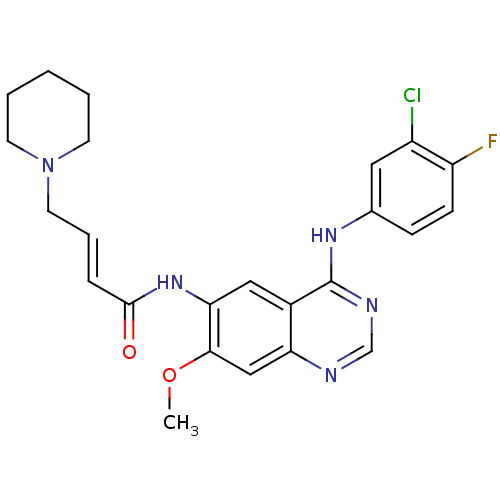
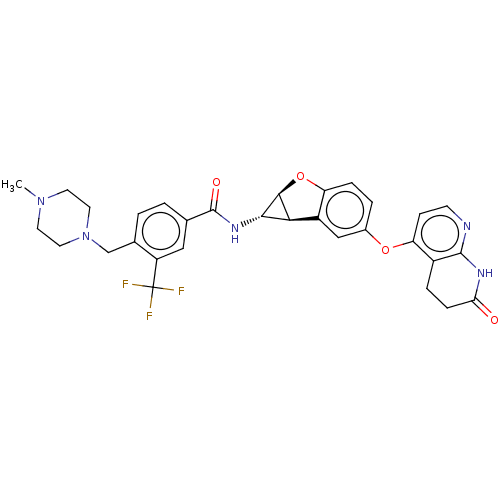
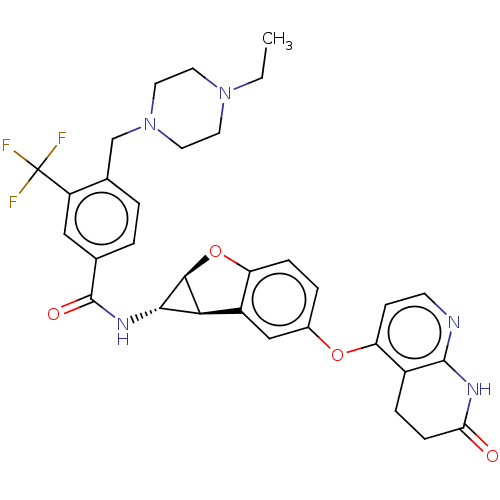
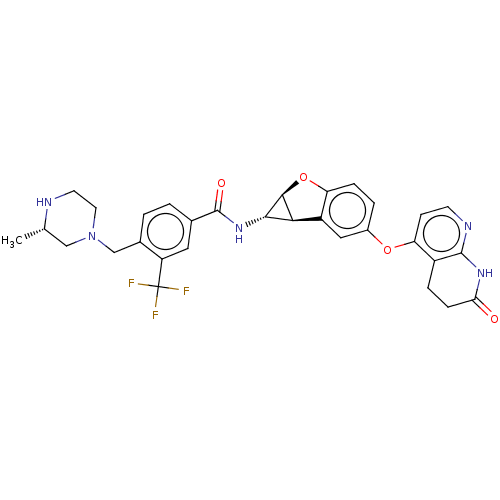
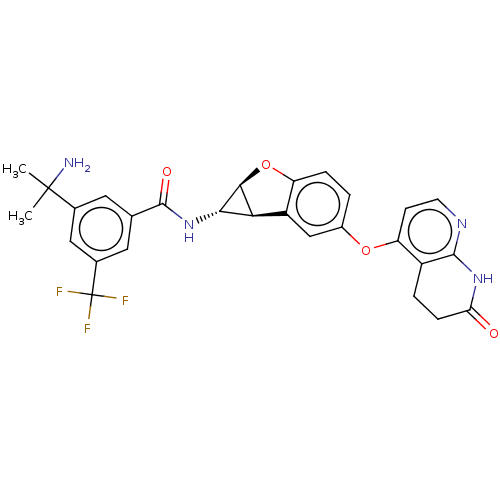
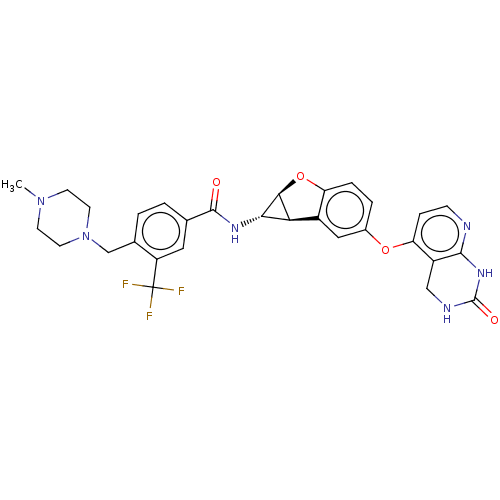
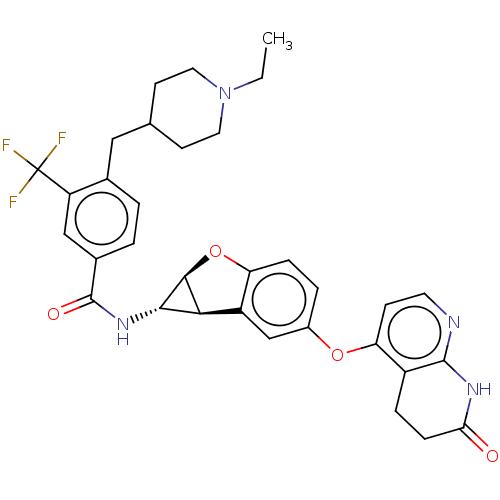
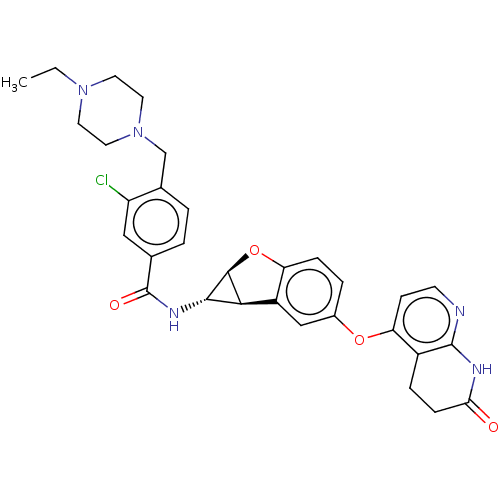
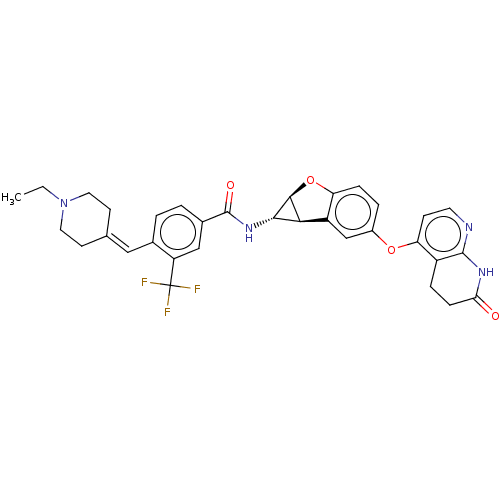
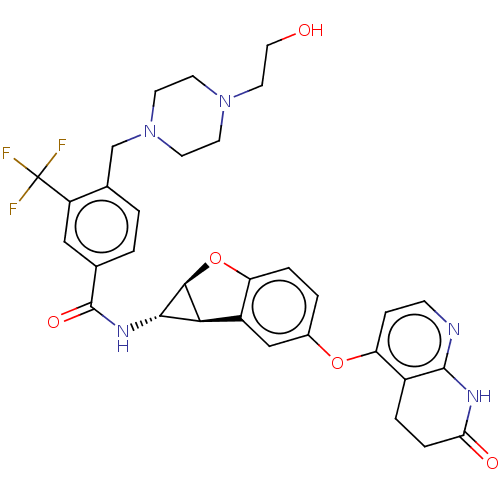
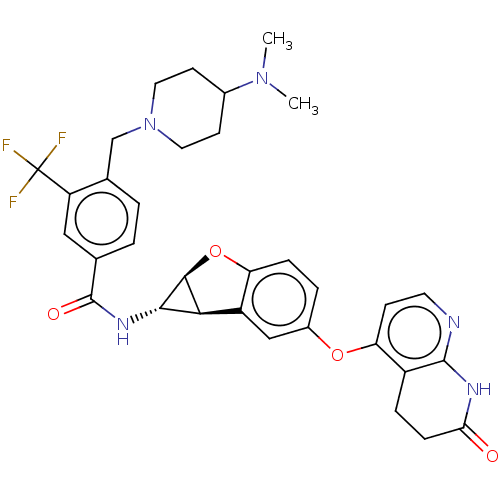
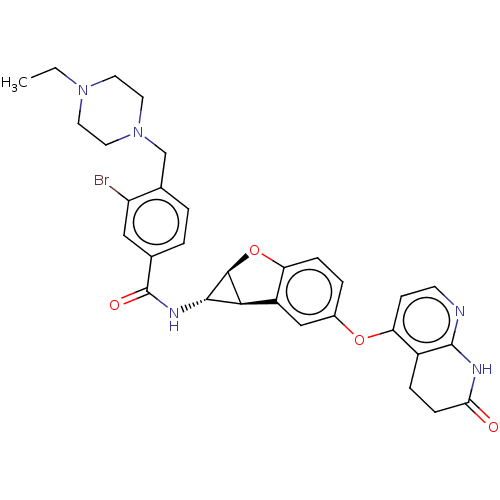
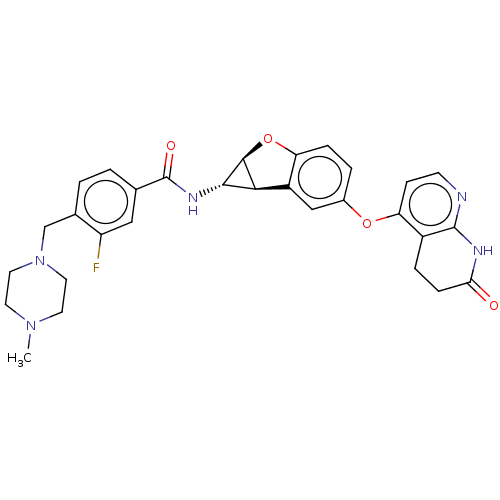
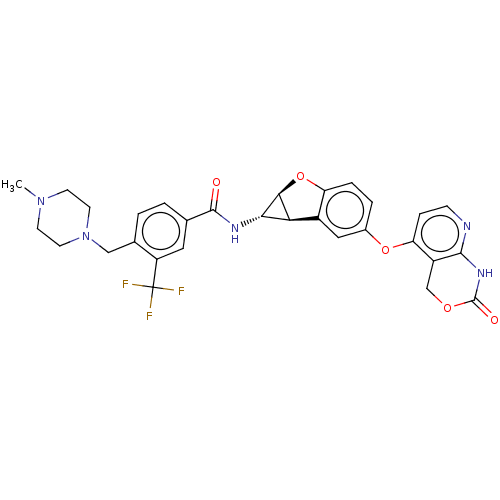
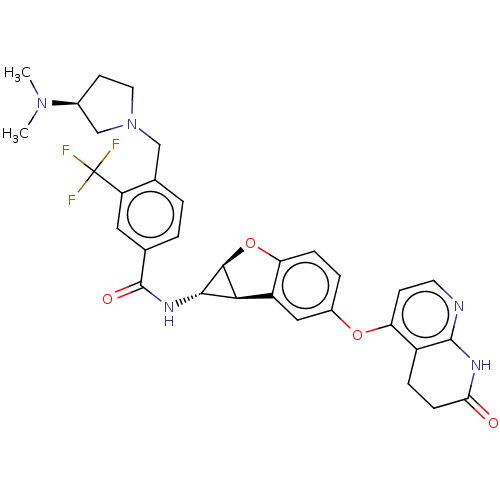
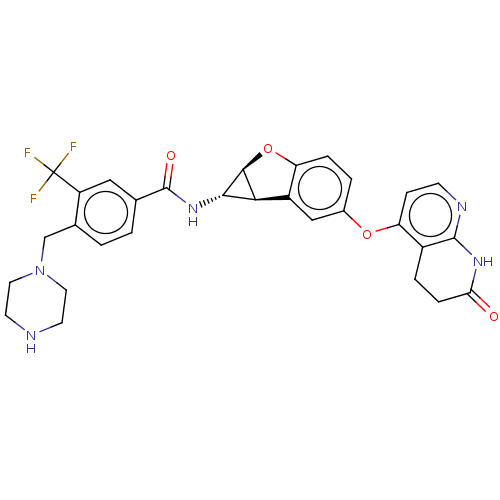
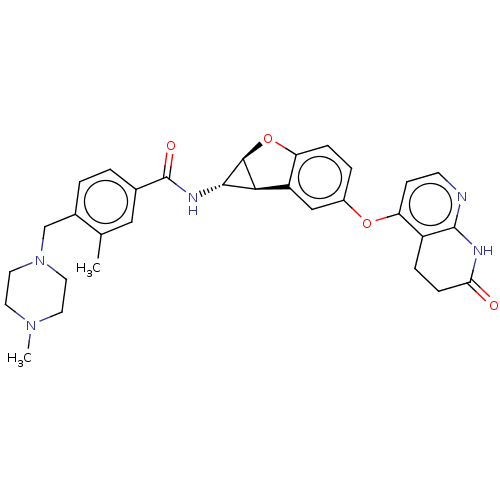
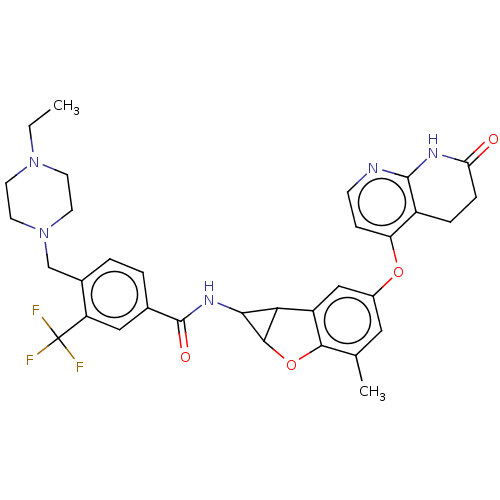
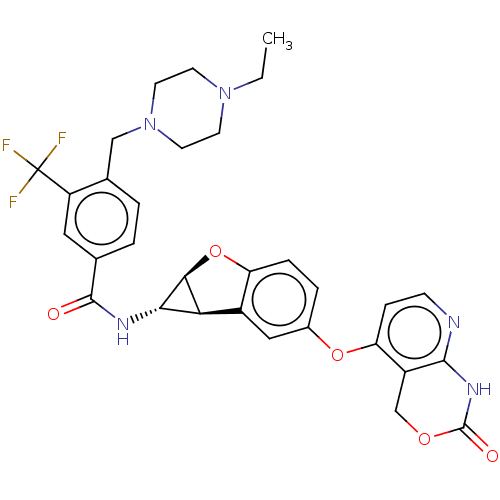
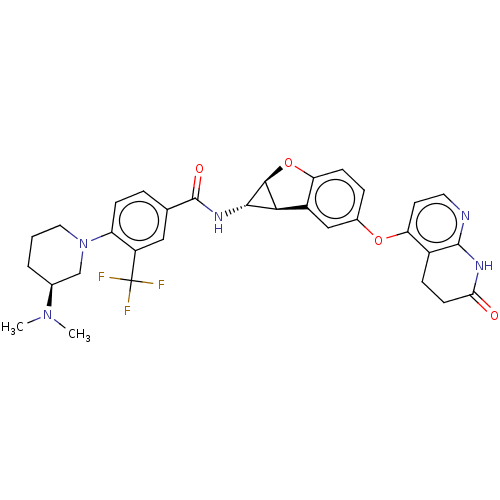
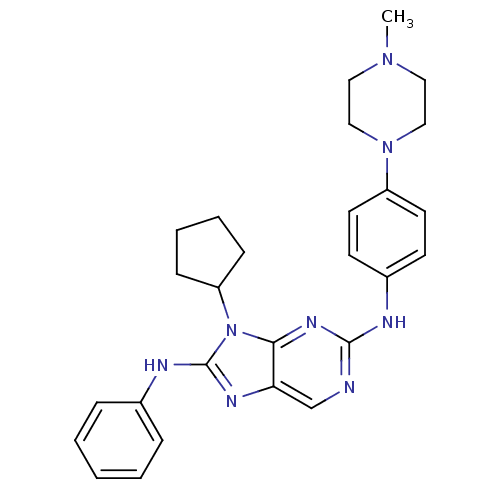
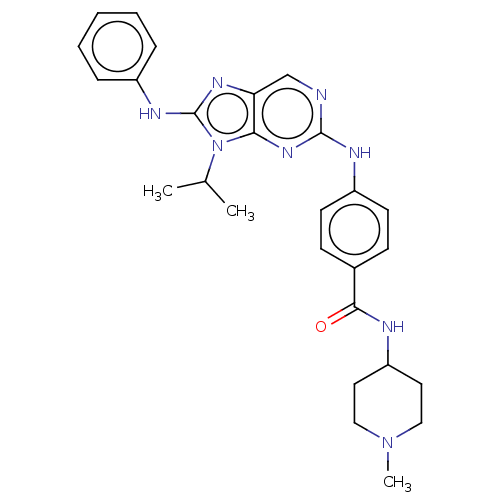
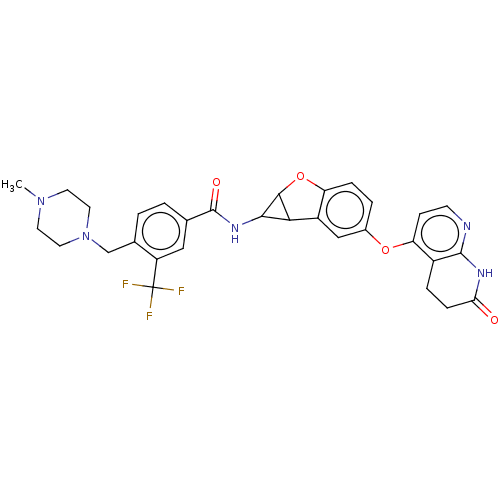
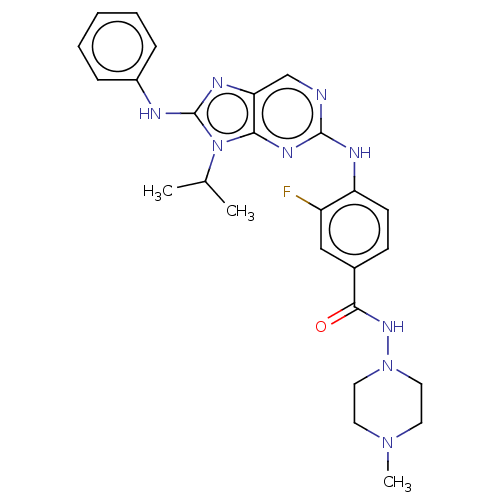
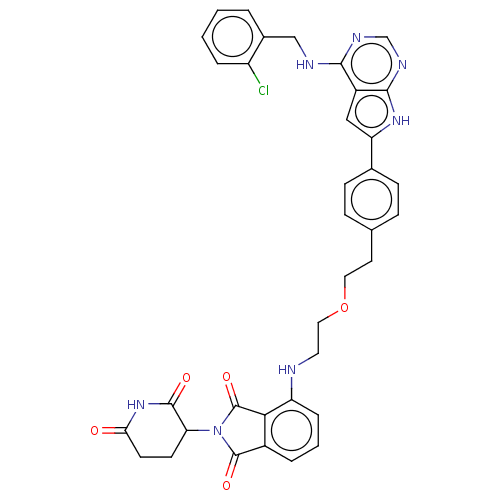
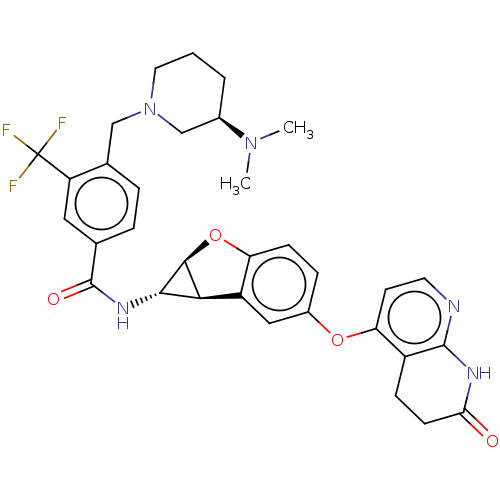
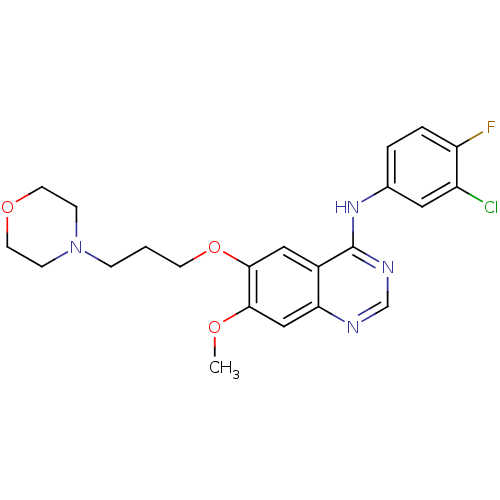
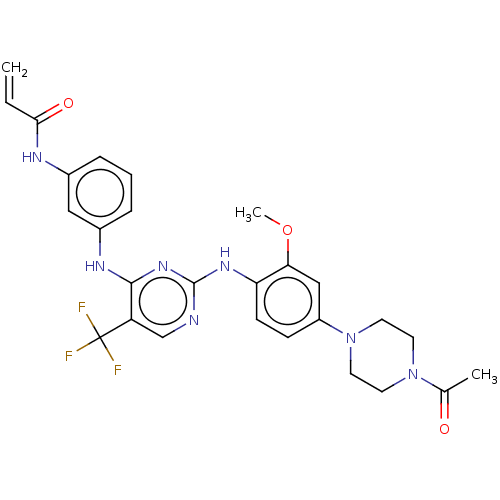
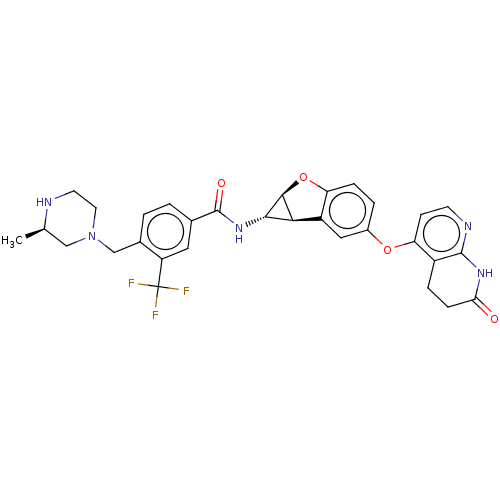
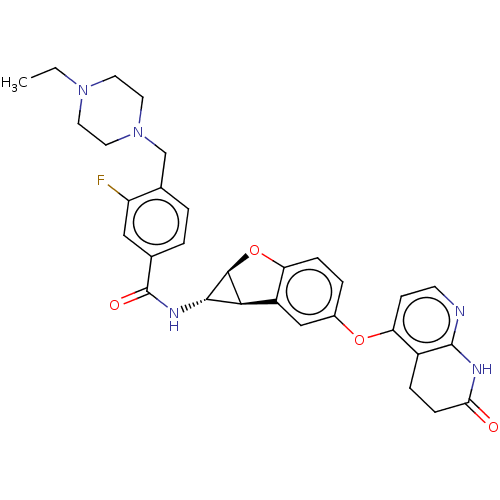
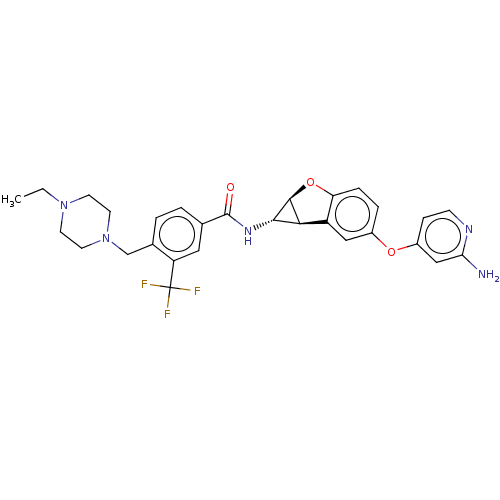
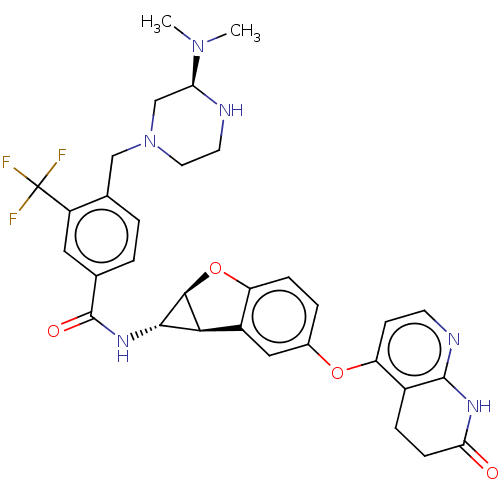
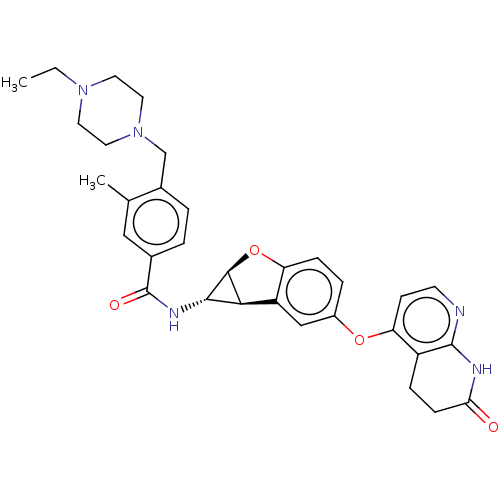
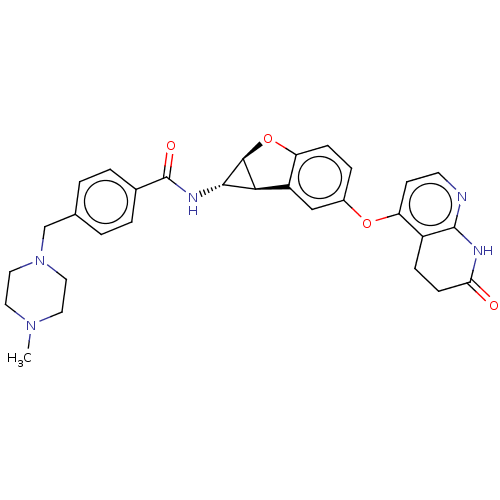
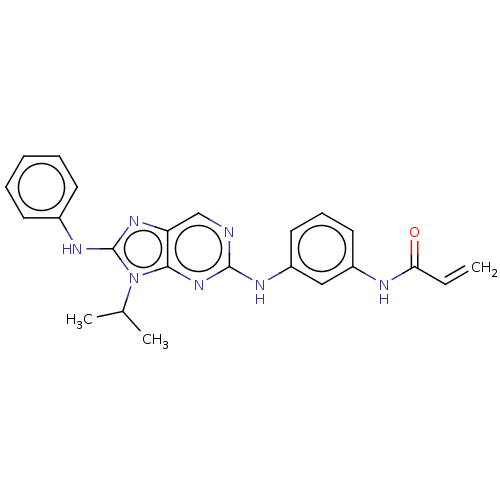
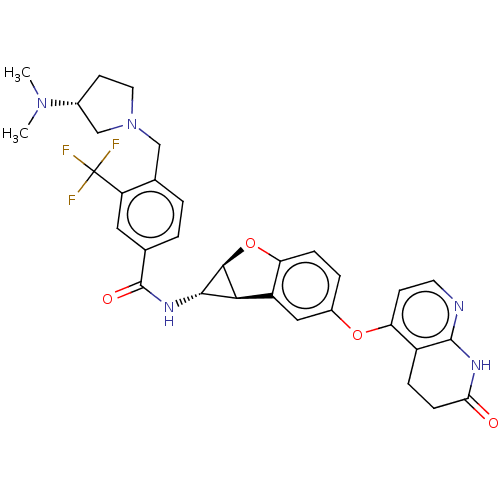
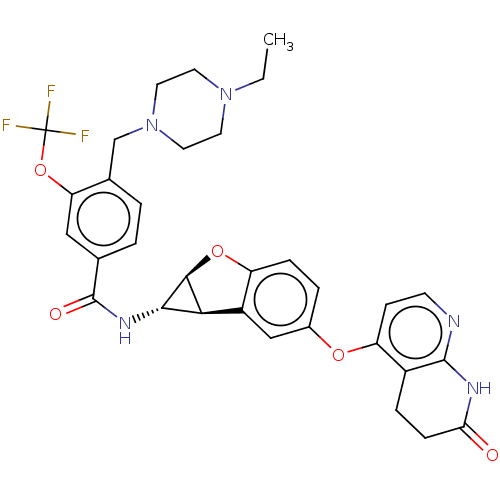
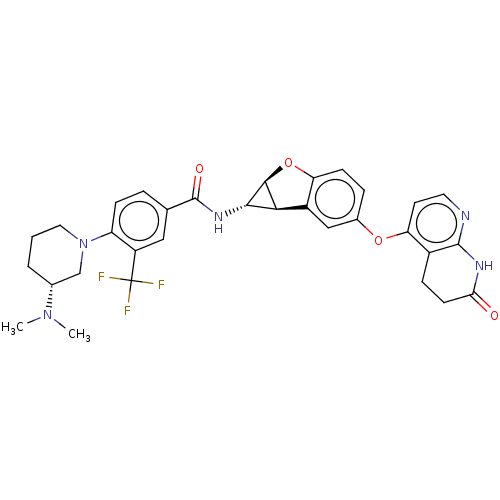
Report error Found 481 Enz. Inhib. hit(s) with Target = 'Epidermal growth factor receptor [L858R]'
Affinity DataIC50: 0.0200nMAssay Description: This assay quantifies the phosphorylation of EGFR at Tyr1068 and was used to measure the inhibitory effect of compounds on the transgenic EGFR del19...More data for this Ligand-Target Pair
Affinity DataIC50: 0.0300nMAssay Description: This assay quantifies the phosphorylation of EGFR at Tyr1068 and was used to measure the inhibitory effect of compounds on the transgenic EGFR del19...More data for this Ligand-Target Pair
Affinity DataIC50: 0.100nMAssay Description:Buffer formulation: a buffer consisted of 50 mM HEPES (pH 7.5), 0.01% BSA, 5 mM MgCl2, 0.1 mM Orthovanadate. After the buffer was formulated, an enzy...More data for this Ligand-Target Pair
Affinity DataIC50: 0.300nMAssay Description:EGFR biochemical activity measurements were carried out using the homogeneous time-resolved fluorescence (HTRF) assay (Cisbio), inhibitors and DMSO n...More data for this Ligand-Target Pair
Affinity DataIC50: 0.400nMAssay Description:The object of this assay was to test the inventive compounds for the kinase inhibitory activity in vitro. In this assay, an isotopic labeling method ...More data for this Ligand-Target Pair
Affinity DataIC50: 0.400nMAssay Description:The object of this assay was to test the inventive compounds for the kinase inhibitory activity in vitro. In this assay, an isotopic labeling method ...More data for this Ligand-Target Pair
Affinity DataIC50: 0.400nMAssay Description:The object of this assay was to test the inventive compounds for the kinase inhibitory activity in vitro. In this assay, an isotopic labeling method ...More data for this Ligand-Target Pair
Affinity DataIC50: 0.400nMAssay Description:All compounds and PROTACs were serially diluted in three-fold increments using 100% DMSO, followed by an intermediate 10-fold dilution using Buffer A...More data for this Ligand-Target Pair
Affinity DataIC50: 0.458nMAssay Description:Kinase Assay Protocol:Bar-coded Corning, low volume NBS, black 384-well plate (Corning Cat. #4514)1. 2.5 μL—4× Test Compound or 100 nL 100×...More data for this Ligand-Target Pair
Affinity DataIC50: 0.510nMAssay Description:The MSA (Mobility Shift Assay) method was used to detect the effects of small molecule inhibitors on the kinase activity of EGFR wild type and its mu...More data for this Ligand-Target Pair
Affinity DataIC50: 0.600nMAssay Description:The object of this assay was to test the inventive compounds for the kinase inhibitory activity in vitro. In this assay, an isotopic labeling method ...More data for this Ligand-Target Pair
Affinity DataIC50: 0.770nMAssay Description:The MSA (Mobility Shift Assay) method was used to detect the effects of small molecule inhibitors on the kinase activity of EGFR wild type and its mu...More data for this Ligand-Target Pair
Affinity DataIC50: 0.800nMAssay Description:EGFR biochemical activity measurements were carried out using the homogeneous time-resolved fluorescence (HTRF) assay (Cisbio), inhibitors and DMSO n...More data for this Ligand-Target Pair

 3D Structure (crystal)
3D Structure (crystal)