Target (1)
Compound (590)
Article Title (3)
Article Author (3)
Assay (96)
Flossdorf, J; Pratorius, HJ; Kula, MR Influence of side-chain structure of aliphatic amino acids on binding to isoleucyl-tRNA synthetase from Escherichia coli MRE 600. Eur J Biochem 66: 147 -55 (1976) Varani, K; Merighi, S; Gessi, S; Klotz, KN; Leung, E; Baraldi, PG; Cacciari, B; Romagnoli, R; Spalluto, G; Borea, PA [(3)H]MRE 3008F20: a novel antagonist radioligand for the pharmacological and biochemical characterization of human A(3) adenosine receptors. Mol Pharmacol 57: 968 -75 (2000) Baraldi, PG; Cacciari, B; Romagnoli, R; Varani, K; Merighi, S; Gessi, S; Borea, PA; Leung, E; Hickey, SL; Spalluto, G Synthesis and preliminary biological evaluation of [3H]-MRE 3008-F20: the first high affinity radioligand antagonist for the human A3 adenosine receptors. Bioorg Med Chem Lett 10: 209 -11 (2000)
Günther, S; Reinke, PY; Fernández-García, Y; Lieske, J; Lane, TJ; Ginn, HM; Koua, FH; Ehrt, C; Ewert, W; Oberthuer, D; Yefanov, O; Meier, S; Lorenzen, K; Krichel, B; Kopicki, J; Gelisio, L; Brehm, W; Dunkel, I; Seychell, B; Gieseler, H; Norton-Baker, B; Escudero-Pérez, B; Domaracky, M; Saouane, S; Tolstikova, A; White, TA; Hänle, A; Groessler, M; Fleckenstein, H; Trost, F; Galchenkova, M; Gevorkov, Y; Li, C; Awel, S; Peck, A; Barthelmess, M; Schlünzen, F; Xavier, PL; Werner, N; Andaleeb, H; Ullah, N; Falke, S; Srinivasan, V; Franca, BA; Schwinzer, M; Brognaro, H; Rogers, C; Melo, D; Zaitsev-Doyle, JJ; Knoska, J; Peña Murillo, GE; Mashhour, AR; Guicking, F; Hennicke, V; Fischer, P; Hakanpää, J; Meyer, J; Gribbon, P; Ellinger, B; Kuzikov, M; Wolf, M; Beccari, AR; Bourenkov, G; Stetten, D; Pompidor, G; Bento, I; Panneerselvam, S; Karpics, I; Schneider, TR; Garcia Alai, MM; Niebling, S; Günther, C; Schmidt, C; Schubert, R; Han, H; Boger, J; Monteiro, DC; Zhang, L; Sun, X; Pletzer-Zelgert, J; Wollenhaupt, J; Feiler, CG; Weiss, MS; Schulz, E; Mehrabi, P; Karničar, K; Usenik, A; Loboda, J; Tidow, H; Chari, A; Hilgenfeld, R; Uetrecht, C; Cox, R; Zaliani, A; Beck, T; Rarey, M; Günther, S; Turk, D; Hinrichs, W; Chapman, HN; Pearson, AR; Betzel, C; Meents, A bioRxiv 2020: (2020) Tomašič, T; Zidar, N; Durcik, M; Ilaš, J; Zega, A; Cruz, CD; Tammela, P; Pál, C; Nyerges, null; Kikelj, D; Mašič, LP US Patent US12258342 (2025) Tomašič, T; Zidar, N; Durcik, M; Ilaš, J; Zega, A; Cruz, CD; Tammela, P; Pál, C; Nyerges, null; Kikelj, D; Mašič, LP US Patent US12258342 (2025)
ChEMBL_29886 (CHEMBL641957) Displacement of [3]-MRE-3008F20 from human Adenosine A3 receptor expressed in CHO cells ChEMBL_29887 (CHEMBL641958) Displacement of [3]-MRE-3008F20 from human Adenosine A3 receptor expressed in CHO cells ChEMBL_32012 (CHEMBL646609) Displacement of [3H]- MRE 308F20 from human Adenosine A3 receptor expressed in CHO cells ChEMBL_375256 (CHEMBL867483) Displacement of [3H]MRE 2029F20 from human adenosine A2B receptor expressed in CHO cells ChEMBL_375257 (CHEMBL867484) Displacement of [3H]MRE 3008F20 from human adenosine A3 receptor expressed in CHO cells ChEBML_31081 Binding affinity of [3H]-MRE 2029-F20 towards human adenosine A2b receptor expressed in CHO cells ChEMBL_32011 (CHEMBL646608) Displacement of [3H]- MRE 308F20 binding from human Adenosine A3 receptor expressed in CHO cells ChEMBL_32020 (CHEMBL649056) Displacement of [3H]-MRE 3008-F20 from Human Adenosine A3 receptor expressed in HEK293 cells ChEMBL_520201 (CHEMBL951844) Displacement of [3H]MRE-2029F20 from human recombinant adenosine A2B receptor expressed in HEK293 cells ChEMBL_29888 (CHEMBL641959) Displacement of [3]-MRE-3008F20 from human Adenosine A3 receptor expressed in CHO cells (95% confidence limits) ChEMBL_757185 (CHEMBL1803409) Displacement of [3H]MRE 3008F20 from human adenosine A3 receptor expressed in CHO cells after 120 mins by scintillation spectrometry ChEMBL_834863 (CHEMBL2073180) Displacement of [3H]MRE-3008-F20 from human adenosine A3 receptor expressed in CHO cells after 120 mins by scintillation counter ChEMBL_794927 (CHEMBL1936206) Displacement of [3H]-MRE-3008-F20 from human adenosine A3 receptor expressed in CHO cells after 120 mins by liquid scintillation counting ChEMBL_593382 (CHEMBL1040510) Agonist activity at TGR5 expressed in human U2-OS cells assessed as increase in MRE/CRE-driven gene expression by luciferase reporter gene assay ChEMBL_608993 (CHEMBL1073135) Agonist activity at human TGR5 receptor expressed in human U2-OS cells assessed as changes in response to cAMP level by MRE/CRE-driven luciferase reporter gene assay ChEMBL_27717 (CHEMBL875784) Displacement of [3H]-DPCPX from human Adenosine A1 receptor expressed in CHO cells; range 269-326 ChEMBL_28053 (CHEMBL643046) Displacement of [3H]DPCPX from human adenosine A1 receptor expressed in CHO cells; range 269-326 ChEMBL_855309 (CHEMBL2161793) Agonist activity at rat GLP1R expressed in HEK293 cells assessed as stimulation of cAMP levels incubated for 6 hrs by multiple response element/cAMP response element (MRE/CRE)-driven reporter gene assay ChEMBL_2031278 (CHEMBL4685436) Agonist activity at human H3 receptor expressed in CHO cells assessed as increase in cAMP accumulation by measuring reduction in forskolin level incubated for 4 hrs by CRE/MRE-luciferase reporter gene assay ChEMBL_2216210 (CHEMBL5129342) Binding affinity to human MMP-1 catalytic domain (100 to 269 residues) expressed in Escherichia coli BL21 Star (DE3) by SPR analysis ChEMBL_2353358 Inhibition of human SIRT5 (34 to 269 residues) using Ac-Leu-Gly-Ser-Lys (Su)-AMC as fluorogenic substrate in presence of NAD+ ChEMBL_2121797 (CHEMBL4830944) Inhibition of human recombinant SIRT5 (34 to 269 residues) using Ac-Leu-Gly-Ser-Lys(Su)-AMC as substrate in presence of NAD+ by fluorescence based analysis ChEMBL_1704679 (CHEMBL4055912) Inhibition of human recombinant MMP-8 catalytic domain (99 to 269 residues) expressed in Escherichia coli using fluorogenic Mca-KPLGL-Dpa-AR-NH2 as substrate by fluorescence spectrophotometric analysis ChEBML_1687482 Inhibition of recombinant human N-terminal His-tagged SIRT5 (34 to 269 residues) expressed in Escherichia coli Transetta(DE3) cells using Benzyl Lys(Succinyl)-AMC substrate after 2 hrs by fluorescence assay ChEMBL_2360798 Inhibition of human recombinant SIRT5 (34 to 269 residues) using Ac-Leu-Gly-Ser-Lys (Su)-AMC as fluorogenic substrate incubated for 2 hrs in presence of NAD+ by fluorescence based analysis ChEMBL_1687482 (CHEMBL4037961) Inhibition of recombinant human N-terminal His-tagged SIRT5 (34 to 269 residues) expressed in Escherichia coli Transetta(DE3) cells using Benzyl Lys(Succinyl)-AMC substrate after 2 hrs by fluorescence assay ChEMBL_2360805 Competitive inhibition of human recombinant SIRT5 (34 to 269 residues) incubated for 2 hrs in presence of 11 uM of fluorogenic substrate Ac-Leu-Gly-Ser-Lys (Su)-AMC by fluorescence based analysis ChEMBL_2360806 Competitive inhibition of human recombinant SIRT5 (34 to 269 residues) incubated for 2 hrs in presence of 33 uM of fluorogenic substrate Ac-Leu-Gly-Ser-Lys (Su)-AMC by fluorescence based analysis ChEMBL_2360807 Competitive inhibition of human recombinant SIRT5 (34 to 269 residues) incubated for 2 hrs in presence of 100 uM of fluorogenic substrate Ac-Leu-Gly-Ser-Lys (Su)-AMC by fluorescence based analysis ChEMBL_2360808 Competitive inhibition of human recombinant SIRT5 (34 to 269 residues) incubated for 2 hrs in presence of 300 uM of fluorogenic substrate Ac-Leu-Gly-Ser-Lys (Su)-AMC by fluorescence based analysis ChEMBL_2121798 (CHEMBL4830945) Inhibition of human recombinant SIRT5 (34 to 269 residues) using Ac-Leu-Gly-Ser-Lys(Su)-AMC as substrate incubated for 1 hr in presence of 400 uM NAD+ by fluorescence based analysis ChEMBL_2121799 (CHEMBL4830946) Inhibition of human recombinant SIRT5 (34 to 269 residues) using Ac-Leu-Gly-Ser-Lys(Su)-AMC as substrate incubated for 1 hr in presence of 200 uM NAD+ by fluorescence based analysis ChEMBL_2121800 (CHEMBL4830947) Inhibition of human recombinant SIRT5 (34 to 269 residues) using Ac-Leu-Gly-Ser-Lys(Su)-AMC as substrate incubated for 1 hr in presence of 100 uM NAD+ by fluorescence based analysis ChEMBL_2121801 (CHEMBL4830948) Inhibition of human recombinant SIRT5 (34 to 269 residues) using Ac-Leu-Gly-Ser-Lys(Su)-AMC as substrate incubated for 1 hr in presence of 50 uM NAD+ by fluorescence based analysis ChEMBL_2121816 (CHEMBL4830963) Inhibition of human recombinant SIRT5 (34 to 269 residues) using Ac-Leu-Gly-Ser-Lys(Su)-AMC as substrate incubated for 1 hr in presence of 800 uM NAD+ by fluorescence based analysis ChEMBL_2360800 Inhibition of human recombinant SIRT5 (34 to 269 residues) using Ac-Leu-Gly-Ser-Lys (Su)-AMC as fluorogenic substrate incubated for 2 hrs in presence of NAD+ at 50 uM by fluorescence based analysis ChEMBL_2360801 Inhibition of human recombinant SIRT5 (34 to 269 residues) using Ac-Leu-Gly-Ser-Lys (Su)-AMC as fluorogenic substrate incubated for 2 hrs in presence of NAD+ at 100 uM by fluorescence based analysis ChEMBL_2360802 Inhibition of human recombinant SIRT5 (34 to 269 residues) using Ac-Leu-Gly-Ser-Lys (Su)-AMC as fluorogenic substrate incubated for 2 hrs in presence of NAD+ at 200 uM by fluorescence based analysis ChEMBL_2360803 Inhibition of human recombinant SIRT5 (34 to 269 residues) using Ac-Leu-Gly-Ser-Lys (Su)-AMC as fluorogenic substrate incubated for 2 hrs in presence of NAD+ at 400 uM by fluorescence based analysis ChEMBL_2360804 Inhibition of human recombinant SIRT5 (34 to 269 residues) using Ac-Leu-Gly-Ser-Lys (Su)-AMC as fluorogenic substrate incubated for 2 hrs in presence of NAD+ at 800 uM by fluorescence based analysis ChEMBL_2121802 (CHEMBL4830949) Inhibition of human recombinant SIRT5 (34 to 269 residues) using 300 uM Ac-Leu-Gly-Ser-Lys(Su)-AMC as substrate incubated for 1 hr in presence of 400 uM NAD+ by fluorescence based analysis ChEMBL_2121803 (CHEMBL4830950) Inhibition of human recombinant SIRT5 (34 to 269 residues) using 100 uM Ac-Leu-Gly-Ser-Lys(Su)-AMC as substrate incubated for 1 hr in presence of 400 uM NAD+ by fluorescence based analysis ChEMBL_2121804 (CHEMBL4830951) Inhibition of human recombinant SIRT5 (34 to 269 residues) using 33 uM Ac-Leu-Gly-Ser-Lys(Su)-AMC as substrate incubated for 1 hr in presence of 400 uM NAD+ by fluorescence based analysis ChEMBL_2121805 (CHEMBL4830952) Inhibition of human recombinant SIRT5 (34 to 269 residues) using 11 uM Ac-Leu-Gly-Ser-Lys(Su)-AMC as substrate incubated for 1 hr in presence of 400 uM NAD+ by fluorescence based analysis ChEMBL_2319786 Inhibition of His-tagged PLK4 (1 to 269 residues) (unknown origin) expressed in Escherichia coli assessed as inhibition constant incubated for 1 hrs using TPSDSLIYDDGLS as substrate in presence [gamma32P]-ATP by TopCount scintillation counter method ChEMBL_2432067 Displacement of (S)-3-(5-chloro-2-hydroxyphenyl)-3-hydroxy-6-(trifluoromethyl)indolin-2-one from biotinylated His6-tagged human IL-1 beta (117 to 269 residues) expressed in Escherichia coli BL21(DE3) by 19F NMR displacement assay Tyrosine Kinase Assay Src, Lck, Flt-1, ZAP, EGFR, FGFR1, and PFGFR-beta were assayed in the Merck research laboratory (Homogeneous proximity tyrosine kinase assays: scintillation proximity assay versus homogeneous time-resolved fluorescence. Anal. Biochem. 1999, 269, 94-104.) Homogenous Time-Resolved Fluorescence (HTRF) Assay MARK3 activity was assayed in vitro using a Cdc25C biotinylated peptide substrate (Cell Signalling Technologies). The phosphopeptide product was quantitated using a Homogenous Time-Resolved Fluorescence (HTRF) assay system (Park et al., 1999, Anal. Biochem. 269:94-104). ChEMBL_2079782 (CHEMBL4735573) Inhibition of recombinant human MMP8 (99 to 269 residues) using Mca-Pro-Leu-Gly-Leu-Dpa-Ala-Arg-NH2 expressed in Escherichia coli expression system as substrate incubated for 15 mins followed by substrate addition and measured after 2 to 4 hrs by fluorescence based assay Radioligand Binding Assay The compounds were evaluated using well established radioligand binding assays protocols (Galli, A. et al., J. Exp. Biol. 1995, 198, 2197-2212; Giros, B. et al., Trends Pharmcol. Sci. 1993, 14, 43-49; Gu, H. et al., J. Biol. Chem. 1994, 269(10), 7124-7130; Shearman, L. P. et al, Am. J. Physiol., 1998, 275(6 Pt 1), C1621-1629; Wolf, W. A. et al., J. Biol. Chem. 1992, 267(29), 20820-20825). The human recombinant transporter proteins dopamine (DAT), norepinephrine (NET) and serotonin (SERT) were selected for the in vitro assays. In Vitro Lipoxygenase (LOX) Inhibitory Assay All the newly synthesized compounds were also evaluated in vitro for their ability to inhibit lipoxygenase enzyme. This was carried out using Abnova lipoxygenase inhibitor screening assay kit (Catalog No. (KA1329). Inhibitors were dissolved in DMSO and were added to the assay in a final volume of 10 μl before initiating with substrate. Three concentrations were prepared (25, 50 and 100 μM) and the concentration that produced 50% enzyme inhibition was determined according to manufacturer instructions [Plačkov et al., Free Radic. Biol. Med., 97:223-235]. Radioligand Binding Assay The compounds were evaluated using well established radioligand binding assays protocols (Galli, A. et al., J. Exp. Biol. 1995, 198, 2197-2212; Giros, B. et al., Trends Pharmcol. Sci. 1993, 14, 43-49; Gu, H. et al., J. Biol. Chem. 1994, 269(10), 7124-7130; Shearman, L. P. et al, Am. J. Physiol., 1998, 275 (6 Pt 1), C1621-1629; Wolf, W. A. et al., J. Biol. Chem. 1992, 267(29), 20820-20825). The human recombinant transporter proteins dopamine (DAT), norepinephrine (NET) and serotonin (SERT) were selected for the in vitro assays. The radioligand binding assays were carried out at 11 different test concentrations 0.1 nm to 1 μM. Receptor Binding Profile of Compound of Examples 1, 2 and 3 Receptor binding is determined for the Compounds of Examples 1, 2 and 3 (corresponding to Formula 1, Formula B and Formula A, respectively). The following literature procedures are used, each of which reference is incorporated herein by reference in their entireties: 5-HT2A: Bryant, H. U. et al. (1996), Life Sci., 15:1259-1268; D2: Hall, D. A. and Strange, P. G. (1997), Brit. J. Pharmacol., 121:731-736; D1: Zhou, Q. Y. et al. (1990), Nature, 347:76-80; SERT: Park, Y. M. et al. (1999), Anal. Biochem., 269:94-104; Mu opiate receptor: Wang, J. B. et al. (1994), FEBS Lett., 338:217-222. In Vitro Pharmacology Assay The monoamine transporters inhibitory activities of selected cycloalkylmethylamines of Formula (I) are reported herein. The compounds were evaluated using well established radioligand binding assays protocols (Galli, A. et al., J. Exp. Biol. 1995, 198, 2197-2212; Giros, B. et al., Trends Pharmcol. Sci. 1993, 14, 43-49; Gu, H. et al., J. Biol. Chem. 1994, 269(10), 7124-7130; Shearman, L. P. et al, Am. J. Physiol., 1998, 275(6 Pt 1), C1621-1629; Wolf, W. A. et al., J. Biol. Chem. 1992, 267(29), 20820-20825). The human recombinant transporter proteins dopamine (DAT), norepinephrine (NET) and serotonin (SERT) were selected for the in vitro assays. The radioligand binding assays were carried out at 11 different test concentrations 0.1 nM to 1 μM. Radioligand Binding Assay The monoamine transporters inhibitory activities of selected compounds cycloalkylmethylamine derivatives comprising Formula (I) are reported herein. The compounds were evaluated using well established radioligand binding assays protocols (Galli, A. et al., J. Exp. Biol. 1995, 198, 2197-2212; Giros, B. et al., Trends Pharmcol. Sci. 1993, 14, 43-49; Gu, H. et al., J. Biol. Chem. 1994, 269(10), 7124-7130; Shearman, L. P. et al, Am. J. Physiol., 1998, 275(6 Pt 1), C1621-1629; Wolf, W. A. et al., J. Biol. Chem. 1992, 267(29), 20820-20825). The human recombinant transporter proteins dopamine (DAT), norepinephrine (NET) and serotonin (SERT) were selected for the in vitro assays. The radioligand binding assays were carried out at 11 different test concentrations 0.1 nm to 1 μM. Fluorogenic Assay Buffer: pH=7.5, 100 mM Tris-HCl, 75 mM NaCl, 2.5 mM CaCl2, 10 mM cysteine, 1% DMSO after all additions. Protein: 0.1 nM Kgp, isolated from culture of Porphyromonas gingivalis, as described in Pike et al. J. Biol. Chem. 1994, 269(1), 406, and Potempa and Nguyen. Current Protocols in Protein Science. 2007, 21.20.1-21.20.27. Fluorogenic substrate: 10 uM Z-His-Glu-Lys-MCA. Time=90 minutes. Temperature=37° C. Each compound: 10 concentrations, starting at either 100 uM or 100 nM, with lower concentrations generated by serial 3-fold dilutions. By testing a range of concentrations for each compound, the concentration required to inhibit the activity of lysine gingipain by 50% (the IC50 ) was determined. Under the described assay conditions, signal-to-noise was excellent, and Z factor was greater than 0.6. Inhibition Assay The specific assay conditions were as follows. Buffer: pH=7.5, 100 mM Tris-HCl, 75 mM NaCl, 2.5 mM CaCl2, 10 mM cysteine, 1% DMSO after all additions. Protein: 0.1 nM Kgp, isolated from culture of Porphyromonas gingivalis, as described in Pike et al. J. Biol. Chem. 1994, 269(1), 406, and Potempa and Nguyen. Current Protocols in Protein Scienc. 2007, 21.20.1-21.20.27. Fluorogenic substrate: 10 uM Z-His-Glu-Lys-MCA. Time=90 minutes. Temperature=37° C. Each compound: 10 concentrations, starting at either 100 uM or 100 nM, with lower concentrations generated by serial 3-fold dilutions. By testing a range of concentrations for each compound, the concentration required to inhibit the activity of lysine gingipain by 50% (the IC50 ) was determined. Under the described assay conditions, signal-to-noise was excellent, and Z factor was greater than 0.6. Enzymatic Assay r-AC protein samples were pre-incubated with various concentrations of test compounds or vehicle control in 100 mM NaH2PO4/citrate buffer pH 4.5, 0.1% Nonidet P-40, 3 mM DTT for 30 min at 37° C. Samples were incubated with 100 μM N-lauroyl ceramide (Nu-Chek Prep, Elysian, Minn.) at 37° for 30 min. The reaction was stopped by addition of a mixture of chloroform/methanol (2:1 vol/vol) containing 1 nmol of heptadecanoic acid (HDA; NuChek Prep). The organic phases were collected, dried under nitrogen, and analyzed by LC/MS in the negative-ion mode using heptadecanoic acid (HDA) as internal standard (m/z=199 for lauric acid, m/z=269 for HDA). HDA was eluted on an XDB Eclipse C18 column isocratically at 2.2 mL/min for 1 min with a solvent mixture of 95% methanol and 5% water, both containing 0.25% acetic acid, and 5 mM ammonium acetate. The column temperature was 50° C. Inhibition Assay To determine the inhibition constants (Ki) of Examples 1-240, compounds were diluted serially in DMSO and added to 50 μL kinase reactions containing 1.5 nM JAK1, 0.2 nM purified JAK2 or 1 nM purified TYK2 enzyme, 100 mM Hepes pH7.2, 0.015% Brij-35, 1.5 μM peptide substrate, 25 μM ATP, 10 mM MgCl2, 4 mM DTT at a final DMSO concentration of 2%. Reactions were incubated at 22° C. in 384-well polypropylene microtiter plates for 30 minutes and then stopped by addition of 25 μL of an EDTA containing solution (100 mM Hepes pH 7.2, 0.015% Brij-35, 150 mM EDTA), resulting in a final EDTA concentration of 50 mM. Ki values were then determined using the Morrison tight binding model. Morrison, J. F., Biochim. Biophys. Acta. 185:269-296 (1969); William, J. W. and Morrison, J. F., Meth. Enzymol., 63:437-467 (1979). JAK1 and TYK2 Inhibition Assay To determine inhibition constants (Ki), compounds were diluted serially in DMSO and added to 50 uL kinase reactions containing 1.5 nM JAK1, 0.2 nM purified JAK2 or 1 nM purified TYK2 enzyme, 100 mM Hepes pH7.2, 0.015% Brij-35, 1.5 uM peptide substrate, 25 uM ATP, 10 mM MgCl2, 4 mM DTT at a final DMSO concentration of 2%. Reactions were incubated at 22 ° C. in 384-well polypropylene microtiter plates for 30 minutes and then stopped by addition of 25 uL of an EDTA containing solution (100 mM Hepes pH 7.2, 0.015% Brij-35, 150 mM EDTA), resulting in a final EDTA concentration of 50 mM. After termination of the kinase reaction, the proportion of phosphorylated product was determined as a fraction of total peptide substrate using the Caliper LabChip 3000 according to the manufacturer's specifications. Ki values were then determined using the Morrison tight binding model (Morrison, J. F., Biochim. Biophys. Acta. 185:269-296 (1969). In Vitro Kinase Assay One or more compounds herein were tested for inhibitory activity of JAK targets according to the following in vitro assay described in Park et al., Analytical Biochemistry 1999, 269, 94-104. The catalytic domains of human JAK1 (a.a. 837-1142), JAK2 (a.a. 828-1132) and JAK3 (a.a. 781-1124) with an N-terminal His tag were expressed using baculovirus in insect cells and purified. The catalytic activity of JAK1, JAK2 or JAK3 was assayed by measuring the phosphorylation of a biotinylated peptide. The phosphorylated peptide was detected by homogenous time resolved fluorescence (HTRF). IC50s of compounds were measured for each kinase in the reactions that contain the enzyme, ATP and 500 nM peptide in 50 mM Tris (pH 7.8) buffer with 100 mM NaCl, 5 mM DTT, and 0.1 mg/mL (0.01%) BSA. The ATP concentration in the reactions was 90 uM for JAK1, 30 uM for JAK2 and 3 uM for JAK3. Reactions were carried out at room temperature for 1 hr and then stopped with 20 uL 45 mM EDTA. Inhibition of Lysine Gingipain The capacities of compounds of the present invention to inhibit the activity of lysine gingipain were measured in a fluorogenic assay similar to those described in Barret Biochemical Journal. 1980, 187(3), 909. The specific assay conditions were as follows. Buffer: pH=7.5, 100 mM Tris-HCl, 75 mM NaCl, 2.5 mM CaCl2, 10 mM cysteine, 1% DMSO after all additions. Protein: 0.1 nM Kgp, isolated from culture of Porphyromonas gingivalis, as described in Pike et al. J. Biol. Chem. 1994, 269(1), 406, and Potempa and Nguyen. Current Protocols in Protein Scienc. 2007, 21.20.1-21.20.27. Fluorogenic substrate: 10 uM Z-His-Glu-Lys-MCA. Time=90 minutes. Temperature=37° C. Each compound: 10 concentrations, starting at either 100 uM or 100 nM, with lower concentrations generated by serial 3-fold dilutions. By testing a range of concentrations for each compound, the concentration required to inhibit the activity of lysine gingipain by 50% (the IC50 ) was determined. Under the described assay conditions, signal-to-noise was excellent, and Z factor was greater than 0.6. JAK2 Inhibition Assay To determine the inhibition constants (Ki), compounds were diluted serially in DMSO and added to 50 kinase reactions containing 0.2 nM purified JAK2 enzyme, 100 mM Hepes pH7.2, 0.015% Brij-35, 1.5 μM peptide substrate, 25 μM ATP, 10 mM MgCl2, 4 mM DTT at a final DMSO concentration of 2%. Reactions were incubated at 22° C. in 384-well polypropylene microtiter plates for 30 minutes and then stopped by addition of 250 μL of an EDTA containing solution (100 mM Hepes pH 7.2, 0.015% Brij-35, 150 mM EDTA), resulting in a final EDTA concentration of 50 mM. After termination of the kinase reaction, the proportion of phosphorylated product was determined as a fraction of total peptide substrate using the Caliper LabChip 3000 according to the manufacturer's specifications. Ki values were then determined using the Morrison tight binding model. Morrison, J. F., Biochim. Biophys. Acta. 185:269-296 (1969); William, J. W. and Morrison, J. F., Meth. Enzymol., 63:437-467 (1979). JAK3 Inhibition Assay To determine inhibition constants (Ki), compounds were diluted serially in DMSO and added to 50 uL kinase reactions containing 5 nM purified JAK3 enzyme, 100 mM Hepes pH7.2, 0.015% Brij-35, 1.5 uM peptide substrate, 5 uM ATP, 10 mM MgCl2, 4 mM DTT at a final DMSO concentration of 2%. Reactions were incubated at 22° C. in 384-well polypropylene microtiter plates for 30 minutes and then stopped by addition of 25 uL of an EDTA containing solution (100 mM Hepes pH 7.2, 0.015% Brij-35, 150 mM EDTA), resulting in a final EDTA concentration of 50 mM. After termination of the kinase reaction, the proportion of phosphorylated product was determined as a fraction of total peptide substrate using the Caliper LabChip 3000 according to the manufacturer's specifications. Ki values were then determined using the Morrison tight binding model (Morrison, J. F., Biochim. Biophys. Acta. 185:269-296 (1969); William, J. W. and Morrison, J. F., Meth. Enzymol., 63:437-467 (1979)). Kinase Assay The compounds of the invention were tested for inhibitory activity of JAK targets according to the following in vitro assay described in Park et al., Analytical Biochemistry 1999, 269, 94-104. The catalytic domains of human JAK1 (a.a. 837-1142), JAK2 (a.a. 828-1132) and JAK3 (a.a. 781-1124) with an N-terminal His tag were expressed using baculovirus in insect cells and purified. The catalytic activity of JAK1, JAK2 or JAK3 was assayed by measuring the phosphorylation of a biotinylated peptide. The phosphorylated peptide was detected by homogenous time resolved fluorescence (HTRF). IC50 was measured for each kinase in the 40 uL reactions that contain the enzyme, ATP and 500 nM peptide in 50 mM Tris (pH 7.8) buffer with 100 mM NaCl, 5 mM DTT, and 0.1 mg/mL (0.01%) BSA. The ATP concentration in the reactions was 90 uM for Jak1, 30 uM for Jak2 and 3 uM for Jak3. Reactions were carried out at room temperature for 1 hr and then stopped with 20 uL 45 mM EDTA. In Vitro Assay Compounds herein were tested for inhibitory activity of JAK targets according to the following in vitro assay described in Park et al., Analytical Biochemistry 1999, 269, 94-104. The catalytic domains of human JAK1 (a.a. 837-1142), JAK2 (a.a. 828-1132) and JAK3 (a.a. 781-1124) with an N-terminal His tag were expressed using baculovirus in insect cells and purified. The catalytic activity of JAK1, JAK2 or JAK3 was assayed by measuring the phosphorylation of a biotinylated peptide. The phosphorylated peptide was detected by homogenous time resolved fluorescence (HTRF). IC50s of compounds were measured for each kinase in the 40 microL reactions that contain the enzyme, ATP and 500 nM peptide in 50 mM Tris (pH 7.8) buffer with 100 mM NaCl, 5 mM DTT, and 0.1 mg/mL (0.01%) BSA. For the 1 mM IC50 measurements, ATP concentration in the reactions was 1 mM. Reactions were carried out at room temperature for 1 hour and then stopped with 20 μL 45 mM EDTA, 300 nM SA-APC, 6 nM Eu-Py20 in assay buffer (Perkin Elmer, Boston, Mass.). Binding to the Europium labeled antibody took place for 40 minutes and HTRF signal was measured on a Fusion plate reader (Perkin Elmer, Boston, Mass.). In Vitro JAK Kinase Assay Compounds herein were tested for inhibitory activity of JAK targets according to the following in vitro assay described in Park et al., Analytical Biochemistry 1999, 269, 94-104. The catalytic domains of human JAK1 (a.a. 837-1142), JAK2 (a.a. 828-1132) and JAK3 (a.a. 781-1124) were expressed using baculovirus in insect cells and purified. The catalytic activity of JAK1, JAK2 or JAK3 was assayed by measuring the phosphorylation of a biotinylated peptide. The phosphorylated peptide was detected by homogenous time resolved fluorescence (HTRF). IC50s of compounds were measured for each kinase in the 40 μL reactions that contain the enzyme, ATP and 500 nM peptide in 50 mM Tris (pH 7.8) buffer with 100 mM NaCl, 5 mM DTT, and 0.1 mg/mL (0.01%) BSA. For the 1 mM IC50 measurements, ATP concentration in the reactions was 1 mM. Reactions were carried out at room temperature for 1 hour and then stopped with 20 μL 45 mM EDTA, 300 nM SA-APC, 6 nM Eu-Py20 in assay buffer (Perkin Elmer, Boston, Mass.). Binding to the Europium labeled antibody took place for 40 minutes and HTRF signal was measured on a PHERA star plate reader (BMG, Cary, N.C.). In Vitro JAK Kinase Assay Compounds herein were tested for inhibitory activity of JAK targets according to the following in vitro assay described in Park et al., Analytical Biochemistry 1999, 269, 94-104. The catalytic domains of human JAK1 (a.a. 837-1142), JAK2 (a.a. 828-1132) and JAK3 (a.a. 781-1124) were expressed using baculovirus in insect cells and purified. The catalytic activity of JAK1, JAK2 or JAK3 was assayed by measuring the phosphorylation of a biotinylated peptide. The phosphorylated peptide was detected by homogenous time resolved fluorescence (HTRF). IC50s of compounds were measured for each kinase in the 40 μL reactions that contain the enzyme, ATP and 500 nM peptide in 50 mM Tris (pH 7.8) buffer with 100 mM NaCl, 5 mM DTT, and 0.1 mg/mL (0.01%) BSA. For the 1 mM IC50 measurements, ATP concentration in the reactions was 1 mM. Reactions were carried out at room temperature for 1 hour and then stopped with 20 μL 45 mM EDTA, 300 nM SA-APC, 6 nM Eu-Py20 in assay buffer (Perkin Elmer, Boston, Mass.). Binding to the Europium labeled antibody took place for 40 minutes and HTRF signal was measured on a PHERA star plate reader (BMG, Cary, N.C.). In Vitro JAK Kinase Assay Compounds herein were tested for inhibitory activity of JAK targets according to the following in vitro assay described in Park et al., Analytical Biochemistry 1999, 269, 94-104. The catalytic domains of human JAK1 (a.a. 837-1142), JAK2 (a.a. 828-1132) and JAK3 (a.a. 781-1124) with an N-terminal His tag were expressed using baculovirus in insect cells and purified. The catalytic activity of JAK1, JAK2 or JAK3 was assayed by measuring the phosphorylation of a biotinylated peptide. The phosphorylated peptide was detected by homogenous time resolved fluorescence (HTRF). IC50s of compounds were measured for each kinase in the 40 microL reactions that contain the enzyme, ATP and 500 nM peptide in 50 mM Tris (pH 7.8) buffer with 100 mM NaCl, 5 mM DTT, and 0.1 mg/mL (0.01%) BSA. For the 1 mM IC50 measurements, ATP concentration in the reactions was 1 mM. Reactions were carried out at room temperature for 1 hour and then stopped with 20 μL 45 mM EDTA, 300 nM SA-APC, 6 nM Eu-Py20 in assay buffer (Perkin Elmer, Boston, Mass.). Binding to the Europium labeled antibody took place for 40 minutes and HTRF signal was measured on a Fusion plate reader (Perkin Elmer, Boston, Mass.). In Vitro JAK Kinase Assay Compounds herein were tested for inhibitory activity of JAK targets according to the following in vitro assay described in Park et al., Analytical Biochemistry 1999, 269, 94-104. The catalytic domains of human JAK1 (a.a. 837-1142), Jak2 (a.a. 828-1132) and Jak3 (a.a. 781-1124) with an N-terminal His tag were expressed using baculovirus in insect cells and purified. The catalytic activity of JAK1, JAK2 or JAK3 was assayed by measuring the phosphorylation of a biotinylated peptide. The phosphorylated peptide was detected by homogenous time resolved fluorescence (HTRF). IC50s of compounds were measured for each kinase in the 40 microL reactions that contain the enzyme, ATP and 500 nM peptide in 50 mM Tris (pH 7.8) buffer with 100 mM NaCl, 5 mM DTT, and 0.1 mg/mL (0.01%) BSA. For the 1 mM IC50 measurements, ATP concentration in the reactions was 1 mM. Reactions were carried out at room temperature for 1 hr and then stopped with 20 μL 45 mM EDTA, 300 nM SA-APC, 6 nM Eu-Py20 in assay buffer (Perkin Elmer, Boston, Mass.). Binding to the Europium labeled antibody took place for 40 minutes and HTRF signal was measured on a Fusion plate reader (Perkin Elmer, Boston, Mass.). In Vitro JAK Kinase Assay Compounds herein were tested for inhibitory activity of JAK targets according to the following in vitro assay described in Park et al., Analytical Biochemistry 1999, 269, 94-104. The catalytic domains of human JAK1 (a.a. 837-1142), Jak2 (a.a. 828-1132) and Jak3 (a.a. 781-1124) with an N-terminal His tag were expressed using baculovirus in insect cells and purified. The catalytic activity of JAK1, JAK2 or JAK3 was assayed by measuring the phosphorylation of a biotinylated peptide. The phosphorylated peptide was detected by homogenous time resolved fluorescence (HTRF). IC50s of compounds were measured for each kinase in the 40 microL reactions that contain the enzyme, ATP and 500 nM peptide in 50 mM Tris (pH 7.8) buffer with 100 mM NaCl, 5 mM DTT, and 0.1 mg/mL (0.01%) BSA. For the 1 mM IC50 measurements, ATP concentration in the reactions was 1 mM. Reactions were carried out at room temperature for 1 hr and then stopped with 20 uL 45 mM EDTA, 300 nM SA-APC, 6 nM Eu-Py20 in assay buffer (Perkin Elmer, Boston, Mass.). Binding to the Europium labeled antibody took place for 40 minutes and HTRF signal was measured on a Fusion plate reader (Perkin Elmer, Boston, Mass.). In Vitro Kinase Assay Compounds herein were tested for inhibitory activity of JAK targets according to the following in vitro assay described in Park et al., Analytical Biochemistry 1999, 269, 94-104. The catalytic domains of human JAK1 (a.a. 837-1142), JAK2 (a.a. 828-1132) and JAK3 (a.a. 781-1124) were expressed using baculovirus in insect cells and purified. The catalytic activity of JAK1, JAK2 or JAK3 was assayed by measuring the phosphorylation of a biotinylated peptide. The phosphorylated peptide was detected by homogenous time resolved fluorescence (HTRF). IC50s of compounds were measured for each kinase in the 40 μL reactions that contain the enzyme, ATP and 500 nM peptide in 50 mM Tris (pH 7.8) buffer with 100 mM NaCl, 5 mM DTT, and 0.1 mg/mL (0.01%) BSA. For the 1 mM IC50 measurements, ATP concentration in the reactions was 1 mM. Reactions were carried out at room temperature for 1 hour and then stopped with 20 μL 45 mM EDTA, 300 nM SA-APC, 6 nM Eu-Py20 in assay buffer (Perkin Elmer, Boston, Mass.). Binding to the Europium labeled antibody took place for 40 minutes and HTRF signal was measured on a PHERA star plate reader (BMG, Cary, N.C.). Kinase Assay Compounds herein were tested for inhibitory activity of JAK targets according to the following in vitro assay described in Park et al., Analytical Biochemistry 1999, 269, 94-104. The catalytic domains of human JAK1 (a.a. 837-1142), JAK2 (a.a. 828-1132) and JAK3 (a.a. 781-1124) with an N-terminal His tag were expressed using baculovirus in insect cells and purified. The catalytic activity of JAK1, JAK2 or JAK3 was assayed by measuring the phosphorylation of a biotinylated peptide. The phosphorylated peptide was detected by homogenous time resolved fluorescence (HTRF). IC50s of compounds were measured for each kinase in the 40 microL reactions that contain the enzyme, ATP and 500 nM peptide in 50 mM Tris (pH 7.8) buffer with 100 mM NaCl, 5 mM DTT, and 0.1 mg/mL (0.01%) BSA. For the 1 mM 1050 measurements, ATP concentration in the reactions was 1 mM. Reactions were carried out at room temperature for 1 hour and then stopped with 20 μL 45 mM EDTA, 300 nM SA-APC, 6 nM Eu-Py20 in assay buffer (Perkin Elmer, Boston, Mass.). Binding to the Europium labeled antibody took place for 40 minutes and HTRF signal was measured on a Fusion plate reader (Perkin Elmer, Boston, Mass.). In Vitro JAK Kinase Assay Compounds herein were tested for inhibitory activity of JAK targets according to the following in vitro assay described in Park et al., Analytical Biochemistry 1999, 269, 94-104. The catalytic domains of human JAK1 (a.a. 837-1142), JAK2 (a.a. 828-1132) and JAK3 (a.a. 781-1124) were expressed using baculovirus in insect cells and purified. The catalytic activity of JAK1, JAK2 or JAK3 was assayed by measuring the phosphorylation of a biotinylated peptide. The phosphorylated peptide was detected by homogenous time resolved fluorescence (HTRF). IC50s of compounds were measured for each kinase in the 40 μL reactions that contain the enzyme, ATP and 500 nM peptide in 50 mM Tris (pH 7.8) buffer with 100 mM NaCl, 5 mM DTT, and 0.1 mg/mL (0.01%) BSA. For the 1 mM IC50 measurements, ATP concentration in the reactions was 1 mM. Reactions were carried out at room temperature for 1 hour and then stopped with 20 μL 45 mM EDTA, 300 nM SA-APC, 6 nM Eu-Py20 in assay buffer (Perkin Elmer, Boston, Mass.). Binding to the Europium labeled antibody took place for 40 minutes and HTRF signal was measured on a PHERA star plate reader (BMG, Cary, N.C.). The data for the JAK1 and/or JAK2 inhibitors were obtained by testing the compounds in the Example A assay at 1 mM ATP. In Vitro JAK Kinase Assay JAK1 inhibitors that can be used for the treatment of cytokine-related diseases or disorders are tested for inhibitory activity of JAK targets according to the following in vitro assay described in Park et al., Analytical Biochemistry 1999, 269, 94-104. The catalytic domains of human JAK1 (a.a. 837-1142), JAK2 (a.a. 828-1132) and JAK3 (a.a. 781-1124) with an N-terminal His tag are expressed using baculovirus in insect cells and purified. The catalytic activity of JAK1, JAK2 or JAK3 was assayed by measuring the phosphorylation of a biotinylated peptide. The phosphorylated peptide was detected by homogenous time resolved fluorescence (HTRF). IC50 s of compounds are measured for each kinase in the 40 μL reactions that contain the enzyme, ATP and 500 nM peptide in 50 mM Tris (pH 7.8) buffer with 100 mM NaCl, 5 mM DTT, and 0.1 mg/mL (0.01%) BSA. For the 1 mM IC50 measurements, ATP concentration in the reactions is 1 mM. Reactions are carried out at room temperature for 1 hour and then stopped with 20 μL 45 mM EDTA, 300 nM SA-APC, 6 nM Eu-Py20 in assay buffer (Perkin Elmer, Boston, MA). Binding to the Europium labeled antibody takes place for 40 minutes and HTRF signal was measured on a Fusion plate reader (Perkin Elmer, Boston, MA). In Vitro JAK Kinase Assay JAK1 inhibitors that can be used for the treatment of cytokine-related diseases or disorders are tested for inhibitory activity of JAK targets according to the following in vitro assay described in Park et al., Analytical Biochemistry 1999, 269, 94-104. The catalytic domains of human JAK1 (a.a. 837-1142), JAK2 (a.a. 828-1132) and JAK3 (a.a. 781-1124) with an N-terminal His tag are expressed using baculovirus in insect cells and purified. The catalytic activity of JAK1, JAK2 or JAK3 was assayed by measuring the phosphorylation of a biotinylated peptide. The phosphorylated peptide was detected by homogenous time resolved fluorescence (HTRF). IC5os of compounds are measured for each kinase in the 40 microL reactions that contain the enzyme, 1 mM ATP and 500 nM peptide in 50 mM Tris (pH 7.8) buffer with 100 mM NaCl, 5 mM DTT, and 0.1 mg/mL (0.01%) BSA. For the 1 mM IC50 measurements, ATP concentration in the reactions is 1 mM. Reactions are carried out at room temperature for 1 hour and then stopped with 20 μL 45 mM EDTA, 300 nM SA-APC, 6 nM Eu-Py20 in assay buffer (Perkin Elmer, Boston, MA). Binding to the Europium labeled antibody takes place for 40 minutes and HTRF signal was measured on a Fusion plate reader (Perkin Elmer, Boston, MA). In Vitro JAK Kinase Assay JAK1 inhibitors that can be used for the treatment of cytokine-related diseases or disorders are tested for inhibitory activity of JAK targets according to the following in vitro assay described in Park et al., Analytical Biochemistry 1999, 269, 94-104. The catalytic domains of human JAK1 (a.a. 837-1142), JAK2 (a.a. 828-1132) and JAK3 (a.a. 781-1124) with an N-terminal His tag are expressed using baculovirus in insect cells and purified. The catalytic activity of JAK1, JAK2 or JAK3 was assayed by measuring the phosphorylation of a biotinylated peptide. The phosphorylated peptide was detected by homogenous time resolved fluorescence (HTRF). ICsos of compounds are measured for each kinase in the 40 microL reactions that contain the enzyme, ATP and 500 nM peptide in 50 mM Tris (pH 7.8) buffer with 100 mM NaCl, 5 mM DTT, and 0.1 mg/mL (0.01%) BSA. For the 1 mM IC50 measurements, ATP concentration in the reactions is 1 mM. Reactions are carried out at room temperature for 1 hour and then stopped with 20 μL 45 mM EDTA, 300 nM SA-APC, 6 nM Eu-Py20 in assay buffer (Perkin Elmer, Boston, Mass.). Binding to the Europium labeled antibody takes place for 40 minutes and HTRF signal was measured on a Fusion plate reader (Perkin Elmer, Boston, Mass.). In Vitro JAK Kinase Assay JAK1 pathway inhibitors that can be used for the treatment of cytokine-related diseases or disorders are tested for inhibitory activity of JAK targets according to the following in vitro assay described in Park et al., Analytical Biochemistry 1999, 269, 94-104. The catalytic domains of human JAK1 (a.a. 837-1142), JAK2 (a.a. 828-1132) and JAK3 (a.a. 781-1124) with an N-terminal His tag are expressed using baculovirus in insect cells and purified. The catalytic activity of JAK1, JAK2 or JAK3 are assayed by measuring the phosphorylation of a biotinylated peptide. The phosphorylated peptide was detected by homogenous time resolved fluorescence (HTRF). IC50s of compounds are measured for each kinase in the 40 microL reactions that contain the enzyme, ATP and 500 nM peptide in 50 mM Tris (pH 7.8) buffer with 100 mM NaCl, 5 mM DTT, and 0.1 mg/mL (0.01%) BSA. For the 1 mM IC50 measurements, ATP concentration in the reactions is 1 mM. Reactions are carried out at room temperature for 1 hour and then stopped with 20 μL 45 mM EDTA, 300 nM SA-APC, 6 nM Eu-Py20 in assay buffer (Perkin Elmer, Boston, MA). Binding to the Europium labeled antibody takes place for 40 minutes and HTRF signal was measured on a Fusion plate reader (Perkin Elmer, Boston, MA). In Vitro JAK Kinase Assay JAK1 pathway inhibitors that can be used for the treatment of cytokine-related diseases or disorders are tested for inhibitory activity of JAK targets according to the following in vitro assay described in Park et al., Analytical Biochemistry 1999, 269, 94-104. The catalytic domains of human JAK1 (a.a. 837-1142), JAK2 (a.a. 828-1132) and JAK3 (a.a. 781-1124) with an N-terminal His tag are expressed using baculovirus in insect cells and purified. The catalytic activity of JAK1, JAK2 or JAK3 was assayed by measuring the phosphorylation of a biotinylated peptide. The phosphorylated peptide was detected by homogenous resolved fluorescence (HTRF). IC50s of compounds are measured for each kinase in the 40 microL reactions that contain the enzyme, ATP and 500 nM peptide in 50 mM Tris (pH 7.8) buffer with 100 mM NaCl, 5 mM DTT, and 0.1 mg/mL (0.01%) BSA. For the 1 mM IC50 measurements, ATP concentration in the reactions is 1 mM. Reactions are carried out at room temperature for 1 hour and then stopped with 20 μL 45 mM EDTA, 300 nM SA-APC, 6 nM Eu-Py20 in assay buffer (Perkin Elmer, Boston, Mass.). Binding to the Europium labeled antibody takes place for 40 minutes and HTRF signal was measured on a Fusion plate reader (Perkin Elmer, Boston, Mass.). In Vitro JAK Kinase Assay JAK1 pathway inhibitors that can be used for the treatment of cytokine-related diseases or disorders are tested for inhibitory activity of JAK targets according to the following in vitro assay described in Park et al., Analytical Biochemistry 1999, 269, 94-104. The catalytic domains of human JAK1 (a.a. 837-1142), JAK2 (a.a. 828-1132) and JAK3 (a.a. 781-1124) with an N-terminal His tag are expressed using baculovirus in insect cells and purified. The catalytic activity of JAK1, JAK2 or JAK3 was assayed by measuring the phosphorylation of a biotinylated peptide. The phosphorylated peptide was detected by homogenous time resolved fluorescence (HTRF). IC50s of compounds are measured for each kinase in the 40 microL reactions that contain the enzyme, ATP and 500 nM peptide in 50 mM Tris (pH 7.8) buffer with 100 mM NaCl, 5 mM DTT, and 0.1 mg/mL (0.01%) BSA. For the 1 mM IC50 measurements, ATP concentration in the reactions is 1 mM. Reactions are carried out at room temperature for 1 hour and then stopped with 20 μL 45 mM EDTA, 300 nM SA-APC, 6 nM Eu-Py20 in assay buffer (Perkin Elmer, Boston, Mass.). Binding to the Europium labeled antibody takes place for 40 minutes and HTRF signal was measured on a Fusion plate reader (Perkin Elmer, Boston, Mass.). In Vitro JAK Kinase Assay JAK1 pathway inhibitors that can be used for the treatment of cytokine-related diseases or disorders are tested for inhibitory activity of JAK targets according to the following in vitro assay described in Park et al., Analytical Biochemistry 1999, 269, 94-104. The catalytic domains of human JAK1 (a.a. 837-1142), JAK2 (a.a. 828-1132) and JAK3 (a.a. 781-1124) with an N-terminal His tag are expressed using baculovirus in insect cells and purified. The catalytic activity of JAK1, JAK2 or JAK3 was assayed by measuring the phosphorylation of a biotinylated peptide. The phosphorylated peptide was detected by homogenous time resolved fluorescence (HTRF). IC50s of compounds are measured for each kinase in the 40 microL reactions that contain the enzyme, ATP and 500 nM peptide in 50 mM Tris (pH 7.8) buffer with 100 mM NaCl, 5 mM DTT, and 0.1 mg/mL (0.01%) BSA. For the 1 mM IC50 measurements, ATP concentration in the reactions is 1 mM. Reactions are carried out at room temperature for 1 hour and then stopped with 20 L 45 mM EDTA, 300 nM SA-APC, 6 nM Eu-Py20 in assay buffer (Perkin Elmer, Boston, Mass.). Binding to the Europium labeled antibody takes place for 40 minutes and HTRF signal was measured on a Fusion plate reader (Perkin Elmer, Boston, Mass.) In Vitro JAK Kinase Assay JAK1 pathway inhibitors that can be used for the treatment of cytokine-related diseases or disorders were tested for inhibitory activity of JAK targets according to the following in vitro assay described in Park et al., Analytical Biochemistry 1999, 269, 94-104. The catalytic domains of human JAK1 (a.a. 837-1142), JAK2 (a.a. 828-1132) and JAK3 (a.a. 781-1124) with an N-terminal His tag were expressed using baculovirus in insect cells and purified. The catalytic activity of JAK1, JAK2 or JAK3 was assayed by measuring the phosphorylation of a biotinylated peptide. The phosphorylated peptide was detected by homogenous time resolved fluorescence (HTRF). IC50s of compounds were measured for each kinase in the 40 microL reactions that contain the enzyme, ATP and 500 nM peptide in 50 mM Tris (pH 7.8) buffer with 100 mM NaCl, 5 mM DTT, and 0.1 mg/mL (0.01%) BSA. For the 1 mM IC50 measurements, ATP concentration in the reactions is 1 mM. Reactions were carried out at room temperature for 1 hour and then stopped with 20 μL 45 mM EDTA, 300 nM SA-APC, 6 nM Eu-Py20 in assay buffer (Perkin Elmer, Boston, MA). Binding to the Europium labeled antibody took place for 40 minutes and HTRF signal was measured on a Fusion plate reader (Perkin Elmer, Boston, MA). In Vitro JAK Kinase Assay Selective JAK1 inhibitors that can be used in combination with a ROCK inhibitor as described herein are tested for inhibitory activity of JAK targets according to the following in vitro assay described in Park et al., Analytical Biochemistry 1999, 269, 94-104. The catalytic domains of human JAK1 (a.a. 837-1142), JAK2 (a.a. 828-1132) and JAK3 (a.a. 781-1124) with an N-terminal His tag are expressed using baculovirus in insect cells and purified. The catalytic activity of JAK1, JAK2, or JAK3 was assayed by measuring the phosphorylation of a biotinylated peptide. The phosphorylated peptide was detected by homogenous time resolved fluorescence (HTRF). IC50s of compounds are measured for each kinase in the 40 μL reactions that contain the enzyme, ATP and 500 nM peptide in 50 mM Tris (pH 7.8) buffer with 100 mM NaCl, 5 mM DTT, and 0.1 mg/mL (0.01%) BSA. For the 1 mM IC50 measurements, ATP concentration in the reactions is 1 mM. Reactions are carried out at room temperature for 1 hour and then stopped with 20 μL 45 mM EDTA, 300 nM SA-APC, 6 nM Eu-Py20 in assay buffer (Perkin Elmer, Boston, MA). Binding to the Europium labeled antibody takes place for 40 minutes and HTRF signal was measured on a Fusion plate reader (Perkin Elmer, Boston, MA). In vitro JAK Kinase Assay JAK1 inhibitors that can be used for the treatment of cytokine-related diseases or disorders are tested for inhibitory activity of JAK targets according to the following in vitro assay described in Park et al., Analytical Biochemistry 1999, 269, 94-104. The catalytic domains of human JAK1 (a.a. 837-1142), JAK2 (a.a. 828-1132) and JAK3 (a.a. 781-1124) with an N-terminal His tag are expressed using baculovirus in insect cells and purified. The catalytic activity of JAK1, JAK2 or JAK3 was assayed by measuring the phosphorylation of a biotinylated peptide. The phosphorylated peptide was detected by homogenous time resolved fluorescence (HTRF). IC50s of compounds are measured for each kinase in the 40 μL reactions that contain the enzyme, ATP and 500 nM peptide in 50 mM Tris (pH 7.8) buffer with 100 mM NaCl, 5 mM DTT, and 0.1 mg/mL (0.01%) BSA. For the 1 mM IC50 measurements, ATP concentration in the reactions is 1 mM. Reactions are carried out at room temperature for 1 hour and then stopped with 20 μL 45 mM EDTA, 300 nM SA-APC, 6 nM Eu-Py20 in assay buffer (Perkin Elmer, Boston, MA). Binding to the Europium labeled antibody takes place for 40 minutes and HTRF signal was measured on a Fusion plate reader (Perkin Elmer, Boston, MA). JAK Enzyme Assay The activity of the isolated recombinant JAK1 and JAK2 kinase domain was measured by monitoring phosphorylation of a peptide derived from JAK3 (Val-Ala-Leu-Val-Asp-Gly-Tyr-Phe-Arg-Leu-Thr-Thr, fluorescently labeled on the N-terminus with 5-carboxyfluorescein) using the Caliper LabChip technology (Caliper Life Sciences, Hopkinton, Mass.). To determine inhibition constants (Ki), compounds were diluted serially in DMSO and added to 50 μL kinase reactions containing purified enzyme (1.5 nM JAK1, or 0.2 nM JAK2), 100 mM HEPES buffer (pH 7.2), 0.015% Brij-35, 1.5 μM peptide substrate, ATP (25 μM), 10 mM MgCl2, 4 mM DTT at a final DMSO concentration of 2%. Reactions were incubated at 22° C. in 384-well polypropylene microtiter plates for 30 minutes and then stopped by addition of 25 μL of an EDTA containing solution (100 mM HEPES buffer (pH 7.2), 0.015% Brij-35, 150 mM EDTA), resulting in a final EDTA concentration of 50 mM. After termination of the kinase reaction, the proportion of phosphorylated product was determined as a fraction of total peptide substrate using the Caliper LabChip 3000 according to the manufacturer's specifications. Ki values were then determined using the Morrison tight binding model (Morrison, J. F., Biochim. Biophys. Acta. 185:269-296 (1969); William, J. W. and Morrison, J. F., Meth. In Vitro JAK Kinase Assay The compounds in Table A were tested for inhibitory activity of JAK targets according to the following in vitro assay described in Park et al., Analytical Biochemistry 1999, 269, 94-104. The catalytic domains of human JAK1 (a.a. 837-1142), JAK2 (a.a. 828-1132) and JAK3 (a.a. 781-1124) were expressed using baculovirus in insect cells and purified. The catalytic activity of JAK1, JAK2 or JAK3 was assayed by measuring the phosphorylation of a biotinylated peptide. The phosphorylated peptide was detected by homogenous time resolved fluorescence (HTRF). IC50s of compounds were measured for each kinase in the 40 μL reactions that contain the enzyme, ATP and 500 nM peptide in 50 mM Tris (pH 7.8) buffer with 100 mM NaCl, 5 mM DTT, and 0.1 mg/mL (0.01%) BSA. For the 1 mM IC50 measurements, ATP concentration in the reactions was 1 mM. Reactions were carried out at room temperature for 1 hour and then stopped with 20 μL 45 mM EDTA, 300 nM SA-APC, 6 nM Eu-Py20 in assay buffer (Perkin Elmer, Boston, Mass.). Binding to the Europium labeled antibody took place for 40 minutes and HTRF signal was measured on a PHERA star plate reader (BMG, Cary, N.C.). The data for the JAK1 and/or JAK2 inhibitors were obtained by testing the compounds in the Example D assay at 1 mM ATP. In Vitro JAK Kinase Assay Compounds herein were tested for inhibitory activity of JAK targets according to the following in vitro assay described in Park et al., Analytical Biochemistry 1999, 269, 94-104. The catalytic domains of human JAK1 (a.a. 837-1142), JAK2 (a.a. 828-1132) and JAK3 (a.a. 781-1124) were expressed using baculovirus in insect cells and purified. The catalytic activity of JAK1, JAK2 or JAK3 was assayed by measuring the phosphorylation of a biotinylated peptide. The phosphorylated peptide was detected by homogenous time resolved fluorescence (HTRF). IC50s of compounds were measured for each kinase in the 40 μL reactions that contain the enzyme, ATP and 500 nM peptide in 50 mM Tris (pH 7.8) buffer with 100 mM NaCl, 5 mM DTT, and 0.1 mg/mL (0.01%) BSA. For the 1 mM IC50 measurements, ATP concentration in the reactions was 1 mM. Reactions were carried out at room temperature for 1 hour and then stopped with 20 μL 45 mM EDTA, 300 nM SA-APC, 6 nM Eu-Py20 in assay buffer (Perkin Elmer, Boston, Mass.). Binding to the Europium labeled antibody took place for 40 minutes and HTRF signal was measured on a PHERA star plate reader (BMG, Cary, N.C.). The Example compounds were each tested in the Example A assay (see Table 1 for data for the compounds of the examples as tested by the assay of Example A at 1 mM ATP). In Vitro JAK Kinase Assay Selective JAK1 inhibitors that can be used in combination with an immunomodulatory agent and steroid for the treatment of hematological diseases or disorders are tested for inhibitory activity of JAK targets according to the following in vitro assay described in Park et al., Analytical Biochemistry 1999, 269, 94-104. The catalytic domains of human JAK1 (a.a. 837-1142), JAK2 (a.a. 828-1132) and JAK3 (a.a. 781-1124) with an N-terminal His tag are expressed using baculovirus in insect cells and purified. The catalytic activity of JAK1, JAK2, or JAK3 was assayed by measuring the phosphorylation of a biotinylated peptide. The phosphorylated peptide was detected by homogenous time resolved fluorescence (HTRF). IC50s of compounds are measured for each kinase in the 40 μL reactions that contain the enzyme, ATP and 500 nM peptide in 50 mM Tris (pH 7.8) buffer with 100 mM NaCl, 5 mM DTT, and 0.1 mg/mL (0.01%) BSA. For the 1 mM IC50 measurements, ATP concentration in the reactions is 1 mM. Reactions are carried out at room temperature for 1 hour and then stopped with 20 μL 45 mM EDTA, 300 nM SA-APC, 6 nM Eu-Py20 in assay buffer (Perkin Elmer, Boston, Mass.). Binding to the Europium labeled antibody takes place for 40 minutes and HTRF signal was measured on a Fusion plate reader (Perkin Elmer, Boston, Mass.). In Vitro JAK Kinase Assay The compounds in Table 1 were tested for inhibitory activity of JAK targets according to the following in vitro assay described in Park et al., Analytical Biochemistry 1999, 269, 94-104. The catalytic domains of human JAK1 (a.a. 837-1142), JAK2 (a.a. 828-1132) and JAK3 (a.a. 781-1124) were expressed using baculovirus in insect cells and purified. The catalytic activity of JAK1, JAK2 or JAK3 was assayed by measuring the phosphorylation of a biotinylated peptide. The phosphorylated peptide was detected by homogenous time resolved fluorescence (HTRF). IC50s of compounds were measured for each kinase in the 40 μL reactions that contain the enzyme, ATP and 500 nM peptide in 50 mM Tris (pH 7.8) buffer with 100 mM NaCl, 5 mM DTT, and 0.1 mg/mL (0.01%) BSA. For the 1 mM IC50 measurements, ATP concentration in the reactions was 1 mM. Reactions were carried out at room temperature for 1 hour and then stopped with 20 μL 45 mM EDTA, 300 nM SA-APC, 6 nM Eu-Py20 in assay buffer (Perkin Elmer, Boston, Mass.). Binding to the Europium labeled antibody took place for 40 minutes and HTRF signal was measured on a PHERA star plate reader (BMG, Cary, N.C.). The data for the JAK1 and/or JAK2 inhibitors were obtained by testing the compounds in the Example J assay at 1 mM ATP. JAK Enzyme Assay The activity of the isolated recombinant JAK1 and JAK2 kinase domain was measured by monitoring phosphorylation of a peptide derived from JAK3 (Val-Ala-Leu-Val-Asp-Gly-Tyr-Phe-Arg-Leu-Thr-Thr, fluorescently labeled on the N-terminus with 5-carboxyfluorescein) using the Caliper LabChip technology (Caliper Life Sciences, Hopkinton, Mass.). To determine inhibition constants (Ki) compounds were diluted serially in DMSO and added to 50 μL kinase reactions containing purified enzyme (1.5 nM JAK1, or 0.2 nM JAK2), 100 mM HEPES buffer (pH 7.2), 0.015% Brij-35, 1.5 μM peptide substrate, ATP (25 μM), 10 mM MgCl2, 4 mM DTT at a final DMSO concentration of 2%. Reactions were incubated at 22° C. in 384-well polypropylene microtiter plates for 30 minutes and then stopped by addition of 25 μL of an EDTA containing solution (100 mM HEPES buffer (pH 7.2), 0.015% Brij-35, 150 mM EDTA), resulting in a final EDTA concentration of 50 mM. After termination of the kinase reaction, the proportion of phosphorylated product was determined as a fraction of total peptide substrate using the Caliper LabChip 3000 according to the manufacturer's specifications. Ki values were then determined using the Morrison tight binding model (Morrison, J. F., Biochim. Biophys. Acta. 185:269-296 (1969); William, J. W. and Morrison, J. F., Meth. Enzymol., 63:437-467 (1979)) modified for ATP-competitive inhibition [Ki=Ki,app/(1+[ATP]/Km,app)]. JAK Enzyme Assays The activity of the isolated recombinant JAK1 and JAK2 kinase domain was measured by monitoring phosphorylation of a peptide derived from JAK3 (Val-Ala-Leu-Val-Asp-Gly-Tyr-Phe-Arg-Leu-Thr-Thr, fluorescently labeled on the N-terminus with 5-carboxyfluorescein) using the Caliper LabChip technology (Caliper Life Sciences, Hopkinton, Mass.). To determine inhibition constants (Ki), compounds were diluted serially in DMSO and added to 50 μL kinase reactions containing purified enzyme (1.5 nM JAK1, or 0.2 nM JAK2), 100 mM HEPES buffer (pH 7.2), 0.015% Brij-35, 1.5 μM peptide substrate, ATP (25 μM), 10 mM MgCl2, 4 mM DTT at a final DMSO concentration of 2%. Reactions were incubated at 22° C. in 384-well polypropylene microtiter plates for 30 minutes and then stopped by addition of 25 μL of an EDTA containing solution (100 mM HEPES buffer (pH 7.2), 0.015% Brij-35, 150 mM EDTA), resulting in a final EDTA concentration of 50 mM. After termination of the kinase reaction, the proportion of phosphorylated product was determined as a fraction of total peptide substrate using the Caliper LabChip 3000 according to the manufacturer's specifications. Ki values were then determined using the Morrison tight binding model (Morrison, J. F., Biochim. Biophys. Acta. 185:269-296 (1969); William, J. W. and Morrison, J. F., Meth. Enzymol., 63:437-467 (1979)) modified for ATP-competitive inhibition [Ki=Ki,app/(1+[ATP]/Km,app)]. GSK3beta Mobility Shift Assay The purpose of GSK3β assay is to evaluate the inhibition (% inhibition, Kiapp and Ki values) of small molecule inhibitors by using a fluorescence-based microfluidic mobility shift assay. GSK3β catalyzes the production of ADP from ATP that accompanies the phosphoryl transfer to the substrate peptide FL-Peptide-15 (5-FAM-KRREILSRRPpSYR-COOH, CPC Scientific, Sunnyvale, Calif.) (SEQ ID NO:2). The mobility shift assay electrophoretically separates the fluorescently labeled peptides (substrate and phosphorylated product) following the kinase reaction. Both substrate and product are measured and the ratio of these values is used to generate % conversion of substrate to product by the LabChip EZ Reader. Active GSK3β (H350L) was purchased from Upstate/Millipore. Typical reaction solutions (50 μL final reaction volume) contained 2% DMSO (±inhibitor), 4 mM MgCl2, 1 mM DTT, 40 μM ATP (ATP Km=9.43 μM), 0.005% Tween-20, 2 μM FL-Peptide-15, and 0.6 nM GSK3β in 25 mM HEPES buffer at pH 7.5. The assay was initiated with the addition of ATP, following 15 minutes pre-incubation of enzyme and inhibitor at room temperature in the reaction mixture. The reaction was stopped after 30 minutes at room temperature by the addition of 50 μL of 80 mM EDTA. The Ki value was determined from the fit of the data to the Morrison tight-binding competitive inhibition equation with the enzyme concentration as a variable. See Morrison, J. F. (1969) Kinetics of the reversible inhibition of enzyme-catalysed reactions by tight-binding inhibitors, Biochimica et biophysica acta 185, 269-286; Murphy, D. J. (2004) Determination of accurate KI values for tight-binding enzyme inhibitors: an in silico study of experimental error and assay design, Analytical biochemistry 327, 61-67. Inhibition Assay Protein Kinase C beta 2 (PKCβII) catalyzes the production of ADP from ATP that accompanies the phosphoryl transfer to the PKC Pseudosubstrate peptide (A→S, RFARKGSLRQKNV). This transfer is coupled to the oxidation of β-NADH through the activities of Pyruvate Kinase (PK) and Lactate Dehydrogenase (LDH). β-NADH conversion to NAD+ is monitored by the decrease in absorbance at 340 nm (e=6.22 cm−1 mM−1) using a Molecular Devices SPECTRA max PLUS spectrophotometer.A typical assay was carried out on a 96-well, clear microtiter plate in a Molecular Devices spectrophotometer for 20 minutes at 30° C. in 0.1 mL of assay buffer containing 50 mM HEPES, pH 7.4, 5 nM PKC, 23 units of pyruvate kinase, 33 units of lactate dehydrogenase, 0.15 mM peptide, 0.1 mM ATP, 1 mM DTT, 4 mM PEP, 8 mM MgCl2, 0.3 mM NADH, 60 mM CaCl2, 10 mg/mL PS, 50 ng/mL PMA, 7.5% DMSO and from about 10,000 nM to 0.169 nM compound inhibitor. Stock solutions of 3-sn-phosphatidyl-L-serine (PS) and phorbol-12-myristate-13-acetate (PMA) were sonicated for 30 seconds just prior to addition to assay buffer and assays were initiated by the addition of 100 μM ATP.Steady-state kinetic parameters for the bi-bi kinase reaction were determined at saturating phospho-acceptor peptide substrate concentration (0.15 mM) by fitting initial velocity data to the Michaelis-Menten equation, v=V max [S]/(K M +[S]) where v is the measured initial velocity, Vmax is the maximal enzyme velocity, [S] is the ATP substrate concentration, and KM is the Michealis constant for ATP. Enzyme turnover values (kcat) were calculated according to kcat=Vmax[E], where [E] is the total enzyme concentration. Enzyme inhibition constants (apparent Ki values) were determined by fitting initial velocities at variable inhibitor concentrations to a model for ATP competitive inhibition based on the Morrison equation). Morrison, J. F., Biochim. Biophys Acta 185: 269-286 (1969). Enzyme Assay AXL enzyme inhibition (% inhibition, Kiapp and Ki values) by small molecule inhibitors was evaluated using a fluorescence-based microfluidic mobility shift assay. AXL catalyzes the production of ADP from ATP that accompanies the phosphoryl transfer to the substrate peptide FL-Peptide-30 (5-FAM-KKKKEEIYFFF-CONH2, CPC Scientific, Sunnyvale, Calif.). The mobility shift assay electrophoretically separates the fluorescently labeled peptides (substrate and phosphorylated product) following the kinase reaction. Both substrate and product are measured and the ratio of these values is used to generate % conversion of substrate to product by the LabChip EZ Reader. Human wild-type receptor tyrosine kinase protein Axl comprising residues 505-811 was produced in-house using the baculoviral expression vector system that incorporated a hexahistidine affinity tag into the protein (LJIC-1916B1.1). The enzyme was preactivated by auto-phosphorylation of 34 uM non-activated enzyme in the presence of 2 mM ATP, 4 mM MgCl2, 50 mM NaCl and 1 mM TCEP in 20 mM HEPES, pH 7.3 at 4° C. for 30 minutes. Typical reaction solutions (50 μL final reaction volume) contained 2% DMSO (±inhibitor), 10 mM MgCl2, 1 mM DTT, 120 μM ATP (ATP Km=70.4 μM), 0.01% Tween-20, 3 μM FL-Peptide-30, and 0.5 nM phosphorylated AXL enzyme in 100 mM HEPES buffer at pH 7.3. The assay was initiated with the addition of ATP, following a fifteen minutes pre-incubation of enzyme and inhibitor at room temperature in the reaction mixture. The reaction was stopped after 30 minutes at 25° C. by the addition of 50 μL of 200 mM EDTA, pH 7.5. The Ki values were determined from the fit of the data to the Morrison tight-binding competitive inhibition equation with the enzyme concentration as a variable. (See, Morrison, J. F. (1969) Kinetics of the reversible inhibition of enzyme-catalysed reactions by tight-binding inhibitors, Biochimica et biophysica acta 185, 269-286; and Murphy, D. J. (2004) Determination of accurate KI values for tight-binding enzyme inhibitors: an in silico study of experimental error and assay design, Analytical biochemistry 327, 61-67.) Inhibition of Lysine Gingipain The capacities of compounds of the present invention to inhibit the activity of lysine gingipain were measured in a fluorogenic assay similar to those described by Barret (Biochemical Journal. 1980, 187(3), 909). The specific assay conditions were as follows. Buffer: pH=7.5, 100 mM Tris-HCl, 75 mM NaCl, 2.5 mM CaCl2), 10 mM cysteine, 1% DMSO after all additions. Protein: 0.1 nM Kgp, isolated from culture of Porphyromonas gingivalis, as described by Pike et al. (J. Biol. Chem. 1994, 269(1), 406), and Potempa and Nguyen (Current Protocols in Protein Science. 2007, 21.20.1-21.20.27). Fluorogenic substrate: 10 μM Z-His-Glu-Lys-MCA. Time=90 minutes. Temperature=37° C. Each compound: 10 concentrations, starting at either 100 μM or 100 nM, with lower concentrations generated by serial 3-fold dilutions. By testing a range of concentrations for each compound, the concentration required to inhibit the activity of lysine gingipain by 50% (the IC50 ) was determined. Under the described assay conditions, the signal-to-noise ratio was excellent, and the Z factor was greater than 0.6. Compounds in Table 1 were tested, as well as the compounds set forth in Table 2 below.The capacities of compounds of the present invention to inhibit the activity of cathepsins B, H, K, L, and S were measured in similar assays. Boc-Leu-Arg-Arg-AMC (20 μM) in sodium acetate buffer (50 mM, pH 5.5) containing DTT (1 mM) and EDTA (2 mM) was used for the Cathepsin B assay. L-Arg-AMC (20 μM) in sodium acetate buffer (50 mM, pH 5.5) containing DTT (1 mM) and EDTA (2 mM) was used for the Cathepsin H assay. Z-Phe-Arg-AMC (10 μM) in HEPES buffer (50 mM, pH 7.4) containing DTT (2.5 mM) and EDTA (1 mM) was used for the Cathepsin K assay. Z-Phe-Arg-AMC (20 μM) in sodium acetate buffer (50 mM, pH 5.5) containing DTT (1 mM) and EDTA (2 mM) was used for the Cathepsin L assay. Z-Leu-Arg-AMC (10 μM) in sodium acetate buffer (25 mM, pH 4.5) containing DTT (2.5 mM) and NaCl (50 mM) was used for the Cathepsin S assay. Ki Determination for Genotypes 1b and 3a NS3 Protease Purified NS3 protease domain (amino acids 1-181) of the genotype 1b and 3a virus were generated as above. The internally quenched fluorogenic depsipeptide substrate Ac-DED(Edans)-EEAbuΨ[COO]ASK(Dabcyl)-NH2 and a synthetic peptide containing the hydrophobic core residues of the NS4A protein cofactor (KKGSVVIVGRIILSGRKK; NS4A peptide) were obtained from Anaspec, Inc. (San Jose, Calif.). Other chemicals and biochemicals were of reagent grade or better and were purchased from standard suppliers.Reactions were run at room temperature in buffer consisting of 50 mM HEPES, 40% glycerol, 0.05% Triton X-100, 10 mM DTT, and 10% DMSO. The final assay solutions contained 50 pM NS3 genotype 1 b protease or 200 pM genotype 3a protease, 20 μM NS4A peptide, and 4 μM substrate (genotype 1b) or 2 μM substrate (genotype 3a). Inhibitor concentrations varied from 100 nM to 5 pM in 3-fold dilutions, and no-inhibitor controls were included.Compound dilutions were made in DMSO at 20× final concentration. Reaction mixtures were prepared in 96-well assay plates. A solution of enzyme and NS4A peptide in assay buffer (25 μL volume with both reagents at 4× final concentration) was mixed with 45 μL assay buffer and 5 μL of either inhibitor or DMSO, and pre-incubated at room temperature for 1 hour. The reaction was started by addition of 25 μL substrate solution at 4× final concentration. Plates were mixed vigorously for 5-10 seconds and reactions were allowed to proceed for 90 minutes, fluorescence was measured every 30 s between 90 and 120 minutes reaction time using a Tecan InfiniTe M1000 or PerkinElmer Envision multimode plate reader with an excitation wavelength of 340 nm and an emission wavelength of 490 nm.Rates were calculated from the progress curves at steady state, in the time frame of 90-120 minutes after addition of substrate. To determine the Ki, rates were plotted as a function of inhibitor concentration, and the data were fit with equation 1 (Morrison, J. F., Biochimica et Biophysica Acta 1969, 185, 269-286) to calculate Ki app using GraphPad Prism 5. Active fraction of enzyme was determined by active site titration with known potent inhibitors. Ki was calculated from Ki app/(1+[[S]/Km]).
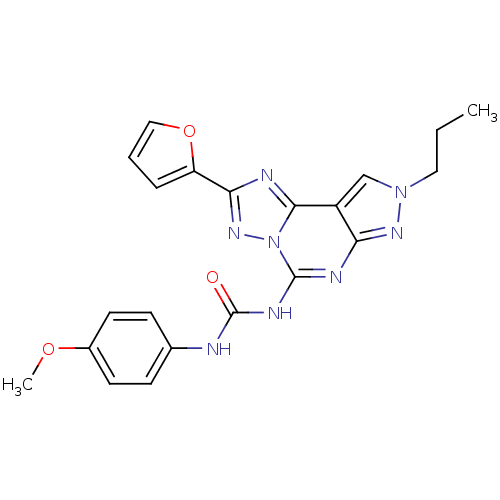 BDBM85618 CHEMBL302765 MRE 3008F20 J1.251.181G
BDBM85618 CHEMBL302765 MRE 3008F20 J1.251.181G US9062078, 269 US9695183, 269 BDBM164139 US9475819, 269 US9637496, 269
US9062078, 269 US9695183, 269 BDBM164139 US9475819, 269 US9637496, 269 US8653263, 269 US11059828, Example 269 US9586962, Example 269 BDBM118616 US11667644, Example 269 USRE48841, Example 269
US8653263, 269 US11059828, Example 269 US9586962, Example 269 BDBM118616 US11667644, Example 269 USRE48841, Example 269 US8987249, 269 US10238633, Example 269 US9713606, 269 BDBM150595
US8987249, 269 US10238633, Example 269 US9713606, 269 BDBM150595 BDBM254363 US10150765, Example 269 US10703749, Example 269 US9464084, 269 US10112937, Example 269
BDBM254363 US10150765, Example 269 US10703749, Example 269 US9464084, 269 US10112937, Example 269 US11369599, Compound 269 BDBM189699 US10213433, Compound 269 US20240043404, Example 269 US9174982, 269
US11369599, Compound 269 BDBM189699 US10213433, Compound 269 US20240043404, Example 269 US9174982, 269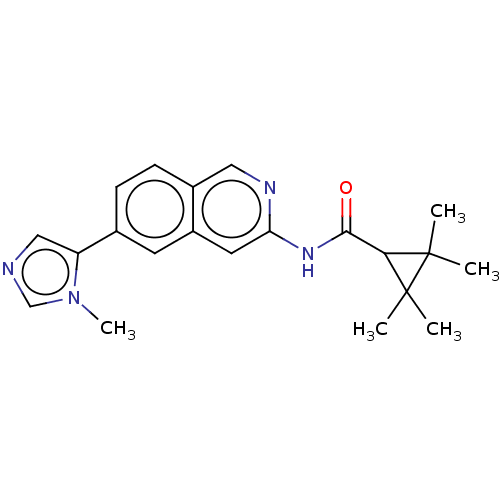 US11673881, Compound 269 US10556885, Compound 269 US11174244, Compound 269 US10508099, Compound 269 BDBM385113 US10287267, Compound 269
US11673881, Compound 269 US10556885, Compound 269 US11174244, Compound 269 US10508099, Compound 269 BDBM385113 US10287267, Compound 269 US10172845, Example 269 US10441581, Example 269 BDBM305253 US11648243, Example 269 US10144734, Example 269
US10172845, Example 269 US10441581, Example 269 BDBM305253 US11648243, Example 269 US10144734, Example 269 US10391089, Compound 269 US9675614, 269 US10307413, Compound 269 US10980794, Cmpd No 269 BDBM179219
US10391089, Compound 269 US9675614, 269 US10307413, Compound 269 US10980794, Cmpd No 269 BDBM179219 US11312704, Compound 269 US11124497, Compound 269 US11524940, Compound 269 US11472793, Compound 269 BDBM509821
US11312704, Compound 269 US11124497, Compound 269 US11524940, Compound 269 US11472793, Compound 269 BDBM509821 BDBM168870 US9079866, 269 US9745328, Compound 269 US9884878, Compound 269
BDBM168870 US9079866, 269 US9745328, Compound 269 US9884878, Compound 269 BDBM268236 US9549932, 269 US11529356, Example 269 US10772893, Example 269
BDBM268236 US9549932, 269 US11529356, Example 269 US10772893, Example 269 US10112931, Example 269 US9688672, Example 269 US9434719, 269 BDBM247667
US10112931, Example 269 US9688672, Example 269 US9434719, 269 BDBM247667 US10245267, Example 269 US9694016, 269 US10709712, Example 269 BDBM77822
US10245267, Example 269 US9694016, 269 US10709712, Example 269 BDBM77822 BDBM145079 US9303025, 269 US8952157, 269
BDBM145079 US9303025, 269 US8952157, 269 BDBM176236 US9688695, 269 US10047103, 269
BDBM176236 US9688695, 269 US10047103, 269 US8633188, 269 BDBM103382 US8546380, 269
US8633188, 269 BDBM103382 US8546380, 269 US9694002, 269 BDBM71147 US9546164, 269
US9694002, 269 BDBM71147 US9546164, 269 BDBM280116 US10028961, Compound 269 US10946023, Compound 269 US10172864, Compound 269
BDBM280116 US10028961, Compound 269 US10946023, Compound 269 US10172864, Compound 269 BDBM365547 US11414410, Example 269 US9868729, Example 269 US10533001, Example 269
BDBM365547 US11414410, Example 269 US9868729, Example 269 US10533001, Example 269 US10329302, Example 269 US11702424, Example 269 US10793579, Example 269 BDBM403116
US10329302, Example 269 US11702424, Example 269 US10793579, Example 269 BDBM403116 US10550105, Example 269 BDBM431324 US10870641, Example 269 US11014913, Example 269
US10550105, Example 269 BDBM431324 US10870641, Example 269 US11014913, Example 269 US9428502, 269 BDBM244659 US10226449, cpd 269 US20250017938, Compound 002-269
US9428502, 269 BDBM244659 US10226449, cpd 269 US20250017938, Compound 002-269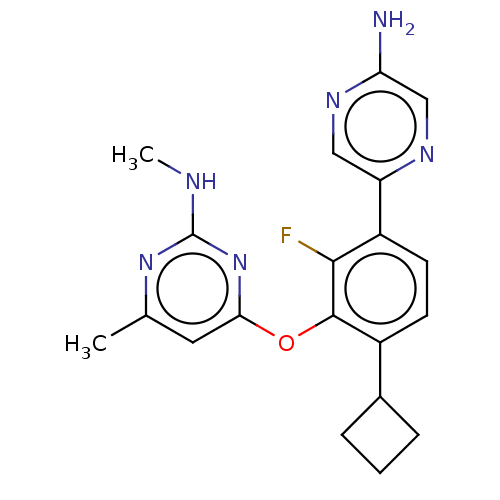 BDBM167188 US9073876, 269 US9732093, Compound 269
BDBM167188 US9073876, 269 US9732093, Compound 269 BDBM167778 US9073922, 269 US9796708, Example 269
BDBM167778 US9073922, 269 US9796708, Example 269 BDBM261048 US9540323, 269 US9540323, example 269
BDBM261048 US9540323, 269 US9540323, example 269 US10155731, Example 269 US9278944, 269 BDBM212578
US10155731, Example 269 US9278944, 269 BDBM212578 US11547697, Compound 269 BDBM130572 US9682141, 269
US11547697, Compound 269 BDBM130572 US9682141, 269 US9556187, Example 269 BDBM198472 US9216999, 269
US9556187, Example 269 BDBM198472 US9216999, 269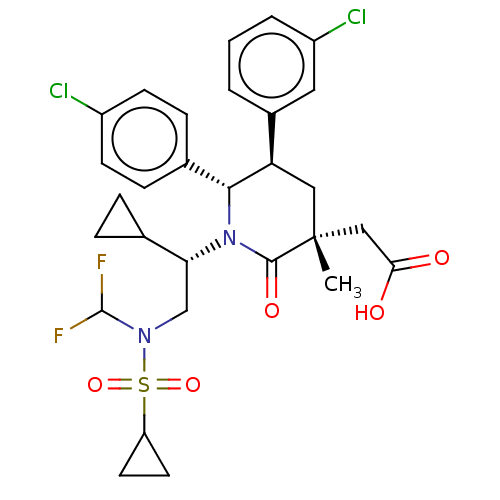 US9593129, Example 269 BDBM215308 US9296736, 269
US9593129, Example 269 BDBM215308 US9296736, 269 US9604984, Example 269 BDBM232723 US9346815, 269
US9604984, Example 269 BDBM232723 US9346815, 269 US9611261, Example 269 BDBM171635 US9085576, 269
US9611261, Example 269 BDBM171635 US9085576, 269 US9682966, 269 BDBM156476 US10118915, Compound 269
US9682966, 269 BDBM156476 US10118915, Compound 269 US9738649, Example 269 BDBM245030 US9422293, 269
US9738649, Example 269 BDBM245030 US9422293, 269 US11312711, Example 3 US9452980, 269 US10501411, Example 269 US11697636, Example 269 BDBM250359
US11312711, Example 3 US9452980, 269 US10501411, Example 269 US11697636, Example 269 BDBM250359 BDBM374149 US10577367, Example 269 US10246456, Example 269
BDBM374149 US10577367, Example 269 US10246456, Example 269 BDBM452130 US11555029, No. 269 US10710986, Example 269
BDBM452130 US11555029, No. 269 US10710986, Example 269 US10023570, Example 269 US10174027, Example 269 BDBM284259
US10023570, Example 269 US10174027, Example 269 BDBM284259 US10478424, Example 269 BDBM273486 US10071079, Example 269
US10478424, Example 269 BDBM273486 US10071079, Example 269 US10562891, Example 269 US11008308, Example 269 BDBM434350
US10562891, Example 269 US11008308, Example 269 BDBM434350 US10947215, Example 269 BDBM298863 US10125118, Example 269
US10947215, Example 269 BDBM298863 US10125118, Example 269 US10947252, Example 269 BDBM486821 US11427601, Example 269
US10947252, Example 269 BDBM486821 US11427601, Example 269 US11285140, Example 269 BDBM400513 US9999619, Example 269
US11285140, Example 269 BDBM400513 US9999619, Example 269 US11292767, Example 269 US10214492, Example 269 BDBM356918
US11292767, Example 269 US10214492, Example 269 BDBM356918 US11352329, COMPD # 269 BDBM461231 US10774053, Compound 269
US11352329, COMPD # 269 BDBM461231 US10774053, Compound 269 US11390631, Example 269 BDBM561732 US12268694, Example 269
US11390631, Example 269 BDBM561732 US12268694, Example 269 US20230279025, Example 269 US11453683, Example 269 BDBM573411
US20230279025, Example 269 US11453683, Example 269 BDBM573411 US20250017938, Compound 001-269 US8871790, 269 BDBM137201
US20250017938, Compound 001-269 US8871790, 269 BDBM137201 US9624228, Example 269 BDBM318312 US10301317, Example 269
US9624228, Example 269 BDBM318312 US10301317, Example 269 US11046698, Compound I-269 US10577373, Compound I-269 US10508120, Compound I-269 BDBM423814
US11046698, Compound I-269 US10577373, Compound I-269 US10508120, Compound I-269 BDBM423814 US11053244, Example 269 US10544143, Example 269 BDBM427831 US10544143, Example 264 US10730877, Example 269
US11053244, Example 269 US10544143, Example 269 BDBM427831 US10544143, Example 264 US10730877, Example 269 US10787452, Compound I-N-269 US10392391, Compound I-N-269 US11370798, Cmpd. # I-N-269 US11117900, Compound I-N-269 BDBM411834 US20230271963, Compound I-N-269
US10787452, Compound I-N-269 US10392391, Compound I-N-269 US11370798, Cmpd. # I-N-269 US11117900, Compound I-N-269 BDBM411834 US20230271963, Compound I-N-269 BDBM428940 US10533010, Example I-269 US11208415, Example I-269
BDBM428940 US10533010, Example I-269 US11208415, Example I-269 BDBM476811 US11345716, Compound III-269 US10870660, Compound III-269
BDBM476811 US11345716, Compound III-269 US10870660, Compound III-269 BDBM71145 US9694002, 269 US9546164, 267 US9546164, 269 US9694002, 267
BDBM71145 US9694002, 269 US9546164, 267 US9546164, 269 US9694002, 267 US10479784, Compound IA-269 BDBM420769 US10961232, Compound IA-269
US10479784, Compound IA-269 BDBM420769 US10961232, Compound IA-269 US10633389, Example 269-1a BDBM439960 US20230279020, Example 269-1a
US10633389, Example 269-1a BDBM439960 US20230279020, Example 269-1a US11352356, Compound I-269 US10730874, Compound I-269 BDBM455260
US11352356, Compound I-269 US10730874, Compound I-269 BDBM455260 US11396508, Compound I-269 US10647713, Compound I-269 BDBM442308
US11396508, Compound I-269 US10647713, Compound I-269 BDBM442308 US20230279020, Example 269-3 US10633389, Example 269-3 BDBM440603
US20230279020, Example 269-3 US10633389, Example 269-3 BDBM440603 US9732060, Compound I-269 US10065941, Compound I-269 BDBM272048
US9732060, Compound I-269 US10065941, Compound I-269 BDBM272048 US9802960, Compound I-269 BDBM338094 US9751854, Compound I-269
US9802960, Compound I-269 BDBM338094 US9751854, Compound I-269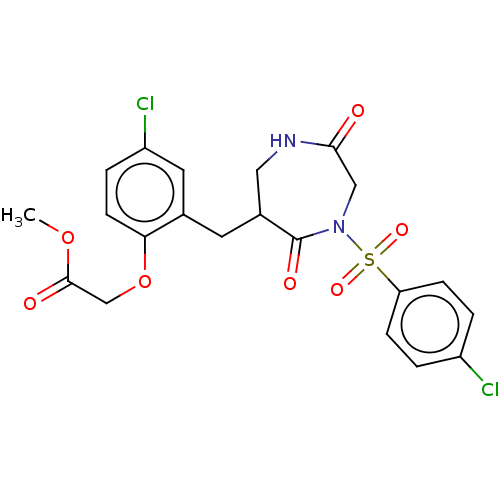 BDBM100752 US8507714, 269
BDBM100752 US8507714, 269 BDBM110741 US8614206, 269
BDBM110741 US8614206, 269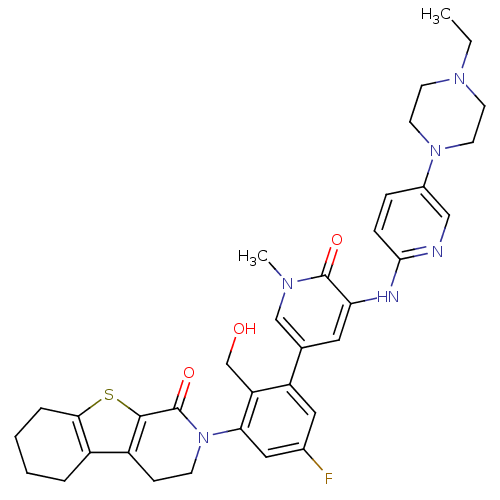 BDBM111957 US8618107, 269
BDBM111957 US8618107, 269 BDBM125421 US8772480, 269
BDBM125421 US8772480, 269 BDBM126459 US8778951, 269
BDBM126459 US8778951, 269 BDBM127491 US8791131, 269
BDBM127491 US8791131, 269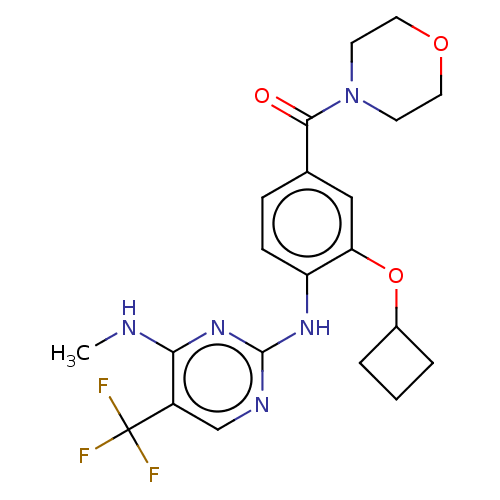 BDBM129166 US8802674, 269
BDBM129166 US8802674, 269 BDBM132979 US8846746, 269
BDBM132979 US8846746, 269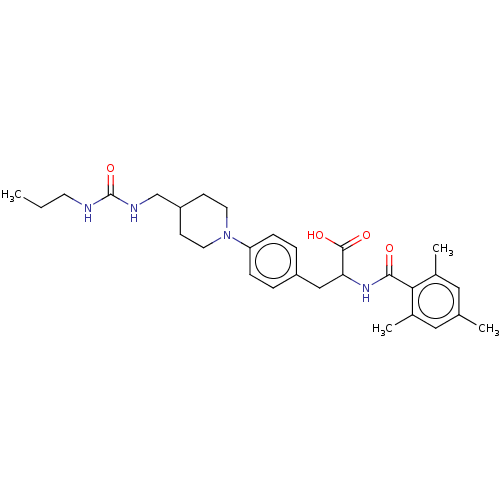 BDBM141994 US8927534, 269
BDBM141994 US8927534, 269 BDBM142813 US8940736, 269
BDBM142813 US8940736, 269 BDBM144068 US8969325, 269
BDBM144068 US8969325, 269 BDBM145485 US8952169, 269
BDBM145485 US8952169, 269 BDBM147431 US8957093, 269
BDBM147431 US8957093, 269 BDBM154253 US9012443, 269
BDBM154253 US9012443, 269 BDBM156837 US9029559, 269
BDBM156837 US9029559, 269 BDBM164802 US9067871, 269
BDBM164802 US9067871, 269 BDBM169919 US9073940, 269
BDBM169919 US9073940, 269 BDBM172609 US9090628, 269
BDBM172609 US9090628, 269 BDBM175724 US9688680, 269
BDBM175724 US9688680, 269 BDBM191499 US9181272, 269
BDBM191499 US9181272, 269 BDBM201838 US9233979, 269
BDBM201838 US9233979, 269 BDBM203090 US9242970, 269
BDBM203090 US9242970, 269 BDBM205800 US9255090, 269
BDBM205800 US9255090, 269 BDBM214206 US9283222, 269
BDBM214206 US9283222, 269 BDBM216069 US9302989, 269
BDBM216069 US9302989, 269 BDBM220680 US9296741, 269
BDBM220680 US9296741, 269 BDBM227318 US9328106, 269
BDBM227318 US9328106, 269 BDBM232448 US9346795, 269
BDBM232448 US9346795, 269 BDBM240554 US9409866, 269
BDBM240554 US9409866, 269 BDBM249038 US9434725, 269
BDBM249038 US9434725, 269 BDBM256087 US9481672, 269
BDBM256087 US9481672, 269 BDBM257800 US9493446, 269
BDBM257800 US9493446, 269 BDBM258834 US9499482, 269
BDBM258834 US9499482, 269 BDBM262417 US9708336, 269
BDBM262417 US9708336, 269 BDBM70576 US9693992, 269
BDBM70576 US9693992, 269 BDBM70771 US9693997, 269
BDBM70771 US9693997, 269 US8481733, 269 BDBM98457
US8481733, 269 BDBM98457 US8501936, 269 BDBM99753
US8501936, 269 BDBM99753 US8592431, 269 BDBM107401
US8592431, 269 BDBM107401 US8604016, 269 BDBM108689
US8604016, 269 BDBM108689 US8623889, 269 BDBM112271
US8623889, 269 BDBM112271 US8637500, 269 BDBM115412
US8637500, 269 BDBM115412 US8637532, 269 BDBM116577
US8637532, 269 BDBM116577 US8722692, 269 BDBM121830
US8722692, 269 BDBM121830 US8759532, 269 BDBM124585
US8759532, 269 BDBM124585 US8772305, 269 BDBM125874
US8772305, 269 BDBM125874 US8796244, 269 BDBM128676
US8796244, 269 BDBM128676 US8841312, 269 BDBM132520
US8841312, 269 BDBM132520 US8846698, 269 BDBM134803
US8846698, 269 BDBM134803 US8846719, 269 BDBM133577
US8846719, 269 BDBM133577 US8853258, 269 BDBM30582
US8853258, 269 BDBM30582 US8871934, 269 BDBM137676
US8871934, 269 BDBM137676 US8912224, 269 BDBM140833
US8912224, 269 BDBM140833 US8957068, 269 BDBM146217
US8957068, 269 BDBM146217 US8962648, 269 BDBM149036
US8962648, 269 BDBM149036 US9012651, 269 BDBM155824
US9012651, 269 BDBM155824 US9023865, 269 BDBM157428
US9023865, 269 BDBM157428 US9023882, 269 BDBM157976
US9023882, 269 BDBM157976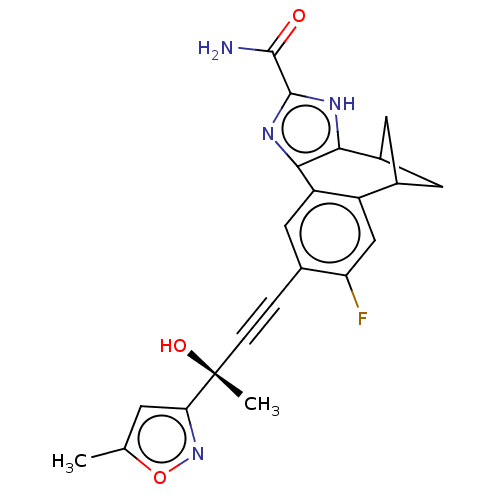 US9034866, 269 BDBM158794
US9034866, 269 BDBM158794 US9085555, 269 BDBM170841
US9085555, 269 BDBM170841 US9120749, 269 BDBM176903
US9120749, 269 BDBM176903 US9145354, 269 BDBM183607
US9145354, 269 BDBM183607 US9145392, 269 BDBM182568
US9145392, 269 BDBM182568 US9169246, 269 BDBM188727
US9169246, 269 BDBM188727 US9169252, 269 BDBM187718
US9169252, 269 BDBM187718 US9169260, 269 BDBM189003
US9169260, 269 BDBM189003 US9187424, 269 BDBM201502
US9187424, 269 BDBM201502 US9221809, 269 BDBM203718
US9221809, 269 BDBM203718 US9226922, 269 BDBM199633
US9226922, 269 BDBM199633 US9242996, 269 BDBM204195
US9242996, 269 BDBM204195 US9260425, 269 BDBM206516
US9260425, 269 BDBM206516 US9260439, 269 BDBM207385
US9260439, 269 BDBM207385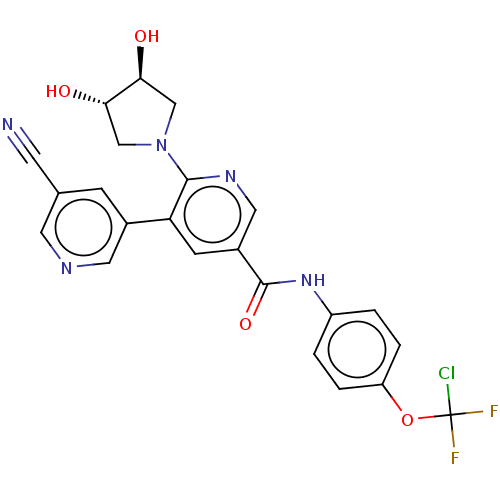 US9278981, 269 BDBM213677
US9278981, 269 BDBM213677 US9290451, 269 BDBM210570
US9290451, 269 BDBM210570 US9321756, 269 BDBM225003
US9321756, 269 BDBM225003 US9328096, 269 BDBM226576
US9328096, 269 BDBM226576 US9334269, 269 BDBM229942
US9334269, 269 BDBM229942 US9340517, 269 BDBM231882
US9340517, 269 BDBM231882 US9353090, 269 BDBM234138
US9353090, 269 BDBM234138 US9394297, 269 BDBM238610
US9394297, 269 BDBM238610 US9434711, 269 BDBM248405
US9434711, 269 BDBM248405 US9452986, 269 BDBM250965
US9452986, 269 BDBM250965 US9458110, 269 BDBM252197
US9458110, 269 BDBM252197 US9458171, 269 BDBM253309
US9458171, 269 BDBM253309 US9663469, 269 BDBM327722
US9663469, 269 BDBM327722 US9675593, 269 BDBM170316
US9675593, 269 BDBM170316 US9682940, 269 BDBM118125
US9682940, 269 BDBM118125 US9682976, 269 BDBM161832
US9682976, 269 BDBM161832 US9695118, 269 BDBM77702
US9695118, 269 BDBM77702 US10988451, Example 273 BDBM449656 US11643400, Example 269 US10703733, Example 269
US10988451, Example 273 BDBM449656 US11643400, Example 269 US10703733, Example 269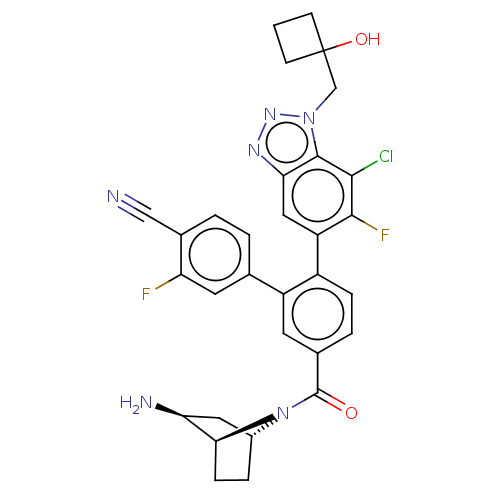 US11510915, Example 269 US10723742, Example 269 BDBM456528 US10723742, Example 257
US11510915, Example 269 US10723742, Example 269 BDBM456528 US10723742, Example 257 US9688643, I-269 US9150546, I-269 US9718790, I-0269 BDBM183117
US9688643, I-269 US9150546, I-269 US9718790, I-0269 BDBM183117 US9790229, Compound 74 BDBM348351 US10577374, Compound 269 US9790229, Compound 269
US9790229, Compound 74 BDBM348351 US10577374, Compound 269 US9790229, Compound 269 (3aR,6aR)-1-(indoline-1- carbonyl)hexahydropyrrolo[3,4- b]pyrrole-5(1H)-carbonitrile US11390584, Example 269 BDBM404617 US10689345, Example 269 US11066365, Example 269 US10343992, Example 269
(3aR,6aR)-1-(indoline-1- carbonyl)hexahydropyrrolo[3,4- b]pyrrole-5(1H)-carbonitrile US11390584, Example 269 BDBM404617 US10689345, Example 269 US11066365, Example 269 US10343992, Example 269 BDBM105759 US8575197, I-269
BDBM105759 US8575197, I-269 BDBM116119 US8633183, E-269
BDBM116119 US8633183, E-269 BDBM280711 US10030020, Example 269
BDBM280711 US10030020, Example 269 BDBM295539 US10112929, Example 269
BDBM295539 US10112929, Example 269 BDBM297153 US10112941, Example 269
BDBM297153 US10112941, Example 269 BDBM326572 US9662327, Compound 269
BDBM326572 US9662327, Compound 269 BDBM337222 US9745291, Compound 269
BDBM337222 US9745291, Compound 269 BDBM339491 US10202339, Compound 269
BDBM339491 US10202339, Compound 269 BDBM340743 US9763922, Example 269
BDBM340743 US9763922, Example 269 BDBM343975 US9777008, Compound 269
BDBM343975 US9777008, Compound 269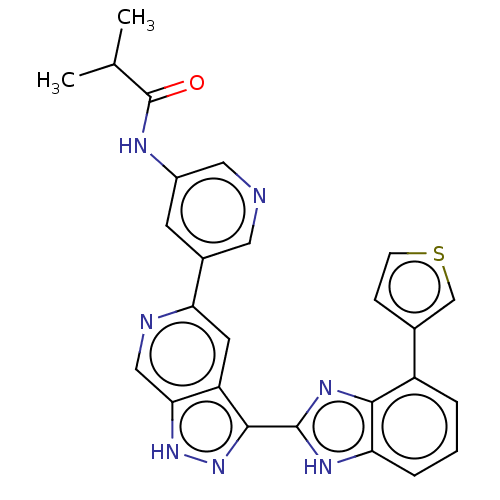 BDBM346315 US10202377, Compound 269
BDBM346315 US10202377, Compound 269 BDBM348061 US9790221, Compound 269
BDBM348061 US9790221, Compound 269 BDBM348546 US9790229, Compound 269
BDBM348546 US9790229, Compound 269 BDBM357513 US10214519, Example 269
BDBM357513 US10214519, Example 269 BDBM361895 US10221177, Compound 269
BDBM361895 US10221177, Compound 269 BDBM364811 US9862730, Example 269
BDBM364811 US9862730, Example 269 BDBM366341 US9873693, Compound 269
BDBM366341 US9873693, Compound 269 BDBM367581 US10227299, Example 269
BDBM367581 US10227299, Example 269 BDBM371162 US10239843, Example 269
BDBM371162 US10239843, Example 269 BDBM372009 US10239870, Example 269
BDBM372009 US10239870, Example 269 BDBM372862 US10246429, Example 269
BDBM372862 US10246429, Example 269 BDBM388120 US10294229, Example 269
BDBM388120 US10294229, Example 269 BDBM390382 US9951086, Example 269
BDBM390382 US9951086, Example 269 BDBM398792 US10323022, Example 269
BDBM398792 US10323022, Example 269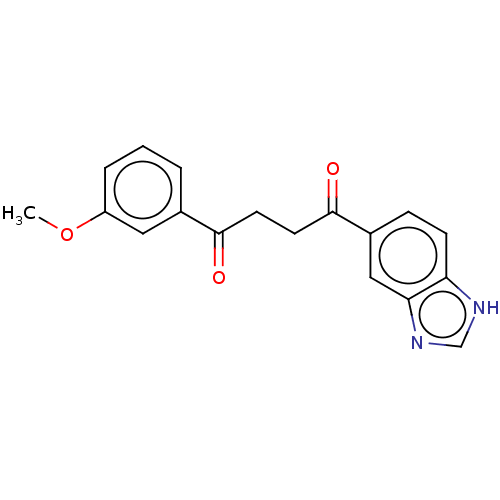 BDBM418349 US10457672, Compound 269
BDBM418349 US10457672, Compound 269 BDBM429592 US10544136, Compound 269
BDBM429592 US10544136, Compound 269 BDBM438014 US10597366, Compound 269
BDBM438014 US10597366, Compound 269 BDBM438702 US10618903, Example 269
BDBM438702 US10618903, Example 269 BDBM450088 US10703755, Example 269
BDBM450088 US10703755, Example 269 BDBM456540 US10723742, Example 269
BDBM456540 US10723742, Example 269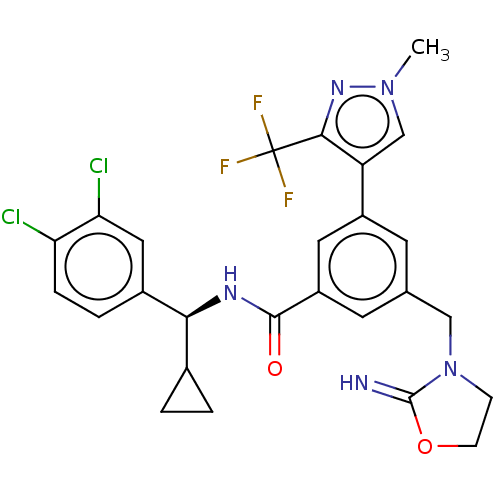 BDBM473538 US10844044, Example 269
BDBM473538 US10844044, Example 269 BDBM477160 US10881652, Example 269
BDBM477160 US10881652, Example 269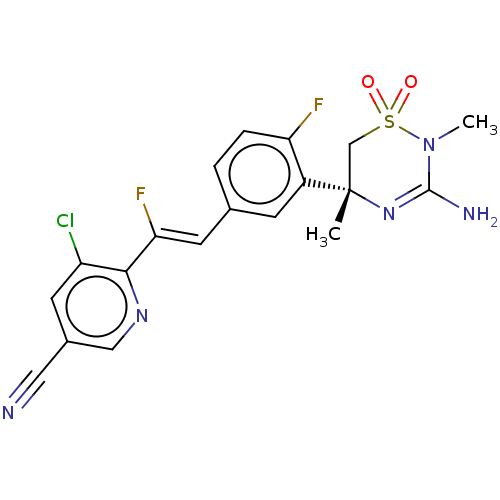 BDBM501350 US11021493, Example 269
BDBM501350 US11021493, Example 269 BDBM516179 US11053226, Example 269
BDBM516179 US11053226, Example 269 BDBM529863 US11203591, Example 269
BDBM529863 US11203591, Example 269 BDBM538361 US11254663, Example 269
BDBM538361 US11254663, Example 269 BDBM538926 US11254668, Example 269
BDBM538926 US11254668, Example 269 BDBM540338 US11261186, Example 269
BDBM540338 US11261186, Example 269 BDBM546996 US11292791, Example 269
BDBM546996 US11292791, Example 269 BDBM583377 US11524959, Compound 269.
BDBM583377 US11524959, Compound 269. BDBM588022 US11535621, Example 269
BDBM588022 US11535621, Example 269 BDBM588641 US11548892, Compound 269
BDBM588641 US11548892, Compound 269 BDBM595894 US11591336, Compound 269
BDBM595894 US11591336, Compound 269 BDBM596711 US11596639, Example 269
BDBM596711 US11596639, Example 269 BDBM607309 US11685745, Example 269
BDBM607309 US11685745, Example 269 BDBM613007 US11725000, Compound 269
BDBM613007 US11725000, Compound 269 BDBM615478 US20230271949, Example 269
BDBM615478 US20230271949, Example 269 BDBM619945 US20230295157, Example 269
BDBM619945 US20230295157, Example 269 BDBM631999 US20230348426, Example 269
BDBM631999 US20230348426, Example 269 BDBM637354 US20230382904, Compound 269
BDBM637354 US20230382904, Compound 269 BDBM641375 US11845723, Example 269
BDBM641375 US11845723, Example 269 BDBM647058 US20240025884, Example 269
BDBM647058 US20240025884, Example 269 BDBM648028 US20240025919, Compound 269
BDBM648028 US20240025919, Compound 269 BDBM659152 US20240092758, Example 269
BDBM659152 US20240092758, Example 269 BDBM660615 US20240092819, Compound 269
BDBM660615 US20240092819, Compound 269 BDBM661660 US20240101572, Example 269
BDBM661660 US20240101572, Example 269 BDBM670325 US11970474, Example 269
BDBM670325 US11970474, Example 269 BDBM677932 US20240174674, Compound 269
BDBM677932 US20240174674, Compound 269 BDBM682518 US20240207300, Compound 269
BDBM682518 US20240207300, Compound 269 BDBM687248 US20240246964, Compound 269
BDBM687248 US20240246964, Compound 269 BDBM689782 US20240262804, Example 269
BDBM689782 US20240262804, Example 269 BDBM695103 US20240294551, Example 269
BDBM695103 US20240294551, Example 269 BDBM716241 US12209085, Example 269
BDBM716241 US12209085, Example 269 BDBM718414 US20250042889, Example 269
BDBM718414 US20250042889, Example 269 BDBM722007 US20250059174, Example 269
BDBM722007 US20250059174, Example 269 BDBM723157 US20250059220, Compound 269
BDBM723157 US20250059220, Compound 269 BDBM723693 US20250064789, Compound 269
BDBM723693 US20250064789, Compound 269 BDBM724379 US20250066350, Example 269
BDBM724379 US20250066350, Example 269 BDBM726955 US20250084074, Example 269
BDBM726955 US20250084074, Example 269 BDBM733966 US20250127903, Compound 269
BDBM733966 US20250127903, Compound 269 BDBM734722 US20250129067, Compound 269
BDBM734722 US20250129067, Compound 269 BDBM745787 US12319655, Example 269
BDBM745787 US12319655, Example 269 BDBM749216 US20250188079, Compound 269
BDBM749216 US20250188079, Compound 269 BDBM749646 US12331033, Example 269
BDBM749646 US12331033, Example 269 BDBM753856 US20250206717, Example 269
BDBM753856 US20250206717, Example 269 BDBM758088 US20250230147, Compound 269
BDBM758088 US20250230147, Compound 269 BDBM761731 US12378224, Example 269
BDBM761731 US12378224, Example 269 US10065950, Example 269 BDBM272425
US10065950, Example 269 BDBM272425 US10072001, Example 269 BDBM277077
US10072001, Example 269 BDBM277077 US10112899, Example 269 BDBM294798
US10112899, Example 269 BDBM294798 US10189854, Compound 269 BDBM332305
US10189854, Compound 269 BDBM332305 US10259811, Example 269 BDBM377251
US10259811, Example 269 BDBM377251 US10280166, Compound 269 BDBM384007
US10280166, Compound 269 BDBM384007 US10336717, Compound 269 BDBM403984
US10336717, Compound 269 BDBM403984 US10377770, Example 269 BDBM410448
US10377770, Example 269 BDBM410448 US10435369, Example 269 BDBM414382
US10435369, Example 269 BDBM414382 US10544143, Example 269 BDBM427836
US10544143, Example 269 BDBM427836 US10562878, Compound 269 BDBM433712
US10562878, Compound 269 BDBM433712 US10660877, Example 269 BDBM443684
US10660877, Example 269 BDBM443684 US10710967, Example 269 BDBM451671
US10710967, Example 269 BDBM451671 US10730863, Example 269 BDBM453877
US10730863, Example 269 BDBM453877 US10781211, Example 269 BDBM464032
US10781211, Example 269 BDBM464032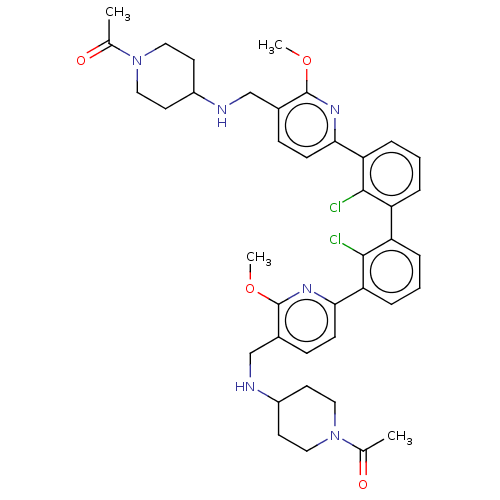 US10899735, No. 269 BDBM479810
US10899735, No. 269 BDBM479810 US10934302, Example 269 BDBM485018
US10934302, Example 269 BDBM485018 US10961200, Compound 269 BDBM488890
US10961200, Compound 269 BDBM488890 US10975056, Example 269 BDBM491316
US10975056, Example 269 BDBM491316 US11034692, Compound 269 BDBM504005
US11034692, Compound 269 BDBM504005 US11225469, Compound 269 BDBM533744
US11225469, Compound 269 BDBM533744 US11286268, Compound 269 BDBM544785
US11286268, Compound 269 BDBM544785 US11420970, Example 269 BDBM568067
US11420970, Example 269 BDBM568067 US11427558, Example 269 BDBM569122
US11427558, Example 269 BDBM569122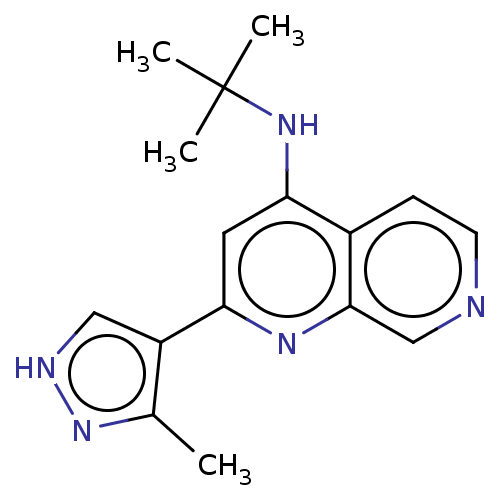 US11458138, Example 269 BDBM574389
US11458138, Example 269 BDBM574389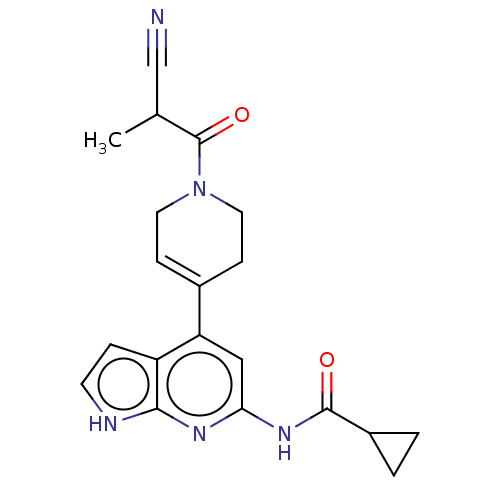 US11524968, Example 269 BDBM584521
US11524968, Example 269 BDBM584521 US11584747, Example 269 BDBM594464
US11584747, Example 269 BDBM594464 US11649255, Example 269 BDBM603296
US11649255, Example 269 BDBM603296 US11654147, Compound 269 BDBM603829
US11654147, Compound 269 BDBM603829 US11773078, Example 269 BDBM621517
US11773078, Example 269 BDBM621517 US11780845, Example 269 BDBM623250
US11780845, Example 269 BDBM623250 US11802133, Example 269 BDBM631332
US11802133, Example 269 BDBM631332 US11814367, Compound 269 BDBM634820
US11814367, Compound 269 BDBM634820 US11834453, Example 269 BDBM638166
US11834453, Example 269 BDBM638166 US11834467, Example 269 BDBM638674
US11834467, Example 269 BDBM638674 US11912686, Compound 269 BDBM654323
US11912686, Compound 269 BDBM654323 US11964953, Example 269 BDBM669004
US11964953, Example 269 BDBM669004 US12128028, Example 269 BDBM701873
US12128028, Example 269 BDBM701873 US12384753, Example 269 BDBM763246
US12384753, Example 269 BDBM763246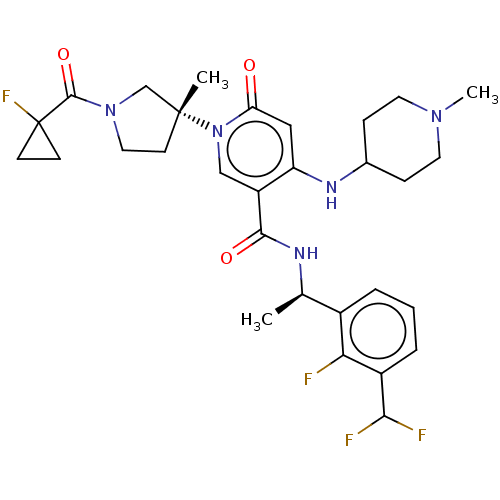 US20230348424, Example 269 BDBM631686
US20230348424, Example 269 BDBM631686 US20230348450, Example 269 BDBM632881
US20230348450, Example 269 BDBM632881 US20240002391, Compound 269 BDBM643433
US20240002391, Compound 269 BDBM643433 US20240025883, Example 269 BDBM646764
US20240025883, Example 269 BDBM646764 US20240043427, Example 269 BDBM650181
US20240043427, Example 269 BDBM650181 US20240116910, Compound 269 BDBM665585
US20240116910, Compound 269 BDBM665585 US20240116946, Example 269 BDBM666236
US20240116946, Example 269 BDBM666236 US20240140931, Compound 269 BDBM670565
US20240140931, Compound 269 BDBM670565 US20240174662, Example 269 BDBM677605
US20240174662, Example 269 BDBM677605 US20240199605, Example 269 BDBM681710
US20240199605, Example 269 BDBM681710 US20240218021, Example 269 BDBM684346
US20240218021, Example 269 BDBM684346 US20240245673, Example 269 BDBM686117
US20240245673, Example 269 BDBM686117 US20240246937, Example 269 BDBM686825
US20240246937, Example 269 BDBM686825 US20240368104, Example 269 BDBM703322
US20240368104, Example 269 BDBM703322 US20250026748, Compound 269 BDBM715105
US20250026748, Compound 269 BDBM715105 US20250074891, Compound 269 BDBM725519
US20250074891, Compound 269 BDBM725519 US20250122174, Example 269 BDBM730686
US20250122174, Example 269 BDBM730686 US20250129078, Compound 269 BDBM735251
US20250129078, Compound 269 BDBM735251 US20250129103, Compound 269 BDBM735681
US20250129103, Compound 269 BDBM735681 US20250145606, Example 269 BDBM738710
US20250145606, Example 269 BDBM738710 US20250145633, Example 269 BDBM739023
US20250145633, Example 269 BDBM739023 US20250170122, Compound 269 BDBM744342
US20250170122, Compound 269 BDBM744342 US20250197382, Compound 269 BDBM752003
US20250197382, Compound 269 BDBM752003 US20250221979, Example 269 BDBM756957
US20250221979, Example 269 BDBM756957 US20250236608, Example 269 BDBM759675
US20250236608, Example 269 BDBM759675 US20250243180, Compound 269 BDBM761277
US20250243180, Compound 269 BDBM761277 US8575197, II-269 BDBM106356
US8575197, II-269 BDBM106356 US8653087, III-269 BDBM117589
US8653087, III-269 BDBM117589 US8735386, I-269 BDBM122799
US8735386, I-269 BDBM122799 US9212130, I-269 BDBM196349
US9212130, I-269 BDBM196349 US9771320, Example 269 BDBM342035
US9771320, Example 269 BDBM342035 US9926282, Example 269 BDBM380274
US9926282, Example 269 BDBM380274 WO2022086828, Example 269 BDBM534533
WO2022086828, Example 269 BDBM534533 (S)-4-((1-benzylpyrrolidin-3-yl)oxy)-2,6-difluoro-3-methyl-N-(thiazol-4-yl)benzenesulfonamide hydrochloride US11299490, Example 269 US10815229, Example 269 US10662184, Example 269 US10246453, Example 269 BDBM374002
(S)-4-((1-benzylpyrrolidin-3-yl)oxy)-2,6-difluoro-3-methyl-N-(thiazol-4-yl)benzenesulfonamide hydrochloride US11299490, Example 269 US10815229, Example 269 US10662184, Example 269 US10246453, Example 269 BDBM374002 BDBM195202 US9212153, 269,Ex. 221
BDBM195202 US9212153, 269,Ex. 221 BDBM246625 US9550763, Compound I-269
BDBM246625 US9550763, Compound I-269 BDBM306107 US10144742, Compound I-269
BDBM306107 US10144742, Compound I-269 BDBM321540 US10183021, Compound I-269
BDBM321540 US10183021, Compound I-269 BDBM323872 US10189809, Compound I-269
BDBM323872 US10189809, Compound I-269 BDBM330707 US9725442, Compound I-269
BDBM330707 US9725442, Compound I-269 BDBM346670 US10202379, Reference Example 269
BDBM346670 US10202379, Reference Example 269 BDBM351168 US9796700, Compound I-269
BDBM351168 US9796700, Compound I-269 BDBM387539 US9938267, Cmpd ID 269
BDBM387539 US9938267, Cmpd ID 269 BDBM405799 US10351547, Compound 2-269
BDBM405799 US10351547, Compound 2-269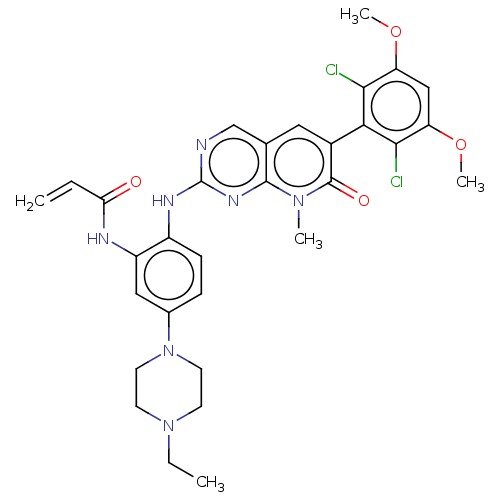 BDBM438588 US10618902, Compound I-269
BDBM438588 US10618902, Compound I-269 BDBM441332 US10640495, Example I-269
BDBM441332 US10640495, Example I-269 BDBM464935 US10793563, Compound I-269
BDBM464935 US10793563, Compound I-269 BDBM465618 US10793568, Compound I-269
BDBM465618 US10793568, Compound I-269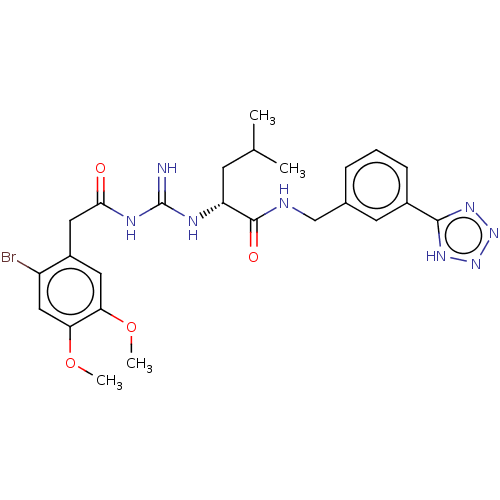 BDBM50055923 US9884814, Compound 269 CHEMBL3321941
BDBM50055923 US9884814, Compound 269 CHEMBL3321941 BDBM529057 US11198695, Example I-269
BDBM529057 US11198695, Example I-269 BDBM529332 US11198695, Example II-269
BDBM529332 US11198695, Example II-269 BDBM554820 US11339144, Compound I-269
BDBM554820 US11339144, Compound I-269 BDBM589528 US11555012, Compound I-269
BDBM589528 US11555012, Compound I-269 BDBM617331 US11753403, Compound I-269
BDBM617331 US11753403, Compound I-269 BDBM640712 US20230399319, Example 2-269
BDBM640712 US20230399319, Example 2-269 BDBM719323 US12221453, Compound I-269
BDBM719323 US12221453, Compound I-269 BDBM727408 US20250090540, Example I-269
BDBM727408 US20250090540, Example I-269 BDBM741404 US20250162989, Compound I-269
BDBM741404 US20250162989, Compound I-269 BDBM750162 US12331046, Compound I-269
BDBM750162 US12331046, Compound I-269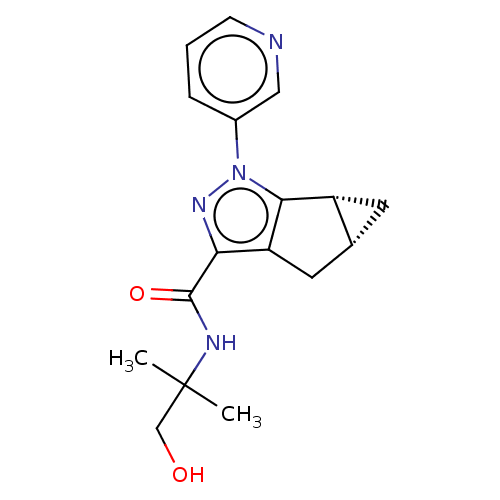 CHEMBL3354975 BDBM50040970 US11214548, Compound 269
CHEMBL3354975 BDBM50040970 US11214548, Compound 269 US10150728, Example I-269 BDBM307056
US10150728, Example I-269 BDBM307056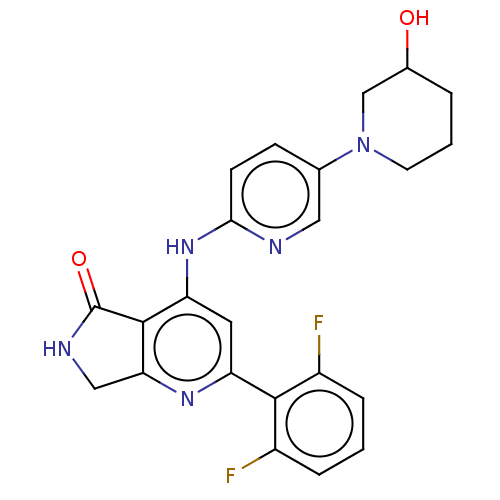 US10196390, Compound I-269 BDBM332980
US10196390, Compound I-269 BDBM332980 US10597377, Compound 2-269 BDBM438264
US10597377, Compound 2-269 BDBM438264 US10696692, Example 269 BDBM50011278 CHEMBL3260356
US10696692, Example 269 BDBM50011278 CHEMBL3260356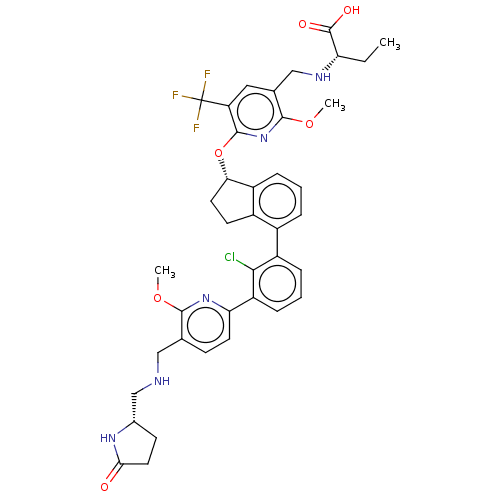 US10774071, Example B-269 BDBM461802
US10774071, Example B-269 BDBM461802 US10919885, Compound No. 269 BDBM483120
US10919885, Compound No. 269 BDBM483120 US11186577, Cmpd. No. 269 BDBM526921
US11186577, Cmpd. No. 269 BDBM526921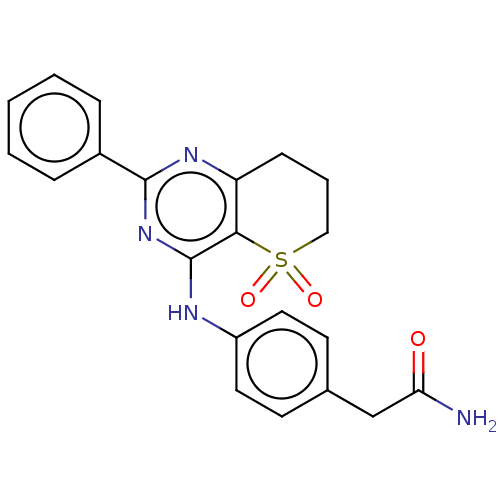 US11401286, Example 269 CHEMBL2385753 BDBM50491738
US11401286, Example 269 CHEMBL2385753 BDBM50491738 US11420958, Ex. No. 269 BDBM566900
US11420958, Ex. No. 269 BDBM566900 US11458149, Compound I-269 BDBM574655
US11458149, Compound I-269 BDBM574655 US11518764, Compound I-269 BDBM582260
US11518764, Compound I-269 BDBM582260 US11685732, Compound I-269 BDBM606625
US11685732, Compound I-269 BDBM606625 US12053473, Example I-269 BDBM689429
US12053473, Example I-269 BDBM689429 US20230390274, Compound A-269 BDBM639066
US20230390274, Compound A-269 BDBM639066 US20240150321, Compound I-269 BDBM673958
US20240150321, Compound I-269 BDBM673958 US20250170126, Compound I-269 BDBM744637
US20250170126, Compound I-269 BDBM744637 US9409866, 268 BDBM240553 US9409866, 269
US9409866, 268 BDBM240553 US9409866, 269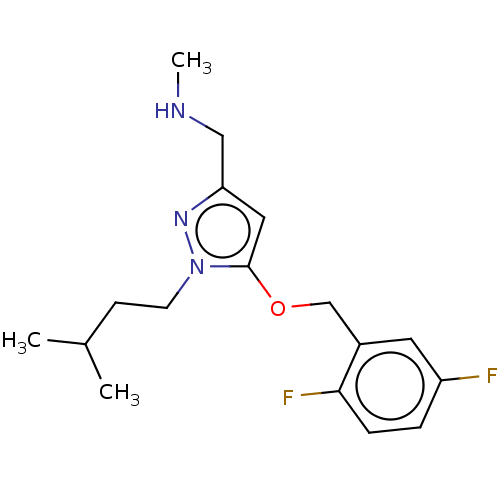 BDBM322097 1-{5-[(2,5-difluorobenzyl)oxy]-1-(3-methylbutyl)-1H-pyrazol-3-yl}-N-methylmethanamine US11576897, Example 269 US10183913, Example 269 US11312688, Example 269
BDBM322097 1-{5-[(2,5-difluorobenzyl)oxy]-1-(3-methylbutyl)-1H-pyrazol-3-yl}-N-methylmethanamine US11576897, Example 269 US10183913, Example 269 US11312688, Example 269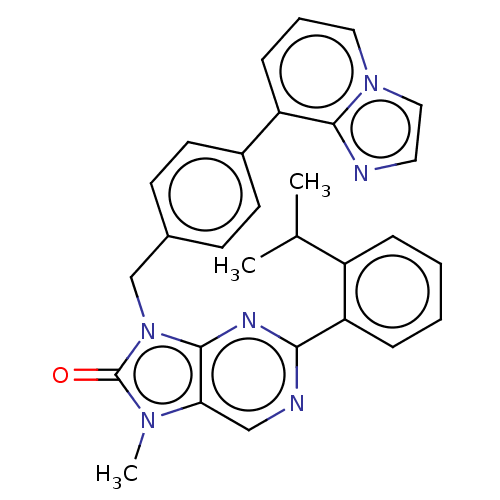 US10399980, Compound I-269 9-(4-(imidazo[1,2-a]pyridin-8-yl)benzyl)-2-(2- US10189841, Compound I-269 US11161848, Compound I-269 BDBM325063
US10399980, Compound I-269 9-(4-(imidazo[1,2-a]pyridin-8-yl)benzyl)-2-(2- US10189841, Compound I-269 US11161848, Compound I-269 BDBM325063 US10508083, Example 269 N-(cis-2-(((cis-4-(3- methoxyphenyl)cyclohexyl)oxy)- methyl)piperidin-3- yl)methanesulfonamide US11292766, Example 269 US10287305, Example 269 BDBM386832
US10508083, Example 269 N-(cis-2-(((cis-4-(3- methoxyphenyl)cyclohexyl)oxy)- methyl)piperidin-3- yl)methanesulfonamide US11292766, Example 269 US10287305, Example 269 BDBM386832 US11111242, Example 269 BDBM316773 2-[(1-acryloylpiperidin-4-yl)amino]-N-butyl-5H-pyrrolo[2,3-b]pyrazine-7-carboxamide US9617258, Example 269 US20250134869, Example 269
US11111242, Example 269 BDBM316773 2-[(1-acryloylpiperidin-4-yl)amino]-N-butyl-5H-pyrrolo[2,3-b]pyrazine-7-carboxamide US9617258, Example 269 US20250134869, Example 269 BDBM579391 US11479560, Title ARN-1-269
BDBM579391 US11479560, Title ARN-1-269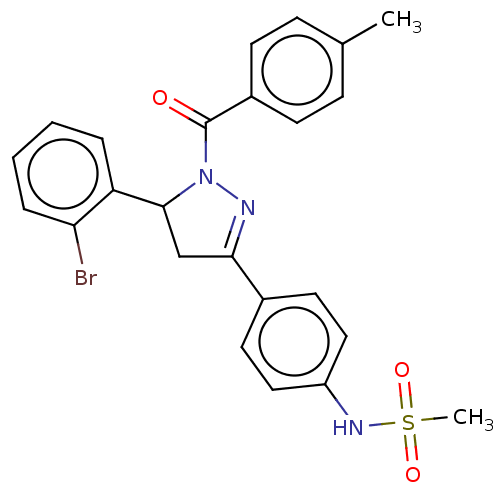 BDBM595517 US11591319, ID lqr-8-269
BDBM595517 US11591319, ID lqr-8-269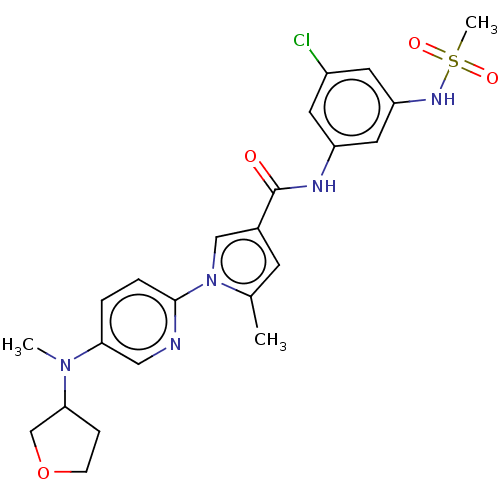 BDBM697487 BDBM697894 BDBM697893 US20240316047, Example 269
BDBM697487 BDBM697894 BDBM697893 US20240316047, Example 269 US11072582, Compound 269 (2S,5R)-1-(4-(2,6-dimethoxypyridin-3-yl)benzoyl)-5-(2-fluorophenyl)pyrrolidine-2-carboxylic acid US10781171, Compound 269 BDBM403991 US10017468, Compound 269
US11072582, Compound 269 (2S,5R)-1-(4-(2,6-dimethoxypyridin-3-yl)benzoyl)-5-(2-fluorophenyl)pyrrolidine-2-carboxylic acid US10781171, Compound 269 BDBM403991 US10017468, Compound 269 US10953005, Example 269 US10555944, Example 269 6-ethoxy-4-(6-(6-(6-oxo-1-propyl-1,6-dihydropyridine-3-carbonyl)-3,6-diazabicyclo[3.1.1]heptan-3-yl)pyridin-3-yl)pyrazolo[1,5-a]pyridine-3-carbonitrile 2,2,2-trifluoroacetate US10112942, Example 269 US10172851, Example 269 BDBM296535 US10137124, Example 269
US10953005, Example 269 US10555944, Example 269 6-ethoxy-4-(6-(6-(6-oxo-1-propyl-1,6-dihydropyridine-3-carbonyl)-3,6-diazabicyclo[3.1.1]heptan-3-yl)pyridin-3-yl)pyrazolo[1,5-a]pyridine-3-carbonitrile 2,2,2-trifluoroacetate US10112942, Example 269 US10172851, Example 269 BDBM296535 US10137124, Example 269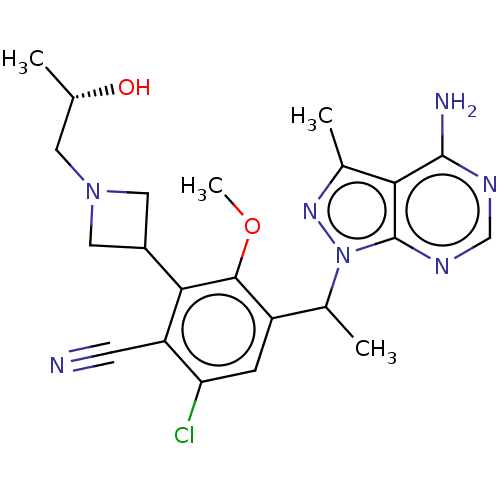 US9707233, 269 US11433071, Example 269 US10092570, Example 361 US10376513, Example 269 BDBM261246 4-[1-(4-Amino-3-methyl-1H-pyrazolo[3,4-d]pyrimidin-1-yl)ethyl]-6-chloro-2-{1-[(2S)-2-hydroxypropyl]azetidin-3-yl}-3-methoxybenzonitrile US10519163, Example 29 US10646492, Example 269 US10675284, Example 29 US11136326, Example 29 US9932341, Example 269 US9730939, Example 269
US9707233, 269 US11433071, Example 269 US10092570, Example 361 US10376513, Example 269 BDBM261246 4-[1-(4-Amino-3-methyl-1H-pyrazolo[3,4-d]pyrimidin-1-yl)ethyl]-6-chloro-2-{1-[(2S)-2-hydroxypropyl]azetidin-3-yl}-3-methoxybenzonitrile US10519163, Example 29 US10646492, Example 269 US10675284, Example 29 US11136326, Example 29 US9932341, Example 269 US9730939, Example 269 BDBM343669 US10442803, Example 269 US9777003, Example 269 US11091484, Example 269 N-(6-Benzyl-1,7-dimethyl-6H-pyrrolo[2,3- e][1,2,4]triazolo[4,3-a]pyridin-4- yl)cyclopropanecarboxamide
BDBM343669 US10442803, Example 269 US9777003, Example 269 US11091484, Example 269 N-(6-Benzyl-1,7-dimethyl-6H-pyrrolo[2,3- e][1,2,4]triazolo[4,3-a]pyridin-4- yl)cyclopropanecarboxamide BDBM191487 US9181272, 257 US9181272, 268 US9181272, 269
BDBM191487 US9181272, 257 US9181272, 268 US9181272, 269 BDBM210117 US9546153, ex. 269 US9546153, ex. 368
BDBM210117 US9546153, ex. 269 US9546153, ex. 368 BDBM450087 US10703755, Example 268 US10703755, Example 269
BDBM450087 US10703755, Example 268 US10703755, Example 269 BDBM629970 US20230340011, Example 269. US20230340011, Example 436.
BDBM629970 US20230340011, Example 269. US20230340011, Example 436. BDBM655109 US11912703, Example 270 US11912703, Example 269
BDBM655109 US11912703, Example 270 US11912703, Example 269 BDBM714073 US20250025443, Compound 269 US20250025443, Example 107
BDBM714073 US20250025443, Compound 269 US20250025443, Example 107 BDBM714235 US20250025443, Compound 405 US20250025443, Example 269
BDBM714235 US20250025443, Compound 405 US20250025443, Example 269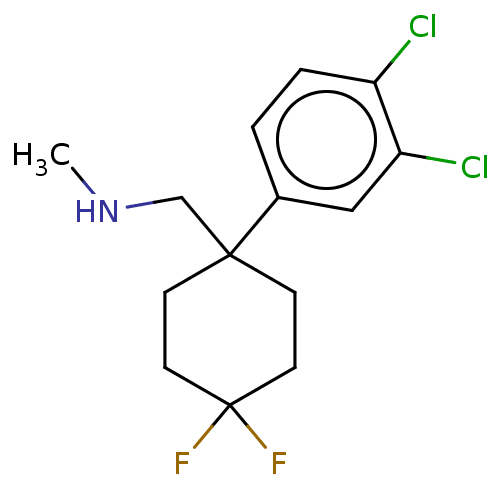 US10562878, Compound 268 US10562878, Compound 269 BDBM433711
US10562878, Compound 268 US10562878, Compound 269 BDBM433711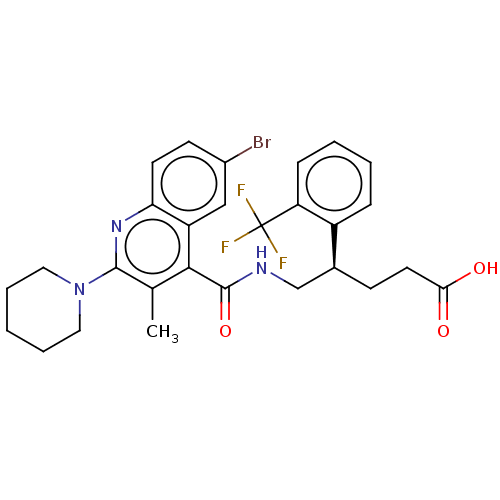 US11149018, Example 269 US11149018, Example 112 BDBM520828
US11149018, Example 269 US11149018, Example 112 BDBM520828 US11254668, Example 269 US11254668, Example 268 BDBM538925
US11254668, Example 269 US11254668, Example 268 BDBM538925 US11292791, Example 267 BDBM546994 US11292791, Example 269
US11292791, Example 267 BDBM546994 US11292791, Example 269 US9303033, J30, Table 37A, Compound 269 BDBM219595
US9303033, J30, Table 37A, Compound 269 BDBM219595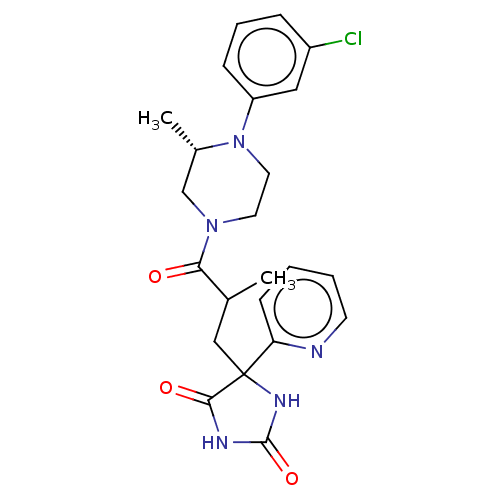 US9926281, Compound 446 BDBM379839 US9926281, Compound 269
US9926281, Compound 446 BDBM379839 US9926281, Compound 269 BDBM276330 US10071988, Example 269 2-[[1-[3-(Difluoromethyl)phenyl]triazol-4-yl]methoxy]-5-fluoro- US10233173, Example 269
BDBM276330 US10071988, Example 269 2-[[1-[3-(Difluoromethyl)phenyl]triazol-4-yl]methoxy]-5-fluoro- US10233173, Example 269 N-((1R,2R)-2-Aminocyclopentyl)-4-oxo-5-(6-phenoxypyridin-3-yl)- 4,5-dihydro-3H-1-thia-3,5,8-triazaacenaphthylene-2-carboxamide US12065446, Example 269 BDBM485435 US11319329, Ex # 269 US10934310, Ex # 269
N-((1R,2R)-2-Aminocyclopentyl)-4-oxo-5-(6-phenoxypyridin-3-yl)- 4,5-dihydro-3H-1-thia-3,5,8-triazaacenaphthylene-2-carboxamide US12065446, Example 269 BDBM485435 US11319329, Ex # 269 US10934310, Ex # 269 BDBM681111 US12012467, Compound DI-791 US12012467, Example 269
BDBM681111 US12012467, Compound DI-791 US12012467, Example 269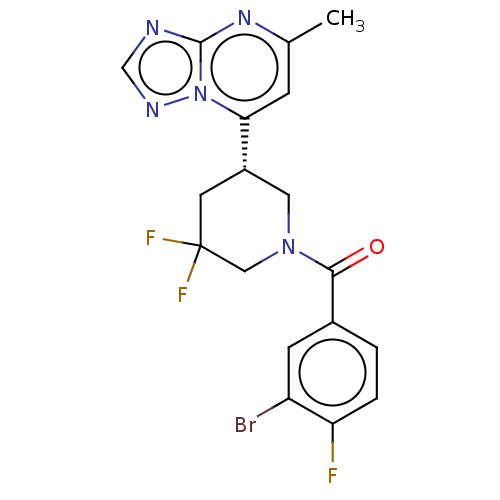 CHEMBL4074744 US11419874, DNS-8254 BDBM50236225 US11186582, Example 269
CHEMBL4074744 US11419874, DNS-8254 BDBM50236225 US11186582, Example 269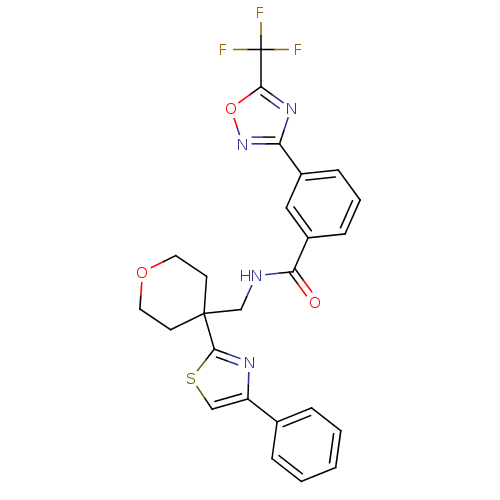 US10011611, TMP269 BDBM50446481 US10722597, Compound TMP-269 CHEMBL3110004
US10011611, TMP269 BDBM50446481 US10722597, Compound TMP-269 CHEMBL3110004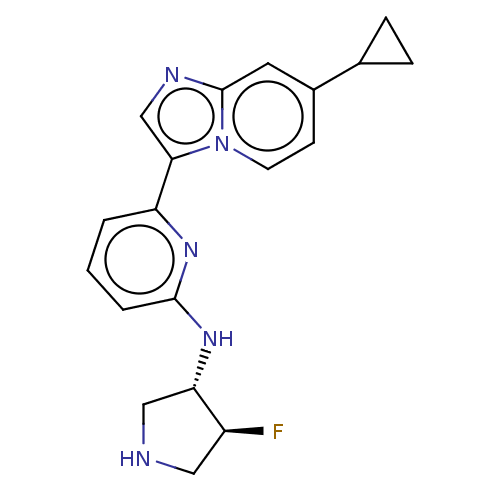 US20230303563, Compound 269 Exemplary Synthetic Procedure #38 BDBM621165
US20230303563, Compound 269 Exemplary Synthetic Procedure #38 BDBM621165 US9855269, Compound 116 US9592235, Example 269 BDBM50011559 CHEMBL3262578
US9855269, Compound 116 US9592235, Example 269 BDBM50011559 CHEMBL3262578 US11046691, Compound 269 1-(2-chlorophenyl)-4-(((1s,3s)-3- hydroxy-1-methylcyclobutyl)amino)-7- (trifluoromethyl)pyrido[2,3- d]pyrimidin-2(1H)-one US11130759, Cpd. No. 269 US11084798, Cpd No 269 BDBM508016
US11046691, Compound 269 1-(2-chlorophenyl)-4-(((1s,3s)-3- hydroxy-1-methylcyclobutyl)amino)-7- (trifluoromethyl)pyrido[2,3- d]pyrimidin-2(1H)-one US11130759, Cpd. No. 269 US11084798, Cpd No 269 BDBM508016 6-(4-(4-fluorophenyl)-1-((1r,3s,5R,7S)-3- hydroxyadamantan-1-yl)-1H-imidazol-5- yl)imidazo[1,2-b]pyridazine-3-carbonitrile US10287295, Example 269' US11352360, Example 269' BDBM283068 US9884868, Example 269'
6-(4-(4-fluorophenyl)-1-((1r,3s,5R,7S)-3- hydroxyadamantan-1-yl)-1H-imidazol-5- yl)imidazo[1,2-b]pyridazine-3-carbonitrile US10287295, Example 269' US11352360, Example 269' BDBM283068 US9884868, Example 269' BDBM239817 US9394311, 100 Roche-Dataset for PDE10A, Compound 269
BDBM239817 US9394311, 100 Roche-Dataset for PDE10A, Compound 269 Roche-Dataset for PDE10A, Compound 712 BDBM120668 US8703768, 269
Roche-Dataset for PDE10A, Compound 712 BDBM120668 US8703768, 269 US10206931, Example 269 (4R)-4-methyl-6-(2- BDBM349461
US10206931, Example 269 (4R)-4-methyl-6-(2- BDBM349461 US9187424, 76 US9187424, 33 US9187424, 269 US9187424, 53 BDBM201269
US9187424, 76 US9187424, 33 US9187424, 269 US9187424, 53 BDBM201269 N-[4-(2,4-difluorophenoxy)-3-(2,6-dimethyl-7- oxofuro[2,3-c]pyridin-4-yl) phenyl]ethanesulfonamide US10023592, Example 269 US10941160, Example 269 BDBM285224
N-[4-(2,4-difluorophenoxy)-3-(2,6-dimethyl-7- oxofuro[2,3-c]pyridin-4-yl) phenyl]ethanesulfonamide US10023592, Example 269 US10941160, Example 269 BDBM285224 BDBM500610 US11078201, Compound I-269 2-(4-(7-((5-(4- methylpiperazin- 1-yl)pyridin- 2-yl)amino)-1- oxoisoindolin- 4-yl)-lH- pyrrolo[2.3- b]pyridin-1- yl)acctamide US11548890, Compound I-269 US11021481, Compound I-269
BDBM500610 US11078201, Compound I-269 2-(4-(7-((5-(4- methylpiperazin- 1-yl)pyridin- 2-yl)amino)-1- oxoisoindolin- 4-yl)-lH- pyrrolo[2.3- b]pyridin-1- yl)acctamide US11548890, Compound I-269 US11021481, Compound I-269 US20230381148, Compound I-269 BDBM593577 US11578066, Compound I-269 1-({5-[5-(difluoromethyl)-1,3,4-oxadiazol-2- yl]-1,3-thiazol-2-yl]methyl)- 1H,2H,3H,4H,5H-pyrido[3,4-b]azepin-2-one US20240269137, Compound I-269
US20230381148, Compound I-269 BDBM593577 US11578066, Compound I-269 1-({5-[5-(difluoromethyl)-1,3,4-oxadiazol-2- yl]-1,3-thiazol-2-yl]methyl)- 1H,2H,3H,4H,5H-pyrido[3,4-b]azepin-2-one US20240269137, Compound I-269 BDBM382560 US10273259, Example 269 US10711020, Example 270 US10273259, Example 270
BDBM382560 US10273259, Example 269 US10711020, Example 270 US10273259, Example 270 US9745291, Compound 45 US9765068, Compound 45 BDBM336999 US9745291, Compound 269
US9745291, Compound 45 US9765068, Compound 45 BDBM336999 US9745291, Compound 269 6-chloro-3-(3-(4-chloro-3,5-dimethylphenoxy)propyl)-1-methyl-N-(phenylsulfonyl)-1H-indole-2-carboxamide US10093640, Example 269 BDBM289061 US10844032, Example 269
6-chloro-3-(3-(4-chloro-3,5-dimethylphenoxy)propyl)-1-methyl-N-(phenylsulfonyl)-1H-indole-2-carboxamide US10093640, Example 269 BDBM289061 US10844032, Example 269 US10927105, Ex 269 1-(4-(3-chloro-5- (pyrimidin-5- yl)benzyl)- piperazine-1- carbonyl)-1H- pyrazole-3- carboxylic acid BDBM483772 US11655217, Example 269
US10927105, Ex 269 1-(4-(3-chloro-5- (pyrimidin-5- yl)benzyl)- piperazine-1- carbonyl)-1H- pyrazole-3- carboxylic acid BDBM483772 US11655217, Example 269 US11077100, Example 269 BDBM510409 (5S,8S)-N- (4-chloro-2,6- difluorobenzyl)- 5-fluoro-8- hydroxy-8- methyl-5,6,7,8- tetrahydroquinoline- 5-carboxamide US11439633, Example 269
US11077100, Example 269 BDBM510409 (5S,8S)-N- (4-chloro-2,6- difluorobenzyl)- 5-fluoro-8- hydroxy-8- methyl-5,6,7,8- tetrahydroquinoline- 5-carboxamide US11439633, Example 269 US10023557, Example 269 N-((5-chloro-1H-pyrrolo[2,3-b]pyridin-3- yl)methyl)-2-((3-chloroquinolin-6- yl)methyl)isonicotinamide US10308637, Example 269 BDBM283622
US10023557, Example 269 N-((5-chloro-1H-pyrrolo[2,3-b]pyridin-3- yl)methyl)-2-((3-chloroquinolin-6- yl)methyl)isonicotinamide US10308637, Example 269 BDBM283622 US11485745, Example 269 US10730889, Example 269 BDBM455676 N-[[6-(quinoxaline-2-carbonyl)- 6-azaspiro[2.5]octan-2- y]]methyl]furo[2,3-c]pyridine-2- carboxamide
US11485745, Example 269 US10730889, Example 269 BDBM455676 N-[[6-(quinoxaline-2-carbonyl)- 6-azaspiro[2.5]octan-2- y]]methyl]furo[2,3-c]pyridine-2- carboxamide (S)-(1-(7-amino-2-(furan-2- US20240217978, Example 269 BDBM683613
(S)-(1-(7-amino-2-(furan-2- US20240217978, Example 269 BDBM683613 BDBM61987 cid_231244 KUC105790N KSC-6-269 2-(4-methoxyphenyl)-1H-indole
BDBM61987 cid_231244 KUC105790N KSC-6-269 2-(4-methoxyphenyl)-1H-indole US11479560, Title WZ-II-270 BDBM579374 US11479560, Title WZ-II-269
US11479560, Title WZ-II-270 BDBM579374 US11479560, Title WZ-II-269 6-(4-Fluoro-3-methyl-phenyl)-1-[(5-methyl-3-pyridyl)methyl]-3H- imidazo[4,5-b]pyridin-2-one US11207298, Example 269 US10617676, Example 269 BDBM436934
6-(4-Fluoro-3-methyl-phenyl)-1-[(5-methyl-3-pyridyl)methyl]-3H- imidazo[4,5-b]pyridin-2-one US11207298, Example 269 US10617676, Example 269 BDBM436934 BDBM292983 US10376514, Example 269 9-(1-Fluoro-2-morpholinoethyl)-6-(4-methoxybenzyl)pyrido[3,2-e][1,2,4]triazolo[1,5-c]pyrimidin-5(6H)-one US10105367, Example 269
BDBM292983 US10376514, Example 269 9-(1-Fluoro-2-morpholinoethyl)-6-(4-methoxybenzyl)pyrido[3,2-e][1,2,4]triazolo[1,5-c]pyrimidin-5(6H)-one US10105367, Example 269 (1R,3S)-3-(3-{[(5-methyl-1,2- oxazol-3-yl)acetyl]amino}-1H- pyrazol-5-yl)cyclopentyl propan-2-ylcarbamate US11014911, Example 269 US11718603, Example 269 BDBM498444
(1R,3S)-3-(3-{[(5-methyl-1,2- oxazol-3-yl)acetyl]amino}-1H- pyrazol-5-yl)cyclopentyl propan-2-ylcarbamate US11014911, Example 269 US11718603, Example 269 BDBM498444 BDBM418771 US10450297, Example 269 N-(2-hydroxyethyl)-4-({4-[({2- [methyl(methylsulfonyl)amino]pyridin-3- yl}methyl)amino]-5- (trifluoromethyl)pyrimidin-2-yl}amino)-N- propylbenzamide (269)
BDBM418771 US10450297, Example 269 N-(2-hydroxyethyl)-4-({4-[({2- [methyl(methylsulfonyl)amino]pyridin-3- yl}methyl)amino]-5- (trifluoromethyl)pyrimidin-2-yl}amino)-N- propylbenzamide (269) BDBM378140 N-((1r,4r)-4-aminocyclohexyl)pyrimidine-2-carboxamide US10266526, Compound 269
BDBM378140 N-((1r,4r)-4-aminocyclohexyl)pyrimidine-2-carboxamide US10266526, Compound 269 (S)-5-(2-Methyl-4-phenoxyphenyl)-4-oxo-N-(pyrrolidin-2-ylmethyl)- 4,5-dihydro-3H-1-thia-3,5,8-triazaacenaphthylene-2-carboxamide; US10822348, Example 269 BDBM467616 US10800792, Example 269
(S)-5-(2-Methyl-4-phenoxyphenyl)-4-oxo-N-(pyrrolidin-2-ylmethyl)- 4,5-dihydro-3H-1-thia-3,5,8-triazaacenaphthylene-2-carboxamide; US10822348, Example 269 BDBM467616 US10800792, Example 269 2-(azepan-1-yl)-4-((4-(2-(4- ethylpiperazin-1-yl)-2-oxoethoxy)phenyl) amino)pyrimido[4,5-d]pyridazin-5(6H)- one US10183944, Example 269 US10647720, Ex. # 269 BDBM322899
2-(azepan-1-yl)-4-((4-(2-(4- ethylpiperazin-1-yl)-2-oxoethoxy)phenyl) amino)pyrimido[4,5-d]pyridazin-5(6H)- one US10183944, Example 269 US10647720, Ex. # 269 BDBM322899 BDBM278047 2-(3-methoxyazetidin-1-yl)-5-(4-((4-(4-(oxetan-3-yl)piperazin-1-yl)phenyl)amino)-1,3,5-triazin-2-yl)benzonitrile US10253019, Example 269 US10040781, Example 269
BDBM278047 2-(3-methoxyazetidin-1-yl)-5-(4-((4-(4-(oxetan-3-yl)piperazin-1-yl)phenyl)amino)-1,3,5-triazin-2-yl)benzonitrile US10253019, Example 269 US10040781, Example 269 BDBM437535 US10617680, Example 269 2-{4-[4-(4-Fluoro-phenyl)-1-methyl-6-oxo-1,6-dihydro- pyridin-3-yl]-pyrazol-1-yl}-4-methoxy-benzoic acid US11020380, Example 269
BDBM437535 US10617680, Example 269 2-{4-[4-(4-Fluoro-phenyl)-1-methyl-6-oxo-1,6-dihydro- pyridin-3-yl]-pyrazol-1-yl}-4-methoxy-benzoic acid US11020380, Example 269 N-(1-(4- cyanobenzyl)piperidin-4- yl)-5-(4-(2-(4- fluorophenyl)propan-2- yl)piperazine-1- carbonyl)picolinamide BDBM328130 US9663496, Compound 269 US10377742, Compound 269 US9663496, Compound 268
N-(1-(4- cyanobenzyl)piperidin-4- yl)-5-(4-(2-(4- fluorophenyl)propan-2- yl)piperazine-1- carbonyl)picolinamide BDBM328130 US9663496, Compound 269 US10377742, Compound 269 US9663496, Compound 268 US10183953, Example 269 ((1S,4R,6R)-6-((5-bromopyridin-2-yl)oxy)-2-azabicyclo[2.2.2]octan-2-yl)(3-fluoro-2-(pyrimidin-2-yl)phenyl)methanone US9611277, Example 269 BDBM315088
US10183953, Example 269 ((1S,4R,6R)-6-((5-bromopyridin-2-yl)oxy)-2-azabicyclo[2.2.2]octan-2-yl)(3-fluoro-2-(pyrimidin-2-yl)phenyl)methanone US9611277, Example 269 BDBM315088 US11634395, Example 269 4-amino-2-methoxy-N-((1S,2R)-2-(8-methylnaphthalen-1-yl)-1- (5-oxo-4,5-dihydro-1,3,4-oxadiazol-2- yl)propyl)benzenesulfonamide US10889555, Example 269 BDBM477499
US11634395, Example 269 4-amino-2-methoxy-N-((1S,2R)-2-(8-methylnaphthalen-1-yl)-1- (5-oxo-4,5-dihydro-1,3,4-oxadiazol-2- yl)propyl)benzenesulfonamide US10889555, Example 269 BDBM477499 (S)-2-((1-(3-fluoro-4- (1H-1,2,4-triazol-3- yl)phenyl)-7,8,9,10- tetrahydro-6-oxa-2,10a- diazacycloocta[cd]inden-4- yl)oxy)propanamide BDBM272964 US10065970, Example 269 US10435414, Example 269
(S)-2-((1-(3-fluoro-4- (1H-1,2,4-triazol-3- yl)phenyl)-7,8,9,10- tetrahydro-6-oxa-2,10a- diazacycloocta[cd]inden-4- yl)oxy)propanamide BDBM272964 US10065970, Example 269 US10435414, Example 269 BDBM85619 CHEMBL138332 1-(3-chlorophenyl)-3-[2-(furan-2-yl)-8-propyl-8H-pyrazolo[4,3-e][1,2,4]triazolo[1,5-c]pyrimidin-5-yl]urea MRE 3010F20
BDBM85619 CHEMBL138332 1-(3-chlorophenyl)-3-[2-(furan-2-yl)-8-propyl-8H-pyrazolo[4,3-e][1,2,4]triazolo[1,5-c]pyrimidin-5-yl]urea MRE 3010F20 US9981909, Example 269 BDBM396642 (S)-3-[4-Chloro-2-(4-fluoro-phenoxy)-phenoxy]-pyrrolidine
US9981909, Example 269 BDBM396642 (S)-3-[4-Chloro-2-(4-fluoro-phenoxy)-phenoxy]-pyrrolidine 3-{4-[(2-methylbenzene)sulfonyl]phenyl}-1- (pyridin-3-ylmethyl)urea BDBM543258 US11279687, Compound 269
3-{4-[(2-methylbenzene)sulfonyl]phenyl}-1- (pyridin-3-ylmethyl)urea BDBM543258 US11279687, Compound 269 BDBM404925 US11370786, Example 269 (R)-1-(2-fluoro-4-(6-(2-(4-(3-(2,2,2- trifluoroethoxy)cyclobutoxy)pyridin- 2-yl)acetamido)pyridazin-3-yl)butyl)- N-methyl-1H-1,2,3-triazole-4- carboxamide US10344025, Example 269
BDBM404925 US11370786, Example 269 (R)-1-(2-fluoro-4-(6-(2-(4-(3-(2,2,2- trifluoroethoxy)cyclobutoxy)pyridin- 2-yl)acetamido)pyridazin-3-yl)butyl)- N-methyl-1H-1,2,3-triazole-4- carboxamide US10344025, Example 269 US11236047, Example 269 BDBM535345 US20230338337, Compound 269 (3R)-6-[1-(azetidin-1-yl)-2- hydroxypropan-2-yl]-3-(4- chlorophenyl)-2-[(5-chloropyridin-2- yl)methyl]-3-methoxy-2,3-dihydro-1H- isoindol-1-one
US11236047, Example 269 BDBM535345 US20230338337, Compound 269 (3R)-6-[1-(azetidin-1-yl)-2- hydroxypropan-2-yl]-3-(4- chlorophenyl)-2-[(5-chloropyridin-2- yl)methyl]-3-methoxy-2,3-dihydro-1H- isoindol-1-one US10822352, Comp No. 269 US10287301, Compound 269 BDBM386340 (2S,4R)-1-(2-(3- acetyl-5-(2-(2- methoxyethylamino) pyrimidin-5-yl)- 1H-indazol-1- yl)acetyl)-N-(6- bromopyridin-2- yl)-4- fluoropyrrolidine-2- carboxamide
US10822352, Comp No. 269 US10287301, Compound 269 BDBM386340 (2S,4R)-1-(2-(3- acetyl-5-(2-(2- methoxyethylamino) pyrimidin-5-yl)- 1H-indazol-1- yl)acetyl)-N-(6- bromopyridin-2- yl)-4- fluoropyrrolidine-2- carboxamide BDBM361112 2-Amino-N-(4′-(N—((R)-pyrrolidin-3-yl)sulfamoyl)-3′-sulfamoyl-2′-(2H-tetrazol-5-yl)-3,4,5,6-tetrahydro-[1,1′-biphenyl]-3-yl)acetamide US10544130, Example 269 US10221163, Example 269
BDBM361112 2-Amino-N-(4′-(N—((R)-pyrrolidin-3-yl)sulfamoyl)-3′-sulfamoyl-2′-(2H-tetrazol-5-yl)-3,4,5,6-tetrahydro-[1,1′-biphenyl]-3-yl)acetamide US10544130, Example 269 US10221163, Example 269 BDBM417652 US10457669, Example 269 2-(6-{5-chloro-2-[(oxan- 4-yl)amino]pyrimidin-4- yl}-1-oxo-2,3-dihydro- 1H-isoindol-2-yl)-N-[1- (2,5-dimethyl-1,3- thiazol-4-yl)ethyl] acetamide US11001575, Example 269
BDBM417652 US10457669, Example 269 2-(6-{5-chloro-2-[(oxan- 4-yl)amino]pyrimidin-4- yl}-1-oxo-2,3-dihydro- 1H-isoindol-2-yl)-N-[1- (2,5-dimethyl-1,3- thiazol-4-yl)ethyl] acetamide US11001575, Example 269 3-(7-Chloro-1H-indazol-5-yl)-5-(difluoromethyl)-2-isopropyl-imidazo[4,5- BDBM551152 US11312712, Example 269
3-(7-Chloro-1H-indazol-5-yl)-5-(difluoromethyl)-2-isopropyl-imidazo[4,5- BDBM551152 US11312712, Example 269 BDBM601009 US11634391, Compound 269 5-(4-methyl-2,3-dihydro-1,4-benzoxazin-7-yl)-1,3,4-oxadiazol-2-ol
BDBM601009 US11634391, Compound 269 5-(4-methyl-2,3-dihydro-1,4-benzoxazin-7-yl)-1,3,4-oxadiazol-2-ol BDBM654659 US11912693, Compound 269 2,2-diethyl-6-[3-(p-tolyl)-1,2,4- oxadiazol-5-yl]chroman-4-one
BDBM654659 US11912693, Compound 269 2,2-diethyl-6-[3-(p-tolyl)-1,2,4- oxadiazol-5-yl]chroman-4-one US10377753, Example 269 BDBM409393 1-(3,3-Difluoroazetidin-1-yl)-2-[6-(m-tolyl)pyrrolo[3,2-b]pyridin-
US10377753, Example 269 BDBM409393 1-(3,3-Difluoroazetidin-1-yl)-2-[6-(m-tolyl)pyrrolo[3,2-b]pyridin- US10508120, Compound I-974 US10577373, Compound I-974 US11414431, Compound I-269 BDBM424519 US11046698, Compound I-974
US10508120, Compound I-974 US10577373, Compound I-974 US11414431, Compound I-269 BDBM424519 US11046698, Compound I-974 US11504367, Example 269 5-(4-((4- cyanophenyl)ethynyl)-3- fluorophenoxy)-1H-1,2,3- triazole-4-carboxylic acid BDBM581171
US11504367, Example 269 5-(4-((4- cyanophenyl)ethynyl)-3- fluorophenoxy)-1H-1,2,3- triazole-4-carboxylic acid BDBM581171 2-methyl-6-oxo-N-{1-[4- (trifluoromethyl)phenyl]cyclobutyl}- 1,6-dihydropyrimidine-4-carboxamide US9815796, Example 269 BDBM355178
2-methyl-6-oxo-N-{1-[4- (trifluoromethyl)phenyl]cyclobutyl}- 1,6-dihydropyrimidine-4-carboxamide US9815796, Example 269 BDBM355178 5-[4-(3,3-Dimethylbutyl)-3-(trifluoromethyl)phenyl]-3,6-dihydro-2H-1,3,4-oxadiazin-2-one BDBM568806 US11427553, Example 269
5-[4-(3,3-Dimethylbutyl)-3-(trifluoromethyl)phenyl]-3,6-dihydro-2H-1,3,4-oxadiazin-2-one BDBM568806 US11427553, Example 269 BDBM585654 6-(2,4-Difluoro-3-methyl-phenyl)-1-(pyridazin-3- ylmethyl)pyrazolo[4,3-b]pyridine US11530210, Example 269
BDBM585654 6-(2,4-Difluoro-3-methyl-phenyl)-1-(pyridazin-3- ylmethyl)pyrazolo[4,3-b]pyridine US11530210, Example 269 US11389456, Ex. No. I-269 5-((3-(pentafluoro-lambda6- sulfaneyl)phenoxy)- 1H-1,2,3-triazole-4- carboxylic acid BDBM560898
US11389456, Ex. No. I-269 5-((3-(pentafluoro-lambda6- sulfaneyl)phenoxy)- 1H-1,2,3-triazole-4- carboxylic acid BDBM560898 ((1-((6-chloropyridin-3- yl)amino)isoquinolin-6- yl)imino)(cyclopropylmeth- yl)(methyl)-lambda6-sulfanone US20240101531, Example 269 BDBM660883
((1-((6-chloropyridin-3- yl)amino)isoquinolin-6- yl)imino)(cyclopropylmeth- yl)(methyl)-lambda6-sulfanone US20240101531, Example 269 BDBM660883 6-(4-(4-cyanophenyl)-5-hydroxy-1H-pyrazol-1-yl)-N-(3-(dimethylamino)propyl)nicotinamide US10407409, Example 269 BDBM413244
6-(4-(4-cyanophenyl)-5-hydroxy-1H-pyrazol-1-yl)-N-(3-(dimethylamino)propyl)nicotinamide US10407409, Example 269 BDBM413244 BDBM351548 US9796704, Entry 269 7-{[(4-cyclopentylmorpholin-2-yl)methyl]oxy}- N-(3,4-dichlorophenyl)-6- (methyloxy)quinazolin-4-amine
BDBM351548 US9796704, Entry 269 7-{[(4-cyclopentylmorpholin-2-yl)methyl]oxy}- N-(3,4-dichlorophenyl)-6- (methyloxy)quinazolin-4-amine BDBM593290 US11578061, Example 269 4-((4,4- difluorocyclohexyl)methoxy)-N- (4-((2R,6S)-2,6- dimethylmorpholino)phenyl)-5- fluoropyrimidin-2-amine
BDBM593290 US11578061, Example 269 4-((4,4- difluorocyclohexyl)methoxy)-N- (4-((2R,6S)-2,6- dimethylmorpholino)phenyl)-5- fluoropyrimidin-2-amine US20240279215, Compound 269 oxazol-5-ylmethyl (4-(2- (4-(oxetan-3- yl)piperazin-1- yl)ethyl)phenyl)carbamate BDBM691808
US20240279215, Compound 269 oxazol-5-ylmethyl (4-(2- (4-(oxetan-3- yl)piperazin-1- yl)ethyl)phenyl)carbamate BDBM691808 (S)-5-chloro-4-((1-(2-fluorophenyl)ethyl)amino)-2-methyl-N-(thiazol-2-yl)benzenesulfonamide BDBM526162 US11174268, Example 269
(S)-5-chloro-4-((1-(2-fluorophenyl)ethyl)amino)-2-methyl-N-(thiazol-2-yl)benzenesulfonamide BDBM526162 US11174268, Example 269 2-Ethoxy-4-{6-[2-(8-fluoro-naphthalen-2-yl)-ethylamino]-pyrimidin-4-yl}- benzoic acid BDBM640148 US11839613, Example 269
2-Ethoxy-4-{6-[2-(8-fluoro-naphthalen-2-yl)-ethylamino]-pyrimidin-4-yl}- benzoic acid BDBM640148 US11839613, Example 269 3-(2-aminobenzo[d]thiazol-4-yl)-6-(2-aminopropylsulfonyl)-2-(2H-tetrazol-5-yl)benzenesulfonamide BDBM368082 US10227331, Example 269
3-(2-aminobenzo[d]thiazol-4-yl)-6-(2-aminopropylsulfonyl)-2-(2H-tetrazol-5-yl)benzenesulfonamide BDBM368082 US10227331, Example 269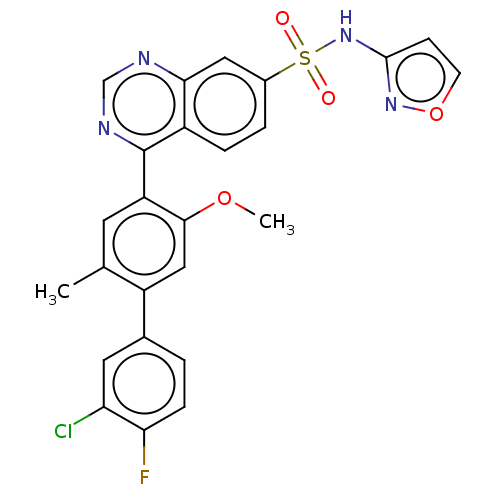 4-(3'-chloro-4'-fluoro-5-methoxy-2-methyl-4-biphenylyl)-N-3-isoxazolyl-7-quinazolinesulfonamide US9776995, Example 269 BDBM343097
4-(3'-chloro-4'-fluoro-5-methoxy-2-methyl-4-biphenylyl)-N-3-isoxazolyl-7-quinazolinesulfonamide US9776995, Example 269 BDBM343097 5-[(3R)-3- aminopiperidin-1-yl]- N-(2- cyanophenyl)pyrazolo [1,5-a]pyrimidine-3- carboxamide trifluoroacetate BDBM402688 US10329294, Example 269
5-[(3R)-3- aminopiperidin-1-yl]- N-(2- cyanophenyl)pyrazolo [1,5-a]pyrimidine-3- carboxamide trifluoroacetate BDBM402688 US10329294, Example 269 BDBM426925 US10544113, No. 269 2-(4-Fluorophenyl)-3-[4-[(3-hydroxy-1-piperidinyl)carbonyl]-2-methylphenyl]-1,3-thiazolidin-4-one
BDBM426925 US10544113, No. 269 2-(4-Fluorophenyl)-3-[4-[(3-hydroxy-1-piperidinyl)carbonyl]-2-methylphenyl]-1,3-thiazolidin-4-one BDBM744974 US20250171431, Compound 269 3-(2-{5-methoxy-6-[(1s,3s)-3- (dimethylamino)cyclobutoxy]- 3-pyridylamino}-4- pyrimidinylamino)-2- quinolinecarbonitrile
BDBM744974 US20250171431, Compound 269 3-(2-{5-methoxy-6-[(1s,3s)-3- (dimethylamino)cyclobutoxy]- 3-pyridylamino}-4- pyrimidinylamino)-2- quinolinecarbonitrile N-((2S,4S)-1-(4-amino-3-benzylbutanoyl)- 2-methylpiperidin-4-yl)-5- cyclopropylisoxazole-3-carboxamide US10577363, Compound 269 BDBM432569
N-((2S,4S)-1-(4-amino-3-benzylbutanoyl)- 2-methylpiperidin-4-yl)-5- cyclopropylisoxazole-3-carboxamide US10577363, Compound 269 BDBM432569 N-(1-(4- cyanobenzyl)piperidin-4- yl)-5-(4-(pyridin-4- yloxy)piperidine-1- carbonyl)picolinamide BDBM328131 US9663496, Compound 269
N-(1-(4- cyanobenzyl)piperidin-4- yl)-5-(4-(pyridin-4- yloxy)piperidine-1- carbonyl)picolinamide BDBM328131 US9663496, Compound 269 US10052306, 269 BDBM238277 2-[5-hydroxy-5-(2- phenylethyl)- octahydrocyclopenta[c] pyrrol-2-yl]-1-(4- phenylphenyl)ethan-1- one
US10052306, 269 BDBM238277 2-[5-hydroxy-5-(2- phenylethyl)- octahydrocyclopenta[c] pyrrol-2-yl]-1-(4- phenylphenyl)ethan-1- one US10130633, Example 269 N-[5-(2-ethylphenyl)-1,3,4- thiadiazol-2-yl]-3-[(morpholin- 4-ylacetyl)amino]-4- (trifluoromethoxy)benzamide BDBM300699
US10130633, Example 269 N-[5-(2-ethylphenyl)-1,3,4- thiadiazol-2-yl]-3-[(morpholin- 4-ylacetyl)amino]-4- (trifluoromethoxy)benzamide BDBM300699 (1R,3S,5R)-2-(2-(3- acetyl-5-(2- methylpyrimidin-5- yl)-7- (trifluoromethyl)-1H- pyrazolo[3,4- c]pyridin-1-yl)acetyl)- N-(6-bromo-3- methylpyridin-2-yl)-5- methyl-2- azabicyclo[3.1.0] hexane-3-carboxamide US11708351, Compound 269 BDBM511932 US11084800, Cpd No. 269
(1R,3S,5R)-2-(2-(3- acetyl-5-(2- methylpyrimidin-5- yl)-7- (trifluoromethyl)-1H- pyrazolo[3,4- c]pyridin-1-yl)acetyl)- N-(6-bromo-3- methylpyridin-2-yl)-5- methyl-2- azabicyclo[3.1.0] hexane-3-carboxamide US11708351, Compound 269 BDBM511932 US11084800, Cpd No. 269 (5-amino-5-(1-(2-((2- morpholinoethyl)amino)-2-oxoethyl)-1H- tetrazol-5-yl)pentyl)boronic acid US11845767, Example 269 BDBM642224
(5-amino-5-(1-(2-((2- morpholinoethyl)amino)-2-oxoethyl)-1H- tetrazol-5-yl)pentyl)boronic acid US11845767, Example 269 BDBM642224 5-((1H-pyrazol-1-yl)methyl)-N-((2,6-dimethoxy-4-(pyridin-3- ylethynyl)phenyl)sulfonyl)-6-methoxypicolinamide BDBM736544 US20250136553, Compound 269
5-((1H-pyrazol-1-yl)methyl)-N-((2,6-dimethoxy-4-(pyridin-3- ylethynyl)phenyl)sulfonyl)-6-methoxypicolinamide BDBM736544 US20250136553, Compound 269 BDBM504860 3-({3- fluoro-4-[5- (trifluoro- methyl)- 1,2,4-oxa- diazol-3- yl]phenyl} methoxy)- 5-methyl- pyridine US11066396, Example 269
BDBM504860 3-({3- fluoro-4-[5- (trifluoro- methyl)- 1,2,4-oxa- diazol-3- yl]phenyl} methoxy)- 5-methyl- pyridine US11066396, Example 269 BDBM657590 N-[1-{4-[2-(aminomethyl)-4-fluorophenyl]thiophen-2-yl}ethyl]-6,7-dimethoxy-2-methylquinazolin-4-amine US20240083857, Example 269
BDBM657590 N-[1-{4-[2-(aminomethyl)-4-fluorophenyl]thiophen-2-yl}ethyl]-6,7-dimethoxy-2-methylquinazolin-4-amine US20240083857, Example 269 US10059713, Example 269 7-(2-fluorobenzenesulfonyl)-4-(2-methylmorpholin-4-yl)-2-(methylsulfanyl)-5H-pyrrolo[3,2-d]pyrimidin-6-amine BDBM271148
US10059713, Example 269 7-(2-fluorobenzenesulfonyl)-4-(2-methylmorpholin-4-yl)-2-(methylsulfanyl)-5H-pyrrolo[3,2-d]pyrimidin-6-amine BDBM271148 US11613531, Example 269 N-(4-(1H-pyrazol-4-yl)phenyl)-2-(3-(3-methoxyphenyl)pyrrolidin-1-yl)pyrimidin-4-amine BDBM598614
US11613531, Example 269 N-(4-(1H-pyrazol-4-yl)phenyl)-2-(3-(3-methoxyphenyl)pyrrolidin-1-yl)pyrimidin-4-amine BDBM598614 US9579320, Example 269 4-(3-(2H-1,2,3-triazol-2-yl)phenylamino)-2-((1R,3S)-3-aminocyclohexylamino)pyrimidine-5-carboxamide (Racemic) BDBM290865
US9579320, Example 269 4-(3-(2H-1,2,3-triazol-2-yl)phenylamino)-2-((1R,3S)-3-aminocyclohexylamino)pyrimidine-5-carboxamide (Racemic) BDBM290865 US9662339, 269 1-[(S)-4-(2,3-Dihydro- [1,4]dioxino[2,3-b]pyridin-3-yl)- benzyl]-4-phenyl-piperidin-4-ol BDBM327186
US9662339, 269 1-[(S)-4-(2,3-Dihydro- [1,4]dioxino[2,3-b]pyridin-3-yl)- benzyl]-4-phenyl-piperidin-4-ol BDBM327186 US9855269, Compound 269 BDBM364026 3-(4-aminopyrido[3,2-d]pyrimidin-6-yl)-5-fluoro-N-[(1-methyl-4-piperidyl)methyl]benzamide
US9855269, Compound 269 BDBM364026 3-(4-aminopyrido[3,2-d]pyrimidin-6-yl)-5-fluoro-N-[(1-methyl-4-piperidyl)methyl]benzamide Preparation of 4-[2-(difluoromethyl)-3,3-difluoro-2-methyl-propanoyl]-3,5-dihydro-2H-pyrido[3,4-f][1,4]oxazepine-9-carbonitrile (Example 268) and 4-[2-(difluoromethyl)-3,3-difluoro-2-methyl-butanoyl]-3,5-dihydro-2H-pyrido[3,4-f][1,4]oxazepine-9-carbonitrile (Example 269) US11203600, Example 269 BDBM530188
Preparation of 4-[2-(difluoromethyl)-3,3-difluoro-2-methyl-propanoyl]-3,5-dihydro-2H-pyrido[3,4-f][1,4]oxazepine-9-carbonitrile (Example 268) and 4-[2-(difluoromethyl)-3,3-difluoro-2-methyl-butanoyl]-3,5-dihydro-2H-pyrido[3,4-f][1,4]oxazepine-9-carbonitrile (Example 269) US11203600, Example 269 BDBM530188 2-(4-(3-amino-1H-pyrazolo [3,4-b]pyridin-5-yl) benzylamino)-N-(2- cyclopropylethyl)-5- (trifluoromethyl) nicotinamide US10030016, Example 269 BDBM282389
2-(4-(3-amino-1H-pyrazolo [3,4-b]pyridin-5-yl) benzylamino)-N-(2- cyclopropylethyl)-5- (trifluoromethyl) nicotinamide US10030016, Example 269 BDBM282389 BDBM349135 Cyclopropanecarboxylic acid {5-[4-(3-dimethylamino- piperidine-1-carbonyl)- phenyl]-[1,2,4]triazolo[1,5- a]pyridin-2-yl}-amide US10206907, Compound 269
BDBM349135 Cyclopropanecarboxylic acid {5-[4-(3-dimethylamino- piperidine-1-carbonyl)- phenyl]-[1,2,4]triazolo[1,5- a]pyridin-2-yl}-amide US10206907, Compound 269 BDBM366017 (2S)-2-cyclohexyl-2-[({2,6- dimethoxy-4-[(2-methyl-3- phenylphenyl)methoxy]phenyl} methyl)amino]ethan-1-ol US9872852, Example 269
BDBM366017 (2S)-2-cyclohexyl-2-[({2,6- dimethoxy-4-[(2-methyl-3- phenylphenyl)methoxy]phenyl} methyl)amino]ethan-1-ol US9872852, Example 269 BDBM611376 (2S)-1-furo[3,2-c]pyridin-2- ylsulfonyl-N-[[3-[4- (trifluoromethyl)phenoxy]- phenyl]methyl]pyrrolidine-2- carboxamide US10626112, Example 269
BDBM611376 (2S)-1-furo[3,2-c]pyridin-2- ylsulfonyl-N-[[3-[4- (trifluoromethyl)phenoxy]- phenyl]methyl]pyrrolidine-2- carboxamide US10626112, Example 269 BDBM732725 US20250115569, Example 269 (R)-5-cyano-N-methyl-N-(2,2,2-trifluoro-1- (3-methyl-4- (trifluoromethyl)phenyl)ethyl)pyridine-3- sulfonamide
BDBM732725 US20250115569, Example 269 (R)-5-cyano-N-methyl-N-(2,2,2-trifluoro-1- (3-methyl-4- (trifluoromethyl)phenyl)ethyl)pyridine-3- sulfonamide 2-(3-Chloro-5- (dimethylamino)phenyl)- N5-(4-cyano-3- fluorophenyl)-6,7- dihydropyrazolo[1,5-a] pyrazine-3,5(4H)- dicarboxamide US9598423, Example 269 BDBM302701
2-(3-Chloro-5- (dimethylamino)phenyl)- N5-(4-cyano-3- fluorophenyl)-6,7- dihydropyrazolo[1,5-a] pyrazine-3,5(4H)- dicarboxamide US9598423, Example 269 BDBM302701 3-(trans-2- ((cyclopropylmethyl)- amino)cyclopropyl)-N- (1-(2,2,2- trifluoroethyl)- piperidin-4- yl)benzamide dihydrochloride(optical isomer,retention time long) US9751885, 269 BDBM338832
3-(trans-2- ((cyclopropylmethyl)- amino)cyclopropyl)-N- (1-(2,2,2- trifluoroethyl)- piperidin-4- yl)benzamide dihydrochloride(optical isomer,retention time long) US9751885, 269 BDBM338832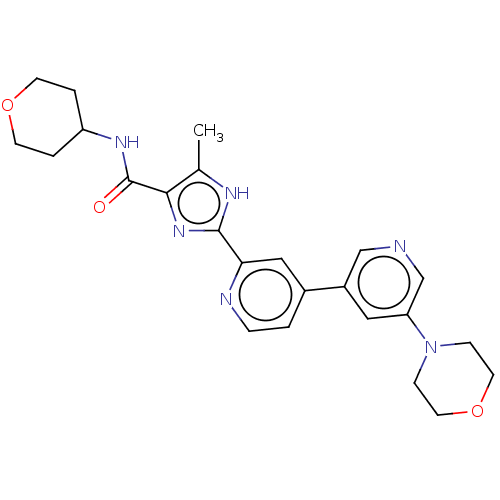 BDBM557217 5-Methyl-2-(5- morpholin-4-yl-3, 4'-bipyridin-2'-yl)- N-(tetrahydro-2H- pyran-4-yl)-1H- US11352340, Example 269
BDBM557217 5-Methyl-2-(5- morpholin-4-yl-3, 4'-bipyridin-2'-yl)- N-(tetrahydro-2H- pyran-4-yl)-1H- US11352340, Example 269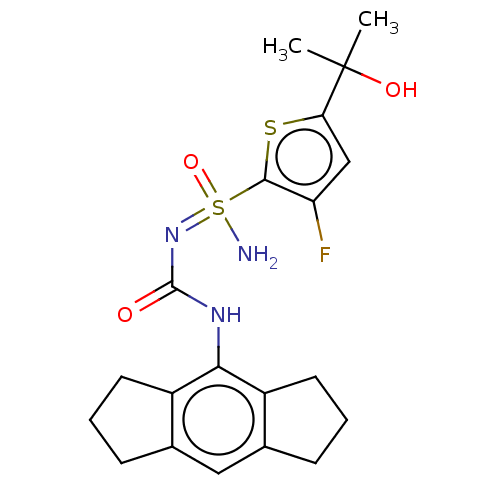 BDBM612696 BDBM612695 US11724992, Example 269 BDBM612580 3-Fluoro-N'-(1,2,3,5,6,7- hexahydro-s-indacen-4- ylcarbamoyl)-5-(2- hydroxypropan-2-yl) thiophene-2- sulfonimidamide
BDBM612696 BDBM612695 US11724992, Example 269 BDBM612580 3-Fluoro-N'-(1,2,3,5,6,7- hexahydro-s-indacen-4- ylcarbamoyl)-5-(2- hydroxypropan-2-yl) thiophene-2- sulfonimidamide N-[4-(4-fluoro-2-methoxyphenyl)pyridin-3-yl]-N-methyl-3-{[2-(sulfamoylamino)ethyl]-sulfonyl}-5-(trifluoromethyl)benzamide US10385022, Example 269 BDBM411273
N-[4-(4-fluoro-2-methoxyphenyl)pyridin-3-yl]-N-methyl-3-{[2-(sulfamoylamino)ethyl]-sulfonyl}-5-(trifluoromethyl)benzamide US10385022, Example 269 BDBM411273 US10174037, Example 269 3-Methyl-6-(methylamino)-1- (1-(4- (trifluoromethyl)phenyl)ethyl)- 1,5-dihydro-4H-pyrazolo[3,4- d]pyrimidin-4-one BDBM320864
US10174037, Example 269 3-Methyl-6-(methylamino)-1- (1-(4- (trifluoromethyl)phenyl)ethyl)- 1,5-dihydro-4H-pyrazolo[3,4- d]pyrimidin-4-one BDBM320864 US10183938, Compound (R)-269 (R)-6-(dimethylamino)-N-(1'-azaspiro[cyclopropane-1,2'-bicyclo[2.2.2]octan]-3'-yl)benzo[b]thiophene-2-carboxamide BDBM322515
US10183938, Compound (R)-269 (R)-6-(dimethylamino)-N-(1'-azaspiro[cyclopropane-1,2'-bicyclo[2.2.2]octan]-3'-yl)benzo[b]thiophene-2-carboxamide BDBM322515 US9656955, Example 269 BDBM308755 trans-4-(4-fluorophenyl)-1-[(1-methyl-1H-imidazol-4-yl)sulfonyl]-N-(pyridin-3-ylmethyl)pyrrolidin-3-amine
US9656955, Example 269 BDBM308755 trans-4-(4-fluorophenyl)-1-[(1-methyl-1H-imidazol-4-yl)sulfonyl]-N-(pyridin-3-ylmethyl)pyrrolidin-3-amine Preparation of 4-[2-(difluoromethyl)-3,3-difluoro-2-methyl-propanoyl]-3,5-dihydro-2H-pyrido[3,4-f][1,4]oxazepine-9-carbonitrile (Example 268) and 4-[2-(difluoromethyl)-3,3-difluoro-2-methyl-butanoyl]-3,5-dihydro-2H-pyrido[3,4-f][1,4]oxazepine-9-carbonitrile (Example 269) US11203600, Example 268 US11203600, Example 269 BDBM530187
Preparation of 4-[2-(difluoromethyl)-3,3-difluoro-2-methyl-propanoyl]-3,5-dihydro-2H-pyrido[3,4-f][1,4]oxazepine-9-carbonitrile (Example 268) and 4-[2-(difluoromethyl)-3,3-difluoro-2-methyl-butanoyl]-3,5-dihydro-2H-pyrido[3,4-f][1,4]oxazepine-9-carbonitrile (Example 269) US11203600, Example 268 US11203600, Example 269 BDBM530187 (4-fluorophenyl)(3-(3-phenyl-1H-pyrazol-5-yl)-5,6-dihydro-[1,2,4]triazolo[4,3-a]pyrazin-7(8H)-yl)methanone BDBM430842 US10544150, Compound 269
(4-fluorophenyl)(3-(3-phenyl-1H-pyrazol-5-yl)-5,6-dihydro-[1,2,4]triazolo[4,3-a]pyrazin-7(8H)-yl)methanone BDBM430842 US10544150, Compound 269 (R)-N-((S)-2-(dimethylamino)-3-(4-hydroxy-2-methylphenyl)propyl)-3-(pyridin-3-yl)-3-(1-(trifluoromethyl)cyclopropyl)propanamide US11634396, Compound 269 BDBM601330
(R)-N-((S)-2-(dimethylamino)-3-(4-hydroxy-2-methylphenyl)propyl)-3-(pyridin-3-yl)-3-(1-(trifluoromethyl)cyclopropyl)propanamide US11634396, Compound 269 BDBM601330 (S)-1,5-dimethyl-N-(6-(5-(methyl- d3)-1,2,4-oxadiazol-3-yl)-2,3- dihydrobenzofuran-3-yl)-1H- pyrazole-4-carboxamide BDBM690880 US12065436, Compound 269
(S)-1,5-dimethyl-N-(6-(5-(methyl- d3)-1,2,4-oxadiazol-3-yl)-2,3- dihydrobenzofuran-3-yl)-1H- pyrazole-4-carboxamide BDBM690880 US12065436, Compound 269 1-(5-(6,7- dimethoxy- quinazolin- 4-yloxy)- 2,4-difluoro- phenyl)- 3-(3-(2- fluoropropan- 2-yl)isoxazol- 5-yl)urea US9730937, Example 269 BDBM333719
1-(5-(6,7- dimethoxy- quinazolin- 4-yloxy)- 2,4-difluoro- phenyl)- 3-(3-(2- fluoropropan- 2-yl)isoxazol- 5-yl)urea US9730937, Example 269 BDBM333719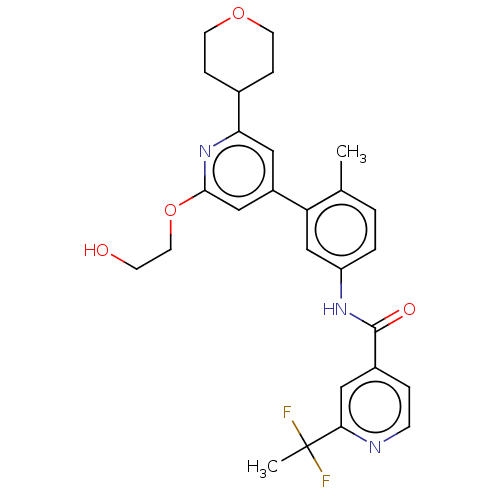 2-(1,1-difluoroethyl)-N- (3-(2-(2- hydroxyethoxy)-6- (tetrahydro-2H-pyran-4- yl)pyridin-4-yl)-4- methylphenyl) isonicotinamide BDBM313643 US10167279, Example 269
2-(1,1-difluoroethyl)-N- (3-(2-(2- hydroxyethoxy)-6- (tetrahydro-2H-pyran-4- yl)pyridin-4-yl)-4- methylphenyl) isonicotinamide BDBM313643 US10167279, Example 269 4-(5-cyano-2-methoxyphenyl)-N-(5-(N- isopropylsulfamoyl)-5,6-dihydro-4H- pyrrolo[3,4-d]thiazol-2-yl)-6- methylnicotinamide US20250230171, Compound 269 BDBM758696
4-(5-cyano-2-methoxyphenyl)-N-(5-(N- isopropylsulfamoyl)-5,6-dihydro-4H- pyrrolo[3,4-d]thiazol-2-yl)-6- methylnicotinamide US20250230171, Compound 269 BDBM758696 BDBM319226 US10173991, Example 269 methyl N-{4-[2- ({[(cyclopropyl- carbamoyl)methyl] carbamoyl} (propane-2- sulfonyl)methyl)- 6-fluoro-1,3- benzothiazol-5-yl] phenyl}carbamate
BDBM319226 US10173991, Example 269 methyl N-{4-[2- ({[(cyclopropyl- carbamoyl)methyl] carbamoyl} (propane-2- sulfonyl)methyl)- 6-fluoro-1,3- benzothiazol-5-yl] phenyl}carbamate BDBM363867 6,7-Dimethoxy-N-((1-methyl-1H-pyrazol-3-yl)methyl)-1-oxo-2-(pentan-3-yl)-1,2-dihydroisoquinoline-4-carboxamide US9856220, Example 269
BDBM363867 6,7-Dimethoxy-N-((1-methyl-1H-pyrazol-3-yl)methyl)-1-oxo-2-(pentan-3-yl)-1,2-dihydroisoquinoline-4-carboxamide US9856220, Example 269 BDBM644291 US11866430, Example 269 8-{4-[(4-chloro-2-hydroxyphenyl)methyl]piperazin-1-yl}-5-methyl-6-oxo-5,6-dihydro-1,5-naphthyridine-2,7-dicarbonitrile
BDBM644291 US11866430, Example 269 8-{4-[(4-chloro-2-hydroxyphenyl)methyl]piperazin-1-yl}-5-methyl-6-oxo-5,6-dihydro-1,5-naphthyridine-2,7-dicarbonitrile BDBM680054 US12011444, Example 269 5-{6-[2-(6-Fluoro-2,4-dimethyl-indol-1-yl)-ethylamino]-pyrimidin-4-yl}-3- trifluoromethyl-thiophene-2-carboxylic acid
BDBM680054 US12011444, Example 269 5-{6-[2-(6-Fluoro-2,4-dimethyl-indol-1-yl)-ethylamino]-pyrimidin-4-yl}-3- trifluoromethyl-thiophene-2-carboxylic acid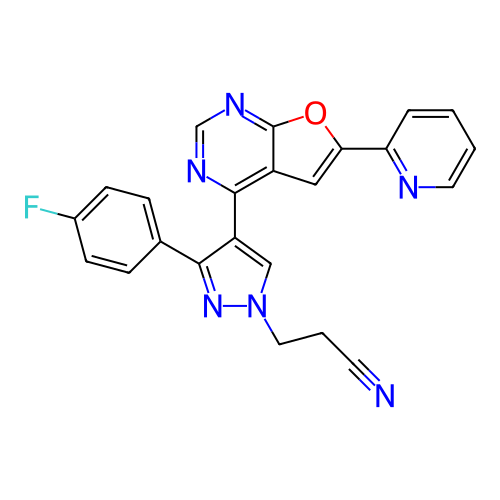 BDBM717448 3-{3-(4-fluorophenyl)-4-[6- (pyridin-2-yl)furo[2,3- d]pyrimidin-4-yl]-1H-pyrazol-1- yl}propanenitrile US20250034159, Example 269
BDBM717448 3-{3-(4-fluorophenyl)-4-[6- (pyridin-2-yl)furo[2,3- d]pyrimidin-4-yl]-1H-pyrazol-1- yl}propanenitrile US20250034159, Example 269 US10428044, Example 269 BDBM415164 Preparation of N-{4-[4-oxo-3-(phenylamino)-4,5,6,7-tetrahydro-1H-indol-2-yl]pyridin-2-yl}pyridine-2-carboxamide
US10428044, Example 269 BDBM415164 Preparation of N-{4-[4-oxo-3-(phenylamino)-4,5,6,7-tetrahydro-1H-indol-2-yl]pyridin-2-yl}pyridine-2-carboxamide US9687479, 269 1-(5-[(5-chlorothiophen-2-yl)methyl]amino-3-(3-fluoropyridin-2-yl)-1H-pyrazol-1-yl)-2,2-dimethylpropan-1-one BDBM163389
US9687479, 269 1-(5-[(5-chlorothiophen-2-yl)methyl]amino-3-(3-fluoropyridin-2-yl)-1H-pyrazol-1-yl)-2,2-dimethylpropan-1-one BDBM163389 (R)-4-((2-(2-chloro-3′-(7-cyano-5-((3-hydroxypyrrolidin-1-yl)methyl)benzo[d]oxazol-2-yl)-2′-methylbiphenyl-3-ylcarbamoyl)-1-methyl-6,7-dihydro-1H-imidazo[4,5-c]pyridin-5(4H)-yl)methyl)-1-methylcyclohexanecarboxylic acid (Peak 1) US10800768, Example 269 US11339149, Example 269 US10308644, Example 268 BDBM395658
(R)-4-((2-(2-chloro-3′-(7-cyano-5-((3-hydroxypyrrolidin-1-yl)methyl)benzo[d]oxazol-2-yl)-2′-methylbiphenyl-3-ylcarbamoyl)-1-methyl-6,7-dihydro-1H-imidazo[4,5-c]pyridin-5(4H)-yl)methyl)-1-methylcyclohexanecarboxylic acid (Peak 1) US10800768, Example 269 US11339149, Example 269 US10308644, Example 268 BDBM395658 3-(4-cyclopropyl-1H-1,2,3-triazol-1-yl)-N-(6-(4-cyclopropyl-4H-1,2,4-triazol-3-yl)pyridin-2-yl)benzamide USRE48711, Example 269 BDBM517658
3-(4-cyclopropyl-1H-1,2,3-triazol-1-yl)-N-(6-(4-cyclopropyl-4H-1,2,4-triazol-3-yl)pyridin-2-yl)benzamide USRE48711, Example 269 BDBM517658 3-{5-[4-(difluoromethoxy)benzyl]-1,3,4-oxadiazol-2-yl}-7-(piperidin-4-yl)pyrazolo[1,5-c]pyrimidin-5(4H)-one hydrochloride BDBM298057 US10118930, Example 269
3-{5-[4-(difluoromethoxy)benzyl]-1,3,4-oxadiazol-2-yl}-7-(piperidin-4-yl)pyrazolo[1,5-c]pyrimidin-5(4H)-one hydrochloride BDBM298057 US10118930, Example 269 4-(1-ethyl-8-oxo-spiro[6,7- dihydro-4H-pyrazolo[3,4- c]azepine-5,4'- tetrahydropyran]-3-yl)butyl thiazole-4-carboxylate US10793580, Example 269 BDBM466172
4-(1-ethyl-8-oxo-spiro[6,7- dihydro-4H-pyrazolo[3,4- c]azepine-5,4'- tetrahydropyran]-3-yl)butyl thiazole-4-carboxylate US10793580, Example 269 BDBM466172 BDBM338674 3-(trans-2-((cyclo- propylmethyl)amino)- cyclopropyl)-N-(1-(2,2,2- trifluoroethyl)piperidin- 4-yl)benzamide dihydrochloride US9751885, 269 US9751885, 271 US9751885, 270 US9751885, 108
BDBM338674 3-(trans-2-((cyclo- propylmethyl)amino)- cyclopropyl)-N-(1-(2,2,2- trifluoroethyl)piperidin- 4-yl)benzamide dihydrochloride US9751885, 269 US9751885, 271 US9751885, 270 US9751885, 108 BDBM667739 US11957687, Compound 269 (E)-N-(2-allyl-6-(prop-1-en- 1-yl)pyridin-4-yl)-2-oxo-6- (trifluoromethyl)-1,2- dihydropyridine-3- carboxamide
BDBM667739 US11957687, Compound 269 (E)-N-(2-allyl-6-(prop-1-en- 1-yl)pyridin-4-yl)-2-oxo-6- (trifluoromethyl)-1,2- dihydropyridine-3- carboxamide BDBM693350 US12071425, Compound 269 4-(2-(4- ((ethyl(methyl)sulfamoyl)amino)-2,3,5- trifluoro-phenoxy)-3-pyridyl)-2- (((3S,5S)-5-fluoro-3- piperidyl)amino)pyrimidine
BDBM693350 US12071425, Compound 269 4-(2-(4- ((ethyl(methyl)sulfamoyl)amino)-2,3,5- trifluoro-phenoxy)-3-pyridyl)-2- (((3S,5S)-5-fluoro-3- piperidyl)amino)pyrimidine BDBM695673 US12084420, Compound 269 N-(tert-butyl)-5-(5$#8243;- (ethylsulfonamido)dispiro[cyclopropane- 1,1'-cyclohexane-4',3$#8243;-indoline]-1$#8243;- carbonyl)thiophene-3-sulfonamide
BDBM695673 US12084420, Compound 269 N-(tert-butyl)-5-(5$#8243;- (ethylsulfonamido)dispiro[cyclopropane- 1,1'-cyclohexane-4',3$#8243;-indoline]-1$#8243;- carbonyl)thiophene-3-sulfonamide Ethyl (3-chloro- 4-fluorophenyl) (2-((ethoxy- carbonyl) amino)-7-(2- methylpyridin- 4-yl)furo[2,3- c]pyridin-3- yl)carbamate BDBM387690 US10294212, No. 269
Ethyl (3-chloro- 4-fluorophenyl) (2-((ethoxy- carbonyl) amino)-7-(2- methylpyridin- 4-yl)furo[2,3- c]pyridin-3- yl)carbamate BDBM387690 US10294212, No. 269 US10336739, Example 269 2-{5-[6-butyl-5- (2,6- dimethoxyphenyl)- 2,4-dihydroxy- pyridin- 3-yl]-1,3,4- oxadiazol- 2-yl}-N-[(4- methoxyphenyl) methyl]acetamide BDBM406655
US10336739, Example 269 2-{5-[6-butyl-5- (2,6- dimethoxyphenyl)- 2,4-dihydroxy- pyridin- 3-yl]-1,3,4- oxadiazol- 2-yl}-N-[(4- methoxyphenyl) methyl]acetamide BDBM406655 US11987588, Compound 269 3-((S)-3-((R)-8-(2-fluoro-5-methylphenylsulfonyl)-1-oxa-8-azaspiro[4.5]decan-3-ylamino)-2-hydroxypropoxy)-N-methylbenzenesulfonamide BDBM675800
US11987588, Compound 269 3-((S)-3-((R)-8-(2-fluoro-5-methylphenylsulfonyl)-1-oxa-8-azaspiro[4.5]decan-3-ylamino)-2-hydroxypropoxy)-N-methylbenzenesulfonamide BDBM675800 US20240174696, Compound 269 BDBM678543 4-(2-Methyl-2,8-diazaspiro[4.5]decan-8-yl)-5-(oxetan-3-yloxy)-2-(pyridin-4-yl)pyrido[3,4-d]pyrimidine
US20240174696, Compound 269 BDBM678543 4-(2-Methyl-2,8-diazaspiro[4.5]decan-8-yl)-5-(oxetan-3-yloxy)-2-(pyridin-4-yl)pyrido[3,4-d]pyrimidine  tert-butyl (2-((2-(4-(2-(5-chloro- 2,4-dimethoxyphenyl)imidazo- [1,2-a]pyridin-7-yl)benzamido)- ethyl)amino)ethyl)carbamate US10508109, Example 269 BDBM423083
tert-butyl (2-((2-(4-(2-(5-chloro- 2,4-dimethoxyphenyl)imidazo- [1,2-a]pyridin-7-yl)benzamido)- ethyl)amino)ethyl)carbamate US10508109, Example 269 BDBM423083 US11065253, Example 269 US10172859, Example 269 US9732094, Example 269 2-[2-Chloro-4-fluoro- 5-(6-morpholin-4-yl- thieno[3,2-d}- pyrimidin-4-yl)- phenyl]-2-(3-methyl- pyrazin-2-yl)- acetamide1H NMR (500 MHz, DMSO-d6) ppm = 8.87 (s,1H), 8.43 (d, J = 2.6, 1H), 8.40 (d, J = 2.7 1H), 7.82-7.78 (m, 1H), 7.74 (d, J = 10.0, 1H), 7.55 (d,J = 1.78), 7.41-7.36 (m, 1H), 6.53 (s, 1H), 5.63(s, 1H), 3.79-3.73 (m, 3H), 3.43-3.38 (m, 4H),2.51 (s, 3H). BDBM315849
US11065253, Example 269 US10172859, Example 269 US9732094, Example 269 2-[2-Chloro-4-fluoro- 5-(6-morpholin-4-yl- thieno[3,2-d}- pyrimidin-4-yl)- phenyl]-2-(3-methyl- pyrazin-2-yl)- acetamide1H NMR (500 MHz, DMSO-d6) ppm = 8.87 (s,1H), 8.43 (d, J = 2.6, 1H), 8.40 (d, J = 2.7 1H), 7.82-7.78 (m, 1H), 7.74 (d, J = 10.0, 1H), 7.55 (d,J = 1.78), 7.41-7.36 (m, 1H), 6.53 (s, 1H), 5.63(s, 1H), 3.79-3.73 (m, 3H), 3.43-3.38 (m, 4H),2.51 (s, 3H). BDBM315849 1-cyclohexyl-N-[4-[2- [[4-(dimethylamino)- cyclohexyl]amino]-8- isopropyl-7-oxo-pter- idin-6-yl]-2-fluoro- phenyl]methanesulfon- amide US12344603, Compound 269 BDBM755000
1-cyclohexyl-N-[4-[2- [[4-(dimethylamino)- cyclohexyl]amino]-8- isopropyl-7-oxo-pter- idin-6-yl]-2-fluoro- phenyl]methanesulfon- amide US12344603, Compound 269 BDBM755000 4-Chloro-3-methyl-1-(5-methyl- thiazol-2-ylmethyl)-1H- pyrazolo[3,4-b]pyridine-5- carboxylic acid (4-trifluoromethyl- phenyl)-amide BDBM381436 US10272074, Example 269
4-Chloro-3-methyl-1-(5-methyl- thiazol-2-ylmethyl)-1H- pyrazolo[3,4-b]pyridine-5- carboxylic acid (4-trifluoromethyl- phenyl)-amide BDBM381436 US10272074, Example 269 N-(4-(1-(4-(5-(difluoromethyl)-1,3,4-oxadiazol-2-yl)benzyl)-1H-pyrazol-4-yl)phenyl)-4,5-dihydro-1H-imidazol-2-amine US20230286970, Compound 269 BDBM618143
N-(4-(1-(4-(5-(difluoromethyl)-1,3,4-oxadiazol-2-yl)benzyl)-1H-pyrazol-4-yl)phenyl)-4,5-dihydro-1H-imidazol-2-amine US20230286970, Compound 269 BDBM618143 US10590140, Example 269 6-(3-(2- (benzyl(methyl)amino) ethyl)phenyl)-2- (ethylamino)-9-methyl- 6,7,8,9-tetrahydro-5H- pyrimido[4,5- e][1,4]diazepin-5-one BDBM436494
US10590140, Example 269 6-(3-(2- (benzyl(methyl)amino) ethyl)phenyl)-2- (ethylamino)-9-methyl- 6,7,8,9-tetrahydro-5H- pyrimido[4,5- e][1,4]diazepin-5-one BDBM436494 US12065436, Compound 163 (S)-1,5-dimethyl-N-(6-(5-methyl- 1,2,4-oxadiazol-3-yl)-2,3- dihydrobenzofuran-3-yl)-1H- pyrazole-4-carboxamide BDBM690774 US12065436, Compound 269
US12065436, Compound 163 (S)-1,5-dimethyl-N-(6-(5-methyl- 1,2,4-oxadiazol-3-yl)-2,3- dihydrobenzofuran-3-yl)-1H- pyrazole-4-carboxamide BDBM690774 US12065436, Compound 269 US12208088, Compound 269 2-(3-(4'-Chloro-1',2'-dihydrospiro[cyclopentane- 1,3'-pyrrolo[2,3-b]pyridin]-3-en-5'-yl)-2- fluorophenyl)-N-(cyanomethyl)isonicotinamide BDBM716031
US12208088, Compound 269 2-(3-(4'-Chloro-1',2'-dihydrospiro[cyclopentane- 1,3'-pyrrolo[2,3-b]pyridin]-3-en-5'-yl)-2- fluorophenyl)-N-(cyanomethyl)isonicotinamide BDBM716031 US12371428, Compound 269 BDBM760581 N-[4-[(6,7-dimethoxy-1,5- naphthyridin-4-yl)oxy]-3- fluorophenyl]-5-(4-fluorophenyl)-6- (methoxymethyl)-1-methyl-4- oxopyridine-3-carboxamide
US12371428, Compound 269 BDBM760581 N-[4-[(6,7-dimethoxy-1,5- naphthyridin-4-yl)oxy]-3- fluorophenyl]-5-(4-fluorophenyl)-6- (methoxymethyl)-1-methyl-4- oxopyridine-3-carboxamide BDBM470884 methyl (4- (((1S,3'R,6'R,7'S,8'E,11'S,12'R )-6-chloro-7'-methoxy-11',12'- dimethyl-13',13'-dioxido-15'- oxo-3,4-dihydro-2H- spiro[naphthalene-1,22'- [20]oxa[13]thia[1,14] diazatetracyclo [14.7.2.0~3,6~.0~19,24~] pentacosa[8,16,18,24]tetraen]- 7'-yl)methyl)-1- piperazinyl)acetate US10821115, Example 269 US11224601, Example 269
BDBM470884 methyl (4- (((1S,3'R,6'R,7'S,8'E,11'S,12'R )-6-chloro-7'-methoxy-11',12'- dimethyl-13',13'-dioxido-15'- oxo-3,4-dihydro-2H- spiro[naphthalene-1,22'- [20]oxa[13]thia[1,14] diazatetracyclo [14.7.2.0~3,6~.0~19,24~] pentacosa[8,16,18,24]tetraen]- 7'-yl)methyl)-1- piperazinyl)acetate US10821115, Example 269 US11224601, Example 269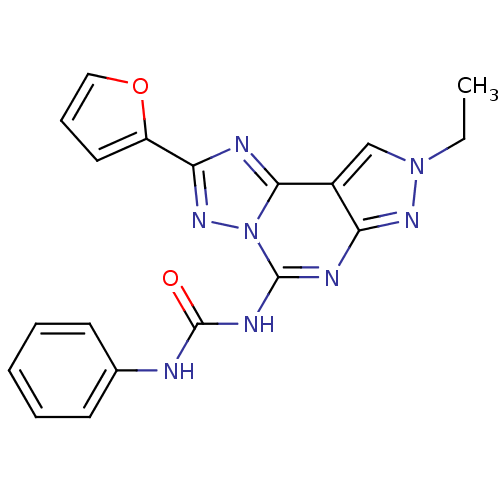 1-(8-Ethyl-2-furan-2-yl-8H-pyrazolo[4,3-e][1,2,4]triazolo[1,5-c]pyrimidin-5-yl)-3-phenyl-urea MRE 3048F20 BDBM50109473 CHEMBL352796 1-(8-ethyl-2-(furan-2-yl)-8H-pyrazolo[4,3-e][1,2,4]triazolo[1,5-c]pyrimidin-5-yl)-3-phenylurea
1-(8-Ethyl-2-furan-2-yl-8H-pyrazolo[4,3-e][1,2,4]triazolo[1,5-c]pyrimidin-5-yl)-3-phenyl-urea MRE 3048F20 BDBM50109473 CHEMBL352796 1-(8-ethyl-2-(furan-2-yl)-8H-pyrazolo[4,3-e][1,2,4]triazolo[1,5-c]pyrimidin-5-yl)-3-phenylurea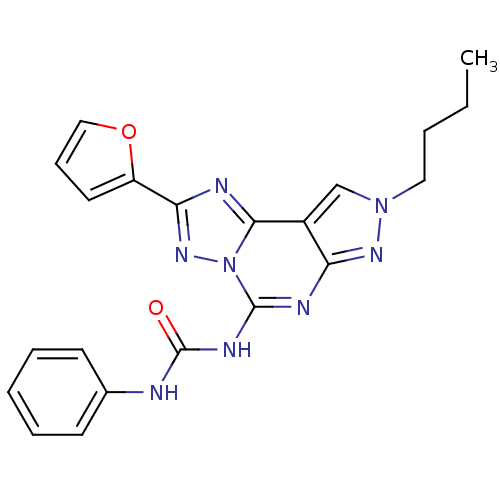 BDBM50109456 MRE 3062F20 CHEMBL349240 1-(8-Butyl-2-furan-2-yl-8H-pyrazolo[4,3-e][1,2,4]triazolo[1,5-c]pyrimidin-5-yl)-3-phenyl-urea 1-(8-butyl-2-(furan-2-yl)-8H-pyrazolo[4,3-e][1,2,4]triazolo[1,5-c]pyrimidin-5-yl)-3-phenylurea
BDBM50109456 MRE 3062F20 CHEMBL349240 1-(8-Butyl-2-furan-2-yl-8H-pyrazolo[4,3-e][1,2,4]triazolo[1,5-c]pyrimidin-5-yl)-3-phenyl-urea 1-(8-butyl-2-(furan-2-yl)-8H-pyrazolo[4,3-e][1,2,4]triazolo[1,5-c]pyrimidin-5-yl)-3-phenylurea CHEMBL168018 MRE 3055F20 1-(2-(furan-2-yl)-8-propyl-8H-pyrazolo[4,3-e][1,2,4]triazolo[1,5-c]pyrimidin-5-yl)-3-phenylurea BDBM50109481 1-(2-furan-2-yl-8-propyl-8H-pyrazolo[4,3-e][1,2,4]triazolo[1,5-c]pyrimidin-5-yl)-3-phenyl-urea
CHEMBL168018 MRE 3055F20 1-(2-(furan-2-yl)-8-propyl-8H-pyrazolo[4,3-e][1,2,4]triazolo[1,5-c]pyrimidin-5-yl)-3-phenylurea BDBM50109481 1-(2-furan-2-yl-8-propyl-8H-pyrazolo[4,3-e][1,2,4]triazolo[1,5-c]pyrimidin-5-yl)-3-phenyl-urea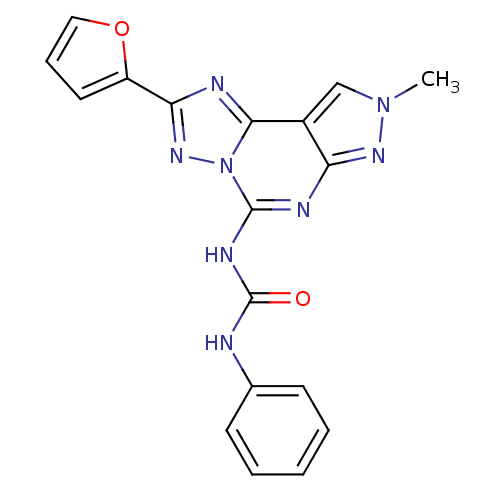 MRE 3046F20 CHEMBL324735 1-(2-Furan-2-yl-8-methyl-8H-pyrazolo[4,3-e][1,2,4]triazolo[1,5-c]pyrimidin-5-yl)-3-phenyl-urea BDBM50109461 1-(2-(furan-2-yl)-8-methyl-8H-pyrazolo[4,3-e][1,2,4]triazolo[1,5-c]pyrimidin-5-yl)-3-phenylurea
MRE 3046F20 CHEMBL324735 1-(2-Furan-2-yl-8-methyl-8H-pyrazolo[4,3-e][1,2,4]triazolo[1,5-c]pyrimidin-5-yl)-3-phenyl-urea BDBM50109461 1-(2-(furan-2-yl)-8-methyl-8H-pyrazolo[4,3-e][1,2,4]triazolo[1,5-c]pyrimidin-5-yl)-3-phenylurea 2-((R)-2-carbamoylpyrrolidin-1-yl)-N-((S)-2-(diethylamino)propyl)-6-(5-(thiophen-2-yl)pyrazolo[1,5-a]pyrimidin-3-yl)isonicotinamide US10100058, Example 269 BDBM292613
2-((R)-2-carbamoylpyrrolidin-1-yl)-N-((S)-2-(diethylamino)propyl)-6-(5-(thiophen-2-yl)pyrazolo[1,5-a]pyrimidin-3-yl)isonicotinamide US10100058, Example 269 BDBM292613 5-[4-amino-5- (trifluoromethyl) pyrrolo[2,1- f][1,2,4]triazin-7-yl]- N-[(3R)-3-(4- chlorophenyl)-3- hydroxypropyl]-2- fluoropyridine-3- carboxamide US11618753, Example 269 BDBM599131
5-[4-amino-5- (trifluoromethyl) pyrrolo[2,1- f][1,2,4]triazin-7-yl]- N-[(3R)-3-(4- chlorophenyl)-3- hydroxypropyl]-2- fluoropyridine-3- carboxamide US11618753, Example 269 BDBM599131 BDBM331434 4-[5-(Dimethyl-1H-1,2,3-triazol-5-yl)-8-[(S)-(2-fluorophenyl)(oxan-4-yl)methyl]-3,8,10-triazatricyclo[7.4.0.02,7]trideca-1(13),2 US9725449, Example 269
BDBM331434 4-[5-(Dimethyl-1H-1,2,3-triazol-5-yl)-8-[(S)-(2-fluorophenyl)(oxan-4-yl)methyl]-3,8,10-triazatricyclo[7.4.0.02,7]trideca-1(13),2 US9725449, Example 269 BDBM345724 1-Cyclopropyl-N-[2-(2-fluoro-6- methyl-phenoxy)-6-(1-methyl-6- oxo-1,6-dihydro-pyridin-3-yl)- pyrimidin-4-yl]- methanesulfonamide US10202360, Example 269
BDBM345724 1-Cyclopropyl-N-[2-(2-fluoro-6- methyl-phenoxy)-6-(1-methyl-6- oxo-1,6-dihydro-pyridin-3-yl)- pyrimidin-4-yl]- methanesulfonamide US10202360, Example 269 BDBM487847 N-(5-cyano-6-(2H-1,2,3-triazol- 2-yl)pyridin-3-yl)-1-(1,5- naphthyridin-4-yl)-5- (trifluoromethyl)-1H-pyrazole-4- carboxamide US10954214, Compound 269
BDBM487847 N-(5-cyano-6-(2H-1,2,3-triazol- 2-yl)pyridin-3-yl)-1-(1,5- naphthyridin-4-yl)-5- (trifluoromethyl)-1H-pyrazole-4- carboxamide US10954214, Compound 269 BDBM630679 ((1s,3s)-3-Hydroxy-3-methylcyclobutyl)(6-(1-methyl-1H-pyrrolo[2,3-b]pyridin-6-yl)-2-azaspiro[3.3]heptan-2-yl)methanone US11802111, Example 269
BDBM630679 ((1s,3s)-3-Hydroxy-3-methylcyclobutyl)(6-(1-methyl-1H-pyrrolo[2,3-b]pyridin-6-yl)-2-azaspiro[3.3]heptan-2-yl)methanone US11802111, Example 269 BDBM639800 2-[[6,7-Dichloro-2-(2-hydroxy acetyl)-10-(1H-pyrazol-4-yl)-3,4-dihydro-1H-pyrazino[1,2-a]indol-9-yl]oxy]acetamide US20230391786, Example 269
BDBM639800 2-[[6,7-Dichloro-2-(2-hydroxy acetyl)-10-(1H-pyrazol-4-yl)-3,4-dihydro-1H-pyrazino[1,2-a]indol-9-yl]oxy]acetamide US20230391786, Example 269 US9593097, Example 269 BDBM299899 3-amino-6-(1-{2- [(1- cyanocyclopropyl) methoxy]-6-[(3- fluoroazetidin-1- yl)carbonyl] pyrimidin- 4-yl}piperidin- 4-yl)pyridine-2- carboxamide
US9593097, Example 269 BDBM299899 3-amino-6-(1-{2- [(1- cyanocyclopropyl) methoxy]-6-[(3- fluoroazetidin-1- yl)carbonyl] pyrimidin- 4-yl}piperidin- 4-yl)pyridine-2- carboxamide US9718828, Example, 269 BDBM267670 (R)-3-(4-amino-3-(4-(4-(trifluoromethyl)pyridin-2-ylcarbamoyl)phenyl)-1H-pyrazolo[3,4-d]pyrimidin-1-yl)-N-ethylpiperidine-1-carboxamide
US9718828, Example, 269 BDBM267670 (R)-3-(4-amino-3-(4-(4-(trifluoromethyl)pyridin-2-ylcarbamoyl)phenyl)-1H-pyrazolo[3,4-d]pyrimidin-1-yl)-N-ethylpiperidine-1-carboxamide US9738655, Example 269 BDBM336047 4-(2-(3-(3-chloro-2- fluorophenyl)isothiazole-5- carbonyl)-5-(4- methoxypiperidin-1-yl)-1,2,3,4- tetrahydroisoquinoline-1- carboxamido)benzoic acid, TFA salt
US9738655, Example 269 BDBM336047 4-(2-(3-(3-chloro-2- fluorophenyl)isothiazole-5- carbonyl)-5-(4- methoxypiperidin-1-yl)-1,2,3,4- tetrahydroisoquinoline-1- carboxamido)benzoic acid, TFA salt (R)-3-(4-(4-methyl-1-propionylpiperidin-4-yl)phenylamino)-5-(3-(3-methyl-2-oxoimidazolidin-1-yl)piperidin-1-yl)pyrazine-2-carboxamide US9656988, Example 269 BDBM309800
(R)-3-(4-(4-methyl-1-propionylpiperidin-4-yl)phenylamino)-5-(3-(3-methyl-2-oxoimidazolidin-1-yl)piperidin-1-yl)pyrazine-2-carboxamide US9656988, Example 269 BDBM309800 BDBM306626 US10144746, Compound 269 ethyl N-[amino[4-({(1R,8S)- 12-(2,4-difluoro-phenyl)-15- oxatetracyclo[6.6.1.02,7.09,14]- pentadeca-2,4,6,9,11,13- hexaen-4-yl]formamido}- methyl)-3,5- difluorophenyl]methylidene]- carbamate
BDBM306626 US10144746, Compound 269 ethyl N-[amino[4-({(1R,8S)- 12-(2,4-difluoro-phenyl)-15- oxatetracyclo[6.6.1.02,7.09,14]- pentadeca-2,4,6,9,11,13- hexaen-4-yl]formamido}- methyl)-3,5- difluorophenyl]methylidene]- carbamate BDBM307998 BDBM307973 US9650358, Example 269 N-{5-chloro-4-[1-(tetrahydro-2H-pyran-4-ylmethyl)-1H-benzimidazol-6-yl]pyridin-2-yl}-3-azabicyclo[3.1.0]hexane-6-carboxamide
BDBM307998 BDBM307973 US9650358, Example 269 N-{5-chloro-4-[1-(tetrahydro-2H-pyran-4-ylmethyl)-1H-benzimidazol-6-yl]pyridin-2-yl}-3-azabicyclo[3.1.0]hexane-6-carboxamide BDBM452947 US10711004, Example 269 7-methyl-1-[[3- [(1S,5S,6R)-3-(2,4- difluorophenyl)-6- bicyclo[3.1.0] hex-2-enyl]-1,2,4- oxadiazol-5-yl] methyl]purin-6-one
BDBM452947 US10711004, Example 269 7-methyl-1-[[3- [(1S,5S,6R)-3-(2,4- difluorophenyl)-6- bicyclo[3.1.0] hex-2-enyl]-1,2,4- oxadiazol-5-yl] methyl]purin-6-one BDBM492128 5-[6-(tert-Butylamino)-4-(difluoromethyl)-3-pyridyl]-N-[(1R)-2-hydroxy-1,2-dimethyl-propyl]-4-[(2S)-2-methylpyrrolidine-1-carbonyl]thiazole-2-carboxamide US10975068, Example 269
BDBM492128 5-[6-(tert-Butylamino)-4-(difluoromethyl)-3-pyridyl]-N-[(1R)-2-hydroxy-1,2-dimethyl-propyl]-4-[(2S)-2-methylpyrrolidine-1-carbonyl]thiazole-2-carboxamide US10975068, Example 269 BDBM597382 ((5R,9S)-2-Methyl-3-(1-methyl-1H-indol-2-yl)-4,5,6,7,8,9- hexahydro-2H-5,9-epiminocycloocta[c]pyrazol-10-yl)(quinolin-6- yl)methanone; US11597728, Example 269
BDBM597382 ((5R,9S)-2-Methyl-3-(1-methyl-1H-indol-2-yl)-4,5,6,7,8,9- hexahydro-2H-5,9-epiminocycloocta[c]pyrazol-10-yl)(quinolin-6- yl)methanone; US11597728, Example 269 N-[4-ethyl-5-(4-fluorophenyl)-1-methyl-1H-pyrazol-3-yl]-6-(4-fluoro-3,5-dimethyl-1H-pyrazol-1-yl)pyrimidin-4-amine BDBM531636 US11208400, Example 269
N-[4-ethyl-5-(4-fluorophenyl)-1-methyl-1H-pyrazol-3-yl]-6-(4-fluoro-3,5-dimethyl-1H-pyrazol-1-yl)pyrimidin-4-amine BDBM531636 US11208400, Example 269 US11149018, Example 269 BDBM520989 (−)-5-[({6-Bromo-3-methyl-2-[(2H10)piperidin-1-yl]quinolin-4-yl}carbonyl)amino]-4-[2-(trifluoromethyl)phenyl]pentanoic acid (Enantiomer 1)
US11149018, Example 269 BDBM520989 (−)-5-[({6-Bromo-3-methyl-2-[(2H10)piperidin-1-yl]quinolin-4-yl}carbonyl)amino]-4-[2-(trifluoromethyl)phenyl]pentanoic acid (Enantiomer 1) US11440913, Example 269 US11440913, Example 270 BDBM571202 2-chloro-5-{2-acetamidoimidazo[1,2- b]pyridazin-6-yl}-N-{1-[2-fluoro-5- (trifluoromethyl)phenyl]ethyl}pyridine- 3-carboxamide
US11440913, Example 269 US11440913, Example 270 BDBM571202 2-chloro-5-{2-acetamidoimidazo[1,2- b]pyridazin-6-yl}-N-{1-[2-fluoro-5- (trifluoromethyl)phenyl]ethyl}pyridine- 3-carboxamide US11731943, Example 269 BDBM613528 methyl N-[4-carbamoyl-1-[4- (cyanomethyl)-1-[[4- (cyclopenten-1-yl)-3-hydroxy- phenyl]methyl]-3-fluoro-4- piperidyl]pyrazol-3-yl]carbamate
US11731943, Example 269 BDBM613528 methyl N-[4-carbamoyl-1-[4- (cyanomethyl)-1-[[4- (cyclopenten-1-yl)-3-hydroxy- phenyl]methyl]-3-fluoro-4- piperidyl]pyrazol-3-yl]carbamate (S)-2-(3-(2-cyclobutyl-1-(4-methyl-4H- 1,2,4-triazol-3-yl)ethyl)phenyl)-6-(((1- methylcyclobutyl)amino)methyl)-4- (trifluoromethyl)isoindolin-1-one US12187709, Compound 269 BDBM710911
(S)-2-(3-(2-cyclobutyl-1-(4-methyl-4H- 1,2,4-triazol-3-yl)ethyl)phenyl)-6-(((1- methylcyclobutyl)amino)methyl)-4- (trifluoromethyl)isoindolin-1-one US12187709, Compound 269 BDBM710911 5-{5-[(2-Fluorophenyl)(oxan-4-yl)methyl]-9-methanesulfonyl-5H-pyrido[3,2-b]indol-3-yl}-1,4-dimethyl-1H-1,2,3-triazole US10112941, Example 116 BDBM297009 US10112941, Example 269
5-{5-[(2-Fluorophenyl)(oxan-4-yl)methyl]-9-methanesulfonyl-5H-pyrido[3,2-b]indol-3-yl}-1,4-dimethyl-1H-1,2,3-triazole US10112941, Example 116 BDBM297009 US10112941, Example 269 6-Bromo-N-[2-[(2R)-2- fluoro-3-hydroxy-3-methyl- butyl]-6-morpholino-1-oxo- isoindolin-5- yl]pyrazolo[1,5- a]pyrimidine-3-carboxamide US10988478, Example 269 BDBM494220
6-Bromo-N-[2-[(2R)-2- fluoro-3-hydroxy-3-methyl- butyl]-6-morpholino-1-oxo- isoindolin-5- yl]pyrazolo[1,5- a]pyrimidine-3-carboxamide US10988478, Example 269 BDBM494220 BDBM344967 1-[1-(2-fluorophenyl)-1H- 1-[1-(2-fluorophenyl)-1H-indazol-4-yl]-3-{1-[(1-hydroxycyclopropyl)carbon-yl]azetidin-3-yl}imidazolidin-2-one US9783527, Example 269
BDBM344967 1-[1-(2-fluorophenyl)-1H- 1-[1-(2-fluorophenyl)-1H-indazol-4-yl]-3-{1-[(1-hydroxycyclopropyl)carbon-yl]azetidin-3-yl}imidazolidin-2-one US9783527, Example 269 BDBM406985 US10336761, Example 269 3-fluoro-N-[5-({[2-(piperazin-1-yl)ethyl]amino}methyl)-2-(pyridin-2-yl)pyrrolo[2,1-f][1,2,4]triazin-4-yl]pyridin-4-amine
BDBM406985 US10336761, Example 269 3-fluoro-N-[5-({[2-(piperazin-1-yl)ethyl]amino}methyl)-2-(pyridin-2-yl)pyrrolo[2,1-f][1,2,4]triazin-4-yl]pyridin-4-amine BDBM569624 US11427567, Example 269 4-(1-(4-(Azetidin-1- ylmethyl)-2-chloro-3- fluorophenyl)-1H- imidazol-4-yl)-N-(1- (methylsulfonyl)piperidin- 4-yl)-5- (trifluoromethyl)pyrimidin- 2-amine
BDBM569624 US11427567, Example 269 4-(1-(4-(Azetidin-1- ylmethyl)-2-chloro-3- fluorophenyl)-1H- imidazol-4-yl)-N-(1- (methylsulfonyl)piperidin- 4-yl)-5- (trifluoromethyl)pyrimidin- 2-amine BDBM673134 US20240150293, Example 269 (1r,4r)-4-(3-chloroanilino)-2′-(2-chloro-4-methoxyphenyl)-2′,3′-dihydrospiro[cyclohexane-1,1′-indene]-4-carboxylic acid, enantiomer 2
BDBM673134 US20240150293, Example 269 (1r,4r)-4-(3-chloroanilino)-2′-(2-chloro-4-methoxyphenyl)-2′,3′-dihydrospiro[cyclohexane-1,1′-indene]-4-carboxylic acid, enantiomer 2 N-(5-(2-(3,3-dimethylazetidin-1- yl)acetamido)-2-methylpyridin- 3-yl)-2-(1-(trifluoromethyl)-1H- pyrazol-4-yl)pyrazolo[5,1- b]thiazole-7-carboxamide BDBM664188 US20240109917, Example 269
N-(5-(2-(3,3-dimethylazetidin-1- yl)acetamido)-2-methylpyridin- 3-yl)-2-(1-(trifluoromethyl)-1H- pyrazol-4-yl)pyrazolo[5,1- b]thiazole-7-carboxamide BDBM664188 US20240109917, Example 269 N-{6-[(3-cyclopropyl-1 H-pyrazol-5-yl)amino]-5-methoxy-1,2-benzoxazol-3-yl}-2,6-dimethoxy-4-(oxan-3-yl)benzene-1-sulfonamide BDBM731056 US20250122183, Example 269
N-{6-[(3-cyclopropyl-1 H-pyrazol-5-yl)amino]-5-methoxy-1,2-benzoxazol-3-yl}-2,6-dimethoxy-4-(oxan-3-yl)benzene-1-sulfonamide BDBM731056 US20250122183, Example 269 US10174016, Example 269 BDBM320102 3-(5-ethyl-1,3- thiazol-2-yl)-5- [(2R)- tetrahydrofuran-2- ylmethoxy]-N- {(1R)-1-[2- (trifluoromethyl) pyrimidin-5- yl]ethyl}benzamide US10202369, Example 278
US10174016, Example 269 BDBM320102 3-(5-ethyl-1,3- thiazol-2-yl)-5- [(2R)- tetrahydrofuran-2- ylmethoxy]-N- {(1R)-1-[2- (trifluoromethyl) pyrimidin-5- yl]ethyl}benzamide US10202369, Example 278 US11964973, Example 270 US11964973, Example 269 (+-)-trans-8-(4-((5-isopropoxypyridin-2-yl)oxy)-3-methoxypiperidin-1-yl)-5-methyl-6-oxo-5,6-dihydro-1,5-naphthyridine-2-carbonitrile BDBM669281
US11964973, Example 270 US11964973, Example 269 (+-)-trans-8-(4-((5-isopropoxypyridin-2-yl)oxy)-3-methoxypiperidin-1-yl)-5-methyl-6-oxo-5,6-dihydro-1,5-naphthyridine-2-carbonitrile BDBM669281 US20240092784, Example 269 (E)-N-Cyclopropyl-7-hydroxy-4-isobutyl-3-(2-(5-methyl-1,3,4-oxadiazol-2-yl)vinyl)-5-oxo-4,5-dihydropyrazolo[1,5-a]pyrimidine-6-carboxamide BDBM659832
US20240092784, Example 269 (E)-N-Cyclopropyl-7-hydroxy-4-isobutyl-3-(2-(5-methyl-1,3,4-oxadiazol-2-yl)vinyl)-5-oxo-4,5-dihydropyrazolo[1,5-a]pyrimidine-6-carboxamide BDBM659832 2-fluoro-N-(6-(4-(4-fluorophenyl)- 1-((1-methyl-1H-pyrazol-3-yl) methyl)-1H-imidazol-5-yl) imidazo[1,2-b]pyridazin-2-yl) isonicotinamide BDBM274694 US9556179, Compound 269
2-fluoro-N-(6-(4-(4-fluorophenyl)- 1-((1-methyl-1H-pyrazol-3-yl) methyl)-1H-imidazol-5-yl) imidazo[1,2-b]pyridazin-2-yl) isonicotinamide BDBM274694 US9556179, Compound 269 BDBM602377 (R)-7-chloro-N-(1-(3-(difluoro(1- isopropylpiperidin-4- yl)methyl)phenyl)ethyl)-6-(1- isopropylpiperidin-4-yl)-2- methylpyrido[2,3-d] pyrimidin-4-amine US11648254, Compound 269
BDBM602377 (R)-7-chloro-N-(1-(3-(difluoro(1- isopropylpiperidin-4- yl)methyl)phenyl)ethyl)-6-(1- isopropylpiperidin-4-yl)-2- methylpyrido[2,3-d] pyrimidin-4-amine US11648254, Compound 269 US10442819, Example 269 BDBM416124 6-[(cis-4- methoxycyclohexyl)carbonyl]- 8-[2-(trifluoromethyl)-5,6- dihydro[1,2,4]triazolo[1,5- a]pyrazin-7(8H)-yl]-6,11- dihydro-5H-pyrido[2,3- b][1,5]benzodiazepine
US10442819, Example 269 BDBM416124 6-[(cis-4- methoxycyclohexyl)carbonyl]- 8-[2-(trifluoromethyl)-5,6- dihydro[1,2,4]triazolo[1,5- a]pyrazin-7(8H)-yl]-6,11- dihydro-5H-pyrido[2,3- b][1,5]benzodiazepine US10899738, Cpd. No 269 rac-1-((1S,2R)-2- (cyano(phenyl)(1-((1-(4- (pyridin-4- ylsulfonyl)phenyl)azetidin-3- yl)methyl)piperidin-4- yl)methyl)cyclopentyl)-3- methylurea BDBM480208
US10899738, Cpd. No 269 rac-1-((1S,2R)-2- (cyano(phenyl)(1-((1-(4- (pyridin-4- ylsulfonyl)phenyl)azetidin-3- yl)methyl)piperidin-4- yl)methyl)cyclopentyl)-3- methylurea BDBM480208 [(2R)-3-(3-ethyl-4-oxo- spiro[6,8-dihydro-5H- pyrazolo[4,3-c]azepine-7,4'- tetrahydropyran]-1-yl)-2- methyl-propyl] 3-(piperidine- 1-carbonyl)benzoate BDBM481474 US10906915, Ex. 269
[(2R)-3-(3-ethyl-4-oxo- spiro[6,8-dihydro-5H- pyrazolo[4,3-c]azepine-7,4'- tetrahydropyran]-1-yl)-2- methyl-propyl] 3-(piperidine- 1-carbonyl)benzoate BDBM481474 US10906915, Ex. 269 Synthesis of 1-((R)-2-((3R,5R,8R,9R,10S,13S,14S,17R)-3-(ethoxymethyl)-3-hydroxy-13-methylhexadecahydro-1H-cyclopenta[a]phenanthren-17-yl)propyl)-1H-pyrazole-4-carbonitrile (269) & 1-((S)-2-((3R,5R,8R,9R,10S,13S,14S,17R)-3-(ethoxymethyl)-3-hydroxy-13-methylhexadecahydro-1H-cyclopenta[a]phenanthren-17-yl)propyl)-1H-pyrazole-4-carbonitrile (270) US20230322846, Example 269 BDBM625780
Synthesis of 1-((R)-2-((3R,5R,8R,9R,10S,13S,14S,17R)-3-(ethoxymethyl)-3-hydroxy-13-methylhexadecahydro-1H-cyclopenta[a]phenanthren-17-yl)propyl)-1H-pyrazole-4-carbonitrile (269) & 1-((S)-2-((3R,5R,8R,9R,10S,13S,14S,17R)-3-(ethoxymethyl)-3-hydroxy-13-methylhexadecahydro-1H-cyclopenta[a]phenanthren-17-yl)propyl)-1H-pyrazole-4-carbonitrile (270) US20230322846, Example 269 BDBM625780 BDBM366672 US9873709, Example 1-269 4-(4-chlorophenyl)-1- [2-[4-[methyl-(1- methyl-4- piperidyl)carbamoyl] anilino]- [1,2,4]triazolo[1,5- a]pyridin-8- yl]piperidine-4- carboxylic acid; formic acid
BDBM366672 US9873709, Example 1-269 4-(4-chlorophenyl)-1- [2-[4-[methyl-(1- methyl-4- piperidyl)carbamoyl] anilino]- [1,2,4]triazolo[1,5- a]pyridin-8- yl]piperidine-4- carboxylic acid; formic acid BDBM448703 {1-{1-[2-fluoro-5- (trifluoromethoxy)benzoyl]piperidin- 4-yl}-3-[3-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)- 1H-pyrrol-1-yl]azatidin-3-yl}acetamide US10695337, Example 269
BDBM448703 {1-{1-[2-fluoro-5- (trifluoromethoxy)benzoyl]piperidin- 4-yl}-3-[3-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)- 1H-pyrrol-1-yl]azatidin-3-yl}acetamide US10695337, Example 269 BDBM553214 N-[5-(6-cyano-4- methylpyridin-3-yl)-4- fluoro-2-[rac-(3R,5S)- 3,4,5-trimethylpiperazin- 1-yl]phenyl]-6-oxo-4- (trifluoromethyl)-1H- pyridine-3-carboxamide US11319299, Example 269
BDBM553214 N-[5-(6-cyano-4- methylpyridin-3-yl)-4- fluoro-2-[rac-(3R,5S)- 3,4,5-trimethylpiperazin- 1-yl]phenyl]-6-oxo-4- (trifluoromethyl)-1H- pyridine-3-carboxamide US11319299, Example 269 BDBM742747 (3S,4R)-4-((5-fluoro-7-(5-(1,1,1- trifluoropropan-2-yl)pyridin-2- yl)pyrrolo[2,1-f][1,2,4]triazin-2- yl)amino)tetrahydro-2H-pyran-3-ol US20250163063, Example 269
BDBM742747 (3S,4R)-4-((5-fluoro-7-(5-(1,1,1- trifluoropropan-2-yl)pyridin-2- yl)pyrrolo[2,1-f][1,2,4]triazin-2- yl)amino)tetrahydro-2H-pyran-3-ol US20250163063, Example 269 N-ethyl-1-((1s,4s)-4-((3- fluoro-4- (trifluoromethyl)pyridin-2- yl)oxy)cyclohexyl)-3-(2- methoxypyrimidin-5-yl)-1H- pyrazolo[4,3-c]pyridin-6- amine US11247990, Example 269 BDBM537444
N-ethyl-1-((1s,4s)-4-((3- fluoro-4- (trifluoromethyl)pyridin-2- yl)oxy)cyclohexyl)-3-(2- methoxypyrimidin-5-yl)-1H- pyrazolo[4,3-c]pyridin-6- amine US11247990, Example 269 BDBM537444 US11345681, Example 269 3-(2-{3-Cyanoimidazo[1,2-a]pyridin-7-yl}ethynyl)-1-[(3S,5R)-5-(methoxymethyl)-1-(prop-2-enoyl)pyrrolidin-3-yl]-5-(methylamino)pyrazole-4-carboxamide BDBM556095
US11345681, Example 269 3-(2-{3-Cyanoimidazo[1,2-a]pyridin-7-yl}ethynyl)-1-[(3S,5R)-5-(methoxymethyl)-1-(prop-2-enoyl)pyrrolidin-3-yl]-5-(methylamino)pyrazole-4-carboxamide BDBM556095 3-((10-Hydroxy-7-((S)-2-(5- methyl-1H-1,2,4-triazol-3- yl)pyrrolidine-1-carbonyl)-7- azaspiro[4.5]decan-10- yl)methyl)-6-phenylpyrimidin- 4(3H)-one BDBM633855 US11807646, Example 269
3-((10-Hydroxy-7-((S)-2-(5- methyl-1H-1,2,4-triazol-3- yl)pyrrolidine-1-carbonyl)-7- azaspiro[4.5]decan-10- yl)methyl)-6-phenylpyrimidin- 4(3H)-one BDBM633855 US11807646, Example 269 BDBM662845 US20240109865, Example 269. 5-[(1R)-1-(3,5-dichloro-2- methyl-4-pyridyl)ethoxy]-6- fluoro-3-[5-fluoro-6-(2- methylsulfonyl-2,6- diazaspiro[3.3]heptan-6-yl)-3- pyridyl]-1H-indazole
BDBM662845 US20240109865, Example 269. 5-[(1R)-1-(3,5-dichloro-2- methyl-4-pyridyl)ethoxy]-6- fluoro-3-[5-fluoro-6-(2- methylsulfonyl-2,6- diazaspiro[3.3]heptan-6-yl)-3- pyridyl]-1H-indazole N-[2-(3,5-dimethyl-1- piperidyl)ethyl]-4-[[8- [1-(4,4,4- trifluorobutanoyl)-3,6- dihydro-2H-pyridin-4- yl]-[1,2,4]triazolo[1,5- a]pyridin-2- yl]amino]benzamide US9873709, Example 2-269 BDBM366973
N-[2-(3,5-dimethyl-1- piperidyl)ethyl]-4-[[8- [1-(4,4,4- trifluorobutanoyl)-3,6- dihydro-2H-pyridin-4- yl]-[1,2,4]triazolo[1,5- a]pyridin-2- yl]amino]benzamide US9873709, Example 2-269 BDBM366973 (3S)-3-[1-[4-[(1S,8R)-5-(2,6- difluorophenyl)-11,11-dimethyl-3,4- diazatricyclo[6.2.1.02,7]undeca-2,4,6- trien-1-yl]pyrimidin-2-yl]-1,2,4-triazol-3- yl]thiolane 1,1-dioxide US11008312, Example 269 BDBM497783
(3S)-3-[1-[4-[(1S,8R)-5-(2,6- difluorophenyl)-11,11-dimethyl-3,4- diazatricyclo[6.2.1.02,7]undeca-2,4,6- trien-1-yl]pyrimidin-2-yl]-1,2,4-triazol-3- yl]thiolane 1,1-dioxide US11008312, Example 269 BDBM497783 BDBM678827 7-[(4R)-4- hydroxy-2- oxopyrrolidin-1- yl]-1-methyl-4-[4- methyl-4-(5- methyl-1,3- benzoxazol-2- yl)piperidin-1-yl]- 2-oxo-1,2- dihydroquinoline- 3-carbonitrile US11998539, Example 269
BDBM678827 7-[(4R)-4- hydroxy-2- oxopyrrolidin-1- yl]-1-methyl-4-[4- methyl-4-(5- methyl-1,3- benzoxazol-2- yl)piperidin-1-yl]- 2-oxo-1,2- dihydroquinoline- 3-carbonitrile US11998539, Example 269 BDBM711413 ((6-(difluoromethoxy)-2-(3'-(5-fluoro-6- (pyrrolidin-1-ylmethyl)pyridin-3-yl)-2'- (fluoromethyl)-2-methyl-[1,1'-biphenyl]-3- yl)benzo[d]oxazol-5-yl)methyl)-L-proline US12187713, Example 269
BDBM711413 ((6-(difluoromethoxy)-2-(3'-(5-fluoro-6- (pyrrolidin-1-ylmethyl)pyridin-3-yl)-2'- (fluoromethyl)-2-methyl-[1,1'-biphenyl]-3- yl)benzo[d]oxazol-5-yl)methyl)-L-proline US12187713, Example 269 US10214537, Example 269 7-(3-((cis)-2,6-dimethyl-4-(methylsulfonyl) piperazin-1-yl)phenyl)-5-(1-(tetrahydro-2H- pyran-4-yl)-1H-pyrazol-5-yl)pyrrolo[2,1-f][1,2,4] triazin-4-amine BDBM358200
US10214537, Example 269 7-(3-((cis)-2,6-dimethyl-4-(methylsulfonyl) piperazin-1-yl)phenyl)-5-(1-(tetrahydro-2H- pyran-4-yl)-1H-pyrazol-5-yl)pyrrolo[2,1-f][1,2,4] triazin-4-amine BDBM358200 US10968216, Example 269 2-{[(2S)-1-(dimethylamino)propan-2-yl]oxy}-5-fluoro-N-(4-fluoro-2-methylphenyl)-4-(3-oxo-5,6,7,8-tetrahydro[1,2,4]triazolo[4,3-a]pyridin-2(3H)-yl)benzamide BDBM490601
US10968216, Example 269 2-{[(2S)-1-(dimethylamino)propan-2-yl]oxy}-5-fluoro-N-(4-fluoro-2-methylphenyl)-4-(3-oxo-5,6,7,8-tetrahydro[1,2,4]triazolo[4,3-a]pyridin-2(3H)-yl)benzamide BDBM490601 (1R,3R)-1-[2,6-difluoro-4-[1- [[(1S,2R)-2- fluorocyclopropyl]methyl] azetidin-3-yl]oxy-phenyl]-2- (2-fluoro-2-methyl-propyl)-3- methyl-1,3,4,9- tetrahydropyrido[3,4-b]indole BDBM731870 US20250114338, Example 269
(1R,3R)-1-[2,6-difluoro-4-[1- [[(1S,2R)-2- fluorocyclopropyl]methyl] azetidin-3-yl]oxy-phenyl]-2- (2-fluoro-2-methyl-propyl)-3- methyl-1,3,4,9- tetrahydropyrido[3,4-b]indole BDBM731870 US20250114338, Example 269 1-[(3S,4S)-1-(2,6- difluorobenzyl)-4- methoxypyrrolidin-3-yl]-3-[1- (2-methylpyridin-4-yl)-1H- pyrazolo[3,4-c]pyridin-5- yl]urea US9884048, Example 269 US9884048, Example 274 US9884048, Example 273 BDBM275390
1-[(3S,4S)-1-(2,6- difluorobenzyl)-4- methoxypyrrolidin-3-yl]-3-[1- (2-methylpyridin-4-yl)-1H- pyrazolo[3,4-c]pyridin-5- yl]urea US9884048, Example 269 US9884048, Example 274 US9884048, Example 273 BDBM275390 4-(1-(6- ((Ethylamino)methyl)-2- methylpyridin-3-yl)-1H- pyrazol-4-yl)-N-(1-((1- methyl-1H-imidazol-4- yl)sulfonyl)piperidin-4- yl)-5- (trifluoromethyl)pyrimidin- 2-amine US11472791, Example 269 BDBM576947
4-(1-(6- ((Ethylamino)methyl)-2- methylpyridin-3-yl)-1H- pyrazol-4-yl)-N-(1-((1- methyl-1H-imidazol-4- yl)sulfonyl)piperidin-4- yl)-5- (trifluoromethyl)pyrimidin- 2-amine US11472791, Example 269 BDBM576947 6-(3-cyanopyrrolo[1,2-b] pyridazin-7-yl)-N-((R)-2- fluoro-3-hydroxy-3- methylbutyl)-4-(((1r,4R)- 4-((5-methyl-1,3,4- oxadiazol-2-yl)amino) cyclohexyl)amino) nicotinamide BDBM407327 US10336762, Compound 269
6-(3-cyanopyrrolo[1,2-b] pyridazin-7-yl)-N-((R)-2- fluoro-3-hydroxy-3- methylbutyl)-4-(((1r,4R)- 4-((5-methyl-1,3,4- oxadiazol-2-yl)amino) cyclohexyl)amino) nicotinamide BDBM407327 US10336762, Compound 269 BDBM505621 (S)-8-chloro-4-((3-chloro-4- fluorophenyl)amino)-6-(((6- (oxetan-3-yl )-4,5,6,7- tetrahydrothieno[2,3-c]pyridin- yl)(1H-1,2,3-triazol-4- yl)methyl)amino)quinoline-3- carbonitrile US11066414, Compound 269
BDBM505621 (S)-8-chloro-4-((3-chloro-4- fluorophenyl)amino)-6-(((6- (oxetan-3-yl )-4,5,6,7- tetrahydrothieno[2,3-c]pyridin- yl)(1H-1,2,3-triazol-4- yl)methyl)amino)quinoline-3- carbonitrile US11066414, Compound 269 BDBM591203 5-(8-amino-7-fluoro-3-((6-isopropyl-7- oxo-5,6,7,8-tetrahydro-4H-pyrazolo[1,5- d][1,4]diazepin-2-yl)amino)isoquinolin- 6-yl)-1-ethyl-1H-pyrazole-3-carbonitrile US11566003, Compound 269
BDBM591203 5-(8-amino-7-fluoro-3-((6-isopropyl-7- oxo-5,6,7,8-tetrahydro-4H-pyrazolo[1,5- d][1,4]diazepin-2-yl)amino)isoquinolin- 6-yl)-1-ethyl-1H-pyrazole-3-carbonitrile US11566003, Compound 269 US20240109900, Example 269 4-(6-((1S,6R,7R)-7-(aminomethyl)-7-(2- fluorophenyl)-3-azabicyclo[4.1.0]heptan-3-yl)-1H- pyrazolo[3,4-b]pyrazin-3-yl)-3-chloro-1- methylpyridin-2(1H)-one BDBM663431
US20240109900, Example 269 4-(6-((1S,6R,7R)-7-(aminomethyl)-7-(2- fluorophenyl)-3-azabicyclo[4.1.0]heptan-3-yl)-1H- pyrazolo[3,4-b]pyrazin-3-yl)-3-chloro-1- methylpyridin-2(1H)-one BDBM663431 4-cyclopropyl-7-(2-((7-isopropyl-2- (oxetan-3-ylmethyl)-1,2,3,4- tetrahydroisoquinolin-6-yl)amino)- 5-(trifluoromethyl)pyrimidin-4-yl)- 3,4-dihydrothieno[2,3- f][1,4]thiazepin-5(2H)-one 1,1- dioxide BDBM738037 US20250144094, Example 269
4-cyclopropyl-7-(2-((7-isopropyl-2- (oxetan-3-ylmethyl)-1,2,3,4- tetrahydroisoquinolin-6-yl)amino)- 5-(trifluoromethyl)pyrimidin-4-yl)- 3,4-dihydrothieno[2,3- f][1,4]thiazepin-5(2H)-one 1,1- dioxide BDBM738037 US20250144094, Example 269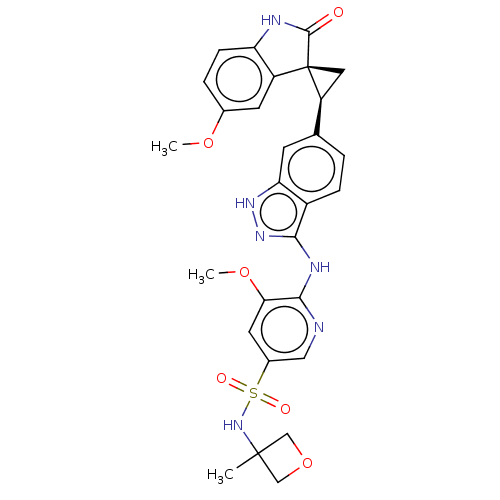 5-methoxy-6-({6-[(1R,2S)-5'-methoxy-2'-oxo-1',2'-dihydrospiro[cyclopropane-1,3'-indol]-2-yl]-1H-indazol-3-yl}amino)-N-(3-methyloxetan-3-yl)pyridine-3-sulfonamide BDBM636140 US20230365537, Example 269
5-methoxy-6-({6-[(1R,2S)-5'-methoxy-2'-oxo-1',2'-dihydrospiro[cyclopropane-1,3'-indol]-2-yl]-1H-indazol-3-yl}amino)-N-(3-methyloxetan-3-yl)pyridine-3-sulfonamide BDBM636140 US20230365537, Example 269 (S)-3-(1,4-dimethyl-1H-benzo[d][1,2,3]triazol-5-yl)-3-(3-(((R)-4-ethyl-1,1-dioxido-3,4-dihydro-2H-benzo[b][1,4,5]oxathiazepin-2-yl)methyl)-4-methylphenyl)-2,2-dimethylpropanoic acid BDBM304965 US10144731, Example 269
(S)-3-(1,4-dimethyl-1H-benzo[d][1,2,3]triazol-5-yl)-3-(3-(((R)-4-ethyl-1,1-dioxido-3,4-dihydro-2H-benzo[b][1,4,5]oxathiazepin-2-yl)methyl)-4-methylphenyl)-2,2-dimethylpropanoic acid BDBM304965 US10144731, Example 269 N-((2S)-1-((2-((4- chlorophenyl)carbamoyl)- 2-((R)-4-isopropyl-2- oxoimidazolidin-1-yl)- 2,3-dihydro-1H-inden- 5-yl)amino)-3,3- dicyclopropyl-1- oxopropan-2-yl)-4- methyl-1,2,5- oxadiazole-3- carboxamide US20250154138, Example 269 BDBM740366
N-((2S)-1-((2-((4- chlorophenyl)carbamoyl)- 2-((R)-4-isopropyl-2- oxoimidazolidin-1-yl)- 2,3-dihydro-1H-inden- 5-yl)amino)-3,3- dicyclopropyl-1- oxopropan-2-yl)-4- methyl-1,2,5- oxadiazole-3- carboxamide US20250154138, Example 269 BDBM740366 7-(5-(5-((1R,5S)-9-acetyl-3-oxa-7,9-diazabicyclo[3.3.1]nonan-7-yl)-1,3,4-thiadiazol-2-yl)-4-((tetrahydro-2H-pyran-4-yl)amino)pyridin-2-yl)pyrrolo[1,2-b]pyridazine-3-carbonitrile BDBM609304 US11702414, Example 269
7-(5-(5-((1R,5S)-9-acetyl-3-oxa-7,9-diazabicyclo[3.3.1]nonan-7-yl)-1,3,4-thiadiazol-2-yl)-4-((tetrahydro-2H-pyran-4-yl)amino)pyridin-2-yl)pyrrolo[1,2-b]pyridazine-3-carbonitrile BDBM609304 US11702414, Example 269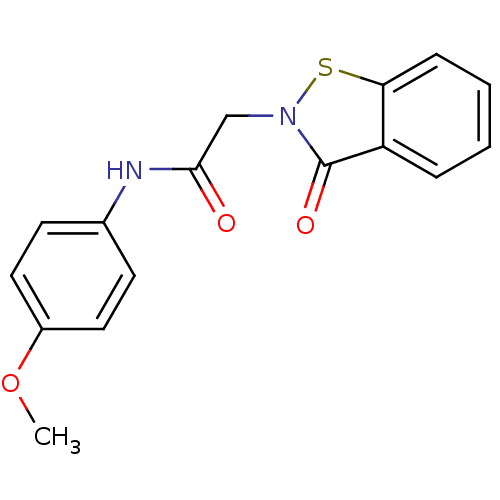 KSC-4-269 KUC104551N N-(4-methoxyphenyl)-2-(3-oxo-1,2-benzothiazol-2-yl)acetamide BDBM61940 N-(4-methoxyphenyl)-2-(3-oxidanylidene-1,2-benzothiazol-2-yl)ethanamide cid_44968082 2-(3-keto-1,2-benzothiazol-2-yl)-N-(4-methoxyphenyl)acetamide
KSC-4-269 KUC104551N N-(4-methoxyphenyl)-2-(3-oxo-1,2-benzothiazol-2-yl)acetamide BDBM61940 N-(4-methoxyphenyl)-2-(3-oxidanylidene-1,2-benzothiazol-2-yl)ethanamide cid_44968082 2-(3-keto-1,2-benzothiazol-2-yl)-N-(4-methoxyphenyl)acetamide US11866450, Compound 269 BDBM644854 Preparation of (14S)-12,12-dimethyl-8-(3-{3-[1-(trifluoromethyl)cyclopropyl]-3,3-dideuterio-propoxy}-1H-pyrazol-1-yl)-2λ6-thia-3,9,11,18,23-pentaazatetracyclo[17.3.1.111,14.05,10]tetracosa-1(22),5(10),6,8,19(23),20-hexaene-2,2,4-trione
US11866450, Compound 269 BDBM644854 Preparation of (14S)-12,12-dimethyl-8-(3-{3-[1-(trifluoromethyl)cyclopropyl]-3,3-dideuterio-propoxy}-1H-pyrazol-1-yl)-2λ6-thia-3,9,11,18,23-pentaazatetracyclo[17.3.1.111,14.05,10]tetracosa-1(22),5(10),6,8,19(23),20-hexaene-2,2,4-trione  US20250019387, Example 269 BDBM713358 (Method 2):(R)-1-(3-((7-(5,7-difluoronaphthalen-1-yl)-8-fluoro-2-((hexahydro-1H-pyrrolizin-7a-yl)methoxy)pyrido[4,3-d]pyrimidin-4-yl)(methyl)amino)pyrrolidin-1-yl)prop-2-en-1-one
US20250019387, Example 269 BDBM713358 (Method 2):(R)-1-(3-((7-(5,7-difluoronaphthalen-1-yl)-8-fluoro-2-((hexahydro-1H-pyrrolizin-7a-yl)methoxy)pyrido[4,3-d]pyrimidin-4-yl)(methyl)amino)pyrrolidin-1-yl)prop-2-en-1-one (S or R)-3-(4-((2S,5R or 2R,5S)-5-(5-amino-9-fluoro-7-methoxy- [1,2,4]triazolo[1,5-c]quinazolin-2-yl)-2-methylpiperidin-1-yl)- 1H-pyrazol-1-yl)-3-methylbutan-2-ol US11312719, Example 269 BDBM551723
(S or R)-3-(4-((2S,5R or 2R,5S)-5-(5-amino-9-fluoro-7-methoxy- [1,2,4]triazolo[1,5-c]quinazolin-2-yl)-2-methylpiperidin-1-yl)- 1H-pyrazol-1-yl)-3-methylbutan-2-ol US11312719, Example 269 BDBM551723 (3R,6R,7S,8E,22S)-6′-Chloro-7-[2-[(3R,4S)-3,4-difluoropyrrolidin-1-yl]ethoxy]-12,12-dimethyl-15,15-dioxo-spiro[11,20-dioxa-15-thia-1,14-diazatetracyclo[14.7.2.03,6.019,24]pentacosa-8,16,18,24-tetraene-22,1′-tetralin]-13-one US11130769, Ex. No. 269 BDBM520572
(3R,6R,7S,8E,22S)-6′-Chloro-7-[2-[(3R,4S)-3,4-difluoropyrrolidin-1-yl]ethoxy]-12,12-dimethyl-15,15-dioxo-spiro[11,20-dioxa-15-thia-1,14-diazatetracyclo[14.7.2.03,6.019,24]pentacosa-8,16,18,24-tetraene-22,1′-tetralin]-13-one US11130769, Ex. No. 269 BDBM520572 US10780090, Compound I-269a US10780090, Compound I-269b BDBM462975 [(1R,2S,4R)-4-{[5-({4-[(7S)-4,7-dihydro-5H-thieno[2,3-c]pyran-7-yl]-5-methyl-2- thienyl}carbonyl)pyrimidin-4-yl]amino}-2-hydroxycyclopentyl]methyl sulfamate US10780090, Compound I-269
US10780090, Compound I-269a US10780090, Compound I-269b BDBM462975 [(1R,2S,4R)-4-{[5-({4-[(7S)-4,7-dihydro-5H-thieno[2,3-c]pyran-7-yl]-5-methyl-2- thienyl}carbonyl)pyrimidin-4-yl]amino}-2-hydroxycyclopentyl]methyl sulfamate US10780090, Compound I-269 US11878965, Example 269 BDBM646278 ((2S,5R)-5-amino-2- methylpiperidin-1-yl)(2-(1- (cyclopropylmethyl)-6-(2-(2,4- dimethylthiazol-5-yl)-4- methylpyridin-3-yl)-1H-pyrrolo[2,3- b]pyridin-2-yl)-7-methoxy-1-methyl- 1H-benzo[d]imidazol-5- yl)methanone
US11878965, Example 269 BDBM646278 ((2S,5R)-5-amino-2- methylpiperidin-1-yl)(2-(1- (cyclopropylmethyl)-6-(2-(2,4- dimethylthiazol-5-yl)-4- methylpyridin-3-yl)-1H-pyrrolo[2,3- b]pyridin-2-yl)-7-methoxy-1-methyl- 1H-benzo[d]imidazol-5- yl)methanone (R)-5-cyano-N-ethyl-N-(2,2,2-trifluoro-1-(3-methyl-4-(trifluoromethyl)phenyl)ethyl)pyridine-3-sulfonamide (Example 268) and (R)-5-cyano-N-methyl-N-(2,2,2-trifluoro-1-(3-methyl-4-(trifluoromethyl)phenyl)ethyl)pyridine-3-sulfonamide (Example 269) BDBM732724 US20250115569, Example 268
(R)-5-cyano-N-ethyl-N-(2,2,2-trifluoro-1-(3-methyl-4-(trifluoromethyl)phenyl)ethyl)pyridine-3-sulfonamide (Example 268) and (R)-5-cyano-N-methyl-N-(2,2,2-trifluoro-1-(3-methyl-4-(trifluoromethyl)phenyl)ethyl)pyridine-3-sulfonamide (Example 269) BDBM732724 US20250115569, Example 268 (R)-4-((2-(2-chloro-3′-(7-cyano-5-((3-hydroxypyrrolidin-1-yl)methyl)benzo[d]oxazol-2-yl)-2′-methylbiphenyl-3-ylcarbamoyl)-1-methyl-6,7-dihydro-1H-imidazo[4,5-c]pyridin-5(4H)-yl)methyl)-1-methylcyclohexanecarboxylic acid (Peak 2) US10308644, Example 269 BDBM395659
(R)-4-((2-(2-chloro-3′-(7-cyano-5-((3-hydroxypyrrolidin-1-yl)methyl)benzo[d]oxazol-2-yl)-2′-methylbiphenyl-3-ylcarbamoyl)-1-methyl-6,7-dihydro-1H-imidazo[4,5-c]pyridin-5(4H)-yl)methyl)-1-methylcyclohexanecarboxylic acid (Peak 2) US10308644, Example 269 BDBM395659 US20250034136, Compound I-269 BDBM717096 N-[2-chloro-4-(trifluoromethyl)phenyl]-2-{6-ethyl-7-[(1S,6S)-5-(5- hydroxy-6-methylpyrimidine-4-carbonyl)-2,5-diazabicyclo[4.2.0]octan- 2-yl]-2-{[(2S)-2-methoxypropyl](methyl)amino}-8-oxo-5H,8H- pyrido[2,3-b]pyrazin-5-yl}acetamide
US20250034136, Compound I-269 BDBM717096 N-[2-chloro-4-(trifluoromethyl)phenyl]-2-{6-ethyl-7-[(1S,6S)-5-(5- hydroxy-6-methylpyrimidine-4-carbonyl)-2,5-diazabicyclo[4.2.0]octan- 2-yl]-2-{[(2S)-2-methoxypropyl](methyl)amino}-8-oxo-5H,8H- pyrido[2,3-b]pyrazin-5-yl}acetamide BDBM705110 US20240376097, Example 269 2-((3-fluoro-1H-pyrrolo[2,3-b] pyridin-5-yl)oxy)-N-((4-((((1r,4r)- 4-hydroxy-4-methylcyclohexyl) methyl)amino)-3-nitrophenyl) sulfonyl)-4-(6-((R)-4-(4-isopropyl- 3-methoxybenzyl)-2-(2- isopropylphenyl)piperazin-1-yl)-2- azaspiro[3.3]heptan-2-yl) benzamide
BDBM705110 US20240376097, Example 269 2-((3-fluoro-1H-pyrrolo[2,3-b] pyridin-5-yl)oxy)-N-((4-((((1r,4r)- 4-hydroxy-4-methylcyclohexyl) methyl)amino)-3-nitrophenyl) sulfonyl)-4-(6-((R)-4-(4-isopropyl- 3-methoxybenzyl)-2-(2- isopropylphenyl)piperazin-1-yl)-2- azaspiro[3.3]heptan-2-yl) benzamide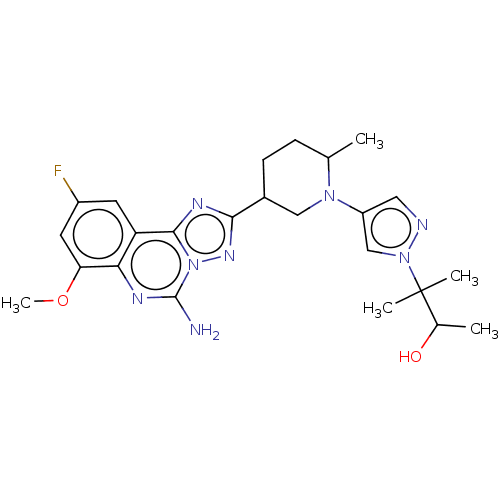 US11312719, Example 268 US11312719, Example 269 (R or S)-3-(4-((2S,5R or 2R,5S)-5-(5-amino-9-fluoro-7-methoxy- [1,2,4]triazolo[1,5-c]quinazolin-2-yl)-2-methylpiperidin-1-yl)- 1H-pyrazol-1-yl)-3-methylbutan-2-ol US11312719, Example 270 US11312719, Example 271 BDBM551722
US11312719, Example 268 US11312719, Example 269 (R or S)-3-(4-((2S,5R or 2R,5S)-5-(5-amino-9-fluoro-7-methoxy- [1,2,4]triazolo[1,5-c]quinazolin-2-yl)-2-methylpiperidin-1-yl)- 1H-pyrazol-1-yl)-3-methylbutan-2-ol US11312719, Example 270 US11312719, Example 271 BDBM551722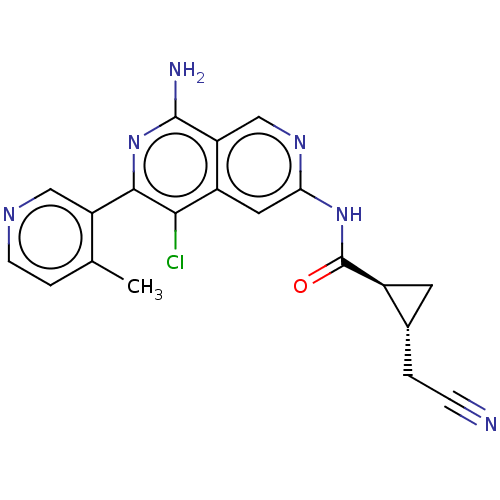 US11034692, Compound 270 (1R,2S)—N-[8-amino-5-chloro-6-(4-methylpyridin-3-yl)-2,7-naphthyridin-3-yl]-2-(cyanomethyl)cyclopropane-1-carboxamide (Compound 269) and (1S,2R)—N-[8-amino-5-chloro-6-(4-methylpyridin-3-yl)-2,7-naphthyridin-3-yl]-2-(cyanomethyl)cyclopropane-1-carboxamide BDBM504006
US11034692, Compound 270 (1R,2S)—N-[8-amino-5-chloro-6-(4-methylpyridin-3-yl)-2,7-naphthyridin-3-yl]-2-(cyanomethyl)cyclopropane-1-carboxamide (Compound 269) and (1S,2R)—N-[8-amino-5-chloro-6-(4-methylpyridin-3-yl)-2,7-naphthyridin-3-yl]-2-(cyanomethyl)cyclopropane-1-carboxamide BDBM504006 5-amino-3-[(5R,7R,8R,12aR,14R,15S,15aR,16S)- 14-(7-amino-3H-[1,2,3]triazolo[4,5-d]pyrimidin- 3-yl)-15-fluoro-2,16-dihydroxy-2,10-dioxido-10- sulfanyloctahydro-12H-5,8-methanofuro[3,2- 1][1,3,9,11,6,2,10]tetraoxathiadiphosphacyclo- tetradecin-7-yl]-3,6-dihydro-7H-[1,2,3]triazolo [4,5-d]pyrimidin-7-one (Diastereomer 1) US11453697, Example 269 BDBM574137 US11453697, Example 270
5-amino-3-[(5R,7R,8R,12aR,14R,15S,15aR,16S)- 14-(7-amino-3H-[1,2,3]triazolo[4,5-d]pyrimidin- 3-yl)-15-fluoro-2,16-dihydroxy-2,10-dioxido-10- sulfanyloctahydro-12H-5,8-methanofuro[3,2- 1][1,3,9,11,6,2,10]tetraoxathiadiphosphacyclo- tetradecin-7-yl]-3,6-dihydro-7H-[1,2,3]triazolo [4,5-d]pyrimidin-7-one (Diastereomer 1) US11453697, Example 269 BDBM574137 US11453697, Example 270 BDBM690323 N-(5-(2-cyclopropyl-2H-tetrazol-5-yl)-2-methylphenyl)-5-(1-((2R,3S)-3-hydroxybutan-2-yl)-3-methyl-1H-pyrazol-4-yl)pyrazolo[1,5-a]pyridine-3-carboxamide or N-(5-(2-cyclopropyl-2H-tetrazol-5-yl)-2-methylphenyl)-5-(1-((2S,3R)-3-hydroxybutan-2-yl)-3-methyl-1H-pyrazol-4-yl)pyrazolo[1,5-a]pyridine-3-carboxamide US20240262826, Compound 269
BDBM690323 N-(5-(2-cyclopropyl-2H-tetrazol-5-yl)-2-methylphenyl)-5-(1-((2R,3S)-3-hydroxybutan-2-yl)-3-methyl-1H-pyrazol-4-yl)pyrazolo[1,5-a]pyridine-3-carboxamide or N-(5-(2-cyclopropyl-2H-tetrazol-5-yl)-2-methylphenyl)-5-(1-((2S,3R)-3-hydroxybutan-2-yl)-3-methyl-1H-pyrazol-4-yl)pyrazolo[1,5-a]pyridine-3-carboxamide US20240262826, Compound 269 US9957264, Example 5-269 BDBM391198 2-((3R,4R or 3S,4S)-4-(3-((4- ((R or S)-1-(tert- butylamino)-2,2,2- trifluoroethyl)phenyl) amino)-4-oxo-4,5-dihydro- 1H-pyrazolo[4,3-c]pyridin-1- yl)-3-fluorotetrahydro-2H- pyran-4-yl)acetonitrile (from I-22A. The mixture was purified by Chiral HPLC with a Venusil Chiral ODH column using hexanes:EtOH (80:20), Tr = 38.0 minutes)
US9957264, Example 5-269 BDBM391198 2-((3R,4R or 3S,4S)-4-(3-((4- ((R or S)-1-(tert- butylamino)-2,2,2- trifluoroethyl)phenyl) amino)-4-oxo-4,5-dihydro- 1H-pyrazolo[4,3-c]pyridin-1- yl)-3-fluorotetrahydro-2H- pyran-4-yl)acetonitrile (from I-22A. The mixture was purified by Chiral HPLC with a Venusil Chiral ODH column using hexanes:EtOH (80:20), Tr = 38.0 minutes)