre-incubated for 5 mins before MAGL substrate addition and further incubated for 10 mins
- GC Enzyme Assay Enzyme activity was determined as the production of fluorescent resorufin from the substrate. The fluorescence was measured (excitation 570 nm, emission 610 nm) in a fluorimeter.
- Millipore Kinase Panel Assay Employing the Milipore panel of purified kinases EXAMPLE 87 (IC50=1 nM) inhibited 98% of purified Syk kinase activity at 50 nM. IC50 values were determined for those kinases that were inhibited by >80% at 300 nM in the Millipore kinase panel.
- ChEBML_161755 Inhibitory concentration against Protein phosphatase 1(10 nM) isolated from rabbit muscle
- ChEBML_62404 Inhibition of [3H]spiperone binding to Dopamine receptor D2 at 0.02 nM
- ChEMBL_124654 (CHEMBL737063) Inhibition of Mitogen-activated protein kinase p38 beta at 1000 nM
- ChEMBL_141126 (CHEMBL882639) Inhibitory activity against Candida albicans (Nmt) assessed as inhibitory concentration (nM)
- ChEMBL_142785 (CHEMBL752160) Inhibition of reuptake of [3H]-NE (20 nM) by norepinephrine transporter
- ChEMBL_147059 (CHEMBL754879) Displacement of [3H]DPDPE (0.63 nM) from Opioid receptor delta 1
- ChEMBL_149012 (CHEMBL758576) Displacement of [3H]DAGO (1.28 nM) from Opioid receptor mu 1
- ChEMBL_202318 (CHEMBL807674) Inhibition of [3H]paroxetine (0.2 nM) binding to 5-HT transporter
- ChEMBL_303180 (CHEMBL829674) Inhibitory constant against dopamine D2 receptor using 0.2 nM [3H]-spiperone
- ChEMBL_306214 (CHEMBL831123) Inhibition of Matrix metalloprotease-12 in presence of 5 nM acetohydroximate
- ChEMBL_306214 (CHEMBL831123) Inhibition of Matrix metalloproteinase-12 in pressence of 5 nM acetohydroximate
- ChEMBL_306336 (CHEMBL828156) Irreversible inhibition of fatty acid amide hydrolase; range=1-3 nM
- ChEMBL_306356 (CHEMBL828175) Irreversible inhibition of fatty acid amide hydrolase; range=. 5-6 nM
- ChEMBL_49585 (CHEMBL661238) Compound was tested for the inhibition of chymotrypsin at 120 nM
- ChEMBL_49586 (CHEMBL661239) Compound was tested for the inhibition of chymotrypsin at 138 nM
- ChEMBL_49587 (CHEMBL661240) Compound was tested for the inhibition of chymotrypsin at 276 nM
- ChEMBL_49588 (CHEMBL661241) Compound was tested for the inhibition of chymotrypsin at 69 nM
- ChEMBL_61584 (CHEMBL675756) Inhibitory concentration against Dopamine receptor D2 (Inactive at >1000 nM concentration)
- ChEMBL_61673 (CHEMBL670051) Inhibition of reuptake of [3H]DA (20 nM) by dopamine transporter
- ChEMBL_62090 (CHEMBL674970) Affinity at dopamine D2 receptor, (For haloperidol Ki(nM)= 1.5+/-1.2)
- IC50 Measurements Enzyme activity was determined as the production of fluorescent 4-MU from the substrate. The fluorescence was measured (excitation 365 nm, emission 445 nm) in an Aviv fluorimeter.
- In Vitro AMSH Activity Assay AMSH activity was measured using 10 nM purified AMSH (E-548B, Boston Biochem) and 80 nM di-ubiquitin substrate DiUbK63TAMRA (UF-310, Boston Biochem). TAMRA fluorescence intensity was monitored using a PHERAstar plate reader with excitation at 540 nm and emission at 590 nm. Reactions were performed in black low-volume 384-well plates and analyzed as described above.
- Kat6b Activity Assay Kat6b was incubated for 30 mins at 22° C. in the presence of different concentrations of test substances (0 μM, and within the range 0.01-20 μM) in assay buffer [25 mM Tris/HCl pH 8, 1 mM EGTA, 2.5 mM Glutathion, 0.02% Chicken Albumin, 0.05% Pluronic F127, 25 mM NaCl, 500 nM H4 peptide and 600 nM Acetyl Coenzym A].The reaction was stopped by addition of Detection Solution (25 mM HEPES pH 7.5, 0.1% BSA, 22 nM SAXL665 (Cisbio #610SAXLE), 100 μM Anacardic Acid (Enzo #ALX-270-381), 1 nM Anti-Histone H4 (ACETYL K8) Antibody (ABCAM #AB15823), 0.5 nM Anti-Rabbit IgG Eu (Perkin Elmer #AD0083).The fluorescence emission at 620 nm and 665 nm after excitation at 330-350 nm was measured in a TR-FRET measuring instrument, for instance a Rubystar or a Pherastar (both from BMG Lab Technologies, Offenburg, Germany) or a Viewlux (Perkin-Elmer). The ratio of the emission at 665 nm and at 622 nm was used as indicator of the amount of acetylated peptide.
- Fluorescence Titration Assay To assess direct binding of inhibitors to oxidized NQO2 (NQO2ox), fluorescence quenching of FAD was monitored with an excitation wavelength of 350 nm and an emission wavelength of 430 nm. NQO2 (775 or 38.75 nM) was titrated with the indicated concentrations of TBB, TBBz, and DMAT.
- Biochemical Assay The enzyme and peptide solution was incubated with compound for 15 minutes at room temp before the reaction was initiated by the addition of ATP. The standard 5l reaction mixture contained 500 μM ATP, 2 μM peptide (STK1 Peptide), 0.75 nM of IRAK4 in reaction buffer (50 mM HEPES, pH 7.0, 0.02% NaN3, 0.01% BSA, 0.1 mM Orthovanadate, 5 mM MgCl2, 0.025% NP-40, 1 mM DTT). After 120 min of incubation at room temperature, 5 μl of Stop and Detect Solution (1:100 Cryptate labeled anti-phosphorylated peptide antibody solution and 125 nM Tracer in a 50 mM HEPES pH 7.0 detection buffer containing sufficient EDTA) was added. The plate was then further incubated for 60 minutes at room temperature and read on Envision 2103 Multilabeled reader (PerkinElmer) with excitation/emission/FRET emission at 340 nm/615 nm/665 nm, respectively. Fluorescence intensities at 615 nm and 665 nm emission wavelengths were expressed as a ratio (665 nm/615 nm).
- Binding Assay DDR1 and DDR2 binding assays were performed using Life Technologies LanthaScreen Europium Kinase Binding assay. The compounds were incubated with 5 nM DDR1 (Carna Biosciences) or 5 nM DDR2 (Life Technologies) for 1 hour at room temperature in white 384-well OptiPlate (PerkinElmer), containing 20 nM or 10 nM Kinase Tracer 178 respectively and 2 nM Europium labelled anti-GST antibody (Life Technologies) in assay buffer (50 mM HEPES pH 7.5, 10 mM MgCl2, 1 mM EGTA and 0.01% BRIJ35).The ratio of fluorescence emission 665 nm/615 nm after excitation at 340 nm was obtained using the Tecan Spark 20M plate reader. IC50 values were determined in GraphPad Prism 7.0 software, using 4 parameter model: log(inhibitor) vs. response. IC50 values were converted in Ki using the Cheng-Prusoff equation (Ki=IC50/(1+[Tracer]/Kd).
- Binding Assay DDR1 and DDR2 binding assays were performed using Life Technologies LanthaScreen™ Europium Kinase Binding assay. The compounds were incubated with 5 nM DDR1 (Carna Biosciences) or 5 nM DDR2 (Life Technologies) for 1 hour at rt in white 384-well OptiPlate (PerkinElmer), containing 20 nM or 10 nM Kinase Tracer 178 respectively and 2 nM Europium labelled anti-GST antibody (Life Technologies) in assay buffer (50 mM HEPES pH 7.5, 10 mM MgCI2, 1 mM EGTA and 0.01% BRIJ35). The ratio of fluorescence emission 665 nm/615 nm after excitation at 340 nm was obtained using the Tecan Spark 20M plate reader. IC50 values were determined in GraphPad Prism 7.0 software, using 4 parameter model: log(inhibitor) vs. response. IC50 values were converted in Ki using the Cheng-Prusoff equation (Ki=IC50/(1+[Tracer]/Kd).
- Evaluation of Inhibitors with Recombinant LC/A Recombinant BoNT LC/A activity was measured in black 96-well microtiter plates by use of a Molecular Devices (Sunnyvale, CA) SpectraMax GeminiEM plate reader. Fluorimeter parameters consisted of excitation@490 nm (slit width =2 nm), an emission@532 nm (slit width =2 nm), and a cut-off filter at 495 nm. Initial rates were measured from the linear region of each assay. IC50 values were determined by using equation that includes initial rates in the presence/absence of inhibitor.
- AlphaScreen Assay Demethylase reactions and AlphaScreen assays were performed as described (Sayegh et al., 2013), with a few exceptions. All demethylase buffers contained 125 μM αKG, 50 μM (NH4)2Fe(SO4)2 and 64 nM C-terminal H3K4me3(1-20)-GGK-biotinylated peptides (Sayegh et al., 2013). The enzymes were used at the following concentrations: 19 nM FLAG-KDM5A, 25 nM FLAG-KDM5B, 8 nM FLAGKDM5C, 29 nM His-FLAG-KDM6A, and 8 nM FLAG-KDM6B. Demethylation reactions contained 0.05% DMSO. The AlphaScreen general IgG (protein A) detection kit from PerkinElmer Life Sciences was used as described (Sayegh et al., 2013). The AlphaScreen optic module on a Pherastar (BMG Labtech) microplate reader was used to record luminescence emission at 570 nm.
- ChEBML_161777 Inhibitory concentration against Protein phosphatase 2A (25 nM) isolated from bovine myocardial tissue
- ChEBML_210583 In vitro concentration of compound required to inhibit 50% of 6 nM thrombin
- ChEBML_225937 Antagonistic activity against RAR alpha in transcriptional activation assay with 32 nM TTNPB
- ChEBML_225941 Antagonistic activity against RAR beta in transcriptional activation assay with 10 nM TTNPB
- ChEBML_225945 Antagonistic activity against RAR gamma in transcriptional activation assay with 3.2 nM TTNPB
- ChEBML_29107 Affinity for adenosine A1 receptor of calf brain using 1 nM [3H]PIA
- ChEBML_44517 In vitro agonist efficacy against PPAR gamma along with 100 nM BRL-49653
- ChEBML_58650 Inhibition of [3H]SCH-23,390 binding to Dopamine receptor D1 at 0.25 nM
- ChEMBL_157586 (CHEMBL769302) Concentration inhibiting 1 nM LTB4-induced aggregation in GP polymorphonuclear (PMN) leukocytes.
- ChEMBL_1623 (CHEMBL616509) Affinity at 5-hydroxytryptamine 1D receptor (For sumatriptan = Ki (nM)-12+/-1.9)
- ChEMBL_195314 (CHEMBL799869) Antagonist activity of TTNPB (10 nM) function at retinoic acid receptor alpha
- ChEMBL_195805 (CHEMBL807717) Antagonist activity of TTNPB (10 nM) function at retinoic acid receptor beta
- ChEMBL_196316 (CHEMBL806259) Antagonist activity of TTNPB (10 nM) function at retinoic acid receptor gamma
- ChEMBL_198189 (CHEMBL799117) Inhibition of reuptake of [3H]5-HT (20 nM) by serotonin transporter
- ChEMBL_201658 (CHEMBL803032) Inhibitory activity against Serotonin transporter using 0.2 nM [3H]paroxetine as radioligand
- ChEMBL_2205 (CHEMBL617030) Affinity at 5-hydroxytryptamine 2 receptor (For ketanserin = Ki(nM)= 0.7+/-0.09)
- ChEMBL_2589 (CHEMBL617457) Inhibitory concentration against 5-hydroxytryptamine 2A receptor (Inactive at >1000 nM concentration)
- ChEMBL_303199 (CHEMBL829825) Inhibitory constant against sigma receptor type 1 using 3 nM [3H]pentazocine
- ChEMBL_303294 (CHEMBL827427) Inhibitory constant against sigma receptor type 2 using 3 nM [3H]ditolylguanidine
- ChEMBL_305584 (CHEMBL828032) Inhibition of HER1 kinase in intact DHER14 cells; Range = 5-15 nM
- ChEMBL_305585 (CHEMBL874430) Inhibition of HER2 kinase in intact CSH12 cells; Range = 25-50 nM
- ChEMBL_306078 (CHEMBL833032) Inhibitory concentration against Nicotinic acetylcholine receptor alpha7; Range is 0.5-8 nM
- ChEMBL_306101 (CHEMBL830876) Inhibitory concentration against Nicotinic acetylcholine receptor alpha7; Range is 100-200 nM
- ChEMBL_306102 (CHEMBL830877) Inhibitory concentration against Nicotinic acetylcholine receptor alpha9; Range is 100-200 nM
- ChEMBL_306126 (CHEMBL833065) Inhibitory concentration against Nicotinic acetylcholine receptor alpha7; Range is 0.3-1.5 nM
- ChEMBL_306143 (CHEMBL831378) Inhibitory concentration against Nicotinic acetylcholine receptor alpha7; Range is 0.3-1.5 nM
- ChEMBL_3141 (CHEMBL617983) Affinity at 5-hydroxytryptamine 3 receptor (For granisetron = Ki (nM)=0.3+/-0.01)
- ChEMBL_320908 (CHEMBL881235) Antagonistic activity against estrogen receptor beta in presence of 0.1 nM estradiol
- ChEMBL_320911 (CHEMBL881238) Antagonistic activity against estrogen receptor alpha in presence of 0.1 nM estradiol
- ChEMBL_49882 (CHEMBL875000) Displacement of 0.1 nM [3H]pCCK-8 from guinea pig pancreatic membranes
- ChEMBL_50384 (CHEMBL662840) Binding affinity for Cytochrome P450 17A1 (17-alpha-hydroxypregnenolone Km=560 nM)
- ChEMBL_50385 (CHEMBL662841) Binding affinity for Cytochrome P450 17A1 (17-alpha-hydroxypregnenolone Km=560 nM)
- ChEMBL_62156 (CHEMBL676559) Inhibitory concentration binding to Dopamine transporter using 0.5 nM [3H]WIN-35428
- ChEMBL_62157 (CHEMBL676560) Inhibitory concentration binding to dopamine transporter using 0.5 nM [3H]WIN-35428
- ChEMBL_69106 (CHEMBL678731) Inhibition of 1 nM alpha melanocortin stimulating hormone (MSH) from frog skin
- ChEMBL_866 (CHEMBL615922) Inhibitory concentration against 5-hydroxytryptamine 1A receptor (Inactive at >1000 nM concentration)
- Fluorophore Displacement Assay Briefly, steady-state fluorescence spectra of ANS binding was monitored by measuring the increase in fluorescence signal between 450550 nm following excitation at 400 nm while for DAUDA binding was measured between 450600 nm following excitation at 335 nm. Spectrophotometer slit widths were set to 5 and 10 nm for the excitation and emission monochromators, respectively. DAUDA (0.025 mM) and ANS (0.25 mM) were titrated into a hIFABP solution at a protein concentration of 0.5 u1 μM in buffer (20 mM MES pH 5.5, 50 mM NaCl) at 20 C
- Homogeneous Time-resolved Fluorescence (HTRF) Assay Enzymes were assayed with retinoblastoma substrate in 384-well plates containing diluted test compounds. Final ATP concentration was 3x the respective enzyme Km. The phosphorylated substrate was analyzed by adding lance europium anti-rabbit IgG and anti-His-allophycocyanin, resulting in fluorescence resonance energy transfer between europium anti-rabbit and allophycocyanin, and quantified by fluorescence intensity ratio 665 nm/615 nm (excited at 340 nm). IC50 values were calculated from net readings at 665 nm, normalized for europium readings at 615 nm.
- Binding Assay Kinetic analysis of NS3 inhibitor binding experiment were performed using the KinTek stopped flow instrument (SF-2005; excitation, 325 nm; and emission, 410 nm) with a programmable shutter to minimized photobleaching.
- Binding Fluorescence Titrations Binding of TLM and the TLM analogs to KasA was quantified by monitoring changes in the intrinsic tryptophan fluroescence of the enzyme using 280-nm excitation and 557-nm emission.
- Binding Assay ERRγ: The arylethene derivative of the present invention was sequentially added to a 384 well plate from a concentration of 10 μM to a final concentration of two-fold dilution. Then, a GST-bound ERR gamma ligand-binding domain (LBD) was added to a final concentration of 5 nM, and a fluorescein-conjugated coactivator PGC1a and a Tb-a-GST antibody were added to 500 nM and 5 nM, respectively. After all reagents were added, a reaction was carried out with gentle shaking at 20° C. for 1 hour, and after the reaction, a binding activity was measured by a TR-FRET method. That is, excitation at 340 nm was performed, each emission value at 495 nm and 520 nm was measured, the result assay was a value measured at 490 nm/a value measured at 520 nm, and an analysis program was Prism 6.
- Fluorescence Polarization Assay For competition-based fluorescence polarization assays, the ability of the test compounds to displace fluorophore-labeled peptides from their respective binding proteins was analyzed as previously described.3 Peptide sequences were: STAT1: 5-carboxyfluorescein-GY(PO3H2)DKPHVL; STAT3: 5-carboxyfluorescein-GY(PO3H2)LPQTV-NH2; STAT4: 5-carboxyfluorescein-GY(PO3H2)LPQNID-OH; STAT5a and STAT5b: 5-carboxyfluorescein-GY(PO3H2)LVLDKW; STAT6: 5-carboxyfluorescein-GY(PO3H2)VPWQDLI-OH; Lck SH2: 5-carboxyfluorescein-GY(PO3H2)EEIP. Due to protein instability, STAT2 was not analyzed. Final concentration of 5-carboxyfluorescein-labeled peptides: 10 nM. Proteins were used at the following final concentrations, which correspond to the Kd-values of the interactions with the respective fluorophore-labeled peptide: STAT1: 420 nM; STAT3: 270 nM; STAT4: 130 nM; STAT5a: 130 nM; STAT5b: 100 nM; STAT6: 310 nM; Lck SH2: 30 nM. Pipetting was carried out partly using a Biomek FX workstation (Beckman-Coulter). Pr
- ChEBML_148090 Displacement of [3H]DAMGO (2 nM ) from opioid receptor mu 1 in human brain
- ChEBML_208850 Binding affinity towards Tachykinin receptor 2 using [125I]iodohistidyl-NKA (0.1 nM)) as radioligand
- ChEBML_49396 Displacement of 0.1 nM [3H]Boc[Nle28,31]-CCK27-33 from guinea pig pancreatic membranes
- ChEMBL_138203 (CHEMBL748432) Inhibition of 0.1 nM [3H]cis-methyldioxolane binding to rat neocortex muscarinic receptor
- ChEMBL_138333 (CHEMBL748213) Inhibition of 0.03 nM [3H]quinuclidinyl benzylate binding to rat neocortex muscarinic receptor
- ChEMBL_1442176 (CHEMBL3375249) Inhibition of recombinant human BChE at 50 nM by stopped flow apparatus method
- ChEMBL_145391 (CHEMBL750081) Inhibition of [3H]naltrindole (0.55 nM) binding from human Opioid receptor kappa 1
- ChEMBL_154437 (CHEMBL756494) Inhibition of HDJ-2 farnesylation in PSN-1 cells with 100 nM GGT1
- ChEMBL_154439 (CHEMBL756496) Inhibition of K-Ras farnesylation in PSN-1 cells with 100 nM GGT1
- ChEMBL_157422 (CHEMBL768878) 50% reduction in myeloperoxidase secretion from human PMNs mediated by 100 nM C5a
- ChEMBL_225937 (CHEMBL874059) Antagonistic activity against RAR alpha in transcriptional activation assay with 32 nM TTNPB
- ChEMBL_225941 (CHEMBL846506) Antagonistic activity against RAR beta in transcriptional activation assay with 10 nM TTNPB
- ChEMBL_225945 (CHEMBL843953) Antagonistic activity against RAR gamma in transcriptional activation assay with 3.2 nM TTNPB
- ChEMBL_226541 (CHEMBL846481) Binding affinity against sigma receptor in bovine cerebellum using 2.0 nM [3H]- haloperidol
- ChEMBL_29619 (CHEMBL639641) Inhibition of 1 nM [3H]- N6- (phenylisopropyl) adenosine binding to Adenosine A1 receptor
- ChEMBL_304333 (CHEMBL839760) Displacement of 5 nM GM-BODIPY from mouse heat shock protein HSP90-alpha
- ChEMBL_305910 (CHEMBL832625) Inhibitory activity against hepatitis C virus NS5B polymerase enzyme; Range=6-9 nM
- ChEMBL_305955 (CHEMBL874549) Inhibitory concentration against Nicotinic acetylcholine receptor alpha-7 Range is 3-5 nM
- ChEMBL_306141 (CHEMBL831376) Inhibitory concentration against Nicotinic acetylcholine receptor alpha4-beta2; Range is 3-5 nM
- ChEMBL_306172 (CHEMBL830967) Inhibitory concentration against Nicotinic acetylcholine receptor alpha3-beta2; Range is 3-5 nM
- ChEMBL_306215 (CHEMBL831124) Inhibitory concentration against Nicotinic acetylcholine receptor alpha3-beta2; Range is 0.5-8 nM
- ChEMBL_306217 (CHEMBL874559) Inhibitory concentration against Nicotinic acetylcholine receptor alpha4-beta2; Range is 0.5-8 nM
- ChEMBL_306274 (CHEMBL827551) Inhibitory concentration against Nicotinic acetylcholine receptor alpha2-beta2; Range is 0.3-1.5 nM
- ChEMBL_306275 (CHEMBL827552) Inhibitory concentration against Nicotinic acetylcholine receptor alpha2-beta4; Range is 0.3-1.5 nM
- ChEMBL_306276 (CHEMBL827553) Inhibitory concentration against Nicotinic acetylcholine receptor alpha3-beta2; Range is 0.3-1.5 nM
- ChEMBL_306279 (CHEMBL828422) Inhibitory concentration against Nicotinic acetylcholine receptor alpha4-beta2; Range is 0.3-1.5 nM
- ChEMBL_306297 (CHEMBL828589) Inhibitory concentration against Nicotinic acetylcholine receptor alpha2-beta4; Range is 0.3-1.5 nM
- ChEMBL_306298 (CHEMBL828590) Inhibitory concentration against Nicotinic acetylcholine receptor alpha4-beta2; Range is 0.3-1.5 nM
- ChEMBL_306418 (CHEMBL828089) Inhibition of EGF Receptor autophosphorylation in A431 cell lysate; Range = 1-10 nM
- ChEMBL_306444 (CHEMBL829088) Inhibition of EGF Receptor autophosphorylation in A431 cell lysate; Range = 10-50 nM
- ChEMBL_306445 (CHEMBL829089) Inhibition of EGF Receptor autophosphorylation in A431 cell lysate; Range = 25-50 nM
- ChEMBL_306459 (CHEMBL829536) Inhibition of EGF Receptor autophosphorylation in A431 cell lysate; Range = 6.7-20 nM
- ChEMBL_306537 (CHEMBL827826) Inhibition of EGF Receptor autophosphorylation in A431 cell lysate; Range = 10000-50000 nM
- ChEMBL_306538 (CHEMBL827827) Inhibition of EGF Receptor autophosphorylation in A431 cell lysate; Range = 3-5 nM
- ChEMBL_306555 (CHEMBL828300) Inhibition of EGF Receptor autophosphorylation in A431 cell lysate; Range = 1-10 nM
- ChEMBL_306556 (CHEMBL828301) Inhibition of EGF Receptor autophosphorylation in A431 cell lysate; Range = 4-10 nM
- ChEMBL_306557 (CHEMBL828302) Inhibition of EGF Receptor autophosphorylation in A431 cell lysate; Range = 5-25 nM
- ChEMBL_306573 (CHEMBL832921) Inhibition of EGF Receptor autophosphorylation in A431 cell lysate; Range = 50-250 nM
- ChEMBL_306574 (CHEMBL832922) Inhibition of EGF Receptor autophosphorylation in A431 cell lysate; Range = 6.7-20 nM
- ChEMBL_306626 (CHEMBL829928) Inhibition of EGF Receptor autophosphorylation in A431 cell lysate; Range = 1000-5000 nM
- ChEMBL_306642 (CHEMBL832979) Inhibition of EGF Receptor autophosphorylation in A431 cell lysate; Range = 1000-10000 nM
- ChEMBL_310145 (CHEMBL838124) Inhibition of Ras farnesylation in H-Ras transformed NIH3T3 cells at 100 nM
- ChEMBL_44517 (CHEMBL658541) In vitro agonist efficacy against PPAR gamma along with 100 nM BRL-49653
- ChEMBL_44520 (CHEMBL656170) In vitro agonistic activity against PPAR gamma along with 100 nM BRL-49653
- ChEMBL_48586 (CHEMBL662466) Inhibition of inositol phosphate production induced by Cholecystokinin type B receptor (0.5 nM)
- ChEMBL_50690 (CHEMBL663077) Inhibition of Cytochrome P450 19A1 against Androstenedione at 0.25 uM (Km=55 nM)
- ChEMBL_60014 (CHEMBL675839) Inhibition of [3H]spiroperidol binding to rat striatal membrane using 0.5 nM ligand.
- ChEMBL_62077 (CHEMBL672376) Displacement of [3H]spiperone (0.5 nM) from rat corpus striatum dopamine D2 receptor
- ChEMBL_62147 (CHEMBL675903) Inhibitory activity against Dopamine transporter using 0.5 nM [3H]WIN-35428 as radioligand
- ChEMBL_86643 (CHEMBL693976) 50% reduction in myeloperoxidase secretion from human PMNs mediated by 100 nM C5a
- ChEMBL_1625308 (CHEMBL3867777) Inhibition of human ABCB1 transfected in HEK293 cells assessed as potentiation of doxorubicin-induced cytotoxicity by measuring doxorubicin IC50 at 50 nM after 72 hrs by CCK8 assay (Rvb = 504.65 +/- 44.94 nM)
- ChEMBL_1625309 (CHEMBL3867778) Inhibition of human ABCB1 transfected in HEK293 cells assessed as potentiation of doxorubicin-induced cytotoxicity by measuring doxorubicin IC50 at 100 nM after 72 hrs by CCK8 assay (Rvb = 504.65 +/- 44.94 nM)
- ChEMBL_1625310 (CHEMBL3867779) Inhibition of human ABCB1 transfected in HEK293 cells assessed as potentiation of doxorubicin-induced cytotoxicity by measuring doxorubicin IC50 at 200 nM after 72 hrs by CCK8 assay (Rvb = 504.65 +/- 44.94 nM)
- ChEMBL_1625311 (CHEMBL3867780) Inhibition of human ABCB1 transfected in HEK293 cells assessed as potentiation of doxorubicin-induced cytotoxicity by measuring doxorubicin IC50 at 500 nM after 72 hrs by CCK8 assay (Rvb = 504.65 +/- 44.94 nM)
- ChEMBL_1625312 (CHEMBL3867781) Inhibition of human ABCB1 transfected in HEK293 cells assessed as potentiation of doxorubicin-induced cytotoxicity by measuring doxorubicin IC50 at 1000 nM after 72 hrs by CCK8 assay (Rvb = 504.65 +/- 44.94 nM)
- ChEMBL_1625317 (CHEMBL3867786) Inhibition of human ABCC1 transfected in HEK293 cells assessed as potentiation of etoposide-induced cytotoxicity by measuring etoposide IC50 at 500 nM after 72 hrs by CCK8 assay (Rvb = 38.54 +/- 5.62 nM)
- ChEMBL_1625318 (CHEMBL3867787) Inhibition of human ABCC1 transfected in HEK293 cells assessed as potentiation of etoposide-induced cytotoxicity by measuring etoposide IC50 at 25000 nM after 72 hrs by CCK8 assay (Rvb = 38.54 +/- 5.62 nM)
- ChEMBL_1625349 (CHEMBL3867818) Inhibition of human ABCB1 expressed in KBV1 cells assessed as potentiation of colchicine-induced cytotoxicity by measuring colchicine IC50 at 50 nM after 72 hrs by MTT assay (Rvb = 487.57 +/- 30.54 nM)
- ChEMBL_1625350 (CHEMBL3867819) Inhibition of human ABCB1 expressed in KBV1 cells assessed as potentiation of colchicine-induced cytotoxicity by measuring colchicine IC50 at 100 nM after 72 hrs by MTT assay (Rvb = 487.57 +/- 30.54 nM)
- ChEMBL_1625351 (CHEMBL3867820) Inhibition of human ABCB1 expressed in KBV1 cells assessed as potentiation of colchicine-induced cytotoxicity by measuring colchicine IC50 at 200 nM after 72 hrs by MTT assay (Rvb = 487.57 +/- 30.54 nM)
- ChEMBL_1625352 (CHEMBL3867821) Inhibition of human ABCB1 expressed in KBV1 cells assessed as potentiation of colchicine-induced cytotoxicity by measuring colchicine IC50 at 500 nM after 72 hrs by MTT assay (Rvb = 487.57 +/- 30.54 nM)
- ChEMBL_1625353 (CHEMBL3867822) Inhibition of human ABCB1 expressed in KBV1 cells assessed as potentiation of colchicine-induced cytotoxicity by measuring colchicine IC50 at 1000 nM after 72 hrs by MTT assay (Rvb = 487.57 +/- 30.54 nM)
- ChEMBL_1625368 (CHEMBL3867837) Inhibition of human ABCB1 expressed in KBV1 cells assessed as potentiation of vincristine-induced cytotoxicity by measuring vincristine IC50 at 50 nM after 72 hrs by MTT assay (Rvb = 277.68 +/- 56.61 nM)
- ChEMBL_1625369 (CHEMBL3867838) Inhibition of human ABCB1 expressed in KBV1 cells assessed as potentiation of vincristine-induced cytotoxicity by measuring vincristine IC50 at 100 nM after 72 hrs by MTT assay (Rvb = 277.68 +/- 56.61 nM)
- ChEMBL_1625370 (CHEMBL3867839) Inhibition of human ABCB1 expressed in KBV1 cells assessed as potentiation of vincristine-induced cytotoxicity by measuring vincristine IC50 at 200 nM after 72 hrs by MTT assay (Rvb = 277.68 +/- 56.61 nM)
- ChEMBL_1625371 (CHEMBL3867840) Inhibition of human ABCB1 expressed in KBV1 cells assessed as potentiation of vincristine-induced cytotoxicity by measuring vincristine IC50 at 500 nM after 72 hrs by MTT assay (Rvb = 277.68 +/- 56.61 nM)
- ChEMBL_1625372 (CHEMBL3867841) Inhibition of human ABCB1 expressed in KBV1 cells assessed as potentiation of vincristine-induced cytotoxicity by measuring vincristine IC50 at 1000 nM after 72 hrs by MTT assay (Rvb = 277.68 +/- 56.61 nM)
- Coactivator Peptide Competitive FP Assay (IC50/EC50) AR functionality was determined by fluorescence polarization (excitation @485 nm, emission @ 530 nm) assay, which measures fluorescently labeled SRC2-3 peptide displacement from DHT-bound AR-LBD.
- Activity Assay A BACE2 activity assay kit from AnaSpec (Fremont, Calif.) was used to determine the potency of the selected compounds by AnaSpec BACE2 fluorescent assay. To each well of a black 96-well micro plate, 50 μl of the β-secretase substrate (Hylite Fluor 488) diluted 1:100 using assay buffer was first added.The compounds were serially diluted (1:2) into DMSO from their 10 mM stock concentrations in DMSO. The serially diluted inhibitors were diluted 1:100 into 1× assay buffer in a 96-well polypropylene plate (assay dilution plate). The final concentration of these compounds tested was 300 nM, 150 nM, 75 nM, 37.5 nM, 18.75 nM, 9.375 nM, 4.688 nM, 2.344 nM, 1.172 nM, 0.586 nM, and 0.293 nM.A volume of 10 μl of the inhibitor from the assay dilution plate, a control inhibitor LY2886721 or inhibitor vehicle was then added to each well of the black 96-well micro plate.The recombinant BACE2 was diluted in an assay buffer and the final BACE2 was 10.6 nM. The enzyme was added in a volume of 40 μl to each well of a black 96-well micro plate.The reagents were mixed gently and the assay plate was placed into a POLARstar fluorescent plate reader with excitation wavelength set at 485 nm and the emission wavelength set at 520 nm. The plate reader was set to take readings every 5 minutes for up to 30 minutes or a 30 minute endpoint reading was used.
- Imidazole Spin Shift Assay In short, the difference between the imidazole-bound low-spin peak at 430 nm and the inhibitorbound high-spin peak at 395 nm was measured as a function of inhibitor concentration.
- Lanthascreen Competitive Binding Assay The assay was performed according to manufacturer protocol. A mixture of nM GST-PPARG-LBD, 5 nM Tb-GST-antibody, 5 nM Fluormone Pan-PPAR Green, and serial dilutions of the experimental compound, beginning at 10 μM downwards, was added to wells of black 384-well low-volume plates (Greiner) to a total volume of 18 μL. All dilutions were made in TR-FRET assay buffer C. DMSO at 2% final concentration was used as a no-ligand control. Experiment was performed in triplicate, and incubated for 2 hours in the dark prior to assay read in Perkin Elmer ViewLux ultra HTS microplate reader. FRET signal was measured by excitation at 340 nm and emission at 520 nm for fluorescein and 490 nm for terbium. Fold change over DMSO was calculated using GraphPad Prism Software (La Jolla, Calif.) by calculating 520 nm/490 nm ratio. Graphs were plotted as fold change of FRET signal for compound treatment over DMSO-only control.
- Test of AKT Kinase Inhibiting Activity After all the reagents were prepared according to the above method, except for the enzyme, the reagents were equilibrated to the room temperature and loaded. a) first, a compound stock solution (10 mM DMSO solution) was diluted with DMSO to obtain a 100 µM compound solution, the compound solution was diluted with the 1× kinase reaction buffer to obtain a 2.5 µM compound working solution (containing 2.5% DMSO). A 2.5% DMSO solution was prepared from the 1× kinase reaction buffer, and the 2.5 µM compound working solution was diluted 7 times with the 2.5% DMSO solution according to a 4-fold gradient to obtain compound working solutions at 8 concentrations (2500 nM, 625 nM, 156 nM, 39 nM, 9.8 nM, 2.4 nM, 0.6 nM, and 0.15 nM). Except for control wells, 4 µL of diluted compound working solution was placed in each reaction well, and 4 µL of previously prepared 2.5% DMSO/kinase buffer was placed in each control well.
- In-Vitro Biochemical Assays Avi-humanTEAD4217-434 (1 nM, produced as described in Hau et al. ChemBioChem 14, 1218, 2013) and LANCE Eu-W1024 Streptavidin (0.5 nM, PerkinElmer) were first pre-incubated for 1 h at room temperature in HEPES (pH 7.4, 50 mM), KCl (100 mM), Tween-20 (0.05%), TCEP (0.25 mM), EDTA (1 mM), and BSA (0.05%)]. N-terminus Cy5 labeled humanYAP60-100 (20 nM) was then added to this preparation. Compounds were dissolved at 10 mM in 100% DMSO and serial dilutions were made in 100% DMSO. The diluted compound solutions were incubated in white 384-well plates (Greiner Bio-One) for 1 h at room temperature with the above described mix. The final DMSO concentration present in the assay was 1%. The fluorescence was measured (50 μs delay between excitation and fluorescence, 75 μs integration time) with a Genios Pro reader (Tecan) and use of an excitation wavelength of 340 nm and emission wavelengths of 620 nm and 665 nm. Data analyses were carried out by using the TR-FRET ratio emission 655 nm/620 nm. The IC50 values were estimated by fitting the data by nonlinear fit regression (GraphPad Prism). In the alternative format, the assay was conducted in the presence of 5 nM His-humanTEAD4217-434, 10 nM N-biotinylated YAP60-100, 0.2 nM anti-His Europium labelled antibody and 10 nM SA-XL665.
- ChEBML_138998 Inhibition of [3H]dihydromorphine (1 nM) binding to mu opioid receptor of rat brain homogenate
- ChEBML_145841 Inhibition of 9 (2 nM) binding to Opioid receptor kappa 1 of rat brain homogenate
- ChEBML_146239 inhibition of 1.0 nM [3H]- DAGO binding to guinea pig brain membrane opioid receptor mu
- ChEBML_159532 Inhibition of 3 nM [3H]R5020 binding to progesterone receptor of human T47D cell cytosol
- ChEBML_201798 Ability to displace 0.4 nM [3H]paroxetine binding to serotonin transporter in rat frontal cortex
- ChEBML_214260 Inhibition of 1 nM AVP-induced cAMP accumulation in cells expressing human vasopressin V2 receptor
- ChEBML_214539 Inhibition of 1 nM AVP-induced calcium mobilisation in cells expressing human vasopressin V1a receptor
- ChEBML_220586 Displacement of [3H]2-BFI (1 nM) from imidazoline receptor I-2 in human brain
- ChEBML_30135 Displacement of [3H]CGS-21680 (Kd, 14.9 nM) from A2A receptor in rat striatal membranes
- ChEBML_30557 Affinity for adenosine A2 receptor at rat striatal membrane using 5 nM [3H]CGS-21680
- ChEBML_30558 Affinity for adenosine A2 receptor at rat striatal membrane using 5 nM [3H]CGS-21680
- ChEBML_30559 Affinity for adenosine A2 receptor at rat striatal membrane using 5 nM [3H]CGS-21680
- ChEBML_31037 Affinity for adenosine A2 receptor at rat striatal membrane using 5 nM [3H]CGS-21680
- ChEBML_32815 Inhibitory activity against rabbit kidney aminopeptidase using 10 nM of [3H]Leu-enkephalin as substrate
- ChEBML_36170 Inhibition of specific binding of [125 I ] Angiotensin-II (0.2 nM) to bovine adrenal cortex
- ChEBML_69892 Apparent affinity by displacement of preincubated [35S]TBPS from rat cortical homogenates at 60 nM
- ChEBML_71109 Displacement of [3H]dexamethasone from human Glucocorticoid receptor (GR); Value ranges from 150-220 nM
- ChEMBL_145729 (CHEMBL755016) Displacement of 0.5 nM [3H]bremazocine from opioid receptor sites in guinea pig membranes
- ChEMBL_145870 (CHEMBL754075) Inhibition of 0.5 nM [3H]bremazocine binding to guinea pig brain membrane opioid receptors
- ChEMBL_147892 (CHEMBL874638) Inhibition of 10 nM 2-MeS-ADP-stimulated phospholipase C in turkey erythrocyte membrane
- ChEMBL_159383 (CHEMBL768816) Inhibition of 3 nM [3H]R5020 binding to progesterone receptor in human T47D cells
- ChEMBL_164464 (CHEMBL770730) Inhibition of specific [3H]-clonidine binding (0.4 nM) to rat brain membranes alpha2 adrenoceptor
- ChEMBL_164466 (CHEMBL770732) Inhibition of specific [3H]-prazosin binding (0.2 nM) to rat brain membranes alpha1 adrenoceptor.
- ChEMBL_164468 (CHEMBL770734) Inhibition of specific [3H]clonidine binding (0.4 nM) to rat brain membranes alpha2 adrenoceptor
- ChEMBL_164469 (CHEMBL770735) Inhibition of specific [3H]clonidine binding (0.4 nM) to rat brain membranes alpha2 adrenoceptor
- ChEMBL_164995 (CHEMBL770198) Inhibition of specific [3H]prazosin binding (0.2 nM) to rat brain membranes alpha1 adrenoceptor.
- ChEMBL_217652 (CHEMBL820187) Binding affinity towards cRARbeta2 receptor by displacing 0.82 nM 3[H]all-trans-RA
- ChEMBL_2600 (CHEMBL617468) Displacement of [3H]ketanserin (0.5 nM) from rat cerebral cortex 5-hydroxytryptamine 2A receptors
- ChEMBL_28851 (CHEMBL643393) Affinity for adenosine A1 receptor at rat cortical receptors using 1 nM [3H]PIA
- ChEMBL_28982 (CHEMBL640784) Affinity for adenosine A1 receptor at rat cortical receptors using 1 nM [3H]PIA
- ChEMBL_29602 (CHEMBL640260) Dissociation constant of [3H]DPCPX binding to adenosine A1 receptor (AR) at 0.05 nM
- ChEMBL_29603 (CHEMBL640261) Dissociation constant of [3H]-DPCPX binding to adenosine A1 receptor (AR) at 0.1 nM
- ChEMBL_29604 (CHEMBL640262) Dissociation constant of [3H]DPCPX binding to adenosine A1 receptor (AR) at 100 nM
- ChEMBL_29605 (CHEMBL640263) Dissociation constant of [3H]DPCPX binding to adenosine A1 receptor (AR) at 200 nM
- ChEMBL_29606 (CHEMBL640264) Dissociation constant of [3H]DPCPX binding to adenosine A1 receptor (AR) at 5 nM
- ChEMBL_29607 (CHEMBL640265) Dissociation constant of [3H]-DPCPX binding to adenosine A1 receptor(AR) at 0.05 nM
- ChEMBL_29724 (CHEMBL646657) Dissociation constant of [3H]DPCPX binding to adenosine A1 receptor(AR) at 0.1 nM
- ChEMBL_29725 (CHEMBL646658) Dissociation constant of [3H]-DPCPX binding to adenosine A1 receptor(AR) at 100 nM
- ChEMBL_29726 (CHEMBL646659) Dissociation constant of [3H]DPCPX binding to adenosine A1 receptor(AR) at 5 nM
- ChEMBL_306316 (CHEMBL827761) Displacement of [3H]NCS-382 (16 nM) from GHB receptor of rat cerebrocortical membranes
- ChEMBL_326588 (CHEMBL863371) Binding affinity to P2Y12 receptor expressed in human platelets from 0.047 to 50 nM
- ChEMBL_34002 (CHEMBL643631) Inhibition of specific [3H]prazosin binding (0.2 nM) to rat brain membranes alpha1 adrenoceptor.
- ChEMBL_49396 (CHEMBL658758) Displacement of 0.1 nM [3H]Boc[Nle28,31]-CCK27-33 from guinea pig pancreatic membranes
- ChEMBL_557836 (CHEMBL956561) Displacement of [3H]CP-55940 from CB1 receptor in mouse brain at 100 nM
- ChEMBL_59878 (CHEMBL673004) Inhibition of [3H]spiroperidol (0.5 nM) binding to dopamine receptor from rat striatal membrane.
- ChEMBL_609658 (CHEMBL1067221) Inhibition of Dengue virus type 2 NS5 RNA methyltransferase at 80 nM enzyme concentration
- ChEMBL_609659 (CHEMBL1067222) Inhibition of Dengue virus type 2 NS5 RNA methyltransferase at 8 nM enzyme concentration
- ChEMBL_71494 (CHEMBL680747) Selectivity for gardos channel; (Gardos channel vs 2000 nM for the cardiac IKs channel).)
- ChEMBL_982327 (CHEMBL2426847) Binding affinity to beta-2 adrenergic receptor (unknown origin) at 1 to 10000 nM
- ChEMBL_99657 (CHEMBL704423) Inhibition of binding of [3H]LTB4 (1 nM) to intact human polymorphonuclear leukocytes (PMNs)
- ChEMBL_99659 (CHEMBL704425) Inhibition of specific binding of LTB4 ( 0.1 nM) to receptors on intact human neutrophils
- ChEMBL_1625397 (CHEMBL3867866) Inhibition of human ABCB1 expressed in NCI-ADR-RES cells assessed as potentiation of colchicine-induced cytotoxicity by measuring colchicine IC50 at 50 nM after 72 hrs by MTT assay (Rvb = 1607.50 +/- 497.42 nM)
- ChEMBL_1625398 (CHEMBL3867867) Inhibition of human ABCB1 expressed in NCI-ADR-RES cells assessed as potentiation of colchicine-induced cytotoxicity by measuring colchicine IC50 at 100 nM after 72 hrs by MTT assay (Rvb = 1607.50 +/- 497.42 nM)
- ChEMBL_1625399 (CHEMBL3867868) Inhibition of human ABCB1 expressed in NCI-ADR-RES cells assessed as potentiation of colchicine-induced cytotoxicity by measuring colchicine IC50 at 200 nM after 72 hrs by MTT assay (Rvb = 1607.50 +/- 497.42 nM)
- ChEMBL_1625400 (CHEMBL3867869) Inhibition of human ABCB1 expressed in NCI-ADR-RES cells assessed as potentiation of colchicine-induced cytotoxicity by measuring colchicine IC50 at 500 nM after 72 hrs by MTT assay (Rvb = 1607.50 +/- 497.42 nM)
- ChEMBL_1625401 (CHEMBL3867870) Inhibition of human ABCB1 expressed in NCI-ADR-RES cells assessed as potentiation of colchicine-induced cytotoxicity by measuring colchicine IC50 at 1000 nM after 72 hrs by MTT assay (Rvb = 1607.50 +/- 497.42 nM)
- ChEMBL_1625411 (CHEMBL3867880) Inhibition of human ABCB1 expressed in NCI-ADR-RES cells assessed as potentiation of vincristine-induced cytotoxicity by measuring vincristine IC50 at 50 nM after 72 hrs by MTT assay (Rvb = 3714.80 +/- 383.58 nM)
- ChEMBL_1625412 (CHEMBL3867881) Inhibition of human ABCB1 expressed in NCI-ADR-RES cells assessed as potentiation of vincristine-induced cytotoxicity by measuring vincristine IC50 at 100 nM after 72 hrs by MTT assay (Rvb = 3714.80 +/- 383.58 nM)
- ChEMBL_1625413 (CHEMBL3867882) Inhibition of human ABCB1 expressed in NCI-ADR-RES cells assessed as potentiation of vincristine-induced cytotoxicity by measuring vincristine IC50 at 200 nM after 72 hrs by MTT assay (Rvb = 3714.80 +/- 383.58 nM)
- ChEMBL_1625414 (CHEMBL3867883) Inhibition of human ABCB1 expressed in NCI-ADR-RES cells assessed as potentiation of vincristine-induced cytotoxicity by measuring vincristine IC50 at 500 nM after 72 hrs by MTT assay (Rvb = 3714.80 +/- 383.58 nM)
- ChEMBL_1625415 (CHEMBL3867884) Inhibition of human ABCB1 expressed in NCI-ADR-RES cells assessed as potentiation of vincristine-induced cytotoxicity by measuring vincristine IC50 at 1000 nM after 72 hrs by MTT assay (Rvb = 3714.80 +/- 383.58 nM)
- Enzyme Inhibition Assay Fluorescent assays were carried out in black 96-well plates. Caspase activity was monitored using a Labsystem Fluoroskan II spectrofluorometer with an excitation wavelength of 405 nm and an emission wavelength of 505 nm. Inhibition IC50 values were calculated based on the inhibition data obtained throughout the inhibitor concentration range from 0.3 nM to 300 uM.
- HTRF2 Assay As the potencies of the HDM2 inhibitors increased, an improved HTRF assay (HTRF2 assay) was developed. All assay conditions remained the same as described above, with the exception of the following changes in reagent concentrations: 0.2 nM GST-hMDM2 (1-188), 0.5 nM biotinylated-p53 (1-83), 0.18 nM SA-XLent, and 100 mM KF.
- Enzymatic Assay The compounds of the invention are assessed for the ability to interact with human LTA4 hydrolase in an enzymatic assay that measures the ability of the enzyme to cleave the peptide bond of arginyl-aminomethylcoumarin (Arg-AMC). LTA4H Enzyme (1 nM final), Arg-AMC substrate (50 M final), and compound are combined in a reaction buffer (50 mM Tris-HCl (pH 7.5), 100 mM KCl, 0.5% bovine serum albumin) at room temperature for 1 h. The formation of product is assessed by measuring the fluorescence of aminomethylcoumarin product (excitation wavelength 380 nm/emission wavelength 460 nm). In general, the preferred potency range (IC50) of compounds in the LTA4H Enzyme assay is between 0.1 nM to 10 M, the more preferred potency range is 0.1 nM to 0.1 M, and the most preferred potency range is 0.1 nM to 10 nM.
- RIPK1 HTRF Binding Assay A solution was prepared containing 0.2 nM Anti GST-Tb (Cisbio, 61GSTTLB), 90.6 nM probe and 1 nM His-GST-TVMV-hRIPK1 (1-324) in FRET Buffer (20 mM HEPES, 10 mM MgCl2, 0.015% Brij-35, 4 mM DTT, 0.05 mg/mL BSA). Using Formulatrix Tempest, the detection antibody/enzyme/probe solution (2 mL) was dispensed into wells of a 1536 plate (Black Low Binding Polystyrene 1536 Plate (Corning, 3724)) containing 10 nL of compounds of interest at appropriate concentration in DMSO. The plate was incubated at rt for 1 h. FRET was measured using the EnVision plate reader (Excitation: 340 nM, Emission: 520 nM/495 nM). Total signal (0% inhibition) was calculated from wells containing 10 nL DMSO only. Blank signal (100% inhibition) calculated from wells containing 10 nL of 15 nM staurosporine and internal controls.
- RIPK1 HTRF Binding Assay A solution was prepared containing 0.2 nM Anti GST-Tb (Cisbio, 61GSTTLB), 90.6 nM probe and 1 nM His-GST-TVMV-hRIPK1(1-324) in FRET Buffer (20 mM HEPES, 10 mM MgCl2, 0.015% Brij-35, 4 mM DTT, 0.05 mg/mL BSA). Using Formulatrix Tempest, the detection antibody/enzyme/probe solution (2 mL) was dispensed into wells of a 1536 plate (Black Low Binding Polystyrene 1536 Plate (Corning, 3724)) containing 10 nL of compounds of interest at appropriate concentration in DMSO. The plate was incubated at rt for 1 h. FRET was measured using the EnVision plate reader (Excitation: 340 nM, Emission: 520 nM/495 nM). Total signal (0% inhibition) was calculated from wells containing 10 nL DMSO only. Blank signal (100% inhibition) calculated from wells containing 10 nL of 15 nM staurosporine and internal controls.
- In Vitro Kinase Assays Biological activities of the MMT3-72 and its active metabolite MMT3-72-M2 were evaluated against JAK1, JAK2, JAK3 and TYK2 using kinase assays (FIG. 3 , Table 1). The compound MMT3-72 showed modest inhibitory activities against JAK1 and JAK2 (199.3 nM and 448.3 nM, respectively) and poor inhibitory activities against JAK3 and TYK2 (6821 nM and 2976 nM, respectively). However, the active metabolite MMT3-72-M2 showed strong inhibitory activities against JAK1 (2.0 nM), JAK2 (16.3 nM), and TYK2 (55.2 nM), but only weak inhibitory activities against JAK3 (701.3 nM). In comparison, fedratinib strongly inhibited JAK1 (10.1 nM) and JAK2 (15.6 nM), but poorly inhibited JAK3 and TYK2. The inhibitory profiles of JAK1, 2, and TYK2 of MMT3-72-M2 may have advantages to treat UC since JAK2/TYK2/IL-12/IL-23 signaling is strongly implicated in UC, while JAK1 isoform has long been identified as potential target in treating IBD as seen in Upadacitinib. In addition, MMT3-72-M2 showed poor inhibitory activities against JAK3 that may also be preferred in treating UC to reduce the unwanted adverse effects. Tofacitinib inhibited JAK3 with an IC50 of 1.6 nM and showed serious adverse effects. JAK3 inhibition has been shown to potentially lead to lymphopenia and thus hypothetically to an increased risk of infection.
- FRET Assay Compounds of the invention were dissolved in DMSO at a concentration of 3 mM with subsequent dilutions in assay buffer (50 mM HEPES PH7.0, 150 mM NaCl, 0.05% BSA, 0.2% Pluronic F-127) such that the assay contained 1% DMSO. In a white 384 shallow well Microplate (Proxiplate-384 Plus, PerkinElmer, 6008280), 150 nL of compound or vehicle (1% DMSO in assay buffer) for the high control (HC) wells and 5 μL of 30 nM ENL Protein (6×HIS ENL YEATS Domain, EpiCypher, 15-0069) were combined and incubated 15 minutes at RT. Low control (LC) wells received 5 μL of assay buffer instead of ENL protein. Then 5 μL of 15 nM H3K9cr peptide (H3 aa1-20, biotinylated; EpiCypher, 12-0099) in assay buffer was added and incubated 30 minutes at RT. Finally a 5 μL mix of 45 nM Anti-6HIS ULight (PerkinElmer, TRF0105) and 1.5 nM Streptavidin-Europium Chelate (PerkinElmer, AD0060) were added and incubated for a further 30 minutes at RT. The TR-FRET signal (665 nm signal/615 nm signal X 10,000) was measured using a PerkinElmer 2104 EnVision (Xenon Flash Lamp excitation, 320 nm±37.5 nm excitation filter, 407 nm cut off dichroic mirror, 615 nm±4.25 (Europium) nm and 665 nm±3.75 nM (ULight) emission filters). Compound concentration response curves were performed in duplicate over the concentration range of 0.15 nM-30 μM. The response at each compound concentration minus the LC value was converted to percent inhibition of the vehicle control group response (HC-LC). The relationship between the % inhibition and the compound concentration was analyzed using a four parameter logistic equation to estimate lower and upper asymptotes, the compound concentration producing 50% inhibition (IC50 value) and the slope at the mid-point location.
- Homogenous Time-Resolved Fluorescence (HTRF) Binding Assay All binding studies were performed in an HTRF assay buffer consisting of dPBS supplemented with 0.1% (withv) bovine serum albumin and 0.05% (v/v) Tween-20. For the PD-1-Ig/PD-L1-His binding assay, inhibitors were pre-incubated with PD-L1-His (10 nM final) for 15 m in 4 μl of assay buffer, followed by addition of PD-1-Ig (20 nM final) in 1 μl of assay buffer and further incubation for 15 m. PD-L1 from either human, cyno, or mouse were used. HTRF detection was achieved using europium crypate-labeled anti-Ig (1 nM final) and allophycocyanin (APC) labeled anti-His (20 nM final). Antibodies were diluted in HTRF detection buffer and 5 μl was dispensed on top of binding reaction. The reaction mixture was allowed to equilibrate for 30 minutes and signal (665 nm/620 nm ratio) was obtained using an EnVision fluorometer. Additional binding assays were established between PD-1-Ig/PD-L2-His (20 & 5 nM, respectively), CD80-His/PD-L1-Ig (100 & 10 nM, respectively) and CD80-His/CTLA4-Ig (10 & 5 nM, respectively). Competition studies between biotinylated SEQ ID NO:71 and human PD-L1-His were performed as follows. Inhibitors were pre-incubated with PD-L1-His (10 nM final) for 60 m in 4 μl of assay buffer followed by addition of biotinylated SEQ ID NO:71 (0.5 nM final) in 1 μl of assay buffer. Binding was allowed to equilibrate for 30 m followed by addition of europium crypated labeled Strepatavidin (2.5 pM final) and APC-labeled anti-His (20 nM final) in 5 μl of HTRF buffer. The reaction was allowed to equilibrate for 30 m and signal (665 nm/620 nm ratio) was obtained using an EnVision fluorometer.
- ABL1 (T315I) Kinase Activity Assay Serially diluting the compound of the present invention from 1 uM initial concentration in three-fold fashion and formulating 10 concentrations (50.8 pM, 152.0 pM, 457.0 pM, 1.37 nM, 4.12 nM, 12.3 nM, 37.0 nM, 111.0 nM, 333.0 nM and 1.0 uM). 5.0 uM Abltide was added into each well and then human T315I mutant enzyme was added. [gamma-33P] ATP was added at room temperature, with final concentration of 1.0 uM, and the reaction was performed for 120 minutes. 20 ul aliquots were transferred onto the ion exchange chromatography paper P81. The paper was thoroughly washed with a 0.75% phosphoric acid solution three times, and then washed with acetone once. Finally, gamma-33P radioactivity was measured.
- In Vitro Binding Screening Assay Equipment: MicroBeta2 LumiJET system, PerkinElmer. Method: Chem-1 cell membranes stably expressing human GnRHR were resuspended in HEPES at pH 7.4. 6 μg of human GnRHR cell membranes were added to each well and incubated with 0.05 nM [125I]-[D-Trp6]-LH-RH and each of the test compounds at 25° C. for 60 min. The compounds were each dissolved in DMSO, with test concentrations being 500 nM, 62.5 nM, 7.81 nM, 0.97 nM, 0.12 nM, 15 pM, and 1.9 pM. 1 pM [D-Trp6]-LH-RH was used in the non-specific binding assay. Cell membranes were collected by filtration under vacuum and washed before liquid scintillation. The binding IC50 and IC90 of the compounds were calculated using Prism.
- Inhibition Assay Assay buffers consist of 20 mM citric acid, 60 mM disodium hydrogen orthophosphate, 1 mM EDTA, 0.1% CHAPS, 4 mM DTT, pH 5.8 for legumain, 50 mM dihydrogen sodium orthophosphate, 1 mM EDTA, 5 mM DTT, pH 6.25 for cathepsin B and cathepsin Land 100 mM Tris, 0.1% CHAPS, 10% sucrose, 10 mM DTT, pH 7.4 for caspase-3. Concentrations of substrates during the measurement were 10 nM (legumain, cathepsin Land caspase-3) and 50 nM (cathepsin B) and concentration of enzymes were 100 nM for cathepsin Land caspase-3, 270 nM for legumain and 360 nM for cathepsin B. Each enzyme was incubated with inhibitor concentrations ranging from 1 nM to 1 mM in the presence of the substrates.
- Fluorescence Polarization-Based Binding Assay Sensitive and quantitative FP-based binding assays were developed and optimized to determine the binding affinities of small-molecule inhibitors to the recombinant Mcl-1, A1/Bfl-1, Bcl-w, Bcl-2, and Bcl-xL proteins. The concentrations of the proteins used in the competitive binding experiments were 10 nM for Mcl-1, 80 nM for Bcl-xL, and 60 nM for Bcl-2. The fluorescent probes, Flu-BID or FAM-BID were fixed at 2 nM for all assays, which binds with Kd values of 2.3 nM, 14.2 nM and 24.1 nM against Mcl-1, Bcl-2 and Bcl-xL respectively. 5 μL of the tested compound in DMSO and 120 μL of protein/probe complex in the assay buffer (20 mM phosphate pH 7.4, 50 mM NaCl, 1 mM EDTA. 0.05% Pluronic F68)s) were added to 96 well black assay plates, incubated at room temperature for 3 h and the polarization values (mP) were measured at an excitation wavelength at 485 nm and an emission wavelength at 530 nm using the plate reader Synergy H1 Hybrid, BioTek.
- ChEBML_145206 Inhibition of opioid receptor kappa by displacing 0.5 nM [3H]bremazocine in guinea pig brain membrane
- ChEBML_145721 Inhibition of total opioid receptor by displacing 0.5 nM [3H]bremazocine in guinea pig brain membrane
- ChEBML_146240 Inhibition of opioid receptor mu by displacing 1 nM [3H]DAGO in guinea pig brain membrane
- ChEBML_146692 Inhibition of 0.5 nM [3H]- Bremazocine binding to Opioid receptor mu 1 of bovine striatum membrane
- ChEBML_196324 Binding affinity towards retinoic acid receptor gamma was determined using [3H]ATRA (5 nM) as radioligand
- ChEBML_217648 Binding affinity towards cRAR-beta-2 receptor by displacing 0.82 nM 3[H]all-trans-RA
- ChEBML_28985 Antagonism of binding of 1 nM [3H]cyclohexyladenosine to adenosine A1 receptors on rat cortical membranes
- ChEBML_29319 Binding affinity towards adenosine A1 receptor in rat cerebral cortical membranes with 1 nM [3H]cyclohexyladenosine
- ChEBML_35496 Inhibition of Aminopeptidase N activity in pig kidney with 10 nM [3H]Leu-enkephalin as substrate
- ChEBML_53962 Ability to inhibit purified Dihydrofolate reductase from human leukemic lymphoblasts was determined spectrophotometrically at 340 nM.
- ChEBML_99661 Inhibition of specific binding of [3H]LTB4 ( 0.1 nM) to LTB4 receptor on intact human neutrophils
- ChEMBL_1505398 (CHEMBL3595281) Inhibition of full-length human ARTD1 using 500 nM NAD substrate after 1 hr incubation
- ChEMBL_1505399 (CHEMBL3595282) Inhibition of full-length human ARTD2 using 500 nM NAD substrate after 3 hrs incubation
- ChEMBL_1505400 (CHEMBL3595283) Inhibition of full-length human ARTD3 using 500 nM NAD substrate after 1 hrs incubation
- ChEMBL_159532 (CHEMBL767565) Inhibition of 3 nM [3H]R5020 binding to progesterone receptor of human T47D cell cytosol
- ChEMBL_1753428 (CHEMBL4188188) Binding affinity to biotinylated-KDM4A (unknown origin) at 300 nM by bio-layer interferometric method
- ChEMBL_201798 (CHEMBL806133) Ability to displace 0.4 nM [3H]paroxetine binding to serotonin transporter in rat frontal cortex
- ChEMBL_209071 (CHEMBL809537) In vitro antagonist activity against Thrombin induced gel-filtered platelet (GFP) aggregation at 0.15 nM.
- ChEMBL_209257 (CHEMBL815698) Inhibition of secretion of radiolabeled serotonin from washed human platelets stimulated by 1 nM thrombin
- ChEMBL_214260 (CHEMBL818586) Inhibition of 1 nM AVP-induced cAMP accumulation in cells expressing human vasopressin V2 receptor
- ChEMBL_214539 (CHEMBL819376) Inhibition of 1 nM AVP-induced calcium mobilisation in cells expressing human vasopressin V1a receptor
- ChEMBL_217986 (CHEMBL824105) Inhibition of apolipoprotein B (apoB) secreted by human hepatoma cells (Hep G2) at 100 nM
- ChEMBL_220590 (CHEMBL841869) Displacement of [3H]2-BFI (1 nM) from imidazoline receptor I-2 in human brain
- ChEMBL_29452 (CHEMBL641037) Binding of 1 nM [3H]CHA to Adenosine A1 receptor of rat cerebral cortical membranes
- ChEMBL_320923 (CHEMBL881350) In vitro binding affinity against rat cannabinoid receptor 1 using 0.5 nM [3H]CP-55940
- ChEMBL_32815 (CHEMBL646467) Inhibitory activity against rabbit kidney aminopeptidase using 10 nM of [3H]Leu-enkephalin as substrate
- ChEMBL_38798 (CHEMBL652339) Evaluated for Adenylyl cyclase activation as % of the maximal stimulation with isoproterenol at 100 nM
- ChEMBL_462940 (CHEMBL929874) Inhibition of VEGF-stimulated VEGFR2 autophosphorylation in HUVEC cells at 50 nM relative to control
- ChEMBL_552365 (CHEMBL1005955) Displacement of [3H]histamine from human histamine H4 receptor expressed in SK-NM-C cells
- ChEMBL_58530 (CHEMBL884446) Ability to displace [3H]spiperone (0.5 nM) from corpus striatum of rat Dopamine receptor D2
- ChEMBL_58793 (CHEMBL667035) Inhibition of 333 nM dopamine stimulated GTP binding in CHO cells expressing human D4 receptor
- ChEMBL_62107 (CHEMBL674985) Ability to displace [3H]spiperone (0.3 nM) from CHO cells of human Dopamine receptor D3
- ChEMBL_62809 (CHEMBL674122) Antagonism of cocaine''s inhibition of [3H]DA uptake at the dose of 200 nM
- ChEMBL_62810 (CHEMBL674123) Antagonism of cocaine''s inhibition of [3H]DA uptake at the dose of 500 nM
- ChEMBL_62811 (CHEMBL871936) Antagonism of cocaine''s inhibition of [3H]DA uptake at the dose of 50 nM
- ChEMBL_643624 (CHEMBL1212488) Displacement of [125I]peptide YY from human recombinant Y1 receptor expressed in SK-NM- cell
- ChEMBL_67167 (CHEMBL681151) In vitro displacement of 0.5 nM [3H]17-beta-estradiol from human Estrogen receptor alpha
- ChEMBL_67329 (CHEMBL678663) In vitro displacement of 0.5 nM [3H]17-beta-estradiol from human Estrogen receptor beta
- ChEMBL_687982 (CHEMBL1291654) Inhibition of GCP2 by top scintillation counter in presence of 30 nM NAA[3]G
- ChEMBL_71102 (CHEMBL679614) Displacement of [3H]dexamethasone from human Glucocorticoid receptor (GR); Value ranges from 10-12 nM
- ChEMBL_71103 (CHEMBL679615) Displacement of [3H]dexamethasone from human Glucocorticoid receptor (GR); Value ranges from 106-131 nM
- ChEMBL_71104 (CHEMBL679616) Displacement of [3H]dexamethasone from human Glucocorticoid receptor (GR); Value ranges from 1273-2947 nM
- ChEMBL_71105 (CHEMBL679617) Displacement of [3H]dexamethasone from human Glucocorticoid receptor (GR); Value ranges from 13-26 nM
- ChEMBL_71106 (CHEMBL679618) Displacement of [3H]dexamethasone from human Glucocorticoid receptor (GR); Value ranges from 133-161 nM
- ChEMBL_71107 (CHEMBL679619) Displacement of [3H]dexamethasone from human Glucocorticoid receptor (GR); Value ranges from 1382-1049 nM
- ChEMBL_71108 (CHEMBL679620) Displacement of [3H]dexamethasone from human Glucocorticoid receptor (GR); Value ranges from 1428-1433 nM
- ChEMBL_71109 (CHEMBL686057) Displacement of [3H]dexamethasone from human Glucocorticoid receptor (GR); Value ranges from 150-220 nM
- ChEMBL_71110 (CHEMBL686058) Displacement of [3H]dexamethasone from human Glucocorticoid receptor (GR); Value ranges from 159-161 nM
- ChEMBL_71111 (CHEMBL686059) Displacement of [3H]dexamethasone from human Glucocorticoid receptor (GR); Value ranges from 17-21 nM
- ChEMBL_71112 (CHEMBL686060) Displacement of [3H]dexamethasone from human Glucocorticoid receptor (GR); Value ranges from 2-2 nM
- ChEMBL_71113 (CHEMBL686061) Displacement of [3H]dexamethasone from human Glucocorticoid receptor (GR); Value ranges from 215-217 nM
- ChEMBL_71114 (CHEMBL686295) Displacement of [3H]dexamethasone from human Glucocorticoid receptor (GR); Value ranges from 23-33 nM
- ChEMBL_71115 (CHEMBL686296) Displacement of [3H]dexamethasone from human Glucocorticoid receptor (GR); Value ranges from 26-67 nM
- ChEMBL_71116 (CHEMBL686297) Displacement of [3H]dexamethasone from human Glucocorticoid receptor (GR); Value ranges from 28-34 nM
- ChEMBL_71117 (CHEMBL686298) Displacement of [3H]dexamethasone from human Glucocorticoid receptor (GR); Value ranges from 294-554 nM
- ChEMBL_71118 (CHEMBL686299) Displacement of [3H]dexamethasone from human Glucocorticoid receptor (GR); Value ranges from 3-3 nM
- ChEMBL_71228 (CHEMBL682527) Displacement of [3H]dexamethasone from human Glucocorticoid receptor (GR); Value ranges from 3-4 nM
- ChEMBL_71229 (CHEMBL682528) Displacement of [3H]dexamethasone from human Glucocorticoid receptor (GR); Value ranges from 366-578 nM
- ChEMBL_71230 (CHEMBL682529) Displacement of [3H]dexamethasone from human Glucocorticoid receptor (GR); Value ranges from 378-814 nM
- ChEMBL_71231 (CHEMBL682530) Displacement of [3H]dexamethasone from human Glucocorticoid receptor (GR); Value ranges from 379-840 nM
- ChEMBL_71232 (CHEMBL680299) Displacement of [3H]dexamethasone from human Glucocorticoid receptor (GR); Value ranges from 39-94 nM
- ChEMBL_71233 (CHEMBL680300) Displacement of [3H]dexamethasone from human Glucocorticoid receptor (GR); Value ranges from 43-50 nM
- ChEMBL_71234 (CHEMBL680301) Displacement of [3H]dexamethasone from human Glucocorticoid receptor (GR); Value ranges from 44-53 nM
- ChEMBL_71235 (CHEMBL680302) Displacement of [3H]dexamethasone from human Glucocorticoid receptor (GR); Value ranges from 5-6 nM
- ChEMBL_71236 (CHEMBL680303) Displacement of [3H]dexamethasone from human Glucocorticoid receptor (GR); Value ranges from 536-560 nM
- ChEMBL_71237 (CHEMBL680304) Displacement of [3H]dexamethasone from human Glucocorticoid receptor (GR); Value ranges from 559-1423 nM
- ChEMBL_71238 (CHEMBL680305) Displacement of [3H]dexamethasone from human Glucocorticoid receptor (GR); Value ranges from 6-7 nM
- ChEMBL_71239 (CHEMBL680306) Displacement of [3H]dexamethasone from human Glucocorticoid receptor (GR); Value ranges from 6-8 nM
- ChEMBL_71240 (CHEMBL680307) Displacement of [3H]dexamethasone from human Glucocorticoid receptor (GR); Value ranges from 61-63 nM
- ChEMBL_71241 (CHEMBL680308) Displacement of [3H]dexamethasone from human Glucocorticoid receptor (GR); Value ranges from 639-673 nM
- ChEMBL_71242 (CHEMBL683338) Displacement of [3H]dexamethasone from human Glucocorticoid receptor (GR); Value ranges from 681-974 nM
- ChEMBL_71243 (CHEMBL683339) Displacement of [3H]dexamethasone from human Glucocorticoid receptor (GR); Value ranges from 70-92 nM
- ChEMBL_71244 (CHEMBL683340) Displacement of [3H]dexamethasone from human Glucocorticoid receptor (GR); Value ranges from 838-988 nM
- ChEMBL_71245 (CHEMBL683341) Displacement of [3H]dexamethasone from human Glucocorticoid receptor (GR); Value ranges from 9-12 nM
- ChEMBL_71246 (CHEMBL683342) Displacement of [3H]dexamethasone from human Glucocorticoid receptor (GR); Value ranges from 949-1169 nM
- ChEMBL_88150 (CHEMBL693235) Stimulation of alkaline phosphatase activity in human endometrial Ishikawa cells with 1 nM E2 estradiol
- ChEMBL_88151 (CHEMBL693236) Stimulation of alkaline phosphatase activity in human endometrial Ishikawa cells with 1 nM E2 estradiol
- ChEMBL_2111476 (CHEMBL4820326) Positive allosteric modulator activity at human CB2R expressed in CHO-K1 cell membrane assessed as CP55940 EC50 for [35S]GTPgammaS binding at 1 nM measured after 90 mins by liquid scintillation counting analysis (Rvb = 1.9 nM)
- ChEMBL_2111477 (CHEMBL4820327) Positive allosteric modulator activity at human CB2R expressed in CHO-K1 cell membrane assessed as CP55940 EC50 for [35S]GTPgammaS binding at 100 nM measured after 90 mins by liquid scintillation counting analysis (Rvb = 1.9 nM)
- TR-FRET Assay Compounds were screened in the TR-FRET assay for JAK2 and c-Src kinase inhibition. Ultra light poly GT (Perkin Elmer) was used as the substrate for JAK2 and c-Src with the ATP concentration of 10 μM and 50 μM, respectively. The Eu-labelled anti-phospho tyrosine antibody (Perkin Elmer) was added at 1 nM and the fluorescence emission at 615 nm and 665 nm was measured with an excitation wavelength of 340 nm.
- Enzymatic Assay FGFR3: In a final reaction volume of 30 uL, FGFR3 (h) (40 ng/ml) is incubated with 50 mM HEPES pH 7.5, 6 mM MnCl2, 1 mM DTT, 0.1 mM Na3VO4, 0.01% Triton-X-100, 500 nM Btn-Flt3 and 25 uM ATP in the presence of compound (1% DMSO final). After incubation for 60 minutes at room temperature the reaction is stopped with 2.27 nM EU-anti P-Tyr, 7 mM EDTA, 31.25 nM SA-XL-665 and 0.02% BSA which is present for 60 minutes at room temperature. Time-Resolved Fluorescence Resonance Energy Transfer (TR-FRET) signal (ex340 nm. Em 620 nm, em 655 nm) is measured afterwards and results are expressed in RFU (Relative Fluorescence Units). In this assay, the inhibitory effect of different compound concentrations (range 10 uM to 0.1 nM) is determined and used to calculate an IC50 (M) and pIC50 (-log IC50) value.
- TR-FRET Assay Compounds were screened in the TR-FRET assay with FGFR1 kinase. 5 ng of FGFR1 [Upstate, USA] kinase was used for assay. The compound was incubated with the kinase for 30 minutes at RT. After the incubation, substrate mix [40 nM Ultra light poly GT (Perkin Elmer, USA) and 13 μM ATP (Sigma)] was added. The above reaction was stopped by the addition of 40 mM EDTA after the 30 mM kinase reaction. The Eu-labelled antiphospho-tyrosine antibody [Perkin Elmer, USA] was added at 0.5 nM and the fluorescence emission at 615 nm/665 nm [excitation at 340 nm] was measured. The compounds were initially screened at 100 nM and 1 μM concentrations. The compounds with >50% inhibition at 100 nM of FGFR1 were taken for the full dose response studies. The final DMSO concentration in the assay was 1%.
- ChEMBL_1622157 (CHEMBL3864509) Inhibition of Smo in mouse Shh Light2 cells assessed as inhibition of SAG-induced Hh pathway activity by measuring SAG-EC50 at 50 nM measured after 36 hrs by Gli-luciferase reporter gene assay (Rvb = 2.93 nM)
- ChEMBL_1622158 (CHEMBL3864510) Inhibition of Smo in mouse Shh Light2 cells assessed as inhibition of SAG-induced Hh pathway activity by measuring SAG-EC50 at 100 nM measured after 36 hrs by Gli-luciferase reporter gene assay (Rvb = 2.93 nM)
- ChEMBL_1622159 (CHEMBL3864511) Inhibition of Smo in mouse Shh Light2 cells assessed as inhibition of SAG-induced Hh pathway activity by measuring SAG-EC50 at 500 nM measured after 36 hrs by Gli-luciferase reporter gene assay (Rvb = 2.93 nM)
- Enzyme Inhibition Assay The IC50s of inhibitor against the human chitinase were determined using the fluorogenic substrate 4MU-NAG3. The fluorescence of the liberated 4MU was quantified using a Perkin Elmer LS2 fluorimeter (excitation 366 nm, emission 445 nm).
- Enzyme Activity (EC50) Analysis Assay In order to confirm the PPARδ activation effect of the example compounds, the following experiment was performed. First, the example compounds were diluted in DMSO to 100× based on the final concentration, then 1/50 was diluted in assay buffer, and 10 μL of each was added to a 384 well plate. Afterwards, GST-conjugated PPARδ ligand-binding domain (LBD) was added to a final concentration of 5 nM, and fluorescien-conjugated C33 coactivator peptide and Tb-α-GST antibody were added to a final concentration of 100 nM and 10 nM, respectively. The final volume of each test system was set to 20 μL, and the test for each concentration was repeated twice, and the binding activity after reaction was measured by TR-FRET method. In other words, after excitation at 340 nm and measurement of emission values at 495 nm and 520 nm, the results were analyzed using the measured value at 490 nm/measured at 520 nm, and the analysis program Prism 6 was used to calculate the EC50 value.
- Homogenous Time-Resolved Fluorescence (HTRF) Binding Assay All binding studies were performed in an HTRF assay buffer consisting of dPBS supplemented with 0.1% (with) bovine serum albumin and 0.05% (v/v) Tween-20. For the h/PD-L1-His binding assay, inhibitors were pre-incubated with PD-L1-His (10 nM final) for 15 m in 4 μl of assay buffer, followed by addition of PD-1-Ig (20 nM final) in 1 μl of assay buffer and further incubation for 15 m. HTRF detection was achieved using europium crypate-labeled anti-Ig (1 nM final) and allophycocyanin (APC) labeled anti-His (20 nM final). Antibodies were diluted in HTRF detection buffer and 5 μl was dispensed on top of the binding reaction. The reaction mixture was allowed to equilibrate for 30 minutes and the resulting signal (665 nm/620 nm ratio) was obtained using an EnVision fluorometer. Additional binding assays were established between the human proteins PD-1-Ig/PD-L2-His (20 & 5 nM, respectively) and CD80-His/PD-L1-Ig (100 & 10 nM, respectively).
- In-Vitro Biochemical TR-FRET Assay Avi-humanTEAD4217-434 (1 nM, produced as described in Hau et al. ChemBioChem 14, 1218, 2013) and LANCE Eu-W1024 Streptavidin (0.5 nM, PerkinElmer) were first pre-incubated for 1 h at room temperature in HEPES (pH 7.4, 50 mM), KCl (100 mM), Tween-20 (0.05%), TCEP (0.25 mM), EDTA (1 mM), and BSA (0.05%)]. N-terminus Cy5 labeled humanYAP60-100 (20 nM) was then added to this preparation. Compounds were dissolved at 10 mM in 100% DMSO and serial dilutions were made in 100% DMSO. The diluted compound solutions were incubated in white 384-well plates (Greiner Bio-One) for 1 h at room temperature with the above described mix. The final DMSO concentration present in the assay was 1%. The fluorescence was measured (50 us delay between excitation and fluorescence, 75 us integration time) with a Genios Pro reader (Tecan) and use of an excitation wavelength of 340 nm and emission wavelengths of 620 nm and 665 nm. Data analyses were carried out by using the TR-FRET ratio emission 655 nm/620 nm.
- LanthaScreen TR-FRET Assay The assay has been run at room temperature on a liquid handling robot. To the assay plates containing 50 mL compound or control solutions, 4.5 uL of solution A (50 mM Tris-HCl pH7.4, 2.0m MDTT, 0.05% Tween20, 0.02 mM Na3VO4) including a generic concentration of 2.0 uM ATP was added per well, followed by 4.5 uL of solution B (0.5% BSA) including a generic concentration of 50 nM poly(EAY) to give 9.05 uL of a reaction volume with final concentrations of 2.0 uM ATP, 50 nM poly(EAY), 25 mM Tris-HCl pH7.4, 1.0 mM DTT, 0.025% Tween20, 0.01 mM Na3VO4, 0.025% BSA as well as specific concentration of the respective enzyme and individual concentrations of divalent cations: (FGFR-3-K650E) 0.2 nM GST-FGR-3-K650E, 3.0 mM MgCl2, (KIT) 36.6 nM GST-KIT, 10 mM MnCl2, (RET) 0.11 nM GST-Xa-RET, 1.0 mM MnCl2, 10 mM MgCl2, (LCK) 3.3 nM His-LCK, 10 mM MnCl2. (KDR) 0.38 nM GST-KDR, 10 mM MgCl2, 1.0 mM MnCl2, (PDGFaV561D) 4.4 nM GST-PDGFRaV561D, 10 mM MnCl2.
- Peptidase Assay In brief, the enzyme (29 nM) was incubated with L-alanine-p-nitroanilide (1 mM) in 50 mM HEPES (pH 7.5), 100 mM KCL, 1% DMSO in the absence or presence of a compound of the invention for 1 hour at ambient temperature. Reaction was terminated by addition of acetic acid (1%). Formation of colored nitro-aniline was measured by the increase in absorbance at 405 nm in a Victor 2 plate reader (Wallac). Spontaneous hydrolysis of the substrate was corrected for by subtracting the absorbance of control incubations without enzyme.In embodiments, the compounds of the invention, when tested in this assay, demonstrated the ability to inhibit peptidase activity at IC50 values of less than 100 μM, in some embodiments less than 1 μM, in some embodiments less than 100 nM, in some embodiments less than 75 nM, in some embodiments less than 50 nM, in some embodiments less than 25 nM, in some embodiments less than 10 nM, in some embodiments less than 5 nM.
- Potency Activity Assay A BACE1 activity assay kit from AnaSpec (Fremont, Calif.) was used to determine the potency of the selected BACE1 inhibitors by AnaSpec BACE1 fluorescent assay. The recombinant β-secretase enzyme was diluted 1:200 in 1× assay buffer (1 part of 1× assay buffer and 1 part of TBS containing 0.1% Tween 20). The enzyme was added in a volume of 40 μl to each well of a black 96-well micro plate.The BACE1 inhibitors were serially diluted into DMSO from their 10 mM stock concentrations in DMSO. The serially diluted inhibitors were diluted 1:100 into 1× assay buffer in a 96-well polypropylene plate (assay dilution plate). The final concentration of the BACE1 inhibitor tested was 300 nM, 100 nM, 30 nM, 10 nM, 3 nM, 1 nM 0.3 nM, and 0.00 nM. Dose responses for inhibitors were run in duplicate.A volume of 10 μl of the inhibitor from the assay dilution plate, a control inhibitor LY2886721 or inhibitor vehicle was then added to each well of the black 96-well micro plate containing BACE1.
- TR-FRET Binding A Compound IC50 values were assessed following a 10-point, half-log10 dilution schema starting at 10 uM compound concentration. The assay was performed in 384-well LV plates (Matrix 4365, Thermo Scientific) and run in the presence and absence of the compound of interest. Each well of 12 uL assay mixture contained 10 mM Tris (pH 7.4), 1 mM DTT, 0.005% Tween-20, 150 mM NaCl, 10% DMSO, 3 nM GST-Mcl-1, 1 nM LanthaScreen Eu-tagged GST antibody (#PV5594, Invitrogen, Life Technologies), and 8 nM fluorescently labeled peptide. Reactions were incubated at 24 °C for 60 min before reading on a Tecan F500 spectrofluorometer with excitation at 340 nm and emission at 620 nm and 665 nm. Subsequently, the ratio of fluorescence emission intensity at 665 nm to 620 nm was calculated for each reaction, and the dose response of the ratio to testing compound concentration was fitted to a select fit model that will provide the best fit quality using automatic parameters to derive IC50 values for each testing compound.
- Binding Assay The assay was run using a fluorescein isothiocyanate-labeled BH3 peptide derived from Bak (FITC-AHx-GQVGRQLAIIGDDINR-NH2) that was purchased from GenScript (Piscataway, N.J.) at >95% purity and used without further purification. 10 nM FITC-Bak peptide and 14 nM recombinant Mcl-1 (residues 172-327) were added to assay buffer (3 mM dithiothreitol, 50 mM NaCl, 20 mM Tris, pH 7.5). For selectivity assays, 40 nM Bcl-2 (residues 1-207A96T,G110R, Δ35-91, replaced with Bcl-xL35-50) or 4 nM Bcl-xL (residues 1-209, loop 45-86 deleted) were incubated with 10 nM FITC-Bak in assay buffer.
- Matriptase Inhibition Enzymatic assays and Ki determination were performed at room temperature in an assay buffer containing 50 mM Tris-HCl, 150 mM NaCl and 500 μg/mL BSA at pH 7.4. To determine which method to use for the evaluation of inhibition, 0.25 nM protease was added to a reaction buffer containing 0 nM, 2.5 nM or 1 mM of inhibitors and 200 μM of a fluorogenic substrate (Boc-Gln-Ala-Arg-AMC). Proteolytic activity was monitored by measuring the release of fluorescence (excitation; 360 nm, emission; 441 nm) in a FLX800 TBE microplate reader (Bio-Tek Instruments, Winooski, Vt., USA).
- ChEBML_146873 Inhibition of [3H]DPDPE binding to guinea pig brain membrane Opioid receptor delta 1 at 1.0 nM
- ChEBML_146877 Inhibition of Opioid receptor delta 1 by displacing 1 nM [3H]DPDPE in guinea pig brain membrane
- ChEBML_162605 In vitro inhibitory concentration required against Protein-tyrosine phosphatase alpha in the presence of 300 nM DTT
- ChEBML_197244 Binding affinity towards Retinoic acid receptor RXR-gamma by displacing 3.4 nM 3[H]-9-cis-RA
- ChEBML_204741 Inhibitory activity against human 5-alpha-reductase type 2 using 18213 3H testosterone 210 nM as substrate
- ChEBML_210874 In vitro inhibitory concentration required against Tyrosine phosphatase SHP-2 in the presence of 300 nM DTT
- ChEBML_221915 Binding affinity towards mu opioid receptor using [3H]- -DAMGO (1.5 nM) as radioligand in rat brain membranes
- ChEBML_221937 Binding affinity towards mu opioid receptor using [3H]- -DAMGO (1.5 nM) as radioligand in rat brain membranes
- ChEBML_29281 Inhibition of adenosine induced negative inotropic activity against guinea-pig atria (Adenosine A1 receptor) at 10 nM
- ChEBML_29628 Inhibition of 1 nM [3H]N-6-(phenylisopropyl)adenosine binding to A1 receptors in rat cortical membranes
- ChEBML_30208 Binding affinity against adrenergic beta 2 receptor versus 1 nM [3H]dihydroalprenolol in rat cerebral cerebellar membranes
- ChEBML_33196 Binding affinity at human Alpha-2A adrenergic receptor in CHO cells by [3H]rauwolscine (1 nM) displacement.
- ChEBML_33659 Binding affinity at human Alpha-2C adrenergic receptor in CHO cells by [3H]rauwolscine (1 nM) displacement.
- ChEBML_33866 Binding affinity at human Alpha-1A adrenergic receptor in CHO cells uby [3H]prazosin (0.25 nM) displacement.
- ChEBML_62870 In vitro binding affinity against dopamine receptor D2 in rat striata; value ranges from 1.8-3.0 nM
- ChEBML_90174 In vitro inhibition of human platelet aggregation induced by alpha-thrombin (at a concentration of 0.15 nM)
- ChEMBL_144336 (CHEMBL751847) Inhibition of [3H]-nicotinic acid (20 nM) binding to nicotinic acid receptor in rat spleen membrane.
- ChEMBL_146285 (CHEMBL756530) In vitro opioid receptor binding competition against 1 nM [3H]- Naltrexone in the absence of NaCl.
- ChEMBL_1505401 (CHEMBL3595284) Inhibition of human ARTD4 (250 to 565) using 500 nM NAD substrate after 2.5 hrs incubation
- ChEMBL_1505402 (CHEMBL3595285) Inhibition of human ARTD7 (460 to 656) using 250 nM NAD substrate after 2 hrs incubation
- ChEMBL_1505403 (CHEMBL3595286) Inhibition of human ARTD10 (809 to 1017) using 500 nM NAD substrate after 2.5 hrs incubation
- ChEMBL_1509403 (CHEMBL3603346) Binding affinity to human serum albumin with excitation at 285 nm by fluorescence emission spectroscopic analysis
- ChEMBL_164465 (CHEMBL770731) Inhibition of specific [3H]-prazosin binding (0.2 nM) to rat brain membranes alpha-1 adrenergic receptor
- ChEMBL_195324 (CHEMBL799719) Binding affinity towards retinoic acid receptor alpha was determined using [3H]ATRA (5 nM) as radioligand
- ChEMBL_195813 (CHEMBL807724) Binding affinity towards retinoic acid receptor beta was determined using [3H]ATRA (5 nM) as radioligand
- ChEMBL_196324 (CHEMBL806267) Binding affinity towards retinoic acid receptor gamma was determined using [3H]ATRA (5 nM) as radioligand
- ChEMBL_207469 (CHEMBL816750) Inhibitory concentration against radioligand [3H]SQ-29,548 (5 nM) binding to TP receptors in human platelets
- ChEMBL_211080 (CHEMBL812861) Tested for the TYR(Me)2 arginine-vasopressin as radioligand at 0.3 nM in A7r5 cells
- ChEMBL_217648 (CHEMBL818520) Binding affinity towards cRAR-beta-2 receptor by displacing 0.82 nM 3[H]all-trans-RA
- ChEMBL_217650 (CHEMBL820185) Binding affinity towards cRAR-beta-2 receptor by displacing 1.1 nM 3[H]-9-cis-RA
- ChEMBL_217653 (CHEMBL820188) Binding affinity towards cRAR-beta-2 receptor by displacing 1.1 nM 3[H]-9-cis-RA
- ChEMBL_2576 (CHEMBL617123) Ability to displace [3H]ketanserin (0.5 nM) from cerebral cortex of rat 5-hydroxytryptamine 2A receptor
- ChEMBL_2812 (CHEMBL875915) Ability to displace [3H]mesulergine (0.5 nM) from SR-3T3 cells of rat 5-HT2C receptor
- ChEMBL_287 (CHEMBL615725) In vitro inhibition of [3H]DPAT (1 nM) binding to 5-HT1A receptor from bovine hippocampus
- ChEMBL_29319 (CHEMBL642307) Binding affinity towards adenosine A1 receptor in rat cerebral cortical membranes with 1 nM [3H]cyclohexyladenosine
- ChEMBL_2956 (CHEMBL617896) Ability to displace [3H]mesulergine (0.5 nM) from CHO cells of human 5-hydroxytryptamine 2C receptor
- ChEMBL_303379 (CHEMBL839696) Inhibitory constant was determined against 5-hydroxytryptamine 1A receptor using 1.2 nM [3H]8-OH-DPAT
- ChEMBL_303523 (CHEMBL839637) Ability to displace [125I]-MCH()0.5 nM from human MCH1R(2.5 uM) expressed in CHO cells
- ChEMBL_303782 (CHEMBL830139) Inhibition of 3 nM [3H]-MLA binding to Nicotinic acetylcholine receptor alpha7 of rat brain membranes
- ChEMBL_303812 (CHEMBL827071) Inhibition of 1 nM [3H]mesulergine binding to Serotonin 5-HT2C receptor from rat brain cortex
- ChEMBL_303813 (CHEMBL827072) Inhibition of 0.4 nM [3H]ketanserin binding to Serotonin 5-HT2A receptor from rat brain cortex
- ChEMBL_303818 (CHEMBL827077) Inhibition of 5 nM [3H]DTBZ binding to Vesicular Monoamine Transporter (VAMT2) of rat vesicle membranes
- ChEMBL_306882 (CHEMBL828688) Inhibition of 100 nM capsaicin effect on intracellular [Ca2+] concentration in HEK293 cells expressing human TRPV1
- ChEMBL_312839 (CHEMBL825050) Antagonist activity against 100 nM quinpirole-stimulated mitogenesis in CHO cells expressing human dopamine D3 receptor
- ChEMBL_312875 (CHEMBL874953) Inhibition of 100 nM NECA-stimulated cAMP production in CHO cells expressing human adenosine A2B receptor
- ChEMBL_320864 (CHEMBL884789) Binding affinity towards human CRTH2 receptor expressed in CHO cells; range 25 nM to 8 uM
- ChEMBL_320936 (CHEMBL882455) In vitro binding affinity towards rat Nicotinic acetylcholine receptor alpha2-beta2 using 0.5 nM [3H]epibatidine
- ChEMBL_320937 (CHEMBL882456) In vitro binding affinity towards rat Nicotinic acetylcholine receptor alpha2-beta4 using 0.5 nM [3H]epibatidine
- ChEMBL_320938 (CHEMBL882457) In vitro binding affinity towards rat Nicotinic acetylcholine receptor alpha3-beta2 using 0.5 nM [3H]epibatidine
- ChEMBL_320939 (CHEMBL882458) In vitro binding affinity towards rat Nicotinic acetylcholine receptor alpha3-beta4 using 0.5 nM [3H]epibatidine
- ChEMBL_320940 (CHEMBL882459) In vitro binding affinity towards rat Nicotinic acetylcholine receptor alpha4-beta2 using 0.5 nM [3H]epibatidine
- ChEMBL_320941 (CHEMBL882460) In vitro binding affinity towards rat Nicotinic acetylcholine receptor alpha4-beta4 using 0.5 nM [3H]epibatidine
- ChEMBL_321012 (CHEMBL871844) Binding affinity towards human Adenosine A1 receptor expressed in CHO cells using 1 nM [3H]DPCPX
- ChEMBL_321017 (CHEMBL883650) Binding affinity towards human Adenosine A2b receptor expressed in HEK293 cells using 5 nM [3H]DPCPX
- ChEMBL_32720 (CHEMBL645992) Displacement of [3H]prazosin (0.3 nM) from rat Alpha-1D adrenergic receptor expressed in CHO cells
- ChEMBL_33584 (CHEMBL647135) Displacement of [3H]prazosin (0.3 nM) from bovine Alpha-1A adrenergic receptor expressed in BHK cells
- ChEMBL_34001 (CHEMBL643630) Inhibition of specific [3H]-prazosin binding (0.2 nM) to rat brain membranes alpha-1 adrenergic receptor
- ChEMBL_34052 (CHEMBL643909) Inhibition of specific [3H]clonidine binding (0.4 nM) to rat brain membranes Alpha-2 adrenergic receptor
- ChEMBL_34053 (CHEMBL643910) Inhibition of specific [3H]clonidine binding (0.4 nM) to rat brain membranes alpha-2 adrenergic receptor
- ChEMBL_34290 (CHEMBL647040) Compound is evaluated for receptor binding affinity using 100 nM concentration on rat cerebral cortical membranes
- ChEMBL_35496 (CHEMBL646406) Inhibition of Aminopeptidase N activity in pig kidney with 10 nM [3H]Leu-enkephalin as substrate
- ChEMBL_36123 (CHEMBL647749) Inhibition of 1.0 nM [3H]mibolerone binding to human androgen receptor of PC3/AR cell lysate
- ChEMBL_38886 (CHEMBL652717) Inhibitory activity against beta adrenergic receptor of rat frontal cortex homogenate using (1.0 nM) [3H]- dihydroalprenolol
- ChEMBL_40267 (CHEMBL656503) Concentration required to inhibit specific binding of [3H]-BK(0.06 nM) to the bradykinin receptor B2
- ChEMBL_40430 (CHEMBL876526) Displacement of [3H]BK (1 nM) from human Bradykinin receptor B2 expressed in Cos-7 cells
- ChEMBL_46141 (CHEMBL660014) Displacement of 0.5 nM [3H](R)-(+)-WIN-55212-2 from Cannabinoid receptor of rat cerebellar membranes
- ChEMBL_461964 (CHEMBL945752) Antagonist activity at human CCR3 receptor assessed as inhibition of chemotaxis in eosinophil at 30 nM
- ChEMBL_567 (CHEMBL615586) In vitro inhibition of [125I]IPAPP (0.25 nM) binding to 5-HT1A receptor from bovine hippocampus
- ChEMBL_58163 (CHEMBL672686) Ability to displace [3H]-SCH- 23390 (0.2 nM) from corpus striatum of rat Dopamine receptor D1
- ChEMBL_585 (CHEMBL615455) In vitro inhibition of [3H]DPAT (1 nM) binding to 5-HT1A receptor from bovine hippocampus
- ChEMBL_58505 (CHEMBL672000) Ability to displace [3H]-SCH- 23390 (0.2 nM) from corpus striatum of rat Dopamine receptor D1
- ChEMBL_587 (CHEMBL615457) In vitro inhibition of [3H]DPAT (1 nM) binding to 5-HT1A receptor from bovine hippocampus
- ChEMBL_619398 (CHEMBL1104156) Inhibition of p70S6K assessed as decrease in NADH absorbance at 340 nm in the presence of
- ChEMBL_619399 (CHEMBL1104157) Inhibition of PKAalpha assessed as decrease in NADH absorbance at 340 nm in the presence of
- ChEMBL_619401 (CHEMBL1104159) Inhibition of ROCK1 assessed as decrease in NADH absorbance at 340 nm in the presence of
- ChEMBL_66541 (CHEMBL677808) In vitro displacement of 3 nM [3H]4-hydroxytamoxifen from estrogen receptor of rat uterine cystol
- ChEMBL_666886 (CHEMBL1262283) Displacement of 20 nM [3H]GSK304649 from Human immunodeficiency virus 1 integrase by scintillation proximity assay
- ChEMBL_693081 (CHEMBL1638241) Inhibition of Mycobacterium smegmatis MC2 155 ATP synthase subunit c-mediated ATP production at 100 nM
- ChEMBL_765233 (CHEMBL1827786) Antagonist activity at Agrobacterium tumefaciens TraR receptor in presence of 100 nM 3-oxo-C8-HSL
- ChEMBL_936870 (CHEMBL2320752) Inhibition of pure alpha-glucosidase (unknown origin) assessed as para-nitrophenol release at 420 nm OD
- ChEMBL_99661 (CHEMBL704427) Inhibition of specific binding of [3H]LTB4 ( 0.1 nM) to LTB4 receptor on intact human neutrophils
- Akt1 Kinase Assay A fluorescence polarization IMAP type assay is used. After a 90 min incubation of reaction mixtures containing enzyme, substrate, and inhibitor, IMAP beads are added and plates are read (lamp filter: 544 nm, emission filter: 615 nm).
- GHS Ligand Binding Assay Briefly, direct ligand binding experiments were performed using fluorescence energy transfer with the purified receptor labeled with Alexa Fluor 350 at its N terminus and a ghrelin peptide labeled with FITC (JMV 4946). Titration experimentswere carried out with protein concentrations in the 10 nM protein concentration range and increasing ligand concentrations. Competition experiments were carried out by adding increasing concentrations of the competing compound to a receptor-JMV 4946 mixture (100 nM concentration range). Fluorescence emission spectra were recorded at 20 °C between 400 and 600 nm on a Cary Eclipse spectrofluorimeter (Varian) with an excitation at 346 or 488 nm. Buffer contributions were systematically subtracted. The FRET ratio corresponds to the ratio of theacceptor-emitted fluorescence at 520 nm from excitation attwo different wavelengths, 346 and 488 nm.
- Lanthascreen Competitive Binding Assay The assay was performed according to manufacturer protocol. A mixture of 5 nM GST-PPARG-LBD, 5 nM Tb-GST-antibody, 5 nM Fluormone Pan- PPAR Green, and serial dilutions of the experimental compound, beginning at 10 μΜ downwards, was added to wells of black 384-well low- volume plates (Greiner) to a total volume of 18 xL. All dilutions were made in TR-FRET assay buffer C. DMSO at 2% final concentration was used as a no-ligand control. Experiment was performed in triplicate, and incubated for 2 hours in the dark prior to assay read in Perkin Elmer ViewLux ultra HTS microplate reader. FRET signal was measured by excitation at 340 nm and emission at 520 nm for fluorescein and 490 nm for terbium.
- Time-Resolved Fluorescence Resonance Energy Transfer (TR-FRET) Binding Assay Probe 1:The assay was carried out in 18 uL low volume plates (Owens Corning) in reaction buffer (50 mM HEPES (NaOH), pH 7.5, 100 mM NaCl, 10 mM MgCl2, 1 mM DTT, 1% Glycerol) using 6.8 nM recombinant, human, C-terminally-His tagged NAMPT, 1 nM Tb-anti-His antibody (Invitrogen, Cat #PV5895), and 200 nM probe (Oregon Green 488-conjugated APO0866; A-1251667.0. Plates were covered, and reactions were carried out for 2-3 hours. Plates were read with Envision (Laser Lantha low volume protocol) after 2 to 3 hours. Excitation was carried out at 337 nm, and the ratio of emission of Oregon Green (520 nm) to terbium (492 nm) was determined and used to calculate IC50 values of test compounds.
- Time-Resolved Fluorescence Resonance Energy Transfer (TR-FRET) Binding Assay Probe 2:The assay was carried out in 18 uL low volume plates (Owens Corning) in reaction buffer (50 mM HEPES (NaOH), pH 7.5, 100 mM NaCl, 10 mM MgCl2, 1 mM DTT, 1% Glycerol) using 6.8 nM recombinant, human, C-terminally-His tagged NAMPT, 1 nM Tb-anti-His antibody (Invitrogen, Cat #PV5895), and 200 nM probe (Oregon Green 488-conjugated APO0866; A-1287128.0. Plates were covered, and reactions were carried out for 2-3 hours. Plates were read with Envision (Laser Lantha low volume protocol) after 2 to 3 hours. Excitation was carried out at 337 nm, and the ratio of emission of Oregon Green (520 nm) to terbium (492 nm) was determined and used to calculate IC50 values of test compounds.
- Homogeneous Time Resolved Fluorescence (HTRF) Assay To determine the potency of compounds for inhibiting nucleotide exchange, various concentrations were incubated with K-Ras G12C (25 nM in reaction, 12.5 nM final). After 18 hours at room temperature, the SOS1 GTP exchange factor (1.67 nM during exchange, 1.25 nM final) was added to initiate nucleotide exchange to GTP (200 μM during exchange, 150 M final). The level of GTP exchange was assessed by addition of a Ras binding domain derived from C-Raf and the HTRF detection antibodies Tb-anti-FLAG and D2-anti-his (Cis-Bio) at 50 nM, 1 nM and 12.5 nM, respectively. After 2 hours, the ratio of 665 nm to 615 nM emission with 320 nM excitation was measured on an Envision plate reader (Perkin Elmer).The final reaction volume was 20l in a ProxiPlate-384F Plus (Perkin Elmer), in buffer containing 20 mM HEPES, 150 mM NaCl, 1 mM MgCl2, 0.1% BSA, 0.03% Tween-20 and 1 mM DTT. K-Ras G12C (residues 2-188) and SOS1 (residues 564-1049) had N-terminal 6-His and the Raf-RBD construct (residues 51-186 of RAFI) had an N-terminal Flag-tag. All constructs were expressed in E. coli and had a Tev cleavage site between the tag and protein of interest that was not used during purification.
- TR-FRET Assay All assay components were dissolved in buffer composition 20 mM Hepes pH 7.5, 150 mM NaCl, 5 mM DTT, 0.005% Tween 20, and 100 ug/ml BSA for BRD4 (1-477 and 44-460). The final concentrations of the bromodomain proteins are 1.6 nM BRD4(44-168), 1 nM BRD4(333-460), and 1 nM BRD4(1-477 or 44-460), and the fluorescent probe molecule is 100 nM, 50 nM, and 7.5 nM respectively. All proteins were biotinylated. A streptavidin labeled with terbium cryptate (Cisbio SA-Tb) was used as detection, and pre-mixed with the bromodomain protein at a final concentration of 0.2 nM. In some instances for BRD4 (44-460), anti-His terbium cryptate was used as a detection. 7.5 nl of dose-responsed test compound or dmso vehicle (0.0375%) was pre-spotted in a black Corning 384 well plate and 10 ul each of bromodomain/detection reagent and fluorescent small molecule solution were added to the plate, and the reaction incubated for 60 min at room temperature. Plates were then read on EnVision plate reader, (λex=340 nm, acceptor λEm=520 nm, and donor λEm=615 nm, LANCE D400 mirror). Time resolved fluorescence intensity measurements were made at both emissions, and the ratio of acceptor/donor was calculated and used for data analysis.
- Time Resolved Fluorescence Energy Transfer (TR-FRET) Assay Assay 2: The test is performed in white 384-well plates (Greiner Bio-One, reference 781207) in a total volume of 60 μL by adding 1 μL of compounds tested at different concentrations diluted in 100% DMSO (1.7% final DMSO concentration) in reaction buffer (PBS, 125 mM NaCl, 0.001% Novexin (consists of carbohydrate polymers), designed to increase the solubility and stability of proteins; Expedeon Ltd., Cambridgeshire, United Kingdom), 0.01% Gelatin, 0.01% 0.2%, Pluronic F-127 (block copolymer from ethylenoxide and propyleneoxide), 1 mM DTT). After addition of 1.25 nM MDM2-biotinylated or 2.5 nM MDM4-biotinylated (internal preparations), and 0.625 nM Europium labeled streptavidin (Perkin Elmer), the solution is pre-incubated for 15 minutes at room temperature, then 10 nM Cy5-p53 peptide (internal preparation) is added before an incubation at room temperature for 15 minutes prior to reading the plate. For measurement of samples, a Victor II microplate reader (Perkin Elmer) is used with the following settings: Excitation 340 nm, Emission Donor 620 nm and Emission Acceptor 665 nm. For selected compounds displaying IC50s between 0.05 and 5 nM on MDM2, a slightly modified assay is used with the following adaptations: 0.1 nM MDM2, 0.1 nM Europium labeled streptavidin and Tecan genios Pro is used as a microplate reader for the fluorescence measurements (p53-MDM2 Assay 2).
- Time Resolved Fluorescence Resonance Energy Transfer (TR-FRET) Assay All assay components were dissolved in buffer composition 20 mM Hepes pH 7.5, 150 mM NaCl, 5 mM DTT, 0.005% Tween 20, and 100 ug/ml BSA for BRD4 (1-477 and 44-460). The final concentrations of the bromodomain proteins are 1.6 nM BRD4(44-168), 1 nM BRD4(333-460), and 1 nM BRD4(1-477 or 44-460), and the fluorescent probe molecule is 100 nM, 50 nM, and 7.5 nM respectively. All proteins were biotinylated. A streptavidin labeled with terbium cryptate (Cisbio SA-Tb) was used as detection, and pre-mixed with the bromodomain protein at a final concentration of 0.2 nM. In some instances for BRD4 (44-460), anti-His terbium cryptate was used as a detection. 7.5 nl of dose-responsed test compound or dmso vehicle (0.0375%) was pre-spotted in a black Corning 384 well plate and 10 ul each of bromodomain/detection reagent and fluorescent small molecule solution were added to the plate, and the reaction incubated for 60 min at room temperature. Plates were then read on EnVision plate reader, (λex=340 nm, acceptor λEm=520 nm, and donor λEm=615 nm, LANCE D400 mirror). Time resolved fluorescence intensity measurements were made at both emissions, and the ratio of acceptor/donor was calculated and used for data analysis.
- Biochemical Assay Increasing concentrations of compounds were added to pre-mixed biotinylated DCAF15 at 200 nM, Bodipy-FL-labelled RRM2 domain in RBM39 at 200 nM, and terbium (Tb)-coupled streptavidin at 2 nM (Invitrogen) in 384-well microplates in a buffer containing 50 mM Tris pH 7.5, 100 mM NaCl, 0.1% pluronic acid and 2% DMSO. Before TR-FRET measurements were conducted, the reactions were incubated for 15 min at room temperature. After excitation of terbium (Tb) fluorescence at 337 nm, emission at 490 nm (Tb) and 520 nm (Bodipy-FL) were recorded with a 70 μs delay to reduce background fluorescence and the reaction was followed over 1 h by recording 60 technical replicates of each data point using a PHERAstar FS microplate reader (BMG Labtech). The TR-FRET signal of each data point was extracted by calculating the 520/490 nm ratio. Data were analyzed with GraphPad Prism 7.
- Biological Assay Recombinant human factor B (expressed in drosophila cells and purified using standard methods) labeled with biotin (10 nM), europium-labeled streptavidin (5 nM) and (+) or (−)-2-((1E,3E,5E)-5-(1-(6-((2-(3-(4-((R)-3-amino-3-phenylpropanoyl)-1-(4-amino-6,7-dimethoxyquinazolin-2-yl)piperazin-2-yl)phenoxy)ethyl)amino)-6-oxohexyl)-3,3-dimethyl-5-sulfoindolin-2-ylidene)penta-1,3-dien-1-yl)-1-ethyl-3,3-dimethyl-5-sulfo-3H-indol-1-ium (Biological Example 2.6, 240 nM activity against factor B when tested using the assay of Biological Example 1) (75 nM) were incubated with test compound at various concentrations up to 2 hours at room temperature in 20 mM Tris/HCl, pH 7.4, 0.005% (v/v) Tween20.The time-gated decrease in fluorescence intensity related to the competition between labeled and unlabeled factor B ligands was recorded at both 620 nm and 665 nm, 70 μs after excitation at 337 nm using a microplate spectrofluorimeter.
- Enzymatic Assay FGFR1: In a final reaction volume of 30 μL, FGFR1 (h) (25 ng/ml) is incubated with 50 mM HEPES pH 7.5, 6 mM MnCl2, 1 mM DTT, 0.1 mM Na3VO4, 0.01% Triton-X-100, 500 nM Btn-Flt3 and 5 μM ATP in the presence of compound (1% DMSO final). After incubation for 60 minutes at room temperature the reaction is stopped with 2.27 nM EU-anti P-Tyr, 7 mM EDTA, 31.25 nM SA-XL-665 and 0.02% BSA which is present for 60 minutes at room temperature. Time-Resolved Fluorescence Resonance Energy Transfer (TR-FRET) signal (ex340 nm. Em 620 nm, em 655 nm) is measured afterwards and results are expressed in RFU (Relative Fluorescence Units). In this assay, the inhibitory effect of different compound concentrations (range 10 μM to 0.1 nM) is determined and used to calculate an IC50 (M) and pIC50 (−log IC50) value.
- Enzymatic Assay FGFR1: In a final reaction volume of 30 μL, FGFR1 (h) (25 ng/ml) was incubated with 50 mM HEPES pH 7.5, 6 mM MnCl2, 1 mM DTT, 0.1 mM Na3VO4, 0.01% Triton-X-100, 500 nM Btn-Flt3 and 5 μM ATP in the presence of compound (1% DMSO final). After incubation for 60 minutes at room temperature the reaction was stopped with 2.27 nM EU-anti P-Tyr, 7 mM EDTA, 31.25 nM SA-XL-665 and 0.02% BSA which was present for 60 minutes at room temperature. Time-Resolved Fluorescence Resonance Energy Transfer (TR-FRET) signal (ex340 nm. Em 620 nm, em 655 nm) was measured afterwards and results are expressed in RFU (Relative Fluorescence Units). In this assay, the inhibitory effect of different compound concentrations (range 10 μM to 0.1 nM) was determined and used to calculate an IC50 (M) and pIC50 (−log IC50) value.
- Enzymatic Assay FGFR2: In a final reaction volume of 30 μL, FGFR2 (h) (150 ng/ml) is incubated with 50 mM HEPES pH 7.5, 6 mM MnCl2, 1 mM DTT, 0.1 mM Na3VO4, 0.01% Triton-X-100, 500 nM Btn-Flt3 and 0.4 μM ATP in the presence of compound (1% DMSO final). After incubation for 60 minutes at room temperature the reaction is stopped with 2.27 nM EU-anti P-Tyr, 7 mM EDTA, 31.25 nM SA-XL-665 and 0.02% BSA which is present for 60 minutes at room temperature. Time-Resolved Fluorescence Resonance Energy Transfer (TR-FRET) signal (ex340 nm. Em 620 nm, em 655 nm) is measured afterwards and results are expressed in (Relative Fluorescence Units). In this assay, the inhibitory effect of different compound concentrations (range 10 μM to 0.1 nM) is determined and used to calculate an IC50 (M) and pIC50 (−log IC50) value.
- Enzymatic Assay FGFR2: In a final reaction volume of 30 μL, FGFR2 (h) (150 ng/ml) was incubated with 50 mM HEPES pH 7.5, 6 mM MnCl2, 1 mM DTT, 0.1 mM Na3VO4, 0.01% Triton-X-100, 500 nM Btn-Flt3 and 0.4 μM ATP in the presence of compound (1% DMSO final). After incubation for 60 minutes at room temperature the reaction was stopped with 2.27 nM EU-anti P-Tyr, 7 mM EDTA, 31.25 nM SA-XL-665 and 0.02% BSA which was present for 60 minutes at room temperature. Time-Resolved Fluorescence Resonance Energy Transfer (TR-FRET) signal (ex340 nm. Em 620 nm, em 655 nm) was measured afterwards and results are expressed in (Relative Fluorescence Units). In this assay, the inhibitory effect of different compound concentrations (range 10 μM to 0.1 nM) was determined and used to calculate an IC50 (M) and pIC50 (−log IC50) value.
- Enzymatic Assay FGFR3: In a final reaction volume of 30 μL, FGFR3 (h) (40 ng/ml) was incubated with 50 mM HEPES pH 7.5, 6 mM MnCl2, 1 mM DTT, 0.1 mM Na3VO4, 0.01% Triton-X-100, 500 nM Btn-Flt3 and 25 μM ATP in the presence of compound (1% DMSO final). After incubation for 60 minutes at room temperature the reaction was stopped with 2.27 nM EU-anti P-Tyr, 7 mM EDTA, 31.25 nM SA-XL-665 and 0.02% BSA which was present for 60 minutes at room temperature. Time-Resolved Fluorescence Resonance Energy Transfer (TR-FRET) signal (ex340 nm. Em 620 nm, em 655 nm) was measured afterwards and results are expressed in RFU (Relative Fluorescence Units). In this assay, the inhibitory effect of different compound concentrations (range 10 μM to 0.1 nM) was determined and used to calculate an IC50 (M) and pIC50 (−log IC50) value.
- Enzymatic Assay FGFR4: In a final reaction volume of 30 μL, FGFR4 (h) (60 ng/ml) is incubated with 50 mM HEPES pH 7.5, 6 mM MnCl2, 1 mM DTT, 0.1 mM Na3VO4, 0.01% Triton-X-100, 500 nM Btn-Flt3 and 5 μM ATP in the presence of compound (1% DMSO final). After incubation for 60 minutes at room temperature the reaction is stopped with 2.27 nM EU-anti P-Tyr, 7 mM EDTA, 31.25 nM SA-XL-665 and 0.02% BSA which is present for 60 minutes at room temperature. Time-Resolved Fluorescence Resonance Energy Transfer (TR-FRET) signal (ex340 nm. Em 620 nm, em 655 nm) is measured afterwards and results are expressed in RFU (Relative Fluorescence Units). In this assay, the inhibitory effect of different compound concentrations (range 10 μM to 0.1 nM) is determined and used to calculate an IC50 (M) and pIC50 (−log IC50) value.
- Enzymatic Assay FGFR4: In a final reaction volume of 30 μL, FGFR4 (h) (60 ng/ml) was incubated with 50 mM HEPES pH 7.5, 6 mM MnCl2, 1 mM DTT, 0.1 mM Na3VO4, 0.01% Triton-X-100, 500 nM Btn-Flt3 and 5 μM ATP in the presence of compound (1% DMSO final). After incubation for 60 minutes at room temperature the reaction was stopped with 2.27 nM EU-anti P-Tyr, 7 mM EDTA, 31.25 nM SA-XL-665 and 0.02% BSA which was present for 60 minutes at room temperature. Time-Resolved Fluorescence Resonance Energy Transfer (TR-FRET) signal (ex340 nm. Em 620 nm, em 655 nm) was measured afterwards and results are expressed in RFU (Relative Fluorescence Units). In this assay, the inhibitory effect of different compound concentrations (range 10 μM to 0.1 nM) was determined and used to calculate an IC50 (M) and pIC50 (−log IC50) value.
- Enzymatic Assay KDR (VEGFR2): In a final reaction volume of 30 μL, KDR (h) (150 ng/ml) is incubated with 50 mM HEPES pH 7.5, 6 mM MnCl2, 1 mM DTT, 0.1 mM Na3VO4, 0.01% Triton-X-100, 500 nM Btn-Flt3 and 3 μM ATP in the presence of compound (1% DMSO final). After incubation for 120 minutes at room temperature the reaction is stopped with 2.27 nM EU-anti P-Tyr, 7 mM EDTA, 31.25 nM SA-XL-665 and 0.02% BSA which is present for 60 minutes at room temperature. Time-Resolved Fluorescence Resonance Energy Transfer (TR-FRET) signal (ex340 nm. Em 620 nm, em 655 nm) is measured afterwards and results are expressed in RFU (Relative Fluorescence Units). In this assay, the inhibitory effect of different compound concentrations (range 10 μM to 0.1 nM) is determined and used to calculate an IC50 (M) and pIC50 (−log IC50) value.
- Enzymatic Assay KDR (VEGFR2): In a final reaction volume of 30 μL, KDR (h) (150 ng/ml) was incubated with 50 mM HEPES pH 7.5, 6 mM MnCl2, 1 mM DTT, 0.1 mM Na3VO4, 0.01% Triton-X-100, 500 nM Btn-Flt3 and 3 μM ATP in the presence of compound (1% DMSO final). After incubation for 120 minutes at room temperature the reaction was stopped with 2.27 nM EU-anti P-Tyr, 7 mM EDTA, 31.25 nM SA-XL-665 and 0.02% BSA which was present for 60 minutes at room temperature. Time-Resolved Fluorescence Resonance Energy Transfer (TR-FRET) signal (ex340 nm. Em 620 nm, em 655 nm) was measured afterwards and results are expressed in RFU (Relative Fluorescence Units). In this assay, the inhibitory effect of different compound concentrations (range 10 μM to 0.1 nM) was determined and used to calculate an IC50 (M) and pIC50 (−log IC50) value.
- FGFR1 (Enzymatic Assay) In a final reaction volume of 30 μL, FGFR1 (h) (25 ng/ml) was incubated with 50 mM HEPES pH 7.5, 6 mM MnCl2, 1 mM DTT, 0.1 mM Na3VO4, 0.01% Triton-X-100, 500 nM Btn-Flt3 and 5 μM ATP in the presence of compound (1% DMSO final). After incubation for 60 minutes at room temperature the reaction was stopped with 2.27 nM EU-anti P-Tyr, 7 mM EDTA, 31.25 nM SA-XL-665 and 0.02% BSA which was present for 60 minutes at room temperature. Time-Resolved Fluorescence Resonance Energy Transfer (TR-FRET) signal (ex340 nm. Em 620 nm, em 655 nm) was measured afterwards and results are expressed in RFU (Relative Fluorescence Units). In this assay, the inhibitory effect of different compound concentrations (range 10 μM to 0.1 nM) was determined and used to calculate an IC50 (M) and pIC50 (−log IC50) value.
- FGFR2 (Enzymatic Assay) In a final reaction volume of 30 μL, FGFR2 (h) (150 ng/ml) was incubated with 50 mM HEPES pH 7.5, 6 mM MnCl2, 1 mM DTT, 0.1 mM Na3VO4, 0.01% Triton-X-100, 500 nM Btn-Flt3 and 0.4 μM ATP in the presence of compound (1% DMSO final). After incubation for 60 minutes at room temperature the reaction was stopped with 2.27 nM EU-anti P-Tyr, 7 mM EDTA, 31.25 nM SA-XL-665 and 0.02% BSA which was present for 60 minutes at room temperature. Time-Resolved Fluorescence Resonance Energy Transfer (TR-FRET) signal (ex340 nm. Em 620 nm, em 655 nm) was measured afterwards and results are expressed in (Relative Fluorescence Units). In this assay, the inhibitory effect of different compound concentrations (range 10 μM to 0.1 nM) was determined and used to calculate an IC50 (M) and pIC50 (−log IC50) value.
- FGFR2 (Enzymatic Assay) In a final reaction volume of 30 μL, FGFR2 (h) (150 ng/ml) was incubated with 50 mM HEPES pH 7.5, 6 mM MnCl2, 1 mM DTT, 0.1 mM Na3VO4, 0.01% Triton-X-100, 500 nM Btn-Flt3 and 0.4 μM ATP in the presence of compound (1% DMSO final). After incubation for 60 minutes at room temperature the reaction was stopped with 2.27 nM EU-anti P-Tyr, 7 mM EDTA, 31.25 nM SA-XL-665 and 0.02% BSA which was present for 60 minutes at room temperature. Time-Resolved Fluorescence Resonance Energy Transfer (TR-FRET) signal (ex340 nm. Em 620 nm, em 655 nm) was measured afterwards and results are expressed in RFU (Relative Fluorescence Units). In this assay, the inhibitory effect of different compound concentrations (range 10 μM to 0.1 nM) was determined and used to calculate an IC50 (M) and pIC50 (−log IC50) value.
- FGFR3 (Enzymatic Assay) In a final reaction volume of 30 μL, FGFR3 (h) (40 ng/ml) was incubated with 50 mM HEPES pH 7.5, 6 mM MnCl2, 1 mM DTT, 0.1 mM Na3VO4, 0.01% Triton-X-100, 500 nM Btn-Flt3 and 25 μM ATP in the presence of compound (1% DMSO final). After incubation for 60 minutes at room temperature the reaction was stopped with 2.27 nM EU-anti P-Tyr, 7 mM EDTA, 31.25 nM SA-XL-665 and 0.02% BSA which was present for 60 minutes at room temperature. Time-Resolved Fluorescence Resonance Energy Transfer (TR-FRET) signal (ex340 nm. Em 620 nm, em 655 nm) was measured afterwards and results are expressed in RFU (Relative Fluorescence Units). In this assay, the inhibitory effect of different compound concentrations (range 10 μM to 0.1 nM) was determined and used to calculate an IC50 (M) and pIC50 (−log IC50) value.
- FGFR4 (Enzymatic Assay) In a final reaction volume of 30 μL, FGFR4 (h) (60 ng/ml) was incubated with 50 mM HEPES pH 7.5, 6 mM MnCl2, 1 mM DTT, 0.1 mM Na3VO4, 0.01% Triton-X-100, 500 nM Btn-Flt3 and 5 μM ATP in the presence of compound (1% DMSO final). After incubation for 60 minutes at room temperature the reaction was stopped with 2.27 nM EU-anti P-Tyr, 7 mM EDTA, 31.25 nM SA-XL-665 and 0.02% BSA which was present for 60 minutes at room temperature. Time-Resolved Fluorescence Resonance Energy Transfer (TR-FRET) signal (ex340 nm. Em 620 nm, em 655 nm) was measured afterwards and results are expressed in RFU (Relative Fluorescence Units). In this assay, the inhibitory effect of different compound concentrations (range 10 μM to 0.1 nM) was determined and used to calculate an IC50 (M) and pIC50 (−log IC50) value.
- FRET Assay For the FRET measurements, the Cerulean-Runt (Runx1 41-190) and Venus-CBFbeta (1-141) fusion proteins were dialyzed into FRET assay buffer. Protein concentrations were determined by UV absorption at 433 nm for Cerulean-Runt and 513 nm for Venus-CBFbeta. Equal molar concentrations of the two proteins were mixed together, diluted to 150 nM with FRET assay buffer containing 0.1% BSA, and incubated for 1 hr. A PHERAstar microplate reader (BMG Labtech, Durham, NC, USA) was used to measure fluorescence (excitation at 433 nm and emission measured at 474 and 525 nm). For IC50 determinations, the ratios of the fluorescence intensities at 525 nm and 474 nm were plotted versus the log of compound concentration, and the resulting curve was fit to a sigmoidal curve using Origin 7.0 (MicroCal, Northampton, MA, USA). Mean values of IC50 originating from two independent measurements performed in duplicate together with the standard deviations are reported.
- In Vitro Assay An in vitro assay showed inhibition of CXCR2-mediated intracellular calcium release by treatment with compound 9 (IC50=586±91 nM). Briefly, human neutrophils were suspended in HBSS− (without Ca2+ and Mg2+) containing 10 mM HEPES and FLIPR Calcium 3 dye (3.1×107 cells in total volume 1.7 mL). Cells were aliquoted (200 μL of the cell suspension per tube, 8 tubes total) and 2 μL of compound 9 (with appropriate dilutions) were added to each of 6 tubes. The tested concentrations of compound 9 were 156 nM, 312 nM, 625 nM, 1250 nM, 2500 nM and 5000 nM. As controls, 2 μL of DMSO (1% final concentration) were added to 2 other tubes. Cells were incubated for 30 min at 37° C. After dye loading, tubes were centrifuged at 6,000 rpm for 1 min, supernatant was removed and the cell pellet was re-suspended in 200 μL of HBSS+ (with Ca2+ and Mg2+) containing 10 mM HEPES.
- Interaction between Compound and Human PD-L1 Protein Example 40: In the experiment, the instrument OctectRED of Fortebio Company based on biofilm interferometry (BLI) technology was used to capture human PD-L1/AVI with SA chips. The concentration of antigen PD-L1/AVI was diluted to 10 μg/mL with running buffer (PBS+0.02% Tween-20+2% DMSO), and the loading time was 300 s; similarly, the analyte was also diluted to the corresponding concentrations (20 nM, 10 nM, 5 nM, 2.5 nM, 1.25 nM, 0.625 nM, 0.3125 nM) with running buffer in gradient manner, and a buffer blank control group was set at the same time. The binding time of human PD-L1/AVI to the analyte was 180 s, and the dissociation time was 300 s; the chip was regenerated with 10 mM glycine HCl, pH 1.7 solution and repeated 5-second pulse for 3 times. The data were fitted to a 1:1 binding model to determine the equilibrium dissociation constant KD.
- KDR (VEGFR2) (Enzymatic Assay) In a final reaction volume of 30 μL, KDR (h) (150 ng/ml) was incubated with 50 mM HEPES pH 7.5, 6 mM MnCl2, 1 mM DTT, 0.1 mM Na3VO4, 0.01% Triton-X-100, 500 nM Btn-Flt3 and 3 μM ATP in the presence of compound (1% DMSO final). After incubation for 120 minutes at room temperature the reaction was stopped with 2.27 nM EU-anti P-Tyr, 7 mM EDTA, 31.25 nM SA-XL-665 and 0.02% BSA which was present for 60 minutes at room temperature. Time-Resolved Fluorescence Resonance Energy Transfer (TR-FRET) signal (ex340 nm. Em 620 nm, em 655 nm) was measured afterwards and results are expressed in RFU (Relative Fluorescence Units). In this assay, the inhibitory effect of different compound concentrations (range 10 μM to 0.1 nM) was determined and used to calculate an IC50 (M) and pIC50 (−log IC50) value.
- KRAS WT Biochemical Assay Biochemical compound potencies are assessed by evaluating inhibition of SOS1-mediated nucleotide exchange in KRAS WT. In this assay, the SOS1-promoted exchange of fluorescently-labeled GDP (BOPIDY-GDP) is monitored by time-resolved fluorescence resonance energy transfer (TR-FRET). Compounds solubilized in DMSO are dispensed as concentration series into 384-well white assay plates. A preformed complex of 0.06 nM biotin-tagged recombinant human KRAS WT, 15 nM GDP and 0.06 nM terbium-labeled streptavidin (CisBIO) prepared in 10 uL/well assay buffer (20 mM HEPES, pH 7.5, 50 mM NaCl, 10 mM MgCl2, 0.01% Tween-20 and 1 mM dithiothreitol) is added and allowed to incubate for 10-minutes at room temperature. The reaction is initiated with the addition of 5 uL 300 nM recombinant human SOS1 and 300 nM BODIPY-GDP in assay buffer. After a 120-minute incubation, the fluorescence is measured with excitation at 337 nm and emission at 490 and 520 nm.
- PI3Kdelta HTRF Binding Assay A solution was prepared containing 0.2 nM Anti GST-Tb (Cisbio, 61GSTTLB), 40 nM probe and 1 nM GST-tagged PIK3C5 in complex with PIK3R1 (Invitrogen #PV5273) in FRET Buffer (20 mM HEPES, 10 mM MgC12, 0.015% Brij-35, 4mM DTT, 0.05 mg/mL BSA). Using Formulatrix Tempest, the detection antibody/enzyme/probe solution (2 mL) was dispensed into wells of a 1536 plate (Black Low Binding Polystyrene 1536 Plate (Corning, 3724)) containing 10 nL of compounds of interest at appropriate concentration in DMSO. The plate was incubated at rt for 1 h. FRET was measured using the EnVision plate reader (Excitation: 340 nM, Emission: 520 nM/495 nM). Total signal (0% inhibition) was calculated from wells containing 10 nL DMSO only. Blank signal (100% inhibition) calculated from wells containing 10 nL of 15 nM staurosporine and internal controls.Preferred compounds have low to no activity against PI3K, preferably compounds have PIK3 activity of 1 pM or greater.
- mPGES1 enzyme assay mPGES1 was diluted with assay buffer and added into plate at 49 μL/well supplemented with 1 μL of compound (the final concentration of each compound at seven gradient concentrations was 10000 nM, 1000 nM, 100 nM, 10 nM, 1 nM, 0.1 nM and 0.01 nM). Each well was added with 3.8 μL of 20 μg/mL PGH precooled on ice after shaking for 30 seconds and incubated for 7 minutes on ice. Then, each well was added with 53.8 μL of 6 mg/mL SnCl2 to quench the reaction. The sample was diluted with dilute buffer at 1:400. 10 μl of diluted sample, 5 μL PGE2-d2 and 5 μL anti-PGE2 Cryptate were added to a black 384-well plate, and incubated at 4° C. overnight. HTRF was determined using Flexstation, and the IC50 of the compounds was obtained by data processing software.
- TR-FRET Assay for ERBB Kinases All compounds and PROTACs were serially diluted in three-fold increments using 100% DMSO, followed by an intermediate 10-fold dilution using Buffer A (50 mM HEPES, pH 7.5, 50 mM NaCl, 10 mM MgCl2, 1 mM DTT, and 0.1% Pluronic F-68). Two microliters of serially diluted compound or PROTAC were then transferred to black 384-well Proxiplates (PerkinElmer, #6008260) using an Integra Viaflo96. Next, 10 uL of protein kinase in Buffer A was added to each well of the assay plate and pre-incubated with compound for 10 minutes. Kinase reactions were then initiated by addition of 5 uL substrate mix containing 3 mM ATP and 30 uM fluorescein-labeled Poly-GluTyr (Thermo Fisher, #PV3610) in Buffer A and allowed to proceed for 10 minutes at room temperature. Reactions were quenched by addition of a 5 uL mixture containing 5 nM LanthaScreen Tb-pY20 Antibody (Thermo Fisher, #PV3552) and 40 mM EDTA in Buffer A. Assay plates were then read using a Synergy2 (Biotek Instruments, Winooski, Vt.) via excitation thru a 340/20 nm bandpass filter and emission collected thru 490/10 nm (donor) and 520/25 nm (acceptor) bandpass filters. The final kinase concentrations used for each 15 uL reaction were as follows: 0.2 nM EGFR Exon20NPG (SignalChem, #E10-132GG), 0.1 nM wild type EGFR (BPS Bioscience, #40187), 0.3 nM EGFR L858R/T790M/C797S (BPS Bioscience, #40351), 0.1 nM EGFR L858R (BPS Bioscience, #40189), 0.4 nM L858R/T790M (BPS Bioscience, #40350), InM EGFR Dell9 (SignalChem, #E10-122JG), 10 nM EGFR Dell9 T790M (SignalChem, #E10-122KG), 0.3 nM Her2 (BPS Bioscience, #40230), 15 nM Her2 InsYVMA (SignalChem, #E27-13BG).
- Enzyme Assay JAK2 (Kd): JAK2 (Invitrogen, PV4210) is used at a final concentration of 5 nM. The binding experiment is performed in 50 mM Hepes pH 7.5, 0.01% Brij-35, 10 mM MgCl2, 1 mM EGTA using 25 nM kinase tracer 236 (Invitrogen, PV5592) and 2 nM Eu-anti-GST (Invitrogen, PV5594) with varying compound concentrations. Detection of tracer is performed according to the manufacturers procedure
- Fluorescence Binding Assays Varying amounts of enzyme were diluted in PBS supplemented with 0.5 g/L pluronic F127 and incubated with 500 nM ML349-FL for 30 minutes at room temperature. The fluorescent polarization from each well was measured using a Tecan F500 plate reader using 485/20 nm excitation and 535/25 nm emission polarization filters. Each concentration was measured in 2 replicates.
- Radioligand Binding Assay The radioligands used were 1 nM (2R,3R,4S,5R)-2-(2-chloro-6-cyclopentylamino-purin-9-yl)-5-hydroxymethyl-tetrahydro-3,4-diol ([3H]CCPA) for hA1, 10 nM (1-(6-amino-9H-purin-9-yl)-1-deoxy-N-ethyl-β-D-ribofuronamide) ([3H]NECA) for hA2a, and 1 nM 2-(1-hexynyl)-N6-methyladenosine [3H] ([3H]HEMADO) for hA3 receptors.
- Kinase Binding Assay Table 6: LanthaScreen Eu-anti-GST Antibody (Invitrogen, PV5594) and DYRK1A (Invitrogen, PV3785) were diluted to make the final concentrations of 6 nM and 15 nM respectively in 1× kinase buffer A, resulting in the preparation of antibody/kinase mixed solution. This antibody/kinase mixed solution was added to the assay plate where the diluted compound was loaded at the concentration of 5 μl/well. At this time, the final concentrations of the antibody and the DYRK1A were 2 nM and 5 nM respectively.Next, kinase tracer 236 solution (Invitrogen, PV5592) was diluted in 1× kinase buffer A to make the concentration of 45 nM. This diluted solution was added to the assay plate at the concentration of 5 μl/well. At this time, the final concentration of kinase tracer 236 was 15 nM and Kd value of Kinase tracer 236 was determined through tracer titration assay.Finally, after reacting at room temperature for 1 hour, fluorescence was measured (Excitation 340 nm, Kinase Tracer Emission 665 nm, LanthaScreen Eu-anti-GST Antibody Emission 620 nm) using Synergy neo (BioTek). Emission ratio (Kinase Tracer Emission Antibody Emission) was calculated based on the measured values, which was presented as a dose-response curve.
- TR-FRET Assay [MEK1 1mM ATP IC50 uM] A BRAF-MEK-ERK cascade assay is used to evaluate the effects of these compounds as inhibitors of the MAP kinase pathway. An enzymatic cascade assay is set up using recombinant human activated BRAF (V599E) kinase (Cat No. 14-557), human full length MEK1 kinase (Cat No. 14-706) and human full length active MAP Kinase 2/ERK2 (Cat No. 14-536) enzymes procured from Upstate. TR-FRET (Time resolved fluorescence resonance energy transfer) detection technology is used for the read out. The assay buffer solution contains 50 mM Tris pH 7.5, 10 mM MgCl2 , 1 mM DTT, 0.01% Tween 20, 0.1 nM activated BRAF, 2 nM inactive MEK1,10 nM inactive ERK2, 1 mM ATP and 500 nM long chain biotin-peptide substrate (LCB- FFKNIVTPRTPPP) in a 384 well format. The kinase reaction is stopped after 90 minutes with 10 mM EDTA and Lance detection mix (2 nM Eu-labeled phospho-serine/threonine antibody (Cat. No. AD0176-Perkin Elmer), 20 nM SA-APC (Cat No. CR130-100-Perkin Elmer) is added. The TR-FRET signal (Excitation at 340 nm, Emission at 615 nm and 665 nm) is read with 50 μs delay time on a Victor3 V fluorimeter.
- hEP2 cAMP Assay Stock solutions of test compounds are made at a concentration of 10 mM in DMSO, and serially diluted in DMSO to concentrations required for inhibition dose response curves (tested concentration range 30 μM-0.4 nM; 30 μM-0.015 nM or 1 μM-0.01 nM).PGE2 (Cayman 14010, stock solution: 75 μM in DMSO) is used as agonist at 75 nM final concentration, corresponding to EC80.Two point five microliters of diluted compounds are transferred into the assay plate. Plate is pre-incubated 45 minutes at room temperature. Subsequently, 2.5 microliters of PGE2 (final conc. 75 nM) are transferred into the assay plate. Plate is incubated 30 minutes at room temperature. Five μl of each donor (anti-cAMP cryptate) and acceptor (cAMP-d2) are added and the plate is incubated another hour at room temperature in the dark and then read using a BMG LABTECH PHERAstar reader (Excitation: 337 nm, Emission: 620 and 665 nm).The obtained Delta F (fluorescence) values (665 nm/620 nM) are converted into % cAMP values using the measurements of the cAMP calibrator provided in the kit. For each compound concentration the percentage of cAMP compared to DMSO control value as average±STDEV (each concentration is measured in duplicate) is calculated.
- hEP4 cAMP Assay Stock solutions of test compounds are made at a concentration of 10 mM in DMSO, and serially diluted in DMSO to concentrations required for inhibition dose response curves (tested concentration range 30 μM-0.4 nM; 30 μM-0.015 nM or 1 μM-0.01 nM).PGE2 (Cayman 14010, stock solution: 6 μM in DMSO) is used as agonist at 6 nM final concentration, corresponding to EC80.Two point five microliters of diluted compounds are transferred into the assay plate. Plate is pre-incubated 45 minutes at room temperature. Subsequently, 2.5 microliters of PGE2 (final conc. 6 nM) are transferred into the assay plate. Plate is incubated 30 minutes at room temperature. Five μl of each donor (anti-cAMP cryptate) and acceptor (cAMP-d2) are added and the plate is incubated another hour at room temperature in the dark and then read using a BMG LABTECH PHERAstar reader (Excitation: 337 nm, Emission: 620 and 665 nm).The obtained Delta F (fluorescence) values (665 nm/620 nM) are converted into % cAMP values using the measurements of the cAMP calibrator provided in the kit. For each compound concentration the percentage of cAMP compared to DMSO control value as average±STDEV (each concentration is measured in duplicate) is calculated.
- Affinity Assays Using Enzyme Activity Assay Buffer 1 (50 mM HEPES, 0.1 M NaCl, pH 7.5) was used to prepare a 0.4 μg/mL rhPSMA solution and a 40 μM solution of the substrate N-Acetyl-Asp-Glu. rhPSMA was mixed with the small molecules to be tested in a 96-well plate, with a constant rhPSMA content of 50 ng/well maintained. Meanwhile, the small molecules were step-wise diluted to final concentrations of 1 μM, 100 nM, 33.3 nM, 11.1 nM, 3.7 nM, 1.2 nM, 0.41 nM, 0.137 nM, 0.045 nM, and 0 nM. In addition, a positive control was set up using PSMA-617. The rhPSMA-small molecules were taken at 40 μL/well and well mixed with the 40 μM solution of the substrate N-Acetyl-Asp-Glu (40 μL/well). The mixtures were incubated at 37° C. in the dark for 1 h, were heated at 70° C. for 5 min to quench the reactions, and were cooled to room temperature. Buffer 2 (0.2 M NaOH, 0.1% beta-Mercaptoethanol) was used to prepare a 15 mM OPA solution. The OPA solution was added to the reaction systems at 80 μL/well and well mixed, and then the plate was incubated at room temperature for 10 min. The mixtures were taken at 100 μL/well and added to a 96-well Flat Black. With the excitation wavelength set to 330 nm and the emission wavelength to 465 nm, the intensity of signals was measured.
- Bim Binding Assays (Mcl-1 TR-FRET Assays) The inhibition of the Mcl-1/Bim interaction was measured using a time-resolved fluorescence resonance energy transfer (TR-FRET) assay. The recombinant human Mcl-1 (C-terminally 6×His tagged Mcl-1 containing residues 171-327) was generated at Amgen Inc (Thousand Oaks, Calif.). A biotinylated peptide derived from human Bim (residues 51-76) was purchased from CPC Scientific (San Jose, Calif.). The TR-FRET assay was conducted in a 384-well white OptiPlate (PerkinElmer, Waltham, Mass.) in a total volume of 40 μL. The reaction mixture contained 0.1 nM Mcl-1 (171-327), 0.05 nM biotin-Bim(51-76), 0.05 nM LANCE Eu-W1024 Anti-6×His (PerkinElmer), 0.072 nM Streptavidin-XLent (Cisbio, Bedford, Mass.), and serially diluted test compounds in the binding buffer of 20 mM HEPES, pH 7.5, 150 mM NaCl, 0.016 mM Brij 35, and 1 mM dithiothreitol. Test compounds were pre-incubated with Mcl-1 (171-327) and biotin-Bim (51-76) for 60 min before addition of the detection mixture (LANCE Eu-W1024 Anti-6×His and Streptavidin-XLent). The reaction plates were further incubated overnight and then were read on an Envision multimode reader (PerkinElmer). Fluorescence signals were measured at 620 nm (40-nm bandwidth) and 665 nm (7.5-nm bandwidth) with a 300 μs delay after excitation at 320 nm (75-nm bandwidth). The signal ratio at 665/620 nm corresponded to the Mcl-1/Bim interaction and was used in all data analyses
- Enzymatic Assay The ability of test compounds to inhibit the activity of JMJD2A was determined in 384-well plate format under the following reaction conditions: 2 nM JMJD2A, 300 nM H3K9me3-biotin labeled peptide (Anaspec cat #64360), 2 μM alpha-ketoglutaric acid in assay buffer of 50 mM HEPES, pH7.3, 0.005% Brij35, 0.5 mM TCEP, 0.2 mg/ml BSA, 50 μM sodium L-ascorbate, and 2 μM ammonium iron(II) sulfate. Reaction product was determined quantitatively by TR-FRET after the addition of detection reagent Phycolink Streptavidin-allophycocyanin (Prozyme) and Europium-anti-di-methylated histone H3 lysine 9 (H3K9me2) antibody (PerkinElmer) in the presence of 5 mM EDTA in LANCE detection buffer (PerkinElmer) at a final concentration of 50 nM and 1 nM, respectively.The assay reaction was initiated by the following: 2 μl of the mixture of 900 nM H3K9me3-biotin labeled peptide and 6 μM alpha-ketoglutaric acid with 2 μl of 11-point serial diluted inhibitor in 3% DMSO were added to each well of plate, followed by the addition of 2 μl of 6 nM JMJD2A to initiate the reaction. The reaction mixture was incubated at room temperature for 30 minutes, and terminated by the addition of 6 μl of 5 mM EDTA in LANCE detection buffer containing 100 nM Phycolink Streptavidin-allophycocyanin and 2 nM Europium-anti-H3K9me2 antibody. Plates were read by EnVisionMultilabel Reader in TR-FRET mode (excitation at 320 nm, emission at 615 nm and 665 nm) after 1 hour incubation at room temperature.
- Enzymatic Assay The ability of test compounds to inhibit the activity of JMJD2C was determined in 384-well plate format under the following reaction conditions: 0.3 nM JMJD2C, 300 nM H3K9me3-biotin labeled peptide (Anaspec cat #64360), 2 μM alpha-ketoglutaric acid in assay buffer of 50 mM HEPES, pH7.3, 0.005% Brij35, 0.5 mM TCEP, 0.2 mg/ml BSA, 50 μM sodium L-ascorbate, and 2 μM ammonium iron(II) sulfate. Reaction product was determined quantitatively by TR-FRET after the addition of detection reagent Phycolink Streptavidin-allophycocyanin (Prozyme) and Europium-anti-di-methylated histone H3 lysine 9 (H3K9me2) antibody (PerkinElmer) in the presence of 5 mM EDTA in LANCE detection buffer (PerkinElmer) at a final concentration of 50 nM and 1 nM, respectively.The assay reaction was initiated by the following: 2 μl of the mixture of 900 nM H3K9me3-biotin labeled peptide and 6 μM alpha-ketoglutaric acid with 2 μl of 11-point serial diluted inhibitor in 3% DMSO were added to each well of the plate, followed by the addition of 2 μl of 0.9 nM JMJD2C to initiate the reaction. The reaction mixture was incubated at room temperature for 30 minutes, and terminated by the addition of 6 μl of 5 mM EDTA in LANCE detection buffer containing 100 nM Phycolink Streptavidin-allophycocyanin and 2 nM Europium-anti-H3K9me2 antibody. Plates were read by EnVisionMultilabel Reader in TR-FRET mode (excitation at 320 nm, emission at 615 nm and 665 nm) after 1 hour incubation at room temperature.
- In Vitro Enzyme Inhibition Assay JMJD2C: The ability of test compounds to inhibit the activity of JMJD2C was determined in 384-well plate format under the following reaction conditions: 0.3 nM JMJD2C, 300 nM H3K9me3-biotin labeled peptide (Anaspec cat #64360), 2 μM alpha-ketoglutaric acid in assay buffer of 50 mM HEPES, pH7.3, 0.005% Brij35, 0.5 mM TCEP, 0.2 mg/ml BSA, 50 μM sodium L-ascorbate, and 2 μM ammonium iron(II) sulfate. Reaction product was determined quantitatively by TR-FRET after the addition of detection reagent Phycolink Streptavidin-allophycocyanin (Prozyme) and Europium-anti-di-methylated histone H3 lysine 9 (H3K9me2) antibody (PerkinElmer) in the presence of 5 mM EDTA in LANCE detection buffer (PerkinElmer) at a final concentration of 50 nM and 1 nM, respectively.The assay reaction was initiated by the following: 2 μl of the mixture of 900 nM H3K9me3-biotin labeled peptide and 6 μM alpha-ketoglutaric acid with 2 μl of 11-point serial diluted inhibitor in 3% DMSO were added to each well of the plate, followed by the addition of 2 μl of 0.9 nM JMJD2C to initiate the reaction. The reaction mixture was incubated at RT for 30 min, and terminated by the addition of 6 μl of 5 mM EDTA in LANCE detection buffer containing 100 nM Phycolink Streptavidin-allophycocyanin and 2 nM Europium-anti-H3K9me2 antibody. Plates were read by EnVisionMultilabel Reader in TR-FRET mode (excitation at 320 nm, emission at 615 nm and 665 nm) after 1 hr incubation at RT.
- In Vitro Enzyme Inhibition Assay Jarid1b: The ability of test compounds to inhibit the activity of Jarid1B was determined in 384-well plate format under the following reaction conditions: 0.8 nM Jarid1B, 300 nM H3K4me3-biotin labeled peptide (Anaspec cat #64357), 2 μM alpha-ketoglutaric acid in assay buffer of 50 mM HEPES, pH7.3, 0.005% Brij35, 0.5 mM TCEP, 0.2 mg/ml BSA, 50 μM sodium L-ascorbate, and 2 μM ammonium iron(II) sulfate. Reaction product was determined quantitatively by TR-FRET after the addition of detection reagent Phycolink Streptavidin-allophycocyanin (Prozyme) and Europium-anti-H3K4me or -H3K4me2 antibody (PerkinElmer) in the presence of 5 mM EDTA in LANCE detection buffer (PerkinElmer) at a final concentration of 25 nM and 1 nM, respectively.The assay reaction was initiated by the following: 2 μl of the mixture of 900 nM H3K4me3-biotin labeled peptide and 6 μM alpha-ketoglutaric acid with 2 μl of 11-point serial diluted inhibitor in 3% DMSO was added to each well of the plate, followed by the addition of 2 μl of 2.4 nM Jarid1B to initiate the reaction. The reaction mixture was incubated at RT for 30 min, and terminated by the addition of 6 μl of 5 mM EDTA in LANCE detection buffer containing 50 nM Phycolink Streptavidin-allophycocyanin and 2 nM Europium-anti-H3K4me/H3K4me2 antibody. Plates were read by EnVisionMultilabel Reader in TR-FRET mode (excitation at 320 nm, emission at 615 nm and 665 nm) after 1 hr incubation at RT.
- Binding Assay ADRB2 IC50 from binding assay(nM), Determined using the Tag-lite Adrenoceptor Beta2 receptor ligand binding assay.
- ChEBML_136063 Compound was tested for Inhibition of [3H]DAMGO (0.25 nM) binding to mu receptor from bovine striatal membranes
- ChEBML_140037 Binding affinity to the rat cardiac muscarinic acetylcholine receptor M2 using 0.3 nM [3H]N-methylscopolamine as radioligand
- ChEBML_144288 Binding ability to compete with [125I]Tyr3-NT (0.15 nM) for human NT receptors cloned in CHO cells.
- ChEBML_144458 Inhibition of neutral endopeptidase activity in rabbit kidney with 20 nM [3H]D-Ala2-Leu-enkephalin as substrate
- ChEBML_145276 Binding affinity towards Opioid receptor mu 1 of guinea pig brain membranes using radioligand 0.25 nM [3H]DAMGO
- ChEBML_195315 Antagonistic activity was evaluated in terms of inhibition of Retinoic acid receptor alpha transactivation by ATRA (50 nM)
- ChEBML_201578 Binding affinity for Sigma opioid receptor type 2 in guinea pig brain homogenate with 4 nM [3H]-(+)-DTG
- ChEBML_204888 Inhibitory activity against rat Steroid 5-alpha-reductase type I using 18213 3H testosterone 210 nM as substrate
- ChEBML_222070 Binding affinity towards mu-opioid receptor by displacement of [3H]NAL at 10 nM from rat brain homogenates
- ChEBML_28843 Ability to inhibit binding of 1 nM [3H]cyclohexyladenosine to adenosine A1 receptor in rat cerebral cortical membranes
- ChEBML_29612 In vitro binding affinity to Adenosine A1 receptor of rat cerebral cortical membranes using 1 nM [3H]PIA
- ChEBML_29616 Inhibition of 1 nM [3H]- N6 -(phenylisopropyl) adenosine binding to Adenosine A1 receptor in rat fat cell membrane
- ChEBML_29631 Inhibition of binding of 1 nM N6-[3H]cyclohexyladenosine to adenosine A1 receptor on rat cerebral cortical membranes.
- ChEBML_49723 Ability to displace 1 nM [3H]pCCK-8 from Cholecystokinin type A receptor in guinea pig pancreatic membranes
- ChEBML_52212 Binding affinity against Cysteinyl leukotriene D4 receptor from guinea pig lung was determined using [3H]LTD4 (0.2 nM)
- ChEBML_66423 Compound was tested for inhibitory activity against FK506 binding protein 12 (FKBP12); Value ranges from 190-900 nM
- ChEBML_75663 Ability to displace specific binding of 0.2 nM [3H]Boc[Nle28,31]-CCK27-33 from guinea pig brain membranes
- ChEMBL_103299 (CHEMBL712253) Anti-estrogenicity for 50% inhibition of the MVLN cell luciferase activity due to 0.1 nM E2 (estradiol)
- ChEMBL_104760 (CHEMBL710992) In vitro binding affinity against melatonin receptor using 2-[125I]iodomelatonin (0.05 nM) and chicken brain membranes
- ChEMBL_1336 (CHEMBL616962) Ability to displace [3H]5-CT (1.5 nM) from HeLa cells of human 5-hydroxytryptamine 1B receptor
- ChEMBL_136061 (CHEMBL746681) Inhibition against mu receptor from displacement studies using 0.5 nM [3H]-DAMGO in rhesus monkey cortex membrane
- ChEMBL_145334 (CHEMBL750422) Binding inhibition of [3H]- -naltrindole (0.15 nM) to membranes in CHO cells expressing Opioid receptor delta 1
- ChEMBL_146344 (CHEMBL753793) Ki value determined against Opioid receptor delta 1 using [3H]DPDPE at the Kd concentration 2.1 nM
- ChEMBL_146612 (CHEMBL750113) Inhibition against delta receptor from displacement studies using 1.5 nM [3H]DPDPE in rhesus monkey cortex membrane
- ChEMBL_1473182 (CHEMBL3418234) Displacement of 0.1 nM [125I]CCL2 from human CCR2 expressing human U2OS cell membrane by scintillation spectrometry
- ChEMBL_147891 (CHEMBL884926) Inhibition of 10 nM 2-MeS-ADP-stimulated phospholipase C in turkey erythrocyte membranes using [3H]inositol
- ChEMBL_148518 (CHEMBL758128) Ki value determined against Opioid receptor mu 1 using [3H]DAMGO at the Kd concentration 0.57 nM
- ChEMBL_148690 (CHEMBL857686) Inhibition of radioligand [3H]DAMGO binding to rat brain Opioid receptor mu 1 using 100 nM DAMGO
- ChEMBL_1509401 (CHEMBL3603344) Binding affinity to human serum albumin with excitation at 280 nm after 2 hrs by spectrofluorimetric analysis
- ChEMBL_1573399 (CHEMBL3804561) Positive allosteric modulation of human mGlu1 receptor assessed as glutamate EC50 at 10 uM (Rvb = 983.4 nM)
- ChEMBL_164467 (CHEMBL770733) Inhibition of specific [3H]-prazosin binding (0.2 nM) to rat brain membranes alpha1 adrenoceptor; 2.1*10e-6
- ChEMBL_1804555 (CHEMBL4303779) hERG binding assays: Displacement of [3H]-Dofetilide (5 nM final) from hERG membranes obtained from HEK293 cells
- ChEMBL_197244 (CHEMBL803366) Binding affinity towards Retinoic acid receptor RXR-gamma by displacing 3.4 nM 3[H]-9-cis-RA
- ChEMBL_201510 (CHEMBL805637) Binding affinity at the Serotonin transporter in rat midbrain by inhibition of 0.5 nM [3H]-paroxetine binding
- ChEMBL_2130177 (CHEMBL4839606) Inhibition of human recombinant CSF1R using ATP/Ulight-TK peptide at 50 nM incubated for 15 mins
- ChEMBL_221915 (CHEMBL842480) Binding affinity towards mu opioid receptor using [3H]- -DAMGO (1.5 nM) as radioligand in rat brain membranes
- ChEMBL_221937 (CHEMBL842476) Binding affinity towards mu opioid receptor using [3H]- -DAMGO (1.5 nM) as radioligand in rat brain membranes
- ChEMBL_223732 (CHEMBL844474) Inhibition of 1 nM [3H]N-6-(phenylisopropyl)adenosine binding to A1 receptors in rat cortical membranes
- ChEMBL_2318 (CHEMBL617525) Ability to displace 0.75 nM [3H]ketanserin in 5-hydroxytryptamine 2 receptor from rat frontal cortex homogenate
- ChEMBL_27613 (CHEMBL643986) Binding affinity against A2 adenosine receptors from rat striatal membranes with 50 nM CPA using [3H]NECA
- ChEMBL_27614 (CHEMBL643987) Binding affinity against Adenosine A2 receptor from rat striatal membrane with 50 nM CPA using [3H]NECA
- ChEMBL_27759 (CHEMBL643364) Binding affinity against Adenosine A2 receptor from rat striatal membranes with 50 nM CPA using [3H]-NECA
- ChEMBL_27905 (CHEMBL642153) Inhibition of [3H]DPCPX binding to human Adenosine A1 receptor expressed in CHO cells at 10000 nM
- ChEMBL_27906 (CHEMBL642154) Inhibition of [3H]DPCPX binding to human Adenosine A1 receptor expressed in CHO cells at 10000 nM
- ChEMBL_2813 (CHEMBL617842) Displacement of [3H]mesulergine (0.5 nM) from rat 5-hydroxytryptamine 2C receptor expressed in SR-3T3 cells
- ChEMBL_288 (CHEMBL615726) In vitro inhibition of [3H]DPAT (1 nM) binding to 5-hydroxytryptamine 1A receptor from bovine hippocampus
- ChEMBL_29281 (CHEMBL640342) Inhibition of adenosine induced negative inotropic activity against guinea-pig atria (Adenosine A1 receptor) at 10 nM
- ChEMBL_29628 (CHEMBL636646) Inhibition of 1 nM [3H]N-6-(phenylisopropyl)adenosine binding to A1 receptors in rat cortical membranes
- ChEMBL_30333 (CHEMBL637928) Inhibition of [125I]AB-MECA (0.15 nM) binding to Adenosine A3 receptor of RBL-2H3 cell membranes
- ChEMBL_30462 (CHEMBL643123) Affinity to Adenosine A3 receptor of rat testis membrane using [3H](R)-PIA with 150 nM DPCPX
- ChEMBL_30605 (CHEMBL642012) Inhibition of [3H]DPCPX binding to human Adenosine A2B receptor expressed in HEK cells at 10000 nM
- ChEMBL_306686 (CHEMBL830020) Inhibition of 100 nM-NECA stimulation of cAMP levels in human adenosine A2b receptor expressing CHO cells
- ChEMBL_30702 (CHEMBL644777) Binding affinity against adenosine A2 receptor from rat striatal membranes with 50 nM CPA using [3H]-NECA
- ChEMBL_30799 (CHEMBL645075) Compound was tested for the inhibition of adenosine deaminase from calf intestine; Range of 0.01-0.001 nM
- ChEMBL_31414 (CHEMBL883238) Displacement of [3H]CGS-21680 from human Adenosine A3 receptor expressed in HEK293 cells at 10000 nM
- ChEMBL_320903 (CHEMBL872402) Binding affinity (low) towards bovine dopamine receptor 1 by using [3H]-SCH- 23390 (0.3 nM) as radioligand
- ChEMBL_320905 (CHEMBL871723) Binding affinity (low) towards porcine dopamine receptor 1 by using [3H]-SCH- 23390 (0.3 nM) as radioligand
- ChEMBL_320990 (CHEMBL872137) Binding affinity towards human Adenosine A3 receptor expressed in HEK293 cells using 0.1 nM [3H]AB-MECA
- ChEMBL_321018 (CHEMBL883651) Binding affinity towards human Adenosine A2a receptor expressed in HEK293 cells using 6 nM [3H]CGS-21680
- ChEMBL_321555 (CHEMBL881969) Inhibitory concentration against human Adenosine A3 receptor expressed in HEK293 cells using 0.1 nM [3H]AB-MECA
- ChEMBL_32528 (CHEMBL641409) Binding affinity at human Alpha-2B adrenergic receptor in CHO cells by [3H]rauwolscine (1 nM) displacement.
- ChEMBL_34197 (CHEMBL649214) Displacement of [3H]prazosin (0.5 nM) from hamster Alpha-1B adrenergic receptor expressed in rat-1 cells
- ChEMBL_34492 (CHEMBL647204) Inhibition of 1 nM [3H]RX-821002 binding to rat whole brain membrane Alpha-2 adrenergic receptors
- ChEMBL_40291 (CHEMBL653423) Inhibition of [3H]BK (1.0 nM) binding to the human bradykinin receptor B2, expressed in CHO cells
- ChEMBL_4173 (CHEMBL619240) Tested for inhibition of 5-HPETE production by rat 5-LO; value ranges from 16-36 nM
- ChEMBL_46620 (CHEMBL657118) Binding affinity against brain Cannabinoid receptor 1 using [3H]CP-55940 as radioligand at 1.1 nM concentration
- ChEMBL_46621 (CHEMBL657119) Binding affinity against brain cannabinoid receptor 1 using [3H]CP-55940 as radioligand at 0.25 nM concentration
- ChEMBL_564 (CHEMBL615584) In vitro inhibition of [3H]5-HT (2 nM) binding to 5-HT1A receptor from bovine hippocampus
- ChEMBL_566 (CHEMBL615585) In vitro inhibition of azido-[125I]-IPAPP (0.25 nM) binding to 5-HT1A receptor from bovine hippocampus
- ChEMBL_568 (CHEMBL884524) In vitro inhibition of [125I]IPAPP (2.5 nM) binding to 5-hydroxytryptamine 1A receptor from bovine hippocampus
- ChEMBL_583 (CHEMBL615453) In vitro inhibition of [3H]5-HT (2 nM) binding to 5-HT1A receptor from bovine hippocampus
- ChEMBL_586 (CHEMBL615456) In vitro inhibition of [3H]DPAT (1 nM) binding to 5-hydroxytryptamine 1A receptor from bovine hippocampus
- ChEMBL_619400 (CHEMBL1104158) Inhibition of GSK3-beta assessed as decrease in NADH absorbance at 340 nm in the presence of
- ChEMBL_63101 (CHEMBL674494) Displacement of [3H]-YM-09151-2 (0.06 nM) from human Dopamine receptor D4 expressed in CHO cells
- ChEMBL_68357 (CHEMBL679499) Inhibition of Flt3/ITD mutant expressed in hematopoietic cells or AML cell lines; range = 30-100 nM
- ChEMBL_70426 (CHEMBL681283) Inhibition of FPP incorporation into biotinylated K-Ras-derived peptide by human farnesyl transferase 5 nM ATP
- ChEMBL_880071 (CHEMBL2213420) Binding affinity to heparin induced human Tau 441 at 50 pM to 500 nM after 30 mins
- ChEMBL_894 (CHEMBL615805) Ability to displace [3H]5-CT (2.0 nM) from HeLa cells of human 5-hydroxytryptamine 1A receptor
- ChEMBL_90174 (CHEMBL697152) In vitro inhibition of human platelet aggregation induced by alpha-thrombin (at a concentration of 0.15 nM)
- Bace-1 Enzymatic Assay These experiments have been preformed using BACE-1 FRET assay kit (PanVera Corporation, Madison, WI), using a multiwell spectrofluorometer instrument capable of 530-545 nm excitation and 570-590 nm emission wavelengths (Wallac Victor2 1420, Perkin-Elmer, Turku, Finland).
- ChEMBL_1752854 (CHEMBL4187614) Inhibition of human PAI-1 assessed as remaining enzyme activity by measuring p-nitroaniline release pre-incubated for 15 mins before 9.8 nM human tPA substrate addition for 15 mins using 67 nM enzyme and chromogenic substrate S-2288 by spectrophotometry
- ChEMBL_1752858 (CHEMBL4187618) Inhibition of human PAI-1 assessed as remaining enzyme activity by measuring p-nitroaniline release pre-incubated for 15 mins before 2 nM Spectrozyme tPA substrate addition for 15 mins using 5 nM enzyme and chromogenic substrate S-2288 by spectrophotometry
- ChEMBL_1752859 (CHEMBL4187619) Inhibition of human PAI-1 assessed as remaining enzyme activity by measuring p-nitroaniline release pre-incubated for 15 mins before 9.8 nM human tPA substrate addition for 15 mins using 24.5 nM enzyme and chromogenic substrate S-2288 by spectrophotometry
- DPP Inhibition Assay The DPP activity resulted in the formation of the fluorescent product amidomethylcoumarin (AMC), which was monitored by excitation at 355 nm and measurement of fluorescent emission at 460 nm. IC50 values reported are means of at least two separate experiments.
- SERT Binding Affinity Assay Conditions for SERT binding assay,Receptor Source Human recombinant (CHO cells)Vehicle 1.0% DMSOIncubation Time 1 hIncubation Temperature 25° C.Incubation Buffer 5 mM Tris-HCl, pH 7.4Ligand 2.0 nM [3H]imipramineNon-Specific Ligand 10.0 μM imipramineKd 1.7 nM
- mu-Calpain Inhibition Assay Assays were initiated by addition of CaCl2, and the increase in fluorescence (ex @370 nm, em @440 nm) was monitored. MDL28170 and buffer with 2% DMSO were used as controls. Ki values were determined according to Dixon methods.
- Biological Activity Assay The KDM1A demethylase enzyme activity can obtained from mammalian cells or tissues expressing KDM1A from an endogenous or recombinant gene and purified or assayed from a whole cell extract. These methods can be used to determine the concentration of the disclosed compounds can inhibit fifty percent of the enzyme activity (IC50). In one aspect, the disclosed compounds exhibit inhibition fifty percent of the KDM1A enzyme activity at a concentration of less than 500 nM, less than 100 nM, less than 50 nM or less than 10 nM.
- Fluorescence Polarization Assay (FPA) Briefly, reaction mixes (100 µL) containing 20 mM HEPES (pH 7.3), 50 mM KCl, 2 mM DTT, 5 mM MgCl2, 20 mM Na2MoO4, 0.01% Triton X-100 with 0.1 mg/mL BSA, 50 nM of recombinant Hsp90α protein, 5 nM of GA-FITC, and varying concentrations of tested compounds or GA were added in a low-binding black 96-well plates. After incubation for 5 h, plates were read on Omega POLARstar Multi-Mode Microplate Reader with excitation at 485 nm and emission at 520 nm.
- In Vitro Assay of Compounds of the Present Invention for Inhibiting Src Kinase Activity Assay After the compound serially diluted with DMSO and a Src recombinant protein were incubated at room temperature for 10 min, the reaction was initiated by the addition of a biotin-labeled TK substrate and ATP. After the mixture was left to stand at room temperature for 40 min Sa-XL 665 and a Cryptate-labeled antibody were added. The fluorescence values at 615 nm and 665 nm were measured, and the ratio of 665 nm to 615 nm was calculated.
- KRASG12D Nucleotide Exchange Assay Recombinant GDP-loaded KRAS G12D (20 nM) was treated with compound at room temperature for 20 minutes in assay buffer (10 mM Hepes pH 7.4, 150 mM NaCl, 5 mM MgCl2, 0.0025% Igepal-CA630, 0.05% BSA, 1 mM DTT, 0.5 nM SA-Tb). BIODIPY-labeled GDP (400 nM) and recombinant SOS (10 nM) were added, and the reaction was incubated for 30 minutes. HTRF signal was measured (PerkinElmer Envision), the signal ratio (λem 520/λem 495) was calculated, and IC50 values were calculated from the dose-response curve.
- Sialidase Assay Briefly, the reaction mixtures containing the substrates and up to 100 nM sialidase were incubated at 37°C and stopped by the addition of 0.5 M Na2CO3 pH 9.0 for the colorimetric assay or 0.25 M glycine pH 10.0 for the fluorimetric assay. Released p-NP was detected spectrophometrically at 405 nm. The fluorescenceassociated with the release of 4-MU was measured at the excitation wavelength of 365 nm and an emission wavelength of 445 nm. These measurements were performed using an EnVision microplate reader (Perkin Elmer, Waltham, Mass.).
- Competition Binding Assays Briefly, 5 mg of a protein mixture of the four cell lines or a single cell line were incubated with compound dilution series in DMSO (3 nM, 10 nM, 30 nM, 100 nM, 300 nM, 1 μM, 3 μM, 30 μM, and DMSO) or single compound dose (5 μM) for 45 min at 4 °C. This preincubation step was followed by incubation with kinobeads (35 μL settled beads). Bound proteins were eluted with LDS sample buffer (NuPAGE,Invitrogen) containing 50 mM DTT. For the calculation of a correction factor, the flowthrough of the DMSO control was incubated with fresh beads for a second time (pulldown of pulldown).
- In Vitro Assay Inhibition of LIMK1 and LIMK2. Inhibition of LIMK1 and LIMK2 was measured with Lanthascreen Eu binding assay (Life Technologies). This TR-FRET assay is based on the competition between a fluorescent tracer and the inhibitor to be tested, allowing the determination of an inhibition constant, Ki. 133 compounds have been tested. A dozen of these compounds have their Ki below 10 nM, and about fifty below 50 nM. These results are very promising, as Ki values of the reference compounds LIMKi3 and LX7101 are 8 and 3.9 nM, respectively, and the Pyr1 compound tested on breast cancer tumors has an IC50 of 50 nM on LIMK1 and 75 nM on LIMK2.
- R132H IDH1 Enzymatic Assay Each test compound (10 mM stock in DMSO) is diluted in DMSO to make a 10-point, 3-fold dilution series. 125 nL of each dilution or DMSO alone is dispensed to a 384-well Greiner Lumitrac 200 assay plate using an Echo Liquid Handler. To each well of the plate is added 20 uL of enzyme in assay buffer or assay buffer alone. Assay buffer consists of 50 mM sodium phosphate, pH 7.0, 50 mM magnesium chloride, 50 mM sodium chloride, and 0.01% (w/v) bovine serum albumin. When present, the R132H mutant IDH1 enzyme is at a working concentration of 1.875 nM (final concentration in assay of 1.5 nM). The assay plate is allowed to incubate for 30 minutes at room temperature and 5 uL of 5× substrate mixture (2.5 uM nicotinamide adenine dinucleotide phosphate, 100 uM adenosine diphosphate, 7.5 mM glyceraldehyde-3-phosphate, 7.5 ug/mL of spinach glyceraldehyde-3-phosphate dehydrogenase, 25 nM phosphoglycerate kinase, and 5 mM alpha-ketoglutarate in assay buffer) is added to all wells. The reaction plate is incubated for 60 minutes followed by addition of 25 uL of Promega Kinase-GLO reagent to all wells and 10-minute incubation.Luminescence is measured using a PerkinElmer Envision plate reader. The percent activity of each dilution is determined as the ratio of background corrected signal to the background corrected signal of wells receiving only DMSO. IC50 values are determined by fitting percent activity data to a four-parameter logistic dose response equation. The IC50 values of the exemplified compounds are included in the tables above in Examples section.Using the above biological assay, all compounds in the examples have IC50 of about 1 nM to about 40,000 nM, or more specifically, about 1 nM to about 20,000 nM, or even more specifically, about 5 nM to about 15,000 nM, or even more specifically, about 5 nM to about 10,000 nM, or even more specifically, about 5 nM to about 5,000 nM, or still more specifically, about 5 nM to about 1,000 nM. Such a result is indicative of the intrinsic activity of the compounds in use as an inhibitor of a mutant IDH1 enzyme.
- Binding Assay ERRα/ERRβ/ERα : In an ERR beta binding assay, GST-bound ERR alpha LBD was used so that a final concentration was 10 nM and a fluorescein-conjugated coactivator PGC1a was 250 nM, and all experiments other than that was the same as the ERR gamma binding assay.In an ER alpha binding assay, a GST-bound ER alpha ligand-binding domain (LBD) was added to a 384 well plate to which the arylethene derivative of the present invention was added to a final concentration of 7.3 nM. Then, a fluorescein-conjugated coactivator PGC1a and a Tb-a-GST antibody were added to 250 nM and 5 nM, respectively, and beta-estradiol as an agonist was added to a final concentration of 4 nM. All subsequent experiments was the same as the ERR gamma binding assay.
- HiRange Homogenous Time-Resolved Fluorescence (HTRF) Assay Briefly, suspensions of cells expressing either the mu, kappa or delta opioid receptors were prepared in buffer containing 0.5 mM isobutyl-methyl xanthine (IBMX). Cells were incubated with varying concentrations of PEG-opioid conjugates and 3 uM forskolin for 30 minutes at room temperature. cAMP was detected following a two-step assay protocol per the manufacturer's instructions and time resolved fluorescence was measured with the following settings: 330 nm excitation; 620 nm and 665 nm emission; 380 nm dichroic mirror. The 665 nm/620 nm ratio is expressed as Delta F % and test compound-related data is expressed as a percentage of average maximum response in wells without forskolin. EC50 values were calculated for each compound from a sigmoidal dose-response plot of concentrations versus maximum response.
- Kinase Binding Assay TrkA binding activity was determined in a TrkA LanthaScreen Eu Kinase Binding Assay. 5 nM His-tagged recombinant human TrkA (6HIS tagged cytoplasmic domain from Invitrogen, Catalog No. PV3144) was incubated with 4 nM Alexa-Fluor Tracer 236 (Invitrogen Cat. No. PV5592), 2 nM biotinylated anti-His (Invitrogen Cat. No. PV6090), and 2 nM europium-labeled Streptavidin (Invitrogen Cat. No. PV5899), in buffer (25 mM MOPS, pH 7.5, 5 mM MgCl2, 0.005% Triton X-100). Three fold serial dilutions of compounds of the invention in DMSO were added to a final percentage of 2% DMSO. After 60-minute incubation at 22° C., the reaction was measured using the EnVision mutlimode plate reader (PerkinElmer) via TR-FRET dual wavelength detection at 615 nM and 665 nM. The percent of control was calculated using a ratiometric emission factor.
- Kinase Binding Assay TrkA binding activity was determined in a TrkA LanthaScreen Eu Kinase Binding Assay. 5 nM His-tagged recombinant human TrkA (6HIS tagged cytoplasmic domain from Invitrogen, Catalog No. PV3144) was incubated with 4 nM Alexa-Fluor Tracer 236 (Invitrogen Cat. No. PV5592), 2 nM biotinylated anti-His (Invitrogen Cat. No. PV6090), and 2 nM europium-labeled Streptavidin (Invitrogen Cat. No. PV5899), in buffer (25 mM MOPS, pH 7.5, 5 mM MgCl2, 0.005% Triton X-100). Three fold serial dilutions of compounds of the invention in DMSO were added to a final percentage of 2% DMSO. After 60-minute incubation at 22° C., the reaction was measured using the EnVision mutlimode plate reader (PerkinElmer) via TR-FRET dual wavelength detection at 615 nM and 665 nM. The percent of control was calculated using a ratiometric emission factor.
- DLK Biochemical Assay Assay Protocol for 1536 MTP format: test compounds were predispensed (60 nl) in test plates; +3 μl DLK solution (10 nM DLK, catalytic domain, Carna Biosiences, 0.6 mM DTT in assay buffer); +2 μl substrate solution (15 nM MKK7, LDB; 60 μM ATP; 0.4 mM DTT in assay buffer); 90 min incubation at RT; +2 μl detection mix (5 nM XL665-labeled anti-His antibody, Cisbio; 0.1 nM Eu-labeled anti-phospho-MKK7 (T275/S277), Cisbio/Millipore in detection buffer); 16 h incubation at 37° C.; endpoint measurement BMG (HTRF). Assay Protocol for 384 low volume MTP format: 1 μl of test compounds were pipetted into test plate; +5 μl DLK solution (10 nM DLK, catalytic domain, Carna Biosiences in assay buffer); +5 μl substrate solution (15 nM MKK7, LDB; 60 μM ATP, in assay buffer); 90 min incubation at RT; +9 μl detection mix (20 nM XL665-labeled anti-His antibody, Cisbio; 0.4 nM Eu-labeled anti-phospho-MKK7 (T275/S277), Cisbio/Millipore in detection buffer); 16 h incubation at 4° C.; endpoint measurement BMG (HTRF).
- Inhibition of Pro Part of ProNGF to hSortilin The pro Sortilin assay was performed in total volume of 20 μl in 50 mM HEPES pH 7.4 assay buffer containing 100 mM NaCl, 2.0 mM CaCl2, 0.1% BSA and 0.1% Tween-20. Varying concentration of compounds where incubated for 15 min at RT with 50 nM of 6his-Sortilin and 6 nM of pro-GST. 4 nM of anti-GST-Eu and 25 nM of anti-His-XL665 were added together with KF (final concentration 200 mM) and, following 150 min incubation at room temperature in the dark. the fluorescence was measured with excitation of 320 nm and dual emission of 665 and 615 nm on the an envision reader (Perkin Elmer). Signal was expressed in terms of HTRF ratio (fluorescence intensity at 665 nm/fluorescence intensity at 615 nm×10,000). Inhibition of binding of proGST to Sortilin was expressed as a percent inhibition of the control response to 20 μM of Neurotensin (100% inhibition) relative to a buffer basal control (0% inhibition). IC50 values were calculated by nonlinear regression using the sigmoid concentration-response (variable slope) using Xlfit 4 (IDBS, UK).
- Inhibition of hProgranulin to hSortilin The progranulin Sortilin assay was performed in total volume of 20 μl in 50 mM Phosphate buffer pH 7.0 assay buffer containing 0.1% BSA. Varying concentration of compounds where incubated for 15 min at RT with 50 nM of 6his-Sortilin and 4 nM of Progranulin. 0.7 nM of anti-progranulin-Eu and 7 nM of anti-HisXL665 were added together with KF (final concentration 200 mM) and, following 120 min incubation at room temperature in the dark, the plate was kept at 4° C. overnight and next day the fluorescence was measured with excitation of 320 nm and dual emission of 665 and 615 nm on the an envision reader (Perkin Elmer). Signal was expressed in terms of HTRF ratio (fluorescence intensity at 665 nm/fluorescence intensity at 615 nm×10,000). Inhibition of binding of progranulin to Sortilin was expressed as a percent inhibition of the control response to 20 μM of Neurotensin (100% inhibition) relative to a buffer basal control (0% inhibition). IC50 values were calculated by nonlinear regression using the sigmoid concentration-response (variable slope) using Xlfit 4 (IDBS, UK).
- KRAS Biochemical Assay KRAS Biochemical Assay—BODIPY-GDP Exchange TR-FRET. Biochemical compound potencies were assessed by evaluating inhibition of SOS1-mediated nucleotide exchange in KRAS G12D. The SOS1-promoted exchange of fluorescently-labeled GDP (BOPIDY-GDP) was monitored by time-resolved fluorescence resonance energy transfer (TR-FRET). Compounds solubilized in DMSO were dispensed as concentration series into 384-well white assay plates. A preformed complex of biotin-tagged recombinant human KRAS (1.5 nM mutant G12D or wild type) and 0.15 nM terbium-labeled streptavidin (CisBIO) prepared in 10 μL/well assay buffer (20 mM HEPES, pH 7.5, 50 mM NaCl, 10 mM MgCl2, 0.01% Tween-20 and 1 mM dithiothreitol) was added and allowed to incubate for 10-minutes. The reaction was initiated with the addition of 5 μL of 3 nM recombinant human SOS1 and 300 nM BODIPY-GDP in assay buffer. After a 60-minute incubation, the fluorescence was measured with excitation at 337 nm and emission at 490 and 520 nm. The TR-FRET ratio was determined as the fluorescence at 520 nm divided by the fluorescence at 490 nm multiplied by 10,000.
- Biological Assay 2 BCL-2 Assay: In the assay 11 different concentrations of each compound (0.1 nM, 0.33 nM, 1.1 nM, 3.8 nM, 13 nM, 44 nM, 0.15 μM, 0.51 μM, 1.7 μM, 5.9 μM and 20 μM) were typically measured as duplicates in the same microtiter plate. For that, 100-fold concentrated DMSO solutions were prepared by serial dilutions (1:3.4) of a 2 mM stock solution in a clear, 384-well microtiter plate (Greiner Bio-One, Frickenhausen, Germany). From there, 50 nl were transferred in a dark test plate (Greiner Bio-One, Frickenhausen, Germany). The assay was initiated by addition of 2 μl of a 2,5-fold concentrated His-BCL-2 solution (usually for a 1 nM end concentration in 5 μl reaction volume) in aqueous assay buffer [50 mM Tris/HCl pH 7, 100 mM sodium chloride (NaCl), 50 mM potassium fluoride (KF), 0.005% Tween-20, 2 mM DTT, 0.1% bovine gamma globulin (BGG)] to the compounds in the assay plate. This was followed by a 10-minute incubation step at 22° C. for pre-equilibration of the putative complex between His-BCL-2 and the compounds. After that, 3 μl of a 1.67-fold concentrated solution (in assay buffer) consisting of Bad BH3-derived peptide (1 nM end concentration) and TR-FRET detection reagents [1.67 nM anti-His-Eu cryptate and 1.67 nM streptavidin-XL665 (both from Cisbio Bioassays, Codolet, France)], were added.
- Biological Assay BCL-XL Assay: In the assay 11 different concentrations of each compound (0.1 nM, 0.33 nM, 1.1 nM, 3.8 nM, 13 nM, 44 nM, 0.15 μM, 0.51 μM, 1.7 μM, 5.9 μM and 20 μM) were typically measured as duplicates in the same microtiter plate. For that, 100-fold concentrated DMSO solutions were prepared by serial dilutions (1:3.4) of a 2 mM stock solution in a clear, 384-well microtiter plate (Greiner Bio-One, Frickenhausen, Germany). From there, 50 nl were transferred in a dark test plate (Greiner Bio-One, Frickenhausen, Germany). The assay was initiated by addition of 2 μl of a 2,5-fold concentrated His-BCL-XL solution (usually for a 1 nM end concentration in 5 μl reaction volume) in aqueous assay buffer [50 mM Tris/HCl pH 7, 100 mM sodium chloride (NaCl), 50 μM potassium fluoride (KF), 0.005% Tween-20, 2 mM DTT, 0.1% bovine gamma globulin (BGG)] to the compounds in the assay plate. This was followed by a 10-minute incubation step at 22° C. for pre-equilibration of the putative complex between His-BCL-XL and the compounds. After that, 3 μl of a 1.67-fold concentrated solution (in assay buffer) consisting of Bad BH3-derived peptide (1 nM end concentration) and TR-FRET detection reagents [1.67 nM anti-His-Eu cryptate and 1.67 nM streptavidin-XL665 (both from Cisbio Bioassays, Codolet, France)], were added.
- Biological Assay MCL-1 Assay: In the assay 11 different concentrations of each compound (0.1 nM, 0.33 nM, 1.1 nM, 3.8 nM, 13 nM, 44 nM, 0.15 μM, 0.51 μM, 1.7 μM, 5.9 μM and 20 μM) were typically measured as duplicates in the same microtiter plate. For that, 100-fold concentrated DMSO solutions were prepared by serial dilutions (1:3.4) of a 2 mM stock solution in a clear, 384-well microtiter plate (Greiner Bio-One, Frickenhausen, Germany). From there, 50 nl were transferred in a dark test plate (Greiner Bio-One, Frickenhausen, Germany). The assay was initiated by addition of 2 μl of a 2,5-fold concentrated MBP-MCL-1 solution (usually for a 1 nM end concentration in 5 μl reaction volume) in aqueous assay buffer [50 mM Tris/HCl pH 7, 100 mM sodium chloride (NaCl), 50 mM potassium fluoride (KF), 0.005% Tween-20, 2 mM DTT, 0.1% bovine gamma globulin (BGG)] to the compounds in the assay plate. This was followed by a 10-minute incubation step at 22° C. for pre-equilibration of the putative complex between MBP-MCL-1 and the compounds. After that, 3 μl of a 1.67-fold concentrated solution (in assay buffer) consisting of Noxa BH3-derived peptide (1 nM end concentration) and TR-FRET detection reagents [1.67 nM anti-MBP-Eu cryptate and 1.67 nM streptavidin-XL665 (both from Cisbio Bioassays, Codolet, France)], were added.
- Enzyme Kinetics - Measuring Inactivation Rates of Inhibitors Inactivation solutions contained a final concentration of 16.55 nM of Cerezyme (Recombinant GCase) in 200 μL of Reaction Buffer (50 mM acetate, 0.2% v/v Triton X-100, 0.3% w/v sodium taurocholate, pH 5.5). The Cerezyme was collected from leftover patient vials and had approximately 8275 nM in the vial. These enzyme Reaction Buffers were brought to 37 C. and spiked with a corresponding inhibitor to make the final inhibitor concentration either 20 nM, 40 nM, 60 nM, 80 nM, 120 nM, 160 nM and 200 nM. Once the inhibitor was added this was considered time=0 minutes. Meanwhile a 96 well plate inside a Biotek Synergy 4 hybrid multi-mode reader at 37 C. was being incubated. In the wells of the plate were waiting a high concentration (3.2 mM) of 2,4-DNP-β-Glc substrate in 180 μL of Reaction Buffer. As the inhibitor inactivation was occurring, 20 μL aliquots were taken from the inactivation solutions and diluted on the plate containing the substrate bringing the final concentration of the substrate to 4 mM. Once all the aliquots were added the solution in the wells was measured for absorbance at 400 nm for 8 minutes. At this time the measurements were paused and the next set of aliquots were taken to see how residual enzyme activity changes over time.
- Inhibitory Activity of Compounds against RIPK1 Enzyme The compound to be tested was dissolved in DMSO to prepare a 10 mM stock solution, diluted 3.16 times with DMSO into a series of concentration gradients, and then MOPS pH 7.0 buffer solution was used to dilute 50 times to prepare a working solution, and mixed well with 36 nM RIPK1 (final concentration), 0.33 mg/ml substrate MBP. Afterwards, 10 mM magnesium ions and 155 μM phosphorus 33 isotope-labeled ATP were added to react, The final concentration of DMSO was 2%. After the reaction was proceeded at room temperature for 2 hours, phosphoric acid was added to terminate. The final reaction system was processed and detected using a liquid scintillation counter. After the test results are subtracted from the blank control and compared with the reading value of the control group to convert to the activity percentage, which was plotted as a curve with the final concentration of the corresponding compound, and four-parameter fitting was used to obtain the IC50 of the compound that inhibits RIPK1 enzyme activity. It can be seen from the experimental results that the exemplary compounds of the present invention possessed high inhibitory activity against RIPK1, with an IC50 value of less than 200 nM (for example, 0.1 nM to 200 nM); IC50 values of some compounds were even less than 100 nM (for example. 0.1 nM to 100 nM) or less than 50 nM (for example, 0.1 nM to 50 nM).
- ChEBML_139646 In vitro 100 nM cortex affinity to displace [3H]- -CD providing a measure of muscarinic m1 receptor agonist affinity
- ChEBML_140336 Tested for the ability to displace [3H]CGS-19,755 (10 nM) binding in rat forebrain membranes for NMDA receptor
- ChEBML_141662 Tested for inhibition against [3H]-SP (substance P) (1 nM) binding to NK1 receptor in guinea pig lung membranes
- ChEBML_141778 Tested for displacement of [125I]-labeled substance P from cloned hNK1 receptor expressed in CHO cells at 100 nM
- ChEBML_146616 Compound was tested for Inhibition of 9 (1 nM) binding to Opioid receptor delta 1 from bovine striatal Membranes
- ChEBML_196320 Ability to displace 3[H](all-E)-retinoic acid (5 nM) from Retinoic acid receptor gamma using transactivation assay
- ChEBML_201887 Binding effinity for sigma receptor using 2 nM of [3H]DTG as radioligand, in membranes from brain minus cerebellum
- ChEBML_220578 Inhibition of 3 nM [3H]clonidine binding to rat kidney membrane imidazoline receptor I-1 in presence of rauwolscine
- ChEBML_42207 Agonist effect on 45 [Ca2+] influx in vanilloid receptor expressing CHO cells relative to maximal capsaicin (300 nM) response
- ChEBML_46144 Concentration required to displace 50% of 0.5 nM [3H](aminoalkyl)indole binding to cannabinoid receptor in rat cerebellum membranes
- ChEBML_48600 Ability to displace 1 nM [3H]pCCK-8 from rat Cholecystokinin type B receptor stably expressing in CHO cells
- ChEBML_58186 Compound was tested for binding affinity against Dopamine receptor D1 using 1 nM [3H]SCH-23390 as the radioligand.
- ChEBML_62243 Binding affinity against Dopamine receptor D2 of rat was determined using 2 nM of [3H]sulpiride in binding assay
- ChEBML_64518 50% inhibitory activity against enkephalinase purified from rat kidney with [3H]D-Ala2-Leu-enkephalin (20 nM) as substrate.
- ChEBML_68127 Tested for inhibition of substrate hydrolysis in the presence of porcine kidney esterase at a concentration of 45 nM
- ChEBML_96622 In vitro inhibitory concentration required against Leukocyte antigen related receptor phosphatase (LAR) in the presence of 300 nM DTT
- ChEMBL_103159 (CHEMBL711304) Compound concentration required to induce transcriptional activation in MVLN cells comparable to 50% of 0.1 nM estradiol response
- ChEMBL_103160 (CHEMBL707870) Compound concentration required to induce transcriptional activation in MVLN cells equal to 50% of 0.1 nM estradiol response
- ChEMBL_1337 (CHEMBL616963) Ability to displace [3H]5-CT (1.5 nM) from HeLa cells of human 5-hydroxytryptamine 1B receptor receptor
- ChEMBL_136088 (CHEMBL745611) Binding affinity for mu opioid receptor in guinea pig brain membranes; using 2 nM [3H]DAMGO as radioligand
- ChEMBL_138749 (CHEMBL747922) In vivo binding affinity against mu opioid receptor was measured by using labeled ligand [3H]naloxone (0.5 nM)
- ChEMBL_142944 (CHEMBL750796) Binding affinity at the Norepinephrine transporter in rat frontal cortex by inhibition of 0.5 nM [3H]nisoxetine binding
- ChEMBL_144288 (CHEMBL754789) Binding ability to compete with [125I]Tyr3-NT (0.15 nM) for human NT receptors cloned in CHO cells.
- ChEMBL_144458 (CHEMBL755105) Inhibition of neutral endopeptidase activity in rabbit kidney with 20 nM [3H]D-Ala2-Leu-enkephalin as substrate
- ChEMBL_145335 (CHEMBL750570) Inhibition of binding of [3H]- -naltrindole (0.15 nM) to membranes from CHO cells expressing Opioid receptor delta 1
- ChEMBL_145383 (CHEMBL750073) Inhibition against kappa receptor from displacement studies using 1.5 nM [3H]U-69593 in rhesus monkey cortex membrane
- ChEMBL_145566 (CHEMBL749570) Ki value determined against Opioid receptor kappa 1 using [3H]U69, 593 at the Kd concentration 0.95 nM
- ChEMBL_145802 (CHEMBL756069) Binding affinity kappa opioid receptor in guinea pig brain membranes, using 1 nM [3H]U-69593 as radioligand
- ChEMBL_145803 (CHEMBL756070) Binding affinity kappa opioid receptor in guinea pig brain membranes; using 1 nM [3H]U-69593 as radioligand
- ChEMBL_146286 (CHEMBL756531) In vitro opioid receptor binding competition against 1 nM [3H]- Naltrexone in the presence of 100 mM NaCl.
- ChEMBL_146868 (CHEMBL752611) Binding affinity for delta opioid receptor in guinea pig brain membranes, using 2 nM [3H]DPDPE as radioligand
- ChEMBL_146869 (CHEMBL749997) Binding affinity for delta opioid receptor in guinea pig brain membranes; using 2 nM [3H]DPDPE as radioligand
- ChEMBL_146898 (CHEMBL751672) Affinity was assessed using competitive binding assay labeled with [3H]DPDPE (6.3 nM) for Opioid receptor delta 1
- ChEMBL_148842 (CHEMBL753959) Affinity was assessed using competitive binding assay labeled with [3H]- DAGO (1.28 nM) for Opioid receptor mu 1
- ChEMBL_1509399 (CHEMBL3603342) Binding affinity to human serum albumin with excitation at 285 nm after 30 mins by fluorescence spectrophotometric analysis
- ChEMBL_1509400 (CHEMBL3603343) Binding affinity to human serum albumin with excitation at 295 nm after 30 mins by fluorescence spectrophotometric analysis
- ChEMBL_151406 (CHEMBL754752) Binding activity of P3 purinoceptor-like protein (P3LP) using radioligand 40 nM [3H]NECA from rat brain membranes
- ChEMBL_153000 (CHEMBL759483) Inhibition constant(Ki) for inhibition of PPIase activity of Escherichia coli parvulin (Conc=4 nM) of Parvulins sfamily
- ChEMBL_1548484 (CHEMBL3756831) Binding affinity to Cyclophilin A (unknown origin) at 0.625 nM to 10 uM by surface plasmon resonance analysis
- ChEMBL_164470 (CHEMBL769917) Inhibition of specific [3H]-clonidine binding (0.4 nM) to rat brain membranes alpha-2 adrenoceptor; 2.8*10e-7
- ChEMBL_1747824 (CHEMBL4182334) Binding affinity to recombinant human chymase assessed as dissociation constant at 12.5 to 200 nM by SPR assay
- ChEMBL_1747827 (CHEMBL4182337) Binding affinity to recombinant human chymase assessed as dissociation constant at 6 to 1000 nM by SPR assay
- ChEMBL_201294 (CHEMBL805784) Binding affinity of compound towards sigma receptor using [3H]DTG (4 nM) ligand in hippocampus rat was determined
- ChEMBL_201427 (CHEMBL810660) Binding affinity for Sigma opioid receptor type 1 in guinea pig brain homogenate with 0.5 nM [3H](+)-PENT
- ChEMBL_201571 (CHEMBL872706) Ki value determined against Sigma opioid receptor type 1 using [(+)-[3H]pentazocine at the Kd concentration 2 nM.
- ChEMBL_201578 (CHEMBL809700) Binding affinity for Sigma opioid receptor type 2 in guinea pig brain homogenate with 4 nM [3H]-(+)-DTG
- ChEMBL_201749 (CHEMBL809883) Inhibition against sigma receptor using displacement of 3 nM [3H]pentazocine in homogenate of guinea pig brain cerebellum
- ChEMBL_204742 (CHEMBL805435) Inhibitory activity against human Steroid 5-alpha-reductase type 2 using 18213 3H testosterone 210 nM as substrate
- ChEMBL_211689 (CHEMBL819860) Effect on tubulin binding and the concentration required to cause 50% decrease in tubulin assembly at 350 nm.
- ChEMBL_216624 (CHEMBL819196) Reversible inhibitory activity toward alpha-chymotrypsin (5 nM concentration) at a pH of 7.8 by progress curve method.
- ChEMBL_2199051 (CHEMBL5111567) Inhibition of human DNA polymerase theta using DNA 16/19 as substrate at 5 nM of enzyme concentration
- ChEMBL_2199052 (CHEMBL5111568) Inhibition of human DNA polymerase theta using DNA 16/19 as substrate at 2 nM of enzyme concentration
- ChEMBL_2199053 (CHEMBL5111569) Inhibition of human DNA polymerase theta using DNA 16/19 as substrate at 1 nM of enzyme concentration
- ChEMBL_2199054 (CHEMBL5111570) Inhibition of human DNA polymerase theta using DNA 16/19 as substrate at 0.5 nM of enzyme concentration
- ChEMBL_221916 (CHEMBL842481) Binding affinity for mu opioid receptor in guinea pig brain membranes, using 2 nM [3H]-DAMGO as radioligand
- ChEMBL_27575 (CHEMBL643497) Binding affinity against human adenosine A1 receptor expressed in CHO cells using [3H]DPCPX; Range = 9.6-15 nM
- ChEMBL_286 (CHEMBL615724) In vitro inhibition of [3H]5-HT (2 nM) binding to 5-hydroxytryptamine 1A receptor from bovine hippocampus
- ChEMBL_29616 (CHEMBL639639) Inhibition of 1 nM [3H]- N6 -(phenylisopropyl) adenosine binding to Adenosine A1 receptor in rat fat cell membrane
- ChEMBL_29618 (CHEMBL639640) Inhibition of 1 nM [3H]- N6- (phenylisopropyl) adenosine binding to A1 adenosine receptor in rat cerebral cortical membranes
- ChEMBL_29620 (CHEMBL639642) Inhibition of 1 nM [3H]- N6-(phenylisopropyl) adenosine binding to Adenosine A1 receptor in rat cerebral cortical membranes
- ChEMBL_29630 (CHEMBL873035) Inhibition of 1 nM [3H]N-6-(phenylisopropyl)adenosine binding to adenosine A1 receptors in rat cortical membranes
- ChEMBL_29631 (CHEMBL636648) Inhibition of binding of 1 nM N6-[3H]cyclohexyladenosine to adenosine A1 receptor on rat cerebral cortical membranes.
- ChEMBL_303374 (CHEMBL839692) Binding affinity towards recombinant human Adenosine A1 receptor was determined using [3H]R-PIA (2.0 nM) as radioligand
- ChEMBL_303423 (CHEMBL840086) Binding affinity towards recombinant human Adenosine A2a receptor was determined using [3H]CGS-21680 (10 nM) as radioligand
- ChEMBL_306397 (CHEMBL828738) Inhibitory concentration against interleukin-8 receptor of human neutrophils by using [125I]IL-8 (0.125 nM) as radioligand
- ChEMBL_30724 (CHEMBL648803) Binding affinity against Adenosine A2 receptor using [3H]NECA with 50 nM CPA in rat striatal brain membranes
- ChEMBL_30755 (CHEMBL649786) Inhibition of [3H]CGS-21680 binding to human Adenosine A2a receptor expressed in HEK293 cells at 10000 nM
- ChEMBL_31249 (CHEMBL642373) Displacement of [125 I]AB-MECA from adenosine A3 receptor in bovine cortical membranes with 20 nM DPCPX
- ChEMBL_312899 (CHEMBL827419) Mitogenic stimulation or antagonism of 30 nM quinpirole-stimulated mitogenesis in CHO cells expressing human dopamine D2 receptor
- ChEMBL_312900 (CHEMBL827420) Mitogenic stimulation or antagonism of 30 nM quinpirole-stimulated mitogenesis in CHO cells expressing human dopamine D3 receptor
- ChEMBL_34811 (CHEMBL648955) Binding affinity towards Angiotensin II receptor, type 1 in rat liver membrane using [125I]-angiotensin II (0.1 nM)
- ChEMBL_36468 (CHEMBL653121) Ability to displace [3H]AII (2 nM) from its specific binding sites in rat adrenal cortical microsome preparation
- ChEMBL_366425 (CHEMBL871378) Displacement of [3H]Histamine from human histamine H4 receptor transfected in SK-N-MC cells at 100 nM
- ChEMBL_366426 (CHEMBL871379) Displacement of [3H]histamine from human histamine H4 receptor transfected in SK-N-MC cells at 300 nM
- ChEMBL_366428 (CHEMBL871381) Displacement of [3H]histamine from human histamine H4 receptor transfected in SK-N-MC cells at 10 nM
- ChEMBL_366429 (CHEMBL871382) Displacement of [3H]histamine from human histamine H4 receptor transfected in SK-N-MC cells at 30 nM
- ChEMBL_42367 (CHEMBL655234) In vitro antagonist effect against 50 nM capsaicin-induced 45 [Ca2+] influx in vanilloid receptor expressing CHO cells
- ChEMBL_43470 (CHEMBL659372) Inhibition of phosphodiesterase from bovine aorta with 1 uM cGMP calcium (10 uM) and calmodulin (15 nM) (insignificant)
- ChEMBL_43471 (CHEMBL659373) Inhibition of phosphodiesterase from bovine aorta with 1 uM cGMP calcium (10 uM) and calmodulin (15 nM) (insignificant)
- ChEMBL_46998 (CHEMBL658967) Binding affinity towards Cannabinoid receptor 2 from mouse spleen membranes using 0.8 nM [3H]CP-55940 as radioligand
- ChEMBL_52212 (CHEMBL666681) Binding affinity against Cysteinyl leukotriene D4 receptor from guinea pig lung was determined using [3H]LTD4 (0.2 nM)
- ChEMBL_565 (CHEMBL833691) In vitro inhibition of [3H]5-HT (2 nM) binding to 5-hydroxytryptamine 1A receptor from bovine hippocampus
- ChEMBL_584 (CHEMBL615454) In vitro inhibition of [3H]5-HT (2 nM) binding to 5-hydroxytryptamine 1A receptor from bovine hippocampus
- ChEMBL_60033 (CHEMBL672129) compound was tested for inhibition of 0.5 nM [3H]spiroperidol binding to dopamine receptor from rat striatal membrane.
- ChEMBL_61488 (CHEMBL672390) Compound was evaluated for inhibition of specifically bound [3H]- cocaine (2.7 nM) against Dopamine transporter in cynomolgus monkeys
- ChEMBL_61834 (CHEMBL673204) Binding affinity at the Dopamine transporter in rat striata by inhibition of 0.5 nM [3H]WIN-35428 binding
- ChEMBL_62928 (CHEMBL673833) Binding affinity of compound towards Dopamine receptor D3 using [3H]spiperone (1.2 nM) ligand in cortex was determined
- ChEMBL_66423 (CHEMBL677280) Compound was tested for inhibitory activity against FK506 binding protein 12 (FKBP12); Value ranges from 190-900 nM
- ChEMBL_70420 (CHEMBL682255) Inhibition of FPP incorporation into biotinylated K-Ras-derived peptide by human farnesyl transferase with 5 nM ATP
- ChEMBL_70422 (CHEMBL682257) Inhibition of FPP incorporation into biotinylated K-Ras-derived peptide by human farnesyl transferase with 5 nM ATP
- ChEMBL_70424 (CHEMBL681281) Inhibition of FPP incorporation into biotinylated K-Ras-derived peptide by human farnesyl transferase with 5 nM ATP
- ChEMBL_70428 (CHEMBL681285) Inhibition of FPP incorporation into biotinylated K-Ras-derived peptide by human farnesyl transferase with 5 nM ATP
- ChEMBL_745145 (CHEMBL1772324) Inhibition of bovine AChE at 30 nM using S-acetylthiocholine as as substrate by Lineweaver-Burk plot analysis
- ChEMBL_75663 (CHEMBL683309) Ability to displace specific binding of 0.2 nM [3H]Boc[Nle28,31]-CCK27-33 from guinea pig brain membranes
- ChEMBL_89797 (CHEMBL698515) Inhibitory activity against human Inosine-5'-monophosphate dehydrogenase 1 (IMPDH type I isoform); Range is 33-37 nM
- ChEMBL_89938 (CHEMBL699564) Inhibitory activity against human Inosine-5'-monophosphate dehydrogenase 2 (IMPDH type II isoform); Range is 6-10 nM
- ChEMBL_956971 (CHEMBL2378319) Binding affinity to human recombinant DAAO at 496 nm spectral modification by spectrophotometric analysis in presence of FAD
- ChEMBL_956972 (CHEMBL2378320) Binding affinity to human recombinant DAAO at 443 nm spectral modification by spectrophotometric analysis in presence of FAD
- ChEMBL_956973 (CHEMBL2378321) Binding affinity to human recombinant DAAO at 498 nm spectral modification by spectrophotometric analysis in presence of FAD
- ChEMBL_956974 (CHEMBL2378322) Binding affinity to human recombinant DAAO at 444 nm spectral modification by spectrophotometric analysis in presence of FAD
- ChEMBL_956975 (CHEMBL2378323) Binding affinity to human recombinant DAAO at 395 nm spectral modification by spectrophotometric analysis in presence of FAD
- ChEMBL_956976 (CHEMBL2378324) Binding affinity to human recombinant DAAO at 362 nm spectral modification by spectrophotometric analysis in presence of FAD
- ChEMBL_956977 (CHEMBL2378325) Binding affinity to human recombinant DAAO at 600 nm spectral modification by spectrophotometric analysis in presence of FAD
- ChEMBL_956978 (CHEMBL2378326) Binding affinity to human recombinant DAAO at 520 nm spectral modification by spectrophotometric analysis in presence of FAD
- ChEMBL_956979 (CHEMBL2378327) Binding affinity to human recombinant DAAO at 440 nm spectral modification by spectrophotometric analysis in presence of FAD
- ChEMBL_956980 (CHEMBL2378328) Binding affinity to human recombinant DAAO at 490 nm spectral modification by spectrophotometric analysis in presence of FAD
- ChEMBL_956981 (CHEMBL2378329) Binding affinity to human recombinant DAAO at 492 nm spectral modification by spectrophotometric analysis in presence of FAD
- ChEMBL_957004 (CHEMBL2378386) Binding affinity to human recombinant DAAO at 442 nm spectral modification by spectrophotometric analysis in presence of FAD
 BDBM356619 US10214478, Compound NM-001
BDBM356619 US10214478, Compound NM-001 BDBM356624 US10214478, Compound NM-008
BDBM356624 US10214478, Compound NM-008 BDBM356628 US10214478, Compound NM-009
BDBM356628 US10214478, Compound NM-009 BDBM356629 US10214478, Compound NM-011
BDBM356629 US10214478, Compound NM-011 BDBM356630 US10214478, Compound NM-012
BDBM356630 US10214478, Compound NM-012 US10214478, Compound NM-002 BDBM356620
US10214478, Compound NM-002 BDBM356620 US10214478, Compound NM-003 BDBM356621
US10214478, Compound NM-003 BDBM356621 US10214478, Compound NM-004 BDBM356622
US10214478, Compound NM-004 BDBM356622 US10214478, Compound NM-005 BDBM356623
US10214478, Compound NM-005 BDBM356623 US10544104, Compound 5d CHEMBL573578 US9765037, Compound 5d BDBM50298225 US11247972, Compound 5d CHEMBL2069955 NM-PP1
US10544104, Compound 5d CHEMBL573578 US9765037, Compound 5d BDBM50298225 US11247972, Compound 5d CHEMBL2069955 NM-PP1