Compound (493)
Article Title (14)
Assay (471)
Zhao, B; Bower, MJ; McDevitt, PJ; Zhao, H; Davis, ST; Johanson, KO; Green, SM; Concha, NO; Zhou, BB Structural basis for Chk1 inhibition by UCN-01. J Biol Chem 277: 46609 -15 (2002) Ye, H; Lv, T; Min, T; Mao, D; Chen, X; Ding, B; Zhang, C HR1405-01, a Safe intravenous NSAID with superior anti-inflammatory and analgesic activities in preclinical trials. Eur J Med Chem 235: (2022) Jiang, Y; Li, X; Hou, J; Huang, Y; Jia, Y; Zou, M; Zhang, J; Wang, X; Xu, W; Zhang, Y Discovery of BC-01, a novel mutual prodrug (hybrid drug) of ubenimex and fluorouracil as anticancer agent. Eur J Med Chem 121: 649 -657 (2016) Komander, D; Kular, GS; Bain, J; Elliott, M; Alessi, DR; Van Aalten, DM Structural basis for UCN-01 (7-hydroxystaurosporine) specificity and PDK1 (3-phosphoinositide-dependent protein kinase-1) inhibition. Biochem J 375: 255 -62 (2003) Solingapuram Sai, KK; Kil, KE; Tu, Z; Chu, W; Finck, BN; Rothfuss, JM; Shoghi, KI; Welch, MJ; Gropler, RJ; Mach, RH Synthesis, radiolabeling and initial in vivo evaluation of [(11)C]KSM-01 for imaging PPAR-a receptors. Bioorg Med Chem Lett 22: 6233 -6 (2012) Banerjee, S; Buhrlage, SJ; Huang, HT; Deng, X; Zhou, W; Wang, J; Traynor, R; Prescott, AR; Alessi, DR; Gray, NS Characterization of WZ4003 and HTH-01-015 as selective inhibitors of the LKB1-tumour-suppressor-activated NUAK kinases. Biochem J 457: 215 -25 PubChem, PC High-throughput multiplex microsphere dose response for inhibitors of toxin protease, specifically Lethal Factor protease, compounds from Cherry Pick 01 PubChem Bioassay (2012) PubChem, PC High-throughput multiplex microsphere dose response for inhibitors of toxin protease, specifically Lethal Factor protease, compounds from Powder Set 01 PubChem Bioassay (2012) PubChem, PC High-throughput multiplex microsphere dose response for inhibitors of toxin protease, specifically Botulinum neurotoxin light chain A protease, compounds from Cherry Pick 01 PubChem Bioassay (2012) PubChem, PC High-throughput multiplex microsphere dose response for inhibitors of toxin protease, specifically Botulinum neurotoxin light chain A protease, compounds from Powder Set 01 PubChem Bioassay (2012) PubChem, PC High-throughput multiplex microsphere dose response for inhibitors of toxin protease, specifically Botulinum neurotoxin light chain F protease, compounds from Cherry Pick 01 PubChem Bioassay (2012) PubChem, PC High-throughput multiplex microsphere dose response for inhibitors of toxin protease, specifically Botulinum neurotoxin light chain F protease, compounds from Powder Set 01 PubChem Bioassay (2012) Jiang, B; Jiang, J; Kaltheuner, IH; Iniguez, AB; Anand, K; Ferguson, FM; Ficarro, SB; Seong, BKA; Greifenberg, AK; Dust, S; Kwiatkowski, NP; Marto, JA; Stegmaier, K; Zhang, T; Geyer, M; Gray, NS Structure-activity relationship study of THZ531 derivatives enables the discovery of BSJ-01-175 as a dual CDK12/13 covalent inhibitor with efficacy in Ewing sarcoma. Eur J Med Chem 221: (2021) Liang, Q; Chen, Y; Yu, K; Chen, C; Zhang, S; Wang, A; Wang, W; Wu, H; Liu, X; Wang, B; Wang, L; Hu, Z; Wang, W; Ren, T; Zhang, S; Liu, Q; Yun, CH; Liu, J Discovery of N-(3-(5-((3-acrylamido-4-(morpholine-4-carbonyl)phenyl)amino)-1-methyl-6-oxo-1,6-dihydropyridin-3-yl)-2-methylphenyl)-4-(tert-butyl)benzamide (CHMFL-BTK-01) as a highly selective irreversible Bruton's tyrosine kinase (BTK) inhibitor. Eur J Med Chem 131: 107 -125 (2017)
ChEMBL_691521 (CHEMBL1634439) Inhibition of Influenza A virus (A/R(duck/Mongolia/54/01-duck/Mongolia/47/01)(H5H1)) neuraminidase after 30 mins by fluorescence analysis ChEMBL_64049 (CHEMBL677409) Inhibitory activity of compound against VanA Enterococcus faecalis MGH-01 ChEMBL_1437608 (CHEMBL3390210) Inhibition of Influenza A virus A/duck/china/QJ/01(H5N1) Neuraminidase ChEBML_1636262 Inhibition of HT-01 probe binding to recombinant mouse DAGLbeta expressed in HEK293T cell membrane proteomes preincubated for 30 mins followed by HT-01 probe addition measured after 30 mins by gel-based competitive activity based protein profiling assay ChEMBL_1636262 (CHEMBL3879160) Inhibition of HT-01 probe binding to recombinant mouse DAGLbeta expressed in HEK293T cell membrane proteomes preincubated for 30 mins followed by HT-01 probe addition measured after 30 mins by gel-based competitive activity based protein profiling assay ChEMBL_1636263 (CHEMBL3879161) Inhibition of HT-01 probe binding to recombinant human DAGLalpha expressed in HEK293T cell membrane proteomes preincubated for 30 mins followed by HT-01 probe addition measured after 30 mins by gel-based competitive activity based protein profiling assay ChEMBL_2274344 Inhibition of ABHD6 in mouse brain membrane using HT-01 probe as substrate incubated for 30 mins by gel based competitive ABPP assay ChEMBL_2277529 Binding affinity to N-terminal His-tagged recombinant ZIKV ZG-01 RdRp (272 to 903 residues) expressed in Escherichia coli rosetta by surface plasmon resonance assay ChEMBL_965196 (CHEMBL2393981) Inhibition of Influenza A virus H1N1 A/duck/China/QJ/01 neuraminidase N1 using 4-MU-NANA as substrate after 5 mins by fluorescence assay ChEBML_1970661 Inhibition of recombinant His-tagged full length human FES expressed in baculovirus expression system using Tyr 01 peptide as substrate after 1 hr by Z'-LYTE assay ChEMBL_1710916 (CHEMBL4120965) Inhibition of Influenza A virus (A/Mississippi/03/01 (H1N1)) neuraminidase activity using 4-MUNANA as substrate measured every min for 60 mins by fluorescence assay ChEMBL_1710917 (CHEMBL4120966) Inhibition of Influenza A virus (A/Mississippi/03/01 (H1N1)) neuraminidase H274Y mutant using 4-MUNANA as substrate measured every min for 60 mins by fluorescence assay ChEMBL_1837923 (CHEMBL4338056) Antibacterial activity against vancomycin-resistant Enterococcus faecalis AUS-RBWH-VRE-01 assessed as reduction in fungal growth incubated for 24 hrs by spectrophotometry based broth microdilution assay ChEBML_1970497 Inhibition of recombinant human full length AURKA expressed in baculovirus expression system using Ser/Thr-01 peptide as substrate after 1 hr in presence of ATP by Z'LYTE assay ChEMBL_1853740 (CHEMBL4354364) Inhibition of recombinant human His-tagged TrkC cytoplasmic domain (510 to 825 residues) expressed in baculovirus using tyr 01 as substrate incubated for 1 hr by Z'-Lyte assay ChEBML_1970725 Inhibition of recombinant full length human His-tagged MATK expressed in Escherichia coli using tyr-01 peptide as substrate incubated for 60 mins in presence of ATP by Z'-LYTE assay ChEBML_1970765 Inhibition of recombinant full length GST-tagged human PKG2 expressed in baculovirus expression system using Ser/Thr 01 as substrate after 1 hr in presence of ATP by Z'-LYTE assay ChEMBL_1783270 (CHEMBL4254787) Inhibition of Influenza A virus (A/Shanghai/01/2014(H7N9)) recombinant neuraminidase R292K mutant expressed in HEK293 cells pre-incubated for 10 mins before 4-MUNANA substrate addition by fluorometry ChEBML_1970650 Inhibition of recombinant human GST-tagged EPHA2 cytoplasmic domain expressed in baculovirus expression system using tyrosine-01 peptide as substrate incubated for 60 mins in presence of ATP by Z'-Lyte assay ChEBML_1970653 Inhibition of recombinant human GST-tagged EPHA5 cytoplasmic domain expressed in baculovirus expression system using tyrosine-01 peptide as substrate incubated for 60 mins in presence of ATP by Z'-Lyte assay ChEMBL_1853738 (CHEMBL4354362) Inhibition of recombinant human His-tagged TrkA catalytic domain (441 to 796 residues) expressed in baculovirus expression system using tyr 01 as substrate incubated for 1 hr by Z'-Lyte assay ChEMBL_1853739 (CHEMBL4354363) Inhibition of recombinant human His-tagged TrkB catalytic domain (526 to 838 residues) expressed in baculovirus expression system using tyr 01 as substrate incubated for 1 hr by Z'-Lyte assay ChEBML_1970466 Inhibition of recombinant full length His-tagged human AURKB expressed in baculovirus expression system using Ser/Thr-01 peptide as substrate incubated for 60 mins in presence of ATP by Z'-LYTE assay ChEBML_1970743 Inhibition of recombinant human His-tagged TrkC cytoplasmic domain (510 to 825 residues) expressed in baculovirus using tyr 01 as substrate incubated for 1 hr in presence of ATP by Z'-Lyte assay ChEBML_1970783 Inhibition of recombinant full length human GST-tagged PRKD2 expressed in baculovirus expression system using serine/threonine-01 peptide as substrate incubated for 60 mins in presence of ATP by Z'-LYTE assay ChEBML_1970784 Inhibition of recombinant full length human GST-tagged PRKG1 expressed in baculovirus expression system using serine/threonine-01 peptide as substrate incubated for 60 mins in presence of ATP by Z'-LYTE assay ChEMBL_2215242 (CHEMBL5128374) Inhibition of Influenza A virus (A/GuangdongSB/01/2009(H1N1)) neuraminidase using 4-MUNANA as substrate preincubated for 1 hr followed by substrate addition and measured after 1 hr by fluorescence assay ChEMBL_1710915 (CHEMBL4120964) Inhibition of Influenza A virus (A/Mississippi/03/01 (H1N1)) neuraminidase activity using 4-MUNANA as substrate preincubated for 60 mins followed by substrate addition measured every min for 60 mins by fluorescence assay ChEBML_1970657 Inhibition of recombinant human His-tagged EPHB4 (561 to 987 residues) catalytic domain expressed in baculovirus expression system using tyrosine-01 peptide as substrate incubated for 60 mins in presence of ATP by Z'-LYTE assay ChEBML_1970742 Inhibition of recombinant human His-tagged NTRK2 cytoplasmic domain (526 to 838 residues) expressed in baculovirus expression system using tyrosine-01 peptide as substrate incubated for 60 mins in presence of ATP by Z'-LYTE assay ChEMBL_1280517 (CHEMBL3096191) Inhibition of influenza A virus A/duck/China/QJ/01(H5N1) neuraminidase using 4-MU-NANA as substrate incubated for 5 mins prior to substrate addition measured after 30 to 60 mins by fluorescence assay ChEMBL_1710918 (CHEMBL4120967) Inhibition of Influenza A virus (A/Mississippi/03/01 (H1N1)) neuraminidase H274Y mutant using 4-MUNANA as substrate preincubated for 60 mins followed by substrate addition measured every min for 60 mins by fluorescence assay ChEMBL_2059828 (CHEMBL4714829) Inhibition of recombinant human His-tagged FGFR1 (308 to 731 residues) cytoplasmic domain expressed in baculovirus expression system using Tyr 01 as substrate incubated for 1 hr in presence of ATP by Z'-Lyte assay ChEMBL_932510 (CHEMBL3074565) Inhibition of influenza A virus (A/duck/China/QJ/01(H5N1)) neuraminidase using 4-MU-NANA as substrate preincubated with enzyme for 5 min prior to substrate addition measured after 30-60 min by fluorescence analysis AKT Kinase Inhibiting Activity Envision model plate reader (Molecular Devices)White 384-well plate (Thermo, Art. No. #264706)Main reagents included in an HTRF kinEASE TK kit (Cisbio, Art. No. #62TKOPEC)TK-biotin substrateStreptavidin-XL665Europium-labeled tyrosine kinase substrate antibody5× enzyme reaction bufferSEBHTRF assay bufferAKT1 (Cama, Art. No. #01-101)AKT2 (Cama, Art. No. #01-102)AKT3 (Invitrogen, Art. No. #PV3185)10 mM ATP (Invitrogen, Art. No. #PV3227)1 M DTT (Sigma, Art. No. #D5545)1 M MgCl2 (Sigma, Art. No. #M8266). Inhibitory Activity Assay BTK: The 2×BTK/Tyr 01 mixture is prepared in 50 mM HEPES pH 7.5, 0.01% BRIJ-35, 10 mM MgCl2, 1 mM EGTA. The final 10 μL Kinase Reaction consists of 1.04-10.4 ng BTK and 2 μM Tyr 01 in 50 mM HEPES pH 7.5, 0.01% BRIJ-35, 10 mM MgCl2, 1 mM EGTA. After the 1 hour Kinase Reaction incubation, 5 μL of a 1:256 dilution of Development Reagent B is added. AlphaLISA-based IL-10 Detection Concentration-dependent effects of HG-9-91-01 analogs on IL-10 release and cellular cytotoxicity in Zymosan A-stimulated BMDCs were measured using AlphaLISA-based IL-10 detection (PerkinElmer) or CellTiterGlo (Promega) as described previously. ChEMBL_1280515 (CHEMBL3096189) Non competitive inhibition of influenza A virus A/duck/China/QJ/01(H5N1) neuraminidase using 4-MU-NANA as substrate incubated for 5 mins prior to substrate addition measured after 30 to 60 mins by Lineweaver-Burk plot analysis Inhibitory Activity Assay KDR: The 2×KDR (VEGFR2)/Tyr 01 mixture is prepared in 50 mM HEPES pH 7.5, 0.01% BRIJ-35, 10 mM MgCl2, 1 mM EGTA. The final 10 μL Kinase Reaction consists of 0.5-11.7 ng KDR (VEGFR2) and 2 μM Tyr 01 in 50 mM HEPES pH 7.5, 0.01% BRIJ-35, 10 mM MgCl2, 1 mM EGTA. After the 1 hour Kinase Reaction incubation, 5 μL of a 1:256 dilution of Development Reagent B is added. ChEMBL_1584616 (CHEMBL3820944) Inhibition of Influenza A virus A/Shanghai/01/2014(H7N9) recombinant wild type neuraminidase transfected in HEK293 cells using MUNANA as substrate assessed as release of 4-methylumbelliferone preincubated for 10 mins followed by substrate addition measured after 15 mins by fluorometric assay Enzyme Assay Aurora B (AurB): The 2×AURKB (Aurora B)/Ser/Thr 01 (ThermoFisher Scientific proprietary) mixture was prepared in 50 mM HEPES pH 7.5, 0.01% BRIJ-35, 10 mM MgCl2, 1 mM EGTA. The final 10 μL Kinase Reaction consisted of 23 nM AURKB (Aurora B), 2 μM Ser/Thr 01 and 75 μM ATP (Km app measured as 81 μM ATP) in 50 mM HEPES pH 7.5, 0.01% BRIJ-35, 10 mM MgCl2, 1 mM EGTA. After the 1 hour Kinase Reaction incubation, 5 μL of a 1:4096 dilution of Development Reagent was added. Caliper-based Mobility Shift Assay IC50‘s for HG-9-91-01 derivatives were measured by Caliper-based mobility shift assay (PerkinElmer). For these experiments, full length His6-MBP-tagged hSIK2 (4 nM) was incubated with HG-9-91-01 derivatives in buffer containing 100 mM HEPES 7.5, 10 mM MgCl2, 2.5 mM DTT, 0.004% Tween20, 0.003% Brij-35, 30 μM ATP and 1.5 μM ProfilerPro FL-Peptide 10 (5-FAMKKKVSRSGLYRSPSMPENLNRPR-COOH, PerkinElmer, Catalog No. 760354) at rt. Reactions were quenched by adding 20 mM EDTA (pH 8) after 1 hr, and percentage of substrate conversion was measured by LabChip EZ Reader II (PerkinElmer). Inhibition Assay Inhibitory activities of compounds against KDR (VEGFR2) were also measured by Invitrogen using Z'-LYTE Method as described above with the following modification. The 2x KDR (VEGFR2)/Tyr 01 mixture is prepared in 50 mM HEPES pH 7.5, 0.01% B RIJ-35, 10 mM MgCl2, 1 mM EGTA. The final 10 μL Kinase Reaction consists of 0.5-11.7 ng KDR (VEGFR2) and 2 μM Tyr 01 in 50 mM HEPES pH 7.5, 0.01% BRIJ-35, 10 mM MgCl2, 1 mM EGTA. After the 1 hour Kinase Reaction incubation, 5 μL of a 1:256 dilution of Development Reagent B is added. Inhibitory Activity Assay KDR (VEGFR2): Inhibitory activities of compounds against KDR (VEGFR2) were also measured by Invitrogen using Z′-LYTE Method as described above with the following modification. The 2× KDR (VEGFR2)/Tyr 01 mixture is prepared in 50 mM HEPES pH 7.5, 0.01% BRIJ-35, 10 mM MgCl2, 1 mM EGTA. The final 10 μL Kinase Reaction consists of 0.5-11.7 ng KDR (VEGFR2) and 2 μM Tyr 01 in 50 mM HEPES pH 7.5, 0.01% BRIJ-35, 10 mM MgCl2, 1 mM EGTA. After the 1 hour Kinase Reaction incubation, 5 μL of a 1:256 dilution of Development Reagent B is added. ChEMBL_2342385 Inhibition of N-terminal His-tagged human DNMT3A (623 to 912 residues)/N-terminal GST-tagged human DNMT3L (160 to 387 residues) co-expressed in baculovirus infected Sf9 cells assessed as DNA methylation using DNA oligonucleotide IDT-01 as substrate preincubated for 15 mins followed by substrate addition in presence of [3H]-SAM and measured after 4 hrs by radioactive methylation assay ChEMBL_2342390 Inhibition of N-terminal His-tagged human DNMT3B (564 to 853 residues)/N-terminal GST-tagged human DNMT3L (160 to 387 residues) co-expressed in baculovirus infected Sf9 cells assessed as DNA methylation using DNA oligonucleotide IDT-01 as substrate preincubated for 15 mins followed by substrate addition in presence of [3H]-SAM and measured after 4 hrs by radioactive methylation assay Binding Assay The binding of SPI-01 to the enzyme-substrate complex was investigated. CSP analysis was carried out on the 40 kDa complex of 15N-labeled full length precursor SUMO-1-GGHSTV (SUMO-1-FL) with unlabeled SENP1-C603S. An equimolar amount of SPI-01 was added to the 1:1 enzyme-substrate complex. The only observed CSP on the 15N-labeled precursor SUMO-1-FL was on the C-terminal residues S99 and V101 (FIGS. 7 and 8) (Song et al., PNAS 101:14373-8 (2004)). This result indicates that SPI-01 binds the enzyme-substrate complex at the interface between SENP and the C-terminal tails of precursor SUMO-FL. X-ray crystal structures showed that the C-terminal tail of precursor SUMO sits in and projects out of the catalytic tunnel of SENPs (Shen et al., Nat. Struct. Mol. Biol. 13:1069-77 (2006)). In the case of SENP1, the region that interacts with the projected C-terminus is predominantly acidic and favors the C-terminus of SUMO-1, which is polar and positively charged, over that of SUMO-2, whose C-terminus is mainly hydrophobic (Shen et al., Nat. Struct. Mol. Biol. 13:1069-77 (2006); and Shen et al., The Biochemical Journal 397:279-88 (2006)). TNAP luminescent HTS assay Sanford-Burnham Center for Chemical Genomics (SBCCG) Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) NIH Molecular Libraries Screening Centers Network (MLSCN) Grant Number: MH077602-01 Alkaline phosphatase (EC 3.1.3.1) (APs) catalyze the hydrolysis of phosphomonoesters, releasing phosphate and alcohol. APs are dimeric enzymes found in the most organism. In human, four isozymes of APs have been identified. Three isozymes are tissue-specific and the fourth one is tissue-nonsepecifc, named TNAP. TNAP deficiency is associated with defective bone mineralization in the form of rickets and osteomalacia. Therefore, there are therapeutic potentials of inhibiting TNAP activity. The goal of this HTS is to identify novel and specific inhibitors of TNAP. TNAP screening was developed and performed at the Sanford-Burnham Center for Chemical Genomics (SBCCG) as part of the Molecular Library Screening Center Network (MLSCN). XO1 submission, MH077602-01, Pharmacological inhibitors o Kinase Assay (ALK) Kinase Assay Protocol:Bar-coded Corning, low volume NBS, black 384-well plate (Corning Cat. #4514) 1. 2.5 uL-4x Test Compound or 100 nL 100x plus 2.4 uL kinase buffer; 2. 5 uL-2x Peptide/Kinase Mixture; 3. 2.5 uL-4xATP Solution; 4. 30-second plate shake; 5. 60-minute Kinase Reaction incubation at room temperature; 6. 5 uL-Development Reagent Solution; 7. 30-second plate shake; 8. 60-minute Development Reaction incubation at room temperature; 9. Read on fluorescence plate reader and analyze the data. Kinase-Specific Assay Conditions:The 2xALK/Tyr 01 mixture is prepared in 50 mM HEPES pH 7.5, 0.01% BRIJ-35, 10 mM MgCl2, 1 mM EGTA. The final 10 uL Kinase Reaction consists of 4.25-96 ng ALK and 2 uM Tyr 01 in 50 mM HEPES pH 7.5, 0.01% BRIJ-35, 10 mM MgCl2, 1 mM EGTA. After the 1 hour Kinase Reaction incubation, 5 uL of a 1:256 dilution of Development Reagent B is added. Kinase Inhibition Assay The buffer for PIM-2 assay was composed of HEPES 50 mM, at pH 7.5, with 1 mM MgCl2, 1 mM DTT, 3 microM Na3VO4, and 0.2 mg/mL BSAFull-length human PIM-2 was expressed and purified as described in Fedorov O, et al., PNAS 2007 104, 51, 20523-28.Assay Conditions (Final Concentrations)Enzyme concentration=1.5 nMAktide substrate (Chemical Abstract Service Registry Number 324029-01-8)=5 microMATP=4 microM33P- -ATP=1 nM. Counter Screen for Placental Alkaline Phosphatase-based Assays Positives Sanford-Burnham Center for Chemical Genomics (SBCCG) Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) NIH Molecular Libraries Screening Centers Network (MLSCN) Grant Number: MH077609-01 This functional assay was developed for detection of compounds inhibiting placental alkaline phosphatase. These compounds would be observed as false positives of assays employing alkaline phosphatase-based detection. This assay was primarily utilized as counter screen for EphA4 hits identified in the screening assay, AID 689 performed at the Sanford-Burnham Center for Chemical Genomics (SBCCG). LATS2 Biochemical Caliper Assay Human LATS2 kinase domain protein was purchased from Carnabio (catalogue number 01-124), which harbors the catalytic domain of amino acids 553-1088, and was co-purified with human His-tagged MOBKL1A (NP_775-739). 5 microlitre of enzyme buffer, 100 nL of compounds and 5 microlitre of localising aid buffer were added into 384-well plates. The final assay reaction mix contains 100 mM HEPES, pH7.5, 0.1% BSA, 0.01% Triton X-100, 1 mM DTT, 10 mM MgCl2, 10 micromolar Sodium Orthovanadate, 10 micromolar Beta-Glycerophosphate, 400 micromolar ATP, 1% DMSO, 1.1 nM LATS2 enzyme (Carnabio, 01-124), 1 micromolar localising aid of FAM-KKLRRTLSVA-COOH (SEQ ID NO: 24) (NanoSyn). The plates were incubated at 25° C. for 3 hours. 40 microlitre of stop buffer with 25 mM EDTA (NanoSyn) was added to each well to terminate the reaction. Localising aids and products were separated electrophoretically using the microfluidic-based Caliper Labchip 3000 Drug Discovery System (Caliper Life Sciences). Plates were read using blue laser excitation and green fluorescence detection and quantified by fluorescence intensity. The IC50 is measured when the effect of the compound reduces the product fluorescence signal by 50%. In Vitro Competitive Activity-Based Protein Profiling Proteomes (human prefrontal cortex or cell membrane fractions) (50 μL, 1.0 mg/mL total protein concentration) were preincubated with varying concentrations of inhibitors at 37° C. After 30 min, FP-Rh or HT-01 (1.0 μL, 50 μM in DMSO) was added and the mixture was incubated for another 30 min at room temperature. Reactions were quenched with SDS loading buffer (15 μL 4×) and run on SDS-PAGE. Following gel imaging, serine hydrolase activity was determined by measuring fluorescent intensity of gel bands corresponding to MAGL using ImageJ 1.49 k software. Kinase Inhibition Assay Method for PIM2 Kinase Inhibition Assay: Dowex Technique i. Kinase Buffer (KB)The buffer for PIM2 assay was composed of HEPES 50 mM, at pH 7.5, with 1 mM MgCl2, 1 mM DTT, 3 uM Na3VO4, and 0.2 mg/mL BSA.Full-length human PIM2 was expressed and purified as described in Fedorov O, et al., PNAS 2007 104, 51, 20523-28 ii. Assay Conditions (Final Concentrations)Enzyme concentration=1.5 nMAktide substrate (Chemical Abstract Service Registry Number 324029-01-8)=5 uM ATP=4 uM. 33P gamma ATP=1 nM iii. Robotized Dowex AssaySee above: same procedure as described for PIM1. PARP1 Assay PARP1 In vitro selectivity assays were conducted as a 10 ul reaction volume in a NUNC Maxisorp 384-well assay plate pre-coated in-house with Histones. 5 ul of Human High specific Activity PARP1 (Trevigen: 4668-100-01) was used at a final reaction concentration of 0.02 units/ml in 1×PARP Buffer (Trevigen: 4671-096-02) with 5 ul of 1×PARP cocktail, which is a mixture of 10×PARP Cocktail (Trevigen: 4671-096-03), 10× Activate DNA (Trevigen: 4671-096-06) and 20×PARP Buffer (as above). The reaction was incubated at room temperature for 60 minutes to allow histones on the coated plate to become PARylated. The wells were then washed with PBS/0.1% Triton X100. PARP1 activity was then detected by measuring the extent of PARylation. Firstly, 10 ul of Streptavidin-HRP (Trevigen: 4800-30-06), diluted 1 in 250 in 1×PARG Assay Buffer (Trevigen: 4680-096-02), was added to each well and incubated at room temperature for 60 minutes. Secondly, following another wash with PBS/0.1% Triton X100, Peroxy Glow Reagents A and B (Trevigen: 4675-096-01 and 4675-096-02) were mixed in equal quantities immediately before use and 100 ul was added to each well. Luminescence signal was then measured immediately. Dose-Response of Allosteric Antagonists for the VLA-4 Integrin Assay Support: 1 X01 MH077638-01 MLSCN Assay for Allosteric Ligands for the VLA-4 Integrin PI: SKLAR, LARRY A University of New Mexico Assay Overview: This is a high throughput flow cytometry cell-based assay to measure dose response of small molecules# allosteric inhibition of integrin alpha-4-beta-1 heterodimer very late antigen (VLA-4). Inhibition of VLA-4 activation affects the affinity and conformation of integrins. Affinity states have been directly evaluated by a peptide ligand derived from the LDV sequence of a native ligand binding region for VLA-4. These dose response assessments report IC50. LATS1 Biochemical Assay: Homogenous Time Resolved Fluorescence (HTRF) Assay The LATS1 biochemical HTRF assay was performed using the HTRF KinEASE-STK S1 kit (CisBio, catalogue number 62ST1PEC) according to manufacturer's instructions. Human LATS1 kinase domain protein was purchased from Carnabio (catalogue number 01-123), which harbors the catalytic domain of amino acids 589-1130, and was co-purified with human His-tagged MOBKL1A (NP_775-739). Compounds were added by ECHO liquid handler (Labcyte) into 384-well plates. Then 5 microlitre of the following solution was added into the wells (50 mM HEPES, 0.01% BSA, 100 nM Orthaovanadate, 1 mM MgCl2, 1 mM DTT, 0.6 ng/microlitre LATS1 enzyme (Carnabio, 01-123), 2 micromolar STK1 localising aid (CisBio)), followed with 5 microlitre of 2 mM ATP in the kinase buffer (50 mM HEPES, 0.01% BSA, 100 nM Orthaovanadate, 1 mM MgCl2, 1 mM DTT) and incubated for 30 minutes at room temperature. 10 microlitre of detection mix (50 mM HEPES, 0.01% BSA, 100 nM Orthaovanadate, 1 mM MgCl2, 1 mM DTT, STK Antibody-Cryptate (CisBio), Streptavidin-XL665 (CisBio), 500 micromolar potassium fluoride, 50 nM EDTA) was added to each well and incubated for 1 hour at room temperature. Plates were read on Pherastar (BMG Labtech) for HTRF (665 nm/620 nm). The IC50 is measured when the effect of the compound reduces the HTRF signal by 50%. In Vitro Competitive Activity-Based Protein Assay Proteomes (mouse brain membrane fraction or cell lysates for mouse assays; human prefrontal cortex or cell membrane fractions for human assays) (50 μL, 1.0 mg/mL total protein concentration) were preincubated with varying concentrations of inhibitors at 37° C. After 30 min, FP Rh or HT-01 (1.0 μL, 50 μM in DMSO) was added and the mixture was incubated for another 30 min at 37° C. Reactions were quenched with SDS loading buffer (15 μL-4×) and run on SDS-PAGE. Following gel imaging, serine hydrolase activity was determined by measuring fluorescent intensity of gel bands corresponding to MAGL and FAAH using ImageJ 1.43u software. In Vitro Competitive Activity-Based Protein Profiling Assay Proteomes (mouse brain membrane fraction or cell lysates for mouse assays; human prefrontal cortex or cell membrane fractions for human assays) (50 μL, 1.0 mg/mL total protein concentration) were preincubated with varying concentrations of inhibitors at 37° C. After 30 min, FP Rh or HT-01 (1.0 μL, 50 μM in DMSO) was added and the mixture was incubated for another 30 min at 37° C. Reactions were quenched with SDS loading buffer (15 μL 4×) and run on SDS-PAGE. Following gel imaging, serine hydrolase activity was determined by measuring fluorescent intensity of gel bands corresponding to MAGL using ImageJ 1.43u software. Mode of action assay-Automated electrophysiology assay of compounds that potentiate KCNQ2 potassium channel BioAssay Type: Confirmatory, Concentration-Response Relationship Observed Source (MLPCN Center Name): Johns Hopkins Ion Channel Center (JHICC) Center Affiliation: Johns Hopkins University, School of Medicine Screening Center PI: Min Li, Ph.D. Assay Provider: Min Li, Ph.D. Network: Molecular Libraries Probe Production Centers Network (MLPCN) Grant Proposal Number: 1 R03 DA027716-01 Grant Proposal PI: Min Li, Ph.D., Johns Hopkins University School of Medicine Assay Implementation: Haibo Yu Ph.D, Bill Shi Ph.D., David Meyers Ph.D., Jia Xu Ph.D. Name: Mode of action assay-Automated electrophysiology assay of compounds that potentiate KCNQ2 potassium channel Description: See the related assay (PubChem AID:2239). Dose response of Retigabine-insensitive compounds that potentiate KCNQ2 potassium channel Name: Dose response of Retigabine-insensitive compounds that potentiate KCNQ2 potassium channel BioAssay Type: Confirmatory, Concentration-Response Relationship Observed Source (MLPCN Center Name): Johns Hopkins Ion Channel Center (JHICC) Center Affiliation: Johns Hopkins University, School of Medicine Screening Center PI: Min Li, Ph.D. Assay Provider: Min Li, Ph.D. Network: Molecular Libraries Probe Production Centers Network (MLPCN) Grant Proposal Number: 1 R03 DA027716-01 Grant Proposal PI: Min Li, Ph.D., Johns Hopkins University School of Medicine Assay Implementation: Haibo Yu Ph.D, Bill Shi Ph.D., David Meyers Ph.D., Jia Xu Ph.D. Name: Dose response of Retigabine-insensitive compounds that potentiate KCNQ2 potassium channel Description: See the related assay (PubChem AID:2239). SAR Analysis for the identification of Selective Antagonists of ERK1/2 Activity in GPR35-Overexpressing U2OS Cells Data Source: Dr. Mary Abood Source Affiliation: Temple University Network: NIH Molecular Libraries Probe Production Centers Network (MLPCN) Grant Proposal Number: 1X01MH085708-01 Assay Provider: Dr. Lawrence Barak, Duke University The aim of this assay was to characterize downstream ERK phosphorylation activity of compounds originally identified in "Image-based HTS for Selective Antagonists of GPR35" (AID 2058). Componds were either acquired from commercial sources or synthesized by the Sanford-Burnham Center for Chemical Genomics. This In-Cell Western assay utilizes a cell line permanently expressing a beta-arrestin GFP biosensor and human GPR35 receptor. Upon agonist-mediated GPCR activation, ERK1/2 phosphorylation occurs as measured by pERK1/2 antibodies. SIK2 Kinase Activity Assay IC50's for selected compounds in Table 5 below were measured by Caliper-based mobility shift assay (PerkinElmer). For these experiments, full length His6-MBP-tagged hSIK2 (4 nM) was incubated with HG-9-91-01 derivatives in buffer 5 containing 100 mM HEPES 7.5, 10 mM MgC2, 2.5 mM DTT, 0.004% Tween20, 0.003% Brij-35, 30 μM ATP and 1.5 μM ProfilerPro FL-Peptide 10 (5-FAMKKKVSRSGLYRSPSMPENLNRPR-COOH, PerkinElmer, Catalog No. 760354) at rt. Reactions were quenched by adding 20 mM EDTA (pH 8) after 1 hr, and percentage of substrate conversion was measured by LabChip EZ Reader II (PerkinElmer). IC50's for SIK2 inhibition were calculated using SmartFit nonlinear regression in Genedata Screener software suite (Genedata). Biological Assays for Inhibition of Rho-Associated Protein Kinase Assays for ROCK inhibition were performed using the following protein constructs: glutathione S-transferase (GST)-tagged human ROCK1 catalytic domain 1-477 from Carna Biosciences (cat #01-109; apparent Km value for ATP is 10 μM) and GST-tagged human ROCK2 catalytic domain 1-553 from Carna Biosciences (Cat#01-110; apparent Km value for ATP is 15 μM). Protein constructs were purified from a baculovirus expression system. The peptide substrate was fluorescent LANCE Ultra ULight-CREBtide: CKRREILSRRPSYRK (PerkinElmer, # TRF0107-D). Kinase reactions were carried out in in a 10 μL volume in 384-well plates: 50 nM ULight-CREBtide substrate, 2 nM constitutively active ROCK1 or ROCK2 kinase, and test compound in DMSO (or DMSO only for controls) were diluted into assay buffer containing 50 mM Tris-HCl (pH=7.5), 10 mM MgCl2, 1 mM EGTA, 0.01% Tween-20, and 2 mM DTT such that the final concentration of DMSO was 0.5%. After a 90 minute incubation at room temperature on a shaker table, the kinase reaction was stopped by addition of 10 mM EDTA, and phosphorylation of the substrate was detected by adding 1 nM LANCE Ultra Europium-anti-phospho-CREB (ser133) antibody (PerkinElmer, # TRF0200-D) and incubating for 60 minutes on a shaker at room temperature. The flurorescence resonance energy transfer (FRET) signals were read and analyzed on an Envision 2103 Multilabel Reader (Perkin Elmer). The concentration of test compound required to inhibit substrate phosphorylation by 50% (the IC50) was calculated by non-linear regression using GraphPad PRIZM. Confirmation Cuvette-Based Assay for Inhibitors of AmpC Beta-Lactamase (assay with detergent) NIH Molecular Libraries Screening Centers Network [MLSCN] NIH Chemical Genomics Center [NCGC] MLSCN Grant: MH079825-01 Assay Provider: Shoichet, Brian K. This aggregation profiling approach exploits the sensitivity of aggregate formation to detergent. Inhibition of b-lactamase is measured in the presence and absence of 0.01% Triton X-100 (Feng 2007). This particular assay is a confirmation of previous qHTS (Inglese, 2006), Pubchem AID 584, assay with presence of 0.01% Triton X-100. For a related assay without detergent, see AID 585. Compounds that inhibit only in the absence of detergent are considered likely promiscuous aggregators. This confirmation assay is a cuvette-based screen of an oxadiazole carbamate series which is an expansion around docking hit Pubchem SID 4244870. Counterscreen for NR2E3 inverse agonists Source (MLPCN Center Name): The Scripps Research Institute Molecular Screening Center Affiliation: The Scripps Research Institute, TSRI Assay Provider: Konstantin Petrukhin, Columbia University Network: Molecular Library Probe Production Centers Network (MLPCN) Grant Proposal Number: 1 R21 NS061718-01 Fast Track Grant Proposal PI: Konstantin Petrukhin, Columbia University External Assay ID: PPARG-NCOR2_IAG_HTRF_1536_3XIC50 DCSRUN Name: Counterscreen for NR2E3 inverse agonists: TR-FRET-based biochemical high throughput dose response assay to identify inverse agonists of the interaction between peroxisome proliferator-activated receptor gamma (PPARg) and nuclear receptor co-repressor 2 (NCOR2). Description: Nuclear receptors are small molecule- and hormone-regulated transcription factors with discrete DNA-binding and ligand-binding domains, and are essential during development and for maintenance of proper cell function in adults. Small pharmacological compounds that bind to the cle In Vitro Competitive Activity-Based Protein Profiling Proteomes (mouse brain membrane fraction or cell lysates for mouse assays; human prefrontal cortex or PC3 cell membrane fractions for human assays) (50 μL, 1.0 or 2.0 mg/mL total protein concentration) were preincubated with varying concentrations of inhibitors at 37° C. After 30 min, FP-Rh or JW912 or HT-01 (1.0 μL, 50 μM in DMSO) was added and the mixture was incubated for another 30 min at 37° C. Reactions were quenched with SDS loading buffer (15 μL-4×) and run on SDS-PAGE. Following gel imaging, serine hydrolase activity was determined by measuring fluorescent intensity of gel bands corresponding to MAGL and FAAH using ImageJ 1.43u software. IC50 data from this assay is shown in Table 1. PIM2 Activity Assay Kinase Buffer (KB) The buffer for PIM2 assay was composed of HEPES 50 mM, at pH 7.5, with 1 mM MgCl2, 1 mM DTT, 3 uM Na3VO4, and 0.2 mg/mL BSA.Full-length human PIM2 was expressed and purified as described in Fedorov O, et al., PNAS 2007 104, 51, 20523-28.ii. Assay Conditions (Final Concentrations)Enzyme concentration=1.5 nMAktide substrate (Chemical Abstract Service Registry Number 324029-01-8)=5 uMATP=4 uM 33P-gamma-ATP=1 nMiii. Robotized Dowex AssayThe test mix consisted of: 1) 3x Enzyme mix (done in Kinase Buffer 3x), 5 uL/well 2) 3x substrate and ATP mix (done in ddH2O), together with 33P-gamma-ATP, 5 uL/well 3) 3x test compounds (diluted into ddH2O-3% DMSO)-5 uL/well. SAR Analysis for the identification of Selective Antagonists of ERK1/2 Activity in GPR35-Overexpressing U2OS Cells - Set 2 Data Source: Dr. Mary Abood Source Affiliation: Temple University Network: NIH Molecular Libraries Probe Production Centers Network (MLPCN) Grant Proposal Number: 1X01MH085708-01 Assay Provider: Dr. Lawrence Barak, Duke University The aim of this assay was to characterize downstream ERK phosphorylation activity of compounds originally identified in "Image-based HTS for Selective Antagonists of GPR35" (AID 2058). Componds were either acquired from commercial sources or synthesized by the Sanford-Burnham Center for Chemical Genomics. This In-Cell Western assay utilizes a cell line permanently expressing a beta-arrestin GFP biosensor and human GPR35 receptor. Upon agonist-mediated GPCR activation, ERK1/2 phosphorylation occurs as measured by pERK1/2 antibodies. Absorbance-based biochemical high throughput dose response assay for activators of procaspase-3 Source (MLPCN Center Name): The Scripps Research Institute Molecular Screening Center (SRIMSC) Affiliation: The Scripps Research Institute, TSRI Assay Provider: Paul Hergenrother, University of Illinois at Urbana-Champaign Network: Molecular Library Probe Production Centers Network (MLPCN) Grant Proposal Number: R01 CA120439-01 Grant Proposal PI: Paul Hergenrother, University of Illinois at Urbana-Champaign External Assay ID: PROCASPASE3_ACT_EPIABS_1536_3XEC50 DRUN Name: Absorbance-based biochemical high throughput dose response assay for activators of procaspase-3. Description: Cancer progression depends upon evasion of the programmed cell death (apoptosis) machinery that normally kills an unneeded or rogue cell (1). Although apoptosis induction using chemotherapeutics is a common anti-cancer treatment, cancer cells often survive because of defects in the pro-apoptotic proteins activated by these drugs (2). The main effector proteins involved in the apopto Confirmation dose response assay for compounds that inhibit/block inward-rectifying potassium ion channel Kir2.1 Data Source: Johns Hopkins Ion Channel Center (JHICC) BioAssay Type: Confirmatory, Confirmatory Screening, Multiple Concentration Activity Observed. Source (MLPCN Center Name): Johns Hopkins Ion Channel Center (JHICC) Center Affiliation: Johns Hopkins University, School of Medicine Screening Center PI: Min Li, Ph.D. Assay Provider: Elena Makhina Ph.D., University of Pittsburgh Network: Molecular Libraries Probe Production Centers Network (MLPCN) Grant Proposal Number: 1 R03 DA026212-01 Grant Proposal PI: Elena Makhina Ph.D., University of Pittsburgh Assay Implementation: Meng Wu Ph.D., Shunyou Long M.S., Haibo Yu Ph.D., Hao-ran Wang Ph.D., Bill Shi Ph.D., David Meyers Ph.D., and Jia Xu Ph.D. Name: Confirmation dose response assay for compounds that inhibit/block inward-rectifying potassium ion channel Kir2.1. Description: See the related essay (PubChem AID: 1672). Dose Response Assay for S1P3 Antagonists Source (MLSCN Center Name): The Scripps Research Institute Molecular Screening Center Center Affiliation: The Scripps Research Institute, TSRI Assay Provider: The Scripps Research Institute, TSRI Network: Molecular Library Screening Center Network (MLSCN) Grant Proposal Number: 1 R03 MH076533-01 The biology of S1P receptor subtypes: Sphingosine 1-phosphate (S1P) influences heart rate [1] [2], coronary artery caliber, endothelial integrity, lung epithelial integrity [3] and lymphocyte recirculation [1] [4]-[6] through five related high affinity G-protein coupled receptors [7]. Inhibition of lymphocyte recirculation by nonselective S1P receptor agonists produces clinical immunosuppression preventing transplant rejection, but is associated with transient bradycardia. Understanding the contribution of individual receptors has been limited by the unavailability of selective agonists or antagonists for the 5 receptor subtypes. Separation of receptor subtype usage for control of endothelial Dose Response Assays for S1P1 Agonists and Agonism Potentiators - Potentiator Assay 60K MLSMR Source (MLSCN Center Name): The Scripps Research Institute Molecular Screening Center Center Affiliation: The Scripps Research Institute, TSRI Assay Provider: The Scripps Research Institute, TSRI Network: Molecular Library Screening Center Network (MLSCN) Grant Proposal Number: 1 R03 MH076533-01 The biology of S1P receptor subtypes Sphingosine 1-phosphate (S1P) influences heart rate (1,2), coronary artery caliber, endothelial integrity, lung epithelial integrity (3) and lymphocyte recirculation (1,4-6) through five related high affinity G-protein coupled receptors (7). Inhibition of lymphocyte recirculation by nonselective S1P receptor agonists produce clinical immunosuppression preventing transplant rejection, but is associated with transient bradycardia. Understanding the contribution of individual receptors has been limited by the unavailability of selective agonists or antagonists for the 5 receptor subtypes. Separation of receptor subtype usage for control of endothelial and Dose response cell-based assay to measure STAT1 inhibition Source (MLPCN Center Name): The Scripps Research Institute Molecular Screening Center Center Affiliation: The Scripps Research Institute (TSRI) Assay Provider: David Frank, Dana Farber Cancer Institute Network: Molecular Libraries Probe Production Centers Network (MLPCN) Grant Proposal Number: 1 X01 MH079826-01 Grant Proposal PI: David Frank, Dana Farber Cancer Institute External Assay ID: STAT1_INH_LUMI_1536_IC50 Name: Dose response cell-based assay to measure STAT1 inhibition Description: Members of the signal transducer and activator of transcription (STAT) family of transcription factors mediate inflammation, cell survival, differentiation, and proliferation (1, 2). In response to stimuli such as growth factors and cytokines (1-3), cytosolic STATs are activated by phosphorylation by the Janus-activated kinases (Jaks), inducing STAT dimerization, nuclear translocation, and binding to specific enhancer elements in target genes (2). Although structurally similar, STAT prote Dose response cell-based assay to measure STAT3 activation Source (MLPCN Center Name): The Scripps Research Institute Molecular Screening Center Center Affiliation: The Scripps Research Institute (TSRI) Assay Provider: David Frank, Dana Farber Cancer Institute Network: Molecular Libraries Probe Production Centers Network (MLPCN) Grant Proposal Number: 1 X01 MH079826-01 Grant Proposal PI: David Frank, Dana Farber Cancer Institute External Assay ID: STAT3_ACT_LUMI_1536_EC50 Name: Dose response cell-based assay to measure STAT3 activation Description: Members of the signal transducer and activator of transcription (STAT) family of transcription factors mediate inflammation, cell survival, differentiation, and proliferation (1, 2). In response to stimuli such as growth factors and cytokines (1-3), cytosolic STATs are activated by phosphorylation by the Janus-activated kinases (Jaks), inducing STAT dimerization, nuclear translocation, and binding to specific enhancer elements in target genes (2). Although structurally similar, STAT protei Dose response cell-based assay to measure STAT3 inhibition Source (MLPCN Center Name): The Scripps Research Institute Molecular Screening Center Center Affiliation: The Scripps Research Institute (TSRI) Assay Provider: David Frank, Dana Farber Cancer Institute Network: Molecular Libraries Probe Production Centers Network (MLPCN) Grant Proposal Number: 1 X01 MH079826-01 Grant Proposal PI: David Frank, Dana Farber Cancer Institute External Assay ID: STAT3_INH_LUMI_1536_IC50 Name: Dose response cell-based assay to measure STAT3 inhibition Description: Members of the signal transducer and activator of transcription (STAT) family of transcription factors mediate inflammation, cell survival, differentiation, and proliferation (1, 2). In response to stimuli such as growth factors and cytokines (1-3), cytosolic STATs are activated by phosphorylation by the Janus-activated kinases (Jaks), inducing STAT dimerization, nuclear translocation, and binding to specific enhancer elements in target genes (2). Although structurally similar, STAT pro JHICC_CHT_Inh_3H uptake_CRC Data Source (MLPCN Center Name): Johns Hopkins Ion Channel Center (JHICC) Center Affiliation: Johns Hopkins University, School of Medicine Screening Center PI: Min Li, Ph.D. Assay Provider: Alicia Ruggiero, Ph.D., Vanderbilt University Medical Center Network: Molecular Libraries Probe Production Centers Network (MLPCN) Grant Proposal Number: 1R03DA028852-01 Grant Proposal PI: Alicia Ruggiero, Ph.D., Vanderbilt University Medical Center Assay Implementation: Meng Wu Ph.D., Xiaofang Huang, M.S., Zhihong Lin, Ph. D., Kaiping Xu, M.S., Shunyou Long, M.S., and Owen McManus, Ph.D. Description: In the brain, the chemical acetylcholine (ACh) exerts powerful modulatory control over arousal, motor and cognitive circuits, and has been found to be deficient in Alzheimer's Disease (AD). The current drugs available to positively impact cognitive deficits in Alzheimer's Disease (AD) and other dementias are the cholinesterase inhibitors. These prevent the breakdown of the neurotransmitter Mode of action - current amplitude concentration response for ztz240, a potentiator of KCNQ2 potassium channels Data Source: Johns Hopkins Ion Channel Center BioAssay Type: Mode of Action, Electrophysiology Source (MLPCN Center Name): Johns Hopkins Ion Channel Center (JHICC) Center Affiliation: Johns Hopkins University, School of Medicine Screening Center PI: Min Li, Ph.D. Assay Provider: Min Li, Ph.D. Network: Molecular Libraries Probe Production Centers Network (MLPCN) Grant Proposal Number: 1 R03 DA027716-01 Grant Proposal PI: Min Li, Ph.D., Johns Hopkins University School of Medicine Assay Implementation: Zhaobing Gao Ph.D., Wei Wang Ph.D., Haibo Yu Ph.D., Meng Wu Ph.D., Shunyou Long M.S., Amy Scott, Beiyan Zou Ph.D., Joseph Babcock, Bill Shi Ph.D., David Meyers Ph.D., Jia Xu Ph.D. Name: Mode of action - current amplitude CRC assay for ztz240, a compound that potentiates KCNQ2 potassium channels Description: See the related primary assay (PubChem AID: 2239). Mode of action - deactivation constant concentration response for ztz240, a potentiator of KCNQ2 potassium channels Data Source: Johns Hopkins Ion Channel Center BioAssay Type: Mode of Action, Electrophysiology Source (MLPCN Center Name): Johns Hopkins Ion Channel Center (JHICC) Center Affiliation: Johns Hopkins University, School of Medicine Screening Center PI: Min Li, Ph.D. Assay Provider: Min Li, Ph.D. Network: Molecular Libraries Probe Production Centers Network (MLPCN) Grant Proposal Number: 1 R03 DA027716-01 Grant Proposal PI: Min Li, Ph.D., Johns Hopkins University School of Medicine Assay Implementation: Zhaobing Gao Ph.D., Wei Wang Ph.D., Haibo Yu Ph.D., Meng Wu Ph.D., Shunyou Long M.S., Amy Scott, Beiyan Zou Ph.D., Joseph Babcock, Bill Shi Ph.D., David Meyers Ph.D., Jia Xu Ph.D. Name: Mode of action - deactivation constant CRC assay for ztz240, a compound that potentiates KCNQ2 potassium channels Description: See the related primary assay (PubChem AID: 2239). S1P3 Agonist Dose-Response Potency Assay Source (MLSCN Center Name): The Scripps Research Institute Molecular Screening Center Center Affiliation: The Scripps Research Institute, TSRI Network: Molecular Library Screening Center Network (MLSCN) Proposal number 1 R03 MH076533-01 External Assay ID: (4.3) S1P3_AG_BLA_1536_EC50 Drun1 Name: S1P3 Agonist Dose-Response Potency Assay The biology of S1P receptor subtypes: Sphingosine 1-phosphate (S1P) influences heart rate [1] [2], coronary artery caliber, endothelial integrity, lung epithelial integrity [3] and lymphocyte recirculation [1] [4]-[6] through five related high affinity G-protein coupled receptors [7]. Inhibition of lymphocyte recirculation by nonselective S1P receptor agonists produces clinical immunosuppression preventing transplant rejection, but is associated with transient bradycardia. Understanding the contribution of individual receptors has been limited by the unavailability of selective agonists or antagonists for the 5 receptor subtypes. Separation of recept sEH_DR_Inh_Infinite200_Fluorescence_01072008 Data Source: Columbia University Molecular Screening Center Source (MLSCN Center Name): Columbia University Molecular Screening Center Center Affiliation: Columbia University Molecular Screening Center Assay Provider: Dr. Bruce D. Hammock, UC, Davis, CA. Network: Molecular Library Screening Center Network (MLSCN) Grant Proposal Number: X01 MH078954-01 Hypertension and vascular inflammation are associated with cardiovascular diseases, the primary cause of death in our society. Because a large proportion of patients are not responding to current therapies, the next generation of drugs will not only need to reduce blood pressure but also treat vascular and renal inflammation as well as reduce smooth muscle cell proliferation, which in turn should also reduce hypertension related organ damage. Using inhibitors developed in the Hammock laboratory, it was shown that the inhibition of soluble epoxide hydrolase (sEH) has therapeutic application in the treatment of hypertension and several in CHOP dose-response primary assay NIH Molecular Libraries Screening Centers Network [MLSCN] Emory Chemical Biology Discovery Center in MLSCN Assay provider: Dr. Randal Kaufman, University of Michigan MLSCN Grant: R03MH084182-01, U54HG003918 Assay Overview Many genetic and environmental diseases result from defective protein folding within the secretory pathway so that aberrantly folded proteins are recognized by the cellular surveillance system and retained within the endoplasmic reticulum (ER). Under conditions of malfolded protein accumulation, the cell activates the Unfolded Protein Response (UPR) to clear the malfolded proteins, and if unsuccessful, initiates a cell death response. Preliminary studies have shown that CHOP is a crucial factor in the apoptotic arm of the UPR; XBP1 activates genes encoding ER protein chaperones and thereby mediates the adaptive UPR response to increase clearance of malfolded proteins. Inhibition of CHOP is hypothesized to enhance survival by preventing UPR programmed cell death. Cathepsin B dose-response confirmation Screening Center: Penn Center for Molecular Discovery Center Affiliation: University of Pennsylvania Network: Molecular Library Screening Center Network (MLSCN) Assay Provider: Scott Diamond, University of Pennsylvania Grant number: MH076406-01 Human liver cathepsin B (EC 3.4.22.1) is a lysosomal cysteine protease. There has been a recent resurgence of interest in cathepsin B due to research showing that proteolysis by this enzyme is required for the entry and replication of the Ebola and SARS viruses in human cells. Thus cathepsin B inhibitors have potential as novel anti-viral agents. Cathepsin B is also implicated in cancer progression. Upregulation and secretion of this enzyme occurs in many types of tumors and correlates positively with their invasive and metastatic capabilities. Cathepsin B facilitates tumor invasion by dissolving extracellular barriers. Inhibitors of cathepsin B thus have been studied as potential anti-cancer agents. A high-throughput screen for cathepsin B Cathepsin G dose-response confirmation Screening Center: Penn Center for Molecular Discovery Center Affiliation: University of Pennsylvania Network: Molecular Library Screening Center Network (MLSCN) Assay Provider: Scott Diamond, University of Pennsylvania Grant number: MH076406-01 Cathepsin G (EC 3.4.21.20) is a chymotrypsin-like serine protease that is secreted from neutrophils. Disregulated cathepsin G activity is implicated in the progression of various chronic inflammatory diseases such as asthma and chronic pulmonary obstructive disease. Thus cathepsin G inhibitors represent useful probes to further elucidate the role of this enzyme in inflammation and may provide a starting point for the development of novel therapeutic agents. A high-throughput screen for cathepsin G inhibitors was designed as an end-point assay monitoring the release of the fluorophore aminomethyl coumarin (AMC) upon enzymatic hydrolysis of an AMC-labeled dipeptide. Primary HTS results have been reported previously (AID 581). Compounds identifi Cathepsin L dose-response confirmation Screening Center: Penn Center for Molecular Discovery Center Affiliation: University of Pennsylvania Network: Molecular Library Screening Center Network (MLSCN) Assay Provider: Scott Diamond, University of Pennsylvania Grant number: MH076406-01 Human liver cathepsin L (EC 3.4.22.15) is a lysosomal cysteine protease. Recent interest in cathepsin L has been generated by research showing that proteolysis by this enzyme is required for the entry and replication of the SARS and Ebola viruses in human cells. Thus cathepsin L inhibitors have potential as novel anti-viral agents. Cathepsin L inhibitors may also be active against Plasmodium falciparum, the parasite responsible for human malaria. Plasmodium contains cathepsin L-like cysteine proteases known as falcipains that appear to promote virulence of the parasite through haemoglobin digestion and erythrocyte invasion. A high-throughput screen for cathepsin L inhibitors was designed as an end-point assay monitoring the release of the fl Cathepsin S dose-response confirmation Screening Center: Penn Center for Molecular Discovery Center Affiliation: University of Pennsylvania Network: Molecular Library Screening Center Network (MLSCN) Assay Provider: Scott Diamond, University of Pennsylvania Grant number: MH076406-01 Human cathepsin S (EC 3.4.22.27) is a lysosomal cysteine protease that is expressed in antigen-presenting cells, especially dendritic cells, B-cells and macrophages. Cathepsin S plays a key role in the processing of antigenic peptides for presentation by MHC Class II molecules on the surface of antigen-presenting cells. Thus inhibitors of cathepsin S may be immunomodulators effective in the treatment of autoimmune diseases. A high-throughput screen for cathepsin S inhibitors was designed as an end-point assay monitoring the release of the fluorophore aminomethyl coumarin (AMC) upon enzymatic hydrolysis of an AMC-labeled dipeptide. Primary HTS results have been reported previously (AID 501). Compounds identified as hits in HTS of mixtures of 1 Chemical Antagonists IAP-family anti-apoptotic proteins Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Screening Centers Network (MLSCN) Grant Proposal Number: MH081277-01 Assay Provider: John C. Reed, Sanford-Burnham Medical Research Institute, San Diego, CA Apoptosis plays an essential role in many aspects of normal development and physiology, becoming dysregulated in myriad diseases characterized by insufficient or excessive cell death. Caspases are intracellular proteases that are suppressed by Inhibitor of Apoptosis Proteins (IAPs), a family of evolutionarily conserved anti-apoptotic proteins. Proteins released from mitochondria (SMAC and HtrA2) can competitively displace IAPs from the Caspases, thus helping to drive apoptosis. It has been shown that only a few residues at the N-terminus of activated SMAC protein (4-mer) are sufficient to affect the release of IAPs from Caspases. Thus, it is plausib Chemical Antagonists of IAP-family anti-apoptotic proteins confirmation Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Screening Centers Network (MLSCN) Grant Proposal Number: MH081277-01 Assay Provider: John C. Reed, Sanford-Burnham Medical Research Institute, San Diego, CA This XIAP dose response assay is developed and performed to confirm hits originally identified in the XIAP HTS binding assay (AID 1018) and to study the structure-activity relationship on analogs of the confirmed hits. Compounds are either acquired from commercial sources or synthesized internally. Apoptosis plays an essential role in many aspects of normal development and physiology, becoming dysregulated in myriad diseases characterized by insufficient or excessive cell death. Caspases are intracellular proteases that are suppressed by Inhibitor of Apoptosis Proteins (IAPs), a family of evolutionarily conserved anti-apoptotic proteins. Proteins Counterscreen for S1P2 Antagonists: Dose Response Cell-Based Screen to Identify Antagonists of CRE-BLA Source (MLSCN Center Name): The Scripps Research Institute Molecular Screening Center Center Affiliation: The Scripps Research Institute (TSRI) Assay Provider: Hugh Rosen, TSRI Network: Molecular Library Screening Center Network (MLSCN) Grant Proposal Number: 1 R03 MH076533-01 Grant Proposal PI: Germana Sanna External Assay ID: CRE _ANT_BLA_384_IC50_Counterscreen_ S1P2_Hits Name: Counterscreen for S1P2 Antagonists: Dose Response Cell-Based Screen to Identify Antagonists of CRE-BLA Description: Sphingosine 1-phosphate (S1P) influences heart rate [1,2], coronary artery caliber, endothelial integrity, lung epithelial integrity [3] and lymphocyte recirculation [2,4] through five related high affinity G-protein coupled receptors [5]. Subtype-selective modulators of S1P receptors will be of broad utility in understanding cell functions in vitro and vascular physiology in vivo, as well as de-convoluting the role of individual subtypes in cellular processes. The S1P receptor 2 (S1P2) Counterscreen for agonists of nuclear receptor subfamily 2, group E, member 3 (NR2E3) Source (MLPCN Center Name): The Scripps Research Institute Molecular Screening Center Affiliation: The Scripps Research Institute, TSRI Assay Provider: Konstantin Petrukhin, Columbia University Network: Molecular Library Probe Production Centers Network (MLPCN) Grant Proposal Number: 1 R21 NS061718-01 Fast Track Grant Proposal PI: Konstantin Petrukhin, Columbia University External Assay ID: PPARG-NCOR2_AG_TR-FRET_1536_3XIC50 CSDRUN Name: Counterscreen for agonists of nuclear receptor subfamily 2, group E, member 3 (NR2E3): TR-FRET-based biochemical high throughput dose response assay to identify agonists of the interaction between peroxisome proliferator-activated receptor gamma (PPARg) and nuclear receptor co-repressor 2 (NCOR2). Description: Nuclear receptors are small molecule- and hormone-regulated transcription factors with discrete DNA-binding and ligand-binding domains, and are essential during development and for maintenance of proper cell function in adults. Small pharmaco Counterscreen for biased ligands (agonists) of the melanocortin 4 receptor (MC4R) Source (MLPCN Center Name): The Scripps Research Institute Molecular Screening Center (SRISMC) Center Affiliation: The Scripps Research Institute, TSRI Assay Provider: Scott DeWire, Trevena Inc Network: Molecular Library Probe Production Center Network (MLPCN) Grant Proposal Number: 1 RC2 MH090877-01 Grant Proposal PI: Scott DeWire, Trevena Inc External Assay ID: OPRM1-OPRD1_AG_LUMI_1536_3XEC50 DCSRUN Name: Counterscreen for biased ligands (agonists) of the melanocortin 4 receptor (MC4R): Luminescence-based cell-based high throughput dose response assay to identify agonists of heterodimerization of the mu 1 (OPRM1) and delta 1 (OPRD1) opioid receptors. Description: Heterotrimeric G-protein coupled receptors (GPCRs) are major targets for disease therapeutics, due in part to their broad tissue distribution, structural diversity, varied modes of action, and disease-associated mutations (1-4). However, it has recently been demonstrated that GPCRs do not only signal in this simplistic f Counterscreen for procaspase-3 activators: absorbance-based biochemical high throughput dose response assay for activators of procaspase-7 Source (MLPCN Center Name): The Scripps Research Institute Molecular Screening Center (SRIMSC) Affiliation: The Scripps Research Institute, TSRI Assay Provider: Paul Hergenrother, University of Illinois at Urbana-Champaign Network: Molecular Library Probe Production Centers Network (MLPCN) Grant Proposal Number: R01 CA120439-01 Grant Proposal PI: Paul Hergenrother, University of Illinois at Urbana-Champaign External Assay ID: PROCASPASE7_ACT_EPIABS_1536_3XEC50 DCSRUN Name: Counterscreen for procaspase-3 activators: absorbance-based biochemical high throughput dose response assay for activators of procaspase-7. Description: Cancer progression depends upon evasion of the programmed cell death (apoptosis) machinery that normally kills an unneeded or rogue cell (1). Although apoptosis induction using chemotherapeutics is a common anti-cancer treatment, cancer cells often survive because of defects in the pro-apoptotic proteins activated by these drugs (2). The Discovery of Novel Allosteric Modulators of the M1 Muscarinic Receptor: PAM Calcium Assay SAR Assay Provider: P. Jeffery Conn Assay Provider Affiliation: Vanderbilt University Grant Title: Discovery of novel allosteric modulators of the M1 Muscarinic receptor Grant Number: 1 R03 MH077606-01 Selective M1 activation is an attractive therapeutic approach for the treatment of cognitive impairment, Alzheimer's disease, schizophrenia and a number of other CNS disorders. Until recently, no highly selective M1 activators existed, and those that claimed to be highly M1 selective were either not centrally penetrant or possessed significant ancillary pharmacology which prohibited their use as probes to study M1 receptor function. We have identified that different M1 PAM chemotypes display different modes of activity on downstream receptor signaling. Thus, all allosteric M1 activation is not equivalent, and additional tool compounds representing diverse chemotypes are required to truly dissect and study M1 function in the CNS. Dosage response for compounds that protect hERG from block by proarrhythmic agents using manual patch clamp Data Source: Johns Hopkins Ion Channel Center BioAssay Type: Mode of action, Concentration-Response Relationship Observed Source (MLPCN Center Name): Johns Hopkins Ion Channel Center (JHICC) Center Affiliation: Johns Hopkins University, School of Medicine Screening Center PI: Min Li, Ph.D. Assay Provider: Dr. Sabina Kupershmidt, Vanderbilt University Medical Center Network: Molecular Libraries Probe Production Centers Network (MLPCN) Grant Proposal Number: 1R03MH084820-01 Grant Proposal PI: Dr. Sabina Kupershmidt, Vanderbilt University Medical Center Assay Implementation: Dr. Sabina Kupershmidt, Vanderbilt University Medical Center Name: Dosage response for compounds that protect hERG from block by proarrhythmic agents using manual patch clamp Description: See the related bioassay (AID: 1511). Principle of the assay Patch clamp is gold standard to measure channel activities. Upon depolarization of cells expressing potassium channels, in this case, human ether-a-go-go-related g Dose Response Assays for S1P1 Agonists and Agonism Potentiators - Agonist Assay 60K MLSMR Source (MLSCN Center Name): The Scripps Research Institute Molecular Screening Center Center Affiliation: The Scripps Research Institute, TSRI Assay Provider: The Scripps Research Institute, TSRI Network: Molecular Library Screening Center Network (MLSCN) Grant Proposal Number: 1 R03 MH076533-01 The biology of S1P receptor subtypes Sphingosine 1-phosphate (S1P) influences heart rate (1,2), coronary artery caliber, endothelial integrity, lung epithelial integrity (3) and lymphocyte recirculation (1,4-6) through five related high affinity G-protein coupled receptors (7). Inhibition of lymphocyte recirculation by nonselective S1P receptor agonists produce clinical immunosuppression preventing transplant rejection, but is associated with transient bradycardia. Understanding the contribution of individual receptors has been limited by the unavailability of selective agonists or antagonists for the 5 receptor subtypes. Separation of receptor subtype usage for control of endothelial and ep Dose Response Assays for S1P1 Agonists and Agonism Potentiators - Parental Cell Line Counter Screen 60K MLSMR Source (MLSCN Center Name): The Scripps Research Institute Molecular Screening Center Center Affiliation: The Scripps Research Institute, TSRI Assay Provider: The Scripps Research Institute, TSRI Network: Molecular Library Screening Center Network (MLSCN) Grant Proposal Number: 1 R03 MH076533-01 The biology of S1P receptor subtypes Sphingosine 1-phosphate (S1P) influences heart rate (1,2), coronary artery caliber, endothelial integrity, lung epithelial integrity (3) and lymphocyte recirculation (1,4-6) through five related high affinity G-protein coupled receptors (7). Inhibition of lymphocyte recirculation by nonselective S1P receptor agonists produce clinical immunosuppression preventing transplant rejection, but is associated with transient bradycardia. Understanding the contribution of individual receptors has been limited by the unavailability of selective agonists or antagonists for the 5 receptor subtypes. Separation of receptor subtype usage for control of endothelial and ep Dose Response Cell Based Assay for Antagonists of the S1P2 Receptor Source (MLSCN Center Name): The Scripps Research Institute Molecular Screening Center Center Affiliation: The Scripps Research Institute (TSRI) Assay Provider: Hugh Rosen, TSRI Network: Molecular Library Screening Center Network (MLSCN) Grant Proposal Number: 1 R03 MH076533-01 Grant Proposal PI: Germana Sanna External Assay ID: S1P2_ANT_BLA_384_IC50 Name: Dose Response Cell Based Assay for Antagonists of the S1P2 Receptor Description: Sphingosine 1-phosphate (S1P) influences heart rate [1,2], coronary artery caliber, endothelial integrity, lung epithelial integrity [3] and lymphocyte recirculation [2,4] through five related high affinity G-protein coupled receptors [5]. Subtype-selective modulators of S1P receptors will be of broad utility in understanding cell functions in vitro and vascular physiology in vivo, as well as de-convoluting the role of individual subtypes in cellular processes. The S1P receptor 2 (S1P2), also known as endothelial differentiation sphingolipid G-pro Dose Response Cell-based Assay for Inhibitors of Wee1 Degradation Source (MLPCN Center Name): The Scripps Research Institute Molecular Screening Center Center Affiliation: The Scripps Research Institute (TSRI) Assay Provider: Nagi Ayad, TSRI Network: Molecular Libraries Probe Production Centers Network (MLPCN) Grant Proposal Number: 1R21NS056991-01 Grant Proposal PI: Nagi Ayad, TSRI External Assay ID: Wee1Degradation_ACT_LUMI_1536_EC50 Name: Dose Response Cell-based Assay for Inhibitors of Wee1 Degradation Description: Cell cycle progression and entry into mitosis are regulated by a highly conserved cellular process known as checkpoint signaling (1-4). The Wee1 nuclear tyrosine kinase functions in this process by regulating the cdc2/cyclinB protein complex. Specifically, Wee1 mediates inhibitory phosphorylation of cdc2, leading to delayed mitosis and cell cycle arrest in cells with DNA damage so that DNA repair and replication can occur (1-4). Wee1 activity is inhibited during mitosis by its phosphorylation and ubiquitination by E3 ligases, and Dose Response cell-based high-throughput screening assay to identify antagonists of galanin receptor 2 (GALR2) Source (MLSCN Center Name): The Scripps Research Institute Molecular Screening Center Affiliation: The Scripps Florida Research Institute, TSRI Assay Provider: Steven Brown, TSRI Network: Molecular Library Screening Center Network (MLSCN) Grant Proposal Number: 1 R21 NS057101-01 Grant Proposal PI: Steven Brown External Assay ID: GALR2_ANT_BLA_1536_IC50 Name: Dose Response cell-based high-throughput screening assay to identify antagonists of galanin receptor 2 (GALR2) Description: Galanin, a 29 amino acid neuropeptide (30 residues in humans), is cleaved from preprogalanin and is involved in many physiological processes including nervous system development, feeding, metabolism and reproduction, and regulation of neurotransmitter and hormone release [1, 2]. The physiologic response to galanin is mediated in part by three G protein-coupled metabotropic 7-transmembrane receptor subtypes, GALR1, GALR2 and GALR3. These receptors are expressed throughout the peripheral and central nervous Dose Response confirmation of Image-Based HTS for Selective Agonists for NTR1 Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Probe Production Centers Network (MLPCN) Grant Proposal Number: 1 R03 MH089653-01 Assay Provider: Dr. Lawrence Barak, Duke University Medical Center Addiction leading to abuse should be treatable by pharmacological approaches, and programs that identify new drugs to treat methamphetamine abuse address an immediate goal of the National Institute on Drug Abuse (NIDA) that new approaches are needed for treating METH addiction. Currently, small molecule drug-like compounds are not available for treating METH abuse. Neurotensin receptor 1 (NTR1) peptide agonists produce behaviors that are exactly opposite to the psychostimulant effects observed with methamphetamine abuse, such as hyperactivity, neurotoxicity, psychotic episodes, and cognitive deficits, and repeated administrations of NTR1 agonists do not l Dose Response confirmation of compounds that inhibit HePTP Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Probe Production Centers Network (MLPCN) Grant Number: XO1 MH077603-01 Assay Provider: Dr. Tomas Mustelin, Sanford-Burnham Medical Research Institute Protein tyrosine phosphatases (PTPs), working with protein tyrosine kinases (PTKs), control the phosphorylation state of many proteins in the signal transduction pathways. HePTP is a tyrosine phosphatase expressed in hematopoietic cells and regulates the MAP kinases Erk and p38. It has been found that HePTP is often dysregualted in the preleukemic disorder myelodysplastic syndrome, as well as in acute myelogeneous leukemia. Small molecule inhibitors of HePTP will be useful as molecular probes for studying the mechanism of signal transduction and MAP kinase regulation, and may have therapeutic potential for the treatment of hematopoietic malignancies. Th Dose response biochemical high throughput screening assay for inhibitors of the p97 ATPase Source (MLPCN Center Name): The Scripps Research Institute Molecular Screening Center Center Affiliation: The Scripps Research Institute (TSRI) Assay Provider: Raymond Deshaies, California Institute of Technology Network: Molecular Libraries Probe Production Centers Network (MLPCN) Grant Proposal Number: 1 R03 MH085687-01 Grant Proposal PI: Raymond Deshaies, California Institute of Technology External Assay ID: p97_INH_Lumi_384_IC50 Name: Dose response biochemical high throughput screening assay for inhibitors of the p97 ATPase Description: Misfolded proteins accumulate in the endoplasmic reticulum (ER) in response to environmental stress (1). To reduce the burden these proteins place on the secretory pathway, eukaryotic cells have evolved a process, known as ER-associated degradation (ERAD), to recognize and eliminate these proteins (1, 2). The highly conserved p97 ATPase functions in ERAD by hydrolyzing ATP needed to export ubiquitinated substrates to the cytosol for degradation Dose response cell-based high throughput screening assay to identify enhancers of beta-glucosidase activity Source (MLSCN Center Name): The Scripps Research Institute Molecular Screening Center Center Affiliation: The Scripps Research Institute, TSRI Assay Provider: Jeffery W Kelly, TSRI Network: Molecular Library Screening Center Network (MLSCN) Grant Proposal Number: MH078940-01 Grant Proposal PI: Jeffery W Kelly External Assay ID: Betaglucosidase_ACT_Fluor_384_EC50 Name: Dose response cell-based high throughput screening assay to identify enhancers of beta-glucosidase activity Description: Gaucher disease (GD) is the most common lysosomal storage disorder caused by a loss of lysosomal glucocerebrosidase (GC) activity, leading to substrate accumulation. Glucosylceramide accumulation in monocyte-macrophage cells leads to hepatomegaly, splenomegaly, anemia, thrombocytopenia, bone lesions, and in severe cases, central nervous system (CNS) involvement [1]. Patients without CNS involvement are classified as type 1, while those with CNS involvement are classified as type 2 or type 3. N370S Dose response cell-based high-throughput screening assay for agonists of the transient receptor potential channel ML3 (TRPML3) Source (MLPCN Center Name): The Scripps Research Institute Molecular Screening Center (SRIMSC) Center Affiliation: The Scripps Research Institute (TSRI) Assay Provider: Stefan Heller, Stanford University Network: Molecular Libraries Probe Production Centers Network (MLPCN) Grant Proposal Number 1 R03 MH083077-01 Grant Proposal PI: Stefan Heller , Stanford University External Assay ID: TRPML3_AG_Calcium_1536_EC50 Name: Dose response cell-based high-throughput screening assay for agonists of the transient receptor potential channel ML3 (TRPML3) Description: Cell signaling pathways that mediate osmosensation, photosensation, and thermosensation depend on a family of diverse transient receptor potential (TRP) cation channels, which are activated by agonist-receptor coupling (1-5). A role for these channels in inner ear hair cell mechanotransduction was gleaned from TRP channel mutations identified in flies, worms, and lower vertebrates with defective balance and impaired sensitivity to Dose response cell-based high-throughput screening assay for agonists of the transient receptor potential channel N1 (TRPN1) Source (MLPCN Center Name): The Scripps Research Institute Molecular Screening Center (SRIMSC) Center Affiliation: The Scripps Research Institute (TSRI) Assay Provider: Stefan Heller, Stanford University Network: Molecular Libraries Probe Production Centers Network (MLPCN) Grant Proposal Number 1 R03 MH083077-01 Grant Proposal PI: Stefan Heller , Stanford University External Assay ID: TRPN1_AG_Calcium_1536_EC50 Name: Dose response cell-based high-throughput screening assay for agonists of the transient receptor potential channel N1 (TRPN1) Description: Cell signaling pathways that mediate osmosensation, photosensation, and thermosensation depend on a family of diverse transient receptor potential (TRP) cation channels, which are activated by agonist-receptor coupling (1-4). A role for these channels in inner ear hair cell mechanotransduction was gleaned from TRP channel mutations identified in flies, worms, and lower vertebrates with defective balance and impaired sensitivity to t Dose response cell-based screening assay for antagonists of neuropeptide Y receptor Y1 (NPY-Y1) Source (MLSCN Center Name): The Scripps Research Institute Molecular Screening Center Affiliation: The Scripps Research Institute, TSRI Assay Provider: Claes Wahlestedt, Scripps Florida Network: Molecular Library Screening Center Network (MLSCN) Grant Proposal Number 1 R21 NS056950-01 Grant Proposal PI: Claes Wahlestedt External Assay ID: NPY-Y1_ANT_CNGC_1536_IC50 Name: Dose response cell-based screening assay for antagonists of neuropeptide Y receptor Y1 (NPY-Y1) Description: Neuropeptide Y (NPY) is a neurotransmitter with physiologic roles including control of feeding behavior, regulation of cortical neural activity, heart neural activity, and emotional regulation. Importantly, NPY is implicated in human diseases such as obesity, depression and alcoholism. NPY mediates its biological effects in part through activation of the Galphai protein coupled receptors (GPCRs) NPY-Y1 and Y2 receptors, which decrease cytosolic cAMP production. Recent studies have implicated these recep Dose response confirmation of the uHTS fluorescent assay for identification of inhibitors of ATG4B Set 2 Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Probe Production Centers Network (MLPCN) Grant Number: 1 R03 MH090871-01 Assay Provider: Dr. John C. Reed, Sanford-Burnham Medical Research Institute, San Diego CA Autophagy is an evolutionarily conserved process whereby cells catabolize damaged proteins and organelles for purposes of generating substrates for sustaining ATP production during times of nutrient deprivation. The autophagic process involves membrane vesicles engulfing cytosol and organelles, delivering their contents to lysosomes for digestion. The genes responsible for autophagy have been identified, largely through genetic analysis of yeast, Saccharomyces cerevisiae, and are conserved in mammals, plants, and essentially all eukaryotes. While autophagy is critical for cell survival in the context of nutrient deprivation, circumstances Dose response confirmation of the uHTS fluorescent assay for identification of inhibitors of ATG4B. Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Probe Production Centers Network (MLPCN) Grant Number: 1 R03 MH090871-01 Assay Provider: Dr. John C. Reed, Sanford-Burnham Medical Research Institute, San Diego CA Autophagy is an evolutionarily conserved process whereby cells catabolize damaged proteins and organelles for purposes of generating substrates for sustaining ATP production during times of nutrient deprivation. The autophagic process involves membrane vesicles engulfing cytosol and organelles, delivering their contents to lysosomes for digestion. The genes responsible for autophagy have been identified, largely through genetic analysis of yeast, Saccharomyces cerevisiae, and are conserved in mammals, plants, and essentially all eukaryotes. While autophagy is critical for cell survival in the context of nutrient deprivation, circumstances Dose response counterscreen assay for STAT3 activators: cell-based high throughput assay to measure STAT1 activation Source (MLPCN Center Name): The Scripps Research Institute Molecular Screening Center Center Affiliation: The Scripps Research Institute (TSRI) Assay Provider: David Frank, Dana Farber Cancer Institute Network: Molecular Libraries Probe Production Centers Network (MLPCN) Grant Proposal Number: 1 X01 MH079826-01 Grant Proposal PI: David Frank, Dana Farber Cancer Institute External Assay ID: STAT1_ACT_LUMI_1536_EC50 (CSDRUN) Name: Dose response counterscreen assay for STAT3 activators: cell-based high throughput assay to measure STAT1 activation Description: Members of the signal transducer and activator of transcription (STAT) family of transcription factors mediate inflammation, cell survival, differentiation, and proliferation (1, 2). In response to stimuli such as growth factors and cytokines (1-3), cytosolic STATs are activated by phosphorylation by the Janus-activated kinases (Jaks), inducing STAT dimerization, nuclear translocation, and binding to specific enhancer element Dose response counterscreen assay for STAT3 inhibitors: cell-based high throughput assay to measure STAT1 inhibition Source (MLPCN Center Name): The Scripps Research Institute Molecular Screening Center Center Affiliation: The Scripps Research Institute (TSRI) Assay Provider: David Frank, Dana-Farber Cancer Institute Network: Molecular Libraries Probe Production Centers Network (MLPCN) Grant Proposal Number: 1 X01 MH079826-01 Grant Proposal PI: David Frank, Dana-Farber Cancer Institute External Assay ID: STAT1_INH_LUMI_1536_IC50 (CSDRUN) Name: Dose response counterscreen assay for STAT3 inhibitors: cell-based high throughput assay to measure STAT1 inhibition Description: Members of the signal transducer and activator of transcription (STAT) family of transcription factors mediate inflammation, cell survival, differentiation, and proliferation (1, 2). In response to stimuli such as growth factors and cytokines (1-3), cytosolic STATs are activated by phosphorylation by the Janus-activated kinases (Jaks), inducing STAT dimerization, nuclear translocation, and binding to specific enhancer element Dose response counterscreen for STAT1 activators: cell-based high throughput assay to measure STAT3 activation Source (MLPCN Center Name): The Scripps Research Institute Molecular Screening Center Center Affiliation: The Scripps Research Institute (TSRI) Assay Provider: David Frank, Dana Farber Cancer Institute Network: Molecular Libraries Probe Production Centers Network (MLPCN) Grant Proposal Number: 1 X01 MH079826-01 Grant Proposal PI: David Frank, Dana Farber Cancer Institute External Assay ID: STAT3_ACT_LUMI_1536_EC50 (CSDRUN) Name: Dose response counterscreen for STAT1 activators: cell-based high throughput assay to measure STAT3 activation Description: Members of the signal transducer and activator of transcription (STAT) family of transcription factors mediate inflammation, cell survival, differentiation, and proliferation (1, 2). In response to stimuli such as growth factors and cytokines (1-3), cytosolic STATs are activated by phosphorylation by the Janus-activated kinases (Jaks), inducing STAT dimerization, nuclear translocation, and binding to specific enhancer elements Dose response counterscreen for STAT1 inhibitors: cell-based high throughput assay to measure STAT3 inhibition Source (MLPCN Center Name): The Scripps Research Institute Molecular Screening Center Center Affiliation: The Scripps Research Institute (TSRI) Assay Provider: David Frank, Dana Farber Cancer Institute Network: Molecular Libraries Probe Production Centers Network (MLPCN) Grant Proposal Number: 1 X01 MH079826-01 Grant Proposal PI: David Frank, Dana Farber Cancer Institute External Assay ID: STAT3_INH_LUMI_1536_IC50 (CSDRUN) Name: Dose response counterscreen for STAT1 inhibitors: cell-based high throughput assay to measure STAT3 inhibition Description: Members of the signal transducer and activator of transcription (STAT) family of transcription factors mediate inflammation, cell survival, differentiation, and proliferation (1, 2). In response to stimuli such as growth factors and cytokines (1-3), cytosolic STATs are activated by phosphorylation by the Janus-activated kinases (Jaks), inducing STAT dimerization, nuclear translocation, and binding to specific enhancer elements Dose response for HTS for Beta-2AR agonists via FAP method from CP1 University of New Mexico Assay Overview: Assay Support: R03 MH093192-01 Project Title: HTS for Non-Canonical Ligands for Beta 2 Adrenergic Receptor Internalization Assay Provider: Jonathan Jarvik, Carnegie Mellon University Screening Center/ PI: UNMCMD/ Larry Sklar Lead Biologist: Yang Wu Chemistry Center/ PI: Vanderbilt Specialty Chemistry Center/Craig Lindsley Chemistry Center Lead: Shaun Stauffer Assay Implementation: Yang Wu, Phillip Tapia, Terry Foutz, Stephanie Chavez, Dominique Perez, Annette Evangelisti, Anna Waller, Cristian Bologa, Mark Carter Assay Background and Significance: G protein-coupled receptors represent the largest family of proteins in the human genome with an estimated number of approximately 800. Because of their central involvement in almost every aspect of human physiology, they also represent the largest target for medical intervention [Lin and Civelli, Annu Med 36 (2004), 204-14]. T Dose response for HTS for Beta-2AR agonists via FAP method from Powderset1 University of New Mexico Assay Overview: Assay Support: R03 MH093192-01 Project Title: HTS for Non-Canonical Ligands for Beta 2 Adrenergic Receptor Internalization Assay Provider: Jonathan Jarvik, Carnegie Mellon University Screening Center/ PI: UNMCMD/ Larry Sklar Lead Biologist: Yang Wu Chemistry Center/ PI: Vanderbilt Specialty Chemistry Center/Craig Lindsley Chemistry Center Lead: Shaun Stauffer Assay Implementation: Yang Wu, Phillip Tapia, Terry Foutz, Stephanie Chavez, Dominique Perez, Annette Evangelisti, Anna Waller, Cristian Bologa, Mark Carter Assay Background and Significance: G protein-coupled receptors represent the largest family of proteins in the human genome with an estimated number of approximately 800. Because of their central involvement in almost every aspect of human physiology, they also represent the largest target for medical intervention [Lin and Civelli, Annu Med 36 (2004), 204-14]. T Dose response for HTS for Beta-2AR agonists via FAP method from Powderset2 University of New Mexico Assay Overview: Assay Support: R03 MH093192-01 Project Title: HTS for Non-Canonical Ligands for Beta 2 Adrenergic Receptor Internalization Assay Provider: Jonathan Jarvik, Carnegie Mellon University Screening Center/ PI: UNMCMD/ Larry Sklar Lead Biologist: Yang Wu Chemistry Center/ PI: Vanderbilt Specialty Chemistry Center/Craig Lindsley Chemistry Center Lead: Shaun Stauffer Assay Implementation: Yang Wu, Phillip Tapia, Terry Foutz, Stephanie Chavez, Dominique Perez, Annette Evangelisti, Anna Waller, Cristian Bologa, Mark Carter Assay Background and Significance: G protein-coupled receptors represent the largest family of proteins in the human genome with an estimated number of approximately 800. Because of their central involvement in almost every aspect of human physiology, they also represent the largest target for medical intervention [Lin and Civelli, Annu Med 36 (2004), 204-14]. T Dose response for HTS for Beta-2AR agonists via FAP method from Powderset3 University of New Mexico Assay Overview: Assay Support: R03 MH093192-01 Project Title: HTS for Non-Canonical Ligands for Beta 2 Adrenergic Receptor Internalization Assay Provider: Jonathan Jarvik, Carnegie Mellon University Screening Center/ PI: UNMCMD/ Larry Sklar Lead Biologist: Yang Wu Chemistry Center/ PI: Vanderbilt Specialty Chemistry Center/Craig Lindsley Chemistry Center Lead: Shaun Stauffer Assay Implementation: Yang Wu, Phillip Tapia, Terry Foutz, Stephanie Chavez, Dominique Perez, Annette Evangelisti, Anna Waller, Cristian Bologa, Mark Carter Assay Background and Significance: G protein-coupled receptors represent the largest family of proteins in the human genome with an estimated number of approximately 800. Because of their central involvement in almost every aspect of human physiology, they also represent the largest target for medical intervention [Lin and Civelli, Annu Med 36 (2004), 204-14]. T Estrogen Receptor (alpha) binding: Dose Response of Primary Screen Assay University of New Mexico Assay Overview: Assay Support: 1X01 MH077627-01 Assay for Ligands of GPR30 and Classical Estrogen Receptors PI: Eric Prossnitz, PhD Assay Development: Megan Dennis Assay Implementation: Megan Dennis, Mark Haynes, PhD Target Team Leader for the Center: Eric Prossnitz, PhD (EProssnitz@salud.unm.edu) Assay Background and Significance: The physiological effects of estrogen are diverse and numerous, with roles in growth, development and homeostasis of numerous tissues. Roles for estrogen in mammalian female reproductive development are among the best defined, but estrogen also plays a part in regulation of skeletal cancer, (cardio)vascular function, the central nervous system as well as in the immune system. Stimulation with estrogen induces many signaling pathways, leading to an array of cellular responses including adhesion, migration, survival, proliferation and angiogenesis in both normal and neoplastic tissues [Edwards et al., 2005]. Effects of estrogen Estrogen Receptor (beta) binding: Dose Response of Primary Screen Assay University of New Mexico Assay Overview: Assay Support: 1X01 MH077627-01 Assay for Ligands of GPR30 and Classical Estrogen Receptors PI: Eric Prossnitz, PhD Assay Development: Megan Dennis Assay Implementation: Megan Dennis, Mark Haynes, PhD Target Team Leader for the Center: Eric Prossnitz, PhD (EProssnitz@salud.unm.edu) Assay Background and Significance: The physiological effects of estrogen are diverse and numerous, with roles in growth, development and homeostasis of numerous tissues. Roles for estrogen in mammalian female reproductive development are among the best defined, but estrogen also plays a part in regulation of skeletal cancer, (cardio)vascular function, the central nervous system as well as in the immune system. Stimulation with estrogen induces many signaling pathways, leading to an array of cellular responses including adhesion, migration, survival, proliferation and angiogenesis in both normal and neoplastic tissues [Edwards et al., 2005]. Effects of estrogen Estrogen Receptor-alpha Coactivator Binding Inhibitors ELISA Secondary Assay NIH Molecular Libraries Screening Centers Network [MLSCN] Emory Chemical Biology Discovery Center in MLSCN Assay provider: John A. Katzenellenbogen, University of Illinois at Urbana-Champaign MLSCN Grant: 1 X01MH78953-01 Assay Overview Estrogens, which are responsible for the growth of many breast cancers, act through the estrogen receptors, ER-alpha and ER-beta, which are ligand-modulated transcription factors and members of the nuclear receptor gene superfamily. ER-alpha and ER-beta are well validated protein targets for various aspects of women's health and breast cancer prevention and treatment. As an essential step in their action as regulators of gene transcription, the ERs recruit various coregulator proteins, both coactivators and corepressors, to the DNA-bound (or protein tethered) ER. These coregulators have various activities: alteration of chromatin architecture, regulation of nucleosome core histone modifications (acetylation, methylation, phosphorylation), and Estrogen Receptor-beta Coactivator Binding Inhibitors Dose Response Confirmation NIH Molecular Libraries Screening Centers Network [MLSCN] Emory Chemical Biology Discovery Center in MLSCN Assay provider: John A. Katzenellenbogen, University of Illinois Urbana-Champaign MLSCN Grant: 1 X01MH78953-01 Assay Overview Estrogens, which are responsible for the growth of many breast cancers, act through the estrogen receptors, ER-alpha and ER-beta, which are ligand-modulated transcription factors and members of the nuclear receptor gene superfamily. ER-alpha and ER-beta are well validated protein targets for various aspects of women's health and breast cancer prevention and treatment. As an essential step in their action as regulators of gene transcription, the ERs recruit various coregulator proteins, both coactivators and corepressors, to the DNA-bound (or protein tethered) ER. These coregulators have various activities: alteration of chromatin architecture, regulation of nucleosome core histone modifications (acetylation, methylation, phosphorylation), and ac Estrogen Receptor-beta Coactivator Binding Inhibitors ELISA Secondary Assay NIH Molecular Libraries Screening Centers Network [MLSCN] Emory Chemical Biology Discovery Center in MLSCN Assay provider: John A. Katzenellenbogen, University of Illinois Urbana-Champaign MLSCN Grant: 1 X01MH78953-01 Assay Overview Estrogens, which are responsible for the growth of many breast cancers, act through the estrogen receptors, ER-alpha and ER-beta, which are ligand-modulated transcription factors and members of the nuclear receptor gene superfamily. ER-alpha and ER-beta are well validated protein targets for various aspects of women's health and breast cancer prevention and treatment. As an essential step in their action as regulators of gene transcription, the ERs recruit various coregulator proteins, both coactivators and corepressors, to the DNA-bound (or protein tethered) ER. These coregulators have various activities: alteration of chromatin architecture, regulation of nucleosome core histone modifications (acetylation, methylation, phosphorylation), and ac Fluorescence-based biochemical high throughput dose response assay for activators of the calcium sensitivity of cardiac Regulated Thin Filaments (RTF) Source (MLPCN Center Name): The Scripps Research Institute Molecular Screening Center (SRIMSC) Affiliation: The Scripps Research Institute, TSRI Assay Provider: James D. Potter, University of Miami School of Medicine Network: Molecular Library Probe Production Centers Network (MLPCN) Grant Proposal Number: 1 R21 NS064821-01 Grant Proposal PI: James D. Potter, University of Miami School of Medicine External Assay ID: RTF_ACT_FLINT_1536_3XEC50 DRUN SENS Name: Fluorescence-based biochemical high throughput dose response assay for activators of the calcium sensitivity of cardiac Regulated Thin Filaments (RTF). Description: Cardiomyopathies are myocardial diseases that often lead to cardiac remodeling to compensate for deficiencies in cardiac output (1). Cardiomyopathies are characterized as having systolic dysfunctions (i.e. reduced ejection fraction) in dilated cardiomyopathy or diastolic dysfunctions (i.e. impaired relaxation) in hypertrophic and restrictive cardiomyopathies (2). The Fluorescence-based biochemical high throughput dose response assay for inhibitors of the calcium sensitivity of cardiac Regulated Thin Filaments (RTF) Source (MLPCN Center Name): The Scripps Research Institute Molecular Screening Center (SRIMSC) Affiliation: The Scripps Research Institute, TSRI Assay Provider: James D. Potter, University of Miami School of Medicine Network: Molecular Library Probe Production Centers Network (MLPCN) Grant Proposal Number 1 R21 NS064821-01 Grant Proposal PI: James D. Potter, University of Miami School of Medicine External Assay ID: RTF_INH_FLINT_1536_3XIC50 DRUN DESENS Name: Fluorescence-based biochemical high throughput dose response assay for inhibitors of the calcium sensitivity of cardiac Regulated Thin Filaments (RTF). Description: Cardiomyopathies are myocardial diseases that often lead to cardiac remodeling to compensate for deficiencies in cardiac output (1). Cardiomyopathies are characterized as having systolic dysfunctions (i.e. reduced ejection fraction) in dilated cardiomyopathy or diastolic dysfunctions (i.e. impaired relaxation) in hypertrophic and restrictive cardiomyopathies (2). Th Fluorescence-based cell-based high-throughput dose response assay for agonists of NPY-Y2. Source (MLPCN Center Name): The Scripps Research Institute Molecular Screening Center (SRIMSC) Center Affiliation: The Scripps Research Institute (TSRI) Assay Provider: Claes Wahlestedt, Scripps Florida Network: Molecular Library Probe Production Centers Network (MLPCN) Grant Proposal Number: 1 R21 NS056950-01 Grant Proposal PI: Claes Wahlestedt External Assay ID: NPY-Y2_AG_CNGC_1536_3XEC50 Name: Fluorescence-based cell-based high-throughput dose response assay for agonists of NPY-Y2. Description: Neuropeptide Y (NPY) is a neurotransmitter with diverse physiologic roles including control of feeding behavior, regulation of cortical neural activity, heart neural activity, and emotional regulation. Importantly, NPY is implicated in human diseases such as obesity, depression and alcoholism. NPY mediates its biological effects in part through activation of the NPY-Y2 receptor, a 381-amino acid Galphai protein coupled receptor (GPCR) which decreases cytosolic cAMP production. NPY Y2 is e Fluorescence-based cell-based high-throughput dose response assay for potentiators or agonists of NPY-Y2. Source (MLPCN Center Name): The Scripps Research Institute Molecular Screening Center (SRIMSC) Center Affiliation: The Scripps Research Institute (TSRI) Assay Provider: Claes Wahlestedt, Scripps Florida Network: Molecular Library Probe Production Centers Network (MLPCN) Grant Proposal Number: 1 R21 NS056950-01 Grant Proposal PI: Claes Wahlestedt External Assay ID: NPY-Y2_POT_CNGC_1536_3XEC50 Name: Fluorescence-based cell-based high-throughput dose response assay for potentiators or agonists of NPY-Y2. Description: Neuropeptide Y (NPY) is a neurotransmitter with diverse physiologic roles including control of feeding behavior, regulation of cortical neural activity, heart neural activity, and emotional regulation. Importantly, NPY is implicated in human diseases such as obesity, depression and alcoholism. NPY mediates its biological effects in part through activation of the NPY-Y2 receptor, a 381-amino acid Galphai protein coupled receptor (GPCR) which decreases cytosolic cAMP produc Fluorescence-based dose response cell-based high-throughput screening assay for agonists of NPY-Y1. Source (MLPCN Center Name): The Scripps Research Institute Molecular Screening Center (SRIMSC) Center Affiliation: The Scripps Research Institute (TSRI) Assay Provider: Claes Wahlestedt, Scripps Florida Network: Molecular Library Probe Production Centers Network (MLPCN) Grant Proposal Number: 1 R21 NS056950-01 Grant Proposal PI: Claes Wahlestedt External Assay ID: NPY-Y1_AG_CNGC_1536_3XEC50 Name: Fluorescence-based dose response cell-based high-throughput screening assay for agonists of NPY-Y1. Description: Neuropeptide Y (NPY) is a neurotransmitter with physiologic roles including control of feeding behavior, regulation of cortical neural activity, heart neural activity, and emotional regulation. Importantly, NPY is implicated in human diseases such as obesity, depression and alcoholism. NPY mediates its biological effects in part through activation of the Galphai protein coupled receptors (GPCRs) NPY-Y1 and Y2 receptors, which decrease cytosolic cAMP production. Recent studies ha Fluorescence-based dose response cell-based high-throughput screening assay for potentiators or agonists of NPY-Y1. Source (MLPCN Center Name): The Scripps Research Institute Molecular Screening Center (SRIMSC) Center Affiliation: The Scripps Research Institute (TSRI) Assay Provider: Claes Wahlestedt, Scripps Florida Network: Molecular Library Probe Production Centers Network (MLPCN) Grant Proposal Number: 1 R21 NS056950-01 Grant Proposal PI: Claes Wahlestedt External Assay ID: NPY-Y1_AG_CNGC_1536_3XEC50 Name: Fluorescence-based dose response cell-based high-throughput screening assay for agonists of NPY-Y1. Description: Neuropeptide Y (NPY) is a neurotransmitter with physiologic roles including control of feeding behavior, regulation of cortical neural activity, heart neural activity, and emotional regulation. Importantly, NPY is implicated in human diseases such as obesity, depression and alcoholism. NPY mediates its biological effects in part through activation of the Galphai protein coupled receptors (GPCRs) NPY-Y1 and Y2 receptors, which decrease cytosolic cAMP production. Recent studies ha Fluorescence-polarization-based biochemical polarscreen dose response binding assay for partial agonists of the peroxisome proliferator-activated receptor gamma (PPARg) Source (MLPCN Center Name): The Scripps Research Institute Molecular Screening Center (SRISMC) Center Affiliation: The Scripps Research Institute, TSRI Assay Provider: Patrick Griffin, TSRI Network: Molecular Library Probe Production Center Network (MLPCN) Grant Proposal Number: MH079861-01 Grant Proposal PI: Patrick Griffin, TSRI External Assay ID: PPARG_AG_FP_0384_3XIC50 POLARSCREEN DRUN Name: Fluorescence-polarization-based biochemical polarscreen dose response binding assay for partial agonists of the peroxisome proliferator-activated receptor gamma (PPARg). Description: Peroxisome proliferator-activated receptors (PPARs) belong to the nuclear receptor superfamily and are lipid sensors functioning as ligand-dependent transcription factors regulating gene expression patterns of diverse biological processes (1, 2). PPARs play a critical role in metabolic processes such as glucose metabolism, lipid metabolism, and have been implicated in anti-atherogenic, anti-inflammatory as well Fluorescent secondary assay for dose-response confirmation of chemical inhibitors of HePTP Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Screening Centers Network (MLSCN) Grant Number: XO1 MH077603-01 Assay Provider: Dr. Tomas Mustelin, Sanford-Burnham Medical Research Institute Protein tyrosine phosphatases (PTPs), working with protein tyrosine kinases (PTKs), control the phosphorylation state of many proteins in the signal transduction pathways. HePTP is a tyrosine phosphatase expressed in hematopoietic cells and regulates the MAP kinases Erk and p38. It has been found that HePTP is often dysregualted in the preleukemic disorder myelodysplastic syndrome, as well as in acute myelogeneous leukemia. Small molecule inhibitors of HePTP will be useful as molecular probes for studying the mechanism of signal transduction and MAP kinase regulation, and may have therapeutic potential for the treatment of hematopoietic malignancies. This flu HTS Discovery of Chemical Inhibitors of HePTP, a Leukemia Target Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Screening Centers Network (MLSCN) Grant Number: XO1 MH077603-01 Assay Provider: Dr. Tomas Mustelin, Sanford-Burnham Medical Research Institute Protein tyrosine phosphatases (PTPs), working with protein tyrosine kinases (PTKs), control the phosphorylation state of many proteins in the signal transduction pathways. HePTP is a tyrosine phosphatase expressed in hematopoietic cells and regulates the MAP kinases Erk and p38. It has been found that HePTP is often dysregualted in the preleukemic disorder myelodysplastic syndrome, as well as in acute myelogeneous leukemia. Small molecule inhibitors of HePTP will be useful as molecular probes for studying the mechanism of signal transduction and MAP kinase regulation, and may have therapeutic potential for the treatment of hematopoietic malignancies. This colorim HTS Image-Based Screen for Selective Agonists of the KOR Receptor Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Probe Production Centers Network (MLPCN) Grant Number: 1X01DA026208-01 Assay Provider: Dr. Lawrance Barak, Duke University, Durham, NC Drug addiction is a disease originating in the central nervous system that produces compulsive behaviors despite the negative consequences that may result. Major addictive drugs of abuse include components of tobacco, opiates, marijuana, ethanol, cocaine, and derivatives of amphetamines. While the addictive behaviors produced by these substances may be generally similar, the drugs act at different receptor sites in the brain. Recent studies have shown that opioid receptors play a role regulating the addictive behaviors of other receptors that interact with illicit and legal substances of abuse. Opioid receptors are composed of multiple subtypes whose contributions to a HTS Image-Based Screen for Selective Antagonists of the KOR Receptor Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Probe Production Centers Network (MLPCN) Grant Number: 1X01DA026208-01 Assay Provider: Dr. Lawrance Barak , Duke University, Durham NC Drug addiction is a disease originating in the central nervous system that produces compulsive behaviors despite the negative consequences that may result. Major addictive drugs of abuse include components of tobacco, opiates, marijuana, ethanol, cocaine, and derivatives of amphetamines. While the addictive behaviors produced by these substances may be generally similar, the drugs act at different receptor sites in the brain. Recent studies have shown that opioid receptors play a role regulating the addictive behaviors of other receptors that interact with illicit and legal substances of abuse. Opioid receptors are composed of multiple subtypes whose contributions to a High Throughput Screen to Identify Compounds that Inhibit Class II HMG-CoA Reductases - Confirmatory Screen Southern Research Molecular Libraries Screening Center (SRMLSC) Southern Research Institute (Birmingham, Alabama) NIH Molecular Libraries Screening Centers Network (MLSCN) Submitted by Dr. Cynthia Stauffacher (Purdue University) Award: 1-R03-MH082373-01 A number of common human pathogens, including Enterococcus faecalis, Streptococcus pneumoniae, and Staphylococcus aureus, are becoming progressively more resistant to antibiotics and pose a serious public health threat, especially to post-surgical and trauma patients. Therefore, the discovery of drugs targeting novel pathways in these pathogens has become increasingly important. The synthesis of isoprenoids, which in these gram-positive pathogenic bacteria (Hedl, 2002) occurs exclusively via the mevalonate pathway, is essential for their survival. The central mevalonate pathway enzyme, 3-hydroxy-3-methylglutaryl coenzyme A reductase (HMGR; EC 1.1.1.34), reduces HMG-CoA to mevalonate using NADPH (Hedl, 2004). Bacterial HMG-CoA redu High Throughput Screening Assay for Hsp70 Inhibitors Burnham Center for Chemical Genomics (BCCG) Burnham Institute for Medical Research (San Diego, CA) NIH Molecular Libraries Screening Centers Network (MLSCN) MLSCN Grant: XO1 MH079863-01 Over-expression of molecular chaperones occurs commonly in cancers and provides protection from a wide variety of cellular stresses, both endogenous and iatrogenic. Molecular chaperones also play important roles in maintaining the activity of several signal-transducing proteins and transcriptions factors involved in malignant transformation. The human genome contains nine Hsp70-family genes. These chaperones include Hsp70 and Hsc70, which are commonly over-expressed in cancers and which confer resistance to myriad cellular stresses, including cytotoxic chemotherapy. This work's aim is to identify chemical probes of Hsp70 through a fluorescence polarization (FP) assay using Fluorescein-labeled ATP. Additional TR-FRET-based assay was developed and utilized as secondary assay in hit confirmation. Identification of Molecular Probes that Activate MRP-1 - Dose Response Southern Research Molecular Libraries Screening Center (SRMLSC) Southern Research Institute (Birmingham, Alabama) NIH Molecular Libraries Screening Centers Network (MLSCN) Award 1X01-MH077620-01: Submitted by Dr. Gary A. Piazza (Southern Research Institute) in collaboration with Sharon Terry (Genetic Alliance) Pseudoxanthoma elasticum (PXE) is a rare genetic disorder (Bergen, 2007), which involves damage to connective tissues that result in multiple manifestations including skin abnormalities, blindness, and cardiovascular complications. Recently, PXE has been found to be caused by loss of function mutations in the MRP-6 (ABCC6) gene (Chassaing, 2005). MRP-6 mutations have also been reported to be associated with a strong increase in the prevalence of premature coronary artery disease (Trip, 2002) MRP-6 belongs to a family of ATP-dependent transmembrane proteins that function to transport solutes across plasma membranes. In addition to their role in protecting tissues from xenobi Image-based HTS for Selective Agonists of GPR55 Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Probe Production Centers Network (MLPCN) Grant Proposal Number: 1X01 DA026205-01 Assay Provider: Dr. Mary Abood, California Pacific Medical Center Research Institute (currently Temple University) The cannabinoid and endocannabinoid system has been implicated in the pathophysiology of drug dependence and addiction disorders. GPR55, an orphan G-Protein Coupled Receptor, has been reported to be a cannabinoid receptor, but its status as such remains unresolved due to conflicting results from pharmacological studies. The goal of the project is to identify small molecule agonists of GPR55, which may aid in the deorphanization efforts of this receptor and ultimately further the understanding of the role of GPR55 in drug addiction. This high content imaging assay utilizes a cell line permanently expressing a In Vitro Hsc70 Dose Response Fluorescence Polarization Assay Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Screening Centers Network (MLSCN) MLSCN Grant: XO1 MH079863-01 Assay Provider: Dr. John C. Reed, Sanford-Burnham Medical Research Institute ) This Hsc70 dose response assay is developed and performed to study the specificity of analogs of hits tested in the "In Vitro Hsp70 Dose Response Fluorescence Polarization Assay for SAR Study" (AID 1072). Over-expression of molecular chaperones occurs commonly in cancers and provides protection from a wide variety of cellular stresses, both endogenous and iatrogenic. Molecular chaperones also play important roles in maintaining the activity of several signal-transducing proteins and transcriptions factors involved in malignant transformation. The human genome contains nine Hsp70-family genes. These chaperones include Hsp70 and Hsc70, which are commonly over-ex Inhibition Assay Assay plates: 96-well MultiScreen 0.65 um filter plates (Millipore Cat. No.: MADVNOB10)Streptavidin coated beads: Streptavidin Sepharose, suspension 5.0 mL, in 50 mM EDTA/PBS diluted (1:100), (Amersham, Cat. No.: 17-5113-01)Compounds: 10 mM in 100% dimethylsulfoxide (DMSO), final conc.: compound 0.003-100 uM in 10% DMSOEnzyme: SYK RPA purified, truncated construct of Spleen Tyrosine Kinase aa 360-635, stock solution 1 mg/mL, MW: 31.2 KDa, final conc.:0.0005 uM. Peptide 1: biotinylated peptide is derived from a naturally occurring phosphor-acceptor consensus sequence (Biotin-EPEGDYEEVLE), special order from QCB, stock solution 20 mM, final conc.: 5.0 uM.ATP: Adenosine-5'-triphosphate 20 mM, (ROCHE Cat. No.: 93202720), final concentration: 20 uM Buffer: HEPES: 2-Hydroxyethyl piperazine-2-ethanesulfonic acid (Sigma, Cat. No.: H-3375)final concentration: 50 mM HEPES pH7.5BSA: Bovine Serum Albumin Fraction V, fatty acid free (Roche Diagnostics GmbH, Cat. No. 9100221). Intein inhibitors as potential Tuberculosis drugs Southern Research's Specialized Biocontainment Screening Center (SRSBSC) Southern Research Institute (Birmingham, Alabama) NIH Molecular Libraries Probe Production Centers Network (MLPCN) Assay Provider: Dr. Henry Paulus, Boston Biomedical Research Institute Award: 1 R03 MH087438-01 Tuberculosis (TB) is globally the most widespread infectious disease. Two billion people, one third of the world's population, are infected with Mycobacterium tuberculosis and 5-10% of these suffer active disease, leading to nearly 3 million deaths annually. The alarming world-wide increase in multidrug-resistant and extensively drug-resistant TB calls for new anti-mycobacterial drugs against new types of targets that are less likely to undergo mutations that lead to drug resistance. This project seeks to identify inhibitors of protein splicing, which is essential for the expression of three genes in M. tuberculosis. Protein splicing inhibitors, by disabling two separate vital functions and at th Late stage assay provider counterscreen for partial agonists of the peroxisome proliferator-activated receptor gamma (PPARg) Source (MLPCN Center Name): The Scripps Research Institute Molecular Screening Center (SRISMC) Center Affiliation: The Scripps Research Institute, TSRI Assay Provider: Patrick Griffin, TSRI Network: Molecular Library Probe Production Center Network (MLPCN) Grant Proposal Number: MH079861-01 Grant Proposal PI: Patrick Griffin, TSRI External Assay ID: PPARA_AG_LUMI_0384_3XEC50 MDRUN Name: Late stage assay provider counterscreen for partial agonists of the peroxisome proliferator-activated receptor gamma (PPARg): Luminescence-based cell-based dose response assay for partial agonists of the peroxisome proliferator-activated receptor alpha (PPARA). Description: Peroxisome proliferator-activated receptors (PPARs) belong to the nuclear receptor superfamily and are lipid sensors functioning as ligand-dependent transcription factors regulating gene expression patterns of diverse biological processes (1, 2). PPARs play a critical role in metabolic processes such as glucose metabolism, lipid Late-stage fluorescence-based counterscreen for antagonists of the G-protein coupled receptor 7 (GPR7) Source (MLPCN Center Name): The Scripps Research Institute Molecular Screening Center (SRIMSC) Affiliation: The Scripps Research Institute, TSRI Assay Provider: Olivier Civelli, University of California, Irvine Network: Molecular Library Probe Production Centers Network (MLPCN) Grant Proposal Number: 1-R03-DA026557-01 Grant Proposal PI: Olivier Civelli External Assay ID: MCHR1_ANT_FLUO8_384_3X%IC50 Name: Late-stage fluorescence-based counterscreen for antagonists of the G-protein coupled receptor 7 (GPR7): cell-based dose response assay to identify antagonists of the melanin-concentrating hormone receptor 1 (MCHR1). Description: Heterotrimeric G-protein coupled receptors (GPCRs) are major targets for disease therapeutics, due in part to their broad tissue distribution, structural diversity, varied modes of action, and disease-associated mutations (1-4). For example, targeting of opiod receptors by opiates such as morphine is a widespread clinical application for GPCR modulation in Luminescence-based cell-based high throughput dose response assay for activators of the Aryl Hydrocarbon Receptor (AHR) Source (MLPCN Center Name): The Scripps Research Institute Molecular Screening Center (SRIMSC) Center Affiliation: The Scripps Research Institute (TSRI) Assay Provider: Michael Denison, University of California, Davis Network: Molecular Libraries Probe Production Centers Network (MLPCN) Grant Proposal Number: 1-X01-DA026558-01 Grant Proposal PI: Michael Denison External Assay ID: AHR_ACT_LUMI_1536_3XEC50 DRUN Name: Luminescence-based cell-based high throughput dose response assay for activators of the Aryl Hydrocarbon Receptor (AHR). Description: Transcription factors are critical regulators of gene expression (1). Under conditions such as environmental stress and exposure to endogenous toxins, transcription factors can rapidly modulate the transcription of genes whose products regulate cell proliferation and metabolism. The aryl hydrocarbon receptor (AHR) is a ligand-activated transcription factor of the basic helix-loop-helix protein superfamily involved in the biological respons Luminescence-based cell-based high throughput dose response assay for activators of the GAA850 frataxin (FXN) promoter Source (MLPCN Center Name): The Scripps Research Institute Molecular Screening Center (SRISMC) Center Affiliation: The Scripps Research Institute, TSRI Assay Provider: Marek Napierala, University of Texas MD Anderson Cancer Center Network: Molecular Library Probe Production Center Network (MLPCN) Grant Proposal Number: 11R21NS064827-01 Grant Proposal PI: Marek Napierala, University of Texas MD Anderson Cancer Center External Assay ID: FXN-GAA850_ACT_LUMI_1536_3XEC50 DRUN Name: Luminescence-based cell-based high throughput dose response assay for activators of the GAA850 frataxin (FXN) promoter. Description: The most frequently inherited early-onset ataxia in Caucasians is Friedreich's ataxia (FRDA), a severe autosomal recessive neurodegenerative disease. FRDA is associated with progressive neurological disability, hypertrophic cardiomyopathy, increased risk of diabetes mellitus, severe disruption of iron-sulfur (Fe-S) cluster biosynthesis, mitochondrial iron overload coupled to Luminescence-based dose response biochemical high throughput screening assay for inhibitors of the Heat Shock Protein 90 (HSP90) Source (MLPCN Center Name): The Scripps Research Institute Molecular Screening Center (SRIMSC) Center Affiliation: The Scripps Research Institute (TSRI) Assay Provider: Robert Matts, Oklahoma State University Network: Molecular Libraries Probe Production Centers Network (MLPCN) Grant Proposal Number: 1 X01 MH083240-01 Grant Proposal PI: Robert Matts, Oklahoma State University External Assay ID: HSP90_INH_LUMI_1536_3XIC50 Name: Luminescence-based dose response biochemical high throughput screening assay for inhibitors of the Heat Shock Protein 90 (HSP90) Description: Hsp90 is a ubiquitously conserved molecular chaperone (1, 2) that assists the folding of client substrates involved in a variety of human disorders characterized by dysregulated cell proliferation (ie, cancer and viral infections), accumulation of protein aggregates (ie, neurodegenerative diseases) and stress-induced apoptotic cell death (ie, multiple sclerosis) (3). Its diverse biology stems in part from interactions Measurement of GPCR-mediated thallium flux through GIRK channels: Dose-Response Testing Assay Provider: Colleen Niswender Assay Provider Affliation: Vanderbilt University Grant Title: Measurement of GPCR-mediated thallium flux through GIRK channels Grant Number: 1 R03 MH076398-01 The aim of this work was to use high throughput screening of a small molecule library to identify compounds that interact with the alpha2C adrenergic receptor. The assay utilized thallium influx through G-protein Inwardly Rectifying K+ (GIRK) channels as a measure of alpha2C activation. Compounds were tested at 10uM final concentration against Human Embryonic Kidney (HEK) 293 cells stably expressing endogenous alpha2C and cDNAs for GIRK 1 and GIRK 2. The Hamamatsu FDSS 6000 plate reader was used to collect kinetic fluorescence intensities during treatment with the test compound. The alpha2C agonist UK-14304 (Tocris) was the positive control, and DMSO, the compound vehicle, was used as the negative control. Measurement of GPCR-mediated thallium flux through GIRK channels: Dose-Response with Rauwolscine Assay Provider: Colleen Niswender Assay Provider Affliation: Vanderbilt University Grant Title: Measurement of GPCR-mediated thallium flux through GIRK channels Grant Number: 1 R03 MH076398-01 The aim of this work was to use high throughput screening of a small molecule library to identify compounds that interact with the alpha2C adrenergic receptor. The assay utilized thallium influx through G-protein Inwardly Rectifying K+ (GIRK) channels as a measure of alpha2C activation. Compounds were tested at 10uM final concentration against Human Embryonic Kidney (HEK) 293 cells stably expressing endogenous alpha2C and cDNAs for GIRK 1 and GIRK 2. The Hamamatsu FDSS 6000 plate reader was used to collect kinetic fluorescence intensities during treatment with the test compound. The alpha2C agonist UK-14304 (Tocris) was the positive control, and DMSO, the compound vehicle, was used as the negative control. Mycobacterium tuberculosis Pantothenate Synthetase Assay Southern Research Molecular Libraries Screening Center (SRMLSC) Southern Research Institute (Birmingham, Alabama) NIH Molecular Libraries Screening Centers Network (MLSCN) Award: 1R03MH076412-01 Multi-drug resistant Mycobacterium tuberculosis is becoming an increased health problem, especially in immunocompromised individuals with HIV. This form of TB is more difficult to treat and as a result has a higher mortality rate. Because of this, the discovery of drugs targeting novel pathways such as the synthesis of pantothenate has become increasingly important. Pantothenate synthetase (PS; EC 6.3.2.1), encoded by the panC gene, catalyzes the essential ATP-dependent condensation of D-pantoate and alpha-alanine to form pantothenate in bacteria, yeast and plants; pantothenate is a key precursor for the biosynthesis of coenzyme A (CoA) and acyl carrier protein (ACP). The activity of PS was measured spectrophotometrically by coupling the formation of AMP to the reactions of myokinase, pyr Mycobacterium tuberculosis Pantothenate Synthetase Secondary Assay Southern Research Molecular Libraries Screening Center (SRMLSC) Southern Research Institute (Birmingham, Alabama) NIH Molecular Libraries Screening Centers Network (MLSCN) Award: 1R03MH076412-01 Multi-drug resistant Mycobacterium tuberculosis is becoming an increased health problem, especially in immunocompromised individuals with HIV. This form of TB is more difficult to treat and as a result has a higher mortality rate. Because of this, the discovery of drugs targeting novel pathways such as the synthesis of pantothenate has become increasingly important. Pantothenate synthetase (PS; EC 6.3.2.1), encoded by the panC gene, catalyzes the essential ATP-dependent condensation of D-pantoate and alpha-alanine to form pantothenate in bacteria, yeast and plants; pantothenate is a key precursor for the biosynthesis of coenzyme A (CoA) and acyl carrier protein (ACP). The activity of PS was measured spectrophotometrically by coupling the formation of AMP to the reactions of myokinase, pyruva QFRET-based biochemical high throughput dose response assay for inhibitors of the Plasmodium falciparum M18 Aspartyl Aminopeptidase (PFM18AAP). Source (MLPCN Center Name): The Scripps Research Institute Molecular Screening Center Affiliation: The Scripps Research Institute, TSRI Assay Provider: John Dalton and Donald Gardiner, Queensland Institute of Medical Research, Australia Network: Molecular Library Probe Production Centers Network (MLPCN) Grant Proposal Number: 1 R03 MH084103-01 Grant Proposal PI: John Dalton and Donald Gardiner, Queensland Institute of Medical Research, Australia External Assay ID: PFM18AAP_INH_QFRET_1536_3XIC50 Name: QFRET-based biochemical high throughput dose response assay for inhibitors of the Plasmodium falciparum M18 Aspartyl Aminopeptidase (PFM18AAP). Description: Aminopeptidases (APs) are metalloproteases that cleave amino-terminal (N-terminal) amino acids during protein synthesis (1, 2) These enzymes are characterized in part by their post-translational removal of leucine, aspartate, proline, methionine, etc from proteins and peptides, in order that proteins are properly regulated, targete QFRET-based biochemical high throughput dose response assay to identify exosite inhibitors of ADAM10. Source (MLPCN Center Name): The Scripps Research Institute Molecular Screening Center (SRIMSC) Center Affiliation: Torrey Pines Institute for Molecular Sciences (TPIMS) Assay Provider: Dmitriy Minond, Torrey Pines Institute for Molecular Sciences (TPIMS) Network: Molecular Libraries Probe Production Centers Network (MLPCN) Grant Proposal Number: 1 R03 DA033985-01 Grant Proposal PI: Dmitriy Minond, Torrey Pines Institute for Molecular Sciences (TPIMS) External Assay ID: ADAM10_INH_QFRET_1536_3XIC50 DRUN Name: QFRET-based biochemical high throughput dose response assay to identify exosite inhibitors of ADAM10. Description: Approximately 20-30% of breast cancer patients have tumors that over-express human epidermal growth factor receptor (HER2), which confers an aggressive tumor phenotype and poor prognosis [1-3]. A Disintegrin and Metalloprotease (ADAM) proteases are responsible for amplification of HER2 signaling due to either cleavage of its extracellular domain or release of HER2 QFRET-based biochemical high throughput dose response assay to identify exosite inhibitors of ADAM17 Source (MLPCN Center Name): The Scripps Research Institute Molecular Screening Center (SRIMSC) Center Affiliation: Torrey Pines Institute for Molecular Sciences (TPIMS) Assay Provider: Dmitriy Minond, Torrey Pines Institute for Molecular Sciences (TPIMS) Network: Molecular Libraries Probe Production Centers Network (MLPCN) Grant Proposal Number: 1 R03 DA033985-01 Grant Proposal PI: Dmitriy Minond, Torrey Pines Institute for Molecular Sciences (TPIMS) External Assay ID: ADAM17_INH_QFRET_1536_3XIC50 INH DRUN Name: QFRET-based biochemical high throughput dose response assay to identify exosite inhibitors of ADAM17. Description: Approximately 20-30% of breast cancer patients have tumors that over-express human epidermal growth factor receptor (HER2), which confers an aggressive tumor phenotype and poor prognosis [1-3]. A Disintegrin and Metalloprotease (ADAM) proteases are responsible for amplification of HER2 signaling due to either cleavage of its extracellular domain or release of H RNA polymerase SAR Screening Center: Penn Center for Molecular Discovery Center Affiliation: University of Pennsylvania Network: Molecular Library Screening Center Network (MLSCN) Assay Provider: Arkady Mustaev, Public Health Research Institute, Newark, NJ Grant number: MH076325-01 DNA-directed RNA polymerase (EC 2.7.7.6) is responsible for bacterial RNA synthesis and as such is essential for bacterial gene expression. Owing to its central role in DNA transcription, the enzyme RNA polymerase is the target of various natural antibiotics. The best known is rifampicin, a potent and broad-spectrum anti-infective agent that is particularly effective against intracellular pathogens, such as Mycobacterium tuberculosis, for which it is one of the most widely used chemotherapeutic agents. However, the emergence of drug-resistant bacteria has become a major public health problem, so the discovery of novel RNA polymerase inhibitors is an important goal. A high-throughput screen was designed to discover novel in RNA polymerase dose-response confirmation Screening Center: Penn Center for Molecular Discovery Center Affiliation: University of Pennsylvania Network: Molecular Library Screening Center Network (MLSCN) Assay Provider: Arkady Mustaev, Public Health Research Institute, Newark, NJ Grant number: MH076325-01 DNA-directed RNA polymerase (EC 2.7.7.6) is responsible for bacterial RNA synthesis and as such is essential for bacterial gene expression. Owing to its central role in DNA transcription, the enzyme RNA polymerase is the target of various natural antibiotics. The best known is rifampicin, a potent and broad-spectrum anti-infective agent that is particularly effective against intracellular pathogens, such as Mycobacterium tuberculosis, for which it is one of the most widely used chemotherapeutic agents. However, the emergence of drug-resistant bacteria has become a major public health problem, so the discovery of novel RNA polymerase inhibitors is an important goal. A high-throughput screen was designed to discover novel in SAR analysis for the identification of translation initiation inhibitors (PABP) Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute(SBMRI, San Diego, CA) Network: NIH Molecular Libraries Probe Production Centers Network (MLPCN) Grant Number: 1R03MH084835-01 Assay Provider: Jerry Pelletier, Ph.D, McGill University, Montreal, Canada Translation is an essential cellular process whose deregulation is associated with alterations in cell growth, cell cycle progression, and cell death responses. The initiation phase of translation is a key target for regulation when cells are exposed to various environmental cues (e.g. insulin, amino acid starvation, mitogenic stimulation, hypoxia, etc). As well, translation initiation control is usurped upon viral infection and is deregulated in many human cancers. Over-expression of certain translation factors can lead to malignant transformation and many of the components of the translational apparatus are over-expressed in human cancers. Several tumor sup SAR analysis for the identification of translation initiation inhibitors (eIF4H) Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBIMR, San Diego, CA) Network: NIH Molecular Libraries Probe Production Centers Network (MLPCN) Grant Number: 1R03MH084835-01 Assay Provider: Jerry Pelletier, Ph.D, McGill University, Montreal, Canada Translation is an essential cellular process whose deregulation is associated with alterations in cell growth, cell cycle progression, and cell death responses. The initiation phase of translation is a key target for regulation when cells are exposed to various environmental cues (e.g. insulin, amino acid starvation, mitogenic stimulation, hypoxia, etc). As well, translation initiation control is usurped upon viral infection and is deregulated in many human cancers. Over-expression of certain translation factors can lead to malignant transformation and many of the components of the translational apparatus are over-expressed in human cancers. Several tumor su SAR analysis of Antagonists of IAP-family anti-apoptotic proteins Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Screening Centers Network (MLSCN) Grant Proposal Number: MH081277-01 Assay Provider: John C. Reed, Sanford-Burnham Medical Research Institute, San Diego, CA This XIAP dose response assay is developed and performed to confirm hits originally identified in the XIAP HTS binding assay (AID 1018) and to study the structure-activity relationship on analogs of the confirmed hits. Compounds are either acquired from commercial sources or synthesized internally. The assay was performed in the assay providers' laboratory. Apoptosis plays an essential role in many aspects of normal development and physiology, becoming dysregulated in myriad diseases characterized by insufficient or excessive cell death. Caspases are intracellular proteases that are suppressed by Inhibitor of Apoptosis Proteins (IAPs), a fami SAR analysis of an In Vitro TNAP Dose Response Luminescent Assay Sanford-Burnham Center for Chemical Genomics (SBCCG) Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) NIH Molecular Libraries Screening Centers Network (MLSCN) Grant Number: MH077602-01 Assay Provider: Dr. Jose Luis Millan, Sanford-Burnham Medical Research Institute, San Diego, CA This TNAP dose response assay is developed and performed for the purpose of SAR study on analogs of hits originally identified in the TNAP luminescent HTS assay (AID 518) Alkaline phosphatase (EC 3.1.3.1) (APs) catalyze the hydrolysis of phosphomonoesters, releasing phosphate and alcohol. APs are dimeric enzymes found in the most organism. In human, four isozymes of APs have been identified. Three isozymes are tissue-specific and the fourth one is tissue-nonsepecifc, named TNAP. TNAP deficiency is associated with defective bone mineralization in the form of rickets and osteomalacia. Therefore, there are therapeutic potentials of inhibiting TNAP activity SAR analysis of compounds that inhibit HePTP - Set 2 Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Probe Production Centers Network (MLSCN) Grant Number: XO1 MH077603-01 Assay Provider: Dr. Tomas Mustelin, Sanford-Burnham Medical Research Institute Protein tyrosine phosphatases (PTPs), working with protein tyrosine kinases (PTKs), control the phosphorylation state of many proteins in the signal transduction pathways. HePTP is a tyrosine phosphatase expressed in hematopoietic cells and regulates the MAP kinases Erk and p38. It has been found that HePTP is often dysregualted in the preleukemic disorder myelodysplastic syndrome, as well as in acute myelogeneous leukemia. Small molecule inhibitors of HePTP will be useful as molecular probes for studying the mechanism of signal transduction and MAP kinase regulation, and may have therapeutic potential for the treatment of hematopoietic malignancies. Th SAR analysis of compounds that potentiate TRAIL-induced apoptosis in PPC-1 cells. Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Production Centers Network (MLPCN) Grant Number: X01 MH083230-01 Assay Provider: Dr. Dmitri Rozanov, Sanford-Burnham Medical Research Institute, San Diego CA This dose response assay is developed and performed to confirm hits originally identified in "uHTS for the identification of compounds that potentiate TRAIL-induced apoptosis of cancer cells" (AID 1443) and to study the structure-activity relationship on analogs of the confirmed hits. Compounds are either acquired from commercial sources or synthesized internally. Cytotoxic chemotherapy induces apoptosis via a pathway involving mitochondria, sometimes referred to as the "intrinsic pathway." An acquired resistance to anticancer drugs commonly results from the accumulation of defects in components of the mitochondrial pathway for apoptosis. Discov SMAD Transcription Factor Inhibitors Dose Response Confirmation NIH Molecular Libraries Screening Centers Network [MLSCN] Emory Chemical Biology Discovery Center in MLSCN Assay provider: F.M. Hoffmann, University of Wisconsin-Madison MLSCN Grant: 1R21NS057002-01 Assay Overview: Transforming growth factor beta (TGF-Beta) regulates a variety of processes in mammalian cells, including proliferation, apoptosis, cell migration and extracellular matrix production. Aberrant increases in TGF-Beta signaling have been implicated in several pathological conditions including cancer and fibrosis. Inhibition of TGF-Beta signaling is an important tool in elucidating the multiple biological functions of TGF-Beta and is of significant interest as a potential therapeutic strategy in fibrotic diseases and several advanced cancers. Smad proteins mediate cellular responses to TGF-Beta. TGF-Beta alters cellular gene expression and cell behavior by binding and activating the Type II and Type I serine kinase receptors on the cell membrane. Activated Type I recep SMAD Transcription Factor Inhibitors Secondary Dose Response Confirmation NIH Molecular Libraries Screening Centers Network [MLSCN] Emory Chemical Biology Discovery Center in MLSCN Assay provider: F.M. Hoffmann, University of Wisconsin-Madison MLSCN Grant: 1R21NS057002-01 Assay Overview: Transforming growth factor beta (TGF-Beta) regulates a variety of processes in mammalian cells, including proliferation, apoptosis, cell migration and extracellular matrix production. Aberrant increases in TGF-Beta signaling have been implicated in several pathological conditions including cancer and fibrosis. Inhibition of TGF-Beta signaling is an important tool in elucidating the multiple biological functions of TGF-Beta and is of significant interest as a potential therapeutic strategy in fibrotic diseases and several advanced cancers. Smad proteins mediate cellular responses to TGF-Beta. TGF-Beta alters cellular gene expression and cell behavior by binding and activating the Type II and Type I serine kinase receptors on the cell membrane. Activated Type I recep Screening for Inhibitors of the Mevalonate Pathway in Streptococcus Pneumoniae - DPM-DC - Secondary Assay Southern Research Molecular Libraries Screening Center (SRMLSC) Southern Research Institute (Birmingham, Alabama) NIH Molecular Libraries Screening Centers Network (MLSCN) Assay Provider: Dr. Thomas S. Leyh, Albert Einstein College of Medicine of Yeshiva University Award: R03 MH078936-01 Streptococcus pneumoniae takes the lives of nearly 4,000 people daily and antibiotic resistant strains are becoming an increasing problem. Because of this, the discovery of drugs targeting novel pathways has become increasingly important. The mevalonate pathway produces isopentenyl diphosphate (the molecular building block of isoprenoids) and is essential for the survival of the pathogen in mouse lung and serum. The biosynthesis of isopentenyl diphosphate involves three consecutive reactions that are catalyzed by the enzymes mevalonate kinase (MK; E.C. 2.7.1.36), phosphomevalonate kinase (PMK; E.C. 2.7.4.2), and diphosphomevalonate decarboxylase (DPM-DC; E.C. 4.1.1.33). DPM-DC catalyzes the ATP- TR-FRET-based biochemical dose response competitive binding lanthascreen assay for partial agonists of the peroxisome proliferator-activated receptor gamma (PPARg) Source (MLPCN Center Name): The Scripps Research Institute Molecular Screening Center (SRISMC) Center Affiliation: The Scripps Research Institute, TSRI Assay Provider: Patrick Griffin, TSRI Network: Molecular Library Probe Production Center Network (MLPCN) Grant Proposal Number: MH079861-01 Grant Proposal PI: Patrick Griffin, TSRI External Assay ID: PPARG_AG_TRFRET_0384_3XIC50 LANTHASCREEN DRUN Name: TR-FRET-based biochemical dose response competitive binding lanthascreen assay for partial agonists of the peroxisome proliferator-activated receptor gamma (PPARg). Description: Peroxisome proliferator-activated receptors (PPARs) belong to the nuclear receptor superfamily and are lipid sensors functioning as ligand-dependent transcription factors regulating gene expression patterns of diverse biological processes (1, 2). PPARs play a critical role in metabolic processes such as glucose metabolism, lipid metabolism, and have been implicated in anti-atherogenic, anti-inflammatory as well TR-FRET-based biochemical high throughput dose response assay to identify NR2E3 inverse agonists Source (MLPCN Center Name): The Scripps Research Institute Molecular Screening Center Affiliation: The Scripps Research Institute, TSRI Assay Provider: Konstantin Petrukhin, Columbia University Network: Molecular Library Probe Production Centers Network (MLPCN) Grant Proposal Number: 1 R21 NS061718-01 Fast Track Grant Proposal PI: Konstantin Petrukhin, Columbia University External Assay ID: NR2E3_IAG_HTRF_1536_3XIC50 DRUN Name: TR-FRET-based biochemical high throughput dose response assay to identify NR2E3 inverse agonists. Description: Nuclear receptors are small molecule- and hormone-regulated transcription factors with discrete DNA-binding and ligand-binding domains, and are essential during development and for maintenance of proper cell function in adults. Small pharmacological compounds that bind to the cleft of the ligand-binding domain could alter receptor conformation and subsequently modify transcription of target genes. Such ligands (agonists and antagonists) have been d TRFRET-based cell-based high throughput dose response assay to identify inhibitors of cell surface Prion Protein (PRPC) Source (MLPCN Center Name): The Scripps Research Institute Molecular Screening Center (SRIMSC) Affiliation: The Scripps Research Institute, TSRI Assay Provider: Corinne Lasmezas, The Scripps Research Institute Molecular Screening Center Network: Molecular Library Probe Production Centers Network (MLPCN) Grant Proposal Number: 1 R03 DA035192-01 Grant Proposal PI: Corinne Lasmezas, The Scripps Research Institute Molecular Screening Center External Assay ID: PRPC_INH_TRFRET_1536_3XIC50 DRUN Name: TRFRET-based cell-based high throughput dose response assay to identify inhibitors of cell surface Prion Protein (PRPC) Description: Prion diseases are lethal infectious neurodegenerative diseases affecting animals and humans [1]. In humans, Creutzfeldt-Jakob Disease (CJD) is either sporadic, affecting mainly people over 60 years, iatrogenic, or genetic with a high penetrance. Prion diseases are characterized by the accumulation, in brain and lymphoid tissue of PrPsc [2], a misfolded, aggrega XBP1 DR counterscreen for CHOP NIH Molecular Libraries Screening Centers Network [MLSCN] Emory Chemical Biology Discovery Center in MLSCN Assay provider: Dr. Randal Kaufman, University of Michigan MLSCN Grant: R03MH084182-01, U54HG003918 Assay Overview Many genetic and environmental diseases result from defective protein folding within the secretory pathway so that aberrantly folded proteins are recognized by the cellular surveillance system and retained within the endoplasmic reticulum (ER). Under conditions of malfolded protein accumulation, the cell activates the Unfolded Protein Response (UPR) to clear the malfolded proteins, and if unsuccessful, initiates a cell death response. Preliminary studies have shown that CHOP is a crucial factor in the apoptotic arm of the UPR; XBP1 activates genes encoding ER protein chaperones and thereby mediates the adaptive UPR response to increase clearance of malfolded proteins. Inhibition of CHOP is hypothesized to enhance survival by preventing UPR programmed cell death. uHTS fluorescence polarization assay for the identification of translation initiation inhibitors (PABP) Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Production Centers Network (MLPCN) Grant Number: 1R03MH084835-01 Assay Provider: Jerry Pelletier, Ph.D, McGill University, Montreal, Canada Translation is an essential cellular process whose deregulation is associated with alterations in cell growth, cell cycle progression, and cell death responses. The initiation phase of translation is a key target for regulation when cells are exposed to various environmental cues (e.g. insulin, amino acid starvation, mitogenic stimulation, hypoxia, etc). As well, translation initiation control is usurped upon viral infection and is deregulated in many human cancers. Over-expression of certain translation factors can lead to malignant transformation and many of the components of the translational apparatus are over-expressed in human cancers. Several tumor suppresso uHTS fluorescence polarization assay for the identification of translation initiation inhibitors (eIF4H) Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Production Centers Network (MLPCN) Grant Number: 1R03MH084835-01 Assay Provider: Jerry Pelletier, Ph.D, McGill University, Montreal, Canada Translation is an essential cellular process whose deregulation is associated with alterations in cell growth, cell cycle progression, and cell death responses. The initiation phase of translation is a key target for regulation when cells are exposed to various environmental cues (e.g. insulin, amino acid starvation, mitogenic stimulation, hypoxia, etc). As well, translation initiation control is usurped upon viral infection and is deregulated in many human cancers. Over-expression of certain translation factors can lead to malignant transformation and many of the components of the translational apparatus are over-expressed in human cancers. Several tumor suppresso 14-3-3 protein interaction modulators Dose Response Confirmation NIH Molecular Libraries Screening Centers Network [MLSCN] Emory Chemical Biology Discovery Center in MLSCN MLSCN Grant: 1 X01MH78953-01 The 14-3-3 proteins are the prototype for a novel class of protein modules that can recognize phosphoserine/threonine (pS/T)-containing motifs in a variety of signaling proteins. To date, 14-3-3 proteins have been reported to bind more than 200 client proteins. Through these interactions, 14-3-3 proteins play important roles in a wide range of vital regulatory processes, such as Bad-induced apoptosis, Raf-1-mediated cell proliferation, and Cdc25-regulated cell cycle progression. In addition to their participation in diverse physiological processes, 14-3-3 proteins have been implicated in a number of clinically important pathological conditions, such as neurodegenerative disorders and cancers. Thus, such studies on the 14-3-3/client-protein interactions may provide tremendous opportunities for therapeutic interventions. Therefore, chemical tools woul AKT counterscreen of confirmed PKD inhibitors-57K library The PKD HTS assay was developed and run at the University of Pittsburgh Molecular Screening Center (PMLSC) as part of the Molecular Library Screening Center Network (MLSCN)(1R03DA24898-01). Protein kinase D (PKD) is a novel family of serine/threonine kinases targeted by diacylglycerol. It regulates many fundamental cell functions including cell proliferation, survival, differentiation and protein trafficking, and plays important roles in pathological conditions such as cardiac hypertrophy and cancer in multiple organ systems. However, the mechanisms underlying these effects of PKD are not clearly understood, and the role of PKD in cancer and other diseases has not been fully defined. This is partly due to the lack of effective pharmacological tools that specifically target PKD in normal cellular processes and in pathological conditions. The immediate goal of this proposal is to demonstrate the feasibility of an IMAP-based HTS fluorescent polarization (FP) assay and its use to identi Absorbance-based biochemical high throughput dose response assay for activators of Methionine sulfoxide reductase A (MsrA) Source (MLPCN Center Name): The Scripps Research Institute Molecular Screening Center (SRIMSC) Affiliation: The Scripps Research Institute, TSRI Assay Provider: Herbert Weissbach, Florida Atlantic University Network: Molecular Library Probe Production Centers Network (MLPCN) Grant Proposal Number: 1R03DA032473-01 Grant Proposal PI: Herbert Weissbach, Florida Atlantic University External Assay ID: MSRA_ACT_ABS_1536_3XEC50 DRUN Name: Absorbance-based biochemical high throughput dose response assay for activators of Methionine sulfoxide reductase A (MsrA). Description: Oxidative damage, resulting from the production of reactive oxygen species (ROS) within cells, is believed to be a major factor in age-related diseases and the aging process. One of the mechanisms by which this damage occurs is via oxidation of methionine residues to methionine sulfoxide (Met(O)) derivatives in cellular proteins, which can lead to protein inactivation (1). These Met(O) species can be repaired/reduced by Activity Assay Inhibitory activity-1 assay: To a solution containing 20 mM Tris-HCl (pH 7.5), 1 mM MgCl2, 100 μM EDTA, 330 μg/ml bovine serum albumin, 50 kU/ml 5′-nucleotidase, 0.1 μCi 3H-cAMP (64 nM cAMP), and PDE10A (H-PDE10A2, Human Phosphodiesterase 10A2, Scottish Biomedical), a test compound was added, and the mixture was reacted at 25° C. for 2 hours. To the reaction solution, QAE-Sephadex (17-0190-01, GE Healthcare Japan Corp.) suspended in 10 mM HEPES-Na (pH 7.0) (hereinafter, also referred to as a “QAE-Sephadex suspension”) was added, and the mixture was shaken for 1 minute and left standing for 5 minutes to obtain a supernatant. To the supernatant, a QAE-Sephadex suspension was further added, and the mixture was shaken for 1 minute and left standing for 5 minutes. Then, the obtained supernatant was transferred to LumaPlate (PerkinElmer, Inc.) and assayed using a radiation counter (TopCount NXT, PerkinElmer, Inc.). Additional SAR compounds tested via Multiplex dose response to identify specific small molecule inhibitors of Ras and Ras-related GTPases specifically Cdc42 wildtype University of New Mexico Assay Overview: Assay Support: NIH I RO3 MH081231-01 HTS to identify specific small molecule inhibitors of Ras and Ras-related GTPases PI: Angela Wandinger-Ness, Ph.D. Co-PI: Larry Sklar, Ph.D. Assay Development: Zurab Surviladze, Ph.D. Assay Implementation: Zurab Surviladze, Danuta Wlodek, Terry Foutz, Mark Carter, Anna Waller Target Team Leader for the Center: Larry Sklar (lsklar@salud.unm.edu) UNM Cheminformatics: Cristian Bologa, Ph.D., Fabiola Miscioscia, Ph.D., Ramona Curpan, Ph.D., Oleg Ursu, Ph.D. Chemistry: University of Kansas Specialized Chemistry Center KU Specialized Chemistry Center PI: Jeff Aube, Ph.D. KU SCC Project Manager: Jennifer E. Golden. Ph.D. KU SCC Chemists on this project: Chad Schroeder, M.S., Denise Simpson, Ph.D., Julica Noeth, B.S. Dose Response Assay Background and Significance: Ras and related small molecular weight GTPases function in the regulation of signaling and cell growth, and collectively serve to control cell proli Additional SAR compounds tested via Multiplex dose response to identify specific small molecule inhibitors of Ras and Ras-related GTPases specifically Rab2 wildtype University of New Mexico Assay Overview: Assay Support: NIH I RO3 MH081231-01 HTS to identify specific small molecule inhibitors of Ras and Ras-related GTPases PI: Angela Wandinger-Ness, Ph.D. Co-PI: Larry Sklar, Ph.D. Assay Development: Zurab Surviladze, Ph.D. Assay Implementation: Zurab Surviladze, Danuta Wlodek, Terry Foutz, Mark Carter, Anna Waller Target Team Leader for the Center: Larry Sklar (lsklar@salud.unm.edu) UNM Cheminformatics: Cristian Bologa, Ph.D., Fabiola Miscioscia, Ph.D., Ramona Curpan, Ph.D., Oleg Ursu, Ph.D. Chemistry: University of Kansas Specialized Chemistry Center KU Specialized Chemistry Center PI: Jeff Aube, Ph.D. KU SCC Project Manager: Jennifer E. Golden. Ph.D. KU SCC Chemists on this project: Chad Schroeder, M.S., Denise Simpson, Ph.D., Julica Noeth, B.S. Dose Response Assay Background and Significance: Ras and related small molecular weight GTPases function in the regulation of signaling and cell growth, and collectively serve to control cell proli Additional SAR compounds tested via Multiplex dose response to identify specific small molecule inhibitors of Ras and Ras-related GTPases specifically Rab7 wildtype University of New Mexico Assay Overview: Assay Support: NIH I RO3 MH081231-01 HTS to identify specific small molecule inhibitors of Ras and Ras-related GTPases PI: Angela Wandinger-Ness, Ph.D. Co-PI: Larry Sklar, Ph.D. Assay Development: Zurab Surviladze, Ph.D. Assay Implementation: Zurab Surviladze, Danuta Wlodek, Terry Foutz, Mark Carter, Anna Waller Target Team Leader for the Center: Larry Sklar (lsklar@salud.unm.edu) UNM Cheminformatics: Cristian Bologa, Ph.D., Fabiola Miscioscia, Ph.D., Ramona Curpan, Ph.D., Oleg Ursu, Ph.D. Chemistry: University of Kansas Specialized Chemistry Center KU Specialized Chemistry Center PI: Jeff Aube, Ph.D. KU SCC Project Manager: Jennifer E. Golden. Ph.D. KU SCC Chemists on this project: Chad Schroeder, M.S., Denise Simpson, Ph.D., Julica Noeth, B.S. Dose Response Assay Background and Significance: Ras and related small molecular weight GTPases function in the regulation of signaling and cell growth, and collectively serve to control cell proli Additional SAR compounds tested via Multiplex dose response to identify specific small molecule inhibitors of Ras and Ras-related GTPases specifically Rac1 wildtype University of New Mexico Assay Overview: Assay Support: NIH I RO3 MH081231-01 HTS to identify specific small molecule inhibitors of Ras and Ras-related GTPases PI: Angela Wandinger-Ness, Ph.D. Co-PI: Larry Sklar, Ph.D. Assay Development: Zurab Surviladze, Ph.D. Assay Implementation: Zurab Surviladze, Danuta Wlodek, Terry Foutz, Mark Carter, Anna Waller Target Team Leader for the Center: Larry Sklar (lsklar@salud.unm.edu) UNM Cheminformatics: Cristian Bologa, Ph.D., Fabiola Miscioscia, Ph.D., Ramona Curpan, Ph.D., Oleg Ursu, Ph.D. Chemistry: University of Kansas Specialized Chemistry Center KU Specialized Chemistry Center PI: Jeff Aube, Ph.D. KU SCC Project Manager: Jennifer E. Golden. Ph.D. KU SCC Chemists on this project: Chad Schroeder, M.S., Denise Simpson, Ph.D., Julica Noeth, B.S. Dose Response Assay Background and Significance: Ras and related small molecular weight GTPases function in the regulation of signaling and cell growth, and collectively serve to control cell proli Additional SAR compounds tested via Multiplex dose response to identify specific small molecule inhibitors of Ras and Ras-related GTPases specifically Ras wildtype University of New Mexico Assay Overview: Assay Support: NIH I RO3 MH081231-01 HTS to identify specific small molecule inhibitors of Ras and Ras-related GTPases PI: Angela Wandinger-Ness, Ph.D. Co-PI: Larry Sklar, Ph.D. Assay Development: Zurab Surviladze, Ph.D. Assay Implementation: Zurab Surviladze, Danuta Wlodek, Terry Foutz, Mark Carter, Anna Waller Target Team Leader for the Center: Larry Sklar (lsklar@salud.unm.edu) UNM Cheminformatics: Cristian Bologa, Ph.D., Fabiola Miscioscia, Ph.D., Ramona Curpan, Ph.D., Oleg Ursu, Ph.D. Chemistry: University of Kansas Specialized Chemistry Center KU Specialized Chemistry Center PI: Jeff Aube, Ph.D. KU SCC Project Manager: Jennifer E. Golden. Ph.D. KU SCC Chemists on this project: Chad Schroeder, M.S., Denise Simpson, Ph.D., Julica Noeth, B.S. Dose Response Assay Background and Significance: Ras and related small molecular weight GTPases function in the regulation of signaling and cell growth, and collectively serve to control cell proli Allosteric Modulators of D1 Receptors: Dose-dependent Assay Assay Provider: Val Watts Assay Provider Affiliation: Purdue University Grant Title: Allosteric Modulators of D1 Receptors Grant Number: 1 X01 MH077619-01 Dopamine receptors have been classified into two large families, the D1-like and the D2-like (Neve et al., 2004). Members of the D1-like receptor family include D1 and D5 dopamine receptors. Activation of D1-like receptors stimulate Gs which in turn activates adenylate cyclase resulting in enhanced cyclic AMP accumulation (Neve et al., 2004). Members of the D2-like receptor family include D2, D3, and D4 dopamine receptors. Activation of D2-like dopamine receptors is often linked to inhibition of drug-stimulated cyclic AMP accumulation (Watts et al., 1999, Watts et al., 1998). The field of dopamine research greatly benefited in the latter part of the last century by the availability of dopamine D2 agonists and antagonists. Indeed, the 'dopamine hypothesis' of schizophrenia resulted directly from the availability of D2 dopami Allosteric Modulators of D1 Receptors: Dose-dependent Counterscreen Assay Provider: Val Watts Assay Provider Affiliation: Purdue University Grant Title: Allosteric Modulators of D1 Receptors Grant Number: 1 X01 MH077619-01 Dopamine receptors have been classified into two large families, the D1-like and the D2-like (Neve et al., 2004). Members of the D1-like receptor family include D1 and D5 dopamine receptors. Activation of D1-like receptors stimulate Gs which in turn activates adenylate cyclase resulting in enhanced cyclic AMP accumulation (Neve et al., 2004). Members of the D2-like receptor family include D2, D3, and D4 dopamine receptors. Activation of D2-like dopamine receptors is often linked to inhibition of drug-stimulated cyclic AMP accumulation (Watts et al., 1999, Watts et al., 1998). The field of dopamine research greatly benefited in the latter part of the last century by the availability of dopamine D2 agonists and antagonists. Indeed, the 'dopamine hypothesis' of schizophrenia resulted directly from the availability of D2 dopami AlphaScreen confirmatory assay for validation of inhibitors of SUMOylation Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Probe Production Centers Network (MLPCN) Grant Number: 1 R03 MH084862-01 Assay Provider: Dr. Yuan Chen, Beckman Research Institute, City Of Hope, CA Protein modification by the SUMO (Small Ubiquitin-like MOdifier) family of proteins is an important post-translational modification that plays an essential role in many functions including gene transcription, cell cycle progression, DNA repair, viral infection, and the development of neurodegenerative diseases (1, 2). Recent proteomic studies have found that approximately 10% of the proteins encoded by the yeast genome are substrates for SUMO modification (3-5). The mechanism of how SUMOylation is involved in these cellular functions remains largely unclear. The inhibitors of SUMOylation would be useful to probe the roles of SUMOylation in cellular regulat BAP1 Enzyme inhibitors Dose Response Confirmation NIH Molecular Libraries Screening Centers Network [MLSCN] Emory Chemical Biology Discovery Center in MLSCN Assay provider: Keith D. Wilkinson, Emory University MLSCN Grant: 1 R03 MH076382-01 Assay Overview: BAP1 (BRCA1 associated protein 1) is a member of the Ubiquitin carboxy-terminal hydrolase (UCH) family of deubiquitinating enzymes(DUB). These proteases reverse the conjugation of ubiquitin to targeted proteins. The importance of ubiquitin conjugation in many cellular processes suggests a critical role of DUBs in normal physiology and potentially in pathological conditions. In order to identify molecular probes for studying DUB function and for targeting DUB for potential therapeutic applications, an enzyme-based kinetic high throughput assay has been developed. The assay monitors the hydrolysis of simple ubiquitin conjugates such as Ub-AMC by cysteine protease activity of BAP1 and the release of free AMC. The amount of the fluorescence intensity of released free AMC is pro Binding Assays (DELFIA) BIR3 domains (10 nM) were incubated with SMAC peptide (10 nM) in assay buffer (50 mM Tris, 120 mM NaCl, 0.1% BSA, 1 mM DTT, 0.05% Triton X100) for one hour at room temperature in the presence of inhibitory compounds. The assay mixture was transferred to a streptavidin coated plate and incubated for one hour at room temperature to allow binding of the biotinylated peptide and associated BIR3 domains to the plate. After several washing steps Eu labeled anti-GST antibody (e.g. Perkin Elmer DELFIA Eu-N1-antiGST AD0250) was added to detect BIR3 domain-SMAC peptide interactions according to Perkin Elmer's instructions. Briefly, the antibody was added (dilution 1:5000 in Perkin Elmer DELFIA Assay Buffer 2013-01) and incubated for one hour. After 3 washing steps using Delfia Washing Buffer (Perkin Elmer DELFIA Wash 2013-05), Enhancement Solution (Perkin Elmer Enhancement Asolution 2013-02) was added and incubation continued for 10 minutes. Biochemical Assay The enzymatic activity of human HPK1 (MAP4K1) was monitored in a biochemical assay in the presence or absence of compounds and using a synthetic peptide substrate. An increase in phosphorylation of the peptide by HPK1 was indicative of its kinase activity.Recombinant HPK1 kinase domain produced via baculovirus infection of insect cells was obtained from Proteros (Proteros Biostructures #PR-0322) and was pre-activated in the presence of 2 mM ATP (Sigma-Aldrich, cat #GE27-2056-01) and 2 mM magnesium chloride for 16 hours at 4° C. The protein reaction mixture was then loaded to a desalting column (Thermo Fisher Scientific, Cat #89889) to remove excess ATP. HPK1 was eluted with buffer containing 20 mM Tris (2-Amino-2-(hydroxymethyl)propane-1,3-diol) pH 8.0, 150 mM NaCl, 2 mM dithiothreitol and 5% glycerol, and was frozen at −80° C. for later use. HPK1 dual phosphorylation was confirmed by mass spectrometry. CDK7 counterscreen of confirmed PKD inhibitors-57K library addition The PKD HTS assay was developed and run at the University of Pittsburgh Molecular Screening Center (PMLSC) as part of the Molecular Library Screening Center Network (MLSCN)(1R03DA24898-01). Protein kinase D (PKD) is a novel family of serine/threonine kinases targeted by diacylglycerol. It regulates many fundamental cell functions including cell proliferation, survival, differentiation and protein trafficking, and plays important roles in pathological conditions such as cardiac hypertrophy and cancer in multiple organ systems. However, the mechanisms underlying these effects of PKD are not clearly understood, and the role of PKD in cancer and other diseases has not been fully defined. This is partly due to the lack of effective pharmacological tools that specifically target PKD in normal cellular processes and in pathological conditions. The immediate goal of this proposal is to demonstrate the feasibility of an IMAP-based HTS fluorescent polarization (FP) assay and its use to identi Cathepsin B mixture HTS dose-response confirmation Screening Center: Penn Center for Molecular Discovery Center Affiliation: University of Pennsylvania Network: Molecular Library Screening Center Network (MLSCN) Assay Provider: Scott Diamond, University of Pennsylvania Grant number: MH076406-01 One of our goals at the Penn Center for Molecular Discovery (PCMD) is to develop capabilities for screening multiple members of target classes, for example cysteine and serine proteases. Many HTS labs focus effort on one target of interest within a class due to resource and time constraints. A few compounds are then tested for selectivity against additional target class members during the hit-to-lead process. Our goal is to test the entire MLSCN compound library against multiple cysteine and serine proteases to obtain a profile of activity against these enzymes classes. This profile may then be used to immediately identify selective compounds during subsequent screening of novel enzyme targets. It may also be possible to identify a subset of t Cathepsin L dose-response testing in the presence of cysteine Screening Center: Penn Center for Molecular Discovery Center Affiliation: University of Pennsylvania Network: Molecular Library Screening Center Network (MLSCN) Assay Provider: Scott Diamond, University of Pennsylvania Grant number: MH076406-01 Human liver cathepsin L (EC 3.4.22.15) is a lysosomal cysteine protease. Recent interest in cathepsin L has been generated by research showing that proteolysis by this enzyme is required for the entry and replication of the SARS and Ebola viruses in human cells. Thus cathepsin L inhibitors have potential as novel anti-viral agents. Cathepsin L inhibitors may also be active against Plasmodium falciparum, the parasite responsible for human malaria. Plasmodium contains cathepsin L-like cysteine proteases known as falcipains that appear to promote virulence of the parasite through haemoglobin digestion and erythrocyte invasion. A high-throughput screen for cathepsin L inhibitors was designed as an end-point assay monitoring the release of the fl Cathepsin L probe #2 dose-response testing Screening Center: Penn Center for Molecular Discovery Center Affiliation: University of Pennsylvania Network: Molecular Library Screening Center Network (MLSCN) Assay Provider: Scott Diamond, University of Pennsylvania Grant number: MH076406-01 Human liver cathepsin L (EC 3.4.22.15) is a lysosomal cysteine protease. Recent interest in cathepsin L has been generated by research showing that proteolysis by this enzyme is required for the entry and replication of the SARS and Ebola viruses in human cells. Thus cathepsin L inhibitors have potential as novel anti-viral agents. Cathepsin L inhibitors may also be active against Plasmodium falciparum, the parasite responsible for human malaria. Plasmodium contains cathepsin L-like cysteine proteases known as falcipains that appear to promote virulence of the parasite through haemoglobin digestion and erythrocyte invasion. A high-throughput screen for cathepsin L inhibitors was designed as an end-point assay monitoring the release of the fl Colorimetric assay for HTS discovery of chemical inhibitors of EphA4 receptor antagonists Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Screening Centers Network (MLSCN) Grant Number: XO1 MH077603-01 Assay Provider: Dr. Elena Pasquale, Sanford-Burnham Medical Research Institute EphA4 is a member of the large Eph family of receptor tyrosine kinases. The signaling ability of EphA4, and the other nine closely related EphA receptors, is activated by binding the six GPI-linked ephrin-A ligands. In addition, EphA4 also binds the three transmembrane ephrin-B ligands, which are the ligands for the other class of Eph receptors, the EphB receptors (EphB1-EphB6). Eph receptor-ephrin interaction requires cell-cell contact because both the receptor and the ligand are membrane-bound. Importantly, signals are generated both through the Eph receptor kinase domain (forward signals) and through signaling molecules associated with the ephrins (reverse Complement C1s ELISA Molecular Library Screening Center Network (MLSCN) Penn Center for Molecular Discovery (PCMD) Assay Provider: Scott L. Diamond, University of Pennsylvania MLSCN Grant: X01-MH076406-01 The classical pathway mediates specific antibody responses. The classical pathway is initiated by the binding of antibodies to cell surface antigens. Subsequent binding of the antibody to complement C1q subunits of C1 result in catalytically active C1s subunits. The two activated C1s subunits are then able to catalyze the assembly of the C3 convertase (complement C4b2a) from complements C2 and C4.(Ref. 1) Assay The high-throughput screen on mixture plates for complement factor C1s inhibitors and single compound IC50 determination of active compounds has been reported earlier (AID 538 and AID 787). In this assay, we use ELISA, which can determine the activity of the classical pathway as a whole, to test the activity of hits from the C1s mixture screen. Materials Human Serum was purchased from Comp Complement factor C1s IC50 from mixture screen Molecular Library Screening Center Network (MLSCN) Penn Center for Molecular Discovery (PCMD) Assay Provider: Dr. Scott L. Diamond, University of Pennsylvania MLSCN Grant: X01-MH076406-01 Complement factor C1s (EC 3.4.21.42) is a trypsin-like serine protease that is activated in one of the first steps in the classical complement cascade. Despite the essential role for the complement cascade in immune defense, unregulated activation leading to acute inflammation and tissue damage has been implicated in many disease states. Under normal conditions the activity of C1s is modulated by its endogenous inhibitor, C1 esterase inhibitor. Pathological conditions lead to excessive activation of C1s; thus a small molecule inhibitor would be useful in the treatment of ischemia-reperfusion injury and other complement-mediated diseases. Assay The high-throughput screen for complement factor C1s inhibitors has been reported earlier (AID 538). The assay consisted of an end-point assay monitoring th Confirmatory Cherry Pick 3 SAR Dose Response Multiplex in TOR pathway GFP-fusion proteins for Saccharomyes cerevisiae, specifically AGP1 University of New Mexico Assay Overview: Assay Support: 1R03 MH086450-01 Project Title: Chemical Screen of TOR pathway GFP fusion proteins in S. cerevisiae Assay Provider: Maggie Werner-Washburne, UNM Screening Center/ PI: UNMCMD/ Larry Sklar Lead Biologist: Jun Chen Chemistry Center/ PI: University of Kansas Specialized Chemistry Center/ Jeff Aube Chemistry Center Lead: Jennifer Golden, Blake Peterson Assay Implementation: Jun Chen, Stephanie Sedillo, Anna Waller, Annette Evangelisti, Cristian Bologa, Oleg Ursu, Mark Carter Assay Background and Significance: The target of rapamycin, TOR, is a ser/thr protein kinase evolutionarily conserved from yeast to man [Wullschleger, et al. 2006]. TOR functions in two distinct protein complexes, TOR complex 1 (TORC1) and TORC2 [Cafferkey, et al. 1993; Stan, et al. 1994]. Curiously, only TOR in TORC1 is bound and inhibited by the lipophilic macrolide rapamycin [Kunz, et al. 1993; Helliwell, et al. 1998; Zhang, et al. 2006]. Although t Confirmatory Cherry Pick 3 SAR Dose Response Multiplex in TOR pathway GFP-fusion proteins for Saccharomyes cerevisiae, specifically CIT2 University of New Mexico Assay Overview: Assay Support: 1R03 MH086450-01 Project Title: Chemical Screen of TOR pathway GFP fusion proteins in S. cerevisiae Assay Provider: Maggie Werner-Washburne, UNM Screening Center/ PI: UNMCMD/ Larry Sklar Lead Biologist: Jun Chen Chemistry Center/ PI: University of Kansas Specialized Chemistry Center/ Jeff Aube Chemistry Center Lead: Jennifer Golden, Blake Peterson Assay Implementation: Jun Chen, Stephanie Sedillo, Anna Waller, Annette Evangelisti, Cristian Bologa, Oleg Ursu, Mark Carter Assay Background and Significance: The target of rapamycin, TOR, is a ser/thr protein kinase evolutionarily conserved from yeast to man [Wullschleger, et al. 2006]. TOR functions in two distinct protein complexes, TOR complex 1 (TORC1) and TORC2 [Cafferkey, et al. 1993; Stan, et al. 1994]. Curiously, only TOR in TORC1 is bound and inhibited by the lipophilic macrolide rapamycin [Kunz, et al. 1993; Helliwell, et al. 1998; Zhang, et al. 2006]. Although th Confirmatory Cherry Pick 3 SAR Dose Response Multiplex in TOR pathway GFP-fusion proteins for Saccharomyes cerevisiae, specifically LAP4 University of New Mexico Assay Overview: Assay Support: 1R03 MH086450-01 Project Title: Chemical Screen of TOR pathway GFP fusion proteins in S. cerevisiae Assay Provider: Maggie Werner-Washburne, UNM Screening Center/ PI: UNMCMD/ Larry Sklar Lead Biologist: Jun Chen Chemistry Center/ PI: University of Kansas Specialized Chemistry Center/ Jeff Aube Chemistry Center Lead: Jennifer Golden, Blake Peterson Assay Implementation: Jun Chen, Stephanie Sedillo, Anna Waller, Annette Evangelisti, Cristian Bologa, Oleg Ursu, Mark Carter Assay Background and Significance: The target of rapamycin, TOR, is a ser/thr protein kinase evolutionarily conserved from yeast to man [Wullschleger, et al. 2006]. TOR functions in two distinct protein complexes, TOR complex 1 (TORC1) and TORC2 [Cafferkey, et al. 1993; Stan, et al. 1994]. Curiously, only TOR in TORC1 is bound and inhibited by the lipophilic macrolide rapamycin [Kunz, et al. 1993; Helliwell, et al. 1998; Zhang, et al. 2006]. Although t Confirmatory Cherry Pick 3 SAR Dose Response Multiplex in TOR pathway GFP-fusion proteins for Saccharomyes cerevisiae, specifically MEP2 University of New Mexico Assay Overview: Assay Support: 1R03 MH086450-01 Project Title: Chemical Screen of TOR pathway GFP fusion proteins in S. cerevisiae Assay Provider: Maggie Werner-Washburne, UNM Screening Center/ PI: UNMCMD/ Larry Sklar Lead Biologist: Jun Chen Chemistry Center/ PI: University of Kansas Specialized Chemistry Center/ Jeff Aube Chemistry Center Lead: Jennifer Golden, Blake Peterson Assay Implementation: Jun Chen, Stephanie Sedillo, Anna Waller, Annette Evangelisti, Cristian Bologa, Oleg Ursu, Mark Carter Assay Background and Significance: The target of rapamycin, TOR, is a ser/thr protein kinase evolutionarily conserved from yeast to man [Wullschleger, et al. 2006]. TOR functions in two distinct protein complexes, TOR complex 1 (TORC1) and TORC2 [Cafferkey, et al. 1993; Stan, et al. 1994]. Curiously, only TOR in TORC1 is bound and inhibited by the lipophilic macrolide rapamycin [Kunz, et al. 1993; Helliwell, et al. 1998; Zhang, et al. 2006]. Although t Confirmatory Cherry Pick 3 SAR Dose Response Multiplex in TOR pathway GFP-fusion proteins for Saccharomyes cerevisiae, specifically RPL19A University of New Mexico Assay Overview: Assay Support: 1R03 MH086450-01 Project Title: Chemical Screen of TOR pathway GFP fusion proteins in S. cerevisiae Assay Provider: Maggie Werner-Washburne, UNM Screening Center/ PI: UNMCMD/ Larry Sklar Lead Biologist: Jun Chen Chemistry Center/ PI: University of Kansas Specialized Chemistry Center/ Jeff Aube Chemistry Center Lead: Jennifer Golden, Blake Peterson Assay Implementation: Jun Chen, Stephanie Sedillo, Anna Waller, Annette Evangelisti, Cristian Bologa, Oleg Ursu, Mark Carter Assay Background and Significance: The target of rapamycin, TOR, is a ser/thr protein kinase evolutionarily conserved from yeast to man [Wullschleger, et al. 2006]. TOR functions in two distinct protein complexes, TOR complex 1 (TORC1) and TORC2 [Cafferkey, et al. 1993; Stan, et al. 1994]. Curiously, only TOR in TORC1 is bound and inhibited by the lipophilic macrolide rapamycin [Kunz, et al. 1993; Helliwell, et al. 1998; Zhang, et al. 2006]. Although t Counter Screen using XIAP-Bir3 of the Chemical Antagonists of IAP-family anti-apoptotic proteins confirmation assay Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Screening Centers Network (MLSCN) Grant Proposal Number: MH081277-01 Assay Provider: John C. Reed, Sanford-Burnham Medical Research Institute, San Diego, CA This dose response assay is developed and performed as a counter screen to compounds in the Chemical Antagonists of IAP-family anti-apoptotic proteins confirmation (AID 1449) and to study the structure-activity relationship on analogs of the confirmed hits. Compounds are either acquired from commercial sources or synthesized internally. Apoptosis plays an essential role in many aspects of normal development and physiology, becoming dysregulated in myriad diseases characterized by insufficient or excessive cell death. Caspases are intracellular proteases that are suppressed by Inhibitor of Apoptosis Proteins (IAPs), a family of evolutionarily conse Counter screen SAR assay for PMM2 inhibitors via a fluorescence intensity assay Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Probe Production Centers Network (MLPCN) Grant Number: R03 MH082386-01 Assay Provider: Dr. Hudson H. Freeze, Sanford-Burnham Medical Research Institute, San Diego, CA Congenital Disorders of Glycosylation (CDGs) are rare genetic disorders in the synthesis of N-linked glycan chains. Mutations in PMM2, encoding phosphomannomutase 2 (PMM2, Man-6-P-> Man-1-P) cause the most common form, CDG-Ia. Patients have a host of problems including hypotonia, variable psychomotor retardation, seizures, peripheral neuropathy, cardiomyopathy, and protein losing enteropathy. There is no therapy for this disorder. A current approach to ameliorate the physiological conditions associated with CDG-Ia is to provide high influx of mannose for patience. We previously developed a HTS assay through the MLSCN to identify inhibitors of phos Counter screen for S1P2 Agonists: Dose Response High Throughput Cell-Based Screen to Identify Activators of CRE-BLA Source (MLSCN Center Name): The Scripps Research Institute Molecular Screening Center Center Affiliation: The Scripps Research Institute (TSRI) Assay Provider: Hugh Rosen, TSRI Grant Proposal Number: 1 R03 MH076533-01 Grant Proposal PI: Germana Sanna External Assay ID: Counterscreen for S1P2 Agonists: Dose Response High Throughput Cell-Based Screen to Identify Activators of CRE-BLA Name: CRE_AG_BLA_384_EC50_%ACT Description: Sphingosine 1-phosphate (S1P) influences heart rate [1,2], coronary artery caliber, endothelial integrity, lung epithelial integrity [3] and lymphocyte recirculation [2,4] through five related high affinity G-protein coupled receptors [5]. Subtype-selective modulators of S1P receptors will be of broad utility in understanding cell functions in vitro and vascular physiology in vivo, as well as de-convoluting the role of individual subtypes in cellular processes. The S1P receptor 2 (S1P2), also known as endothelial differentiation sphingolipid G-protein-coup Counterscreen assay for PERK inhibitors: Dose response cell-based high throughput screening assay to measure inhibition of PERK at 6 hours Source (MLPCN Center Name): The Scripps Research Institute Molecular Screening Center (SRIMSC) Center Affiliation: The Scripps Research Institute (TSRI) Assay Provider: David Ron, New York University Network: Molecular Libraries Probe Production Centers Network (MLPCN) Grant Proposal Number R03 MH082370-01 Grant Proposal PI: David Ron, New York University External Assay ID: PERK_INH_SEAP_1536_EC50 6HR CS Name: Counterscreen assay for PERK inhibitors: Dose response cell-based high throughput screening assay to measure inhibition of PERK at 6 hours Description: The endoplasmic reticulum (ER) is essential for protein synthesis and folding (1, 2). In response to environmental stress, misfolded proteins accumulate in the ER lumen, leading to activation of a set of signaling cascades known as the unfolded protein response (UPR). In eukaryotic cells the UPR reduces the burden of misfolded proteins by inhibiting protein synthesis and increasing clearance of misfolded proteins (3, 4). The Counterscreen assay for inhibitors of Wee1 degradation: dose response cell-based assay to identify inhibitors of cyclin B degradation Source (MLPCN Center Name): The Scripps Research Institute Molecular Screening Center Center Affiliation: The Scripps Research Institute (TSRI) Assay Provider: Nagi Ayad, TSRI Network: Molecular Libraries Probe Production Centers Network (MLPCN) Grant Proposal Number: 1R21NS056991-01 Grant Proposal PI: Nagi Ayad, TSRI External Assay ID: cyclinBDegradation_ACT_LUMI_1536_EC50 Name: Counterscreen assay for inhibitors of Wee1 degradation: dose response cell-based assay to identify inhibitors of cyclin B degradation Description: Cell cycle progression and entry into mitosis are regulated by a highly conserved cellular process known as checkpoint signaling (1-4). The Wee1 nuclear tyrosine kinase functions in this process by regulating the cdc2/cyclin B protein complex. Specifically, Wee1 mediates inhibitory phosphorylation of cdc2, leading to delayed mitosis and cell cycle arrest in cells with DNA damage so that DNA repair and replication can occur (1-4). Wee1 activity is inhibited du Counterscreen for S1P2 Agonists: Dose Response High Throughput Cell-Based Screen to Identify Activators of CRE-BLA: S1P2 Purchased Analogues Source (MLSCN Center Name): The Scripps Research Institute Molecular Screening Center Center Affiliation: The Scripps Research Institute (TSRI) Assay Provider: Hugh Rosen, TSRI Grant Proposal Number: 1 R03 MH076533-01 Grant Proposal PI: Germana Sanna External Assay ID: CRE_AG_BLA_384_EC50_S1P2_Purchased_Analogues Name: Counterscreen for S1P2 Agonists: Dose Response High Throughput Cell-Based Screen to Identify Activators of CRE-BLA: S1P2 Purchased Analogues Description: Sphingosine 1-phosphate (S1P) influences heart rate [1,2], coronary artery caliber, endothelial integrity, lung epithelial integrity [3] and lymphocyte recirculation [2,4] through five related high affinity G-protein coupled receptors [5]. Subtype-selective modulators of S1P receptors will be of broad utility in understanding cell functions in vitro and vascular physiology in vivo, as well as de-convoluting the role of individual subtypes in cellular processes. The S1P receptor 2 (S1P2), also known as endothelial Counterscreen for activators of the nuclear receptor Steroidogenic Factor 1 (SF-1): A cell-based dose-response assay for inhibition of the RAR-related orphan receptor A (RORA) Source (MLSCN Center Name): The Scripps Research Institute Molecular Screening Center Center Affiliation: The Scripps Research Institute, TSRI Assay Provider: Orphagen Pharmaceuticals, San Diego, CA Network: Molecular Library Screening Center Network (MLSCN) Grant Proposal Number: 1X01-MH077624-01 External Assay ID: RORA_AG_LUMI_1536_CS_EC50 Name: Counterscreen for activators of the nuclear receptor Steroidogenic Factor 1 (SF-1): A cell-based dose-response assay for inhibition of the RAR-related orphan receptor A (RORA) Description: Nuclear receptors are a family of small molecule and hormone-regulated transcription factors that share conserved DNA-binding and ligand-binding domains. Small pharmacological compounds able to bind to the cleft of the ligand-binding domain could alter its conformation and subsequently modify transcription of target genes. Such ligand agonists and/or antagonists have already been successfully designed for 23 nuclear receptors among the 48 previousl Counterscreen for exosite inhibitors of ADAM10: QFRET-based biochemical high throughput dose response assay to identify inhibitors of ADAM17 Source (MLPCN Center Name): The Scripps Research Institute Molecular Screening Center (SRIMSC) Center Affiliation: Torrey Pines Institute for Molecular Sciences (TPIMS) Assay Provider: Dmitriy Minond, Torrey Pines Institute for Molecular Sciences (TPIMS) Network: Molecular Libraries Probe Production Centers Network (MLPCN) Grant Proposal Number: 1 R03 DA033985-01 Grant Proposal PI: Dmitriy Minond, Torrey Pines Institute for Molecular Sciences (TPIMS) External Assay ID: ADAM17_INH_QFRET_1536_3IC50 DCSRUN for ADAM10_INH Name: Counterscreen for exosite inhibitors of ADAM10: QFRET-based biochemical high throughput dose response assay to identify inhibitors of ADAM17 Description: Approximately 20-30% of breast cancer patients have tumors that over-express human epidermal growth factor receptor (HER2), which confers an aggressive tumor phenotype and poor prognosis [1-3]. A Disintegrin and Metalloprotease (ADAM) proteases are responsible for amplification of HER2 signaling due to either c Counterscreen for exosite inhibitors of ADAM17: Fluorescence resonance energy transfer (FRET)-based biochemical high throughput dose response assay to identify inhibitors of ADAM10 Source (MLPCN Center Name): The Scripps Research Institute Molecular Screening Center (SRIMSC) Center Affiliation: Torrey Pines Institute for Molecular Sciences (TPIMS) Assay Provider: Dmitriy Minond, Torrey Pines Institute for Molecular Sciences (TPIMS) Network: Molecular Libraries Probe Production Centers Network (MLPCN) Grant Proposal Number: 1 R03 DA033985-01 Grant Proposal PI: Dmitriy Minond, Torrey Pines Institute for Molecular Sciences (TPIMS) External Assay ID: ADAM10_INH_QFRET_1536_3XIC50 INH DCSRUN for ADAM17 INH Name: Counterscreen for exosite inhibitors of ADAM17: Fluorescence resonance energy transfer (FRET)-based biochemical high throughput dose response assay to identify inhibitors of ADAM10. Description: Approximately 20-30% of breast cancer patients have tumors that over-express human epidermal growth factor receptor (HER2), which confers an aggressive tumor phenotype and poor prognosis [1-3]. A Disintegrin and Metalloprotease (ADAM) proteases are responsible for a Counterscreen for inhibitors of EBNA-1: fluorescence polarization-based biochemical high throughput dose response assay to identify inhibitors of the Epstein-Barr virus-encoded protein, ZTA Source (MLPCN Center Name): The Scripps Research Institute Molecular Screening Center (SRIMSC) Center Affiliation: The Scripps Research Institute (TSRI) Assay Provider: Paul Lieberman, Wistar Institute Network: Molecular Libraries Probe Production Centers Network (MLPCN) Grant Proposal Number: I R21 NS063906-01 Grant Proposal PI: Paul Lieberman, Wistar Institute External Assay ID: ZTA_INH_FP_1536_3XIC50 EBNA DRUN CS Name: Counterscreen for inhibitors of EBNA-1: fluorescence polarization-based biochemical high throughput dose response assay to identify inhibitors of the Epstein-Barr virus-encoded protein, ZTA. Description: During each cell cycle in eukaryotes, the genome must be completely replicated and this replication must begin at the correct time and site (initiation site or origin) (1). Pathogenic viruses often take advantage of this cellular precision to maintain replication of their own genome. The Epstein-Barr virus (EBV) is an orally-transmitted herpesvirus associated with Counterscreen for inhibitors of M1 and M17 aminopeptidases: QFRET-based biochemical high throughput dose response assay for inhibitors of the Cathepsin L proteinase (CTSL1). Source (MLPCN Center Name): The Scripps Research Institute Molecular Screening Center Affiliation: The Scripps Research Institute, TSRI Assay Provider: John Dalton and Donald Gardiner, Queensland Institute of Medical Research, Australia Network: Molecular Library Probe Production Centers Network (MLPCN) Grant Proposal Number: 1 R03 MH084103-01 Grant Proposal PI: John Dalton and Donald Gardiner, Queensland Institute of Medical Research, Australia External Assay ID: CTSL1_INH_QFRET_1536_3XIC50 CSDRUN Name: Counterscreen for inhibitors of M1 and M17 aminopeptidases: QFRET-based biochemical high throughput dose response assay for inhibitors of the Cathepsin L proteinase (CTSL1). Description: Aminopeptidases (APs) are metalloproteases that cleave amino-terminal (N-terminal) amino acids during protein synthesis (1, 2) These enzymes are characterized in part by their post-translational removal of leucine, aspartate, proline, methionine, etc from proteins and peptides, in order that protei Counterscreen for inhibitors of MCL1: fluorescence polarization-based biochemical high throughput dose response assay for inhibitors of BCL2-related protein, long isoform (BCLXL). Source (MLPCN Center Name): The Scripps Research Institute Molecular Screening Center Affiliation: The Scripps Research Institute, TSRI Assay Provider: Michael Cardone, Eutropics Network: Molecular Library Probe Production Centers Network (MLPCN) Grant Proposal Number: 1 R43 CA135915-01 Fast Track Grant Proposal PI: Michael Cardone, Eutropics External Assay ID: BCLXLBIM_INH_FP_1536_3XIC50 Name: Counterscreen for inhibitors of MCL1: fluorescence polarization-based biochemical high throughput dose response assay for inhibitors of BCL2-related protein, long isoform (BCLXL). Description: Cancer initialization and survival depends upon evasion of the programmed cell death (apoptosis) machinery that normally kills an unneeded or rogue cell (1). Although an effective mechanism for anti-cancer chemotherapeutics is apoptosis induction, cancer cells develop resistance to the pro-apoptotic proteins activated by these drugs (2). Multiple myeloma (MM) and chronic lymphoblastic leukemia (CLL) ar Counterscreen for inhibitors of gld-1: Fluorescence polarization-based biochemical high throughput dose response assay for inhibitors of the HIV Rev protein-RRE RNA interaction Source (MLPCN Center Name): The Scripps Research Institute Molecular Screening Center (SRIMSC) Affiliation: The Scripps Research Institute, TSRI Assay Provider: James R. Williamson, TSRI Network: Molecular Library Probe Production Centers Network (MLPCN) Grant Proposal Number: 1 R21 NS056951-01 Grant Proposal PI: James R. Williamson External Assay ID: HIVREVRRE_INH_FRET_1536_3XIC50 GLD1 DCSRUN Name: Counterscreen for inhibitors of gld-1: Fluorescence polarization-based biochemical high throughput dose response assay for inhibitors of the HIV Rev protein-RRE RNA interaction. Description: Post-transcriptional control of gene expression, such as alternative splicing and tissue-specific silencing, allow for great protein diversity (1). The elements in the mRNA 3' untranslated regions (UTRs) influence the expression of genes involved in proliferation and differentiation of stem cells and germ cells (2). These elements are critical during spermatogenesis and oogenesis in hermaphrodi Counterscreen for inhibitors of tRNA 2'-phosphotransferase (TPT1): fluorescence polarization-based biochemical high throughput dose response assay to identify inhibitors of RNAse T1. Source (MLPCN Center Name): The Scripps Research Institute Molecular Screening Center (SRIMSC) Center Affiliation: The Scripps Research Institute (TSRI) Assay Provider: Heather Harding, New York University School of Medicine Network: Molecular Libraries Probe Production Centers Network (MLPCN) Grant Proposal Number: 1-R03-DA026554-01 Grant Proposal PI: Heather Harding, New York University School of Medicine External Assay ID: RNASETI _INH_FP_1536_3XIC50 Name: Counterscreen for inhibitors of tRNA 2'-phosphotransferase (TPT1): fluorescence polarization-based biochemical high throughput dose response assay to identify inhibitors of RNAse T1. Description: The process of transcription converts the DNA sequences found in genes into mRNA (1, 2). This process is coupled to the subsequent removal of mRNA introns by splicing, which additionally serves to increase proteome complexity (3, 4). Intron splicing also occurs for transfer RNA (tRNA), which functions in the delivery of amino acid Counterscreen for inhibitors of the Retinoic Acid Receptor-related orphan receptor A (RORA): A cell-based dose-response assay for inhibition of the Steroidogenic Factor 1 (SF-1) Source (MLSCN Center Name): The Scripps Research Institute Molecular Screening Center Center Affiliation: The Scripps Research Institute, TSRI Assay Provider: Orphagen Pharmaceuticals, San Diego, CA Network: Molecular Library Screening Center Network (MLSCN) Proposal number 1X01-MH077624-01 External Assay ID: SF1_INH_Lumi_1536_CS_IC50 Name: Counterscreen for inhibitors of the Retinoic Acid Receptor-related orphan receptor A (RORA): A cell-based dose-response assay for inhibition of the Steroidogenic Factor 1 (SF-1) Description: Nuclear receptors are a family of small molecule and hormone-regulated transcription factors that share conserved DNA-binding and ligand-binding domains. Small pharmacological compounds able to bind to the cleft of the ligand-binding domain could alter its conformation and subsequently modify transcription of target genes. Such ligand agonists and/or antagonists have already been successfully designed for 23 nuclear receptors among the 48 previously ident Counterscreen for inhibitors of the nuclear receptor Steroidogenic Factor 1 (SF-1): A cell-based dose-response assay for inhibition of the RAR-related orphan receptor A (RORA) Source (MLSCN Center Name): The Scripps Research Institute Molecular Screening Center Center Affiliation: The Scripps Research Institute, TSRI Assay Provider: Orphagen Pharmaceuticals, San Diego, CA Network: Molecular Library Screening Center Network (MLSCN) Grant Proposal number 1X01-MH077624-01 External Assay ID: RORA_INH_Lumi_1536_CS_IC50 Name: Counterscreen for inhibitors of the nuclear receptor Steroidogenic Factor 1 (SF-1): A cell-based dose-response assay for inhibition of the RAR-related orphan receptor A (RORA) Description: Nuclear receptors are a family of small molecule and hormone-regulated transcription factors that share conserved DNA-binding and ligand-binding domains. Small pharmacological compounds able to bind to the cleft of the ligand-binding domain could alter its conformation and subsequently modify transcription of target genes. Such ligand agonists and/or antagonists have already been successfully designed for 23 nuclear receptors among the 48 previously Discovery of novel allosteric modulators of the M1 muscarinic receptor: Agonist Activity against Muscarinic Panel Assay Provider: P. Jeffrey Conn Assay Provider Affiliation: Vanderbilt University Grant Title: Discovery of novel allosteric modulators of the M1 muscarinic receptor Grant Number: 1 R03 MH077606-01 The M1 muscarinic receptor is thought to be an important therapeutic target in schizophrenia. A cell-based fluorometric calcium assay was developed for high throughput screening. This assay was used to identify compounds with high selectivity for the M1 receptor subtype that act at an allosteric site on the receptor, thus providing increased specificity for this single receptor subtype. It is anticipated that these compounds will provide important tools for the study of muscarinic receptor function in the CNS. Agents that enhance cholinergic transmission or activate muscarinic acetylcholine receptors (mAChRs) have been developed to ameliorate the loss of cognitive function in patients with Alzheimer's Disease (AD). While cholinergic agents have been partially successful in improving c Discovery of novel allosteric modulators of the M1 muscarinic receptor: Agonist NMS binding at M1 Assay Provider: P. Jeffrey Conn Assay Provider Affiliation: Vanderbilt University Grant Title: Discovery of novel allosteric modulators of the M1 muscarinic receptor Grant Number: 1 R03 MH077606-01 The M1 muscarinic receptor is thought to be an important therapeutic target in schizophrenia. A cell-based fluorometric calcium assay was developed for high throughput screening. This assay was used to identify compounds with high selectivity for the M1 receptor subtype that act at an allosteric site on the receptor, thus providing increased specificity for this single receptor subtype. It is anticipated that these compounds will provide important tools for the study of muscarinic receptor function in the CNS. Agents that enhance cholinergic transmission or activate muscarinic acetylcholine receptors (mAChRs) have been developed to ameliorate the loss of cognitive function in patients with Alzheimer's Disease (AD). While cholinergic agents have been partially successful in improving c Discovery of novel allosteric modulators of the M1 muscarinic receptor: Agonist NMS competition at M2 Assay Provider: P. Jeffrey Conn Assay Provider Affiliation: Vanderbilt University Grant Title: Discovery of novel allosteric modulators of the M1 muscarinic receptor Grant Number: 1 R03 MH077606-01 The M1 muscarinic receptor is thought to be an important therapeutic target in schizophrenia. A cell-based fluorometric calcium assay was developed for high throughput screening. This assay was used to identify compounds with high selectivity for the M1 receptor subtype that act at an allosteric site on the receptor, thus providing increased specificity for this single receptor subtype. It is anticipated that these compounds will provide important tools for the study of muscarinic receptor function in the CNS. Agents that enhance cholinergic transmission or activate muscarinic acetylcholine receptors (mAChRs) have been developed to ameliorate the loss of cognitive function in patients with Alzheimer's Disease (AD). While cholinergic agents have been partially successful in improving c Discovery of novel allosteric modulators of the M1 muscarinic receptor: Agonist NMS competition at M3 Assay Provider: P. Jeffrey Conn Assay Provider Affiliation: Vanderbilt University Grant Title: Discovery of novel allosteric modulators of the M1 muscarinic receptor Grant Number: 1 R03 MH077606-01 The M1 muscarinic receptor is thought to be an important therapeutic target in schizophrenia. A cell-based fluorometric calcium assay was developed for high throughput screening. This assay was used to identify compounds with high selectivity for the M1 receptor subtype that act at an allosteric site on the receptor, thus providing increased specificity for this single receptor subtype. It is anticipated that these compounds will provide important tools for the study of muscarinic receptor function in the CNS. Agents that enhance cholinergic transmission or activate muscarinic acetylcholine receptors (mAChRs) have been developed to ameliorate the loss of cognitive function in patients with Alzheimer's Disease (AD). While cholinergic agents have been partially successful in improving c Discovery of novel allosteric modulators of the M1 muscarinic receptor: Agonist NMS competition at M4 Assay Provider: P. Jeffrey Conn Assay Provider Affiliation: Vanderbilt University Grant Title: Discovery of novel allosteric modulators of the M1 muscarinic receptor Grant Number: 1 R03 MH077606-01 The M1 muscarinic receptor is thought to be an important therapeutic target in schizophrenia. A cell-based fluorometric calcium assay was developed for high throughput screening. This assay was used to identify compounds with high selectivity for the M1 receptor subtype that act at an allosteric site on the receptor, thus providing increased specificity for this single receptor subtype. It is anticipated that these compounds will provide important tools for the study of muscarinic receptor function in the CNS. Agents that enhance cholinergic transmission or activate muscarinic acetylcholine receptors (mAChRs) have been developed to ameliorate the loss of cognitive function in patients with Alzheimer's Disease (AD). While cholinergic agents have been partially successful in improving c Dose Response Cell Based Assay for Agonists of the S1P2 Receptor Source (MLSCN Center Name): The Scripps Research Institute Molecular Screening Center Center Affiliation: The Scripps Research Institute (TSRI) Assay Provider: Hugh Rosen, TSRI Grant Proposal Number: 1 R03 MH076533-01 Grant Proposal PI: Germana Sanna External Assay ID: S1P2_AG_BLA_384_EC50 Name: Dose Response Cell Based Assay for Agonists of the S1P2 Receptor Description: Sphingosine 1-phosphate (S1P) influences heart rate [1, 2], coronary artery caliber, endothelial integrity, lung epithelial integrity [3] and lymphocyte recirculation [2, 4] through five related high affinity G-protein coupled receptors [5]. Subtype-selective modulators of S1P receptors will be of broad utility in understanding cell functions in vitro and vascular physiology in vivo, as well as de-convoluting the role of individual subtypes in cellular processes. The S1P receptor 2 (S1P2), also known as endothelial differentiation sphingolipid G-protein-coupled receptor 5 (EDG5), signals through Gi, Gq and G1 Dose Response Cell Based Assay for Agonists of the S1P2 Receptor of Purchased Analogues Source (MLSCN Center Name): The Scripps Research Institute Molecular Screening Center Center Affiliation: The Scripps Research Institute (TSRI) Assay Provider: Hugh Rosen, TSRI Grant Proposal Number: 1 R03 MH076533-01 Grant Proposal PI: Germana Sanna External Assay ID: S1P2_AG_BLA_384_EC50_Purchased_Analogues Name: Dose Response Cell Based Assay for Agonists of the S1P2 Receptor of Purchased Analogues Description: Sphingosine 1-phosphate (S1P) influences heart rate [1, 2], coronary artery caliber, endothelial integrity, lung epithelial integrity [3] and lymphocyte recirculation [2, 4] through five related high affinity G-protein coupled receptors [5]. Subtype-selective modulators of S1P receptors will be of broad utility in understanding cell functions in vitro and vascular physiology in vivo, as well as de-convoluting the role of individual subtypes in cellular processes. The S1P receptor 2 (S1P2), also known as endothelial differentiation sphingolipid G-protein-coupled recepto Dose Response Confirmation Screen via Multiplex HTS Assay for Inhibitors of MEK Kinase PB1 Domains, specifically MEK5 binding to MEK Kinase 2 Mutant University of New Mexico Assay Overview: Assay Support: 1R03MH084830-01 Project Title: TR-FRET HTS Assay for Inhibitors of MEKK2-MEK5 PB1 Domain Interaction PI: Kazuhiro Nakamura, Ph.D Center PI: Larry Sklar, Ph.D Assay Implementatiion: Zurab Surviladze Ph.D, Mark Haynes Ph.D, Anna Waller Ph.D, Mark Carter MS Assay Background and Significance: PB1 (Phox/Bem1p) domains function as protein-protein interaction sites by forming PB1-PB1 domain heterodimers (Moscat, et al. 2006). There are at least 20 human PB1 domain-containing proteins. Different PB1 domains contribute to the formation of specific protein complexes critical for biological responses including proliferation, apoptosis, cell polarity, and angiogenesis. These proteins include the mitogen/extracellular signal regulated kinase kinases MEKK2 and MEKK3 (MAP3Ks), as well as the downstream regulated kinase MEK5 (a MAP2K), which solely govern the ERK5 MAPK pathway involved in angiogenesis, cell growth and inhibition of apo Dose Response Confirmation for Small Molecule Inhibitors of Epstein-Barr Virus NIH Molecular Libraries Screening Centers Network [MLSCN] Emory Chemical Biology Discovery Center in MLSCN Assay provider: Theodore Jardetzky; Northwestern University MLSCN Grant: 1R21NS059415-01 Epstein-Barr virus (EBV), or human herpes virus 4 (HHV-4), is a member of the larger herpesvirus family that consists of three subfamilies (##, ##, ##). Epstein-Barr virus (EBV) is an extremely prevalent human herpesvirus. Disease syndromes in humans caused by EBV reflect the cell types that EBV infects, which are primarily of lymphoid or epithelial origin. Infection of both cell types is associated with a variety of proliferative disorders and cancers. Current treatments for such malignancies are traditional chemotherapy, radiation-therapy, surgery, and if possible, restoration of immune system function. Like all herpesviruses, EBV has mechanisms to evade the immune system and maintain latency in the human host. A treatment directed specifically against EBV could significantly improve patie Dose Response Confirmation for Small Molecule Inhibitors of Eukaryotic Translation Initiation Dose Response Confirmation for Small Molecule Inhibitors of Eukaryotic Translation Initiation NIH Molecular Libraries Screening Centers Network [MLSCN] Emory Chemical Biology Discovery Center in MLSCN Assay provider: Dr. Jerry Pelletier, McGill UNIVERSITY MLSCN Grant: 1 R03 MH081216-01 Title: Dose Response Confirmation for Small Molecule Inhibitors of Eukaryotic Translation Initiation Assay Overview The recruitment of the 40S ribosomal subunit and associated factors (43S pre-initiation complex) to the mRNA during translation initiation is highly regulated by eukaryotic initiation factor (eIF) 4F. This complex consists of three subunits: (i) eIF4E, the cap-binding protein responsible for binding of the complex to the mRNA cap structure; (ii) eIF4A, an RNA helicase thought to be required to unwind 5' proximal local mRNA secondary structure to facilitate access of the 43S ribosomal complex to the mRNA template; and (iii) eIF4G, a modular scaffold that mediates mRNA binding to the Dose Response Confirmation for small molecular inhibitors for p47phox, a regulatory protein of NADPH oxidases (Noxs) NIH Molecular Libraries Screening Centers Network [MLSCN] Emory Chemical Biology Discovery Center in MLSCN Assay provider: Susan Smith, Emory University MLSCN Grant: MH083234-01 Oxidative stress (the excess production of cellular oxidizing substances) is a central component in many diseases. Reactive oxygen species (ROS) produce oxidative stress that plays a central role in inflammation in general, and in the tissue damage and abnormal cell growth and fibrosis associated with many diseases. ROS-associated diseases often represent chronic conditions, and are frequently associated with tissue damage, fibrosis, and in some cases probable genetic damage. Numerous examples of ROS-associated diseases have been identified, and include diseases and pathologies of the cardiovascular system, nervous system, endocrine system, respiratory system, excretory system, and others. Nox enzymes provide most of the ROS in these conditions. Noxs, particularly Nox2 and Nox1, confer the major source of oxi Dose Response Confirmation via Multiplex HTS Assay for Inhibitors of MEK Kinase PB1 Domains, specifically MEK5 binding to MEK Kinase 2 Wildtype University of New Mexico Assay Overview: Assay Support: 1R03MH084830-01 Project Title: TR-FRET HTS Assay for Inhibitors of MEKK2-MEK5 PB1 Domain Interaction PI: Kazuhiro Nakamura, Ph.D Center PI: Larry Sklar, Ph.D Assay Implementatiion: Zurab Surviladze Ph.D, Mark Haynes Ph.D, Anna Waller Ph.D, Mark Carter MS Assay Background and Significance: PB1 (Phox/Bem1p) domains function as protein-protein interaction sites by forming PB1-PB1 domain heterodimers (Moscat, et al. 2006). There are at least 20 human PB1 domain-containing proteins. Different PB1 domains contribute to the formation of specific protein complexes critical for biological responses including proliferation, apoptosis, cell polarity, and angiogenesis. These proteins include the mitogen/extracellular signal regulated kinase kinases MEKK2 and MEKK3 (MAP3Ks), as well as the downstream regulated kinase MEK5 (a MAP2K), which solely govern the ERK5 MAPK pathway involved in angiogenesis, cell growth and inhibition of apo Dose Response cell-based high-throughput screening assay to identify agonists of galanin receptor 2 (GALR2) Source (MLSCN Center Name): The Scripps Research Institute Molecular Screening Center Affiliation: The Scripps Florida Research Institute, TSRI Assay Provider: Steven Brown, TSRI Network: Molecular Library Screening Center Network (MLSCN) Grant Proposal Number: 1 R21 NS057101-01 Grant Proposal PI: Steven Brown External Assay ID: GALR2_AG_BLA_1536_EC50 Name: Dose Response cell-based high-throughput screening assay to identify agonists of galanin receptor 2 (GALR2) Description: Galanin, a 29 amino acid neuropeptide (30 residues in humans), is cleaved from preprogalanin and is involved in many physiological processes including nervous system development, feeding, metabolism and reproduction, and regulation of neurotransmitter and hormone release [1, 2]. The physiologic response to galanin is mediated in part by three G protein-coupled metabotropic 7-transmembrane receptor subtypes, GALR1, GALR2 and GALR3. These receptors are expressed throughout the peripheral and central nervous sys Dose Response concentration confirmation of uHTS hits from a small molecule activators of human intestinal alkaline phosphatase via a luminescent assay - Set 2 Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Production Centers Network (MLPCN) Grant Proposal Number: X01-MH077602-01 Assay Provider Dr. Jose Luis Millan, Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Alkaline phosphatase (EC 3.1.3.1) (APs) catalyze the hydrolysis of phosphomonoesters, releasing inorganic phosphate and alcohol. APs are dimeric enzymes found in most organisms. In human, four isozymes of APs have been identified. One isozyme is tissue-nonspecific (designated TNAP) and three other isozymes are tissue-specific and named according to the tissue of their predominant expression: intestinal (IAP), placental (PLAP) and germ cell (GCAP) alkaline phosphatases. IAP expression is largely restricted to the gut, especially to the epithelial cells (enterocytes) of the small intestinal mucosa. The exact biological function of Dose Response concentration confirmation of uHTS hits from a small molecule activators of human intestinal alkaline phosphatase via a luminescent assay Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Production Centers Network (MLPCN) Grant Proposal Number: X01-MH077602-01 Assay Provider Dr. Jose Luis Millan, Sanford-Burnham Medical Research Institute(SBMRI, San Diego, CA) Alkaline phosphatase (EC 3.1.3.1) (APs) catalyze the hydrolysis of phosphomonoesters, releasing inorganic phosphate and alcohol. APs are dimeric enzymes found in most organisms. In human, four isozymes of APs have been identified. One isozyme is tissue-nonspecific (designated TNAP) and three other isozymes are tissue-specific and named according to the tissue of their predominant expression: intestinal (IAP), placental (PLAP) and germ cell (GCAP) alkaline phosphatases. IAP expression is largely restricted to the gut, especially to the epithelial cells (enterocytes) of the small intestinal mucosa. The exact biological function of Dose Response concentration confirmation of uHTS hits from a small molecule activators of mouse intestinal alkaline phosphatase via a luminescent assay Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Production Centers Network (MLPCN) Grant Proposal Number: X01-MH077602-01 Alkaline phosphatase (EC 3.1.3.1) (APs) catalyze the hydrolysis of phosphomonoesters, releasing inorganic phosphate and alcohol. APs are dimeric enzymes found in most organisms. In human, four isozymes of APs have been identified. One isozyme is tissue-nonspecific (designated TNAP) and three other isozymes are tissue-specific and named according to the tissue of their predominant expression: intestinal (IAP), placental (PLAP) and germ cell (GCAP) alkaline phosphatases. IAP expression is largely restricted to the gut, especially to the epithelial cells (enterocytes) of the small intestinal mucosa. The exact biological function of IAP is unknown. The goal of this HTS is to confirm hits in "uHTS Luminescent assay for identificatio Dose Response confirmation of HTS hits from an HePTP Fluorescent Assay using OMFP substrate - Set 2 Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Probe Production Centers Network (MLPCN) Grant Number: XO1 MH077603-01 Assay Provider: Dr. Tomas Mustelin, Sanford-Burnham Medical Research Institute Protein tyrosine phosphatases (PTPs), working with protein tyrosine kinases (PTKs), control the phosphorylation state of many proteins in the signal transduction pathways. HePTP is a tyrosine phosphatase expressed in hematopoietic cells and regulates the MAP kinases Erk and p38. It has been found that HePTP is often dysregualted in the preleukemic disorder myelodysplastic syndrome, as well as in acute myelogeneous leukemia. Small molecule inhibitors of HePTP will be useful as molecular probes for studying the mechanism of signal transduction and MAP kinase regulation, and may have therapeutic potential for the treatment of hematopoietic malignancies. Th Dose Response confirmation of Inhibitors of Mdm2/MdmX interaction in luminescent format Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego CA) Network: NIH Molecular Libraries Probe Production Centers Network (MLPCN) Grant Number: R03 MH089489-01 Assay Provider: Dr. Geoffrey M. Wahl, Salk Institute for Biological Studies, San Diego, CA A wild type but attenuated p53 is retained in approximately 50% of human tumors, and reactivation of p53 in such tumors is an attractive chemotherapeutic strategy. p53 activity is restricted in vivo by mdm2 and mdmx, and knockout of either of these proteins is embryonic lethal in a p53-dependent manner (1, 2). Both proteins bind to p53 via a hydrophobic N-terminal pocket and block p53-dependent transcription of genes required for tumor suppression. Efforts to reactivate p53 with small molecules have focused on inhibition of the mdm2/p53 interaction, which leads to increased p53 levels and activity. However, recent reports indicate that targetin Dose Response confirmation of UBC13 Polyubiquitin Inhibitors using a Bfl-1 counterscreen Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego CA) Network: NIH Molecular Libraries Probe Production Centers Network (MLPCN) Grant Number: 1R03 MH085677-01 Assay Provider: Dr. John C. Reed, Sanford-Burnham Medical Research Institute, San Diego CA Tumor Necrosis Factor Receptor-Associated Factors (TRAFs) are a family of adapter proteins that bind an unusual ubiquitin-conjugating enzyme, Ubc13, which produces polyubiquitin chains linked at lysine 63 of ubiquitin. These lysine 63-linked ubiquitin polymers trigger changes in protein activity. Ubiquitination by Ubc13 of TRAFs and the various protein kinases to which TRAFs bind is recognized as a critical step in signaling by TNFRs, TLRs, NLRs, and T-cell and B-cell antigen receptors (TCR/BCR) during innate and acquired immune responses. Since aberrant signaling by these receptor systems is linked to a wide variety of autoimmune, inflammato Dose Response confirmation of inhibitors of Sentrin-specific proteases (SENPs) using a Caspase-3 Selectivity assay Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Probe Production Centers Network(MLPCN) Grant Proposal Number: 1R21 NS061758-01 fast track Assay Provider: Dr. Guy Salvesen, Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Modification of proteins by SUMO is a dynamic and reversible process. SUMOylation/deSUMOylation cycle regulates SUMOs function. Sentrin-specific proteases (SENPs) are involved in both the maturation of SUMO precursors (endopeptidase cleavage) and deconjugation of the targets (isopeptidase cleavage) [1-3]. There are seven SENPs (1, 2, 3, 5, 6, 7, 8) in humans, and several of these have been characterized as SUMO (or Nedd8) specific enzymes. SENP8 is not a SUMO protease, instead it functions on a small ubiquitin related protein Nedd8. The objective of this project is to generate small molecule inhibitors specific for Dose Response confirmation of inhibitors of Sentrin-specific proteases (SENPs) using a Luminescent Interference Counterscreen assay Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Production Centers Network(MLPCN)Grant Proposal Number: 1R21 NS061758-01 fast track Assay Provider: Dr. Guy Salvesen, Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Modification of proteins by SUMO is a dynamic and reversible process. SUMOylation/deSUMOylation cycle regulates SUMOs function. Sentrin-specific proteases (SENPs) are involved in both the maturation of SUMO precursors (endopeptidase cleavage) and deconjugation of the targets (isopeptidase cleavage) [1-3]. There are seven SENPs (1, 2, 3, 5, 6, 7, 8) in humans, and several of these have been characterized as SUMO (or Nedd8) specific enzymes. SENP8 is not a SUMO protease, instead it functions on a small ubiquitin related protein Nedd8. The objective of this project is to generate small molecule inhibitors specific for SENP8 Dose Response confirmation of uHTS activators of Human Intestinal Alkaline Phosphatase using Mouse Intestinal Alkaline Phosphatase Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Production Centers Network (MLPCN) Grant Proposal Number: X01-MH077602-01 Alkaline phosphatase (EC 3.1.3.1) (APs) catalyze the hydrolysis of phosphomonoesters, releasing inorganic phosphate and alcohol. APs are dimeric enzymes found in most organisms. In human, four isozymes of APs have been identified. One isozyme is tissue-nonspecific (designated TNAP) and three other isozymes are tissue-specific and named according to the tissue of their predominant expression: intestinal (IAP), placental (PLAP) and germ cell (GCAP) alkaline phosphatases. IAP expression is largely restricted to the gut, especially to the epithelial cells (enterocytes) of the small intestinal mucosa. The exact biological function of IAP is unknown. The goal of this MLPCN probe project is to identify novel and specific activators of Dose Response confirmation of uHTS activators of Human Intestinal Alkaline Phosphatase using Placental Alkaline Phosphatase Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Production Centers Network (MLPCN) Grant Proposal Number: X01-MH077602-01 Assay Provider Dr. Jose Luis Millan, Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Alkaline phosphatase (EC 3.1.3.1) (APs) catalyze the hydrolysis of phosphomonoesters, releasing inorganic phosphate and alcohol. APs are dimeric enzymes found in most organisms. In human, four isozymes of APs have been identified. One isozyme is tissue-nonspecific (designated TNAP) and three other isozymes are tissue-specific and named according to the tissue of their predominant expression: intestinal (IAP), placental (PLAP) and germ cell (GCAP) alkaline phosphatases. IAP expression is largely restricted to the gut, especially to the epithelial cells (enterocytes) of the small intestinal mucosa. The exact biological function of Dose Response confirmation of uHTS activators of Human Intestinal Alkaline Phosphatase using Tissue Nonspecific Alkaline Phosphatase. Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Production Centers Network (MLPCN) Grant Proposal Number: X01-MH077602-01 Assay Provider Dr. Jose Luis Millan, Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Alkaline phosphatase (EC 3.1.3.1) (APs) catalyze the hydrolysis of phosphomonoesters, releasing inorganic phosphate and alcohol. APs are dimeric enzymes found in most organisms. In human, four isozymes of APs have been identified. One isozyme is tissue-nonspecific (designated TNAP) and three other isozymes are tissue-specific and named according to the tissue of their predominant expression: intestinal (IAP), placental (PLAP) and germ cell (GCAP) alkaline phosphatases. IAP expression is largely restricted to the gut, especially to the epithelial cells (enterocytes) of the small intestinal mucosa. The exact biological function of Dose Response confirmation of uHTS activators of Mouse Intestinal Alkaline Phosphatase using Human Intestinal Alkaline Phosphatase Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Production Centers Network (MLPCN) Grant Proposal Number: X01-MH077602-01 Assay Provider Dr. Jose Luis Millan, Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Alkaline phosphatase (EC 3.1.3.1) (APs) catalyze the hydrolysis of phosphomonoesters, releasing inorganic phosphate and alcohol. APs are dimeric enzymes found in most organisms. In human, four isozymes of APs have been identified. One isozyme is tissue-nonspecific (designated TNAP) and three other isozymes are tissue-specific and named according to the tissue of their predominant expression: intestinal (IAP), placental (PLAP) and germ cell (GCAP) alkaline phosphatases. IAP expression is largely restricted to the gut, especially to the epithelial cells (enterocytes) of the small intestinal mucosa. The exact biological function of Dose Response confirmation of uHTS activators of Mouse Intestinal Alkaline Phosphatase using Placental Alkaline Phosphatase Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Production Centers Network (MLPCN) Grant Proposal Number: X01-MH077602-01 Alkaline phosphatase (EC 3.1.3.1) (APs) catalyze the hydrolysis of phosphomonoesters, releasing inorganic phosphate and alcohol. APs are dimeric enzymes found in most organisms. In human, four isozymes of APs have been identified. One isozyme is tissue-nonspecific (designated TNAP) and three other isozymes are tissue-specific and named according to the tissue of their predominant expression: intestinal (IAP), placental (PLAP) and germ cell (GCAP) alkaline phosphatases. IAP expression is largely restricted to the gut, especially to the epithelial cells (enterocytes) of the small intestinal mucosa. The exact biological function of IAP is unknown. The goal of this HTS is to confirm hits in "uHTS Luminescent assay for identificatio Dose Response confirmation of uHTS activators of Mouse Intestinal Alkaline Phosphatase using Tissue Nonspecific Alkaline Phosphatase. Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Production Centers Network (MLPCN) Grant Proposal Number: X01-MH077602-01 Alkaline phosphatase (EC 3.1.3.1) (APs) catalyze the hydrolysis of phosphomonoesters, releasing inorganic phosphate and alcohol. APs are dimeric enzymes found in most organisms. In human, four isozymes of APs have been identified. One isozyme is tissue-nonspecific (designated TNAP) and three other isozymes are tissue-specific and named according to the tissue of their predominant expression: intestinal (IAP), placental (PLAP) and germ cell (GCAP) alkaline phosphatases. IAP expression is largely restricted to the gut, especially to the epithelial cells (enterocytes) of the small intestinal mucosa. The exact biological function of IAP is unknown. The goal of this HTS is to confirm hits in "uHTS Luminescent assay for identificatio Dose Response confirmation of uHTS for inhibitors of Sentrin-specific protease 6 (SENP6) using a Luminescent assay Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Probe Production Centers Network(MLPCN) Grant Proposal Number: 1R21 NS061758-01 fast track Assay Provider: Dr. Guy Salvesen, Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Modification of proteins by SUMO is a dynamic and reversible process. SUMOylation/deSUMOylation cycle regulates SUMOs function. Sentrin-specific proteases (SENPs) are involved in both the maturation of SUMO precursors (endopeptidase cleavage) and deconjugation of the targets (isopeptidase cleavage) [1-3]. There are seven SENPs (1, 2, 3, 5, 6, 7, 8) in humans, and several of these have been characterized as SUMO (or Nedd8) specific enzymes. The objective of this project is to generate small molecule inhibitors specific for SENP6 (the deSUMOylating enzyme). 1536-well chemiluminescent screening assay utilizes RLRGG- Dose Response confirmation of uHTS for inhibitors of Sentrin-specific protease 8 (SENP8) using a Luminescent assay Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Production Centers Network(MLPCN)Grant Proposal Number: 1R21 NS061758-01 fast track Assay Provider: Dr. Guy Salvesen, Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Modification of proteins by SUMO is a dynamic and reversible process. SUMOylation/deSUMOylation cycle regulates SUMOs function. Sentrin-specific proteases (SENPs) are involved in both the maturation of SUMO precursors (endopeptidase cleavage) and deconjugation of the targets (isopeptidase cleavage) [1-3]. There are seven SENPs (1, 2, 3, 5, 6, 7, 8) in humans, and several of these have been characterized as SUMO (or Nedd8) specific enzymes. SENP8 is not a SUMO protease, instead it functions on a small ubiquitin related protein Nedd8. The objective of this project is to generate small molecule inhibitors specific for SENP8 Dose Response confirmation of uHTS for the identification of UBC13 Polyubiquitin Inhibitors via a TR-FRET Assay Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego CA) Network: NIH Molecular Libraries Probe Production Centers Network (MLPCN) Grant Number: 1R03 MH085677-01 Assay Provider: Dr. John C. Reed, Sanford-Burnham Medical Research Institute, San Diego CA Tumor Necrosis Factor Receptor-Associated Factors (TRAFs) are a family of adapter proteins that bind an unusual ubiquitin-conjugating enzyme, Ubc13, which produces polyubiquitin chains linked at lysine 63 of ubiquitin. These lysine 63-linked ubiquitin polymers trigger changes in protein activity. Ubiquitination by Ubc13 of TRAFs and the various protein kinases to which TRAFs bind is recognized as a critical step in signaling by TNFRs, TLRs, NLRs, and T-cell and B-cell antigen receptors (TCR/BCR) during innate and acquired immune responses. Since aberrant signaling by these receptor systems is linked to a wide variety of autoimmune, inflammato Dose Response confirmation of uHTS for the identification of UBC13 Polyubiquitin Inhibitors via a TR-FRET Assay reconfirm Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego CA) Network: NIH Molecular Libraries Probe Production Centers Network (MLPCN) Grant Number: 1R03 MH085677-01 Assay Provider: Dr. John C. Reed, Sanford-Burnham Medical Research Institute, San Diego CA Tumor Necrosis Factor Receptor-Associated Factors (TRAFs) are a family of adapter proteins that bind an unusual ubiquitin-conjugating enzyme, Ubc13, which produces polyubiquitin chains linked at lysine 63 of ubiquitin. These lysine 63-linked ubiquitin polymers trigger changes in protein activity. Ubiquitination by Ubc13 of TRAFs and the various protein kinases to which TRAFs bind is recognized as a critical step in signaling by TNFRs, TLRs, NLRs, and T-cell and B-cell antigen receptors (TCR/BCR) during innate and acquired immune responses. Since aberrant signaling by these receptor systems is linked to a wide variety of autoimmune, inflammato Dose Response confirmation of uHTS hits from a small molecule inhibitors of LYP via a fluorescence intensity assay - Set 2 Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Probe Production Centers Network (MLPCN) Grant Number: 1R21NS056945-01 Assay Provider: Dr. Nunzio Bottini, La Jolla Institute for Allergy and Immunology, La Jolla, CA LYP, is a lymphocyte specific protein tyrosine phosphatase that plays a critical regulatory role in T cell receptor signaling. The PTPN22 gene encodes this phosphatase. A single-nucleotide polymorphism in PTPN22 is associated with a number of autoimmune disorders, including type 1 diabetes, rheumatoid arthritis, juvenile rheumatoid arthritis, systemic lupus erythematosus and Grave's disease. The autoimmunity-predisposing allele is a gain-of-function mutant suggesting that a specific small-molecule inhibitor could eliminate its effect. This biochemical assay employs a fluorescent readout based on the enzyme's ability to liberate Dose Response confirmation of uHTS hits from a small molecule inhibitors of LYP via a fluorescence intensity assay using pCAP substrate Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Probe Production Centers Network (MLPCN) Grant Number: 1R21NS056945-01 Assay Provider: Dr. Nunzio Bottini, La Jolla Institute for Allergy and Immunology, La Jolla, CA LYP, is a lymphocyte specific protein tyrosine phosphatase that plays a critical regulatory role in T cell receptor signaling. The PTPN22 gene encodes this phosphatase. A single-nucleotide polymorphism in PTPN22 is associated with a number of autoimmune disorders, including type 1 diabetes, rheumatoid arthritis, juvenile rheumatoid arthritis, systemic lupus erythematosus and Grave's disease. The autoimmunity-predisposing allele is a gain-of-function mutant suggesting that a specific small-molecule inhibitor could eliminate its effect. Finding specific inhibitors of protein phosphatases has proven extremely difficult. The goal of th Dose Response confirmation of uHTS hits from a small molecule inhibitors of human intestinal alkaline phosphatase via a luminescent assay - Set 2 Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute(SBMRI, San Diego, CA) Network: NIH Molecular Libraries Probe Production Centers Network (MLPCN) Grant Proposal Number: X01-MH077602-01 Assay Provider Dr. Jose Luis Millan, Sanford-Burnham Medical Research Institute(SBMRI, San Diego, CA) Alkaline phosphatase (EC 3.1.3.1) (APs) catalyze the hydrolysis of phosphomonoesters, releasing inorganic phosphate and alcohol. APs are dimeric enzymes found in most organisms. In human, four isozymes of APs have been identified. One isozyme is tissue-nonspecific (designated TNAP) and three other isozymes are tissue-specific and named according to the tissue of their predominant expression: intestinal (IAP), placental (PLAP) and germ cell (GCAP) alkaline phosphatases. IAP expression is largely restricted to the gut, especially to the epithelial cells (enterocytes) of the small intestinal mucosa. The exact biological functio Dose Response confirmation of uHTS hits from a small molecule inhibitors of human intestinal alkaline phosphatase via a luminescent assay Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Production Centers Network (MLPCN) Grant Proposal Number: X01-MH077602-01 Assay Provider Dr. Jose Luis Millan, Sanford-Burnham Medical Research Institute(SBMRI, San Diego, CA) Alkaline phosphatase (EC 3.1.3.1) (APs) catalyze the hydrolysis of phosphomonoesters, releasing inorganic phosphate and alcohol. APs are dimeric enzymes found in most organisms. In human, four isozymes of APs have been identified. One isozyme is tissue-nonspecific (designated TNAP) and three other isozymes are tissue-specific and named according to the tissue of their predominant expression: intestinal (IAP), placental (PLAP) and germ cell (GCAP) alkaline phosphatases. IAP expression is largely restricted to the gut, especially to the epithelial cells (enterocytes) of the small intestinal mucosa. The exact biological function of Dose Response confirmation of uHTS hits from a small molecule inhibitors of mouse intestinal alkaline phosphatase via a luminescent assay Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Production Centers Network (MLPCN) Grant Proposal Number: X01-MH077602-01 Assay Provider Dr. Jose Luis Millan, Sanford-Burnham Medical Research Institute(SBMRI, San Diego, CA) Alkaline phosphatase (EC 3.1.3.1) (APs) catalyze the hydrolysis of phosphomonoesters, releasing inorganic phosphate and alcohol. APs are dimeric enzymes found in most organisms. In human, four isozymes of APs have been identified. One isozyme is tissue-nonspecific (designated TNAP) and three other isozymes are tissue-specific and named according to the tissue of their predominant expression: intestinal (IAP), placental (PLAP) and germ cell (GCAP) alkaline phosphatases. IAP expression is largely restricted to the gut, especially to the epithelial cells (enterocytes) of the small intestinal mucosa. The exact biological function of Dose Response confirmation of uHTS inhibitors of Human Intestinal Alkaline Phosphatase using Mouse Intestinal Alkaline Phosphatase Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Production Centers Network (MLPCN) Grant Proposal Number: X01-MH077602-01 Assay Provider Dr. Jose Luis Millan, Sanford-Burnham Medical Research Institute(SBMRI, San Diego, CA) Alkaline phosphatase (EC 3.1.3.1) (APs) catalyze the hydrolysis of phosphomonoesters, releasing inorganic phosphate and alcohol. APs are dimeric enzymes found in most organisms. In human, four isozymes of APs have been identified. One isozyme is tissue-nonspecific (designated TNAP) and three other isozymes are tissue-specific and named according to the tissue of their predominant expression: intestinal (IAP), placental (PLAP) and germ cell (GCAP) alkaline phosphatases. IAP expression is largely restricted to the gut, especially to the epithelial cells (enterocytes) of the small intestinal mucosa. The exact biological function of Dose Response confirmation of uHTS inhibitors of Human Intestinal Alkaline Phosphatase using Placental Alkaline Phosphatase Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute(SBMRI, San Diego, CA) Network: NIH Molecular Libraries Probe Production Centers Network (MLPCN) Grant Proposal Number: X01-MH077602-01 Assay Provider Dr. Jose Luis Millan, Sanford-Burnham Medical Research Institute, San Diego, CA) Alkaline phosphatase (EC 3.1.3.1) (APs) catalyze the hydrolysis of phosphomonoesters, releasing inorganic phosphate and alcohol. APs are dimeric enzymes found in most organisms. In human, four isozymes of APs have been identified. One isozyme is tissue-nonspecific (designated TNAP) and three other isozymes are tissue-specific and named according to the tissue of their predominant expression: intestinal (IAP), placental (PLAP) and germ cell (GCAP) alkaline phosphatases. IAP expression is largely restricted to the gut, especially to the epithelial cells (enterocytes) of the small intestinal mucosa. The exact biological function of I Dose Response confirmation of uHTS inhibitors of Human Intestinal Alkaline Phosphatase using Tissue Nonspecific Alkaline Phosphatase. Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute(SBMRI, San Diego, CA) Network: NIH Molecular Libraries Probe Production Centers Network (MLPCN) Grant Proposal Number: X01-MH077602-01 Assay Provider Dr. Jose Luis Millan, Sanford-Burnham Medical Research Institute(SBMRI, San Diego, CA) Alkaline phosphatase (EC 3.1.3.1) (APs) catalyze the hydrolysis of phosphomonoesters, releasing inorganic phosphate and alcohol. APs are dimeric enzymes found in most organisms. In human, four isozymes of APs have been identified. One isozyme is tissue-nonspecific (designated TNAP) and three other isozymes are tissue-specific and named according to the tissue of their predominant expression: intestinal (IAP), placental (PLAP) and germ cell (GCAP) alkaline phosphatases. IAP expression is largely restricted to the gut, especially to the epithelial cells (enterocytes) of the small intestinal mucosa. The exact biological functio Dose Response confirmation of uHTS inhibitors of Mouse Intestinal Alkaline Phosphatase using Human Intestinal Alkaline Phosphatase Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute(SBMRI, San Diego, CA) Network: NIH Molecular Libraries Probe Production Centers Network (MLPCN) Grant Proposal Number: X01-MH077602-01 Assay Provider Dr. Jose Luis Millan, Sanford-Burnham Medical Research Institute(SBMRI, San Diego, CA) Alkaline phosphatase (EC 3.1.3.1) (APs) catalyze the hydrolysis of phosphomonoesters, releasing inorganic phosphate and alcohol. APs are dimeric enzymes found in most organisms. In human, four isozymes of APs have been identified. One isozyme is tissue-nonspecific (designated TNAP) and three other isozymes are tissue-specific and named according to the tissue of their predominant expression: intestinal (IAP), placental (PLAP) and germ cell (GCAP) alkaline phosphatases. IAP expression is largely restricted to the gut, especially to the epithelial cells (enterocytes) of the small intestinal mucosa. The exact biological functio Dose Response confirmation of uHTS inhibitors of Mouse Intestinal Alkaline Phosphatase using Placental Alkaline Phosphatase Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Production Centers Network (MLPCN) Grant Proposal Number: X01-MH077602-01 Assay Provider Dr. Jose Luis Millan, Sanford-Burnham Medical Research Institute(SBMRI, San Diego, CA) Alkaline phosphatase (EC 3.1.3.1) (APs) catalyze the hydrolysis of phosphomonoesters, releasing inorganic phosphate and alcohol. APs are dimeric enzymes found in most organisms. In human, four isozymes of APs have been identified. One isozyme is tissue-nonspecific (designated TNAP) and three other isozymes are tissue-specific and named according to the tissue of their predominant expression: intestinal (IAP), placental (PLAP) and germ cell (GCAP) alkaline phosphatases. IAP expression is largely restricted to the gut, especially to the epithelial cells (enterocytes) of the small intestinal mucosa. The exact biological function of Dose Response confirmation of uHTS inhibitors of Mouse Intestinal Alkaline Phosphatase using Tissue Nonspecific Alkaline Phosphatase. Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Production Centers Network (MLPCN) Grant Proposal Number: X01-MH077602-01 Assay Provider Dr. Jose Luis Millan, Sanford-Burnham Medical Research Institute(SBMRI, San Diego, CA) Alkaline phosphatase (EC 3.1.3.1) (APs) catalyze the hydrolysis of phosphomonoesters, releasing inorganic phosphate and alcohol. APs are dimeric enzymes found in most organisms. In human, four isozymes of APs have been identified. One isozyme is tissue-nonspecific (designated TNAP) and three other isozymes are tissue-specific and named according to the tissue of their predominant expression: intestinal (IAP), placental (PLAP) and germ cell (GCAP) alkaline phosphatases. IAP expression is largely restricted to the gut, especially to the epithelial cells (enterocytes) of the small intestinal mucosa. The exact biological function of Dose Response confirmation of uHTS small molecule inhibitors of Plasmodium falciparum Glucose-6-phosphate dehydrogenase via a fluorescence intensity assay Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego CA) Network: NIH Molecular Libraries Probe Production Centers Network (MLPCN) Grant Number: 1R21AI082434-01 Assay Provider: Lars Bode, Ph.D., University of California San Diego, San Diego, CA Tropical malaria caused by the protozoan parasite Plasmodium falciparum is responsible for up to three million deaths annually. Due to increasing regional distribution and resistances against the clinically used antimalarials, novel antimalarial drugs - which have new mechanisms of action and are suitable for combination therapies - are urgently required. Plasmodium falciparum Glucose-6-phosphate dehydrogenase (PfGluPho) is a potential novel target for antimalarial drug design. Glucose-6-Phosphate Dehydrogenase (G6PD) reaction is the first and rate-limiting step in the pentose phosphate pathway (PPP), catalyzed by a bifunctional enzyme Plasmodium fal Dose Response of Developing T Cell Immune Modulators with powder sourced compounds University of New Mexico Assay Overview: Assay Support: 1 X01 MH085707-01 Project Title: HTS for developing T Cell Immune Modulators Assay Provider: Inkyu Hwang, PhD The Scripps Research Institute (La Jolla) Lead Biologist: Mark K Haynes, PhD Chemistry Center/ PI: Craig Lindsley Specialized Chemistry Center: The Vanderbilt Specialized Chemistry Center Accelerated Probe Development Assay Implementation: Mark K. Haynes PhD, Chelin Hu MS, Anna Waller PhD, Mark Carter MS Assay Background and Significance: When naive T cells encounter antigen presenting cells (APC) displaying cognate MHC-peptide complexes (pMHCs), the T cell and the APC form a highly organized structure at the contact site termed the 'immunological synapse', which is critical not only for prolonged and stable T/APC contact but also for the efficient and organized T cell signaling required for cell cycle progression and development of effector functions. Several membrane proteins and their ligand partners Dose Response of SAR compounds via Multiplex HTS Assay for Inhibitors of MEK Kinase PB1 Domains, specifically MEK5 binding to MEK Kinase 2 Wildtype University of New Mexico Assay Overview: Assay Support: 1R03MH084830-01 Project Title: TR-FRET HTS Assay for Inhibitors of MEKK2-MEK5 PB1 Domain Interaction PI: Kazuhiro Nakamura, Ph.D Center PI: Larry Sklar, Ph.D Assay Implementatiion: Zurab Surviladze Ph.D, Mark Haynes Ph.D, Anna Waller Ph.D, Mark Carter MS Assay Background and Significance: PB1 (Phox/Bem1p) domains function as protein-protein interaction sites by forming PB1-PB1 domain heterodimers (Moscat, et al. 2006). There are at least 20 human PB1 domain-containing proteins. Different PB1 domains contribute to the formation of specific protein complexes critical for biological responses including proliferation, apoptosis, cell polarity, and angiogenesis. These proteins include the mitogen/extracellular signal regulated kinase kinases MEKK2 and MEKK3 (MAP3Ks), as well as the downstream regulated kinase MEK5 (a MAP2K), which solely govern the ERK5 MAPK pathway involved in angiogenesis, cell growth and inhibition of apo Dose Response orthogonal assay utilizing the direct end-point detection of NADPH for uHTS small molecule inhibitors of Plasmodium falciparum Glucose-6-phosphate dehydrogenase Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego CA) Network: NIH Molecular Libraries Probe Production Centers Network (MLPCN) Grant Number: 1R21AI082434-01 Assay Provider: Lars Bode, Ph.D., University of California San Diego, San Diego, CA Tropical malaria caused by the protozoan parasite Plasmodium falciparum is responsible for up to three million deaths annually. Due to increasing regional distribution and resistances against the clinically used antimalarials, novel antimalarial drugs - which have new mechanisms of action and are suitable for combination therapies - are urgently required. Plasmodium falciparum Glucose-6-phosphate dehydrogenase (PfGluPho) is a potential novel target for antimalarial drug design. Glucose-6-Phosphate Dehydrogenase (G6PD) reaction is the first and rate-limiting step in the pentose phosphate pathway (PPP), catalyzed by a bifunctional enzyme Plasmodium fal Dose Response orthogonal kinetic assay utilizing the direct detection of NADPH for uHTS small molecule inhibitors of Plasmodium falciparum Glucose-6-phosphate dehydrogenase Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego CA) Network: NIH Molecular Libraries Probe Production Centers Network (MLPCN) Grant Number: 1R21AI082434-01 Assay Provider: Lars Bode, Ph.D., University of California San Diego, San Diego, CA Tropical malaria caused by the protozoan parasite Plasmodium falciparum is responsible for up to three million deaths annually. Due to increasing regional distribution and resistances against the clinically used antimalarials, novel antimalarial drugs - which have new mechanisms of action and are suitable for combination therapies - are urgently required. Plasmodium falciparum Glucose-6-phosphate dehydrogenase (PfGluPho) is a potential novel target for antimalarial drug design. Glucose-6-Phosphate Dehydrogenase (G6PD) reaction is the first and rate-limiting step in the pentose phosphate pathway (PPP), catalyzed by a bifunctional enzyme Plasmodium fal Dose Response selectivity of inhibitors of STriatal-Enriched Phosphatase (STEP) in the Lymphoid Phosphatase (PTPN22) Inhibition Assay Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Probe Production Centers Network (MLPCN) Grant Number: 1R03MH095532-01 Assay Provider: Dr. Lutz Tautz, Sanford-Burnham Medical Research Institute, San Diego CA Disturbance of the dynamic balance between protein tyrosine phosphorylation and dephosphorylation is crucial for the development of many serious conditions, including cancer, diabetes, and autoimmune disorders. This is the first time that tyrosine phosphatase inhibitors are being proposed to improve cognitive function in Alzheimer's disease (AD). STriatal-Enriched Phosphatase (STEP) is a brain-specific protein tyrosine phosphatase that is highly expressed in regions where consolidation of memory occurs and regulates the internalization of NMDARs. Our recent work demonstrates that STEP is elevated in the prefrontal cortex of human AD patients Dose Response selectivity of inhibitors of STriatal-Enriched Phosphatase (STEP) in the dual-specificity protein-tyrosine phosphatase VHR Inhibition Assay Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Probe Production Centers Network (MLPCN) Grant Number: 1R03MH095532-01 Assay Provider: Dr. Lutz Tautz, Sanford-Burnham Medical Research Institute, San Diego CA Disturbance of the dynamic balance between protein tyrosine phosphorylation and dephosphorylation is crucial for the development of many serious conditions, including cancer, diabetes, and autoimmune disorders. This is the first time that tyrosine phosphatase inhibitors are being proposed to improve cognitive function in Alzheimer's disease (AD). STriatal-Enriched Phosphatase (STEP) is a brain-specific protein tyrosine phosphatase that is highly expressed in regions where consolidation of memory occurs and regulates the internalization of NMDARs. Our recent work demonstrates that STEP is elevated in the prefrontal cortex of human AD patients Dose Response selectivity of inhibitors of Striatal-Enriched Phosphatase (STEP) in a SHP2 (PTPN11) Inhibition Assay Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Probe Production Centers Network (MLPCN) Grant Number: 1R03MH095532-01 Assay Provider: Dr. Lutz Tautz, Sanford-Burnham Medical Research Institute, San Diego CA Disturbance of the dynamic balance between protein tyrosine phosphorylation and dephosphorylation is crucial for the development of many serious conditions, including cancer, diabetes, and autoimmune disorders. This is the first time that tyrosine phosphatase inhibitors are being proposed to improve cognitive function in Alzheimer's disease (AD). STriatal-Enriched Phosphatase (STEP) is a brain-specific protein tyrosine phosphatase that is highly expressed in regions where consolidation of memory occurs and regulates the internalization of NMDARs. Our recent work demonstrates that STEP is elevated in the prefrontal cortex of human AD patients Dose Response validation of uHTS RPN11 inhibitor hits using a Thrombin Fluorescence Polarization assay Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford Burnham Medical Research Institute (SBMRI, La Jolla, CA) Network: NIH Molecular Libraries Probe Production Centers Network (MLPCN) Grant Number: 1 R03 MH094180-01 Assay Provider: Dr. Raymond Deshaies, California Institute of Technology, Pasadena, CA Protein modification by the attachment of ubiquitin to cellular proteins is a key mechanism in regulating many cellular and physiological processes. Ubiquitin is covalently attached via an enzymatic cascade to target proteins through an isopeptide bond between the C-terminus of ubiquitin and a lysine residue of the acceptor substrate [1]. Assembly of a chain of >=4 ubiquitins linked together via Lys48 of ubiquitin marks cellular proteins for degradation by the 26S proteasome [2-3]. The 26S proteasome is a 2.5 megadalton macromolecular protein complex that comprises two distinct subparticles: the 19S cap regulatory particle (RP) and the 20S core Dose response assay for compounds that inhibit KCNQ2 potassium channels on automated electrophysiological assay II Source (MLPCN Center Name): Johns Hopkins Ion Channel Center (JHICC_KCNQ2_Inh_HFCP_IWS_CRC) Center Affiliation: Johns Hopkins University, School of Medicine Screening Center PI: Min Li, Ph.D. Assay Provider: Min Li, Ph.D. Network: Molecular Libraries Probe Production Centers Network (MLPCN) Grant Proposal Number: 1 R03 DA027716-01 Grant Proposal PI: Min Li, Ph.D., Johns Hopkins University School of Medicine Assay Implementation: Haibo Yu Ph.D., Kaiping Xu, Shunyou Long M.S, David Meyers Ph.D., Meng Wu Ph.D., Owen McManus Ph.D. Name: Dose response assay for compounds that inhibit KCNQ2 potassium channels on automated electrophysiological assay II Description: Voltage-gated potassium (K) channels are critical for neuronal function in excitable tissues such as brain and heart. They are also found in non-excitable tissues important for other functions such as hormone secretion, oxygen-sensing and immune responses. There are more than 100 genes in the human genome encoding different but Dose response assay for dual activators of procaspase-3 and procaspase-7: Absorbance-based biochemical high throughput screening assay to identify activators of procaspase-7 Source (MLPCN Center Name): The Scripps Research Institute Molecular Screening Center (SRIMSC) Affiliation: The Scripps Research Institute, TSRI Assay Provider: Paul Hergenrother, University of Illinois at Urbana-Champaign Network: Molecular Library Probe Production Centers Network (MLPCN) Grant Proposal Number: R01 CA120439-01 Grant Proposal PI: Paul Hergenrother, University of Illinois at Urbana-Champaign External Assay ID: PROCASPASE7_ACT_EPIABS_1536_3XEC50 DUAL Name: Dose response assay for dual activators of procaspase-3 and procaspase-7: Absorbance-based biochemical high throughput screening assay to identify activators of procaspase-7. Description: Cancer progression depends upon evasion of the programmed cell death (apoptosis) machinery that normally kills an unneeded or rogue cell (1). Although apoptosis induction using chemotherapeutics is a common anti-cancer treatment, cancer cells often survive because of defects in the pro-apoptotic proteins activated by these drugs ( Dose response biochemical High Throughput Screening assay for agonists of the steroid receptor coactivator 1 (SRC-1) recruitment by the peroxisome proliferator-activated receptor gamma (PPARgamma) Source (MLSCN Center Name): The Scripps Research Institute Molecular Screening Center Center Affiliation: The Scripps Research Institute (TSRI) Assay Provider: Pat Griffin, TSRI Network: Molecular Library Screening Center Network (MLSCN) Grant Proposal Number: 1 X01 MH079861-01 Grant Proposal PI: Patrick Griffin, Scripps Florida External Assay ID: PPARgSRC1_AG_TRFRET_1536_EC50 Name: Dose response biochemical High Throughput Screening assay for agonists of the steroid receptor coactivator 1 (SRC-1) recruitment by the peroxisome proliferator-activated receptor gamma (PPARgamma) Description: Peroxisome proliferator-activated receptors (PPARs) belong to the nuclear receptor superfamily and are lipid sensors functioning as ligand-dependent transcription factors regulating gene expression patterns of diverse biological processes [1, 2]. PPARs play a critical role in metabolic processes such as glucose metabolism, lipid metabolism, and have been implicated in anti-atherogenic, anti-inf Dose response biochemical High Throughput Screening assay for agonists of the steroid receptor coactivator 2 (SRC-2) recruitment by the peroxisome proliferator-activated receptor gamma (PPARgamma) Source (MLSCN Center Name): The Scripps Research Institute Molecular Screening Center Center Affiliation: The Scripps Research Institute (TSRI) Assay Provider: Pat Griffin, TSRI Network: Molecular Library Screening Center Network (MLSCN) Grant Proposal Number: 1 X01 MH079861-01 Grant Proposal PI: Pat Griffin, TSRI External Assay ID: PPARgSRC2_AG_TRFRET_1536_EC50 Name: Dose response biochemical High Throughput Screening assay for agonists of the steroid receptor coactivator 2 (SRC-2) recruitment by the peroxisome proliferator-activated receptor gamma (PPARgamma) Description: Peroxisome proliferator-activated receptors (PPARs) belong to the nuclear receptor superfamily and are lipid sensors functioning as ligand-dependent transcription factors regulating gene expression patterns of diverse biological processes [1, 2]. PPARs play a critical role in metabolic processes such as glucose metabolism, lipid metabolism, and have been implicated in anti-atherogenic, anti-inflammatory as we Dose response biochemical High Throughput Screening assay for agonists of the steroid receptor coactivator 3 (SRC-3) recruitment by the peroxisome proliferator-activated receptor gamma (PPARgamma) Source (MLSCN Center Name): The Scripps Research Institute Molecular Screening Center http://molscreen.florida.scripps.edu/ Center Affiliation: The Scripps Research Institute, TSRI Assay Provider: Pat Griffin, TSRI Network: Molecular Library Screening Center Network (MLSCN) Grant Proposal Number: 1 X01 MH079861-01 Grant Proposal PI: Pat Griffin, TSRI External Assay ID: PPARgSRC3_AG_TRFRET_1536_EC50 Name: Dose response biochemical High Throughput Screening assay for agonists of the steroid receptor coactivator 3 (SRC-3) recruitment by the peroxisome proliferator-activated receptor gamma (PPARgamma) Description: Peroxisome proliferator-activated receptors (PPARs) belong to the nuclear receptor superfamily and are lipid sensors functioning as ligand-dependent transcription factors regulating gene expression patterns of diverse biological processes [1, 2]. PPARs play a critical role in metabolic processes such as glucose metabolism, lipid metabolism, and have been implicated in anti Dose response biochemical assay for autofluorescent inhibitors of Matrix Metalloproteinase 13 (MMP13) activity External Assay ID: MMP13_INH_deltaRFU_ 1536_IC50 Source (MLSCN Center Name): The Scripps Research Institute Molecular Screening Center Center Affiliation: The Scripps Research Institute, TSRI Assay Provider: Florida Atlantic University Network: Molecular Library Screening Center Network (MLSCN) Proposal Number: 1 X01 MH078948-01 Name: Dose response biochemical assay for autofluorescent inhibitors of Matrix Metalloproteinase 13 (MMP13) activity Description: Osteoarthritis (OA) is an age-related debilitating disease affecting more than 80% of people over the age of 75, caused by the destruction of articular cartilage (1). The major components of the cartilage extra cellular matrix (ECM) are type II collagen and the chondroitin sulfate proteoglycan, aggrecan (2). Proteases with potential roles in OA include MMP-1, MMP-13, ADAMTS-4, and ADAMTS-5 (3). MMP-13 is believed to be the more prominent collagenase in OA (3,4). Initial clinical trials with MMP inhibitors targeting active site wer Dose response biochemical assay to identify inhibitors of the HIV Rev - RRE RNA interaction (disruption of protein-RNA interaction) Source (MLSCN Center Name): The Scripps Research Institute Molecular Screening Center Center Affiliation: The Scripps Research Institute, TSRI Assay Provider: James R. Williamson, TSRI Network: Molecular Library Screening Center Network (MLSCN) Grant Proposal Number: 1 X01 MH078935-01 Grant Proposal PI: James R. Williamson External Assay ID: HIVREVRRE_INH_FRET_1536_ IC50 Name: Dose response biochemical assay to identify inhibitors of the HIV Rev - RRE RNA interaction (disruption of protein-RNA interaction) Description: Rev is a small basic protein that is critical for HIV replication (1). Early in infection, before synthesis of significant amounts of Rev, mRNA transcripts are processed by a default pathway that fully splices both introns. Export of these mRNAs to the cytoplasm for translation produces a set of small regulatory proteins, including Tat and Rev. Rev binds to the Rev-Responsive Element (RRE) on the viral mRNA (2), which results in efficient export of singly spliced Dose response cell-based high throughput screening assay for antagonists of the Sphingosine 1-Phosphate Receptor 3 (S1P3) Source (MLPCN Center Name): The Scripps Research Institute Molecular Screening Center (SRIMSC) Center Affiliation: The Scripps Research Institute (TSRI) Assay Provider: Hugh Rosen, TSRI Network: Molecular Library Probe Production Centers Network (MLPCN) Grant Proposal Number: 1 R03 MH076533-01 Grant Proposal PI: Germana Sanna, TSRI External Assay ID: S1P3_ANT_BLA_384_IC50 Name: Dose response cell-based high throughput screening assay for antagonists of the Sphingosine 1-Phosphate Receptor 3 (S1P3) Description: Sphingosine 1-phosphate (S1P) influences heart rate [1, 2], coronary artery caliber, endothelial integrity, lung epithelial integrity [3] and lymphocyte recirculation [1, 4-6] through five related high affinity G-protein coupled receptors [7, 8] which are involved in diverse biology. For example, S1P1 activation is sufficient to control lymphocyte numbers, with no discernable role in sinus rhythm control, whereas S1P3 regulates sinus rhythm but not lymphocyte recirculation. Dose response cell-based high-throughput screening assay to measure PERK inhibition Source (MLPCN Center Name): The Scripps Research Institute Molecular Screening Center (SRIMSC) Center Affiliation: The Scripps Research Institute (TSRI) Assay Provider: David Ron, New York University Network: Molecular Libraries Probe Production Centers Network (MLPCN) Grant Proposal Number R03 MH082370-01 Grant Proposal PI: David Ron, New York University External Assay ID: PERK_INH_SEAP_1536_EC50 Name: Dose response cell-based high-throughput screening assay to measure PERK inhibition Description: The endoplasmic reticulum (ER) is essential for protein synthesis and folding (1, 2). In response to environmental stress, misfolded proteins accumulate in the ER lumen, leading to activation of a set of signaling cascades known as the unfolded protein response (UPR). In eukaryotic cells the UPR reduces the burden of misfolded proteins by inhibiting protein synthesis and increasing clearance of misfolded proteins (3, 4). The RNA-dependent protein kinase (PKR)-like ER kinase (PERK) define Dose response confirmation of uHTS RPN11 inhibitor hits in a Fluorescence Polarization assay Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford Burnham Medical Research Institute (SBMRI, La Jolla, CA) Network: NIH Molecular Libraries Probe Production Centers Network (MLPCN) Grant Number: 1 R03 MH094180-01 Assay Provider: Dr. Raymond Deshaies, California Institute of Technology, Pasadena, CA Protein modification by the attachment of ubiquitin to cellular proteins is a key mechanism in regulating many cellular and physiological processes. Ubiquitin is covalently attached via an enzymatic cascade to target proteins through an isopeptide bond between the C-terminus of ubiquitin and a lysine residue of the acceptor substrate [1]. Assembly of a chain of >=4 ubiquitins linked together via Lys48 of ubiquitin marks cellular proteins for degradation by the 26S proteasome [2-3]. The 26S proteasome is a 2.5 megadalton macromolecular protein complex that comprises two distinct subparticles: the 19S cap regulatory particle (RP) and the 20S core Dose response confirmation of uHTS antagonist hits from Gli-SUFU in a luminescent reporter assay Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Production Centers Network(MLPCN) Grant Number: 1R03MH094195-01 Assay Provider: James Chen, Ph.D.,Stanford University, Stanford California The Hh pathway plays a critical role in the patterning of certain embryonic tissues and contributes to their neoplastic transformation later in life. Hh signaling regulates cerebellar patterning by promoting the proliferation of neuronal precursor cells, and constitutive Hh target gene expression can lead to medulloblastoma, the most common pediatric brain tumor(1). Hh signaling is normally initiated by the binding of Hh ligands to the twelve-pass transmembrane protein Ptch1(2) inducing its exit from the cilium, leading to Smo accumulation and activation within the antenna-like organelle. Activated Smo then shifts the balance between repressor and activator forms Dose response confirmation of uHTS small molecule inhibitors of Striatal-Enriched Phosphatase via a fluorescence intensity assay Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Probe Production Centers Network (MLPCN) Grant Number: 1R03MH095532-01 Assay Provider: Dr. Lutz Tautz, Sanford-Burnham Medical Research Institute, San Diego CA Disturbance of the dynamic balance between protein tyrosine phosphorylation and dephosphorylation is crucial for the development of many serious conditions, including cancer, diabetes, and autoimmune disorders. This is the first time that tyrosine phosphatase inhibitors are being proposed to improve cognitive function in Alzheimer's disease (AD). STriatal-Enriched Phosphatase (STEP) is a brain-specific protein tyrosine phosphatase that is highly expressed in regions where consolidation of memory occurs and regulates the internalization of NMDARs. Our recent work demonstrates that STEP is elevated in the prefrontal cortex of human AD patients Dose response counterscreen assay for neuropeptide Y receptor Y1 (NPY-Y1): Cell-based high throughput assay to measure NPY-Y2 antagonism Source (MLSCN Center Name): The Scripps Research Institute Molecular Screening Center Affiliation: The Scripps Research Institute, TSRI Assay Provider: Claes Wahlestedt, Scripps Florida Network: Molecular Library Screening Center Network (MLSCN) Grant Proposal Number 1 R21 NS056950-01 Grant Proposal PI: Claes Wahlestedt External Assay ID: NPY-Y2_ANT_CNGC_1536_IC50 (CS) Name: Dose response counterscreen assay for neuropeptide Y receptor Y1 (NPY-Y1): Cell-based high throughput assay to measure NPY-Y2 antagonism Description: Neuropeptide Y (NPY) is a neurotransmitter with physiologic roles including control of feeding behavior, regulation of cortical neural activity, heart neural activity, and emotional regulation. Importantly, NPY is implicated in human diseases such as obesity, depression and alcoholism. NPY mediates its biological effects in part through activation of the Galphai protein coupled receptors (GPCRs) NPY-Y1 and Y2 receptors, which decrease cytosolic cAMP product Dose response counterscreen for agonists of galanin receptor 2 (GalR2): a cell-based high-throughput screening assay for activators of beta-lactamase activity Source (MLSCN Center Name): The Scripps Research Institute Molecular Screening Center Affiliation: The Scripps Florida Research Institute (TSRI) Assay Provider: Steven Brown, TSRI Network: Molecular Library Screening Center Network (MLSCN) Grant Proposal Number: 1 R21 NS057101-01 Grant Proposal PI: Steven Brown External Assay ID: NFAT_ACT_BLA_1536_EC50 Name: Dose response counterscreen for agonists of galanin receptor 2 (GalR2): a cell-based high-throughput screening assay for activators of beta-lactamase activity Description: Galanin, a 29 amino acid neuropeptide (30 residues in humans), is cleaved from preprogalanin and is involved in many physiological processes including nervous system development, feeding, metabolism and reproduction, and regulation of neurotransmitter and hormone release [1, 2]. The physiologic response to galanin is mediated in part by three G protein-coupled metabotropic 7-transmembrane receptor subtypes, GALR1, GALR2 and GALR3. These receptors are expre Dose response counterscreen for antagonists of galanin receptor 2 (GalR2): a cell-based high-throughput screening assay for inhibitors of beta-lactamase activity Source (MLSCN Center Name): The Scripps Research Institute Molecular Screening Center Affiliation: The Scripps Research Institute (TSRI) Assay Provider: Steven Brown, TSRI Network: Molecular Library Screening Center Network (MLSCN) Grant Proposal Number: 1 R21 NS057101-01 Grant Proposal PI: Steven Brown External Assay ID: NFAT_INH_BLA_1536_ IC50 Name: Dose response counterscreen for antagonists of galanin receptor 2 (GalR2): a cell-based high-throughput screening assay for inhibitors of beta-lactamase activity Description: Galanin, a 29 amino acid neuropeptide (30 residues in humans), is cleaved from preprogalanin and is involved in many physiological processes including nervous system development, feeding, metabolism and reproduction, and regulation of neurotransmitter and hormone release [1, 2]. The physiologic response to galanin is mediated in part by three G protein-coupled metabotropic 7-transmembrane receptor subtypes, GALR1, GALR2 and GALR3. These receptors are expressed t Dose response counterscreen for dual activators of procaspase-3 and procaspase-7: Absorbance-based biochemical high throughput screening assay to identify activators of procaspase-3 Source (MLPCN Center Name): The Scripps Research Institute Molecular Screening Center (SRIMSC) Affiliation: The Scripps Research Institute, TSRI Assay Provider: Paul Hergenrother, University of Illinois at Urbana-Champaign Network: Molecular Library Probe Production Centers Network (MLPCN) Grant Proposal Number: R01 CA120439-01 Grant Proposal PI: Paul Hergenrother, University of Illinois at Urbana-Champaign External Assay ID: PROCASPASE3_ACT_EPIABS_1536_3XEC50 DUAL Name: Dose response counterscreen for dual activators of procaspase-3 and procaspase-7: Absorbance-based biochemical high throughput screening assay to identify activators of procaspase-3. Description: Cancer progression depends upon evasion of the programmed cell death (apoptosis) machinery that normally kills an unneeded or rogue cell (1). Although apoptosis induction using chemotherapeutics is a common anti-cancer treatment, cancer cells often survive because of defects in the pro-apoptotic proteins activated by these Dose response counterscreen for neuropeptide Y receptor Y2 (NPY-Y2): Cell-based high throughput assay to measure NPY-Y1 antagonism Source (MLSCN Center Name): The Scripps Research Institute Molecular Screening Center Affiliation: The Scripps Research Institute, TSRI Assay Provider: Claes Wahlestedt, Scripps Florida Network: Molecular Library Screening Center Network (MLSCN) Grant Proposal Number 1 R21 NS056950-01 Grant Proposal PI: Claes Wahlestedt External Assay ID: NPY-Y1_ANT_CNGC_1536_IC50 (CS) Name: Dose response counterscreen for neuropeptide Y receptor Y2 (NPY-Y2): Cell-based high throughput assay to measure NPY-Y1 antagonism Description: Neuropeptide Y (NPY) is a neurotransmitter with diverse physiologic roles including control of feeding behavior, regulation of cortical neural activity, heart neural activity, and emotional regulation. Importantly, NPY is implicated in human diseases such as obesity, depression and alcoholism. NPY mediates its biological effects in part through activation of the NPY-Y2 receptor, a 381-amino acid Galphai protein coupled receptor (GPCR) which decreases cytosolic cAMP Dose response counterscreen of uHTS hits for ATG4B inhibitors in a Phospholipase A2 assay Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Probe Production Centers Network (MLPCN) Grant Number: 1 R03 MH090871-01 Assay Provider: Dr. John C. Reed, Sanford-Burnham Medical Research Institute, San Diego CA This assay is a counterscreen for the primary screen (AID504462) which looked for inhibitors of Autophagin 1 (ATG4B). In screening for compounds that inhibit ATG4B, the High Throughput Screening (HTS) assay utilized a cleavable form of Phospholipase A2 (PLA2), which is expressed as a fusion protein with the Autophagin substrate LC3/ATG8 appended to its N-terminus, as the substrate for the primary enzymatic reaction. The addition of sequences to the N-terminus of PLA2 inhibits the activity of this enzyme. Cleavage by proteases removing the N-terminal extension then restores enzyme activity, constituting the basis for a protease assay. The sub Dose response counterscreen of uHTS hits for ATG4B inhibitors in a Phospholipase A2 assay Set 2 Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Probe Production Centers Network (MLPCN) Grant Number: 1 R03 MH090871-01 Assay Provider: Dr. John C. Reed, Sanford-Burnham Medical Research Institute, San Diego CA This assay is a counterscreen for the primary screen (AID504462) which looked for inhibitors of Autophagin 1 (ATG4B). In screening for compounds that inhibit ATG4B, the High Throughput Screening (HTS) assay utilized a cleavable form of Phospholipase A2 (PLA2), which is expressed as a fusion protein with the Autophagin substrate LC3/ATG8 appended to its N-terminus, as the substrate for the primary enzymatic reaction. The addition of sequences to the N-terminus of PLA2 inhibits the activity of this enzyme. Cleavage by proteases removing the N-terminal extension then restores enzyme activity, constituting the basis for a protease assay. The sub Dose response orthogonal assay of uHTS small molecule inhibitors of Striatal-Enriched Phosphatase via a colorimetric intensity assay. Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Probe Production Centers Network (MLPCN) Grant Number: 1R03MH095532-01 Assay Provider: Dr. Lutz Tautz, Sanford-Burnham Medical Research Institute, San Diego CA Disturbance of the dynamic balance between protein tyrosine phosphorylation and dephosphorylation is crucial for the development of many serious conditions, including cancer, diabetes, and autoimmune disorders. This is the first time that tyrosine phosphatase inhibitors are being proposed to improve cognitive function in Alzheimer's disease (AD). STriatal-Enriched Phosphatase (STEP) is a brain-specific protein tyrosine phosphatase that is highly expressed in regions where consolidation of memory occurs and regulates the internalization of NMDARs. Our recent work demonstrates that STEP is elevated in the prefrontal cortex of human AD patients Dose responses of compounds that activate the Choline Transporter (CHT) - 10 point CRC Data Source (MLPCN Center Name): Johns Hopkins Ion Channel Center (JHICC) Center Affiliation: Johns Hopkins University, School of Medicine Screening Center PI: Min Li, Ph.D. Assay Provider: Alicia Ruggiero, Ph.D., Vanderbilt University Medical Center Network: Molecular Libraries Probe Production Centers Network (MLPCN) Grant Proposal Number: 1R03DA028852-01 Grant Proposal PI: Alicia Ruggiero, Ph.D., Vanderbilt University Medical Center Assay Implementation: Zhihong Lin Ph. D., Xiaofang Huang M.S., Shunyou Long M.S., Owen McManus Ph.D., and Meng Wu Ph.D. Description: In the brain, the chemical acetylcholine (ACh) exerts powerful modulatory control over arousal, motor and cognitive circuits, and has been found to be deficient in Alzheimer's Disease (AD). The current drugs available to positively impact cognitive deficits in Alzheimer's Disease (AD) and other dementias are the cholinesterase inhibitors. These prevent the breakdown of the neurotransmitter acetylcholine (ACh), an Dose-response biochemical assay for antagonists of the interaction between the Eph receptor B4 (EphB4) and its ligand ephrin-B2 via TNYL-RAW peptide Source (MLSCN Center Name): The Scripps Research Institute Molecular Screening Center Center Affiliation: The Scripps Research Institute, TSRI Assay Provider: Peter Kuhn, TSRI Network: Molecular Library Screening Center Network (MLSCN) Grant Proposal Number: 1 X01 MH079857-01 Grant Proposal PI: Peter Kuhn External Assay ID: EphB4TNYLRAW_INH_FP_1536_IC50 Name: Dose-response biochemical assay for antagonists of the interaction between the Eph receptor B4 (EphB4) and its ligand ephrin-B2 via TNYL-RAW peptide Description: The erythropoietin-producing hepatocellular (Eph) receptor family is the largest family of receptor tyrosine kinases identified to date, with 16 structurally similar family members(1). The Eph family plays important roles in both the developing and adult tissues, and is involved in biological processes such as tissue patterning, vascular system development, axonal guidance, and neuronal development (2-4). During vascular development, the Eph receptor B4 (EphB4) is p Dose-response cell-based assay for activators of the Retinoic Acid Receptor-related orphan receptor A (RORA) Source (MLSCN Center Name): The Scripps Research Institute Molecular Screening Center Center Affiliation: The Scripps Research Institute, TSRI Assay Provider: Orphagen Pharmaceuticals, San Diego, CA Network: Molecular Library Screening Center Network (MLSCN) Grant Proposal Number: 1X01-MH077624-01 External Assay ID: RORA_AG_LUMI_1536_EC50 Name: Dose-response cell-based assay for activators of the Retinoic Acid Receptor-related orphan receptor A (RORA) Description: Nuclear receptors are a family of small molecule and hormone-regulated transcription factors that share conserved DNA-binding and ligand-binding domains. Small pharmacological compounds able to bind to the cleft of the ligand-binding domain could alter its conformation and subsequently modify transcription of target genes. Such ligand agonists and/or antagonists have already been successfully designed for 23 nuclear receptors among the 48 previously identified in the human genome [1-3]. RORA is one of three related orp Dose-response cell-based assay for activators of the nuclear receptor Steroidogenic Factor 1 (SF-1) Source (MLSCN Center Name): The Scripps Research Institute Molecular Screening Center Center Affiliation: The Scripps Research Institute, TSRI Assay Provider: Orphagen Pharmaceuticals, San Diego, CA Network: Molecular Library Screening Center Network (MLSCN) Grant Proposal Number: 1X01-MH077624-01 External Assay ID: SF1_AG_LUMI_1536_EC50 Nuclear receptors are a family of small molecule and hormone-regulated transcription factors that share conserved DNA-binding and ligand-binding domains. Small pharmacological compounds able to bind to the cleft of the ligand-binding domain could alter its conformation and subsequently modify transcription of target genes. Such ligand agonists and/or antagonists have already been successfully designed for 23 nuclear receptors among the 48 previously identified in the human genome [1-3]. The nuclear receptor SF-1 (steroidogenic factor-1) belongs to the class of "unexplored" orphan nuclear receptors that have been poorly investigated at a pharmacolog Dose-response cell-based assay for inhibitors of the Retinoic Acid Receptor-related orphan receptor A (RORA) Source (MLSCN Center Name): The Scripps Research Institute Molecular Screening Center Center Affiliation: The Scripps Research Institute, TSRI Assay Provider: Orphagen Pharmaceuticals, San Diego, CA Network: Molecular Library Screening Center Network (MLSCN) Proposal number 1X01-MH077624-01 External Assay ID: RORA_INH_Lumi_1536_IC50 Name: Dose-response cell-based assay for inhibitors of the Retinoic Acid Receptor-related orphan receptor A (RORA) Description: Nuclear receptors are a family of small molecule and hormone-regulated transcription factors that share conserved DNA-binding and ligand-binding domains. Small pharmacological compounds able to bind to the cleft of the ligand-binding domain could alter its conformation and subsequently modify transcription of target genes. Such ligand agonists and/or antagonists have already been successfully designed for 23 nuclear receptors among the 48 previously identified in the human genome [1-3]. RORA is one of three related orphan nu Dose-response cell-based assay for inhibitors of the nuclear receptor Steroidogenic Factor 1 (SF-1) Source (MLSCN Center Name): The Scripps Research Institute Molecular Screening Center Center Affiliation: The Scripps Research Institute, TSRI Assay Provider: Orphagen Pharmaceuticals, San Diego, CA Network: Molecular Library Screening Center Network (MLSCN) Grant proposal number 1X01-MH077624-01 External Assay ID: SF-1_INH_Lumi_1536_IC50 Name: Dose-response cell-based assay for inhibitors of the nuclear receptor Steroidogenic Factor 1 (SF-1) Description: Nuclear receptors are a family of small molecule and hormone-regulated transcription factors that share conserved DNA-binding and ligand-binding domains. Small pharmacological compounds able to bind to the cleft of the ligand-binding domain could alter its conformation and subsequently modify transcription of target genes. Such ligand agonists and/or antagonists have already been successfully designed for 23 nuclear receptors among the 48 previously identified in the human genome [1-3]. The nuclear receptor SF-1 (steroidogenic Dose-response confirmation of microRNA-mediated mRNA deadenylation inhibitors by fluoresence polarization assay Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Production Centers Network(MLPCN) Grant Number: R03 MH094198-01 Assay Provider: Kalle Gehring, Ph.D., McGill University The regulation of gene expression is a mechanism that allows cells to respond to growth and proliferation stimuli, stress, and nutrient availability. It is managed at multiple levels: mRNA expression, mRNA translation initiation and mRNA decay (1). MicroRNAs (miRNAs) are endogenous small RNAs that post-transcriptionally regulate gene expression to control a wide range of biological processes including cell growth, division and differentiation, as well as metabolism and development. The poly(A) tail of mRNA is bound by several molecules of the poly(A)-binding protein (PABP), an abundant cytoplasmic protein in eukaryotes that promotes translation (2). PABPC1 is a multi-domain protein t Dose-response primary assay and counterscreen assay for HTS small molecule inhibitors of CHOP to regulate the unfolded protein response to ER stress NIH Molecular Libraries Screening Centers Network [MLSCN] Emory Chemical Biology Discovery Center in MLSCN Assay provider: Dr. Randal Kaufman, University of Michigan MLSCN Grant: R03MH084182-01, U54HG003918 Assay Overview Many genetic and environmental diseases result from defective protein folding within the secretory pathway so that aberrantly folded proteins are recognized by the cellular surveillance system and retained within the endoplasmic reticulum (ER). Under conditions of malfolded protein accumulation, the cell activates the Unfolded Protein Response (UPR) to clear the malfolded proteins, and if unsuccessful, initiates a cell death response. Preliminary studies have shown that CHOP is a crucial factor in the apoptotic arm of the UPR; XBP1 activates genes encoding ER protein chaperones and thereby mediates the adaptive UPR response to increase clearance of malfolded proteins. Inhibition of CHOP is hypothesized to enhance survival by preventing UPR programmed cell death. E3 Ligase dose-response_384 Molecular Library Screening Center Network (MLSCN) Penn Center for Molecular Discovery (PCMD) Assay Provider: Brent Stockwell, Columbia University MLSCN Grant: R03MH082369-01 The E3 ligases are involved in regulating other proteins by covalent ligation to the 76 amino acid protein ubiquitin. This post-translational modification can result in altered conformation, altered activity, or degradation of the substrate protein. Thus, E3 ligases are effectors of a major means of post-translational modification of proteins in many species, including mammals. The dipeptide boronic acid bortezomib is a potent proteasome inhibitor, has selective anticancer activity in tumor cells and in mice and was recently approved for clinical use in multiple myeloma. MDM2 E3 ligase is involved in numerous types of human cancer. Selective E3 ligase inhibitors would be preferable as they would be more selective and less toxic. Inhibitors of the MDM2-UBCH5 interaction should disrupt the E3 ligase activity of M E3 Ligase_Mutant_Dose Response Molecular Library Screening Center Network (MLSCN) Penn Center for Molecular Discovery (PCMD) Assay Provider: Brent Stockwell, Columbia University MLSCN Grant: R03MH082369-01 The E3 ligases are involved in regulating other proteins by covalent ligation to the 76 amino acid protein ubiquitin. This post-translational modification can result in altered conformation, altered activity, or degradation of the substrate protein. Thus, E3 ligases are effectors of a major means of post-translational modification of proteins in many species, including mammals. The dipeptide boronic acid bortezomib is a potent proteasome inhibitor, has selective anticancer activity in tumor cells and in mice and was recently approved for clinical use in multiple myeloma. MDM2 E3 ligase is involved in numerous types of human cancer. Selective E3 ligase inhibitors would be preferable as they would be more selective and less toxic. Inhibitors of the MDM2-UBCH5 interaction should disrupt the E3 ligase activity of M E3 Ligase_WT_Dose Response Molecular Library Screening Center Network (MLSCN) Penn Center for Molecular Discovery (PCMD) Assay Provider: Brent Stockwell, Columbia University MLSCN Grant: R03MH082369-01 The E3 ligases are involved in regulating other proteins by covalent ligation to the 76 amino acid protein ubiquitin. This post-translational modification can result in altered conformation, altered activity, or degradation of the substrate protein. Thus, E3 ligases are effectors of a major means of post-translational modification of proteins in many species, including mammals. The dipeptide boronic acid bortezomib is a potent proteasome inhibitor, has selective anticancer activity in tumor cells and in mice and was recently approved for clinical use in multiple myeloma. MDM2 E3 ligase is involved in numerous types of human cancer. Selective E3 ligase inhibitors would be preferable as they would be more selective and less toxic. Inhibitors of the MDM2-UBCH5 interaction should disrupt the E3 ligase activity of M Epi-absorbance-based counterscreen for selective VIM-2 inhibitors: dose response biochemical high throughput screening assay to identify inhibitors of IMP-1 metallo-beta-lactamase. Source (MLPCN Center Name): The Scripps Research Institute Molecular Screening Center (SRIMSC) Center Affiliation: The Scripps Research Institute (TSRI) Assay Provider: Peter Hodder, TSRI Network: Molecular Libraries Probe Production Centers Network (MLPCN) Grant Proposal Number: 1 R21 NS059451-01 Fast Track Grant Proposal PI: Peter Hodder, TSRI External Assay ID: IMP1NITRO_INH_EPIABS_1536_3XIC50 Name: Epi-absorbance-based counterscreen for selective VIM-2 inhibitors: dose response biochemical high throughput screening assay to identify inhibitors of IMP-1 metallo-beta-lactamase. Description: The emergence of gram-negative bacteria that exhibit multi-drug resistance, combined with the paucity of new antibiotics, poses a public health challenge (1). The production of bacterial beta-lactamase enzymes, in particular, is a common mechanism of drug resistance (2-4). The beta-lactamases evolved from bacteria with resistance to naturally-occurring beta-lactams or penams (5), agents which Epi-absorbance-based dose response biochemical high throughput screening assay for selective inhibitors of VIM-2 metallo-beta-lactamase Source (MLPCN Center Name): The Scripps Research Institute Molecular Screening Center (SRIMSC) Center Affiliation: The Scripps Research Institute (TSRI) Assay Provider: Peter Hodder, TSRI Network: Molecular Libraries Probe Production Centers Network (MLPCN) Grant Proposal Number: 1 R21 NS059451-01 Fast Track Grant Proposal PI: Peter Hodder, TSRI External Assay ID: VIM2NITRO_INH_EPIABS_1536_3XIC50 Name: Epi-absorbance-based dose response biochemical high throughput screening assay for selective inhibitors of VIM-2 metallo-beta-lactamase Description: The emergence of gram-negative bacteria that exhibit multi-drug resistance, combined with the paucity of new antibiotics, poses a public health challenge (1). The production of bacterial beta-lactamase enzymes, in particular, is a common mechanism of drug resistance (2-4). The beta-lactamases evolved from bacteria with resistance to naturally-occurring beta-lactams or penams (5), agents which inhibit the transpeptidase involved in cell Estrogen Receptor-alpha Coactivator Binding Inhibitors Dose Response Confirmation NIH Molecular Libraries Screening Centers Network [MLSCN] Emory Chemical Biology Discovery Center in MLSCN Assay provider: John A. Katzenellenbogen, University of Illinois at Urbana-Champaign MLSCN Grant: 1 X01MH78953-01 Title: HTS for Estrogen Receptor-alpha Coactivator Binding inhibitors Assay Overview Estrogens, which are responsible for the growth of many breast cancers, act through the estrogen receptors, ER-alpha and ER-beta, which are ligand-modulated transcription factors and members of the nuclear receptor gene superfamily. ER-alpha and ER-beta are well validated protein targets for various aspects of women's health and breast cancer prevention and treatment. As an essential step in their action as regulators of gene transcription, the ERs recruit various coregulator proteins, both coactivators and corepressors, to the DNA-bound (or protein tethered) ER. These coregulators have various activities: alteration of chromatin architecture, regulation of nucleosome core FRET-based counterscreen for selective VIM-2 inhibitors: dose response biochemical high throughput screening assay to identify epi-absorbance assay artifacts. Source (MLPCN Center Name): The Scripps Research Institute Molecular Screening Center (SRIMSC) Center Affiliation: The Scripps Research Institute (TSRI) Assay Provider: Peter Hodder, TSRI Network: Molecular Libraries Probe Production Centers Network (MLPCN) Grant Proposal Number: 1 R21 NS059451-01 Fast Track Grant Proposal PI: Peter Hodder, TSRI External Assay ID: VIM2CCF2_INH_FRET_1536_3XIC50 Name: FRET-based counterscreen for selective VIM-2 inhibitors: dose response biochemical high throughput screening assay to identify epi-absorbance assay artifacts. Description: The emergence of gram-negative bacteria that exhibit multi-drug resistance, combined with the paucity of new antibiotics, poses a public health challenge (1). The production of bacterial beta-lactamase enzymes, in particular, is a common mechanism of drug resistance (2-4). The beta-lactamases evolved from bacteria with resistance to naturally-occurring beta-lactams or penams (5), agents which inhibit the transpeptidas FRET-based counterscreen for selective VIM-2 inhibitors: dose response biochemical high throughput screening assay to identify inhibitors of IMP-1 metallo-beta-lactamase. Source (MLPCN Center Name): The Scripps Research Institute Molecular Screening Center (SRIMSC) Center Affiliation: The Scripps Research Institute (TSRI) Assay Provider: Peter Hodder, TSRI Network: Molecular Libraries Probe Production Centers Network (MLPCN) Grant Proposal Number: 1 R21 NS059451-01 Fast Track Grant Proposal PI: Peter Hodder, TSRI External Assay ID: IMP1CCF2_INH_FRET_1536_3X%IC50 Name: FRET-based counterscreen for selective VIM-2 inhibitors: dose response biochemical high throughput screening assay to identify inhibitors of IMP-1 metallo-beta-lactamase. Description: The emergence of gram-negative bacteria that exhibit multi-drug resistance, combined with the paucity of new antibiotics, poses a public health challenge (1). The production of bacterial beta-lactamase enzymes, in particular, is a common mechanism of drug resistance (2-4). The beta-lactamases evolved from bacteria with resistance to naturally-occurring beta-lactams or penams (5), agents which inhibit the Factor XIIa Dose Response Confirmation Molecular Library Screening Center Network (MLSCN) Penn Center for Molecular Discovery (PCMD) Assay Provider: Scott L. Diamond, University of Pennsylvania MLSCN Grant: X01-MH076406-01 Target Factor XII (FXII) is a 80 kDa zymogen found at a concentration of 0.375 uM in plasma, and upon activation by kallikrein at R353, a disulfide-linked two chain molecule called factor XIIa alpha (FXIIa) is generated. FXIIa is also capable of autoactivation by binding to negatively charged surfaces (1). Kallikrein can also cleave other scissile bonds in FXIIa alpha outside of the catalytic domain at R334, R343, and R353, generating FXIIa beta, a 30 kDa enzyme that is no longer able to bind to surfaces, and which activates prekallikrein (PK) to kallikrein, using high molecular weight kininogen (HK) as a cofactor (2, 3). FXIIa is irreversibly inhibited by C-1 inhibitor (C1INH), a 105 kDa plasma SERPIN (4-6). FXII, PK, HK, C1INH, and factor XI (FXI) have been traditionally placed within the i Fluorescence Polarization Dose Response Assay for TR3-Based Bcl-B Inhibitors Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA). Network: NIH Molecular Libraries Screening Centers Network (MLSCN) Grant Number: XO1 MH077632-01 Assay Provider: Dr. John C. Reed, Sanford-Burnham Medical Research Institute This dose response assay is developed and performed for the purpose of confirming hits originally identified in Fluorescence Polarization Screen Assay for Bcl-B Phenotype Converters (AID 1240). Bcl-B is an anti-apoptotic member of the Bcl-2 family that is prominently expressed in plasma and multiple myeloma cells. TR3 (NR4A1; HMR; NP10; GFRP1; NAK1; NUR77; NGFIB) is an orphan member of the steroid/thyroid/retinoid nuclear receptor superfamily that translocates from cellular nuclei to mitochondria upon exposure to various pro-apoptotic stimuli. At mitochondria, TR3 binds to Bcl-B and converts it into a pro-apoptotic protein. A specific 9-amino Fluorescence counterscreen assay for TRPML3 agonists: dose response cell-based high-throughput screening assay to identify agonists of the transient receptor potential channel N1 (TRPN1) Source (MLPCN Center Name): The Scripps Research Institute Molecular Screening Center (SRIMSC) Center Affiliation: The Scripps Research Institute (TSRI) Assay Provider: Stefan Heller, Stanford University Network: Molecular Libraries Probe Production Centers Network (MLPCN) Grant Proposal Number 1 R03 MH083077-01 Grant Proposal PI: Stefan Heller , Stanford University External Assay ID: TRPN1_AG_Calcium_1536_3XEC50 CS Name: Fluorescence counterscreen assay for TRPML3 agonists: dose response cell-based high-throughput screening assay to identify agonists of the transient receptor potential channel N1 (TRPN1) Description: Cell signaling pathways that mediate osmosensation, photosensation, and thermosensation depend on a family of diverse transient receptor potential (TRP) cation channels, which are activated by agonist-receptor coupling (1-5). A role for these channels in inner ear hair cell mechanotransduction was gleaned from TRP channel mutations identified in flies, worms, and low Fluorescence counterscreen assay for TRPN1 agonists: dose response cell-based high-throughput screening assay to identify agonists of the transient receptor potential channel ML3 (TRPML3) Source (MLPCN Center Name): The Scripps Research Institute Molecular Screening Center (SRIMSC) Center Affiliation: The Scripps Research Institute (TSRI) Assay Provider: Stefan Heller, Stanford University Network: Molecular Libraries Probe Production Centers Network (MLPCN) Grant Proposal Number 1 R03 MH083077-01 Grant Proposal PI: Stefan Heller , Stanford University External Assay ID: TRPML3_AG_FLUO8_1536_3XEC50 Counterscreen Name: Fluorescence counterscreen assay for TRPN1 agonists: dose response cell-based high-throughput screening assay to identify agonists of the transient receptor potential channel ML3 (TRPML3) Description: Cell signaling pathways that mediate osmosensation, photosensation, and thermosensation depend on a family of diverse transient receptor potential (TRP) cation channels, which are activated by agonist-receptor coupling (1-4). A role for these channels in inner ear hair cell mechanotransduction was gleaned from TRP channel mutations identified in flies, wo Fluorescence counterscreen for potentiators or agonists of NPY-Y1: Cell-based high-throughput dose response assay for potentiators or agonists of NPY-Y2. Source (MLPCN Center Name): The Scripps Research Institute Molecular Screening Center (SRIMSC) Center Affiliation: The Scripps Research Institute (TSRI) Assay Provider: Claes Wahlestedt, Scripps Florida Network: Molecular Library Probe Production Centers Network (MLPCN) Grant Proposal Number: 1 R21 NS056950-01 Grant Proposal PI: Claes Wahlestedt External Assay ID: NPY-Y2_POT_CNGC_1536_3XEC50 CSDRUN Name: Fluorescence counterscreen for potentiators or agonists of NPY-Y1: Cell-based high-throughput dose response assay for potentiators or agonists of NPY-Y2. Description: Neuropeptide Y (NPY) is a neurotransmitter with physiologic roles including control of feeding behavior, regulation of cortical neural activity, heart neural activity, and emotional regulation. Importantly, NPY is implicated in human diseases such as obesity, depression and alcoholism. NPY mediates its biological effects in part through activation of the Galphai protein coupled receptors (GPCRs) NPY-Y1 and Y2 receptor Fluorescence polarization assay for PKD inhibitiors-interference assay-57K HTS campaign The PKD HTS assay was developed and run at the University of Pittsburgh Molecular Screening Center (PMLSC) as part of the Molecular Library Screening Center Network (MLSCN)(1R03DA24898-01). Protein kinase D (PKD) is a novel family of serine/threonine kinases targeted by diacylglycerol. It regulates many fundamental cell functions including cell proliferation, survival, differentiation and protein trafficking, and plays important roles in pathological conditions such as cardiac hypertrophy and cancer in multiple organ systems. However, the mechanisms underlying these effects of PKD are not clearly understood, and the role of PKD in cancer and other diseases has not been fully defined. This is partly due to the lack of effective pharmacological tools that specifically target PKD in normal cellular processes and in pathological conditions. The immediate goal of this proposal is to demonstrate the feasibility of an IMAP-based fluorescent polarization (FP) assay for high throughput scree Fluorescence polarization-based biochemical high throughput dose response assay for inhibitors of GLD-1 protein - TGE RNA interaction Source (MLPCN Center Name): The Scripps Research Institute Molecular Screening Center (SRIMSC) Affiliation: The Scripps Research Institute, TSRI Assay Provider: James R. Williamson, TSRI Network: Molecular Library Probe Production Centers Network (MLPCN) Grant Proposal Number: 1 R21 NS056951-01 Grant Proposal PI: James R. Williamson External Assay ID: GLD1_INH_FP_1536_3XIC50 DRUN Name: Fluorescence polarization-based biochemical high throughput dose response assay for inhibitors of gld-1 protein - TGE RNA interaction. Description: Post-transcriptional control of gene expression, such as alternative splicing and tissue-specific silencing, allow for great protein diversity (1). The elements in the mRNA 3' untranslated regions (UTRs) influence the expression of genes involved in proliferation and differentiation of stem cells and germ cells (2). These elements are critical during spermatogenesis and oogenesis in hermaphrodite Caenorhabditis elegans nematodes (3). gld-1 (defective Fluorescence polarization-based biochemical high throughput dose response assay for inhibitors of myeloid cell leukemia sequence 1 (MCL1) interactions with BIM-BH3 peptide. Source (MLPCN Center Name): The Scripps Research Institute Molecular Screening Center Affiliation: The Scripps Research Institute, TSRI Assay Provider: Michael Cardone, Eutropics Network: Molecular Library Probe Production Centers Network (MLPCN) Grant Proposal Number: 1 R43 CA135915-01 Fast Track Grant Proposal PI: Michael Cardone, Eutropics External Assay ID: MCL1BIM_INH_FP_1536_3XIC50 Name: Fluorescence polarization-based biochemical high throughput dose response assay for inhibitors of myeloid cell leukemia sequence 1 (MCL1) interactions with BIM-BH3 peptide. Description: Cancer initialization and survival depends upon evasion of the programmed cell death (apoptosis) machinery that normally kills an unneeded or rogue cell (1). Although an effective mechanism for anti-cancer chemotherapeutics is apoptosis induction, cancer cells develop resistance to the pro-apoptotic proteins activated by these drugs (2). Multiple myeloma (MM) and chronic lymphoblastic leukemia (CLL) are two we Fluorescence polarization-based biochemical high throughput dose response assay for inhibitors of the Epstein-Barr virus nuclear antigen 1 (EBNA-1) Source (MLPCN Center Name): The Scripps Research Institute Molecular Screening Center (SRIMSC) Center Affiliation: The Scripps Research Institute (TSRI) Assay Provider: Paul Lieberman, Wistar Institute Network: Molecular Libraries Probe Production Centers Network (MLPCN) Grant Proposal Number: I R21 NS063906-01 Grant Proposal PI: Paul Lieberman, Wistar Institute External Assay ID: EBNA1_INH_FP_1536_3XIC50 Name: Fluorescence polarization-based biochemical high throughput dose response assay for inhibitors of the Epstein-Barr virus nuclear antigen 1 (EBNA-1). Description: During each cell cycle in eukaryotes, the genome must be completely replicated and this replication must begin at the correct time and site (initiation site or origin) (1). Pathogenic viruses often take advantage of this cellular precision to maintain replication of their own genome. The Epstein-Barr virus (EBV) is an orally-transmitted herpesvirus associated with numerous human neoplasms (2) that infects over 90% o Fluorescence polarization-based biochemical high throughput dose response assay to identify inhibitors of tRNA 2'-phosphotransferase (TPT1). Source (MLPCN Center Name): The Scripps Research Institute Molecular Screening Center (SRIMSC) Center Affiliation: The Scripps Research Institute (TSRI) Assay Provider: Heather Harding, New York University School of Medicine Network: Molecular Libraries Probe Production Centers Network (MLPCN) Grant Proposal Number: 1-R03-DA026554-01 Grant Proposal PI: Heather Harding, New York University School of Medicine External Assay ID: TPT1_INH_FP_1536_3XIC50 Name: Fluorescence polarization-based biochemical high throughput dose response assay to identify inhibitors of tRNA 2'-phosphotransferase (TPT1). Description: The process of transcription converts the DNA sequences found in genes into mRNA (1, 2). This process is coupled to the subsequent removal of mRNA introns by splicing, which additionally serves to increase proteome complexity (3, 4). Intron splicing also occurs for transfer RNA (tRNA), which functions in the delivery of amino acids to the growing peptide chain as directed by Fluorescence-based biochemical dose response assay to identify inhibitors of Protein Arginine Deiminase 4 (PAD4) Source (MLPCN Center Name): The Scripps Research Institute Molecular Screening Center (SRIMSC) Affiliation: The Scripps Research Institute, TSRI Assay Provider: Paul Thompson, University of South Carolina Network: Molecular Library Probe Production Centers Network (MLPCN) Grant Proposal Number: R01 GM079357-01 Grant Proposal PI: Paul Thompson External Assay ID: PAD4_INH_FLUO_2X%IC50 Name: Fluorescence-based biochemical dose response assay to identify inhibitors of Protein Arginine Deiminase 4 (PAD4). Description: Rheumatoid Arthritis (RA) is a chronic and progressive autoimmune disorder that affects about one percent of the US population (1). Existing therapies treat the symptoms of the disease but not the underlying cause, and are associated with numerous side effects (2). The activity of Protein Arginine Deiminase 4 (PAD4), one of four known active PAD isozymes, is increased in RA; where it is thought to generate a subset of antigens that the immune system recognizes as foreign ( Fluorescence-based biochemical high throughput dose response assay for inhibitors of the Hepatitis C Virus non-structural protein 3 helicase (NS3). Source (MLPCN Center Name): The Scripps Research Institute Molecular Screening Center (SRIMSC) Center Affiliation: The Scripps Research Institute (TSRI) Assay Provider: David Frick, New York Medical College Network: Molecular Libraries Probe Production Centers Network (MLPCN) Grant Proposal Number: 1 R03 MH085690-01 Grant Proposal PI: David Frick, New York Medical College External Assay ID: NS3DNA_INH_FLINT_1536_3XIC50 Name: Fluorescence-based biochemical high throughput dose response assay for inhibitors of the Hepatitis C Virus non-structural protein 3 helicase (NS3). Description: The flavivirus Hepatitis C virus (HCV) is a major cause of liver failure and hepatocellular cancer, with about 170 million people infected worldwide (1). The HCV has a small RNA genome that is directly translated by the infected host cell into a single precursor polyprotein that is processed by enzymatic cleavage into 10 proteins of diverse function. The non-structural proteins include p7, NS2, NS3, NS4 Fluorescence-based counterscreen for agonists of NPY-Y1: cell-based high-throughput dose response assay for agonists of NPY-Y2. Source (MLPCN Center Name): The Scripps Research Institute Molecular Screening Center (SRIMSC) Center Affiliation: The Scripps Research Institute (TSRI) Assay Provider: Claes Wahlestedt, Scripps Florida Network: Molecular Library Probe Production Centers Network (MLPCN) Grant Proposal Number 1 R21 NS056950-01 Grant Proposal PI: Claes Wahlestedt External Assay ID: NPY-Y2_AG_CNGC_1536_3XEC50 CSDRUN Name: Fluorescence-based counterscreen for agonists of NPY-Y1: cell-based high-throughput dose response assay for agonists of NPY-Y2. Description: Neuropeptide Y (NPY) is a neurotransmitter with physiologic roles including control of feeding behavior, regulation of cortical neural activity, heart neural activity, and emotional regulation. Importantly, NPY is implicated in human diseases such as obesity, depression and alcoholism. NPY mediates its biological effects in part through activation of the Galphai protein coupled receptors (GPCRs) NPY-Y1 and Y2 receptors, which decrease cytosolic Fluorescence-based counterscreen for agonists of NPY-Y2: cell-based high-throughput dose response assay for agonists of NPY-Y1. Source (MLPCN Center Name): The Scripps Research Institute Molecular Screening Center (SRIMSC) Center Affiliation: The Scripps Research Institute (TSRI) Assay Provider: Claes Wahlestedt, Scripps Florida Network: Molecular Library Probe Production Centers Network (MLPCN) Grant Proposal Number: 1 R21 NS056950-01 Grant Proposal PI: Claes Wahlestedt External Assay ID: NPY-Y1_AG_CNGC_1536_3XEC50 CSDRUN Name: Fluorescence-based counterscreen for agonists of NPY-Y2: cell-based high-throughput dose response assay for agonists of NPY-Y1. Description: Neuropeptide Y (NPY) is a neurotransmitter with diverse physiologic roles including control of feeding behavior, regulation of cortical neural activity, heart neural activity, and emotional regulation. Importantly, NPY is implicated in human diseases such as obesity, depression and alcoholism. NPY mediates its biological effects in part through activation of the NPY-Y2 receptor, a 381-amino acid Galphai protein coupled receptor (GPCR) which dec Fluorescence-based counterscreen for potentiators or agonists of NPY-Y2: cell-based high-throughput dose response assay for potentiators or agonists of NPY-Y1. Source (MLPCN Center Name): The Scripps Research Institute Molecular Screening Center (SRIMSC) Center Affiliation: The Scripps Research Institute (TSRI) Assay Provider: Claes Wahlestedt, Scripps Florida Network: Molecular Library Probe Production Centers Network (MLPCN) Grant Proposal Number: 1 R21 NS056950-01 Grant Proposal PI: Claes Wahlestedt External Assay ID: NPY-Y1_POT_CNGC_1536_3XEC50 CSDRUN Name: Fluorescence-based counterscreen for potentiators or agonists of NPY-Y2: cell-based high-throughput dose response assay for potentiators or agonists of NPY-Y1. Description: Neuropeptide Y (NPY) is a neurotransmitter with diverse physiologic roles including control of feeding behavior, regulation of cortical neural activity, heart neural activity, and emotional regulation. Importantly, NPY is implicated in human diseases such as obesity, depression and alcoholism. NPY mediates its biological effects in part through activation of the NPY-Y2 receptor, a 381-amino acid Galphai protein Fluorescence-based dose response cell-based high throughput screening assay to identify antagonists of the G-protein coupled receptor 7 (GPR7). Source (MLPCN Center Name): The Scripps Research Institute Molecular Screening Center (SRIMSC) Affiliation: The Scripps Research Institute, TSRI Assay Provider: Olivier Civelli, University of California, Irvine Network: Molecular Library Probe Production Centers Network (MLPCN) Grant Proposal Number: 1-R03-DA026557-01 Grant Proposal PI: Olivier Civelli External Assay ID: GPR7_ANT_FLUO8_1536_3X%IC50 Name: Fluorescence-based dose response cell-based high throughput screening assay to identify antagonists of the G-protein coupled receptor 7 (GPR7). Description: Heterotrimeric G-protein coupled receptors (GPCRs) are major targets for disease therapeutics, due in part to their broad tissue distribution, structural diversity, varied modes of action, and disease-associated mutations (1-4). For example, targeting of opiod receptors by opiates such as morphine is a widespread clinical application for GPCR modulation in pain management. The recently de-orphanized GPR7 (5) is localized predom Formylpeptide Receptor (FPRL1) Ligand Structure Activity Relationship (SAR) Analysis : Dose Response Assay University of New Mexico Assay Overview: Assay Support: NIH 1R03MH076381-01 Assay for Formylpeptide Receptor Family Ligands PI: Bruce S. Edwards, Ph.D. Assay Background and Significance Formyl peptide receptors. The G-protein coupled formylpeptide receptor (FPR) was one of the originating members of the chemoattractant receptor superfamily (Le et al., 2002a; Oppenheim et al., 1991). N-formylated peptides such as fMLF are high affinity FPR ligands that trigger a variety of biologic activities in myeloid cells, including chemokinesis, chemotaxis, cytokine production and superoxide generation (He et al., 2003; Le et al., 2001b; Murphy, 1994; Murphy, 1996; Tiffany et al., 2001). Since such peptides are derived from bacterial or mitochondrial proteins (Carp, 1982; Marasco et al., 1984; Schiffmann et al., 1975a; Schiffmann et al., 1975b), it has been proposed that a primary FPR function is to promote trafficking of phagocytic myeloid cells to sites of infection and tissue damage where HTS TR-FRET-based dose response confirmatory assay for Siah-1 Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Probe Production Centers Network (MLPCN) Grant Number: 1 R03 MH086475-01 Assay Provider: Dr. John C. Reed, Sanford-Burnham Medical Research Institute, San Diego CA Proteasomal degradation typically requires post-translational modification of target proteins with K48-linked polyubiquitin chains. This process of protein proteolysis plays a key role in normal cellular function. The E3 ubiquitin ligase, Siah-1, facilitates the transfer of ubiquitin to its substrate proteins destined for degradation by way of its RING domain. Siah-1 is a member of a family of highly conserved RING domain proteins, which regulate a variety of cellular functions, including cell cycle arrest, tumor suppression, and apoptosis through the beta-catenin degradation pathway. Siah-1 has also been identified as a p53-inducible gene, HTS Dose response counterscreen for assays utilizing the enzyme, b-galactosidase Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Production Centers Network (MLPCN) Grant Number: 1X01DA026208-01 Assay Provider: Dr. Lawrance Barak , Duke University, Durham NC b-galactosidase (b-gal), a hydrolase enzyme that catalyzes the hydrolysis of b-galactosides to monosaccharides is utilized in many different screening technologies involving enzyme reaction coupling and reporter assays, for example DiscoverX b-Arrestin GPCR assays such as the KOR1 Agonist or Antagonist. This assay was developed and performed as a counterscreen for screening assays that utilize b-gal and a reaction that it catalyzes. By detecting inhibitors and activators of this enzyme, it is possible to attribute activity not to the primary assay in question, but rather to interaction with the method of detection. References Fowler et al. (1970). "The amino acid sequ HTS Dose response counterscreen for assays utilizing the enzyme, beta-galactosidase - Set 2 Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute(SBMRI, San Diego, CA) Network: NIH Molecular Libraries Probe Production Centers Network (MLPCN) Grant Number: 1X01DA026208-01 Assay Provider: Dr. Lawrance Barak , Duke University, Durham, NC b-galactosidase (b-gal), a hydrolase enzyme that catalyzes the hydrolysis of b-galactosides to monosaccharides is utilized in many different screening technologies involving enzyme reaction coupling and reporter assays, for example DiscoverX b-Arrestin GPCR assays such as the APJ Agonist or Antagonist. This assay was developed and performed as a counterscreen for screening assays that utilize b-gal and a reaction that it catalyzes. By detecting inhibitors and activators of this enzyme, it is possible to attribute activity not to the primary assay in question, but rather to interaction with the method of detection. References Fowler et al. (1970). "The amino aci HTS Dose response counterscreen for assays utilizing the enzyme, beta-galactosidase - Set 3 Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute(SBMRI, San Diego, CA) Network: NIH Molecular Libraries Probe Production Centers Network (MLPCN) Grant Number: 1X01DA026208-01 Assay Provider: Dr. Lawrance Barak , Duke University, Durham, NC b-galactosidase (b-gal), a hydrolase enzyme that catalyzes the hydrolysis of b-galactosides to monosaccharides is utilized in many different screening technologies involving enzyme reaction coupling and reporter assays. This assay was developed and performed as a counterscreen for the primary assay originally identified as "uHTS identification of small molecule antagonists of the CCR6 receptor via a luminescent beta-arrestin assay", AID 493098. By detecting inhibitors and activators of this enzyme, it is possible to attribute activity not to the primary assay in question, but rather to interaction with the method of detection. References Fowler et al. (1970). HTS HePTP Fluorescent Assay using OMFP substrate for In Vitro dose response studies Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Screening Centers Network (MLSCN) Grant Number: XO1 MH077603-01 Assay Provider: Dr. Tomas Mustelin, Sanford-Burnham Medical Research Institute Protein tyrosine phosphatases (PTPs), working with protein tyrosine kinases (PTKs), control the phosphorylation state of many proteins in the signal transduction pathways. HePTP is a tyrosine phosphatase expressed in hematopoietic cells and regulates the MAP kinases Erk and p38. It has been found that HePTP is often dysregualted in the preleukemic disorder myelodysplastic syndrome, as well as in acute myelogeneous leukemia. Small molecule inhibitors of HePTP will be useful as molecular probes for studying the mechanism of signal transduction and MAP kinase regulation, and may have therapeutic potential for the treatment of hematopoietic malignancies. This bioc HTS Image-Based Screen for Agonists of the DOR Receptor Data Source: Sanford-Burnham Center for Chemical Genomics Source Affiliation: Sanford-Burnham Medical Research Institute Network: NIH Molecular Libraries Probe Production Centers Network Grant Number: 1X01DA026208-01 Assay Provider: Dr. Lawrance Barak, Duke University, Durham, NC Drug addiction is a disease that involves the G-protein coupled receptors in the central nervous system resulting in compulsive or abnormal behavior. Recent studies have shown that opioid receptors play a role in regulating other receptors that interact with drug and other substance abuse. The opioid receptors are composed of multiple of multiple receptor subtypes whose contribution the addictive behaviors are not fully delineated. The aim of this assay is to identify small molecule agonists of the human delta opioid receptor (DOR). The dose response assay is developed and performed to evaluate selectivity of hits originally identified in a uHTS luminescent beta-arrestin assay for agonists of the KOR re HTS Image-Based Screen for Antagonists of the DOR Receptor Data Source: Sanford-Burnham Center for Chemical Genomics Source Affiliation: Sanford-Burnham Medical Research Institute Network: NIH Molecular Libraries Probe Production Centers Network Grant Number: 1X01DA026208-01 Assay Provider: Dr. Lawrance Barak, Duke University, Durham, NC Drug addiction is a disease that involves the G-protein coupled receptors in the central nervous system resulting in compulsive or abnormal behavior. Recent studies have shown that opioid receptors play a role in regulating other receptors that interact with drug and other substance abuse. The opioid receptors are composed of multiple of multiple receptor subtypes whose contribution the addictive behaviors are not fully delineated. The aim of this assay is to identify small molecule antagonists of the human delta opioid receptor (DOR). The dose response assay is developed and performed to evaluate selectivity of hits originally identified in a uHTS luminescent beta-arrestin assay for agonists of the KOR High throughput discovery of novel modulators of ROMK K+ channel activity: Analog Library Testing Assay Provider: Jerod Denton Assay Provider Affiliation: Vanderbilt University Grant Title: High throughput discovery of novel modulators of ROMK K+ channel activity Grant Number: R21 NS057041-01 The Renal Outer Medullary Potassium channel (ROMK, Kir1.1) is expressed in the renal tubule where it critically regulates fluid and electrolyte homeostasis (1). An emerging body of evidence suggests that ROMK could be a target for a novel class loop diuretic that lowers blood pressure while preserving plasma potassium levels (2). Furthermore, homozygous loss-of-function mutations in the gene encoding ROMK (KCNJ1) cause antenatal Bartter syndrome, a severe salt and water wasting disease in infants (3). ROMK is thus an important pharmacological target for the management of disease. Its actual therapeutic value and drugability, however, are unknown due to the lack of small-molecule probes targeting the channel. The discovery of ROMK modulators will provide important new tools for studying High-throughput multiplex microsphere dose response for inhibitors of toxin protease, specifically Botulinum neurotoxin light chain A protease, compounds from Cherry Pick 01 University of New Mexico Assay Overview: Assay Support: 1 R03 MH093184-01A1 Project Title: High-throughput multiplex microsphere screening for toxin protease inhibitors Assay Provider: Steven Graves Ph.D. Screening Center/PI: UNMCMD/ Larry Sklar Ph.D. Lead Biologist: Bruce Edwards Ph.D., Screening Operations Team: Jingshu Zhu, Mark Carter MS, Kristine Gouveia MS, Matthew Garcia Chemistry Center PI: Craig W. Lindsley Chemistry Lead: Kyle Emmitte Specialized Chemistry Center: Vanderbilt Specialized Chemistry Center For Accelerated Probe Development Proteases regulate many biological pathways that include: coagulation, immune system activation, metastasis, and viral life cycles. Within the larger set of proteases, pharmaceutical development for the proteases of the two-part bacterial toxins of Clostridium botulinum and Bacillus anthracis is of great interest due to their role in natural disease and biothreat scenarios (1-4). Botulinum Neurotoxin A Light Chain (BoNTALC) is also High-throughput multiplex microsphere dose response for inhibitors of toxin protease, specifically Botulinum neurotoxin light chain A protease, compounds from Powder Set 01 University of New Mexico Assay Overview: Assay Support: 1 R03 MH093184-01A1 Project Title: High-throughput multiplex microsphere screening for toxin protease inhibitors Assay Provider: Steven Graves Ph.D. Screening Center/PI: UNMCMD/ Larry Sklar Ph.D. Lead Biologist: Bruce Edwards Ph.D., Screening Operations Team: Jingshu Zhu, Mark Carter MS, Kristine Gouveia MS, Matthew Garcia Chemistry Center PI: Craig W. Lindsley Chemistry Lead: Kyle Emmitte Specialized Chemistry Center: Vanderbilt Specialized Chemistry Center For Accelerated Probe Development Proteases regulate many biological pathways that include: coagulation, immune system activation, metastasis, and viral life cycles. Within the larger set of proteases, pharmaceutical development for the proteases of the two-part bacterial toxins of Clostridium botulinum and Bacillus anthracis is of great interest due to their role in natural disease and biothreat scenarios (1-4). Botulinum Neurotoxin A Light Chain (BoNTALC) is also High-throughput multiplex microsphere dose response for inhibitors of toxin protease, specifically Botulinum neurotoxin light chain F protease, compounds from Cherry Pick 01 University of New Mexico Assay Overview: Assay Support: 1 R03 MH093184-01A1 Project Title: High-throughput multiplex microsphere screening for toxin protease inhibitors Assay Provider: Steven Graves Ph.D. Screening Center/PI: UNMCMD/ Larry Sklar Ph.D. Lead Biologist: Bruce Edwards Ph.D., Screening Operations Team: Jingshu Zhu, Mark Carter MS, Kristine Gouveia MS, Matthew Garcia Chemistry Center PI: Craig W. Lindsley Chemistry Lead: Kyle Emmitte Specialized Chemistry Center: Vanderbilt Specialized Chemistry Center For Accelerated Probe Development Proteases regulate many biological pathways that include: coagulation, immune system activation, metastasis, and viral life cycles. Within the larger set of proteases, pharmaceutical development for the proteases of the two-part bacterial toxins of Clostridium botulinum and Bacillus anthracis is of great interest due to their role in natural disease and biothreat scenarios (1-4). Botulinum Neurotoxin A Light Chain (BoNTALC) is also High-throughput multiplex microsphere dose response for inhibitors of toxin protease, specifically Botulinum neurotoxin light chain F protease, compounds from Powder Set 01 University of New Mexico Assay Overview: Assay Support: 1 R03 MH093184-01A1 Project Title: High-throughput multiplex microsphere screening for toxin protease inhibitors Assay Provider: Steven Graves Ph.D. Screening Center/PI: UNMCMD/ Larry Sklar Ph.D. Lead Biologist: Bruce Edwards Ph.D., Screening Operations Team: Jingshu Zhu, Mark Carter MS, Kristine Gouveia MS, Matthew Garcia Chemistry Center PI: Craig W. Lindsley Chemistry Lead: Kyle Emmitte Specialized Chemistry Center: Vanderbilt Specialized Chemistry Center For Accelerated Probe Development Proteases regulate many biological pathways that include: coagulation, immune system activation, metastasis, and viral life cycles. Within the larger set of proteases, pharmaceutical development for the proteases of the two-part bacterial toxins of Clostridium botulinum and Bacillus anthracis is of great interest due to their role in natural disease and biothreat scenarios (1-4). Botulinum Neurotoxin A Light Chain (BoNTALC) is also High-throughput multiplex microsphere dose response for inhibitors of toxin protease, specifically Lethal Factor protease, compounds from Cherry Pick 01 University of New Mexico Assay Overview: Assay Support: 1 R03 MH093184-01A1 Project Title: High-throughput multiplex microsphere screening for toxin protease inhibitors Assay Provider: Steven Graves Ph.D. Screening Center/PI: UNMCMD/ Larry Sklar Ph.D. Lead Biologist: Bruce Edwards Ph.D., Screening Operations Team: Jingshu Zhu, Mark Carter MS, Kristine Gouveia MS, Matthew Garcia Chemistry Center PI: Craig W. Lindsley Chemistry Lead: Kyle Emmitte Specialized Chemistry Center: Vanderbilt Specialized Chemistry Center For Accelerated Probe Development Proteases regulate many biological pathways that include: coagulation, immune system activation, metastasis, and viral life cycles. Within the larger set of proteases, pharmaceutical development for the proteases of the two-part bacterial toxins of Clostridium botulinum and Bacillus anthracis is of great interest due to their role in natural disease and biothreat scenarios (1-4). Botulinum Neurotoxin A Light Chain (BoNTALC) is also High-throughput multiplex microsphere dose response for inhibitors of toxin protease, specifically Lethal Factor protease, compounds from Powder Set 01 University of New Mexico Assay Overview: Assay Support: 1 R03 MH093184-01A1 Project Title: High-throughput multiplex microsphere screening for toxin protease inhibitors Assay Provider: Steven Graves Ph.D. Screening Center/PI: UNMCMD/ Larry Sklar Ph.D. Lead Biologist: Bruce Edwards Ph.D., Screening Operations Team: Jingshu Zhu, Mark Carter MS, Kristine Gouveia MS, Matthew Garcia Chemistry Center PI: Craig W. Lindsley Chemistry Lead: Kyle Emmitte Specialized Chemistry Center: Vanderbilt Specialized Chemistry Center For Accelerated Probe Development Proteases regulate many biological pathways that include: coagulation, immune system activation, metastasis, and viral life cycles. Within the larger set of proteases, pharmaceutical development for the proteases of the two-part bacterial toxins of Clostridium botulinum and Bacillus anthracis is of great interest due to their role in natural disease and biothreat scenarios (1-4). Botulinum Neurotoxin A Light Chain (BoNTALC) is also Homogeneous Time-Resolved Fluorescence Resonance Energy Transfer (HTRF) Assay Data Source: Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute Network: NIH Molecular Libraries Production Centers Network (MLPCN) Grant Number: 1 R03 MH085675-01 Assay Provider: Dr Ilya Bezprozvanny, UT Southwestern Medical Center , Dallas, TX Chronic pain (neuropathic pain, inflammatory pain, cancer pain) is a major health problem. Opiate-based drugs, such as morphine and morphine derivatives, are the primary standard of care for the treatment of chronic pain. Unfortunately, patients develop tolerance to opiates due to desensitization of the opiate receptor. Thus, alternative anti-nociceptive ("pain killing") pathways need to be explored for treatment of chronic pain. The N-type voltage-gated Ca2+ channels (CaV2.2s) in dorsal root ganglia neurons is a well validated target for chronic pain (1, 2). We previously demonstrated the interaction between CaV2.2 and the first PDZ domain of molecular Human Glucose-6-Phosphate Dehydrogenase Dose Response Selectivity Assay for Inhibitors of Plasmodium falciparum Glucose-6-Phosphate Dehydrogenase Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego CA) Network: NIH Molecular Libraries Probe Production Centers Network (MLPCN) Grant Number: 1R21AI082434-01 Assay Provider: Lars Bode, Ph.D., University of California San Diego, San Diego, CA Tropical malaria caused by the protozoan parasite Plasmodium falciparum is responsible for up to three million deaths annually. Due to increasing regional distribution and resistances against the clinically used antimalarials, novel antimalarial drugs - which have new mechanisms of action and are suitable for combination therapies - are urgently required. Plasmodium falciparum Glucose-6-phosphate dehydrogenase (PfGluPho) is a potential novel target for antimalarial drug design. Glucose-6-Phosphate Dehydrogenase (G6PD) reaction is the first and rate-limiting step in the pentose phosphate pathway (PPP), catalyzed by a bifunctional enzyme Plasmodium fal Identification of Novel Modulators of Cl- dependent Transport Process via HTS: Dose-Dependent Assay 2 with KCC2 Vanderbilt Screening Center for GPCRs, Ion Channels and Transporters Assay Provider: Eric Delpire Assay Provider Affliation: Vanderbilt University Grant Title: Identification of Novel Modulators of Cl- dependent Transport Process via HTS Grant Number: R21NS053658-01 Cation-chloride cotransporters such as K-Cl cotransport and Na-K-2Cl cotransport play major roles in a variety of physiological settings, including the modulation of GABAergic synaptic transmission. For instance, KCC2, a neuronal-specific K-Cl cotransporter is up-regulated in the brain during postnatal development, and is responsible for lowering the intracellular Cl- concentration in neurons, thus promoting GABA inhibition. Reduction in KCC2 expression results in brain hyperexcitability, as demonstrated by animal models. Furthermore, KCC2 expression is decreased in brain tissue isolated from epileptic patients. There are very few pharmacological agents that affect K-Cl cotransporters. First, there are no specific inhibi Identification of Novel Modulators of Cl- dependent Transport Process via HTS: Rubidium Flux Vanderbilt Screening Center for GPCRs, Ion Channels and Transporters Assay Provider: Eric Delpire Assay Provider Affliation: Vanderbilt University Grant Title: Identification of Novel Modulators of Cl- dependent Transport Process via HTS Grant Number: R21NS053658-01 Cation-chloride cotransporters such as K-Cl cotransport and Na-K-2Cl cotransport play major roles in a variety of physiological settings, including the modulation of GABAergic synaptic transmission. For instance, KCC2, a neuronal-specific K-Cl cotransporter is up-regulated in the brain during postnatal development, and is responsible for lowering the intracellular Cl- concentration in neurons, thus promoting GABA inhibition. Reduction in KCC2 expression results in brain hyperexcitability, as demonstrated by animal models. Furthermore, KCC2 expression is decreased in brain tissue isolated from epileptic patients. There are very few pharmacological agents that affect K-Cl cotransporters. First, there are no specific inhibi Identification of Novel Modulators of Cl- dependent Transport Process via HTS; Dose-dependent Assay with KCC2 Vanderbilt Screening Center for GPCRs, Ion Channels and Transporters Assay Provider: Eric Delpire Assay Provider Affliation: Vanderbilt University Grant Title: Identification of Novel Modulators of Cl- dependent Transport Process via HTS Grant Number: R21NS053658-01 Cation-chloride cotransporters such as K-Cl cotransport and Na-K-2Cl cotransport play major roles in a variety of physiological settings, including the modulation of GABAergic synaptic transmission. For instance, KCC2, a neuronal-specific K-Cl cotransporter is up-regulated in the brain during postnatal development, and is responsible for lowering the intracellular Cl- concentration in neurons, thus promoting GABA inhibition. Reduction in KCC2 expression results in brain hyperexcitability, as demonstrated by animal models. Furthermore, KCC2 expression is decreased in brain tissue isolated from epileptic patients. There are very few pharmacological agents that affect K-Cl cotransporters. First, there are no specific inhibi Identification of SV40 T antigen inhibitors: A route to novel anti-viral reagents A biochemical assay using the ADP-Hunter methodology, purified TAg, and ATP to identify compounds that inhibit the ATPase activity of Tag Southern Research's Specialized Biocontainment Screening Center (SRSBSC) Southern Research Institute (Birmingham, Alabama) NIH Molecular Libraries Probe Centers Network (MLPCN) Assay Provider: Dr. Jeffery Brodsky, University of Pittsburgh Grant number: 1R03MH084077-01 Assay Rationale and Summary: The oncogenic virus Simian Virus 40 (SV40) is a well-characterized model system to examine the underlying mechanisms of growth control and cancer. SV40 is also closely related to two viruses, JC and BK virus, which infect humans, and result in morbidity and mortality in immuno-compromised patients. Although it is controversial whether SV40 also causes disease in humans, the virus has been found in specific tumors but not in the surrounding tissue. Unlike BKV and JCV, SV40 grows relatively well in mammalian cell culture systems; moreover, ~10% of the p Identification of SV40 T antigen inhibitors: Cytotoxicity screen of selected hits Southern Research's Specialized Biocontainment Screening Center (SRSBSC) Southern Research Institute (Birmingham, Alabama) NIH Molecular Libraries Probe Production Centers Network (MLPCN) Assay Provider: Dr. Jeffery Brodsky, University of Pittsburgh Award: R03MH084077-01 Cytoxicity Assay Rationale and Summary: The oncogenic virus Simian Virus 40 (SV40) is a well-characterized model system to examine the underlying mechanisms of growth control and cancer. SV40 is also closely related to two viruses, JC and BK virus, which infect humans, and result in morbidity and mortality in immuno-compromised patients. Although it is controversial whether SV40 also causes disease in humans, the virus has been found in specific tumors but not in the surrounding tissue. Unlike BKV and JCV, SV40 grows relatively well in mammalian cell culture systems; moreover, ~10% of the population examined in one study has antibodies against SV40, probably because the first polio vaccines were contaminated with Image-Based HTS for Selective Antagonists for GPR55 Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Probe Production Centers Network (MLPCN) Grant Proposal Number: 1X01 DA026205-01 Assay Provider: Dr. Mary Abood, California Pacific Medical Center Research Institute The cannabinoid and endocannabinoid system has been implicated in the pathophysiology of drug dependence and addiction disorders. GPR55, an orphan G-Protein Coupled Receptor, has been reported to be a cannabinoid receptor, but its status as such remains unresolved due to conflicting results from pharmacological studies. The goal of this project is to identify small molecule antagonists of GPR55, which may aid in the deorphanization efforts of this receptor and ultimately further the understanding of the role of GPR55 in drug addiction. This high content imaging assay utilizes a cell line permanently expressing a beta-arrestin GFP biosens Image-Based HTS for Selective Antagonists of GPR35 Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Probe Production Centers Network (MLPCN) Grant Proposal Number: 1X01MH085708-01 Assay Provider: Dr. Lawrence Barak, Duke University Addictive behavior stems from abnormal signaling activities in the brain. Thus identification of compounds blocking this modified signaling activity may lead to treatments for addictive behavior. GPR35, a to-date uncharacterized orphan G-Protein Coupled Receptor, is thought to play a role in addiction and has homology to other known receptors of abuse. The goal of this project is to identify small molecule antagonists of GPR35, which may aid in characterization of this receptor and ultimately further the understanding of the role of GPR35 in addictive behavior. This high content imaging assay utilizes a cell line permanently expressing a beta-arrestin GFP biosensor and th In Vitro Bfl-1 Dose Response Fluorescence Polarization Assay for SAR Study Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Screening Centers Network (MLSCN) Grant Number: XO1 MH077632-01 Assay Provider: Dr. John C. Reed, Sanford-Burnham Medical Research Institute This Bfl-1 dose response assay is developed and performed for the purpose of SAR study on analogs of hits originally identified in the Bfl-1 fluorescence polarization HTS assay (AID 432). Bfl-1, also known as A1 in mice is an anti-apoptotic and NF-kB-inducible member of the Bcl-2 protein family involved in regulation of apoptosis. Due to difficulties with accomplishing targeted gene ablation in mouse models, the endogenous functions of Bfl-1 are largely unknown. Chemical inhibitors of Bfl-1 can be used as research tools for neutralizing Bfl-1 in human and mouse cells. In Vitro Hsp70 Dose Response Fluorescence Polarization Assay for SAR Study Sanford-Burnham Center for Chemical Genomics (SBCCG) Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) NIH Molecular Libraries Screening Centers Network (MLSCN) MLSCN Grant: XO1 MH079863-01 Assay Provider: Dr. John C. Reed, Sanford-Burnham Medical Research Institute This Hsp70 dose response assay is developed and performed for the purpose of SAR study on analogs of hits originally identified in the fluorescence polarization HTS assay for Hsp70 Inhibitors (AID 583). Over-expression of molecular chaperones occurs commonly in cancers and provides protection from a wide variety of cellular stresses, both endogenous and iatrogenic. Molecular chaperones also play important roles in maintaining the activity of several signal-transducing proteins and transcriptions factors involved in malignant transformation. The human genome contains nine Hsp70-family genes. These chaperones include Hsp70 and Hsc70, which are commonly over-expressed in cancers and which confer resistance In Vitro MKP-3 Dose Response Assay for SAR Study Data Source: Burnham Center for Chemical Genomics (BCCG) Source Affiliation: Burnham Institute for Medical Research (BIMR, La Jolla, CA) Network: NIH Molecular Libraries Screening Centers Network (MLSCN) Grant Number: XO1 MH076390-01 Assay Provider: Dr. John Lazo, University of Pittsburg This MKP-3 dose response assay is developed and performed for the purpose of SAR study on analogs of hits originally identified in the MKP-3 in vitro HTS assay (AID 425) MKP-3 (mitogen-activated protein kinase phosphatase-3; EC 3.1.3.48, EC 3.1.3.16), a dual specificity phosphatase negatively regulates ERK1/2 by catalyzing the removal of a phosphoryl group from Thr(P) and Tyr(P) in the activation loop consensus motif -pTXpY. MKP-3 screening was performed using a biochemical assay developed at the laboratory of Prof. John Lazo (University of Pittsburg). The assay was optimized and run at the Burnham Center for Chemical Genomics (BCCG) as part of the Molecular Library Screening Center Network (MLSCN Inhibitors of T-Type Calcium Channel Assay Provider: Xinmin Xie Assay Provider Affiliation: Bioscience Division, SRI International, Menlo Park, CA Grant Title: HTS Assay for Cav3 T-Type Channels using FLIPR Grant Number: NS050771-01 T-type Ca2+ channels are also called low voltage-activated channels because they open at voltages near the resting membrane potential of most cells. In many types of neurons, Ca2+ influx through T-type channels triggers low-threshold spikes, which in turn trigger a burst of action potentials mediated by Na+ channels (1). Burst firing is thought to play an important role in the synchronized activity of the thalamus observed in absence epilepsy, and also in a wider range of neurological disorders characterized by thalamocortical dysrhythmia (2). Prominent T-currents are also observed in dorsal root ganglion neurons, with subsets of nociceptors expressing more T-current than high voltage-activated Ca2+ currents (3). Considerable evidence supports the notion that a T-channel antagonist would Inhibitors of T-Type Calcium Channels (SynthLib1) Assay Provider: Xinmin Xie Assay Provider Affiliation: Bioscience Division, SRI International, Menlo Park, CA Grant Title: HTS Assay for Cav3 T-Type Channels using FLIPR Grant Number: NS050771-01 T-type Ca2+ channels are also called low voltage-activated channels because they open at voltages near the resting membrane potential of most cells. In many types of neurons, Ca2+ influx through T-type channels triggers low-threshold spikes, which in turn trigger a burst of action potentials mediated by Na+ channels (1). Burst firing is thought to play an important role in the synchronized activity of the thalamus observed in absence epilepsy, and also in a wider range of neurological disorders characterized by thalamocortical dysrhythmia (2). Prominent T-currents are also observed in dorsal root ganglion neurons, with subsets of nociceptors expressing more T-current than high voltage-activated Ca2+ currents (3). Considerable evidence supports the notion that a T-channel antagonist would Inhibitors of T-Type Calcium Channels (SynthLib2) Assay Provider: Xinmin Xie Assay Provider Affiliation: Bioscience Division, SRI International, Menlo Park, CA Grant Title: HTS Assay for Cav3 T-Type Channels using FLIPR Grant Number: NS050771-01 T-type Ca2+ channels are also called low voltage-activated channels because they open at voltages near the resting membrane potential of most cells. In many types of neurons, Ca2+ influx through T-type channels triggers low-threshold spikes, which in turn trigger a burst of action potentials mediated by Na+ channels (1). Burst firing is thought to play an important role in the synchronized activity of the thalamus observed in absence epilepsy, and also in a wider range of neurological disorders characterized by thalamocortical dysrhythmia (2). Prominent T-currents are also observed in dorsal root ganglion neurons, with subsets of nociceptors expressing more T-current than high voltage-activated Ca2+ currents (3). Considerable evidence supports the notion that a T-channel antagonist would Inhibitors of T-Type Calcium Channels Assay Provider: Xinmin Xie Assay Provider Affiliation: Bioscience Division, SRI International, Menlo Park, CA Grant Title: HTS Assay for Cav3 T-Type Channels using FLIPR Grant Number: NS050771-01 T-type Ca2+ channels are also called low voltage-activated channels because they open at voltages near the resting membrane potential of most cells. In many types of neurons, Ca2+ influx through T-type channels triggers low-threshold spikes, which in turn trigger a burst of action potentials mediated by Na+ channels (1). Burst firing is thought to play an important role in the synchronized activity of the thalamus observed in absence epilepsy, and also in a wider range of neurological disorders characterized by thalamocortical dysrhythmia (2). Prominent T-currents are also observed in dorsal root ganglion neurons, with subsets of nociceptors expressing more T-current than high voltage-activated Ca2+ currents (3). Considerable evidence supports the notion that a T-channel antagonist would Kallikrein 5 1536 HTS Dose Response Confirmation Molecular Library Screening Center Network (MLSCN) Penn Center for Molecular Discovery (PCMD) Assay Provider: Scott L. Diamond, University of Pennsylvania MLSCN Grant: X01-MH076406-01 Target Human kallikrein 5 (hK5) is a member of the human tissue kallikrein family, which contains 15 kallikrein-like serine proteases (1). It is synthesized as a 293 amino acid zymogen, and loses a 29 amino acid signal peptide upon secretion, followed by cleavage at the Arg66-Ile67 bond, which releases a 37 amino acid activation peptide, resulting in a 237 amino acid mature enzyme (2). hK5 generally exhibits a trypsin-like specificity for P1-Arg over P1-Lys residues, and has been observed to digest different extracellular matrix and plasma proteins (3, 4). Protein inhibitors include alpha2-antiplasmin, antithrombin III and alpha2-macroglobulin (4). In addition, Zn2+ and other divalent cations strongly inhibit hK5 (5). hK5 is highly expressed in skin tissue, specifically the outermost layer of LYP1 Fluorescent Assay using OMFP substrate for In Vitro dose response studies. Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Probe Production Centers Network (MLPCN) Grant Number: 1R21NS056945-01 Assay Provider: Dr. Nunzio Bottini, La Jolla Institute for Allergy and Immunology, La Jolla, CA LYP, is a lymphocyte specific protein tyrosine phosphatase that plays a critical regulatory role in T cell receptor signaling. The PTPN22 gene encodes this phosphatase. A single-nucleotide polymorphism in PTPN22 is associated with a number of autoimmune disorders, including type 1 diabetes, rheumatoid arthritis, juvenile rheumatoid arthritis, systemic lupus erythematosus and Grave's disease. The autoimmunity-predisposing allele is a gain-of-function mutant suggesting that a specific small-molecule inhibitor could eliminate its effect. This biochemical assay employs a fluorescent readout based on the enzyme's ability to liberate Late stage assay provider counterscreen results from the probe development effort to identify inhibitors of kruppel-like factor 5 (KLF5) Source (MLPCN Center Name): The Scripps Research Institute Molecular Screening Center Affiliation: The Scripps Research Institute, TSRI Assay Provider: Vincent Yang, Emory University Network: Molecular Library Probe Production Centers Network (MLPCN) Grant Proposal Number: 1-R03-DA026215-01 Grant Proposal PI: Vincent Yang External Assay ID: KLF5-WESTERN-BLOT_INH_LUMI_3XIC50 MDCSRUN SAR_Round 1 Name: Late stage assay provider counterscreen results from the probe development effort to identify inhibitors of kruppel-like factor 5 (KLF5): chemiluminescence-based western blot assay for inhibitors of KLF5 protein levels. Description: Transcription factors are essential regulators of transcription that bind DNA to control both the rate and frequency of gene expression (1). Many diseases of cell homeostasis are associated with aberrant transcription factor activity (2). Colon cancer, in particular, is a disease of uncontrolled proliferation of the epithelial cells that line the intestinal Late stage counterscreen for the probe development effort to identify selective agonists of the Transient Receptor Potential Channels 3 (TRPML3) Source (MLPCN Center Name): The Scripps Research Institute Molecular Screening Center (SRIMSC) Center Affiliation: The Scripps Research Institute (TSRI) Assay Provider: Stefan Heller, Stanford University Network: Molecular Libraries Probe Production Centers Network (MLPCN) Grant Proposal Number 1 R03 MH083077-01 Grant Proposal PI: Stefan Heller, Stanford University External Assay ID: TRPN1_AG_FLUO8_1536_3XEC50_LATE STAGE CS DRUN Name: Late stage counterscreen for the probe development effort to identify selective agonists of the Transient Receptor Potential Channels 3 (TRPML3): fluorescence-based cell-based dose response assay for TRPN1 agonists. Description: Cell signaling pathways that mediate osmosensation, photosensation, and thermosensation depend on a family of diverse transient receptor potential (TRP) cation channels, which are activated by agonist-receptor coupling (1-5). A role for these channels in inner ear hair cell mechanotransduction was gleaned from TRP channel muta Late stage counterscreen results from the probe development effort to identify STAT1 inhibitors: luminescence-based cell-based dose response assay for STAT3 inhibitors Source (MLPCN Center Name): The Scripps Research Institute Molecular Screening Center Center Affiliation: The Scripps Research Institute (TSRI) Assay Provider: David Frank, Dana Farber Cancer Institute Network: Molecular Libraries Probe Production Centers Network (MLPCN) Grant Proposal Number: 1 X01 MH079826-01 Grant Proposal PI: David Frank, Dana Farber Cancer Institute External Assay ID: STAT3_INH_LUMI_1536_IC50 MDCSRUN SAR_ROUND_0 Name: Late stage counterscreen results from the probe development effort to identify STAT1 inhibitors: luminescence-based cell-based dose response assay for STAT3 inhibitors. Description: Members of the signal transducer and activator of transcription (STAT) family of transcription factors mediate inflammation, cell survival, differentiation, and proliferation (1, 2). In response to stimuli such as growth factors and cytokines (1-3), cytosolic STATs are activated by phosphorylation by the Janus-activated kinases (Jaks), inducing STAT dimerization, nuc Late stage counterscreen results from the probe development effort to identify activators of signal transducer and activator of transcription 1 (STAT1) Source (MLPCN Center Name): The Scripps Research Institute Molecular Screening Center Center Affiliation: The Scripps Research Institute (TSRI) Assay Provider: David Frank, Dana Farber Cancer Institute Network: Molecular Libraries Probe Production Centers Network (MLPCN) Grant Proposal Number: 1 X01 MH079826-01 Grant Proposal PI: David Frank, Dana Farber Cancer Institute External Assay ID: STAT3_ACT_LUMI_1536_EC50 MDCSRUN SAR_ROUND_0 Name: Late stage counterscreen results from the probe development effort to identify activators of signal transducer and activator of transcription 1 (STAT1): luminescence-based cell-based dose response assay for STAT3 activators. Description: Members of the signal transducer and activator of transcription (STAT) family of transcription factors mediate inflammation, cell survival, differentiation, and proliferation (1, 2). In response to stimuli such as growth factors and cytokines (1-3), cytosolic STATs are activated by phosphorylation by the Janus-a Late stage results from the probe development effort to identify STAT1 inhibitors: luminescence-based cell-based dose response assay for STAT1 inhibitors Source (MLPCN Center Name): The Scripps Research Institute Molecular Screening Center Center Affiliation: The Scripps Research Institute (TSRI) Assay Provider: David Frank, Dana Farber Cancer Institute Network: Molecular Libraries Probe Production Centers Network (MLPCN) Grant Proposal Number: 1 X01 MH079826-01 Grant Proposal PI: David Frank, Dana Farber Cancer Institute External Assay ID: STAT1_INH_LUMI_1536_IC50 MDRUN SAR_ ROUND_0 Name: Late stage results from the probe development effort to identify STAT1 inhibitors: luminescence-based cell-based dose response assay for STAT1 inhibitors. Description: Members of the signal transducer and activator of transcription (STAT) family of transcription factors mediate inflammation, cell survival, differentiation, and proliferation (1, 2). In response to stimuli such as growth factors and cytokines (1-3), cytosolic STATs are activated by phosphorylation by the Janus-activated kinases (Jaks), inducing STAT dimerization, nuclear translocat Late stage results from the probe development effort to identify activators of signal transducer and activator of transcription 1 (STAT1) Source (MLPCN Center Name): The Scripps Research Institute Molecular Screening Center Center Affiliation: The Scripps Research Institute (TSRI) Assay Provider: David Frank, Dana Farber Cancer Institute Network: Molecular Libraries Probe Production Centers Network (MLPCN) Grant Proposal Number: 1 X01 MH079826-01 Grant Proposal PI: David Frank, Dana Farber Cancer Institute External Assay ID: STAT1_ACT_LUMI_1536_EC50 MDRUN SAR_ROUND_0 Name: Late stage results from the probe development effort to identify activators of signal transducer and activator of transcription 1 (STAT1): luminescence-based cell-based dose response assay for STAT1 activators. Description: Members of the signal transducer and activator of transcription (STAT) family of transcription factors mediate inflammation, cell survival, differentiation, and proliferation (1, 2). In response to stimuli such as growth factors and cytokines (1-3), cytosolic STATs are activated by phosphorylation by the Janus-activated kinases Late stage results from the probe development effort to identify selective agonists of the Transient Receptor Potential Channels 3 (TRPML3) Source (MLPCN Center Name): The Scripps Research Institute Molecular Screening Center (SRIMSC) Center Affiliation: The Scripps Research Institute (TSRI) Assay Provider: Stefan Heller, Stanford University Network: Molecular Libraries Probe Production Centers Network (MLPCN) Grant Proposal Number: 1 R03 MH083077-01 Grant Proposal PI: Stefan Heller, Stanford University External Assay ID: TRPML3-SELECTIVE_AG_FLUO8_1536_3XEC50_LATE STAGE Name: Late stage results from the probe development effort to identify selective agonists of the Transient Receptor Potential Channels 3 (TRPML3): fluorescence-based cell-based dose response assay for TRPML3 agonists. Description: Cell signaling pathways that mediate osmosensation, photosensation, and thermosensation depend on a family of diverse transient receptor potential (TRP) cation channels, which are activated by agonist-receptor coupling (1-5). A role for these channels in inner ear hair cell mechanotransduction was gleaned from TRP channel muta Late-stage fluorescence-based dose response cell-based screening assay to identify antagonists of the G-protein coupled receptor 7 (GPR7): Intracellular calcium release Source (MLPCN Center Name): The Scripps Research Institute Molecular Screening Center (SRIMSC) Affiliation: The Scripps Research Institute, TSRI Assay Provider: Olivier Civelli, University of California, Irvine Network: Molecular Library Probe Production Centers Network (MLPCN) Grant Proposal Number: 1-R03-DA026557-01 Grant Proposal PI: Olivier Civelli External Assay ID: GPR7_ANT_FLUO8_96_IC50 Name: Late-stage fluorescence-based dose response cell-based screening assay to identify antagonists of the G-protein coupled receptor 7 (GPR7): Intracellular calcium release. Description: Heterotrimeric G-protein coupled receptors (GPCRs) are major targets for disease therapeutics, due in part to their broad tissue distribution, structural diversity, varied modes of action, and disease-associated mutations (1-4). For example, targeting of opiod receptors by opiates such as morphine is a widespread clinical application for GPCR modulation in pain management. The recently de-orphanized GPR7 (5 Luminescence counterscreen assay for p97 inhibitors: Dose response biochemical high throughput screening assay to identify inhibitors of the C522A mutant p97 ATPase. Source (MLPCN Center Name): The Scripps Research Institute Molecular Screening Center Center Affiliation: The Scripps Research Institute (TSRI) Assay Provider: Raymond Deshaies, California Institute of Technology Network: Molecular Libraries Probe Production Centers Network (MLPCN) Grant Proposal Number: 1 R03 MH085687-01 Grant Proposal PI: Raymond Deshaies, California Institute of Technology External Assay ID: P97C522A_INH_LUMI_384_IC50 CS Name: Luminescence counterscreen assay for p97 inhibitors: Dose response biochemical high throughput screening assay to identify inhibitors of the C522A mutant p97 ATPase. Description: Misfolded proteins accumulate in the endoplasmic reticulum (ER) in response to environmental stress (1). To reduce the burden these proteins place on the secretory pathway, eukaryotic cells have evolved a process, known as ER-associated degradation (ERAD), to recognize and eliminate these proteins (1, 2). The highly conserved p97 ATPase functions in ERAD by hydr Luminescence counterscreen assay for p97 inhibitors: dose response biochemical high throughput screening assay to identify inhibitors of the C522A mutant p97 ATPase: synthesized compounds. Source (MLPCN Center Name): The Scripps Research Institute Molecular Screening Center (SRIMSC) Center Affiliation: The Scripps Research Institute (TSRI) Assay Provider: Raymond Deshaies, California Institute of Technology Network: Molecular Libraries Probe Production Centers Network (MLPCN) Grant Proposal Number: 1 R03 MH085687-01 Grant Proposal PI: Raymond Deshaies, California Institute of Technology External Assay ID: p97C552A_INH _LUMI_384_IC50_SAR_CS ROUND1 Name: Luminescence counterscreen assay for p97 inhibitors: dose response biochemical high throughput screening assay to identify inhibitors of the C522A mutant p97 ATPase: synthesized compounds. Description: Misfolded proteins accumulate in the endoplasmic reticulum (ER) in response to environmental stress (1). To reduce the burden these proteins place on the secretory pathway, eukaryotic cells have evolved a process, known as ER-associated degradation (ERAD), to recognize and eliminate these proteins (1, 2). The highly con Luminescence-based biochemical high throughput dose response assay for inhibitors of the interaction of the lipase co-activator protein, abhydrolase domain Source (MLPCN Center Name): The Scripps Research Institute Molecular Screening Center (SRIMSC) Affiliation: The Scripps Research Institute, TSRI Assay Provider: James Granneman, Wayne State University Network: Molecular Library Probe Production Centers Network (MLPCN) Grant Proposal Number: 1 R21 NS061634-01 Grant Proposal PI: James Granneman, Wayne State University External Assay ID: ABHD5-MLDP_INH_LUMI_1536_3XIC50 2K Name: Luminescence-based biochemical high throughput dose response assay for inhibitors of the interaction of the lipase co-activator protein, abhydrolase domain containing 5 (ABHD5) with perilipin-5 (MLDP; PLIN5) (2K validation set). Description: Adipocytes are important regulators of vertebrate energy stores, in part through the storage of dietary fat (triglyceride) that is mobilized via lipolysis during fasting states for use by tissues such as heart and skeletal muscle (1, 2). However, in chronic conditions of overnutrition, such as obesity and lipid storage diso Luminescence-based biochemical high throughput dose response assay for inhibitors of the interaction of the lipase co-activator protein, abhydrolase domain Source (MLPCN Center Name): The Scripps Research Institute Molecular Screening Center (SRIMSC) Affiliation: The Scripps Research Institute, TSRI Assay Provider: James Granneman, Wayne State University Network: Molecular Library Probe Production Centers Network (MLPCN) Grant Proposal Number: 1 R21 NS061634-01 Grant Proposal PI: James Granneman, Wayne State University External Assay ID: ABHD5-PLIN1_INH_LUMI_1536_3XIC50 2K Name: Luminescence-based biochemical high throughput dose response assay for inhibitors of the interaction of the lipase co-activator protein, abhydrolase domain containing 5 (ABHD5) with perilipin-1 (PLIN1) (2K validation set). Description: Adipocytes are important regulators of vertebrate energy stores, in part through the storage of dietary fat (triglyceride) that is mobilized via lipolysis during fasting states for use by tissues such as heart and skeletal muscle (1, 2). However, in chronic conditions of overnutrition, such as obesity and lipid storage disorders Luminescence-based cell-based assay provider high throughput dose response assay for partial agonists of the peroxisome proliferator-activated receptor gamma (PPARg) Source (MLPCN Center Name): The Scripps Research Institute Molecular Screening Center (SRISMC) Center Affiliation: The Scripps Research Institute, TSRI Assay Provider: Patrick Griffin, TSRI Network: Molecular Library Probe Production Center Network (MLPCN) Grant Proposal Number: MH079861-01 Grant Proposal PI: Patrick Griffin, TSRI External Assay ID: PPARG_AG_LUMI_0384_3XEC50 DRUN Name: Luminescence-based cell-based assay provider high throughput dose response assay for partial agonists of the peroxisome proliferator-activated receptor gamma (PPARg). Description: Peroxisome proliferator-activated receptors (PPARs) belong to the nuclear receptor superfamily and are lipid sensors functioning as ligand-dependent transcription factors regulating gene expression patterns of diverse bHeres what I wrote for the intro.iological processes (1, 2). PPARs play a critical role in metabolic processes such as glucose metabolism, lipid metabolism, and have been implicated in anti-atherogenic, anti- Luminescence-based cell-based high throughput dose response assay for biased ligands (agonists) of the melanocortin 4 receptor (MC4R) Source (MLPCN Center Name): The Scripps Research Institute Molecular Screening Center (SRISMC) Center Affiliation: The Scripps Research Institute, TSRI Assay Provider: Scott DeWire, Trevena Inc Network: Molecular Library Probe Production Center Network (MLPCN) Grant Proposal Number: 1 RC2 MH090877-01 Grant Proposal PI: Scott DeWire, Trevena Inc External Assay ID: MC4R_AG_LUMI_1536_3XEC50 DRUN Name: Luminescence-based cell-based high throughput dose response assay for biased ligands (agonists) of the melanocortin 4 receptor (MC4R). Description: Heterotrimeric G-protein coupled receptors (GPCRs) are major targets for disease therapeutics, due in part to their broad tissue distribution, structural diversity, varied modes of action, and disease-associated mutations (1-4). However, it has recently been demonstrated that GPCRs do not only signal in this simplistic fashion, but rather activate a network of downstream effects comprised of parallel signal transduction pathways. GPCR ligands Luminescence-based counterscreen for activators of the Aryl Hydrocarbon Receptor (AHR): cell-based high throughput dose response screening assay for activators of the Pregnane X Receptor (PXR) Source (MLPCN Center Name): The Scripps Research Institute Molecular Screening Center (SRIMSC) Center Affiliation: The Scripps Research Institute (TSRI) Assay Provider: Michael Denison, University of California, Davis Network: Molecular Libraries Probe Production Centers Network (MLPCN) Grant Proposal Number: 1-X01-DA026558-01 Grant Proposal PI: Michael Denison External Assay ID: PXR_ACT_LUMI_1536_3XEC50 DCSRUN Name: Luminescence-based counterscreen for activators of the Aryl Hydrocarbon Receptor (AHR): cell-based high throughput dose response screening assay for activators of the Pregnane X Receptor (PXR). Description: Transcription factors are critical regulators of gene expression (1). Under conditions such as environmental stress and exposure to endogenous toxins, transcription factors can rapidly modulate the transcription of genes whose products regulate cell proliferation and metabolism. The aryl hydrocarbon receptor (AHR) is a ligand-activated transcription factor of the ba Luminescent HTS for small molecule inhibitors of MT1-MMP transcription Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Screening Centers Network (MLSCN) Grant Number: XO1 MH78949-01 Assay Provider: Dr. Alex Strongin, Sanford-Burnham Medical Research Institute The sustained presence of matrix metalloproteinases (MMPs) in a tumor environment is a characteristic of many cancer types. The expression of the MT1-MMP mRNA and the MT1-MMP protein closely correlates with increased tumor volume, tumor invasiveness, and the incidence of local and distant metastases. Tumorigenic MT1-MMP is effective in both its active and inactive states. MT1-MMP protects malignant cells against the host immune surveillance thus making tumor cells resistant to the anti-tumor immunity mechanisms. Proteolytically active MT1-MMP is trafficked to centrosomes. Through the proteolysis of centrosomal proteins MT1-MMP promotes mitotic spindle aberrati Luminescent assay for HTS discovery of chemical inhibitors of placental alkaline phosphatase confirmation Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Screening Centers Network (MLSCN) Grant Proposal Number: MH077602-01 Assay Provider: Dr. Jose Luis Millan, Sanford-Burnham Medical Research Institute, San Diego, CA. This PLAP dose response assay is developed and performed to confirm hits originally identified in the PLAP Luminescent HTS assay (AID 690) and to study the structure-activity relationship on analogs of the confirmed hits. Compounds are either acquired from commercial sources or synthesized internally. Alkaline phosphatase (EC 3.1.3.1) (APs) catalyze the hydrolysis of phosphomonoesters, releasing phosphate and alcohol. APs are dimeric enzymes found in most organisms. In human, four isozymes of APs have been identified: three isozymes are tissue-specific and the fourth one is tissue-nonspecific. Placental alkaline phosphatase (PLAP) is high Luminescent assay for identification of inhibitors of human intestinal alkaline phosphatase Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Screening Centers Network (MLSCN) Grant Proposal Number: XO1 MH077602-01 Alkaline phosphatase (EC 3.1.3.1) (APs) catalyze the hydrolysis of phosphomonoesters, releasing inorganic phosphate and alcohol. APs are dimeric enzymes found in most organisms. In human, four isozymes of APs have been identified. One isozyme is tissue-nonspecific (designated TNAP) and three other isozymes are tissue-specific and named according to the tissue of their predominant expression: intestinal (IAP), placental (PLAP) and germ cell (GCAP) alkaline phosphatases. IAP expression is largely restricted to the gut, especially to the epithelial cells (enterocytes) of the small intestinal mucosa. IAP is inhibited by a number of inhibitors (1). They include L-phenylalanine, (2, 3), L-tryptophan (4), L-leucine and phenylalanine-g MKP-3 in vitro HTS assay Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Screening Centers Network (MLSCN) Grant Number: XO1 MH076390-01 Assay Provider: Dr. John Lazo, University of Pittsburg MKP-3 (mitogen-activated protein kinase phosphatase-3; EC 3.1.3.48, EC 3.1.3.16), a dual specificity phosphatase negatively regulates ERK1/2 by catalyzing the removal of a phosphoryl group from Thr(P) and Tyr(P) in the activation loop consensus motif -pTXpY. MKP-3 screening was performed using a biochemical assay developed at the laboratory of Prof. John Lazo (University of Pittsburg). The assay was optimized and run at the Sanford-Burnham Center for Chemical Genomics (SBCCG) as part of the Molecular Library Screening Center Network (MLSCN). Enzyme activity and its inhibition by screened compounds was measured in an end-point assay based on hydrolysis of 3-O-methylfluorescein phosphate Modulation of the Metabotropic Glutamate Receptor mGluR4: Selectivity at mGluR1 Assigned Assay Grant Number: NS053536-01 Screening Center Name & PI: Vanderbilt Screening Center for GPCRs, Ion Channels and Transporters, C. David Weaver Chemistry Center Name & PI: Vanderbilt Specialized Chemistry Center for Accelerated Probe Development, Craig W. Lindsley Assay Submitter & Institution: Colleen M. Niswender, Vanderbilt University The primary pathophysiological change giving rise to the symptoms of Parkinson's disease (PD) is a loss of the dopaminergic neurons in the substantia nigra pars compacta (SNc) that are involved in modulating the function of basal ganglia (BG) nuclei. Unfortunately, traditional therapies for treatment of PD based on dopamine replacement strategies eventually fail in most patients and are associated with numerous side effects. A great deal of effort has been focused on developing a detailed understanding of the circuitry and function of the BG to develop novel, nondopaminergic, approaches for restoring normal BG function in PD patients. Modulation of the Metabotropic Glutamate Receptor mGluR4: Selectivity at mGluR2 Assigned Assay Grant Number: NS053536-01 Screening Center Name & PI: Vanderbilt Screening Center for GPCRs, Ion Channels and Transporters, C. David Weaver Chemistry Center Name & PI: Vanderbilt Specialized Chemistry Center for Accelerated Probe Development, Craig W. Lindsley Assay Submitter & Institution: Colleen M. Niswender, Vanderbilt University The primary pathophysiological change giving rise to the symptoms of Parkinson's disease (PD) is a loss of the dopaminergic neurons in the substantia nigra pars compacta (SNc) that are involved in modulating the function of basal ganglia (BG) nuclei. Unfortunately, traditional therapies for treatment of PD based on dopamine replacement strategies eventually fail in most patients and are associated with numerous side effects. A great deal of effort has been focused on developing a detailed understanding of the circuitry and function of the BG to develop novel, nondopaminergic, approaches for restoring normal BG function in PD patients. Modulation of the Metabotropic Glutamate Receptor mGluR4: Selectivity at mGluR7 Assigned Assay Grant Number: NS053536-01 Screening Center Name & PI: Vanderbilt Screening Center for GPCRs, Ion Channels and Transporters, C. David Weaver Chemistry Center Name & PI: Vanderbilt Specialized Chemistry Center for Accelerated Probe Development, Craig W. Lindsley Assay Submitter & Institution: Colleen M. Niswender, Vanderbilt University The primary pathophysiological change giving rise to the symptoms of Parkinson's disease (PD) is a loss of the dopaminergic neurons in the substantia nigra pars compacta (SNc) that are involved in modulating the function of basal ganglia (BG) nuclei. Unfortunately, traditional therapies for treatment of PD based on dopamine replacement strategies eventually fail in most patients and are associated with numerous side effects. A great deal of effort has been focused on developing a detailed understanding of the circuitry and function of the BG to develop novel, nondopaminergic, approaches for restoring normal BG function in PD patients. Modulation of the Metabotropic Glutamate Receptor mGluR4: Selectivity at mGluR8 Assigned Assay Grant Number: NS053536-01 Screening Center Name & PI: Vanderbilt Screening Center for GPCRs, Ion Channels and Transporters, C. David Weaver Chemistry Center Name & PI: Vanderbilt Specialized Chemistry Center for Accelerated Probe Development, Craig W. Lindsley Assay Submitter & Institution: Colleen M. Niswender, Vanderbilt University The primary pathophysiological change giving rise to the symptoms of Parkinson's disease (PD) is a loss of the dopaminergic neurons in the substantia nigra pars compacta (SNc) that are involved in modulating the function of basal ganglia (BG) nuclei. Unfortunately, traditional therapies for treatment of PD based on dopamine replacement strategies eventually fail in most patients and are associated with numerous side effects. A great deal of effort has been focused on developing a detailed understanding of the circuitry and function of the BG to develop novel, nondopaminergic, approaches for restoring normal BG function in PD patients. Multiplex dose response to identify specific small molecule inhibitors of Ras and Ras-related GTPases specifically Cdc42 activated mutant University of New Mexico Assay Overview: Assay Support: NIH I RO3 MH081231-01 HTS to identify specific small molecule inhibitors of Ras and Ras-related GTPases PI: Angela Wandinger-Ness, Ph.D. Co-PI: Larry Sklar, Ph.D. Assay Development: Zurab Surviladze, Ph.D. Assay Implementation: Zurab Surviladze, Danuta Wlodek, Terry Foutz, Mark Carter, Anna Waller Target Team Leader for the Center: Larry Sklar (lsklar@salud.unm.edu) Dose Response Assay Background and Significance: Ras and related small molecular weight GTPases function in the regulation of signaling and cell growth, and collectively serve to control cell proliferation, differentiation and apoptosis [Tekai et al. 2001; Wennerberg et al. 2005]. The Ras-related GTPases are divided into four subfamilies with the Rab proteins regulating membrane transport, Rho proteins (including Rac and Cdc 42) regulating cytoskeletal rearrangements and responses to signaling, Arf/Sar proteins regulating membrane and microtubule dynamics as well Multiplex dose response to identify specific small molecule inhibitors of Ras and Ras-related GTPases specifically Rab2 wildtype University of New Mexico Assay Overview: Assay Support: NIH I RO3 MH081231-01 HTS to identify specific small molecule inhibitors of Ras and Ras-related GTPases PI: Angela Wandinger-Ness, Ph.D. Co-PI: Larry Sklar, Ph.D. Assay Development: Zurab Surviladze, Ph.D. Assay Implementation: Zurab Surviladze, Danuta Wlodek, Terry Foutz, Mark Carter, Anna Waller Target Team Leader for the Center: Larry Sklar (lsklar@salud.unm.edu) Dose Response Assay Background and Significance: Ras and related small molecular weight GTPases function in the regulation of signaling and cell growth, and collectively serve to control cell proliferation, differentiation and apoptosis [Takai et al. 2001; Wennerberg et al. 2005]. The Ras-related GTPases are divided into four subfamilies with the Rab proteins regulating membrane transport, Rho proteins (including Rac and Cdc 42) regulating cytoskeletal rearrangements and responses to signaling, Arf/Sar proteins regulating membrane and microtubule dynamics as well Multiplex dose response to identify specific small molecule inhibitors of Ras and Ras-related GTPases specifically Rac activated mutant University of New Mexico Assay Overview: Assay Support: NIH I RO3 MH081231-01 HTS to identify specific small molecule inhibitors of Ras and Ras-related GTPases PI: Angela Wandinger-Ness, Ph.D. Co-PI: Larry Sklar, Ph.D. Assay Development: Zurab Surviladze, Ph.D. Assay Implementation: Zurab Surviladze, Danuta Wlodek, Terry Foutz, Mark Carter, Anna Waller Target Team Leader for the Center: Larry Sklar (lsklar@salud.unm.edu) Dose Response Assay Background and Significance: Ras and related small molecular weight GTPases function in the regulation of signaling and cell growth, and collectively serve to control cell proliferation, differentiation and apoptosis [Takai et al. 2001; Wennerberg et al. 2005]. The Ras-related GTPases are divided into four subfamilies with the Rab proteins regulating membrane transport, Rho proteins (including Rac and Cdc 42) regulating cytoskeletal rearrangements and responses to signaling, Arf/Sar proteins regulating membrane and microtubule dynamics as well Multiplexed dose response to identify specific small molecule inhibitors of Ras and Ras-related GTPases specifically Rac wildtype University of New Mexico Assay Overview: Assay Support: NIH I RO3 MH081231-01 HTS to identify specific small molecule inhibitors of Ras and Ras-related GTPases PI: Angela Wandinger-Ness, Ph.D. Co-PI: Larry Sklar, Ph.D. Assay Development: Zurab Surviladze, Ph.D. Assay Implementation: Zurab Surviladze, Danuta Wlodek, Terry Foutz, Mark Carter, Anna Waller Target Team Leader for the Center: Larry Sklar (lsklar@salud.unm.edu) Dose Response Assay Background and Significance: Ras and related small molecular weight GTPases function in the regulation of signaling and cell growth, and collectively serve to control cell proliferation, differentiation and apoptosis [Takai et al. 2001; Wennerberg et al. 2005]. The Ras-related GTPases are divided into four subfamilies with the Rab proteins regulating membrane transport, Rho proteins (including Rac and Cdc 42) regulating cytoskeletal rearrangements and responses to signaling, Arf/Sar proteins regulating membrane and microtubule dynamics as well Multiplexed dose response to identify specific small molecule inhibitors of Ras and Ras-related GTPases specifically Ras wildtype University of New Mexico Assay Overview: Assay Support: NIH I RO3 MH081231-01 HTS to identify specific small molecule inhibitors of Ras and Ras-related GTPases PI: Angela Wandinger-Ness, Ph.D. Co-PI: Larry Sklar, Ph.D. Assay Development: Zurab Surviladze, Ph.D. Assay Implementation: Zurab Surviladze, Danuta Wlodek, Terry Foutz, Mark Carter, Anna Waller Target Team Leader for the Center: Larry Sklar (lsklar@salud.unm.edu) Dose Response Assay Background and Significance: Ras and related small molecular weight GTPases function in the regulation of signaling and cell growth, and collectively serve to control cell proliferation, differentiation and apoptosis [Tekai et al. 2001; Wennerberg et al. 2005]. The Ras-related GTPases are divided into four subfamilies with the Rab proteins regulating membrane transport, Rho proteins (including Rac and Cdc 42) regulating cytoskeletal rearrangements and responses to signaling, Arf/Sar proteins regulating membrane and microtubule dynamics as well Name: High Throughput Screen to Identify Compounds that increase expression of NF-kB in Human Neuronal Cells - Dose Response Southern Research Molecular Libraries Screening Center (SRMLSC) Southern Research Institute (Birmingham, Alabama) NIH Molecular Libraries Screening Centers Network (MLSCN) Award: R03 MH082367-01, Submitted by Dr. Maurizio Grimaldi (Neuropharmacology Laboratory, Southern Research Institute) The pharmacological treatment of neurodegenerative disorders has been a disappointment when compared to the successes obtained in stroke, other neurological diseases like seizures, and in mental health diseases. It has to be said that the pathogenesis of neurodegenerative disorders and their early diagnosis represent a definite obstacle to effective intervention. Nuclear factor kB (NF-kB) is a key cellular signaling factor in the central nervous system. Although NF-kB signaling pathways have been extensively investigated in cancer and in immunological diseases, NF-kB role in the central nervous system physiology and pathology in non inflammatory disorders of the brain is still unclear. NF-kB has Oxadiazole SAR compounds tested by Multiplex dose response to identify specific small molecule inhibitors of Ras and Ras-related GTPases specifically Cdc42 activated mutant University of New Mexico Assay Overview: Assay Support: NIH I RO3 MH081231-01 HTS to identify specific small molecule inhibitors of Ras and Ras-related GTPases PI: Angela Wandinger-Ness, Ph.D. Co-PI: Larry Sklar, Ph.D. Assay Development: Zurab Surviladze, Ph.D. Assay Implementation: Zurab Surviladze, Danuta Wlodek, Terry Foutz, Mark Carter, Anna Waller Target Team Leader for the Center: Larry Sklar (lsklar@salud.unm.edu) Chemistry: University of Kansas Specialized Chemistry Center Target Team Leader for Chemistry: Jennifer Golden Dose Response Assay Background and Significance: Ras and related small molecular weight GTPases function in the regulation of signaling and cell growth, and collectively serve to control cell proliferation, differentiation and apoptosis [Tekai et al. 2001; Wennerberg et al. 2005]. The Ras-related GTPases are divided into four subfamilies with the Rab proteins regulating membrane transport, Rho proteins (including Rac and Cdc 42) regulating cytoskeletal r Oxadiazole SAR compounds tested via Multiplex dose response to identify specific small molecule inhibitors of Ras and Ras-related GTPases specifically Cdc42 wildtype University of New Mexico Assay Overview: Assay Support: NIH I RO3 MH081231-01 HTS to identify specific small molecule inhibitors of Ras and Ras-related GTPases PI: Angela Wandinger-Ness, Ph.D. Co-PI: Larry Sklar, Ph.D. Assay Development: Zurab Surviladze, Ph.D. Assay Implementation: Zurab Surviladze, Danuta Wlodek, Terry Foutz, Mark Carter, Anna Waller Target Team Leader for the Center: Larry Sklar (lsklar@salud.unm.edu) UNM Cheminformatics: Cristian Bologa, Ph.D., Fabiola Miscioscia, Ph.D., Ramona Curpan, Ph.D., Oleg Ursu, Ph.D. Chemistry: University of Kansas Specialized Chemistry Center KU Specialized Chemistry Center PI: Jeff Aube, Ph.D. KU SCC Project Manager: Jennifer E. Golden. Ph.D. KU SCC Chemists on this project: Chad Schroeder, M.S., Denise Simpson, Ph.D., Julica Noeth, B.S. Dose Response Assay Background and Significance: Ras and related small molecular weight GTPases function in the regulation of signaling and cell growth, and collectively serve to control cell proli Oxadiazole SAR compounds tested via Multiplex dose response to identify specific small molecule inhibitors of Ras and Ras-related GTPases specifically Rab2 wildtype University of New Mexico Assay Overview: Assay Support: NIH I RO3 MH081231-01 HTS to identify specific small molecule inhibitors of Ras and Ras-related GTPases PI: Angela Wandinger-Ness, Ph.D. Co-PI: Larry Sklar, Ph.D. Assay Development: Zurab Surviladze, Ph.D. Assay Implementation: Zurab Surviladze, Danuta Wlodek, Terry Foutz, Mark Carter, Anna Waller Target Team Leader for the Center: Larry Sklar (lsklar@salud.unm.edu) UNM Cheminformatics: Cristian Bologa, Ph.D., Fabiola Miscioscia, Ph.D., Ramona Curpan, Ph.D., Oleg Ursu, Ph.D. Chemistry: University of Kansas Specialized Chemistry Center KU Specialized Chemistry Center PI: Jeff Aube, Ph.D. KU SCC Project Manager: Jennifer E. Golden. Ph.D. KU SCC Chemists on this project: Chad Schroeder, M.S., Denise Simpson, Ph.D., Julica Noeth, B.S. Dose Response Assay Background and Significance: Ras and related small molecular weight GTPases function in the regulation of signaling and cell growth, and collectively serve to control cell proli Oxadiazole SAR compounds tested via Multiplex dose response to identify specific small molecule inhibitors of Ras and Ras-related GTPases specifically Rab7 wildtype University of New Mexico Assay Overview: Assay Support: NIH I RO3 MH081231-01 HTS to identify specific small molecule inhibitors of Ras and Ras-related GTPases PI: Angela Wandinger-Ness, Ph.D. Co-PI: Larry Sklar, Ph.D. Assay Development: Zurab Surviladze, Ph.D. Assay Implementation: Zurab Surviladze, Danuta Wlodek, Terry Foutz, Mark Carter, Anna Waller Target Team Leader for the Center: Larry Sklar (lsklar@salud.unm.edu) UNM Cheminformatics: Cristian Bologa, Ph.D., Fabiola Miscioscia, Ph.D., Ramona Curpan, Ph.D., Oleg Ursu, Ph.D. Chemistry: University of Kansas Specialized Chemistry Center KU Specialized Chemistry Center PI: Jeff Aube, Ph.D. KU SCC Project Manager: Jennifer E. Golden. Ph.D. KU SCC Chemists on this project: Chad Schroeder, M.S., Denise Simpson, Ph.D., Julica Noeth, B.S. Dose Response Assay Background and Significance: Ras and related small molecular weight GTPases function in the regulation of signaling and cell growth, and collectively serve to control cell proli Oxadiazole SAR compounds tested via Multiplex dose response to identify specific small molecule inhibitors of Ras and Ras-related GTPases specifically Rac1 wildtype University of New Mexico Assay Overview: Assay Support: NIH I RO3 MH081231-01 HTS to identify specific small molecule inhibitors of Ras and Ras-related GTPases PI: Angela Wandinger-Ness, Ph.D. Co-PI: Larry Sklar, Ph.D. Assay Development: Zurab Surviladze, Ph.D. Assay Implementation: Zurab Surviladze, Danuta Wlodek, Terry Foutz, Mark Carter, Anna Waller Target Team Leader for the Center: Larry Sklar (lsklar@salud.unm.edu) UNM Cheminformatics: Cristian Bologa, Ph.D., Fabiola Miscioscia, Ph.D., Ramona Curpan, Ph.D., Oleg Ursu, Ph.D. Chemistry: University of Kansas Specialized Chemistry Center KU Specialized Chemistry Center PI: Jeff Aube, Ph.D. KU SCC Project Manager: Jennifer E. Golden. Ph.D. KU SCC Chemists on this project: Chad Schroeder, M.S., Denise Simpson, Ph.D., Julica Noeth, B.S. Dose Response Assay Background and Significance: Ras and related small molecular weight GTPases function in the regulation of signaling and cell growth, and collectively serve to control cell proli Oxadiazole SAR compounds tested via Multiplex dose response to identify specific small molecule inhibitors of Ras and Ras-related GTPases specifically Ras wildtype University of New Mexico Assay Overview: Assay Support: NIH I RO3 MH081231-01 HTS to identify specific small molecule inhibitors of Ras and Ras-related GTPases PI: Angela Wandinger-Ness, Ph.D. Co-PI: Larry Sklar, Ph.D. Assay Development: Zurab Surviladze, Ph.D. Assay Implementation: Zurab Surviladze, Danuta Wlodek, Terry Foutz, Mark Carter, Anna Waller Target Team Leader for the Center: Larry Sklar (lsklar@salud.unm.edu) UNM Cheminformatics: Cristian Bologa, Ph.D., Fabiola Miscioscia, Ph.D., Ramona Curpan, Ph.D., Oleg Ursu, Ph.D. Chemistry: University of Kansas Specialized Chemistry Center KU Specialized Chemistry Center PI: Jeff Aube, Ph.D. KU SCC Project Manager: Jennifer E. Golden. Ph.D. KU SCC Chemists on this project: Chad Schroeder, M.S., Denise Simpson, Ph.D., Julica Noeth, B.S. Dose Response Assay Background and Significance: Ras and related small molecular weight GTPases function in the regulation of signaling and cell growth, and collectively serve to control cell proli PLK1 counterscreening assay for identified PKD inhibitors The PKD HTS assay was developed and run at the University of Pittsburgh Molecular Screening Center (PMLSC) as part of the Molecular Library Screening Center Network (MLSCN)(1R03DA24898-01). Protein kinase D (PKD) is a novel family of serine/threonine kinases targeted by diacylglycerol. It regulates many fundamental cell functions including cell proliferation, survival, differentiation and protein trafficking, and plays important roles in pathological conditions such as cardiac hypertrophy and cancer in multiple organ systems. However, the mechanisms underlying these effects of PKD are not clearly understood, and the role of PKD in cancer and other diseases has not been fully defined. This is partly due to the lack of effective pharmacological tools that specifically target PKD in normal cellular processes and in pathological conditions. The immediate goal of this proposal is to demonstrate the feasibility of an IMAP-based HTS fluorescent polarization (FP) assay and its use to identi Profiling Assay to determine GST-GSH interactions in multiplex bead-based assays University of New Mexico Assay Overview: Assay Support: NIH I RO3 MH081231-01 HTS to identify specific small molecule inhibitors of Ras and Ras-related GTPases PI: Angela Wandinger-Ness, Ph.D. Co-PI: Larry Sklar, Ph.D. Assay Development: Zurab Surviladze, Ph.D. Assay Implementation: Zurab Surviladze, Danuta Wlodek, Terry Foutz, Mark Carter, Anna Waller Profiling Assay Background and Significance: The objective of the HTS associated with this counterscreen was to identify small molecule regulators of Ras and Ras-related GTPases (see Summary Report and PubChem AIDs 1333, 1334, 1335, 1336, 1337, 1339, 1340, 1341). The primary HTS assay was a no-wash fluorescent GTP-binding assay adapted to multiplexed, high-throughput measurements whereby multiple GTPases were simultaneously screened against the MLSCN library. The specificity is based on the observation that individual GTPases including wt and activated forms exhibit measurably distinct affinities for Bodipy-FI-GTP vs GTP. The assay Pyrazoline SAR compounds tested via Multiplex dose response to identify specific small molecule inhibitors of Ras and Ras-related GTPases specifically Rab2 wildtype University of New Mexico Assay Overview: Assay Support: NIH I RO3 MH081231-01 HTS to identify specific small molecule inhibitors of Ras and Ras-related GTPases PI: Angela Wandinger-Ness, Ph.D. Co-PI: Larry Sklar, Ph.D. Assay Development: Zurab Surviladze, Ph.D. Assay Implementation: Zurab Surviladze, Danuta Wlodek, Terry Foutz, Mark Carter, Anna Waller Target Team Leader for the Center: Larry Sklar (lsklar@salud.unm.edu) UNM Cheminformatics: Cristian Bologa, Ph.D., Fabiola Miscioscia, Ph.D., Ramona Curpan, Ph.D., Oleg Ursu, Ph.D. Chemistry: University of Kansas Specialized Chemistry Center KU Specialized Chemistry Center PI: Jeff Aube, Ph.D. KU SCC Project Manager: Jennifer E. Golden. Ph.D. KU SCC Chemists on this project: Chad Schroeder, M.S., Denise Simpson, Ph.D., Julica Noeth, B.S. Dose Response Assay Background and Significance: Ras and related small molecular weight GTPases function in the regulation of signaling and cell growth, and collectively serve to control cell proli Pyrazoline SAR compounds tested via Multiplex dose response to identify specific small molecule inhibitors of Ras and Ras-related GTPases specifically Rab7 wildtype University of New Mexico Assay Overview: Assay Support: NIH I RO3 MH081231-01 HTS to identify specific small molecule inhibitors of Ras and Ras-related GTPases PI: Angela Wandinger-Ness, Ph.D. Co-PI: Larry Sklar, Ph.D. Assay Development: Zurab Surviladze, Ph.D. Assay Implementation: Zurab Surviladze, Danuta Wlodek, Terry Foutz, Mark Carter, Anna Waller Target Team Leader for the Center: Larry Sklar (lsklar@salud.unm.edu) UNM Cheminformatics: Cristian Bologa, Ph.D., Fabiola Miscioscia, Ph.D., Ramona Curpan, Ph.D., Oleg Ursu, Ph.D. Chemistry: University of Kansas Specialized Chemistry Center KU Specialized Chemistry Center PI: Jeff Aube, Ph.D. KU SCC Project Manager: Jennifer E. Golden. Ph.D. KU SCC Chemists on this project: Chad Schroeder, M.S., Denise Simpson, Ph.D., Julica Noeth, B.S. Dose Response Assay Background and Significance: Ras and related small molecular weight GTPases function in the regulation of signaling and cell growth, and collectively serve to control cell proli Pyrazoline SAR compounds tested via Multiplex dose response to identify specific small molecule inhibitors of Ras and Ras-related GTPases specifically Ras wildtype University of New Mexico Assay Overview: Assay Support: NIH I RO3 MH081231-01 HTS to identify specific small molecule inhibitors of Ras and Ras-related GTPases PI: Angela Wandinger-Ness, Ph.D. Co-PI: Larry Sklar, Ph.D. Assay Development: Zurab Surviladze, Ph.D. Assay Implementation: Zurab Surviladze, Danuta Wlodek, Terry Foutz, Mark Carter, Anna Waller Target Team Leader for the Center: Larry Sklar (lsklar@salud.unm.edu) UNM Cheminformatics: Cristian Bologa, Ph.D., Fabiola Miscioscia, Ph.D., Ramona Curpan, Ph.D., Oleg Ursu, Ph.D. Chemistry: University of Kansas Specialized Chemistry Center KU Specialized Chemistry Center PI: Jeff Aube, Ph.D. KU SCC Project Manager: Jennifer E. Golden. Ph.D. KU SCC Chemists on this project: Chad Schroeder, M.S., Denise Simpson, Ph.D., Julica Noeth, B.S. Dose Response Assay Background and Significance: Ras and related small molecular weight GTPases function in the regulation of signaling and cell growth, and collectively serve to control cell prol QFRET-based counterscreen for inhibitors of PFM18AAP: biochemical high throughput dose response assay for inhibitors of the Cathepsin L proteinase (CTSL1). Source (MLPCN Center Name): The Scripps Research Institute Molecular Screening Center Affiliation: The Scripps Research Institute, TSRI Assay Provider: John Dalton and Donald Gardiner, Queensland Institute of Medical Research, Australia Network: Molecular Library Probe Production Centers Network (MLPCN) Grant Proposal Number: 1 R03 MH084103-01 Grant Proposal PI: John Dalton and Donald Gardiner, Queensland Institute of Medical Research, Australia External Assay ID: CTSL1_INH_QFRET_1536_3XIC50 Name: QFRET-based counterscreen for inhibitors of PFM18AAP: biochemical high throughput dose response assay for inhibitors of the Cathepsin L proteinase (CTSL1). Description: Aminopeptidases (APs) are metalloproteases that cleave amino-terminal (N-terminal) amino acids during protein synthesis (1, 2) . These enzymes are characterized in part by their post-translational removal of leucine, aspartate, proline, methionine, etc from proteins and peptides, in order that proteins are properly regulat Rml C and D dose-response confirmation Molecular Library Screening Center Network (MLSCN) Penn Center for Molecular Discovery (PCMD) Assay Provider: Michael McNeil, Colorado State University, Fort Collins, CO MLSCN Grant: DA024889-01 This screen is for compounds that have the potential to be developed into new drugs against tuberculosis (TB) because they inhibit the enzymes required for the formation of the cell wall of the tuberculosis bacterium. New drugs are needed because the rate of cure with the present drugs is very slow, and prevalence of Mycobacterium tuberculosis resistance to present drugs is increasing. Recently, an increase in co-infection of HIV and M. tuberculosis has occurred, and treatment with present drugs results in harmful HIV/TB drug interactions. To identify potential anti-TB agents, we focused on two enzymes that act sequentially in the formation of dTDP-rhamnose (dTDP-Rha), a biosynthetic precursor required for TB cell wall formation and found to be essential for the growth of M. smegmatis and M S1P3 Dose Response Assay Counterscreen for 5-Hydroxytryptamine(Serotonin) Receptor Subtype 1E Antagonists External Assay ID: S1P3_IC50_CS_5H1E_Antagonists Name: S1P3 Dose Response Assay Counterscreen for 5-Hydroxytryptamine(Serotonin) Receptor Subtype 1E Antagonists Source (MLSCN Center Name): The Scripps Research Institute Molecular Screening Center Center Affiliation: The Scripps Research Institute, TSRI Assay Provider: The Scripps Research Institute Molecular Screening Center Network: Molecular Library Screening Center Network (MLSCN) Grant Proposal Number: 1 R03 MH076345-01 Description: The neurotransmitter, serotonin (5HT, 5-hydroxytryptamine) is important in a large number of neurological behaviors including of mood [1], appetite [2], cognition [3], pain [4], and memory [5]. The serotonin receptors are a large group of GPCRs which are divided into families, 5HT1-7, based upon functionality and pharmacology [6]. The 5HT1 family which includes at least 6 members, 5HT1A-F, is indicated in anxiety, depression and cognition [7]. All of the 5HT1 receptors activate gi/o g-proteins to inhi SAR Analysis for the identification of selective inhibits of the transient receptor potential cation channel C4 (TRPC4): Automated Electrophysiology Data Source: Johns Hopkins Ion Channel Center BioAssay Type: Confirmatory, Concentration-Response Relationship Observed Source (MLPCN Center Name): Johns Hopkins Ion Channel Center (JHICC) Center Affiliation: Johns Hopkins University, School of Medicine Screening Center PI: Min Li, Ph.D. Assay Provider: Michael Zhu, Ph.D., Ohio State University Network: Molecular Libraries Probe Production Centers Network (MLPCN) Grant Proposal Number: 1 R21 NS056942-01 Grant Proposal PI: Michael Zhu, Ph.D., Ohio State University Assay Implementation: Jie Shi, Melissa Miller, Shunyou Long M.S., David Meyers Ph.D., Meng Wu Ph.D., Owen McManus Ph.D. Name: SAR Analysis for the identification of selective inhibitors of the transient receptor potential cation channel C4 (TRPC4): Automated Electrophysiology Description: The inhibitory effects of test compounds on TRPC4 channels were evaluated in electrophysiologcal assays using a QPatch16 automated electrophysiology instrument. Whole cell recordings were SAR Analysis of Agonists of the DOR Receptor using an Image-Based Assay - Set 2 Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Production Centers Network(MLPCN) Grant Number: 1X01DA026208-01 Assay Provider: Dr. Lawrance Barak, Duke University, Durham NC Drug addiction is a disease that involves the G-protein coupled receptors in the central nervous system resulting in compulsive or abnormal behavior. Recent studies have shown that opioid receptors play a role in regulating other receptors that interact with drug and other substance abuse. The opioid receptors are composed of multiple of multiple receptor subtypes whose contribution the addictive behaviors are not fully delineated. The aim of this assay is to identify small molecule agonists of the human delta opioid receptor (DOR). This dose response assay is developed and performed to confirm hits originally identified in "uHTS identification of small molecule agonists of SAR Analysis of Agonists of the DOR Receptor using an Image-Based Assay - Set 3 Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute(SBMRI, San Diego, CA) Network: NIH Molecular Libraries Production Centers Network(MLPCN) Grant Number: 1X01DA026208-01 Assay Provider: Dr. Lawrance Barak, Duke University, Durham NC Drug addiction is a disease that involves the G-protein coupled receptors in the central nervous system resulting in compulsive or abnormal behavior. Recent studies have shown that opioid receptors play a role in regulating other receptors that interact with drug and other substance abuse. The opioid receptors are composed of multiple of multiple receptor subtypes whose contribution the addictive behaviors are not fully delineated. The aim of this assay is to identify small molecule agonists of the human delta opioid receptor (DOR). This dose response assay is developed and performed to confirm hits originally identified in "uHTS identification of small molecule agonists of SAR Analysis of Agonists of the DOR Receptor using an Image-Based Assay Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Production Centers Network(MLPCN) Grant Number: 1X01DA026208-01 Assay Provider: Dr. Lawrance Barak, Duke University, Durham NC Drug addiction is a disease that involves the G-protein coupled receptors in the central nervous system resulting in compulsive or abnormal behavior. Recent studies have shown that opioid receptors play a role in regulating other receptors that interact with drug and other substance abuse. The opioid receptors are composed of multiple of multiple receptor subtypes whose contribution the addictive behaviors are not fully delineated. The aim of this assay is to identify small molecule agonists of the human delta opioid receptor (DOR). This dose response assay is developed and performed to confirm hits originally identified in "uHTS identification of small molecule agonists of SAR Analysis of Agonists of the MOR Receptor using an Image-Based Assay - Set 2 Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Production Centers Network(MLPCN) Grant Number: 1X01DA026208-01 Assay Provider: Dr. Lawrance Barak, Duke University, Durham NC Drug addiction is a disease that involves the G-protein coupled receptors in the central nervous system resulting in compulsive or abnormal behavior. Recent studies have shown that opioid receptors play a role in regulating other receptors that interact with drug and other substance abuse. The opioid receptors are composed of multiple of multiple receptor subtypes whose contribution the addictive behaviors are not fully delineated. The aim of this assay is to identify small molecule agonists of the human mu opioid receptor (MOR). This dose response assay is developed and performed to confirm hits originally identified in "uHTS identification of small molecule agonists of th SAR Analysis of Agonists of the MOR Receptor using an Image-Based Assay - Set 3 Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Production Centers Network(MLPCN) Grant Number: 1X01DA026208-01 Assay Provider: Dr. Lawrance Barak, Duke University, Durham NC Drug addiction is a disease that involves the G-protein coupled receptors in the central nervous system resulting in compulsive or abnormal behavior. Recent studies have shown that opioid receptors play a role in regulating other receptors that interact with drug and other substance abuse. The opioid receptors are composed of multiple of multiple receptor subtypes whose contribution the addictive behaviors are not fully delineated. The aim of this assay is to identify small molecule agonists of the human mu opioid receptor (MOR). This dose response assay is developed and performed to confirm hits originally identified in "uHTS identification of small molecule agonists of th SAR Analysis of Agonists of the MOR Receptor using an Image-Based Assay Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Production Centers Network(MLPCN) Grant Number: 1X01DA026208-01 Assay Provider: Dr. Lawrance Barak, Duke University, Durham NC Drug addiction is a disease that involves the G-protein coupled receptors in the central nervous system resulting in compulsive or abnormal behavior. Recent studies have shown that opioid receptors play a role in regulating other receptors that interact with drug and other substance abuse. The opioid receptors are composed of multiple of multiple receptor subtypes whose contribution the addictive behaviors are not fully delineated. The aim of this assay is to identify small molecule agonists of the human mu opioid receptor (MOR). This dose response assay is developed and performed to confirm hits originally identified in "uHTS identification of small molecule agonists of th SAR Analysis of Antagonists of the DOR Receptor using an Image-Based Assay - Set 3 Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Probe Production Centers Network(MLPCN) Grant Number: 1X01DA026208-01 Assay Provider: Dr. Lawrance Barak, Duke University, Durham NC Drug addiction is a disease that involves the G-protein coupled receptors in the central nervous system resulting in compulsive or abnormal behavior. Recent studies have shown that opioid receptors play a role in regulating other receptors that interact with drug and other substance abuse. The opioid receptors are composed of multiple of multiple receptor subtypes whose contribution the addictive behaviors are not fully delineated. The aim of this assay is to identify small molecule antagonists of the human delta opioid receptor (DOR). This dose response assay is developed and performed to confirm hits originally identified in "uHTS identification of small molecule ago SAR Analysis of Antagonists of the DOR Receptor using an Image-Based Assay Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Production Centers Network(MLPCN) Grant Number: 1X01DA026208-01 Assay Provider: Dr. Lawrance Barak, Duke University, Durham NC Drug addiction is a disease that involves the G-protein coupled receptors in the central nervous system resulting in compulsive or abnormal behavior. Recent studies have shown that opioid receptors play a role in regulating other receptors that interact with drug and other substance abuse. The opioid receptors are composed of multiple of multiple receptor subtypes whose contribution the addictive behaviors are not fully delineated. The aim of this assay is to identify small molecule antagonists of the human delta opioid receptor (DOR). This dose response assay is developed and performed to confirm hits originally identified in "uHTS identification of small molecule agonists SAR Analysis of Antagonists of the MOR Receptor using an Image-Based Assay - Set 3 Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Probe Production Centers Network(MLPCN) Grant Number: 1X01DA026208-01 Assay Provider: Dr. Lawrance Barak, Duke University, Durham NC Drug addiction is a disease that involves the G-protein coupled receptors in the central nervous system resulting in compulsive or abnormal behavior. Recent studies have shown that opioid receptors play a role in regulating other receptors that interact with drug and other substance abuse. The opioid receptors are composed of multiple of multiple receptor subtypes whose contribution the addictive behaviors are not fully delineated. The aim of this assay is to identify small molecule antagonists of the human mu opioid receptor (MOR). This dose response assay is developed and performed to confirm hits originally identified in "uHTS identification of small molecule agoni SAR Analysis of Selective Antagonists of GPR55 using an Image-Based Assay - Set 4 Data Source: Sanford-Burnham Center for Chemical Genomics(SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Probe Production Centers Network (MLPCN) Grant Number: 1X01 DA026205-01 Assay Provider: Dr. Mary Abood, (Temple University, formerly at California Pacific Medical Center Research Institute) The cannabinoid and endocannabinoid system has been implicated in the pathophysiology of drug dependence and addiction disorders. GPR55, an orphan G-Protein Coupled Receptor, has been reported to be a cannabinoid receptor, but its status as such remains unresolved due to conflicting results from pharmacological studies. The goal of the project is to identify small molecule antagonists of GPR55, which may aid in the deorphanization efforts of this receptor and ultimately further the understanding of the role of GPR55 in drug addiction. This high content imaging assay utilizes a cell line permanently expressing SAR Analysis of Selective Antagonists of GPR55 using an Image-Based Assay Data Source: Sanford-Burnham Center for Chemical Genomics(SSBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute(SBMRI, San Diego, CA) Network: NIH Molecular Libraries Probe Production Centers Network (MLPCN) Grant Number: 1X01 MH085708-01 Assay Provider: Dr. Lawrence Barak, Duke University, Durham, NC Addictive behavior stems from abnormal signaling activities in the brain. Thus identification of compounds blocking this modified signaling activity may lead to treatments for addictive behavior. GPR35, a to-date uncharacterized orphan G-Protein Coupled Receptor, is thought to play a role in addiction and has homology to other known receptors of abuse. This high-content imaging assay was used as a counter screen for hits originally identified in a high-content screen for antagonists of the GPR35 receptor "Image-based HTS for Selective Antagonists of GPR35" (AID 2058) and to study the structure-activity relationship on analogs of the confirmed hits. Compounds are ei SAR Analysis of small molecule inhibitors of Sentrin-specific protease 8 (SENP8) using a Luminescent assay - Set 2 Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Production Centers Network(MLPCN)Grant Proposal Number: 1R21 NS061758-01 fast track Assay Provider: Dr. Guy Salvesen, Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Modification of proteins by SUMO is a dynamic and reversible process. SUMOylation/deSUMOylation cycle regulates SUMOs function. Sentrin-specific proteases (SENPs) are involved in both the maturation of SUMO precursors (endopeptidase cleavage) and deconjugation of the targets (isopeptidase cleavage) [1-3]. There are seven SENPs (1, 2, 3, 5, 6, 7, 8) in humans, and several of these have been characterized as SUMO (or Nedd8) specific enzymes. SENP8 is not a SUMO protease, instead it functions on a small ubiquitin related protein Nedd8. The objective of this project is to generate small molecule inhibitors specific for SENP8 SAR Confirmatory Dose Response LIBS Assay for Allosteric Ligands of the VLA-4 Integrin University of New Mexico Assay Overview: Assay Support: 1 R01 HL081062-01 Project Title: HTS for Identification of VLA-4 Allosteric Modulators (MLPCN) PI: Larry Sklar Screening Center / PI: UNM Center for Molecular Discovery / Larry Sklar Chemistry Center / PI: Vanderbilt Specialized Chemistry Center / Jeff Aube Assay Implementation: Peter Simons, Yang Wu, Susan Young, Terry Foutz, Stephanie Sedillo, Anna Waller, Annette Evangelisti, Mark Carter, Oleg Ursu, Cristian Bologa Assay Background and Significance: We have developed a novel ligand induced binding site (LIBS) mAb assay for integrin HTS using phycoerythrin (PE) labeled HUTS-21 mAb. The novel assay is a result of an R01 (HL081062) which has taken advantage of compounds identified through an earlier project X01MH077638 (entitled "MLSCN Assay for Allosteric Regulators of the VLA-4 Integrin") that screened a portion of the MLSMR for allosteric regulators of VLA-4 (AIDs 528, 529). The approach described here uses an assay SAR LYP1 Fluorescent Assay using OMFP substrate for In Vitro dose response studies - Set 2 Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Probe Production Centers Network (MLPCN) Grant Number: 1R21NS056945-01 Assay Provider: Dr. Nunzio Bottini, La Jolla Institute for Allergy and Immunology CA LYP, is a lymphocyte specific protein tyrosine phosphatase that plays a critical regulatory role in T cell receptor signaling. The PTPN22 gene encodes this phosphatase. A single-nucleotide polymorphism in PTPN22 is associated with a number of autoimmune disorders, including type 1 diabetes, rheumatoid arthritis, juvenile rheumatoid arthritis, systemic lupus erythematosus and Grave's disease. The autoimmunity-predisposing allele is a gain-of-function mutant suggesting that a specific small-molecule inhibitor could eliminate its effect. This biochemical assay employs a fluorescent readout based on the enzyme's ability to liberate methyl-fluo SAR LYP1 Fluorescent Assay using OMFP substrate for In Vitro dose response studies - Set 3 Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Probe Production Centers Network (MLPCN) Grant Number: 1R21NS056945-01 Assay Provider: Dr. Nunzio Bottini, La Jolla Institute for Allergy and Immunology, La Jolla, CA LYP, is a lymphocyte specific protein tyrosine phosphatase that plays a critical regulatory role in T cell receptor signaling. The PTPN22 gene encodes this phosphatase. A single-nucleotide polymorphism in PTPN22 is associated with a number of autoimmune disorders, including type 1 diabetes, rheumatoid arthritis, juvenile rheumatoid arthritis, systemic lupus erythematosus and Grave's disease. The autoimmunity-predisposing allele is a gain-of-function mutant suggesting that a specific small-molecule inhibitor could eliminate its effect. This biochemical assay employs a fluorescent readout based on the enzyme's ability to liberate SAR LYP1 Fluorescent Assay using OMFP substrate for In Vitro dose response studies - Set 4 Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Probe Production Centers Network (MLPCN) Grant Number: 1R21NS056945-01 Assay Provider: Dr. Nunzio Bottini , La Jolla Institute for Allergy and Immunology CA LYP, is a lymphocyte specific protein tyrosine phosphatase that plays a critical regulatory role in T cell receptor signaling. The PTPN22 gene encodes this phosphatase. A single-nucleotide polymorphism in PTPN22 is associated with a number of autoimmune disorders, including type 1 diabetes, rheumatoid arthritis, juvenile rheumatoid arthritis, systemic lupus erythematosus and Grave's disease. The autoimmunity-predisposing allele is a gain-of-function mutant suggesting that a specific small-molecule inhibitor could eliminate its effect. This biochemical assay employs a fluorescent readout based on the enzyme's ability to liberate methyl-fl SAR LYP1 Fluorescent Assay using OMFP substrate for In Vitro dose response studies Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Probe Production Centers Network (MLPCN) Grant Number: 1R21NS056945-01 Assay Provider: Dr. Nunzio Bottini, La Jolla Institute for Allergy and Immunology, La Jolla, CA LYP, is a lymphocyte specific protein tyrosine phosphatase that plays a critical regulatory role in T cell receptor signaling. The PTPN22 gene encodes this phosphatase. A single-nucleotide polymorphism in PTPN22 is associated with a number of autoimmune disorders, including type 1 diabetes, rheumatoid arthritis, juvenile rheumatoid arthritis, systemic lupus erythematosus and Grave's disease. The autoimmunity-predisposing allele is a gain-of-function mutant suggesting that a specific small-molecule inhibitor could eliminate its effect. This biochemical assay employs a fluorescent readout based on the enzyme's ability to liberate SAR LYP1 Fluorescent Assay using pCAP substrate for In Vitro dose response studies - Set 2 Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute(SBMRI, San Diego, CA) Network: NIH Molecular Libraries Probe Production Centers Network (MLPCN) Grant Number: 1R21NS056945-01 Assay Provider: Dr. Nunzio Bottini, La Jolla Institute for Allergy and Immunology LYP, is a lymphocyte specific protein tyrosine phosphatase that plays a critical regulatory role in T cell receptor signaling. The PTPN22 gene encodes this phosphatase. A single-nucleotide polymorphism in PTPN22 is associated with a number of autoimmune disorders, including type 1 diabetes, rheumatoid arthritis, juvenile rheumatoid arthritis, systemic lupus erythematosus and Grave's disease. The autoimmunity-predisposing allele is a gain-of-function mutant suggesting that a specific small-molecule inhibitor could eliminate its effect. Finding specific inhibitors of protein phosphatases has proven extremely difficult. The goal of this project is to SAR LYP1 Fluorescent Assay using pCAP substrate for In Vitro dose response studies - Set 3 Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute(SBMRI, San Diego, CA) Network: NIH Molecular Libraries Probe Production Centers Network (MLPCN) Grant Number: 1R21NS056945-01 Assay Provider: Dr. Nunzio Bottini, La Jolla Institute for Allergy and Immunology, La Jolla, CA LYP, is a lymphocyte specific protein tyrosine phosphatase that plays a critical regulatory role in T cell receptor signaling. The PTPN22 gene encodes this phosphatase. A single-nucleotide polymorphism in PTPN22 is associated with a number of autoimmune disorders, including type 1 diabetes, rheumatoid arthritis, juvenile rheumatoid arthritis, systemic lupus erythematosus and Grave's disease. The autoimmunity-predisposing allele is a gain-of-function mutant suggesting that a specific small-molecule inhibitor could eliminate its effect. Finding specific inhibitors of protein phosphatases has proven extremely difficult. The goal of this SAR LYP1 Fluorescent Assay using pCAP substrate for In Vitro dose response studies - Set 4 Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Probe Production Centers Network(MLPCN) Grant Number: 1R21NS056945-01 Assay Provider: Dr. Nunzio Bottini, La Jolla Institute for Allergy and Immunology, La Jolla, CA LYP, is a lymphocyte specific protein tyrosine phosphatase that plays a critical regulatory role in T cell receptor signaling. The PTPN22 gene encodes this phosphatase. A single-nucleotide polymorphism in PTPN22 is associated with a number of autoimmune disorders, including type 1 diabetes, rheumatoid arthritis, juvenile rheumatoid arthritis, systemic lupus erythematosus and Grave's disease. The autoimmunity-predisposing allele is a gain-of-function mutant suggesting that a specific small-molecule inhibitor could eliminate its effect. Finding specific inhibitors of protein phosphatases has proven extremely difficult. The goal of this SAR LYP1 Fluorescent Assay using pCAP substrate for In Vitro dose response studies Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Probe Production Centers Network (MLPCN) Grant Number: 1R21NS056945-01 Assay Provider: Dr. Nunzio Bottini, La Jolla Institute for Allergy and Immunology, La Jolla, CA LYP, is a lymphocyte specific protein tyrosine phosphatase that plays a critical regulatory role in T cell receptor signaling. The PTPN22 gene encodes this phosphatase. A single-nucleotide polymorphism in PTPN22 is associated with a number of autoimmune disorders, including type 1 diabetes, rheumatoid arthritis, juvenile rheumatoid arthritis, systemic lupus erythematosus and Grave's disease. The autoimmunity-predisposing allele is a gain-of-function mutant suggesting that a specific small-molecule inhibitor could eliminate its effect. Finding specific inhibitors of protein phosphatases has proven extremely difficult. The goal of thi SAR Selectivity Analysis of small molecule inhibitors of PEST using pCAP in a fluorescence assay Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Probe Production Centers Network(MLPCN) Grant Number: 1R21NS056945-01 Assay Provider: Dr. Nunzio Bottini, La Jolla Institute for Allergy and Immunology, La Jolla, CA PTP-PEST, encoded by PTPN12 gene, is a protein tyrosine phosphatase that contains a C-terminal PEST motif, which serves as a protein-protein interaction domain. This PTP dephosphorylates c-ABL, and may play a role in oncogenesis. This is a selectivity counterscreen to aid with identification of specific inhibitors of LYP phosphatase (AID 2135). This assay uses a phosphorylated coumarin amino acid (pCAP) moiety attached to a 14-mer peptide, which is a substrate for LYP. This dose response assay is developed and performed to assess selectivity of hits originally found in "uHTS identification of small molecule inhibitors of LYP via a fluo SAR analysis of Agonists of the GPR35 Receptor using an Image-Based Assay - Set 3 Data Source: Sanford-Burnham Center for Chemical Genomics Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI) Network: NIH Molecular Libraries Production Centers Network (MLPCN) Grant Number: 1X01 DA026205-01 Assay Provider: Dr. Mary Abood, (Temple University, formerly at California Pacific Medical Center Research Institute) Addictive behavior stems from abnormal signaling activities in the brain. Thus identification of compounds blocking this modified signaling activity may lead to treatments for addictive behavior. GPR35, a to-date uncharacterized orphan G-Protein Coupled Receptor, is thought to play a role in addiction and has homology to other known receptors of abuse. This dose response assay is developed and performed to evaluate selectivity of hits originally identified in "Image-Based HTS for Selective Agonists for GPR55" (AID 1961) and to study the structure-activity relationship. Compounds are either acquired from commercial sources or synthesized i SAR analysis of Agonists of the GPR35 Receptor using an Image-Based Assay - Set 4 Data Source: Sanford-Burnham Center for Chemical Genomics Source Affiliation: Sanford-Burnham Medical Research Institute Network: NIH Molecular Libraries Probe Production Centers Network (MLPCN) Grant Number: 1X01 DA026205-01 Assay Provider: Dr. Mary Abood, (Temple University, formerly at California Pacific Medical Center Research Institute) Addictive behavior stems from abnormal signaling activities in the brain. Thus identification of compounds blocking this modified signaling activity may lead to treatments for addictive behavior. GPR35, a to-date uncharacterized orphan G-Protein Coupled Receptor, is thought to play a role in addiction and has homology to other known receptors of abuse. This high-content imaging assay is developed and performed to evaluate selectivity of hits originally identified in "Image-based HTS for Selective Antagonists of GPR55" (AID 2013) and to study the structure-activity relationship on analogs of the confirmed hits. Compounds are either acquired fr SAR analysis of Agonists of the Kappa Opioid Receptor (KOR) using an Image-Based Assay - Set 2 Data Source: Sanford-Burnham Center for Chemical Genomics Source Affiliation: Sanford-Burnham Medical Research Institute Network: NIH Molecular Libraries Production Centers Network Grant Number: 1X01DA026208-01 Assay Provider: Dr. Lawrance Barak, Duke University, Durham NC Drug addiction is a disease that involves the G-protein coupled receptors in the central nervous system resulting in compulsive or abnormal behavior. Recent studies have shown that opioid receptors play a role in regulating other receptors that interact with drug and other substance abuse. The opioid receptors are composed of multiple of multiple receptor subtypes whose contribution the addictive behaviors are not fully delineated. The aim of this assay is to identify small molecule agonists of the human kappa opioid receptor (KOR). This dose response assay is developed and performed to confirm hits originally identified in "uHTS identification of small molecule agonists of the kappa opioid receptor via a lum SAR analysis of Agonists of the Kappa Opioid Receptor (KOR) using an Image-Based Assay Data Source: Sanford-Burnham Center for Chemical Genomics Source Affiliation: Sanford-Burnham Medical Research Institute Network: NIH Molecular Libraries Production Centers Network Grant Number: 1X01DA026208-01 Assay Provider: Dr. Lawrance Barak, Duke University, Durham NC Drug addiction is a disease that involves the G-protein coupled receptors in the central nervous system resulting in compulsive or abnormal behavior. Recent studies have shown that opioid receptors play a role in regulating other receptors that interact with drug and other substance abuse. The opioid receptors are composed of multiple of multiple receptor subtypes whose contribution the addictive behaviors are not fully delineated. The aim of this assay is to identify small molecule agonists of the human kappa opioid receptor (KOR). This dose response assay is developed and performed to confirm hits originally identified in "uHTS identification of small molecule agonists of the kappa opioid receptor via a lum SAR analysis of Agonists of the Kappa Opioid Receptor (KOR) using an Image-Based Assay-Set 3 Data Source: Sanford-Burnham Center for Chemical Genomics Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Production Centers Network Grant Number: 1X01DA026208-01 Assay Provider: Dr. Lawrance Barak, Duke University, Durham NC Drug addiction is a disease that involves the G-protein coupled receptors in the central nervous system resulting in compulsive or abnormal behavior. Recent studies have shown that opioid receptors play a role in regulating other receptors that interact with drug and other substance abuse. The opioid receptors are composed of multiple of multiple receptor subtypes whose contribution the addictive behaviors are not fully delineated. The aim of this assay is to identify small molecule agonists of the human kappa opioid receptor (KOR). This dose response assay is developed and performed to confirm hits originally identified in "uHTS identification of small molecule agonists of the kappa op SAR analysis of Antagonists of IAP-family anti-apoptotic proteins - Set 2 Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Production Centers Network(MLPCN)Grant Proposal Number: MH081277-01 Assay Provider: John C. Reed, Sanford-Burnham Medical Research Institute, San Diego, CA This XIAP dose response assay is developed and performed to confirm hits originally identified in the XIAP HTS binding assay (AID 1018) and to study the structure-activity relationship on analogs of the confirmed hits. Compounds are either acquired from commercial sources or synthesized internally. The assay was performed in the assay providers' laboratory. Apoptosis plays an essential role in many aspects of normal development and physiology, becoming dysregulated in myriad diseases characterized by insufficient or excessive cell death. Caspases are intracellular proteases that are suppressed by Inhibitor of Apoptosis Proteins (IAPs), a famil SAR analysis of Antagonists of XIAP-Bir3 domain of IAP-family anti-apoptotic proteins - Set 2 Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Production Centers Network(MLPCN)Grant Proposal Number: MH081277-01 Assay Provider: John C. Reed, Sanford-Burnham Medical Research Institute San Diego, CA This dose response assay is developed and performed as a counter screen to compounds in the Chemical Antagonists of IAP-family anti-apoptotic proteins confirmation (AID 1449) and to study the structure-activity relationship on analogs of the confirmed hits. Compounds are either acquired from commercial sources or synthesized internally. This assay was performed in the assay providers' laboratory. Apoptosis plays an essential role in many aspects of normal development and physiology, becoming dysregulated in myriad diseases characterized by insufficient or excessive cell death. Caspases are intracellular proteases that are suppressed by Inhibit SAR analysis of Antagonists of XIAP-Bir3 domain of IAP-family anti-apoptotic proteins Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Screening Centers Network (MLSCN) Grant Proposal Number: MH081277-01 Assay Provider: John C. Reed, Sanford-Burnham Medical Research Institute, San Diego, CA This dose response assay is developed and performed as a counter screen to compounds in the Chemical Antagonists of IAP-family anti-apoptotic proteins confirmation (AID 1449) and to study the structure-activity relationship on analogs of the confirmed hits. Compounds are either acquired from commercial sources or synthesized internally. This assay was performed in the assay providers' laboratory. Apoptosis plays an essential role in many aspects of normal development and physiology, becoming dysregulated in myriad diseases characterized by insufficient or excessive cell death. Caspases are intracellular proteases that are suppressed by Inhibi SAR analysis of Antagonists of the GPR35 Receptor using an Image-Based Assay - Set 4 Data Source: Sanford-Burnham Center for Chemical Genomics Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Production Centers Network (MLPCN) Grant Number: 1X01 DA026205-01 Assay Provider: Dr. Mary Abood, (Temple University, formerly at California Pacific Medical Center Research Institute) Addictive behavior stems from abnormal signaling activities in the brain. Thus identification of compounds blocking this modified signaling activity may lead to treatments for addictive behavior. GPR35, a to-date uncharacterized orphan G-Protein Coupled Receptor, is thought to play a role in addiction and has homology to other known receptors of abuse. This high-content imaging assay is developed and performed to evaluate selectivity of hits originally identified in a high-content screen for agonists of the GPR55 receptor, "Image-Based HTS for Selective Agonists of GPR55"(AID 1961) and to study the structure-activity relatio SAR analysis of Antagonists of the GPR35 Receptor using an Image-Based Assay Data Source: Sanford-Burnham Center for Chemical Genomics Source Affiliation: Sanford-Burnham Medical Research Institute Network: NIH Molecular Libraries Production Centers Network (MLPCN) Grant Number: 1X01 DA026205-01 Assay Provider: Dr. Mary Abood, (Temple University, formerly at California Pacific Medical Center Research Institute) Addictive behavior stems from abnormal signaling activities in the brain. Thus identification of compounds blocking this modified signaling activity may lead to treatments for addictive behavior. GPR35, a to-date uncharacterized orphan G-Protein Coupled Receptor, is thought to play a role in addiction and has homology to other known receptors of abuse. This high-content imaging assay is developed and performed to evaluate selectivity of hits originally identified in a high-content screen for agonists of the GPR55 receptor, "Image-Based HTS for Selective Agonists of GPR55"(AID 1961) and to study the structure-activity relationship on analogs of th SAR analysis of Antagonists of the Kappa Opioid Receptor (KOR) using an Image-Based Assay - Set 2 Data Source: Sanford-Burnham Center for Chemical Genomics Source Affiliation: Sanford-Burnham Medical Research Institute Network: NIH Molecular Libraries Production Centers Network Grant Number: 1X01DA026208-01 Assay Provider: Dr. Lawrance Barak, Duke University, Durham NC Drug addiction is a disease that involves the G-protein coupled receptors in the central nervous system resulting in compulsive or abnormal behavior. Recent studies have shown that opioid receptors play a role in regulating other receptors that interact with drug and other substance abuse. The opioid receptors are composed of multiple of multiple receptor subtypes whose contribution the addictive behaviors are not fully delineated. The aim of this assay is to identify small molecule antagonists of the human kappa opioid receptor (KOR). This dose response assay is developed and performed to confirm hits originally identified in "uHTS identification of small molecule agonists of the kappa opioid receptor via a SAR analysis of Antagonists of the Kappa Opioid Receptor (KOR) using an Image-Based Assay - Set 3 Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Probe Production Centers Network Grant Number: 1X01DA026208-01 Assay Provider: Dr. Lawrance Barak, Duke University, Durham NC Drug addiction is a disease that involves the G-protein coupled receptors in the central nervous system resulting in compulsive or abnormal behavior. Recent studies have shown that opioid receptors play a role in regulating other receptors that interact with drug and other substance abuse. The opioid receptors are composed of multiple of multiple receptor subtypes whose contribution the addictive behaviors are not fully delineated. The aim of this assay is to identify small molecule antagonists of the human kappa opioid receptor (KOR). This dose response assay is developed and performed to confirm hits originally identified in "uHTS identification of small molecule agonists SAR analysis of Antagonists of the Kappa Opioid Receptor (KOR) using an Image-Based Assay - Set 4 Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Probe Production Centers Network Grant Number: 1X01DA026208-01 Assay Provider: Dr. Lawrance Barak, Duke University, Durham NC Drug addiction is a disease that involves the G-protein coupled receptors in the central nervous system resulting in compulsive or abnormal behavior. Recent studies have shown that opioid receptors play a role in regulating other receptors that interact with drug and other substance abuse. The opioid receptors are composed of multiple of multiple receptor subtypes whose contribution the addictive behaviors are not fully delineated. The aim of this assay is to identify small molecule antagonists of the human kappa opioid receptor (KOR). This dose response assay is developed and performed to confirm hits originally identified in "uHTS identification of small molecule agonists SAR analysis of Antagonists of the Kappa Opioid Receptor (KOR) using an Image-Based Assay Data Source: Sanford-Burnham Center for Chemical Genomics Source Affiliation: Sanford-Burnham Medical Research Institute Network: NIH Molecular Libraries Production Centers Network Grant Number: 1X01DA026208-01 Assay Provider: Dr. Lawrance Barak, Duke University, Durham NC Drug addiction is a disease that involves the G-protein coupled receptors in the central nervous system resulting in compulsive or abnormal behavior. Recent studies have shown that opioid receptors play a role in regulating other receptors that interact with drug and other substance abuse. The opioid receptors are composed of multiple of multiple receptor subtypes whose contribution the addictive behaviors are not fully delineated. The aim of this assay is to identify small molecule antagonists of the human kappa opioid receptor (KOR). This dose response assay is developed and performed to confirm hits originally identified in "uHTS identification of small molecule agonists of the kappa opioid receptor via a SAR analysis of GM-Tri-DAP induced IL-8 secretion in MCF-7/NOD1 cells - Set 2 Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Screening Centers Network (MLSCN) Grant Number: 1 R03 MH084844-01 Assay Provider: Dr. John C. Reed, Sanford-Burnham Medical Research Institute The modulation of immune response activity is one of the major goals in the development of novel therapeutics for auto-immune and inflammatory diseases. The innate system resides at the intersection of the pathways of microbial recognition, inflammation, and cell death, thereby offering various therapeutic targets. In this context, NOD1 and NOD2 are of particular interest, since they recognize distinct structures derived from bacterial peptidoglycans and directly activate NF-kB, a central regulator of immune response, inflammation, and apoptosis. The NOD proteins participate in the signaling events triggered by host recognition of specific motifs (mostly, murope SAR analysis of Muramyl dipeptide (MDP) induced IL-8 secretion in MCF-7/NOD2 cells - Set 2 Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Screening Centers Network (MLSCN) Grant Number: 1 R03 MH084844-01 Assay Provider: Dr. John C. Reed, Sanford-Burnham Medical Research Institute The modulation of immune response activity is one of the major goals in the development of novel therapeutics for auto-immune and inflammatory diseases. The innate system resides at the intersection of the pathways of microbial recognition, inflammation, and cell death, thereby offering various therapeutic targets. In this context, NOD1 and NOD2 are of particular interest, since they recognize distinct structures derived from bacterial peptidoglycans and directly activate NF-kB, a central regulator of immune response, inflammation, and apoptosis. The NOD proteins participate in the signaling events triggered by host recognition of specific motifs (mostly, murope SAR analysis of NF-kB dependent luciferase using DAP as an inducer Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Probe Production Centers Network (MLPCN) Grant Number: 1 R03 MH084844-01 Assay Provider: Dr. John C. Reed, Sanford-Burnham Medical Research Institute, San Diego CA The modulation of immune response activity is one of the major goals in the development of novel therapeutics for auto-immune and inflammatory diseases. The innate system resides at the intersection of the pathways of microbial recognition, inflammation, and cell death, thereby offering various therapeutic targets. In this context, NOD1 and NOD2 are of particular interest, since they recognize distinct structures derived from bacterial peptidoglycans and directly activate NF-kB, a central regulator of immune response, inflammation, and apoptosis. Mutations in the NOD1 and NOD2 genes are associated with a number of human inflammatory diso SAR analysis of NF-kappaB dependent luciferase using DAP as an inducer - Set 2 Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Probe Production Centers Network (MLPCN) Grant Number: 1 R03 MH084844-01 Assay Provider: Dr. John C. Reed, Sanford-Burnham Medical Research Institute, San Diego CA The modulation of immune response activity is one of the major goals in the development of novel therapeutics for auto-immune and inflammatory diseases. The innate system resides at the intersection of the pathways of microbial recognition, inflammation, and cell death, thereby offering various therapeutic targets. In this context, NOD1 and NOD2 are of particular interest, since they recognize distinct structures derived from bacterial peptidoglycans and directly activate NF-kB, a central regulator of immune response, inflammation, and apoptosis. Mutations in the NOD1 and NOD2 genes are associated with a number of human inflammatory diso SAR analysis of NF-kappaB dependent luciferase using Doxorucibin as an inducer - Set 2 Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego CA) Network: NIH Molecular Libraries Production Centers Network (MLPCN) Grant Number: 1 R03 MH084844-01 Assay Provider: Dr. John C. Reed, Sanford-Burnham Medical Research Institute, San Diego CA The modulation of immune response activity is one of the major goals in the development of novel therapeutics for auto-immune and inflammatory diseases. The innate system resides at the intersection of the pathways of microbial recognition, inflammation, and cell death, thereby offering various therapeutic targets. In this context, NOD1 and NOD2 are of particular interest, since they recognize distinct structures derived from bacterial peptidoglycans and directly activate NF-kB, a central regulator of immune response, inflammation, and apoptosis. Mutations in the NOD1 and NOD2 genes are associated with a number of human inflammatory disorders, SAR analysis of NF-kappaB dependent luciferase using PMA/Ionomycin as an inducer - 2 Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Probe Production Centers Network (MLPCN) Grant Number: 1 R03 MH084844-01 Assay Provider: Dr. John C. Reed, Sanford-Burnham Medical Research Institute, San Diego CA The modulation of immune response activity is one of the major goals in the development of novel therapeutics for auto-immune and inflammatory diseases. The innate system resides at the intersection of the pathways of microbial recognition, inflammation, and cell death, thereby offering various therapeutic targets. In this context, NOD1 and NOD2 are of particular interest, since they recognize distinct structures derived from bacterial peptidoglycans and directly activate NF-kB, a central regulator of immune response, inflammation, and apoptosis. NF-kB pathway activated by antigen receptors is critical for acquired (as opposed to innate) SAR analysis of Tumor necrosis factor alpha (TNF-alpha) induced IL-8 secretion in MCF-7/NOD1 cells - Set 2 Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Screening Centers Network (MLSCN) Grant Number: 1 R03 MH084844-01 Assay Provider: Dr. John C. Reed, Sanford-Burnham Medical Research Institute The modulation of immune response activity is one of the major goals in the development of novel therapeutics for auto-immune and inflammatory diseases. The innate system resides at the intersection of the pathways of microbial recognition, inflammation, and cell death, thereby offering various therapeutic targets. In this context, NOD1 and NOD2 are of particular interest, since they recognize distinct structures derived from bacterial peptidoglycans and directly activate NF-kB, a central regulator of immune response, inflammation, and apoptosis. The NOD proteins participate in the signaling events triggered by host recognition of specific motifs (mostly, murope SAR analysis of agonists of the Cannabinoid Receptor 2 using an Image-Based Assay - Set 2 Data Source: Sanford-Burnham Center for Chemical Genomics Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Production Centers Network (MLPCN) Grant Number: 1X01 DA026205-01 Assay Provider: Dr. Mary Abood, Temple University, formerly at California Pacific Medical Center Research Institute Drug addiction is a disease that involves the G-protein coupled receptors in the central nervous system resulting in compulsive or abnormal behavior. Recent studies have shown that opioid receptors play a role in regulating other receptors that interact with drug and other substance abuse. The cannabinoid receptors (type 1 and 2) are members of the G-protein coupled receptor family and have been found to be involved in alterations in mood and cognition, as experienced by marijuana users. The specific aim of this assay is to identify small molecule agonists of the human cannabinoid receptor type 2 (CB2). This dose response assay SAR analysis of antagonists of the Cannabinoid Receptor 1 using an Image-Based Assay - Set 3 Data Source: Sanford-Burnham Center for Chemical Genomics Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Production Centers Network (MLPCN) Grant Number: 1X01 DA026205-01 Assay Provider: Dr. Mary Abood, Temple University, formerly at California Pacific Medical Center Research Institute Drug addiction is a disease that involves the G-protein coupled receptors in the central nervous system resulting in compulsive or abnormal behavior. Recent studies have shown that opioid receptors play a role in regulating other receptors that interact with drug and other substance abuse. The cannabinoid receptors (type 1 and 2) are members of the G-protein coupled receptor family and have been found to be involved in alterations in mood and cognition, as experienced by marijuana users. The specific aim of this assay is to identify small molecule antagonists of the human cannabinoid receptor type 1 (CB1). This dose response a SAR analysis of antagonists of the Cannabinoid Receptor 1 using an Image-Based Assay - Set 4 Data Source: Sanford-Burnham Center for Chemical Genomics Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego) Network: NIH Molecular Libraries Probe Production Centers Network (MLPCN) Grant Number: 1X01 DA026205-01 Assay Provider: Dr. Mary Abood, Temple University, formerly at California Pacific Medical Center Research Institute Drug addiction is a disease that involves the G-protein coupled receptors in the central nervous system resulting in compulsive or abnormal behavior. Recent studies have shown that opioid receptors play a role in regulating other receptors that interact with drug and other substance abuse. The cannabinoid receptors (type 1 and 2) are members of the G-protein coupled receptor family and have been found to be involved in alterations in mood and cognition, as experienced by marijuana users. The specific aim of this assay is to identify small molecule antagonists of the human cannabinoid receptor type 1 (CB1). The dose response ass SAR analysis of antagonists of the Cannabinoid Receptor 2 using an Image-Based Assay - Set 2 Data Source: Sanford-Burnham Center for Chemical Genomics Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Production Centers Network (MLPCN) Grant Number: 1X01 DA026205-01 Assay Provider: Dr. Mary Abood, Temple University, formerly at California Pacific Medical Center Research Institute Drug addiction is a disease that involves the G-protein coupled receptors in the central nervous system resulting in compulsive or abnormal behavior. Recent studies have shown that opioid receptors play a role in regulating other receptors that interact with drug and other substance abuse. The cannabinoid receptors (type 1 and 2) are members of the G-protein coupled receptor family and have been found to be involved in alterations in mood and cognition, as experienced by marijuana users. The specific aim of this assay is to identify small molecule antagonists of the human cannabinoid receptor type 2 (CB2). The dose response ass SAR analysis of compounds that inhibit Human Immunodeficiency Virus Fusion. Data Source: Burnham Center for Chemical Genomics (BCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Production Centers Network (MLPCN) Grant Number: 1R21NS059403-01 Assay Provider: Dr. Miriam Gochin, Touro University-California, Vallejo, CA The fusion-active conformation of the envelope protein gp41 of HIV-1 consists of an N-terminal trimeric a-helical coiled coil domain, and three anti-parallel C-terminal helices which fold down the grooves of the coiled coil to form a six-helix bundle. Disruption of the six-helix bundle is considered to be a key component of an effective non-peptide fusion inhibitor. This structure forms as a result of a conformational change in gp41, triggered by gp120 and co-receptor binding to host cell receptors. Prevention of six-helix bundle formation has been recognized as an important mechanism for viral fusion inhibition A metallopeptide-based fluorescence assay has been develope SAR analysis of compounds that inhibit NOD1 - Set 3 Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Probe Production Centers Network (MLPCN) Grant Number: 1 R03 MH084844-01 Assay Provider: Dr. John C. Reed, Sanford-Burnham Medical Research Institute, San Diego CA The modulation of immune response activity is one of the major goals in the development of novel therapeutics for auto-immune and inflammatory diseases. The innate system resides at the intersection of the pathways of microbial recognition, inflammation, and cell death, thereby offering various therapeutic targets. In this context, NOD1 and NOD2 are of particular interest, since they recognize distinct structures derived from bacterial peptidoglycans and directly activate NF-kB, a central regulator of immune response, inflammation, and apoptosis. Mutations in the NOD1 and NOD2 genes are associated with a number of human inflammatory diso SAR analysis of compounds that inhibit NOD1 revised Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Probe Production Centers Network (MLPCN) Grant Number: 1 R03 MH084844-01 Assay Provider: Dr. John C. Reed, Sanford-Burnham Medical Research Institute, San Diego CA The modulation of immune response activity is one of the major goals in the development of novel therapeutics for auto-immune and inflammatory diseases. The innate system resides at the intersection of the pathways of microbial recognition, inflammation, and cell death, thereby offering various therapeutic targets. In this context, NOD1 and NOD2 are of particular interest, since they recognize distinct structures derived from bacterial peptidoglycans and directly activate NF-kB, a central regulator of immune response, inflammation, and apoptosis. Mutations in the NOD1 and NOD2 genes are associated with a number of human inflammatory diso SAR analysis of compounds that inhibit NOD2 - Set 3 Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Probe Production Centers Network (MLPCN) Grant Number: 1 R03 MH084844-01 Assay Provider: Dr. John C. Reed, Sanford-Burnham Medical Research Institute, San Diego CA The modulation of immune response activity is one of the major goals in the development of novel therapeutics for auto-immune and inflammatory diseases. The innate system resides at the intersection of the pathways of microbial recognition, inflammation, and cell death, thereby offering various therapeutic targets. In this context, NOD1 and NOD2 are of particular interest, since they recognize distinct structures derived from bacterial peptidoglycans and directly activate NF-kB, a central regulator of immune response, inflammation, and apoptosis. Mutations in the NOD1 and NOD2 genes are associated with a number of human inflammatory disor SAR analysis of compounds that inhibit NOD2 revised Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Probe Production Centers Network (MLPCN) Grant Number: 1 R03 MH084844-01 Assay Provider: Dr. John C. Reed, Sanford-Burnham Medical Research Institute, San Diego CA The modulation of immune response activity is one of the major goals in the development of novel therapeutics for auto-immune and inflammatory diseases. The innate system resides at the intersection of the pathways of microbial recognition, inflammation, and cell death, thereby offering various therapeutic targets. In this context, NOD1 and NOD2 are of particular interest, since they recognize distinct structures derived from bacterial peptidoglycans and directly activate NF-kB, a central regulator of immune response, inflammation, and apoptosis. Mutations in the NOD1 and NOD2 genes are associated with a number of human inflammatory disor SAR analysis of compounds that potentiate TRAIL-induced apoptosis in MDA-MB-435 cells. Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Production Centers Network (MLPCN) Grant Number: X01 MH083230-01 Assay Provider: Dr. Dmitri Rozanov, Sanford-Burnham Medical Research Institute, San Diego CA This assay was developed and performed to confirm hits originally identified in "uHTS for the identification of compounds that potentiate TRAIL-induced apoptosis of cancer cells" (AID 1443) and to study the structure-activity relationship on analogs of the confirmed hits. Compounds are either acquired from commercial sources or synthesized internally. The TRAIL-resistant cell line, MDA-MB-435 is used, because we would like to determine if compounds can potentiate TRAIL-mediated cytotoxicity not only in TRAIL-sensitive PPC-1 carcinoma cells(AIDs 1443 and 1624) but also in TRAIL-resistant cells. Cytotoxic chemotherapy induces apoptosis via a pat SAR analysis of inhibitors of TNFa specific NF-kB induction - Set 3 Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Probe Production Centers Network (MLPCN) Grant Number: 1 R03 MH084844-01 Assay Provider: Dr. John C. Reed, Sanford-Burnham Medical Research Institute, San Diego CA Multiple cellular stimuli acting through various pathways lead to NF-kB induction. The assay described below uses tumor necrosis factor alpha (TNFa), a canonical NF-kB inducer, and is designed for identification of hits specific to TNFa-modulated pathways. We utilized this assay to assess selectivity of hits emerging from the primary screening of the library in NOD1- and NOD2-specific assays (AIDs 1578 and 1566). The HEK-293-T NF-kB-Luc cell line designed for luminescent detection of NF-kB induction was utilized in this assay. This dose response assay is developed and performed to confirm hits originally identified in "uHTS luminescence a SAR analysis of inhibitors of TNFa specific NF-kB induction revised Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Production Centers Network (MLPCN) Grant Number: 1 R03 MH084844-01 Assay Provider: Dr. John C. Reed, Sanford-Burnham Medical Research Institute, San Diego CA Multiple cellular stimuli acting through various pathways lead to NF-kB induction. The assay described below uses tumor necrosis factor alpha (TNFa), a canonical NF-kB inducer, and is designed for identification of hits specific to TNFa-modulated pathways. We utilized this assay to assess selectivity of hits emerging from the primary screening of the library in NOD1- and NOD2-specific assays (AIDs 1578 and 1566). The HEK-293-T NF-kB-Luc cell line designed for luminescent detection of NF-kB induction was utilized in this assay. This dose response assay is developed and performed to confirm hits originally identified in "uHTS luminescence assay f SAR analysis of small molecule agonists of the kappa opioid receptor via a luminescent beta-arrestin assay - Set 2 Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Production Centers Network (MLPCN) Grant Number: 1X01DA026208-01 Assay Provider: Dr. Lawrance Barak , Duke University, Durham NC Drug addiction is a disease originating in the central nervous system that produces compulsive behaviors despite the negative consequences that may result. Major addictive drugs of abuse include components of tobacco, opiates, marijuana, ethanol, cocaine, and derivatives of amphetamines. While the addictive behaviors produced by these substances may be generally similar, the drugs act at different receptor sites in the brain. Recent studies have shown that opioid receptors play a role regulating the addictive behaviors of other receptors that interact with illicit and legal substances of abuse. Opioid receptors are composed of multiple subtypes whose contributions to addicti SAR analysis of small molecule agonists of the kappa opioid receptor via a luminescent beta-arrestin assay - Set 3 Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Production Centers Network (MLPCN) Grant Number: 1X01DA026208-01 Assay Provider: Dr. Lawrance Barak , Duke University, Durham NC Drug addiction is a disease originating in the central nervous system that produces compulsive behaviors despite the negative consequences that may result. Major addictive drugs of abuse include components of tobacco, opiates, marijuana, ethanol, cocaine, and derivatives of amphetamines. While the addictive behaviors produced by these substances may be generally similar, the drugs act at different receptor sites in the brain. Recent studies have shown that opioid receptors play a role regulating the addictive behaviors of other receptors that interact with illicit and legal substances of abuse. Opioid receptors are composed of multiple subtypes whose contributions to addicti SAR analysis of small molecule agonists of the kappa opioid receptor via a luminescent beta-arrestin assay Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Production Centers Network (MLPCN) Grant Number: 1X01DA026208-01 Assay Provider: Dr. Lawrance Barak , Duke University, Durham NC Drug addiction is a disease originating in the central nervous system that produces compulsive behaviors despite the negative consequences that may result. Major addictive drugs of abuse include components of tobacco, opiates, marijuana, ethanol, cocaine, and derivatives of amphetamines. While the addictive behaviors produced by these substances may be generally similar, the drugs act at different receptor sites in the brain. Recent studies have shown that opioid receptors play a role regulating the addictive behaviors of other receptors that interact with illicit and legal substances of abuse. Opioid receptors are composed of multiple subtypes whose contributions to addicti SAR analysis of small molecule antagonists of the kappa opioid receptor via a luminescent beta-arrestin assay - Set 3 Data Source: Burnham Center for Chemical Genomics (BCCG) Source Affiliation: Burnham Institute for Medical Research (BIMR, La Jolla, CA) Network: NIH Molecular Libraries Probe Production Centers Network (MLPCN) Grant Number: 1X01DA026208-01 Assay Provider: Dr. Lawrance Barak , Duke University, Durham NC Drug addiction is a disease originating in the central nervous system that produces compulsive behaviors despite the negative consequences that may result. Major addictive drugs of abuse include components of tobacco, opiates, marijuana, ethanol, cocaine, and derivatives of amphetamines. While the addictive behaviors produced by these substances may be generally similar, the drugs act at different receptor sites in the brain. Recent studies have shown that opioid receptors play a role regulating the addictive behaviors of other receptors that interact with illicit and legal substances of abuse. Opioid receptors are composed of multiple subtypes whose contributions to addictive behavio SAR analysis of small molecule antagonists of the kappa opioid receptor via a luminescent beta-arrestin assay - Set 4 Data Source: Burnham Center for Chemical Genomics (BCCG) Source Affiliation: Burnham Institute for Medical Research (BIMR, La Jolla, CA) Network: NIH Molecular Libraries Probe Production Centers Network (MLPCN) Grant Number: 1X01DA026208-01 Assay Provider: Dr. Lawrance Barak , Duke University, Durham NC Drug addiction is a disease originating in the central nervous system that produces compulsive behaviors despite the negative consequences that may result. Major addictive drugs of abuse include components of tobacco, opiates, marijuana, ethanol, cocaine, and derivatives of amphetamines. While the addictive behaviors produced by these substances may be generally similar, the drugs act at different receptor sites in the brain. Recent studies have shown that opioid receptors play a role regulating the addictive behaviors of other receptors that interact with illicit and legal substances of abuse. Opioid receptors are composed of multiple subtypes whose contributions to addictive behavio SAR analysis of small molecule antagonists of the kappa opioid receptor via a luminescent beta-arrestin assay - Set 5 Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Probe Production Centers Network (MLPCN) Grant Number: 1X01DA026208-01 Assay Provider: Dr. Lawrance Barak , Duke University, Durham NC Drug addiction is a disease originating in the central nervous system that produces compulsive behaviors despite the negative consequences that may result. Major addictive drugs of abuse include components of tobacco, opiates, marijuana, ethanol, cocaine, and derivatives of amphetamines. While the addictive behaviors produced by these substances may be generally similar, the drugs act at different receptor sites in the brain. Recent studies have shown that opioid receptors play a role regulating the addictive behaviors of other receptors that interact with illicit and legal substances of abuse. Opioid receptors are composed of multiple subtypes whose contributions to ad SAR analysis of small molecule antagonists of the kappa opioid receptor via a luminescent beta-arrestin assay - Set 6 Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Probe Production Centers Network(MLPCN)Grant Number: 1X01DA026208-01 Assay Provider: Dr. Lawrance Barak , Duke University, Durham NC Drug addiction is a disease originating in the central nervous system that produces compulsive behaviors despite the negative consequences that may result. Major addictive drugs of abuse include components of tobacco, opiates, marijuana, ethanol, cocaine, and derivatives of amphetamines. While the addictive behaviors produced by these substances may be generally similar, the drugs act at different receptor sites in the brain. Recent studies have shown that opioid receptors play a role regulating the addictive behaviors of other receptors that interact with illicit and legal substances of abuse. Opioid receptors are composed of multiple subtypes whose contributions to addi SAR analysis of small molecule antagonists of the kappa opioid receptor via a luminescent beta-arrestin assay Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Production Centers Network (MLPCN) Grant Number: 1X01DA026208-01 Assay Provider: Dr. Lawrance Barak , Duke University, Durham NC Drug addiction is a disease originating in the central nervous system that produces compulsive behaviors despite the negative consequences that may result. Major addictive drugs of abuse include components of tobacco, opiates, marijuana, ethanol, cocaine, and derivatives of amphetamines. While the addictive behaviors produced by these substances may be generally similar, the drugs act at different receptor sites in the brain. Recent studies have shown that opioid receptors play a role regulating the addictive behaviors of other receptors that interact with illicit and legal substances of abuse. Opioid receptors are composed of multiple subtypes whose contributions to addictiv SAR analysis of small molecule inhibitors of Mint-PDZ and N-type Ca2+ channel carboxyl-terminal peptide association using HTRF - Set 2 Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Production Centers Network (MLPCN) Grant Number: 1 R03 MH085675-01 Assay Provider: Dr Ilya Bezprozvanny, UT Southwestern Medical Center , Dallas, TX Chronic pain (neuropathic pain, inflammatory pain, cancer pain) is a major health problem. Opiate-based drugs, such as morphine and morphine derivatives, are the primary standard of care for the treatment of chronic pain. Unfortunately, patients develop tolerance to opiates due to desensitization of the opiate receptor. Thus, alternative anti-nociceptive ("pain killing") pathways need to be explored for treatment of chronic pain. The N-type voltage-gated Ca2+ channels (CaV2.2s) in dorsal root ganglia neurons is a well validated target for chronic pain (1, 2). We previously demonstrated the interaction between CaV2.2 and the first PDZ domain of molecula SAR analysis of small molecule inhibitors of Mint-PDZ and N-type Ca2+ channel carboxyl-terminal peptide association using HTRF Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Production Centers Network (MLPCN) Grant Number: 1 R03 MH085675-01 Assay Provider: Dr Ilya Bezprozvanny, UT Southwestern Medical Center , Dallas, TX Chronic pain (neuropathic pain, inflammatory pain, cancer pain) is a major health problem. Opiate-based drugs, such as morphine and morphine derivatives, are the primary standard of care for the treatment of chronic pain. Unfortunately, patients develop tolerance to opiates due to desensitization of the opiate receptor. Thus, alternative anti-nociceptive ("pain killing") pathways need to be explored for treatment of chronic pain. The N-type voltage-gated Ca2+ channels (CaV2.2s) in dorsal root ganglia neurons is a well validated target for chronic pain (1, 2). We previously demonstrated the interaction between CaV2.2 and the first PDZ domain of molecula SAR assay for compounds activating TNAP in the absence of phosphate acceptor performed in a luminescent assay Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Screening Centers Network (MLSCN) Grant Proposal Number: 1R03 MH082385-01 This TNAP dose response assay is developed and performed to confirm hits originally identified in the TNAP luminescent HTS assay (AID 1001) and to study the structure-activity relationship on analogs of the confirmed hits. Compounds are either acquired from commercial sources or synthesized internally. Alkaline phosphatases (EC 3.1.3.1) (APs) catalyze the hydrolysis of phosphomonoesters, releasing phosphate and alcohol. APs are dimeric enzymes found in the most organisms. In human, four isozymes of APs have been identified. Three isozymes are tissue-specific and the fourth one is tissue-non specific, named TNAP. TNAP deficiency is associated with defective bone mineralization in the form of rickets and osteomalacia. Therefore, t SAR assay for compounds activating TNAP in the presence of 100 mM DEA performed in a luminescence assay Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Screening Centers Network (MLSCN) Grant Proposal Number: 1R03 MH082385-01 This TNAP dose response assay is developed and performed to confirm hits originally identified in the TNAP luminescent HTS assay (AID 1001) and to study the structure-activity relationship on analogs of the confirmed hits. Compounds are either acquired from commercial sources or synthesized internally. Alkaline phosphatases (EC 3.1.3.1) (APs) catalyze the hydrolysis of phosphomonoesters, releasing phosphate and alcohol. APs are dimeric enzymes found in the most organisms. In human, four isozymes of APs have been identified. Three isozymes are tissue-specific and the fourth one is tissue-non specific, named TNAP. TNAP deficiency is associated with defective bone mineralization in the form of rickets and osteomalacia. Therefore, t SAR assay for compounds inhibiting TNAP in the absence of phosphate acceptor performed in a luminescent assay Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Screening Centers Network (MLSCN) Grant Number: MH077602-01 Assay Provider: Dr. Jose Luis Millan, Sanford-Burnham Medical Research Institute, San Diego, CA This TNAP dose response assay is developed and performed for the purpose of SAR study on analogs of hits originally identified in the TNAP luminescent HTS assay (AID 518) Alkaline phosphatase (EC 3.1.3.1) (APs) catalyze the hydrolysis of phosphomonoesters, releasing phosphate and alcohol. APs are dimeric enzymes found in the most organism. In human, four isozymes of APs have been identified. Three isozymes are tissue-specific and the fourth one is tissue-nonsepecifc, named TNAP. TNAP overexpression is associated with excessive calcification observed in different tissues. Therefore, there are therapeutic potentials of in SAR assay for compounds that inhibit PHOSPHO1 Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Probe Production Centers Network (MLPCN) Grant Number: 1 R03 MH084086-01 Assay Provider: Dr. Jose Luis Milan, Sanford-Burnham Medical Research Institute, San Diego CA Mineralization of cartilage and bone occurs by a series of physicochemical and biochemical processes that together facilitate the deposition of hydroxyapatite (HA) in specific areas of the extracellular matrix (ECM). Experimental evidence has pointed to the presence of HA crystals along collagen fibrils in the ECM and also within the lumen of chondroblast- and osteoblast-derived matrix vesicles (MVs). Dr. Milan's working model is that bone mineralization is first initiated within the lumen of MVs. In a second step, HA crystals grow beyond the confines of the MVs and become exposed to the extracellular milieu where they continue to TR-FRET secondary assay for HTS discovery of chemical inhibitors of Hsp70 Sanford-Burnham Center for Chemical Genomics (SBCCG) Sanford-Burnham Medical Research Institute (San Diego, CA) NIH Molecular Libraries Screening Centers Network (MLSCN) MLSCN Grant: XO1 MH079863-01 Over-expression of molecular chaperones occurs commonly in cancers and provides protection from a wide variety of cellular stresses, both endogenous and iatrogenic. Molecular chaperones also play important roles in maintaining the activity of several signal-transducing proteins and transcriptions factors involved in malignant transformation. The human genome contains nine Hsp70-family genes. These chaperones include Hsp70 and Hsc70, which are commonly over-expressed in cancers and which confer resistance to myriad cellular stresses, including cytotoxic chemotherapy. The current assay was developed at the Sanford-Burnham Center for Chemical Genomics (SBCCG), based on Fluorescen-12-ATP binding to GST-Hsp70 in the presence of Terbium-labeled anti-GST antibody. The assay is aimed to support TR-FRET-based biochemical high throughput dose response assay for agonists of nuclear receptor subfamily 2, group E, member 3 (NR2E3) Source (MLPCN Center Name): The Scripps Research Institute Molecular Screening Center Affiliation: The Scripps Research Institute, TSRI Assay Provider: Konstantin Petrukhin, Columbia University Network: Molecular Library Probe Production Centers Network (MLPCN) Grant Proposal Number: 1 R21 NS061718-01 Fast Track Grant Proposal PI: Konstantin Petrukhin, Columbia University External Assay ID: NR2E3_AG_TR-FRET_1536_3XIC50 Name: TR-FRET-based biochemical high throughput dose response assay for agonists of nuclear receptor subfamily 2, group E, member 3 (NR2E3). Description: Nuclear receptors are small molecule- and hormone-regulated transcription factors with discrete DNA-binding and ligand-binding domains, and are essential during development and for maintenance of proper cell function in adults. Small pharmacological compounds that bind to the cleft of the ligand-binding domain could alter receptor conformation and subsequently modify transcription of target genes. Such ligands (agon TRFRET-based biochemical high throughput dose response assay to identify inhibitors of 5-meCpG-binding domain protein 2 (MBD2)-DBD binding to methylated oligonucleotide Source (MLPCN Center Name): The Scripps Research Institute Molecular Screening Center Affiliation: The Scripps Research Institute, TSRI Assay Provider: Bill Nelson Network: Molecular Library Probe Production Centers Network (MLPCN) Grant Proposal Number: 1 R03 MH098712-01 Grant Proposal PI: Bill Nelson External Assay ID: MBD2-CPGDNA_INH_TRFRET_1536_3XIC50 DRUN Name: TRFRET-based biochemical high throughput dose response assay to identify inhibitors of 5-meCpG-binding domain protein 2 (MBD2)-DBD binding to methylated oligonucleotide. Description: Of all the somatic genome changes that accumulate during the pathogenesis of human cancers, only changes in DNA methylation appear to occur consistently (virtually all cases), to arise early (first appearing in preneoplastic lesions), and to be potentially reversible (the DNA sequence remains intact) (1-4). One such change in DNA methylation, increased CpG dinucleotide methylation at CpG islands encompassing the transcriptional regulatory r TRFRET-based cell-based high throughput dose response assay for biased ligands (antagonists) of the melanocortin 4 receptor (MC4R) Source (MLPCN Center Name): The Scripps Research Institute Molecular Screening Center (SRISMC) Center Affiliation: The Scripps Research Institute, TSRI Assay Provider: Scott DeWire, Trevena Inc Network: Molecular Library Probe Production Center Network (MLPCN) Grant Proposal Number: 1 RC2 MH090877-01 Grant Proposal PI: Scott DeWire, Trevena Inc External Assay ID: MC4R_ANT_TRFRET_1536_3XIC50 DRUN Name: TRFRET-based cell-based high throughput dose response assay for biased ligands (antagonists) of the melanocortin 4 receptor (MC4R). Description: Heterotrimeric G-protein coupled receptors (GPCRs) are major targets for disease therapeutics, due in part to their broad tissue distribution, structural diversity, varied modes of action, and disease-associated mutations (1-4). However, it has recently been demonstrated that GPCRs do not only signal in this simplistic fashion, but rather activate a network of downstream effects comprised of parallel signal transduction pathways. GPCR ligands uHTS HTRF assay for identification of inhibitors of SUMOylation Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Probe Production Centers Network (MLPCN) Grant Number: 1 R03 MH084862-01 Assay Provider: Dr. Yuan Chen, Beckman Research Institute, City Of Hope, CA Protein modification by the SUMO (Small Ubiquitin-like MOdifier) family of proteins is an important post-translational modification that plays an essential role in many functions including gene transcription, cell cycle progression, DNA repair, viral infection, and the development of neurodegenerative diseases (1, 2). Recent proteomic studies have found that approximately 10% of the proteins encoded by the yeast genome are substrates for SUMO modification (3-5). The mechanism of how SUMOylation is involved in these cellular functions remains largely unclear. The inhibitors of SUMOylation would be useful to probe the roles of SUMOylation in cellular regulati uHTS Homogeneous Terbium Time-Resolved Fluorescence Resonance Energy Transfer (HTRF) Assay Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Probe Production Centers Network (MLPCN) Grant Number: 1 R03 MH085675-01 Assay Provider: Dr Ilya Bezprozvanny, UT Southwestern Medical Center , Dallas, TX Chronic pain (neuropathic pain, inflammatory pain, cancer pain) is a major health problem. Opiate-based drugs, such as morphine and morphine derivatives, are the primary standard of care for the treatment of chronic pain. Unfortunately, patients develop tolerance to opiates due to desensitization of the opiate receptor. Thus, alternative anti-nociceptive ("pain killing") pathways need to be explored for treatment of chronic pain. The N-type voltage-gated Ca2+ channels (CaV2.2s) in dorsal root ganglia neurons is a well validated target for chronic pain (1, 2). We previously demonstrated the interaction between CaV2.2 and the first PDZ domain of mo uHTS identification of compounds activating TNAP in the absence of phosphate acceptor performed in luminescent assay Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Screening Centers Network (MLSCN) Grant Proposal Number: 1R03 MH082385-01 Alkaline phosphatases (EC 3.1.3.1) (APs) catalyze the hydrolysis of phosphomonoesters, releasing phosphate and alcohol. APs are dimeric enzymes found in the most organisms. In human, four isozymes of APs have been identified. Three isozymes are tissue-specific and the fourth one is tissue-non specific, named TNAP. TNAP deficiency is associated with defective bone mineralization in the form of rickets and osteomalacia. Therefore, there is therapeutic potential of modulating TNAP activity. The goal of this HTS is to identify novel and specific activators of TNAP. The only known to date class of alkaline phosphatases activators are amino-containing alcohols, such as diethanolamine (DEA), that act as phosphoacceptor substrate and exh uHTS identification of small molecule antagonists of the kappa opioid receptor via a luminescent beta-arrestin assay Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Production Centers Network (MLPCN) Grant Number: 1X01DA026208-01 Assay Provider: Dr. Lawrance Barak , Duke University, Durham NC Drug addiction is a disease originating in the central nervous system that produces compulsive behaviors despite the negative consequences that may result. Major addictive drugs of abuse include components of tobacco, opiates, marijuana, ethanol, cocaine, and derivatives of amphetamines. While the addictive behaviors produced by these substances may be generally similar, the drugs act at different receptor sites in the brain. Recent studies have shown that opioid receptors play a role regulating the addictive behaviors of other receptors that interact with illicit and legal substances of abuse. Opioid receptors are composed of multiple subtypes whose contributions to addictiv uHTS identification of small molecule inhibitors of LYP via a fluorescence intensity assay Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Production Centers Network (MLPCN) Grant Number: 1R21NS056945-01 Assay Provider: Dr. Nunzio Bottini, San Diego Institute for Allergy and Immunology CA LYP, is a lymphocyte specific protein tyrosine phosphatase that plays a critical regulatory role in T cell receptor signaling. The PTPN22 gene encodes this phosphatase. A single-nucleotide polymorphism in PTPN22 is associated with a number of autoimmune disorders, including type 1 diabetes, rheumatoid arthritis, juvenile rheumatoid arthritis, systemic lupus erythematosus and Grave's disease. The autoimmunity-predisposing allele is a gain-of-function mutant suggesting that a specific small-molecule inhibitor could eliminate its effect. Finding specific inhibitors of protein phosphatases has proven extremely difficult. The goal of this project is to f uHTS luminescence assay for the identification of compounds that inhibit NOD1 Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Probe Production Centers Network (MLPCN) Grant Number: 1 R03 MH084844-01 Assay Provider: Dr. John C. Reed, Sanford-Burnham Medical Research Institute, San Diego CA The modulation of immune response activity is one of the major goals in the development of novel therapeutics for auto-immune and inflammatory diseases. The innate system resides at the intersection of the pathways of microbial recognition, inflammation, and cell death, thereby offering various therapeutic targets. In this context, NOD1 and NOD2 are of particular interest, since they recognize distinct structures derived from bacterial peptidoglycans and directly activate NF-kB, a central regulator of immune response, inflammation, and apoptosis. Mutations in the NOD1 and NOD2 genes are associated with a number of human inflammatory disor uHTS luminescence assay for the identification of compounds that inhibit NOD2 in MDP treated cells Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, CA) Network: NIH Molecular Libraries Probe Production Centers Network (MLPCN) Grant Number: 1 R03 MH084844-01 Assay Provider: Dr. John C. Reed, Sanford-Burnham Medical Research Institute, San Diego, CA The modulation of immune response activity is one of the major goals in the development of novel therapeutics for auto-immune and inflammatory diseases. The innate system resides at the intersection of the pathways of microbial recognition, inflammation, and cell death, thereby offering various therapeutic targets. In this context, NOD1 and NOD2 are of particular interest, since they recognize distinct structures derived from bacterial peptidoglycans and directly activate NF-kB, a central regulator of immune response, inflammation, and apoptosis. Mutations in the NOD1 and NOD2 genes are associated with a number of human inflammatory dis
 BDBM634329 US20230357271, Example a-01-01
BDBM634329 US20230357271, Example a-01-01 BDBM647579 US20240025893, Example e-01-01
BDBM647579 US20240025893, Example e-01-01 US20240025893, Example b-01-01 BDBM647381
US20240025893, Example b-01-01 BDBM647381 US20240025893, Example c-01-01 BDBM647534
US20240025893, Example c-01-01 BDBM647534 US20240025893, Example d-01-01 BDBM647558
US20240025893, Example d-01-01 BDBM647558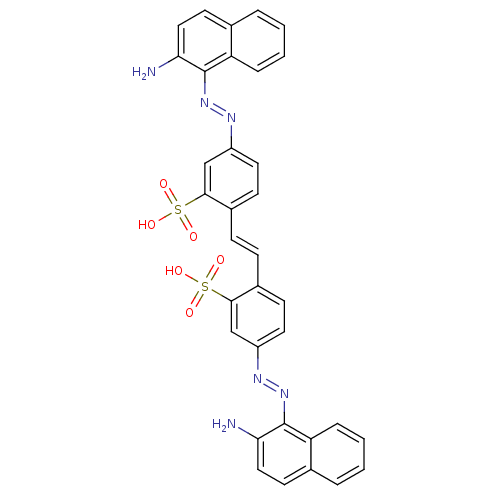 US11041859, Code SPI-01 US9791447, Compound SPI-01 BDBM103826 SPI-01
US11041859, Code SPI-01 US9791447, Compound SPI-01 BDBM103826 SPI-01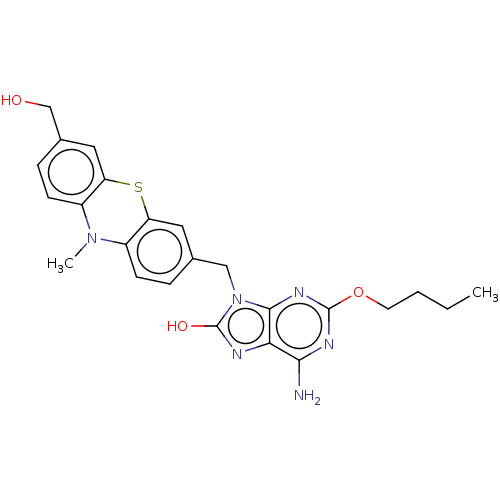 US10457681, Compound Ia-01 US10981914, Compound Ia-01 BDBM419193 US10487084, Compound Ia-01
US10457681, Compound Ia-01 US10981914, Compound Ia-01 BDBM419193 US10487084, Compound Ia-01 BDBM476173 US11364220, Compound 01 US10874634, Cmpd No. 01
BDBM476173 US11364220, Compound 01 US10874634, Cmpd No. 01 BDBM111832 US8623906, JHE-01-129B US9174969, JHE-01-129B
BDBM111832 US8623906, JHE-01-129B US9174969, JHE-01-129B BDBM111889 US9174969, JHE-01-134A US8623906, JHE-01-134A
BDBM111889 US9174969, JHE-01-134A US8623906, JHE-01-134A US8623906, JHE-01-155B US9174969, JHE-01-155B BDBM111860
US8623906, JHE-01-155B US9174969, JHE-01-155B BDBM111860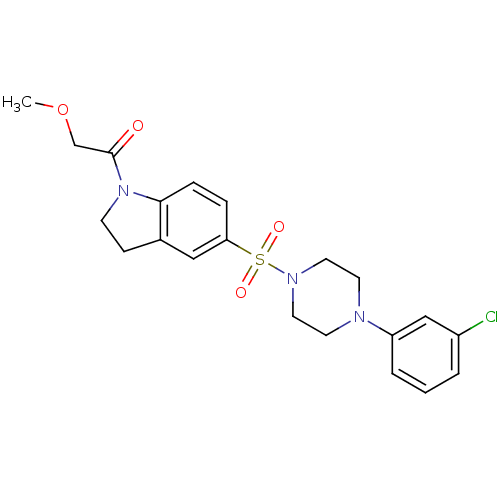 US8623906, JHE-01-169 US9174969, JHE-01-169 BDBM111881
US8623906, JHE-01-169 US9174969, JHE-01-169 BDBM111881 US9174969, JHE-01-137 US8623906, JHE-01-137 BDBM111837
US9174969, JHE-01-137 US8623906, JHE-01-137 BDBM111837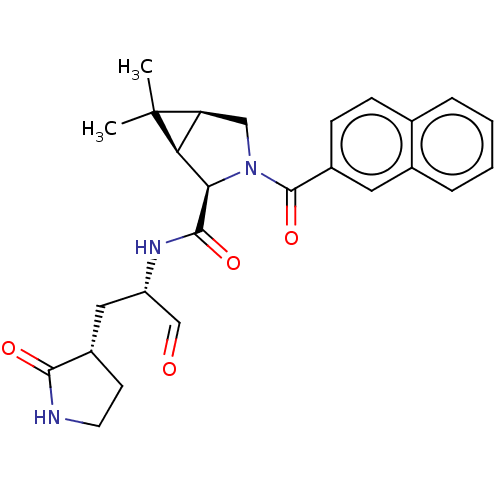 BDBM448224 MI-01
BDBM448224 MI-01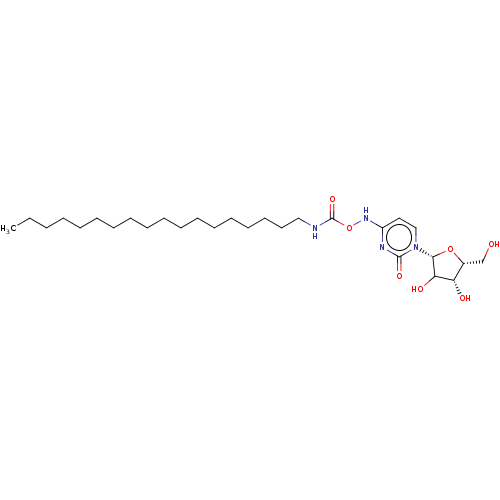 BDBM476315 US10874683, Example EIDD-02357-01 US10874683, Example EIDD-02474-01
BDBM476315 US10874683, Example EIDD-02357-01 US10874683, Example EIDD-02474-01 BDBM634493 US20230357271, Example a-01-26 US20230357271, Example a-01-05
BDBM634493 US20230357271, Example a-01-26 US20230357271, Example a-01-05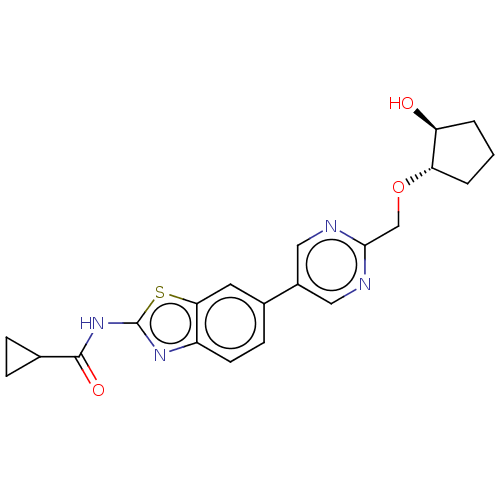 BDBM647362 US20240025893, Example a-01-01 US20240025893, Example a-04-02
BDBM647362 US20240025893, Example a-01-01 US20240025893, Example a-04-02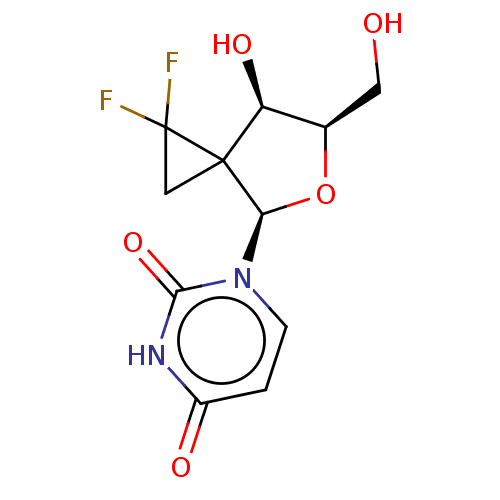 US10874683, Example EIDD-02339-01 BDBM476312 US10874683, Example EIDD-02340-01
US10874683, Example EIDD-02339-01 BDBM476312 US10874683, Example EIDD-02340-01 US20230357271, Example a-01-19 US20230357271, Example a-01-41 BDBM634521
US20230357271, Example a-01-19 US20230357271, Example a-01-41 BDBM634521 AB00980327-01 cid_3370791 BDBM200240
AB00980327-01 cid_3370791 BDBM200240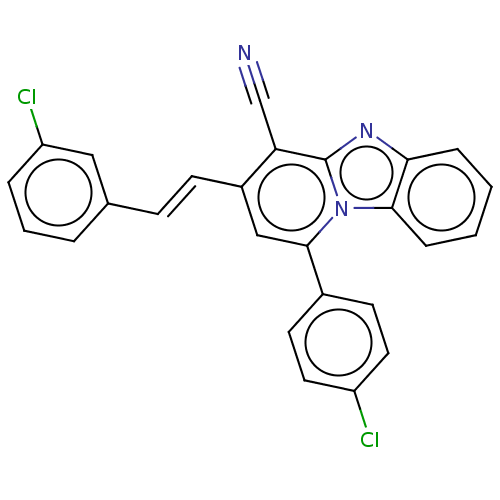 AB00980391-01 cid_5829212 BDBM200241
AB00980391-01 cid_5829212 BDBM200241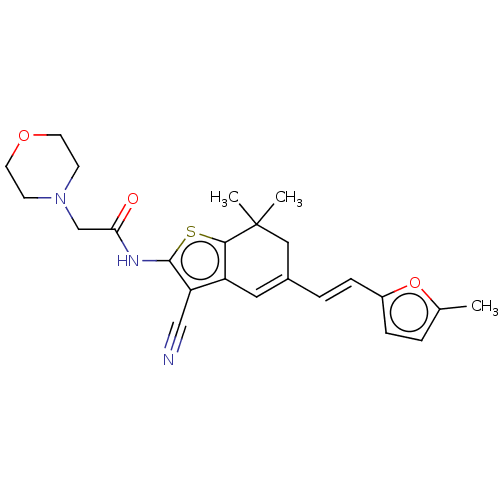 AB00984190-01 BDBM200243 cid_16377116
AB00984190-01 BDBM200243 cid_16377116 AB00984490-01 cid_73673797 BDBM200244
AB00984490-01 cid_73673797 BDBM200244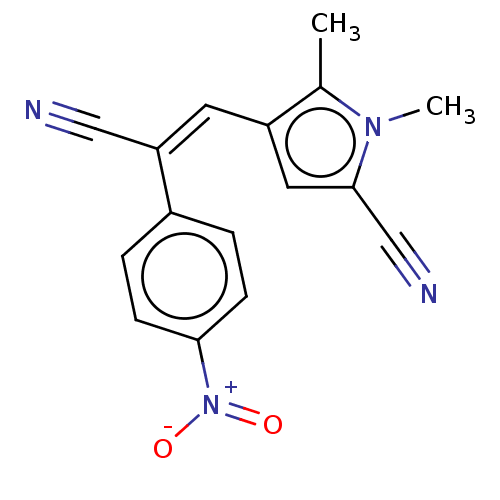 AB00984491-01 BDBM200245 cid_73673798
AB00984491-01 BDBM200245 cid_73673798 AB00984504-01 cid_17399291 BDBM200242
AB00984504-01 cid_17399291 BDBM200242 AB00985077-01 BDBM200267 cid_17551481
AB00985077-01 BDBM200267 cid_17551481 AB00985143-01 BDBM200269 cid_17549910
AB00985143-01 BDBM200269 cid_17549910 AB00985144-01 BDBM200270 cid_17549911
AB00985144-01 BDBM200270 cid_17549911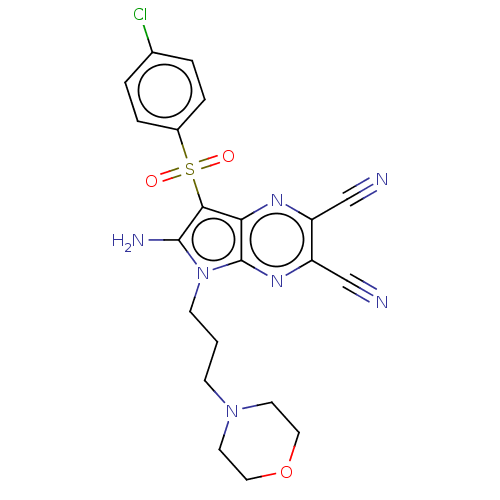 AB00985146-01 cid_17549916 BDBM200268
AB00985146-01 cid_17549916 BDBM200268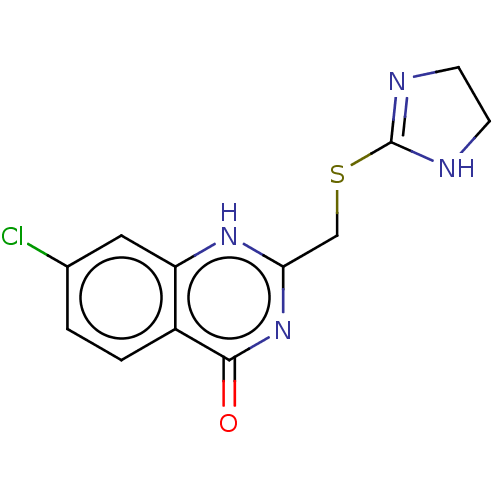 AB00986286-01 cid_17417823 BDBM200265
AB00986286-01 cid_17417823 BDBM200265 AB00987484-01 BDBM200258 cid_73673950
AB00987484-01 BDBM200258 cid_73673950 AB00988196-01 cid_17449592 BDBM200262
AB00988196-01 cid_17449592 BDBM200262 AB00988331-01 BDBM200263 cid_73673967
AB00988331-01 BDBM200263 cid_73673967 AB00988524-01 cid_25529738 BDBM200257
AB00988524-01 cid_25529738 BDBM200257 AB00998676-01 BDBM200254 cid_42923401
AB00998676-01 BDBM200254 cid_42923401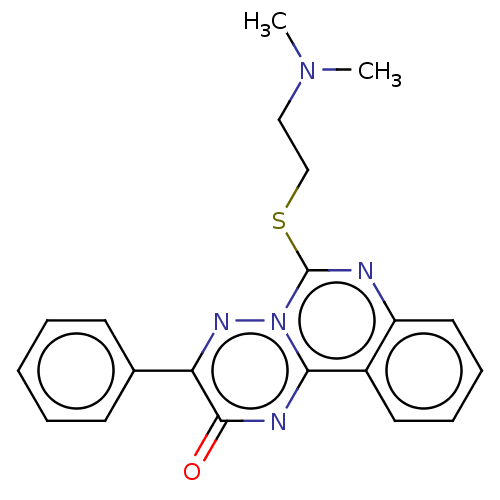 AB01000690-01 cid_45917772 BDBM200252
AB01000690-01 cid_45917772 BDBM200252 AB01002383-01 BDBM200286 cid_40135047
AB01002383-01 BDBM200286 cid_40135047 AB01002949-01 BDBM200281 cid_31583282
AB01002949-01 BDBM200281 cid_31583282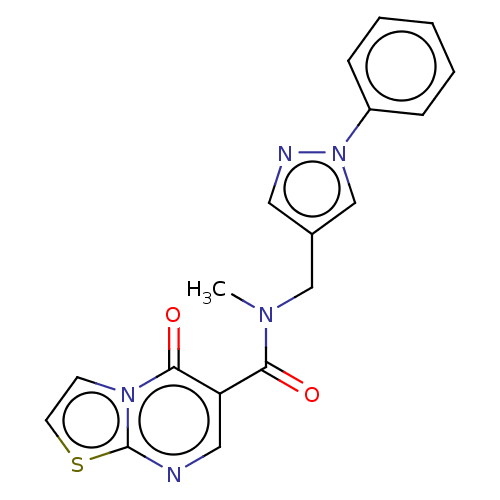 AB01003053-01 BDBM200277 cid_42920332
AB01003053-01 BDBM200277 cid_42920332 AB01003479-01 cid_45913338 BDBM200232
AB01003479-01 cid_45913338 BDBM200232 AB01003538-01 BDBM200288 cid_45896766
AB01003538-01 BDBM200288 cid_45896766 AB01003563-01 cid_45894740 BDBM200285
AB01003563-01 cid_45894740 BDBM200285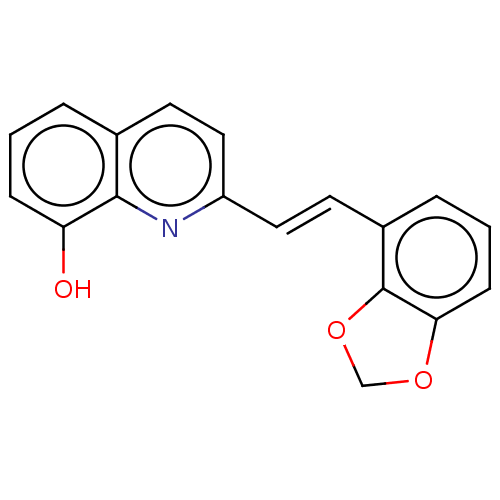 AB01003735-01 BDBM200276 cid_45906904
AB01003735-01 BDBM200276 cid_45906904 AB01003935-01 cid_7844607 BDBM200283
AB01003935-01 cid_7844607 BDBM200283 AB01004165-01 cid_45843625 BDBM200275
AB01004165-01 cid_45843625 BDBM200275 AB01004260-01 BDBM200279 cid_45839325
AB01004260-01 BDBM200279 cid_45839325 AB01005713-01 cid_22618062 BDBM200284
AB01005713-01 cid_22618062 BDBM200284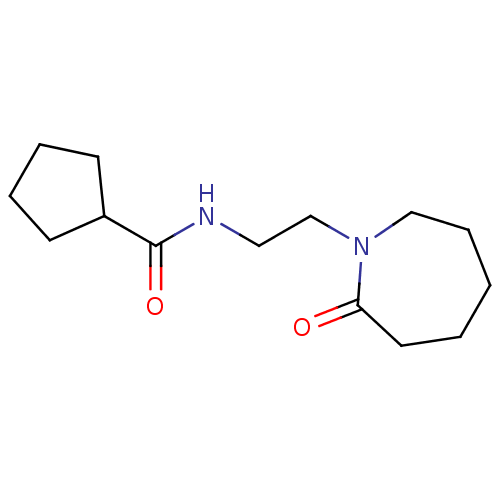 AB01006655-01 cid_45887585 BDBM200273
AB01006655-01 cid_45887585 BDBM200273 BDBM115575 US8633183, C-01
BDBM115575 US8633183, C-01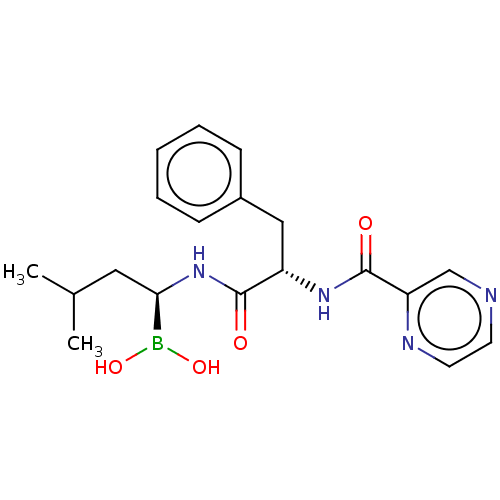 BDBM200228 cid_24871309 AB01275474-01
BDBM200228 cid_24871309 AB01275474-01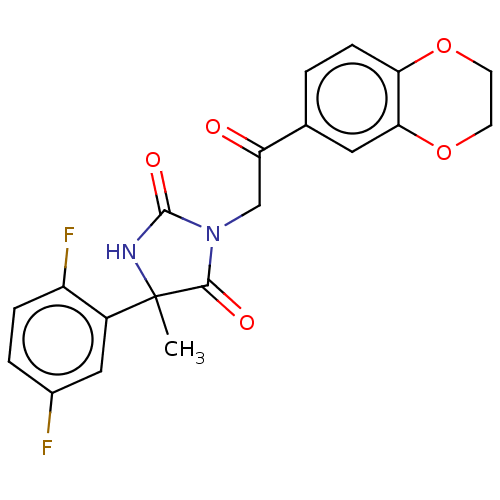 BDBM200247 AB00984167-01 cid_16376379
BDBM200247 AB00984167-01 cid_16376379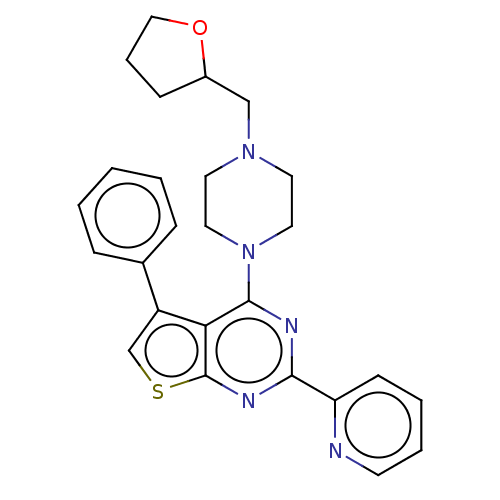 BDBM200248 cid_42909519 AB01001062-01
BDBM200248 cid_42909519 AB01001062-01 BDBM200249 AB01000507-01 cid_41402338
BDBM200249 AB01000507-01 cid_41402338 BDBM200260 AB00987552-01 cid_9418044
BDBM200260 AB00987552-01 cid_9418044 BDBM200271 cid_17549909 AB00985142-01
BDBM200271 cid_17549909 AB00985142-01 BDBM200278 AB01004299-01 cid_40161147
BDBM200278 AB01004299-01 cid_40161147 BDBM200280 AB01004181-01 cid_45898217
BDBM200280 AB01004181-01 cid_45898217 BDBM200282 cid_18076193 AB01005317-01
BDBM200282 cid_18076193 AB01005317-01 BDBM200289 AB01003541-01 cid_33987850
BDBM200289 AB01003541-01 cid_33987850 BDBM200290 AB01003484-01 cid_6086067
BDBM200290 AB01003484-01 cid_6086067 BDBM200291 cid_25976507 AB01002406-01
BDBM200291 cid_25976507 AB01002406-01 BDBM260199 US9522914, A-01
BDBM260199 US9522914, A-01 BDBM260290 US9522914, C-01
BDBM260290 US9522914, C-01 BDBM50430256 CHEMBL391409 NPGI-01
BDBM50430256 CHEMBL391409 NPGI-01 BDBM651982 US11905244, Compound 01
BDBM651982 US11905244, Compound 01 BDBM688353 US20240189315, Example 01
BDBM688353 US20240189315, Example 01 BDBM714655 US20250026741, Compound #01
BDBM714655 US20250026741, Compound #01 BDBM724602 US20250066377, Example 01
BDBM724602 US20250066377, Example 01 BDBM757306 US12357603, Example 01
BDBM757306 US12357603, Example 01 US11746103, EX-01 BDBM614151
US11746103, EX-01 BDBM614151 US12178809, Example 01 BDBM709273
US12178809, Example 01 BDBM709273 US20230295130, Compound 01 BDBM619541
US20230295130, Compound 01 BDBM619541 US20240368106, Example 01 BDBM703335
US20240368106, Example 01 BDBM703335 US8629135, SW-01 BDBM112774
US8629135, SW-01 BDBM112774 US9522914, B-01 BDBM260248
US9522914, B-01 BDBM260248 US9522914, D-01 BDBM260312
US9522914, D-01 BDBM260312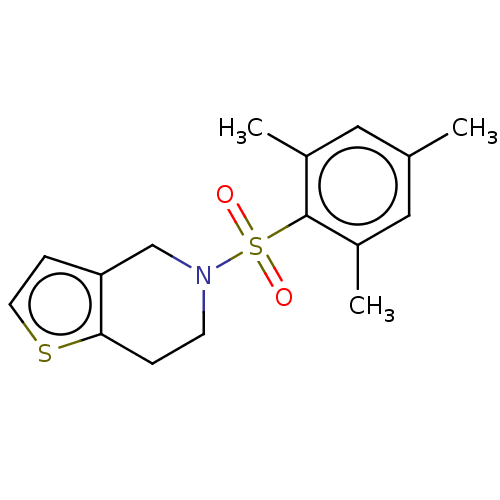 cid_17520970 BDBM200266 AB00986657-01
cid_17520970 BDBM200266 AB00986657-01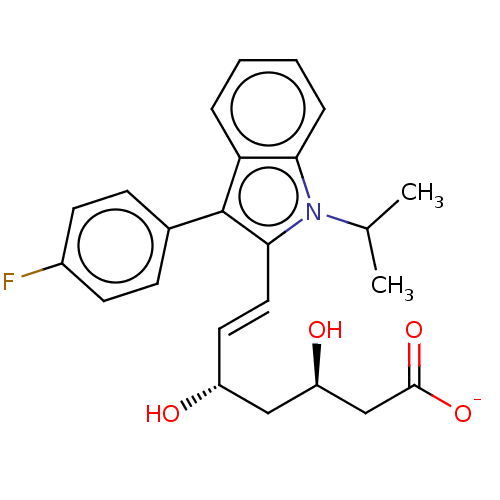 cid_23663976 AB01274723-01 BDBM200236
cid_23663976 AB01274723-01 BDBM200236 cid_2585136 BDBM200259 AB00987441-01
cid_2585136 BDBM200259 AB00987441-01 cid_2697355 BDBM200246 AB00984213-01
cid_2697355 BDBM200246 AB00984213-01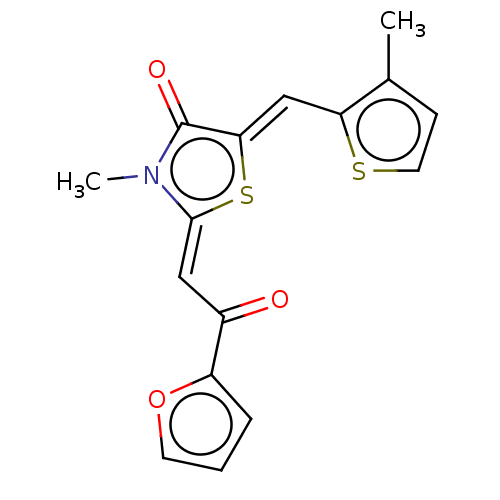 cid_39977303 AB00987062-01 BDBM200261
cid_39977303 AB00987062-01 BDBM200261 cid_40087336 AB00998495-01 BDBM200255
cid_40087336 AB00998495-01 BDBM200255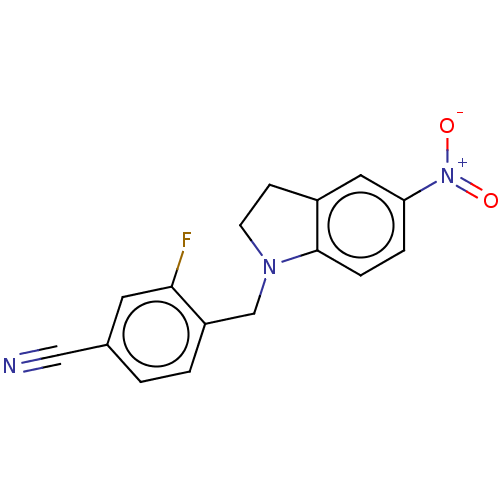 cid_42067840 AB01004372-01 BDBM200231
cid_42067840 AB01004372-01 BDBM200231 cid_45836084 BDBM200251 AB01000998-01
cid_45836084 BDBM200251 AB01000998-01 cid_45867314 AB01003476-01 BDBM200287
cid_45867314 AB01003476-01 BDBM200287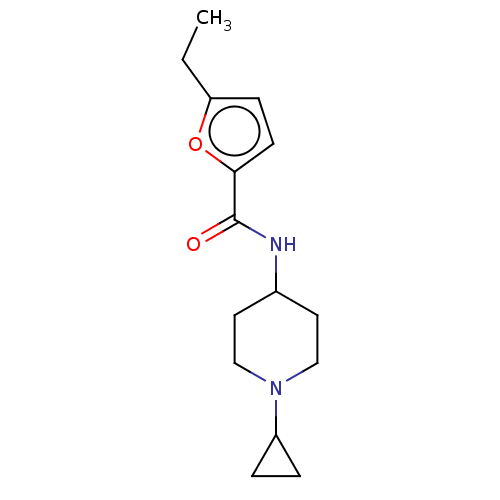 cid_45880860 BDBM200274 AB01004196-01
cid_45880860 BDBM200274 AB01004196-01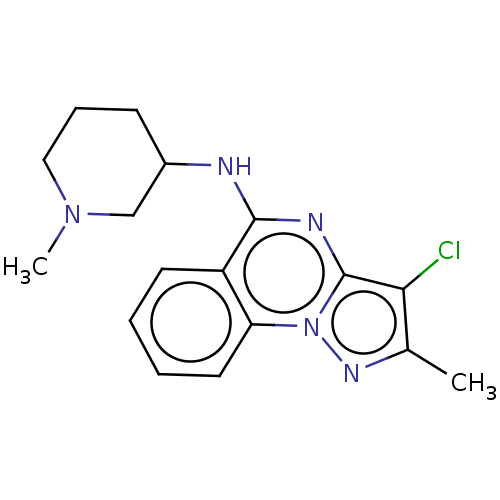 cid_45917090 AB01000744-01 BDBM200250
cid_45917090 AB01000744-01 BDBM200250 cid_4815244 AB00985720-01 BDBM200264
cid_4815244 AB00985720-01 BDBM200264 cid_640701 AB00999694-01 BDBM200253
cid_640701 AB00999694-01 BDBM200253 cid_73673937 AB00988986-01 BDBM200256
cid_73673937 AB00988986-01 BDBM200256 cid_73673997 AB00984802-01 BDBM200230
cid_73673997 AB00984802-01 BDBM200230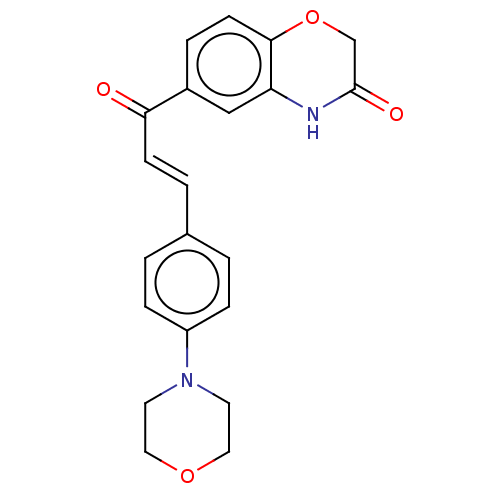 cid_8425524 AB00985701-01 BDBM200272
cid_8425524 AB00985701-01 BDBM200272 US12162836, Compound HMC-C-01-A BDBM637669 US11834414, Compound HMC-C-01-A
US12162836, Compound HMC-C-01-A BDBM637669 US11834414, Compound HMC-C-01-A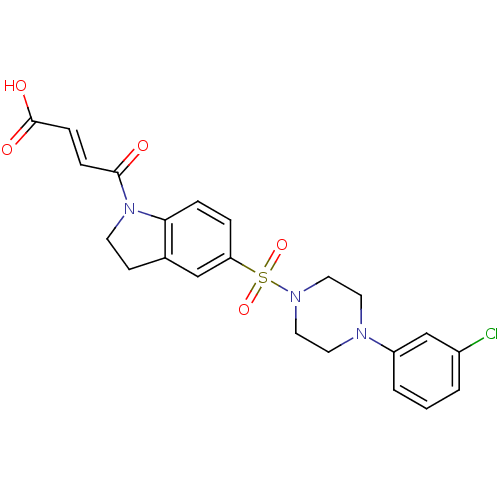 BDBM111826 US8623906, JHE-01-129A
BDBM111826 US8623906, JHE-01-129A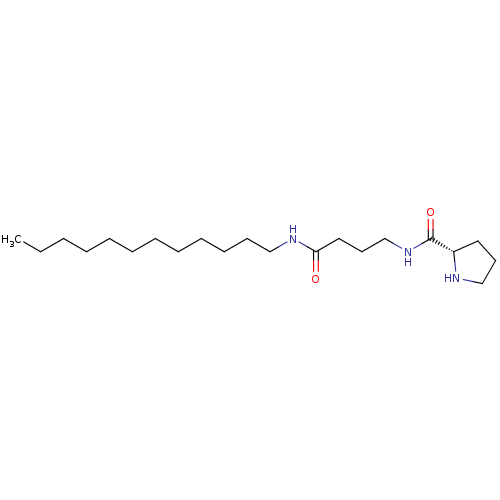 BDBM193289 US9193762, RS-42-01
BDBM193289 US9193762, RS-42-01 BDBM313750 US10167282, Compound TBAP-01
BDBM313750 US10167282, Compound TBAP-01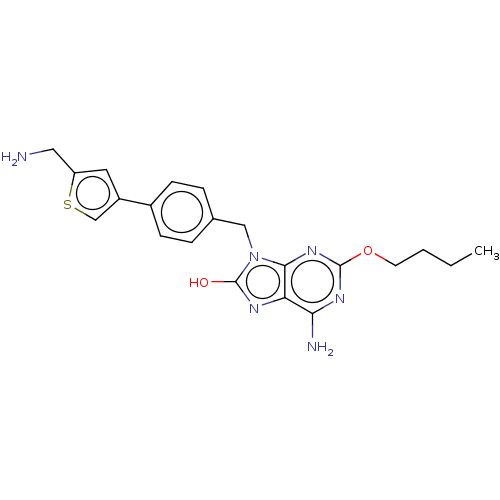 BDBM420040 US10487084, Compound Ib-01
BDBM420040 US10487084, Compound Ib-01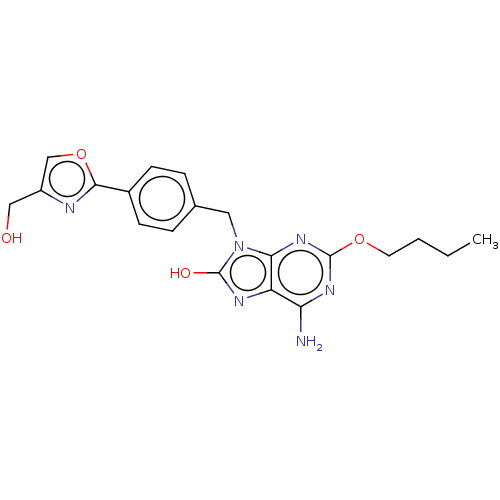 BDBM420042 US10487084, Compound Ic-01
BDBM420042 US10487084, Compound Ic-01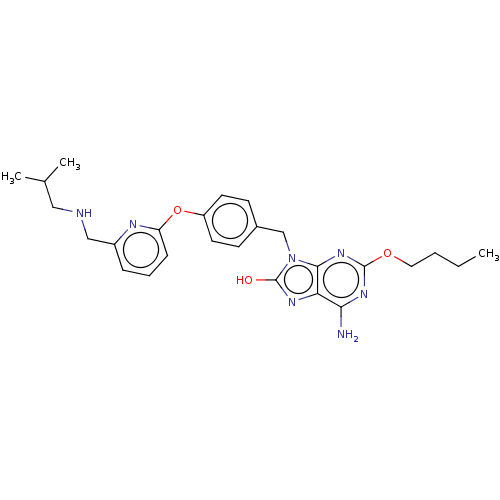 BDBM423499 US10508115, Compound Ic-01
BDBM423499 US10508115, Compound Ic-01 BDBM423507 US10508115, Compound Id-01
BDBM423507 US10508115, Compound Id-01 BDBM423535 US10508115, Compound Ig-01
BDBM423535 US10508115, Compound Ig-01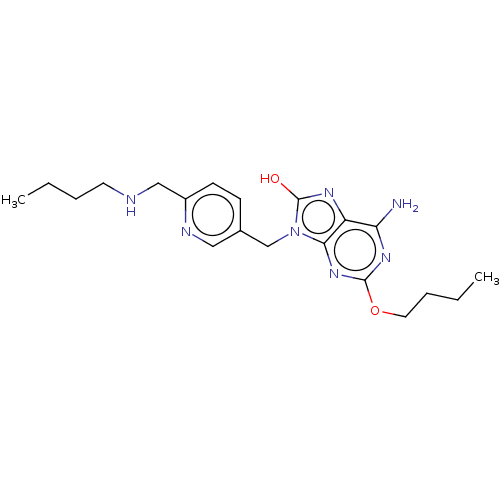 BDBM483337 US10919895, Compound Ic-01
BDBM483337 US10919895, Compound Ic-01 BDBM50421724 PROCINOLOL SD-2124-01
BDBM50421724 PROCINOLOL SD-2124-01 BDBM605422 US11672788, Compound GNTbm-01
BDBM605422 US11672788, Compound GNTbm-01 BDBM650937 US20240043470, Compound 1-01
BDBM650937 US20240043470, Compound 1-01 BDBM651145 US20240043470, Compound 3-01
BDBM651145 US20240043470, Compound 3-01 BDBM651193 US20240043470, Compound 4-01
BDBM651193 US20240043470, Compound 4-01 BDBM688152 US20240246941, Compound SJ001008068-01
BDBM688152 US20240246941, Compound SJ001008068-01 BDBM688154 US20240246941, Compound SJ001008069-01
BDBM688154 US20240246941, Compound SJ001008069-01 BDBM688157 US20240246941, Compound SJ000311270-01
BDBM688157 US20240246941, Compound SJ000311270-01 BDBM688159 US20240246941, Compound SJ000311286-01
BDBM688159 US20240246941, Compound SJ000311286-01 BDBM693147 US12071408, Example III-01
BDBM693147 US12071408, Example III-01 BDBM707299 US12162836, Compound NASMP-01
BDBM707299 US12162836, Compound NASMP-01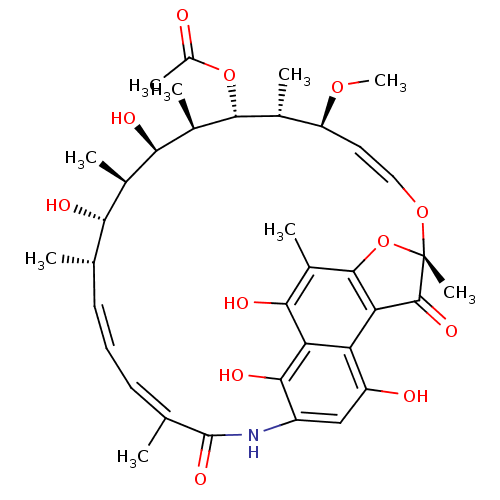 CB-01-11 BDBM50391000 RIFAMYCIN
CB-01-11 BDBM50391000 RIFAMYCIN CHEMBL1255966 NCG-C00093990-01 BDBM50394088
CHEMBL1255966 NCG-C00093990-01 BDBM50394088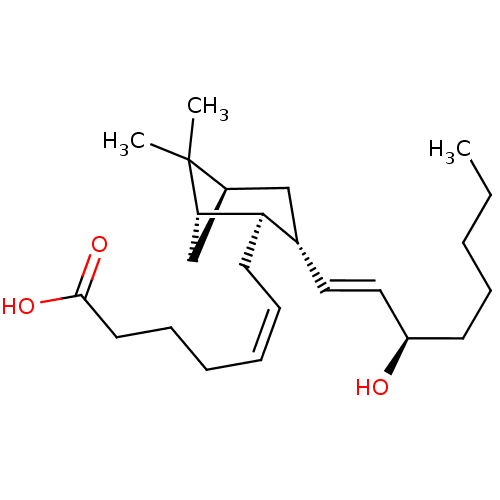 PTA2 BDBM82223 CAS_71111-01-8
PTA2 BDBM82223 CAS_71111-01-8 US10407423, Compound 2-01 BDBM413394
US10407423, Compound 2-01 BDBM413394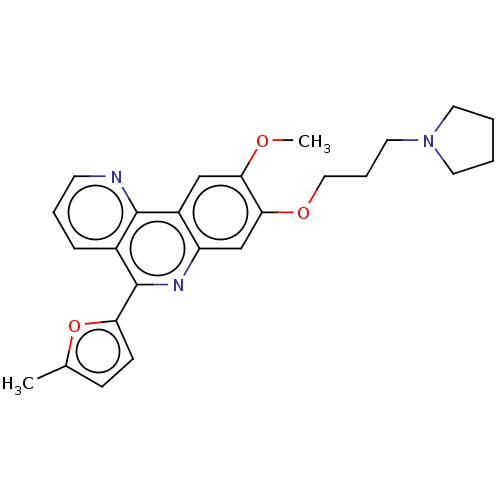 US10407423, Compound 3-01 BDBM413407
US10407423, Compound 3-01 BDBM413407 US10508115, Compound IIa-01 BDBM423536
US10508115, Compound IIa-01 BDBM423536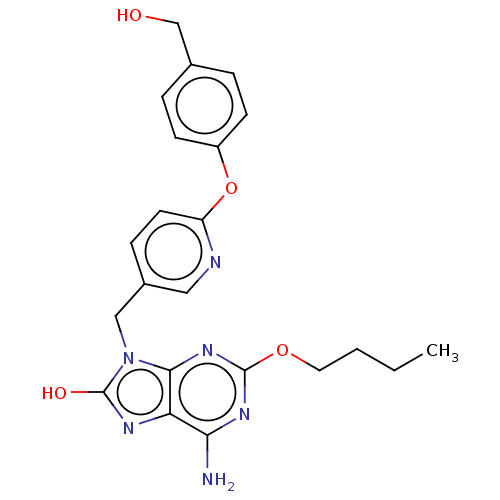 US10508115, Compound Ib-01 BDBM423482
US10508115, Compound Ib-01 BDBM423482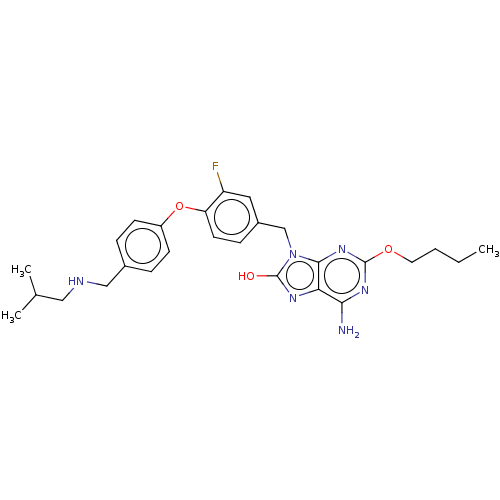 US10508115, Compound Ie-01 BDBM423515
US10508115, Compound Ie-01 BDBM423515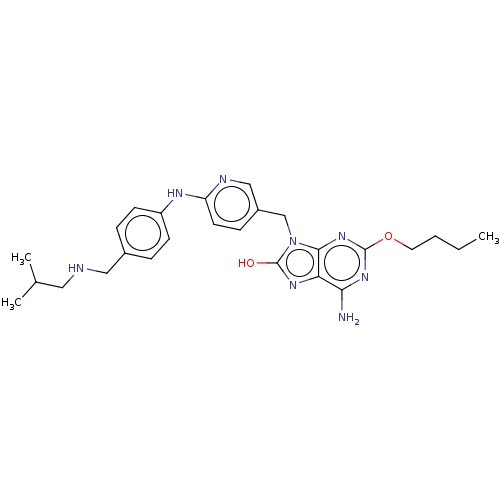 US10508115, Compound If-01 BDBM423525
US10508115, Compound If-01 BDBM423525 US10919895, Compound Ia-01 BDBM483317
US10919895, Compound Ia-01 BDBM483317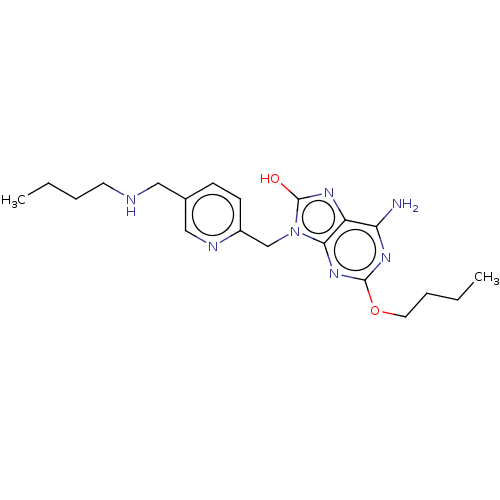 US10919895, Compound Ib-01 BDBM483329
US10919895, Compound Ib-01 BDBM483329 US20230295130, Example T-01 BDBM619377
US20230295130, Example T-01 BDBM619377 US20230322715, Compound ALK-01 BDBM624902
US20230322715, Compound ALK-01 BDBM624902 US20240246941, Compound SJ001008065-01 BDBM688158
US20240246941, Compound SJ001008065-01 BDBM688158 US20240246941, Compound SJ001008066-01 BDBM688153
US20240246941, Compound SJ001008066-01 BDBM688153 US20240246941, Compound SJ001008067-01 BDBM688160
US20240246941, Compound SJ001008067-01 BDBM688160 US20240246941, Compound SJ001009807-01 BDBM688161
US20240246941, Compound SJ001009807-01 BDBM688161 US9174969, JHE-01-129A BDBM189409
US9174969, JHE-01-129A BDBM189409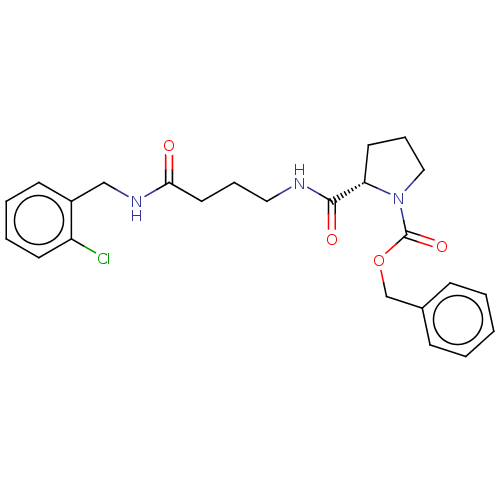 US9193762, RS-37-01 BDBM193282
US9193762, RS-37-01 BDBM193282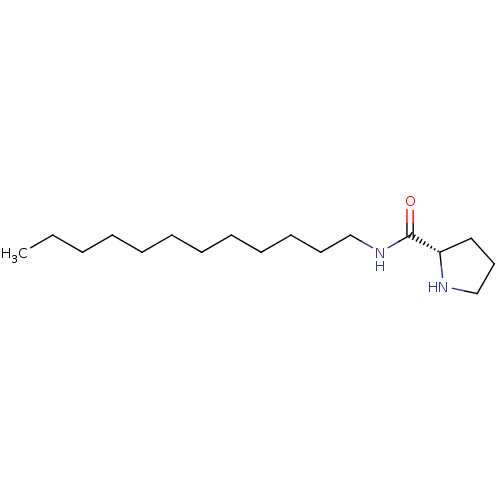 US9193762, RS-47-01 BDBM193277
US9193762, RS-47-01 BDBM193277 US9193762, RS-48-01 BDBM193279
US9193762, RS-48-01 BDBM193279 US9249124, RK2-015-01 BDBM209961
US9249124, RK2-015-01 BDBM209961 US9249124, RK2-017-01 BDBM209951
US9249124, RK2-017-01 BDBM209951 US9249124, RK2-046-01 BDBM209962
US9249124, RK2-046-01 BDBM209962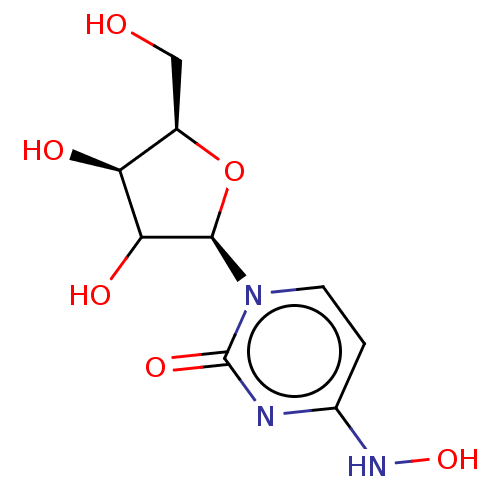 US10874683, Example EIDD-01931-04 US10874683, Example EIDD-02345-01 US10874683, Example EIDD-02416-01 US10874683, Example 00106 US10874683, Example EIDD-02261-01 BDBM476299 US10874683, Example 56.1 US10874683, Example EIDD-02200-01
US10874683, Example EIDD-01931-04 US10874683, Example EIDD-02345-01 US10874683, Example EIDD-02416-01 US10874683, Example 00106 US10874683, Example EIDD-02261-01 BDBM476299 US10874683, Example 56.1 US10874683, Example EIDD-02200-01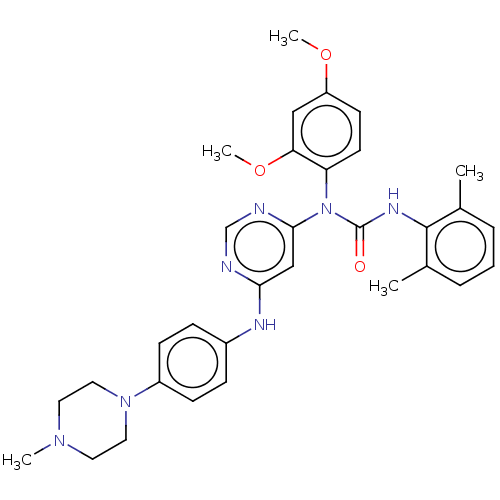 BDBM192712 HG-9-91-01 (1)
BDBM192712 HG-9-91-01 (1)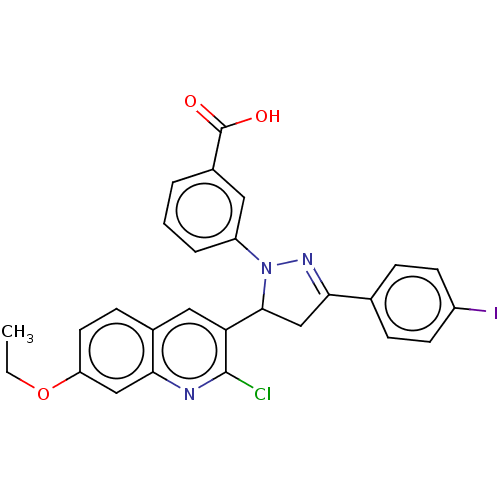 BDBM461290 US10774063, Compound NG-01-02
BDBM461290 US10774063, Compound NG-01-02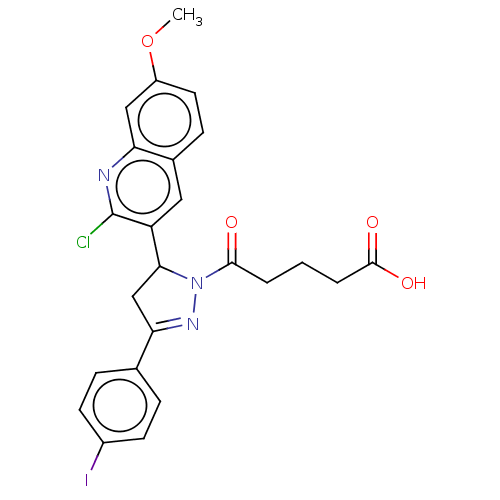 BDBM461292 US10774063, Compound NG-01-24
BDBM461292 US10774063, Compound NG-01-24 BDBM461293 US10774063, Compound NG-01-25
BDBM461293 US10774063, Compound NG-01-25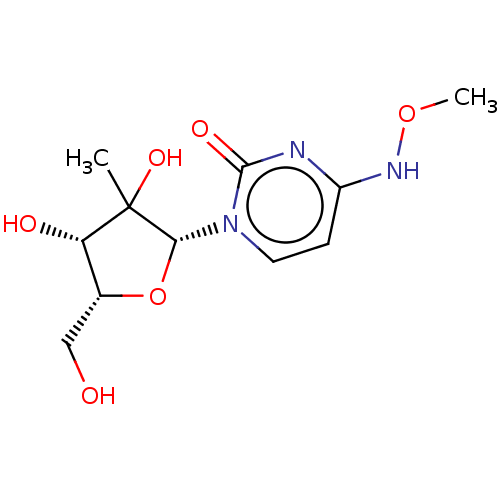 BDBM476306 US10874683, Example EIDD-02054-01
BDBM476306 US10874683, Example EIDD-02054-01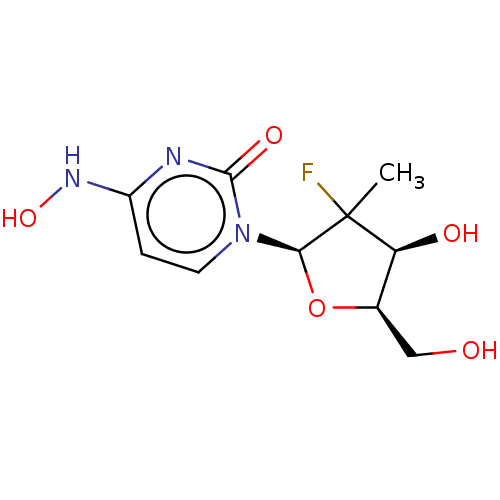 BDBM476307 US10874683, Example EIDD-02080-01
BDBM476307 US10874683, Example EIDD-02080-01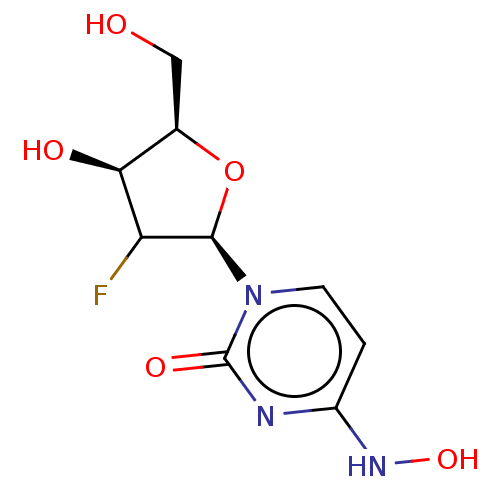 BDBM476308 US10874683, Example EIDD-02085-01
BDBM476308 US10874683, Example EIDD-02085-01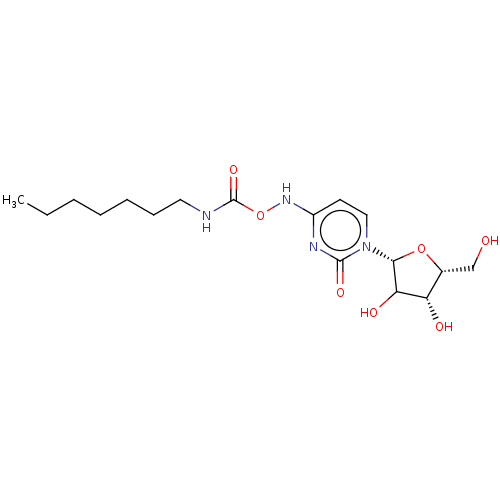 BDBM476309 US10874683, Example EIDD-02107-01
BDBM476309 US10874683, Example EIDD-02107-01 BDBM476313 US10874683, Example EIDD-02340-01
BDBM476313 US10874683, Example EIDD-02340-01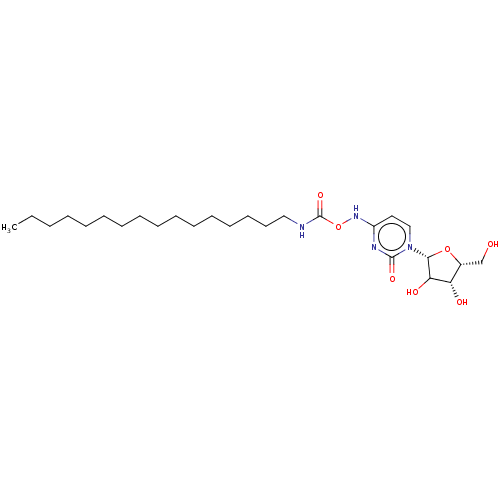 BDBM476314 US10874683, Example EIDD-02356-01
BDBM476314 US10874683, Example EIDD-02356-01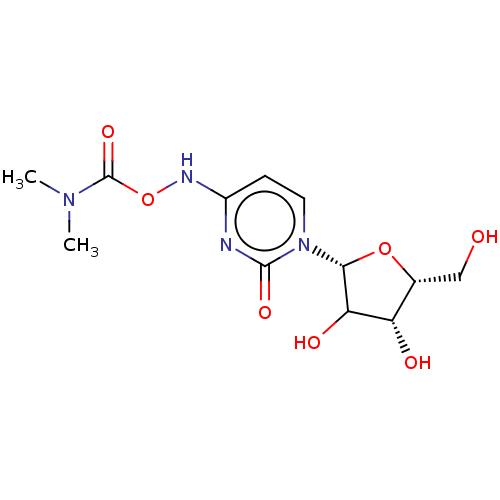 BDBM476319 US10874683, Example EIDD-02423-01
BDBM476319 US10874683, Example EIDD-02423-01 BDBM476321 US10874683, Example EIDD-02475-01
BDBM476321 US10874683, Example EIDD-02475-01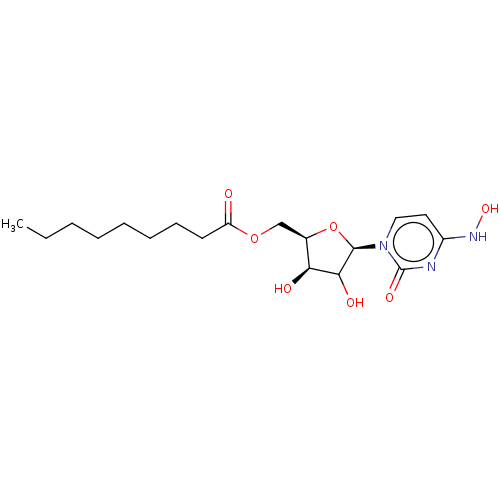 BDBM476323 US10874683, Example EIDD-02476-01
BDBM476323 US10874683, Example EIDD-02476-01 BDBM476325 US10874683, Example EIDD-02416-01
BDBM476325 US10874683, Example EIDD-02416-01 BDBM476326 US10874683, Example EIDD-02345-01
BDBM476326 US10874683, Example EIDD-02345-01 BDBM476327 US10874683, Example EIDD-02261-01
BDBM476327 US10874683, Example EIDD-02261-01 BDBM476328 US10874683, Example EIDD-02427-01
BDBM476328 US10874683, Example EIDD-02427-01 BDBM476329 US10874683, Example EIDD-02207-01
BDBM476329 US10874683, Example EIDD-02207-01 BDBM476334 US10874683, Example EIDD-02200-01
BDBM476334 US10874683, Example EIDD-02200-01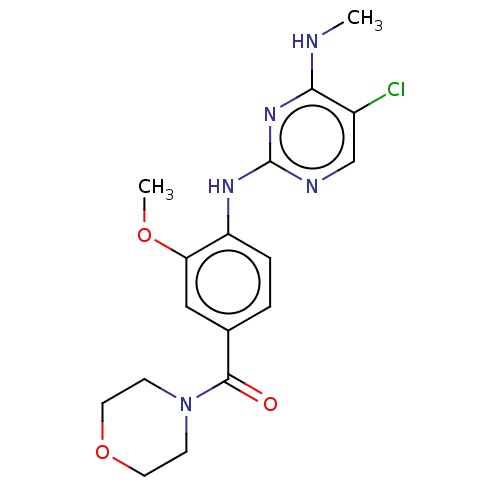 BDBM482160 BDBM50396041 HG-10-102-01
BDBM482160 BDBM50396041 HG-10-102-01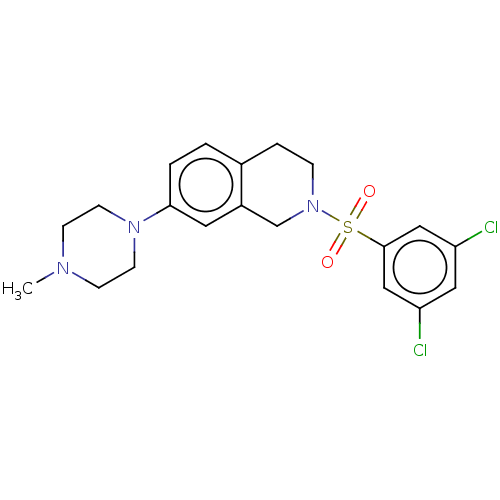 BDBM488140 US10954217, Example KTL-01-257
BDBM488140 US10954217, Example KTL-01-257 BDBM50421953 CHEMBL1256916 L-687384 NCGC00094067-01
BDBM50421953 CHEMBL1256916 L-687384 NCGC00094067-01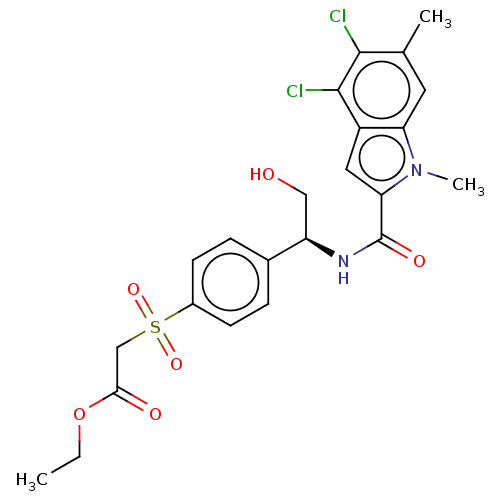 BDBM50519131 CHEMBL4518579 US11304929, Example 01-002
BDBM50519131 CHEMBL4518579 US11304929, Example 01-002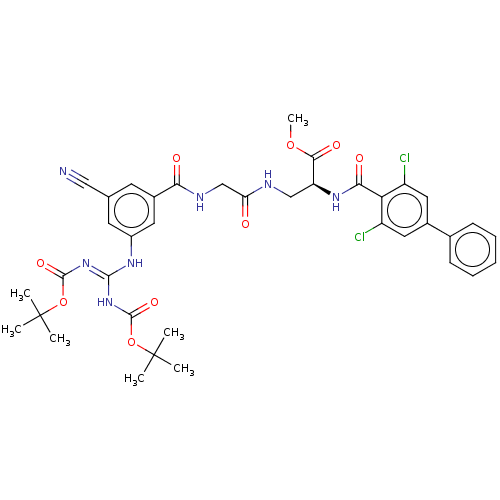 BDBM585089 US11530180, Compound SU15210-0206-01
BDBM585089 US11530180, Compound SU15210-0206-01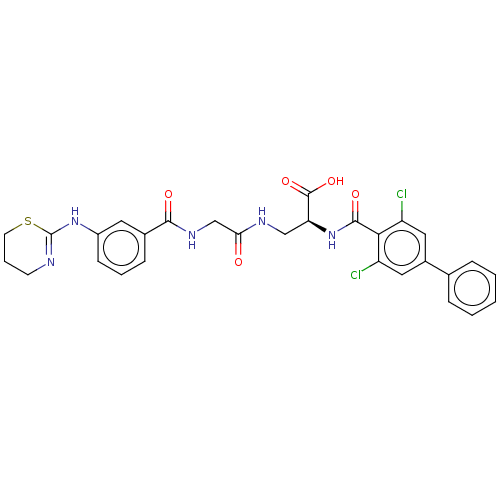 BDBM585090 US11530180, Compound SU15210-0230-01
BDBM585090 US11530180, Compound SU15210-0230-01 BDBM601768 US11643396, Example SG2-005-01
BDBM601768 US11643396, Example SG2-005-01 BDBM601769 US11643396, Example SG2-015-01
BDBM601769 US11643396, Example SG2-015-01 BDBM601783 US11643396, Example RJ1-027-01
BDBM601783 US11643396, Example RJ1-027-01 BDBM601785 US11643396, Example RJ1-036-01
BDBM601785 US11643396, Example RJ1-036-01 BDBM601786 US11643396, Example RJ1-040-01
BDBM601786 US11643396, Example RJ1-040-01 BDBM601789 US11643396, Example SG2-029-01
BDBM601789 US11643396, Example SG2-029-01 BDBM601802 US11643396, Example RJ1-045-01
BDBM601802 US11643396, Example RJ1-045-01 BDBM601804 US11643396, Example RJ1-053-01
BDBM601804 US11643396, Example RJ1-053-01 BDBM601805 US11643396, Example RJ1-057-01
BDBM601805 US11643396, Example RJ1-057-01 BDBM601810 US11643396, Example SG2-064-01
BDBM601810 US11643396, Example SG2-064-01 BDBM601811 US11643396, Example SG2-065-01
BDBM601811 US11643396, Example SG2-065-01 BDBM601812 US11643396, Example SG2-069-01
BDBM601812 US11643396, Example SG2-069-01 BDBM601909 US11643396, Example SG4-039-01
BDBM601909 US11643396, Example SG4-039-01 BDBM602172 US11643417, Ex. No. 2-01
BDBM602172 US11643417, Ex. No. 2-01 BDBM634330 US20230357271, Example a-01-21
BDBM634330 US20230357271, Example a-01-21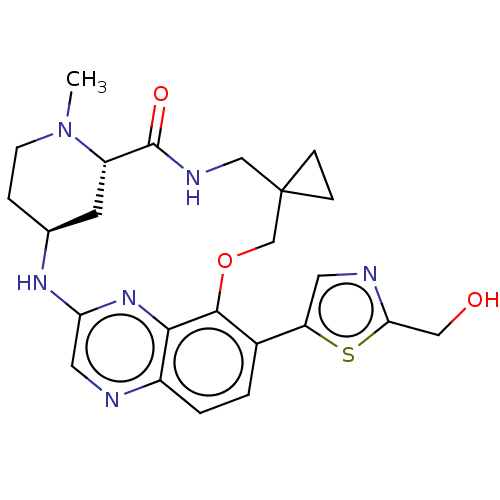 BDBM634349 US20230357271, Example a-01-02
BDBM634349 US20230357271, Example a-01-02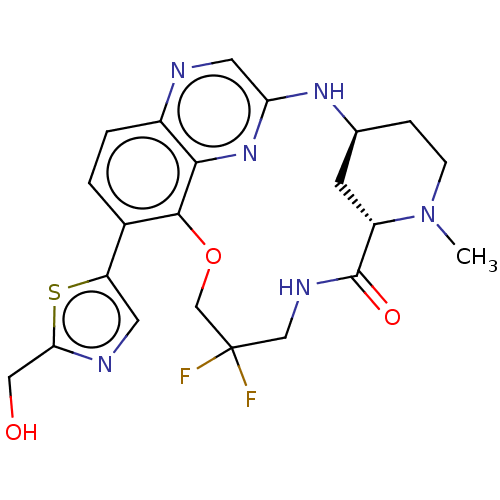 BDBM634353 US20230357271, Example a-01-22
BDBM634353 US20230357271, Example a-01-22 BDBM634384 US20230357271, Example a-01-03
BDBM634384 US20230357271, Example a-01-03 BDBM634449 US20230357271, Example a-01-23
BDBM634449 US20230357271, Example a-01-23 BDBM634492 US20230357271, Example a-01-24
BDBM634492 US20230357271, Example a-01-24 BDBM634494 US20230357271, Example a-01-25
BDBM634494 US20230357271, Example a-01-25 BDBM634495 US20230357271, Example a-01-06
BDBM634495 US20230357271, Example a-01-06 BDBM634496 US20230357271, Example a-01-26
BDBM634496 US20230357271, Example a-01-26 BDBM634503 US20230357271, Example a-01-10
BDBM634503 US20230357271, Example a-01-10 BDBM634504 US20230357271, Example a-01-30
BDBM634504 US20230357271, Example a-01-30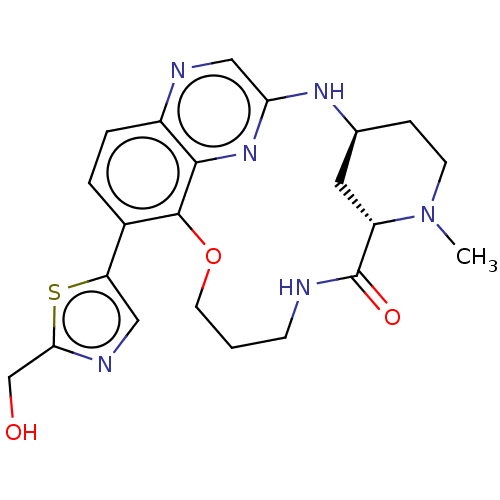 BDBM634505 US20230357271, Example a-01-11
BDBM634505 US20230357271, Example a-01-11 BDBM634506 US20230357271, Example a-01-31
BDBM634506 US20230357271, Example a-01-31 BDBM634507 US20230357271, Example a-01-12
BDBM634507 US20230357271, Example a-01-12 BDBM634508 US20230357271, Example a-01-32
BDBM634508 US20230357271, Example a-01-32 BDBM634509 US20230357271, Example a-01-13
BDBM634509 US20230357271, Example a-01-13 BDBM634510 US20230357271, Example a-01-33
BDBM634510 US20230357271, Example a-01-33 BDBM634511 US20230357271, Example a-01-14
BDBM634511 US20230357271, Example a-01-14 BDBM634512 US20230357271, Example a-01-34
BDBM634512 US20230357271, Example a-01-34 BDBM634514 US20230357271, Example a-01-35
BDBM634514 US20230357271, Example a-01-35 BDBM634516 US20230357271, Example a-01-36
BDBM634516 US20230357271, Example a-01-36 BDBM634525 US20230357271, Example a-01-41
BDBM634525 US20230357271, Example a-01-41 BDBM634527 US20230357271, Example a-01-42
BDBM634527 US20230357271, Example a-01-42 BDBM634529 US20230357271, Example a-01-44
BDBM634529 US20230357271, Example a-01-44 BDBM634531 US20230357271, Example a-01-45
BDBM634531 US20230357271, Example a-01-45 BDBM634533 US20230357271, Example a-02-01
BDBM634533 US20230357271, Example a-02-01 BDBM647363 US20240025893, Example a-01-02
BDBM647363 US20240025893, Example a-01-02 BDBM647364 US20240025893, Example a-01-03
BDBM647364 US20240025893, Example a-01-03 BDBM647365 US20240025893, Example a-01-04
BDBM647365 US20240025893, Example a-01-04 BDBM647366 US20240025893, Example a-01-05
BDBM647366 US20240025893, Example a-01-05 BDBM647371 US20240025893, Example a-01-10
BDBM647371 US20240025893, Example a-01-10 BDBM647372 US20240025893, Example a-01-11
BDBM647372 US20240025893, Example a-01-11 BDBM647373 US20240025893, Example a-01-12
BDBM647373 US20240025893, Example a-01-12 BDBM647374 US20240025893, Example a-02-01
BDBM647374 US20240025893, Example a-02-01 BDBM647375 US20240025893, Example a-03-01
BDBM647375 US20240025893, Example a-03-01 BDBM647376 US20240025893, Example a-04-01
BDBM647376 US20240025893, Example a-04-01 BDBM647380 US20240025893, Example a-05-01
BDBM647380 US20240025893, Example a-05-01 BDBM647382 US20240025893, Example b-01-02
BDBM647382 US20240025893, Example b-01-02 BDBM647475 US20240025893, Example b-04-01
BDBM647475 US20240025893, Example b-04-01 BDBM647541 US20240025893, Example c-03-01
BDBM647541 US20240025893, Example c-03-01 BDBM647550 US20240025893, Example c-04-01
BDBM647550 US20240025893, Example c-04-01 BDBM647553 US20240025893, Example c-05-01
BDBM647553 US20240025893, Example c-05-01 BDBM647554 US20240025893, Example c-06-01
BDBM647554 US20240025893, Example c-06-01 BDBM647557 US20240025893, Example c-07-01
BDBM647557 US20240025893, Example c-07-01 BDBM647560 US20240025893, Example d-01-03
BDBM647560 US20240025893, Example d-01-03 BDBM647561 US20240025893, Example d-01-04
BDBM647561 US20240025893, Example d-01-04 BDBM647562 US20240025893, Example d-01-05
BDBM647562 US20240025893, Example d-01-05 BDBM647563 US20240025893, Example d-01-06
BDBM647563 US20240025893, Example d-01-06 BDBM647564 US20240025893, Example d-01-07
BDBM647564 US20240025893, Example d-01-07 BDBM647573 US20240025893, Example d-04-01
BDBM647573 US20240025893, Example d-04-01 BDBM647580 US20240025893, Example e-01-02
BDBM647580 US20240025893, Example e-01-02 BDBM647581 US20240025893, Example e-01-03
BDBM647581 US20240025893, Example e-01-03 BDBM647582 US20240025893, Example e-02-01
BDBM647582 US20240025893, Example e-02-01 BDBM706968 US20240400537, Example EX-C-01
BDBM706968 US20240400537, Example EX-C-01 BDBM732853 US20250115578, Example 01 US20250115578, KH01
BDBM732853 US20250115578, Example 01 US20250115578, KH01 CAS_84-01-5 chlorproethazine NSC_65750 BDBM86724
CAS_84-01-5 chlorproethazine NSC_65750 BDBM86724 NCGC00094126-01 BDBM50366534 L-745870 CHEMBL555670
NCGC00094126-01 BDBM50366534 L-745870 CHEMBL555670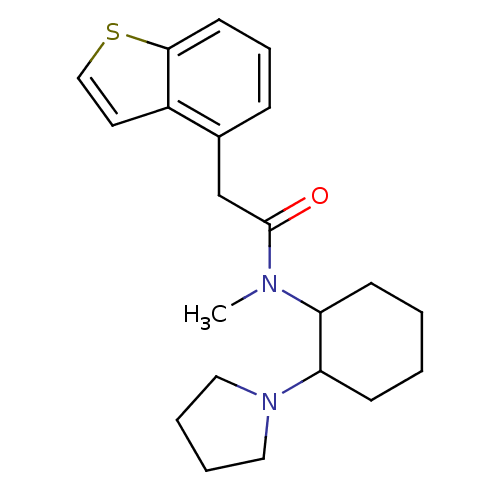 NSC_122115 BDBM81936 PD117302 CAS_111728-01-9
NSC_122115 BDBM81936 PD117302 CAS_111728-01-9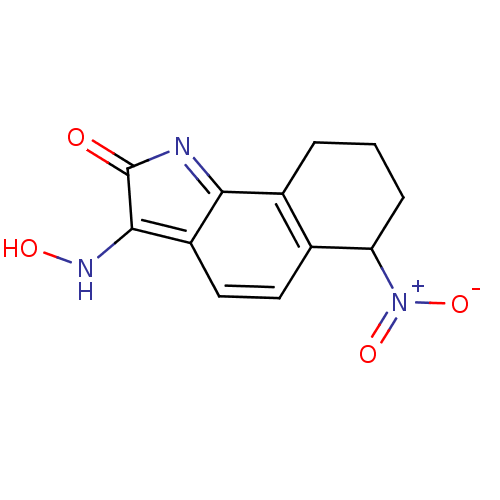 NSC_5311098 CAS_136623-01-3 BDBM85215 NS102
NSC_5311098 CAS_136623-01-3 BDBM85215 NS102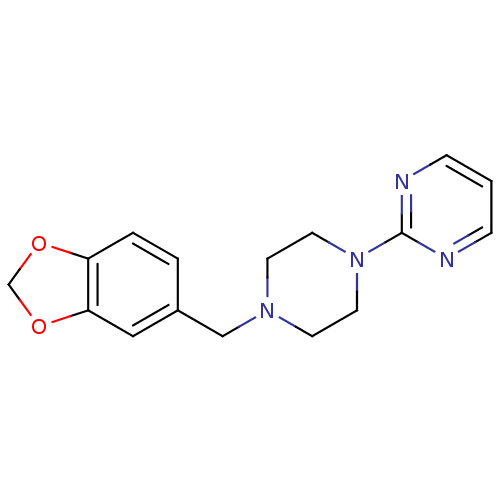 Piribedil NSC_4850 CAS_3605-01-4 BDBM85092
Piribedil NSC_4850 CAS_3605-01-4 BDBM85092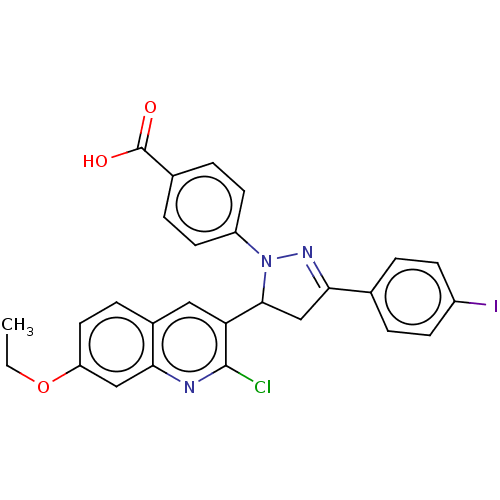 US10774063, Compound NG-01-04 BDBM461291
US10774063, Compound NG-01-04 BDBM461291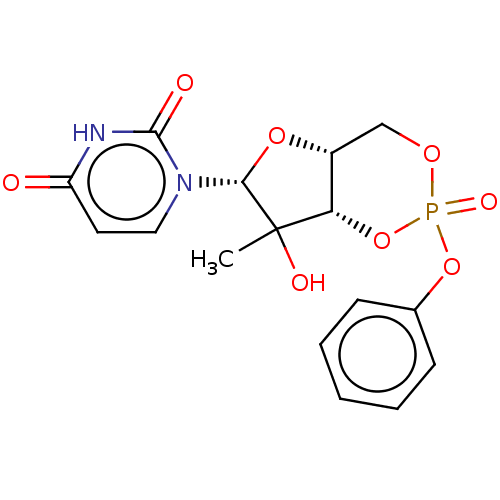 US10874683, Example EIDD-01872-01 BDBM476333
US10874683, Example EIDD-01872-01 BDBM476333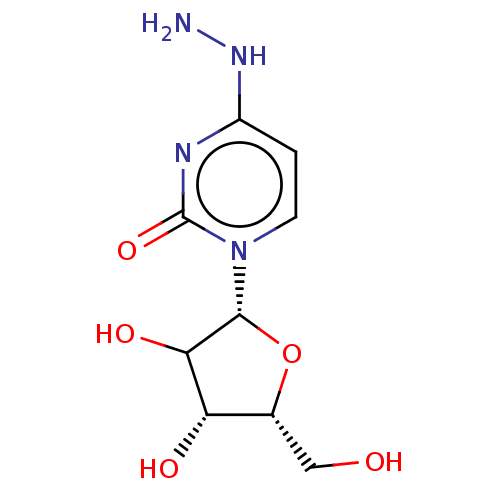 US10874683, Example EIDD-01910-01 BDBM476311
US10874683, Example EIDD-01910-01 BDBM476311 US10874683, Example EIDD-02053-01 BDBM476305
US10874683, Example EIDD-02053-01 BDBM476305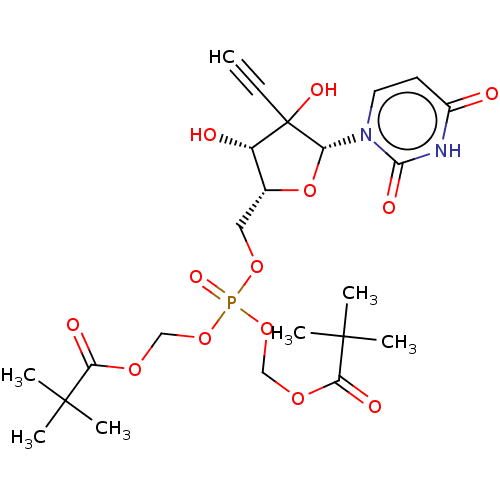 US10874683, Example EIDD-02290-01 BDBM476335
US10874683, Example EIDD-02290-01 BDBM476335 US10874683, Example EIDD-02422-01 BDBM476316
US10874683, Example EIDD-02422-01 BDBM476316 US10874683, Example EIDD-02474-01 BDBM476320
US10874683, Example EIDD-02474-01 BDBM476320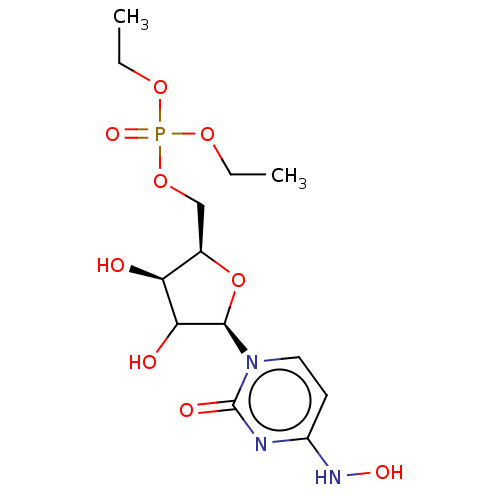 US10874683, Example EIDD-02503-01 BDBM476331
US10874683, Example EIDD-02503-01 BDBM476331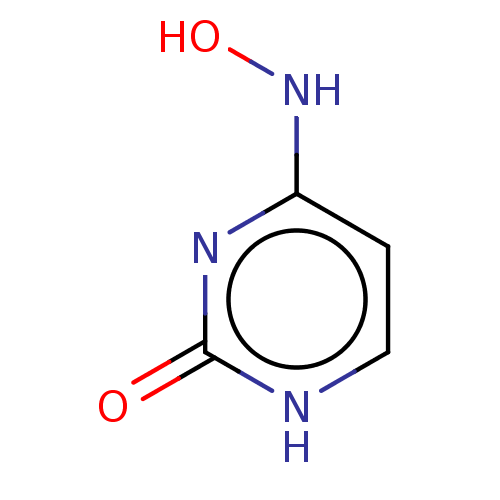 US10874683, Example EIDD-02504-01 BDBM476324
US10874683, Example EIDD-02504-01 BDBM476324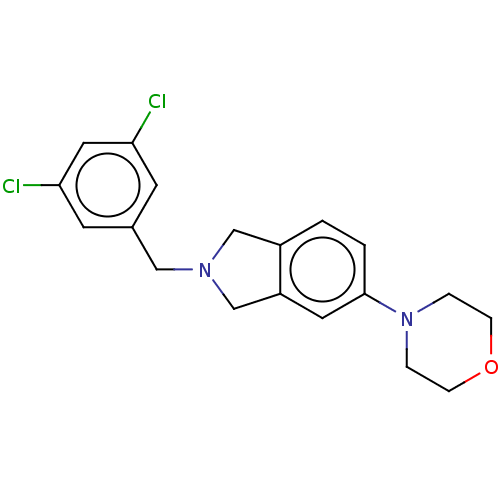 US10954217, Example KTL-01-121 BDBM488121
US10954217, Example KTL-01-121 BDBM488121 US10954217, Example KTL-01-184 BDBM488144
US10954217, Example KTL-01-184 BDBM488144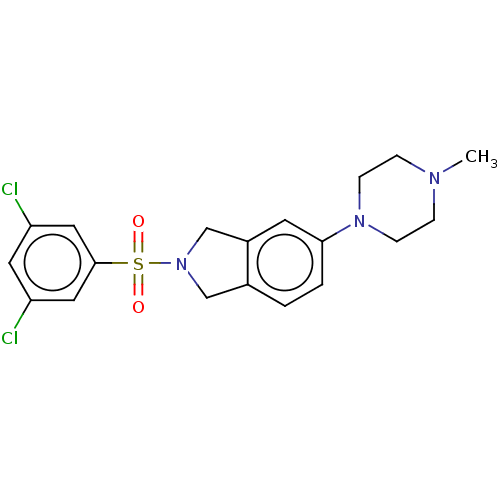 US10954217, Example KTL-01-253 BDBM488127
US10954217, Example KTL-01-253 BDBM488127 US10954217, Example KTL-01-264 BDBM488142
US10954217, Example KTL-01-264 BDBM488142 US10954217, Example KTL-01-276 BDBM488128
US10954217, Example KTL-01-276 BDBM488128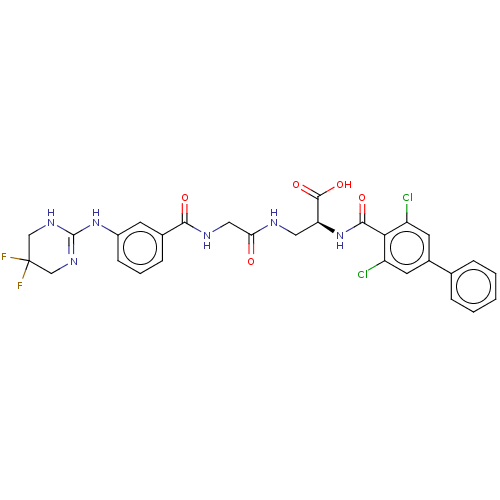 US11530180, Compound SU15210-0238-01 BDBM585091
US11530180, Compound SU15210-0238-01 BDBM585091 US11643396, Example RJ1-024-01 BDBM601782
US11643396, Example RJ1-024-01 BDBM601782 US11643396, Example RJ1-041-01 BDBM601787
US11643396, Example RJ1-041-01 BDBM601787 US11643396, Example RJ1-051-01 BDBM601803
US11643396, Example RJ1-051-01 BDBM601803 US11643396, Example RJ1-060-01 BDBM601806
US11643396, Example RJ1-060-01 BDBM601806 US11643396, Example RJ1-064-01 BDBM601807
US11643396, Example RJ1-064-01 BDBM601807 US11643396, Example RJ1-066-01 BDBM601808
US11643396, Example RJ1-066-01 BDBM601808 US11643396, Example SG2-054-01 BDBM601791
US11643396, Example SG2-054-01 BDBM601791 US11643396, Example SG2-055-01 BDBM601792
US11643396, Example SG2-055-01 BDBM601792 US11643396, Example SG2-060-01 BDBM601794
US11643396, Example SG2-060-01 BDBM601794 US11643396, Example SG2-063-01 BDBM601809
US11643396, Example SG2-063-01 BDBM601809 US11643396, Example SG2-070-01 BDBM601813
US11643396, Example SG2-070-01 BDBM601813 US11643396, Example SG2-071-01 BDBM601814
US11643396, Example SG2-071-01 BDBM601814 US11643396, Example SG2-072-01 BDBM601815
US11643396, Example SG2-072-01 BDBM601815 US11643396, Example SG2-081-01 BDBM601816
US11643396, Example SG2-081-01 BDBM601816 US11643396, Example SG2-085-01 BDBM601817
US11643396, Example SG2-085-01 BDBM601817 US11643396, Example SG2-086-01 BDBM601838
US11643396, Example SG2-086-01 BDBM601838 US11643396, Example SG2-087-01 BDBM601818
US11643396, Example SG2-087-01 BDBM601818 US11643396, Example SG2-088-01 BDBM601819
US11643396, Example SG2-088-01 BDBM601819 US11643396, Example SG2-142-01 BDBM601842
US11643396, Example SG2-142-01 BDBM601842 US11643396, Example SG3-087-01 BDBM601868
US11643396, Example SG3-087-01 BDBM601868 US11834414, Compound CHMSA-01-A BDBM637675
US11834414, Compound CHMSA-01-A BDBM637675 US20230322715, Compound TR-YTH-01 BDBM624881
US20230322715, Compound TR-YTH-01 BDBM624881 US20230357271, Example a-01-04 BDBM634491
US20230357271, Example a-01-04 BDBM634491 US20230357271, Example a-01-07 BDBM634497
US20230357271, Example a-01-07 BDBM634497 US20230357271, Example a-01-08 BDBM634499
US20230357271, Example a-01-08 BDBM634499 US20230357271, Example a-01-09 BDBM634501
US20230357271, Example a-01-09 BDBM634501 US20230357271, Example a-01-15 BDBM634513
US20230357271, Example a-01-15 BDBM634513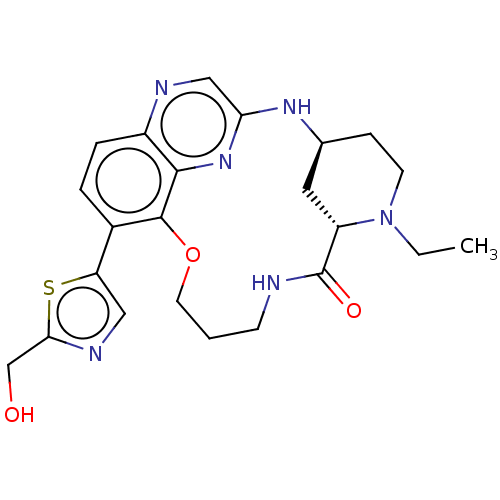 US20230357271, Example a-01-16 BDBM634515
US20230357271, Example a-01-16 BDBM634515 US20230357271, Example a-01-17 BDBM634517
US20230357271, Example a-01-17 BDBM634517 US20230357271, Example a-01-18 BDBM634519
US20230357271, Example a-01-18 BDBM634519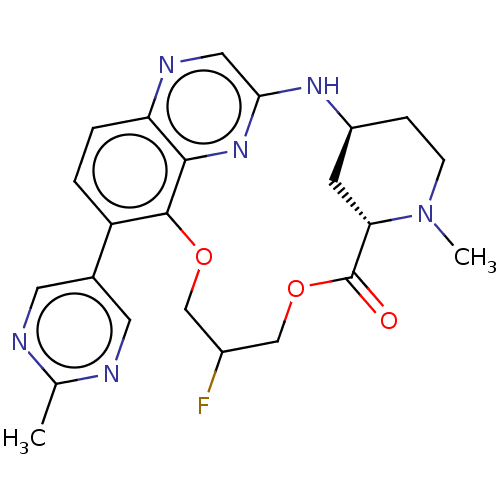 US20230357271, Example a-01-20 BDBM634523
US20230357271, Example a-01-20 BDBM634523 US20230357271, Example a-01-27 BDBM634498
US20230357271, Example a-01-27 BDBM634498 US20230357271, Example a-01-28 BDBM634500
US20230357271, Example a-01-28 BDBM634500 US20230357271, Example a-01-29 BDBM634502
US20230357271, Example a-01-29 BDBM634502 US20230357271, Example a-01-37 BDBM634518
US20230357271, Example a-01-37 BDBM634518 US20230357271, Example a-01-38 BDBM634520
US20230357271, Example a-01-38 BDBM634520 US20230357271, Example a-01-39 BDBM634522
US20230357271, Example a-01-39 BDBM634522 US20230357271, Example a-01-40 BDBM634524
US20230357271, Example a-01-40 BDBM634524 US20240025893, Example a-01-06 BDBM647367
US20240025893, Example a-01-06 BDBM647367 US20240025893, Example a-01-07 BDBM647368
US20240025893, Example a-01-07 BDBM647368 US20240025893, Example a-01-08 BDBM647369
US20240025893, Example a-01-08 BDBM647369 US20240025893, Example a-01-09 BDBM647370
US20240025893, Example a-01-09 BDBM647370 US20240025893, Example b-02-01 BDBM647383
US20240025893, Example b-02-01 BDBM647383 US20240025893, Example b-03-01 BDBM647466
US20240025893, Example b-03-01 BDBM647466 US20240025893, Example b-05-01 BDBM647523
US20240025893, Example b-05-01 BDBM647523 US20240025893, Example b-07-01 BDBM647532
US20240025893, Example b-07-01 BDBM647532 US20240025893, Example b-08-01 BDBM647533
US20240025893, Example b-08-01 BDBM647533 US20240025893, Example c-02-01 BDBM647535
US20240025893, Example c-02-01 BDBM647535 US20240025893, Example d-01-02 BDBM647559
US20240025893, Example d-01-02 BDBM647559 US20240025893, Example d-01-08 BDBM647565
US20240025893, Example d-01-08 BDBM647565 US20240025893, Example d-03-01 BDBM647567
US20240025893, Example d-03-01 BDBM647567 US20240025893, Example e-03-01 BDBM647609
US20240025893, Example e-03-01 BDBM647609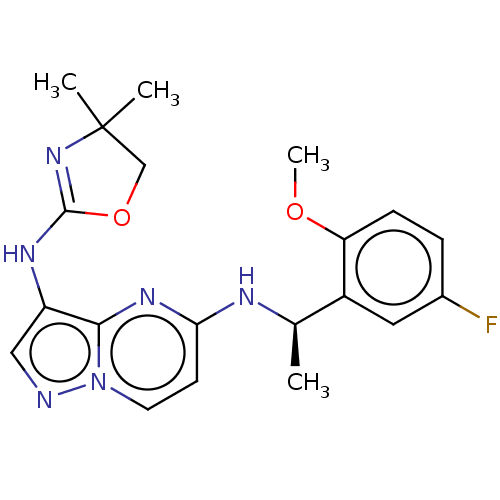 US20240025908, Example 1 BDBM647633 T-01
US20240025908, Example 1 BDBM647633 T-01 US9403797, PAPC-A-01 BDBM50401614 CHEMBL2203843
US9403797, PAPC-A-01 BDBM50401614 CHEMBL2203843 BDBM192713 HG-9-91-01 derivative, 2
BDBM192713 HG-9-91-01 derivative, 2 BDBM192717 HG-9-91-01 derivative, 6
BDBM192717 HG-9-91-01 derivative, 6 BDBM192718 HG-9-91-01 derivative, 7
BDBM192718 HG-9-91-01 derivative, 7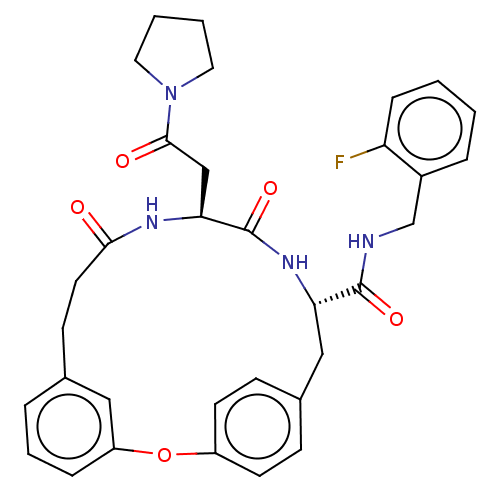 BDBM50587103 CHEMBL5086113 US20240043470, Compound 2-01-B
BDBM50587103 CHEMBL5086113 US20240043470, Compound 2-01-B BDBM701962 CBX-01 US12129248, Compound ACV-56
BDBM701962 CBX-01 US12129248, Compound ACV-56 BDBM706135 SB-FAP-01 US20240382629, Example i
BDBM706135 SB-FAP-01 US20240382629, Example i BDBM710640 US12187701, Compound IHK-01-013-2
BDBM710640 US12187701, Compound IHK-01-013-2 BRD-K01182818-001-01-0 cid_44489856 BDBM75317
BRD-K01182818-001-01-0 cid_44489856 BDBM75317 Salvinorin A CAS_83729-01-5 NSC_128563 BDBM86548
Salvinorin A CAS_83729-01-5 NSC_128563 BDBM86548 US11643396, Example SG2-033-01-1 BDBM601790
US11643396, Example SG2-033-01-1 BDBM601790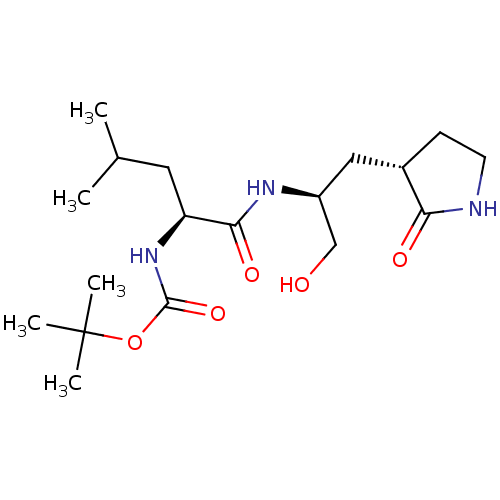 WO2006061714-ID-01 BDBM423459 WO2006061714, Example 1
WO2006061714-ID-01 BDBM423459 WO2006061714, Example 1 cid_44489345 BDBM75315 BRD-K85028121-001-01-6
cid_44489345 BDBM75315 BRD-K85028121-001-01-6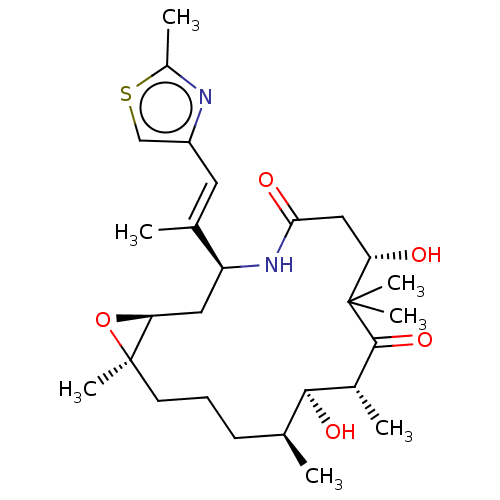 BDBM50564768 NSC-747973 Ixempra kit BMS 247550-01 BMS-247550-01 Ixempra BMS-247550 CHEBI:63605 Azaepothilone b Ixabepilone
BDBM50564768 NSC-747973 Ixempra kit BMS 247550-01 BMS-247550-01 Ixempra BMS-247550 CHEBI:63605 Azaepothilone b Ixabepilone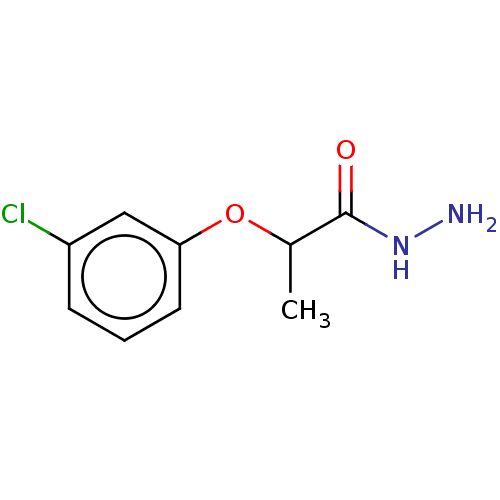 2‐(3‐Chlorophenoxy)propanehydrazide (01) BDBM163684
2‐(3‐Chlorophenoxy)propanehydrazide (01) BDBM163684 BDBM476183 US10865384, Compound IM-01 US10865384, Example 2f
BDBM476183 US10865384, Compound IM-01 US10865384, Example 2f BDBM688162 US20240246941, Compound SJ000311280 US20240246941, Compound SJ000311280-01
BDBM688162 US20240246941, Compound SJ000311280 US20240246941, Compound SJ000311280-01 BDBM688163 US20240246941, Compound SJ000311281 US20240246941, Compound SJ000311281-01
BDBM688163 US20240246941, Compound SJ000311281 US20240246941, Compound SJ000311281-01 US10954217, Example KTL-01-Table 3.A15 BDBM488133
US10954217, Example KTL-01-Table 3.A15 BDBM488133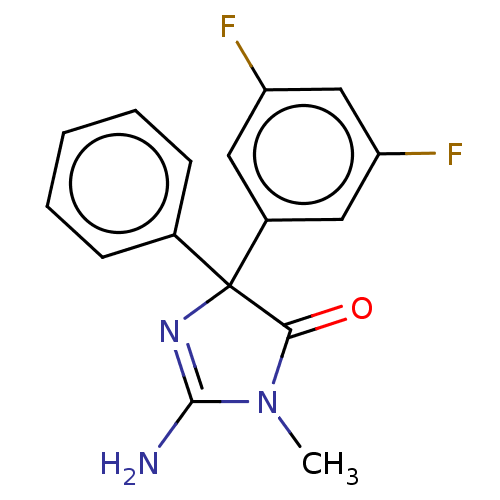 US11142505, FAH-01 BDBM512557 US11091444, Compound FAH-1
US11142505, FAH-01 BDBM512557 US11091444, Compound FAH-1 US20240246941, Compound SJ000311284-01 US20240246941, Compound SJ000311284 BDBM688164
US20240246941, Compound SJ000311284-01 US20240246941, Compound SJ000311284 BDBM688164 BDBM357739 AG-01-128/(I-202) US10214530, Example 18
BDBM357739 AG-01-128/(I-202) US10214530, Example 18 BDBM680895 US12012467, Example 134 US12012467, Compound ZBB-01-75
BDBM680895 US12012467, Example 134 US12012467, Compound ZBB-01-75 BDBM680922 US12012467, Example 154 US12012467, Compound ZBB-01-220
BDBM680922 US12012467, Example 154 US12012467, Compound ZBB-01-220 BDBM680923 US12012467, Example 155 US12012467, Compound ZBB-01-221
BDBM680923 US12012467, Example 155 US12012467, Compound ZBB-01-221 BDBM680924 US12012467, Example 156 US12012467, Compound ZBB-01-222
BDBM680924 US12012467, Example 156 US12012467, Compound ZBB-01-222 BDBM680925 US12012467, Compound ZBB-01-258 US12012467, Example 157
BDBM680925 US12012467, Compound ZBB-01-258 US12012467, Example 157 BDBM724208 US20250066338, Ref Comp B WO2018109607, Ex 4A-01
BDBM724208 US20250066338, Ref Comp B WO2018109607, Ex 4A-01 MW-01-148/(I-302) US10214530, Example 55 BDBM357788
MW-01-148/(I-302) US10214530, Example 55 BDBM357788 MW-01-157/(I-207) US10214530, Example 25 BDBM357736
MW-01-157/(I-207) US10214530, Example 25 BDBM357736 MW-01-162/(I-303) US10214530, Example 56 BDBM357789
MW-01-162/(I-303) US10214530, Example 56 BDBM357789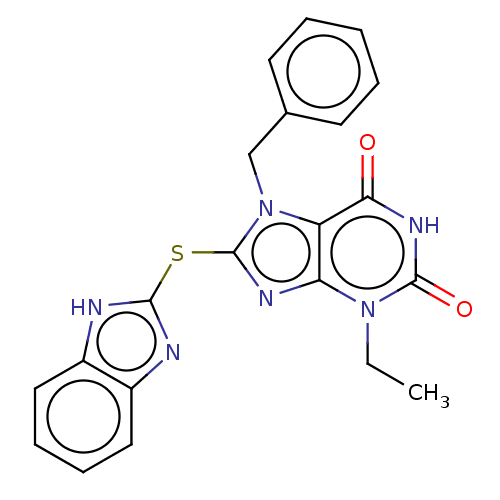 US10214530, Example 52 BDBM357743 MW-01-139/(I-46)
US10214530, Example 52 BDBM357743 MW-01-139/(I-46)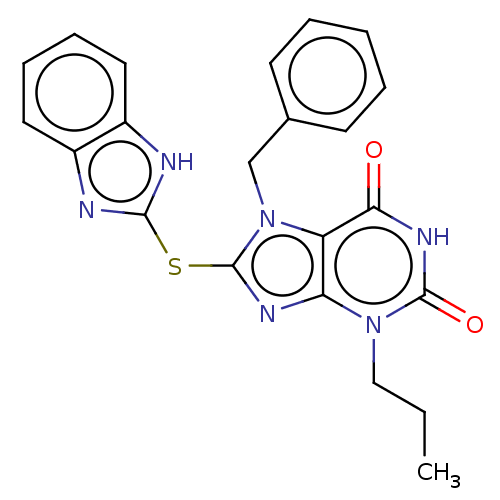 US10214530, Example 53 BDBM357741 MW-01-153/(I-47)
US10214530, Example 53 BDBM357741 MW-01-153/(I-47) US10508115, Compound Ia-01 US10508115, Compound Ia-05 BDBM423474
US10508115, Compound Ia-01 US10508115, Compound Ia-05 BDBM423474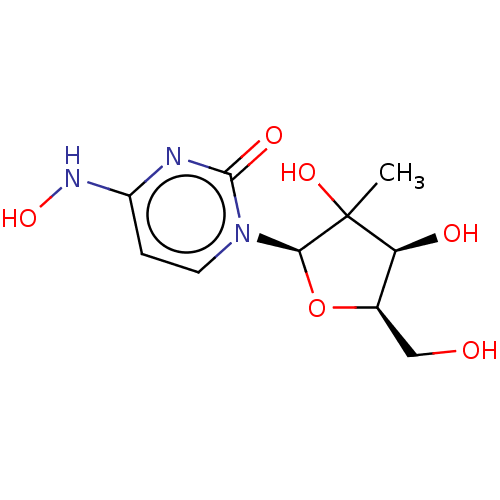 US10874683, Example EIDD-02053-01 BDBM476303 US10874683, Example 56.5
US10874683, Example EIDD-02053-01 BDBM476303 US10874683, Example 56.5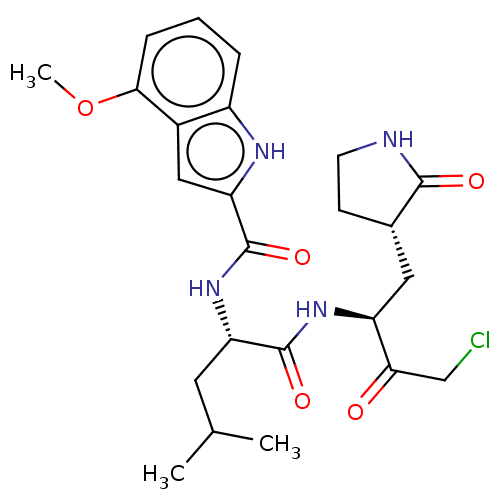 US11753373, Compound A-5-c WO2005113580-Ex-01 BDBM423408
US11753373, Compound A-5-c WO2005113580-Ex-01 BDBM423408 US12012467, Compound ZBB-01-160 BDBM680921 US12012467, Example 153
US12012467, Compound ZBB-01-160 BDBM680921 US12012467, Example 153 US12012467, Compound ZBB-01-161 BDBM680893 US12012467, Example 132
US12012467, Compound ZBB-01-161 BDBM680893 US12012467, Example 132 US12012467, Compound ZBB-01-73 BDBM680894 US12012467, Example 133
US12012467, Compound ZBB-01-73 BDBM680894 US12012467, Example 133 US12012467, Example 135 US12012467, Compound ZBB-01-111 BDBM680896
US12012467, Example 135 US12012467, Compound ZBB-01-111 BDBM680896 US12012467, Example 136 BDBM680897 US12012467, Compound ZBB-01-98
US12012467, Example 136 BDBM680897 US12012467, Compound ZBB-01-98 US20240246941, Compound SJ000311286-1 BDBM688095 US20240246941, Compound SJ000311286-01
US20240246941, Compound SJ000311286-1 BDBM688095 US20240246941, Compound SJ000311286-01 US20240279174, Compound JH-LPH-01 US20240279174, Compound AZ1 BDBM691401
US20240279174, Compound JH-LPH-01 US20240279174, Compound AZ1 BDBM691401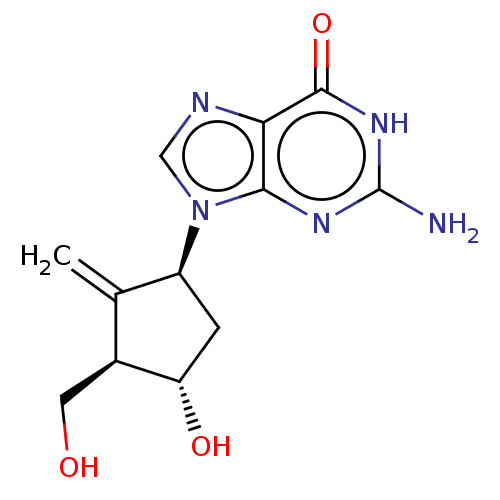 Baraclude Entecavir BDBM50248008 CHEBI:473990 BMS-200475-01 SQ-34676
Baraclude Entecavir BDBM50248008 CHEBI:473990 BMS-200475-01 SQ-34676 ERLOTINIB HYDROCHLORIDE Erlotinib OSI-774 CP-358774-01 BDBM50311470 CHEMBL1079742
ERLOTINIB HYDROCHLORIDE Erlotinib OSI-774 CP-358774-01 BDBM50311470 CHEMBL1079742 N-[4-(Benzyloxy)phenyl]imidodicarbonimidic diamide hydrochloride (B1-01) BDBM233025
N-[4-(Benzyloxy)phenyl]imidodicarbonimidic diamide hydrochloride (B1-01) BDBM233025 TLN-4601 BDBM50481798 Amo-01 Diazepinomicin ECO-4601 BU-4664L
TLN-4601 BDBM50481798 Amo-01 Diazepinomicin ECO-4601 BU-4664L US10774063, Compound TDRL-551 BDBM461287 US10774063, Compound NG-01-25
US10774063, Compound TDRL-551 BDBM461287 US10774063, Compound NG-01-25 US12012467, Compound ZBB-01-74-1 BDBM680920 US12012467, Example 152
US12012467, Compound ZBB-01-74-1 BDBM680920 US12012467, Example 152 BMS 708163 BMS-70816301 BMS-708163 BMS-708163-01 Avagacestat BDBM50458169
BMS 708163 BMS-70816301 BMS-708163 BMS-708163-01 Avagacestat BDBM50458169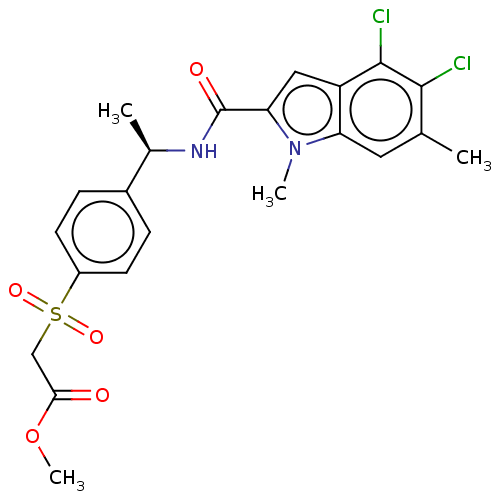 methyl 2-{4-[(1R)-1-[(4,5-dichloro-1,6-dimethyl-1H-indol-2- yl)formamido]ethyl]benzenesulfonyl}acetate US11304929, Example 01-001 US11304929, Example 01-022 BDBM549419
methyl 2-{4-[(1R)-1-[(4,5-dichloro-1,6-dimethyl-1H-indol-2- yl)formamido]ethyl]benzenesulfonyl}acetate US11304929, Example 01-001 US11304929, Example 01-022 BDBM549419 US11304929, Example 01-018 BDBM549067 US11304929, Example 01-024 propan-2-yl 2-{4-[(1R)-1-[(4-chloro-1,6-dimethyl-1H-indol-2- yl)formamido]ethyl]benzenesulfonyl}acetate
US11304929, Example 01-018 BDBM549067 US11304929, Example 01-024 propan-2-yl 2-{4-[(1R)-1-[(4-chloro-1,6-dimethyl-1H-indol-2- yl)formamido]ethyl]benzenesulfonyl}acetate 4-methoxy-2-(4-methoxyphenyl)quinazoline BRD-K05542495-001-01-1 BDBM65746 cid_946273
4-methoxy-2-(4-methoxyphenyl)quinazoline BRD-K05542495-001-01-1 BDBM65746 cid_946273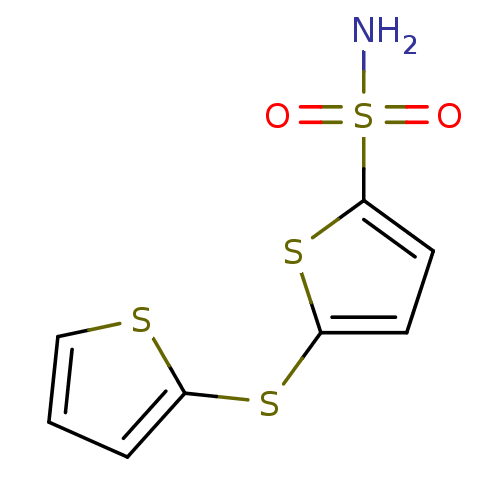 5-(thiophen-2-ylsulfanyl)thiophene-2-sulfonamide 0049-01 LeadQuest Compound 2 BDBM13068
5-(thiophen-2-ylsulfanyl)thiophene-2-sulfonamide 0049-01 LeadQuest Compound 2 BDBM13068 BDBM65740 BRD-K80942107-001-01-1 cid_1597037 4-phenoxy-2-(4-propoxyphenyl)quinazoline
BDBM65740 BRD-K80942107-001-01-1 cid_1597037 4-phenoxy-2-(4-propoxyphenyl)quinazoline cid_1634732 4-(4-methoxyphenoxy)-2-(4-propoxyphenyl)quinazoline BRD-K42568865-001-01-3 BDBM65744
cid_1634732 4-(4-methoxyphenoxy)-2-(4-propoxyphenyl)quinazoline BRD-K42568865-001-01-3 BDBM65744 cid_616867 4-methoxy-2-phenyl-quinazoline BRD-K31596841-001-01-6 4-methoxy-2-phenylquinazoline BDBM65743
cid_616867 4-methoxy-2-phenyl-quinazoline BRD-K31596841-001-01-6 4-methoxy-2-phenylquinazoline BDBM65743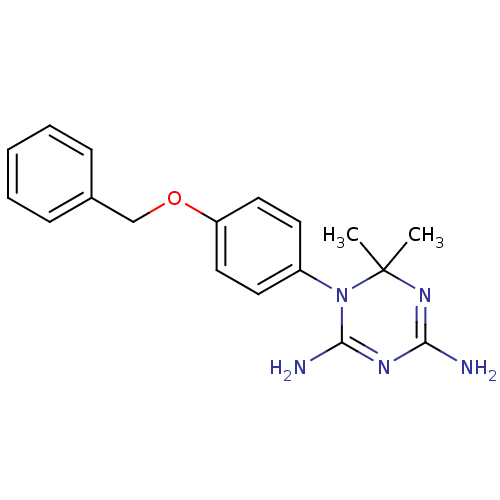 1-(4-(Benzyloxy)phenyl)-6,6-dimethyl-1,6-dihydro-1,3,5-triazine-2,4-diamine hydrochloride (B2-01) CHEMBL7191 BDBM50405043
1-(4-(Benzyloxy)phenyl)-6,6-dimethyl-1,6-dihydro-1,3,5-triazine-2,4-diamine hydrochloride (B2-01) CHEMBL7191 BDBM50405043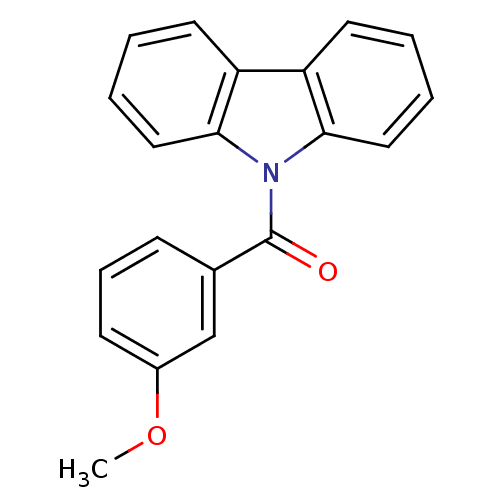 9-carbazolyl-(3-methoxyphenyl)methanone BRD-K58694191-001-01-3 carbazol-9-yl-(3-methoxyphenyl)methanone BDBM54882 cid_24102740
9-carbazolyl-(3-methoxyphenyl)methanone BRD-K58694191-001-01-3 carbazol-9-yl-(3-methoxyphenyl)methanone BDBM54882 cid_24102740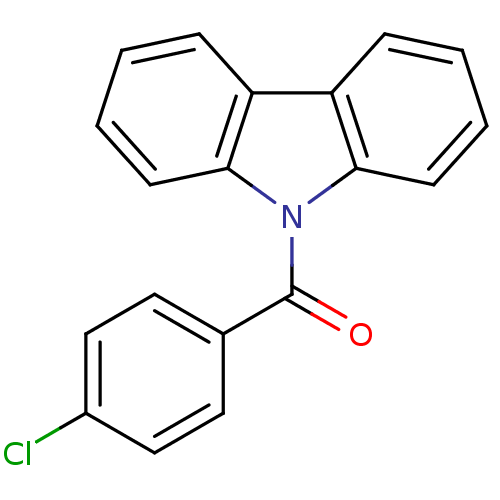 9-carbazolyl-(4-chlorophenyl)methanone BDBM54876 BRD-K47719676-001-01-6 carbazol-9-yl-(4-chlorophenyl)methanone cid_1848658
9-carbazolyl-(4-chlorophenyl)methanone BDBM54876 BRD-K47719676-001-01-6 carbazol-9-yl-(4-chlorophenyl)methanone cid_1848658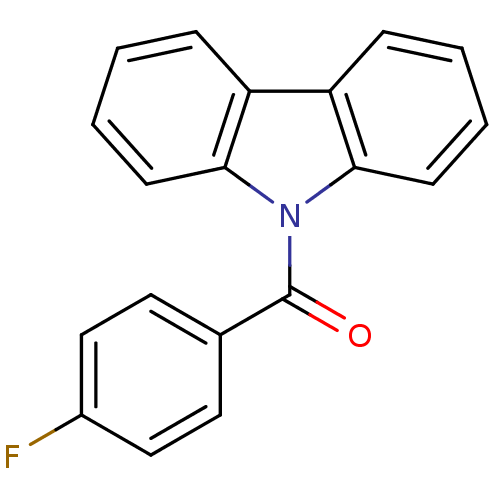 BDBM54865 BRD-K19704225-001-01-8 9-carbazolyl-(4-fluorophenyl)methanone carbazol-9-yl-(4-fluorophenyl)methanone cid_8852543
BDBM54865 BRD-K19704225-001-01-8 9-carbazolyl-(4-fluorophenyl)methanone carbazol-9-yl-(4-fluorophenyl)methanone cid_8852543 BDBM233018 N-{4-[2-(1,3-Dioxo-1,3-dihydro-2H-isoindol-2-yl)ethoxy]phenyl}imidodicarbonimidic diamide hydrochloride (A1-01)
BDBM233018 N-{4-[2-(1,3-Dioxo-1,3-dihydro-2H-isoindol-2-yl)ethoxy]phenyl}imidodicarbonimidic diamide hydrochloride (A1-01) US20240368133, Compound A-01 2-[[4-(1,3-benzo- thiazol-2-yl)piper- azin-1-yl]methyl]- benzoic acid BDBM703524
US20240368133, Compound A-01 2-[[4-(1,3-benzo- thiazol-2-yl)piper- azin-1-yl]methyl]- benzoic acid BDBM703524 BDBM13070 0114-01 LeadQuest Compound 4 4-(3-methyl-5-oxo-4,5-dihydro-1H-pyrazol-1-yl)benzene-1-sulfonamide
BDBM13070 0114-01 LeadQuest Compound 4 4-(3-methyl-5-oxo-4,5-dihydro-1H-pyrazol-1-yl)benzene-1-sulfonamide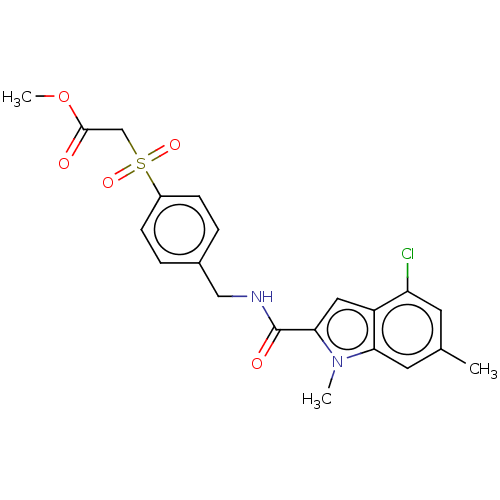 BDBM549522 methyl 2-(4-{[(4-chloro-1,6-dimethyl-1H-indol-2- yl)formamido]methyl}benzenesulfonyl)acetate US11304929, Example 01-030
BDBM549522 methyl 2-(4-{[(4-chloro-1,6-dimethyl-1H-indol-2- yl)formamido]methyl}benzenesulfonyl)acetate US11304929, Example 01-030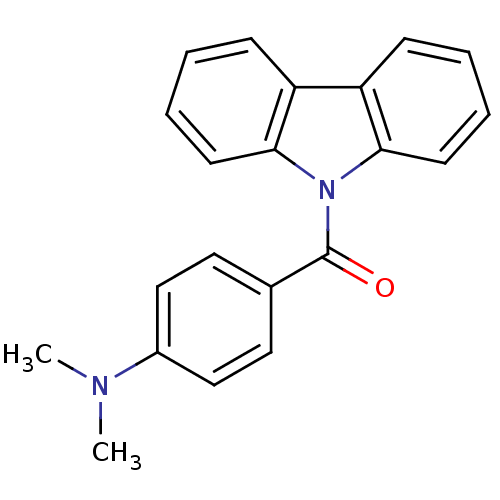 BRD-K84127109-001-01-3 cid_44247470 carbazol-9-yl-[4-(dimethylamino)phenyl]methanone 9-carbazolyl-[4-(dimethylamino)phenyl]methanone BDBM54893
BRD-K84127109-001-01-3 cid_44247470 carbazol-9-yl-[4-(dimethylamino)phenyl]methanone 9-carbazolyl-[4-(dimethylamino)phenyl]methanone BDBM54893 BRD-K85868893-001-01-1 cid_44247472 carbazol-9-yl-[2-(trifluoromethyl)phenyl]methanone 9-carbazolyl-[2-(trifluoromethyl)phenyl]methanone BDBM54894
BRD-K85868893-001-01-1 cid_44247472 carbazol-9-yl-[2-(trifluoromethyl)phenyl]methanone 9-carbazolyl-[2-(trifluoromethyl)phenyl]methanone BDBM54894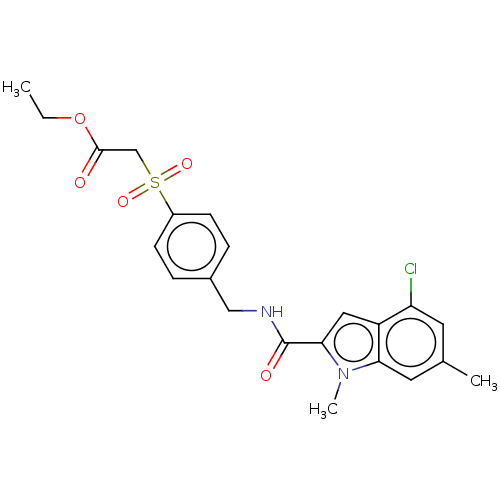 ethyl 2-(4-{[(4-chloro-1,6-dimethyl-1H-indol-2- yl)formamido]methyl}benzenesulfonyl)acetate BDBM549520 US11304929, Example 01-032
ethyl 2-(4-{[(4-chloro-1,6-dimethyl-1H-indol-2- yl)formamido]methyl}benzenesulfonyl)acetate BDBM549520 US11304929, Example 01-032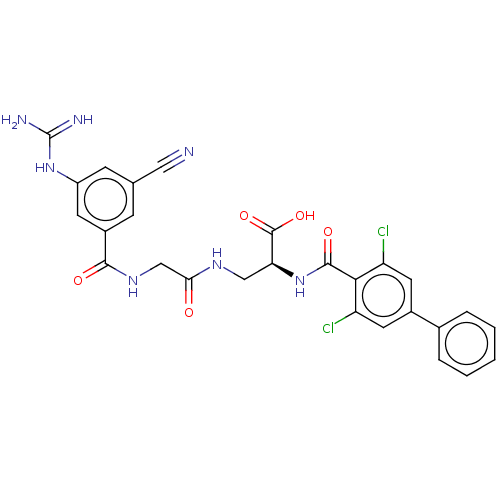 (S)-3-(2-(3-cyano-5 guanidinobenzamido)acetamido)-2-(3,5-dichlorobiphenyl-4-ylcarboxamido)propanoic acid BDBM585088 US11530180, Compound SU15210-0205-01
(S)-3-(2-(3-cyano-5 guanidinobenzamido)acetamido)-2-(3,5-dichlorobiphenyl-4-ylcarboxamido)propanoic acid BDBM585088 US11530180, Compound SU15210-0205-01 BDBM233021 2-(2-(4-(4,6-Diamino-2,2-dimethyl-1,3,5-triazin-1(2H)-yl)phenoxy)ethyl)isoindoline-1,3-dione hydrochloride (A2-01)
BDBM233021 2-(2-(4-(4,6-Diamino-2,2-dimethyl-1,3,5-triazin-1(2H)-yl)phenoxy)ethyl)isoindoline-1,3-dione hydrochloride (A2-01)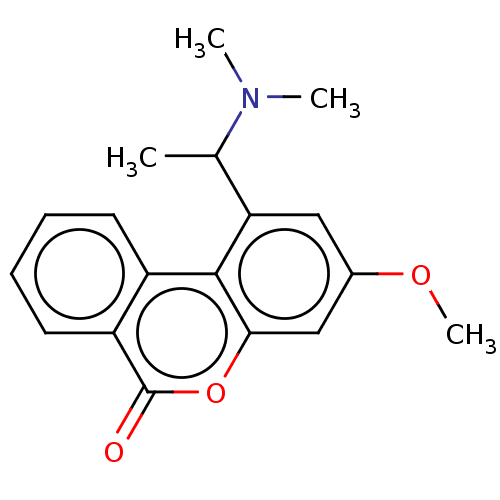 BDBM294222 US9586925, Compound FAR-01-1 US9586925, EXAMPLE XIII (-)-1-(1-(dimethylamino)ethyl)-3-methoxy-6H-benzo[c]chromen-6-one
BDBM294222 US9586925, Compound FAR-01-1 US9586925, EXAMPLE XIII (-)-1-(1-(dimethylamino)ethyl)-3-methoxy-6H-benzo[c]chromen-6-one BDBM294224 (-)-1-(1-(dimethylamino)ethyl)-3-hydroxy-6H-benzo[c]chromen-6-one US9586925, Compound FAR-01-1OH US9586925, EXAMPLE XVII
BDBM294224 (-)-1-(1-(dimethylamino)ethyl)-3-hydroxy-6H-benzo[c]chromen-6-one US9586925, Compound FAR-01-1OH US9586925, EXAMPLE XVII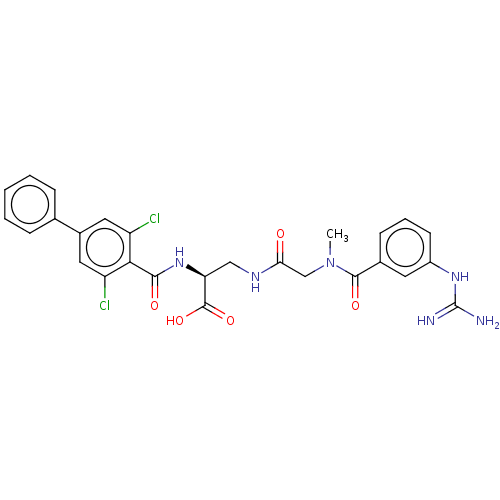 BDBM585086 (S)-2-(3,5-dichlorobiphenyl-4-ylcarboxamido)-3-(2-(3 guanidino-N-methylbenzamido) acetamido)propanoic acid US11530180, Compound SU15210-0202-01
BDBM585086 (S)-2-(3,5-dichlorobiphenyl-4-ylcarboxamido)-3-(2-(3 guanidino-N-methylbenzamido) acetamido)propanoic acid US11530180, Compound SU15210-0202-01 US9586925, EXAMPLE XI BDBM294221 US9586925, Compound FAR-01-2 (-)-3-(1-(dimethylamino)ethyl)-1-methoxy-6H-benzo[c]chromen-6-one
US9586925, EXAMPLE XI BDBM294221 US9586925, Compound FAR-01-2 (-)-3-(1-(dimethylamino)ethyl)-1-methoxy-6H-benzo[c]chromen-6-one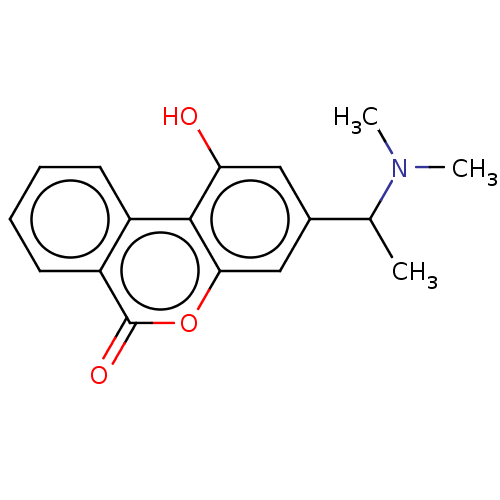 US9586925, EXAMPLE XV BDBM294223 (-)-3-(1-(dimethylamino)ethyl)-1-hydroxy-6H-benzo[c]chromen-6-one US9586925, Compound FAR-01-2OH
US9586925, EXAMPLE XV BDBM294223 (-)-3-(1-(dimethylamino)ethyl)-1-hydroxy-6H-benzo[c]chromen-6-one US9586925, Compound FAR-01-2OH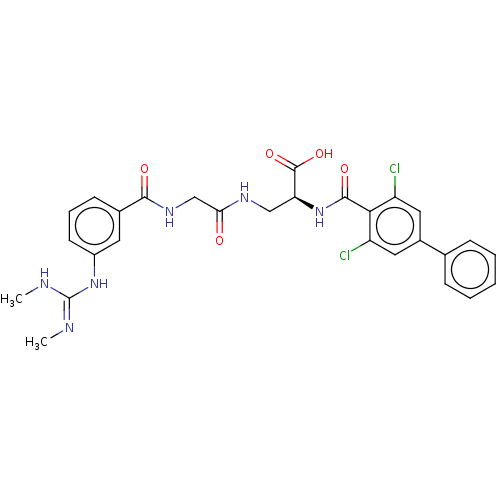 (S,E)-2-(3,5-dichlorobiphenyl-4-ylcarboxamido)-3-(2-(3-(2,3-dimethylguanidino)benzamido)acetamido)propanoic acid US11530180, Compound SU15210-0193-01 BDBM585084
(S,E)-2-(3,5-dichlorobiphenyl-4-ylcarboxamido)-3-(2-(3-(2,3-dimethylguanidino)benzamido)acetamido)propanoic acid US11530180, Compound SU15210-0193-01 BDBM585084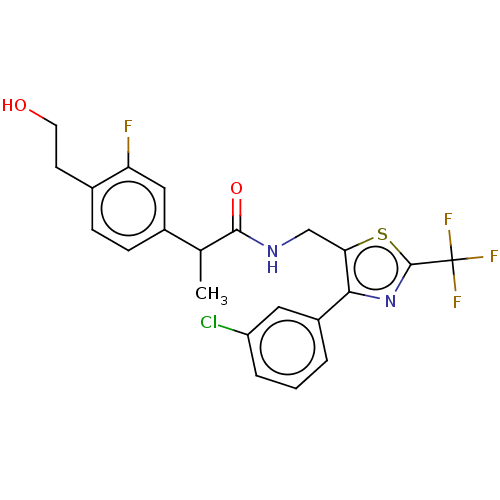 BDBM342115 N-((4-(3-chlorophenyl)-2-(trifluoromethyl)thiazol-5-yl)methyl)-2-(3-fluoro-4-(2-hydroxyethyl)phenyl)-propanamide US9771359, EX-01
BDBM342115 N-((4-(3-chlorophenyl)-2-(trifluoromethyl)thiazol-5-yl)methyl)-2-(3-fluoro-4-(2-hydroxyethyl)phenyl)-propanamide US9771359, EX-01 BDBM54862 4-[9-carbazolyl(oxo)methyl]benzonitrile 4-carbazol-9-ylcarbonylbenzenecarbonitrile BRD-K11704073-001-01-9 4-(carbazole-9-carbonyl)benzonitrile cid_44247467
BDBM54862 4-[9-carbazolyl(oxo)methyl]benzonitrile 4-carbazol-9-ylcarbonylbenzenecarbonitrile BRD-K11704073-001-01-9 4-(carbazole-9-carbonyl)benzonitrile cid_44247467 BDBM549448 US11304929, Example 01-010 methyl 2-{4-[(1R)-1-({3,5-dimethyl-3H-benzo[e]indol-2- yl}formamido)ethyl]benzenesulfonyl}acetate
BDBM549448 US11304929, Example 01-010 methyl 2-{4-[(1R)-1-({3,5-dimethyl-3H-benzo[e]indol-2- yl}formamido)ethyl]benzenesulfonyl}acetate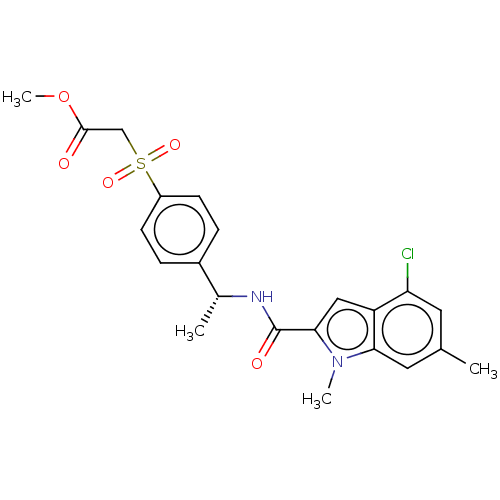 BDBM549458 US11304929, Example 01-012 methyl 2-{4-[(1R)-1-[(4-chloro-1,6-dimethyl-1H-indol-2- yl)formamido]ethyl]benzenesulfonyl}acetate
BDBM549458 US11304929, Example 01-012 methyl 2-{4-[(1R)-1-[(4-chloro-1,6-dimethyl-1H-indol-2- yl)formamido]ethyl]benzenesulfonyl}acetate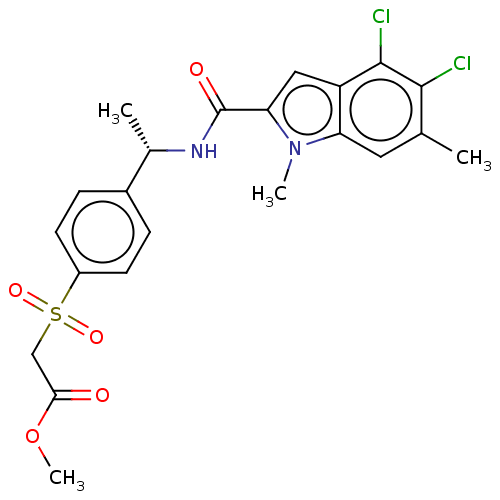 BDBM549466 US11304929, Example 01-017 methyl 2-{4-[(1S)-1-[(4,5-dichloro-1,6-dimethyl-1H-indol-2- yl)formamido]ethyl]benzenesulfonyl}acetate
BDBM549466 US11304929, Example 01-017 methyl 2-{4-[(1S)-1-[(4,5-dichloro-1,6-dimethyl-1H-indol-2- yl)formamido]ethyl]benzenesulfonyl}acetate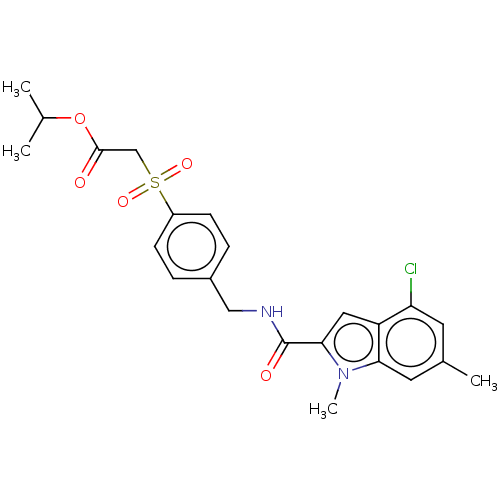 BDBM549519 US11304929, Example 01-031 propan-2-yl 2-(4-{[(4-chloro-1,6-dimethyl-1H-indol-2- yl)formamido]methyl}benzenesulfonyl)acetate
BDBM549519 US11304929, Example 01-031 propan-2-yl 2-(4-{[(4-chloro-1,6-dimethyl-1H-indol-2- yl)formamido]methyl}benzenesulfonyl)acetate US11304929, Example 01-015 BDBM549459 methyl 2-{4-[(1R)-1-[(4-chloro-1,6,7-trimethyl-1H-indol-2- yl)formamido]ethyl]benzenesulfonyl}acetate
US11304929, Example 01-015 BDBM549459 methyl 2-{4-[(1R)-1-[(4-chloro-1,6,7-trimethyl-1H-indol-2- yl)formamido]ethyl]benzenesulfonyl}acetate US11304929, Example 01-019 BDBM549488 methyl 2-{4-[(1R)-1-[(5-chloro-1-methyl-1H-indol-2- yl)formamido]ethyl]benzenesulfonyl}acetate
US11304929, Example 01-019 BDBM549488 methyl 2-{4-[(1R)-1-[(5-chloro-1-methyl-1H-indol-2- yl)formamido]ethyl]benzenesulfonyl}acetate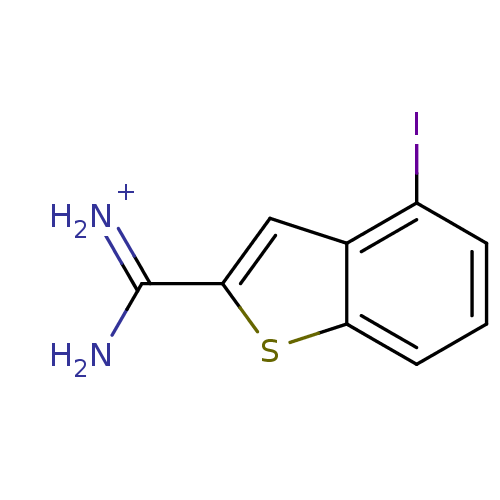 [amino(4-iodo-1-benzothiophen-2-yl)methylidene]azanium APC-6860 4-Iodobenzo[b]thiophene-2-carboxamidine BDBM14169 CRA-6860 CA-01
[amino(4-iodo-1-benzothiophen-2-yl)methylidene]azanium APC-6860 4-Iodobenzo[b]thiophene-2-carboxamidine BDBM14169 CRA-6860 CA-01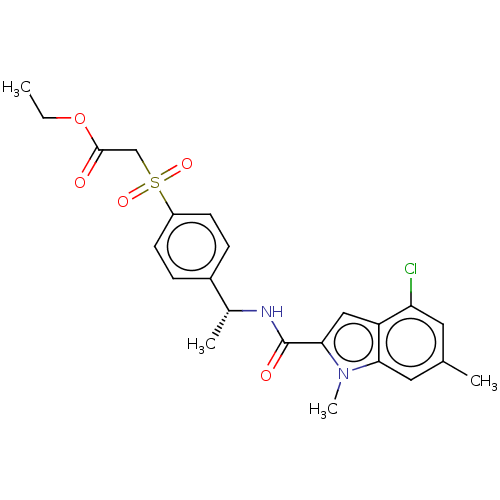 ethyl 2-{4-[(1R)-1-[(4-chloro-1,6-dimethyl-1H-indol-2- yl)formamido]ethyl]benzenesulfonyl}acetate BDBM549430 US11304929, Example 01-004
ethyl 2-{4-[(1R)-1-[(4-chloro-1,6-dimethyl-1H-indol-2- yl)formamido]ethyl]benzenesulfonyl}acetate BDBM549430 US11304929, Example 01-004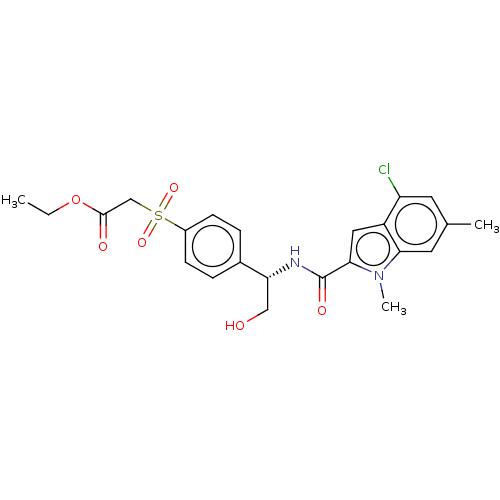 BDBM549432 ethyl 2-{4-[(1S)-1-[(4-chloro-1,6-dimethyl-1H-indol-2-yl)formamido]-2- hydroxyethyl]benzenesulfonyl}acetate US11304929, Example 01-005
BDBM549432 ethyl 2-{4-[(1S)-1-[(4-chloro-1,6-dimethyl-1H-indol-2-yl)formamido]-2- hydroxyethyl]benzenesulfonyl}acetate US11304929, Example 01-005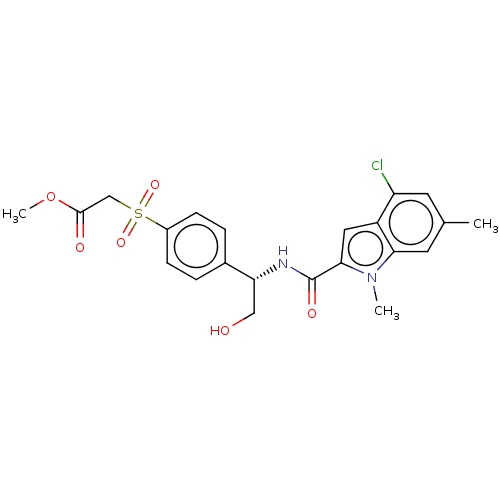 BDBM549454 US11304929, Example 01-013 methyl 2-{4-[(1S)-1-[(4-chloro-1,6-dimethyl-1H-indol-2-yl)formamido]-2- hydroxyethyl]benzenesulfonyl}acetate
BDBM549454 US11304929, Example 01-013 methyl 2-{4-[(1S)-1-[(4-chloro-1,6-dimethyl-1H-indol-2-yl)formamido]-2- hydroxyethyl]benzenesulfonyl}acetate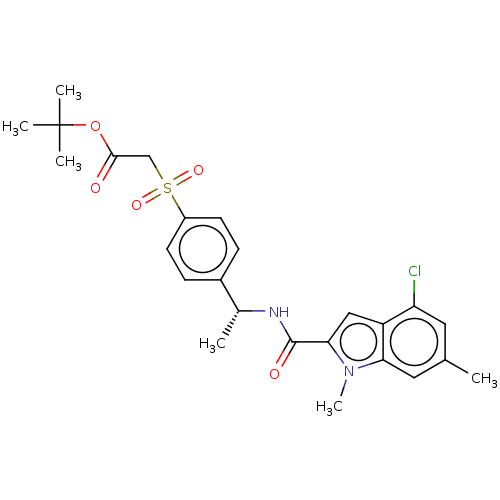 BDBM549455 US11304929, Example 01-014 tert-butyl 2-{4-[(1R)-1-[(4-chloro-1,6-dimethyl-1H-indol-2- yl)formamido]ethyl]benzenesulfonyl}acetate
BDBM549455 US11304929, Example 01-014 tert-butyl 2-{4-[(1R)-1-[(4-chloro-1,6-dimethyl-1H-indol-2- yl)formamido]ethyl]benzenesulfonyl}acetate BDBM549481 US11304929, Example 01-022 (13C)methyl 2-{4-[(1R)-1-[(4,5-dichloro-1,6-dimethyl-1H-indol-2- yl)formamido]ethyl]benzenesulfonyl}acetate
BDBM549481 US11304929, Example 01-022 (13C)methyl 2-{4-[(1R)-1-[(4,5-dichloro-1,6-dimethyl-1H-indol-2- yl)formamido]ethyl]benzenesulfonyl}acetate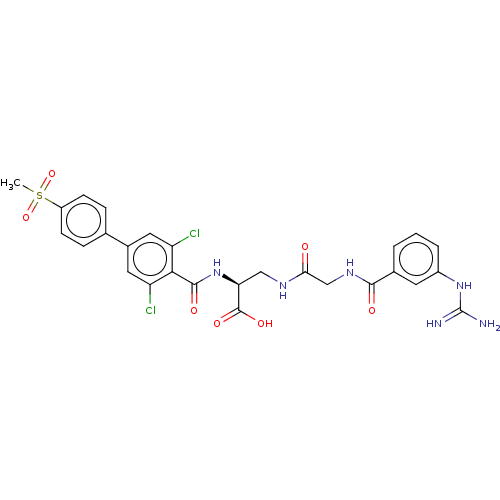 BDBM585079 US11530180, Compound SU15210-0110-01 (S)-2-(3,5-dichloro-4′-(methylsulfonyl)biphenyl-4-ylcarboxamido)-3-(2-(3 guanidinobenzamido)acetamido)propanoic acid
BDBM585079 US11530180, Compound SU15210-0110-01 (S)-2-(3,5-dichloro-4′-(methylsulfonyl)biphenyl-4-ylcarboxamido)-3-(2-(3 guanidinobenzamido)acetamido)propanoic acid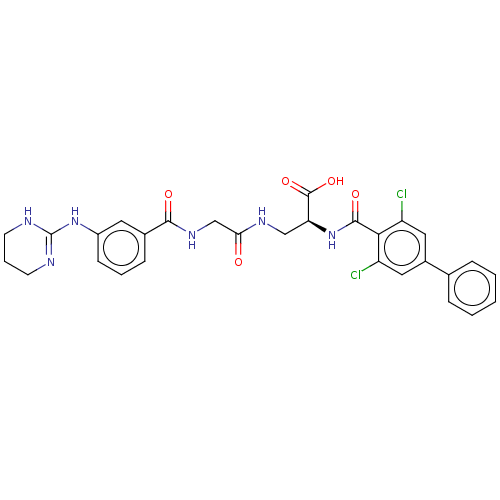 BDBM585080 US11530180, Compound SU15210-0147-01 (S)-2-(3,5-dichlorobiphenyl-4-ylcarboxamido)-3-(2-(3-(1,4,5,6-tetrahydropyrimidin-2-ylamino)benzamido)acetamido)propanoic acid
BDBM585080 US11530180, Compound SU15210-0147-01 (S)-2-(3,5-dichlorobiphenyl-4-ylcarboxamido)-3-(2-(3-(1,4,5,6-tetrahydropyrimidin-2-ylamino)benzamido)acetamido)propanoic acid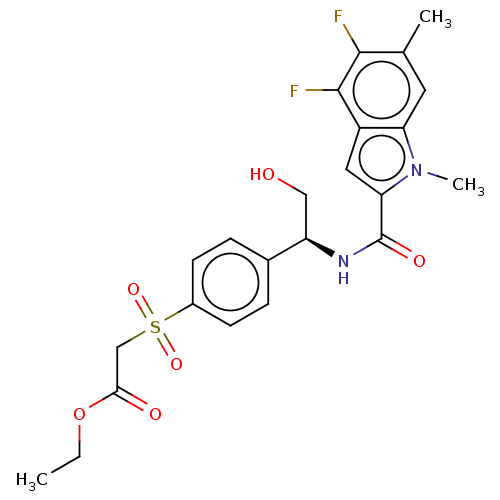 US11304929, Example 01-028 ethyl 2-{4-[(1S)-1-[(4,5-difluoro-1,6-dimethyl-1H-indol-2-yl)formamido]-2- hydroxyethyl]benzenesulfonyl}acetate BDBM549391
US11304929, Example 01-028 ethyl 2-{4-[(1S)-1-[(4,5-difluoro-1,6-dimethyl-1H-indol-2-yl)formamido]-2- hydroxyethyl]benzenesulfonyl}acetate BDBM549391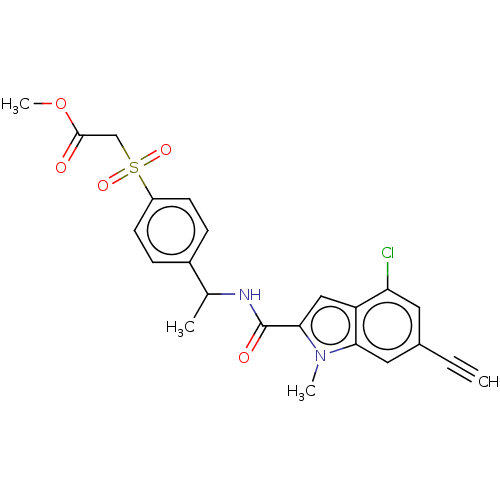 methyl 2-(4-{1-[(4-chloro-6-ethynyl-1-methyl-1H-indol-2- yl)formamido]ethyl}benzenesulfonyl)acetate BDBM549440 US11304929, Example 01-008
methyl 2-(4-{1-[(4-chloro-6-ethynyl-1-methyl-1H-indol-2- yl)formamido]ethyl}benzenesulfonyl)acetate BDBM549440 US11304929, Example 01-008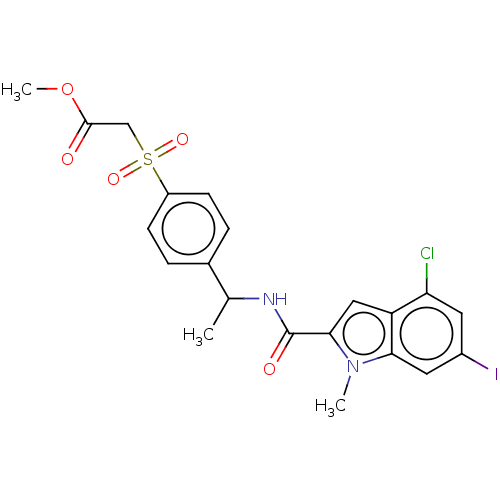 methyl 2-(4-{1-[(4-chloro-6-iodo-1-methyl-1H-indol-2- yl)formamido]ethyl}benzenesulfonyl)acetate BDBM549478 US11304929, Example 01-020
methyl 2-(4-{1-[(4-chloro-6-iodo-1-methyl-1H-indol-2- yl)formamido]ethyl}benzenesulfonyl)acetate BDBM549478 US11304929, Example 01-020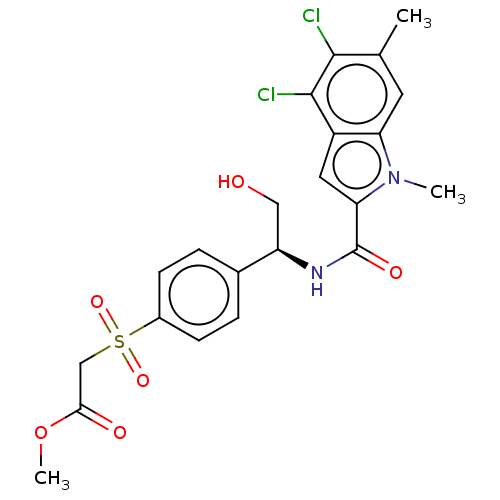 methyl 2-{4-[(1S)-1-[(4,5-dichloro-1,6-dimethyl-1H-indol-2-yl)formamido]-2- hydroxyethyl]benzenesulfonyl}acetate US11304929, Example 01-007 BDBM549438
methyl 2-{4-[(1S)-1-[(4,5-dichloro-1,6-dimethyl-1H-indol-2-yl)formamido]-2- hydroxyethyl]benzenesulfonyl}acetate US11304929, Example 01-007 BDBM549438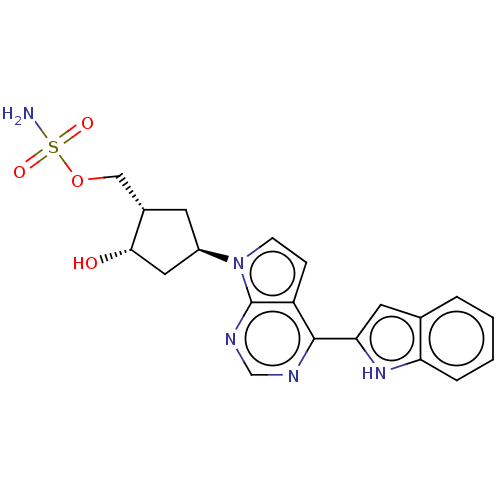 ((1S,2S,4R)-4-(4-(1H-indol-2-yl)-7H-pyrrolo[2,3- d]pyrimidin-7-yl)-2-hydroxycyclopentyl)methyl sulfamate US9593121, 1-01 BDBM299236
((1S,2S,4R)-4-(4-(1H-indol-2-yl)-7H-pyrrolo[2,3- d]pyrimidin-7-yl)-2-hydroxycyclopentyl)methyl sulfamate US9593121, 1-01 BDBM299236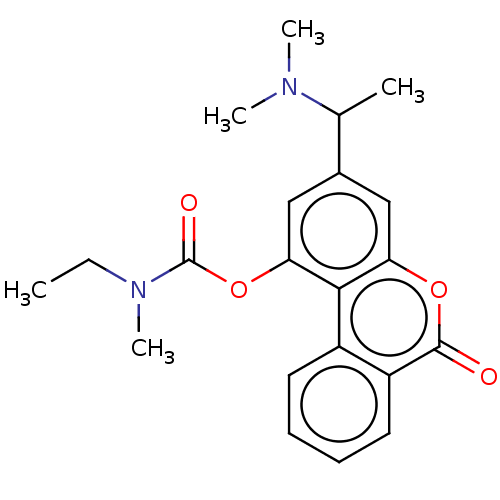 BDBM294225 US9586925, EXAMPLE XIX US9586925, Compound FAR-01-2CA (-)-3-(1-(dimethylamino)ethyl)-6-oxo-6H-benzo[c]chromen-1-yl ethyl(methyl)carbamate
BDBM294225 US9586925, EXAMPLE XIX US9586925, Compound FAR-01-2CA (-)-3-(1-(dimethylamino)ethyl)-6-oxo-6H-benzo[c]chromen-1-yl ethyl(methyl)carbamate BDBM294226 US9586925, EXAMPLE XXI US9586925, Compound FAR-01-1CA (-)-1-(1-(dimethylamino)ethyl)-6-oxo-6H-benzo[c]chromen-3-yl ethyl(methyl)carbamate
BDBM294226 US9586925, EXAMPLE XXI US9586925, Compound FAR-01-1CA (-)-1-(1-(dimethylamino)ethyl)-6-oxo-6H-benzo[c]chromen-3-yl ethyl(methyl)carbamate DN-18001AF APO-066 L1 PL-1 CHEBI:68554 Deferiprone CP-20 BDBM50525976 Ferriprox L-1 CP20 PL1 DN-180-01-AF APO-66
DN-18001AF APO-066 L1 PL-1 CHEBI:68554 Deferiprone CP-20 BDBM50525976 Ferriprox L-1 CP20 PL1 DN-180-01-AF APO-66 US11304929, Example 01-024 BDBM549489 propan-2-yl 2-{4-[(1R)-1-[(4-chloro-1,6-dimethyl-1H-indol-2- yl)formamido]ethyl]benzenesulfonyl}acetate
US11304929, Example 01-024 BDBM549489 propan-2-yl 2-{4-[(1R)-1-[(4-chloro-1,6-dimethyl-1H-indol-2- yl)formamido]ethyl]benzenesulfonyl}acetate US11304929, Example 01-037 BDBM549139 propan-2-yl 2-{4-[(1R)-1-[(4,5-dichloro-1,6-dimethyl-1H-indol-2- yl)formamido]ethyl]benzenesulfonyl}acetate
US11304929, Example 01-037 BDBM549139 propan-2-yl 2-{4-[(1R)-1-[(4,5-dichloro-1,6-dimethyl-1H-indol-2- yl)formamido]ethyl]benzenesulfonyl}acetate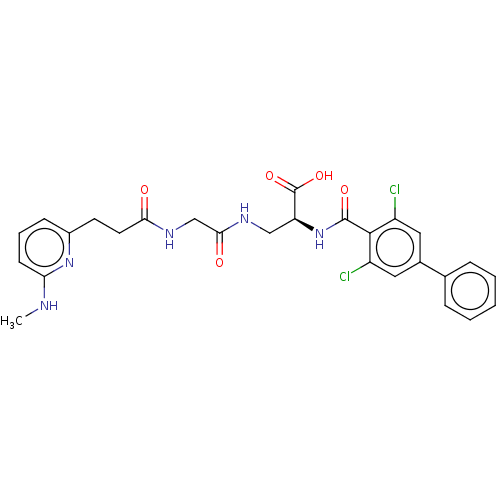 US11530180, Compound SU15210-0152-01 (S)-2-(3,5-dichlorobiphenyl-4-ylcarboxamido)-3-(2-(3-(6-(methylamino)pyridin-2-yl)propanamido)acetamido)propanoic acid BDBM585083
US11530180, Compound SU15210-0152-01 (S)-2-(3,5-dichlorobiphenyl-4-ylcarboxamido)-3-(2-(3-(6-(methylamino)pyridin-2-yl)propanamido)acetamido)propanoic acid BDBM585083 (2S)-2-(2,6-dichloro-4-morpholinobenzamido)-3-(2-(3-(5-fluoro-1,4,5,6-tetrahydropyrimidin-2-ylamino)benzamido)acetamido)propanoic acid US11530180, Compound SU15210-0248-01 BDBM585093
(2S)-2-(2,6-dichloro-4-morpholinobenzamido)-3-(2-(3-(5-fluoro-1,4,5,6-tetrahydropyrimidin-2-ylamino)benzamido)acetamido)propanoic acid US11530180, Compound SU15210-0248-01 BDBM585093 2-((4-(6-(Benzyloxy)pyridin-2-yl)piperidin-1-yl)methyl)-1-methyl-1H-benzo[d]imidazole-6-carboxylic acid US10208019, Example 9A-01 BDBM349716
2-((4-(6-(Benzyloxy)pyridin-2-yl)piperidin-1-yl)methyl)-1-methyl-1H-benzo[d]imidazole-6-carboxylic acid US10208019, Example 9A-01 BDBM349716 BDBM549461 US11304929, Example 01-016 ethyl 2-{4-[(1R)-1-({4,5-dichloro-1,6-dimethyl-1H-pyrrolo[2,3-b]pyridin-2- yl}formamido)ethyl]benzenesulfonyl}acetate
BDBM549461 US11304929, Example 01-016 ethyl 2-{4-[(1R)-1-({4,5-dichloro-1,6-dimethyl-1H-pyrrolo[2,3-b]pyridin-2- yl}formamido)ethyl]benzenesulfonyl}acetate BDBM549475 US11304929, Example 01-021 propan-2-yl 2-{4-[(1S)-1-[(4,5-dichloro-1,6-dimethyl-1H-indol-2-yl)formamido]- 2-hydroxyethyl]benzenesulfonyl}acetate
BDBM549475 US11304929, Example 01-021 propan-2-yl 2-{4-[(1S)-1-[(4,5-dichloro-1,6-dimethyl-1H-indol-2-yl)formamido]- 2-hydroxyethyl]benzenesulfonyl}acetate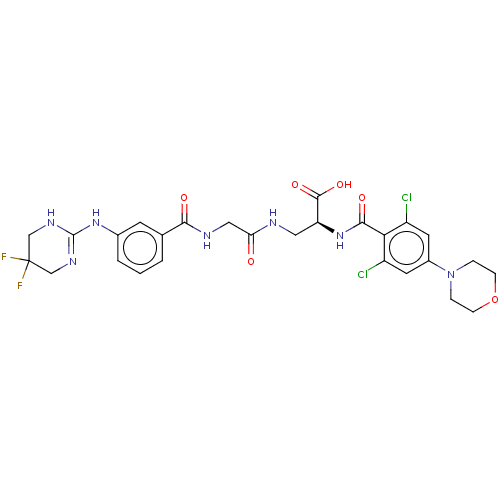 BDBM585096 US11530180, Compound SU15210-0271-01 (S)-2-(2,6-dichloro-4-morpholinobenzamido)-3-(2-(3-(5,5-difluoro-1,4,5,6-tetrahydropyrimidin-2-ylamino)benzamido)acetamido)propanoic acid
BDBM585096 US11530180, Compound SU15210-0271-01 (S)-2-(2,6-dichloro-4-morpholinobenzamido)-3-(2-(3-(5,5-difluoro-1,4,5,6-tetrahydropyrimidin-2-ylamino)benzamido)acetamido)propanoic acid US11304929, Example 01-029 methyl 2-({6-[(1R)-1-[(4-chloro-1,6-dimethyl-1H-indol-2- yl)formamido]ethyl]pyridin-3-yl}sulfonyl)acetate BDBM549507
US11304929, Example 01-029 methyl 2-({6-[(1R)-1-[(4-chloro-1,6-dimethyl-1H-indol-2- yl)formamido]ethyl]pyridin-3-yl}sulfonyl)acetate BDBM549507 US11304929, Example 01-038 propan-2-yl 2-{4-[(1S)-1-[(4-chloro-1,6-dimethyl-1H-indol-2-yl)formamido]-2- hydroxyethyl]benzenesulfonyl}acetate BDBM549349
US11304929, Example 01-038 propan-2-yl 2-{4-[(1S)-1-[(4-chloro-1,6-dimethyl-1H-indol-2-yl)formamido]-2- hydroxyethyl]benzenesulfonyl}acetate BDBM549349 US11304929, Example 01-039 BDBM549370 propan-2-yl 2-{4-[(1S)-1-[(4,5-difluoro-1,6-dimethyl-1H-indol-2-yl)formamido]-2- hydroxyethyl]benzenesulfonyl}acetate
US11304929, Example 01-039 BDBM549370 propan-2-yl 2-{4-[(1S)-1-[(4,5-difluoro-1,6-dimethyl-1H-indol-2-yl)formamido]-2- hydroxyethyl]benzenesulfonyl}acetate cis-3-(5-cyclopropoxy-3- methylpyridin-2-yl)-8- (3,5-difluorophenyl)-8- (dimethylamino)-1,3- diazaspiro[4.5]decan-2- one BDBM706933 US20240400537, Example EX-I-01
cis-3-(5-cyclopropoxy-3- methylpyridin-2-yl)-8- (3,5-difluorophenyl)-8- (dimethylamino)-1,3- diazaspiro[4.5]decan-2- one BDBM706933 US20240400537, Example EX-I-01 methyl 2-{4-[(1R)-1-({4,5-dichloro-1,6-dimethyl-1H-pyrrolo[2,3-b]pyridin-2- yl}formamido)ethyl]benzenesulfonyl}acetate US11304929, Example 01-006 BDBM549437
methyl 2-{4-[(1R)-1-({4,5-dichloro-1,6-dimethyl-1H-pyrrolo[2,3-b]pyridin-2- yl}formamido)ethyl]benzenesulfonyl}acetate US11304929, Example 01-006 BDBM549437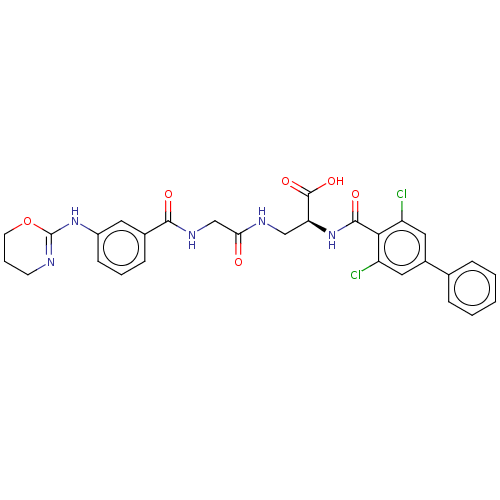 (S)-2-(3,5-dichlorobiphenyl-4-ylcarboxamido)-3-(2-(3-(5,6-dihydro-4H-1,3-oxazin-2-ylamino)benzamido)acetamido)propanoic acid BDBM585087 US11530180, Compound SU15210-0203-01
(S)-2-(3,5-dichlorobiphenyl-4-ylcarboxamido)-3-(2-(3-(5,6-dihydro-4H-1,3-oxazin-2-ylamino)benzamido)acetamido)propanoic acid BDBM585087 US11530180, Compound SU15210-0203-01 BDBM549485 US11304929, Example 01-023 methyl 2-{4-[(1S)-1-({4,5-dichloro-1,6-dimethyl-1H-pyrrolo[2,3-b]pyridin-2- yl}formamido)-2-hydroxyethyl]benzenesulfonyl}acetate
BDBM549485 US11304929, Example 01-023 methyl 2-{4-[(1S)-1-({4,5-dichloro-1,6-dimethyl-1H-pyrrolo[2,3-b]pyridin-2- yl}formamido)-2-hydroxyethyl]benzenesulfonyl}acetate N-(5-(2-((((1S,2S)-2-Hydroxycyclopentyl)oxy)methyl)pyrimidin-5-yl)-4-methoxypyrazolo[1,5-a]pyridin-2-yl)cyclopropanecarboxamide BDBM647610 US20240025893, Example e-04-01
N-(5-(2-((((1S,2S)-2-Hydroxycyclopentyl)oxy)methyl)pyrimidin-5-yl)-4-methoxypyrazolo[1,5-a]pyridin-2-yl)cyclopropanecarboxamide BDBM647610 US20240025893, Example e-04-01 US11304929, Example 01-033 BDBM549051 propan-2-yl 2-{4-[(1R)-1-[(4-chloro-5-fluoro-1,6-dimethyl-1H-indol-2- yl)formamido]ethyl]benzenesulfonyl}acetate
US11304929, Example 01-033 BDBM549051 propan-2-yl 2-{4-[(1R)-1-[(4-chloro-5-fluoro-1,6-dimethyl-1H-indol-2- yl)formamido]ethyl]benzenesulfonyl}acetate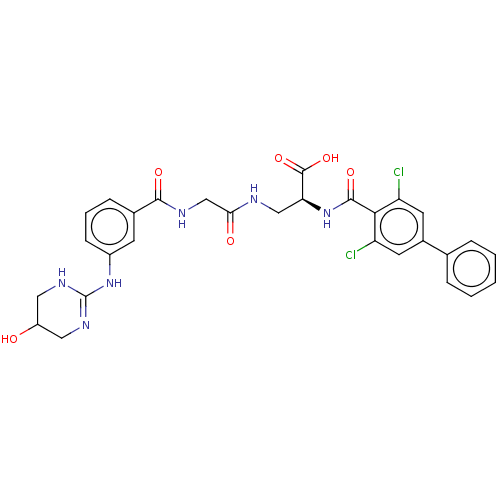 US11530180, Compound SU15210-0150-01 (2S)-2-(3,5-dichlorobiphenyl-4-yl carboxamido)-3-(243-(5-hydroxy-1,4,5,6-tetrahydropyrimidin-2-yl amino)benzamido)acetamido)propanoic acid BDBM585081
US11530180, Compound SU15210-0150-01 (2S)-2-(3,5-dichlorobiphenyl-4-yl carboxamido)-3-(243-(5-hydroxy-1,4,5,6-tetrahydropyrimidin-2-yl amino)benzamido)acetamido)propanoic acid BDBM585081 carbazol-9-yl-[4-(trifluoromethoxy)phenyl]methanone 9-carbazolyl-[4-(trifluoromethoxy)phenyl]methanone carbazol-9-yl-[4-(trifluoromethyloxy)phenyl]methanone BRD-K83315615-001-01-2 BDBM54892 cid_44247468
carbazol-9-yl-[4-(trifluoromethoxy)phenyl]methanone 9-carbazolyl-[4-(trifluoromethoxy)phenyl]methanone carbazol-9-yl-[4-(trifluoromethyloxy)phenyl]methanone BRD-K83315615-001-01-2 BDBM54892 cid_44247468 propan-2-yl 2-{4-[(1R)-1-({4-chloro-1,6-dimethyl-1H-pyrrolo[2,3-b]pyridin-2- yl}formamido)ethyl]benzenesulfonyl}acetate US11304929, Example 01-036 BDBM543835
propan-2-yl 2-{4-[(1R)-1-({4-chloro-1,6-dimethyl-1H-pyrrolo[2,3-b]pyridin-2- yl}formamido)ethyl]benzenesulfonyl}acetate US11304929, Example 01-036 BDBM543835 propan-2-yl 2-{4-[(1S)-1-[(4-chloro-5-fluoro-1,6-dimethyl-1H-indol-2- yl)formamido]-2-hydroxyethyl]benzenesulfonyl}acetate BDBM549112 US11304929, Example 01-035
propan-2-yl 2-{4-[(1S)-1-[(4-chloro-5-fluoro-1,6-dimethyl-1H-indol-2- yl)formamido]-2-hydroxyethyl]benzenesulfonyl}acetate BDBM549112 US11304929, Example 01-035 Ammonium 2-((4-(6-((4-Methylbenzyl)oxy)pyridin-2-yl)piperidin-1-yl)methyl)-1-(2-methoxyethyl)-1H-benzo[d]imidazole-6-carboxylate BDBM349712 US10208019, Example 8A-01
Ammonium 2-((4-(6-((4-Methylbenzyl)oxy)pyridin-2-yl)piperidin-1-yl)methyl)-1-(2-methoxyethyl)-1H-benzo[d]imidazole-6-carboxylate BDBM349712 US10208019, Example 8A-01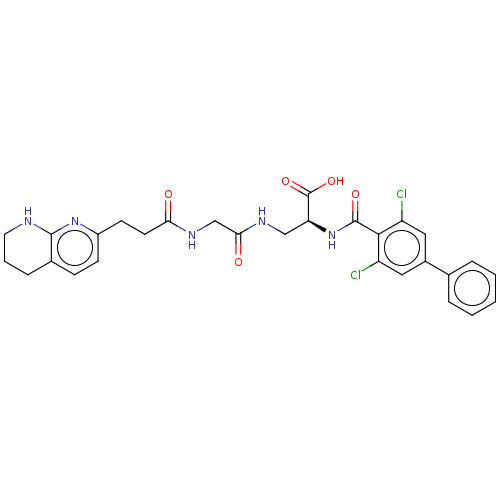 US11530180, Compound SU15210-0151-01 (S)-2-(3,5-dichloro-[1,1′-biphenyl]-4-carboxamido)-3-(2-(3-(5,6,7,8-tetrahydro-1,8-naphthyridin-2-yl)propanamido)acetamido)propanoic acid BDBM585082
US11530180, Compound SU15210-0151-01 (S)-2-(3,5-dichloro-[1,1′-biphenyl]-4-carboxamido)-3-(2-(3-(5,6,7,8-tetrahydro-1,8-naphthyridin-2-yl)propanamido)acetamido)propanoic acid BDBM585082 propan-2-yl 2-{4-[(1S)-1-({4-chloro-1,6-dimethyl-1H-pyrrolo[2,3-b]pyridin-2- yl}formamido)-2-hydroxyethyl]benzenesulfonyl}acetate BDBM549078 US11304929, Example 01-034
propan-2-yl 2-{4-[(1S)-1-({4-chloro-1,6-dimethyl-1H-pyrrolo[2,3-b]pyridin-2- yl}formamido)-2-hydroxyethyl]benzenesulfonyl}acetate BDBM549078 US11304929, Example 01-034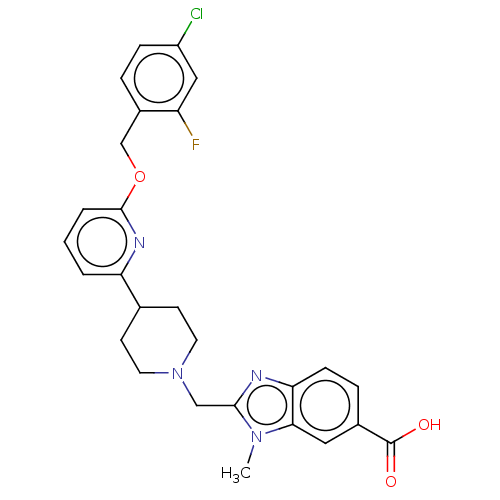 2-((4-(6-((4-Chloro-2-fluorobenzyl)oxy)pyridin-2-yl)piperidin-1-yl)methyl)-1-methyl-1H-benzo[d]imidazole-6-carboxylic acid BDBM349654 US10208019, Example 1A-01
2-((4-(6-((4-Chloro-2-fluorobenzyl)oxy)pyridin-2-yl)piperidin-1-yl)methyl)-1-methyl-1H-benzo[d]imidazole-6-carboxylic acid BDBM349654 US10208019, Example 1A-01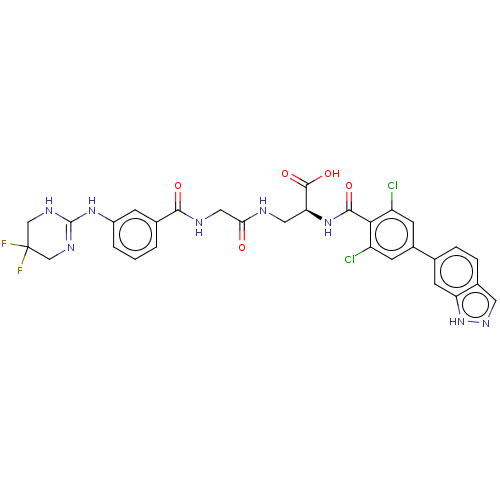 BDBM585097 (S)-2-(2,6-dichloro-4-(1H-indazol-6-yl)benzamido)-3-(2-(3-(5,5-difluoro-1,4,5,6-tetrahydropyrimidin-2-ylamino)benzamido)acetamido)propanoic acid US11530180, Compound SU15210-0272-01
BDBM585097 (S)-2-(2,6-dichloro-4-(1H-indazol-6-yl)benzamido)-3-(2-(3-(5,5-difluoro-1,4,5,6-tetrahydropyrimidin-2-ylamino)benzamido)acetamido)propanoic acid US11530180, Compound SU15210-0272-01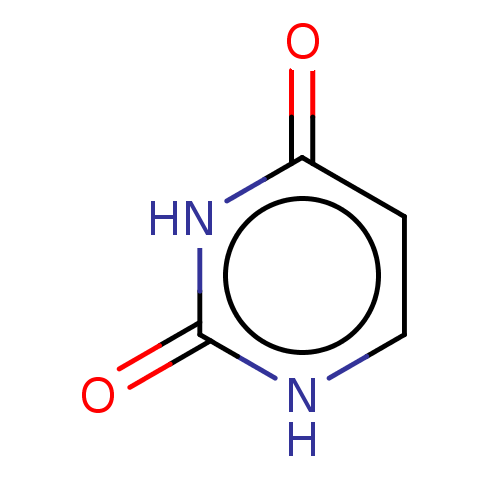 BMS-205603-01 Lamivudine impurity e SQ-7726 NSC-3970 BDBM50549809 CHEBI:17568 Fluorouracil specified compound c Hybar x SQ-8493 Lamivudine impurity e rs SQ-6201 Uracil Pyrod
BMS-205603-01 Lamivudine impurity e SQ-7726 NSC-3970 BDBM50549809 CHEBI:17568 Fluorouracil specified compound c Hybar x SQ-8493 Lamivudine impurity e rs SQ-6201 Uracil Pyrod US11304929, Example 01-003 methyl 2-{4-[(1R)-1-{[4-chloro-1-methyl-6-(1-methyl-1H-pyrazol-4-yl)-1H-indol- 2-yl]formamido}ethyl]benzenesulfonyl}acetate BDBM549427
US11304929, Example 01-003 methyl 2-{4-[(1R)-1-{[4-chloro-1-methyl-6-(1-methyl-1H-pyrazol-4-yl)-1H-indol- 2-yl]formamido}ethyl]benzenesulfonyl}acetate BDBM549427 US11304929, Example 01-009 BDBM549441 ethyl 2-{4-[(1R)-1-{[4-chloro-1-methyl-6-(1-methyl-1H-pyrazol-4-yl)-1H-indol-2- yl]formamido}ethyl]benzenesulfonyl}acetate
US11304929, Example 01-009 BDBM549441 ethyl 2-{4-[(1R)-1-{[4-chloro-1-methyl-6-(1-methyl-1H-pyrazol-4-yl)-1H-indol-2- yl]formamido}ethyl]benzenesulfonyl}acetate US11530180, Compound SU15210-0241-01 (2S)-2-(2,6-dichloro-4-(1H-indazol-6-yl)benzamido)-3-(2-(3-(5-fluoro-1,4,5,6-tetrahydropyrimidin-2-ylamino)benzamido)acetamido)propanoic acid BDBM585092
US11530180, Compound SU15210-0241-01 (2S)-2-(2,6-dichloro-4-(1H-indazol-6-yl)benzamido)-3-(2-(3-(5-fluoro-1,4,5,6-tetrahydropyrimidin-2-ylamino)benzamido)acetamido)propanoic acid BDBM585092 US20240025893, Example d-05-01 3-((Dimethylamino)methyl)-N-(6-(2-((((1S,2S)-2-hydroxycyclopentyl)oxy)methyl)pyrimidin-5-yl)benzo[d]thiazol-2-yl)cyclobutane-1-carboxamide BDBM647577
US20240025893, Example d-05-01 3-((Dimethylamino)methyl)-N-(6-(2-((((1S,2S)-2-hydroxycyclopentyl)oxy)methyl)pyrimidin-5-yl)benzo[d]thiazol-2-yl)cyclobutane-1-carboxamide BDBM647577 propan-2-yl 2-{4-[(1R)-1-{[4-chloro-1-methyl-6-(4-methylpiperazin-1-yl)-1H- indol-2-yl]formamido}ethyl]benzenesulfonyl}acetate BDBM549514 US11304929, Example 01-041
propan-2-yl 2-{4-[(1R)-1-{[4-chloro-1-methyl-6-(4-methylpiperazin-1-yl)-1H- indol-2-yl]formamido}ethyl]benzenesulfonyl}acetate BDBM549514 US11304929, Example 01-041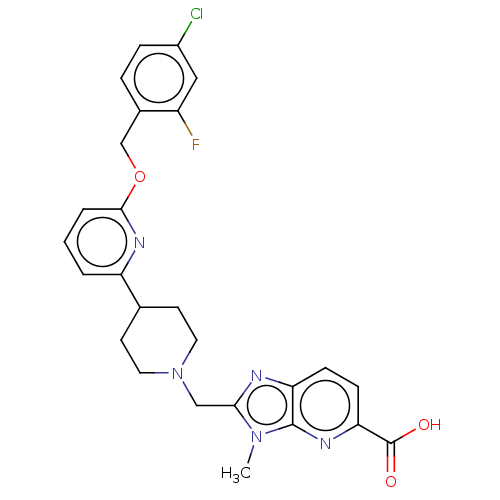 2-((4-(6-((4-Chloro-2-fluorobenzyl)oxy)pyridin-2-yl)piperidin-1-yl)methyl)-3-methyl-3H-imidazo[4,5-b]pyridine-5-carboxylic acid US10208019, Example 2A-01 BDBM349684
2-((4-(6-((4-Chloro-2-fluorobenzyl)oxy)pyridin-2-yl)piperidin-1-yl)methyl)-3-methyl-3H-imidazo[4,5-b]pyridine-5-carboxylic acid US10208019, Example 2A-01 BDBM349684 2-[(4-{6-[(4-Chloro-2-fluorobenzyl)oxy]pyridin-2-yl}piperidin-1-yl)methyl]-1-(1,3-oxazol-5-ylmethyl)-1H-benzimidazole-6-carboxylic acid BDBM349731 US10208019, Example 10A-01
2-[(4-{6-[(4-Chloro-2-fluorobenzyl)oxy]pyridin-2-yl}piperidin-1-yl)methyl]-1-(1,3-oxazol-5-ylmethyl)-1H-benzimidazole-6-carboxylic acid BDBM349731 US10208019, Example 10A-01 2-[(4-{6-[(4-cyano-2-fluorobenzyl)oxy]pyridin-2-yl}piperidin-1-yl)methyl]-1-[(2S)-oxetan-2-ylmethyl]-1H-benzimidazole-6-carboxylic acid US10208019, Example 4A-01 BDBM349708
2-[(4-{6-[(4-cyano-2-fluorobenzyl)oxy]pyridin-2-yl}piperidin-1-yl)methyl]-1-[(2S)-oxetan-2-ylmethyl]-1H-benzimidazole-6-carboxylic acid US10208019, Example 4A-01 BDBM349708 BDBM349711 US10208019, Example 7A-01 2-[(4-{6-[(4-cyano-2-fluorobenzyl)oxy]pyridin-2-yl}piperidin-1-yl)methyl]-1-(1,3-oxazol-2-ylmethyl)-1H-benzimidazole-6-carboxylic acid
BDBM349711 US10208019, Example 7A-01 2-[(4-{6-[(4-cyano-2-fluorobenzyl)oxy]pyridin-2-yl}piperidin-1-yl)methyl]-1-(1,3-oxazol-2-ylmethyl)-1H-benzimidazole-6-carboxylic acid US10208019, Example 3A-01 BDBM349707 2-[(4-{6-[(4-chloro-2-fluorobenzyl)oxy]pyridin-2-yl}piperidin-1-yl)methyl]-1-[(2S)-oxetan-2-ylmethyl]-1H-benzimidazole-6-carboxylic acid
US10208019, Example 3A-01 BDBM349707 2-[(4-{6-[(4-chloro-2-fluorobenzyl)oxy]pyridin-2-yl}piperidin-1-yl)methyl]-1-[(2S)-oxetan-2-ylmethyl]-1H-benzimidazole-6-carboxylic acid US10208019, Example 5A-01 2-[(4-{6-[(4-Cyano-2-fluorobenzyl)oxy]pyridin-2-yl}piperazin-1-yl)methyl]-1-[(2S)-oxetan-2-ylmethyl]-1H-benzimidazole-6-carboxylic acid BDBM349709
US10208019, Example 5A-01 2-[(4-{6-[(4-Cyano-2-fluorobenzyl)oxy]pyridin-2-yl}piperazin-1-yl)methyl]-1-[(2S)-oxetan-2-ylmethyl]-1H-benzimidazole-6-carboxylic acid BDBM349709 propan-2-yl 2-{4-[(1S)-1-{[4-chloro-1-methyl-6-(4-methylpiperazin-1-yI)-1H- indol-2-yl]formamido}-2-hydroxyethyl]benzenesulfonyl}acetate BDBM549512 US11304929, Example 01-040
propan-2-yl 2-{4-[(1S)-1-{[4-chloro-1-methyl-6-(4-methylpiperazin-1-yI)-1H- indol-2-yl]formamido}-2-hydroxyethyl]benzenesulfonyl}acetate BDBM549512 US11304929, Example 01-040 US10208019, Example 14A-02 US10208019, Example 14A-01 BDBM349899 2-[(4-{6-[(4-cyanobenzyl)oxy]pyridin-2-yl}piperidin-1-yl)methyl]-1-(tetrahydrofuran-2-ylmethyl)-1H-benzimidazole-6-carboxylic acid
US10208019, Example 14A-02 US10208019, Example 14A-01 BDBM349899 2-[(4-{6-[(4-cyanobenzyl)oxy]pyridin-2-yl}piperidin-1-yl)methyl]-1-(tetrahydrofuran-2-ylmethyl)-1H-benzimidazole-6-carboxylic acid US11304929, Example 01-011 propan-2-yl 2-{4-[(1R)-1-{[4-chloro-1-methyl-6-(1-methyl-1H-pyrazol-4-yl)-1H- indol-2-yl]formamido}ethyl]benzenesulfonyl}acetate BDBM549449
US11304929, Example 01-011 propan-2-yl 2-{4-[(1R)-1-{[4-chloro-1-methyl-6-(1-methyl-1H-pyrazol-4-yl)-1H- indol-2-yl]formamido}ethyl]benzenesulfonyl}acetate BDBM549449 US20240025893, Example d-02-01 (1s,3s)-3-(tert-Butoxy)-N-(6-(2-((((1S,2S)-2-hydroxycyclopentyl)oxy)methyl)pyrimidin-5-yl)benzo[d]thiazol-2-yl)cyclobutane-1-carboxamide BDBM647566
US20240025893, Example d-02-01 (1s,3s)-3-(tert-Butoxy)-N-(6-(2-((((1S,2S)-2-hydroxycyclopentyl)oxy)methyl)pyrimidin-5-yl)benzo[d]thiazol-2-yl)cyclobutane-1-carboxamide BDBM647566 BDBM349710 US10208019, Example 6A-01 2-[(4-{6-[(4-cyano-2-fluorobenzyl)oxy]pyridin-2-yl}piperidin-1-yl)methyl]-3-[(2S)-oxetan-2-ylmethyl]-3H-imidazo[4,5-b]pyridine-5-carboxylic acid
BDBM349710 US10208019, Example 6A-01 2-[(4-{6-[(4-cyano-2-fluorobenzyl)oxy]pyridin-2-yl}piperidin-1-yl)methyl]-3-[(2S)-oxetan-2-ylmethyl]-3H-imidazo[4,5-b]pyridine-5-carboxylic acid BDBM349895 2-{[4-{6-[(4-Chloro-2-fluorobenzyl)oxy]pyridin-2-yl}-2-methylpiperidin-1-yl]methyl}-1-(2-methoxyethyl)-1H-benzimidazole-6-carboxylic acid US10208019, Example 13A-01 US10208019, Example 13A-04
BDBM349895 2-{[4-{6-[(4-Chloro-2-fluorobenzyl)oxy]pyridin-2-yl}-2-methylpiperidin-1-yl]methyl}-1-(2-methoxyethyl)-1H-benzimidazole-6-carboxylic acid US10208019, Example 13A-01 US10208019, Example 13A-04 BDBM744715 (2R,4S)-N-((S)-1-(((R)-2-amino-6,7-dihydro-5H-cyclopenta[b]pyridin-5-yl)amino)-1-oxopropan-2-yl)-4-(4-fluorobenzyl) pyrrolidine-2-carboxamide US20250171414, Compound L001-IS-01
BDBM744715 (2R,4S)-N-((S)-1-(((R)-2-amino-6,7-dihydro-5H-cyclopenta[b]pyridin-5-yl)amino)-1-oxopropan-2-yl)-4-(4-fluorobenzyl) pyrrolidine-2-carboxamide US20250171414, Compound L001-IS-01 BRD-K70771662-001-01-3 4-chloro-N-(3-cyano-4-phenylsulfanylphenyl)benzamide 4-chloro-N-[3-cyano-4-(phenylthio)phenyl]benzamide BDBM65747 4-chloranyl-N-(3-cyano-4-phenylsulfanyl-phenyl)benzamide cid_1471787
BRD-K70771662-001-01-3 4-chloro-N-(3-cyano-4-phenylsulfanylphenyl)benzamide 4-chloro-N-[3-cyano-4-(phenylthio)phenyl]benzamide BDBM65747 4-chloranyl-N-(3-cyano-4-phenylsulfanyl-phenyl)benzamide cid_1471787 ethyl 2-{4-[(1R)-1-[(5-chloro-1-methyl-6-{1-[2-(morpholin-4-yl)ethyl]-1H-pyrazol- 4-yl}-1H-indol-2-yl)formamido]ethyl]benzenesulfonyl}acetate US11304929, Example 01-025 BDBM549504
ethyl 2-{4-[(1R)-1-[(5-chloro-1-methyl-6-{1-[2-(morpholin-4-yl)ethyl]-1H-pyrazol- 4-yl}-1H-indol-2-yl)formamido]ethyl]benzenesulfonyl}acetate US11304929, Example 01-025 BDBM549504 methyl 2-{4-[(1R)-1-[(5-chloro-1-methyl-6-{1-[2-(morpholin-4-yl)ethyl]-1H- pyrazol-4-yl}-1H-indol-2-yl)formamido]ethyl]benzenesulfonyl}acetate US11304929, Example 01-027 BDBM549503
methyl 2-{4-[(1R)-1-[(5-chloro-1-methyl-6-{1-[2-(morpholin-4-yl)ethyl]-1H- pyrazol-4-yl}-1H-indol-2-yl)formamido]ethyl]benzenesulfonyl}acetate US11304929, Example 01-027 BDBM549503 BDBM349810 2-[(4-{6-[(4-chloro-2-fluorobenzyl)oxy]pyridin-2-yl}piperidin-1-yl)methyl]-1-[2-(1-methyl-1H-imidazol-4-yl)ethyl]-1H-benzimidazole-6-carboxylic acid US10208019, Example 11A-01
BDBM349810 2-[(4-{6-[(4-chloro-2-fluorobenzyl)oxy]pyridin-2-yl}piperidin-1-yl)methyl]-1-[2-(1-methyl-1H-imidazol-4-yl)ethyl]-1H-benzimidazole-6-carboxylic acid US10208019, Example 11A-01 BDBM549510 propan-2-yl 2-{4-[(1R)-1-[(5-chloro-1-methyl-6-{1-[2-(morpholin-4-yl)ethyl]-1H- pyrazol-4-yl}-1H-indol-2-yl)formamido]ethyl]benzenesulfonyl}acetate US11304929, Example 01-026
BDBM549510 propan-2-yl 2-{4-[(1R)-1-[(5-chloro-1-methyl-6-{1-[2-(morpholin-4-yl)ethyl]-1H- pyrazol-4-yl}-1H-indol-2-yl)formamido]ethyl]benzenesulfonyl}acetate US11304929, Example 01-026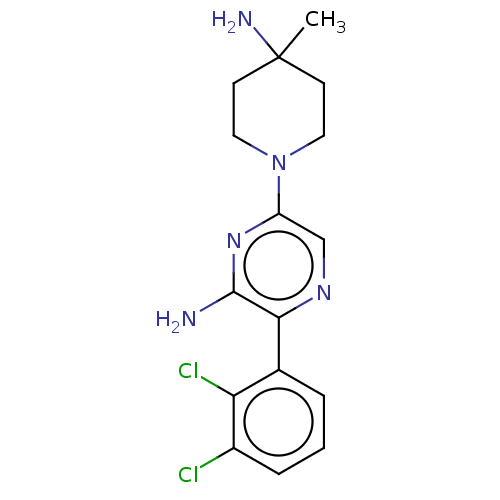 US10093646, Compound 1 US12281118, Example SHP099 US10301278, Example 00003 US11702392, Compound SHP-099 US10336774, Example 01 US11401259, Compound 1 US12053470, Compound 1 US10774065, Example 00003 BDBM38019 US20240270753, Compound SHP099 US10858359, SHP099 US20230348467, Compound SHP-099
US10093646, Compound 1 US12281118, Example SHP099 US10301278, Example 00003 US11702392, Compound SHP-099 US10336774, Example 01 US11401259, Compound 1 US12053470, Compound 1 US10774065, Example 00003 BDBM38019 US20240270753, Compound SHP099 US10858359, SHP099 US20230348467, Compound SHP-099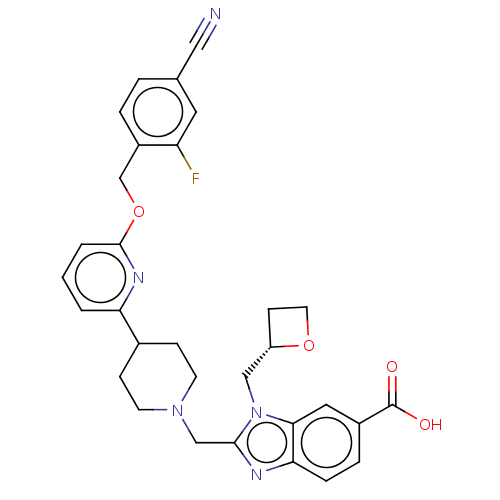 2-{[4-(6-{[(4-cyano-2-fluorophenyl)(methyl-d2)]oxy}pyridin-2-yl)piperidin-1-yl]methyl}-1-[(2S)-oxetan-2-ylmethyl]-1H-benzimidazole-6-carboxylic acid US10208019, Example 4A-01 US10208019, Example 1A-09 BDBM349662
2-{[4-(6-{[(4-cyano-2-fluorophenyl)(methyl-d2)]oxy}pyridin-2-yl)piperidin-1-yl]methyl}-1-[(2S)-oxetan-2-ylmethyl]-1H-benzimidazole-6-carboxylic acid US10208019, Example 4A-01 US10208019, Example 1A-09 BDBM349662 BDBM284394 US10174027, Example 404 US10023570, Example 404 6-(1-methyl-1H-pyraz 01-4- yl)-4-(6-(4-((tetrahydro- 2H-pyran-2- yl)methyl)piperazin-1- yl)pyridin-3- yl)pyrazolo[1,5-a]pyridine- 3-carbonitrile
BDBM284394 US10174027, Example 404 US10023570, Example 404 6-(1-methyl-1H-pyraz 01-4- yl)-4-(6-(4-((tetrahydro- 2H-pyran-2- yl)methyl)piperazin-1- yl)pyridin-3- yl)pyrazolo[1,5-a]pyridine- 3-carbonitrile BDBM349884 US10208019, Example 12A-01 2-((4-(6-((4-Chloro-2-fluorobenzyl)oxy)pyridin-2-yl)-2-(trifluoromethyl)piperazin-1-yl)methyl)-1-(2-methoxyethyl)-1H-benzo[d]imidazole-6-carboxylic acid US10208019, Example 12A-02
BDBM349884 US10208019, Example 12A-01 2-((4-(6-((4-Chloro-2-fluorobenzyl)oxy)pyridin-2-yl)-2-(trifluoromethyl)piperazin-1-yl)methyl)-1-(2-methoxyethyl)-1H-benzo[d]imidazole-6-carboxylic acid US10208019, Example 12A-02 (benzyl (2 S)-2-[[2,6-dichloro-4-(1,1-di oxo-1,4-thiazinan-4-yl)benzoyl]amino]-3-[[2-[[3-[(5-fluoro-1,4,5,6-tetrahydropyrimidin-2-yl)amino]benzoyl]amino]acetyl]amino]propanoate US11530180, Compound SU15210-0253-01 BDBM585094
(benzyl (2 S)-2-[[2,6-dichloro-4-(1,1-di oxo-1,4-thiazinan-4-yl)benzoyl]amino]-3-[[2-[[3-[(5-fluoro-1,4,5,6-tetrahydropyrimidin-2-yl)amino]benzoyl]amino]acetyl]amino]propanoate US11530180, Compound SU15210-0253-01 BDBM585094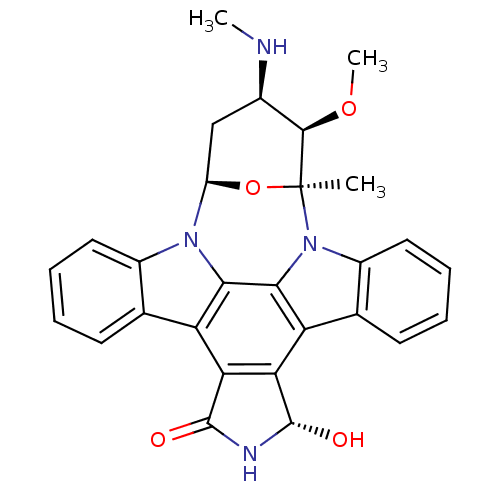 18-hydroxy-3-methoxy-2-methyl-4-methylamino-(2R,3S,4S,6S)-29-oxa-1,7,17-triazaoctacyclo[12.12.2.12,6.07,28.08,13.015,19.020,27.021,26]nonacosa-8(13),9,11,14(28),15(19),20(27),21(26),22,24-nonaen-16-one BDBM50280450 CHEMBL574737 UCN-01 UCN-02
18-hydroxy-3-methoxy-2-methyl-4-methylamino-(2R,3S,4S,6S)-29-oxa-1,7,17-triazaoctacyclo[12.12.2.12,6.07,28.08,13.015,19.020,27.021,26]nonacosa-8(13),9,11,14(28),15(19),20(27),21(26),22,24-nonaen-16-one BDBM50280450 CHEMBL574737 UCN-01 UCN-02 3-chloro-N-[3-cyano-4-[(4-methoxyphenyl)thio]phenyl]benzamide 3-chloranyl-N-[3-cyano-4-(4-methoxyphenyl)sulfanyl-phenyl]benzamide BDBM65738 cid_2769230 3-chloro-N-[3-cyano-4-(4-methoxyphenyl)sulfanylphenyl]benzamide BRD-K36087356-001-01-8
3-chloro-N-[3-cyano-4-[(4-methoxyphenyl)thio]phenyl]benzamide 3-chloranyl-N-[3-cyano-4-(4-methoxyphenyl)sulfanyl-phenyl]benzamide BDBM65738 cid_2769230 3-chloro-N-[3-cyano-4-(4-methoxyphenyl)sulfanylphenyl]benzamide BRD-K36087356-001-01-8 cid_1489476 BDBM65739 2-chloro-N-[3-cyano-4-(4-methoxyphenyl)sulfonylphenyl]benzamide 2-chloranyl-N-[3-cyano-4-(4-methoxyphenyl)sulfonyl-phenyl]benzamide 2-chloro-N-[3-cyano-4-(4-methoxyphenyl)sulfonyl-phenyl]benzamide BRD-K68340261-001-01-6
cid_1489476 BDBM65739 2-chloro-N-[3-cyano-4-(4-methoxyphenyl)sulfonylphenyl]benzamide 2-chloranyl-N-[3-cyano-4-(4-methoxyphenyl)sulfonyl-phenyl]benzamide 2-chloro-N-[3-cyano-4-(4-methoxyphenyl)sulfonyl-phenyl]benzamide BRD-K68340261-001-01-6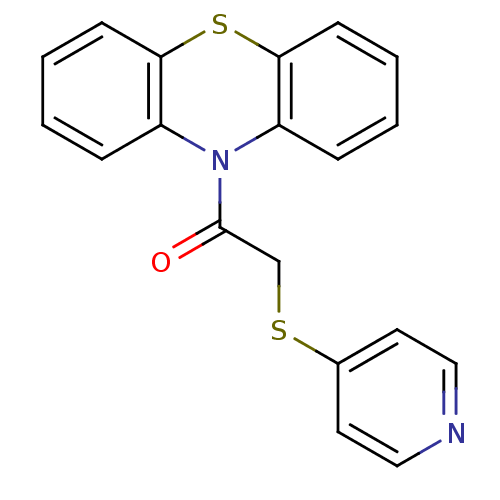 1-phenothiazin-10-yl-2-(4-pyridylthio)ethanone 1-(10-phenothiazinyl)-2-(pyridin-4-ylthio)ethanone BRD-K50117178-001-01-0 BDBM54879 1-phenothiazin-10-yl-2-pyridin-4-ylsulfanyl-ethanone 1-phenothiazin-10-yl-2-pyridin-4-ylsulfanylethanone cid_44247469
1-phenothiazin-10-yl-2-(4-pyridylthio)ethanone 1-(10-phenothiazinyl)-2-(pyridin-4-ylthio)ethanone BRD-K50117178-001-01-0 BDBM54879 1-phenothiazin-10-yl-2-pyridin-4-ylsulfanyl-ethanone 1-phenothiazin-10-yl-2-pyridin-4-ylsulfanylethanone cid_44247469 BDBM54884 1-phenothiazin-10-yl-2-thiophen-2-ylsulfanyl-ethanone BRD-K59611824-001-01-1 1-(10-phenothiazinyl)-2-(thiophen-2-ylthio)ethanone 1-phenothiazin-10-yl-2-(2-thienylthio)ethanone 1-phenothiazin-10-yl-2-thiophen-2-ylsulfanylethanone cid_1230713
BDBM54884 1-phenothiazin-10-yl-2-thiophen-2-ylsulfanyl-ethanone BRD-K59611824-001-01-1 1-(10-phenothiazinyl)-2-(thiophen-2-ylthio)ethanone 1-phenothiazin-10-yl-2-(2-thienylthio)ethanone 1-phenothiazin-10-yl-2-thiophen-2-ylsulfanylethanone cid_1230713 BRD-K28023767-001-01-6 2-[(4-chlorophenyl)thio]-1-phenothiazin-10-yl-ethanone 2-[(4-chlorophenyl)thio]-1-(10-phenothiazinyl)ethanone cid_3390924 2-(4-chlorophenyl)sulfanyl-1-phenothiazin-10-yl-ethanone 2-(4-chlorophenyl)sulfanyl-1-phenothiazin-10-ylethanone BDBM54870
BRD-K28023767-001-01-6 2-[(4-chlorophenyl)thio]-1-phenothiazin-10-yl-ethanone 2-[(4-chlorophenyl)thio]-1-(10-phenothiazinyl)ethanone cid_3390924 2-(4-chlorophenyl)sulfanyl-1-phenothiazin-10-yl-ethanone 2-(4-chlorophenyl)sulfanyl-1-phenothiazin-10-ylethanone BDBM54870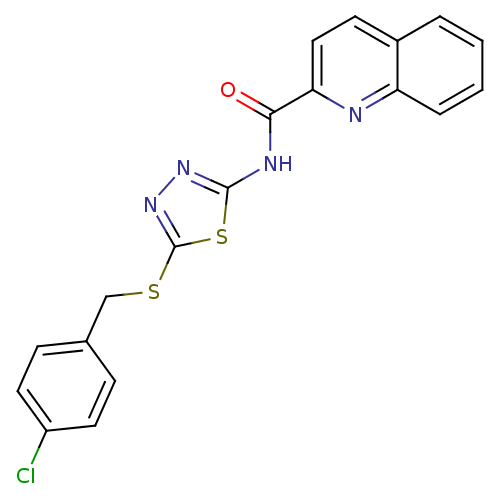 BRD-K57435385-001-01-8 N-[5-[(4-chlorophenyl)methylthio]-1,3,4-thiadiazol-2-yl]-2-quinolinecarboxamide N-[5-[(4-chlorophenyl)methylsulfanyl]-1,3,4-thiadiazol-2-yl]quinoline-2-carboxamide N-[5-[(4-chlorobenzyl)thio]-1,3,4-thiadiazol-2-yl]quinaldamide cid_2091582 BDBM68336
BRD-K57435385-001-01-8 N-[5-[(4-chlorophenyl)methylthio]-1,3,4-thiadiazol-2-yl]-2-quinolinecarboxamide N-[5-[(4-chlorophenyl)methylsulfanyl]-1,3,4-thiadiazol-2-yl]quinoline-2-carboxamide N-[5-[(4-chlorobenzyl)thio]-1,3,4-thiadiazol-2-yl]quinaldamide cid_2091582 BDBM68336 BRD-K92790413-001-01-9 BDBM54897 2-(4-methoxyphenyl)sulfanyl-1-phenothiazin-10-ylethanone cid_8392433 2-[(4-methoxyphenyl)thio]-1-(10-phenothiazinyl)ethanone 2-[(4-methoxyphenyl)thio]-1-phenothiazin-10-yl-ethanone 2-(4-methoxyphenyl)sulfanyl-1-phenothiazin-10-yl-ethanone
BRD-K92790413-001-01-9 BDBM54897 2-(4-methoxyphenyl)sulfanyl-1-phenothiazin-10-ylethanone cid_8392433 2-[(4-methoxyphenyl)thio]-1-(10-phenothiazinyl)ethanone 2-[(4-methoxyphenyl)thio]-1-phenothiazin-10-yl-ethanone 2-(4-methoxyphenyl)sulfanyl-1-phenothiazin-10-yl-ethanone N-[3-cyano-4-(4-methylphenyl)sulfanyl-phenyl]-4-fluoranyl-benzamide BRD-K43069600-001-01-1 N-[3-cyano-4-[(4-methylphenyl)thio]phenyl]-4-fluorobenzamide N-[3-cyano-4-(p-tolylthio)phenyl]-4-fluoro-benzamide N-[3-cyano-4-(4-methylphenyl)sulfanylphenyl]-4-fluorobenzamide BDBM65745 cid_2769158
N-[3-cyano-4-(4-methylphenyl)sulfanyl-phenyl]-4-fluoranyl-benzamide BRD-K43069600-001-01-1 N-[3-cyano-4-[(4-methylphenyl)thio]phenyl]-4-fluorobenzamide N-[3-cyano-4-(p-tolylthio)phenyl]-4-fluoro-benzamide N-[3-cyano-4-(4-methylphenyl)sulfanylphenyl]-4-fluorobenzamide BDBM65745 cid_2769158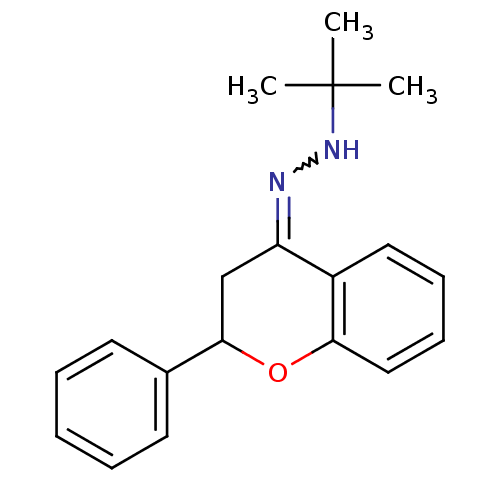 2-methyl-N-[(E)-(2-phenyl-2,3-dihydrochromen-4-ylidene)amino]propan-2-amine BDBM54852 BRD-A92251157-003-01-0 cid_16240206 2-methyl-N-[(E)-(2-phenyl-3,4-dihydro-2H-1-benzopyran-4-ylidene)amino]-2-propanamine tert-butyl-[(E)-(2-phenylchroman-4-ylidene)amino]amine tert-butyl-[(2-phenylchroman-4-ylidene)amino]amine;hydrochloride
2-methyl-N-[(E)-(2-phenyl-2,3-dihydrochromen-4-ylidene)amino]propan-2-amine BDBM54852 BRD-A92251157-003-01-0 cid_16240206 2-methyl-N-[(E)-(2-phenyl-3,4-dihydro-2H-1-benzopyran-4-ylidene)amino]-2-propanamine tert-butyl-[(E)-(2-phenylchroman-4-ylidene)amino]amine tert-butyl-[(2-phenylchroman-4-ylidene)amino]amine;hydrochloride (5Z)-1-methyl-5-[[1-(2-naphthyl)pyrrol-2-yl]methylene]-2-thioxo-hexahydropyrimidine-4,6-quinone (5Z)-1-methyl-5-[[1-(2-naphthalenyl)-2-pyrrolyl]methylidene]-2-sulfanylidene-1,3-diazinane-4,6-dione BDBM54869 (5Z)-1-methyl-5-[(1-naphthalen-2-ylpyrrol-2-yl)methylidene]-2-sulfanylidene-1,3-diazinane-4,6-dione cid_5823856 BRD-K26980645-001-01-7
(5Z)-1-methyl-5-[[1-(2-naphthyl)pyrrol-2-yl]methylene]-2-thioxo-hexahydropyrimidine-4,6-quinone (5Z)-1-methyl-5-[[1-(2-naphthalenyl)-2-pyrrolyl]methylidene]-2-sulfanylidene-1,3-diazinane-4,6-dione BDBM54869 (5Z)-1-methyl-5-[(1-naphthalen-2-ylpyrrol-2-yl)methylidene]-2-sulfanylidene-1,3-diazinane-4,6-dione cid_5823856 BRD-K26980645-001-01-7 4-(dimethylamino)-6-methyl-6,10,12a-tris(oxidanyl)-1,3,11,12-tetrakis(oxidanylidene)-4a,5,5a,11a-tetrahydro-4H-tetracene-2-carboxamide 4-(dimethylamino)-6,10,12a-trihydroxy-1,3,11,12-tetraketo-6-methyl-4a,5,5a,11a-tetrahydro-4H-tetracene-2-carboxamide BRD-A88208128-003-01-2 cid_44247471 4-(dimethylamino)-6,10,12a-trihydroxy-6-methyl-1,3,11,12-tetraoxo-4a,5,5a,11a-tetrahydro-4H-tetracene-2-carboxamide BDBM54850
4-(dimethylamino)-6-methyl-6,10,12a-tris(oxidanyl)-1,3,11,12-tetrakis(oxidanylidene)-4a,5,5a,11a-tetrahydro-4H-tetracene-2-carboxamide 4-(dimethylamino)-6,10,12a-trihydroxy-1,3,11,12-tetraketo-6-methyl-4a,5,5a,11a-tetrahydro-4H-tetracene-2-carboxamide BRD-A88208128-003-01-2 cid_44247471 4-(dimethylamino)-6,10,12a-trihydroxy-6-methyl-1,3,11,12-tetraoxo-4a,5,5a,11a-tetrahydro-4H-tetracene-2-carboxamide BDBM54850 US11420970, Example 156 (R)-1-((R)-3-amino-1-(4-((6-amino-9H-purin-9-yl)methyl)-6-(2,4,5-trifluorophenyl)pyridin-3-yl)piperidin-3-yl)-2,2-difluoroethan-1-01 and (S)-1-((R)-3-amino-1-(4-((6-amino-9H-purin-9-yl) methyl)-6-(2,4,5-trifluorophenyl)pyridin-3-yl)piperidin-3-yl)-2,2-difluoroethan-1-ol BDBM567955
US11420970, Example 156 (R)-1-((R)-3-amino-1-(4-((6-amino-9H-purin-9-yl)methyl)-6-(2,4,5-trifluorophenyl)pyridin-3-yl)piperidin-3-yl)-2,2-difluoroethan-1-01 and (S)-1-((R)-3-amino-1-(4-((6-amino-9H-purin-9-yl) methyl)-6-(2,4,5-trifluorophenyl)pyridin-3-yl)piperidin-3-yl)-2,2-difluoroethan-1-ol BDBM567955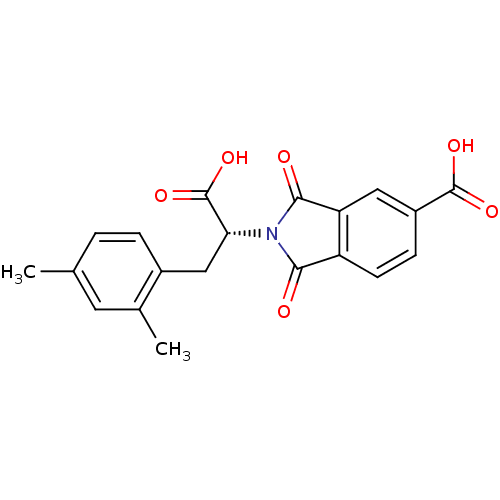 2-[(1R)-1-carboxy-2-(2,4-dimethylphenyl)ethyl]-1,3-dioxoisoindole-5-carboxylic acid BDBM41852 cid_16752644 2-[(1R)-1-carboxy-2-(2,4-dimethylphenyl)ethyl]-1,3-diketo-isoindoline-5-carboxylic acid NCGC00161794-01 2-[(2R)-3-(2,4-dimethylphenyl)-1-oxidanyl-1-oxidanylidene-propan-2-yl]-1,3-bis(oxidanylidene)isoindole-5-carboxylic acid 2-[(1R)-1-carboxy-2-(2,4-dimethylphenyl)ethyl]-1,3-dioxo-5-isoindolecarboxylic acid
2-[(1R)-1-carboxy-2-(2,4-dimethylphenyl)ethyl]-1,3-dioxoisoindole-5-carboxylic acid BDBM41852 cid_16752644 2-[(1R)-1-carboxy-2-(2,4-dimethylphenyl)ethyl]-1,3-diketo-isoindoline-5-carboxylic acid NCGC00161794-01 2-[(2R)-3-(2,4-dimethylphenyl)-1-oxidanyl-1-oxidanylidene-propan-2-yl]-1,3-bis(oxidanylidene)isoindole-5-carboxylic acid 2-[(1R)-1-carboxy-2-(2,4-dimethylphenyl)ethyl]-1,3-dioxo-5-isoindolecarboxylic acid 2-[(1R)-1-carboxy-2-(4-phenylphenyl)ethyl]-1,3-dioxoisoindole-5-carboxylic acid 2-[(1R)-1-carboxy-2-(4-phenylphenyl)ethyl]-1,3-diketo-isoindoline-5-carboxylic acid BDBM41854 NCGC00161802-01 cid_16752652 2-[(1R)-1-carboxy-2-(4-phenylphenyl)ethyl]-1,3-dioxo-5-isoindolecarboxylic acid 1,3-bis(oxidanylidene)-2-[(2R)-1-oxidanyl-1-oxidanylidene-3-(4-phenylphenyl)propan-2-yl]isoindole-5-carboxylic acid
2-[(1R)-1-carboxy-2-(4-phenylphenyl)ethyl]-1,3-dioxoisoindole-5-carboxylic acid 2-[(1R)-1-carboxy-2-(4-phenylphenyl)ethyl]-1,3-diketo-isoindoline-5-carboxylic acid BDBM41854 NCGC00161802-01 cid_16752652 2-[(1R)-1-carboxy-2-(4-phenylphenyl)ethyl]-1,3-dioxo-5-isoindolecarboxylic acid 1,3-bis(oxidanylidene)-2-[(2R)-1-oxidanyl-1-oxidanylidene-3-(4-phenylphenyl)propan-2-yl]isoindole-5-carboxylic acid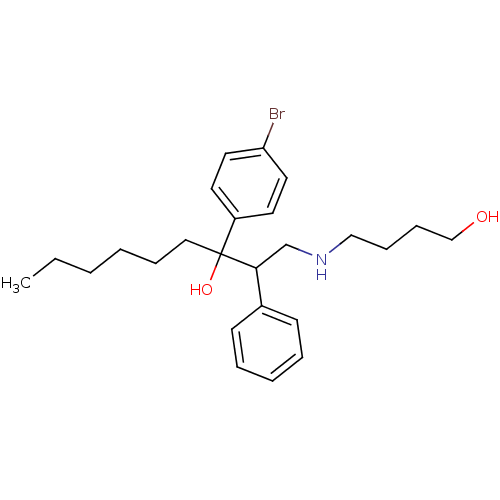 3-(4-bromophenyl)-1-(4-hydroxybutylamino)-2-phenyl-3-nonanol BDBM54851 3-(4-bromophenyl)-1-(4-hydroxybutylamino)-2-phenyl-nonan-3-ol;hydrochloride cid_16193797 3-(4-bromophenyl)-1-(4-hydroxybutylamino)-2-phenyl-nonan-3-ol 3-(4-bromophenyl)-1-(4-oxidanylbutylamino)-2-phenyl-nonan-3-ol BRD-A91513005-001-01-2 3-(4-bromophenyl)-1-(4-hydroxybutylamino)-2-phenylnonan-3-ol
3-(4-bromophenyl)-1-(4-hydroxybutylamino)-2-phenyl-3-nonanol BDBM54851 3-(4-bromophenyl)-1-(4-hydroxybutylamino)-2-phenyl-nonan-3-ol;hydrochloride cid_16193797 3-(4-bromophenyl)-1-(4-hydroxybutylamino)-2-phenyl-nonan-3-ol 3-(4-bromophenyl)-1-(4-oxidanylbutylamino)-2-phenyl-nonan-3-ol BRD-A91513005-001-01-2 3-(4-bromophenyl)-1-(4-hydroxybutylamino)-2-phenylnonan-3-ol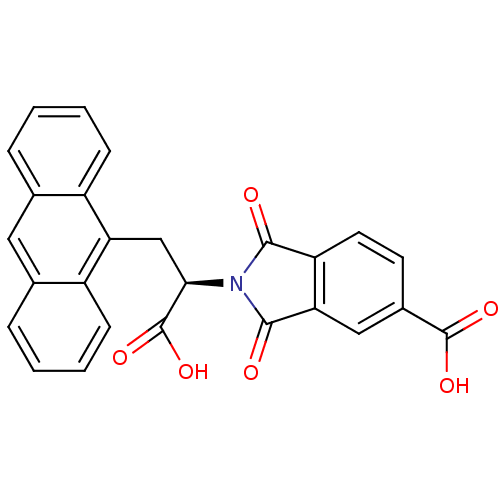 cid_16752655 NCGC00161805-01 BDBM41855 2-[(1R)-2-(9-anthracenyl)-1-carboxyethyl]-1,3-dioxo-5-isoindolecarboxylic acid 2-[(1R)-2-anthracen-9-yl-1-carboxyethyl]-1,3-dioxoisoindole-5-carboxylic acid 2-[(1R)-2-(9-anthryl)-1-carboxy-ethyl]-1,3-diketo-isoindoline-5-carboxylic acid 2-[(2R)-3-anthracen-9-yl-1-oxidanyl-1-oxidanylidene-propan-2-yl]-1,3-bis(oxidanylidene)isoindole-5-carboxylic acid
cid_16752655 NCGC00161805-01 BDBM41855 2-[(1R)-2-(9-anthracenyl)-1-carboxyethyl]-1,3-dioxo-5-isoindolecarboxylic acid 2-[(1R)-2-anthracen-9-yl-1-carboxyethyl]-1,3-dioxoisoindole-5-carboxylic acid 2-[(1R)-2-(9-anthryl)-1-carboxy-ethyl]-1,3-diketo-isoindoline-5-carboxylic acid 2-[(2R)-3-anthracen-9-yl-1-oxidanyl-1-oxidanylidene-propan-2-yl]-1,3-bis(oxidanylidene)isoindole-5-carboxylic acid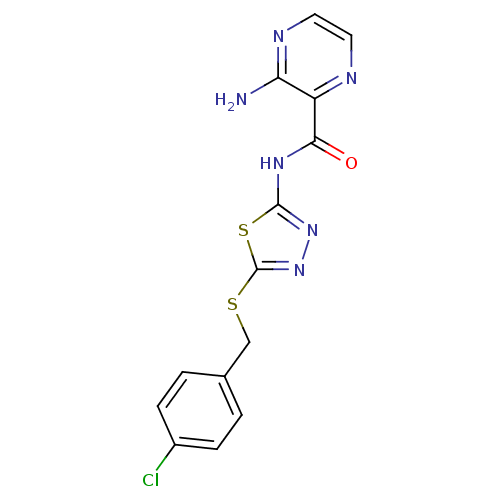 cid_25475077 3-amino-N-[5-[(4-chlorobenzyl)thio]-1,3,4-thiadiazol-2-yl]pyrazinamide 3-amino-N-[5-[(4-chlorophenyl)methylsulfanyl]-1,3,4-thiadiazol-2-yl]pyrazine-2-carboxamide 3-amino-N-[5-[(4-chlorophenyl)methylthio]-1,3,4-thiadiazol-2-yl]-2-pyrazinecarboxamide 3-azanyl-N-[5-[(4-chlorophenyl)methylsulfanyl]-1,3,4-thiadiazol-2-yl]pyrazine-2-carboxamide BDBM68334 BRD-K38089706-001-01-9
cid_25475077 3-amino-N-[5-[(4-chlorobenzyl)thio]-1,3,4-thiadiazol-2-yl]pyrazinamide 3-amino-N-[5-[(4-chlorophenyl)methylsulfanyl]-1,3,4-thiadiazol-2-yl]pyrazine-2-carboxamide 3-amino-N-[5-[(4-chlorophenyl)methylthio]-1,3,4-thiadiazol-2-yl]-2-pyrazinecarboxamide 3-azanyl-N-[5-[(4-chlorophenyl)methylsulfanyl]-1,3,4-thiadiazol-2-yl]pyrazine-2-carboxamide BDBM68334 BRD-K38089706-001-01-9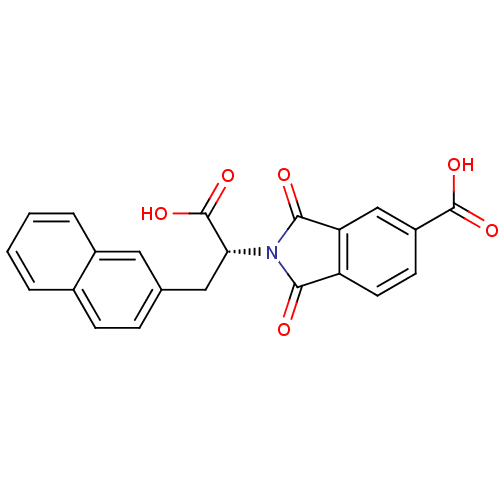 2-[(1R)-1-carboxy-2-(2-naphthalenyl)ethyl]-1,3-dioxo-5-isoindolecarboxylic acid BDBM41851 2-[(1R)-1-carboxy-2-(2-naphthyl)ethyl]-1,3-diketo-isoindoline-5-carboxylic acid 2-[(1R)-1-carboxy-2-naphthalen-2-ylethyl]-1,3-dioxoisoindole-5-carboxylic acid 2-[(2R)-3-naphthalen-2-yl-1-oxidanyl-1-oxidanylidene-propan-2-yl]-1,3-bis(oxidanylidene)isoindole-5-carboxylic acid cid_16752641 NCGC00161791-01
2-[(1R)-1-carboxy-2-(2-naphthalenyl)ethyl]-1,3-dioxo-5-isoindolecarboxylic acid BDBM41851 2-[(1R)-1-carboxy-2-(2-naphthyl)ethyl]-1,3-diketo-isoindoline-5-carboxylic acid 2-[(1R)-1-carboxy-2-naphthalen-2-ylethyl]-1,3-dioxoisoindole-5-carboxylic acid 2-[(2R)-3-naphthalen-2-yl-1-oxidanyl-1-oxidanylidene-propan-2-yl]-1,3-bis(oxidanylidene)isoindole-5-carboxylic acid cid_16752641 NCGC00161791-01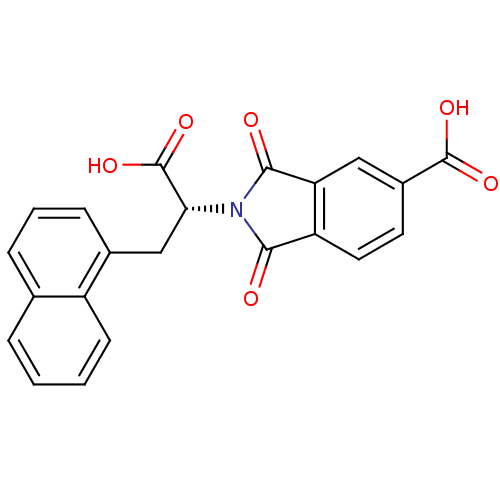 BDBM41850 2-[(1R)-1-carboxy-2-(1-naphthalenyl)ethyl]-1,3-dioxo-5-isoindolecarboxylic acid 2-[(1R)-1-carboxy-2-(1-naphthyl)ethyl]-1,3-diketo-isoindoline-5-carboxylic acid cid_16752640 2-[(2R)-3-naphthalen-1-yl-1-oxidanyl-1-oxidanylidene-propan-2-yl]-1,3-bis(oxidanylidene)isoindole-5-carboxylic acid NCGC00161790-01 2-[(1R)-1-carboxy-2-naphthalen-1-ylethyl]-1,3-dioxoisoindole-5-carboxylic acid
BDBM41850 2-[(1R)-1-carboxy-2-(1-naphthalenyl)ethyl]-1,3-dioxo-5-isoindolecarboxylic acid 2-[(1R)-1-carboxy-2-(1-naphthyl)ethyl]-1,3-diketo-isoindoline-5-carboxylic acid cid_16752640 2-[(2R)-3-naphthalen-1-yl-1-oxidanyl-1-oxidanylidene-propan-2-yl]-1,3-bis(oxidanylidene)isoindole-5-carboxylic acid NCGC00161790-01 2-[(1R)-1-carboxy-2-naphthalen-1-ylethyl]-1,3-dioxoisoindole-5-carboxylic acid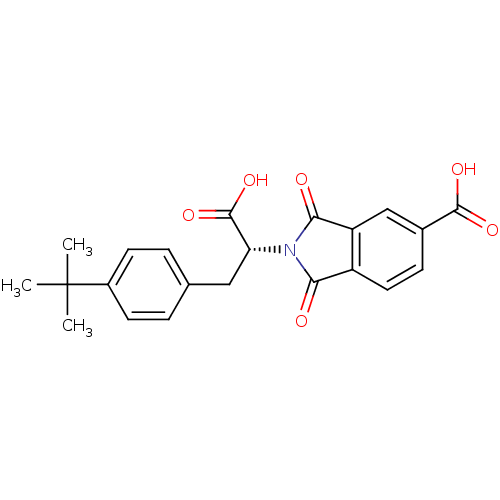 BDBM41853 2-[(2R)-3-(4-tert-butylphenyl)-1-oxidanyl-1-oxidanylidene-propan-2-yl]-1,3-bis(oxidanylidene)isoindole-5-carboxylic acid 2-[(1R)-2-(4-tert-butylphenyl)-1-carboxyethyl]-1,3-dioxo-5-isoindolecarboxylic acid NCGC00161801-01 cid_16752651 2-[(1R)-2-(4-tert-butylphenyl)-1-carboxy-ethyl]-1,3-diketo-isoindoline-5-carboxylic acid 2-[(1R)-2-(4-tert-butylphenyl)-1-carboxyethyl]-1,3-dioxoisoindole-5-carboxylic acid
BDBM41853 2-[(2R)-3-(4-tert-butylphenyl)-1-oxidanyl-1-oxidanylidene-propan-2-yl]-1,3-bis(oxidanylidene)isoindole-5-carboxylic acid 2-[(1R)-2-(4-tert-butylphenyl)-1-carboxyethyl]-1,3-dioxo-5-isoindolecarboxylic acid NCGC00161801-01 cid_16752651 2-[(1R)-2-(4-tert-butylphenyl)-1-carboxy-ethyl]-1,3-diketo-isoindoline-5-carboxylic acid 2-[(1R)-2-(4-tert-butylphenyl)-1-carboxyethyl]-1,3-dioxoisoindole-5-carboxylic acid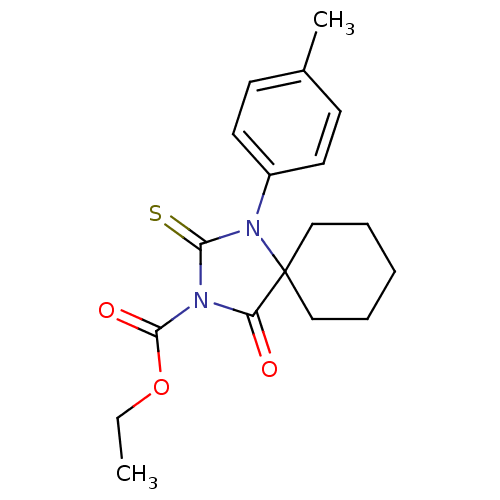 ethyl 1-(4-methylphenyl)-4-oxidanylidene-2-sulfanylidene-1,3-diazaspiro[4.5]decane-3-carboxylate 4-keto-1-(p-tolyl)-2-thioxo-1,3-diazaspiro[4.5]decane-3-carboxylic acid ethyl ester BRD-K97464451-001-01-9 1-(4-methylphenyl)-4-oxo-2-sulfanylidene-1,3-diazaspiro[4.5]decane-3-carboxylic acid ethyl ester ethyl 1-(4-methylphenyl)-4-oxo-2-sulfanylidene-1,3-diazaspiro[4.5]decane-3-carboxylate cid_3889161 BDBM68335
ethyl 1-(4-methylphenyl)-4-oxidanylidene-2-sulfanylidene-1,3-diazaspiro[4.5]decane-3-carboxylate 4-keto-1-(p-tolyl)-2-thioxo-1,3-diazaspiro[4.5]decane-3-carboxylic acid ethyl ester BRD-K97464451-001-01-9 1-(4-methylphenyl)-4-oxo-2-sulfanylidene-1,3-diazaspiro[4.5]decane-3-carboxylic acid ethyl ester ethyl 1-(4-methylphenyl)-4-oxo-2-sulfanylidene-1,3-diazaspiro[4.5]decane-3-carboxylate cid_3889161 BDBM68335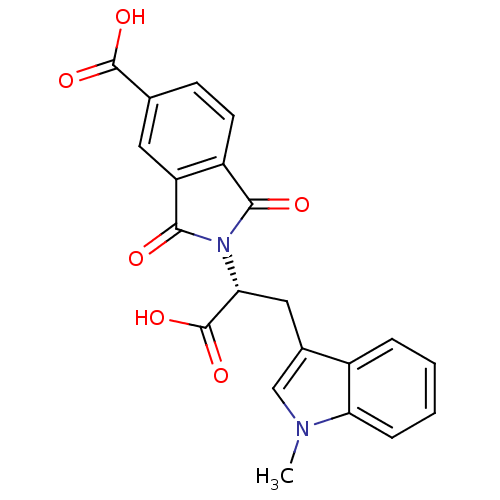 2-[(1R)-1-carboxy-2-(1-methylindol-3-yl)ethyl]-1,3-diketo-isoindoline-5-carboxylic acid cid_16752638 2-[(2R)-3-(1-methylindol-3-yl)-1-oxidanyl-1-oxidanylidene-propan-2-yl]-1,3-bis(oxidanylidene)isoindole-5-carboxylic acid NCGC00161788-01 2-[(1R)-1-carboxy-2-(1-methylindol-3-yl)ethyl]-1,3-dioxoisoindole-5-carboxylic acid 2-[(1R)-1-carboxy-2-(1-methyl-3-indolyl)ethyl]-1,3-dioxo-5-isoindolecarboxylic acid BDBM41849
2-[(1R)-1-carboxy-2-(1-methylindol-3-yl)ethyl]-1,3-diketo-isoindoline-5-carboxylic acid cid_16752638 2-[(2R)-3-(1-methylindol-3-yl)-1-oxidanyl-1-oxidanylidene-propan-2-yl]-1,3-bis(oxidanylidene)isoindole-5-carboxylic acid NCGC00161788-01 2-[(1R)-1-carboxy-2-(1-methylindol-3-yl)ethyl]-1,3-dioxoisoindole-5-carboxylic acid 2-[(1R)-1-carboxy-2-(1-methyl-3-indolyl)ethyl]-1,3-dioxo-5-isoindolecarboxylic acid BDBM41849 cid_16752637 2-[(1R)-1-carboxy-2-(1H-indol-3-yl)ethyl]-1,3-dioxo-5-isoindolecarboxylic acid NCGC00161787-01 2-[(1R)-1-carboxy-2-(1H-indol-3-yl)ethyl]-1,3-diketo-isoindoline-5-carboxylic acid 2-[(1R)-1-carboxy-2-(1H-indol-3-yl)ethyl]-1,3-dioxoisoindole-5-carboxylic acid 2-[(2R)-3-(1H-indol-3-yl)-1-oxidanyl-1-oxidanylidene-propan-2-yl]-1,3-bis(oxidanylidene)isoindole-5-carboxylic acid BDBM41848
cid_16752637 2-[(1R)-1-carboxy-2-(1H-indol-3-yl)ethyl]-1,3-dioxo-5-isoindolecarboxylic acid NCGC00161787-01 2-[(1R)-1-carboxy-2-(1H-indol-3-yl)ethyl]-1,3-diketo-isoindoline-5-carboxylic acid 2-[(1R)-1-carboxy-2-(1H-indol-3-yl)ethyl]-1,3-dioxoisoindole-5-carboxylic acid 2-[(2R)-3-(1H-indol-3-yl)-1-oxidanyl-1-oxidanylidene-propan-2-yl]-1,3-bis(oxidanylidene)isoindole-5-carboxylic acid BDBM41848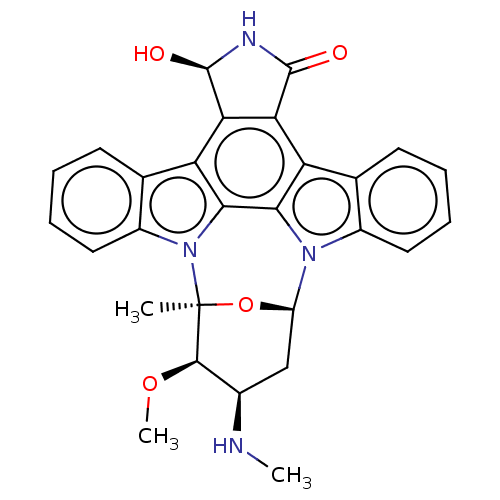 (5S,6R,7R,9R,16R)-16-hydroxy-6-methoxy-5-methyl-7-(methylamino)-6,7,8,9,15,16-hexahydro-5H,14H-5,9-epoxy-4b,9a,15-triazadibenzo[b,h]cyclonona[1,2,3,4-jkl]cyclopenta[e]-as-indacen-14-one (2S,3R,4R,6R,18R)-18-hydroxy-3-methoxy-2-methyl-4-(methylamino)-29-oxa-1,7,17-triazaoctacyclo[12.12.2.1^{2,6}.0^{7,28}.0^{8,13}.0^{15,19}.0^{20,27}.0^{21,26}]nonacosa-8,10,12,14,19,21,23,25,27-nonaen-16-one 7-Hydroxystaurosporine BDBM17054 UCN-01
(5S,6R,7R,9R,16R)-16-hydroxy-6-methoxy-5-methyl-7-(methylamino)-6,7,8,9,15,16-hexahydro-5H,14H-5,9-epoxy-4b,9a,15-triazadibenzo[b,h]cyclonona[1,2,3,4-jkl]cyclopenta[e]-as-indacen-14-one (2S,3R,4R,6R,18R)-18-hydroxy-3-methoxy-2-methyl-4-(methylamino)-29-oxa-1,7,17-triazaoctacyclo[12.12.2.1^{2,6}.0^{7,28}.0^{8,13}.0^{15,19}.0^{20,27}.0^{21,26}]nonacosa-8,10,12,14,19,21,23,25,27-nonaen-16-one 7-Hydroxystaurosporine BDBM17054 UCN-01 BRD-K32172653-001-01-2 N-[(2S,3S)-5-[(2R)-1-hydroxypropan-2-yl]-3-methyl-2-[[methyl(thiophen-2-ylsulfonyl)amino]methyl]-6-oxo-2,3,4,7-tetrahydro-1,5-benzoxazonin-9-yl]-2-phenylacetamide N-[(2S,3S)-3-methyl-2-[[methyl(thiophen-2-ylsulfonyl)amino]methyl]-6-oxidanylidene-5-[(2R)-1-oxidanylpropan-2-yl]-2,3,4,7-tetrahydro-1,5-benzoxazonin-9-yl]-2-phenyl-ethanamide cid_44485194 BDBM75312 N-[(2S,3S)-5-[(1R)-2-hydroxy-1-methyl-ethyl]-6-keto-3-methyl-2-[[methyl(2-thienylsulfonyl)amino]methyl]-2,3,4,7-tetrahydro-1,5-benzoxazonin-9-yl]-2-phenyl-acetamide
BRD-K32172653-001-01-2 N-[(2S,3S)-5-[(2R)-1-hydroxypropan-2-yl]-3-methyl-2-[[methyl(thiophen-2-ylsulfonyl)amino]methyl]-6-oxo-2,3,4,7-tetrahydro-1,5-benzoxazonin-9-yl]-2-phenylacetamide N-[(2S,3S)-3-methyl-2-[[methyl(thiophen-2-ylsulfonyl)amino]methyl]-6-oxidanylidene-5-[(2R)-1-oxidanylpropan-2-yl]-2,3,4,7-tetrahydro-1,5-benzoxazonin-9-yl]-2-phenyl-ethanamide cid_44485194 BDBM75312 N-[(2S,3S)-5-[(1R)-2-hydroxy-1-methyl-ethyl]-6-keto-3-methyl-2-[[methyl(2-thienylsulfonyl)amino]methyl]-2,3,4,7-tetrahydro-1,5-benzoxazonin-9-yl]-2-phenyl-acetamide 1-(1,3-benzodioxol-5-yl)-3-[(2S,3R)-2-[[cyclopropylmethyl(methyl)amino]methyl]-5-[(1R)-2-hydroxy-1-methyl-ethyl]-6-keto-3-methyl-2,3,4,7-tetrahydro-1,5-benzoxazonin-9-yl]urea 1-(1,3-benzodioxol-5-yl)-3-[(2S,3R)-2-[[cyclopropylmethyl(methyl)amino]methyl]-5-[(2R)-1-hydroxypropan-2-yl]-3-methyl-6-oxo-2,3,4,7-tetrahydro-1,5-benzoxazonin-9-yl]urea BRD-K49995760-001-01-7 1-(1,3-benzodioxol-5-yl)-3-[(2S,3R)-2-[[cyclopropylmethyl(methyl)amino]methyl]-3-methyl-6-oxidanylidene-5-[(2R)-1-oxidanylpropan-2-yl]-2,3,4,7-tetrahydro-1,5-benzoxazonin-9-yl]urea cid_44489252 BDBM75314
1-(1,3-benzodioxol-5-yl)-3-[(2S,3R)-2-[[cyclopropylmethyl(methyl)amino]methyl]-5-[(1R)-2-hydroxy-1-methyl-ethyl]-6-keto-3-methyl-2,3,4,7-tetrahydro-1,5-benzoxazonin-9-yl]urea 1-(1,3-benzodioxol-5-yl)-3-[(2S,3R)-2-[[cyclopropylmethyl(methyl)amino]methyl]-5-[(2R)-1-hydroxypropan-2-yl]-3-methyl-6-oxo-2,3,4,7-tetrahydro-1,5-benzoxazonin-9-yl]urea BRD-K49995760-001-01-7 1-(1,3-benzodioxol-5-yl)-3-[(2S,3R)-2-[[cyclopropylmethyl(methyl)amino]methyl]-3-methyl-6-oxidanylidene-5-[(2R)-1-oxidanylpropan-2-yl]-2,3,4,7-tetrahydro-1,5-benzoxazonin-9-yl]urea cid_44489252 BDBM75314 BDBM75321 N-[[(3S,9R,10R)-3,10-dimethyl-16-(methylsulfonylamino)-13-oxidanylidene-12-[(2S)-1-oxidanylpropan-2-yl]-2,8-dioxa-12-azabicyclo[12.4.0]octadeca-1(14),15,17-trien-9-yl]methyl]-N-methyl-2-phenyl-ethanamide N-[[(3S,9R,10R)-12-[(1S)-2-hydroxy-1-methyl-ethyl]-13-keto-16-(methanesulfonamido)-3,10-dimethyl-2,8-dioxa-12-azabicyclo[12.4.0]octadeca-1(14),15,17-trien-9-yl]methyl]-N-methyl-2-phenyl-acetamide BRD-K41135617-001-01-3 N-[[(3S,9R,10R)-12-[(2S)-1-hydroxypropan-2-yl]-16-(methanesulfonamido)-3,10-dimethyl-13-oxo-2,8-dioxa-12-azabicyclo[12.4.0]octadeca-1(14),15,17-trien-9-yl]methyl]-N-methyl-2-phenylacetamide cid_44495024
BDBM75321 N-[[(3S,9R,10R)-3,10-dimethyl-16-(methylsulfonylamino)-13-oxidanylidene-12-[(2S)-1-oxidanylpropan-2-yl]-2,8-dioxa-12-azabicyclo[12.4.0]octadeca-1(14),15,17-trien-9-yl]methyl]-N-methyl-2-phenyl-ethanamide N-[[(3S,9R,10R)-12-[(1S)-2-hydroxy-1-methyl-ethyl]-13-keto-16-(methanesulfonamido)-3,10-dimethyl-2,8-dioxa-12-azabicyclo[12.4.0]octadeca-1(14),15,17-trien-9-yl]methyl]-N-methyl-2-phenyl-acetamide BRD-K41135617-001-01-3 N-[[(3S,9R,10R)-12-[(2S)-1-hydroxypropan-2-yl]-16-(methanesulfonamido)-3,10-dimethyl-13-oxo-2,8-dioxa-12-azabicyclo[12.4.0]octadeca-1(14),15,17-trien-9-yl]methyl]-N-methyl-2-phenylacetamide cid_44495024 BDBM75320 cid_44494902 4-(dimethylamino)-N-[(3R,9S,10S)-12-[(1R)-2-hydroxy-1-methyl-ethyl]-13-keto-3,10-dimethyl-9-[[methyl(piperonyl)amino]methyl]-2,8-dioxa-12-azabicyclo[12.4.0]octadeca-1(14),15,17-trien-16-yl]butyramide N-[(3R,9S,10S)-9-[[1,3-benzodioxol-5-ylmethyl(methyl)amino]methyl]-12-[(2R)-1-hydroxypropan-2-yl]-3,10-dimethyl-13-oxo-2,8-dioxa-12-azabicyclo[12.4.0]octadeca-1(14),15,17-trien-16-yl]-4-(dimethylamino)butanamide N-[(3R,9S,10S)-9-[[1,3-benzodioxol-5-ylmethyl(methyl)amino]methyl]-3,10-dimethyl-13-oxidanylidene-12-[(2R)-1-oxidanylpropan-2-yl]-2,8-dioxa-12-azabicyclo[12.4.0]octadeca-1(14),15,17-trien-16-yl]-4-(dimethylamino)butanamide BRD-K94081657-001-01-6
BDBM75320 cid_44494902 4-(dimethylamino)-N-[(3R,9S,10S)-12-[(1R)-2-hydroxy-1-methyl-ethyl]-13-keto-3,10-dimethyl-9-[[methyl(piperonyl)amino]methyl]-2,8-dioxa-12-azabicyclo[12.4.0]octadeca-1(14),15,17-trien-16-yl]butyramide N-[(3R,9S,10S)-9-[[1,3-benzodioxol-5-ylmethyl(methyl)amino]methyl]-12-[(2R)-1-hydroxypropan-2-yl]-3,10-dimethyl-13-oxo-2,8-dioxa-12-azabicyclo[12.4.0]octadeca-1(14),15,17-trien-16-yl]-4-(dimethylamino)butanamide N-[(3R,9S,10S)-9-[[1,3-benzodioxol-5-ylmethyl(methyl)amino]methyl]-3,10-dimethyl-13-oxidanylidene-12-[(2R)-1-oxidanylpropan-2-yl]-2,8-dioxa-12-azabicyclo[12.4.0]octadeca-1(14),15,17-trien-16-yl]-4-(dimethylamino)butanamide BRD-K94081657-001-01-6 1-[[(2R,3R)-9-(dimethylamino)-5-[(2R)-1-hydroxypropan-2-yl]-3-methyl-6-oxo-2,3,4,7-tetrahydro-1,5-benzoxazonin-2-yl]methyl]-3-(3,5-dimethyl-4-isoxazolyl)-1-methylurea 1-[[(2R,3R)-9-(dimethylamino)-5-[(1R)-2-hydroxy-1-methyl-ethyl]-6-keto-3-methyl-2,3,4,7-tetrahydro-1,5-benzoxazonin-2-yl]methyl]-3-(3,5-dimethylisoxazol-4-yl)-1-methyl-urea 1-[[(2R,3R)-9-(dimethylamino)-5-[(2R)-1-hydroxypropan-2-yl]-3-methyl-6-oxo-2,3,4,7-tetrahydro-1,5-benzoxazonin-2-yl]methyl]-3-(3,5-dimethyl-1,2-oxazol-4-yl)-1-methylurea BDBM75318 cid_44494069 1-[[(2R,3R)-9-(dimethylamino)-3-methyl-6-oxidanylidene-5-[(2R)-1-oxidanylpropan-2-yl]-2,3,4,7-tetrahydro-1,5-benzoxazonin-2-yl]methyl]-3-(3,5-dimethyl-1,2-oxazol-4-yl)-1-methyl-urea BRD-K72792753-001-01-7
1-[[(2R,3R)-9-(dimethylamino)-5-[(2R)-1-hydroxypropan-2-yl]-3-methyl-6-oxo-2,3,4,7-tetrahydro-1,5-benzoxazonin-2-yl]methyl]-3-(3,5-dimethyl-4-isoxazolyl)-1-methylurea 1-[[(2R,3R)-9-(dimethylamino)-5-[(1R)-2-hydroxy-1-methyl-ethyl]-6-keto-3-methyl-2,3,4,7-tetrahydro-1,5-benzoxazonin-2-yl]methyl]-3-(3,5-dimethylisoxazol-4-yl)-1-methyl-urea 1-[[(2R,3R)-9-(dimethylamino)-5-[(2R)-1-hydroxypropan-2-yl]-3-methyl-6-oxo-2,3,4,7-tetrahydro-1,5-benzoxazonin-2-yl]methyl]-3-(3,5-dimethyl-1,2-oxazol-4-yl)-1-methylurea BDBM75318 cid_44494069 1-[[(2R,3R)-9-(dimethylamino)-3-methyl-6-oxidanylidene-5-[(2R)-1-oxidanylpropan-2-yl]-2,3,4,7-tetrahydro-1,5-benzoxazonin-2-yl]methyl]-3-(3,5-dimethyl-1,2-oxazol-4-yl)-1-methyl-urea BRD-K72792753-001-01-7 BRD-K25694548-001-01-3 N-[(2R,3S)-5-[(1S)-2-hydroxy-1-methyl-ethyl]-6-keto-3-methyl-2-[[methyl(phenylcarbamoyl)amino]methyl]-3,4-dihydro-2H-1,5-benzoxazocin-10-yl]-4-thiazol-2-yl-benzamide BDBM75319 cid_44494900 N-[(2R,3S)-3-methyl-2-[[methyl(phenylcarbamoyl)amino]methyl]-6-oxidanylidene-5-[(2S)-1-oxidanylpropan-2-yl]-3,4-dihydro-2H-1,5-benzoxazocin-10-yl]-4-(1,3-thiazol-2-yl)benzamide N-[(2R,3S)-5-[(2S)-1-hydroxypropan-2-yl]-3-methyl-2-[[methyl(phenylcarbamoyl)amino]methyl]-6-oxo-3,4-dihydro-2H-1,5-benzoxazocin-10-yl]-4-(1,3-thiazol-2-yl)benzamide N-[(2R,3S)-2-[[[anilino(oxo)methyl]-methylamino]methyl]-5-[(2S)-1-hydroxypropan-2-yl]-3-methyl-6-oxo-3,4-dihydro-2H-1,5-benzoxazocin-10-yl]-4-(2-thiazolyl)benzamide
BRD-K25694548-001-01-3 N-[(2R,3S)-5-[(1S)-2-hydroxy-1-methyl-ethyl]-6-keto-3-methyl-2-[[methyl(phenylcarbamoyl)amino]methyl]-3,4-dihydro-2H-1,5-benzoxazocin-10-yl]-4-thiazol-2-yl-benzamide BDBM75319 cid_44494900 N-[(2R,3S)-3-methyl-2-[[methyl(phenylcarbamoyl)amino]methyl]-6-oxidanylidene-5-[(2S)-1-oxidanylpropan-2-yl]-3,4-dihydro-2H-1,5-benzoxazocin-10-yl]-4-(1,3-thiazol-2-yl)benzamide N-[(2R,3S)-5-[(2S)-1-hydroxypropan-2-yl]-3-methyl-2-[[methyl(phenylcarbamoyl)amino]methyl]-6-oxo-3,4-dihydro-2H-1,5-benzoxazocin-10-yl]-4-(1,3-thiazol-2-yl)benzamide N-[(2R,3S)-2-[[[anilino(oxo)methyl]-methylamino]methyl]-5-[(2S)-1-hydroxypropan-2-yl]-3-methyl-6-oxo-3,4-dihydro-2H-1,5-benzoxazocin-10-yl]-4-(2-thiazolyl)benzamide BRD-K56287340-001-01-8 cid_44485648 N-[(2S,3R)-2-[[cyclopropylmethyl(methyl)amino]methyl]-5-[(1R)-2-hydroxy-1-methyl-ethyl]-6-keto-3-methyl-3,4-dihydro-2H-1,5-benzoxazocin-10-yl]-4-thiazol-2-yl-benzamide N-[(2S,3R)-2-[[cyclopropylmethyl(methyl)amino]methyl]-5-[(2R)-1-hydroxypropan-2-yl]-3-methyl-6-oxo-3,4-dihydro-2H-1,5-benzoxazocin-10-yl]-4-(2-thiazolyl)benzamide BDBM75313 N-[(2S,3R)-2-[[cyclopropylmethyl(methyl)amino]methyl]-3-methyl-6-oxidanylidene-5-[(2R)-1-oxidanylpropan-2-yl]-3,4-dihydro-2H-1,5-benzoxazocin-10-yl]-4-(1,3-thiazol-2-yl)benzamide N-[(2S,3R)-2-[[cyclopropylmethyl(methyl)amino]methyl]-5-[(2R)-1-hydroxypropan-2-yl]-3-methyl-6-oxo-3,4-dihydro-2H-1,5-benzoxazocin-10-yl]-4-(1,3-thiazol-2-yl)benzamide
BRD-K56287340-001-01-8 cid_44485648 N-[(2S,3R)-2-[[cyclopropylmethyl(methyl)amino]methyl]-5-[(1R)-2-hydroxy-1-methyl-ethyl]-6-keto-3-methyl-3,4-dihydro-2H-1,5-benzoxazocin-10-yl]-4-thiazol-2-yl-benzamide N-[(2S,3R)-2-[[cyclopropylmethyl(methyl)amino]methyl]-5-[(2R)-1-hydroxypropan-2-yl]-3-methyl-6-oxo-3,4-dihydro-2H-1,5-benzoxazocin-10-yl]-4-(2-thiazolyl)benzamide BDBM75313 N-[(2S,3R)-2-[[cyclopropylmethyl(methyl)amino]methyl]-3-methyl-6-oxidanylidene-5-[(2R)-1-oxidanylpropan-2-yl]-3,4-dihydro-2H-1,5-benzoxazocin-10-yl]-4-(1,3-thiazol-2-yl)benzamide N-[(2S,3R)-2-[[cyclopropylmethyl(methyl)amino]methyl]-5-[(2R)-1-hydroxypropan-2-yl]-3-methyl-6-oxo-3,4-dihydro-2H-1,5-benzoxazocin-10-yl]-4-(1,3-thiazol-2-yl)benzamide BRD-K94479191-001-01-5 methyl N-[[(8R,9S)-6-[(2S)-1-hydroxypropan-2-yl]-8-methyl-5-oxo-10-oxa-1,6,13,14-tetrazabicyclo[10.2.1]pentadeca-12(15),13-dien-9-yl]methyl]-N-methylcarbamate methyl N-methyl-N-[[(8R,9S)-8-methyl-5-oxidanylidene-6-[(2S)-1-oxidanylpropan-2-yl]-10-oxa-1,6,13,14-tetrazabicyclo[10.2.1]pentadeca-12(15),13-dien-9-yl]methyl]carbamate N-[[(8R,9S)-6-[(1S)-2-hydroxy-1-methyl-ethyl]-5-keto-8-methyl-10-oxa-1,6,13,14-tetrazabicyclo[10.2.1]pentadeca-12(15),13-dien-9-yl]methyl]-N-methyl-carbamic acid methyl ester N-[[(8R,9S)-6-[(2S)-1-hydroxypropan-2-yl]-8-methyl-5-oxo-10-oxa-1,6,13,14-tetrazabicyclo[10.2.1]pentadeca-12(15),13-dien-9-yl]methyl]-N-methylcarbamic acid methyl ester BDBM75316 cid_44489808
BRD-K94479191-001-01-5 methyl N-[[(8R,9S)-6-[(2S)-1-hydroxypropan-2-yl]-8-methyl-5-oxo-10-oxa-1,6,13,14-tetrazabicyclo[10.2.1]pentadeca-12(15),13-dien-9-yl]methyl]-N-methylcarbamate methyl N-methyl-N-[[(8R,9S)-8-methyl-5-oxidanylidene-6-[(2S)-1-oxidanylpropan-2-yl]-10-oxa-1,6,13,14-tetrazabicyclo[10.2.1]pentadeca-12(15),13-dien-9-yl]methyl]carbamate N-[[(8R,9S)-6-[(1S)-2-hydroxy-1-methyl-ethyl]-5-keto-8-methyl-10-oxa-1,6,13,14-tetrazabicyclo[10.2.1]pentadeca-12(15),13-dien-9-yl]methyl]-N-methyl-carbamic acid methyl ester N-[[(8R,9S)-6-[(2S)-1-hydroxypropan-2-yl]-8-methyl-5-oxo-10-oxa-1,6,13,14-tetrazabicyclo[10.2.1]pentadeca-12(15),13-dien-9-yl]methyl]-N-methylcarbamic acid methyl ester BDBM75316 cid_44489808