Target (25)
Compound (175)
Article Title (31)
Assay (192)
Saitoh, T; Kuramochi, K; Imai, T; Takata, K; Takehara, M; Kobayashi, S; Sakaguchi, K; Sugawara, F Podophyllotoxin directly binds a hinge domain in E2 of HPV and inhibits an E2/E7 interaction in vitro. Bioorg Med Chem 16: 5815 -25 (2008) Abdel-Magid, AF Prostaglandin e2 synthase-1 inhibitors as potential treatment for osteoarthritis: patent highlight. ACS Med Chem Lett 3: 703 -704 (2012) Friesen, RW; Mancini, JA Microsomal prostaglandin E2 synthase-1 (mPGES-1): a novel anti-inflammatory therapeutic target. J Med Chem 51: 4059 -67 (2008) Wiegard, A; Hanekamp, W; Griessbach, K; Fabian, J; Lehr, M Pyrrole alkanoic acid derivatives as nuisance inhibitors of microsomal prostaglandin E2 synthase-1. Eur J Med Chem 48: 153 -63 (2012) Koeberle, A; Laufer, SA; Werz, O Design and Development of Microsomal Prostaglandin E2 Synthase-1 Inhibitors: Challenges and Future Directions. J Med Chem 59: 5970 -86 (2016) Riendeau, D; Aspiotis, R; Ethier, D; Gareau, Y; Grimm, EL; Guay, J; Guiral, S; Juteau, H; Mancini, JA; Méthot, N; Rubin, J; Friesen, RW Inhibitors of the inducible microsomal prostaglandin E2 synthase (mPGES-1) derived from MK-886. Bioorg Med Chem Lett 15: 3352 -5 (2005) Lim, C; Lee, M; Park, EJ; Cho, R; Park, HJ; Lee, SJ; Cho, H; Lee, SK; Kim, S Sulfonamide derivatives of styrylheterocycles as a potent inhibitor of COX-2-mediated prostaglandin E2 production. Bioorg Med Chem Lett 20: 6938 -41 (2010) Zhou, L; Jeong, IH; Xue, S; Xue, M; Wang, L; Li, S; Liu, R; Jeong, GH; Wang, X; Cai, J; Yin, J; Huang, B Inhibition of the Ubiquitin Transfer Cascade by a Peptidomimetic Foldamer Mimicking the E2 N-Terminal Helix. J Med Chem 66: 491 -502 (2023) Tran, TD; Park, H; Kim, HP; Ecker, GF; Thai, KM Inhibitory activity of prostaglandin E2 production by the synthetic 2'-hydroxychalcone analogues: Synthesis and SAR study. Bioorg Med Chem Lett 19: 1650 -3 (2009) Hajduk, PJ; Dinges, J; Miknis, GF; Merlock, M; Middleton, T; Kempf, DJ; Egan, DA; Walter, KA; Robins, TS; Shuker, SB; Holzman, TF; Fesik, SW NMR-based discovery of lead inhibitors that block DNA binding of the human papillomavirus E2 protein. J Med Chem 40: 3144 -50 (1997) Koeberle, A; Zettl, H; Greiner, C; Wurglics, M; Schubert-Zsilavecz, M; Werz, O Pirinixic acid derivatives as novel dual inhibitors of microsomal prostaglandin E2 synthase-1 and 5-lipoxygenase. J Med Chem 51: 8068 -76 (2008) Nakao, Y; Oku, N; Matsunaga, S; Fusetani, N Cyclotheonamides E2 and E3, new potent serine protease inhibitors from the marine sponge of the genus Theonella. J Nat Prod 61: 667 -70 (1998) Cameron, KO; Lefker, BA; Ke, HZ; Li, M; Zawistoski, MP; Tjoa, CM; Wright, AS; DeNinno, SL; Paralkar, VM; Owen, TA; Yu, L; Thompson, DD Discovery of CP-533536: an EP2 receptor selective prostaglandin E2 (PGE2) agonist that induces local bone formation. Bioorg Med Chem Lett 19: 2075 -8 (2009) Greiner, C; Zettl, H; Koeberle, A; Pergola, C; Northoff, H; Schubert-Zsilavecz, M; Werz, O Identification of 2-mercaptohexanoic acids as dual inhibitors of 5-lipoxygenase and microsomal prostaglandin E2 synthase-1. Bioorg Med Chem 19: 3394 -401 (2011) Waltenberger, B; Wiechmann, K; Bauer, J; Markt, P; Noha, SM; Wolber, G; Rollinger, JM; Werz, O; Schuster, D; Stuppner, H Pharmacophore modeling and virtual screening for novel acidic inhibitors of microsomal prostaglandin E2 synthase-1 (mPGES-1). J Med Chem 54: 3163 -74 (2011) Fox, BM; Beck, HP; Roveto, PM; Kayser, F; Cheng, Q; Dou, H; Williamson, T; Treanor, J; Liu, H; Jin, L; Xu, G; Ma, J; Wang, S; Olson, SH A selective prostaglandin E2 receptor subtype 2 (EP2) antagonist increases the macrophage-mediated clearance of amyloid-beta plaques. J Med Chem 58: 5256 -73 (2015) Choi, D; Piao, YL; Wu, Y; Cho, H Control of the intracellular levels of prostaglandin E2 through inhibition of the 15-hydroxyprostaglandin dehydrogenase for wound healing. Bioorg Med Chem 21: 4477 -84 (2013) Shiro, T; Takahashi, H; Kakiguchi, K; Inoue, Y; Masuda, K; Nagata, H; Tobe, M Synthesis and SAR study of imidazoquinolines as a novel structural class of microsomal prostaglandin E2 synthase-1 inhibitors. Bioorg Med Chem Lett 22: 285 -8 (2011) Huang, H; Feng, X; Xiao, Z; Liu, L; Li, H; Ma, L; Lu, Y; Ju, J; She, Z; Lin, Y Azaphilones and p-terphenyls from the mangrove endophytic fungus Penicillium chermesinum (ZH4-E2) isolated from the South China Sea. J Nat Prod 74: 997 -1002 (2011) He, S; Li, C; Liu, Y; Lai, L Discovery of highly potent microsomal prostaglandin e2 synthase 1 inhibitors using the active conformation structural model and virtual screen. J Med Chem 56: 3296 -309 (2013) Svouraki, A; Garscha, U; Kouloura, E; Pace, S; Pergola, C; Krauth, V; Rossi, A; Sautebin, L; Halabalaki, M; Werz, O; Gaboriaud-Kolar, N; Skaltsounis, AL Evaluation of Dual 5-Lipoxygenase/Microsomal Prostaglandin E2 Synthase-1 Inhibitory Effect of Natural and Synthetic Acronychia-Type Isoprenylated Acetophenones. J Nat Prod 80: 699 -706 (2017) Lauro, G; Strocchia, M; Terracciano, S; Bruno, I; Fischer, K; Pergola, C; Werz, O; Riccio, R; Bifulco, G Exploration of the dihydropyrimidine scaffold for the development of new potential anti-inflammatory agents blocking prostaglandin E2 synthase-1 enzyme (mPGES-1). Eur J Med Chem 80: 407 -15 (2014) Mohd Aluwi, MF; Rullah, K; Yamin, BM; Leong, SW; Abdul Bahari, MN; Lim, SJ; Mohd Faudzi, SM; Jalil, J; Abas, F; Mohd Fauzi, N; Ismail, NH; Jantan, I; Lam, KW Synthesis of unsymmetrical monocarbonyl curcumin analogues with potent inhibition on prostaglandin E2 production in LPS-induced murine and human macrophages cell lines. Bioorg Med Chem Lett 26: 2531 -8 (2016) Hanke, T; Dehm, F; Liening, S; Popella, SD; Maczewsky, J; Pillong, M; Kunze, J; Weinigel, C; Barz, D; Kaiser, A; Wurglics, M; Lämmerhofer, M; Schneider, G; Sautebin, L; Schubert-Zsilavecz, M; Werz, O Aminothiazole-featured pirinixic acid derivatives as dual 5-lipoxygenase and microsomal prostaglandin E2 synthase-1 inhibitors with improved potency and efficiency in vivo. J Med Chem 56: 9031 -44 (2013) Liedtke, AJ; Keck, PR; Lehmann, F; Koeberle, A; Werz, O; Laufer, SA Arylpyrrolizines as inhibitors of microsomal prostaglandin E2 synthase-1 (mPGES-1) or as dual inhibitors of mPGES-1 and 5-lipoxygenase (5-LOX). J Med Chem 52: 4968 -72 (2009) Bäurle, S; Nagel, J; Peters, O; Bräuer, N; Ter Laak, A; Preusse, C; Rottmann, A; Heldmann, D; Bothe, U; Blume, T; Zorn, L; Walter, D; Zollner, TM; Steinmeyer, A; Langer, G Identification of a Benzimidazolecarboxylic Acid Derivative (BAY 1316957) as a Potent and Selective Human Prostaglandin E2 Receptor Subtype 4 (hEP4-R) Antagonist for the Treatment of Endometriosis. J Med Chem 62: 2541 -2563 (2019) Hieke, M; Greiner, C; Dittrich, M; Reisen, F; Schneider, G; Schubert-Zsilavecz, M; Werz, O Discovery and biological evaluation of a novel class of dual microsomal prostaglandin E2 synthase-1/5-lipoxygenase inhibitors based on 2-[(4,6-diphenethoxypyrimidin-2-yl)thio]hexanoic acid. J Med Chem 54: 4490 -507 (2011) De Simone, R; Chini, MG; Bruno, I; Riccio, R; Mueller, D; Werz, O; Bifulco, G Structure-based discovery of inhibitors of microsomal prostaglandin E2 synthase-1, 5-lipoxygenase and 5-lipoxygenase-activating protein: promising hits for the development of new anti-inflammatory agents. J Med Chem 54: 1565 -75 (2011) Blouin, M; Han, Y; Burch, J; Farand, J; Mellon, C; Gaudreault, M; Wrona, M; Lévesque, JF; Denis, D; Mathieu, MC; Stocco, R; Vigneault, E; Therien, A; Clark, P; Rowland, S; Xu, D; O'Neill, G; Ducharme, Y; Friesen, R The discovery of 4-{1-[({2,5-dimethyl-4-[4-(trifluoromethyl)benzyl]-3-thienyl}carbonyl)amino]cyclopropyl}benzoic acid (MK-2894), a potent and selective prostaglandin E2 subtype 4 receptor antagonist. J Med Chem 53: 2227 -38 (2010) Sharif, NA; Xu, SX; Williams, GW; Crider, JY; Griffin, BW; Davis, TL Pharmacology of [3H]prostaglandin E1/[3H]prostaglandin E2 and [3H]prostaglandin F2alpha binding to EP3 and FP prostaglandin receptor binding sites in bovine corpus luteum: characterization and correlation with functional data. J Pharmacol Exp Ther 286: 1094 -102 (1998) Lister, MD; Glaser, KB; Ulevitch, RJ; Dennis, EA Inhibition studies on the membrane-associated phospholipase A2 in vitro and prostaglandin E2 production in vivo of the macrophage-like P388D1 cell. Effects of manoalide, 7,7-dimethyl-5,8-eicosadienoic acid, and p-bromophenacyl bromide. J Biol Chem 264: 8520 -8 (1989)
ChEMBL_1910802 (CHEMBL4413248) Inhibition of E2-activated human PDHK2 ChEMBL_582704 (CHEMBL1051801) Antagonist activity against prostaglandin E2 receptor ChEMBL_1700822 (CHEMBL4051804) Inhibition of human placental cytosolic 17beta-HSD2 using [3H]-E2/E2 substrate and NAD+ after 20 mins by HPLC based radio-detection method ChEMBL_305014 (CHEMBL829449) Inhibitory concentration against human prostaglandin E2 synthase (mPGES-1) ChEMBL_429971 (CHEMBL917701) Displacement of [3H]E2 from ERalpha ligand binding domain ChEMBL_429972 (CHEMBL917702) Displacement of [3H]E2 from ERbeta ligand binding domain ChEMBL_450189 (CHEMBL900460) Displacement of [3H]E2 from human ERalpha ligand binding domain ChEMBL_450190 (CHEMBL900461) Displacement of [3H]E2 from human ERbeta ligand binding domain ChEMBL_66867 (CHEMBL675206) Displacement of [3H]-estradiol (E2) from sheep uterine estrogen receptor ChEMBL_803011 (CHEMBL1954168) Displacement of [3H]E2 from rat ERbeta1 after 90 mins ChEMBL_460516 (CHEMBL926596) Inhibition of human COX1-mediated conversion of arachidonic acid to prostaglandin-E2 ChEMBL_532974 (CHEMBL989644) Binding affinity to HPV1a recombinant E2 protein by surface plasmon resonance method ChEMBL_532978 (CHEMBL989648) Binding affinity to HPV11 recombinant E2 protein by surface plasmon resonance method ChEMBL_532980 (CHEMBL989650) Binding affinity to HPV16 recombinant E2 protein by surface plasmon resonance method ChEMBL_72863 (CHEMBL683959) Binding affinity of second enantiomer (E2) against human glucagon receptor was determined ChEMBL_310691 (CHEMBL837410) Agonist activity as alkaline phosphatase induction in Ishikawa endometrial cells compared to E2 ChEMBL_312806 (CHEMBL837766) Inhibition of 10e-9 M E2 stimulated MCF-7 breast cancer cell proliferation ChEMBL_619036 (CHEMBL1102078) Binding affinity to ubiquitin E2 variant domain of human Tsg101 by fluorescent anisotropy ChEMBL_637922 (CHEMBL1166108) Displacement of [3H]E2 from human ERalpha after 16 hrs by scintillation counting ChEMBL_637923 (CHEMBL1166109) Displacement of [3H]E2 from human ERbeta after 16 hrs by scintillation counting ChEMBL_700705 (CHEMBL1645744) Inhibition of [3H]E2 binding to human placental 17beta-HSD2 after 20 mins ChEBML_159933 The compound was evaluated for prostaglandin E2 inhibition using recombinant Prostaglandin G/H synthase 2 ChEMBL_1497878 (CHEMBL3582785) Displacement of [3H]-E2 from estrogen receptor-alpha (unknown origin) by scintillation counting analysis ChEMBL_1497879 (CHEMBL3582786) Displacement of [3H]-E2 from estrogen receptor-beta (unknown origin) by scintillation counting analysis ChEMBL_225530 (CHEMBL848481) In vitro inhibitory effect on production of prostaglandin E2 (PGE2) in rat synovial cells. ChEMBL_496057 (CHEMBL998487) Inhibition of COX1 assessed as inhibition of arachidonic acid conversion to prostaglandin-E2 release ChEMBL_532975 (CHEMBL989645) Binding affinity to HPV1a recombinant E2 protein activator domain by surface plasmon resonance assay ChEMBL_532976 (CHEMBL989646) Binding affinity to HPV1a recombinant E2 protein hinge domain by surface plasmon resonance assay ChEMBL_544743 (CHEMBL1010180) Inhibition of human platelet COX1 assessed as effect on prostaglandin E2 production by ELISA ChEMBL_653105 (CHEMBL1226308) Displacement of E2-Alexa633 from GFP-tagged GPR30 expressed in COS7 cells by FACS ChEMBL_653106 (CHEMBL1226309) Displacement of E2-Alexa633 from GFP-tagged ERalpha expressed in COS7 cells by FACS ChEMBL_653107 (CHEMBL1226310) Displacement of E2-Alexa633 from GFP-tagged ERbeta expressed in COS7 cells by FACS ChEMBL_67504 (CHEMBL682288) Inhibition of [3H]E2 binding to estrogen receptor alpha in MCF-7 cell lysate ChEMBL_100268 (CHEMBL712011) Agonist activity in transcriptional activation assay in MCF-7-2a cells compared to estradiol E2 ChEMBL_1284702 (CHEMBL3107402) Inhibition of COX-2 in human whole blood assessed as prostaglandin E2 production by RIA ChEMBL_158938 (CHEMBL767826) The compound was evaluated for prostaglandin E2 inhibition using recombinant Prostaglandin G/H synthase 1 ChEMBL_159933 (CHEMBL769658) The compound was evaluated for prostaglandin E2 inhibition using recombinant Prostaglandin G/H synthase 2 ChEMBL_2168580 (CHEMBL5053639) Displacement of fluorescent-labeled E2 from LBD of ERalpha (unknown origin) by TR-FRET assay ChEMBL_2168581 (CHEMBL5053640) Displacement of fluorescent-labeled E2 from LBD of ERbeta (unknown origin) by TR-FRET assay ChEMBL_532977 (CHEMBL989647) Binding affinity to HPV1a recombinant E2 protein DNA binding domain by surface plasmon resonance assay ChEMBL_787297 (CHEMBL1919075) Inhibition of human placental microsomal 17beta-HSD2 using [3H]E2 as substrate by HPLC analysis ChEMBL_838278 (CHEMBL2076226) TP_TRANSPORTER: increase in dihydrofluorescein intracellular accumulation (dihydrofluorescein: 1 uM) in SK-E2 cells (expressing BSEP) ChEMBL_838294 (CHEMBL2076242) TP_TRANSPORTER: increase in bodipy intracellular accumulation (Bodipy: 0.2 uM) in SK-E2 cells (expressing BSEP) ChEMBL_88150 (CHEMBL693235) Stimulation of alkaline phosphatase activity in human endometrial Ishikawa cells with 1 nM E2 estradiol ChEMBL_88151 (CHEMBL693236) Stimulation of alkaline phosphatase activity in human endometrial Ishikawa cells with 1 nM E2 estradiol ChEMBL_1832421 (CHEMBL4332429) Binding affinity to CD81 (unknown origin) assessed as disruption of interaction between CD81 the HCV envelope protein E2 by measuring Kd for CD81/E2 interaction at 5 uM by surface plasmon resonance assay (Rvb = 1.3 nM) Inhibition of 17beta-HSD1 Tritiated E1 was incubated with 17beta-HSD1, cofactor, and inhibitor. The amount of labeled E2 formed was quantified by HPLC. Detection and quantification of the steroids were performed using a radioflow detector (Berthold Technologies, Bad Wildbad). The conversion rate was calculated: %conversion = (%E2/ (%E2 +%E1) x 100). Each value was calculated from at least three independent experiments. ChEMBL_1750754 (CHEMBL4185514) Inhibition of NEDD8 (unknown origin) assessed as decrease in formation of E2-UBL thioester reaction product ChEMBL_1750756 (CHEMBL4185516) Inhibition of SAE (unknown origin) assessed as decrease in formation of E2-UBL thioester reaction product ChEMBL_1750757 (CHEMBL4185517) Inhibition of ATG7 (unknown origin) assessed as decrease in formation of E2-UBL thioester reaction product ChEMBL_1750758 (CHEMBL4185518) Inhibition of UBA6 (unknown origin) assessed as decrease in formation of E2-UBL thioester reaction product ChEMBL_2031808 (CHEMBL4685966) Displacement of fluorescent-labelled E2 from recombinant human GST-tagged ERalpha by LanthaScreen TR-FRET assay ChEMBL_2031809 (CHEMBL4685967) Displacement of fluorescent-labelled E2 from recombinant human GST-tagged ERbeta by LanthaScreen TR-FRET assay ChEMBL_633223 (CHEMBL1119430) Antagonist activity at ERbeta expressed in yeast assessed as inhibition of E2-induced alpha-galactosidase activity ChEMBL_633226 (CHEMBL1119433) Antagonist activity at ERalpha expressed in yeast assessed as inhibition of E2-induced alpha-galactosidase activity ChEMBL_715249 (CHEMBL1664450) Displacement of [3H]E2 from human placental 17beta-HSD2 after 10 mins by fluid scintillation counting ChEMBL_103299 (CHEMBL712253) Anti-estrogenicity for 50% inhibition of the MVLN cell luciferase activity due to 0.1 nM E2 (estradiol) ChEMBL_1439099 (CHEMBL3384914) Displacement of [3H]E2 from human placental microsomal 17beta-HSD2 after 20 mins by cell-free assay ChEMBL_1486828 (CHEMBL3535467) Noncompetitive inhibition of MRP4-mediated E2-17betaG transport in human MRP4 expressing HEK293 cells by Dixon plot ChEMBL_1489317 (CHEMBL3533496) Noncompetitive inhibition of MRP1-mediated E2-17betaG transport in human MRP1 expressing HEK293 cells by Dixon plot ChEMBL_2074723 (CHEMBL4730257) Displacement of [3H]-17beta-E2 from human ERbeta LBD by solid phase competitive radio ligand binding assay ChEMBL_2212559 (CHEMBL5125508) Displacement of 3H-E2 from ERalpha in human MCF7 incubated for 2 hrs by competition binding assay ChEMBL_313095 (CHEMBL836811) Inhibition of 10e-9 M E2 stimulated transcriptional activation in ER+MCF-7/2a breast cancer cells ChEMBL_313096 (CHEMBL836812) Inhibition of 10e-9 M E2 stimulated transcriptional activation in ER+MCF-7/2a breast cancer cells ChEMBL_940755 (CHEMBL2330135) Displacement of [3H]E2 from human recombinant ERbeta receptor after overnight incubation by liquid scintillation counting analysis ChEMBL_940756 (CHEMBL2330136) Displacement of [3H]E2 from human recombinant ERalpha receptor after overnight incubation by liquid scintillation counting analysis ChEMBL_544744 (CHEMBL1010181) Inhibition of human recombinant COX2 expressed in Sf21 cells assessed as effect on prostaglandin E2 production by ELISA ChEMBL_775981 (CHEMBL1912979) Displacement of [3H]E2 from estrogen receptor in JW rabbit uterus after 2 hrs by liquid scintillation counting ChEMBL_823597 (CHEMBL2044598) Inhibition of human placenta microsomal 17betaHSD2 using [3H]E2 as substrate after 20 mins by radioflow detection assay ChEMBL_143041 (CHEMBL750290) Affinity for rat Tachykinin receptor 1 determined in displacement screening by using radioligand 3,4-[3H]-(L-Pro e2) SP. ChEMBL_1561162 (CHEMBL3777355) Displacement of [3H]-E2 from human ER-alpha incubated for 16 to 20 hrs by liquid scintillation counting analysis ChEMBL_1561163 (CHEMBL3777356) Displacement of [3H]-E2 from human ER-beta incubated for 16 to 20 hrs by liquid scintillation counting analysis ChEMBL_1781487 (CHEMBL4253004) Displacement of [3H]E2 from human recombinant ERalpha assessed as receptor binding after 45 mins by scintillation counting method ChEMBL_322407 (CHEMBL871883) Tested for prostaglandin E2 production as a function of COX-2 inhibition using endotoxin-treated murine RAW 264.7 macrophages ChEBML_1684948 Antagonist activity at rat EP1 receptor expressed in African green monkey COS1 cells assessed as inhibition of prostaglandin-E2-induced increase in intracellular calcium level preincubated for 60 secs followed by prostaglandin-E2 addition measured for 60 secs by Fluo 4-AM dye based fluorescence assay Estrogen Receptor Binding Assay. Arzoxifene and its analogues were assayed in the standard ER competitive radioligand binding assay, using full length human recombinant ER-alpha and ER-beta with [3H]-estradiol. IC50 values were calculated from binding of the sample expressed as a percentage relative to E2 (50 nM, 100%). Relative binding affinity (RBA; relative to E2) was calculated from IC50 (E2)/IC50 (sample). The samples were assayed in triplicate at least five concentrations. ChEMBL_1501406 (CHEMBL3588435) Antagonist activity at human EP2 receptor overexpressed in human ECV304 cells assessed as inhibition of prostaglandin E2-induced cAMP production ChEMBL_1501407 (CHEMBL3588436) Antagonist activity at rat EP2 receptor overexpressed in human ECV304 cells assessed as inhibition of prostaglandin E2-induced cAMP production ChEMBL_1501408 (CHEMBL3588437) Antagonist activity at mouse EP2 receptor overexpressed in human ECV304 cells assessed as inhibition of prostaglandin E2-induced cAMP production ChEMBL_1619745 (CHEMBL3861914) Inhibition of purified human placental microsomal 17beta-HSD2 using [3H]-E2 substrate and NAD+ by HPLC based radioactive displacement assay ChEMBL_1861100 (CHEMBL4361956) Inhibition of mouse liver homogenate 17beta-HSD2 using [3H]-E2 as substrate in presence of NAD+ by radio-HPLC analysis ChEMBL_1861111 (CHEMBL4361967) Inhibition of rat liver homogenate 17beta-HSD2 using [3H]-E2 as substrate in presence of NAD+ by radio-HPLC analysis ChEMBL_1863754 (CHEMBL4364729) Inhibition of mouse liver homogenate 17beta-HSD2 using [3H]-E2 as substrate in presence of NAD+ by radio-HPLC analysis ChEMBL_1898529 (CHEMBL4400644) Inhibition of mouse liver homogenate 17beta-HSD2 using [3H]-E2 as substrate in presence of NAD+ by radio-HPLC analysis ChEMBL_81576 (CHEMBL689336) Dissociation constant after binding to human papillomavirus E2 DNA-binding domain (DBD) by observing the changes in [15N]-HSQC spectra. Binding Assay Recombinant ER-α or ER-β ligand binding domain (LBD) was combined with [3H]E2 (PerkinElmer, Waltham, Mass.) in buffer A (10 mM Tris, pH 7.4, 1.5 mM disodium EDTA, 0.25 M sucrose, 10 mM sodium molybdate, 1 mM PMSF) to determine the equilibrium dissociation constant (Kd) of [3H]E2. Protein was incubated with increasing concentrations of [3H]E2 with and without a high concentration of unlabeled E2 at 4° C. for 18h in order to determine total and non-specific binding. Non-specific binding was then subtracted from total binding to determine specific binding. Ligand binding curves were analyzed by nonlinear regression with one site saturation to determine the Kd of E2 (ER-α: 0.65 nM; ER-β: 1.83 nM). In addition, the concentration of [3H]E2 required to saturate ER-α and ER-β LBD was determined to be 1-3 nM.Increasing concentrations of two β-SERMs (14m and 12u) (range: 10−11 to 10−6 M) were incubated with [3H]E2 (1-2 nM) and ER LBD using the conditions described above. Following incubation, plates were harvested with GF/B filters on the Unifilter-96 Harvester (PerkinElmer) and washed three times with ice-cold buffer B (50 mM Tris, pH 7.2). The filter plates were dried at room temperature, then Microscint-O cocktail was added to each well and the filter plates were sealed with TopSeal-A. Radioactivity was counted in a TopCount NXT Microplate Scintillation Counter using the settings for [3H] in Microscint cocktail (PerkinElmer).The specific binding of [3H]E2 at each concentration of compound was determined by subtracting the nonspecific binding of [3H]E2 (determined by incubating with 10−6 M unlabeled E2) and expressing it as a percentage of the specific binding in the absence of compound. The concentration of compound that reduced the specific binding of [3H]E2 by 50% (IC50) was determined by computer-fitting the data with SigmaPlot and non-linear regression with the four parameter logistic curve. The equilibrium binding constant (Ki) of each compound was then calculated by: Ki=Kd×IC50/(Kd+L), where Kd is the equilibrium dissociation constant of [3H]E2, and L is the concentration of [3H]E2. ChEMBL_1541061 (CHEMBL3744292) Inhibition of 17beta-HSD2 in human placental microsomal fraction using E2/NAD+ as substrate/cofactor after 20 mins by HPLC analysis ChEMBL_1826276 (CHEMBL4326150) Displacement of [3H]prostaglandin E2 from human EP2 receptor expressed in HEK293 cell membranes after 60 mins by liquid scintillation counting ChEMBL_1861097 (CHEMBL4361953) Inhibition of human placental microsomal fraction 17beta-HSD2 using [3H]-E2 as substrate in presence of NAD+ by radio-HPLC analysis ChEMBL_1863749 (CHEMBL4364724) Inhibition of human placental microsomal fraction 17beta-HSD2 using [3H]-E2 as substrate in presence of NAD+ by radio-HPLC analysis ChEMBL_1898526 (CHEMBL4400641) Inhibition of human placental microsomal fraction 17beta-HSD2 using [3H]-E2 as substrate in presence of NAD+ by radio-HPLC analysis ChEMBL_938945 (CHEMBL2328434) Inhibition of 17beta-HSD2 in human MDA-MB-231 cells using [3H]E2 as substrate after 6 hrs by HPLC analysis ChEMBL_1539819 (CHEMBL3736957) Antagonist activity at ERalpha in human T47D-KBLuc cells assessed as inhibition of E2-induced transcriptional activity by luciferase reporter gene assay ChEMBL_1649547 (CHEMBL3998681) Inhibition of recombinant human PDK1 in presence of E1 protein and PDC core E2/E3BP incubated for 10 mins by titration assay ChEMBL_1649549 (CHEMBL3998683) Inhibition of recombinant human PDK3 in presence of E1 protein and PDC core E2/E3BP incubated for 10 mins by titration assay ChEMBL_1649550 (CHEMBL3998684) Inhibition of recombinant human PDK4 in presence of E1 protein and PDC core E2/E3BP incubated for 10 mins by titration assay ChEMBL_1861108 (CHEMBL4361964) Inhibition of 17beta-HSD2 in human MDA-MB-231 cells using [3H]-E2 as substrate after 6 hrs by radio-HPLC analysis ChEMBL_794210 (CHEMBL1932035) Inhibition of human microsomal 17beta-HSD2 in cell-free system using [2,4,6,7-3H]E2 as substrate after 20 mins by radioflow detector ChEMBL_938947 (CHEMBL2328436) Inhibition of 17beta-HSD1 in human placental cytosolic fraction using [3H]E1 as substrate assessed as formation of E2 by HPLC analysis ChEMBL_938948 (CHEMBL2328437) Inhibition of 17beta-HSD2 in human placental microsomal fraction using [3H]E2 as substrate assessed as formation of E1 by HPLC analysis ChEMBL_1471515 (CHEMBL3420078) Inhibition of 17 beta-HSD1 in human T47D cells assessed as inhibition of transformation of [14C]E1 to [14C]E2 after overnight incubation ChEMBL_1524156 (CHEMBL3631241) Inhibition of human placental microsomal 17beta HSD2 using unlabeled- and labelled [2,4,6,7-3H]-E2 as substrate incubated for 20 mins by HPLC analysis ChEMBL_1524162 (CHEMBL3631247) Inhibition of mouse liver microsomal 17beta HSD2 using unlabeled- and labelled [2,4,6,7-3H]-E2 as substrate incubated for 20 mins by HPLC analysis ChEMBL_1524167 (CHEMBL3631252) Inhibition of rat liver microsomal 17beta HSD2 using unlabeled- and labelled [2,4,6,7-3H]-E2 as substrate incubated for 20 mins by HPLC analysis ChEMBL_1898536 (CHEMBL4400651) Inhibition of 17beta-HSD2 in human using MDA-MB-231 cells using [3H]-E2 as substrate after 6 hrs by cell-based assay ChEMBL_2324328 Inhibition of human placental microsomal fraction 17beta-HSD2 using [3H]E2 as substrate measured after 20 mins in presence of NAD+ by HPLC analysis ChEMBL_736715 (CHEMBL1695096) Inhibition of Prostaglandin E2 synthase-1 in IL1-beta stimulated microsomal fraction of human A549 cell assessed as PGE2 level by RP-HPLC ChEMBL_768581 (CHEMBL1832600) Inhibition of 17beta-HSD1 in human T47D cells expressing estrogen receptor assessed as conversion of [14C]-E1 to [14C]-E2 after 24 hrs ChEBML_1696129 Displacement of [3H]E2 from GST-fused ERbeta-LBD (unknown origin) expressed in Escherichia coli BL21 incubated for 1 hr by liquid scintillation counting method ChEMBL_1282732 (CHEMBL3102029) Antagonist activity at ERbeta (unknown origin) assessed as inhibition of E2-induced receptor activation after 22 hrs by cell-based luciferase reporter gene assay ChEMBL_1646099 (CHEMBL3995155) Inhibition of human placental microsomal 17beta-HSD2 using [2,4,6,7-3H]-E2 as substrate after 20 mins in presence of NAD+ by RP-HPLC method ChEMBL_1668111 (CHEMBL4017999) Antagonist activity at CMX-GAL4N fused human ERalpha expressed in HEK293 cells assessed as reduction in E2 induced response by luciferase reporter gene assay ChEMBL_1769949 (CHEMBL4222061) Antagonist activity at ERalpha (unknown origin) expressed in human MCF7 cells assessed as inhibition of E2-induced response by dual luciferase reporter gene assay ChEMBL_2168592 (CHEMBL5053651) Antagonist activity at ERalpha (unknown origin) expressed in human U2OS cells assessed as inhibition of E2-induced transactivation by dual luciferase reporter gene assay ChEMBL_2168593 (CHEMBL5053652) Antagonist activity at ERbeta (unknown origin) expressed in human U2OS cells assessed as inhibition of E2-induced transactivation by dual luciferase reporter gene assay ChEMBL_2173335 (CHEMBL5058469) Inhibition of human placental microsomal fraction 17beta-HSD2 using [3H]-E2 as substrate in presence of NAD+ measured after 20 mins by HPLC method ChEMBL_2217339 (CHEMBL5130471) Inhibition of human placental microsomal fraction 17beta-HS2 using [3H]-E2 as substrate in presence of NAD+ measured after 10 mins by HPLC method ChEMBL_1287299 (CHEMBL3111007) Inhibition of 17-beta HSD1 in human T47D cells assessed as transformation of [14C]E1 to [14C]E2 after 24 hrs by thin layer chromatography ChEMBL_1687824 (CHEMBL4038394) Inhibition of BRD4 (unknown origin) expressed in human H1299 cells co-expressing HPV16-LCR/E2 after 24 hrs by Bright Glo-luciferase reporter gene assay ChEMBL_1696128 (CHEMBL4047018) Displacement of [3H]E2 from GST-fused ERalpha-LBD (unknown origin) expressed in Escherichia coli BL21 incubated for 1 hr by liquid scintillation counting method ChEMBL_1696129 (CHEMBL4047019) Displacement of [3H]E2 from GST-fused ERbeta-LBD (unknown origin) expressed in Escherichia coli BL21 incubated for 1 hr by liquid scintillation counting method ChEMBL_1789617 (CHEMBL4261351) Inhibition of mouse microsomal fraction 17beta-HSD2 using [3H]-E2 as substrate after 20 mins in presence of NAD+ by radio-flow detector based analysis ChEMBL_1826278 (CHEMBL4326152) Displacement of [3H]prostaglandin E2 from recombinant human full length EP1 receptor expressed in Chem-1 cell membranes after 60 mins by liquid scintillation counting ChEMBL_1826279 (CHEMBL4326153) Displacement of [3H]prostaglandin E2 from recombinant human full length EP3 receptor expressed in Chem-1 cell membranes after 60 mins by liquid scintillation counting ChEMBL_1826280 (CHEMBL4326154) Displacement of [3H]prostaglandin E2 from recombinant human full length EP4 receptor expressed in Chem-1 cell membranes after 60 mins by liquid scintillation counting ChEMBL_2017938 (CHEMBL4671516) Displacement of [3H]prostaglandin E2 from full-length recombinant human EP4 receptor expressed in HEK293 cell membranes measured after 120 mins by scintillation counting method ChEMBL_2017939 (CHEMBL4671517) Displacement of [3H]prostaglandin E2 from full-length recombinant human EP3 receptor expressed in HEK293 cell membranes measured after 120 mins by scintillation counting method ChEMBL_2017940 (CHEMBL4671518) Displacement of [3H]prostaglandin E2 from full-length recombinant human EP2 receptor expressed in HEK293 cell membranes measured after 120 mins by scintillation counting method ChEMBL_2017941 (CHEMBL4671519) Displacement of [3H]prostaglandin E2 from full-length recombinant human EP1 receptor expressed in HEK293 cell membranes measured after 60 mins by scintillation counting method Competitive Radioligand-Binding Assay Estrogen receptor (ERβ) binding affinity of the NRBAs was also determined using an in vitro competitive radioligand-binding assay with [3H]-estradiol ([3H]-E2, PerkinElmer), a high affinity ligand for both ERα and ERβ. The equilibrium dissociation constant (Kd) of [3H]-E2 was determined by incubating increasing concentrations of [3H]-E2 (0.01 to 10 nM) with bacterially expressed ERα or ERβ ligand binding domain (LBD) at 4° C. for 18 hours (h). Non-specific binding was determined by adding 1000 nM E2 to the incubation mixture. It was determined that the minimum concentration of [3H]-E2 required to saturate ERα and ERβ binding sites in the incubation mixture was 1 nM, respectively. The binding affinity of the NRBAs was determined under identical conditions by incubating increasing concentrations (3×10−2 to 1,000 nM) of ligand with isolated ER LBD and 1 nM [3H]-E2. Following incubation, bound and free [3H]-E2 were separated by using vacuum filtration with the Harvester (PerkinElmer). Briefly, the incubation mixture was filtered through a high affinity protein binding filter, and washed several times to remove any unbound radioactivity. The filter plate was air dried and sealed on the bottom. Scintillation cocktail was added to each well and the top of the plate was sealed. Radioactivity was counted in a TopCount NXT Microplate Scintillation Counter. ChEMBL_1539818 (CHEMBL3736956) Antagonist activity at luciferase-fused ERalpha in human HEK293 cells expressing eYFP assessed as reduction of E2-induced estrogenic activity after 2 hrs by BRET assay ChEMBL_1717011 (CHEMBL4132011) Antagonist activity at ER in human Ishikawa cells assessed as inhibition of E2-induced alkaline phosphatase induction after 72 hrs by PNPP substrate based spectophotometric method ChEMBL_1789612 (CHEMBL4261346) Inhibition of human placental microsomal fraction 17beta-HSD2 using [3H]-E2 as substrate after 20 mins in presence of NAD+ by radio-flow detector based analysis ChEMBL_1846277 (CHEMBL4346818) Inhibition of recombinant CDK2/Cyclin E2 (unknown origin) expressed in baculovirus expression system using biotinylated-histone H1 as substrate measured after 60 mins by HTRF assay Inhibition of 17beta-HSD2 Tritiated E2 was incubated with 17beta-HSD2, cofactor, and inhibitor. The amount of labeled E1 formed was quantified by HPLC. Detection and quantification of the steroids were performed using a radioflow detector (Berthold Technologies, Bad Wildbad). The conversion rate was calculated: %conversion = (%E1/ (%E2 +%E1) x 100). Each value was calculated from at least three independent experiments. ChEBML_1684873 Inhibition of recombinant CYP1A1 (unknown origin) expressed in Escherichia coli DH5alphaF'Iq coexpressing human NADPH-P450 reductase using E2-d4 as substrate after 10 mins in presence of NADP+ ChEMBL_1337650 (CHEMBL3242838) Inhibition of steroid sulfatase in human MCF7 cells assessed as conversion of [3H]E1S to [3H]E1 and [3H]E2 after 5 hrs by liquid scintillation counting ChEMBL_1649548 (CHEMBL3998682) Inhibition of N-terminal His6-SUMO tagged recombinant human PDK2 in presence of E1 protein and PDC core E2/E3BP incubated for 10 mins by titration assay ChEMBL_1791556 (CHEMBL4263475) Antagonist activity at full length ERalpha (unknown origin) assessed as reduction in E2 induced response after 24 hrs by cell based ERE-driven luciferase reporter gene assay ChEMBL_1791557 (CHEMBL4263476) Antagonist activity at full length ERbeta (unknown origin) assessed as reduction in E2 induced response after 24 hrs by cell based ERE-driven luciferase reporter gene assay ChEMBL_1336447 (CHEMBL3242591) Inhibition of LPS-induced COX-2 in peritoneal macrophage of C57BL/J6 mouse assessed as prostaglandin E2 formation preincubated for 1 hr followed by LPS challenge by radioimmunoassay ChEMBL_1670395 (CHEMBL4020283) Inhibition of BRD4 in human H1299 cells assessed as decrease in HPV LCR-E2-EP400-mediated transcriptional repression after 24 hrs by Bright-Glo luciferase reporter gene assay ChEMBL_1684873 (CHEMBL4035352) Inhibition of recombinant CYP1A1 (unknown origin) expressed in Escherichia coli DH5alphaF'Iq coexpressing human NADPH-P450 reductase using E2-d4 as substrate after 10 mins in presence of NADP+ ChEMBL_1684874 (CHEMBL4035353) Inhibition of recombinant CYP1B1 (unknown origin) expressed in Escherichia coli DH5alphaF'Iq coexpressing human NADPH-P450 reductase using E2-d4 as substrate after 10 mins in presence of NADP+ ChEMBL_1891475 (CHEMBL4393302) Antagonist activity at FLAG-tagged ERalpha (unknown origin) expressed in HEK293 cells assessed as reduction in E2-induced ER-alpha-mediated transcriptional activity by luciferase reporter gene assay ChEMBL_2031821 (CHEMBL4685979) Antagonist activity at ERalpha (unknown origin) expressed in human U2OS cells assessed as inhibition of E2-induced transactivation measured after 21 hrs by dual luciferase reporter gene assay ChEMBL_2031822 (CHEMBL4685980) Antagonist activity at ERbeta (unknown origin) expressed in human U2OS cells assessed as inhibition of E2-induced transactivation measured after 21 hrs by dual luciferase reporter gene assay ChEMBL_1891729 (CHEMBL4393556) Inhibition of ovine COX1 assessed as reduction in prostaglandin E2 production incubated for 10 mins before arachidonic acid addition and measured after 25 mins by LC-MS/MS analysis ChEMBL_1984040 (CHEMBL4617446) Antagonist activity at human Gal4-fused ERalpha expressed in HEK293 cells assessed as inhibition of E2-induced transcriptional activation after 24 hrs by luciferase/beta-galactosidase reporter gene assay ChEMBL_1984042 (CHEMBL4617448) Antagonist activity at human Gal4-fused ERbeta expressed in HEK293 cells assessed as inhibition of E2-induced transcriptional activation after 24 hrs by luciferase/beta-galactosidase reporter gene assay ChEMBL_1646098 (CHEMBL3995154) Inhibition of human placental cytosolic 17beta-HSD1 assessed as reduction in activation of [2,4,6,7-3H]-E1 substrate to E2 after 10 mins in presence of NADH by RP-HPLC method ChEMBL_1891730 (CHEMBL4393557) Inhibition of COX2 (unknown origin) assessed as reduction in prostaglandin E2 production incubated for 10 mins before arachidonic acid addition and measured after 25 mins by LC-MS/MS analysis ChEMBL_2381114 Inhibition of recombinant 17beta-HSD10 (unknown origin) using E2 as substrate and NAD+ as cofactor preincubated for 5 mins followed by substrate addition and measured after 20 mins by fluorometric assay ChEMBL_2469668 Antagonist activity at C-terminal RLucII-fused human ER-alpha expressed in HEK293T cells co-expressing CoA-YFP-fused LXXLL coactivator motif from NCOA2 in presence of E2 by BRET assay ChEMBL_1619749 (CHEMBL3861918) Inhibition of N-terminal 6His-tagged human human HSD17B14 expressed in Escherichia coli BL21 (DE3) pLysS using E2 substrate and NAD+ incubated for 2 hrs using purified enzyme by fluorimetric assay ChEMBL_1739715 (CHEMBL4155465) Binding affinity to recombinant 17beta-HSD14 (unknown origin) expressed in Escherichia coli BL21 pLysS strain using E2 as substrate in presence of NAD measured continuously for 15 mins by fluorimetric method ChEMBL_1759878 (CHEMBL4194886) Binding affinity to BRD4 in human H1299 cells stably expressing E2 and HPV16-LCR luciferase reporter incubated for 24 hrs by Bright-Glo luciferase reporter gene assay based BRD4 engagement assay ChEMBL_1993615 (CHEMBL4627510) Antagonist activity at ERbeta (unknown origin) expressed in human HeLa cells assessed as inhibition of E2-induced transcriptional activity measured after 24 hrs by luciferase reporter gene assay based Schild plot analysis ChEMBL_2060600 (CHEMBL4715601) Inhibition of recombinant N-terminal His-tagged human PTP1B (E2 to N321) expressed in Escherichia coli expression system assessed by dephosphorylation of substrate pNPP measured for 4 mins by plate reader analysis ChEMBL_2381116 Mixed type inhibition of recombinant 17beta-HSD10 (unknown origin) using E2 as substrate and NAD+ as cofactor preincubated for 5 mins followed by substrate addition and measured after 20 mins by fluorometric assay Competitive Binding Assay Competitive binding of estrogen to GPR30 and ER alpha were conducted using COS-7 cells transfected with GPR30-GFP or ER alpha-GFP and fluorescent estrogen, E2-Alexa633 labeled as a reporter. ChEMBL_1284733 (CHEMBL3107542) Inhibition of COX-1 in mouse J774 cells using arachidonic acid as substrate assessed as inhibition of prostaglandin E2 production preincubated for 15 mins followed by substrate addition measured after 30 mins by RIA ChEMBL_1284732 (CHEMBL3107541) Inhibition of LPS-induced COX-2 in mouse J774 cells using arachidonic acid as substrate assessed as inhibition of prostaglandin E2 production preincubated for 15 mins followed by substrate addition measured after 30 mins by RIA ChEMBL_1792852 (CHEMBL4264771) Inhibition of recombinant mouse full-length 17beta-HSD10 (2 to 261 residues) assessed as reduction in transformation of [14C]-E2 to E1 after 48 hrs in presence of 1 mM NAD+ cofactor by liquid scintillation counting method ChEMBL_2251629 (CHEMBL5165839) Inhibition of recombinant N-terminal GST/His6-fusion tagged human CDK2/Cyclin E2 expressed in Sf9 insect cells incubated for 20 mins followed by [33P]gamma-ATP addition and measured after 2 hrs by radiometric HotSpot Kinase assay ChEMBL_1667760 (CHEMBL4017648) Antagonist activity at rat EP1 receptor expressed in African green monkey COS1 cells assessed as decrease in prostaglandin E2-induced increase in intracellular calcium level incubated for 90 mins measured for 60 secs by Fluo 4-AM dye based fluorescence assay Binding Assay The ER binding affinity of the compounds was determined using an in vitro competitive radioligand binding assay was [2,4,6,7-3H(N)]-Estradiol ([3H]E2), a natural high affinity ER ligand, and bacterially expressed GST fusion ER-alpha or ER-beta ligand binding domain (LBD) protein. Competition-Based Ligand Binding Assay. Ligand binding was determined using a scintillation proximity assay with streptavidin-coated polyvinyltoluene scintillation beads (Amersham) and biotinylated receptor. Receptor-bound [3H]-E2 was determined by scintillation counting (Perkin-Elmer). The IC50 values were calculated using a four-parameter logistic equation. Estrone Detection Assay for Evaluation of HSD17beta13 Activity The liquid chromatography/mass spectrometry (LC/MS) estrone detection assay monitors the conversion of estradiol to estrone by HSD17B13. This assay was undertaken in a 96wp format (Eppendorf deep well Plate 96/500) in an 80 µl reaction volume containing: 4 µM of Estradiol (E2; Cayman; #10006315), 6 mM NAD+ (Sigma; #N0623) and 30 nM HSD17B13 enzyme (in-house; E. coli expressed His-tagged, purified, soluble protein) in a reaction containing 1 M potassium phosphate buffer pH 7.4, with 0.5% vehicle (DMSO). Reactions were incubated for 2 hours at 26.5° C., and estradiol (E2) conversion to estrone (E1) was quantitated by LC-MS/MS based analyte detection for both E2 and E1 using LCMS grade reagents.Reactions were terminated by the addition of two volumes of acetonitrile (MeCN; LCMS grade; CAS# 75/05/8) containing deuterated (D4)-E1 used as internal standard (Clear Synth; #CS-T-54273; 500 ng/mL final concentration). Samples were applied to pre-prepared Bond Elut-C18 extraction cartridges (3 mL; Agilent; #12102028), washed and eluted in MeCN. Eluates were dried under nitrogen and re-suspended in 60% methanol (LCMS grade methanol; CAS# 67/56/1) before submission for analysis. Aqueous linearity for E2 and E1 were included for quantification. Binding Assay The method employed was adapted from the scientific literature and described in detail by Osbourn et al. (1993, Biochemistry, 32, 6229-6236). Recombinant human ERalpha and ERR proteins were purified from transfected Sf9-cells. The in vitro assays involved the use of either ERalpha or ERbeta proteins and [3H]E2, at a fixed concentration of 0.5 nM, as the labeled ligand. Recombinant human ERalpha or ERbeta proteins were dissolved in binding buffer (10 mM Tris-HCL, pH 7.5, 10% glycerol, 1 mM DTT, 1 mg/ml BSA) and duplicate aliquots were then incubated with [3H]E2 at a final concentration of 0.5 nM, together with a vehicle control (0.4% DMSO), or the same amount of vehicle containing increasing concentrations of unlabeled steroid ligands as competitors. After incubation for 2 h at 25 C., the unbound ligands were removed and the amounts of [3H]E2 bound to either ERalpha or ERbeta proteins were measured. Inhibition Assay The method employed was adapted from the scientific literature and described in detail by Osboum et al. (1993, Biochemistry, 32, 6229-6236). Recombinant human ERalpha and ERbeta proteins were purified from transfected Sf9-cells. The in vitro assays involved the use of either ERalpha or ERbeta proteins and [3H]E2, at a fixed concentration of 0.5 nM, as the labeled ligand. Recombinant human ERalpha or ERbeta proteins were dissolved in binding buffer (10 mM Tris-HCL, pH 7.5, 10% glycerol, 1 mM DTT, 1 mg/ml BSA) and duplicate aliquots were then incubated with [3H]E2 at a final concentration of 0.5 nM, together with a vehicle control (0.4% DMSO), or the same amount of vehicle containing increasing concentrations of unlabeled steroid ligands as competitors. After incubation for 2 h at 25 C., the unbound ligands were removed and the amounts of [3H]E2 bound to either ERalpha or ERbeta proteins were measured. Evaluation of HSD17f313 Activity Assay Table 3: The liquid chromatography/mass spectrometry (LC/MS) estrone detection assay monitors the conversion of estradiol to estrone by HSD17B13. This assay was undertaken in a 96 wp format (Eppendorf deep well Plate 96/500) in an 80 μl reaction volume containing: 4 μM of Estradiol (E2; Cayman; Ser. No. 10/006,315), 6 mM NAD+(Sigma; #N0623) and 30 nM HSD17B13 enzyme (in-house; E. coli expressed His-tagged, purified, soluble protein) in a reaction containing 1M potassium phosphate buffer pH 7.4, with 0.5% vehicle (DMSO). Reactions were incubated for 2 hours at 26.5° C., and estradiol (E2) conversion to estrone (E1) was quantitated by LC-MS/MS based analyte detection for both E2 and E1 using LCMS grade reagents.Reactions were terminated by the addition of two volumes of acetonitrile (MeCN; LCMS grade; CAS #75/05/8) containing deuterated (D4)-E1 used as internal standard (Clear Synth; #CS-T-54273; 500 ng/mL final concentration). Samples were applied to pre-prepared Bond Elut-C18 extraction cartridges (3 mL; Agilent; Ser. No. 12/102,028), washed and eluted in MeCN. Eluates were dried under nitrogen and re-suspended in 60% methanol (LCMS grade methanol; CAS #67/56/1) before submission for analysis. Aqueous linearity for E2 and E1 were included for quantification. Pseudotyped HCV particle (HCVpp) reporter assay Plasmids expressing HCV E1 and E2 envelope proteins of GT1a H77 strain (Proc Natl Acad Sci USA 1997 94:8738-43) or GT1b Con1 strain (Science 1999 285:110-3) were constructed by cloning the nucleic acids encoding the last 60 amino acids of HCV core protein and all of the HCV E1 and E2 proteins into pcDNA3.1(+) vector. Plasmid pVSV-G expressing the glycoprotein G of the vesicular stomatitis virus (VSV G) is from Clontech (cat #631530). The HIV packaging construct expressing the firefly luciferase reporter gene was modified based on the envelope defective pNL.4.3.Luc-R .E vector (Virology 1995 206:935-44) by further deleting part of the HIV envelope protein. Enzyme Inhibition on the Conversion of E2 to E1 by 17beta-HSD1 For steady-state kinetic study of hybrid inhibitors, a Fluorolog 3 instrument was used to monitor the fluorescent signal of NADPH formed during estradiol oxidation, through which the substrate conversion is monitored. The excitation wavelength was 340 nm, and the emission wavelength was 460 nm. A standard curve monitoring the increase of fluorescence (cps) caused by an increase of NADPH revealed that the formation of 1 uM NADPH corresponds to a 316,000 cps increase. The Km for E2 in the absence of EM-1745, and four apparent Km values in the presence of 0.5, 2, 4, and 7 nM EM-1745 were obtained. Ki was further determined by a plot of Km vs. the inhibitor concentration. In Vitro Assay The method employed was adapted from the scientific literature and described in detail by Osbourn et al. (1993, Biochemistry, 32, 6229-6236). Recombinant human ERα and ERβ proteins were purified from transfected Sf9-cells. The in vitro assays involved the use of either ERα or ERβ proteins and [3H]E2, at a fixed concentration of 0.5 nM, as the labeled ligand. Recombinant human ERα or ERβ proteins were dissolved in binding buffer (10 mM Tris-HCL, pH 7.5, 10% glycerol, 1 mM DTT, 1 mg/ml BSA) and duplicate aliquots were then incubated with [3H]E2 at a final concentration of 0.5 nM, together with a vehicle control (0.4% DMSO), or the same amount of vehicle containing increasing concentrations of unlabeled steroid ligands as competitors. After incubation for 2 h at 25° C., the unbound ligands were removed and the amounts of [3H]E2 bound to either ERα or ERβ proteins were measured. The average amounts of [3H]E2 bound to either ERα or ERβ proteins at each concentration of competitor were used to make inhibition curves. IC50 values were subsequently determined by a non-linear, least squares regression analysis. Inhibition constants (Ki) were calculated using the equation of Cheng and Prusoff (Cheng et al., 1973, Biochem. Pharmacol., 22, 3099-3108), using the measured IC50 of the tested compounds, the concentration of radioligand employed in the assay, and the historical values for the Kd of the radioligand, which were established as 0.2 nM and 0.13 nM for ERα and ERβ, respectively. Biochemical Assay Through the Molecular Library Probe Production Center Network (MLPCN), at least 300,000 compounds were screened using a TR-FRET method, an ALPHASCREEN method or both, and tested for their ability to inhibit SUMOylation of a target protein via SUMO E1 or SUMO E2. The assays were based on SUMOylation of the target protein RanGAP1, which is a protein that is efficiently SUMOylated with only the SUMO E1 and E2 enzymes, and does not use E3 ligases. A fluorescence resonance energy transfer (FRET) assay was the primary assay followed by a chemoluminescence-based secondary assay using ALPHA screen to eliminate false positive hits. Then, the hits were screened by a poly-ubiquitination assay using ubiquitin, ubiquitin E1, Ubc5 and Apc11 to eliminate inhibitors not specific to SUMOylation. The screening identified a potent family of SUMOylation inhibitors based on a tricyclic scaffold. Estrone Detection Assay for Evaluation of HSD17I313 Activity Reactions were terminated by the addition of two volumes of acetonitrile (MeCN; LCMS grade; CAS #75/05/8) containing deuterated (D4)-E1 used as internal standard (Clear Synth; #CS-T-54273; 500 ng/mL final concentration). Samples were applied to pre-prepared Bond Elut-C18 extraction cartridges (3 mL; Agilent; Ser. No. 12/102,028), washed and eluted in MeCN. Eluates were dried under nitrogen and re-suspended in 60% methanol (LCMS grade methanol; CAS #67/56/1) before submission for analysis. Aqueous linearity for E2 and E1 were included for quantification.[0357]Analysis of samples was undertaken on a XBridge BEH C18 column (Waters; #186003033) using 0.1% Diethyl Amine in MeCN (mobile phase A; DEA CAS #109-89-7) and 0.1% Diethyl Amine in milli-Q water (mobile phase B) in a 3 min gradient allowing 25% B. Analytes were detected in negative mode using MRM analysis, with E2 having a RT of 1.85 min and E1 having a RT of 2 min. In Vitro Enzyme Activity and Drug Binding Ub-charging of E2 enzyme was performed for 90 min at 30° C. in a reaction containing 0.1 μM GST-E1, 3.3 μM E2/UbcH7, 10 μM FLAG-Ub and 0.188 μM PINK1 (all R&D systems). In a separate reaction 0.75 μM Parkin (Ubiquigent) was preincubated with 20 μM compound 4 (or equal volume of DMSO). Both reactions were diluted in Ub buffer (final concentration: 20 mM HEPES pH 7.2, 10 mM MgCl2, 0.1 mM EGTA, 500 μM TCEP and 10% ATP regeneration system (20 mM HEPES pH 7.6, 10 mM ATP, 300 mM phosphocreatine, 10 mM MgCl2, 10% glycerol, 1.5 mg/mL creatine phosphokinase (all Sigma Aldrich))). 1 U Apyrase (Sigma) was added per 9 μL reaction and both reactions were combined and incubated for an additional 30 min. 100 μL of 1×LDS buffer was added per 10 μL reaction and samples were split for −/+DTT (20 mM final). hPGDH Inhibitor Screening Biochemical Assay The in vitro biochemical assay can be performed in white, 384 plates in total 20 μl reaction volume consisting of 10 nM of 15-PGDH/HPGD (R&D System #5660-DH), 15 μM Prostaglandin E2 (Sigma, Cat #P5640-10MG) and 0.25 mM β-Nicotinamide adenine dinucleotide sodium salt (Sigma, Cat #N0632-5G) made in reaction buffer (50 mM Tris-HCl, pH 7.5, 0.01% Tween 20) at 10-point dose response curve for test/tool compounds. Briefly, 5 μl (4×) of compounds solution and 5 μl (final concentration, 10 nM) of enzyme solution is added to white 384 well plates and incubated for 10 mins at 37° C. 5 μl (4×) of Prostaglandin E2 and 5 μl (4×) of β-Nicotinamide adenine dinucleotide sodium salt is added to the wells and incubated for 10 mins at rt. Fluorescence is recorded at ex/em=340 nm/485 nm. The percentage (%) inhibition of enzyme activity was determined relative to positive control (1% DMSO) and IC50 was calculated using GraphPad prism software (four parameter-variable slope equation). Enzymatic Assay (Inhibition of Type 1 17beta-HSD) The enzymatic reaction was performed in the reaction buffer containing substrate, [14C]-estrone, and the test inhibitors. After the reaction, radiolabeled steroids were extracted from the reaction mixture, and solvent was evaporated to dryness. Steroids were dissolved and separated on TLC plates. Radioactivity signals were detected and quantified using a PhosphoImager (Sunny Vale, CA). The percentage of transformation of [14C]-E1 into [14C]-E2 was calculated. The IC50 values were calculated using an unweighted iterative least-squares method for four-parameter logistic curve fitting (DE50 program, CHUL Research Center, Quebec). Biochemical Assay A hydroxyprostaglandin dehydrogenase inhibition screening biochemical assay can be performed to assess the synthesized inhibitors provided herein. Provided herein is an exemplary biochemical assay for hPGDH inhibitor screening.The in vitro biochemical assay can be performed in white, 384 plates in total 20 μl reaction volume consisting of 10 nM of 15-PGDH/HPGD (R&D System #5660-DH), 15 μM Prostaglandin E2 (Sigma, Cat #P5640-10MG) and 0.25 mM (3-Nicotinamide adenine dinucleotide sodium salt (Sigma, Cat #N0632-5G) made in reaction buffer (50 mM Tris-HCl, pH 7.5, 0.01% Tween 20) at 10-point dose response curve for test/tool compounds. Briefly, 5 μl (4×) of compounds solution and 5 μl (final concentration, 10 nM) of enzyme solution is added to white 384 well plates and incubated for 10 mins at 37° C. 5 μl (4×) of Prostaglandin E2 and 5 μl (4×) of β-Nicotinamide adenine dinucleotide sodium salt is added to the wells and incubated for 10 mins at room temperature. Fluorescence is recorded at ex/em=340 nm/485 nm. The percentage (%) inhibition of enzyme activity was determined relative to positive control (1% DMSO) and IC50 was calculated using GraphPad prism software (four parameter-variable slope equation). Cell Response Assay The rat EP1 receptor-expressing cells were washed with the assay buffer. 100 μL of a fluorescent calcium indicator (Calcium kit II, Fluo 4 (Dojindo Laboratories).The intracellular calcium concentration was measured as a fluorescent signal using FDSS (registered trademark) 7000 (manufactured by Hamamatsu Photonics K. K.). 50 μL of each test compound (final concentrations: 1 nM to 10 μM) was added to each well after 20 seconds from initiating the reading of the fluorescent signal, and the fluorescence signal was measured for 60 seconds. Then, 50 μL of a prostaglandin E2 buffer solution was added to each well (final concentration 10 nM) and the fluorescence signal was measured for 60 seconds. Evaluation of HSD17B2 Activity Assay Table 2: The fluorescence based detection assay monitors the conversion of the co-factor NAD+ to NADH, which occurs coincident with the conversion of estradiol to estrone by HSD17B2. These assays were performed in 384 well plates (Greiner V-shape PP-microplate). The 20 μl reaction volume contained: 0.7 μM estradiol (E2); 6 mM NAD+ (Sigma); 250 nM HSD17B2 enzyme (Origene; Cat #TP303293); 0.25 M potassium phosphate buffer pH 7.4, 0.75% vehicle (DMSO). Reactions were incubated for 2 hours at 37° C., and the reaction was stopped by freezing the reaction plates at −80° C. Zero time samples were frozen immediately.The conversion of NAD+ to NADH was quantitated using NAD-Glo kits (Promega; Cat #G9062) according to the manufacturers' instructions. A volume of 15 μL of enzyme reaction mixture was added to 15 μL of reconstituted Glo reagent, and the plates were incubated for 30 minutes at room temperature. Luminescence was quantitated on a Tecan Spark Reader®. A standard curve of NADH (0.1-50 μL) in potassium phosphate buffer pH 7.4/1% DMSO was run on each plate.Activity of the enzyme in the absence of E2 was used to evaluate specificity of conversion. Enzyme activity in the presence of test samples was expressed as a percentage of the uninhibited enzyme in the presence of 1% DMSO vehicle only, and was plotted versus inhibitor concentration. Non-linear regression was performed using a four-parameter logistic model and GraphPad Prism software. Cell Response Assay The rat EP1 receptor-expressing cells were washed with the assay buffer. 100 μL of a fluorescent calcium indicator (Fluo-4 NW Calcium Assay Kit (Molecular Probes). The intracellular calcium concentration was measured as a fluorescent signal using FlexStation (registered trademark) (manufactured by Molecular Devices). 50 μL of each test compound that had been diluted with the assay buffer (final concentrations: 1 nM to 10 μM) was added to each well after 20 seconds from initiating the reading of the fluorescent signal, and the fluorescence signal was measured for 60 seconds. Then, 50 μL of a prostaglandin E2 buffer solution was added to each well (final concentration 10 nM) and the fluorescence signal was measured for 60 seconds. PDK Inhibition Assay To determine the IC50 for PDK inhibitors, a mixture containing 0.05-0.2 μM PDK, 6μM E1, with or without 0.5 μM of the PDC core E2/E3BP, and various amounts of inhibitor was incubated at 25 °C for 10 min in a buffer of 20 mM Tris-Cl, pH 7.5, 10 mM KCl, 5 mM MgCl2, 2 mM DTT, 0.02% (v/v) Tween 20, and 0.1 mg/ml bovine serumalbumin before the addition of 50 μM ATP to initiate the reaction. All inhibition titrations were performed at 10 dose points ranging from 31.6 to 1 mM in a 3.162-fold dilution series, with each inhibitor concentration tested in duplicate. The remaining steps were described previously [Wynn et al., J. Biol. Chem., 283:25305-25315]. Inhibitory Activity Assay Microsomes were prepared from COS-1 cells transiently transfected with a plasmid containing human mPGES-1 cDNA, and used as mPGES-1 enzyme. The mPGES-1 enzyme was diluted with a sodium phosphate buffer (pH 7.2) containing reduced glutathione (2.5 mM) and EDTA (1 mM), DMSO or a DMSO solution of a test compound (final concentration of DMSO was 1%) was added to the enzyme, and the mixture was preincubated at 4° C. for 15 minutes. Then, PGH2 as the substrate was added at a final concentration of 1 μM to start the enzymatic reaction, and after incubation at 4° C. for 4 minutes, a solution of ferric chloride (25 mM) and citric acid (50 mM) was added to terminate the enzymatic reaction. Generated PGE2 was measured by using Prostaglandin E2 Express EIA Kit (Cayman Chemical). Binding Assay Measurement of EP2 receptor binding action was performed in accordance with the method of Abramovitz et al. (Biochimica et Biophysica Acta, 1483, 285 (2000)). A test compound dissolved in dimethyl sulfoxide (final concentration: 1.0 (V/V) %) and [3H]prostaglandin E2 (NET-428, manufactured by PerkinElmer) (final concentration: 10 nM) were added to a buffer solution (10 mM MES-KOH (pH 6.0), 10 mM MgCl2, 1 mM EDTA) in which 10 μg of a membrane fraction of HEK293 cells expressing human EP2 receptor (ES-562-M, manufactured by Euroscreen) was suspended, and then incubated at 30° C. for 60 minutes. The membrane fraction was recovered on glass fiber filter paper (GF/B, manufactured by Whatman) using a cell harvester (M30R, manufactured by Brandel), and after washing with a buffer solution (10 mM MES-KOH (pH 6.0), 10 mM MgCl2), radioactivity was measured with a liquid scintillation analyzer (2000CA, manufactured by Packard). Pharmaceutical Assay To determine the HECT E3 ligase selectivity of the compounds, a panel of biochemical HECT E3 ligase autoubiquitinylation assays was employed (Smurf-1, Smurf-2, WWP1, WWP2, ITCH, Nedd4, Nedd4L and E6AP). The conjugation of ubiquitin to a protein substrate is a multistep process. In an initial ATP-requiring step, a thioester bond is formed between the carboxyl terminus of ubiquitin and an internal cystein residue of the ubiquitin-activating enzyme (E1). Activated ubiquitin is then transferred to a specific cystein residue of an ubiquitin-conjugating enzyme (E2). E2s donate ubiquitin to a HECT E3 ligase (E3) from which it is transferred to the substrate protein. HECT E3 ligases can auto-ubiquitinylate. This event is detected in the TR-FRET (Time-Resolved Fluorescence Resonance Energy Transfer) assay used in this panel. The reaction mix contains E1, E2, tagged-E3, biotin-conjugated ubiquitin, the compound and ATP in a suitable buffer and is incubated for 45 minutes to allow auto-ubiquitinylation of the E3 ligase. To measure the extent of ubiquitinylated E3 ligase by TR-FRET, the donor fluorophore Europium cryptate (Eu3+ cryptate), conjugated to streptavidin which subsequently binds to biotinylated ubiquitin, and the modified allophycocyanin XL665 (HTRF primary acceptor fluorophore) coupled to a tag-specific antibody (HA, His or GST), which recognizes the respective E3 ligase fusion proteins, are added after the reaction is complete. When these two fluorophores are brought together by a biomolecular interaction (in this case ubiquitinylation of the E3 ligase), a portion of the energy captured by the Cryptate during excitation is released through fluorescence emission at 620 nm, while the remaining energy is transferred to XL665. This energy is then released by XL665 as specific fluorescence at 665 nm. Light at 665 nm is emitted only through FRET with Europium. Because Europium Cryptate is present in the assay, light at 620 nm is detected even when the biomolecular interaction does not bring XL665 within close proximity. In Vitro PDHK1 Activity Assay The inhibitory potency of compounds of PDHK1 activity was determined in an enzymatic assay. Pyruvate dehydrogenase complex (PDHc) is an enzyme that catalyzes the reaction of pyruvate and a lipoamide to give the acetylated dihydrolipoamide and carbon dioxide. PDHc has three subunits E1, E2 and E3. E1 has 3 serine phosphorylation sites S293, S232, S300. If E1 was phosphorylated, PDH will be inactivated. PDHK can phosphorylate PDH protein (also named PDHE1) in PDH complex (PDC) to inactivate it. In the PDHK1 enzymatic assays, the final reaction mixture contains 1 nM of PDHK1 (in house purification from Escherichia coli), 25 nM of PDHE1 (in house purification from Escherichia coli) and 20 μM of ATP, and the experiment was reacted at 30° C. for 45 minutes. After adding detection reagent AlphaScreen Histidine (Nickel Chelate, from Perkin Elmer), and anti-phosphorylation s-293 antibody (from Abcam), Anti-Rabbit IgG Alpha (from Perkin Elmer)), the plate was incubated at 30° C. for 60 min before measuring the light change using a luminometer (Envision). Biochemical Assay For the biochemical assay panel, 50 nl of the test compounds, reference compounds and buffer/DMSO control are transferred to the respective wells of a 384-well white GREINER "SMALL VOLUME" PS plate. The assay panel is run at room temperature on a Biomek FX liquid handling workstation. To the assay plates containing 50 nl compound or control solutions in 90% DMSO, 4.5 ul of E3 ligase solution were added per well, followed by 4.5 ul of the pre-incubated E1/E2/Ub mix or the pre-diluted ubiquitin (LOW control). Plates are shaken vigorously after each addition. In this assay the compound concentrations range from 3 nM to 10 uM in an 8-point dose-response curve. After 45 min of incubation the ubiquitinylation reactions were stopped by adding 4.5 ul 2 mM NEM, immediately followed by 4.5 ul of a detection solution including the XL665-labeled antibody and the streptavidin-coupled europium to give a total volume of 18 ul. After an incubation time of 45 min in the dark, the plates are transferred into the Pherastar fluorescence reader to measure the TR-FRET signal. Detection of the Antagonism of the Human Prostaglandin D Receptor Signal 5. 1 Detection PrincipleBinding of prostaglandin D2 to the human PGD receptor induces activation of membrane-bound adenylate cyclases and leads to the formation of cAMP. In the presence of the phosphodiesterase inhibitor IBMX, the cAMP which has accumulated as a result of this stimulation and is released by cell lysis is employed in a competitive detection method. In this test, the cAMP present in the lysate competes with a fluorescently labelled cAMP (cAMP-d2) for binding to an anti-cAMP antibody labelled with an Eu cryptate.The absence of cellular cAMP leads to a maximum signal owing to this cAMP-d2 molecule binding to the antibody. Excitation of the cAMP-d2 molecule at 337 nm leads to a fluorescence resonance energy transfer (FRET) to the Eu cryptate molecules of the anti-cAMP antibody (labelled therewith), followed by a long-lasting emission signal at 665 nm (and also at 620 nM). The two signals are measured in a suitable measuring device in a time-resolved manner, i.e. once the background fluorescence has subsided. Any increase of the low FRET signal owing to prostglandin E2 administration (measured as change in the well ratio=emission665nm/emission620nm*10 000) indicates the action of antagonists.5.2. Detection Method5.2.1. Test for Antagonism (Figures Per Well of a 384-Well Plate):4 μl of a cAMP-d2/cell suspension (625 000 cells/ml) were added to a test plate with the substance solutions (0.05 μl; 100% DMSO, concentration range 0.8 nM-16.5 μM) already charged. After 20 minutes of pre-incubation at room temperature (RT), 2 μl of a 3×PGD2 solution (6 nM, in PBS-IBMX) were added and the mixture was incubated in the presence of the agonist for a further 30 min at RT (volume: 6 μl). The reaction was then stopped by addition of 2 μl of lysis buffer and the mixture was incubated at RT for a further 20 min prior to the actual measurement (volume: 8 μl). HBV Replication Inhibition Assay HBV replication inhibition by the compounds of this invention could be determined in cells infected or transfected with HBV, or cells with stably integrated HBV, such as HepG2.2.15 cells (Sells et al. 1987). In this example, HepG2.2.15 cells were maintained in cell culture medium containing 10% fetal bovine serum (FBS), Geneticin, L-glutamine, penicillin and streptomycin. HepG2.2.15 cells could be seeded in 96-well plates at a density of 40,000 cells/well and be treated with serially diluted compounds at a final DMSO concentration of 0.5% either alone or in combination by adding drugs in a checker box format. Cells were incubated with compounds for three days, after which medium was removed and fresh medium containing compounds was added to cells and incubated for another three days. At day 6, supernatant was removed and treated with DNase at 37° C. for 60 minutes, followed by enzyme inactivation at 75° C. for 15 minutes. Encapsidated HBV DNA was released from the virions and covalently linked HBV polymerase by incubating in lysis buffer (Affymetrix QS0010) containing 2.5 μg proteinase K at 50° C. for 40 minutes. HBV DNA was denatured by addition of 0.2 M NaOH and detected using a branched DNA (BDNA) QuantiGene assay kit according to manufacturer recommendation (Affymetrix). HBV DNA levels could also be quantified using qPCR, based on amplification of encapsidated HBV DNA extraction with QuickExtraction Solution (Epicentre Biotechnologies) and amplification of HBV DNA using HBV specific PCR probes that can hybridize to HBV DNA and a fluorescently labeled probe for quantitation. In addition, cell viability of HepG2.2.15 cells incubated with test compounds alone or in combination was determined by using CellTitre-Glo reagent according to the manufacturer protocol (Promega). The mean background signal from wells containing only culture medium was subtracted from all other samples, and percent inhibition at each compound concentration was calculated by normalizing to signals from HepG2.2.15 cells treated with 0.5% DMSO using equation E1.% inhibition=(DMSOave−Xi)/DMSOave×100% E1:where DMSOave is the mean signal calculated from the wells that were treated with DMSO control (0% inhibition control) and Xi is the signal measured from the individual wells. EC50 values, effective concentrations that achieved 50% inhibitory effect, were determined by non-linear fitting using Graphpad Prism software (San Diego, Calif.) and equation E2Y=Y min+(Y max−Y min)/(1+10(Log EC50−X)×HillSlope) E2:where Y represents percent inhibition values and X represents the logarithm of compound concentrations.
 US9878991, Compound E2 BDBM236119 US9365563, E2
US9878991, Compound E2 BDBM236119 US9365563, E2 BDBM286270 US11780853, Example E2 US9518060, Example E2 US11220518, Ex. No. E2
BDBM286270 US11780853, Example E2 US9518060, Example E2 US11220518, Ex. No. E2 US10584127, Compound E2-1.2 US10294230, Compound E2-1.2 BDBM388295 US11136328, Compound E2-1.2 US11752155, Compound E2-1.2B'
US10584127, Compound E2-1.2 US10294230, Compound E2-1.2 BDBM388295 US11136328, Compound E2-1.2 US11752155, Compound E2-1.2B' BDBM168073 US9073931, E2
BDBM168073 US9073931, E2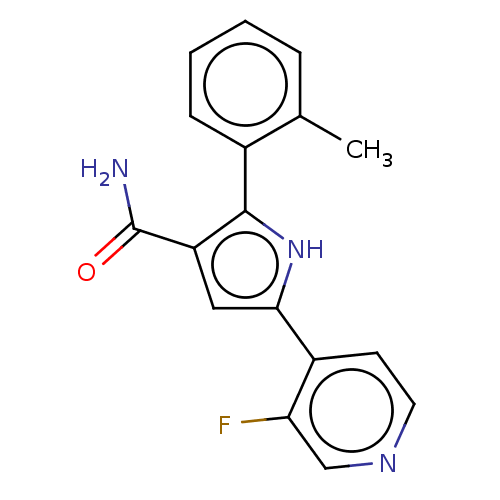 BDBM187245 US9670191, E2
BDBM187245 US9670191, E2 BDBM229618 US9340506, E2
BDBM229618 US9340506, E2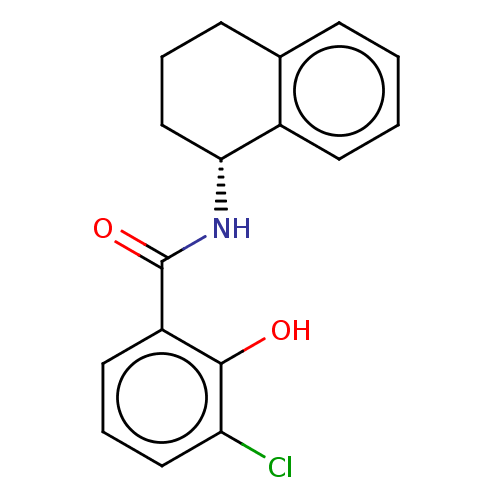 BDBM253797 US9459250, E2
BDBM253797 US9459250, E2 BDBM36490 E2-NH3+
BDBM36490 E2-NH3+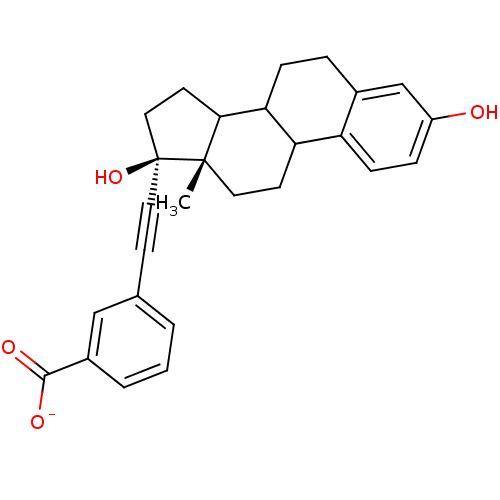 BDBM36491 E2-COO-
BDBM36491 E2-COO-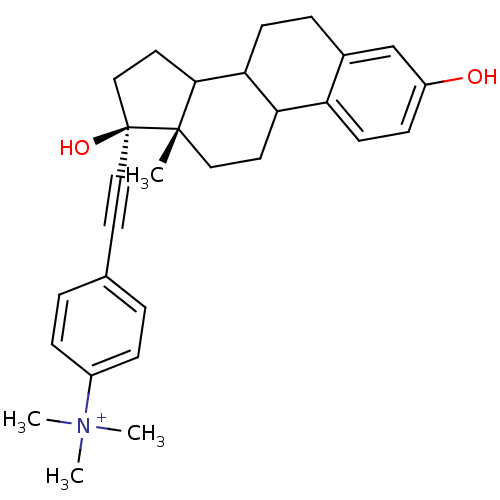 BDBM36492 E2-NMe3+
BDBM36492 E2-NMe3+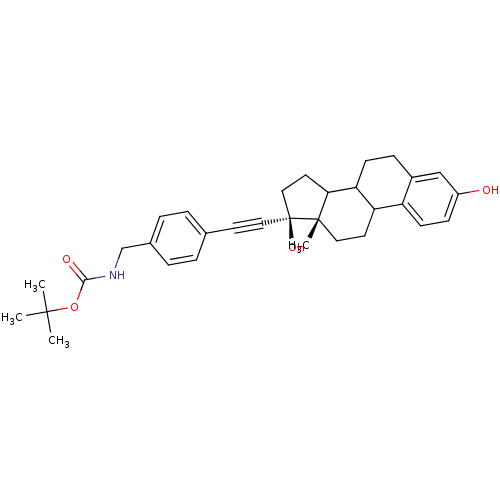 BDBM36493 E2-NB
BDBM36493 E2-NB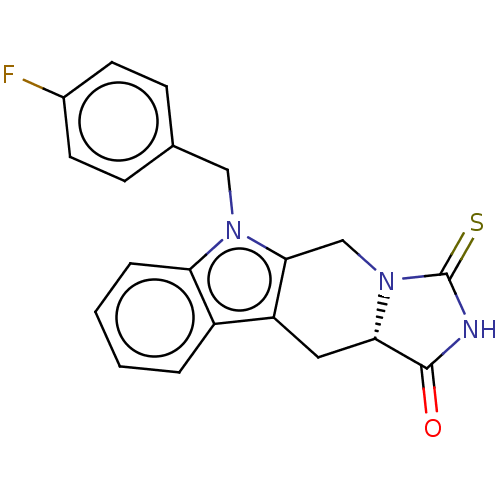 BDBM380539 US9926318, E2
BDBM380539 US9926318, E2 US8999981, E2 BDBM153932
US8999981, E2 BDBM153932 US9416126, E2 BDBM239259
US9416126, E2 BDBM239259 BDBM519243 US11116757, No. E2
BDBM519243 US11116757, No. E2 BDBM567414 US11420968, Example E2
BDBM567414 US11420968, Example E2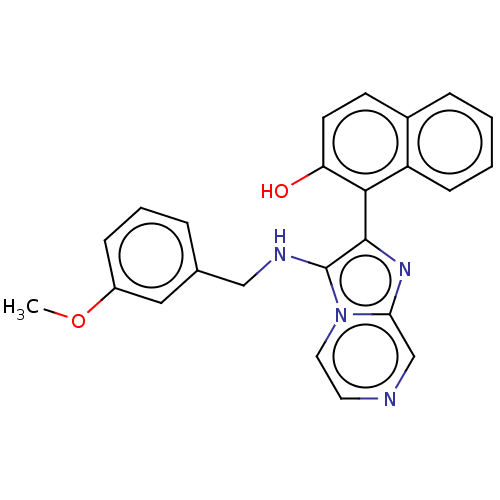 BDBM585372 US11530209, Compound E2
BDBM585372 US11530209, Compound E2 BDBM703001 US20240366813, Compound E2
BDBM703001 US20240366813, Compound E2 BDBM720105 US20250051321, Compound E2
BDBM720105 US20250051321, Compound E2 BDBM748526 US20250188030, Compound E2
BDBM748526 US20250188030, Compound E2 CHEMBL505589 BDBM50259922 Cyclotheonamide E2
CHEMBL505589 BDBM50259922 Cyclotheonamide E2 US10570077, Compound E2 BDBM435383
US10570077, Compound E2 BDBM435383 US11291655, No E2 BDBM546414
US11291655, No E2 BDBM546414 BDBM361983 US10221182, Compound E2-1.5
BDBM361983 US10221182, Compound E2-1.5 BDBM361993 US10221182, Compound E2-1.4
BDBM361993 US10221182, Compound E2-1.4 BDBM362002 US10221182, Compound E2-1.7
BDBM362002 US10221182, Compound E2-1.7 BDBM362004 US10221182, Compound E2-1.16
BDBM362004 US10221182, Compound E2-1.16 BDBM433562 US10562878, Compound 170 E2
BDBM433562 US10562878, Compound 170 E2 BDBM433567 US10562878, Compound 172 E2
BDBM433567 US10562878, Compound 172 E2 BDBM433614 US10562878, Compound 134 E2
BDBM433614 US10562878, Compound 134 E2 BDBM433619 US10562878, Compound 165 E2
BDBM433619 US10562878, Compound 165 E2 BDBM433621 US10562878, Compound 166 E2
BDBM433621 US10562878, Compound 166 E2 BDBM433623 US10562878, Compound 173 E2
BDBM433623 US10562878, Compound 173 E2 BDBM433628 US10562878, Compound 152 E2
BDBM433628 US10562878, Compound 152 E2 BDBM433630 US10562878, Compound 153 E2
BDBM433630 US10562878, Compound 153 E2 BDBM433649 US10562878, Compound 184 E2
BDBM433649 US10562878, Compound 184 E2 BDBM433651 US10562878, Compound 187 E2
BDBM433651 US10562878, Compound 187 E2 BDBM433730 US10562878, Compound 205 E2
BDBM433730 US10562878, Compound 205 E2 BDBM433737 US10562878, Compound 147 E2
BDBM433737 US10562878, Compound 147 E2 BDBM433739 US10562878, Compound 148 E2
BDBM433739 US10562878, Compound 148 E2 BDBM433749 US10562878, Compound 164 E2
BDBM433749 US10562878, Compound 164 E2 BDBM433772 US10562878, Compound 140 E2
BDBM433772 US10562878, Compound 140 E2 BDBM433774 US10562878, Compound 139 E2
BDBM433774 US10562878, Compound 139 E2 BDBM433777 US10562878, Compound 20 E2
BDBM433777 US10562878, Compound 20 E2 BDBM433786 US10562878, Compound 155 E2
BDBM433786 US10562878, Compound 155 E2 BDBM433788 US10562878, Compound 136 E2
BDBM433788 US10562878, Compound 136 E2 BDBM433792 US10562878, Compound 21 E2
BDBM433792 US10562878, Compound 21 E2 BDBM433806 US10562878, Compound 22 E2
BDBM433806 US10562878, Compound 22 E2 BDBM433815 US10562878, Compound 154 E2
BDBM433815 US10562878, Compound 154 E2 BDBM433841 US10562878, Compound 234 E2
BDBM433841 US10562878, Compound 234 E2 BDBM433881 US10562878, Compound 142 E2
BDBM433881 US10562878, Compound 142 E2 BDBM433893 US10562878, Compound 145 E2
BDBM433893 US10562878, Compound 145 E2 BDBM433895 US10562878, Compound 146 E2
BDBM433895 US10562878, Compound 146 E2 BDBM433907 US10562878, Compound 161 E2
BDBM433907 US10562878, Compound 161 E2 BDBM433913 US10562878, Compound 143 E2
BDBM433913 US10562878, Compound 143 E2 BDBM433915 US10562878, Compound 144 E2
BDBM433915 US10562878, Compound 144 E2 BDBM433917 US10562878, Compound 159 E2
BDBM433917 US10562878, Compound 159 E2 BDBM433919 US10562878, Compound 160 E2
BDBM433919 US10562878, Compound 160 E2 BDBM595470 US11591313, Compound 038-E2
BDBM595470 US11591313, Compound 038-E2 BDBM595473 US11591313, Compound 039-E2
BDBM595473 US11591313, Compound 039-E2 BDBM617052 US11752155, Compound E2-22.2B'
BDBM617052 US11752155, Compound E2-22.2B' BDBM617053 US11752155, Compound E2-22.4B'
BDBM617053 US11752155, Compound E2-22.4B' BDBM617054 US11752155, Compound E2-22.6B'
BDBM617054 US11752155, Compound E2-22.6B'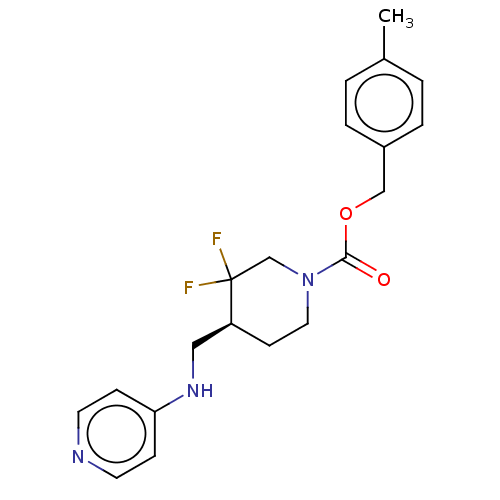 BDBM617055 US11752155, Compound E2-38.2B'
BDBM617055 US11752155, Compound E2-38.2B' BDBM701606 US12128028, Example 2 (E2)
BDBM701606 US12128028, Example 2 (E2) US10221182, Compound E2-1.3 BDBM361990
US10221182, Compound E2-1.3 BDBM361990 US10221182, Compound E2-1.6 BDBM362000
US10221182, Compound E2-1.6 BDBM362000 US10562878, Compound 133 E2 BDBM433612
US10562878, Compound 133 E2 BDBM433612 US10562878, Compound 138 E2 BDBM433790
US10562878, Compound 138 E2 BDBM433790 US10562878, Compound 141 E2 BDBM433879
US10562878, Compound 141 E2 BDBM433879 US10562878, Compound 156 E2 BDBM433802
US10562878, Compound 156 E2 BDBM433802 US10562878, Compound 157 E2 BDBM433889
US10562878, Compound 157 E2 BDBM433889 US10562878, Compound 158 E2 BDBM433891
US10562878, Compound 158 E2 BDBM433891 US10562878, Compound 163 E2 BDBM433745
US10562878, Compound 163 E2 BDBM433745 US10562878, Compound 185 E2 BDBM433658
US10562878, Compound 185 E2 BDBM433658 US10562878, Compound 188 E2 BDBM433667
US10562878, Compound 188 E2 BDBM433667 US10562878, Compound 195 E2 BDBM433750
US10562878, Compound 195 E2 BDBM433750 US10562878, Compound 206 E2 BDBM433735
US10562878, Compound 206 E2 BDBM433735 US10562878, Compound 231 E2 BDBM433821
US10562878, Compound 231 E2 BDBM433821 US10562878, Compound 60 E2 BDBM433828
US10562878, Compound 60 E2 BDBM433828 US10562878, Compound 61 E2 BDBM433832
US10562878, Compound 61 E2 BDBM433832 US10562878, Compound 62 E2 BDBM433837
US10562878, Compound 62 E2 BDBM433837 US10562878, Compound 64 E2 BDBM433844
US10562878, Compound 64 E2 BDBM433844 US11591313, Compound 040-E2 BDBM595475
US11591313, Compound 040-E2 BDBM595475 US20240246927, Patent ID E2 BDBM686266
US20240246927, Patent ID E2 BDBM686266 US10562878, Compound 163 E2 US10562878, Compound 163 E1 US10562878, Compound 148 E1 US10562878, Compound 147 E2 US10562878, Compound 147 E1 US10562878, Compound 148 E2 US10562878, Compound 164 E1 US10562878, Compound 164 E2 BDBM433736
US10562878, Compound 163 E2 US10562878, Compound 163 E1 US10562878, Compound 148 E1 US10562878, Compound 147 E2 US10562878, Compound 147 E1 US10562878, Compound 148 E2 US10562878, Compound 164 E1 US10562878, Compound 164 E2 BDBM433736 17beta-estradiol (E2) 17α-ethinylestradiol Ovocyclin US9561238, E2 [2,4,6,7-3H]-17beta-estradiol CHEMBL135 (1S,10R,11S,14S,15S)-15-methyltetracyclo[8.7.0.0^{2,7}.0^{11,15}]heptadeca-2,4,6-triene-5,14-diol US9034854, E2 CS336 [3H]]estradiol [2,4,6,7-3H]-E2 ESTRADIOL US9422324, E2 BDBM17292 [3H]-estradiol US9040509, E2 17 beta-Estradiol Estradiol-17 alpha
17beta-estradiol (E2) 17α-ethinylestradiol Ovocyclin US9561238, E2 [2,4,6,7-3H]-17beta-estradiol CHEMBL135 (1S,10R,11S,14S,15S)-15-methyltetracyclo[8.7.0.0^{2,7}.0^{11,15}]heptadeca-2,4,6-triene-5,14-diol US9034854, E2 CS336 [3H]]estradiol [2,4,6,7-3H]-E2 ESTRADIOL US9422324, E2 BDBM17292 [3H]-estradiol US9040509, E2 17 beta-Estradiol Estradiol-17 alpha US10562878, Compound 144 E1 US10562878, Compound 159 E1 US10562878, Compound 159 E2 US10562878, Compound 143 E2 BDBM433912 US10562878, Compound 143 E1 US10562878, Compound 144 E2
US10562878, Compound 144 E1 US10562878, Compound 159 E1 US10562878, Compound 159 E2 US10562878, Compound 143 E2 BDBM433912 US10562878, Compound 143 E1 US10562878, Compound 144 E2 US10562878, Compound 153 E1 BDBM433613 US10562878, Compound 134 E2 US10562878, Compound 152 E2 US10562878, Compound 134 E1 US10562878, Compound 152 E1 US10562878, Compound 153 E2
US10562878, Compound 153 E1 BDBM433613 US10562878, Compound 134 E2 US10562878, Compound 152 E2 US10562878, Compound 134 E1 US10562878, Compound 152 E1 US10562878, Compound 153 E2 BDBM433633 US10562878, Compound cis 121 E2
BDBM433633 US10562878, Compound cis 121 E2 BDBM433708 US10562878, Compound trans 124 E2
BDBM433708 US10562878, Compound trans 124 E2 BDBM433717 US10562878, Compound trans 125 E2
BDBM433717 US10562878, Compound trans 125 E2 BDBM559876 US11375716, Formula Ia-iii-e2
BDBM559876 US11375716, Formula Ia-iii-e2 BDBM559878 US11375716, Formula Ia-xiv-e2
BDBM559878 US11375716, Formula Ia-xiv-e2 US10562878, Compound trans 121 E2 BDBM433635
US10562878, Compound trans 121 E2 BDBM433635 US11375716, Formula Ia-ii-e2 BDBM559874
US11375716, Formula Ia-ii-e2 BDBM559874 (E)-N-(2-aminophenyl)-3-(2-cinnamylthiazol-4-yl)acrylamide BDBM375766 US10301323, Compound E2 US9908899, Compound E2
(E)-N-(2-aminophenyl)-3-(2-cinnamylthiazol-4-yl)acrylamide BDBM375766 US10301323, Compound E2 US9908899, Compound E2 BDBM433771 US10562878, Compound 140 E2 US10562878, Compound 139 E1 US10562878, Compound 140 E1 US10562878, Compound 139 E2
BDBM433771 US10562878, Compound 140 E2 US10562878, Compound 139 E1 US10562878, Compound 140 E1 US10562878, Compound 139 E2 BDBM433892 US10562878, Compound 146 E1 US10562878, Compound 145 E1 US10562878, Compound 146 E2 US10562878, Compound 145 E2
BDBM433892 US10562878, Compound 146 E1 US10562878, Compound 145 E1 US10562878, Compound 146 E2 US10562878, Compound 145 E2 US10562878, Compound 141 E1 US10562878, Compound 142 E2 US10562878, Compound 141 E2 BDBM433878 US10562878, Compound 142 E1
US10562878, Compound 141 E1 US10562878, Compound 142 E2 US10562878, Compound 141 E2 BDBM433878 US10562878, Compound 142 E1 US10562878, Compound 156 E1 BDBM433785 US10562878, Compound 155 E1 US10562878, Compound 156 E2 US10562878, Compound 155 E2
US10562878, Compound 156 E1 BDBM433785 US10562878, Compound 155 E1 US10562878, Compound 156 E2 US10562878, Compound 155 E2 US10562878, Compound 157 E2 US10562878, Compound 157 E1 US10562878, Compound 158 E2 US10562878, Compound 158 E1 BDBM433888
US10562878, Compound 157 E2 US10562878, Compound 157 E1 US10562878, Compound 158 E2 US10562878, Compound 158 E1 BDBM433888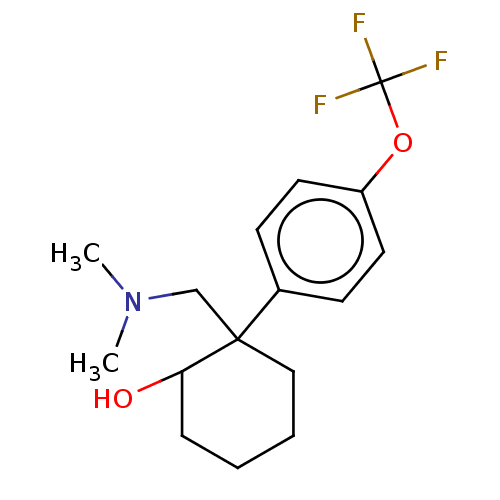 US10562878, Compound 162 E1 BDBM433905 US10562878, Compound 161 E1 US10562878, Compound 161 E2 US10562878, Compound 162 E2
US10562878, Compound 162 E1 BDBM433905 US10562878, Compound 161 E1 US10562878, Compound 161 E2 US10562878, Compound 162 E2 US10562878, Compound 166 E1 BDBM433618 US10562878, Compound 165 E1 US10562878, Compound 166 E2 US10562878, Compound 165 E2
US10562878, Compound 166 E1 BDBM433618 US10562878, Compound 165 E1 US10562878, Compound 166 E2 US10562878, Compound 165 E2 US10562878, Compound 185 E1 US10562878, Compound 184 E1 US10562878, Compound 185 E2 US10562878, Compound 184 E2 BDBM433648
US10562878, Compound 185 E1 US10562878, Compound 184 E1 US10562878, Compound 185 E2 US10562878, Compound 184 E2 BDBM433648 BDBM218873 US9303033, E2, Table 6A, Compound 12
BDBM218873 US9303033, E2, Table 6A, Compound 12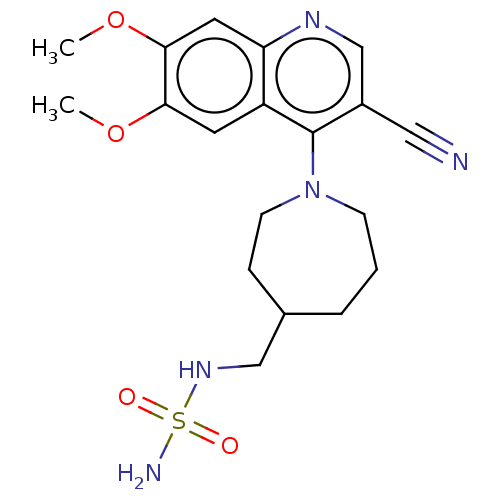 US11591313, Compound 039 BDBM595471 US11591313, Compound 039-E2
US11591313, Compound 039 BDBM595471 US11591313, Compound 039-E2 US10562878, Compound 202 US10562878, Compound 188 E1 BDBM433650 US10562878, Compound 187 E2 US10562878, Compound 187 E1 US10562878, Compound 188 E2
US10562878, Compound 202 US10562878, Compound 188 E1 BDBM433650 US10562878, Compound 187 E2 US10562878, Compound 187 E1 US10562878, Compound 188 E2 US10562878, Compound trans 121 E1 US10562878, Compound trans 121 E2 BDBM433632 US10562878, Compound cis 121 E2 US10562878, Compound cis 121 E1
US10562878, Compound trans 121 E1 US10562878, Compound trans 121 E2 BDBM433632 US10562878, Compound cis 121 E2 US10562878, Compound cis 121 E1 BDBM361982 US10221182, Compound E2-1.5 US10221182, Compound E1-1.5
BDBM361982 US10221182, Compound E2-1.5 US10221182, Compound E1-1.5 BDBM361992 US10221182, Compound E2-1.4 US10221182, Compound E1-1.4
BDBM361992 US10221182, Compound E2-1.4 US10221182, Compound E1-1.4 BDBM362003 US10221182, Compound E1-1.16 US10221182, Compound E2-1.16
BDBM362003 US10221182, Compound E1-1.16 US10221182, Compound E2-1.16 BDBM433561 US10562878, Compound 170 E1 US10562878, Compound 170 E2
BDBM433561 US10562878, Compound 170 E1 US10562878, Compound 170 E2 BDBM433566 US10562878, Compound 172 E2 US10562878, Compound 172 E1
BDBM433566 US10562878, Compound 172 E2 US10562878, Compound 172 E1 BDBM433787 US10562878, Compound 136 E1 US10562878, Compound 136 E2
BDBM433787 US10562878, Compound 136 E1 US10562878, Compound 136 E2 BDBM433814 US10562878, Compound 154 E2 US10562878, Compound 154 E1
BDBM433814 US10562878, Compound 154 E2 US10562878, Compound 154 E1 BDBM433820 US10562878, Compound 231 E2 US10562878, Compound 231 E1
BDBM433820 US10562878, Compound 231 E2 US10562878, Compound 231 E1 BDBM433830 US10562878, Compound 59 E2 US10562878, Compound 59 E1
BDBM433830 US10562878, Compound 59 E2 US10562878, Compound 59 E1 BDBM433840 US10562878, Compound 234 E2 US10562878, Compound 234 E1
BDBM433840 US10562878, Compound 234 E2 US10562878, Compound 234 E1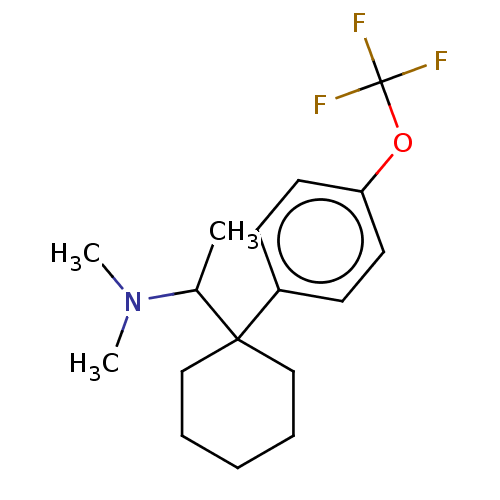 BDBM433842 US10562878, Compound 63 E2 US10562878, Compound 63 E1
BDBM433842 US10562878, Compound 63 E2 US10562878, Compound 63 E1 BDBM433918 US10562878, Compound 160 E2 US10562878, Compound 160 E1
BDBM433918 US10562878, Compound 160 E2 US10562878, Compound 160 E1 US10221182, Compound E1-1.3 US10221182, Compound E2-1.3 BDBM361989
US10221182, Compound E1-1.3 US10221182, Compound E2-1.3 BDBM361989 US10221182, Compound E2-1.6 BDBM361999 US10221182, Compound E1-1.6
US10221182, Compound E2-1.6 BDBM361999 US10221182, Compound E1-1.6 US10221182, Compound E2-1.7 US10221182, Compound E1-1.7 BDBM362001
US10221182, Compound E2-1.7 US10221182, Compound E1-1.7 BDBM362001 US10562878, Compound 133 E2 BDBM433611 US10562878, Compound 133 E1
US10562878, Compound 133 E2 BDBM433611 US10562878, Compound 133 E1 US10562878, Compound 138 E2 US10562878, Compound 138 E1 BDBM433789
US10562878, Compound 138 E2 US10562878, Compound 138 E1 BDBM433789 US10562878, Compound 169 E1 US10562878, Compound 169 E2 BDBM433625
US10562878, Compound 169 E1 US10562878, Compound 169 E2 BDBM433625 US10562878, Compound 193 E1 US10562878, Compound 193 E2 BDBM433793
US10562878, Compound 193 E1 US10562878, Compound 193 E2 BDBM433793 US10562878, Compound 205 E1 US10562878, Compound 205 E2 BDBM433729
US10562878, Compound 205 E1 US10562878, Compound 205 E2 BDBM433729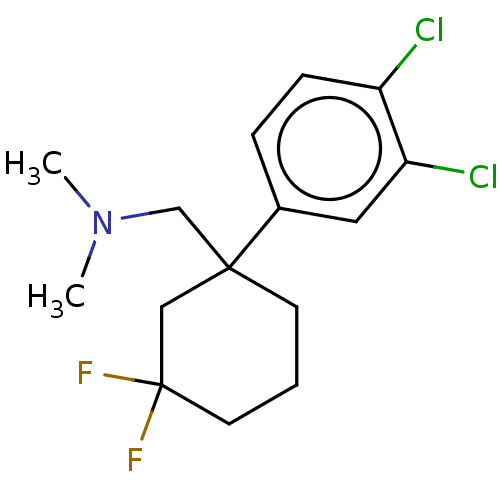 US10562878, Compound 206 E2 US10562878, Compound 206 E1 BDBM433734
US10562878, Compound 206 E2 US10562878, Compound 206 E1 BDBM433734 US10562878, Compound 60 E2 US10562878, Compound 60 E1 BDBM433827
US10562878, Compound 60 E2 US10562878, Compound 60 E1 BDBM433827 US10562878, Compound 61 E2 US10562878, Compound 61 E1 BDBM433829
US10562878, Compound 61 E2 US10562878, Compound 61 E1 BDBM433829 US10562878, Compound 62 E1 US10562878, Compound 62 E2 BDBM433836
US10562878, Compound 62 E1 US10562878, Compound 62 E2 BDBM433836 US10562878, Compound 64 E2 US10562878, Compound 64 E1 BDBM433843
US10562878, Compound 64 E2 US10562878, Compound 64 E1 BDBM433843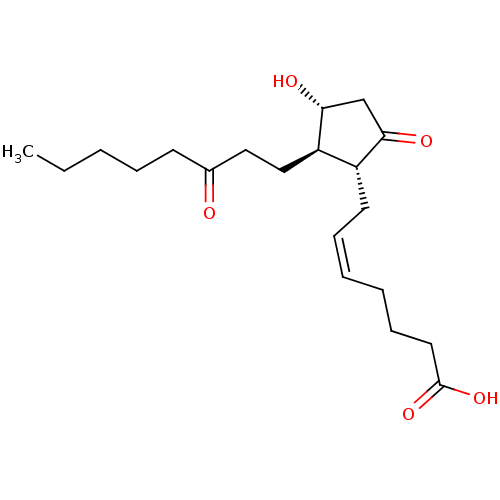 BDBM82093 PGE2 13,14-dihydro CAS_363-23-5 13,14-DIHYDRO-15-KETO PROSTAGLANDIN E2 13,14-dihydro-15-oxo-prostaglandin E2 PGE2, 13,14-dihydro15-oxo
BDBM82093 PGE2 13,14-dihydro CAS_363-23-5 13,14-DIHYDRO-15-KETO PROSTAGLANDIN E2 13,14-dihydro-15-oxo-prostaglandin E2 PGE2, 13,14-dihydro15-oxo 17-PHENYL TRINOR PROSTAGLANDIN E2 PGE2,17-Phenyl BDBM82094 CAS_38315-43-4
17-PHENYL TRINOR PROSTAGLANDIN E2 PGE2,17-Phenyl BDBM82094 CAS_38315-43-4 BDBM433775 US10562878, Compound 20 US10562878, Compound 20 E2 US10562878, Compound 20 E1
BDBM433775 US10562878, Compound 20 US10562878, Compound 20 E2 US10562878, Compound 20 E1 BDBM433784 US10562878, Compound 22 US10562878, Compound 22 E1 US10562878, Compound 22 E2
BDBM433784 US10562878, Compound 22 US10562878, Compound 22 E1 US10562878, Compound 22 E2 US10562878, Compound 173 E1 US10562878, Compound 173 US10562878, Compound 173 E2 BDBM433615
US10562878, Compound 173 E1 US10562878, Compound 173 US10562878, Compound 173 E2 BDBM433615 US10562878, Compound 21 E1 BDBM433778 US10562878, Compound 21 US10562878, Compound 21 E2
US10562878, Compound 21 E1 BDBM433778 US10562878, Compound 21 US10562878, Compound 21 E2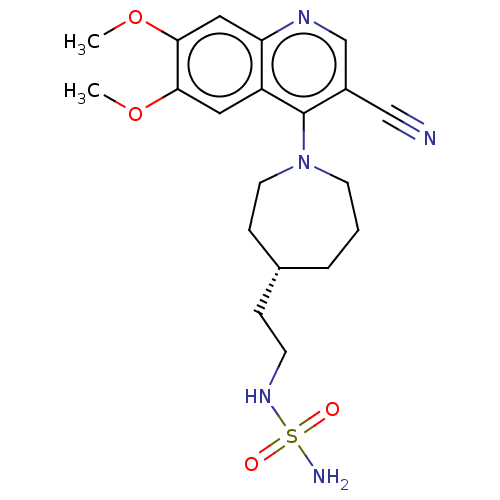 US11591313, Compound 038-E1 BDBM595468 US11591313, Compound 038 US11591313, Compound 038-E2
US11591313, Compound 038-E1 BDBM595468 US11591313, Compound 038 US11591313, Compound 038-E2 BDBM50337905 CHEMBL1684051 US10562878, Compound 19 E2 1-(1-(4-(trifluoromethoxy)phenyl)cyclohexyl)ethanamine
BDBM50337905 CHEMBL1684051 US10562878, Compound 19 E2 1-(1-(4-(trifluoromethoxy)phenyl)cyclohexyl)ethanamine CHEMBL1684049 BDBM50337903 1-(1-(naphthalen-1-yl)cyclohexyl)ethanamine US10562878, Compound 16 E2
CHEMBL1684049 BDBM50337903 1-(1-(naphthalen-1-yl)cyclohexyl)ethanamine US10562878, Compound 16 E2 BDBM50337914 CHEMBL1684043 US10562878, Compound 17 E2 (+/-)-1-(1-(3,4-dichlorophenyl)cyclohexyl)propan-1-amine
BDBM50337914 CHEMBL1684043 US10562878, Compound 17 E2 (+/-)-1-(1-(3,4-dichlorophenyl)cyclohexyl)propan-1-amine Carboxylic acid deriv. E2 BDBM11794 2-{[(3-chlorophenyl)carbamoyl][(2-chlorophenyl)methyl]amino}propanoic acid
Carboxylic acid deriv. E2 BDBM11794 2-{[(3-chlorophenyl)carbamoyl][(2-chlorophenyl)methyl]amino}propanoic acid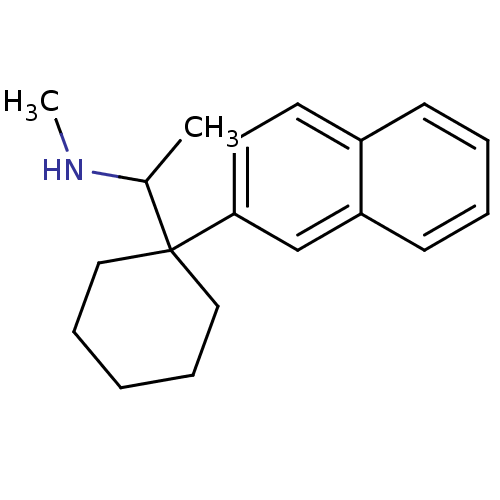 N-methyl-1-(1-(naphthalen-2-yl)cyclohexyl)ethanamine BDBM50337912 US10562878, Compound 58 E2 CHEMBL1684056
N-methyl-1-(1-(naphthalen-2-yl)cyclohexyl)ethanamine BDBM50337912 US10562878, Compound 58 E2 CHEMBL1684056 US10562878, Compound cis 124 BDBM433706 US10562878, Compound trans 124 E1 US10562878, Compound trans 124 E2
US10562878, Compound cis 124 BDBM433706 US10562878, Compound trans 124 E1 US10562878, Compound trans 124 E2 US10562878, Compound cis 125 BDBM433715 US10562878, Compound trans 125 E1 US10562878, Compound trans 125 E2
US10562878, Compound cis 125 BDBM433715 US10562878, Compound trans 125 E1 US10562878, Compound trans 125 E2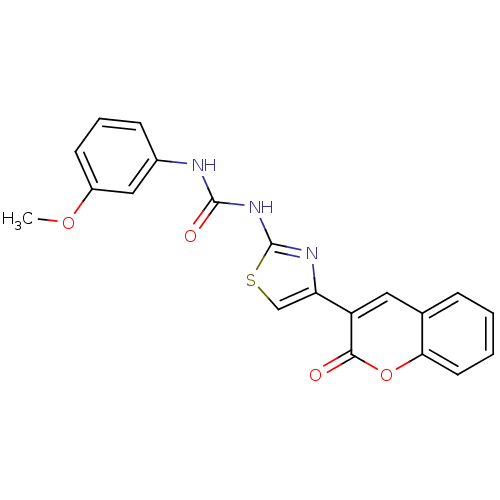 BDBM152438 1-(3-Methoxyphenyl)-3-(4-(2-oxo-2H-chromen-3-yl)thiazol-2-yl)urea (e2)
BDBM152438 1-(3-Methoxyphenyl)-3-(4-(2-oxo-2H-chromen-3-yl)thiazol-2-yl)urea (e2)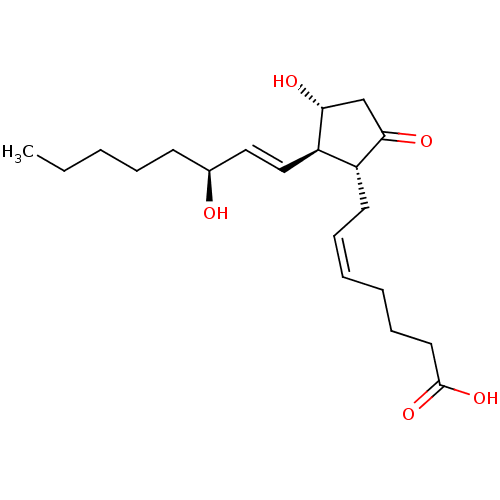 BDBM35847 [3H]Prostaglandin E2 PGE2 CHEMBL548 DINOPROSTONE (15S)-prostaglandin E2 prostaglandin E2 [3H]PGE2 (E,Z)-(1R,2R,3R)-7-(3-Hydroxy-2-((3S)-(3-hydroxy-1-octenyl))-5-oxocyclopentyl)-5-heptenoic acid [3H]Dinoprostone (5Z,11alpha,13E,15S)-11,15-dihydroxy-9-oxoprosta-5,13-dien-1-oic acid (5Z,13E,15S)-11alpha,15-dihydroxy-9-oxoprosta-5,13-dien-1-oic acid
BDBM35847 [3H]Prostaglandin E2 PGE2 CHEMBL548 DINOPROSTONE (15S)-prostaglandin E2 prostaglandin E2 [3H]PGE2 (E,Z)-(1R,2R,3R)-7-(3-Hydroxy-2-((3S)-(3-hydroxy-1-octenyl))-5-oxocyclopentyl)-5-heptenoic acid [3H]Dinoprostone (5Z,11alpha,13E,15S)-11,15-dihydroxy-9-oxoprosta-5,13-dien-1-oic acid (5Z,13E,15S)-11alpha,15-dihydroxy-9-oxoprosta-5,13-dien-1-oic acid BDBM387142 US9938276, E2 2-((3aR, 7aR)-6-(2- propylbenzoyl)octahydro-1H-pyrrolo[2,3- c]pyridin-1-yl)isonicotinonitrile
BDBM387142 US9938276, E2 2-((3aR, 7aR)-6-(2- propylbenzoyl)octahydro-1H-pyrrolo[2,3- c]pyridin-1-yl)isonicotinonitrile US10457675, Example 88 US10065948, 88 BDBM272152 E2-(abs)-N-hydroxy-1-phenyl-2,3-dihydro-1H-indene- 1-carboxamide
US10457675, Example 88 US10065948, 88 BDBM272152 E2-(abs)-N-hydroxy-1-phenyl-2,3-dihydro-1H-indene- 1-carboxamide US10562878, Compound 2 E2 (+/-)-1-(1-(3,4-dichlorophenyl)cyclohexyl)ethanamine CHEMBL1684042 1-(1-(3,4-dichlorophenyl)cyclohexyl)ethanamine BDBM50337909
US10562878, Compound 2 E2 (+/-)-1-(1-(3,4-dichlorophenyl)cyclohexyl)ethanamine CHEMBL1684042 1-(1-(3,4-dichlorophenyl)cyclohexyl)ethanamine BDBM50337909 US11447505, Example E2 5-methyl-4-(1-piperidyl)- 3-(1H-pyrazol-4-yl)-1H- pyrrolo[2,3-b]pyridine BDBM572413
US11447505, Example E2 5-methyl-4-(1-piperidyl)- 3-(1H-pyrazol-4-yl)-1H- pyrrolo[2,3-b]pyridine BDBM572413 (+/-)-1-(1-(naphthalen-2-yl)cyclohexyl)ethanamine 1-(1-(naphthalen-2-yl)cyclohexyl)ethanamine BDBM50337904 CHEMBL1684050 US10562878, Compound 13 E2
(+/-)-1-(1-(naphthalen-2-yl)cyclohexyl)ethanamine 1-(1-(naphthalen-2-yl)cyclohexyl)ethanamine BDBM50337904 CHEMBL1684050 US10562878, Compound 13 E2 US10065948, 32 E2-(abs)-5-(3-Fluoro-2-methylphenyl)-N-hydroxy-1- phenyl-4,5,6,7-tetrahydro-1H-indazole-5-carboxamide BDBM272096
US10065948, 32 E2-(abs)-5-(3-Fluoro-2-methylphenyl)-N-hydroxy-1- phenyl-4,5,6,7-tetrahydro-1H-indazole-5-carboxamide BDBM272096 1-[4-(3-fluorophenoxy)-6-(trifluoromethyl)pyrimidin-2-yl]-4-(pyrrolidin-1-ylmethyl)piperidin-4-ol (E2) US9920053, Example 2 BDBM376872
1-[4-(3-fluorophenoxy)-6-(trifluoromethyl)pyrimidin-2-yl]-4-(pyrrolidin-1-ylmethyl)piperidin-4-ol (E2) US9920053, Example 2 BDBM376872 BDBM272098 E2-(abs)-5-(3-Fluoro-2-methylphenyl)-2-(2- fluorophenyl)-N-hydroxy-4,5,6,7-tetrahydro-2H- indazole-5-carboxamide US10065948, 34
BDBM272098 E2-(abs)-5-(3-Fluoro-2-methylphenyl)-2-(2- fluorophenyl)-N-hydroxy-4,5,6,7-tetrahydro-2H- indazole-5-carboxamide US10065948, 34 US10065948, 38 BDBM272102 E2-(abs)-5-(3-Fluoro-2-methylphenyl)-N-hydroxy-1- (o-tolyl)-4,5,6,7-tetrahydro-1H-indazole-5- carboxamide
US10065948, 38 BDBM272102 E2-(abs)-5-(3-Fluoro-2-methylphenyl)-N-hydroxy-1- (o-tolyl)-4,5,6,7-tetrahydro-1H-indazole-5- carboxamide US10065948, 85 E2-(abs)-6-(3-Fluoro-2-methylphenyl)-N-hydroxy-2- (2,2,2-trifluoroethyl)-2,4,5,6,7,8- hexahydrocyclohepta[c]pyrazole-6-carboxamide BDBM272149
US10065948, 85 E2-(abs)-6-(3-Fluoro-2-methylphenyl)-N-hydroxy-2- (2,2,2-trifluoroethyl)-2,4,5,6,7,8- hexahydrocyclohepta[c]pyrazole-6-carboxamide BDBM272149 Compound 11 16beta-alkyl-E2 BDBM17294 (13S,14S,15S)-15-methyl-13-nonyltetracyclo[8.7.0.0^{2,7}.0^{11,15}]heptadeca-2,4,6-triene-5,14-diol
Compound 11 16beta-alkyl-E2 BDBM17294 (13S,14S,15S)-15-methyl-13-nonyltetracyclo[8.7.0.0^{2,7}.0^{11,15}]heptadeca-2,4,6-triene-5,14-diol BDBM272129 E2-(abs)-5-(3-Fluoro-2-methylphenyl)-2-(3- fluorophenyl)-N-hydroxy-5,6-dihydro-4H- cyclopenta[d]thiazole-5-carboxamide US10065948, 65
BDBM272129 E2-(abs)-5-(3-Fluoro-2-methylphenyl)-2-(3- fluorophenyl)-N-hydroxy-5,6-dihydro-4H- cyclopenta[d]thiazole-5-carboxamide US10065948, 65 BDBM751305 US20250195530, Example E2 8-(2-chloro-4- (2-(4- methylpiperazin-1- yl)ethoxy)phenyl)- 9-(3- chlorobenzyl)- 6-(1- methylcyclopropoxy)- 9H- purine
BDBM751305 US20250195530, Example E2 8-(2-chloro-4- (2-(4- methylpiperazin-1- yl)ethoxy)phenyl)- 9-(3- chlorobenzyl)- 6-(1- methylcyclopropoxy)- 9H- purine E2-(abs)-5-(3-Fluoro-2-methylphenyl)-2-(2- fluorophenyl)-N-hydroxy-5,6-dihydro-4H- cyclopenta[d]thiazole-5-carboxamide US10065948, 62 BDBM272126
E2-(abs)-5-(3-Fluoro-2-methylphenyl)-2-(2- fluorophenyl)-N-hydroxy-5,6-dihydro-4H- cyclopenta[d]thiazole-5-carboxamide US10065948, 62 BDBM272126 BDBM556745 1-(3-((2-tert-Butyl-4-chloro-5-methylphenoxy)methyl)-6,7-dihydro-1H-pyrazolo[4,3-c]pyridin-5(4H)-yl)ethanone US11352330, Example E2
BDBM556745 1-(3-((2-tert-Butyl-4-chloro-5-methylphenoxy)methyl)-6,7-dihydro-1H-pyrazolo[4,3-c]pyridin-5(4H)-yl)ethanone US11352330, Example E2 US10065948, 75 BDBM272139 E2-(abs)-5-(3-Fluoro-2-methylphenyl)-2-(4-fluoro-2- methylphenyl)-N-hydroxy-5,6-dihydro-4H- cyclopenta[d]thiazole-5-carboxamide
US10065948, 75 BDBM272139 E2-(abs)-5-(3-Fluoro-2-methylphenyl)-2-(4-fluoro-2- methylphenyl)-N-hydroxy-5,6-dihydro-4H- cyclopenta[d]thiazole-5-carboxamide US10065948, 81 BDBM272145 E2-(abs)-5-(3-fluoro-2-methylphenyl)-N-hydroxy-3- phenyl-5,6-dihydro-4H-cyclopenta[d]isothiazole-5- carboxamide US10457675, Example 81
US10065948, 81 BDBM272145 E2-(abs)-5-(3-fluoro-2-methylphenyl)-N-hydroxy-3- phenyl-5,6-dihydro-4H-cyclopenta[d]isothiazole-5- carboxamide US10457675, Example 81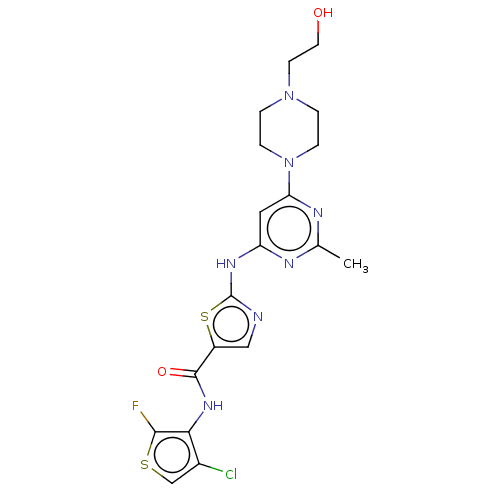 BDBM632967 N-(4-chloro-2-fluorothiophen-3-yl)-2- ((6-(4-(2-hydroxyethyl)piperazin-1-yl)- 2-methylpyrimidin-4-yl)amino)thiazole- 5-carboxamide US20230348453, Compound E2
BDBM632967 N-(4-chloro-2-fluorothiophen-3-yl)-2- ((6-(4-(2-hydroxyethyl)piperazin-1-yl)- 2-methylpyrimidin-4-yl)amino)thiazole- 5-carboxamide US20230348453, Compound E2 US10065948, 77 BDBM272141 E2-(abs)-2-(1,5-dimethyl-1H-pyrazol-4-yl)-5-(3- fluoro-2-methylphenyl)-N-hydroxy-5,6-dihydro-4H- cyclopenta[d]thiazole-5-carboxamide
US10065948, 77 BDBM272141 E2-(abs)-2-(1,5-dimethyl-1H-pyrazol-4-yl)-5-(3- fluoro-2-methylphenyl)-N-hydroxy-5,6-dihydro-4H- cyclopenta[d]thiazole-5-carboxamide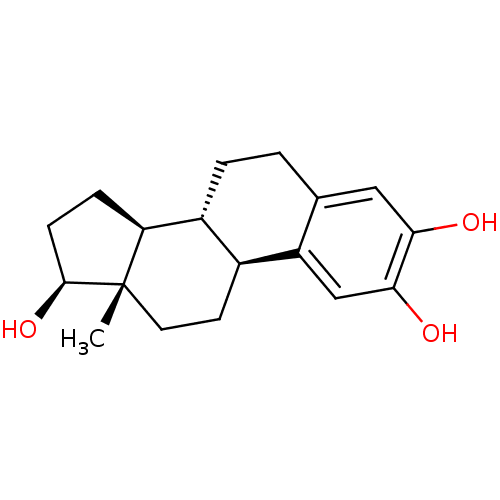 2-OH-estradiol 2-hydroxy-17beta-estradiol cid_247304 (17beta)-estra-1,3,5(10)-triene-2,3,17-triol US10864248, Compound 2 2-OH-E2 BDBM50262140 CHEMBL467987 estra-1,3,5(10)-triene-2,3,17beta-triol
2-OH-estradiol 2-hydroxy-17beta-estradiol cid_247304 (17beta)-estra-1,3,5(10)-triene-2,3,17-triol US10864248, Compound 2 2-OH-E2 BDBM50262140 CHEMBL467987 estra-1,3,5(10)-triene-2,3,17beta-triol 2-((4-(7-(((2S,5R)-5-(Ethylsulfonamido)tetrahydro- 2H-pyran-2-yl)methyl)-2,7-diazaspiro[3.5]nonan-2- yl)pyrimidin-5-yl)oxy)-5-fluoro-N-((E2)-2- hydroxycyclobutyl)-N-isopropylbenzamide US11919901, Compound 257 BDBM656860
2-((4-(7-(((2S,5R)-5-(Ethylsulfonamido)tetrahydro- 2H-pyran-2-yl)methyl)-2,7-diazaspiro[3.5]nonan-2- yl)pyrimidin-5-yl)oxy)-5-fluoro-N-((E2)-2- hydroxycyclobutyl)-N-isopropylbenzamide US11919901, Compound 257 BDBM656860 [5-(6-amino-9H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl]methyl 10-[(1S,10R,11S,14S,15S)-5,14-dihydroxy-15-methyltetracyclo[8.7.0.0^{2,7}.0^{11,15}]heptadeca-2,4,6-trien-13-yl]decanoate BDBM17291 E2-adenosine hybrid compound, 9
[5-(6-amino-9H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl]methyl 10-[(1S,10R,11S,14S,15S)-5,14-dihydroxy-15-methyltetracyclo[8.7.0.0^{2,7}.0^{11,15}]heptadeca-2,4,6-trien-13-yl]decanoate BDBM17291 E2-adenosine hybrid compound, 9 [5-(6-amino-9H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl]methyl 8-[(1S,10R,11S,14S,15S)-5,14-dihydroxy-15-methyltetracyclo[8.7.0.0^{2,7}.0^{11,15}]heptadeca-2,4,6-trien-13-yl]octanoate E2-adenosine hybrid compound, 7 BDBM17288
[5-(6-amino-9H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl]methyl 8-[(1S,10R,11S,14S,15S)-5,14-dihydroxy-15-methyltetracyclo[8.7.0.0^{2,7}.0^{11,15}]heptadeca-2,4,6-trien-13-yl]octanoate E2-adenosine hybrid compound, 7 BDBM17288 [5-(6-amino-9H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl]methyl 9-[(1S,10R,11S,14S,15S)-5,14-dihydroxy-15-methyltetracyclo[8.7.0.0^{2,7}.0^{11,15}]heptadeca-2,4,6-trien-13-yl]nonanoate BDBM17290 EM-1745 E2-adenosine hybrid compound, 8 EM1745
[5-(6-amino-9H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl]methyl 9-[(1S,10R,11S,14S,15S)-5,14-dihydroxy-15-methyltetracyclo[8.7.0.0^{2,7}.0^{11,15}]heptadeca-2,4,6-trien-13-yl]nonanoate BDBM17290 EM-1745 E2-adenosine hybrid compound, 8 EM1745 BDBM713106 US20250019387, Example 43 (Method 3-E2): 1-((1R,6S,7R)-3-(7-(8-chloronaphthalen-1-yl)-2-(((S)-1-methylpyrrolidin-2-yl)methoxy)-5,6,7,8-tetrahydropyrido[3,4-d]pyrimidin-4-yl)-3-azabicyclo[4.1.0]heptan-7-yl)prop-2-en-1-one and 1-((1S,6R,7S)-3-(7-(8-chloronaphthalen-1-yl)-2-(((S)-1-methylpyrrolidin-2-yl)methoxy)-5,6,7,8-tetrahydropyrido[3,4-d]pyrimidin-4-yl)-3-azabicyclo[4.1.0]heptan-7-yl)prop-2-en-1-one
BDBM713106 US20250019387, Example 43 (Method 3-E2): 1-((1R,6S,7R)-3-(7-(8-chloronaphthalen-1-yl)-2-(((S)-1-methylpyrrolidin-2-yl)methoxy)-5,6,7,8-tetrahydropyrido[3,4-d]pyrimidin-4-yl)-3-azabicyclo[4.1.0]heptan-7-yl)prop-2-en-1-one and 1-((1S,6R,7S)-3-(7-(8-chloronaphthalen-1-yl)-2-(((S)-1-methylpyrrolidin-2-yl)methoxy)-5,6,7,8-tetrahydropyrido[3,4-d]pyrimidin-4-yl)-3-azabicyclo[4.1.0]heptan-7-yl)prop-2-en-1-one