Compound (183)
Article Title (75)
Article Author (68)
Assay (31)
Wang, J; Knapp, S; Pyne, NJ; Pyne, S; Elkins, JM Crystal Structure of Sphingosine Kinase 1 with PF-543. ACS Med Chem Lett 5: 1329 -33 (2014) Munchhof, MJ; Li, Q; Shavnya, A; Borzillo, GV; Boyden, TL; Jones, CS; LaGreca, SD; Martinez-Alsina, L; Patel, N; Pelletier, K; Reiter, LA; Robbins, MD; Tkalcevic, GT Discovery of PF-04449913, a Potent and Orally Bioavailable Inhibitor of Smoothened. ACS Med Chem Lett 3: 106 -111 (2012) Cheng, H; Orr, STM; Bailey, S; Brooun, A; Chen, P; Deal, JG; Deng, YL; Edwards, MP; Gallego, GM; Grodsky, N; Huang, B; Jalaie, M; Kaiser, S; Kania, RS; Kephart, SE; Lafontaine, J; Ornelas, MA; Pairish, M; Planken, S; Shen, H; Sutton, S; Zehnder, L; Almaden, CD; Bagrodia, S; Falk, MD; Gukasyan, HJ; Ho, C; Kang, X; Kosa, RE; Liu, L; Spilker, ME; Timofeevski, S; Visswanathan, R; Wang, Z; Meng, F; Ren, S; Shao, L; Xu, F; Kath, JC Structure-Based Drug Design and Synthesis of PI3Kα-Selective Inhibitor (PF-06843195). J Med Chem 64: 644 -661 (2021) Filipski, KJ; Edmonds, DJ; Garnsey, MR; Smaltz, DJ; Coffman, K; Futatsugi, K; Lee, J; O'Neil, SV; Wright, A; Nason, D; Gosset, JR; Orozco, CC; Blackler, D; Fakhoury, G; Gutierrez, JA; Perez, S; Ross, T; Stock, I; Tesz, G; Dullea, R Design of Next-Generation DGAT2 Inhibitor PF-07202954 with Longer Predicted Half-Life. ACS Med Chem Lett 14: 1427 -1433 (2023) Xue, CB; Wang, A; Han, Q; Zhang, Y; Cao, G; Feng, H; Huang, T; Zheng, C; Xia, M; Zhang, K; Kong, L; Glenn, J; Anand, R; Meloni, D; Robinson, DJ; Shao, L; Storace, L; Li, M; Hughes, RO; Devraj, R; Morton, PA; Rogier, DJ; Covington, M; Scherle, P; Diamond, S; Emm, T; Yeleswaram, S; Contel, N; Vaddi, K; Newton, R; Hollis, G; Metcalf, B Discovery of INCB8761/PF-4136309, a Potent, Selective, and Orally Bioavailable CCR2 Antagonist. ACS Med Chem Lett 2: 913 -918 (2011) Gopalsamy, A; Aulabaugh, AE; Barakat, A; Beaumont, KC; Cabral, S; Canterbury, DP; Casimiro-Garcia, A; Chang, JS; Chen, MZ; Choi, C; Dow, RL; Fadeyi, OO; Feng, X; France, SP; Howard, RM; Janz, JM; Jasti, J; Jasuja, R; Jones, LH; King-Ahmad, A; Knee, KM; Kohrt, JT; Limberakis, C; Liras, S; Martinez, CA; McClure, KF; Narayanan, A; Narula, J; Novak, JJ; O'Connell, TN; Parikh, MD; Piotrowski, DW; Plotnikova, O; Robinson, RP; Sahasrabudhe, PV; Sharma, R; Thuma, BA; Vasa, D; Wei, L; Wenzel, AZ; Withka, JM; Xiao, J; Yayla, HG PF-07059013: A Noncovalent Modulator of Hemoglobin for Treatment of Sickle Cell Disease. J Med Chem 64: 326 -342 (2021) Johnson, DS; Stiff, C; Lazerwith, SE; Kesten, SR; Fay, LK; Morris, M; Beidler, D; Liimatta, MB; Smith, SE; Dudley, DT; Sadagopan, N; Bhattachar, SN; Kesten, SJ; Nomanbhoy, TK; Cravatt, BF; Ahn, K Discovery of PF-04457845: A Highly Potent, Orally Bioavailable, and Selective Urea FAAH Inhibitor. ACS Med Chem Lett 2: 91 -96 (2011) Dow, RL; Li, JC; Pence, MP; Gibbs, EM; LaPerle, JL; Litchfield, J; Piotrowski, DW; Munchhof, MJ; Manion, TB; Zavadoski, WJ; Walker, GS; McPherson, RK; Tapley, S; Sugarman, E; Guzman-Perez, A; DaSilva-Jardine, P Discovery of PF-04620110, a Potent, Selective, and Orally Bioavailable Inhibitor of DGAT-1. ACS Med Chem Lett 2: 407 -412 (2011) Freeman-Cook, KD; Hoffman, RL; Behenna, DC; Boras, B; Carelli, J; Diehl, W; Ferre, RA; He, YA; Hui, A; Huang, B; Huser, N; Jones, R; Kephart, SE; Lapek, J; McTigue, M; Miller, N; Murray, BW; Nagata, A; Nguyen, L; Niessen, S; Ninkovic, S; O'Doherty, I; Ornelas, MA; Solowiej, J; Sutton, SC; Tran, K; Tseng, E; Visswanathan, R; Xu, M; Zehnder, L; Zhang, Q; Zhang, C; Dann, S Discovery of PF-06873600, a CDK2/4/6 Inhibitor for the Treatment of Cancer. J Med Chem 64: 9056 -9077 (2021) Pujala, B; Ramachandran, SA; Sonawane, M; Kamble, MM; Panpatil, D; Adhikari, S; Soni, S; Subbareddy, V; Shinde, BU; Nayak, AK; Bansal, C; Gupta, A; Mukherjee, K; Agarwal, AK; Guerrero, J; Herrera, FJ; Bernales, S; Guha, M; Chakravarty, S; Pham, SM; Rai, R Discovery of MDV6058 (PF-06952229), a selective and potent TGFβR1 inhibitor: Design, synthesis and optimization. Bioorg Med Chem Lett 75: (2022) Gerstenberger, BS; Ambler, C; Arnold, EP; Banker, ME; Brown, MF; Clark, JD; Dermenci, A; Dowty, ME; Fensome, A; Fish, S; Hayward, MM; Hegen, M; Hollingshead, BD; Knafels, JD; Lin, DW; Lin, TH; Owen, DR; Saiah, E; Sharma, R; Vajdos, FF; Xing, L; Yang, X; Yang, X; Wright, SW Discovery of Tyrosine Kinase 2 (TYK2) Inhibitor (PF-06826647) for the Treatment of Autoimmune Diseases. J Med Chem 63: 13561 -13577 (2020) Wright, SW; Farley, KA; Han, S; Knafels, JD; Lee, KL In Retrospect: Root-Cause Analysis of Structure-Activity Relationships in IRAK4 Inhibitor Zimlovisertib (PF-06650833). ACS Med Chem Lett 15: 540 -545 (2024) Stepan, AF; Tran, TP; Helal, CJ; Brown, MS; Chang, C; O'Connor, RE; De Vivo, M; Doran, SD; Fisher, EL; Jenkinson, S; Karanian, D; Kormos, BL; Sharma, R; Walker, GS; Wright, AS; Yang, EX; Brodney, MA; Wager, TT; Verhoest, PR; Obach, RS Late-Stage Microsomal Oxidation Reduces Drug-Drug Interaction and Identifies Phosphodiesterase 2A Inhibitor PF-06815189. ACS Med Chem Lett 9: 68 -72 (2018) Ryckmans, T; Aubdool, AA; Bodkin, JV; Cox, P; Brain, SD; Dupont, T; Fairman, E; Hashizume, Y; Ishii, N; Kato, T; Kitching, L; Newman, J; Omoto, K; Rawson, D; Strover, J Design and pharmacological evaluation of PF-4840154, a non-electrophilic reference agonist of the TrpA1 channel. Bioorg Med Chem Lett 21: 4857 -9 (2011) Zheng, C; Cao, G; Xia, M; Feng, H; Glenn, J; Anand, R; Zhang, K; Huang, T; Wang, A; Kong, L; Li, M; Galya, L; Hughes, RO; Devraj, R; Morton, PA; Rogier, DJ; Covington, M; Baribaud, F; Shin, N; Scherle, P; Diamond, S; Yeleswaram, S; Vaddi, K; Newton, R; Hollis, G; Friedman, S; Metcalf, B; Xue, CB Discovery of INCB10820/PF-4178903, a potent, selective, and orally bioavailable dual CCR2 and CCR5 antagonist. Bioorg Med Chem Lett 21: 1442 -6 (2011) Johnson, DS; Choi, C; Fay, LK; Favor, DA; Repine, JT; White, AD; Akunne, HC; Fitzgerald, L; Nicholls, K; Snyder, BJ; Whetzel, SZ; Zhang, L; Serpa, KA Discovery of PF-00217830: aryl piperazine napthyridinones as D2 partial agonists for schizophrenia and bipolar disorder. Bioorg Med Chem Lett 21: 2621 -5 (2011) Bhattacharya, SK; Andrews, K; Beveridge, R; Cameron, KO; Chen, C; Dunn, M; Fernando, D; Gao, H; Hepworth, D; Jackson, VM; Khot, V; Kong, J; Kosa, RE; Lapham, K; Loria, PM; Londregan, AT; McClure, KF; Orr, ST; Patel, J; Rose, C; Saenz, J; Stock, IA; Storer, G; VanVolkenburg, M; Vrieze, D; Wang, G; Xiao, J; Zhang, Y Discovery of PF-5190457, a Potent, Selective, and Orally Bioavailable Ghrelin Receptor Inverse Agonist Clinical Candidate. ACS Med Chem Lett 5: 474 -9 (2014) Cheng, H; Li, C; Bailey, S; Baxi, SM; Goulet, L; Guo, L; Hoffman, J; Jiang, Y; Johnson, TO; Johnson, TW; Knighton, DR; Li, J; Liu, KK; Liu, Z; Marx, MA; Walls, M; Wells, PA; Yin, MJ; Zhu, J; Zientek, M Discovery of the Highly Potent PI3K/mTOR Dual Inhibitor PF-04979064 through Structure-Based Drug Design. ACS Med Chem Lett 4: 91 -7 (2013) Siu, M; Johnson, TO; Wang, Y; Nair, SK; Taylor, WD; Cripps, SJ; Matthews, JJ; Edwards, MP; Pauly, TA; Ermolieff, J; Castro, A; Hosea, NA; LaPaglia, A; Fanjul, AN; Vogel, JE N-(Pyridin-2-yl) arylsulfonamide inhibitors of 11beta-hydroxysteroid dehydrogenase type 1: Discovery of PF-915275. Bioorg Med Chem Lett 19: 3493 -7 (2009) Zou, HY; Li, Q; Lee, JH; Arango, ME; Burgess, K; Qiu, M; Engstrom, LD; Yamazaki, S; Parker, M; Timofeevski, S; Cui, JJ; McTigue, M; Los, G; Bender, SL; Smeal, T; Christensen, JG Sensitivity of selected human tumor models to PF-04217903, a novel selective c-Met kinase inhibitor. Mol Cancer Ther 11: 1036 -47 Blasina, A; Hallin, J; Chen, E; Arango, ME; Kraynov, E; Register, J; Grant, S; Ninkovic, S; Chen, P; Nichols, T; O'Connor, P; Anderes, K Breaching the DNA damage checkpoint via PF-00477736, a novel small-molecule inhibitor of checkpoint kinase 1. Mol Cancer Ther 7: 2394 -404 Pettersson, M; Johnson, DS; Humphrey, JM; Am Ende, CW; Butler, TW; Dorff, PH; Efremov, IV; Evrard, E; Green, ME; Helal, CJ; Kauffman, GW; Mullins, PB; Navaratnam, T; O'Donnell, CJ; O'Sullivan, TJ; Patel, NC; Stepan, AF; Stiff, CM; Subramanyam, C; Trapa, P; Tran, TP; Vetelino, BC; Yang, E; Xie, L; Pustilnik, LR; Steyn, SJ; Wood, KM; Bales, KR; Hajos-Korcsok, E; Verhoest, PR Discovery of Clinical Candidate PF-06648671: A Potent γ-Secretase Modulator for the Treatment of Alzheimer's Disease. J Med Chem 67: 10248 -10262 Garnsey, MR; Smith, AC; Polivkova, J; Arons, AL; Bai, G; Blakemore, C; Boehm, M; Buzon, LM; Campion, SN; Cerny, M; Chang, SC; Coffman, K; Farley, KA; Fonseca, KR; Ford, KK; Garren, J; Kong, JX; Koos, MRM; Kung, DW; Lian, Y; Li, MM; Li, Q; Martinez-Alsina, LA; O'Connor, R; Ogilvie, K; Omoto, K; Raymer, B; Reese, MR; Ryder, T; Samp, L; Stevens, KA; Widlicka, DW; Yang, Q; Zhu, K; Fortin, JP; Sammons, MF Discovery of the Potent and Selective MC4R Antagonist PF-07258669 for the Potential Treatment of Appetite Loss. J Med Chem 66: 3195 -3211 (2023) Dow, RL; Andrews, MP; Li, JC; Michael Gibbs, E; Guzman-Perez, A; Laperle, JL; Li, Q; Mather, D; Munchhof, MJ; Niosi, M; Patel, L; Perreault, C; Tapley, S; Zavadoski, WJ Defining the key pharmacophore elements of PF-04620110: discovery of a potent, orally-active, neutral DGAT-1 inhibitor. Bioorg Med Chem 21: 5081 -97 (2013) Hall, J; Brault, A; Vincent, F; Weng, S; Wang, H; Dumlao, D; Aulabaugh, A; Aivazian, D; Castro, D; Chen, M; Culp, J; Dower, K; Gardner, J; Hawrylik, S; Golenbock, D; Hepworth, D; Horn, M; Jones, L; Jones, P; Latz, E; Li, J; Lin, LL; Lin, W; Lin, D; Lovering, F; Niljanskul, N; Nistler, R; Pierce, B; Plotnikova, O; Schmitt, D; Shanker, S; Smith, J; Snyder, W; Subashi, T; Trujillo, J; Tyminski, E; Wang, G; Wong, J; Lefker, B; Dakin, L; Leach, K Discovery of PF-06928215 as a high affinity inhibitor of cGAS enabled by a novel fluorescence polarization assay. PLoS ONE 12: 1 -16 (2017) af Forselles, KJ; Root, J; Clarke, T; Davey, D; Aughton, K; Dack, K; Pullen, N In vitro and in vivo characterization of PF-04418948, a novel, potent and selective prostaglandin EP₂ receptor antagonist. Br J Pharmacol 164: 1847 -56 Murray, BW; Guo, C; Piraino, J; Westwick, JK; Zhang, C; Lamerdin, J; Dagostino, E; Knighton, D; Loi, CM; Zager, M; Kraynov, E; Popoff, I; Christensen, JG; Martinez, R; Kephart, SE; Marakovits, J; Karlicek, S; Bergqvist, S; Smeal, T Small-molecule p21-activated kinase inhibitor PF-3758309 is a potent inhibitor of oncogenic signaling and tumor growth. Proc Natl Acad Sci U S A 107: 9446 -51 Wager, TT; Pettersen, BA; Schmidt, AW; Spracklin, DK; Mente, S; Butler, TW; Howard, H; Lettiere, DJ; Rubitski, DM; Wong, DF; Nedza, FM; Nelson, FR; Rollema, H; Raggon, JW; Aubrecht, J; Freeman, JK; Marcek, JM; Cianfrogna, J; Cook, KW; James, LC; Chatman, LA; Iredale, PA; Banker, MJ; Homiski, ML; Munzner, JB; Chandrasekaran, RY Discovery of two clinical histamine H(3) receptor antagonists: trans-N-ethyl-3-fluoro-3-[3-fluoro-4-(pyrrolidinylmethyl)phenyl]cyclobutanecarboxamide (PF-03654746) and trans-3-fluoro-3-[3-fluoro-4-(pyrrolidin-1-ylmethyl)phenyl]-N-(2-methylpropyl)cyclobutanecarboxamide (PF-03654764). J Med Chem 54: 7602 -20 (2011) Zou, HY; Li, Q; Engstrom, LD; West, M; Appleman, V; Wong, KA; McTigue, M; Deng, YL; Liu, W; Brooun, A; Timofeevski, S; McDonnell, SR; Jiang, P; Falk, MD; Lappin, PB; Affolter, T; Nichols, T; Hu, W; Lam, J; Johnson, TW; Smeal, T; Charest, A; Fantin, VR PF-06463922 is a potent and selective next-generation ROS1/ALK inhibitor capable of blocking crizotinib-resistant ROS1 mutations. Proc Natl Acad Sci U S A 112: 3493 -8 Skerratt, SE; Andrews, M; Bagal, SK; Bilsland, J; Brown, D; Bungay, PJ; Cole, S; Gibson, KR; Jones, R; Morao, I; Nedderman, A; Omoto, K; Robinson, C; Ryckmans, T; Skinner, K; Stupple, P; Waldron, G The Discovery of a Potent, Selective, and Peripherally Restricted Pan-Trk Inhibitor (PF-06273340) for the Treatment of Pain. J Med Chem 59: 10084 -10099 (2016) Mowbray, CE; Bell, AS; Clarke, NP; Collins, M; Jones, RM; Lane, CA; Liu, WL; Newman, SD; Paradowski, M; Schenck, EJ; Selby, MD; Swain, NA; Williams, DH Challenges of drug discovery in novel target space. The discovery and evaluation of PF-3893787: a novel histamine H4 receptor antagonist. Bioorg Med Chem Lett 21: 6596 -602 (2011) Arhancet, GB; Walker, DP; Metz, S; Fobian, YM; Heasley, SE; Carter, JS; Springer, JR; Jones, DE; Hayes, MJ; Shaffer, AF; Jerome, GM; Baratta, MT; Zweifel, B; Moore, WM; Masferrer, JL; Vazquez, ML Discovery and SAR of PF-4693627, a potent, selective and orally bioavailable mPGES-1 inhibitor for the potential treatment of inflammation. Bioorg Med Chem Lett 23: 1114 -9 (2013) Futatsugi, K; Cabral, S; Kung, DW; Huard, K; Lee, E; Boehm, M; Bauman, J; Clark, RW; Coffey, SB; Crowley, C; Dechert-Schmitt, AM; Dowling, MS; Dullea, R; Gosset, JR; Kalgutkar, AS; Kou, K; Li, Q; Lian, Y; Loria, PM; Londregan, AT; Niosi, M; Orozco, C; Pettersen, JC; Pfefferkorn, JA; Polivkova, J; Ross, TT; Sharma, R; Stock, IA; Tesz, G; Wisniewska, H; Goodwin, B; Price, DA Discovery of Ervogastat (PF-06865571): A Potent and Selective Inhibitor of Diacylglycerol Acyltransferase 2 for the Treatment of Non-alcoholic Steatohepatitis. J Med Chem 65: 15000 -15013 (2022) Carr&aagrovr;, G; Rizzi, A; Guerrini, R; Barnes, TA; McDonald, J; Hebbes, CP; Mela, F; Kenigs, VA; Marzola, G; Rizzi, D; Gavioli, E; Zucchini, S; Regoli, D; Morari, M; Salvadori, S; Rowbotham, DJ; Lambert, DG; Kapusta, DR; Calo', G [(pF)Phe4,Arg14,Lys15]N/OFQ-NH2 (UFP-102), a highly potent and selective agonist of the nociceptin/orphanin FQ receptor. J Pharmacol Exp Ther 312: 1114 -23 (2005) Brodney, MA; Auperin, DD; Becker, SL; Bronk, BS; Brown, TM; Coffman, KJ; Finley, JE; Hicks, CD; Karmilowicz, MJ; Lanz, TA; Liston, D; Liu, X; Martin, BA; Nelson, RB; Nolan, CE; Oborski, CE; Parker, CP; Richter, KE; Pozdnyakov, N; Sahagan, BG; Schachter, JB; Sokolowski, SA; Tate, B; Wood, DE; Wood, KM; Van Deusen, JW; Zhang, L Design, synthesis, and in vivo characterization of a novel series of tetralin amino imidazoles as γ-secretase inhibitors: discovery of PF-3084014. Bioorg Med Chem Lett 21: 2637 -40 (2011) Futatsugi, K; Smith, AC; Tu, M; Raymer, B; Ahn, K; Coffey, SB; Dowling, MS; Fernando, DP; Gutierrez, JA; Huard, K; Jasti, J; Kalgutkar, AS; Knafels, JD; Pandit, J; Parris, KD; Perez, S; Pfefferkorn, JA; Price, DA; Ryder, T; Shavnya, A; Stock, IA; Tsai, AS; Tesz, GJ; Thuma, BA; Weng, Y; Wisniewska, HM; Xing, G; Zhou, J; Magee, TV Discovery of PF-06835919: A Potent Inhibitor of Ketohexokinase (KHK) for the Treatment of Metabolic Disorders Driven by the Overconsumption of Fructose. J Med Chem 63: 13546 -13560 (2020) Ren, L; Moreno, D; Baer, BR; Barbour, P; Bettendorf, T; Bouhana, K; Brown, K; Brown, SA; Fell, JB; Hartley, DP; Hicken, EJ; Laird, ER; Lee, P; McCown, J; Otten, JN; Prigaro, B; Wallace, R; Kahn, D Identification of the Clinical Candidate PF-07284890 (ARRY-461), a Highly Potent and Brain Penetrant BRAF Inhibitor for the Treatment of Cancer. J Med Chem 67: 13019 -13032 Butler, CR; Popiolek, M; McAllister, LA; LaChapelle, EA; Kramer, M; Beck, EM; Mente, S; Brodney, MA; Brown, M; Gilbert, A; Helal, C; Ogilvie, K; Starr, J; Uccello, D; Grimwood, S; Edgerton, J; Garst-Orozco, J; Kozak, R; Lotarski, S; Rossi, A; Smith, D; O'Connor, R; Lazzaro, J; Steppan, C; Steyn, SJ Design and Synthesis of Clinical Candidate PF-06852231 (CVL-231): A Brain Penetrant, Selective, Positive Allosteric Modulator of the M4 Muscarinic Acetylcholine Receptor. J Med Chem 67: 10831 -10847 Johnson, PS; Ryckmans, T; Bryans, J; Beal, DM; Dack, KN; Feeder, N; Harrison, A; Lewis, M; Mason, HJ; Mills, J; Newman, J; Pasquinet, C; Rawson, DJ; Roberts, LR; Russell, R; Spark, D; Stobie, A; Underwood, TJ; Ward, R; Wheeler, S Discovery of PF-184563, a potent and selective V1a antagonist for the treatment of dysmenorrhoea. The influence of compound flexibility on microsomal stability. Bioorg Med Chem Lett 21: 5684 -7 (2011) Ahn, K; Smith, SE; Liimatta, MB; Beidler, D; Sadagopan, N; Dudley, DT; Young, T; Wren, P; Zhang, Y; Swaney, S; Van Becelaere, K; Blankman, JL; Nomura, DK; Bhattachar, SN; Stiff, C; Nomanbhoy, TK; Weerapana, E; Johnson, DS; Cravatt, BF Mechanistic and pharmacological characterization of PF-04457845: a highly potent and selective fatty acid amide hydrolase inhibitor that reduces inflammatory and noninflammatory pain. J Pharmacol Exp Ther 338: 114 -24 O'Neill, BT; Beck, EM; Butler, CR; Nolan, CE; Gonzales, C; Zhang, L; Doran, SD; Lapham, K; Buzon, LM; Dutra, JK; Barreiro, G; Hou, X; Martinez-Alsina, LA; Rogers, BN; Villalobos, A; Murray, JC; Ogilvie, K; LaChapelle, EA; Chang, C; Lanyon, LF; Steppan, CM; Robshaw, A; Hales, K; Boucher, GG; Pandher, K; Houle, C; Ambroise, CW; Karanian, D; Riddell, D; Bales, KR; Brodney, MA Design and Synthesis of Clinical Candidate PF-06751979: A Potent, Brain Penetrant, β-Site Amyloid Precursor Protein Cleaving Enzyme 1 (BACE1) Inhibitor Lacking Hypopigmentation. J Med Chem 61: 4476 -4504 (2018) Dounay, AB; Anderson, M; Bechle, BM; Evrard, E; Gan, X; Kim, JY; McAllister, LA; Pandit, J; Rong, S; Salafia, MA; Tuttle, JB; Zawadzke, LE; Verhoest, PR PF-04859989 as a template for structure-based drug design: identification of new pyrazole series of irreversible KAT II inhibitors with improved lipophilic efficiency. Bioorg Med Chem Lett 23: 1961 -6 (2013) Andrews, MD; Fish, PV; Blagg, J; Brabham, TK; Brennan, PE; Bridgeland, A; Brown, AD; Bungay, PJ; Conlon, KM; Edmunds, NJ; af Forselles, K; Gibbons, CP; Green, MP; Hanton, G; Holbrook, M; Jessiman, AS; McIntosh, K; McMurray, G; Nichols, CL; Root, JA; Storer, RI; Sutton, MR; Ward, RV; Westbrook, D; Whitlock, GA Pyrimido[4,5-d]azepines as potent and selective 5-HT2C receptor agonists: design, synthesis, and evaluation of PF-3246799 as a treatment for urinary incontinence. Bioorg Med Chem Lett 21: 2715 -20 (2011) Andaloussi, M; Lim, HD; van der Meer, T; Sijm, M; Poulie, CB; de Esch, IJ; Leurs, R; Smits, RA A novel series of histamine H4 receptor antagonists based on the pyrido[3,2-d]pyrimidine scaffold: comparison of hERG binding and target residence time with PF-3893787. Bioorg Med Chem Lett 23: 2663 -70 (2013) Verhoest, PR; Basak, AS; Parikh, V; Hayward, M; Kauffman, GW; Paradis, V; McHardy, SF; McLean, S; Grimwood, S; Schmidt, AW; Vanase-Frawley, M; Freeman, J; Van Deusen, J; Cox, L; Wong, D; Liras, S Design and discovery of a selective small molecule¿ opioid antagonist (2-methyl-N-((2'-(pyrrolidin-1-ylsulfonyl)biphenyl-4-yl)methyl)propan-1-amine, PF-4455242). J Med Chem 54: 5868 -77 (2011) Li, JJ; Iula, DM; Nguyen, MN; Hu, LY; Dettling, D; Johnson, TR; Du, DY; Shanmugasundaram, V; Van Camp, JA; Wang, Z; Harter, WG; Yue, WS; Boys, ML; Wade, KJ; Drummond, EM; Samas, BM; Lefker, BA; Hoge, GS; Lovdahl, MJ; Asbill, J; Carroll, M; Meade, MA; Ciotti, SM; Krieger-Burke, T Rational design and synthesis of 4-((1R,2R)-2-hydroxycyclohexyl)-2(trifluoromethyl)benzonitrile (PF-998425), a novel, nonsteroidal androgen receptor antagonist devoid of phototoxicity for dermatological indications. J Med Chem 51: 7010 -4 (2008) Cui, JJ; Tran-Dubé, M; Shen, H; Nambu, M; Kung, PP; Pairish, M; Jia, L; Meng, J; Funk, L; Botrous, I; McTigue, M; Grodsky, N; Ryan, K; Padrique, E; Alton, G; Timofeevski, S; Yamazaki, S; Li, Q; Zou, H; Christensen, J; Mroczkowski, B; Bender, S; Kania, RS; Edwards, MP Structure based drug design of crizotinib (PF-02341066), a potent and selective dual inhibitor of mesenchymal-epithelial transition factor (c-MET) kinase and anaplastic lymphoma kinase (ALK). J Med Chem 54: 6342 -63 (2011) Jones, P; Storer, RI; Sabnis, YA; Wakenhut, FM; Whitlock, GA; England, KS; Mukaiyama, T; Dehnhardt, CM; Coe, JW; Kortum, SW; Chrencik, JE; Brown, DG; Jones, RM; Murphy, JR; Yeoh, T; Morgan, P; Kilty, I Design and Synthesis of a Pan-Janus Kinase Inhibitor Clinical Candidate (PF-06263276) Suitable for Inhaled and Topical Delivery for the Treatment of Inflammatory Diseases of the Lungs and Skin. J Med Chem 60: 767 -786 (2017) Vazquez, ML; Kaila, N; Strohbach, JW; Trzupek, JD; Brown, MF; Flanagan, ME; Mitton-Fry, MJ; Johnson, TA; TenBrink, RE; Arnold, EP; Basak, A; Heasley, SE; Kwon, S; Langille, J; Parikh, MD; Griffin, SH; Casavant, JM; Duclos, BA; Fenwick, AE; Harris, TM; Han, S; Caspers, N; Dowty, ME; Yang, X; Banker, ME; Hegen, M; Symanowicz, PT; Li, L; Wang, L; Lin, TH; Jussif, J; Clark, JD; Telliez, JB; Robinson, RP; Unwalla, R Identification of N-{cis-3-[Methyl(7H-pyrrolo[2,3-d]pyrimidin-4-yl)amino]cyclobutyl}propane-1-sulfonamide (PF-04965842): A Selective JAK1 Clinical Candidate for the Treatment of Autoimmune Diseases. J Med Chem 61: 1130 -1152 (2018) Poel, TJ; Thomas, RC; Adams, WJ; Aristoff, PA; Barbachyn, MR; Boyer, FE; Brieland, J; Brideau, R; Brodfuehrer, J; Brown, AP; Choy, AL; Dermyer, M; Dority, M; Ford, CW; Gadwood, RC; Hanna, D; Hongliang, C; Huband, MD; Huber, C; Kelly, R; Kim, JY; Martin, JP; Pagano, PJ; Ross, D; Skerlos, L; Sulavik, MC; Zhu, T; Zurenko, GE; Prasad, JV Antibacterial oxazolidinones possessing a novel C-5 side chain. (5R)-trans-3-[3-Fluoro-4- (1-oxotetrahydrothiopyran-4-yl)phenyl]-2- oxooxazolidine-5-carboxylic acid amide (PF-00422602), a new lead compound. J Med Chem 50: 5886 -9 (2007) Walsky, RL; Obach, RS; Hyland, R; Kang, P; Zhou, S; West, M; Geoghegan, KF; Helal, CJ; Walker, GS; Goosen, TC; Zientek, MA Selective mechanism-based inactivation of CYP3A4 by CYP3cide (PF-04981517) and its utility as an in vitro tool for delineating the relative roles of CYP3A4 versus CYP3A5 in the metabolism of drugs. Drug Metab Dispos 40: 1686 -97 (2012) Henderson, JL; Kormos, BL; Hayward, MM; Coffman, KJ; Jasti, J; Kurumbail, RG; Wager, TT; Verhoest, PR; Noell, GS; Chen, Y; Needle, E; Berger, Z; Steyn, SJ; Houle, C; Hirst, WD; Galatsis, P Discovery and preclinical profiling of 3-[4-(morpholin-4-yl)-7H-pyrrolo[2,3-d]pyrimidin-5-yl]benzonitrile (PF-06447475), a highly potent, selective, brain penetrant, and in vivo active LRRK2 kinase inhibitor. J Med Chem 58: 419 -32 (2015) Ruggeri, RB; Buckbinder, L; Bagley, SW; Carpino, PA; Conn, EL; Dowling, MS; Fernando, DP; Jiao, W; Kung, DW; Orr, ST; Qi, Y; Rocke, BN; Smith, A; Warmus, JS; Zhang, Y; Bowles, D; Widlicka, DW; Eng, H; Ryder, T; Sharma, R; Wolford, A; Okerberg, C; Walters, K; Maurer, TS; Zhang, Y; Bonin, PD; Spath, SN; Xing, G; Hepworth, D; Ahn, K; Kalgutkar, AS Discovery of 2-(6-(5-Chloro-2-methoxyphenyl)-4-oxo-2-thioxo-3,4-dihydropyrimidin-1(2H)-yl)acetamide (PF-06282999): A Highly Selective Mechanism-Based Myeloperoxidase Inhibitor for the Treatment of Cardiovascular Diseases. J Med Chem 58: 8513 -28 (2015) Crosignani, S; Bingham, P; Bottemanne, P; Cannelle, H; Cauwenberghs, S; Cordonnier, M; Dalvie, D; Deroose, F; Feng, JL; Gomes, B; Greasley, S; Kaiser, SE; Kraus, M; Négrerie, M; Maegley, K; Miller, N; Murray, BW; Schneider, M; Soloweij, J; Stewart, AE; Tumang, J; Torti, VR; Van Den Eynde, B; Wythes, M Discovery of a Novel and Selective Indoleamine 2,3-Dioxygenase (IDO-1) Inhibitor 3-(5-Fluoro-1H-indol-3-yl)pyrrolidine-2,5-dione (EOS200271/PF-06840003) and Its Characterization as a Potential Clinical Candidate. J Med Chem 60: 9617 -9629 (2017) Meyers, MJ; Arhancet, GB; Hockerman, SL; Chen, X; Long, SA; Mahoney, MW; Rico, JR; Garland, DJ; Blinn, JR; Collins, JT; Yang, S; Huang, HC; McGee, KF; Wendling, JM; Dietz, JD; Payne, MA; Homer, BL; Heron, MI; Reitz, DB; Hu, X Discovery of (3S,3aR)-2-(3-chloro-4-cyanophenyl)-3-cyclopentyl-3,3a,4,5-tetrahydro-2H-benzo[g]indazole-7-carboxylic acid (PF-3882845), an orally efficacious mineralocorticoid receptor (MR) antagonist for hypertension and nephropathy. J Med Chem 53: 5979 -6002 (2010) Ryder, TF; Calabrese, MF; Walker, GS; Cameron, KO; Reyes, AR; Borzilleri, KA; Delmore, J; Miller, R; Kurumbail, RG; Ward, J; Kung, DW; Brown, JA; Edmonds, DJ; Eng, H; Wolford, AC; Kalgutkar, AS Acyl Glucuronide Metabolites of 6-Chloro-5-[4-(1-hydroxycyclobutyl)phenyl]-1 H-indole-3-carboxylic Acid (PF-06409577) and Related Indole-3-carboxylic Acid Derivatives are Direct Activators of Adenosine Monophosphate-Activated Protein Kinase (AMPK). J Med Chem 61: 7273 -7288 (2018) Cameron, KO; Kung, DW; Kalgutkar, AS; Kurumbail, RG; Miller, R; Salatto, CT; Ward, J; Withka, JM; Bhattacharya, SK; Boehm, M; Borzilleri, KA; Brown, JA; Calabrese, M; Caspers, NL; Cokorinos, E; Conn, EL; Dowling, MS; Edmonds, DJ; Eng, H; Fernando, DP; Frisbie, R; Hepworth, D; Landro, J; Mao, Y; Rajamohan, F; Reyes, AR; Rose, CR; Ryder, T; Shavnya, A; Smith, AC; Tu, M; Wolford, AC; Xiao, J Discovery and Preclinical Characterization of 6-Chloro-5-[4-(1-hydroxycyclobutyl)phenyl]-1H-indole-3-carboxylic Acid (PF-06409577), a Direct Activator of Adenosine Monophosphate-activated Protein Kinase (AMPK), for the Potential Treatment of Diabetic Nephropathy. J Med Chem 59: 8068 -81 (2016) Hughes, RO; Rogier, DJ; Devraj, R; Zheng, C; Cao, G; Feng, H; Xia, M; Anand, R; Xing, L; Glenn, J; Zhang, K; Covington, M; Morton, PA; Hutzler, JM; Davis, JW; Scherle, P; Baribaud, F; Bahinski, A; Mo, ZL; Newton, R; Metcalf, B; Xue, CB Discovery of ((1S,3R)-1-isopropyl-3-((3S,4S)-3-methoxy-tetrahydro-2H-pyran-4-ylamino)cyclopentyl)(4-(5-(trifluoromethyl)pyridazin-3-yl)piperazin-1-yl)methanone, PF-4254196, a CCR2 antagonist with an improved cardiovascular profile. Bioorg Med Chem Lett 21: 2626 -30 (2011) Dow, RL; Carpino, PA; Hadcock, JR; Black, SC; Iredale, PA; DaSilva-Jardine, P; Schneider, SR; Paight, ES; Griffith, DA; Scott, DO; Oamppound39Connor, RE; Nduaka, CI Discovery of 2-(2-chlorophenyl)-3-(4-chlorophenyl)-7-(2,2-difluoropropyl)-6,7-dihydro-2H-pyrazolo[3,4-f][1,4]oxazepin-8(5H)-one (PF-514273), a novel, bicyclic lactam-based cannabinoid-1 receptor antagonist for the treatment of obesity. J Med Chem 52: 2652 -5 (2009) Swain, NA; Batchelor, D; Beaudoin, S; Bechle, BM; Bradley, PA; Brown, AD; Brown, B; Butcher, KJ; Butt, RP; Chapman, ML; Denton, S; Ellis, D; Galan, SRG; Gaulier, SM; Greener, BS; de Groot, MJ; Glossop, MS; Gurrell, IK; Hannam, J; Johnson, MS; Lin, Z; Markworth, CJ; Marron, BE; Millan, DS; Nakagawa, S; Pike, A; Printzenhoff, D; Rawson, DJ; Ransley, SJ; Reister, SM; Sasaki, K; Storer, RI; Stupple, PA; West, CW Discovery of Clinical Candidate 4-[2-(5-Amino-1H-pyrazol-4-yl)-4-chlorophenoxy]-5-chloro-2-fluoro-N-1,3-thiazol-4-ylbenzenesulfonamide (PF-05089771): Design and Optimization of Diaryl Ether Aryl Sulfonamides as Selective Inhibitors of Na J Med Chem 60: 7029 -7042 (2017) Stepan, AF; Claffey, MM; Reese, MR; Balan, G; Barreiro, G; Barricklow, J; Bohanon, MJ; Boscoe, BP; Cappon, GD; Chenard, LK; Cianfrogna, J; Chen, L; Coffman, KJ; Drozda, SE; Dunetz, JR; Ghosh, S; Hou, X; Houle, C; Karki, K; Lazzaro, JT; Mancuso, JY; Marcek, JM; Miller, EL; Moen, MA; O'Neil, S; Sakurada, I; Skaddan, M; Parikh, V; Smith, DL; Trapa, P; Tuttle, JB; Verhoest, PR; Walker, DP; Won, A; Wright, AS; Whritenour, J; Zasadny, K; Zaleska, MM; Zhang, L; Shaffer, CL Discovery and Characterization of (R)-6-Neopentyl-2-(pyridin-2-ylmethoxy)-6,7-dihydropyrimido[2,1-c][1,4]oxazin-4(9H)-one (PF-06462894), an Alkyne-Lacking Metabotropic Glutamate Receptor 5 Negative Allosteric Modulator Profiled in both Rat and Nonhuman Primates. J Med Chem 60: 7764 -7780 (2017) Davoren, JE; Lee, CW; Garnsey, M; Brodney, MA; Cordes, J; Dlugolenski, K; Edgerton, JR; Harris, AR; Helal, CJ; Jenkinson, S; Kauffman, GW; Kenakin, TP; Lazzaro, JT; Lotarski, SM; Mao, Y; Nason, DM; Northcott, C; Nottebaum, L; O'Neil, SV; Pettersen, B; Popiolek, M; Reinhart, V; Salomon-Ferrer, R; Steyn, SJ; Webb, D; Zhang, L; Grimwood, S Discovery of the Potent and Selective M1 PAM-Agonist N-[(3R,4S)-3-Hydroxytetrahydro-2H-pyran-4-yl]-5-methyl-4-[4-(1,3-thiazol-4-yl)benzyl]pyridine-2-carboxamide (PF-06767832): Evaluation of Efficacy and Cholinergic Side Effects. J Med Chem 59: 6313 -28 (2016) Thorarensen, A; Dowty, ME; Banker, ME; Juba, B; Jussif, J; Lin, T; Vincent, F; Czerwinski, RM; Casimiro-Garcia, A; Unwalla, R; Trujillo, JI; Liang, S; Balbo, P; Che, Y; Gilbert, AM; Brown, MF; Hayward, M; Montgomery, J; Leung, L; Yang, X; Soucy, S; Hegen, M; Coe, J; Langille, J; Vajdos, F; Chrencik, J; Telliez, JB Design of a Janus Kinase 3 (JAK3) Specific Inhibitor 1-((2S,5R)-5-((7H-Pyrrolo[2,3-d]pyrimidin-4-yl)amino)-2-methylpiperidin-1-yl)prop-2-en-1-one (PF-06651600) Allowing for the Interrogation of JAK3 Signaling in Humans. J Med Chem 60: 1971 -1993 (2017) Lee, KL; Ambler, CM; Anderson, DR; Boscoe, BP; Bree, AG; Brodfuehrer, JI; Chang, JS; Choi, C; Chung, S; Curran, KJ; Day, JE; Dehnhardt, CM; Dower, K; Drozda, SE; Frisbie, RK; Gavrin, LK; Goldberg, JA; Han, S; Hegen, M; Hepworth, D; Hope, HR; Kamtekar, S; Kilty, IC; Lee, A; Lin, LL; Lovering, FE; Lowe, MD; Mathias, JP; Morgan, HM; Murphy, EA; Papaioannou, N; Patny, A; Pierce, BS; Rao, VR; Saiah, E; Samardjiev, IJ; Samas, BM; Shen, MWH; Shin, JH; Soutter, HH; Strohbach, JW; Symanowicz, PT; Thomason, JR; Trzupek, JD; Vargas, R; Vincent, F; Yan, J; Zapf, CW; Wright, SW Discovery of Clinical Candidate 1-{[(2S,3S,4S)-3-Ethyl-4-fluoro-5-oxopyrrolidin-2-yl]methoxy}-7-methoxyisoquinoline-6-carboxamide (PF-06650833), a Potent, Selective Inhibitor of Interleukin-1 Receptor Associated Kinase 4 (IRAK4), by Fragment-Based Drug Design. J Med Chem 60: 5521 -5542 (2017) Pfefferkorn, JA; Choi, C; Larsen, SD; Auerbach, B; Hutchings, R; Park, W; Askew, V; Dillon, L; Hanselman, JC; Lin, Z; Lu, GH; Robertson, A; Sekerke, C; Harris, MS; Pavlovsky, A; Bainbridge, G; Caspers, N; Kowala, M; Tait, BD Substituted pyrazoles as hepatoselective HMG-CoA reductase inhibitors: discovery of (3R,5R)-7-[2-(4-fluoro-phenyl)-4-isopropyl-5-(4-methyl-benzylcarbamoyl)-2H-pyrazol-3-yl]-3,5-dihydroxyheptanoic acid (PF-3052334) as a candidate for the treatment of hypercholesterolemia. J Med Chem 51: 31 -45 (2008) Johnson, TW; Richardson, PF; Bailey, S; Brooun, A; Burke, BJ; Collins, MR; Cui, JJ; Deal, JG; Deng, YL; Dinh, D; Engstrom, LD; He, M; Hoffman, J; Hoffman, RL; Huang, Q; Kania, RS; Kath, JC; Lam, H; Lam, JL; Le, PT; Lingardo, L; Liu, W; McTigue, M; Palmer, CL; Sach, NW; Smeal, T; Smith, GL; Stewart, AE; Timofeevski, S; Zhu, H; Zhu, J; Zou, HY; Edwards, MP Discovery of (10R)-7-amino-12-fluoro-2,10,16-trimethyl-15-oxo-10,15,16,17-tetrahydro-2H-8,4-(metheno)pyrazolo[4,3-h][2,5,11]-benzoxadiazacyclotetradecine-3-carbonitrile (PF-06463922), a macrocyclic inhibitor of anaplastic lymphoma kinase (ALK) and c-ros oncogene 1 (ROS1) with preclinical brain expo J Med Chem 57: 4720 -44 (2014) Li, H; Tatlock, J; Linton, A; Gonzalez, J; Jewell, T; Patel, L; Ludlum, S; Drowns, M; Rahavendran, SV; Skor, H; Hunter, R; Shi, ST; Herlihy, KJ; Parge, H; Hickey, M; Yu, X; Chau, F; Nonomiya, J; Lewis, C Discovery of (R)-6-cyclopentyl-6-(2-(2,6-diethylpyridin-4-yl)ethyl)-3-((5,7-dimethyl-[1,2,4]triazolo[1,5-a]pyrimidin-2-yl)methyl)-4-hydroxy-5,6-dihydropyran-2-one (PF-00868554) as a potent and orally available hepatitis C virus polymerase inhibitor. J Med Chem 52: 1255 -8 (2010) Fensome, A; Ambler, CM; Arnold, E; Banker, ME; Brown, MF; Chrencik, J; Clark, JD; Dowty, ME; Efremov, IV; Flick, A; Gerstenberger, BS; Gopalsamy, A; Hayward, MM; Hegen, M; Hollingshead, BD; Jussif, J; Knafels, JD; Limburg, DC; Lin, D; Lin, TH; Pierce, BS; Saiah, E; Sharma, R; Symanowicz, PT; Telliez, JB; Trujillo, JI; Vajdos, FF; Vincent, F; Wan, ZK; Xing, L; Yang, X; Yang, X; Zhang, L Dual Inhibition of TYK2 and JAK1 for the Treatment of Autoimmune Diseases: Discovery of (( S)-2,2-Difluorocyclopropyl)((1 R,5 S)-3-(2-((1-methyl-1 H-pyrazol-4-yl)amino)pyrimidin-4-yl)-3,8-diazabicyclo[3.2.1]octan-8-yl)methanone (PF-06700841). J Med Chem 61: 8597 -8612 (2018) Verhoest, PR; Fonseca, KR; Hou, X; Proulx-Lafrance, C; Corman, M; Helal, CJ; Claffey, MM; Tuttle, JB; Coffman, KJ; Liu, S; Nelson, F; Kleiman, RJ; Menniti, FS; Schmidt, CJ; Vanase-Frawley, M; Liras, S Design and discovery of 6-[(3S,4S)-4-methyl-1-(pyrimidin-2-ylmethyl)pyrrolidin-3-yl]-1-(tetrahydro-2H-pyran-4-yl)-1,5-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one (PF-04447943), a selective brain penetrant PDE9A inhibitor for the treatment of cognitive disorders. J Med Chem 55: 9045 -54 (2012) Galan, SR; Jones, RM; Smith, NN; Dorr, PK; Westby, M; Perruccio, F; Rodrigues, D An imidazopiperidine series of CCR5 antagonists for the treatment of HIV: the discovery of N-{(1S)-1-(3-fluorophenyl)-3-[(3-endo)-3-(5-isobutyryl-2-methyl-4,5,6,7-tetrahydro-1H-imidazo[4,5-c]pyridin-1-yl)-8-azabicyclo[3.2.1]oct-8-yl]propyl}acetamide (PF-232798). J Med Chem 54: 67 -77 (2011) Cheng, H; Nair, SK; Murray, BW; Almaden, C; Bailey, S; Baxi, S; Behenna, D; Cho-Schultz, S; Dalvie, D; Dinh, DM; Edwards, MP; Feng, JL; Ferre, RA; Gajiwala, KS; Hemkens, MD; Jackson-Fisher, A; Jalaie, M; Johnson, TO; Kania, RS; Kephart, S; Lafontaine, J; Lunney, B; Liu, KK; Liu, Z; Matthews, J; Nagata, A; Niessen, S; Ornelas, MA; Orr, ST; Pairish, M; Planken, S; Ren, S; Richter, D; Ryan, K; Sach, N; Shen, H; Smeal, T; Solowiej, J; Sutton, S; Tran, K; Tseng, E; Vernier, W; Walls, M; Wang, S; Weinrich, SL; Xin, S; Xu, H; Yin, MJ; Zientek, M; Zhou, R; Kath, JC Discovery of 1-{(3R,4R)-3-[({5-Chloro-2-[(1-methyl-1H-pyrazol-4-yl)amino]-7H-pyrrolo[2,3-d]pyrimidin-4-yl}oxy)methyl]-4-methoxypyrrolidin-1-yl}prop-2-en-1-one (PF-06459988), a Potent, WT Sparing, Irreversible Inhibitor of T790M-Containing EGFR Mutants. J Med Chem 59: 2005 -24 (2016) Verhoest, PR; Chapin, DS; Corman, M; Fonseca, K; Harms, JF; Hou, X; Marr, ES; Menniti, FS; Nelson, F; Oamppound39Connor, R; Pandit, J; Proulx-Lafrance, C; Schmidt, AW; Schmidt, CJ; Suiciak, JA; Liras, S Discovery of a Novel Class of Phosphodiesterase 10A Inhibitors and Identification of Clinical Candidate 2-[4-(1-Methyl-4-pyridin-4-yl-1H-pyrazol-3-yl)-phenoxymethyl]-quinoline (PF-2545920) for the Treatment of Schizophrenia (dagger) dagger Coordinates of the PDE10A crystal structures have been depo J Med Chem 52: 5188 -96 (2009) Cui, JJ; Shen, H; Tran-Dubé, M; Nambu, M; McTigue, M; Grodsky, N; Ryan, K; Yamazaki, S; Aguirre, S; Parker, M; Li, Q; Zou, H; Christensen, J Lessons from (S)-6-(1-(6-(1-methyl-1H-pyrazol-4-yl)-[1,2,4]triazolo[4,3-b]pyridazin-3-yl)ethyl)quinoline (PF-04254644), an inhibitor of receptor tyrosine kinase c-Met with high protein kinase selectivity but broad phosphodiesterase family inhibition leading to myocardial degeneration in rats. J Med Chem 56: 6651 -65 (2013) Cui, JJ; McTigue, M; Nambu, M; Tran-Dubé, M; Pairish, M; Shen, H; Jia, L; Cheng, H; Hoffman, J; Le, P; Jalaie, M; Goetz, GH; Ryan, K; Grodsky, N; Deng, YL; Parker, M; Timofeevski, S; Murray, BW; Yamazaki, S; Aguirre, S; Li, Q; Zou, H; Christensen, J Discovery of a novel class of exquisitely selective mesenchymal-epithelial transition factor (c-MET) protein kinase inhibitors and identification of the clinical candidate 2-(4-(1-(quinolin-6-ylmethyl)-1H-[1,2,3]triazolo[4,5-b]pyrazin-6-yl)-1H-pyrazol-1-yl)ethanol (PF-04217903) for the treatment of J Med Chem 55: 8091 -109 (2012) Planken, S; Behenna, DC; Nair, SK; Johnson, TO; Nagata, A; Almaden, C; Bailey, S; Ballard, TE; Bernier, L; Cheng, H; Cho-Schultz, S; Dalvie, D; Deal, JG; Dinh, DM; Edwards, MP; Ferre, RA; Gajiwala, KS; Hemkens, M; Kania, RS; Kath, JC; Matthews, J; Murray, BW; Niessen, S; Orr, ST; Pairish, M; Sach, NW; Shen, H; Shi, M; Solowiej, J; Tran, K; Tseng, E; Vicini, P; Wang, Y; Weinrich, SL; Zhou, R; Zientek, M; Liu, L; Luo, Y; Xin, S; Zhang, C; Lafontaine, J Discovery of N-((3R,4R)-4-Fluoro-1-(6-((3-methoxy-1-methyl-1H-pyrazol-4-yl)amino)-9-methyl-9H-purin-2-yl)pyrrolidine-3-yl)acrylamide (PF-06747775) through Structure-Based Drug Design: A High Affinity Irreversible Inhibitor Targeting Oncogenic EGFR Mutants with Selectivity over Wild-Type EGFR. J Med Chem 60: 3002 -3019 (2017)
Loughnan, ML; Nicke, A; Jones, A; Adams, DJ; Alewood, PF; Lewis, RJ J Med Chem 47: 1234 -41 (2004) De Bruyn, T; van Westen, GJ; Ijzerman, AP; Stieger, B; de Witte, P; Augustijns, PF; Annaert, PP Mol Pharmacol 83: 1257 -67 (2013) Akritopoulou-Zanze, I; Albert, DH; Bousquet, PF; Cunha, GA; Harris, CM; Moskey, M; Dinges, J; Stewart, KD; Sowin, TJ Bioorg Med Chem Lett 17: 3136 -40 (2007) Thalji, RK; Raha, K; Andreotti, D; Checchia, A; Cui, H; Meneghelli, G; Profeta, R; Tonelli, F; Tommasi, S; Bakshi, T; Donovan, BT; Howells, A; Jain, S; Nixon, C; Quinque, G; McCloskey, L; Bax, BD; Neu, M; Chan, PF; Stavenger, RA Bioorg Med Chem Lett 29: 1407 -1412 (2019) Chou, O; Juang, YP; Chao, TL; Tsai, SF; Chiu, PF; Chiou, CT; Tsai, KC; Chang, SY; Liang, PH; Wong, CH J Nat Prod 86: 1428 -1436 (2023) Cirillo, PF; Asojo, OA; Khire, U; Lee, Y; Mootien, S; Hegan, P; Sutherland, AG; Peterson-Roth, E; Ledizet, M; Koski, RA; Anthony, KG ACS Med Chem Lett 11: 1843 -1847 (2020) Salsi, E; Bayden, AS; Spyrakis, F; Amadasi, A; Campanini, B; Bettati, S; Dodatko, T; Cozzini, P; Kellogg, GE; Cook, PF; Roderick, SL; Mozzarelli, A J Med Chem 53: 345 -56 (2010) Martin, FM; Beckett, RP; Bellamy, CL; Courtney, PF; Davies, SJ; Drummond, AH; Dodd, R; Pratt, LM; Patel, SR; Ricketts, ML; Todd, RS; Tuffnell, AR; Ward, JW; Whittaker, M Bioorg Med Chem Lett 9: 2887 -92 (1999) Vandeveer, GH; Arduini, RM; Baker, DP; Barry, K; Bohnert, T; Bowden-Verhoek, JK; Conlon, P; Cullen, PF; Guan, B; Jenkins, TJ; Liao, SY; Lin, L; Liu, YT; Marcotte, D; Mertsching, E; Metrick, CM; Negrou, E; Powell, N; Scott, D; Silvian, LF; Hopkins, BT Bioorg Med Chem Lett 80: (2023) Cozzi, NV; Daley, PF Bioorg Med Chem Lett 26: 959 -64 (2016) Shen, S; Doubleday, PF; Weerawarna, PM; Zhu, W; Kelleher, NL; Silverman, RB ACS Med Chem Lett 11: 1949 -1955 (2020) Borthwick, AD; Crame, AJ; Ertl, PF; Exall, AM; Haley, TM; Hart, GJ; Mason, AM; Pennell, AM; Singh, OM; Weingarten, GG; Woolven, JM J Med Chem 45: 1 -18 (2001) Nolan, KA; Zhao, H; Faulder, PF; Frenkel, AD; Timson, DJ; Siegel, D; Ross, D; Burke, TR; Stratford, IJ; Bryce, RA J Med Chem 50: 6316 -25 (2007) Fitzpatrick, PF; Chadegani, F; Zhang, S; Dougherty, V Biochemistry 56: 869 -875 (2017) Markovich, KM; Tantishaiyakul, V; Hamada, A; Miller, DD; Romstedt, KJ; Shams, G; Shin, Y; Fraundorfer, PF; Doyle, K; Feller, DR J Med Chem 35: 466 -79 (1992) Li, ZH; Liu, XQ; Geng, PF; Suo, FZ; Ma, JL; Yu, B; Zhao, TQ; Zhou, ZQ; Huang, CX; Zheng, YC; Liu, HM ACS Med Chem Lett 8: 384 -389 (2017) Perez, C; Barkley-Levenson, AM; Dick, BL; Glatt, PF; Martinez, Y; Siegel, D; Momper, JD; Palmer, AA; Cohen, SM J Med Chem 62: 1609 -1625 (2019) Ehlert, FJ; Griffin, MT; Glidden, PF J Pharmacol Exp Ther 279: 1335 -44 (1996) Zuo, C; Xu, YS; He, PF; Zhang, WJ Eur J Med Chem 261: Yoshigae, Y; Sridar, C; Kent, UM; Hollenberg, PF Drug Metab Dispos 41: 858 -69 (2013) Nara, S; Tanaka, R; Eishima, J; Hara, M; Takahashi, Y; Otaki, S; Foglesong, RJ; Hughes, PF; Turkington, S; Kanda, Y J Med Chem 46: 2467 -73 (2003) Shook, BC; Rassnick, S; Wallace, N; Crooke, J; Ault, M; Chakravarty, D; Barbay, JK; Wang, A; Powell, MT; Leonard, K; Alford, V; Scannevin, RH; Carroll, K; Lampron, L; Westover, L; Lim, HK; Russell, R; Branum, S; Wells, KM; Damon, S; Youells, S; Li, X; Beauchamp, DA; Rhodes, K; Jackson, PF J Med Chem 55: 1402 -17 (2012) Schotte, A; Janssen, PF; Gommeren, W; Luyten, WH; Van Gompel, P; Lesage, AS; De Loore, K; Leysen, JE Psychopharmacology (Berl) 124: 57 -73 (1996) Monkovic, I; Willner, D; Adam, MA; Brown, M; Crenshaw, RR; Fuller, CE; Juby, PF; Luke, GM; Matiskella, JA; Montzka, TA J Med Chem 31: 1548 -58 (1988) Ares, JJ; Kador, PF; Miller, DD J Med Chem 29: 2384 -9 (1986) Kelly, PF; Ashwell, S; Thomson, B; Collis, A; Davis, J; Walker, D; Lu, W US Patent US11311527 (2022) Miller, WH; Ali, FE; Bondinell, WE; Callahan, JF; Calvo, RR; Eggleston, DS; Haltiwanger, RC; Huffman, WF; Hwang, SM; Jakas, DR; Keenan, RM; Koster, PF; Ku, TW; Kwon, C; Newlander, KA; Nichols, AJ; Parker, MF; Samanen, JM; Southall, LS; Takata, DT Bioorg Med Chem Lett 6: 2481 -2486 (1996) Swarbrick, ME; Beswick, PJ; Gleave, RJ; Green, RH; Bingham, S; Bountra, C; Carter, MC; Chambers, LJ; Chessell, IP; Clayton, NM; Collins, SD; Corfield, JA; Hartley, CD; Kleanthous, S; Lambeth, PF; Lucas, FS; Mathews, N; Naylor, A; Page, LW; Payne, JJ; Pegg, NA; Price, HS; Skidmore, J; Stevens, AJ; Stocker, R; Stratton, SC; Stuart, AJ; Wiseman, JO Bioorg Med Chem Lett 19: 4504 -8 (2009) Lamie, PF; Ali, WAM; Bazgier, V; Rárová, L Eur J Med Chem 123: 803 -813 (2016) Xu, J; Zhang, X; Huang, F; Li, G; Leadlay, PF J Nat Prod 84: 1579 -1586 (2021) Allen, LF; Eiseman, IA; Fry, DW; Lenehan, PF Semin Oncol 30: 65 -78 Bisacchi, GS; Ahmad, S; Alam, M; Ashfaq, A; Barrish, J; Cheng, PT; Greytok, J; Hermsmeier, M; Lin, PF; Merchant, Z; Skoog, M; Spergel, S; Zahler, R Bioorg Med Chem Lett 5: 459 -464 (1995) Grabowska, K; Puszko, AK; Lipinski, PF; Laskowska, AK; Wilenska, B; Witkowska, E; Perret, GY; Misicka, A Bioorg Med Chem 25: 597 -602 (2017) Chen, T; Zhuo, LS; Liu, PF; Fang, WR; Li, YM; Huang, W Eur J Med Chem 192: (2020) Zhang, TJ; Zhang, Y; Zhang, ZH; Wang, ZR; Zhang, X; Hu, SS; Lu, PF; Guo, S; Meng, FH Bioorg Med Chem Lett 60: (2022) Dutta, AS; Gormley, JJ; McLachlan, PF; Major, JS J Med Chem 33: 2552 -60 (1990) Clark, MA; Acharya, RA; Arico-Muendel, CC; Belyanskaya, SL; Benjamin, DR; Carlson, NR; Centrella, PA; Chiu, CH; Creaser, SP; Cuozzo, JW; Davie, CP; Ding, Y; Franklin, GJ; Franzen, KD; Gefter, ML; Hale, SP; Hansen, NJ; Israel, DI; Jiang, J; Kavarana, MJ; Kelley, MS; Kollmann, CS; Li, F; Lind, K; Mataruse, S; Medeiros, PF; Messer, JA; Myers, P; O'Keefe, H; Oliff, MC; Rise, CE; Satz, AL; Skinner, SR; Svendsen, JL; Tang, L; van Vloten, K; Wagner, RW; Yao, G; Zhao, B; Morgan, BA Nat Chem Biol 5: 647 -54 (2009) Kim, CU; Misco, PF; Luh, BY; Terry, B; Bisacchi, G; Mansuri, MM Bioorg Med Chem Lett 3: 1571 -1576 (1993) Lowe, JA; Archer, RL; Chapin, DS; Chen, JB; Helweg, D; Johnson, JL; Koe, BK; Lebel, LA; Moore, PF; Nielsen, JA J Med Chem 34: 624 -8 (1991) Knudsen, LB; Nielsen, PF; Huusfeldt, PO; Johansen, NL; Madsen, K; Pedersen, FZ; Thøgersen, H; Wilken, M; Agersø, H J Med Chem 43: 1664 -9 (2000) DOLENTE, C; HEWINGS, DS; HUNZIKER, D; KRUMMENACHER, D; PETTAZZONI, PF; WICHMANN, J US Patent US20240174621 (2024) Wang, L; Bharti, na; Kumar, R; Pavlov, PF; Winblad, B Eur J Med Chem 209: (2021) Guerreiro, PS; Estácio, SG; Antunes, F; Fernandes, AS; Pinheiro, PF; Costa, JG; Castro, M; Miranda, JP; Guedes, RC; Oliveira, NG Chem Biol Drug Des 88: 915 -925 (2016) Prati, F; Buonfiglio, R; Furlotti, G; Cavarischia, C; Mangano, G; Picollo, R; Oggianu, L; di Matteo, A; Olivieri, S; Bovi, G; Porceddu, PF; Reggiani, A; Garrone, B; Di Giorgio, FP; Ombrato, R ACS Med Chem Lett 11: 825 -831 (2020) Yan, XQ; Wang, ZC; Qi, PF; Li, G; Zhu, HL Eur J Med Chem 177: 425 -447 (2019) Burtea, A; Collins, MR; Del Bel, ML; Greasley, SE; Johnson, E; Johnson, TW; Kumpf, RA; Kung, P; Montgomery, TP; Ninkovic, S; Rescourio, GC; Richardson, PF; Scales, SA; Sutton, SC US Patent US20250122183 (2025) Sherwin, PF; McMullan, PC; Covey, DF J Med Chem 32: 651 -8 (1989) Qian, XK; Zhang, J; Song, PF; Zhao, YS; Ma, HY; Jin, Q; Wang, DD; Guan, XQ; Li, SY; Bao, X; Zou, LW Bioorg Med Chem 40: (2021) Galemmo, RA; Maduskuie, TP; Dominguez, C; Rossi, KA; Knabb, RM; Wexler, RR; Stouten, PF Bioorg Med Chem Lett 8: 2705 -10 (1999) Vinick, FJ; Saccomano, NA; Koe, BK; Nielsen, JA; Williams, IH; Thadeio, PF; Jung, S; Meltz, M; Johnson, J; Lebel, LA J Med Chem 34: 86 -9 (1991) Saccomano, NA; Vinick, FJ; Koe, BK; Nielsen, JA; Whalen, WM; Meltz, M; Phillips, D; Thadieo, PF; Jung, S; Chapin, DS J Med Chem 34: 291 -8 (1991) Chang, CT; Edwards, MW; Torrence, PF; Mertes, MP J Med Chem 22: 1137 -9 (1979) Zhang, J; Zhang, QY; Tu, PF; Xu, FC; Liang, H J Nat Prod 81: 364 -370 (2018) Jacobsen, EJ; TenBrink, RE; Stelzer, LS; Belonga, KL; Carter, DB; Im, HK; Im, WB; Sethy, VH; Tang, AH; VonVoigtlander, PF; Petke, JD J Med Chem 39: 158 -75 (1996) Merchant, KM; Gill, GS; Harris, DW; Huff, RM; Eaton, MJ; Lookingland, K; Lutzke, BS; Mccall, RB; Piercey, MF; Schreur, PJ; Sethy, VH; Smith, MW; Svensson, KA; Tang, AH; Vonvoigtlander, PF; Tenbrink, RE J Pharmacol Exp Ther 279: 1392 -403 (1996) Chen, Z; Wang, ZC; Yan, XQ; Wang, PF; Lu, XY; Chen, LW; Zhu, HL; Zhang, HW Bioorg Med Chem Lett 25: 1947 -51 (2015) Gummadi, VR; Rajagopalan, S; Looi, CY; Paydar, M; Renukappa, GA; Ainan, BR; Krishnamurthy, NR; Panigrahi, SK; Mahasweta, K; Raghuramachandran, S; Rajappa, M; Ramanathan, A; Lakshminarasimhan, A; Ramachandra, M; Wong, PF; Mustafa, MR; Nanduri, S; Hosahalli, S Bioorg Med Chem Lett 23: 4911 -8 (2013) Zhang, WT; Ruan, JL; Wu, PF; Jiang, FC; Zhang, LN; Fang, W; Chen, XL; Wang, Y; Cao, BS; Chen, GY; Zhu, YJ; Gu, J; Chen, JG J Med Chem 52: 718 -25 (2009) Cai, L; Xiong, PF; Li, T; Li, C; Wu, ZX; Hong, YL; Wang, JT; Zhang, MY; Yang, XQ; Xu, QQ; Shi, H; Luo, QC; Li, R; Liu, MM Eur J Med Chem 271: Li, PH; Zeng, P; Chen, SB; Yao, PF; Mai, YW; Tan, JH; Ou, TM; Huang, SL; Li, D; Gu, LQ; Huang, ZS J Med Chem 59: 238 -52 (2016) Taylor, JW; Jin, QK; Sbacchi, M; Wang, L; Belfiore, P; Garnier, M; Kazantzis, A; Kapurniotu, A; Zaratin, PF; Scheideler, MA J Med Chem 45: 1108 -21 (2002) Ni, WW; Liu, Q; Ren, SZ; Li, WY; Yi, LL; Jing, H; Sheng, LX; Wan, Q; Zhong, PF; Fang, HL; Ouyang, H; Xiao, ZP; Zhu, HL Bioorg Med Chem 26: 4145 -4152 (2018) Freeman, JP; Michalson, ET; D'Andrea, SV; Baczynskyj, L; Von Voigtlander, PF; Lahti, RA; Smith, MW; Lawson, CF; Scahill, TA; Mizsak, SA J Med Chem 34: 1891 -6 (1991) Amirnasr, A; Verdijk, RM; van Kuijk, PF; Taal, W; Sleijfer, S; Wiemer, EAC PLoS ONE 12: Troxler, T; Enz, A; Hoyer, D; Langenegger, D; Neumann, P; Pfäffli, P; Schoeffter, P; Hurth, K Bioorg Med Chem Lett 18: 979 -82 (2008) KOBAYAKAWA, Y; OSHIMA, T; ITO, S; SCHÖPF, P US Patent US20240317759 (2024) Sloman, DL; Chessari, G; Schöpf, P; Howard, S; Kawai, Y; Shibata, K; Asakura, H; Uno, T; Sagara, T; Nakamura, M; Kobayakawa, Y; Bennett, DJ; Bharathan, I; Graham, TH; Han, Y; Hussain, Z; Ma, X; Mandal, M; Otte, RD; Palani, A; Swaminathan, U; Uehling, M; Ye, Y; Chau, R; Christian, AH; Gathiaka, S; Henderson, TJ; Hennessy, ET; Hoover, AJ; Kawamura, S; Kolaj, I; Lyons, TW; Mitcheltree, MJ; Sather, A US Patent US20240239788 (2024) Silva, LR; da Silva Santos-Júnior, PF; de Andrade Brandão, J; Anderson, L; Bassi, ÊJ; Xavier de Araújo-Júnior, J; Cardoso, SH; da Silva-Júnior, EF Bioorg Med Chem 28: (2020)
ChEMBL_1515039 (CHEMBL3616251) Inhibition of human MGAT3 expressed in HEK293 cells assessed as effect on incorporation of [1,3-14C] glycerol into TAG by TLC method in presence of DGAT1 inhibitor PF-04620110 and DGAT2 inhibitor PF-06424439 ChEMBL_2035328 (CHEMBL4689486) Inhibition pf human plasmin using Mes-Darg-Phe-Arg-AMC as substrate by fluorescence assay ChEMBL_1634420 (CHEMBL3877212) Inhibition of PF-06658607 binding to BTK in human Ramos cells after 1 hr by gel-based ABPP assay ChEMBL_1634421 (CHEMBL3877213) Inhibition of PF-06422899 binding to EGFR in human A431 cells after 1 hr by gel-based ABPP assay ChEMBL_2254306 (CHEMBL5168516) Displacement of [3H]PF-06883365 from FAP-tagged human GLP-1R expressed in CHO cells assessed as inhibition constant by radioligand binding assay ChEBML_1687029 Agonist activity at mGlu4 assessed as decrease in PF-induced presynaptic calcium amplitude in rat coronal slices by Fluo-4FF-AM dye based fluorescence assay ChEMBL_1687029 (CHEMBL4037508) Agonist activity at mGlu4 assessed as decrease in PF-induced presynaptic calcium amplitude in rat coronal slices by Fluo-4FF-AM dye based fluorescence assay ChEMBL_1634422 (CHEMBL3877214) Inhibition of PF-06658607 binding to recombinant C-terminal FLAG-tagged FAM213A (unknown origin) expressed in HEK293T cells after 1 hr by gel-based ABPP assay ChEMBL_1634423 (CHEMBL3877215) Inhibition of PF-06422899 binding to recombinant C-terminal FLAG-tagged DUS2L (unknown origin) expressed in HEK293T cells after 1 hr by gel-based ABPP assay ChEMBL_1748624 (CHEMBL4183134) Displacement of [3H]-PF-6475886 from recombinant human full length FL-tagged BACE1 expressed in HEK293 cell membranes after 30 mins by parallel scintillation proximity assay ChEMBL_1748625 (CHEMBL4183135) Displacement of [3H]-PF-6475886 from recombinant human full length Myc-DDK-tagged BACE2 expressed in HEK293 cell membranes after 30 mins by parallel scintillation proximity assay ChEMBL_2347444 Inhibition of MPS1 in human MDA-MB-468 cells assessed as decrease in phosphorylation of histone H3 at Ser10 incubated for 2 hrs in presence of PF-2771 by ELISA ChEMBL_1742691 (CHEMBL4158441) Antagonist activity at human TRPV4 expressed in BHK/AC9 or HEK MSR2 cells assessed as inhibition of PF-04674114-induced Ca2+ flux pre-incubated for 10 mins before agonist addition by FLIPR assay FRET protease activity assay PF-00835231 was evaluated against 3CLpro from a variety of other coronaviruses representing alpha, beta and gamma groups of Coronaviridae, using biochemical F rster Resonance Energy Transfer (FRET) protease activity assays. TR-FRET Competition Assay Assay buffer was 20 mM HEPES, pH 7.5, 10 mM MgCl2, 0.01% BSA, 1 mM DTT, 0.0005% Tween 20, and 2% DMSO. Inactivation kinetic reactions were performed by preparing 15 μL of a 1.33Χ solution of (final concentrations) 2 nM Eu-Ab, 1-8 nM kinase (optimal concentrations of each kinase were empirically determined) and a variable concentration of PF- 06651600, and pre-incubating this for a variable amount of time (detailed below). This was then combined this with 5 μL of 4X solution of the validated probe (150 nM, final concentration). For all kinases, the following experiments were performed: (A) [PF-06651500] = 0, 4.9, 14.8, 44.4, 133.3, and 400 nM; pre-incubation time = 2 h. (B) [PF-06651500] = 0, 0.5, 1.0, 2.0, 4.0, and 8.0 μM; pre-incubation time = 120 s. For JAK3, the following additional experiments were performed: [PF-06651500] = 0, 0.66, 1.98, 5.93, 17.8, 53.3, 160, 480 nM; pre-incubation time of (C) 30 s, (D) 60 s, and (E) 1.5 h. The assays were read using an EnVi Biacore SPR Assay In running buffer (0.01 M HEPES pH 7.4, 0.15 M NaCl, 0.005% Surfactant P20, mM MgCl2, and 1% DMSO)N-terminally GST-fused purified FAK enzyme was captured on both spot 1 and 2. Spot 1 was subsequently blocked by loading with 30 nM PF-562,271 at the beginning of each cycle. Concentration series' of the test compounds were injected over the spots at 25° C. Biacore SPR Assay Binding parameters of compounds were determined using a Biacore S51 sensor. An anti-GST antibody was immobilized onto a CM5 chip by primary amine-coupling in accordance with the manufacturer's recommendations.In running buffer (0.01 M HEPES pH 7.4, 0.15 M NaCl, 0.005% Surfactant P20, mM MgCl2, and 1% DMSO)N-terminally GST-fused purified FAK enzyme was captured on both spot 1 and 2. Spot 1 was subsequently blocked by loading with 30 nM PF-562,271 at the beginning of each cycle. Concentration series' of the test compounds were injected over the spots at 25 C. The specific binding was calculated as difference between spot 2 and 1 signals followed by solvent correction. Biochemical Assay PI3KA_FL: Genes encoding for full length p110alpha and p85alpha subunits of PI3Kalpha complex were subcloned from existing constructs into pFASTBAC Dual vector (Life Technologies, Carlsbad, Calif.) using standard cloning procedures. Gene encoding p110alpha subunit was subcloned into polyhedrine promoter while gene encoding p85alpha subunit was subcloned into p10 promoter. Additionally, sequence encoding for histidine tag and Tobacco Etch Virus ("TEV") cleavage site preceded p110alpha ORF (Open Reading Frame). The biochemical assays of kinase activity of full-length PI3Kalpha (full-length p110alpha/p85a) or truncated PI3Kalpha (p110alpha/iSH2 p85a) were conducted using a fluorescence polarization format similar to the procedure of Yuan J., et al., (2011) PF-04691502, a Potent and Selective Oral Inhibitor of PI3K and mTOR Kinases with Antitumor Activity, Mol Cancer Ther. 10, 2189-2199. The enzymatic reactions were conducted in 50 uL volumes in 96-well plates. GABAA (α1β2γ2) Receptor Current using Patch-Clamp Technique A whole-cell patch-clamp technique was used to study the allosteric regulation effect of the compound of the present invention on the GABAA (α1β2γ2) receptor.HEK293 cell lines stably expressing the GABAA (α1β2γ2) receptor were used. The GABAA (α1β2γ2) receptor gene information was as follows: GABA-α1: NM_000806; GABA-β2: NM_021911; GABA-γ2: NM_198904. The voltage stimulus of GABA receptor current recorded by a whole-cell patch-clamp technique was as follows: when whole-cell sealing was formed, the cell membrane voltage was clamped at −70 mV. The peak value of current was recorded after sequentially spraying test compounds from low concentration to high concentration and 100 μM GABA onto the cell surface in a Gap-free mode. The mode of administration of test compounds was as follows: for each concentration, the test compound was administered 1-2 times; the cells were washed with extracellular fluid for 1 min before detection was performed on the test compound at another concentration; and finally, 100 μM GABA was given as the control. The experimental data was collected by an EPC-10 amplifier (HEKA) and stored in PatchMaster (HEKA) software. A microelectrode puller was used to pull capillary glass tubes into a recording electrode. A microelectrode manipulator was manipulated under an inverted microscope to contact the recording electrode with cells, and negative pressure suction was applied to form a GΩ seal. After the GΩ seal was formed, rapid capacitance compensation (pF) was conducted, and then negative pressure was continued to break cell membranes, forming a whole-cell recording mode. Then slow capacitance compensation was conducted, and the membrane capacitance (pF) and series resistance were recorded. Biochemical Assay PI3KA_Act: Genes encoding for full length p110alpha and p85alpha nSH-iSH2=niSH2 (p85a aminoacids 322-600) subunits of PI3Kalpha complex were subcloned from existing constructs into pFASTBAC Dual vector (Life Technologies, Carlsbad, Calif.) using standard cloning procedures. Gene encoding p110alpha subunit was subcloned into polyhedrine promoter while gene encoding p85alpha niSH2 domains was subcloned into p10 promoter. Additionally, Human Rhinovirus 3C Protease (HRV 3C) site was introduced between nSH2 and iSH2, replacing aminoacids 431-438 of p85alpha with LEVLFQGP HRV 3C recognition sequence, using standard QuickChange mutagenesis protocol (Agilent Technologies, CA). Recombinant baculovirus was generated using Bac-to-Bac protocol (Life Technologies, Carlsbad, Calif.). The biochemical assays of kinase activity of full-length PI3Kalpha (full-length p110alpha/p85a) or truncated PI3Kalpha (p110alpha/iSH2 p85a) were conducted using a fluorescence polarization format similar to the procedure of Yuan J., et al., (2011) "PF-04691502, a Potent and Selective Oral Inhibitor of PI3K and mTOR Kinases with Antitumor Activity, Mol Cancer Ther. 10, 2189-2199. hERG Kd Inhibition Assay Compounds were dissolved in DMSO to a stock concentration of 10 mM and tested in 10-dose IC50 mode, in triplicate, with a 3-fold serial dilution starting at 100 uM. Control compound E-4031 was tested in 10-dose IC50 mode with 3-fold serial dilution starting at 1 uM. The assay is based on the competition of fluorescently labeled Tracer binding to the membrane preparation containing 1x Predictor hERG Membrane with 1 nM Predictor hERG Tracer Red in a buffer with the composition of 25 mM Hepes, pH 7.5, 15 mM KCl, 1 mM MgCl2, 0.05% PF-127, and 1% DMSO. Compounds in DMSO were added into the membrane mixture by using sonication, the tracer was added and gently mixed in the dark. The fluorescence was measured after 4 hours incubation at room temperature. The measurement parameters are as follows: Ex=531 nm FP and Em=595 nm P and S. FS-3 Assay Autotaxin Inhibitor Screening Kits are available from Echelon Biosciences, Logan, Utah, USA [http://echelon-inc.com/, accessed 6 Oct. 2011]. Using the methods of Gierse et al [A novel autotaxin inhibitor reduces lysophosphatidic acid levels in plasma and the site of inflammation, Journal of Pharmacology and Experimental Therapeutics, vol 334(1), 310-317 (2010). The FS-3 assay to identify ATX inhibitors was preformed as follows: 3 ul of standard inhibitor (referred to as PF-8380 in Gierse et al above) and test compounds were added to an assay plate. To each assay well, containing test compounds or standard, 24 ul of human Autotaxin enzyme (2 nM) was added. The assay plate was then centrifuged at 1000 rpm for 1 minute and allowed to incubate at 37° C. for 30 minutes. Following the incubation period each plate was read in a fluorescence plate reader (Spectra Max M5: excitation: 494 nm and emission: 520 nm) and IC50 values were derived from inhibition of FS-3 fluorescence (as described above). Mpro Enzyme Activity Assay Representative Compounds of the Disclosure, PF-00835231, and PF-07321332 were tested for their capability to inhibit the SARS-CoV-2 main protease Mpro by using a biochemical FRET-based Mpro enzyme activity assay. Representative compounds of Formula I′ and II′ were also tested. See PCT/US2021/046311. Briefly, recombinant Mpro protein (Ser1-Gln306; with proven proteolytic activity) was purchased from Biosynth Carbosynth (Staad, Switzerland). An EDANS- and Dabcyl-labeled peptide was purchased from Life Technologies GmbH (Darmstadt, Germany), and served as substrate peptide for Mpro proteolytic cleavage allowing fluorescence resonance energy transfer (FRET) read-out. Due to the Mpro-mediated cleavage of the substrate peptide, the EDANS fluorescence (λexc.=336 nm; λem.=490 nm) becomes dequenched (from disappearing Dabcyl) and increases with increasing Mpro activity. The assay buffer was 20 mM Tris buffer supplemented with 100 mM NaCl, and 1 mM EDTA, adjusted to pH 7.3 with 1N HCl. The test compounds were diluted from 20 mM stocks in DMSO; the stock of the substrate peptide was 250 μM in aqua bidest. The catalytic activity of the recombinant Mpro enzyme was 20 U/mg. It was checked in advance that neither the assay buffer nor the Mpro protein by itself emit fluorescence at 490 nm under 336 nm excitation. The basal emission of the uncleaved substrate peptide was subtracted from all results by baseline correction. The enzyme assay was carried out in black U-form half-area 96-wells. Each assay sample was finally composed of 0.4 μL substrate peptide stock (3× ad 20 μL assay buffer to yield finally 2 μM; 100 pmol), 0.1 μL Mpro enzyme (20 mU in assay buffer ad 20 μL) and 20 μL of 3× (in assay buffer) test compound dilution, resulting in a final sample volume of 60 μL. The final test compound concentrations were: 10 μM for compound fast-screening, and 0-200 μM for IC50 determinations. Initially, Mpro enzyme and test compound was added and mixed in 96-well and pre-incubated for 30 min in the dark with 200 rpm swiveling at room temperature. Subsequently, the reaction was started by addition of the substrate peptide, and followed by a fluorescence kinetic (λexc.=336 nm/λem.=500 nm/CutOff=435 nm; 30 min with 2 min increment by using a SpectraMax M5 multiwell plate reader (Molecular Devices, San Jose, CA, USA). Inhibitory Activity against 3CLPpro The inhibitory activity of the compounds against SARS-CoV-2 3CLpro was determined by fluorescence resonance energy transfer technique. A suitable amount of the above compounds was respectively weighed, and formulated in DMSO to give solutions over suitable concentration gradients. 5 μL of the formulated solution and 91 μL of Assay Reagent (Assay Buffer: 2019-nCoV Mpro/3CLpro=90:1, purchased from Shanghai Beyotime Biotechnology Co., Ltd.) were added to a 96-well black plate, mixed uniformly, and incubated for 10 min at 37° C. in the dark. 4 μL of Substrate (100 μM Dabcyl-KTSAVLQSGFRKME-Edans, purchased from Shanghai Beyotime Biotechnology Co., Ltd.) was rapidly added to each well, and mixed uniformly. After incubation for 5 min at 37° C. in the dark, the signal gradually became stable. The fluorescence in 5-30 min was detected on a multifunctional plate reader (Thermo Fisher Technology Co., LTD., Varioskan Flash) and the inhibition percentage of the sample was calculated. The excitation wavelength was 340 nm and the emission wavelength was 490 nm. The Assay Reagent free of the compound was used as a control with 100% enzyme activity, the Assay Buffer free of SARS-CoV-2 Mpro/3CLpro was used as a blank control, and S-216722 (Shandong Xuanshuo Medical Technology Co., Ltd.) and PF-07321332 (Jinan Jianfeng Chemical Co., Ltd.) were used as positive controls. The rest of the treatment method was the same. SARS-CoV-2 Coronavirus 3C Protease FRET Assay The proteolytic activity of the main protease, 3CLpro, of SARS-CoV-2 was monitored using a continuous fluorescence resonance energy transfer (FRET) assay. The SARS-CoV-23CLpro assay measures the activity of full length SARS-CoV-23CL protease to cleave a synthetic fluorogenic substrate peptide with the following sequence: Dabcyl-KTSAVLQ-SGFRKME-Edans modelled on a consensus peptide (V. Grum-Tokars et al. Evaluating the 3C-like protease activity of SARS-coronavirus: recommendations for standardized assays for drug discovery. Virus Research 133 (2008) 63 73). The fluorescence of the cleaved Edans peptide (excitation 340 nm / emission 490 nm) is measured using a fluorescence intensity protocol on a Flexstation reader (Molecular Devices). The fluorescent signal is reduced in the present of PF-835231, a potent inhibitor of SARS-CoV-23CLpro. The assay reaction buffer contained 20 mM Tris-HCl (pH 7.3), 100 nM NaCl, 1 mM EDTA and 25 µM peptide substrate. Enzyme reactions were initiated with the addition of 15 nM SARS-CoV-23CL protease and allowed to proceed for 60 minutes at 23 oC. Percent inhibition or activity was calculated based on control wells containing no compound (0% inhibition/100% activity) and a control compound (100% inhibition/0% activity). IC50 values were generated using a four-parameter fit model using ABASE software (IDBS). Ki values were fit to the Morrison equation with the enzyme concentration parameter fixed to 15 nM, the Km parameter fixed to 14 µM and the substrate concentration parameter fixed to 25 µM using ABASE software (IDBS). SARS-CoV-2 Coronavirus 3C Protease FRET Assay The proteolytic activity of the main protease, 3CLpro, of SARS-CoV-2 was monitored using a continuous fluorescence resonance energy transfer (FRET) assay. The SARS-CoV-2 3CLpro assay measures the activity of full-length SARS-CoV-2 3CL protease to cleave a synthetic fluorogenic substrate peptide with the following sequence: Dabcyl-KTSAVLQ-SGFRKME-Edans modelled on a consensus peptide (V. Grum-Tokars et al. Evaluating the 3C-like protease activity of SARS-coronavirus: recommendations for standardized assays for drug discovery. Virus Research 133 (2008) 63-73). The fluorescence of the cleaved Edans peptide (excitation 340 nm/emission 490 nm) is measured using a fluorescence intensity protocol on a Flexstation reader (Molecular Devices). The fluorescent signal is reduced in the present of PF-835231, a potent inhibitor of SARS-CoV-2 3CLpro. The assay reaction buffer contained 20 mM Tris-HCl (pH 7.3), 100 nM NaCl, 1 mM EDTA and 25 μM peptide substrate. Enzyme reactions were initiated with the addition of 15 nM SARS-CoV-2 3CL protease and allowed to proceed for 60 minutes at 23° C. Percent inhibition or activity was calculated based on control wells containing no compound (0% inhibition/100% activity) and a control compound (100% inhibition/0% activity). IC50 values were generated using a four-parameter fit model using ABASE software (IDBS). Ki values were fit to the Morrison equation with the enzyme concentration parameter fixed to 15 nM, the Km parameter fixed to 14 μM and the substrate concentration parameter fixed to 25 μM using ABASE software (IDBS). Biochemical Assay The biochemical assays of kinase activity of full-length PI3Kα (full-length p110α/p85α) or truncated PI3Kα (p110α/iSH2 p85α) were conducted using a fluorescence polarization format similar to the procedure of Yuan J., et al., (2011) PF-04691502, a Potent and Selective Oral Inhibitor of PI3K and mTOR Kinases with Antitumor Activity, Mol Cancer Ther. 10, 2189-2199. The enzymatic reactions were conducted in 50 μL volumes in 96-well plates. The reactions contained human recombinant PI3Kα (2 nM full-length p110α/p85α or 0.5 nM p110α/iSH2 p85) and 30 μM phosphatidylinositol 4,5-bisphosphate (PIP2) (Avanti Polar Lipids, Inc., Alabaster, Ala.) and were sonicated for 1 minute prior to adding PI3Kα enzyme (PI3KA_Act or PI3KA_FL), DMSO or test compound (12-point 3-fold serial dilution, 3 μM top dose, 2% DMSO final concentration), 5 mM MgCl2, 50 mM HEPES pH 7.4, 150 mM NaCl, 1 mM DTT, and 0.05% 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS). The reactions were initiated by the addition of ATP (41 μM, Km-level, for full-length p110α/p85 or 1 mM ATP for p110α/iSH2 p85), following a 15-min preincubation. The reactions were incubated for 30 min at room temperature, stopped with EDTA pH 8 (10 mM final concentration). In a detection plate, 15 μL of detector/probe mixture, containing 480 nM GST-Grp1PH domain protein (University of Dundee, Dundee, UK) and 12 nM carboxytetramethylrhodamine (TAMRA)-tagged fluorescent phosphatidylinositol (3,4,5)-triphosphate (PIP3) (Echelon Biosciences, Inc., Salt Lake City, Utah) in assay buffer, was mixed with 15 μL of kinase reaction mixture. The plate was shaken for 30 minutes and fluorescence polarization values were measured on an LJL Analyst HT plate reader (Molecular Devices, Sunnyvale, Calif.). Electrophysiology Assay HEK293 cells were cultured in DMEM with 4.5 g L-1 glucose, L-glutamine, and sodium pyruvate (Mediatech) containing 10% (v/v) FBS (Axenia BioLogix) and 1% (v/v) penicillin-streptomycin, at 37° C. and with 5% CO2. Cells were lifted with trypsin-EDTA (Life Technologies) and passaged to 6-well plates (Warner Instruments) 3-4 d before recording. Transient transfection was performed with Lipofectamine 2000 (Thermo Fisher Scientific) 2 days before recording. The plasmids of human Kir6.2 and SUR1 were the gift from Dr. Show-Ling Shyng (Oregon Health and Science University), and we fused mCherry fluorescent protein to the C-terminus of Kir6.2. The vector ratio for co-transfection of Kir6.2 to SUR1 was 1:10. Before recording, cells were lifted with trypsin-EDTA, kept in modified Tyrode's saline (140 mM NaCl, 5 mM KCl, 10 mM HEPES, 2 mM CaCl2), 1 mM MgCl2, 10 mM glucose, pH 7.2 ˜ 7.3 with HCl), and were used within 8 hours. For recording, an aliquot of cells was transferred to a recording chamber on a Nikon-TE2000 Inverted Scope (Nikon Instruments), and transfection was confirmed with fluorescent microscopy. The pipette solution contained: 145 mM KCl, 1 mM MgCl2, 5 mM EGTA, 2 mM CaCl2), 20 mM HEPES, 0.3 mM K2-ATP and 0.3 mM K2-ADP. Patch borosilicate pipettes (Sutter Instrument) were pulled from a Sutter P-97 puller with resistances of 2-3 MΩ. Data were acquired using a Axopatch 200B amplifier controlled by Clampex 10.2 via Digidata 1550 Å (Axon Instruments), sampled at 10 kHz, filtered at 2 kHz. Membrane capacitance was around 15 pF. Rs was around 5 MΩ. The membrane potential was held at −80 mV and a ramp to +80 mV (1 mV/ms) was applied every second. Bath was switched to 150 mM KCl, 10 mM HEPES, 2 mM CaCl2), and the chemical to be tested was dissolved in it and puffed with VC3-8xP pressurized perfusion system (ALA Science). Fluorescence Polarization Assay Assays were performed at room temperature in 384-well microtiter plates with an incubation volume of 20.2 μL. Solutions of test compounds were prepared in DMSO and serially diluted with DMSO to yield 8 μL of each of 10 solutions differing by 3-fold in concentration, at 32 serial dilutions per plate. 100% inhibition is determined using a known PDE9 inhibitor, such as 1-(2-chlorophenyl)-6-[(2R)-3,3,3-trifluoro-2-methylpropyl]-1,5-dihydro-4H-pyrazolo[3,4-d]pyrimidine-4-one (BAY 73-6691), (6-[(3S,4S)-4-methyl-1-(pyrimidin-2-ylmethyl)pyrrolidin-3-yl]-1-(tetrahydro-2H-pyran-4-yl)-1,5-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one (PF-04447943). 0% of inhibition is determined by using DMSO (1% final concentrations). A Labcyte Echo 555 (Labcyte, Sunnyvale, CA) is used to dispense 200 nL from each well of the titration plate to the 384 well assay plate. Human PDE9A2 membrane preps were diluted to 1 ng/ml. FAM-labeled cGMP substrate (Molecular Devices, Sunnyvale, CA) was at a concentration of 100 nM (Km of PDE9 for cGMP is 70-170 nM) in the assay buffer (10 mM Tris HCl, pH 7.2, 10 mM MgCl2, 0.05% NaN3 0.01% Tween-20, and 1 mM DTT). PDE9 enzyme mix and compounds were mixed and incubated at room temperature for 30 min. Following which, FAMcGMP substrate was added, shaken and incubated for an additional 60 min at room temperature. The final concentration of human PDE9 membrane preparations were 0.5 ng/ml. The final concentration of FAM-cGMP was 50 nM. After the incubation period, the enzymatic reaction was stopped by addition of binding solution (IMAP-FP, Molecular Devices, comprised of 80% Solution A, 20% Solution B and a 1:600 dilution of binding reagent) to each well. The plates were shaken then incubated at room temperature for 1 h prior to determining the fluorescence polarization (mP) using a Perkin Elmer EnVision™ plate reader (Waltham, MA). SARS-CoV-2 Coronavirus 3C Protease FRET Assay The proteolytic activity of the main protease, 3CLpro, of SARS-CoV-2 was monitored using a continuous fluorescence resonance energy transfer (FRET) assay. The SARS-CoV-2 3CLpro assay measures the activity of full-length SARS-CoV-2 3CL protease to cleave a synthetic fluorogenic substrate peptide with the following sequence: Dabcyl-KTSAVLQ-SGFRKME-Edans modelled on a consensus peptide (V. Grum-Tokars et al. Evaluating the 3C-like protease activity of SARS-coronavirus: recommendations for standardized assays for drug discovery. Virus Research 133 (2008) 63-73). The fluorescence of the cleaved Edans peptide (excitation 340 nm / emission 490 nm) is measured using a fluorescence intensity protocol on a Flexstation reader (Molecular Devices). The fluorescent signal is reduced in the present of PF-835231, a potent inhibitor of SARS-CoV-2 3CLpro. The assay reaction buffer contained 20 mM Tris-HCI (pH 7.3), 100 nM NaCI, 1 mM EDTA and 25 μM peptide substrate. Enzyme reactions were initiated with the addition of 15 nM SARS-CoV-2 3CL protease and allowed to proceed for 60 minutes at 23 °C. Percent inhibition or activity was calculated based on control wells containing no compound (0% inhibition/100% activity) and a control compound (100% inhibition/0% activity). ICso values were generated using a four-parameter fit model using ABASE software (I DBS). Ki values were fit to the Morrison equation with the enzyme concentration parameter fixed to 15 nM, the Km parameter fixed to 14 μM and the substrate concentration parameter fixed to 25 μM using ABASE software (I DBS).Proteolytic activity of SARS-CoV-2 Coronavirus 3CL protease is measured using a continuous fluorescence resonance energy transfer assay. The SARS-CoV-2 3CLpro FRET assay measures the protease catalyzed cleavage of TAMRA-SITSAVLQSGFRKMK-(DABCYL)-OH to TAMRA - SITSAVLQ and SGFRKMK(DABCYL)-OH. The fluorescence of the cleaved TAMRA (ex. 558 nm / em.581 nm) peptide was measured using a TECAN SAFI RE fluorescence plate reader over the course of 10 min. Typical reaction solutions contained 20 mM HEPES (pH 7.0), 1 mM EDTA, 4.0 mM FRET substrate, 4% DMSO and 0.005% Tween-20. Assays were initiated with the addition of 25 nM SARS 3CLpro (nucleotide sequence 9985-10902 of the Urbani strain of SARS coronavirus complete genome sequence (NCBI accession number AY278741)). Fluorescence polarization assay Rhesus PDE9A2 was amplified from rhesus whole brain cDNA (Biochain Institute) essentially as described in Hutson, et al. Neuropharmacology (2011) 61(4):665-676. HEK 293 or CHO cells over-expressing rhesus or rat PDE9A2 (created by DiscoverX from mRNA Genbank accession # NM_138543) respectively were lysed in 20 mM HEPES, 1 mM EDTA buffer with protease inhibitors (Roche, Indianapolis, Ind.). After brief homogenization, cells were pelleted via centrifugation at 75,000×g for 20 min at 4° C. The pellets were re-suspended, centrifuged and re-pelleted again in the same manner. The membrane fraction was collected in 20 mM HEPES, 1 mM MgCl2 with protease inhibitors. Human PDE9 (PDE9A2, GenBank Accession No. NM_001001567), full length with N-terminal GST tag, was purchased from BPS Bioscience. The fluorescence polarization assay for cyclic nucleotide phosphodiesterases was performed using an IMAP FP kit supplied by Molecular Devices, Sunnyvale, Calif. (product # R8139). IMAP technology has been applied previously to phosphodiesterase assays (Huang, W., et al., J. Biomol Screen, 2002, 7: 215). Assays were performed at room temperature in 384-well microtiter plates with an incubation volume of 20.2 μL. Solutions of test compounds were prepared in DMSO and serially diluted with DMSO to yield 8 μL of each of 10 solutions differing by 3-fold in concentration, at 32 serial dilutions per plate. 100% inhibition is determined using a known PDE9 inhibitor, such as 1-(2-chlorophenyl)-6-[(2R)-3,3,3-trifluoro-2-methylpropyl]-1,5-dihydro-4H-pyrazolo[3,4-d]pyrimidine-4-one (BAY 73-6691) (Wunder et al, Mol. Pharmacol., 2005, 68(6): 1775-81), (6-[(3S,4S)-4-methyl-1-(pyrimidin-2-ylmethyl)pyrrolidin-3-yl]-1-(tetrahydro-2H-pyran-4-yl)-1,5-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one (PF-04447943) (Wager et al., ACS Chemical Neuroscience, 2010, 1:435-449). 0% of inhibition is determined by using DMSO (1% final concentrations). A Labcyte Echo 555 (Labcyte, Sunnyvale, Calif.) is used to dispense 200 nL from each well of the titration plate to the 384 well assay plate. Rhesus membrane preps were diluted to 4 ng/ml, rat membrane preps diluted to 13 ng/ml, and human PDE9A2 diluted to 1 ng/ml. FAM-labeled cGMP substrate (Molecular Devices, Sunnyvale, Calif.) was at a concentration of 100 nM (Km of PDE9 for cGMP is 70-170 nM) in the assay buffer (10 mM Tris HCl, pH 7.2, 10 mM MgCl2, 0.05% NaN3 0.01% Tween-20, and 1 mM DTT). PDE9 enzyme mix and compounds were mixed and incubated at room temperature for 30 min. Following which, FAMcGMP substrate was added, shaken and incubated for an additional 60 min at room temperature. The final concentrations of rhesus and rat membrane preparations were 2 ng/ml and 6.5 ng/ml, respectively, while the human PDE9 was used at a final concentration of 0.5 ng/ml. The final concentration of FAM-cGMP was 50 nM. After the incubation period, the enzymatic reaction was stopped by addition of binding solution (IMAP-FP, Molecular Devices, comprised of 80% Solution A, 20% Solution B and a 1:600 dilution of binding reagent) to each well. The plates were shaken then incubated at room temperature for 1 h prior to determining the fluorescence polarization (mP) using a Perkin Elmer EnVision plate reader (Waltham, Mass.).
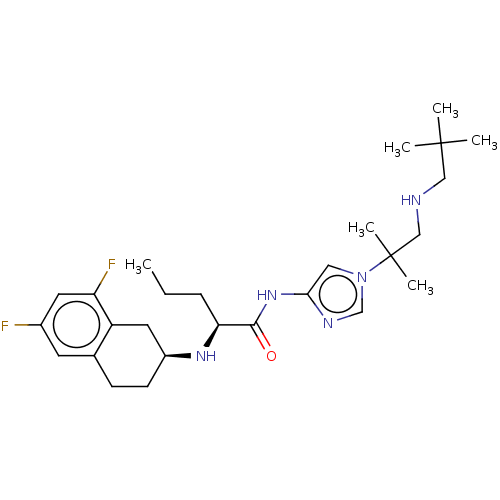 Nirogacestat PF-03084014 PF-3084014 PF 03084014 PF 3084014 BDBM50458159
Nirogacestat PF-03084014 PF-3084014 PF 03084014 PF 3084014 BDBM50458159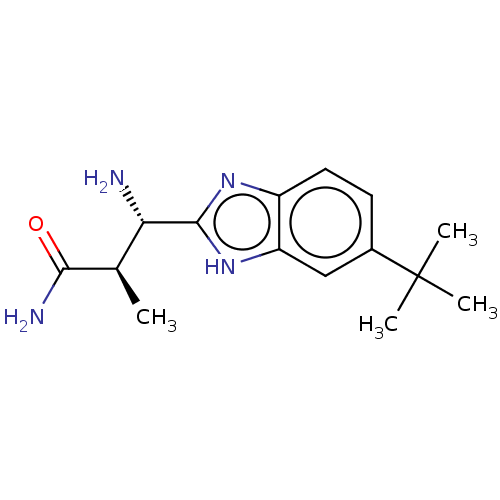 BDBM50507839 PF-6305591 Pf-06305591
BDBM50507839 PF-6305591 Pf-06305591 PF-00215937 BDBM84797 PF-00215955
PF-00215937 BDBM84797 PF-00215955 BDBM50457459 Pf-00489791 PF-489,791 UK-489,791
BDBM50457459 Pf-00489791 PF-489,791 UK-489,791 CHEMBL587528 GNF-Pf-1521 BDBM50040883 GNF-Pf-472
CHEMBL587528 GNF-Pf-1521 BDBM50040883 GNF-Pf-472 BDBM694370 US20250188102, Compound PF-07321332 US12077605, Compound PF-07321332
BDBM694370 US20250188102, Compound PF-07321332 US12077605, Compound PF-07321332 JAK-I BDBM50611955 PF-03394197 JAKI OCLACITINIB PF 03394197
JAK-I BDBM50611955 PF-03394197 JAKI OCLACITINIB PF 03394197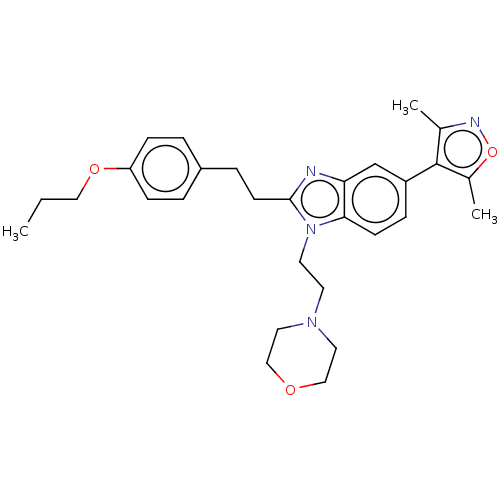 BDBM188517 PF-CBP1
BDBM188517 PF-CBP1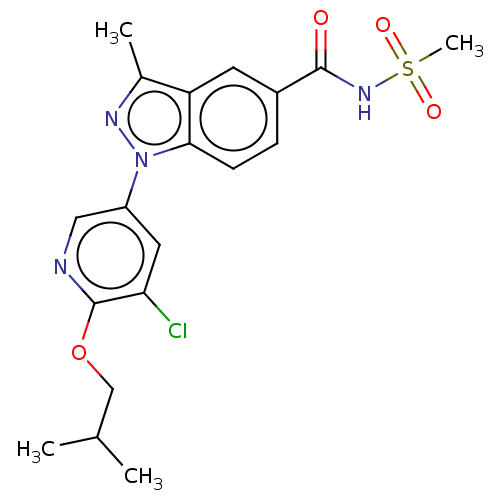 BDBM50466964 Pf-05241328
BDBM50466964 Pf-05241328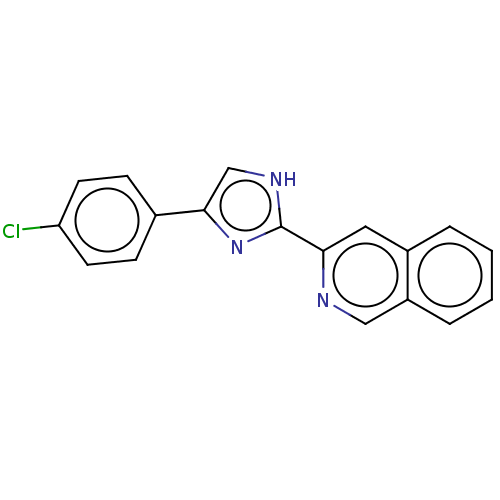 BDBM84796 PF-00215924
BDBM84796 PF-00215924 PF-00215937 BDBM84798
PF-00215937 BDBM84798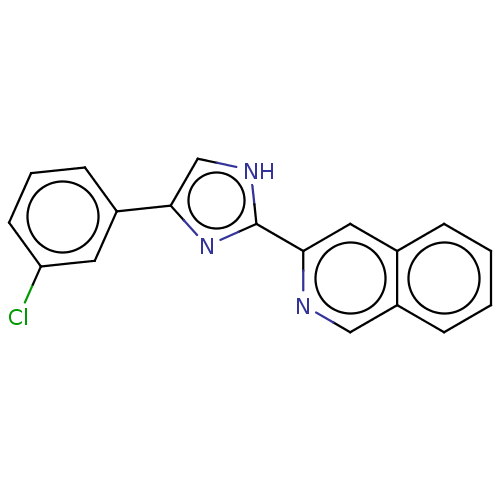 PF-00240292 BDBM84800
PF-00240292 BDBM84800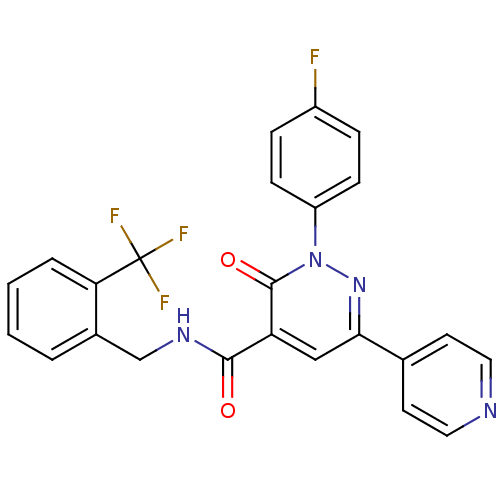 PF-00416121 BDBM84801
PF-00416121 BDBM84801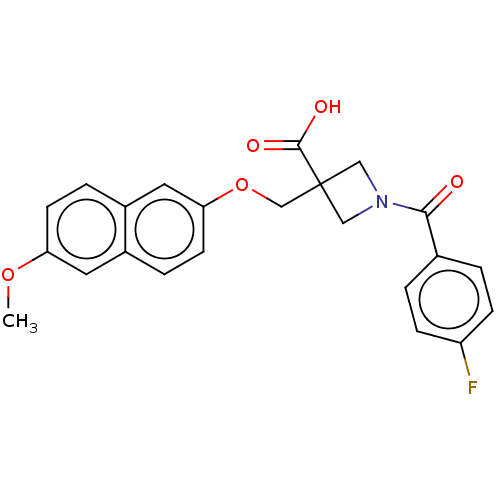 PF-04418948 BDBM50403531
PF-04418948 BDBM50403531 PF-04764755 BDBM84799
PF-04764755 BDBM84799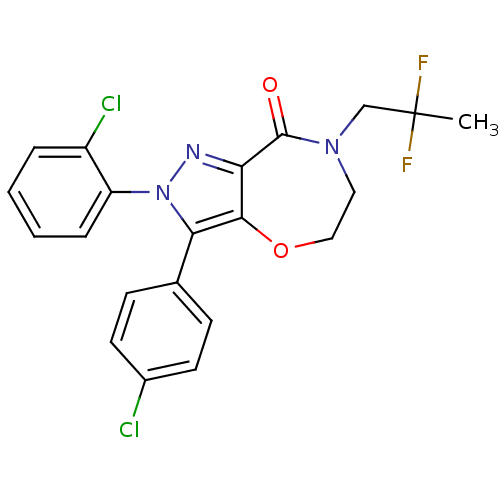 PF-514273 BDBM29075
PF-514273 BDBM29075 Pf-07059013 BDBM50565929
Pf-07059013 BDBM50565929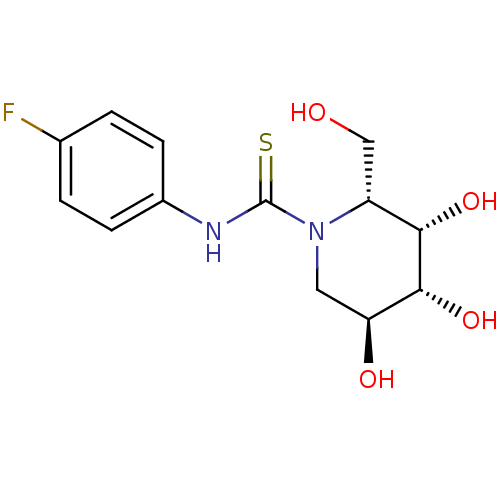 BDBM113759 DGJ-pF PhT
BDBM113759 DGJ-pF PhT BDBM113760 pF PhIM-DGJ
BDBM113760 pF PhIM-DGJ BDBM50550246 Pf-06273340 PF06273340
BDBM50550246 Pf-06273340 PF06273340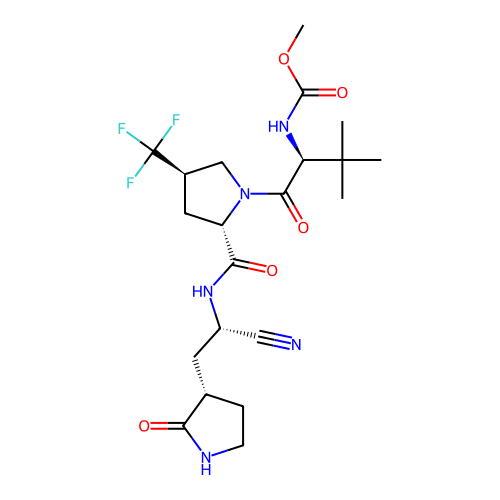 BDBM50645473 Ibuzatrelvir Pf-07817883
BDBM50645473 Ibuzatrelvir Pf-07817883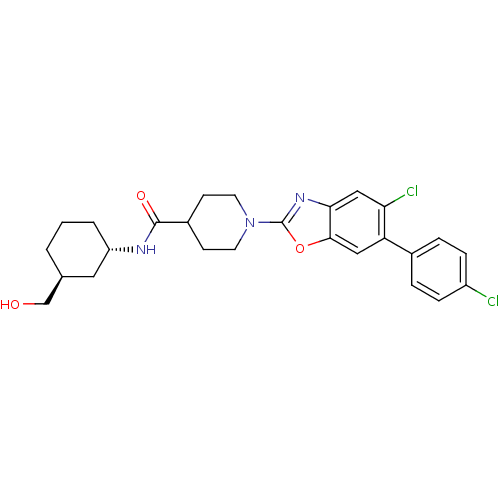 CHEMBL2325079 BDBM50426967 PF-4693627
CHEMBL2325079 BDBM50426967 PF-4693627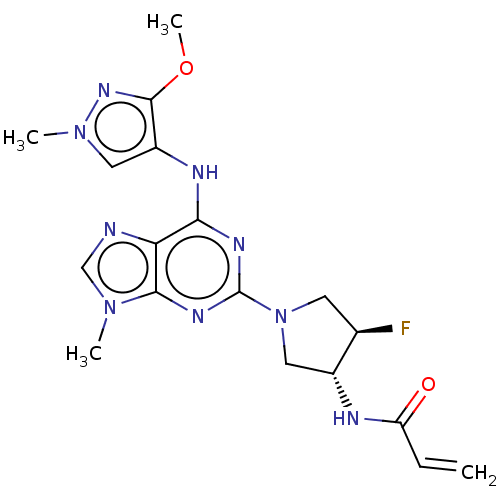 PF-06747775 BDBM50450870 Mavelertinib
PF-06747775 BDBM50450870 Mavelertinib PF-431396 BDBM50373223 CHEMBL541649
PF-431396 BDBM50373223 CHEMBL541649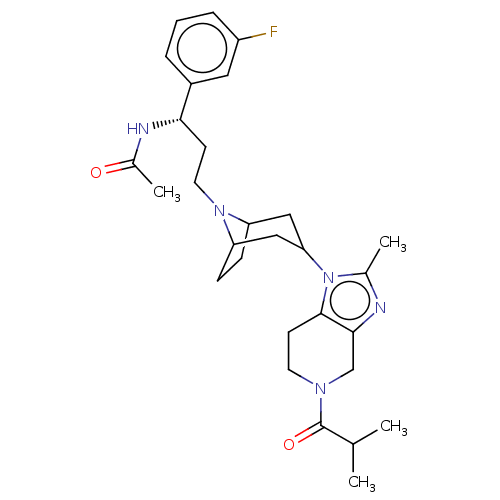 US10167299, PF-232798The BDBM314060
US10167299, PF-232798The BDBM314060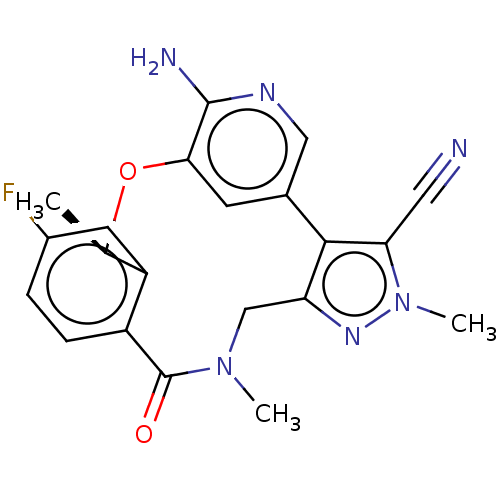 US12129258, Example Lorlatinib CHEMBL3286830 US10780082, Compound PF-06463922 BDBM50018830 US10543199, Compound PF-06463922 US12338251, Compound Lorlatinib US11517561, Compound PF-06463922
US12129258, Example Lorlatinib CHEMBL3286830 US10780082, Compound PF-06463922 BDBM50018830 US10543199, Compound PF-06463922 US12338251, Compound Lorlatinib US11517561, Compound PF-06463922 BDBM50198437 GNF-Pf-5524 CHEMBL577016
BDBM50198437 GNF-Pf-5524 CHEMBL577016 BDBM50212588 CHEMBL602986 GNF-Pf-75
BDBM50212588 CHEMBL602986 GNF-Pf-75 BDBM50239983 CHEMBL15134 GNF-Pf-5411
BDBM50239983 CHEMBL15134 GNF-Pf-5411 BDBM50264645 GNF-Pf-3892 CHEMBL597863
BDBM50264645 GNF-Pf-3892 CHEMBL597863 BDBM50348091 CHEMBL579363 GNF-PF-1973
BDBM50348091 CHEMBL579363 GNF-PF-1973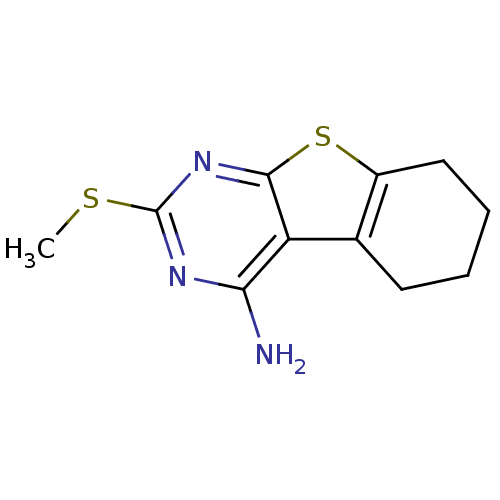 BDBM50354361 CHEMBL575039 GNF-PF-2787
BDBM50354361 CHEMBL575039 GNF-PF-2787 BDBM50393483 GNF-Pf-2995 CHEMBL581454
BDBM50393483 GNF-Pf-2995 CHEMBL581454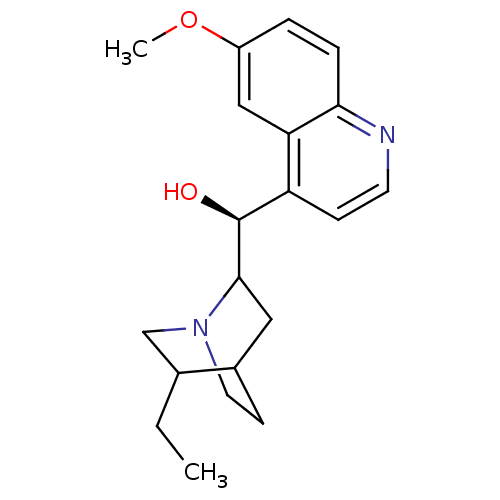 BDBM50407157 DIHYDROQUINIDINE GNF-Pf-5606
BDBM50407157 DIHYDROQUINIDINE GNF-Pf-5606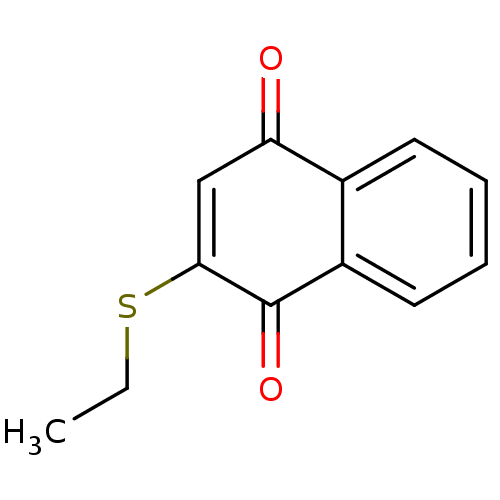 BDBM50430488 GNF-Pf-2144 CHEMBL531383
BDBM50430488 GNF-Pf-2144 CHEMBL531383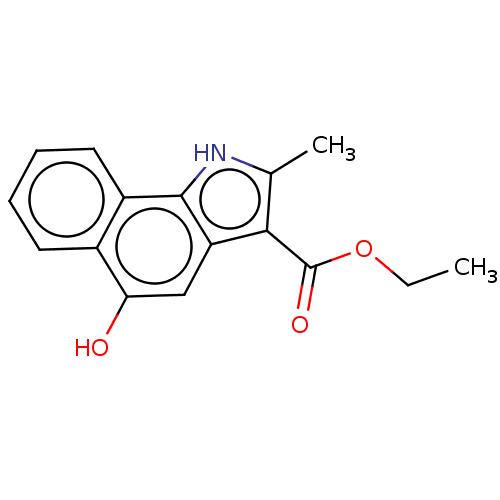 BDBM50468657 GNF-Pf-3537 CHEMBL601110
BDBM50468657 GNF-Pf-3537 CHEMBL601110 BDBM50528977 CHEMBL579408 GNF-Pf-4596
BDBM50528977 CHEMBL579408 GNF-Pf-4596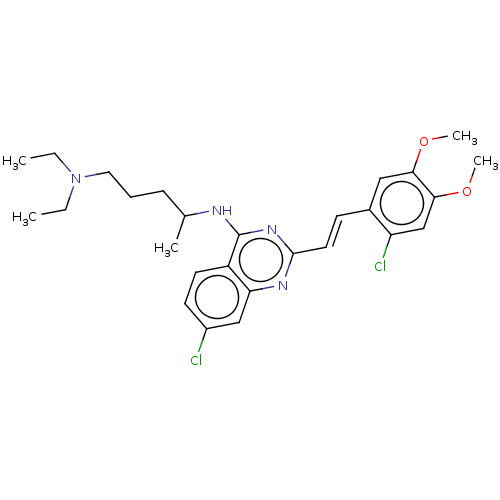 BDBM50529754 GNF-Pf-2789 CHEMBL579353
BDBM50529754 GNF-Pf-2789 CHEMBL579353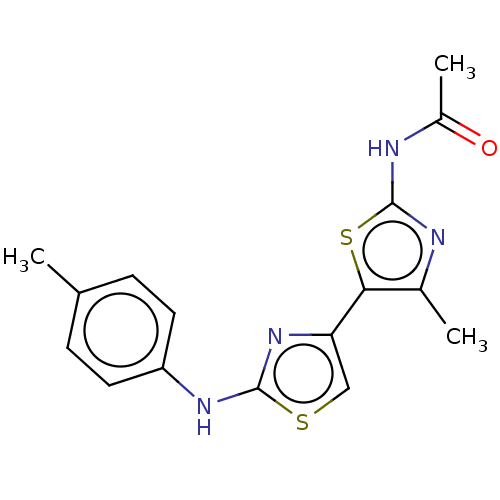 BDBM50536322 GNF-Pf-4167 CHEMBL604514
BDBM50536322 GNF-Pf-4167 CHEMBL604514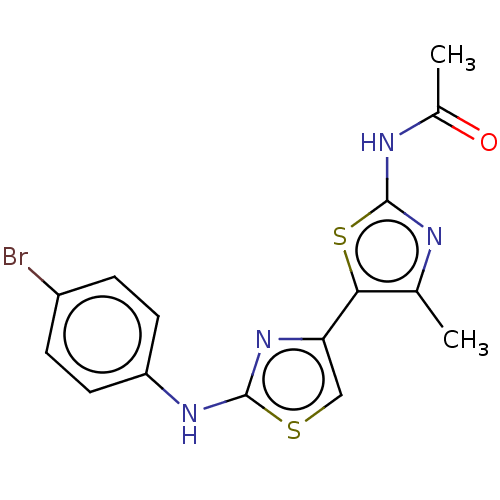 BDBM50536344 CHEMBL600947 GNF-Pf-2911
BDBM50536344 CHEMBL600947 GNF-Pf-2911 BDBM50572276 GNF-Pf-2080 CHEMBL578495
BDBM50572276 GNF-Pf-2080 CHEMBL578495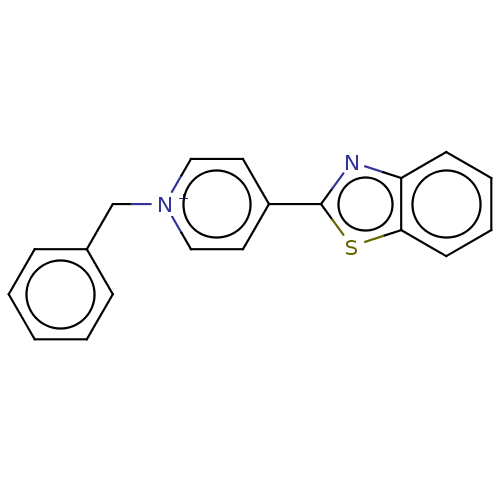 BDBM50595064 CHEMBL582490 GNF-Pf-3382
BDBM50595064 CHEMBL582490 GNF-Pf-3382 BDBM50598657 GNF-Pf-1339 CHEMBL600353
BDBM50598657 GNF-Pf-1339 CHEMBL600353 BDBM50649404 Abrocitinib Cibinqo Pf-04965842
BDBM50649404 Abrocitinib Cibinqo Pf-04965842 BDBM762514 US12378231, Example PF-04457845
BDBM762514 US12378231, Example PF-04457845 CHEMBL240752 GNF-Pf-344 BDBM50477567
CHEMBL240752 GNF-Pf-344 BDBM50477567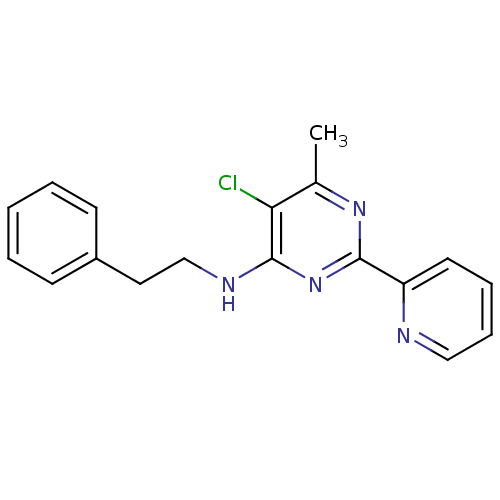 CHEMBL472879 GNF-Pf-359 BDBM50433375
CHEMBL472879 GNF-Pf-359 BDBM50433375 CHEMBL477328 GNF-Pf-3071 BDBM50496217
CHEMBL477328 GNF-Pf-3071 BDBM50496217 CHEMBL528724 BDBM50019984 GNF-Pf-1241
CHEMBL528724 BDBM50019984 GNF-Pf-1241 CHEMBL576209 GNF-Pf-5153 BDBM50561140
CHEMBL576209 GNF-Pf-5153 BDBM50561140 CHEMBL578702 GNF-Pf-2242 BDBM50458278
CHEMBL578702 GNF-Pf-2242 BDBM50458278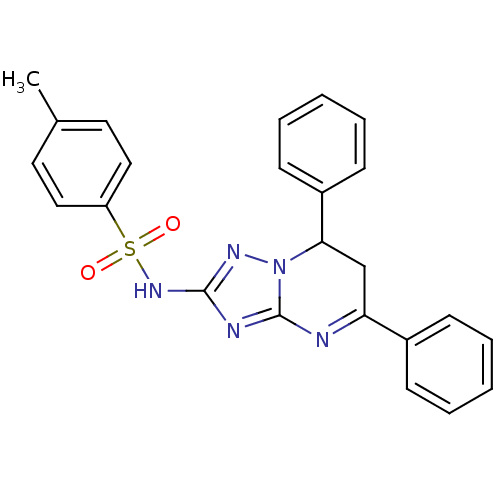 CHEMBL582739 GNF-Pf-590 BDBM50420173
CHEMBL582739 GNF-Pf-590 BDBM50420173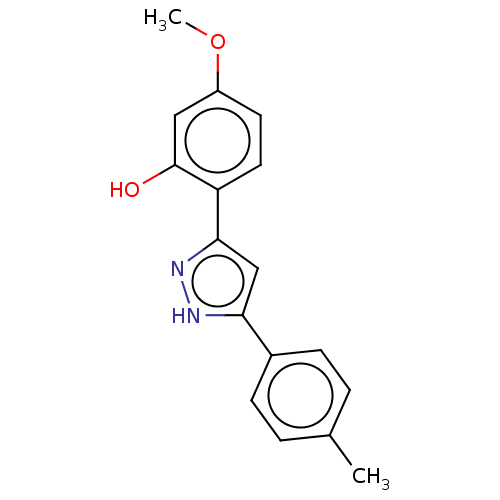 CHEMBL584668 GNF-Pf-4068 BDBM50148010
CHEMBL584668 GNF-Pf-4068 BDBM50148010 CHEMBL585793 BDBM50406805 GNF-Pf-4574
CHEMBL585793 BDBM50406805 GNF-Pf-4574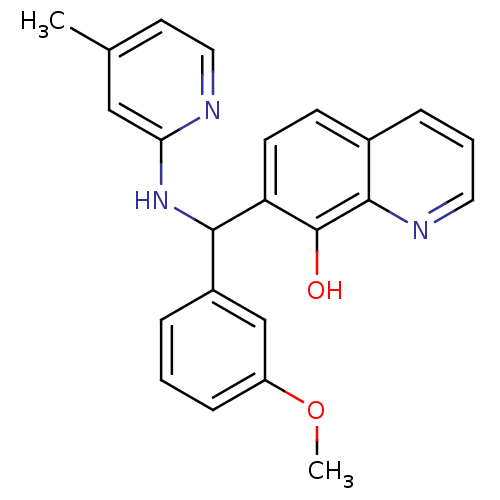 CHEMBL598057 GNF-Pf-4324 BDBM50447146
CHEMBL598057 GNF-Pf-4324 BDBM50447146 CHEMBL599894 GNF-Pf-3800 BDBM50598131
CHEMBL599894 GNF-Pf-3800 BDBM50598131 CHEMBL601773 GNF-Pf-3462 BDBM50390992
CHEMBL601773 GNF-Pf-3462 BDBM50390992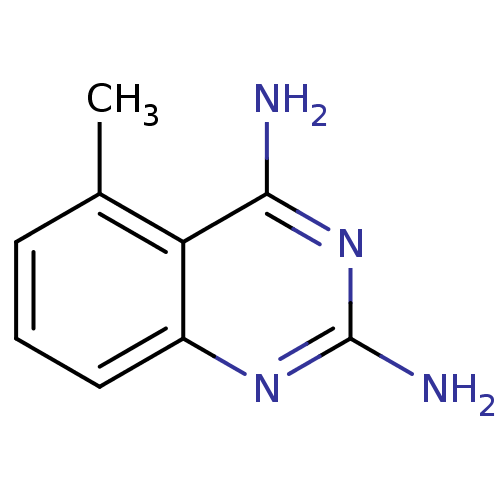 CHEMBL7077 BDBM50404679 GNF-Pf-690
CHEMBL7077 BDBM50404679 GNF-Pf-690 CINCHORINE GNF-PF-3189 BDBM50370411
CINCHORINE GNF-PF-3189 BDBM50370411 GNF-PF-1829 BDBM50356521 CHEMBL579169
GNF-PF-1829 BDBM50356521 CHEMBL579169 GNF-PF-2549 CHEMBL388886 BDBM50371109
GNF-PF-2549 CHEMBL388886 BDBM50371109 GNF-PF-2559 BDBM50378389 CHEMBL583584
GNF-PF-2559 BDBM50378389 CHEMBL583584 GNF-PF-5069 BDBM50347948 CHEMBL601821
GNF-PF-5069 BDBM50347948 CHEMBL601821 GNF-Pf-100 BDBM50393279 CHEMBL600512
GNF-Pf-100 BDBM50393279 CHEMBL600512 GNF-Pf-1115 CHEMBL584646 BDBM50430175
GNF-Pf-1115 CHEMBL584646 BDBM50430175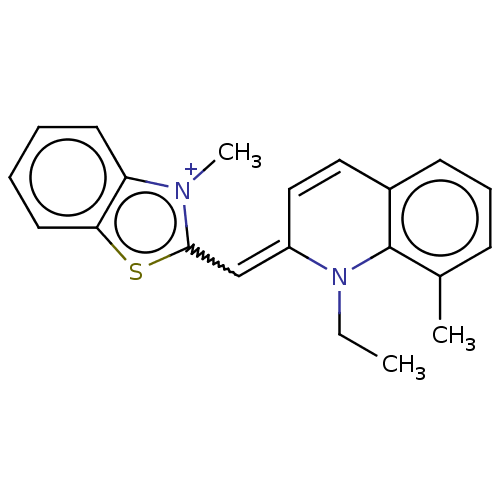 GNF-Pf-1442 CHEMBL193627 BDBM50232763
GNF-Pf-1442 CHEMBL193627 BDBM50232763 GNF-Pf-2723 BDBM50461989 CHEMBL599901
GNF-Pf-2723 BDBM50461989 CHEMBL599901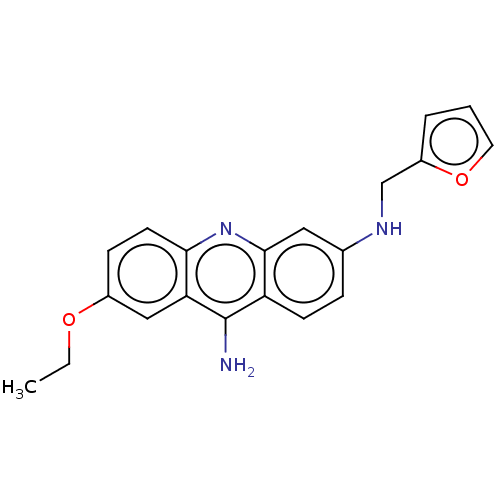 GNF-Pf-3043 CHEMBL602783 BDBM50267913
GNF-Pf-3043 CHEMBL602783 BDBM50267913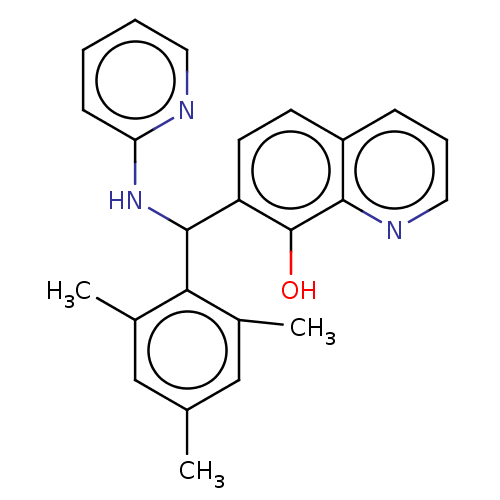 GNF-Pf-3313 BDBM50561129 CHEMBL600752
GNF-Pf-3313 BDBM50561129 CHEMBL600752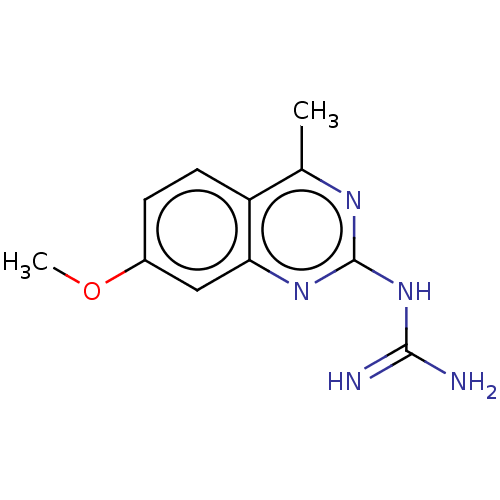 GNF-Pf-3346 BDBM50238837 CHEMBL581893
GNF-Pf-3346 BDBM50238837 CHEMBL581893 GNF-Pf-3356 BDBM50504518 CHEMBL579731
GNF-Pf-3356 BDBM50504518 CHEMBL579731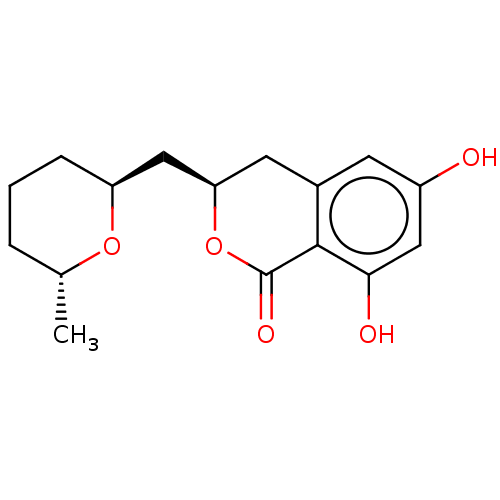 GNF-Pf-5378 CHEMBL578530 BDBM50457621
GNF-Pf-5378 CHEMBL578530 BDBM50457621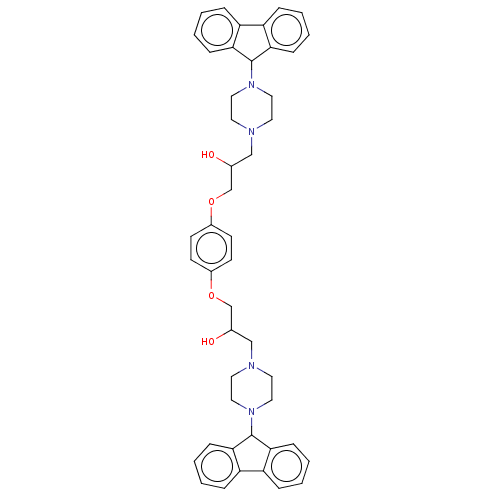 GNF-Pf-721 CHEMBL600902 BDBM50500171
GNF-Pf-721 CHEMBL600902 BDBM50500171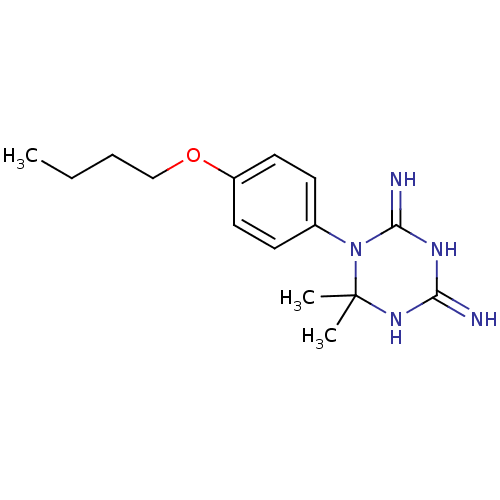 GNF-Pf-784 CHEMBL119315 BDBM50407729
GNF-Pf-784 CHEMBL119315 BDBM50407729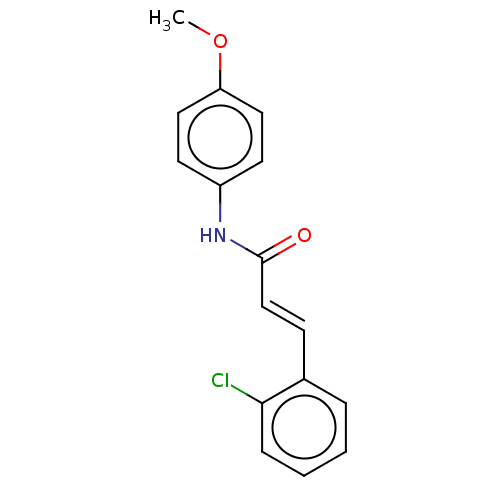 GNF-Pf-930 BDBM50117998 CHEMBL585432
GNF-Pf-930 BDBM50117998 CHEMBL585432 Pf-07284892 ARRY-558 BDBM50640216
Pf-07284892 ARRY-558 BDBM50640216 US12331038, Compound PF-06835919 BDBM749703
US12331038, Compound PF-06835919 BDBM749703 BDBM50479298 GNF-Pf-4421 CHEMBL206540 MMV007116
BDBM50479298 GNF-Pf-4421 CHEMBL206540 MMV007116 BDBM50518923 GNF-Pf-2217 CHEMBL579300 MMV665786
BDBM50518923 GNF-Pf-2217 CHEMBL579300 MMV665786 BDBM50646325 GMI-1070 PF-06460031 Rivipansel
BDBM50646325 GMI-1070 PF-06460031 Rivipansel BDBM726690 PF-07220060 US20250084095, Reference Cpd
BDBM726690 PF-07220060 US20250084095, Reference Cpd CHEMBL2331664 BDBM50428107 US8791131, 257 PF-04979064
CHEMBL2331664 BDBM50428107 US8791131, 257 PF-04979064 CHEMBL598685 SJ000111021 GNF-Pf-4035 BDBM50406806
CHEMBL598685 SJ000111021 GNF-Pf-4035 BDBM50406806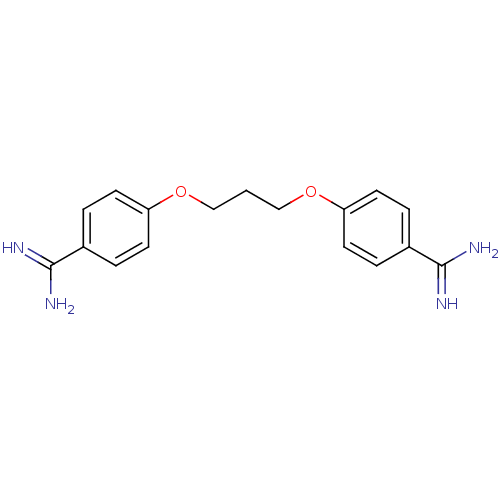 GNF-PF-3839 BDBM50368092 PROPAMIDINE CHLORIDE
GNF-PF-3839 BDBM50368092 PROPAMIDINE CHLORIDE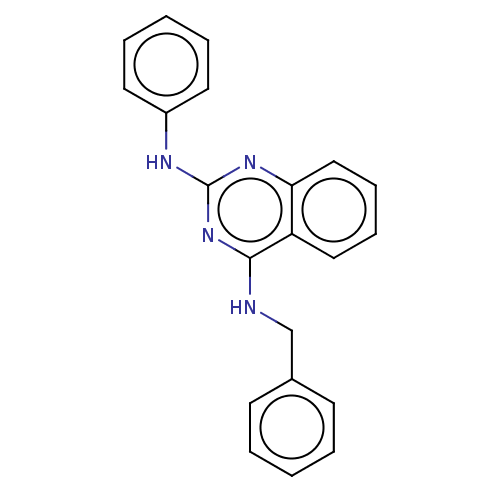 GNF-Pf-4180 MMV006169 BDBM50518231 CHEMBL597248
GNF-Pf-4180 MMV006169 BDBM50518231 CHEMBL597248 Pf-06852231 Cvl-231 BDBM50635592 Emraclidine
Pf-06852231 Cvl-231 BDBM50635592 Emraclidine US10882859, Reference compound PF-06263276 BDBM477065
US10882859, Reference compound PF-06263276 BDBM477065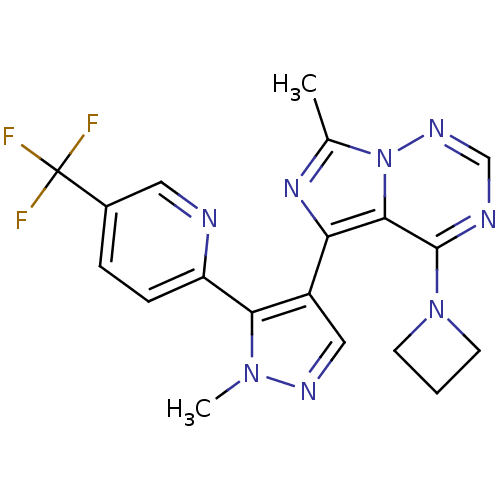 US11419874, PF-05180999 BDBM107767 US8598155, 2
US11419874, PF-05180999 BDBM107767 US8598155, 2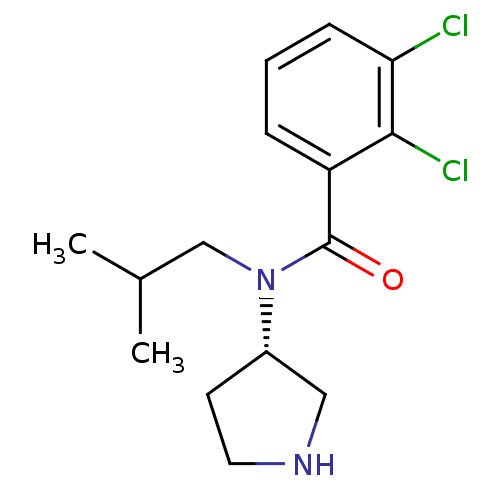 PF-184298 (S)-2,3-dichloro-N-isobutyl-N-(pyrrolidin-3-yl)benzamide BDBM50310656 CHEMBL1080787 PF-18298
PF-184298 (S)-2,3-dichloro-N-isobutyl-N-(pyrrolidin-3-yl)benzamide BDBM50310656 CHEMBL1080787 PF-18298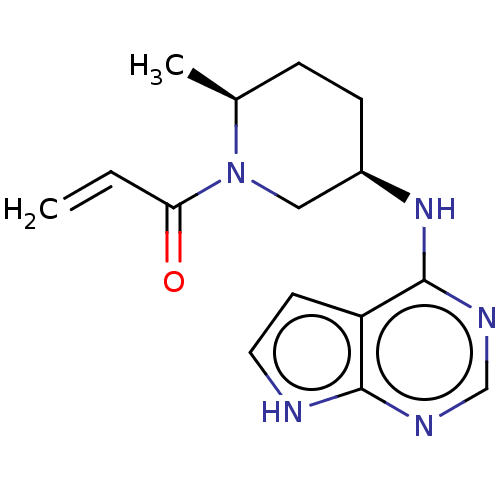 US20250134869, Example 5 BDBM209866 US11111242, Example 5 PF-06651600 US20230348487, Example PF-06651600 US9617258, Example 5
US20250134869, Example 5 BDBM209866 US11111242, Example 5 PF-06651600 US20230348487, Example PF-06651600 US9617258, Example 5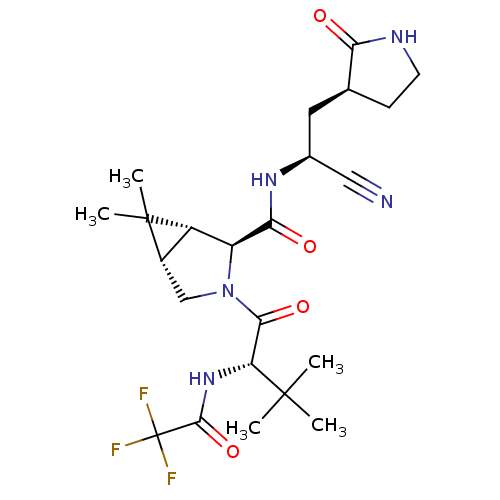 CVD-0018409 science.abl4784, 6 BDBM496902 US11351149, Example 13 PF-07321332 US11753373, Compound D-1-d US20230391736, Example PF-07321332 WO2021250648, Example 13 US20240208970, Compound PF-07321332
CVD-0018409 science.abl4784, 6 BDBM496902 US11351149, Example 13 PF-07321332 US11753373, Compound D-1-d US20230391736, Example PF-07321332 WO2021250648, Example 13 US20240208970, Compound PF-07321332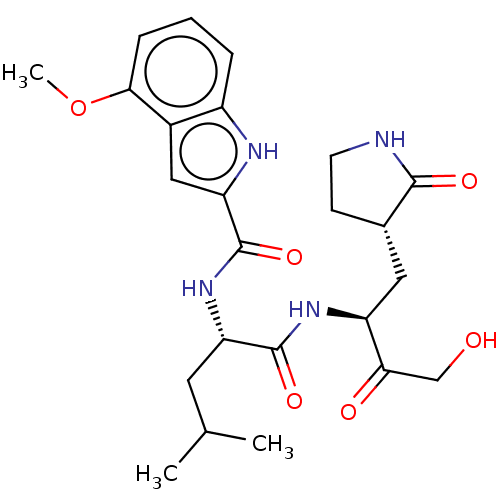 WO2005113580-Ex-02 BDBM420298 PF-00835231 US11524940, Compound 741 US11753373, Compound A-5-d CVD-0006356 WO2021205298, Compound 1 US12077605, Compound PF-00835231 cmdc.202100576, 6b PF-0835231
WO2005113580-Ex-02 BDBM420298 PF-00835231 US11524940, Compound 741 US11753373, Compound A-5-d CVD-0006356 WO2021205298, Compound 1 US12077605, Compound PF-00835231 cmdc.202100576, 6b PF-0835231 BDBM50434370 CHEMBL593252 GNF-Pf-3564 Thionin (14)
BDBM50434370 CHEMBL593252 GNF-Pf-3564 Thionin (14) BDBM676548 PF-03531814 US20240166643, Comparative Example 1
BDBM676548 PF-03531814 US20240166643, Comparative Example 1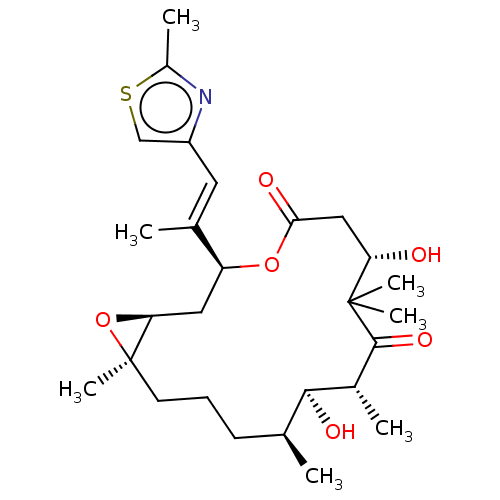 CHEBI:31550 Patupilone GNF-Pf-193 BDBM50103627
CHEBI:31550 Patupilone GNF-Pf-193 BDBM50103627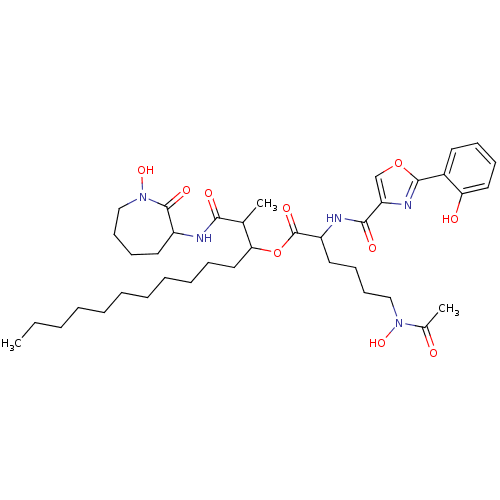 CHEMBL499519 GNF-PF-5618 BDBM50241913 nocardimicin B
CHEMBL499519 GNF-PF-5618 BDBM50241913 nocardimicin B CHEMBL598279 BDBM50575014 MMV000963 GNF-Pf-3570 SJ000171111
CHEMBL598279 BDBM50575014 MMV000963 GNF-Pf-3570 SJ000171111 GNF-Pf-1199 CHEBI:16562 BDBM50226209 CHEMBL347645
GNF-Pf-1199 CHEBI:16562 BDBM50226209 CHEMBL347645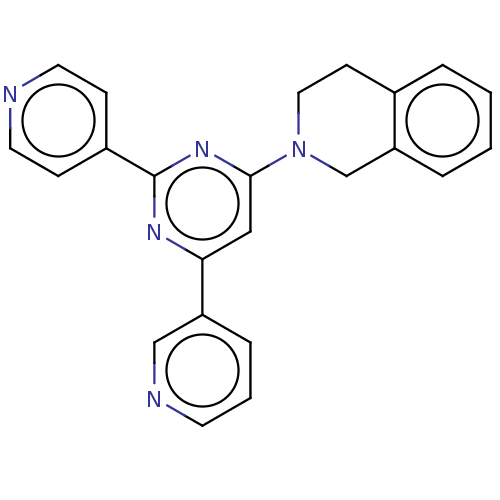 GNF-Pf-1447 CHEMBL548646 BDBM50536193 TCMDC-125419
GNF-Pf-1447 CHEMBL548646 BDBM50536193 TCMDC-125419 GNF-Pf-4091 BDBM50631333 TCMDC-123919 CHEMBL581588
GNF-Pf-4091 BDBM50631333 TCMDC-123919 CHEMBL581588 PF-03531549 BDBM676549 US20240166643, Comparative Example 2
PF-03531549 BDBM676549 US20240166643, Comparative Example 2 BDBM50044616 SAM-531 PF-05212365 Cerlapirdine WAY-262531
BDBM50044616 SAM-531 PF-05212365 Cerlapirdine WAY-262531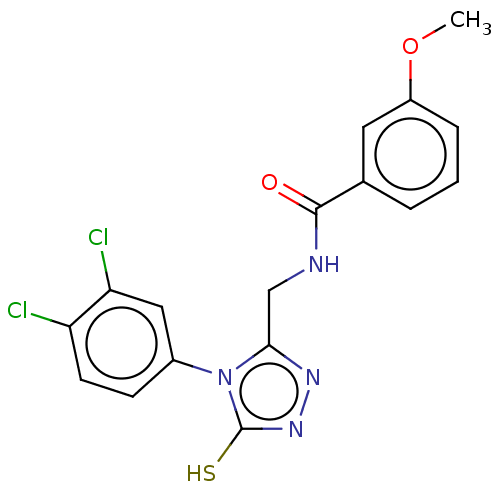 CHEMBL579923 SJ000233075 GNF-Pf-4873 BDBM50035868 TCMDC-123816
CHEMBL579923 SJ000233075 GNF-Pf-4873 BDBM50035868 TCMDC-123816 GNF-Pf-2233 TCMDC-125776 CHEMBL537778 BDBM50601909 MMV007224
GNF-Pf-2233 TCMDC-125776 CHEMBL537778 BDBM50601909 MMV007224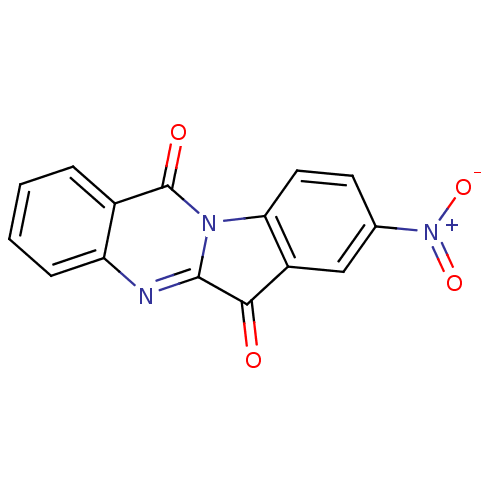 GNF-Pf-3777 US10669273, Compound 5i CHEMBL432537 BDBM50442991
GNF-Pf-3777 US10669273, Compound 5i CHEMBL432537 BDBM50442991 TCMDC-123459 BDBM50134735 GNF-Pf-2870 CHEMBL531018 SJ000111341
TCMDC-123459 BDBM50134735 GNF-Pf-2870 CHEMBL531018 SJ000111341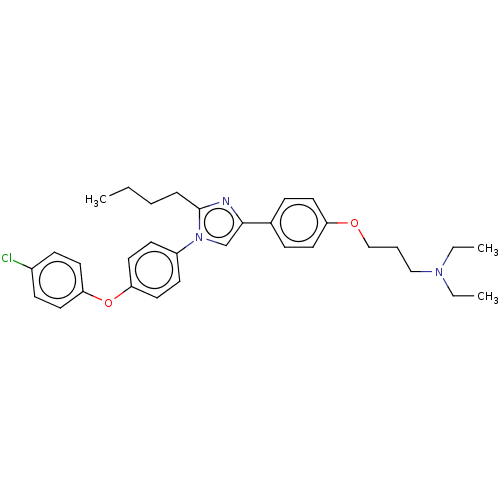 TTP448 US11524942, Compound I Azeliragon PF-04494700 BDBM50249580
TTP448 US11524942, Compound I Azeliragon PF-04494700 BDBM50249580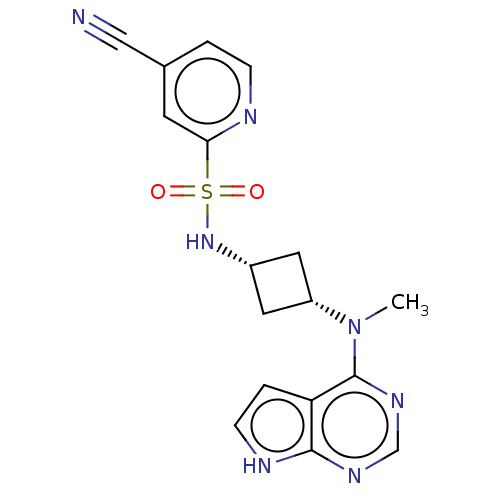 US10966980, Example 8 US9035074, 8 PF-02384554 BDBM159756
US10966980, Example 8 US9035074, 8 PF-02384554 BDBM159756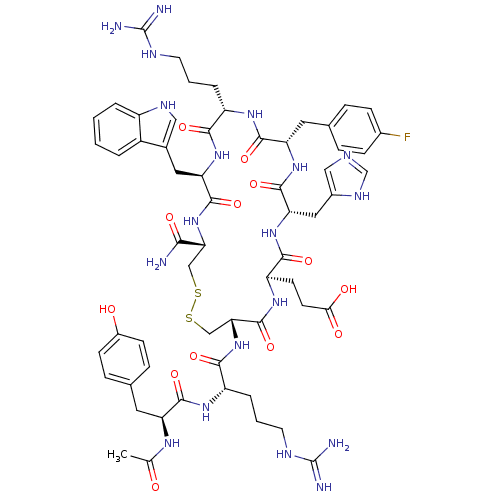 Ac-YR[CEH(pF-dF)RWC]-NH2 BDBM50165927 CHEMBL407809
Ac-YR[CEH(pF-dF)RWC]-NH2 BDBM50165927 CHEMBL407809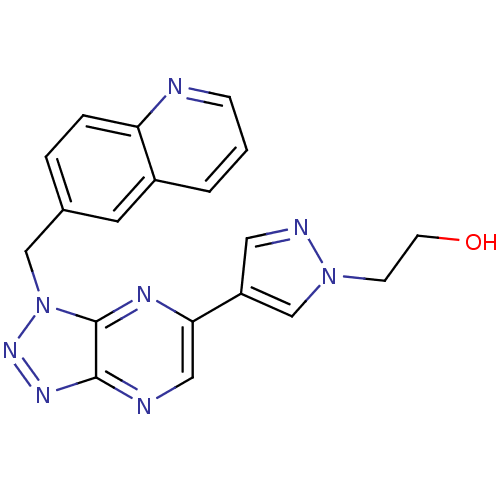 CHEMBL2170804 CHEMBL2001019 US9062045, Comparator No. 3 (PF-04217903) BDBM50396934
CHEMBL2170804 CHEMBL2001019 US9062045, Comparator No. 3 (PF-04217903) BDBM50396934 CHEMBL444219 Ac-His-DPhe(pF)-Arg-Trp-NH2 BDBM50252898
CHEMBL444219 Ac-His-DPhe(pF)-Arg-Trp-NH2 BDBM50252898 IDN-6556 Emricasan BDBM50461533 PF-03491390 VAY785 VAY-785
IDN-6556 Emricasan BDBM50461533 PF-03491390 VAY785 VAY-785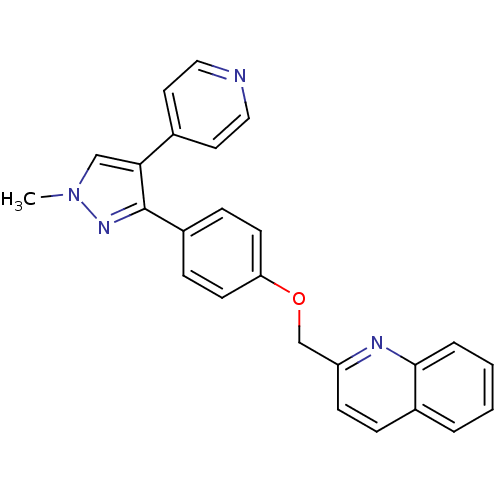 PF-2545920 US9138494, MP-10 BDBM31592 substituted pyrazole, 9
PF-2545920 US9138494, MP-10 BDBM31592 substituted pyrazole, 9 SJ000110765 MMV665943 TCMDC-123486 CHEMBL548209 GNF-Pf-4694 BDBM50626501
SJ000110765 MMV665943 TCMDC-123486 CHEMBL548209 GNF-Pf-4694 BDBM50626501 GNF-Pf-3515 1-(4-methylquinazolin-2-yl)guanidine CHEMBL602365 BDBM50335552
GNF-Pf-3515 1-(4-methylquinazolin-2-yl)guanidine CHEMBL602365 BDBM50335552 GNF-Pf-4337 CHEMBL277148 2-Phenyl-1H-quinolin-4-one BDBM50044964
GNF-Pf-4337 CHEMBL277148 2-Phenyl-1H-quinolin-4-one BDBM50044964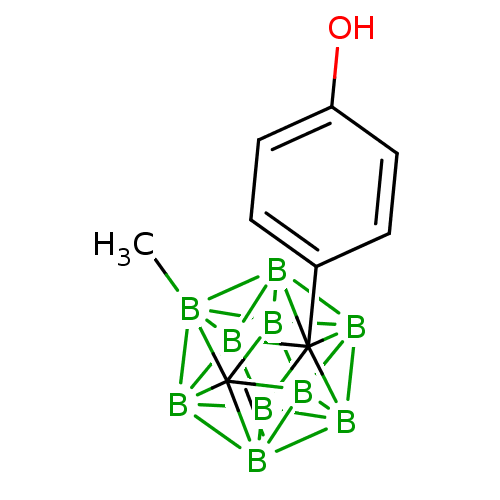 Rucaparib camsylate CO-338 C0-338 BDBM50076330 PF-1367338-BW Rubraca
Rucaparib camsylate CO-338 C0-338 BDBM50076330 PF-1367338-BW Rubraca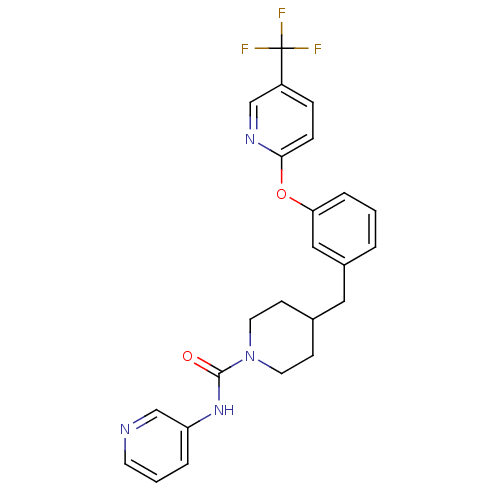 US20250170100, Compound Comparator 2 PF-3845, 10 US20240308985, Comparator TableD2.2 BDBM81356
US20250170100, Compound Comparator 2 PF-3845, 10 US20240308985, Comparator TableD2.2 BDBM81356 N-phenyl-4-(quinolin-3-ylmethyl)piperidine-1-carboxamide PF-750 BDBM26740
N-phenyl-4-(quinolin-3-ylmethyl)piperidine-1-carboxamide PF-750 BDBM26740 BDBM50066492 N*6*-(3,4,5-Trimethoxy-benzyl)-quinazoline-2,4,6-triamine CHEMBL113999 GNF-Pf-607
BDBM50066492 N*6*-(3,4,5-Trimethoxy-benzyl)-quinazoline-2,4,6-triamine CHEMBL113999 GNF-Pf-607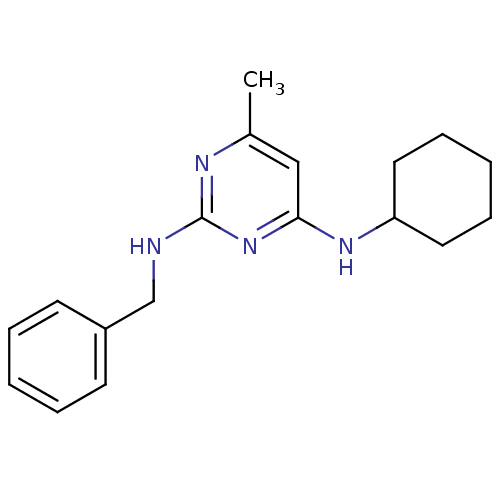 BDBM50304834 GNF-Pf-2094 N2-benzyl-N4-cyclohexyl-6-methylpyrimidine-2,4-diamine CHEMBL578928
BDBM50304834 GNF-Pf-2094 N2-benzyl-N4-cyclohexyl-6-methylpyrimidine-2,4-diamine CHEMBL578928 CHEMBL1095047 BDBM50317209 PF-877423 (2R)-N-(adamantan-2-yl)-1-ethylpyrrolidine-2-carboxamide
CHEMBL1095047 BDBM50317209 PF-877423 (2R)-N-(adamantan-2-yl)-1-ethylpyrrolidine-2-carboxamide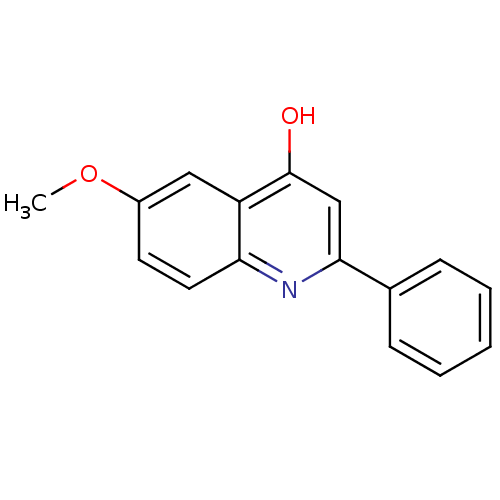 CHEMBL14120 GNF-PF-117 6-Methoxy-2-phenyl-1H-quinolin-4-one BDBM50041130
CHEMBL14120 GNF-PF-117 6-Methoxy-2-phenyl-1H-quinolin-4-one BDBM50041130 GNF-Pf-2700 1-(8-ethoxy-4-methylquinazolin-2-yl)guanidine BDBM50322849 CHEMBL602578
GNF-Pf-2700 1-(8-ethoxy-4-methylquinazolin-2-yl)guanidine BDBM50322849 CHEMBL602578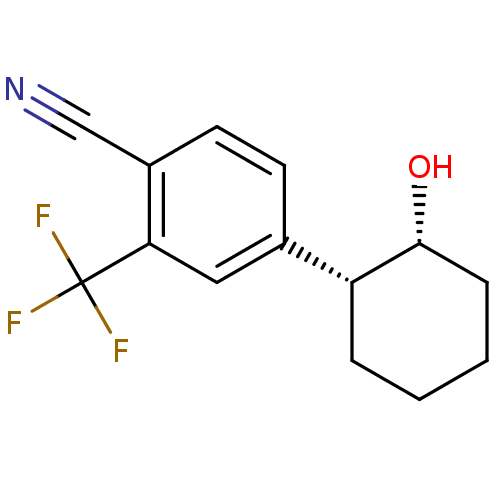 arylcyclohexanol, (-)-6a PF-0998425 4-[(1R,2R)-2-hydroxycyclohexyl]-2-(trifluoromethyl)benzonitrile BDBM25436
arylcyclohexanol, (-)-6a PF-0998425 4-[(1R,2R)-2-hydroxycyclohexyl]-2-(trifluoromethyl)benzonitrile BDBM25436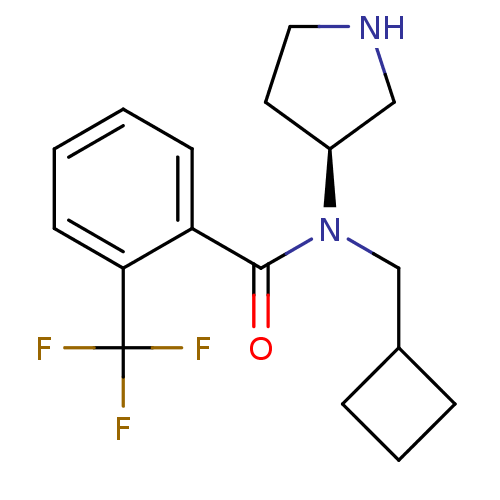 BDBM50297077 CHEMBL552338 PF-3409409 (S)-N-(cyclobutylmethyl)-N-(pyrrolidin-3-yl)-2-(trifluoromethyl)benzamide
BDBM50297077 CHEMBL552338 PF-3409409 (S)-N-(cyclobutylmethyl)-N-(pyrrolidin-3-yl)-2-(trifluoromethyl)benzamide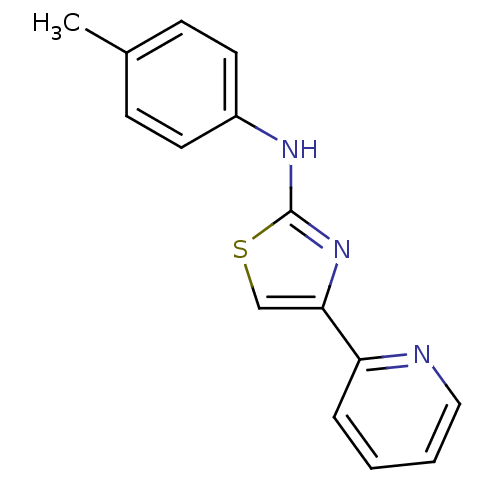 CHEMBL470514 GNF-Pf-3677 4-(pyridin-2-yl)-N-p-tolylthiazol-2-amine BDBM50249555
CHEMBL470514 GNF-Pf-3677 4-(pyridin-2-yl)-N-p-tolylthiazol-2-amine BDBM50249555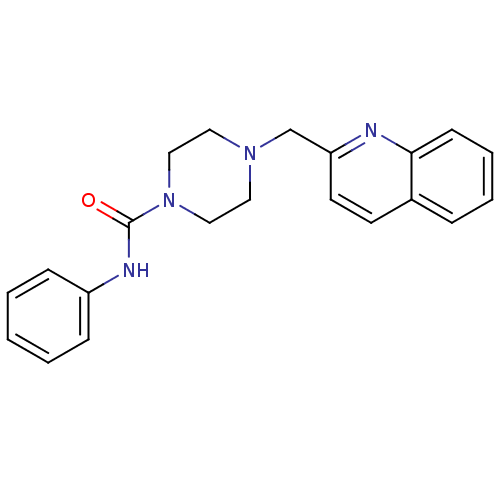 N-phenyl-4-(quinolin-2-ylmethyl)piperazine-1-carboxamide US9169224, 19 PF-622 BDBM26741
N-phenyl-4-(quinolin-2-ylmethyl)piperazine-1-carboxamide US9169224, 19 PF-622 BDBM26741 P-638 BDBM50277158 GNF-Pf-2016 3123L CHEBI:17939 TCMDC-123493 CL-13900 Puromycin
P-638 BDBM50277158 GNF-Pf-2016 3123L CHEBI:17939 TCMDC-123493 CL-13900 Puromycin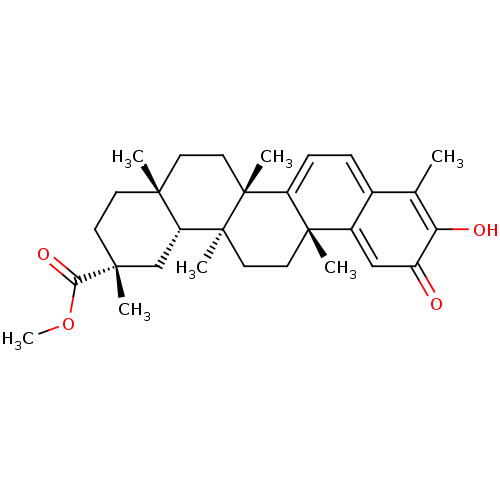 PRISTIMERIN GNF-Pf-476 Pristimerine BDBM50481947 CHEBI:8416 US11660306, Example Pristimerin acs.jmedchem.1c00409_ST.332
PRISTIMERIN GNF-Pf-476 Pristimerine BDBM50481947 CHEBI:8416 US11660306, Example Pristimerin acs.jmedchem.1c00409_ST.332 BDBM50191291 5-(4-(dimethylamino)benzylidene)-2-(4-hydroxyphenylimino)thiazolidin-4-one CHEMBL211589 GNF-PF-1788
BDBM50191291 5-(4-(dimethylamino)benzylidene)-2-(4-hydroxyphenylimino)thiazolidin-4-one CHEMBL211589 GNF-PF-1788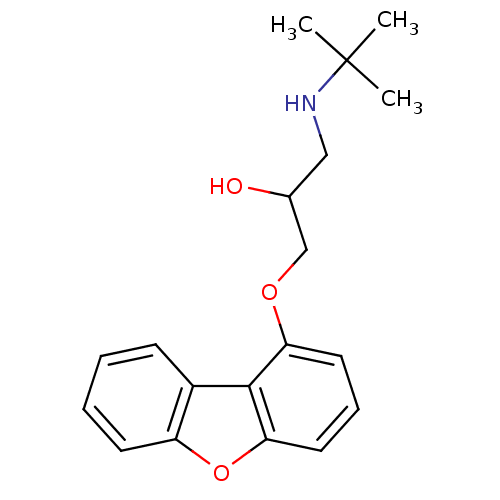 BDBM50274012 CHEMBL513104 GNF-PF-1694 1-tert-Butylamino-3-(dibenzofuran-1-yloxy)-propan-2-ol
BDBM50274012 CHEMBL513104 GNF-PF-1694 1-tert-Butylamino-3-(dibenzofuran-1-yloxy)-propan-2-ol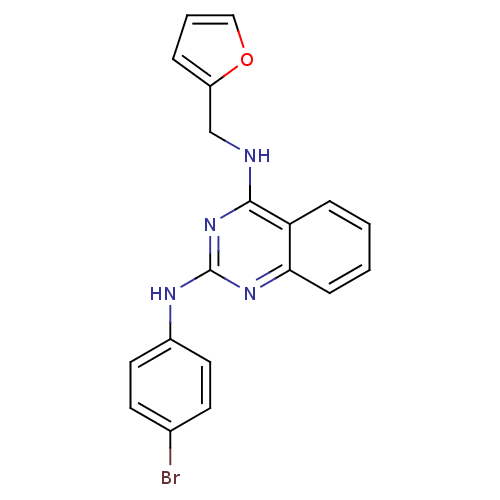 N2-(4-bromophenyl)-N4-(furan-2-ylmethyl)quinazoline-2,4-diamine CHEMBL379386 BDBM50187063 GNF-PF-2722
N2-(4-bromophenyl)-N4-(furan-2-ylmethyl)quinazoline-2,4-diamine CHEMBL379386 BDBM50187063 GNF-PF-2722 2-(3-(4-cyanophenoxy)-2,4-dicyclopropyl-1H-pyrrol-1-yl)-N-methylacetamide CHEMBL1083754 BDBM50318934 PF-02367982
2-(3-(4-cyanophenoxy)-2,4-dicyclopropyl-1H-pyrrol-1-yl)-N-methylacetamide CHEMBL1083754 BDBM50318934 PF-02367982 BDBM50216181 PF-429242 CHEMBL233611 (R)-N-(2-methoxyphenethyl)-4-((diethylamino)methyl)-N-(pyrrolidin-3-yl)benzamide
BDBM50216181 PF-429242 CHEMBL233611 (R)-N-(2-methoxyphenethyl)-4-((diethylamino)methyl)-N-(pyrrolidin-3-yl)benzamide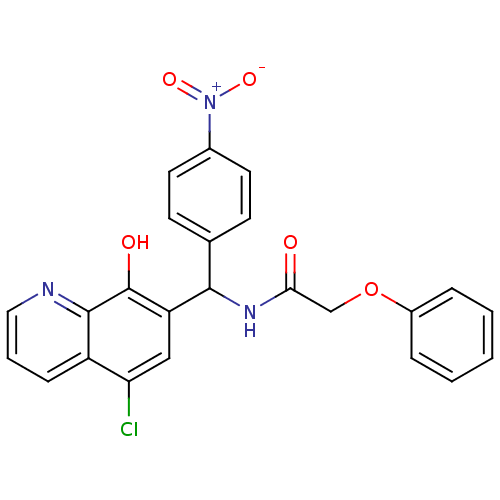 BDBM50256061 GNF-Pf-4399 CHEMBL482160 N-((5-chloro-8-hydroxyquinolin-7-yl)(4-nitrophenyl)methyl)-2-phenoxyacetamide
BDBM50256061 GNF-Pf-4399 CHEMBL482160 N-((5-chloro-8-hydroxyquinolin-7-yl)(4-nitrophenyl)methyl)-2-phenoxyacetamide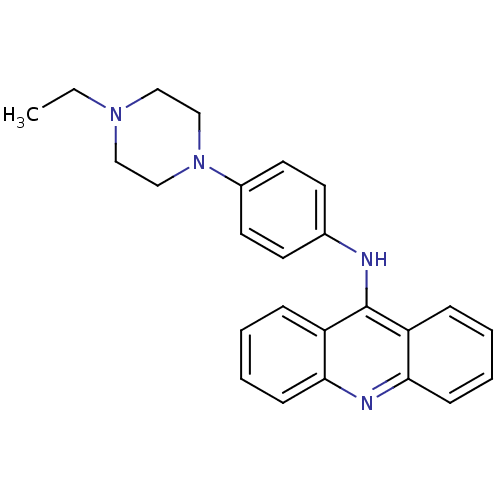 CHEMBL217366 TCMDC-124255 N-(4-(4-ethylpiperazin-1-yl)phenyl)acridin-9-amine GNF-PF-3878 BDBM50196122
CHEMBL217366 TCMDC-124255 N-(4-(4-ethylpiperazin-1-yl)phenyl)acridin-9-amine GNF-PF-3878 BDBM50196122 CHEMBL481505 N-((5-chloro-8-hydroxyquinolin-7-yl)(4-methoxyphenyl)methyl)-2-phenoxyacetamide BDBM50256059 GNF-PF-3875
CHEMBL481505 N-((5-chloro-8-hydroxyquinolin-7-yl)(4-methoxyphenyl)methyl)-2-phenoxyacetamide BDBM50256059 GNF-PF-3875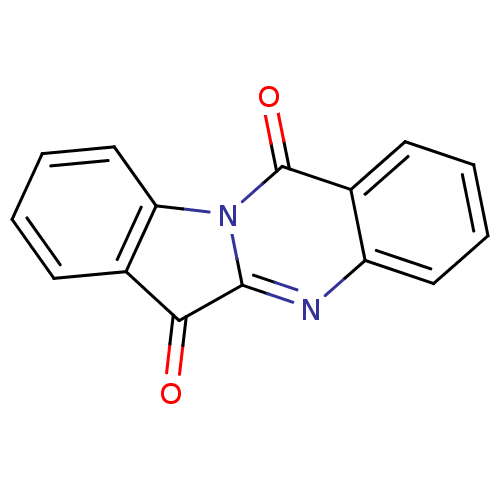 Indolo[2,1-b]quinazoline-6,12-dione CHEMBL306946 GNF-PF-2691 BDBM50240612 TRYPTANTHRIN US10669273, Compound 5a TCMDC-125859
Indolo[2,1-b]quinazoline-6,12-dione CHEMBL306946 GNF-PF-2691 BDBM50240612 TRYPTANTHRIN US10669273, Compound 5a TCMDC-125859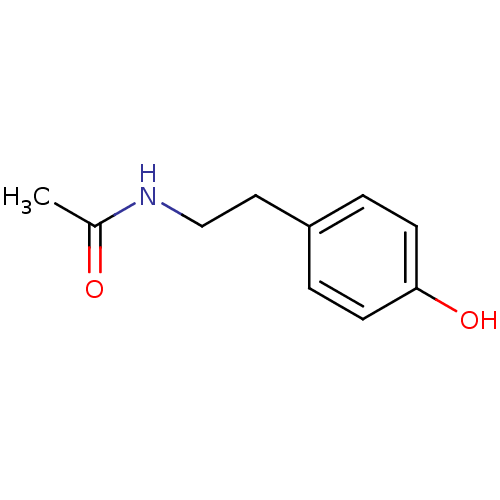 N-[2-(4-Hydroxy-phenyl)-ethyl]-acetamide BDBM50136842 CHEMBL152117 N-[2-(4-hydroxyphenyl)ethylacetamide GNF-Pf-5230
N-[2-(4-Hydroxy-phenyl)-ethyl]-acetamide BDBM50136842 CHEMBL152117 N-[2-(4-hydroxyphenyl)ethylacetamide GNF-Pf-5230 8-chloro-N-(3-morpholinopropyl)-5H-pyrimido[5,4-b]indol-4-amine BDBM50322743 cid_661700 GNF-Pf-4478 CHEMBL597857
8-chloro-N-(3-morpholinopropyl)-5H-pyrimido[5,4-b]indol-4-amine BDBM50322743 cid_661700 GNF-Pf-4478 CHEMBL597857 CHEBI:49960 ZD-64 ZD-6474 NSC-760766 Zactima GNF-PF-2188 NSC-744325 Caprelsa VANDETANIB BDBM50595124 ZD6474
CHEBI:49960 ZD-64 ZD-6474 NSC-760766 Zactima GNF-PF-2188 NSC-744325 Caprelsa VANDETANIB BDBM50595124 ZD6474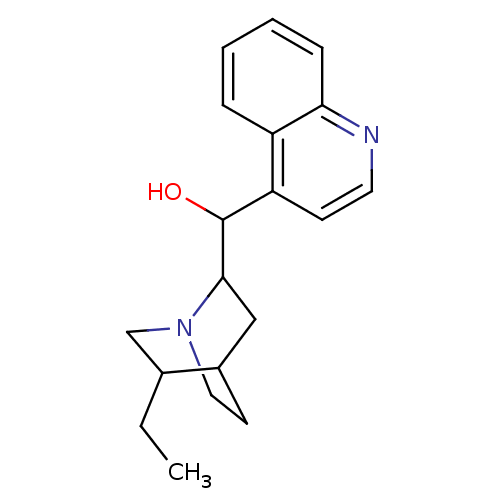 CHEMBL428695 (5-Ethyl-1-aza-bicyclo[2.2.2]oct-2-yl)-quinolin-4-yl-methanol GNF-Pf-4292 BDBM50047001
CHEMBL428695 (5-Ethyl-1-aza-bicyclo[2.2.2]oct-2-yl)-quinolin-4-yl-methanol GNF-Pf-4292 BDBM50047001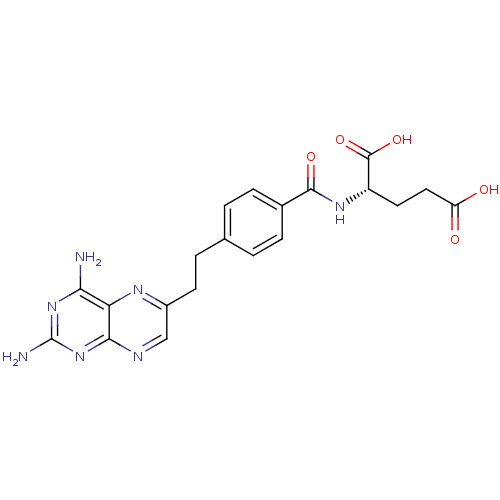 GNF-PF-173 BDBM50016461 CHEMBL293263 (S)-2-{4-[2-(2,4-Diamino-pteridin-6-yl)-ethyl]-benzoylamino}-pentanedioic acid
GNF-PF-173 BDBM50016461 CHEMBL293263 (S)-2-{4-[2-(2,4-Diamino-pteridin-6-yl)-ethyl]-benzoylamino}-pentanedioic acid GNF-PF-5188 8-hexyl-3,5,5-trimethyl-2,3,4,5-tetrahydro-1H-chromeno[3,4-c]pyridin-10-ol CHEMBL525381 BDBM50265648
GNF-PF-5188 8-hexyl-3,5,5-trimethyl-2,3,4,5-tetrahydro-1H-chromeno[3,4-c]pyridin-10-ol CHEMBL525381 BDBM50265648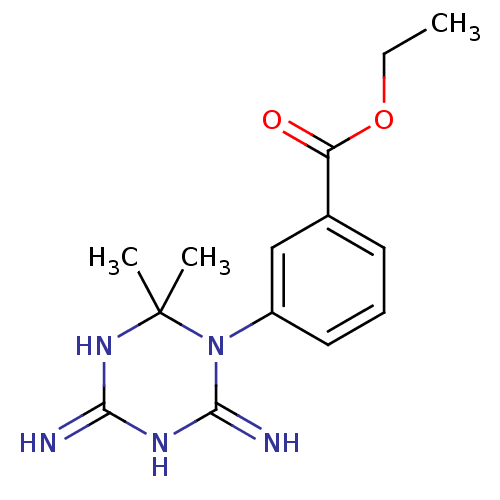 3-(4,6-Diamino-2,2-dimethyl-2H-[1,3,5]triazin-1-yl)-benzoic acid ethyl ester BDBM50291770 CHEMBL268356 GNF-PF-785
3-(4,6-Diamino-2,2-dimethyl-2H-[1,3,5]triazin-1-yl)-benzoic acid ethyl ester BDBM50291770 CHEMBL268356 GNF-PF-785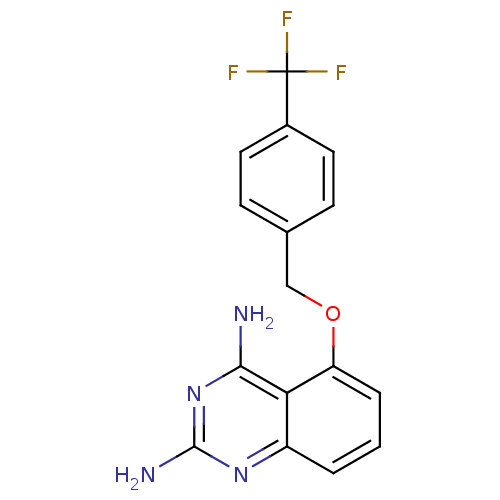 GNF-Pf-1166 5-(4-Trifluoromethyl-benzyloxy)-quinazoline-2,4-diamine 5-(4-(trifluoromethyl)benzyloxy)quinazoline-2,4-diamine BDBM50130952 CHEMBL84108
GNF-Pf-1166 5-(4-Trifluoromethyl-benzyloxy)-quinazoline-2,4-diamine 5-(4-(trifluoromethyl)benzyloxy)quinazoline-2,4-diamine BDBM50130952 CHEMBL84108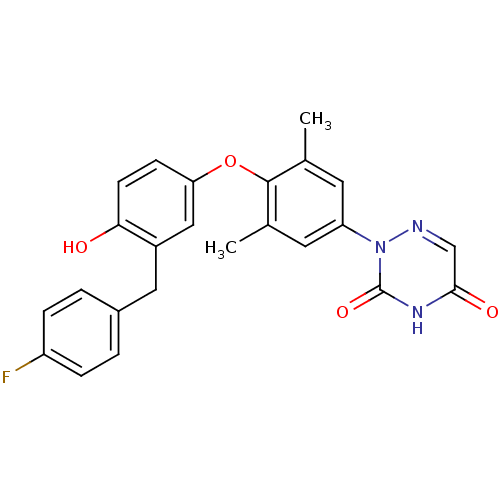 PF-00277343 2-(4-(3-(4-fluorobenzyl)-4-hydroxyphenoxy)-3,5-dimethylphenyl)-1,2,4-triazine-3,5(2H,4H)-dione BDBM50304781 CHEMBL594025
PF-00277343 2-(4-(3-(4-fluorobenzyl)-4-hydroxyphenoxy)-3,5-dimethylphenyl)-1,2,4-triazine-3,5(2H,4H)-dione BDBM50304781 CHEMBL594025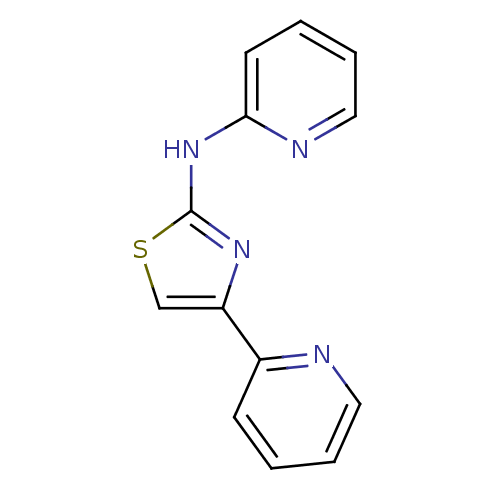 CHEMBL489770 N-(4-(pyridin-2-yl)thiazol-2-yl)pyridin-2-amine US9034574, I GNF-PF-4773 TCMDC-124275 BDBM50264020
CHEMBL489770 N-(4-(pyridin-2-yl)thiazol-2-yl)pyridin-2-amine US9034574, I GNF-PF-4773 TCMDC-124275 BDBM50264020 GNF-Pf-1931 TCMDC-123993 5-(6-Ethyl-thieno[2,3-d]pyrimidin-4-ylsulfanyl)-[1,3,4]thiadiazol-2-ylamine BDBM50098485 CHEMBL6229
GNF-Pf-1931 TCMDC-123993 5-(6-Ethyl-thieno[2,3-d]pyrimidin-4-ylsulfanyl)-[1,3,4]thiadiazol-2-ylamine BDBM50098485 CHEMBL6229 Pyrido[1,2-a:3,4-b']diindol-5-ium,12,13-dihydro-13-oxo-, chloride CHEMBL258765 NSC-622398 BDBM50229977 GNF-PF-1458
Pyrido[1,2-a:3,4-b']diindol-5-ium,12,13-dihydro-13-oxo-, chloride CHEMBL258765 NSC-622398 BDBM50229977 GNF-PF-1458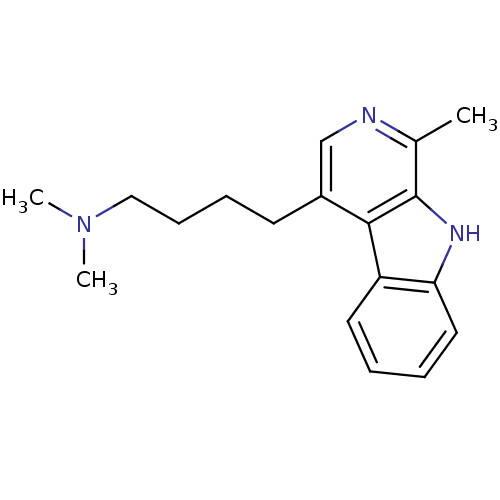 BDBM50330491 GNF-Pf-3207 N,N-dimethyl-4-(1-methyl-9H-pyrido[3,4-b]indol-4-yl)butan-1-aminium CHEMBL602178
BDBM50330491 GNF-Pf-3207 N,N-dimethyl-4-(1-methyl-9H-pyrido[3,4-b]indol-4-yl)butan-1-aminium CHEMBL602178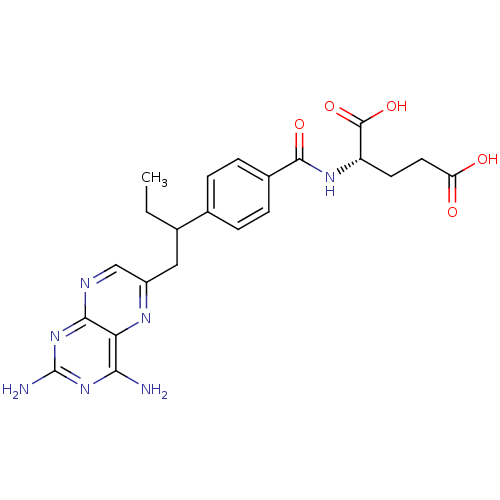 CHEMBL296373 TCMDC-137768 Edatrexate (S)-2-{4-[1-(2,4-Diamino-pteridin-6-ylmethyl)-propyl]-benzoylamino}-pentanedioic acid GNF-Pf-63 BDBM50016460
CHEMBL296373 TCMDC-137768 Edatrexate (S)-2-{4-[1-(2,4-Diamino-pteridin-6-ylmethyl)-propyl]-benzoylamino}-pentanedioic acid GNF-Pf-63 BDBM50016460 N1,N1-Dimethyl-N4-(5-methyl-[1,2,4]triazolo[1,5-a]pyrimidin-7-yl)-benzene-1,4-diamine GNF-Pf-2905 BDBM50345412 CHEMBL582694
N1,N1-Dimethyl-N4-(5-methyl-[1,2,4]triazolo[1,5-a]pyrimidin-7-yl)-benzene-1,4-diamine GNF-Pf-2905 BDBM50345412 CHEMBL582694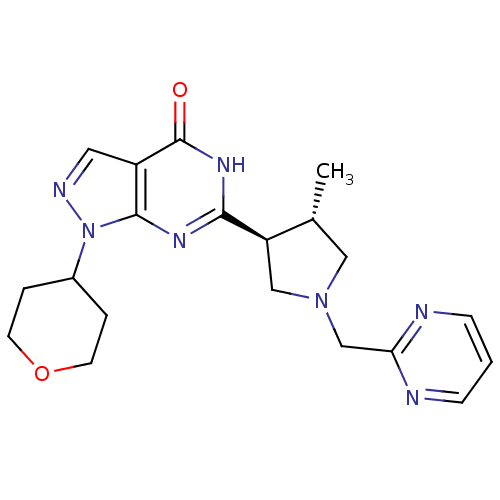 US11608342, Reference compound disclosed in WO2008/139293 WO2008/139293, AF27873 US10513524, Reference compound disclosed in WO2008/139293 BDBM50397838 CHEMBL2179105 PF-04447943
US11608342, Reference compound disclosed in WO2008/139293 WO2008/139293, AF27873 US10513524, Reference compound disclosed in WO2008/139293 BDBM50397838 CHEMBL2179105 PF-04447943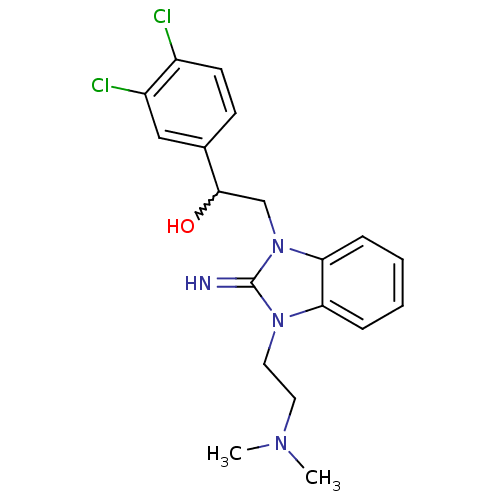 1-(3,4-dichlorophenyl)-2-(3-(2-(dimethylamino)ethyl)-2-imino-2,3-dihydrobenzo[d]imidazol-1-yl)ethanol CHEMBL395209 GNF-PF-2112 BDBM50201545
1-(3,4-dichlorophenyl)-2-(3-(2-(dimethylamino)ethyl)-2-imino-2,3-dihydrobenzo[d]imidazol-1-yl)ethanol CHEMBL395209 GNF-PF-2112 BDBM50201545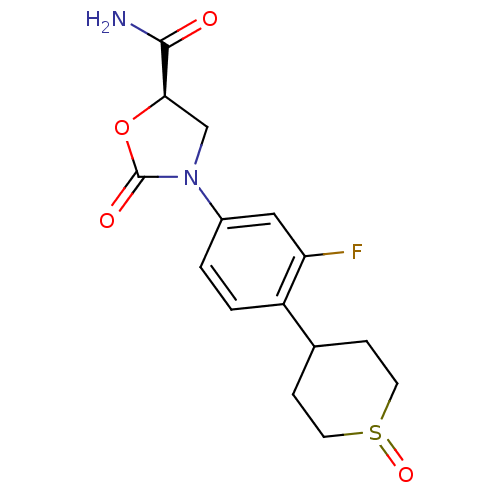 PF-00422602 (5R)-3-[3-fluoro-4-(1-oxidotetrahydro-2H-thiopyran-4-yl)phenyl]-2-oxo-1,3-oxazolidine-5-carboxamide CHEMBL437955 BDBM50226483
PF-00422602 (5R)-3-[3-fluoro-4-(1-oxidotetrahydro-2H-thiopyran-4-yl)phenyl]-2-oxo-1,3-oxazolidine-5-carboxamide CHEMBL437955 BDBM50226483 (E)-N-[2-(4-Ethyl-piperazin-1-yl)-4-methyl-quinolin-6-yl]-3-(4-methoxy-phenyl)-acrylamide GNF-Pf-1550 BDBM50172447 CHEMBL196573
(E)-N-[2-(4-Ethyl-piperazin-1-yl)-4-methyl-quinolin-6-yl]-3-(4-methoxy-phenyl)-acrylamide GNF-Pf-1550 BDBM50172447 CHEMBL196573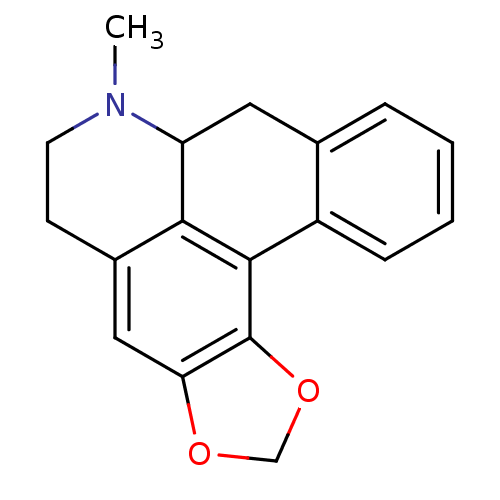 GNF-Pf-4466 BDBM50284797 cid_235224 7-Methyl-6,7,7a,8-tetrahydro-5H-benzo[g][1,3]dioxolo[4',5':4,5]benzo[1,2,3-de]quinoline CHEMBL36654
GNF-Pf-4466 BDBM50284797 cid_235224 7-Methyl-6,7,7a,8-tetrahydro-5H-benzo[g][1,3]dioxolo[4',5':4,5]benzo[1,2,3-de]quinoline CHEMBL36654 CHEMBL599883 GNF-Pf-4070 3-methoxy-3'-(methylthio)-5,5'-bi(1,2,4-triazine) BDBM50335540 3-Methoxy-5-(3-methylthio-1,2,4-triazin-5-yl-)-1,2,4-triazine
CHEMBL599883 GNF-Pf-4070 3-methoxy-3'-(methylthio)-5,5'-bi(1,2,4-triazine) BDBM50335540 3-Methoxy-5-(3-methylthio-1,2,4-triazin-5-yl-)-1,2,4-triazine GNF-PF-2857 CHEMBL217665 [4-(4-methylpiperazin-1-yl)phenyl]quinolin-4-ylamine N-(4-(4-methylpiperazin-1-yl)phenyl)quinolin-4-amine BDBM50196110
GNF-PF-2857 CHEMBL217665 [4-(4-methylpiperazin-1-yl)phenyl]quinolin-4-ylamine N-(4-(4-methylpiperazin-1-yl)phenyl)quinolin-4-amine BDBM50196110 3-(4-bromophenyl)-5H-indeno[1,2-c]pyridazin-5-one CHEMBL338094 GNF-Pf-3108 BDBM50031071 3-(4-Bromo-phenyl)-indeno[1,2-c]pyridazin-5-one
3-(4-bromophenyl)-5H-indeno[1,2-c]pyridazin-5-one CHEMBL338094 GNF-Pf-3108 BDBM50031071 3-(4-Bromo-phenyl)-indeno[1,2-c]pyridazin-5-one CHEMBL2163795 CHEMBL274189 PALMATINE CHEMBL1270849 BDBM50292332 2,3,9,10-tetramethoxy-5,6-dihydro-isoquino[3,2-a]isoquinolinylium 2,3,9,10-Tetramethoxy-5,6-dihydro-isoquino[3,2-a]isoquinolinylium; chloride GNF-Pf-4086
CHEMBL2163795 CHEMBL274189 PALMATINE CHEMBL1270849 BDBM50292332 2,3,9,10-tetramethoxy-5,6-dihydro-isoquino[3,2-a]isoquinolinylium 2,3,9,10-Tetramethoxy-5,6-dihydro-isoquino[3,2-a]isoquinolinylium; chloride GNF-Pf-4086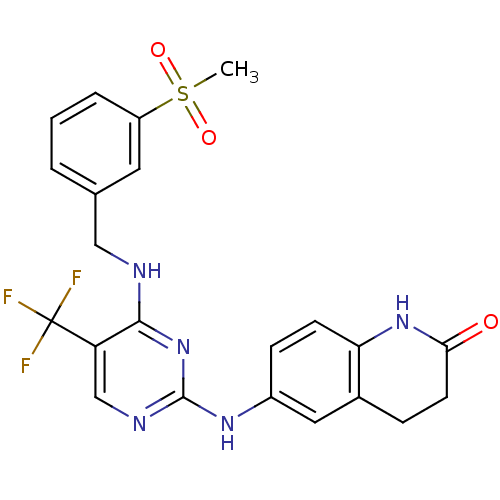 BDBM50269948 US10266549, Example 32 PF-228 6-(4-(3-(methylsulfonyl)benzylamino)-5-(trifluoromethyl)pyrimidin-2-ylamino)-3,4-dihydroquinolin-2(1H)-one CHEMBL514554 D3RKN_2 US10774092, Example 32
BDBM50269948 US10266549, Example 32 PF-228 6-(4-(3-(methylsulfonyl)benzylamino)-5-(trifluoromethyl)pyrimidin-2-ylamino)-3,4-dihydroquinolin-2(1H)-one CHEMBL514554 D3RKN_2 US10774092, Example 32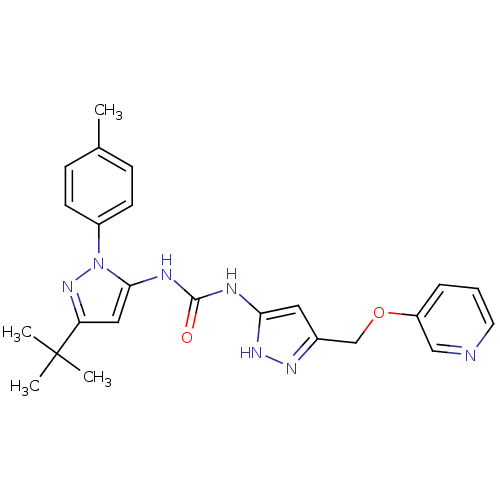 PF-4618433 CHEMBL1084269 1-(3-tert-butyl-1-p-tolyl-1H-pyrazol-5-yl)-3-(3-((pyridin-3-yloxy)methyl)-1H-pyrazol-5-yl)urea BDBM50318872
PF-4618433 CHEMBL1084269 1-(3-tert-butyl-1-p-tolyl-1H-pyrazol-5-yl)-3-(3-((pyridin-3-yloxy)methyl)-1H-pyrazol-5-yl)urea BDBM50318872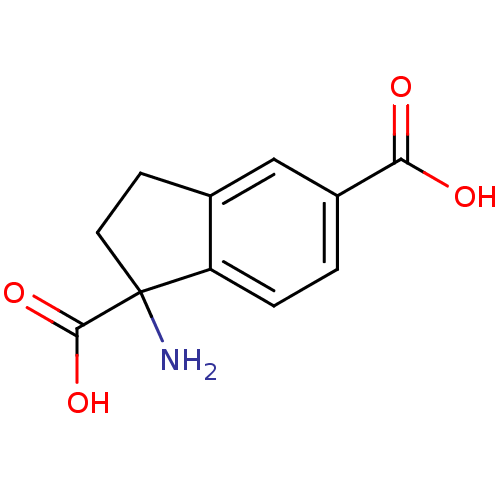 1-Amino-indan-1,5-dicarboxylic acid BDBM50030630 GNF-Pf-1401 (RS)-1-aminoindan-1,5-dicarboxylic acid AIDC CHEMBL313938 AIDA 1-Amino-indan-1,5-dicarboxylic acid(RS-AIDA)
1-Amino-indan-1,5-dicarboxylic acid BDBM50030630 GNF-Pf-1401 (RS)-1-aminoindan-1,5-dicarboxylic acid AIDC CHEMBL313938 AIDA 1-Amino-indan-1,5-dicarboxylic acid(RS-AIDA) GNF-Pf-3427 Acridin-9-yl-[4-(4-methyl-piperazin-1-yl)-phenyl]-amine BDBM50196135 CHEMBL106525 N-(4-(4-methylpiperazin-1-yl)phenyl)acridin-9-amine TCMDC-123912
GNF-Pf-3427 Acridin-9-yl-[4-(4-methyl-piperazin-1-yl)-phenyl]-amine BDBM50196135 CHEMBL106525 N-(4-(4-methylpiperazin-1-yl)phenyl)acridin-9-amine TCMDC-123912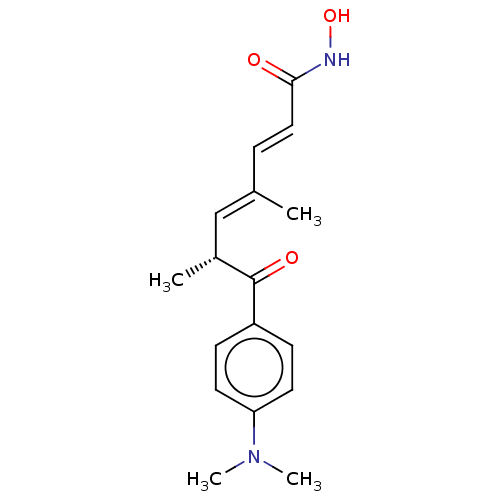 US10011611, Trichostatin A GNF-Pf-1011 CHEBI:46024 Trichostatin A (TSA) US20230322747, Compound trichostatin A BDBM50005711 Trichostatin A US9265734, TSA US11535598, Compound Trichostatin A US10722597, Compound Trichostatin TRICHOSTATIN
US10011611, Trichostatin A GNF-Pf-1011 CHEBI:46024 Trichostatin A (TSA) US20230322747, Compound trichostatin A BDBM50005711 Trichostatin A US9265734, TSA US11535598, Compound Trichostatin A US10722597, Compound Trichostatin TRICHOSTATIN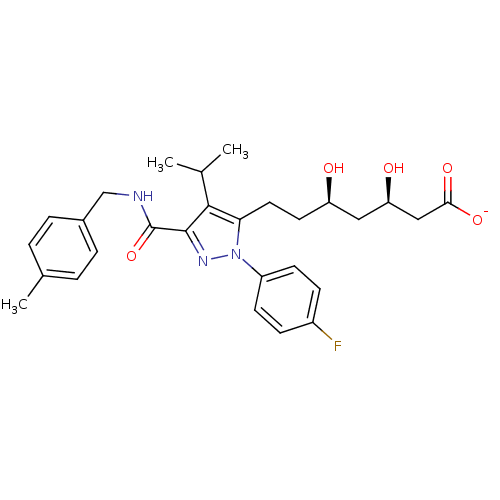 PF-3052334 pyrazole-based inhibitor, 35 BDBM20692 sodium (3R,5R)-7-[1-(4-fluorophenyl)-3-{[(4-methylphenyl)methyl]carbamoyl}-4-(propan-2-yl)-1H-pyrazol-5-yl]-3,5-dihydroxyheptanoate
PF-3052334 pyrazole-based inhibitor, 35 BDBM20692 sodium (3R,5R)-7-[1-(4-fluorophenyl)-3-{[(4-methylphenyl)methyl]carbamoyl}-4-(propan-2-yl)-1H-pyrazol-5-yl]-3,5-dihydroxyheptanoate 6-(1,3-Dioxo-1H,3H-benzo[de]isoquinolin-2-yl)-hexanoic acid hydroxyamide GNF-Pf-2024 6-(1,3-dioxo-1H-benzo[de]isoquinolin-2(3H)-yl)-N-hydroxyhexanamide Scriptaid, 9 Scriptaid BDBM50328678 CHEMBL96051
6-(1,3-Dioxo-1H,3H-benzo[de]isoquinolin-2-yl)-hexanoic acid hydroxyamide GNF-Pf-2024 6-(1,3-dioxo-1H-benzo[de]isoquinolin-2(3H)-yl)-N-hydroxyhexanamide Scriptaid, 9 Scriptaid BDBM50328678 CHEMBL96051 GNF-PF-5152 1-[2-(2-Hydroxy-3-piperidin-1-yl-propoxy)-phenyl]-3-phenyl-propan-1-one 1-(2-(2-hydroxy-3-(piperidin-1-yl)propoxy)phenyl)-3-phenylpropan-1-one BDBM50135012 CHEMBL319952
GNF-PF-5152 1-[2-(2-Hydroxy-3-piperidin-1-yl-propoxy)-phenyl]-3-phenyl-propan-1-one 1-(2-(2-hydroxy-3-(piperidin-1-yl)propoxy)phenyl)-3-phenylpropan-1-one BDBM50135012 CHEMBL319952 PF-06877900 US11472809, Example 14 US10144738, Example 14 2-((1r,3s)-3-Hydroxy-1-(4-(6-(1-methyl-1H-pyrazol-4-yl)pyrazolo[1,5-a]pyrazin-4-yl)-1H-pyrazol-1-yl)cyclobutyl)acetonitrile (trans isomer) BDBM305815
PF-06877900 US11472809, Example 14 US10144738, Example 14 2-((1r,3s)-3-Hydroxy-1-(4-(6-(1-methyl-1H-pyrazol-4-yl)pyrazolo[1,5-a]pyrazin-4-yl)-1H-pyrazol-1-yl)cyclobutyl)acetonitrile (trans isomer) BDBM305815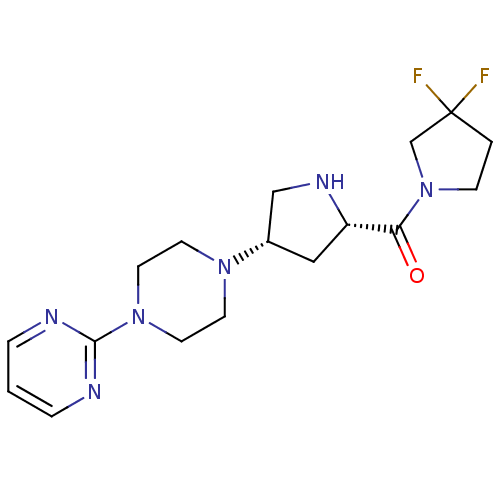 BDBM50249479 CHEMBL515387 cis-(3,3-Difluoro-pyrrolidin-1-yl)-[(2S,4S)-(4-(4-pyrimidin-2-yl-piperazin-1-yl)-pyrrolidin-2-yl]-methanone PF-00734200 2-(4-{(3S,5S)-5-[(3,3-difluoropyrrolidin-1-yl)carbonyl]pyrrolidin-3-yl}piperazin-1-yl)pyrimidine
BDBM50249479 CHEMBL515387 cis-(3,3-Difluoro-pyrrolidin-1-yl)-[(2S,4S)-(4-(4-pyrimidin-2-yl-piperazin-1-yl)-pyrrolidin-2-yl]-methanone PF-00734200 2-(4-{(3S,5S)-5-[(3,3-difluoropyrrolidin-1-yl)carbonyl]pyrrolidin-3-yl}piperazin-1-yl)pyrimidine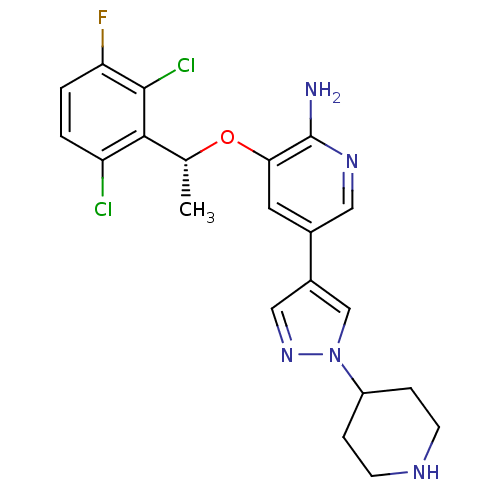 US12129258, Example Crizotinib BDBM50306682 US10370379, Crizotinib (R)-3-(1-(2,6-dichloro-3-fluorophenyl)ethoxy)-5-(1-(piperidin-4-yl)-1H-pyrazol-4-yl)pyridin-2-amine CHEMBL601719 US11059827, Compound Crizotinib US9199944, Crizotinib PF-2341066 3-[(1R)-1-(2,6-dichloro-3-fluorophenyl)ethoxy]-5-(1-piperidin-4-yl-1H-pyrazol-4-yl)pyridin-2-amine US10543199, Compound Crizotinib US9126941, PF-2341066 US11517561, Compound Crizotinib CRIZOTINIB 3-(2,6-dichloro-3-fluorobenzyloxy)-5-(1-(piperidin-4-yl)-1H-pyrazol-4-yl)pyridin-2-amine US10780082, Compound Crizotinib US9226923, Crizotinib
US12129258, Example Crizotinib BDBM50306682 US10370379, Crizotinib (R)-3-(1-(2,6-dichloro-3-fluorophenyl)ethoxy)-5-(1-(piperidin-4-yl)-1H-pyrazol-4-yl)pyridin-2-amine CHEMBL601719 US11059827, Compound Crizotinib US9199944, Crizotinib PF-2341066 3-[(1R)-1-(2,6-dichloro-3-fluorophenyl)ethoxy]-5-(1-piperidin-4-yl-1H-pyrazol-4-yl)pyridin-2-amine US10543199, Compound Crizotinib US9126941, PF-2341066 US11517561, Compound Crizotinib CRIZOTINIB 3-(2,6-dichloro-3-fluorobenzyloxy)-5-(1-(piperidin-4-yl)-1H-pyrazol-4-yl)pyridin-2-amine US10780082, Compound Crizotinib US9226923, Crizotinib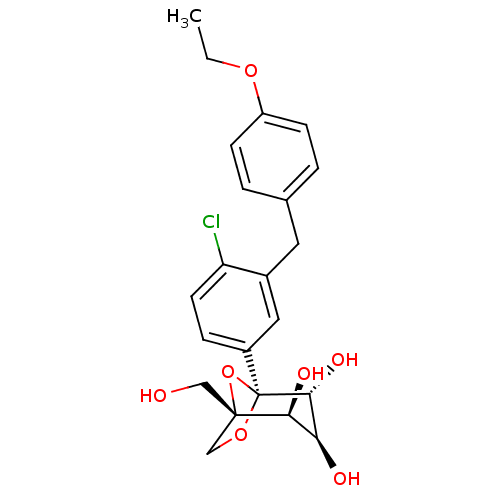 CHEMBL1770248 5-(4-chloro-3-(4-ethoxybenzyl)phenyl)-1-hydroxymethyl-6,8-dioxabicyclo(3.2.1)octane-2,3,4-triol ertugliflozin PF-04971729 BDBM50342885 Steglatro (1S,2S,3S,4R,5S)-5-[4-Chloro-3-(4-ethoxybenzyl)phenyl]-1-(hydroxymethyl)-6,8-dioxa-bicyclo[3.2.1]octane-2,3,4-triol
CHEMBL1770248 5-(4-chloro-3-(4-ethoxybenzyl)phenyl)-1-hydroxymethyl-6,8-dioxabicyclo(3.2.1)octane-2,3,4-triol ertugliflozin PF-04971729 BDBM50342885 Steglatro (1S,2S,3S,4R,5S)-5-[4-Chloro-3-(4-ethoxybenzyl)phenyl]-1-(hydroxymethyl)-6,8-dioxa-bicyclo[3.2.1]octane-2,3,4-triol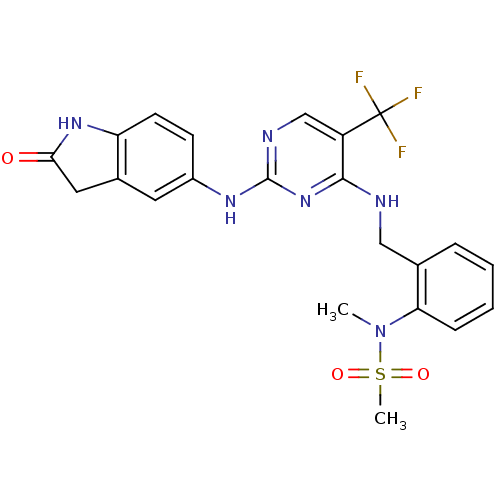 CHEMBL472212 D3RKN_6 N-methyl-N-(2-((2-(2-oxoindolin-5-ylamino)-5-(trifluoromethyl)pyrimidin-4-ylamino)methyl)phenyl)methanesulfonamide N-methyl-N-{2-[({2-[(2-oxo-2,3-dihydro-1H-indol-5-yl)amino]-5-(trifluoromethyl)pyrimidin-4-yl}amino)methyl]phenyl}methanesulfonamide PF-431396 BDBM50246060 CHEMBL541649
CHEMBL472212 D3RKN_6 N-methyl-N-(2-((2-(2-oxoindolin-5-ylamino)-5-(trifluoromethyl)pyrimidin-4-ylamino)methyl)phenyl)methanesulfonamide N-methyl-N-{2-[({2-[(2-oxo-2,3-dihydro-1H-indol-5-yl)amino]-5-(trifluoromethyl)pyrimidin-4-yl}amino)methyl]phenyl}methanesulfonamide PF-431396 BDBM50246060 CHEMBL541649 CHEMBL486232 BDBM50243232 GNF-PF-5434 N~2~-(MORPHOLIN-4-YLCARBONYL)-N~1~-[(1S,2E)-1-(2-PHENYLETHYL)-3-(PHENYLSULFONYL)PROP-2-ENYL]-D-LEUCINAMIDE N-((S)-4-methyl-1-oxo-1-((S)-5-phenyl-1-(phenylsulfonyl)pent-1-en-3-ylamino)pentan-2-yl)morpholine-4-carboxamide
CHEMBL486232 BDBM50243232 GNF-PF-5434 N~2~-(MORPHOLIN-4-YLCARBONYL)-N~1~-[(1S,2E)-1-(2-PHENYLETHYL)-3-(PHENYLSULFONYL)PROP-2-ENYL]-D-LEUCINAMIDE N-((S)-4-methyl-1-oxo-1-((S)-5-phenyl-1-(phenylsulfonyl)pent-1-en-3-ylamino)pentan-2-yl)morpholine-4-carboxamide N-methyl-N-{3-[({2-[(2-oxo-2,3-dihydro-1H-indol-5-yl)amino]-5-(trifluoromethyl)pyrimidin-4-yl}amino)methyl]pyridin-2-yl}methanesulfonamide CHEMBL2430359 CHEMBL1084546 BDBM50318884 PF-562271 N-methyl-N-(3-((2-(2-oxoindolin-5-ylamino)-5-(trifluoromethyl)pyrimidin-4-ylamino)methyl)pyridin-2-yl)methanesulfonamide
N-methyl-N-{3-[({2-[(2-oxo-2,3-dihydro-1H-indol-5-yl)amino]-5-(trifluoromethyl)pyrimidin-4-yl}amino)methyl]pyridin-2-yl}methanesulfonamide CHEMBL2430359 CHEMBL1084546 BDBM50318884 PF-562271 N-methyl-N-(3-((2-(2-oxoindolin-5-ylamino)-5-(trifluoromethyl)pyrimidin-4-ylamino)methyl)pyridin-2-yl)methanesulfonamide (6R)-6-cyclopentyl-6-[2-(2,6-diethylpyridin-4-yl)ethyl]-3-[(5,7-dimethyl[1,2,4]triazolo[1,5-a]pyrimidin-2-yl)methyl]-4-hydroxy-5,6-dihydro-2H-pyran-2-one CHEMBL490672 (R)-6-cyclopentyl-6-[2-(2,6-diethylpyridin-4-yl)ethyl]-3-[(5,7-dimethyl[1,2,4]triazolo[1,5-a]pyrimidin-2-yl)methyl]-4-hydroxy-5,6-dihydro-2Hpyran-2-one PF-00868554 BDBM50293131
(6R)-6-cyclopentyl-6-[2-(2,6-diethylpyridin-4-yl)ethyl]-3-[(5,7-dimethyl[1,2,4]triazolo[1,5-a]pyrimidin-2-yl)methyl]-4-hydroxy-5,6-dihydro-2H-pyran-2-one CHEMBL490672 (R)-6-cyclopentyl-6-[2-(2,6-diethylpyridin-4-yl)ethyl]-3-[(5,7-dimethyl[1,2,4]triazolo[1,5-a]pyrimidin-2-yl)methyl]-4-hydroxy-5,6-dihydro-2Hpyran-2-one PF-00868554 BDBM50293131 Rythmol PROPAFENONE 1-(2-(2-hydroxy-3-(propylamino)propoxy)phenyl)-3-phenylpropan-1-one 1-[2-(2-Hydroxy-3-propylamino-propoxy)-phenyl]-3-phenyl-propan-1-one GNF-Pf-4594 CHEMBL631 1N-[2-(5-dimethylaminomethyl-2-furylmethylsulfanyl)ethyl]-1N-methyl-2-nitro-(Z)-1-ethene-1,1-diamine BDBM50067133 1-[2-(2-Hydroxy-3-propylamino-propoxy)-phenyl]-3-phenyl-propan-1-one (propafenone)
Rythmol PROPAFENONE 1-(2-(2-hydroxy-3-(propylamino)propoxy)phenyl)-3-phenylpropan-1-one 1-[2-(2-Hydroxy-3-propylamino-propoxy)-phenyl]-3-phenyl-propan-1-one GNF-Pf-4594 CHEMBL631 1N-[2-(5-dimethylaminomethyl-2-furylmethylsulfanyl)ethyl]-1N-methyl-2-nitro-(Z)-1-ethene-1,1-diamine BDBM50067133 1-[2-(2-Hydroxy-3-propylamino-propoxy)-phenyl]-3-phenyl-propan-1-one (propafenone) (4-Bromo-2-fluoro-phenyl)-[6-methoxy-7-(1-methyl-piperidin-4-ylmethoxy)-quinazolin-4-yl]-amine N-(4-bromo-2-fluorophenyl)-6-methoxy-7-[(1-methyl-4-piperidinyl)methoxy]-4-quinazolinamine BDBM50340915 ZD-64 GNF-Pf-2188 ZD-6474 Zactima CHEMBL24828 4-BROMO-2-FLUORO-N-[(4E)-6-METHOXY-7-[(1-METHYLPIPERIDIN-4-YL)METHOXY]QUINAZOLIN-4(1H)-YLIDENE]ANILINE N-(4-bromo-2-fluorophenyl)-6-methoxy-7-((1-methylpiperidin-4-yl)methoxy)quinazolin-4-amine
(4-Bromo-2-fluoro-phenyl)-[6-methoxy-7-(1-methyl-piperidin-4-ylmethoxy)-quinazolin-4-yl]-amine N-(4-bromo-2-fluorophenyl)-6-methoxy-7-[(1-methyl-4-piperidinyl)methoxy]-4-quinazolinamine BDBM50340915 ZD-64 GNF-Pf-2188 ZD-6474 Zactima CHEMBL24828 4-BROMO-2-FLUORO-N-[(4E)-6-METHOXY-7-[(1-METHYLPIPERIDIN-4-YL)METHOXY]QUINAZOLIN-4(1H)-YLIDENE]ANILINE N-(4-bromo-2-fluorophenyl)-6-methoxy-7-((1-methylpiperidin-4-yl)methoxy)quinazolin-4-amine 1-[10-(4-amino-2-methyl-1-quinoliniumyl)decyl]-2-methyl-4-quinoliniumamine dichloride Dequqlinium 1,1'-(Deca-4,6-diyne-1,10-diyl)bis(4-amino-2-methylquinolinium) Diiodide Hydrate 1-[10-(4-amino-2-methyl-1-quinoliniumyl)decyl]-2-methyl-4-quinoliniumamine(dequalinium) 1-[10-(4-amino-2-methyl-1-quinoliniumyl)decyl]-2-methyl-4-quinoliniumamine GNF-Pf-5483 BDBM50048403 Dequalinium chloride 1-[10-(4-amino-2-methyl-1-quinoliniumyl)decyl]-2-methyl-4-quinoliniumamine(Deq) CHEMBL121663 1-[10-(4-amino-2-methyl-1-quinoliniumyl)decyl]-2-methyl-4-quinoliniumamine dichloride ; Dequalinium dequalinium DQ+
1-[10-(4-amino-2-methyl-1-quinoliniumyl)decyl]-2-methyl-4-quinoliniumamine dichloride Dequqlinium 1,1'-(Deca-4,6-diyne-1,10-diyl)bis(4-amino-2-methylquinolinium) Diiodide Hydrate 1-[10-(4-amino-2-methyl-1-quinoliniumyl)decyl]-2-methyl-4-quinoliniumamine(dequalinium) 1-[10-(4-amino-2-methyl-1-quinoliniumyl)decyl]-2-methyl-4-quinoliniumamine GNF-Pf-5483 BDBM50048403 Dequalinium chloride 1-[10-(4-amino-2-methyl-1-quinoliniumyl)decyl]-2-methyl-4-quinoliniumamine(Deq) CHEMBL121663 1-[10-(4-amino-2-methyl-1-quinoliniumyl)decyl]-2-methyl-4-quinoliniumamine dichloride ; Dequalinium dequalinium DQ+ (2,8-Bis-trifluoromethyl-quinolin-4-yl)-piperidin-2-yl-methanol med.21724, Compound Mefloquine rac-mefloquine (2,8-Bis-trifluoromethyl-quinolin-4-yl)-piperidin-2-yl-methanol; (Mefloquine or MF) 2,8-di(trifluoromethyl)-4-quinolyl-hexahydro-2-pyridinylmethanol(Mefloquine) GNF-Pf-5544 Lariam melfoquine (2,8-bis(trifluoromethyl)quinolin-4-yl)(piperidin-2-yl)methanol 2-[(2,8-Bis-trifluoromethyl-quinolin-4-yl)-hydroxy-methyl]-piperidinium BDBM50022889 MEFLOQUINE rel-(2,8-bis(trifluoromethyl)quinolin-4-yl)(piperidin-2-yl)methanol (2,8-Bis-trifluoromethyl-quinolin-4-yl)-(1-methyl-piperidin-2-yl)-methanol (S)-(2,8-bis(trifluoromethyl)quinolin-4-yl)((R)-piperidin-2-yl)methanol (S)-(2,8-Bis-trifluoromethyl-quinolin-4-yl)-(R)-piperidin-2-yl-methanol [2,8-bis(trifluoromethyl)quinolin-4-yl]-(2-piperidyl)methanol (2,8-Bis-trifluoromethyl-quinolin-4-yl)-piperidin-2-yl-methanol( Mefloquine) CHEMBL416956 (S)-(2,8-Bis-trifluoromethyl-quinolin-4-yl)-piperidin-2-yl-methanol
(2,8-Bis-trifluoromethyl-quinolin-4-yl)-piperidin-2-yl-methanol med.21724, Compound Mefloquine rac-mefloquine (2,8-Bis-trifluoromethyl-quinolin-4-yl)-piperidin-2-yl-methanol; (Mefloquine or MF) 2,8-di(trifluoromethyl)-4-quinolyl-hexahydro-2-pyridinylmethanol(Mefloquine) GNF-Pf-5544 Lariam melfoquine (2,8-bis(trifluoromethyl)quinolin-4-yl)(piperidin-2-yl)methanol 2-[(2,8-Bis-trifluoromethyl-quinolin-4-yl)-hydroxy-methyl]-piperidinium BDBM50022889 MEFLOQUINE rel-(2,8-bis(trifluoromethyl)quinolin-4-yl)(piperidin-2-yl)methanol (2,8-Bis-trifluoromethyl-quinolin-4-yl)-(1-methyl-piperidin-2-yl)-methanol (S)-(2,8-bis(trifluoromethyl)quinolin-4-yl)((R)-piperidin-2-yl)methanol (S)-(2,8-Bis-trifluoromethyl-quinolin-4-yl)-(R)-piperidin-2-yl-methanol [2,8-bis(trifluoromethyl)quinolin-4-yl]-(2-piperidyl)methanol (2,8-Bis-trifluoromethyl-quinolin-4-yl)-piperidin-2-yl-methanol( Mefloquine) CHEMBL416956 (S)-(2,8-Bis-trifluoromethyl-quinolin-4-yl)-piperidin-2-yl-methanol