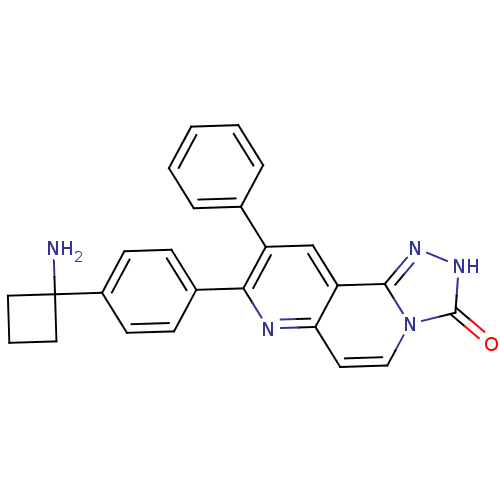
 CHEMBL1079175 US10550114, Compound MK-2206 8-(4-(1-aminocyclobutyl)phenyl)-9-phenyl-[1,2,4]triazolo[3,4-f][1,6]naphthyridin-3(2H)-one MK-2206 BDBM50313650
CHEMBL1079175 US10550114, Compound MK-2206 8-(4-(1-aminocyclobutyl)phenyl)-9-phenyl-[1,2,4]triazolo[3,4-f][1,6]naphthyridin-3(2H)-one MK-2206 BDBM50313650 BDBM102776 US8541572, 2206
BDBM102776 US8541572, 2206 BDBM266310 US9718790, I-2206
BDBM266310 US9718790, I-2206 BDBM736020 US20250129104, Compound 2206
BDBM736020 US20250129104, Compound 2206 MK-52 MK-51 CHEMBL2092821 BDBM50451497
MK-52 MK-51 CHEMBL2092821 BDBM50451497 BDBM50485492 MK-5172 MK-5172 MONOHYDRATE Grazoprevir monohydrate Grazoprevir MK-5172 ANHYDROUS
BDBM50485492 MK-5172 MK-5172 MONOHYDRATE Grazoprevir monohydrate Grazoprevir MK-5172 ANHYDROUS MK-2461 CHEMBL1802916 BDBM50347659 US20250129067, Compound MK 2461
MK-2461 CHEMBL1802916 BDBM50347659 US20250129067, Compound MK 2461 BDBM103838 MK-5172
BDBM103838 MK-5172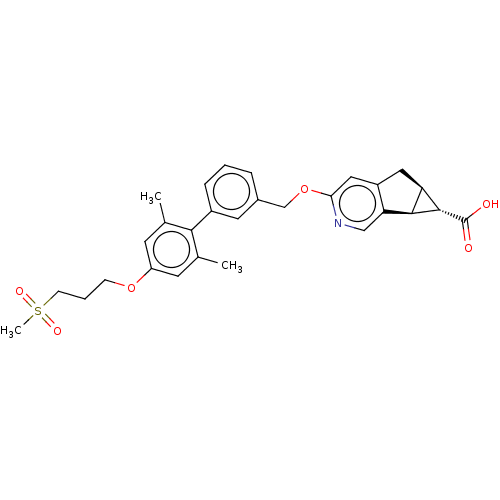 BDBM50566358 Mk-8666
BDBM50566358 Mk-8666 MK-8189 BDBM50616544
MK-8189 BDBM50616544 Mk-0752 BDBM50458158
Mk-0752 BDBM50458158 MK-24(S)-S(O)(NH)Ph BDBM50214607 CHEMBL541423 MK-24
MK-24(S)-S(O)(NH)Ph BDBM50214607 CHEMBL541423 MK-24 BDBM50360601 MK-0893 CHEMBL1933349
BDBM50360601 MK-0893 CHEMBL1933349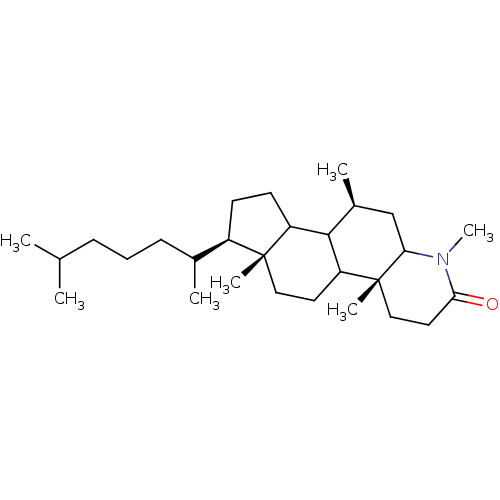 BDBM50368906 MK-386 CHEMBL25448
BDBM50368906 MK-386 CHEMBL25448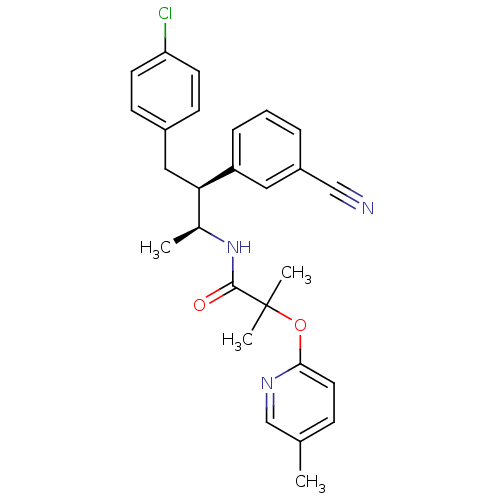 BDBM50373120 MK-0364 CHEMBL260977
BDBM50373120 MK-0364 CHEMBL260977 BDBM50381914 CHEMBL2023109 MK-7725
BDBM50381914 CHEMBL2023109 MK-7725 BDBM50423686 CHEMBL506044 MK-0668
BDBM50423686 CHEMBL506044 MK-0668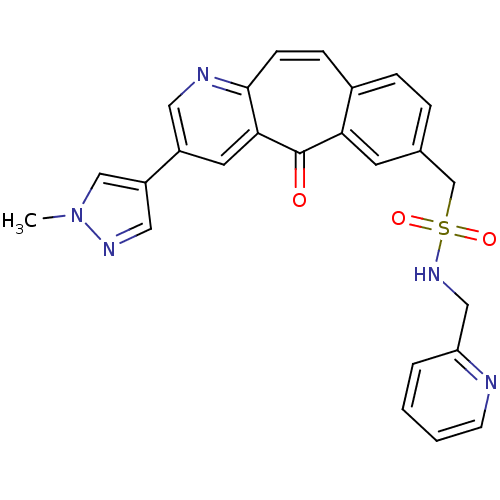 BDBM50427138 MK-8033 CHEMBL2323775
BDBM50427138 MK-8033 CHEMBL2323775 BDBM50479471 MK-1107 CHEMBL491019
BDBM50479471 MK-1107 CHEMBL491019 CHEMBL253364 BDBM50230827 MK-9
CHEMBL253364 BDBM50230827 MK-9 CHEMBL489586 BDBM50479470 MK-4965
CHEMBL489586 BDBM50479470 MK-4965 MK 0893 BDBM50168081 CHEMBL3799802
MK 0893 BDBM50168081 CHEMBL3799802 MK-0617 CHEMBL566154 BDBM50423695
MK-0617 CHEMBL566154 BDBM50423695 MK-1220 CHEMBL1672609 BDBM50485489
MK-1220 CHEMBL1672609 BDBM50485489 MK-50 CHEMBL2113644 BDBM50451496
MK-50 CHEMBL2113644 BDBM50451496 MK-55 BDBM50474237 CHEMBL313379
MK-55 BDBM50474237 CHEMBL313379 MK-7445 BDBM50484630 CHEMBL1939502
MK-7445 BDBM50484630 CHEMBL1939502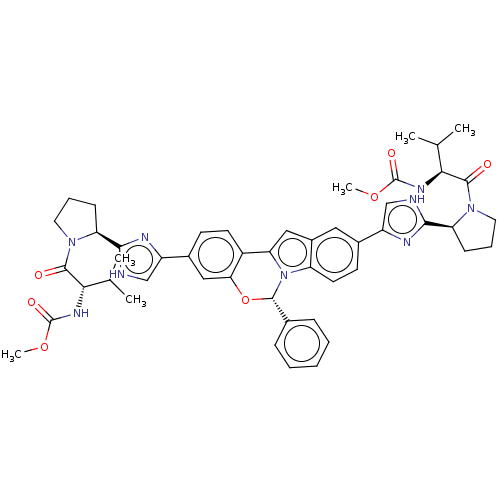 MK-8742 BDBM50531952 Elbasvir
MK-8742 BDBM50531952 Elbasvir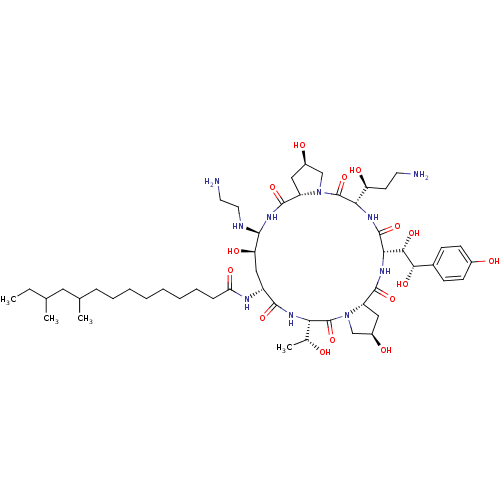 MK-991 BDBM50366679 CHEMBL1793852
MK-991 BDBM50366679 CHEMBL1793852 Mk-2748 BDBM50642717 MK2748
Mk-2748 BDBM50642717 MK2748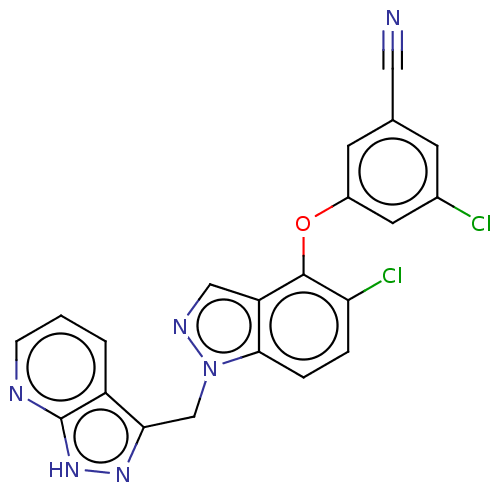 Mk-6186 Mk6186 BDBM50484632
Mk-6186 Mk6186 BDBM50484632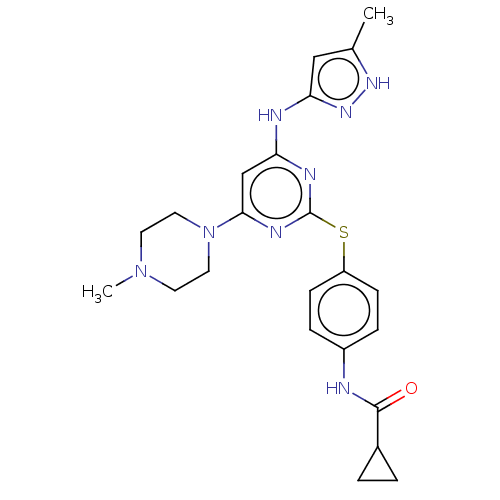 BDBM50004205 TOZASERTIB MK-045 MK-0457 US9249124, VX680 VX-68 VX-680
BDBM50004205 TOZASERTIB MK-045 MK-0457 US9249124, VX680 VX-68 VX-680 BDBM565698 US11407768, Compound MK-8722
BDBM565698 US11407768, Compound MK-8722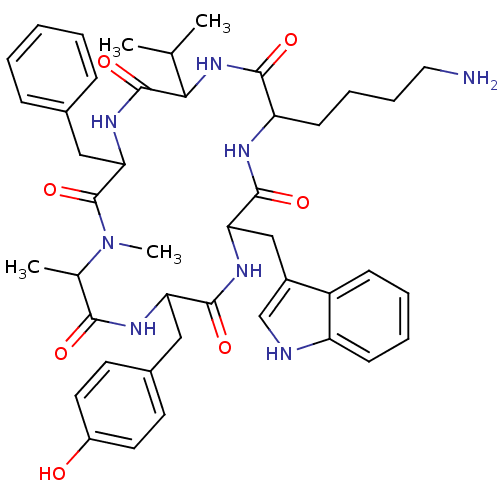 BDBM81766 MK 678 NSC_3086456 CAS_3086456
BDBM81766 MK 678 NSC_3086456 CAS_3086456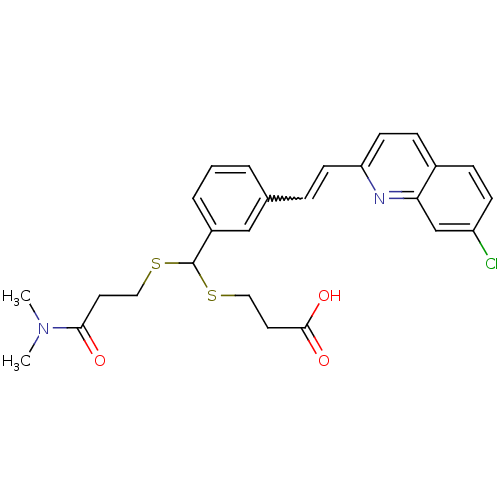 BDBM86671 MK 571 NSC_60719 CAS_60719
BDBM86671 MK 571 NSC_60719 CAS_60719 CHEMBL481611 BDBM50255753 MK-0822 Odanacatib
CHEMBL481611 BDBM50255753 MK-0822 Odanacatib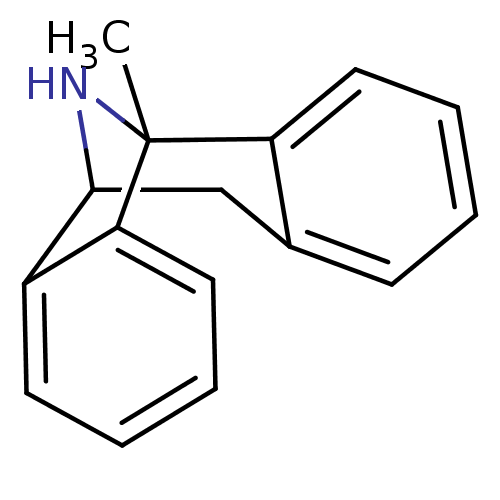 MK-801 BDBM86153 CAS_180081 NSC_180081
MK-801 BDBM86153 CAS_180081 NSC_180081 Pifeltro Doravirine MK-1439 BDBM50508293
Pifeltro Doravirine MK-1439 BDBM50508293 BDBM50213266 CHEBI:471744 MK-0787 Imipenem
BDBM50213266 CHEBI:471744 MK-0787 Imipenem Birabresib OTX-015 MK-8628 BDBM50092312
Birabresib OTX-015 MK-8628 BDBM50092312 MK-4618 BDBM50146154 KRP-114V Vibegron
MK-4618 BDBM50146154 KRP-114V Vibegron MK-0518 Isentress Raltegravir Potassium MK0518 POTASSIUM Raltegravir monopotassium salt BDBM50480673 Isentress hd MK-0518 POTASSIUM
MK-0518 Isentress Raltegravir Potassium MK0518 POTASSIUM Raltegravir monopotassium salt BDBM50480673 Isentress hd MK-0518 POTASSIUM CAS_123679 MK-912 L-657,743 NSC_123679 BDBM81811
CAS_123679 MK-912 L-657,743 NSC_123679 BDBM81811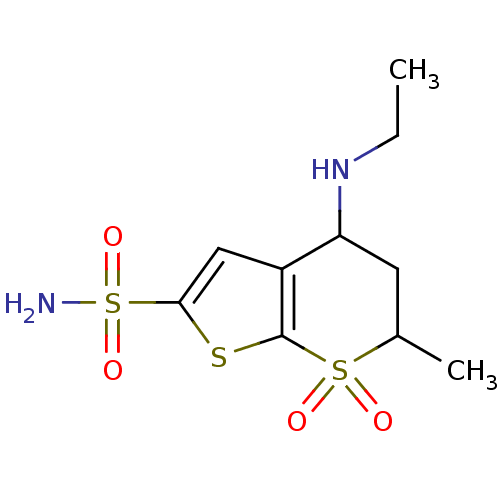 US10172837, Dorzolamide MK-507 Dorzolamide CHEMBL269001 BDBM50043906
US10172837, Dorzolamide MK-507 Dorzolamide CHEMBL269001 BDBM50043906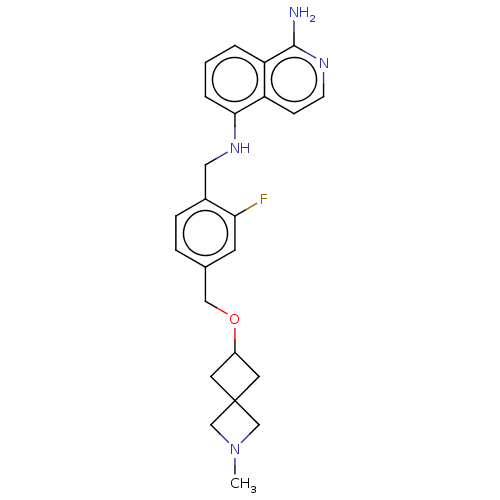 US20240059691, Example 2206 BDBM652626 5-N-[[2-fluoro-4-[(2-methyl-2-azaspiro[3.3]heptan-6- yl)oxymethyl]phenyl]methyl]isoquinoline-1,5-diamine
US20240059691, Example 2206 BDBM652626 5-N-[[2-fluoro-4-[(2-methyl-2-azaspiro[3.3]heptan-6- yl)oxymethyl]phenyl]methyl]isoquinoline-1,5-diamine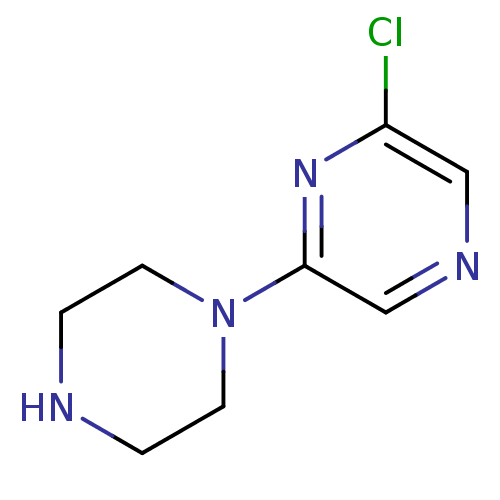 6'-Chloro-3,4,5,6-tetrahydro-2H-[1,2']bipyrazinyl L-630,571 MK 212 2-chloro-6-piperazin-1-ylpyrazine CHEMBL269521 6'-Chloro-3,4,5,6-tetrahydro-2H-[1,2']bipyrazinyl(MK-212) BDBM50017452 MK-212
6'-Chloro-3,4,5,6-tetrahydro-2H-[1,2']bipyrazinyl L-630,571 MK 212 2-chloro-6-piperazin-1-ylpyrazine CHEMBL269521 6'-Chloro-3,4,5,6-tetrahydro-2H-[1,2']bipyrazinyl(MK-212) BDBM50017452 MK-212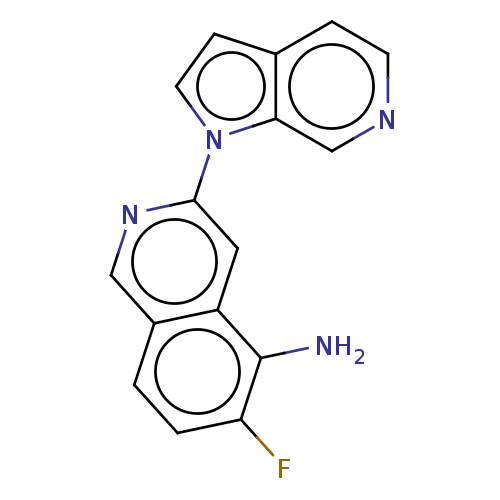 BDBM281567 US20240165276, Compound MK-6420 US10022461, Compound 18
BDBM281567 US20240165276, Compound MK-6420 US10022461, Compound 18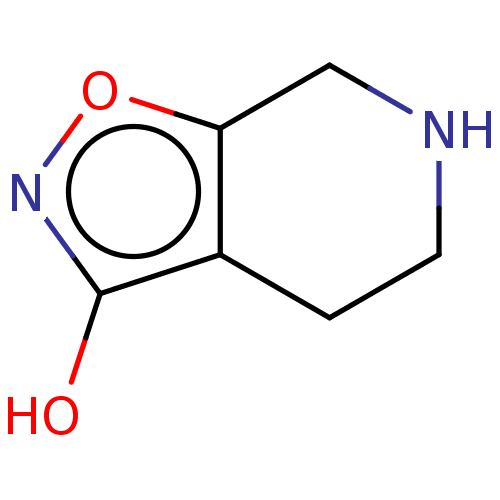 BDBM50224809 CHEBI:34373 MK-0928 Gaboxadol LU-02030
BDBM50224809 CHEBI:34373 MK-0928 Gaboxadol LU-02030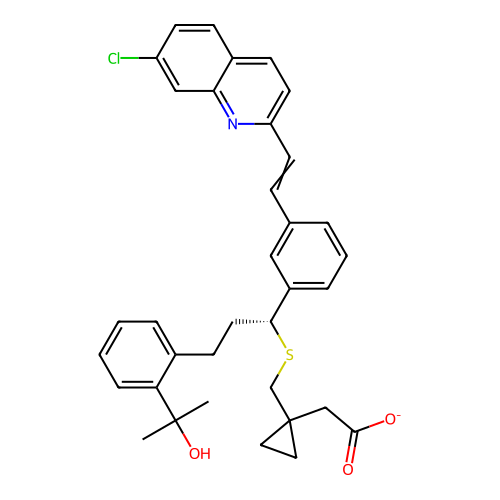 CHEBI:6993 BDBM50239015 Montelukast Sodium Singulair MK-476
CHEBI:6993 BDBM50239015 Montelukast Sodium Singulair MK-476 MK-0826 BDBM50248009 Invanz CHEBI:60070 Ertapenem sodium
MK-0826 BDBM50248009 Invanz CHEBI:60070 Ertapenem sodium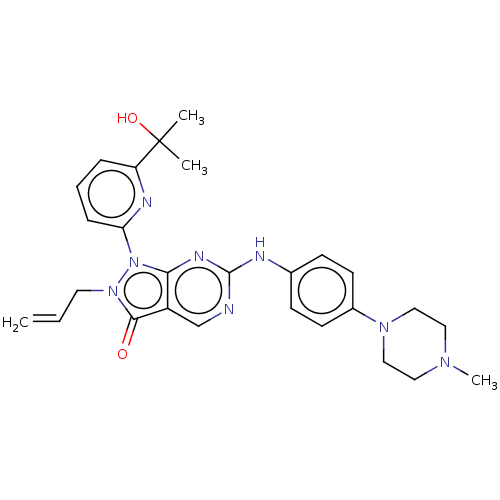 US12220415, Compound AZD1775 US20240182482, Compound MK-1775 US11248006, Compound AZD1775 BDBM50240826 US11479555, Example AZD1775 AZD-1775 US11124518, Example AZD1775 MK-1775
US12220415, Compound AZD1775 US20240182482, Compound MK-1775 US11248006, Compound AZD1775 BDBM50240826 US11479555, Example AZD1775 AZD-1775 US11124518, Example AZD1775 MK-1775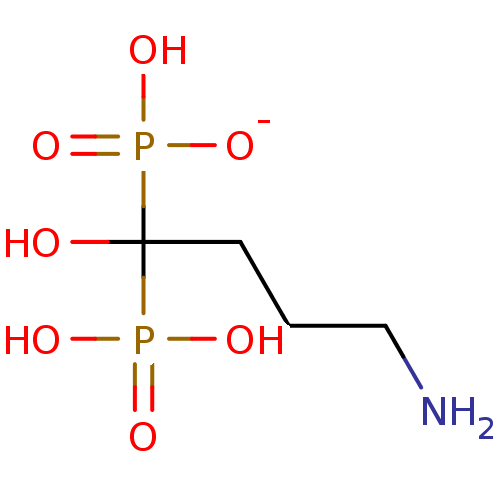 MK-217 Alendronate Sodium BDBM50247920 Binosto Fosamax G-704650
MK-217 Alendronate Sodium BDBM50247920 Binosto Fosamax G-704650 MK-458 NAXAGOLIDE HYDROCHLORIDE BDBM50446931 L-647339 Naxagolide HCl
MK-458 NAXAGOLIDE HYDROCHLORIDE BDBM50446931 L-647339 Naxagolide HCl MK-8931 SCH 900931 BDBM50580216 SCH-900931 SCH900931 VERUBECESTAT
MK-8931 SCH 900931 BDBM50580216 SCH-900931 SCH900931 VERUBECESTAT MK-6721 NMED-160 Z-160 Z160 BDBM50461289 NP-118809
MK-6721 NMED-160 Z-160 Z160 BDBM50461289 NP-118809 MK-8228 Letermovir Prevymis AIC246 AIC 246 AIC-246 BDBM50614677
MK-8228 Letermovir Prevymis AIC246 AIC 246 AIC-246 BDBM50614677 BDBM50067593 Crixivan MK-639 CHEBI:44032 L-735524 Indinavir US10806794, Compound Indinavir
BDBM50067593 Crixivan MK-639 CHEBI:44032 L-735524 Indinavir US10806794, Compound Indinavir BDBM50111186 MK-27 CHEMBL9075 (S)-2-Benzylamino-5-guanidino-pentanoic acid amide
BDBM50111186 MK-27 CHEMBL9075 (S)-2-Benzylamino-5-guanidino-pentanoic acid amide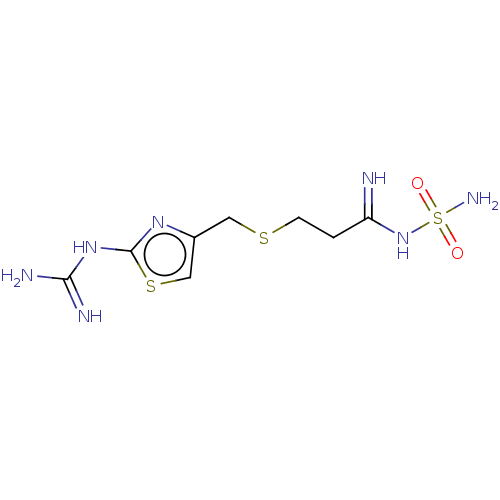 MK-208 Famotidine Pepcid Pepcid Rpd Pepcid Ac Fluxid CHEBI:4975 BDBM50103514
MK-208 Famotidine Pepcid Pepcid Rpd Pepcid Ac Fluxid CHEBI:4975 BDBM50103514 CHEBI:474180 L-743,872 Caspofungin Acetate Cancidas Caspofungin MK-991 BDBM50478215 L-743872
CHEBI:474180 L-743,872 Caspofungin Acetate Cancidas Caspofungin MK-991 BDBM50478215 L-743872 MK-42 BDBM50111176 (S)-4-Allyloxycarbonyl-4-[(naphthalen-2-ylmethyl)-amino]-butyl-ammonium
MK-42 BDBM50111176 (S)-4-Allyloxycarbonyl-4-[(naphthalen-2-ylmethyl)-amino]-butyl-ammonium BDBM50092127 CHEMBL331559 MK-852 8-Acetylamino-22-(4-aminomethyl-benzyl)-5-carbamoylmethyl-16-carboxymethyl-1,1-dimethyl-4,7,15,18,21,24-hexaoxo-icosahydro-2,10,11-trithia-3a,6,14,17,20,23-hexaaza-cyclopentacyclotricosene-13-carboxylic acid (MK-0852)
BDBM50092127 CHEMBL331559 MK-852 8-Acetylamino-22-(4-aminomethyl-benzyl)-5-carbamoylmethyl-16-carboxymethyl-1,1-dimethyl-4,7,15,18,21,24-hexaoxo-icosahydro-2,10,11-trithia-3a,6,14,17,20,23-hexaaza-cyclopentacyclotricosene-13-carboxylic acid (MK-0852) CHEMBL9257 BDBM50111191 (S)-2-(3,4-Dimethoxy-benzylamino)-5-guanidino-pentanoic acid amide MK-25
CHEMBL9257 BDBM50111191 (S)-2-(3,4-Dimethoxy-benzylamino)-5-guanidino-pentanoic acid amide MK-25 CHEMBL274352 BDBM50111187 MK-18 (S)-5-Guanidino-2-[(naphthalen-2-ylmethyl)-amino]-pentanoic acid amide
CHEMBL274352 BDBM50111187 MK-18 (S)-5-Guanidino-2-[(naphthalen-2-ylmethyl)-amino]-pentanoic acid amide MK-20 BDBM50111196 (R)-5-Guanidino-2-[(naphthalen-2-ylmethyl)-amino]-pentanoic acid amide CHEMBL266055
MK-20 BDBM50111196 (R)-5-Guanidino-2-[(naphthalen-2-ylmethyl)-amino]-pentanoic acid amide CHEMBL266055 MK-3102 US10155775, Omarigliptin OMARIGLIPTIN US10822319, Compound Omarigliptin US10479798, Compound MK3102 BDBM50003020 US20240115577, Compound 9
MK-3102 US10155775, Omarigliptin OMARIGLIPTIN US10822319, Compound Omarigliptin US10479798, Compound MK3102 BDBM50003020 US20240115577, Compound 9 MK-43 (S)-5-Guanidino-2-[(naphthalen-2-ylmethyl)-amino]-pentanoic acid allyl ester BDBM50111195
MK-43 (S)-5-Guanidino-2-[(naphthalen-2-ylmethyl)-amino]-pentanoic acid allyl ester BDBM50111195 US20250101023, Example Dinaciclib Dinaciclib US20230416221, Compound Dinaciclib SCH-727965 US11643396, Example Dinaciclib BDBM50139171 MK-7965
US20250101023, Example Dinaciclib Dinaciclib US20230416221, Compound Dinaciclib SCH-727965 US11643396, Example Dinaciclib BDBM50139171 MK-7965 (R)-2-[(1H-Indol-3-ylmethyl)-amino]-3-naphthalen-2-yl-propionamide BDBM50111188 CHEMBL9219 MK-15
(R)-2-[(1H-Indol-3-ylmethyl)-amino]-3-naphthalen-2-yl-propionamide BDBM50111188 CHEMBL9219 MK-15 (S)-2-(2-Chloro-3,4-dimethoxy-benzylamino)-5-guanidino-pentanoic acid amide BDBM50111204 MK-26 CHEMBL8929
(S)-2-(2-Chloro-3,4-dimethoxy-benzylamino)-5-guanidino-pentanoic acid amide BDBM50111204 MK-26 CHEMBL8929 (S)-2-[(1H-Indol-3-ylmethyl)-amino]-3-naphthalen-2-yl-propionamide CHEMBL428480 BDBM50111197 MK-16
(S)-2-[(1H-Indol-3-ylmethyl)-amino]-3-naphthalen-2-yl-propionamide CHEMBL428480 BDBM50111197 MK-16 BDBM50111193 CHEMBL9138 (R)-3-(1H-Indol-3-yl)-2-[(naphthalen-2-ylmethyl)-amino]-propionamide MK-14
BDBM50111193 CHEMBL9138 (R)-3-(1H-Indol-3-yl)-2-[(naphthalen-2-ylmethyl)-amino]-propionamide MK-14 CHEMBL273475 5-(S)-Guanidino-2-[(1H-indol-3-ylmethyl)-amino]-pentanoic acid amide BDBM50111198 MK-17
CHEMBL273475 5-(S)-Guanidino-2-[(1H-indol-3-ylmethyl)-amino]-pentanoic acid amide BDBM50111198 MK-17 CHEMBL8730 5-(R)-Guanidino-2-[(1H-indol-3-ylmethyl)-amino]-pentanoic acid amide BDBM50111200 MK-19
CHEMBL8730 5-(R)-Guanidino-2-[(1H-indol-3-ylmethyl)-amino]-pentanoic acid amide BDBM50111200 MK-19 MK-11 (S)-3-(1H-Indol-3-yl)-2-[(naphthalen-2-ylmethyl)-amino]-propionamide BDBM50111202 CHEMBL9274
MK-11 (S)-3-(1H-Indol-3-yl)-2-[(naphthalen-2-ylmethyl)-amino]-propionamide BDBM50111202 CHEMBL9274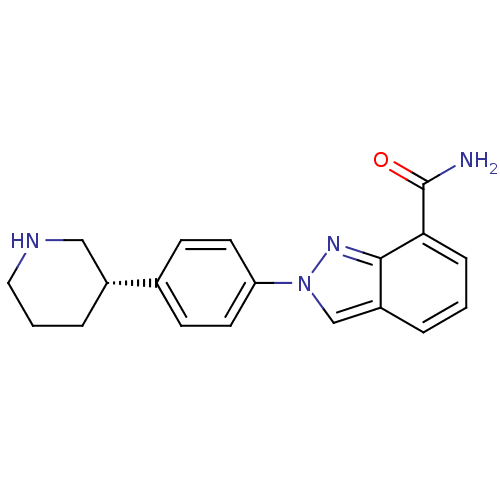 MK-4827 CHEMBL1094636 (S)-2-(4-(piperidin-3-yl)phenyl)-2H-indazole-7-carboxamide Niraparib BDBM50316226
MK-4827 CHEMBL1094636 (S)-2-(4-(piperidin-3-yl)phenyl)-2H-indazole-7-carboxamide Niraparib BDBM50316226 PL 100 (PHARMACEUTICAL) MK-8122 PL-100 Ppl-100 PPL 100 Tmb-607 BDBM50482335 MX-100
PL 100 (PHARMACEUTICAL) MK-8122 PL-100 Ppl-100 PPL 100 Tmb-607 BDBM50482335 MX-100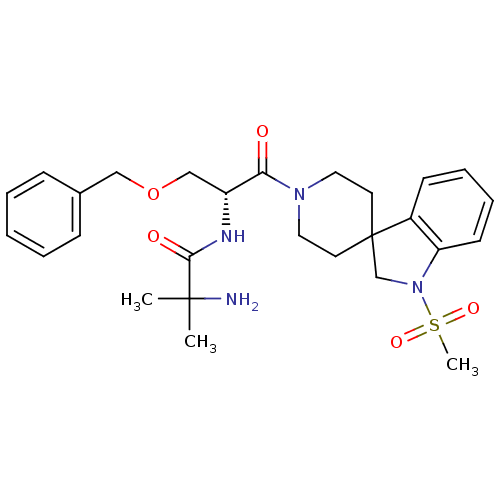 1N-[1-benzyloxymethyl-2-[1-methylsulfonylspiro[2,3-dihydro-1H-indole-3,4'-(hexahydropyridine)]-1-yl]-2-oxo-(1R)-ethyl]-2-amino-2-methylpropanamide [3a(MK-0677)] CHEMBL13817 1-{[(2R)-3-(benzyloxy)-1-{1-methanesulfonyl-1,2-dihydrospiro[indole-3,4'-piperidine]-1'-yl}-1-oxopropan-2-yl]carbamoyl}-1-methylethan-1-aminium 1N-[1-benzyloxymethyl-2-[1-methylsulfonylspiro[2,3-dihydro-1H-indole-3,4'-(hexahydropyridine)]-1-yl]-2-oxo-(1R)-ethyl]-2-amino-2-methylpropanamide BDBM50049478 MK-0677 1N-[1-benzyloxymethyl-2-[1-methylsulfonylspiro[2,3-dihydro-1H-indole-3,4'-(hexahydropyridine)]-1-yl]-2-oxo-(1R)-ethyl]-2-amino-2-methylpropanamide(MK-0677) MK 0677 MK-677
1N-[1-benzyloxymethyl-2-[1-methylsulfonylspiro[2,3-dihydro-1H-indole-3,4'-(hexahydropyridine)]-1-yl]-2-oxo-(1R)-ethyl]-2-amino-2-methylpropanamide [3a(MK-0677)] CHEMBL13817 1-{[(2R)-3-(benzyloxy)-1-{1-methanesulfonyl-1,2-dihydrospiro[indole-3,4'-piperidine]-1'-yl}-1-oxopropan-2-yl]carbamoyl}-1-methylethan-1-aminium 1N-[1-benzyloxymethyl-2-[1-methylsulfonylspiro[2,3-dihydro-1H-indole-3,4'-(hexahydropyridine)]-1-yl]-2-oxo-(1R)-ethyl]-2-amino-2-methylpropanamide BDBM50049478 MK-0677 1N-[1-benzyloxymethyl-2-[1-methylsulfonylspiro[2,3-dihydro-1H-indole-3,4'-(hexahydropyridine)]-1-yl]-2-oxo-(1R)-ethyl]-2-amino-2-methylpropanamide(MK-0677) MK 0677 MK-677 1-methyl-16-azatetracyclo[7.6.1.02,7.010,15]hexadeca-2(7),3,5,10(15),11,13-hexaene(MK-801) (+)-1-methyl-16-azatetracyclo[7.6.1.02,7.010,15]hexadeca-2(7),3,5,10(15),11,13-hexaene (+/-) MK-8011-methyl-(9R,1R)-16-azatetracyclo[7.6.1.02,7.010,15]hexadeca-2(7),3,5,10(15),11,13-hexaene (5S,10R)-(+)-5-methyl-10,11-dihydro-5Hdibenzo[a,d]cyclohepten-5,10-imine MK-801 1-methyl-16-azatetracyclo[7.6.1.02,7.010,15]hexadeca-2(7),3,5,10(15),11,13-hexaene (+/-)-1-methyl-16-azatetracyclo[7.6.1.02,7.010,15]hexadeca-2(7),3,5,10(15),11,13-hexaene (1S,9R)-1-methyl-16-azatetracyclo[7.6.1.0^{2,7}.0^{10,15}]hexadeca-2(7),3,5,10(15),11,13-hexaene BDBM50000663 MK-801 (Dizocilpine) (-)-MK801 US11944616, Compound Dizocilpine (Dizocilpine)1-methyl-16-azatetracyclo[7.6.1.02,7.010,15]hexadeca-2(7),3,5,10(15),11,13-hexaene (1S,9R)-1-methyl-16-azatetracyclo[7.6.1.0^{2,7}.0^{10,15}]hexadeca-2,4,6,10(15),11,13-hexaene MK-801,(+) (5S,10S)-(+)-5-methyl-10,11-dihydro-5Hdibenzo[a,d]cyclohepten-5,10-imine 1-methyl-16-azatetracyclo[7.6.1.02,7.010,15]hexadeca-2(7),3,5,10,12,14-hexaene CHEMBL284237 MK-801,(-) 1-methyl-16-azatetracyclo[7.6.1.02,7.010,15]hexadeca-2,4,6,10(15),11,13-hexaene (-)-1-methyl-16-azatetracyclo[7.6.1.02,7.010,15]hexadeca-2(7),3,5,10(15),11,13-hexaene (MK-801)1-methyl-16-azatetracyclo[7.6.1.02,7.010,15]hexadeca-2(7),3,5,10(15),11,13-hexaene dizocilpine (+)-MK-801 (+)MK-801 (+/-)-MK801 10,11-Dihydro-5-methyl-5H-dibenzo[a,d]cyclohepten-5,10-imine.(MK-801)
1-methyl-16-azatetracyclo[7.6.1.02,7.010,15]hexadeca-2(7),3,5,10(15),11,13-hexaene(MK-801) (+)-1-methyl-16-azatetracyclo[7.6.1.02,7.010,15]hexadeca-2(7),3,5,10(15),11,13-hexaene (+/-) MK-8011-methyl-(9R,1R)-16-azatetracyclo[7.6.1.02,7.010,15]hexadeca-2(7),3,5,10(15),11,13-hexaene (5S,10R)-(+)-5-methyl-10,11-dihydro-5Hdibenzo[a,d]cyclohepten-5,10-imine MK-801 1-methyl-16-azatetracyclo[7.6.1.02,7.010,15]hexadeca-2(7),3,5,10(15),11,13-hexaene (+/-)-1-methyl-16-azatetracyclo[7.6.1.02,7.010,15]hexadeca-2(7),3,5,10(15),11,13-hexaene (1S,9R)-1-methyl-16-azatetracyclo[7.6.1.0^{2,7}.0^{10,15}]hexadeca-2(7),3,5,10(15),11,13-hexaene BDBM50000663 MK-801 (Dizocilpine) (-)-MK801 US11944616, Compound Dizocilpine (Dizocilpine)1-methyl-16-azatetracyclo[7.6.1.02,7.010,15]hexadeca-2(7),3,5,10(15),11,13-hexaene (1S,9R)-1-methyl-16-azatetracyclo[7.6.1.0^{2,7}.0^{10,15}]hexadeca-2,4,6,10(15),11,13-hexaene MK-801,(+) (5S,10S)-(+)-5-methyl-10,11-dihydro-5Hdibenzo[a,d]cyclohepten-5,10-imine 1-methyl-16-azatetracyclo[7.6.1.02,7.010,15]hexadeca-2(7),3,5,10,12,14-hexaene CHEMBL284237 MK-801,(-) 1-methyl-16-azatetracyclo[7.6.1.02,7.010,15]hexadeca-2,4,6,10(15),11,13-hexaene (-)-1-methyl-16-azatetracyclo[7.6.1.02,7.010,15]hexadeca-2(7),3,5,10(15),11,13-hexaene (MK-801)1-methyl-16-azatetracyclo[7.6.1.02,7.010,15]hexadeca-2(7),3,5,10(15),11,13-hexaene dizocilpine (+)-MK-801 (+)MK-801 (+/-)-MK801 10,11-Dihydro-5-methyl-5H-dibenzo[a,d]cyclohepten-5,10-imine.(MK-801) (S)-5-Guanidino-2-[(1,2,3,4-tetrahydro-naphthalen-2-ylmethyl)-amino]-pentanoic acid amide BDBM50111185 CHEMBL8676 MK-28
(S)-5-Guanidino-2-[(1,2,3,4-tetrahydro-naphthalen-2-ylmethyl)-amino]-pentanoic acid amide BDBM50111185 CHEMBL8676 MK-28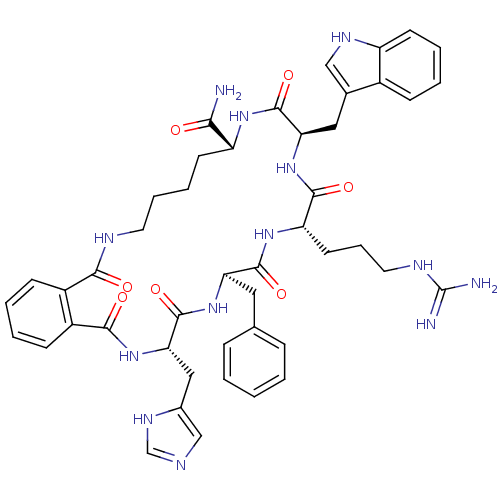 BDBM50114085 CHEMBL314401 (O)C-(C6H4)-C(O)-c[His-D-Phe-Arg-Trp-Lys]-NH2) (MK-6)
BDBM50114085 CHEMBL314401 (O)C-(C6H4)-C(O)-c[His-D-Phe-Arg-Trp-Lys]-NH2) (MK-6) BDBM50114084 CHEMBL439188 (O)C-(C6H4)-C(O)-c[His-D-Nal(2')-Arg-Trp-Lys]-NH2) (MK-7)
BDBM50114084 CHEMBL439188 (O)C-(C6H4)-C(O)-c[His-D-Nal(2')-Arg-Trp-Lys]-NH2) (MK-7)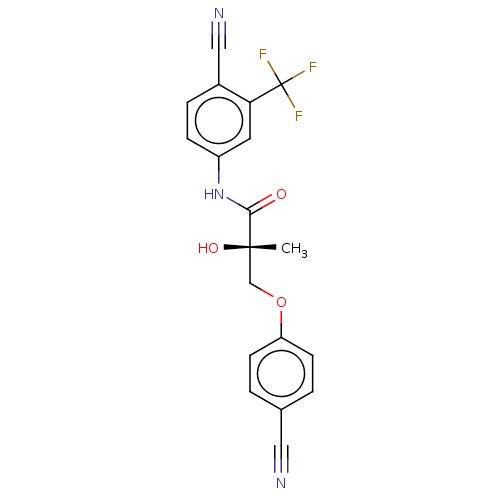 US10806720, Compound Enobosarm-(agonist) BDBM50529668 US11648234, Compound Enobosarm-(agonist) Ostarine MK-2866 Gtx-024 US11230523, Compound Enobosarm-(agonist) Enobosarm
US10806720, Compound Enobosarm-(agonist) BDBM50529668 US11648234, Compound Enobosarm-(agonist) Ostarine MK-2866 Gtx-024 US11230523, Compound Enobosarm-(agonist) Enobosarm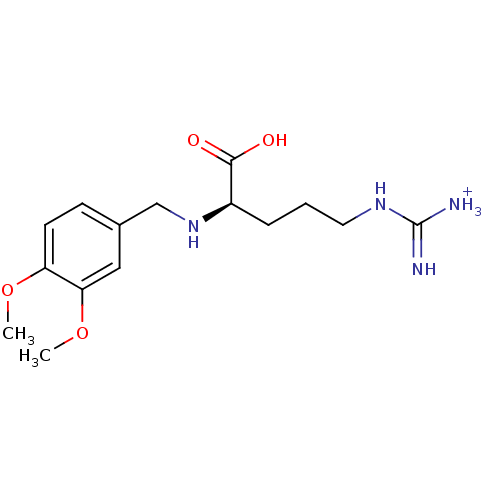 BDBM50111181 MK-33 N-Alkylaminoacid derivative (2R)-5-[(E)-[amino(azaniumyl)methylidene]amino]-2-{[(3,4-dimethoxyphenyl)methyl]amino}pentanoic acid
BDBM50111181 MK-33 N-Alkylaminoacid derivative (2R)-5-[(E)-[amino(azaniumyl)methylidene]amino]-2-{[(3,4-dimethoxyphenyl)methyl]amino}pentanoic acid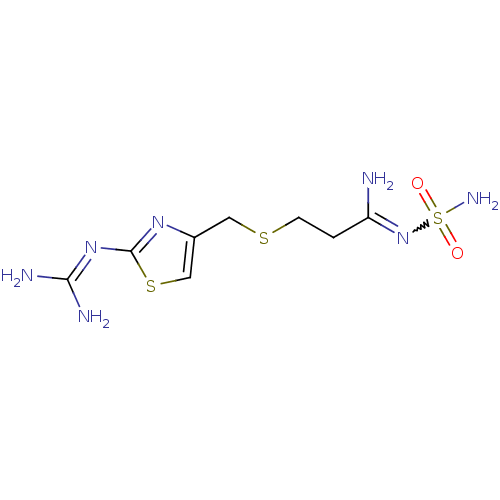 Famotidine YM 11170 Pepcid cid_5702160 3-[({2-[(diaminomethylidene)amino]-1,3-thiazol-4-yl}methyl)sulfanyl]-N'-sulfamoylpropanimidamide MK 208 BDBM22891
Famotidine YM 11170 Pepcid cid_5702160 3-[({2-[(diaminomethylidene)amino]-1,3-thiazol-4-yl}methyl)sulfanyl]-N'-sulfamoylpropanimidamide MK 208 BDBM22891 MK-31 BDBM50111182 N-Alkylaminoacid derivative (2R)-5-[(E)-[amino(azaniumyl)methylidene]amino]-2-[(naphthalen-2-ylmethyl)amino]pentanoic acid
MK-31 BDBM50111182 N-Alkylaminoacid derivative (2R)-5-[(E)-[amino(azaniumyl)methylidene]amino]-2-[(naphthalen-2-ylmethyl)amino]pentanoic acid MK-55 5-Guanidino-2-[(naphthalen-2-ylmethyl)-amino]-pentanoic acid (3-guanidino-propyl)-amide N-Alkylaminoacid derivative CHEMBL313379 BDBM50111206
MK-55 5-Guanidino-2-[(naphthalen-2-ylmethyl)-amino]-pentanoic acid (3-guanidino-propyl)-amide N-Alkylaminoacid derivative CHEMBL313379 BDBM50111206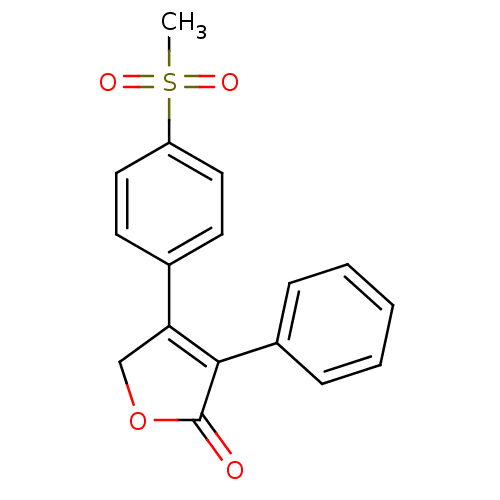 US11786535, Compound Rofecoxib Rofecoxib BDBM22369 US11478464, Compound Rofecoxib 4-(4-methanesulfonylphenyl)-3-phenyl-2,5-dihydrofuran-2-one CHEMBL122 MK 0966
US11786535, Compound Rofecoxib Rofecoxib BDBM22369 US11478464, Compound Rofecoxib 4-(4-methanesulfonylphenyl)-3-phenyl-2,5-dihydrofuran-2-one CHEMBL122 MK 0966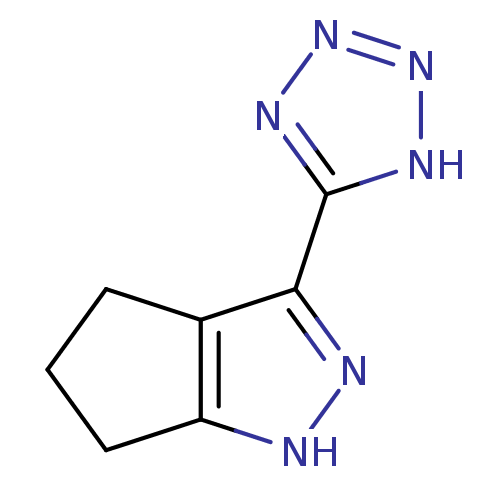 BDBM50273099 3-(1H-tetrazol-5-yl)-1,4,5,6-tetrahydrocyclopenta[c]pyrazole 3-(2H-tetrazol-5-yl)-1,4,5,6-tetrahydro-cyclopentapyrazole CHEMBL456145 MK-0354
BDBM50273099 3-(1H-tetrazol-5-yl)-1,4,5,6-tetrahydrocyclopenta[c]pyrazole 3-(2H-tetrazol-5-yl)-1,4,5,6-tetrahydro-cyclopentapyrazole CHEMBL456145 MK-0354 MK-32 N-Alkylaminoacid derivative (2R)-5-[(E)-[amino(azaniumyl)methylidene]amino]-2-[(1H-indol-3-ylmethyl)amino]pentanoic acid BDBM50111205
MK-32 N-Alkylaminoacid derivative (2R)-5-[(E)-[amino(azaniumyl)methylidene]amino]-2-[(1H-indol-3-ylmethyl)amino]pentanoic acid BDBM50111205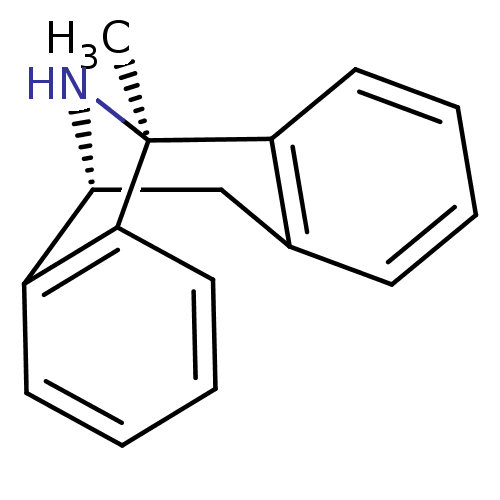 (1S,9R)-1-methyl-16-azatetracyclo[7.6.1.0^{2,7}.0^{10,15}]hexadeca-2,4,6,10(15),11,13-hexaene (5S,10S)-(+)-5-methyl-10,11-dihydro-5Hdibenzo[a,d]cyclohepten-5,10-imine 1-methyl-16-azatetracyclo[7.6.1.02,7.010,15]hexadeca-2(7),3,5,10(15),11,13-hexaene(MK-801) (+)-1-methyl-16-azatetracyclo[7.6.1.02,7.010,15]hexadeca-2(7),3,5,10(15),11,13-hexaene 1-methyl-16-azatetracyclo[7.6.1.02,7.010,15]hexadeca-2(7),3,5,10,12,14-hexaene CHEMBL284237 1-methyl-16-azatetracyclo[7.6.1.02,7.010,15]hexadeca-2,4,6,10(15),11,13-hexaene (-)-1-methyl-16-azatetracyclo[7.6.1.02,7.010,15]hexadeca-2(7),3,5,10(15),11,13-hexaene (MK-801)1-methyl-16-azatetracyclo[7.6.1.02,7.010,15]hexadeca-2(7),3,5,10(15),11,13-hexaene dizocilpine (+)-MK-801 (+)MK-801 (+/-) MK-8011-methyl-(9R,1R)-16-azatetracyclo[7.6.1.02,7.010,15]hexadeca-2(7),3,5,10(15),11,13-hexaene (5S,10R)-(+)-5-methyl-10,11-dihydro-5Hdibenzo[a,d]cyclohepten-5,10-imine MK-801 1-methyl-16-azatetracyclo[7.6.1.02,7.010,15]hexadeca-2(7),3,5,10(15),11,13-hexaene BDBM50344263 (+/-)-1-methyl-16-azatetracyclo[7.6.1.02,7.010,15]hexadeca-2(7),3,5,10(15),11,13-hexaene (+/-)-MK801 (1S,9R)-1-methyl-16-azatetracyclo[7.6.1.0^{2,7}.0^{10,15}]hexadeca-2(7),3,5,10(15),11,13-hexaene MK-801 (Dizocilpine) (-)-MK801 (Dizocilpine)1-methyl-16-azatetracyclo[7.6.1.02,7.010,15]hexadeca-2(7),3,5,10(15),11,13-hexaene 10,11-Dihydro-5-methyl-5H-dibenzo[a,d]cyclohepten-5,10-imine.(MK-801)
(1S,9R)-1-methyl-16-azatetracyclo[7.6.1.0^{2,7}.0^{10,15}]hexadeca-2,4,6,10(15),11,13-hexaene (5S,10S)-(+)-5-methyl-10,11-dihydro-5Hdibenzo[a,d]cyclohepten-5,10-imine 1-methyl-16-azatetracyclo[7.6.1.02,7.010,15]hexadeca-2(7),3,5,10(15),11,13-hexaene(MK-801) (+)-1-methyl-16-azatetracyclo[7.6.1.02,7.010,15]hexadeca-2(7),3,5,10(15),11,13-hexaene 1-methyl-16-azatetracyclo[7.6.1.02,7.010,15]hexadeca-2(7),3,5,10,12,14-hexaene CHEMBL284237 1-methyl-16-azatetracyclo[7.6.1.02,7.010,15]hexadeca-2,4,6,10(15),11,13-hexaene (-)-1-methyl-16-azatetracyclo[7.6.1.02,7.010,15]hexadeca-2(7),3,5,10(15),11,13-hexaene (MK-801)1-methyl-16-azatetracyclo[7.6.1.02,7.010,15]hexadeca-2(7),3,5,10(15),11,13-hexaene dizocilpine (+)-MK-801 (+)MK-801 (+/-) MK-8011-methyl-(9R,1R)-16-azatetracyclo[7.6.1.02,7.010,15]hexadeca-2(7),3,5,10(15),11,13-hexaene (5S,10R)-(+)-5-methyl-10,11-dihydro-5Hdibenzo[a,d]cyclohepten-5,10-imine MK-801 1-methyl-16-azatetracyclo[7.6.1.02,7.010,15]hexadeca-2(7),3,5,10(15),11,13-hexaene BDBM50344263 (+/-)-1-methyl-16-azatetracyclo[7.6.1.02,7.010,15]hexadeca-2(7),3,5,10(15),11,13-hexaene (+/-)-MK801 (1S,9R)-1-methyl-16-azatetracyclo[7.6.1.0^{2,7}.0^{10,15}]hexadeca-2(7),3,5,10(15),11,13-hexaene MK-801 (Dizocilpine) (-)-MK801 (Dizocilpine)1-methyl-16-azatetracyclo[7.6.1.02,7.010,15]hexadeca-2(7),3,5,10(15),11,13-hexaene 10,11-Dihydro-5-methyl-5H-dibenzo[a,d]cyclohepten-5,10-imine.(MK-801)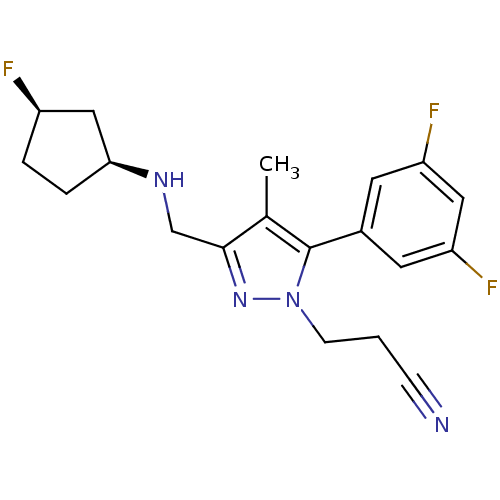 3-[5-(3,5-difluorophenyl)-3-({[(1S,3R)-3-fluorocyclopentyl]amino}-methyl)-4-methyl-1H-pyrazol-1-yl]propanenitrile CHEMBL560667 MK-1925 BDBM50296579
3-[5-(3,5-difluorophenyl)-3-({[(1S,3R)-3-fluorocyclopentyl]amino}-methyl)-4-methyl-1H-pyrazol-1-yl]propanenitrile CHEMBL560667 MK-1925 BDBM50296579 4'-(2-Ethyl-5,7-dimethyl-imidazo[4,5-b]pyridin-3-ylmethyl)-biphenyl-2-sulfonic acid benzoylamide CHEMBL293511 MK-996 L-158282 BDBM50038189
4'-(2-Ethyl-5,7-dimethyl-imidazo[4,5-b]pyridin-3-ylmethyl)-biphenyl-2-sulfonic acid benzoylamide CHEMBL293511 MK-996 L-158282 BDBM50038189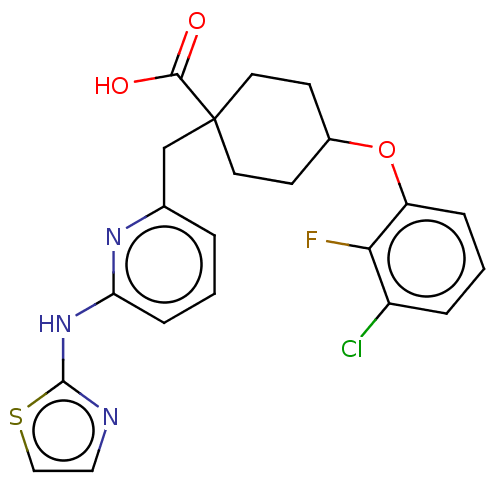 4-(3-chloro-2-fluorophenoxy)-1-[[6-(1,3-thiazol-2-ylamino)pyridin-2-yl]methyl]cyclohexane-1-carboxylic acid BDBM209862 MK-5108
4-(3-chloro-2-fluorophenoxy)-1-[[6-(1,3-thiazol-2-ylamino)pyridin-2-yl]methyl]cyclohexane-1-carboxylic acid BDBM209862 MK-5108 BDBM50111184 N-Alkylaminoacid derivative (2R)-5-[(E)-[amino(azaniumyl)methylidene]amino]-2-{[(2-chloro-3,4-dimethoxyphenyl)methyl]amino}pentanoic acid MK-34
BDBM50111184 N-Alkylaminoacid derivative (2R)-5-[(E)-[amino(azaniumyl)methylidene]amino]-2-{[(2-chloro-3,4-dimethoxyphenyl)methyl]amino}pentanoic acid MK-34 BDBM50111175 N-Alkylaminoacid derivative (2S)-5-[(E)-[amino(azaniumyl)methylidene]amino]-N-(naphthalen-2-yl)-2-[(naphthalen-2-ylmethyl)amino]pentanamide MK-56
BDBM50111175 N-Alkylaminoacid derivative (2S)-5-[(E)-[amino(azaniumyl)methylidene]amino]-N-(naphthalen-2-yl)-2-[(naphthalen-2-ylmethyl)amino]pentanamide MK-56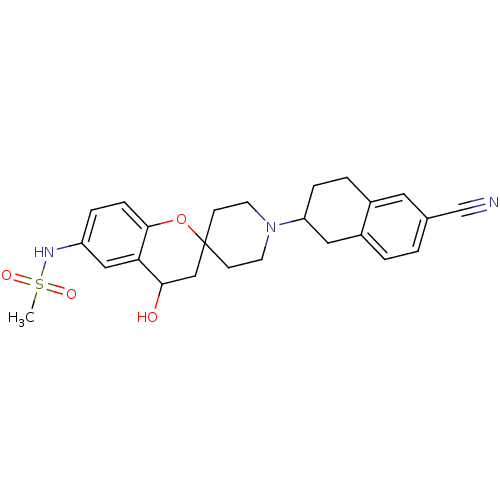 BDBM50128003 N-[1'-(6-cyano-1,2,3,4-tetrahydro-2-naphthalenyl)-4-hydroxyspiro[3,4-dihydro-2H-chromene-2,4'-(hexahydropyridine)]-6-yl]methanesulfonamide CHEMBL52627 MK-499
BDBM50128003 N-[1'-(6-cyano-1,2,3,4-tetrahydro-2-naphthalenyl)-4-hydroxyspiro[3,4-dihydro-2H-chromene-2,4'-(hexahydropyridine)]-6-yl]methanesulfonamide CHEMBL52627 MK-499 CHEMBL9048 (2S,4S)-4-Hydroxy-1-naphthalen-2-ylmethyl-pyrrolidine-2-carboxylic acid [2-(1H-imidazol-4-yl)-ethyl]-amide BDBM50111189 MK-69
CHEMBL9048 (2S,4S)-4-Hydroxy-1-naphthalen-2-ylmethyl-pyrrolidine-2-carboxylic acid [2-(1H-imidazol-4-yl)-ethyl]-amide BDBM50111189 MK-69 MK-54 BDBM50111183 N-Alkylaminoacid derivative (2S)-5-[(E)-[amino(azaniumyl)methylidene]amino]-N-(naphthalen-1-yl)-2-[(naphthalen-2-ylmethyl)amino]pentanamide
MK-54 BDBM50111183 N-Alkylaminoacid derivative (2S)-5-[(E)-[amino(azaniumyl)methylidene]amino]-N-(naphthalen-1-yl)-2-[(naphthalen-2-ylmethyl)amino]pentanamide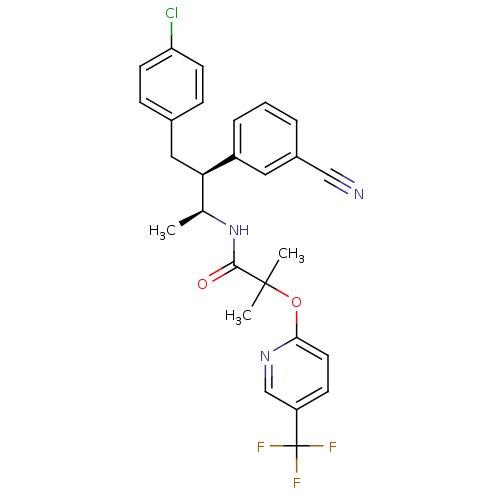 N-[(1S,2S)-3-(4-chlorophenyl)-2-(3-cyanophenyl)-1-methylpropyl]-2-methyl-2-{[5-(trifluoromethyl)pyridin-2-yl]oxy}propanamide CHEMBL220360 Taranbant N-((2S,3S)-4-(4-chlorophenyl)-3-(3-cyanophenyl)butan-2-yl)-2-methyl-2-(5-(trifluoromethyl)pyridin-2-yloxy)propanamide MK-0634 BDBM50200841 MK-0364 TARANABANT
N-[(1S,2S)-3-(4-chlorophenyl)-2-(3-cyanophenyl)-1-methylpropyl]-2-methyl-2-{[5-(trifluoromethyl)pyridin-2-yl]oxy}propanamide CHEMBL220360 Taranbant N-((2S,3S)-4-(4-chlorophenyl)-3-(3-cyanophenyl)butan-2-yl)-2-methyl-2-(5-(trifluoromethyl)pyridin-2-yloxy)propanamide MK-0634 BDBM50200841 MK-0364 TARANABANT (R)-5-Guanidino-2-[(naphthalen-2-ylmethyl)-amino]-pentanoic acid [(S)-1-carbamoyl-2-(1H-indol-3-yl)-ethyl]-amide CHEMBL447178 BDBM50111179 MK-30A
(R)-5-Guanidino-2-[(naphthalen-2-ylmethyl)-amino]-pentanoic acid [(S)-1-carbamoyl-2-(1H-indol-3-yl)-ethyl]-amide CHEMBL447178 BDBM50111179 MK-30A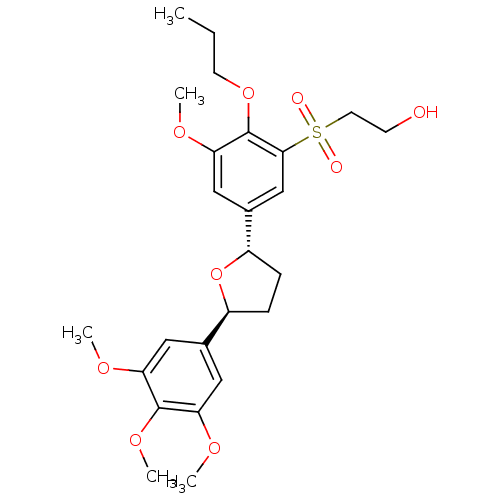 BDBM50002823 CHEMBL299944 L-680573 MK-287 2-{3-Methoxy-2-propoxy-5-[(2S,5S)-5-(3,4,5-trimethoxy-phenyl)-tetrahydro-furan-2-yl]-benzenesulfonyl}-ethanol
BDBM50002823 CHEMBL299944 L-680573 MK-287 2-{3-Methoxy-2-propoxy-5-[(2S,5S)-5-(3,4,5-trimethoxy-phenyl)-tetrahydro-furan-2-yl]-benzenesulfonyl}-ethanol BDBM50111192 MK-53 (2S)-5-[(E)-[amino(azaniumyl)methylidene]amino]-N-(1H-indol-3-ylmethyl)-2-[(naphthalen-2-ylmethyl)amino]pentanamide N-Alkylaminoacid derivative
BDBM50111192 MK-53 (2S)-5-[(E)-[amino(azaniumyl)methylidene]amino]-N-(1H-indol-3-ylmethyl)-2-[(naphthalen-2-ylmethyl)amino]pentanamide N-Alkylaminoacid derivative MK-29A BDBM50111180 CHEMBL8720 (S)-5-Guanidino-2-[(naphthalen-2-ylmethyl)-amino]-pentanoic acid [(S)-1-carbamoyl-2-(1H-indol-3-yl)-ethyl]-amide
MK-29A BDBM50111180 CHEMBL8720 (S)-5-Guanidino-2-[(naphthalen-2-ylmethyl)-amino]-pentanoic acid [(S)-1-carbamoyl-2-(1H-indol-3-yl)-ethyl]-amide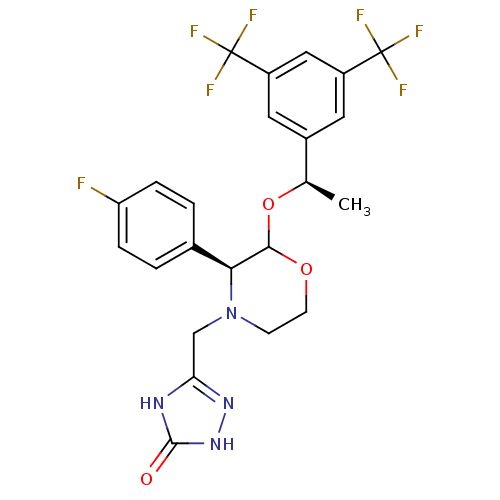 CHEMBL135613 MK-869 BDBM50106711 5-[2-[1-(3,5-Bis-trifluoromethyl-phenyl)-ethoxy]-3-(4-fluoro-phenyl)-morpholin-4-ylmethyl]-2,4-dihydro-[1,2,4]triazol-3-one
CHEMBL135613 MK-869 BDBM50106711 5-[2-[1-(3,5-Bis-trifluoromethyl-phenyl)-ethoxy]-3-(4-fluoro-phenyl)-morpholin-4-ylmethyl]-2,4-dihydro-[1,2,4]triazol-3-one CHEMBL267007 MK-29B (S)-5-Guanidino-2-[(1H-indol-3-ylmethyl)-amino]-pentanoic acid [(S)-1-carbamoyl-2-(1H-indol-3-yl)-ethyl]-amide BDBM50111190
CHEMBL267007 MK-29B (S)-5-Guanidino-2-[(1H-indol-3-ylmethyl)-amino]-pentanoic acid [(S)-1-carbamoyl-2-(1H-indol-3-yl)-ethyl]-amide BDBM50111190 MK-10 22-Benzyl-19-(3-guanidino-propyl)-16-(1H-indol-3-ylmethyl)-4,7,15,18,21,24-hexaoxo-docosahydro-3a,8,14,17,20,23-hexaaza-cyclopentacyclotricosene-13-carboxylic acid amide BDBM50114081
MK-10 22-Benzyl-19-(3-guanidino-propyl)-16-(1H-indol-3-ylmethyl)-4,7,15,18,21,24-hexaoxo-docosahydro-3a,8,14,17,20,23-hexaaza-cyclopentacyclotricosene-13-carboxylic acid amide BDBM50114081 MK-70 BDBM50111178 N-Alkylaminoacid derivative (2S,4S)-N-{3-[(E)-[amino(azaniumyl)methylidene]amino]propyl}-4-hydroxy-1-(naphthalen-2-ylmethyl)pyrrolidine-2-carboxamide
MK-70 BDBM50111178 N-Alkylaminoacid derivative (2S,4S)-N-{3-[(E)-[amino(azaniumyl)methylidene]amino]propyl}-4-hydroxy-1-(naphthalen-2-ylmethyl)pyrrolidine-2-carboxamide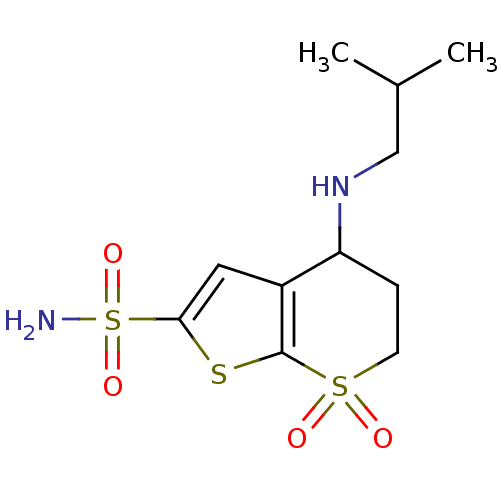 4-Isobutylamino-7,7-dioxo-4,5,6,7-tetrahydro-7lambda*6*-thieno[2,3-b]thiopyran-2-sulfonic acid amide MK-417 BDBM50017725 MK-927 CHEMBL545013 4-Isobutylamino-7,7-dioxo-4,5,6,7-tetrahydro-7lambda*6*-thieno[2,3-b]thiopyran-2-sulfonic acid amide; hydrochloride (S)-4-Isobutylamino-7,7-dioxo-4,5,6,7-tetrahydro-7lambda*6*-thieno[2,3-b]thiopyran-2-sulfonic acid amide
4-Isobutylamino-7,7-dioxo-4,5,6,7-tetrahydro-7lambda*6*-thieno[2,3-b]thiopyran-2-sulfonic acid amide MK-417 BDBM50017725 MK-927 CHEMBL545013 4-Isobutylamino-7,7-dioxo-4,5,6,7-tetrahydro-7lambda*6*-thieno[2,3-b]thiopyran-2-sulfonic acid amide; hydrochloride (S)-4-Isobutylamino-7,7-dioxo-4,5,6,7-tetrahydro-7lambda*6*-thieno[2,3-b]thiopyran-2-sulfonic acid amide 4-Isobutylamino-7,7-dioxo-4,5,6,7-tetrahydro-7lambda*6*-thieno[2,3-b]thiopyran-2-sulfonic acid amide MK-417 BDBM50041029 CHEMBL417975 MK-927 4-Isopropylamino-7,7-dioxo-4,5,6,7-tetrahydro-7lambda*6*-thieno[2,3-b]thiopyran-2-sulfonic acid amide CHEMBL1204135 (S)-4-Isobutylamino-7,7-dioxo-4,5,6,7-tetrahydro-7lambda*6*-thieno[2,3-b]thiopyran-2-sulfonic acid amide
4-Isobutylamino-7,7-dioxo-4,5,6,7-tetrahydro-7lambda*6*-thieno[2,3-b]thiopyran-2-sulfonic acid amide MK-417 BDBM50041029 CHEMBL417975 MK-927 4-Isopropylamino-7,7-dioxo-4,5,6,7-tetrahydro-7lambda*6*-thieno[2,3-b]thiopyran-2-sulfonic acid amide CHEMBL1204135 (S)-4-Isobutylamino-7,7-dioxo-4,5,6,7-tetrahydro-7lambda*6*-thieno[2,3-b]thiopyran-2-sulfonic acid amide BDBM24064 L-706,000 [35S]-MK499 N-[(4R)-1'-[(2R)-6-cyano-1,2,3,4-tetrahydronaphthalen-2-yl]-3,4-dihydrospiro[1-benzopyran-2,4'-piperidine]-4-ol]methanesulfonamide L-706000 MK-499
BDBM24064 L-706,000 [35S]-MK499 N-[(4R)-1'-[(2R)-6-cyano-1,2,3,4-tetrahydronaphthalen-2-yl]-3,4-dihydrospiro[1-benzopyran-2,4'-piperidine]-4-ol]methanesulfonamide L-706000 MK-499 [(7R)-4-(5-Chloro-1,3-benzoxazol-2-yl)-7-methyl-1,4-diazepan-1-yl][5-methyl-2-(2H-1,2,3-triazol-2-yl)phenyl]methanone BDBM50318701 CHEMBL1083659 MK-4305
[(7R)-4-(5-Chloro-1,3-benzoxazol-2-yl)-7-methyl-1,4-diazepan-1-yl][5-methyl-2-(2H-1,2,3-triazol-2-yl)phenyl]methanone BDBM50318701 CHEMBL1083659 MK-4305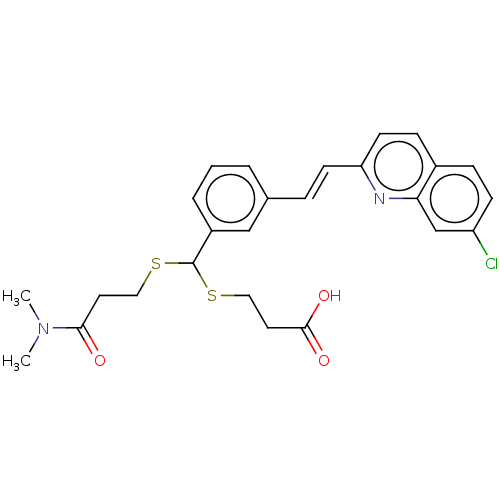 CHEMBL15177 3-[{3-[2-(7-Chloro-quinolin-2-yl)-vinyl]-phenyl}-(2-dimethylcarbamoyl-ethylsulfanyl)-methylsulfanyl]-propionic acid(MK-571, L-660711) BDBM50001285 MK 571 (E)-3-((3-(2-(7-chloroquinolin-2-yl)vinyl)phenyl)(3-(dimethylamino)-3-oxopropylthio)methylthio)propanoic acid 3-((3-(2-(7-chloroquinolin-2-yl)vinyl)phenyl)(3-(dimethylamino)-3-oxopropylthio)methylthio)propanoic acid 3-[{3-[2-(7-Chloro-quinolin-2-yl)-vinyl]-phenyl}-(2-dimethylcarbamoyl-ethylsulfanyl)-methylsulfanyl]-propionic acid(MK-571) MK-571 3-(((3-(2-(7-chloroquinoline-2-yl)ethenyl)phenyl)((3-dimethylamino-3-oxopropyl)thio)methyl)thio)propanoic acid 3-[{3-[(E)-2-(7-Chloro-quinolin-2-yl)-vinyl]-phenyl}-(2-dimethylcarbamoyl-ethylsulfanyl)-methylsulfanyl]-propionic acid 3-[{3-[(E)-2-(7-Chloro-quinolin-2-yl)-vinyl]-phenyl}-(2-diethylcarbamoyl-ethylsulfanyl)-methylsulfanyl]-propionic acid L-660711
CHEMBL15177 3-[{3-[2-(7-Chloro-quinolin-2-yl)-vinyl]-phenyl}-(2-dimethylcarbamoyl-ethylsulfanyl)-methylsulfanyl]-propionic acid(MK-571, L-660711) BDBM50001285 MK 571 (E)-3-((3-(2-(7-chloroquinolin-2-yl)vinyl)phenyl)(3-(dimethylamino)-3-oxopropylthio)methylthio)propanoic acid 3-((3-(2-(7-chloroquinolin-2-yl)vinyl)phenyl)(3-(dimethylamino)-3-oxopropylthio)methylthio)propanoic acid 3-[{3-[2-(7-Chloro-quinolin-2-yl)-vinyl]-phenyl}-(2-dimethylcarbamoyl-ethylsulfanyl)-methylsulfanyl]-propionic acid(MK-571) MK-571 3-(((3-(2-(7-chloroquinoline-2-yl)ethenyl)phenyl)((3-dimethylamino-3-oxopropyl)thio)methyl)thio)propanoic acid 3-[{3-[(E)-2-(7-Chloro-quinolin-2-yl)-vinyl]-phenyl}-(2-dimethylcarbamoyl-ethylsulfanyl)-methylsulfanyl]-propionic acid 3-[{3-[(E)-2-(7-Chloro-quinolin-2-yl)-vinyl]-phenyl}-(2-diethylcarbamoyl-ethylsulfanyl)-methylsulfanyl]-propionic acid L-660711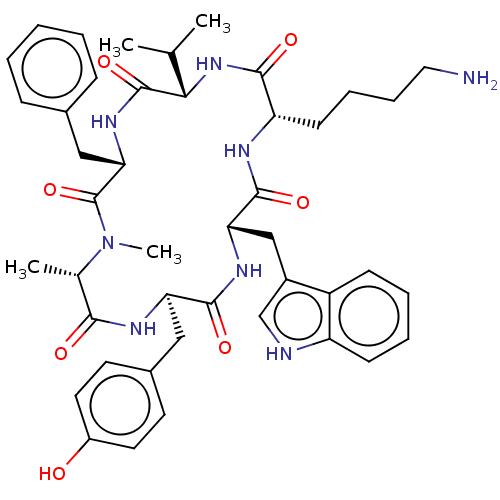 MK-678 BDBM50068063 (12S,15R,18S)-9-(4-Amino-butyl)-3-benzyl-15-(4-hydroxy-benzyl)-12-(1H-indol-3-ylmethyl)-6-isopropyl-1,18-dimethyl-1,4,7,10,13,16hexaaza-cyclooctadecane-2,5,8,11,14,17-hexaone CHEMBL2370925
MK-678 BDBM50068063 (12S,15R,18S)-9-(4-Amino-butyl)-3-benzyl-15-(4-hydroxy-benzyl)-12-(1H-indol-3-ylmethyl)-6-isopropyl-1,18-dimethyl-1,4,7,10,13,16hexaaza-cyclooctadecane-2,5,8,11,14,17-hexaone CHEMBL2370925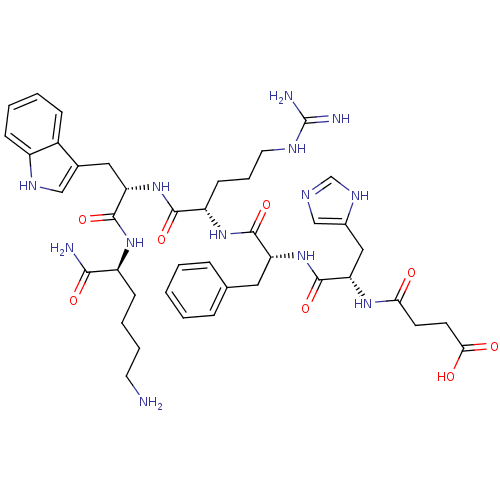 BDBM50114086 MK-11 CHEMBL433413 N-[1-(1-{1-[1-(5-Amino-1-carbamoyl-pentylcarbamoyl)-2-(1H-indol-3-yl)-ethylcarbamoyl]-4-guanidino-butylcarbamoyl}-2-phenyl-ethylcarbamoyl)-2-(3H-imidazol-4-yl)-ethyl]-succinamic acid
BDBM50114086 MK-11 CHEMBL433413 N-[1-(1-{1-[1-(5-Amino-1-carbamoyl-pentylcarbamoyl)-2-(1H-indol-3-yl)-ethylcarbamoyl]-4-guanidino-butylcarbamoyl}-2-phenyl-ethylcarbamoyl)-2-(3H-imidazol-4-yl)-ethyl]-succinamic acid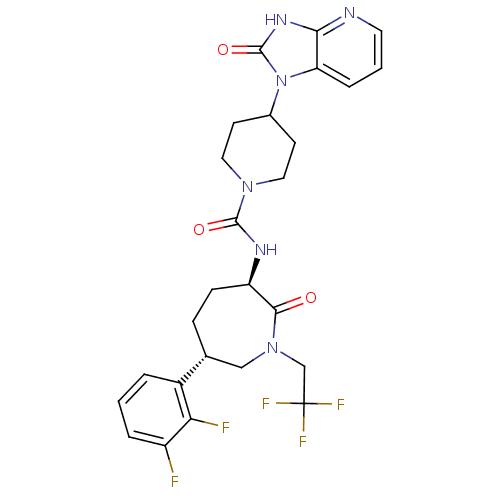 N-[(3R,6S)-6-(2,3-difluorophenyl)-2-oxo-1-(2,2,2-trifluoroethyl)azepan-3-yl]-4-(2-oxo-2,3-dihydro-1H-imidazo[4,5-b]pyridin-1-yl)piperidine-1-carboxamide BDBM50224431 CHEMBL236593 Telcagepant MK-0974
N-[(3R,6S)-6-(2,3-difluorophenyl)-2-oxo-1-(2,2,2-trifluoroethyl)azepan-3-yl]-4-(2-oxo-2,3-dihydro-1H-imidazo[4,5-b]pyridin-1-yl)piperidine-1-carboxamide BDBM50224431 CHEMBL236593 Telcagepant MK-0974 BDBM50013920 MK-679 3-[{3-[2-(7-Chloro-quinolin-2-yl)-vinyl]-phenyl}-(2-dimethylcarbamoyl-ethylsulfanyl)-methylsulfanyl]-propionic acid 3-[(R)-{3-[(E)-2-(7-Chloro-quinolin-2-yl)-vinyl]-phenyl}-(2-dimethylcarbamoyl-ethylsulfanyl)-methylsulfanyl]-propionic acid CHEMBL280481 VERLUKAST
BDBM50013920 MK-679 3-[{3-[2-(7-Chloro-quinolin-2-yl)-vinyl]-phenyl}-(2-dimethylcarbamoyl-ethylsulfanyl)-methylsulfanyl]-propionic acid 3-[(R)-{3-[(E)-2-(7-Chloro-quinolin-2-yl)-vinyl]-phenyl}-(2-dimethylcarbamoyl-ethylsulfanyl)-methylsulfanyl]-propionic acid CHEMBL280481 VERLUKAST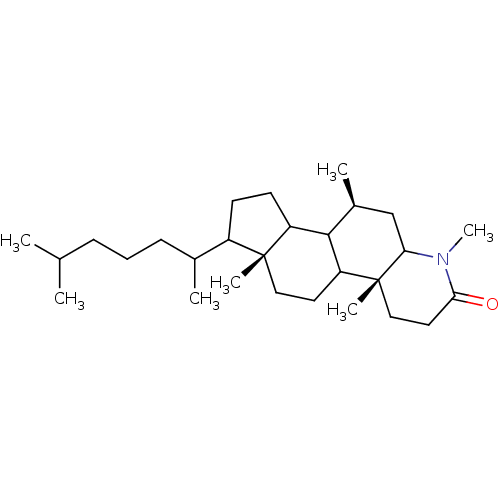 BDBM50032771 MK-386 CHEMBL25448 (4aR,6aR,7R,10S)-7-(1,5-Dimethyl-hexyl)-1,4a,6a,10-tetramethyl-hexadecahydro-indeno[5,4-f]quinolin-2-one (4aR,6aR,10S)-7-(1,5-Dimethyl-hexyl)-1,4a,6a,10-tetramethyl-hexadecahydro-indeno[5,4-f]quinolin-2-one
BDBM50032771 MK-386 CHEMBL25448 (4aR,6aR,7R,10S)-7-(1,5-Dimethyl-hexyl)-1,4a,6a,10-tetramethyl-hexadecahydro-indeno[5,4-f]quinolin-2-one (4aR,6aR,10S)-7-(1,5-Dimethyl-hexyl)-1,4a,6a,10-tetramethyl-hexadecahydro-indeno[5,4-f]quinolin-2-one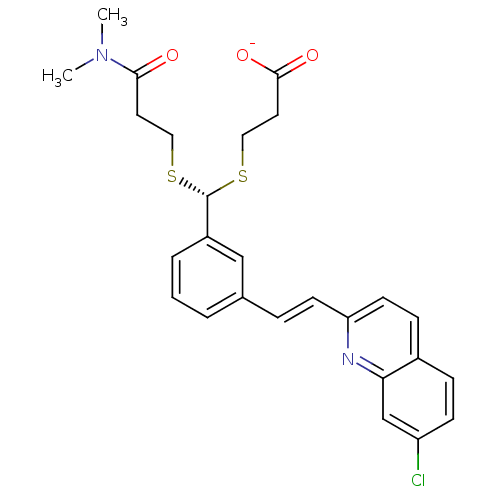 Sodium;3-[(R)-{3-[(E)-2-(7-chloro-quinolin-2-yl)-vinyl]-phenyl}-(2-dimethylcarbamoyl-ethylsulfanyl)-methylsulfanyl]-propionate MK-679 BDBM50052029 Sodium; 3-[(R)-{3-[(E)-2-(7-chloro-quinolin-2-yl)-vinyl]-phenyl}-(2-dimethylcarbamoyl-ethylsulfanyl)-methylsulfanyl]-propionate L-668019 CHEMBL89340
Sodium;3-[(R)-{3-[(E)-2-(7-chloro-quinolin-2-yl)-vinyl]-phenyl}-(2-dimethylcarbamoyl-ethylsulfanyl)-methylsulfanyl]-propionate MK-679 BDBM50052029 Sodium; 3-[(R)-{3-[(E)-2-(7-chloro-quinolin-2-yl)-vinyl]-phenyl}-(2-dimethylcarbamoyl-ethylsulfanyl)-methylsulfanyl]-propionate L-668019 CHEMBL89340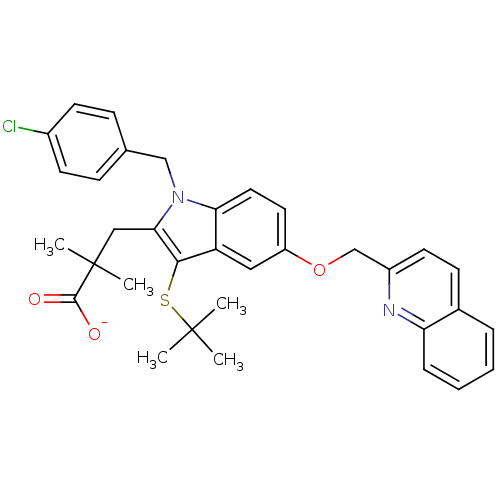 3-[3-tert-Butylsulfanyl-1-(4-chloro-benzyl)-5-(quinolin-2-ylmethoxy)-1H-indol-2-yl]-2,2-dimethyl-propionic acid Sodium; 3-[3-tert-butylsulfanyl-1-(4-chloro-benzyl)-5-(quinolin-2-ylmethoxy)-1H-indol-2-yl]-2,2-dimethyl-propionate MK-591 BDBM50052018 CHEMBL313489
3-[3-tert-Butylsulfanyl-1-(4-chloro-benzyl)-5-(quinolin-2-ylmethoxy)-1H-indol-2-yl]-2,2-dimethyl-propionic acid Sodium; 3-[3-tert-butylsulfanyl-1-(4-chloro-benzyl)-5-(quinolin-2-ylmethoxy)-1H-indol-2-yl]-2,2-dimethyl-propionate MK-591 BDBM50052018 CHEMBL313489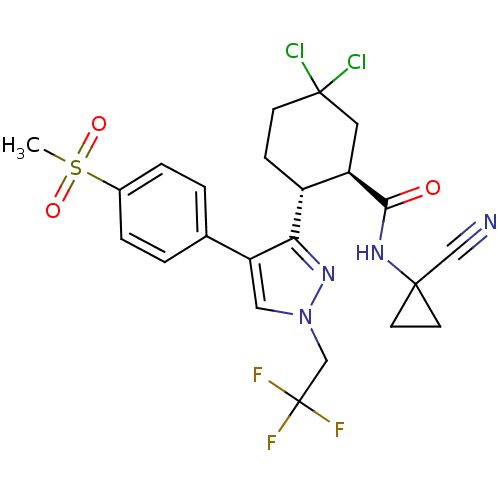 CHEMBL523352 (1R,2R)-5,5-Dichloro-N-(1-cyanocyclopropyl)-2-[4-[4-(methylsulfonyl)phenyl]-1-(2,2,2-trifluoroethyl)-1H-pyrazol-3-yl]cyclohexanecarboxamide (1R,2R)-5,5-dichloro-N-(1-cyanocyclopropyl)-2-(4-(4-(methylsulfonyl)phenyl)-1-(2,2,2-trifluoroethyl)-1H-pyrazol-3-yl)cyclohexanecarboxamide BDBM50253098 MK-1256
CHEMBL523352 (1R,2R)-5,5-Dichloro-N-(1-cyanocyclopropyl)-2-[4-[4-(methylsulfonyl)phenyl]-1-(2,2,2-trifluoroethyl)-1H-pyrazol-3-yl]cyclohexanecarboxamide (1R,2R)-5,5-dichloro-N-(1-cyanocyclopropyl)-2-(4-(4-(methylsulfonyl)phenyl)-1-(2,2,2-trifluoroethyl)-1H-pyrazol-3-yl)cyclohexanecarboxamide BDBM50253098 MK-1256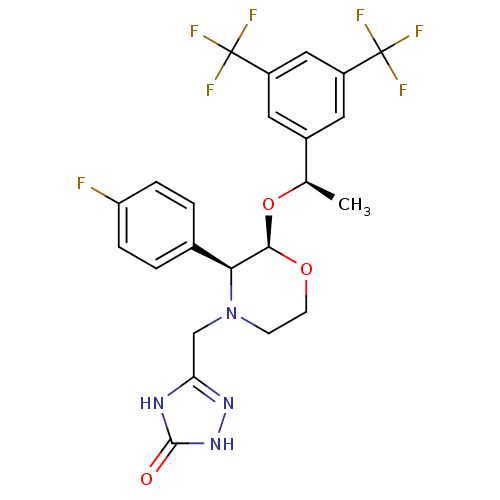 US10100030, Aprepitant 5-[(2R,3S)-2-[(R)-1-(3,5-Bis-trifluoromethyl-phenyl)-ethoxy]-3-(4-fluoro-phenyl)-morpholin-4-ylmethyl]-2,4-dihydro-[1,2,4]triazol-3-one MK-0869 US10011568, Aprepitant MK-869 3-[2-{1-[3,5-di(trifluoromethyl)phenyl]ethoxy}-3-(4-fluorophenyl)-1,4-oxazinan-4-ylmethyl]-4,5-dihydro-1H-1,2,4-triazol-5-one APREPITANT CHEMBL1471 Emend 5-(((2R,3S)-2-((R)-1-(3,5-bis(trifluoromethyl)phenyl)ethoxy)-3-(4-fluorophenyl)morpholino)methyl)-2H-1,2,4-triazol-3(4H)-one 5-[2-[1-(3,5-Bis-trifluoromethyl-phenyl)-ethoxy]-3-(4-fluoro-phenyl)-morpholin-4-ylmethyl]-2,4-dihydro-[1,2,4]triazol-3-one BDBM50220136
US10100030, Aprepitant 5-[(2R,3S)-2-[(R)-1-(3,5-Bis-trifluoromethyl-phenyl)-ethoxy]-3-(4-fluoro-phenyl)-morpholin-4-ylmethyl]-2,4-dihydro-[1,2,4]triazol-3-one MK-0869 US10011568, Aprepitant MK-869 3-[2-{1-[3,5-di(trifluoromethyl)phenyl]ethoxy}-3-(4-fluorophenyl)-1,4-oxazinan-4-ylmethyl]-4,5-dihydro-1H-1,2,4-triazol-5-one APREPITANT CHEMBL1471 Emend 5-(((2R,3S)-2-((R)-1-(3,5-bis(trifluoromethyl)phenyl)ethoxy)-3-(4-fluorophenyl)morpholino)methyl)-2H-1,2,4-triazol-3(4H)-one 5-[2-[1-(3,5-Bis-trifluoromethyl-phenyl)-ethoxy]-3-(4-fluoro-phenyl)-morpholin-4-ylmethyl]-2,4-dihydro-[1,2,4]triazol-3-one BDBM50220136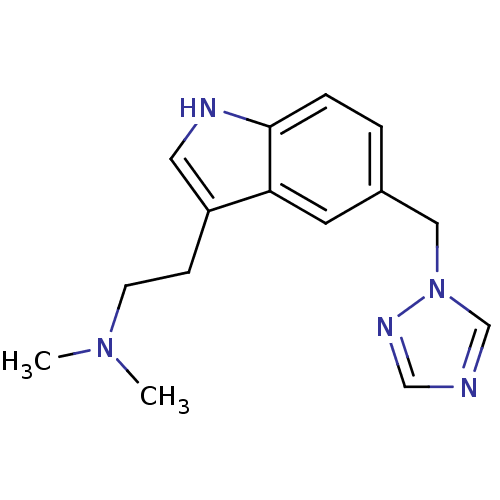 CHEMBL905 N,N-dimethyl-5-(1H-1,2,4-triazol-1-ylmethyl)-1H-indole-3-ethanamine BDBM50033437 MK 462 free base risatriptan N,N-dimethyl-2-[5-(1H-1,2,4-triazol-1-ylmethyl)-1H-indol-3-yl]ethanamine N,N-dimethyl-2-[5-(1,2,4-triazol-1-ylmethyl)-1H-indol-3-yl]-ethanamine RIZATRIPTAN
CHEMBL905 N,N-dimethyl-5-(1H-1,2,4-triazol-1-ylmethyl)-1H-indole-3-ethanamine BDBM50033437 MK 462 free base risatriptan N,N-dimethyl-2-[5-(1H-1,2,4-triazol-1-ylmethyl)-1H-indol-3-yl]ethanamine N,N-dimethyl-2-[5-(1,2,4-triazol-1-ylmethyl)-1H-indol-3-yl]-ethanamine RIZATRIPTAN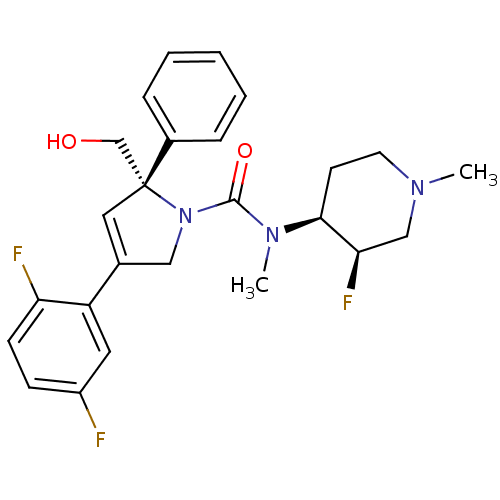 BDBM50252149 CHEMBL481931 MK-0731 (S)-4-(2,5-difluorophenyl)-N-((3R,4S)-3-fluoro-1-methylpiperidin-4-yl)-2-(hydroxymethyl)-N-methyl-2-phenyl-2H-pyrrole-1(5H)-carboxamide (2S)-4-(2,5-difluorophenyl)-N-[(3R,4S)-3-fluoro-1-methylpiperidin-4-yl]-2-(hydroxymethyl)-N-methyl-2-phenyl-2,5-dihydro-1H-pyrrole-1-carboxamide
BDBM50252149 CHEMBL481931 MK-0731 (S)-4-(2,5-difluorophenyl)-N-((3R,4S)-3-fluoro-1-methylpiperidin-4-yl)-2-(hydroxymethyl)-N-methyl-2-phenyl-2H-pyrrole-1(5H)-carboxamide (2S)-4-(2,5-difluorophenyl)-N-[(3R,4S)-3-fluoro-1-methylpiperidin-4-yl]-2-(hydroxymethyl)-N-methyl-2-phenyl-2,5-dihydro-1H-pyrrole-1-carboxamide Sitagliptin (13) US10479798, Compound MK0431 (1R)-3-oxo-3-[3-(trifluoroethyl)-5,6-dihydro[1,2,4]triazolo[4,3-a]pyrazin-7(8H)-yl]-1-(2,4,5-trifluorobenzyl)propylamine Sitagliptin CHEMBL393336 Triazolopiperazine Analogue 1 MK-0431 BDBM11162 (3R)-3-amino-1-[3-(trifluoromethyl)-5H,6H,7H,8H-[1,2,4]triazolo[3,4-a]pyrazin-7-yl]-4-(2,4,5-trifluorophenyl)butan-1-one hydrochloride
Sitagliptin (13) US10479798, Compound MK0431 (1R)-3-oxo-3-[3-(trifluoroethyl)-5,6-dihydro[1,2,4]triazolo[4,3-a]pyrazin-7(8H)-yl]-1-(2,4,5-trifluorobenzyl)propylamine Sitagliptin CHEMBL393336 Triazolopiperazine Analogue 1 MK-0431 BDBM11162 (3R)-3-amino-1-[3-(trifluoromethyl)-5H,6H,7H,8H-[1,2,4]triazolo[3,4-a]pyrazin-7-yl]-4-(2,4,5-trifluorophenyl)butan-1-one hydrochloride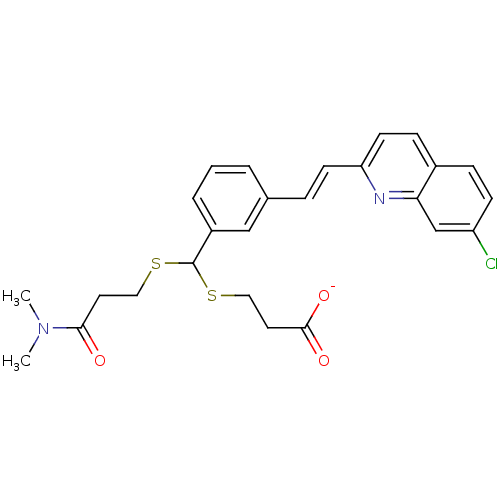 Sodium; 3-[{3-[(E)-2-(7-chloro-quinolin-2-yl)-vinyl]-phenyl}-(2-dimethylcarbamoyl-ethylsulfanyl)-methylsulfanyl]-propionate L-660771 BDBM50052019 Sodium;3-[{3-[(E)-2-(7-chloro-quinolin-2-yl)-vinyl]-phenyl}-(2-dimethylcarbamoyl-ethylsulfanyl)-methylsulfanyl]-propionate CHEMBL89768 MK-571 sodium (E)-3-((3-(2-(7-chloroquinolin-2-yl)vinyl)phenyl)(3-(dimethylamino)-3-oxopropylthio)methylthio)propanoate
Sodium; 3-[{3-[(E)-2-(7-chloro-quinolin-2-yl)-vinyl]-phenyl}-(2-dimethylcarbamoyl-ethylsulfanyl)-methylsulfanyl]-propionate L-660771 BDBM50052019 Sodium;3-[{3-[(E)-2-(7-chloro-quinolin-2-yl)-vinyl]-phenyl}-(2-dimethylcarbamoyl-ethylsulfanyl)-methylsulfanyl]-propionate CHEMBL89768 MK-571 sodium (E)-3-((3-(2-(7-chloroquinolin-2-yl)vinyl)phenyl)(3-(dimethylamino)-3-oxopropylthio)methylthio)propanoate CHEMBL599872 (1R,21S,24S)-21-tert-butyl-N-((1R,2R)-1-{[(cyclopropylsulfonyl)amino]carbonyl}-2-ethylcyclopropyl)-16,16-dimethyl-3,19,22-trioxo-2,18-dioxa-4,20,23-triazatetracyclo[21.2.1.14,70.06,11]heptacosa-6,8,10-triene-24-carboxamide BDBM50326055 MK-7009 (1R,21S,24S)-21-tert-Butyl-N-((1R,2R)-1-{[(cyclopropylsulfonyl)amino]carbonyl}-2-ethylcyclopropyl)-16,16-dimethyl-3,19,22-trioxo-2,18-dioxa-4,20,23-triazatetracyclo[21.2.1.14,7.06,11]-heptacosa-6,8,10-triene-24-carboxamide
CHEMBL599872 (1R,21S,24S)-21-tert-butyl-N-((1R,2R)-1-{[(cyclopropylsulfonyl)amino]carbonyl}-2-ethylcyclopropyl)-16,16-dimethyl-3,19,22-trioxo-2,18-dioxa-4,20,23-triazatetracyclo[21.2.1.14,70.06,11]heptacosa-6,8,10-triene-24-carboxamide BDBM50326055 MK-7009 (1R,21S,24S)-21-tert-Butyl-N-((1R,2R)-1-{[(cyclopropylsulfonyl)amino]carbonyl}-2-ethylcyclopropyl)-16,16-dimethyl-3,19,22-trioxo-2,18-dioxa-4,20,23-triazatetracyclo[21.2.1.14,7.06,11]-heptacosa-6,8,10-triene-24-carboxamide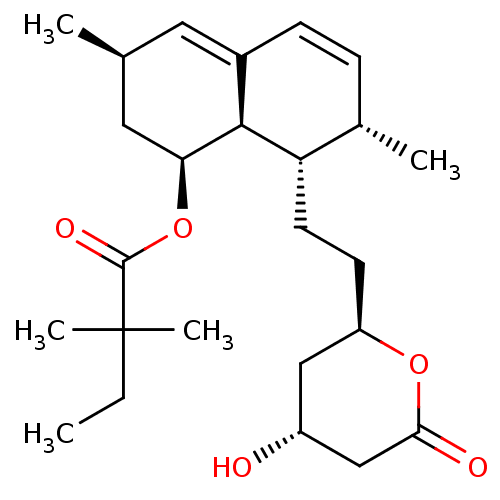 cid_54454 MK-733 SIMVASTATIN LACTONE Simvastatin (1S,3R,7S,8S,8aR)-8-{2-[(2R,4R)-4-hydroxy-6-oxotetrahydro-2H-pyran-2-yl]ethyl}-3,7-dimethyl-1,2,3,7,8,8a-hexahydronaphthalen-1-yl 2,2-dimethylbutanoate Zocor BDBM50139181 2,2-dimethylbutyric acid, 8-ester with (4R,6R)-6-(2-((1S,2S,6R,8S,8aR)-1,2,6,7,8,8a-hexahydro-8-hydroxy-2,6-dimethyl-1-naphthyl)ethyl)tetrahydro-4-hydroxy-2H-pyran-2-one
cid_54454 MK-733 SIMVASTATIN LACTONE Simvastatin (1S,3R,7S,8S,8aR)-8-{2-[(2R,4R)-4-hydroxy-6-oxotetrahydro-2H-pyran-2-yl]ethyl}-3,7-dimethyl-1,2,3,7,8,8a-hexahydronaphthalen-1-yl 2,2-dimethylbutanoate Zocor BDBM50139181 2,2-dimethylbutyric acid, 8-ester with (4R,6R)-6-(2-((1S,2S,6R,8S,8aR)-1,2,6,7,8,8a-hexahydro-8-hydroxy-2,6-dimethyl-1-naphthyl)ethyl)tetrahydro-4-hydroxy-2H-pyran-2-one 3-[1-(4-chlorobenzyl)-3-t-butyl-thio-5-isopropylindol-2-yl]-2,2-dimethylpropanoic acid cid_3651377 3-(1-(4-chlorobenzyl)-3-(tert-butylthio)-5-isopropyl-1H-indol-2-yl)-2,2-dimethylpropanoic acid CHEMBL29097 MK886 MK-886 3-(3-(tert-butylthio)-1-(4-chlorobenzyl)-5-isopropyl-1H-indol-2-yl)-2,2-dimethylpropanoic acid 3-[3-tert-Butylsulfanyl-1-(4-chloro-benzyl)-5-isopropyl-1H-indol-2-yl]-2,2-dimethyl-propionic acid BDBM50006805
3-[1-(4-chlorobenzyl)-3-t-butyl-thio-5-isopropylindol-2-yl]-2,2-dimethylpropanoic acid cid_3651377 3-(1-(4-chlorobenzyl)-3-(tert-butylthio)-5-isopropyl-1H-indol-2-yl)-2,2-dimethylpropanoic acid CHEMBL29097 MK886 MK-886 3-(3-(tert-butylthio)-1-(4-chlorobenzyl)-5-isopropyl-1H-indol-2-yl)-2,2-dimethylpropanoic acid 3-[3-tert-Butylsulfanyl-1-(4-chloro-benzyl)-5-isopropyl-1H-indol-2-yl]-2,2-dimethyl-propionic acid BDBM50006805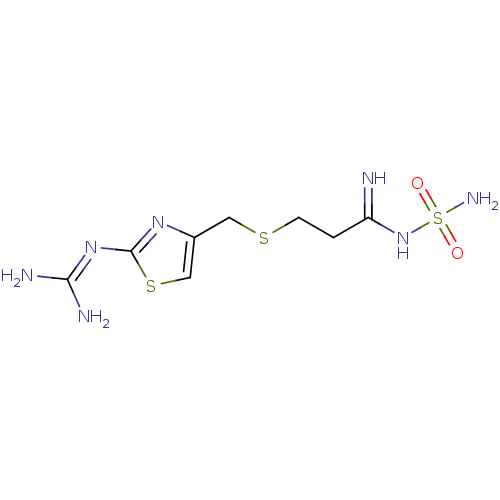 3-(2-Guanidino-thiazol-4-ylmethylsulfanyl)-N-sulfonylamino-propionamidine BDBM50036754 Pepcid Pepcid ac (1Z)-N'-(aminosulfonyl)-3-[({2-[(diaminomethylene)amino]-1,3-thiazol-4-yl}methyl)thio]propanimidamide N-sulfomoyl-3-(2-Guanidino-2H-1lambda*4*-thiazol-4-ylmethylsulfanyl)-propionamidine (1Z)-N'-(aminosulfonyl)-3-[({2-[(diaminomethylene)amino]-1,3-thiazol-4-yl}methyl)thio]propanimidamide (Famotidine) N'-(aminosulfonyl)-3-(((2-((diaminomethylene)amino)-4-thiazolyl)methyl)thio)propanimidamide Pepcid ac (geltab) MK-208 CHEMBL902 Pepcid rpd Fluxid FAMOTIDINE
3-(2-Guanidino-thiazol-4-ylmethylsulfanyl)-N-sulfonylamino-propionamidine BDBM50036754 Pepcid Pepcid ac (1Z)-N'-(aminosulfonyl)-3-[({2-[(diaminomethylene)amino]-1,3-thiazol-4-yl}methyl)thio]propanimidamide N-sulfomoyl-3-(2-Guanidino-2H-1lambda*4*-thiazol-4-ylmethylsulfanyl)-propionamidine (1Z)-N'-(aminosulfonyl)-3-[({2-[(diaminomethylene)amino]-1,3-thiazol-4-yl}methyl)thio]propanimidamide (Famotidine) N'-(aminosulfonyl)-3-(((2-((diaminomethylene)amino)-4-thiazolyl)methyl)thio)propanimidamide Pepcid ac (geltab) MK-208 CHEMBL902 Pepcid rpd Fluxid FAMOTIDINE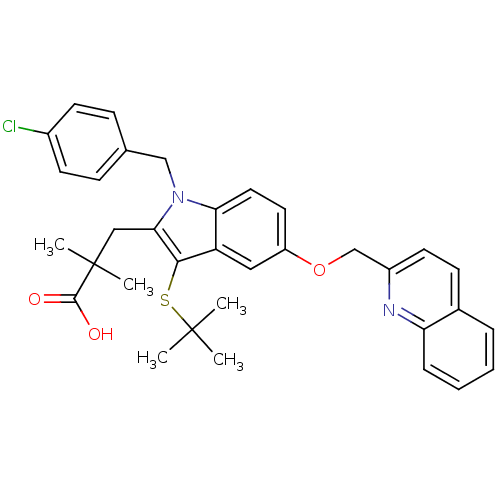 2-[3-tert-Butylsulfanyl-1-(4-chloro-benzyl)-5-(quinolin-2-ylmethoxy)-1H-indol-2-yl]-2-methyl-propionic acid 3-[3-tert-Butylsulfanyl-1-(4-chloro-benzyl)-5-(quinolin-2-ylmethoxy)-1H-indol-2-yl]-2,2-dimethyl-propionic acid CHEMBL16596 3-(1-(4-chlorobenzyl)-3-(tert-butylthio)-5-(quinolin-2-ylmethoxy)-1H-indol-2-yl)-2,2-dimethylpropanoic acid 3-(3-(tert-butylthio)-1-(4-chlorobenzyl)-5-(quinolin-2-ylmethoxy)-1H-indol-2-yl)-2,2-dimethylpropanoic acid MK-591 BDBM50029559 L-686708 3-[3-(TERT-BUTYLTHIO)-1-(4-CHLOROBENZYL)-5-(QUINOLIN-2-YLMETHOXY)-1H-INDOL-2-YL]-2,2-DIMETHYLPROPANOIC ACID
2-[3-tert-Butylsulfanyl-1-(4-chloro-benzyl)-5-(quinolin-2-ylmethoxy)-1H-indol-2-yl]-2-methyl-propionic acid 3-[3-tert-Butylsulfanyl-1-(4-chloro-benzyl)-5-(quinolin-2-ylmethoxy)-1H-indol-2-yl]-2,2-dimethyl-propionic acid CHEMBL16596 3-(1-(4-chlorobenzyl)-3-(tert-butylthio)-5-(quinolin-2-ylmethoxy)-1H-indol-2-yl)-2,2-dimethylpropanoic acid 3-(3-(tert-butylthio)-1-(4-chlorobenzyl)-5-(quinolin-2-ylmethoxy)-1H-indol-2-yl)-2,2-dimethylpropanoic acid MK-591 BDBM50029559 L-686708 3-[3-(TERT-BUTYLTHIO)-1-(4-CHLOROBENZYL)-5-(QUINOLIN-2-YLMETHOXY)-1H-INDOL-2-YL]-2,2-DIMETHYLPROPANOIC ACID L-364718 1H-Indole-2-carboxylic acid (1-methyl-2-oxo-5-phenyl-2,3-dihydro-1H-benzo[e][1,4]diazepin-3-yl)-amide(devazepide) DEVAZEPIDE 1H-Indole-2-carboxylic acid (1-methyl-2-oxo-5-phenyl-2,3-dihydro-1H-benzo[e][1,4]diazepin-3-yl)-amide 1H-Indole-2-carboxylic acid (1-methyl-2-oxo-5-phenyl-2,3-dihydro-1H-benzo[e][1,4]diazepin-3-yl)-amide (L-364,718 ((S)-devazepide) 1H-Indole-2-carboxylic acid (1-methyl-2-oxo-5-phenyl-2,3-dihydro-1H-benzo[e][1,4]diazepin-3-yl)-amide (MK-329, L-364,718) CHEMBL9506 1H-Indole-2-carboxylic acid ((S)-1-methyl-2-oxo-5-phenyl-2,3-dihydro-1H-benzo[e][1,4]diazepin-3-yl)-amide 1H-Indole-2-carboxylic acid (1-methyl-2-oxo-5-phenyl-2,3-dihydro-1H-benzo[e][1,4]diazepin-3-yl)-amide: 0.1C4H10O. 0.15CH2Cl2 MK-329 1H-Indole-2-carboxylic acid (1-methyl-2-oxo-5-phenyl-2,3-dihydro-1H-benzo[e][1,4]diazepin-3-yl)-amide(L-364718) (Z)-N-(1-methyl-2-oxo-5-phenyl-2,3-dihydro-1H-benzo[e][1,4]diazepin-3-yl)-1H-indole-2-carboxamide L-364,718 (S)-1H-Indole-2-carboxylic acid (1-methyl-2-oxo-5-phenyl-2,3-dihydro-1H-benzo[e][1,4]diazepin-3-yl)-amide BDBM50005463 (S)-N-(1-methyl-2-oxo-5-phenyl-2,3-dihydro-1H-benzo[e][1,4]diazepin-3-yl)-1H-indole-2-carboxamide CCK antagonist synthetic 18 (R)-1H-Indole-2-carboxylic acid (1-methyl-2-oxo-5-phenyl-2,3-dihydro-1H-benzo[e][1,4]diazepin-3-yl)-amide: 0.1C4H10O 1H-Indole-2-carboxylic acid (1-methyl-2-oxo-5-phenyl-2,3-dihydro-1H-benzo[e][1,4]diazepin-3-yl)-amide(Devazepide or (R) L364718) CCK antagonist synthetic 17
L-364718 1H-Indole-2-carboxylic acid (1-methyl-2-oxo-5-phenyl-2,3-dihydro-1H-benzo[e][1,4]diazepin-3-yl)-amide(devazepide) DEVAZEPIDE 1H-Indole-2-carboxylic acid (1-methyl-2-oxo-5-phenyl-2,3-dihydro-1H-benzo[e][1,4]diazepin-3-yl)-amide 1H-Indole-2-carboxylic acid (1-methyl-2-oxo-5-phenyl-2,3-dihydro-1H-benzo[e][1,4]diazepin-3-yl)-amide (L-364,718 ((S)-devazepide) 1H-Indole-2-carboxylic acid (1-methyl-2-oxo-5-phenyl-2,3-dihydro-1H-benzo[e][1,4]diazepin-3-yl)-amide (MK-329, L-364,718) CHEMBL9506 1H-Indole-2-carboxylic acid ((S)-1-methyl-2-oxo-5-phenyl-2,3-dihydro-1H-benzo[e][1,4]diazepin-3-yl)-amide 1H-Indole-2-carboxylic acid (1-methyl-2-oxo-5-phenyl-2,3-dihydro-1H-benzo[e][1,4]diazepin-3-yl)-amide: 0.1C4H10O. 0.15CH2Cl2 MK-329 1H-Indole-2-carboxylic acid (1-methyl-2-oxo-5-phenyl-2,3-dihydro-1H-benzo[e][1,4]diazepin-3-yl)-amide(L-364718) (Z)-N-(1-methyl-2-oxo-5-phenyl-2,3-dihydro-1H-benzo[e][1,4]diazepin-3-yl)-1H-indole-2-carboxamide L-364,718 (S)-1H-Indole-2-carboxylic acid (1-methyl-2-oxo-5-phenyl-2,3-dihydro-1H-benzo[e][1,4]diazepin-3-yl)-amide BDBM50005463 (S)-N-(1-methyl-2-oxo-5-phenyl-2,3-dihydro-1H-benzo[e][1,4]diazepin-3-yl)-1H-indole-2-carboxamide CCK antagonist synthetic 18 (R)-1H-Indole-2-carboxylic acid (1-methyl-2-oxo-5-phenyl-2,3-dihydro-1H-benzo[e][1,4]diazepin-3-yl)-amide: 0.1C4H10O 1H-Indole-2-carboxylic acid (1-methyl-2-oxo-5-phenyl-2,3-dihydro-1H-benzo[e][1,4]diazepin-3-yl)-amide(Devazepide or (R) L364718) CCK antagonist synthetic 17 1-Ethyl-6-fluoro-4-oxo-7-piperazin-1-yl-1,4,4a,8a-tetrahydro-quinoline-3-carboxylic acid (norfloxacin) Chibroxin (norfloxacin)1-Ethyl-6-fluoro-4-oxo-7-piperazin-1-yl-1,4-dihydro-quinoline-3-carboxylic acid 1-Ethyl-6-fluoro-4-oxo-7-piperazin-1-yl-1,4-dihydro-quinoline-3-carboxylic acid(Norfloxacin) CHEMBL9 1-ethyl-6-fluoro-7-hexahydro-1-pyrazinyl-4-oxo-1,4-dihydro-3-quinolinecarboxylic acid 1-Ethyl-6-fluoro-4-oxo-7-piperazin-1-yl-1,4-dihydro-quinoline-3-carboxylic acid (Norfloxacin) 1-Ethyl-6-fluoro-4-oxo-7-piperazin-1-yl-1,4-dihydro-quinoline-3-carboxylic acid(1-norfloxacin) NORFLOXACIN Noroxin 1-ethyl-6-fluoro-4-oxo-7-(piperazin-1-yl)-1,4-dihydroquinoline-3-carboxylic acid (NFLX)1-Ethyl-6-fluoro-4-oxo-7-piperazin-1-yl-1,4-dihydro-quinoline-3-carboxylic acid 1-Ethyl-6-fluoro-4-oxo-7-piperazin-1-yl-1,4-dihydro-quinoline-3-carboxylic acid BDBM50045000 MK-366
1-Ethyl-6-fluoro-4-oxo-7-piperazin-1-yl-1,4,4a,8a-tetrahydro-quinoline-3-carboxylic acid (norfloxacin) Chibroxin (norfloxacin)1-Ethyl-6-fluoro-4-oxo-7-piperazin-1-yl-1,4-dihydro-quinoline-3-carboxylic acid 1-Ethyl-6-fluoro-4-oxo-7-piperazin-1-yl-1,4-dihydro-quinoline-3-carboxylic acid(Norfloxacin) CHEMBL9 1-ethyl-6-fluoro-7-hexahydro-1-pyrazinyl-4-oxo-1,4-dihydro-3-quinolinecarboxylic acid 1-Ethyl-6-fluoro-4-oxo-7-piperazin-1-yl-1,4-dihydro-quinoline-3-carboxylic acid (Norfloxacin) 1-Ethyl-6-fluoro-4-oxo-7-piperazin-1-yl-1,4-dihydro-quinoline-3-carboxylic acid(1-norfloxacin) NORFLOXACIN Noroxin 1-ethyl-6-fluoro-4-oxo-7-(piperazin-1-yl)-1,4-dihydroquinoline-3-carboxylic acid (NFLX)1-Ethyl-6-fluoro-4-oxo-7-piperazin-1-yl-1,4-dihydro-quinoline-3-carboxylic acid 1-Ethyl-6-fluoro-4-oxo-7-piperazin-1-yl-1,4-dihydro-quinoline-3-carboxylic acid BDBM50045000 MK-366 Proscar BDBM50334788 (4aR,6aS,7S)-4a,6a-Dimethyl-2-oxo-2,4a,4b,5,6,6a,7,8,9,9a,9b,10,11,11a-tetradecahydro-1H-indeno[5,4-f]quinoline-7-carboxylic acid tert-butylamide (4aR,6aS)-4a,6a-Dimethyl-2-oxo-2,4a,4b,5,6,6a,7,8,9,9a,9b,10,11,11a-tetradecahydro-1H-indeno[5,4-f]quinoline-7-carboxylic acid tert-butylamide CHEMBL710 FINASTERIDE 4a,6a-Dimethyl-2-oxo-2,4a,4b,5,6,6a,7,8,9,9a,9b,10,11,11a-tetradecahydro-1H-indeno[5,4-f]quinoline-7-carboxylic acid tert-butylamide(Finasteride) MK-906 (R)-4a,6a-Dimethyl-2-oxo-2,4a,4b,5,6,6a,7,8,9,9a,9b,10,11,11a-tetradecahydro-1H-indeno[5,4-f]quinoline-7-carboxylic acid tert-butylamide (4aR,4bS,6aS,7S,9aS,9bS,11aR)-N-tert-butyl-4a,6a-dimethyl-2-oxo-2,4a,4b,5,6,6a,7,8,9,9a,9b,10,11,11a-tetradecahydro-1H-indeno[5,4-f]quinoline-7-carboxamide Propecia (4aR,6aS,7S,11aR)-4a,6a-Dimethyl-2-oxo-2,4a,4b,5,6,6a,7,8,9,9a,9b,10,11,11a-tetradecahydro-1H-indeno[5,4-f]quinoline-7-carboxylic acid tert-butylamide US9061023, Finasteride (4aR,4bS,6aS,7S,9aS,9bS,11aR)-4a,6a-Dimethyl-2-oxo-2,4a,4b,5,6,6a,7,8,9,9a,9b,10,11,11a-tetradecahydro-1H-indeno[5,4-f]quinoline-7-carboxylic acid tert-butylamide (4aR,4bS,6aS,9aS,9bS,11aR)-4a,6a-Dimethyl-2-oxo-2,4a,4b,5,6,6a,7,8,9,9a,9b,10,11,11a-tetradecahydro-1H-indeno[5,4-f]quinoline-7-carboxylic acid tert-butylamide (4aR,6aS,11aR)-4a,6a-Dimethyl-2-oxo-hexadecahydro-indeno[5,4-f]quinoline-7-carboxylic acid tert-butylamide 4a,6a-Dimethyl-2-oxo-2,4a,4b,5,6,6a,7,8,9,9a,9b,10,11,11a-tetradecahydro-1H-indeno[5,4-f]quinoline-7-carboxylic acid tert-butylamide (17beta-(N-tert-butylcarbamoyl)-4-aza-5alpha-androst-1-en-3-one 4a,6a,9a-Trimethyl-2-oxo-2,4a,4b,5,6,6a,7,8,9,9a,9b,10,11,11a-tetradecahydro-1H-indeno[5,4-f]quinoline-7-carboxylic acid (Finasteride)
Proscar BDBM50334788 (4aR,6aS,7S)-4a,6a-Dimethyl-2-oxo-2,4a,4b,5,6,6a,7,8,9,9a,9b,10,11,11a-tetradecahydro-1H-indeno[5,4-f]quinoline-7-carboxylic acid tert-butylamide (4aR,6aS)-4a,6a-Dimethyl-2-oxo-2,4a,4b,5,6,6a,7,8,9,9a,9b,10,11,11a-tetradecahydro-1H-indeno[5,4-f]quinoline-7-carboxylic acid tert-butylamide CHEMBL710 FINASTERIDE 4a,6a-Dimethyl-2-oxo-2,4a,4b,5,6,6a,7,8,9,9a,9b,10,11,11a-tetradecahydro-1H-indeno[5,4-f]quinoline-7-carboxylic acid tert-butylamide(Finasteride) MK-906 (R)-4a,6a-Dimethyl-2-oxo-2,4a,4b,5,6,6a,7,8,9,9a,9b,10,11,11a-tetradecahydro-1H-indeno[5,4-f]quinoline-7-carboxylic acid tert-butylamide (4aR,4bS,6aS,7S,9aS,9bS,11aR)-N-tert-butyl-4a,6a-dimethyl-2-oxo-2,4a,4b,5,6,6a,7,8,9,9a,9b,10,11,11a-tetradecahydro-1H-indeno[5,4-f]quinoline-7-carboxamide Propecia (4aR,6aS,7S,11aR)-4a,6a-Dimethyl-2-oxo-2,4a,4b,5,6,6a,7,8,9,9a,9b,10,11,11a-tetradecahydro-1H-indeno[5,4-f]quinoline-7-carboxylic acid tert-butylamide US9061023, Finasteride (4aR,4bS,6aS,7S,9aS,9bS,11aR)-4a,6a-Dimethyl-2-oxo-2,4a,4b,5,6,6a,7,8,9,9a,9b,10,11,11a-tetradecahydro-1H-indeno[5,4-f]quinoline-7-carboxylic acid tert-butylamide (4aR,4bS,6aS,9aS,9bS,11aR)-4a,6a-Dimethyl-2-oxo-2,4a,4b,5,6,6a,7,8,9,9a,9b,10,11,11a-tetradecahydro-1H-indeno[5,4-f]quinoline-7-carboxylic acid tert-butylamide (4aR,6aS,11aR)-4a,6a-Dimethyl-2-oxo-hexadecahydro-indeno[5,4-f]quinoline-7-carboxylic acid tert-butylamide 4a,6a-Dimethyl-2-oxo-2,4a,4b,5,6,6a,7,8,9,9a,9b,10,11,11a-tetradecahydro-1H-indeno[5,4-f]quinoline-7-carboxylic acid tert-butylamide (17beta-(N-tert-butylcarbamoyl)-4-aza-5alpha-androst-1-en-3-one 4a,6a,9a-Trimethyl-2-oxo-2,4a,4b,5,6,6a,7,8,9,9a,9b,10,11,11a-tetradecahydro-1H-indeno[5,4-f]quinoline-7-carboxylic acid (Finasteride)
- Beshore, DC; N Di Marco, C; Chang, RK; Greshock, TJ; Ma, L; Wittmann, M; Seager, MA; Koeplinger, KA; Thompson, CD; Fuerst, J; Hartman, GD; Bilodeau, MT; Ray, WJ; Kuduk, SD MK-7622: A First-in-Class M ACS Med Chem Lett 9: 652-656 (2018)
- Bungard, CJ; Hartman, GD; Manikowski, JJ; Perkins, JJ; Bai, C; Brandish, PE; Euler, DH; Hershey, JC; Schmidt, A; Fang, Y; Norcross, RT; Rushmore, TH; Thompson, CD; Meissner, RS Discovery of selective glucocorticoid receptor modulator MK-5932. Bioorg Med Chem 19: 7374-86 (2011)
- Lai, MT; Munshi, V; Touch, S; Tynebor, RM; Tucker, TJ; McKenna, PM; Williams, TM; DiStefano, DJ; Hazuda, DJ; Miller, MD Antiviral activity of MK-4965, a novel nonnucleoside reverse transcriptase inhibitor. Antimicrob Agents Chemother 53: 2424-31 (2009)
- Chang, M; Park, SR; Kim, J; Jang, M; Park, JH; Park, JE; Park, HG; Suh, YG; Jeong, YS; Park, YH; Kim, HD Silicon switch approach in TRPV1 antagonist MK-056 and its analogues. Bioorg Med Chem 18: 111-6 (2010)
- Isabel, E; Bateman, KP; Chauret, N; Cromlish, W; Desmarais, S; Duong, le T; Falgueyret, JP; Gauthier, JY; Lamontagne, S; Lau, CK; Léger, S; LeRiche, T; Lévesque, JF; Li, CS; Massé, F; McKay, DJ; Mellon, C; Nicoll-Griffith, DA; Oballa, RM; Percival, MD; Riendeau, D; Robichaud, J; Rodan, GA; Rodan, SB; Seto, C; Thérien, M; Truong, VL; Wesolowski, G; Young, RN; Zamboni, R; Black, WC The discovery of MK-0674, an orally bioavailable cathepsin K inhibitor. Bioorg Med Chem Lett 20: 887-92 (2010)
- Bebbington, D; Binch, H; Charrier, JD; Everitt, S; Fraysse, D; Golec, J; Kay, D; Knegtel, R; Mak, C; Mazzei, F; Miller, A; Mortimore, M; O'Donnell, M; Patel, S; Pierard, F; Pinder, J; Pollard, J; Ramaya, S; Robinson, D; Rutherford, A; Studley, J; Westcott, J The discovery of the potent aurora inhibitor MK-0457 (VX-680). Bioorg Med Chem Lett 19: 3586-92 (2009)
- Wolkenberg, SE; Nolt, MB; Bilodeau, MT; Trotter, BW; Manley, PJ; Kett, NR; Nanda, KK; Wu, Z; Cato, MJ; Kane, SA; Kiss, L; Spencer, RH; Wang, J; Lynch, JJ; Regan, CP; Stump, GL; Li, B; White, R; Yeh, S; Dinsmore, CJ; Lindsley, CW; Hartman, GD Discovery of MK-1832, a Kv1.5 inhibitor with improved selectivity and pharmacokinetics. Bioorg Med Chem Lett 27: 1062-1069 (2017)
- Blizzard, TA; Chen, H; Kim, S; Wu, J; Bodner, R; Gude, C; Imbriglio, J; Young, K; Park, YW; Ogawa, A; Raghoobar, S; Hairston, N; Painter, RE; Wisniewski, D; Scapin, G; Fitzgerald, P; Sharma, N; Lu, J; Ha, S; Hermes, J; Hammond, ML Discovery of MK-7655, aß-lactamase inhibitor for combination with Primaxin®. Bioorg Med Chem Lett 24: 780-5 (2014)
- Rudd, MT; Manley, PJ; Hanney, B; Meng, Z; Shu, Y; de Leon, P; Frie, JL; Han, Y; Wai, JM; Yang, ZQ; Perkins, JJ; Hurzy, DM; Manikowski, JJ; Zhu, H; Bungard, CJ; Converso, A; Meissner, RS; Cosden, ML; Hayashi, I; Ma, L; O'Brien, J; Uebele, VN; Schachter, JB; Bhandari, N; Ward, GJ; Fillgrove, KL; Lu, B; Liang, Y; Dubost, DC; Puri, V; Eddins, DM; Vardigan, JD; Drolet, RE; Kern, JT; Uslaner, JM Discovery of MK-8768, a Potent and Selective mGluR2 Negative Allosteric Modulator. ACS Med Chem Lett 14: 1088-1094 (2023)
- Velcicky, J; Feifel, R; Hawtin, S; Heng, R; Huppertz, C; Koch, G; Kroemer, M; Moebitz, H; Revesz, L; Scheufler, C; Schlapbach, A Novel 3-aminopyrazole inhibitors of MK-2 discovered by scaffold hopping strategy. Bioorg Med Chem Lett 20: 1293-7 (2010)
- Gauthier, JY; Chauret, N; Cromlish, W; Desmarais, S; Duong, le T; Falgueyret, JP; Kimmel, DB; Lamontagne, S; Léger, S; LeRiche, T; Li, CS; Massé, F; McKay, DJ; Nicoll-Griffith, DA; Oballa, RM; Palmer, JT; Percival, MD; Riendeau, D; Robichaud, J; Rodan, GA; Rodan, SB; Seto, C; Thérien, M; Truong, VL; Venuti, MC; Wesolowski, G; Young, RN; Zamboni, R; Black, WC The discovery of odanacatib (MK-0822), a selective inhibitor of cathepsin K. Bioorg Med Chem Lett 18: 923-8 (2008)
- Anderson, DR; Hegde, S; Reinhard, E; Gomez, L; Vernier, WF; Lee, L; Liu, S; Sambandam, A; Snider, PA; Masih, L Aminocyanopyridine inhibitors of mitogen activated protein kinase-activated protein kinase 2 (MK-2). Bioorg Med Chem Lett 15: 1587-90 (2005)
- Harper, S; McCauley, JA; Rudd, MT; Ferrara, M; DiFilippo, M; Crescenzi, B; Koch, U; Petrocchi, A; Holloway, MK; Butcher, JW; Romano, JJ; Bush, KJ; Gilbert, KF; McIntyre, CJ; Nguyen, KT; Nizi, E; Carroll, SS; Ludmerer, SW; Burlein, C; DiMuzio, JM; Graham, DJ; McHale, CM; Stahlhut, MW; Olsen, DB; Monteagudo, E; Cianetti, S; Giuliano, C; Pucci, V; Trainor, N; Fandozzi, CM; Rowley, M; Coleman, PJ; Vacca, JP; Summa, V; Liverton, NJ Discovery of MK-5172, a Macrocyclic Hepatitis C Virus NS3/4a Protease Inhibitor. ACS Med Chem Lett 3: 332-6 (2012)
- Boga, SB; Deng, Y; Zhu, L; Nan, Y; Cooper, AB; Shipps, GW; Doll, R; Shih, NY; Zhu, H; Sun, R; Wang, T; Paliwal, S; Tsui, HC; Gao, X; Yao, X; Desai, J; Wang, J; Alhassan, AB; Kelly, J; Patel, M; Muppalla, K; Gudipati, S; Zhang, LK; Buevich, A; Hesk, D; Carr, D; Dayananth, P; Black, S; Mei, H; Cox, K; Sherborne, B; Hruza, AW; Xiao, L; Jin, W; Long, B; Liu, G; Taylor, SA; Kirschmeier, P; Windsor, WT; Bishop, R; Samatar, AA MK-8353: Discovery of an Orally Bioavailable Dual Mechanism ERK Inhibitor for Oncology. ACS Med Chem Lett 9: 761-767 (2018)
- Li, L; Berthelette, C; Chateauneuf, A; Ouellet, M; Sturino, CF; Wang, Z Potent and selective 5-LO inhibitor bearing benzothiophene pharmacophore: discovery of MK-5286. Bioorg Med Chem Lett 20: 7440-3 (2010)
- Anderson, DR; Meyers, MJ; Vernier, WF; Mahoney, MW; Kurumbail, RG; Caspers, N; Poda, GI; Schindler, JF; Reitz, DB; Mourey, RJ Pyrrolopyridine inhibitors of mitogen-activated protein kinase-activated protein kinase 2 (MK-2). J Med Chem 50: 2647-54 (2007)
- PubChem, PC Screening for Inhibitors of the Mevalonate Pathway in Streptococcus Pneumoniae - MK Dose Response PubChem Bioassay (2007)
- Brockunier, L; Stelmach, J; Guo, J; Spencer, T; Rosauer, K; Bansal, A; Cai, SJ; Chen, N; Cummings, J; Huang, L; Johnson, T; Levesque, S; Luo, L; Maloney, K; Metzger, J; Mortko, C; Ortega, K; Pai, LY; Pereira, A; Salituro, G; Shang, J; Shepherd, C; Sherrie Xu, S; Yang, Q; Cui, J; Roy, S; Parmee, E; Raghavan, S Soluble guanylate cyclase stimulators for the treatment of hypertension: Discovery of MK-2947. Bioorg Med Chem Lett 30: (2020)
- Liu, P; Hamill, TG; Chioda, M; Chobanian, H; Fung, S; Guo, Y; Chang, L; Bakshi, R; Hong, Q; Dellureficio, J; Lin, LS; Abbadie, C; Alexander, J; Jin, H; Mandala, S; Shiao, LL; Li, W; Sanabria, S; Williams, D; Zeng, Z; Hajdu, R; Jochnowitz, N; Rosenbach, M; Karanam, B; Madeira, M; Salituro, G; Powell, J; Xu, L; Terebetski, JL; Leone, JF; Miller, P; Cook, J; Holahan, M; Joshi, A; O'Malley, S; Purcell, M; Posavec, D; Chen, TB; Riffel, K; Williams, M; Hargreaves, R; Sullivan, KA; Nargund, RP; DeVita, RJ Discovery of MK-3168: A PET Tracer for Imaging Brain Fatty Acid Amide Hydrolase. ACS Med Chem Lett 4: 509-13 (2013)
- Gallant, M; Beaulieu, C; Berthelette, C; Colucci, J; Crackower, MA; Dalton, C; Denis, D; Ducharme, Y; Friesen, RW; Guay, D; Gervais, FG; Hamel, M; Houle, R; Krawczyk, CM; Kosjek, B; Lau, S; Leblanc, Y; Lee, EE; Levesque, JF; Mellon, C; Molinaro, C; Mullet, W; O'Neill, GP; O'Shea, P; Sawyer, N; Sillaots, S; Simard, D; Slipetz, D; Stocco, R; Sørensen, D; Truong, VL; Wong, E; Wu, J; Zaghdane, H; Wang, Z Discovery of MK-7246, a selective CRTH2 antagonist for the treatment of respiratory diseases. Bioorg Med Chem Lett 21: 288-93 (2010)
- Jiang, J; Ding, FX; Zhou, X; Bateman, TJ; Dong, S; Gu, X; Keh deJesus, R; Pio, B; Tang, H; Chobanian, HR; Levorse, D; Hu, M; Thomas-Fowlkes, B; Margulis, M; Koehler, M; Weinglass, A; Gibson, J; Houle, K; Yudkovitz, J; Hampton, C; Pai, LY; Samuel, K; Cutarelli, T; Sullivan, K; Parmee, ER; Davies, I; Pasternak, A Discovery of MK-8153, a Potent and Selective ROMK Inhibitor and Novel Diuretic/Natriuretic. J Med Chem 64: 7691-7701 (2021)
- Feng, D; Biftu, T; Romero, FA; Kekec, A; Dropinski, J; Kassick, A; Xu, S; Kurtz, MM; Gollapudi, A; Shao, Q; Yang, X; Lu, K; Zhou, G; Kemp, D; Myers, RW; Guan, HP; Trujillo, ME; Li, C; Weber, A; Sebhat, IK Discovery of MK-8722: A Systemic, Direct Pan-Activator of AMP-Activated Protein Kinase. ACS Med Chem Lett 9: 39-44 (2018)
- Kuduk, SD; Skudlarek, JW; DiMarco, CN; Bruno, JG; Pausch, MH; O'Brien, JA; Cabalu, TD; Stevens, J; Brunner, J; Tannenbaum, PL; Garson, SL; Savitz, AT; Harrell, CM; Gotter, AL; Winrow, CJ; Renger, JJ; Coleman, PJ Identification of MK-8133: An orexin-2 selective receptor antagonist with favorable development properties. Bioorg Med Chem Lett 25: 2488-92 (2015)
- Riendeau, D; Aspiotis, R; Ethier, D; Gareau, Y; Grimm, EL; Guay, J; Guiral, S; Juteau, H; Mancini, JA; Méthot, N; Rubin, J; Friesen, RW Inhibitors of the inducible microsomal prostaglandin E2 synthase (mPGES-1) derived from MK-886. Bioorg Med Chem Lett 15: 3352-5 (2005)
- Liverton, NJ; Carroll, SS; Dimuzio, J; Fandozzi, C; Graham, DJ; Hazuda, D; Holloway, MK; Ludmerer, SW; McCauley, JA; McIntyre, CJ; Olsen, DB; Rudd, MT; Stahlhut, M; Vacca, JP MK-7009, a potent and selective inhibitor of hepatitis C virus NS3/4A protease. Antimicrob Agents Chemother 54: 305-11 (2009)
- Bungard, CJ; Williams, PD; Ballard, JE; Bennett, DJ; Beaulieu, C; Bahnck-Teets, C; Carroll, SS; Chang, RK; Dubost, DC; Fay, JF; Diamond, TL; Greshock, TJ; Hao, L; Holloway, MK; Felock, PJ; Gesell, JJ; Su, HP; Manikowski, JJ; McKay, DJ; Miller, M; Min, X; Molinaro, C; Moradei, OM; Nantermet, PG; Nadeau, C; Sanchez, RI; Satyanarayana, T; Shipe, WD; Singh, SK; Truong, VL; Vijayasaradhi, S; Wiscount, CM; Vacca, JP; Crane, SN; McCauley, JA Discovery of MK-8718, an HIV Protease Inhibitor Containing a Novel Morpholine Aspartate Binding Group. ACS Med Chem Lett 7: 702-7 (2016)
- Selnick, HG; Hess, JF; Tang, C; Liu, K; Schachter, JB; Ballard, JE; Marcus, J; Klein, DJ; Wang, X; Pearson, M; Savage, MJ; Kaul, R; Li, TS; Vocadlo, DJ; Zhou, Y; Zhu, Y; Mu, C; Wang, Y; Wei, Z; Bai, C; Duffy, JL; McEachern, EJ Discovery of MK-8719, a Potent O-GlcNAcase Inhibitor as a Potential Treatment for Tauopathies. J Med Chem 62: 10062-10097 (2019)
- Riendeau, D; Percival, MD; Brideau, C; Charleson, S; Dubé, D; Ethier, D; Falgueyret, JP; Friesen, RW; Gordon, R; Greig, G; Guay, J; Mancini, J; Ouellet, M; Wong, E; Xu, L; Boyce, S; Visco, D; Girard, Y; Prasit, P; Zamboni, R; Rodger, IW; Gresser, M; Ford-Hutchinson, AW; Young, RN; Chan, CC Etoricoxib (MK-0663): preclinical profile and comparison with other agents that selectively inhibit cyclooxygenase-2. J Pharmacol Exp Ther 296: 558-66 (2001)
- Bell, IM; Stump, CA; Gallicchio, SN; Staas, DD; Zartman, CB; Moore, EL; Sain, N; Urban, M; Bruno, JG; Calamari, A; Kemmerer, AL; Mosser, SD; Fandozzi, C; White, RB; Zrada, MM; Selnick, HG; Graham, SL; Vacca, JP; Kane, SA; Salvatore, CA MK-8825: a potent and selective CGRP receptor antagonist with good oral activity in rats. Bioorg Med Chem Lett 22: 3941-5 (2012)
- Paone, DV; Nguyen, DN; Shaw, AW; Burgey, CS; Potteiger, CM; Deng, JZ; Mosser, SD; Salvatore, CA; Yu, S; Roller, S; Kane, SA; Selnick, HG; Vacca, JP; Williams, TM Orally bioavailable imidazoazepanes as calcitonin gene-related peptide (CGRP) receptor antagonists: discovery of MK-2918. Bioorg Med Chem Lett 21: 2683-6 (2011)
- Layton, ME; Kern, JC; Hartingh, TJ; Shipe, WD; Raheem, I; Kandebo, M; Hayes, RP; Huszar, S; Eddins, D; Ma, B; Fuerst, J; Wollenberg, GK; Li, J; Fritzen, J; McGaughey, GB; Uslaner, JM; Smith, SM; Coleman, PJ; Cox, CD Discovery of MK-8189, a Highly Potent and Selective PDE10A Inhibitor for the Treatment of Schizophrenia. J Med Chem 66: 1157-1171 (2023)
- Huang, X; Brubaker, J; Zhou, W; Biju, PJ; Xiao, L; Shao, N; Huang, Y; Dong, L; Liu, Z; Bitar, R; Buevich, A; Jung, J; Peterson, SL; Butcher, JW; Close, J; Martinez, M; MacCoss, RN; Zhang, H; Crawford, S; McCormick, KD; Aslanian, R; Nargund, R; Correll, C; Gervais, F; Qiu, H; Yang, X; Garlisi, C; Rindgen, D; Maloney, KM; Siliphaivanh, P; Palani, A Discovery of MK-8318, a Potent and Selective CRTh2 Receptor Antagonist for the Treatment of Asthma. ACS Med Chem Lett 9: 679-684 (2018)
- Nair, AG; Zeng, Q; Selyutin, O; Rosenblum, SB; Jiang, Y; Yang, DY; Keertikar, K; Zhou, G; Dwyer, M; Kim, SH; Shankar, B; Yu, W; Tong, L; Chen, L; Mazzola, R; Caldwell, J; Tang, H; Agrawal, S; Liu, R; Kong, R; Ingravallo, P; Xia, E; Zhai, Y; Nomeir, A; Asante-Appiah, E; Kozlowski, JA MK-8325: A silyl proline-containing NS5A inhibitor with pan-genotype activity for treatment of HCV. Bioorg Med Chem Lett 28: 1954-1957 (2018)
- Nicoll-Griffith, DA; Yergey, JA; Trimble, LA; Silva, JM; Li, C; Chauret, N; Gauthier, JY; Grimm, E; Léger, S; Roy, P; Thérien, M; Wang, Z; Prasit, P; Zamboni, R; Young, RN; Brideau, C; Chan, CC; Mancini, J; Riendeau, D Synthesis, characterization, and activity of metabolites derived from the cyclooxygenase-2 inhibitor rofecoxib (MK-0966, Vioxx). Bioorg Med Chem Lett 10: 2683-6 (2000)
- Shah, SK; He, S; Guo, L; Truong, Q; Qi, H; Du, W; Lai, Z; Liu, J; Jian, T; Hong, Q; Dobbelaar, P; Ye, Z; Sherer, E; Feng, Z; Yu, Y; Wong, F; Samuel, K; Madiera, M; Karanam, BV; Reddy, VB; Mitelman, S; Tong, SX; Chicchi, GG; Tsao, KL; Trusca, D; Feng, Y; Wu, M; Shao, Q; Trujillo, ME; Eiermann, GJ; Li, C; Pachanski, M; Fernandez, G; Nelson, D; Bunting, P; Morissette, P; Volksdorf, S; Kerr, J; Zhang, BB; Howard, AD; Zhou, YP; Pasternak, A; Nargund, RP; Hagmann, WK Discovery of MK-1421, a Potent, Selective sstr3 Antagonist, as a Development Candidate for Type 2 Diabetes. ACS Med Chem Lett 6: 513-7 (2015)
- Roecker, AJ; Reger, TS; Mattern, MC; Mercer, SP; Bergman, JM; Schreier, JD; Cube, RV; Cox, CD; Li, D; Lemaire, W; Bruno, JG; Harrell, CM; Garson, SL; Gotter, AL; Fox, SV; Stevens, J; Tannenbaum, PL; Prueksaritanont, T; Cabalu, TD; Cui, D; Stellabott, J; Hartman, GD; Young, SD; Winrow, CJ; Renger, JJ; Coleman, PJ Discovery of MK-3697: a selective orexin 2 receptor antagonist (2-SORA) for the treatment of insomnia. Bioorg Med Chem Lett 24: 4884-90 (2014)
- Chobanian, HR; Guo, Y; Liu, P; Chioda, MD; Fung, S; Lanza, TJ; Chang, L; Bakshi, RK; Dellureficio, JP; Hong, Q; McLaughlin, M; Belyk, KM; Krska, SW; Makarewicz, AK; Martel, EJ; Leone, JF; Frey, L; Karanam, B; Madeira, M; Alvaro, R; Shuman, J; Salituro, G; Terebetski, JL; Jochnowitz, N; Mistry, S; McGowan, E; Hajdu, R; Rosenbach, M; Abbadie, C; Alexander, JP; Shiao, LL; Sullivan, KM; Nargund, RP; Wyvratt, MJ; Lin, LS; DeVita, RJ Discovery of MK-4409, a Novel Oxazole FAAH Inhibitor for the Treatment of Inflammatory and Neuropathic Pain. ACS Med Chem Lett 5: 717-21 (2014)
- Sebhat, IK; Franklin, C; Lo, MM; Chen, D; Jewell, JP; Miller, R; Pang, J; Palyha, O; Kan, Y; Kelly, TM; Guan, XM; Marsh, DJ; Kosinski, JA; Metzger, JM; Lyons, K; Dragovic, J; Guzzo, PR; Henderson, AJ; Reitman, ML; Nargund, RP; Wyvratt, MJ Discovery of MK-5046, a Potent, Selective Bombesin Receptor Subtype-3 Agonist for the Treatment of Obesity ACS Med Chem Lett 2: 43-47 (2011)
- Chobanian, HR; Guo, Y; Liu, P; Chioda, M; Lanza, TJ; Chang, L; Kelly, TM; Kan, Y; Palyha, O; Guan, XM; Marsh, DJ; Metzger, JM; Gorski, JN; Raustad, K; Wang, SP; Strack, AM; Miller, R; Pang, J; Madeira, M; Lyons, K; Dragovic, J; Reitman, ML; Nargund, RP; Lin, LS Discovery of MK-7725, A Potent, Selective Bombesin Receptor Subtype-3 Agonist for the Treatment of Obesity. ACS Med Chem Lett 3: 252-256 (2012)
- Lovering, F; Kirincich, S; Wang, W; Combs, K; Resnick, L; Sabalski, JE; Butera, J; Liu, J; Parris, K; Telliez, JB Identification and SAR of squarate inhibitors of mitogen activated protein kinase-activated protein kinase 2 (MK-2). Bioorg Med Chem 17: 3342-51 (2009)
- Kobayashi, K; Tsujita, T; Ito, H; Ozaki, S; Tani, T; Ishii, Y; Okuda, S; Tadano, K; Fukuroda, T; Ohta, H; Okamoto, O Identification of MK-1925: a selective, orally active and brain-penetrable opioid receptor-like 1 (ORL1) antagonist. Bioorg Med Chem Lett 19: 4729-32 (2009)
- Dorsey, BD; McDonough, C; McDaniel, SL; Levin, RB; Newton, CL; Hoffman, JM; Darke, PL; Zugay-Murphy, JA; Emini, EA; Schleif, WA; Olsen, DB; Stahlhut, MW; Rutkowski, CA; Kuo, LC; Lin, JH; Chen, IW; Michelson, SR; Holloway, MK; Huff, JR; Vacca, JP Identification of MK-944a: a second clinical candidate from the hydroxylaminepentanamide isostere series of HIV protease inhibitors. J Med Chem 43: 3386-99 (2000)
- He, S; Ye, Z; Truong, Q; Shah, S; Du, W; Guo, L; Dobbelaar, PH; Lai, Z; Liu, J; Jian, T; Qi, H; Bakshi, RK; Hong, Q; Dellureficio, J; Pasternak, A; Feng, Z; deJesus, R; Yang, L; Reibarkh, M; Bradley, SA; Holmes, MA; Ball, RG; Ruck, RT; Huffman, MA; Wong, F; Samuel, K; Reddy, VB; Mitelman, S; Tong, SX; Chicchi, GG; Tsao, KL; Trusca, D; Wu, M; Shao, Q; Trujillo, ME; Eiermann, GJ; Li, C; Zhang, BB; Howard, AD; Zhou, YP; Nargund, RP; Hagmann, WK The Discovery of MK-4256, a Potent SSTR3 Antagonist as a Potential Treatment of Type 2 Diabetes. ACS Med Chem Lett 3: 484-489 (2012)
- Bell, IM; Gallicchio, SN; Stump, CA; Bruno, JG; Fan, H; Gantert, LT; Hostetler, ED; Kemmerer, AL; McWherter, M; Moore, EL; Mosser, SD; Purcell, ML; Riffel, K; Salvatore, CA; Sanabria-Bohórquez, S; Staas, DD; White, RB; Williams, M; Zartman, CB; Cook, JJ; Hargreaves, RJ; Kane, SA; Graham, SL; Selnick, HG [(11)C]MK-4232: The First Positron Emission Tomography Tracer for the Calcitonin Gene-Related Peptide Receptor. ACS Med Chem Lett 4: 863-8 (2013)
- Chang, W; Altman, MD; Lesburg, CA; Perera, SA; Piesvaux, JA; Schroeder, GK; Wyss, DF; Cemerski, S; Chen, Y; DiNunzio, E; Haidle, AM; Ho, T; Kariv, I; Knemeyer, I; Kopinja, JE; Lacey, BM; Laskey, J; Lim, J; Long, BJ; Ma, Y; Maddess, ML; Pan, BS; Presland, JP; Spooner, E; Steinhuebel, D; Truong, Q; Zhang, Z; Fu, J; Addona, GH; Northrup, AB; Parmee, E; Tata, JR; Bennett, DJ; Cumming, JN; Siu, T; Trotter, BW Discovery of MK-1454: A Potent Cyclic Dinucleotide Stimulator of Interferon Genes Agonist for the Treatment of Cancer. J Med Chem 65: 5675-5689 (2022)
- Palani, A; Nawrocki, AR; Orvieto, F; Bianchi, E; Mandić, E; Pessi, A; Huang, C; Deng, Q; Toussaint, N; Walsh, E; Reddy, V; Ashley, E; He, H; Mumick, S; Hawes, B; Marsh, D; Erion, M; Nargund, R; Carrington, PE Discovery of MK-1462: GLP-1 and Glucagon Receptor Dual Agonist for the Treatment of Obesity and Diabetes. ACS Med Chem Lett 13: 1248-1254 (2022)
- Tang, H; Zhu, Y; Teumelsan, N; Walsh, SP; Shahripour, A; Priest, BT; Swensen, AM; Felix, JP; Brochu, RM; Bailey, T; Thomas-Fowlkes, B; Pai, LY; Hampton, C; Corona, A; Hernandez, M; Metzger, J; Forrest, M; Zhou, X; Owens, K; Tong, V; Parmee, E; Roy, S; Kaczorowski, GJ; Yang, L; Alonso-Galicia, M; Garcia, ML; Pasternak, A Discovery of MK-7145, an Oral Small Molecule ROMK Inhibitor for the Treatment of Hypertension and Heart Failure. ACS Med Chem Lett 7: 697-701 (2016)
- Cox, CD; Hostetler, ED; Flores, BA; Evelhoch, JL; Fan, H; Gantert, L; Holahan, M; Eng, W; Joshi, A; McGaughey, G; Meng, X; Purcell, M; Raheem, IT; Riffel, K; Yan, Y; Renger, JJ; Smith, SM; Coleman, PJ Discovery of [¹¹C]MK-8193 as a PET tracer to measure target engagement of phosphodiesterase 10A (PDE10A) inhibitors. Bioorg Med Chem Lett 25: 4893-8 (2015)
- Biftu, T; Sinha-Roy, R; Chen, P; Qian, X; Feng, D; Kuethe, JT; Scapin, G; Gao, YD; Yan, Y; Krueger, D; Bak, A; Eiermann, G; He, J; Cox, J; Hicks, J; Lyons, K; He, H; Salituro, G; Tong, S; Patel, S; Doss, G; Petrov, A; Wu, J; Xu, SS; Sewall, C; Zhang, X; Zhang, B; Thornberry, NA; Weber, AE Omarigliptin (MK-3102): a novel long-acting DPP-4 inhibitor for once-weekly treatment of type 2 diabetes. J Med Chem 57: 3205-12 (2014)
- Sahoo, SP; Graham, DW; Acton, J; Biftu, T; Bugianesi, RL; Girotra, NN; Kuo, C; Ponpipom, MM; Doebber, TW; Wu, MS; Hwang, S; Lam, M; MacIntyre, D; Bach, TJ; Luell, S; Meurer, R; Davies, P; Alberts, AW; Chabala, JC Synthesis and biological activity of MK 287 (L-680,573): a potent, specific and orally active paf receptor antagonist Bioorg Med Chem Lett 1: 327-332 (1991)
- Vachal, P; Duffy, JL; Campeau, LC; Amin, RP; Mitra, K; Murphy, BA; Shao, PP; Sinclair, PJ; Ye, F; Katipally, R; Lu, Z; Ondeyka, D; Chen, YH; Zhao, K; Sun, W; Tyagarajan, S; Bao, J; Wang, SP; Cote, J; Lipardi, C; Metzger, D; Leung, D; Hartmann, G; Wollenberg, GK; Liu, J; Tan, L; Xu, Y; Chen, Q; Liu, G; Blaustein, RO; Johns, DG Invention of MK-8262, a Cholesteryl Ester Transfer Protein (CETP) Inhibitor Backup to Anacetrapib with Best-in-Class Properties. J Med Chem 64: 13215-13258 (2021)
- Chen, H; Blizzard, TA; Kim, S; Wu, J; Young, K; Park, YW; Ogawa, AM; Raghoobar, S; Painter, RE; Wisniewski, D; Hairston, N; Fitzgerald, P; Sharma, N; Scapin, G; Lu, J; Hermes, J; Hammond, ML Side chain SAR of bicyclicß-lactamase inhibitors (BLIs). 2. N-Alkylated and open chain analogs of MK-8712. Bioorg Med Chem Lett 21: 4267-70 (2011)
- Gallant, M; Aspiotis, R; Day, S; Dias, R; Dubé, D; Dubé, L; Friesen, RW; Girard, M; Guay, D; Hamel, P; Huang, Z; Lacombe, P; Laliberté, S; Lévesque, JF; Liu, S; Macdonald, D; Mancini, J; Nicholson, DW; Styhler, A; Townson, K; Waters, K; Young, RN; Girard, Y Discovery of MK-0952, a selective PDE4 inhibitor for the treatment of long-term memory loss and mild cognitive impairment. Bioorg Med Chem Lett 20: 6387-93 (2010)
- Kattar, SD; Gulati, A; Margrey, KA; Keylor, MH; Ardolino, M; Yan, X; Johnson, R; Palte, RL; McMinn, SE; Nogle, L; Su, J; Xiao, D; Piesvaux, J; Lee, S; Hegde, LG; Woodhouse, JD; Faltus, R; Moy, LY; Xiong, T; Ciaccio, PJ; Pearson, K; Patel, M; Otte, KM; Leyns, CEG; Kennedy, ME; Bennett, DJ; DiMauro, EF; Fell, MJ; Fuller, PH Discovery of MK-1468: A Potent, Kinome-Selective, Brain-Penetrant Amidoisoquinoline LRRK2 Inhibitor for the Potential Treatment of Parkinson's Disease. J Med Chem 66: 14912-14927 (2023)
- Neelamkavil, SF; Stamford, AW; Kowalski, T; Biswas, D; Boyle, C; Chackalamannil, S; Xia, Y; Jayne, C; Neustadt, B; Hao, J; Liu, H; Dai, X; Baker, H; Hawes, B; O'Neill, K; Tang, H; Greenlee, WJ Discovery of MK-8282 as a Potent G-Protein-Coupled Receptor 119 Agonist for the Treatment of Type 2 Diabetes. ACS Med Chem Lett 9: 457-461 (2018)
- Robichaud, J; Black, WC; Thérien, M; Paquet, J; Oballa, RM; Bayly, CI; McKay, DJ; Wang, Q; Isabel, E; Léger, S; Mellon, C; Kimmel, DB; Wesolowski, G; Percival, MD; Massé, F; Desmarais, S; Falgueyret, JP; Crane, SN Identification of a nonbasic, nitrile-containing cathepsin K inhibitor (MK-1256) that is efficacious in a monkey model of osteoporosis. J Med Chem 51: 6410-20 (2008)
- Trujillo, JI; Meyers, MJ; Anderson, DR; Hegde, S; Mahoney, MW; Vernier, WF; Buchler, IP; Wu, KK; Yang, S; Yang, S; Hartmann, SJ; Reitz, DB Novel tetrahydro-beta-carboline-1-carboxylic acids as inhibitors of mitogen activated protein kinase-activated protein kinase 2 (MK-2). Bioorg Med Chem Lett 17: 4657-63 (2007)
- Lin, S; Malkani, S; Lombardo, M; Yang, L; Mills, SG; Chapman, K; Thompson, JE; Zhang, WX; Wang, R; Cubbon, RM; O'Neill, EA; Hale, JJ Design, synthesis, and biological evaluation of aminopyrazine derivatives as inhibitors of mitogen-activated protein kinase-activated protein kinase 2 (MK-2). Bioorg Med Chem Lett 25: 5402-8 (2015)
- Labelle, M; Belley, M; Gareau, Y; Gauthier, JY; Guay, D; Gordon, R; Grossman, SG; Jones, TR; Leblanc, Y; McAuliffe, M; McFarlane, C; Masson, P; Metters, KM; Ouimet, N; Patrick, DH; Piechuta, H; Rochette, C; Sawyer, N; Xiang, YB; Pickett, CB Discovery of MK-0476, a potent and orally active leukotriene D4 receptor antagonist devoid of peroxisomal enxyme induction Bioorg Med Chem Lett 5: 283-288 (1995)
- Harrison, ST; Mulhearn, J; Wolkenberg, SE; Miller, PJ; O'Malley, SS; Zeng, Z; Williams, DL; Hostetler, ED; Sanabria-Bohórquez, S; Gammage, L; Fan, H; Sur, C; Culberson, JC; Hargreaves, RJ; Cook, JJ; Hartman, GD; Barrow, JC Synthesis and Evaluation of 5-Fluoro-2-aryloxazolo[5,4-b]pyridines as β-Amyloid PET Ligands and Identification of MK-3328. ACS Med Chem Lett 2: 498-502 (2011)
- Prasit, P; Wang, Z; Brideau, C; Chan, CC; Charleson, S; Cromlish, W; Ethier, D; Evans, JF; Ford-Hutchinson, AW; Gauthier, JY; Gordon, R; Guay, J; Gresser, M; Kargman, S; Kennedy, B; Leblanc, Y; Léger, S; Mancini, J; O'Neill, GP; Ouellet, M; Percival, MD; Perrier, H; Riendeau, D; Rodger, I; Zamboni, R The discovery of rofecoxib, [MK 966, Vioxx, 4-(4'-methylsulfonylphenyl)-3-phenyl-2(5H)-furanone], an orally active cyclooxygenase-2-inhibitor. Bioorg Med Chem Lett 9: 1773-8 (1999)
- Chakravarty, PK; Naylor, EM; Chen, A; Chang, RS; Chen, TB; Faust, KA; Lotti, VJ; Kivlighn, SD; Gable, RA; Zingaro, GJ A highly potent, orally active imidazo[4,5-b]pyridine biphenylacylsulfonamide (MK-996; L-159,282): a new AT1-selective angiotensin II receptor antagonist. J Med Chem 37: 4068-72 (1995)
- Chauret, N; Yergey, JA; Brideau, C; Friesen, RW; Mancini, J; Riendeau, D; Silva, J; Styhler, A; Trimble, LA; Nicoll-Griffith, DA In vitro metabolism considerations, including activity testing of metabolites, in the discovery and selection of the COX-2 inhibitor etoricoxib (MK-0663). Bioorg Med Chem Lett 11: 1059-62 (2001)
- Alexandre, FR; Rahali, R; Rahali, H; Guillon, S; Convard, T; Fillgrove, K; Lai, MT; Meillon, JC; Xu, M; Small, J; Dousson, CB; Raheem, IT Synthesis and Antiviral Evaluation of Carbocyclic Nucleoside Analogs of Nucleoside Reverse Transcriptase Translocation Inhibitor MK-8591 (4'-Ethynyl-2-fluoro-2'-deoxyadenosine). J Med Chem 61: 9218-9228 (2018)
- Bakshi, RK; Patel, GF; Rasmusson, GH; Baginsky, WF; Cimis, G; Ellsworth, K; Chang, B; Bull, H; Tolman, RL; Harris, GS 4,7 beta-Dimethyl-4-azacholestan-3-one (MK-386) and related 4-azasteroids as selective inhibitors of human type 1 5 alpha-reductase. J Med Chem 37: 3871-4 (1994)
- Lin, S; Lombardo, M; Malkani, S; Hale, JJ; Mills, SG; Chapman, K; Thompson, JE; Zhang, WX; Wang, R; Cubbon, RM; O'Neill, EA; Luell, S; Carballo-Jane, E; Yang, L Novel 1-(2-aminopyrazin-3-yl)methyl-2-thioureas as potent inhibitors of mitogen-activated protein kinase-activated protein kinase 2 (MK-2). Bioorg Med Chem Lett 19: 3238-42 (2009)
- Oballa, RM; Belair, L; Black, WC; Bleasby, K; Chan, CC; Desroches, C; Du, X; Gordon, R; Guay, J; Guiral, S; Hafey, MJ; Hamelin, E; Huang, Z; Kennedy, B; Lachance, N; Landry, F; Li, CS; Mancini, J; Normandin, D; Pocai, A; Powell, DA; Ramtohul, YK; Skorey, K; Sørensen, D; Sturkenboom, W; Styhler, A; Waddleton, DM; Wang, H; Wong, S; Xu, L; Zhang, L Development of a liver-targeted stearoyl-CoA desaturase (SCD) inhibitor (MK-8245) to establish a therapeutic window for the treatment of diabetes and dyslipidemia. J Med Chem 54: 5082-96 (2011)
- Katz, JD; Jewell, JP; Guerin, DJ; Lim, J; Dinsmore, CJ; Deshmukh, SV; Pan, BS; Marshall, CG; Lu, W; Altman, MD; Dahlberg, WK; Davis, L; Falcone, D; Gabarda, AE; Hang, G; Hatch, H; Holmes, R; Kunii, K; Lumb, KJ; Lutterbach, B; Mathvink, R; Nazef, N; Patel, SB; Qu, X; Reilly, JF; Rickert, KW; Rosenstein, C; Soisson, SM; Spencer, KB; Szewczak, AA; Walker, D; Wang, W; Young, J; Zeng, Q Discovery of a 5H-benzo[4,5]cyclohepta[1,2-b]pyridin-5-one (MK-2461) inhibitor of c-Met kinase for the treatment of cancer. J Med Chem 54: 4092-108 (2011)
- Chan, CC; Boyce, S; Brideau, C; Charleson, S; Cromlish, W; Ethier, D; Evans, J; Ford-Hutchinson, AW; Forrest, MJ; Gauthier, JY; Gordon, R; Gresser, M; Guay, J; Kargman, S; Kennedy, B; Leblanc, Y; Leger, S; Mancini, J; O'Neill, GP; Ouellet, M; Patrick, D; Percival, MD; Perrier, H; Prasit, P; Rodger, I Rofecoxib [Vioxx, MK-0966; 4-(4'-methylsulfonylphenyl)-3-phenyl-2-(5H)-furanone]: a potent and orally active cyclooxygenase-2 inhibitor. Pharmacological and biochemical profiles. J Pharmacol Exp Ther 290: 551-60 (1999)
- Boatman, PD; Lauring, B; Schrader, TO; Kasem, M; Johnson, BR; Skinner, P; Jung, JK; Xu, J; Cherrier, MC; Webb, PJ; Semple, G; Sage, CR; Knudsen, J; Chen, R; Luo, WL; Caro, L; Cote, J; Lai, E; Wagner, J; Taggart, AK; Carballo-Jane, E; Hammond, M; Colletti, SL; Tata, JR; Connolly, DT; Waters, MG; Richman, JG (1aR,5aR)1a,3,5,5a-Tetrahydro-1H-2,3-diaza-cyclopropa[a]pentalene-4-carboxylic acid (MK-1903): a potent GPR109a agonist that lowers free fatty acids in humans. J Med Chem 55: 3644-66 (2012)
- Guay, D; Boulet, L; Friesen, RW; Girard, M; Hamel, P; Huang, Z; Laliberté, F; Laliberté, S; Mancini, JA; Muise, E; Pon, D; Styhler, A Optimization and structure-activity relationship of a series of 1-phenyl-1,8-naphthyridin-4-one-3-carboxamides: identification of MK-0873, a potent and effective PDE4 inhibitor. Bioorg Med Chem Lett 18: 5554-8 (2008)
- Shen, HC; Ding, FX; Raghavan, S; Deng, Q; Luell, S; Forrest, MJ; Carballo-Jane, E; Wilsie, LC; Krsmanovic, ML; Taggart, AK; Wu, KK; Wu, TJ; Cheng, K; Ren, N; Cai, TQ; Chen, Q; Wang, J; Wolff, MS; Tong, X; Holt, TG; Waters, MG; Hammond, ML; Tata, JR; Colletti, SL Discovery of a biaryl cyclohexene carboxylic acid (MK-6892): a potent and selective high affinity niacin receptor full agonist with reduced flushing profiles in animals as a preclinical candidate. J Med Chem 53: 2666-70 (2010)
- Sturino, CF; O'Neill, G; Lachance, N; Boyd, M; Berthelette, C; Labelle, M; Li, L; Roy, B; Scheigetz, J; Tsou, N; Aubin, Y; Bateman, KP; Chauret, N; Day, SH; Lévesque, JF; Seto, C; Silva, JH; Trimble, LA; Carriere, MC; Denis, D; Greig, G; Kargman, S; Lamontagne, S; Mathieu, MC; Sawyer, N; Slipetz, D; Abraham, WM; Jones, T; McAuliffe, M; Piechuta, H; Nicoll-Griffith, DA; Wang, Z; Zamboni, R; Young, RN; Metters, KM Discovery of a potent and selective prostaglandin D2 receptor antagonist, [(3R)-4-(4-chloro-benzyl)-7-fluoro-5-(methylsulfonyl)-1,2,3,4-tetrahydrocyclopenta[b]indol-3-yl]-acetic acid (MK-0524). J Med Chem 50: 794-806 (2007)
- Blouin, M; Han, Y; Burch, J; Farand, J; Mellon, C; Gaudreault, M; Wrona, M; Lévesque, JF; Denis, D; Mathieu, MC; Stocco, R; Vigneault, E; Therien, A; Clark, P; Rowland, S; Xu, D; O'Neill, G; Ducharme, Y; Friesen, R The discovery of 4-{1-[({2,5-dimethyl-4-[4-(trifluoromethyl)benzyl]-3-thienyl}carbonyl)amino]cyclopropyl}benzoic acid (MK-2894), a potent and selective prostaglandin E2 subtype 4 receptor antagonist. J Med Chem 53: 2227-38 (2010)
- Semple, G; Skinner, PJ; Gharbaoui, T; Shin, YJ; Jung, JK; Cherrier, MC; Webb, PJ; Tamura, SY; Boatman, PD; Sage, CR; Schrader, TO; Chen, R; Colletti, SL; Tata, JR; Waters, MG; Cheng, K; Taggart, AK; Cai, TQ; Carballo-Jane, E; Behan, DP; Connolly, DT; Richman, JG 3-(1H-tetrazol-5-yl)-1,4,5,6-tetrahydro-cyclopentapyrazole (MK-0354): a partial agonist of the nicotinic acid receptor, G-protein coupled receptor 109a, with antilipolytic but no vasodilatory activity in mice. J Med Chem 51: 5101-8 (2008)
- Jones, P; Altamura, S; Boueres, J; Ferrigno, F; Fonsi, M; Giomini, C; Lamartina, S; Monteagudo, E; Ontoria, JM; Orsale, MV; Palumbi, MC; Pesci, S; Roscilli, G; Scarpelli, R; Schultz-Fademrecht, C; Toniatti, C; Rowley, M Discovery of 2-{4-[(3S)-piperidin-3-yl]phenyl}-2H-indazole-7-carboxamide (MK-4827): a novel oral poly(ADP-ribose)polymerase (PARP) inhibitor efficacious in BRCA-1 and -2 mutant tumors. J Med Chem 52: 7170-85 (2009)
- Lin, LS; Lanza, T; Jewell, JP; Liu, P; Jones, C; Kieczykowski, GR; Treonze, K; Si, Q; Manior, S; Koo, G; Tong, X; Wang, J; Schuelke, A; Pivnichny, J; Wang, R; Raab, C; Vincent, S; Davies, P; Maccoss, M; Mumford, RA; Hagmann, WK Discovery of N-{N-[(3-cyanophenyl)sulfonyl]-4(R)-cyclobutylamino-(L)-prolyl}-4-[(3',5'-dichloroisonicotinoyl) amino]-(L)-phenylalanine (MK-0668), an extremely potent and orally active antagonist of very late antigen-4. J Med Chem 52: 3449-52 (2009)
- Venkatraman, S; Lebsack, AD; Alves, K; Gardner, MF; James, J; Lingham, RB; Maniar, S; Mumford, RA; Si, Q; Stock, N; Treonze, KM; Wang, B; Zunic, J; Munoz, B Discovery of N-{N-[(3-cyanobenzene) sulfonyl]-4(R)-(3,3-difluoropiperidin-1-yl)-(l)-prolyl}-4-[(3',5'-dichloro-isonicotinoyl) amino]-(l)-phenylalanine (MK-0617), a highly potent and orally active VLA-4 antagonist. Bioorg Med Chem Lett 19: 5803-6 (2009)
- Lin, LS; Ha, S; Ball, RG; Tsou, NN; Castonguay, LA; Doss, GA; Fong, TM; Shen, CP; Xiao, JC; Goulet, MT; Hagmann, WK Conformational analysis and receptor docking of N-[(1S,2S)-3-(4-chlorophenyl)-2-(3-cyanophenyl)-1-methylpropyl]-2-methyl-2-{[5-(trifluoromethyl)pyridin-2-yl]oxy}propanamide (taranabant, MK-0364), a novel, acyclic cannabinoid-1 receptor inverse agonist. J Med Chem 51: 2108-14 (2008)
- Tucker, TJ; Sisko, JT; Tynebor, RM; Williams, TM; Felock, PJ; Flynn, JA; Lai, MT; Liang, Y; McGaughey, G; Liu, M; Miller, M; Moyer, G; Munshi, V; Perlow-Poehnelt, R; Prasad, S; Reid, JC; Sanchez, R; Torrent, M; Vacca, JP; Wan, BL; Yan, Y Discovery of 3-{5-[(6-amino-1H-pyrazolo[3,4-b]pyridine-3-yl)methoxy]-2-chlorophenoxy}-5-chlorobenzonitrile (MK-4965): a potent, orally bioavailable HIV-1 non-nucleoside reverse transcriptase inhibitor with improved potency against key mutant viruses. J Med Chem 51: 6503-11 (2008)
- Lin, LS; Lanza, TJ; Jewell, JP; Liu, P; Shah, SK; Qi, H; Tong, X; Wang, J; Xu, SS; Fong, TM; Shen, CP; Lao, J; Xiao, JC; Shearman, LP; Stribling, DS; Rosko, K; Strack, A; Marsh, DJ; Feng, Y; Kumar, S; Samuel, K; Yin, W; Van der Ploeg, LH; Goulet, MT; Hagmann, WK Discovery of N-[(1S,2S)-3-(4-Chlorophenyl)-2- (3-cyanophenyl)-1-methylpropyl]-2-methyl-2- {[5-(trifluoromethyl)pyridin-2-yl]oxy}propanamide (MK-0364), a novel, acyclic cannabinoid-1 receptor inverse agonist for the treatment of obesity. J Med Chem 49: 7584-7 (2006)
- Xiong, Y; Guo, J; Candelore, MR; Liang, R; Miller, C; Dallas-Yang, Q; Jiang, G; McCann, PE; Qureshi, SA; Tong, X; Xu, SS; Shang, J; Vincent, SH; Tota, LM; Wright, MJ; Yang, X; Zhang, BB; Tata, JR; Parmee, ER Discovery of a novel glucagon receptor antagonist N-[(4-{(1S)-1-[3-(3, 5-dichlorophenyl)-5-(6-methoxynaphthalen-2-yl)-1H-pyrazol-1-yl]ethyl}phenyl)carbonyl]-ß-alanine (MK-0893) for the treatment of type II diabetes. J Med Chem 55: 6137-48 (2012)
- Cox, CD; Breslin, MJ; Whitman, DB; Schreier, JD; McGaughey, GB; Bogusky, MJ; Roecker, AJ; Mercer, SP; Bednar, RA; Lemaire, W; Bruno, JG; Reiss, DR; Harrell, CM; Murphy, KL; Garson, SL; Doran, SM; Prueksaritanont, T; Anderson, WB; Tang, C; Roller, S; Cabalu, TD; Cui, D; Hartman, GD; Young, SD; Koblan, KS; Winrow, CJ; Renger, JJ; Coleman, PJ Discovery of the dual orexin receptor antagonist [(7R)-4-(5-chloro-1,3-benzoxazol-2-yl)-7-methyl-1,4-diazepan-1-yl][5-methyl-2-(2H-1,2,3-triazol-2-yl)phenyl]methanone (MK-4305) for the treatment of insomnia. J Med Chem 53: 5320-32 (2010)
- Linders, JT; Monn, JA; Mattson, MV; George, C; Jacobson, AE; Rice, KC Synthesis and binding properties of MK-801 isothiocyanates; (+)-3-isothiocyanato-5-methyl-10,11-dihydro-5H-dibenzo[a,d]cyclohepten- 5,10-imine hydrochloride: a new, potent and selective electrophilic affinity ligand for the NMDA receptor-coupled phencyclidine binding site. J Med Chem 36: 2499-507 (1993)
- Narjes, F; Crescenzi, B; Ferrara, M; Habermann, J; Colarusso, S; Ferreira, Mdel R; Stansfield, I; Mackay, AC; Conte, I; Ercolani, C; Zaramella, S; Palumbi, MC; Meuleman, P; Leroux-Roels, G; Giuliano, C; Fiore, F; Di Marco, S; Baiocco, P; Koch, U; Migliaccio, G; Altamura, S; Laufer, R; De Francesco, R; Rowley, M Discovery of (7R)-14-cyclohexyl-7-{[2-(dimethylamino)ethyl](methyl) amino}-7,8-dihydro-6H-indolo[1,2-e][1,5]benzoxazocine-11-carboxylic acid (MK-3281), a potent and orally bioavailable finger-loop inhibitor of the hepatitis C virus NS5B polymerase. J Med Chem 54: 289-301 (2011)
- Debenham, JS; Madsen-Duggan, C; Clements, MJ; Walsh, TF; Kuethe, JT; Reibarkh, M; Salowe, SP; Sonatore, LM; Hajdu, R; Milligan, JA; Visco, DM; Zhou, D; Lingham, RB; Stickens, D; DeMartino, JA; Tong, X; Wolff, M; Pang, J; Miller, RR; Sherer, EC; Hale, JJ Discovery of N-[Bis(4-methoxyphenyl)methyl]-4-hydroxy-2-(pyridazin-3-yl)pyrimidine-5-carboxamide (MK-8617), an Orally Active Pan-Inhibitor of Hypoxia-Inducible Factor Prolyl Hydroxylase 1-3 (HIF PHD1-3) for the Treatment of Anemia. J Med Chem 59: 11039-11049 (2016)
- Lan, P; Romero, FA; Wodka, D; Kassick, AJ; Dang, Q; Gibson, T; Cashion, D; Zhou, G; Chen, Y; Zhang, X; Zhang, A; Li, Y; Trujillo, ME; Shao, Q; Wu, M; Xu, S; He, H; MacKenna, D; Staunton, J; Chapman, KT; Weber, A; Sebhat, IK; Makara, GM Hit-to-Lead Optimization and Discovery of 5-((5-([1,1'-Biphenyl]-4-yl)-6-chloro-1H-benzo[d]imidazol-2-yl)oxy)-2-methylbenzoic Acid (MK-3903): A Novel Class of Benzimidazole-Based Activators of AMP-Activated Protein Kinase. J Med Chem 60: 9040-9052 (2017)
- Yan, L; Huo, P; Debenham, JS; Madsen-Duggan, CB; Lao, J; Chen, RZ; Xiao, JC; Shen, CP; Stribling, DS; Shearman, LP; Strack, AM; Tsou, N; Ball, RG; Wang, J; Tong, X; Bateman, TJ; Reddy, VB; Fong, TM; Hale, JJ Discovery of N-[(4R)-6-(4-chlorophenyl)-7-(2,4-dichlorophenyl)-2,2-dimethyl-3,4-dihydro-2H-pyrano[2,3-b]pyridin-4-yl]-5-methyl-1H-pyrazole-3-carboxamide (MK-5596) as a novel cannabinoid-1 receptor (CB1R) inverse agonist for the treatment of obesity. J Med Chem 53: 4028-37 (2010)
- Kinzel, O; Alfieri, A; Altamura, S; Brunetti, M; Bufali, S; Colaceci, F; Ferrigno, F; Filocamo, G; Fonsi, M; Gallinari, P; Malancona, S; Hernando, JI; Monteagudo, E; Orsale, MV; Palumbi, MC; Pucci, V; Rowley, M; Sasso, R; Scarpelli, R; Steinkühler, C; Jones, P Identification of MK-5710 ((8aS)-8a-methyl-1,3-dioxo-2-[(1S,2R)-2-phenylcyclo- propyl]-N-(1-phenyl-1H-pyrazol-5-yl)hexahydro-imidazo[1,5-a]pyrazine-7(1H)-carboxamide), a potent smoothened antagonist for use in Hedgehog pathway dependent malignancies, part 2. Bioorg Med Chem Lett 21: 4429-35 (2011)
- Malancona, S; Altamura, S; Filocamo, G; Kinzel, O; Hernando, JI; Rowley, M; Scarpelli, R; Steinkühler, C; Jones, P Identification of MK-5710 ((8aS)-8a-methyl-1,3-dioxo-2-[(1S,2R)-2-phenylcyclopropyl]-N-(1-phenyl-1H-pyrazol-5-yl)hexahydroimid azo[1,5-a]pyrazine-7(1H)-carboxamide), a potent smoothened antagonist for use in Hedgehog pathway dependent malignancies, part 1. Bioorg Med Chem Lett 21: 4422-8 (2011)
- Cox, CD; Coleman, PJ; Breslin, MJ; Whitman, DB; Garbaccio, RM; Fraley, ME; Buser, CA; Walsh, ES; Hamilton, K; Schaber, MD; Lobell, RB; Tao, W; Davide, JP; Diehl, RE; Abrams, MT; South, VJ; Huber, HE; Torrent, M; Prueksaritanont, T; Li, C; Slaughter, DE; Mahan, E; Fernandez-Metzler, C; Yan, Y; Kuo, LC; Kohl, NE; Hartman, GD Kinesin spindle protein (KSP) inhibitors. 9. Discovery of (2S)-4-(2,5-difluorophenyl)-n-[(3R,4S)-3-fluoro-1-methylpiperidin-4-yl]-2-(hydroxymethyl)-N-methyl-2-phenyl-2,5-dihydro-1H-pyrrole-1-carboxamide (MK-0731) for the treatment of taxane-refractory cancer. J Med Chem 51: 4239-52 (2008)
- Acton, JJ; Akiyama, TE; Chang, CH; Colwell, L; Debenham, S; Doebber, T; Einstein, M; Liu, K; McCann, ME; Moller, DE; Muise, ES; Tan, Y; Thompson, JR; Wong, KK; Wu, M; Xu, L; Meinke, PT; Berger, JP; Wood, HB Discovery of (2R)-2-(3-{3-[(4-Methoxyphenyl)carbonyl]-2-methyl-6-(trifluoromethoxy)-1H-indol-1-yl}phenoxy)butanoic acid (MK-0533): a novel selective peroxisome proliferator-activated receptor gamma modulator for the treatment of type 2 diabetes mellitus with a reduced potential to increase plasma a J Med Chem 52: 3846-54 (2009)
- Northrup, AB; Katcher, MH; Altman, MD; Chenard, M; Daniels, MH; Deshmukh, SV; Falcone, D; Guerin, DJ; Hatch, H; Li, C; Lu, W; Lutterbach, B; Allison, TJ; Patel, SB; Reilly, JF; Reutershan, M; Rickert, KW; Rosenstein, C; Soisson, SM; Szewczak, AA; Walker, D; Wilson, K; Young, JR; Pan, BS; Dinsmore, CJ Discovery of 1-[3-(1-methyl-1H-pyrazol-4-yl)-5-oxo-5H-benzo[4,5]cyclohepta[1,2-b]pyridin-7-yl]-N-(pyridin-2-ylmethyl)methanesulfonamide (MK-8033): A Specific c-Met/Ron dual kinase inhibitor with preferential affinity for the activated state of c-Met. J Med Chem 56: 2294-310 (2013)
- Paone, DV; Shaw, AW; Nguyen, DN; Burgey, CS; Deng, JZ; Kane, SA; Koblan, KS; Salvatore, CA; Mosser, SD; Johnston, VK; Wong, BK; Miller-Stein, CM; Hershey, JC; Graham, SL; Vacca, JP; Williams, TM Potent, orally bioavailable calcitonin gene-related peptide receptor antagonists for the treatment of migraine: discovery of N-[(3R,6S)-6-(2,3-difluorophenyl)-2-oxo-1- (2,2,2-trifluoroethyl)azepan-3-yl]-4- (2-oxo-2,3-dihydro-1H-imidazo[4,5-b]pyridin- 1-yl)piperidine-1-carboxamide (MK-0974). J Med Chem 50: 5564-7 (2007)
- H., FP; Anmol, G; D., KS; H., KM; A., MK; Xin, Y WIPO WO2022093881 (2022)
- Noha, RM; Abdelhameid, MK; Ismail, MM; Mohammed, MR; Salwa, E Eur J Med Chem 209: (2021)
- Boy, KM; Guernon, JM; Zuev, DS; Xu, L; Zhang, Y; Shi, J; Marcin, LR; Higgins, MA; Wu, YJ; Krishnananthan, S; Li, J; Trehan, A; Smith, D; Toyn, JH; Meredith, JE; Burton, CR; Kimura, SR; Zvyaga, T; Zhuo, X; Lentz, KA; Grace, JE; Denton, R; Morrison, JS; Mathur, A; Albright, CF; Ahlijanian, MK; Olson, RE; Thompson, LA; Macor, JE ACS Med Chem Lett 10: 312-317 (2019)
- Murph, MM; Jiang, GW; Altman, MK; Jia, W; Nguyen, DT; Fambrough, JM; Hardman, WJ; Nguyen, HT; Tran, SK; Alshamrani, AA; Madan, D; Zhang, J; Prestwich, GD Bioorg Med Chem 23: 5999-6013 (2015)
- Ameriks, MK; Rech, JC; Savall, BM US Patent US10047092 (2018)
- O'Brien, PM; Sliskovic, DR; Anderson, MK; Bousley, RF; Krause, BR; Stanfield, RL Bioorg Med Chem Lett 5: 289-294 (1995)
- Kumar, K; Man-Un Ung, P; Wang, P; Wang, H; Li, H; Andrews, MK; Stewart, AF; Schlessinger, A; DeVita, RJ Eur J Med Chem 157: 1005-1016 (2018)
- Li, K; McGee, LR; Fisher, B; Sudom, A; Liu, J; Rubenstein, SM; Anwer, MK; Cushing, TD; Shin, Y; Ayres, M; Lee, F; Eksterowicz, J; Faulder, P; Waszkowycz, B; Plotnikova, O; Farrelly, E; Xiao, SH; Chen, G; Wang, Z Bioorg Med Chem Lett 23: 1238-44 (2013)
- Azim, MK; Ahmed, W; Khan, IA; Rao, NA; Khan, KM Bioorg Med Chem Lett 18: 3011-5 (2008)
- Chianelli, D; Rucker, PV; Roland, J; Tully, DC; Nelson, J; Liu, X; Bursulaya, B; Hernandez, ED; Wu, J; Prashad, M; Schlama, T; Liu, Y; Chu, A; Schmeits, J; Huang, DJ; Hill, R; Bao, D; Zoll, J; Kim, Y; Groessl, T; McNamara, P; Liu, B; Richmond, W; Sancho-Martinez, I; Phimister, A; Seidel, HM; Badman, MK; Joseph, SB; Laffitte, B; Molteni, V J Med Chem 63: 3868-3880 (2020)
- Bakthavatchalam, R; Basu, MK; Behera, AK; Venkateshappa, C; Hewson, CA; Kadnur, SV; Kalindjian, SB; Kulkarni, B; Saxena, R; Suresh, J; Viswanathan, V; Zainuddin, M; Dharshinis, AP; Kristam, R US Patent US9969687 (2018)
- Zhou, HJ; Aujay, MA; Bennett, MK; Dajee, M; Demo, SD; Fang, Y; Ho, MN; Jiang, J; Kirk, CJ; Laidig, GJ; Lewis, ER; Lu, Y; Muchamuel, T; Parlati, F; Ring, E; Shenk, KD; Shields, J; Shwonek, PJ; Stanton, T; Sun, CM; Sylvain, C; Woo, TM; Yang, J J Med Chem 52: 3028-38 (2009)
- Sliedregt, LA; Rensen, PC; Rump, ET; van Santbrink, PJ; Bijsterbosch, MK; Valentijn, AR; van der Marel, GA; van Boom, JH; van Berkel, TJ; Biessen, EA J Med Chem 42: 609-18 (1999)
- Wendeler, M; Lee, HF; Bermingham, A; Miller, JT; Chertov, O; Bona, MK; Baichoo, NS; Ehteshami, M; Beutler, J; Oamppound39Keefe, BR; Götte, M; Kvaratskhelia, M; Le Grice, S ACS Chem Biol 3: 635-44 (2008)
- Jadhav, RD; Kadam, KS; Kandre, S; Guha, T; Reddy, MM; Brahma, MK; Deshmukh, NJ; Dixit, A; Doshi, L; Potdar, N; Enose, AA; Vishwakarma, RA; Sivaramakrishnan, H; Srinivasan, S; Nemmani, KV; Gupte, A; Gangopadhyay, AK; Sharma, R Eur J Med Chem 54: 324-42 (2012)
- Campbell, MK; Czirr, E; Harish, R US Patent US11957671 (2024)
- Bellenie, BR; Carter, MK; Cheung, KM; Davis, OA; Hoelder, S; Lloyd, MG; Varela Rodriguez, A; Woodward, H; Innocenti, P US Patent US12110286 (2024)
- Sawyer, JS; Beight, DW; Smith, EC; Snyder, DW; Chastain, MK; Tielking, RL; Hartley, LW; Carlson, DG J Med Chem 48: 893-6 (2005)
- Ouertatani-Sakouhi, H; El-Turk, F; Fauvet, B; Cho, MK; Pinar Karpinar, D; Le Roy, D; Dewor, M; Roger, T; Bernhagen, J; Calandra, T; Zweckstetter, M; Lashuel, HA J Biol Chem 285: 26581-98 (2010)
- Christensen, MK; Erichsen, KD; Olesen, UH; Tjørnelund, J; Fristrup, P; Thougaard, A; Nielsen, SJ; Sehested, M; Jensen, PB; Loza, E; Kalvinsh, I; Garten, A; Kiess, W; Björkling, F J Med Chem 56: 9071-88 (2013)
- Volgraf, M; Lumb, JP; Brastianos, HC; Carr, G; Chung, MK; Münzel, M; Mauk, AG; Andersen, RJ; Trauner, D Nat Chem Biol 4: 535-7 (2008)
- Clark, MK; Scott, SA; Wojtkowiak, J; Chirco, R; Mathieu, P; Reiners, JJ; Mattingly, RR; Borch, RF; Gibbs, RA J Med Chem 50: 3274-82 (2007)
- Clements MK
- Shahripour, AB; Plummer, MS; Lunney, EA; Sawyer, TK; Stankovic, CJ; Connolly, MK; Rubin, JR; Walker, NP; Brady, KD; Allen, HJ; Talanian, RV; Wong, WW; Humblet, C Bioorg Med Chem Lett 11: 2779-82 (2001)
- ANDERSON, ED; ARONOW, SD; BOYLES, NA; DAHLGREN, MK; FENG, S; GERASYUTO, AI; HICKEY, ER; IRVIN, TC; KESICKI, EA; KLIPPEL-GIESE, A; KNIGHT, JL; KOLAKOWSKI, GR; KUMAR, M; LONG, KF; MAYNE, CG; MCELLIGOTT, DL; MCLEAN, JA; PUCA, L; RAVI, KK; SEVERANCE, DL; WELCH, MB; WIDJAJA, T US Patent US20230286960 (2023)
- Dahlgren, MK; Garcia, AB; Hare, AA; Tirado-Rives, J; Leng, L; Bucala, R; Jorgensen, WL J Med Chem 55: 10148-59 (2012)
- Wadkins, RM; Hyatt, JL; Wei, X; Yoon, KJ; Wierdl, M; Edwards, CC; Morton, CL; Obenauer, JC; Damodaran, K; Beroza, P; Danks, MK; Potter, PM J Med Chem 48: 2906-15 (2005)
- Leftheris, K; Kline, T; Natarajan, S; DeVirgilio, MK; Cho, YH; Pluscec, J; Ricca, C; Robinson, S; Seizinger, BR; Manne, V; Meyers, CA Bioorg Med Chem Lett 4: 887-892 (1994)
- Ramesh, C; Nayak, TK; Burai, R; Dennis, MK; Hathaway, HJ; Sklar, LA; Prossnitz, ER; Arterburn, JB J Med Chem 53: 1004-14 (2010)
- Fan, C; Clay, MD; Deyholos, MK; Vederas, JC Bioorg Med Chem 18: 2141-2151 (2010)
- Yeh, VS; Beno, DW; Brodjian, S; Brune, ME; Cullen, SC; Dayton, BD; Dhaon, MK; Falls, HD; Gao, J; Grihalde, N; Hajduk, P; Hansen, TM; Judd, AS; King, AJ; Klix, RC; Larson, KJ; Lau, YY; Marsh, KC; Mittelstadt, SW; Plata, D; Rozema, MJ; Segreti, JA; Stoner, EJ; Voorbach, MJ; Wang, X; Xin, X; Zhao, G; Collins, CA; Cox, BF; Reilly, RM; Kym, PR; Souers, AJ J Med Chem 55: 1751-7 (2012)
- Springer, RH; Dimmitt, MK; Novinson, T; O'Brien, DE; Robins, RK; Simon, LN; Miller, JP J Med Chem 19: 291-6 (1976)
- Tian, X; Switzer, AG; Derose, SA; Mishra, RK; Solinsky, MG; Mumin, RN; Ebetino, FH; Jayasinghe, LR; Webster, ME; Colson, AO; Crossdoersen, D; Pinney, BB; Farmer, JA; Dowty, ME; Obringer, CM; Cruze, CA; Burklow, ML; Suchanek, PM; Dong, L; Dirr, MK; Sheldon, RJ; Wos, JA J Med Chem 51: 6055-66 (2008)
- Thomas, AA; Hunt, KW; Volgraf, M; Watts, RJ; Liu, X; Vigers, G; Smith, D; Sammond, D; Tang, TP; Rhodes, SP; Metcalf, AT; Brown, KD; Otten, JN; Burkard, M; Cox, AA; Do, MK; Dutcher, D; Rana, S; DeLisle, RK; Regal, K; Wright, AD; Groneberg, R; Scearce-Levie, K; Siu, M; Purkey, HE; Lyssikatos, JP; Gunawardana, IW J Med Chem 57: 878-902 (2014)
- Woo, PW; Kostlan, CR; Sircar, JC; Dong, MK; Gilbertsen, RB J Med Chem 35: 1451-7 (1992)
- Halland, N; Schmidt, F; Weiss, T; Li, Z; Czech, J; Saas, J; Ding-Pfennigdorff, D; Dreyer, MK; Strübing, C; Nazare, M J Med Chem 65: 1567-1584 (2022)
- Elsayed, MK; Burayk, S US Patent US11478464 (2022)
- Fenwick MK
- Gannon, MK; Holt, JJ; Bennett, SM; Wetzel, BR; Loo, TW; Bartlett, MC; Clarke, DM; Sawada, GA; Higgins, JW; Tombline, G; Raub, TJ; Detty, MR J Med Chem 52: 3328-41 (2009)
- Bum-Erdene, K; Liu, D; Xu, D; Ghozayel, MK; Meroueh, SO ACS Med Chem Lett 12: 60-66 (2021)
- Muddana, HS; Fenley, AT; Mobley, DL; Gilson, MK J Comput Aided Mol Des 28: 305-17 (2014)
- Burnett, GL; Yang, YC; Aggen, JB; Pitzen, J; Gliedt, MK; Semko, CM; Marquez, A; Evans, JW; Wang, G; Won, WS; Tomlinson, ACA; Kiss, G; Tzitzilonis, C; Thottumkara, AP; Cregg, J; Mellem, KT; Choi, JS; Lee, JC; Zhao, Y; Lee, BJ; Meyerowitz, JG; Knox, JE; Jiang, J; Wang, Z; Wildes, D; Wang, Z; Singh, M; Smith, JAM; Gill, AL J Med Chem 66: 149-169 (2023)
- Blanchard, S; Soh, CK; Lee, CP; Poulsen, A; Bonday, Z; Goh, KL; Goh, KC; Goh, MK; Pasha, MK; Wang, H; Williams, M; Wood, JM; Ethirajulu, K; Dymock, BW Bioorg Med Chem Lett 22: 2880-4 (2012)
- Christopoulos, A; Grant, MK; Ayoubzadeh, N; Kim, ON; Sauerberg, P; Jeppesen, L; El-Fakahany, EE J Pharmacol Exp Ther 298: 1260-8 (2001)
- Carpenter, AJ; Al-Barazanji, KA; Barvian, KK; Bishop, MJ; Britt, CS; Cooper, JP; Goetz, AS; Grizzle, MK; Hertzog, DL; Ignar, DM; Morgan, RO; Peckham, GE; Speake, JD; Swain, WR Bioorg Med Chem Lett 16: 4994-5000 (2006)
- Premkumar, L; Kurth, F; Duprez, W; Grøftehauge, MK; King, GJ; Halili, MA; Heras, B; Martin, JL J Biol Chem 289: 19869-80 (2014)
- Baviskar, AT; Banerjee, UC; Gupta, M; Singh, R; Kumar, S; Gupta, MK; Kumar, S; Raut, SK; Khullar, M; Singh, S; Kumar, R Bioorg Med Chem 21: 5782-93 (2013)
- Korzhnev, DM; Hadden, MK J Med Chem 59: 9321-9336 (2016)
- Jude, KM; Banerjee, AL; Haldar, MK; Manokaran, S; Roy, B; Mallik, S; Srivastava, DK; Christianson, DW J Am Chem Soc 128: 3011-8 (2006)
- Meyers, MJ; Pelc, M; Kamtekar, S; Day, J; Poda, GI; Hall, MK; Michener, ML; Reitz, BA; Mathis, KJ; Pierce, BS; Parikh, MD; Mischke, DA; Long, SA; Parlow, JJ; Anderson, DR; Thorarensen, A Bioorg Med Chem Lett 20: 1543-7 (2010)
- Aik, W; Demetriades, M; Hamdan, MK; Bagg, EA; Yeoh, KK; Lejeune, C; Zhang, Z; McDonough, MA; Schofield, CJ J Med Chem 56: 3680-8 (2013)
- Ali, SM; Tedford, CE; Gregory, R; Handley, MK; Yates, SL; Hirth, WW; Phillips, JG J Med Chem 42: 903-9 (1999)
- Lerch, ML; Harper, MK; Faulkner, DJ J Nat Prod 66: 667-70 (2003)
- Hao, J; Beck, J; Zhou, X; Lackner, GL; Johnston, R; Reinhard, M; Goldsmith, P; Hollinshead, S; Dehlinger, V; Filla, SA; Wang, XS; Richardson, J; Posada, M; Mohutsky, M; Schober, D; Katner, JS; Chen, Q; Hu, B; Remick, DM; Coates, DA; Mathes, BM; Hawk, MK; Svensson, KA; Hembre, E J Med Chem 65: 3786-3797 (2022)
- Penning, TD; Chandrakumar, NS; Desai, BN; Djuric, SW; Gasiecki, AF; Malecha, JW; Miyashiro, JM; Russell, MA; Askonas, LJ; Gierse, JK; Harding, EI; Highkin, MK; Kachur, JF; Kim, SH; Villani-Price, D; Pyla, EY; Ghoreishi-Haack, NS; Smith, WG Bioorg Med Chem Lett 13: 1137-9 (2003)
- Hilinski, MK; Mrozowski, RM; Clark, DE; Lannigan, DA Bioorg Med Chem Lett 22: 3244-7 (2012)
- Himmelbauer, MK; Xin, Z; Jones, JH; Enyedy, I; King, K; Marcotte, DJ; Murugan, P; Santoro, JC; Hesson, T; Spilker, K; Johnson, JL; Luzzio, MJ; Gilfillan, R; de Turiso, FG J Med Chem 62: 10740-10756 (2019)
- Hu, Y; Ho, MK; Chan, KH; New, DC; Wong, YH Bioorg Med Chem Lett 20: 2582-5 (2010)
- Williams, PD; Perlow, DS; Payne, LS; Holloway, MK; Siegl, PK; Schorn, TW; Lynch, RJ; Doyle, JJ; Strouse, JF; Vlasuk, GP J Med Chem 34: 887-900 (1991)
- Hu, MK; Wu, LJ; Hsiao, G; Yen, MH J Med Chem 45: 2277-82 (2002)
- Hussain MK
- Kim, ND; Yoon, J; Kim, JH; Lee, JT; Chon, YS; Hwang, MK; Ha, I; Song, WJ Bioorg Med Chem Lett 16: 3772-6 (2006)
- Ibrahim, MK; Taghour, MS; Metwaly, AM; Belal, A; Mehany, ABM; Elhendawy, MA; Radwan, MM; Yassin, AM; El-Deeb, NM; Hafez, EE; ElSohly, MA; Eissa, IH Eur J Med Chem 155: 117-134 (2018)
- Masci, D; Hind, C; Islam, MK; Toscani, A; Clifford, M; Coluccia, A; Conforti, I; Touitou, M; Memdouh, S; Wei, X; La Regina, G; Silvestri, R; Sutton, JM; Castagnolo, D Eur J Med Chem 178: 500-514 (2019)
- Koh, DW; Coyle, DL; Mehta, N; Ramsinghani, S; Kim, H; Slama, JT; Jacobson, MK J Med Chem 46: 4322-32 (2003)
- Yu, BZ; Kaimal, R; Bai, S; El Sayed, KA; Tatulian, SA; Apitz, RJ; Jain, MK; Deng, R; Berg, OG J Nat Prod 72: 24-8 (2009)
- Dezube, M; Sugg, EE; Birkemo, LS; Croom, DK; Dougherty, RW; Ervin, GN; Grizzle, MK; James, MK; Johnson, MF; Mosher, JT J Med Chem 38: 3384-90 (1995)
- Isabel, E; Black, WC; Bayly, CI; Grimm, EL; Janes, MK; McKay, DJ; Nicholson, DW; Rasper, DM; Renaud, J; Roy, S; Tam, J; Thornberry, NA; Vaillancourt, JP; Xanthoudakis, S; Zamboni, R Bioorg Med Chem Lett 13: 2137-40 (2003)
- Shirai, F; Mizutani, A; Yashiroda, Y; Tsumura, T; Kano, Y; Muramatsu, Y; Chikada, T; Yuki, H; Niwa, H; Sato, S; Washizuka, K; Koda, Y; Mazaki, Y; Jang, MK; Yoshida, H; Nagamori, A; Okue, M; Watanabe, T; Kitamura, K; Shitara, E; Honma, T; Umehara, T; Shirouzu, M; Fukami, T; Seimiya, H; Yoshida, M; Koyama, H J Med Chem 63: 4183-4204 (2020)
- Shankaraiah, N; Nekkanti, S; Chudasama, KJ; Senwar, KR; Sharma, P; Jeengar, MK; Naidu, VG; Srinivasulu, V; Srinivasulu, G; Kamal, A Bioorg Med Chem Lett 24: 5413-7 (2015)
- Jeon, MK; Lee, KM; Kim, IH; Jang, YK; Kang, SK; Lee, JM; Jung, KY; Kumar, JA; Rhee, SD; Jung, WH; Song, JS; Bae, MA; Kim, KR; Ahn, JH Bioorg Med Chem Lett 24: 4281-5 (2014)
- Golebiowski, A; Whitehouse, D; Beckett, RP; Van Zandt, M; Ji, MK; Ryder, TR; Jagdmann, E; Andreoli, M; Lee, Y; Sheeler, R; Conway, B; Olczak, J; Mazur, M; Czestkowski, W; Piotrowska, W; Cousido-Siah, A; Ruiz, FX; Mitschler, A; Podjarny, A; Schroeter, H Bioorg Med Chem Lett 23: 4837-41 (2013)
- Lee, J; Jin, MK; Kang, SU; Kim, SY; Lee, J; Shin, M; Hwang, J; Cho, S; Choi, YS; Choi, HK; Kim, SE; Suh, YG; Lee, YS; Kim, YH; Ha, HJ; Toth, A; Pearce, LV; Tran, R; Szabo, T; Welter, JD; Lundberg, DJ; Wang, Y; Lazar, J; Pavlyukovets, VA; Morgan, MA; Blumberg, PM Bioorg Med Chem Lett 15: 4143-50 (2005)
- Peoples, AJ; Hughes, D; Ling, LL; Millett, W; Nitti, A; Spoering, A; Steadman, VA; Chiva, JC; Lazarides, L; Jones, MK; Poullennec, KG; Lewis, K; Epstein, S US Patent US9402878 (2016)
- Bruncko, M; Oost, TK; Belli, BA; Ding, H; Joseph, MK; Kunzer, A; Martineau, D; McClellan, WJ; Mitten, M; Ng, SC; Nimmer, PM; Oltersdorf, T; Park, CM; Petros, AM; Shoemaker, AR; Song, X; Wang, X; Wendt, MD; Zhang, H; Fesik, SW; Rosenberg, SH; Elmore, SW J Med Chem 50: 641-62 (2007)
- Kim, J; Kwon, J; Lee, D; Jo, S; Park, DS; Choi, J; Park, E; Hwang, JY; Ko, Y; Choi, I; Ju, MK; Ahn, J; Kim, J; Han, SJ; Kim, TH; Cechetto, J; Nam, J; Ahn, S; Sommer, P; Liuzzi, M; Lee, J Bioorg Med Chem Lett 24: 5473-7 (2015)
- Pettit, GR; Grealish, MP; Jung, MK; Hamel, E; Pettit, RK; Chapuis, JC; Schmidt, JM J Med Chem 45: 2534-42 (2002)
- Kang MK
- Barile, E; Marconi, GD; De, SK; Baggio, C; Gambini, L; Salem, AF; Kashyap, MK; Castro, JE; Kipps, TJ; Pellecchia, M ACS Chem Biol 12: 444-455 (2017)
- Jain, KS; Bariwal, JB; Kathiravan, MK; Phoujdar, MS; Sahne, RS; Chauhan, BS; Shah, AK; Yadav, MR Bioorg Med Chem 16: 4759-800 (2008)
- Maity, P; Chatterjee, J; Patil, KT; Arora, S; Katiyar, MK; Kumar, M; Samarbakhsh, A; Joshi, G; Bhutani, P; Chugh, M; Gavande, NS; Kumar, R J Med Chem 66: 3135-3172 (2023)
- Kim, E; Park, CS; Han, T; Bae, MH; Chong, W; Lee, CH; Shin, YA; Ahn, BN; Kim, MK; Shin, CY; Son, MH; Kim, JK; Moon, HS; Shim, HJ; Kim, EJ; Kim, SH; Lim, JI; Lee, CH Bioorg Med Chem Lett 18: 4993-6 (2008)
- Mascitti, V; Maurer, TS; Robinson, RP; Bian, J; Boustany-Kari, CM; Brandt, T; Collman, BM; Kalgutkar, AS; Klenotic, MK; Leininger, MT; Lowe, A; Maguire, RJ; Masterson, VM; Miao, Z; Mukaiyama, E; Patel, JD; Pettersen, JC; Préville, C; Samas, B; She, L; Sobol, Z; Steppan, CM; Stevens, BD; Thuma, BA; Tugnait, M; Zeng, D; Zhu, T J Med Chem 54: 2952-60 (2011)
- Kim, Y; Kim, J; Kim, S; Ki, Y; Seo, SH; Tae, J; Ko, MK; Jang, HS; Lim, EJ; Song, C; Cho, Y; Koh, HY; Chong, Y; Choo, IH; Keum, G; Min, SJ; Choo, H Eur J Med Chem 85: 629-37 (2014)
- Krapf, MK; Gallus, J; Wiese, M J Med Chem 60: 4474-4495 (2017)
- Khandazhinskaya, AL; Shirokova, EA; Skoblov, YS; Victorova, LS; Goryunova, LY; Beabealashvilli, RS; Pronyaeva, TR; Fedyuk, NV; Zolin, VV; Pokrovsky, AG; Kukhanova, MK J Med Chem 45: 1284-91 (2002)
- Davis, PD; Elliott, LH; Harris, W; Hill, CH; Hurst, SA; Keech, E; Kumar, MK; Lawton, G; Nixon, JS; Wilkinson, SE J Med Chem 35: 994-1001 (1992)
- Alam, A; Pal, C; Goyal, M; Kundu, MK; Kumar, R; Iqbal, MS; Dey, S; Bindu, S; Sarkar, S; Pal, U; Maiti, NC; Adhikari, S; Bandyopadhyay, U Bioorg Med Chem 19: 7365-73 (2011)
- Nagarajan, SR; Devadas, B; Malecha, JW; Lu, HF; Ruminski, PG; Rico, JG; Rogers, TE; Marrufo, LD; Collins, JT; Kleine, HP; Lantz, MK; Zhu, J; Green, NF; Russell, MA; Landis, BH; Miller, LM; Meyer, DM; Duffin, TD; Engleman, VW; Finn, MB; Freeman, SK; Griggs, DW; Williams, ML; Nickols, MA; Pegg, JA; Shannon, KE; Steininger, C; Westlin, MM; Nickols, GA; Keene, JL Bioorg Med Chem 15: 3783-800 (2007)
- Dresser, MJ; Xiao, G; Leabman, MK; Gray, AT; Giacomini, KM Pharm Res 19: 1244-7 (2002)
- Ko, S; Lee, MK; Shin, D; Park, H Bioorg Med Chem 17: 7769-74 (2009)
- Poláchová, E; Bach, K; Heuten, E; Stanchev, S; Tichá, A; Lampe, P; Majer, P; Langer, T; Lemberg, MK; Stříšovský, K J Med Chem 66: 251-265 (2023)
- Hill, JE; Linder, MK; Davies, KS; Sawada, GA; Morgan, J; Ohulchanskyy, TY; Detty, MR J Med Chem 57: 8622-34 (2014)
- Woodrow, MD; Ballantine, SP; Barker, MD; Clarke, BJ; Dawson, J; Dean, TW; Delves, CJ; Evans, B; Gough, SL; Guntrip, SB; Holman, S; Holmes, DS; Kranz, M; Lindvaal, MK; Lucas, FS; Neu, M; Ranshaw, LE; Solanke, YE; Somers, DO; Ward, P; Wiseman, JO Bioorg Med Chem Lett 19: 5261-5 (2009)
- Burger, MT; Nishiguchi, G; Han, W; Lan, J; Simmons, R; Atallah, G; Ding, Y; Tamez, V; Zhang, Y; Mathur, M; Muller, K; Bellamacina, C; Lindvall, MK; Zang, R; Huh, K; Feucht, P; Zavorotinskaya, T; Dai, Y; Basham, S; Chan, J; Ginn, E; Aycinena, A; Holash, J; Castillo, J; Langowski, JL; Wang, Y; Chen, MY; Lambert, A; Fritsch, C; Kauffmann, A; Pfister, E; Vanasse, KG; Garcia, PD J Med Chem 58: 8373-86 (2015)
- Siegel, D; Hui, HC; Doerffler, E; Clarke, MO; Chun, K; Zhang, L; Neville, S; Carra, E; Lew, W; Ross, B; Wang, Q; Wolfe, L; Jordan, R; Soloveva, V; Knox, J; Perry, J; Perron, M; Stray, KM; Barauskas, O; Feng, JY; Xu, Y; Lee, G; Rheingold, AL; Ray, AS; Bannister, R; Strickley, R; Swaminathan, S; Lee, WA; Bavari, S; Cihlar, T; Lo, MK; Warren, TK; Mackman, RL J Med Chem 60: 1648-1661 (2017)
- Liu, J; Shikhman, AR; Lotz, MK; Wong, CH Chem Biol 8: 701-11 (2001)
- Scott, JD; DeMong, DE; Greshock, TJ; Basu, K; Dai, X; Harris, J; Hruza, A; Li, SW; Lin, SI; Liu, H; Macala, MK; Hu, Z; Mei, H; Zhang, H; Walsh, P; Poirier, M; Shi, ZC; Xiao, L; Agnihotri, G; Baptista, MA; Columbus, J; Fell, MJ; Hyde, LA; Kuvelkar, R; Lin, Y; Mirescu, C; Morrow, JA; Yin, Z; Zhang, X; Zhou, X; Chang, RK; Embrey, MW; Sanders, JM; Tiscia, HE; Drolet, RE; Kern, JT; Sur, SM; Renger, JJ; Bilodeau, MT; Kennedy, ME; Parker, EM; Stamford, AW; Nargund, R; McCauley, JA; Miller, MW J Med Chem 60: 2983-2992 (2017)
- Haile, PA; Casillas, LN; Votta, BJ; Wang, GZ; Charnley, AK; Dong, X; Bury, MJ; Romano, JJ; Mehlmann, JF; King, BW; Erhard, KF; Hanning, CR; Lipshutz, DB; Desai, BM; Capriotti, CA; Schaeffer, MC; Berger, SB; Mahajan, MK; Reilly, MA; Nagilla, R; Rivera, EJ; Sun, HH; Kenna, JK; Beal, AM; Ouellette, MT; Kelly, M; Stemp, G; Convery, MA; Vossenkämper, A; MacDonald, TT; Gough, PJ; Bertin, J; Marquis, RW J Med Chem 62: 6482-6494 (2019)
- Mahapatra, MK; Kumar, R; Kumar, M Bioorg Chem 71: 1-9 (2017)
- Hopper, DW; Vera, MD; How, D; Sabatini, J; Xiang, JS; Ipek, M; Thomason, J; Hu, Y; Feyfant, E; Wang, Q; Georgiadis, KE; Reifenberg, E; Sheldon, RT; Keohan, CC; Majumdar, MK; Morris, EA; Skotnicki, J; Sum, PE Bioorg Med Chem Lett 19: 2487-91 (2009)
- Zentner, I; Sierra, LJ; Maciunas, L; Vinnik, A; Fedichev, P; Mankowski, MK; Ptak, RG; Martín-García, J; Cocklin, S Bioorg Med Chem Lett 23: 1132-5 (2013)
- Mann, MK; Zepeda-Velázquez, CA; González-Álvarez, H; Dong, A; Kiyota, T; Aman, AM; Loppnau, P; Li, Y; Wilson, B; Arrowsmith, CH; Al-Awar, R; Harding, RJ; Schapira, M J Med Chem 64: 15017-15036 (2021)
- Manthey, MK; Huang, DT; Bubb, WA; Christopherson, RI J Med Chem 41: 4550-5 (1998)
- Selness, SR; Devraj, RV; Devadas, B; Walker, JK; Boehm, TL; Durley, RC; Shieh, H; Xing, L; Rucker, PV; Jerome, KD; Benson, AG; Marrufo, LD; Madsen, HM; Hitchcock, J; Owen, TJ; Christie, L; Promo, MA; Hickory, BS; Alvira, E; Naing, W; Blevis-Bal, R; Messing, D; Yang, J; Mao, MK; Yalamanchili, G; Vonder Embse, R; Hirsch, J; Saabye, M; Bonar, S; Webb, E; Anderson, G; Monahan, JB Bioorg Med Chem Lett 21: 4066-71 (2011)
- Su, DS; Lim, JL; Markowitz, MK; Wan, BL; Murphy, KL; Reiss, DR; Harrell, CM; O'Malley, SS; Ransom, RW; Chang, RS; Pettibone, DJ; Tang, C; Prueksaritanont, T; Freidinger, RM; Bock, MG Bioorg Med Chem Lett 17: 3006-9 (2007)
- Ahn, K; Johnson, DS; Mileni, M; Beidler, D; Long, JZ; McKinney, MK; Weerapana, E; Sadagopan, N; Liimatta, M; Smith, SE; Lazerwith, S; Stiff, C; Kamtekar, S; Bhattacharya, K; Zhang, Y; Swaney, S; Van Becelaere, K; Stevens, RC; Cravatt, BF Chem Biol 16: 411-20 (2009)
- Zhou, HJ; Wang, J; Yao, B; Wong, S; Djakovic, S; Kumar, B; Rice, J; Valle, E; Soriano, F; Menon, MK; Madriaga, A; Kiss von Soly, S; Kumar, A; Parlati, F; Yakes, FM; Shawver, L; Le Moigne, R; Anderson, DJ; Rolfe, M; Wustrow, D J Med Chem 58: 9480-97 (2015)
- Press, NJ; Taylor, RJ; Fullerton, JD; Tranter, P; McCarthy, C; Keller, TH; Brown, L; Cheung, R; Christie, J; Haberthuer, S; Hatto, JD; Keenan, M; Mercer, MK; Press, NE; Sahri, H; Tuffnell, AR; Tweed, M; Fozard, JR Bioorg Med Chem Lett 15: 3081-5 (2005)
- Groutas, WC; Abrams, WR; Theodorakis, MC; Kasper, AM; Rude, SA; Badger, RC; Ocain, TD; Miller, KE; Moi, MK; Brubaker, MJ J Med Chem 28: 204-9 (1985)
- Nambu, M; Covel, JA; Kapoor, M; Li, X; Moloney, MK; Numa, MM; Soltow, QA; Trzoss, M; Webb, P; Webb, RR; Mutz, M Bioorg Med Chem Lett 27: 2465-2471 (2017)
- Dumas, J; Hatoum-Mokdad, H; Sibley, R; Riedl, B; Scott, WJ; Monahan, MK; Lowinger, TB; Brennan, C; Natero, R; Turner, T; Johnson, JS; Schoenleber, R; Bhargava, A; Wilhelm, SM; Housley, TJ; Ranges, GE; Shrikhande, A Bioorg Med Chem Lett 10: 2051-4 (2000)
- Astrand, A; Grimster, N; Kawatkar, S; Kettle, JG; Nilsson, MK; Ruston, L; Su, Q; Vasbinder, M; Winter-Holt, JJ; Woessner, RD; Chuaqui, CE; McCabe, J US Patent US10167276 (2019)
- Zhao, L; O'Reilly, MK; Shultz, MD; Chmielewski, J Bioorg Med Chem Lett 13: 1175-7 (2003)
- Dinavahi, SS; Gowda, R; Bazewicz, CG; Battu, MB; Lin, JM; Chitren, RJ; Pandey, MK; Amin, S; Robertson, GP; Gowda, K Eur J Med Chem 187: (2020)
- Parai, MK; Huggins, DJ; Cao, H; Nalam, MN; Ali, A; Schiffer, CA; Tidor, B; Rana, TM J Med Chem 55: 6328-41 (2012)
- Witter, DJ; Biftu, T; Biju, P; Bogen, SL; Hong, Q; Huang, C; Huang, X; Li, B; Park, MK; Sloman, DL US Patent US10399972 (2019)
- William, AD; Lee, AC; Goh, KC; Blanchard, S; Poulsen, A; Teo, EL; Nagaraj, H; Lee, CP; Wang, H; Williams, M; Sun, ET; Hu, C; Jayaraman, R; Pasha, MK; Ethirajulu, K; Wood, JM; Dymock, BW J Med Chem 55: 169-96 (2012)
- Rivara, M; Patel, MK; Amori, L; Zuliani, V Bioorg Med Chem Lett 22: 6401-4 (2012)
- Topczewski, JJ; Lodge, AM; Yasapala, SN; Payne, MK; Keshavarzi, PM; Quinn, DM Bioorg Med Chem Lett 23: 5786-9 (2013)
- Wang, M; Winneroski, LL; Ardecky, RJ; Babine, RE; Brooks, DA; Etgen, GJ; Hutchison, DR; Kauffman, RF; Kunkel, A; Mais, DE; Montrose-Rafizadeh, C; Ogilvie, KM; Oldham, BA; Peters, MK; Rito, CJ; Rungta, DK; Tripp, AE; Wilson, SB; Xu, Y; Zink, RW; McCarthy, JR Bioorg Med Chem Lett 14: 6113-6 (2004)
- Karaj, E; Sindi, SH; Kuganesan, N; Koranne, RA; Knoff, JR; James, AW; Fu, Y; Kotsull, LN; Pflum, MK; Shah, Z; Taylor, WR; Tillekeratne, LMV J Med Chem 65: 14764-14791 (2022)
- Banerjee, A; Pawar, MY; Patil, S; Yadav, PS; Kadam, PA; Kattige, VG; Deshpande, DS; Pednekar, PV; Pisat, MK; Gharat, LA Bioorg Med Chem Lett 24: 4838-44 (2014)
- Kumar, CBP; Raghu, MS; Prathibha, BS; Prashanth, MK; Kanthimathi, G; Kumar, KY; Parashuram, L; Alharthi, FA Bioorg Med Chem Lett 44: (2021)
- Li, G; Tao, ZF; Tong, Y; Przytulinska, MK; Kovar, P; Merta, P; Chen, Z; Zhang, H; Sowin, T; Rosenberg, SH; Lin, NH Bioorg Med Chem Lett 17: 6499-504 (2007)
- Purohit, MK; Scovell, I; Neschadim, A; Katsman, Y; Branch, DR; Kotra, LP Bioorg Med Chem Lett 23: 2324-7 (2013)
- Rahnasto, MK; Raunio, HA; Wittekindt, C; Salminen, KA; Leppänen, J; Juvonen, RO; Poso, A; Lahtela-Kakkonen, MK Bioorg Med Chem 19: 7186-93 (2011)
- Vu, CB; Luke, GP; Kawahata, N; Shakespeare, WC; Wang, Y; Sundaramoorthi, R; Metcalf, CA; Keenan, TP; Pradeepan, S; Corpuz, E; Merry, T; Bohacek, RS; Dalgarno, DC; Narula, SS; van Schravendijk, MR; Ram, MK; Adams, S; Liou, S; Keats, JA; Violette, SM; Guan, W; Weigele, M; Sawyer, TK Bioorg Med Chem Lett 13: 3071-4 (2003)
- Lee, KL; Foley, MA; Chen, L; Behnke, ML; Lovering, FE; Kirincich, SJ; Wang, W; Shim, J; Tam, S; Shen, MW; Khor, S; Xu, X; Goodwin, DG; Ramarao, MK; Nickerson-Nutter, C; Donahue, F; Ku, MS; Clark, JD; McKew, JC J Med Chem 50: 1380-400 (2007)
- Czechtizky, W; Dedio, J; Desai, B; Dixon, K; Farrant, E; Feng, Q; Morgan, T; Parry, DM; Ramjee, MK; Selway, CN; Schmidt, T; Tarver, GJ; Wright, AG ACS Med Chem Lett 4: 768-72 (2013)
- Castro-Falcón, G; Seiler, GS; Demir, Ö; Rathinaswamy, MK; Hamelin, D; Hoffmann, RM; Makowski, SL; Letzel, AC; Field, SJ; Burke, JE; Amaro, RE; Hughes, CC J Med Chem 61: 10463-10472 (2018)
- Rauf, MK; Talib, A; Badshah, A; Zaib, S; Shoaib, K; Shahid, M; Flörke, U; Imtiaz-ud-Din, na; Iqbal, J Eur J Med Chem 70: 487-96 (2013)
- Lele, AC; Raju, A; Khambete, MP; Ray, MK; Rajan, MG; Arkile, MA; Jadhav, NJ; Sarkar, D; Degani, MS ACS Med Chem Lett 6: 1140-4 (2015)
- Manzoor, S; Angeli, A; Zara, S; Carradori, S; Rahman, MA; Raza, MK; Supuran, CT; Hoda, N Eur J Med Chem 243: (2022)
- Jackson, JJ; Shibuya, GM; Ravishankar, B; Adusumilli, L; Bradford, D; Brockstedt, DG; Bucher, C; Bui, M; Cho, C; Colas, C; Cutler, G; Dukes, A; Han, X; Hu, DX; Jacobson, S; Kassner, PD; Katibah, GE; Ko, MYM; Kolhatkar, U; Leger, PR; Ma, A; Marshall, L; Maung, J; Ng, AA; Okano, A; Pookot, D; Poon, D; Ramana, C; Reilly, MK; Robles, O; Schwarz, JB; Shakhmin, AA; Shunatona, HP; Sreenivasan, R; Tivitmahaisoon, P; Xu, M; Zaw, T; Wustrow, DJ; Zibinsky, M J Med Chem 65: 12895-12924 (2022)
- Shishodia, S; Nuñez, R; Strohmier, BP; Bursch, KL; Goetz, CJ; Olp, MD; Jensen, DR; Fenske, TG; Ordonez-Rubiano, SC; Blau, ME; Roach, MK; Peterson, FC; Volkman, BF; Dykhuizen, EC; Smith, BC J Med Chem 65: 13714-13735 (2022)
- Archibald, SC; Head, JC; Linsley, JM; Porter, JR; Robinson, MK; Shock, A; Warrellow, GJ Bioorg Med Chem Lett 10: 993-5 (2000)
- Rogacki, MK; Pitta, E; Balabon, O; Huss, S; Lopez-Roman, EM; Argyrou, A; Blanco-Ruano, D; Cacho, M; Vande Velde, CML; Augustyns, K; Ballell, L; Barros, D; Bates, RH; Cunningham, F; Van der Veken, P J Med Chem 61: 11221-11249 (2018)
- Gryder, BE; Akbashev, MJ; Rood, MK; Raftery, ED; Meyers, WM; Dillard, P; Khan, S; Oyelere, AK ACS Chem Biol 8: 2550-60 (2013)
- Nguyen, TT; Ding, D; Wolter, WR; Pérez, RL; Champion, MM; Mahasenan, KV; Hesek, D; Lee, M; Schroeder, VA; Jones, JI; Lastochkin, E; Rose, MK; Peterson, CE; Suckow, MA; Mobashery, S; Chang, M J Med Chem 61: 8825-8837 (2018)
- Kozikowski, AP; Saiah, MK; Bergmann, JS; Johnson, KM J Med Chem 37: 3440-2 (1994)
- Romagnoli, R; Oliva, P; Salvador, MK; Camacho, ME; Padroni, C; Brancale, A; Ferla, S; Hamel, E; Ronca, R; Grillo, E; Bortolozzi, R; Rruga, F; Mariotto, E; Viola, G Eur J Med Chem 181: (2019)
- Sanders MK
- Parihar, HS; Suryanarayanan, A; Ma, C; Joshi, P; Venkataraman, P; Schulte, MK; Kirschbaum, KS Bioorg Med Chem Lett 11: 2133-6 (2001)
- Baumhover, NJ; Martin, ME; Parameswarappa, SG; Kloepping, KC; O'Dorisio, MS; Pigge, FC; Schultz, MK Bioorg Med Chem Lett 21: 5757-61 (2011)
- Pierre, F; Chua, PC; O'Brien, SE; Siddiqui-Jain, A; Bourbon, P; Haddach, M; Michaux, J; Nagasawa, J; Schwaebe, MK; Stefan, E; Vialettes, A; Whitten, JP; Chen, TK; Darjania, L; Stansfield, R; Anderes, K; Bliesath, J; Drygin, D; Ho, C; Omori, M; Proffitt, C; Streiner, N; Trent, K; Rice, WG; Ryckman, DM J Med Chem 54: 635-54 (2011)
- Crosignani, S; Page, P; Missotten, M; Colovray, V; Cleva, C; Arrighi, JF; Atherall, J; Macritchie, J; Martin, T; Humbert, Y; Gaudet, M; Pupowicz, D; Maio, M; Pittet, PA; Golzio, L; Giachetti, C; Rocha, C; Bernardinelli, G; Filinchuk, Y; Scheer, A; Schwarz, MK; Chollet, A J Med Chem 51: 2227-2243 (2008)
- Dart, ML; Machleidt, T; Jost, E; Schwinn, MK; Robers, MB; Shi, C; Kirkland, TA; Killoran, MP; Wilkinson, JM; Hartnett, JR; Zimmerman, K; Wood, KV ACS Med Chem Lett 9: 546-551 (2018)
- Baxter, EW; Conway, KA; Kennis, L; Bischoff, F; Mercken, MH; Winter, HL; Reynolds, CH; Tounge, BA; Luo, C; Scott, MK; Huang, Y; Braeken, M; Pieters, SM; Berthelot, DJ; Masure, S; Bruinzeel, WD; Jordan, AD; Parker, MH; Boyd, RE; Qu, J; Alexander, RS; Brenneman, DE; Reitz, AB J Med Chem 50: 4261-4 (2007)
- Bae, IH; Kim, HS; You, Y; Chough, C; Choe, W; Seon, MK; Lee, SG; Keum, G; Jang, SK; Moon Kim, B Eur J Med Chem 101: 163-78 (2015)
- Goodwin, NC; Mabon, R; Harrison, BA; Shadoan, MK; Almstead, ZY; Xie, Y; Healy, J; Buhring, LM; DaCosta, CM; Bardenhagen, J; Mseeh, F; Liu, Q; Nouraldeen, A; Wilson, AG; Kimball, SD; Powell, DR; Rawlins, DB J Med Chem 52: 6201-4 (2009)
- Stoermer, D; Vitharana, D; Hin, N; Delahanty, G; Duvall, B; Ferraris, DV; Grella, BS; Hoover, R; Rojas, C; Shanholtz, MK; Smith, KP; Stathis, M; Wu, Y; Wozniak, KM; Slusher, BS; Tsukamoto, T J Med Chem 55: 5922-32 (2012)
- Mukku, VJ; Edrada, RA; Schmitz, FJ; Shanks, MK; Chaudhuri, B; Fabbro, D J Nat Prod 66: 686-9 (2003)
- Tolentino, KT; Mashinson, V; Sharma, MK; Chhonker, YS; Murry, DJ; Hopkins, CR Eur J Med Chem 244: (2022)
- Sliskovic, DR; Blankley, CJ; Krause, BR; Newton, RS; Picard, JA; Roark, WH; Roth, BD; Sekerke, C; Shaw, MK; Stanfield, RL J Med Chem 35: 2095-103 (1992)
- Zaraei, SO; Al-Ach, NN; Anbar, HS; El-Gamal, R; Tarazi, H; Tokatly, RT; Kalla, RR; Munther, MA; Wahba, MM; Alshihabi, AM; Shehata, MK; Sbenati, RM; Shahin, AI; El-Awady, R; Al-Tel, TH; El-Gamal, MI Eur J Med Chem 238: (2022)
- Xu, R; Sim, MK; Go, ML J Med Chem 41: 3220-31 (1998)
- Morningstar, ML; Roth, T; Farnsworth, DW; Smith, MK; Watson, K; Buckheit, RW; Das, K; Zhang, W; Arnold, E; Julias, JG; Hughes, SH; Michejda, CJ J Med Chem 50: 4003-4015 (2007)
- Peterson, EA; Boezio, AA; Andrews, PS; Boezio, CM; Bush, TL; Cheng, AC; Choquette, D; Coats, JR; Colletti, AE; Copeland, KW; DuPont, M; Graceffa, R; Grubinska, B; Kim, JL; Lewis, RT; Liu, J; Mullady, EL; Potashman, MH; Romero, K; Shaffer, PL; Stanton, MK; Stellwagen, JC; Teffera, Y; Yi, S; Cai, T; La, DS Bioorg Med Chem Lett 22: 4967-74 (2012)
- Hu, J; Hu, K; Liu, T; Stern, MK; Mistry, R; Challiss, RA; Costanzi, S; Wess, J J Biol Chem 288: 34777-90 (2013)
- Ellis, CD; Oppong, KA; Laufersweiler, MC; O'Neil, SV; Soper, DL; Wang, Y; Wos, JA; Fancher, AN; Lu, W; Suchanek, MK; Wang, RL; De, B; Demuth, TP Bioorg Med Chem Lett 16: 4728-32 (2006)
- Gim, HJ; Li, H; Jeong, JH; Lee, SJ; Sung, MK; Song, MY; Park, BH; Oh, SJ; Ryu, JH; Jeon, R Bioorg Med Chem 23: 3322-36 (2015)
- Xiao, J; Free, RB; Barnaeva, E; Conroy, JL; Doyle, T; Miller, B; Bryant-Genevier, M; Taylor, MK; Hu, X; Dulcey, AE; Southall, N; Ferrer, M; Titus, S; Zheng, W; Sibley, DR; Marugan, JJ J Med Chem 57: 3450-63 (2014)
- Dhorma, LP; Teli, MK; Nangunuri, BG; Venkanna, A; Ragam, R; Maturi, A; Mirzaei, A; Vo, DK; Maeng, HJ; Kim, MH Eur J Med Chem 227: (2022)
- Liu, LZ; Ma, T; Zhou, J; Long Hu, Z; Jun Zhang, X; Zhen Zhang, H; Zeng, M; Liu, J; Li, L; Jiang, Y; Zou, Z; Wang, F; Zhang, L; Xu, J; Wang, J; Xiao, F; Fang, X; Zou, H; Efanov, AM; Thomas, MK; Lin, HV; Chen, J Bioorg Med Chem Lett 30: (2020)
- Travis, S; Green, KD; Thamban Chandrika, N; Pang, AH; Frantom, PA; Tsodikov, OV; Garneau-Tsodikova, S; Thompson, MK RSC Med Chem 14: 947-956 (2023)
- Thorson, MK; Puerta, DT; Cohen, SM; Barrios, AM Bioorg Med Chem Lett 24: 4019-22 (2014)
- Wu, H; Zink, N; Carter, LP; Mehta, AK; Hernandez, RJ; Ticku, MK; Lamb, R; France, CP; Coop, A J Pharmacol Exp Ther 305: 675-9 (2003)
- Vaarla, K; Kesharwani, RK; Santosh, K; Vedula, RR; Kotamraju, S; Toopurani, MK Bioorg Med Chem Lett 25: 5797-803 (2015)
- Tripathi, A; Choubey, PK; Sharma, P; Seth, A; Tripathi, PN; Tripathi, MK; Prajapati, SK; Krishnamurthy, S; Shrivastava, SK Eur J Med Chem 183: (2019)
- Sze, KH; Lam, WH; Zhang, H; Ke, YH; Tse, MK; Woo, PC; Lau, SK; Lau, CC; Cai, JP; Tung, ET; Lo, RK; Xu, S; Kao, RY; Hao, Q; Yuen, KY Cell Chem Biol 24: 182-194 (2017)
- Urlam, MK; Pireddu, R; Ge, Y; Zhang, X; Sun, Y; Lawrence, HR; Guida, WC; Sebti, SM; Lawrence, NJ Medchemcomm 4: 932-941 (2013)
- Vadnere, MK; Maggiora, L; Mertes, MP J Med Chem 29: 1714-20 (1986)
- Duncia, JV; Santella, JB; Higley, CA; VanAtten, MK; Weber, PC; Alexander, RS; Kettner, CA; Pruitt, JR; Liauw, AY; Quan, ML; Knabb, RM; Wexler, RR Bioorg Med Chem Lett 8: 775-80 (1999)
- Wallace, BD; Roberts, AB; Pollet, RM; Ingle, JD; Biernat, KA; Pellock, SJ; Venkatesh, MK; Guthrie, L; O'Neal, SK; Robinson, SJ; Dollinger, M; Figueroa, E; McShane, SR; Cohen, RD; Jin, J; Frye, SV; Zamboni, WC; Pepe-Ranney, C; Mani, S; Kelly, L; Redinbo, MR Chem Biol 22: 1238-49 (2015)
- Luo, G; Chen, L; Kostich, WA; Hamman, B; Allen, J; Easton, A; Bourin, C; Gulianello, M; Lippy, J; Nara, S; Pattipati, SN; Dandapani, K; Dokania, M; Vattikundala, P; Sharma, V; Elavazhagan, S; Verma, MK; Lal Das, M; Wagh, S; Balakrishnan, A; Johnson, BM; Santone, KS; Thalody, G; Denton, R; Saminathan, H; Holenarsipur, VK; Kumar, A; Rao, A; Putlur, SP; Sarvasiddhi, SK; Shankar, G; Louis, JV; Ramarao, M; Conway, CM; Li, YW; Pieschl, R; Tian, Y; Hong, Y; Bristow, L; Albright, CF; Bronson, JJ; Macor, JE; Dzierba, CD J Med Chem 65: 4534-4564 (2022)
- Vasudevan, A; Wodka, D; Verzal, MK; Souers, AJ; Gao, J; Brodjian, S; Fry, D; Dayton, B; Marsh, KC; Hernandez, LE; Ogiela, CA; Collins, CA; Kym, PR Bioorg Med Chem Lett 14: 4879-82 (2004)
- Uwaydah, IM; Waddle, MK; Rogers, ME J Med Chem 22: 889-90 (1979)
- Buck, IM; Black, JW; Cooke, T; Dunstone, DJ; Gaffen, JD; Griffin, EP; Harper, EA; Hull, RA; Kalindjian, SB; Lilley, EJ; Linney, ID; Low, CM; McDonald, IM; Pether, MJ; Roberts, SP; Shankley, NP; Shaxted, ME; Steel, KI; Sykes, DA; Tozer, MJ; Watt, GF; Walker, MK; Wright, L; Wright, PT J Med Chem 48: 6803-12 (2005)
- Wambua, MK; Nalawansha, DA; Negmeldin, AT; Pflum, MK J Med Chem 57: 642-50 (2014)
- Wang, R; Chai, WM; Yang, Q; Wei, MK; Peng, Y Bioorg Med Chem 24: 4620-4625 (2016)
- Ishida, K; Simizu, S; Teruya, T; Wierzba, MK; Osada, H Bioorg Med Chem Lett 14: 2505-9 (2004)
- Munk, SA; Lai, RK; Burke, JE; Arasasingham, PN; Kharlamb, AB; Manlapaz, CA; Padillo, EU; Wijono, MK; Hasson, DW; Wheeler, LA; Garst, ME J Med Chem 39: 1193-5 (1996)
- Thamm, S; Willwacher, MK; Aspnes, GE; Bretschneider, T; Brown, NF; Buschbom-Helmke, S; Fox, T; Gargano, EM; Grabowski, D; Hoenke, C; Matera, D; Mueck, K; Peters, S; Reindl, S; Riether, D; Schmid, M; Tautermann, CS; Teitelbaum, AM; Trünkle, C; Veser, T; Winter, M; Wortmann, L J Med Chem 66: 2832-2850 (2023)
- Seganish, WM; McElroy, WT; Brumfield, S; Herr, RJ; Yet, L; Yang, J; Harding, III, JP; Ho, GD; Tulshian, D; Yu, W; Wong, MK; Lavey, B; Kozlowski, JA US Patent US9598440 (2017)
- Peng, X; Lanter, JC; Y-P Chen, A; Brand, MA; Wozniak, MK; Hoekman, S; Longin, O; Regeling, H; Zonneveld, W; P L Bell, R; Koenig, G; Hurst, RS; Blain, JF; Burnett, DA Bioorg Med Chem Lett 80: (2023)
- Sun, WX; Han, HW; Yang, MK; Wen, ZL; Wang, YS; Fu, JY; Lu, YT; Wang, MY; Bao, JX; Lu, GH; Qi, JL; Wang, XM; Lin, HY; Yang, YH Bioorg Med Chem 27: (2019)
- Yau, MK; Suen, JY; Xu, W; Lim, J; Liu, L; Adams, MN; He, Y; Hooper, JD; Reid, RC; Fairlie, DP ACS Med Chem Lett 7: 105-10 (2016)
- Hevener, KE; Yun, MK; Qi, J; Kerr, ID; Babaoglu, K; Hurdle, JG; Balakrishna, K; White, SW; Lee, RE J Med Chem 53: 166-77 (2010)
- Odell, LR; Abdel-Hamid, MK; Hill, TA; Chau, N; Young, KA; Deane, FM; Sakoff, JA; Andersson, S; Daniel, JA; Robinson, PJ; McCluskey, A J Med Chem 60: 349-361 (2017)
- Ahn, S; Basavana Gowda, MK; Lee, M; Masagalli, JN; Mailar, K; Choi, WJ; Noh, M Eur J Med Chem 187: (2020)
- Kozikowski, AP; Eddine Saiah, MK; Johnson, KM; Bergmann, JS J Med Chem 38: 3086-93 (1995)
- Moussa, BA; El-Zaher, AA; El-Ashrey, MK; Fouad, MA Eur J Med Chem 148: 477-486 (2018)
- Thomas, AA; Hunt, KW; Newhouse, B; Watts, RJ; Liu, X; Vigers, G; Smith, D; Rhodes, SP; Brown, KD; Otten, JN; Burkard, M; Cox, AA; Geck Do, MK; Dutcher, D; Rana, S; DeLisle, RK; Regal, K; Wright, AD; Groneberg, R; Liao, J; Scearce-Levie, K; Siu, M; Purkey, HE; Lyssikatos, JP J Med Chem 57: 10112-29 (2014)
- Hristova-Kazmierski, MK; Horan, P; Davis, P; Yamamura, HI; Kramer, T; Horvath, R; Kazmierski, WM; Porreca, F; Hruby, VJ Bioorg Med Chem Lett 3: 831-834 (1993)
- Pätsi, HT; Kilpeläinen, TP; Auno, S; Dillemuth, PMJ; Arja, K; Lahtela-Kakkonen, MK; Myöhänen, TT; Wallén, EAA ACS Med Chem Lett 12: 1578-1584 (2021)
- Wu, RS; Wolpert-DeFilippes, MK; Quinn, FR J Med Chem 23: 256-61 (1980)
- Cristancho Ortiz, CJ; de Freitas Silva, M; Pruccoli, L; Fonseca Nadur, N; de Azevedo, LL; Kümmerle, AE; Guedes, IA; Dardenne, LE; Leomil Coelho, LF; Guimarães, MJ; da Silva, FMR; Castro, N; Gontijo, VS; Rojas, VCT; de Oliveira, MK; Vilela, FC; Giusti-Paiva, A; Barbosa, G; Lima, LM; Pinheiro, GB; Veras, LG; Mortari, MR; Tarozzi, A; Viegas, C RSC Med Chem 13: 568-584 (2022)
- Abd El-Gaber, MK; Hassan, HY; Mahfouz, NM; Farag, HH; Bekhit, AA Eur J Med Chem 93: 481-91 (2015)
- Peluso, S; de L Ufret, M; O'Reilly, MK; Imperiali, B Chem Biol 9: 1323-8 (2002)
- Ray2010 Assay 59 MK-801 forebrain .
- Ray2010 Assay 60 MK-801 cortex .
- Ray2010 Assay 62 MK-801 hippocampus .
- Ray2010 Assay 63 MK-801 striatum .
- Ray2010 Assay 66 MK-801 midbrain .
- Ray2010 Assay 67 MK-801 cerebellum .
- Ray2010 Assay 64 MK-801 Thalamus + hypothalamus .
- Ray2010 Assay 65 MK-801 spinal cord .
- Ray2010 Assay 68 MK-801 basal forebrain .
- ChEMBL_1855070 (CHEMBL4355799) Displacement of MK-0499 from human ERG
- ChEMBL_2246303 (CHEMBL5160513) Inhibition of MK-499 binding to hERG
- ChEMBL_1366317 (CHEMBL3295963) Displacement of [35S]MK-499 from human ERG
- ChEMBL_1538961 (CHEMBL3737860) Displacement of [35S]-MK-0499 from human ERG
- ChEMBL_1798125 (CHEMBL4270242) Inhibition of MK-499 binding to human ERG
- ChEMBL_1924512 (CHEMBL4427468) Displacement of [35S]MK-499 from human ERG
- ChEMBL_686861 (CHEMBL1291160) Displacement of labeled MK-499 from human ERG
- ChEMBL_745160 (CHEMBL1772391) Displacement of [35S]MK-0499 from human ERG
- ChEMBL_103915 (CHEMBL711650) Inhibition of EGF-dependent mouse keratinocyte MK cell proliferation
- ChEMBL_1498874 (CHEMBL3583954) Inhibition of MK-0499 binding to Nav1.5 (unknown origin)
- ChEMBL_1498875 (CHEMBL3583955) Inhibition of MK-0499 binding to Cav1.2 (unknown origin)
- ChEMBL_154227 (CHEMBL763473) Inhibition of MK 801 binding to NMDA, PCP receptor
- ChEMBL_563071 (CHEMBL1011901) Inhibition of human ERG by MK-0499 binding assay
- ChEMBL_879794 (CHEMBL2210522) Displacement of radiolabeled MK-499 from human ERG channel
- ChEMBL_1335137 (CHEMBL3239686) Inhibition of human ERG by MK-499 displacement binding analysis
- ChEMBL_1447890 (CHEMBL3378637) Inhibition of human ERG by MK-499 displacement binding analysis
- ChEMBL_1884189 (CHEMBL4385771) Displacement of [3H]MK-801 from NMDA receptor (unknown origin)
- ChEMBL_2201494 (CHEMBL5114202) Displacement of [3H]MK-801 from NMDA receptor (unknown origin)
- ChEMBL_594565 (CHEMBL1037060) Displacement of [3H]MK-912 from human adrenergic alpha2A receptor
- ChEMBL_461764 (CHEMBL927770) Displacement of [35S]MK-499 from ERG expressed in HEK293 cells
- ChEMBL_487380 (CHEMBL1021907) Displacement of MK-499 from human ERG channel in HEK293 cells
- ChEMBL_563165 (CHEMBL964269) Displacement of [35S]MK-0677 from human growth hormone secretagogue receptor
- ChEMBL_632336 (CHEMBL1112964) Displacement of labeled MK-499 from human ERG in HEK293 cells
- ChEMBL_72508 (CHEMBL685173) Inhibitory concentration (biotinylated probe) is evaluated for [35S]- MK-0677 binding
- ChEMBL_2161344 (CHEMBL5046094) Inhibition of [35S]MK-499 binding to hERG expressed in HEK293 cells
- ChEMBL_33509 (CHEMBL647006) Inhibitory constant against Alpha-2B adrenergic receptor using [3H]MK-912 radioligand
- ChEMBL_454215 (CHEMBL903398) Displacement of [3H]MK-912 from adrenergic alpha2A receptor in HT29 cells
- ChEMBL_536741 (CHEMBL984535) Displacement of [35S]MK-499 from human ERG expressed in HEK293 cells
- ChEMBL_597859 (CHEMBL1039122) Displacement of [35S]MK-499 from human ERG expressed in HEK293 cells
- ChEMBL_699709 (CHEMBL1648352) Displacement of radiolabeled MK-499 from human ERG expressed in HEK293 cells
- ChEMBL_790372 (CHEMBL1925825) Displacement of [35S]-MK-499 from human ERG expressed in HEK cells
- ChEMBL_2134017 (CHEMBL4843627) Inhibition of [S35]-MK-0499 binding to human ERG expressed in HEK293 cells
- ChEMBL_33053 (CHEMBL647294) Inhibition of [3H]- MK-912 binding against human recombinant Alpha-2A adrenergic receptor
- ChEMBL_33078 (CHEMBL647725) Inhibition of [3H]- MK-912 binding against human recombinant Alpha-2A adrenergic receptor
- ChEMBL_336038 (CHEMBL865929) Displacement of labelled MK-499 from cloned channel hERG expressed in HEK cells
- ChEMBL_357922 (CHEMBL871230) Displacement of labeled MK-499 from cloned hERG channel expressed in HEK cells
- ChEMBL_41016 (CHEMBL655034) Compound was tested for inhibition of Beta-lactamase from Pseudomonas aeruginosa MK-1184
- ChEMBL_425254 (CHEMBL856827) Displacement of [35S]MK-499 from hERG potassium channel expressed in HEK293 cells
- ChEMBL_1473546 (CHEMBL3420425) Displacement of radio-labeled MK-499 from ERG channel in human embryonic kidney cells
- ChEMBL_1852493 (CHEMBL4353117) Displacement of [3H]-MK-801 from NMDA receptor (unknown origin) by scintillation counting method
- ChEMBL_950641 (CHEMBL2352644) Displacement of [3H]MK-801 from NMDA receptor complex (unknown origin) after 40 mins
- ChEMBL_1512245 (CHEMBL3611641) Binding affinity to human ERG expressed in HEK cells by MK-499 radioligand displacement assay
- ChEMBL_2161353 (CHEMBL5046103) Inhibition of [35S]MK-499 binding to hERG expressed in HEK293 cells at 60 uM
- ChEMBL_32915 (CHEMBL645903) Compound was tested for binding affinity using [3H]MK-912 against Alpha-2 adrenergic receptor
- ChEMBL_340159 (CHEMBL865594) Displacement of [35S]-labeled MK-499 from cloned human ERG receptor expressed in HEK cells
- ChEMBL_72512 (CHEMBL685177) Binding Affinity against Growth hormone secretagogue receptor of swine using [35S]MK-0677 as radioligand
- ChEMBL_2062193 (CHEMBL4717446) Inhibition of human ERG expressed in HEK293 cells assessed as reduction in S35-MK-0499 binding
- ChEMBL_2238202 (CHEMBL5152098) Binding affinity to NMDAR (unknown origin) assessed as inhibition constant by [3H]MK-801 binding assay
- ChEMBL_2268622 Inhibition of human recombinant LSD1/CoREST using ART(mK)QTARKSTGGKAPRKQLAGGK-biotin as substrate incubated for 15 mins
- ChEMBL_2349019 Displacement of [35S]MK-0677 from human GHSR1a expressed in HEK293 cell membrane by filter binding assay
- ChEMBL_572595 (CHEMBL1025211) Displacement of radiolabeled MK-499 from human ERG expressed in HEK293 cells coexpressing IKr channel protein
- ChEBML_154220 Compound was tested for its affinity towards PCP receptor in rat brain membrane [3H]MK-801 as radioligand
- ChEMBL_33067 (CHEMBL647966) Binding affinity towards recombinant human alpha-2A adrenergic receptor was determined using [3H]MK-912 as radioligand
- ChEMBL_1659450 (CHEMBL4009062) Displacement of [3H] MK-912 from human adrenergic alpha 2A receptor expressed in HT29 cells after 60 mins
- ChEMBL_226537 (CHEMBL846477) In vitro binding affinity at PCP binding site of sigma-opioid receptors using (+)-[3H]-MK-801 as radioligand
- ChEMBL_306231 (CHEMBL831139) Inhibitory activity against alpha-2A adrenergic receptor by using [3H]MK-912 as radioligand expressed in COS-1 cells
- ChEMBL_306290 (CHEMBL828432) In vivo inhibitory concentration against displacement of 35[S] MK-499 binding to hERG channel expressed in HEK293 cells
- ChEMBL_306311 (CHEMBL827704) In vitro inhibitory concentration against displacement of 35[S] MK-499 binding to hERG channel expressed in HEK293 cells
- ChEMBL_33037 (CHEMBL648052) Compound was evaluated for inhibition of binding of [3H]MK-91 to Alpha-2 adrenergic receptor in bovine pineal
- ChEMBL_306656 (CHEMBL831429) Inhibitory activity against constitutively activated human Alpha-2A adrenergic receptor transiently expressed in COS-1 cells using [3H]MK-912
- ChEMBL_33630 (CHEMBL648088) Compound was evaluated for inhibition of binding of [3H]MK-91 to Alpha-2 adrenergic receptor in porcine alpha2-clone
- ChEMBL_72509 (CHEMBL685174) Compound was evaluated for binding affinity using [35S]MK-0677 (800-1100 Ci/mmol), as radioligand having specific high activity
- ChEMBL_140406 (CHEMBL746732) Inhibition of [3H]-MK-801 binding to a N-methyl-D-aspartic acid(NMDA) receptor in glycine-sensitive rat cortical membranes.
- ChEMBL_215102 (CHEMBL820996) In vitro displacement of [35S]-MK- 499 from HEK 293 cells stably transfected with hERG voltage-gated potassium channel subunit Kv11.1
- ChEMBL_2459006 Inhibition of C-terminal His-tagged LSD1 (unknown origin) expressed in Escherichia coli BL21 (DE3) using ART(mK)QTARKSTGGKAPRKQLAGGK-Biotin as substrate
- ChEMBL_72513 (CHEMBL685178) Binding affinity towards human Growth hormone secretagogue receptor type 1 using competitive binding assay with radiolabeled [35S]-MK-0677 expressed as IC50
- ChEMBL_819403 (CHEMBL2033070) Antagonist activity at integrin alpha5beta1 in human A375M cells assessed as inhibition of cell adhesion in presence of Mg2+ and MK-0429
- ChEMBL_1507833 (CHEMBL3598769) Displacement of [3H]-(+)-MK-801 from phencyclidine binding site of NMDA receptor in human frontal cortex after 22 hrs by scintillation counting analysis
- ChEMBL_816921 (CHEMBL2026264) Displacement of [3H]-MK-912 from human cloned adrenergic alpha2A receptor expressed in insect Sf9 membranes after 60 mins by liquid scintillation counting
- ChEMBL_72514 (CHEMBL685179) Binding affinity to cloned human growth hormone secretagogue receptor type 1 in a competitive binding assay with [35S]MK-0677 as a radiolabeled ligand
- ChEMBL_819486 (CHEMBL2033291) Antagonist activity at integrin alpha5beta1 in human A375M cells assessed as inhibition of cell adhesion to fibronectin in presence of Mg2+ and MK-0429
- ChEMBL_140711 (CHEMBL751881) Functional antagonism at the NMDA receptor-ion channel complex was demonstarted by the ability to inhibit the binding of the channel-blocking agent [3H](+)-MK-801
- ChEMBL_140710 (CHEMBL751880) Functional antagonism at the N-methyl-D-aspartate glutamate receptor 1 was demonstarted by the ability to inhibit the binding of the channel-blocking agent [3H](+)-MK-801
- ChEMBL_141414 (CHEMBL751708) Compound was tested for inhibition of [3H]MK-801 binding to N-methyl-D-aspartate glutamate receptor at NR2B subunit in high affinity fraction of porcine brain membranes
- ChEMBL_141415 (CHEMBL751709) Compound was tested for inhibition of [3H]MK-801 binding to N-methyl-D-aspartate glutamate receptor lacking NR2B subunit in low affinity fraction of porcine brain membranes
- ChEMBL_2427364 Inhibition of N-terminal GST-tagged LSD1 (171 to 852 residues) (unknown origin) expressed in Escherichia coli using ART(mK)QTARKSTGGKAPRKQLAGGK-Biotin as substrate measured for 20 mins by spectrophotometric analysis
- ChEBML_1624157 Inhibition of ABCC2 (unknown origin) expressed in MDCK2 cells assessed as inhibition of calcein-AM efflux measured after 30 mins in presence of ABCB1 inhibitor GF120918 and ABCC1 inhibitor MK-571 by flow cytometry
- ChEMBL_2328669 Inhibition of recombinant human LSD1 (172 to 852 residues)/His-tagged human CoREST (286 to 482 residues) expressed in Escherichia coli BL21 (DE3) cells using ART(mK)QTARKSTGGKAPRKQLAGGK-Biotin as substrate assessed as increase in H3K4 methylation incubated for 40 mins
- ChEMBL_2105757 (CHEMBL4814432) Inhibition of N-terminal His-SUMO tagged human LSD1 (172 to 852 residues) expressed in Escherichia coli BL21 (DE3) using ART(mK)QTARKSTGGKAPRKQLAGGK-Biotin as substrate preincubated for 60 mins followed by substrate addition and measured after 15 mins by HTRF assay
- Screening for Inhibitors of the Mevalonate Pathway in Streptococcus Pneumoniae - MK Dose Response Southern Research Molecular Libraries Screening Center (SRMLSC) Southern Research Institute (Birmingham, Alabama) NIH Molecular Libraries Screening Centers Network (MLSCN) Assay Provider: Dr. Thomas S. Leyh, Albert Einstein College of medicine of Yeshiva University Streptococcus pneumonia (SP) takes the lives of nearly 4,000 people daily and antibiotic resistant strains are becoming an increasing problem. Because of this, the discovery of drugs targeting novel pathways such as the mevalonate pathway has become increasingly important. The pathway produces isopentyl diphosphate (the molecular building block of isoprenoids) and is essential for the survival of the pathogen in mouse lung and serum. The mevalonate pathway is comprised of three consecutive reactions that are catalyzed by the enzymes mevalonate kinase (MK; E.C. 2.7.1.36), phosphomevalonate kinase (PMK; E.C. 2.7.4.2), and diphosphomevalonate decarboxylase (PDM-DC; E.C. 4.1.1.33). MK catalyzes the ATP dependent conversion o
- ChEMBL_2105758 (CHEMBL4814433) Inhibition of N-terminal His-SUMO tagged human LSD1 (172 to 852 residues)/His-tagged CoREST (286 to 482 residues) expressed in Escherichia coli BL21 (DE3) using ART(mK)QTARKSTGGKAPRKQLAGGK-Biotin as substrate preincubated for 60 mins followed by substrate addition and measured after 15 mins by HTRF assay
- [3H] MK-801 Binding Assay Assays were conducted as described in Moskal et al. (Moskal, J. R., Kuo, A. G., Weiss, C., Wood, P. L., O'Connor Hanson, A., Kelso, S., Harris, R. B., Disterhoft, J. F., 2005. GLYX-13: a monoclonal antibody-derived peptide that acts as an N-methyl-D-aspartate receptor modulator. Neuropharmacology. 49, 1077-87) The potentiation of [3H]MK-801 binding (5 nM; 22.5 Ci/mmol) to well washed rat cortical membranes (200 μg) was measured under non-equilibrium conditions (15 min @ 25° C.) in the presence of increasing concentrations of test compounds and 50 μM glutamate. Zero levels were determined in the absence of any glycine ligand and in the presence of 30 μM 5.7 DCKA. Maximal stimulation was measured in the presence of 1 mM glycine, and 500 μM glutamate was present in all samples.
- Omnia Assay Briefly, a 1.25× stock of MK-2 enzyme from Invitrogen (PV3317), a 5× stock of ATP (AS001A), and ST3-Sox peptide substrate (KNZ1031C) were prepared in 1× kinase reaction buffer consisting of 20 mM Tris, pH 7.5, 5 mM MgCl2, 1 mM EGTA, 5 mM β-glycerophosphate, 5% glycerol (10× stock, KB002A) and 0.2 mM DTT. Compound potency assays were initiated by adding a 0.5 μL volume of 100% DMSO and serially diluted compounds prepared in 100% DMSO to a Corning (#3574) 384-well, white, non-binding surface microtiter plate (Corning, N.Y.) followed immediately by 10 μL of the ST3-Sox peptide and ATP substrate solution. Kinase reactions were started with the addition of 40 μL of MK-2 enzyme and monitored every 71 seconds for 30-240 minutes at λex360/λem485 in a Synergy2, Synergy4 or Synergy H4 plate reader from BioTek (Winooski, Vt.). At the conclusion of each assay, progress curves from each well were examined for linear reaction kinetics and fit statistics (R2, 95% confidence interval, absolute sum of squares). Initial velocity (0 minutes to +30 minutes) from each reaction was estimated from the slope of a plot of relative fluorescence units vs time (minutes) and normalized to the no enzyme and no inhibitor control groups for % Inhibition. The resulting % Inhibition values were then plotted against inhibitor concentration to estimate IC50 from log[Inhibitor] vs Response, Variable Slope model in GraphPad Prism from GraphPad Software (San Diego, Calif.). Potency results for the compounds tested are shown in Table A in the column entitled MK2 IC50. [Reagent] used:[MK-2]=0.4 nM, [ATP]=1.0 mM and [ST3-Sox]=10 μM (ATP appKM=8-10 μM)
- Inhibition Assay Thoroughly mixed test compounds, enzyme and reaction buffer, preincubated the mixture for 15 minutes at 37° C. and then primed reaction by adding substrate, successfully detecting fluorescence value at 460 nm for 5 minutes. At the same time, set the blank control group without substrate and solvent control group with DMSO replacing test compound, as well as positive control group of Vildagliptin (LAF-237) and Sitagliptin (MK-0431) [Bioorg. Med. Chem. Lett., 2005, 15, 4770-4773]. All final reaction volumes were 100 μL. Each concentration of each sample consisted of parallel wells in triplate.
- MK-2 IC50 Value Determination The phosphorylation of HSP27 peptide by MAPKAPK2 was measured using an anion exchange resin capture assay method. The reaction was carried out in reaction buffer containing enzyme and test compound in the presence of the HSP-peptide with [gamma-33P]ATP /ATP. After the reaction was terminated and [gamma-33P]ATP was removed from solution by the addition of AG 1X8 ion exchange resin. An aliquot was removed from the quenched reaction mixture and added to a 96-well plate. Microscint-40 (Packard) was added and the amount of phosphorylated-peptide was determined.
- FLAP Binding Assay (Test A) Compounds were tested in a competition binding assay using 3H-MK591 as tracer. (Preparation of MK-591 is described in Bioorg. Med. Chem. Lett. 1999, 9, 2391). A 100,000×g pellet from COS-7 cells stably transfected with a plasmid expressing human ALOX5AP was the source of FLAP. Membrane pellets were resuspended in buffer (100 mM Tris-HCl, 0.05% Tween-20, 140 mM NaCl, 2 mM EDTA, 0.5 mM DTT, 5% Glycerol, pH 7.5) to give a final protein concentration of 12 mg/mL (2 μg/well). To perform assays, 1.4 μL compounds were dispensed into 96-well plates in 3-fold dilution series in triplicate. 84 μL radioligand (25000 CPM, 2 nM final concentration in assay) was then added followed by 84 μL membrane suspension and incubation at rt for 60 min. Following filtration, filter plates were dried 12 h at RT (or 50° C. for 1 hour). 50 μL scintillant was then added, the filterplates were sealed and radioactivity was measured in a microbeta counter. Specific binding was defined as total binding minus non-specific binding. Total binding was defined as 3H-MK591 bound to membranes in the absence of competitor, non-specific binding was defined as 3H-MK591 in the presence of 0.1 mM MK-591. IC50 values were determined by plotting % inhibition versus log compound concentration and using a one site dose response model.
- MK-499 Filter Binding Assay Drug cardiac arrhythmia is an important safety concern for pharmaceutical development and health regulatory authorities. Blockade of heterologously-expressed human ether-a-go-go gene (hERG) channel prolongs the duration of the cardiac action potential leading to a long QT interval that can lead to sudden death (De Ponti, F.; et al Drug Safety 2002, 25, pp. 263-286). It is important to have compounds devoid of hERG channel activity as measured by an in vitro assay. Affinity of compounds for the hERG channel was evaluated in radioligand competition experiments using HEK293 cells that were stably transfected with the hERG channel and radiolabeled ligand, MK-499 a potent antiarrhythmic. This assay correlates well with QT prolongation in vivo (Jamieson, C.; et al., J. Med. Chem. 2006, 49, pp. 5029-5046). 25 μL Target membranes (in assay buffer: 10 mM HEPES/NaOH, pH 7.4, 70 mM NaCl, 60 mM KCl, 2 mM MgCl2, 1 mM CaCl2) purified from a HEK293 cell line expressing the human Ether- -go-go Related Gene (hERG) ion channel, 1 μL test compound in 10 mM DMSO and 25 μL (6,000 cpm/well; in assay buffer) [35S]MK-0499 radioligand (Merck/Perkin Elmer) were added to the assay plate (Axygen; 384 Deep well Diamond Plate , clear). After incubation of the binding reaction at room temperature (RT) for 90 min 50 μL of the assay were transferred to a Multiscreen HTS 384 FC filter plate (Millipore), which had been pre-wetted with 20 μL 0.01% PEI/0.01% Triton X-100 for at least 30 min at RT. Then, 30 μL wash buffer (10 mM HEPES/NaOH, pH 7.4, 130 mM NaCl, 2 mM MgCl2, 1 mM CaCl2) equilibrated to RT were added to each well of the assay plate and subsequently transferred to the filter plate. The assay mixture was aspirated through the filter plate using a Biotek ELx405 washer. The filter plate was washed twice with 100 μl cold wash buffer per wash and well and then dried in a drying oven for at least 75 min at 55° C. Afterwards, the bottom of each filter plate was heat sealed with a solid foil seal, then 10 μL of Microscint 0 (Perkin Elmer) were added to each well of the filter plate and finally, the top of each filter plate was sealed with a clear seal. The plates were stored for at least 20 min in a MicroBeta2 reader (Perkin Elmer) before they are counted (60 sec/well).
- Screening for Inhibitors of the Mevalonate Pathway in Streptococcus Pneumoniae - DPM-DC - Secondary Assay Southern Research Molecular Libraries Screening Center (SRMLSC) Southern Research Institute (Birmingham, Alabama) NIH Molecular Libraries Screening Centers Network (MLSCN) Assay Provider: Dr. Thomas S. Leyh, Albert Einstein College of Medicine of Yeshiva University Award: R03 MH078936-01 Streptococcus pneumoniae takes the lives of nearly 4,000 people daily and antibiotic resistant strains are becoming an increasing problem. Because of this, the discovery of drugs targeting novel pathways has become increasingly important. The mevalonate pathway produces isopentenyl diphosphate (the molecular building block of isoprenoids) and is essential for the survival of the pathogen in mouse lung and serum. The biosynthesis of isopentenyl diphosphate involves three consecutive reactions that are catalyzed by the enzymes mevalonate kinase (MK; E.C. 2.7.1.36), phosphomevalonate kinase (PMK; E.C. 2.7.4.2), and diphosphomevalonate decarboxylase (DPM-DC; E.C. 4.1.1.33). DPM-DC catalyzes the ATP-
- Screening for Inhibitors of the Mevalonate Pathway in Streptococcus Pneumoniae - DPM-DC Dose Response Southern Research Molecular Libraries Screening Center (SRMLSC) Southern Research Institute (Birmingham, Alabama) NIH Molecular Libraries Screening Centers Network (MLSCN) Assay Provider: Dr. Thomas S. Leyh, Albert Einstein College of Medicine of Yeshiva University Streptococcus pneumoniae takes the lives of nearly 4,000 people daily and antibiotic resistant strains are becoming an increasing problem. Because of this, the discovery of drugs targeting novel pathways has become increasingly important. The mevalonate pathway produces isopentenyl diphosphate (the molecular building block of isoprenoids) and is essential for the survival of the pathogen in mouse lung and serum. The biosynthesis of isopentenyl diphosphate involves three consecutive reactions that are catalyzed by the enzymes mevalonate kinase (MK; E.C. 2.7.1.36), phosphomevalonate kinase (PMK; E.C. 2.7.4.2), and diphosphomevalonate decarboxylase (DPM-DC; E.C. 4.1.1.33). DPM-DC catalyzes the ATP-dependent conversion of (R
- Radioligand Binding Assay To perform the competition binding assay, thawed membrane homogenate was added to each well of a 96-well plate (20 ug/well). The experimental compounds were serially diluted in 100% DMSO and added to each row of the assay plate to achieve desired compound concentrations, keeping the DMSO concentration in the assay plate at 1.33% of the final reaction volume. Next, 3H Ro 25-6981 (4 nM) was added to the assay plate. After incubation for 1 hr at room temperature, the membrane bound radioligand was harvested on to GF/B filter plates (treated with 0.5% PEI for 1 hr at room temperature). The filter plates were dried at 50° C. for 20 mins, incubated with microscint 20 for 10 minutes and finally, the counts were read on TopCount (Perkin Elmer). Non-specific binding was determined using MK-0657 (the preparation of this compound is described as example 1 in WO 2004 108705 (40 uM).
- Radioligand Binding Assay To perform the competition binding assay, thawed membrane homogenate was added to each well of a 96-well plate (20 μg/well). The experimental compounds were serially diluted in 100% DMSO and added to each row of the assay plate to achieve desired compound concentrations, keeping the DMSO concentration in the assay plate at 1.33% of the final reaction volume. Next, 3H Ro 25-6981 (4 nM) was added to the assay plate. After incubation for 1 hr at room temperature, the membrane bound radioligand was harvested on to GF/B filter plates (treated with 0.5% PEI for 1 hr at room temperature). The filter plates were dried at 50° C. for 20 mins, incubated with microscint 20 for 10 minutes and finally, the counts were read on TopCount (Perkin Elmer). Non-specific binding was determined using MK-0657 (the preparation of this compound is described as example 1 in WO 2004 108705 (40 μM). CPM values were converted to % inhibition and the concentration response curves were plotted using custom made software.
- Radioligand Binding Assay Human CGRP receptors (consisting of CRLR and RAMP1) expressed in insect Sf21 cell membrane homogenates were re-suspended in the binding buffer (10 mM HEPES, pH 7.4, 5 mM MgCl2, 0.2% BSA) to a final assay concentration of 0.6 protein per well. Saturation isotherms were determined by the addition of various concentrations of 3H-telcagepant (Ho et al, The Lancet, 2008, 372, 2115) (in a total reaction volume of 250 μL) for 60 min at room temperature. At the end of the incubation, membranes were filtered onto a unifilter, a 96-well white microplate with bonded GF/B filter pre-incubated with 0.5% PEI, with a Tomtec cell harvester and washed 5 times with distilled water. Non-specific binding (NSB) was measured in the presence of 10 nM MK-3207 hydrochloride (CAS No. 957116-20-0). Radioactivity on the filter was counted (1 min) on a microbeta counter after addition of 50 μL of scintillation fluid. For inhibition experiments, membranes were incubated with 0.5 nM 3H-telcagepant and 10 concentrations of the inhibitory compound (0.001-10 IC50 values were derived from the inhibition curve and the affinity constant (Ki) values were calculated using the Cheng-Prussoff equation (Cheng et al, Biochem. Pharmacol. 1973, 22, 3099-3108).
- Radioligand Binding Assay Human CGRP receptors expressed (consisting of CRLR and RAMP1) in insect Sf21 cell membrane homogenates were re-suspended in the binding buffer (10 mM HEPES, pH 7.4, 5 mM MgCl2, 0.2% BSA) to a final assay concentration of 0.6 μg protein per well. Saturation isotherms were determined by the addition of various concentrations of 3H-telcagepant (Ho et al, The Lancet, 2008, 372, 2115) (in a total reaction volume of 250 μL) for 60 min at rt. At the end of the incubation, membranes were filtered onto a unifilter, a 96-well white microplate with bonded GF/B filter pre-incubated with 0.5% PEI, with a Tomtec cell harvester and washed 5 times with distilled water. Non-specific binding (NSB) was measured in the presence of 10 nM MK-3207 hydrochloride (CAS No. 957116-20-0). Radioactivity on the filter was counted (1 min) on a microbeta counter after addition of 50 μL of scintillation fluid. For inhibition experiments, membranes were incubated with 0.5 nM 3H-telcagepant and 10 concentrations of the inhibitory compound (0.001-10 μM). IC50 values were derived from the inhibition curve and the affinity constant (Ki) values were calculated using the Cheng-Prussoff equation (Cheng et al, Biochem. Pharmacol. 1973, 22, 3099-3108). The pKi values (where pKi=−log10 Ki) of certain compounds of the invention are tabulated below.
- Radioligand binding assay Radioligand binding assay. Human CGRP receptors expressed (consisting of CRLR and RAMP1) in insect Sf21 cell membrane homogenates were re-suspended in the binding buffer (10 mM HEPES, pH 7.4, 5 mM MgCl2, 0.2% BSA) to a final assay concentration of 0.6 μg protein per well. Saturation isotherms were determined by the addition of various concentrations of 3H-telcagepant (Ho et al, The Lancet, 2008, 372, 2115) (in a total reaction volume of 250 μL) for 60 min at rt. At the end of the incubation, membranes were filtered onto a unifilter, a 96-well white microplate with bonded GF/B filter pre-incubated with 0.5% PEI, with a Tomtec cell harvester and washed 5 times with distilled water. Non-specific binding (NSB) was measured in the presence of 10 nM MK-3207 hydrochloride (CAS No. 957116-20-0). Radioactivity on the filter was counted (1 min) on a microbeta counter after addition of 50 μL of scintillation fluid. For inhibition experiments, membranes were incubated with 0.5 nM 3H-telcagepant and 10 concentrations of the inhibitory compound (0.001-10 μM). IC50 values were derived from the inhibition curve and the affinity constant (Ki) values were calculated using the Cheng-Prussoff equation (Cheng et al, Biochem. Pharmacol. 1973, 22, 3099-3108).
- Recombinant CDK8 Protein-Cyclin C Complex In Vitro Capability of compounds of the present invention to bind to CDK8 protein was detected using LanthaScreen (ThermoFisher) assay. We detected FRET signal proportional to the amount of CDK8-bound fluorescently-labeled ligand (Tracer 236) that competes with inhibitor for ATP binding site. We carried out the measurements in a 15 μl reaction volume using a 384-well plate (Corning, #CLS4513). Enzyme CDK8/Cyclin C (ThermoFisher, #PR7261B) was mixed with antibodies Anti-His-tag-Biotin (ThermoFisher, #PV6090), Streptavidin-Eu (ThermoFisher, #PV6025), the resulting mixture was added to plate wells (5 MKπ/well). Final concentrations of substances were as follows: CDK8/Cyclin C 5 nM, Streptavidin-Eu 3 nM, Anti-His-tag-Biotin 3 nM. Staurosporine (S4400, Sigma) was used as a reference inhibitor, and 0.1% solution of dimethyl sulphoxide (DMSO) in reaction buffer was used as a blank, the reaction buffer comprised 250 mM HEPES (pH 7.5), 50 mM MgCl2, 5 mM EGTA, and 0.05% Brij-35.The inhibitors and controls in question were added to corresponding wells (5 μl/well). The plate was incubated at room temperature for 20 minutes. After incubating, 5 μl/well of tracer solution Alexa Fluor-647 (Kinase Tracer 236, ThermoFisher, #PV5592)) was added to the wells. Final concentration of tracer was 10 nM. Reaction buffer, instead of tracer solution, was used as a negative control. The plate was incubated for 40 minutes at 25 C., TR-FRET signal was then measured according to the manufacturer's guidelines on SPARK20 plate reader (Tecan, Switzerland), and the value was converted to the amount of kinase-bound tracer.
- Binding Assay To prepare cell membranes with human α1- and α2-adrenergic receptors, CHO cells stably overexpressing α1- and α2-adrenergic receptors are lysed and then subjected to differential centrifugation. After lysis in binding buffer (50 mM tris(hydroxymethyl)aminomethane/1 N hydrochloric acid, 5 mM magnesium chloride, pH 7.4) using an Ultra Turrax (Jahnke & Kunkel, Werk), the homogenate is centrifuged at 1000 g and at 4° C. for 10 min. The resulting sediment is discarded and the supernatant is centrifuged at 20 000 g and at 4° C. for 30 min. The supernatant is discarded and the sediment is resuspended in binding buffer and stored at −70° C. until the binding test. For the binding test the radioligands 3H-MK-912 (2.2-3.2 TBq/mmol, PerkinElmer) (0.4 nM for α2C-adrRez and 1 nM for α2A-adrRez), 0.25 nM 3H-prazosin (α1AC-adrRez; 2.6-3.3 TBq/mmol, PerkinElmer), 0.25 nM 3H-rauwolscine (α2B-adrRez, 2.6-3.2 TBq/mmol, PerkinElmer) are incubated for 60 minutes with 5-20 μg cell membranes in binding buffer (total test volume 0.2 ml) in the presence of the test substances at 30° C. in 96-well filter plates (FC/B glass fibre, Multiscreen Millipore). The incubating is terminated by aspiration of the unbound radioactivity and the plates are then washed with binding buffer and subsequently dried at 40° C. for 1 hour. Liquid scintillator (Ultima Gold, PerkinElmer) is then added and the radioactivity that remained on the plates is measured in a liquid scintillation counter (Microbeta, Wallac). Non-specific binding is defined as radioactivity in the presence of 1-10 μM WB-4101 (α2C-adrRez and α2A-adrRez), prazosin (α2B-adrRez and α1AC-adrRez) (all from Sigma) and is generally <25% of the bound total radioactivity.
- Biological Assay (HTS) NMDA receptors are ion channels that are highly permeable to Ca2+ ions, rendering it possible to monitor NMDA receptor function using cell-based calcium flux assay. In this assay, co-agonists glutamate and glycine are added to cells heterologously expressing human GluN1/GluN2B NMDA receptors to initiate cellular Ca2+ influx. The time course of the changes in intracellular calcium is measured using a fluorescent dye and a FLIPR (Fluorometric Imaging Plate Reader) device. Twenty four hours before measurements, the expression of the NMDA receptors in the stable cell line is induced with Tet-On inducible system in the presence of a non-selective NMDA receptor blocker. On the day of the experiment, cell culture media is carefully washed and the cells are loaded with Calcium 5 Dye Kit (Molecular Devices) in dye loading buffer containing 137 mM NaCl, 4 mM KCl, 2 mM CaCl2), 1.5 mM MgCl2, 10 mM HEPES and 5 mM D-glucose; pH 7.4. After 1 h incubation at the room temperature, the dye is washed away with the assay buffer (150 mM, 3 mM, 2 mM CaCl2), 0.01 mM EDTA, 10 mM HEPES and 5 mM D-glucose; pH 7.4) In the FLIPR TETRA reader, various concentrations of the test compounds are added to the cells for 5 min while fluorescence is monitored to detect potential agonist activity. Next, co-agonists, glutamate and glycine are added for another 5 minutes. The concentration of glutamate corresponding to EC40 is used to maximize the assay's signal window and ability to detect NMDA receptor antagonists and negative allosteric modulators. A saturating concentration (10 μM) of glycine is also present in the assay. A non-selective NMDA receptor antagonist, (+)MK-801 is used as a positive control for antagonist activity. The fluorescent signal in the presence of test compounds is quantified and normalized to the signal defined by the appropriate control wells.
- Biological Assay (standard) NMDA receptors are ion channels that are highly permeable to Ca2+ ions, rendering it possible to monitor NMDA receptor function using cell-based calcium flux assay. In this assay, co-agonists glutamate and glycine are added to cells heterologously expressing human GluN1/GluN2B NMDA receptors to initiate cellular Ca2+ influx. The time course of the changes in intracellular calcium is measured using a fluorescent dye and a FLIPR (Fluorometric Imaging Plate Reader) device.Twenty four hours before measurements, the expression of the NMDA receptors in the stable cell line is induced with Tet-On inducible system in the presence of a non-selective NMDA receptor blocker. On the day of the experiment, cell culture media is carefully washed and the cells are loaded with Calcium 5 Dye Kit (Molecular Devices) in dye loading buffer containing 137 mM NaCl, 4 mM KCl, 2 mM CaCl2), 0.5 mM MgCl2, 10 mM HEPES and 5 mM D-glucose; pH 7.4. After 1 h incubation at the room temperature, the dye is washed away with the assay buffer (137 mM NaCl, 4 mM KCl, 2 mM CaCl2), 0.01 mM EDTA, 10 mM HEPES and 5 mM D-glucose; pH 7.4) In the FLIPR TETRA reader, various concentrations of the test compounds are added to the cells for 5 min while fluorescence is monitored to detect potential agonist activity. Next, co-agonists, glutamate and glycine are added for another 5 minutes. The concentration of glutamate corresponding to ~EC40 used to maximize the assay's signal window and ability to detect NMDA receptor antagonists and negative allosteric modulators. A saturating concentration (10 μM) of glycine is also present in the assay. A non-selective NMDA receptor antagonist, (+)MK-801 is used as a positive control for antagonist activity. The fluorescent signal in the presence of test compounds is quantified and normalized to the signal defined by the appropriate control wells.
- NMDA receptor FLIPR assay NMDA receptors are ion channels that are highly permeable to Ca2+ ions, rendering it possible to monitor NMDA receptor function using cell-based calcium flux assay. In this assay, co-agonists glutamate and glycine are added to cells heterologously expressing human GluN1/GluN2B NMDA receptors to initiate cellular Ca2+ influx. The time course of the changes in intracellular calcium is measured using a fluorescent dye and a FLIPR (Fluorometric Imaging Plate Reader) device.Twenty four hours before measurements, the expression of the NMDA receptors in the stable cell line is induced with Tet-On inducible system in the presence of a non-selective NMDA receptor blocker. On the day of the experiment, cell culture media is carefully washed and the cells are loaded with Calcium 5 Dye Kit (Molecular Devices) in dye loading buffer containing 149 mM NaCl, 4 mM KCl, 2 mM CaCl2, and 1.5 mM MgCl2, 10 mM HEPES and 5 mM D-glucose; pH 7.4. After 1 h incubation at the room temperature, the dye is washed away with the assay buffer (149 mM NaCl (standard assay) or 150 mM (HTS assay), 4 mM KCl (standard assay) or 3 mM (HTS assay), 2 mM CaCl2, 0.01 mM EDTA, 10 mM HEPES and 5 mM D-glucose; pH 7.4) In the FLIPR TETRA reader, various concentrations of the test compounds are added to the cells for 5 min while fluorescence is monitored to detect potential agonist activity. Next, co-agonists, glutamate and glycine are added for another 5 minutes. The concentration of glutamate corresponding to EC80 (standard assay) or EC40 (HTS assay) is used to maximize the assay's signal window or the ability to detect NMDA receptor antagonists and negative allosteric modulators, respectively. A saturating concentration (10 μM) of glycine is also present in the assay. A non-selective NMDA receptor antagonist, (+)MK-801 is used as a positive control for antagonist activity. The fluorescent signal in the presence of test compounds is quantified and normalized to the signal defined by the appropriate control wells.
- NR2B standard assay NMDA receptors are ion channels that are highly permeable to Ca2+ ions, rendering it possible to monitor NMDA receptor function using cell-based calcium flux assay. In this assay, co-agonists glutamate and glycine are added to cells heterologously expressing human GluN1/GluN2B NMDA receptors to initiate cellular Ca2+ influx. The time course of the changes in intracellular calcium is measured using a fluorescent dye and a FLIPR (Fluorometric Imaging Plate Reader) device.Twenty four hours before measurements, the expression of the NMDA receptors in the stable cell line is induced with Tet-On inducible system in the presence of a non-selective NMDA receptor blocker. On the day of the experiment, cell culture media is carefully washed and the cells are loaded with Calcium 5 Dye Kit (Molecular Devices) in dye loading buffer containing 149 mM NaCl, 4 mM KCl, 2 mM CaCl2, and 1.5 mM MgCl2, 10 mM HEPES and 5 mM D-glucose; pH 7.4. After 1 h incubation at the room temperature, the dye is washed away with the assay buffer (149 mM NaCl (standard assay) or 150 mM (HTS assay), 4 mM KCl (standard assay) or 3 mM (HTS assay), 2 mM CaCl2, 0.01 mM EDTA, 10 mM HEPES and 5 mM D-glucose; pH 7.4) In the FLIPR TETRA reader, various concentrations of the test compounds are added to the cells for 5 min while fluorescence is monitored to detect potential agonist activity. Next, co-agonists, glutamate and glycine are added for another 5 minutes. The concentration of glutamate corresponding to EC80 (standard assay) or EC40 (HTS assay) is used to maximize the assay's signal window or the ability to detect NMDA receptor antagonists and negative allosteric modulators, respectively. A saturating concentration (10 μM) of glycine is also present in the assay. A non-selective NMDA receptor antagonist, (+)MK-801 is used as a positive control for antagonist activity. The fluorescent signal in the presence of test compounds is quantified and normalized to the signal defined by the appropriate control wells.
- Effect of Compounds of Formula (I) on Cloned Human GluN1/GluN2B Ion Channels Expressed in Mammalian Cells NMDA receptors are ion channels that are highly permeable to Ca2+ ions, rendering it possible to monitor NMDA receptor function using cell-based calcium flux assay. In this assay, co-agonists glutamate and glycine are added to cells heterologously expressing human GluN1/GluN2B NMDA receptors to initiate cellular Ca2+ influx. The time course of the changes in intracellular calcium is measured using a fluorescent dye and a FLIPR (Fluorometric Imaging Plate Reader) device.Twenty four hours before measurements, the expression of the NMDA receptors in the stable cell line is induced with Tet-On inducible system in the presence of a non-selective NMDA receptor blocker. On the day of the experiment, cell culture media is carefully washed and the cells are loaded with Calcium 5 Dye Kit (Molecular Devices) in dye loading buffer containing 137 mM NaCl, 4 mM KCl, 2 mM CaCl2, 0.5 mM MgCl2 (standard assay) or 1.5 mM MgCl2 (HTS assay), 10 mM HEPES and 5 mM D-glucose; pH 7.4. After 1 h incubation at the room temperature, the dye is washed away with the assay buffer (137 mM NaCl (standard assay) or 150 mM (HTS assay), 4 mM KCl (standard assay) or 3 mM (HTS assay), 2 mM CaCl2, 0.01 mM EDTA, 10 mM HEPES and 5 mM D-glucose; pH 7.4) In the FLIPR TETRA reader, various concentrations of the test compounds are added to the cells for 5 min while fluorescence is monitored to detect potential agonist activity. Next, co-agonists, glutamate and glycine are added for another 5 minutes. The concentration of glutamate corresponding to EC40 (standard assay) or EC40 (HTS assay) is used to maximize the assay's signal window and ability to detect NMDA receptor antagonists and negative allosteric modulators. A saturating concentration (10 μM) of glycine is also present in the assay. A non-selective NMDA receptor antagonist, (+)MK-801 is used as a positive control for antagonist activity. The fluorescent signal in the presence of test compounds is quantified and normalized to the signal defined by the appropriate control wells.
- Effects of Test Articles on Cloned Human NR1/GuN2B Ion Channels Expressed in Mammalian Cells NMDA receptors are ion channels that are highly permeable to Ca2+ ions, rendering it possible to monitor NMDA receptor function using cell-based calcium flux assay. In this assay, co-agonists glutamate and glycine are added to cells heterologously expressing human GluN1/GluN2B NMDA receptors to initiate cellular Ca2+ influx. The time course of the changes in intracellular calcium is measured using a fluorescent dye and a FLIPR (Fluorometric Imaging Plate Reader) device.Twenty-four hours before measurements, the expression of the NMDA receptors in the stable cell line is induced with Tet-On inducible system in the presence of a non-selective NMDA receptor blocker. On the day of the experiment, cell culture media is carefully washed, and the cells are loaded with Calcium 5 Dye Kit (Molecular Devices) in dye loading buffer containing 137 mM NaCl, 4 mM KCl, 2 mM CaCl2, 0.5 mM MgCl2 (standard assay) or 1.5 mM MgCl2 (HTS assay), 10 mM HEPES and 5 mM D-glucose; pH 7.4. After 1 h incubation at the room temperature, the dye is washed away with the assay buffer (137 mM NaCl (standard assay) or 150 mM (HTS assay), 4 mM KCl (standard assay) or 3 mM (HTS assay), 2 mM CaCl2), 0.01 mM EDTA, 10 mM HEPES and 5 mM D-glucose; pH 7.4) In the FLIPR TETRA reader, various concentrations of the test compounds are added to the cells for 5 min while fluorescence is monitored to detect potential agonist activity. Next, co-agonists, glutamate and glycine are added for another 5 minutes. The concentration of glutamate corresponding to EC40 (standard assay) or EC40 (HTS assay) is used to maximize the assay's signal window and ability to detect NMDA receptor antagonists and negative allosteric modulators. A saturating concentration (10 μM) of glycine is also present in the assay. A non-selective NMDA receptor antagonist, (+)MK-801 is used as a positive control for antagonist activity. The fluorescent signal in the presence of test compounds is quantified and normalized to the signal defined by the appropriate control wells.
- HTS cell-based calcium flux assay NMDA receptors are ion channels that are highly permeable to Ca2+ ions, rendering it possible to monitor NMDA receptor function using cell-based calcium flux assay. In this assay, co-agonists glutamate and glycine are added to cells heterologously expressing human GluN1/GluN2B NMDA receptors to initiate cellular Ca2+ influx. The time course of the changes in intracellular calcium is measured using a fluorescent dye and a FLIPR (Fluorometric Imaging Plate Reader) device.Twenty four hours before measurements, the expression of the NMDA receptors in the stable cell line is induced with Tet-On inducible system in the presence of a non-selective NMDA receptor blocker. On the day of the experiment, cell culture media is carefully washed and the cells are loaded with Calcium 5 Dye Kit (Molecular Devices) in dye loading buffer containing 137 mM NaCl, 4 mM KCl, 2 mM CaCl2, 0.5 mM MgCl2 (standard assay) or 1.5 mM MgCl2 (HTS assay), 10 mM HEPES and 5 mM D-glucose; pH 7.4. After 1 h incubation at the room temperature, the dye is washed away with the assay buffer (137 mM NaCl (standard assay) or 150 mM (HTS assay), 4 mM KCl (standard assay) or 3 mM (HTS assay), 2 mM CaCl2, 0.01 mM EDTA, 10 mM HEPES and 5 mM D-glucose; pH 7.4) In the FLIPR TETRA reader, various concentrations of the test compounds are added to the cells for 5 min while fluorescence is monitored to detect potential agonist activity. Next, co-agonists, glutamate and glycine are added for another 5 minutes. The concentration of glutamate corresponding to EC40 (HTS assay) is used to maximize the assay's signal window and ability to detect NMDA receptor antagonists and negative allosteric modulators. A saturating concentration (10 μM) of glycine is also present in the assay. A non-selective NMDA receptor antagonist, (+)MK-801 is used as a positive control for antagonist activity. The fluorescent signal in the presence of test compounds is quantified and normalized to the signal defined by the appropriate control wells.
- Standard cell-based calcium flux assay NMDA receptors are ion channels that are highly permeable to Ca2+ ions, rendering it possible to monitor NMDA receptor function using cell-based calcium flux assay. In this assay, co-agonists glutamate and glycine are added to cells heterologously expressing human GluN1/GluN2B NMDA receptors to initiate cellular Ca2+ influx. The time course of the changes in intracellular calcium is measured using a fluorescent dye and a FLIPR (Fluorometric Imaging Plate Reader) device.Twenty four hours before measurements, the expression of the NMDA receptors in the stable cell line is induced with Tet-On inducible system in the presence of a non-selective NMDA receptor blocker. On the day of the experiment, cell culture media is carefully washed and the cells are loaded with Calcium 5 Dye Kit (Molecular Devices) in dye loading buffer containing 137 mM NaCl, 4 mM KCl, 2 mM CaCl2, 0.5 mM MgCl2 (standard assay) or 1.5 mM MgCl2 (HTS assay), 10 mM HEPES and 5 mM D-glucose; pH 7.4. After 1 h incubation at the room temperature, the dye is washed away with the assay buffer (137 mM NaCl (standard assay) or 150 mM (HTS assay), 4 mM KCl (standard assay) or 3 mM (HTS assay), 2 mM CaCl2, 0.01 mM EDTA, 10 mM HEPES and 5 mM D-glucose; pH 7.4) In the FLIPR TETRA reader, various concentrations of the test compounds are added to the cells for 5 min while fluorescence is monitored to detect potential agonist activity. Next, co-agonists, glutamate and glycine are added for another 5 minutes. The concentration of glutamate corresponding to EC40 (standard assay) is used to maximize the assay's signal window and ability to detect NMDA receptor antagonists and negative allosteric modulators. A saturating concentration (10 μM) of glycine is also present in the assay. A non-selective NMDA receptor antagonist, (+)MK-801 is used as a positive control for antagonist activity. The fluorescent signal in the presence of test compounds is quantified and normalized to the signal defined by the appropriate control wells.
- Inhibition of Specific Binding to the Rat NR1/NR2B Receptor The assay depends on the binding of a tracer to the GluN2B subunit-containing NMDA receptors and the ability of the test compounds to displace such binding. 3-[3H] 1-(azetidin-1-yl)-2-[6-(4-fluoro-3-methyl-phenyl)pyrrolo[3,2-b]pyridin-1-yl]ethanone is a high-affinity GluN2B-selective antagonist, which binds to the Ifenprodil binding site located at the interphase between GluN1 and GluN2B subunits. Alternatively, The assay measures binding affinity for ligands that compete for the Ifenprodil binding site in the native NMDA receptors from adult rat cortical membranes.In brief, rat adult cortex is homogenized in the assay buffer (50 mM Tris; pH 7.4). The resulting cortical membranes containing native NMDA receptors are purified by centrifugation and extensively washed, then re-suspended in the assay buffer. The test compounds, tracer and membranes are mixed together and incubated with shaking for 2 hours at room temperature to reach binding equilibrium. Non-specific binding of the tracer is determined by pre-incubation of brain membranes with 10 μM of CP 101,606. Following the incubation, the bound and unbound tracer is separated by filtration with cell harvester and GF/B filter plates (PerkinElmer) soaked with polyethylenimine.The extent of binding is measured by counting [3H] radioactivity retained on the filters plates with liquid scintillator counter. Binding affinity (equilibrium dissociation constant Ki) for the test compounds is determined by fitting experimental data with the following model log EC50=log(10^ log Ki*(1+[Radioligand]/HotKd)) and Y=Bottom+(Top-Bottom)/(1+10^(X-Log EC50)) where [Radioligand] is the concentration of the tracer, HotKdNM is the equilibrium dissociation constant of the tracer, Top and Bottom are the curve plateaus in the units of Y axis.HNR2BC: Effects of Test Articles on Cloned Human NR1/NR2B Ion Channels Expressed in Mammalian CellsNMDA receptors are ion channels that are highly permeable to Ca2+ ions, rendering it possible to monitor NMDA receptor function using cell-based calcium flux assay. In this assay, co-agonists glutamate and glycine are added to cells heterologously expressing human GluN1/GluN2B NMDA receptors to initiate cellular Ca2+ influx. The time course of the changes in intracellular calcium is measured using a fluorescent dye and a FLIPR (Fluorometric Imaging Plate Reader) device.Twenty four hours before measurements, the expression of the NMDA receptors in the stable cell line is induced with Tet-On inducible system in the presence of a non-selective NMDA receptor blocker. On the day of the experiment, cell culture media is carefully washed and the cells are loaded with Calcium 5 Dye Kit (Molecular Devices) in dye loading buffer containing 137 mM NaCl, 4 mM KCl, 2 mM CaCl2, 0.5 mM MgCl2, 10 mM HEPES and 5 mM D-glucose; pH 7.4. After 1 h incubation at the room temperature, the dye is washed away with the assay buffer (137 mM NaCl, 4 mM KCl, 2 mM CaCl2, 0.01 mM EDTA, 10 mM HEPES and 5 mM D-glucose; pH 7.4) In the FLIPR TETRA reader, various concentrations of the test compounds are added to the cells for 5 min while fluorescence is monitored to detect potential agonist activity. Next, co-agonists, glutamate and glycine are added for another 5 minutes. The concentration of glutamate corresponding to EC80 is used to maximize the assay's signal window and ability to detect NMDA receptor antagonists and negative allosteric modulators. A saturating concentration (10 μM) of glycine is also present in the assay. A non-selective NMDA receptor antagonist, (+)MK-801 is used as a positive control for antagonist activity. The fluorescent signal in the presence of test compounds is quantified and normalized to the signal defined by the appropriate control wells.
 CHEMBL1079175 US10550114, Compound MK-2206 8-(4-(1-aminocyclobutyl)phenyl)-9-phenyl-[1,2,4]triazolo[3,4-f][1,6]naphthyridin-3(2H)-one MK-2206 BDBM50313650
CHEMBL1079175 US10550114, Compound MK-2206 8-(4-(1-aminocyclobutyl)phenyl)-9-phenyl-[1,2,4]triazolo[3,4-f][1,6]naphthyridin-3(2H)-one MK-2206 BDBM50313650 BDBM102776 US8541572, 2206
BDBM102776 US8541572, 2206 BDBM266310 US9718790, I-2206
BDBM266310 US9718790, I-2206 BDBM736020 US20250129104, Compound 2206
BDBM736020 US20250129104, Compound 2206 MK-52 MK-51 CHEMBL2092821 BDBM50451497
MK-52 MK-51 CHEMBL2092821 BDBM50451497 BDBM50485492 MK-5172 MK-5172 MONOHYDRATE Grazoprevir monohydrate Grazoprevir MK-5172 ANHYDROUS
BDBM50485492 MK-5172 MK-5172 MONOHYDRATE Grazoprevir monohydrate Grazoprevir MK-5172 ANHYDROUS MK-2461 CHEMBL1802916 BDBM50347659 US20250129067, Compound MK 2461
MK-2461 CHEMBL1802916 BDBM50347659 US20250129067, Compound MK 2461 BDBM103838 MK-5172
BDBM103838 MK-5172 BDBM50566358 Mk-8666
BDBM50566358 Mk-8666 MK-8189 BDBM50616544
MK-8189 BDBM50616544 Mk-0752 BDBM50458158
Mk-0752 BDBM50458158 MK-24(S)-S(O)(NH)Ph BDBM50214607 CHEMBL541423 MK-24
MK-24(S)-S(O)(NH)Ph BDBM50214607 CHEMBL541423 MK-24 BDBM50360601 MK-0893 CHEMBL1933349
BDBM50360601 MK-0893 CHEMBL1933349 BDBM50368906 MK-386 CHEMBL25448
BDBM50368906 MK-386 CHEMBL25448 BDBM50373120 MK-0364 CHEMBL260977
BDBM50373120 MK-0364 CHEMBL260977 BDBM50381914 CHEMBL2023109 MK-7725
BDBM50381914 CHEMBL2023109 MK-7725 BDBM50423686 CHEMBL506044 MK-0668
BDBM50423686 CHEMBL506044 MK-0668 BDBM50427138 MK-8033 CHEMBL2323775
BDBM50427138 MK-8033 CHEMBL2323775 BDBM50479471 MK-1107 CHEMBL491019
BDBM50479471 MK-1107 CHEMBL491019 CHEMBL253364 BDBM50230827 MK-9
CHEMBL253364 BDBM50230827 MK-9 CHEMBL489586 BDBM50479470 MK-4965
CHEMBL489586 BDBM50479470 MK-4965 MK 0893 BDBM50168081 CHEMBL3799802
MK 0893 BDBM50168081 CHEMBL3799802 MK-0617 CHEMBL566154 BDBM50423695
MK-0617 CHEMBL566154 BDBM50423695 MK-1220 CHEMBL1672609 BDBM50485489
MK-1220 CHEMBL1672609 BDBM50485489 MK-50 CHEMBL2113644 BDBM50451496
MK-50 CHEMBL2113644 BDBM50451496 MK-55 BDBM50474237 CHEMBL313379
MK-55 BDBM50474237 CHEMBL313379 MK-7445 BDBM50484630 CHEMBL1939502
MK-7445 BDBM50484630 CHEMBL1939502 MK-8742 BDBM50531952 Elbasvir
MK-8742 BDBM50531952 Elbasvir MK-991 BDBM50366679 CHEMBL1793852
MK-991 BDBM50366679 CHEMBL1793852 Mk-2748 BDBM50642717 MK2748
Mk-2748 BDBM50642717 MK2748 Mk-6186 Mk6186 BDBM50484632
Mk-6186 Mk6186 BDBM50484632 BDBM50004205 TOZASERTIB MK-045 MK-0457 US9249124, VX680 VX-68 VX-680
BDBM50004205 TOZASERTIB MK-045 MK-0457 US9249124, VX680 VX-68 VX-680 BDBM565698 US11407768, Compound MK-8722
BDBM565698 US11407768, Compound MK-8722 BDBM81766 MK 678 NSC_3086456 CAS_3086456
BDBM81766 MK 678 NSC_3086456 CAS_3086456 BDBM86671 MK 571 NSC_60719 CAS_60719
BDBM86671 MK 571 NSC_60719 CAS_60719 CHEMBL481611 BDBM50255753 MK-0822 Odanacatib
CHEMBL481611 BDBM50255753 MK-0822 Odanacatib MK-801 BDBM86153 CAS_180081 NSC_180081
MK-801 BDBM86153 CAS_180081 NSC_180081 Pifeltro Doravirine MK-1439 BDBM50508293
Pifeltro Doravirine MK-1439 BDBM50508293 BDBM50213266 CHEBI:471744 MK-0787 Imipenem
BDBM50213266 CHEBI:471744 MK-0787 Imipenem Birabresib OTX-015 MK-8628 BDBM50092312
Birabresib OTX-015 MK-8628 BDBM50092312 MK-4618 BDBM50146154 KRP-114V Vibegron
MK-4618 BDBM50146154 KRP-114V Vibegron MK-0518 Isentress Raltegravir Potassium MK0518 POTASSIUM Raltegravir monopotassium salt BDBM50480673 Isentress hd MK-0518 POTASSIUM
MK-0518 Isentress Raltegravir Potassium MK0518 POTASSIUM Raltegravir monopotassium salt BDBM50480673 Isentress hd MK-0518 POTASSIUM CAS_123679 MK-912 L-657,743 NSC_123679 BDBM81811
CAS_123679 MK-912 L-657,743 NSC_123679 BDBM81811 US10172837, Dorzolamide MK-507 Dorzolamide CHEMBL269001 BDBM50043906
US10172837, Dorzolamide MK-507 Dorzolamide CHEMBL269001 BDBM50043906 US20240059691, Example 2206 BDBM652626 5-N-[[2-fluoro-4-[(2-methyl-2-azaspiro[3.3]heptan-6- yl)oxymethyl]phenyl]methyl]isoquinoline-1,5-diamine
US20240059691, Example 2206 BDBM652626 5-N-[[2-fluoro-4-[(2-methyl-2-azaspiro[3.3]heptan-6- yl)oxymethyl]phenyl]methyl]isoquinoline-1,5-diamine 6'-Chloro-3,4,5,6-tetrahydro-2H-[1,2']bipyrazinyl L-630,571 MK 212 2-chloro-6-piperazin-1-ylpyrazine CHEMBL269521 6'-Chloro-3,4,5,6-tetrahydro-2H-[1,2']bipyrazinyl(MK-212) BDBM50017452 MK-212
6'-Chloro-3,4,5,6-tetrahydro-2H-[1,2']bipyrazinyl L-630,571 MK 212 2-chloro-6-piperazin-1-ylpyrazine CHEMBL269521 6'-Chloro-3,4,5,6-tetrahydro-2H-[1,2']bipyrazinyl(MK-212) BDBM50017452 MK-212 BDBM281567 US20240165276, Compound MK-6420 US10022461, Compound 18
BDBM281567 US20240165276, Compound MK-6420 US10022461, Compound 18 BDBM50224809 CHEBI:34373 MK-0928 Gaboxadol LU-02030
BDBM50224809 CHEBI:34373 MK-0928 Gaboxadol LU-02030 CHEBI:6993 BDBM50239015 Montelukast Sodium Singulair MK-476
CHEBI:6993 BDBM50239015 Montelukast Sodium Singulair MK-476 MK-0826 BDBM50248009 Invanz CHEBI:60070 Ertapenem sodium
MK-0826 BDBM50248009 Invanz CHEBI:60070 Ertapenem sodium US12220415, Compound AZD1775 US20240182482, Compound MK-1775 US11248006, Compound AZD1775 BDBM50240826 US11479555, Example AZD1775 AZD-1775 US11124518, Example AZD1775 MK-1775
US12220415, Compound AZD1775 US20240182482, Compound MK-1775 US11248006, Compound AZD1775 BDBM50240826 US11479555, Example AZD1775 AZD-1775 US11124518, Example AZD1775 MK-1775 MK-217 Alendronate Sodium BDBM50247920 Binosto Fosamax G-704650
MK-217 Alendronate Sodium BDBM50247920 Binosto Fosamax G-704650 MK-458 NAXAGOLIDE HYDROCHLORIDE BDBM50446931 L-647339 Naxagolide HCl
MK-458 NAXAGOLIDE HYDROCHLORIDE BDBM50446931 L-647339 Naxagolide HCl MK-8931 SCH 900931 BDBM50580216 SCH-900931 SCH900931 VERUBECESTAT
MK-8931 SCH 900931 BDBM50580216 SCH-900931 SCH900931 VERUBECESTAT MK-6721 NMED-160 Z-160 Z160 BDBM50461289 NP-118809
MK-6721 NMED-160 Z-160 Z160 BDBM50461289 NP-118809 MK-8228 Letermovir Prevymis AIC246 AIC 246 AIC-246 BDBM50614677
MK-8228 Letermovir Prevymis AIC246 AIC 246 AIC-246 BDBM50614677 BDBM50067593 Crixivan MK-639 CHEBI:44032 L-735524 Indinavir US10806794, Compound Indinavir
BDBM50067593 Crixivan MK-639 CHEBI:44032 L-735524 Indinavir US10806794, Compound Indinavir BDBM50111186 MK-27 CHEMBL9075 (S)-2-Benzylamino-5-guanidino-pentanoic acid amide
BDBM50111186 MK-27 CHEMBL9075 (S)-2-Benzylamino-5-guanidino-pentanoic acid amide MK-208 Famotidine Pepcid Pepcid Rpd Pepcid Ac Fluxid CHEBI:4975 BDBM50103514
MK-208 Famotidine Pepcid Pepcid Rpd Pepcid Ac Fluxid CHEBI:4975 BDBM50103514 CHEBI:474180 L-743,872 Caspofungin Acetate Cancidas Caspofungin MK-991 BDBM50478215 L-743872
CHEBI:474180 L-743,872 Caspofungin Acetate Cancidas Caspofungin MK-991 BDBM50478215 L-743872 MK-42 BDBM50111176 (S)-4-Allyloxycarbonyl-4-[(naphthalen-2-ylmethyl)-amino]-butyl-ammonium
MK-42 BDBM50111176 (S)-4-Allyloxycarbonyl-4-[(naphthalen-2-ylmethyl)-amino]-butyl-ammonium BDBM50092127 CHEMBL331559 MK-852 8-Acetylamino-22-(4-aminomethyl-benzyl)-5-carbamoylmethyl-16-carboxymethyl-1,1-dimethyl-4,7,15,18,21,24-hexaoxo-icosahydro-2,10,11-trithia-3a,6,14,17,20,23-hexaaza-cyclopentacyclotricosene-13-carboxylic acid (MK-0852)
BDBM50092127 CHEMBL331559 MK-852 8-Acetylamino-22-(4-aminomethyl-benzyl)-5-carbamoylmethyl-16-carboxymethyl-1,1-dimethyl-4,7,15,18,21,24-hexaoxo-icosahydro-2,10,11-trithia-3a,6,14,17,20,23-hexaaza-cyclopentacyclotricosene-13-carboxylic acid (MK-0852) CHEMBL9257 BDBM50111191 (S)-2-(3,4-Dimethoxy-benzylamino)-5-guanidino-pentanoic acid amide MK-25
CHEMBL9257 BDBM50111191 (S)-2-(3,4-Dimethoxy-benzylamino)-5-guanidino-pentanoic acid amide MK-25 CHEMBL274352 BDBM50111187 MK-18 (S)-5-Guanidino-2-[(naphthalen-2-ylmethyl)-amino]-pentanoic acid amide
CHEMBL274352 BDBM50111187 MK-18 (S)-5-Guanidino-2-[(naphthalen-2-ylmethyl)-amino]-pentanoic acid amide MK-20 BDBM50111196 (R)-5-Guanidino-2-[(naphthalen-2-ylmethyl)-amino]-pentanoic acid amide CHEMBL266055
MK-20 BDBM50111196 (R)-5-Guanidino-2-[(naphthalen-2-ylmethyl)-amino]-pentanoic acid amide CHEMBL266055 MK-3102 US10155775, Omarigliptin OMARIGLIPTIN US10822319, Compound Omarigliptin US10479798, Compound MK3102 BDBM50003020 US20240115577, Compound 9
MK-3102 US10155775, Omarigliptin OMARIGLIPTIN US10822319, Compound Omarigliptin US10479798, Compound MK3102 BDBM50003020 US20240115577, Compound 9 MK-43 (S)-5-Guanidino-2-[(naphthalen-2-ylmethyl)-amino]-pentanoic acid allyl ester BDBM50111195
MK-43 (S)-5-Guanidino-2-[(naphthalen-2-ylmethyl)-amino]-pentanoic acid allyl ester BDBM50111195 US20250101023, Example Dinaciclib Dinaciclib US20230416221, Compound Dinaciclib SCH-727965 US11643396, Example Dinaciclib BDBM50139171 MK-7965
US20250101023, Example Dinaciclib Dinaciclib US20230416221, Compound Dinaciclib SCH-727965 US11643396, Example Dinaciclib BDBM50139171 MK-7965 (R)-2-[(1H-Indol-3-ylmethyl)-amino]-3-naphthalen-2-yl-propionamide BDBM50111188 CHEMBL9219 MK-15
(R)-2-[(1H-Indol-3-ylmethyl)-amino]-3-naphthalen-2-yl-propionamide BDBM50111188 CHEMBL9219 MK-15 (S)-2-(2-Chloro-3,4-dimethoxy-benzylamino)-5-guanidino-pentanoic acid amide BDBM50111204 MK-26 CHEMBL8929
(S)-2-(2-Chloro-3,4-dimethoxy-benzylamino)-5-guanidino-pentanoic acid amide BDBM50111204 MK-26 CHEMBL8929 (S)-2-[(1H-Indol-3-ylmethyl)-amino]-3-naphthalen-2-yl-propionamide CHEMBL428480 BDBM50111197 MK-16
(S)-2-[(1H-Indol-3-ylmethyl)-amino]-3-naphthalen-2-yl-propionamide CHEMBL428480 BDBM50111197 MK-16 BDBM50111193 CHEMBL9138 (R)-3-(1H-Indol-3-yl)-2-[(naphthalen-2-ylmethyl)-amino]-propionamide MK-14
BDBM50111193 CHEMBL9138 (R)-3-(1H-Indol-3-yl)-2-[(naphthalen-2-ylmethyl)-amino]-propionamide MK-14 CHEMBL273475 5-(S)-Guanidino-2-[(1H-indol-3-ylmethyl)-amino]-pentanoic acid amide BDBM50111198 MK-17
CHEMBL273475 5-(S)-Guanidino-2-[(1H-indol-3-ylmethyl)-amino]-pentanoic acid amide BDBM50111198 MK-17 CHEMBL8730 5-(R)-Guanidino-2-[(1H-indol-3-ylmethyl)-amino]-pentanoic acid amide BDBM50111200 MK-19
CHEMBL8730 5-(R)-Guanidino-2-[(1H-indol-3-ylmethyl)-amino]-pentanoic acid amide BDBM50111200 MK-19 MK-11 (S)-3-(1H-Indol-3-yl)-2-[(naphthalen-2-ylmethyl)-amino]-propionamide BDBM50111202 CHEMBL9274
MK-11 (S)-3-(1H-Indol-3-yl)-2-[(naphthalen-2-ylmethyl)-amino]-propionamide BDBM50111202 CHEMBL9274 MK-4827 CHEMBL1094636 (S)-2-(4-(piperidin-3-yl)phenyl)-2H-indazole-7-carboxamide Niraparib BDBM50316226
MK-4827 CHEMBL1094636 (S)-2-(4-(piperidin-3-yl)phenyl)-2H-indazole-7-carboxamide Niraparib BDBM50316226 PL 100 (PHARMACEUTICAL) MK-8122 PL-100 Ppl-100 PPL 100 Tmb-607 BDBM50482335 MX-100
PL 100 (PHARMACEUTICAL) MK-8122 PL-100 Ppl-100 PPL 100 Tmb-607 BDBM50482335 MX-100 1N-[1-benzyloxymethyl-2-[1-methylsulfonylspiro[2,3-dihydro-1H-indole-3,4'-(hexahydropyridine)]-1-yl]-2-oxo-(1R)-ethyl]-2-amino-2-methylpropanamide [3a(MK-0677)] CHEMBL13817 1-{[(2R)-3-(benzyloxy)-1-{1-methanesulfonyl-1,2-dihydrospiro[indole-3,4'-piperidine]-1'-yl}-1-oxopropan-2-yl]carbamoyl}-1-methylethan-1-aminium 1N-[1-benzyloxymethyl-2-[1-methylsulfonylspiro[2,3-dihydro-1H-indole-3,4'-(hexahydropyridine)]-1-yl]-2-oxo-(1R)-ethyl]-2-amino-2-methylpropanamide BDBM50049478 MK-0677 1N-[1-benzyloxymethyl-2-[1-methylsulfonylspiro[2,3-dihydro-1H-indole-3,4'-(hexahydropyridine)]-1-yl]-2-oxo-(1R)-ethyl]-2-amino-2-methylpropanamide(MK-0677) MK 0677 MK-677
1N-[1-benzyloxymethyl-2-[1-methylsulfonylspiro[2,3-dihydro-1H-indole-3,4'-(hexahydropyridine)]-1-yl]-2-oxo-(1R)-ethyl]-2-amino-2-methylpropanamide [3a(MK-0677)] CHEMBL13817 1-{[(2R)-3-(benzyloxy)-1-{1-methanesulfonyl-1,2-dihydrospiro[indole-3,4'-piperidine]-1'-yl}-1-oxopropan-2-yl]carbamoyl}-1-methylethan-1-aminium 1N-[1-benzyloxymethyl-2-[1-methylsulfonylspiro[2,3-dihydro-1H-indole-3,4'-(hexahydropyridine)]-1-yl]-2-oxo-(1R)-ethyl]-2-amino-2-methylpropanamide BDBM50049478 MK-0677 1N-[1-benzyloxymethyl-2-[1-methylsulfonylspiro[2,3-dihydro-1H-indole-3,4'-(hexahydropyridine)]-1-yl]-2-oxo-(1R)-ethyl]-2-amino-2-methylpropanamide(MK-0677) MK 0677 MK-677 1-methyl-16-azatetracyclo[7.6.1.02,7.010,15]hexadeca-2(7),3,5,10(15),11,13-hexaene(MK-801) (+)-1-methyl-16-azatetracyclo[7.6.1.02,7.010,15]hexadeca-2(7),3,5,10(15),11,13-hexaene (+/-) MK-8011-methyl-(9R,1R)-16-azatetracyclo[7.6.1.02,7.010,15]hexadeca-2(7),3,5,10(15),11,13-hexaene (5S,10R)-(+)-5-methyl-10,11-dihydro-5Hdibenzo[a,d]cyclohepten-5,10-imine MK-801 1-methyl-16-azatetracyclo[7.6.1.02,7.010,15]hexadeca-2(7),3,5,10(15),11,13-hexaene (+/-)-1-methyl-16-azatetracyclo[7.6.1.02,7.010,15]hexadeca-2(7),3,5,10(15),11,13-hexaene (1S,9R)-1-methyl-16-azatetracyclo[7.6.1.0^{2,7}.0^{10,15}]hexadeca-2(7),3,5,10(15),11,13-hexaene BDBM50000663 MK-801 (Dizocilpine) (-)-MK801 US11944616, Compound Dizocilpine (Dizocilpine)1-methyl-16-azatetracyclo[7.6.1.02,7.010,15]hexadeca-2(7),3,5,10(15),11,13-hexaene (1S,9R)-1-methyl-16-azatetracyclo[7.6.1.0^{2,7}.0^{10,15}]hexadeca-2,4,6,10(15),11,13-hexaene MK-801,(+) (5S,10S)-(+)-5-methyl-10,11-dihydro-5Hdibenzo[a,d]cyclohepten-5,10-imine 1-methyl-16-azatetracyclo[7.6.1.02,7.010,15]hexadeca-2(7),3,5,10,12,14-hexaene CHEMBL284237 MK-801,(-) 1-methyl-16-azatetracyclo[7.6.1.02,7.010,15]hexadeca-2,4,6,10(15),11,13-hexaene (-)-1-methyl-16-azatetracyclo[7.6.1.02,7.010,15]hexadeca-2(7),3,5,10(15),11,13-hexaene (MK-801)1-methyl-16-azatetracyclo[7.6.1.02,7.010,15]hexadeca-2(7),3,5,10(15),11,13-hexaene dizocilpine (+)-MK-801 (+)MK-801 (+/-)-MK801 10,11-Dihydro-5-methyl-5H-dibenzo[a,d]cyclohepten-5,10-imine.(MK-801)
1-methyl-16-azatetracyclo[7.6.1.02,7.010,15]hexadeca-2(7),3,5,10(15),11,13-hexaene(MK-801) (+)-1-methyl-16-azatetracyclo[7.6.1.02,7.010,15]hexadeca-2(7),3,5,10(15),11,13-hexaene (+/-) MK-8011-methyl-(9R,1R)-16-azatetracyclo[7.6.1.02,7.010,15]hexadeca-2(7),3,5,10(15),11,13-hexaene (5S,10R)-(+)-5-methyl-10,11-dihydro-5Hdibenzo[a,d]cyclohepten-5,10-imine MK-801 1-methyl-16-azatetracyclo[7.6.1.02,7.010,15]hexadeca-2(7),3,5,10(15),11,13-hexaene (+/-)-1-methyl-16-azatetracyclo[7.6.1.02,7.010,15]hexadeca-2(7),3,5,10(15),11,13-hexaene (1S,9R)-1-methyl-16-azatetracyclo[7.6.1.0^{2,7}.0^{10,15}]hexadeca-2(7),3,5,10(15),11,13-hexaene BDBM50000663 MK-801 (Dizocilpine) (-)-MK801 US11944616, Compound Dizocilpine (Dizocilpine)1-methyl-16-azatetracyclo[7.6.1.02,7.010,15]hexadeca-2(7),3,5,10(15),11,13-hexaene (1S,9R)-1-methyl-16-azatetracyclo[7.6.1.0^{2,7}.0^{10,15}]hexadeca-2,4,6,10(15),11,13-hexaene MK-801,(+) (5S,10S)-(+)-5-methyl-10,11-dihydro-5Hdibenzo[a,d]cyclohepten-5,10-imine 1-methyl-16-azatetracyclo[7.6.1.02,7.010,15]hexadeca-2(7),3,5,10,12,14-hexaene CHEMBL284237 MK-801,(-) 1-methyl-16-azatetracyclo[7.6.1.02,7.010,15]hexadeca-2,4,6,10(15),11,13-hexaene (-)-1-methyl-16-azatetracyclo[7.6.1.02,7.010,15]hexadeca-2(7),3,5,10(15),11,13-hexaene (MK-801)1-methyl-16-azatetracyclo[7.6.1.02,7.010,15]hexadeca-2(7),3,5,10(15),11,13-hexaene dizocilpine (+)-MK-801 (+)MK-801 (+/-)-MK801 10,11-Dihydro-5-methyl-5H-dibenzo[a,d]cyclohepten-5,10-imine.(MK-801) (S)-5-Guanidino-2-[(1,2,3,4-tetrahydro-naphthalen-2-ylmethyl)-amino]-pentanoic acid amide BDBM50111185 CHEMBL8676 MK-28
(S)-5-Guanidino-2-[(1,2,3,4-tetrahydro-naphthalen-2-ylmethyl)-amino]-pentanoic acid amide BDBM50111185 CHEMBL8676 MK-28 BDBM50114085 CHEMBL314401 (O)C-(C6H4)-C(O)-c[His-D-Phe-Arg-Trp-Lys]-NH2) (MK-6)
BDBM50114085 CHEMBL314401 (O)C-(C6H4)-C(O)-c[His-D-Phe-Arg-Trp-Lys]-NH2) (MK-6) BDBM50114084 CHEMBL439188 (O)C-(C6H4)-C(O)-c[His-D-Nal(2')-Arg-Trp-Lys]-NH2) (MK-7)
BDBM50114084 CHEMBL439188 (O)C-(C6H4)-C(O)-c[His-D-Nal(2')-Arg-Trp-Lys]-NH2) (MK-7) US10806720, Compound Enobosarm-(agonist) BDBM50529668 US11648234, Compound Enobosarm-(agonist) Ostarine MK-2866 Gtx-024 US11230523, Compound Enobosarm-(agonist) Enobosarm
US10806720, Compound Enobosarm-(agonist) BDBM50529668 US11648234, Compound Enobosarm-(agonist) Ostarine MK-2866 Gtx-024 US11230523, Compound Enobosarm-(agonist) Enobosarm BDBM50111181 MK-33 N-Alkylaminoacid derivative (2R)-5-[(E)-[amino(azaniumyl)methylidene]amino]-2-{[(3,4-dimethoxyphenyl)methyl]amino}pentanoic acid
BDBM50111181 MK-33 N-Alkylaminoacid derivative (2R)-5-[(E)-[amino(azaniumyl)methylidene]amino]-2-{[(3,4-dimethoxyphenyl)methyl]amino}pentanoic acid Famotidine YM 11170 Pepcid cid_5702160 3-[({2-[(diaminomethylidene)amino]-1,3-thiazol-4-yl}methyl)sulfanyl]-N'-sulfamoylpropanimidamide MK 208 BDBM22891
Famotidine YM 11170 Pepcid cid_5702160 3-[({2-[(diaminomethylidene)amino]-1,3-thiazol-4-yl}methyl)sulfanyl]-N'-sulfamoylpropanimidamide MK 208 BDBM22891 MK-31 BDBM50111182 N-Alkylaminoacid derivative (2R)-5-[(E)-[amino(azaniumyl)methylidene]amino]-2-[(naphthalen-2-ylmethyl)amino]pentanoic acid
MK-31 BDBM50111182 N-Alkylaminoacid derivative (2R)-5-[(E)-[amino(azaniumyl)methylidene]amino]-2-[(naphthalen-2-ylmethyl)amino]pentanoic acid MK-55 5-Guanidino-2-[(naphthalen-2-ylmethyl)-amino]-pentanoic acid (3-guanidino-propyl)-amide N-Alkylaminoacid derivative CHEMBL313379 BDBM50111206
MK-55 5-Guanidino-2-[(naphthalen-2-ylmethyl)-amino]-pentanoic acid (3-guanidino-propyl)-amide N-Alkylaminoacid derivative CHEMBL313379 BDBM50111206 US11786535, Compound Rofecoxib Rofecoxib BDBM22369 US11478464, Compound Rofecoxib 4-(4-methanesulfonylphenyl)-3-phenyl-2,5-dihydrofuran-2-one CHEMBL122 MK 0966
US11786535, Compound Rofecoxib Rofecoxib BDBM22369 US11478464, Compound Rofecoxib 4-(4-methanesulfonylphenyl)-3-phenyl-2,5-dihydrofuran-2-one CHEMBL122 MK 0966 BDBM50273099 3-(1H-tetrazol-5-yl)-1,4,5,6-tetrahydrocyclopenta[c]pyrazole 3-(2H-tetrazol-5-yl)-1,4,5,6-tetrahydro-cyclopentapyrazole CHEMBL456145 MK-0354
BDBM50273099 3-(1H-tetrazol-5-yl)-1,4,5,6-tetrahydrocyclopenta[c]pyrazole 3-(2H-tetrazol-5-yl)-1,4,5,6-tetrahydro-cyclopentapyrazole CHEMBL456145 MK-0354 MK-32 N-Alkylaminoacid derivative (2R)-5-[(E)-[amino(azaniumyl)methylidene]amino]-2-[(1H-indol-3-ylmethyl)amino]pentanoic acid BDBM50111205
MK-32 N-Alkylaminoacid derivative (2R)-5-[(E)-[amino(azaniumyl)methylidene]amino]-2-[(1H-indol-3-ylmethyl)amino]pentanoic acid BDBM50111205 (1S,9R)-1-methyl-16-azatetracyclo[7.6.1.0^{2,7}.0^{10,15}]hexadeca-2,4,6,10(15),11,13-hexaene (5S,10S)-(+)-5-methyl-10,11-dihydro-5Hdibenzo[a,d]cyclohepten-5,10-imine 1-methyl-16-azatetracyclo[7.6.1.02,7.010,15]hexadeca-2(7),3,5,10(15),11,13-hexaene(MK-801) (+)-1-methyl-16-azatetracyclo[7.6.1.02,7.010,15]hexadeca-2(7),3,5,10(15),11,13-hexaene 1-methyl-16-azatetracyclo[7.6.1.02,7.010,15]hexadeca-2(7),3,5,10,12,14-hexaene CHEMBL284237 1-methyl-16-azatetracyclo[7.6.1.02,7.010,15]hexadeca-2,4,6,10(15),11,13-hexaene (-)-1-methyl-16-azatetracyclo[7.6.1.02,7.010,15]hexadeca-2(7),3,5,10(15),11,13-hexaene (MK-801)1-methyl-16-azatetracyclo[7.6.1.02,7.010,15]hexadeca-2(7),3,5,10(15),11,13-hexaene dizocilpine (+)-MK-801 (+)MK-801 (+/-) MK-8011-methyl-(9R,1R)-16-azatetracyclo[7.6.1.02,7.010,15]hexadeca-2(7),3,5,10(15),11,13-hexaene (5S,10R)-(+)-5-methyl-10,11-dihydro-5Hdibenzo[a,d]cyclohepten-5,10-imine MK-801 1-methyl-16-azatetracyclo[7.6.1.02,7.010,15]hexadeca-2(7),3,5,10(15),11,13-hexaene BDBM50344263 (+/-)-1-methyl-16-azatetracyclo[7.6.1.02,7.010,15]hexadeca-2(7),3,5,10(15),11,13-hexaene (+/-)-MK801 (1S,9R)-1-methyl-16-azatetracyclo[7.6.1.0^{2,7}.0^{10,15}]hexadeca-2(7),3,5,10(15),11,13-hexaene MK-801 (Dizocilpine) (-)-MK801 (Dizocilpine)1-methyl-16-azatetracyclo[7.6.1.02,7.010,15]hexadeca-2(7),3,5,10(15),11,13-hexaene 10,11-Dihydro-5-methyl-5H-dibenzo[a,d]cyclohepten-5,10-imine.(MK-801)
(1S,9R)-1-methyl-16-azatetracyclo[7.6.1.0^{2,7}.0^{10,15}]hexadeca-2,4,6,10(15),11,13-hexaene (5S,10S)-(+)-5-methyl-10,11-dihydro-5Hdibenzo[a,d]cyclohepten-5,10-imine 1-methyl-16-azatetracyclo[7.6.1.02,7.010,15]hexadeca-2(7),3,5,10(15),11,13-hexaene(MK-801) (+)-1-methyl-16-azatetracyclo[7.6.1.02,7.010,15]hexadeca-2(7),3,5,10(15),11,13-hexaene 1-methyl-16-azatetracyclo[7.6.1.02,7.010,15]hexadeca-2(7),3,5,10,12,14-hexaene CHEMBL284237 1-methyl-16-azatetracyclo[7.6.1.02,7.010,15]hexadeca-2,4,6,10(15),11,13-hexaene (-)-1-methyl-16-azatetracyclo[7.6.1.02,7.010,15]hexadeca-2(7),3,5,10(15),11,13-hexaene (MK-801)1-methyl-16-azatetracyclo[7.6.1.02,7.010,15]hexadeca-2(7),3,5,10(15),11,13-hexaene dizocilpine (+)-MK-801 (+)MK-801 (+/-) MK-8011-methyl-(9R,1R)-16-azatetracyclo[7.6.1.02,7.010,15]hexadeca-2(7),3,5,10(15),11,13-hexaene (5S,10R)-(+)-5-methyl-10,11-dihydro-5Hdibenzo[a,d]cyclohepten-5,10-imine MK-801 1-methyl-16-azatetracyclo[7.6.1.02,7.010,15]hexadeca-2(7),3,5,10(15),11,13-hexaene BDBM50344263 (+/-)-1-methyl-16-azatetracyclo[7.6.1.02,7.010,15]hexadeca-2(7),3,5,10(15),11,13-hexaene (+/-)-MK801 (1S,9R)-1-methyl-16-azatetracyclo[7.6.1.0^{2,7}.0^{10,15}]hexadeca-2(7),3,5,10(15),11,13-hexaene MK-801 (Dizocilpine) (-)-MK801 (Dizocilpine)1-methyl-16-azatetracyclo[7.6.1.02,7.010,15]hexadeca-2(7),3,5,10(15),11,13-hexaene 10,11-Dihydro-5-methyl-5H-dibenzo[a,d]cyclohepten-5,10-imine.(MK-801) 3-[5-(3,5-difluorophenyl)-3-({[(1S,3R)-3-fluorocyclopentyl]amino}-methyl)-4-methyl-1H-pyrazol-1-yl]propanenitrile CHEMBL560667 MK-1925 BDBM50296579
3-[5-(3,5-difluorophenyl)-3-({[(1S,3R)-3-fluorocyclopentyl]amino}-methyl)-4-methyl-1H-pyrazol-1-yl]propanenitrile CHEMBL560667 MK-1925 BDBM50296579 4'-(2-Ethyl-5,7-dimethyl-imidazo[4,5-b]pyridin-3-ylmethyl)-biphenyl-2-sulfonic acid benzoylamide CHEMBL293511 MK-996 L-158282 BDBM50038189
4'-(2-Ethyl-5,7-dimethyl-imidazo[4,5-b]pyridin-3-ylmethyl)-biphenyl-2-sulfonic acid benzoylamide CHEMBL293511 MK-996 L-158282 BDBM50038189 4-(3-chloro-2-fluorophenoxy)-1-[[6-(1,3-thiazol-2-ylamino)pyridin-2-yl]methyl]cyclohexane-1-carboxylic acid BDBM209862 MK-5108
4-(3-chloro-2-fluorophenoxy)-1-[[6-(1,3-thiazol-2-ylamino)pyridin-2-yl]methyl]cyclohexane-1-carboxylic acid BDBM209862 MK-5108 BDBM50111184 N-Alkylaminoacid derivative (2R)-5-[(E)-[amino(azaniumyl)methylidene]amino]-2-{[(2-chloro-3,4-dimethoxyphenyl)methyl]amino}pentanoic acid MK-34
BDBM50111184 N-Alkylaminoacid derivative (2R)-5-[(E)-[amino(azaniumyl)methylidene]amino]-2-{[(2-chloro-3,4-dimethoxyphenyl)methyl]amino}pentanoic acid MK-34 BDBM50111175 N-Alkylaminoacid derivative (2S)-5-[(E)-[amino(azaniumyl)methylidene]amino]-N-(naphthalen-2-yl)-2-[(naphthalen-2-ylmethyl)amino]pentanamide MK-56
BDBM50111175 N-Alkylaminoacid derivative (2S)-5-[(E)-[amino(azaniumyl)methylidene]amino]-N-(naphthalen-2-yl)-2-[(naphthalen-2-ylmethyl)amino]pentanamide MK-56 BDBM50128003 N-[1'-(6-cyano-1,2,3,4-tetrahydro-2-naphthalenyl)-4-hydroxyspiro[3,4-dihydro-2H-chromene-2,4'-(hexahydropyridine)]-6-yl]methanesulfonamide CHEMBL52627 MK-499
BDBM50128003 N-[1'-(6-cyano-1,2,3,4-tetrahydro-2-naphthalenyl)-4-hydroxyspiro[3,4-dihydro-2H-chromene-2,4'-(hexahydropyridine)]-6-yl]methanesulfonamide CHEMBL52627 MK-499 CHEMBL9048 (2S,4S)-4-Hydroxy-1-naphthalen-2-ylmethyl-pyrrolidine-2-carboxylic acid [2-(1H-imidazol-4-yl)-ethyl]-amide BDBM50111189 MK-69
CHEMBL9048 (2S,4S)-4-Hydroxy-1-naphthalen-2-ylmethyl-pyrrolidine-2-carboxylic acid [2-(1H-imidazol-4-yl)-ethyl]-amide BDBM50111189 MK-69 MK-54 BDBM50111183 N-Alkylaminoacid derivative (2S)-5-[(E)-[amino(azaniumyl)methylidene]amino]-N-(naphthalen-1-yl)-2-[(naphthalen-2-ylmethyl)amino]pentanamide
MK-54 BDBM50111183 N-Alkylaminoacid derivative (2S)-5-[(E)-[amino(azaniumyl)methylidene]amino]-N-(naphthalen-1-yl)-2-[(naphthalen-2-ylmethyl)amino]pentanamide N-[(1S,2S)-3-(4-chlorophenyl)-2-(3-cyanophenyl)-1-methylpropyl]-2-methyl-2-{[5-(trifluoromethyl)pyridin-2-yl]oxy}propanamide CHEMBL220360 Taranbant N-((2S,3S)-4-(4-chlorophenyl)-3-(3-cyanophenyl)butan-2-yl)-2-methyl-2-(5-(trifluoromethyl)pyridin-2-yloxy)propanamide MK-0634 BDBM50200841 MK-0364 TARANABANT
N-[(1S,2S)-3-(4-chlorophenyl)-2-(3-cyanophenyl)-1-methylpropyl]-2-methyl-2-{[5-(trifluoromethyl)pyridin-2-yl]oxy}propanamide CHEMBL220360 Taranbant N-((2S,3S)-4-(4-chlorophenyl)-3-(3-cyanophenyl)butan-2-yl)-2-methyl-2-(5-(trifluoromethyl)pyridin-2-yloxy)propanamide MK-0634 BDBM50200841 MK-0364 TARANABANT (R)-5-Guanidino-2-[(naphthalen-2-ylmethyl)-amino]-pentanoic acid [(S)-1-carbamoyl-2-(1H-indol-3-yl)-ethyl]-amide CHEMBL447178 BDBM50111179 MK-30A
(R)-5-Guanidino-2-[(naphthalen-2-ylmethyl)-amino]-pentanoic acid [(S)-1-carbamoyl-2-(1H-indol-3-yl)-ethyl]-amide CHEMBL447178 BDBM50111179 MK-30A BDBM50002823 CHEMBL299944 L-680573 MK-287 2-{3-Methoxy-2-propoxy-5-[(2S,5S)-5-(3,4,5-trimethoxy-phenyl)-tetrahydro-furan-2-yl]-benzenesulfonyl}-ethanol
BDBM50002823 CHEMBL299944 L-680573 MK-287 2-{3-Methoxy-2-propoxy-5-[(2S,5S)-5-(3,4,5-trimethoxy-phenyl)-tetrahydro-furan-2-yl]-benzenesulfonyl}-ethanol BDBM50111192 MK-53 (2S)-5-[(E)-[amino(azaniumyl)methylidene]amino]-N-(1H-indol-3-ylmethyl)-2-[(naphthalen-2-ylmethyl)amino]pentanamide N-Alkylaminoacid derivative
BDBM50111192 MK-53 (2S)-5-[(E)-[amino(azaniumyl)methylidene]amino]-N-(1H-indol-3-ylmethyl)-2-[(naphthalen-2-ylmethyl)amino]pentanamide N-Alkylaminoacid derivative MK-29A BDBM50111180 CHEMBL8720 (S)-5-Guanidino-2-[(naphthalen-2-ylmethyl)-amino]-pentanoic acid [(S)-1-carbamoyl-2-(1H-indol-3-yl)-ethyl]-amide
MK-29A BDBM50111180 CHEMBL8720 (S)-5-Guanidino-2-[(naphthalen-2-ylmethyl)-amino]-pentanoic acid [(S)-1-carbamoyl-2-(1H-indol-3-yl)-ethyl]-amide CHEMBL135613 MK-869 BDBM50106711 5-[2-[1-(3,5-Bis-trifluoromethyl-phenyl)-ethoxy]-3-(4-fluoro-phenyl)-morpholin-4-ylmethyl]-2,4-dihydro-[1,2,4]triazol-3-one
CHEMBL135613 MK-869 BDBM50106711 5-[2-[1-(3,5-Bis-trifluoromethyl-phenyl)-ethoxy]-3-(4-fluoro-phenyl)-morpholin-4-ylmethyl]-2,4-dihydro-[1,2,4]triazol-3-one CHEMBL267007 MK-29B (S)-5-Guanidino-2-[(1H-indol-3-ylmethyl)-amino]-pentanoic acid [(S)-1-carbamoyl-2-(1H-indol-3-yl)-ethyl]-amide BDBM50111190
CHEMBL267007 MK-29B (S)-5-Guanidino-2-[(1H-indol-3-ylmethyl)-amino]-pentanoic acid [(S)-1-carbamoyl-2-(1H-indol-3-yl)-ethyl]-amide BDBM50111190 MK-10 22-Benzyl-19-(3-guanidino-propyl)-16-(1H-indol-3-ylmethyl)-4,7,15,18,21,24-hexaoxo-docosahydro-3a,8,14,17,20,23-hexaaza-cyclopentacyclotricosene-13-carboxylic acid amide BDBM50114081
MK-10 22-Benzyl-19-(3-guanidino-propyl)-16-(1H-indol-3-ylmethyl)-4,7,15,18,21,24-hexaoxo-docosahydro-3a,8,14,17,20,23-hexaaza-cyclopentacyclotricosene-13-carboxylic acid amide BDBM50114081 MK-70 BDBM50111178 N-Alkylaminoacid derivative (2S,4S)-N-{3-[(E)-[amino(azaniumyl)methylidene]amino]propyl}-4-hydroxy-1-(naphthalen-2-ylmethyl)pyrrolidine-2-carboxamide
MK-70 BDBM50111178 N-Alkylaminoacid derivative (2S,4S)-N-{3-[(E)-[amino(azaniumyl)methylidene]amino]propyl}-4-hydroxy-1-(naphthalen-2-ylmethyl)pyrrolidine-2-carboxamide 4-Isobutylamino-7,7-dioxo-4,5,6,7-tetrahydro-7lambda*6*-thieno[2,3-b]thiopyran-2-sulfonic acid amide MK-417 BDBM50017725 MK-927 CHEMBL545013 4-Isobutylamino-7,7-dioxo-4,5,6,7-tetrahydro-7lambda*6*-thieno[2,3-b]thiopyran-2-sulfonic acid amide; hydrochloride (S)-4-Isobutylamino-7,7-dioxo-4,5,6,7-tetrahydro-7lambda*6*-thieno[2,3-b]thiopyran-2-sulfonic acid amide
4-Isobutylamino-7,7-dioxo-4,5,6,7-tetrahydro-7lambda*6*-thieno[2,3-b]thiopyran-2-sulfonic acid amide MK-417 BDBM50017725 MK-927 CHEMBL545013 4-Isobutylamino-7,7-dioxo-4,5,6,7-tetrahydro-7lambda*6*-thieno[2,3-b]thiopyran-2-sulfonic acid amide; hydrochloride (S)-4-Isobutylamino-7,7-dioxo-4,5,6,7-tetrahydro-7lambda*6*-thieno[2,3-b]thiopyran-2-sulfonic acid amide 4-Isobutylamino-7,7-dioxo-4,5,6,7-tetrahydro-7lambda*6*-thieno[2,3-b]thiopyran-2-sulfonic acid amide MK-417 BDBM50041029 CHEMBL417975 MK-927 4-Isopropylamino-7,7-dioxo-4,5,6,7-tetrahydro-7lambda*6*-thieno[2,3-b]thiopyran-2-sulfonic acid amide CHEMBL1204135 (S)-4-Isobutylamino-7,7-dioxo-4,5,6,7-tetrahydro-7lambda*6*-thieno[2,3-b]thiopyran-2-sulfonic acid amide
4-Isobutylamino-7,7-dioxo-4,5,6,7-tetrahydro-7lambda*6*-thieno[2,3-b]thiopyran-2-sulfonic acid amide MK-417 BDBM50041029 CHEMBL417975 MK-927 4-Isopropylamino-7,7-dioxo-4,5,6,7-tetrahydro-7lambda*6*-thieno[2,3-b]thiopyran-2-sulfonic acid amide CHEMBL1204135 (S)-4-Isobutylamino-7,7-dioxo-4,5,6,7-tetrahydro-7lambda*6*-thieno[2,3-b]thiopyran-2-sulfonic acid amide BDBM24064 L-706,000 [35S]-MK499 N-[(4R)-1'-[(2R)-6-cyano-1,2,3,4-tetrahydronaphthalen-2-yl]-3,4-dihydrospiro[1-benzopyran-2,4'-piperidine]-4-ol]methanesulfonamide L-706000 MK-499
BDBM24064 L-706,000 [35S]-MK499 N-[(4R)-1'-[(2R)-6-cyano-1,2,3,4-tetrahydronaphthalen-2-yl]-3,4-dihydrospiro[1-benzopyran-2,4'-piperidine]-4-ol]methanesulfonamide L-706000 MK-499 [(7R)-4-(5-Chloro-1,3-benzoxazol-2-yl)-7-methyl-1,4-diazepan-1-yl][5-methyl-2-(2H-1,2,3-triazol-2-yl)phenyl]methanone BDBM50318701 CHEMBL1083659 MK-4305
[(7R)-4-(5-Chloro-1,3-benzoxazol-2-yl)-7-methyl-1,4-diazepan-1-yl][5-methyl-2-(2H-1,2,3-triazol-2-yl)phenyl]methanone BDBM50318701 CHEMBL1083659 MK-4305 CHEMBL15177 3-[{3-[2-(7-Chloro-quinolin-2-yl)-vinyl]-phenyl}-(2-dimethylcarbamoyl-ethylsulfanyl)-methylsulfanyl]-propionic acid(MK-571, L-660711) BDBM50001285 MK 571 (E)-3-((3-(2-(7-chloroquinolin-2-yl)vinyl)phenyl)(3-(dimethylamino)-3-oxopropylthio)methylthio)propanoic acid 3-((3-(2-(7-chloroquinolin-2-yl)vinyl)phenyl)(3-(dimethylamino)-3-oxopropylthio)methylthio)propanoic acid 3-[{3-[2-(7-Chloro-quinolin-2-yl)-vinyl]-phenyl}-(2-dimethylcarbamoyl-ethylsulfanyl)-methylsulfanyl]-propionic acid(MK-571) MK-571 3-(((3-(2-(7-chloroquinoline-2-yl)ethenyl)phenyl)((3-dimethylamino-3-oxopropyl)thio)methyl)thio)propanoic acid 3-[{3-[(E)-2-(7-Chloro-quinolin-2-yl)-vinyl]-phenyl}-(2-dimethylcarbamoyl-ethylsulfanyl)-methylsulfanyl]-propionic acid 3-[{3-[(E)-2-(7-Chloro-quinolin-2-yl)-vinyl]-phenyl}-(2-diethylcarbamoyl-ethylsulfanyl)-methylsulfanyl]-propionic acid L-660711
CHEMBL15177 3-[{3-[2-(7-Chloro-quinolin-2-yl)-vinyl]-phenyl}-(2-dimethylcarbamoyl-ethylsulfanyl)-methylsulfanyl]-propionic acid(MK-571, L-660711) BDBM50001285 MK 571 (E)-3-((3-(2-(7-chloroquinolin-2-yl)vinyl)phenyl)(3-(dimethylamino)-3-oxopropylthio)methylthio)propanoic acid 3-((3-(2-(7-chloroquinolin-2-yl)vinyl)phenyl)(3-(dimethylamino)-3-oxopropylthio)methylthio)propanoic acid 3-[{3-[2-(7-Chloro-quinolin-2-yl)-vinyl]-phenyl}-(2-dimethylcarbamoyl-ethylsulfanyl)-methylsulfanyl]-propionic acid(MK-571) MK-571 3-(((3-(2-(7-chloroquinoline-2-yl)ethenyl)phenyl)((3-dimethylamino-3-oxopropyl)thio)methyl)thio)propanoic acid 3-[{3-[(E)-2-(7-Chloro-quinolin-2-yl)-vinyl]-phenyl}-(2-dimethylcarbamoyl-ethylsulfanyl)-methylsulfanyl]-propionic acid 3-[{3-[(E)-2-(7-Chloro-quinolin-2-yl)-vinyl]-phenyl}-(2-diethylcarbamoyl-ethylsulfanyl)-methylsulfanyl]-propionic acid L-660711 MK-678 BDBM50068063 (12S,15R,18S)-9-(4-Amino-butyl)-3-benzyl-15-(4-hydroxy-benzyl)-12-(1H-indol-3-ylmethyl)-6-isopropyl-1,18-dimethyl-1,4,7,10,13,16hexaaza-cyclooctadecane-2,5,8,11,14,17-hexaone CHEMBL2370925
MK-678 BDBM50068063 (12S,15R,18S)-9-(4-Amino-butyl)-3-benzyl-15-(4-hydroxy-benzyl)-12-(1H-indol-3-ylmethyl)-6-isopropyl-1,18-dimethyl-1,4,7,10,13,16hexaaza-cyclooctadecane-2,5,8,11,14,17-hexaone CHEMBL2370925 BDBM50114086 MK-11 CHEMBL433413 N-[1-(1-{1-[1-(5-Amino-1-carbamoyl-pentylcarbamoyl)-2-(1H-indol-3-yl)-ethylcarbamoyl]-4-guanidino-butylcarbamoyl}-2-phenyl-ethylcarbamoyl)-2-(3H-imidazol-4-yl)-ethyl]-succinamic acid
BDBM50114086 MK-11 CHEMBL433413 N-[1-(1-{1-[1-(5-Amino-1-carbamoyl-pentylcarbamoyl)-2-(1H-indol-3-yl)-ethylcarbamoyl]-4-guanidino-butylcarbamoyl}-2-phenyl-ethylcarbamoyl)-2-(3H-imidazol-4-yl)-ethyl]-succinamic acid N-[(3R,6S)-6-(2,3-difluorophenyl)-2-oxo-1-(2,2,2-trifluoroethyl)azepan-3-yl]-4-(2-oxo-2,3-dihydro-1H-imidazo[4,5-b]pyridin-1-yl)piperidine-1-carboxamide BDBM50224431 CHEMBL236593 Telcagepant MK-0974
N-[(3R,6S)-6-(2,3-difluorophenyl)-2-oxo-1-(2,2,2-trifluoroethyl)azepan-3-yl]-4-(2-oxo-2,3-dihydro-1H-imidazo[4,5-b]pyridin-1-yl)piperidine-1-carboxamide BDBM50224431 CHEMBL236593 Telcagepant MK-0974 BDBM50013920 MK-679 3-[{3-[2-(7-Chloro-quinolin-2-yl)-vinyl]-phenyl}-(2-dimethylcarbamoyl-ethylsulfanyl)-methylsulfanyl]-propionic acid 3-[(R)-{3-[(E)-2-(7-Chloro-quinolin-2-yl)-vinyl]-phenyl}-(2-dimethylcarbamoyl-ethylsulfanyl)-methylsulfanyl]-propionic acid CHEMBL280481 VERLUKAST
BDBM50013920 MK-679 3-[{3-[2-(7-Chloro-quinolin-2-yl)-vinyl]-phenyl}-(2-dimethylcarbamoyl-ethylsulfanyl)-methylsulfanyl]-propionic acid 3-[(R)-{3-[(E)-2-(7-Chloro-quinolin-2-yl)-vinyl]-phenyl}-(2-dimethylcarbamoyl-ethylsulfanyl)-methylsulfanyl]-propionic acid CHEMBL280481 VERLUKAST BDBM50032771 MK-386 CHEMBL25448 (4aR,6aR,7R,10S)-7-(1,5-Dimethyl-hexyl)-1,4a,6a,10-tetramethyl-hexadecahydro-indeno[5,4-f]quinolin-2-one (4aR,6aR,10S)-7-(1,5-Dimethyl-hexyl)-1,4a,6a,10-tetramethyl-hexadecahydro-indeno[5,4-f]quinolin-2-one
BDBM50032771 MK-386 CHEMBL25448 (4aR,6aR,7R,10S)-7-(1,5-Dimethyl-hexyl)-1,4a,6a,10-tetramethyl-hexadecahydro-indeno[5,4-f]quinolin-2-one (4aR,6aR,10S)-7-(1,5-Dimethyl-hexyl)-1,4a,6a,10-tetramethyl-hexadecahydro-indeno[5,4-f]quinolin-2-one Sodium;3-[(R)-{3-[(E)-2-(7-chloro-quinolin-2-yl)-vinyl]-phenyl}-(2-dimethylcarbamoyl-ethylsulfanyl)-methylsulfanyl]-propionate MK-679 BDBM50052029 Sodium; 3-[(R)-{3-[(E)-2-(7-chloro-quinolin-2-yl)-vinyl]-phenyl}-(2-dimethylcarbamoyl-ethylsulfanyl)-methylsulfanyl]-propionate L-668019 CHEMBL89340
Sodium;3-[(R)-{3-[(E)-2-(7-chloro-quinolin-2-yl)-vinyl]-phenyl}-(2-dimethylcarbamoyl-ethylsulfanyl)-methylsulfanyl]-propionate MK-679 BDBM50052029 Sodium; 3-[(R)-{3-[(E)-2-(7-chloro-quinolin-2-yl)-vinyl]-phenyl}-(2-dimethylcarbamoyl-ethylsulfanyl)-methylsulfanyl]-propionate L-668019 CHEMBL89340 3-[3-tert-Butylsulfanyl-1-(4-chloro-benzyl)-5-(quinolin-2-ylmethoxy)-1H-indol-2-yl]-2,2-dimethyl-propionic acid Sodium; 3-[3-tert-butylsulfanyl-1-(4-chloro-benzyl)-5-(quinolin-2-ylmethoxy)-1H-indol-2-yl]-2,2-dimethyl-propionate MK-591 BDBM50052018 CHEMBL313489
3-[3-tert-Butylsulfanyl-1-(4-chloro-benzyl)-5-(quinolin-2-ylmethoxy)-1H-indol-2-yl]-2,2-dimethyl-propionic acid Sodium; 3-[3-tert-butylsulfanyl-1-(4-chloro-benzyl)-5-(quinolin-2-ylmethoxy)-1H-indol-2-yl]-2,2-dimethyl-propionate MK-591 BDBM50052018 CHEMBL313489 CHEMBL523352 (1R,2R)-5,5-Dichloro-N-(1-cyanocyclopropyl)-2-[4-[4-(methylsulfonyl)phenyl]-1-(2,2,2-trifluoroethyl)-1H-pyrazol-3-yl]cyclohexanecarboxamide (1R,2R)-5,5-dichloro-N-(1-cyanocyclopropyl)-2-(4-(4-(methylsulfonyl)phenyl)-1-(2,2,2-trifluoroethyl)-1H-pyrazol-3-yl)cyclohexanecarboxamide BDBM50253098 MK-1256
CHEMBL523352 (1R,2R)-5,5-Dichloro-N-(1-cyanocyclopropyl)-2-[4-[4-(methylsulfonyl)phenyl]-1-(2,2,2-trifluoroethyl)-1H-pyrazol-3-yl]cyclohexanecarboxamide (1R,2R)-5,5-dichloro-N-(1-cyanocyclopropyl)-2-(4-(4-(methylsulfonyl)phenyl)-1-(2,2,2-trifluoroethyl)-1H-pyrazol-3-yl)cyclohexanecarboxamide BDBM50253098 MK-1256 US10100030, Aprepitant 5-[(2R,3S)-2-[(R)-1-(3,5-Bis-trifluoromethyl-phenyl)-ethoxy]-3-(4-fluoro-phenyl)-morpholin-4-ylmethyl]-2,4-dihydro-[1,2,4]triazol-3-one MK-0869 US10011568, Aprepitant MK-869 3-[2-{1-[3,5-di(trifluoromethyl)phenyl]ethoxy}-3-(4-fluorophenyl)-1,4-oxazinan-4-ylmethyl]-4,5-dihydro-1H-1,2,4-triazol-5-one APREPITANT CHEMBL1471 Emend 5-(((2R,3S)-2-((R)-1-(3,5-bis(trifluoromethyl)phenyl)ethoxy)-3-(4-fluorophenyl)morpholino)methyl)-2H-1,2,4-triazol-3(4H)-one 5-[2-[1-(3,5-Bis-trifluoromethyl-phenyl)-ethoxy]-3-(4-fluoro-phenyl)-morpholin-4-ylmethyl]-2,4-dihydro-[1,2,4]triazol-3-one BDBM50220136
US10100030, Aprepitant 5-[(2R,3S)-2-[(R)-1-(3,5-Bis-trifluoromethyl-phenyl)-ethoxy]-3-(4-fluoro-phenyl)-morpholin-4-ylmethyl]-2,4-dihydro-[1,2,4]triazol-3-one MK-0869 US10011568, Aprepitant MK-869 3-[2-{1-[3,5-di(trifluoromethyl)phenyl]ethoxy}-3-(4-fluorophenyl)-1,4-oxazinan-4-ylmethyl]-4,5-dihydro-1H-1,2,4-triazol-5-one APREPITANT CHEMBL1471 Emend 5-(((2R,3S)-2-((R)-1-(3,5-bis(trifluoromethyl)phenyl)ethoxy)-3-(4-fluorophenyl)morpholino)methyl)-2H-1,2,4-triazol-3(4H)-one 5-[2-[1-(3,5-Bis-trifluoromethyl-phenyl)-ethoxy]-3-(4-fluoro-phenyl)-morpholin-4-ylmethyl]-2,4-dihydro-[1,2,4]triazol-3-one BDBM50220136 CHEMBL905 N,N-dimethyl-5-(1H-1,2,4-triazol-1-ylmethyl)-1H-indole-3-ethanamine BDBM50033437 MK 462 free base risatriptan N,N-dimethyl-2-[5-(1H-1,2,4-triazol-1-ylmethyl)-1H-indol-3-yl]ethanamine N,N-dimethyl-2-[5-(1,2,4-triazol-1-ylmethyl)-1H-indol-3-yl]-ethanamine RIZATRIPTAN
CHEMBL905 N,N-dimethyl-5-(1H-1,2,4-triazol-1-ylmethyl)-1H-indole-3-ethanamine BDBM50033437 MK 462 free base risatriptan N,N-dimethyl-2-[5-(1H-1,2,4-triazol-1-ylmethyl)-1H-indol-3-yl]ethanamine N,N-dimethyl-2-[5-(1,2,4-triazol-1-ylmethyl)-1H-indol-3-yl]-ethanamine RIZATRIPTAN BDBM50252149 CHEMBL481931 MK-0731 (S)-4-(2,5-difluorophenyl)-N-((3R,4S)-3-fluoro-1-methylpiperidin-4-yl)-2-(hydroxymethyl)-N-methyl-2-phenyl-2H-pyrrole-1(5H)-carboxamide (2S)-4-(2,5-difluorophenyl)-N-[(3R,4S)-3-fluoro-1-methylpiperidin-4-yl]-2-(hydroxymethyl)-N-methyl-2-phenyl-2,5-dihydro-1H-pyrrole-1-carboxamide
BDBM50252149 CHEMBL481931 MK-0731 (S)-4-(2,5-difluorophenyl)-N-((3R,4S)-3-fluoro-1-methylpiperidin-4-yl)-2-(hydroxymethyl)-N-methyl-2-phenyl-2H-pyrrole-1(5H)-carboxamide (2S)-4-(2,5-difluorophenyl)-N-[(3R,4S)-3-fluoro-1-methylpiperidin-4-yl]-2-(hydroxymethyl)-N-methyl-2-phenyl-2,5-dihydro-1H-pyrrole-1-carboxamide Sitagliptin (13) US10479798, Compound MK0431 (1R)-3-oxo-3-[3-(trifluoroethyl)-5,6-dihydro[1,2,4]triazolo[4,3-a]pyrazin-7(8H)-yl]-1-(2,4,5-trifluorobenzyl)propylamine Sitagliptin CHEMBL393336 Triazolopiperazine Analogue 1 MK-0431 BDBM11162 (3R)-3-amino-1-[3-(trifluoromethyl)-5H,6H,7H,8H-[1,2,4]triazolo[3,4-a]pyrazin-7-yl]-4-(2,4,5-trifluorophenyl)butan-1-one hydrochloride
Sitagliptin (13) US10479798, Compound MK0431 (1R)-3-oxo-3-[3-(trifluoroethyl)-5,6-dihydro[1,2,4]triazolo[4,3-a]pyrazin-7(8H)-yl]-1-(2,4,5-trifluorobenzyl)propylamine Sitagliptin CHEMBL393336 Triazolopiperazine Analogue 1 MK-0431 BDBM11162 (3R)-3-amino-1-[3-(trifluoromethyl)-5H,6H,7H,8H-[1,2,4]triazolo[3,4-a]pyrazin-7-yl]-4-(2,4,5-trifluorophenyl)butan-1-one hydrochloride Sodium; 3-[{3-[(E)-2-(7-chloro-quinolin-2-yl)-vinyl]-phenyl}-(2-dimethylcarbamoyl-ethylsulfanyl)-methylsulfanyl]-propionate L-660771 BDBM50052019 Sodium;3-[{3-[(E)-2-(7-chloro-quinolin-2-yl)-vinyl]-phenyl}-(2-dimethylcarbamoyl-ethylsulfanyl)-methylsulfanyl]-propionate CHEMBL89768 MK-571 sodium (E)-3-((3-(2-(7-chloroquinolin-2-yl)vinyl)phenyl)(3-(dimethylamino)-3-oxopropylthio)methylthio)propanoate
Sodium; 3-[{3-[(E)-2-(7-chloro-quinolin-2-yl)-vinyl]-phenyl}-(2-dimethylcarbamoyl-ethylsulfanyl)-methylsulfanyl]-propionate L-660771 BDBM50052019 Sodium;3-[{3-[(E)-2-(7-chloro-quinolin-2-yl)-vinyl]-phenyl}-(2-dimethylcarbamoyl-ethylsulfanyl)-methylsulfanyl]-propionate CHEMBL89768 MK-571 sodium (E)-3-((3-(2-(7-chloroquinolin-2-yl)vinyl)phenyl)(3-(dimethylamino)-3-oxopropylthio)methylthio)propanoate CHEMBL599872 (1R,21S,24S)-21-tert-butyl-N-((1R,2R)-1-{[(cyclopropylsulfonyl)amino]carbonyl}-2-ethylcyclopropyl)-16,16-dimethyl-3,19,22-trioxo-2,18-dioxa-4,20,23-triazatetracyclo[21.2.1.14,70.06,11]heptacosa-6,8,10-triene-24-carboxamide BDBM50326055 MK-7009 (1R,21S,24S)-21-tert-Butyl-N-((1R,2R)-1-{[(cyclopropylsulfonyl)amino]carbonyl}-2-ethylcyclopropyl)-16,16-dimethyl-3,19,22-trioxo-2,18-dioxa-4,20,23-triazatetracyclo[21.2.1.14,7.06,11]-heptacosa-6,8,10-triene-24-carboxamide
CHEMBL599872 (1R,21S,24S)-21-tert-butyl-N-((1R,2R)-1-{[(cyclopropylsulfonyl)amino]carbonyl}-2-ethylcyclopropyl)-16,16-dimethyl-3,19,22-trioxo-2,18-dioxa-4,20,23-triazatetracyclo[21.2.1.14,70.06,11]heptacosa-6,8,10-triene-24-carboxamide BDBM50326055 MK-7009 (1R,21S,24S)-21-tert-Butyl-N-((1R,2R)-1-{[(cyclopropylsulfonyl)amino]carbonyl}-2-ethylcyclopropyl)-16,16-dimethyl-3,19,22-trioxo-2,18-dioxa-4,20,23-triazatetracyclo[21.2.1.14,7.06,11]-heptacosa-6,8,10-triene-24-carboxamide cid_54454 MK-733 SIMVASTATIN LACTONE Simvastatin (1S,3R,7S,8S,8aR)-8-{2-[(2R,4R)-4-hydroxy-6-oxotetrahydro-2H-pyran-2-yl]ethyl}-3,7-dimethyl-1,2,3,7,8,8a-hexahydronaphthalen-1-yl 2,2-dimethylbutanoate Zocor BDBM50139181 2,2-dimethylbutyric acid, 8-ester with (4R,6R)-6-(2-((1S,2S,6R,8S,8aR)-1,2,6,7,8,8a-hexahydro-8-hydroxy-2,6-dimethyl-1-naphthyl)ethyl)tetrahydro-4-hydroxy-2H-pyran-2-one
cid_54454 MK-733 SIMVASTATIN LACTONE Simvastatin (1S,3R,7S,8S,8aR)-8-{2-[(2R,4R)-4-hydroxy-6-oxotetrahydro-2H-pyran-2-yl]ethyl}-3,7-dimethyl-1,2,3,7,8,8a-hexahydronaphthalen-1-yl 2,2-dimethylbutanoate Zocor BDBM50139181 2,2-dimethylbutyric acid, 8-ester with (4R,6R)-6-(2-((1S,2S,6R,8S,8aR)-1,2,6,7,8,8a-hexahydro-8-hydroxy-2,6-dimethyl-1-naphthyl)ethyl)tetrahydro-4-hydroxy-2H-pyran-2-one 3-[1-(4-chlorobenzyl)-3-t-butyl-thio-5-isopropylindol-2-yl]-2,2-dimethylpropanoic acid cid_3651377 3-(1-(4-chlorobenzyl)-3-(tert-butylthio)-5-isopropyl-1H-indol-2-yl)-2,2-dimethylpropanoic acid CHEMBL29097 MK886 MK-886 3-(3-(tert-butylthio)-1-(4-chlorobenzyl)-5-isopropyl-1H-indol-2-yl)-2,2-dimethylpropanoic acid 3-[3-tert-Butylsulfanyl-1-(4-chloro-benzyl)-5-isopropyl-1H-indol-2-yl]-2,2-dimethyl-propionic acid BDBM50006805
3-[1-(4-chlorobenzyl)-3-t-butyl-thio-5-isopropylindol-2-yl]-2,2-dimethylpropanoic acid cid_3651377 3-(1-(4-chlorobenzyl)-3-(tert-butylthio)-5-isopropyl-1H-indol-2-yl)-2,2-dimethylpropanoic acid CHEMBL29097 MK886 MK-886 3-(3-(tert-butylthio)-1-(4-chlorobenzyl)-5-isopropyl-1H-indol-2-yl)-2,2-dimethylpropanoic acid 3-[3-tert-Butylsulfanyl-1-(4-chloro-benzyl)-5-isopropyl-1H-indol-2-yl]-2,2-dimethyl-propionic acid BDBM50006805 3-(2-Guanidino-thiazol-4-ylmethylsulfanyl)-N-sulfonylamino-propionamidine BDBM50036754 Pepcid Pepcid ac (1Z)-N'-(aminosulfonyl)-3-[({2-[(diaminomethylene)amino]-1,3-thiazol-4-yl}methyl)thio]propanimidamide N-sulfomoyl-3-(2-Guanidino-2H-1lambda*4*-thiazol-4-ylmethylsulfanyl)-propionamidine (1Z)-N'-(aminosulfonyl)-3-[({2-[(diaminomethylene)amino]-1,3-thiazol-4-yl}methyl)thio]propanimidamide (Famotidine) N'-(aminosulfonyl)-3-(((2-((diaminomethylene)amino)-4-thiazolyl)methyl)thio)propanimidamide Pepcid ac (geltab) MK-208 CHEMBL902 Pepcid rpd Fluxid FAMOTIDINE
3-(2-Guanidino-thiazol-4-ylmethylsulfanyl)-N-sulfonylamino-propionamidine BDBM50036754 Pepcid Pepcid ac (1Z)-N'-(aminosulfonyl)-3-[({2-[(diaminomethylene)amino]-1,3-thiazol-4-yl}methyl)thio]propanimidamide N-sulfomoyl-3-(2-Guanidino-2H-1lambda*4*-thiazol-4-ylmethylsulfanyl)-propionamidine (1Z)-N'-(aminosulfonyl)-3-[({2-[(diaminomethylene)amino]-1,3-thiazol-4-yl}methyl)thio]propanimidamide (Famotidine) N'-(aminosulfonyl)-3-(((2-((diaminomethylene)amino)-4-thiazolyl)methyl)thio)propanimidamide Pepcid ac (geltab) MK-208 CHEMBL902 Pepcid rpd Fluxid FAMOTIDINE 2-[3-tert-Butylsulfanyl-1-(4-chloro-benzyl)-5-(quinolin-2-ylmethoxy)-1H-indol-2-yl]-2-methyl-propionic acid 3-[3-tert-Butylsulfanyl-1-(4-chloro-benzyl)-5-(quinolin-2-ylmethoxy)-1H-indol-2-yl]-2,2-dimethyl-propionic acid CHEMBL16596 3-(1-(4-chlorobenzyl)-3-(tert-butylthio)-5-(quinolin-2-ylmethoxy)-1H-indol-2-yl)-2,2-dimethylpropanoic acid 3-(3-(tert-butylthio)-1-(4-chlorobenzyl)-5-(quinolin-2-ylmethoxy)-1H-indol-2-yl)-2,2-dimethylpropanoic acid MK-591 BDBM50029559 L-686708 3-[3-(TERT-BUTYLTHIO)-1-(4-CHLOROBENZYL)-5-(QUINOLIN-2-YLMETHOXY)-1H-INDOL-2-YL]-2,2-DIMETHYLPROPANOIC ACID
2-[3-tert-Butylsulfanyl-1-(4-chloro-benzyl)-5-(quinolin-2-ylmethoxy)-1H-indol-2-yl]-2-methyl-propionic acid 3-[3-tert-Butylsulfanyl-1-(4-chloro-benzyl)-5-(quinolin-2-ylmethoxy)-1H-indol-2-yl]-2,2-dimethyl-propionic acid CHEMBL16596 3-(1-(4-chlorobenzyl)-3-(tert-butylthio)-5-(quinolin-2-ylmethoxy)-1H-indol-2-yl)-2,2-dimethylpropanoic acid 3-(3-(tert-butylthio)-1-(4-chlorobenzyl)-5-(quinolin-2-ylmethoxy)-1H-indol-2-yl)-2,2-dimethylpropanoic acid MK-591 BDBM50029559 L-686708 3-[3-(TERT-BUTYLTHIO)-1-(4-CHLOROBENZYL)-5-(QUINOLIN-2-YLMETHOXY)-1H-INDOL-2-YL]-2,2-DIMETHYLPROPANOIC ACID L-364718 1H-Indole-2-carboxylic acid (1-methyl-2-oxo-5-phenyl-2,3-dihydro-1H-benzo[e][1,4]diazepin-3-yl)-amide(devazepide) DEVAZEPIDE 1H-Indole-2-carboxylic acid (1-methyl-2-oxo-5-phenyl-2,3-dihydro-1H-benzo[e][1,4]diazepin-3-yl)-amide 1H-Indole-2-carboxylic acid (1-methyl-2-oxo-5-phenyl-2,3-dihydro-1H-benzo[e][1,4]diazepin-3-yl)-amide (L-364,718 ((S)-devazepide) 1H-Indole-2-carboxylic acid (1-methyl-2-oxo-5-phenyl-2,3-dihydro-1H-benzo[e][1,4]diazepin-3-yl)-amide (MK-329, L-364,718) CHEMBL9506 1H-Indole-2-carboxylic acid ((S)-1-methyl-2-oxo-5-phenyl-2,3-dihydro-1H-benzo[e][1,4]diazepin-3-yl)-amide 1H-Indole-2-carboxylic acid (1-methyl-2-oxo-5-phenyl-2,3-dihydro-1H-benzo[e][1,4]diazepin-3-yl)-amide: 0.1C4H10O. 0.15CH2Cl2 MK-329 1H-Indole-2-carboxylic acid (1-methyl-2-oxo-5-phenyl-2,3-dihydro-1H-benzo[e][1,4]diazepin-3-yl)-amide(L-364718) (Z)-N-(1-methyl-2-oxo-5-phenyl-2,3-dihydro-1H-benzo[e][1,4]diazepin-3-yl)-1H-indole-2-carboxamide L-364,718 (S)-1H-Indole-2-carboxylic acid (1-methyl-2-oxo-5-phenyl-2,3-dihydro-1H-benzo[e][1,4]diazepin-3-yl)-amide BDBM50005463 (S)-N-(1-methyl-2-oxo-5-phenyl-2,3-dihydro-1H-benzo[e][1,4]diazepin-3-yl)-1H-indole-2-carboxamide CCK antagonist synthetic 18 (R)-1H-Indole-2-carboxylic acid (1-methyl-2-oxo-5-phenyl-2,3-dihydro-1H-benzo[e][1,4]diazepin-3-yl)-amide: 0.1C4H10O 1H-Indole-2-carboxylic acid (1-methyl-2-oxo-5-phenyl-2,3-dihydro-1H-benzo[e][1,4]diazepin-3-yl)-amide(Devazepide or (R) L364718) CCK antagonist synthetic 17
L-364718 1H-Indole-2-carboxylic acid (1-methyl-2-oxo-5-phenyl-2,3-dihydro-1H-benzo[e][1,4]diazepin-3-yl)-amide(devazepide) DEVAZEPIDE 1H-Indole-2-carboxylic acid (1-methyl-2-oxo-5-phenyl-2,3-dihydro-1H-benzo[e][1,4]diazepin-3-yl)-amide 1H-Indole-2-carboxylic acid (1-methyl-2-oxo-5-phenyl-2,3-dihydro-1H-benzo[e][1,4]diazepin-3-yl)-amide (L-364,718 ((S)-devazepide) 1H-Indole-2-carboxylic acid (1-methyl-2-oxo-5-phenyl-2,3-dihydro-1H-benzo[e][1,4]diazepin-3-yl)-amide (MK-329, L-364,718) CHEMBL9506 1H-Indole-2-carboxylic acid ((S)-1-methyl-2-oxo-5-phenyl-2,3-dihydro-1H-benzo[e][1,4]diazepin-3-yl)-amide 1H-Indole-2-carboxylic acid (1-methyl-2-oxo-5-phenyl-2,3-dihydro-1H-benzo[e][1,4]diazepin-3-yl)-amide: 0.1C4H10O. 0.15CH2Cl2 MK-329 1H-Indole-2-carboxylic acid (1-methyl-2-oxo-5-phenyl-2,3-dihydro-1H-benzo[e][1,4]diazepin-3-yl)-amide(L-364718) (Z)-N-(1-methyl-2-oxo-5-phenyl-2,3-dihydro-1H-benzo[e][1,4]diazepin-3-yl)-1H-indole-2-carboxamide L-364,718 (S)-1H-Indole-2-carboxylic acid (1-methyl-2-oxo-5-phenyl-2,3-dihydro-1H-benzo[e][1,4]diazepin-3-yl)-amide BDBM50005463 (S)-N-(1-methyl-2-oxo-5-phenyl-2,3-dihydro-1H-benzo[e][1,4]diazepin-3-yl)-1H-indole-2-carboxamide CCK antagonist synthetic 18 (R)-1H-Indole-2-carboxylic acid (1-methyl-2-oxo-5-phenyl-2,3-dihydro-1H-benzo[e][1,4]diazepin-3-yl)-amide: 0.1C4H10O 1H-Indole-2-carboxylic acid (1-methyl-2-oxo-5-phenyl-2,3-dihydro-1H-benzo[e][1,4]diazepin-3-yl)-amide(Devazepide or (R) L364718) CCK antagonist synthetic 17 1-Ethyl-6-fluoro-4-oxo-7-piperazin-1-yl-1,4,4a,8a-tetrahydro-quinoline-3-carboxylic acid (norfloxacin) Chibroxin (norfloxacin)1-Ethyl-6-fluoro-4-oxo-7-piperazin-1-yl-1,4-dihydro-quinoline-3-carboxylic acid 1-Ethyl-6-fluoro-4-oxo-7-piperazin-1-yl-1,4-dihydro-quinoline-3-carboxylic acid(Norfloxacin) CHEMBL9 1-ethyl-6-fluoro-7-hexahydro-1-pyrazinyl-4-oxo-1,4-dihydro-3-quinolinecarboxylic acid 1-Ethyl-6-fluoro-4-oxo-7-piperazin-1-yl-1,4-dihydro-quinoline-3-carboxylic acid (Norfloxacin) 1-Ethyl-6-fluoro-4-oxo-7-piperazin-1-yl-1,4-dihydro-quinoline-3-carboxylic acid(1-norfloxacin) NORFLOXACIN Noroxin 1-ethyl-6-fluoro-4-oxo-7-(piperazin-1-yl)-1,4-dihydroquinoline-3-carboxylic acid (NFLX)1-Ethyl-6-fluoro-4-oxo-7-piperazin-1-yl-1,4-dihydro-quinoline-3-carboxylic acid 1-Ethyl-6-fluoro-4-oxo-7-piperazin-1-yl-1,4-dihydro-quinoline-3-carboxylic acid BDBM50045000 MK-366
1-Ethyl-6-fluoro-4-oxo-7-piperazin-1-yl-1,4,4a,8a-tetrahydro-quinoline-3-carboxylic acid (norfloxacin) Chibroxin (norfloxacin)1-Ethyl-6-fluoro-4-oxo-7-piperazin-1-yl-1,4-dihydro-quinoline-3-carboxylic acid 1-Ethyl-6-fluoro-4-oxo-7-piperazin-1-yl-1,4-dihydro-quinoline-3-carboxylic acid(Norfloxacin) CHEMBL9 1-ethyl-6-fluoro-7-hexahydro-1-pyrazinyl-4-oxo-1,4-dihydro-3-quinolinecarboxylic acid 1-Ethyl-6-fluoro-4-oxo-7-piperazin-1-yl-1,4-dihydro-quinoline-3-carboxylic acid (Norfloxacin) 1-Ethyl-6-fluoro-4-oxo-7-piperazin-1-yl-1,4-dihydro-quinoline-3-carboxylic acid(1-norfloxacin) NORFLOXACIN Noroxin 1-ethyl-6-fluoro-4-oxo-7-(piperazin-1-yl)-1,4-dihydroquinoline-3-carboxylic acid (NFLX)1-Ethyl-6-fluoro-4-oxo-7-piperazin-1-yl-1,4-dihydro-quinoline-3-carboxylic acid 1-Ethyl-6-fluoro-4-oxo-7-piperazin-1-yl-1,4-dihydro-quinoline-3-carboxylic acid BDBM50045000 MK-366 Proscar BDBM50334788 (4aR,6aS,7S)-4a,6a-Dimethyl-2-oxo-2,4a,4b,5,6,6a,7,8,9,9a,9b,10,11,11a-tetradecahydro-1H-indeno[5,4-f]quinoline-7-carboxylic acid tert-butylamide (4aR,6aS)-4a,6a-Dimethyl-2-oxo-2,4a,4b,5,6,6a,7,8,9,9a,9b,10,11,11a-tetradecahydro-1H-indeno[5,4-f]quinoline-7-carboxylic acid tert-butylamide CHEMBL710 FINASTERIDE 4a,6a-Dimethyl-2-oxo-2,4a,4b,5,6,6a,7,8,9,9a,9b,10,11,11a-tetradecahydro-1H-indeno[5,4-f]quinoline-7-carboxylic acid tert-butylamide(Finasteride) MK-906 (R)-4a,6a-Dimethyl-2-oxo-2,4a,4b,5,6,6a,7,8,9,9a,9b,10,11,11a-tetradecahydro-1H-indeno[5,4-f]quinoline-7-carboxylic acid tert-butylamide (4aR,4bS,6aS,7S,9aS,9bS,11aR)-N-tert-butyl-4a,6a-dimethyl-2-oxo-2,4a,4b,5,6,6a,7,8,9,9a,9b,10,11,11a-tetradecahydro-1H-indeno[5,4-f]quinoline-7-carboxamide Propecia (4aR,6aS,7S,11aR)-4a,6a-Dimethyl-2-oxo-2,4a,4b,5,6,6a,7,8,9,9a,9b,10,11,11a-tetradecahydro-1H-indeno[5,4-f]quinoline-7-carboxylic acid tert-butylamide US9061023, Finasteride (4aR,4bS,6aS,7S,9aS,9bS,11aR)-4a,6a-Dimethyl-2-oxo-2,4a,4b,5,6,6a,7,8,9,9a,9b,10,11,11a-tetradecahydro-1H-indeno[5,4-f]quinoline-7-carboxylic acid tert-butylamide (4aR,4bS,6aS,9aS,9bS,11aR)-4a,6a-Dimethyl-2-oxo-2,4a,4b,5,6,6a,7,8,9,9a,9b,10,11,11a-tetradecahydro-1H-indeno[5,4-f]quinoline-7-carboxylic acid tert-butylamide (4aR,6aS,11aR)-4a,6a-Dimethyl-2-oxo-hexadecahydro-indeno[5,4-f]quinoline-7-carboxylic acid tert-butylamide 4a,6a-Dimethyl-2-oxo-2,4a,4b,5,6,6a,7,8,9,9a,9b,10,11,11a-tetradecahydro-1H-indeno[5,4-f]quinoline-7-carboxylic acid tert-butylamide (17beta-(N-tert-butylcarbamoyl)-4-aza-5alpha-androst-1-en-3-one 4a,6a,9a-Trimethyl-2-oxo-2,4a,4b,5,6,6a,7,8,9,9a,9b,10,11,11a-tetradecahydro-1H-indeno[5,4-f]quinoline-7-carboxylic acid (Finasteride)
Proscar BDBM50334788 (4aR,6aS,7S)-4a,6a-Dimethyl-2-oxo-2,4a,4b,5,6,6a,7,8,9,9a,9b,10,11,11a-tetradecahydro-1H-indeno[5,4-f]quinoline-7-carboxylic acid tert-butylamide (4aR,6aS)-4a,6a-Dimethyl-2-oxo-2,4a,4b,5,6,6a,7,8,9,9a,9b,10,11,11a-tetradecahydro-1H-indeno[5,4-f]quinoline-7-carboxylic acid tert-butylamide CHEMBL710 FINASTERIDE 4a,6a-Dimethyl-2-oxo-2,4a,4b,5,6,6a,7,8,9,9a,9b,10,11,11a-tetradecahydro-1H-indeno[5,4-f]quinoline-7-carboxylic acid tert-butylamide(Finasteride) MK-906 (R)-4a,6a-Dimethyl-2-oxo-2,4a,4b,5,6,6a,7,8,9,9a,9b,10,11,11a-tetradecahydro-1H-indeno[5,4-f]quinoline-7-carboxylic acid tert-butylamide (4aR,4bS,6aS,7S,9aS,9bS,11aR)-N-tert-butyl-4a,6a-dimethyl-2-oxo-2,4a,4b,5,6,6a,7,8,9,9a,9b,10,11,11a-tetradecahydro-1H-indeno[5,4-f]quinoline-7-carboxamide Propecia (4aR,6aS,7S,11aR)-4a,6a-Dimethyl-2-oxo-2,4a,4b,5,6,6a,7,8,9,9a,9b,10,11,11a-tetradecahydro-1H-indeno[5,4-f]quinoline-7-carboxylic acid tert-butylamide US9061023, Finasteride (4aR,4bS,6aS,7S,9aS,9bS,11aR)-4a,6a-Dimethyl-2-oxo-2,4a,4b,5,6,6a,7,8,9,9a,9b,10,11,11a-tetradecahydro-1H-indeno[5,4-f]quinoline-7-carboxylic acid tert-butylamide (4aR,4bS,6aS,9aS,9bS,11aR)-4a,6a-Dimethyl-2-oxo-2,4a,4b,5,6,6a,7,8,9,9a,9b,10,11,11a-tetradecahydro-1H-indeno[5,4-f]quinoline-7-carboxylic acid tert-butylamide (4aR,6aS,11aR)-4a,6a-Dimethyl-2-oxo-hexadecahydro-indeno[5,4-f]quinoline-7-carboxylic acid tert-butylamide 4a,6a-Dimethyl-2-oxo-2,4a,4b,5,6,6a,7,8,9,9a,9b,10,11,11a-tetradecahydro-1H-indeno[5,4-f]quinoline-7-carboxylic acid tert-butylamide (17beta-(N-tert-butylcarbamoyl)-4-aza-5alpha-androst-1-en-3-one 4a,6a,9a-Trimethyl-2-oxo-2,4a,4b,5,6,6a,7,8,9,9a,9b,10,11,11a-tetradecahydro-1H-indeno[5,4-f]quinoline-7-carboxylic acid (Finasteride)