Compound (286)
Article Title (3)
Article Author (104)
Assay (55)
Aihara, Y; Yoshida, A; Furuta, T; Wakimoto, T; Akizawa, T; Konishi, M; Kan, T Regioselective synthesis of methylated epigallocatechin gallate via nitrobenzenesulfonyl (Ns) protecting group. Bioorg Med Chem Lett 19: 4171 -4 (2009) Varming, T; Christophersen, P; Møller, A; Peters, D; Axelsson, O; Nielsen, E& Synthesis and biological activity of the neuronal calcium channel blocker 2-amino-1-(2,5-dimethoxyphenyl)-5-trifluoromethyl benzimidazole (NS-649) Bioorg Med Chem Lett 6: 245 -248 (1996) Wätjen, F; Bigge, CF; Jensen, LH; Boxer, PA; Lescosky, LJ; Nielsen, E&; Malone, TC; Campbell, GW; Coughenour, LL; Rock, DM; Drejer, J; Marcoux, FW NS 257 (1,2,3,6,7,8-hexahydro-3(hydroxyimino)-N,N,7-trimethyl-2-oxobenzo[2,1-b:3,4-c']dipyrrole-5-sulfonamide) is a potent, systemically active ampa receptor antagonist Bioorg Med Chem Lett 4: 371 -376 (1994)
Abutaleb NS Agarwal, NS; Rich, DH J Med Chem 29: 2519 -24 (1987) Srour, AM; Ahmed, NS; Abd El-Karim, SS; Anwar, MM; El-Hallouty, SM Bioorg Med Chem 28: (2020) Khan, MIH; Sawyer, BJ; Akins, NS; Le, HV Eur J Med Chem 243: (2022) Stepanov, AI; Astrat'ev, AA; Sheremetev, AB; Lagutina, NK; Palysaeva, NV; Tyurin, AY; Aleksandrova, NS; Sadchikova, NP; Suponitsky, KY; Atamanenko, OP; Konyushkin, LD; Semenov, RV; Firgang, SI; Kiselyov, AS; Semenova, MN; Semenov, VV Eur J Med Chem 94: 237 -51 (2015) Huang, YM; Alharbi, NS; Sun, B; Shantharam, CS; Rakesh, KP; Qin, HL Eur J Med Chem 181: (2019) Bahadur, A; Iqbal, S; Muneer, S; Alsaab, HO; Awwad, NS; Ibrahium, HA Bioorg Med Chem Lett 57: (2022) Wendeler, M; Lee, HF; Bermingham, A; Miller, JT; Chertov, O; Bona, MK; Baichoo, NS; Ehteshami, M; Beutler, J; Oamppound39Keefe, BR; Götte, M; Kvaratskhelia, M; Le Grice, S ACS Chem Biol 3: 635 -44 (2008) Dounay, AB; Barta, NS; Bikker, JA; Borosky, SA; Campbell, BM; Crawford, T; Denny, L; Evans, LM; Gray, DL; Lee, P; Lenoir, EA; Xu, W Bioorg Med Chem Lett 19: 1159 -63 (2009) Gupta, P; Mehrotra, S; Sharma, A; Chugh, M; Pandey, R; Kaushik, A; Khurana, S; Srivastava, N; Srivastava, T; Deshmukh, A; Panda, A; Aggarwal, P; Bhavesh, NS; Bhatnagar, RK; Mohmmed, A; Gupta, D; Malhotra, P J Med Chem 60: 8298 -8308 (2017) Large, JM; Birchall, K; Bouloc, NS; Merritt, AT; Smiljanic-Hurley, E; Tsagris, DJ; Wheldon, MC; Ansell, KH; Coombs, PJ; Kettleborough, CA; Whalley, D; Stewart, LB; Bowyer, PW; Baker, DA; Osborne, SA Bioorg Med Chem Lett 29: (2019) Mylari, BL; Oates, PJ; Beebe, DA; Brackett, NS; Coutcher, JB; Dina, MS; Zembrowski, WJ J Med Chem 44: 2695 -700 (2001) CUTSHALL, NS; HADD, MA; KESHIPEDDY, SK; LEMUS, RH; METZ, M US Patent US20250066383 (2025) Bonifazi, A; Yano, H; Ellenberger, MP; Muller, L; Kumar, V; Zou, MF; Cai, NS; Guerrero, AM; Woods, AS; Shi, L; Newman, AH J Med Chem 60: 2890 -2907 (2017) Penning, TD; Chandrakumar, NS; Desai, BN; Djuric, SW; Gasiecki, AF; Liang, CD; Miyashiro, JM; Russell, MA; Askonas, LJ; Gierse, JK; Harding, EI; Highkin, MK; Kachur, JF; Kim, SH; Villani-Price, D; Pyla, EY; Ghoreishi-Haack, NS; Smith, WG Bioorg Med Chem Lett 12: 3383 -6 (2002) Lee, S; Lee, H; Yi, KY; Lee, BH; Yoo, SE; Lee, K; Cho, NS Bioorg Med Chem Lett 15: 2998 -3001 (2005) Ahn, SK; Auh, J; Choi, NS; Han, CK; Kim, T; Pae, K; Shin, YJ; Han, D; Han, CK US Patent US9464044 (2016) Poudel, YB; He, L; Cox, M; Zhang, Q; Johnson, WL; Cong, Q; Cheng, H; Chowdari, NS; Tarby, C; Donnell, AF; Broekema, M; O'Malley, DP; Zhang, Y; A M Subbaiah, M; Kumar, BV; Subramani, L; Wang, B; Li, YX; Sivaprakasam, P; Critton, D; Mulligan, D; Sandhu, B; Xie, C; Ramakrishnan, R; Nagar, J; Dudhgaonkar, S; Oderinde, MS; Murtaza, A; Schieven, GL; Mathur, A; Gavai, AV; Vite, G; Gangwar, S ACS Med Chem Lett 15: 181 -188 (2024) Kottirsch, G; Zerwes, HG; Cook, NS; Tapparelli, C Bioorg Med Chem Lett 7: 727 -732 (1997) Cutshall, NS; O'Day, C; Prezhdo, M Bioorg Med Chem Lett 15: 3374 -9 (2005) Zyk, NY; Ber, AP; Nimenko, EA; Shafikov, RR; Evteev, SA; Petrov, SA; Uspenskaya, AA; Dashkova, NS; Ivanenkov, YA; Skvortsov, DA; Beloglazkina, EK; Majouga, AG; Machulkin, AE Bioorg Med Chem Lett 71: (2022) Mun, J; Jabbar, AA; Devi, NS; Yin, S; Wang, Y; Tan, C; Culver, D; Snyder, JP; Van Meir, EG; Goodman, MM J Med Chem 55: 6738 -50 (2012) Kastrinsky, DB; Sangodkar, J; Zaware, N; Izadmehr, S; Dhawan, NS; Narla, G; Ohlmeyer, M Bioorg Med Chem 23: 6528 -34 (2015) Mamontova, EM; Zakharenko, AL; Zakharova, OD; Dyrkheeva, NS; Volcho, KP; Reynisson, J; Arabshahi, HJ; Salakhutdinov, NF; Lavrik, OI Bioorg Med Chem 28: (2020) Quibell, M; Watts, JP; Flinn, NS US Patent US8552202 (2013) Tal-Gan, Y; Freeman, NS; Klein, S; Levitzki, A; Gilon, C Bioorg Med Chem 18: 2976 -85 (2010) Gandhi NS Garton, NS; Barker, MD; Davis, RP; Douault, C; Hooper-Greenhill, E; Jones, E; Lewis, HD; Liddle, J; Lugo, D; McCleary, S; Preston, AGS; Ramirez-Molina, C; Neu, M; Shipley, TJ; Somers, DO; Watson, RJ; Wilson, DM Bioorg Med Chem Lett 26: 4606 -4612 (2016) Maity, P; Chatterjee, J; Patil, KT; Arora, S; Katiyar, MK; Kumar, M; Samarbakhsh, A; Joshi, G; Bhutani, P; Chugh, M; Gavande, NS; Kumar, R J Med Chem 66: 3135 -3172 (2023) Eastwood, P; Esteve, C; González, J; Fonquerna, S; Aiguadé, J; Carranco, Is; Doménech, T; Aparici, Ms; Miralpeix, M; Albertí, J; Córdoba, M; Fernández, R; Pont, Ms; Godessart, Ns; Prats, N; Loza, Ms; Cadavid, Ms; Nueda, A ACS Med Chem Lett 2: 213 -218 (2011) Noncovich, A; Priest, C; Ung, J; Patron, AP; Servant, G; Brust, P; Servant, N; Faber, N; Liu, H; Gonsalves, NS; Ditschun, TL Bioorg Med Chem Lett 27: 3931 -3938 (2017) Chandra, KM; Goud, NS; Arifuddin, M; Alvala, M; Alvala, R; Angeli, A; Supuran, CT Bioorg Med Chem Lett 39: (2021) Gray, NS; Wodicka, L; Thunnissen, AM; Norman, TC; Kwon, S; Espinoza, FH; Morgan, DO; Barnes, G; LeClerc, S; Meijer, L; Kim, SH; Lockhart, DJ; Schultz, PG Science 281: 533 -538 (1998) El Massry, AM; Asal, AM; Khattab, SN; Haiba, NS; Awney, HA; Helmy, M; Langer, V; Amer, A Bioorg Med Chem 20: 2624 -37 (2012) Letavic, MA; Axt, MZ; Barberia, JT; Carty, TJ; Danley, DE; Geoghegan, KF; Halim, NS; Hoth, LR; Kamath, AV; Laird, ER; Lopresti-Morrow, LL; McClure, KF; Mitchell, PG; Natarajan, V; Noe, MC; Pandit, J; Reeves, L; Schulte, GK; Snow, SL; Sweeney, FJ; Tan, DH; Yu, CH Bioorg Med Chem Lett 12: 1387 -90 (2002) Hamilton, NM; Dawson, M; Fairweather, EE; Hamilton, NS; Hitchin, JR; James, DI; Jones, SD; Jordan, AM; Lyons, AJ; Small, HF; Thomson, GJ; Waddell, ID; Ogilvie, DJ J Med Chem 55: 4431 -45 (2012) Perry, MWD; Björhall, K; Bonn, B; Carlsson, J; Chen, Y; Eriksson, A; Fredlund, L; Hao, H; Holden, NS; Karabelas, K; Lindmark, H; Liu, F; Pemberton, N; Petersen, J; Rodrigo Blomqvist, S; Smith, RW; Svensson, T; Terstiege, I; Tyrchan, C; Yang, W; Zhao, S; Öster, L J Med Chem 60: 5057 -5071 (2017) Ilyinsky, NS; Shchyolkina, AK; Borisova, OF; Mamaeva, OK; Zvereva, MI; Azhibek, DM; Livshits, MA; Mitkevich, VA; Balzarini, J; Sinkevich, YB; Luzikov, YN; Dezhenkova, LG; Kolotova, ES; Shtil, AA; Shchekotikhin, AE; Kaluzhny, DN Eur J Med Chem 85: 605 -14 (2014) Ibrahim, HS; Abou-Seri, SM; Ismail, NS; Elaasser, MM; Aly, MH; Abdel-Aziz, HA Eur J Med Chem 108: 415 -22 (2016) Bennett, JM; Campbell, AD; Campbell, AJ; Carr, MG; Dunsdon, RM; Greening, JR; Hurst, DN; Jennings, NS; Jones, PS; Jordan, S; Kay, PB; O'Brien, MA; King-Underwood, J; Raynham, TM; Wilkinson, CS; Wilkinson, TC; Wilson, FX Bioorg Med Chem Lett 11: 355 -7 (2001) Nørager, NG; Poulsen, MH; Jensen, AG; Jeppesen, NS; Kristensen, AS; Strømgaard, K J Med Chem 57: 4940 -9 (2014) Yoon, HR; Balupuri, A; Lee, J; Lee, C; Son, DH; Jeoung, RG; Kim, KA; Choi, S; Kang, NS Bioorg Med Chem Lett 101: (2024) Hudelson, MG; Ketkar, NS; Holder, LB; Carlson, TJ; Peng, CC; Waldher, BJ; Jones, JP J Med Chem 51: 648 -54 (2008) Aneja, B; Khan, NS; Khan, P; Queen, A; Hussain, A; Rehman, MT; Alajmi, MF; El-Seedi, HR; Ali, S; Hassan, MI; Abid, M Eur J Med Chem 163: 840 -852 (2019) Devadas, B; Freeman, SK; McWherter, CA; Kishore, NS; Lodge, JK; Jackson-Machelski, E; Gordon, JI; Sikorski, JA J Med Chem 41: 996 -1000 (1998) Liu, G; Henry, KJ; Szczepankiewicz, BG; Winn, M; Kozmina, NS; Boyd, SA; Wasicak, J; von Geldern, TW; Wu-Wong, JR; Chiou, WJ; Dixon, DB; Nguyen, B; Marsh, KC; Opgenorth, TJ J Med Chem 41: 3261 -75 (1998) Tamagnan, G; Baldwin, RM; Kula, NS; Baldessarini, RJ; Innis, RB Bioorg Med Chem Lett 10: 1113 -5 (2000) Pradhan, NS; Patil, NS; Walavalkar, RR; Kulkarni, NS; Pawar, SB; Pawar, TS US Patent US9359341 (2016) Levy, DE; Schulman, H; Paraselli, BR; Kumar, NS; Dabbugoddu, B; Balasubramanyam, C US Patent US11325908 (2022) Khan, MM; Simizu, S; Lai, NS; Kawatani, M; Shimizu, T; Osada, H ACS Chem Biol 6: 245 -51 (2011) Sampson, D; Zhu, XY; Eyunni, SV; Etukala, JR; Ofori, E; Bricker, B; Lamango, NS; Setola, V; Roth, BL; Ablordeppey, SY Bioorg Med Chem 22: 3105 -14 (2014) Barnes, L; Blaber, H; Brooks, DTK; Byers, L; Buckley, D; Byron, ZC; Chilvers, RG; Cochrane, L; Cooney, E; Damian, HA; Francis, L; Fu He, D; Grace, JMJ; Green, HJ; Hogarth, EJP; Jusu, L; Killalea, CE; King, O; Lambert, J; Lee, ZJ; Lima, NS; Long, CL; Mackinnon, ML; Mahdy, S; Matthews-Wright, J; Millward, MJ; Meehan, MF; Merrett, C; Morrison, L; Parke, HRI; Payne, C; Payne, L; Pike, C; Seal, A; Senior, AJ; Smith, KM; Stanelyte, K; Stillibrand, J; Szpara, R; Taday, FFH; Threadgould, AM; Trainor, RJ; Waters, J; Williams, O; Wong, CKW; Wood, K; Barton, N; Gruszka, A; Henley, Z; Rowedder, JE; Cookson, R; Jones, KL; Nadin, A; Smith, IE; Macdonald, SJF; Nortcliffe, A J Med Chem 62: 10402 -10422 (2019) Boger, J; Payne, LS; Perlow, DS; Lohr, NS; Poe, M; Blaine, EH; Ulm, EH; Schorn, TW; LaMont, BI; Lin, TY J Med Chem 28: 1779 -90 (1986) McGovern-Gooch, KR; Mahajani, NS; Garagozzo, A; Schramm, AJ; Hannah, LG; Sieburg, MA; Chisholm, JD; Hougland, JL Biochemistry 56: 919 -931 (2017) Letavic, MA; Bonaventure, P; Carruthers, NI; Dugovic, C; Koudriakova, T; Lord, B; Lovenberg, TW; Ly, KS; Mani, NS; Nepomuceno, D; Pippel, DJ; Rizzolio, M; Shelton, JE; Shah, CR; Shireman, BT; Young, LK; Yun, S J Med Chem 58: 5620 -36 (2015) Kessler, RM; Votaw, JR; de Paulis, T; Bingham, DR; Ansari, MS; Mason, NS; Holburn, G; Schmidt, DE; Votaw, DB; Manning, RG Synapse 15: 169 -76 (1993) Sharples, CG; Karig, G; Simpson, GL; Spencer, JA; Wright, E; Millar, NS; Wonnacott, S; Gallagher, T J Med Chem 45: 3235 -45 (2002) Hobson, AD; Harris, CM; van der Kam, EL; Turner, SC; Abibi, A; Aguirre, AL; Bousquet, P; Kebede, T; Konopacki, DB; Gintant, G; Kim, Y; Larson, K; Maull, JW; Moore, NS; Shi, D; Shrestha, A; Tang, X; Zhang, P; Sarris, KK J Med Chem 58: 9154 -70 (2015) Balaji, S; Karthikeyan, C; Moorthy, NS; Trivedi, P Bioorg Med Chem Lett 14: 6089 -94 (2004) Murthy, NS; Bakeris, T; Kavarana, MJ; Hamilton, DS; Lan, Y; Creighton, DJ J Med Chem 37: 2161 -6 (1994) Sirivolu, VR; Vernekar, SK; Ilina, T; Myshakina, NS; Parniak, MA; Wang, Z J Med Chem 56: 8765 -80 (2013) Parlow, JJ; Case, BL; Dice, TA; Fenton, RL; Hayes, MJ; Jones, DE; Neumann, WL; Wood, RS; Lachance, RM; Girard, TJ; Nicholson, NS; Clare, M; Stegeman, RA; Stevens, AM; Stallings, WC; Kurumbail, RG; South, MS J Med Chem 46: 4050 -62 (2003) Qiao, L; Baumann, CA; Crysler, CS; Ninan, NS; Abad, MC; Spurlino, JC; Desjarlais, RL; Kervinen, J; Neeper, MP; Bayoumy, SS; Williams, R; Deckman, IC; Dasgupta, M; Reed, RL; Huebert, ND; Tomczuk, BE; Moriarty, KJ Bioorg Med Chem Lett 16: 123 -8 (2006) Ahmed-Belkacem, R; Hausdorff, M; Delpal, A; Sutto-Ortiz, P; Colmant, AMG; Touret, F; Ogando, NS; Snijder, EJ; Canard, B; Coutard, B; Vasseur, JJ; Decroly, E; Debart, F J Med Chem 65: 6231 -6249 (2022) Pabla NS Pagadala, NS; Bjorndahl, TC; Joyce, M; Wishart, DS; Syed, K; Landi, A Bioorg Med Chem 25: 5875 -5888 (2017) Park, CM; Kim, SY; Park, WK; Park, NS; Seong, CM Bioorg Med Chem Lett 18: 3844 -7 (2008) Pradhan, NS; Patil, NS; Walavalkar, RR; Kulkarni, NS; Pawar, SB; Pawar, TS US Patent US9359341 (2016) Pradhan, NS; Patil, NS; Walavalkar, RR; Kulkarni, NS; Pawar, SB; Pawar, TS US Patent US9359341 (2016) Bryan, MC; Do, S; Katsumoto, T; Liang, J; Rajapaksa, NS; Kiefer, Jr., JR; Fu, L US Patent US10899772 (2021) Haviv, F; Fitzpatrick, TD; Swenson, RE; Nichols, CJ; Mort, NA; Bush, EN; Diaz, G; Bammert, G; Nguyen, A; Rhutasel, NS J Med Chem 36: 363 -9 (1993) Heimbrook, DC; Saari, WS; Balishin, NL; Fisher, TW; Friedman, A; Kiefer, DM; Rotberg, NS; Wallen, JW; Oliff, A J Med Chem 34: 2102 -7 (1991) Londregan, AT; Piotrowski, DW; Futatsugi, K; Warmus, JS; Boehm, M; Carpino, PA; Chin, JE; Janssen, AM; Roush, NS; Buxton, J; Hinchey, T Bioorg Med Chem Lett 23: 1407 -11 (2013) Molteni, V; He, X; Nabakka, J; Yang, K; Kreusch, A; Gordon, P; Bursulaya, B; Warner, I; Shin, T; Biorac, T; Ryder, NS; Goldberg, R; Doughty, J; He, Y Bioorg Med Chem Lett 14: 1477 -81 (2004) STOCK, NS; CHEN, AC; BRAVO, YM; JACINTHO, JD; BACCEI, JM; STEARNS, BA; CLARK, RC US Patent US20240041837 (2024) Thomas, ST; Yang, X; Sampson, NS Bioorg Med Chem Lett 21: 2216 -9 (2011) Epperson, JR; Deskus, JA; Gentile, AJ; Iben, LG; Ryan, E; Sarbin, NS Bioorg Med Chem Lett 14: 1023 -6 (2004) Gordon, DE; Jang, GM; Bouhaddou, M; Xu, J; Obernier, K; White, KM; O'Meara, MJ; Rezelj, VV; Guo, JZ; Swaney, DL; Tummino, TA; Hüttenhain, R; Kaake, RM; Richards, AL; Tutuncuoglu, B; Foussard, H; Batra, J; Haas, K; Modak, M; Kim, M; Haas, P; Polacco, BJ; Braberg, H; Fabius, JM; Eckhardt, M; Soucheray, M; Bennett, MJ; Cakir, M; McGregor, MJ; Li, Q; Meyer, B; Roesch, F; Vallet, T; Mac Kain, A; Miorin, L; Moreno, E; Naing, ZZC; Zhou, Y; Peng, S; Shi, Y; Zhang, Z; Shen, W; Kirby, IT; Melnyk, JE; Chorba, JS; Lou, K; Dai, SA; Barrio-Hernandez, I; Memon, D; Hernandez-Armenta, C; Lyu, J; Mathy, CJP; Perica, T; Pilla, KB; Ganesan, SJ; Saltzberg, DJ; Rakesh, R; Liu, X; Rosenthal, SB; Calviello, L; Venkataramanan, S; Liboy-Lugo, J; Lin, Y; Huang, XP; Liu, Y; Wankowicz, SA; Bohn, M; Safari, M; Ugur, FS; Koh, C; Savar, NS; Tran, QD; Shengjuler, D; Fletcher, SJ; O'Neal, MC; Cai, Y; Chang, JCJ; Broadhurst, DJ; Klippsten, S; Sharp, PP; Wenzell, NA; Kuzuoglu-Ozturk, D; Wang, HY; Trenker, R; Young, JM; Cavero, DA; Hiatt, J; Roth, TL; Rathore, U; Subramanian, A; Noack, J; Hubert, M; Stroud, RM; Frankel, AD; Rosenberg, OS; Verba, KA; Agard, DA; Ott, M; Emerman, M; Jura, N; von Zastrow, M; Verdin, E; Ashworth, A; Schwartz, O; d'Enfert, C; Mukherjee, S; Jacobson, M; Malik, HS; Fujimori, DG; Ideker, T; Craik, CS; Floor, SN; Fraser, JS; Gross, JD; Sali, A; Roth, BL; Ruggero, D; Taunton, J; Kortemme, T; Beltrao, P; Vignuzzi, M; García-Sastre, A; Shokat, KM; Shoichet, BK; Krogan, NJ Nature 583: 459 -468 (2020) Bhuniya, D; Umrani, D; Dave, B; Salunke, D; Kukreja, G; Gundu, J; Naykodi, M; Shaikh, NS; Shitole, P; Kurhade, S; De, S; Majumdar, S; Reddy, SB; Tambe, S; Shejul, Y; Chugh, A; Palle, VP; Mookhtiar, KA; Cully, D; Vacca, J; Chakravarty, PK; Nargund, RP; Wright, SD; Graziano, MP; Singh, SB; Roy, S; Cai, TQ Bioorg Med Chem Lett 21: 3596 -602 (2011) Sirisoma, NS; Ratra, GS; Tomizawa, M; Casida, JE Bioorg Med Chem Lett 11: 2979 -81 (2001) Stock, NS; Bain, G; Zunic, J; Li, Y; Ziff, J; Roppe, J; Santini, A; Darlington, J; Prodanovich, P; King, CD; Baccei, C; Lee, C; Rong, H; Chapman, C; Broadhead, A; Lorrain, D; Correa, L; Hutchinson, JH; Evans, JF; Prasit, P J Med Chem 54: 8013 -29 (2011) Azizeh, BY; Van Tine, BA; Sturm, NS; Hutzler, AM; David, C; Trivedi, D; Hruby, VJ Bioorg Med Chem Lett 5: 1849 -1852 (1995) Goodnow, RA; Hicks, A; Sidduri, A; Kowalczyk, A; Dominique, R; Qiao, Q; Lou, JP; Gillespie, P; Fotouhi, N; Tilley, J; Cohen, N; Choudhry, S; Cavallo, G; Tannu, SA; Ventre, JD; Lavelle, D; Tare, NS; Oh, H; Lamb, M; Kurylko, G; Hamid, R; Wright, MB; Pamidimukkala, A; Egan, T; Gubler, U; Hoffman, AF; Wei, X; Li, YL; O'Neil, J; Marcano, R; Pozzani, K; Molinaro, T; Santiago, J; Singer, L; Hargaden, M; Moore, D; Catala, AR; Chao, LC; Hermann, G; Venkat, R; Mancebo, H; Renzetti, LM J Med Chem 53: 3502 -16 (2010) Lipchock, JM; Hendrickson, HP; Douglas, BB; Bird, KE; Ginther, PS; Rivalta, I; Ten, NS; Batista, VS; Loria, JP Biochemistry 56: 96 -106 (2017) Marson, CM; Matthews, CJ; Yiannaki, E; Atkinson, SJ; Soden, PE; Shukla, L; Lamadema, N; Thomas, NS J Med Chem 56: 6156 -74 (2013) Tibon, NS; Ng, CH; Cheong, SL Eur J Med Chem 188: (2020) Ellis, ES; Byrne, C; Murphy, OE; Tilford, NS; Baxter, GS Br J Pharmacol 114: 400 -4 (1995) Tulsi, NS; Downey, AM; Cairo, CW Bioorg Med Chem 18: 8679 -86 (2010) Wang NS Senger, S; Convery, MA; Chan, C; Watson, NS Bioorg Med Chem Lett 16: 5731 -5 (2006) Jiang, W; Tucci, FC; Chen, CW; Arellano, M; Tran, JA; White, NS; Marinkovic, D; Pontillo, J; Fleck, BA; Wen, J; Saunders, J; Madan, A; Foster, AC; Chen, C Bioorg Med Chem Lett 16: 4674 -8 (2006) Antczak, MI; Zhang, Y; Wang, C; Doran, J; Naidoo, J; Voruganti, S; Williams, NS; Markowitz, SD; Ready, JM J Med Chem 60: 3979 -4001 (2017) Friedman, MM; Cox, P; Frank, KE; Hoemann, MZ; Osuma, A; Wilson, NS; Xu, X; Cusack, K; Huntley, R; Herold, JM US Patent US10280184 (2019) Xia NS Pidugu, VR; Yarla, NS; Pedada, SR; Kalle, AM; Satya, AK Bioorg Med Chem 24: 5611 -5617 (2016) Mohammadi-Khanaposhtani, M; Saeedi, M; Zafarghandi, NS; Mahdavi, M; Sabourian, R; Razkenari, EK; Alinezhad, H; Khanavi, M; Foroumadi, A; Shafiee, A; Akbarzadeh, T Eur J Med Chem 92: 799 -806 (2015) Zaryanova, EV; Lozinskaya, NA; Beznos, OV; Volkova, MS; Chesnokova, NB; Zefirov, NS Bioorg Med Chem Lett 27: 3787 -3793 (2017) Zechner, M; Castro Jaramillo, CA; Zubler, NS; Taddio, MF; Mu, L; Altmann, KH; Krämer, SD J Med Chem 66: 6782 -6797 (2023) Lamie, PF; El-Kalaawy, AM; Abdel Latif, NS; Rashed, LA; Philoppes, JN Eur J Med Chem 214: (2021) Tawfik, HO; El-Moselhy, TF; El-Din, NS; El-Hamamsy, MH Bioorg Med Chem 27: (2019) Warda, ET; Shehata, IA; El-Ashmawy, MB; El-Gohary, NS Bioorg Med Chem 28: (2020) El-Sayed, NS; Nam, YW; Egorova, PA; Nguyen, HM; Orfali, R; Rahman, MA; Yang, G; Wulff, H; Bezprozvanny, I; Parang, K; Zhang, M J Med Chem 65: 303 -322 (2022) Penning, TD; Russell, MA; Chen, BB; Chen, HY; Liang, CD; Mahoney, MW; Malecha, JW; Miyashiro, JM; Yu, SS; Askonas, LJ; Gierse, JK; Harding, EI; Highkin, MK; Kachur, JF; Kim, SH; Villani-Price, D; Pyla, EY; Ghoreishi-Haack, NS; Smith, WG J Med Chem 45: 3482 -90 (2002) Ivanova, JN; Nocentini, A; Ta Rs, K; Leita Ns, JN; Dvinskis, E; Kazaks, A; Domračeva, I; Supuran, CT; Žalubovskis, R J Med Chem 66: 5703 -5718 (2023)
ChEMBL_2272495 Binding affinity to recombinant human NSD3 PWWP1 domain (247 to 398 residues) by SPR analysis ChEMBL_961297 (CHEMBL2390473) Activation of human AMPK alpha2 (1 to 398) expressed in African green monkey COS7 cells ChEMBL_2211767 (CHEMBL5124716) Inhibition of GST-tagged NSD3-PWWP1 domain (247 to 398 residues) (unknown origin) by TR-FRET assay ChEMBL_2476104 Binding affinity to human NSD3 PWWP1 domain (247 to 398 residues) assessed as dissociation constant by SPR analysis ChEMBL_2476105 Inhibition of GST-tagged NSD3 (247 to 398 residues) (unknown origin) incubated for 60 mins by TR-FRET assay ChEMBL_2051300 (CHEMBL4705999) Binding affinity to NSD3 PWWP1 domain (263 to 398 residues) (unknown origin) expressed in Escherichia coli BL21(DE3) by SPR method ChEMBL_2272496 Antagonist activity against recombinant human GST-tagged NSD3 PWWP1 domain (247 to 398 residues) incubated for 60 mins by TR-FRET assay ChEMBL_1898487 (CHEMBL4400602) Inhibition of N-terminal GST-fused human FGFR1 cytoplasmic domain (398 to 822(end) amino acids) using CSKtide substrate incubated for 90 mins ChEMBL_2069443 (CHEMBL4724696) Inhibition of recombinant human GST-tagged ERK5 (1 to 398 assessed as phosphorylated peptide substrate incubated for 60 mins by TR-FRET based biochemical assay ChEMBL_1649227 (CHEMBL3998361) Inhibition of human N-terminal GST-tagged FGFR1 cytoplasmic domain (398 to 822 residues) expressed in baculovirus expression system using CSKtide as substrate after 90 mins ChEMBL_2353761 Antagonist activity against recombinant human GST-tagged NSD3 PWWP1 domain (247 to 398 residues) expressed in Escherichia coli BL21(DE3) incubated for 60 mins by TR-FRET assay ChEMBL_1799440 (CHEMBL4271732) Inhibition of human N-terminal GST-tagged FGFR1 (398 to 822 residues) cytoplasmic domain expressed in baculovirus expression system after 60 mins by off-chip mobility shift assay ChEMBL_2128597 (CHEMBL4838026) Inhibition of human N-terminal GST fusion tagged FGFR1 cytoplasmic domain (398 to 822 residues) expressed in baculovirus infected Sf21 insect cells in presence of ATP at Km concentration ChEMBL_2434809 Binding affinity to CM5 chip immobilized N-terminal GST-tagged NSD3 PWWP1 domain (263 to 398 residues) (unknown origin) expressed in Escherichia coli BL21(DE3) assessed as dissociation constant by SPR analysis ChEMBL_1823032 (CHEMBL4322796) Binding affinity to recombinant N-terminal His-FLAG-GST-tagged MAP3K11 (unknown origin) (99 to 398 residues) expressed in baculovirus infected Sf9 insect cells incubated for 1 hr by TR-FRET assay ChEMBL_2213923 (CHEMBL5127055) Inhibition of N-terminal GST-tagged recombinant wild type human FGFR1 (398 to 822 residues) expressed in baculovirus infected Sf21 cells incubated for 1 hr in presence of ATP by caliper mobility shift assay ChEMBL_1914929 (CHEMBL4417512) Inhibition of N-terminal GST-tagged human FGFR1 cytoplasmic domain (398-822 AA) expressed in baculovirus using FAM-labelled peptide as substrate pre-incubated for 10 mins followed by substrate addition by mobility shift assay ChEMBL_2069349 (CHEMBL4724602) Inhibition of human N-terminal GST-tagged FGFR1 (398 to end residues) expressed in baculovirus expression system using CSKtide as substrate incubated for 30 mins in presence of ATP by off-chip mobility shift assay ChEMBL_2282176 Inhibition of human MCT4 in human SNU-398 cells assessed as inhibition of lactate efflux preincubated for 30 mins followed by D(+)glucose and measured after 4 hrs by dialysis based UHPLC-ESI-Q-Orbitrap-MS analysis ChEMBL_1645072 (CHEMBL3994001) Inhibition of recombinant human GST-tagged FGFR1 cytoplasmic domain (398 to 822 residues) expressed in baculovirus expression system preincubated for 10 mins followed by FAM-labeled peptide substrate addition measured after 1 hr by mobility shift assay ChEMBL_2282175 Inhibition of human MCT4 in human SNU-398 cells assessed as inhibition of radioactive lactate efflux preincubated for 30 mins followed by D(+)glucose and measured after 4 hrs by dialysis based UHPLC-ESI-Q-Orbitrap-MS analysis ChEBML_1717792 Inhibition of N-terminal GST tagged human FGFR1 (398 to 822 end residues) cytoplasmic domain expressed in baculovirus expression system preincubated for 10 mins followed by fluorescein amidite-labelled peptide substrate addition measured after 1 hr by mobility shift assay ChEBML_1970665 Inhibition of recombinant N-terminal His-tagged human FGFR3 K650E mutant (398 to 806 residues) cytoplasmic domain expressed in baculovirus expression system using Tyr 04 peptide as substrate after 1 hr in presence of ATP by Z'-LYTE assay ChEMBL_1995346 (CHEMBL4629241) Inhibition of N-terminal GST-tagged human FGFR1 cytoplasmic domain (398-822 end amino acids residues) expressed in baculovirus expression system preincubated for 30 to 120 mins followed by incubation with substrate and ATP for 30 mins by off-chip mobility shift assay ChEMBL_2491644 Inhibition of recombinant N-terminal GST tagged FGFR1 (398 to 822 residues) (unknown origin) expressed in Sf21 cells using biotinylated PYK2 peptide (biotin-AGAGSIESDIYAEIPDETC-NH2) as substrate preincubated for 30 mins followed by substrate addition and measured after 1 hr by AlphaScreen assay ChEMBL_1989429 (CHEMBL4623164) Inhibition of N-terminal GST fused human recombinant FGFR1 cytoplasmic domain (398 to 822 (end) residues) expressed in baculovirus expression system using FAM-labeled peptide and ATP as substrate preincubated for 10 mins followed by substrate addition measured after 1 hr by mobility shift assay ChEMBL_2251914 (CHEMBL5166124) Inhibition of human recombinant N-terminal GST-tagged ERK5 (1 to 398 residues) expressed in Escherichia coli using biotinylated peptide Ahx-PPGDYSTTPGGTLFSTTPGGTRI as substrate preincubated for 15 mins followed by substrate addition measured after 60 mins in presence of ATP by TR-FRET method ChEMBL_1832017 (CHEMBL4332025) Inhibition of His-tagged MAP2K5 activated N-terminal GST-tagged recombinant human ERK5 (1 to 398 residues) expressed in Escherichia coli using biotin-Ahx-PPGDYSTTPGGTLFSTTPGGTRI peptide as substrate preincubated for 15 mins followed by substrate addition and measured after 60 mins in presence of ATP by TR-FRET assay ChEMBL_2184175 (CHEMBL5096257) Inhibition of recombinant N-terminal GST-tagged human FGFR1 (398 to 822 residues) expressed in baculovirus expression system using IGF-1Rtide peptide as substrate preincubated for 10 mins followed by substrate addition and measured after 1 hrs in the presence of ATP by ADP-Glo reagent based luminescence assay In Vitro Assessment of Peptide Substrate Cleavage by BoNTA-LC Reactions between recombinant BoNTA-LC and fluorescent peptide substrate were carried out in 96-well microplates. Reaction progress was measured continuously by increase in fluorescence at Ex =398 nm, Em=485 nm as the cleavage of the substrate relieved the quenching of DACIA fluorescence by the 2,4-dinitrophenyl-lysine. The initial velocity values for enzymatic cleavage of peptide substrate were plotted against inhibitor concentration. The points were fit by non-linear regression analysis using the graphing program Prism (GraphPad). IC50 is the inhibitor concentration that achieves half-maximal enzyme inhibition. cAMP Assay CHO cells expressing human CB1 or CB2 receptors are seeded 17-24 hours prior to the experiment 50,000 cells per well in a black 96 well plate with flat clear bottom (Corning Costar #3904) in DMEM (Invitrogen No. 31331), 1×HT supplement, with 10% fetal calf serum and incubated at 5% CO2 and 37° C. in a humidified incubator. The growth medium was exchanged with Krebs Ringer Bicarbonate buffer with 1 mM IBMX and incubated at 30° C. for 30 min. Compounds were added to a final assay volume of 100 μl and incubated for 30 min at 30° C. Using the cAMP-Nano-TRF detection kit the assay (Roche Diagnostics) was stopped by the addition of 50 μl lysis reagent (Tris, NaCl, 1.5% Triton X100, 2.5% NP40, 10% NaN3) and 50 μl detection solutions (20 μM mAb Alexa700-cAMP 1:1, and 48 μM Ruthenium-2-AHA-cAMP) and shaken for 2 h at room temperature. The time-resolved energy transfer is measured by a TRF reader (Evotec Technologies GmbH), equipped with a ND:YAG laser as excitation source. The plate is measured twice with the excitation at 355 nm and at the emission with a delay of 100 ns and a gate of 100 ns, total exposure time 10 s at 730 (bandwidth 30 nm) or 645 nm (bandwidth 75 nm), respectively. cAMP Assay CHO cells expressing human CB1 or CB2 receptors are seeded 17-24 hours prior to the experiment 50.000 cells per well in a black 96 well plate with flat clear bottom (Corning Costar #3904) in DMEM (Invitrogen No. 31331), 1×HT supplement, with 10% fetal calf serum and incubated at 5% CO2 and 37° C. in a humidified incubator. The growth medium was exchanged with Krebs Ringer Bicarbonate buffer with 1 mM IBMX and incubated at 30° C. for 30 min. Compounds were added to a final assay volume of 100 μl and incubated for 30 min at 30° C. Using the cAMP-Nano-TRF detection kit the assay (Roche Diagnostics) was stopped by the addition of 50 μl lysis reagent (Tris, NaCl, 1.5% Triton X100, 2.5% NP40, 10% NaN3) and 50 μl detection solutions (20 μM mAb Alexa700-cAMP 1:1, and 48 μM Ruthenium-2-AHA-cAMP) and shaken for 2 h at room temperature. The time-resolved energy transfer is measured by a TRF reader (Evotec Technologies GmbH), equipped with a ND:YAG laser as excitation source. The plate is measured twice with the excitation at 355 nm and at the emission with a delay of 100 ns and a gate of 100 ns, total exposure time 10s at 730 (bandwidth 30 nm) or 645 nm (bandwidth 75 nm), respectively. cAMP Assay CHO cells expressing human CB1 or CB2 receptors are seeded 17-24 hours prior to the experiment 50.000 cells per well in a black 96 well plate with flat clear bottom (Corning Costar #3904) in DMEM (Invitrogen No. 31331), 1.times.HT supplement, with 10% fetal calf serum and incubated at 5% CO2 and 37° C. in a humidified incubator. The growth medium was exchanged with Krebs Ringer Bicarbonate buffer with 1 mM IBMX and incubated at 30.degree. C. for 30 min Compounds were added to a final assay volume of 100 ul and incubated for 30 min at 30° C. Using the cAMP-Nano-TRF detection kit the assay (Roche Diagnostics) was stopped by the addition of 50 ul lysis reagent (Tris, NaCl, 1.5% Triton X100, 2.5% NP40, 10% NaN.sub.3) and 50 ul detection solutions (20 uM mAb Alexa700-cAMP 1:1, and 48 uM Ruthenium-2-AHA-cAMP) and shaken for 2 h at room temperature. The time-resolved energy transfer is measured by a TRF reader (Evotec Technologies GmbH), equipped with a ND:YAG laser as excitation source. The plate is measured twice with the excitation at 355 nm and at the emission with a delay of 100 ns and a gate of 100 ns, total exposure time 10 s at 730 (bandwidth 30 nm) or 645 nm (bandwidth 75 nm), respectively. The FRET signal is calculated. Assay of the Citric Acid Cycle Activation NaCT-CHO and pME-CHO were plated at 20,000 cells/well into white CulturPlate -96 (PerkinElmer) two days before the assay. Prior to assay incubation, the cultured plates were washed twice with washing buffer, 10 mM HEPES-Tris(pH7.4) containing 140 mM choline chloride, 2 mM KCl, 1 mM CaCl2, 1 mM MgCl2. The compounds to be tested were dissolved and diluted in DMSO (Wako Pure Chemical industries) to 1,000 times of a final concentration, and further diluted to two times as high as the final concentration in assay buffer, 10 mM HEPES-Tris(pH7.4) containing 140 mM NaCl, 2 mM KCl, 1 mM CaCl2, 1 mM MgCl2. The range of final concentrations was properly determined based on the test compounds activity. Each 25 μL compound solution was added to well and subsequently 25 μL radio-labeled substrate solution containing 0.4 mM (0.4 MBq/mL) [1,5-14C]-citric acid (PerkinElmer) in the assay buffer was added. After 1 hour incubation at 37° C., the reaction mixture was discarded and washed three times with pre-chilled washing buffer and then 0.1 mL MicroScint 20 (PerkinElmer) was added to well. The plate was sealed with TopSeal-A (PerkinElmer) and the radioactivity was measured using a TopCount (PerkinElmer). Non-specific activity (NS cpm) and total radio activity (Total cpm) were determined by counting of pME-CHO plated wells and NaCT-CHO plated wells without compounds, respectively. Diffusion of [14C] CO2 was able to be estimated from residual radioactivity (R cpm) by an equation (Total−R)/(Total−NS)×100(%). The difference of Total and R was disappeared in the presence of 0.1 μM antimycin A. EC50 values were calculated by regression analysis using SAS Statistical Analysis System (SAS institute Japan Ltd. Release 9.1). cAMP Assay CHO cells expressing human CB1 or CB2 receptors are seeded 17-24 hours prior to the experiment 50.000 cells per well in a black 96 well plate with flat clear bottom (Corning Costar #3904) in DMEM (Invitrogen No. 31331), 1×HT supplement, with 10% fetal calf serum and incubated at 5% CO2 and 37 °C. in a humidified incubator. The growth medium was exchanged with Krebs Ringer Bicarbonate buffer with 1 mM IBMX and incubated at 30 °C. for 30 min. Compounds were added to a final assay volume of 100 μl and incubated for 30 min at 30 °C. Using the cAMP-Nano-TRF detection kit the assay (Roche Diagnostics) was stopped by the addition of 50 μl lysis reagent (Tris, NaCl, 1.5% Triton X100, 2.5% NP40, 10% NaN3) and 50 μl detection solutions (20 μM mAb Alexa700-cAMP 1:1, and 48 μM Ruthenium-2-AHA-cAMP) and shaken for 2 h at room temperature. The time-resolved energy transfer is measured by a TRF reader (Evotec Technologies GmbH), equipped with a ND:YAG laser as excitation source. The plate is measured twice with the excitation at 355 nm and at the emission with a delay of 100 ns and a gate of 100 ns, total exposure time 10 s at 730 (bandwidth 30 nm) or 645 nm (bandwidth 75 nm), respectively. The FRET signal is calculated as follows: FRET=T730-Alexa730-P(T645-B645) with P=Ru730-B730/Ru645-B645, where T730 is the test well measured at 730 nM, T645 is the test well measured at 645 nm, B730 and B645 are the buffer controls at 730 nm and 645 nm, respectively. cAMP content is determined from the function of a standard curve spanning from 10 μM to 0.13 nM cAMP. cAMP Assay CHO cells expressing human CB1 or CB2 receptors are seeded 17-24 hours prior to the experiment 50.000 cells per well in a black 96 well plate with flat clear bottom (Corning Costar #3904) in DMEM (Invitrogen No. 31331), 1×HT supplement, with 10% fetal calf serum and incubated at 5% CO2 and 37° C. in a humidified incubator. The growth medium was exchanged with Krebs Ringer Bicarbonate buffer with 1 mM IBMX and incubated at 30° C. for 30 min. Compounds were added to a final assay volume of 100 μl and incubated for 30 min at 30° C. Using the cAMP-Nano-TRF detection kit the assay (Roche Diagnostics) was stopped by the addition of 50 μl lysis reagent (Tris, NaCl, 1.5% Triton X100, 2.5% NP40, 10% NaN3) and 50 μl detection solutions (20 μM mAb Alexa700-cAMP 1:1, and 48 μM Ruthenium-2-AHA-cAMP) and shaken for 2 h at room temperature. The time-resolved energy transfer is measured by a TRF reader (Evotec Technologies GmbH), equipped with a ND:YAG laser as excitation source. The plate is measured twice with the excitation at 355 nm and at the emission with a delay of 100 ns and a gate of 100 ns, total exposure time 10 s at 730 (bandwidth 30 nm) or 645 nm (bandwidth 75 nm), respectively. The FRET signal is calculated as follows: FRET=T730-Alexa730-P(T645-B645) with P=Ru730-B730/Ru645-B645, where T730 is the test well measured at 730 nM, T645 is the test well measured at 645 nm, B730 and B645 are the buffer controls at 730 nm and 645 nm, respectively. cAMP content is determined from the function of a standard curve spanning from 10 μM to 0.13 nM cAMP. cAMP Assay CHO cells expressing human CB1 or CB2 receptors are seeded 17-24 hours prior to the experiment 50.000 cells per well in a black 96 well plate with flat clear bottom (Corning Costar #3904) in DMEM (Invitrogen No. 31331), 1×HT supplement, with 10% fetal calf serum and incubated at 5% CO2 and 37° C. in a humidified incubator. The growth medium was exchanged with Krebs Ringer Bicarbonate buffer with 1 mM IBMX and incubated at 30° C. for 30 min. Compounds were added to a final assay volume of 100 μl and incubated for 30 min at 30° C. Using the cAMP-Nano-TRF detection kit the assay (Roche Diagnostics) was stopped by the addition of 50 μl lysis reagent (Tris, NaCl, 1.5% Triton X100, 2.5% NP40, 10% NaN3) and 50 μl detection solutions (20 μM mAb Alexa700-cAMP 1:1, and 48 μM Ruthenium-2-AHA-cAMP) and shaken for 2 h at room temperature. The time-resolved energy transfer is measured by a TRF reader (Evotec Technologies GmbH), equipped with a ND:YAG laser as excitation source. The plate is measured twice with the excitation at 355 nm and at the emission with a delay of 100 ns and a gate of 100 ns, total exposure time 10 s at 730 (bandwidth 30 nm) or 645 nm (bandwidth 75 nm), respectively. The FRET signal is calculated as follows: FRET=T730-Alexa730-P(T645-B645) with P=Ru730-B730/Ru645-B645, where T730 is the test well measured at 730 nM, T645 is the test well measured at 645 nm, B730 and B645 are the buffer controls at 730 nm and 645 nm, respectively. cAMP content is determined from the function of a standard curve spanning from 10 μM to 0.13 nM cAMP. CYP450 Enzyme Induction Study A final incubation system of 500 μL contained 50 μL of liver microsomes (protein concentration: 0.2 mg/mL, Corning), 1 μL of mixed specific substrates of CYP450 (CYP1A2, CYP2B6, CYP2C9, CYP2C19, CYP2D6, CYP3A4), 398 μL of PBS (pH 7.4), 1 μL of specific positive inhibitor (positive control) or test compound (in acetonitrile) and 50 μL of NADPH+MgCl2. Samples were prepared in duplicate of 0.5 mL for each CYP450 subtype. For each tube, the 450 μL mixed solution of substrates and enzyme and the NADPH solution were separately pre-incubated at 37° C. for 5 min. The 50 μL mixed solution of NADPH+MgCl2 was added for reaction. At 30 min, 50 μL of the mixture was taken and 300 μL of glacial acetonitrile containing an internal standard was added to terminate the reaction. Additionally, 2 blanks of 500 μL each were prepared in parallel without adding NADPH as the negative control group. cAMP Assay CHO cells expressing human CB1 or CB2 receptors are seeded 17-24 hours prior to the experiment 50.000 cells per well in a black 96 well plate with flat clear bottom (Corning Costar #3904) in DMEM (Invitrogen No. 31331), 1xHT supplement, with 10% fetal calf serum and incubated at 5% CO2 and 37° C. in a humidified incubator. The growth medium was exchanged with Krebs Ringer Bicarbonate buffer with 1 mM IBMX and incubated at 30° C. for 30 min. Compounds were added to a final assay volume of 100 ul and incubated for 30 min at 30° C. Using the cAMP-Nano-TRF detection kit the assay (Roche Diagnostics) was stopped by the addition of 50 ul lysis reagent (Tris, NaCl, 1.5% Triton X100, 2.5% NP40, 10% NaN3) and 50 ul detection solutions (20 uM mAb Alexa700-cAMP 1:1, and 48 uM Ruthenium-2-AHA-cAMP) and shaken for 2 h at room temperature. The time-resolved energy transfer is measured by a TRF reader (Evotec Technologies GmbH), equipped with a ND:YAG laser as excitation source. The plate is measured twice with the excitation at 355 nm and at the emission with a delay of 100 ns and a gate of 100 ns, total exposure time 10 s at 730 (bandwidth 30 nm) or 645 nm (bandwidth 75 nm), respectively. The FRET signal is calculated as follows: FRET=T730-Alexa730-P (T645-B645) with P=Ru730-B730/Ru645-B645, where T730 is the test well measured at 730 nM, T645 is the test well measured at 645 nm, B730 and B645 are the buffer controls at 730 nm and 645 nm, respectively. cAMP content is determined from the function of a standard curve spanning from 10 uM to 0.13 nM cAMP. EC50 values were determined using Activity Base analysis (ID Business Solution, Limited). The EC50 values for a wide range of cannabinoid agonists generated from this assay were in agreement with the values published in the scientific literature. cAMP Assay CHO cells expressing human CB1 or CB2 receptors are seeded 17-24 hours prior to the experiment 50.000 cells per well in a black 96 well plate with flat clear bottom (Corning Costar #3904) in DMEM (Invitrogen No. 31331), lx HT supplement, with 10% fetal calf serum and incubated at 5% CO2 and 37° C. in a humidified incubator. The growth medium was exchanged with Krebs Ringer Bicarbonate buffer with 1 mM IBMX and incubated at 30° C. for 30 min. Compounds were added to a final assay volume of 100 ul and incubated for 30 min at 30° C. Using the cAMP-Nano-TRF detection kit the assay (Roche Diagnostics) was stopped by the addition of 50 ul lysis reagent (Tris, NaCl, 1.5% Triton X100, 2.5% NP40, 10% NaN3) and 50 ul detection solutions (20 uM mAb Alexa700-cAMP 1:1, and 48 uM Ruthenium-2-AHA-cAMP) and shaken for 2 h at room temperature. The time-resolved energy transfer is measured by a TRF reader (Evotec Technologies GmbH), equipped with a ND:YAG laser as excitation source. The plate is measured twice with the excitation at 355 nm and at the emission with a delay of 100 ns and a gate of 100 ns, total exposure time 10 s at 730 (bandwidth 30 nm) or 645 nm (bandwidth 75 nm), respectively. The FRET signal is calculated as follows: FRET=T730-Alexa730-P(T645-B645) with P=Ru730-B730/Ru645-B645, where T730 is the test well measured at 730 nM, T645 is the test well measured at 645 nm, B730 and B645 are the buffer controls at 730 nm and 645 nm, respectively. cAMP content is determined from the function of a standard curve spanning from 10 uM to 0.13 nM cAMP. EC50 values were determined using Activity Base analysis (ID Business Solution, Limited). The EC50 values for a wide range of cannabinoid agonists generated from this assay were in agreement with the values published in the scientific literature. cAMP Assay CHO-dhfr(minus) cells expressing human GPBAR1 receptors are seeded 17-24 hours prior to the experiment 50.000 cells per well in a black 96 well plate with flat clear bottom (Corning Costar #3904) in DMEM (Invitrogen No. 31331), lx HT supplement, with 10% fetal calf serum and incubated at 5% CO2 and 37° C. in a humidified incubator. The growth medium was exchanged with Krebs Ringer Bicarbonate buffer with 1 mM IBMX and incubated at 30° C. for 30 min Compounds were added to a final assay volume of 100 μl and incubated for 30 min at 30° C. The assay was stopped by the addition of 50 μl lysis reagent (Tris, NaCl, 1.5% Triton X100, 2.5% NP40, 10% NaN3) and 50 detection solutions (20 μM mAb Alexa700-cAMP 1:1, and 48 μM Ruthenium-2-AHA-cAMP) and shaked for 2 h at room temperature. The time-resolved energy transfer is measured by a TRF reader (Evotec Technologies GmbH, Hamburg Germany), equipped with a ND:YAG laser as excitation source. The plate is measured twice with the excitation at 355 nm and at the emission with a delay of 100 ns and a gate of 100 ns, total exposure time 10 s at 730 (bandwidth 30 nm) or 645 nm (bandwidth 75 nm), respectively. The measured signal at 730 nm has to be corrected for the ruthenium background, the direct excitation of Alexa and the buffer control. The FRET signal is calculated as follows: FRET=T730−Alexa730−P(T645−B645) with P=Ru730−B730/Ru645−B645, where T730 is the test well measured at 730 nM, T645 is the test well measured at 645 nm, B730 and B645 are the buffer controls at 730 nm and 645 nm, respectively. cAMP content is determined from the function of a standard curve spanning from 10 μM to 0.13 nM cAMP. CYP450 Enzyme Inhibition Assay 500 μL of a final incubation system contains 50 μL of human liver microsomes (protein concentration: 0.2 mg/mL, Corning), 1 μL of mixed CYP450 specific substrates (CYP1A2, CYP 2B6, CYP 2C9, CYP2C19, CYP 2D6, and CYP 3A4), 398 μL PBS buffer (pH7.4), 1 μL specific positive inhibitor (positive control group) or the test compound (formulated with acetonitrile), and 50 μL NADPH+MgCl2. Duplicate incubation systems of 0.5 mL each were formulated for each CYP450 subtype. A total volume of 450 μL of a uniformly mixed solution of the substrate and the enzyme was formulated in each tube, and the solution and NADPH were pre-incubated at 37° C. for 5 minutes, respectively. Then 50 μL of the mixed solution of NADPH+MgCl2 was added for reaction. 50 μL of the reaction solution was taken out at 30 minutes, and the reaction was terminated with 300 μL of ice acetonitrile containing an internal standard. In addition, two control groups of 500 μL each without NADPH were prepared in parallel as a negative control group. cAMP Assay CHO cells expressing human CB1 or CB2 receptors are seeded 17-24 hours prior to the experiment 50.000 cells per well in a black 96 well plate with flat clear bottom (Corning Costar #3904) in DMEM (Invitrogen No. 31331), 1xHT supplement, with 10% fetal calf serum and incubated at 5% CO2 and 37° C. in a humidified incubator. The growth medium was exchanged with Krebs Ringer Bicarbonate buffer with 1 mM IBMX and incubated at 30° C. for 30 min. Compounds were added to a final assay volume of 100 ul and incubated for 30 min at 30° C. Using the cAMP-Nano-TRF detection kit the assay (Roche Diagnostics) was stopped by the addition of 50 ul lysis reagent (Tris, NaCl, 1.5% Triton X100, 2.5% NP40, 10% NaN3) and 50 ul detection solutions (20 uM mAb Alexa700-cAMP 1:1, and 48 uM Ruthenium-2-AHA-cAMP) and shaken for 2 h at room temperature. The time-resolved energy transfer is measured by a TRF reader (Evotec Technologies GmbH), equipped with a ND:YAG laser as excitation source. The plate is measured twice with the excitation at 355 nm and at the emission with a delay of 100 ns and a gate of 100 ns, total exposure time 10 s at 730 (bandwidth 30 nm) or 645 nm (bandwidth 75 nm), respectively. The FRET signal is calculated as follows: FRET=T730-Alexa730-P(T645-B645) with P=Ru730-B730/Ru645-B645, where T730 is the test well measured at 730 nM, T645 is the test well measured at 645 nm, B730 and B645 are the buffer controls at 730 nm and 645 nm, respectively. cAMP content is determined from the function of a standard curve spanning from 10 uM to 0.13 nM cAMP. EC50 values were determined using Activity Base analysis (ID Business Solution, Limited). The EC50 values for a wide range of cannabinoid agonists generated from this assay were in agreement with the values published in the scientific literature. The compounds of the invention are CB2 receptor agonists with EC50 below 1 uM and selectivity versus CB1 in the corresponding assay of at least 10 fold. CYP450 Enzyme Induction Study 500 μL of a final incubation system contains 50 μL of liver microsomes (protein concentration: 0.2 mg/mL), 1 μL of mixed CYP450 specific substrates (CYP1A2, CYP 2B6, CYP 2C9, CYP2C19, CYP 2D6, and CYP 3A4), 398 μL PBS buffer (PH7.4), 1 μL specific positive inhibitor (positive control group) or the test compound (formulated with acetonitrile), and 50 μL NADPH+MgCl2. Duplicate incubation systems of 0.5 mL each were formulated for each CYP450 subtype. A total volume of 450 μL of a uniformly mixed solution of the substrate and the enzyme was formulated in each tube, and the solution and NADPH were pre-incubated at 37° C. for 5 minutes, respectively. Then 50 μL of the mixed solution of NADPH+MgCl2 was added for reaction. 50 μL of the reaction solution was taken out at 30 minutes, and the reaction was terminated with 300 μL of ice acetonitrile containing an internal standard. In addition, two control groups of 500 μL each without NADPH were prepared in parallel as a negative control group. cAMP Assay CHO cells expressing human CB1 or CB2 receptors are seeded 17-24 hours prior to the experiment 50.000 cells per well in a black 96 well plate with flat clear bottom (Corning Costar #3904) in DMEM (Invitrogen No. 31331), 1x HT supplement, with 10% fetal calf serum and incubated at 5% CO2 and 37° C. in a humidified incubator. The growth medium was exchanged with Krebs Ringer Bicarbonate buffer with 1 mM IBMX and incubated at 30° C. for 30 min. Compounds were added to a final assay volume of 100 ul and incubated for 30 min at 30° C. Using the cAMP-Nano-TRF detection kit the assay (Roche Diagnostics) was stopped by the addition of 50 ul lysis reagent (Tris, NaCl, 1.5% Triton X100, 2.5% NP40, 10% NaN3) and 50 ul detection solutions (20 uM mAb Alexa700-cAMP 1:1, and 48 uM Ruthenium-2-AHA-cAMP) and shaken for 2 h at room temperature. The time-resolved energy transfer is measured by a TRF reader (Evotec Technologies GmbH), equipped with a ND:YAG laser as excitation source. The plate is measured twice with the excitation at 355 nm and at the emission with a delay of 100 ns and a gate of 100 ns, total exposure time 10 s at 730 (bandwidth 30 nm) or 645 nm (bandwidth 75 nm), respectively. The FRET signal is calculated as follows: FRET=T730-Alexa730-P(T645-B645) with P=Ru730-B730/Ru645-B645, where T730 is the test well measured at 730 nM, T645 is the test well measured at 645 nm, B730 and B645 are the buffer controls at 730 nm and 645 nm, respectively. cAMP content is determined from the function of a standard curve spanning from 10 uM to 0.13 nM cAMP. EC50 values were determined using Activity Base analysis (ID Business Solution, Limited). The EC50 values for a wide range of cannabinoid agonists generated from this assay were in agreement with the values published in the scientific literature. The compounds of the invention are CB2 receptor agonists with EC50 below 1 uM and selectivity versus CB1 in the corresponding assay of at least 10 fold. Particular compound of the invention are CB2 receptor agonists with EC50 below 0.05 uM and selectivity versus CB1 in the corresponding assay of at least 500 fold. cAMP Assay CHO cells expressing human CB1 or CB2 receptors are seeded 17-24 hours prior to the experiment 50.000 cells per well in a black 96 well plate with flat clear bottom (Corning Costar #3904) in DMEM (Invitrogen No. 31331), 1×HT supplement, with 10% fetal calf serum and incubated at 5% CO2 and 37° C. in a humidified incubator. The growth medium was exchanged with Krebs Ringer Bicarbonate buffer with 1 mM IBMX and incubated at 30° C. for 30 min. Compounds were added to a final assay volume of 100 μl and incubated for 30 min at 30° C. Using the cAMP-Nano-TRF detection kit the assay (Roche Diagnostics) was stopped by the addition of 50 μl lysis reagent (Tris, NaCl, 1.5% Triton X100, 2.5% NP40, 10% NaN3) and 50 μl detection solutions (20 μM mAb Alexa700-cAMP 1:1, and 48 μM Ruthenium-2-AHA-cAMP) and shaken for 2 h at room temperature. The time-resolved energy transfer is measured by a TRF reader (Evotec Technologies GmbH), equipped with a ND:YAG laser as excitation source. The plate is measured twice with the excitation at 355 nm and at the emission with a delay of 100 ns and a gate of 100 ns, total exposure time 10 s at 730 (bandwidth 30 nm) or 645 nm (bandwidth 75 nm), respectively. The FRET signal is calculated as follows: FRET=T730-Alexa730-P(T645-B645) with P=Ru730-B730/Ru645-B645, where T730 is the test well measured at 730 nM, T645 is the test well measured at 645 nm, B730 and B645 are the buffer controls at 730 nm and 645 nm, respectively. cAMP content is determined from the function of a standard curve spanning from 10 μM to 0.13 nM cAMP.EC50 values were determined using Activity Base analysis (ID Business Solution, Limited). The EC50 values for a wide range of cannabinoid agonists generated from this assay were in agreement with the values published in the scientific literature.The compounds of the invention are CB2 receptor agonists with EC50 below 1 μM and selectivity versus CB1 in the corresponding assay of at least 10 fold. Particular compound of the invention are CB2 receptor agonists with EC50 below 0.05 μM and selectivity versus CB1 in the corresponding assay of at least 500 fold. Assessing Antiviral Activity Against Influenza To assess their antiviral activity, some compounds were tested against murine adapted human influenza (PR8) in vitro. Canine MDCK cells were grown to confluency ( 1.0×10{circumflex over ( )}4 cells/well) in 96-well plate format in Eagle's Minimal Essential Medium (EMEM) supplemented with 10% fetal bovine serum (FBS) and 100 U/ml each of penicillin and streptomycin. Wells were washed in 1×PBS and infected with an PR8 variant expressing mCherry downstream and separated by a 2A autocleavage site from the NS-1 protein at a multiplicity of 0.01 infectious unit (IU) per cell in serum free EMEM. Assays were performed in triplicate. One hour later, virus containing medium in the cells was replaced with fresh complete medium containing the indicated compounds at 25, 12.5, 6.25, 3.13, 1.56, 0.78, 0.39 μM or the carrier in which the compounds are dissolved (DMSO) and supplemented with 2.5 μg/ml TPCK trypsin. Final concentration of DMSO was 0.5% in each treatment. Virus yield in the culture was determined at 3 days post infection by quantification of fluorescent (mCherry positive) cells in each well by fluorescent microscopy. Results were plotted using CDD Vault (CDD Vault was developed by Collaborative Drug Discovery, Inc., 1633 Bayshore Hwy, Suite 342, Burlingame, Calif. 94010) in order to calculate IC50s. Activity Assay The EGLN-1 (or EGLN-3) enzyme activity is determined using mass spectrometry (matrix-assisted laser desorption ionization, time-of-flight MS, MALDI-TOF MS. Recombinant human EGLN-1-179/426 is prepared as described above and in the Supplemental Data. Full-length recombinant human EGLN-3 is prepared in a similar way, however it is necessary to use the His-MBP-TVMV-EGLN-3 fusion for the assay due to the instability of the cleaved protein. For both enzymes, the HIF-1α peptide corresponding to residues 556-574 is used as substrate. The reaction is conducted in a total volume of 50 uL containing TrisCl (5 mM, pH 7.5), ascorbate (120 uM), 2-oxoglutarate (3.2 uM), HIF-1α (8.6 uM), and bovine serum albumin (0.01%). The enzyme, quantity predetermined to hydroxylate 20% of substrate in 20 minutes, is added to start the reaction. Where inhibitors are used, compounds are prepared in dimethyl sulfoxide at 10-fold final assay concentration. After 20 minutes at room temperature, the reaction is stopped by transferring 10 uL of reaction mixture to 50 uL of a mass spectrometry matrix solution (α-cyano-4-hydroxycinnamic acid, 5 mg/mL in 50% acetonitrile/0.1% TFA, 5 mM NH4PO4). Two microliters of the mixture is spotted onto a MALDI-TOF MS target plate for analysis with an Applied Biosystems (Foster City, Calif.) 4700 Proteomics Analyzer MALDI-TOF MS equipped with a Nd:YAG laser (355 nm, 3 ns pulse width, 200 Hz repetition rate). Activity Assay The EGLN-1 (or EGLN-3) enzyme activity is determined using mass spectrometry (matrix-assisted laser desorption ionization, time-of-flight MS, MALDI-TOF MS- for assay details, see reference (Greis et al., 2006). Recombinant human EGLN-1-179/426 is prepared as described above and in the Supplemental Data. Full-length recombinant human EGLN-3 is prepared in a similar way, however it is necessary to use the His-MBP-TVMVEGLN-3 fusion for the assay due to the instability of the cleaved protein. For both enzymes, the HIF-1α peptide corresponding to residues 556-574 (DLDLEALAPYIPADDDFQL) (SEQ ID NO. 1) is used as substrate. The reaction is conducted in a total volume of 50 uL containing TrisCl (5 mM, pH 7.5), ascorbate (120 μM), 2-oxoglutarate (3.2 μM), HIF-1α (8.6 μM), and bovine serum albumin (0.01%). The enzyme, quantity predetermined to hydroxylate 20% of substrate in 20 minutes, is added to start the reaction. Where inhibitors are used, compounds are prepared in dimethyl sulfoxide at 10-fold final assay concentration. After 20 minutes at room temperature, the reaction is stopped by transferring 10 μL of reaction mixture to 50 μL of a mass spectrometry matrix solution (α-cyano-4-hydroxycinnamic acid, 5 mg/mL in 50% acetonitrile/0.1% TFA, 5 mM NH4PO4). Two microliters of the mixture is spotted onto a MALDI-TOF MS target plate for analysis with an Applied Biosystems (Foster City, Calif.) 4700 Proteomics Analyzer MALDI-TOF MS equipped with a Nd:YAG laser (355 nm, 3 ns pulse width, 200 Hz repetition rate). cAMP Assay The following mixtures and buffer solutions were prepared: (a) Buffer 1: HBSS (Mediatech Cat#21-023-CV) with 5 mM HEPES (1 mM stock, Gibco BRL Cat#15630-056) and 0.1% BSA (7.5% stock, Invitrogen Cat#15260-037); (b) Buffer 2: 0.5 mM IBMX (200 mM stock in DMSO, Sigma 15879) in Buffer 1; (c) 1 uM cAMP Standard (50 uM stock, Perkin Elmer Cat#AD0262) diluted in Buffer 2 and serially diluted in Buffer 2, 12 doses @1/2 dilutions resulting in a dose range of 1 uM to 0.5 nM; (d) d2 labelled cAMP (CisBio HTRF Detection Kit Cat #62AM4PEB reconstituted with 6 ml dH2O) diluted 1/20 with lysis buffer (CisBio HTRF Detection Kit Cat #62AM4PEB); (e) anti-cAMP (CisBio HTRF Detection Kit Cat #62AM4PEB reconstituted with 5 ml dH2O) diluted 1/20 with lysis buffer (CisBio HTRF Detection Kit Cat #62AM4PEB); and (f) Forskolin (Sigma Cat#F6886, 10 mM in DMSO) diluted first in DMSO to 1 mM and then to 1.5 uM in Buffer 2. A FLEXDROP (Perkin Elmer) was cleaned with ethanol then water, and primed with Buffer 2. A 384 well V bottom polypropylene plate containing d2 labelled cAMP and a second 384 well V bottom polypropylene plate containing anti-cAMP was prepared (50 ul per well). Media as "dumped~ from the cell plate and 30 uL Buffer 1 was added to each well using a Multidrop. The content of the cell plate was again "dumped" and 10 uL Buffer 2 was added to each well using a Flexdrop. 12.5 nL test compound dilutions or control compound dilutions (10 mM to 0.5 uM) were added to the cell plate using an ECHO 555 (Labcyte). The cell plate was mixed (Speed 6, Lab-Line Instruments Titer Plate Shaker) and centrifugated (1000 RPMs, 1 min). Using the Flexdrop, 2 ul additions were made into the cell plate: Buffer 2 was added to Column 24; and, 1.5 uM Forskolin was added to columns 1 through 23. Final volume of the cell plate was 12 ul with 250 nM Forskolin in all wells except column 12, and serial dilutions of test compound or control ranging from 10 uM to 0.5 nM. The cell plate was again mixed (speed 6) and centrifugated (1000 RPMs, 1 min). The cell plate was incubated for 30 minutes at room temperature (~27° C.). The contents of row P were removed and the cAMP standard dilutions were added in duplicate to Row P (P1-12 and P13-24). After incubation, 6 uL d2 labelled cAMP and 6 uL of Anti-cAMP were added to all wells of the cell plate using a BioMek FX (Beckman Coulter). The cell plate was again mixed (speed 6) and centrifugated (1000 RPMs, 1 min) and was incubated for 60 minutes in the dark at room Temp (~27° C.). After this final incubation, the cell plate was read in HTRF mode (fluorescence at 665 nm and 620 nm) on an Envision plate Reader (Perkin Elmer). The Envision reader outputs a ratio of channel 1/channel 2 fluorescence 10,000 (Normalized signal (NS)). Amount of cAMP in nM was calculated for each well (based on NS) from a cAMP standard curve located on each plate (at P1-12 and P13-24). Activity Assay The EGLN-1 (or EGLN-3) enzyme activity is determined using mass spectrometry (matrix-assisted laser desorption ionization, time-of-flight MS, MALDI-TOF MS. Recombinant human EGLN-1-179/426 is prepared as described above and in the Supplemental Data. Full-length recombinant human EGLN-3 is prepared in a similar way, however it is necessary to use the His-MBP-TVMV-EGLN-3 fusion for the assay due to the instability of the cleaved protein. For both enzymes, the HIF-1α peptide corresponding to residues 556-574 is used as substrate. The reaction is conducted in a total volume of 50 μL containing TrisCl (5 mM, pH 7.5), ascorbate (120 μM), 2-oxoglutarate (3.2 μM), HIF-1α (8.6 μM), and bovine serum albumin (0.01%). The enzyme, quantity predetermined to hydroxylate 20% of substrate in 20 minutes, is added to start the reaction. Where inhibitors are used, compounds are prepared in dimethyl sulfoxide at 10-fold final assay concentration. After 20 minutes at room temperature, the reaction is stopped by transferring 10 μL of reaction mixture to 50 μL of a mass spectrometry matrix solution (α-cyano-4-hydroxycinnamic acid, 5 mg/mL in 50% acetonitrile/0.1% TFA, 5 mM NH4PO4). Two microliters of the mixture is spotted onto a MALDI-TOF MS target plate for analysis with an Applied Biosystems (Foster City, Calif.) 4700 Proteomics Analyzer MALDI-TOF MS equipped with a Nd:YAG laser (355 nm, 3 ns pulse width, 200 Hz repetition rate). Hydroxylated peptide product is identified from substrate by the gain of 16 Da. Data defined as percent conversion of substrate to product is analyzed in GraphPad Prism 4 to calculate IC50 values. EGLN-1 Activity Assay The EGLN-1 (or EGLN-3) enzyme activity is determined using mass spectrometry (matrix-assisted laser desorption ionization, time-of-flight MS, MALDI-TOF MS for assay details, see reference (Greis et al., 2006). Recombinant human EGLN-1-179/426 is prepared as described above and in the Supplemental Data. Full-length recombinant human EGLN-3 is prepared in a similar way, however it is necessary to use the His-MBP-TVMV EGLN-3 fusion for the assay due to the instability of the cleaved protein. For both enzymes, the HIF-1C. peptide corresponding to residues 556-574 (DLDLEALAPYIPAD DDFQL) is used as substrate. The reaction is conducted in a total volume of 50 uL containing TrisCl (5 mM, pH 7.5), ascorbate (120 uM), 2-oxoglutarate (3.2 uM), HIF-1C. (8.6 uM), and bovine serum albumin (0.01%). The enzyme, quantity predetermined to hydroxylate 20% of substrate in 20 minutes, is added to start the reaction. Where inhibitors are used, compounds are prepared in dimethyl Sulfoxide at 10-fold final assay concentration. After 20 minutes at room temperature, the reaction is stopped by transferring 10 LIL of reaction mixture to 50 LL of a mass spectrometry matrix Solution (C-cyano-4-hydroxycinnamic acid, 5 mg/mL in 50% acetonitrile/0.1% TFA, 5 mM NHPO). Two micro liters of the mixture is spotted onto a MALDI-TOF MS target plate for analysis with an Applied Biosystems (Foster City, Calif.) 4700 Proteomics Analyzer MALDI-TOF MS equipped with a Nd:YAG laser (355 nm, 3 ns pulse width, 200 HZ repetition rate). Hydroxylated peptide product is identified from substrate by the gain of 16 Da. Data defined as percent conversion of Substrate to product is analyzed in GraphPad Prism 4 to calculate ICso values. EGLN-1 Activity Assay The EGLN-1 (or EGLN-3) enzyme activity is determined using mass spectrometry (matrix-assisted laser desorption ionization, time-of-flight MS, MALDI-TOF MS- for assay details, see reference (Greis et al., 2006). Recombinant human EGLN-1-179/426 is prepared as described above and in the Supplemental Data. Full-length recombinant human EGLN-3 is prepared in a similar way, however it is necessary to use the His-MBP-TVMVEGLN-3 fusion for the assay due to the instability of the cleaved protein. For both enzymes, the HIF-1α peptide corresponding to residues 556-574 (DLDLEALAPYIPADDDFQL) (SEQ ID NO. 1) is used as substrate. The reaction is conducted in a total volume of 50 uL containing TrisCl (5 mM, pH 7.5), ascorbate (120 μM), 2-oxoglutarate (3.2 μM), HIF-1α (8.6 μM), and bovine serum albumin (0.01%). The enzyme, quantity predetermined to hydroxylate 20% of substrate in 20 minutes, is added to start the reaction. Where inhibitors are used, compounds are prepared in dimethyl sulfoxide at 10-fold final assay concentration. After 20 minutes at room temperature, the reaction is stopped by transferring 10 μL of reaction mixture to 50 μL of a mass spectrometry matrix solution (α-cyano-4-hydroxycinnamic acid, 5 mg/mL in 50% acetonitrile/0.1% TFA, 5 mM NH4PO4). Two microliters of the mixture is spotted onto a MALDI-TOF MS target plate for analysis with an Applied Biosystems (Foster City, Calif.) 4700 Proteomics Analyzer MALDI-TOF MS equipped with a Nd:YAG laser (355 nm, 3 ns pulse width, 200 Hz repetition rate). Hydroxylated peptide product is identified from substrate by the gain of 16 Da. Data defined as percent conversion of substrate to product is analyzed in GraphPad Prism 4 to calculate IC50 values. EGLN-1 Activity Assay The EGLN-1 (or EGLN-3) enzyme activity is determined using mass spectrometry (matrix-assisted laser desorption ionization, time-of-flight MS, MALDI-TOF MS-for assay details, see reference (Greis et. al., 2006). Recombinant human EGLN-1-179/426 is prepared as described above and in the Supplemental Data. Full-length recombinant human EGLN-3 is prepared in a similar way, however it is necessary to use the His-MBP-TVMVEGLN-3 fusion for the assay due to the instability of the cleaved protein. For both enzymes, the HIF-1α peptide corresponding to residues 556-574 (DLDLEALAPYIPADDDFQL) (SEQ ID NO. 1) is used as substrate. The reaction is conducted in a total volume of 50 uL containing TrisCl (5 mM, pH 7.5), ascorbate (120 μM), 2-oxoglutarate (3.2 μM), HIF-1α (8.6 μM), and bovine serum albumin (0.01%). The enzyme, quantity predetermined to hydroxylate 20% of substrate in 20 minutes, is added to start the reaction. Where inhibitors are used, compounds are prepared in dimethyl sulfoxide at 10-fold final assay concentration. After 20 minutes at room temperature, the reaction is stopped by transferring 10 μL of reaction mixture to 50 μL of a mass spectrometry matrix solution (α-cyano-4-hydroxycinnamic acid, 5 mg/mL in 50% acetonitrile/0.1% TFA, 5 mM NH4PO4). Two microliters of the mixture is spotted onto a MALDI-TOF MS target plate for analysis with an Applied Biosystems (Foster City, Calif.) 4700 Proteomics Analyzer MALDI-TOF MS equipped with a Nd:YAG laser (355 nm, 3 ns pulse width, 200 Hz repetition rate), Hydroxylated peptide product is identified from substrate by the gain of 16 Da. Data defined as percent conversion of substrate to product is analyzed in GraphPad Prism 4 to calculate IC50 values. Biochemical Assay Buffer pH7.4: Prepare 1 (M) KH2PO4 and 1 (M) K2HPO4. Titrate 1(M) K2HPO4 with 1 (M) KH2PO4 to obtain pH 7.40. Dilute this buffer 10 fold in Water (30 ml buffer+270 ml of water) to obtain 100 mM phosphate buffer. Adjust pH to 7.40±0.02 using 5(N) HCl or 5(N) NaOH. NADPH Regeneration System (NRS): Prepare a solution containing 13 mM NADP, 33 mM Glucose-6-phosphate, 33 mM MgCl2and 4 U/ml Glucose-6-phosphate dehydrogenase in buffer. Liver Microsome (LM) suspension: Thaw LM vial on ice, then mix 1.0 ml LM (20 mg/ml) with 19 ml buffer [final LM Conc: 1 mg/ml] LM+NRS suspension: Mix 5.0 ml NRS with 20 ml LM suspension [final LM Conc: 0.8 mg/ml] System suitability standard: a synthesized compound having Mol wt 686.2 used as System suitability standard. Dissolve this compound in ice-cold acetonitrile to obtain concentration of 0.1 μg/ml and store at 4° C. Compound Dilution: Compound Stock: 10 mM in DMSO Sub stock (100 μM): 4 μl of 10 mM Compound Stock+398 μl Acetonitrile. Working plate (2 μM): 10 μl of 100 μM Sub stock+490 μl buffer Assay Procedure Incubate all plastic materials including tips at 37° C. overnight. Incubate LM suspension and NRS at 37° C. for 15 min before use. Add 40 μl buffer to the wells of blank plate. Add 40 μl compound (from working plate) to 0, 5, 10, 20, 30 and 60 min plates. Initiate reaction by adding 40 μl of LM+NRS suspension in each plate. Terminate reaction by adding 240 μl ice-cold acetonitrile containing system suitability standard at designated time points. For T=0 add 240 μl ice-cold acetonitrile containing system suitability standard before LM+NRS addition. Centrifuge (3500 rpm, 20 min and 15° C.) the plates. Mix 110 μl supernatant with 110 μl water and quantitate amount of Compound in the solution using LC-MS/MS.
 US10172845, Example 398 US10441581, Example 398 US11648243, Example 398 US10881652, Example 398 US10144734, Example 398 BDBM305381
US10172845, Example 398 US10441581, Example 398 US11648243, Example 398 US10881652, Example 398 US10144734, Example 398 BDBM305381 BDBM78453 US10245267, Example 398 US9694016, 398 US10709712, Example 398
BDBM78453 US10245267, Example 398 US9694016, 398 US10709712, Example 398 US9745328, Compound 398 US9079866, 398 BDBM168997 US9884878, Compound 398
US9745328, Compound 398 US9079866, 398 BDBM168997 US9884878, Compound 398 BDBM71326 US9546164, 398 US9694002, 398
BDBM71326 US9546164, 398 US9694002, 398 US8952157, 398 US9303025, 398 BDBM145207
US8952157, 398 US9303025, 398 BDBM145207 BDBM171772 US9611261, Example 398 US9085576, 398
BDBM171772 US9611261, Example 398 US9085576, 398 BDBM232852 US9604984, Example 398 US9346815, 398
BDBM232852 US9604984, Example 398 US9346815, 398 US11547697, Compound 398 US9682141, 398 BDBM138090
US11547697, Compound 398 US9682141, 398 BDBM138090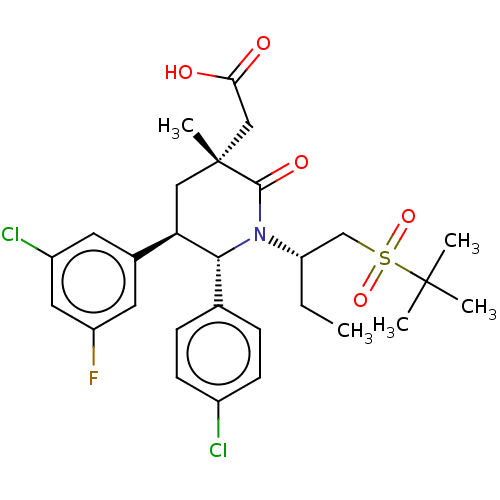 US9593129, Example 398 US9296736, 398 BDBM215410
US9593129, Example 398 US9296736, 398 BDBM215410 US9682966, 398 US10118915, Compound 398 BDBM156625
US9682966, 398 US10118915, Compound 398 BDBM156625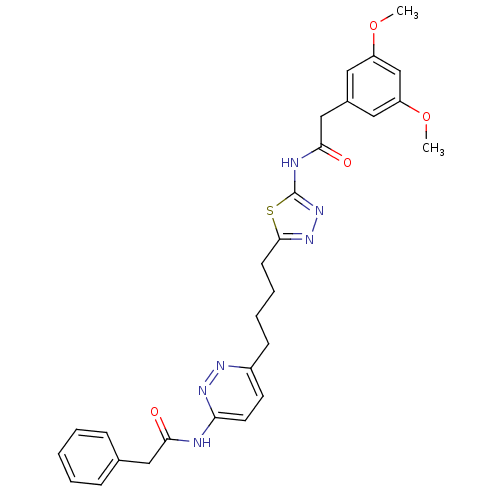 BDBM108818 US8604016, 398 US9938267, Cmpd ID 398
BDBM108818 US8604016, 398 US9938267, Cmpd ID 398 BDBM175633 US9688672, Example 398 US10112931, Example 398
BDBM175633 US9688672, Example 398 US10112931, Example 398 BDBM273613 US10478424, Example 398 US10071079, Example 398
BDBM273613 US10478424, Example 398 US10071079, Example 398 BDBM374256 US10577367, Example 398 US10246456, Example 398
BDBM374256 US10577367, Example 398 US10246456, Example 398 BDBM452260 US11555029, No. 398 US10710986, Example 398
BDBM452260 US11555029, No. 398 US10710986, Example 398 US10028961, Compound 398 US10946023, Compound 398 BDBM280239
US10028961, Compound 398 US10946023, Compound 398 BDBM280239 US10125118, Example 398 US10947215, Example 398 BDBM298992
US10125118, Example 398 US10947215, Example 398 BDBM298992 US10544130, Example 398 US10221163, Example 398 BDBM361241
US10544130, Example 398 US10221163, Example 398 BDBM361241 US10562891, Example 398 BDBM434484 US11008308, Example 398
US10562891, Example 398 BDBM434484 US11008308, Example 398 US10711020, Example 398 US10273259, Example 398 BDBM382689
US10711020, Example 398 US10273259, Example 398 BDBM382689 US10947252, Example 398 BDBM486931 US11427601, Example 398
US10947252, Example 398 BDBM486931 US11427601, Example 398 US11014913, Example 398 US10870641, Example 398 BDBM477154
US11014913, Example 398 US10870641, Example 398 BDBM477154 US11236047, Example 398 BDBM535724 US20230338337, Compound 398
US11236047, Example 398 BDBM535724 US20230338337, Compound 398 US11453683, Example 398 BDBM573567 US20230279025, Example 398
US11453683, Example 398 BDBM573567 US20230279025, Example 398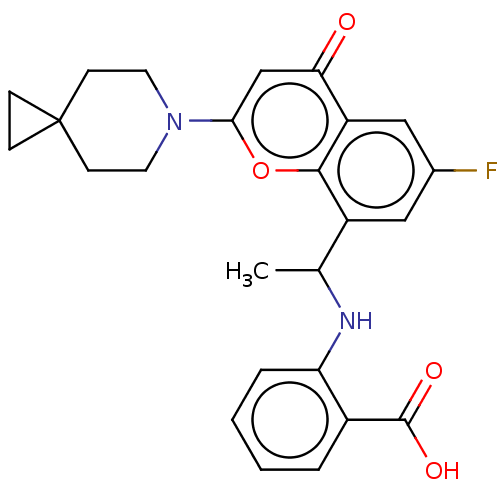 US20230286960, Example 398 BDBM602724 US11649227, Example 398
US20230286960, Example 398 BDBM602724 US11649227, Example 398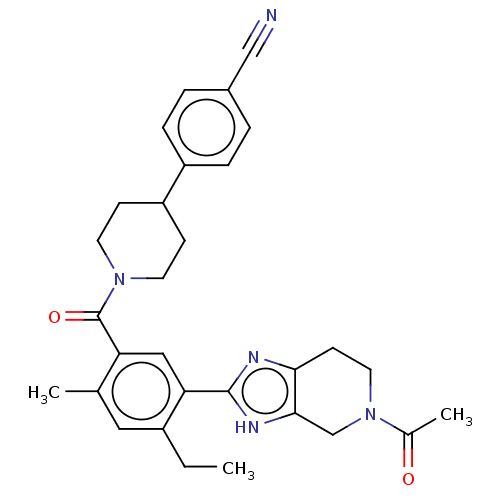 US8871790, 398 BDBM137312 US20250017938, Compound 001-398
US8871790, 398 BDBM137312 US20250017938, Compound 001-398 US11046698, Compound I-398 US10508120, Compound I-398 US10577373, Compound I-398 BDBM423943
US11046698, Compound I-398 US10508120, Compound I-398 US10577373, Compound I-398 BDBM423943 BDBM338215 US9802960, Compound I-398 US9751854, Compound I-398
BDBM338215 US9802960, Compound I-398 US9751854, Compound I-398 BDBM565830 US12221453, Compound I-398 US11414431, Compound I-398
BDBM565830 US12221453, Compound I-398 US11414431, Compound I-398 US10633389, Example 398-1a US20230279020, Example 398-1a BDBM440060
US10633389, Example 398-1a US20230279020, Example 398-1a BDBM440060 US11396508, Compound I-398 BDBM442437 US10647713, Compound I-398
US11396508, Compound I-398 BDBM442437 US10647713, Compound I-398 US11548890, Compound I-398 BDBM502542 US11028085, Compound I-398
US11548890, Compound I-398 BDBM502542 US11028085, Compound I-398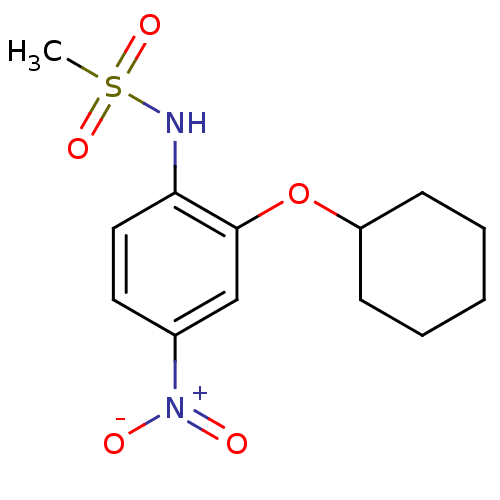 N-(2-Cyclohexyloxy-4-nitro-phenyl)-methanesulfonamide N-[2-(cyclohexyloxy)-4-nitrophenyl]methanesulfonamide N-(2-Cyclohexyloxy-4-nitro-phenyl)-methanesulfonamide(NS-398) N-[2-(cyclohexyloxy)-4-nitrophenyl]methane sulfonamide N-(2-(cyclohexyloxy)-4-nitrophenyl)methanesulfonamide N-{2-(cyclohexyloxy)-4-[hydroxy(oxido)amino]phenyl}methanesulfonamide NS-398 CHEMBL7162 NS398 BDBM50029593
N-(2-Cyclohexyloxy-4-nitro-phenyl)-methanesulfonamide N-[2-(cyclohexyloxy)-4-nitrophenyl]methanesulfonamide N-(2-Cyclohexyloxy-4-nitro-phenyl)-methanesulfonamide(NS-398) N-[2-(cyclohexyloxy)-4-nitrophenyl]methane sulfonamide N-(2-(cyclohexyloxy)-4-nitrophenyl)methanesulfonamide N-{2-(cyclohexyloxy)-4-[hydroxy(oxido)amino]phenyl}methanesulfonamide NS-398 CHEMBL7162 NS398 BDBM50029593 BDBM121959 US8722692, 398
BDBM121959 US8722692, 398 BDBM137805 US8871934, 398
BDBM137805 US8871934, 398 BDBM146533 US8957068, 398
BDBM146533 US8957068, 398 BDBM154505 US9012443, 398
BDBM154505 US9012443, 398 BDBM170046 US9073940, 398
BDBM170046 US9073940, 398 BDBM172336 US9675593, 398
BDBM172336 US9675593, 398 BDBM182697 US9145392, 398
BDBM182697 US9145392, 398 BDBM187855 US9169252, 398
BDBM187855 US9169252, 398 BDBM189129 US9169260, 398
BDBM189129 US9169260, 398 BDBM201966 US9233979, 398
BDBM201966 US9233979, 398 BDBM203215 US9242970, 398
BDBM203215 US9242970, 398 BDBM226705 US9328096, 398
BDBM226705 US9328096, 398 BDBM248506 US9434711, 398
BDBM248506 US9434711, 398 BDBM251094 US9452986, 398
BDBM251094 US9452986, 398 BDBM88340 US9695118, 398
BDBM88340 US9695118, 398 BDBM97852 US8481536, 398
BDBM97852 US8481536, 398 US8501936, 398 BDBM99879
US8501936, 398 BDBM99879 US8507533, 398 BDBM100845
US8507533, 398 BDBM100845 US8637500, 398 BDBM115515
US8637500, 398 BDBM115515 US8637532, 398 BDBM116694
US8637532, 398 BDBM116694 US8841312, 398 BDBM132648
US8841312, 398 BDBM132648 US8846698, 398 BDBM134917
US8846698, 398 BDBM134917 US8952169, 398 BDBM145600
US8952169, 398 BDBM145600 US9023865, 398 BDBM157557
US9023865, 398 BDBM157557 US9029559, 398 BDBM156956
US9029559, 398 BDBM156956 US9034866, 398 BDBM158923
US9034866, 398 BDBM158923 US9085555, 398 BDBM170968
US9085555, 398 BDBM170968 US9120749, 398 BDBM177032
US9120749, 398 BDBM177032 US9125913, 398 BDBM178826
US9125913, 398 BDBM178826 US9163007, 398 BDBM185891
US9163007, 398 BDBM185891 US9226922, 398 BDBM199762
US9226922, 398 BDBM199762 US9242996, 398 BDBM204319
US9242996, 398 BDBM204319 US9255090, 398 BDBM205929
US9255090, 398 BDBM205929 US9260425, 398 BDBM206639
US9260425, 398 BDBM206639 US9260439, 398 BDBM207514
US9260439, 398 BDBM207514 US9283222, 398 BDBM214335
US9283222, 398 BDBM214335 US9302989, 398 BDBM217103
US9302989, 398 BDBM217103 US9328106, 398 BDBM227445
US9328106, 398 BDBM227445 US9340517, 398 BDBM232010
US9340517, 398 BDBM232010 US9458171, 398 BDBM253438
US9458171, 398 BDBM253438 US9493446, 398 BDBM257929
US9493446, 398 BDBM257929 US9688643, I-398 US9150546, I-398 US9718790, I-0397 BDBM183228
US9688643, I-398 US9150546, I-398 US9718790, I-0397 BDBM183228 BDBM50000215 NS-49
BDBM50000215 NS-49 BDBM161583 US9108983, Example 398
BDBM161583 US9108983, Example 398 BDBM296296 US10112937, Example 398
BDBM296296 US10112937, Example 398 BDBM316440 US10172864, Compound 398
BDBM316440 US10172864, Compound 398 BDBM380403 US9926282, Example 398
BDBM380403 US9926282, Example 398 BDBM410574 US10377770, Example 398
BDBM410574 US10377770, Example 398 BDBM431453 US10550105, Example 398
BDBM431453 US10550105, Example 398 BDBM479939 US10899735, No. 398
BDBM479939 US10899735, No. 398 BDBM494347 US10988478, Example 398
BDBM494347 US10988478, Example 398 BDBM512803 acs.jmedchem.1c00409_ST.398
BDBM512803 acs.jmedchem.1c00409_ST.398 BDBM540462 US11261186, Example 398
BDBM540462 US11261186, Example 398 BDBM544912 US11286268, Compound 398
BDBM544912 US11286268, Compound 398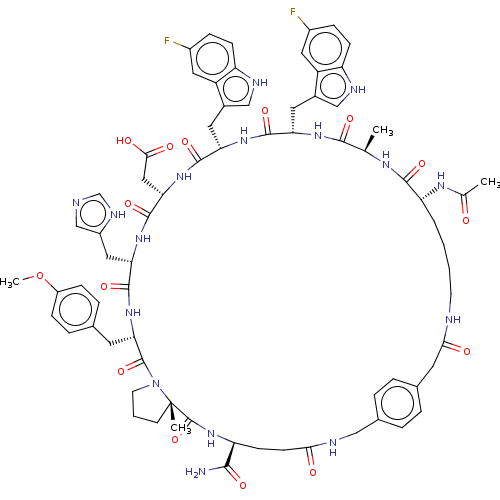 BDBM586311 US11530244, Compound 398
BDBM586311 US11530244, Compound 398 BDBM588795 US11548892, Compound 398
BDBM588795 US11548892, Compound 398 BDBM595931 US11591336, Compound 398
BDBM595931 US11591336, Compound 398 BDBM596628 US11596639, Example 398
BDBM596628 US11596639, Example 398 BDBM603425 US11649255, Example 398
BDBM603425 US11649255, Example 398 BDBM613135 US11725000, Compound 398
BDBM613135 US11725000, Compound 398 BDBM615607 US20230271949, Example 398
BDBM615607 US20230271949, Example 398 BDBM618407 US20230286970, Compound 398
BDBM618407 US20230286970, Compound 398 BDBM622825 US20230312605, Compound 398
BDBM622825 US20230312605, Compound 398 BDBM623379 US11780845, Example 398
BDBM623379 US11780845, Example 398 BDBM632128 US20230348426, Example 398
BDBM632128 US20230348426, Example 398 BDBM634949 US11814367, Compound 398
BDBM634949 US11814367, Compound 398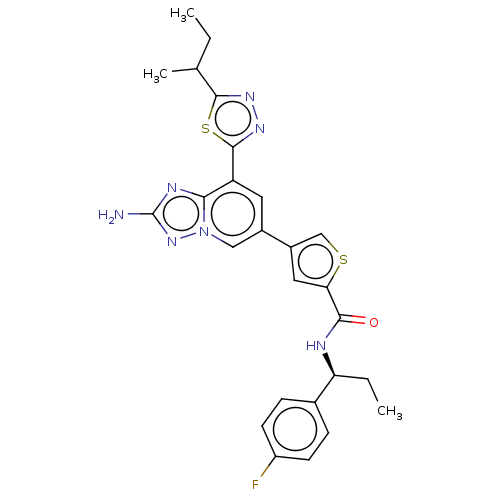 BDBM637483 US20230382904, Compound 398
BDBM637483 US20230382904, Compound 398 BDBM641504 US11845723, Example 398
BDBM641504 US11845723, Example 398 BDBM647187 US20240025884, Example 398
BDBM647187 US20240025884, Example 398 BDBM650303 US20240043427, Example 398
BDBM650303 US20240043427, Example 398 BDBM655363 US11912703, Example 398
BDBM655363 US11912703, Example 398 BDBM695232 US20240294551, Example 398
BDBM695232 US20240294551, Example 398 BDBM697617 US20240316047, Example 398
BDBM697617 US20240316047, Example 398 BDBM718541 US20250042889, Example 398
BDBM718541 US20250042889, Example 398 BDBM723819 US20250064789, Compound 398
BDBM723819 US20250064789, Compound 398 BDBM735374 US20250129078, Compound 398
BDBM735374 US20250129078, Compound 398 BDBM739149 US20250145633, Example 398
BDBM739149 US20250145633, Example 398 BDBM745916 US12319655, Example 398
BDBM745916 US12319655, Example 398 US10307413, Compound 398 BDBM394754
US10307413, Compound 398 BDBM394754 US10660877, Example 398 BDBM443810
US10660877, Example 398 BDBM443810 US10730863, Example 398 BDBM453958
US10730863, Example 398 BDBM453958 US10961200, Compound 398 BDBM489019
US10961200, Compound 398 BDBM489019 US10975056, Example 398 BDBM491424
US10975056, Example 398 BDBM491424 US11186582, Example 398 BDBM527524
US11186582, Example 398 BDBM527524 US11242361, Compound 398 BDBM536692
US11242361, Compound 398 BDBM536692 US11254663, Example 398 BDBM538478
US11254663, Example 398 BDBM538478 US11292791, Example 398 BDBM547125
US11292791, Example 398 BDBM547125 US11524968, Example 398 BDBM584649
US11524968, Example 398 BDBM584649 US11773078, Example 398 BDBM621646
US11773078, Example 398 BDBM621646 US11834453, Example 398 BDBM638295
US11834453, Example 398 BDBM638295 US12378224, Example 398 BDBM761776
US12378224, Example 398 BDBM761776 US12384753, Example 398 BDBM763374
US12384753, Example 398 BDBM763374 US20230340011, Example 398. BDBM630098
US20230340011, Example 398. BDBM630098 US20240116946, Example 398 BDBM666365
US20240116946, Example 398 BDBM666365 US20240174662, Example 398 BDBM677734
US20240174662, Example 398 BDBM677734 US20240218021, Example 398 BDBM683911
US20240218021, Example 398 BDBM683911 US20240246937, Example 398 BDBM686955
US20240246937, Example 398 BDBM686955 US20240246964, Compound 398 BDBM687377
US20240246964, Compound 398 BDBM687377 US20250026748, Compound 398 BDBM715234
US20250026748, Compound 398 BDBM715234 US20250059220, Compound 398 BDBM723286
US20250059220, Compound 398 BDBM723286 US20250197382, Compound 398 BDBM752132
US20250197382, Compound 398 BDBM752132 US8575197, I-398 BDBM105888
US8575197, I-398 BDBM105888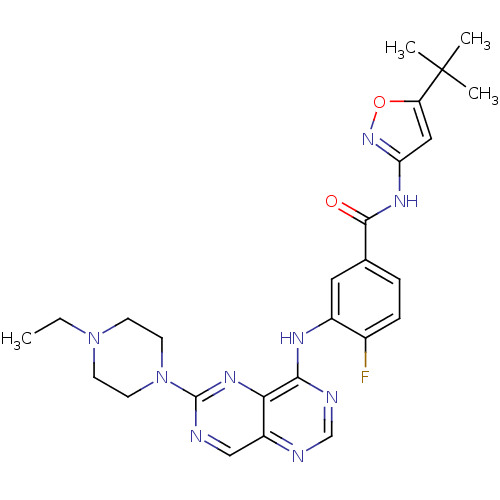 US8653087, III-398 BDBM117718
US8653087, III-398 BDBM117718 US9546153, ex. 398 BDBM210385
US9546153, ex. 398 BDBM210385 US9662327, Compound 398 BDBM326813
US9662327, Compound 398 BDBM326813 US9718825, Example 398 BDBM268970
US9718825, Example 398 BDBM268970 US9862730, Example 398 BDBM364940
US9862730, Example 398 BDBM364940 BDBM146302 US8957068, 397 US8957068, 398
BDBM146302 US8957068, 397 US8957068, 398 BDBM195380 US9212153, 464,Ex. 398
BDBM195380 US9212153, 464,Ex. 398 BDBM246744 US9550763, Compound I-398
BDBM246744 US9550763, Compound I-398 BDBM346787 US10202379, Reference Example 398
BDBM346787 US10202379, Reference Example 398 BDBM441440 US10640495, Example I-398
BDBM441440 US10640495, Example I-398 BDBM465063 US10793563, Compound I-398
BDBM465063 US10793563, Compound I-398 BDBM567028 US11420958, Ex. No. 398
BDBM567028 US11420958, Ex. No. 398 BDBM674087 US20240150321, Compound I-398
BDBM674087 US20240150321, Compound I-398 BDBM728767 US20250092056, Compound I-398
BDBM728767 US20250092056, Compound I-398 BDBM742128 US20250163057, Compound I-398
BDBM742128 US20250163057, Compound I-398 US10144742, Compound I-398 BDBM306236
US10144742, Compound I-398 BDBM306236 US10150728, Example I-398 BDBM307185
US10150728, Example I-398 BDBM307185 US10183021, Compound I-398 BDBM321624
US10183021, Compound I-398 BDBM321624 US10919885, Compound No. 398 BDBM483249
US10919885, Compound No. 398 BDBM483249 US11339144, Compound I-398 BDBM554929
US11339144, Compound I-398 BDBM554929 US11518764, Compound I-398 BDBM582385
US11518764, Compound I-398 BDBM582385 US11555012, Compound I-398 BDBM589657
US11555012, Compound I-398 BDBM589657 US20250019387, Table 1a.398 BDBM713671
US20250019387, Table 1a.398 BDBM713671 US20250090540, Example I-398 BDBM727537
US20250090540, Example I-398 BDBM727537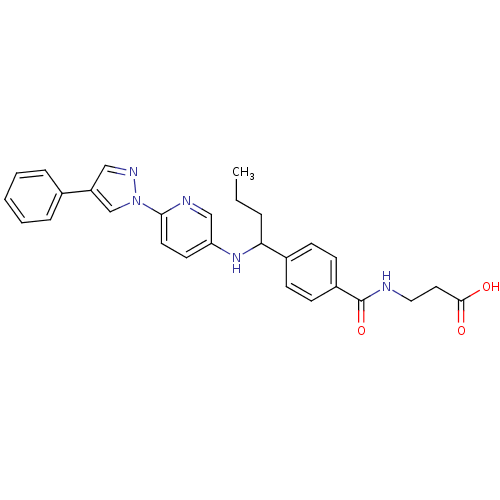 US8507533, 398 BDBM100844 US8507533, 160
US8507533, 398 BDBM100844 US8507533, 160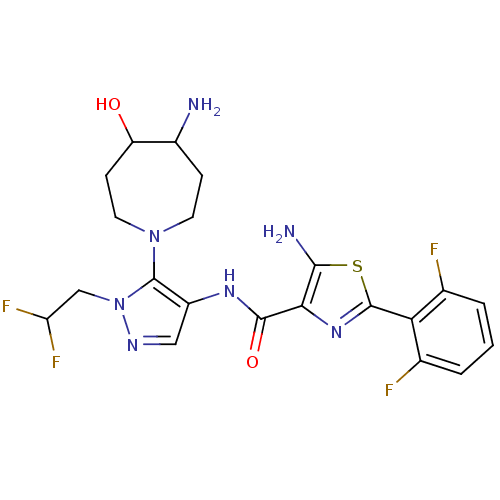 US8614206, 398 US8614206, 399 BDBM110847
US8614206, 398 US8614206, 399 BDBM110847 US9212153, 398,Ex. 337 BDBM195318
US9212153, 398,Ex. 337 BDBM195318 BDBM247795 US9434719, 398 as TFA salt
BDBM247795 US9434719, 398 as TFA salt BDBM508081 US11046691, Compound 398 US11130759, Cpd. No. 398 4-[(2,2-difluoroethyl)amino]-7-methyl- 1-phenylpyrido[2,3-d]pyrimidin-2(1H)- one US11084798, Cpd No 398
BDBM508081 US11046691, Compound 398 US11130759, Cpd. No. 398 4-[(2,2-difluoroethyl)amino]-7-methyl- 1-phenylpyrido[2,3-d]pyrimidin-2(1H)- one US11084798, Cpd No 398 US10017468, Compound 398 US11072582, Compound 398 US10781171, Compound 398 (2S,5R)-5-(2,3-difluorophenyl)-1-(2'-methoxy-[1,1'-biphenyl]-4-carbonyl)pyrrolidine-2-carboxylic acid BDBM404228
US10017468, Compound 398 US11072582, Compound 398 US10781171, Compound 398 (2S,5R)-5-(2,3-difluorophenyl)-1-(2'-methoxy-[1,1'-biphenyl]-4-carbonyl)pyrrolidine-2-carboxylic acid BDBM404228 US10137124, Example 398 US10112942, Example 398 US10172851, Example 398 BDBM296664 US10555944, Example 398 6-(2-hydroxy- 2- methylpropoxy)- 4-(6-(6- ((1s,3s)-3- methoxycyclo- butane-1- carbonyl)-3,6- diazabicyclo[3. 1.1]heptan-3- yl)pyridin-3- yl)pyrazolo[1,5- a]pyridine-3- carbonitrile US10953005, Example 398
US10137124, Example 398 US10112942, Example 398 US10172851, Example 398 BDBM296664 US10555944, Example 398 6-(2-hydroxy- 2- methylpropoxy)- 4-(6-(6- ((1s,3s)-3- methoxycyclo- butane-1- carbonyl)-3,6- diazabicyclo[3. 1.1]heptan-3- yl)pyridin-3- yl)pyrazolo[1,5- a]pyridine-3- carbonitrile US10953005, Example 398 US10189841, Compound I-398 US10399980, Compound I-398 2-(2-isopropylphenyl)-9-(4-(2-methylpiperidine-1- BDBM325192
US10189841, Compound I-398 US10399980, Compound I-398 2-(2-isopropylphenyl)-9-(4-(2-methylpiperidine-1- BDBM325192 BDBM268363 US9549932, 398 N-methyl-5-[2-(morpholin-4-yl)-8- (1H-pyrazol-5-yl)-1,7- naphthyridin-4-yl]pyridine-2- carboxamide US10772893, Example 398 US11529356, Example 398
BDBM268363 US9549932, 398 N-methyl-5-[2-(morpholin-4-yl)-8- (1H-pyrazol-5-yl)-1,7- naphthyridin-4-yl]pyridine-2- carboxamide US10772893, Example 398 US11529356, Example 398 BDBM539053 US11254668, Example 399 US11254668, Example 398
BDBM539053 US11254668, Example 399 US11254668, Example 398 BDBM700377 US20240336593, Example 140 US20250206717, Example 398
BDBM700377 US20240336593, Example 140 US20250206717, Example 398 US11584747, Example 398 US11584747, Example 399 BDBM594580
US11584747, Example 398 US11584747, Example 399 BDBM594580 US20250025443, Compound 398 BDBM714017 US20250025443, Example 50
US20250025443, Compound 398 BDBM714017 US20250025443, Example 50 US20250025443, Compound 748 BDBM714364 US20250025443, Example 398
US20250025443, Compound 748 BDBM714364 US20250025443, Example 398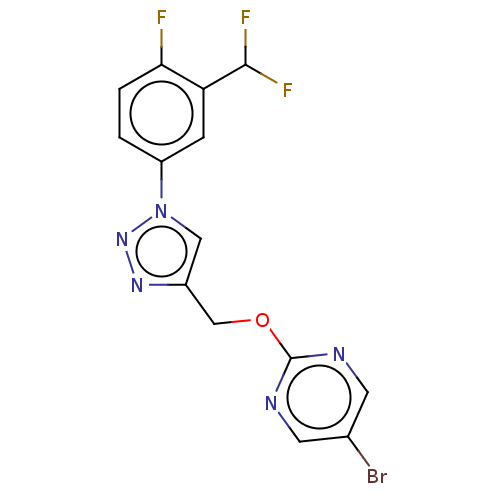 5-Bromo-2-[[1-[3-(difluoromethyl)-4-fluoro-phenyl]triazol-4- US10071988, Example 398 BDBM276459 US10233173, Example 398
5-Bromo-2-[[1-[3-(difluoromethyl)-4-fluoro-phenyl]triazol-4- US10071988, Example 398 BDBM276459 US10233173, Example 398 4-Oxo-5-(4-phenoxyphenyl)-N-((1R,2S)-2- propionamidocyclopentyl)-4,5-dihydro-3H-1-thia-3,5,8- triazaacenaphthylene-2-carboxamide US12065446, Example 398 BDBM485561 US10934310, Ex # 398 US11319329, Ex # 398
4-Oxo-5-(4-phenoxyphenyl)-N-((1R,2S)-2- propionamidocyclopentyl)-4,5-dihydro-3H-1-thia-3,5,8- triazaacenaphthylene-2-carboxamide US12065446, Example 398 BDBM485561 US10934310, Ex # 398 US11319329, Ex # 398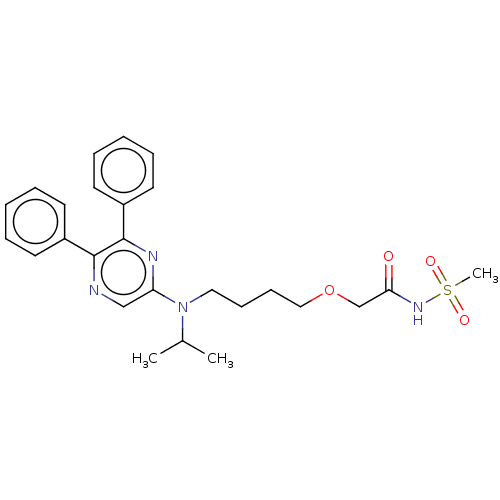 ACT-293987 BDBM50235383 Uptravi Selexipag NS-304
ACT-293987 BDBM50235383 Uptravi Selexipag NS-304 US10793563, Compound I-329 BDBM464996 US10793563, Compound I-398
US10793563, Compound I-329 BDBM464996 US10793563, Compound I-398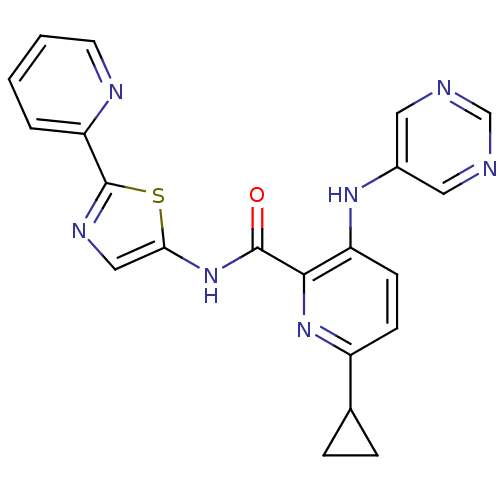 BDBM50397998 CHEMBL2180800 US8470820, 62 Roche-Dataset for PDE10A, Compound 398
BDBM50397998 CHEMBL2180800 US8470820, 62 Roche-Dataset for PDE10A, Compound 398 US20230340011, Example 398. BDBM630005 US20230340011, Example 304. US20230340011, Example 305.
US20230340011, Example 398. BDBM630005 US20230340011, Example 304. US20230340011, Example 305. US10472354, Example 398 US10174016, Example 398 BDBM320231 US10202369, Example 398 3-{[(5R)-3-methyl-2- oxo-1,3-oxazolidin- 5-yl]methoxy}-5-(5- methyl-1,3-thiazol- 2-yl)-N-{(1R)-1-[2- (trifluoromethyl) pyrimidin-5- yl]ethyl}benzamide
US10472354, Example 398 US10174016, Example 398 BDBM320231 US10202369, Example 398 3-{[(5R)-3-methyl-2- oxo-1,3-oxazolidin- 5-yl]methoxy}-5-(5- methyl-1,3-thiazol- 2-yl)-N-{(1R)-1-[2- (trifluoromethyl) pyrimidin-5- yl]ethyl}benzamide US20240217978, Example 398 (S)-2-((7-amino-2-(furan-2- BDBM683742
US20240217978, Example 398 (S)-2-((7-amino-2-(furan-2- BDBM683742 (R/S)-3-Methyl-6-[2-methyl-3-(trifluoromethyl)phenyl]-1-(oxetan-2- ylmethyl)imidazo[4,5-b]pyridin-2-one US10617676, Example 398 BDBM437062 US11207298, Example 398
(R/S)-3-Methyl-6-[2-methyl-3-(trifluoromethyl)phenyl]-1-(oxetan-2- ylmethyl)imidazo[4,5-b]pyridin-2-one US10617676, Example 398 BDBM437062 US11207298, Example 398 BDBM483856 1,1,1,3,3,3- hexafluoropropan-2- yl 4-(7-cyclobutyl- N-methyl-5,6,7,8- tetrahydroimidazo- [l,2-a]pyrazine- 2-carboxamido)-4- methylpiperidine-1- carboxylate US11655217, Example 398 US10927105, Ex 398
BDBM483856 1,1,1,3,3,3- hexafluoropropan-2- yl 4-(7-cyclobutyl- N-methyl-5,6,7,8- tetrahydroimidazo- [l,2-a]pyrazine- 2-carboxamido)-4- methylpiperidine-1- carboxylate US11655217, Example 398 US10927105, Ex 398 N~2~-acetyl-N-[3-({4-[({3- [methyl(methylsulfonyl)amino]pyridin-2- yl}methyl)amino]-5- (trifluoromethyl)pyrimidin-2- yl}amino)phenyl]glycinamide (398) BDBM418898 US10450297, Example 398
N~2~-acetyl-N-[3-({4-[({3- [methyl(methylsulfonyl)amino]pyridin-2- yl}methyl)amino]-5- (trifluoromethyl)pyrimidin-2- yl}amino)phenyl]glycinamide (398) BDBM418898 US10450297, Example 398 US11485745, Example 398 BDBM455799 US10730889, Example 398 N-[[6-[4- (trifluoromethyl]pyridine-3- carbonyl]-6-azaspiro[2.5]octan- 2-yl]methyl]-1,3- dihydropyrrolo[3,4-c]pyridine-2- carboxamide
US11485745, Example 398 BDBM455799 US10730889, Example 398 N-[[6-[4- (trifluoromethyl]pyridine-3- carbonyl]-6-azaspiro[2.5]octan- 2-yl]methyl]-1,3- dihydropyrrolo[3,4-c]pyridine-2- carboxamide 2-(2-isopropylphenyl)-9-(4-(2-methylpiperidine-1- US11161848, Compound I-398 BDBM522840
2-(2-isopropylphenyl)-9-(4-(2-methylpiperidine-1- US11161848, Compound I-398 BDBM522840 BDBM661012 N-(6-chloropyridin-3-yl)-3- methylisoquinolin-1-amine US20240101531, Example 398
BDBM661012 N-(6-chloropyridin-3-yl)-3- methylisoquinolin-1-amine US20240101531, Example 398 US10266526, Compound 398 N-((1r,4r)-4-aminocyclohexyl)-5-methylthiazole-2-carboxamide BDBM378267
US10266526, Compound 398 N-((1r,4r)-4-aminocyclohexyl)-5-methylthiazole-2-carboxamide BDBM378267 US11053244, Example 406 US10544143, Example 398 BDBM427964 US10730877, Example 501 US10544143, Example 406
US11053244, Example 406 US10544143, Example 398 BDBM427964 US10730877, Example 501 US10544143, Example 406 5-[2-(2,4-difluoroanilino)-5- methylsulfonylphenyl]-1,3- dimethylpyridin-2-one BDBM486188 US10941160, Example 398
5-[2-(2,4-difluoroanilino)-5- methylsulfonylphenyl]-1,3- dimethylpyridin-2-one BDBM486188 US10941160, Example 398 BDBM284388 US10174027, Example 398 US10023570, Example 398 6-(1-methyl-1H-pyrazol-4- yl)-4-(6-(4-(2- (trifluoromethoxy)ethyl) piperazin-1-yl)pyridin-3- yl)pyrazolo[1,5-a]pyridine- 3-carbonitrile
BDBM284388 US10174027, Example 398 US10023570, Example 398 6-(1-methyl-1H-pyrazol-4- yl)-4-(6-(4-(2- (trifluoromethoxy)ethyl) piperazin-1-yl)pyridin-3- yl)pyrazolo[1,5-a]pyridine- 3-carbonitrile 2-{6-[5-chloro-2- (ethylamino)pyrimidin- 4-yl]-1-oxo-2,3- dihydro-1H- isoindol-2-yl}-N- [(1S)-2-hydroxy-1-(3- methylphenyl) ethyl]acetamide US10457669, Example 398 BDBM417766 US11001575, Example 398
2-{6-[5-chloro-2- (ethylamino)pyrimidin- 4-yl]-1-oxo-2,3- dihydro-1H- isoindol-2-yl}-N- [(1S)-2-hydroxy-1-(3- methylphenyl) ethyl]acetamide US10457669, Example 398 BDBM417766 US11001575, Example 398 5-[2-(2,4-difluoroanilino)-5- methylsulfonylphenyl]-1,3- dimethylpyridin-2-one BDBM486188 US10023592, Example 398 BDBM285344
5-[2-(2,4-difluoroanilino)-5- methylsulfonylphenyl]-1,3- dimethylpyridin-2-one BDBM486188 US10023592, Example 398 BDBM285344 BDBM386464 US10287301, Compound 398 (2S,4R)-1-(2-(3- acetyl-5-(2- (hydroxymethyl) pyrimidin-5-yl)- 1H-indazol-1- yl)acetyl)-N-(6- bromopyrazin-2- yl)-4- fluoropyrrolidine- 2-carboxamide US10822352, Comp No. 398
BDBM386464 US10287301, Compound 398 (2S,4R)-1-(2-(3- acetyl-5-(2- (hydroxymethyl) pyrimidin-5-yl)- 1H-indazol-1- yl)acetyl)-N-(6- bromopyrazin-2- yl)-4- fluoropyrrolidine- 2-carboxamide US10822352, Comp No. 398 BDBM405054 1-[2-fluoro-4-(4-{2-[2-fluoro-5- (trifluoromethoxy)phenyl]acetamido}- 1H-1,2,3-triazol-1-yl)butyl]-N- (pyridin-2-ylmethyl)-1H-1,2,3- triazole-4-carboxamide US11370786, Example 398 US10344025, Example 398
BDBM405054 1-[2-fluoro-4-(4-{2-[2-fluoro-5- (trifluoromethoxy)phenyl]acetamido}- 1H-1,2,3-triazol-1-yl)butyl]-N- (pyridin-2-ylmethyl)-1H-1,2,3- triazole-4-carboxamide US11370786, Example 398 US10344025, Example 398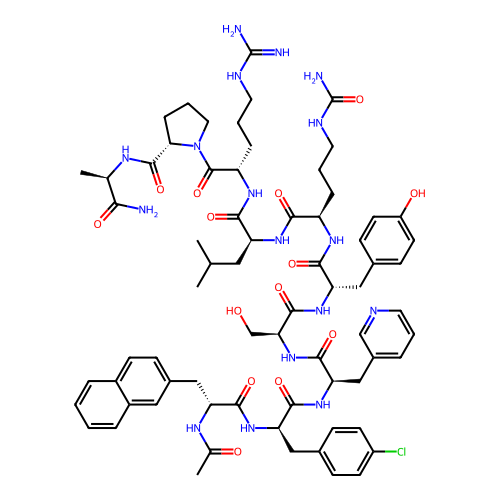 BDBM50248241 Cetrorelix Acetate D-20761 CHEBI:31387 Cetrotide NS-75A SB-075 Acetate Cetrorelix
BDBM50248241 Cetrorelix Acetate D-20761 CHEBI:31387 Cetrotide NS-75A SB-075 Acetate Cetrorelix 3-(1-isopropyl-1H-indol-5-yl)-5-(2- methoxyphenyl)-1,2,4-oxadiazole US11912693, Compound 398 BDBM654785
3-(1-isopropyl-1H-indol-5-yl)-5-(2- methoxyphenyl)-1,2,4-oxadiazole US11912693, Compound 398 BDBM654785 3-(5-Methylpyridin-2-yl)-2-(1H-tetrazol-5-yl)benzenesulfonamide, TFA Salt BDBM262546 US9708336, 398
3-(5-Methylpyridin-2-yl)-2-(1H-tetrazol-5-yl)benzenesulfonamide, TFA Salt BDBM262546 US9708336, 398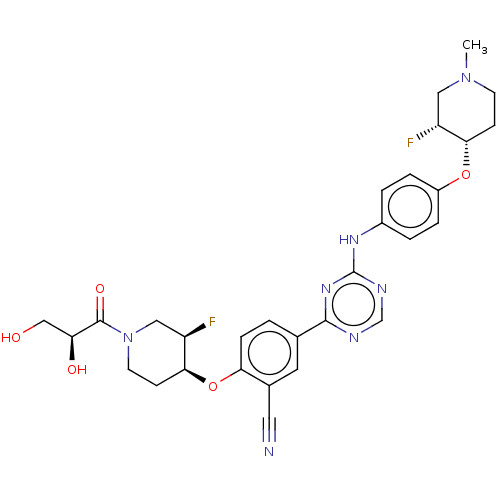 2-(((3R,4S)-1-((S)-2,3-dihydroxypropanoyl)-3-fluoropiperidin-4-yl)oxy)-5-(4-((4-(((3R,4S)-3-fluoro-1-methylpiperidin-4-yl)oxy)phenyl)amino)-1,3,5-triazin-2-yl)benzonitrile US10040781, Example 398 BDBM278176 US10253019, Example 398
2-(((3R,4S)-1-((S)-2,3-dihydroxypropanoyl)-3-fluoropiperidin-4-yl)oxy)-5-(4-((4-(((3R,4S)-3-fluoro-1-methylpiperidin-4-yl)oxy)phenyl)amino)-1,3,5-triazin-2-yl)benzonitrile US10040781, Example 398 BDBM278176 US10253019, Example 398 1-Methyl-1H-pyrrolo[2,3- b]pyridine-5-carboxylic acid (4- chloro-phenyl)-amide US10272074, Example 398 BDBM381563
1-Methyl-1H-pyrrolo[2,3- b]pyridine-5-carboxylic acid (4- chloro-phenyl)-amide US10272074, Example 398 BDBM381563 5-[2-(4-Fluorophenyl)-6-(trifluoromethyl)pyrrolo[2,3-b]pyridin-1-yl]indolin-2- US11312712, Example 398 BDBM551279
5-[2-(4-Fluorophenyl)-6-(trifluoromethyl)pyrrolo[2,3-b]pyridin-1-yl]indolin-2- US11312712, Example 398 BDBM551279 BDBM164181 US9687479, 398 N-benzyl-1-[(2-methoxyphenyl)carbonyl]-3-(thiophen-2-yl)-1H-1,2,4-triazol-5-amine
BDBM164181 US9687479, 398 N-benzyl-1-[(2-methoxyphenyl)carbonyl]-3-(thiophen-2-yl)-1H-1,2,4-triazol-5-amine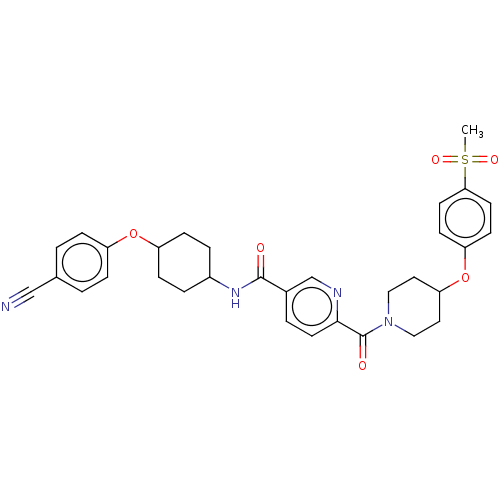 N-(4-(4-cyanophenoxy) cyclohexyl)-6-(4-(4- (methylsulfonyl) phenoxy)piperidine-1- carbonyl)nicotinamide BDBM328260 US9663496, Compound 398
N-(4-(4-cyanophenoxy) cyclohexyl)-6-(4-(4- (methylsulfonyl) phenoxy)piperidine-1- carbonyl)nicotinamide BDBM328260 US9663496, Compound 398 US10377742, Compound 398 N-(4-(4-cyanophenoxy)cyclohexyl)-6-(4-(4- (methylsulfonyl)phenoxy)piperidine-1- carbonyl)nicotinamide BDBM409372
US10377742, Compound 398 N-(4-(4-cyanophenoxy)cyclohexyl)-6-(4-(4- (methylsulfonyl)phenoxy)piperidine-1- carbonyl)nicotinamide BDBM409372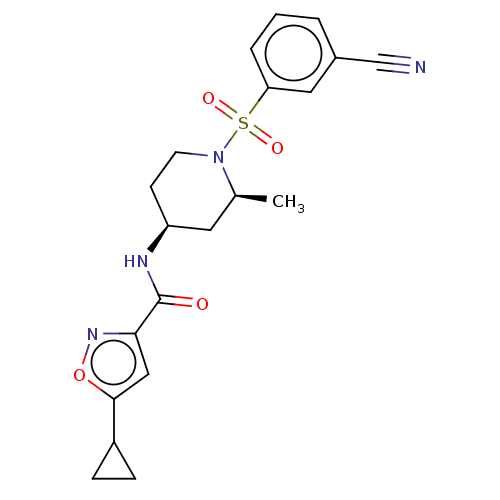 BDBM432698 N-((2S,4S)-1-((3-cyanophenyl)sulfonyl)-2- methylpiperidin-4-yl)-5- cyclopropylisoxazole-3-carboxamide US10577363, Compound 398
BDBM432698 N-((2S,4S)-1-((3-cyanophenyl)sulfonyl)-2- methylpiperidin-4-yl)-5- cyclopropylisoxazole-3-carboxamide US10577363, Compound 398 BDBM409533 US10377753, Example 398 2-[6-(5-Ethyl-2-thienyl)-3-fluoro-pyrrolo[3,2-b]pyridin-1-yl]-1-(3-
BDBM409533 US10377753, Example 398 2-[6-(5-Ethyl-2-thienyl)-3-fluoro-pyrrolo[3,2-b]pyridin-1-yl]-1-(3- BDBM691937 US20240279215, Compound 398 oxazol-5-ylmethyl (4-((2- (dimethylcarbamoyl)-2- azaspiro[3.3]heptan-6- yl)methyl)-3- fluorophenyl)carbamate
BDBM691937 US20240279215, Compound 398 oxazol-5-ylmethyl (4-((2- (dimethylcarbamoyl)-2- azaspiro[3.3]heptan-6- yl)methyl)-3- fluorophenyl)carbamate BDBM745096 4-(3-{p-[4-(8-chloro-3- quinolylamino)-2- pyrimidinylamino]phenoxy} propyl)-1lambda6,4-thiazinane-1,1- dione US20250171431, Compound 398
BDBM745096 4-(3-{p-[4-(8-chloro-3- quinolylamino)-2- pyrimidinylamino]phenoxy} propyl)-1lambda6,4-thiazinane-1,1- dione US20250171431, Compound 398 US9579320, Example 398 BDBM290967 (R)-2-(1-amino-3-hydroxy-1-oxo-2-ylamino)-4-(3-methoxyphenylamino)pyrimidine-5-carboxamide
US9579320, Example 398 BDBM290967 (R)-2-(1-amino-3-hydroxy-1-oxo-2-ylamino)-4-(3-methoxyphenylamino)pyrimidine-5-carboxamide BDBM512053 (1R,3S,5R)-2-(2-(3- acetyl-5-(2- methylpyrimidin-5- yl)-1H-pyrazolo[3,4- c]pyridin-1-yl)acetyl)- N-(6-bromo-3- methylpyridin-2-yl)-5- (2-oxopropyl)-2- azabicyclo[3.1.0]hexane- 3-carboxamide US11084800, Cpd No. 398 US11708351, Compound 398
BDBM512053 (1R,3S,5R)-2-(2-(3- acetyl-5-(2- methylpyrimidin-5- yl)-1H-pyrazolo[3,4- c]pyridin-1-yl)acetyl)- N-(6-bromo-3- methylpyridin-2-yl)-5- (2-oxopropyl)-2- azabicyclo[3.1.0]hexane- 3-carboxamide US11084800, Cpd No. 398 US11708351, Compound 398 3-{4-[3-chloro-2-(morpholin-4-yl)pyridine-4- sulfonyl]phenyl}-1-(pyridin-3-ylmethyl)urea BDBM543387 US11279687, Compound 398
3-{4-[3-chloro-2-(morpholin-4-yl)pyridine-4- sulfonyl]phenyl}-1-(pyridin-3-ylmethyl)urea BDBM543387 US11279687, Compound 398 BDBM657719 N-[1-{5-[2-(aminomethyl)-4-chlorophenyl]thiophen-2-yl}ethyl]-6,7-dimethoxy-2-methylquinazolin-4-amine US20240083857, Example 398
BDBM657719 N-[1-{5-[2-(aminomethyl)-4-chlorophenyl]thiophen-2-yl}ethyl]-6,7-dimethoxy-2-methylquinazolin-4-amine US20240083857, Example 398 N-[(3S)-3-(4- chlorophenyl)-3- hydroxypropyl]- 5-{2- acetamidoimidazo [1,2-b]pyridazin- 6-yl}-2- methylbenzamide BDBM571323 US11440913, Example 398
N-[(3S)-3-(4- chlorophenyl)-3- hydroxypropyl]- 5-{2- acetamidoimidazo [1,2-b]pyridazin- 6-yl}-2- methylbenzamide BDBM571323 US11440913, Example 398 (7-(2-Fluoro-3-methylbenzyl)-2-azaspiro[3.5]nonan-2-yl)((1s,3s)-3-hydroxy-3-methylcyclobutyl)methanone US11802111, Example 398 BDBM630808
(7-(2-Fluoro-3-methylbenzyl)-2-azaspiro[3.5]nonan-2-yl)((1s,3s)-3-hydroxy-3-methylcyclobutyl)methanone US11802111, Example 398 BDBM630808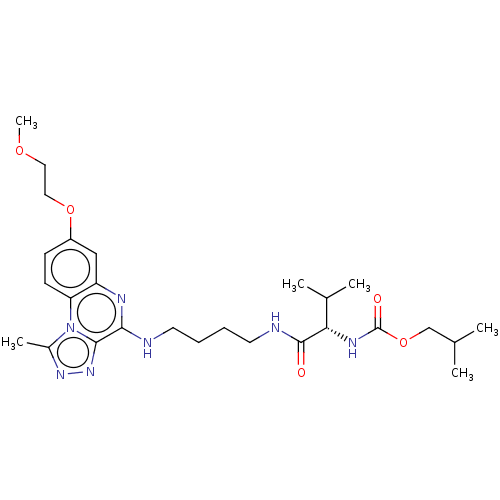 BDBM502479 (S)-(1-{4-[7- (2-methoxy- ethoxy)-1- methyl- [1,2,4]tri- azolo[4,3- a]quinoxaline- 4-ylamino]- butylcarbamo- US11028090, Example 398
BDBM502479 (S)-(1-{4-[7- (2-methoxy- ethoxy)-1- methyl- [1,2,4]tri- azolo[4,3- a]quinoxaline- 4-ylamino]- butylcarbamo- US11028090, Example 398 US10173991, Example 398 3-[2- ({[(cyclopropyl- carbamoyl)methyl] carbamoyl}(2- methoxyethane- sulfonyl)methyl)- 1,3-benzothiazol- 5-yl]-N,N- dimethyl- benzamide BDBM319352
US10173991, Example 398 3-[2- ({[(cyclopropyl- carbamoyl)methyl] carbamoyl}(2- methoxyethane- sulfonyl)methyl)- 1,3-benzothiazol- 5-yl]-N,N- dimethyl- benzamide BDBM319352 US11724992, Example 398 (S) or (R)-N'- ((1,2,3,5,6,7- hexahydro-s- indacen-4-yl) carbamoyl)- 1-methyl- 1H- indazole-5- sulfonimi- damide BDBM612708
US11724992, Example 398 (S) or (R)-N'- ((1,2,3,5,6,7- hexahydro-s- indacen-4-yl) carbamoyl)- 1-methyl- 1H- indazole-5- sulfonimi- damide BDBM612708 US9776995, Example 398 4-(2-chloro-3'-fluoro-5-methoxy-4-biphenylyl)-N-3-isoxazolyl-3-oxo-2,3-dihydro-7-isoquinolinesulfonamide BDBM343165
US9776995, Example 398 4-(2-chloro-3'-fluoro-5-methoxy-4-biphenylyl)-N-3-isoxazolyl-3-oxo-2,3-dihydro-7-isoquinolinesulfonamide BDBM343165 3-((2S)-2-hydroxy-3-(8-(2-methoxyphenylsulfonyl)-1-oxa-8-azaspiro[4.5]decan-3-ylamino)propoxy)-N-methylbenzenesulfonamide US11987588, Compound 398 BDBM675929
3-((2S)-2-hydroxy-3-(8-(2-methoxyphenylsulfonyl)-1-oxa-8-azaspiro[4.5]decan-3-ylamino)propoxy)-N-methylbenzenesulfonamide US11987588, Compound 398 BDBM675929 BDBM355307 N-[(1R)-1-{3-fluoro-4- [(trifluoromethyl)sulfanyl]phenyl}eth- yl]-6-oxo-2-phenyl-1,6- dihydropyrimidine-4-carboxamide US9815796, Example 398
BDBM355307 N-[(1R)-1-{3-fluoro-4- [(trifluoromethyl)sulfanyl]phenyl}eth- yl]-6-oxo-2-phenyl-1,6- dihydropyrimidine-4-carboxamide US9815796, Example 398 BDBM563572 US11407771, Compound 398 N-[4-(2-{2-[3-(5-Cyclopropyl-2- fluoro-phenyl)-ureido]-thiazol-5- yl}-ethyl)-pyridin-2-yl]- acetamide
BDBM563572 US11407771, Compound 398 N-[4-(2-{2-[3-(5-Cyclopropyl-2- fluoro-phenyl)-ureido]-thiazol-5- yl}-ethyl)-pyridin-2-yl]- acetamide BDBM667619 US11957687, Compound 398 2-oxo-N-((R)-((R)- tetrahydro-2H-pyran-3- yl)(p-tolyl)methyl)-6- (trifluoromethyl)-1,2- dihydropyridine-3- carboxamide
BDBM667619 US11957687, Compound 398 2-oxo-N-((R)-((R)- tetrahydro-2H-pyran-3- yl)(p-tolyl)methyl)-6- (trifluoromethyl)-1,2- dihydropyridine-3- carboxamide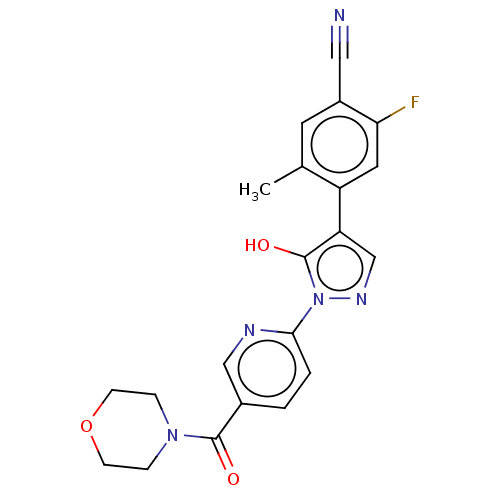 US10407409, Example 398 2-fluoro-4-(5-hydroxy-1-(5-(morpholine-4-carbonyl)pyridin-2-yl)-1H-pyrazol-4-yl)-5-methylbenzonitrile BDBM413146
US10407409, Example 398 2-fluoro-4-(5-hydroxy-1-(5-(morpholine-4-carbonyl)pyridin-2-yl)-1H-pyrazol-4-yl)-5-methylbenzonitrile BDBM413146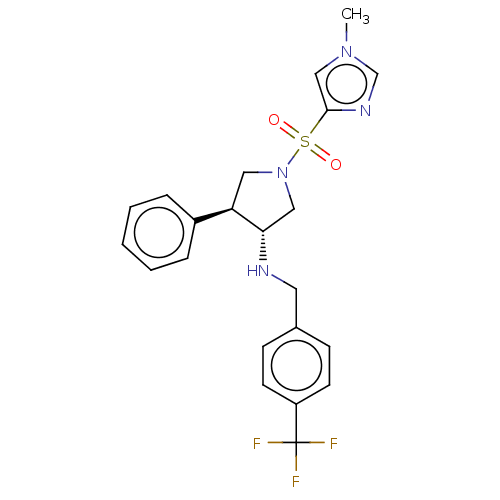 US9656955, Example 398 BDBM308884 (3R,4S)-1-[(1-methyl-1H-imidazol-4-yl)sulfonyl]-4-phenyl-N-[4-(trifluoromethyl)benzyl]pyrrolidin-3-amine
US9656955, Example 398 BDBM308884 (3R,4S)-1-[(1-methyl-1H-imidazol-4-yl)sulfonyl]-4-phenyl-N-[4-(trifluoromethyl)benzyl]pyrrolidin-3-amine 2-Ethylamino-6-fluoro-4-{6-[2-(4-methoxy-2-methyl-indol-1-yl)-ethylamino]-pyrimidin- 4-yl}-benzoic acid BDBM680183 US12011444, Example 398
2-Ethylamino-6-fluoro-4-{6-[2-(4-methoxy-2-methyl-indol-1-yl)-ethylamino]-pyrimidin- 4-yl}-benzoic acid BDBM680183 US12011444, Example 398 Preparation of 1-(cyclopropylmethyl)-3-{4-[4-oxo-3-(phenylamino)-4,5,6,7-tetrahydro-1H-indol-2-yl]pyridin-2-yl}thiourea BDBM415291 US10428044, Example 398
Preparation of 1-(cyclopropylmethyl)-3-{4-[4-oxo-3-(phenylamino)-4,5,6,7-tetrahydro-1H-indol-2-yl]pyridin-2-yl}thiourea BDBM415291 US10428044, Example 398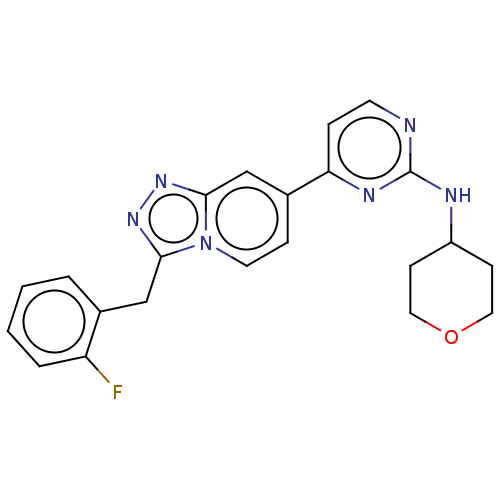 US9670208, Example 398 4-(3-(2-fluorobenzyl)-[1,2,4]triazolo[4,3-a]pyridin-7-yl)-N-(tetrahydro-2H-pyran-4-yl)pyrimidin-2-amine BDBM193772
US9670208, Example 398 4-(3-(2-fluorobenzyl)-[1,2,4]triazolo[4,3-a]pyridin-7-yl)-N-(tetrahydro-2H-pyran-4-yl)pyrimidin-2-amine BDBM193772 (R)-N-(5-(5-ethyl-1,2,4-oxadiazol-3- yl)-2,3-dihydro-1H-inden-1-yl)-3- methyl-1H-pyrazole-4-carboxamide BDBM691009 US12065436, Compound 398
(R)-N-(5-(5-ethyl-1,2,4-oxadiazol-3- yl)-2,3-dihydro-1H-inden-1-yl)-3- methyl-1H-pyrazole-4-carboxamide BDBM691009 US12065436, Compound 398 BDBM333848 1-(3-tert-butyl- 1-(4- methoxy- phenyl)- 1H-pyrazol-5- yl)- 3-(3-(6,7- dimethoxy- quinazolin- 4-yloxy) phenyl) urea US9730937, Example 398
BDBM333848 1-(3-tert-butyl- 1-(4- methoxy- phenyl)- 1H-pyrazol-5- yl)- 3-(3-(6,7- dimethoxy- quinazolin- 4-yloxy) phenyl) urea US9730937, Example 398 (1r,4r)-2′-[3-(benzenesulfonyl)propyl]-4-(3-chloroanilino)-2′,3′-dihydrospiro[cyclohexane-1,1′-indene]-4-carboxylic acid BDBM673252 US20240150293, Example 398
(1r,4r)-2′-[3-(benzenesulfonyl)propyl]-4-(3-chloroanilino)-2′,3′-dihydrospiro[cyclohexane-1,1′-indene]-4-carboxylic acid BDBM673252 US20240150293, Example 398 3-{[(2S)-2- (methoxymethyl) pyrrolidin-1-yl] carbonyl}-2- methyl-7- (piperidin-4-yl) pyrazolo[1,5-a] pyrimidin-5(4H)- one hydrochloride BDBM298195 US10118930, Example 398
3-{[(2S)-2- (methoxymethyl) pyrrolidin-1-yl] carbonyl}-2- methyl-7- (piperidin-4-yl) pyrazolo[1,5-a] pyrimidin-5(4H)- one hydrochloride BDBM298195 US10118930, Example 398 4-chloro-2-fluoro-N-(6-(4-(4- fluorophenyl)-1,2-dimethyl- 1H-imidazol-5-yl) imidazo[1,2-b]pyridazin-2-yl) benzamide US9556179, Compound 398 BDBM274821
4-chloro-2-fluoro-N-(6-(4-(4- fluorophenyl)-1,2-dimethyl- 1H-imidazol-5-yl) imidazo[1,2-b]pyridazin-2-yl) benzamide US9556179, Compound 398 BDBM274821 BDBM388248 US10294229, Example 398 1-(5-(3-(1-acetylazetidin-3-yl)isoxazol-5-yl)-4-(isopropylamino)pyridin-2-yl)-1H-pyrazolo[3,4-b]pyridine-5-carbonitrile
BDBM388248 US10294229, Example 398 1-(5-(3-(1-acetylazetidin-3-yl)isoxazol-5-yl)-4-(isopropylamino)pyridin-2-yl)-1H-pyrazolo[3,4-b]pyridine-5-carbonitrile BDBM416254 US10442819, Example 398 3-{[8-(1-methyl-1H-pyrazol- 4-yl)-5,11-dihydro-6H- pyrido[2,3- b][1,5]benzodiazepin-6- yl]carbonyl}cyclobutanone O-ethyloxime
BDBM416254 US10442819, Example 398 3-{[8-(1-methyl-1H-pyrazol- 4-yl)-5,11-dihydro-6H- pyrido[2,3- b][1,5]benzodiazepin-6- yl]carbonyl}cyclobutanone O-ethyloxime BDBM421282 5-(8-Amino-6-(trifluoromethyl)imidazo[1,2- a]pyrazin-3-yl)-2-fluoro-N-(4- (hydroxymethyl)bicyclo[2.1.1]hexan-1- yl)benzenesulfonamide US10479795, Example 398
BDBM421282 5-(8-Amino-6-(trifluoromethyl)imidazo[1,2- a]pyrazin-3-yl)-2-fluoro-N-(4- (hydroxymethyl)bicyclo[2.1.1]hexan-1- yl)benzenesulfonamide US10479795, Example 398 BDBM537573 US11247990, Example 398 4-(6-(ethylamino)-1- ((1s,4s)-4-(2- fluorophenoxy)cyclohexyl)- 1H-pyrazolo[4,3-c]pyridin- 3-yl)-1-methylpyridin- 2(1H)-one
BDBM537573 US11247990, Example 398 4-(6-(ethylamino)-1- ((1s,4s)-4-(2- fluorophenoxy)cyclohexyl)- 1H-pyrazolo[4,3-c]pyridin- 3-yl)-1-methylpyridin- 2(1H)-one US11964973, Example 398 trans-8-(4-((5-Isopropoxypyridin-2-yl)oxy)-3-ethoxypiperidin-1-yl)-5-methyl-6-oxo-5,6-dihydro-1,5-naphthyridine-2-carbonitrile BDBM669400
US11964973, Example 398 trans-8-(4-((5-Isopropoxypyridin-2-yl)oxy)-3-ethoxypiperidin-1-yl)-5-methyl-6-oxo-5,6-dihydro-1,5-naphthyridine-2-carbonitrile BDBM669400 (2S)-N-[[2-(dimethylamino)-6- [2-hydroxy-4- (trifluoromethyl)phenyl]- pyrimidin-4-yl]methyl]-1-(5- fluorobenzofuran-2- yl)sulfonyl-azetidine-2- carboxamide BDBM611504 US10626112, Example 398
(2S)-N-[[2-(dimethylamino)-6- [2-hydroxy-4- (trifluoromethyl)phenyl]- pyrimidin-4-yl]methyl]-1-(5- fluorobenzofuran-2- yl)sulfonyl-azetidine-2- carboxamide BDBM611504 US10626112, Example 398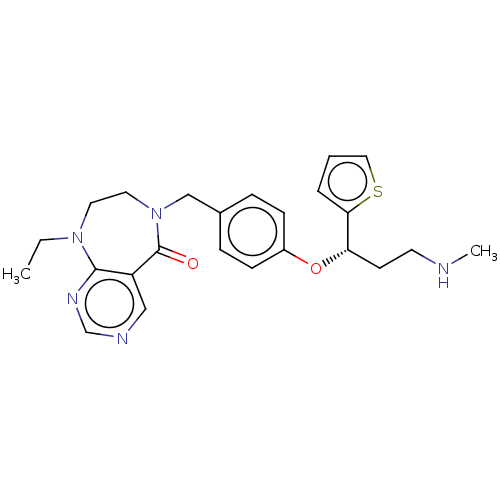 BDBM436622 (S)-9-ethyl-6-(4-(3- (methylamino)-1- (thiophen-2- yl)propoxy)benzyl)- 6,7,8,9-tetrahydro- 5H-pyrimido[4,5- e][1,4]diazepin-5-one US10590140, Example 398
BDBM436622 (S)-9-ethyl-6-(4-(3- (methylamino)-1- (thiophen-2- yl)propoxy)benzyl)- 6,7,8,9-tetrahydro- 5H-pyrimido[4,5- e][1,4]diazepin-5-one US10590140, Example 398 BDBM664315 N-(5-(2-(3,3-dimethylazetidin-1- yl)acetamido)-2-methylpyridin- 3-yl)-2-(4-methylpyrimidin-5- yl)pyrazolo[5,1-b]thiazole-7- carboxamide US20240109917, Example 398
BDBM664315 N-(5-(2-(3,3-dimethylazetidin-1- yl)acetamido)-2-methylpyridin- 3-yl)-2-(4-methylpyrimidin-5- yl)pyrazolo[5,1-b]thiazole-7- carboxamide US20240109917, Example 398 BDBM742875 (3S,4R)-4-((7-(4-(1- (difluoromethyl)cyclopropyl)phenyl)- 5-fluoropyrrolo[2,1-f][1,2,4]triazin-2- yl)amino)tetrahydro-2H-pyran-3-ol US20250163063, Example 398
BDBM742875 (3S,4R)-4-((7-(4-(1- (difluoromethyl)cyclopropyl)phenyl)- 5-fluoropyrrolo[2,1-f][1,2,4]triazin-2- yl)amino)tetrahydro-2H-pyran-3-ol US20250163063, Example 398 (3-fluoro-2-(5-fluoropyrimidin-2-yl)phenyl)((1S,2R,4R)-2-((5-(trifluoromethyl)pyrazin-2-yl)amino)-7-azabicyclo[2.2.1]heptan-7-yl)methanone US9637496, 398 BDBM326681
(3-fluoro-2-(5-fluoropyrimidin-2-yl)phenyl)((1S,2R,4R)-2-((5-(trifluoromethyl)pyrazin-2-yl)amino)-7-azabicyclo[2.2.1]heptan-7-yl)methanone US9637496, 398 BDBM326681 (5-Cyclopropyl-1-methyl-1H-pyrazol-4-yl)((5R,8S)-2-methyl-3- (2,3,4-trifluorophenyl)-2,4,5,6,7,8-hexahydro-5,8- epiminocyclohepta[c]pyrazol-9-yl)methanone; US11597728, Example 398 BDBM597510
(5-Cyclopropyl-1-methyl-1H-pyrazol-4-yl)((5R,8S)-2-methyl-3- (2,3,4-trifluorophenyl)-2,4,5,6,7,8-hexahydro-5,8- epiminocyclohepta[c]pyrazol-9-yl)methanone; US11597728, Example 398 BDBM597510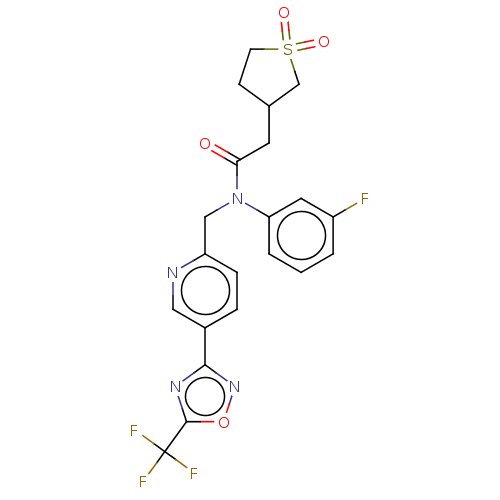 2-(1,1- dioxido- tetrahydro- thiophen- 3-yl)-N- (3-fluoro- phenyl)-N- ({5-[5- (trifluoro- methyl)- 1,2,4-oxa- diazol-3- yl]pyridin- 2-yl} US11066396, Example 398 BDBM505000
2-(1,1- dioxido- tetrahydro- thiophen- 3-yl)-N- (3-fluoro- phenyl)-N- ({5-[5- (trifluoro- methyl)- 1,2,4-oxa- diazol-3- yl]pyridin- 2-yl} US11066396, Example 398 BDBM505000 4-chloro-5-({4-[4-(5-fluoro-2-methoxyphenyl)-1H-pyrrolo[2,3-b]pyridin-2-yl]piperidin-1-yl}methyl)-1,3-thiazol-2-amine US9796708, Example 398 BDBM353166
4-chloro-5-({4-[4-(5-fluoro-2-methoxyphenyl)-1H-pyrrolo[2,3-b]pyridin-2-yl]piperidin-1-yl}methyl)-1,3-thiazol-2-amine US9796708, Example 398 BDBM353166 BDBM659961 US20240092784, Example 398 N-Cyclopropyl-7-hydroxy-4-neopentyl-5-oxo-1-(2,2,2-trifluoroethyl)-2,3,4,5-tetrahydro-1H-pyrrolo[2',3':3,4]pyrazolo[1,5-a]pyrimidine-6-carboxamide
BDBM659961 US20240092784, Example 398 N-Cyclopropyl-7-hydroxy-4-neopentyl-5-oxo-1-(2,2,2-trifluoroethyl)-2,3,4,5-tetrahydro-1H-pyrrolo[2',3':3,4]pyrazolo[1,5-a]pyrimidine-6-carboxamide 5-[4-(Difluoromethyl)-6-[[(1S)-1-(trifluoromethyl)propyl]amino]-3-pyridyl]-N-[(1S,3S)-3-hydroxycyclopentyl]-4-[(2S)-2-methylpyrrolidine-1-carbonyl]thiazole-2-carboxamide BDBM492257 US10975068, Example 398
5-[4-(Difluoromethyl)-6-[[(1S)-1-(trifluoromethyl)propyl]amino]-3-pyridyl]-N-[(1S,3S)-3-hydroxycyclopentyl]-4-[(2S)-2-methylpyrrolidine-1-carbonyl]thiazole-2-carboxamide BDBM492257 US10975068, Example 398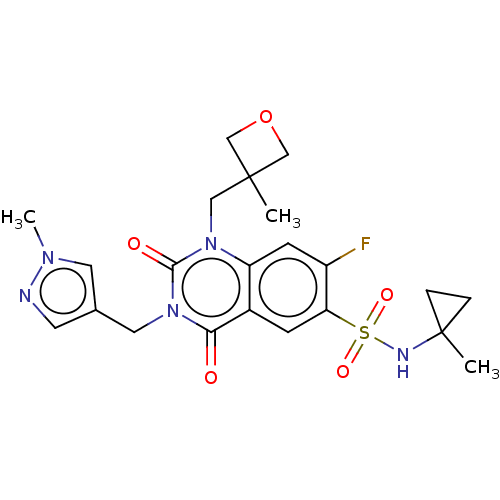 BDBM371291 7-Fluoro-3-((1-methyl-1H-pyrazol-4-yl)methyl)-N-(1-methylcyclopropyl)-1-((3-methyloxetan-3-yl)methyl)-2,4-dioxo-1,2,3,4-tetrahydroquinazoline-6-sulfonamide US10239843, Example 398
BDBM371291 7-Fluoro-3-((1-methyl-1H-pyrazol-4-yl)methyl)-N-(1-methylcyclopropyl)-1-((3-methyloxetan-3-yl)methyl)-2,4-dioxo-1,2,3,4-tetrahydroquinazoline-6-sulfonamide US10239843, Example 398 BDBM531765 6-(4-chloro-3,5-dimethyl-1H-pyrazol-1-yl)-N-{1-(cyclopropylmethyl)-3-[4-(difluoromethoxy)phenyl]-4-methyl-1H-pyrazol-5-yl}pyrimidin-4-amine US11208400, Example 398
BDBM531765 6-(4-chloro-3,5-dimethyl-1H-pyrazol-1-yl)-N-{1-(cyclopropylmethyl)-3-[4-(difluoromethoxy)phenyl]-4-methyl-1H-pyrazol-5-yl}pyrimidin-4-amine US11208400, Example 398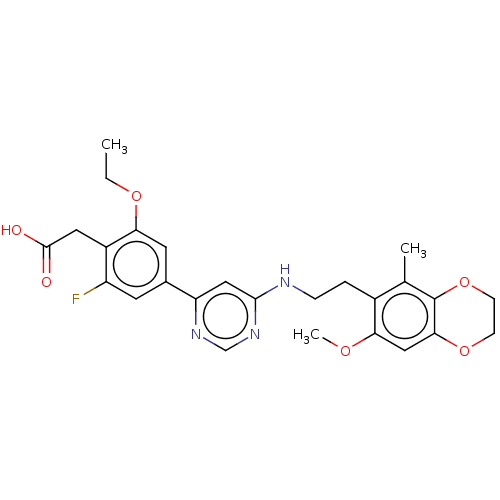 BDBM640275 (2-Ethoxy-6-fluoro-4-{6-[2-(7-methoxy-5-methyl-2,3-dihydro-benzo[1,4]dioxin-6- yl)-ethylamino]-pyrimidin-4-yl}-phenyl)-acetic acid US11839613, Example 398
BDBM640275 (2-Ethoxy-6-fluoro-4-{6-[2-(7-methoxy-5-methyl-2,3-dihydro-benzo[1,4]dioxin-6- yl)-ethylamino]-pyrimidin-4-yl}-phenyl)-acetic acid US11839613, Example 398 BDBM669399 US11964973, Example 398 trans-8-(4-((5-Isopropoxypyridin-2-yl)oxy)-3-ethoxypiperidin-1-yl)-5-methyl-6-oxo-5,6-dihydro-1,5-naphthyridine-2-carbonitrile US11964973, Example 397
BDBM669399 US11964973, Example 398 trans-8-(4-((5-Isopropoxypyridin-2-yl)oxy)-3-ethoxypiperidin-1-yl)-5-methyl-6-oxo-5,6-dihydro-1,5-naphthyridine-2-carbonitrile US11964973, Example 397 BDBM678956 1-methyl-4-[4-(5-methyl-1,3-benzoxazol-2-yl)piperidin-1-yl]-7-[methyl(oxetan-3-yl)amino]-2-oxo-1,2-dihydroquinoline-3-carbonitrile US11998539, Example 398
BDBM678956 1-methyl-4-[4-(5-methyl-1,3-benzoxazol-2-yl)piperidin-1-yl]-7-[methyl(oxetan-3-yl)amino]-2-oxo-1,2-dihydroquinoline-3-carbonitrile US11998539, Example 398 US10703755, Example 398 BDBM450198 9-({1-[(3S)-1-Azabicyclo[2.2.2]oct-3-ylmethyl]-1H-pyrazol-4-yl}methyl)-2-ethoxy-8-(3-fluoropyridin-3-yl)-6-methyl-9H-purine
US10703755, Example 398 BDBM450198 9-({1-[(3S)-1-Azabicyclo[2.2.2]oct-3-ylmethyl]-1H-pyrazol-4-yl}methyl)-2-ethoxy-8-(3-fluoropyridin-3-yl)-6-methyl-9H-purine US20250230171, Compound 398 2'-chloro-N-(5-(5-(difluoromethyl)-2- fluorobenzoyl)-5,6-dihydro-4H- pyrrolo[3,4-d]thiazol-2-yl)-5'-methoxy- 6-methyl-[4,4'-bipyridine]-3- carboxamide BDBM759049
US20250230171, Compound 398 2'-chloro-N-(5-(5-(difluoromethyl)-2- fluorobenzoyl)-5,6-dihydro-4H- pyrrolo[3,4-d]thiazol-2-yl)-5'-methoxy- 6-methyl-[4,4'-bipyridine]-3- carboxamide BDBM759049 BDBM315975 US10172859, Example 398 US11065253, Example 398 US9732094, Example 398 1-Ethyl-6-{[4-fluoro- 3-(7-morpholin-4-yl- quinazolin-4-yl)- phenyl]hydroxy- methyl}-1H-pyridin-2- one1H NMR (500 MHz, DMSO-d6) ppm = 9.10 (s,1H), 7.63-7.56 (m, 2H), 7.54 (dd, J = 9.4, 2.5, 1H),7.52-7.43 (m, 2H), 7.41 (dd, J = 9.1, 6.9, 1H),7.20 (d, J = 2.4, 1H), 6.58 (d, J = 1H), 6.35 (dd,J = 9.1, 1.4, 1H), 6.31 (dd, J = 7.1, 1.4, 1H), 5.91 (d,J = 5.0, 1H), 4.05-3.88 (m, 2H), 3.81-3.74 (m,4H), 3.48-3.39 (m, 4H), 0.95 (t, J = 6.9, 3H).
BDBM315975 US10172859, Example 398 US11065253, Example 398 US9732094, Example 398 1-Ethyl-6-{[4-fluoro- 3-(7-morpholin-4-yl- quinazolin-4-yl)- phenyl]hydroxy- methyl}-1H-pyridin-2- one1H NMR (500 MHz, DMSO-d6) ppm = 9.10 (s,1H), 7.63-7.56 (m, 2H), 7.54 (dd, J = 9.4, 2.5, 1H),7.52-7.43 (m, 2H), 7.41 (dd, J = 9.1, 6.9, 1H),7.20 (d, J = 2.4, 1H), 6.58 (d, J = 1H), 6.35 (dd,J = 9.1, 1.4, 1H), 6.31 (dd, J = 7.1, 1.4, 1H), 5.91 (d,J = 5.0, 1H), 4.05-3.88 (m, 2H), 3.81-3.74 (m,4H), 3.48-3.39 (m, 4H), 0.95 (t, J = 6.9, 3H). (S)-6-(azetidin-1-ylmethyl)-2-(3-(3,3- difluoro-1-(fluoro(4-methyl-4H-1,2,4- triazol-3-yl)methyl)cyclobutyl)phenyl)-4- (trifluoromethyl)isoindolin-1-one US20250230147, Compound 398 BDBM758217
(S)-6-(azetidin-1-ylmethyl)-2-(3-(3,3- difluoro-1-(fluoro(4-methyl-4H-1,2,4- triazol-3-yl)methyl)cyclobutyl)phenyl)-4- (trifluoromethyl)isoindolin-1-one US20250230147, Compound 398 BDBM758217 BDBM407456 US10336762, Compound 398 (R)-4-((4- carbamoylbicyclo[2.2.2] octan-1-yl)amino)-6-(3- cyanopyrrolo[1,2- b]pyridazin-7-yl)-N-(2- fluoro-3-hydroxy-3-methyl- butyl)nicotinamide
BDBM407456 US10336762, Compound 398 (R)-4-((4- carbamoylbicyclo[2.2.2] octan-1-yl)amino)-6-(3- cyanopyrrolo[1,2- b]pyridazin-7-yl)-N-(2- fluoro-3-hydroxy-3-methyl- butyl)nicotinamide N-[(6-amino-2,4-dimethyl- pyridin-3-yl)methyl]-6- fluoro-11- (4-fluorophenyl)-13-methyl-15- oxatetracyclo[6.6.1.02,7.09,14] pentadeca-2,4,6,9(14),10,12- hexaene-4-carboxamide US10144746, Compound 398 BDBM306706
N-[(6-amino-2,4-dimethyl- pyridin-3-yl)methyl]-6- fluoro-11- (4-fluorophenyl)-13-methyl-15- oxatetracyclo[6.6.1.02,7.09,14] pentadeca-2,4,6,9(14),10,12- hexaene-4-carboxamide US10144746, Compound 398 BDBM306706 US11648254, Compound 398 BDBM602506 (R)-1-(4-((1-(3-(difluoromethyl)-2- fluorophenyl)ethyl)amino)-2-methyl-7- ((oxetan-3- ylmethyl)amino)pyrido[2,3-d]pyrimidin- 6-yl)cyclopropane-1-carbonitrile
US11648254, Compound 398 BDBM602506 (R)-1-(4-((1-(3-(difluoromethyl)-2- fluorophenyl)ethyl)amino)-2-methyl-7- ((oxetan-3- ylmethyl)amino)pyrido[2,3-d]pyrimidin- 6-yl)cyclopropane-1-carbonitrile BDBM282518 (S)-3-(4-(4-amino-7-(2- (dimethylamino)ethyl)-7H- pyrrolo[2,3-d]pyrimidin- 5-yl)benzylamino)-6-cyano- N-(1-(3,4-difluorophenyl) ethyl)pyrazine-2-carboxamide US10030016, Example 398
BDBM282518 (S)-3-(4-(4-amino-7-(2- (dimethylamino)ethyl)-7H- pyrrolo[2,3-d]pyrimidin- 5-yl)benzylamino)-6-cyano- N-(1-(3,4-difluorophenyl) ethyl)pyrazine-2-carboxamide US10030016, Example 398 BDBM422494 US10508104, Example 398 4-[4-({1-[(1R)-1-(3,5- difluorophenyl)-2- methoxyethyl]-6-hydroxy- 4-oxo-2-[(propan-2- yIoxy)methyl]-1,4- dihydropyrimidin-5- yl}sulfonyl)phenyl] pyridine-3-carbonitrile
BDBM422494 US10508104, Example 398 4-[4-({1-[(1R)-1-(3,5- difluorophenyl)-2- methoxyethyl]-6-hydroxy- 4-oxo-2-[(propan-2- yIoxy)methyl]-1,4- dihydropyrimidin-5- yl}sulfonyl)phenyl] pyridine-3-carbonitrile BDBM711039 2-(3-((S)-(4-methyl-4H-1,2,4-triazol-3- yl)((1r,3S)-3-(trifluoromethoxy)- cyclobutyl)methyl)phenyl)-6-(((1- methylcyclobutyl)amino)methyl)-4- (trifluoromethyl)isoindolin-1-one US12187709, Compound 398
BDBM711039 2-(3-((S)-(4-methyl-4H-1,2,4-triazol-3- yl)((1r,3S)-3-(trifluoromethoxy)- cyclobutyl)methyl)phenyl)-6-(((1- methylcyclobutyl)amino)methyl)-4- (trifluoromethyl)isoindolin-1-one US12187709, Compound 398 US10954214, Compound 398 N-(5-chloro-6-(2H-1,2,3-triazol- 2-yl)pyridin-3-yl)-1-(7- (difluoromethyl)thieno[2,3- c]pyridin-4-yl)-5- (trifluoromethyl)-1H-pyrazole-4- carboxamide BDBM487976
US10954214, Compound 398 N-(5-chloro-6-(2H-1,2,3-triazol- 2-yl)pyridin-3-yl)-1-(7- (difluoromethyl)thieno[2,3- c]pyridin-4-yl)-5- (trifluoromethyl)-1H-pyrazole-4- carboxamide BDBM487976 US9718828, Example, 398 BDBM267792 4-{8-amino-3-[(3R,6S)-6-methyl-1-propanoylpiperidin-3-yl]imidazo[1,5-a]pyrazin-1-yl}-3-fluoro-N-[4-(trifluoromethyl)pyridin-2-yl]benzamide
US9718828, Example, 398 BDBM267792 4-{8-amino-3-[(3R,6S)-6-methyl-1-propanoylpiperidin-3-yl]imidazo[1,5-a]pyrazin-1-yl}-3-fluoro-N-[4-(trifluoromethyl)pyridin-2-yl]benzamide 7-[[5-(4-hydroxy-1- piperidyl)-2- pyridyl]amino]-4- imidazo[1,2- a]pyrazin-3- yl-isoindolin-1-one US11078201, Compound I-398 US11021481, Compound I-133 BDBM500474 US11548890, Compound I-133
7-[[5-(4-hydroxy-1- piperidyl)-2- pyridyl]amino]-4- imidazo[1,2- a]pyrazin-3- yl-isoindolin-1-one US11078201, Compound I-398 US11021481, Compound I-133 BDBM500474 US11548890, Compound I-133 BDBM599256 5-[4-amino-5-(trifluoromethyl) pyrrolo[2,1-f][1,2,4]triazin-7-yl]- 2-chloro-4-fluoro-N-[(3R,4S)-4- fluoro-1-(3-methylbutanoyl) pyrrolidin-3-yl]benzamide US11618753, Example 398
BDBM599256 5-[4-amino-5-(trifluoromethyl) pyrrolo[2,1-f][1,2,4]triazin-7-yl]- 2-chloro-4-fluoro-N-[(3R,4S)-4- fluoro-1-(3-methylbutanoyl) pyrrolidin-3-yl]benzamide US11618753, Example 398 N-[(1R,2S)-2- hydroxy- cyclopentyl]- 2-{[(2R)-1- methoxypropan-2- yl]oxy}-6-[4-(1H- pyrazolo[3,4- b]pyridin-3- yl)piperidin-1- yl]pyrimidine-4- carboxamide US9593097, Example 398 BDBM300018
N-[(1R,2S)-2- hydroxy- cyclopentyl]- 2-{[(2R)-1- methoxypropan-2- yl]oxy}-6-[4-(1H- pyrazolo[3,4- b]pyridin-3- yl)piperidin-1- yl]pyrimidine-4- carboxamide US9593097, Example 398 BDBM300018 1-[4-[2-[4-[4- (cyclopropylmethyl) piperazine-1- carbonyl]anilino]- [1,2,4]triazolo[1,5- a]pyridin-8-yl]-3,6- dihydro-2H-pyridin-1- yl]-4,4,4-trifluoro- butan-1-one US9873709, Example 2-398 BDBM367099
1-[4-[2-[4-[4- (cyclopropylmethyl) piperazine-1- carbonyl]anilino]- [1,2,4]triazolo[1,5- a]pyridin-8-yl]-3,6- dihydro-2H-pyridin-1- yl]-4,4,4-trifluoro- butan-1-one US9873709, Example 2-398 BDBM367099 BDBM297273 US10112941, Example 398 US10112941, Example 399 2-{5-[(S)-(4-Chlorophenyl)(oxan-4-yl)methyl]-3-(dimethyl-1H-1,2,3-triazol-5-yl)-5H-pyrido[3,2-b]indol-7-yl}propan-2-ol
BDBM297273 US10112941, Example 398 US10112941, Example 399 2-{5-[(S)-(4-Chlorophenyl)(oxan-4-yl)methyl]-3-(dimethyl-1H-1,2,3-triazol-5-yl)-5H-pyrido[3,2-b]indol-7-yl}propan-2-ol 5-(1-(tetrahydro-2H-pyran-4-yl)-1H- pyrazol-5-yl)-7-(3-(1-(((tetrahydro-2H- pyran-4-yl)amino)methyl)cyclopropyl) phenyl)pyrrolo[2,1-f][1,2,4]triazin-4- amine US10214537, Example 398 BDBM358329
5-(1-(tetrahydro-2H-pyran-4-yl)-1H- pyrazol-5-yl)-7-(3-(1-(((tetrahydro-2H- pyran-4-yl)amino)methyl)cyclopropyl) phenyl)pyrrolo[2,1-f][1,2,4]triazin-4- amine US10214537, Example 398 BDBM358329 7-((5-(4- hydroxypipen din-1-yl)pyridin-2- yl)amino)-4- (imidazo[1,5- a]pyrazin-3- yl)isoindolin- 1-one BDBM500736 US11078201, Compound I-125 US11021481, Compound I-398 US11548890, Compound I-125
7-((5-(4- hydroxypipen din-1-yl)pyridin-2- yl)amino)-4- (imidazo[1,5- a]pyrazin-3- yl)isoindolin- 1-one BDBM500736 US11078201, Compound I-125 US11021481, Compound I-398 US11548890, Compound I-125 BDBM498573 US11718603, Example 399 US11014911, Example 398 US11014911, Example 399 (1R,3S)-3-(3-{[(6- methylpyridin-3- yl)acetyl]amino}-1H-pyrazol-5- yl)cyclopentyl [(2$#958;)-1,1,1- trifluorobutan-2-yl]carbamate - Isomer A
BDBM498573 US11718603, Example 399 US11014911, Example 398 US11014911, Example 399 (1R,3S)-3-(3-{[(6- methylpyridin-3- yl)acetyl]amino}-1H-pyrazol-5- yl)cyclopentyl [(2$#958;)-1,1,1- trifluorobutan-2-yl]carbamate - Isomer A BDBM553324 US11319299, Example 398 4-(difluoromethyl)-N-[4- fluoro-2-[rac-(3R)-3,4- dimethylpiperazin-1-yl]- 5-[2-[rac-(2R)-2- methylmorpholin-4- yl]pyrimidin-5- yl]phenyl]-6-oxo-1H- pyridine-3-carboxamide
BDBM553324 US11319299, Example 398 4-(difluoromethyl)-N-[4- fluoro-2-[rac-(3R)-3,4- dimethylpiperazin-1-yl]- 5-[2-[rac-(2R)-2- methylmorpholin-4- yl]pyrimidin-5- yl]phenyl]-6-oxo-1H- pyridine-3-carboxamide BDBM569753 US11427567, Example 398 4-(1-(2-Chloro-4-((4- ethylpiperazin-1- yl)methyl)phenyl)-1H- imidazol-4-yl)-N- ((3R,4R)-3-fluoro-1- (methylsulfonyl)piperidin- 4-yl)-5- (trifluoromethyl)pyrimidin- 2-amine
BDBM569753 US11427567, Example 398 4-(1-(2-Chloro-4-((4- ethylpiperazin-1- yl)methyl)phenyl)-1H- imidazol-4-yl)-N- ((3R,4R)-3-fluoro-1- (methylsulfonyl)piperidin- 4-yl)-5- (trifluoromethyl)pyrimidin- 2-amine US11034692, Compound 398 BDBM504132 (1S,2S)-N-(8-amino-6-(4-methyl-6- (1H-pyrazol-1-yl)pyridin-3-yl)-2,7- naphthyridin-3-yl)-2-(1-methyl-1H- pyrazol-4-yl)cyclopropane-1- carboxamide
US11034692, Compound 398 BDBM504132 (1S,2S)-N-(8-amino-6-(4-methyl-6- (1H-pyrazol-1-yl)pyridin-3-yl)-2,7- naphthyridin-3-yl)-2-(1-methyl-1H- pyrazol-4-yl)cyclopropane-1- carboxamide 1-(3-(((S)-2,2-dimethyl-1,3- dioxolan-4-yl)methoxy)-4- methyl-1-phenyl-1H-pyrazol- 5-yl)-3-((3S,4R)-1-(2- methoxyethyl)-4-(3,4,5- trifluorophenyl)pyrrolidin-3- yl)urea US10323022, Example 398 BDBM398921
1-(3-(((S)-2,2-dimethyl-1,3- dioxolan-4-yl)methoxy)-4- methyl-1-phenyl-1H-pyrazol- 5-yl)-3-((3S,4R)-1-(2- methoxyethyl)-4-(3,4,5- trifluorophenyl)pyrrolidin-3- yl)urea US10323022, Example 398 BDBM398921 methyl rac-((1S,2R)-2- (cyano(1-((3-methyl-1-(4- ((1-methyl-1H-pyrazol-4- yl)sulfonyl)phenyl)azetidin- 3-yl)methyl)piperidin-4- yl)(phenyl)methyl)cyclopentyl) carbamate BDBM480333 US10899738, Cpd. No 398
methyl rac-((1S,2R)-2- (cyano(1-((3-methyl-1-(4- ((1-methyl-1H-pyrazol-4- yl)sulfonyl)phenyl)azetidin- 3-yl)methyl)piperidin-4- yl)(phenyl)methyl)cyclopentyl) carbamate BDBM480333 US10899738, Cpd. No 398 ((1S,6R,7R)-3-(6-((3-chloro-2-(1H-pyrazol-1- yl)pyridin-4-yl)thio)pyrido[2,3-b]pyrazin-2-yl)-7-(2- fluorophenyl)-3-azabicyclo[4.1.0]heptan-7- yl)methanamine US20240109900, Example 398 BDBM663560
((1S,6R,7R)-3-(6-((3-chloro-2-(1H-pyrazol-1- yl)pyridin-4-yl)thio)pyrido[2,3-b]pyrazin-2-yl)-7-(2- fluorophenyl)-3-azabicyclo[4.1.0]heptan-7- yl)methanamine US20240109900, Example 398 BDBM663560 US11034692, Compound 398 BDBM504078 US11034692, Compound 345 trans-N-(8-amino-6-(4-methyl-6-(1H- pyrazol-1-yl)pyridin-3-yl)-2,7- naphthyridin-3-yl)-2-(1-methyl-1H- pyrazol-4-yl)cyclopropane-1-carboxamide
US11034692, Compound 398 BDBM504078 US11034692, Compound 345 trans-N-(8-amino-6-(4-methyl-6-(1H- pyrazol-1-yl)pyridin-3-yl)-2,7- naphthyridin-3-yl)-2-(1-methyl-1H- pyrazol-4-yl)cyclopropane-1-carboxamide BDBM467740 US10800792, Example 398 (R,E)-N-(1-(2-Cyano-4,4-dimethylpent-2-enoyl)piperidin-3-yl)-5- (*R)-(2-methyl-4-phenoxyphenyl)-4-oxo-4,5-dihydro-3H-1-thia- 3,5,8-triazaacenaphthylene-2-carboxamide; US10822348, Example 880
BDBM467740 US10800792, Example 398 (R,E)-N-(1-(2-Cyano-4,4-dimethylpent-2-enoyl)piperidin-3-yl)-5- (*R)-(2-methyl-4-phenoxyphenyl)-4-oxo-4,5-dihydro-3H-1-thia- 3,5,8-triazaacenaphthylene-2-carboxamide; US10822348, Example 880 BDBM505753 tert-butyl 4-(4-(((8-chloro-4-((3- chloro-4-fluorophenyl)amino)-3- cyanoquinolin-6-yl)amino)(1- methyl-1H-1,2,3-triazol-4- yl)methyl)-1H-1,2,3-triazol-1- yl)piperidine-1-carboxyl ate US11066414, Compound 398
BDBM505753 tert-butyl 4-(4-(((8-chloro-4-((3- chloro-4-fluorophenyl)amino)-3- cyanoquinolin-6-yl)amino)(1- methyl-1H-1,2,3-triazol-4- yl)methyl)-1H-1,2,3-triazol-1- yl)piperidine-1-carboxyl ate US11066414, Compound 398 BDBM583506 2-{5-[(1R,4R,7R)-7-amino-2- azabicyclo[2.2.1]heptane-2- carbonyl]-7-methoxy-1-methyl- 1H-1,3-benzodiazol-2-yl}-1- (cyclopropylmethyl)-N-[(1,1- dioxidotetrahydrothiophen-3- yl)methyl]-1H-indole-6- carboxamide US11524959, Compound 398.
BDBM583506 2-{5-[(1R,4R,7R)-7-amino-2- azabicyclo[2.2.1]heptane-2- carbonyl]-7-methoxy-1-methyl- 1H-1,3-benzodiazol-2-yl}-1- (cyclopropylmethyl)-N-[(1,1- dioxidotetrahydrothiophen-3- yl)methyl]-1H-indole-6- carboxamide US11524959, Compound 398. (3S,6S,9S,12S,18S,27S,30S,34S)-6-benzyl-18-[(4-chlorophenyl)methyl]-3,9-diisobutyl-4,7,16,19,22,25,30,31-octamethyl-27-[(1S)-1-methylpropyl]-12-[2-(p-tolyl)ethyl]-34-(pyrrolidine-1-carbonyl)-1,4,7,10,13,16,19,22,25,28,31-undecazacyclotetratriacontane-2,5,8,11,14,17,20,23,26,29,32-undecone US20240148821, Compound 398 BDBM671367
(3S,6S,9S,12S,18S,27S,30S,34S)-6-benzyl-18-[(4-chlorophenyl)methyl]-3,9-diisobutyl-4,7,16,19,22,25,30,31-octamethyl-27-[(1S)-1-methylpropyl]-12-[2-(p-tolyl)ethyl]-34-(pyrrolidine-1-carbonyl)-1,4,7,10,13,16,19,22,25,28,31-undecazacyclotetratriacontane-2,5,8,11,14,17,20,23,26,29,32-undecone US20240148821, Compound 398 BDBM671367 (S)-3-((1R,3R)-1-(2,6- difluoro-4-((1-(3- fluoropropyl)azetidin-3- yl)amino)phenyl)-5-fluoro-3- methyl-3,4-dihydro-1H- pyrido[3,4-b]indol-2(9H)-yl)- 2-fluoro-2-methylpropan-1-ol BDBM732079 US20250114338, Example 398
(S)-3-((1R,3R)-1-(2,6- difluoro-4-((1-(3- fluoropropyl)azetidin-3- yl)amino)phenyl)-5-fluoro-3- methyl-3,4-dihydro-1H- pyrido[3,4-b]indol-2(9H)-yl)- 2-fluoro-2-methylpropan-1-ol BDBM732079 US20250114338, Example 398 US12310975, Example 398 2-{[6-({5-chloro-2-[4- ({1-[2-(2,6- dioxopiperidin-3-yl)-1,3- dioxo-2,3-dihydro-1H- isoindol-4-yl]pyrrolidin- 2-yl}methyl)piperazin-1- yl]pyrimidin-4- yl}amino)-2-oxo-1- (propan-2-yl)-1,2- dihydroquinolin-3- yl]oxy}-N- methylacetamide BDBM743856
US12310975, Example 398 2-{[6-({5-chloro-2-[4- ({1-[2-(2,6- dioxopiperidin-3-yl)-1,3- dioxo-2,3-dihydro-1H- isoindol-4-yl]pyrrolidin- 2-yl}methyl)piperazin-1- yl]pyrimidin-4- yl}amino)-2-oxo-1- (propan-2-yl)-1,2- dihydroquinolin-3- yl]oxy}-N- methylacetamide BDBM743856 BDBM759801 (1S,5S)—N-(4-(1-ethyl-3-(4-fluorophenyl)-1H-pyrazol-4-yl)-7-methoxypyrido[3,2-d]pyrimidin-6-yl)-3-methyl-3-azabicyclo[3.1.0]hexane-1-carboxamide and (1R,5R)—N-(4-(1-ethyl-3-(4-fluorophenyl)-1H-pyrazol-4-yl)-7-methoxypyrido[3,2-d]pyrimidin-6-yl)-3-methyl-3-azabicyclo[3.1.0]hexane-1-carboxamide US20250236608, Example 398
BDBM759801 (1S,5S)—N-(4-(1-ethyl-3-(4-fluorophenyl)-1H-pyrazol-4-yl)-7-methoxypyrido[3,2-d]pyrimidin-6-yl)-3-methyl-3-azabicyclo[3.1.0]hexane-1-carboxamide and (1R,5R)—N-(4-(1-ethyl-3-(4-fluorophenyl)-1H-pyrazol-4-yl)-7-methoxypyrido[3,2-d]pyrimidin-6-yl)-3-methyl-3-azabicyclo[3.1.0]hexane-1-carboxamide US20250236608, Example 398