 BDBM733127 US20250115590, Compound OC-1
BDBM733127 US20250115590, Compound OC-1 BDBM733128 US20250115590, Compound OC-2
BDBM733128 US20250115590, Compound OC-2 BDBM733129 US20250115590, Compound OC-3
BDBM733129 US20250115590, Compound OC-3 BDBM733134 US20250115590, Compound OC-8
BDBM733134 US20250115590, Compound OC-8 BDBM733135 US20250115590, Compound OC-9
BDBM733135 US20250115590, Compound OC-9 BDBM733136 US20250115590, Compound OC-10
BDBM733136 US20250115590, Compound OC-10 BDBM733137 US20250115590, Compound OC-11
BDBM733137 US20250115590, Compound OC-11 BDBM733138 US20250115590, Compound OC-12
BDBM733138 US20250115590, Compound OC-12 BDBM733139 US20250115590, Compound OC-13
BDBM733139 US20250115590, Compound OC-13 BDBM733140 US20250115590, Compound OC-14
BDBM733140 US20250115590, Compound OC-14 US20250115590, Compound OC-4 BDBM733130
US20250115590, Compound OC-4 BDBM733130 US20250115590, Compound OC-5 BDBM733131
US20250115590, Compound OC-5 BDBM733131 US20250115590, Compound OC-6 BDBM733132
US20250115590, Compound OC-6 BDBM733132 US20250115590, Compound OC-7 BDBM733133
US20250115590, Compound OC-7 BDBM733133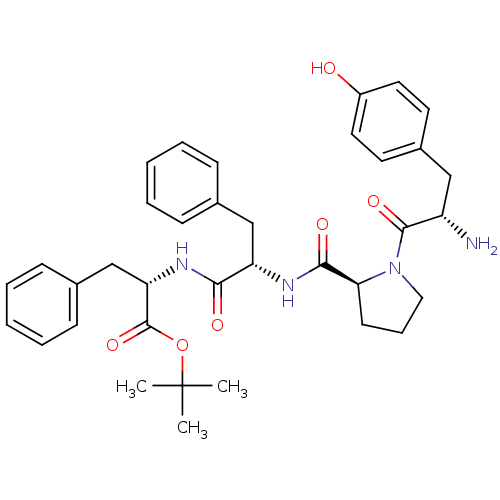 Tyr-Pro-Phe-Phe-OC(CH3)3 BDBM50271442 CHEMBL500195
Tyr-Pro-Phe-Phe-OC(CH3)3 BDBM50271442 CHEMBL500195 BDBM50090934 CHEMBL317303 L-Arg NO2-L-Dbu-OC(CH3)3
BDBM50090934 CHEMBL317303 L-Arg NO2-L-Dbu-OC(CH3)3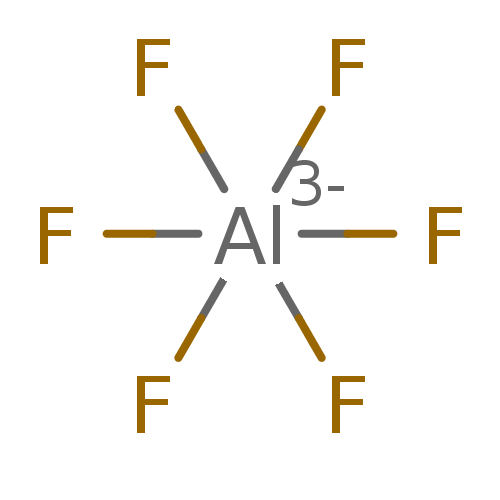 BDBM50164079 CHEMBL181624 hexafluoridoaluminate(3-) (OC-6-11)-hexafluoroaluminate(3-) [AlF6](3-) hexafluoroaluminate(3-) AlF6(3-)
BDBM50164079 CHEMBL181624 hexafluoridoaluminate(3-) (OC-6-11)-hexafluoroaluminate(3-) [AlF6](3-) hexafluoroaluminate(3-) AlF6(3-)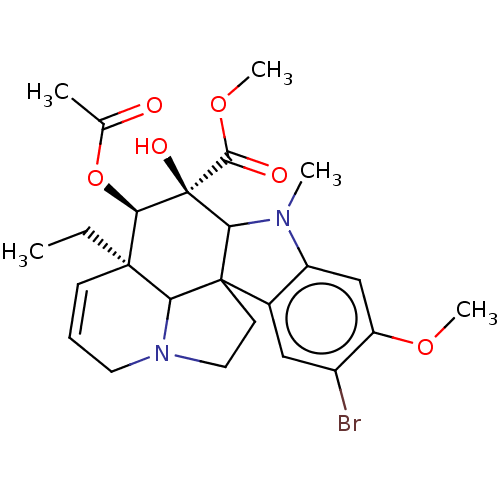 cid_6618799 SMR000306514 4-Acetoxy-9-bromo-3a-ethyl-5-hydroxy-8-methoxy-6-methyl-3a,4,5,5a,6,11,12,12b-oc BDBM56683 MLS000727817
cid_6618799 SMR000306514 4-Acetoxy-9-bromo-3a-ethyl-5-hydroxy-8-methoxy-6-methyl-3a,4,5,5a,6,11,12,12b-oc BDBM56683 MLS000727817 BDBM683092 US20240216357, Example 37 4-(6-(3-amino-8-a- zabicyclo[3.2.1]oc- tane-8-yl)-4- hydroxy-3- (3-hydroxy- 4-methoxyphenyl) pyridin-2-yl)-2- fluorobenzonitrile
BDBM683092 US20240216357, Example 37 4-(6-(3-amino-8-a- zabicyclo[3.2.1]oc- tane-8-yl)-4- hydroxy-3- (3-hydroxy- 4-methoxyphenyl) pyridin-2-yl)-2- fluorobenzonitrile endo-8-[7-(5- chloro-3- methoxyquinoxalin- 6-yl)-5H- pyrrolo[2,3- b]pyrazin-3-yl]-8- azabicyclo[3.2.1]oc- tan-3-amine, hydrochloride US11466016, Example 107 BDBM575974
endo-8-[7-(5- chloro-3- methoxyquinoxalin- 6-yl)-5H- pyrrolo[2,3- b]pyrazin-3-yl]-8- azabicyclo[3.2.1]oc- tan-3-amine, hydrochloride US11466016, Example 107 BDBM575974 4-(6-(3-amino-8-a- zabicyclo[3.2.1]oc- tane-8-yl)-3-(3- fluoro-4-methoxy- phenyl)-4-hydroxy- pyridin-2-yl)-2- fluorobenzonitrile BDBM683070 US20240216357, Example 14
4-(6-(3-amino-8-a- zabicyclo[3.2.1]oc- tane-8-yl)-3-(3- fluoro-4-methoxy- phenyl)-4-hydroxy- pyridin-2-yl)-2- fluorobenzonitrile BDBM683070 US20240216357, Example 14 4-(2-((1-((dimethyl- amino)methyl)cyclo- propyl)methoxy)-6,8- difluoro-4-(6- methoxy-3,8- diazabicyclo[3.2.1]oc- tan-3-yl)quinazolin- 7-yl)naphthalen-2-ol US20240239788, Example 25 BDBM685226
4-(2-((1-((dimethyl- amino)methyl)cyclo- propyl)methoxy)-6,8- difluoro-4-(6- methoxy-3,8- diazabicyclo[3.2.1]oc- tan-3-yl)quinazolin- 7-yl)naphthalen-2-ol US20240239788, Example 25 BDBM685226 US11466016, Example 113 {6-[endo-3-amino- 8-aza- bicyclo[3.2.1]oc- tan-8-yl]-3-(5- chloro-3- methoxyquinoxalin- 6-yl)-1H- pyrazolo[3,4- b]pyrazin-5- yl}methanol BDBM575979
US11466016, Example 113 {6-[endo-3-amino- 8-aza- bicyclo[3.2.1]oc- tan-8-yl]-3-(5- chloro-3- methoxyquinoxalin- 6-yl)-1H- pyrazolo[3,4- b]pyrazin-5- yl}methanol BDBM575979 (1R,2S,3S,5S)-8-[7- (5-chloro-3- methoxyquinoxalin- 6-yl)-5H- pyrrolo[2,3- b]pyrazin-3-yl]-2- fluoro-8- azabicyclo[3.2.1]oc- tan-3-amine, hydrochloride BDBM575973 US11466016, Example 106
(1R,2S,3S,5S)-8-[7- (5-chloro-3- methoxyquinoxalin- 6-yl)-5H- pyrrolo[2,3- b]pyrazin-3-yl]-2- fluoro-8- azabicyclo[3.2.1]oc- tan-3-amine, hydrochloride BDBM575973 US11466016, Example 106 2-(5-{3-[endo-3-amino- 8-azabicyclo[3.2.1]oc- tan-8-yl]-5H- pyrrolo[2,3-b]pyrazin- 7-yl}-4-chloro- 2-methyl-2H-indazol-3- yl)acetonitrile US11466016, Example 61 BDBM575933
2-(5-{3-[endo-3-amino- 8-azabicyclo[3.2.1]oc- tan-8-yl]-5H- pyrrolo[2,3-b]pyrazin- 7-yl}-4-chloro- 2-methyl-2H-indazol-3- yl)acetonitrile US11466016, Example 61 BDBM575933 4-(6-(3-amino-8-a- zabicyclo[3.2.1]oc- tane-8-yl)-3-(5- fluoro-3- methylben- zo[d]isoxazol-6-yl)- 4-hydroxypyridin- 2-yl)-2-fluoroben- zonitrile BDBM683080 US20240216357, Example 24
4-(6-(3-amino-8-a- zabicyclo[3.2.1]oc- tane-8-yl)-3-(5- fluoro-3- methylben- zo[d]isoxazol-6-yl)- 4-hydroxypyridin- 2-yl)-2-fluoroben- zonitrile BDBM683080 US20240216357, Example 24 US11466016, Example 60 2-(5-{3-[endo-3-amino- 8-azabicyclo[3.2.1]oc- tan-8-yl]-5H-pyrrolo [2,3-b]pyrazin-7-yl}-4- chloro-2H-indazol-2-yl)- N-tert-butylacetamide BDBM575932
US11466016, Example 60 2-(5-{3-[endo-3-amino- 8-azabicyclo[3.2.1]oc- tan-8-yl]-5H-pyrrolo [2,3-b]pyrazin-7-yl}-4- chloro-2H-indazol-2-yl)- N-tert-butylacetamide BDBM575932 US11466016, Example 116 BDBM575981 {6-[endo-3-amino- 3-methyl-8- azabicyclo[3.2.1]oc- tan-8-yl]-3-(3,4- dichloro-2-methyl- 2H-indazol-5-yl)- 1H-pyrazolo[3,4- b]pyrazin-5- yl}methanol
US11466016, Example 116 BDBM575981 {6-[endo-3-amino- 3-methyl-8- azabicyclo[3.2.1]oc- tan-8-yl]-3-(3,4- dichloro-2-methyl- 2H-indazol-5-yl)- 1H-pyrazolo[3,4- b]pyrazin-5- yl}methanol BDBM685297 2-(3-(6-chloro-2-((1- ((dimethylamino)meth- yl)cyclopro- pyl)methoxy)- 8-fluoro-7-(3-hydroxy- naphthalen-1-yl) quinazolin-4-yl)-3,8- diazabicyclo[3.2.1]oc- tan-1-yl)acetonitrile US20240239788, Example 36
BDBM685297 2-(3-(6-chloro-2-((1- ((dimethylamino)meth- yl)cyclopro- pyl)methoxy)- 8-fluoro-7-(3-hydroxy- naphthalen-1-yl) quinazolin-4-yl)-3,8- diazabicyclo[3.2.1]oc- tan-1-yl)acetonitrile US20240239788, Example 36 US11466016, Example 104 BDBM575971 (1R,2S,3S,5S)-8-[7- (3,4-dichloro-2- methyl-2H-indazol- 5-yl)-5H- pyrrolo[2,3- b]pyrazin-3-yl]-2- fluoro-8- azabicyclo[3.2.1]oc- tan-3-amine, hydrochloride
US11466016, Example 104 BDBM575971 (1R,2S,3S,5S)-8-[7- (3,4-dichloro-2- methyl-2H-indazol- 5-yl)-5H- pyrrolo[2,3- b]pyrazin-3-yl]-2- fluoro-8- azabicyclo[3.2.1]oc- tan-3-amine, hydrochloride {6-[(1S,2S,3S,5R)- 3-amino-2-fluoro-8- azabicyclo[3.2.1]oc- tan-8-yl]-3-(5- chloro-3- methoxyquinoxalin- 6-yl)-1H- pyrazolo[3,4- b]pyrazin-5- yl}methanol BDBM575976 US11466016, Example 109
{6-[(1S,2S,3S,5R)- 3-amino-2-fluoro-8- azabicyclo[3.2.1]oc- tan-8-yl]-3-(5- chloro-3- methoxyquinoxalin- 6-yl)-1H- pyrazolo[3,4- b]pyrazin-5- yl}methanol BDBM575976 US11466016, Example 109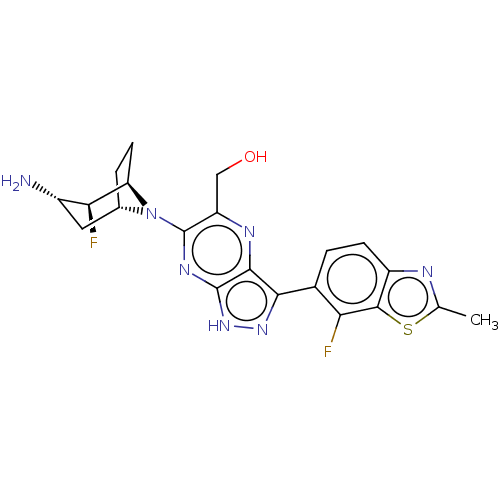 BDBM575966 US11466016, Example 98 {6-[(1R,2S,3S,5S)- 3-amino-2-fluoro-8- azabicyclo[3.2.1]oc- tan-8-yl]-3-(7- chloro-2-methyl- 1,3-benzothiazol-6- yl)-1H-pyrazolo [3,4-b]pyrazin- 5-yl}methanol
BDBM575966 US11466016, Example 98 {6-[(1R,2S,3S,5S)- 3-amino-2-fluoro-8- azabicyclo[3.2.1]oc- tan-8-yl]-3-(7- chloro-2-methyl- 1,3-benzothiazol-6- yl)-1H-pyrazolo [3,4-b]pyrazin- 5-yl}methanol BDBM575983 US11466016, Example 118 {6-[(1R,2S,3S,5S)- 3-amino-2-fluoro-8- azabicyclo[3.2.1]oc- tan-8-yl]-3-[5- chloro-3- (dimethylamino)qui- noxalin-6-yl]-1H- pyrazolo[3,4- b]pyrazin-5- yl}methanol
BDBM575983 US11466016, Example 118 {6-[(1R,2S,3S,5S)- 3-amino-2-fluoro-8- azabicyclo[3.2.1]oc- tan-8-yl]-3-[5- chloro-3- (dimethylamino)qui- noxalin-6-yl]-1H- pyrazolo[3,4- b]pyrazin-5- yl}methanol US11466016, Example 100 {6-[(1R,2S,3S,5S)- 3-amino-2-fluoro-8- azabicyclo[3.2.1]oc- tan-8-yl]-3-(5- chloro-3-methoxy- 2-methylquinolin- 6-yl)-1H-pyrazolo [3,4-b]pyrazin- 5-yl}methanol BDBM575967
US11466016, Example 100 {6-[(1R,2S,3S,5S)- 3-amino-2-fluoro-8- azabicyclo[3.2.1]oc- tan-8-yl]-3-(5- chloro-3-methoxy- 2-methylquinolin- 6-yl)-1H-pyrazolo [3,4-b]pyrazin- 5-yl}methanol BDBM575967 US11466016, Example 102 {6-[(1S,2R,3S,5R)- 3-amino-2-fluoro-8- azabicyclo[3.2.1]oc- tan-8-yl]-3-(3,4- dichloro-2-methyl- 2H-indazol-5-yl)- 1H-pyrazolo[3,4- b]pyrazin-5- yl}methanol BDBM575969
US11466016, Example 102 {6-[(1S,2R,3S,5R)- 3-amino-2-fluoro-8- azabicyclo[3.2.1]oc- tan-8-yl]-3-(3,4- dichloro-2-methyl- 2H-indazol-5-yl)- 1H-pyrazolo[3,4- b]pyrazin-5- yl}methanol BDBM575969 US11466016, Example 103 {6-[(1R,2S,3S,5S)- 3-amino-2-fluoro-8- azabicyclo[3.2.1]oc- tan-8-yl]-3-(3,4- dichloro-2-ethyl-2H- indazol-5-yl)-1H- pyrazolo[3,4- b]pyrazin-5- yl}methanol BDBM575970
US11466016, Example 103 {6-[(1R,2S,3S,5S)- 3-amino-2-fluoro-8- azabicyclo[3.2.1]oc- tan-8-yl]-3-(3,4- dichloro-2-ethyl-2H- indazol-5-yl)-1H- pyrazolo[3,4- b]pyrazin-5- yl}methanol BDBM575970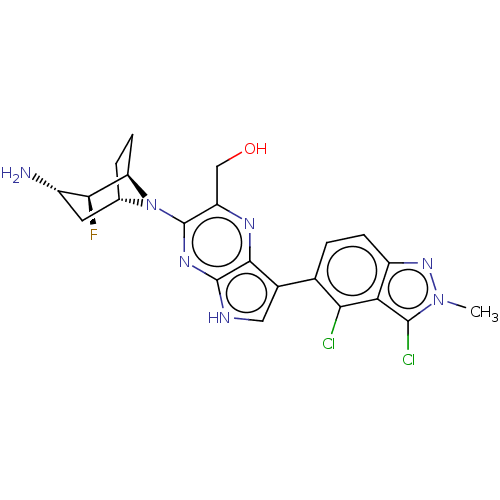 US11466016, Example 122 {3- [(1R,2S,3S,5S)-3- amino-2-fluoro-8- azabicyclo[3.2.1]oc- tan-8-yl]-7-(3,4- dichloro-2-methyl- 2H-indazol-5-yl)- 5H-pyrrolo[2,3- b]pyrazin-2- yl}methanol BDBM575988
US11466016, Example 122 {3- [(1R,2S,3S,5S)-3- amino-2-fluoro-8- azabicyclo[3.2.1]oc- tan-8-yl]-7-(3,4- dichloro-2-methyl- 2H-indazol-5-yl)- 5H-pyrrolo[2,3- b]pyrazin-2- yl}methanol BDBM575988 US11466016, Example 108 BDBM575975 {6-[(1R,2S,3S,5S)- 3-amino-2-fluoro-8- azabicyclo[3.2.1]oc- tan-8-yl]-3-(3- chloro-2,4-dimeth- yl-2H-indazol-5- yl)-1H-pyrazolo [3,4-b]pyrazin- 5-yl}methanol
US11466016, Example 108 BDBM575975 {6-[(1R,2S,3S,5S)- 3-amino-2-fluoro-8- azabicyclo[3.2.1]oc- tan-8-yl]-3-(3- chloro-2,4-dimeth- yl-2H-indazol-5- yl)-1H-pyrazolo [3,4-b]pyrazin- 5-yl}methanol BDBM575972 US11466016, Example 105 {6-[(1R,2S,3S,5S)- 3-amino-2-fluoro-8- azabicyclo[3.2.1]oc- tan-8-yl]-3-(3- bromo-4-fluoro-2- methyl-2H-indazol- 5-yl)-1H- pyrazolo[3,4- b]pyrazin-5- yl}methanol
BDBM575972 US11466016, Example 105 {6-[(1R,2S,3S,5S)- 3-amino-2-fluoro-8- azabicyclo[3.2.1]oc- tan-8-yl]-3-(3- bromo-4-fluoro-2- methyl-2H-indazol- 5-yl)-1H- pyrazolo[3,4- b]pyrazin-5- yl}methanol US11466016, Example 101 {6-[(1R,2S,3S,5S)- 3-amino-2-fluoro-8- azabicyclo[3.2.1]oc- tan-8-yl]-3-[4- chloro-2-(propan-2- yl)-2H-indazol-5- yl]-1H-pyrazolo [3,4-b]pyrazin- 5-yl}methanol BDBM575968
US11466016, Example 101 {6-[(1R,2S,3S,5S)- 3-amino-2-fluoro-8- azabicyclo[3.2.1]oc- tan-8-yl]-3-[4- chloro-2-(propan-2- yl)-2H-indazol-5- yl]-1H-pyrazolo [3,4-b]pyrazin- 5-yl}methanol BDBM575968
- Yim, CB; Dijkgraaf, I; Merkx, R; Versluis, C; Eek, A; Mulder, GE; Rijkers, DT; Boerman, OC; Liskamp, RM J Med Chem 53: 3944-53 (2010)
- Jiang, J; Lin, P; Hoang, M; Chang, L; Tan, C; Feighner, S; Palyha, OC; Hreniuk, DL; Pan, J; Sailer, AW; Morin, NR; MacNeil, DJ; Howard, AD; Van der Ploeg, LH; Goulet, MT; DeVita, RJ Bioorg Med Chem Lett 16: 5275-9 (2006)
- ChEMBL_708717 (CHEMBL1667059) Inhibition of methicillin-resistant Staphylococcus aureus OC 3726 PBP 2a
- ChEMBL_553829 (CHEMBL966192) Inhibition of bocillin FL binding to methicillin-resistant Staphylococcus aureus OC 3726 penicillin-binding protein 2a
- Solid Phase Integrin αVβ6 Binding Assay Microplates were coated with recombinant human integrin αVβ6 (2 μg/mL) in PBS (100 μL/well 25 OC, overnight). The coating solution was removed, washed with wash buffer (0.05% Tween 20; 0.5 mM MnCl2; in 1×TBS). Plate was blocked with 200 μL/well of Block Buffer (1% BSA; 5% sucrose; 0.5 mM MnCl2; in 1×TBS) at 37° C. for 2 h. Dilutions of testing compounds and recombinant TGFβ1 LAP (0.67 μg/mL) in binding buffer (0.05% BSA; 2.5% sucrose; 0.5 mM MnCl2; in 1×TBS) were added. The plate was incubated for 2 hours at 25° C., washed, and incubated for 1 hour with Biotin-Anti-hLAP. Bound antibody was detected by peroxidase-conjugated streptavidin. The IC50 values for testing compounds were calculated by a four-parameter logistic regression.
- Two-electrode Voltage Clamp Electrophysiology Two-electrode voltage clamp (TEVC) recordings were performed on Xenopus oocytes at room temperature 3-6 days postinjection using an OC-725C TEVC amplifier (Warner Instruments, Hamden, CT). Glass electrodes had a tip resistance of 0.5-2.5 megaohms and were pulled from thin walled glass capillary tubes (World Precision Instruments, Hertfordshire, UK) using a PC-10 puller (Narishige, East Meadow, NY). Voltage and current electrodes were filled with 0.3 and 3 M KCl, respectively. During recordings, oocytes were placed in a recording chamber and perfused with the extracellular recording solution comprised of 90 mM NaCl, 1 mM KCl, 10 mM HEPES, 0.5 mM BaCl2, and 0.01 mM EDTA (pH 7.4 with NaOH). Current responses were recorded at a holding potential of -40 and -60 mV for GluN1/N2 and GluN1/N3 receptors, respectively. Compounds were dissolved in extracellular recording solution and applied to the oocyte by gravity-driven perfusion using a computer-controlled 8-modular valve positioner (Digital MVP, Hamilton, Reno, NV).
- fluorescent peptide assay The assays were carried out with 20 nM enzyme (except for MPI3, for which 10 nM enzyme was used) and 10 μM substrate at 37oC with continuous shaking. All the analyses were carried out in triplicate. The substrate (DABCYL-Lys-Thr-Ser-Ala-Val-Leu-Gln-Ser-Gly-Phe-Arg-Lys-Met-Glu-EDANS) was purchased from Bachem and stored as 1 mM solution in 100% DMSO. Enzyme activity was monitored by fluorescence with excitation at 336 nm and emission at 455 nm wavelength. The dilution buffer (used for enzyme and substrate dilution) is 10 mMNaxHyPO4,10 mM NaCl, 0.5 mM EDTA, pH 7.6. Final composition of the assay buffer is 10 mM NaxHyPO4, 10 mM NaCl, 0.5 mM EDTA, 2 μM DTT (coming from enzyme stock solution), pH 7.6 with 1.25% DMSO. All the inhibitors were stored as 10 mMin 100%DMSO solutions in -20 oC freezer.
- GyraseB Assay Initially, equimolar quantities of about 0.5µM each of E. coli gyraseA and S. aureus gyraseB were incubated for a period of 45 min with salmon sperm DNA (Sigma) in 50mM Tris (pH 7.5), 75mM ammonium acetate buffer for the reconstitution of the hybrid topoisomerases at 4 OC. Later the assay was performed in a 96-well microtiter plate as mentioned earlier (7). The assay buffer includes 50 mM Tris (pH 7.5), 75 mM ammonium acetate, 5% w/v glycerol, 0.5mM ΕDTA, 6 mM magnesiumchloride, 0.001% Triton X-100, 1 mMdithiothreitol, DNA of 2 μg/mL (~3 μM base pairs), 250µM ATP, 2.2nM of E. coligyrase A and S. aureusgyraseB. Reactions were performed with various drug concentration ranges for the calculation of IC50, with a negative moxifloxacin and positive novobiocin control as standards. The reaction was allowed to proceed for 60 min and was quenched by addition of 20uL of malachite green reagent (POMG-25H, Bioassay systems, USA) subsequently absorbance was read at 650nm after 20 mi
- MraY Assay Park's nucleotide-N-C6-dansyl (2 mM stock solution, 1.88 μL), MgCl2 (0.5 M, 5 μL), KCl (2 M, 5 μL), Triton X-100 (0.5%, 5.63 μL), Tris buffer (pH 8.0, 50 mM), neryl phosphate (0.1 M, 2.25 μL), and inhibitor molecule (0-100 μg/mL in Tris buffer) were placed in a 500 μL Eppendorf tube. To a stirred reaction mixture, P-60 (10 μL) was added (total volume of reaction mixture: 50 μL adjust with Tris buffer). The reaction mixture was incubated for 2 h at room temperature (26 oC) and quenched with CHCl3 (100 μL). Two phases were mixed via vortex and centrifuged at 25,000 xg for 10 min. The upper aqueous phase was assayed via reverse-phase HPLC. The water phase (10 μL) was injected into HPLC (solvent: CH3CN/0.05 M aq. NH4HCO3=25:75; UV: 350 nm; flow rate: 0.5 mL/min; column: Kinetex 5 μm C8, 100 Å, 150×4.60 mm), and the area of the peak for lipid I-neryl derivative was quantified to obtain the IC50 value.
- MraY Assay Park's nucleotide-N -C6-dansyl (2 mM stock solution, 1.88 μL), MgCl2 (0.5 M, 5 L), KCI (2 M, 5 μL), Triton X-100 (0.5%, 5.63 μL), Tris buffer (pH 8.0, 50 mM), neryl phosphate (0.1 M, 2.25 μL), and inhibitor molecue (0-100 μg/mL in Tris buffer) were placed in a 500 μL Eppendorf tube. To a stirred reaction mixture, P-60 (10 μL) was added (total volume of reaction mixture: 50 μL adjust with Tris buffer). The reaction mixture was incubated for 2 h at room temperature (26 oC) and quenched with CHCl3 (100 μL). Two phases were mixed via vortex and centrifuged at 25,000 xg for 10 min. The upper aqueous phase was assayed via reverse-phase HPLC. The water phase (10 μL) was injected into HPLC (solvent: CH3CN/0.05 M aq. NH4HCO3=25:75; UV: 350 nm; flow rate: 0.5 mL/min; column: Kinetex 5 μm C8, 100 Å, 150 ×4.60 mm), and the area of the peak for lipid I-neryl derivative was quantified to obtain the IC50 value.
- SARS-CoV-2 Coronavirus 3C Protease FRET Assay The proteolytic activity of the main protease, 3CLpro, of SARS-CoV-2 was monitored using a continuous fluorescence resonance energy transfer (FRET) assay. The SARS-CoV-23CLpro assay measures the activity of full length SARS-CoV-23CL protease to cleave a synthetic fluorogenic substrate peptide with the following sequence: Dabcyl-KTSAVLQ-SGFRKME-Edans modelled on a consensus peptide (V. Grum-Tokars et al. Evaluating the 3C-like protease activity of SARS-coronavirus: recommendations for standardized assays for drug discovery. Virus Research 133 (2008) 63 73). The fluorescence of the cleaved Edans peptide (excitation 340 nm / emission 490 nm) is measured using a fluorescence intensity protocol on a Flexstation reader (Molecular Devices). The fluorescent signal is reduced in the present of PF-835231, a potent inhibitor of SARS-CoV-23CLpro. The assay reaction buffer contained 20 mM Tris-HCl (pH 7.3), 100 nM NaCl, 1 mM EDTA and 25 µM peptide substrate. Enzyme reactions were initiated with the addition of 15 nM SARS-CoV-23CL protease and allowed to proceed for 60 minutes at 23 oC. Percent inhibition or activity was calculated based on control wells containing no compound (0% inhibition/100% activity) and a control compound (100% inhibition/0% activity). IC50 values were generated using a four-parameter fit model using ABASE software (IDBS). Ki values were fit to the Morrison equation with the enzyme concentration parameter fixed to 15 nM, the Km parameter fixed to 14 µM and the substrate concentration parameter fixed to 25 µM using ABASE software (IDBS).
- Chamber Assay of CFTR-mediated short-circuit currents ssing chamber experiments were performed using human bronchial epithelial (HBE) cells derived from CF subjects heterozygous for F508del and a minimal function CFTR mutation (F508del/MF-HBE) and cultured as previously described (Neuberger T, Burton B, Clark H, Van Goor F Methods Mol Biol 2011:741:39-54). After four days the apical media was removed, and the cells were grown at an air liquid interface for >14 days prior to use. This resulted in a monolayer of fully differentiated columnar cells that were ciliated, features that are characteristic of human bronchial airway epithelia. To isolate the CFTR-mediated short-circuit (ISC) current, F508del/MF-HBE grown on Costar Snapwell cell culture inserts were mounted in an Ussing chamber and the transepithelial ISC was measured under voltage-clamp recording conditions (Vhold= 0 mV) at 37 oC. The basolateral solution contained (in mM) 145 NaCl, 0.83 K2HPO4, 3.3 KH2PO4, 1.2 MgCl2, 1.2 CaCl2, 10 Glucose, 10 HEPES (pH adjusted to 7.4 with NaOH) and the apical solution contained (in mM) 145 NaGluconate, 1.2 MgCl2, 1.2 CaCl2, 10 glucose, 10 HEPES (pH adjusted to 7.4 with NaOH) and 30 µM amiloride to block the epithelial sodium channel. Forskolin (20 µM) was added to the apical surface to activate CFTR, followed by apical addition of a CFTR inhibitor cocktail consisting of BPO, GlyH-101 and CFTR inhibitor 172 (each at 20 µM final assay concentration) to specifically isolate CFTR currents. The CFTR-mediated ISC (µA/cm2) for each condition was determined from the peak forskolin response to the steady-state current following inhibition.
- PHH Natural Infection Assay Detailed procedures regarding primary human hepatocyte (PHH) HBV natural infection assay are described as below. One tube of frozen PHH (10 million cells) is thawed in 37 oC water bath and then transferred to 20 mL of PHH thawing medium (Sigma, InVitroGRO HT Medium, Cat. S03319) with gently mixing. The cells were then centrifuged at 80 g/min for 5 min, the supernatant was discarded and the tube was refilled with 25 mL of PHH plating medium (Sigma , InVitroGRO CP Medium, Cat. S03317). The tube was shaken very gently to re-suspend all cells, and then 50 µl of cells were transferred to each well 384-well collagen I coated plate withappropriate liquid handling equipment, e.g. Integra VI AFL0384 or Agilent Bravo. The cells were then cultured for 24 hours in a cell incubator. For HBV infection, after PHH attachment on the culture plate, the plating medium was removed and replenished with PHH culture medium containing HBV virus. The PHH culture medium was prepared with Dulbecco's Modified Eagle Medium (DMEM)/F12 (1: 1 in volume ratio) containing 10% fetal bovine serum (Gibco, Cat.10099141), 5 ng/mL human epidermal growth factor (Gibco, Cat.PHG0311L), 20 ng/mL dexamethasone (Sigma, Cat.D4902-100mg), 250 ng/mL human recombinant insulin (Gibco, Cat.41400045) and 100 U/mL penicillin. HBV virus at 200 genome equivalent (GE) per cell with 4% PEG8000 (Sigma, Cat.P1458) containing culture medium were added to the PHH culture medium for infection. The cells were then cultured for 24 hours in cell incubator. Then the cell culture supernatant was removed. The HBV-infected PHH were cultured with sandwich culture method with PHH culture medium containing 1% DMSO and 0.25 mg/mL matrix gel for 72 hours. The supernatant was then refreshed with PHH culture medium containing different concentrations of testing compounds for two times with 72-hour interval. At the end of treatment, the supernatant was collected for viral markers measurements, including HBsAg, HBeAg, HBV DNA and cytotoxicity. HBsAg and HBeAg were detected using alphalisa method using their specific antibodies. For HBV DNA detection, HBV DNA Quantitative Fluorescence Diagnostic Kit (Sansure Biotech Inc.) was used following the manufacture s protocol.
- PHH Natural Infection Assay Detailed procedures regarding primary human hepatocyte (PHH) HBV natural infection assay are described as below. One tube of frozen PHH (10 million cells) is thawed in 37 oC water bath and then transferred to 20 mL of PHH thawing medium (Sigma , InVitroGRO HT Medium, Cat. S03319) with gently mixing. The cells were then centrifuged at 80 g/min for 5 min, the supernatant was discarded and the tube was refilled with 25 mL of PHH plating medium (Sigma , InVitroGRO CP Medium, Cat. S03317). The tube was shaken very gently to re-suspend all cells.50 μl of cells were transferred to each well 384-well collagen I coated plate with appropriate liquid handling equipment, e.g. Integra VIAFLO384 or Agilent Bravo. The cells were then cultured for 24 hours in a cell incubator. For HBV infection, after PHH attachment on the culture plate, the plating medium was removed and replenished with PHH culture medium containing HBV virus. The PHH culture medium was prepared with Dulbecco's Modified Eagle Medium (DMEM)/F12 (1: 1 in volume ratio) containing 10% fetal bovine serum (Gibco, Cat.10099141), 5 ng/mL human epidermal growth factor (Gibco, Cat.PHG0311L), 20 ng/mL dexamethasone (Sigma, Cat.D4902-100mg), 250 ng/mL human recombinant insulin (Gibco, Cat.41400045) and 100 U/mL penicillin. HBV virus at 200 genome equivalent (GE) per cell with 4% PEG8000 (Sigma, Cat.P1458) containing culture medium were added to the PHH culture medium for infection. The cells were then cultured for 24 hours in cell incubator. Then the cell culture supernatant was removed. The HBV-infected PHH were cultured with sandwich culture method with PHH culture medium containing 1% DMSO and 0.25 mg/mL matrix gel for 72 hours. The supernatant was then refreshed with PHH culture medium containing different concentrations of testing compounds for two times with 72-hour interval. At the end of treatment, the supernatant was collected for viral markers measurements, including HBsAg, HBeAg, HBV DNA and cytotoxicity. HBsAg and HBeAg were detected using alphalisa method using their specific antibodies. For HBV DNA detection, HBV DNA Quantitative Fluorescence Diagnostic Kit (Sansure Biotech Inc.) was used following the manufactures protocol. Cytotoxicity was determined using Cell Counting Kit-8 (CCK8, Dojindo Molecular Technologies, Inc.).
- Test for Evaluating Rat MC4 Receptor Activation, Using Cells Expressing Rat MC4 Receptor Experiment Method(1) Construction of Rat MC4 Receptor-Expressing VectorA rat MC4 receptor gene (GenBank Accession No.: NM_013099.2) was introduced into an expression vector pcDNA 3.1/V5-His TOPO (registered trademark) (Thermo Fisher Scientific Inc.).(2) Construction of Cells Transiently Expressing Rat MC4 ReceptorAn expression vector for a rat MC4 receptor was introduced into FreeStyle 293-F cell (Thermo Fisher Scientific Inc.). For the introduction, electroporation was employed. That is, 1×107 FreeStyle 293-F cell were suspended in 80 μL of an electroporation buffer (Thermo Fisher Scientific Inc.), and 20 μg of the expression vector was added thereto. The resultant was put into a cuvette (OC-100 Processing Assembly, MaxCyte, Inc.) and electroporated with MaxCyte STX (registered trademark) (MaxCyte, Inc.). The cells were cultured over one day, suspended in a Cell Banker (registered trademark) 1 (JUJI FIELD Inc.), and stored frozen until use.(3) Measurement of Amount of cAMP ProducedMeasurement was carried out in accordance with the attached instructions, using a LANCE (registered trademark) Ultra cAMP Kit (PerkinElmer, Inc.). That is, after dissolution in DMSO, the test compound (a final concentration of 1 pM to 30 μM) diluted with an assay buffer (Hank's balanced salt solution, 5 mM HEPES, 0.5 mM IBMX, 0.1% bovine serum albumin, pH 7.4), or α-MSH (Bachem Inc., a final concentration of 1 pM to 30 μM) was added to OptiPlate-384 (PerkinElmer, Inc.). Furthermore, a suspension of cells transiently expressing the rat MC4 receptor, that had been prepared using the assay buffer, was added thereto at 1,000 cells/well, followed by being left to stand at room temperature for about 1 hour. Thereafter, an Eu-cAMP tracer solution and an ULight -anti-cAMP solution were added thereto, followed by being left to stand at room temperature for about 1 hour. The amount of cAMP was calculated using EnVision (registered trademark) (PerkinElmer Inc.).For the agonistic activity, an efficacy (EC50 (μM)) was calculated by a non-linear regression method with a Sigmoid-Emax model, by defining the maximum reaction with α-MSH as 100% and the reaction with the vehicle alone as 0%, respectively.
- GOAT activity assay GOAT activity was assessed using a time-resolved fluorescence energy transfer (TR-FRET) assay in a 384-well format. His-tag human GOAT enzyme was in the form of a cell membrane preparation from sf9 cells infected with hGOAT-V5-His baculovirus. Varying concentrations of test compound with final DMSO concentration kept to 0.5% were added to membrane solution. Human GOAT membrane activity was established in a buffer having final concentration 0.25 mg/mL in 50 mM MOPS, pH7.5; 50 mM KCl; 0.1 mg/mL BSA; 50 μM CHAPS; and 2 mM EDTA. Substrate solution consisting of biotinylated ghrelin peptide (final concentration 100 nM), octanoyl coA (final concentration 2 μM) and palmitoyl CoA (final concentration 50 μM) was added to initiate the reaction. Plates were sealed, centrifuged for 1 minute at 2000 rpm, then incubated at 30° C. for 80 minutes with gentle shaking on an Eppendorf mix plate. Reaction termination and detection mix consisting of chicken anti-active ghrelin antibody (final concentration of 10 nM), Europium W1024-labeled streptavidin (final concentration of 4 nM), GOAT anti-chicken Dylight (final concentration of 12.5 nM), and GS[DAP-oc]-FL-amide inhibitor (final concentration of 1 μM) was added before further incubation for 40 minutes at 30° C. The plate was then read on an Envision in HTRF mode with excitation filter UV (TRF) 340 and first emission filter of APC 665 and a second emission filter of Europium 615. HTRF readings were acquired as per instrument defined LANCE-DELFIA protocol with a delay and window times of 50 μs for both; number of sequential windows: 1; time between flashes: 2000 μs between each of 100 flashes and 10 flashes for the second detector. The HTRF ratio was calculated directly by the instrument as the ratio of 665 window/615 window. Percent inhibition was calculated as 100−(100×(U−NC)/(PC−NC)) where U was the unknown value HTRF ratio (test compound value), NC was the negative control (100% inhibition value generated from a potent inhibitor), and PC was the positive control (100% activity generated from 0.5% DMSO vehicle). IC50 values were generated in GraphPad Prism (Version 4.03) using non-linear regression curve fit and sigmoidal dose response variable slope analysis.
- PHH Natural Infection Assay Detailed procedures regarding primary human hepatocyte (PHH) HBV natural infection assay are described as below. One tube of frozen PHH (10 million cells) is thawed in 37 oC water bath and then transferred to 20 mL of PHH thawing medium (Sigma , InVitroGRO HT Medium, Cat. S03319) with gently mixing. The cells were then centrifuged at 80 g/min for 5 min, the supernatant was discarded and the tube was refilled with 25 mL of PHH plating medium (Sigma , InVitroGRO CP Medium, Cat. S03317). The tube was shaken very gently to re-suspend all cells.50 μl of cells were transferred to each well 384-well collagen I coated plate with appropriate liquid handling equipment, e.g. Integra VIAFLO384 or Agilent Bravo. The cells were then cultured for 24 hours in a cell incubator. For HBV infection, after PHH attachment on the culture plate, the plating medium was removed and replenished with PHH culture medium containing HBV virus. The PHH culture medium was prepared with Dulbecco's Modified Eagle Medium (DMEM)/F12 (1: 1 in volume ratio) containing 10% fetal bovine serum (Gibco, Cat.10099141), 5 ng/mL human epidermal growth factor (Gibco, Cat.PHG0311L), 20 ng/mL dexamethasone (Sigma, Cat.D4902-100mg), 250 ng/mL human recombinant insulin (Gibco, Cat.41400045) and 100 U/mL penicillin. HBV virus at 200 genome equivalent (GE) per cell with 4% PEG8000 (Sigma, Cat.P1458) containing culture medium were added to the PHH culture medium for infection. The cells were then cultured for 24 hours in cell incubator. Then the cell culture supernatant was removed. The HBV-infected PHH were cultured with sandwich culture method with PHH culture medium containing 1% DMSO and 0.25 mg/mL matrix gel for 72 hours. The supernatant was then refreshed with PHH culture medium containing different concentrations of testing compounds for two times with 72-hour interval. At the end of treatment, the supernatant was collected for viral markers measurements, including HBsAg, HBeAg, HBV DNA and cytotoxicity. HBsAg and HBeAg were detected using alphalisa method using their specific antibodies. For HBV DNA detection, HBV DNA Quantitative Fluorescence Diagnostic Kit (Sansure Biotech Inc.) was used following the manufacture s protocol. Cytotoxicity was determined using Cell Counting Kit-8 (CCK8, Dojindo Molecular Technologies, Inc.).
- Test for Evaluating Human MC Receptor Activation, Using Cells Expressing Human MC4, MC1, or MC3 Receptor Experiment Method(1) Construction of Human MC Receptor-Expressing VectorA human MC4 receptor gene (GenBank Accession No.: NM_005912.2), a human MC1 receptor gene (GenBank Accession No.: NM_002386.3), or a human MC3 receptor gene (GenBank Accession No.: NM_019888.3) was introduced into an expression vector pcDNA 3.1/V5-His TOPO (registered trademark) (Thermo Fisher Scientific Inc.).(2) Construction of Cells Transiently Expressing Human MC ReceptorAn expression vector for a human MC4, MC1, or MC3 receptor was introduced into FreeStyle 293-F cell (Thermo Fisher Scientific Inc., product number: R790-07). For the introduction, electroporation was employed. That is, 1×107 FreeStyle 293-F cell were suspended in 80 μL of an electroporation buffer (Thermo Fisher Scientific Inc., product number: B201-100), and 20 μg of the expression vector was added thereto. The resultant was put into a cuvette (OC-100 Processing Assembly, MaxCyte, Inc.) and electroporated with MaxCyte SIX (registered trademark) (MaxCyte, Inc.). The cells were cultured over one day, suspended in a Cell Banker (registered trademark) 1 (JUJI FIELD Inc.), product number: BLC-1), and stored frozen until their use.(3) Measurement of Amount of cAMP ProducedMeasurement was carried out by using a LANCE (registered trademark) Ultra cAMP Kit (PerkinElmer, Inc.) in accordance with the attached instructions. That is, after dissolution in DMSO, the test compound (a final concentration of 1 pM to 30 μM) diluted with an assay buffer (Hank's balanced salt solution, 5 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), 0.5 mM 3-isobutyl-1-methylxanthine (IBMX), 0.1% bovine serum albumin, pH 7.4), or α-MSH (Bachem Inc., a final concentration of 1 pM to 30 μM) was added to OptiPlate-384 (PerkinElmer, Inc.). Furthermore, a suspension of cells transiently expressing the human MC4, MC1, or MC3 receptor prepared by using the assay buffer was added thereto at 1,000 cells/well, followed by being left to stand at room temperature for about 1 hour. Thereafter, an Eu-cAMP tracer solution and an ULight -anti-cAMP solution were added thereto, followed by being left to stand at room temperature for about 1 hour. The amount of cAMP was calculated using EnVision (registered trademark) (PerkinElmer Inc.).For the agonistic activity, an efficacy (EC50 (μM)) was calculated by a non-linear regression method with a Sigmoid-Emax model, by defining the maximum reaction with α-MSH as 100% and the reaction with the vehicle alone as 0%, respectively.
 BDBM733127 US20250115590, Compound OC-1
BDBM733127 US20250115590, Compound OC-1 BDBM733128 US20250115590, Compound OC-2
BDBM733128 US20250115590, Compound OC-2 BDBM733129 US20250115590, Compound OC-3
BDBM733129 US20250115590, Compound OC-3 BDBM733134 US20250115590, Compound OC-8
BDBM733134 US20250115590, Compound OC-8 BDBM733135 US20250115590, Compound OC-9
BDBM733135 US20250115590, Compound OC-9 BDBM733136 US20250115590, Compound OC-10
BDBM733136 US20250115590, Compound OC-10 BDBM733137 US20250115590, Compound OC-11
BDBM733137 US20250115590, Compound OC-11 BDBM733138 US20250115590, Compound OC-12
BDBM733138 US20250115590, Compound OC-12 BDBM733139 US20250115590, Compound OC-13
BDBM733139 US20250115590, Compound OC-13 BDBM733140 US20250115590, Compound OC-14
BDBM733140 US20250115590, Compound OC-14 US20250115590, Compound OC-4 BDBM733130
US20250115590, Compound OC-4 BDBM733130 US20250115590, Compound OC-5 BDBM733131
US20250115590, Compound OC-5 BDBM733131 US20250115590, Compound OC-6 BDBM733132
US20250115590, Compound OC-6 BDBM733132 US20250115590, Compound OC-7 BDBM733133
US20250115590, Compound OC-7 BDBM733133 Tyr-Pro-Phe-Phe-OC(CH3)3 BDBM50271442 CHEMBL500195
Tyr-Pro-Phe-Phe-OC(CH3)3 BDBM50271442 CHEMBL500195 BDBM50090934 CHEMBL317303 L-Arg NO2-L-Dbu-OC(CH3)3
BDBM50090934 CHEMBL317303 L-Arg NO2-L-Dbu-OC(CH3)3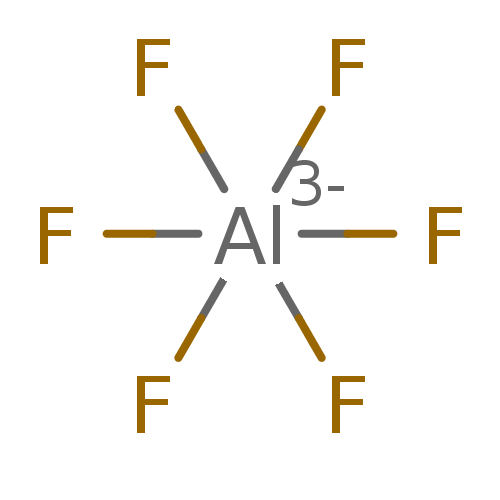 BDBM50164079 CHEMBL181624 hexafluoridoaluminate(3-) (OC-6-11)-hexafluoroaluminate(3-) [AlF6](3-) hexafluoroaluminate(3-) AlF6(3-)
BDBM50164079 CHEMBL181624 hexafluoridoaluminate(3-) (OC-6-11)-hexafluoroaluminate(3-) [AlF6](3-) hexafluoroaluminate(3-) AlF6(3-) cid_6618799 SMR000306514 4-Acetoxy-9-bromo-3a-ethyl-5-hydroxy-8-methoxy-6-methyl-3a,4,5,5a,6,11,12,12b-oc BDBM56683 MLS000727817
cid_6618799 SMR000306514 4-Acetoxy-9-bromo-3a-ethyl-5-hydroxy-8-methoxy-6-methyl-3a,4,5,5a,6,11,12,12b-oc BDBM56683 MLS000727817 BDBM683092 US20240216357, Example 37 4-(6-(3-amino-8-a- zabicyclo[3.2.1]oc- tane-8-yl)-4- hydroxy-3- (3-hydroxy- 4-methoxyphenyl) pyridin-2-yl)-2- fluorobenzonitrile
BDBM683092 US20240216357, Example 37 4-(6-(3-amino-8-a- zabicyclo[3.2.1]oc- tane-8-yl)-4- hydroxy-3- (3-hydroxy- 4-methoxyphenyl) pyridin-2-yl)-2- fluorobenzonitrile endo-8-[7-(5- chloro-3- methoxyquinoxalin- 6-yl)-5H- pyrrolo[2,3- b]pyrazin-3-yl]-8- azabicyclo[3.2.1]oc- tan-3-amine, hydrochloride US11466016, Example 107 BDBM575974
endo-8-[7-(5- chloro-3- methoxyquinoxalin- 6-yl)-5H- pyrrolo[2,3- b]pyrazin-3-yl]-8- azabicyclo[3.2.1]oc- tan-3-amine, hydrochloride US11466016, Example 107 BDBM575974 4-(6-(3-amino-8-a- zabicyclo[3.2.1]oc- tane-8-yl)-3-(3- fluoro-4-methoxy- phenyl)-4-hydroxy- pyridin-2-yl)-2- fluorobenzonitrile BDBM683070 US20240216357, Example 14
4-(6-(3-amino-8-a- zabicyclo[3.2.1]oc- tane-8-yl)-3-(3- fluoro-4-methoxy- phenyl)-4-hydroxy- pyridin-2-yl)-2- fluorobenzonitrile BDBM683070 US20240216357, Example 14 4-(2-((1-((dimethyl- amino)methyl)cyclo- propyl)methoxy)-6,8- difluoro-4-(6- methoxy-3,8- diazabicyclo[3.2.1]oc- tan-3-yl)quinazolin- 7-yl)naphthalen-2-ol US20240239788, Example 25 BDBM685226
4-(2-((1-((dimethyl- amino)methyl)cyclo- propyl)methoxy)-6,8- difluoro-4-(6- methoxy-3,8- diazabicyclo[3.2.1]oc- tan-3-yl)quinazolin- 7-yl)naphthalen-2-ol US20240239788, Example 25 BDBM685226 US11466016, Example 113 {6-[endo-3-amino- 8-aza- bicyclo[3.2.1]oc- tan-8-yl]-3-(5- chloro-3- methoxyquinoxalin- 6-yl)-1H- pyrazolo[3,4- b]pyrazin-5- yl}methanol BDBM575979
US11466016, Example 113 {6-[endo-3-amino- 8-aza- bicyclo[3.2.1]oc- tan-8-yl]-3-(5- chloro-3- methoxyquinoxalin- 6-yl)-1H- pyrazolo[3,4- b]pyrazin-5- yl}methanol BDBM575979 (1R,2S,3S,5S)-8-[7- (5-chloro-3- methoxyquinoxalin- 6-yl)-5H- pyrrolo[2,3- b]pyrazin-3-yl]-2- fluoro-8- azabicyclo[3.2.1]oc- tan-3-amine, hydrochloride BDBM575973 US11466016, Example 106
(1R,2S,3S,5S)-8-[7- (5-chloro-3- methoxyquinoxalin- 6-yl)-5H- pyrrolo[2,3- b]pyrazin-3-yl]-2- fluoro-8- azabicyclo[3.2.1]oc- tan-3-amine, hydrochloride BDBM575973 US11466016, Example 106 2-(5-{3-[endo-3-amino- 8-azabicyclo[3.2.1]oc- tan-8-yl]-5H- pyrrolo[2,3-b]pyrazin- 7-yl}-4-chloro- 2-methyl-2H-indazol-3- yl)acetonitrile US11466016, Example 61 BDBM575933
2-(5-{3-[endo-3-amino- 8-azabicyclo[3.2.1]oc- tan-8-yl]-5H- pyrrolo[2,3-b]pyrazin- 7-yl}-4-chloro- 2-methyl-2H-indazol-3- yl)acetonitrile US11466016, Example 61 BDBM575933 4-(6-(3-amino-8-a- zabicyclo[3.2.1]oc- tane-8-yl)-3-(5- fluoro-3- methylben- zo[d]isoxazol-6-yl)- 4-hydroxypyridin- 2-yl)-2-fluoroben- zonitrile BDBM683080 US20240216357, Example 24
4-(6-(3-amino-8-a- zabicyclo[3.2.1]oc- tane-8-yl)-3-(5- fluoro-3- methylben- zo[d]isoxazol-6-yl)- 4-hydroxypyridin- 2-yl)-2-fluoroben- zonitrile BDBM683080 US20240216357, Example 24 US11466016, Example 60 2-(5-{3-[endo-3-amino- 8-azabicyclo[3.2.1]oc- tan-8-yl]-5H-pyrrolo [2,3-b]pyrazin-7-yl}-4- chloro-2H-indazol-2-yl)- N-tert-butylacetamide BDBM575932
US11466016, Example 60 2-(5-{3-[endo-3-amino- 8-azabicyclo[3.2.1]oc- tan-8-yl]-5H-pyrrolo [2,3-b]pyrazin-7-yl}-4- chloro-2H-indazol-2-yl)- N-tert-butylacetamide BDBM575932 US11466016, Example 116 BDBM575981 {6-[endo-3-amino- 3-methyl-8- azabicyclo[3.2.1]oc- tan-8-yl]-3-(3,4- dichloro-2-methyl- 2H-indazol-5-yl)- 1H-pyrazolo[3,4- b]pyrazin-5- yl}methanol
US11466016, Example 116 BDBM575981 {6-[endo-3-amino- 3-methyl-8- azabicyclo[3.2.1]oc- tan-8-yl]-3-(3,4- dichloro-2-methyl- 2H-indazol-5-yl)- 1H-pyrazolo[3,4- b]pyrazin-5- yl}methanol BDBM685297 2-(3-(6-chloro-2-((1- ((dimethylamino)meth- yl)cyclopro- pyl)methoxy)- 8-fluoro-7-(3-hydroxy- naphthalen-1-yl) quinazolin-4-yl)-3,8- diazabicyclo[3.2.1]oc- tan-1-yl)acetonitrile US20240239788, Example 36
BDBM685297 2-(3-(6-chloro-2-((1- ((dimethylamino)meth- yl)cyclopro- pyl)methoxy)- 8-fluoro-7-(3-hydroxy- naphthalen-1-yl) quinazolin-4-yl)-3,8- diazabicyclo[3.2.1]oc- tan-1-yl)acetonitrile US20240239788, Example 36 US11466016, Example 104 BDBM575971 (1R,2S,3S,5S)-8-[7- (3,4-dichloro-2- methyl-2H-indazol- 5-yl)-5H- pyrrolo[2,3- b]pyrazin-3-yl]-2- fluoro-8- azabicyclo[3.2.1]oc- tan-3-amine, hydrochloride
US11466016, Example 104 BDBM575971 (1R,2S,3S,5S)-8-[7- (3,4-dichloro-2- methyl-2H-indazol- 5-yl)-5H- pyrrolo[2,3- b]pyrazin-3-yl]-2- fluoro-8- azabicyclo[3.2.1]oc- tan-3-amine, hydrochloride {6-[(1S,2S,3S,5R)- 3-amino-2-fluoro-8- azabicyclo[3.2.1]oc- tan-8-yl]-3-(5- chloro-3- methoxyquinoxalin- 6-yl)-1H- pyrazolo[3,4- b]pyrazin-5- yl}methanol BDBM575976 US11466016, Example 109
{6-[(1S,2S,3S,5R)- 3-amino-2-fluoro-8- azabicyclo[3.2.1]oc- tan-8-yl]-3-(5- chloro-3- methoxyquinoxalin- 6-yl)-1H- pyrazolo[3,4- b]pyrazin-5- yl}methanol BDBM575976 US11466016, Example 109 BDBM575966 US11466016, Example 98 {6-[(1R,2S,3S,5S)- 3-amino-2-fluoro-8- azabicyclo[3.2.1]oc- tan-8-yl]-3-(7- chloro-2-methyl- 1,3-benzothiazol-6- yl)-1H-pyrazolo [3,4-b]pyrazin- 5-yl}methanol
BDBM575966 US11466016, Example 98 {6-[(1R,2S,3S,5S)- 3-amino-2-fluoro-8- azabicyclo[3.2.1]oc- tan-8-yl]-3-(7- chloro-2-methyl- 1,3-benzothiazol-6- yl)-1H-pyrazolo [3,4-b]pyrazin- 5-yl}methanol BDBM575983 US11466016, Example 118 {6-[(1R,2S,3S,5S)- 3-amino-2-fluoro-8- azabicyclo[3.2.1]oc- tan-8-yl]-3-[5- chloro-3- (dimethylamino)qui- noxalin-6-yl]-1H- pyrazolo[3,4- b]pyrazin-5- yl}methanol
BDBM575983 US11466016, Example 118 {6-[(1R,2S,3S,5S)- 3-amino-2-fluoro-8- azabicyclo[3.2.1]oc- tan-8-yl]-3-[5- chloro-3- (dimethylamino)qui- noxalin-6-yl]-1H- pyrazolo[3,4- b]pyrazin-5- yl}methanol US11466016, Example 100 {6-[(1R,2S,3S,5S)- 3-amino-2-fluoro-8- azabicyclo[3.2.1]oc- tan-8-yl]-3-(5- chloro-3-methoxy- 2-methylquinolin- 6-yl)-1H-pyrazolo [3,4-b]pyrazin- 5-yl}methanol BDBM575967
US11466016, Example 100 {6-[(1R,2S,3S,5S)- 3-amino-2-fluoro-8- azabicyclo[3.2.1]oc- tan-8-yl]-3-(5- chloro-3-methoxy- 2-methylquinolin- 6-yl)-1H-pyrazolo [3,4-b]pyrazin- 5-yl}methanol BDBM575967 US11466016, Example 102 {6-[(1S,2R,3S,5R)- 3-amino-2-fluoro-8- azabicyclo[3.2.1]oc- tan-8-yl]-3-(3,4- dichloro-2-methyl- 2H-indazol-5-yl)- 1H-pyrazolo[3,4- b]pyrazin-5- yl}methanol BDBM575969
US11466016, Example 102 {6-[(1S,2R,3S,5R)- 3-amino-2-fluoro-8- azabicyclo[3.2.1]oc- tan-8-yl]-3-(3,4- dichloro-2-methyl- 2H-indazol-5-yl)- 1H-pyrazolo[3,4- b]pyrazin-5- yl}methanol BDBM575969 US11466016, Example 103 {6-[(1R,2S,3S,5S)- 3-amino-2-fluoro-8- azabicyclo[3.2.1]oc- tan-8-yl]-3-(3,4- dichloro-2-ethyl-2H- indazol-5-yl)-1H- pyrazolo[3,4- b]pyrazin-5- yl}methanol BDBM575970
US11466016, Example 103 {6-[(1R,2S,3S,5S)- 3-amino-2-fluoro-8- azabicyclo[3.2.1]oc- tan-8-yl]-3-(3,4- dichloro-2-ethyl-2H- indazol-5-yl)-1H- pyrazolo[3,4- b]pyrazin-5- yl}methanol BDBM575970 US11466016, Example 122 {3- [(1R,2S,3S,5S)-3- amino-2-fluoro-8- azabicyclo[3.2.1]oc- tan-8-yl]-7-(3,4- dichloro-2-methyl- 2H-indazol-5-yl)- 5H-pyrrolo[2,3- b]pyrazin-2- yl}methanol BDBM575988
US11466016, Example 122 {3- [(1R,2S,3S,5S)-3- amino-2-fluoro-8- azabicyclo[3.2.1]oc- tan-8-yl]-7-(3,4- dichloro-2-methyl- 2H-indazol-5-yl)- 5H-pyrrolo[2,3- b]pyrazin-2- yl}methanol BDBM575988 US11466016, Example 108 BDBM575975 {6-[(1R,2S,3S,5S)- 3-amino-2-fluoro-8- azabicyclo[3.2.1]oc- tan-8-yl]-3-(3- chloro-2,4-dimeth- yl-2H-indazol-5- yl)-1H-pyrazolo [3,4-b]pyrazin- 5-yl}methanol
US11466016, Example 108 BDBM575975 {6-[(1R,2S,3S,5S)- 3-amino-2-fluoro-8- azabicyclo[3.2.1]oc- tan-8-yl]-3-(3- chloro-2,4-dimeth- yl-2H-indazol-5- yl)-1H-pyrazolo [3,4-b]pyrazin- 5-yl}methanol BDBM575972 US11466016, Example 105 {6-[(1R,2S,3S,5S)- 3-amino-2-fluoro-8- azabicyclo[3.2.1]oc- tan-8-yl]-3-(3- bromo-4-fluoro-2- methyl-2H-indazol- 5-yl)-1H- pyrazolo[3,4- b]pyrazin-5- yl}methanol
BDBM575972 US11466016, Example 105 {6-[(1R,2S,3S,5S)- 3-amino-2-fluoro-8- azabicyclo[3.2.1]oc- tan-8-yl]-3-(3- bromo-4-fluoro-2- methyl-2H-indazol- 5-yl)-1H- pyrazolo[3,4- b]pyrazin-5- yl}methanol US11466016, Example 101 {6-[(1R,2S,3S,5S)- 3-amino-2-fluoro-8- azabicyclo[3.2.1]oc- tan-8-yl]-3-[4- chloro-2-(propan-2- yl)-2H-indazol-5- yl]-1H-pyrazolo [3,4-b]pyrazin- 5-yl}methanol BDBM575968
US11466016, Example 101 {6-[(1R,2S,3S,5S)- 3-amino-2-fluoro-8- azabicyclo[3.2.1]oc- tan-8-yl]-3-[4- chloro-2-(propan-2- yl)-2H-indazol-5- yl]-1H-pyrazolo [3,4-b]pyrazin- 5-yl}methanol BDBM575968